- 1Key Laboratory of Stress Physiology and Ecology of Gansu Province, Northwest Institute of Eco-Environment and Resources, Chinese Academy of Sciences, Lanzhou, China
- 2University of Chinese Academy of Sciences, Beijing, China
- 3Desert Animal Adaptations and Husbandry, Wyler Department of Dryland Agriculture, Jacob Blaustein Institutes for Desert Research, Ben-Gurion University of the Negev, Beer Sheva, Israel
- 4Key Laboratory of Extreme Environmental Microbial Resources and Engineering, Lanzhou, China
- 5Ningxia Feed Engineering Technology Research Center, Ningxia University, Yinchuan, China
- 6Shandong Huakun Rural Revitalization Institute Co., Ltd., Jinan, China
- 7Yellow River Estuary Tan Sheep Institute of Industrial Technology, Dongying, China
Grape pomace (GP), a by-product in wine production, is nutritious and can be used as a feed ingredient for ruminants; however, its role in shaping sheep gastrointestinal tract (GIT) microbiota is unclear. We conducted a controlled trial using a randomized block design with 10 Tan lambs fed a control diet (CD) and 10 Tan lambs fed a pelleted diet containing 8% GP (dry matter basis) for 46 days. Rumen, jejunum, cecum, and colon bacterial and archaeal composition were identified by 16S rRNA gene sequencing. Dry matter intake (DMI) was greater (p < 0.05) in the GP than CD group; however, there was no difference in average daily gain (ADG, p < 0.05) and feed conversion ratio (FCR, p < 0.05) between the two groups. The GP group had a greater abundance of Prevotella 1 and Prevotella 7 in the rumen; of Sharpe, Ruminococcaceae 2, and [Ruminococcus] gauvreauii group in the jejunum; of Ruminococcaceae UCG-014 and Romboutsia in the cecum, and Prevotella UCG-001 in the colon; but lesser Rikenellaceae RC9 gut group in the rumen and cecum, and Ruminococcaceae UCG-005 and Ruminococcaceae UCG-010 in the colon than the CD group. The pathways of carbohydrate metabolism, such as L-rhamnose degradation in the rumen, starch and glycogen degradation in the jejunum, galactose degradation in the cecum, and mixed acid fermentation and mannan degradation in the colon were up-graded; whereas, the pathways of tricarboxylic acid (TCA) cycle VIII, and pyruvate fermentation to acetone in the rumen and colon were down-graded with GP. The archaeal incomplete reductive TCA cycle was enriched in the rumen, jejunum, and colon; whereas, the methanogenesis from H2 and CO2, the cofactors of methanogenesis, including coenzyme M, coenzyme B, and factor 420 biosynthesis were decreased in the colon. The study concluded that a diet including GP at 8% DM did not affect ADG or FCR in Tan lambs. However, there were some potential benefits, such as enhancing propionate production by microbiota and pathways in the GIT, promoting B-vitamin production in the rumen, facilitating starch degradation and amino acid biosynthesis in the jejunum, and reducing methanogenesis in the colon.
1. Introduction
In ruminant production, feed typically accounts for 60–70% of total production expenditures (Arthur et al., 2004; Karisa et al., 2014). Therefore, saving feed costs by the use of by-products as a feed source for livestock could increase profits substantially. Grapes are cultivated widely to produce wine, the most popular beverage in the world. According to the Food and Agriculture Organization (FAO), there are 75,866 km2 of land worldwide devoted to vineyards, with an annual yield of 74 million tons of grapes, of which approximately 75% is used for winemaking (Beres et al., 2017). In the production of grape juice, whole grape bunches are pressed, and grape pomace (GP), including seeds, skins and stems, remains (Yu and Ahmedna, 2013). The GP, approximately 20% of the total mass, is generally discarded, which can cause serious environmental problems when accumulated.
GP is rich in tannins and other polyphenols. Tannins are high molecular weight polyphenolic compounds that possess antimicrobial, anti-parasitic, anti-viral, antioxidant, anti-inflammatory, and immunomodulatory properties (Besharati et al., 2022). In addition, tannins can increase nitrogen utilization in the gut, improve ruminal fermentation, and decrease methane production in ruminants. However, at high intakes, tannins are considered as anti-nutritional since they bind to proteins, carbohydrates, and other substances, consequently reducing fiber digestibility. GP can be fed to ruminants at approximately 10% of the dry matter intake (Chikwanha et al., 2019; Gao et al., 2019). Previous studies reported that GP could affect nutrient utilization (Abarghuei et al., 2010), enhance meat and milk quality and antioxidant capacity (Ianni and Martino, 2020; Molosse et al., 2021), and reduce methane (CH4) emission (Moate et al., 2014; Russo et al., 2017). In addition, GP can improve rumen fermentation, increase propionate concentration to boost growth performance, and decrease methane emission (Khiaosa-ard et al., 2015; Foiklang et al., 2016a). However, at high intakes (usually >20% DM), GP can be detrimental to rumen fermentation and reduce the digestibilities of dry matter (DM), neutral detergent fiber (NDF) and acid detergent fiber (ADF), and the production of volatile fatty acid (VFAs) (Besharati and Taghizadeh, 2009; Ishida et al., 2015).
The mammalian digestive tract is colonized by vast numbers of microorganisms, including bacteria, fungi, archaea, protozoa, and viruses, which outnumber the cells in the body by hundreds of times and are considered the ‘second genome’ of the animal (Zhu et al., 2010). The rumen is a unique organ in ruminants, accounting for approximately 75% of the total volume of the sheep’s stomach (Membrive, 2016). The microbes in the rumen can ferment complex cellulose, starch, proteins, and lipids, producing VFAs, which provide up to 70% of the host’s energy needs, and can synthesize microbial proteins and vitamin B complexes (Mizrahi et al., 2021). Microbes in the lower gut can also degrade feed and produce CH4. The jejunum is the main site of digestion and absorption of protein and carbohydrates, while microbial fermentation in the hindgut can degrade up to 30% of cellulose and hemicellulose in ruminants (O’Hara et al., 2020). In addition, archaea in the gastrointestinal tract (GIT) produce large amounts of CH4. Enteric CH4 from ruminants accounts for 73% of livestock emissions (Rodrigues, 2016), and the CH4 emission is the equivalent of 2–12% of the digestible energy intake of the host (Johnson and Johnson, 1995). The rumen is the primary site of methane production in sheep, while the hindgut contributes only 6–14% of methane production (Immig, 1996).
The effect of GP on rumen and feces microbiota has been studied (Kafantaris et al., 2017; Biscarini et al., 2018), but the results have been equivocal. GP may potentially enhance the abundance of beneficial bacteria and suppress methane emission (Ross et al., 2013), or have no impact on the composition and function of rumen microbiota (Buffa et al., 2020). There has been no comprehensive study on the potential shaping effects of feed supplemented with GP on the GIT microbiome. To fill this gap, we examined the effect of GP on bacteria and archaea in different segments of the GIT of Tan lambs. We hypothesized that adding GP (8%) would enhance sheep growth, have a positive impact on gastrointestinal microbe fermentation, and reduce methane production.
2. Materials and methods
The protocol of the study and all procedures on the lambs complied with animal research and welfare recommendations and were approved by the Academic Committee of the Northwestern Institute of Eco-Environment Resources, Chinese Academy of Sciences (protocol number: CAS201810082).
2.1. Experimental design, diets, and management
Twenty castrated, 3-month-old Tan lambs of similar body condition and weight (22 ± 1.7 kg) were divided randomly into two groups (a separate sheepfold for each group) of 10 lambs each: one group was fed a control diet (CD) and one group was fed a diet containing 8% GP by DM. The diets were offered twice daily, at 8:00 and 18:00, with free access to water and lick mineral blocks for 46 d. The GP was bought from a winery at the eastern foot of the Helan Mountain, and after drying, the feed was prepared by the Feed Engineering Technology Research Center of Ningxia University. The two diets were iso-nitrogenous, with the same energy yield and were prepared according to the Feeding Standards for sheep and goats (Ministry of Agriculture of China, 2005; Table 1). The lambs were weighed before morning feed was offered on days 0 and 47 to calculate average daily gain (ADG). All feed offered and feed remains before the feed was offered each morning were weighed to determine daily dry matter intake (DMI). Feed conversion ratio (FCR) was calculated as the ratio between DMI (g/d) and ADG (g/d).
2.2. Sample collection and feed composition
After the feeding trial, 5 lambs were selected randomly from each group and transported to a nearby abattoir for traditional halal slaughter. The rumen, jejunum, cecum, and colon were separated by ligation with cotton thread to prevent the contents from moving between adjoining sections, their contents were collected with sterile forceps into 2 mL sterile lyophilization tubes, stored in liquid nitrogen, and brought to the laboratory for storage at-80°C.
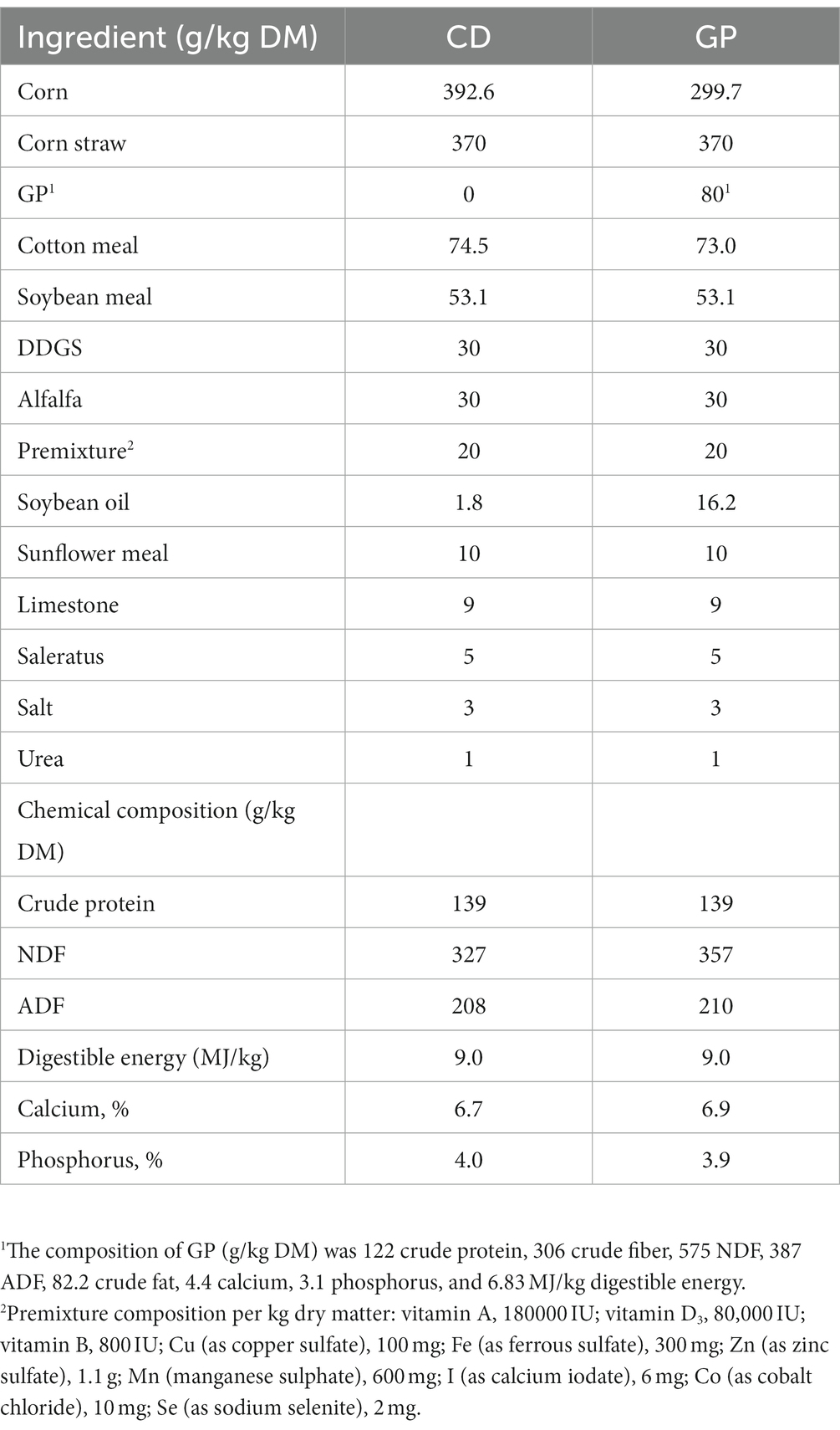
The chemical composition of GP and feeds was determined following the Association of Official Analytical Chemists (AOAC International, 2007), and included DM (934.01), ether extract (945.16), crude protein (990.03, Kjeldahl, N × 6.25), calcium (927.02), and phosphorus (965.17). The NDF (assayed with a heat-stable amylase) and ADF were measured using the filter bag technique with a fiber analyzer (A2000; ANKOM Technology, NY, USA), as described by Van Soest et al. (1991) (Table 2).
