
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 02 October 2023
Sec. Biology of Archaea
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1254891
This article is part of the Research Topic Molecular Biology of Archaea - 2022 View all 19 articles
CRISPR (clustered regularly interspaced short palindromic repeats)-Cas systems are widely distributed among bacteria and archaea. In this study, we demonstrate the successful utilization of the type I-D CRISPR-Cas system for genetic engineering in the thermoacidophilic archaeon Sulfolobus acidocaldarius. Given its extreme growth conditions characterized by a temperature of 75°C and pH 3, an uracil auxotrophic selection system was previously established, providing a basis for our investigations. We developed a novel plasmid specifically designed for genome editing, which incorporates a mini-CRISPR array that can be induced using xylose, resulting in targeted DNA cleavage. Additionally, we integrated a gene encoding the β-galactosidase of Saccharolobus solfataricus into the plasmid, enabling blue-white screening and facilitating the mutant screening process. Through the introduction of donor DNA containing genomic modifications into the plasmid, we successfully generated deletion mutants and point mutations in the genome of S. acidocaldarius. Exploiting the PAM (protospacer adjacent motif) dependence of type I systems, we experimentally confirmed the functionality of three different PAMs (CCA, GTA, and TCA) through a self-targeting assessment assay and the gene deletion of upsE. Our findings elucidate the application of the endogenous Type I-D CRISPR-Cas system for genetic engineering in S. acidocaldarius, thus expanding its genetic toolbox.
The CRISPR-Cas system is an RNA-guided prokaryotic defense system to protect bacterial and archaeal cells from foreign DNA, such as virus invasion or conjugative plasmids (Barrangou et al., 2007; Brouns et al., 2008; Hale et al., 2009; Garneau et al., 2010; Marraffini and Sontheimer, 2010; Westra et al., 2012; Elmore et al., 2015). This self-defense mechanism consists of various steps (McGinn and Marraffini, 2019). First, the infected cell acquires a piece of foreign DNA and incorporates it into its own genome between specific clustered regularly interspaced short palindromic repeats (CRISPR). The integrated sequences are called spacers, which function as a memory from past survived infections (Fineran and Charpentier, 2012). Depending on the species, there are several of these clusters, which are accompanied by specific genes encoding for CRISPR-associated (Cas) proteins. CRISPR systems are divided into 2 classes, 6 types, and 33 subtypes and several variants, according to the properties of the Cas proteins (Makarova et al., 2015). Class 1 systems (type I, III, and IV) consist of a ribonucleoprotein (RNP) effector complex that is composed of several Cas proteins and bound crRNA (CRISPR RNA) during interference (Liu and Doudna, 2020). In comparison, class 2 systems (type II, V, and VI) only utilize one multidomain Cas protein, which interacts with crRNA for interference (Makarova et al., 2020; Nidhi et al., 2021). Approximately 47% of analyzed bacterial and archaeal genomes contain CRISPR systems, which, however, are much more prevalent in archaea (87%) than in bacteria (50%). Type I systems are the most dominant form of CRISPR systems, present in 64 and 60% of archaea and bacteria, respectively (Makarova et al., 2011, 2013). Type I and II systems interfere with invading DNA (Sinkunas et al., 2013), whereas type III systems, for example, interact with DNA and RNA in a transcriptional-dependent fashion (Samai et al., 2015).
In Sulfolobales, most CRISPR systems include type I-A, I-D, type III-B, and III-D (Vestergaard et al., 2014). Most research studies regarding CRISPR systems in Sulfolobales were performed in Saccharolobus solfataricus and Sulfolobus islandicus, showing the roles of different Cas proteins during CRISPR activity and the necessity of a protospacer adjacent motif (PAM) for type I systems (Peng et al., 2013), properties of protospacer and crRNA for interference (Manica et al., 2013; Mousaei et al., 2016), and the degradation properties of the type I-D system (Lin et al., 2020). For a more detailed insight into the different aspects of CRISPR-Cas systems in Sulfolobales, we refer to the reviews of the study mentioned in the reference (Garrett et al., 2011, 2015; Cannone et al., 2013; Manica and Schleper, 2013; Zhang and White, 2013; Zink et al., 2020).
After exploring various aspects of CRISPR-Cas systems in Sulfolobales, the endogenous CRISPR type I and III systems in S. islandicus were utilized for genome editing (Li et al., 2016). In this approach, the endogenous CRISPR-Cas system was primed using a specific spacer incorporated into a mini-CRISPR array integrated into a plasmid. By targeting a wild-type sequence, the CRISPR system induced DNA damage at the desired genomic site. Notably, the plasmid itself was not targeted due to the absence of a PAM sequence. The resulting DNA damage was subsequently repaired by cellular DNA repair mechanisms, thereby enabling the introduction of genetic modifications through homologous recombination with a provided repair fragment (Yang et al., 2020).
Furthermore, Schleper et al. demonstrated the potential of CRISPR systems in Sulfolobales by utilizing the type III system for RNA interference assays in Sa. solfataricus. Their studies highlighted the versatility and applicability of CRISPR systems in this context (Zebec et al., 2016; Zink et al., 2021).
S. acidocaldarius exhibits CRISPR-Cas type I-D and III-D systems (Figure 1) (Makarova et al., 2020). The type I-D system is a unique type I system, as it does not have the Cas8 scaffold protein, but the Cas10 protein, which originates from a type III system, leads to the hypothesis of an evolutionary link between type I and III systems (Lin et al., 2020). The cluster contains the signature protein from the type I system, Cas3′ (helicase domain), without the endonuclease domain (Cas3″), as well as a Cas10-like large subunit, Cas10d from the type III system, together with Csc1 (Cas5) and the backbone Csc2 (Cas7) (Makarova et al., 2011). These Cas genes are associated with CRISPR clusters with different numbers of spacer sequences throughout the genome, with the type I-D genes associated with cluster 1 and cluster 2 (Figure 1).

Figure 1. CRISPR loci in S. acidocaldarius. CRISPR type I-D and III-D systems are allocated on different loci of the genome. The Cas proteins (orange and blue, respectively) are associated with CRISPR clusters with different repeat sequences and different quantities of spacers. Furthermore, there are unclassified Type III system and other Cas proteins (white) that probably do not form a functional Cas cluster.
To distinguish the foreign DNA from inherent DNA, type I and II systems need specific motifs that can be found next to the targeting sites (protospacer), called protospacer adjacent motif (PAM) (Mojica et al., 2009; Fischer et al., 2012). These PAMs differ in length and sequence between species and are located next to the protospacer at the 5′ end in type I systems and 3′ end in type II systems (Gleditzsch et al., 2019). These sequences can be discovered by analyzing the adjacent motifs of the protospacers of previous infections in the endogenous CRISPR clusters via bioinformatics tools. It is shown in S. islandicus LAL14/1 that the type I-D system is able to cleave dsDNA using the PAM GTN, similar to other type I systems, and also ssDNA by a ruler-like mechanism that is similar to type III systems without the need for a PAM (Lin et al., 2020). Previously, Lillestøl et al. proposed CCN, GTN, and TCN as PAMs for different Sulfolobales species (Lillestøl et al., 2009).
