- 1Spice and Beverage Research Institute, Chinese Academy of Tropical Agricultural Science, Wanning, China
- 2College of Agricultural Resources and Environmental Sciences, Henan Agricultural University, Zhengzhou, China
- 3Sanya Research Institute, Chinese Academy of Tropical Agricultural Science, Sanya, China
- 4Jiangsu Provincial Key Lab of Solid Organic Waste Utilization, Jiangsu Collaborative Innovation Center of Solid Organic Wastes, Educational Ministry Engineering Center of Resource-Saving Fertilizers, Nanjing Agricultural University, Nanjing, China
Coffee is an important cash crop worldwide, but it has been plagued by serious continuous planting obstacles. Intercropping with Areca catechu could alleviate the continuous planting obstacle of coffee due to the diverse root secretions of Areca catechu. However, the mechanism of Areca catechu root secretion in alleviating coffee continuous planting obstacle is still unclear. The changes of coffee rhizosphere soil microbial compositions and functions were explored by adding simulated root secretions of Areca catechu, the primary intercropping plant species (i.e., amino acids, plant hormone, organic acids, phenolic acids, flavonoids and sugars) in current study. The results showed that the addition of coffee root exudates altered soil physicochemical properties, with significantly increasing the availability of potassium and organic matter contents as well as promoting soil enzyme activity. However, the addition of plant hormone, organic acids, or phenolic acids led to a decrease in the Shannon index of bacterial communities in continuously planted coffee rhizosphere soil (RS-CP). The inclusion of phenolic acids specifically caused the decrease of fungal Shannon index. Plant hormone, flavonoids, phenolic acids, and sugars increased the relative abundance of beneficial bacteria with reduced bacterial pathogens. Flavonoids and organic acids increased the relative abundance of potential fungal pathogen Fusarium. The polyphenol oxidase, dehydrogenase, urease, catalase, and pH were highly linked with bacterial community structure. Moreover, catalase, pH, and soil-available potassium were the main determinants of fungal communities. In conclusion, this study highlight that the addition of plant hormone, phenolic acids, and sugars could enhance enzyme activity, and promote synergistic interactions among microorganisms by enhancing the physicochemical properties of RS-CP, maintaining the soil functions in coffee continuous planting soil, which contribute to alleviate the obstacles associated with continuous coffee cultivation.
1. Introduction
As a major cash crop worldwide, coffee plays a crucial role in international agricultural trade (Higashi, 2019). However, the sustainability of coffee production is at risk due to continuous cropping obstacles (CCO) which can cause a decline in coffee quality and production over time (Xiong et al., 2015; Zhao et al., 2018; Su et al., 2022). The CCO phenomenon is closely associated with deteriorating soil properties, the accumulation of allelopathic substances, and microbial imbalances (Xiong et al., 2018; Zhao et al., 2021; Li Q. et al., 2022). It is therefore imperative to investigate the interactions among root secretion, soil properties, and microbiome to maintaining the health of coffee rhizosphere soil of coffee rhizosphere soil and mitigate the negative effects of CCO on coffee production.
Continuous cultivation of coffee can decrease soil organic carbon content and porosity, proliferation of pests and diseases in the topsoil of coffee plantations, and reduced plant adaptability (Ajayi et al., 2021; Rezende et al., 2021; Ayalew et al., 2022). Under long-term continuous cultivation, the root exudates of coffee, such as phenolic compounds, can potentially have toxic effects on certain microorganisms, resulting in the changes of microbial community structure (Jurburg et al., 2020). Previous research has highlighted the significance of soil physic-chemical properties remediation and soil microbiome structure regulation as key factors in alleviating CCO (Xiong et al., 2015; Cheng et al., 2020; Zeng et al., 2020; Liu Q. et al., 2022; Wang et al., 2022). Root secretions also appear to play a critical role in regulating CCO, with intercropping crop groups increasing the diversity of root exudates and optimizing soil nutrients in intercropping ecosystems, thereby promoting positive effects on soil microbial diversity and stability (Zeng et al., 2020). Intercropping systems can alleviate CCO of crops through the improvement of soil bacteria metabolism, soil enzyme activity, and the alleviation of autotoxicity via root metabolite interactions (Cheng et al., 2020; Liu Q. et al., 2022). Consequently, the intercropping system is deemed to offer stable services for crop growth by regulating the soil microbiome and reducing allelochemicals (Guo et al., 2020). Areca catechu is a tropical cash crop that is extensively intercropping with coffee to increase the productivity per unit area in several coffee plantations (Sujatha et al., 2011). The research team were observed that coffee plants intercropped with Areca catechu exhibited more vigorous growth compared to those grown solely in previous study. However, further studies need to be conducted to investigate the mechanism of intercropping with Areca catechu as a means of alleviating coffee CCO.
It is well established that plants play a crucial role in regulating the biological and abiotic conditions of their rhizosphere soil through root secretions, which in turn can affect plant growth by modifying rhizosphere environment (Hu et al., 2018; Zhao et al., 2021; Li M. et al., 2022). Research has revealed that root secretions can act as selective agents that potentially influence the composition of the rhizosphere microbiome (Zhalnina et al., 2018; Zhou et al., 2020). Root exudates aid in nutrient acquisition by modulating the rhizosphere microbiome and facilitating plant–soil feedback to help plants fend off pathogens (Hu et al., 2018). Root exudates mediate the proliferation of specific bacterial communities primarily by targeting plant genetics and physiology to modify the composition of soil microbial communities (Fu et al., 2021; Kawasaki et al., 2021). For example, phloridzin could attract specific pathogenic microorganisms that cause sequential crop diseases (Hofmann et al., 2009); the addition of various organic acids leads to a reduction in soil microbial community richness and diversity index, ultimately leading to specific changes in microbial community structure (Ma et al., 2019); Phenolic acids can decrease soil ecosystem stability by reducing beneficial microbial communities and increasing harmful microbial communities (Huang et al., 2019). Plants are capable of adjusting the composition of their secretions at different stages of growth (Upadhyay et al., 2022). Despite the existence of various types of secretions, the effects of different secretions on soil physicochemical properties and microbial community composition remain unclear.
In this study, we conducted a field-controlled experiment to investigate the effects of primary root exudates from Areca catechu on the physicochemical properties, enzyme activity, microbial community, and their interactions in the rhizosphere soil of coffee plants. We aim to answer the following scientific questions: (1) Investigate which type of Areca catechu root exudate has a greater impact on the coffee rhizosphere microbiota; (2) Reveale the main driving factors of Areca catechu root exudate regulating coffee rhizosphere microorganisms.
2. Materials and methods
2.1. Experiment site
The experiment was conducted in the planting resource garden of the Spice Beverage Research Institute located in Hainan Province, China (110°6′ E, 18°24′ N). The soil in the experimental site showed a soil organic carbon (SOM) content of 164.2 g/kg, an alkali-hydrolyzed nitrogen content (AN) of 98.5 g/kg, a soil-available phosphorus content (AP) of 250.2 mg/kg, and a soil-available potassium content (AHK) of 84.1 mg/kg.
2.2. Experimental material
The soil was collected from a coffee plantation with a long history of monocropping coffee. The topsoil was excavated, and the coffee roots were dug out. The roots were gently shaken to remove non-rhizosphere soil, and then the roots with rhizosphere soil were placed in a bag and vigorously shaken to obtain the rhizosphere soil of the coffee. A sieve (<2 mm) was used to remove plant debris and fully homogenize it for the experiment.
According to previous studies, root exudates of Areca catechu were analyzed and determined by using liquid chromatography-mass spectrometry technology. From 6 different categories, 20 compounds with relatively high content were selected based on accessibility and relative abundance to represent the main root exudate composition of Areca catechu for subsequent experiments.
2.3. Experimental design
Eight treatments were added with different types of root secretions: sterile water (CK), 20% sterile methanol solution (KCK), 20% sterile methanol solution dissolved organic acids (OA), 20% sterile methanol solution dissolved amino acids (AMA), 20% sterile methanol solution dissolved phenolic acids (PA), 20% sterile methanol solution dissolved flavonoids (FLA), 20% sterile methanol solution dissolves sugars (SUG), 20% sterile methanol solution dissolves plant hormone (AUX). Each treatment was repeated six times (Supplementary Table S1).
A randomized block group design was used in the experiment. Each treatment was replicated six times, with each replicate consisting of 600 g of soil placed in large containers. The packaged soil was placed in coffee fields to ensure uniform environmental conditions. Before each addition, the prepared exogenous secretion solutions are adjusted to neutral pH using sodium hydroxide to eliminate the strong interference of soil pH on microbial communities. Subsequently, the solutions were added to the packaged soil in the coffee fields. The daily input of carbon from other crops in sandy soil is used to estimate the amount of root exudates secreted by Areca catechus: 0.05–0.1 mg C/g per day (Trofymow et al., 1987; Iijima et al., 2000; Baudoin et al., 2003), as few studies have reported the daily carbon input from Areca catechu tree roots into the soil. The exogenous secretion added per unit soil was 0.075 mg C/g/d. In each simulated root exudate, the carbon content of each carbon source material in the solution was equal (e.g., the carbon content of citric acid, succinic acid, and tartaric acid accounted for 1/3 of the total carbon content in each added solution, excluding the carbon content in methanol). After 6 weeks of cultivation, destructive sampling was conducted, and the soil samples were stored at −80°C.
2.4. Analysis of soil physic-chemical properties and soil enzyme activities
Soil pH was measured with a pH meter (FE28, China; soil and water ratio 1:2.5). soil-available phosphorus (AP), soil-available potassium (AK), alkali-hydrolyzed nitrogen (AHN), and soil organic matter (SOM) were separately measured by the molybdenum blue, flame photometry, potassium persulfate oxidation, and dichromate oxidation method, respectively (Lu, 1999). The concentrations of soil urease (S-UE), soil dehydrogenase (S-DHA), soil cellulase (S-CL), soil catalase (S-CAT), peroxidase (S-POD), soil polyphenol oxidase (S-PPO), soil acid phosphatase (S-ACP) and soil alkaline phosphatase (S-ALP) were determined by micro method kits from Suzhou Grace Biotechnology Co., Ltd. with the use of an enzyme marker (SynergyH1, USA).
2.5. Soil DNA extraction and sequencing
Soil microbial DNA was extracted three times from 0.5 g fresh soil by using the EZNA® Soil DNA Extraction Kit (Omega, USA). The 0.8% agarose gels was used to check the purity and quality of genomic DNA. The DNA was extracted using sequences: 338F (5′-ACTCCT ACGGGAGGCAGCA-3′) and 806R (5′-GGACTACHVGGGT WTCTAAT-3′) bacterial primers with barcode tags (Song et al., 2020). Fungal internal transcribed spacer (ITS1) genes were amplified using the ITS5F (5′-GGAAGTAAAAGTCGTAACAAGG-3′) and ITS1R (5′- GCTGCGTTCTTCATCGATGC-3′) barcode primers, with the amplification program according to the reference description (Zhou et al., 2021). The PCR amplification products were quantified by fluorescence with the Quant-iT PicoGreen dsDNA Assay Kit and a Microplate reader (BioTek, FLx800) and were used for quantification. Each sample was mixed in the corresponding ratio based on the sequencing volume requirement of each sample, based on the results of the fluorescence quantification. Sequencing libraries were prepared using the Illumina TruSeq Nano DNA LT Library Prep Kit, which was used for sequencing, followed by double end sequencing (Paired-end) of DNA fragments from the community using the Illumina platform.
