
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 09 October 2023
Sec. Microbiological Chemistry and Geomicrobiology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1260585
Shrimp rearing generate organic waste that is trapped in the pond sediment. In excess, these wastes may impair aquaculture ecosystem and shrimps’ health. To promote the biological oxidation of accumulated organic waste, the pond is drained and dried at the end of each production cycle. However, this practice is not always conducive to maintaining microbial decomposition activities in sediments. Shrimp production in New Caledonia is no exception to this problem of pollution of pond bottoms. One promising way of treating this waste would be bioremediation, using a native halophyte plant and its microbiota. Thus, this study explored the nutrient removal potential of Suaeda australis and its microbiota on sediments from four shrimp farms. Suaeda australis was grown in an experimental greenhouse for 6 months. In order to mimic the drying out of the sediments, pots containing only sediments were left to dry in the open air without halophytes. An analysis of the chemical composition and active microbiota was carried out initially and after 6 months in the sediments of the halophyte cultures and in the dry sediments for each farm, respectively. In the initial state, the chemical parameters and the microbial diversity of the sediment varied considerably from one farm to another. Growing Suaeda australis reduced the nitrogen, phosphorus and sulfur content in all type of sediment. However, this reduction varied significantly from one sediment to another. The rhizosphere of Suaeda australis is mainly composed of micro-organisms belonging to the Alphaproteobacteria class. However, the families recruited from this class vary depending on the farm in question. Depending on the sediment, the variation in microbiota leads to different putative biochemical functions. For two of the farms, a similar reduction in nitrogen concentration was observed in both dry and cultivated sediments. This suggests that certain initial chemical characteristics of the sediments influence the nutrient removal efficiency of Suaeda australis. Our study therefore highlights the need to control the pH of sediments before cultivation or in dry sediments in order to ensure optimal microbial decomposition of organic waste and nutrient cycling.
In New Caledonia, the Pacific blue shrimp Penaeus stylirostris is farmed mainly on a semi-intensive basis. On this South Pacific island, the shrimps are farmed in earthen basins of between 3 and 12 hectares dug directly into the saltpans (Goarant et al., 2004; Della Patrona and Brun, 2009). During the rearing, the uneaten feed but also feces, shrimp’s exoskeleton, and dead phytoplankton tend to accumulate at the bottom of the pond (Boyd, 1995; Páez-Osuna et al., 1997; Funge-Smith and Briggs, 1998; Avnimelech and Ritvo, 2003). In fact, only 46.7% of the nitrogen and 7.4% of the phosphorus in the pellets are assimilated by shrimp in semi-intensive farming (Páez-Osuna et al., 1997), leading to the accumulation of huge quantities of organic waste in the pond sediments. Excessive accumulation of organic waste in the sediment during rearing leads to eutrophication of the pond ecosystem, which can encourage the development of diseases in shrimp, resulting in high mortality (Funge-Smith and Briggs, 1998). Indeed, the anaerobic decomposition of the accumulated organic matter leads to the release of hydrogen sulfide (H2S) and ammonia (NH4+), which are toxic for the pond ecosystem. In New Caledonia, at the end of each production cycle, the ponds are usually completely drained to discard the polluted water and then dried in the sun for several months (Boyd, 1995; Yang et al., 2017). The aim of this drying period is to accelerate the aerobic microbial decomposition of the organic waste accumulated in the sediment (Boyd and Pippopinyo, 1994). Thus, this preparation and regeneration of the aquaculture pond between two rearing cycles is therefore a vital step to ensure the success of the next rearing (Della Patrona and Brun, 2009). However, excessive pond drying period may on the contrary limit the microbial decomposition as water stress is reported to narrow the microbial activities (Schimel, 2018). Otherwise, it was reported that for an optimal organic matter degradation, the optimum range of shrimp pond sediment moisture content should be maintained between 10 and 20% and the pH around 7.5–8 (Boyd and Pippopinyo, 1994; Della Patrona and Brun, 2009). As a result, the drying period used by shrimp farmers may not always be effective if the physical and chemical parameters that encourage optimal microbial life and activity are not taken into account. In New Caledonia, ponds can dry out for up to 6 months, which represents a considerable amount of time without economic production for farmers. This can be explained by the seasonal nature of shrimp farming, which means that some farmers only have one production cycle a year (Goarant et al., 2004). However, since few years, New Caledonia shrimp farming face important production decrease, from a peak at 2500 t in 2004 to less than 1,500 t nowadays leading to a negative economic impact (FAO data base “Fisheries and Aquaculture”). It is therefore necessary to find new ways of improving shrimp farming production.
In New Caledonia, salt tolerant plants called halophytes such as glasswort (Sarcocornia quinqueflora), austral seablite (Suaeda australis) and sea purslane (Sesuvium portulacastrum) grow naturally at the vicinity of the shrimp farms (e.g., on the dikes of earthen-ponds) (Colette et al., 2022). In addition, halophytic species rapidly colonize pond sediments when they are abandoned or left empty for several months (Della Patrona, personal communication). Thus, the cultivation of halophytes has recently been explored as a means of improving the quality of pond bottoms by reducing the organic waste accumulated in the sediments at the end of a shrimp rearing cycle (Colette et al., 2022, 2023). The integration of plant cultivation into aquaculture farming systems to limit eutrophication of the aquaculture ecosystem use waste product derived from aquaculture activities to produce plant biomass, and therefore reduce waste concentrations in farming system (e.g., water or sediment) (Miranda et al., 2008; Mariscal-Lagarda et al., 2014; Mariscal-Lagarda and Páez-Osuna, 2014; Yaobin et al., 2019). The nutrient removal of organic matter accumulated in sediments cannot rely solely on plant nutrition. In fact, microbial transformation is crucial for plant nutrition, because in the soil, most nutrients such as N, P and S are bound to organic molecules and are poorly bioavailable to plants (Thies and Grossman, 2006; Jacoby et al., 2017). Microbial communities play a key role in the decomposition of organic matter and the biogeochemical cycling of nutrients in ecosystems (Allison et al., 2010; Luo et al., 2017). Thus, to evaluate the nutrient removal effectiveness of shrimp sediments by halophytes, the microbiota associated with the rhizosphere of plant species must be taken into account (Li et al., 2022). In a previous study, we have demonstrated that the microbiota associated with the rhizosphere of halophytes growing in shrimp sediments varied according to halophyte species (Sarcocornia quinqueflora, Atriplex jubata, and Suaeda australis) (Colette et al., 2023). In addition, microbial guilds selected by the plants were differently involved in functions linked to the N, C and S biogeochemical cycles in the sediment. However, this previous study was conducted using only one type of shrimp pond sediment. It would therefore be interesting to assess whether the nutrient removal capacity of halophyte species also varies according to the different types of shrimp pond sediment. To do this, we chose to focus on Suaeda australis, as this species with a deep root system proved to be effective at eliminating nitrogen (Colette et al., 2022, 2023). This study therefore aimed to assess (i) the impact of Suaeda australis and its associated microbiota on both sediment chemistry and microbial communities, (ii) and whether the effect of Suaeda australis and its microbiota on the biochemical parameters of sediments varied according to the shrimp farm sediments. We also aimed to determine whether the initial characteristics (chemical composition and active microbial communities) of the sediments influenced the effectiveness of Suaeda australis and its associated rhizosphere microbiota in removing sediment nutrient. To answer these questions, we used four different shrimp farm sediments whose chemical composition we compared in the initial state and either after six months of Suaeda australis culture or after six months of drying. The active microbial communities of the sediment and halophyte rhizosphere were also explored by sequencing the cDNA of the V4 region of the 16S RNA gene (Cristescu, 2019; Wood et al., 2020).
The experimental greenhouse is located on a shrimp farm (Aigue Marine) in Boulouparis, New Caledonia, bordering Saint-Vincent Bay. The experiment extended from September (2021) to February (2022) in the same meteorological and watering conditions as in our previous study described in (Colette et al., 2023). Sediments from four different shrimp farms were collected with a medium-sized excavator at the end of the shrimp rearing, on the first days of the drying period of the ponds. In order to keep the names of the shrimp farms anonymous, we will refer to them as A, D, F, and P. The sediments collected were transported to the greenhouse and stored for a few days before being poured into 42 L pots. Two-month-old seedlings of Suaeda australis were planted on these sediments. The seedlings were obtained from germinations of seeds from mother plants grown in another experimental greenhouse in New Caledonia (Colette et al., 2023). Then, to avoid drastic change, the young seedlings were transplanted with a part of their initial growth substrate in the 42 L pots filled with sediment. To ensure minimum survival of the halophytes, 3 to 4 young seedlings per pot were planted in the 42 L pots. For each sediment from each farm (A, D, F and P), 9 pots were used for the cultivation of S. australis.
The old plants were grown for 6 months in the greenhouse and irrigated daily using an automatic sprinkler system. We chose a cultivation period of 6 months for our experimentation as it is the duration of pond sediment drying practiced by shrimp farmers in New Caledonia (Goarant et al., 2004). Twice a week, S. australis were watered with lagoon seawater used for the shrimp farm activities. For each sediment from each farm (A, D, F and P), 3 pots of 42 L were also used for a dry treatment condition. This consisted of pots containing only pond sediment, placed outside the greenhouse without watering for the 6 months of the experiment to mimic the effect of drying out the pond (dry conditions). During the drying period used by shrimp farmers, the bottom of the emptied ponds is dried in the sun (Boyd, 1995; Della Patrona and Brun, 2009). Thus, the pots were exposed to the same weather conditions as the sediments of the emptied ponds and subjected to the natural drying period.
Sampling was carried out at the beginning (D0) and at the end of the 6 months experiment. For each farm (A, D, F and P), the sediment was sampled in the 42 L pots on D0 (before the culture of S. australis) and at the end of the experiment in the dry condition (Dry) and in sediment with S. australis cultivation. For the chemical analyses, sediment of each farm (A, D, F and P), were sampled in triplicates in different 42 L pots from each modality (D0, dry, S. australis). To ensure sampling homogeneity, each sediment samples consisted of a pool of 6 samples from different pots, homogenized in a clean bucket and then stored in aluminum trays and 50 mL tubes. The collected sediment in aluminum tray was then oven-dried at 35°C for several days for analysis of the pH, total and available forms of phosphorus, total sulfur and organic carbon. The other part of the sediment collected in the 50 mL tubes were stored at −20°C for the analysis of available nitrogen forms (NO3− and NH4+).
Several publications have used cDNA metabarcoding and proved that RNA is a useful tool to identify living organisms and to perform biological survey and monitoring (Laroche et al., 2018; Amarasiri et al., 2021; Miyata et al., 2021; Veilleux et al., 2021). Indeed, the high turnover of RNA molecules in the environment (from days to weeks) compared to DNA (from months to years) reflect better the metabolically active lineages at the sampling time. Thus, in our research, we opted to extract RNA to investigate the active microbiota in the rhizosphere and sediment, minimizing the chances of detecting microorganisms that may be inactive or dead in the samples. In order to explore the active microbial diversity in the sediment, the top 2–3 cm of sediment from the 42 L pots were also collected aseptically using RNA/DNA free gloves and spatula. For each farm (A, D, F, and P), the sampling was carried out at the same time as sampling for chemical analyses at D0 and in the Dry and S. australis conditions. We collected sample in triplicate and one replicate consist of 3 to 4 sediment samples from the same pot. Then, sampled sediment was transferred into RNA/DNA free 15 mL tubes. The collecting sediment were stored at 4°C during transport to the laboratory and then frozen at - 80°C until further processing.
The Laboratory of Analytics Means (LAMA/ISO 9001, Noumea, New Caledonia) performed the analyses of cation exchange capacity (CEC), calcium carbonate (CaCO3), pH, organic carbon (C.org), nitrate (NO3−), ammonium (NH4+), total and available forms of phosphorus and total sulfur. The CEC was measured by the cobaltihexamine chloride methods (ISO 23470) and the CaCO3 with a Bernard calcimeter. Sediment pH was measured with pH electrod SCHOTT Blue Line in soil/distilled water ratio of 1:2.5. The nitrate (NO3−) and ammoniums (NH4+) were extracted from the sediment with KCl solution at 1 N. The nitrate and ammonium concentrations were evaluated by colorimetric method based on the Griess reaction (ISO 14256-2:2005) and Nessler method (ISO 14256-2:2005) respectively. Total organic carbon was determined using the Walkley and Black method (Pétard, 1993). Total phosphorus was measured by Murphey and Riley method (Murphy and Riley, 1962) whereas available phosphorus forms by Olsen method (Olsen, 1954). Total sulfur was determined after alkaline fusion by ICP-OES. A non-parametric test of Kruskal-Wallis followed by a Dunn’s test were performed with R software to show statistically significant differences of sediment chemistry between the experimental conditions.