In this study, we used the endogenous type I CRISPR system of S. acidocaldarius to generate deletion mutants and introduce point mutations in previously characterized genes. Using this as proof of concept, we were able to expand the genetic toolbox of this thermoacidophilic model organism.
For all experiments, Sulfolobus acidocaldarius MW001 was used as the background strain and aerobically grown in Brock standard medium (Brock et al., 1972) supplemented with 0.1% (w/v) N-Z-Amine and 0.2% (w/v) dextrin at pH of 3 and 75°C. For inducing protein expression from plasmids containing a xylose-inducible promoter, dextrin was replaced with 0.2% xylose. For complementation of the uracil auxotrophy, 20 μg/ml of uracil was added to the medium.
To cultivate S. acidocaldarius on a plate, Brock medium that was concentrated two times supplemented with 6 mM CaCl2, 20 mM MgCl2, 0.2% (w/v) N-Z-Amine, and 0.4% (w/v) dextrin was mixed in equal amount with freshly boiled 1.4% (w/v) Gelrite (Carl Roth, Karlsruhe, German). Cultures on plates were incubated for 5–7 days at 75°C in humidified containers. To remove plasmids from cells, they were streaked out on the second selection plates, which contained an additional 10 μg/ml uracil and 100 μg/ml 5-FOA.
S. acidocaldarius strain MW001 was grown in 50 ml of Brock medium supplemented with NZ-Amine, dextrin, and uracil. Upon reaching an optical density (OD600) of 0.5–0.7, a portion of the culture was transferred to 50 ml of fresh medium and harvested at an optical density OD600 of 0.2–0.3. The culture was cooled down on ice and then centrifuged for 15 min at 4000 × g at 4°C and washed three times with 30 ml and one last time with 1 ml of ice-cold 20 mM sucrose. The cells were resuspended in 20 mM sucrose to reach a final theoretical optical density of 20 and divided into a portion of 50 μl and immediately used for transformation or stored at −80°C without freezing in liquid nitrogen.
To prevent restriction by the SuaI restriction system, plasmids were methylated prior to their transformation into S. acidocaldarius. For this purpose, E. coli ER1821 cells, containing the additional plasmid pM.EsaBC4I (New England Biolabs, Frankfurt am Main, Germany), were transformed with the obtained plasmids. The methylated plasmids were, then, purified and electroporated into competent MW001 cells, using a Gene Pulser Xcell (Bio-Rad, München, Germany) with a constant time protocol at 1.5 kV, 25 μF, 600 Ω in 1-mm cuvettes. Cells were regenerated for 30 min at 75°C in 450 μl Brock medium without pH adjustment. To recover the transformants, 50 μl of cell suspension was inoculated in 5 ml of Brock medium supplemented with 0.2% (w/v) xylose and 0.1% (w/v) dextrin and incubated for 2 days at 75°C. From these cultures, 150 μl were plated per selection plate.
To obtain a base vector for using the endogenous CRISPR-Cas system, an expression plasmid named pSVA13134 was designed. To that end, lacI/lacZ was cloned into the NcoI/NotI cloning site of pSVAxylFX-Stop (van der Kolk et al., 2020) using primers 11670/11671 for amplifying the plasmid and 11672/11673 for amplifying lacI/lacZ (Table 1). The primers contained overlapping regions at each end, which consisted of the cluster 1 repeat (GTAATAACGACAAGAAACTAAAAC) and SapI restriction site. In between the repeats was the lacI/lacZ cluster. Cloning was performed using in vivo assembly resulting in pSVAxylFX-CRISPR. The lacS (sso3019) gene of Sa. solfataricus, encoding a β-galactosidase, was also added using NdeI and NheI with the primers 11642/11643 resulting in pSVA13133.
Primers 12042/12049 were used to delete additional ApaI and XhoI sites and 12050/12051 to move an ApaI restriction site to another position, using T4 PNK cloning, to expand the usable multiple cloning site of the resulting plasmid pSVA13134.
Repair fragments for homologous recombination, containing genetic modifications, were then cloned into the ApaI/NdeI restriction sites using the primers, as shown in Table 1. For that, an upstream and a downstream fragment of upsE were amplified from genomic S. acidocaldarius DNA, and both PCR products ligated, using overlap extension PCR, resulting in pSVA13271 (for upsE KO) and pSVA13280 (Walker A mutation K232A) (Table 2). Spacers for targeted CRISPR activity were generated by annealing the forward and reverse primers at 98°C for 10 s followed by 50°C for 10 s. The primer contained SapI restriction sites, parts of the cluster 1 repeat, and target sequence for the CRISPR system.
For the self-assessment protocol, the protospacer of the first spacer of CRISPR cluster 1 (AGAAAATATCTCAAGGAGGGCGAGGAAGTATGCGAAAG) was cloned into the SapI restriction site using FX cloning with primers 13606/13607 for the non-PAM control and primers 13608-13613 for the different tested PAMs CCA, GTA, and TCA, respectively (Table 1).
Potential candidates were first selected by blue-white screening, spraying X-Gal (25mg/ml in DMF) diluted in a 1:4 ratio with dextrin [20% (w/v)] on visible colonies. Afterward, the plates were incubated for up to 4 h until colonies turned blue. To verify the genotype of potential mutants, blue colonies were lysed for 2 min in 10 μl 0.2 M NaOH. To prevent DNA denaturation, 60 μl of 0.2 M Tris (pH 7.8) was added, as well as 60 μl of ddH2O added to dilute the sample. After vortexing, 1 μl was used as a template for a 20 μl PCR reaction using the Phusion polymerase. MW001 DNA was used as a wild-type control. For analysis of deletion mutants, plasmid DNA was used as a negative control to ensure that the signal was due to genetic alteration, not from plasmid amplification. After the analysis of gel electrophoresis, PCR products were sequenced (Eurofins Genomics Europe).
UV treatment was carried out following the protocol described by Fröls et al. (2008). In total, 10 ml of culture with an optical density (OD) of 0.2 to 0.3 was exposed to 75 J/m2 of UV light at 254 nm using a UV Crosslinker device (Spectroline, Westbury, NY). The cultures were, then, incubated at 75°C for 3 h. To determine the number of aggregated cells after UV exposure, the cell culture was diluted to OD 0.2, and 5 μl spotted onto a microscope slide coated with a thin layer of 1% (w/v) agarose in Brock minimal medium. After drying the cell suspension, a coverslip was added, and pictures were taken in three fields per sample under a phase contrast microscope. The number of free and aggregated cells (≥3) was counted using the ImageJ cell counter (NIH, Bethesda, MD).
Wagner et al. established a genetic toolbox for S. acidocaldarius in 2012 based on a uracil auxotrophic strain MW001 in combination with a number of plasmids usable for the construction of deletion mutants, or mutants in which genes were either mutated genomically or tags were added to the gene of interest (Wagner et al., 2012). Moreover, using this system, genes were placed ectopically into the genome for genetic modifications (Wagner et al., 2012). To this end, plasmids are integrated into the genomic DNA after transformation via homologous recombination and can be excised after specific selection using 5-FOA and uracil, leading to alteration of the genomic DNA.