2.6. Bioinformatics analysis
Microbiome bioinformatics were performed with QIIME 22019.4 (Bolyen et al., 2019) with slight modification according to the official tutorials.1 Briefly, raw sequence data were demultiplexed using the demux plugin following by primers cutting with cutadapt plugin (Martin, 2011). Sequences were then quality filtered, denoised, merged and chimera removed using the DADA2 plugin (Callahan et al., 2016). Non-singleton amplicon sequence variants (OTUs) were aligned with maff and used to construct a phylogeny with fasttree2 (Price et al., 2010). Taxonomy was assigned to OTUs using the classify-sklearn naïve Bayes taxonomy classifier in feature-classifier plugin (Bokulich et al., 2018) against the Greengenes 13_8 99% OTUs reference sequences (Mcdonald et al., 2012). Alpha-diversity metrics Shannon, beta diversity metrics weighted UniFrac (Lozupone et al., 2007) unweighted UniFrac (Lozupone and Knight, 2005), Jaccard distance, and Bray–Curtis dissimilarity were estimated using the diversity plugin with samples were rarefied to sequences per sample.
2.7. Statistical analysis
All statistical analyses were performed in R 4.2.0 and visualized with ggplot2. Principal coordinate analysis (PCoA) based on Bray-Curtis distance was used to explore differences of several root secretions on bacterial and fungal community structure, respectively. Soil physicochemical properties, soil enzyme activity, soil microbial diversity indices, relative abundance of specific microbial taxa, and relative abundance of soil microbial functional groups were analyzed using one-way ANOVA to determine differences among the different root secretions addition. The statistical significance (p < 0.05) was calculated using Tukey test. Correlations between soil properties, soil enzyme activity and soil microbial community diversity were calculated and analyzed using Spearman correlation matrices. FAPROTAX (Liang et al., 2020) and FUNGuild (Jiang et al., 2021) are commonly used to predict the functions of bacterial and fungal communities.
3. Results
3.1. Changes of soil physical and chemical properties and enzyme activities
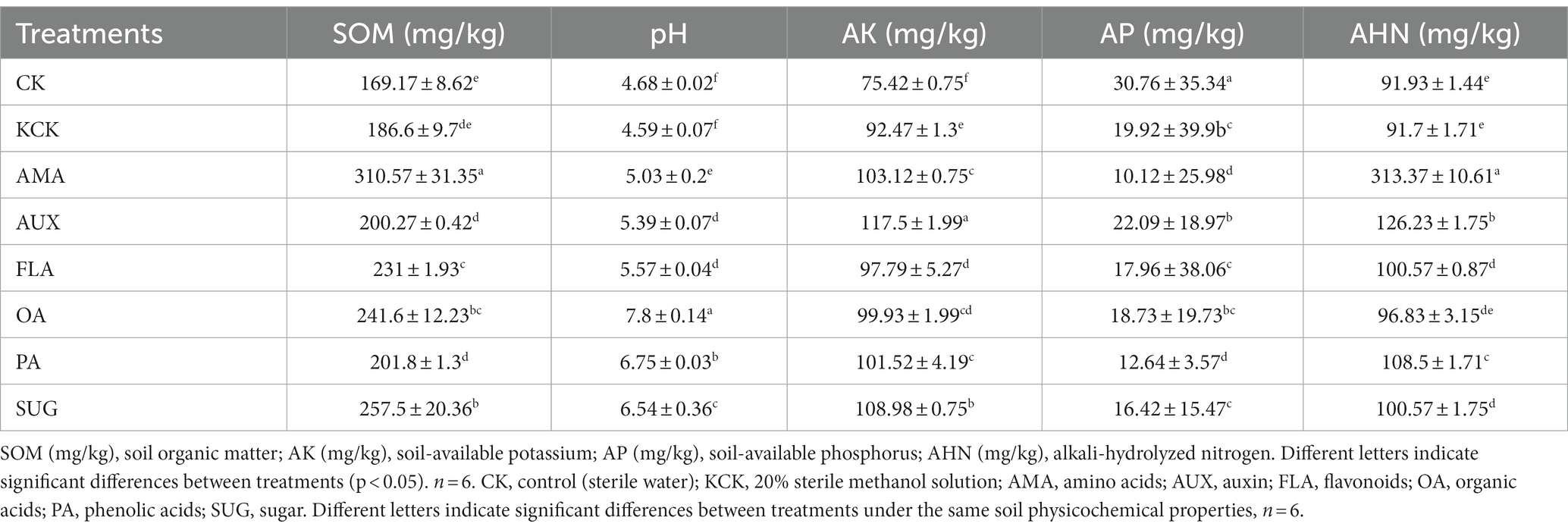
One-way ANOVA analysis revealed significant effects of exogenous secretion addition on soil properties and enzyme activities. There were significant changes in soil-available potassium and phosphorus between the CK treatment and the KCK treatment. Soil-available potassium showed a significant increase of 22.61%, while soil-available phosphorus exhibited a significant decrease of 35.24%. Compared to the control treatment (KCK), the addition of AMA significantly increased soil organic matter, alkali-hydrolyzed nitrogen, and soil-available potassium by 66.44, 241.73, and 11.52%, respectively. The soil-available potassium and alkali-hydrolyzed nitrogen levels were significantly increased by 27.06 and 37.65%, respectively, under the AUX addition treatment. Treatment with FLA significantly increased organic matter by 23.79%. The addition of OA significantly increased organic matter and pH by 29.48 and 69.93%, respectively. In the PA treatment, pH and alkali-hydrolyzed nitrogen were significantly higher by 47.06 and 18.32%, respectively. The addition of SUG significantly increased organic matter, pH, and alkali-hydrolyzed nitrogen levels. A significant increase of 38.00, 42.48, and 17.85% was observed in organic matter, pH, and soil-available potassium of the SUG treatment. However, the soil-available phosphorus content was significantly reduced by 49.20 and 36.54% in the AMA and PA, treatments, respectively (all p < 0.05, Table 1).
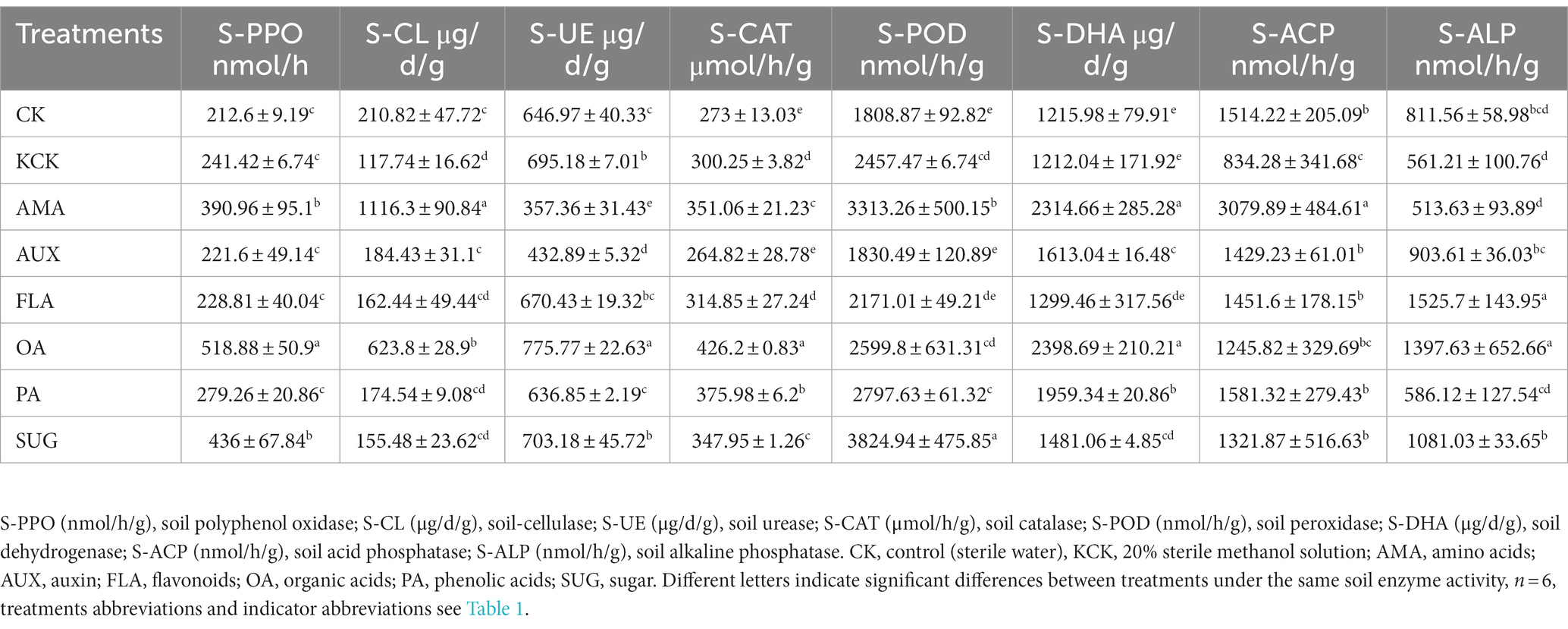
There were significant differences in the activities of cellulase, urease, hydrogen peroxide enzyme, peroxidase, and acid phosphatase between the CK treatment and the KCK treatment. The activity of urease, hydrogen peroxide enzyme, and peroxidase in the KCK treatment showed a significant increase of 6.90, 9.08, and 26.39% respectively, while the activities of cellulase and acid phosphatase exhibited a significant decrease of 79.10 and 81.50%, respectively. Compared to the KCK treatment, the activities of polyphenol oxidase, dehydrogenase, and peroxidase were significantly increased by 83.90, 90.35, and 83.17%, respectively, under the AMA addition treatment. The addition of OA increased the activities of soil polyphenol oxidase, peroxidase, and dehydrogenase by 144.07, 56.12, and 97.26%, respectively. In the PA treatment, catalase, peroxidase, and dehydrogenase activities were significantly increased by 37.72, 54.66, and 61.13%, respectively. The SUG addition significantly increased polyphenol oxidase and peroxidase activities by 105.08 and 111.45%, respectively (all p < 0.05, Table 2).