For each sediment sample, RNA was extracted using RNA PowerSoil Total RNA Isolation Kit (MoBio Laboratories, Inc.) and then reverse-transcripted into complementary DNA (cDNA) as described in our previous study (Colette et al., 2023) using Second Strand cDNA Synthesis Kit (Invitrogen). All cDNAs were sent to MrDNA (Shallowater, Texas, United States) where PCR using the 515f-806R primers couple (Caporaso et al., 2011), barcode indexing and sequencing of the V4 hypervariable region of the reverse-transcripted procaryotic 16S rRNA molecule were carried out. The sequencing was done with an average of 20 k raw reads per sample.
The amplicon analysis was performed with DADA2 version 1.6 package1 on R software as described in Colette et al. (2023). The chimeras were removed using the consensus method, and the taxonomy was assigned using the Silva 138 SSU Ref NR99 database (Quast et al., 2012). Sequences with no affiliation or affiliated to the Eukaryota, Mitochondria or Chloroplasts were removed from the ASV table, prior to further analysis.
The alpha diversity of each sediment sample was calculated on R software with the microeco (v0.20.0) package (Liu et al., 2021). Then, data were normalized with the Counts Per Million (CPM) as described in Callac et al. (2022, 2023). The beta diversity was investigated by a PCoA (principal coordinate analysis) plot using the Bray distance with the microeco package on R software. Then, a permutation test (PERMANOVA) was performed to highlight Bray distances significantly different between farms (A, D, F and P) and modalities (D0, S. australis and Dry). Venn diagrams were then made to exhibit both shared and specific ASVs between the farm sediments for each of the three different modalities (D0, dry and S. australis). The Venn diagrams were built using the open-source component for the web environment Jvenn2. For each modality, stacked bar charts of the relative abundance of microbial communities were made to display the composition of the sediment microbial communities according to the farms sampled. Stacked bar plot were performed on R software with microeco and ggplot (v3.4.2) packages. For the S. australis modality, we performed a Functional Annotation of the Prokaryotic Taxa to predict the putative functions of the specific sedimentary microbial communities on each farm. The functional annotation was based on FAPROTAX v1.2.4 database and was done on R software with microeco package.
At the beginning of the experiment (D0), the chemical parameters of the sediments varied from farm to farm (Table 1). With the exception of sediment from farm D, which had a pH of 7.1, the pH of farms A, F and P was around 8 (Table 1). Sediment CEC values ranged from 23.7 to 30.4, with the highest value observed on farm D.
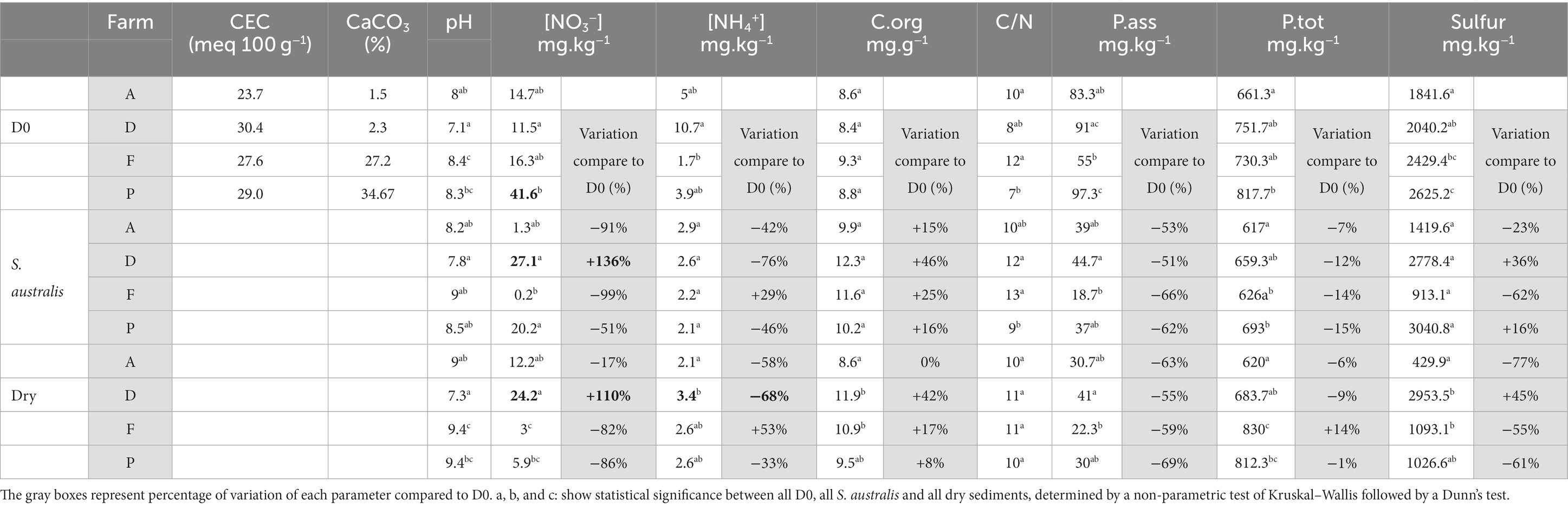
Table 1. Average of sediment chemical parameters across four shrimp farms named A, D, F, and P, at the beginning of the experiment (D0) and after 150 days of S. australis cultivation or in dry sediment.
Farms A and D both had a lower percentage of calcium carbonate (CaCO3) in their sediments (1.5 to 2.3%) than farms F and P (27.2 to 34.67%). The C/N ratio of farm sediments ranged from 7 to 12 (Table 1). Sediment organic carbon concentration did not vary significantly between farms, unlike NO3−, NH4+, phosphorus and sulfur concentrations. Sediments from farm P had significantly higher concentrations of sulfur, total and available phosphorus, and NO3− (Table 1). Sediments from farms A and D had significantly lower sulfur concentrations than those from farms F and P.
After 150 days of S. australis cultivation, sediment pH was still significantly lower in farm D than in the other farms (Table 1). For farms A, F and P, S. australis cultivation reduced the NO3− concentrations by 50 to 99% compared with D0. For farm D only, S. australis cultivation increased sediment NO3− concentration by 136% (Table 1). For all farms, S. australis cultivation increased the concentration of organic carbon in sediment compared to D0 and decreased the concentrations of phosphorus (total and available forms). Farm D showed the greatest increase in organic carbon (+ 46%) compared with the others farms (+ 15 to 25%). The greatest reduction in phosphorus under S. australis cultivation was found in the sediments of the farms F and P (Table 1). In terms of sulfur concentration, S. australis cultivation increased concentrations on farms D (+36%) and P (+16%) whereas it decreased on farms A (−23%) and F (−62%).
Sediment drying resulted in a pH increase of one unit compared with D0 in all farms except farm D (Table 1). Sediment pH values were around 9 on farms A, F, and P, and 7.3 on farm D (Table 1). NO3− concentration was reduced by 80% in farms F and P and by 17% in farm A. Conversely, NO3− and total sulfur concentration in dried sediments from farm D increased by 110 and 45%, respectively, compared to D0 (Table 1). The dried sediments from farm D showed higher NO3− and total sulfur concentrations than those from the other farms, which were not detected at D0 (Table 1). For all farms, sediment drying reduced the concentration of available forms of phosphorus compared to D0, within a range quite similar to that of the S. australis cultivation (Table 1). Sediment organic carbon increased in dry sediments from farms D, F, and P compared to D0, but the increase was smaller than under S. australis cultivation.
On D0, farms F and P had higher values of richness indices (Chao1 and observed ASV) and evenness indices of Shannon compared to the farms A and D (Table 2). After 150 days of S. australis cultivation, the average of all alpha diversity indices decreased in farms F and P compared with D0, while they increased in farm A and F (Table 2). The highest values of alpha diversity indices (Chao1, Observed ASV, Shannon and Simpson) with S. australis cultivation were found in farm A sediment. For all farms, average of richness indices decreased in the dry sediment compared with D0. In addition, for all farms, the richness indices values in dry sediments were lower than in sediment with S. australis cultivation (Table 2). In the dry sediments, the highest of Chao1 and Observed ASV values were found in farm P.
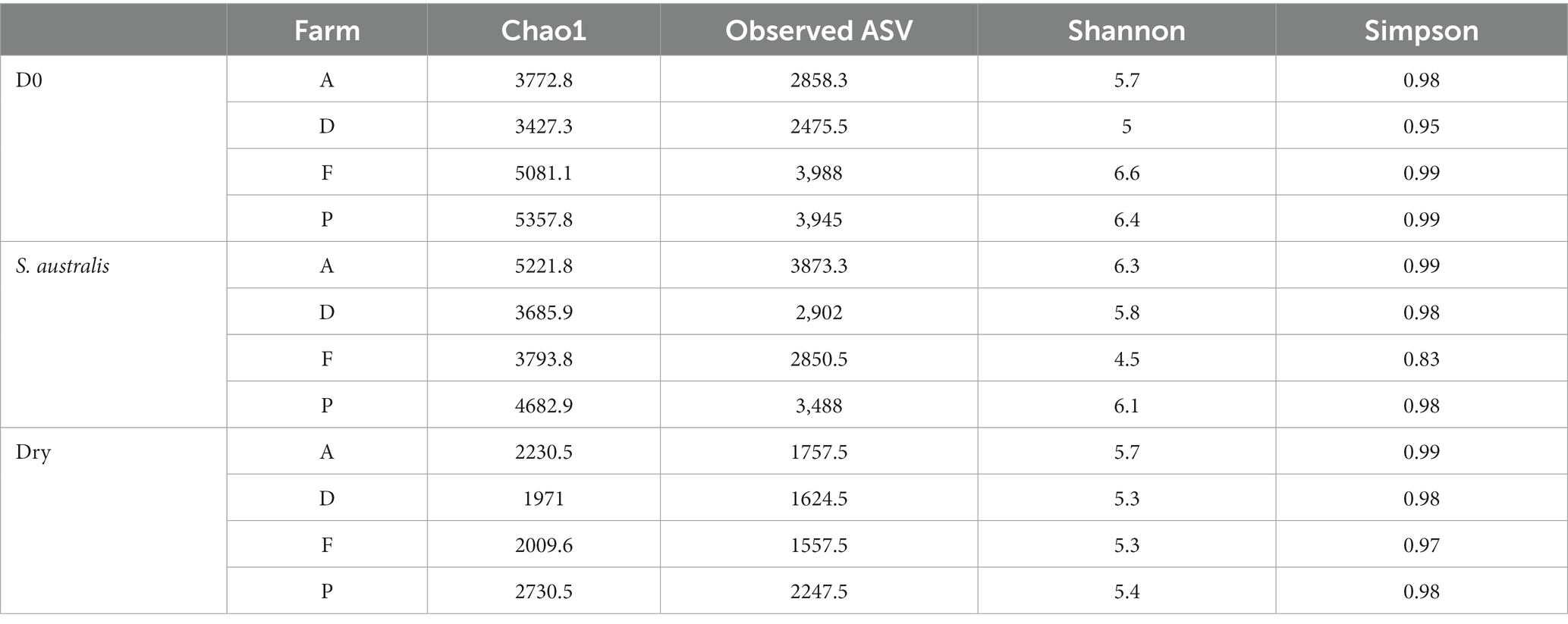
Table 2. Comparison of average alpha diversity indices of richness (Chao1, observed ASV) and evenness (Shannon and Simpson) between the different shrimp farms sediment at the beginning of the experiment (D0) and after 150 days of S. australis cultivation or in dry sediment.
Our dataset was composed of 20,467 ASVs. The PCoA diagram displayed that the sediment microbiota differed between D0, dry and S. australis, as the points representing these different modalities are clearly separated in the graph (Figure 1). In addition, a differential test of distances among groups performed with a PERMANOVA evidenced that those three modalities were statistically different with a value of p at 0.001 (Table 3).
