- 1Department of Veterinary and Animal Sciences, University of Copenhagen, Copenhagen, Denmark
- 2Department of Aquatic Pathology, Can Tho University, Can Tho, Vietnam
- 3Department of Aquatic Product Processing, Can Tho University, Can Tho, Vietnam
Introduction: Motile Aeromonas septicemia (MAS) is a burden for striped catfish (Pangasius hypophthalmus) farmers in Vietnam. MAS can be caused by several species of Aeromonas but Aeromonas hydrophila is seen as the leading cause of MAS in aquaculture, but recent reports suggest that A. dhakensis is also causing MAS.
Methods: Here we investigated the bacterial etiology of MAS and compared the genomic features of A. hydrophila and A. dhakensis. We collected 86 isolates from diseased striped catfish fingerlings over 5 years from eight provinces in Vietnam. Species identification was done using PCR, MALDI-TOF and whole genome sequence (WGS). The MICs of commonly used antimicrobials was established. Thirty presumed A. hydrophila isolates were sequenced for species confirmation and genomic comparison. A phylogenetic analysis was conducted using publicly available sequences and sequences from this study.
Results: A total of 25/30 isolates were A. dhakensis sequence type (ST) 656 and 5/30 isolates were A. hydrophila ST 251. Our isolates and all publicly available A. hydrophila isolates from Vietnam belonged to ST 251 and differed with <200 single nucleotide polymorphisms (SNP). Similarly, all A. dhakensis isolates from Vietnam belonged to ST 656 and differed with <100 SNPs. The tet(A) gene was found in 1/5 A. hydrophila and 19/25 A. dhakensis. All A. hydrophila had an MIC ≤2 mg/L while 19/25 A. dhakensis had MIC ≥8 mg/L for oxytetracycline. The floR gene was only found in A. dhakensis (14/25) which showed a MIC ≥8 mg/L for florfenicol. Key virulence genes, i.e., aerA/act, ahh1 and hlyA were present in all genomes, while ast was only present in A. dhakensis.
Discussion: This study confirms previous findings where A. dhakensis was the dominating pathogen causing MAS and that the importance of A. hydrophila has likely been overestimated. The differences in antimicrobial susceptibility between the two species could indicate a need for targeted antimicrobial treatment plans. The lipopolysaccharide regions and outer membrane proteins did not significantly differ in their immunogenic potentials, but it remains to be determined with in vivo experiments whether there is a difference in the efficacy of available vaccines against A. hydrophila and A. dhakensis.
1. Introduction
Aeromonas spp. are Gram-negative bacteria belonging to the family Aeromonadaceae. They are commonly found in aquatic environments but can also be isolated from a variety of other sources (Janda and Abbott, 2010; Khor et al., 2018). Today, there are 36 species of Aeromonas and many are capable of causing disease in animals and humans. Aeromonas spp. are important pathogens in aquaculture, i.e., Aeromonas hydrophila, Aeromonas caviae, Aeromonas veronii, Aeromonas dhakensis and Aeromonas salmonicida all cause diseases in global aquaculture productions (Fernández-Bravo and Figueras, 2020). A. salmonicida infects cold-water fish whereas the other mentioned Aeromonas spp. are mesophilic and motile bacteria infecting warm-water farmed fish and less often humans (Fernández-Bravo and Figueras, 2020).
Vietnam is one of the world’s leading exporters of fish, ranking as the third biggest exporter of aquatic animal products in 2020 after China and Norway. In 2020, the value of exported aquaculture products from Vietnam reached USD 8.5 billion (FAO, 2022). Frozen striped catfish (Pangasius hypophthalmus) fillets are undoubtedly the most important exported product and Vietnam is the leading producer and exporter of farmed striped catfish, e.g., to China and the United States of America (FAO, 2022). Striped catfish is mainly cultured in the Mekong Delta in southern Vietnam where the production has intensified and grown fast during the last decade. The vast majority of striped catfish farmers experience one or often more disease outbreaks during the 9-month production cycle (Hoa et al., 2021). The most frequently occurring disease in striped catfish farming is motile Aeromonas septicemia which causes big economical losses for the aquaculture sector, not only in Vietnam but globally (Stratev and Odeyemi, 2017). Motile Aeromonas septicemia is caused by motile Aeromonas spp., mainly A. hydrophila, A. caviae, A. veronii and A. dhakensis, but variations in the prevalence of each species occur depending on the geographical region and cultured aquatic species (Stratev and Odeyemi, 2017; Azzam-Sayuti et al., 2021; Saharia et al., 2021). The symptoms of motile Aeromonas septicemia include anorexia, hemorrhage of internal organs and red coloring of the skin due do hemorrhage (Nielsen et al., 2001).
A. hydrophila has been considered the most predominant species causing motile Aeromonas septicemia and was detected in 11% of larvae, in 30% of fry and 30% of apparently healthy striped catfish fingerlings (Hoa et al., 2021). However, already in the beginning of the 2000’s, it was questioned whether the importance of A. hydrophila as a disease causing pathogen had been overestimated (Nielsen et al., 2001). With new and more precise molecular identification methods, new species of Aeromonas diverged and isolates previously characterized as A. hydrophila were re-identified as other or new Aeromonas spp. (Beaz-Hidalgo et al., 2013). In 2002, A. dhakensis was described for the first time under the name Aeromonas hydrophila subsp. dhakensis when isolates from children with diarrhea in Bangladesh showed distinctively different biochemical properties compared to A. hydrophila (Huys et al., 2002). Later, A. hydrophila subsp. dhakensis was elevated to the rank of species based on phylogenetic and multilocus phylogenetic analysis, and it is today considered a distinct species of Aeromonas, called A. dhakensis (Beaz-Hidalgo et al., 2013). A. dhakensis is described as both a human clinical pathogen and a pathogen in aquaculture. A. dhakensis was recently described as the most frequently isolated Aeromonas spp. (43%) from cultured freshwater fish farming in Malaysia, followed by A. veronii (22%) and A. hydrophila (20%) (Azzam-Sayuti et al., 2021). A study analyzing samples collected between 2013–2019 from striped catfish in the Mekong Delta area in Vietnam reports that 75% of the suspected A. hydrophila isolates were identified as A. dhakensis using different identification methods (Bartie et al., 2023).
A. dhakensis continues to be misidentified as A. hydrophila due to the application of phenotypic identification methods not able to differentiate between the two species (Chen et al., 2016). Matrix-Assisted Laser Desorption/Ionization Time Of Flight (MALDI-TOF) is a method which technically would be able to identify A. dhakensis at species. However, the current commercially available MALDI-TOF databases does not include A. dhakensis, but includes A. hydrophila, A. jandaei, A. punctata (caviae), A. sobria and A. salmonicida. Thus the current databases will give the closest matching bacterial species, i.e., A. hydrophila or only identify the isolate on genus level. The most accurate methods identifying A. dhakensis is by sequencing the housekeeping genes (rpoB, rpoD or gyrB) and whole genome sequence analysis (Chen et al., 2016).
Due to the relatively new taxonomy of A. dhakensis as a separate species and the lack of accurate identification methods in diagnostic laboratories, the real prevalence of A. dhakensis as a disease-causing pathogen in striped catfish is unknown. Thus the significance of A. hydrophila is likely overestimated, at least in Vietnamese striped catfish aquaculture (Chen et al., 2016; Lau et al., 2020; Bartie et al., 2023). The objective of this study was to conduct a comprehensive comparative analysis of the virulence in silico, prevalence, and genetic characteristics of A. dhakensis and A. hydrophila isolates obtained between 2017 and 2021 from striped catfish fingerlings showing symptoms of motile Aeromonas septicemia in eight different provinces of the Mekong Delta. By investigating key virulence factors, antimicrobial resistance profiles, and genomic traits, we aimed to elucidate the unique contributions of each species to the occurrence and severity of motile Aeromonas septicemia in striped catfish fingerlings. Through this comparative analysis, we have provided valuable insights into the epidemiology these emerging pathogens, ultimately contributing to the sustainable aquaculture practices and the health of striped catfish production in Vietnam.
2. Materials and methods
2.1. Specimens and bacterial isolation
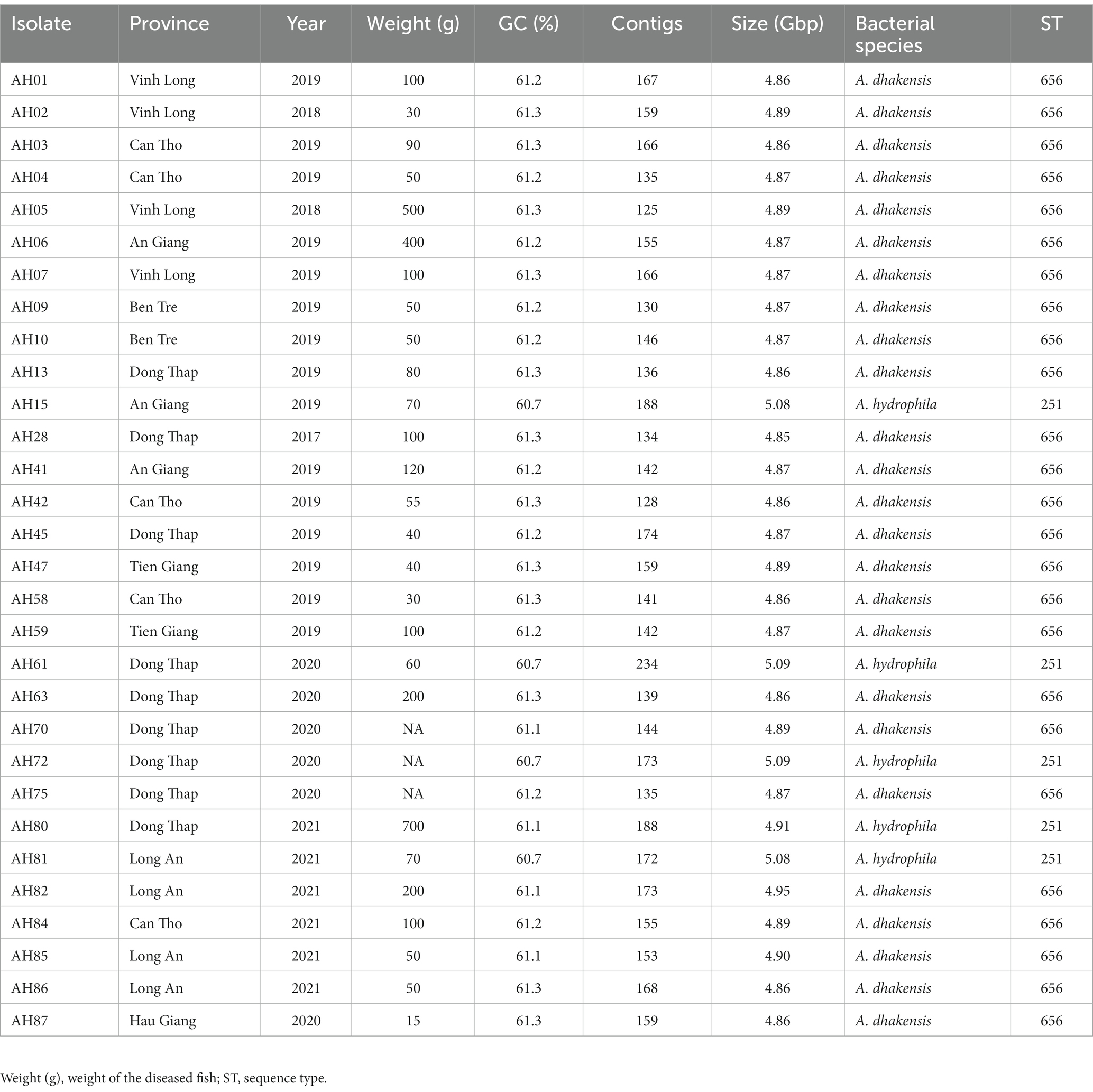
We collected diseased P. hypophthalmus fingerlings (size 10-100 g) and larger fish (size 100-800 g) from grow-out farms showing clinical signs of motile Aeromonas septicemia, i.e., pin-point hemorrhagic red spots scattered in the whole body, exophthalmia, pink-red fluid in the abdomen and enlarged spleen, liver and kidneys. Fish were anesthetized with Aqui-S® (Bayer, Vietnam) and the skin was disinfected with alcohol 70% before performing necropsy. Samples from the liver and kidneys were taken using sterile inoculation loops that were cultured onto tryptic soya agar plates (TSA, Merck, Germany) and incubated at 28°C for 24 h at Can Tho University, Vietnam. Representative colony isolates on TSA plates were re-streaked onto new TSA plates to ensure purity. Isolates were initially characterized by morphology, Gram staining, catalase, oxidase, and O/F test and subsequent identified by API 20E kit (bioMérieux, France). A total of 86 bacterial isolates were collected from commercial striped catfish farms (pond sizes ranged from 0.5–1.2 ha and water depth from 4–5.5 m) located in eight different provinces in the Mekong Delta in southern Vietnam, i.e., An Giang (15 strains), Ben Tre (5 strains), Can Tho (13 strains), Dong Thap (39 strains), Hau Giang (1 strain), Long An (5 strains), Tien Giang (4 strains) and Vinh Long (4 strains) provinces. The sample locations are shown in Figure 1. Bacterial specimens were collected between 2017 and 2021 by the Department of Aquatic Pathology, College of Aquaculture and Fisheries at Can Tho University. Isolates were stored in tryptic soya broth (TSB, Merck, Germany) containing 25% glycerol and kept at –80°C before shipment to the University of Copenhagen, Denmark, for further characterization.
