- 1Facultad de Medicina y Facultad de Ciencias de la Vida, Instituto de Ciencias Biomédicas, Universidad Andrés Bello, Santiago, Chile
- 2Laboratorio de Microbiología, Departamento de Bioquímica y Biología Molecular, Facultad de Ciencias Químicas y Farmacéuticas, Universidad de Chile, Santiago, Chile
- 3Facultad de Medicina Veterinaria y Agronomía, Universidad de Las Américas, Santiago, Chile
- 4Núcleo de Investigaciones Aplicadas en Ciencias Veterinarias y Agronómicas, Facultad de Medicina Veterinaria y Agronomía, Universidad de Las Américas, Santiago, Chile
- 5Departamento de Ciencias Químicas y Biológicas, Universidad Bernardo O'Higgins, Santiago, Chile
The type VI secretion system (T6SS) is a contact-dependent contractile multiprotein apparatus widely distributed in Gram-negative bacteria. These systems can deliver different effector proteins into target bacterial and/or eukaryotic cells, contributing to the environmental fitness and virulence of many bacterial pathogens. Salmonella harbors five different T6SSs encoded in different genomic islands. The T6SS encoded in Salmonella Pathogenicity Island 6 (SPI-6) contributes to Salmonella competition with the host microbiota and its interaction with infected host cells. Despite its relevance, information regarding the total number of effector proteins encoded within SPI-6 and its distribution among different Salmonella enterica serotypes is limited. In this work, we performed bioinformatic and comparative genomics analyses of the SPI-6 T6SS gene cluster to expand our knowledge regarding the T6SS effector repertoire and the global distribution of these effectors in Salmonella. The analysis of a curated dataset of 60 Salmonella enterica genomes from the Secret6 database revealed the presence of 23 new putative T6SS effector/immunity protein (E/I) modules. These effectors were concentrated in the variable regions 1 to 3 (VR1-3) of the SPI-6 T6SS gene cluster. VR1-2 were enriched in candidate effectors with predicted peptidoglycan hydrolase activity, while VR3 was enriched in candidate effectors of the Rhs family with C-terminal extensions with predicted DNase, RNase, deaminase, or ADP-ribosyltransferase activity. A global analysis of known and candidate effector proteins in Salmonella enterica genomes from the NCBI database revealed that T6SS effector proteins are differentially distributed among Salmonella serotypes. While some effectors are present in over 200 serotypes, others are found in less than a dozen. A hierarchical clustering analysis identified Salmonella serotypes with distinct profiles of T6SS effectors and candidate effectors, highlighting the diversity of T6SS effector repertoires in Salmonella enterica. The existence of different repertoires of effector proteins suggests that different effector protein combinations may have a differential impact on the environmental fitness and pathogenic potential of these strains.
Introduction
The type VI secretion system (T6SS) is a multiprotein nanomachine composed of 13 structural components and various accessory proteins that deliver protein effectors into target cells through a contractile mechanism (Cherrak et al., 2019; Coulthurst, 2019). The T6SS needle, composed of an inner tube (made of a stack of Hcp hexamer rings) and comprising a trimer of VgrG and a PAAR protein, is wrapped into a contractile sheath formed by the polymerization of TssB/TssC subunits. These are assembled into an extended, metastable conformation (Silverman et al., 2013; Cherrak et al., 2019). Contraction of the sheath upon contact with a target cell or sensing cell envelope damage propels the needle toward the target cell (Brackmann et al., 2017). T6SS effector proteins are classified as either cargo or specialized effectors. Cargo effectors are delivered by non-covalent interaction with some core components (Coulthurst, 2019), while specialized effectors are additional domains of either VgrG, Hcp, or PAAR proteins (Durand et al., 2014; Whitney et al., 2014; Diniz and Coulthurst, 2015; Ma et al., 2017; Pissaridou et al., 2018).
The extensive repertoire of effector proteins makes the T6SS a highly versatile machine that can target prokaryotic or eukaryotic cells (Coulthurst, 2019; Monjarás Feria and Valvano, 2020). Among the antibacterial effector proteins, some target the peptidic or glycosidic bonds of the peptidoglycan (Ma and Mekalanos, 2010; Russell et al., 2012; Srikannathasan et al., 2013; Whitney et al., 2013; Berni et al., 2019; Wood et al., 2019), or the FtsZ cell division ring (Ting et al., 2018). These antibacterial effectors are encoded in bi-cistronic elements with immunity proteins (E/I pairs) that bind tightly and specifically to their cognate effector preventing self-intoxication and killing of sibling cells (Russell et al., 2012). Other T6SS effectors are eukaryote-specific, such as those targeting the actin or microtubule cytoskeleton networks (Monjarás Feria and Valvano, 2020), and others (known as trans-kingdom effectors) can target both bacterial and eukaryotic cells (Jiang et al., 2014). These effectors include those targeting conserved molecules (NAD+ and NADP+) and macromolecules (DNA, phospholipids) or forming pores in membranes (Whitney et al., 2015; Tang et al., 2018; Ahmad et al., 2019).
Many enteric pathogens (e.g., Salmonella, Shigella, and Vibrio) use the T6SS to colonize the intestinal tract of infected hosts (Sana et al., 2016; Chassaing and Cascales, 2018), while some strains of the gut commensal Bacteroides fragilis use their T6SSs only for competition against other Bacteroidales species (Coyne and Comstock, 2019). The T6SS is, therefore, a key player in bacterial warfare.
The Salmonella genus includes more than 2,600 serotypes distributed between species S. enterica and S. bongori (Issenhuth-Jeanjean et al., 2014), which differ in clinical signs and host range (Uzzau et al., 2000). Serotypes are defined based on variations in the somatic, flagellar and capsular antigens, according to the Kauffmann-White-Le Minor serotyping scheme (Grimont and Weill, 2007; Issenhuth-Jeanjean et al., 2014). Worldwide, Salmonella infections are responsible for 95.1 million cases of gastroenteritis per year (GBD 2017 Non-Typhoidal Salmonella Invasive Disease Collaborators, 2019). In addition, the World Health Organization (WHO) has also included Salmonella as a high-priority pathogen due to the emergence of strains with high levels of fluoroquinolone resistance (GBD 2017 Non-Typhoidal Salmonella Invasive Disease Collaborators, 2019). In Salmonella, 5 T6SS gene clusters have been identified within Salmonella Pathogenicity Islands (SPIs) SPI-6, SPI-19, SPI-20, SPI-21, and SPI-22 (Blondel et al., 2009; Fookes et al., 2011). These T6SSs are distributed in 4 different evolutionary lineages: T6SSSPI-6 belongs to subtype i3, T6SSSPI-19 to subtype i1, T6SSSPI-22 to subtype i4a, and both T6SSSPI-20 and T6SSSPI-21 belong to subtype i2 (Bao et al., 2019). Besides their distinct evolutionary origin, these five T6SS gene clusters are differentially distributed among distinct serotypes, subspecies, and species of Salmonella (Blondel et al., 2009).
Notably, most of these T6SSs have been shown to contribute to the virulence and pathogenesis of different Salmonella serotypes (Blondel et al., 2010; Mulder et al., 2012; Pezoa et al., 2013, 2014; Sana et al., 2016; Xian et al., 2020; Hespanhol et al., 2022; Sibinelli-Sousa et al., 2022). One of the most studied and widely distributed T6SS corresponds to that encoded in SPI-6. Depending on the serotype, the SPI-6 T6SS gene cluster comprises a region of ~35 to 50 kb encoding ~30 to 45 ORFs, including each of the 13 T6SS core components. The genetic architecture of the SPI-6 T6SS gene cluster is highly conserved among serotypes; nonetheless, there are structural differences restricted to three variable regions of the island (herein referred to as VR1, VR2, and VR3, Figure 1) (Blondel et al., 2009). In S. Typhimurium and S. Dublin, 9 SPI-6 T6SS effector proteins have been described to date (Russell et al., 2012; Benz et al., 2013; Whitney et al., 2013; Koskiniemi et al., 2014; Sana et al., 2016; Sibinelli-Sousa et al., 2020; Amaya et al., 2022; Jurėnas et al., 2022; Lorente-Cobo et al., 2022), most of which are encoded within these variable regions (Figure 1; Table 1).

Figure 1. Schematic representation of selected SPI-6 T6SS gene clusters. The figure shows an alignment of the SPI-6 T6SS gene cluster of S. Dublin CT_02021853, S. Typhi CT18 and S. Typhimurium 14028s. The location of variable regions 1–3 is shown. ORFs encoding previously described T6SS effectors and cognate immunity proteins are shown in red and green, respectively. ORFs encoding T6SS core components are shown in blue. Grayscale represents the percentage of identity between nucleotide sequences.

Table 1. T6SS effectors and cognate immunity proteins encoded in SPI-6 previously identified in Salmonella enterica.
