
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 06 November 2023
Sec. Aquatic Microbiology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1259653
Ciliates serve as excellent indicators for water quality monitoring. However, their utilization is hindered by various taxonomic confusions. The ciliate genus Lacrymaria Bory de Saint-Vincent, 1824 is commonly found in different aquatic habitats, but its taxonomy has been sparsely investigated using state-of-the-art methods. This study investigated two new Lacrymaria species from Nanhui Wetland, Shanghai, China, using living observation, protargol staining, and molecular phylogeny methods. Lacrymaria songi sp. nov. is 180–340 × 20–25 μm in size and possesses 12–16 somatic kineties, 1 terminal contractile vacuole, 2 macronuclear nodules, and 2 types of rod-shaped extrusomes. Lacrymaria dragescoi sp. nov. is distinguished from its congeners by its cell size of 210–400 × 25–35 μm, 14–17 somatic kineties, 1 terminal contractile vacuole, 1 macronucleus, and 2 types of rod-shaped extrusomes. Phylogenetic analyses based on SSU rRNA gene sequences indicate that Lacrymariidae is monophyletic but Lacrymaria is not. Additionally, a brief review of the genus Lacrymaria is provided in this study. We suggest that L. bulbosa Alekperov, 1984, L. lanceolata Kahl, 1930, and L. ovata Burkovsky, 1970 be removed from the genus and propose Phialina lanceolata nov. comb. and Phialina ovata nov. comb. for the latter two.
ZooBank registration: Present work: urn:lsid:zoobank.org:pub:CDFB1EBD-80BD-4533-B391-CEE89F62EDC4 Lacrymaria songi sp. nov.: urn:lsid:zoobank.org:act:417E7C2D-DAEC-4711-90BB-64AB3CD2F7D5 Lacrymaria dragescoi sp. nov.: urn:lsid:zoobank.org:act:8778D6B0-1F2E-473C-BE19-3F685391A40D.
Ciliates are excellent indicators for water quality monitoring and play a vital role in the aquatic microbial food web (Wang et al., 2022; Weisse and Montagnes, 2022). Lacrymaria ciliates are common raptorial microorganisms found in aquatic habitats worldwide (Kahl, 1930; Dragesco, 1960, 1965; Rajter et al., 2019; Wang et al., 2019). They can be easily identified by the bubble-like head located at the front end of their body, which is covered by short oblique kineties (Lynn, 2008). The family Lacrymariidae de Fromentel 1876 includes four genera, namely, Lacrymaria Bory de Saint-Vincent, 1824, Pelagolacrymaria Foissner, 1999, Phialina Bory de Saint-Vincent, 1824, and Phialinides Foissner, 1988 (Lynn, 2008). In contrast to other well-studied haptorians, such as pleurostomatids and spathidiids, research on the Lacrymariidae is limited, and its phylogeny remains unresolved (Foissner, 1988; Foissner and Xu, 2007; Rajter et al., 2019; Pan et al., 2020; Wu et al., 2021, 2022; Chi et al., 2022; Zhang G. et al., 2022a,b,c).
Lacrymaria Bory de Saint-Vincent, 1824 is the largest and oldest genus of the family Lacrymariidae. It is distinguished from its relatives by the presence of a retractable neck (Foissner, 1983). However, for a long time, its closest related genus Phialina was considered as its synonym, which results in the affiliation of most Lacrymaria species needing to be re-considered. Several Lacrymaria species have already been transferred to Phialina, Lagynus, or Pelagolacrymaria in recent studies (Supplementary Table S1; Foissner, 1983, 1987; Song and Wilbert, 1989; Sola et al., 1990; Foissner et al., 1995, 1999, 2002; Wang et al., 2019; Jiang et al., 2023). Since the descriptions of most Lacrymaria species are rough and superficial, the species delimitation is understudied. Recent phylogenetic analysis of Haptoria based on either single gene locus or multiple gene loci has shown that Lacrymaria is not monophyletic (Wu et al., 2017; Huang et al., 2018; Rajter et al., 2019; Wang et al., 2019). However, this conclusion is not confident because only 3 out of 53 nominal Lacrymaria species have molecular information, and the DNA sequence that detached from Lacrymaria in gene trees is not reported along with morphometrics (Huang et al., 2018; Rajter et al., 2019).
Recent studies on haptorian ciliates in China have revealed a high diversity of the order Pleurostomatida (Liu et al., 2017; Pan et al., 2020; Wu et al., 2021, 2022; Zhang G. et al., 2022a,b,c, 2023). However, little attention has been given to the family Lacrymariidae. A project on ciliate fauna conducted in Changjiang Estuary has led to the discovery of various new or rarely known ciliates (Chen et al., 2022; Han et al., 2022; He et al., 2022; Zhang Z. et al., 2022a,b). As a new contribution, we investigated the phylogeny and taxonomy of two new Lacrymaria species, namely, L. songi sp. nov. and L. dragescoi sp. nov., using integrative methods including live observation, silver staining, and DNA sequencing. Additionally, we provide a brief review of the genus Lacrymaria.
Lacrymaria songi sp. nov. and Lacrymaria dragescoi sp. nov. were both collected on 28 September 2022 from two adjacent sites of Nanhui Wetland (N30°53′27.56″, E121°58′38.78″), Shanghai, China (Figure 1). For the habitat of L. songi sp. nov., the water temperature was 23.6°C, the pH was 7.43, the concentration of dissolved oxygen (DO) was 4.22 mg/L, and the salinity measured in the Petri dish was 17‰; for the habitat of L. dragescoi sp. nov., the water temperature was 23.6°C, the pH was 7.61, the DO was 5.47 mg/L, and the salinity measured in the Petri dish was 20‰. All environmental parameters, except salinity, were measured in situ, and the salinity was measured when Lacrymaria species were detected in the raw cultures.
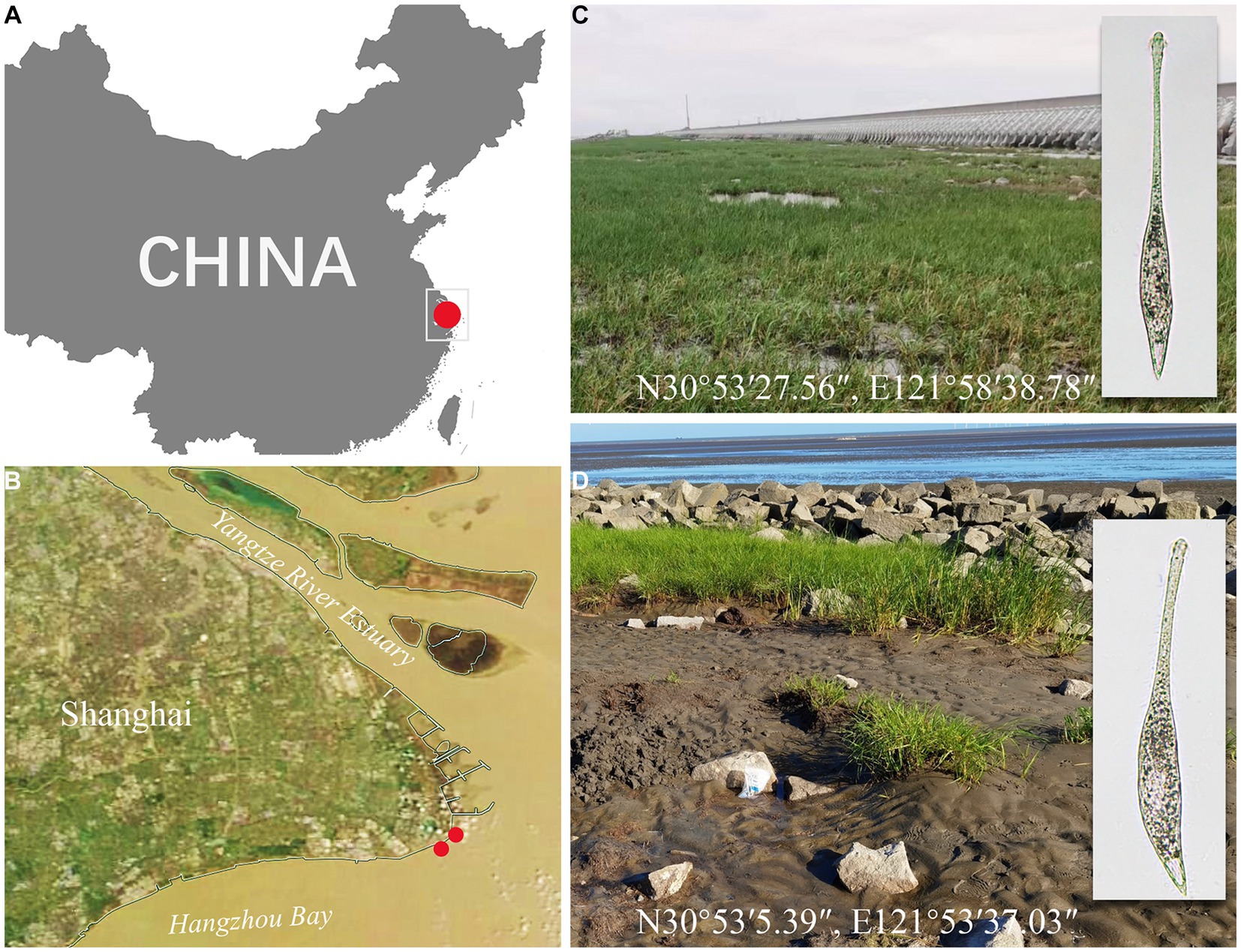
Figure 1. Sampling sites (A–D). (A) A part of the map of China showing the location of Shanghai. (B) A satellite image of Shanghai showing the location of Nanhui Wetland. (C) Photograph of the sampling site in a tidal flat of Nanhui Wetland (N30°53′27.56″, E121°58′38.78″) from where Lacrymaria songi sp. nov. was collected. (D) Photograph of the sampling site in the tidal flat of Nanhui Wetland (N30°53′5.39″, E121°53′37.03″) from where Lacrymaria dragescoi sp. nov. was collected.
After the samples were transported to the laboratory, raw cultures were immediately established in Petri dishes with rice grains to enrich the growth of bacteria, serving as food for the ciliates. L. songi sp. nov. and L. dragescoi sp. nov. were detected after 3 weeks.
Live cells were observed using bright field and differential interference contrast microscopy (Olympus BX53) at magnifications of 100–1,000. The ciliary pattern was revealed using the protargol preparation method (Wilbert, 1975). The protargol reagent was manually synthesized following the method described by Pan et al. (2013). Counts and measurements of stained specimens were performed at a magnification of 1,000, and drawings were made at the same magnification with the aid of a camera lucida. Terminology and systematics are explained by Lynn (2008) and Vd’ačný et al. (2011).