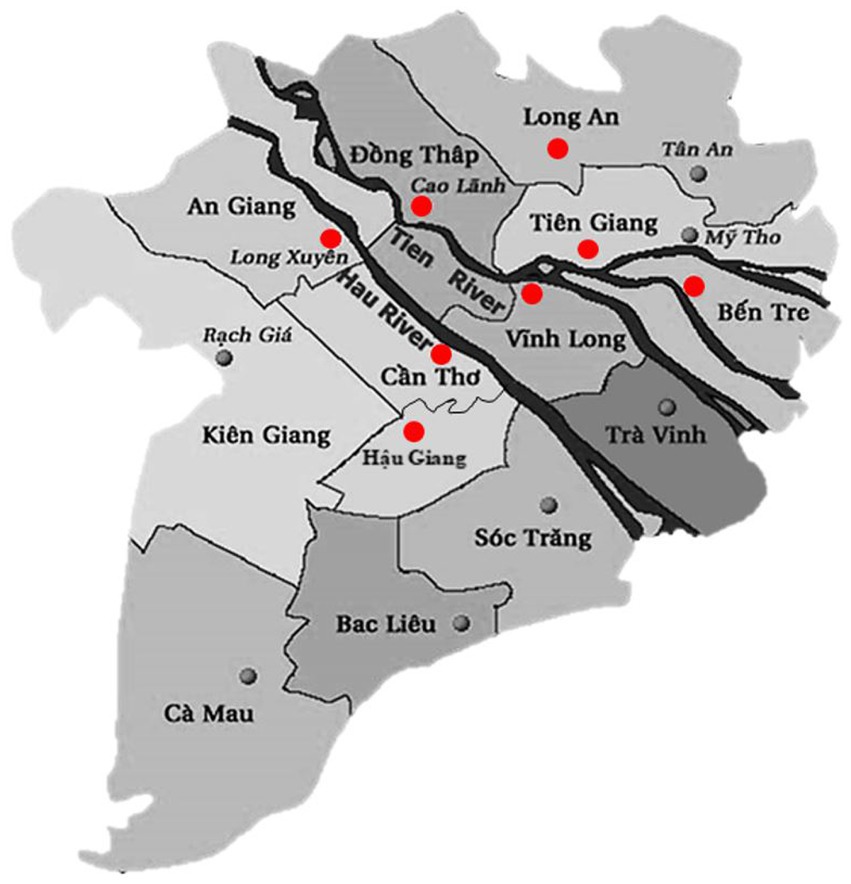
Figure 1. Map showing the location of provinces (red dots) where strains were collected. Province borders are marked with thin black lines. The main river branches of the Mekong Delta are marked with thick black color.
2.2. Bacterial species identification
At the University of Copenhagen material from each vial with a bacterial isolate was streaked onto agar plates with blood agar base and 5% sterile bovine blood and incubated overnight at 28°C to confirm the purity. The species of all 86 isolates was determined using A. hydrophila species specific PCR primers: aerolysin aer gene 5’CCA AGG GGT CTG TGG CGA CA 3′ (forward read) and 5′ TTT CAC CGG TAA CAG GAT TG 3′ (reverse read) with expected size of 209 bp amplicons (Pollard et al., 1990). To confirm results from the PCR, fresh bacterial colonies from all isolates were cultured on agar plates and their identity confirmed by using the automated VITEK® MS (bioMérieux, France) MALDI-TOF mass spectrometry.
2.3. Antimicrobial susceptibility testing
The minimum inhibitory concentration (MIC) of each isolate was determined by microbroth dilution. Each bacterial strain was streaked onto agar plates with blood agar base and 5% sterile bovine blood and incubated overnight at 37°C. Tubes with 5 mL demineralized water and tubes with 11 mL Mueller-Hinton (Oxoid) broth were prepared and incubated overnight to reduce the risk of contamination. Characteristic colonies (1 to 3) were suspended in the tubes containing demineralized water. Using a nephelometer, the bacterial concentration was measured to 0.5 McFarland Standard. Now 10 μL of the suspension was pipetted into the tube with Mueller-Hinton broth and vortexed. The Mueller-Hinton broth was poured onto an empty Petri dish and gently mixed on the table. A multi-channel pipette was used to suspend 50 μL in each well on Sensititre plates. The MIC testing was performed using the commercial Sensititre kit (AVIAN1F) from ThermoScientific™ containing the following antimicrobials: enrofloxacin, gentamicin, ceftiofur, neomycin, erythromycin, oxytetracycline, tetracycline, amoxicillin, spectinomycin, sulphadimethoxine, trimethoprim/sulfamethoxazole, florfenicol, sulphathiazole, penicillin, streptomycin, novobiocin, tylosin tartrate, and clindamycin. Escherichia coli ATCC25922 was used as control strain. Results were interpreted using the Thermo Scientific™ Sensititre™ SWIN™ Software System. There are no clinical breakpoints established for neither A. hydrophila nor A. dhakensis in aquaculture and therefore the epidemiological cutoff values for enrofloxacin, gentamicin, oxytetracycline and florfenicol, established by Clinical and Laboratory Standards Institute (CLSI) guidelines VET04 third edition (Clinical and Laboratory Standards Institute, 2017), for A. hydrophila were used for the relevant antimicrobials.
2.4. Whole genome sequencing
A total of 30 isolates were selected for whole genome sequencing based on results from the PCR and MALDI-TOF species identification. Only isolates confirmed as A. hydrophila from the MALDI-TOF analysis were chosen. Sampling year and location were considered so that isolates from all sampling years and provinces were represented. Isolates were cultured on blood agar and incubated at 37°C for 24 h. A pure bacterial colony was transferred to Luria broth (Oxoid) and incubated at 37°C for 13 h. Genomic DNA was extracted using the bacterial DNA extraction kit Maxwell® RSC Cultured Cells DNA Kit (Promega Corporation, Madison, WI, United States). Quality parameters of the extracted DNA was assessed using NanoDrop™ One (Thermo Fisher Scientific, Waltham, MA, United States) and the DNA concentration using Qubit 2.0 Fluorometer (Invitrogen, Thermo Fisher Scientific, Waltham, MA, United States). Whole genome sequencing was performed with Illumina MiSeq (Illumina Inc., San Diego, CA, United States) at Statens Serum Institut (Copenhagen, Denmark) using the Nextera XT DNA Library Preparation Kit (96 samples) (Illumina Inc., San Diego, CA, United States), generating minimum sequence coverage of 50X.
2.5. Bioinformatic analysis
Raw sequence reads were processed for quality control using FastQC version v0.11.9. The raw reads are available at the European Nucleotide Archive (ENA) under the project number PRJEB59357. Trimming raw sequences was done with Trimmomatic version 0.3 (Bolger et al., 2014). Cleaned reads were assembled using the SPAdes 3.9 assembly tool (Nurk et al., 2013). For quality control of the assembled genomes, we used QUAST version 5.2 (Mikheenko et al., 2018). The number of contigs in each genome ranged from 125–234 contigs and the total size of the genomes was 4.85–5.09 Gbp. The assembled genomes were then subjected to further analyses. Confirmation of species identity was performed using Kraken2 (Wood and Salzberg, 2014) and KmerFinder version 3.2 (Hasman et al., 2014; Larsen et al., 2014; Clausen et al., 2018).
Multi-locus sequence typing (MLST) was done using PubMLST (Jolley et al., 2018). Screening for antimicrobial resistance genes was done in the Comprehensive Antibiotic Resistance Database Resistance Gene Identifier (CARD RGI) (Alcock et al., 2020). The selection criteria was set to perfect and strict hits. We screened the draft genomes for virulence genes in the Virulence Factor Data Base (VFDB) using the VFanalyzer tool (Liu et al., 2019) and blasted the genomes against seven additional virulence genes relevant to Aeromonas spp. downloaded from NCBI (ahpA, alt, dns, elastase, gcaT, lip and ser) using MyDbFinder.1 The threshold for ID% was set to 90% and the minimum length to 60%. To detect plasmids, we uploaded the assembled genomes to PlasmidFinder 2.1 in CGE (Camacho et al., 2009; Carattoli et al., 2014) using the Enterobactericeae database set at 95% threshold for minimum identity and minimum 60% for coverage.
The sequences were then analyzed for single nucleotide polymorphism along with publically available genomes. A search for all available A. hydrophila and A. dhakensis sequences was conducted in National Center for Biotechnology Information (NCBI), and all sequences, where at least the isolation source was available, were used in the phylogenetic analysis. Additionally, the collection date and geographical location was included in the metadata when available. Single-nucleotide variants were called by using Snippy version 4.6.02 under the following parameters: mapping quality of 60, a minimum base quality of 13, a minimum read coverage of four, and a 75% concordance at a locus. Since species confirmatory results showed that among the 30 sequenced assumed A. hydrophila most strains (25 strains) were in fact A. dhakensis, two separate phylogenetic analyses were performed with A. dhakensis (GeneBank: CP084351.1) and A. hydrophila (GeneBank: CP005966.1) used as references during variant calling and alignment. The core genome single-nucleotide variants were aligned with Snippy-core version 4.1.0 for phylogeny inference.
