- 1Department of Bacteriology, Noguchi Memorial Institute for Medical Research, College of Health Sciences, University of Ghana, Accra, Ghana
- 2Veterinary Services Department, Ministry of Food and Agriculture, Accra, Ghana
- 3Department of Immunology, Noguchi Memorial Institute for Medical Research, College of Health Sciences, University of Ghana, Accra, Ghana
- 4National Food Institute, Research Group for Global Capacity Building, WHO Collaborating Centre for Antimicrobial Resistance in Foodborne Pathogens and Genomics, FAO Reference Laboratory for Antimicrobial Resistance, European Union Reference Laboratory for Antimicrobial Resistance, Technical University of Denmark, Kongens Lyngby, Denmark
Introduction: Enterococcus spp. have gradually evolved from commensals to causing life-threatening hospital-acquired infections globally due to their inherent antimicrobial resistance ability and virulence potential. Enterococcus spp. recovered from livestock and raw meat samples were characterized using antimicrobial susceptibility testing and whole-genome sequencing.
Materials and methods: Isolates were confirmed using the MALDI-ToF mass spectrometer, and antimicrobial susceptibility was determined using the Kirby-Bauer disk diffusion method. Whole genome sequencing was performed on isolates resistant to two or more antibiotics. Bioinformatics analysis was performed to determine sequence types, resistance and virulence gene content and evolutionary relationships between isolates from meat and livestock samples, and other enterococci genomes curated by PATRIC. eBURST analysis was used to assign genomes to clonal complexes.
Results: Enterococcus spp. were predominantly E. faecalis (96/236; 41%) and E. faecium (89/236; 38%). Overall, isolates showed resistance to erythromycin (78/236; 33%), tetracycline (71/236; 30%), ciprofloxacin (20/236; 8%), chloramphenicol (12/236; 5%), linezolid (7/236; 3%), ampicillin (4/236; 2%) and vancomycin (1/236, 0.4%). Resistance to two or more antimicrobial agents was detected among 17% (n = 40) Enterococcus spp. Resistance genes for streptogramins [lsa(A), lsa(E), msr(C)], aminoglycosides [aac(6′)-Ii, aph(3′)-III, ant(6)-Ia, aac(6′)-aph(2″), str], amphenicol [cat], macrolides [erm(B), erm(T), msr(C)], tetracyclines [tet(M), tet(L), tet(S)] and lincosamides [lsa(A), lsa(E), lnu(B)] were detected among the isolates. Genes for biofilm formation, adhesins, sex pheromones, cytolysins, hyaluronidase, oxidative stress resistance, quorum-sensing and anti-phagocytic activity were also identified. Potential plasmids with replicon sequences (rep1, rep2, repUS43, repUS47, rep9a, rep9b) and other mobile genetic elements (Tn917, cn_5536_ISEnfa1, Tn6009, ISEnfa1, ISEfa10) were detected. Clinically relevant E. faecium ST32 and ST416 clones were identified in meat samples.
Conclusion: The occurrence of antimicrobial-resistant Enterococcus spp. in livestock and raw meat samples, carrying multiple resistance and virulence genes, including known clones associated with hospital-acquired infections, underscores the critical need for employing robust tools like whole genome sequencing. Such tools provide detailed data essential for ongoing surveillance efforts aimed at addressing the challenge of antimicrobial resistance with a focus on one health.
1 Introduction
Enterococcus spp. exist as commensals in the gut of warm-blooded animals and humans but possess virulence genes, and are recognized as opportunistic pathogens that can cause a variety of hospital-acquired infections, such as urinary tract and intra-abdominal infections, bacteremia, and endocarditis (Iweriebor et al., 2015; Ngbede et al., 2017). E. faecium and E. faecalis are commonly associated with opportunistic infections, with other species being rarely pathogenic (Beukers et al., 2017). Enterococci have notable resistance to adverse environmental conditions, hence the capacity to colonize different ecological niches and spread within the food chain through contaminated animals and foods (Klibi et al., 2013). The inherent high antimicrobial resistance (AMR) ability of Enterococcus spp., and their ability to genetically acquire and transmit antimicrobial drug resistant determinants among themselves and other bacteria in the environment, presents a significant challenge for therapeutic measures (Iweriebor et al., 2015).
One common phenomenon that increases AMR among bacterial species including Enterococci is the use of antibiotics in animal production, which subsequently creates ideal conditions for evolution and selection of resistant strains (Iweriebor et al., 2015; Ngbede et al., 2017). Several studies have documented the widespread use of antibiotics in animal husbandry in Ghana, with majority of farmers using antibiotics for the purposes of treatment or prophylaxis, and at a lesser extent for growth stimulation (Donkor et al., 2012; Osei Sekyere, 2014; Boamah et al., 2016). Although Donkor et al. (2012) reports that there is no significant difference in the frequency of antimicrobial usage among different animals (cattle, goat, sheep, pig, poultry), Osei Sekyere (2014) indicates that the frequency of antimicrobial usage is influenced by the size or financial status of farms and mixed farming practices. In an effort to reduce costs and maintain profits, farmers in Ghana disregard the recommended waiting period after administering antibiotics to food animals by selling animal products like eggs and meat for human consumption (Boamah et al., 2016; Nkansa, 2020). This causes persistence of antibiotic residues in animal products, thereby exposing microorganisms in animals and humans to sub-inhibitory concentrations of antibiotic, leading to the emergence and spread of antibiotic-resistant bacterial strains through food or other environmental pathways (Iweriebor et al., 2015; Boamah et al., 2016; McEwen and Collignon, 2018; Collignon and McEwen, 2019). Unavailability of regulations regarding the use of antimicrobials in animals, limited involvement of veterinarians in the administration of antibiotics, and the unavailability of optimal storage conditions for antibiotics are among factors identified to be influencing the extensive and inappropriate use of antimicrobials in animal husbandry in the country (Donkor et al., 2012; Osei Sekyere, 2014; Boamah et al., 2016; Nkansa, 2020).
Enterococci are one of two indicator bacteria used to study the extent of AMR in populations owing to their proven resistance and virulence transmissibility as demonstrated in surveillance programs implemented in developed countries (Bager, 2000; European Food Safety Authority, European Centre for Disease Prevention and Control, 2013). Although the multidrug resistance potential of Enterococcus spp. has been demonstrated in other studies across Africa (Klibi et al., 2013; Iweriebor et al., 2015; Ngbede et al., 2017; Katakweba et al., 2018), data on Enterococci of human, animal and environmental origin in Ghana is scarce. Reports (Obiri-Danso et al., 2003, 2005; Ayum and Gifty, 2010; Labi et al., 2015; Quansah et al., 2018, 2020; Akrong et al., 2019; Akita et al., 2021) of isolated Enterococcus spp. from human clinical sources and environmental sources in Ghana have consistently lacked genomic characterization. Given that there are clear reports of inappropriate use of antibiotics on farms in Ghana, the limited application of genomic characterization obscures understanding of the genetic basis of ensuing antibiotic resistance, including the evolution and transmission of resistance genes, and the dynamics of resistance in bacterial populations. The application of whole genome sequencing (WGS) offers extensive genomic information and is valuable for characterizing bacterial isolates and conducting genomic surveillance of AMR in bacteria. This study employed WGS and antimicrobial susceptibility testing to examine Enterococcus spp. recovered from archived raw meat and livestock samples collected in Ghana.
2 Materials and methods
2.1 Study site and isolates
Samples used in the study were obtained from three livestock farms and three slaughterhouses in southern Ghana independent of each other, from 2018 to 2019 and archived at the Bacteriology Department, Noguchi Memorial Institute for Medical Research. Livestock samples consisted of fecal swabs collected from cattle, goats, pigs, poultry and sheep. Meat samples were collected from the thigh, brisket and flank/mid-loin of carcasses at slaughterhouses using sterile swabs and stored in brain heart infusion broth. Carcasses sampled included beef, chevon and mutton. One milliliter of each archived broth sample was pre-enriched with 9 mL of trypticase soy broth and incubated overnight at 37°C. A loopful of pre-enriched samples was then cultured on Bile Esculin Azide agar (Merck, Germany) and incubated for 18–24 h at 37°C. Presumptive Enterococcus spp. were seen as small transparent colonies with black halos. Presumptive Enterococcus spp. were confirmed using MALDI-TOF mass spectrometer (Bruker, Billerica, MA, USA) following subculture on nutrient agar (Oxoid, Basingstoke, Hants, United Kingdom).
Antimicrobial susceptibility was determined using the Kirby-Bauer disk diffusion method and interpreted with reference to the Clinical and Laboratory Standards Institute (CLSI) guideline (CLSI, 2021). Antibiotics used included ampicillin (10ug), vancomycin (30ug), erythromycin (15ug), tetracycline (30ug), ciprofloxacin (5ug), chloramphenicol (30ug) and linezolid (30ug) from Oxoid (Basingstoke, Hants, UK). The reference strain S. aureus ATCC 25923 was applied as quality control.
2.2 Whole-genome sequencing and analysis
Genomic DNA was extracted from Enterococcus spp. resistant to two or more antimicrobial agents using QIAamp® DNA mini kit (QIAGEN Inc. GmbH, Holden, Germany), with reference to the manufacturer’s protocol. Libraries of DNA were prepared using the Illumina® DNA Prep (M) Tagmentation kit (Illumina Inc. San Diego, CA 92122, United States) followed by multiplexing and 300 bp paired-end sequencing on the Illumina Miseq platform (Illumina, San Diego, CA, United States). Fastq files generated were assessed for quality using Fastqc (v0.11.9) and trimmed with Trimmomatic (v.0.39) at a minimum quality threshold of ≥ Q20 (Andrews, 2010; Bolger et al., 2014). Reads that met the quality threshold (Q20 or higher) after trimming, were subjected to de-novo assembly using Unicycler (v0.4.8). De-novo assembly was performed using the following parameters: minimum contig size of 200 bp, number of contigs <400, and minimum coverage of 20x. All assembled sequence data were submitted to GenBank and assigned accession numbers under Bio project PRJNA851427.
Assembled genomes were analyzed using online tools hosted by Center for Genomic Epidemiology.1 KmerFinder (v3.2; Hasman et al., 2014) was applied to confirm the identity of isolates sequenced. Kraken (v2.1.3; Wood et al., 2019) and Bracken (v2.6.1; Lu et al., 2017) were used to assess contamination. Resistance determinants were predicted using ResFinder (v4.1; Bortolaia et al., 2020), virulence genes by VirulenceFinder (v2.0; Joensen et al., 2014), plasmids and other mobile genetic elements by PlasmidFinder (v2.1; Carattoli et al., 2014) and MobileElementFinder (v1.0.3; Johansson et al., 2021) respectively. Sequence types were determined using the MLST option (v2.0.9; Larsen et al., 2012); eBURST analysis from PubMLST (https://pubmlst.org/bigsdb?db=pubmlst_efaecalis_seqdef&page=plugin&name=BURST&scheme_id=1) was used to assign genomes to clonal complexes based on their MLST profiles (Feil et al., 2004; Leavis et al., 2006; Jolley et al., 2018).
