
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol., 06 October 2023
Sec. Virology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1261651
The endosomal sorting complex required for transport (ESCRT) is an essential molecular machinery in eukaryotic cells that facilitates the invagination of endosomal membranes, leading to the formation of multivesicular bodies (MVBs). It participates in various cellular processes, including lipid bilayer remodeling, cytoplasmic separation, autophagy, membrane fission and re-modeling, plasma membrane repair, as well as the invasion, budding, and release of certain enveloped viruses. The ESCRT complex consists of five complexes, ESCRT-0 to ESCRT-III and VPS4, along with several accessory proteins. ESCRT-0 to ESCRT-II form soluble complexes that shuttle between the cytoplasm and membranes, mainly responsible for recruiting and transporting membrane proteins and viral particles, as well as recruiting ESCRT-III for membrane neck scission. ESCRT-III, a soluble monomer, directly participates in vesicle scission and release, while VPS4 hydrolyzes ATP to provide energy for ESCRT-III complex disassembly, enabling recycling. Studies have confirmed the hijacking of ESCRT complexes by enveloped viruses to facilitate their entry, replication, and budding. Recent research has focused on the interaction between various components of the ESCRT complex and different viruses. In this review, we discuss how different viruses hijack specific ESCRT regulatory proteins to impact the viral life cycle, aiming to explore commonalities in the interaction between viruses and the ESCRT system.
The Endosomal Sorting Complexes Required for Transport (ESCRT) system is a peripheral membrane protein complex composed of ESCRT-0, ESCRT-I, ESCRT-II, ESCRT-III, VPS4-VTA1, and several accessory proteins including ALIX (Johnson et al., 2018; Zhang et al., 2021). They are recruited to distinct cellular locations to perform key steps in essential processes such as protein degradation, cell division, and membrane sealing (Migliano et al., 2022). The ESCRT system was initially discovered in yeast cells (Rothman and Stevens, 1986) and subsequently found to exist in insects, mammalian cells, and even archaea. It is evolutionarily conserved in all eukaryotes and plays important roles in cytoplasmic separation (Spitzer et al., 2006), cellular autophagy (Rusten and Stenmark, 2009; Isono, 2021), other membrane fission and remodeling (Schoneberg et al., 2017), as well as replication and budding processes of certain enveloped and non-enveloped viruses (Wirblich et al., 2006; Broniarczyk et al., 2014; Ahmed et al., 2019; Rose et al., 2020), among other biological activities. Although proteins and complexes involved in the ESCRT pathway must work collaboratively, they are recruited sequentially and carry out distinct functions. Early-acting factors such as ESCRT-I and ALIX recognize and assist in remodeling the appropriate membrane, concentrating vesicular cargo, and recruiting late-acting factors (Hurley, 2010).
The core structure of ESCRT is illustrated in Figure 1. The ESCRT-0 complex, a heterodimer, primarily recognizes ubiquitinated substrates. In eukaryotes, it consists of hepatocyte growth factor-regulated tyrosine kinase substrate (HRS) and signals transducing adaptor molecule (STAM1/2), whereas in yeast cells, it is composed of VPS27 and HSE1. These subunits form a composite helix through the GAT (GGA and Tom1) domain in a 1:1 ratio (Asao et al., 1997; Prag et al., 2007). As a regulatory factor in one of the essential intracellular vesicular transport pathways, ESCRT-0 recognizes, recruits, and binds to membrane proteins on the cell membrane. It then associates with ESCRT-I and ESCRT-II complexes to form vesicles for intracellular transport and degradation of substances. Therefore, ESCRT-0 plays a critical role in regulating vesicular transport pathways in the cytoplasm.
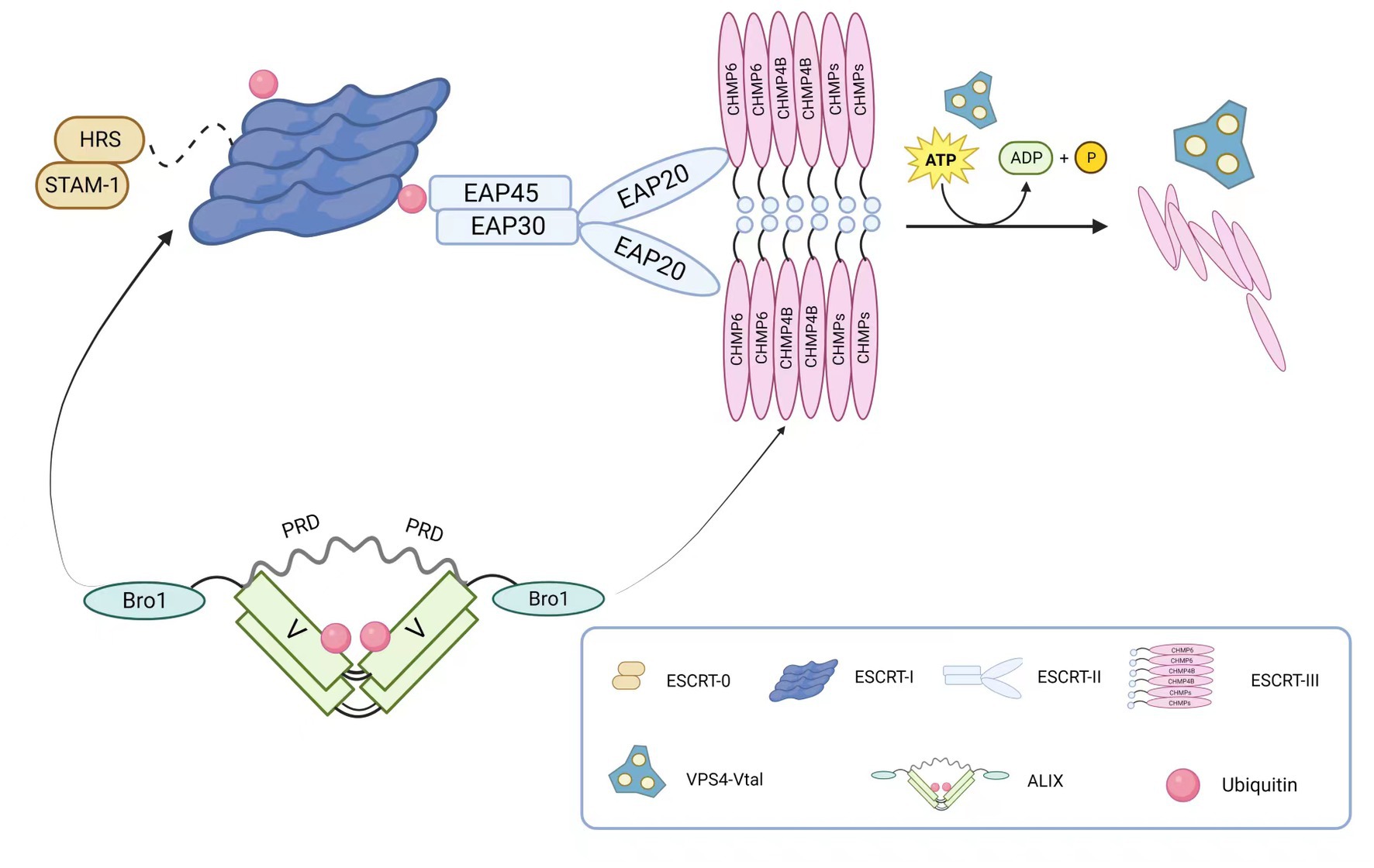
Figure 1. The main components of the ESCRT complex. ESCRT-0 is a dimer composed of HRS and STAM-1, primarily responsible for recognizing ubiquitinated substrates. ESCRT-I connects to ESCRT-0’s HRS through TSG101. ESCRT-II consists of one EAP45, one EAP30, and two EAP20, forming a “Y” shaped structure. The linkage between ESCRT-I and ESCRT-II is achieved through the connection of VPS28 and EAP45. ESCRT-III assembles into a tight fila-mentous structure on the cell membrane, and ESCRT-II and ESCRT-III recruit CHMP6 through EAP20. The ESCRT-associated protein ALIX is composed of the N-terminal Bro1 domain and the C-terminal proline-rich domain (PRD), located on either side of the “V” shaped central region. Through its Bro1 domain, ALIX recruits TSG101 and CHMP4B. Membrane neck constriction and ESCRT-III disassembly are catalyzed by the ATPase VPS4. The VPS4 complex hydrolyzes ATP to provide energy for thinning the bud neck and for disassembling and recycling ESCRT-III aggre-gates through the central pore.
ESCRT-I, structurally forming an elongated heterotetramer, was the first discovered ESCRT structure. It consists of VPS23/TSG101, VPS28, VPS37, and MVB12/UBAP1 (Ubiquitin Associated Protein 1) in a 1:1:1:1 ratio. On one hand, the UEV domain of VPS23/TSG101 interacts with ESCRT-0 and ubiquitinated membrane proteins. ESCRT-I can bind two ubiquitin moieties on the endosome due to the association of UBAP1 and yeast MVB12 with ubiquitin. On the other hand, VPS28 interacts with the GLUE domain of ESCRT-II protein EAP45, thereby interacting with the ESCRT-II complex. Additionally, ESCRT-I and Alix can recruit ESCRT-III components, which mediate the final scission step during cell division. Moreover, ESCRT-I is involved in regulating various cellular processes such as viral infection (Morita et al., 2007) and apoptosis (Morita et al., 2007; Kaul and Chakrabarti, 2017).
As a key factor in intracellular membrane transport and degradation pathways, ESCRT-II participates in forming and dissociating intracellular vesicles in the endolysosomal system. In yeast cells, this complex consists of VPS36, VPS25, and VPS22, while in mammals, it is composed of the endosomal sorting complex required for transport-associated proteins (EAP). The Y-shaped structure of ESCRT-II is formed by EAP30, EAP45, and EA20 in a 1:1:2 ratio (Alam et al., 2006). Similar to ESCRT-I, ESCRT-II has only one ubiquitin-binding domain, which limits its ability to recruit ubiquitinated proteins. However, it contains domains that interact with ubiquitin (Slagsvold et al., 2005), ESCRT-I (Teo et al., 2006), and specific membrane components. Therefore, it does not directly participate in sorting ubiquitinated proteins but functions after cargo selection by ESCRT-0. Additionally, the SNF8 protein (VPS22) of ESCRT-II interacts with calcium transporters, thereby participating in receptor-mediated calcium signaling in various cells and tissues (Carrasquillo et al., 2012).
In contrast to early ESCRT complexes (ESCRT-0, -I, and -II) that form stable protein complexes in the cytoplasm, the ESCRT-III complex assembles only transiently on endosomes. ESCRT-III belongs to the CHMP family, soluble monomers that are widely present in eukaryotes and participate in intracellular membrane transport and degradation pathways. It also acts as a membrane remodeling machinery, providing the driving force for membrane constriction at the neck and subsequent scission (Fabrikant et al., 2009; Wollert and Hurley, 2010). The ESCRT-III complex consists of 12 proteins, including CHMP1-7 and IST1, and their recruitment and assembly are regulated by early ESCRT complexes (ESCRT-0, -I, and -II). In most cases, ESCRT-III exists as filaments in cells, assembling on membranes to form tight helical structures. It mediates the rupture and sealing of various cellular membranes, including those between the nuclear envelope and plasma membrane, and is released from the membrane in the final stage through the transient ATP-dependent reaction with other ESCRT proteins, facilitated by Vps4. Furthermore, several studies have shown that the ESCRT-III complex plays important roles in various biological processes, including cell division (Harker-Kirschneck et al., 2022; Hurtig et al., 2023), immune regulation (Li et al., 2022), and neurodegenerative diseases (Lee et al., 2007a; Han et al., 2012).
The AAA-ATPase complex Vacuolar protein sorting 4 (VPS4) consists of the type I AAA-ATPase Vps4 and its cofactor Vta1. By harnessing the energy provided by ATP hydrolysis, it disassembles the ESCRT-III complex on endosomal membranes, enabling recycling. Upon completion of ESCRT-III disassembly, the Vps4 complex dissociates into its inactive protomers. Therefore, the Vps4 complex terminates each round of MVB cargo sorting and vesicle formation (Xiao et al., 2007; Shestakova et al., 2010). Two types of VPS4 complexes, VPS4A and VPS4B, have been identified in mammals, and they play important roles in cell biology and disease, including cell division (Morita et al., 2010), membrane fusion and fission (Finken-Eigen et al., 1997; Migliano and Teis, 2018), and viral infection (Kieffer et al., 2008; Broniarczyk et al., 2017) among other biological processes. Nearly all viruses that utilize ESCRT budding require the recruitment of VPS4, indicating its critical role in viral budding (Hurley and Hanson, 2010).
ALIX protein is a multifunctional cellular protein with a structure depicted in Figure 1. It is primarily composed of three domains: Bro1, V, and PRD (proline-rich domain). Each domain mediates interactions with different cellular and protein partners (Bissig and Gruenberg, 2014). The interaction between AAA-ATPase complex Vacuolar protein sorting 4 (VPS4) and apoptosis-linked gene 2 (ALG-2)-interacting protein X (ALIX) is a potential determinant of ESCRT-I and ESCRT-II function, promoting certain ESCRT-mediated activities through direct interactions with ESCRT-I and ESCRT-III. ALIX can also associate with various effector and adaptor proteins, such as TSG101 (Strack et al., 2003; von Schwedler et al., 2003), tyrosine kinases (Schmidt et al., 2005), and CEP55 (centrosomal protein 55; Carlton et al., 2008), and these interactions are crucial for many viral budding processes. Furthermore, ALIX exhibits diverse functions, including promoting exocytosis (Fan et al., 2019), cell migration (Xie et al., 2023), and apoptosis (Strappazzon et al., 2010).
The molecular mechanism underlying membrane invagination is illustrated in Figure 2. Initially, ESCRT-0 recognizes and enriches ubiquitinated substrates to recruit ESCRT-I and initiate the budding process. Ubiquitination serves as a critical entry point for the ESCRT pathway and plays a crucial role in retroviral budding (Strack et al., 2000). Ubiquitinated cargo molecules are sorted into MVBs, where proteins such as TSG101, UBAP1, EAP45, and ALIX contain ubiquitin-binding sites. ESCRT-I and ESCRT-II then interact with each other to induce inward membrane invagination and form initial buds. Subsequently, ESCRT-III cuts the neck of the buds, releasing small vesicles into the lumen of the endosome, completing the budding process. Finally, the VPS4-VTA1 complex, powered by ATP hydrolysis, dissociates the ESCRT-III complexes from the endosomal membrane, separating the ESCRT structures and allowing for recycling. Once MVBs fuse with lysosomes, the intraluminal vesicles (ILVs) are degraded within the lysosomal lumen. Therefore, the ESCRT system plays a crucial role in sorting substances and downregulating surface receptors, while also participating in the precise regulation of cell signaling (Taelman et al., 2010; Sigismund et al., 2012). During this process, upstream ESCRT mechanisms or other recruiting factors, such as viral Gag proteins, initiate membrane deformation and allow downstream ESCRT machinery to assemble in the absence of pre-existing membrane necks or pores. ESCRT-III components are recruited by ESCRT-I/-II and/or Bro domain proteins (Christ et al., 2016) and preferentially aggregate on curved membranes (Lee et al., 2015; de Franceschi et al., 2019; Bertin et al., 2020; Nepal et al., 2020). The following sections will describe several important functions of the ESCRT complex.