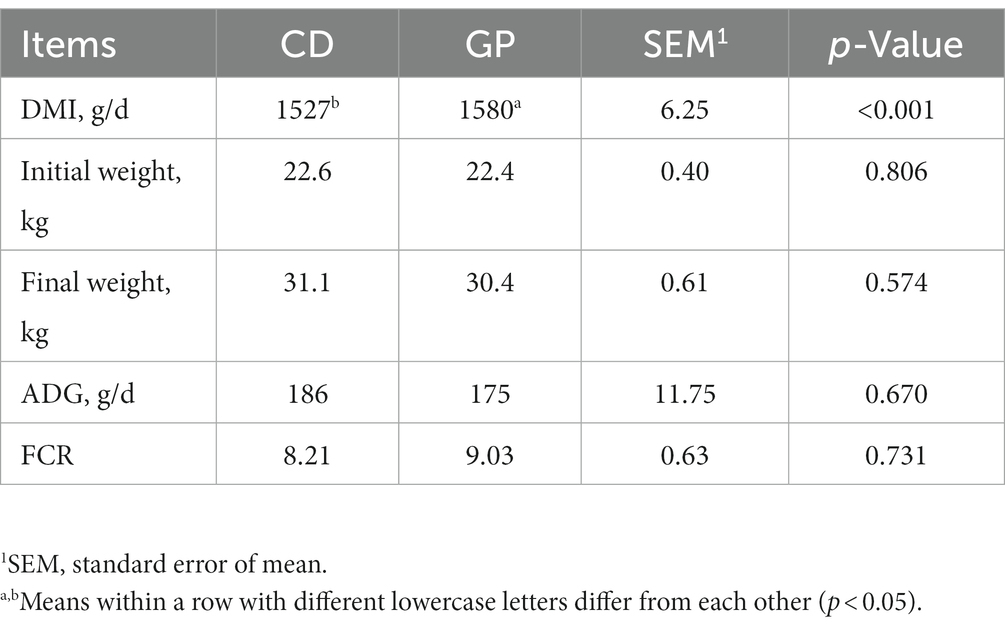
Table 2. Dry matter intake (DMI), body weights, average daily gain (ADG) and feed conversion ratio (FCR) in lambs consuming control (CD) and grape pomace (GP) diets.
2.3. DNA extraction, amplification, and sequencing
The cetyltrimethylammonium bromide method was used to extract DNA from each sample, and subsequent determination of DNA purity and concentration used agarose gel electrophoresis. The polymerase chain reaction (PCR) was done with the V3-V4 region of bacteria and the V4-V5 region of archaea as targets. After PCR product mixing and purification, PCR products were extracted using the library constructed by the TruSeq® DNA PCR-Free Sample Preparation Kit, and after Qubit and real-time PCR quantification, the library was sequenced using NovaSeq6000.
2.4. Bioinformatics analysis
The QIIME 2 (Bolyen et al., 2019) was used for microbiome bioinformatics. The q2-demux plugin was used to demultiplex and quality filter the raw sequence data; the feature table was clustered by 97% similarity using Vsearch (Rognes et al., 2016). All the operational taxonomic units (OTUs) were aligned using the mafft (Katoh et al., 2002), and were used to construct a phylogenic tree using fasttree2 (Price et al., 2010). Taxonomy was assigned to OTUs using the q2-feature-classifier (Bokulich et al., 2018), classify-sklearn naïve Bayes taxonomy classifier against the Silva 138 99% reference sequences (McDonald et al., 2012) to classify and annotate OTUs. Filtering feature tables included total-frequency-based filtering by data volume, species-based filtering of tables and sequences. Alpha and beta diversities, microbial composition and MetaStat analysis of difference species calculation and visualization used the R package microeco (Liu et al., 2020); and functional prediction used phylogenetic investigation of communities by reconstruction of unobserved states (PICRUSt2) (Douglas et al., 2020) based on the metabolic pathways from all domains of life (MetaCyc) database (Caspi et al., 2020).
2.5. Statistical analyses
The ADG, DMI and FCR were compared between dietary groups using a one-way ANOVA (SPSS 26, Chicago, IL, USA). Alpha diversity indices were compared between groups using the Wilcox test, beta diversity was analyzed using multivariate analysis of variance (PERMANOVA), differences in the relative abundances of the rumen, jejunum cecum, and colonic microbiota at the genus level were tested by MetaStat analysis (Paulson et al., 2011), for different site differential pathway analysis of treatments used the Welch’s t-test test, correlations between differential microbiota and MetaCyc pathways used the “corr.test” function in R4.2.2, and false discovery rate (FDR)-corrected Spearman’s correlation test and visualized used the Pheatmap R package. A level of p < 0.05 was accepted as significant and 0.05 < p < 0.10 as tended to be significant.
3. Results
3.1. Effect of grape pomace on average daily gain, dry matter intake and feed conversion ratio in Tan lambs
The DMI was greater (p < 0.05) in the GP than CD group, but there was no difference (p > 0.05) between the two groups in initial and final body weights, ADG and FCR (Table 2).
3.2. Grape pomace altered gastrointestinal bacterial community structure
A total of 3,441,427 and 3,460,722 paired-end sequencing reads were obtained from four different parts of the GIT of the lambs using 16S rRNA gene sequencing for bacterial V3-V4 region and archaeal V4-V5 region, and 8,253 sequences and 6,430 OTUs were obtained by quality control and Vsearch clustering. In addition, 2055 and 297 OTUs were obtained by total-frequency-based filtering (samples with a total frequency of less than 31,488 and all features with a total abundance of less than ten were filtered), contingency-based filtering (filtered the feature present in at least two samples), and taxonomy-based filtering of tables (removed all features that contain either mitochondria or chloroplast, excluded the archaea at kingdom-level annotation in bacterial results and the bacteria in archaeal results) for subsequent analyses.
The first set of questions examined the feasibility of sequencing. The construction of the rarefaction curve (Figure 1A) demonstrated that the curve tended to plateau and, therefore, the sequencing data were large enough to reflect most of the microbial diversity information in the samples. Next, we analyzed the effect of diet and GIT position on alpha diversity (Figure 1B). The Chao1 index was lesser (p = 0.008) and the Shannon index tended to be lesser (p = 0.056) in the rumen of the GP than in CD lambs. In the GIT, the Shannon (p = 0.0005) and Chao1 indices (p = 0.031) in the rumen were greater than in the jejunum, and both were greater than in the cecum (Shannon: p = 0.031, Chao1: p = 0.006) and colon (Shannon: p = 0.006). The Shannon index of the jejunum was lesser than the cecum (Shannon: p < 0.001) and colon (Shannon: p < 0.001, Chao1: p = 0.046), and Chao1 index of the cecum was lesser than the colon (p = 0.046). We compared the beta diversity of microorganisms in CD and GP groups in different GIT sites; they were different (p = 0.001) and the figure separated the rumen, jejunum, and hindgut at the OTU level (Figure 1C). Only in the rumen (p = 0.051) and cecum (p = 0.096) did the bacterial composition tend to differ between the CD and GP groups (Figures 1D,E).
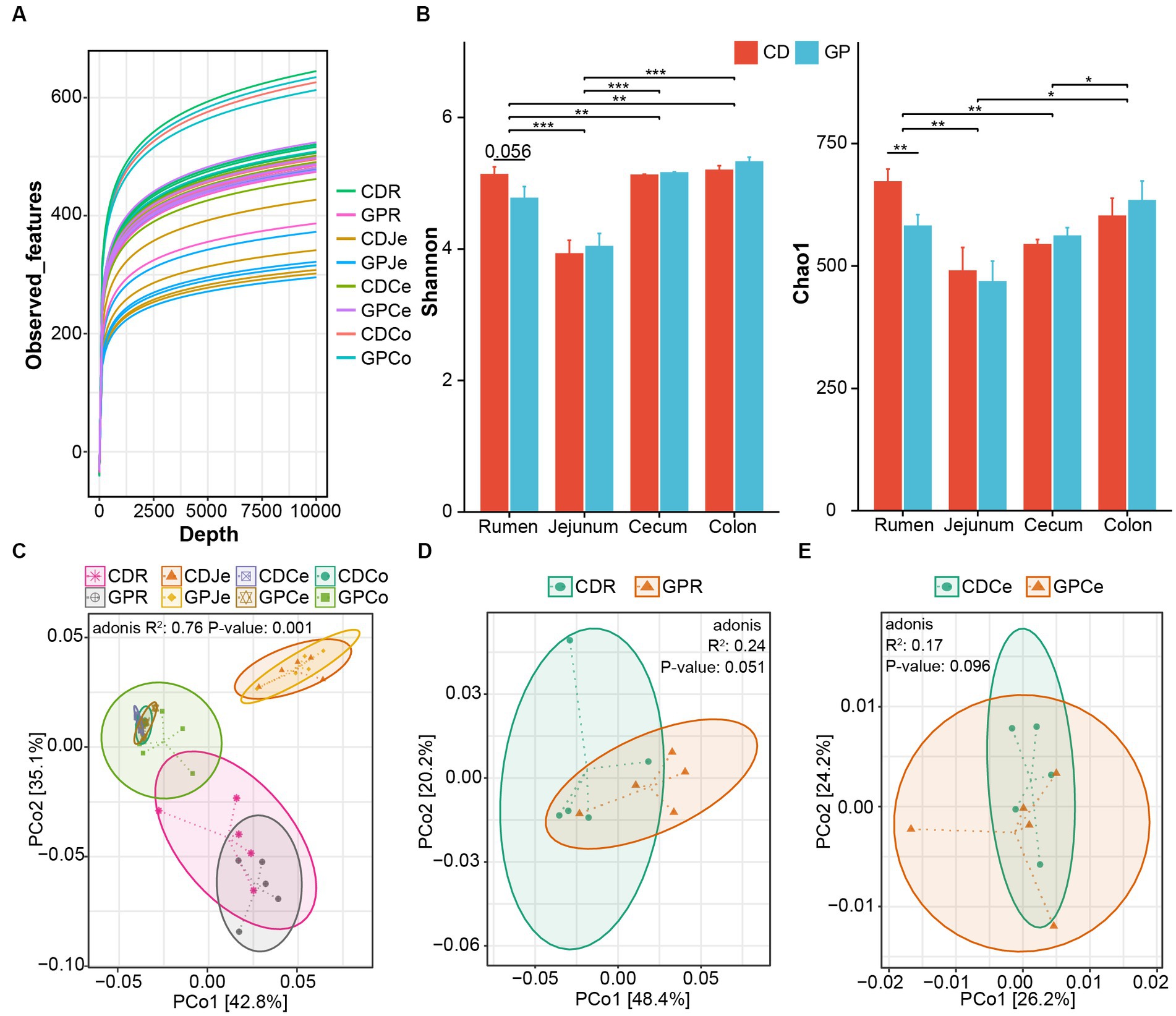
Figure 1. Effect of control (CD) and grape pomace (GP) diets on the bacterial community structure of the gastrointestinal tract (GIT). (A) The rarefaction curve calculated by observed features; (B) Alpha diversity representing species richness and evenness using Chao1 and Shannon indices of CD and GP in GIT; Principal-coordinate analysis (PCoA) of the GIT (C), rumen (D) and cecum (E) bacteria community at OTUs level by PERMANOVA analysis based on weight uniFrac distances. CDR, control diets rumen; GPR, GP rumen; CDJe, control diets jejunum; GPJe, GP jejunum; CDCe, control diets cecum; GPCe, GP cecum; CDCo, control diets colon; GPCo, GP colon. Data are presented as mean ± standard error of mean (SEM) (n = 5). ***p < 0.001, **0.001 < p < 0.01, *0.01 < p < 0.05.