3.2. Changes in diversity indices of bacterial and fungal communities upon addition of Areca catechu main root secretions
The one-way ANOVA analysis revealed significant effects of different root secretion additions on soil microbial Shannon index. There were no significant differences in Shannon index of soil bacteria and fungi communities between the CK and KCK treatments. In the bacterial community, compared to CK, the Shannon index was significantly reduced by 25.18, 26.44, and 25.50% in the AUX, OA, and PA treatments, respectively. Additionally, compared to KCK, the AUX treatment resulted in a significant reduction of 22.20% in Shannon index. In the fungal community, the PA treatment exhibited a significant reduction of 38.38% in Shannon index compared to CK, and the FLA and PA treatments exhibited significant reductions of 15.03 and 22.54%, respectively, compared to KCK (all p < 0.05, Figure 1).
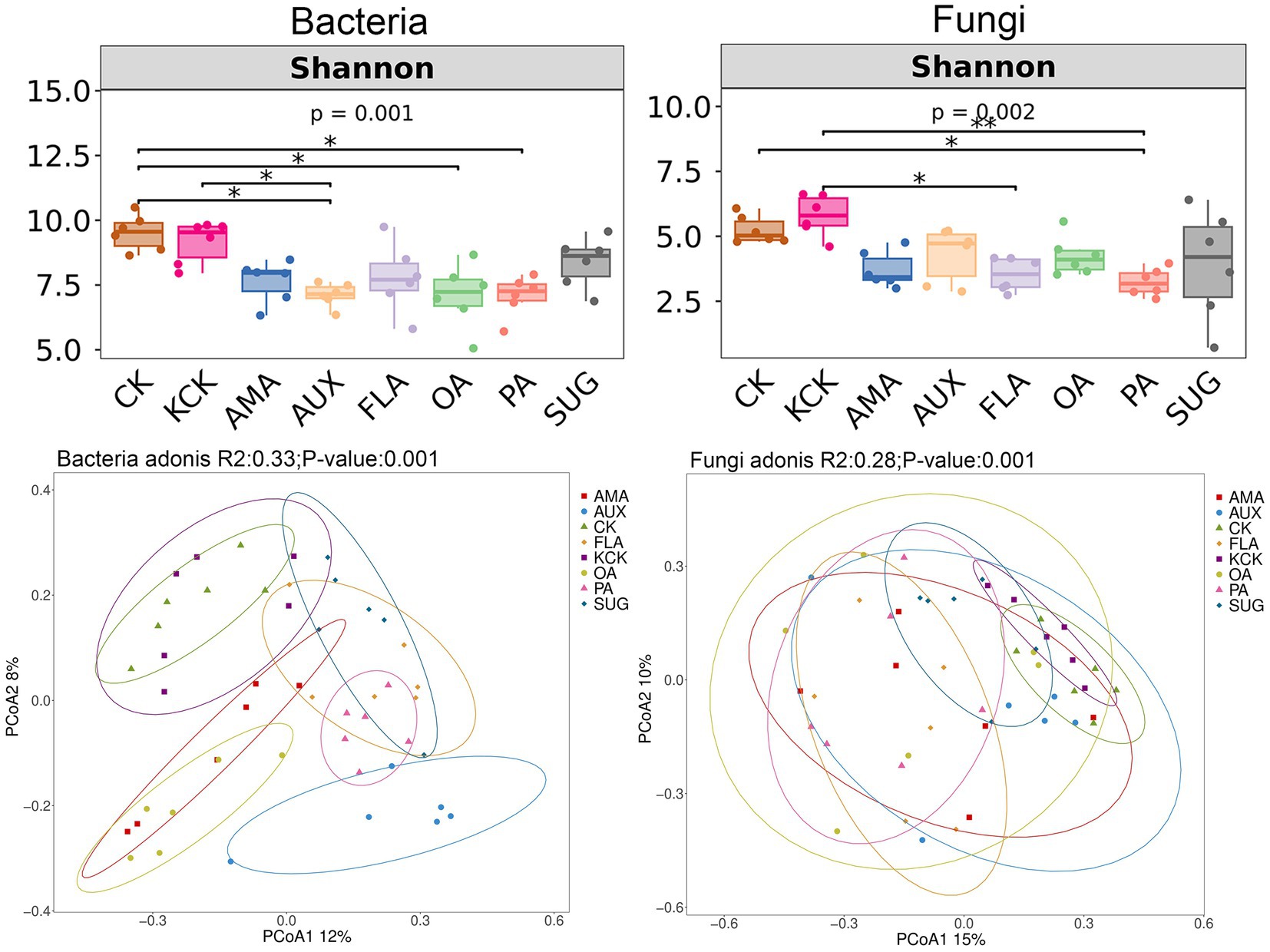
Figure 1. Diversity of bacterial and fungal communities in rhizosphere soil. n = 6. CK, control (sterile water); KCK, 20% sterile methanol solution; AMA, amino acids; AUX, auxin; FLA, flavonoids; OA, organic acids; PA, phenolic acids; SUG, sugar. * Correlation is significant at the 0.05 level; ** Correlation is significant at the 0.01 level; and *** Correlation is significant at the 0.001 level.
The bacterial community Shannon index significantly and negatively correlated with S-PPO (p < 0.05), S-DHA (p < 0.01), pH (p < 0.01), and AHN (p < 0.01). Similarly, the fungal community Shannon index was significantly and negatively correlated with S-CAT (p < 0.05), S-DHA (p < 0.01), pH (p < 0.01), SOM (p < 0.05), and AHN (p < 0.01). Soil microbial diversity indices were also significantly and negatively correlated with soil AP (p < 0.05) (Supplementary Table S3).
Principal Coordinate Analysis (PCoA) was used to exploring the dissimilarity in community composition between treatments. There were no significant differences in bacterial and fungal community structure between the CK and KCK treatments. The addition of FLA, AMA, OA, AUX, and PA treatments had a significant effect on the bacterial community structure when compared to KCK. There was no significant difference in the bacterial community structure between the SUG addition treatment and KCK treatment. Similarly, the fungal community structures under the PA and FLA addition treatments were significantly different from KCK, while there was no significant difference in the fungal community structures between the AMA, AUX, OA, SUG, and KCK treatments (all p < 0.05, Figure 1).
3.3. Effect of Areca catechu main root secretions addition on the major bacterial communities
In the CK and KCK treatments, the major bacterial communities. (relative abundance >1%) were Mycobacterium, Burkholderia-Caballeronia-Paraburkholderia, Acidothermus, Sphingomonas, Bacillus, Methylobacterium, Bradyrhizobium, and JG30-KF-AS9, and there was no significant difference in the relative abundance of these bacterial genera between the two treatments. Compared to KCK, the addition of AMA significantly reduced Bacillus and Bradyrhizobium by 72.47 and 97.00%, respectively (p < 0.001). The addition of AUX increased Burkholderia-Caballeronia-Paraburkholderia by 3866.61% (p < 0.001), while significantly reducing Mycobacterium, Acidothermus, Bacillus, and Bradyrhizobium by 67.22% (p < 0.01), 83.54% (p < 0.01), 56.24% (p < 0.01), and 90.66% (p < 0.001), respectively. In the FLA treatment, Burkholderia-Caballeronia-Paraburkholderia was significantly increased by 1513.02% (p < 0.001), while Mycobacterium was significantly decreased by 80.80% (p < 0.01). The addition of OA significantly reduced Acidothermus, Bacillus, Bradyrhizobium, and JG30-KF-AS9 by 95.59% (p < 0.01), 74.91% (p < 0.01), 67.98% (p < 0.05), and 92.83% (p < 0.05), respectively. In the PA treatment, Methylobacterium was significantly increased by 131.51% (p < 0.001), while Mycobacterium, Acidothermus, and Bacillus were significantly reduced by 96.51% (p < 0.01), 90.91% (p < 0.01), and 34.68% (p < 0.05), respectively. The addition of SUG significantly decreased Mycobacterium by 85.56% (p < 0.01) while increasing Sphingomonas by 126.12% (p < 0.001). Hyphomicrobium became the dominant genus in all treatments, except the KCK treatment, and significantly increased by 91.99% (p < 0.001) and 104.24% (p < 0.001) in the PA and SUG treatments, respectively (Figure 2; Supplementary Table S2).
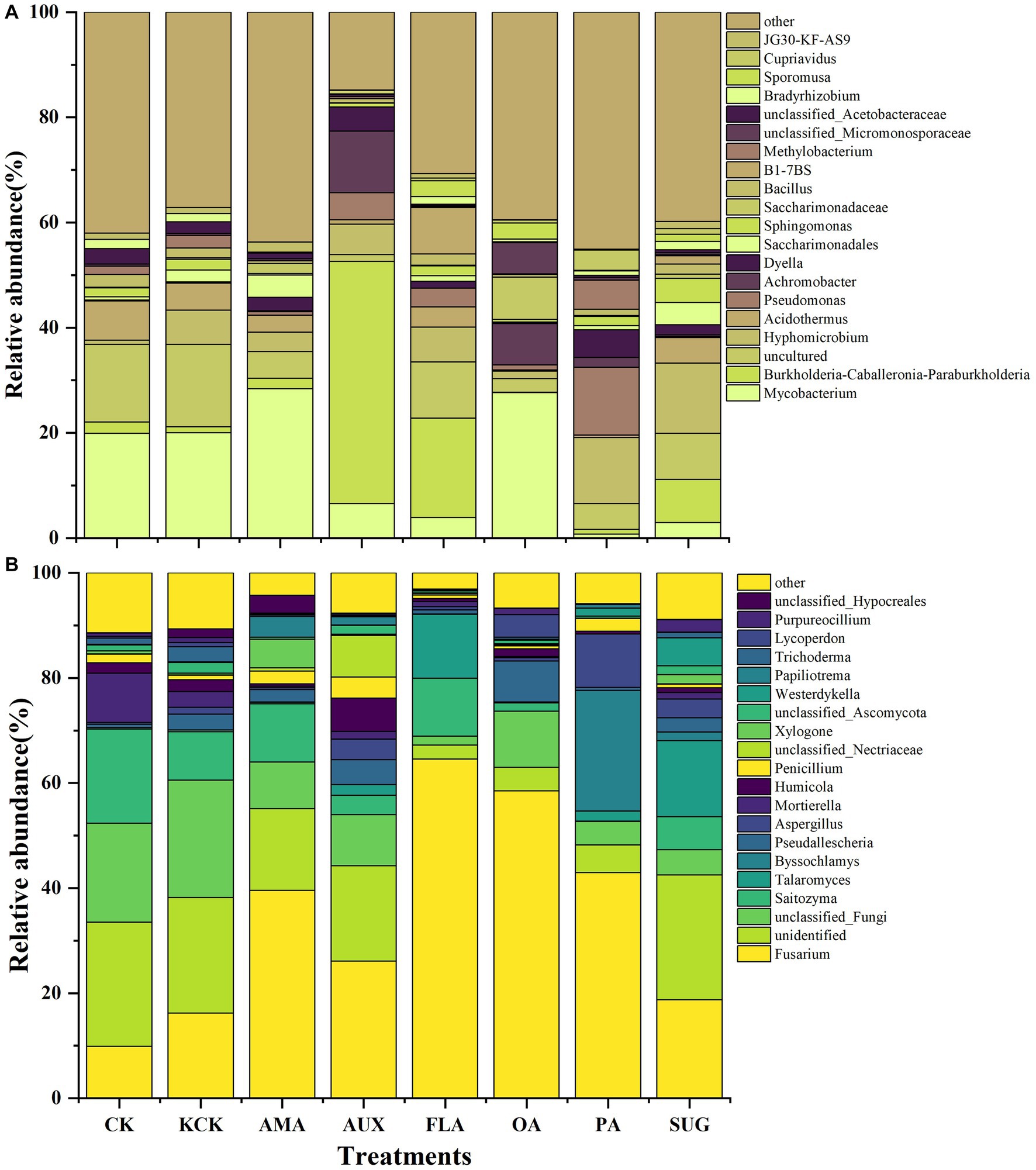
Figure 2. Relative abundance of top 20 genera of bacterial (A) and fungal (B) microbial communities in rhizosphere soil. n = 6. CK, control (sterile water); KCK, 20% sterile methanol solution; AMA, amino acids; AUX, auxin; FLA, flavonoids; OA, organic acids; PA, phenolic acids; SUG, sugar.