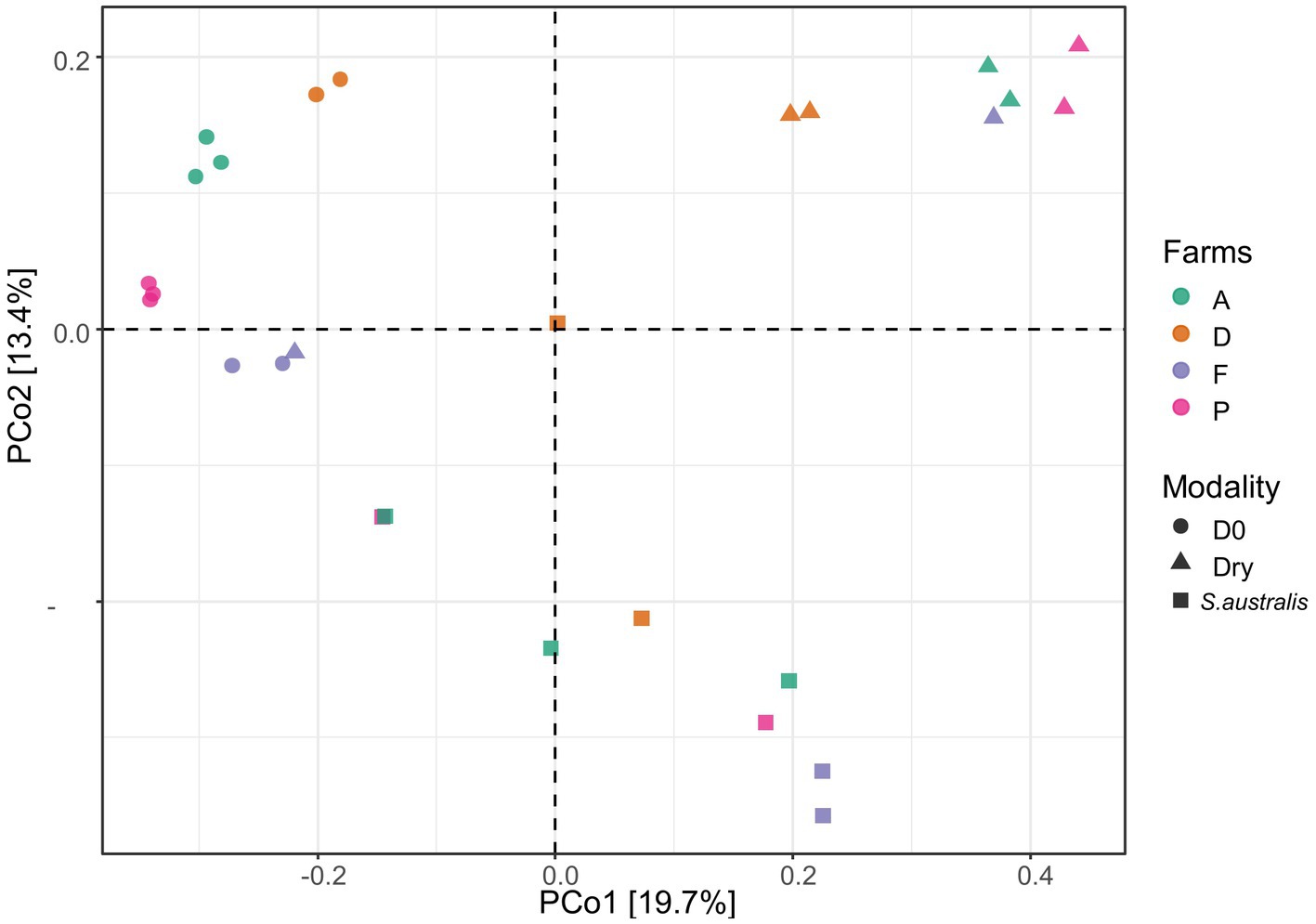
Figure 1. Principle coordinate analysis (PCoA) plot, of the microbial diversity in the sediments from the farms A, D, F, and P following the different modalities (D0, dry, S. australis), made using the Bray-Curtis distance.
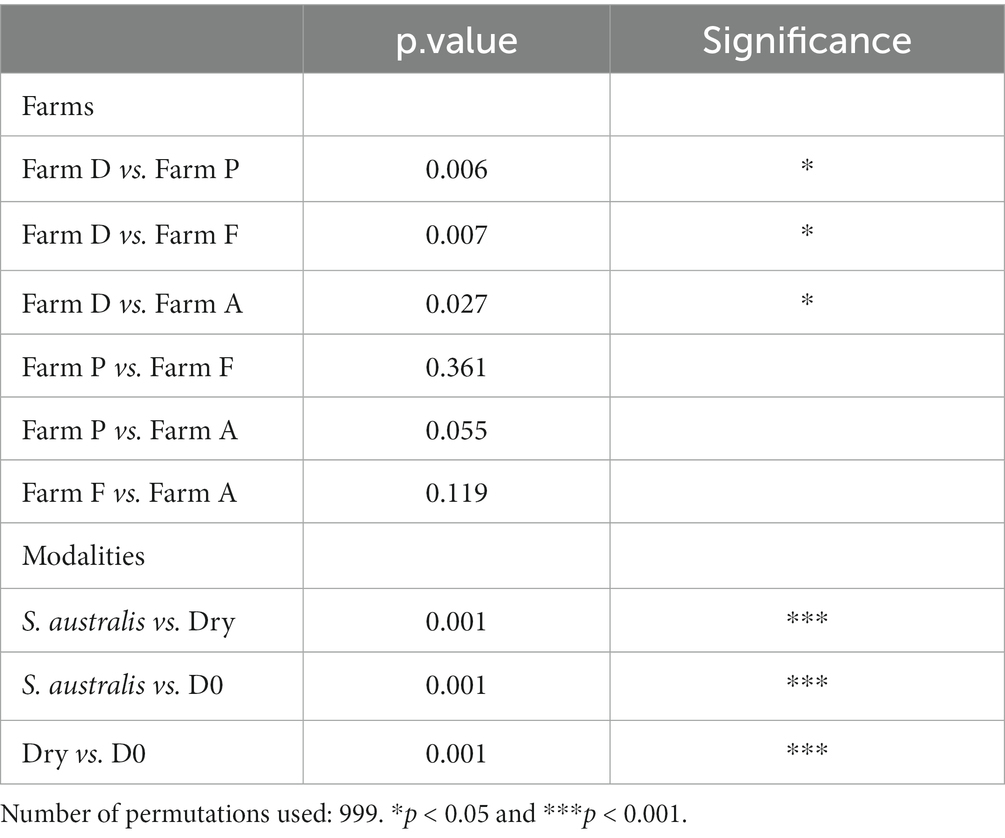
Table 3. Results of the permutation test (PERMANOVA) based on Bray–Curtis measure for pairwise comparison of farms (A, D, F, and P) and modalities (D0, S. australis and Dry).
All modalities confounded, the PERMANOVA also evidenced that farm D sediment microbiota was significantly different (value of p < 0.05) from farms A, F, and P (Table 3). This can be evidenced in the plot for the modalities D0 where samples from farm D were distant from farms F and P (Figure 1). Furthermore, when considering the dry modality in the PCoA diagram, sediment samples from farm D were isolated from other farms.
For the S. australis modality, sediment samples were randomly distributed across the graph (Figure 1). For this modality, there was more distance between replicates of sediment samples, highlighting a greater variability of microbiota within sediments from the same farm. For farm F only, replicates of sediment microbiota samples with the S. australis modality were close to each other (Figure 1).
Venn diagram showed that at the beginning of the experiment (D0), microbiota of the four farms shared 404 ASVs (Figure 2A). At D0, the farms F and P shared 504 ASVs and fewer ASVs with farm A (67 and 98 ASVs respectively), while farms A and D together shared 280 ASVs.
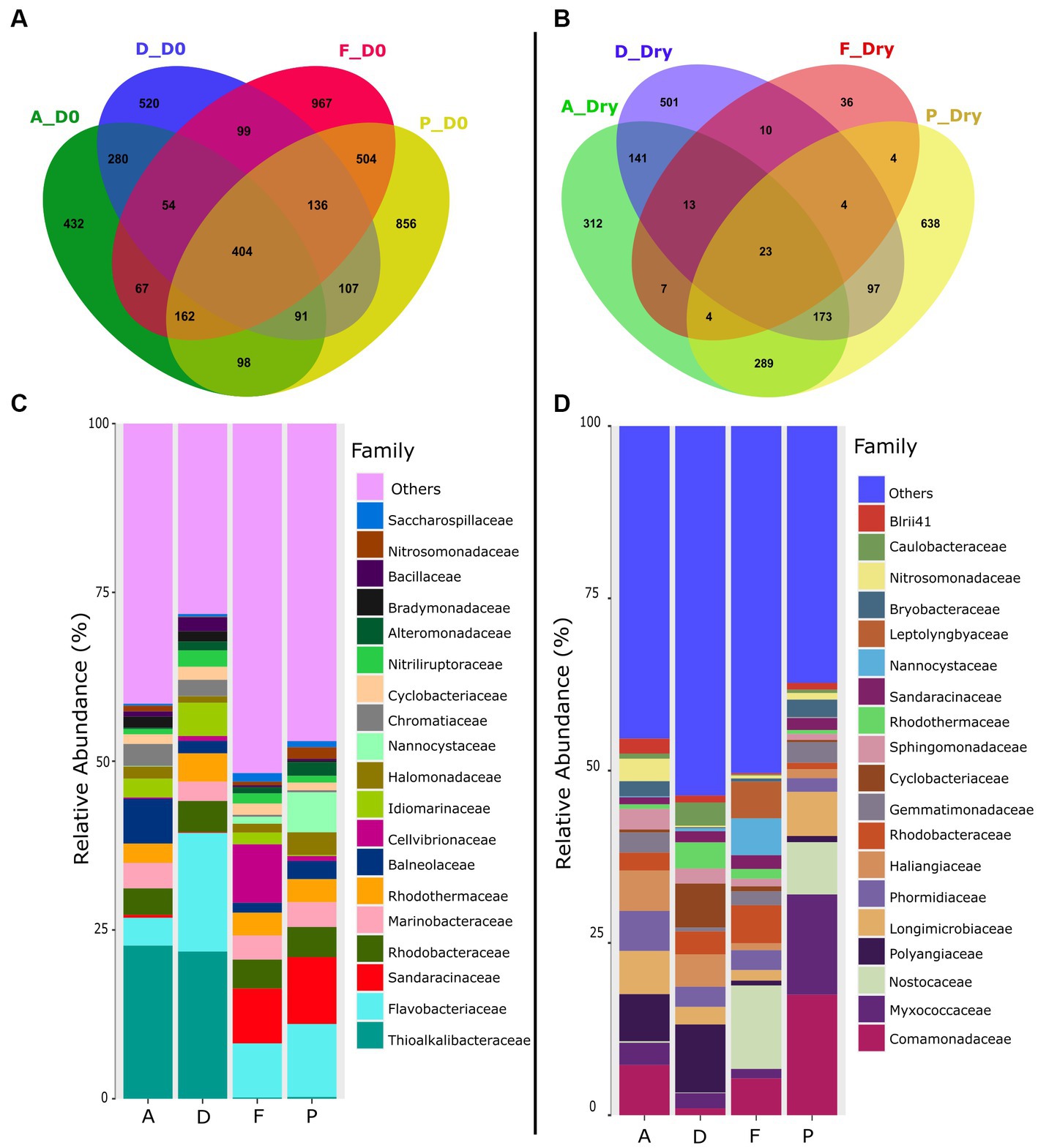
Figure 2. Venn diagrams of shared and specific ASVs in the sediment of farms A, D, F and P, in (A) the sediment at the beginning of the experiment (D0), (B) in the dry sediment. The relative abundance of the specific ASVs at the bacterial family level found in each farms sediments was represented in (C) at the beginning of the experiment and in (D) dry sediment.
On D0, Thioalkalibacteraceae was the main bacterial family found in the sediments of the farms A and D with a relative abundance of about 22% whereas this family accounted for less than 0.27% in the farms F and P (Figure 2C and Supplementary Table S1). The Chromatiaceae family was also mainly found in the farms A and D sediments with relative abundance around 3% whereas it was less than 0.3% in the other farms. Furthermore, on D0, microbiota of the farms F and P was composed of Sandaracinaceae (between 8 and 9%) whereas this family was less than 0.5% in the farms A and D.
For the four farms on D0, the active microbiota was composed of members of the Flavobacteriaceae, Marinobacteraecae, Rhodobactacteraceae, and Rhodothermaceae families. The microbiota of the farm F was also composed of Cellvibrionaceae (8.65%) which was represented less than 1% of the microbial abundance in the sediments of the other farms (Figure 2C and Supplementary Table S1). The microbiota of the farm P was also composed by Nannocystaceae (6%), which encompassed less than 1% in the other farms.
In dry conditions, however, the Venn diagram showed that only 23 ASVs were shared between the four farms (Figure 2B). In the dry sediment, among the 19 main families identified, only 5 were present at D0 (Nannocystaceae, Sandaracinaceae, Rhodotermaceae, Cyclobacteraceae, Rhodobacteraceae, Figure 2D). Polyangiaceae and Haliangiaceae families were present in the farm A (6.8%) and D (9.9%) but accounted for less than 1% in the farms F and P (Figure 2D and Supplementary Table S2). The Nostocaceae family was present in sediments of the farms F and P; but was absent in farms A and D. Comamonadaceae family was found in sediments of the farms A (7%), farms F (5%) and farm P (17%) but represent only 1% in farm D. The Nitrosomonadaceae family was found in farm A with a relative abundance of 3% but was less than 1% in other farms. The dry sediment of the farm P was enriched in Myxococcaceae (14.5%) compared to others farms (less than 3%). Farm F was composed of Leptolyngyaceae (5%) and Nannocystaceae (5%) whereas the relative abundance of those families were less than 0.4% in the dry sediments (Figure 2D and Supplementary Table S2).