Putative recombinogenic regions were detected and masked using Gubbins version 2.4.1 (Croucher et al., 2015). A maximum-likelihood phylogenetic tree was build using RAxML version 8.2.12 and the generalized time-reversible model with 200 bootstraps (Stamatakis, 2014). The final trees were rooted on the reference genomes and the trees were annotated and visualized with iTOL version 3 (Letunic and Bork, 2021). To support these results we constructed single nucleotide polymorphism matrices.
All of the 30 genomes were annotated using Prokka version 1.14.5 (Seemann, 2014) and the output master annotation files (.gff) were used to run a pan genome pipeline using Roary version 3.13.0 (Page et al., 2015). The pan genome was visualized in Phandango (Hadfield et al., 2018). The unique gene regions were first extracted and then analyzed in VRprofile (Li et al., 2018). Annotation was also done with Rapid Annotations using Subsystems Technology (RAST) to compare genes coding for capsular and extracellular polysaccharides (Aziz et al., 2008; Overbeek et al., 2014; Brettin et al., 2015). Since the currently used vaccine against motile Aeromonas septicemia is from inactivated bacteria cultures (PHARMAQ AS, Norway), we decided to further investigate any differences between the LPS and the outer membrane proteins of A. dhakensis and A. hydrophila. The analysis can provide information of potential species level differences in these regions that may have an influence on antigenic ability and vaccine efficacy. The sequences of six outer membrane proteins from A. hydrophila were downloaded from NCBI (GenBank: HQ326181.1, DQ177328.1, OM912661.1, HF546053.1, OM912660.1 and JQ349084.1) and combined into one multifasta file. This multifasta file was used as the reference in MyDbFinder version 2.0 (see footnote 1) with the threshold for %ID set to 90% and minimum length set to 60%. We then blasted the reference file against 5 genomes of each species. The similarities of the lipopolysaccharide (LPS) regions of each species were compared by downloading the A. hydrophila LPS region sequenced by Jimenez et al. (2008) and comparing 5 genomes of each species to the downloaded sequences using the Gview server3 The LPS sequences were downloaded from NCBI (GenBank: EU296246.1, EU296247.1 and EU296248.1).
3. Results
3.1. Species identification
All isolates were Gram-negative, catalase positive, oxidase positive, O/F positive and rod-shaped. The PCR analysis showed that many isolates lacked the 209 base pairs size amplicon band indicative of A. hydrophila. We therefore decided to run all isolates through MALDI-TOF to confirm the species. From the MALDI-TOF run, 83/85 isolates were identified as the genus Aeromonas. On species level, 10/85 isolates were identified as A. hydrophila, 2/85 had “no match” and 73/85 isolates were identified only at genus level as Aeromonas spp. Looking closer at the MALDI-TOF results to determine which Aeromonas spp. was the closest match, 2/85 isolates matched with A. punctata, 2/85 matched A. veronii, 68/85 matched closest with A. hydrophila and 11/85 isolates were only identified on genus level (Figure 2). Based on these results and considering sampling year and sampling location, a total of 30 isolates confirmed as A. hydrophila at species level were chosen for whole genome sequencing.
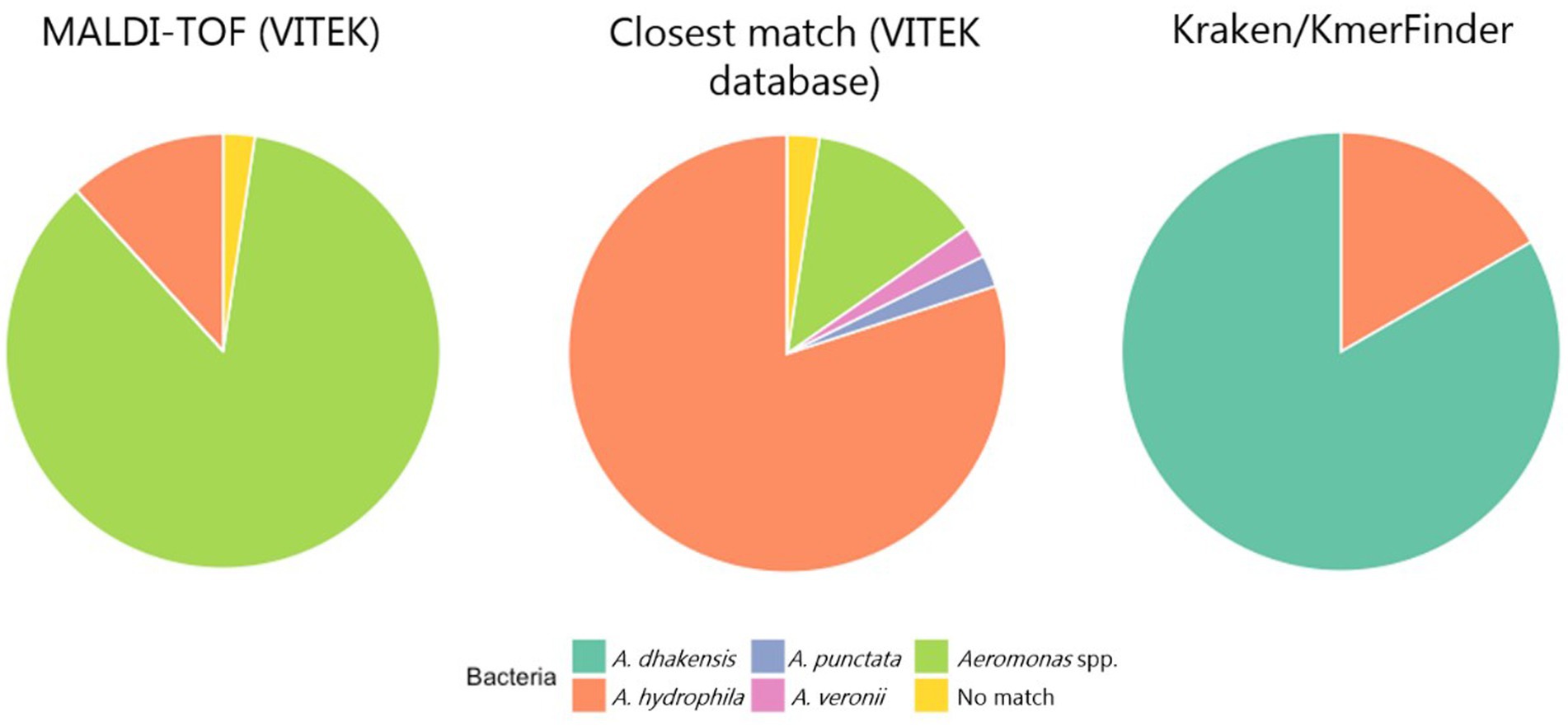
Figure 2. The proportion of identified Aeromonas spp. based on MALDI-TOF, looking for the closest matching species in the VITEK database and analysis of the sequenced genomes using KmerFinder/Kraken2.
3.2. Genomic characteristics of Aeromonas dhakensis and Aeromonas hydrophila
The 30 sequenced isolates were from the following provinces: An Giang (3), Ben Tre (2), Can Tho (5), Dong Thap (9), Hau Giang (1), Hiep Thanh Com (1), Long An (4), Tien Giang (2) and Vinh Long (4). Years of isolation of the strains included: 2017 (1), 2018 (2), 2019 (15), 2020 (6) and 2021 (6). The genome GC content ranged from 60.7–61.3%. After sequencing the selected isolates, the first analysis performed was species identification based on the k-mers in the DNA sequence data using KmerFinder. KmerFinder identified 5/30 genomes as A. hydrophila and 25/30 isolates as A. dhakensis. These results were confirmed by the results from Kraken. Multi-locus sequence typing grouped all A. dhakensis isolates into sequence type (ST) 656 and all A. hydrophila genomes into ST 251 (Table 1). The genomes of the A. hydrophila isolates were larger than the genomes of A. dhakensis as they were ≥ 4.91 Gbp and the A. dhakensis genomes were all <4.91 Gbp. The GC content of A. dhakensis was ≥61.1%, while the GC content of A. hydrophila was ≤61.1%. Details about the 30 sequenced isolates and their genomes can be seen in Table 1. The sequences have been deposited in ENA with the project accession number PRJEB59357.
3.3. Phylogenetic analysis
Two separate phylogenetic trees were constructed for A. hydrophila and A. dhakensis. Figure 3 shows the tree constructed using all A. hydrophila strains sequenced in this study and 16 publicly available A. hydrophila genomes downloaded from ENA. The reference strain was A. hydrophila (ST 251) isolated from channel catfish in the United States. The single nucleotide polymorphism (SNP) analysis revealed that one of the strains from ENA had over 200,000 SNPs comparing it to all other strains. Assembly and species analysis of the strain in KmerFinder and pubMLST revealed that this strain is in fact A. dhakensis ST 335. The species of all other isolates were confirmed as A. hydrophila with all strains originating from striped catfish in Vietnam. There were nine different sequence types and one unknown sequence type among the publicly available genomes. The SNP matrix (Supplementary Table S1) shows how these strains from Vietnam group and differ in less than 200 SNPs, including the strains sequenced in this study. All strains have ST 251, also known as virulent A. hydrophila. One channel catfish isolate from China (SRR21285549) closely resembles the Vietnamese strains with less than 140 SNPs and also belongs to ST 251. This strain and all the Vietnamese strains closely resemble the reference strain with less than 140 SNPs which supports the results from the sequence type analysis as all of these genomes are ST 251.