For the fungal community, the dominant genera were Fusarium, Saitozyma, Mortierella, Trichoderma, and Penicillium in CK and KCK treatments, and there was no significant difference in the relative abundance of these bacterial genera between the two treatments. Compared with KCK, the Mortierella treated by AMA was significantly reduced by 92.29% (p < 0.001); Saitozym and Mortierella were significantly reduced by 60.18% (p < 0.05) and 49.34% (p < 0.01) as a result of AUX treatment; Fusarium under FLA treatment was significantly increased by 298.75% (p < 0.001), Trichoderma and Mortierella were significantly decreased by 97.00% (p < 0.05) and 75.73% (p < 0.01); Fusarium treated with OA was significantly increased by 261.24% (p < 0.001), Saitozyma and Mortierella were significantly decreased by 83.06 and 99.40% (p < 0.001); Saitozyma, Mortierella was significantly reduced by 99.49 and 99.81% (p < 0.001) in the PA treatment group; Mortierella significantly reduced by 57.86% (p < 0.001) in treatment SUG (Figure 2; Supplementary Table S2).
Compared to the control group (KCK), the addition of Areca catechu main root secretions in coffee rhizosphere soil demonstrated inhibitory effects on potential pathogenic genera, such as Mycobacterium and Acidothermus, within the bacterial community. Furthermore, the inclusion of AUX, FLA, PA, and SUG led to a significant increase in the relative abundance of two potential beneficial genera, namely Burkholderia-Caballeronia-Paraburkholderia and Sphingomonas. Notably, the PA and SUG treatments resulted in the emergence of a new dominant potential beneficial genus, Hyphomicrobium. Within the fungal community, the FLA and OA treatments significantly elevated the relative abundance of the potential pathogenic genus Fusarium. Additionally, the overall relative abundance of the potential beneficial genus Mortierella experienced a significant reduction across all treatments (Supplementary Tables S2, S3).
3.4. Correlation of soil physicochemical properties and enzyme activities with microbial communities
Significant positive correlations were found between soil SOM and S-PPO (R = 0.60, p < 0.001), S-CAT (R = 0.56, p < 0.01), S-POD (R = 0.56, p < 0.001) and S-DHA (R = 0.61, p < 0.001), and significant negative correlations were found with S-UE (R = 0.01, p < 0.05). pH and S-PPO (R = 0.64, p < 0.001), S-CAT (R = 0.72, p < 0.001), S-POD (R = 0.39, p < 0.01) and S-DHA (R = 0.53, p < 0.001). AP exhibited significant negative correlations with several indicators including S-PPO (R = −0.55, p < 0.01), S-CL (R = −0.23, p < 0.01), S-CAT (R = −0.62, p < 0.001), S-POD (R = −0.73, p < 0.001), S-DHA (R = −0.45, p < 0.01), and S-ACP (R = −0.28, p < 0.01). The inter-root soil bacterial community genus level structure had a significant response to soil S-PPO, S-UE, S-CAT, S-DHA, and pH, and the fungal genus level structure had a significant response to soil S-CAT, pH, and AK (Figure 3). This study demonstrates that there is a close correlation between soil physicochemical factors as well as between physicochemical factors and soil microbial community structure.
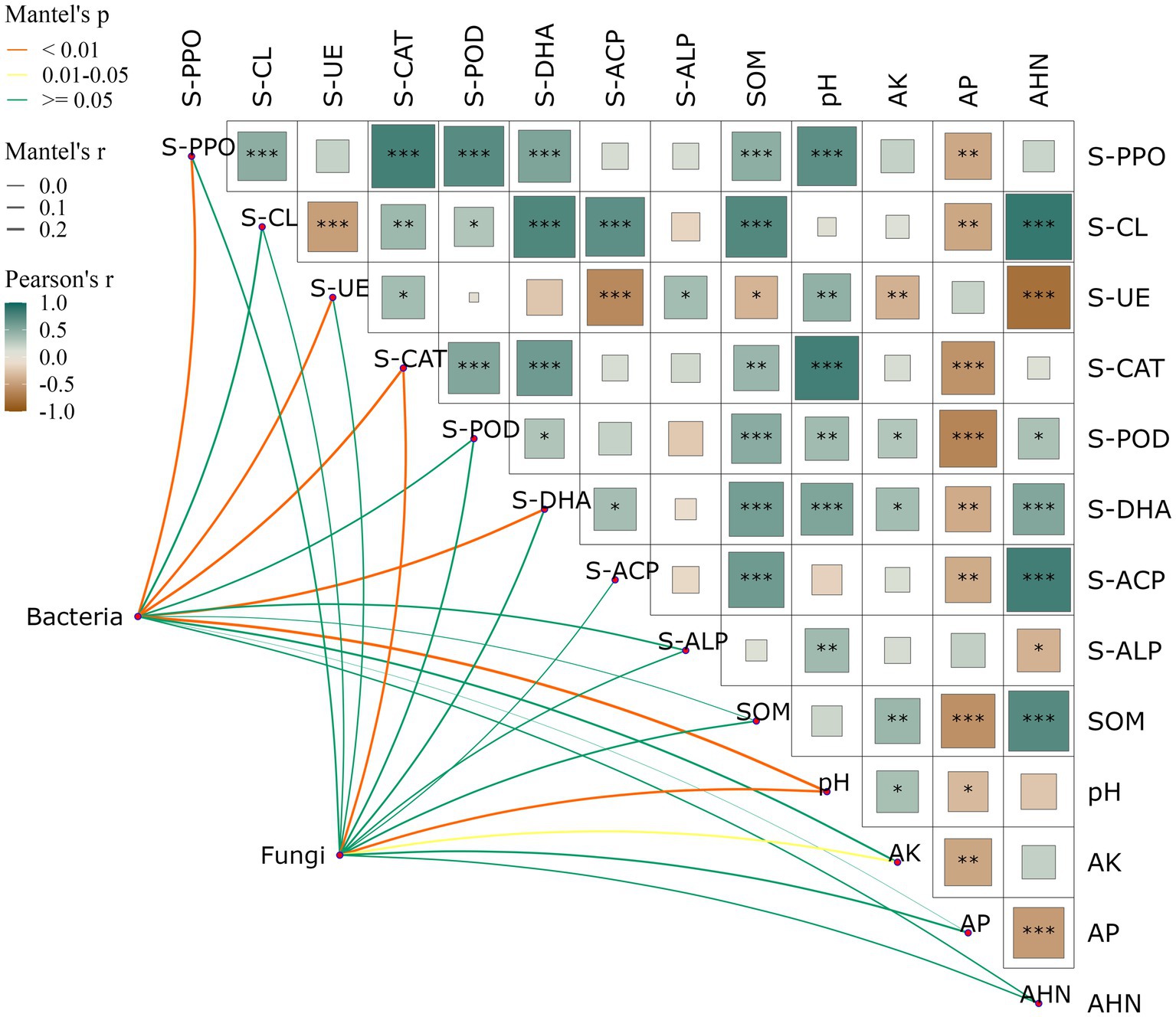
Figure 3. Effects of soil physic-chemical and enzyme activities on relative abundance of bacterial (A) and fungi (B) communities at the genus level. Heatmap showing the correlation between soil properties, network diagram shows the correlation between soil microbial community and soil properties. Red represents positive correlation, blue represents negative correlation, the darker the color, the stronger the correlation. The symbol “*” indicates statistical significance at p < 0.05, “**” indicates p < 0.01, and “***” indicates p < 0.001.
There was a strong correlation between soil physicochemical factors and enzyme activities and the dominant genus of rhizosphere bacteria (Figure 4). For the relative abundance of dominant genera in the bacterial community, there was a significant positive correlation between Mycobacterium and S-CL (R = 0.32), and a significant negative correlation with pH (R = 0.35), AK (R = 0.42), and AHN (R = 0.31); Burkholderia-Caballeronia-Paraburkholderia was negatively correlated with S-PPO (R = 0.47), S-CAT (R = 0.64), S-POD (R = 0.35) and pH (R = 0.31), and positively correlated with AHN (R = 0.29); Hypomicrobium has a significant negative correlation with AK (R = 0.45); Acidothermus was negatively correlated with S-PPO (R = 0.39), S-CAT (R = 0.40), S-DHA (R = 0.48), pH (R = 0.51) and AHN (R = 0.32); Sphingomonas showed significant negative correlations with S-DHA (R = 0.55) and AHN (R = 0.50); For the relative abundance of dominant genera in fungal communities, there was a significant positive correlation between Fusarium and S-CAT (R = 0.34), SOM (R = 0.30), pH (R = 0.31), and a significant negative correlation with AP (R = 0.30). Mortierella and S-PPO (R = 0.35), S-CAT (R = 0.54), S-POD (R = 0.35), S-DHA (R = 0.49), SOM (R = 0.45), pH (R = 0.57), AK (R = 0.31), AHN (R = 0.44) were significantly negatively correlated with each other and positively correlated with AP (R = 0.62).