3.3. Composition and biomarkers of gastrointestinal bacteria in different treatments
We analyzed the overlap and differences of the OTUs between the CD and GP groups. The number of OTUs in the GP group was lesser in the rumen and jejunum, but greater in the cecum and colon than in the CD group. Upset plots also display the zonal changes in bacterial OTUs at different sites and similarities in adjacent sites (Figure 2A). The dominant phyla (Figure 2B) were Firmicutes (CD: 52.7%, GP: 43.0%) and Bacteroidetes (CD: 35.2%, GP: 41.3%) in the rumen; Firmicutes (CD: 63.8%, GP: 60.4%), Actinobacteria (CD: 23.7%, GP: 29.4%) and Bacteroidetes (CD: 7.43%, GP: 6.22%) in the jejunum; Firmicutes (CD: 66.9%, GP: 69.1%) and Bacteroidetes (CD: 27.9%, GP: 26.4%) in the cecum; and Firmicutes (CD: 68.7, GP: 64.3%) and Bacteroidetes (CD: 26.6%, GP: 28.6%) in the colon. The dominant genera (Figure 2C) were Prevotella 1 (CD: 8.57%, GP: 20.3%), Rikenellaceae RC9 gut group (CD: 10.0%, GP: 5.68%) in the rumen; Bifidobacterium (CD: 13.4%, GP:18.0%), Romboutsia (CD: 15.4%, GP:7.84%), Paeniclostridium (CD: 11.42%, GP: 5.76%) in the jejunum; Ruminococcaceae UCG-005 (CD: 11.35%, GP:10.50%), Rikenellaceae RC9 gut group (CD: 9.92%, GP:7.88%) in the cecum; and Ruminococcaceae UCG-005 (CD: 10.62%, GP:6.69%), Rikenellaceae RC9 gut group (CD: 6.97%, GP: 6.34%), in the colon.
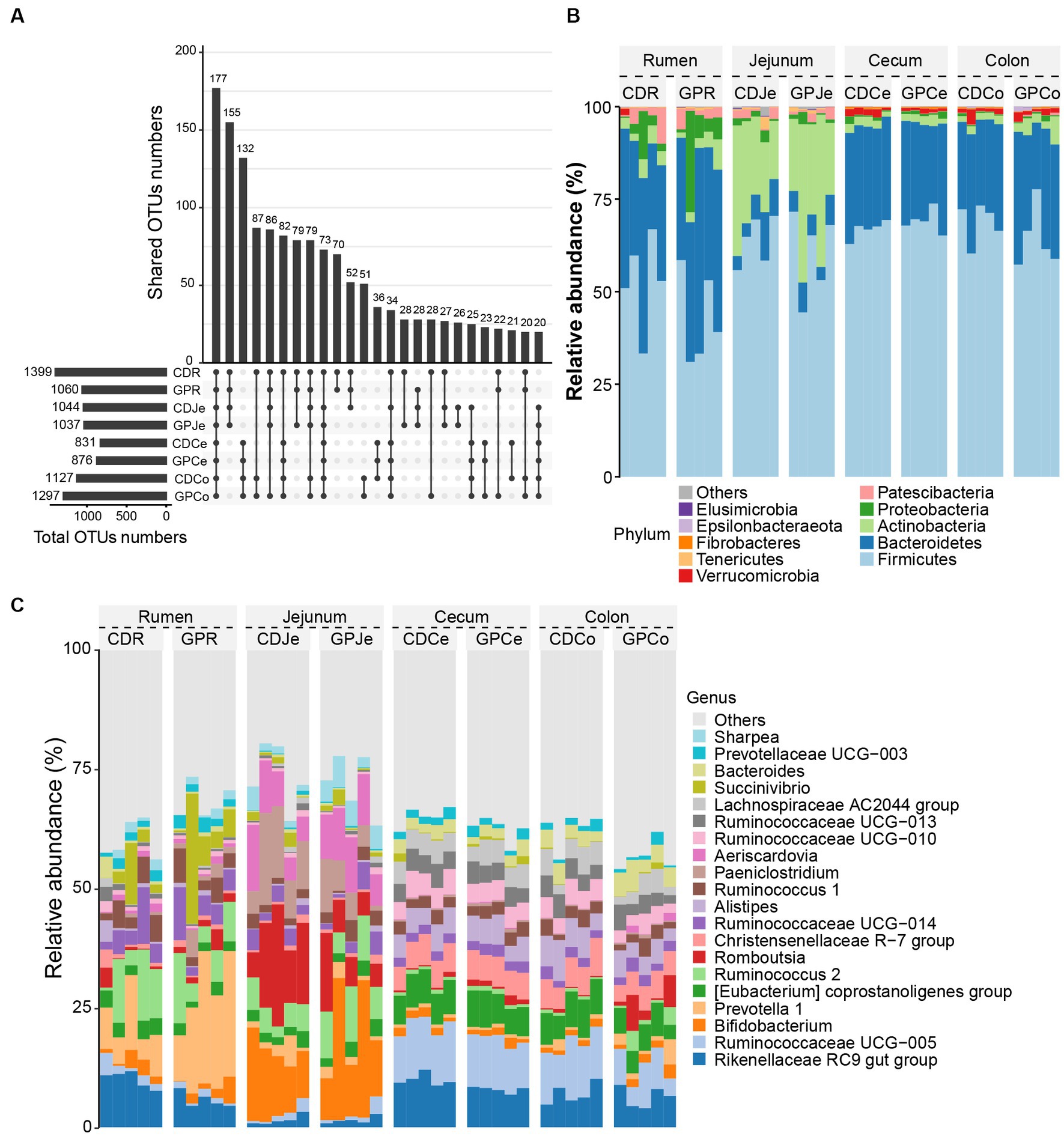
Figure 2. Composition of the gastrointestinal tract bacteria in each group. (A) The upset diagram in different treatments for different sites; (B) Top 10 bacteria at phylum level; (C) Top 20 bacteria at the genus level. CDR, control diet rumen; GPR, grape pomace diet rumen, CDJe, control diet jejunum; GPK, GP jejunum; CDCe, control diets cecum; GPCe, GP cecum; CDCo, control diets colon; GPCo, GP colon.
We screened for biomarkers at the four sites of the GP group. MetaStat analysis (Figure 3) revealed that there were differences between GP and CD in the GIT. Compared to the CD group, the relative abundances of Prevotella 1 (p = 0.01), Prevotella 7 (p = 0.008), Paeniclostridium (p = 0.030), and Clostridium sensu stricto 1 (p = 0.009) increased in the rumen of the GP group, while Rikenellaceae RC9 gut group (p = 0.02) and [Eubacterium] coprostanoligenes group (p = 0.008) decreased. Prevotella 1 displayed the greatest variation in the rumen. The abundance of several bacteria in the jejunum increased in the GP group, including Ruminococcus 2 (p = 0.041), Sharpea (p = 0.023), [Ruminococcus] gauvreauii group (p = 0.016), Olsenella (p = 0.037) and Mogibacterium (p = 0.008). The abundance of Ruminococcaceae UCG-014 (p = 0.030), Romboutsia (p = 0.039), Lachnoclostridium 10 (p = 0.020) and Paeniclostridium (p = 0.046) were greater in the cecum, and the abundance of Rikenellaceae RC9 gut group (p = 0.018), Akkermansia (p = 0.023) were lesser in the GP than CD group. In the colon of the GP group, similar trends were exhibited in that the abundance of Ruminococcaceae UCG-005 (p = 0.028), Ruminococcaceae UCG-010 (p = 0.049), Ruminococcaceae UCG-009 (p = 0.006), Ruminococcaceae UCG-002 (p = 0.006) and Oscillibacter (p = 0.043) were lesser, but of Prevotellaceae UCG-001 (p = 0.049) was greater than in the CD group.
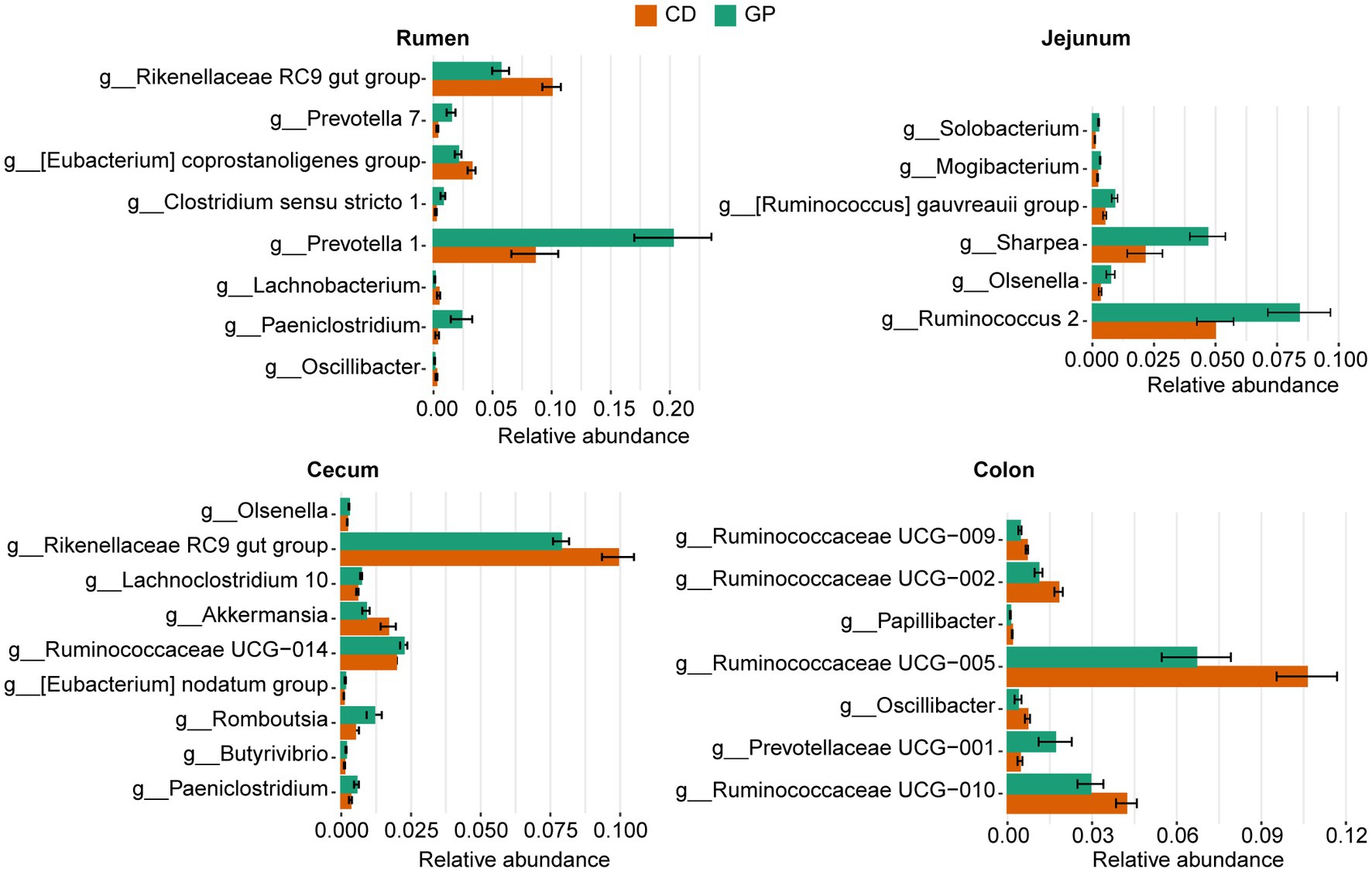
Figure 3. Bacteria at the genus level with significant differences in relative abundances (>0.1%) between control (CD) and grape pomace (GP) groups in the rumen, jejunum, cecum, and colon. Data are presented as mean ± standard deviation (SD) (n = 5). The bars are arranged in descending order of value of p.
3.4. PICRUSt2 predicted significant differences in Metacyc pathways
The functions in gastrointestinal bacteria following GP feed were predicted using PICRUSt2, in which the sequences and abundances were input, and gene family and pathway abundances were output. The functional predictions based on the MetaCyc database of metabolic pathways and enzymes obtained 388 pathways.
The differential pathways (>0.1%) after treatment were analyzed using Welch’s t-test in each region (Figure 4), with the most differential pathways in the colon and least in the cecum. In the rumen, the GP up-regulated the ASPASN-PWY (superpathway of L-aspartate and L-asparagine biosynthesis), PWY − 5,154 (L-arginine biosynthesis III) involved in vitamin biosynthesis, amino acid biosynthesis: PANTO−PWY (phosphopantothenate biosynthesis I), PWY − 6,892 (thiazole biosynthesis I) and carbohydrate degradation: and RHAMCAT−PWY (L-rhamnose degradation I); and down-regulated sugar metabolism PWY − 6,588 (pyruvate fermentation to acetone), REDCITCYC (TCA cycle VIII), PENTOSE−P − PWY (pentose phosphate pathway), PRPP−PWY (superpathway of histidine, purine, and pyrimidine biosynthesis), and PWY − 5,345 (superpathway of L-methionine biosynthesis). In the jejunum, the GP group up-graded the pathways in polysaccharide degradation: GLYCOCAT-PWY (glycogen degradation I), PWY-6737 (starch degradation V); but down-graded 11 pathways of amino acid metabolism: branched amino acid biosynthesis (ILEUSYN-PWY, ILEUSYN-PWY, PWY-5103, VALSYN-PWY, BRANCHED-CHAIN-AA-SYN-PWY), arginine (ARGSYNBSUB-PWY), serine (SER-GLYSYN-PWY) e.g., folate biosynthesis (PWY-7539, PWY-6147) and thiamine biosynthesis (PWY-6892). In the cecum, the pathways of PWY-6317 (galactose degradation I), PWY0-781 (aspartate superpathway) were up-graded, but vitamin biosynthesis (PYRIDOXSYN-PWY, PWY0-845) was down-regulated. The GP up-regulated seven amino acid metabolism pathways (HOMOSER-METSYN-PWY, P4-PWY, PWY0-781, PWY-5347, PWY-6630, PWY-6628), mannan degradation (PWY-7456), and mixed acid fermentation (TFERMENTATION-PWY); and down-regulated 11 pathways linked with amino acid metabolism such as L-isoleucine biosynthesis (PWY-5101, ILEUSYN-PWY, PWY-5103), L-valine biosynthesis (VALSYN-PWY), L-lysine biosynthesis (PWY-2942); carbohydrate metabolism: TCA cycle VIII (REDCITCYC), nicotinamide adenine dinucleotide biosynthesis I (PYRIDNUCSYN-PWY), pyruvate fermentation to acetone (PWY-6588); cofactor, carrier, and vitamin biosynthesis (COA-PWY, 1CMET2-PWY, PYRIDNUCSYN-PWY) and nucleoside and nucleotide biosynthesis (PWY-5686, PWY-6122, PWY-6277, PWY-7208, PWY-7219) in the colon.