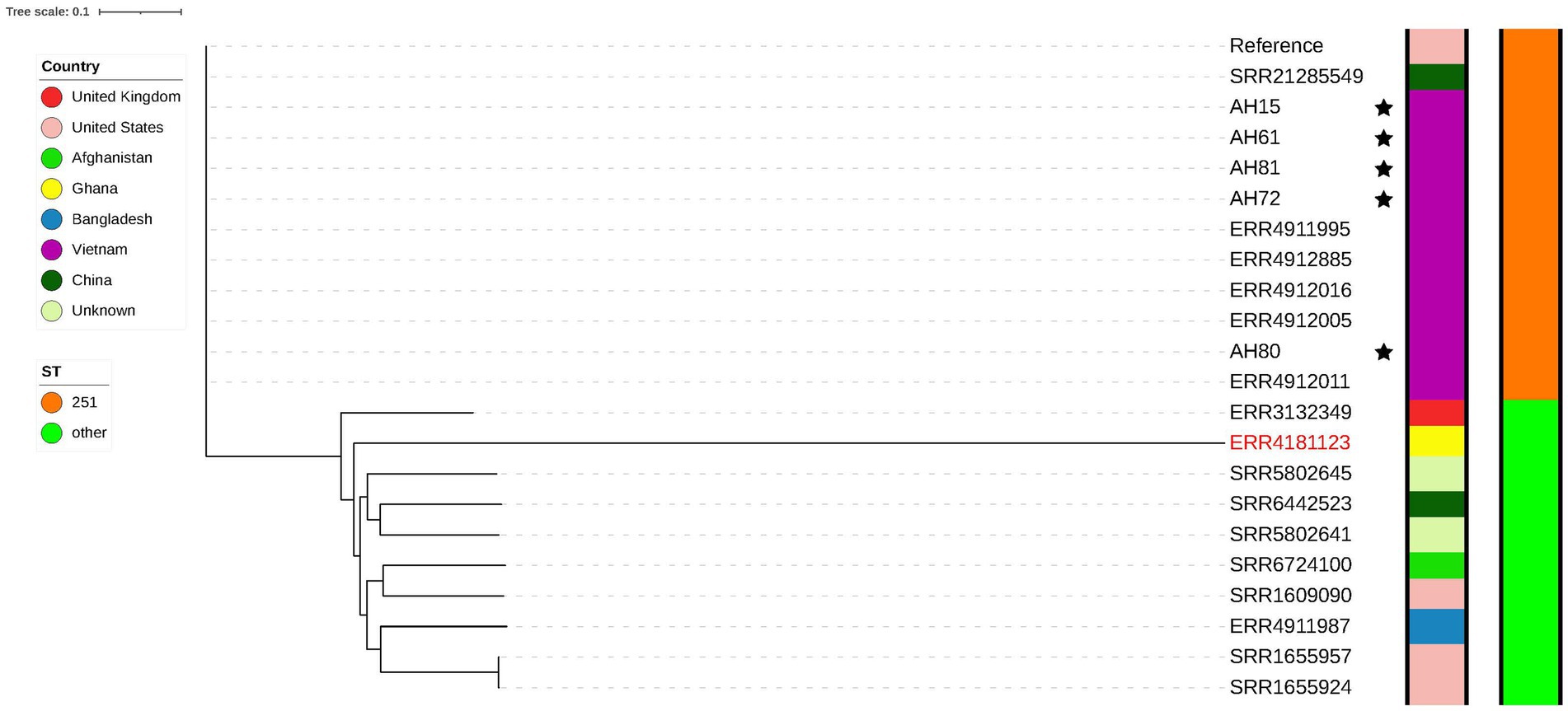
Figure 3. Phylogenetic tree showing strains of A. hydrophila. The genomes of the five strains sequenced in this study are marked with a star. A genome later confirmed as A. dhakensis is marked in a red colored font.
The phylogenetic tree composed of A. dhakensis genomes seen in Figure 4 was generated using the 25 sequenced A. dhakensis genomes and 20 publicly available A. dhakensis genomes accessed from ENA. The reference strain was isolated from a human clinical sample from China. The sequence types and species of all publicly available A. dhakensis genomes were confirmed using KmerFinder and pubMLST. Our isolates had a maximum difference of 108 SNPs and all belonged to ST 656. One isolate (ST 337) originating from a striped catfish in Malaysia did not group with the other isolates from the same region. The rest of the isolates from Southeast Asia (all from Vietnam) belonged to ST 656 with a difference in SNPs between any isolate being maximum 98 SNPs. Among the publicly available A. dhakensis genomes there was a variation in the sequence types with isolates representing eight different sequence types and two unknown sequence types. The number of SNPs between the isolates sequenced for this study and isolates from outside of Vietnam was 65,000–68,000 SNPs. The publicly available A. dhakensis isolates had a difference of 80,000 to 94,000 SNPs when compared to our Vietnamese isolates. The SNP matrices are available in the Supplementary Tables S1, S2.

Figure 4. Phylogenetic tree showing isolates of A. dhakensis. The genomes sequenced in this study are marked with a star.
3.4. Antimicrobial susceptibility, antimicrobial resistance genes and plasmids
All A. hydrophila isolates had an MIC ≤2 mg/L for tetracycline and oxytetracycline. Four out of five isolates were considered wild type for tetracyclines (≤ 0.25 mg/L). Most A. dhakensis isolates (19/25) had an MIC ≥8 mg/L for tetracycline and oxytetracycline. Six A. dhakensis isolates were wild type for tetracyclines. All A. hydrophila isolates had an MIC ≤1 mg/L for florfenicol while 11/25 A. dhakensis isolates had an MIC ≤1 mg/L and were wild type for florfenicol, the rest of the A. dhakensis isolates (14/25) had an MIC ≥8 mg/L and. The MIC for sulphathiazole was ≤128 mg/L for all A. hydrophila isolates and only two A. dhakensis isolates had an MIC <256 mg/L. All A. hydrophila were wild type (≤ 0.12 mg/L) for enrofloxacin, while 20/25 of the A. dhakensis isolates were non-wild type for enrofloxacin. All isolates, both A. dhakensis and A. hydrophila, were wild type for gentamicin (MIC ≤2 mg/L). The MIC values of the most commonly used antimicrobials in Vietnamese aquaculture production are summarized in Figure 5. The MIC values of all isolates (n = 86) for all tested antimicrobials can be found in the Supplementary Table S3.
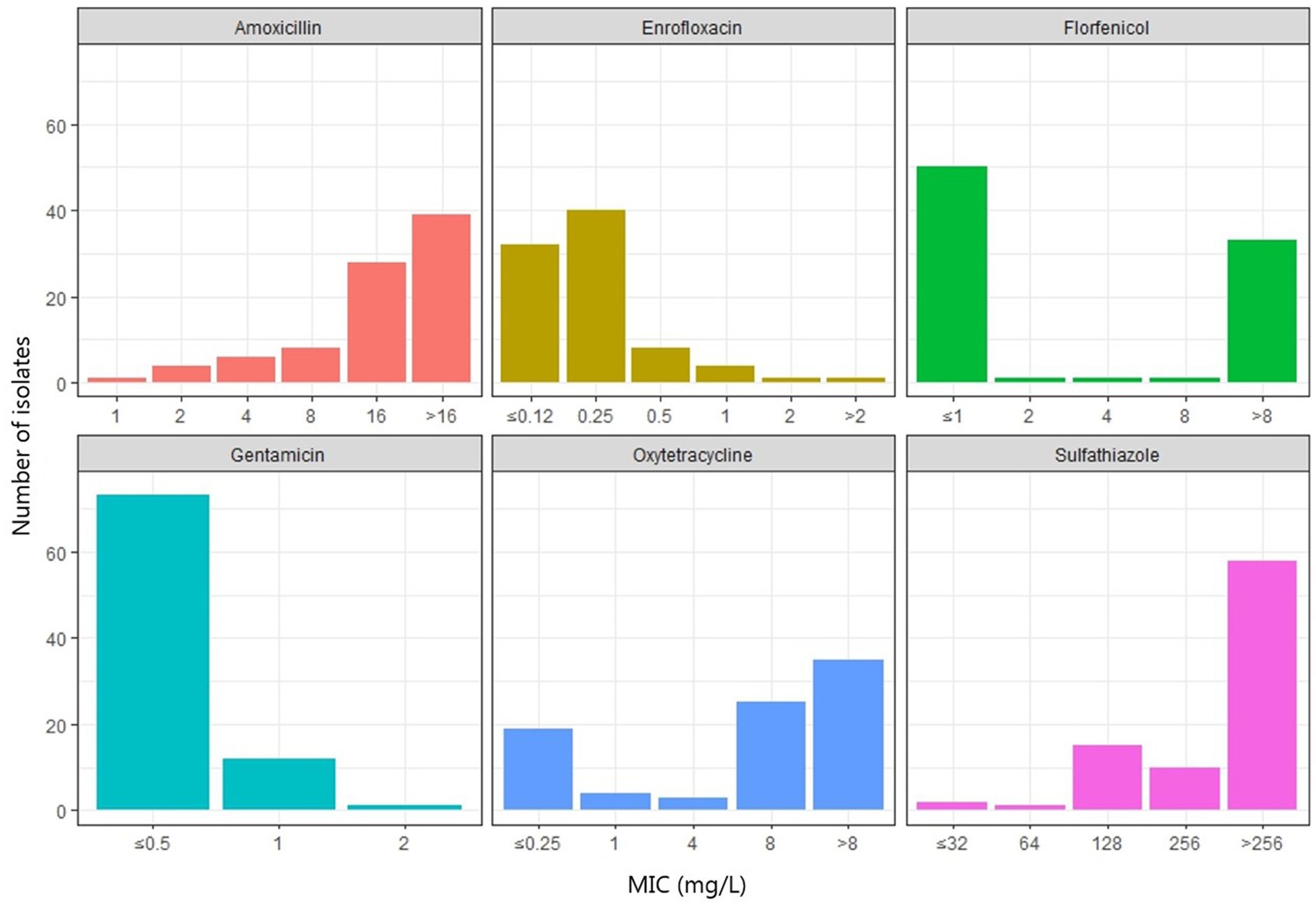
Figure 5. Minimum inhibitory concentrations (mg/L) of six antimicrobials commonly used for treatment of motile Aeromonas septicemia in striped catfish production in Vietnam.
There were no plasmids detected in any of the genomes. We found the sulfonamide resistance genes sul1 (70%) and sul2 (63%), the quinolone resistance genes QnrS2 (62%) and aac(6′)-Ib-cr (7%). Beta-lactam resistance encoding genes included ampH (97%) and imiH (3%), cphA2 (83%), cphA4 (17%) and cphA3 (3%). The cphA genes can also code for carbapenemase resistance. The trimethoprim resistance genes dfrA1 and dfr22 were found in 70 and 3% of the isolates, respectively. The tetracycline resistance gene tet(A) was present in 67% of the genomes. Resistance genes to phenicols; floR (47%) and rifampin, arr-2 (7%) were also found. All of the A. dhakensis genomes carried the cphA2 resistance gene but no other cphA genes, while all the A. hydrophila genomes carried cphA4 or cphA3. Apart from the genome of isolate AH80 that carried the sul1 gene, none of the other A. hydrophila genomes carried genes coding for sulfonamide resistance. Twenty A. dhakensis isolates carried sul1 and 19 isolates carried sul2. The floR gene was only found in A. dhakensis. Apart from isolate AH80, genes encoding resistance to trimethoprim, tetracycline and phenicol were not present in the A. hydrophila isolates. A heatmap for antimicrobial resistance genes is provided in Figure 6.
