- 1Biotechnology and Germplasm Resources Institute, Yunnan Academy of Agricultural Sciences, Kunming, China
- 2School of Agriculture, Yunnan University, Kunming, China
Rice blast, caused by Magnaporthe oryzae, is a major threat to global rice production causing significant crop losses and impacting grain quality. The annual loss of rice production due to this disease ranges from 10% to 30%. The use of biologically controlled strains, instead of chemical pesticides, to control plant diseases has become a research hotspot. In this study, an antagonistic endophytic bacterial strain was isolated from the roots of Oryza officinalis using the traditional isolation and culture methods. A phylogenetic tree based on 16S RNA and whole-genome sequencing identified isolate G5 as a strain of Bacillus subtilis. This isolate displayed strong antagonistic effects against different physiological strains of M. oryzae. After co-culture in LB medium for 7 days, the inhibition rates of the mycelial growth of four strains of M. oryzae, ZB15, WH97, Guy11, and T-39800E were 98.07 ± 0.0034%, 98.59 ± 0.0051%, 99.16 ± 0.0012%, and 98.69 ± 0.0065%, respectively. Isolate G5 significantly inhibited the formation of conidia of M. oryzae, with an inhibition rate of 97% at an OD600 of 2. Isolate G5 was able to provide 66.81% protection against rice blast under potted conditions. Whole-genome sequencing revealed that the genome size of isolate G5 was 4,065,878 bp, including 4,182 coding genes. Using the anti-SMASH software, 14 secondary metabolite synthesis gene clusters were predicted to encode antifungal substances, such as fengycin, surfactin, and bacilysin. The G5 isolate also contained genes related to plant growth promotion. These findings provide a theoretical basis for expounding the biocontrol mechanisms of this strain and suggest further development of biogenic agents that could effectively inhibit rice blast pathogen growth and reduce crop damage, while being environmentally friendly, conducive to ecological development, and a sustainable alternative to chemical pesticides. This study also enriches the relevant research on endophytes of wild rice, which proves that wild rice is a valuable microbial resource bank.
1. Introduction
There are 2.5 to 3.5 billion people around the world who depend on rice (Oryza sativa L.) for nutrition; its demand is still growing in some low-income regions (Asibi et al., 2019). As a result of the ascomycete fungus Magnaporthe oryzae, rice blast is one of the most devastating rice diseases in the world (Rossman et al., 1990; Komatsu et al., 2016). Rice blast causes an average yield loss of 10% to 30% per year, which represents a global yield loss of approximately 157 million tons annually. Several outbreaks have been reported. An estimated 800,000 tons of rice were lost in Japan during the rice blast epidemic in 1953 (Wang et al., 2014). Rice blast diseases result in the loss of approximately 564,000 tons of rice each year in eastern India alone (Devanna et al., 2022).
Rice blast is a non-organ selective disease. It is possible for M. oryzae to infect rice plants at any developmental stage, including the leaves, stems, nodes, panicles, and roots, reducing market value, and affecting food security (Li et al., 2007). Currently, the most important measures for controlling rice blast in rice production are planting disease-resistant varieties and controlling the use of chemical pesticides. These measures have been effective in reducing the impact of rice blast on crop yield; however, they also have limitations. Owing to the high number of different strains and rapid evolution leading to variation in M. oryzae, the general disease-resistant varieties lose their resistance after 3–5 years (Dean et al., 2005). The extensive use of chemicals not only increases the drug resistance of pathogenic bacteria and accelerates their mutation but also leads to environmental pollution (Karthikeyan and Gnanamanickam, 2008). A biocontrol agent is increasingly popular because of the fact that it is eco-friendly, has a low residual effect, is highly selectable, and can control an organism for an extended period of time. Therefore, it is important to implement integrated disease management strategies that reasonably apply the necessary agricultural, biological, physical, chemical, and other comprehensive technical measures to economically, safely, and effectively eliminate or control diseases and increase production and income (Ons et al., 2020). Compared with traditional control methods, integrated control can better realize the benign, healthy, and sustainable development of the agricultural economy.
Plant endophytes, including fungi, bacteria, and actinomycetes, which colonize plant organs, tissues, and intercellular spaces, have a stable living space inside the healthy tissues of the host plant (Sturz and Nowak, 2000). These are critical microbial resources whose urgent development has been proven beneficial (Tan and Zou, 2001). Bacteria are dominant components of the plant microbiome and are important endophyte components (Afzal et al., 2019). The probiotic effects of endophytic bacteria on plants include (1) promoting plant nutrient absorption, such as nitrogen, phosphorus, and ions; (2) regulating plant growth and development by regulating plant hormones (auxin, cytokinin, ethylene, etc.) (Davison, 1988); and (3) helping plants resist stresses, including biotic and abiotic stresses.
Endophytic bacteria have direct probiotic effects on plants such as promoting plant nutrient absorption and regulating plant growth and development (Bevivino et al., 1998). Endophytic bacteria indirectly mediate resistance to pathogenic bacteria through a series of biochemical pathways. These biochemical pathways include the secretion of antibiotics and cell wall-degrading enzymes, reduction of ethylene levels in plants, induction of systemic resistance in plants, reduction of the concentration of ions needed by pathogens, and synthesis of volatile organic compounds that inhibit pathogenic bacteria (Santoyo et al., 2016; Glick, 2020).
One of the three wild rice species in China, Oryza officinalis Wall., grows mostly in hills or woodlands and warm, rainy, humid environments. O. officinalis Wall. has strong growth potential; its biomass is approximately 20 times that of cultivated rice (Kiran et al., 2013). It has well-developed vascular bundles, barren tolerance, shading, photooxidation, and a strong ability to absorb mineral elements simultaneously (Cheng et al., 2005). O. officinalis Wall. is known to have endured a variety of harsh conditions and natural calamities over a long period of time, making it a valuable genetic resource for rice breeding (Tan et al., 2004; Devanna et al., 2014). It also harbors a special and effective microbial community ecosystem, which is an ideal source of endophytes for the biocontrol of rice diseases (Tian et al., 2023). Moreover, compared with cultivated rice endophytes and rhizosphere bacteria, there are few studies on wild rice.
Bacillus spp. are the most widely used biocontrol bacteria in research and production. A broad range of ecological niches are inhabited by bacteria from this group, such as soil, water, and air, as well as on the surfaces and rhizosphere of plants, in the gastrointestinal tract of animals, and in many extreme environments (Wang et al., 2022). Bacillus species exhibit a wide array of secondary metabolisms and possess the capacity to synthesize various structurally distinct antagonistic substances, thereby contributing to their extensive bacteriostatic spectrum within the field of biotechnology (Djordje et al., 2018). Another unique feature arises from their spore-forming ability, which enables them to grow under unfavorable environmental conditions, such as high temperature, drought, ultraviolet light, and other stresses (Siddikee et al., 2010). In addition, their reproductive ability and colonization speed are superior to those of other biocontrol bacteria (Dutta et al., 2014). There are many examples of successful biocontrol strains derived from Bacillus spp. B. thuringiensis is regarded as the most effective bioinsecticide, showing a good biocontrol effect on most insects of the orders Diptera, Lepidoptera, and Coleoptera (Khan et al., 2022). B. cereus HS24, B. tequilensis GYLH001, and B. velezensis ZW-10 show strong antimicrobial effects against M. oryzae (Li et al., 2018; Huang et al., 2019; Chen et al., 2020). B. subtilis L1-21 performs well as a biocontrol agent for Botrytis cinerea after tomato harvests (Bu et al., 2021) and endophytic B. subtilis Lu144 effectively reduces the incidence of mulberry bacterial wilt (Ji et al., 2010). Wei et al. (2011) demonstrated the ability of B. amyloliquefaciens to reduce Ralstonia solanacearum infections in potato plants.
Most of the beneficial bacteria with biocontrol effects have come from cultivated rice or its rhizosphere soil; however, there are few reports of bacteria isolated from wild rice. The objective of this study was to isolate, screen, and identify biocontrol bacteria in wild rice and to conduct a comprehensive investigation of its potential control effects on M. oryzae. Seven microbial strains were isolated from healthy root samples of O. officinalis Wall. G5 was the only strain that inhibited M. oryzae strongly. An analysis of the strain's morphology and its 16S rRNA sequence identified it as B. subtilis. The antagonistic effects of G5 on the four different strains of M. oryzae were determined using the plate-confrontation method. Both in vitro and field trials were conducted to evaluate the biocontrol effects against M. oryzae. Furthermore, we explored the inhibitory mechanisms of B. subtilis G5 and the host defense responses against M. oryzae. Moreover, the genome of this strain was studied, including the genes involved in antifungal production.
2. Materials and methods
2.1. Strain, media, and cultural conditions
M. oryzae strains, WH97 and Guy11, which are widely used in research, ZB15, which is prevalent in southwestern China, and T-39800E, identified in our previous study, were grown on potato dextrose agar plates at 28°C. The isolated endophytic bacteria were inoculated onto Luria–Bertani (LB) agar plates (Sangong Co., Ltd., Shanghai, China) at 28°C.
2.2. Isolation and selection of endophytic bacteria against M. oryzae from healthy O. officinalis wall
Healthy O. officinalis Wall. plants were collected from the Biotechnology and Germplasm Resources Institute of the Yunnan Academy of Agricultural Sciences, Yunnan Province, China. Root samples from wild rice plants were thoroughly washed with tap water to remove airborne counterparts and then soaked in 75% (v/v) ethanol for 1 min to kill common bacteria and some fungi. To further eliminate microorganisms on the surface of plant tissues, all samples were soaked in 1% (v/v) NaClO for 5 min, followed by rinsing five times with sterile water under aseptic conditions for the purpose of depleting epiphytic microorganisms. The surface moisture was removed using sterilized filter paper. The dried tissue was, then, placed in a sterilized mortar and ground; 5 ml of sterile water was added to the ground tissue. After serial dilutions of the suspension, 1 ml aliquots were evenly spread on LB agar plates and incubated at 37°C for 2 to 3 days. Single colonies were selected and purified from LB agar plates by repeated streaking.