Using the CSI phylogeny tool, evolutionary relationships between E. faecalis and E. faecium isolates from this study and other African Enterococci genomes (curated by Bacterial and Viral Bioinformatics Resource Center),2 were analyzed and inferred from single nucleotide polymorphisms (SNPs), by comparing assembled sequences to the reference genomes E. faecium SRR24 (NZ_CP038996.1) and E. faecalis V583 (226185.9). Close genetic relatedness was determined based on a pairwise SNP difference below 10 between the genomes (Weterings et al., 2015). The maximum-likelihood phylogenetic tree generated was annotated in Interactive Tree of Life (iTOL; Letunic and Bork, 2021). The accession numbers of downloaded genomes have been included in the Supplementary materials.
2.3 Statistical analysis
Statistical analysis was performed to assess the difference in Enterococcus spp. isolation from the respective samples and to also assess the correlation between resistance phenotypes and their respective genotype using the chi square test of independence and logistic regression. A significance level of α = 0.05 or lower was employed to determine statistical significance. Analyses were carried out using Stata version 16.
3 Results
3.1 Distribution of Enterococcus species recovered
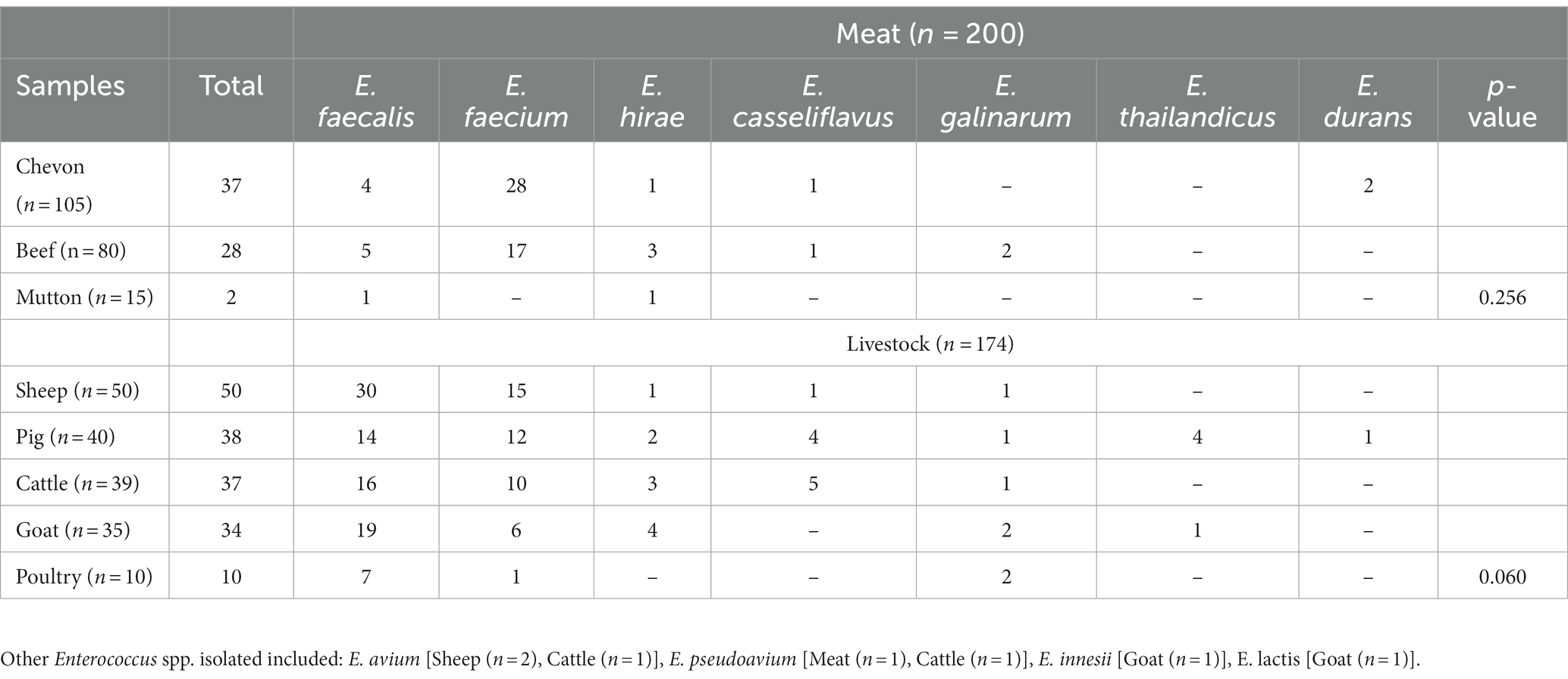
In total, the 374 samples tested originated from raw meat (n = 200) and livestock (n = 174). Meat samples included chevon (n = 105), beef (n = 80) and mutton (n = 15) whereas livestock samples comprised of sheep (n = 50), pigs (n = 40), cattle (n = 39), goats (n = 35) and poultry (n = 10; Table 1). Out of the 374 samples, 236 (63%) Enterococcus spp. were recovered, with E. faecalis (n = 96; 41%) and E. faecium (n = 89; 38%) predominating. E. hirae, E. casseliflavus, E. galinarum, E. thailandicus, among others were also isolated in this study (Table 1). The isolation of Enterococcus spp. from the different samples was not statistically significant (Table 1).
3.2 Antimicrobial susceptibility profile of Enterococcus species
Enterococcus spp. recovered were resistant to erythromycin (78/236; 33%), tetracycline (71/236; 30%), ciprofloxacin (20/236; 8%), chloramphenicol (12/236; 5%), linezolid (7/236;3%), ampicillin (4/236; 2%) and vancomycin (1/236; 0.4%; Table 2). Resistance to two or more antimicrobial agents was detected among 17% (n = 40) Enterococcus spp., with E. faecium and E. faecalis accounting for 98% (39/40) of this population. These isolates were resistant to erythromycin (38/40; 95%), tetracycline (39/40; 98%), ciprofloxacin (10/40; 25%), chloramphenicol (8/40; 20%), linezolid (1/40; 2.5%) and ampicillin (1/40; 2.5%). Use of tetracycline (2/3; 67%), penicillin (2/3; 67%) and colistin (1/3; 33%) was recorded in the livestock farms sampled.
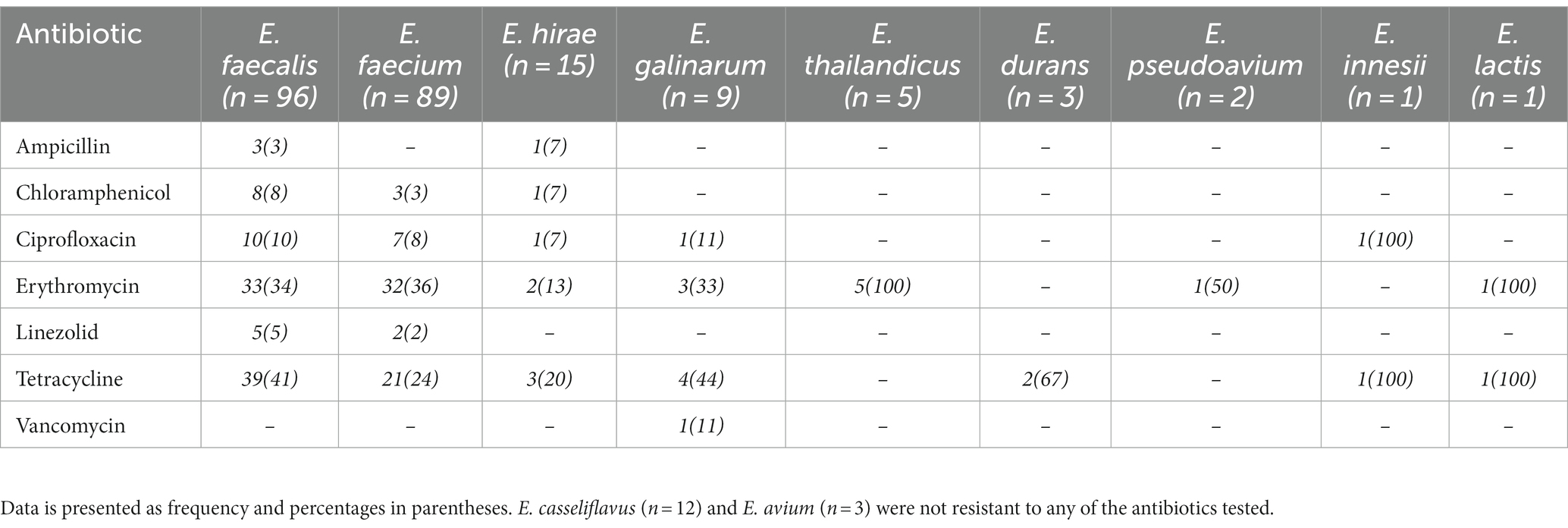
Table 2. Antimicrobial resistance profile of Enterococcus spp. recovered from livestock and raw meat samples.
3.3 Genomic characteristics of Enterococcus species
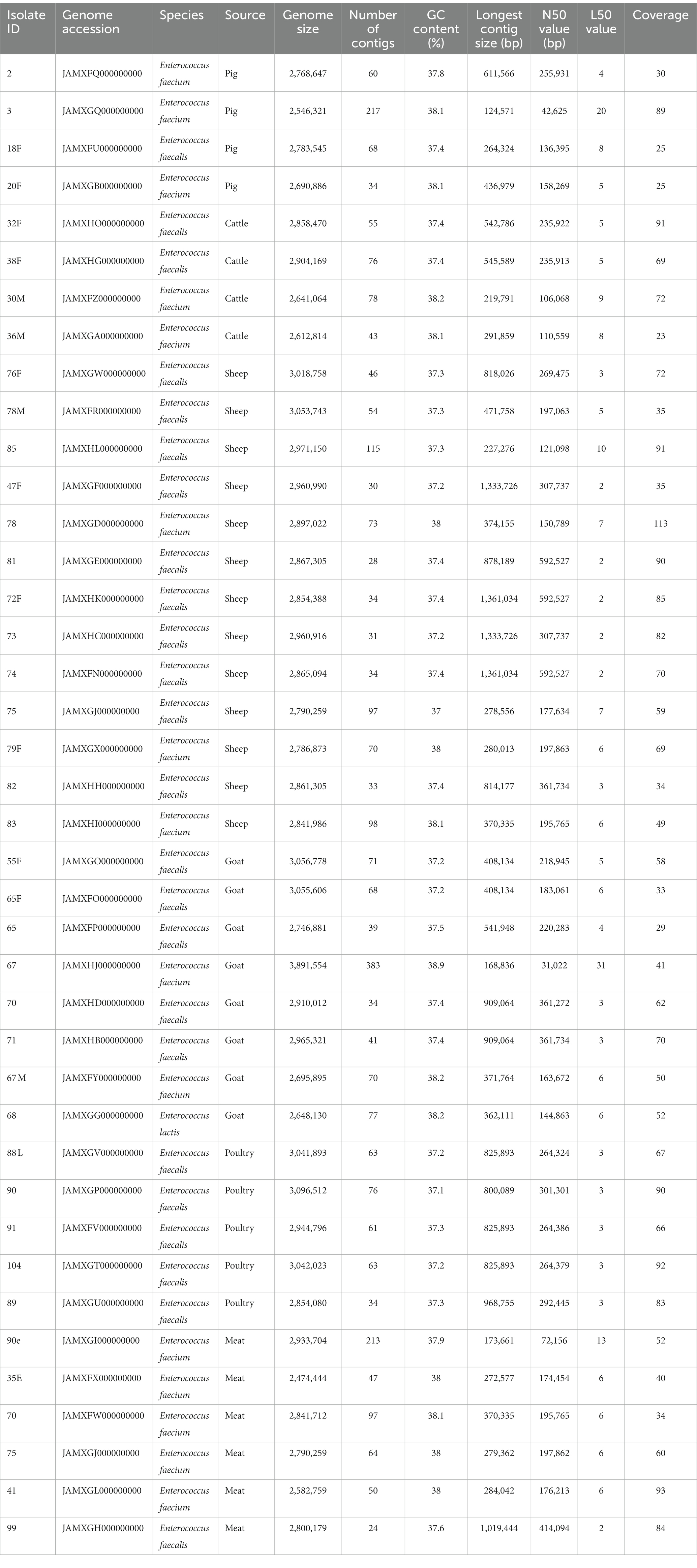
A summary of the genome characteristics of Enterococcus spp. sequenced is presented in Table 3. All assembled genomes passed QC with Q scores >20, minimum coverage of 20, number of contigs <400 and minimum contig size of 200 bp. The assembled genomes ranged in size from 2.5–3.1 Mb, with average GC content of 37.6.
