
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 19 October 2023
Sec. Microbe and Virus Interactions with Plants
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1254246
This article is part of the Research Topic Studying Plant Virus-Host Interactions: Elucidating the Natural Resistance Mechanism View all 6 articles
Thrips palmi (Thysanoptera: Thripidae) is a major agricultural pest infesting over 200 plant species. Along with direct injury caused by feeding, T. palmi spreads several orthotospoviruses. Groundnut bud necrosis orthotospovirus (GBNV, family Tospoviridae, genus Orthotospovirus) is the predominant orthotospovirus in Asia, vectored by T. palmi. It is responsible for almost 89 million USD losses in Asia annually. Several transcripts of T. palmi related to innate immune response, receptor binding, cell signaling, cellular trafficking, viral replication, and apoptosis are responsive to the infection of orthotospoviruses in thrips. Expression of T. palmi tyrosine kinase Btk29A isoform X1 (Btk29A) and collagen alpha-1(III) chain-like (COL3A1) are significantly regulated post-GBNV and capsicum chlorosis orthotospovirus infection. In the present study, T. palmi Btk29A and COL3A1 were silenced and the effect on virus titer and fitness was assessed. The expression of Btk29A and COL3A1 was significantly reduced by 3.62 and 3.15-fold, respectively, 24 h post-dsRNA exposure. Oral administration of Btk29A and COL3A1 dsRNAs induced 60 and 50.9% mortality in T. palmi. The GBNV concentration in T. palmi significantly dropped post-silencing Btk29A. In contrast, the silencing of COL3A1 led to an increase in GBNV concentration in T. palmi compared to the untreated control. To the best of our knowledge, this is the first report on the effect of silencing Btk29A and COL3A1 on the fitness and GBNV titer in T. palmi.
The melon thrips, Thrips palmi Karny (Thysanoptera: Thripidae), is a major pest of vegetables, legumes, and ornamental plants. T. palmi was first identified in Sumatra (Karny, 1925) and is currently reported from the rest of Asia, Australia, the Pacific, North and South America, and West Africa (Cannon et al., 2007). It is considered an A1 quarantine pest by EPPO [The European and Mediterranean Plant Protection Organization (EPPO), 2018]. Larvae and adults of T. palmi rasp the tender plant parts and suck the cellular content. White silvery scarring develops on the affected plant parts that gradually turn brownish. In addition to direct injury caused by feeding, T. palmi transmits many orthotospoviruses (Riley et al., 2011). T. palmi-transmitted groundnut bud necrosis orthotospovirus (GBNV) causes more than USD 80 million losses annually in Asia (Reddy et al., 1995). GBNV can cause yield losses of up to 100% in tomatoes (Venkata Ramana et al., 2011). Outbreaks of watermelon bud necrosis orthotospovirus (WBNV) transmitted by T. palmi resulted in the failure of watermelon cultivation in southern India (Singh and Krishnareddy, 1995). The economic losses caused by T. palmi during 1994–2004 were around £ 16.9–19.6 million (MacLeod et al., 2004), though the exact figures on worldwide economic impact are not available. The application of insecticides is the most common option to protect crops from thrips and orthotospoviruses. However, thrips develop quick resistance to most of the common classes of insecticides due to their high fecundity, short generation period, and wide host range (Bao and Sonoda, 2012; Bao et al., 2014; Shi et al., 2020, 2021).
Lately, advancements in genome-assisted pest management techniques paved the way for inventive pest control methods. In particular, RNA interference (RNAi) has been extensively used to control chewing pests by delivering dsRNA through various techniques (Yu et al., 2013; Mamta and Rajam, 2017; Vogel et al., 2019; Adeyinka et al., 2020; Christiaens et al., 2020; Zhu and Palli, 2020; Hough et al., 2022). However, limited knowledge of the gene functions of target pests restrains the scope of genome-assisted insect resistance programs. RNAi has been successfully utilized to study the gene function of Frankliniella occidentalis and T. tabaci (Badillo-Vargas et al., 2015; Han et al., 2019; Singh et al., 2019; Andongma et al., 2020; Wu et al., 2022; Zhang et al., 2022a,b). Silencing of hsp70, TLR3, and TOB1 induced significant mortality and also inhibited chili leaf curl virus (ChiLCV) transmission by Bemisia tabaci adults (Chakraborty and Ghosh, 2022; Thesnim et al., 2023). Aphb and CP19 silencing affected fitness of aphid species (Mao and Zeng, 2012; Shang et al., 2020). RNAi of Nilaparvata lugens Nldl and Nljag in nymphs produced lethal or teratogenic effects (Yang et al., 2022). Little attempts were made to study the gene functions of T. palmi. In our previous study, T. palmi transcripts related to innate immune response, receptor binding, cell signaling, cellular trafficking, viral replication, and apoptosis were responsive to the acquisition of GBNV (Mahanta et al., 2022). Silencing of T. palmi UHRF1BP1 and PFAS using modified anti-sense oligos showed a decline in GBNV titer in T. palmi (Priti et al., 2022). Besides, T. palmi tyrosine kinase Btk29A isoform X1 (Btk29A) and collagen alpha-1(III) chain-like (COL3A1) were significantly regulated in response to GBNV and capsicum chlorosis orthotospovirus (CaCV) infection (Widana Gamage et al., 2018; Mahanta et al., 2022). Btk29A is involved in ATP binding and cellularization of Drosophila embryos (Thomas and Wieschaus, 2004). Dysfunctional Btk causes selective apoptosis and decreases HIV-1 production in HLM-1 cells (Guendel et al., 2015) indicating that Btk positively regulates productive virus infection either by activating the infected cells or enhancing the release of the virus from the infected cells. Collagens are structural proteins of the extracellular matrix and basal membrane and are constituents of viral biofilm (VB) development of human T-cell leukemia virus type 1 (HTLV-1; Millen et al., 2019). HTLV-1 infects T- cells through cell-to-cell transmission by polarized budding into synaptic clefts and cell surface transfer of VBs. Knockout of collagen impairs the transfer of viral protein from infected to acceptor cells. Type III collagen is also involved in cell adhesion, migration, and differentiation through its interaction with cell surface receptor integrins (Kim et al., 2005). In the present study, T. palmi Btk29A and COL3A1 were silenced to study the effect on thrips fitness and orthotospovirus titer in thrips.
The study employed a homogenous population of T. palmi derived from a single adult female. The isofemale line has been maintained on eggplant since 2018. The identity of the population was further verified by morphometric keys (Bhatti, 1980; Cluever and Smith, 2017) and mitochondrial cytochrome oxidase subunit I (mtCOI) nucleotide sequencing.
The initial GBNV inoculum was collected from a pure culture maintained at the Advanced Center for Plant Virology, Indian Agricultural Research Institute (IARI), New Delhi. Sap inoculation of healthy cowpea plants at the two-leaf stage was done with GBNV under insect-proof conditions as described by Ghosh et al. (2021). The presence of GBNV infection in cowpea was verified using RT-PCR using GBNV-specific primers AG109F-AG110R (Table 1).