For the usage of the CRISPR-Cas system for genetic engineering, we designed plasmids based on the expression vector pSVAxylFX-CRISPR, which replicates in S. acidocaldarius does not integrate into the genome and is based on the plasmid pRN1 (Berkner et al., 2007). It derives from pSVAxylFX-Stop (van der Kolk et al., 2020) and contains 2 CRISPR repeats of cluster 1 (GTAATAACGACAAGAAACTAAAAC), which are downstream of a D-xylose-inducible promoter Pxyl/Psaci_1938. Additionally, lacSSso from Sa. solfataricus was integrated into pSVAxylFX-CRISPR, to allow for blue–white screening in S. acidocaldarius using X-gal and verify successful transformation. The final vector pSVA13134, which was the base for all plasmids used for genetic manipulations (Figure 2A), also contains a multiple cloning site that is suitable for inserting the repair fragment. The spacer/target sequence can be ordered as a primer pair and cloned into pSVA13134 by restriction with SapI (Figure 2B). Spacer primers are designed by searching for a 37 nt protospacer sequence in the target area, which needs to be flanked by a PAM at the 5′ end (Figure 2C). Selection of positive E. coli clones is accomplished with blue–white screening because of the presence of the lacI/lacZ cassette in between the SapI restriction sites (Figure 2A) (Geertsma, 2013).
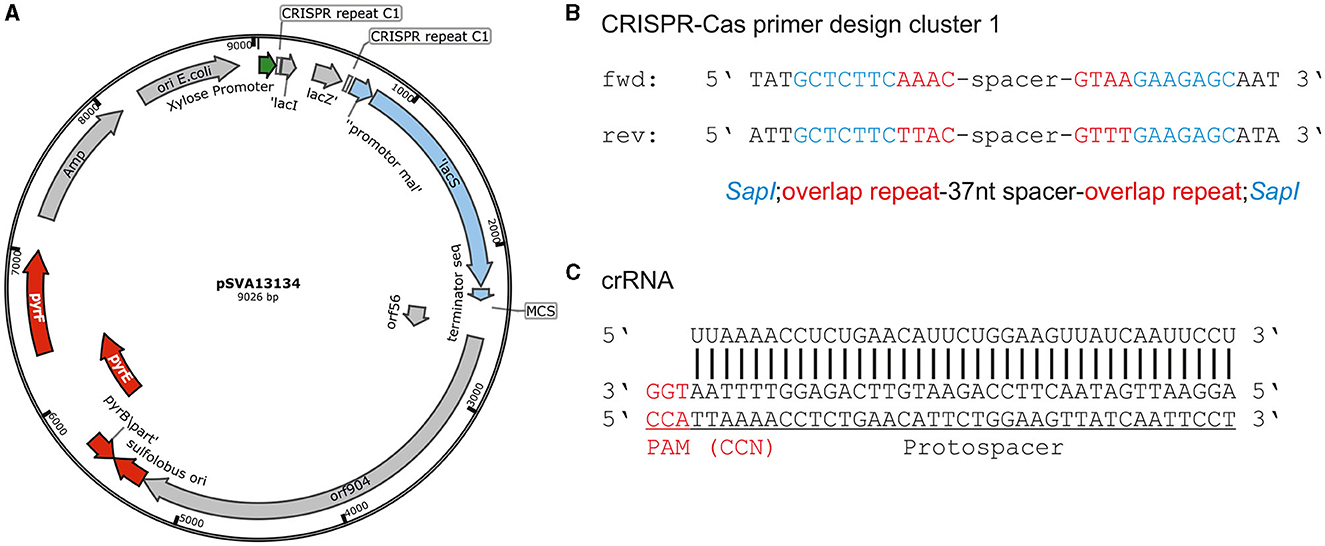
Figure 2. CRISPR base vector and CRISPR RNA design. (A) Plasmid map of base vector pSVA13134 containing xylose-inducible promoter Pxyl/Psaci_1938, repeats of CRISPR cluster 1 (C1), and lacSSso for blue-white screening in S. acidocaldarius under a maltose-inducible promoter. (B) Primer design for cloning of spacer sequence onto pSVA13134 using FX-cloning. Primer consists of SapI restriction site, few nucleotides from overlap of cluster 1 repeats, and a specific spacer depending on the target sequence. The total length is ~65 nt. (C) Protospacer localization downstream of PAM (CCA) and CRISPR RNA binding to the target site for the deletion of upsE.
The CRISPR vectors used in this study are expression vectors, containing a S. acidocaldarius ORI. Therefore, a standard electroporation protocol for expression vectors was used, where transformed cells were plated on the first selection plates after 30 min of recovery at 75°C. However, we did not obtain any colonies with the CRISPR plasmids using this standard protocol. Therefore, we introduced an additional recovery step in the liquid medium after electroporation, which was similar to the lactose selection system in Sa. solfataricus PBL2025 (Albers and Driessen, 2008). Different recovery periods of 1, 2, and 3 d in Brock medium were tested, containing different carbon sources (D-xylose, sucrose, and dextrin). As the plasmid mini-CRISPR array is under the control of a D-xylose-inducible promoter Pxyl/Psaci_1938, induction of the CRISPR array is tested on plates, as well as in liquid medium for the 1–3 days of recovery step. Ultimately, positive genetically modified colonies only formed after 2 days of induction in Brock-NZ-Amine-D-xylose and plating on the first selection plates (Brock-NZ-Amine-dextrin) (Figure 3). No other combination yielded any positive colonies on plates.