The VR1 is located downstream of gene tssC and encodes the E/I modules Tae2/Tai2 and Tae4/Tai4. Tae2 and Tae4 are peptidoglycan hydrolases able to cleave the DD-crosslinks between D-mDAP and D-alanine or the covalent link between D-Glu and mDAP of the tetrapeptide stem, respectively, thus contributing to interbacterial competition and mice colonization (Russell et al., 2012; Sana et al., 2016). VR2 is located downstream of gene tssM and encodes many proteins of unknown function and two E/I modules with peptidoglycan hydrolase activity: Tge2/Tgi2P is predicted to have N-acetylglucosaminidase activity (Whitney et al., 2013), while Tlde1/Tldi shows L,D carboxypeptidase activity against the peptide stems of the peptidoglycan layer (Sibinelli-Sousa et al., 2020; Lorente-Cobo et al., 2022). Finally, the VR3 is located downstream of gene tssI and encodes a variable number of Rhs elements, some of them harboring endonuclease domains such as HNHc (DNase) and Ntox47 (RNase), and an ART domain (ADP-ribosyltransferase) linked to the C-terminal of these Rhs proteins (Koskiniemi et al., 2014; Amaya et al., 2022; Jurėnas et al., 2022).
Most of our knowledge regarding the presence and distribution of SPI-6 T6SS effector proteins comes from studies using reference strains of a limited number of serotypes (e.g., S. Typhimurium and S. Dublin) (Russell et al., 2012; Benz et al., 2013; Whitney et al., 2013; Koskiniemi et al., 2014; Sana et al., 2016; Sibinelli-Sousa et al., 2020; Amaya et al., 2022; Jurėnas et al., 2022; Lorente-Cobo et al., 2022). In this study, we performed a bioinformatic prediction analysis searching for putative T6SS effectors in a dataset of 60 genomes covering 37 S. enterica serotypes retrieved from the curated Secret6 database. Our analysis identified 23 new putative antibacterial effectors encoded in E/I modules within the 3 VRs of the SPI-6 T6SS gene cluster. These candidates include 5 effectors with putative peptidoglycan hydrolase activity, 16 effectors with potential nuclease activity and 2 effectors targeting the bacterial translation machinery. Finally, we expanded our analysis to include all available Salmonella genomes deposited in the NCBI database and determined the global distribution of these new putative effectors. A hierarchical clustering analysis identified that some effectors are conserved in most Salmonella serotypes. In contrast, most other effectors are differentially distributed in different serotypes. The presence of different sets of T6SS effectors suggests that distinct repertoires of these proteins may have a differential impact on the pathogenicity and environmental adaptation of Salmonella serotypes.
Materials and methods
Identification of candidate SPI-6 T6SS effectors
First, we searched the Secret6 database1 for Salmonella genomes encoding the minimal 13 core components of a T6SS and identified a total of 60 genomes that met this requirement. Then, to identify putative T6SS effectors encoded within SPI-6 of Salmonella, each ORF of this island was analyzed with the Bastion6 pipeline (Wang et al., 2018) excluding the 13 T6SS core components. ORFs presenting a Bastion6 score ≥ 0.7 were considered as candidate T6SS effectors. Each Bastion6 prediction was further analyzed with tools implemented in the Operon-Mapper web server (Taboada et al., 2018) to determine if it was likely part of a bi-cistronic unit also encoding a putative immunity protein [i.e., a small protein with potential signal peptides (SignalP 6.0) and/or transmembrane domains (TMHMM 2.0)]. Conserved functional domains and motifs in the candidate T6SS effectors were identified using the PROSITE, NCBI-CDD, Motif-finder, and Pfam databases (Kanehisa et al., 2002; Sigrist et al., 2013; Finn et al., 2014; Lu et al., 2019) implemented in the GenomeNet2 search engine. An e-value cutoff score of 0.01 was used. Finally, a biochemical functional prediction for each putative effector and immunity protein identified was performed by HMM homology searches using the HHpred HMM-HMM comparison tool (Zimmermann et al., 2017). It is worth mentioning that most genes (ORFs) identified do not have formal names, making extremely difficult referring to them using conventional genetic nomenclature. Thus, in figures and tables we will refer to ORFs encoding effectors and immunity proteins according to the corresponding protein name (in the case of those previously reported in the literature) or the functional domains present in the predicted proteins (in the case of ORFs encoding new candidate effectors and immunity proteins).
Hierarchical clustering analysis of the new SPI-6 T6SS effectors
For hierarchical clustering analysis, a presence/absence matrix of each T6SS effector and candidate effector was constructed for each bacterial genome by means of BLASTn analyses and manual curation of the data. A 90% identity and 90% sequence coverage threshold was used to select positive matches. The matrix generated was uploaded as a csv file to the online server MORPHEUS2 using default parameters (i.e., one minus Pearson’s correlation, average linkage method).
Salmonella 16S rDNA phylogenetic analyses
The 16S rDNA sequences were obtained from the 60 Salmonella genomes previously analyzed. The sequences were concatenated and aligned with ClustalW using the Molecular Evolutionary Genetics Analysis (MEGA) software version 7.0 (Kumar et al., 2016). A phylogenetic tree was built from the alignments obtained from MEGA by performing a bootstrap test of phylogeny (1,000 replications) using the maximum-likelihood method with a Jones-Taylor-Thornton correction model.
Sequence and phylogenetic analyses
The DNA sequence encoding each T6SS effector identified in this study was subjected to BLASTn analyses to find orthologs in all Salmonella genome sequences deposited in the NCBI database (October 2022). For selection of positive matches, a 90% identity and 90% sequence coverage threshold was used. Conservation of sequences was determined by multiple sequence alignments using T-Coffee Expresso (Notredame et al., 2000), MAFFT (Katoh et al., 2017), and ESPript 3 (Robert and Gouet, 2014). Comparative genomic analysis of SPI-6 T6SS gene clusters was performed using Mauve (Darling et al., 2004) and EasyFig v2.2.5 (Sullivan et al., 2011). Nucleotide sequences were analyzed using Artemis version 18 (Rutherford et al., 2000).
Results
Analysis of a curated dataset of Salmonella genomes reveals 23 new putative E/I modules encoded within the SPI-6 T6SS gene cluster
To identify new T6SS effectors with high confidence, we first screened the SPI-6 T6SS gene clusters of a dataset of 60 Salmonella enterica genomes from the Secret6 curated database (Zhang et al., 2023). This database includes 60 strains covering 37 Salmonella serotypes (Supplementary Table S1). Each ORF within SPI-6 T6SS gene clusters was analyzed based on four criteria: (i) identification of candidate effectors through Bastion6 analysis (a bioinformatic tool that predicts T6SS effectors based on amino acid sequence, evolutionary information, and physicochemical properties); (ii) identification of putative immunity proteins by detection of signal peptides (SignalP 6.0), transmembrane domains (TMHMM 2.0) and operon prediction (Operon-mapper; Taboada et al., 2018); (iii) identification of conserved functional domains associated with bona fide T6SS effectors (INTERPROSCAN, PROSITE, NCBI-CDD, MOTIF, and Pfam) and (iv) functional biochemical prediction using the HHpred HMM-HMM server. In addition, we analyzed these gene clusters to identify potential unannotated ORFs which could encode putative effectors and cognate immunity proteins.
Our analysis identified 23 new putative effector proteins and cognate immunity proteins (Table 2). These candidates included both cargo and specialized effector proteins with diverse predicted biochemical functions, including peptidoglycan hydrolases (5), DNases (8), RNases (6), deaminases (1), ADP-ribosyltransferases (2) and hybrid DNases/RNases (1) (Table 2). In addition, our analysis showed that the repertoire of E/I modules in SPI-6 vary considerably between closely related strains (Figure 2). Of note, comparative genomic analyses revealed that each identified E/I module is encoded within one of the 3 VRs previously described (Blondel et al., 2009). One E/I module is encoded within VR1, four within VR2, and 18 are encoded within VR3 (Figure 3).

Table 2. New putative T6SS effectors and cognate immunity proteins encoded in SPI-6 of Salmonella enterica.