T. palmi differential gene expression data in response to GBNV and CaCV (Widana Gamage et al., 2018; Mahanta et al., 2022) were analyzed for the selection of potential gene targets in response to orthotospovirus infection. The expression of the Btk29A and COL3A1 in T. palmi was highly abundant post-GBNV and CaCV acquisition. Therefore, T. palmi Btk29A and COL3A1 were targeted for silencing using the cognate dsRNA. The available sequences of Btk29A and COL3A1 were retrieved from NCBI and aligned in MEGAX (Kumar et al., 2018) to find the conserved regions. SiRNA Wizard web tool (accessed on 10-01-2022)1 was used to predict putative siRNAs from the conserved regions of T. palmi Btk29A and COL3A1. The region identified for dsRNA design was subjected to evaluation for off-target effects in humans, mice, birds, butterflies, bees, ants, and plants using the NCBI BLASTn tool.2 The region in both genes that showed the maximum number of putative siRNAs without having any off-target effect was selected for dsRNA synthesis. The selected regions were 1,166–1,444 for Btk29A and 213–469 for COL3A1 genes.
The dsRNA stretches were amplified using primer pairs listed in Table 1. The PCR conditions were optimized using a gradient PCR. Total DNA was isolated from T. palmi using a CTAB extraction buffer as described by Jangra and Ghosh (2022) and used for amplification in a thermal cycler. A 25 μL PCR mixture comprised of 1X PCR buffer (Thermo Fisher Scientific, United States), 0.4 μM each forward and reverse primer (GCC Biotech, India), 0.26 mM dNTP mix (Thermo Fisher Scientific), ~ 60 ng template DNA, and 2 U of DreamTaq DNA polymerase (Thermo Fisher Scientific) were used. PCR was performed in a T100 thermocycler (Bio-Rad, United States) with an initial denaturation at 94°C for 5 min, 35 cycles of denaturation at 94°C for 40 s, annealing at 55°C for 40 s, extension at 72°C for 40 s, followed by a final extension at 72°C for 10 min. The PCR products were resolved on a 2% agarose gel, stained with GoodView (BR Biochem, India), and visualized under a gel documentation system (MaestroGen Inc., Taiwan). The amplified PCR products were sequenced for confirmation purposes.
The PCR amplicons were cloned between two T7 promoters in the L4440 expression vector (Addgene, United States) and transformed into RNase III deficient E. coli HT115 cells (University of Minnesota, United States). The presence of the desired inserts was confirmed by colony PCR and restriction digestion. Production of dsRNA was induced by adding 0.8 M isopropyl-β-D-1-thiogalactopyranoside (IPTG, HiMedia, India) to the culture of recombinant E. coli HT115 cells and incubating overnight at 37°C in a shaking incubator (Gnanasekaran et al., 2019, 2021). Total RNA was extracted from HT115 cells using Trizol reagent (Invitrogen, United States) according to the manufacturer’s protocol. DsRNA from the total RNA was purified using 1 U of DNase I (RNase-free, Thermo Fisher Scientific) and 1 U of RNase A (DNase and protease-free, Thermo Fisher Scientific) for 1 h at 37°C in the presence of 500 mM sodium chloride. The enzymes were inactivated by chloroform extraction as described by Chakraborty and Ghosh (2022). The quality and concentration of the purified dsRNAs were assessed in a spectrophotometer (Nano-300, Genetix Biotech Asia, India) and visualized on a 2% agarose gel. A dsRNA targeting Bemisia tabaci transducer of erbB2.1 (BtTOB1; Thesnim et al., 2023) and not specific to T. palmi based on nucleotide homology screening was used as a negative control.
The purified Btk29A and COL3A1 dsRNAs were administered to T. palmi orally using an artificial feeding setup as reported by Priti et al. (2022). Briefly, a diet was prepared by mixing 50 mg/mL pine pollen (Lost Empire Herbs, United States) extract with 1% sucrose solution and 0.0001% methylene blue tracker dye (Bio Basic, United States). The diet was supplemented with 3 μg/μL dsRNA (total volume 300 μL). The diet with dsRNA was filled into UV-sterilized detachable caps of 2 mL microcentrifuge tubes and covered with stretched parafilm. Around 20 adults or larvae were taken in the perforated microcentrifuge tubes. The cap filled with diet was closed gently. T. palmi were allowed to feed for 24 h at 28 ± 1°C temperature, 60 ± 10% relative humidity, and 16 h light–8 h dark. In the control sets, a diet without dsRNA and a diet with BtTOB1 dsRNA (non-specific to T. palmi) were supplied. Several such replicates were maintained to get enough dsRNA-treated and untreated thrips to assess the mortality, expression of the target genes, and virus titer in thrips.
The expression of T. palmi Btk29A and COL3A1 was assessed 24 h post-dsRNA feeding by RT-qPCR. β-tubulin was taken as the endogenous control. Table 1 lists the primer pairs used in RT-qPCR. Five surviving T. palmi in three biological replicates for each gene under study were considered for estimating the mRNA expression level. Total RNA was extracted using NucleoSpin RNA XS (Macherey-Nagel, Germany) according to the manufacturer’s protocol. Total RNA was quantified in a Nano-300 Micro Spectrophotometer (Genetix Biotech Asia), and cDNA was synthesized using FIREScript RT cDNA synthesis kit (Solis BioDyne, Tartu, Estonia). One μg RNA template, 5 μM oligo dT primers, 500 μM dNTP mix, 2 μL of 1 X reaction buffer, 10 U FIREScript RT, and 1 U RiboGrip RNase inhibitor were used in 20 μL reaction mixture. cDNA was synthesized in a T100 thermocycler (BioRad, United States) by reverse transcription at 50°C for 60 min, and enzyme inactivation at 85°C for 5 min. The qPCR assay was performed in an Insta Q48M real-time PCR (Himedia, India). The 20 μL qPCR mixture included 1X GoTaq qPCR Master Mix (Promega, United States), 300 nM CXR passive reference dye, 0.25 μM of each forward and reverse primer, and 2 μL of template cDNA. The following thermal cycling was followed: initial denaturation at 94°C for 5 min, 40 cycles of 94°C for 40 s, 56°C for 40 s, and 72°C for 40 s. A melting curve analysis was carried out after each reaction to assess the specificity of the amplicons. Three biological and two technical replicates were used in the RT-qPCR. A non-template reaction is used as a negative control. The relative expression of Btk29A and COL3A1 in dsRNA-fed T. palmi was measured following the 2−ΔΔCT method (Livak and Schmittgen, 2001). The statistical analysis was performed and graphs were generated in Microsoft Excel 2019. Expression of T. palmi Btk29A and COL3A1 in BtTOB1 dsRNA-fed thrips served as the negative control.
T. palmi individuals that fed on the artificial diet were identified by a blue tinge in their abdomen and were exclusively considered to assess the effect of dsRNA on survival. The mortality of T. palmi was recorded 24 h post-dsRNA feeding. The whole experiment was replicated nine times for each treatment. The mean mortality in dsRNA-exposed T. palmi was recorded and compared with the thrips fed on the dsRNA-free diet and diet with BtTOB1 dsRNA. Tukey’s test was performed with XLSTAT 2014.5.03 to differentiate means across categories with a 95% confidence interval. The morphological changes, if any, post-dsRNA feeding were also checked under a stereo microscope (M205 FA, Leica, Germany) and captured in a Leica DFC425 C.