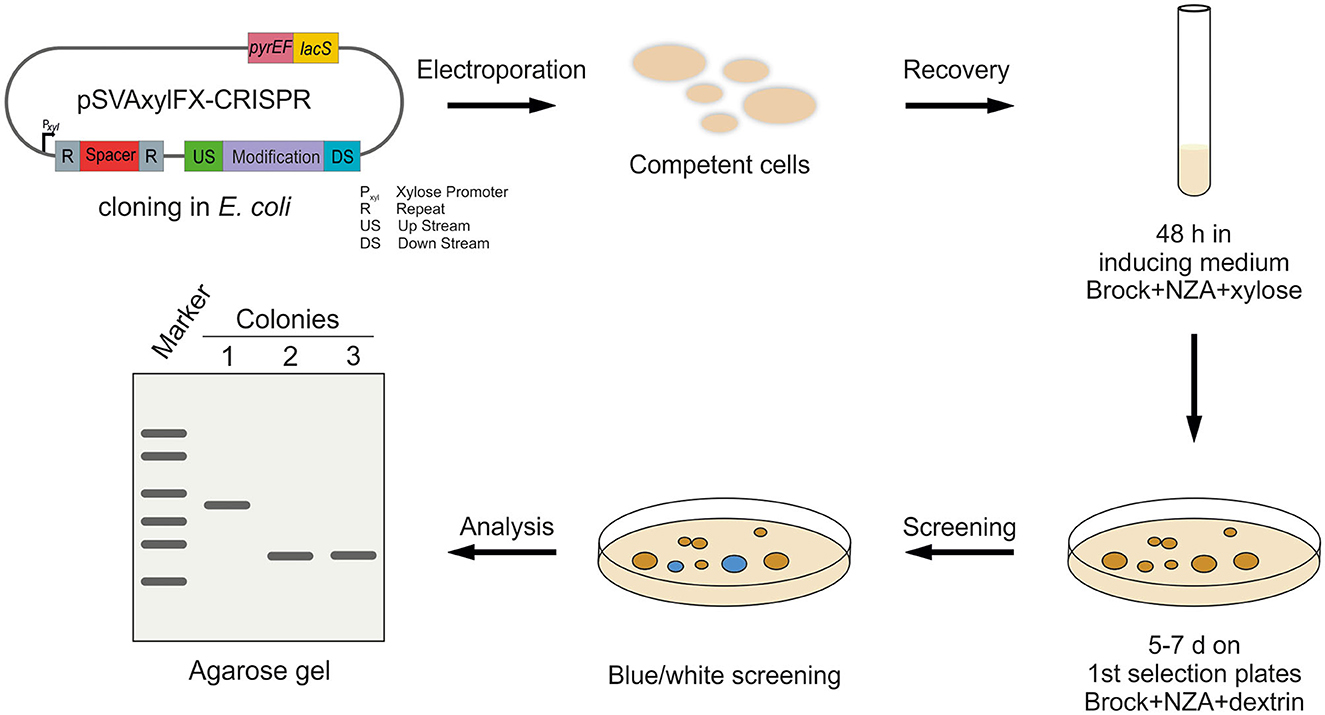
Figure 3. Workflow for CRISPR-based gene modification in S. acidocaldarius. After cloning of the CRISPR plasmids in E. coli, the methylated plasmid is transformed into competent S. acidocaldarius MW001 cells. Afterward, transformants are regenerated for 48 h in standard Brock medium supplemented with NZ-Amine and D-xylose for induction of the CRISPR system. Cells are, then, transferred to the first selection plates (NZ-Amine and dextrin) and incubated at 75°C for 5–7 days. After confirming plasmid presence using blue-white screening, blue colonies are further analyzed using PCR and agarose gel separation.
In general, transformation in S. acidocaldarius always yields a certain amount of false positive colonies on the first selection plates, probably due to uracil cross-feeding from lysed cells. Therefore, transformants were diluted in a 1:100 ratio after transformation (50 μl transformants in 5 ml inducing medium for 2 days to prevent a high amount of background colonies, resulting in a ratio, that allowed for consistent colony formation).
For S. acidocaldarius, Sa. solfataricus, and S. islandicus, several PAM sequences have been predicted bioinformatically by analyzing the targets of the protospacer sequences of CRISPR arrays from different Sulfolobales (Lillestøl et al., 2009). Three commonly found motifs were 5′-CCN-3′, 5′-GTN-3′, and 5′-TCN-3′. To verify the activity of the endogenous CRISPR type I-D system in S. acidocaldarius, a plasmid self-targeting assessment was performed. To that end, a plasmid containing a target sequence of the endogenous CRISPR-Cas system was transformed into S. acidocaldarius MW001. In the case of a functional CRISPR-Cas system and PAM sequence, the CRISPR-Cas RNP complex is able to cleave the plasmid, which also harbors a selection cassette, leading to the formation of fewer colonies compared with a non-target control (Figure 4A). Therefore, the protospacer sequence corresponding to the last acquired spacer of the CRISPR cluster 1 (AGAAAATATCTCAAGGAGGGCGAGGAAGTATGCGAAAG) was used as a target (Figure 4B). The sequence was flanked at the 5′-end by the PAMs CCA, GTA, or TCA in the CRISPR expression cassette, which was designed to mimic the sequence and arrangement of the native type I-D array in S. acidocaldarius. As a negative control, no PAM was inserted at the 5′-end of the target, which was just flanked by the native cluster 1 repeat sequence (AAC) (Figure 4A).
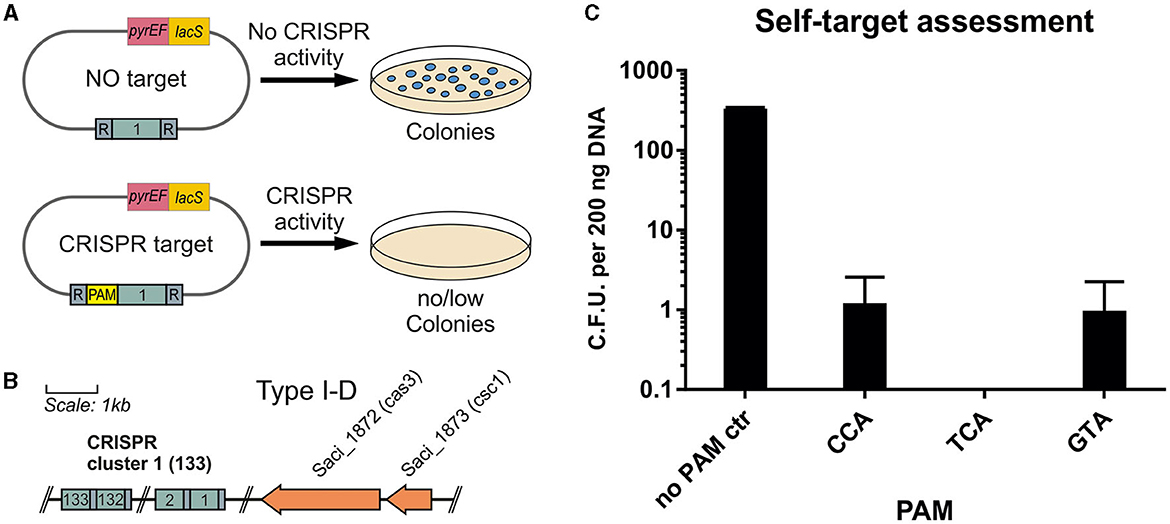
Figure 4. Self-targeting assessment of the endogenous CRISPR type I system. (A) Plasmids containing the corresponding target sequence of the first spacer from the CRISPR cluster 1 are used. (B) To verify an active system and functional cleavage capability, the target sequence is cloned onto plasmid pSVA13134 with and without a flanking PAM sequence. In the case of a non-functional PAM and CRISPR-Cas system, the plasmid is not cleaved, and colonies are able to form. In the case of a functional system, the plasmid is destroyed, and colonies are unable to form on the selective plates. (C) Colonies per 200 ng of plasmid DNA formed in the self-target assessment assay. As a control, the target is in between the native cluster 1 repeats leading to a no PAM control (AAC). Additionally, CCA, TCA, and GTA are used as PAMs accompanying the target at the 5′-end, leading to a possible recognition of the CRISPR-Cas complex. All 3 PAMs showed a high percentage of cleavage with 0 to 3 colonies forming per replicate. The average of three biological replicates is shown.
We showed that the presence of the target sequence for the first spacer of cluster 1 totally abolished the presence of the plasmid when using TCA as PAM. After blue-white screening to verify the presence of the plasmid, there was nearly full clearance for all tested PAMs with ~1 colony per 200 ng plasmid DNA for CCA and GTA PAMs (Figure 4C). The no PAM control yielded, on average, 303 colonies, showing a clearance effect for the used PAMs.