At the family level, the specific microbiota was varying between the farms (A, D, F and P) (Figures 3A,B). In the farm A, specific microbiota was composed of Balneolaceae (10%), Micromonosporaceae (5%) and Bradymonadaceae (3%). Specific microbiota of farm D was mainly composed of Terasakiellaceae (36%) family which was absent in the other farms (Figure 3B and Supplementary Table S3). In the farm F, the specific microbiota contained members Oscillatoriaceae family (7%) which was totally absent in the other sediments, and Sphingomonadaceae (3%) which was less than 1% t in the other farms. The specific microbiota of the farm F was also composed by lineages related to the Haliangiaceae (6%) and A4b (Fermentative organism found inside of anammox granules) (5%) which accounted for less than 2% in the specific microbiota of the others farms. The specific microbiota of the farm P was composed of Nodosilineaceae (8%) in greater proportion than in other farms, and of Nannocystaceae (5%) which was also present in the farm F (4%) and in lesser proportion (< 1.5%) in the farms A and D. The specific microbiota of the farm P also included the Geothermobacteraceae family (6%), which was absent from the other farms.
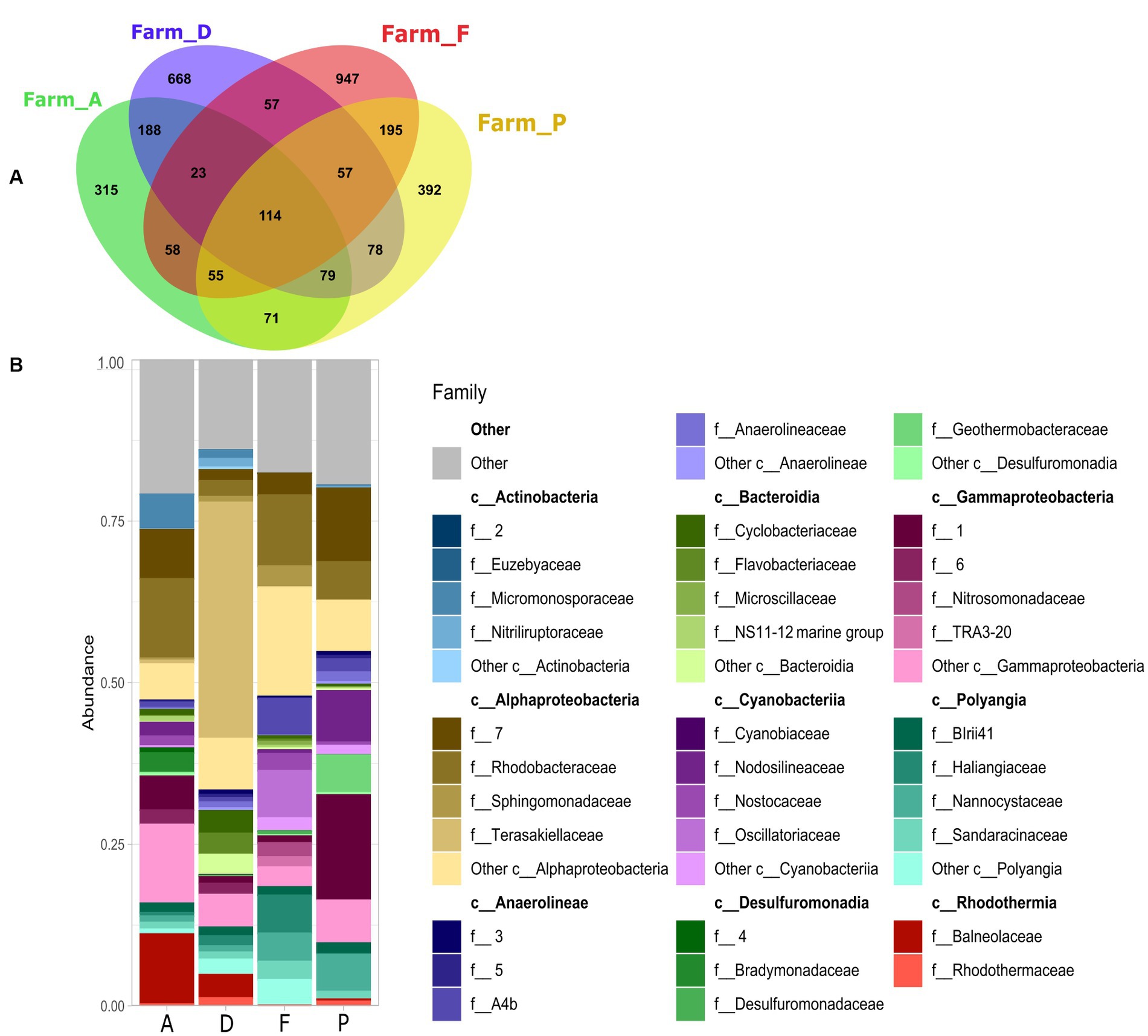
Figure 3. (A) Venn diagram of shared and specific ASVs in the sediments with S. australis from the farms A, D, F, and P. (B) Stacked bar plot represent relative abundance of the 10 main bacterial classes and the 5 main families per class, of the specific ASVs found in each farm.
Rhodobacteraceae was found in the specific microbiota of all farms, with relative abundance of 3% in the farm D, of approximately 12% in the farms A and F and about 6% in the farm P (Figure 3B and Supplementary Table S3).
In the farm D, the putative functions of the specific microbiota of the sediment with S. australis were significantly and positively correlated with the sulfur cycle (sulfide and sulfur oxidation), ammonia oxidation and aromatics degradation (Figure 4). For farm A sediment microbiota, correlations were significantly positive with hydrocarbon degradation and methanol oxidation.
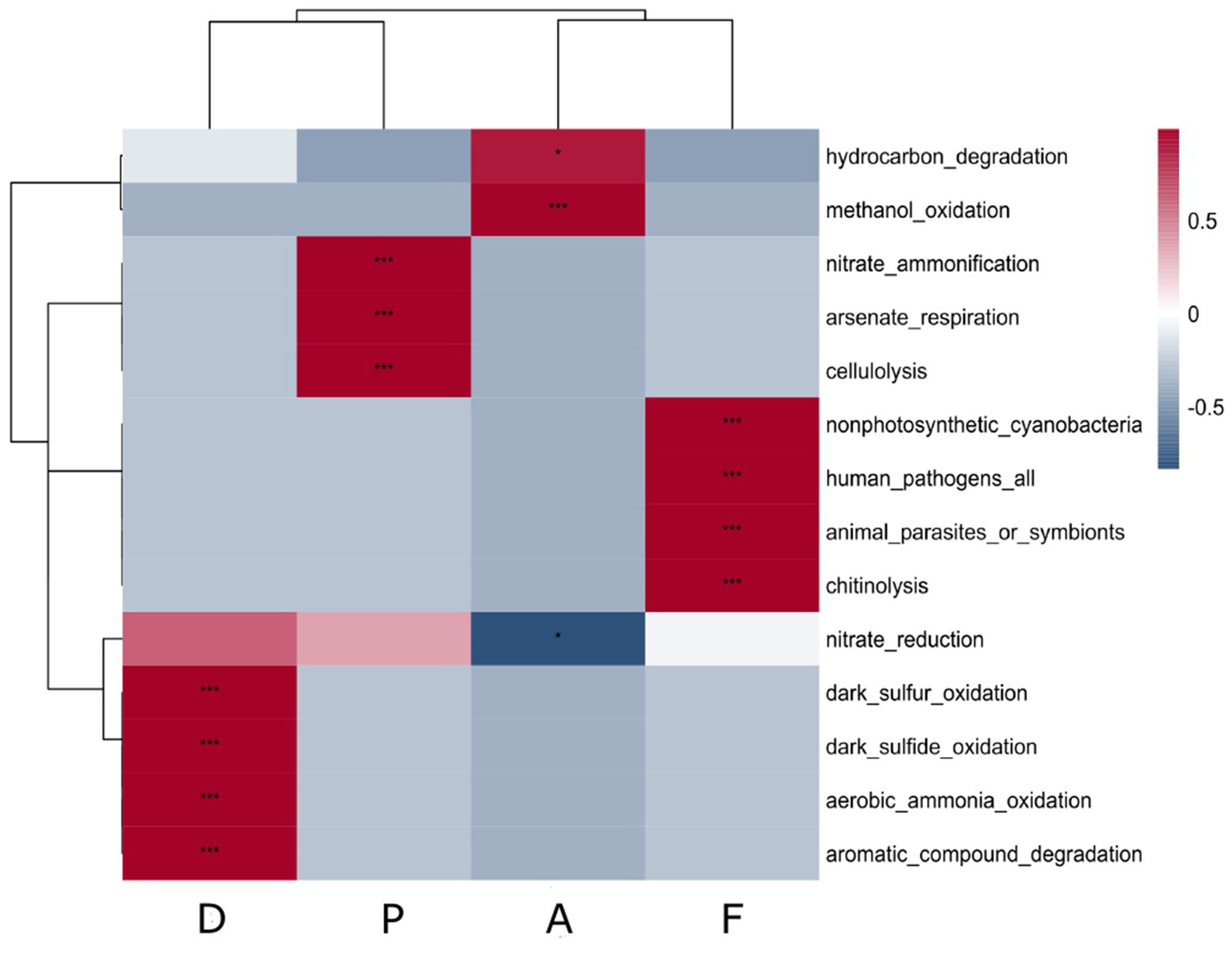
Figure 4. Correlation heatmap of putative ecological function associated with the specific microbiota of S. australis according to the farm sediment. Heatmap color gradient is linked to Pearson correlation coefficient intensity with in red the positive correlation and in blue the negative correlation. Significant correlations between ecological function and farms are indicated by an asterisk (*).
In the farm P, the putative functions were significantly positively correlated with nitrate ammonification, arsenate respiration and cellulolysis. Other lysis functions, such as chitinolysis, were also found as putative functions in farm F (Figure 4). However, in the sediment microbiota from Farm F, significant positive correlations were also found.
We could observe that at D0 and for all the farms, the Gammaproteobacteria was the main class that dominated the sediment microbiota (39%) followed by the Bacteroidia (15%) (Figure 5A). However, in the dry conditions (Figure 5B), the abundance of those two bacterial classes decreased by a factor two. In the dry conditions the relative abundance of the Polyangia increased compared to D0 (8% compared to 15%) and the Cyanobacteria rise in great proportion (14%) (Figure 5B). The Myxococcia and Anaerolineae found in the dry sediment were absent at D0. In the rhizosphere of S. australis, the relative abundance of Alphaproteobacteria had significantly increased compared to the others conditions and constituted the main bacterial class of the rhizosphere microbiota (32%) (Figure 5C). Cyanobacteria, Polyangia, Bacteroidia and Myxococcia were also present in the S. australis rhizosphere but in a lesser proportion than in the dry sediment.
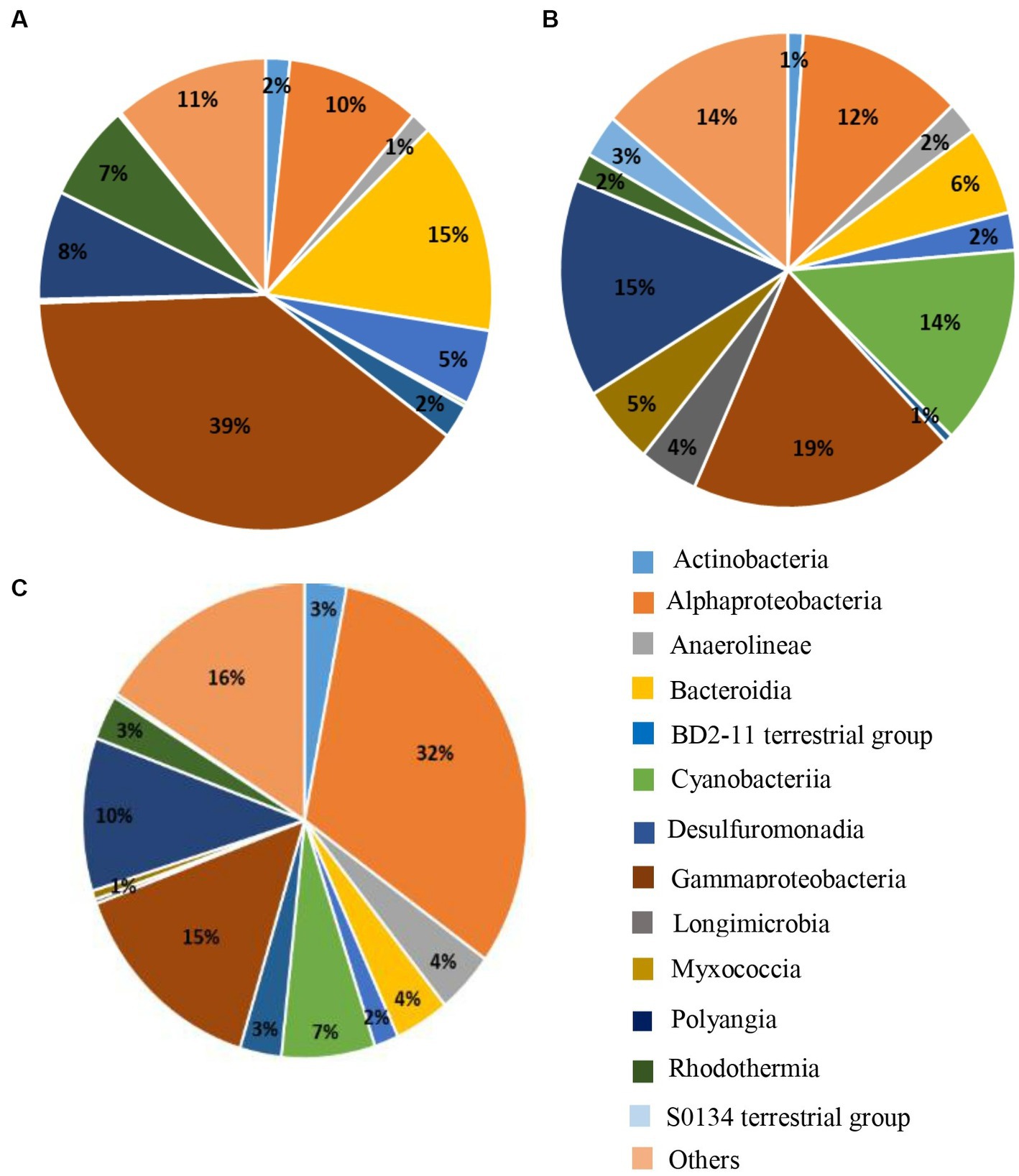
Figure 5. Pie charts of the 15 main bacterial classes representing at least 84% of the relative abundance found in the microbiota of the sediment at (A) D0, (B) in the dry sediment, and (C) in the rhizosphere of S. australis, all farms combined.