Five cells of each species were isolated from the raw cultures using sterile micropipettes and washed at least five times with filtered (0.22 μm) habitat water to remove contaminants. Genomic DNA was extracted by a DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany) using one-quarter of the volume recommended by the manufacturer’s instructions as described by Shao et al. (2023). The SSU rRNA gene was amplified by PCR using the primers 18S-F (5’-AAC CTG GTT GAT CCT GCC AGT-3′) and 5.8S-R (5′- TAC TGA TAT GCT TAA GTT CAG CGG-3′) (Medlin et al., 1988; Sogin, 1989). The cycling parameters were as follows: an initial denaturation of 3 min at 95°C, followed by 30 cycles of 30 s at 95°C, 20 s at 56°C, and 1.5 min at 72°C, with a final extension of 5 min at 72°C. The PCR products were, then, purified, cloned, and sequenced, following the method described by Chen et al. (2022). The sequencing data were assembled using SeqMan v7.1 (DNAStar), and sequence similarities were calculated using BioEdit v.7.2.5 (Hall, 1999).
The SSU rRNA gene sequences of Lacrymaria songi sp. nov. and L. dragescoi sp. nov. were aligned with 52 other sequences downloaded from GenBank, including three metopids as outgroup taxa, namely, Clevelandella panesthiae (KC139719), Metopus palaeformis (AY007450), and Nyctotherus ovalis (AJ222678). The alignment was performed using the MUSCLE algorithm on the Web Server Guidance1 with default settings (Sela et al., 2015). Maximum likelihood (ML) analyses were conducted using RAxML-HPC2 (Stamatakis, 2014) on XSEDE v.8.2.11 on the CIPRES Science Gateway2 under the GTRGAMMA model with 1,000 bootstraps. Bayesian inference (BI) analysis was performed using MrBayes v.3.2.7 (Ronquist et al., 2012) on the same platform under the GTR + I + Γmodel, which was selected by jModelTest 2 via the Akaike Information Criterion (Darriba et al., 2012). Markov chain Monte Carlo simulations were run for 1,000,000 generations, and trees were sampled every 100 generations with a burn-in of 2,500 trees (25%). The tree topology was visualized using Figtree v1.4.4 (Rambaut, 2018).
The support of the dataset for competing phylogenetic hypotheses was evaluated using the approximately unbiased (AU) test to test the monophyly of the genus Lacrymaria (Shimodaira and Hasegawa, 2001). The site-wise likelihoods for the resulting constrained topology and the non-constrained ML topology were calculated using RAxML v.8.2.11 under a partitioned GTR + GAMMA model (Yang, 1996; Stamatakis, 2014). The same model was used to estimate the site likelihoods for those trees prior to conducting the AU test. The scores of each constraint tree were compared with the unconstrained ML result using the AU test option implemented in CONSEL (Shimodaira and Hasegawa, 2001).
Subclass Haptoria Corliss, 1974.
Family Lacrymariidae de Fromentel, 1876.
Genus Lacrymaria Bory de Saint-Vincent, 1824.
Size: approximately 180–340 × 20–25 μm in vivo. Body shape: highly variable depending on the state of contraction, ranging from a vase-shaped body in the contracted state to fusiform to clavate in the extended state. Neck: flexible, occupying half of the body length and up to two-thirds of body length when swimming, and neck beating 92 times/min when preying. Extrusomes have two types: type I—approximately 10 μm long, rod-shaped, mostly arranged in bundles, scattered in main body, and attached to oral bulge; type II—approximately 4 μm long, rod-shaped, scattered in main body and 12–16 somatic kineties. Single terminally located contractile vacuole. Two macronuclear nodules. Brackish habitat.
A muddy tidal flat of Nanhui Wetland (N30°53′27.56″, E121°58′38.78″), Shanghai, China.
A protargol slide (registration no. TJ2022090805-1) with the holotype circled in black ink and one paratype slide (TJ2022090805-2) are deposited in the Laboratory of Protozoology, Ocean University of China.
The species is named in honor of Prof. Weibo Song, Ocean University of China, in recognition of his outstanding contribution to Ciliatology.
The SSU rRNA gene sequence of L. songi sp. nov. has been deposited in GenBank (accession no. OR689566) with 1,641 bp long and GC content of 42.41%.
Cell: highly contractile, when fully extended cell size in vivo approximately 180–340 × 20–25 μm and length:width ratio of 11:1 (Figures 2A, 3A–C) and when contracted, cell size approximately 65–102 × 28–40 μm and length:width ratio of 3:1 (Figures 2D, 3D). Body shape fusiform to clavate with flexible neck, occupying half of body length, and up to two-thirds of body length when swimming. The posterior end tapered and tail-like when free swimming but vase-shaped with neck retracting into trunk and the posterior end broadly tapered when contracted (Figures 2A,D,H, 3A,E,J,K).
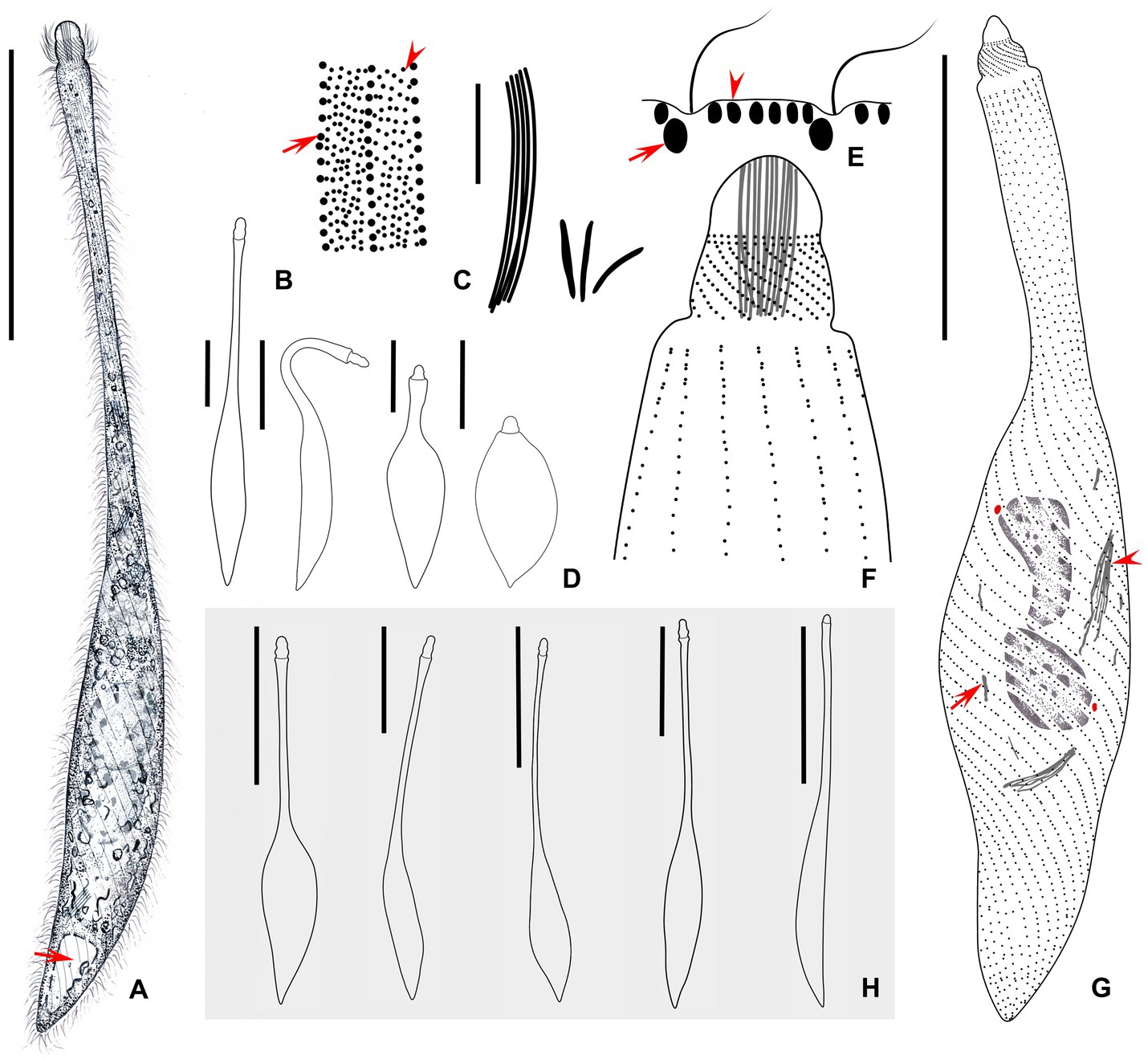
Figure 2. (A–H) Morphology of Lacrymaria songi sp. nov. from life (A,B,D,E,H) and after protargol staining (C,F,H). (A) Typical extended individual arrow points to the contractile vacuole. (B) Two types of cortical granules; type I is marked by an arrowhead and type II is marked by an arrow. (C) Two types of extrusomes. (D) Shape variants in one individual. (E) Arrangement of cortical granules on the cell surface; type I is marked by an arrowhead and type II is marked by an arrow. (F) The anterior end of the ciliary pattern. (G) Ciliary pattern of the holotype specimen; arrowhead points to a long type of extrusome, arrow points to a short type of extrusome, and red dots indicate micronuclei. (H) Shape variants in different individuals. Scale bars: 100 μm in (A,H); 5 μm in (C); 50 μm in (D,G).