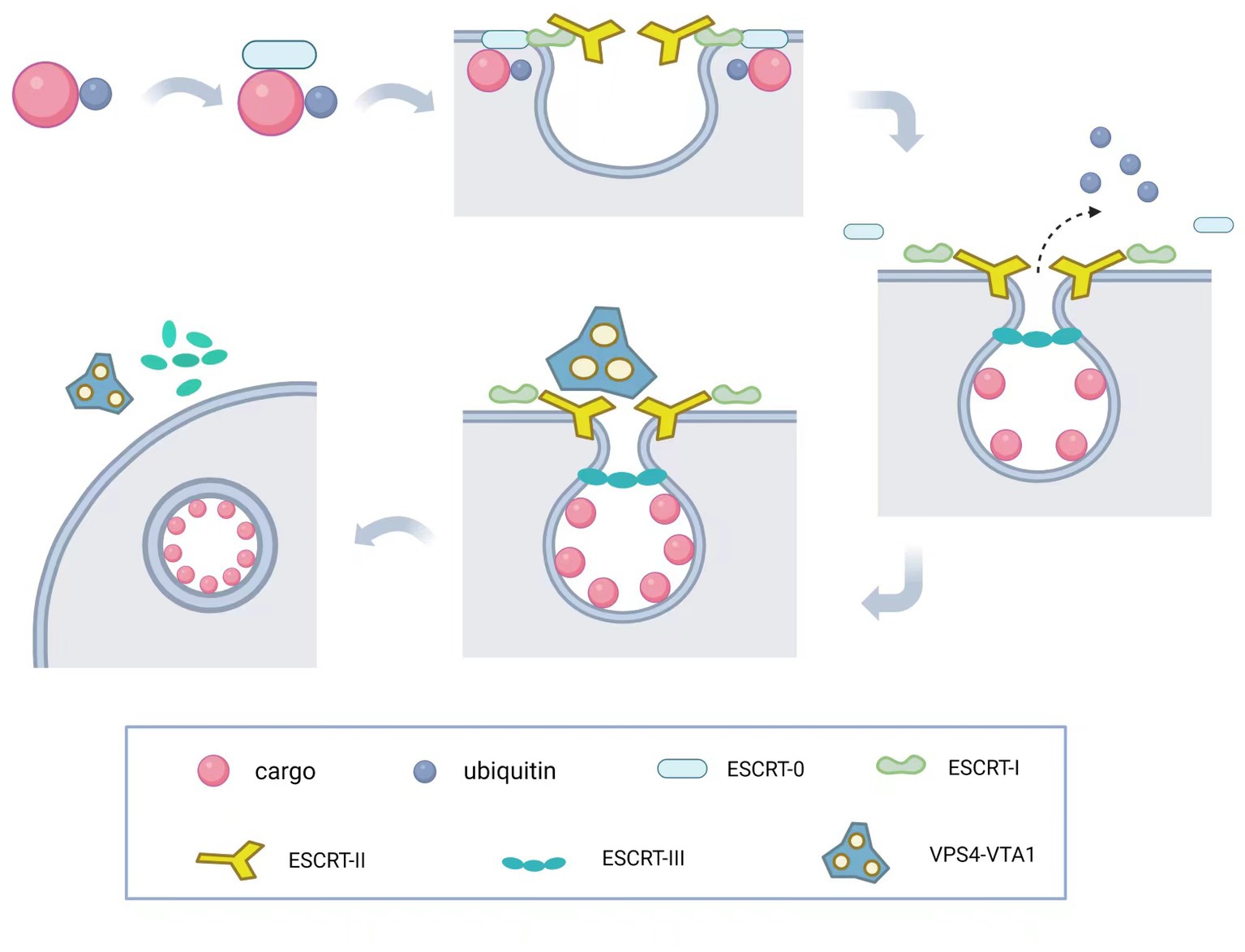
Figure 2. Molecular mechanism of ESCRT complex. The ESCRT-mediated budding process involves a series of coordinated steps. First, ESCRT-0 recognizes and enriches ubiquitinated substrates to recruit ESCRT-I and initiate its budding process. Then, ESCRT-I and ESCRT-II interact to induce invagination of the endosomal membrane, forming the initial bud. Subsequently, ESCRT-III cleaves the bud neck to release the cargo into the endosomal lumen, completing the budding process. Finally, the VPS4-VTA1 complex uses ATP hydrolysis to provide energy, dissociating the ESCRT-III complex from the endosomal membrane and allowing the recycling of ESCRT machinery and the endosomal membrane to occur.
Degradation and recycling of intracellular substances are fundamental mechanisms for maintaining cellular homeostasis and responding to adverse external conditions. Most cells employ several distinct degradation systems: the ubiquitin-proteasome degradation system, endocytosis, autophagy, and lysosomal degradation system (Lefebvre et al., 2018). Among these, autophagy is a conserved cellular mechanism in all eukaryotes, through which organelles, proteins, and invading pathogens can be sequestered and degraded within vesicles (Marshall and Vierstra, 2018). Within cells, various types of autophagic pathways coexist, relying on different combinations of cellular and molecular mechanisms, but ultimately delivering cargo for degradation in lysosomes (Rusten and Stenmark, 2009). The ESCRT machinery is capable of participating in different types of autophagic processes to degrade cytoplasmic components, such as macroautophagy and microautophagy. The distinction between the two lies in the fact that, in macroautophagy, autophagic substrates are transported to vesicles called autophagosomes, while in microautophagy, substrates are directly engulfed by invaginations of the vacuolar membrane.
The formation of autophagosomes requires the induction, transport, and fusion of membrane-bound organelles. Previous studies have shown that disruption of the ESCRT machinery in mammals leads to autophagosome accumulation, indicating the important role of ESCRT in regulating autophagy (Filimonenko et al., 2007; Lee et al., 2007a). The relationship between ESCRT and autophagy is linked through shared molecular mechanisms involved in autophagosome formation and fusion. The ESCRT machinery rescues mildly damaged lysosomes and interacts with autophagy-related proteins (ATGs) to form a bridge between endocytic and macroautophagy processes, facilitating the formation of the endo-lysosomal system and cell death. ESCRT components have been proposed as potential regulators of autophagosome membrane closure (Hurley, 2015; Knorr et al., 2015). Proper autophagosome membrane closure requires ESCRT-III component CHMP2A and AAA-ATPase vacuolar protein sorting-associated protein 4 (VPS4) activity (Takahashi et al., 2018). Upon maturation, autophagosomes fuse with late endosomes/lysosomes for degradation, and this process involves ESCRT proteins such as ALIX (Petiot and Sadoul, 2009) and VPS4. Additionally, certain components of the ESCRT complex participate in regulating different steps of the autophagic pathway. For instance, the ESCRT-III protein SNF7 plays a role in autophagosome formation and membrane sealing (Schaefer et al., 2020), while VPS4 promotes the fusion of autophagosomes with late endosomes, facilitating the progression of autophagy (Teis et al., 2010; Tang et al., 2015). Therefore, the ESCRT complex plays a crucial role in the autophagic pathway and is closely associated with the mechanisms of autophagy.
ESCRT is an evolutionarily conserved membrane remodeling complex with the unique ability to catalyze membrane constriction within necks. Among them, ESCRT-III, in collaboration with the AAA ATPase VPS4, plays a central role in the main membrane remodeling and scission functions of the ESCRT machinery (Stoten and Carlton, 2018). ESCRT-III exerts its membrane fission catalytic ability on almost all cellular membranes. It catalyzes the formation of ILVs within endosomal membranes, drives membrane scission from the cell surface, facilitates nuclear envelope reformation, and mediates autophagosome closure (Schoneberg et al., 2017). It is recruited to various distinct sites of action through interactions with multiple adaptor proteins. VPS4, on the other hand, is recruited to the membrane to facilitate the disassembly of ESCRT-III components (Yang et al., 2015). Cells utilize this system for reverse membrane fission during processes such as protein degradation, cell division, and retrovirus release. Retroviruses recruit ESCRT-III to bud sites to ensure cell exit. In this process, the ESCRT complex, through interaction with viral particle proteins, forms a protein membrane fission machinery that promotes viral particle release and aids in the evolution of the L domain (Scheffer et al., 2014).
Not all organisms require the complete ESCRT complex, but ESCRT-III and VPS4 appear to be indispensable, with CHMP2, CHMP3, CHMP4, and VPS4 considered essential components in membrane remodeling. However, even in the presence of ESCRT-III proteins, membrane fission is halted when AAA ATPase VPS4 is absent. Furthermore, ESCRT demonstrates critical activity in many other cellular membrane remodeling events, such as enveloped virus budding, cytokinesis abscission, exosome biogenesis (Larios et al., 2020), plasma membrane repair, immune synapse formation, and nuclear membrane homeostasis (Christ et al., 2017).
The ESCRT machinery was initially discovered through genetic studies in yeast, where mutations led to improper sorting of proteins into vesicles, functionally equivalent to lysosomes (Alfred and Vaccari, 2016). These mutant strains, known as “E-vps mutants,” exhibited enlarged pre-vacuolar endosome-like compartments containing undegraded proteins (Haguenauer-Tsapis, 2012). Subsequently, it was found that most class E VPS genes act sequentially, concentrating transport cargoes and incorporating them into late endosomes (also known as multivesicular bodies or MVBs), which eventually fuse with lysosomes for degradation (Katzmann et al., 2001). During the sorting process, various complexes of ESCRT are recruited from the cytoplasm to the inner membrane of endosomes through specific subunit interactions. These complexes facilitate the sequestration of intracellular waste, proteins, and other molecules into MVBs through the formation of endosomes. Subsequently, certain MVBs can fuse with lysosomes, directing the enclosed material into lysosomes for degradation.
ESCRT complexes play crucial roles in this process, with the ubiquitination of cargoes providing a critical signal for initial cargo binding by ESCRT-0 (Urbe, 2005). The ESCRT-0 subunits HRS and STAM, ESCRT-I subunit TSG101, and ESCRT-II subunit Vps36 all contain ubiquitin-binding domains that interact with ubiquitinated cargoes. Specifically, ESCRT-0, ESCRT-I, and ESCRT-II subunits facilitate the aggregation and recognition of membrane receptors by interacting with specific signaling molecules on the membrane. Subsequently, ESCRT-III and VPS4-VTA1 subunits act on the outer membrane of the MVB, forming a cone-shaped structure that ultimately drives MVB formation and separation of internal vesicles (Huotari and Helenius, 2011). With the assistance of ESCRT complexes, the VPS4 complex dissociates ESCRT structures from the limiting membrane, and after fusion of the MVB with lysosomes, ILVs are degraded within the lysosomal lumen. Additionally, several accessory proteins participate in the regulation of this process, such as ALIX and VPS27, which interact with different subunits of the ESCRT complex, thereby modulating endosomal sorting (Bilodeau et al., 2003; Murrow et al., 2015). Thus, the ESCRT pathway also plays a critical role in the sorting and downregulation of cell surface receptors, and it is involved in fine-tuning cell signaling pathways (Raiborg and Stenmark, 2009).
ESCRT was initially discovered as a regulator of intracellular cargo sorting (Katzmann et al., 2001; Babst et al., 2002). Recent studies have shown that ESCRT is also involved in a series of critical events during cell division, such as gametogenesis, chromosome segregation, and cytokinesis (Mathieu et al., 2022). The final stage of cell division is abscission, which involves the severing of the narrow membrane bridge between the two daughter cells (Steigemann and Gerlich, 2009). ESCRT is essential for cell division, and its role in cytokinesis can be traced back to ancient bacteria, suggesting that the ancestral function of these proteins was to promote membrane remodeling for division and was selected throughout evolution to perform various other cellular biological functions. In mammalian cell lines, the ESCRT-III complex mediates abscission, with the bridging protein Cep55 considered a key regulatory factor in abscission. This is due to its recruitment of ESCRT-III to the site of abscission, termed the midbody (MB; Tedeschi et al., 2020). Cep55 recruits ESCRT-III subcomplexes, subsequently facilitating membrane constriction and rupture of the intercellular bridge (Carlton and Martin-Serrano, 2007; Mierzwa and Gerlich, 2014; Addi et al., 2018). During this process, ESCRT-associated components are sequentially recruited to the membrane to mediate the scission event (Wollert et al., 2009b) and exhibit membrane-severing activity in vitro (Wollert et al., 2009a). The charged multivesicular body protein 1B (CHMP1B) of ESCRT-III interacts with the microtubule-severing enzyme spastin, which is crucial for cell division (Yang et al., 2009). The ESCRT-I subunit tumor susceptibility gene 101 (TSG101) and the ESCRT-III subunit charged multivesicular body protein 4B (CHMP4B) are sequentially recruited to the central region of the intercellular bridge, forming a series of cortical rings. In the late stage of cell division, CHMP4B is acutely recruited to the constricted site where abscission occurs, and VPS4 follows CHMP4B to that location, triggering immediate cell separation (Carlton and Martin-Serrano, 2007). ESCRT-III and VPS4 are spatially and temporally associated with the abscission event, indicating their direct involvement in membrane scission during cytokinesis.
During cell division, ESCRT interacts with structures such as the centrosome, chromosomes, and the cytoskeletal framework, participating in the regulation of cytokinesis, chromosome segregation, and recombination, as well as gametogenesis and embryonic development. Moreover, ESCRT may be closely associated with cell fate determination during cell division, including cell fate transitions, stem cell differentiation, and tumor development (Valentine et al., 2014; Wilson et al., 2020). In conclusion, the role of ESCRT in cell division is gradually being unraveled, and it holds significant implications for a comprehensive understanding of the mechanisms underlying cell division regulation and related diseases.
Cell apoptosis is a programmed cell death and the most extensively studied form of regulated cell death. It is characterized by sequential activation of the cysteine-aspartic protease caspases (Green, 2019). This process involves chromatin fragmentation and marginalization, as well as the generation and membrane budding of apoptotic bodies (Elmore, 2007). During cell apoptosis, cells release various cell factors and vesicles containing cellular components and nuclear fragments referred to as apoptotic bodies. The ESCRT complex can participate in the formation and clearance of apoptotic bodies. Specifically, the ESCRT-0 and ESCRT-III complexes interact with vesicle-associated proteins on the cell membrane, inducing membrane curvature and eventually leading to vesicle formation and release. Subsequently, the ESCRT-III complex assembles within the cell, forming a contractile structure that can engulf and release the apoptotic bodies to the extracellular space. Under normal circumstances, the binding of TSG101 with ALIX prevents cell apoptosis, but disruption of this process may occur due to dysregulation of cellular solute Ca2+ and subsequent upregulation of ALG-2 (Yang et al., 2022). Furthermore, the ESCRT-III complex can facilitate the release of mitochondrial content through extracellular vesicles, thereby participating in the process of apoptosis. ESCRT-III plays a crucial role in repairing damaged membranes involved in various types of regulated cell death, such as necroptosis, pyroptosis, and ferroptosis. Inhibition of the ESCRT-III mechanism through genetic depletion of its core components increases susceptibility to cell death induced by anticancer drugs, indicating that ESCRT-III could be a potential target to overcome drug resistance during cancer treatment. Therefore, the ESCRT complex plays a significant role in regulating the release and clearance of cellular components during cell apoptosis, maintaining normal physiological processes within the organism (Yang et al., 2022).
Furthermore, cells undergoing necroptosis do not always lead to cell death. The known repair mechanisms of ESCRT involve vesiculation or shedding of damaged membranes and play a critical balancing role in sorting and downregulating activated cell surface receptors (Raiborg and Stenmark, 2009) and repairing damaged membranes to maintain membrane integrity (Christ et al., 2017). This delays the cell death pathway, including cell apoptosis, necroptosis, pyroptosis, and ferroptosis (Jimenez et al., 2014; Gong et al., 2017; Ruhl et al., 2018). Cell apoptosis, necroptosis, and pyroptosis are all passive “suicidal” forms of molecular processes that result in the formation of several nanometer-sized membrane pores and catastrophic cell rupture (Espiritu, 2021). In summary, ESCRT provides time for dying cells, and ESCRT-dependent membrane repair negatively regulates cell death: ESCRT-I and -III are involved in cell apoptosis, while ESCRT-III mediates necroptosis, pyroptosis, and ferroptosis (Table 1).
The ESCRT machinery is not only essential for cellular processes such as membrane repair but is also hijacked by certain viruses during different stages of replication, particularly during the budding phase. The interaction between certain ESCRT components and viral proteins is primarily mediated by short tetrapeptide motifs known as Late assembly (L) domains. These motifs mediate the recruitment and interaction of class E proteins to facilitate virus budding, envelopment of retroviruses (e.g., HIV), and other RNA viruses (e.g., filoviruses, orthomyxoviruses, and paramyxoviruses), redirecting cellular ESCRT proteins to the cytoplasmic membrane for viral particle release and egress from infected cells.