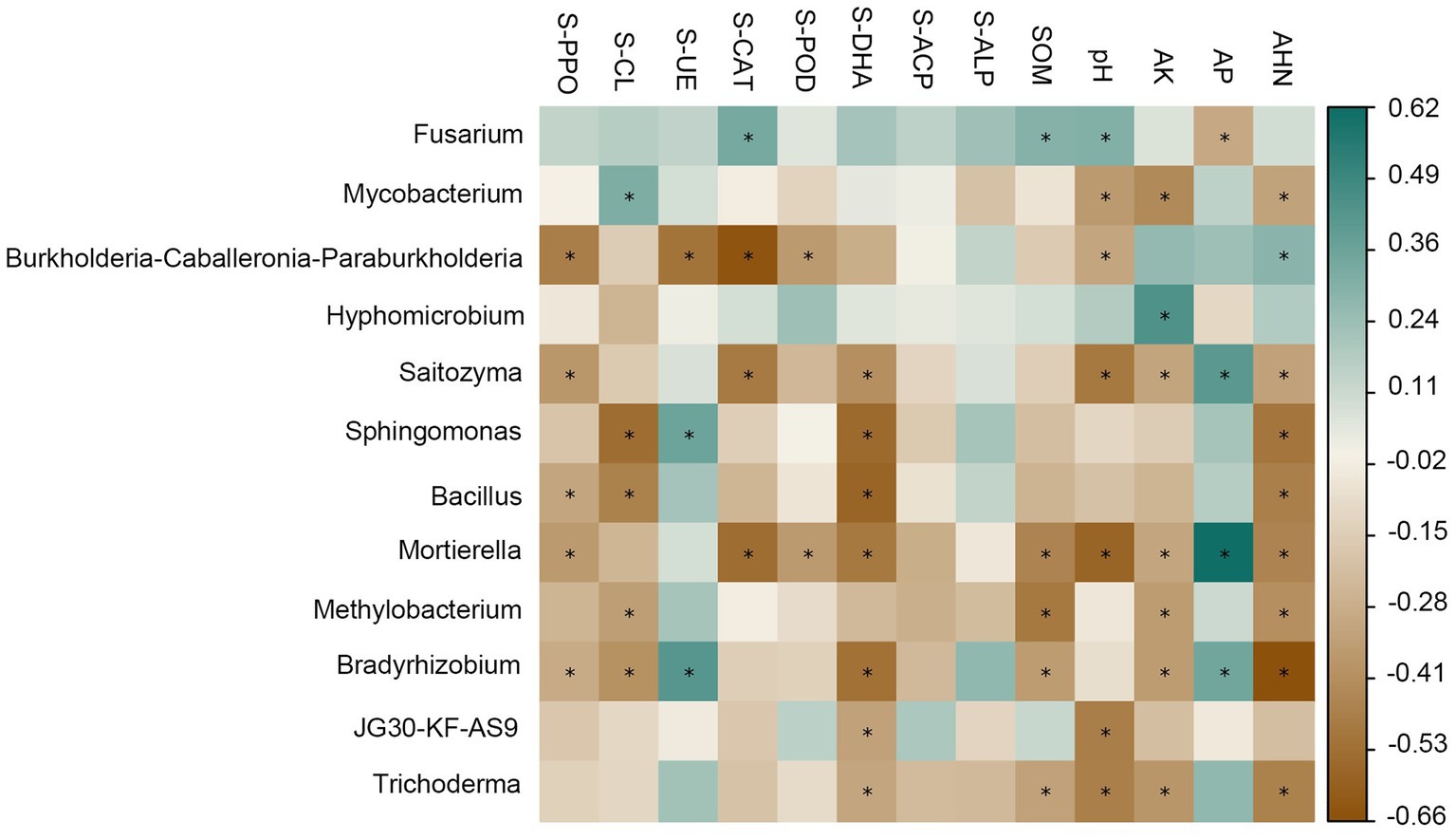
Figure 4. Relationships among soil physic-chemical, enzyme activities and dominant genera relative abundance of bacterial and fungal communities at the genus level. Cyan represents positive correlation, brown represents negative correlation, the darker the color, the stronger the correlation. The symbol “*” indicates statistical significance at p < 0.05.
3.5. Functional predictions
The functional prediction of the soil bacterial community revealed that the main gene functional groups in each treatment were chemoheterotrophy and aerobic_chemoheterotrophy, which accounted for 66.39% of the total. There were no significant changes in the percentages of these functional groups after the addition of exogenous secretions. In the case of the fungal community, there were no differences between the functional communities of the fungi under the CK and KCK treatments. The Undefined Saprotroph also significantly increased by 310.21% in the PA treatment. The addition of various compounds did not significantly alter the relative abundance of the Plant Pathogen and Fungal Parasite functional groups. However, Animal Pathogen, Endophyte, Wood Saprotroph, and Lichen Parasite showed a significant increase in relative abundance in the OA treatment, by 72.90, 35.86, 64.99, and 125.78%, respectively, (all p < 0.05; Figure 5; Supplementary Table S4).
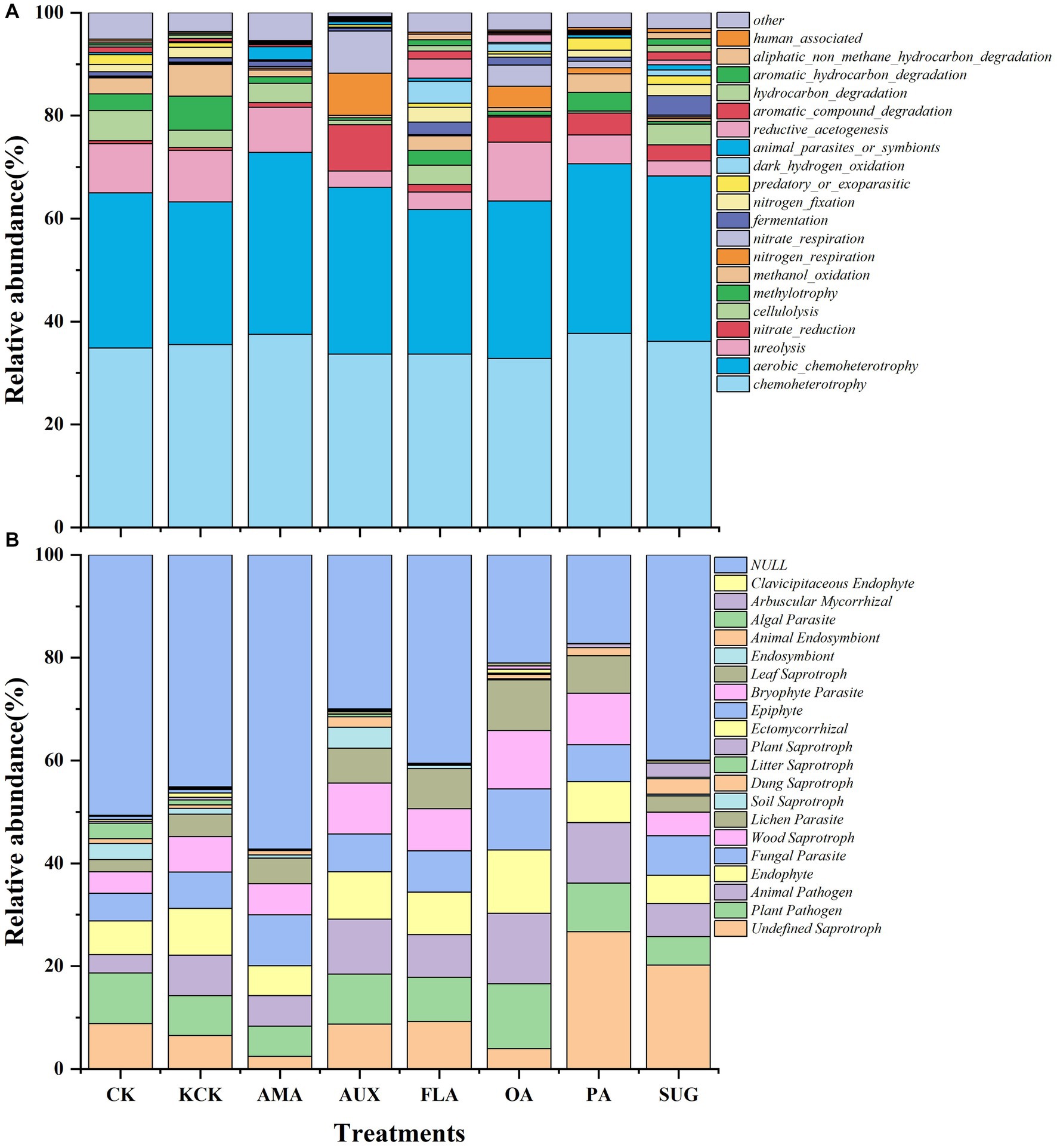
Figure 5. Effects of different root secretions on relative abundance of the top 20 functional genes in soil bacteria (A) and fungi (B) communities. n = 6. CK, control (sterile water); KCK, 20% sterile methanol solution; AMA, amino acids; AUX, auxin; FLA, flavonoids; OA, organic acids; PA, phenolic acids; SUG, sugar.
Based on the functional annotations of the samples in the database and the abundance information, a heat map was created to show the top 20 functions in terms of abundance and their abundance information in each treatment (Figure 6). Among the bacterial functional groups, the cluster analysis revealed that CK, KCK, PA, AMA, AUX, and OA treatments had similar functional community composition, while treatments FLA and SUG were similar but had higher abundance of gene functions such as automatic_compound_degradation, hydrocarbon_degradation, and automatic_hydrocarbon_degradation. Nitrate_reduction, methylotrophy, and methanol_oxidation genes had higher relative abundance in other treatments. In the fungal functional groups, the functional community composition of treatments CK, KCK, and SUG were grouped together, while the other treatments were grouped together.