The eggs of T. palmi were collected using the artificial oviposition setup reported by Jangra et al. (2020). The eggs were incubated at 28°C on wet tissue paper. The newly hatched first instar larvae (L1) were collected using a Camel hairbrush and allowed to feed on a diet mixed with dsRNA for 2 h as outlined above. T. palmi fed on the dsRNA-free diet and diet with BtTOB1 dsRNA were used as negative controls. The dsRNA-fed and non-fed L1 were allowed to acquire GBNV as reported by Ghosh et al. (2021). In brief, T. palmi L1s were released for a 12-h acquisition access period (AAP) on detached GBNV-infected cowpea leaf at 28 ± 1°C and 60 ± 10% relative humidity. Five live L1s at three biological replicates were collected in a microcentrifuge tube post-GBNV acquisition and used to quantify the virus copies. The GBNV titer acquired by dsRNA-fed and non-fed T. palmi larvae were estimated by absolute quantification by RT-qPCR using the primer pair AG335F-AG336R (Table 1). Total RNA extraction and cDNA synthesis was performed as outlined above. One μg RNA template, 5 μM random primers, 500 μM dNTP mix, 2 μL of 1 X reaction buffer, 10 U FIREScript RT, and 1 U RiboGrip RNase inhibitor were used in 20 μL reaction mixture. The cDNA was synthesized in a T100 thermal cycler with primer annealing at 25°C for 10 min, reverse transcription at 50°C for 60 min and enzyme inactivation at 85°C for 5 min. 10 μL of 1X GoTaq qPCR Master Mix (Promega), 300 nM CRX reference dye, 0.25 μM of each forward and reverse primer, and 2 μL template cDNA were used in 20 μL qPCR reaction mixture. Thermal cycling was performed as initial denaturation at 95°C for 5 min, 35 cycles at 95°C for 25 s, 54°C for 25 s, and 72°C for 30 s. A melting curve analysis was performed to ensure the reaction’s specificity. Three biological and two technical replicates were used for each treatment. A standard curve (y = −3.3251x + 28.141, efficiency = 99.86%) of GBNV using primer pair AG335F-AG336R reported by Priti et al. (2022) was used for the quantification of GBNV copies in T. palmi.
The viral titer was estimated by fitting the mean CT values in the standard curve. The following formula was used to calculate the virus copy number in Microsoft Excel 2019. N = (x × 6.022 × 1023)/ (n × 340 × 109), where N = number of viral copies, x = amount of amplicon in ng, and n = length of linearized plasmid.
One-Way ANOVA was performed to determine the mean differences in virus copies among treatments at p ≤ 0.05 in XLSTAT 2014.5.03. The effect of Btk29A and COL3A1 dsRNAs feeding on survival and mRNA expression at this stage (L1) was also recorded as mentioned above.
T. palmi adults had quadrangular heads with seven segmented antennae. At the top of the head, three brick-red ocelli were present in a triangular formation. A pair of interocellar setae originated outside the ocellar triangle. Besides, the nucleotide sequence of a 595 bp portion of mtCOI was amplified using primer pairs LCO 1490 and HCO 2198 (Folmer et al., 1994) and was 100% identical with T. palmi. The sequence can be retrieved from GenBank with the accession number OP223495.
RT-PCR with GBNV-specific primers produced ~1767 bp desired amplicon from all the sap-inoculated plants. The nucleotide sequence was found to be more than 97% identical to other GBNV isolates. The sequence can be retrieved from GenBank with the accession number MN566913.
A conserved 279 nt-long (1,166 to 1,444) fragment of the Btk29A gene (1.538 nt) and a 257 nt-long (213 to 469 nt) fragment of the COL3A1 gene (7.063 nt) of T. palmi were preferred for dsRNA designing (Figure 1). The dsRNA stretches were unique to T. palmi and showed no homology with other non-target organisms such as Homo sapiens (taxid:9606), mice (taxid:10088), Lepidoptera (taxid:7088), Aves (taxid:8782), Hymenoptera (taxid:7399), Formicidae (taxid:36668), and plants (taxid:3193) in BLASTn analysis. In siRNA Wizard, the 279 nt-long T. palmi Btk29A fragment produced 3 putative siRNAs of 21 nt-long. Similarly, the 257 nt-long COL3A1 fragment predicted 13 putative siRNAs of 21 nt-long (Figure 1). A conserved 167 nt (896–1,062) stretch of B. tabaci TOB1 was used to synthesize BtTOB1 dsRNA and considered as a negative control. The BtTOB1 dsRNA was specific to B. tabaci (Thesnim et al., 2023) and showed no homology to T. palmi.
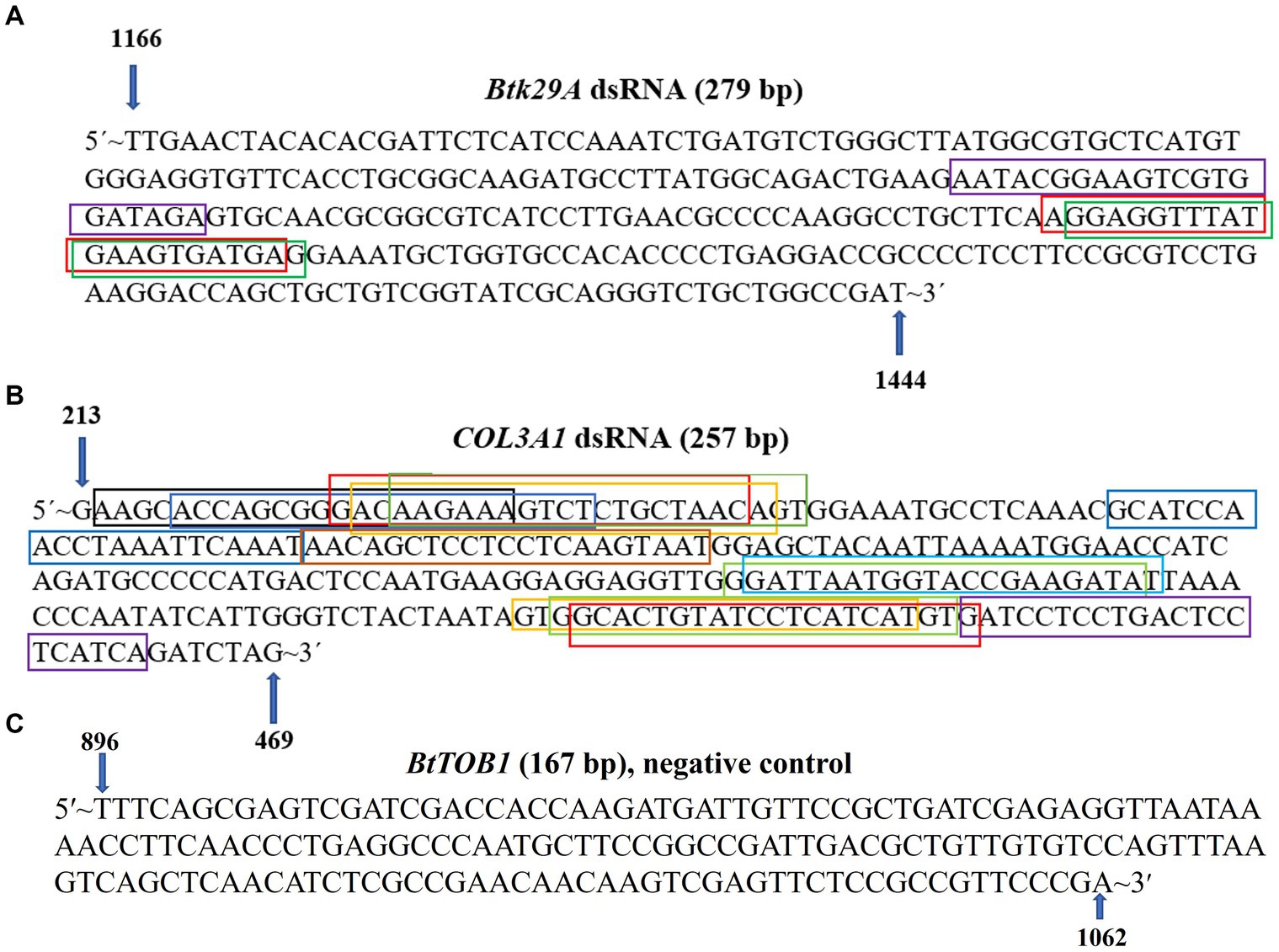
Figure 1. Design of Btk29A and COL3A1 dsRNA. (A) A 279 nt conserved region of Thrips palmi Btk29A was selected for designing Btk29A dsRNA. (B) A 257 nt conserved region of T. palmi COL3A1 was selected for designing COL3A1 dsRNA. The putative siRNAs were marked in different colored boxes. (C) A 167 nt conserved region of Bemisia tabaci TOB1, not specific to T. palmi was used to design the negative control dsRNA.