To determine the potential antagonistic potential of these isolates, a preliminary screening was conducted using the dual-culture method, as previously described (Ferreira et al., 1991). Plates inoculated only with M. oryzae Guy11, which is widely used in research, served as controls. The surface area of the plates was measured by taking photographs with a Canon EOS 77D camera (Canon, Tokyo, Japan) after 7 days of incubation at 28°C. To evaluate the inhibitory effect, the percentage of inhibited growth area was calculated using the formula [(Sc – St)/Sc) × 100], where Sc and St represent the growth area of M. oryzae on the control and treated plates, respectively. Each isolate was tested three times with three replicates each. After screening with the above method, strain G5 was isolated and stored at −80°C in glycerol (30%, v/v) stock. As previously described, scanning electron microscopy (SEM; Quanta FEG650, FEI, Hillsboro, USA) (De et al., 2008) was used to observe the morphology and ultrastructure of M. oryzae in a dual-culture experiment to assess the antifungal activity of G5′.
2.3. Identification of strain G5
The morphological characteristics of strain G5 were examined on LB agar. Genomic DNA extraction from strain G5 was performed using the TaKaRa MiniBEST Bacteria Genomic DNA Extraction Kit Ver.3.0, following the guidelines provided by the manufacturer (TaKaRa, Dalian, China), in order to determine the 16S rRNA gene sequence. The primer sequence is 27F:5′-AGAGTTTGATCCTGGCTCAG-3′; 1492R 5′-GGTTACCTTGTTACGACTT-3′. The 30 μl PCR mixture contained 15 μl of 2 × Phanta Max Master Mix, 1 μl of each 10 μM primer, 4 μl of genomic DNA, and 9 μl of double distilled water. The PCR program was as follows: denaturation at 94°C for 3 min, 30 cycles or 94°C for 15 s, 55°C for 15 s, 68°C for 1.5 min, and final extension at 68°C for 10 min. As a result of PCR product sequencing by Tsingke Biotechnology (Kunming, China), the sequences gained access to the GenBank databases for BLAST searches.
2.4. Effects of G5 on conidial germination and appressorium formation
To obtain conidial precipitates, 1 ml of Guy11-GFP conidial suspension was centrifuged at 3,000 rpm/min for 3 min in a 1.5-ml centrifuge tube. In the control group, 1 ml of sterile water was added to each centrifuge tube to suspend the conidial precipitates; 1 ml of G5 at different concentrations (OD600 = 1, OD600 = 1.5, OD600 = 2) was added to each centrifuge tube to suspend the conidial precipitates. One droplet (20 μl) of conidial suspension from each treatment group was placed on a hydrophobic microscope cover glass with wet filter paper and incubated at 28°C in darkness. Conidial germination and appressorium formation were observed under the upright fluorescent microscope (Leica DM2500, Wetzlar, Germany) after 16 h of incubation. In each replicate, we measured the percentage of conidial germination and appressorium formation. There were three replicates of each treatment, with more than 50 conidia assessed for each treatment. The experiment was repeated three times.
2.5. Biocontrol assays
2.5.1. Antifungal activity on detached rice leaves
The rice leaf experiment included three groups: prevention, treatment, and control. Leaves from Nipponbare plants at the 5-leaf stage, measuring 5 cm and exhibiting normal growth, were immersed in Petri dishes containing a solution of 6-benzylaminopurine (1 mg/ml). The application of three delicate punctures per leaf segment facilitated the penetration of the treatment agents. Three replicates with ten rice plants each were used in each treatment. Prevention group: 5 μl of the cell-free G5 culture filtrate was applied to punctured leaves, and the leaves were incubated at 28°C in the dark. After 24 h, all puncture sites were inoculated with 5 μl droplets of the conidial suspension of M. oryzae, Guy11 (1 × 105 spores/ml). Under alternating light and dark conditions, the leaves were incubated at 28°C for 6 days. Treatment group: The same method as the prevention group was used, but leaves were first inoculated with a spore suspension of M. oryzae, and then, the cell-free G5 culture filtrate was added. Control group: Sterile water was used instead of the cell-free G5 culture filtrate; all other steps were the same. After 6 days of incubation, the lesion diameters were compared.
2.5.2. Antifungal activity in greenhouse
The biocontrol effect of B. subtilis G5 on rice blast was studied using leaf-spraying inoculation with Nipponbare plants. Each treatment consisted of three replicates with 10 rice plants per replicate. In the prevention experiment, rice seedlings at the 3-leaf stage were subjected to a treatment involving the application of 50 ml of the cell-free G5 culture filtrate. Subsequently, after 7 days, all plants were sprayed with 50 ml conidial suspension of Guy11, containing 1 × 105 spores/ml. The plants were, then, placed in a growth chamber with a temperature of 28°C, humidity of 80%, and subjected to a dark period of 24 h, followed by a light/dark cycle of 12 h each. In the control group, the same procedures were followed, with the exception that the leaves were inoculated with 50 ml of distilled water, followed by the application of 50 ml of the Guy11 conidial suspension (1 × 105 spores/ml).
The disease index was assessed 7 days after spray inoculation. We calculated disease index and biocontrol efficacy as follows: Disease index = [Σ (the number of diseased plants in each disease rating × the number of plants at the corresponding rating)/(total number of plants investigated × the highest disease rating)] × 100%.
Biocontrol efficacy = [(relative disease index of control treatment – disease index of treatment)/disease index of control] × 100%.
2.6. Whole-genome sequencing of G5
A NucleoBond® HMW DNA kit (740160.20; Macherey-Nagel, Düren, Germany) was used for high-quality genome extraction of samples according to the manufacturer's instructions. DNA concentration and purity were determined via Qubit4.0 (Q33226; Thermo Fisher Scientific, Waltham, MA, USA) and Nanodrop (SMA4000; Taiwan, China). DNA integrity was assessed using 0.75% agarose gel electrophoresis. gDNA was separated into two samples. One sample was randomly selected to build a library with an insertion of 300 bp. The library was processed using the NovaSeq 6000 platform (Illumina, San Diego, CA, USA) and the paired-end 150 bp sequencing strategy, which allowed for the determination of diversity and detection of minor variants, but has the disadvantage of short reads that do not permit the reconstruction of complete haplotypes. The other sample was subjected to end-repair, 3′ adenylated, adapters, and motor protein ligations. The product was purified using Agencourt AMPure XP Beads (A63881; Beckman Coulter, Brea, CA, USA). Finally, fragments larger than 1 kb were screened using single-molecule nanopore DNA sequencing on a MinION Flow Cell (R9.4.1; Oxford Nanopore Technologies, Oxford, UK). Reads longer than the Illumina raw reads were filtered and assembled using Canu software version 1.3 with the default parameters. The genomic sequences were proofread using NextPolish (v1.4.1) and Pilon (v1.18). The assembled sequences were deposited in the NCBI (BioProject ID: PRJNA997266). Gene prediction and annotation were performed using Prokka (version 1.10) and the National Center for Biotechnology Information NR database. Functional annotation was performed on protein-coding genes using the cluster of NCBI non-redundant (NR) protein sequences, Swiss-Prot, Orthologous Groups of Proteins (COG), and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases. Secondary metabolite gene clusters were predicted using antiSMASH v4.1.0 as previously described (Kai et al., 2017). Sequencing was performed by Sangon Biotech Co. Ltd. (Shanghai, China).
2.7. Statistical analysis
Statistical analyses were performed using SPSS software version 26 (SPSS, Chicago, IL, United States). All treatments were performed in triplicate. Data are expressed as the mean ± standard deviation (SD). The means of the different treatment groups were determined using one-way ANOVA with Duncan's multiple range test to determine whether there were significant differences between the treatment groups. A p-value of < 0.05 was considered to be statistically significant.
3. Results
3.1. Isolation of endophytic actinobacterial antagonists against M. oryzae
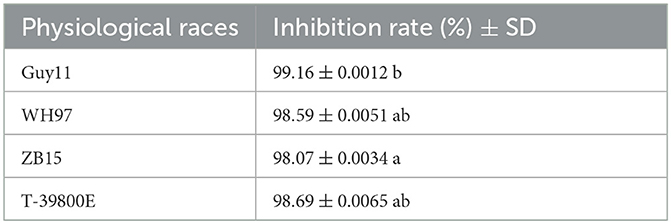
Seven endophytic strains were isolated from the roots of healthy O. officinalis plants. Among them, one strain, named G5, showed the strongest antagonistic activity against M. oryzae Guy11 (Figures 1A, B). Strain G5 showed strong inhibitory activity against all tested strains: WH97, Guy11, ZB15, and T-39800E. The highest percentage inhibition (99.16 ± 0.0012%) was observed for G5 against Guy11, and the lowest recorded (98.07 ± 0.0034%) was observed against ZB15 (Figure 1D). G5 also had an inhibitory effect on WH97 (Figure 1C) and T-39800E (Figure 1E), with inhibition rates of 98.59 ± 0.0051% and 98.69 ± 0.0065%, respectively (Table 1). These results suggest that strain G5 has a strong inhibitory effect on different strains of M. oryzae, although the inhibition rate of the G5 strain against different strains is slightly different.
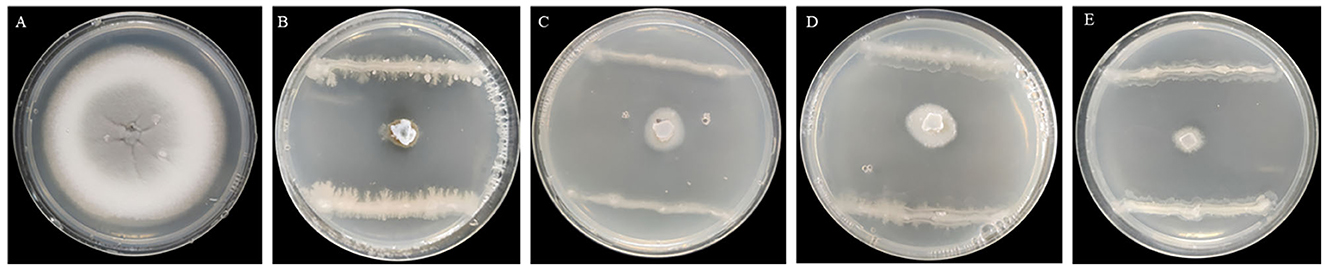
Figure 1. Inhibition effect of G5 on mycelial growth of M. oryzae of different physiological races Guy11 (B), WH97 (C), ZB15 (D), T-39800E (E), and CK (A).