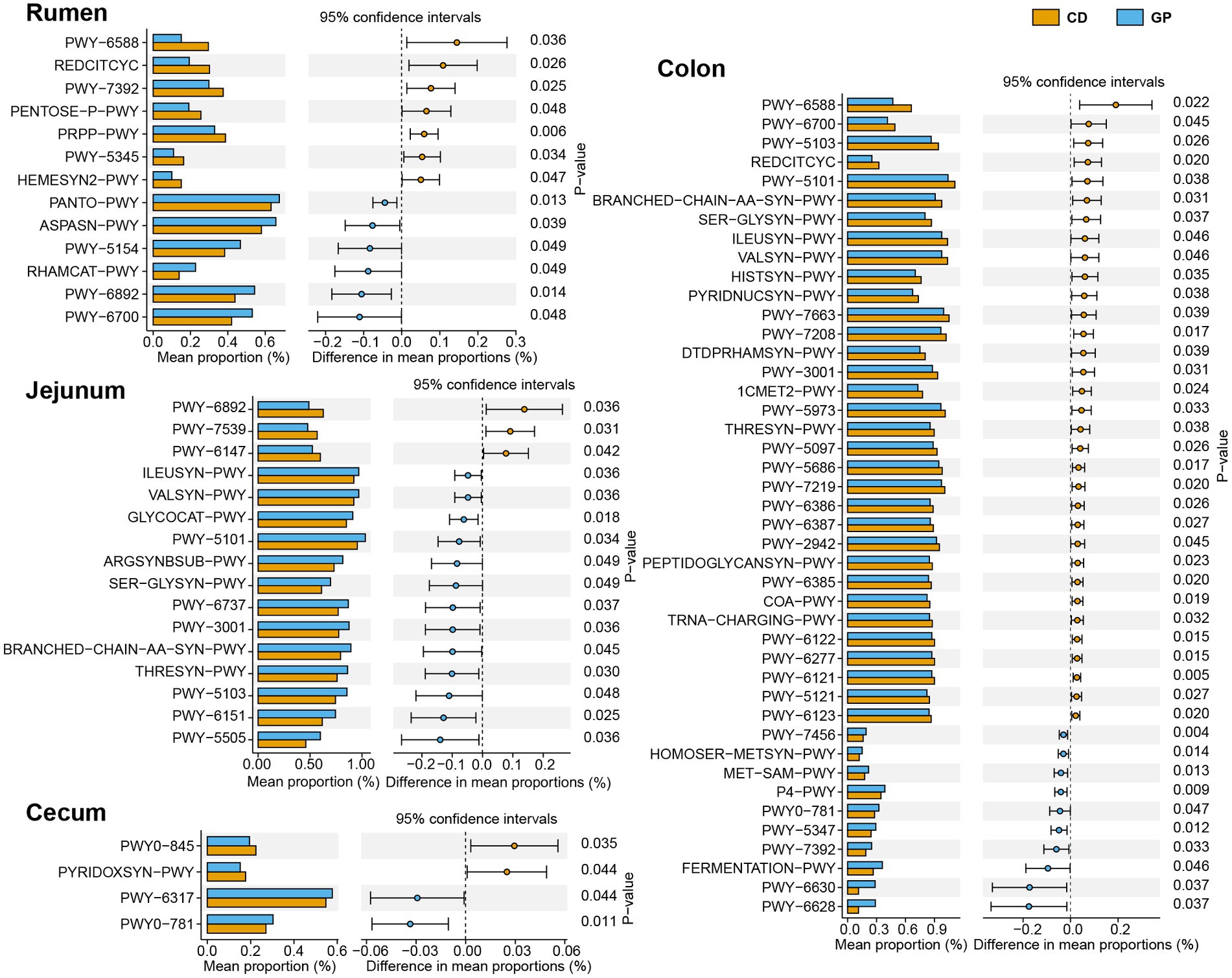
Figure 4. Significantly different MetaCyc pathways in rumen, jejunum, cecum, and colon. The error bars on the columns represent the standard error of mean (SEM).
3.5. Correlation of bacteria at the genus level and MetaCyc pathways
There were strong correlations between most of the bacteria and pathways in different sites (Figure 5). In general, there was a positive correlation between up-graded pathways and the genera that were most abundant in CD; whereas, the less abundant genera in CD displayed negative correlations. In general, there were positive correlations between up-graded pathways and the genera that were most abundant in CD, while, the least abundant genera in CD were correlated negatively. For rumen bacteria, the GP-altered microbes exhibited a functional distinction. In GP lambs, the relative abundances of the rumen Prevotella 1, Prevotella 7, Paeniclostridium, and Clostridium sensu stricto 1 were correlated positively with the up-regulated pathways of L-arginine biosynthesis, queuosine biosynthesis, peptidases, and inhibitors, glycerophospholipid metabolism, and phosphopantothenate biosynthesis, and negatively with the down-regulated pathways. There were positive correlations between the low abundant bacteria in CD, including the Rikenellaceae RC9 gut group, [Eubacterium]coprostanoligenes group, and Oscillibacter with pyruvate fermentation to acetone, TCA cycle Vlll, and the down-regulated pathways. However, there were negative correlations between these bacteria and the up-regulated pathways. In the jejunum, the six significantly different bacteria species were all greater in the GP than CD group. In the jejunum, the GP group had six bacteria species with greater abundance than in the CD group; those genera were correlated positively with amino acid biosynthesis, starch and glycogen digestion. Solobacterium, [Ruminococcus] gauvreauii group and Mogibacterium were correlated positively with most up-regulated pathways. The main changed microorganisms of jejunum were correlated negatively with folate biosynthesis (PWY-7539, PWY-6147) and thiamine biosynthesis (PWY-6892). In the cecum, the Rikenellaceae RC9 gut group was correlated positively with vitamin biosynthesis (PYRIDOXSYN-PWY, PWY0-845), but negatively with the up-graded pathways (galactose degradation and aspartate superpathway). Romboutsia, Olsenella, Paeniclostridium, Butyrivibrio were correlated positively with vitamin biosynthesis (PYRIDOXSYN-PWY, PWY0-845), and negatively with the down-graded pathways. In the colon, the genus Prevotellaceae UCG-001 was correlated positively with MET-SAM-PWY (superpathway of S-adenosyl-L-methionine biosynthesis), Prevotellaceae UCG-001 was correlated negatively with the down-graded pathways, while Ruminococcaceae UCG-002, Ruminococcaceae UCG-010, and Papillibacter were correlated positively with most down-regulated pathways. Ruminococcaceae UCG-005 and Ruminococcaceae UCG-09 were correlated positively with isoleucine (ILEUSYN-PWY, PWY-5101), valine (VALSYN-PWY), lysine (PWY-2942, PWY-5097), histidine (HISTSYN-PWY), and threonine (HRESYN-PWY) biosynthesis.
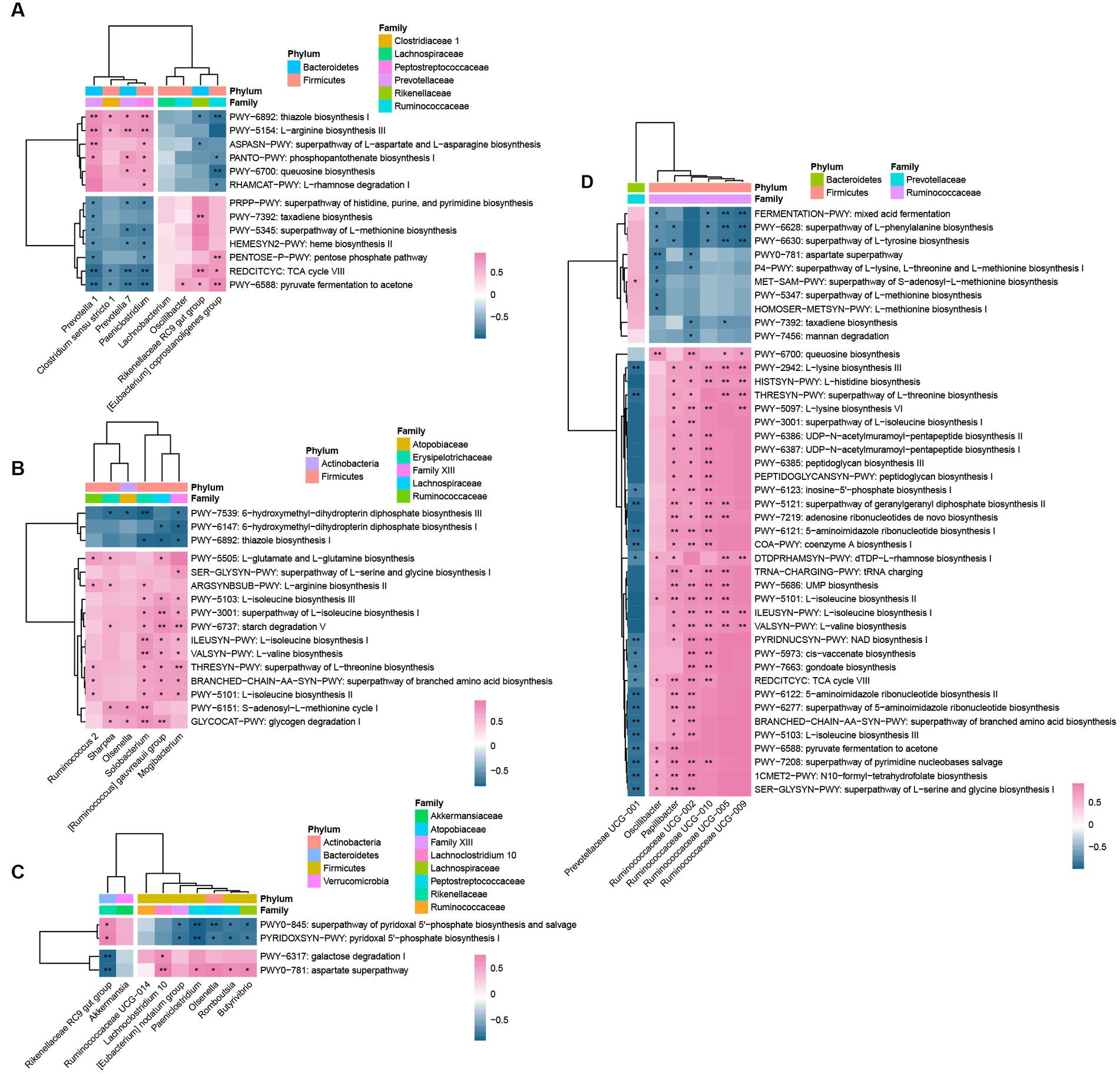
Figure 5. Spearman correlations between genera and pathways in rumen (A), jejunum (B), cecum (C), and colon (D). For each heatmap, rows correspond to bacteria at the genus level, columns correspond to MetaCyc pathways. Red and blue represent the positive and negative correlations, respectively. The intensity of the colors denotes the degree of correlation between the genera abundances and the pathways. **p < 0.01, * p < 0.05.
3.6. The difference in gastrointestinal archaea diversity and biomarkers
Alpha and beta diversities of archaea were used to characterize species diversity within and between GIT segments. Sequencing analysis of archaea from different segments of the GIT revealed that the Shannon index (p = 0.056) tended to be greater in the rumen of GP than CD (Supplementary Figure S1A); however, the Chao1 index did not differ between dietary treatments in the different sites of the GIT. The beta diversity of each site (Supplementary Figure S1B) also displayed differences (p = 0.001), with the GP diets altering the beta diversity of the jejunum (p = 0.006) and colon (p = 0.008) at the OTUs level (Supplementary Figures S1C,D).