Many enveloped viruses utilize the ESCRT pathway to catalyze membrane scission, leading to the release or budding of viral particles (Votteler and Sundquist, 2013). This process is dependent on specific late-domain structures of the respective viruses. All viruses that bud in an ESCRT-dependent manner require the membrane-remodeling functions provided by ESCRT-III and VPS4, but their requirements for upstream factors may vary. For instance, HIV-1 recruits the ESCRT pathway through specific motifs on the viral Gag protein (Morita and Sundquist, 2004), Marburg virus and Ebola virus VP40 interact with ESCRT-associated proteins such as Alix and TSG101 through their L domains to facilitate budding (Timmins et al., 2003; Urata et al., 2007). Enveloped virus infection begins with the attachment of the virus to the host cell’s plasma membrane, followed by viral entry, replication, and expression within the host cell. Finally, mature virus particles are released from the host cell, initiating a new infectious cycle. Studies have revealed the hijacking of the ESCRT system by HIV for its budding, and subsequent research has demonstrated the hijacking of the ESCRT system by various viruses, mediating different steps of the virus life cycle in different cells, thereby facilitating virus proliferation, budding, and dissemination. The ESCRT system has become an integral component of enveloped virus infections (Votteler and Sundquist, 2013; Meng and Lever, 2021). In the following, we summarize the reported interactions between various types of enveloped viruses and different ESCRT components, aiming to enhance our understanding of the interplay between ESCRT complexes and viral infections. Additionally, we aim to provide new research directions and contribute to the development of therapeutic strategies for future studies.
Some envelope RNA viruses utilize the ESCRT pathway to facilitate budding and release from host cells. By interacting with ESCRT proteins, these viruses can redirect intracellular proteins to the cell membrane, promoting viral particle budding and separation. The involvement of ESCRT plays a critical regulatory role in the infection, replication, and spread of enveloped RNA viruses.
Avian paramyxoviruses, including Newcastle disease virus (NDV), are single-stranded RNA viruses that can cause highly contagious diseases in various avian species. The M protein of paramyxoviruses has been identified as a nucleocapsid-associated protein that plays a crucial role in the virus life cycle (Pentecost et al., 2015). During early infection, the M protein typically enters the cell nucleus through its nuclear localization signal and transiently accumulates in the nucleolus. Previous studies have shown that this M protein aggregation, observed in respiratory syncytial virus (RSV), NDV, and measles virus (MeV), can affect the transcription, synthesis, and post-translational modifications of host proteins (Yu et al., 2016). The M protein contains conserved peptides known as the L domain, which can utilize the ESCRT complex to facilitate paramyxovirus release. However, single amino acid mutations in this domain can result in severe budding defects in paramyxoviruses (Duan et al., 2014). A novel late domain sequence, FPIV, was found in the parainfluenza virus 5 (SV5) matrix protein (Schmitt et al., 2005). Studies have shown that mutation of the FPIV-like late domain sequences dramatically impairs the budding process of SV5 (Schmitt et al., 2005) and MuV (Li et al., 2009). A similar sequence motif, FPIV, is present in the NDV M protein, suggesting that this motif could play an essential role in the replication and budding process of NDV. Previous research has demonstrated the critical significance of the FPIV motif within the NDV M protein, with residues F and P playing a pivotal role in NDV replication and budding. Furthermore, the FPIV motif’s recruitment of cellular multivesicular body (MVB) machinery is implicated in NDV budding (Duan et al., 2014). As a core component of the ESCRT-III complex, CHMP4B has been identified as a key factor linking the FPIV L domain and the ESCRT machinery. Previous studies by Li et al. (2013) confirmed that the binding between NDV M protein and CHMP4 promotes virus replication. Similarly, the study by Pei et al. (Pei et al., 2023) demonstrated that the ESCRT-III complex CHMP4s directly interact with NDV M protein through the FPIV L domain, independently of TSG101, ESCRT-II, or ALIX as assembly factors. In summary, critical amino acid mutations in the FPIV L domain prevent effective interaction between NDV M protein and CHMP4B, resulting in the stalling of viral sub-particles during membrane budding and leading to defects in NDV release and growth. However, further research is needed to elucidate the detailed mechanisms underlying the interaction between NDV and the ESCRT system, as well as whether other ESCRT components are utilized to facilitate virus propagation.
Sendai virus (SeV), belonging to the Paramyxoviridae family and Respirovirus genus, is a pneumotropic virus in rodents and one of the most extensively studied members of the Respirovirus genus (Henrickson, 2003). Its structure contains a multifunctional accessory protein called the C protein, which enhances the virulence of the virus by stimulating viral replication (Oda et al., 2021). It is considered that the C proteins are not involved in virus assembly, since they are essentially nonstructural components that are abundantly expressed in infected cells but are also present in trace amounts in mature virions (Sakaguchi et al., 2005). The C proteins are categorized as nonessential accessory proteins but contribute greatly to virus replication in vitro and are indispensable for the in vivo multiplication and pathogenesis of the infection they cause (Kurotani et al., 1998). It is thought that the C protein is involved in virus budding because of the low efficiency of release of progeny virions from C-knockout virus-infected cells and because of the requirement of the C protein for efficient release of virus-like particles. In addition, the C-protein knockout [4C(−)] virus produced largely noninfectious progeny virions with a highly anomalous morphology. Excessive accumulation of viral proteins and genomic RNAs was also observed in 4C(−) virus-infected cells. These results suggest that the late step of budding is abrogated in the absence of the C protein (Hasan et al., 2000). Oda et al. conducted virus infection experiments using SeV generated with the Y3: Bro1 complex crystal structure and analyzed the impact of the interaction between SeV’s C protein and Alix, a component of the ESCRT complex, on SeV budding. Their study revealed that a decrease in the affinity between the C protein and Alix significantly reduced the infectivity of the virus, indicating that the binding of the C protein to Alix enhances SeV budding (Kurotani et al., 1998). Furthermore, some studies have suggested that Alix also regulates cellular immune responses and inflammatory reactions, which may be associated with the immune response and inflammation triggered by SeV infection, although the specific mechanisms remain unclear (Yan et al., 2014; Chen and Yan, 2015; Chen et al., 2021). While the role of the ESCRT system in other viruses has been extensively studied, there is currently a lack of comprehensive research on the interaction between the Sendai virus and the ESCRT system.
Human parainfluenza virus type 1 (HPIV1) is a common respiratory pathogen and a single-stranded, negative-sense, non-segmented RNA virus belonging to the Paramyxoviridae family. The viral genome consists of 15,600 nucleotides and encodes six proteins from six genes. Among them, the C protein serves as an important virulence factor in HPIV1. It can inhibit apoptosis, influence host gene transcription, and regulate the production and signaling of type I interferon (Van Cleve et al., 2006; Bartlett et al., 2008; Boonyaratanakornkit et al., 2009). The C protein of HPIV1 interacts with the Bro1 domain of Alix at a specific site, which is also essential for the interaction between Alix and CHMP4B. The C protein is ubiquitinated and subject to proteasome-mediated degradation, but its interaction with Alix’s Bro1 domain protects it from degradation. Studies have shown that SeV shares a high degree of homology with the C protein of HPIV1 (Andrejeva et al., 2004). However, the major difference between them is that SeV also expresses another accessory protein called the V protein, which has inhibitory effects on the host’s innate antiviral response. In contrast, HPIV1 does not express the V protein, and the C protein is the only known viral antagonist of innate immune responses. Boonyaratanakornkit et al. (2013) proposed that during HPIV1 infection, Alix binds to the C protein and recruits it to the cytoplasmic surface of late endosomes. Subsequently, CHMP4 may recruit the Alix-C core shell to a common site on the cytoplasmic surface of late endosomes, which serves as the source for virus assembly and budding.
Nipah virus (NIV) is a deadly zoonotic virus belonging to the Paramyxoviridae family, capable of causing severe respiratory and neurological diseases in humans and animals. The M protein of NIV is considered to play a central role in virus assembly and budding (Patch et al., 2007; Harrison et al., 2010). When expressed alone, the M protein is released from the cells as virus-like particles, and its budding is independent of the ESCRT pathway (Patch et al., 2008). The C protein of NIV acts as an accessory factor to counteract innate immunity, enhancing the budding of the M protein through the ESCRT pathway. Research by Park et al. (Park et al., 2016) indicates that NIV-C directly interacts with the ESCRT factor TSG101, which is essential for NIV-C to enhance budding and efficiently release infectious Nipah virus. Further studies are being conducted to gain a better understanding of the interaction mechanisms between NIV and the ESCRT system.
Human Immunodeficiency Virus (HIV) is a single-stranded RNA virus belonging to the Retroviridae family. It primarily infects human immune cells, leading to immune system impairment. HIV-1 Gag is synthesized in the cytoplasm and subsequently directed to the plasma membrane, where virus assembly takes place (Lippincott-Schwartz et al., 2017). The formation of HIV viral particles occurs through the multimerization of the Gag precursor on the plasma membrane, and the budding of the virus is mediated by the interaction between Gag and various ESCRT (Endosomal Sorting Complex Required for Transport) proteins. Early components of the cellular ESCRT system, ESCRT-I (TSG101/VPS23) and ALIX mediate HIV-1 budding, both of which interact with the P6 domain of Gag. Within this domain, there are PS/TAP and YPXnL motifs that simultaneously promote virus budding, with the PS/TAP motif binding to ESCRT-I’s TSG101 and the YPXnL motif binding to ESCRT-III-associated ALIX protein. Both pathways require downstream ESCRT-III and VPS4 ATPase (Zavitz et al., 2008; Meng and Lever, 2013; Sette et al., 2013). Additionally, this domain can interact with ESCRT-II’s EAP20, directing HIV particles to the cell membrane and facilitating particle release (Pincetic et al., 2008; Medina et al., 2011; Ghoujal et al., 2012). However, Langelier et al. suggested that HIV-1 release is ESCRT-II-independent since siRNA depletion of ESCRT-II subunit EAP20 did not significantly impact virus release and infectivity. Nevertheless, they did report detrimental effects on virus production at later time points following ESCRT-II knockdown (Langelier et al., 2006). Furthermore, the Gag protein interacts with multiple members of the ESCRT-III complex, including CHMP2, CHMP3, CHMP4, and CHMP6, leading to the formation of membrane helical structures within the cell and aiding in virus particle formation and release (Baumgaertel et al., 2011; Cashikar et al., 2014; Johnson et al., 2019).
Following this, Janvier et al. (2011) investigated the additional role of the ESCRT machinery in HIV-1 release. Their research demonstrated that BST-2/tetherin, acting as a restriction factor, hinders HIV release by tethering mature virus particles to the plasma membrane. As a crucial component of ESCRT-0, HRS facilitates Vpu protein-induced downregulation and degradation of BST-2, thereby aiding in efficient HIV release. Physiologically, HRS is believed to initiate ESCRT-mediated multivesicular body (MVB) formation, with the PSAP motif in HRS recruiting the ESCRT machinery by interacting with ESCRT-I component TSG101 (Bache et al., 2003). The assembly and efficient release of HIV-1 requires highly coordinated interactions between virus-encoded proteins and cellular key components. In summary, we propose that ESCRT-0 and ESCRT-II may be dispensable for HIV budding, while Alix is indispensable. Among the ESCRT-III subunits, only CHMP4 and CHMP2 appear to be essential. Loss of ESCRT function inhibits virus release, resulting in accumulated arrested buds on the surface of infected cells.
The Flaviviridae family comprises enveloped, single-stranded positive-sense RNA viruses, including the Hepatitis C virus (HCV), which is the causative agent of chronic hepatitis. It encodes a large polyprotein precursor of approximately 3,000 amino acid residues, which is processed by viral proteases and cellular signalizes to generate three structural proteins (core, E1, and E2) and seven non-structural proteins (p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B; Scheel and Rice, 2013). The non-structural proteins NS3, NS4A, NS4B, NS5A, and NS5B form the viral replication machinery, while p7 and NS2 are crucial for the production of infectious viruses (Jones et al., 2007; Jirasko et al., 2010). NS5A is a membrane-associated RNA-binding phosphoprotein that also participates in the assembly and maturation of infectious HCV particles (Appel et al., 2008; Tellinghuisen and Foss, 2008).
Enveloped viruses typically hijack the ESCRT system through late-domain motifs or ubiquitination to bud from the plasma membrane. However, some viruses, such as HCV, lack identifiable late-domain motifs despite lacking late structural domains, suggesting that their association with ESCRT proteins may have diverse and complex mechanisms. On one hand, the K2 ubiquitination of NS63 lysine residues mediates its interaction with the HRS ubiquitin-interacting motif (UIM) and its binding to HCV assembly, thus compensating for the apparent deficiency of the late domain. On the other hand, HCV proteins involved in virus assembly, including the core and NS2, are ubiquitinated, and the recognition of ubiquitinated NS2 and potentially other viral proteins by HRS UIM might play a role in ESCRT recruitment and HCV assembly, providing a potential mechanism by which HCV proteins could be recruited to ESCRT complexes (Shirakura et al., 2007; Welbourn et al., 2009; Barouch-Bentov et al., 2016). Subsequently, Ariumi et al. (2011) demonstrated the binding of the HCV core to CHMP4B, indicating that the core possesses a novel motif required for HCV production. According to reports by Blanchard et al., the aspartic acid at position 111 of the core protein is crucial for virus assembly. Subsequently, they introduced mutations in the core protein sequence of the Dj clone by incorporating the L domain sequence. Specifically, they changed the aspartic acid at position 111 to proline-alanine (PTDP to PTAP) through site-directed mutagenesis. The results indicated that this amino acid mutation at this position impedes the formation of HCV core particles (Blanchard et al., 2003).
HRS serves as a critical factor in HCV envelopment, while ESCRT-III and VPS4/VTA1 factors are hijacked by the HCV core and NS5A, respectively, to mediate viral assembly. The essential role of VPS4 and the ESCRT-III complex in HCV production has been demonstrated by Corless et al. (Corless et al., 2010). Deng et al. proposed that the Hepatitis C Virus (HCV) induces the Reactive Oxygen Species (ROS)/JNK (c-Jun N-terminal kinase)/Itch (HECT-type E3 ubiquitin ligase) signaling pathway, which enhances the polyubiquitination of VPS4A. This leads to an augmented interaction between VPS4A and CHMP1B, thereby promoting VPS4A ATPase activity and subsequently facilitating the release of HCV particles (Deng et al., 2022). Furthermore, integrated approaches combining proteomics, RNA interference, viral genetics, and biophysics have validated the recruitment of ESCRT components by HCV proteins to facilitate viral envelopment (Ariumi et al., 2011; Barouch-Bentov et al., 2016). ESCRT-0 component HRS has been identified as crucial for the release of HCV via exosome secretion by Tamai et al. (Tamai et al., 2012). Subsequently, the study by Barouch-Bentov et al. (Barouch-Bentov et al., 2016) revealed that HCV hijacks HRS as an entry point into the ESCRT pathway to promote viral envelopment, highlighting the role of ubiquitinated NS2 residues in HRS recruitment and HCV assembly. The ubiquitination of viral proteins was found to serve as a signal for HRS binding and HCV assembly, thereby compensating for the absence of late structural domains. Additionally, HRS interacts with the viral core protein and envelope glycoproteins, facilitating their incorporation into newly formed viral particles. This interaction is crucial for the generation of infectious viral particles and the dissemination of HCV infection.
Porcine Reproductive and Respiratory Syndrome (PRRS) is an acute, contact-transmitted infectious disease that can cause abortion, stillbirth, weak-born piglets, mummified fetuses, and acute respiratory distress in pigs of all ages. The causative agent, Porcine Reproductive and Respiratory Syndrome Virus (PRRSV) is an enveloped, non-segmented, positive-sense RNA virus belonging to the Arteriviridae family and the genus Arterivirus (Lefkowitz et al., 2018). PRRSV infection is initiated by an interaction between the virus and host cell receptors/factors attached to the plasma membrane (Su et al., 2021). Upon internalization, PRRSV utilizes host machinery for its translation, transcription, and replication (Pasternak et al., 2006). Subsequently, the PRRSV nucleocapsid enters the endoplasmic reticulum (ER) or Golgi apparatus (GA), leading to the formation of enveloped viral particles. Finally, PRRSV viral particles are released through exocytosis, establishing a new cycle of infection. The PRRSV N protein is involved in the formation of the viral envelope by covalent and non-covalent interactions, enveloping the viral genome (Dokland, 2010). Additionally, it interacts with various host cell factors to modulate viral infection.