3.2. Identification of endophytic bacterium G5
The colony characteristics of bacteria cultured under certain conditions, including surface, size, color, edge, texture, and shape, have a certain stability, which is important for identifying the types of bacteria. Gram staining is a common method for observing the morphology of bacteria, the results of which can be used to initially classify bacteria. Therefore, preliminary morphological observations were performed on strain G5. On LB medium, G5 colonies showed typical characteristics of Bacillus sp., such as being dry and round with irregular protrusions near the margin (Supplementary Figure 1). Based on SEM images, the cells were short rod-shaped structures approximately 0.63–0.72 μm in width and 1.40–1.46 μm in length (Figure 2A). Gram staining of these cells was positive (Supplementary Figure 1). In addition, BLAST analysis of the 16s rRNA genes showed that G5 (GenBank database accession number: OQ874691) shared 99% identity with Bacillus subtilis (MZ895428.1 and MZ895403.1). Based on phylogenetic analysis, strain G5 clustered closely with Bacillus spp. Furthermore, strain G5 and B. subtilis clustered together (Figure 2B).
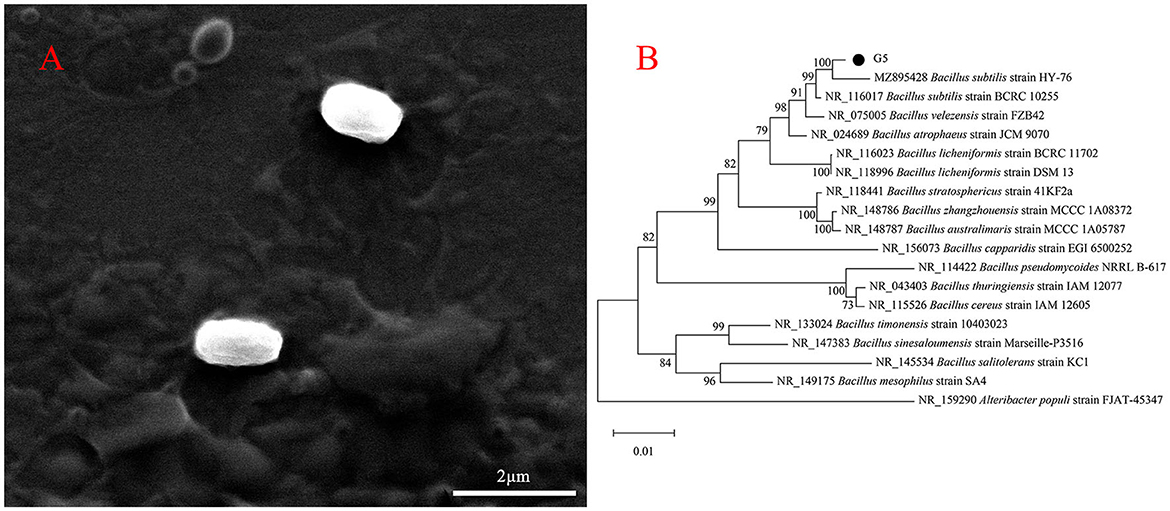
Figure 2. Morphological characteristics of G5 were visualized by SEM (A). Neighbor-joining phylogenetic tree showing the position of B. subtilis G5 isolate with other species of Bacillus spp. and related taxa based on 16S rDNA gene sequences. Bootstrap values (expressed as percentages of 1,000 replications) are indicated at tree branch points (B).
3.3. The morphological and ultrastructural changes in m. oryzae after confrontation with G5
Hyphal structures of M. oryzae were observed using SEM (Figure 3). It was observed that the untreated hyphae of M. oryzae grew normally, showed a straight, uniform appearance, and had a well-developed tube-like structure (Figure 3A). Conversely, abnormalities were observed in fungal hyphae co-cultured with G5 including deformities (Figure 3B), irregular distortions (Figures 3E, F), and inflated (Figure 3D) and shrunken structures (Figure 3C). These phenomena suggest that some substances produced by strain G5 can inhibit the growth of mycelia and destroy the mycelial cell membrane, resulting in mycelial shrinkage.
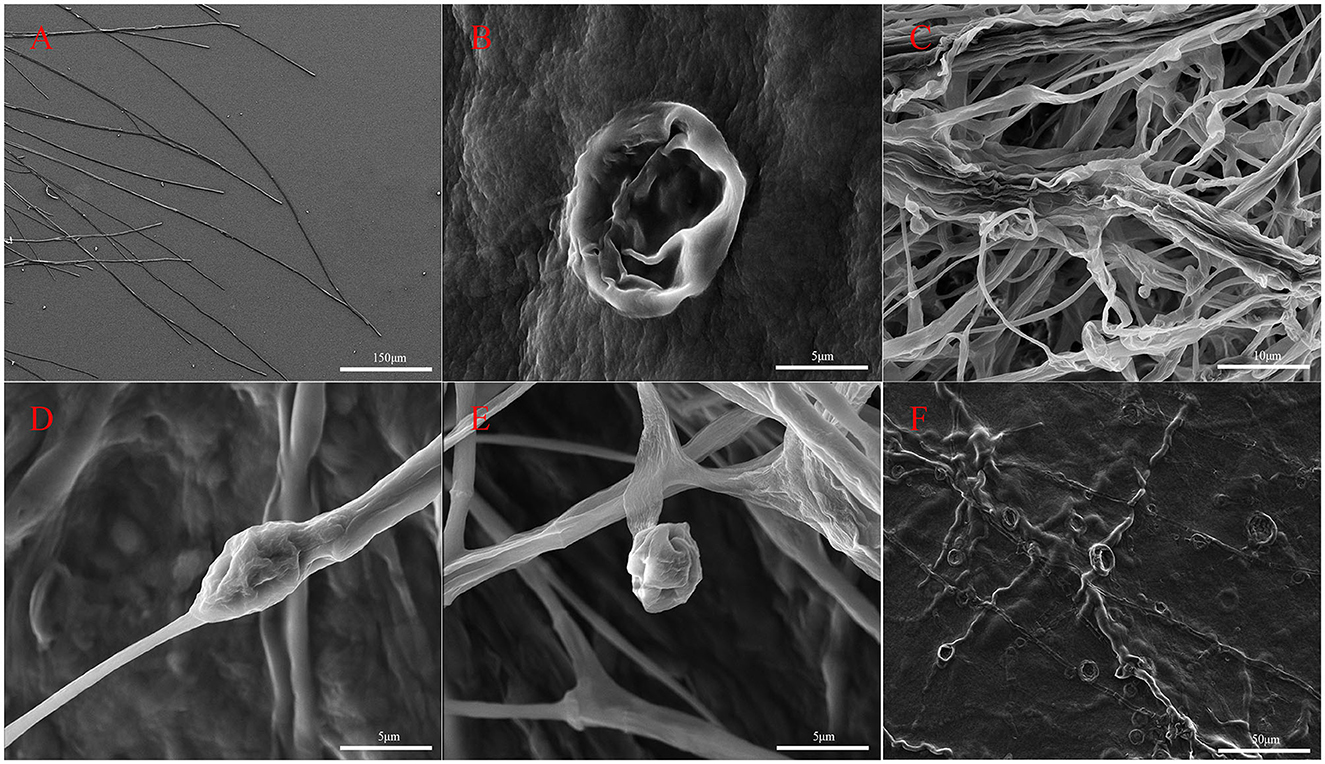
Figure 3. Scanning electron micrographs of M. oryzae mycelia control group (A) and treated with strain G5 (remaining pictures) after 7 days. M. oryzae mycelia treated with strain G5 were wrinkled (B), twisted (E, F), and locally inflated (C), and hyphal tips were deformed (D).
3.4. Effects of G5 on conidial germination and appressorium formation
For M. oryzae, successful infection of rice depends on both conidia germination and the formation of appressoria. Therefore, we studied the antagonistic effects of the strain G5 on these two processes of M. oryzae. G5 did not strongly inhibit conidial germination. At 16 hpi (hour post-infection), none of the G5 concentrations inhibited conidial germination. At all concentrations, OD600 = 1, OD600 = 1.5, and OD600 = 2, G5 allowed 99.33%, 98%, and 98.67% of the conidia to germinate, respectively. When the conidia were treated with G5 at OD600 = 2 and OD600 = 1.5, only 3.00% (Figure 4E) and 6.33% (Figure 4D) of the conidia formed appressoria, respectively. G5 at a lower concentration (OD600 = 1) did not completely impair appressorium formation; 23% of the conidia formed appressoria (Figure 4C). These results show that the inhibition of G5 on M. oryzae is mainly achieved by inhibiting the formation of the appressorium, with almost no inhibitory effect on conidial germination (Figure 4F). We speculated that G5 interferes with the infection mechanism of M. oryzae.