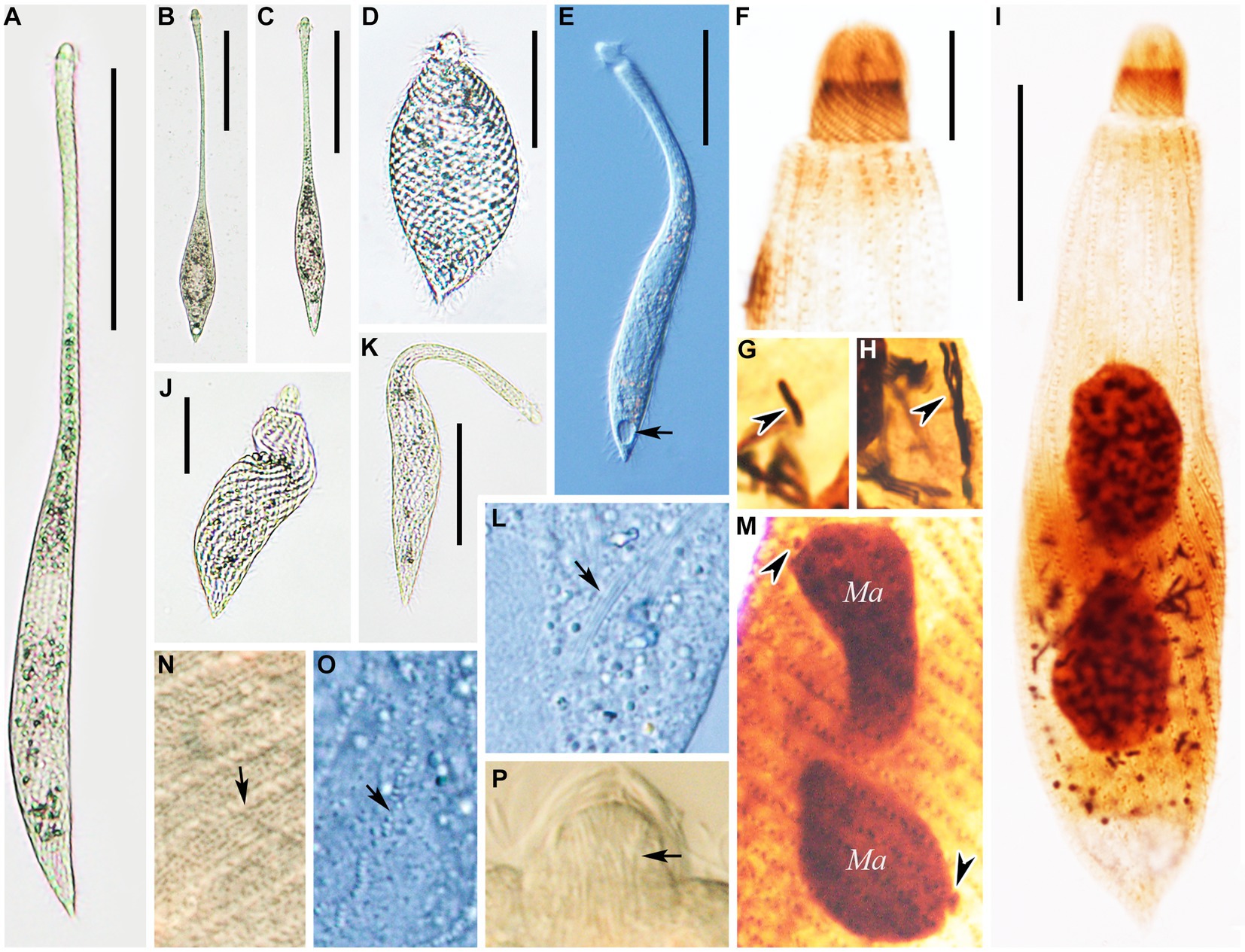
Figure 3. Photomicrographs of Lacrymaria songi sp. nov. from life (A–E,I–K,M–O) and after protargol staining (F–H,L). (A) A representative individual. (B,C) Different free-swimming individuals show shape variants. (D) A completely contracted individual. (E,J,K) Individuals in different contraction states; arrow in (E) points to the contractile vacuole. (F) Details of the anterior portion showing the anterior somatic kineties. (G) Details of the cytoplasm; arrowhead points to a short extrusome. (H) Details of the cytoplasm; arrowhead points to long extrusomes. (I) A representative specimen showing ciliature and nuclear apparatus. (L) Details of the cytoplasm; arrow points to extrusomes. (M) Details of the cytoplasm; arrowheads point to micronuclei. (N,O) Details of cytoplasm in the middle of the body; arrows point to two different cortical granules. (P) Head structure arrows point to extrusomes. Ma, macronuclear nodules. Scale bars: 100 μm in (A–C); 30 μm in (D); 50 μm in (E,H,J); 23 μm in (I); 12 μm in (F).
Two ovoidal macronuclear nodules centrally located with a filament connected to each other, each approximately 12–20 × 8–13 μm in vivo and approximately 10–32 × 6–21 μm after protargol staining (Figures 2G, 3I,M and Table 1). Two micronuclei detected only in 1 out of 30 stained individuals, respectively, located at subapical of each macronuclear nodule (Figure 2G, 3M). However, micronucleus not detected in live cells. Single contractile vacuole terminally located, variable in shape, ranging from rounded to obovate, approximately 11 × 17 μm during diastole, pulsating every 5 min (Figure 3E). Two types of extrusomes: type I approximately 10 μm long, rod-shaped, straight or slightly curved, mostly arranged in bundles, scattered in main body, and attached to oral bulge; type II approximately 4 μm long, rod-shaped, straight or slightly curved, scattered in main body (Figures 2C,F, 3G,H,L,P). Both types of extrusomes easily detected after protargol staining but only type I detectable in vivo. Two types of colorless cortical granules: type I dot-like, approximately 0.4 μm in vivo, densely arranged in five or six rows between kineties in peripheral region of cortex, this character may vary slightly with body contraction; type II dot-like to oval-shaped, approximately 0.8 μm in vivo, only distributed along somatic kineties deep in cortex (Figures 2B,E, 3N,O). Cytoplasm colorless or grayish, containing numerous globular granules (<4 μm in diameter) in trunk, rendering neck hyaline and trunk opaque (Figure 3L). Locomotion usually by swimming fast with neck swinging; when preying, neck extends forward and backward and retracts rapidly, beating approximately 92 times per minute, whereas trunk moves in a small range (Supplementary Video S1).
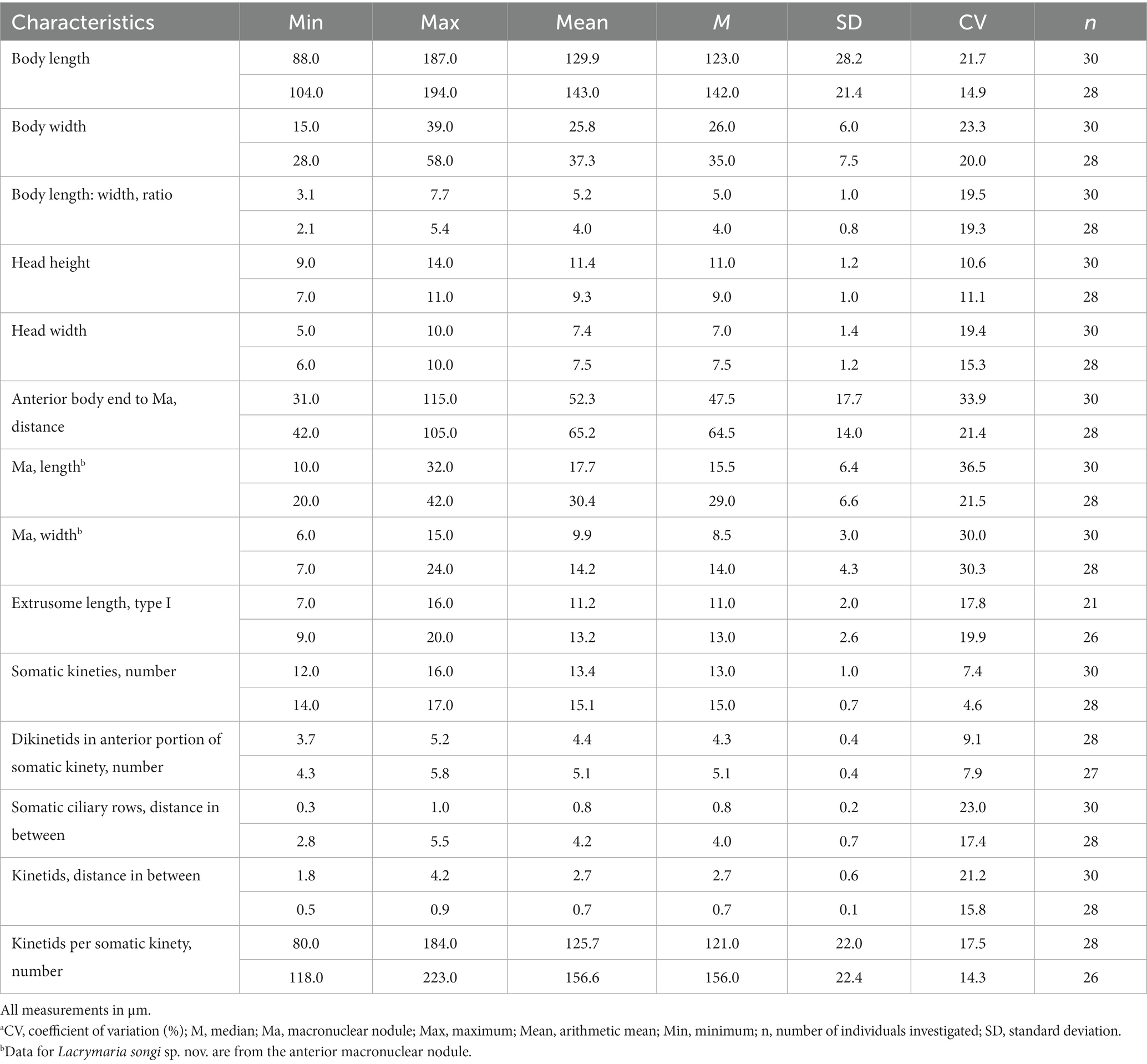
Table 1. Morphometric characteristics of Lacrymaria songi sp. nov. (the upper line) and L. dragescoi sp. nov. (the lower line) based on protargol stained specimens.a
Somatic cilia approximately 9 μm long, densely arranged in 12–16 (13 on average) somatic kineties. Somatic kineties slightly spiral in vivo when cell extended but broadly spiral in contracted individuals and protargol preparations (Figures 3A,D,I). Each kinety composed of 3–6 (4.4 on average) dorsal brush dikinetids anteriorly (Figures 2F,G, 3F), and 80–184 somatic monokinetids posteriorly with some dikinetids irregularly interspersed (Figure 2F and Table 1). Head kineties densely spirally arranged, with cilia approximately 10 μm long. Circumoral kinety is composed of approximately 28 circumoral dikinetids (Figures 2E,F).
Syn. Lacrymaria olor sensu Dragesco, 1966, pr. p. Figure 11b.
Size: approximately 210–400 × 25–35 μm in vivo. Body shape: highly variable depending on the state of contraction, ranging from a vase-shaped body in the contracted state to fusiform to clavate in the extended state. Neck: flexible, occupying half of the body length, and accounting for two-thirds of the body length when swimming, and neck beating 30 times/min when preying. Extrusomes have two types; type I—approximately 13 μm long, rod-shaped, mostly arranged in bundles, scattered in main body and attached to oral bulge; type II—approximately 3 μm long, rod-shaped, scattered in main body, and 14–17 somatic kineties. Single terminally located contractile vacuole. One macronuclear nodule. Brackish habitat.
A tidal flat of Nanhui Wetland (N30°53′5.39″, E121°53′37.03″), Shanghai, China.
A protargol slide (registration no. TJ2022090807-1) with the holotype circled in black ink and one paratype slide (TJ2022090807-2) are deposited in the Laboratory of Protozoology, Ocean University of China, Qingdao, Shandong, China.
The species is named in honor of Prof. Jean Dragesco, in recognition of his contributions to Ciliatology.
The SSU rRNA gene sequence of L. dragescoi sp. nov. has been deposited in GenBank (accession no. OR689567) with 1,642 bp long and GC content of 42.75%.
Cells: highly contractile; when fully extended, cell size of approximately 210–400× 25–35 μm in vivo and length:width ratio of 10:1 (Figures 4A, 5A,B); when contracted, cell size of approximately 100–170 × 36–44 μm and length:width ratio of 3:1 (Figures 4C, 5E). Body shape fusiform to clavate with flexible neck, occupying half of the body length and accounting for two-thirds of the body length when swimming. The posterior end tapered and tail-like when free swimming but vase-shaped with neck retracting into trunk, and posterior end sharply rounded when contracted (Figures 4A–C, 5A,D,E).