PCR with primer pair AG507F-AG508R specific to Btk29A yielded an amplicon of 279 bp (accession no. OP346052). Whereas a 257 bp (accession no. OP345816) fragment of COL3A1 was amplified in PCR with the primer pair AG188F-AG189R. On a 2% agarose gel, the Btk29A and COL3A1 dsRNAs purified from the total RNA of recombinant E. coli HT115 cells generated ~279 and ~ 257 bp bands, respectively (Figure 2). The concentration of the purified dsRNA was 621.5 ng/ μL for Btk29A and 531.0 ng/ μL for COL31A. It was 779.9 ng/μL in the case of BtTOB1 dsRNA.

Figure 2. Synthesis and delivery of dsRNA. (A) Purified Btk29A dsRNA (1) and COL3A1 dsRNA (2), BtTOB1 dsRNA (3) used as a negative control, M1, M3: 100 bp plus DNA ladder, M2: 1 kb plus DNA ladder. (B) Adult T. palmi fed on artificial diet for 24 h indicated by blue tinge on abdomen (red arrow).
The expression of T. palmi Btk29A and COL3A1 was downregulated post-dsRNA feeding. The mean logarithmic expression of T. palmi Btk29A was estimated as −5.66 in the larval stage and 0.13 in the adult stage under controlled conditions. It was 2.53 and 0.39 in larval and adult stages, respectively for T. palmi COL3A1. A 3.62-fold downregulation of T. palmi Btk29A mRNA was recorded 24 h post-feeding on a diet containing Btk29A dsRNA in contrast to T. palmi feeding on a diet without dsRNA (Figure 3). Similarly, a 3.15-fold reduction in T. palmi COL3A1 mRNA expression level was recorded in T. palmi fed on COL3A1 dsRNA. Expression of T. palmi Btk29A and COL3A1 post-exposure to BtTOB1 dsRNA was statistically non-significant with untreated control. There was no significant difference in the expression of the endogenous control, β-tubulin, between dsRNA-fed and non-fed T. palmi, indicating that the Btk29A and COL3A1 dsRNAs had a specific action on the target mRNAs. The primer pairs for Btk29A, COL3A1, and β-tubulin produced no secondary peaks in the RT-qPCR melting curve analysis, indicating that the reactions were specific.
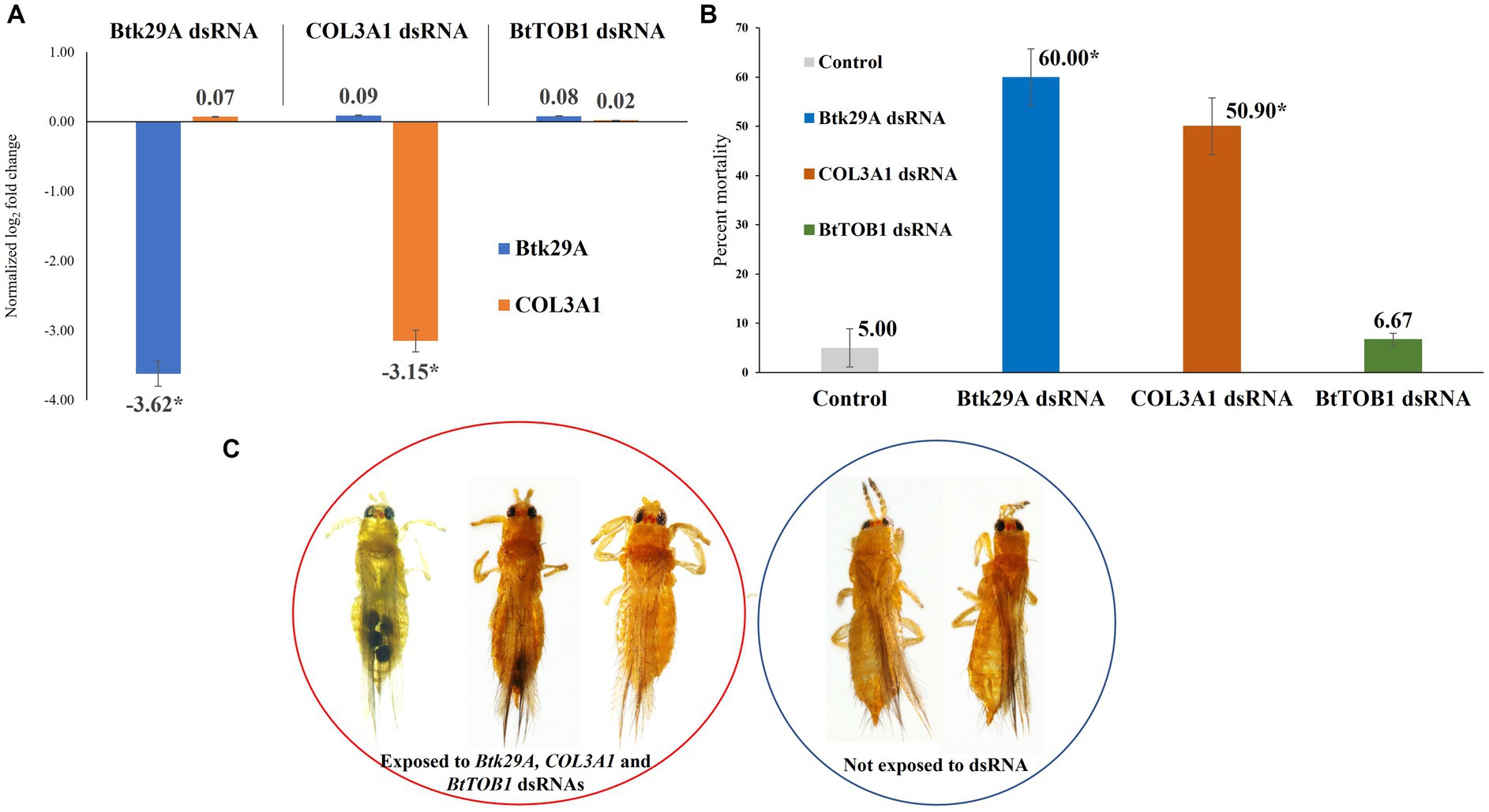
Figure 3. Effect of silencing Btk29A and COL3A1 on survival of T. palmi. (A) Normalized relative expression of T. palmi Btk29A and COL3A1 mRNA post-dsRNA exposure. (B) Percent mortality of adult T. palmi 24 h post-Btk29A and COL3A1 dsRNA feeding at 3 μg/mL. The error bars are the standard error of the mean (SEM). Mean denoted by an asterisk (*) indicates a significant difference (p < 0.05). (C) Btk29A and COL3A1 dsRNA-exposed and unexposed T. palmi. No morphological deformities were recorded post-dsRNA exposure.