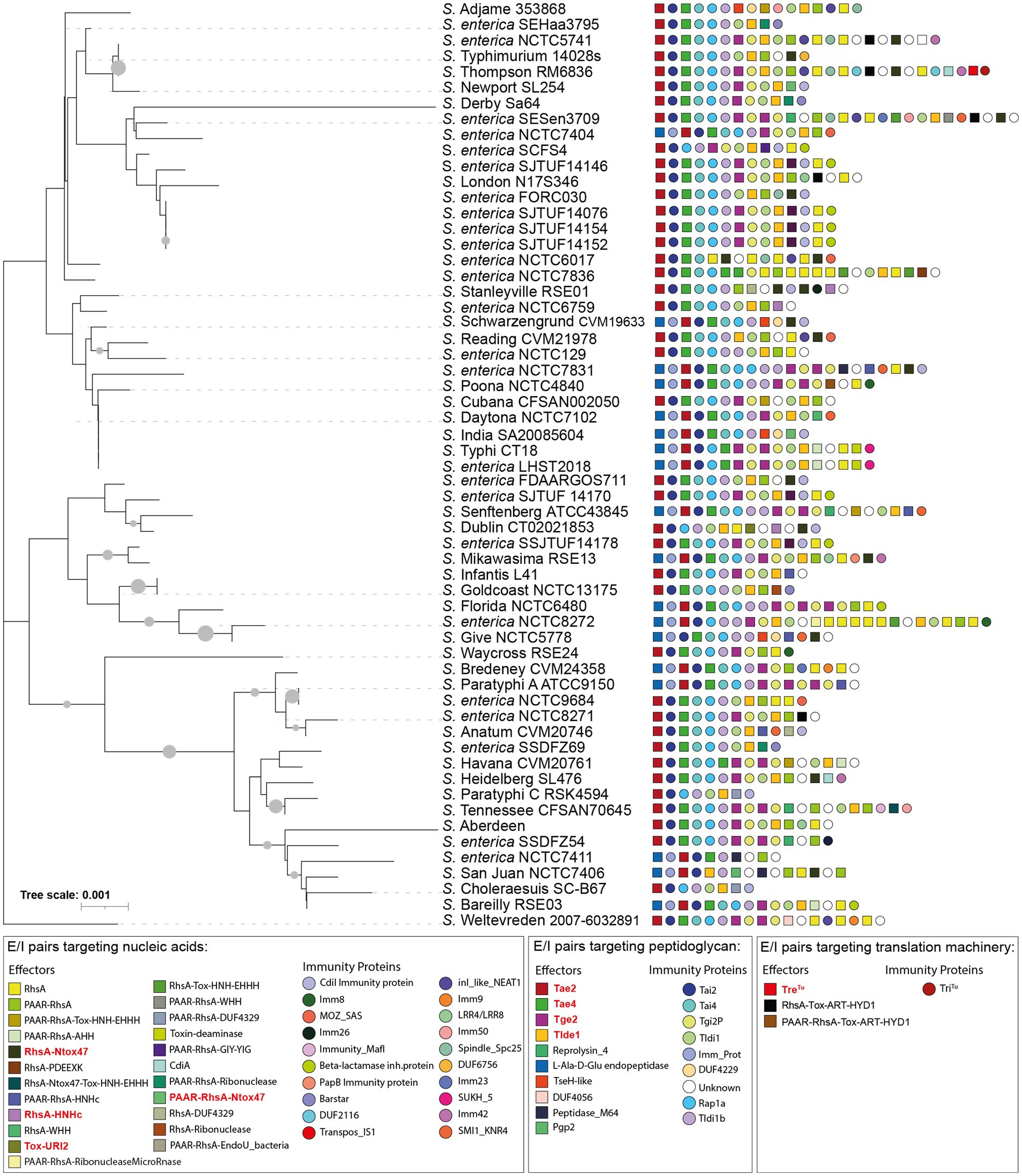
Figure 2. 16S rDNA phylogeny and T6SS E/I module composition of Salmonella enterica SPI-6. Concatenated 16S rDNA nucleotide sequences from 60 Salmonella genomes deposited in Secret6 database were aligned with ClustalW using MEGA version 7.0. Next, a maximum-likelihood phylogenetic tree was built from the alignment using a bootstrap test of phylogeny (1,000 replications) with a Jones-Taylor-Thornton correction model. In the figure, we refer to ORFs encoding effectors and immunity proteins according to the corresponding protein name (in the case of those previously reported in the literature) or the functional domains present in the predicted proteins (in the case of ORFs encoding new candidate effectors and immunity proteins). Squares and circles next to each strain name correspond to ORFs encoding an effector or an immunity protein, respectively. Different colors represent confirmed or predicted functions, as indicated in the figure.
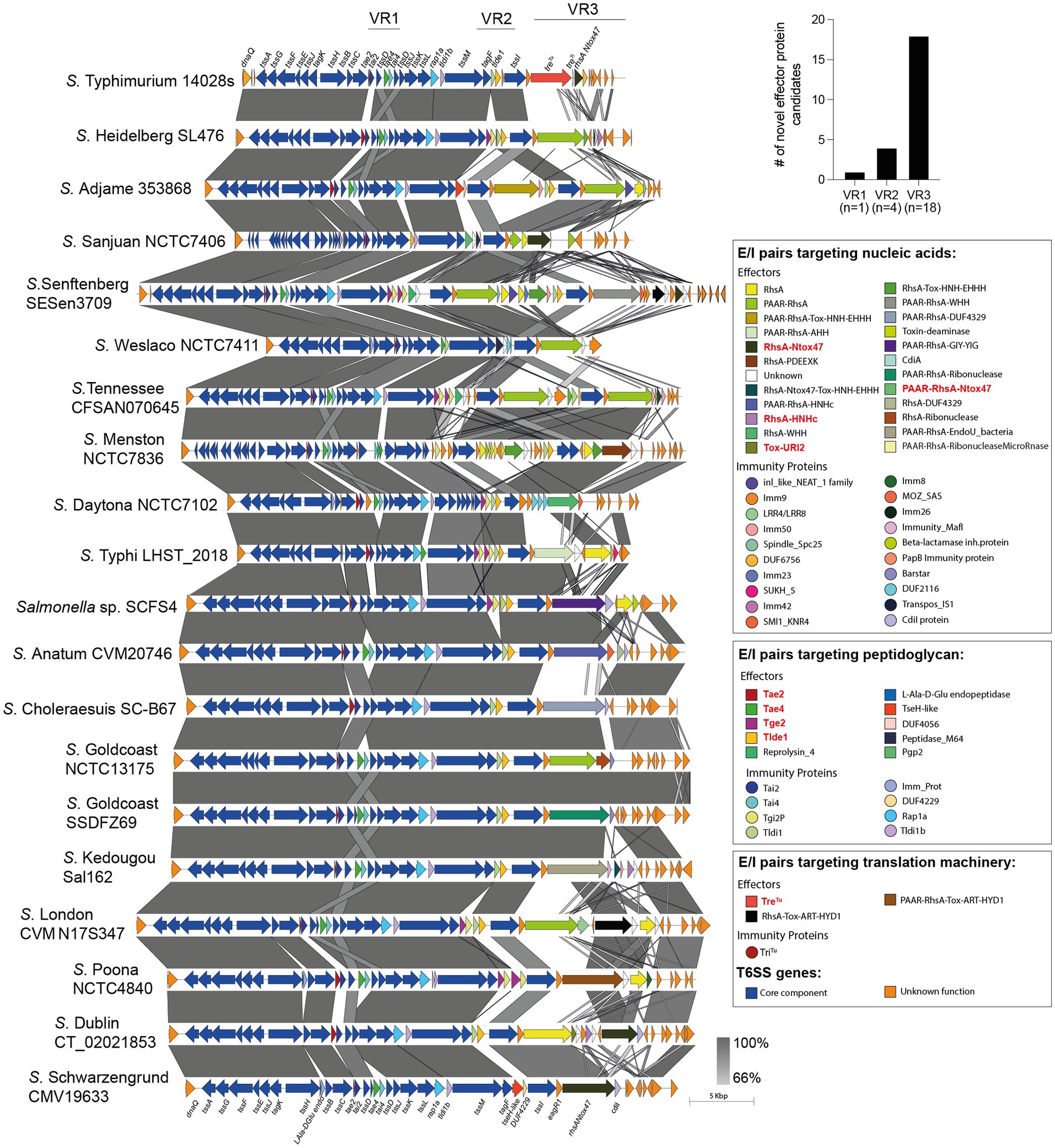
Figure 3. Comparative genomic analysis of SPI-6 T6SS gene clusters in representative Salmonella serotypes reveals new effector encoding genes. The location of variable regions 1–3 is shown. In the figure, we refer to ORFs encoding effectors and immunity proteins according to the corresponding protein name (in the case of those previously reported in the literature) or the functional domains present in the predicted proteins (in the case of ORFs encoding new candidate effectors and immunity proteins). ORFs encoding T6SS core components are shown in blue. ORFs encoding E/I modules are presented in different colors according to the confirmed or predicted functions, as indicated in the figure. Grayscale represents the percentage of identity between nucleotide sequences. Previously described Salmonella T6SS effectors are highlighted in red.
Putative T6SS cargo effectors with predicted peptidoglycan hydrolase activity are confined to VR1 and VR2
Our bioinformatic analysis identified 5 predicted T6SS cargo effectors with putative peptidoglycan hydrolase activity (Table 2; Figure 4). One effector corresponds to an unannotated ORF encoded within VR1. This ORF was identified in 30% (18/60) of the genomes analyzed, is located between genes tssH and tssB (ELZ70_17805 and ELZ70_17795 ORFs in S. Bareilly strain RSE03) and is predicted to encode a 32 amino acids protein with a putative L-Ala-D-Glu-endopeptidase protein domain (Figure 4). This ORF is predicted to be co-transcribed with a downstream unannotated ORF that encodes a 146 amino acids protein with a periplasmic-targeting signal peptide (Table 2), suggesting that this latter ORF encodes the cognate immunity protein of the new candidate effector.