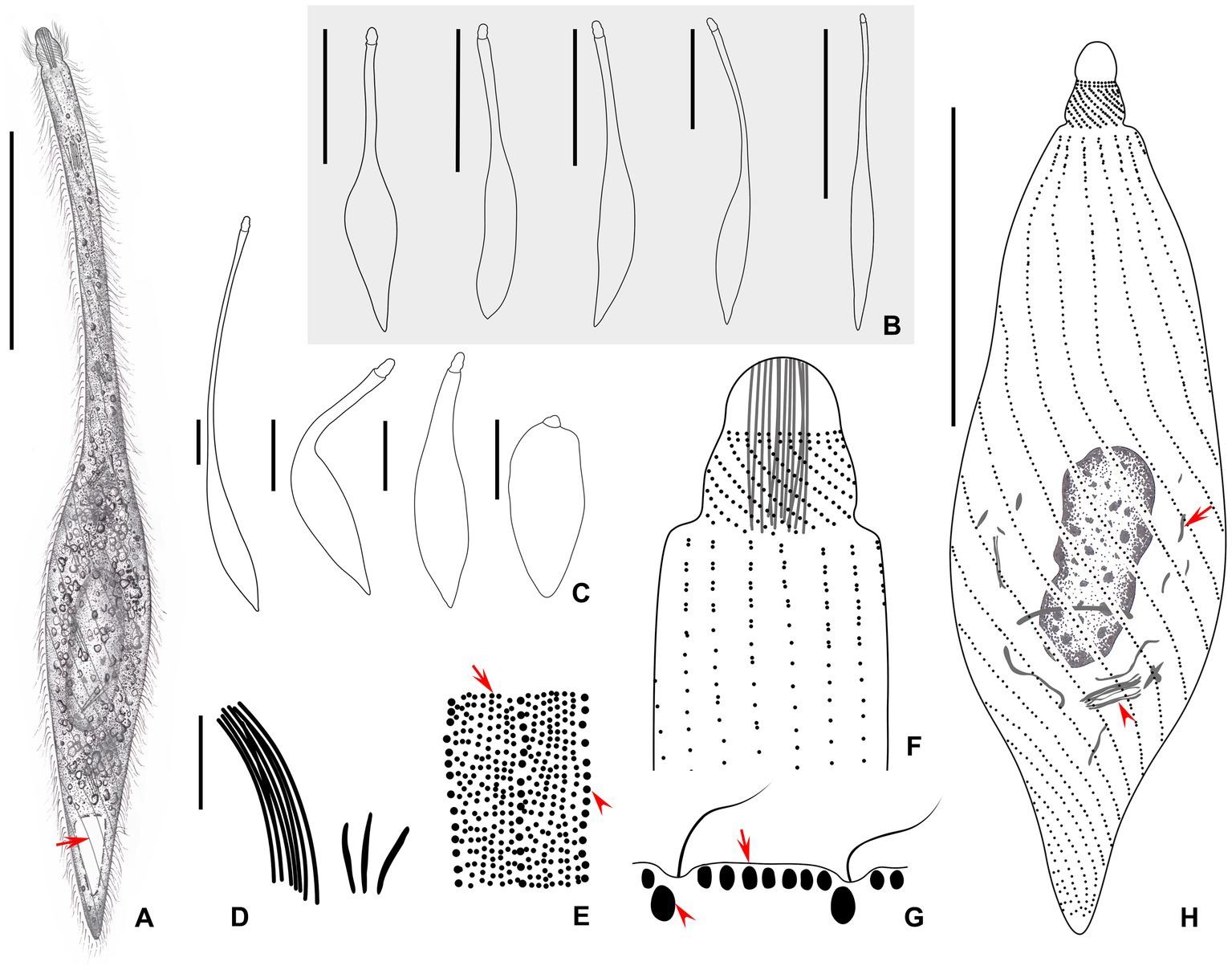
Figure 4. (A–H) Morphology of Lacrymaria dragescoi sp. nov. from life (A–C,E,G) and after protargol staining (D,F,H). (A) A typical extended individual; arrow points to the contractile vacuole. (B) Shape variants between different individuals. (C) Shape variants of the same individual. (D) Two types of extrusomes. (E) Two types of cortical granules; type I is indicated by red arrow and type II is indicated by red arrowheads. (F) Anterior end of ciliary pattern showing the head kineties and somatic kineties. (G) Arrangements of the cortical granules on the cell surface; type I is indicated by red arrow and type II is indicated by red arrowheads. (H) Ciliary pattern of the holotype specimen; arrowhead points to the long type of extrusomes and arrow points to the short type of extrusomes. Scale bars: 100 μm in (A,B); 50 μm in (C); 5 μm in (D); 25 μm in (H).
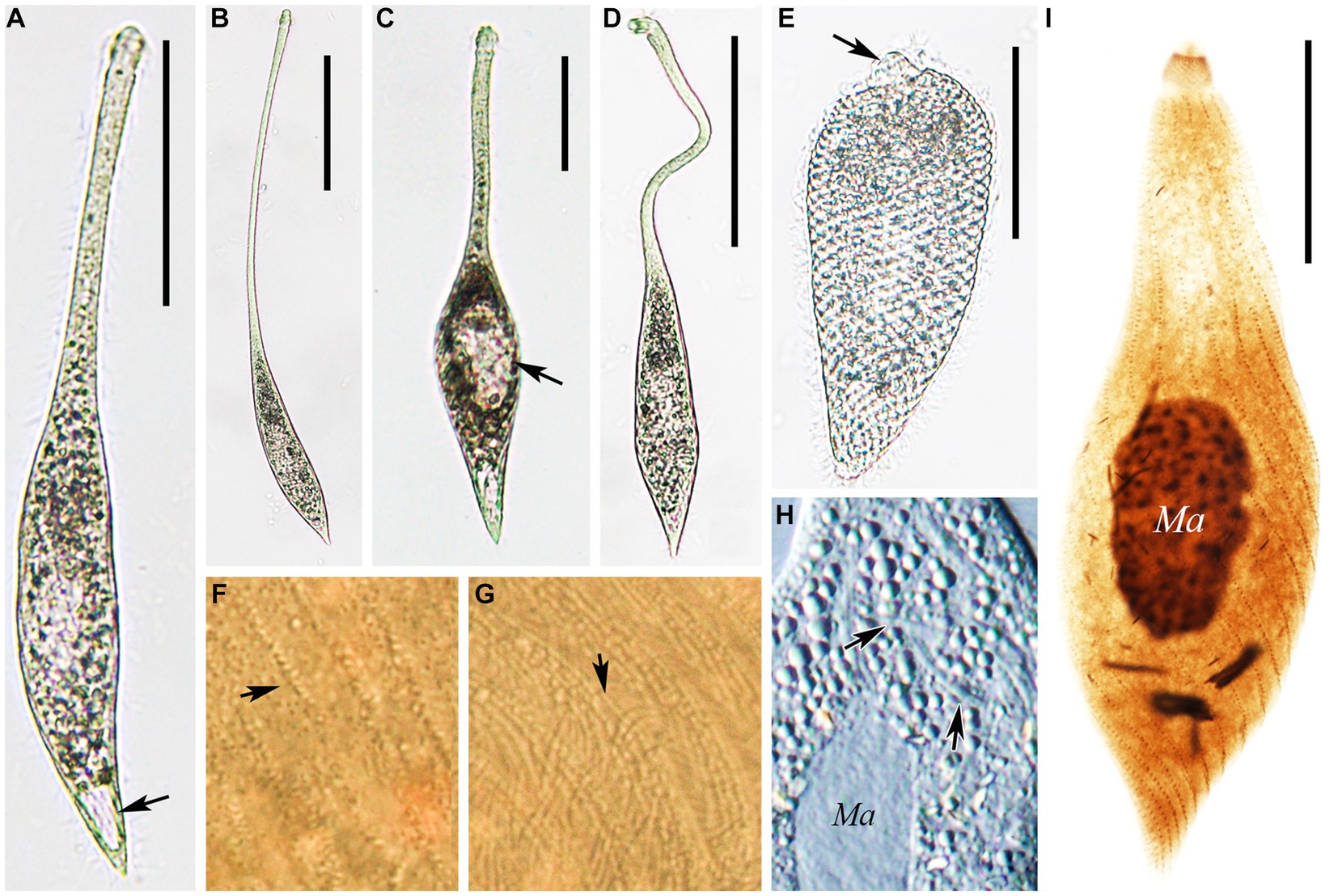
Figure 5. Photomicrographs of Lacrymaria dragescoi sp. nov. from life (A–H) and after protargol staining (I). (A) A representative individual; arrow points to the contractile vacuole. (B) An extended individual. (C) An individual that is seeking prey; arrow points to a large food vacuole. (D) An individual with a neck somewhat extended for searching for food. (E) A completely contracted individual, arrowhead infers to the head. (F,G) Details of the cell surface in the middle of the body; arrows point to the two different cortical granules. (H) Details of the cytoplasm; arrows point to extrusomes. (I) The holotype specimen showing the ciliature and nuclear apparatus. Ma, macronucleus. Scale bars: 100 μm in (A–D); 50 μm in (E); 30 μm in (I).
Nuclear apparatus centrally located, comprising one oval-shaped macronucleus, approximately 17–25 × 6–9 μm in vivo and approximately 20–40 × 7–24 μm after protargol staining (Figures 4H, 5H,I and Table 1). Micronucleus undetected in vivo or after protargol staining. Single contractile vacuole terminally located, variable in shape, ranging from rounded to obovate, approximately 17 × 12 μm during diastole, and pulsating every 3 min (Figures 4A, 5A). Two types of extrusomes: type I approximately 13 μm long, rod-shaped, straight or slightly curved, mostly arranged in bundles, scattered in main body, and attached to oral bulge; type II approximately 3 μm in size, rod-shaped, straight or slightly curved, scattered in main body (Figures 4D,F,H, 5H,I). Both types of extrusomes easily detected after protargol staining but only type I detectable in vivo. Two types of cortical granules: type I dot-like, approximately 0.4 μm in vivo, in the peripheral region of cortex densely arranged in seven or eight rows between kineties, this character may vary slightly with body contraction; type II dot-like to oval-shaped, approximately 0.7 μm in vivo, only distributed along somatic kineties deep in cortex; both types of cortical granules colorless (Figures 4E,G, 5F,G). Cytoplasm colorless or grayish, containing numerous globular granules (< 2.6 μm in diameter) in trunk, rendering neck hyaline and trunk opaque (Figure 5H). Locomotion usually by swimming fast with neck swinging; when preying, neck extends forward and backward and retracts rapidly, beating approximately 30 times per minute, whereas trunk moves in a small range (Supplementary Video S2).