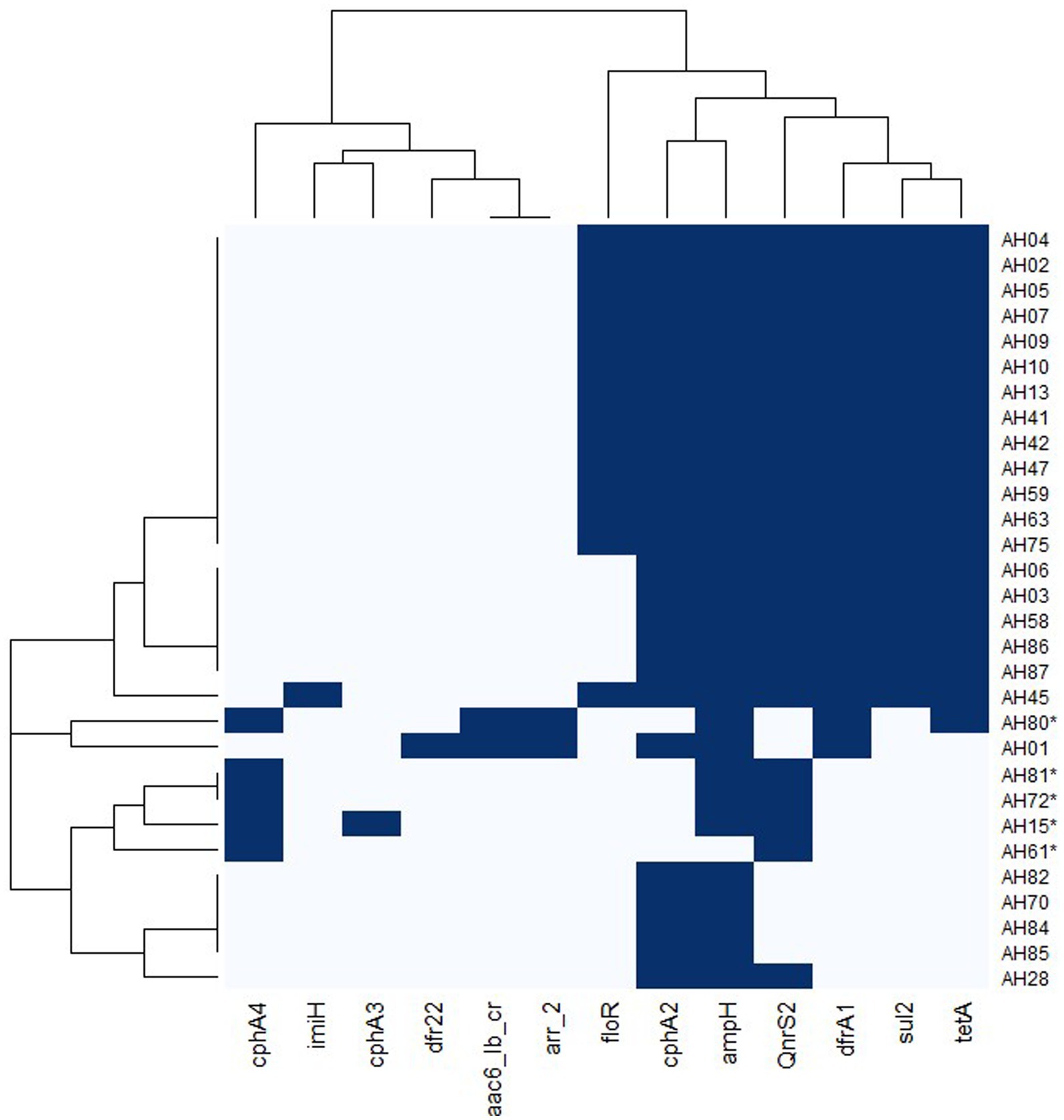
Figure 6. Heatmap of antimicrobial resistance genes in A. dhakensis and A. hydrophila. Dark blue color indicates presence of a gene and light blue color indicates absence of the gene. The A. hydrophila isolates are marked with an asterisk.
3.5. Virulence factors and genes in Aeromonas dhakensis and Aeromonas hydrophila
All A. hydrophila genomes carried virulence genes coding for Flp type IV pili. None of the A. dhakensis genomes carried any genes coding for this type of pili. All genomes carried genes coding for mannose sensitive hemagglutinin (Msh) pilus, polar flagella, Tap type IV pili and type I fimbriae. For type I fimbria, all genomes carried fimC, fimD and fimF. Only some genomes carried fimA and fimE. The type 2 and type 6 secretions systems (T2SS and T6SS) were present in all genomes, while the type 3 secretion system (T3SS) was absent in all genomes. The aerolysin aerA/cytotoxic enterotoxin act, extracellular hemolysin AHH1 gene ahh1 and the hemolysin HlyA gene hlyA were present in all genomes. The gene coding for heat-stable cytotonic enterotoxin, ast, was only present in A. dhakensis genomes. The detailed results from the virulence gene analysis can be found in the Supplementary Table S4. The presence of seven genes (ahpA, alt, dns, elastase, gcaT, lip and ser) relevant for virulence in Aeromonas spp. was investigated in all genomes. The ahpA gene was present in all A. dhakensis, but none of the A. hydrophila genomes. The alt, dns, elastase and lip genes were present in all genomes. The ser gene was present in all A. hydrophila, but none of the A. dhakensis genomes.
3.6. Comparison genomics of Aeromonas hydrophila and Aeromonas dhakensis
3.6.1. Mobile genetic elements and phages
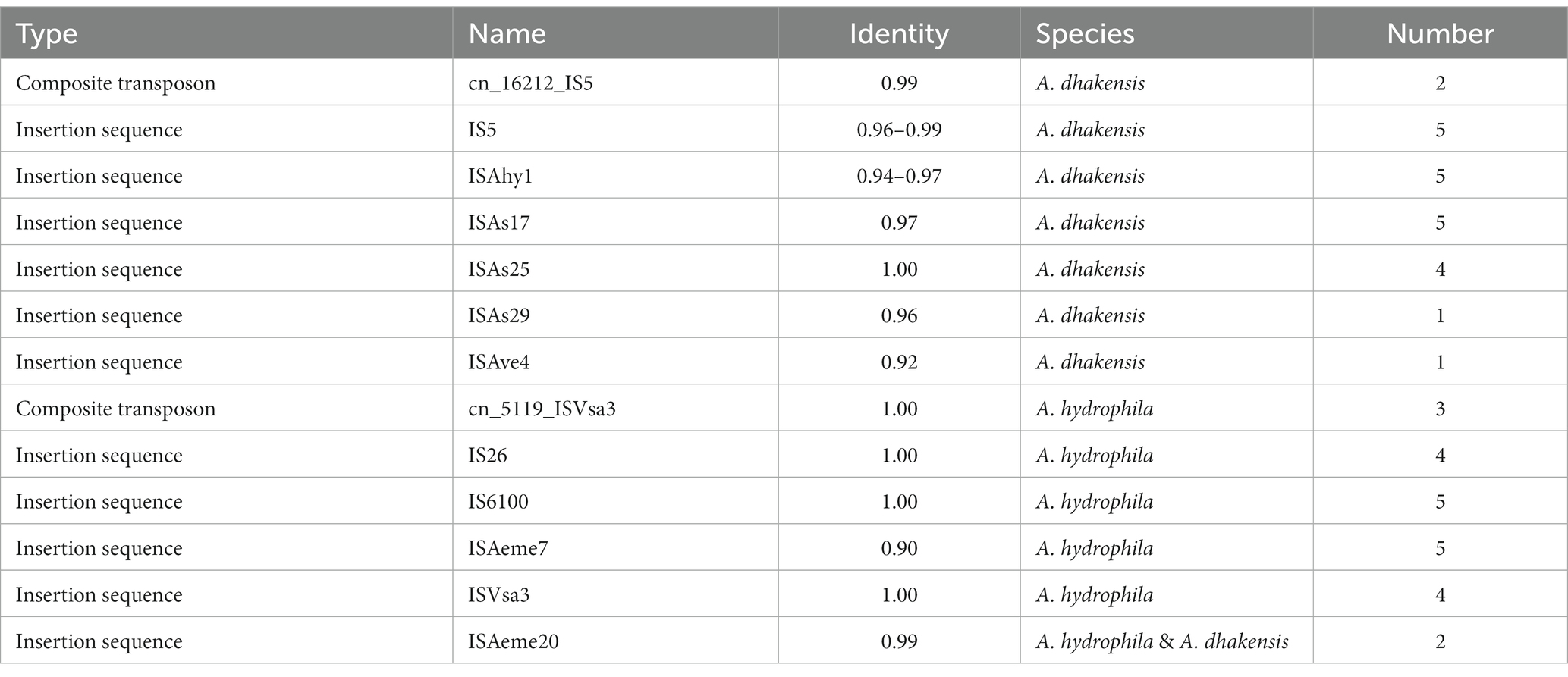
The results from MobileElementFinder showed that insertion sequences were present in all genomes. The results from this analysis are summarized in Table 2. In A. dhakensis, the insertion sequences IS5, ISAhy1 and ISAs17 were present in all genomes. In A. hydrophila, the insertions sequences IS6100 and ISAeme7 were found in all five genomes. Only one insertion sequence, ISAeme20, was present in both species. Two types of composite transposons were found in the genomes. The transposon cn_16212_IS5 was found in two A. dhakensis genomes and cn_5119_ISVsa3 was found in three A. hydrophila genomes.
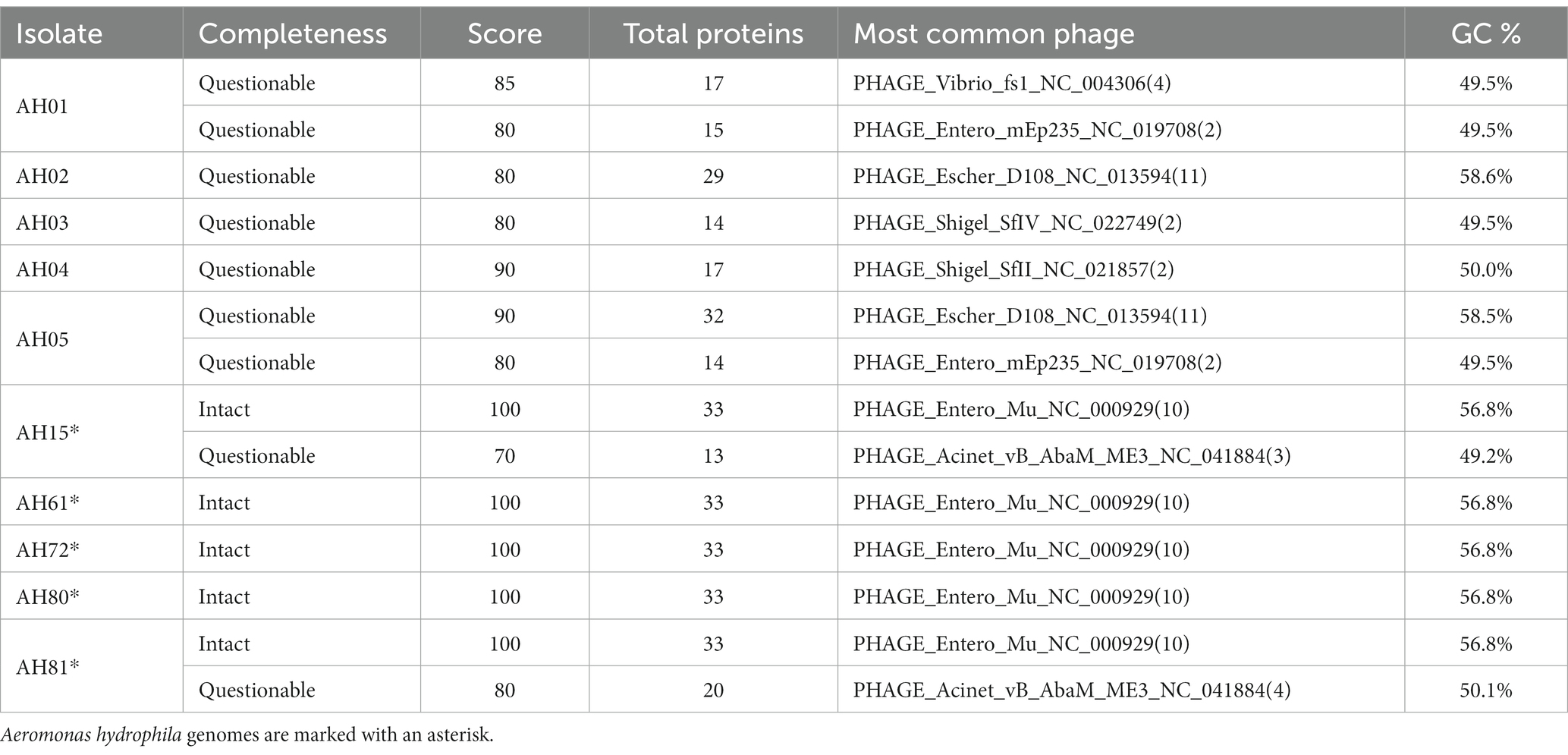
The results from PHASTER showed that A. dhakensis only carried prophage regions of questionable completeness while A. hydrophila had intact prophage regions in every genome. In the A. dhakensis genomes there were five different prophage regions while there were two different prophage regions present in the A. hydrophila genomes. Table 3 summarizes the prophage region findings.