To characterize the archaeal composition, the relative abundances of the top 10 phyla and genera were charted. At the phylum level (Supplementary Figure S2A), in the rumen and jejunum, Euryarchaeota (CDR:95.4%, GPR:91.1%, CDJe:99.1%, GPJe:97.2%) dominated, followed by Halobacterota (CDR:0.16%, GPR: 5.05%, CDJe:0.69%, GPJe:2.47%) and Thermoplasmatota (CDR:4.56%, GPR: 3.32%, CDJe:0.23%, GPJe:0.34%), while in the cecum and colon, Halobacterota (CDCe:46.8%, GPCe:42.5%, CDCo:16.1%, GPCo:56.5%) and Euryarchaeota (CDCe:53.0%, GPCe:56.8%, CDCo:83.8%, GPCo:42.9%) dominated. At the genus level (Supplementary Figure S2B), Methanobrevibacter (CDR:95.2%, GPR: 91.3, CDJe:99.7%, GPJe:97.2%) dominated in the rumen and jejunum, followed by Methanocorpusculum (CDR:0.10%, GPR:4.80%, CDJe:0.67%, GPJe:2.46%) and Methanosphaera (CDR:0.90%, GPR:2.60%, CDJe:4.20%, GPJe:6.17%); and in the cecum and colon, Methanocorpusculum (CDCe:95.2%, GPCe:91.3, CDCo:99.7%, GPCo: 97.2%) and Methanobrevibacter (CDCe:52.4%, GPCe:56.0%, CDCo:83.3%, GPCo: 42.0%) dominated, followed by Methanocorpusculum (CDCe:46.8%, GPCe:42.5%, CDCo:15.9%, GPCo:52.3%).
The Metastats analysis detected the change of archaea in different sites in the GIT at the genus level (Supplementary Figure S2C). The dominant genera were Methanobrevibacter and Methanocorpusculum, but the change in the cecum was from other genera. Methanobrevibacter was lesser in the rumen (p = 0.01), jejunum (p = 0.06) and colon of GP (p = 0.007), but greater in the cecum (p = 0.007) in the CD than GP group. Moreover, the abundance of Methanocorpusculum was greater in the rumen (p = 0.03), jejunum (p = 0.06) and colon (p = 0.009) and tended to be lesser in the cecum (p = 0.09) in the GP than CD group.
We used PICRUSt2 to predict changes in archaeal function in different GIT sites by the MetaCyc database, and 262 pathways were obtained. The relative abundances of pathways greater than 0.1% were tested for differences between dietary groups at each GIT site. The pathways of the jejunum and colon were distinct, whereas the cecum did not show any difference (Figure 6). The incomplete reductive TCA cycle (P42-PWY) was up-regulated, while phosphopantothenate biosynthesis III (PWY-6654) was down-regulated in the rumen of the GP group. In the jejunum in the GP group, only the pathway of incomplete reductive TCA cycle (P42-PWY) was up-regulated, while 14 amino acid metabolism pathways, including arginine biosynthesis (PWY-7400, ARGSYN-PWY, ARGSYNBSUB-PWY), isoleucine biosynthesis (PWY-5104, ILEUSYN-PWY, PWY-5103, PWY-3001), and lysine biosynthesis (PWY-2942, PWY-5097), were down-regulated. In the colon, the GP group had a greater enrichment of pathways in the carbohydrate metabolism as incomplete reductive TCA cycle (P42-PWY), guanosine diphosphate mannose biosynthesis (PWY-5659), superpathway of guanosine diphosphate mannose-derived O-antigen building blocks biosynthesis (PWY-7323), and 5 purine nucleotide biosynthesis, including superpathway of guanosine nucleotides de novo biosynthesis I (PWY-7228), superpathway of guanosine nucleotides de novo biosynthesis II (PWY-6125), adenosine deoxyribonucleotides de novo biosynthesis II (PWY-7220), guanosine deoxyribonucleotides de novo biosynthesis II (PWY-7222), and superpathway of adenosine nucleotides de novo biosynthesis II (PWY-6126). Furthermore, methanogenesis from H2 and CO2 (methanogenesis-PWY), 15 amino acid biosynthesis such as lysine (PWY-2942, PWY-5097), arginine (PWY-7400, ARGSYN-PWY, ARGSYNBSUB-PWY), methionine (HSERMETANA-PWY), isoleucine (PWY-3001, PWY-5103, ILEUSYN-PWY, PWY-5104, PWY-5101), six co-factor, carrier, and vitamin biosynthesis: factor 420 biosynthesis (PWY-5198), co-enzyme M biosynthesis I (P261-PWY), co-enzyme B biosynthesis (P241-PWY), thiamin salvage II (PWY-6897), flavin biosynthesis II (PWY-6167), co-enzyme A biosynthesis I (COA-PWY); and pyruvate fermentation to isobutanol (PWY-7111) were down-regulated.
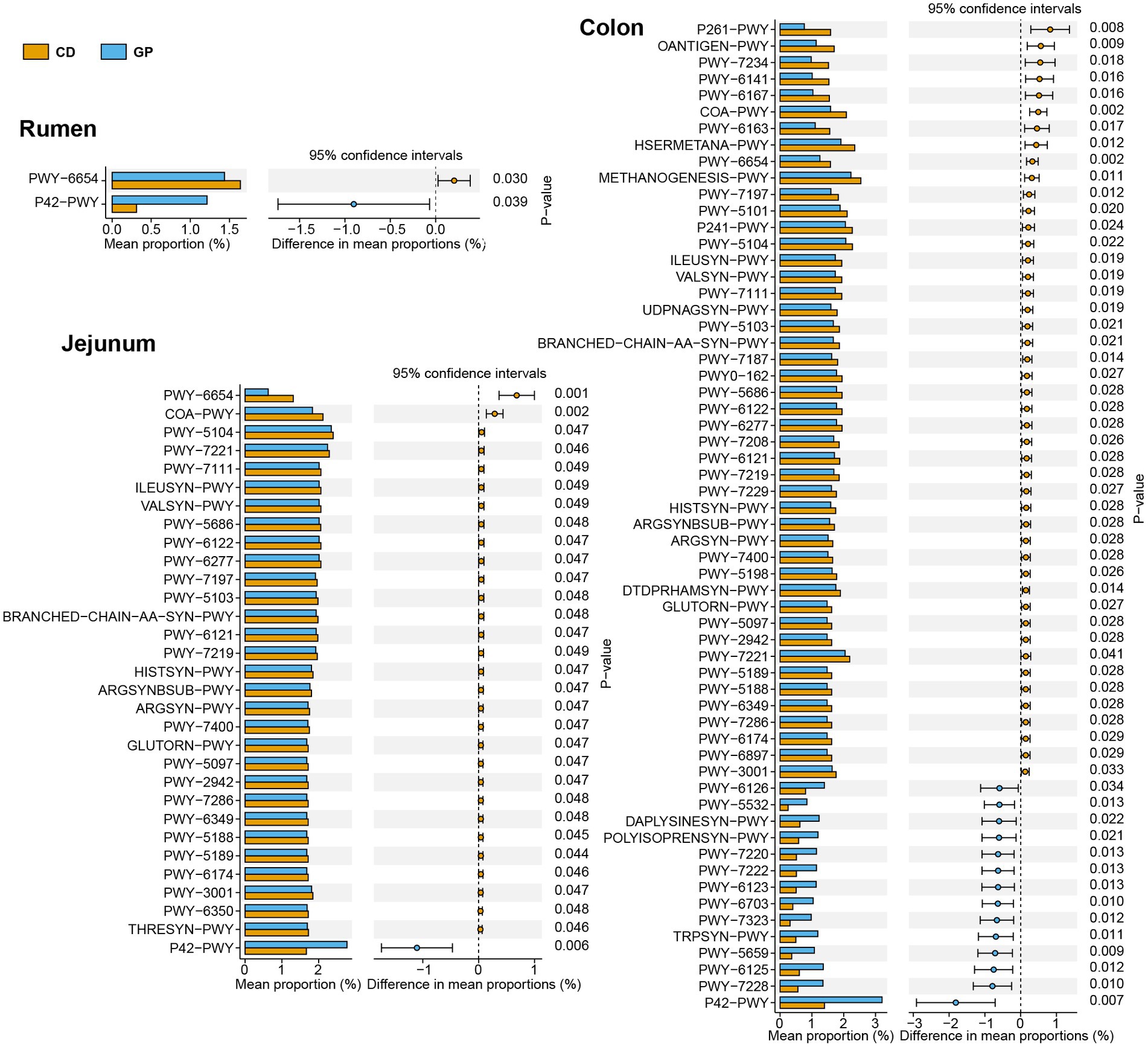
Figure 6. Different archaeal prediction pathways at different regions in Tan lambs after consuming grape pomace. The error bars on the columns represent the standard error of mean (SEM).
4. Discussion
Dietary composition is the main factor influencing gastrointestinal microbiology. In the present study, GP reduced the alpha diversity and altered the community structure of rumen bacteria, and the community structure of jejunal and colonic archaea. Host metabolism was affected by altering the relative abundances of some bacteria (Prevotella, Rikenellaceae RC9 gut group, and Ruminococcus) and archaea (Methanobrevibacter and Methanocorpusculum). The GP induced alterations in the bacteria associated with fermentation in the rumen and colon and changes in their metabolic functions and fermentation patterns, while inhibiting the metabolic activity of colonic archaea.
Under iso-energetic and iso-nitrogenous conditions, replacing 8% of corn with GP and increasing soybean oil in the diet resulted in higher DMI. The high energy soybean oil (16.2 g at 34.7 kJ DE/g) was added to compensate for the low energy GP (80 g at 6.8 kJ DE/g), so that the diets would be iso-energetic and iso-nitrogenous. This increase in intake in the GP diet could be attributed to the potential improvement in palatability due to the soybean oil (Macleod and Buchanan-Smith, 1972). Similar to a previous study, a dietary GP of approximately 10% resulted in an increased DMI in sheep (Zhao et al., 2018). Comparison of the two iso-energetic and iso-nitrogenous diets, a diet with 8% GP did not improve growth performance or FCR, and, consequently our first hypothesis was not supported. However, the present results are consistent with other studies that reported diets with less than 20% GP did not affect FCR in sheep (e.g., Chikwanha et al., 2019). The FCR in the Tan lambs in the present study was relatively high, 8.21 and 9.03; but similar results have been reported in other studies. For example, Liu et al. (2021) reported a FCR of 9.1 with ad libitum intake and a range between 8.1 and 10.3 with different intakes for Tan sheep. Tan sheep are hardy and well adapted to a dry, cold and windy environment, but FCR is poor.
Broadly consistent with previous studies (Zhang et al., 2018; Xie et al., 2021), bacteria and archaea of the GIT displayed spatial heterogeneity in community structure and composition, as he GIT microbiome differed among rumen, small intestine, and hindgut. The alpha diversity of rumen bacteria decreased, while there were marked changes in the bacterial community structure with the consumption of GP. These results differed from those of Rolinec et al. (2023) who reported that the consumption of a diet with 1 and 2% GP increased alpha diversity among rumen bacteria and stimulated higher levels of methanogen abundance. The difference among studies may be attributed to differing levels of dietary GP. It is possible that the polyphenols in GP inhibit bacteria. Polyphenols have a stronger bactericidal effect on gram-positive bacteria than on gram-negative bacteria due to the structure of their cell walls and the arrangement of their outer membranes. These polyphenols modify the membrane’s permeability and integrity of the cell walls of gram-negative bacteria in the rumen (Vasta et al., 2019).
In the present study, although the dominant bacterial species in the rumen and colon differed, their bacteria and functions were similar. Prevotella 1 was the dominant bacterial genus in the rumen of sheep (Henderson et al., 2015; Li et al., 2020). This genus degrades carbohydrates (starch, hemicellulose, and pectin) and proteins to produce succinic and acetic acids (Krieg et al., 2010; Accetto and Avguštin, 2019). The association of Prevotella with propionate production has been reported previously (Strobel, 1992; Purushe et al., 2010; Betancur-Murillo et al., 2023). Propionate is the major substrate for hepatic gluconeogenesis in ruminants, with an energy utilization efficiency of 109% (Orskov, 2012), competes for hydrogen in propionate production by Prevotella, and also reduces the production of methane (Denman et al., 2015). Ruminococcaceae are the dominant fiber fermentation bacteria in the GIT of sheep, producing mainly acetate (Peng et al., 2021; Ma et al., 2022). The reduction in fiber-degrading bacteria may be due to the binding of tannins to the fibers or their inhibitory effect on microbial activity. Tannins can prevent microorganisms from attaching to the fiber, and gram-positive specialized fibrolytic bacteria are very susceptible to tannins (McSweeney et al., 2001). Previous research demonstrated that distiller’s dried grains with solubles (DDGS) and tannin-rich diets reduced the abundance of rumen Ruminoccoccaceae and increased the abundance of Prevotella (Khiaosa-ard et al., 2015; Pannell et al., 2022), as was observed in the present study.