Somatic cilia approximately 8 μm long, densely arranged in 14–17 (15 on average) somatic kineties. Somatic kineties slightly spiral in vivo when cell extended but broadly spiral in contracted individuals and protargol preparations (Figures 4H, 5A,E,I). Each kinety composed of 3–6 (5.1 on average) dorsal brush dikinetids anteriorly and 118–223 somatic monokinetids posteriorly with some dikinetids irregularly interspersed (Figures 4F,H, 5I and Table 1). Head kineties densely spirally arranged, with cilia approximately 8 μm long. Circumoral kinety composed of approximately 30 circumoral dikinetids (Figures 4F,I).
The nucleotide similarities of the SSU rRNA gene sequences between Lacrymaria species range from 94.45 to 99.62%. L. songi sp. nov. differs from congeners except for L. dragescoi sp. nov. by 25–66 nucleotides, with sequence identities ranging from 95.69 to 98.43%. L. dragescoi sp. nov. differs from congeners except for L. songi sp. nov. by 6–60 nucleotides, with sequence identities ranging from 96.08 to 99.62% (Figure 6).
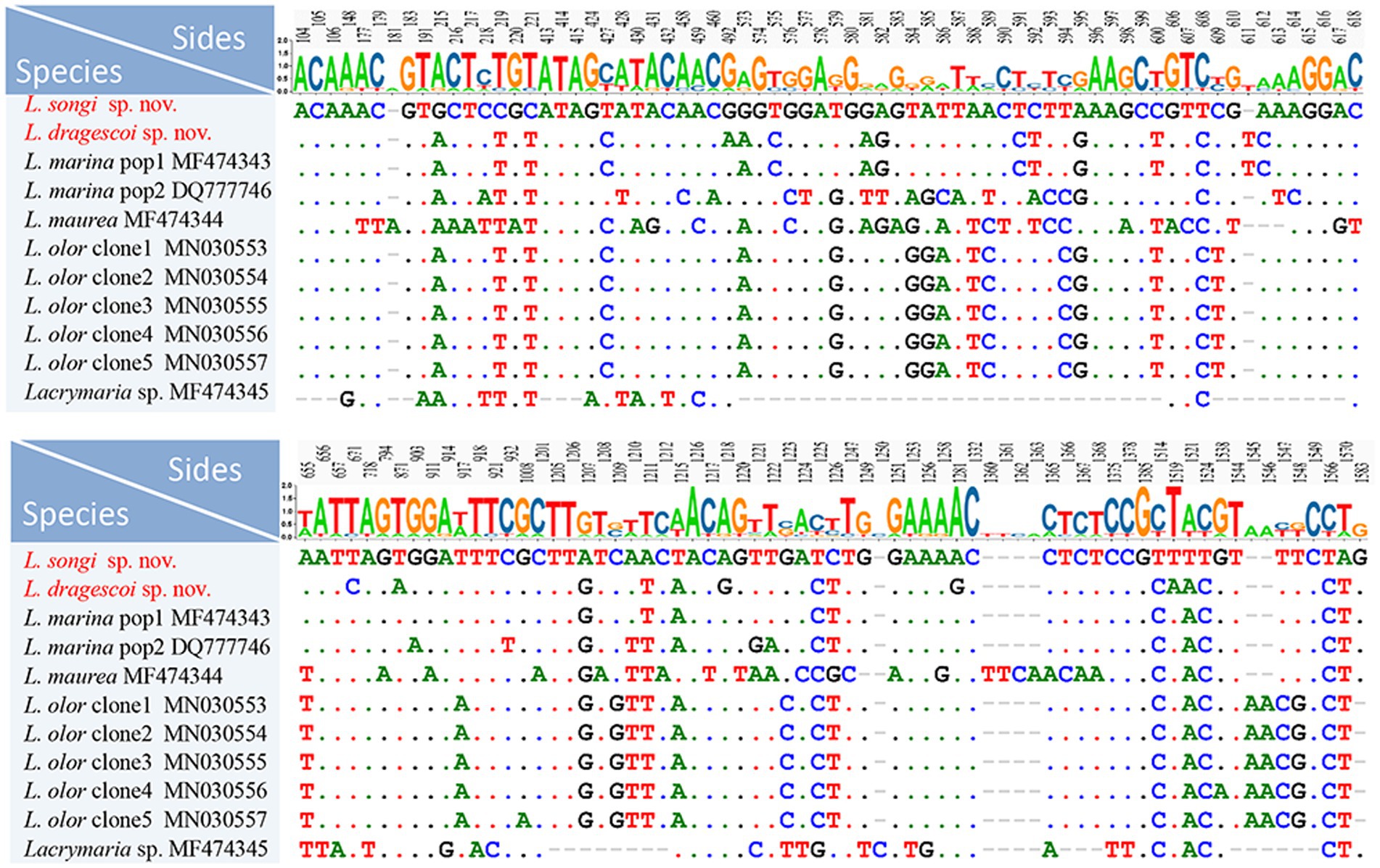
Figure 6. Nucleotide differences between Lacrymaria songi sp. nov., Lacrymaria dragescoi sp. nov., and other Lacrymaria species. Two new sequences in our present study are the first two rows. Numbers indicate the position of nucleotides. Missing sites are indicated by dashes (−).
The topologies of the ML and BI trees were basically congruent with varying levels of support; therefore, only the ML tree is presented in Figure 7. As shown in the ML tree, Lacrymaria songi sp. nov. and L. dragescoi sp. nov. fall in the core of Lacrymaria, and the family Lacrymariidae is recovered as a monophyletic group (Figure 7). The genus Lacrymaria is non-monophyletic with Lacrymaria sp. (MF474345) groups with Phialina. The AU test also refutes the monophyly of Lacrymaria (AU > 0.05). In the ML tree, L. dragescoi sp. nov. groups with Lacrymaria marina pop1 (MF474343) with high support (ML/BI, 100%/0.99), forming a sister clade to L. songi sp. nov. Then, they depict a monophyletic group that is sister to a clade formed with very weak support by Lacrymaria olor clone 1–5 (MN30553–MN30557) and Lacrymariidae (LN869967).
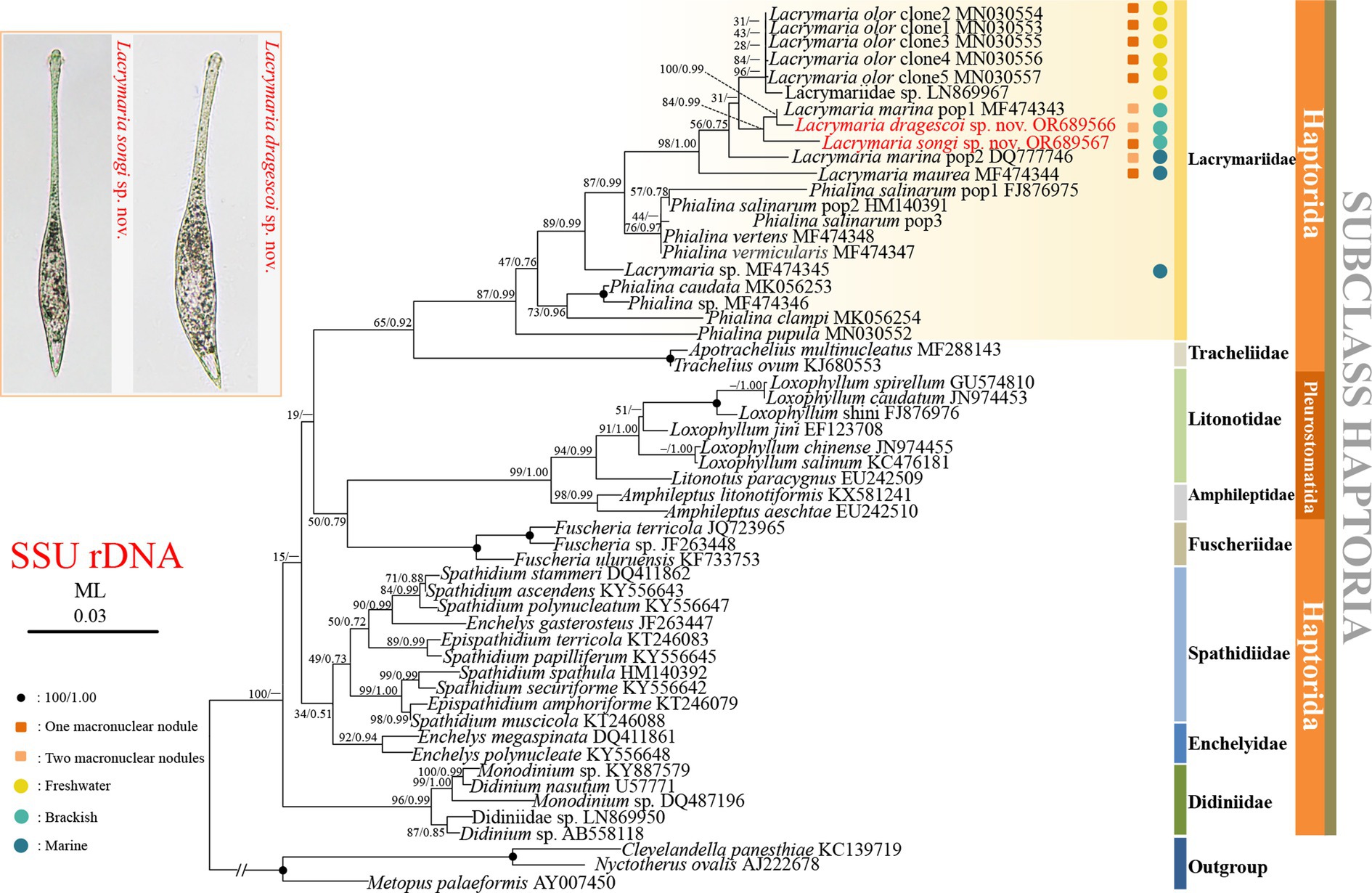
Figure 7. Phylogenetic tree based on SSU rRNA gene sequences, displaying the phylogenetic positions of Lacrymaria songi sp. nov. and L. dragescoi sp. nov. (red). Numbers near branches denote bootstrap values for maximum likelihood (ML) and posterior probabilities for Bayesian inference (BI). “–” indicates the disagreement between ML and BI trees. GenBank accession numbers are provided after species names. The scale bar corresponds to three substitutions per 100 nucleotide positions.
Lacrymaria is easily recognized by its long, contractile, and flexible neck. However, the history of Lacrymaria is marked by confusion. Both Lacrymaria and its relative Phialina were originally defined based on misinterpreted oral features. Phialina has also experienced abandonment and re-activation (Bory de Saint-Vincent, 1824; Foissner, 1983). Currently, there are approximately 53 nominal species within the genus Lacrymaria (Supplementary Table S1). However, only 12 species have been investigated through live observation and silver staining. Furthermore, those Lacrymaria species without infraciliature data have mostly not been rediscovered since their original reports raised questions about their validation and affiliations.