T. palmi fitness was significantly affected post-exposure to Btk29A and COL3A1 dsRNAs under controlled laboratory settings. Feeding on Btk29A dsRNA at a concentration of 3 μg/mL for 24 h induced 60% mortality in T. palmi adults (Figure 3). Whereas the mean mortality of COL3A1 dsRNA-fed T. palmi was 50.9% compared to 5% in T. palmi fed on the diet without dsRNA. At a 95% confidence level, the mean mortality for Btk29A and COL3A1 dsRNA treatments was significant, with p-values of 0.024 and 0.013, respectively, compared to the untreated control. There was no discernible difference in mortality between T. palmi treated with BtTOB1 dsRNA and the untreated control. No immediate morphological deformities were recorded in T. palmi adults post-Btk29A and COL3A1 dsRNA exposure (Figure 3).
The GBNV titer in early instar larvae of T. palmi was affected post-silencing Btk29A and COL3A1. Silencing of Btk29A induced a 9.53-fold decrease in the GBNV titer in T. palmi. The mean GBNV copy in T. palmi that were fed on the diet without dsRNA was 2.14 × 109. In contrast, the mean GBNV copy acquired by T. palmi fed on the diet mixed with Btk29A dsRNA was 2.24 × 108. Surprisingly, the silencing of T. palmi COL3A1 led to a slight increase in the mean GBNV copy acquired by T. palmi. The mean GBNV copy in COL3A1 dsRNA-fed T. palmi was 2.47 × 109, i.e., 0.87-fold higher than the untreated control (Figure 4). At this stage, the expression of T. palmi Btk29A and COL3A1 was downregulated by 5.23 and 2.04-fold, respectively, in dsRNA-fed T. palmi larvae as compared to larvae fed on the diet without dsRNA. There was no significant difference in GBNV titer following BtTOB1 dsRNA exposure when compared to the untreated control.
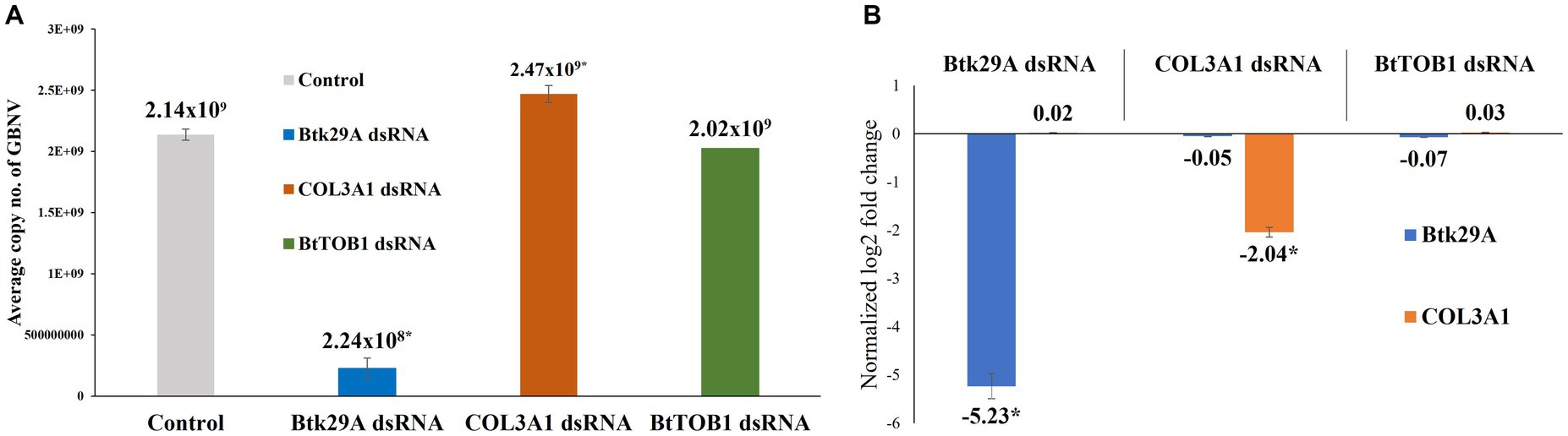
Figure 4. Effect of silencing T. palmi Btk29A and COL3A1 on groundnut bud necrosis orthotospovirus (GBNV) titer in T. palmi. (A) Mean GBNV copy numbers acquired by Btk29A and COL3A1 dsRNA-exposed T. palmi larvae. (B) Expression of T. palmi Btk29A and COL3A1 mRNA in larval stage post-dsRNA exposure. The error bars are the standard error of the mean (SEM). Mean denoted by an asterisk (*) indicates a significant difference (p < 0.05).
RNAi approaches are currently less documented for members of the insect order Thysanoptera, and the function of the thrips genes in different biological and molecular processes remains unclear (Rajesh et al., 2023). The current work is the first to examine the effect of silencing T. palmi genes employing dsRNA. In response to GBNV acquisition, numerous T. palmi transcripts are linked with innate immune response, receptor binding, cell signaling, cellular trafficking, viral replication, and cellular apoptosis (Jagdale and Ghosh, 2019; Mahanta et al., 2022). Response of more than a hundred genes of T. palmi is conserved to GBNV and CaCV infection (Mahanta et al., 2022). The expression of T. palmi Btk29A and COL3A1 were significantly regulated in response to orthotospovirus acquisition (Widana Gamage et al., 2018; Mahanta et al., 2022). In the present study, T. palmi Btk29A and COL3A1 were silenced using exogenous uptake of dsRNA, and the resulting effects were assessed.
DsRNA was administered to T. palmi orally mixed with an artificial diet. In our previous study, a setup was optimized for T. palmi to deliver antisense oligos (Priti et al., 2022). The same setup was used in the current study for the oral delivery of dsRNA molecules. Oral dsRNA delivery has previously been described in T. tabaci and F. occidentalis (Jahani et al., 2018; Singh et al., 2019; Andongma et al., 2020). Different artificial diets such as Luria-Bertani and Tryptone-Soy-Broth and Yeast-based diets were used for delivery of dsRNA in F. occidentalis (Whitten et al., 2016; Andongma et al., 2020). However, these diets were not suitable for T. palmi (Priti et al., 2022). In the current research, an extract of pine pollen and sucrose was used that has been optimized for T. palmi. In this setup, T. palmi thrives for more than 12 days. A blue dye was blended with the diet that was clear through the thrips cuticle thereby confirming the active feeding by thrips. Previously the stability of dsRNA in gut juice and hemolymph was reported in thrips species (Jahani et al., 2018). No degradation of V-ATPase-B dsRNA (1 μg/μl) was reported in F. occidentalis at 0, 1, 2, and 3 days post-oral delivery. Besides oral delivery, leaf-disk, microinjection, and plastid-mediated delivery were reported in F. occidentalis (Badillo-Vargas et al., 2015; Han et al., 2019; Gao et al., 2020; Wu et al., 2022).