The aim of this study was to evaluate the impact of Suaeda australis and its associated active microbiota on sediment chemistry and microbial communities depending on the sediment composition of four shrimp farms (A, D, F, and P). For that we have compared the change of sediment chemistry and active microbial communities from each farm at D0 and in the presence or not of halophyte cultivation. In the absence of halophytes, the sediment was left drying outside the greenhouse along with the experiment to mimic the pond drying periods practiced by shrimp farmers. By comparing these two conditions (with and without halophytes), we were able to highlight the changes caused by plants and their rhizosphere microbiota, and how these vary between the sediments of shrimp farms.
At the beginning of the experiment (D0), chemical parameters of sediment varied between the farms (Table 1). These variations can be easily explained primarily by differences in aquaculture practices on the farms during the rearing cycles as shrimp density, feeding rate, fertilization (N, P) and liming strategy (CaCO3). These parameters can strongly influence the concentration of nutrients in the pond sediment, such as nitrogen, carbon, phosphorus and sulfur (Boyd, 1995; Avnimelech and Ritvo, 2003). The C:N ratio found in sediments at the beginning of the experiment was varying between 7 and 12, and these values were within the range of those reported in others studies of aquaculture pond soils (Munsiri et al., 1995; Smith, 1996). These C:N ratio values were recorded in the case of rapid biological mineralization of the soil, which means that overall, in our study, the organic matter (feed pellets, feces) in the sediments at D0 must have been easily decomposed by the microorganisms (Brust, 2019). In our study, the farms F and P were chemically distinct from farms A and D with higher concentration of CaCO3 in their sediments at the end of the rearing (Table 1). Farms F and P also showed the highest values of alpha diversity richness indices (Chao1, observed ASV) on D0, compared to farms A and D (Table 2). It would be therefore interesting to study the effect of liming on the microbial communities in shrimp pond sediments to explore if this is a factor that could have directly influenced the differences in alpha diversity indices between the studied farms. In fact, in terrestrial soil, previous studies have reported that a proper application of CaCO3 have enhanced the microbial alpha diversity indices (Shannon, Simpson) in acid paddy-field (Guo et al., 2019) and microbial richness indices (Observed ASV, ACE) in sugarcane field (Pang et al., 2019). In shrimp ponds, the main source of sediment calcium carbonate comes from liming which is a common practice to improve the pH and alkalinity of aquaculture ponds (Boyd, 1995; Zhou et al., 2019). It was reported that sediment pH has a pronounced effect on microbial organic matter decomposition. Indeed, Li et al. (2015), reported that below a pH value of 7.5, liming of shrimp pond sediments is necessary to increase the microbial respiration rate and therefore organic decomposition during the drying period. Thus, from the start of the experiment, the pH values found in the sediments of farm D may not be conducive to microbial decomposition of organic matter, unlike the other farms (Table 1).
At the beginning of the experiment, the four shrimp farms sediment were all characterized by the occurrence of Marinobacteraecae, Rhodobactacteraceae, and Rhodothermaceae. Flavobacteriaceae also occurred in all the shrimp farm sediment but with a higher relative abundance in farms D and F than in farms P and A. Flavobacteria family was commonly found in freshwater, marine and terrestrial environments (Rosenberg, 2014). They are chemoorganotrophic bacteria found associated with algae cells, fish or also organic detritus. The Rhodobacteraceae, Flavobacteraceae, Marinobacteraceae families were previously found as dominant taxa families in the shrimp larval rearing water of New Caledonia (Callac et al., 2022, 2023) suggesting that these taxa were usually detected in shrimp rearing. In addition, Rhodobacteraceae and Flavobacteraceae were also found, respectively, as microbial biomarker of shrimp gut and pond sediment samples (Zhou et al., 2021) suggesting that those taxa were commonly associated with shrimp farming ecosystems.
Farms A and D were characterized by the dominance of Thioalkalibacteraceae in their sediment whereas this family was absent in farms F and P (Figure 2A). This family is known to use thiosulfate, elementary sulfur and sulfide as electrons donors (Boden, 2017). The sediment microbiota of those two farms were also composed of Chromatiaceae family which was also reported to be involved in sulfide and elemental sulfur oxidation (Imhoff, 2014). The sediments from these two farms had the lowest total sulfur concentration compared with farms F and P, suggesting a potentially important role for Thioalkalibacteraceae in the elimination of sulfur from these sediments.
Farms F and P sediment were mainly characterized by the occurrence of Sandaracinaceae family from the Myxobacteria phylum. Members of this bacterial phylum are reported to have the ability to lyse living cells of other microorganisms by lytic enzyme and to be cellulose-decomposers (Mohr et al., 2012; Mohr, 2018). The Nannocystaceae was another bacterial family belonging to Myxobacteria phylum and was particularly found in farm P sediment but was absent in farms A and D. This family was reported to be capable of degrading complex macromolecules and lysing microorganisms (Garcia and Müller, 2014). Because of their lytic capacity, Myxobacteria are considered as an important micro-predators playing a key role in structuring and regulating soil microbial communities (Shimkets et al., 2006; Phillips et al., 2022). The presence of this taxon was strongly influenced by bacterial communities and was previously reported to be significantly positively correlated to alpha diversity indices of Chao1 and Observed OTU (Dai et al., 2021) and of ACE, Shannon and Simpson (Wang et al., 2020). Thus, in our study the higher alpha diversity indices (observed ASV, Chao1 and Shannon) may have promoted the occurrence of the Myxobacteria in farms P and F by given them a broader choice of potential prey microorganisms. The microbiota of the farm F had the particularity to be composed of Cellvibrionaceae family whereas this family was nearly absent from the others farms. A widespread trait among strains of this family was reported to have the ability to use complex polysaccharides as substrates (Spring et al., 2015). This family was identified as highly productive of carbohydrate-active enzymes that can be involved in lignocellulose degradation (Leadbeater et al., 2021). Thus, a characteristic of the microbiota of the farms F and P at the beginning of the experiment was the occurrence of bacteria with lytic and complex organic matter degradation capabilities.
After the 150 days of experimentation in pots, the sediment microbiota has evolved differently from D0 regarding the dry or S. australis modalities (Figure 1 and Table 3). Drying had significantly decreased the alpha diversity indices in the sediment of each farm compared to D0 (Table 2). Regarding the drying effect on the microbial diversity, it had been widely reported that prolonged drought has a significant impact on the abundance, structure and activities of the soil microbiome (Bogati and Walczak, 2022). In the soil, water is a solvent and a transport medium of microbial substrates so it has a direct influence on the ability of bacteria to acquire soil substrates. In addition, soil water can also directly influences the physiological state of bacteria (Chen et al., 2007; Schimel, 2018). Myxobacteria was a phylum highly represented in the dry sediments with 5 families: Nannocystaceae, Myxococcaceae, Haliangicaeae, Polyangiaceae, Sandaracinaeceae, out of the 19 main families observed (Figure 2D). In dry sediments, the Myxobacteria phylum represented by Myxococcia and Polyangia classes, had higher relative abundance than in D0 (Figure 5B). Their significant occurrence in dry sediments can be explained by the fact that Myxobacteria are spore-forming micro-organisms that can become dormant under stressful environmental conditions like dryness (Dawid, 2000; Mohr, 2018). The families related to Myxobacteria was varying within farms sediment with Haliangiaceae and Polyangiaceae specifically related to farms A and D, Nannocystaceae to farm F and Myxococcaceae to farm P (Figure 2D). Several Cyanobacteria families were also found in the microbiota in the dry sediment of each farms. However, the relative abundance of the different families also varied between the farms. Thus, Nostocaceae were present in farms F and P but in lesser extent in farms A and D, the Leptolyngbyaceae were only found in the farm F and the Phormidiaceae were found in all farms. Cyanobacteria families were not evidenced at the beginning of the experiment; those bacteria are known as nitrogen-fixing bacteria. They are able to thrive in hostile environments as dryland areas and are pioneers in many nutrient-poor substrates (Hakkoum et al., 2021).
In the farms F and P, sediment drying had significantly reduced the NO3− concentration and total sulfur compared to the beginning of the experiment; while in farm D, sediment drying has considerably increased the NO3− and sulfur concentrations compared to the beginning of the experiment. For farm A the reduction of NO3− levels was much lower. It would therefore appear that sediment drying did not have the same effect on the biogeochemical cycle, depending on the farm sediments (Table 1). In dry conditions, the sediment pH of the farm D was still very different from the others farms (Table 1). Furthermore, in dry sediment, the pH difference between farm D sediment and others farms was greater than at D0. The lower pH values of farm D could therefore explain the significant differences observed in the sediment microbiota of this farm compared to the others (Table 3) as pH was also reported as a main driving factor structuring soil bacterial community (Zhalnina et al., 2015; Tripathi et al., 2018; Wang et al., 2019). The initial composition of the microbial communities may have had a key role in sediment biogeochemical cycles (C, N, S). Indeed, the farms F and P for which the sediment microbiotas were similar at D0 (Figure 2C), had the same range of NO3− and NH4+, total phosphorus and sulfur concentration in their dry sediment compared to the others farms (Table 1).
In dry sediments, farm D was distinguished from the others by a significantly lower relative abundance of Comamonadaceae, whereas this family was among the abundant in dry sediments from the other farms (Figure 2D). Specifically, members of this family related to the Acidovorax, Brachymonas, Comamonas, Hydrogenophaga genera, were reported to be able to denitrify and use nitrates (Willems, 2014). In several studies, the denitrifying bacteria found were mainly members of the Comamonadaceae family (Khan et al., 2002; Wang and Chu, 2016; Chu and Wang, 2017). Thus, occurrence of these family may have favored the reduction of nitrogen level in farms A, F and P compared to the beginning of the experiment. In farm A, we also found Nitrosomonadaceae, which are ammonia oxidizers, with some genera able to oxidize ammonia to nitrite and other genera to oxidizing nitrite to nitrate (Prosser et al., 2014). Thus, the occurrence of this family may explain the higher NO3− concentration in the dry sediment of the farm A compared to farms F and P. However, this family was nearly absent in the dry sediment of the farms D, F, and P (Figure 2C). It may also be possible that some soil biogeochemical processes of denitrification, nitrification, sulfur oxidation and reduction may have occurred largely before our sampling time as we used RNA, which has a shorter lifespan and higher turn-over in the environment than the DNA molecule. Thus, some prokaryotes may not be identified because they were no more “active” in the dry sediment after 6 months (Cristescu, 2019; Wood et al., 2020). Indeed, we also did not evidenced prokaryotes linked to sulfur cycle in dry sediment while sulfur level was reduced in farms A, F and P. After 6 months of drying and based on the sediment microbiota solely, we also could not explain the increase of sulfur and NO3− concentrations in farm D sediment.
The growth of Suaeda australis in sediment had two opposite effects on alpha diversity according to the farms. In farms A and D, the alpha diversity index had increased compared to D0 whereas it decreased in farms F and P while they had the highest alpha diversity indices at the start of the experiment (Table 2). A previous study has also reported that cottons roots have different effect on alpha diversity of the rhizosphere bacterial community in two types of soils (Qiao et al., 2017). They showed that in nutrient-rich soils, bacterial diversity in the rhizosphere was lower, because some bulk soil microorganisms were unable to adapt to variations in chemical and physical properties of the soil and to adapt to variation of root exudates. In our study, the decrease in alpha diversity may reflect the plant’s selection of microorganisms from the bulk soil into its rhizosphere. In contrast, in sediments with lower alpha diversity (farms A and D), the release of root exudate may have enhanced the diversity of microorganisms.