Among the ESCRT proteins that are crucial for PRRSV infection, TSG101 has been identified as the most upstream factor. It has been reported that TSG101 is involved in the replication of classical swine fever virus and the assembly and release of various viruses (Liu et al., 2021). As a novel host factor interacting with PRRSV N protein, TSG101 facilitates the formation of PRRSV viral particles and is involved in viral particle assembly and budding, rather than an attachment, internalization, RNA replication, and N protein translation during the infection process (Sang et al., 2012; Zhang et al., 2022). The study by Zhang et al. (2022) confirmed that inhibiting TSG101 expression or function significantly affects PRRSV replication and infectivity. Furthermore, PRRSV infection leads to the aberrant relocation and dysregulation of TSG101, affecting the balance of the ESCRT pathway and membrane trafficking within the cells.
Although significant progress has been made in elucidating the PRRSV lifecycle, a comprehensive understanding of PRRSV assembly is still lacking. The mechanisms of other identified ESCRT components involved in PRRSV infection remain unclear. Further research is needed to explore the interaction mechanisms between PRRSV and the ESCRT system to better understand the PRRSV lifecycle and viral proliferation process. Moreover, considering the importance of TSG101 in various viral infections, it holds promise as a broad-spectrum antiviral target for development.
Ebola virus (EBOV) is a single-stranded negative-sense enveloped RNA virus belonging to the family Filoviridae, which can cause a severe hemorrhagic fever syndrome with a high fatality rate. The viral matrix protein VP40 is a major component of EBOV virions and is essential for filamentous particle formation (Timmins et al., 2001; Bavari et al., 2002). Oligomerization of VP40 on the plasma membrane is required for particle assembly and virus budding (Panchal et al., 2003). The late (L) budding domain of VP40 recruits hosts ESCRT (endosomal sorting complex required for transport) and ESCRT-related proteins to facilitate membrane scission, thereby mediating efficient virus-cell separation. This structural protein plays a crucial role in the late stages of virus particle assembly and release. Additionally, ESCRT-III is also critical for EBOV release. Previous studies have indicated the involvement of TSG101, a component of the ESCRT-I complex, in EBOV budding (Martin-Serrano et al., 2001). TSG101 recruitment to the plasma membrane, away from its normal functional site in endosomes, has been shown to occur through binding with EBOV VP40 (Martin-Serrano et al., 2001; Panchal et al., 2003). The interaction between the two requires specific amino acid motifs or the late (L) domain (Freed, 2002), which is responsible for the binding of TSG101 to the PTAP motif of VP40. In the final step of the vacuolar protein sorting (VPS) pathway, the ATPase activity of VPS4 generates the energy necessary for the disassembly of protein complexes, allowing for subsequent rounds of sorting. Dominant-negative VPS4 mutants lacking ATPase activity have been shown to inhibit EBOV release (Garrus et al., 2001; Licata et al., 2003). EBOV VP40 also redirects other ESCRT-I proteins from endosomes to the plasma membrane, although these interactions have not been fully characterized. Furthermore, the host ESCRT protein ALIX has been extensively implicated in the budding of retroviruses such as HIV-1 and EIAV (Martin-Serrano and Marsh, 2007; Lee et al., 2007b; Zhai et al., 2011), as well as in the egress of other RNA viruses (Irie et al., 2007). Han et al. (2015) first proposed a role for ALIX in EBOV particle budding and identified the domains of ALIX that mediate its interaction with the required VP40. Specifically, EBOV VP40 recruits host ALIX through the YPx(n)L/I motif, which can act as an alternative to the L domain in promoting virus release. These findings are of significant importance for a comprehensive understanding of the intricate virus-host interactions required for efficient EBOV egress and dissemination.
Marburg virus (MARV) is a filamentous virus capable of inducing severe hemorrhagic fever in both humans and non-human primates, with a mortality rate reaching up to 90%. Comprising seven distinct proteins, the filamentous MARV particles manifest a complex architecture. Among these constituents, the nucleoprotein (NP) encases the non-segmented negative-sense RNA genome. Coordinating with viral proteins VP35, L, VP30, and VP24, NP assembles into an elongated helical nucleocapsid (Muhlberger et al., 1998; Bharat et al., 2011). A surrounding layer of matrix protein VP40 encompasses this nucleocapsid, linking it to the viral envelope, which additionally features the incorporation of the homotrimeric glycoprotein (GP; Feldmann et al., 2001). Both matrix protein VP40 and nucleocapsid protein NP of MARV harbor specific late-domain motifs. The PSAP late-domain motif within NP plays a pivotal role in recruiting the ESCRT-I protein TSG101. Previous investigations have revealed that altering the PSAP late-domain motif within NP compromises its interaction with Tsg101, consequently impeding the liberation of MARV-specific virus-like particles (VLPs; Dolnik et al., 2010). Insights from Dolnik et al. underline the significance of the PSAP motif within NP for the efficient trafficking and envelopment of the nucleocapsid. This motif not only engages Tsg101 but also initiates the recruitment of IQGAP1, a factor involved in the regulation of the actin cytoskeleton. Notably, mutations within the PSAP motif of recombinant rMARV culminate in faulty transport processes, leading to the intracellular accumulation of the nucleocapsid and attenuating the virus’s capacity for robust intercellular dissemination (Dolnik et al., 2014).
The Arenaviridae family stands as a dynamically evolving and diverse collective of viruses. It encompasses an array of enveloped viruses characterized by a bipartite negative-sense RNA genome, encoding four distinct viral proteins, and marked by a non-lytic life cycle. This genome comprises two individual single-stranded RNA entities: a larger segment encoding the viral polymerase (L) and a petite zinc finger motif protein (Z), as well as a minor segment encoding both the viral nucleoprotein (NP) and the glycoprotein precursor (GPC; Southern, 1996). The Z protein, functioning as the pivotal viral matrix protein, is the prime propellant behind the budding of viral particles. In isolation, its expression can assemble VLPs (Hoenen et al., 2007). Earlier research has indicated that the proficiency of the Z protein’s budding activity correlates with the presence of late-domain motifs or aligns with the reliance on the ESCRT pathway (Perez et al., 2003; Cruz Casabona et al., 2009).
Notwithstanding the existence of diverse late-domain motifs within certain arenaviruses, the extent to which virus budding hinges on each motif appears to diverge among different viruses. For instance, the prototypic arenavirus lymphocytic choriomeningitis virus (LCMV) exclusively features a sole PPxY motif within its Z protein, a critical determinant driving the budding process (Perez et al., 2003). In contrast, the highly pathogenic Old World arenavirus Lassa virus (LASV) Z protein comprises dual PTAP and PPxY motifs. Although both motifs contribute to LASV particle release, it has been documented that the PPxY motif holds sway (Strecker et al., 2003). The research underscores the ability of diverse enveloped viruses, including members of the Arenaviridae family like LASV, to enlist ESCRT components, thereby facilitating the release of viral particles (Bieniasz, 2006; Chen and Lamb, 2008). Intriguingly, RNA interference (siRNA)-based experiments have unveiled that depletion of key factors such as HRS, TSG101, VPS22, and VPS24 substantially curbs cellular entry by LASV and LCMV (Pasqual et al., 2011). Furthermore, insights from Shtanko et al.’s investigation spotlight that ALIX/AIP1’s Bro1 domain interfaces with both NP and Z proteins. The indispensable role of ALIX is underscored in facilitating the incorporation of NP into Z-induced VLPs, as illuminated by their study (Shtanko et al., 2011).
Rabies virus (RABV) is a well-established zoonotic pathogen belonging to the Lyssavirus genus within the Rhabdoviridae family. Characterized by a single-stranded negative-sense RNA genome, this virus encodes a set of five essential viral proteins: nucleoprotein (N), phosphoprotein (P), matrix protein (M), glycoprotein (G), and large protein (L; Finke and Conzelmann, 2005). In the context of rhabdovirus infections, the role of the M protein is paramount—it is pivotal in the processes of budding and the formation of distinctive bullet-shaped viral particles (Mebatsion et al., 1999). Detailed proteomic analyses have unveiled the presence of ESCRT-associated factors, including CHMP4B, HSP40, Alix, TSG101, and CHMP2A, within purified RABV viral particles (Finke and Conzelmann, 2005). A recent investigation conducted by Itakura et al. has shed further light on the intricate web of interactions governing the virus’s lifecycle. Specifically, their research has illuminated the pivotal involvement of the ESCRT-I protein, TSG101, in the later stages of this lifecycle. TSG101 has emerged as a key binding partner for the RABV M protein. The concerted effort between the L domain of TSG101 and the L structure domain of RABV M is instrumental in orchestrating the efficient assembly, budding, and distinctive bullet-shaped morphology that characterize RABV (Itakura et al., 2023).
Infectious hematopoietic necrosis virus (IHNV), much like RABV, resides within the Rhabdoviridae family and boasts a negative-sense single-stranded RNA genome. This genomic structure encodes a cluster of six distinctive proteins: nucleoprotein (N), polymerase-associated phosphoprotein (P), matrix protein (M), unique glycoprotein (G), large RNA-dependent RNA polymerase (L), and a concise intergenic region nestled between the G and L genes (Morzunov et al., 1995; Assenberg et al., 2010; Kim and Kim, 2011). Throughout viral replication, the M protein kickstarts the intricate choreography of viral particle assembly and budding (Dancho et al., 2009). The G protein’s budding domain, in turn, orchestrates either the curvature of membranes at the budding site or the strategic aggregation of nucleocapsids onto the plasma membrane. This dual approach seamlessly promotes the liberation of viruses (Robison and Whitt, 2000). Pioneering the exploration of IHNV budding, Chen et al. (2019) stumbled upon the influential role of VPS4. A remarkable impact on IHNV budding emerged in response to siRNA-induced VPS4B attenuation. In stark contrast, when VPS4A was downregulated, IHNV budding seemed relatively unaltered. This body of evidence underscores the pivotal involvement of VPS4B in IHNV budding while suggesting a less pivotal role for VPS4A in IHNV’s emancipation. In parallel, Chen et al. unveiled a network of interactions between IHNV’s M, G, and L proteins and Nedd4, Tsg101, and Alix. Collectively, these discoveries underscore the paramount importance of interactions between virus proteins bearing the L domain and their corresponding E-class proteins in IHNV budding. Furthermore, the host factors at play exert their influential touch upon the intricate ballet of IHNV’s budding journey.
Classical swine fever (CSF), caused by classical swine fever virus (CSFV), is a highly contagious disease of swine with high morbidity and mortality (Moennig, 2015). CSFV is an enveloped, positive-stranded RNA virus that belongs to the genus Pestivirus within the family Flaviviridae (Meyers et al., 1996). The viral genome is a single-stranded +RNA of approximately 12.3 kb with a structure of a single open reading frame (ORF) surrounded by two untranslated regions (UTRs). The ORF encodes a polyprotein that is cleaved into four structural (capsid protein C, envelope glycoproteins Erns, E1, and E2) and eight nonstructural proteins (Npro, p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B; Zhou, 2019; Fan et al., 2021). After CSFV enters the host cells, a series of reactions occur with intracellular components that regulate different stages of the viral life cycle, including entry, trafficking, replication, assembly, and release (Ning et al., 2016; Zheng et al., 2020). ESCRT-I complex Tsg101 protein participates in clathrin-mediated endocytosis of CSFV and is also involved in CSFV trafficking. Tsg101 assists the virus in entering the host cell through the late endosome (Rab7 and Rab9) and finally reaching the lysosome (Lamp-1). Interestingly, Tsg101 is also involved in the viral replication process by interacting with nonstructural proteins 4B and 5B of CSFV (Liu et al., 2021).
Liu et al. investigated the Tsg101, VPS25, CHMP4B, and CHMP7 proteins that play essential roles in viral trafficking and Clathrin-mediated viral entry. CHMP4B and CHMP7 interact with Rab5 to transport CSFV virions from early endosomes to late endosomes. They first demonstrate that HRS (ESCRT-0), VPS28 (ESCRT-I), VPS25 (ESCRT-II), and adaptor protein ALIX play important roles in the formation of virus replication complexes (VRC) together with CHMP2B/4B/7 (ESCRT-III), and VPS4A. Tsg101 and CHMP4B are associated with LAMP-1 in the lysosomes, probably leading to uncoating and release of nucleic acid for genome replication in the endoplasmic reticulum area (Liu et al., 2022).
In addition to enveloped RNA viruses, the ESCRT complex is also involved in the assembly and release processes of enveloped DNA viruses. For instance, Hepatitis B virus (HBV) and Herpes simplex virus type 1 (HSV-1).
HBV is the major pathogen responsible for human liver disease. It is an enveloped, DNA-containing pararetrovirus that replicates primarily through reverse transcription of its pregenomic RNA (pgRNA; Stieler and Prange, 2014). Despite the success of global HBV vaccination programs, a definitive cure for HBV infection has yet to be found (Ringelhan et al., 2015; Thomas et al., 2015). Persistent HBV infection can lead to the development of liver cirrhosis, liver damage, and hepatocellular carcinoma (Ganem and Prince, 2004). When the nucleocapsid of HBV reaches genomic maturation in the cytoplasm, two distinct outcomes can occur: one involves encapsidation of the viral particles and their release through the ESCRT system, while the other involves the return of the viral RC DNA genome to the cell nucleus for further amplification of covalently closed circular DNA (Tuttleman et al., 1986).
During HBV infection, all components of ESCRT-0 are essential for HBV replication. The core antigen of HBV (HBcAg) interacts with ESCRT-0, facilitating the assembly and release of HBV particles. Specifically, the HBcAg protein forms a complex with the HRS subunit of ESCRT-0, which promotes the intracellular aggregation and assembly of HBV core proteins, ultimately leading to the release of HBV particles. Additionally, the surface antigen of HBV (HBsAg) can interact with the ESCRT complex, facilitating the release of HBV particles. Studies by Chou et al. (Chou et al., 2015) have shown that both downregulation and overexpression of HRS result in the inhibition of HBV RNA transcription, DNA replication, and virus particle production, partly through promoting naked capsid secretion.
Due to the absence of the P (S/T)AP-like late-domain motif in HBV structural proteins, which is known to interact with TSG101, it was previously believed that TSG101 was dispensable for HBV export. However, recent research findings have demonstrated that the knockdown of TSG101 reduces extracellular HBV DNA levels and results in the accumulation of viral capsids within cells. This supports the essential role of TSG101 in the process of HBV extracellular capsid release (Zheng et al., 2023). Although the absence of ESCRT-II does not impair the capsid assembly reaction, it selectively impairs the formation and/or stability of nuclear capsids. In cells depleted of ESCRT-II, the levels of enveloped pgRNA are significantly reduced, indicating that ESCRT-II guides steps in capsid formation that accompany replicative capacities, such as facilitating RNA transport and capsid envelopment. Stieler et al. performed knockdown experiments targeting ESCRT-II components EAP20, EAP30, and EAP45, and found that depletion of ESCRT-II components dramatically reduced virus budding. Therefore, depletion of ESCRT-II components EAP20, EAP30, and EAP45 not only inhibits the production and/or release of enveloped virus particles but also impairs the formation of intracellular nuclear capsids. HBV budding on cellular membranes also requires the involvement of ESCRT-III and the VPS4 complex in membrane scission and separation (Stieler and Prange, 2014).