3.4 Antimicrobial resistance determinants
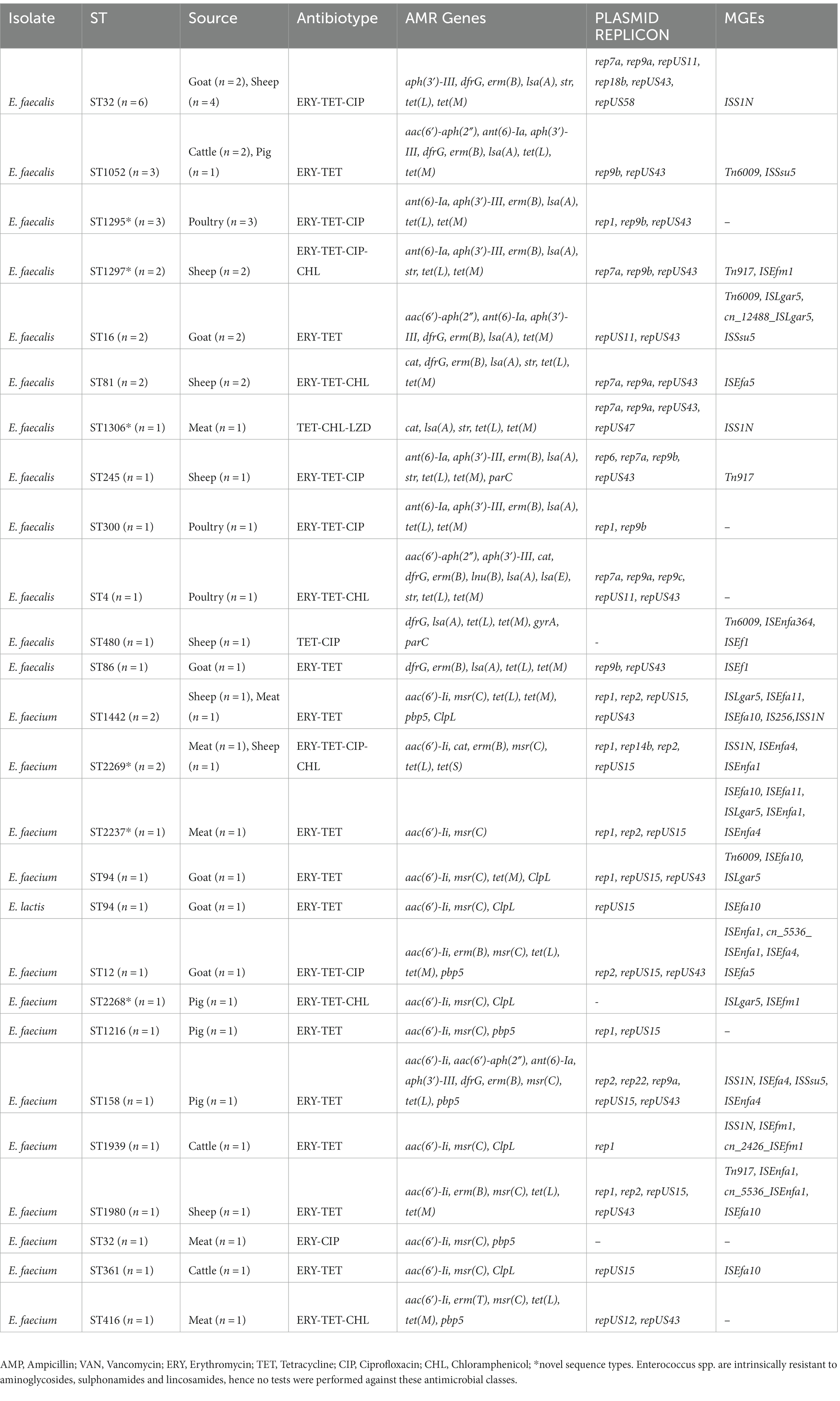
Antimicrobial resistance genes conferring resistance to 9 different antimicrobial classes were detected among E. faecium (8/9; 89%), E. faecalis (8/9; 89%) and E. lactis (3/9; 33%) (Table 4). Genes encoding resistance to tetracyclines [tet(M), tet(L), tet(S)], lincosamides [lsa(A), lsa(E), lnu(B)], macrolides [erm(B), erm(T), msr(C)], aminoglycosides [aac(6′)-Ii, aph(3′)-III, ant(6)-Ia, aac(6′)-aph(2″), str], streptogramins [lsa(A), lsa(E), msr(C)], trimethoprim [dfrG] and amphenicol [cat] were detected among isolates (Table 4). Chromosomal point mutations were observed in E. faecalis genomes (2/24; 8.3%) in the quinolone resistance determinant regions (QRDRs) of the DNA gyrase (gyrA p.S83Y) and DNA topoisomerase IV genes (parC p.S80I, parC p.S80R). Similarly, E. faecium genomes (7/16; 44%) exhibited multiple point mutations in the pbp5 gene which encodes ampicillin resistance (Table 4). The ClpL gene encoding resistance to heat was identified in 7/40 (18%) of Enterococcus spp. genomes. With a value of p of <0.001, chi-square test indicated a significant association between resistance phenotype and the presence of associated gene or genotype. Logistic regression also showed a three-fold increase in the odds of detecting associated genes for each unit increase in the different resistance phenotypes observed. Thus, a significant relationship between the type of resistance phenotype and genotype was observed.
3.5 Virulence factors
Virulence genes were detected in all isolates sequenced. The genes present in E. faecalis isolates include biofilm formation-associated pili genes [ebpA, ebpB, ebpC, SrtA], adhesins [ace, efaAfs, agg], sex pheromones [cad, camE, cCF10, cOB1], cytolysins [cylB, cylL, cylM], hyaluronidase [hylA, hylB], gelatinase [gelE], oxidative stress resistance [tpx], quorum-sensing [fsrB] and anti-phagocytic activity [ElrA]. A narrower spectrum of virulence genes was detected in E. faecium and E. lactis genomes; acm and efaAfm all encode adhesins.
3.6 Mobile genetic elements
Majority of the isolates (39/40; 98%) sequenced possessed mobile genetic elements including integrative conjugative elements, plasmids, insertion sequences, unit and composite transposons (Table 4). A total of 16 different replicon plasmid sequences were identified in the study, including repUS43 (n = 29), rep7a (n = 13), rep1 (n = 12), repUS15 (n = 12), repUS11 (n = 11) and rep9a (n = 11).
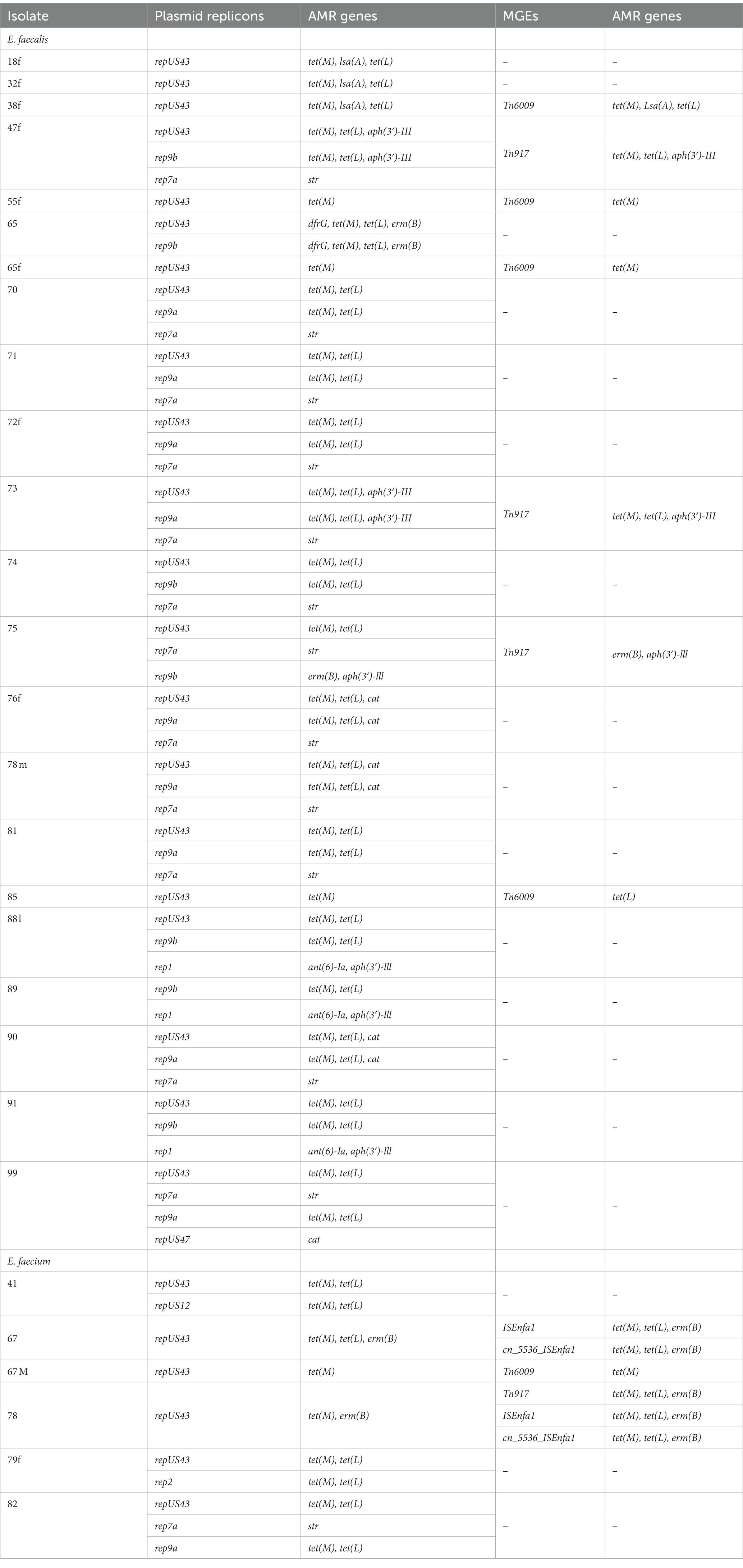
AMR genes (28/40; 70%) were found to occur on the same contig as plasmid replicons. Of note, tet(M), tet(L) and erm(B) genes were borne on repUS43, rep9a and rep9b plasmid replicons whereas aph(3′)-III and ant(6)-Ia genes were carried by rep1 plasmid and str gene, rep7a plasmid (Table 5). Meanwhile, AMR genes encoding tetracycline, macrolide and aminoglycoside resistance were also identified to be associated with other mobile genetic elements such as transposons (Tn917, cn_5536_ISEnfa1), integrative conjugative element (Tn6009) and insertion sequences (ISEnfa1, ISEfa10) in 10/40 (25%) of the isolates (Table 5). The occurrence of virulence genes on plasmids or other mobile genetic elements was observed in 3/40 (8%) of isolates.