3.6.2. Comparison of outer membrane proteins and lipopolysaccharide regions
We blasted the sequenced genomes against a reference sequence of six outer membrane proteins (ompA, ompC, ompTS, ompK, omp38 and omp48,) from A. hydrophila. A. hydrophila and A. dhakensis genomes shared outer membrane proteins. The proteins ompA and ompTS were present in all A. hydrophila genomes and all A. dhakensis genomes. The ompC (25/25) and omp48 (12/25) proteins were only present in A. dhakensis genomes. The omp38 protein was present in all A. hydrophila genomes (5/5) but was not present in any of the A. dhakensis genomes. The protein ompK was not present in either species. The complete results from the outer membrane protein analysis can be found in Supplementary Table S5. We also looked at differences between the genomes in RAST. When comparing the genes coding for capsular and extracellular polysaccharides the only difference between A. dhakensis compared to A. hydrophila were the genes coding for rhamnose containing glycans and dTDP-rhamnose synthesis.
All five A. hydrophila genomes from this study and five A. dhakensis genomes were compared to the LPS region displayed by A. hydrophila AH-3 (Jimenez et al., 2008). In both species, the genes kdkA, waaC, waaA, wahF, waaE, waaF and coaD were similar to the LPS region in isolate AH-3. The region containing the hldD, wahA, waaL, wahB, wahC and wahE varied between the two species (Supplementary Figure S1).
3.6.3. Pan genome analysis and analysis of species-specific coding sequences
The pan genome analysis reviled a core genome consisting of 2,799 genes shared by both A. hydrophila and A. dhakensis. A. dhakensis genomes had 1,276 unique genes while A. hydrophila genomes had 1,357 unique genes (Figure 7). Analyzing the unique gene regions in VRprofile reviled that the unique region of A. dhakensis consisted of 28 genes related to virulence factors, 13 genes related to antimicrobial resistance, two genes related to the type VI secretion system and type VI secretion effectors, 18 prophage related genes, two integron genes, 10 insertions sequence elements, 15 pathogenicity island genes and 12 antimicrobial resistance island genes. In A. hydrophila, the unique genes consisted of 52 genes related to virulence factors, 13 genes related to antimicrobial resistance, 12 prophage related genes, two genes related to the type VI secretion system and one type VI secretion effector, two integron genes, 12 insertion sequence element genes, 31 pathogenicity island genes and 26 antimicrobial resistance island genes (Figure 8).
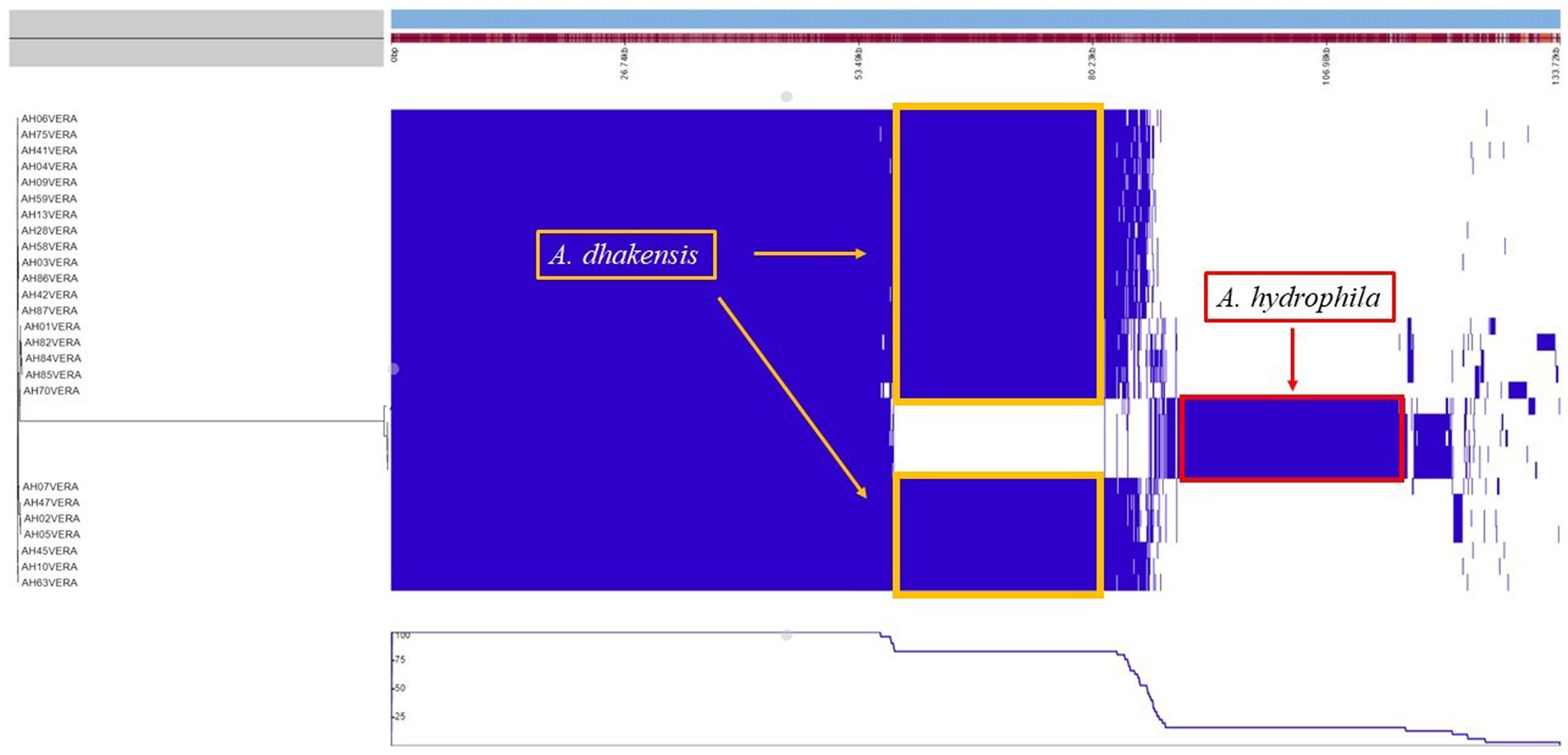
Figure 7. Pan genome analysis of A. hydrophila and A. dhakensis. The unique regions for each species are marked with yellow (A. dhakensis) and red (A. hydrophila) color.
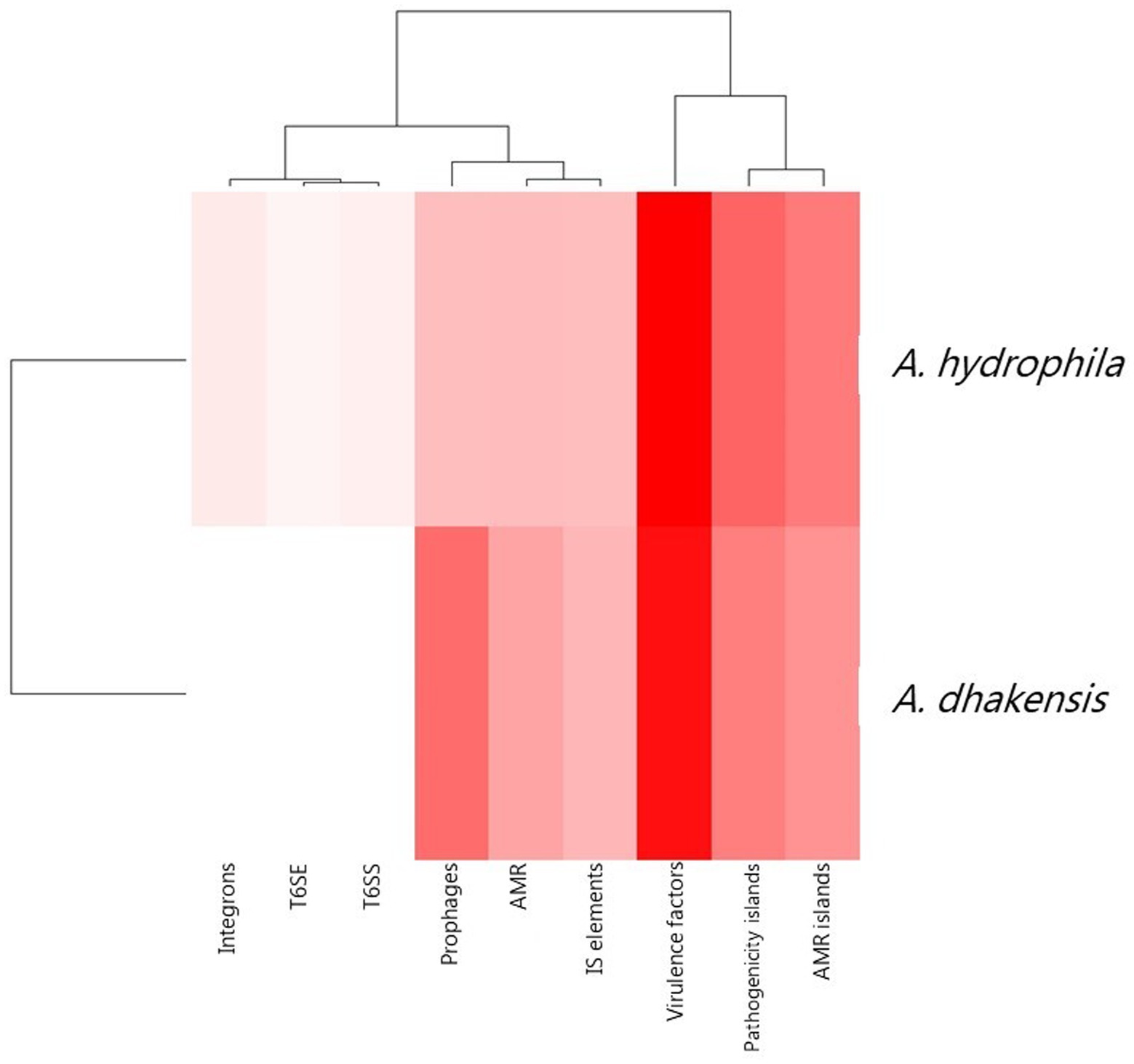
Figure 8. Unique attributes of A. dhakensis and A. hydrophila genomes. AMR (antimicrobial resistance), AMR islands (antimicrobial resistance islands), IS elements (insertions sequence elements), T6SS (type VI secretion systems), T6SE (type VI secretion effectors). A dark red color indicates higher prevalence of the genomic feature and a lighter color indicates lower prevalence of the feature.