The Alphaprotebacteria dominated the specific rhizosphere microbiota of Suaeda australis (Figure 5C). Bacteria of this class have the ability to interact with plants as pathogens, symbionts or non-symbionts (Pini et al., 2011). Other studies have also found that Alphaproteobacteria was the dominant class in the rhizosphere soil (Jorquera et al., 2016). Thus, Suaeda australis may had promoted the occurrence of Alphaproteobacteria in the rhizosphere. However, at the family level, Suaeda australis rhizosphere specific microbial communities’ composition and abundance were varying according to the farms (Figure 3B). For instance, farm D was marked by the predominance of nitrogen-fixing Terasakiellaceae (Alphaproteobacteria class) which was absent in others farms sediment (Figure 3B; Weiler et al., 2018; Espín et al., 2021). In the farms A and F, the rhizosphere microbiota of Suaeda australis had higher relative abundance of Rhodobacteraceae family (Alphaproteobacteria class) compared to the farms D and P. The Rhodobacteraceae family was also reported to be deeply involved in sulfur and carbon biogeochemical cycles (Pujalte et al., 2014). The higher occurrence of this family in the farms A and F may have favored the reduction of sulfur level in sediment compared to D0 (Table 1). A previous study has reported that members of Rhodobacteraceae family are key player in hydrocarbon degradation in Mexico beach sand (Kostka et al., 2011). Thus, higher abundance of taxa from this family in farm A may explained its significant link to hydrocarbon degradation function (Figure 4).
Functions linked to biopolymer lysis were found significantly enriched in the farms F and P with celluloysis and chitinolysis functions. The chitinolysis function is relevant because chitin makes up the shrimp exoskeleton and this biopolymer accumulates in the sediments during shrimp rearing as the shrimps molt or die (Philip and Antony, 2006; Della Patrona and Brun, 2009). Those functions may be linked to the higher abundance of several families related to Myxobacteria (Haliangiaceae, Nannocystaceae, Sandaracinaceae) in farms F and P (Mohr, 2018). Myxobacteria were also found in farms A and D but in a lesser abundance. However, the occurrence of Myxobacteria in the specific rhizosphere microbiota may not be due to the plant influence as those families were found without plant occurrence on D0 and in dry sediment.
In farm F, the specific microbiota of Suaeda australis was significantly correlated with pathogens functions (human pathogens, animal parasites or symbionts) which may be explained by the presence of Alphaproteobacteria from the Sphingomonadaceae family. In fact, some genera of Sphingomonadaceae are known as plant pathogens or to cause human infection (Glaeser and Kämpfer, 2014). The functional assignment of prokaryotic taxa with FAPROTAX is based on information on taxa using standard references (Louca et al., 2016a,b). The FAPROTAX tool is relevant for predicting the functions of prokaryotes related to biogeochemical cycles. It can be applied to the analysis of the microbiome (terrestrial, human, animal), however, it is not specialized in the microbiome of shrimp sediment (Sansupa et al., 2021).
The Desulfuromonadia class was evidenced in the rhizosphere of Suaeda australis in farms A and P. It belongs to the Desulfurobacterota phylum that encompasses sulfate-reducing and fermentative taxa (Murphy et al., 2021). However, the abundance of the families affiliated to this phylum varied within farms, with Geothermobacteraceae and Bradymonadaceae found only in farms P and A, respectively. This evidenced that sediment types had influence the structuration of rhizosphere microbial composition. Previous study has also reported differences in the composition of the rhizosphere microbial communities of Arabidopsis thaliana, lettuces plants or also cotton grown in different types of terrestrial soil (Lundberg et al., 2012; Schreiter et al., 2014; Qiao et al., 2017). In their study, Qiao et al. (2017) reported that the soil background microorganisms were the main cause of the variation of the microbial community in the rhizosphere between different soils. We have highlighted in this study, that at the beginning of the experiment, the microbial diversity and composition varied between the four farms. Thus, as the rhizosphere communities were a subset of soil microbial communities, differences in the initial pool of soil prokaryotic taxa between farms may result in different recruitment of taxa by Suaeda australis. Also, variation of rhizosphere microbial communities between farms may also result from the growth and nutrition of Suaeda australis that could influence the chemical parameters of the sediments. Indeed, we have evidenced that under Suaeda australis cultivation, the pH, nitrogen and sulfur concentrations varied in different ways between the farms (Table 1). For instance, Marschner et al. (2004), evidenced that variation of soil pH value of one unit, can significantly affect the structure of the bacterial communities of Sudan grass rhizosphere, grown in sandy loam soil. In addition, variations in the amount of root exudation according to the farms sediments could also be a factor that influencing the microbial composition of the Suaeda australis rhizosphere (Neumann et al., 2014; Maurer et al., 2021).
Cultivation of Suaeda australis in the sediments seemed effective in removing nitrogen, sulfur and phosphorus but this varied according to the farms (Table 1). The nitrogen concentration was reduced in farms A, F and P whereas the total sulfur was reduced only in farms A and F. For all the farms, the cultivation of Suaeda australis has reduced both the total and available forms of phosphorus (Table 1). In all farms, the cultivation of Suaeda australis had increased the concentration of organic carbon in the sediment compared to D0 (Table 1). This increase may be due to plant litter or to the exudates released by the roots (Yan et al., 2018).
The sediment of the farm D was the only one for which the NO3− level had increased significantly under both Suaeda australis cultivation and sediment drying compared to D0 (Table 1). In contrast, in other sediments from shrimp farms, nitrogen levels were reduced by 55–99% under Suaeda australis cultivation. In addition, for farm D, the practice of drying or growing halophytes increased total sulfur and organic carbon concentrations by at least 36 and 40% respectively, which was significantly higher than the concentrations observed in the others farms (Table 1). Thus, theses experimental conditions have selected microbial communities that appeared to be not effective for sediment nitrogen, carbon and sulfur removal. Farm D also stood out from the others for its lower pH values, higher ammonia concentration and lower alpha diversity index (Tables 1, 2). The lower pH value observed in farm D at the initial state and in dry conditions compared to the other farms may have not favor nutrient removal by rhizosphere microbial communities. It was reported that soil acidity reduces the intrinsic activities of microbial communities (Kemmitt et al., 2006), this could explain the significantly increased of organic carbon and nitrogen in the dry sediment and with the halophytes (Table 1). Under the cultivation of Suaeda australis the pH of the sediment of the farm D increased but it was still lower than the other farms (Table 1). Thus for farm D, liming the sediment accumulation beforehand halophyte cultivation or drying period may be a way to increase the sediment pH and to improve the organic matter decomposition by microbial communities (Li et al., 2015). In addition to control sediment acidity, the liming application in shrimp sediment can also be beneficial to improve the sediment texture as the porosity (Della Patrona and Brun, 2009). This sediment parameter could also shape the microbial diversity and composition (Chau et al., 2011; Seaton et al., 2020).
For the other farms, sediment drying appears more efficient than Suaeda australis cultivation for the reduction of total sulfur. This can be explained by the fact that in the greenhouse, pots containing halophytes were watered twice a week with seawater, which is a source of sulfate for the sedimentary matrix. In addition, plant residues can be sources of organic sulfur for the sediments (Schoenau and Malhi, 2015). It would be therefore interesting to determine the different forms of sulfur in the sediments, such as the quantity of sulfur in its organic form that is available to the plant, or the quantity of bound sulfur, for a better understanding of the effect of halophyte cultivation on the sulfur cycle (Zhou et al., 2009). For all the farms, the available forms of phosphorus were reduced almost as much in the dry sediment as in the halophyte rhizosphere. This may be explained as phosphorus is an essential nutrient for microorganisms growth (Oliverio et al., 2020) and therefore microbial communities have used the pool of available sediment phosphorus for their nutrient requirement. This highlighted a pivotal role of the prokaryotes in the phosphorus cycle.
In the farm A, the nitrogen removal under the halophyte cultivation was significantly higher than in the dry sediment. Thus, for this farm, Suaeda australis nutrition and its associated microbiota was more effective than in dry condition to reduce nitrogen level accumulated in the sediment. However, in the farms F and P, the practice of drying alone, had greatly reduced the level of nitrogen in the sediments (Table 1), which meant that for these two farms, the microbial community alone was almost as effective (farm F) or even more effective (farm P) than the cultivation of halophytes and its microbiota. Those two farms had similar microbial communities at the beginning of the experiment, higher alpha diversity indices and higher CaCO3 concentration in their sediments (Figure 2C and Tables 1, 2). Those parameters may have favored a better nutrients removal in the sediment with and without halophyte cultivation. However, for those two farms F and P, the reduction of the total phosphorus was significantly higher under halophyte cultivation compared to dry sediment. This higher decrease under Suaeda australis cultivation may be linked to the phosphorus nutrition for the plant. In the soil, mineralization of organic P is catalyzed by hydrolytic enzymes such as phosphatase that can be released by plant or soil microorganisms (Marschner et al., 2011; Nannipieri et al., 2011). The mineralization of organic phosphorus is thereby controlled by the plant and microbial demand (Richardson and Simpson, 2011). Thus, a higher mineralization of sediment phosphorus to meet the plant needs could have explained the greater reduction of total phosphorus in the Suaeda australis rhizosphere compared to the dry sediment. This could be the case particularly for the farm F, where the total pool of phosphorus available to the plant was significantly lower than for all the other farms (Table 1). Thus, the effectiveness of nutrient removal by halophyte in shrimp sediment seemed to be strongly influenced by the chemical characteristics of the sediments at the initial state. The aquaculture sediments are formed on the initial soil of the pond as soon as aquaculture activities begin, and then evolve with successive rearing and zootechnical practices (Munsiri et al., 1995). It was also reported that in New Caledonia, the pond sediment pH and CaCO3 concentration may change with the aging of farms (Lemonnier et al., 2021). Thus, the background of farms aquaculture practices, farms age or type of original pond soil may explain variation of sediment chemical parameters between farms.
First, our results showed that the active microbial diversity of sediments varied between shrimp farms, which has so far never been explored before in New Caledonia. The initial CaCO3 concentration in the sediments appeared to have significantly influenced the microbial diversity, with the higher alpha diversity indices and microbiota similarities found in sediments with the higher CaCO3 concentrations.
The difference in microbial and chemical composition between the farms led to different specific microbiota in the Suaeda australis rhizosphere, recruited from the bulk sediments, as well as in the selection of various putative microbial function in the sediment. Alphaproteobacteria class was promoted in Suaeda australis rhizosphere but the related family composition and abundance varied according to the farm sediments. Thus, the initial microbial composition and chemical characteristic of the sediment seemed to have strongly influenced the efficiency of nutrient removal by Suaeda australis. In fact, the significant efficient nitrogen removal without halophyte cultivation in the farms F and P sediments meant that in those farms, the microbial communities played an important and efficient role in the nitrogen cycle. The pH had also emerged as a key factor in the success of nutrient removal and thus in nitrogen removal under halophyte cultivation or sediment drying. In addition, the sediment microbial community of this farm was more different from the other farms. Therefore, the initial chemical parameters of these aquaculture sediments should probably be improved to ensure better nutrient removal efficiency; liming may be a solution to improve pH, texture and neutralize acidity in the sediments.
In conclusion, this study demonstrated the impact of Suaeda australis cultivation on nitrogen, sulfur, and phosphorus removal in shrimp pond sediments; however, the removal efficiency varied depending on the sediment of the shrimp ponds. Thus, the initial chemical and microbial composition of the sediment must be studied in order to select the most appropriate method for nutrient removal (halophyte, drying, lime application, or a combination of several methods). To go further, it would be interesting to repeat this experiment but at the pond scale; and also to compare the nutrient removal efficiency among different halophyte species or a mixture of halophyte species within pond sediment. Thus, ultimately some halophyte species could be rather efficient for the nutrient removal of a type of sediment than another.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: NCBI BioProject (https://www.ncbi.nlm.nih.gov/bioproject/), PRJNA996981.
MC: Conceptualization, Investigation, Methodology, Software, Writing – original draft, Writing – review & editing, Data curation, Formal analysis, Visualization. LG: Supervision, Validation, Writing – review & editing. LD: Conceptualization, Funding acquisition, Investigation, Methodology, Supervision, Validation, Writing – review & editing. DA: Methodology, Writing – review & editing. NC: Data curation, Funding acquisition, Investigation, Software, Supervision, Validation, Visualization, Writing – review & editing, Formal analysis, Methodology.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by Agence Rurale of New Caledonia and by the Government of New Caledonia (PhD. research grant). Funding for sequencing analysis were entirely supported by Ifremer unit from the Department of Biological Resources and Environment (RBE).