3.7 Multi-locus sequence typing
Eighteen E. faecalis isolates belong to ST32 (n = 6), ST1052 (n = 3), ST16 (n = 2), ST81 (n = 2), including two novel sequence types ST1295 (n = 3) and ST1297 (n = 2; Table 4). The remaining isolates had diverse sequence types (Table 4). E. faecium isolates belong to ST2269 (n = 2), ST1442 (n = 2), ST94 (n = 1), ST12 (n = 1), ST1216 (n = 1), ST158 (n = 1), ST1939 (n = 1), ST1980 (n = 1), ST32 (n = 1), ST361 (n = 1), ST2268 (n = 1), ST2237 (n = 1) and ST416 (n = 1). E. lactis ST94 (n = 1) was detected. E. faecium clones (ST1442 and ST2269) were identified in both meat and livestock samples. E. faecium isolates (2/15) identified belong to ST32 and ST416, which are part of the CC17 clonal complex known for causing hospital-acquired infections globally.
3.8 Phylogenetic analysis
The pairwise distance matrix of SNPs showed close genetic relatedness between majority of the E. faecalis isolates from livestock in this study (Figure 1). Clustering of E. faecalis isolates was mostly observed between isolates from the same farm (Figure 2). E. faecalis isolates recovered in this study also showed close genetic relatedness with the South African livestock genomes irrespective of the livestock type (Figures 1, 2). Two isolates from sheep (76F and 78 M) in this study were found clustering together with South African human clinical isolates, with a pairwise SNP difference below 10 (Figures 1, 2). Similar patterns in the distribution of plasmid replicons, AMR and virulence genes were seen among E. faecalis genomes occurring in the same clade (Figure 2).
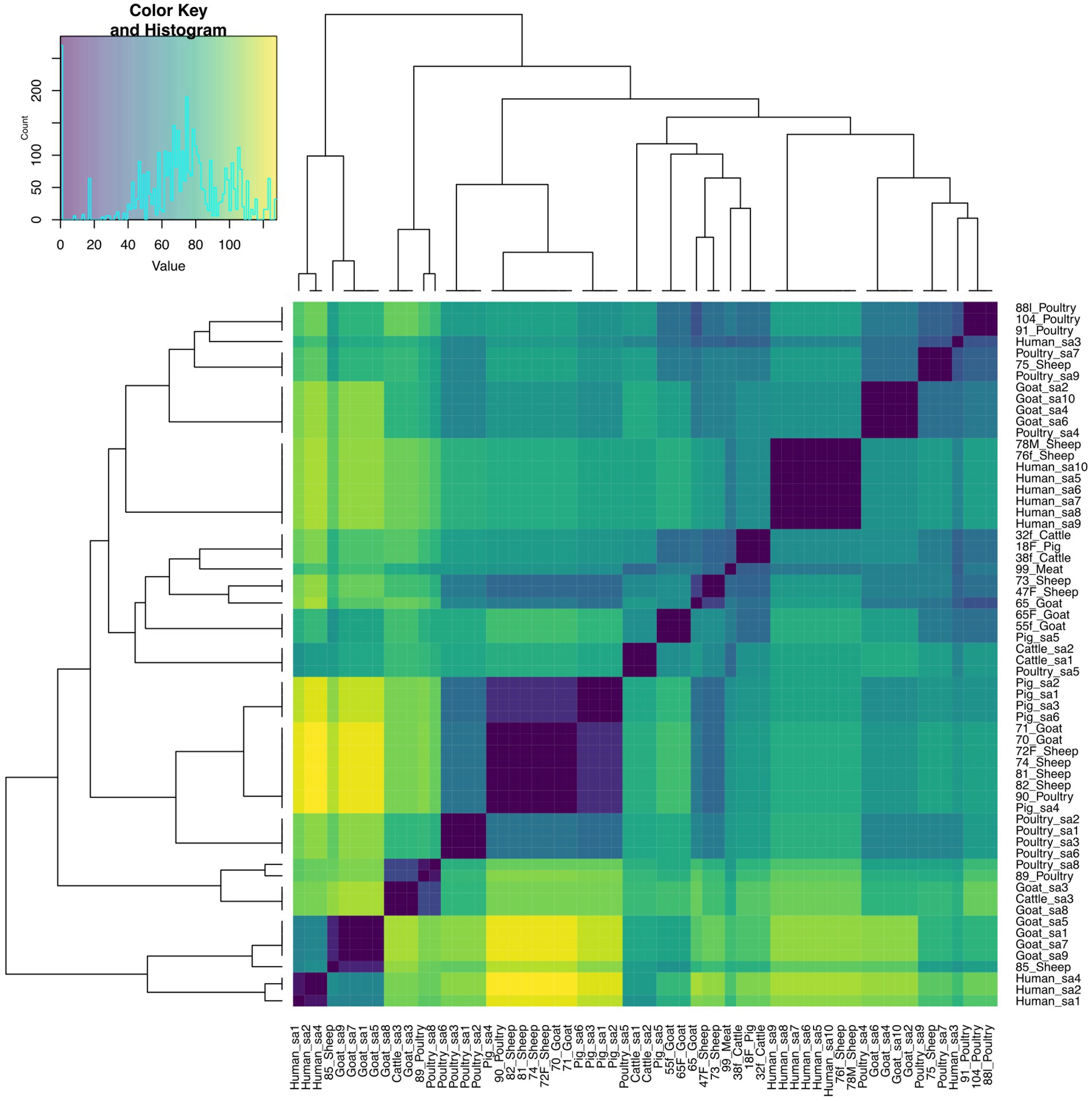
Figure 1. Heatmap and dendrogram showing pairwise SNP distance matrix and relationship among E. faecalis (n = 24) isolates in this study and other South African E. faecalis (n = 38) genomes downloaded from BV-BRC, labeled with the suffix “sa.” Isolates in this study included: Cattle = 2; Goat = 5; Meat = 1; Pig = 1; Poultry = 5 and Sheep = 10. The diagram shows the genetic variation between the isolates in the study. The pairwise distance matrix of SNPs showed close genetic relatedness between majority of the E. faecalis isolates from livestock in this study, with SNP difference below 10. Complete matrix data with pairwise distances recorded has been included in Supplementary material. Heatmap and dendrogram were created with gplots (v.3.1.3) and dendextend (v.3.1.3) packages, respectively, in R.
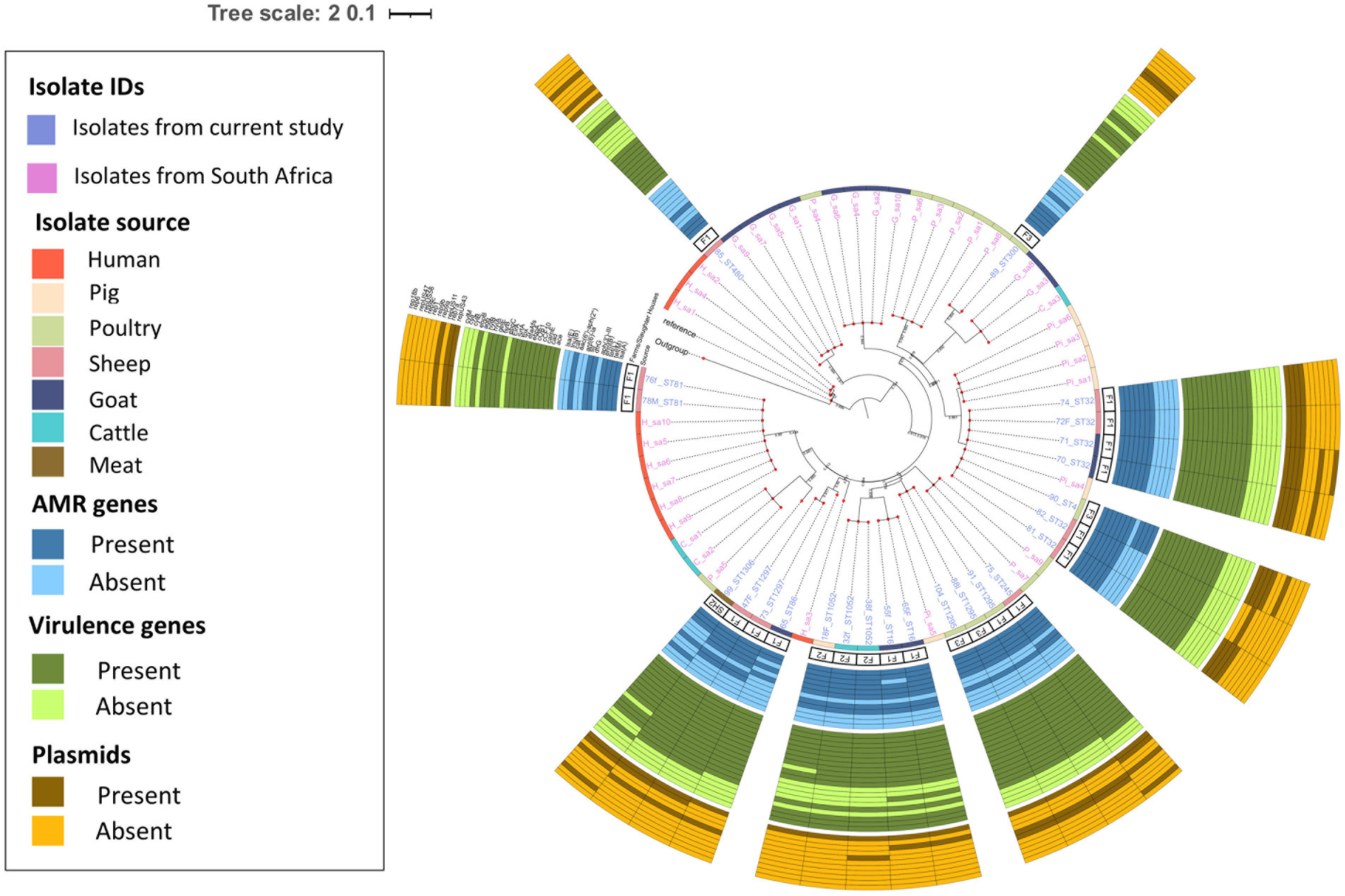
Figure 2. Population structure of Enterococcus faecalis isolates. Phylogeny of 24 study isolates and 38 isolates from South Africa downloaded from BV-BRC. The genomes were compared using E. faecalis V583 strain (226185.9) as reference. SNP-based maximum likelihood tree was constructed using CSI Phylogeny and visualized in iTOL. This phylogenetic tree shows the evolutionary relationship between E. faecalis isolates in this study and other E. faecalis genomes from South Africa. Clustering between E. faecalis isolates from livestock was mostly observed among isolates in this study and other genomes from South Africa. For each isolate in this study, the source, farm (F)/slaughter house (SH), the sequence types, AMR, virulence genes and plasmids distribution are shown.