HSV-1 is a structurally complex enveloped double-stranded DNA virus belonging to the Herpesviridae family. It is a human pathogen that can cause diseases such as oral herpes, keratitis, and encephalitis. Its capsid can package together with the DNA genome in the cell nucleus (Dai and Zhou, 2018). The viral capsid undergoes primary packaging at the inner nuclear membrane (Bigalke and Heldwein, 2017) and then undergoes final secondary packaging within the lumen of cellular compartments in the cytoplasm (Hollinshead et al., 2012). During this process, it utilizes its viral-encoded proteins and the cellular ESCRT mechanism to drive its envelope assembly. ESCRT-III and VPS4 are essential for HSV-1 to accomplish cytoplasmic secondary packaging and play important roles in the wrapping process of HSV-1 at the inner nuclear membrane (Crump et al., 2007; Pawliczek and Crump, 2009).
Generally, viruses recruit one or more early ESCRT components to access ESCRT-III, with the most common being ALIX and the ESCRT-I complex (Votteler and Sundquist, 2013). However, unlike many other members of the enveloped virus family, HSV-1 appears to not require the ESCRT-I subunit TSG101 or the protein ALIX containing the Bro1 domain for recruitment and activation of ESCRT-III. Both of them are dispensable for HSV-1 envelopment even when simultaneously depleted (Pawliczek and Crump, 2009). Its structural components may directly interact with ESCRT-II to control ESCRT-III assembly. Following this, Jenna Barnes and colleagues proposed a hypothesis that due to the lack of ALIX and ESCRT-I involvement, the most likely remaining cellular proteins used by HSV-1 to assemble the ESCRT-III machinery are the ESCRT-II complex and Bro1 family members other than ALIX. Knockdown experiments targeting the critical EAP25/VPS1 subunit of ESCRT-II and the remaining known ESCRT-related Bro1 domain proteins, HD-PTP and BROX, revealed no impact on HSV-1 replication (Barnes and Wilson, 2020).
Considering the aforementioned findings, it is reasonable to hypothesize that ESCRT-III may be directly recruited by proteins present in the capsid or envelope of HSV-1. HSV-1 appears to redundantly utilize multiple cellular ESCRT components for ESCRT-III recruitment, or the virus may completely bypass the requirement for Bro1 domain proteins, ESCRT-I, and ESCRT-II. In the latter scenario, the structural proteins within HSV-1 particles, whether internal or external to the capsid, might have evolved to directly recruit and trigger the assembly of ESCRT-III subunits at sites of envelope constriction and scission (Crump, 2018; Barnes and Wilson, 2019). The interaction between HSV-1 and the ESCRT system may involve the participation of other components, and further investigation is needed to elucidate the specific mechanisms and their implications.
Autographa California Multiple Nuclear Polyhedrovirus (AcMNPV) is the most intensively studied baculovirus and is the type species of the virus family Baculoviridae. Baculoviruses are enveloped, insect-specific double-stranded DNA (dsDNA) viruses that replicate in the nuclei of infected cells. During the infection cycle, baculoviruses produce two phenotypes of enveloped virions: occlusion-derived virions (ODV) and budded virions (BV). The ESCRT pathway is conserved in sequenced insect genomes and the expression levels of certain components of ESCRT-I, -II, -III, Vps4, and Alix were significantly upregulated upon AcMNPV infection (Chen et al., 2014; Li and Blissard, 2015).
Prior studies revealed that efficient entry and egress of AcMNPV BV are dependent on functional Vps4 (Li and Blissard, 2012). Since Vps4 is required for recycling ESCRT III and represents a terminal step in the ESCRT pathway, this suggests that other components of the ESCRT pathway may be specifically involved in entry and egress. However, In the current study, Yue et al. found that functional ESCRT-I and ESCRT-III complexes were required for efficient entry and transport of AcMNPV budded virions early in infection. The components of ESCRT-III (but not ESCRT-I) are also necessary for the efficient nuclear egress of progeny nucleocapsids. In addition, Yue et al. also found that several baculovirus cores or conserved proteins (Ac11, Ac76, Ac78, GP41, Ac93, Ac103, Ac142, and Ac146) interact with Vps4 and components of ESCRT-III. These viral proteins may form an “egress complex” that is involved in recruiting ESCRT-III components to a virus-egress domain on the nuclear membrane (Yue et al., 2018).
ESCRT plays a crucial role in various cellular processes, including the budding of enveloped viruses, regulation of innate immune responses, and degradation of damaged or unwanted cellular components. Although significant progress has been made in understanding the molecular mechanisms of ESCRT-mediated virus budding, much remains to be explored. For instance, the molecular mechanisms underlying the regulation of ESCRT components’ expression and activity require further investigation. Moreover, the precise roles of ESCRT in virus replication and pathogenesis need to be elucidated. An enhanced understanding of how the ESCRT system is hijacked by viruses can greatly contribute to our knowledge of normal cellular physiological functions.
In the future, research on ESCRT function in virus-host interactions will provide new insights into the development of antiviral therapies. A better understanding of the ESCRT complex’s role in innate immune response regulation will lead to the development of novel approaches to treating viral infections. Moreover, the identification and characterization of novel ESCRT-interacting viral proteins and their interaction mechanisms will expand our knowledge of virus-host interactions. Finally, the development of new technologies, such as single-particle cryo-electron microscopy, will allow us to determine the three-dimensional structure of ESCRT and provide a more detailed understanding of its molecular mechanisms. Overall, further research on ESCRT and its interactions with viruses will help to develop new strategies to combat viral infections.
CW: Writing – original draft. YC: Writing – review & editing. SH: Writing – review & editing. XL: Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation (32202767), the Earmarked Fund for China Agriculture Research System (CARS-40), and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Addi, C., Bai, J., and Echard, A. (2018). Actin, microtubule, septin and ESCRT filament remodeling during late steps of cytokinesis. Curr. Opin. Cell Biol. 50, 27–34. doi: 10.1016/j.ceb.2018.01.007
Ahmed, I., Akram, Z., Iqbal, H. M. N., and Munn, A. L. (2019). The regulation of endosomal sorting complex required for transport and accessory proteins in multivesicular body sorting and enveloped viral budding - an overview. Int. J. Biol. Macromol. 127, 1–11. doi: 10.1016/j.ijbiomac.2019.01.015
Alam, S. L., Langelier, C., Whitby, F. G., Koirala, S., Robinson, H., Hill, C. P., et al. (2006). Structural basis for ubiquitin recognition by the human ESCRT-II EAP45 GLUE domain. Nat. Struct. Mol. Biol. 13, 1029–1030. doi: 10.1038/nsmb1160
Alfred, V., and Vaccari, T. (2016). When membranes need an ESCRT: endosomal sorting and membrane remodelling in health and disease. Swiss Med. Wkly. 146:w14347. doi: 10.4414/smw.2016.14347
Andrejeva, J., Childs, K. S., Young, D. F., Carlos, T. S., Stock, N., Goodbourn, S., et al. (2004). The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc. Natl. Acad. Sci. U. S. A. 101, 17264–17269. doi: 10.1073/pnas.0407639101
Appel, N., Zayas, M., Miller, S., Krijnse-Locker, J., Schaller, T., Friebe, P., et al. (2008). Essential role of domain III of nonstructural protein 5A for hepatitis C virus infectious particle assembly. PLoS Pathog. 4:e1000035. doi: 10.1371/journal.ppat.1000035
Ariumi, Y., Kuroki, M., Maki, M., Ikeda, M., Dansako, H., Wakita, T., et al. (2011). The ESCRT system is required for hepatitis C virus production. PLoS One 6, –e14517. doi: 10.1371/journal.pone.0014517
Asao, H., Sasaki, Y., Arita, T., Tanaka, N., Endo, K., Kasai, H., et al. (1997). Hrs is associated with STAM, a signal-transducing adaptor molecule. Its suppressive effect on cytokine-induced cell growth. J. Biol. Chem. 272, 32785–32791. doi: 10.1074/jbc.272.52.32785
Assenberg, R., Delmas, O., Morin, B., Graham, S. C., De Lamballerie, X., Laubert, C., et al. (2010). Genomics and structure/function studies of Rhabdoviridae proteins involved in replication and transcription. Antivir. Res. 87, 149–161. doi: 10.1016/j.antiviral.2010.02.322
Babst, M., Katzmann, D. J., Snyder, W. B., Wendland, B., and Emr, S. D. (2002). Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev. Cell 3, 283–289. doi: 10.1016/S1534-5807(02)00219-8
Bache, K. G., Brech, A., Mehlum, A., and Stenmark, H. (2003). Hrs regulates multivesicular body formation via ESCRT recruitment to endosomes. J. Cell Biol. 162, 435–442. doi: 10.1083/jcb.200302131
Barnes, J., and Wilson, D. W. (2019). Seeking closure: how do herpesviruses recruit the cellular ESCRT apparatus? J. Virol. 93, e00392–e00319. doi: 10.1128/jvi.00392-19
Barnes, J., and Wilson, D. W. (2020). The ESCRT-II subunit EAP20/VPS25 and the Bro1 domain proteins HD-PTP and BROX are individually dispensable for herpes simplex virus 1 replication. J. Virol. 94, e01641–e01619. doi: 10.1128/jvi.01641-19
Barouch-Bentov, R., Neveu, G., Xiao, F., Beer, M., Bekerman, E., Schor, S., et al. (2016). Hepatitis C virus proteins interact with the endosomal sorting complex required for transport (ESCRT) machinery via ubiquitination to facilitate viral envelopment. MBio 7, e01456–e01416. doi: 10.1128/mBio.01456-16
Bartlett, E. J., Cruz, A.-M., Esker, J., Castano, A., Schomacker, H., Surman, S. R., et al. (2008). Human parainfluenza virus type 1 C proteins are nonessential proteins that inhibit the host interferon and apoptotic responses and are required for efficient replication in nonhuman primates. J. Virol. 82, 8965–8977. doi: 10.1128/JVI.00853-08
Baumgaertel, V., Ivanchenko, S., Dupont, A., Sergeev, M., Wiseman, P. W., Kraeusslich, H.-G., et al. (2011). Live-cell visualization of dynamics of HIV budding site interactions with an ESCRT component. Nat. Cell Biol. 13, 469–U277. doi: 10.1038/ncb2215
Bavari, S., Bosio, C. M., Wiegand, E., Ruthel, G., Will, A. B., Geisbert, T. W., et al. (2002). Lipid raft microdomains: a gateway for compartmentalized trafficking of Ebola and Marburg viruses. J. Exp. Med. 195, 593–602. doi: 10.1084/jem.20011500
Bertin, A., de Franceschi, N., de la Mora, E., Maity, S., Alqabandi, M., Miguet, N., et al. (2020). Human ESCRT-III polymers assemble on positively curved membranes and induce helical membrane tube formation. Nat. Commun. 11:2663. doi: 10.1038/s41467-020-16368-5
Bharat, T. A. M., Riches, J. D., Kolesnikova, L., Welsch, S., Kraehling, V., Davey, N., et al. (2011). Cryo-Electron tomography of Marburg virus particles and their morphogenesis within infected cells. PLoS Biol. 9:e1001196. doi: 10.1371/journal.pbio.1001196
Bieniasz, P. D. (2006). Late budding domains and host proteins in enveloped virus release. Virology 344, 55–63. doi: 10.1016/j.virol.2005.09.044
Bigalke, J. M., and Heldwein, E. E. (2017). Have NEC coat, will travel: structural basis of membrane budding during nuclear egress in herpesviruses. Adv Virus Res. 97, 107–141. doi: 10.1016/bs.aivir.2016.07.002
Bilodeau, P. S., Winistorfer, S. C., Kearney, W. R., Robertson, A. D., and Piper, R. C. (2003). Vps27-Hse1 and ESCRT-I complexes cooperate to increase efficiency of sorting ubiquitinated proteins at the endosome. J. Cell Biol. 163, 237–243. doi: 10.1083/jcb.200305007
Bissig, C., and Gruenberg, J. (2014). ALIX and the multivesicular endosome: ALIX in wonderland. Trends Cell Biol. 24, 19–25. doi: 10.1016/j.tcb.2013.10.009
Blanchard, E., Hourioux, C., Brand, D., Ait-Goughoulte, M., Moreau, A., Trassard, S., et al. (2003). Hepatitis C virus-like particle budding: role of the core protein and importance of its asp(111). J. Virol. 77, 10131–10138. doi: 10.1128/JVI.77.18.10131-10138.2003
Boonyaratanakornkit, J. B., Bartlett, E. J., Amaro-Carambot, E., Collins, P. L., Murphy, B. R., and Schmidt, A. C. (2009). The C proteins of human parainfluenza virus type 1 (HPIV1) control the transcription of a broad Array of cellular genes that would otherwise respond to HPIV1 infection. J. Virol. 83, 1892–1910. doi: 10.1128/JVI.01373-08
Boonyaratanakornkit, J., Schomacker, H., Collins, P., and Schmidt, A. (2013). Alix serves as an adaptor that allows human parainfluenza virus type 1 to interact with the host cell ESCRT system. PLoS One 8:e59462. doi: 10.1371/journal.pone.0059462
Broniarczyk, J., Bergant, M., Goździcka-Józefiak, A., and Banks, L. (2014). Human papillomavirus infection requires the TSG101 component of the ESCRT machinery. Virology 460-461, 83–90. doi: 10.1016/j.virol.2014.05.005
Broniarczyk, J., Pim, D., Massimi, P., Bergant, M., Gozdzicka-Jozefiak, A., Crump, C., et al. (2017). The VPS4 component of the ESCRT machinery plays an essential role in HPV infectious entry and capsid disassembly. Sci. Rep. 7:45159. doi: 10.1038/srep45159
Carlton, J. G., Agromayor, M., and Martin-Serrano, J. (2008). Differential requirements for Alix and ESCRT-III in cytokinesis and HIV-1 release. Proc. Natl. Acad. Sci. U. S. A. 105, 10541–10546. doi: 10.1073/pnas.0802008105
Carlton, J. G., and Martin-Serrano, J. (2007). Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science 316, 1908–1912. doi: 10.1126/science.1143422
Carrasquillo, R., Tian, D., Krishna, S., Pollak, M. R., Greka, A., and Schloendorff, J. (2012). SNF8, a member of the ESCRT-II complex, interacts with TRPC6 and enhances its channel activity. BMC Cell Biol. 13. doi: 10.1186/1471-2121-13-33
Cashikar, A. G., Shim, S., Roth, R., Maldazys, M. R., Heuser, J. E., and Hanson, P. I. (2014). Structure of cellular ESCRT-III spirals and their relationship to HIV budding. elife 3:e02184. doi: 10.7554/eLife.02184
Chen, B. J., and Lamb, R. A. (2008). Mechanisms for enveloped virus budding: can some viruses do without an ESCRT? Virology 372, 221–232. doi: 10.1016/j.virol.2007.11.008
Chen, Y., Li, J., Zhou, Y., Feng, Y., Guan, X., Li, D., et al. (2019). The role of infectious hematopoietic necrosis virus (IHNV) proteins in recruiting the ESCRT pathway through three ways in the host cells of fish during IHNV budding. Fish Shellfish Immunol. 92, 833–841. doi: 10.1016/j.fsi.2019.07,011