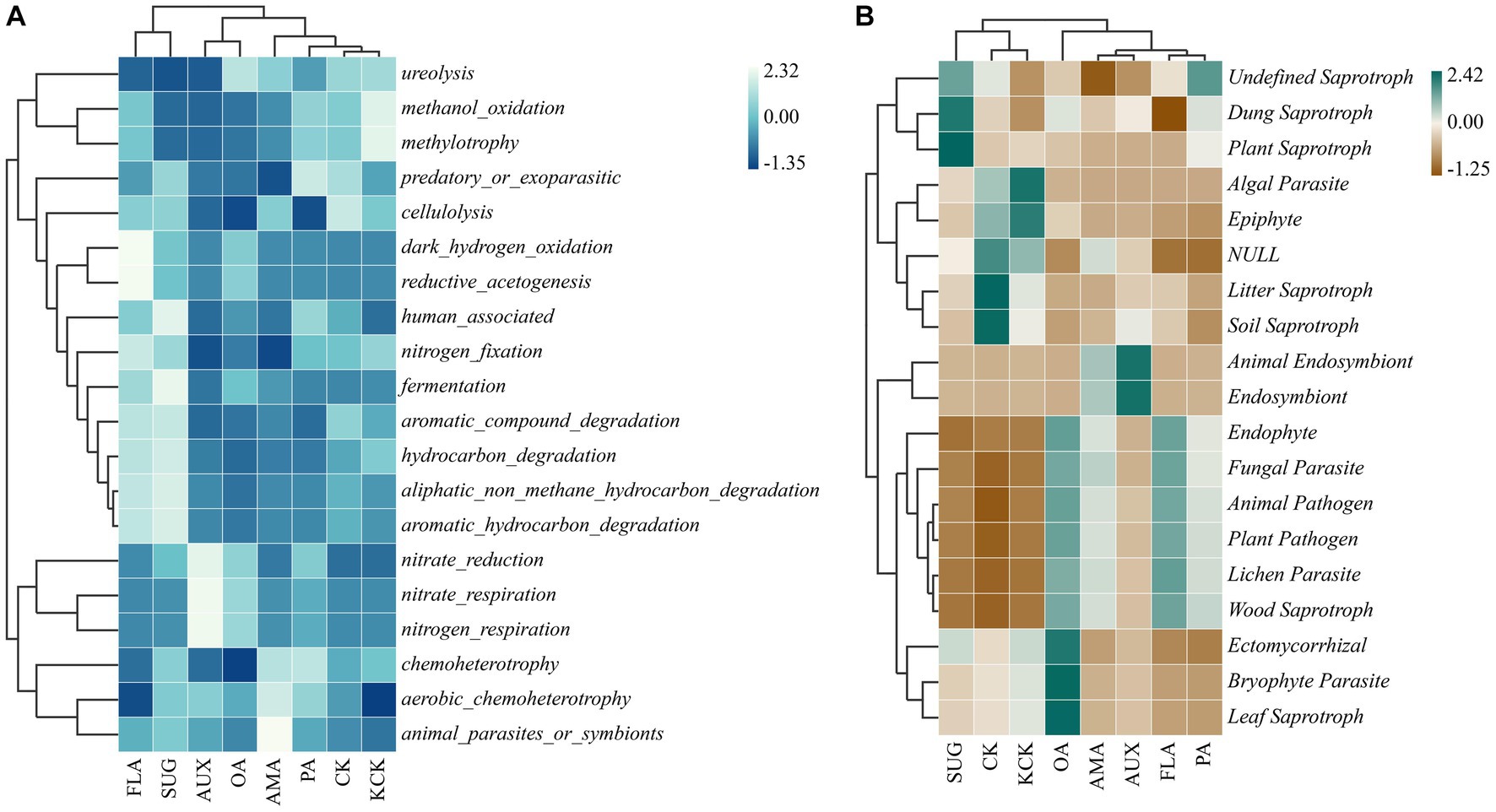
Figure 6. Clustering heat map of the functional diversity of rhizosphere microbial genes, bacterial (A) and fungal (B). n = 6. CK, control (sterile water); KCK, 20% sterile methanol solution; AMA, amino acids; AUX, auxin; FLA, flavonoids; OA, organic acids; PA, phenolic acids; SUG, sugar.
4. Discussion
4.1. Effects of different root secretions on soil physic-chemical properties and enzyme activities
Soil enzymes play an important role in the degradation and transformation of soil nutrients such as alkali-hydrolyzed nitrogen, and also participate in other physiological and biochemical processes. The activity and diversity of soil enzymes can have a significant impact on the soil physico-chemical properties, such as nutrient cycling, soil structure, and organic matter decomposition (Jing et al., 2020). On one hand, the findings of this study indicate that the addition of compounds significantly increased the activities of S-PPO, S-DHA, S-POD, and S-CAT, suggesting that the root secretions of Areca catechu mainly affect redox enzymes (Table 2). Specifically, S-PPO can accelerate the soil humification process, while S-DHA, S-POD, and S-CAT have been found to play crucial roles in soil bio-oxidation (Tan et al., 2017; Telesiński et al., 2019; Li Q. et al., 2022). The four soil enzyme activities mentioned above exhibited a significant positive correlation with soil organic matter (SOM) and pH in this study (Figure 3) supporting the notion that oxidoreductases in lichen can catalyze the formation of alkali-hydrolyzed nitrogen (Beckett et al., 2013). The present study demonstrates that crop root secretions, an important driver of microbial activity and a steady source of soil organic carbon, exert a robust regulatory effect on both soil physicochemical and enzymatic activities (Yan et al., 2023). Through a series of biochemical reactions, various types of secretions are transformed into the nutrients necessary for organisms, providing diverse sources of carbon for the soil microbiome. In the present study, the addition of different root secretions led to significant increases in pH, soil organic matter (SOM), available potassium (AK), and alkali-hydrolyzable nitrogen (AHN), as confirmed by the results presented in Table 1. Interestingly, the noticeable decrease in available phosphorus (AP) observed in this study could be related to the increased activity of catalase. Prior research has indicated that catalase is a key factor that inhibits the conversion of soil organic phosphorus to effective phosphorus (Strickland et al., 2015). Under the absence of exogenous phosphorus interference, the increase in catalase activity led to a significant reduction in soil effective phosphorus content, as demonstrated in this study (Wang et al., 2012). The results of this study suggest that acid phosphatase activity was not affected significantly by the addition of root secretions, which may correspond to a low rate of available phosphorus (AP) accumulation in the soil (Liu et al., 2023).
4.2. Effect of different root secretions on the diversity of soil microbial communities
As a crucial mediator of interaction between crop and soil microbial communities, the alterations in both the type and quantity of root secretions could markedly impact the diversity of soil microbial communities (Zhalnina et al., 2018). Root secretions have complex and diverse effects on microbial diversity, potentially simultaneously influencing multiple microbial groups. However, specific compounds in root secretions, such as plant hormones, have been indicated to directly impact microbial growth and metabolism, thereby influencing microbial diversity (Vives-Peris et al., 2020; Zhao et al., 2021). Some flavonoid and organic acid compounds found in root secretions are only mineralized by specific microorganisms, while most soil microorganisms are unable to utilize them, thus affecting soil microbial diversity (Qu and Wang, 2008; Eilers et al., 2010).
The results from Supplementary Table S4 in the Supplementary Material demonstrated that soil microbial diversity was inversely correlated with soil properties, following the application of AUX, OA, and PA. This suggests that the reduction in bacterial and fungal diversity could be attributed to changes in soil properties resulting from these treatments. While the addition of AUX, OA, and PA had a beneficial effect on AK, AHN, and pH in the rhizosphere soil of this study, the ensuing increase in nitrogen and pH levels may have exceeded the tolerable range of certain microbial communities, thereby inducing adverse changes in their living environment that inhibited their growth. Specifically, the pronounced alterations in the soil environment could hinder the growth of microbial populations that had adapted to the original living conditions (Zheng et al., 2022). The rise in soil nitrogen content accelerated the loss of rare species and led to a decline in microbial diversity. While an increase in other nutrients may help certain microorganisms withstand the negative impacts caused by changes in soil nutrient conditions, the overall effect was a decrease in microbial diversity (Wang et al., 2018).
Root secretions exhibit notable spatiotemporal variability during various physiological stages of crops, and the impacts of distinct components on the assembly of microbial structures may differ significantly (Zhalnina et al., 2018). Previous research has demonstrated that the composition of the rhizosphere microbial community is influenced by the composition of root secretions. The significant differences observed in microbial community structure among FLA, PA, and CK in this study were in line with previous findings (Cadot et al., 2021; Figure 1). The selectivity of microorganisms for the consumption of root secretions influences the aggregation of rhizosphere microorganisms. Certain secretions encourage the colonization of specific microorganisms, while others inhibit microbial growth. As such, different types of secretions can have varying impacts on the diversity of rhizosphere microorganisms (Sasse et al., 2018; Siczek et al., 2018; Bao et al., 2022). In this study, FLA and PA may have divergent impacts on the abundance of dominant species, consequently leading to alterations in microbial diversity (Figures 1, 2).
4.3. Effect of root secretions on the abundance of dominant microbiome
The reduction in microbial community abundance and diversity observed in response to compounds addition may be attributed to the inhibition of harmful microorganisms and stabilization of microbial structure (Salem et al., 2022). This study found that the addition of compounds leads to alterations in both the diversity and relative abundance of dominant species of soil microbial communities. The interaction between plant root exudates and specific microbial communities is complex and diverse. They can attract or repel microorganisms, modulate symbiotic and antagonistic relationships, thereby influencing the health and growth of plants (Zhang et al., 2022). Specific root secretions can attract beneficial bacteria and reduce the proliferation of pathogenic bacteria, contributing to these changes (Liu A. et al., 2022). Terpenoids and polyketides present in root secretions have been found to suppress phytopathogenic mycobacteria while facilitating the recruitment of specific bacteria to rhizosphere soils (Strickland et al., 2015). Chemical substances in plant root exudates can serve as signaling molecules to attract beneficial microorganisms and establish symbiotic relationships. Among them, phenolic and flavonoid compounds have been shown to enhance the expression of nodulation genes in nitrogen-fixing bacteria (Hirsch et al., 2003).These signaling molecules can trigger chemical reactions in rhizobial bacteria, leading to the formation of root nodules and the establishment of symbiotic relationships with plant roots (Xie et al., 2019).
Therefore, the addition of compounds is more likely to impact bacterial communities rather than fungal communities, as demonstrated in Figure 3 where fungal communities were found to be insensitive to the addition of compounds. Our analysis revealed that the relative abundance of dominant bacterial genera had significant negative correlations with soil physicochemical properties such as S-PPO, S-UE, S-CAT, S-DHA, pH, and AK. This finding suggests that the microbial community in soil that has undergone continuous coffee cropping exhibits sensitivity to changes in soil physicochemical properties and enzyme activities brought about by root secretion addition, which is consistent with the results of previous studies (Liu et al., 2019; Figure 4). The addition of root secretions promotes the interaction between microorganisms, and regulates the structure of soil microbial community by reducing the intensity of competition between microbial communities (Eldridge et al., 2017). In addition, the addition of root secretions also enhances the recruitment of microorganisms compatible with root secretions and reduces the survival space of pre-existing dominant species. This may explain the observed reduction in the abundance of dominant genera (Santoyo, 2022). The results observed in this study demonstrate how the addition of primary exudates from Areca catechu roots promotes the transition of soil microbial community structure from imbalance to stability.
4.4. Effect of different compounds on functional genes
The structure and function of soil microorganisms are considered to be inseparable, thus the difference of soil microbial communities under compounds addition may change the functional type of these microbial communities (Xie et al., 2022). Soil microbial community function may be influenced by the addition of simulated compounds, and microbial substrate preferences for root metabolites (amino acids and organic acids) can positively aid in the selection of specific functional taxa (Mundra et al., 2022). Chemoheterotrophy and Aerobic chemoheterotrophy can participate in the carbon cycle and accelerate the decomposition of alkali-hydrolyzed nitrogen (Yu et al., 2021). However, The exogenous secretions did not significantly affect the percentage of chemoheterotrophic functional genes in this study (Figure 5).
In this study, the CK and KCK treatments showed a higher proportion of potential pathogenic functional groups such as Plant Pathogen and Fungal Parasite. These findings suggest that a high relative abundance of pathotrophic fungal functional groups in coffee rhizosphere soils may be the primary cause of CCO (Liu et al., 2019). It is worth noting that the relative abundance of functional groups such as Plant Pathogen, Fungal Parasite, Animal Pathogen, Endophyte, Wood Saprotroph, and Lichen Parasite did not show significant changes after the addition of various compounds (except for organic acid compounds). This indicates that prior to the addition of compounds, the fungal functional groups present in coffee rhizosphere soils were more complex, and the addition of compound compounds led to a decrease in fungal functional group complexity (Figure 5). These results suggest that the potential risk of coffee disease can be mitigated by the addition of secretions from Areca catechu.