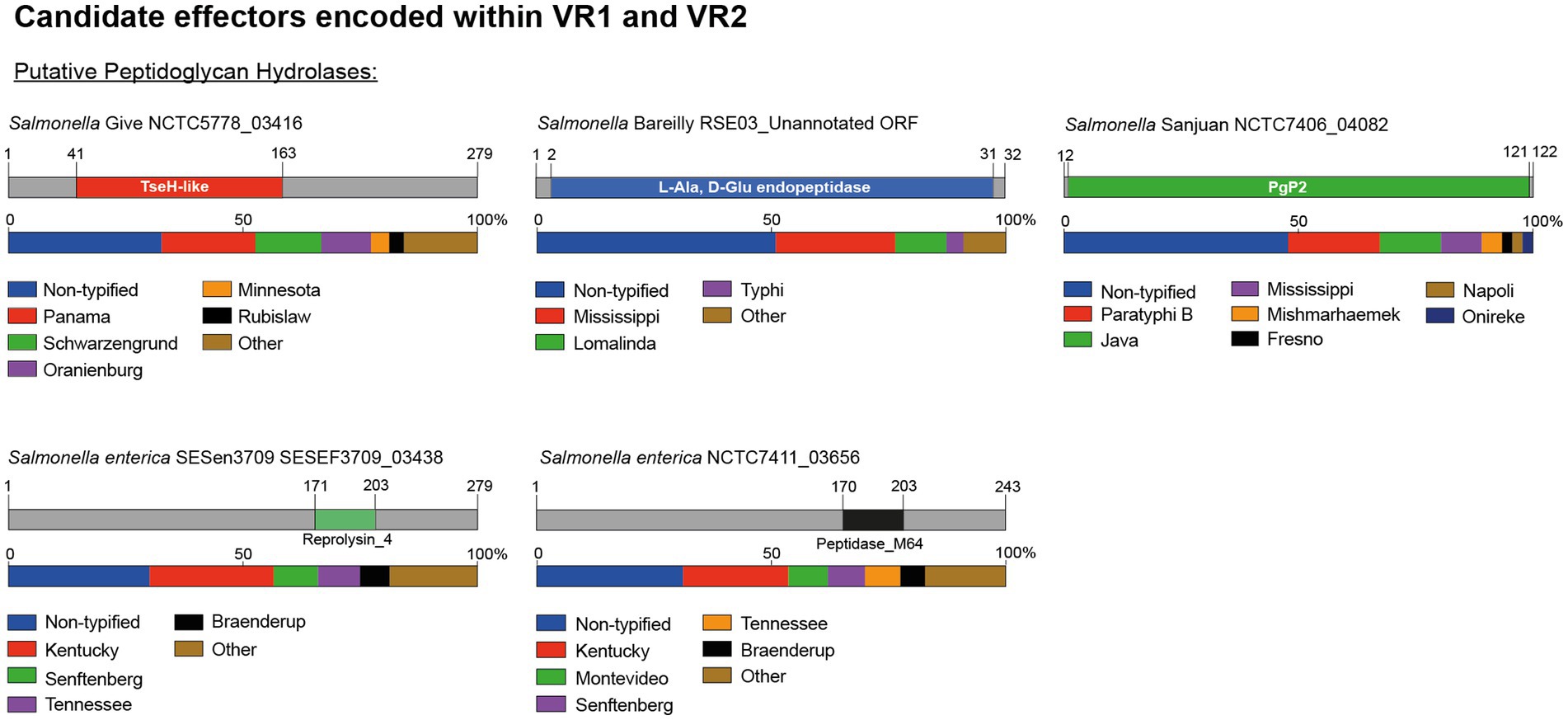
Figure 4. The variable regions 1 and 2 of the SPI-6 T6SS gene cluster encode 5 new putative effectors. Schematic representation and distribution of new putative effectors among Salmonella genomes. Predicted functional domains are show in different colors. Homologs for each candidate effector were identified by BLASTn analyses, as described in Materials and Methods.
In addition, our analysis identified four putative E/I modules encoded in VR2. The first putative effector (NCTC7406_04082 in S. Sanjuan strain NCTC7406) is a 122 amino acid protein that harbors a predicted PgP2 protein domain with putative L,D transpeptidase activity (Table 2; Figure 4). NCTC7406_04082 is part of a bi-cistronic unit with NCTC7406_04081. This latter ORF encodes a 147 amino acid protein with a signal peptide targeting the periplasmic space that may correspond to its cognate immunity protein (Table 2). The second VR2 candidate effector (G9X22_18260 in S. Adjame strain 353868) is a 279 amino acids protein that harbors a putative amidase domain similar to the NlpC/P60 endopeptidase domain of the TseH T6SS effector of Vibrio cholerae (Altindis et al., 2015). This candidate effector is also encoded next to a putative immunity protein of 86 amino acids harboring a DUF4229 protein domain and 2 transmembrane helices that may target this protein to the periplasmic space (Table 2).
The third candidate effector (SESEF3709_03438 in S. enterica strain SESen3709) is a 243 amino acids protein that harbors a Reprolysin_4 domain with putative amidase activity (Figure 4). SESEF3709_03438 is predicted to be part of a bi-cistronic unit with SESEF3709_03437, that encodes a putative cognate immunity protein with a signal peptide for periplasmic targeting.
The final candidate effector of VR2 corresponds to a 243 amino acid protein with a predicted M64 peptidase domain (NCTC7411_03656 in S. enterica strain NCTC7411) (Table 2; Figure 4). Our analysis also revealed that NCTC7411_03656 is likely to be part of bi-cistronic unit with their respective putative immunity protein gene (NCTC7411_03655 in S. enterica strain NCTC7411) (Table 2). In other serotypes, the putative immunity protein gene encodes a protein of 84–144 amino acids harboring a transmembrane domain that targets this protein to the periplasmic space (Table 2).
Putative T6SS specialized effectors with polymorphic nuclease and ADP-ribosyltransferase toxin domains associated to Rhs proteins are restricted to the VR3
Our analysis revealed the presence of 18 candidate effectors encoded within the VR3 of SPI-6, including 16 in the Rhs family of proteins, 1 RNase and 1 deaminase. The size of the Rhs proteins ranged from 500 to 1,500 amino acids harboring different nuclease and ADP-ribosyltransferases domains (Table 2; Figure 5).

Figure 5. The variable region 3 of the SPI-6 T6SS gene cluster encodes 18 new putative effectors. Schematic representation and distribution of new putative effectors among Salmonella genomes. Predicted functional domains are shown in different colors. Homologs for each candidate effector were identified by BLASTn analyses, as described in Materials and Methods.
Eight of the 16 Rhs proteins harbored distinct C-terminal DNase domains, including domains of the HNH/ENDO VII superfamily of nucleases (IPR028048) such as WHH (IPR032869), Tox-HNH-EHH5H (IPR028048) or AHH (IPR032871), and nuclease domains of the GIY-YIG (IPR000305) and PDEEXK (IPR009362) families (Figure 5). In addition, 4 of these 8 candidates also harbored N-terminal PAAR motifs (IPR008727) (Figure 5). The presence of PAAR motifs suggests that these candidates correspond to specialized effector proteins. Each of these candidates were also predicted to be encoded in bi-cistronic units with ORFs encoding their respective immunity protein. Several of these proteins harbored domains previously found in cognate immunity proteins of bacterial toxin systems such as Imm50 (IPR028957), SMI1_KNR4 (PF09346) and CdI (IPR041256), among others (Table 2).
Our bioinformatics analyses also predicted 5 Rhs effectors with C-terminal extensions harboring different RNase protein domains (Table 2; Figure 5). These include Rhs proteins with Guanine-specific ribonuclease N1/T1/U2 (IPR000026), EndoU (IPR029501), and DUF4329 (IPR025479) domains. In addition, three of these proteins also harbored N-terminal PAAR motifs (IPR008727). The gene encoding each of these proteins was also predicted to be co-transcribed with genes encoding putative immunity proteins (Table 2). Remarkably, our analysis also identified a hybrid Rhs effector with predicted C-terminal RNase (Ntox47 domain) and DNase (Tox-HNH-EHHH) domains (CS349_18795 in S. Tennessee strain CFSAN070645). The gene encoding this protein is also predicted to be part of bi-cistronic unit with an ORF encoding a 129 amino acid protein with an Imm50 (IPR028957) domain. We also identified two putative Rhs effectors with a TOX-ART-HYD1 (pfam15633) ADP-ribosyltransferase domain, one of which also includes an N-terminal PAAR motif (NCTC4840_03667 in S. Poona strain NCTC4840). This protein shares 32% identity with STM0291, a recently described Rhs effector with an ART protein domain of S. Typhimurium named TreTu (type VI ribosyltranferase effector targeting EF-Tu; Jurėnas et al., 2022). The low percentage of sequence identity (Supplementary Figure S1) suggests that this could be a divergent STM0291 homolog.
Finally, in VR3 we identified a putative effector with the CdiA RNase domain (IPR041620) not associated to Rhs elements (SEHA_RS26915 in S. Heidelberg SL476) (Table 2; Figure 5). In addition, we also identified a candidate effector harboring potential adenosine deaminase activity (STY_RS01505 in S. Typhi CT18). This effector is a small 86 amino acid protein with a TOX-deaminase domain of the BURPS668_1122 family (IPR032721) found in polymorphic toxin systems (Table 2; Figure 5). The gene encoding this effector is predicted to be co-transcribed with an ORF encoding a putative immunity protein with a SUKH_5 (PF14567) domain (Table 2; Figure 5).