Chen, X., Wei, Q., Sun, H., Zhang, X., Yang, C., Tao, Y., et al. (2021). Exosomes derived from human umbilical cord mesenchymal stem cells regulate macrophage polarization to attenuate systemic lupus erythematosus-associated diffuse alveolar hemorrhage in mice. Int. J. Stem Cells 14, 331–340. doi: 10.15283/ijsc20156
Chen, J.-L., and Yan, H. (2015). Dicaine represses apoptosis-linked gene 2-interacting protein X expression to induce airway epithelial barrier dysfunction. Mol. Med. Rep. 12, 238–242. doi: 10.3892/mmr.2015.3433
Chen, Y. R., Zhong, S., Fei, Z., Gao, S., Zhang, S., Li, Z., et al. (2014). Transcriptome responses of the host Trichoplusia ni to infection by the baculovirus Autographa californica multiple nucleopolyhedrovirus. J. Virol. 88, 13781–13797. doi: 10.1128/JVI.02243-14
Chou, S.-F., Tsai, M.-L., Huang, J.-Y., Chang, Y.-S., and Shih, C. (2015). The dual role of an ESCRT-0 component HGS in HBV transcription and naked capsid secretion. PLoS Pathog. 11:e1005123. doi: 10.1371/journal.ppat.1005123
Christ, L., Raiborg, C., Wenzel, E. M., Campsteijn, C., and Stenmark, H. (2017). Cellular functions and molecular mechanisms of the ESCRT membrane-scission machinery. Trends Biochem. Sci. 42, 42–56. doi: 10.1016/j.tibs.2016.08.016
Christ, L., Wenzel, E. M., Liestol, K., Raiborg, C., Campsteijn, C., and Stenmark, H. (2016). ALIX and ESCRT-I/II function as parallel ESCRT-III recruiters in cytokinetic abscission. J. Cell Biol. 212, 499–513. doi: 10.1083/jcb.201507009
Corless, L., Crump, C. M., Griffin, S. D. C., and Harris, M. (2010). Vps4 and the ESCRT-III complex are required for the release of infectious hepatitis C virus particles. J. Gen. Virol. 91, 362–372. doi: 10.1099/vir.0.017285-0
Crump, C. (2018). Virus assembly and egress of HSV. Adv Exp Med Biol. 1045, 23–44. doi: 10.1007/978-981-10-7230-7_2
Crump, C. A., Yates, C., and Minson, T. (2007). Herpes simplex virus type 1 cytoplasmic envelopment requires functional Vps4. J. Virol. 81, 7380–7387. doi: 10.1128/JVI.00222-07
Cruz Casabona, J., Levingston Macleod, J. M., Eugenia Loureiro, M., Gomez, G. A., and Lopez, N. (2009). The RING domain and the L79 residue of Z protein are involved in both the Rescue of Nucleocapsids and the incorporation of glycoproteins into infectious chimeric arenavirus-like particles. J. Virol. 83, 7029–7039. doi: 10.1128/JVI.00329-09
Dai, X., and Zhou, Z. H. (2018). Structure of the herpes simplex virus 1 capsid with associated tegument protein complexes. Science 360:eaao7298. doi: 10.1126/science.aao7298
Dancho, B., McKenzie, M. O., Connor, J. H., and Lyles, D. S. (2009). Vesicular stomatitis virus matrix protein mutations that affect association with host membranes and viral Nucleocapsids. J. Biol. Chem. 284, 4500–4509. doi: 10.1074/jbc.M808136200
de Franceschi, N., Alqabandi, M., Weissenhorn, W., and Bassereau, P. (2019). Dynamic and sequential protein reconstitution on negatively curved membranes by Giant vesicles fusion. Bio-Protocol 9:e3294. doi: 10.21769/BioProtoc.3294
Deng, L., Liang, Y., Ariffianto, A., Matsui, C., Abe, T., Muramatsu, M., et al. (2022). Hepatitis C virus-induced ROS/JNK signaling pathway activates the E3 ubiquitin ligase itch to promote the release of HCV particles via Polyubiquitylation of VPS4A. J. Virol. 96, e0181–21. doi: 10.1128/jvi.01811-21
Dokland, T. (2010). The structural biology of PRRSV. Virus Res. 154, 86–97. doi: 10.1016/j.virusres.2010.07.029
Dolnik, O., Kolesnikova, L., Stevermann, L., and Becker, S. (2010). Tsg101 is recruited by a late domain of the Nucleocapsid protein to support budding of Marburg virus-like particles. J. Virol. 84, 7847–7856. doi: 10.1128/JVI.00476-10
Dolnik, O., Kolesnikova, L., Welsch, S., Strecker, T., Schudt, G., and Becker, S. (2014). Interaction with Tsg101 is necessary for the efficient transport and release of Nucleocapsids in Marburg virus-infected cells. PLoS Pathog. 10. doi: 10.1371/journal.ppat.1004463
Duan, Z., Hu, Z., Zhu, J., Xu, H., Chen, J., Liu, H., et al. (2014). Mutations in the FPIV motif of Newcastle disease virus matrix protein attenuate virus replication and reduce virus budding. Arch. Virol. 159, 1813–1819. doi: 10.1007/s00705-014-1998-2
Elmore, S. (2007). Apoptosis: a review of programmed cell death. Toxicol. Pathol. 35, 495–516. doi: 10.1080/01926230701320337
Espiritu, R. A. (2021). Repairing plasma membrane damage in regulated necrotic cell death. Mol. Biol. Rep. 48, 2751–2759. doi: 10.1007/s11033-021-06252-w
Fabrikant, G., Lata, S., Riches, J. D., Briggs, J. A. G., Weissenhorn, W., and Kozlov, M. M. (2009). Computational model of membrane fission catalyzed by ESCRT-III. PLoS Comput. Biol. 5:e1000575. doi: 10.1371/journal.pcbi.1000575
Fan, W., Guo, J., Gao, B., Zhang, W., Ling, L., Xu, T., et al. (2019). Flotillin-mediated endocytosis and ALIX-syntenin-1-mediated exocytosis protect the cell membrane from damage caused by necroptosis. Sci. Signal. 12:eaaw3423. doi: 10.1126/scisignal.aaw3423
Fan, J., Liao, Y., Zhang, M., Liu, C., Li, Z., Li, Y., et al. (2021). Anti-classical swine fever virus strategies. Microorganisms 9:761. doi: 10.3390/microorganisms9040761
Feldmann, H., Volchkov, V. E., Volchkova, V. A., Stroher, U., and Klenk, H. D. (2001). Biosynthesis and role of filoviral glycoproteins. J. Gen. Virol. 82, 2839–2848. doi: 10.1099/0022-1317-82-12-2839
Filimonenko, M., Stuffers, S., Raiborg, C., Yamamoto, A., Malerod, L., Fisher, E. M. C., et al. (2007). Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J. Cell Biol. 179, 485–500. doi: 10.1083/jcb.200702115
Finke, S., and Conzelmann, K. K. (2005). Replication strategies of rabies virus. Virus Res. 111, 120–131. doi: 10.1016/j.virusres.2005.04.004
Finken-Eigen, M., Roehricht, R. A., and Koehrer, K. (1997). The VPS4 gene is involved in protein transport out of a yeast pre-vacuolar endosome-like compartment. Curr. Genet. 31, 469–480. doi: 10.1007/s002940050232
Freed, E. O. (2002). Viral late domains. J. Virol. 76, 4679–4687. doi: 10.1128/JVI.76.10.4679-4687.2002
Ganem, D., and Prince, A. M. (2004). Mechanisms of disease: hepatitis B virus infection - natural history and clinical consequences. N. Engl. J. Med. 350, 1118–1129. doi: 10.1056/NEJMra031087
Garrus, J. E., von Schwedler, U. K., Pornillos, O. W., Morham, S. G., Zavitz, K. H., Wang, H. E., et al. (2001). Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cells 107, 55–65. doi: 10.1016/s0092-8674(01)00506-2
Ghoujal, B., Milev, M. P., Ajamian, L., Abel, K., and Mouland, A. J. (2012). ESCRT-II's involvement in HIV-1 genomic RNA trafficking and assembly. Biol. Cell. 104, 706–721. doi: 10.1111/boc.201200021
Gong, Y.-N., Guy, C., Olauson, H., Becker, J. U., Yang, M., Fitzgerald, P., et al. (2017). ESCRT-III acts downstream of MLKL to regulate Necroptotic cell death and its consequences. Cells 169, 286–300. doi: 10.1016/j.cell.2017.03.020
Green, D. R. (2019). The coming decade of cell death research: five riddles. Cells 177, 1094–1107. doi: 10.1016/j.cell.2019.04.024
Haguenauer-Tsapis, R. (2012). An MBoC favorite: morphological classification of the yeast vacuolar protein-sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol. Biol. Cell 23:2622. doi: 10.1091/mbc.E12-03-0184
Han, Z., Madara, J. J., Liu, Y., Liu, W., Ruthel, G., Freedman, B. D., et al. (2015). ALIX rescues budding of a double PTAP/PPEY L-domain deletion mutant of Ebola VP40: a role for ALIX in Ebola virus egress. J. Infect. Dis. 212, S138–S145. doi: 10.1093/infdis/jiu838
Han, J.-H., Ryu, H.-H., Jun, M.-H., Jang, D.-J., and Lee, J.-A. (2012). The functional analysis of the CHMP2B missense mutation associated with neurodegenerative diseases in the endo-lysosomal pathway. Biochem. Biophys. Res. Commun. 421, 544–549. doi: 10.1016/j.bbrc.2012.04.041
Harker-Kirschneck, L., Hafner, A. E., Yao, T., Vanhille-Campos, C., Jiang, X., Pulschen, A., et al. (2022). Physical mechanisms of ESCRT-III-driven cell division. Proc. Natl. Acad. Sci. U. S. A. 119:e2107763119. doi: 10.1073/pnas.2107763119
Harrison, M. S., Sakaguchi, T., and Schmitt, A. P. (2010). Paramyxovirus assembly and budding: building particles that transmit infections. Int. J. Biochem. Cell Biol. 42, 1416–1429. doi: 10.1016/j.biocel.2010.04.005
Hasan, M. K., Kato, A., Muranaka, M., Yamaguchi, R., Sakai, Y., Hatano, I., et al. (2000). Versatility of the accessory C proteins of Sendai virus: contribution to virus assembly as an additional role. J. Virol. 74, 5619–5628. doi: 10.1128/JVI.74.12.5619-5628.2000
Henrickson, K. J. (2003). Parainfluenza viruses. Clin. Microbiol. Rev. 16:242-+. doi: 10.1128/cmr.16.2.242-264.2003
Hoenen, T., Kolesnikova, L., and Becker, S. (2007). Recent advances in filovirus- and arenavirus-like particles. Futur. Virol. 2, 193–203. doi: 10.2217/17460794.2.2.193
Hollinshead, M., Johns, H. L., Sayers, C. L., Gonzalez-Lopez, C., Smith, G. L., and Elliott, G. (2012). Endocytic tubules regulated by Rab GTPases 5 and 11 are used for envelopment of herpes simplex virus. EMBO J. 31, 4204–4220. doi: 10.1038/emboj.2012.262
Huotari, J., and Helenius, A. (2011). Endosome maturation. EMBO J. 30, 3481–3500. doi: 10.1038/emboj.2011.286
Hurley, J. H. (2010). The ESCRT complexes. Crit. Rev. Biochem. Mol. Biol. 45, 463–487. doi: 10.3109/10409238.2010.502516
Hurley, J. H., and Hanson, P. I. (2010). Membrane budding and scission by the ESCRT machinery: it's all in the neck. Nat. Rev. Mol. Cell Biol. 11, 556–566. doi: 10.1038/nrm2937
Hurtig, F., Burgers, T. C. Q., Cezanne, A., Jiang, X., Mol, F. N., Traparic, J., et al. (2023). The patterned assembly and stepwise Vps4-mediated disassembly of composite ESCRT-III polymers drives archaeal cell division. Sci. Adv. 9:eade5224. doi: 10.1126/sciadv.ade5224
Irie, T., Shimazu, Y., Yoshida, T., and Sakaguchi, T. (2007). The YLDL sequence within Sendai virus m protein is critical for budding of virus-like particles and interacts with Alix/AIP1 independently of C protein. J. Virol. 81, 2263–2273. doi: 10.1128/JVI.02218-06
Isono, E. (2021). ESCRT is a great sealer: non-endosomal function of the ESCRT machinery in membrane repair and autophagy. Plant Cell Physiol. 62, 766–774. doi: 10.1093/pcp/pcab045
Itakura, Y., Tabata, K., Saito, T., Intaruck, K., Kawaguchi, N., Kishimoto, M., et al. (2023). Morphogenesis of bullet-shaped rabies virus particles regulated by TSG101. J. Virol. 97:e0043823. doi: 10.1128/jvi.00438-23
Janvier, K., Pelchen-Matthews, A., Renaud, J.-B., Caillet, M., Marsh, M., and Berlioz-Torrent, C. (2011). The ESCRT-0 component HRS is required for HIV-1 Vpu-mediated BST-2/Tetherin Down-regulation. PLoS Pathog. 7:e1001265. doi: 10.1371/journal.ppat.1001265
Jimenez, A. J., Maiuri, P., Lafaurie-Janvore, J., Divoux, S., Piel, M., and Perez, F. (2014). ESCRT machinery is required for plasma membrane repair. Science 343:1247136. doi: 10.1126/science.1247136
Jirasko, V., Montserret, R., Lee, J. Y., Gouttenoire, J., Moradpour, D., Penin, F., et al. (2010). Structural and functional studies of nonstructural protein 2 of the hepatitis C virus reveal its key role as organizer of Virion assembly. PLoS Pathog. 6:e1001233. doi: 10.1371/journal.ppat.1001233
Johnson, D. S., Bleck, M., and Simon, S. M. (2018). Timing of ESCRT-III protein recruitment and membrane scission during HIV-1 assembly. elife 7:e36221. doi: 10.7554/eLife.36221
Johnson, D. S., Bleck, M., and Simon, S. M. (2019). Recruitment dynamics of Escrt-III proteins during HIV-1 gag assembly and plasma membrane scission. Biophys. J. 116:373A. doi: 10.1016/j.bpj.2018.11.2029
Jones, C. T., Murray, C. L., Eastman, D. K., Tassello, J., and Rice, C. M. (2007). Hepatitis C virus p7 and NS2 proteins are essential for production of infectious virus. J. Virol. 81, 8374–8383. doi: 10.1128/JVI.00690-07
Katzmann, D. J., Babst, M., and Emr, S. D. (2001). Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cells 106, 145–155. doi: 10.1016/S0092-8674(01)00434-2
Kaul, Z., and Chakrabarti, O. (2017). Tumor susceptibility gene 101 regulates predisposition to apoptosis via ESCRT machinery accessory proteins. Mol. Biol. Cell 28, 2106–2122. doi: 10.1091/mbc.E16-12-0855
Kieffer, C., Skalicky, J. J., Morita, E., De Domenico, I., Ward, D. M., Kaplan, J., et al. (2008). Two distinct modes of ESCRT-III recognition are required for VPS4 functions in lysosomal protein targeting and HIV-1 budding. Dev. Cell 15, 62–73. doi: 10.1016/j.devcel.2008.05.014
Kim, M. S., and Kim, K. H. (2011). Protection of olive flounder, Paralichthys olivaceus, against viral hemorrhagic septicemia virus (VHSV) by immunization with NV gene-knockout recombinant VHSV. Aquaculture 314, 39–43. doi: 10.1016/j.aquaculture.2011.01.050
Knorr, R. L., Lipowsky, R., and Dimova, R. (2015). Autophagosome closure requires membrane scission. Autophagy 11, 2134–2137. doi: 10.1080/15548627.2015.1091552
Kurotani, A., Kiyotani, K., Kato, A., Shioda, T., Sakai, Y., Mizumoto, K., et al. (1998). Sendai virus C proteins are categorically nonessential gene products but silencing their expression severely impairs viral replication and pathogenesis. Genes Cells 3, 111–124. doi: 10.1046/j.1365-2443.1998.00170.x
Langelier, C., von Schwedler, U. K., Fisher, R. D., De Domenico, I., White, P. L., Hill, C. P., et al. (2006). Human ESCRT-II complex and its role in human immunodeficiency virus type 1 release. J. Virol. 80, 9465–9480. doi: 10.1128/JVI.01049-06
Larios, J., Mercier, V., Roux, A., and Gruenberg, J. (2020). ALIX- and ESCRT-III-dependent sorting of tetraspanins to exosomes. J. Cell Biol. 219:e201904113. doi: 10.1083/jcb.201904113
Lee, J.-A., Beigneux, A., Ahmad, S. T., Young, S. G., and Gao, F.-B. (2007a). ESCRT-III dysfunction causes autophagosome accumulation and neurodegeneration. Curr. Biol. 17, 1561–1567. doi: 10.1016/j.cub.2007.07.029
Lee, S., Joshi, A., Nagashima, K., Freed, E. O., and Hurley, J. H. (2007b). Structural basis for viral late-domain binding to Alix. Nat. Struct. Mol. Biol. 14, 194–199. doi: 10.1038/nsmb1203
Lee, I.-H., Kai, H., Carlson, L.-A., Groves, J. T., and Hurley, J. H. (2015). Negative membrane curvature catalyzes nucleation of endosomal sorting complex required for transport (ESCRT)-III assembly. Proc. Natl. Acad. Sci. U. S. A. 112, 15892–15897. doi: 10.1073/pnas.1518765113
Lefebvre, C., Legouis, R., and Culetto, E. (2018). ESCRT and autophagies: endosomal functions and beyond. Semin. Cell Dev. Biol. 74, 21–28. doi: 10.1016/j.semcdb.2017.08.014
Lefkowitz, E. J., Dempsey, D. M., Hendrickson, R. C., Orton, R. J., Siddell, S. G., and Smith, D. B. (2018). Virus taxonomy: the database of the international committee on taxonomy of viruses (ICTV). Nucleic Acids Res. 46, D708–D717. doi: 10.1093/nar/gkx932