The up-regulated pathways in the rumen and colon were all related to glycolysis. L-rhamnose, a part of complex pectin polysaccharides, produced pyruvate upon degradation and increased propionate production in in vivo and in vitro studies (Hosseini et al., 2011). Mannan, a major component of plant hemicellulose (Moreira and Filho, 2008), promotes the glycolytic pathway, that is, the mixed acid (from glycolysis) fermentation to lactate, succinate, and acetate. The plant structural carbohydrates and hemicellulose are rich in pentose, and down-regulating the pentose phosphate pathway decreases acetate. Down-regulation of the TCA cycle inhibits acetyl-co-enzyme A conversion to reduce further the precursors for acetate and butyrate synthesis (Ungerfeld, 2020). Furthermore, thiamin diphosphate and phosphopantothenate, also known as vitamin B1 and B5, were up-regulated pathways, and, therefore, GP promotes the synthesis of B-vitamins in the rumen. The microbial synthesis of vitamin B in the rumen meets the vitamin requirement of the host and is a co-factor for various enzymes involved in the metabolism of fatty acids, carbohydrates, and amino acids. Thiamin diphosphate is an essential co-factor for a variety of enzymes, such as transketolase, pyruvate dehydrogenase, pyruvate decarboxylase, and α-ketoglutarate dehydrogenase (Lawhorn et al., 2004). Phosphopantothenate is an essential precursor for the synthesis of co-enzyme A, a co-factor in many intermediate metabolic pathways (Begley et al., 2001).
The jejunum digests and metabolizes starch and absorbs nutrients. In the GP group, the dominant genera belonged mainly to Firmicutes, which up-regulated bacterial starch, glycogen degradation, and amino acid biosynthesis. Sharpea and [Ruminococcus] gauvreauii group enhance the degradation of starch and glycogen. Sharpea ferments starch and melibiose to produce lactic acid and CO2 (Morita et al., 2008); lactic acid reduces gut pH and inhibits the proliferation of pathogenic bacteria, and, consequently, Sharpea is beneficial for the host’s health (Xu et al., 2018; Ma et al., 2020). Ruminococcus 2 (family Ruminoccoccaceae) ferments starch to produce propionate (Tong et al., 2018; Chen et al., 2021), while [Ruminococcus] gauvreauii group, (Lachnospiraceae) ferments glucose into acetic acid (Domingo et al., 2008). Mogibacterium ferments starch and produces phenylacetate, but does not ferment carbohydrates (Schleifer, 2009), and its abundance increased when fed a high grain diet (Liu et al., 2015). The up-regulation of several amino acid synthesis pathways in the jejunum may be related to the polyphenols in GP, which bind to proteins, slowing down the degradation in the rumen and increasing protein metabolism in the small intestine (Guerra-Rivas et al., 2017).
The ruminant hindgut, which includes the cecum and colon, also ferments carbohydrates. However, differences in cecal bacteria and archaea between CD and GP were minor. The abundance of Rikenellaceae RC9 gut group decreased, but of Ruminococcaceae UCG-014, Romboutsia and Lachnoclostridium increased in the GP group. GP enriched the galactose degradation and aspartate superpathway. It was reported that the abundance of Ruminococcaceae UCG-014 and Olsenella were correlated positively with feed efficiency in the rumen (Elolimy et al., 2020; McLoughlin et al., 2020). There was an increased abundance of these two bacteria in the cecum in the GP lambs, which may have promoted carbohydrate fermentation and improved feed efficiency. Olsenella was the only Actinobacterium with substantial variation, increased in both jejunum and cecum, and was correlated positively with glycogen and galactose depletion. Olsenella ferments starch and glycogen substrates to produce lactic, acetic, formic, and succinic acids (Göker et al., 2010; Kraatz et al., 2011). Paeniclostridium increased in both the rumen and cecum, and correlated with altered pathways in the rumen. Although this bacterium is potentially pro-inflammatory, as it produces hemorrhagic toxins that cause loss of gastrointestinal tissue and body disease (Lee et al., 2020), it was reported to be the predominant bacterium in the rumen of sheep (Zhao et al., 2023). Paeniclostridium has not been found in in vivo studies in which GP or polyphenols was fed to sheep. Some potential roles of Paeniclostridium were discovered in the bovine intestine, where the glycolytic pathways were up-regulated by the increased abundance of Paeniclostridium and Romboutsia (Sim et al., 2022). There appears to be a relationship between Paeniclostridium, Romboutsia, or Clostridium sensu stricto 1 and the fermentation process as they are the dominant bacteria in avocado oil processing waste feedstocks and dark fermentative hydrogen production (Yang and Wang, 2019; Ijoma et al., 2022). Clostridium sensu stricto 1 can be fermented to produce butyric acid (Wang et al., 2017).
Rikenellaceae RC9 gut group and [Eubacterium] coprostanoligenes group are the main bacteria that decreased in relative abundances with GP. Rikenellaceae RC9 gut group is associated with rumen cellulose degradation, and it was reported that the ruminal abundance decreased in sheep fed high grain diets and diets with low NDF (Zened et al., 2013; Zhang et al., 2017). The [Eubacterium] coprostanoligenes group reduces cholesterol to coprostanol and ferments carbohydrates to produce acetic, formic and succinic acids (Freier et al., 1994). The ruminal abundance increased in animals fed a high energy, high lignin diet (Khatoon et al., 2022), and these bacteria were correlated positively with the TCA cycle, pentose phosphate pathway, and pyruvate fermentation to acetone. GP reduced the abundance of the Rikenellaceae RC9 gut group and the [Eubacterium] coprostanoligenes group, which may affect carbohydrate metabolism. We, therefore, propose that GP increases the abundance of propionate-producing and starch-fermenting bacteria and pathways, and decreases cellulose metabolism, thereby altering gastrointestinal fermentation, which supports our second hypothesis. Previous studies demonstrated that GP promotes propionate production and reduces acetate concentration (Foiklang et al., 2016a,b). The cashew nut shell liquid is rich in polyphenols, as is grape pomace. It has a comparable effect on rumen fermentation and reduces methane emissions (via hydrogenotrophic methanogenesis) by directly inhibiting hydrogenotrophic methanogenesis and reducing the copy number of the mcrA gene, fiber degradation, and the resulting H2 production (Shinkai et al., 2012).
Archaea are the only methane producers in the GIT (Janssen and Kirs, 2008). Four main types of methanogenic pathways have been identified: the pathway from H2 and CO2, the aceticlastic pathway from acetate, methanogenesis from methylated compounds, and methanogenesis from methoxylated aromatic compounds. The H2 and CO2 and methyltropic pathways are the main sources of soluble methane production in the gut (Beauchemin et al., 2020). The rumen and hindgut are the main sites of methane production, where methanogens play an essential role. In the GIT (except cecum), the abundance of Methanobrevibacter decreased, of Methanocorpusculum increased, and the pathways of incomplete reductive TCA cycle were up-regulated. The differences in archaea and pathways between CD and GP were particularly noticeable in the colon. Consistent with findings of earlier studies (Henderson et al., 2015), the most common genus of archaea was Methanobrevibacter, which can convert H2 or acetate to produce methane (Leahy et al., 2013). Methanocorpusculum occurs in the hindgut and feces of cattle and sheep and is associated with methane production (Ngetich et al., 2022), but the mechanism of action warrants further study. GP can inhibit methane production, due to the polyphenols, such as tannins, which inhibit methanogens and protozoa, and reduce fiber degradation and H2 production (Moate et al., 2014). In the current study, GP decreased the relative abundance and function of Methanobrevibacter in the GIT, which would support our third hypothesis. Methanogens are incapable of a complete oxidative or reductive TCA cycle, and incomplete cycles produce biosynthetic intermediates (Goodchild et al., 2004). However, in response to anaerobic or microaerophilic growth conditions, these incomplete cycles can still convert pyruvate into necessary biosynthetic intermediates (Wood et al., 2004). In the colon, the pathways of methanogenesis from H2 and CO2, the essential co-factors (co-enzyme M, co-enzyme B, and factor 420) of methanogenesis were down-regulated. All catabolic processes in methanogenesis require the reduction of the methylated form of co-enzyme M to methane by co-enzyme B, and factor 420 also acts as an electron carrier (Deppenmeier, 2002). GP lowers methane production by increasing propionate-producing microbes and pathways.
5. Conclusion
The current study demonstrated that a diet containing 8% dietary GP by dry weight increased DMI but did not affect ADG and FCR in Tan lambs. The GP enhanced the abundance of Prevotella 1, Ruminococcus 2 and Sharpea, decreased the acetate-producing Ruminococcaceae and methane-producing Methanobrevibacter, stimulated starch metabolism, mixed acid fermentation, and intestinal amino acid and rumen B-vitamins biosynthesis, and down-regulated colonic methanogenesis. Overall, GIT microbiome altered fermentation to promote propionate production and degradation of starch, decrease cellulose fermentation and methanogenesis. In spite of these positive effects on the energy balance of the lambs, the ADG and FCR were not improved with a dietary supplement of 8% GP. Future studies with different GP levels are warranted. It is possible that levels of less than 8% GP would be beneficial for the entire GIT, reduce overall methane production and improve lamb performance.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA877028.
Ethics statement
The animal studies were approved by the Academic Committee of the Northwestern Institute of Eco-Environment Resources, Chinese Academy of Sciences. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
XC: Investigation, Software, Writing — original draft, Visualization. XD: Investigation, Formal analysis, Writing — review & editing. YL: Investigation, Formal analysis, Writing — review & editing. AD: Writing — review & editing. XW: Investigation, Software, Writing — review & editing. KJ: Investigation, Software, Writing — review & editing. QG: Methodology, Resources, Writing — review & editing. GX: Methodology, Resources, Writing — review & editing. HC: Methodology, Resources, Writing — review & editing. GY: Conceptualization, Resources, Writing — review & editing, Project administration and Funding acquisition.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was funded by the grants from the Strategic Priority Research Program (A) of Chinese Academy of Sciences (Grant No. XDA26040301), the Construction of Rural Revitalization Industry Technology Innovation Center and Demonstration Base in the Yellow River Delta (Grant No. E141050101), and the Key Research and Development Program of in Gansu Province (Grant No. 22ZD6NA037).