4. Discussion
A. dhakensis ST656 was the dominating species causing motile Aeromonas septicemia in the Mekong Delta in Vietnam during the studied time period. All confirmed A. hydrophila isolates in our study were from 2019, 2020 and 2021. Another study conducted in the Mekong Delta with isolates collected earlier (2013–2015) showed that A. dhakensis was more frequently isolated than A. hydrophila already at that time (Bartie et al., 2023). Whether there has been a shift from A. hydrophila towards A. dhakensis or if A. dhakensis has been the main causative agent of motile Aeromonas septicemia all along is unclear as current confirmed isolates (next-generation sequencing MLST and/or WGS) of both species only date back to 2013. Our results corroborate previous findings of A. dhakensis being the dominating causative agent of motile Aeromonas septicemia in striped catfish farming in the Mekong Delta. Knowledge about which of the two Aeromonas species is causing motile Aeromonas septicemia may be important if there are species-specific differences in their susceptibility to antimicrobial treatment, their virulence and immunogenicity.
The approved vaccine by PHARMAQ AS, Norway, to prevent motile Aeromonas septicemia in Vietnam contains formalin-inactivated cultures of A. hydrophila serotypes A and B. It is administered through intraperitoneal injection of fish (PHARMAQ, 2018). Since this vaccine is based on A. hydrophila strains it is important to know if the vaccine is equally efficient and effective in preventing disease and transmission of motile Aeromonas septicemia caused by A. dhakensis. We compared structures of the cell wall, that are relevant for the pathogenesis, to look for differences between A. hydrophila compared to A. dhakensis (Jimenez et al., 2008). Analysing the lipopolysaccharide region, we saw that the genes kdkA, waaC, waaA, wahF, waaE, waaF and coaD were identical in A. hydrophila and A. dhakensis, while there were variations in the region containing the genes hldD, wahA, waaL, wahB, wahC and wahE. There were differences in the presence of outer membrane proteins. Only two out of six outer membrane proteins were shared by both species. The genes coding for the other outer membrane proteins were present in either A. dhakensis or A. hydrophila. There were differences between A. hydrophila and A. dhakensis when it came to outer membrane structures relevant to antigen recognition in the host. To establish whether the only current approved vaccine is equally efficient and effective in preventing motile Aeromonas septicemia caused by A. dhakensis as A. hydrophila, experimental infection studies comparing the vaccine’s potential in preventing disease caused by the two bacterial species are needed.
Until the early 2000’s, A. dhakensis was not a distinct species (Huys et al., 2002). With modern molecular-based detection methods it is now possible to distinguish A. dhakensis from A. hydrophila. Whole genome sequencing and multilocus phylogenetic analysis are to date the most accurate methods of identification (Chen et al., 2016; Du et al., 2021), but such methods are not routinely used for diagnostic purposes in Vietnam and most other countries. The PCR primers initially used to identify the A. hydrophila is based on an article published in 1990 (Pollard et al., 1990), before A. dhakensis was identified as a distinct species. If these PCR primers are used in diagnostic laboratories to identify A. dhakensis, this species will routinely be misidentified. The same applies for the MALDI-TOF commercial databases as it does not include. A. dhakensis. Instead, the MALDI-TOF will identify the species on genus level only, or find the closest matching bacterial species, which in our case was A. hydrophila. Thus, the use of such existing identification methods will overestimate the true burden of A. hydrophila and the burden of A. dhakensis will be underestimated. There is a need to develop a species-specific PCR method that can differentiate A. dhakensis from A. hydrophila which can be used in laboratories for research and diagnostic purposes.
In Vietnamese fish production, the most commonly used antimicrobials are phenicols (florfenicol), tetracyclines (oxytetracycline) and sulfonamides (Luu et al., 2021). In striped catfish farming the most commonly used antimicrobial is florfenicol (Pham et al., 2023) and the aquaculture sector is said to be a genetic hotspot for gene transfer, including antimicrobial resistance genes (Watts et al., 2017). It was therefore no surprise to see a high prevalence (47–70%) of antimicrobial resistance genes for these three antimicrobial groups commonly used in the production. However, we noted differences between the A. dhakensis and A. hydrophila isolates regarding the presence of antimicrobial resistance genes and MIC values to the tested antimicrobials. The florfenicol resistance gene, floR, was only found in A. dhakensis (14/25) and these isolates had an MIC ≥8 mg/L for florfenicol while the A. hydrophila isolates had MIC values ≤1 mg/L for florfenicol. None of the A. hydrophila genomes carried the sul2 gene. All of the A. dhakensis genomes carried the cphA4 or cphA3 gene, while A. hydrophila genomes carried cphA2 instead. All of the isolates with an MIC ≥8 mg/L for tetracycline also carried the tetracycline resistance gene tet(A).
We found differences in the antimicrobial susceptibility between A. dhakensis and A. hydrophila with higher MIC values for florfenicol and tetracyclines seen in A. dhakensis. Our findings indicate that the disease-causing organism could impact the choice of which antimicrobial to use. However, in our study we only tested five confirmed A. hydrophila isolates in the MIC analysis. Any differences in the antimicrobial susceptibility between the two Aeromonas spp. isolated from striped catfish and practical implications for treatment needs further studies. Pharmacokinetic studies to support development of antimicrobial treatment guidelines for motile Aeromonas septicemia are also needed.
A study comparing Aeromonas spp. from fish and human clinical isolates found that A. dhakensis carried more virulence genes compared to other Aeromonas spp. (A. caviae, A. veronii and A. hydrophila) (Wu et al., 2019). In our genome analysis, we found differences between the virulence genes present in the A. dhakensis genomes compared to the A. hydrophila genomes. But unlike reported by Wu et al. (2019), when we compared the unique genes from both bacterial species, A. hydrophila had more unique virulence genes than A. dhakensis. The five A. hydrophila genomes carried virulence genes coding for Flp type IV pili. None of the A. dhakensis genomes carried any genes coding for this type of pili. The ahpA gene was present in all A. dhakensis, but none of the A. hydrophila genomes. This protease has previously been associated with pathogenic strains of Aeromonas spp. (Zhu et al., 2007). The serine protease gene ser was present in all A. hydrophila, but none of the A. dhakensis genomes. The gene coding for heatstable cytotonic enterotoxin, ast, was only present in A. dhakensis genomes. The enterotoxin Act is one of the most important virulence factors in A. dhakensis (Rasmussen-Ivey et al., 2016) and the gene coding for this enterotoxin was present in all genomes, including A. hydrophila genomes. The T2SS is known to secrete the enterotoxin Act in Aeromonas spp. and is, together with T6SS, common in aquatic isolates (Rasmussen-Ivey et al., 2016). However, a difference in the geographical spread of T6SS has been suggested where a complete set of core genes has been observed in A. hydrophila isolates from Asia whereas isolates from the United States only carry remnants of this secretion system (Rasmussen-Ivey et al., 2016). The T6SS plays an important role in the bacteria’s evasion of the host immune system. This secretions system can translocate proteins and toxins into the cytosol of other cells (Wang et al., 2023). There was no difference in the protein secretions systems used by the two bacterial species and the T2SS and T6SS were found in all genomes, suggesting that these secretion systems play and important part in the pathogenicity of both A. hydrophila and A. dhakensis in Vietnamese striped catfish production.
5. Conclusion
Our study confirms that the dominating species causing outbreaks of motile Aeromonas septicemia in striped catfish fingerlings in the Mekong Delta from 2017 to 2021 was A. dhakensis ST656. A. hydrophila ST251 was also isolated from diseased striped catfish fingerlings. The true burden of A. dhakensis as an aquatic pathogen has been masked due to diagnostic methods unable of differentiating A. dhakensis from A. hydrophila. As vaccines are important preventive measures to reduce disease and needs for using antimicrobials, studies comparing the current vaccine’s efficiency and effectiveness in preventing infection with both A. dhakensis and A. hydrophila are needed.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
VE: Writing – original draft, Writing – review & editing. LK: Writing – review & editing. YH: Writing – review & editing. TD: Writing – review & editing. TP: Writing – review & editing. AD: Writing – review & editing.
Funding
VE was partly supported through a PhD scholarship from the Sino-Danish Center, a University Partnership between Denmark and China. Financial support to collect and store isolates in Vietnam was received from KMP-Singapore.