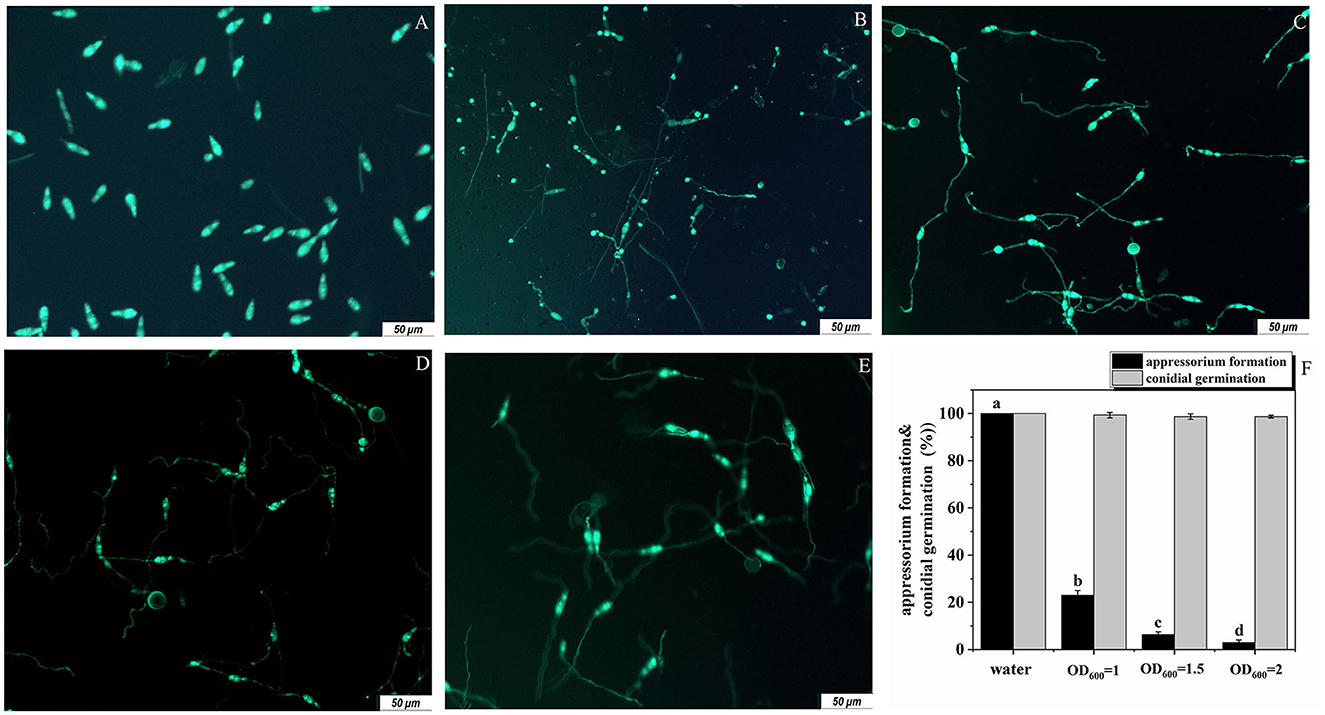
Figure 4. Effect of B. subtilis G5 at various concentrations on conidial germination and appressorium formation at 16 hpi. Conidia of strain Guy11-GFP (A) were treated with sterile water (B) and G5 at the concentrations indicated. Conidium development was observed with sterile water and G5 OD600 = 1 (C), OD600 = 1.5 (D), and OD600 = 2 (E). Conidia examined by differential interference fluorescence microscope. The scale bar indicates 50 μm, conidial germination and appressorium formation with sterile water and G5 at various concentrations. Data from three biological replicates were used to calculate the mean and standard deviation. Mean separation letters indicate statistical significance (P < 0.05) based on a one-way analysis of variance followed by the LSD test (F).
3.5. Biocontrol assays
3.5.1. Antifungal activity on detached rice leaves
The disease control efficacy of G5 on the detached rice leaves against rice blast is shown in Figure 5. Compared with the control group (11.42 ± 2.87 mm), the length of leaf lesion of the prevention group was significantly smaller (1.25 ± 0.42 mm; Figure 5A); the length of leaf lesion was also smaller in the treatment group (4.08 ± 0.80 mm). These results indicate that G5 displays a significant preventive effect that was greater than its curative effect (Figure 5B).
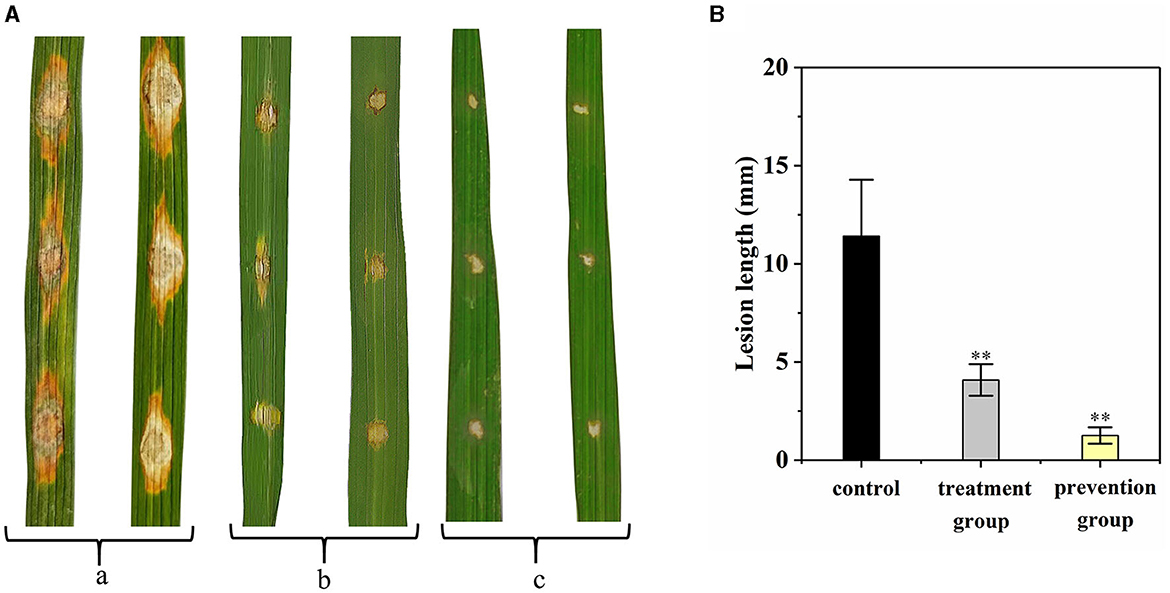
Figure 5. Disease control efficacy on detached rice leaves of G5 against rice blast. (a) Control group. (b) Treatment group. (c) Prevention group. The length of leaf lesions in vitro was analyzed (A). Independent samples t-test analyzed data. The symbols ** indicate significant differences at P < 0.01 (B).
3.5.2. Antifungal activity in greenhouse
We investigated whether G5 confers resistance to rice against rice blast in a greenhouse. Before 7 days of Guy11 inoculation, an inhibitory effect of G5 was observed in rice blast infection. Treating rice plants with G5 strains significantly reduced rice blast. The severity of disease in the control rice plants was observed to be significant, characterized by the formation of large circular or oval brown spots and dense disease spots. In comparison with the disease index of 85.04% observed in rice treated with sterile water, the disease index of rice treated with G5 was significantly lower at 28.23%, indicating a notable biocontrol effect of 66.81% (Figures 6A, B). Additionally, the treatment group exhibited a reduced leaf area covered by diseased spots, accompanied by the limited presence of necrotic spots. In conclusion, G5 has a positive control effect on rice blast.
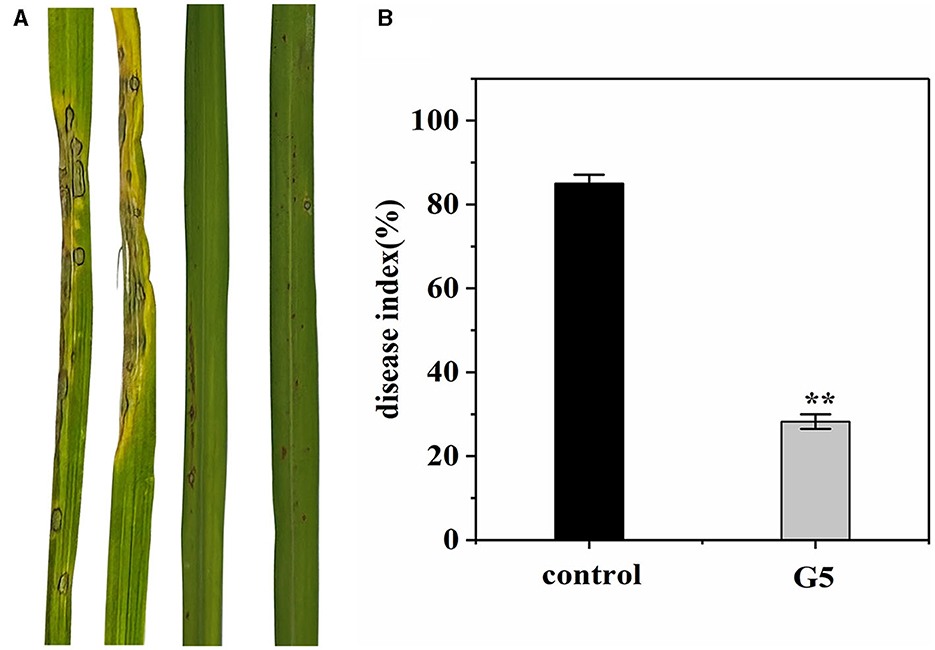
Figure 6. Effect of G5 on resistance against rice blast under greenhouse conditions. The severity of devastating symptoms on the leaves of G5-inoculated rice compared with control under greenhouse conditions (A). According to the disease classification, the disease index of G5-infected and control-infected rice was calculated (B). Bar charts were plotted with mean ± SD from 30 plants, respectively. Independent samples t-test analyzed data. The symbols ** indicate significant differences at P < 0.01.
3.6. Genome sequence assembly and general features of G5
Sequencing and analyzing the whole genome of G5 were completed. Accordingly, the G5 genome comprised a singular circular chromosome measuring 4,065,878 base pairs, exhibiting an average GC content of 43.82%. The genome was found to contain 30 rRNA, 86 tRNA, and 29 sRNA genes, as shown in Figure 7A. Furthermore, all 4,182 predicted open reading frames underwent annotation analysis by comparing with the Nr, Swiss-Prot, COG, and KEGG databases, resulting in the annotation of 3,977 candidate genes. Supplementary Figure 2 displays the functional annotations of the genome. COG analysis further categorized 3,015 genes into 26 functional groups, as presented in Figure 7A. The results revealed four main functional gene classes: amino acid transport and metabolism (251 genes), carbohydrate transport and metabolism (258 genes), transcription (256 genes), and cell wall/membrane/envelope biogenesis (181 genes), which represented 31.83% of the genes predicted in the COG analysis. Additionally, other gene clusters associated with inorganic ion transport and metabolism (173 genes), translation, ribosomal structure, and biogenesis (165 genes), energy production and conversion (164 genes), replication, recombination, and repair (108 genes), and coenzyme transport and metabolism (121 genes) accounted for 20.23% of the predicted genes. Moreover, 24.7% of the predicted genes were associated with general function prediction only; unknown functions were poorly characterized. It is worth noting that 54 genes involved in defense mechanisms were annotated, of which 25 encoded the ABC-type multidrug transport system, 10 encoded the resistance protein ABC-type antimicrobial peptide transport system, five encoded the resistance protein beta-lactamase, four encoded the Na+-driven multidrug efflux pump, two encoded the multidrug resistance efflux pump, two encoded the microcin C7 resistance protein, one encoded streptogramin lyase, and one encoded the vancomycin resistance protein. These genes may be involved in the antibacterial function of G5 through the synthesis and transport of antibiotics.