Genome-wide analysis of the distribution of SPI-6 T6SS effectors and candidate effectors in Salmonella
Identifying new putative T6SS effectors encoded within VR1-3 of SPI-6 encouraged us to determine the presence and distribution of the genes encoding these proteins across Salmonella enterica. The nucleotide sequence corresponding to each effector and candidate effector was used in BLASTn searches examining publicly available Salmonella enterica genome sequences deposited in the NCBI database, and the distribution of each effector protein was determined (Supplementary Table S2).
The analysis of the 9 T6SS effector proteins previously reported in the literature (i.e., Tae2, Tae4, Tge2, Tlde1, RhsA-HNHc, RhsA-Ntox47, PAAR-RhsA-Ntox47, TreTu and Tox-URI2) and the 23 candidate effectors described in this study showed that they are widely and differentially distributed among Salmonella genomes (Supplementary Table S2). Interestingly, we identified these effectors and candidates effector in many non-typified Salmonella strains (Figure 6A).
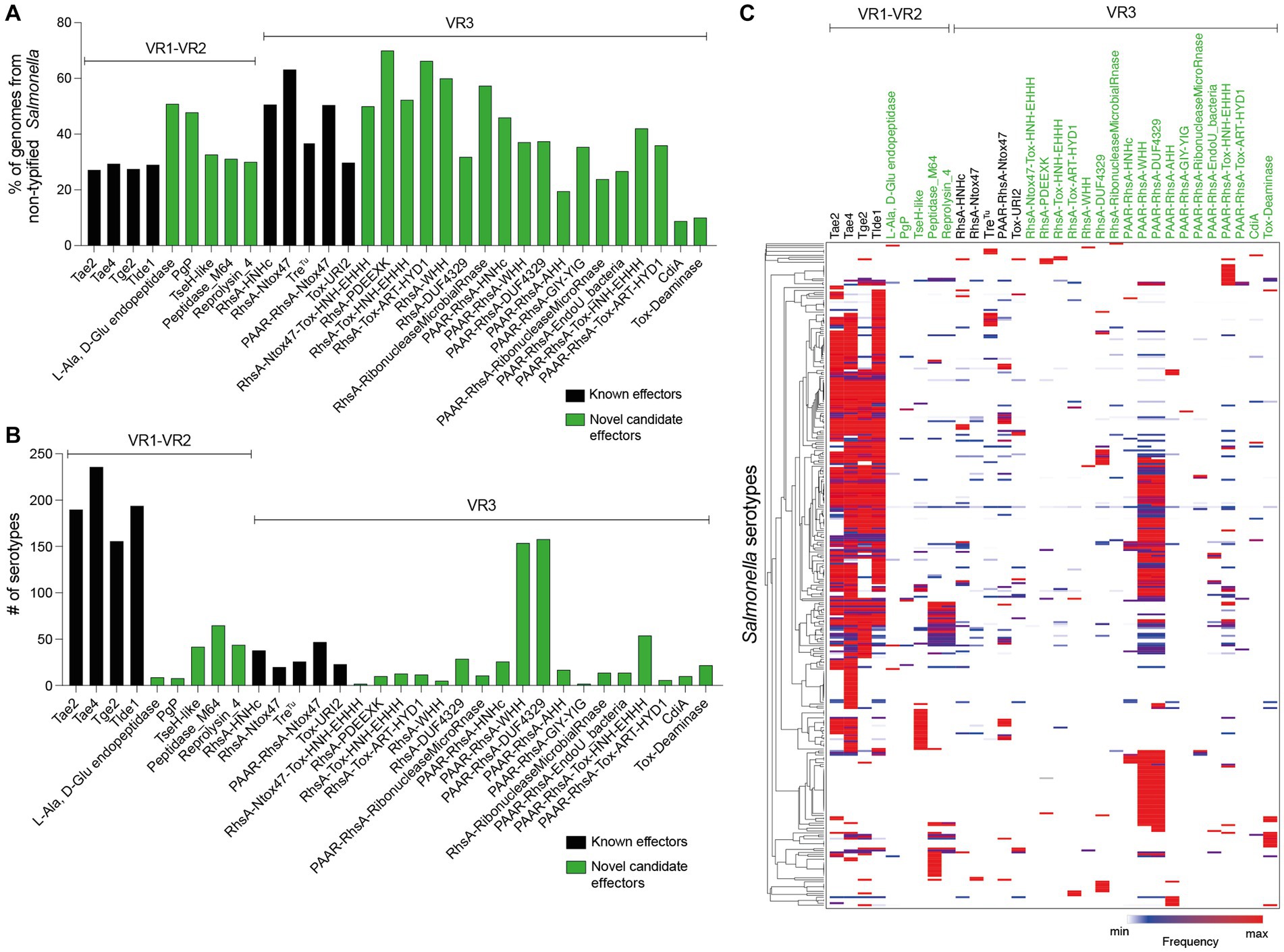
Figure 6. Prevalence of ORFs encoding T6SS effectors and candidate effectors in SPI-6. In the figure, we refer to ORFs encoding effectors according to the corresponding protein name (in the case of those previously reported in the literature) or the functional domains present in the predicted proteins (in the case of ORFs encoding new candidate effectors). Distribution of ORFs encoding T6SS effectors and candidate effectors in non-typified (A) and serotyped (B) Salmonella strains. (C) Prevalence of ORFs encoding T6SS effectors and candidate effectors in the genome of 340 Salmonella serotypes. A hierarchical clustering analysis was performed using MORPHEUS, as described in Materials and Methods. Color code in the heatmap indicates the presence of a given ORF (frequency) among all analyzed strains of a particular Salmonella serotype.
Some effector and candidate effectors were more widespread across different serotypes than others (Figure 6B). Within VR1 and VR2, the previously reported effectors Tae2, Tae4, Tge2, and Tlde1 were identified across 150–240 serotypes, while the five candidate effector proteins identified in this study were found in 5–50 distinct serotypes. A different scenario was observed for effectors and candidate effectors encoded within VR3. In this case, the previously reported effectors were identified in less than 50 serotypes, while some new candidate effectors, such as PAAR-RhsA-WHH and PAAR-RhsA-DUF4329, were identified in over 150 serotypes. The distribution of each candidate effector in different Salmonella serotypes is highlighted in Figures 4, 5.
Finally, we performed a hierarchical clustering analysis to gain further insight into the distribution of effector and candidate effectors identified in 340 Salmonella genomes (Supplementary Table S3). As shown in Figure 6C, the four bona fide effectors encoded within VR1-2 (Tae2, Tae4, Tge2, and Tlde1) were the most conserved across the genomes of 113 different Salmonella serotypes. Nevertheless, these effectors are absent in the genome of 78 Salmonella serotypes, all of which include the genes encoding candidate effectors PAAR-RhsA-WHH and PAAR-RhsA-DUF4329 located within VR3. Furthermore, these candidate effectors are also distributed in the genome of 73 other Salmonella serotypes, suggesting that they play important roles in the biology of this pathogen.
Discussion
The T6SS has emerged as an important virulence and environmental fitness factor for Salmonella (Blondel et al., 2010; Mulder et al., 2012; Pezoa et al., 2013, 2014; Sana et al., 2016; Xian et al., 2020; Hespanhol et al., 2022; Sibinelli-Sousa et al., 2022). However, information regarding the complexity and diversity of effector proteins for each distinct Salmonella T6SS is still lacking. In this context, even though the T6SS encoded in SPI-6 has been shown to contribute to host colonization by S. Typhimurium and S. Dublin (Mulder et al., 2012; Pezoa et al., 2013, 2014; Sana et al., 2016) and to interbacterial competition of S. Typhimurium against the intestinal microbiota (Sibinelli-Sousa et al., 2022), only 9 effector proteins have been identified and characterized so far (Russell et al., 2012; Benz et al., 2013; Whitney et al., 2013; Koskiniemi et al., 2014; Sana et al., 2016; Sibinelli-Sousa et al., 2020; Amaya et al., 2022; Jurėnas et al., 2022; Lorente-Cobo et al., 2022).