In the present study, oral administration of dsRNAs reduced T. palmi Btk29A and COL3A1 mRNA expression levels by 3.62 and 3.15-fold, respectively, at 24 h. In T. palmi, dsRNA targeting B. tabaci TOB1 (nonspecific to T. palmi) was used as a negative control in T. palmi. There was no significant alteration in the expression of T. palmi Btk29A and COL3A1 post-BtTOB1 dsRNA treatment. Further, there was no significant difference in the expression of the endogenous control gene, β-tubulin between dsRNA-exposed and non-exposed T. palmi, indicating that the dsRNA had a specific effect on the target genes. In previous reports, a 25% decrease in the V-ATPase-B mRNA levels was reported in F. occidentalis 3 days post microinjection of V-ATPase-B dsRNA (Badillo-Vargas et al., 2015). Silencing of F. occidentalis TLR6, apoLp, COPE, SAMM50, and hsp11.6 was also achieved using dsRNA (Han et al., 2019; Yuan et al., 2022). In T. tabaci, oral delivery of SNF7 and AQP dsRNAs caused 16.4 and 14.47-fold reductions in respective mRNA levels (Singh et al., 2019). However, the downregulation of T. palmi mRNA in the present study was relatively lower as compared to other thrips species reported previously. The dsRNA concentration, exposure time, target gene expression, delivery method, thrips species, and the defense mechanism of the host might be the reasons behind the variation in the magnitude of silencing (Ramkumar et al., 2021). Silencing of T. palmi Btk29A and COL3A1 negatively influenced had a detrimental impact on T. palmi fitness. Adult T. palmi showed up to 60% mortality 24 h post-dsRNA exposure when compared to T. palmi fed a diet without dsRNA and a diet containing BtTOB1 dsRNA. Btk29A and COL3A1 are involved in a series of molecular and biological processes in T. palmi and depletion of Btk29A and COL3A1 mRNA might interrupt the essential processes for the survival of T. palmi. Increased mortality and decreased fertility were previously reported in V-ATPase-B dsRNA-injected F. occidentalis (Badillo-Vargas et al., 2015). Silencing of α-tubulin also caused high mortality in F. occidentalis larvae (Whitten et al., 2016). High mortality was also reported in adults of F. occidentalis post-silencing of TLR6, apoLp, and COPE (Han et al., 2019). In T. tabaci, up to 72% mortality was reported post-silencing of SNF7 and AQP (Singh et al., 2019). Besides the dsRNA-mediated mRNA depletion, disruption of T. palmi UHRF1BP1 and PFAS expression using modified antisense oligos resulted in 93.33% mortality. Morphological deformities were recorded post-silencing of UHRF1BP1 and PFAS (Priti et al., 2022). However, in the present study, no such morphological deformities were recorded in adults of T. palmi 24 h post-oral delivery. Either the exposure was too short to produce any obvious immediate effects on the structural shape or Btk29A and COL3A1 might not be associated with morphogenesis of T. palmi at all.
Silencing of T. palmi Btk29A and COL3A1 also altered the orthotospovirus titer in T. palmi larvae. Orthotospoviruses are transmitted by T. palmi in a persistent-propagative mode (Ghosh et al., 2017, 2019). It is known that the virus can only be acquired by the early instar larvae, with the emerging adults becoming virulent (Ghosh et al., 2021). Therefore, the orthotospovirus titer in T. palmi was assessed in the larval stage. Silencing of T. palmi Btk29A reduced the virus copies in larval T. palmi by 9.53-fold. Btk29A belongs to the non-receptor tyrosine kinase family that participates in biological processes such as protein tyrosine kinase activity, ATP binding, protein phosphorylation, and intracellular signal transduction (Kumar et al., 2011). In human lung epithelial cells, Btk29A participates in host cell receptor tyrosine kinase signaling to promote influenza virus RNA synthesis, viral ribonucleoprotein (vRNP) nuclear export, and virus release. Other tyrosine kinases have also been found to be associated with virus replication in humans (Kumar et al., 2011). Inhibiting Btk kinase activity induces HIV-1-infected HLM-1 cells to undergo selective apoptosis (Guendel et al., 2015). The decrease of GBNV copies post-silencing of T. palmi Btk29A strongly suggests its involvement in virus replication in insect cells. In contrast, silencing of T. palmi COL3A1 induced a small but significant increase in GBNV copies in larval T. palmi. COL3A1 is associated with the innate immune response against several diseases in humans (Thomas et al., 2007; Kuivaniemi and Tromp, 2019). Collagen is also involved in transfer of viral protein from infected to acceptor cells (Millen et al., 2019). Besides cell adhesion, migration, type III collagen interacts with cell surface receptor integrins (Kim et al., 2005). However, its role in antiviral resistance has never been explored. The depletion of COL3A1 mRNA probably aided the virus to evade the innate immune response which led to an increase in virus copies in T. palmi.
The current study is the first to validate that silencing Btk29A and COL3A1 affects the fitness and orthotospovirus titer in T. palmi. Btk29A and COL3A1 would be novel genetic targets for sustainable thrips management and help elucidate the thrips-orthotospovirus relationships.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Ethical review and approval were not required for the study on animals in accordance with the local legislation and institutional requirements.
VR: Data curation, Formal analysis, Investigation, Validation, Writing – original draft. SJ: Data curation, Formal analysis, Investigation, Software, Writing – original draft. AG: Conceptualization, Funding acquisition, Methodology, Project administration, Resources, Visualization, Writing – review & editing.
The fellowship of VR was supported by the Indian Council of Agricultural Research, New Delhi.
The authors are thankful to the advisories of Vinay Kalia, Debjani Dey, and MS Saharan (IARI). The assistance provided by IARI, New Delhi is thankfully acknowledged.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Adeyinka, O. S., Riaz, S., Toufiq, N., Yousaf, I., Bhatti, M. U., Batcho, A., et al. (2020). Advances in exogenous RNA delivery techniques for RNAi-mediated pest control. Mol. Biol. Rep. 47, 6309–6319. doi: 10.1007/s11033-020-05666-2
Andongma, A. A., Greig, C., Dyson, P. J., Flynn, N., and Whitten, M. M. (2020). Optimization of dietary RNA interference delivery to western flower thrips Frankliniella occidentalis and onion thrips Thrips tabaci. Arch. Insect Biochem. Physiol. 103:e21645. doi: 10.1002/arch.21645
Badillo-Vargas, I. E., Rotenberg, D., Schneweis, B. A., and Whitfield, A. E. (2015). RNA interference tools for the western flower thrips, Frankliniella occidentalis. J. Insect Physiol. 76, 36–46. doi: 10.1016/j.jinsphys.2015.03.009
Bao, W. X., Narai, Y., Nakano, A., Kaneda, T., Murai, T., and Sonoda, S. (2014). Spinosad resistance of melon thrips, Thrips palmi, is conferred by G275E mutation in α6 subunit of nicotinic acetylcholine receptor and cytochrome P450 detoxification. Pestic. Biochem. Physiol. 112, 51–55. doi: 10.1016/j.pestbp.2014.04.013
Bao, W. X., and Sonoda, S. (2012). Resistance to cypermethrin in melon thrips, Thrips palmi (Thysanoptera: Thripidae), is conferred by reduced sensitivity of the sodium channel and CYP450-mediated detoxification. Appl. Entomol. Zool. 47, 443–448. doi: 10.1007/s13355-012-0141-7
Bhatti, J. S. (1980). Species of the genus Thrips from India (Thysanoptera). Syst. Entomol. 5, 109–166. doi: 10.1111/j.1365-3113.1980.tb00404.x
Cannon, R. J. C., Matthews, L., and Collins, D. W. (2007). A review of the pest status and control options for Thrips palmi. Crop Prot. 26, 1089–1098. doi: 10.1016/j.cropro.2006.10.023
Chakraborty, P., and Ghosh, A. (2022). Topical spray of dsRNA induces mortality and inhibits chilli leaf curl virus transmission by Bemisia tabaci Asia II 1. Cells 11:833. doi: 10.3390/cells11050833
Christiaens, O., Niu, J., and Nji Tizi Taning, C. (2020). RNAi in insects: a revolution in fundamental research and pest control applications. Insects 11:415. doi: 10.3390/insects11070415
Cluever, J. D., and Smith, H. A. (2017). A photo-based key of thrips (Thysanoptera) associated with horticultural crops in Florida. Fla. Entomol. 100, 454–467. doi: 10.1653/024.100.0208
Folmer, O., Black, M., Hoeh, W., Lutz, R., and Vrijenhoek, R. (1994). DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 3, 294–299.