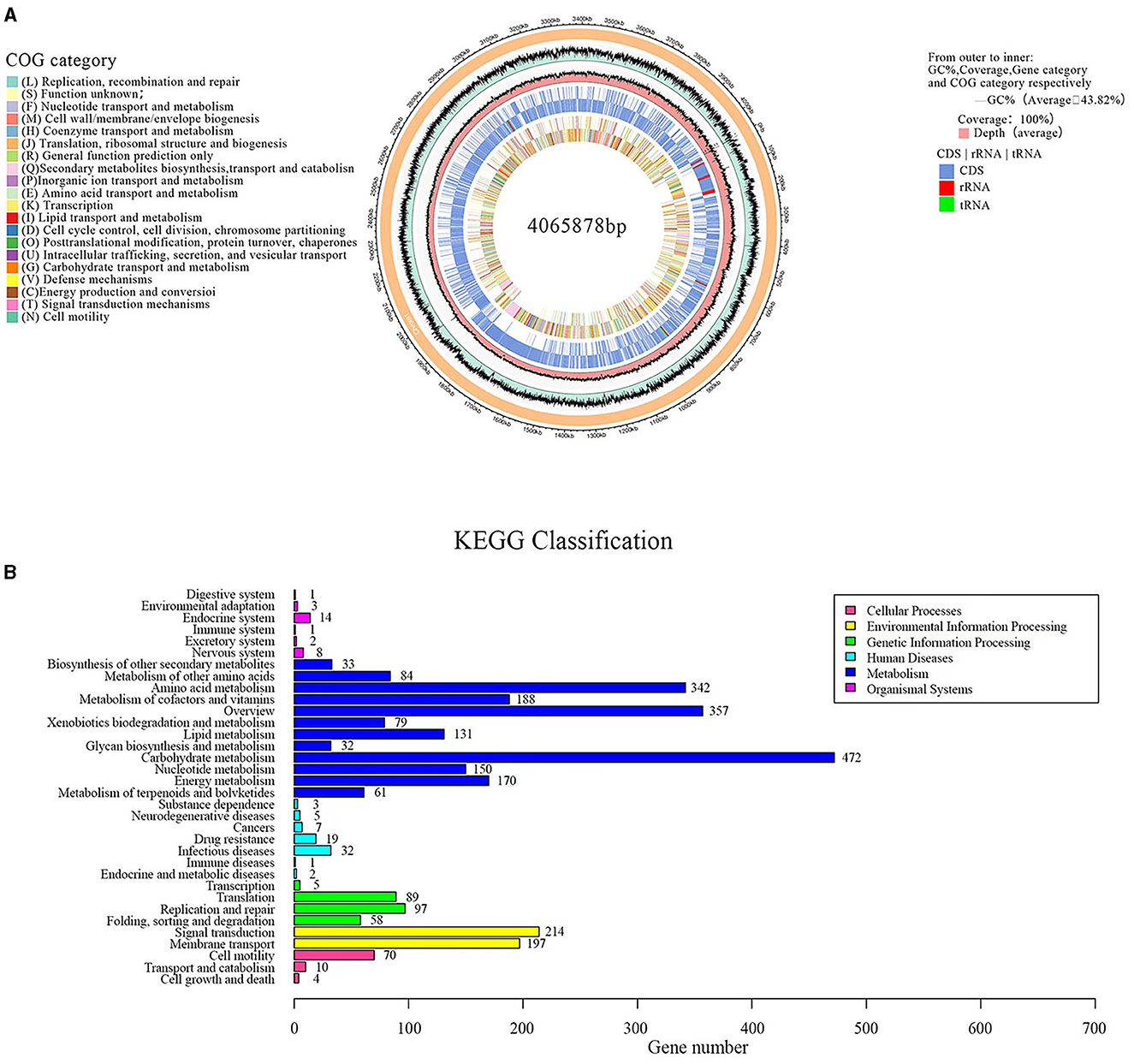
Figure 7. Circular genome map of endophytic B. subtilis G5 (A). From outside to inside, circle 1, the size of the complete genome, each scale represents 20 kb; circle 2, G + C content; circle 3, coverage; circles 4 and 5, gene category on the + and – strands, CDS in light blue, tRNA in red, tRNA in green; circles 6 and 7, the predicted protein-coding genes on the + and – strands, different colors represent different COG function classification. KEGG pathway classification (B). The vertical axis is the name of the metabolic pathway involved, and the horizontal axis is the number of genes annotated to that pathway.
A total of 2,250 genes were mapped to five KEGG branches, including cellular processes, environmental information processing, genetic information processing, metabolism, and organismal systems. Among these, a high proportion of the annotated genes was assigned to metabolism, especially the pathways belonging to carbohydrate metabolism (474 genes), overview maps (357 genes), and amino acid metabolism (342 genes) (Figure 7B). Similar to the COG annotations, carbohydrate metabolism was emphasized in the KEGG pathways. In addition, 35 genes were annotated for the biosynthesis of secondary metabolites, such as penicillin and cephalosporin (EC: 3.5.2.6), and 61 genes were annotated for terpenoid and polyketide metabolism, including terpenoid backbone biosynthesis (EC: 2.7.1.148; EC: 2.7.7.60), non-ribosomal peptide structure, and butanoate metabolism (EC: 2.3.3.10; EC: 2.3.1.9). These genes have been speculated to be closely related to the production of G5 antibacterial substances.
3.7. Genetic basis for pathogen inhibition
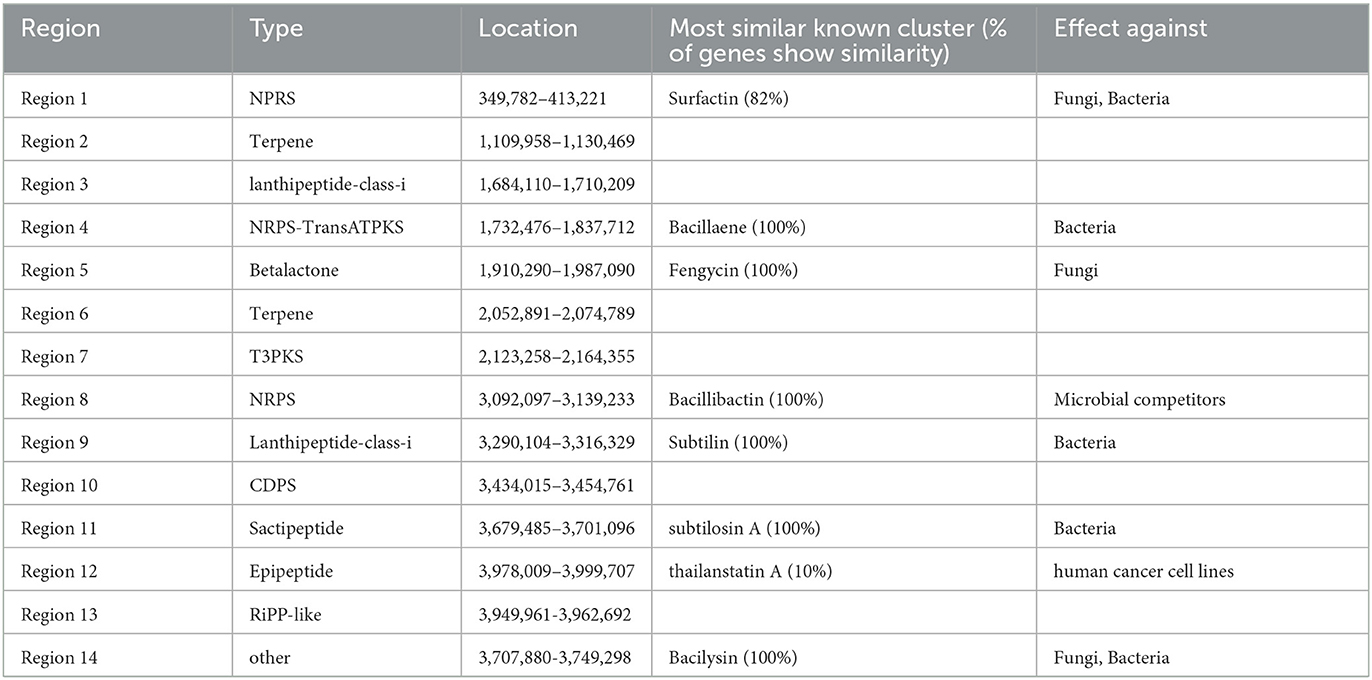
The secondary metabolites of the G5 genome were analyzed using anti-SMASH. Fourteen secondary metabolite gene clusters were predicted (Table 2), including eight highly similar antibiotic synthesis gene clusters: two cyclic lipopeptides (surfactin and fengycin), one polyketide antibiotic (bacillaene), one siderophore (bacillibactin), one dipeptide (bacilysin), one lanthipeptide (subtilin), one thiopeptide (subtilosin A), and one antibiotic gene cluster (thailanstatin A). Fengycin has strong antifungal activity against filamentous fungi; surfactin and bacilysin also exhibit antifungal activities (Ongena and Jacques, 2008). Therefore, the observed inhibitory effect of the G5 strain on M. oryzae may have been the result of the combined action of these three metabolites. B. subtilis G5 also had four gene clusters with unknown functions, including two terpene clusters, one tRNA-dependent cyclodipeptide synthase cluster, one type III polyketide synthase (PKS) cluster, one lanthipeptide class-I cluster, and one RiPP (ribosomally synthesized and post-translationally modified peptides)-like cluster, suggesting that some B. subtilis G5 gene clusters may synthesize new antibacterial substances. The simultaneous production of these antimicrobial metabolites may exhibit synergistic properties in combating pathogens. These metabolites serve as the foundation for pathogen suppression by this endophytic bacterium. Supplementary genes associated with pathogen suppression are presented in Table 2.