The self-target assessment indicated a functional CRISPR-type I-D system in S. acidocaldarius. The expression of endogenous Cas proteins under the native promoter system was sufficient to generate full clearance of transformed plasmids. Through this method, PAMs can easily be tested, as demonstrated previously, e.g., in Pyrococcus furiosus (Elmore et al., 2015). Notably, Lillestøl predicted CCN as a functional PAM for Sa. solfataricus and S islandicus as no valid PAM was predicted for Sulfolobus acidocaldarius (Lillestøl et al., 2009). Our results demonstrated that the three PAMs published by Lillestøl et al. have a similar impact on the clearing of targeted DNA.
After showing the activity of the CRISPR system and functional PAMs through the self-targeting assessment, we used upsE (saci_1494) as a target to test whether we can obtain a gene deletion using the system. UpsE encodes for the UV pili assembly ATPase (Fröls et al., 2008; Ajon et al., 2011; Wagner et al., 2012; van Wolferen et al., 2013). We chose it as a target as the successful deletion can be additionally verified via UV aggregation assays (Fröls et al., 2008). Therefore, we screened for PAM sequences in the sequence of upsE on the sense strand and used 37 nt downstream of it as a protospacer (Figure 5). The spacer sequence was cloned in between the CRISPR repeats of cluster 1 in the artificial CRISPR array on pSVA13271. This plasmid harbors a repair fragment for homologous recombination, which consists of the 500 bp upstream and 500 bp downstream of upsE and is derived from pSVA13134. We tested three different PAMs CCA, GTA, and TCA using the plasmids pSVA13272, pSVA13273, and pSVA13274, respectively. For transformation, 200 ng of plasmid was transformed, and the cells were incubated for 48 h in Brock medium with NZ-Amine and 0.2% (w/v) D-xylose. D-xylose induces the transcription of the CRISPR array on the plasmid, due to a D-xylose-inducible promoter, leading to the production of crRNA (CRISPR RNA), which then forms an RNP complex with the endogenous CRISPR-Cas proteins (Figure 5). After the formation of colonies on the first selection plates, initial screening of the presence of the plasmid was performed using X-Gal (5-bromo-4-chloro-indolyl-β-D-galactopyranoside) as the gene for the β galactosidase (lacS) is encoded on the plasmid. PCR analysis and subsequent sequencing confirmed that all blue colonies were indeed clones in which the upsE gene was deleted. There was no difference in either of the used PAMs CCA, GTA, and TCA (Figure 6A).
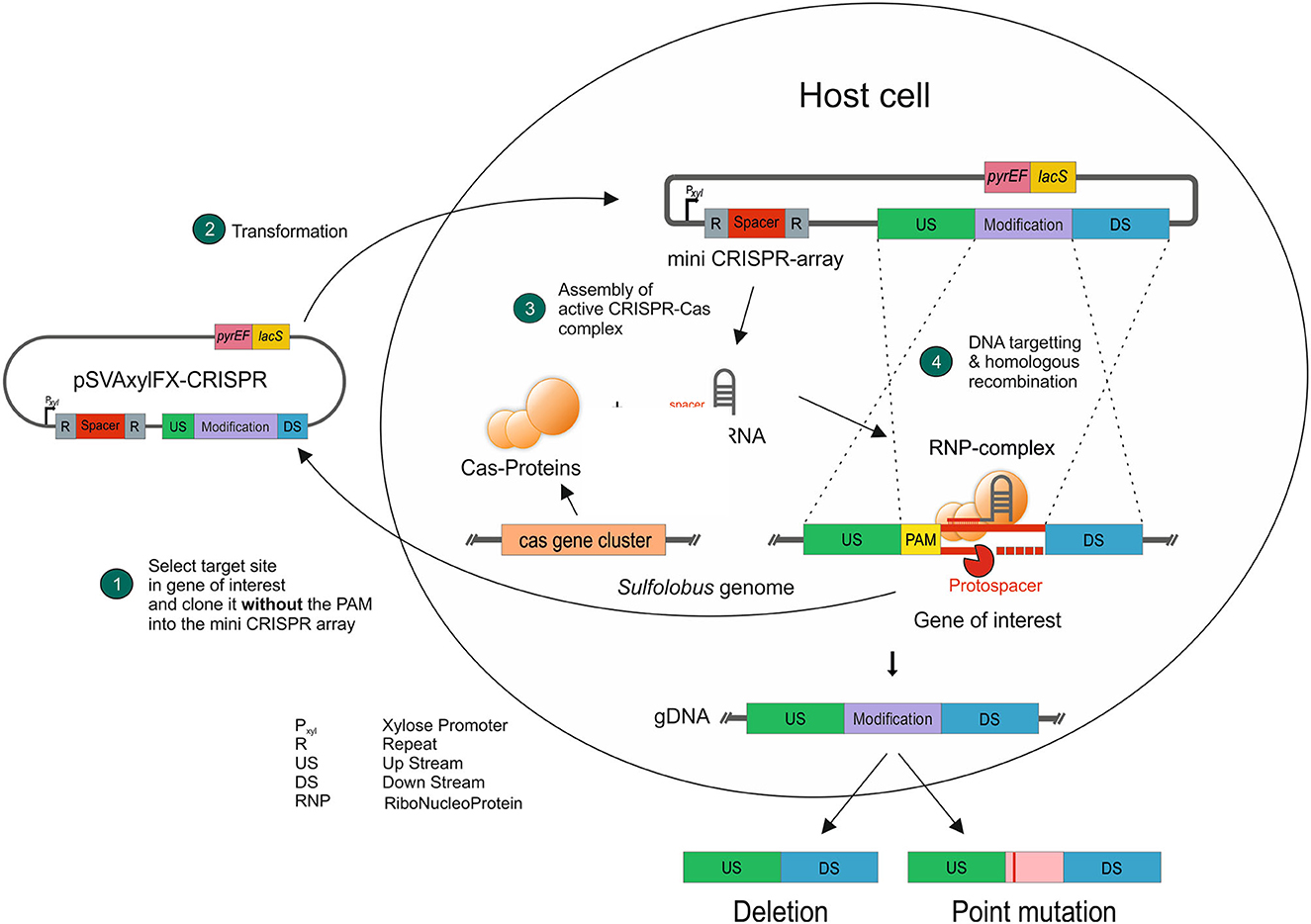
Figure 5. Concept of using the endogenous CRISPR-Cas system presented in this study. A target sequence next to a PAM is cloned into the CRISPR base vector, containing the genetic modification, without the PAM to avoid self-targeting (1). After transformation (2) and induction of the CRISPR array, the endogenous Cas proteins form a ribonucleoprotein complex (3), which scans the genomic DNA for the respective protospacer and then cleaves it. The genetically modified DNA sequence is implemented through DNA repair mechanisms via homologous recombination (4).