Conflict of interest
HC is employed by Shandong Huakun Rural Revitalization Institute Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1264840/full#supplementary-material
References
Abarghuei, M. J., Rouzbehan, Y., and Alipour, D. (2010). The influence of the grape pomace on the ruminal parameters of sheep. Livest. Sci. 132, 73–79. doi: 10.1016/j.livsci.2010.05.002
Accetto, T., and Avguštin, G. (2019). The diverse and extensive plant polysaccharide degradative apparatuses of the rumen and hindgut Prevotella species: A factor in their ubiquity? Syst. Appl. Microbiol. 42, 107–116. doi: 10.1016/j.syapm.2018.10.001
AOAC International (2007). Official methods of analysis of AOAC international, 18th Ed. methods 991.42 and 993.19. Gaithersburg, MD: AOAC International
Arthur, P. F., Archer, J. A., and Herd, R. M. (2004). Feed intake and efficiency in beef cattle: overview of recent Australian research and challenges for the future. Aust. J. Exp. Agric. 44, 361–369. doi: 10.1071/EA02162
Beauchemin, K. A., Ungerfeld, E. M., Eckard, R. J., and Wang, M. (2020). Review: fifty years of research on rumen methanogenesis: lessons learned and future challenges for mitigation. Animal 14, s2–s16. doi: 10.1017/S1751731119003100
Begley, T. P., Kinsland, C., and Strauss, E. (2001). The biosynthesis of coenzyme A in bacteria. Vitam. Horm. 61, 157–171. doi: 10.1016/s0083-6729(01)61005-7
Beres, C., Costa, G. N. S., Cabezudo, I., da Silva-James, N. K., Teles, A. S. C., Cruz, A. P. G., et al. (2017). Towards integral utilization of grape pomace from winemaking process: a review. Waste Manag. 68, 581–594. doi: 10.1016/j.wasman.2017.07.017
Besharati, M., Maggiolino, A., Palangi, V., Kaya, A., Jabbar, M., Eseceli, H., et al. (2022). Tannin in ruminant nutrition: review. Molecules 27:8273. doi: 10.3390/molecules27238273
Besharati, M., and Taghizadeh, A. (2009). Evaluation of dried grape by-product as a tanniniferous tropical feedstuff. Anim. Feed Sci. Technol. 152, 198–203. doi: 10.1016/j.anifeedsci.2009.04.011
Betancur-Murillo, C. L., Aguilar-Marín, S. B., and Jovel, J. (2023). Prevotella: a key player in ruminal metabolism. Microorganisms 11:1. doi: 10.3390/microorganisms11010001
Biscarini, F., Palazzo, F., Castellani, F., Masetti, G., Grotta, L., Cichelli, A., et al. (2018). Rumen microbiome in dairy calves fed copper and grape-pomace dietary supplementations: composition and predicted functional profile. PLoS One 13:e0205670. doi: 10.1371/journal.pone.0205670
Bokulich, N. A., Kaehler, B. D., Rideout, J. R., Dillon, M., Bolyen, E., Knight, R., et al. (2018). Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6:90. doi: 10.1186/s40168-018-0470-z
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Buffa, G., Mangia, N. P., Cesarani, A., Licastro, D., Sorbolini, S., Pulina, G., et al. (2020). Agroindustrial by-products from tomato, grape and myrtle given at low dosage to lactating dairy ewes: effects on rumen parameters and microbiota. Ital. J. Anim. Sci. 19, 1462–1471. doi: 10.1080/1828051X.2020.1848465
Caspi, R., Billington, R., Keseler, I. M., Kothari, A., Krummenacker, M., Midford, P. E., et al. (2020). The MetaCyc database of metabolic pathways and enzymes - a 2019 update. Nucleic Acids Res. 48, D445–D453. doi: 10.1093/nar/gkz862
Chen, X., Su, X., Li, J., Yang, Y., Wang, P., Yan, F., et al. (2021). Real-time monitoring of ruminal microbiota reveals their roles in dairy goats during subacute ruminal acidosis. npj Biofilms Microbomes 7:45. doi: 10.1038/s41522-021-00215-6
Chikwanha, O. C., Muchenje, V., Nolte, J. E., Dugan, M. E. R., and Mapiye, C. (2019). Grape pomace (Vitis vinifera L. cv. Pinotage) supplementation in lamb diets: effects on growth performance, carcass and meat quality. Meat Sci. 147, 6–12. doi: 10.1016/j.meatsci.2018.08.017
Denman, S. E., Martinez Fernandez, G., Shinkai, T., Mitsumori, M., and McSweeney, C. S. (2015). Metagenomic analysis of the rumen microbial community following inhibition of methane formation by a halogenated methane analog. Front. Microbiol. 6:1087. doi: 10.3389/fmicb.2015.01087
Deppenmeier, U. (2002). The unique biochemistry of methanogenesis. Prog. Nucl. Res. Molec. Biol. 71, 223–283. doi: 10.1016/s0079-6603(02)71045-3
Domingo, M.-C., Huletsky, A., Boissinot, M., Bernard, K. A., Picard, F. J., and Bergeron, M. G. (2008). Ruminococcus gauvreauii sp. nov., a glycopeptide-resistant species isolated from a human faecal specimen. Int. J. Syst. Evol. Microbiol. 58, 1393–1397. doi: 10.1099/ijs.0.65259-0
Douglas, G. M., Maffei, V. J., Zaneveld, J. R., Yurgel, S. N., Brown, J. R., Taylor, C. M., et al. (2020). PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 38, 685–688. doi: 10.1038/s41587-020-0548-6
Elolimy, A., Alharthi, A., Zeineldin, M., Parys, C., and Loor, J. J. (2020). Residual feed intake divergence during the preweaning period is associated with unique hindgut microbiome and metabolome profiles in neonatal Holstein heifer calves. J. Anim. Sci. Biotechnol. 11:13. doi: 10.1186/s40104-019-0406-x
Foiklang, S., Wanapat, M., and Norrapoke, T. (2016a). Effect of grape pomace powder, mangosteen peel powder and monensin on nutrient digestibility, rumen fermentation, nitrogen balance and microbial protein synthesis in dairy steers. Asian Australas. J. Anim. Sci. 29, 1416–1423. doi: 10.5713/ajas.15.0689
Foiklang, S., Wanapat, M., and Nor-rapoke, T. (2016b). In vitro rumen fermentation and digestibility of buffaloes as influenced by grape pomace powder and urea treated rice straw supplementation. Anim. Sci. J. 87, 370–377. doi: 10.1111/asj.12428
Freier, T. A., Beitz, D. C., Li, L., and Hartman, P. A. (1994). Characterization of Eubacterium coprostanoligenes sp. nov., a cholesterol-reducing anaerobe. Int. J. Syst. Bacteriol. 44, 137–142. doi: 10.1099/00207713-44-1-137
Gao, X., Tang, F., Zhang, F., Jia, C., Yang, Z., Liu, C., et al. (2019). Effects of the supplementation of distillers’ grape residues on ruminal degradability, whole tract digestibility and nitrogen metabolism in sheep. Arch. Anim. Nutr. 73, 384–398. doi: 10.1080/1745039X.2019.1590142
Göker, M., Held, B., Lucas, S., Nolan, M., Yasawong, M., Glavina del Rio, T., et al. (2010). Complete genome sequence of Olsenella uli type strain (VPI D76D-27CT). Stand. Genomic Sci. 3, 76–84. doi: 10.4056/sigs.1082860
Goodchild, A., Raftery, M., Saunders, N. F., Guilhaus, M., and Cavicchioli, R. (2004). Biology of the cold adapted archaeon, Methanococcoides burtonii determined by proteomics using liquid chromatography-tandem mass spectrometry. J. Proteome Res. 3, 1164–1176. doi: 10.1021/pr0498988
Guerra-Rivas, C., Gallardo, B., Mantecón, Á. R., del Álamo-Sanza, M., and Manso, T. (2017). Evaluation of grape pomace from red wine by-product as feed for sheep. J. Sci. Food Agric. 97, 1885–1893. doi: 10.1002/jsfa.7991
Henderson, G., Cox, F., Ganesh, S., Jonker, A., Young, W., Global Rumen Census Collaborators, et al. (2015). Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Sci. Rep. 5:14567. doi: 10.1038/srep14567
Hosseini, E., Grootaert, C., Verstraete, W., and Van de Wiele, T. (2011). Propionate as a health-promoting microbial metabolite in the human gut. Nutr. Rev. 69, 245–258. doi: 10.1111/j.1753-4887.2011.00388.x
Ianni, A., and Martino, G. (2020). Dietary grape pomace supplementation in dairy cows: effect on nutritional quality of milk and its derived dairy products. Foods 9:168. doi: 10.3390/foods9020168
Ijoma, G. N., Ogola, H. J. O., Rashama, C., and Matambo, T. (2022). Metagenomic insights into the impacts of phytochemicals on bacterial and archaeal community structure and biogas production patterns during anaerobic digestion of avocado oil processing waste feedstocks. Biomass Conv. Bioref., 1–20. doi: 10.1007/s13399-022-03516-8
Immig, I. (1996). The rumen and hindgut as source of ruminant methanogenesis. Environ. Monit. Assess. 42, 57–72. doi: 10.1007/BF00394042
Ishida, K., Kishi, Y., Oishi, K., Hirooka, H., and Kumagai, H. (2015). Effects of feeding polyphenol-rich winery wastes on digestibility, nitrogen utilization, ruminal fermentation, antioxidant status and oxidative stress in wethers. Anim. Sci. J. 86, 260–269. doi: 10.1111/asj.12280
Janssen, P. H., and Kirs, M. (2008). Structure of the archaeal community of the rumen. Appl. Environ. Microbiol. 74, 3619–3625. doi: 10.1128/aem.02812-07
Johnson, K. A., and Johnson, D. E. (1995). Methane emissions from cattle. J. Anim. Sci. 73, 2483–2492. doi: 10.2527/1995.7382483x
Kafantaris, I., Kotsampasi, B., Christodoulou, V., Kokka, E., Kouka, P., Terzopoulou, Z., et al. (2017). Grape pomace improves antioxidant capacity and faecal microflora of lambs. J. Anim. Physiol. Anim. Nutr. 101, e108–e121. doi: 10.1111/jpn.12569
Karisa, B., Moore, S., and Plastow, G. (2014). Analysis of biological networks and biological pathways associated with residual feed intake in beef cattle. Anim. Sci. J. 85, 374–387. doi: 10.1111/asj.12159
Katoh, K., Misawa, K., Kuma, K., and Miyata, T. (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast fourier transform. Nucleic Acids Res. 30, 3059–3066. doi: 10.1093/nar/gkf436
Khatoon, M., Patel, S. H., Pandit, R. J., Jakhesara, S. J., Rank, D. N., Joshi, C. G., et al. (2022). Rumen and fecal microbial profiles in cattle fed high lignin diets using metagenome analysis. Anaerobe 73:102508. doi: 10.1016/j.anaerobe.2021.102508
Khiaosa-ard, R., Metzler-Zebeli, B. U., Ahmed, S., Muro-Reyes, A., Deckardt, K., Chizzola, R., et al. (2015). Fortification of dried distillers grains plus solubles with grape seed meal in the diet modulates methane mitigation and rumen microbiota in Rusitec. J. Dairy Sci. 98, 2611–2626. doi: 10.3168/jds.2014-8751
Kraatz, M., Wallace, R. J., and Svensson, L. (2011). Olsenella umbonata sp. nov., a microaerotolerant anaerobic lactic acid bacterium from the sheep rumen and pig jejunum, and emended descriptions of Olsenella, Olsenella uli and Olsenella profusa. Int. J. Syst. Evol. Microbiol. 61, 795–803. doi: 10.1099/ijs.0.022954-0
Krieg, N. R., Ludwig, W., Euzéby, J., and Whitman, W. B. (2010). “Phylum XIV. Bacteroidetes phyl. Nov” in Bergey’s manual® of systematic bacteriology: volume four the bacteroidetes, spirochaetes, tenericutes (mollicutes), acidobacteria, fibrobacteres, fusobacteria, dictyoglomi, gemmatimonadetes, lentisphaerae, verrucomicrobia, chlamydiae, and planctomycetes. eds. N. R. Krieg, J. T. Staley, D. R. Brown, B. P. Hedlund, B. J. Paster, and N. L. Ward, et al. (New York, NY: Springer New York), 25–469.
Lawhorn, B. G., Mehl, R. A., and Begley, T. P. (2004). Biosynthesis of the thiamin pyrimidine: the reconstitution of a remarkable rearrangement reaction. Org. Biomol. Chem. 2, 2538–2546. doi: 10.1039/B405429F
Leahy, S. C., Kelly, W. J., Ronimus, R. S., Wedlock, N., Altermann, E., and Attwood, G. T. (2013). Genome sequencing of rumen bacteria and archaea and its application to methane mitigation strategies. Animal 7, 235–243. doi: 10.1017/S1751731113000700
Lee, H., Beilhartz, G. L., Kucharska, I., Raman, S., Cui, H., Lam, M. H. Y., et al. (2020). Recognition of semaphorin proteins by P. sordellii lethal toxin reveals principles of receptor specificity in clostridial toxins. Cells 182, 345–356.e16. doi: 10.1016/j.cell.2020.06.005
Li, Z., Mu, C., Xu, Y., Shen, J., and Zhu, W. (2020). Changes in the solid-, liquid-, and epithelium-associated bacterial communities in the rumen of Hu lambs in response to dietary urea supplementation. Front. Microbiol. 11:244. doi: 10.3389/fmicb.2020.00244
Liu, J., Bian, G., Zhu, W., and Mao, S. (2015). High-grain feeding causes strong shifts in ruminal epithelial bacterial community and expression of toll-like receptor genes in goats. Front. Microbiol. 6:167. doi: 10.3389/fmicb.2015.00167
Liu, C., Cui, Y., Li, X., and Yao, M. (2020). Microeco: an R package for data mining in microbial community ecology. FEMS Microbiol. Ecol. 97:fiaa255. doi: 10.1093/femsec/fiaa255
Liu, H., Yang, G., Degen, A., Ji, K., Jiao, D., Liang, Y., et al. (2021). Effect of feed level and supplementary rumen protected lysine and methionine on growth performance, rumen fermentation, blood metabolites and nitrogen balance in growing Tan lambs fed low protein diets. Anim. Feed. Sci. Technol. 279:115024. doi: 10.1016/j.anifeedsci.2021.115024
Ma, Y., Deng, X., Yang, X., Wang, J., Li, T., Hua, G., et al. (2022). Characteristics of bacterial microbiota in different intestinal segments of Aohan fine-wool sheep. Front. Microbiol. 13:874536. doi: 10.3389/fmicb.2022.874536
Ma, C., Gao, Q., Zhang, W., Zhu, Q., Tang, W., Blachier, F., et al. (2020). Supplementing synbiotic in sows’ diets modifies beneficially blood parameters and colonic microbiota composition and metabolic activity in suckling piglets. Front. Vet. Sci. 7:575685. doi: 10.3389/fvets.2020.575685
Macleod, G. K., and Buchanan-Smith, J. G. (1972). Digestibility of hydrogenated tallow, saturated fatty acids and soybean oil-supplemented diets by sheep. J. Anim. Sci. 35, 890–895. doi: 10.2527/jas1972.354890x
McDonald, D., Price, M. N., Goodrich, J., Nawrocki, E. P., DeSantis, T. Z., Probst, A., et al. (2012). An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6, 610–618. doi: 10.1038/ismej.2011.139
McLoughlin, S., Spillane, C., Claffey, N., Smith, P. E., O’Rourke, T., Diskin, M. G., et al. (2020). Rumen microbiome composition is altered in sheep divergent in feed efficiency. Front. Microbiol. 11:1981. doi: 10.3389/fmicb.2020.01981
McSweeney, C. S., Palmer, B., McNeill, D. M., and Krause, D. O. (2001). Microbial interactions with tannins: nutritional consequences for ruminants. Anim. Feed Sci. Technol. 91, 83–93. doi: 10.1016/S0377-8401(01)00232-2
Membrive, C. M. B. (2016). “Anatomy and physiology of the rumen” in Rumenology. eds. D. D. Millen, M. D. B. Arrigoni, and R. D. L. Pacheco (Cham: Springer International Publishing)
Ministry of Agriculture of China. (2005). Feeding standard of meat-producing sheep and goats (standard NY/T 816–2004). Chinese Agricultural Press Beijing, China.