In this study, by means of bioinformatic and comparative genomic analyses, we identified a subset of 23 new SPI-6 T6SS candidate effectors, including peptidoglycan hydrolases, DNases, RNases, deaminases, and ADP-ribosyltransferases. Despite being well conserved, the SPI-6 T6SS gene cluster encodes a variable number of ORFs of unknown function restricted to three variable regions (VR1-3), that include the T6SS effectors previously identified in this species (Blondel et al., 2009). Notably, our analysis showed that every new T6SS effector identified is encoded within one of these variable regions. An interesting observation was that all predicted peptidoglycan targeting effectors are confined to VR1 and VR2. The reason behind this observation remains unclear; however, it is possible that VR1 and VR2 are hot-spots for gene recombination during Salmonella evolution, but the lack of mobile genetic elements surrounding these regions does not support this hypothesis. Importantly, in addition to the 4 peptidoglycan targeting effectors reported so far (Tae2, Tae4, Tge2, and Tlde1) (Russell et al., 2012; Benz et al., 2013; Whitney et al., 2013; Sana et al., 2016; Sibinelli-Sousa et al., 2020; Lorente-Cobo et al., 2022), we identified 5 candidate effectors encoded in VR1 and VR2 that presumably degrade peptidoglycan, indicating that this macromolecule is a common target site for Salmonella T6SS effectors. Of note, the unannotated ORF encoded in VR1 is the first putative effector that likely cleaves the link between L-Ala and D-Glu of the peptidoglycan peptide stems reported in Salmonella and shares homology to the peptidoglycan hydrolase ChiX of Serratia marcescens (30% identity and 45.2% similarity at amino acid sequence level) (Owen et al., 2018). This finding expands the peptidoglycan target sites exploited by Salmonella T6SS effectors against competing bacteria. On the other hand, the PgP2 and TseH-like candidate effectors are predicted to have redundant peptidoglycan degrading functions with other Salmonella T6SS effectors. PgP2 is predicted to have the same L,D transpeptidase exchange activity reported for Tlde1 (Sibinelli-Sousa et al., 2020; Lorente-Cobo et al., 2022), replacing D-Ala by a non-canonical D-amino acid preventing the normal crosslink between mDAP and D-Ala. In addition, the TseH-like candidate effector is a NlpC/P60 endopeptidase family protein (Xu et al., 2010; Altindis et al., 2015; Squeglia et al., 2019) predicted to cleave the covalent link between D-Glu and mDAP, as reported for Tae4 (Benz et al., 2013). These redundant functions suggests that the peptide stems are the main peptidoglycan target sites of Salmonella T6SS effectors, as only one identified effector targets the glycoside bonds in this macromolecule corresponds to Tge2 (Whitney et al., 2013). Remarkably, most serotypes encode combinations of T6SS effectors predicted to have hydrolytic activity toward different regions of the peptidoglycan structure. We hypothesize that this assortment of seemingly redundant effectors may improve the efficiency of the bacterial killing process.
The Reprolysin_4 domain found in some candidate effectors is present in zinc-binding metallo-peptidases harboring the binding motif HExxGHxxGxxH of family M12B peptidases. Of note, this motif is also present in the T6SS antibacterial effector SED_RS06335 with putative peptidoglycan hydrolase activity encoded in SPI-19 of Salmonella Dublin CT_02021853 (Amaya et al., 2022). The last candidate effector targeting the peptidoglycan identified in our study harbors the Peptidase_M64 protein domain that is also present in the IgA proteinase of Clostridium ramosum (Kosowska et al., 2002), recently reclassified as Thomasclavelia ramosa (Lawson et al., 2023). The putative immunity proteins of Reprolysin_4 and Peptidase_M64 have a signal peptide and a transmembrane domain, respectively. This suggests that both candidate effectors target the bacterial periplasm.
On the other hand, the VR3 of the SPI-6 T6SS gene cluster encodes a wide variety of effector proteins including domains found in DNases, RNases, deaminases and ADP-ribosyltransferases. Interestingly, most of these domains are fused to the C-terminal of Rhs proteins contributing to diversify the molecular targets of T6SSs in Salmonella. This was not unexpected since we have previously shown that the VR3 of SPI-6 encodes a variable number of Rhs elements (Blondel et al., 2009; Amaya et al., 2022) and many Rhs proteins have C-terminal polymorphic endonuclease domains associated with T6SS effectors in Salmonella and other bacteria (Zhang et al., 2012; Koskiniemi et al., 2014; Amaya et al., 2022). It is known that Rhs proteins have YD-peptide repeats, which fold into a large β-cage structure that surrounds and protects the C-terminal toxin domain increasing T6SS secretion efficiency (Donato et al., 2020; Jurėnas et al., 2021; Günther et al., 2022). This could explain why many T6SS effectors are associated to these elements.
Altogether, our work expands the repertoire of Salmonella T6SS effectors and provides evidence that the SPI-6 T6SS gene cluster harbors a great diversity of antibacterial effectors encoded in three variable regions. One interesting finding of our study is that peptidoglycan hydrolyzing effectors restricted to VR1 and VR2 are highly conserved in Salmonella genomes, while effectors targeting nucleic acids and the translation machinery encoded in VR3 are broadly distributed in Salmonella serotypes. This suggests that different repertoires of effectors could have an impact on the pathogenic potential and environmental fitness of these bacteria. Importantly, although this study increases the number of putative Salmonella antibacterial effectors against competing bacteria, we could not rule out that those targeting nucleic acids encoded in VR3 may also affect eukaryotic cells. This is an important knowledge gap, since no T6SS effector protein identified to date in Salmonella has been confirmed to target eukaryotic organisms, despite the clear contribution of Salmonella T6SSs to intracellular replication, survival and cytotoxicity inside the host immune cells (Mulder et al., 2012; Blondel et al., 2013; Schroll et al., 2019). Further research is required to address this issue. Finally, we are currently performing experimental work to confirm that each of the 23 candidates identified in our study correspond to bona fide T6SS effector proteins.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
CB, FA, PB, CS, and DP: conceptualization, formal analysis, validation, writing-original draft preparation, writing review and editing, resources, project administration, and funding acquisition. CB and DP: methodology, investigation, and visualization. CS and DP: supervision. All authors contributed to the article and approved the submitted version.
Funding
DP was supported by Fondo Concursable Proyectos de Investigación Regulares UDLA 2023 DI-13/23. CS was supported by FONDECYT grant 1212075. CB was supported by FONDECYT grant 1201805, ECOS-ANID ECOS200037 and HHMI-Gulbenkian International Research Scholar Grant #55008749. FA was supported by CONICYT/ANID fellowship 21191925.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1252344/full#supplementary-material
Footnotes
References
Ahmad, S., Wang, B., Walker, M. D., Tran, H.-K. R., Stogios, P. J., Savchenko, A., et al. (2019). An interbacterial toxin inhibits target cell growth by synthesizing (p)ppApp. Nature 575, 674–678. doi: 10.1038/s41586-019-1735-9
Altindis, E., Dong, T., Catalano, C., and Mekalanos, J. (2015). Secretome analysis of Vibrio cholerae type VI secretion system reveals a new effector–immunity pair. mBio 6:e00075. doi: 10.1128/mBio.00075-15
Amaya, F. A., Blondel, C. J., Barros-Infante, M. F., Rivera, D., Moreno-Switt, A. I., Santiviago, C. A., et al. (2022). Identification of type VI secretion systems effector proteins that contribute to interbacterial competition in Salmonella Dublin. Front. Microbiol. 13:811932. doi: 10.3389/fmicb.2022.811932
Bao, H., Zhao, J.-H., Zhu, S., Wang, S., Zhang, J., Wang, X.-Y., et al. (2019). Genetic diversity and evolutionary features of type VI secretion systems in Salmonella. Future Microbiol. 14, 139–154. doi: 10.2217/fmb-2018-0260
Benz, J., Reinstein, J., and Meinhart, A. (2013). Structural insights into the effector - immunity system Tae4/Tai4 from Salmonella typhimurium. PLoS One 8:e67362. doi: 10.1371/journal.pone.0067362
Berni, B., Soscia, C., Djermoun, S., Ize, B., and Bleves, S. (2019). A type VI secretion system trans-kingdom effector is required for the delivery of a novel antibacterial toxin in Pseudomonas aeruginosa. Front. Microbiol. 10:1218. doi: 10.3389/fmicb.2019.01218
Blondel, C. J., Jiménez, J. C., Contreras, I., and Santiviago, C. A. (2009). Comparative genomic analysis uncovers 3 novel loci encoding type six secretion systems differentially distributed in Salmonella serotypes. BMC Genomics 10:354. doi: 10.1186/1471-2164-10-354
Blondel, C. J., Jiménez, J. C., Leiva, L. E., Alvarez, S. A., Pinto, B. I., Contreras, F., et al. (2013). The type VI secretion system encoded in Salmonella Pathogenicity Island 19 is required for Salmonella enterica serotype Gallinarum survival within infected macrophages. Infect. Immun. 81, 1207–1220. doi: 10.1128/iai.01165-12
Blondel, C. J., Yang, H.-J., Castro, B., Chiang, S., Toro, C. S., Zaldívar, M., et al. (2010). Contribution of the type VI secretion system encoded in SPI-19 to chicken colonization by Salmonella enterica serotypes Gallinarum and Enteritidis. PLoS One 5:e11724. doi: 10.1371/journal.pone.0011724
Brackmann, M., Nazarov, S., Wang, J., and Basler, M. (2017). Using force to punch holes: mechanics of contractile nanomachines. Trends Cell Biol. 27, 623–632. doi: 10.1016/j.tcb.2017.05.00
Chassaing, B., and Cascales, E. (2018). Antibacterial weapons: targeted destruction in the microbiota. Trends Microbiol. 26, 329–338. doi: 10.1016/j.tim.2018.01.006
Cherrak, Y., Flaugnatti, N., Durand, E., Journet, L., and Cascales, E. (2019). Structure and activity of the type VI secretion system. Microbiol. Spectr. 7:10.1128/microbiolspec.PSIB-0031-2019. doi: 10.1128/microbiolspec.PSIB-0031-2019
Coulthurst, S. (2019). The type VI secretion system: a versatile bacterial weapon. Microbiology 165, 503–515. doi: 10.1099/mic.0.000789
Coyne, M. J., and Comstock, L. E. (2019). Type VI secretion systems and the gut microbiota. Microbiol. Spectr. 7:10.1128/microbiolspec.PSIB-0009-2018. doi: 10.1128/microbiolspec.PSIB-0009-2018
Darling, A. C. E., Mau, B., Blattner, F. R., and Perna, N. T. (2004). Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403. doi: 10.1101/gr.2289704
Diniz, J. A., and Coulthurst, S. J. (2015). Intraspecies competition in Serratia marcescens is mediated by type VI-secreted Rhs effectors and a conserved effector-associated accessory protein. J. Bacteriol. 197, 2350–2360. doi: 10.1128/jb.00199-15
Donato, S. L., Beck, C. M., Garza-Sánchez, F., Jensen, S. J., Ruhe, Z. C., Cunningham, D. A., et al. (2020). The β-encapsulation cage of rearrangement hotspot (Rhs) effectors is required for type VI secretion. Proc. Natl. Acad. Sci. U. S. A. 117, 33540–33548. doi: 10.1073/pnas.1919350117
Durand, E., Cambillau, C., Cascales, E., and Journet, L. (2014). VgrG, Tae, Tle, and beyond: the versatile arsenal of type VI secretion effectors. Trends Microbiol. 22, 498–507. doi: 10.1016/j.tim.2014.06.004
Finn, R. D., Bateman, A., Clements, J., Coggill, P., Eberhardt, R. Y., Eddy, S. R., et al. (2014). Pfam: the protein families database. Nucleic Acids Res. 42, D222–D230. doi: 10.1093/nar/gkt1223
Fookes, M., Schroeder, G. N., Langridge, G. C., Blondel, C. J., Mammina, C., Connor, T. R., et al. (2011). Salmonella bongori provides insights into the evolution of the salmonellae. PLoS Pathog. 7:e1002191. doi: 10.1371/journal.ppat.1002191
GBD 2017 Non-Typhoidal Salmonella Invasive Disease Collaborators (2019). The global burden of non-typhoidal Salmonella invasive disease: a systematic analysis for the global burden of disease study 2017. Lancet Infect. Dis. 19, 1312–1324. doi: 10.1016/s1473-3099(19)30418-9
Grimont, P. A. D., and Weill, F.-X. (2007). Antigenic formulae of the Salmonella serovars. 9 WHO Collaborating Centre for Reference and Research on Salmonella, Institut Pasteur, Paris.