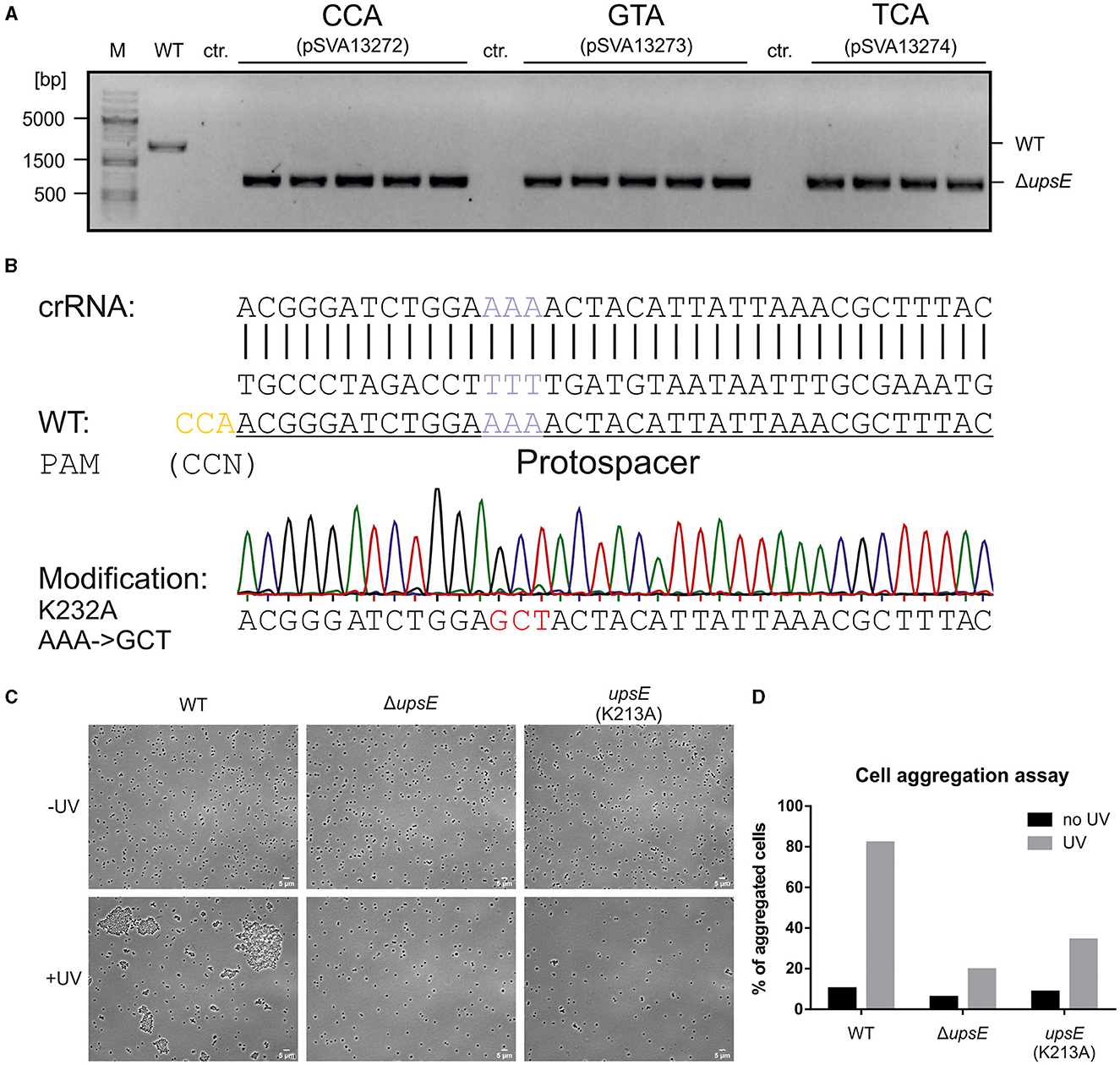
Figure 6. Constructing a deletion mutant and point mutations using the CRISPR plasmid. (A) Verification of ΔupsE deletion mutants using PCR. All three tested PAMs CCA, GTA, and TCA yielded positive signals for upsE deletion. MW001 (WT) DNA was used as a negative control. Control (ctr.) is the PCR reaction with the respective plasmid for the three PAMs alone, to ensure that the signal is due to genomic alteration and not due to the CRISPR plasmid. (B) The sequence of point mutation target site (purple) and the modification sequence (red) leading to an amino acid exchange from lysine to alanine at position 232 (K232A). The PAM CCA (yellow) is identified 12 bp upstream of the target triplet and the following 37 nt are used as a spacer for a targeted CRISPR cleavage. The mutation is verified using Sanger sequencing, shown here as a chromatograph. (C) Cell aggregation assay for the phenotypic analysis of the upsE deletion and point mutation mutants showing light microscopy phase contrast images of the control samples on the top row and the UV-treated cultures on the bottom row. (D) Analysis of cell aggregation assay. Shown are% of aggregated cells after UV treatment. The means correspond to three samples per biological triplicates of MW001 (WT), MW1301 (ΔupsE), and MW1304 (upsE K232A). Compared with the WT control, aggregation is impaired in both mutants.
We were also able to cure the cells of the plasmid after verifying the genotype by putting the cells on the second selection plates containing 5-FOA, which was metabolized to a cytotoxic compound by the pyrEF gene, forcing the plasmid out of the cell (Wagner et al., 2012).
To further expand other possibilities for CRISPR-based genome editing, we tried to introduce a point mutation in the Walker A motif of upsE. In the case of successful genome alteration, the newly generated mutant should not be able to form aggregates upon UV induction similar to the deletion mutant. Therefore, we wanted to mutate a lysine residue at position 232 to an alanine (K232A) (Figure 6B), to abolish the ATPase function (delToro et al., 2016). For this, the target protospacer sequence needs to be spanning over the mutational site so that the crRNA only hybridizes with the WT sequence while abolishing targeting the mutation (Figure 6B). For upsE, a PAM CCA is localized 12 nt upstream of the target site, which is used. The modification was, then, put into a 1 kb repair fragment into the MCS of pSVA13134, containing the previously described point mutation. After following our established protocol, we were able to generate a Walker A mutant after the first transformation, named S. acidocaldarius MW1304, by only screening five clones, showing very high efficiency.
To verify the genetic edition, a UV aggregation assay was performed, showing impaired aggregation for both ΔupsE MW1301 and the Walker A mutant MW1304 (Figure 6C), as previously described (Fröls et al., 2008; Ajon et al., 2011; Wagner et al., 2012; van Wolferen et al., 2013). In contrast to the deletion mutant, there is still some aggregation of the Walker A mutant, but much less compared with the MW001 control, showing that the obtained mutants behave as expected (Figure 6D).
In this study, we have successfully demonstrated the utility of the endogenous type I CRISPR system in S. acidocaldarius as a versatile genetic tool for generating gene deletion mutants and introducing single codon changes within the S. acidocaldarius genome, similar to the previous findings in S. islandicus (Li et al., 2016). By testing various potential protospacer adjacent motifs (PAMs), we have expanded the range of available PAM sites, offering more options for targeted genetic modifications.
Although the cloning process for our method involves an additional step compared with the well-established “pop-in/pop-out” approach (Wagner et al., 2012), the identification of desired mutants can be confirmed on the first selection plate. Consequently, the acquisition of mutants in S. acidocaldarius is significantly accelerated compared with the previous methods. This expedited process is attributed not only to the increased yield of mutants, thereby reducing the screening period, but also to the fact that mutant screening is accomplished during the initial selection plates, ~10 days post-transformation. In contrast, the previous method necessitated screening after the second selection, which occurred ~16 days after transformation. It is worth noting that the second selection is still required to eliminate the plasmid from the cells, but this step is performed after confirming the genotype of the colony of interest.