Acknowledgments
We would like to acknowledge and thank the staff at Can Tho University who helped to collect the bacterial isolates, and all the striped catfish fish farmers who contributed in the An Giang, Ben Tre, Can Tho, Dong Thap, Hau Giang, Long An, Tien Giang and Vinh Long provinces.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1254781/full#supplementary-material
Footnotes
References
Alcock, B. P., Raphenya, A. R., Lau, T. T. Y., Tsang, K. K., Bouchard, M., Edalatmand, A., et al. (2020). CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 48, D517–D525. doi: 10.1093/nar/gkz935
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Azzam-Sayuti, M., Ina-Salwany, M. Y., Zamri-Saad, M., Yusof, M. T., Annas, S., Najihah, M. Y., et al. (2021). The prevalence, putative virulence genes and antibiotic resistance profiles of Aeromonas spp. isolated from cultured freshwater fishes in peninsular Malaysia. Aquaculture 540:736719. doi: 10.1016/j.aquaculture.2021.736719
Bartie, K. L., Ngô, T. P. H., Bekaert, M., Hoang Oanh, D. T., Hoare, R., Adams, A., et al. (2023). Aeromonas hydrophila ST251 and Aeromonas dhakensis are major emerging pathogens of striped catfish in Vietnam. Front. Microbiol. 13:1067235. doi: 10.3389/fmicb.2022.1067235
Beaz-Hidalgo, R., Martínez-Murcia, A., and Figueras, M. J. (2013). Reclassification of Aeromonas hydrophila subsp. dhakensis Huys et al. 2002 and Aeromonas aquariorum Martínez-Murcia et al. 2008 as Aeromonas dhakensis sp. nov. comb nov. and emendation of the species Aeromonas hydrophila. Syst. Appl. Microbiol. 36, 171–176. doi: 10.1016/j.syapm.2012.12.007
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Brettin, T., Davis, J. J., Disz, T., Edwards, R. A., Gerdes, S., Olsen, G. J., et al. (2015). RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 5:8365. doi: 10.1038/srep08365
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421
Carattoli, A., Zankari, E., García-Fernández, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using plasmid finder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Chen, P. L., Lamy, B., and Ko, W. C. (2016). Aeromonas dhakensis, an increasingly recognized human pathogen. Front. Microbiol. 7, 1–8. doi: 10.3389/fmicb.2016.00793
Clausen, P. T. L. C., Aarestrup, F. M., and Lund, O. (2018). Rapid and precise alignment of raw reads against redundant databases with KMA. BMC Bioinformatics 19:307. doi: 10.1186/s12859-018-2336-6
Clinical and Laboratory Standards Institute (2017) Performance standards for antimicrobial susceptibility testing. 950 West Valley road, suite 2500, Wayne, Pennsylvania, USA Available at: www.clsi.org
Croucher, N. J., Page, A. J., Connor, T. R., Delaney, A. J., Keane, J. A., Bentley, S. D., et al. (2015). Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 43:e15. doi: 10.1093/nar/gku1196
Du, X., Wang, M., Zhou, H., Li, Z., Xu, J., Li, Z., et al. (2021). Comparison of the multiple platforms to identify various Aeromonas species. Front. Microbiol. 11, 1–8. doi: 10.3389/fmicb.2020.625961
FAO (2022) World fisheries and aquaculture, FAO: Rome, 2022. Available at: https://www.fao.org/3/ca9229en/online/ca9229en.html#chapter-1_1
Fernández-Bravo, A., and Figueras, M. J. (2020). An update on the genus Aeromonas: taxonomy, epidemiology, and pathogenicity. Microorganisms 8:129. doi: 10.3390/microorganisms8010129
Hadfield, J., Croucher, N. J., Goater, R. J., Abudahab, K., Aanensen, D. M., and Harris, S. R. (2018). Phandango: an interactive viewer for bacterial population genomics. Bioinformatics 34, 292–293. doi: 10.1093/bioinformatics/btx610
Hasman, H., Saputra, D., Sicheritz-Ponten, T., Lund, O., Svendsen, C. A., Frimodt-Møller, N., et al. (2014). Rapid whole-genome sequencing for detection and characterization of microorganisms directly from clinical samples. J. Clin. Microbiol. 52, 139–146. doi: 10.1128/JCM.02452-13
Hoa, T. T. T., Boerlage, A. S., Duyen, T. T. M., Thy, D. T. M., Hang, N. T. T., Humphry, R. W., et al. (2021). Nursing stages of striped catfish (Pangasianodon hypophthalmus) in Vietnam: pathogens, diseases and husbandry practices. Aquaculture 533:736114. doi: 10.1016/j.aquaculture.2020.736114
Huys, G., Kämpfer, P., Albert, M. J., Kühn, I., Denys, R., and Swings, J. (2002). Aeromonas hydrophila subsp. dhakensis subsp. nov., isolated from children with diarrhoea in Bangladesh, and extended description of Aeromonas hydrophila subsp. hydrophila (Chester 1901) Stanier 1943 (approved lists 1980). Int. J. Syst. Evol. Microbiol. 52, 705–712. doi: 10.1099/00207713-52-3-705
Janda, J. M., and Abbott, S. L. (2010). The genus Aeromonas: taxonomy, pathogenicity, and infection. Clin. Microbiol. Rev. 23, 35–73. doi: 10.1128/CMR.00039-09
Jimenez, N., Canals, R., Lacasta, A., Kondakova, A. N., Lindner, B., Knirel, Y. A., et al. (2008). Molecular analysis of three Aeromonas hydrophila AH-3 (serotype O34) lipopolysaccharide core biosynthesis gene clusters. J. Bacteriol. 190, 3176–3184. doi: 10.1128/JB.01874-07
Jolley, K. A., Bray, J. E., and Maiden, M. C. J. (2018). Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 3:124. doi: 10.12688/wellcomeopenres.14826.1
Khor, W. C., et al. (2018). Comparison of clinical isolates of Aeromonas from Singapore and Malaysia with regard to molecular identification, virulence, and antimicrobial profiles. Microb. Drug Resist. 24, 469–478. doi: 10.1089/mdr.2017.0083
Larsen, M. V., Cosentino, S., Lukjancenko, O., Saputra, D., Rasmussen, S., Hasman, H., et al. (2014). Benchmarking of methods for genomic taxonomy. J. Clin. Microbiol. 52, 1529–1539. doi: 10.1128/JCM.02981-13
Lau, T. T. V., Tan, J. A. M. A., Puthucheary, S. D., Puah, S. M., and Chua, K. H. (2020). Genetic relatedness and novel sequence types of clinical Aeromonas dhakensis from Malaysia. Braz. J. Microbiol. 51, 909–918. doi: 10.1007/s42770-020-00239-8
Letunic, I., and Bork, P. (2021). Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
Li, J., Tai, C., Deng, Z., Zhong, W., He, Y., and Ou, H. Y. (2018). VRprofile: gene-cluster-detection-based profiling of virulence and antibiotic resistance traits encoded within genome sequences of pathogenic bacteria. Brief. Bioinform. 19, bbw141–bbw574. doi: 10.1093/bib/bbw141
Liu, B., Zheng, D., Jin, Q., Chen, L., and Yang, J. (2019). VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 47, D687–D692. doi: 10.1093/nar/gky1080
Luu, Q. H., Nguyen, T. B. T., Nguyen, T. L. A., do, T. T. T., Dao, T. H. T., and Padungtod, P. (2021). Antibiotics use in fish and shrimp farms in Vietnam. Aquac. Rep. 20:100711. doi: 10.1016/j.aqrep.2021.100711
Mikheenko, A., Prjibelski, A., Saveliev, V., Antipov, D., and Gurevich, A. (2018). Versatile genome assembly evaluation with QUAST-LG. Bioinformatics 34, i142–i150. doi: 10.1093/bioinformatics/bty266
Nielsen, M. E., Høi, L., Schmidt, A. S., Qian, D., Shimada, T., Shen, J. Y., et al. (2001). Is Aeromonas hydrophila the dominant motile Aeromonas species that causes disease outbreaks in aquaculture production in the Zhejiang Province of China? Dis. Aquat. Org. 46, 23–29. doi: 10.3354/dao046023
Nurk, S., Bankevich, A., Antipov, D., Gurevich, A., Korobeynikov, A., Lapidus, A., et al. (2013). “Assembling genomes and Mini-metagenomes from highly chimeric reads” in BT-research in computational molecular biology. eds. M. Deng, et al. (Berlin, Heidelberg: Springer Berlin Heidelberg), 158–170.
Overbeek, R., Olson, R., Pusch, G. D., Olsen, G. J., Davis, J. J., Disz, T., et al. (2014). The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res. 42, D206–D214. doi: 10.1093/nar/gkt1226
Page, A. J., Cummins, C. A., Hunt, M., Wong, V. K., Reuter, S., Holden, M. T. G., et al. (2015). Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693. doi: 10.1093/bioinformatics/btv421
Pham, Q. V., Nguyen, Q. T., Devreese, M., Croubels, S., Dang, T. H. O., Dalsgaard, A., et al. (2023). Pharmacokinetics and depletion of florfenicol in striped catfish Pangasianodon hypophthalmus after oral administration. Fish. Sci. 89, 357–365. doi: 10.1007/s12562-023-01682-w
PHARMAQ (2018) PHARMAQ products-VACCINE PACKAGE INSERT. Available at: https://www.pharmaq.com/en/pharmaq/our-products/?country=Vietnam&category=all (Accessed August 15, 2023).
Pollard, D. R., Johnson, W. M., Lior, H., Tyler, S. D., and Rozee, K. R. (1990). Detection of the Aerolysin gene in Aeromonas hydrophila by the polymerase chain reaction. J. Clin. Microbiol. 28, 2477–2481. doi: 10.1128/JCM.28.11.2477-2481.1990
Rasmussen-Ivey, C. R., Figueras, M. J., McGarey, D., and Liles, M. R. (2016). Virulence factors of Aeromonas hydrophila: in the wake of reclassification. Front. Microbiol. 7, 1–10. doi: 10.3389/fmicb.2016.01337
Rasmussen-Ivey, C. R., Hossain, M. J., Odom, S. E., Terhune, J. S., Hemstreet, W. G., Shoemaker, C. A., et al. (2016). Classification of a hypervirulent aeromonas hydrophila pathotype responsible for epidemic outbreaks in warm-water fishes. Front. Microbiol. 7, 1–16. doi: 10.3389/fmicb.2016.01615
Saharia, P. K., Hussain, I. A., Pokhrel, H., Kalita, B., Borah, G., and Yasmin, R. (2021). Prevalence of motile Aeromonas Septicaemia (MAS) in fish culture systems of the Central Brahmaputra Valley zone of Assam, India. Aquac. Res. 52, 1201–1214. doi: 10.1111/are.14979
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics (Oxford, England) 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Stratev, D., and Odeyemi, O. A. (2017). An overview of motile Aeromonas septicaemia management. Aquac. Int. 25, 1095–1105. doi: 10.1007/s10499-016-0100-3
Wang, Y., Zeng, M., Xia, L., Valerie Olovo, C., Su, Z., and Zhang, Y. (2023). Bacterial strategies for immune systems — role of the type VI secretion system. Int. Immunopharmacol. 114:109550. doi: 10.1016/j.intimp.2022.109550
Watts, J. E., Schreier, H. J., Lanska, L., and Hale, M. S. (2017). The rising tide of antimicrobial resistance in aquaculture: sources, sinks and solutions. Mar. Drugs 15:158. doi: 10.3390/md15060158
Wood, D. E., and Salzberg, S. L. (2014). Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 15:R46. doi: 10.1186/gb-2014-15-3-r46
Wu, C. J., Ko, W. C., Lee, N. Y., Su, S. L., Li, C. W., Li, M. C., et al. (2019). Aeromonas isolates from fish and patients in Tainan City, Taiwan: genotypic and phenotypic characteristics. Appl. Environ. Microbiol. 85, e01360–e01319. doi: 10.1128/AEM.01360-19
Keywords: Aeromonas dhakensis, motile Aeromonas septicemia (MAS), striped catfish (Pangasianodon hypophthalmus), comparative genomics, aquaculture, Aeromonas hydrophila
Citation: Erickson VI, Khoi LM, Hounmanou YMG, Dung TT, Phu TM and Dalsgaard A (2023) Comparative genomic analysis of Aeromonas dhakensis and Aeromonas hydrophila from diseased striped catfish fingerlings cultured in Vietnam. Front. Microbiol. 14:1254781. doi: 10.3389/fmicb.2023.1254781
Edited by:
Juan M. Tomas, University of Barcelona, SpainReviewed by:
Ulisses Padua Pereira, State University of Londrina, BrazilAna Fernández-Bravo, University of Rovira i Virgili, Spain
Copyright © 2023 Erickson, Khoi, Hounmanou, Dung, Phu and Dalsgaard. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anders Dalsgaard, YWRhbEBzdW5kLmt1LmRr
†These authors have contributed equally to this work
 Vera Irene Erickson
Vera Irene Erickson Le Minh Khoi2†
Le Minh Khoi2† Yaovi Mahuton Gildas Hounmanou
Yaovi Mahuton Gildas Hounmanou Tran Minh Phu
Tran Minh Phu Anders Dalsgaard
Anders Dalsgaard