3.8. Genetic basis for plant growth promotion
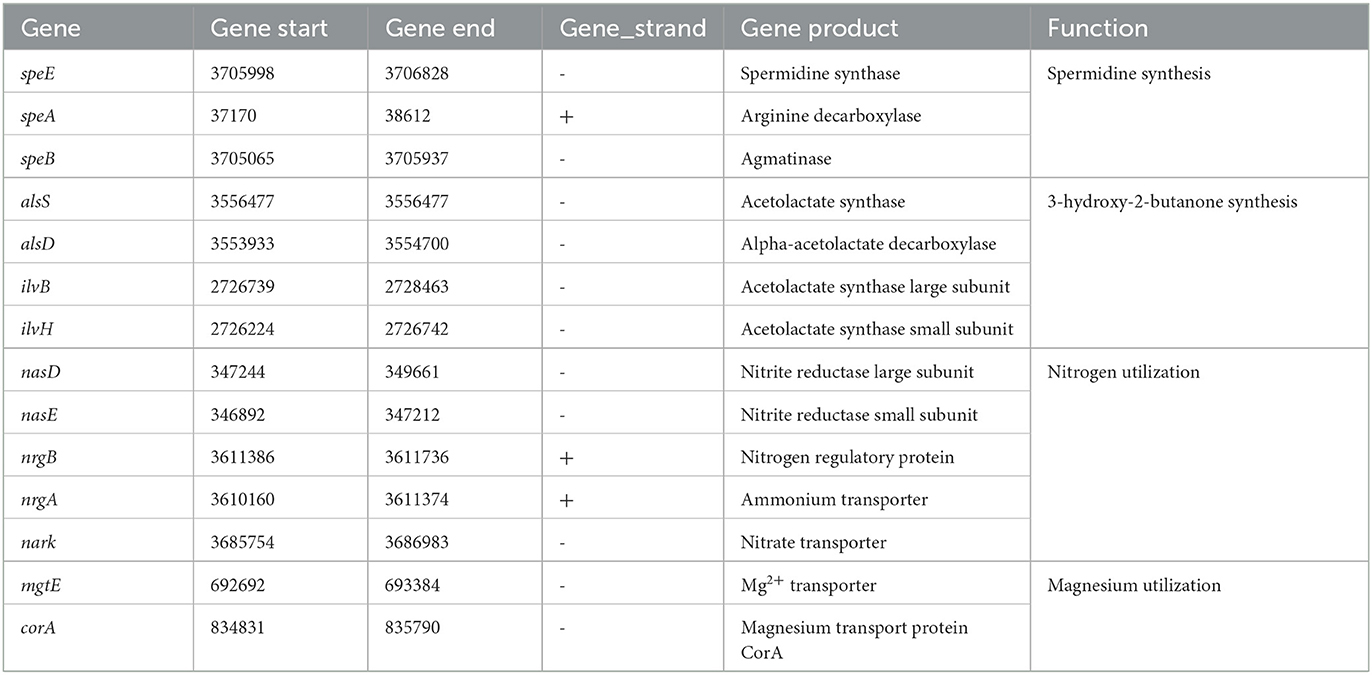
In the G5 genome, numerous genes/gene clusters linked to the promotion of plant growth were detected. These encompass genes involved in siderophore synthesis, 3-hydroxy-2-butanone synthesis, and nutrient utilization. Additionally, genes accountable for spermidine synthesis, such as spermidine synthase (speE), arginine decarboxylase (speA), and agmatinase (speB), were identified. Spermidine is a polyamine that promotes plant growth by inducing expansin-related gene expression and inhibiting ethylene synthesis. Siderophores help plants by delivering iron or inhibiting the growth of phytopathogenic fungi by competing for iron (Arguelles-Arias et al., 2009). The genome analysis revealed the presence of genes associated with the synthesis of the volatile bio-promoting metabolite 3-hydroxy-2-butanone, such as acetolactate decarboxylase (alsD), acetolactate synthase large subunit (ilvB), and acetolactate synthase small subunit (ilvH). This metabolite has been shown to enhance plant growth by increasing plant fresh weight. Furthermore, the identification of genes involved in nitrogen and magnesium utilization, such as nitrite reductase large subunit (nasD), nitrite reductase small subunit (nasE), nitrogen regulatory protein (nrgB), ammonium transporter (nrgA), nitrate transporter (nark), and Mg2+ transporter (mgtE), was also accomplished. It is worth noting that these genes have the potential to augment nutrient accessibility. Moreover, Table 3 provides a comprehensive summary of additional genes that have been identified in relation to the promotion of plant growth.
4. Discussion
Plant roots create a favorable environment for endophytes, offering shelter and nourishment. This enables the endophytes to establish symbiotic connections with host plants. Nutrients discharged through root exudation serve as a continuous source of nutrients for endophytes, supporting essential metabolic functions and overall plant health, even under challenging conditions. Indeed, the pivotal role of endophytic communities in enhancing wild rice adaptability to harsh conditions is well documented (Urs et al., 2013; Zhu et al., 2022). These microorganisms bolster the plant's resilience against diverse stresses, such as drought, salinity, heavy metals, and pathogens.
In our study, the isolation of B. subtilis from wild rice roots revealed its potential to substantially enhance nutrient uptake and root development, thus contributing to the overall plant wellbeing. The incorporation of B. subtilis into wild-type rice roots yielded notable benefits. Ameliorating nutrient absorption can significantly enhance growth and concurrently provide biocontrol against rice blast disease. Visual representations elucidate the dynamics of bacterial interactions with host plants (Figure 8). There have also been previous reports on the positive interplay between B. subtilis and host plants. B. subtilis, a prominent plant growth-promoting rhizobacterium, is an example of this in the field (Earl et al., 2008; Todorova and Kozhuharova, 2010). Its multifaceted mechanisms include nutrient enhancement, hormone modulation, and production of antimicrobial agents. Moreover, its spore-forming capability provides resilience against environmental challenges, making it a promising candidate for sustainable agriculture.
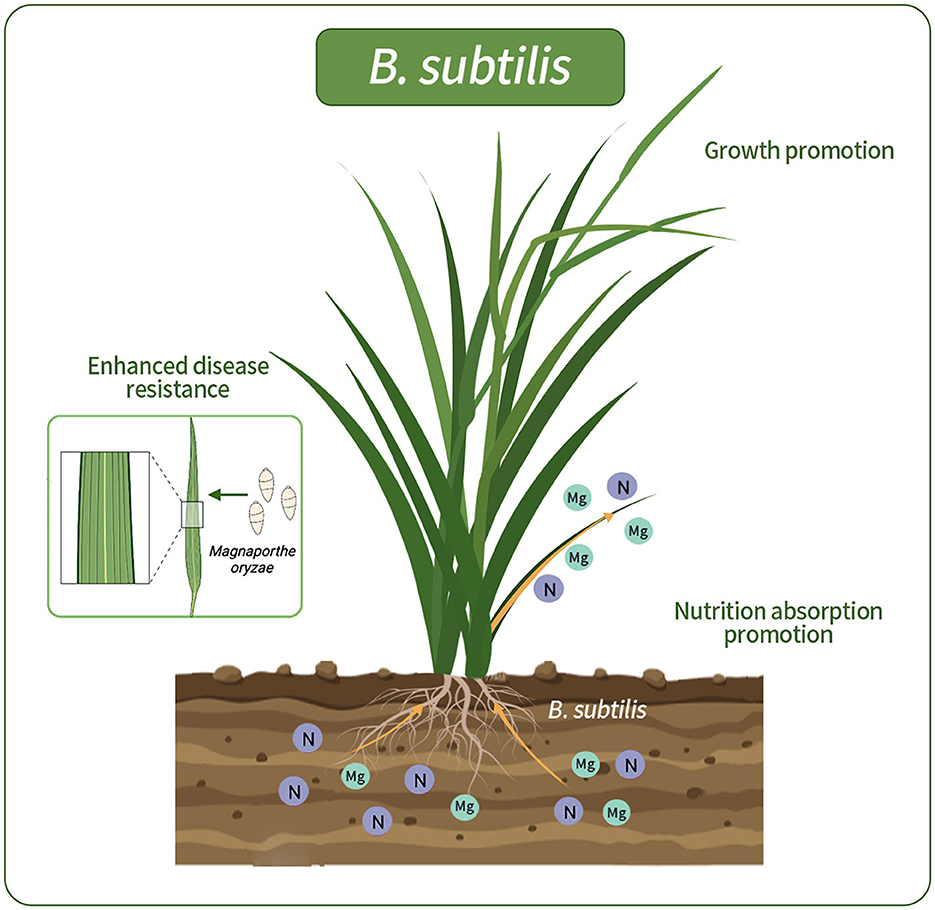
Figure 8. Schematic representations of rice colonized by G5. G5 promoted the absorption of N and Mg elements, promoted rice growth, and enhanced the resistance to rice blast.
The exploration of endophytes in wild rice has revealed diverse species with growth promoting and antagonistic attributes. These findings underscore the significance of wild rice as a repository of valuable microbial resources that are essential for eco-friendly farming methods and the preservation of crop genetic diversity. B. subtilis, a versatile bacterium, has been demonstrated to be a growth promoter and pathogen controller (Ryu et al., 2003; Hayat et al., 2010; Fan et al., 2017). Its array of benefits includes eliciting systemic resistance and withstanding adverse conditions. B. subtilis strains exhibit a spectrum of effects, ranging from iron mobilization to ethylene modulation (Xie et al., 2014; Freitas et al., 2015; Woo et al., 2020). Variants such as GBO3, OKB105, and GOT9 show strain-specific advantages, ranging from iron migration to drought tolerance. Notably, B. subtilis JA has demonstrated the potential as a biocontrol agent via volatile inhibition (Chen et al., 2008), whereas B. subtilis SS12.9 displays iturin-based biocontrol against Xanthomonas oryzae pv. Oryzae (Beri et al., 2012).