E. faecium isolates from livestock sources exhibited greater genetic diversity compared to those from South Africa (Figure 3). E. faecium isolates from meat and livestock were mostly genetically divergent (Figure 4). However, one E. faecium isolate from meat (isolate 70) showed close genetic relatedness to an isolate from sheep (isolate 83) in the livestock group, with a SNP difference below 10 (Figures 3, 4).
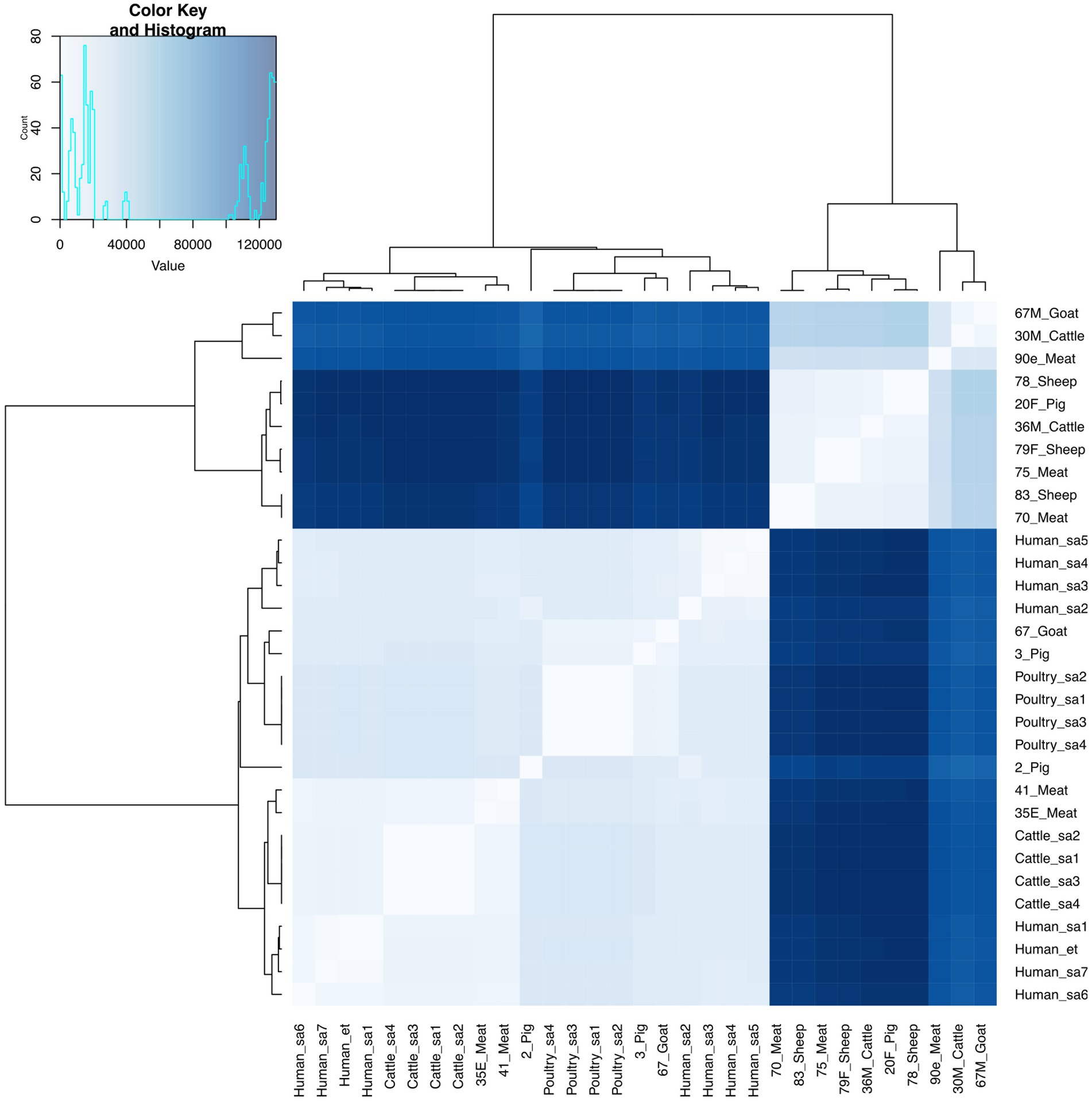
Figure 3. Heatmap and dendrogram showing pairwise SNP distance matrix and relationship among E. faecium (n = 15) isolates in this study and other E. faecium (n = 16) genomes downloaded from BV-BRC, labeled with the suffix “sa” and “et”. Isolates in this study included: Cattle = 2; Goat = 2; Meat = 5; Pig = 3 and Sheep =3. The diagram shows the genetic variation between the isolates in the study. The pairwise distance matrix of SNPs showed significant genetic diversity between E. faecium isolates, with SNP difference greater than 10. Complete matrix data with pairwise distances recorded has been included in Supplementary material. Heatmap and dendrogram were created with gplots (v.3.1.3) and dendextend (v.3.1.3) packages, respectively, in R.
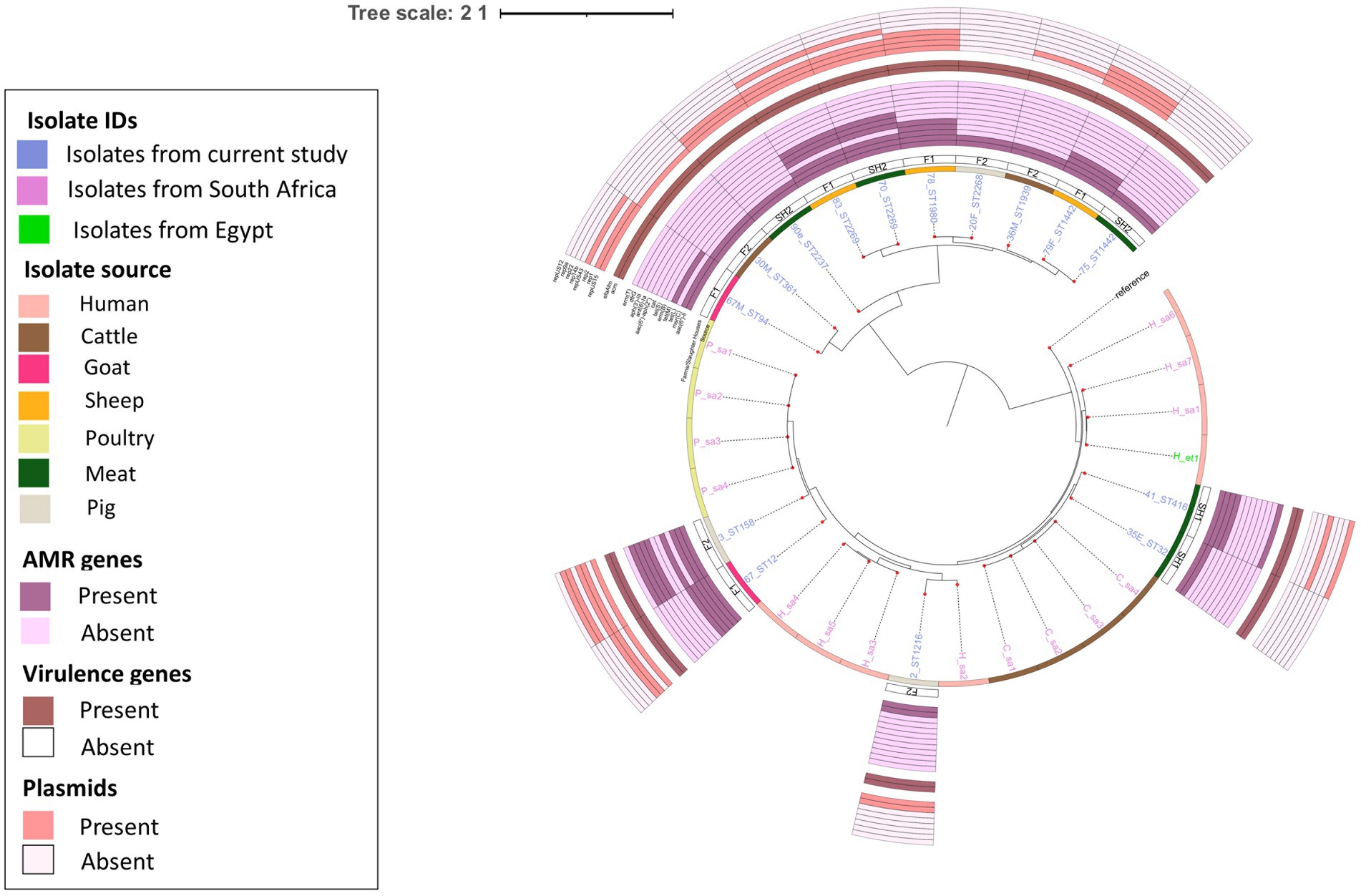
Figure 4. Population structure of E. faecium isolates. Phylogeny of 15 study isolates and 16 isolates from South Africa downloaded from BV-BRC. The genomes were compared using E. faecium SRR24 (NZ CP038996.1) as a reference. SNP-based maximum likelihood tree was constructed using CSI Phylogeny and visualized in iTOL. This phylogenetic tree shows the evolutionary relationship between E. faecalis isolates in this study and other E. faecalis genomes from South Africa. Limited genetic similarity between E. faecium isolates from meat and livestock was observed. For each isolate in this study, the source, farm (F)/slaughter house (SH), the sequence types, AMR, virulence genes and plasmids distribution are shown.