These advancements in the application of the endogenous type I-D CRISPR system in S. acidocaldarius offer a significant improvement in both efficiency and speed, thus facilitating genetic manipulations and expanding the genetic engineering capabilities in this organism.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
JB: Investigation, Methodology, Project administration, Validation, Visualization, Writing—original draft, Writing—review and editing. AR: Investigation, Methodology, Project administration, Supervision, Writing—review and editing. BW: Investigation, Writing—review and editing. AW: Conceptualization, Investigation, Writing—review and editing. BS: Funding acquisition, Resources, Supervision, Writing—review and editing. S-VA: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing—review and editing.
JB and AR were supported by the BMBF 031B0848C. AW was supported by the BMBF 031L0078A and 031B0848A. S-VA and BS acknowledge the funding of the HotAcidFACTORY Sulfolobus acidocaldarius as novel thermoacidophilic bio-factory project within the BMBF funding initiative Mikrobielle Biofabriken für die industrielle Bioökonomie—Neuartige Plattformorganismen für innovative Produkte und nachhaltige Bioprozesse. We acknowledge support by the Open Access Publication Fund of the University of Freiburg.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Ajon, M., Fröls, S., van Wolferen, M., Stoecker, K., Teichmann, D., Driessen, A. J. M., et al. (2011). UV-inducible DNA exchange in hyperthermophilic archaea mediated by type IV pili. Mol. Microbiol. 82, 807–817. doi: 10.1111/j.1365-2958.2011.07861.x
Albers, S.-V., and Driessen, A. J. M. (2008). Conditions for gene disruption by homologous recombination of exogenous DNA into the Sulfolobus solfataricus genome. Archaea 2, 145–149. doi: 10.1155/2008/948014
Barrangou, R., Fremaux, C., Deveau, H., Richards, M., Boyaval, P., Moineau, S., et al. (2007). CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712. doi: 10.1126/science.1138140
Berkner, S., Grogan, D., Albers, S.-V., and Lipps, G. (2007). Small multicopy, non-integrative shuttle vectors based on the plasmid pRN1 for Sulfolobus acidocaldarius and Sulfolobus solfataricus, model organisms of the (cren-) archaea. Nucleic Acids Res. 35, e88. doi: 10.1093/nar/gkm449
Brock, T. D., Brock, K. M., Belly, R. T., and Weiss, R. L. (1972). Sulfolobus: a new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Archiv. Mikrobiol. 84, 54–68.
Brouns, S. J. J., Jore, M. M., Lundgren, M., Westra, E. R., Slijkhuis, R. J. H., Snijders, A. P. L., et al. (2008). Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321, 960–964. doi: 10.1126/science.1159689
Cannone, G., Webber-Birungi, M., and Spagnolo, L. (2013). Electron microscopy studies of Type III CRISPR machines in Sulfolobus solfataricus. Biochem. Soc. Transact. 41, 1427–1430. doi: 10.1042/BST20130166
delToro, D., Ortiz, D., Ordyan, M., Sippy, J., Oh, C.-S., Keller, N., et al. (2016). Walker-A motif acts to coordinate ATP Hydrolysis with motor output in viral DNA packaging. J. Mol. Biol. 428, 2709–2729. doi: 10.1016/j.jmb.2016.04.029
Elmore, J., Deighan, T., Westpheling, J., Terns, R. M., and Terns, M. P. (2015). DNA targeting by the type I-G and type I-A CRISPR-Cas systems of pyrococcus furiosus. Nucleic Acids Res. 43, 10353–10363. doi: 10.1093/nar/gkv1140
Fineran, P. C., and Charpentier, E. (2012). Memory of viral infections by CRISPR-Cas adaptive immune systems: acquisition of new information. Virology 434, 202–209. doi: 10.1016/j.virol.2012.10.003
Fischer, S., Maier, L.-K., Stoll, B., Brendel, J., Fischer, E., Pfeiffer, F., et al. (2012). An archaeal immune system can detect multiple protospacer adjacent motifs (PAMs) to target invader DNA. J. Biologic. Chemistr. 287, 33351–33363. doi: 10.1074/jbc.M112.377002
Fröls, S., Ajon, M., Wagner, M., Teichmann, D., Zolghadr, B., Folea, M., et al. (2008). UV-inducible cellular aggregation of the hyperthermophilic archaeon Sulfolobus solfataricus is mediated by pili formation. Mol. Microbiol. 70, 938–952. doi: 10.1111/j.1365-2958.2008.06459.x
Garneau, J. E., Dupuis, M.-È., Villion, M., Romero, D. A., Barrangou, R., Boyaval, P., et al. (2010). The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468, 67–71. doi: 10.1038/nature09523
Garrett, R. A., Shah, S. A., Erdmann, S., Liu, G., Mousaei, M., León-Sobrino, C., et al. (2015). CRISPR-cas adaptive immune systems of the sulfolobales: unravelling their complexity and diversity. Life 5, 783–817. doi: 10.3390/life5010783
Garrett, R. A., Shah, S. A., Vestergaard, G., Deng, L., Gudbergsdottir, S., Kenchappa, C. S., et al. (2011). CRISPR-based immune systems of the Sulfolobales: complexity and diversity. Biochemic. Soc. Transact. 39, 51–57. doi: 10.1042/BST0390051
Geertsma, E. R. (2013). FX cloning: a versatile high-throughput cloning system for characterization of enzyme variants. Methods Mol. Biol. 978, 133–148. doi: 10.1007/978-1-62703-293-3_10
Gleditzsch, D., Pausch, P., Müller-Esparza, H., Özcan, A., Guo, X., Bange, G., et al. (2019). PAM identification by CRISPR-Cas effector complexes: diversified mechanisms and structures. RNA Biol. 16, 504–517. doi: 10.1080/15476286.2018.1504546
Hale, C. R., Zhao, P., Olson, S., Duff, M. O., Graveley, B. R., Wells, L., et al. (2009). RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell 139:945–956. doi: 10.1016/j.cell.2009.07.040
Li, Y., Pan, S., Zhang, Y., Ren, M., Feng, M., Peng, N., et al. (2016). Harnessing Type I and Type III CRISPR-Cas systems for genome editing. Nucleic Acids Res. 44, e34. doi: 10.1093/nar/gkv1044
Lillestøl, R. K., Shah, S. A., Brügger, K., Redder, P., Phan, H., Christiansen, J., et al. (2009). CRISPR families of the crenarchaeal genus Sulfolobus: bidirectional transcription and dynamic properties. Mol. Microbiol. 72, 259–272. doi: 10.1111/j.1365-2958.2009.06641.x
Lin, J., Fuglsang, A., Kjeldsen, A. L., Sun, K., Bhoobalan-Chitty, Y., and Peng, X. (2020). DNA targeting by subtype I-D CRISPR-Cas shows type I and type III features. Nucleic Acids Res. 48, 10470–10478. doi: 10.1093/nar/gkaa749