5. Conclusion
This study reveals that adding primary exudates from Areca catechu roots can enhance soil fertility and enzyme activity in coffee rhizosphere soils, resulting in significant changes in the structure and function of soil microbial communities. Specifically, the addition of flavonoids, plant hormone, phenolic acids, and sugars can inhibit the growth of bacterial pathogens in continuously cultivated soils by promoting soil nutrient enrichment and increasing soil enzyme activity, thus reducing the risk of coffee diseases. The addition of organic acids and flavonoids may increase the relative abundance of pathogenic genera like Fusarium and lead to a significant increase in the relative abundance of pathogenic fungal functional groups. The inclusion of plant hormone, phenolic acids, and sugars could promote the proliferation and relative dynamic balance of non-pathogenic bacteria in continuously cropped coffee rhizosphere soils, which ultimately contributes to overcoming challenges associated with continuous cropping in coffee rhizosphere soils.
Data availability statement
The data presented in the study are deposited in the Mendeley Data repository, accession number doi: 10.17632/tgfz6h2bxh.1.
Author contributions
SZ: Conceptualization, Data curation, Formal analysis, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. AZ: Methodology, Writing – review & editing. QZ: Conceptualization, Funding acquisition, Methodology, Project administration, Resources, Supervision, Writing – review & editing. YD: Formal analysis, Methodology, Project administration, Supervision, Writing – review & editing. LS: Formal analysis, Supervision, Writing – review & editing. YS: Formal analysis, Project administration, Supervision, Writing – review & editing. FZ: Supervision, Writing – review & editing. DH: Supervision, Writing – review & editing. WX: Formal analysis, Supervision, Visualization, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by the Chinese Academy of Tropical Agricultural Sciences for Science and Technology Innovation Team of National Tropical Agricultural Science Center (No. CATASCXTD202312), the Natural Science Foundation of Hainan (421RC653, 321MS093, 321QN327), the National Natural Science Foundation of China (No. 32101847).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1257164/full#supplementary-material
Footnotes
References
Ajayi, A. E., Tassinari, D., Araujo-Junior, C. F., Faloye, O. T., Abayomi, F., Dias Junior, M. D. S., et al. (2021). Long-term no tillage management impact on soil hydro-physical properties in coffee cultivation. Geoderma 404:115306. doi: 10.1016/j.geoderma.2021.115306
Ayalew, B., Hylander, K., Zewdie, B., Shimales, T., Adugna, G., Mendesil, E., et al. (2022). The impact of shade tree species identity on coffee pests and diseases. Agric. Ecosyst. Environ. 340:108152. doi: 10.1016/j.agee.2022.108152
Bao, L., Liu, Y., Ding, Y., Shang, J., Wei, Y., Tan, Y., et al. (2022). Interactions between phenolic acids and microorganisms in rhizospheric soil from continuous cropping of panax notoginseng. Front. Microbiol. 13:791603. doi: 10.3389/fmicb.2022.791603
Baudoin, E., Benizri, E., and Guckert, A. (2003). Impact of artificial root exudates on the bacterial community structure in bulk soil and maize rhizosphere. Soil Biol. Biochem. 35, 1183–1192. doi: 10.1016/S0038-0717(03)00179-2
Beckett, R. P., Zavarzina, A. G., and Liers, C. (2013). Oxidoreductases and cellulases in lichens: possible roles in lichen biology and soil organic matter turnover. Fungal Biol. 117, 431–438. doi: 10.1016/j.funbio.2013.04.007
Bokulich, N. A., Kaehler, B. D., Rideout, J. R., Dillon, M., Bolyen, E., Knight, R., et al. (2018). Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6:90. doi: 10.1186/s40168-018-0470-z
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Cadot, S., Guan, H., Bigalke, M., Walser, J., Jander, G., Erb, M., et al. (2021). Specific and conserved patterns of microbiota-structuring by maize benzoxazinoids in the field. Microbiome 9:103. doi: 10.1186/s40168-021-01049-2
Callahan, B. J., Mcmurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Cheng, F., Ali, M., Liu, C., Deng, R., and Cheng, Z. (2020). Garlic allelochemical diallyl disulfide alleviates autotoxicity in the root exudates caused by long-term continuous cropping of tomato. J. Agric. Food Chem. 68, 11684–11693. doi: 10.1021/acs.jafc.0c03894
Eilers, K. G., Lauber, C. L., Knight, R., and Fierer, N. (2010). Shifts in bacterial community structure associated with inputs of low molecular weight carbon compounds to soil. Soil Biol. Biochem. 42, 896–903. doi: 10.1016/j.soilbio.2010.02.003
Eldridge, D. J., Delgado Baquerizo, M., Travers, S. K., Val, J., Oliver, I., Hamonts, K., et al. (2017). Competition drives the response of soil microbial diversity to increased grazing by vertebrate herbivores. Ecology 98, 1922–1931. doi: 10.1002/ecy.1879
Fu, R., Feng, H., Dini-Andreote, F., Wang, Z., Bo, C., Cao, W., et al. (2021). Modulation of the tomato rhizosphere microbiome via changes in root exudation mediated by the ethylene receptor NR. Microorganisms 9:2456. doi: 10.3390/microorganisms9122456
Guo, Y., Lv, J., Zhao, Q., Dong, Y., and Dong, K. (2020). Cinnamic acid increased the incidence of fusarium wilt by increasing the pathogenicity of fusarium oxysporum and reducing the physiological and biochemical resistance of faba bean, which was alleviated by intercropping with wheat. Front. Plant Sci. 11:608389. doi: 10.3389/fpls.2020.608389
Higashi, Y. (2019). Coffee and endothelial function: a coffee paradox? Nutrients 11:2104. doi: 10.3390/nu11092104
Hirsch, A. M., Bauer, W. D., Bird, D. M., Cullimore, J., Tyler, B., and Yoder, J. I. (2003). Molecular signals and receptors: controlling rhizosphere interactions between plants and other organisms. Ecology 84, 858–868. doi: 10.1890/0012-9658(2003)084[0858:MSARCR]2.0.CO;2
Hofmann, A., Wittenmayer, L., Arnold, G., Schieber, A., and Merbach, W. (2009). Root exudation of phloridzin by apple seedlings (malus x domestica borkh.) with symptoms of apple replant disease. J. Appl. Bot. Food Qual. 2, 185–189. doi: 10.1614/IPSM-08-126.1
Hu, L., Robert, C., Cadot, S., Zhang, X., Ye, M., Li, B., et al. (2018). Root exudate metabolites drive plant-soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nat. Commun. 9:2738. doi: 10.1038/s41467-018-05122-7
Huang, A. C., Jiang, T., Liu, Y., Bai, Y., Reed, J., Qu, B., et al. (2019). A specialized metabolic network selectively modulates arabidopsis root microbiota. Science 364:eaau6389. doi: 10.1126/science.aau6389
Iijima, M., Griffiths, B., and Bengough, A. G. (2000). Sloughing of cap cells and carbon exudation from maize seedling roots in compacted sand. New Phytol. 145, 477–482. doi: 10.1046/j.1469-8137.2000.00595.x
Jiang, S., Xing, Y., Liu, G., Hu, C., Wang, X., Yan, G., et al. (2021). Changes in soil bacterial and fungal community composition and functional groups during the succession of boreal forests. Soil Biol. Biochem. 161:108393. doi: 10.1016/j.soilbio.2021.108393
Jing, Y., Zhang, Y., Han, I., Wang, P., Mei, Q., and Huang, Y. (2020). Effects of different straw biochars on soil organic carbon, nitrogen, available phosphorus, and enzyme activity in paddy soil. Sci. Rep. 10:8837. doi: 10.1038/s41598-020-65796-2
Jurburg, S. D., Shek, K. L., and Mcguire, K. (2020). Soil microbial composition varies in response to coffee agroecosystem management. FEMS Microbiol. Ecol. 96:fiaa164. doi: 10.1093/femsec/fiaa164
Kawasaki, A., Dennis, P. G., Forstner, C., Raghavendra, A. K. H., Mathesius, U., Richardson, A. E., et al. (2021). Manipulating exudate composition from root apices shapes the microbiome throughout the root system. Plant Physiol. 187, 2279–2295. doi: 10.1093/plphys/kiab337
Li, Q., Jia, W., Zhang, Q., and Cheng, X. (2022). Localized plant-soil-microbe interactions regulate spatial variations of soil oxidase activities within afforested systems in a subtropical area. Geoderma 406:115499. doi: 10.1016/j.geoderma.2021.115499
Li, M., Song, Z., Li, Z., Qiao, R., Zhang, P., Ding, C., et al. (2022). Populus root exudates are associated with rhizosphere microbial communities and symbiotic patterns. Front. Microbiol. 13:1042944. doi: 10.3389/fmicb.2022.1042944
Liang, S., Deng, J., Jiang, Y., Wu, S., Zhou, Y., and Zhu, W. (2020). Functional distribution of bacterial community under different land use patterns based on faprotax function prediction. Pol. J. Environ. Stud. 29, 1245–1261. doi: 10.15244/pjoes/108510
Liu, J., Li, F. Y., Liu, J., Wang, S., Liu, H., Ding, Y., et al. (2023). Grazing promotes soil phosphorus cycling by enhancing soil microbial functional genes for phosphorus transformation in plant rhizosphere in a semi-arid natural grassland. Geoderma 430:116303. doi: 10.1016/j.geoderma.2022.116303
Liu, H., Pan, F., Han, X., Song, F., Zhang, Z., Yan, J., et al. (2019). Response of soil fungal community structure to long-term continuous soybean cropping. Front. Microbiol. 9:3316. doi: 10.3389/fmicb.2018.03316
Liu, A., Wang, W., Chen, X., Zheng, X., Fu, W., Wang, G., et al. (2022). Phytoremediation of DEHP and heavy metals co-contaminated soil by rice assisted with a PGPR consortium: insights into the regulation of ion homeostasis, improvement of photosynthesis and enrichment of beneficial bacteria in rhizosphere soil. Environ. Pollut. 314:120303. doi: 10.1016/j.envpol.2022.120303
Liu, Q., Zhang, L., Wang, L., Wu, Q., Li, K., and Guo, X. (2022). Autotoxin affects the rhizosphere microbial community structure by influencing the secretory characteristics of grapevine roots. Front. Microbiol. 13:953424. doi: 10.3389/fmicb.2022.953424
Lozupone, C. A., Hamady, M., Kelley, S. T., and Knight, R. (2007). Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 73, 1576–1585. doi: 10.1128/AEM.01996-06
Lozupone, C., and Knight, R. (2005). Unifrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71, 8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005
Lu, R.K. (1999). Soil and agro-chemistry analytical methods. Beijing: China Agricultural Science & Technology Press.