4 Discussion
This study revealed a substantial diversity among the isolates collected from both meat and livestock samples. While E. faecalis and E. faecium were the most prevalent species detected among the samples, no statistically significant differences were noted in the distribution of Enterococcus spp. This lack of differentiation among the Enterococcus spp. recovered from the various sources, suggests that these isolates may not exhibit strong niche-specific adaptions based on the particular source or animal host, contrary to reports elsewhere (Zaheer et al., 2020). The variation in the distribution of Enterococcus spp. among livestock across different geographic regions can be attributed to differences in dietary patterns, which impact the composition of gut commensal bacteria (Klibi et al., 2013; Ngbede et al., 2017). Similar to our findings, other studies (Golob et al., 2019; Habib et al., 2022) have also reported consistent contamination of retail meat by Enterococcus spp. and this has been linked to fecal contamination. Furthermore, the presence of antimicrobial resistance genes in these Enterococci isolates, including those related to clinically important antimicrobials like aminoglycosides, streptogramin A, macrolides, and tetracycline, suggests a narrowing of available treatment options (beta-lactam monotherapy or beta-lactam in combination with an aminoglycoside or glycopeptide) for opportunistic Enterococcus infections including those caused by vancomycin resistant strains (Arias et al., 2010; O’Driscoll and Crank, 2015). Among the isolates resistant to two or more antimicrobial agents, resistance to erythromycin, tetracycline, ciprofloxacin and chloramphenicol was common. The incidence of erythromycin and tetracycline resistance in Enterococcus spp. has been reported in other studies in Africa, and has been associated with the extensive use and misuse of these antimicrobials in animal production (Iweriebor et al., 2015; Ngbede et al., 2017; Molechan et al., 2019). The increased levels of resistance recorded against tetracycline could be attributed to the use of tetracycline as recorded in the livestock farms sampled for this study. Nonetheless, erythromycin resistance might also be due to cross resistance associated with another macrolide, tylosin documented to be used among livestock in Ghana (Jackson et al., 2004; Akansale et al., 2019). Resistance to erythromycin among Enterococcus spp. was found to be mediated by erythromycin ribosomal methylation (erm) genes which encode for the modification of ribosomal target through methylation and msr(C) genes which mediate the extrusion of the antibiotic (Munita and Arias, 2016). Moreover, the detection of tet(L), tet(S) and tet(M) genes indicate that tetracycline resistance was due to ribosomal target protection and efflux-mediated mechanisms (Munita and Arias, 2016). Chloramphenicol resistance was mediated by the chloramphenicol acetyltransferase (cat) gene responsible for inactivation of chloramphenicol antibiotic, while ciprofloxacin resistance was encoded by gyrA and parC mutations in the subunits of the DNA gyrase and topoisomerase IV enzymes (Munita and Arias, 2016).
Furthermore, the presence of ClpL gene suggests exposure of these Enterococcus spp. to heat and other adverse environmental conditions. These environmental stress factors have been demonstrated to select for resistant bacteria and influence susceptibility to a range of antibiotics (Huang et al., 2021). The finding of low resistance to ampicillin, linezolid and vancomycin is in line with other reports (Mannu et al., 2003; Bortolaia et al., 2016). Although linezolid resistance is considered uncommon due to its limited use, findings show that the use of other antimicrobials such as macrolides may select for resistance against linezolid (Tyson et al., 2018).
All three livestock farms sampled in this study used antibiotics including tetracycline, penicillin and colistin, similar to other studies in the country where antibiotic use has been described as inappropriate and unregulated (Donkor et al., 2012; Osei Sekyere, 2014; Boamah et al., 2016; Nkansa, 2020). Enterococci notably acquire AMR genes through plasmids and transposons, chromosomal exchange or mutation (Iweriebor et al., 2015; Torres et al., 2018). Mobile genetic elements including integrative conjugative elements, plasmid replicons, insertion sequences and transposons were identified among the isolates confirming the potential of these isolates to acquire and transfer AMR determinants between like-species and other pathogens. Co-occurrence between plasmid replicons (rep1, rep2, repUS43, repUS47, rep9a, rep9b) and AMR genes on the same contig was observed in 70% (28/40) of the isolates. Tetracycline resistance genes [tet(M),tet(L)] and macrolide resistance gene [erm(B)] more frequently co-existed with repUS43, rep9a and rep9b plasmid replicons. The association between tet(M) and tet(L) and repUS43 and rep9b was also reported in a similar study (Fatoba et al., 2022). Aminoglycoside resistance genes [aph(3′)-III, ant(6)-Ia] tended to be more frequently associated with rep1 plasmid, while another aminoglycoside resistance gene (str) often occurred on the same contig with rep7a plasmid replicon. Other mobile genetic elements commonly associated with AMR genes encoding resistance to tetracycline, macrolides and aminoglycosides included transposons (Tn917, cn_5536_ISEnfa1), integrative conjugative element (Tn6009) and insertion sequences (ISEnfa1, ISEfa10). Macrolide resistance gene [erm(B)] was frequently associated with Tn917 consistent with reports elsewhere (Torres et al., 2018). The abundance of plasmid replicon genes and other mobile genetic elements detected in this study confirms the plasticity of the genomes of Enterococcus spp. and supports the potential for horizontal dissemination of the AMR genes (Fatoba et al., 2022). The dissemination of these genes is intensified by clonal expansion which results when horizontally transferred AMR genes become chromosomally integrated (Waddington et al., 2022).
MSLT analysis revealed diverse clones including novel sequence types for both E. faecium and E. faecalis isolates. Novel sequence types for both E. faecalis and E. faecium harbored genes for virulence and resistance with demonstrated phenotypic resistance. The most common African clone, E. faecalis ST16, (Jolley et al., 2018) was found among the isolates (n = 2) in this study. The other clones (ST4, ST16, ST32, ST300) detected, have also been reported in Nigeria and South Africa from similar sources (Jolley et al., 2018). Clonal relationship was observed in four E. faecium isolates [ST1442 (n = 2), ST2269 (n = 2)] from meat and livestock samples. E. faecium ST32 and ST416 recovered from meat clustered with sequence types belonging to the hospital-adapted CC17, known to cause hospital-acquired and clinical infections worldwide (Leavis et al., 2006). Majority of isolates belonging to CC17 are characterized by resistance to quinolones and ampicillin, and the presence of enterococcal surface protein (Leavis et al., 2006; Top et al., 2008). E. faecium isolates (ST32 and ST416) showed resistance to ciprofloxacin but were both susceptible to ampicillin phenotypically, although mutations in the pbp5 gene was detected for both isolates.
The pathogenicity of bacterial species requires not only antibiotic resistance but also the possession of specific virulence factors. The adherence of bacterial cells to host tissues is a critical stage in the development of infection or forming biofilms (Stępień-Pyśniak et al., 2019). In this study, both acm and efaAfm virulence genes were detected in all E. faecium isolates. These genes have also been documented in clinically-derived E. faecium isolates in the United States and Malaysia (Nallapareddy et al., 2008; Soheili et al., 2014). Acm gene, known for encoding an adhesin responsible for E. faecium attachment to collagen, plays a significant role in the competitiveness of this species within clinical environments and is crucial for survival, colonization and infection (Nallapareddy et al., 2008). While the precise function of efaAfm is yet to be confirmed, it is hypothesized to be involved in cell wall adherence (Soheili et al., 2014; Stępień-Pyśniak et al., 2019). Consistent with other reports (Jahansepas et al., 2018; Farman et al., 2019; Trościańczyk et al., 2022), a broader spectrum of virulence genes was identified in E. faecalis than E. faecium, suggesting that E. faecalis is more virulent than E. faecium. Genes mediating adherence to collagen (ace), biotic and abiotic surfaces (efaAfs) and expression of pili on cell surface (ebpA, ebpB, ebpC, SrtA) were all identified in E. faecalis isolates, facilitating cell adhesion and biofilm formation. Genes encoding sex pheromones (cad, camE, cCF10, cOB1) identified in all E. faecalis isolates are noted for promoting biofilm formation and regulation by inducing conjugation between enterococcal cells and mediating the transfer of pheromone-responsive plasmids, which may contain virulence genes (Stępień-Pyśniak et al., 2019; Yoon et al., 2020; Trościańczyk et al., 2022). Furthermore, genes encoding cytolysin (cylB, cylL, cylM), gelatinase (gelE), and hyaluronidase (hylA, hylB), which are known for their ability to damage cells were detected in E. faecalis isolates (Golińska et al., 2013; Stępień-Pyśniak et al., 2019; Lang et al., 2020). Although studies suggest that hyaluronidase encoded by hyl is specific for E. faecium it has been reported in E. faecalis isolates in other studies (Golob et al., 2019; Kiruthiga et al., 2020). ElrA and tpx genes, which are associated with evading host’s immune defenses were also detected, possibly indicating an evolved mechanism in these E. faecalis isolates for immune system evasion. Similar virulence factors have been isolated from E. faecalis isolates from food, animal, and clinical sources in Africa, Europe and Asia (Iweriebor et al., 2015, 2016; Beshiru et al., 2017; Jahansepas et al., 2018; Farman et al., 2019; Stępień-Pyśniak et al., 2019; Yoon et al., 2020). The ST16, ST4 and ST480 E. faecalis identified in this study, have also been reported in cases of clinical infection in other geographical regions (Watanabe et al., 2009;Olsen et al., 2012; Poulsen et al., 2012; Farman et al., 2019) suggesting the widespread nature of these clones. Similar to findings in this study, E. faecalis ST480 identified in Saudi Arabia carried virulence genes encoding sex pheromones [cOB1], biofillm formation-associated pili genes [ebpA, ebpB, SrtA] and adhesins [ace, efaAfs], and were resistant to erythromycin, ciprofloxacin and tetracycline (Farman et al., 2019). Similarly, E. faecalis ST16 isolates were mostly resistant to erythromycin, ciprofloxacin and tetracycline (Farman et al., 2019), and harbored virulence genes encoding cytolysin [cylB, cylL, cylM] and adhesins [efaAfs, ace] (Olsen et al., 2012; Poulsen et al., 2012; Farman et al., 2019). On the other hand, E. faecalis ST4 isolate in this study was resistant to erythromycin, tetracycline and chloramphenicol, and had similar virulence genes encoding biofilm formation [gelE] and adhesins [efaAfs] as E. faecalis ST4 isolated in Japan (Watanabe et al., 2009).
Phylogenetic analysis revealed close genetic relatedness among E. faecalis isolates from livestock sources in this present study and other livestock isolates from South Africa. In contrast to Mbanga et al. (2021), close genetic relatedness was observed among two sheep enterococci isolates and human clinical isolates obtained from South Africa. The enterococci isolates from sheep (76F and 78 M) harbored resistance genes for amphenicols, macrolides, tetracyclines, aminoglycosides, streptogramins and lincosamides, with multiple virulence factors as well. The identified evolutionary relationship between E. faecalis isolates from livestock in this study and those from human clinical sources in South Africa suggests E. faecalis originating from animals could potentially serve as a reservoir for human-associated Enterococcus species. Meanwhile, E. faecium isolates from livestock and meat sources in this study showed significant genetic differences. Livestock-derived E. faecium genomes obtained from South Africa showed some genetic similarity, albeit genetically diverse from isolates in this study. The alignment of E. faecium isolates to geographically distinct clusters is similar to a report by (Freitas et al., 2020), and indicates that these E. faecium populations evolved independently in distinct host environments and geographic regions. Similarly, the rare occurrence of genetic similarity between meat- and livestock-derived Enterococcus spp. may result from slaughter houses sourcing livestock from diverse farms in different geographical settings.