Liu, T. Y., and Doudna, J. A. (2020). Chemistry of Class 1 CRISPR-Cas effectors: binding, editing, and regulation. J. Biologic. Chemistr. 295, 14473–14487. doi: 10.1074/jbc.REV120.007034
Makarova, K. S., Haft, D. H., Barrangou, R., Brouns, S. J. J., Charpentier, E., Horvath, P., et al. (2011). Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 9, 467–477. doi: 10.1038/nrmicro2577
Makarova, K. S., Wolf, Y. I., Alkhnbashi, O. S., Costa, F., Shah, S. A., Saunders, S. J., et al. (2015). An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 13, 722–736. doi: 10.1038/nrmicro3569
Makarova, K. S., Wolf, Y. I., Iranzo, J., Shmakov, S. A., Alkhnbashi, O. S., Brouns, S. J. J., et al. (2020). Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants. Nat. Rev. Microbiology 18, 67–83. doi: 10.1038/s41579-019-0299-x
Makarova, K. S., Wolf, Y. I., and Koonin, E. V. (2013). Comparative genomics of defense systems in archaea and bacteria. Nucleic Acids Res. 41, 4360–4377. doi: 10.1093/nar/gkt157
Manica, A., and Schleper, C. (2013). CRISPR-mediated defense mechanisms in the hyperthermophilic archaeal genus Sulfolobus. RNA biology 10, 671–678. doi: 10.4161/rna.24154
Manica, A., Zebec, Z., Steinkellner, J., and Schleper, C. (2013). Unexpectedly broad target recognition of the CRISPR-mediated virus defence system in the archaeon Sulfolobus solfataricus. Nucleic Acids Res. 41, 10509–10517. doi: 10.1093/nar/gkt767
Marraffini, L. A., and Sontheimer, E. J. (2010). CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat. Rev. Genetics 11, 181–190. doi: 10.1038/nrg2749
McGinn, J., and Marraffini, L. A. (2019). Molecular mechanisms of CRISPR-Cas spacer acquisition. Nat. Rev. Microbiology 17, 7–12. doi: 10.1038/s41579-018-0071-7
Mojica F. J. M. Díez-Villaseñor C. García-Martínez J. Almendros C. (2009). Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 155, 733–740. doi: 10.1099/mic.0.023960-0
Mousaei, M., Deng, L., She, Q., and Garrett, R. A. (2016). Major and minor crRNA annealing sites facilitate low stringency DNA protospacer binding prior to Type I-A CRISPR-Cas interference in sulfolobus. RNA Biol. 13, 1166–1173. doi: 10.1080/15476286.2016.1229735
Nidhi, S., Anand, U., Oleksak, P., Tripathi, P., Lal, J. A., Thomas, G., et al. (2021). Novel CRISPR-Cas systems: an updated review of the current achievements, applications, and future research perspectives. Int. J. Mol. Sci. 22, 7327. doi: 10.3390/ijms22073327
Peng, W., Li, H., Hallstrøm, S., Peng, N., Liang, Y. X., and She, Q. (2013). Genetic determinants of PAM-dependent DNA targeting and pre-crRNA processing in Sulfolobus islandicus. RNA Biol. 10, 738–748. doi: 10.4161/rna.23798
Samai, P., Pyenson, N., Jiang, W., Goldberg, G. W., Hatoum-Aslan, A., and Marraffini, L. A. (2015). Co-transcriptional DNA and RNA cleavage during Type III CRISPR-Cas immunity. Cell 161, 1164–1174. doi: 10.1016/j.cell.2015.04.027
Sinkunas, T., Gasiunas, G., Waghmare, S. P., Dickman, M. J., Barrangou, R., Horvath, P., et al. (2013). In vitro reconstitution of Cascade-mediated CRISPR immunity in Streptococcus thermophilus. EMBO J. 32, 385–394. doi: 10.1038/emboj.2012.352
van der Kolk, N., Wagner, A., Wagner, M., Waßmer, B., Siebers, B., and Albers, S.-V. (2020). Identification of XylR, the activator of arabinose/xylose inducible regulon in sulfolobus acidocaldarius and its application for homologous protein expression. Front. Microbiol. 11, 1066. doi: 10.3389/fmicb.2020.01066
van Wolferen, M., Ajon, M., Driessen, A. J. M., and Albers, S.-V. (2013). Molecular analysis of the UV-inducible pili operon from Sulfolobus acidocaldarius. Microbiol. Open 2, 928–937. doi: 10.1002/mbo3.128
Vestergaard, G., Garrett, R. A., and Shah, S. A. (2014). CRISPR adaptive immune systems of archaea. RNA Biol. 11, 156–167. doi: 10.4161/rna.27990
Wagner, M., van Wolferen, M., Wagner, A., Lassak, K., Meyer, B. H., Reimann, J., et al. (2012). Versatile genetic tool box for the crenarchaeote sulfolobus acidocaldarius. Front. Microbiol. 3, 214. doi: 10.3389/fmicb.2012.00214
Westra, E. R., van Erp, P. B. G., Künne, T., Wong, S. P., Staals, R. H. J., Seegers, C. L. C., et al. (2012). CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol. Cell 46, 595–605. doi: 10.1016/j.molcel.2012.03.018
Yang, H., Ren, S., Yu, S., Pan, H., Li, T., Ge, S., et al. (2020). Methods favoring homology-directed repair choice in response to CRISPR/Cas9 induced-double strand breaks. Int. J. Mol. Sci. 21, 8461. doi: 10.3390/ijms21186461
Zebec, Z., Zink, I. A., Kerou, M., and Schleper, C. (2016). Efficient CRISPR-mediated post-transcriptional gene silencing in a hyperthermophilic archaeon using multiplexed crRNA expression. G3 6, 3161–3168. doi: 10.1534/g3.116.032482
Zhang, J., and White, M. F. (2013). Hot and crispy: CRISPR-Cas systems in the hyperthermophile Sulfolobus solfataricus. Biochem. Soc. Transact. 41, 1422–1426. doi: 10.1042/BST20130031
Zink, I. A., Fouqueau, T., Tarrason Risa, G., Werner, F., Baum, B., Bläsi, U., et al. (2021). Comparative CRISPR type III-based knockdown of essential genes in hyperthermophilic Sulfolobales and the evasion of lethal gene silencing. RNA Biol. 18, 421–434. doi: 10.1080/15476286.2020.1813411
Keywords: archaea, genetic tools, deletion mutant, genetic engineering, type I-D CRISPR system, protospacer adjacent motif
Citation: Bost J, Recalde A, Waßmer B, Wagner A, Siebers B and Albers S-V (2023) Application of the endogenous CRISPR-Cas type I-D system for genetic engineering in the thermoacidophilic archaeon Sulfolobus acidocaldarius. Front. Microbiol. 14:1254891. doi: 10.3389/fmicb.2023.1254891
Received: 07 July 2023; Accepted: 11 September 2023;
Published: 02 October 2023.
Edited by:
Michel Geovanni Santiago-Martínez, University of Connecticut, United StatesReviewed by:
Changyi Zhang, University of Illinois at Urbana-Champaign, United StatesCopyright © 2023 Bost, Recalde, Waßmer, Wagner, Siebers and Albers. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sonja-Verena Albers, c29uamEuYWxiZXJzQGJpb2xvZ2llLnVuaS1mcmVpYnVyZy5kZQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.