The present study corroborated the antagonistic potential of the G5 strain against M. oryzae using multiple approaches, including SEM, plate confrontation, and biocontrol assays. The unique morphological alterations in M. oryzae hyphae induced by G5 treatment underscore its distinct mechanism of action (Sha et al., 2020; Chen et al., 2021). Disruption of appressorium formation emerged as a key facet of the effects of G5 on M. oryzae, in contrast to previous studies (Chen et al., 2019; Liu et al., 2021). In their study, antagonistic bacteria inhibited both conidial germination and appressorium formation of M. oryzae. This underscores the pivotal role of the appressorium in M. oryzae infection and highlights the potential for G5 to mitigate rice blast by impeding infection structure formation.
The G5 genome hosts intriguing antibiotic synthesis genes, such as surfactin, fengycin, and bacillaene, which are known for their antagonistic properties (Heerklotz and Seelig, 2007; Erega et al., 2021). These compounds confer broad-spectrum antibacterial and antifungal properties, suggesting application potential for G5. Additionally, the novel identified gene clusters indicate unexplored antibacterial substances. The G5 strain also contains genes related to plant growth promotion, such as those linked to spermidine synthesis and acetoin production (Zhang et al., 2015; Nascimento et al., 2019). These attributes indicate their potential to bolster plant development and systemic resistance, which warrant further investigation. The diverse biosynthetic abilities of B. subtilis warrant attention and customization for specific applications. It is essential to recognize the disparity between B. subtilis strains and optimize the fermentation conditions or conduct genetic modifications to maximize efficacy.
5. Conclusion
In summary, we successfully isolated the endophytic antagonistic bacterium G5 from O. officinalis Wall. and identified it as a strain of B. subtilis through morphological and genetic analyses. G5 exhibits potent biocontrol effects against M. oryzae via distinct mechanisms. Its genome holds promise for understanding pathogen inhibition and microbial inoculum development. Harnessing the potential of B. subtilis G5 will require tailored approaches and optimization to maximize its role in agriculture.
Data availability statement
The data presented in this study are deposited in the National Center for Biotechnology Information (NCBI) GenBank (https://www.ncbi.nlm.nih.gov/genbank/) under accession number OQ874691. The assembled sequences were deposited in the NCBI (BioProject ID: PRJNA997266).
Author contributions
L-YL: Writing—original draft. Z-XX: Data curation, Writing—original draft. J-LL: Conceptualization, Writing—original draft. D-ZY: Methodology, Writing—review and editing. LL: Investigation, Writing—review and editing. LC: Resources, Writing—review and editing. Q-FZ: Project administration, Writing—review and editing. F-YY: Formal analysis, Writing—review and editing. R-XL: Data curation, Writing—original draft. Z-QC: Conceptualization, Writing—review and editing. S-QX: Resources, Writing—review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Central Guidance on Local Science and Technology Development Fund of Yunnan Province (202207AB110012), the Yunnan Youth Top Talent Project (YNWR-QNBJ-2018-284), and the Scientific and technological talents and platform plan (2019HB034).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1264000/full#supplementary-material
References
Afzal, L., Shinwari, Z. K., Sikandar, and Shahzad, S. (2019). Plant beneficial endophytic bacteria: mechanisms, diversity, host range and genetic determinants. Microbiol. Res. 221, 36–49. doi: 10.1016/j.micres.2019.02.001
Arguelles-Arias, A., Ongena, M., Halimi, B., Lara, Y., Brans, A., Joris, B., et al. (2009). Bacillus amyloliquefaciens ga1 as a source of potent antibiotics and other secondary metabolites for biocontrol of plant pathogens. Microb. Cell Fact. 8, 63. doi: 10.1186/1475-2859-8-63
Asibi, A. E., Chai, Q., and Coulter, J. A. (2019). Rice blast: a disease with implications for global food security. Agronomy. 9, 451. doi: 10.3390/agronomy9080451
Beri, T., Koji, M., Stankovi, S., Topisirovi, L., Degrassi, G., Myers, M., et al. (2012). Antimicrobial activity of bacillus sp. Natural isolates and their potential use in the biocontrol of phytopathogenic bacteria. Food Technol. Biotechnol. 50, 25–31. doi: 10.2147/OTT.S23874
Bevivino, A., Sarrocco, S., Dalmastri, C., Tabacchioni, S., Cantale, C., and Chiarini, L. (1998). Characterization of a free-living maize-rhizosphere population of burkholderia cepacia: effect of seed treatment on disease suppression and growth promotion of maize. Fems Microbiol. Ecol. 27, 225–237. doi: 10.1111/j.1574-6941.1998.tb00539.x
Bu, S. W., Munir, S., He, P. F., Li, Y. M., Wu, Y. X., Li, X. Y., et al. (2021). Bacillus subtilis L1-21 as a biocontrol agent for post harvest gray mold of tomato caused by Botrytis cinerea. Biol. Control. 157, 104568. doi: 10.1016/j.biocontrol.2021.104568
Chen, H., Xiao, X., Wang, J., Wu, L., Zheng, Z., and Yu, Z. (2008). Antagonistic effects of volatiles generated by Bacillus subtilis on spore germination and hyphal growth of the plant pathogen, Botrytis cinerea. Biotechnol. Lett. 30, 919–923. doi: 10.1007/s10529-007-9626-9
Chen, W. Q., Zhao, L., Li, H., Dong, Y. L., Xu, H., Guan, Y., et al. (2019). The isolation of the antagonistic strain Bacillus australimaris CQ07 and the exploration of the pathogenic inhibition mechanism of Magnaporthe oryzae. PLoS ONE. 14, e220410. doi: 10.1371/journal.pone.0220410
Chen, Z., Zhao, L., Chen, W. Q., Dong, Y. L., Yang, C., Li, C., et al. (2020). Isolation and evaluation of Bacillus velezensis ZW-10 as a potential biological control agent against Magnaporthe oryzae. Biotechnol. Biotechnol. Equip. 34, 714–724. doi: 10.1080/13102818.2020.1803766
Chen, Z., Zhao, L., Dong, Y. L., Chen, W. Q., Li, C. L., Gao, X. L., et al. (2021). The antagonistic mechanism of Bacillus velezensis ZW10 against rice blast disease: evaluation of ZW10 as a potential biopesticide. PLoS ONE 16, e0256807. doi: 10.1371/journal.pone.0256807
Cheng, Z. Q., Huang, X. Q., Zhang, Y. Z., Qian, J., Yang, M. Z., Cheng-Jun, W. U., et al. (2005). Diversity in the content of some nutritional components in husked seeds of three wild rice species and rice varieties in Yunnan province of China. J. Integr. Plant Biol. 47, 11. doi: 10.1111/j.1744-7909.2005.00130.x
Davison, J. (1988). Plant Beneficial Bacteria. Nat. Biotechnol. 6, 282–286. doi: 10.1038/nbt0388-282
De, J., Ramaiah, N., and Vardanyan, L. (2008). Detoxification of toxic heavy metals by marine bacteria highly resistant to mercury. Mar. Biotechnol. 10, 471–477. doi: 10.1007/s10126-008-9083-z
Dean, R. A., Talbot, N. J., Ebbole, D. J., Farman, M. L., Mitchell, T. K., Orbach, M. J., et al. (2005). The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 434, 980–986. doi: 10.1038/nature03449
Devanna, B. N., Jain, P., Solanke, A. U., Das, A., Thakur, S., Singh, P. K., et al. (2022). Understanding the dynamics of blast resistance in rice-Magnaporthe oryzae interactions. J. Fungi. 8, 584. doi: 10.3390/jof8060584
Devanna, N. B., Joshitha, V., and Sharma, T. R. (2014). The blast resistance gene Pi54of cloned from Oryza officinalis interacts with avr-pi54 through its novel NBS-LRR domains. PLoS ONE 9, e104840. doi: 10.1371/journal.pone.0104840
Djordje, F., Ivica, D., Tanja, B., Jelena, L., and Slavi, A. S. (2018). Biological control of plant pathogens by bacillus species. J. Biotechnol. 285, 44–55. doi: 10.1016/j.jbiotec.2018.07.044
Dutta, D., Puzari, K., Godoi, R., and Dutta, P. (2014). Endophytes: exploitation as a tool in plant protection. Braz. Arch. Biol. Technol. 57, 621–629. doi: 10.1590/S1516-8913201402043
Earl, A. M., Losick, R., and Kolter, R. (2008). Ecology and genomics of Bacillus subtilis. Trends Microbiol. 16, 269–275. doi: 10.1016/j.tim.2008.03.004
Erega, A., Stefanic, P., Dogsa, I., Danevcic, T., Simunovic, K., Klancnik, A., et al. (2021). Bacillaene mediates the inhibitory effect of bacillus subtilis on campylobacter jejuni biofilms. Appl. Environ. Microbiol. 87, e295520. doi: 10.1128/AEM.02955-20
Fan, H. Y., Zhang, Z. W., Li, Y., Zhang, X., Duan, Y. M., and Wang, Q. (2017). Biocontrol of bacterial fruit blotch by Bacillus subtilis 9407 via surfactin-mediated antibacterial activity and colonization. Front. Microbiol. 8, 1973. doi: 10.3389/fmicb.2017.01973
Ferreira, J., Matthee, F. N., and Thomas, A. C. (1991). Biological control of eutypa lata on grapevine by an antagonistic strain of Bacillus subtilis. Phytopathology. 81, 283–287. doi: 10.1094/Phyto-81-283
Freitas, M. N. A., Medeiros, F. H. V., Carvalho, S. P., Guilherme, L. R. G., Teixeira, W. D., Zhang, H. M., et al. (2015). Augmenting iron accumulation in cassava by the beneficial soil bacterium Bacillus subilis (GBO3). Front. Plant Sci. 6, 596. doi: 10.3389/fpls.2015.00596
Glick, B. R. (2020). Beneficial Plant-Bacterial Interactions: Beneficial Plant-Bacterial Interactions.