The authors thank M. Bull Michel from Aigue Marine aquaculture farm for his support for the project. Thanks go to LAMA Platform (IRD), Noumea, New-Caledonia (France) for sediment chemical analysis and MrDNA lab, Shallowater (Texas), for sequencing analysis. Authors would also thank to the LEAD Ifremer for the financial contribution.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1260585/full#supplementary-material
Allison, S. D., Weintraub, M. N., Gartner, T. B., and Waldrop, M. P. (2010). “Evolutionary-economic principles as regulators of soil enzyme production and ecosystem function” in Soil enzymology. eds. G. Shukla and A. Varma, Soil Biology (Berlin, Heidelberg: Springer Berlin Heidelberg), 229–243.
Amarasiri, M., Furukawa, T., Nakajima, F., and Sei, K. (2021). Pathogens and disease vectors/hosts monitoring in aquatic environments: potential of using eDNA/eRNA based approach. Sci. Total Environ. 796:148810. doi: 10.1016/j.scitotenv.2021.148810
Avnimelech, Y., and Ritvo, G. (2003). Shrimp and fish pond soils: processes and management. Aquaculture 220, 549–567. doi: 10.1016/S0044-8486(02)00641-5
Boden, R. (2017). Reclassification of Halothiobacillus hydrothermalis and Halothiobacillus halophilus to Guyparkeria gen. Nov. in the Thioalkalibacteraceae fam. Nov., with emended descriptions of the genus Halothiobacillus and family Halothiobacillaceae. Int. J. Syst. Evol. Microbiol. 67, 3919–3928. doi: 10.1099/ijsem.0.002222
Bogati, K., and Walczak, M. (2022). The impact of drought stress on soil microbial community, enzyme activities and plants. Agronomy 12:189. doi: 10.3390/agronomy12010189
Boyd, C. E., and Pippopinyo, S. (1994). Factors affecting respiration in dry pond bottom soils. Aquaculture 120, 283–293. doi: 10.1016/0044-8486(94)90085-X
Brust, G. E. (2019). Chapter 9 - Management strategies for organic vegetable fertility. Safety Pract. Organ. Food, 193–212. doi: 10.1016/B978-0-12-812060-6.00009-X
Callac, N., Boulo, V., Giraud, C., Beauvais, M., Ansquer, D., Ballan, V., et al. (2022). Microbiota of the rearing water of Penaeus stylirostris larvae influenced by lagoon seawater and specific key microbial lineages of larval stage and survival. Microbiol Spectr 10, e04241–e04222. doi: 10.1128/spectrum.04241-22
Callac, N., Giraud, C., Boulo, V., Wabete, N., and Pham, D. (2023). Microbial biomarker detection in shrimp larvae rearing water as putative bio-surveillance proxies in shrimp aquaculture. PeerJ 11:e15201. doi: 10.7717/peerj.15201
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Lozupone, C. A., Turnbaugh, P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U. S. A. 108, 4516–4522. doi: 10.1073/pnas.1000080107
Chau, J. F., Bagtzoglou, A. C., and Willig, M. R. (2011). The effect of soil texture on richness and diversity of bacterial communities. Environ. Forensic 12, 333–341. doi: 10.1080/15275922.2011.622348
Chen, M.-M., Zhu, Y.-G., Su, Y.-H., Chen, B.-D., Fu, B.-J., and Marschner, P. (2007). Effects of soil moisture and plant interactions on the soil microbial community structure. Eur. J. Soil Biol. 43, 31–38. doi: 10.1016/j.ejsobi.2006.05.001
Chu, L., and Wang, J. (2017). Denitrification of groundwater using a biodegradable polymer as a carbon source: long-term performance and microbial diversity. RSC Adv. 7, 53454–53462. doi: 10.1039/C7RA11151G
Colette, M., Guentas, L., Gunkel-Grillon, P., Callac, N., and Della Patrona, L. (2022). Is halophyte species growing in the vicinity of the shrimp ponds a promising Agri-aquaculture system for shrimp ponds remediation in New Caledonia? Mar. Pollut. Bull. 177:113563. doi: 10.1016/j.marpolbul.2022.113563
Colette, M., Guentas, L., Patrona, L. D., Ansquer, D., and Callac, N. (2023). Dynamic of active microbial diversity in rhizosphere sediments of halophytes used for bioremediation of earthen shrimp ponds. Environ. Microb. 18:58. doi: 10.1186/s40793-023-00512-x
Cristescu, M. E. (2019). Can environmental RNA revolutionize biodiversity science? Trends Ecol. Evol. 34, 694–697. doi: 10.1016/j.tree.2019.05.003
Dai, W., Wang, N., Wang, W., Ye, X., Cui, Z., Wang, J., et al. (2021). Microbial community structure drives predatory Myxobacteria distribution under different compost manures. Review. doi: 10.21203/rs.3.rs-524148/v1
Dawid, W. (2000). Biology and global distribution of myxobacteria in soils. FEMS Microbiol. Rev. 24, 403–427. doi: 10.1111/j.1574-6976.2000.tb00548.x
Della Patrona, L., and Brun, P. (2009). Elevage de la Crevette Bleue en Nouvelle-Calédonie. Litopenaeus Stylirostris. Bases biologiques et zootechnie. Available at: https://archimer.ifremer.fr/doc/00251/36229/
Espín, Y., Menchén, A., Moreno, J. L., Sanz, D., Álvarez-Ortí, M., Fernández, J. A., et al. (2021). Water and sediment bacterial communities in a small Mediterranean, oxygen-stratified, saline Lake (Lake Alboraj, SE Spain). Appl. Sci. 11:6309. doi: 10.3390/app11146309
Funge-Smith, S. J., and Briggs, M. R. P. (1998). Nutrient budgets in intensive shrimp ponds: implications for sustainability. Aquaculture 164, 117–133. doi: 10.1016/S0044-8486(98)00181-1
Garcia, R., and Müller, R. (2014). “The family Polyangiaceae” in The prokaryotes. eds. E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin, Heidelberg: Springer Berlin Heidelberg), 247–279.
Glaeser, S. P., and Kämpfer, P. (2014). “The family Sphingomonadaceae” in The prokaryotes. eds. E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin, Heidelberg: Springer Berlin Heidelberg), 641–707.
Goarant, C., Harache, Y., Herbland, A., and Mugnier, C. (2004). Styli 2003. Trente ans de crevetticulture en Nouvelle-Calédonie in Nouméa-Koné. Available at: https://archimer.ifremer.fr/doc/00716/82825/ (Accessed September 29, 2023).
Guo, A., Ding, L., Tang, Z., Zhao, Z., and Duan, G. (2019). Microbial response to CaCO3 application in an acid soil in southern China. J. Environ. Sci. 79, 321–329. doi: 10.1016/j.jes.2018.12.007
Hakkoum, Z., Minaoui, F., Douma, M., Mouhri, K., and Loudiki, M. (2021). Impact of human disturbances on soil cyanobacteria diversity and distribution in suburban arid area of Marrakesh, Morocco. Ecol Process 10:42. doi: 10.1186/s13717-021-00303-7
Imhoff, J. F. (2014). “The family Chromatiaceae” in The prokaryotes. eds. E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin, Heidelberg: Springer Berlin Heidelberg), 151–178.
Jacoby, R., Peukert, M., Succurro, A., Koprivova, A., and Kopriva, S. (2017). The Role of Soil Microorganisms in Plant Mineral Nutrition—Current Knowledge and Future Directions. Front. Plant Sci. 8:1617. doi: 10.3389/fpls.2017.01617
Jorquera, M. A., Maruyama, F., Ogram, A. V., Navarrete, O. U., Lagos, L. M., Inostroza, N. G., et al. (2016). Rhizobacterial community structures associated with native plants grown in Chilean extreme environments. Microb. Ecol. 72, 633–646. doi: 10.1007/s00248-016-0813-x
Kemmitt, S., Wright, D., Goulding, K., and Jones, D. (2006). pH regulation of carbon and nitrogen dynamics in two agricultural soils. Soil Biol. Biochem. 38, 898–911. doi: 10.1016/j.soilbio.2005.08.006
Khan, S. T., Horiba, Y., Yamamoto, M., and Hiraishi, A. (2002). Members of the family Comamonadaceae as primary poly (3-Hydroxybutyrate-co-3-Hydroxyvalerate)-degrading Denitrifiers in activated sludge as revealed by a Polyphasic approach. Appl. Environ. Microbiol. 68, 3206–3214. doi: 10.1128/AEM.68.7.3206-3214.2002
Kostka, J. E., Prakash, O., Overholt, W. A., Green, S. J., Freyer, G., Canion, A., et al. (2011). Hydrocarbon-degrading Bacteria and the bacterial community response in Gulf of Mexico Beach sands impacted by the Deepwater horizon oil spill. Appl. Environ. Microbiol. 77, 7962–7974. doi: 10.1128/AEM.05402-11
Laroche, O., Wood, S. A., Tremblay, L. A., Ellis, J. I., Lear, G., and Pochon, X. (2018). A cross-taxa study using environmental DNA/RNA metabarcoding to measure biological impacts of offshore oil and gas drilling and production operations. Mar. Pollut. Bull. 127, 97–107. doi: 10.1016/j.marpolbul.2017.11.042
Leadbeater, D. R., Oates, N. C., Bennett, J. P., Li, Y., Dowle, A. A., Taylor, J. D., et al. (2021). Mechanistic strategies of microbial communities regulating lignocellulose deconstruction in a UK salt marsh. Microbiome 9:48. doi: 10.1186/s40168-020-00964-0
Lemonnier, H., Lopez, E., Schembri, F., and Antypas, F. (2021). Acidification des sédiments des bassins d’élevage de la crevette en Nouvelle-Calédonie. IFREMER/RBE/Pacifique/LEAD Scientific and Technical Report.
Li, K., Chen, D., Huang, Z., Fu, J., Chen, Y., Xue, T., et al. (2022). Effects of shrimp-vegetable rotation on microbial diversity and community structure in pond sediment. Pol. J. Environ. Stud. 31, 2651–2663. doi: 10.15244/pjoes/144097
Li, L., Queiroz, J. F., and Boyd, C. E. (2015). Pond bottom dry-out and liming, part 1. Available at: https://www.globalseafood.org/advocate/pond-bottom-dryout-liming-part-1/
Liu, C., Cui, Y., Li, X., and Yao, M. (2021). Microeco: an R package for data mining in microbial community ecology. FEMS Microbiol. Ecol. 97:fiaa255. doi: 10.1093/femsec/fiaa255
Louca, S., Jacques, S. M. S., Pires, A. P. F., Leal, J. S., Srivastava, D. S., Parfrey, L. W., et al. (2016a). High taxonomic variability despite stable functional structure across microbial communities. Nat Ecol Evol 1:0015. doi: 10.1038/s41559-016-0015
Louca, S., Parfrey, L. W., and Doebeli, M. (2016b). Decoupling function and taxonomy in the global ocean microbiome. Science 353, 1272–1277. doi: 10.1126/science.aaf4507
Lundberg, D. S., Lebeis, S. L., Paredes, S. H., Yourstone, S., Gehring, J., Malfatti, S., et al. (2012). Defining the core Arabidopsis thaliana root microbiome. Nature 488, 86–90. doi: 10.1038/nature11237
Luo, L., Meng, H., and Gu, J.-D. (2017). Microbial extracellular enzymes in biogeochemical cycling of ecosystems. J. Environ. Manag. 197, 539–549. doi: 10.1016/j.jenvman.2017.04.023
Mariscal-Lagarda, M. M., and Páez-Osuna, F. (2014). Mass balances of nitrogen and phosphorus in an integrated culture of shrimp (Litopenaeus vannamei) and tomato (Lycopersicon esculentum mill) with low salinity groundwater: a short communication. Aquac. Eng. 58, 107–112. doi: 10.1016/j.aquaeng.2013.12.003
Mariscal-Lagarda, M. M., Páez-Osuna, F., Esquer-Méndez, J. L., Guerrero-Monroy, I., Del Vivar, A.-R., Brito-Solano, K. Y., et al. (2014). Water quality in an intergrated culture of white shrimp (Litopenaeus vannamei)-tomato (Lycopersicon esculentum) using low salinity groundwater in Sonora, Mexico. Ex. Agric. 50, 306–319. doi: 10.1017/S0014479713000690
Marschner, P., Crowley, D., and Rengel, Z. (2011). Rhizosphere interactions between microorganisms and plants govern iron and phosphorus acquisition along the root axis – model and research methods. Soil Biol. Biochem. 43, 883–894. doi: 10.1016/j.soilbio.2011.01.005
Marschner, P., Crowley, D., and Yang, C. H. (2004). Development of specific rhizosphere bacterial communities in relation to plant species, nutrition and soil type. Plant Soil 261, 199–208. doi: 10.1023/B:PLSO.0000035569.80747.c5
Maurer, D., Malique, F., Alfarraj, S., Albasher, G., Horn, M. A., Butterbach-Bahl, K., et al. (2021). Interactive regulation of root exudation and rhizosphere denitrification by plant metabolite content and soil properties. Plant Soil 467, 107–127. doi: 10.1007/s11104-021-05069-7
Miranda, F. R., Lima, R. N., Crisóstomo, L. A., and Santana, M. G. S. (2008). Reuse of inland low-salinity shrimp farm effluent for melon irrigation. Aquac. Eng. 39, 1–5. doi: 10.1016/j.aquaeng.2008.04.001
Miyata, K., Inoue, Y., Amano, Y., Nishioka, T., Yamane, M., Kawaguchi, T., et al. (2021). Fish environmental RNA enables precise ecological surveys with high positive predictivity. Ecol. Indic. 128:107796. doi: 10.1016/j.ecolind.2021.107796
Mohr, K. (2018). Diversity of Myxobacteria—we only see the tip of the iceberg. Microorganisms 6:84. doi: 10.3390/microorganisms6030084
Mohr, K. I., Garcia, R. O., Gerth, K., Irschik, H., and Müller, R. (2012). Sandaracinus amylolyticus gen. Nov., sp. nov., a starch-degrading soil myxobacterium, and description of Sandaracinaceae fam. Nov. Int. J. Syst. Evol. Microbiol. 62, 1191–1198. doi: 10.1099/ijs.0.033696-0
Munsiri, P., Boyd, C. E., and Hajek, B. F. (1995). Physical and chemical characteristics of bottom soil profiles in ponds at Auburn, Alabama, USA and a proposed system for describing pond soil horizons. J World Aquaculture Soc 26, 346–377. doi: 10.1111/j.1749-7345.1995.tb00831.x
Murphy, J., and Riley, J. P. (1962). A modified single solution method for the determination of phosphate in natural waters. Analytica Chimica Acta 27, 31–36. doi: 10.1016/S0003-2670(00)88444-5
Murphy, C. L., Biggerstaff, J., Eichhorn, A., Ewing, E., Shahan, R., Soriano, D., et al. (2021). Genomic characterization of three novel Desulfobacterota classes expand the metabolic and phylogenetic diversity of the phylum. Environ. Microbiol. 23, 4326–4343. doi: 10.1111/1462-2920.15614
Nannipieri, P., Giagnoni, L., Landi, L., and Renella, G. (2011). “Role of phosphatase enzymes in soil” in Phosphorus in action. eds. E. Bünemann, A. Oberson, and E. Frossard, Soil Biology (Berlin, Heidelberg: Springer Berlin Heidelberg), 215–243.