Lacrymaria bulbosa Alekperov, 1984, L. lanceolata Kahl, 1930, and L. ovata Burkovsky, 1970 do not possess a contractile neck (Kahl, 1930; Gelei, 1954; Burkovsky, 1970a; Alekperov, 1984). This indicates that they should be removed from Lacrymaria. There are three genera of Lacrymaria with an acontractile neck, namely, Pelagolacrymaria Foissner et al., 1999, Phialina Bory de St. Vincent, 1824, and Phialinides Foissner, 1988. In terms of ciliary patterns, L. lanceolata and L. ovata lack a monokinetid circle and a dikinetid circle between the head and the trunk. Therefore, they should be assigned to the genus Phialina as new combinations, i.e., Phialina lanceolata nov. comb. and Phialina ovata nov. comb. However, the affiliation of L. bulbosa cannot be determined presently due to its unknown ciliary pattern.
Lacrymaria sapropelica Kahl, 1927 and L. urnula Kahl, 1930 both have a furrow encircling the neck-like region, which indicates that they should belong to the family Lagynusidae (Jiang et al., 2023). Since their ciliary patterns have not been investigated, further investigation is needed to determine their exact affiliations, particularly through protargol staining and SSU rRNA gene sequencing.
The molecular phylogeny of Lacrymaria was initially investigated by sequencing the SSU rRNA gene of L. marina Kahl, 1933 (Gao et al., 2008). Subsequently, Rossi et al. (2016), Huang et al. (2018), and Rajter et al. (2019) sequenced nine new SSU rRNA gene sequences of the genus and found that Lacrymaria was likely a non-monophyletic genus. However, none of those sequences were reported with the morphological data, which cast doubt on these results.
Previous studies indicate that the following characteristics can be used for the circumscription of Lacrymaria species: the number of somatic kineties, the number of macronuclear nodules, the number and position of micronuclei, the number and position of contractile vacuoles, and characteristics of the extrusomes (Foissner, 1983; Dragesco and Dragesco-Kernéis, 1986; Song and Wilbert, 1989; Foissner et al., 1995; Rajter et al., 2019; Wang et al., 2019).
Lacrymaria olor (Müller, 1786) Bory de Saint-Vincent, 1824 resembles L. songi sp. nov. in body size, the number of somatic kineties, and the shape of the posterior end (Foissner et al., 1995). However, L. olor can be clearly distinguished from L. songi sp. nov by the location of the micronucleus (a micronucleus located between the two macronuclear nodules vs. two micronuclei located at the subapical of each macronuclear nodule) and habitat (freshwater vs. brackish water).
In terms of body length and shape, four species should be compared with Lacrymaria songi sp. nov., namely, L. clavarioides Alekperov, 1984, L. inflata Vuxanovici, 1959, L. maurea Dragesco, 1965, and L. metabolica Bünger, 1908 (Table 2). Among them, only L. inflata has a terminally located contractile vacuole, which is similar to L. songi sp. nov., but the shape of the posterior end (round vs. pointed) and the habitat of L. inflata (freshwater vs. brackish water) are different from those of L. songi sp. nov. Compared with L. songi sp. nov., L. clavarioides has more somatic kineties (20–25 vs. 12–16) and lives in a freshwater habitat (vs. brackish water), L. maurea has a rather short neck (vs. occupying up to two-thirds of the body length when swimming), and L. metabolica has a round-shaped tail (vs. pointed) and lives in freshwater (vs. brackish water) (Kahl, 1930; Vuxanovici, 1959; Dragesco, 1965; Alekperov, 1984).
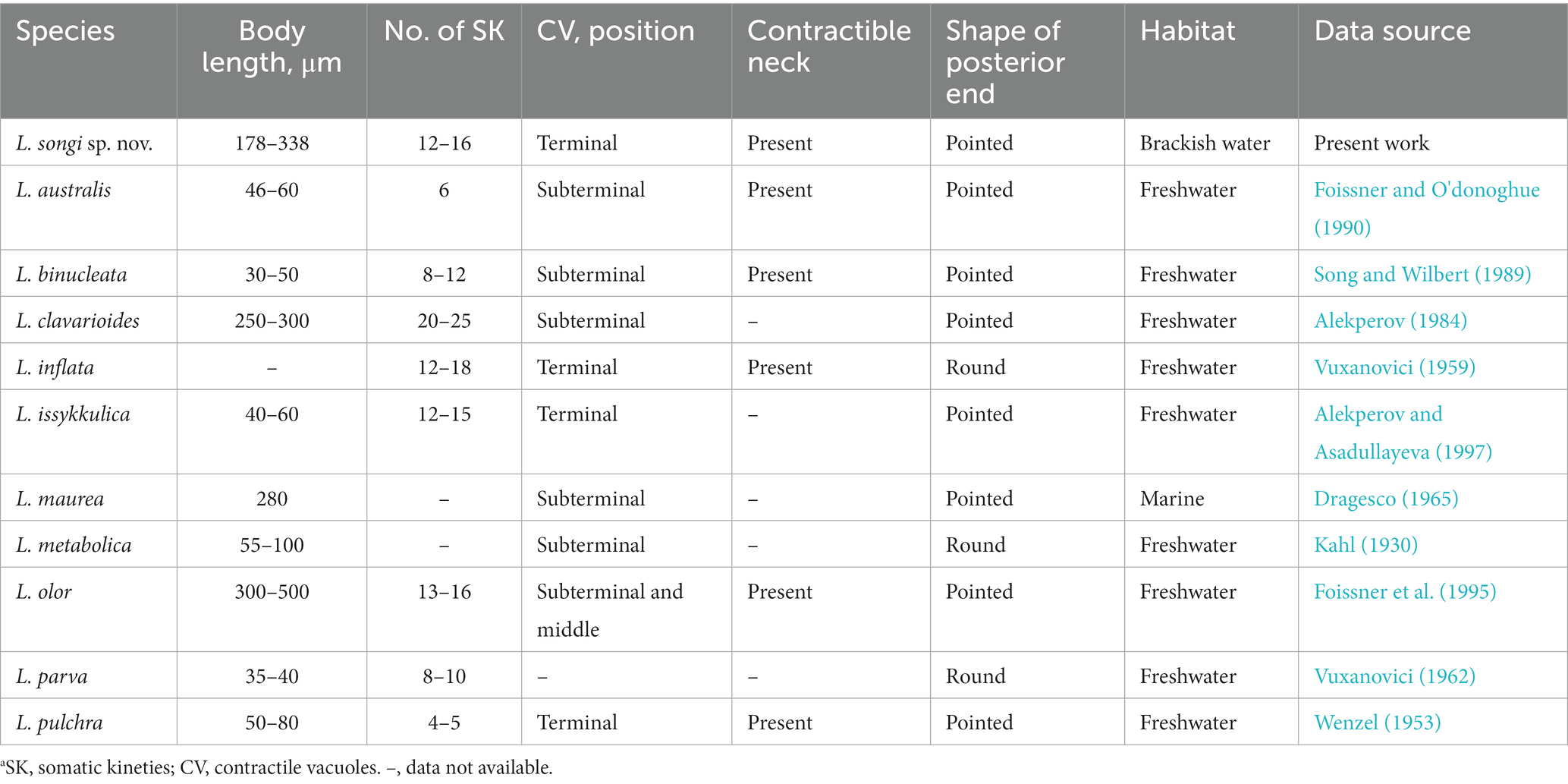
Table 2. Comparison of Lacrymaria songi sp. nov. with congeners that possess two macronuclear nodules.a
There are five more species possessing two macronuclear nodules, namely, Lacrymaria australis Foissner, 1990, L. binucleata Song and Wilbert, 1989, L. issykkulica Alekperov, 1997, L. parva Vuxanovici, 1962, and L. pulchra Wenzel, 1953. They can be separated from L. songi sp. nov. by the body size, the number of somatic kineties, and the position of contractile vacuoles (for details, refer to Table 2; Kahl, 1930; Wenzel, 1953; Vuxanovici, 1962; Song and Wilbert, 1989; Foissner and O'donoghue, 1990; Alekperov and Asadullayeva, 1997).
In terms of body length and shape, a single contractile vacuole, and a single macronucleus, 14 species should be compared with Lacrymaria dragescoi sp. nov. These species are L. acuminata Vuxanovici, 1962, L. acuta Kahl, 1933, L. affinis Bock, 1952, L. delamarci Dragesco, 1960, L. elongata Vuxanovici, 1963, L. filiformis (Maskell, 1886) Foissner, 1983, L. foliacea Vuxanovici, 1962, L. lagynus Gelei, 1954, L. marina Kahl, 1933, L. rotundata Dragesco, 1960, L. salinarum Kahl, 1928, L. trichocystus Dragesco, 1960, L. versatilis (Quennerstedt, 1865) Borror, 1963, and L. vitrea Vuxanovici, 1959.
Lacrymaria dragescoi sp. nov. closely resembles L. marina Kahl, 1933 regarding the general morphology, such as body size, body shape, habitat, and characteristics of extrusomes. However, L. dragescoi sp. nov. can be clearly distinguished from L. marina by the number of somatic kineties (14–17 vs. 19–23; on average 15 vs. 20) (Table 3 and Song and Packroff, 1997).