Ma, H., Li, X., Hou, S., Peng, D., Wang, Y., Xu, F., et al. (2019). The activation and extraction systems using organic acids and lentinus edodes to remediate cadmium contaminated soil. Environ. Pollut. 255:113252. doi: 10.1016/j.envpol.2019.113252
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10–12. doi: 10.14806/EJ.17.1.200
Mcdonald, D., Price, M. N., Goodrich, J., Nawrocki, E. P., Desantis, T. Z., Probst, A., et al. (2012). An improved greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6, 610–618. doi: 10.1038/ismej.2011.139
Mundra, S., Kauserud, H., Okland, T., Nordbakken, J. F., Ransedokken, Y., and Kjønaas, O. J. (2022). Shift in tree species changes the belowground biota of boreal forests. New Phytol. 234, 2073–2087. doi: 10.1111/nph.18109
Price, M. N., Dehal, P. S., and Arkin, A. P. (2010). FastTree 2-approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490
Qu, X. H., and Wang, J. G. (2008). Effect of amendments with different phenolic acids on soil microbial biomass, activity, and community diversity. Appl. Soil Ecol. 39, 172–179. doi: 10.1016/j.apsoil.2007.12.007
Rezende, M. Q., Venzon, M., Dos Santos, P. S., Cardoso, I. M., and Janssen, A. (2021). Extrafloral nectary-bearing leguminous trees enhance pest control and increase fruit weight in associated coffee plants. Agric. Ecosyst. Environ. 319:107538. doi: 10.1016/j.agee.2021.107538
Salem, M. A., Wang, J. Y., and Al-Babili, S. (2022). Metabolomics of plant root exudates: from sample preparation to data analysis. Front. Plant Sci. 13:1062982. doi: 10.3389/fpls.2022.1062982
Santoyo, G. (2022). How plants recruit their microbiome? New insights into beneficial interactions. J. Adv. Res. 40, 45–58. doi: 10.1016/j.jare.2021.11.020
Sasse, J., Martinoia, E., and Northen, T. (2018). Feed your friends: do plant exudates shape the root microbiome? Trends Plant Sci. 23, 25–41. doi: 10.1016/j.tplants.2017.09.003
Siczek, A., Frąc, M., Wielbo, J., and Kidaj, D. (2018). Benefits of flavonoids and straw mulch application on soil microbial activity in pea rhizosphere. Int. J. Environ. Sci. Technol. 15, 755–764. doi: 10.1007/s13762-017-1434-8
Song, Y., Li, X., Yao, S., Yang, X., and Jiang, X. (2020). Correlations between soil metabolomics and bacterial community structures in the pepper rhizosphere under plastic greenhouse cultivation. Sci. Total Environ. 728:138439. doi: 10.1016/j.scitotenv.2020.138439
Strickland, M. S., Mcculley, R. L., Nelson, J. A., and Bradford, M. A. (2015). Compositional differences in simulated root exudates elicit a limited functional and compositional response in soil microbial communities. Front. Microbiol. 6:817. doi: 10.3389/fmicb.2015.00817
Su, Y., Zi, H., Wei, X., Hu, B., Deng, X., Chen, Y., et al. (2022). Application of manure rather than plant-origin organic fertilizers alters the fungal community in continuous cropping tobacco soil. Front. Microbiol. 13:818956. doi: 10.3389/fmicb.2022.818956
Sujatha, S., Bhat, R., Kannan, C., and Balasimha, D. (2011). Impact of intercropping of medicinal and aromatic plants with organic farming approach on resource use efficiency in arecanut (Areca catechu L.) plantation in India. Ind. Crop. Prod. 33, 78–83. doi: 10.1016/j.indcrop.2010.09.001
Tan, X., Wang, Z., Lu, G., He, W., Wei, G., Huang, F., et al. (2017). Kinetics of soil dehydrogenase in response to exogenous cd toxicity. J. Hazard. Mater. 329, 299–309. doi: 10.1016/j.jhazmat.2017.01.055
Telesiński, A., Krzyśko-Bupicka, T., Cybulska, K., Pawłowska, B., Biczak, R., Nieg, M., et al. (2019). Comparison of oxidoreductive enzyme activities in three coal tar creosote-contaminated soils. Soil Res. 57:814. doi: 10.1071/SR19040
Trofymow, J. A., Coleman, D. C., and Cambardella, C. (1987). Rates of rhizodeposition and ammonium depletion in the rhizosphere of axenic oat roots. Plant Soil 97, 333–344. doi: 10.1007/BF02383223
Upadhyay, S. K., Srivastava, A. K., Rajput, V. D., Chauhan, P. K., Bhojiya, A. A., Jain, D., et al. (2022). Root exudates: mechanistic insight of plant growth promoting rhizobacteria for sustainable crop production. Front. Microbiol. 13:916488. doi: 10.3389/fmicb.2022.916488
Vives-Peris, V., de Ollas, C., Gómez-Cadenas, A., and Pérez-Clemente, R. M. (2020). Root exudates: from plant to rhizosphere and beyond. Plant Cell Rep. 39, 3–17. doi: 10.1007/s00299-019-02447-5
Wang, J., Li, X., Zhang, J., Yao, T., Wei, D., Wang, Y., et al. (2012). Effect of root exudates on beneficial microorganisms—evidence from a continuous soybean monoculture. Plant Ecol. 213, 1883–1892. doi: 10.1007/s11258-012-0088-3
Wang, C., Liu, D., and Bai, E. (2018). Decreasing soil microbial diversity is associated with decreasing microbial biomass under nitrogen addition. Soil Biol. Biochem. 120, 126–133. doi: 10.1016/j.soilbio.2018.02.003
Wang, R., Liu, J., Jiang, W., Ji, P., and Li, Y. (2022). Metabolomics and microbiomics reveal impacts of rhizosphere metabolites on alfalfa continuous cropping. Front. Microbiol. 13:833968. doi: 10.3389/fmicb.2022.833968
Xie, H., Chen, Z., Feng, X., Wang, M., Luo, Y., Wang, Y., et al. (2022). L-theanine exuded from camellia sinensis roots regulates element cycling in soil by shaping the rhizosphere microbiome assembly. Sci. Total Environ. 837:155801. doi: 10.1016/j.scitotenv.2022.155801
Xie, X. G., Zhang, F. M., Yang, T., Chen, Y., Li, X. G., and Dai, C. C. (2019). Endophytic fungus drives nodulation and N (2) fixation attributable to specific root exudates. MBio. 10, e719–e728. doi: 10.1128/mBio.00728-19
Xiong, W., Jousset, A., Guo, S., Karlsson, I., Zhao, Q., Wu, H., et al. (2018). Soil protist communities form a dynamic hub in the soil microbiome. ISME J. 12, 634–638. doi: 10.1038/ismej.2017.171
Xiong, W., Zhao, Q., Zhao, J., Xun, W., Li, R., Zhang, R., et al. (2015). Different continuous cropping spans significantly affect microbial community membership and structure in a vanilla-grown soil as revealed by deep pyrosequencing. Microb. Ecol. 70, 209–218. doi: 10.1007/s00248-014-0516-0
Yan, S., Yin, L., Dijkstra, F. A., Wang, P., and Cheng, W. (2023). Priming effect on soil carbon decomposition by root exudate surrogates: a meta-analysis. Soil Biol. Biochem. 178:108955. doi: 10.1016/j.soilbio.2023.108955
Yu, Y., Liu, L., Wang, J., Zhang, Y., and Xiao, C. (2021). Effects of warming on the bacterial community and its function in a temperate steppe. Sci. Total Environ. 792:148409. doi: 10.1016/j.scitotenv.2021.148409
Zeng, J., Liu, J., Lu, C., Ou, X., Luo, K., Li, C., et al. (2020). Intercropping with turmeric or ginger reduce the continuous cropping obstacles that affect pogostemon cablin (patchouli). Front. Microbiol. 11:579719. doi: 10.3389/fmicb.2020.579719
Zhalnina, K., Louie, K. B., Hao, Z., Mansoori, N., Da Rocha, U. N., Shi, S., et al. (2018). Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat. Microbiol. 3, 470–480. doi: 10.1038/s41564-018-0129-3
Zhang, W., Luo, X., Mei, Y., Yang, Q., Zhang, A., Chen, M., et al. (2022). Priming of rhizobial nodulation signaling in the mycosphere accelerates nodulation of legume hosts. New Phytol. 235, 1212–1230. doi: 10.1111/nph.18192
Zhao, Q., Xiong, W., Xing, Y., Sun, Y., Lin, X., and Dong, Y. (2018). Long-term coffee monoculture alters soil chemical properties and microbial communities. Sci. Rep. 8, 6111–6116. doi: 10.1038/s41598-018-24537-2
Zhao, M., Zhao, J., Yuan, J., Hale, L., Wen, T., Huang, Q., et al. (2021). Root exudates drive soil-microbe-nutrient feedbacks in response to plant growth. Plant Cell Environ. 44, 613–628. doi: 10.1111/pce.13928
Zheng, X., Cao, H., Liu, B., Zhang, M., Zhang, C., Chen, P., et al. (2022). Effects of mercury contamination on microbial diversity of different kinds of soil. Microorganisms 10:977. doi: 10.3390/microorganisms10050977
Zhou, Y., Coventry, D. R., Gupta, V. V. S. R., Fuentes, D., Merchant, A., Kaiser, B. N., et al. (2020). The preceding root system drives the composition and function of the rhizosphere microbiome. Genome Biol. 21:89. doi: 10.1186/s13059-020-01999-0
Keywords: continuous cropping obstacle, soil physic-chemical properties, soil enzyme activity, microbial community structure, functional prediction
Citation: Zhao S, Zhang A, Zhao Q, Dong Y, Su L, Sun Y, Zhu F, Hua D and Xiong W (2023) The impact of main Areca Catechu root exudates on soil microbial community structure and function in coffee plantation soils. Front. Microbiol. 14:1257164. doi: 10.3389/fmicb.2023.1257164
Edited by:
Priyanka Chandra, Central Soil Salinity Research Institute (ICAR), IndiaReviewed by:
Hongmiao Wu, Anhui Agricultural University, ChinaHongchen Jiang, China University of Geosciences Wuhan, China
Copyright © 2023 Zhao, Zhang, Zhao, Dong, Su, Sun, Zhu, Hua and Xiong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ang Zhang, YW5nemhhbmdfaGVudUAxNjMuY29t; Qingyun Zhao, cWluZ3l1bl8wMjJAY2F0YXMuY24=
 Shaoguan Zhao
Shaoguan Zhao Ang Zhang1*
Ang Zhang1* Yunping Dong
Yunping Dong Lanxi Su
Lanxi Su Wu Xiong
Wu Xiong