Neumann, G., Bott, S., Ohler, M. A., Mock, H.-P., Lippmann, R., Grosch, R., et al. (2014). Root exudation and root development of lettuce (Lactuca sativa L. cv. Tizian) as affected by different soils. Front. Microbiol. 5:2. doi: 10.3389/fmicb.2014.00002
Oliverio, A. M., Bissett, A., McGuire, K., Saltonstall, K., Turner, B. L., and Fierer, N. (2020). The role of phosphorus limitation in shaping soil bacterial communities and their metabolic capabilities. MBio 11, e01718–e01720. doi: 10.1128/mBio.01718-20
Olsen, S. R. (1954). Estimation of available phosphorus in soils by extraction with sodium bicarbonate. US Department of Agriculture.
Páez-Osuna, F., Guerrero-Galván, S. R., Ruiz-Fernández, A. C., and Espinoza-Angulo, R. (1997). Fluxes and mass balances of nutrients in a semi-intensive shrimp farm in North-Western Mexico. Mar. Pollut. Bull. 34, 290–297. doi: 10.1016/S0025-326X(96)00133-6
Pang, Z., Tayyab, M., Kong, C., Hu, C., Zhu, Z., Wei, X., et al. (2019). Liming positively modulates microbial community composition and function of sugarcane fields. Agronomy 9:808. doi: 10.3390/agronomy9120808
Pétard, J. (1993). Les méthodes d’analyse. Analyse de sols. Available at: https://horizon.documentation.ird.fr/exl-doc/pleins_textes/divers10-09/010005303.pdf
Philip, R., and Antony, S. (2006). Bioremediation in shrimp culture systems. Available at: https://www.researchgate.net/publication/48410828_Bioremediation_in_Shrimp_Culture_Systems
Phillips, K. E., Akbar, S., and Stevens, D. C. (2022). Concepts and conjectures concerning predatory performance of myxobacteria. Front. Microbiol. 13:1031346. doi: 10.3389/fmicb.2022.1031346
Pini, F., Galardini, M., Bazzicalupo, M., and Mengoni, A. (2011). Plant-Bacteria association and Symbiosis: are there common genomic traits in Alphaproteobacteria? Genes 2, 1017–1032. doi: 10.3390/genes2041017
Prosser, J. I., Head, I. M., and Stein, L. Y. (2014). “The family Nitrosomonadaceae” in The prokaryotes. eds. E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin, Heidelberg: Springer Berlin Heidelberg), 901–918.
Pujalte, M. J., Lucena, T., Ruvira, M. A., Arahal, D. R., and Macián, M. C. (2014). “The family Rhodobacteraceae” in The prokaryotes. eds. E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin, Heidelberg: Springer Berlin Heidelberg), 439–512.
Qiao, Q., Wang, F., Zhang, J., Chen, Y., Zhang, C., Liu, G., et al. (2017). The variation in the rhizosphere microbiome of cotton with soil type, genotype and developmental stage. Sci. Rep. 7:3940. doi: 10.1038/s41598-017-04213-7
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2012). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Richardson, A. E., and Simpson, R. J. (2011). Soil microorganisms mediating phosphorus availability update on microbial phosphorus. Plant Physiol. 156, 989–996. doi: 10.1104/pp.111.175448
Sansupa, C., Wahdan, S. F. M., Hossen, S., Disayathanoowat, T., Wubet, T., and Purahong, W. (2021). Can we use functional annotation of prokaryotic taxa (FAPROTAX) to assign the ecological functions of soil Bacteria? Appl. Sci. 11:688. doi: 10.3390/app11020688
Schimel, J. P. (2018). Life in dry soils: effects of drought on soil microbial communities and processes. Annu. Rev. Ecol. Evol. Syst. 49, 409–432. doi: 10.1146/annurev-ecolsys-110617-062614
Schoenau, J. J., and Malhi, S. S. (2015). “Sulfur Forms and Cycling Processes in Soil and Their Relationship to Sulfur Fertility,” in Agronomy Monographs, ed. J. Jez (Madison, WI, USA: American Society of Agronomy, Crop Science Society of America, Soil Science Society of America), 1–10.
Schreiter, S., Ding, G.-C., Heuer, H., Neumann, G., Sandmann, M., Grosch, R., et al. (2014). Effect of the soil type on the microbiome in the rhizosphere of field-grown lettuce. Front. Microbiol. 5:144. doi: 10.3389/fmicb.2014.00144
Seaton, F. M., George, P. B. L., Lebron, I., Jones, D. L., Creer, S., and Robinson, D. A. (2020). Soil textural heterogeneity impacts bacterial but not fungal diversity. Soil Biol. Biochem. 144:107766. doi: 10.1016/j.soilbio.2020.107766
Shimkets, L. J., Dworkin, M., and Reichenbach, H. (2006). “The Myxobacteria” in The prokaryotes. eds. M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (New York, NY: Springer New York), 31–115.
Smith, P. T. (1996). Physical and chemical characteristics of sediments from prawn farms and mangrove habitats on the Clarence River, Australia. Aquaculture 146, 47–83. doi: 10.1016/S0044-8486(96)01361-0
Spring, S., Scheuner, C., Göker, M., and Klenk, H.-P. (2015). A taxonomic framework for emerging groups of ecologically important marine gammaproteobacteria based on the reconstruction of evolutionary relationships using genome-scale data. Front. Microbiol. 6:281. doi: 10.3389/fmicb.2015.00281
Thies, J., and Grossman, J. (2006). “The Soil Habitat and Soil Ecology,” in Biological Approaches to Sustainable Soil Systems Books in Soils, Plants, and the Environment. ed. N. Uphoff (CRC Press), 59–78.
Tripathi, B. M., Stegen, J. C., Kim, M., Dong, K., Adams, J. M., and Lee, Y. K. (2018). Soil pH mediates the balance between stochastic and deterministic assembly of bacteria. ISME J. 12, 1072–1083. doi: 10.1038/s41396-018-0082-4
Veilleux, H. D., Misutka, M. D., and Glover, C. N. (2021). Environmental DNA and environmental RNA: current and prospective applications for biological monitoring. Sci. Total Environ. 782:146891. doi: 10.1016/j.scitotenv.2021.146891
Wang, J., and Chu, L. (2016). Biological nitrate removal from water and wastewater by solid-phase denitrification process. Biotechnol. Adv. 34, 1103–1112. doi: 10.1016/j.biotechadv.2016.07.001
Wang, W., Luo, X., Ye, X., Chen, Y., Wang, H., Wang, L., et al. (2020). Predatory Myxococcales are widely distributed in and closely correlated with the bacterial community structure of agricultural land. Appl. Soil Ecol. 146:103365. doi: 10.1016/j.apsoil.2019.103365
Wang, C., Zhou, X., Guo, D., Zhao, J., Yan, L., Feng, G., et al. (2019). Soil pH is the primary factor driving the distribution and function of microorganisms in farmland soils in northeastern China. Ann. Microbiol. 69, 1461–1473. doi: 10.1007/s13213-019-01529-9
Weiler, B. A., Verhoeven, J. T. P., and Dufour, S. C. (2018). Bacterial communities in tissues and surficial mucus of the cold-water coral Paragorgia arborea. Front. Mar. Sci. 5:378. doi: 10.3389/fmars.2018.00378
Willems, A. (2014). “The family Comamonadaceae” in The prokaryotes. eds. E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin, Heidelberg: Springer Berlin Heidelberg), 777–851.
Wood, S. A., Biessy, L., Latchford, J. L., Zaiko, A., von Ammon, U., Audrezet, F., et al. (2020). Release and degradation of environmental DNA and RNA in a marine system. Sci. Total Environ. 704:135314. doi: 10.1016/j.scitotenv.2019.135314
Yan, J., Wang, L., Hu, Y., Tsang, Y. F., Zhang, Y., Wu, J., et al. (2018). Plant litter composition selects different soil microbial structures and in turn drives different litter decomposition pattern and soil carbon sequestration capability. Geoderma 319, 194–203. doi: 10.1016/j.geoderma.2018.01.009
Yang, P., Lai, D. Y. F., Jin, B., Bastviken, D., Tan, L., and Tong, C. (2017). Dynamics of dissolved nutrients in the aquaculture shrimp ponds of the Min River estuary, China: concentrations, fluxes and environmental loads. Sci. Total Environ. 603-604, 256–267. doi: 10.1016/j.scitotenv.2017.06.074
Yaobin, L., Lin, Q., Fengbo, L., Xiyue, Z., Chunchun, X., Long, J., et al. (2019). Impact of Rice-catfish/shrimp co-culture on nutrients fluxes across sediment-water Interface in intensive aquaculture ponds. Rice Sci. 26, 416–424. doi: 10.1016/j.rsci.2019.06.001
Zhalnina, K., Dias, R., De Quadros, P. D., Davis-Richardson, A., Camargo, F. A. O., Clark, I. M., et al. (2015). Soil pH determines microbial diversity and composition in the park grass experiment. Microb. Ecol. 69, 395–406. doi: 10.1007/s00248-014-0530-2
Zhou, C., An, S., Deng, Z., Yin, D., Zhi, Y., Sun, Z., et al. (2009). Sulfur storage changed by exotic Spartina alterniflora in coastal saltmarshes of China. Ecol. Eng. 35, 536–543. doi: 10.1016/j.ecoleng.2008.01.004
Zhou, W., Han, G., Liu, M., and Li, X. (2019). Effects of soil pH and texture on soil carbon and nitrogen in soil profiles under different land uses in Mun River basin, Northeast Thailand. PeerJ 7:e7880. doi: 10.7717/peerj.7880
Keywords: halophyte, earthen pond sediment, active rhizosphere microbiota, nutrient removal, metabarcoding
Citation: Colette M, Guentas L, Della Patrona L, Ansquer D and Callac N (2023) Suaeda australis and its associated rhizosphere microbiota: a comparison of the nutrient removal potential between different shrimp farm sediments in New Caledonia. Front. Microbiol. 14:1260585. doi: 10.3389/fmicb.2023.1260585
Received: 18 July 2023; Accepted: 25 September 2023;
Published: 09 October 2023.
Edited by:
Sukhwan Yoon, Korea Advanced Institute of Science and Technology (KAIST), Republic of KoreaReviewed by:
Grégoire Michoud, Swiss Federal Institute of Technology Lausanne, SwitzerlandCopyright © 2023 Colette, Guentas, Della Patrona, Ansquer and Callac. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marie Colette, bWFyaWUuY29sZXR0ZUBldHVkaWFudC51bmMubmM=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.