Li, Z., and Blissard, G. W. (2012). Cellular VPS4 is required for efficient entry and egress of budded virions of Autographa californica multiple nucleopolyhedrovirus. J. Virol. 86, 459–472. doi: 10.1128/JVI.06049-11
Li, Z., and Blissard, G. (2015). The vacuolar protein sorting genes in insects: a comparative genome view. Insect Biochem. Mol. Biol. 62, 211–225. doi: 10.1016/j.ibmb.2014.11.007
Li, X., Li, X., Cao, H., Wang, Y., and Zheng, S. J. (2013). Engagement of new castle disease virus (NDV) matrix (M) protein with charged multivesicular body protein (CHMP) 4 facilitates viral replication. Virus Res. 171, 80–88. doi: 10.1016/j.virusres.2012.10.033
Li, Z., Mo, F., Wang, Y., Li, W., Chen, Y., Liu, J., et al. (2022). Enhancing Gasdermin-induced tumor pyroptosis through preventing ESCRT-dependent cell membrane repair augments antitumor immune response. Nat. Commun. 13:6321. doi: 10.1038/s41467-022-34036-8
Li, M., Schmitt, P. T., Li, Z., McCrory, T. S., He, B., and Schmitt, A. P. (2009). Mumps virus matrix, fusion, and nucleocapsid proteins cooperate for efficient production of virus-like particles. J. Virol. 83, 7261–7272. doi: 10.1128/JVI.00421-09
Licata, J. M., Simpson-Holley, M., Wright, N. T., Han, Z. Y., Paragas, J., and Harty, R. N. (2003). Overlapping motifs (PTAP and PPEY) within the Ebola virus VP40 protein function independently as late budding domains: involvement of host proteins TSG101 and VPS-4. J. Virol. 77, 1812–1819. doi: 10.1128/JVI.77.3.1812-1819.2003
Lippincott-Schwartz, J., Freed, E. O., and van Engelenburg, S. B. (2017). A consensus view of ESCRT-mediated human immunodeficiency virus type 1 abscission. Annu Rev Virol. 4, 309–325. doi: 10.1146/annurev-virology-101416-041840
Liu, C.-C., Liu, Y.-Y., Cheng, Y., Zhang, Y.-N., Zhang, J., Liang, X.-D., et al. (2021). The ESCRT-I subunit Tsg101 plays novel dual roles in entry and replication of classical swine fever virus. J. Virol. 95, e01928–e01920. doi: 10.1128/jvi.01928-20
Liu, C. C., Liu, Y. Y., Zhou, J. F., Chen, X., Chen, H., Hu, J. H., et al. (2022). Cellular ESCRT components are recruited to regulate the endocytic trafficking and RNA replication compartment assembly during classical swine fever virus infection. PLoS Pathog. 18:e1010294. doi: 10.1371/journal.ppat.1010294
Marshall, R. S., and Vierstra, R. D. (2018). Autophagy: the master of bulk and selective recycling. Annu Rev Plant Biol. 69, 173–208. doi: 10.1146/annurev-arplant-042817-040606
Martin-Serrano, J., and Marsh, M. (2007). ALIX catches HIV. Cell Host Microbe 1, 5–7. doi: 10.1016/j.chom.2007.02.006
Martin-Serrano, J., Zang, T., and Bieniasz, P. D. (2001). HIV-I and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 7, 1313–1319. doi: 10.1038/nm1201-1313
Mathieu, J., Michel-Hissier, P., Boucherit, V., and Huynh, J.-R. (2022). The deubiquitinase USP8 targets ESCRT-III to promote incomplete cell division. Science 376, 818–823. doi: 10.1126/science.abg2653
Mebatsion, T., Weiland, F., and Conzelmann, K. K. (1999). Matrix protein of rabies virus is responsible for the assembly and budding of bullet-shaped particles and interacts with the transmembrane spike glycoprotein G. J. Virol. 73, 242–250. doi: 10.1128/JVI.73.1.242-250.1999
Medina, G. N., Ehrlich, L. S., Chen, M. H., Khan, M. B., Powell, M. D., and Carter, C. A. (2011). Sprouty 2 binds ESCRT-II factor Eap20 and facilitates HIV-1 gag release. J. Virol. 85, 7353–7362. doi: 10.1128/JVI.00141-11
Meng, B., and Lever, A. M. L. (2013). Wrapping up the bad news - HIV assembly and release. Retrovirology 10:5. doi: 10.1186/1742-4690-10-5
Meng, B., and Lever, A. M. L. (2021). The interplay between ESCRT and viral factors in the enveloped virus life cycle. Viruses 13:324. doi: 10.3390/v13020324
Meyers, G., Thiel, H. J., and Rümenapf, T. (1996). Classical swine fever virus: recovery of infectious viruses from cDNA constructs and generation of recombinant cytopathogenic defective interfering particles. J. Virol. 70, 1588–1595. doi: 10.1128/jvi.70.3.1588-1595.1996
Mierzwa, B., and Gerlich, D. W. (2014). Cytokinetic abscission: molecular mechanisms and temporal control. Dev. Cell 31, 525–538. doi: 10.1016/j.devcel.2014.11.006
Migliano, S. M., and Teis, D. (2018). ESCRT and membrane protein ubiquitination. Prog Mol Subcell Biol. 57, 107–135. doi: 10.1007/978-3-319-96704-2_4
Migliano, S. M., Wenzel, E. M., and Stenmark, H. (2022). Biophysical and molecular mechanisms of ESCRT functions, and their implications for disease. Curr. Opin. Cell Biol. 75:102062. doi: 10.1016/j.ceb.2022.01.007
Moennig, V. (2015). The control of classical swine fever in wild boar. Front. Microbiol. 6:1211. doi: 10.3389/fmicb.2015.01211
Morita, E., Colf, L. A., Karren, M. A., Sandrin, V., Rodesch, C. K., and Sundquist, W. I. (2010). Human ESCRT-III and VPS4 proteins are required for centrosome and spindle maintenance. Proc. Natl. Acad. Sci. U. S. A. 107, 12889–12894. doi: 10.1073/pnas.1005938107
Morita, E., Sandrin, V., Alam, S. L., Eckert, D. M., Gygi, S. P., and Sundquist, W. I. (2007). Identification of human MVB12 proteins as ESCRT-I subunits that function in HIV budding. Cell Host Microbe 2, 41–53. doi: 10.1016/j.chom.2007.06.003
Morita, E., and Sundquist, W. I. (2004). Retrovirus budding. Annu. Rev. Cell Dev. Biol. 20, 395–425. doi: 10.1146/annurev.cellbio.20.010403.102350
Morzunov, S. P., Winton, J. R., and Nichol, S. T. (1995). The complete genome structure and phylogenetic relationship of infectious hematopoietic necrosis virus. Virus Res. 38, 175–192. doi: 10.1016/0168-1702(95)00056-v
Muhlberger, E., Lotfering, B., Klenk, H. D., and Becker, S. (1998). Three of the four nucleocapsid proteins of Marburg virus, NP, VP35, and L, are sufficient to mediate replication and transcription of Marburg virus-specific monocistronic minigenomes. J. Virol. 72, 8756–8764. doi: 10.1128/JVI.72.11.8756-8764.1998
Murrow, L., Malhotra, R., and Debnath, J. (2015). ATG12-ATG3 interacts with Alix to promote basal autophagic flux and late endosome function. Nat. Cell Biol. 17, 300–310. doi: 10.1038/ncb3112
Nepal, B., Sepehri, A., and Lazaridis, T. (2020). Mechanisms of negative membrane curvature sensing and generation by ESCRT III subunit Snf7. Protein Sci. 29, 1473–1485. doi: 10.1002/pro.3851
Ning, P., Gao, L., Zhou, Y., Hu, C., Lin, Z., Gong, C., et al. (2016). Caveolin-1-mediated endocytic pathway is involved in classical swine fever virus Shimen infection of porcine alveolar macrophages. Vet. Microbiol. 195, 81–86. doi: 10.1016/j.vetmic.2016.09.016
Oda, K., Matoba, Y., Sugiyama, M., and Sakaguchi, T. (2021). Structural insight into the interaction of Sendai virus C protein with Alix to stimulate viral budding. J. Virol. 95, e00815–21. doi: 10.1128/JVI.00815-21
Panchal, R. G., Ruthel, G., Kenny, T. A., Kallstrom, G. H., Lane, D., Badie, S. S., et al. (2003). In vivo oligomerization and raft localization of Ebola virus protein VP40 during vesicular budding. Proc. Natl. Acad. Sci. U. S. A. 100, 15936–15941. doi: 10.1073/pnas.2533915100
Park, A., Yun, T., Vigant, F., Pernet, O., Won, S. T., Dawes, B. E., et al. (2016). Nipah virus C protein recruits Tsg101 to promote the efficient release of virus in an ESCRT-dependent pathway. PLoS Pathog. 12:e1005659. doi: 10.1371/journal.ppat.1005659
Pasqual, G., Rojek, J. M., Masin, M., Chatton, J.-Y., and Kunz, S. (2011). Old World arenaviruses enter the host cell via the multivesicular body and depend on the endosomal sorting complex required for transport. PLoS Pathog. 7:e1002232. doi: 10.1371/annotation/952387a7-96a0-44f6-98a6-2d7c7b472b0a
Pasternak, A. O., Spaan, W. J. M., and Snijder, E. J. (2006). Nidovirus transcription: how to make sense…? J. Gen. Virol. 87, 1403–1421. doi: 10.1099/vir.0.81611-0
Patch, J. R., Crameri, G., Wang, L.-F., Eaton, B. T., and Broder, C. C. (2007). Quantitative analysis of Nipah virus proteins released as virus-like particles reveals central role for the matrix protein. Virol. J. 4:1. doi: 10.1186/1743-422x-4-1
Patch, J. R., Han, Z., McCarthy, S. E., Yan, L., Wang, L.-F., Harty, R. N., et al. (2008). The YPLGVG sequence of the Nipah virus matrix protein is required for budding. Virol. J. 5:137. doi: 10.1186/1743-422x-5-137
Pawliczek, T., and Crump, C. M. (2009). Herpes simplex virus type 1 production requires a functional ESCRT-III complex but is independent of TSG101 and ALIX expression. J. Virol. 83, 11254–11264. doi: 10.1128/JVI.00574-09
Pei, Y., Xue, J., Teng, Q., Feng, D., Huang, M., Liang, R., et al. (2023). Mutation of phenylalanine 23 of Newcastle disease virus matrix protein inhibits virus release by disrupting the interaction between the FPIV L-domain and charged multivesicular body protein 4B. Microbiol. Spect. e0411622. doi: 10.1128/spectrum.04116-22
Pentecost, M., Vashisht, A. A., Lester, T., Voros, T., Beaty, S. M., Park, A., et al. (2015). Evidence for ubiquitin-regulated nuclear and subnuclear trafficking among Paramyxovirinae matrix proteins. PLoS Pathog. 11:e1004739. doi: 10.1371/journal.ppat.1004739
Perez, M., Craven, R. C., and de la Torre, J. C. (2003). The small RING finger protein Z drives arenavirus budding: implications for antiviral strategies. Proc. Natl. Acad. Sci. U. S. A. 100, 12978–12983. doi: 10.1073/pnas.2133782100
Petiot, A., and Sadoul, R. (2009). Autophagy discriminates between Alix and ESCRTs. Autophagy 5, 106–107. doi: 10.4161/auto.5.1.7277
Pincetic, A., Medina, G., Carter, C., and Leis, J. (2008). Avian sarcoma virus and human immunodeficiency virus, type 1 use different subsets of ESCRT proteins to facilitate the budding process. J. Biol. Chem. 283, 29822–29830. doi: 10.1074/jbc.M804157200
Prag, G., Watson, H., Kim, Y. C., Beach, B. M., Ghirlando, R., Hummer, G., et al. (2007). The Vps27/Hse1 complex is a GAT domain-based scaffold for ubiquitin-dependent sorting. Dev. Cell 12, 973–986. doi: 10.1016/j.devcel.2007.04.013
Raiborg, C., and Stenmark, H. (2009). The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature 458, 445–452. doi: 10.1038/nature07961
Ringelhan, M., O'Connor, T., Protzer, U., and Heikenwalder, M. (2015). The direct and indirect roles of HBV in liver cancer: prospective markers for HCC screening and potential therapeutic targets. J. Pathol. 235, 355–367. doi: 10.1002/path.4434
Robison, C. S., and Whitt, M. A. (2000). The membrane-proximal stem region of vesicular stomatitis virus G protein confers efficient virus assembly. J. Virol. 74, 2239–2246. doi: 10.1128/JVI.74.5.2239-2246.2000
Rose, K. M., Hirsch, V. M., and Bouamr, F. (2020). Budding of a retrovirus: some assemblies required. Viruses 12. doi: 10.3390/v12101188
Rothman, J. H., and Stevens, T. H. (1986). Protein sorting in yeast: mutants defective in vacuole biogenesis mislocalize vacuolar proteins into the late secretory pathway. Cells 47, 1041–1051. doi: 10.1016/0092-8674(86)90819-6
Ruhl, S., Shkarina, K., Demarco, B., Heilig, R., Santos, J. C., and Broz, P. (2018). ESCRT-dependent membrane repair negatively regulates pyroptosis downstream of GSDMD activation. Science 362, 956–960. doi: 10.1126/science.aar7607
Rusten, T. E., and Stenmark, H. (2009). How do ESCRT proteins control autophagy? J. Cell Sci. 122, 2179–2183. doi: 10.1242/jcs.050021
Sakaguchi, T., Kato, A., Sugahara, F., Shimazu, Y., Inoue, M., Kiyotani, K., et al. (2005). AIP1/Alix is a binding partner of Sendai virus C protein and facilitates virus budding. J. Virol. 79, 8933–8941. doi: 10.1128/JVI.79.14.8933-8941.2005
Sang, Y., Shi, J., Sang, W., Rowland, R. R. R., and Blecha, F. (2012). Replication-competent recombinant porcine reproductive and respiratory syndrome (PRRS) viruses expressing Indicator proteins and antiviral cytokines. Viruses 4, 102–116. doi: 10.3390/v4010102
Schaefer, J. A., Schessner, J. P., Bircham, P. W., Tsuji, T., Funaya, C., Pajonk, O., et al. (2020). ESCRT machinery mediates selective microautophagy of endoplasmic reticulum in yeast. EMBO J. 39:e102586. doi: 10.15252/embj.2019102586
Scheel, T. K. H., and Rice, C. M. (2013). Understanding the hepatitis C virus life cycle paves the way for highly effective therapies. Nat. Med. 19, 837–849. doi: 10.1038/nm.3248
Scheffer, L. L., Sreetama, S. C., Sharma, N., Medikayala, S., Brown, K. J., Defour, A., et al. (2014). Mechanism of Ca2+−triggered ESCRT assembly and regulation of cell membrane repair. Nat. Commun. 5:5646. doi: 10.1038/ncomms6646
Schmidt, M. H. H., Dikic, I., and Bogler, O. (2005). Src phosphorylation of Alix/AIP1 modulates its interaction with binding partners and antagonizes its activities. J. Biol. Chem. 280, 3414–3425. doi: 10.1074/jbc.M409839200
Schmitt, A. P., Leser, G. P., Morita, E., Sundquist, W. I., and Lamb, R. A. (2005). Evidence for a new viral late-domain core sequence, FPIV, necessary for budding of a paramyxovirus. J. Virol. 79, 2988–2997. doi: 10.1128/JVI.79.5.2988-2997.2005
Schoneberg, J., Lee, I.-H., Iwasa, J. H., and Hurley, J. H. (2017). Reverse-topology membrane scission by the ESCRT proteins. Nat. Rev. Mol. Cell Biol. 18, 5–17. doi: 10.1038/nrm.2016.121
Sette, P., Nagashima, K., Piper, R. C., and Bouamr, F. (2013). Ubiquitin conjugation to gag is essential for ESCRT-mediated HIV-1 budding. Retrovirology 10:79. doi: 10.1186/1742-4690-10-79
Shestakova, A., Hanono, A., Drosner, S., Curtiss, M., Davies, B. A., Katzmann, D. J., et al. (2010). Assembly of the AAA ATPase Vps4 on ESCRT-III. Mol. Biol. Cell 21, 1059–1071. doi: 10.1091/mbc.E09-07-0572
Shirakura, M., Murakami, K., Ichimura, T., Suzuki, R., Shimoji, T., Fukuda, K., et al. (2007). E6AP ubiquitin ligase mediates ubiquitylation and degradation of hepatitis C virus core protein. J. Virol. 81, 1174–1185. doi: 10.1128/JVI.01684-06
Shtanko, O., Watanabe, S., Jasenosky, L. D., Watanabe, T., and Kawaoka, Y. (2011). ALIX/AIP1 is required for NP incorporation into Mopeia virus Z-induced virus-like particles. J. Virol. 85, 3631–3641. doi: 10.1128/JVI.01984-10
Sigismund, S., Confalonieri, S., Ciliberto, A., Polo, S., Scita, G., and Di Fiore, P. P. (2012). Endocytosis and signaling: cell logistics shape the eukaryotic cell plan. Physiol. Rev. 92, 273–366. doi: 10.1152/physrev.00005.2011
Slagsvold, T., Aasland, R., Hirano, S., Bache, K. G., Raiborg, C., Trambaiolo, D., et al. (2005). Eap45 in mammalian ESCRT-II binds ubiquitin via a phosphoinositide-interacting GLUE domain. J. Biol. Chem. 280, 19600–19606. doi: 10.1074/jbc.M501510200
Southern, P.J. (1996). Arenaviridae: The viruses and their replication. Fundamental virology, Third edition.