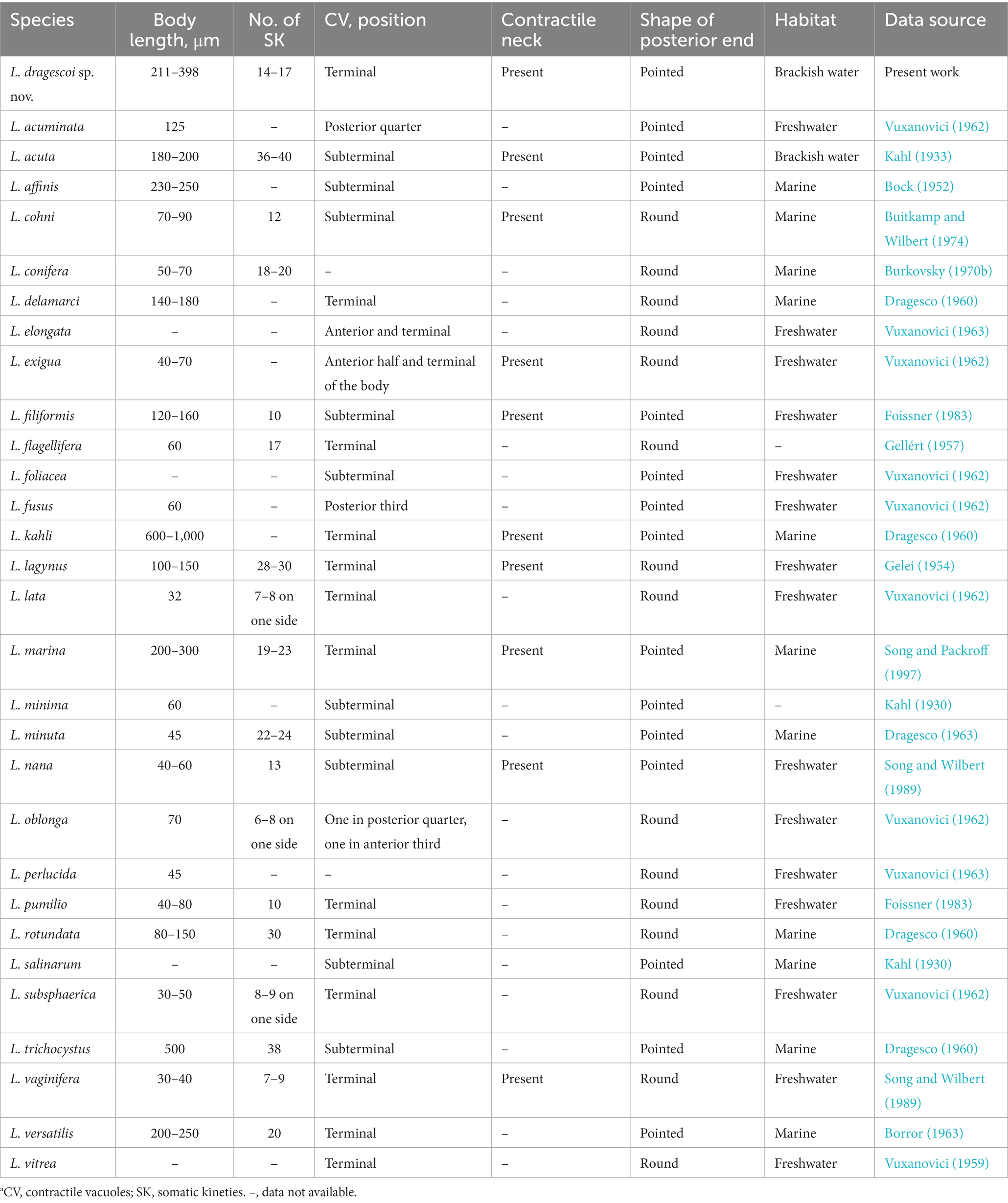
Table 3. Comparison of Lacrymaria dragescoi sp. nov. with congeners that possesses single macronucleus.a
Lacrymaria delamarci, L. lagynus, L. rotundata, L. vitrea, and L. versatilis have a single terminally located contractile vacuole, which is the same as L. dragescoi sp. nov. However, the tail shape of the former four species is round (vs. pointed in L. dragescoi sp. nov.), and L. lagynus and L. rotundata are clearly distinguished from L. dragescoi sp. nov. by the number of somatic kineties (28–30, 30 vs. 14–17). Moreover, L. vitrea differs from L. dragescoi sp. nov. by the habitat (freshwater vs. brackish water) (Gelei, 1954; Vuxanovici, 1959; Dragesco, 1960; Borror, 1963). Unlike L. dragescoi sp. nov., L. versatilis has a wider neck when extended (two-thirds of body width vs. less than one-third of body width in L. dragescoi sp. nov.), which clearly distinguishes them (Borror, 1963).
Other similar congeners with two macronuclear nodules can be distinguished from Lacrymaria dragescoi sp. nov. by the location or the number of contractile vacuoles, the habitat, and the number of somatic kineties (for details, refer to Table 3).
Lacrymaria olor sensu Dragesco, 1966, pr. p. (Figure 11b) resembles L. dragescoi sp. nov. in habitat and most morphological characteristics (Dragesco, 1966). However, the new species is smaller (211–398 μm long vs. 300–500 μm long) and has fewer somatic kineties (14–17 vs. 16–20). Since these differences cannot clearly separate them, we tentatively assign L. olor sensu Dragesco, 1966, pr. p. (Figure 11b) as a synonym of L. dragescoi sp. nov. (Dragesco, 1966).
With the addition of Lacrymaria songi sp. nov. and L. dragescoi sp. nov., the family Lacrymariidae is still monophyletic and the genus Lacrymaria is non-monophyletic, which is consistent with previous studies (Huang et al., 2018; Rajter et al., 2019; Wang et al., 2019).
Lacrymaria songi sp. nov. and L. dragescoi sp. nov. are both depicted in the core of Lacrymaria (Figure 7). L. dragescoi groups with L. marina population 1 and then clusters with L. songi sp. nov., which corresponds well with their morphological characteristics. With the addition of two new sequences, however, two populations of L. marina did not cluster together. Although the morphology of the two L. marina populations was not reported yet, we found that they are from different habitats, i.e., L. marina population 1 was collected from brackish water, while L. marina population 2 was collected from marine water. Therefore, both molecular data and the habitat imply that the two populations of L. marina are different species. Concerning the brackish habitat, L. marina population 1 is likely misidentified.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
JT: Investigation, Visualization, Writing – original draft, Writing – review & editing. GZ: Investigation, Validation, Writing – review & editing. JG: Writing – review & editing. LL: Writing – review & editing. JJ: Conceptualization, Supervision, Writing – review & editing. HP: Conceptualization, Funding acquisition, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the Natural Science Foundation of China (project number: 32170533).
The authors are grateful to Guanhua Wang, a master’s student at Shanghai Ocean University, for her help with sampling.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1259653/full#supplementary-material
Alekperov, I. K. (1984). New species of ciliates (Gymnostomata) from water bodies of Azerbaijan. Zool. Zhurnal 63, 1417–1420.
Alekperov, I. K., and Asadullayeva, E. (1997). New and little-known ciliates (orders Nassulida-Oligotrichida) from the Caspian Sea Apsheronian coast. Communication 2. Zool. Zhurnal 76, 1411–1417.
Bock, K. J. (1952). Über einige holo-und spirotriche Ciliaten aus den marinen Sandgebieten der Kieler Bucht. Zool. Anz. 149, 107–115.
Borror, A. C. (1963). Morphology and ecology of some uncommon ciliates from Alligator Harbor, Florida. Am. Microsc. Soc. 82, 125–131. doi: 10.2307/3223987
Bory de Saint-Vincent, J.B. (1824). Encyclopédie méthodique. Histoire Naturelle des zoophytes, ou animaux rayonnés, fainsant suite a l’histoire naturelle des vers de bruguière. Aggase, Paris, Tome second.
Buitkamp, U., and Wilbert, N. (1974). Morphologie und Taxonomie einiger Ciliaten eines kanadischen Prariebodens. Acta Protozool. 13, 201–210.
Burkovsky, I. (1970a). The ciliates of the mesopsammon of the Kandalaksha Gulg (White Sea). I. Acta Protozool. 7, 475–489.
Burkovsky, I. (1970b). The ciliates of the mesopsammon of the Kandalaksha Gulg (White Sea). II. Acta Protozool. 8, 47–64.
Chen, L., Ren, Y., Han, K., Stoeck, T., Jiang, J., and Pan, H. (2022). Re-description and molecular phylogeny of the free-swimming peritrichs Hastatella radians Erlanger, 1890 and H. Aesculacantha Jarocki & Jakubowska, 1927 (Ciliophora, Peritrichia) from China. Eur. J. Protistol. 84:125891. doi: 10.1016/j.ejop.2022.125891
Chi, Y., Liu, Y. J., Ma, H. G., Wang, Y., Liu, R., Al-Farraj, S. A., et al. (2022). Taxonomy and molecular phylogeny of two poorly known ciliate genera, Balantidion and Acropisthium (Protista: Ciliophora: Litostomatea), including a new species of Balantidion. Eur. J. Protistol. 85:125906. doi: 10.1016/j.ejop.2022.125906
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9:772. doi: 10.1038/nmeth.2109
Dragesco, J. (1960). Ciliés mésopsammiques littoraux. Systématique, morphologie, écologie. Trav. Stat. Biol. Roscoff 12, 1–356.
Dragesco, J. (1963). Compléments a la connaissance des ciliés mésopsammiques de Roscoff. Cah. Biol. Mar. 4, 251–275.
Dragesco, J., and Dragesco-Kernéis, A. (1986). Ciliés libres de l'Afrique intertropicale: introduction à la connaissance et à l'étude des Ciliés. Faune Tropicale 26, 1–559.
Foissner, W. (1983). Taxonomische Studien über die Ciliaten des Großglocknergebietes (Hohe Tauern, Österreich): I. Familien Holophryidae, Prorodontidae, Plagiocampidae, Colepidae, Enchelyidae und Lacrymariidae nov. fam. Ann. Nat. Hist. Mus. Wien Ser. B Bot. Zool. 84, 49–85.
Foissner, W. (1987). Miscellanea nomenclatorica ciliatea (Protozoa: Ciliophora). Arch. Protistenkd. 133, 219–235. doi: 10.1016/S0003-9365(87)80054-4
Foissner, W. (1988). Gemeinsame Arten in der terricolen Ciliatenfauna (Protozoa: Ciliophora) von Australien und Afrika. Stapfia 17, 85–133.
Foissner, W., Agatha, S., and Berger, H. (2002). Soil ciliates (Protozoa, Ciliophora) from Namibia (Southwest Africa), with emphasis on two contrasting environments, the Etosha region and the Namib Desert. Denisia 5, 1–1459.
Foissner, W., Berger, H., Blatterer, H., and Kohmann, F. (1995). Taxonomische und ökologische revision der Ciliaten des Saprobiensystems–band IV: Gymnostomatea, Loxodes, Suctoria. Informationsberichte Bayer. Landesamtes Wasserwirtschaft 1, 1–540.
Foissner, W., Berger, H., and Schaumburg, J. (1999). Identification and ecology of limnetic plankton ciliates. Informationsberichte Bayer 3, 1–793.
Foissner, W., and O'Donoghue, P. J. (1990). Morphology and infraciliature of some freshwater ciliates (Protozoa: Ciliophora) from Western and South Australia. Invertebr. Syst. 3, 661–696. doi: 10.1071/IT9890661
Foissner, W., and Xu, K. (2007). Monograph of the Spathidiida (Ciliophora, Haptoria). Vol. I: Protospathidiidae, Arcuospathidiidae, Apertospathulidae. (Dordrecht: Springer).
Gao, S., Song, W., Ma, H., Clamp, J., Yi, Z., Al-Rasheid, K., et al. (2008). Phylogeny of six genera of the subclass Haptoria (Ciliophora, Litostomatea) inferred from sequences of the gene coding for small subunit ribosomal RNA. J. Eukaryot. Microbiol. 55, 562–566. doi: 10.1111/j.1550-7408.2008.00360.x
Gelei, J. (1954). Über die Lebensgemeinschaft einiger temporärer Tümpel auf einer Bergwiese im Börzsönygebirge (Oberungarn). Acta Biol. Acad. Sci. Hung. 5, 259–343.
Gellért, J. (1957). Néhány hazai lomblevelü és tülevelü erdö talajának ciliáta-faunája Ciliatenfauna im Humus einiger ungarischen Laub-und Nadelholzwälder. Ann. Inst. biol. Tihany 24, 11–34.