Gao, Y., Kim, M. J., Kim, J. H., Jeong, I. H., Clark, J. M., and Lee, S. H. (2020). Transcriptomic identification and characterization of genes responding to sublethal doses of three different insecticides in the western flower thrips, Frankliniella occidentalis. Pestic. Biochem. Physiol. 167:104596. doi: 10.1016/j.pestbp.2020.104596
Ghosh, A., Basavaraj, Y. B., Jangra, S., and Das, A. (2019). Exposure to watermelon bud necrosis virus and groundnut bud necrosis virus alters the life history traits of their vector, Thrips palmi (Thysanoptera: Thripidae). Arch. Virol. 164, 2799–2804. doi: 10.1007/s00705-019-04381-z
Ghosh, A., Dey, D., Mandal, B., and Jain, R. K. (2017). “Thrips as the vectors of tospoviruses in Indian agriculture” in A century of plant virology in India. eds. B. Mandal, G. Rao, V. Baranwal, and R. Jain (Singapore: Springer), 537–561.
Ghosh, A., Mandal, B., and Dietzgen, R. G. (2021). Progression of watermelon bud necrosis virus infection in its vector, Thrips palmi. Cells 10:392. doi: 10.3390/cells10020392
Gnanasekaran, P., Gupta, N., Ponnusamy, K., and Chakraborty, S. (2021). Geminivirus Betasatellite-encoded βC1 protein exhibits novel ATP hydrolysis activity that influences its DNA-binding activity and viral pathogenesis. J. Virol. 95:e0047521. doi: 10.1128/JVI.00475-21
Gnanasekaran, P., Ponnusamy, K., and Chakraborty, S. (2019). A geminivirus betasatellite encoded βC1 protein interacts with PsbP and subverts PsbP-mediated antiviral defence in plants. Mol. Plant Pathol. 20, 943–960. doi: 10.1111/mpp.12804
Guendel, I., Iordanskiy, S., Sampey, G. C., Van Duyne, R., Calvert, V., Petricoin, E., et al. (2015). Role of Bruton’s tyrosine kinase inhibitors in HIV-1-infected cells. J. Neuro-Oncol. 21, 257–275. doi: 10.1007/s13365-015-0323-5
Han, S. H., Kim, J. H., Kim, K., and Lee, S. H. (2019). Selection of lethal genes for ingestion RNA interference against western flower thrips, Frankliniella occidentalis, via leaf disc-mediated dsRNA delivery. Pestic. Biochem. Physiol. 161, 47–53. doi: 10.1016/j.pestbp.2019.07.014
Hough, J., Howard, J. D., Brown, S., Portwood, D. E., Kilby, P. M., and Dickman, M. J. (2022). Strategies for the production of dsRNA biocontrols as alternatives to chemical pesticides. Front. Bioeng. Biotechnol. 10:980592. doi: 10.3389/fbioe.2022.980592
Jagdale, S. S., and Ghosh, A. (2019). In silico analyses of molecular interactions between groundnut bud necrosis virus and its vector, Thrips palmi. VirusDisease 30, 245–251. doi: 10.1007/s13337-019-00521-w
Jahani, M., Christiaens, O., and Smagghe, G. (2018). Analysis of artificial diet to deliver dsRNA in the western flower thrips (Frankliniella occidentalis). IOBC-WPRS Bulletin 131, 41–50.
Jangra, S., Dhall, H., Aggarwal, S., Mandal, B., Jain, R. K., and Ghosh, A. (2020). An observation on the embryonic development in Thrips palmi (Thysanoptera: Thripidae) eggs obtained by an artificial oviposition setup. J. Asia Pac. Entomol. 23, 492–497. doi: 10.1016/j.aspen.2020.03.012
Jangra, S., and Ghosh, A. (2022). Rapid and zero-cost DNA extraction from soft-bodied insects for routine PCR-based applications. PLoS One 17:e0271312. doi: 10.1371/journal.pone.0271312
Kim, J. K., Xu, Y., Xu, X., Keene, D. R., Gurusiddappa, S., Liang, X., et al. (2005). A novel binding site in collagen type III for integrins alpha1beta1 and alpha2beta1. J. Biol. Chem. 280, 32512–32520. doi: 10.1074/jbc.M502431200
Kuivaniemi, H., and Tromp, G. (2019). Type III collagen (COL3A1): gene and protein structure, tissue distribution, and associated diseases. Gene 707, 151–171. doi: 10.1016/j.gene.2019.05.003
Kumar, N., Liang, Y., Parslow, T. G., and Liang, Y. (2011). Receptor tyrosine kinase inhibitors block multiple steps of influenza a virus replication. J. Virol. 85, 2818–2827. doi: 10.1128/JVI.01969-10
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Karny, H. H. (1925). The Thysanoptera found on tobacco in Java and Sumatra. Bulletin van het Deli Proefstation te Medan-Sumatra, 23, 1–55.
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
MacLeod, A., Head, J., and Gaunt, A. (2004). An assessment of the potential economic impact of Thrips palmi on horticulture in England and the significance of a successful eradication campaign. Crop Prot. 23, 601–610. doi: 10.1016/j.cropro.2003.11.010
Mahanta, D. K., Jangra, S., Ghosh, A., Sharma, P. K., Iquebal, M. A., Jaiswal, S., et al. (2022). Groundnut bud necrosis virus modulates the expression of innate immune, endocytosis, and cuticle development-associated genes to circulate and propagate in its vector, Thrips palmi. Front. Microbiol. 13:773238. doi: 10.3389/fmicb.2022.773238
Mamta, B., and Rajam, M. V. (2017). RNAi technology: a new platform for crop pest control. Physiol. Mol. Biol. Plants 23, 487–501. doi: 10.1007/s12298-017-0443-x
Mao, J., and Zeng, F. (2012). Feeding-based RNA Intereference of a gap gene is lethal to the pea aphid, Acyrthosiphon pisum. PLoS One 7:e48718. doi: 10.1371/journal.pone.0048718
Millen, S., Gross, C., Donhauser, N., Mann, M. C., Péloponèse, J. M. Jr., and Thoma-Kress, A. K. (2019). Collagen IV (COL4A1, COL4A2), a component of the viral biofilm, is induced by the HTLV-1 Oncoprotein tax and impacts virus transmission. Front. Microbiol. 10:2439. doi: 10.3389/fmicb.2019.02439
Priti,, Mukherjee, S. K., and Ghosh, A. (2022). Silencing of Thrips palmi UHRF1BP1 and PFAS using antisense Oligos induces mortality and reduces Tospovirus titer in its vector. Pathogens 11:1319. doi: 10.3390/pathogens11111319
Rajesh, V., Rakesh, V., Jangra, S., and Ghosh, A. (2023). RNA interference in thrips vectors: a step forward toward sustainable management. Indian J. Entomol. 1–10. doi: 10.55446/IJE.2023.1113
Ramkumar, G., Asokan, R., Prasannakumar, N. R., Kariyanna, B., Karthi, S., Alwahibi, M. S., et al. (2021). RNA interference suppression of v-ATPase B and juvenile hormone binding protein genes through topically applied dsRNA on Tomato leaves: developing biopesticides to control the south American pinworm, Tuta absoluta (Lepidoptera: Gelechiidae). Front. Physiol. 12. doi: 10.3389/fphys.2021.742871
Reddy, D. V. R., Buiel, A. A. M., Satyanarayana, T., Dwivedi, S. L., Reddy, A. S., Ratna, A. S., et al. (1995). Recent studies on Peanut bud necrosis disease. ICRISAT Asia Centre, 1995:3–7.