To the best of our knowledge, this is the first study reporting on WGS of Enterococcus spp. in Ghana. The study demonstrated the presence of AMR Enterococcus spp. from livestock and raw meat samples in Ghana, and highlighted the potential of these bacteria to serve as reservoirs for resistant genes and further transmission to other pathogenic bacteria. The study also described the mechanisms of resistance exhibited by Enterococcus spp. against important antimicrobial agents. Genomic analysis also detected hospital-acquired strains and other clinically relevant Enterococcus spp. Phylogenetic analysis revealed evolutionary relationship among various livestock and meat isolates as well as demonstrated the potential of these isolates evolving to cause infections in humans. The use of WGS in this study has provided granular data for AMR surveillance from a one-health perspective to help consolidate efforts to strengthen public health surveillance systems in Ghana.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Ethics statement
The animal study was approved by Ethical clearance for this study was obtained from the institutional review board (IRB) and Animal Care and Use Committee at Noguchi Memorial Institute for Medical Research (FWA00001824). The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
GA: Data curation, Formal analysis, Investigation, Methodology, Software, Writing – original draft, Writing – review & editing. ED: Data curation, Formal analysis, Investigation, Methodology, Software, Writing – review & editing, Conceptualization, Funding acquisition, Resources, Supervision, Validation, Visualization. CO-N: Data curation, Formal analysis, Investigation, Methodology, Software, Writing – review & editing. FAO: Methodology, Writing – review & editing. QM: Methodology, Writing – review & editing. PN: Funding acquisition, Writing – review & editing. BA: Writing – review & editing, Methodology. RH: Writing – review & editing, Funding acquisition, Project administration, Resources, Validation, Visualization. BE: Funding acquisition, Project administration, Resources, Validation, Visualization, Writing – review & editing, Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Supervision, Writing – original draft.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the University of Ghana Research Fund [URF/9/ ILG-067/2015–2016] and a grant from the International Society for Infectious Disease Research. Whole-genome sequencing of the Enterococcus spp. was funded by SeqAfrica. The SeqAfrica project is funded by the Department of Health and Social Care’s Fleming Fund using UK aid. The views expressed in this publication are those of the authors and not necessarily those of the UK Department of Health and Social Care or its Management Agent, Mott MacDonald.
Acknowledgments
The authors are grateful to the Directors of the livestock farms, the slaughterhouse workers, and the Veterinary Services Directorate of the Ministry of Food and Agriculture for their co-operation and technical assistance.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1254896/full#supplementary-material
Footnotes
References
Akansale, R., Adzitey, F., and Teye, G.A. (2019). Knowledge of farmers in antibiotic usage and investigation of antibiotic residues in meats in Sunyani municipality, Ghana ARTICLE INFO ABSTRACT, J food safe & hyg. Available at: http://jfsh.tums.ac.ir.
Akita, L. G., Laudien, J., Biney, C., and Akrong, M. O. (2021). A baseline study of spatial variability of bacteria (total coliform, E. Coli, and Enterococcus spp.) as biomarkers of pollution in ten tropical Atlantic beaches: concern for environmental and public health. Environ. Sci. Pollut. Res. 28, 50941–50965. doi: 10.1007/s11356-021-15432-x
Akrong, M. O., Amu-Mensah, F. K., Amu-Mensah, M. A., Darko, H., Addico, G. N. D., and Ampofo, J. A. (2019). Seasonal analysis of bacteriological quality of drinking water sources in communities surrounding Lake Bosomtwe in the Ashanti region of Ghana. Appl Water Sci 9, 1–6. doi: 10.1007/s13201-019-0959-z
Andrews, S. (2010). ‘FastQC: A quality control tool for high throughput sequence data ’. Babraham Bioinformatics, Babraham Institute: Cambridge, United Kingdom.
Arias, C. A., Contreras, G. A., and Murray, B. E. (2010). ‘Management of multidrug-resistant enterococcal infections’, clinical microbiology and infection. US: Blackwell Publishing Ltd., pp. 555–562.
Ayum, G., and Gifty, A. (2010). Microbial quality of chevon and mutton sold in tamale Metropolis of northern Ghana. J. Appl. Sci. Environ. Manag. 14, 53–55. doi: 10.4314/jasem.v14i4.63257
Bager, F. (2000). DANMAP: monitoring antimicrobial resistance in Denmark. Int. J. Antimicrob. Agents 14, 271–274. doi: 10.1016/S0924-8579(00)00135-7
Beshiru, A., Igbinosa, I. H., Omeje, F. I., Ogofure, A. G., Eyong, M. M., and Igbinosa, E. O. (2017). Multi-antibiotic resistant and putative virulence gene signatures in Enterococcus species isolated from pig farms environment. Microb. Pathog. 104, 90–96. doi: 10.1016/j.micpath.2017.01.020
Beukers, A. G., Zaheer, R., Goji, N., Amoako, K. K., Chaves, A. V., Ward, M. P., et al. (2017). Comparative genomics of Enterococcus spp. isolated from bovine feces. BMC Microbiol. 17:52. doi: 10.1186/s12866-017-0962-1
Boamah, V. E., Agyare, C., Odoi, H., and Dalsgaard, A. (2016). Antibiotic practices and factors influencing the use of antibiotics in selected poultry farms in Ghana. J Antimicrob Agents 2:120. doi: 10.4172/2472-1212.1000120
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bortolaia, V., Espinosa-Gongora, C., and Guardabassi, L. (2016). Human health risks associated with antimicrobial-resistant enterococci and Staphylococcus aureus on poultry meat. CMI 22, 130–140. doi: 10.1016/j.cmi.2015.12.003
Bortolaia, V., Kaas, R. S., Ruppe, E., Roberts, M. C., Schwarz, S., Cattoir, V., et al. (2020). ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 75, 3491–3500. doi: 10.1093/jac/dkaa345
Carattoli, A., Zankari, E., García-Fernández, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using plasmidfinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
CLSI. (2021). M100 performance standards for antimicrobial susceptibility testing a CLSI supplement for global application. 31st Edition. Available at: www.clsi.org.
Collignon, P. J., and McEwen, S. A. (2019). One health-its importance in helping to better control antimicrobial resistance. Tropical Med. Infectious Dis. 4:22. doi: 10.3390/tropicalmed4010022
Donkor, E. S., Newman, M. J., and Yeboah-Manu, D. (2012). Epidemiological aspects of non-human antibiotic usage and resistance: implications for the control of antibiotic resistance in Ghana. Tropical Med. Int. Health 17, 462–468. doi: 10.1111/j.1365-3156.2012.02955.x
European Food Safety Authority, European Centre for Disease Prevention and Control (2013). The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2011. EFSA J. 11:3196. doi: 10.2903/j.efsa.2013.3196
Farman, M., Yasir, M., al-Hindi, R. R., Farraj, S. A., Jiman-Fatani, A. A., Alawi, M., et al. (2019). Genomic analysis of multidrug-resistant clinical Enterococcus faecalis isolates for antimicrobial resistance genes and virulence factors from the western region of Saudi Arabia. ARIC 8, 1–11. doi: 10.1186/s13756-019-0508-4
Fatoba, D. O., Amoako, D. G., Akebe, A. L. K., Ismail, A., and Essack, S. Y. (2022). Genomic analysis of antibiotic-resistant Enterococcus spp. reveals novel enterococci strains and the spread of plasmid-borne Tet(M), Tet(L) and Erm(B) genes from chicken litter to agricultural soil in South Africa. J. Environ. Manag. 302:114101. doi: 10.1016/j.jenvman.2021.114101
Feil, E. J., Li, B. C., Aanensen, D. M., Hanage, W. P., and Spratt, B. G. (2004). eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186, 1518–1530. doi: 10.1128/JB.186.5.1518-1530.2004
Freitas, A. R., Tedim, A. P., Duarte, B., Elghaieb, H., Abbassi, M. S., Hassen, A., et al. (2020). Linezolid-resistant (Tn6246::fexB-poxtA) Enterococcus faecium strains colonizing humans and bovines on different continents: similarity without epidemiological link. J. Antimicrob. Chemother. 75, 2416–2423. doi: 10.1093/jac/dkaa227
Golińska, E., Tomusiak, A., Gosiewski, T., Więcek, G., Machul, A., Mikołajczyk, D., et al. (2013). Virulence factors of Enterococcus strains isolated from patients with inflammatory bowel disease. World J. Gastroenterol. 19, 3562–3572. doi: 10.3748/wjg.v19.i23.3562
Golob, M., Pate, M., Kušar, D., Dermota, U., Avberšek, J., Papić, B., et al. (2019). Antimicrobial resistance and virulence genes in Enterococcus faecium and Enterococcus faecalis from humans and retail red meat. Biomed. Res. Int. 2019, 1–12. doi: 10.1155/2019/2815279
Habib, I., Ghazawi, A., Lakshmi, G. B., Mohamed, M. Y. I., Li, D., Khan, M., et al. (2022). Emergence and genomic characterization of the first reported optrA-carrying linezolid-resistant enterococci isolated from retail broiler meat in the United Arab Emirates. Foods. 11:3190. doi: 10.3390/foods11203190
Hasman, H., Saputra, D., Sicheritz-Ponten, T., Lund, O., Svendsen, C. A., Frimodt-Møller, N., et al. (2014). Rapid whole-genome sequencing for detection and characterization of microorganisms directly from clinical samples. J. Clin. Microbiol. 52, 139–146. doi: 10.1128/JCM.02452-13
Huang, L., Ahmed, S., Gu, Y., Huang, J., An, B., Wu, C., et al. (2021). The effects of natural products and environmental conditions on antimicrobial resistance. Mol. Ther. 26:4277. doi: 10.3390/molecules26144277
Iweriebor, B. C., Obi, L. C., and Okoh, A. I. (2015). Virulence and antimicrobial resistance factors of Enterococcusspp. Isolated from fecal samples from piggery farms in eastern cape, South Africa ecological and evolutionary microbiology. BMC Microbiol. 15:136. doi: 10.1186/s12866-015-0468-7
Iweriebor, B. C., Obi, L. C., and Okoh, A. I. (2016). Macrolide, glycopeptide resistance and virulence genes in Enterococcus species isolates from dairy cattle. J. Med. Microbiol. 65, 641–648. doi: 10.1099/jmm.0.000275
Jackson, C. R., Fedorka-Cray, P. J., Barrett, J. B., and Ladely, S. R. (2004). Effects of tylosin use on erythromycin resistance in enterococci isolated from swine. Appl. Environ. Microbiol. 70, 4205–4210. doi: 10.1128/AEM.70.7.4205-4210.2004