Hayat, R., Ali, S., Amara, U., Khalid, R., and Ahmed, I. (2010). Soil beneficial bacteria and their role in plant growth promotion: a review. Ann. Microbiol. 60, 579–598. doi: 10.1007/s13213-010-0117-1
Heerklotz, H., and Seelig, J. (2007). Leakage and lysis of lipid membranes induced by the lipopeptide surfactin. Eur. Biophys. J. 36, 305–314. doi: 10.1007/s00249-006-0091-5
Huang, W., Liu, X., Zhou, X., Wang, X., and Liu, H. (2019). Calcium signaling is suppressed in Magnaporthe oryzae conidia by Bacillus cereus HS24. Phytopathology 110, 309–316. doi: 10.1094/PHYTO-08-18-0311-R
Ji, X. L., Lu, G. B., Gai, Y. P., Zheng, C. C., and Mu, Z. M. (2010). Biological control against bacterial wilt and colonization of mulberry by an endophytic Bacillus subtilis strain. FEMS Microbiol. Ecol. 65, 565–573. doi: 10.1111/j.1574-6941.2008.00543.x
Kai, B., Thomas, W., Chevrette, M. G., Xiaowen, L., Schwalen, C. J., et al. (2017). Antismash 4.0-improvements in chemistry prediction and gene cluster boundary identification. Nucl. Acids Res. 45, 36–41. doi: 10.1093/nar/gkx319
Karthikeyan, V., and Gnanamanickam, S. S. (2008). Biological control of setaria blast (Magnaporthe grisea) with bacterial strains. Crop Prot. 27, 263–267. doi: 10.1016/j.cropro.2007.05.013
Khan, A. R., Mustafa, A., Hyder, S., Valipour, M., Rizvi, Z. F., Gondal, A. S., et al. (2022). Bacillus spp. as bioagents: uses and application for sustainable agriculture. Biology 11, 1763. doi: 10.3390/biology11121763
Kiran, T. V., Rao, Y. V., Subrahmanyam, D., Rani, N. S., Bhadana, V. P., Rao, P. R., et al. (2013). Variation in leaf photosynthetic characteristics in wild rice species. Photosynthetica 51, 350–358. doi: 10.1007/s11099-013-0032-3
Komatsu, K., Urayama, S. L., Katoh, Y., Fuji, S. I., Hase, S., Fukuhara, T., et al. (2016). Detection of Magnaporthe oryzae chryso virus 1 in Japan and establishment of a rapid, sensitive and direct diagnostic method based on reverse transcription loop-mediated isothermal amplification. Arch. Virol. 161, 317–326. doi: 10.1007/s00705-015-2666-x
Li, H., Guan, Y., Dong, Y., Zhao, L., Rong, S., Chen, W., et al. (2018). Isolation and evaluation of endophytic Bacillus tequilensis GYLH001 with potential application for biological control of Magnaporthe oryzae. PLoS ONE 13, e203505. doi: 10.1371/journal.pone.0203505
Li, Y. B., Wu, C. J., Jiang, G. H., Wang, L. Q., and He, Y. Q. (2007). Dynamic analyses of rice blast resistance for the assessment of genetic and environmental effects. Plant Breed. 126, 541–547. doi: 10.1111/j.1439-0523.2007.01409.x
Liu, X. Y., Bao, T. T., Zheng, L., Kgosi, V. T., Liu, H. Y., and Liu, H. X. (2021). Cell wall integrity in Magnaporthe oryzae is weakened by proteins secreted by Bacillus licheniformis BLO6. Biol. Control. 157, 104582. doi: 10.1016/j.biocontrol.2021.104582
Nascimento, F. X., Hernandez, A. G., Glick, B. R., and Rossi, M. J. (2019). Plant growth-promoting activities and genomic analysis of the stress-resistant Bacillus megaterium STB1, a bacterium of agricultural and biotechnological interest. Biotechnol. Rep. 25, e00406. doi: 10.1016/j.btre.2019.e00406
Ongena, M., and Jacques, P. (2008). Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends Microbiol. 16, 115–25. doi: 10.1016/j.tim.2007.12.009
Ons, L., Bylemans, D., Thevissen, K., and Cammue, B. P. A. (2020). Combining biocontrol agents with chemical fungicides for integrated plant fungal disease control. Microorganisms 8, 1930. doi: 10.3390/microorganisms8121930
Rossman, A. Y., Howard, R. J., and Valent, B. (1990). Pyricularia grisea, the correct name for the rice blast disease fungus. Mycologia 4, 509–512. doi: 10.1080/00275514.1990.12025916
Ryu, C. M., Farag, M. A., Hu, C. H., Reddy, M. S., Wei, H. X., and Pare, P. W. (2003). Bacterial volatiles promote growth in Arabidopsis. Proc. Natl. Acad. Sci. USA. 100, 4927–4932. doi: 10.1073/pnas.0730845100
Santoyo, G., Moreno-Hagelsieb, G., Del Carmen Orozco-Mosqueda, M., and Glick, B. R. (2016). Plant growth-promoting bacterial endophytes. Microbiol. Res. 183, 92–99. doi: 10.1016/j.micres.2015.11.008
Sha, Y., Zeng, Q., and Sui, S. (2020). Screening and application of Bacillus strains isolated from nonrhizospheric rice soil for the biocontrol of rice blast. Plant Pathol. J.36, 231–243. doi: 10.5423/PPJ.OA.02.2020.0028
Siddikee, M. A., Chauhan, P. S., and Anandham, H. (2010). Isolation, characterization, and use for plant growth promotion under salt stress, of acc deaminase-producing halotolerant bacteria derived from coastal soil. J. Microbiol. Biotechnol. 20, 1577–1584. doi: 10.4014/jmb.1007.07011
Sturz, A. V., and Nowak, J. (2000). Endophytic communities of rhizobacteria and the strategies required to create yield enhancing associations with crops. Agric. Ecosyst. Environ. Appl. Soil Ecol. 15, 183–190. doi: 10.1016/S0929-1393(00)00094-9
Tan, G. X., Ren, X., Weng, Q. M., Shi, Z. Y., Zhu, L. L., and He, G. C. (2004). Mapping of a new resistance gene to bacterial blight in rice line introgressed from Oryza officinalis. Acta Genetica Sinica. 31, 724–729.
Tan, R. X., and Zou, W. X. (2001). Endophytes: a rich source of functional metabolites. Nat. Prod. Rep. 18, 448–459. doi: 10.1039/b100918o
Tian, Q., Gong, Y., Liu, S., Ji, M., Tang, R., Kong, D., et al. (2023). Endophytic bacterial communities in wild rice (Oryza officinalis) and their plant growth-promoting effects on perennial rice. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1184489
Todorova, S., and Kozhuharova, L. (2010). Characteristics and antimicrobial activity of Bacillus subtilis strains isolated from soil. World J. Microbiol. Biotechnol. 26, 1207–1216. doi: 10.1007/s11274-009-0290-1
Urs, L., Yi, D., Aline, B., Magnus, R., Hajirezaei, M. R., Stefanie, D. H., et al. (2013). Host-related metabolic cues affect colonization strategies of a root endophyte. Proc. Nail. Acad. Sci.U. S. A. 110, 13965–13970. doi: 10.1073/pnas.1301653110
Wang, S. Y., Herrera-Balandrano, D. D., Wang, Y. X., Shi, X. C., Chen, X., Jin, Y., et al. (2022). Biocontrol ability of the bacillus amyloliquefaciens group, b. Amyloliquefaciens, b. Velezensis, b. Nakamurai, and b. Siamensis, for the management of fungal postharvest diseases: a review. J. Agric. Food Chem. 23, 70. doi: 10.1021/acs.jafc.2c01745
Wang, X., Lee, S., Wang, J., Ma, J., and Ji, Y. (2014). Current advances on genetic resistance to rice blast disease. Rice-Germplasm. Genet. Improv. 23, 195–217. doi: 10.5772/56824
Wei, Z., Yang, X., Yin, S., Shen, Q., Ran, W., and Xu, Y. (2011). Efficacy of bacillus-fortified organic fertiliser in controlling bacterial wilt of tomato in the field. Agric. Ecosyst. Environ,. Appl. Soil Ecol. 48, 152–159. doi: 10.1016/j.apsoil.2011.03.013
Woo, 0., Kim, H., Kim, J., Keum, H. L., Lee, K., Sul, W. J., et al. (2020). Bacillus subtilis strain GOT9 confers enhanced tolerance to drought and salt stresses in Arabidopsis thaliana and Brassica campestris. Plant Physiol. Biochem. 148, 359–367. doi: 10.1016/j.plaphy.2020.01.032
Xie, S. S., Wu, H. J., Zang, H. Y., Wu, L. M., Zhu, Q. Q., and Gao, X. W. (2014). Plant growth promotion by spermidine-producing Bacillus subtilis OKB105. Mol. Plant-Microbe Int. 27, 655–663. doi: 10.1094/MPMI-01-14-0010-R
Zhang, N., Yang, D. Q., Wang, D. D., Miao, Y. Z., Shao, J. H., Zhou, X., et al. (2015). Whole transcriptomic analysis of the plant-beneficial rhizobacterium Bacillus amyloliquefaciens SQR9 during enhanced biofilm formation regulated by maizeroot exudates. BMC Genomics. 16, 685. doi: 10.1186/s12864-015-1825-5
Keywords: endophytic bacteria, rice blast, Bacillus subtilis, disease resistance, genome
Citation: Lei L-Y, Xiong Z-X, Li J-L, Yang D-Z, Li L, Chen L, Zhong Q-F, Yin F-Y, Li R-X, Cheng Z-Q and Xiao S-Q (2023) Biological control of Magnaporthe oryzae using natively isolated Bacillus subtilis G5 from Oryza officinalis roots. Front. Microbiol. 14:1264000. doi: 10.3389/fmicb.2023.1264000
Received: 20 July 2023; Accepted: 15 September 2023;
Published: 09 October 2023.
Edited by:
Francisca Suárez-Estrella, University of Almeria, SpainReviewed by:
Orlando Borras-Hidalgo, Qilu University of Technology, ChinaAnjali Chandrol Solanki, Mansarovar Global University, India
Copyright © 2023 Lei, Xiong, Li, Yang, Li, Chen, Zhong, Yin, Li, Cheng and Xiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Su-Qin Xiao, eGlhb3N1cWluMjI3QDEyNi5jb20=
 Ling-Yun Lei
Ling-Yun Lei Zi-Xuan Xiong1,2
Zi-Xuan Xiong1,2 Jin-Lu Li
Jin-Lu Li De-Zheng Yang
De-Zheng Yang Liu Li
Liu Li Rong-Xin Li
Rong-Xin Li