Mizrahi, I., Wallace, R. J., and Moraïs, S. (2021). The rumen microbiome: balancing food security and environmental impacts. Nat. Rev. Microbiol. 19, 553–566. doi: 10.1038/s41579-021-00543-6
Moate, P. J., Williams, S. R. O., Torok, V. A., Hannah, M. C., Ribaux, B. E., Tavendale, M. H., et al. (2014). Grape marc reduces methane emissions when fed to dairy cows. J. Dairy Sci. 97, 5073–5087. doi: 10.3168/jds.2013-7588
Molosse, V. L., Deolindo, G. L., Cécere, B. G., Marcon, H., da Rosa, G., Vedovatto, M., et al. (2021). Effect of dietary supplementation with grape residue flour on weight gain, metabolic profile, leukogram, proteinogram and antioxidant response in suckling lambs. Res. Vet. Sci. 139, 112–120. doi: 10.1016/j.rvsc.2021.07.004
Moreira, L. R., and Filho, E. X. (2008). An overview of mannan structure and mannan-degrading enzyme systems. Appl. Microbiol. Biotechnol. 79, 165–178. doi: 10.1007/s00253-008-1423-4
Morita, H., Shiratori, C., Murakami, M., Takami, H., Toh, H., Kato, Y., et al. (2008). Sharpea azabuensis gen. nov., sp. nov., a gram-positive, strictly anaerobic bacterium isolated from the faeces of thoroughbred horses. Int. J. Syst. Evol. Microbiol. 58, 2682–2686. doi: 10.1099/ijs.0.65543-0
Ngetich, D. K., Bett, R. C., Gachuiri, C. K., and Kibegwa, F. M. (2022). Diversity of gut methanogens and functional enzymes associated with methane metabolism in smallholder dairy cattle. Arch. Microbiol. 204:608. doi: 10.1007/s00203-022-03187-z
O’Hara, E., Neves, A. L. A., Song, Y., and Guan, L. L. (2020). The role of the gut microbiome in cattle production and health: driver or passenger? Annu. Rev. Anim. Biosci. 8, 199–220. doi: 10.1146/annurev-animal-021419-083952
Pannell, D., Kouakou, B., Terrill, T. H., Ogunade, I. M., Estrada-Reyes, Z. M., Bryant, V., et al. (2022). Adding dried distillers grains with solubles influences the rumen microbiome of meat goats fed lespedeza or alfalfa-based diets. Small Rumin. Res. 214:106747. doi: 10.1016/j.smallrumres.2022.106747
Paulson, J. N., Pop, M., and Bravo, H. C. (2011). Metastats: an improved statistical method for analysis of metagenomic data. Genome Biol. 12:P17. doi: 10.1186/gb-2011-12-s1-p17
Peng, X., Wilken, S. E., Lankiewicz, T. S., Gilmore, S. P., Brown, J. L., Henske, J. K., et al. (2021). Genomic and functional analyses of fungal and bacterial consortia that enable lignocellulose breakdown in goat gut microbiomes. Nat. Microbiol. 6, 499–511. doi: 10.1038/s41564-020-00861-0
Price, M. N., Dehal, P. S., and Arkin, A. P. (2010). FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490
Purushe, J., Fouts, D. E., Morrison, M., White, B. A., Mackie, R. I., Coutinho, P. M., et al. (2010). Comparative genome analysis of Prevotella ruminicola and Prevotella bryantii: insights into their environmental niche. Microb. Ecol. 60, 721–729. doi: 10.1007/s00248-010-9692-8
Rodrigues, P. H. M. (2016). “Control and manipulation of ruminal fermentation” in Rumenology. eds. D. D. Millen, M. D. B. Arrigoni, and R. D. L. Pacheco (Cham: Springer International Publishing)
Rognes, T., Flouri, T., Nichols, B., Quince, C., and Mahé, F. (2016). VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. doi: 10.7717/peerj.2584
Rolinec, M., Medo, J., Gábor, M., Miluchová, M., Šimko, M., Gálik, B., et al. (2023). Effect of grape pomace intake on the rumen bacterial community of sheep. Diversity 15:234. doi: 10.3390/d15020234
Ross, E. M., Moate, P. J., Marett, L., Cocks, B. G., and Hayes, B. J. (2013). Investigating the effect of two methane-mitigating diets on the rumen microbiome using massively parallel sequencing. J. Dairy Sci. 96, 6030–6046. doi: 10.3168/jds.2013-6766
Russo, V. M., Jacobs, J. L., Hannah, M. C., Moate, P. J., Dunshea, F. R., and Leury, B. J. (2017). In vitro evaluation of the methane mitigation potential of a range of grape marc products. Anim. Prod. Sci. 57, 1437–1444. doi: 10.1071/AN16495
Schleifer, K.-H. (2009). Phylum XIII. Firmicutes Gibbons and Murray 1978, 5 Firmacutes sic Gibbons and Murray 1978, 5, Bergey’s manual® of systematic bacteriology: volume three the firmicutes, eds. P. VosDe, G. M. Garrity, D. Jones, N. R. Krieg, W. Ludwig, and F. A. Rainey, et al. (New York, NY: Springer New York), 19–1317.
Shinkai, T., Enishi, O., Mitsumori, M., Higuchi, K., Kobayashi, Y., Takenaka, A., et al. (2012). Mitigation of methane production from cattle by feeding cashew nut shell liquid. J. Dairy Sci. 95, 5308–5316. doi: 10.3168/jds.2012-5554
Sim, S., Lee, H., Yoon, S., Seon, H., Park, C., and Kim, M. (2022). The impact of different diets and genders on fecal microbiota in Hanwoo cattle. J. Anim. Sci. Technol. 64, 897–910. doi: 10.5187/jast.2022.e71
Strobel, H. J. (1992). Vitamin B12-dependent propionate production by the ruminal bacterium Prevotella ruminicola 23. Appl. Environ. Microbiol. 58, 2331–2333. doi: 10.1128/aem.58.7.2331-2333.1992
Tong, J., Zhang, H., Yang, D., Zhang, Y., Xiong, B., and Jiang, L. (2018). Illumina sequencing analysis of the ruminal microbiota in high-yield and low-yield lactating dairy cows. PLoS One 13:e0198225. doi: 10.1371/journal.pone.0198225
Ungerfeld, E. M. (2020). Metabolic hydrogen flows in rumen fermentation: principles and possibilities of interventions. Front. Microbiol. 11:589. doi: 10.3389/fmicb.2020.00589
Van Soest, P. J., Robertson, J. B., and Lewis, B. A. (1991). Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J. Dairy Sci. 74, 3583–3597. doi: 10.3168/jds.S0022-0302(91)78551-2
Vasta, V., Daghio, M., Cappucci, A., Buccioni, A., Serra, A., Viti, C., et al. (2019). Invited review: plant polyphenols and rumen microbiota responsible for fatty acid biohydrogenation, fiber digestion, and methane emission: experimental evidence and methodological approaches. J. Dairy Sci. 102, 3781–3804. doi: 10.3168/jds.2018-14985
Wang, Y., Xu, L., Liu, J., Zhu, W., and Mao, S. (2017). A high grain diet dynamically shifted the composition of mucosa-associated microbiota and induced mucosal injuries in the colon of sheep. Front. Microbiol. 8:2080. doi: 10.3389/fmicb.2017.02080
Wood, A. P., Aurikko, J. P., and Kelly, D. P. (2004). A challenge for 21st century molecular biology and biochemistry: what are the causes of obligate autotrophy and methanotrophy? FEMS Microbiol. Rev. 28, 335–352. doi: 10.1016/j.femsre.2003.12.001
Xie, F., Jin, W., Si, H., Yuan, Y., Tao, Y., Liu, J., et al. (2021). An integrated gene catalog and over 10,000 metagenome-assembled genomes from the gastrointestinal microbiome of ruminants. Microbiome 9:137. doi: 10.1186/s40168-021-01078-x
Xu, J., Li, Y., Yang, Z., Li, C., Liang, H., Wu, Z., et al. (2018). Yeast probiotics shape the gut microbiome and improve the health of early-weaned piglets. Front. Microbiol. 9:2011. doi: 10.3389/fmicb.2018.02011
Yang, G., and Wang, J. (2019). Changes in microbial community structure during dark fermentative hydrogen production. Int. J. Hydrog. Energy 44, 25542–25550. doi: 10.1016/j.ijhydene.2019.08.039
Yu, J., and Ahmedna, M. (2013). Functional components of grape pomace: their composition, biological properties and potential applications. Int. J. Food Sci. Technol. 48, 221–237. doi: 10.1111/j.1365-2621.2012.03197.x
Zened, A., Combes, S., Cauquil, L., Mariette, J., Klopp, C., Bouchez, O., et al. (2013). Microbial ecology of the rumen evaluated by 454 GS FLX pyrosequencing is affected by starch and oil supplementation of diets. FEMS Microbiol. Ecol. 83, 504–514. doi: 10.1111/1574-6941.12011
Zhang, H., Shao, M., Huang, H., Wang, S., Ma, L., Wang, H., et al. (2018). The dynamic distribution of small-tail Han sheep microbiota across different intestinal segments. Front. Microbiol. 9:32. doi: 10.3389/fmicb.2018.00032
Zhang, J., Shi, H., Wang, Y., Li, S., Cao, Z., Ji, S., et al. (2017). Effect of dietary forage to concentrate ratios on dynamic profile changes and interactions of ruminal microbiota and metabolites in Holstein heifers. Front. Microbiol. 8:2206. doi: 10.3389/fmicb.2017.02206
Zhao, W., Abdelsattar, M. M., Wang, X., Zhang, N., and Chai, J. (2023). In vitro modulation of rumen fermentation by microbiota from the recombination of rumen fluid and solid phases. Microbiol. Spectr. 11, e03387–e03322. doi: 10.1128/spectrum.03387-22
Zhao, J. X., Li, Q., Zhang, R. X., Liu, W. Z., Ren, Y. S., Zhang, C. X., et al. (2018). Effect of dietary grape pomace on growth performance, meat quality and antioxidant activity in ram lambs. Anim. Feed Sci. Technol. 236, 76–85. doi: 10.1016/j.anifeedsci.2017.12.004
Keywords: wine by-product, rumen, hindgut, microbiome, methanogenesis
Citation: Cheng X, Du X, Liang Y, Degen AA, Wu X, Ji K, Gao Q, Xin G, Cong H and Yang G (2023) Effect of grape pomace supplement on growth performance, gastrointestinal microbiota, and methane production in Tan lambs. Front. Microbiol. 14:1264840. doi: 10.3389/fmicb.2023.1264840
Edited by:
Anusorn Cherdthong, Khon Kaen University, ThailandReviewed by:
Chalong Wachirapakorn, Khon Kaen University, ThailandChaichana Suriyapha, Khon Kaen University, Thailand
Copyright © 2023 Cheng, Du, Liang, Degen, Wu, Ji, Gao, Xin, Cong and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guo Yang, eWFuZ2d1b0BsemIuYWMuY24=
 Xindong Cheng
Xindong Cheng Xia Du
Xia Du Yanping Liang
Yanping Liang Abraham Allan Degen
Abraham Allan Degen Xiukun Wu
Xiukun Wu Kaixi Ji
Kaixi Ji Qiaoxian Gao5
Qiaoxian Gao5 Guo Yang
Guo Yang