Riley, D. G., Joseph, S. V., Srinivasan, R., and Diffie, S. (2011). Thrips vectors of tospoviruses. J. Integrat. Pest Manag. 2, I1–I10. doi: 10.1603/IPM10020
Shang, F., Ding, Y., Ye, C., Yang, L., Chang, Y., Xie, J., et al. (2020). Evaluation of a cuticle protein gene as a potential RNAi target in aphids. Pest Manag. Sci. 76, 134–140. doi: 10.1002/ps.5599
Shi, P., Guo, S. K., Gao, Y. F., Cao, L. J., Gong, Y. J., Chen, J. C., et al. (2020). Variable resistance to spinetoram in populations of Thrips palmi across a small area unconnected to genetic similarity. Evol. Appl. 13, 2234–2245. doi: 10.1111/eva.12996
Shi, P., Guo, S. K., Gao, Y. F., Chen, J. C., Gong, Y. J., Tang, M. Q., et al. (2021). Association between susceptibility of Thrips palmi to spinetoram and frequency of G275E mutation provides basis for molecular quantification of field-evolved resistance. J. Econ. Entomol. 114, 339–347. doi: 10.1093/jee/toaa314
Singh, S., Gupta, M., Pandher, S., Kaur, G., Goel, N., Rathore, P., et al. (2019). RNA sequencing, selection of reference genes and demonstration of feeding RNAi in Thrips tabaci (Lind.) (Thysanoptera: Thripidae). BMC Mol. Biol. 20, 6–21. doi: 10.1186/s12867-019-0123-1
Singh, S. J., and Krishnareddy, M. (1995). Watermelon bud necrosis: a new tospovirus disease. Tospoviruses and Thrips of Floral and Vegetable Crops 431, 68–77.
The European and Mediterranean Plant Protection Organization (EPPO) . (2018). Thrips palmi. Available at: https://onlinelibrary.wiley.com/doi/10.1111/epp.12545 [Accessed June 1, 2018].
Thesnim, P., Jangra, S., Kumar, M., and Ghosh, A. (2023). Effect of silencing Bemisia tabaci TLR3 and TOB1 on fitness and begomovirus transmission. Front. Plant Sci. 14:1136262. doi: 10.3389/fpls.2023.1136262
Thomas, A. H., Edelman, E. R., and Stultz, C. M. (2007). Collagen fragments modulate innate immunity. Exp. Biol. Med. 232, 406–411. doi: 10.3181/00379727-232-2320406
Thomas, J. H., and Wieschaus, E. (2004). src64 and tec29 are required for microfilament contraction during Drosophila cellularization. Development 131, 863–871. doi: 10.1242/dev.00989
Venkata Ramana, C., Venkata Rao, P., Prasada Rao, R. D. V. J., Kumar, S. S., Reddy, I. P., and Reddy, Y. N. (2011). Genetic analysis for Peanut Bud Necrosis Virus (PBNV) resistance in tomato (Lycopersicon Esculentum Mill.). Acta Hortic. 914, 459–463. doi: 10.17660/ActaHortic.2011.914.88
Vogel, E., Santos, D., Mingels, L., Verdonckt, T. W., and Broeck, J. V. (2019). RNA interference in insects: protecting beneficials and controlling pests. Front. Physiol. 9:1912. doi: 10.3389/fphys.2018.01912
Whitten, M. M., Facey, P. D., Del Sol, R., Fernández-Martínez, L. T., Evans, M. C., Mitchell, J. J., et al. (2016). Symbiont-mediated RNA interference in insects. Proc. R. Soc. B Biol. Sci. 283:20160042. doi: 10.1098/rspb.2016.0042
Widana Gamage, S. M. K., Rotenberg, D., Schneweis, D. J., Tsai, C. W., and Dietzgen, R. G. (2018). Transcriptome-wide responses of adult melon thrips (Thrips palmi) associated with capsicum chlorosis virus infection. PLoS One 13:e0208538. doi: 10.1371/journal.pone.0208538
Wu, M., Dong, Y., Zhang, Q., Li, S., Chang, L., Loiacono, F. V., et al. (2022). Efficient control of western flower thrips by plastid-mediated RNA interference. Proc. Natl. Acad. Sci. 119:e2120081119. doi: 10.1073/pnas.2120081119
Yang, X., Liu, S., Lu, W., Du, M., Qiao, Z., Liang, Z., et al. (2022). Delta and jagged are candidate target genes of RNAi biopesticides for the control of Nilaparvata lugens. Front. Bioeng. Biotechnol. 10. doi: 10.3389/fbioe.2022.1023729
Yu, N., Christiaens, O., Liu, J., Niu, J., Cappelle, K., Caccia, S., et al. (2013). Delivery of dsRNA for RNAi in insects: an overview and future directions. Insect Sci. 20, 4–14. doi: 10.1111/j.1744-7917.2012.01534.x
Yuan, J. W., Song, H. X., Chang, Y. W., Yang, F., Xie, H. F., Gong, W. R., et al. (2022). Identification, expression analysis and functional verification of two genes encoding small heat shock proteins in the western flower thrips, Frankliniella occidentalis (Pergande). Int. J. Biol. Macromol. 211, 74–84. doi: 10.1016/j.ijbiomac.2022.05.056
Zhang, T., Liu, L., Jia, Y., Zhi, J., Yue, W., Li, D., et al. (2022a). Induced resistance combined with RNA interference attenuates the Counteradaptation of the Western flower Thrips. Int. J. Mol. Sci. 23:10886. doi: 10.3390/ijms231810886
Zhang, T., Zhi, J. R., Li, D. Y., Liu, L., and Zeng, G. (2022b). Effect of different double-stranded RNA feeding solutions on the RNA interference of V-ATPase-B in Frankliniella occidentalis. Entomol. Exp. Appl. 170, 427–436. doi: 10.1111/eea.13153
Keywords: melon thrips, GBNV, groundnut bud necrosis orthotospovirus, RNAi, dsRNA, virus-vector relationship
Citation: Rajesh V, Jangra S and Ghosh A (2023) Effect of silencing Thrips palmi Btk29A and COL3A1 on fitness and virus acquisition. Front. Microbiol. 14:1254246. doi: 10.3389/fmicb.2023.1254246
Received: 06 July 2023; Accepted: 29 September 2023;
Published: 19 October 2023.
Edited by:
Ying Wen Huang, National Chung Hsing University, TaiwanReviewed by:
Ved Prakash, Kansas State University, United StatesCopyright © 2023 Rajesh, Jangra and Ghosh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Amalendu Ghosh, YW1hbDRlbnRvQGdtYWlsLmNvbQ==
†Present address: Sumit Jangra,Tropical Research and Education Center, Institute of Food and Agricultural Sciences, University of Florida, Homestead, FL, United States
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.