Günther, P., Quentin, D., Ahmad, S., Sachar, K., Gatsogiannis, C., Whitney, J. C., et al. (2022). Structure of a bacterial Rhs effector exported by the type VI secretion system. PLoS Pathog. 18:e1010182. doi: 10.1371/journal.ppat.1010182
Hespanhol, J. T., Sanchez-Limache, D. E., Nicastro, G. G., Mead, L., Llontop, E. E., Chagas-Santos, G., et al. (2022). Antibacterial T6SS effectors with a VRR-Nuc domain are structure-specific nucleases. eLife 11:e82437. doi: 10.7554/eLife.82437
Issenhuth-Jeanjean, S., Roggentin, P., Mikoleit, M., Guibourdenche, M., de Pinna, E., Nair, S., et al. (2014). Supplement 2008–2010 (no. 48) to the white–Kauffmann–Le minor scheme. Res. Microbiol. 165, 526–530. doi: 10.1016/j.resmic.2014.07.004
Jiang, F., Waterfield, N. R., Yang, J., Yang, G., and Jin, Q. (2014). A Pseudomonas aeruginosa type VI secretion phospholipase D effector targets both prokaryotic and eukaryotic cells. Cell Host Microbe 15, 600–610. doi: 10.1016/j.chom.2014.04.010
Jurėnas, D., Rey, M., Byrne, D., Chamot-Rooke, J., Terradot, L., and Cascales, E. (2022). Salmonella antibacterial Rhs polymorphic toxin inhibits translation through ADP-ribosylation of EF-Tu P-loop. Nucleic Acids Res. 50, 13114–13127. doi: 10.1093/nar/gkac1162
Jurėnas, D., Rosa, L. T., Rey, M., Chamot-Rooke, J., Fronzes, R., and Cascales, E. (2021). Mounting, structure and autocleavage of a type VI secretion-associated Rhs polymorphic toxin. Nat. Commun. 12:6998. doi: 10.1038/s41467-021-27388-0
Kanehisa, M., Goto, S., Kawashima, S., and Nakaya, A. (2002). The KEGG databases at GenomeNet. Nucleic Acids Res. 30, 42–46. doi: 10.1093/nar/30.1.42
Katoh, K., Rozewicki, J., and Yamada, K. D. (2017). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20, 1160–1166. doi: 10.1093/bib/bbx108
Koskiniemi, S., Garza-Sánchez, F., Sandegren, L., Webb, J. S., Braaten, B. A., Poole, S. J., et al. (2014). Selection of orphan Rhs toxin expression in evolved Salmonella enterica serovar Typhimurium. PLoS Genet. 10:e1004255. doi: 10.1371/journal.pgen.1004255
Kosowska, K., Reinholdt, J., Rasmussen, L. K., Sabat, A., Potempa, J., Kilian, M., et al. (2002). The Clostridium ramosum IgA proteinase represents a novel type of metalloendopeptidase. J. Biol. Chem. 277, 11987–11994. doi: 10.1074/jbc.M110883200
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lawson, P. A., Saavedra-Perez, L., and Sankaranarayanan, K. (2023). Reclassification of Clostridium cocleatum, Clostridium ramosum, clostridium spiroforme and Clostridium saccharogumia as Thomasclavelia cocleata gen. Nov., comb. nov., Thomasclavelia ramosa comb. nov., gen. Nov., Thomasclavelia spiroformis comb. nov. and Thomasclavelia saccharogumia comb. nov. Int. J. Syst. Evol. Microbiol. 73:10.1099/ijsem.0.005694. doi: 10.1099/ijsem.0.005694
Lorente-Cobo, N., Sibinelli-Sousa, S., Biboy, J., Vollmer, W., Bayer-Santos, E., and Prehna, G. (2022). Molecular characterization of the type VI secretion system effector Tlde1a reveals a structurally altered LD-transpeptidase fold. J. Biol. Chem. 298:102556. doi: 10.1016/j.jbc.2022.102556
Lu, S., Wang, J., Chitsaz, F., Derbyshire, M. K., Geer, R. C., Gonzales, N. R., et al. (2019). CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 48, D265–D268. doi: 10.1093/nar/gkz991
Ma, A. T., and Mekalanos, J. J. (2010). In vivo actin cross-linking induced by Vibrio cholerae type VI secretion system is associated with intestinal inflammation. Proc. Natl. Acad. Sci. 107, 4365–4370. doi: 10.1073/pnas.0915156107
Ma, J., Sun, M., Dong, W., Pan, Z., Lu, C., and Yao, H. (2017). PAAR-Rhs proteins harbor various C-terminal toxins to diversify the antibacterial pathways of type VI secretion systems. Environ. Microbiol. 19, 345–360. doi: 10.1111/1462-2920.13621
Monjarás Feria, J., and Valvano, M. A. (2020). An overview of anti-eukaryotic T6SS effectors. Front. Cell. Infect. Microbiol. 10:584751. doi: 10.3389/fcimb.2020.584751
Mulder, D. T., Cooper, C. A., and Coombes, B. K. (2012). Type VI secretion system-associated gene clusters contribute to pathogenesis of Salmonella enterica serovar Typhimurium. Infect. Immun. 80, 1996–2007. doi: 10.1128/iai.06205-11
Notredame, C., Higgins, D. G., and Heringa, J. (2000). T-coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302, 205–217. doi: 10.1006/jmbi.2000.4042
Owen, R. A., Fyfe, P. K., Lodge, A., Biboy, J., Vollmer, W., Hunter, W. N., et al. (2018). Structure and activity of ChiX: a peptidoglycan hydrolase required for chitinase secretion by Serratia marcescens. Biochem. J. 475, 415–428. doi: 10.1042/BCJ20170633
Pezoa, D., Blondel, C. J., Silva, C. A., Yang, H.-J., Andrews-Polymenis, H., Santiviago, C. A., et al. (2014). Only one of the two type VI secretion systems encoded in the Salmonella enterica serotype Dublin genome is involved in colonization of the avian and murine hosts. Vet. Res. 45:2. doi: 10.1186/1297-9716-45-2
Pezoa, D., Yang, H.-J., Blondel, C. J., Santiviago, C. A., Andrews-Polymenis, H. L., and Contreras, I. (2013). The type VI secretion system encoded in SPI-6 plays a role in gastrointestinal colonization and systemic spread of Salmonella enterica serovar Typhimurium in the chicken. PLoS One 8:e63917. doi: 10.1371/journal.pone.0063917
Pissaridou, P., Allsopp, L. P., Wettstadt, S., Howard, S. A., Mavridou, D. A. I., and Filloux, A. (2018). The Pseudomonas aeruginosa T6SS-VgrG1b spike is topped by a PAAR protein eliciting DNA damage to bacterial competitors. Proc. Natl. Acad. Sci. 115, 12519–12524. doi: 10.1073/pnas.1814181115
Robert, X., and Gouet, P. (2014). Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324. doi: 10.1093/nar/gku316
Russell, A. B., Singh, P., Brittnacher, M., Bui, N. K., Hood, R. D., Carl, M. A., et al. (2012). A widespread bacterial type VI secretion effector superfamily identified using a heuristic approach. Cell Host Microbe 11, 538–549. doi: 10.1016/j.chom.2012.04.007
Rutherford, K., Parkhill, J., Crook, J., Horsnell, T., Rice, P., Rajandream, M.-A., et al. (2000). Artemis: sequence visualization and annotation. Bioinformatics 16, 944–945. doi: 10.1093/bioinformatics/16.10.944
Sana, T. G., Flaugnatti, N., Lugo, K. A., Lam, L. H., Jacobson, A., Baylot, V., et al. (2016). Salmonella Typhimurium utilizes a T6SS-mediated antibacterial weapon to establish in the host gut. Proc. Natl. Acad. Sci. 113, E5044–E5051. doi: 10.1073/pnas.1608858113