Spitzer, C., Schellmann, S., Sabovljevic, A., Shahriari, M., Keshavaiah, C., Bechtold, N., et al. (2006). The Arabidopsis elch mutant reveals functions of an ESCRT component in cytokinesis. Development 133, 4679–4689. doi: 10.1242/dev.02654
Steigemann, P., and Gerlich, D. W. (2009). Cytokinetic abscission: cellular dynamics at the midbody. Trends Cell Biol. 19, 606–616. doi: 10.1016/j.tcb.2009.07.008
Stieler, J. T., and Prange, R. (2014). Involvement of ESCRT-II in hepatitis B virus morphogenesis. PLoS One 9:e91279. doi: 10.1371/journal.pone.0091279
Stoten, C. L., and Carlton, J. G. (2018). ESCRT-dependent control of membrane remodelling during cell division. Semin. Cell Dev. Biol. 74, 50–65. doi: 10.1016/j.semcdb.2017.08.035
Strack, B., Calistri, A., Accola, M. A., Palu, G., and Gottlinger, H. G. (2000). A role for ubiquitin ligase recruitment in retrovirus release. Proc. Natl. Acad. Sci. U. S. A. 97, 13063–13068. doi: 10.1073/pnas.97.24.13063
Strack, B., Calistri, A., Craig, S., Popova, E., and Gottlinger, H. G. (2003). AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cells 114, 689–699. doi: 10.1016/S0092-8674(03)00653-6
Strappazzon, F., Torch, S., Chatellard-Causse, C., Petiot, A., Thibert, C., Blot, B., et al. (2010). Alix is involved in caspase 9 activation during calcium-induced apoptosis. Biochem. Biophys. Res. Commun. 397, 64–69. doi: 10.1016/j.bbrc.2010.05.062
Strecker, T., Eichler, R., ter Meulen, J., Weissenhorn, W., Klenk, H. D., Garten, W., et al. (2003). Lassa virus Z protein is a matrix protein sufficient for the release of virus-like particles. J. Virol. 77, 10700–10705. doi: 10.1128/JVI.77.19.10700-10705.2003
Su, C.-M., Rowland, R. R. R., and Yoo, D. (2021). Recent advances in PRRS virus receptors and the targeting of receptor-ligand for control. Vaccine 9:354. doi: 10.3390/vaccines9040354
Taelman, V. F., Dobrowolski, R., Plouhinec, J.-L., Fuentealba, L. C., Vorwald, P. P., Gumper, I., et al. (2010). Wnt signaling requires sequestration of glycogen synthase kinase 3 inside multivesicular endosomes. Cells 143, 1136–1148. doi: 10.1016/j.cell.2010.11.034
Takahashi, Y., He, H., Tang, Z., Hattori, T., Liu, Y., Young, M. M., et al. (2018). An autophagy assay reveals the ESCRT-III component CHMP2A as a regulator of phagophore closure. Nat. Commun. 9:2855. doi: 10.1038/s41467-018-05254-w
Tamai, K., Shiina, M., Tanaka, N., Nakano, T., Yamamoto, A., Kondo, Y., et al. (2012). Regulation of hepatitis C virus secretion by the Hrs-dependent exosomal pathway. Virology 422, 377–385. doi: 10.1016/j.virol.2011.11.009
Tang, S., Henne, W. M., Borbat, P. P., Buchkovich, N. J., Freed, J. H., Mao, Y., et al. (2015). Structural basis for activation, assembly and membrane binding of ESCRT-III Snf7 filaments. elife 4:e12548. doi: 10.7554/eLife.12548
Tedeschi, A., Almagro, J., Renshaw, M. J., Messal, H. A., Behrens, A., and Petronczki, M. (2020). Cep55 promotes cytokinesis of neural progenitors but is dispensable for most mammalian cell divisions. Nat. Commun. 11:1746. doi: 10.1038/s41467-020-15359-w
Teis, D., Saksena, S., Judson, B. L., and Emr, S. D. (2010). ESCRT-II coordinates the assembly of ESCRT-III filaments for cargo sorting and multivesicular body vesicle formation. EMBO J. 29, 871–883. doi: 10.1038/emboj.2009.408
Tellinghuisen, T. L., and Foss, K. L. (2008). Regulation of hepatitis C virion production via phosphorylation of the NS5A protein. PLoS Pathog. 4:e1000032. doi: 10.1371/journal.ppat.1000032
Teo, H. L., Gill, D. J., Sun, J., Perisic, O., Veprintsev, D. B., Vallis, Y., et al. (2006). ESCRT-I core and ESCRT-II GLUE domain structures reveal role for GLUE in linking to ESCRT-I and membranes. Cells 125, 99–111. doi: 10.1016/j.cell.2006.01.047
Thomas, E., Yoneda, M., and Schiff, E. R. (2015). Viral hepatitis: past and future of HBV and HDV. Cold Spring Harb. Perspect. Med. 5:a021345. doi: 10.1101/cshperspect.a021345
Timmins, J., Schoehn, G., Ricard-Blum, S., Scianimanico, S., Vernet, T., Ruigrok, R. W., et al. (2003). Ebola virus matrix protein VP40 interaction with human cellular factors Tsg101 and Nedd4. J. Mol. Biol. 326, 493–502. doi: 10.1016/S0022-2836(02)01406-7
Timmins, J., Scianimanico, S., Schoehn, G., and Weissenhorn, W. (2001). Vesicular release of Ebola virus matrix protein VP40. Virology 283, 1–6. doi: 10.1006/viro.2001.0860
Tuttleman, J. S., Pourcel, C., and Summers, J. (1986). Formation of the pool of covalently closed circular viral DNA in hepadnavirus-infected cells. Cells 47, 451–460. doi: 10.1016/0092-8674(86)90602-1
Urata, S., Noda, T., Kawaoka, Y., Morikawa, S., Yokosawa, H., and Yasuda, J. (2007). Interaction of Tsg101 with Marburg virus VP40 depends on the PPPY motif, but not the PT/SAP motif as in the case of Ebola virus, and Tsg101 plays a critical role in the budding of Marburg virus-like particles induced by VP40, NP, and GP. J. Virol. 81, 4895–4899. doi: 10.1128/JVI.02829-06
Urbe, S. (2005). Ubiquitin and endocytic protein sorting. Essays Biochem. 41, 81–98. doi: 10.1042/eb0410081
Valentine, M., Hogan, J., and Collier, S. (2014). The Drosophila Chmp1 protein determines wing cell fate through regulation of epidermal growth factor receptor signaling. Dev. Dyn. 243, 977–987. doi: 10.1002/dvdy.24140
Van Cleve, W., Amaro-Carambot, E., Surman, S. R., Bekisz, J., Collins, P. L., Zoon, K. C., et al. (2006). Attenuating mutations in the P/C gene of human parainfluenza virus type 1 (HPIV1) vaccine candidates abrogate the inhibition of both induction and signaling of type I interferon (IFN) by wild-type HPIV1. Virology 352, 61–73. doi: 10.1016/j.virol.2006.04.011
von Schwedler, U. K., Stuchell, M., Muller, B., Ward, D. M., Chung, H. Y., Morita, E., et al. (2003). The protein network of HIV budding. Cells 114, 701–713. doi: 10.1016/S0092-8674(03)00714-1
Votteler, J., and Sundquist, W. I. (2013). Virus budding and the ESCRT pathway. Cell Host Microbe 14, 232–241. doi: 10.1016/j.chom.2013.08.012
Welbourn, S., Jirasko, V., Breton, V., Reiss, S., Penin, F., Bartenschlager, R., et al. (2009). Investigation of a role for lysine residues in non-structural proteins 2 and 2/3 of the hepatitis C virus for their degradation and virus assembly. J. Gen. Virol. 90, 1071–1080. doi: 10.1099/vir.0.009944-0
Wilson, C., Kavaler, J., and Ahmad, S. T. (2020). Expression of a human variant of CHMP2B linked to neurodegeneration in Drosophila external sensory organs leads to cell fate transformations associated with increased notch activity. Dev. Neurobiol. 80, 85–97. doi: 10.1002/dneu.22722
Wirblich, C., Bhattacharya, B., and Roy, P. (2006). Nonstructural protein 3 of bluetongue virus assists virus release by recruiting ESCRT-I protein Tsg101. J. Virol. 80, 460–473. doi: 10.1128/JVI.80.1.460-473.2006
Wollert, T., and Hurley, J. H. (2010). Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature 464, 864–U873. doi: 10.1038/nature08849
Wollert, T., Wunder, C., Lippincott-Schwartz, J., and Hurley, J. H. (2009a). Membrane scission by the ESCRT-III complex. Nature 458, 172–177. doi: 10.1038/nature07836
Wollert, T., Yang, D., Ren, X., Lee, H. H., Im, Y. J., and Hurley, J. H. (2009b). The ESCRT machinery at a glance. J. Cell Sci. 122, 2163–2166. doi: 10.1242/jcs.029884
Xiao, J., Xia, H., Yoshino-Koh, K., Zhou, J., and Xu, Z. (2007). Structural characterization of the ATPase reaction cycle of endosomal AAA protein Vps4. J. Mol. Biol. 374, 655–670. doi: 10.1016/j.jmb.2007.09.067
Xie, Q.-H., Wang, W.-M., Yang, J.-G., Xia, H.-F., Xiao, B.-L., Chen, G.-H., et al. (2023). ALIX promotes cell migration and invasion of head and neck squamous cell carcinoma by regulating the expression of MMP9, MMP14, VEGF-C. Arch. Oral Biol. 151:105696. doi: 10.1016/j.archoralbio.2023.105696
Yan, H., Yi, H., Xia, L., Zhan, Z., He, W., Cao, J., et al. (2014). Staphylococcal enterotoxin B suppresses Alix and compromises intestinal epithelial barrier functions. J. Biomed. Sci. 21:29. doi: 10.1186/1423-0127-21-29
Yang, D., Rismanchi, N., Renvoise, B., Lippincott-Schwartz, J., Blackstone, C., and Hurley, J. (2009). Structural basis for midbody targeting of spastin by the ESCRT-III protein CHMP1B. FASEB J. 23, 503–512. doi: 10.1096/fj.08-118422
Yang, B., Stjepanovic, G., Shen, Q., Martin, A., and Hurley, J. H. (2015). Vps4 disassembles an ESCRT-III filament by global unfolding and processive translocation. Nat. Struct. Mol. Biol. 22, 492–U488. doi: 10.1038/nsmb.3015
Yang, Y., Wang, M., Zhang, Y.-Y., Zhao, S.-Z., and Gu, S. (2022). The endosomal sorting complex required for transport repairs the membrane to delay cell death. Front. Oncol. 12:1007446. doi: 10.3389/fonc.2022.1007446
Yu, X., Shahriari, S., Li, H.-M., and Ghildyal, R. (2016). Measles virus matrix protein inhibits host cell transcription. PLoS One 11:e0161360. doi: 10.1371/journal.pone.0161360
Yue, Q., Yu, Q., Yang, Q., Xu, Y., Guo, Y., Blissard, G. W., et al. (2018). Distinct roles of cellular ESCRT-I and ESCRT-III proteins in efficient entry and egress of budded Virions of Autographa californica multiple Nucleopolyhedrovirus. J. Virol. 92, e01636–17. doi: 10.1128/JVI.01636-17
Zavitz, K., Morham, S., and Wettstein, D.A. (2008). TSG101-GAG interaction and use thereof. United States Patent Appl. Publ. within the TVPP US20030663407.
Zhai, Q., Landesman, M. B., Chung, H.-Y., Dierkers, A., Jeffries, C. M., Trewhella, J., et al. (2011). Activation of the retroviral budding factor ALIX. J. Virol. 85, 9222–9226. doi: 10.1128/JVI.02653-10
Zhang, L., Ju, Y., Chen, S., and Ren, L. (2021). Recent Progress on exosomes in RNA virus infection. Viruses 13:v13020256. doi: 10.3390/v13020256
Zhang, L., Li, R., Geng, R., Wang, L., Chen, X.-X., Qiao, S., et al. (2022). Tumor susceptibility gene 101 (TSG101) contributes to Virion formation of porcine reproductive and respiratory syndrome virus via interaction with the Nucleocapsid (N) protein along with the early secretory pathway. J. Virol. 96:e0000522. doi: 10.1128/jvi.00005-22
Zheng, G., Li, L. F., Zhang, Y., Qu, L., Wang, W., Li, M., et al. (2020). MERTK is a host factor that promotes classical swine fever virus entry and antagonizes innate immune response in PK-15 cells. Emerg. Microbes Infect. 9, 571–581. doi: 10.1080/22221751.2020.1738278
Zheng, Y., Wang, M., Li, S., Bu, Y., Xu, Z., Zhu, G., et al. (2023). Hepatitis B virus hijacks TSG101 to facilitate egress via multiple vesicle bodies. PLoS Pathog. 19:e1011382. doi: 10.1371/journal.ppat.1011382
Keywords: ESCRT, MVBS, enveloped virus, viral entry, viral budding, viral replication
Citation: Wang C, Chen Y, Hu S and Liu X (2023) Insights into the function of ESCRT and its role in enveloped virus infection. Front. Microbiol. 14:1261651. doi: 10.3389/fmicb.2023.1261651
Received: 19 July 2023; Accepted: 20 September 2023;
Published: 06 October 2023.
Edited by:
Kohji Moriishi, University of Yamanashi, JapanReviewed by:
Eiji Morita, Hirosaki University, JapanCopyright © 2023 Wang, Chen, Hu and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiufan Liu, eGZsaXVAeXp1LmVkdS5jbg==; Shunlin Hu, c2xodUB5enUuZWR1LmNu
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.