Jahansepas, A., Aghazadeh, M., Rezaee, M. A., Hasani, A., Sharifi, Y., Aghazadeh, T., et al. (2018). Occurrence of Enterococcus faecalis and Enterococcus faecium in various clinical infections: detection of their drug resistance and virulence determinants. Microb. Drug Resist. 24, 76–82. doi: 10.1089/mdr.2017.0049
Joensen, K. G., Scheutz, F., Lund, O., Hasman, H., Kaas, R. S., Nielsen, E. M., et al. (2014). Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Microbiol. 52, 1501–1510. doi: 10.1128/JCM.03617-13
Johansson, M. H. K., Bortolaia, V., Tansirichaiya, S., Aarestrup, F. M., Roberts, A. P., and Petersen, T. N. (2021). Detection of mobile genetic elements associated with antibiotic resistance in Salmonella enterica using a newly developed web tool: MobileElementFinder. J. Antimicrob. Chemother. 76, 101–109. doi: 10.1093/JAC/DKAA390
Jolley, K. A., Bray, J. E., and Maiden, M. C. J. (2018). Open-access bacterial population genomics: BIGSdb software, the PubMLST.Org website and their applications. Wellcome Open Res. 3:124. doi: 10.12688/wellcomeopenres.14826.1
Katakweba, A. A. S., Muhairwa, A. P., Lupindu, A. M., Damborg, P., Rosenkrantz, J. T., Minga, U. M., et al. (2018). First report on a randomized investigation of antimicrobial resistance in fecal Indicator Bacteria from livestock, poultry, and humans in Tanzania. Microb. Drug Resist. 24, 260–268. doi: 10.1089/mdr.2016.0297
Kiruthiga, A., Padmavathy, K., Shabana, P., Naveenkumar, V., Gnanadesikan, S., and Malaiyan, J. (2020). Improved detection of esp, hyl, asa1, gelE, cylA virulence genes among clinical isolates of enterococci. Biomed Central Res. Notes 13, 1–7. doi: 10.1186/s13104-020-05018-0
Klibi, N., Said, L. B., Jouini, A., Slama, K. B., López, M., Sallem, R. B., et al. (2013). Species distribution, antibiotic resistance and virulence traits in enterococci from meat in Tunisia. Meat Sci. 93, 675–680. doi: 10.1016/j.meatsci.2012.11.020
Labi, A. K., Yawson, A. E., Ganyaglo, G. Y., and Newman, M. J. (2015). Prevalence and associated risk factors of asymptomatic bacteriuria in ante-Natal clients in a large teaching Hospital in Ghana. Ghana Med. J. 49, 154–158. doi: 10.4314/gmj.v49i3.5
Lang, S., Demir, M., Duan, Y., Martin, A., and Schnabl, B. (2020). Cytolysin-positive Enterococcus faecalis is not increased in patients with non-alcoholic steatohepatitis. Liver Int. 40, 860–865. doi: 10.1111/liv.14377
Larsen, M. V., Cosentino, S., Rasmussen, S., Friis, C., Hasman, H., Marvig, R. L., et al. (2012). Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 50, 1355–1361. doi: 10.1128/JCM.06094-11
Leavis, H. L., Bonten, M. J., and Willems, R. J. (2006). Identification of high-risk enterococcal clonal complexes: global dispersion and antibiotic resistance. Curr. Opin. Microbiol. 9, 454–460. doi: 10.1016/j.mib.2006.07.001
Letunic, I., and Bork, P. (2021). Interactive tree of life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
Lu, J., Breitwieser, F. P., Thielen, P., and Salzberg, S. L. (2017). Bracken: estimating species abundance in metagenomics data. PeerJ Comput Sci 3:e104. doi: 10.7717/peerj-cs.104
Mannu, L., Paba, A., Daga, E., Comunian, R., Zanetti, S., Duprè, I., et al. (2003). Comparison of the incidence of virulence determinants and antibiotic resistance between Enterococcus faecium strains of dairy, animal and clinical origin. Int. J. Food Microbiol. 88, 291–304. doi: 10.1016/S0168-1605(03)00191-0
Mbanga, J., Amoako, D. G., Abia, A. L. K., Allam, M., Ismail, A., and Essack, S. Y. (2021). Genomic analysis of Enterococcus spp. isolated from a wastewater treatment plant and its associated waters in Umgungundlovu District, South Africa. Front. Microbiol. 12:648454. doi: 10.3389/fmicb.2021.648454
McEwen, S. A., and Collignon, P. J. (2018). Antimicrobial resistance: a one health perspective. Microbiol. Spectrum 6, 521–547. doi: 10.1128/microbiolspec.arba-0009-2017
Molechan, C., Amoako, D. G., Abia, A. L. K., Somboro, A. M., Bester, L. A., and Essack, S. Y. (2019). Molecular epidemiology of antibiotic-resistant Enterococcus spp. from the farm-to-fork continuum in intensive poultry production in KwaZulu-Natal, South Africa. Sci. Total Environ. 692, 868–878. doi: 10.1016/j.scitotenv.2019.07.324
Nallapareddy, S. R., Singh, K. V., Okhuysen, P. C., and Murray, B. E. (2008). A functional collagen adhesin gene, acm, in clinical isolates of Enterococcus faecium correlates with the recent success of this emerging nosocomial pathogen. Infect. Immun. 76, 4110–4119. doi: 10.1128/IAI.00375-08
Ngbede, E. O., Raji, M. A., Kwanashie, C. N., and Kwaga, J. K. P. (2017). Antimicrobial resistance and virulence profile of enterococci isolated from poultry and cattle sources in Nigeria. Trop. Anim. Health Prod. 49, 451–458. doi: 10.1007/s11250-016-1212-5
O’Driscoll, T., and Crank, C. W. (2015). Vancomycin-resistant enterococcal infections: epidemiology, clinical manifestations, and optimal management. Infec. Drug Resist. 8, 217–230. doi: 10.2147/IDR.S54125
Obiri-Danso, K., Okore-Hanson, A., and Jones, K. (2003). The microbiological quality of drinking water sold on the streets in Kumasi, Ghana. Lett. Appl. Microbiol. 37, 334–339. doi: 10.1046/j.1472-765X.2003.01403.x
Obiri-Danso, K., Weobong, C. A. A., and Jones, K. (2005). Aspects of health-related microbiology of the subin, an urban river in Kumasi, Ghana. J. Water Health 3, 69–76. doi: 10.2166/wh.2005.0007
Olsen, R. H., Schønheyder, H. C., Christensen, H., and Bisgaard, M. (2012). Enterococcus faecalis of human and poultry origin share virulence genes supporting the zoonotic potential of E. faecalis. Zoonoses Public Health 59, 256–263. doi: 10.1111/j.1863-2378.2011.01442.x
Osei Sekyere, J. (2014). Antibiotic types and handling practices in disease management among pig farms in Ashanti region, Ghana. J. Vet. Med. 1, 2014, –8. doi: 10.1155/2014/531952
Poulsen, L. L., Bisgaard, M., Son, N. T., Trung, N. V., An, H. M., and Dalsgaard, A. (2012). Enterococcus faecalis clones in poultry and in humans with urinary tract infections, Vietnam. Emerg. Infect. Dis. 18, 1096–1100. doi: 10.3201/eid1807.111754
Quansah, J. K., Escalante, C., Kunadu, A., Saalia, F., and Chen, J. (2020). Pre- and post-harvest practices of Urban leafy green vegetable farmers in Accra, Ghana and their association with microbial quality of vegetables produced. Agriculture (Basel) 10:18. doi: 10.3390/agriculture10010018
Quansah, J. K., Kunadu, A. P. H., Saalia, F. K., Díaz-Pérez, J., and Chen, J. (2018). Microbial quality of leafy green vegetables grown or sold in Accra metropolis, Ghana. Food Control 86, 302–309. doi: 10.1016/j.foodcont.2017.11.001
Soheili, S., Ghafourian, S., Sekawi, Z., Neela, V., Sadeghifard, N., Ramli, R., et al. (2014). Wide distribution of virulence genes among Enterococcus faecium and Enterococcus faecalis clinical isolates. ScientificWorldJournal. 2014, 1–6. doi: 10.1155/2014/623174
Stępień-Pyśniak, D., Hauschild, T., Kosikowska, U., Dec, M., and Urban-Chmiel, R. (2019). Biofilm formation capacity and presence of virulence factors among commensal Enterococcus spp. from wild birds. Sci. Rep. 9:11204. doi: 10.1038/s41598-019-47602-w
Top, J., Willems, R., van der Velden, S., Asbroek, M., and Bonten, M. (2008). Emergence of clonal complex 17 Enterococcus faecium in the Netherlands. J. Clin. Microbiol. 46, 214–219. doi: 10.1128/JCM.01351-07
Torres, C., Alonso, C. A., Ruiz-Ripa, L., León-Sampedro, R., del Campo, R., and Coque, T. M. (2018). Antimicrobial resistance in Enterococcus spp. of animal origin. Microbiol. Spectrum 6, 1–41. doi: 10.1128/microbiolspec.arba-0032-2018
Trościańczyk, A., Nowakiewicz, A., Osińska, M., Łagowski, D., Gnat, S., and Chudzik-Rząd, B. (2022). Comparative characteristics of sequence types, genotypes and virulence of multidrug-resistant Enterococcus faecium isolated from various hosts in eastern Poland. Spread of clonal complex 17 in humans and animals. Res. Microbiol. 173:103925. doi: 10.1016/j.resmic.2022.103925
Tyson, G. H., Sabo, J. L., Hoffmann, M., Hsu, C. H., Mukherjee, S., Hernandez, J., et al. (2018). Novel linezolid resistance plasmids in Enterococcus from food animals in the USA. J. Antimicrob. Chemother. 73, 3254–3258. doi: 10.1093/jac/dky369
Waddington, C., Carey, M. E., Boinett, C. J., Higginson, E., Veeraraghavan, B., and Baker, S. (2022). Exploiting genomics to mitigate the public health impact of antimicrobial resistance. Genome Med BioMed Central Ltd. 14:15. doi: 10.1186/s13073-022-01020-2
Watanabe, S., Kobayashi, N., Quinones, D., Nagashima, S., Uehara, N., and Watanabe, N. (2009). Genetic diversity of enterococci harboring the high-level gentamicin resistance gene aac(6 0)-Ie-aph(200)-Ia or aph(2 00)-Ie in a Japanese hospital. Microb. Drug Resist. 15, 185–194. doi: 10.1089/mdr.2009.0917
Weterings, V., Zhou, K., Rossen, J. W., van Stenis, D., Thewessen, E., Kluytmans, J., et al. (2015). An outbreak of colistin-resistant Klebsiella pneumoniae carbapenemase-producing Klebsiella pneumoniae in the Netherlands (July to December 2013), with inter-institutional spread. Eur. J. Clin. Microbiol. Infect. Dis. 34, 1647–1655. doi: 10.1007/s10096-015-2401-2
Wood, D. E., Lu, J., and Langmead, B. (2019). Improved metagenomic analysis with kraken 2. Genome Biol. 20:257. doi: 10.1186/s13059-019-1891-0
Yoon, S., Son, S. H., Kim, Y. B., Seo, K. W., and Lee, Y. J. (2020). Molecular characteristics of optrA-carrying Enterococcus faecalis from chicken meat in South Korea. J. American Assoc. Instructors Invest. Poultry Husbandry 99, 6990–6996. doi: 10.1016/j.psj.2020.08.062
Keywords: Enterococcus spp., antimicrobial resistance, whole-genome sequencing, livestock, raw meat, Africa
Citation: Amuasi GR, Dsani E, Owusu-Nyantakyi C, Owusu FA, Mohktar Q, Nilsson P, Adu B, Hendriksen RS and Egyir B (2023) Enterococcus species: insights into antimicrobial resistance and whole-genome features of isolates recovered from livestock and raw meat in Ghana. Front. Microbiol. 14:1254896. doi: 10.3389/fmicb.2023.1254896
Edited by:
Shangshang Qin, Zhengzhou University, ChinaReviewed by:
Nikolaos Strepis, Erasmus Medical Center, NetherlandsTeresa Semedo-Lemsaddek, University of Lisbon, Portugal
Copyright © 2023 Amuasi, Dsani, Owusu-Nyantakyi, Owusu, Mohktar, Nilsson, Adu, Hendriksen and Egyir. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Beverly Egyir, YmVneWlyQG5vZ3VjaGkudWcuZWR1Lmdo; YmV2ZXJseWVneWlyQGdtYWlsLmNvbQ==
 Grebstad Rabbi Amuasi1
Grebstad Rabbi Amuasi1 Christian Owusu-Nyantakyi
Christian Owusu-Nyantakyi Rene S. Hendriksen
Rene S. Hendriksen Beverly Egyir
Beverly Egyir