Schroll, C., Huang, K., Ahmed, S., Kristensen, B. M., Pors, S. E., Jelsbak, L., et al. (2019). The SPI-19 encoded type-six secretion-systems (T6SS) of Salmonella enterica serovars Gallinarum and Dublin play different roles during infection. Vet. Microbiol. 230, 23–31. doi: 10.1016/j.vetmic.2019.01.006
Sibinelli-Sousa, S., de Araújo-Silva, A. L., Hespanhol, J. T., and Bayer-Santos, E. (2022). Revisiting the steps of Salmonella gut infection with a focus on antagonistic interbacterial interactions. FEBS J. 289, 4192–4211. doi: 10.1111/febs.16211
Sibinelli-Sousa, S., Hespanhol, J. T., Nicastro, G. G., Matsuyama, B. Y., Mesnage, S., Patel, A., et al. (2020). A family of T6SS antibacterial effectors related to L,D-transpeptidases targets the peptidoglycan. Cell Rep. 31:107813. doi: 10.1016/j.celrep.2020.107813
Sigrist, C. J. A., de Castro, E., Cerutti, L., Cuche, B. A., Hulo, N., Bridge, A., et al. (2013). New and continuing developments at PROSITE. Nucleic Acids Res. 41, D344–D347. doi: 10.1093/nar/gks1067
Silverman, J. M., Agnello, D. M., Zheng, H., Andrews, B. T., Li, M., Catalano, C. E., et al. (2013). Haemolysin coregulated protein is an exported receptor and chaperone of type VI secretion substrates. Mol. Cell 51, 584–593. doi: 10.1016/j.molcel.2013.07.025
Squeglia, F., Moreira, M., Ruggiero, A., and Berisio, R. (2019). The cell wall hydrolytic NlpC/P60 endopeptidases in mycobacterial cytokinesis: a structural perspective. Cells 8:609. doi: 10.3390/cells8060609
Srikannathasan, V., English, G., Bui, N. K., Trunk, K., O’Rourke, P. E. F., Rao, V. A., et al. (2013). Structural basis for type VI secreted peptidoglycan DL-endopeptidase function, specificity and neutralization in Serratia marcescens. Acta Crystallogr. Sect. D Biol. Crystallogr. 69, 2468–2482. doi: 10.1107/s0907444913022725
Sullivan, M. J., Petty, N. K., and Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Taboada, B., Estrada, K., Ciria, R., and Merino, E. (2018). Operon-mapper: a web server for precise operon identification in bacterial and archaeal genomes. Bioinformatics 34, 4118–4120. doi: 10.1093/bioinformatics/bty496
Tang, J. Y., Bullen, N. P., Ahmad, S., and Whitney, J. C. (2018). Diverse NADase effector families mediate interbacterial antagonism via the type VI secretion system. J. Biol. Chem. 293, 1504–1514. doi: 10.1074/jbc.ra117.000178
Ting, S.-Y., Bosch, D. E., Mangiameli, S. M., Radey, M. C., Huang, S., Park, Y.-J., et al. (2018). Bifunctional immunity proteins protect bacteria against FtsZ-targeting ADP-ribosylating toxins. Cells 175, 1380–1392.e14. doi: 10.1016/j.cell.2018.09.037
Uzzau, S., Brown, D. J., Wallis, T., Rubino, S., Leori, G., Bernard, S., et al. (2000). Host adapted serotypes of Salmonella enterica. Epidemiol. Amp. Infect. 125, 229–255. doi: 10.1017/s0950268899004379
Wang, J., Yang, B., Leier, A., Marquez-Lago, T. T., Hayashida, M., Rocker, A., et al. (2018). Bastion6: a bioinformatics approach for accurate prediction of type VI secreted effectors. Bioinformatics 34, 2546–2555. doi: 10.1093/bioinformatics/bty155
Whitney, J. C., Beck, C. M., Goo, Y. A., Russell, A. B., Harding, B. N., Leon, J. A. D., et al. (2014). Genetically distinct pathways guide effector export through the type VI secretion system. Mol. Microbiol. 92, 529–542. doi: 10.1111/mmi.12571
Whitney, J. C., Chou, S., Russell, A. B., Biboy, J., Gardiner, T. E., Ferrin, M. A., et al. (2013). Identification, structure, and function of a novel type VI secretion peptidoglycan glycoside hydrolase effector-immunity pair. J. Biol. Chem. 288, 26616–26624. doi: 10.1074/jbc.m113.488320
Whitney, J. C., Quentin, D., Sawai, S., LeRoux, M., Harding, B. N., Ledvina, H. E., et al. (2015). An interbacterial NAD(P)+ glycohydrolase toxin requires elongation factor Tu for delivery to target cells. Cells 163, 607–619. doi: 10.1016/j.cell.2015.09.027
Wood, T. E., Howard, S. A., Förster, A., Nolan, L. M., Manoli, E., Bullen, N. P., et al. (2019). The Pseudomonas aeruginosa T6SS delivers a periplasmic toxin that disrupts bacterial cell morphology. Cell Rep. 29, 187–201.e7. doi: 10.1016/j.celrep.2019.08.094
Xian, H., Yuan, Y., Yin, C., Wang, Z., Ji, R., Chu, C., et al. (2020). The SPI-19 encoded T6SS is required for Salmonella Pullorum survival within avian macrophages and initial colonization in chicken dependent on inhibition of host immune response. Vet. Microbiol. 250:108867. doi: 10.1016/j.vetmic.2020.108867
Xu, Q., Abdubek, P., Astakhova, T., Axelrod, H. L., Bakolitsa, C., Cai, X., et al. (2010). Structure of the γ-D-glutamyl-L-diamino acid endopeptidase YkfC from Bacillus cereus in complex with L-ala-γ-D-Glu: insights into substrate recognition by NlpC/P60 cysteine peptidases. Acta crystallographica. Sect. F Struct. Biol. Crystallizat. Commun. 66, 1354–1364. doi: 10.1107/S1744309110021214
Zhang, D., de Souza, R. F., Anantharaman, V., Iyer, L. M., and Aravind, L. (2012). Polymorphic toxin systems: comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol. Direct 7:18. doi: 10.1186/1745-6150-7-18
Zhang, J., Guan, J., Wang, M., Li, G., Djordjevic, M., Tai, C., et al. (2023). SecReT6 update: a comprehensive resource of bacterial type VI secretion systems. Sci. China Life Sci. 66, 626–634. doi: 10.1007/s11427-022-2172-x
Keywords: Salmonella, T6SS, SPI-6, effector, immunity protein
Citation: Blondel CJ, Amaya FA, Bustamante P, Santiviago CA and Pezoa D (2023) Identification and distribution of new candidate T6SS effectors encoded in Salmonella Pathogenicity Island 6. Front. Microbiol. 14:1252344. doi: 10.3389/fmicb.2023.1252344
Edited by:
Sébastien Holbert, INRA Centre Val de Loire, FranceReviewed by:
Qiuhe Lu, Cleveland Clinic, United StatesLuke Peter Allsopp, Imperial College London, United Kingdom
Fernando Navarro-Garcia, National Polytechnic Institute of Mexico (CINVESTAV), Mexico
Copyright © 2023 Blondel, Amaya, Bustamante, Santiviago and Pezoa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Carlos A. Santiviago, Y3NhbnRpdmlhZ29AY2lxLnVjaGlsZS5jbA==; David Pezoa, ZHBlem9hQHVkbGEuY2w=
†These authors have contributed equally to this work and share first authorship
 Carlos J. Blondel
Carlos J. Blondel Fernando A. Amaya
Fernando A. Amaya Paloma Bustamante
Paloma Bustamante Carlos A. Santiviago
Carlos A. Santiviago David Pezoa
David Pezoa