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Han, K., Pan, H., and Jiang, J. (2022). Taxonomy and SSU rRNA gene-based phylogeny of two new Euplotes species from China: E. Chongmingensis n. sp. and E. Paramieti n. sp. (Protista, Ciliophora). BMC Microbiol. 22:133. doi: 10.1186/s12866-022-02543-9
He, J., Jiang, J., Agatha, S., and Pan, H. (2022). Taxonomy and phylogeny of the freshwater tintinnid Tintinnopsis tubuformis Chiang, 1956 (Ciliophora, Oligotrichea) and a proposed synonymization of T. longa nom. Corr. Chiang, 1956. J. Eukaryot. Microbiol. 69:e12918. doi: 10.1111/jeu.12918
Huang, J. B., Zhang, T., Zhang, Q., Li, Y., Warren, A., Pan, H., et al. (2018). Further insights into the highly derived haptorids (Ciliophora, Litostomatea): phylogeny based on multigene data. Zool. Scr. 47, 231–242. doi: 10.1111/zsc.12269
Jiang, L., Wang, C., Al-Farraj, S. A., Hines, H. N., and Hu, X. (2023). Morphological and molecular examination of the ciliate family Lagynusidae (Protista, Ciliophora, Prostomatea) with descriptions of two new genera and two new species from China. Mar. Life Sci. Technol. 5, 178–195. doi: 10.1007/s42995-023-00174-1
Kahl, A. (1930). Urtiere oder Protozoa. I. Wimpertiere oder Ciliate (Infusoria). Allgemeiner Teil Prostomata 18, 1–180.
Kahl, A. (1933). “Ciliata libera et ectocommensalia” in Die Tierwelt der Nord- und Ostsee Lief. eds. G. Grimpe and E. Wagler, vol. 23 (Leipzig: Akademische Verlagsgesellschaft), 29–146.
Liu, W., Jiang, J., Xu, Y., Pan, X., Qu, Z., Luo, X., et al. (2017). Diversity of free-living marine ciliates (Alveolata, Ciliophora): faunal studies in coastal waters of China during the years 2011–2016. Eur. J. Protistol. 61, 424–438. doi: 10.1016/j.ejop.2017.04.007
Lynn, D.H. (2008). The ciliated protozoa: Characterization, classification, and guide to the literature, 3rd Springer: Dordrecht.
Medlin, L., Elwood, H. J., Stickel, S., and Sogin, M. L. (1988). The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene 71, 491–499. doi: 10.1016/0378-1119(88)90066-2
Pan, X., Bourland, W. A., and Song, W. (2013). Protargol synthesis: an in-house protocol. J. Eukaryot. Microbiol. 60, 609–614. doi: 10.1111/jeu.12067
Pan, H., Zhang, Q., Dong, J., and Jiang, J. (2020). Morphology and phylogeny of two novel pleurostomatids (Ciliophora, Litostomatea), establishing a new genus. J. Eukaryot. Microbiol. 67, 252–262. doi: 10.1111/jeu.12779
Rajter, Ľ., Bourland, W., and Vďačný, P. (2019). Morpho-molecular characterization of the litostomatean predatory ciliate Phialina pupula (Müller, 1773) Foissner, 1983 (Haptoria, Lacrymariidae). Acta Protozool. 58, 53–68. doi: 10.4467/16890027AP.19.004.10835
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Rossi, A., Boscaro, V., Carducci, D., Serra, V., Modeo, L., Verni, F., et al. (2016). Ciliate communities and hidden biodiversity in freshwater biotopes of the Pistoia province (Tuscany, Italy). Eur. J. Protistol. 53, 11–19. doi: 10.1016/j.ejop.2015.12.005
Sela, I., Ashkenazy, H., Katoh, K., and Pupko, T. (2015). GUIDANCE2: accurate detection of unreliable alignment regions accounting for the uncertainty of multiple parameters. Nucleic Acids Res. 43, W7–W14. doi: 10.1093/nar/gkv318
Shao, C., Lyu, J., Li, T., Wang, J., and Lian, C. (2023). Morphology, morphogenesis and molecular phylogeny of a novel saline soil ciliate, Urosoma quadrinucleatum n. sp.(Ciliophora, Hypotrichia). Eur. J. Protistol. 88:125970. doi: 10.1016/j.ejop.2023.125970
Shimodaira, H., and Hasegawa, M. (2001). CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics 17, 1246–1247. doi: 10.1093/bioinformatics/17.12.1246
Sogin, M. L. (1989). Evolution of eukaryotic microorganisms and their small subunit ribosomal RNAs. Am. Zool. 29, 487–499. doi: 10.1093/icb/29.2.487
Sola, A. I., Guinea, A., Longas, J. F., and Frenandez-Galiano, D. (1990). Buccal and somatic infraciliature of Lagynus elegans (Engelmann, 1862) Quennersted, 1867 (Ciliophora, Prostomatea): remarks on its systematic position. J. Protozool. 37, 328–333. doi: 10.1111/j.1550-7408.1990.tb01154.x
Song, W., and Packroff, G. (1997). Taxonomische Untersuchungen an marinen Ciliaten aus China mit Beschreibungen von zwei neuen Arten, Strombidium globosaneum nov. spec. and S. platum nov. spec.(Protozoa, Ciliophora). Arch. Protistenkd. 147, 331–360. doi: 10.1016/S0003-9365(97)80059-0
Song, W., and Wilbert, N. (1989). Taxonomische Untersuchungen an Aufwuchsciliaten (Protozoa, Ciliophora) im Poppelsdorfer Weiher, Bonn. Lauterbornia 3, 2–221.
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Vd’ačný, P., Bourland, W. A., Orsi, W., Epstein, S. S., and Foissner, W. (2011). Phylogeny and classification of the Litostomatea (Protista, Ciliophora), with emphasis on free-living taxa and the 18S rRNA gene. Mol. Phylogenet. Evol. 59, 510–522. doi: 10.1016/j.ympev.2011.02.016
Vuxanovici, A. (1959). Contributii la studiul unor infuzori holotrichi. Studii Cerc. Biol. (Biol. Anim.) 11, 307–335.
Vuxanovici, A. (1962). Contributii la sistematica ciliatelor (Nota III). Studii Cerc. Biol. (Biol. Anim.) 14, 549–573.
Vuxanovici, A. (1963). Contributii la sistematica ciliatelor (Nota IV). Studii Cerc. Biol. (Biol. Anim.) 15, 65–93.
Wang, Z., Chi, Y., Li, T., Song, W. Y., Wang, Y., Wu, T., et al. (2022). Biodiversity of freshwater ciliates (Protista, Ciliophora) in the Lake Weishan wetland, China: the state of the art. Mar. Life Sci. Tech. 4, 429–451. doi: 10.1007/s42995-022-00154-x
Wang, Y., Ji, D., and Yin, J. (2019). Morphology and phylogeny of two Phialina species (Ciliophora, Haptoria) from northern China. Eur. J. Protistol. 67, 46–58. doi: 10.1016/j.ejop.2018.10.002
Weisse, T., and Montagnes, D. J. (2022). Ecology of planktonic ciliates in a changing world: concepts, methods, and challenges. J. Eukaryot. Microbiol. 69:e12879. doi: 10.1111/jeu.12879
Wilbert, N. (1975). Eine verbesserte technik der protargolimpra gnation fur ciliaten. Mikrokosmos 64, 171–179.
Wu, L., Jiao, X., Shen, Z., Yi, Z., Li, J., Warren, A., et al. (2017). New taxa refresh the phylogeny and classification of pleurostomatid ciliates (Ciliophora, Litostomatea). Zool. Scr. 46, 245–253. doi: 10.1111/zsc.12193
Wu, L., Li, J., Warren, A., and Lin, X. (2021). Species diversity of the pleurostomatid ciliate genus Amphileptus (Ciliophora, Haptoria), with notes on the taxonomy and molecular phylogeny of three species. Front. Mar. Sci. 8:642767. doi: 10.3389/fmars.2021.642767
Wu, L., Li, J., Warren, A., and Lin, X. (2022). Taxonomy and systematics of a new pleurostomatid ciliate, Pseudolitonotus spirelis gen. Et sp. n. (Protozoa, Ciliophora, Haptoria). Mar. Life Sci. Technol. 4, 31–41. doi: 10.1007/s42995-021-00123-w
Yang, Z. (1996). Among-site rate variation and its impact on phylogenetic analyses. Trends Ecol. Evol. 11, 367–372. doi: 10.1016/0169-5347(96)10041-0
Zhang, G., Chi, Y., Wang, Z., Wang, Y., Liu, R., Warren, A., et al. (2022a). Taxonomic and phylogenetic studies on two new freshwater Amphileptus species (Ciliophora, Pleurostomatida), from Lake Weishan, northern China. Eur. J. Protistol. 82:125854. doi: 10.1016/j.ejop.2021.125854
Zhang, G., Li, Y., Gong, R., Qiao, Y., Al-Farraj, S. A., Pan, H., et al. (2023). Taxonomy and molecular phylogeny of pleurostomatid ciliates from China with a description of two new species. Protist 174:125975. doi: 10.1016/j.protis.2023.125975
Zhang, G., Sheng, Y., Liu, Y., Cao, X., al-Farraj, S. A., Vďačný, P., et al. (2022b). Integrative studies on three new freshwater Amphileptus species (Ciliophora, Pleurostomatida) discovered in northern China. Mar. Life Sci. Technol. 4, 452–470. doi: 10.1007/s42995-022-00143-0
Zhang, G., Zhao, Y., Chi, Y., Warren, A., Pan, H., and Song, W. (2022c). Updating the phylogeny and taxonomy of pleurostomatid ciliates (Protista: Ciliophora) with establishment of a new family, a new genus and two new species. Zool. J. Linnean Soc. 196, 105–123. doi: 10.1093/zoolinnean/zlac028
Zhang, Z., Berger, H., Pan, H., and Jiang, J. (2022a). Two hypotrichs (Ciliophora, Hypotricha) from China: morphology and SSU rDNA sequence of Holosticha aestuarina nov. spec. And H. Muuiensis Kim et al., 2017. Eur. J. Protistol 86:125931. doi: 10.1016/j.ejop.2022.125931
Zhang, Z., Hines, H. N., Pan, H., and Jiang, J. (2022b). The description of a new brackish water ciliate species from China, Trachelostyla aestuarina n. sp., with a species key and biogeographic investigation for Trachelostyla (Ciliophora, Sporadotrichida). Front. Mar. Sci. 9:1056587. doi: 10.3389/fmars.2022.1056587
Keywords: Lacrymariidae, new combination, new species, Changjiang Estuary, SSU rRNA gene, taxonomy
Citation: Tang J, Zhang G, Guo J, Luo L, Jiang J and Pan H (2023) A new contribution to the raptorial ciliate genus Lacrymaria (Protista: Ciliophora): a brief review and comprehensive descriptions of two new species from Changjiang Estuary. Front. Microbiol. 14:1259653. doi: 10.3389/fmicb.2023.1259653
Received: 16 July 2023; Accepted: 03 October 2023;
Published: 06 November 2023.
Edited by:
Zhenzhen Yi, South China Normal University, ChinaReviewed by:
Daode Ji, Yantai Nanshan University, ChinaCopyright © 2023 Tang, Zhang, Guo, Luo, Jiang and Pan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongbo Pan, aGJwYW5Ac2hvdS5lZHUuY24=; Jiamei Jiang, am0tamlhbmdAc2hvdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.