
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 01 December 2023
Sec. Microbial Physiology and Metabolism
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1252155
 Mizuki Kanno1,2
Mizuki Kanno1,2 Takuya Shiota2
Takuya Shiota2 So Ueno2
So Ueno2 Minato Takahara2
Minato Takahara2 Keisuke Haneda3
Keisuke Haneda3 Yuhei O. Tahara4
Yuhei O. Tahara4 Masaki Shintani1,2,3,5,6
Masaki Shintani1,2,3,5,6 Ryoma Nakao7
Ryoma Nakao7 Makoto Miyata4
Makoto Miyata4 Kazuhide Kimbara1,2,3
Kazuhide Kimbara1,2,3 Hiroyuki Futamata1,2,3,5
Hiroyuki Futamata1,2,3,5 Yosuke Tashiro1,2,3,8*
Yosuke Tashiro1,2,3,8*Membrane vesicles (MVs) are small spherical structures (20–400 nm) produced by most bacteria and have important biological functions including toxin delivery, signal transfer, biofilm formation, and immunomodulation of the host. Although MV formation is enhanced in biofilms of a wide range of bacterial species, the underlying mechanisms are not fully understood. An opportunistic pathogen, Pseudomonas aeruginosa, causes chronic infections that can be difficult to treat due to biofilm formation. Since MVs are abundant in biofilms, can transport virulence factors to the host, and have inflammation-inducing functions, the mechanisms of enhanced MV formation in biofilms needs to be elucidated to effectively treat infections. In this study, we evaluated the characteristics of MVs in P. aeruginosa PAO1 biofilms, and identified factors that contribute to enhanced MV formation. Vesiculation was significantly enhanced in the static culture; MVs were connected to filamentous substances in the biofilm, and separation between the outer and inner membranes and curvature of the membrane were observed in biofilm cells. By screening a transposon mutant library (8,023 mutants) for alterations in MV formation in biofilms, 66 mutants were identified as low-vesiculation strains (2/3 decrease relative to wild type), whereas no mutant was obtained that produced more MVs (twofold increase). Some transposons were inserted into genes related to biofilm formation, including flagellar motility (flg, fli, and mot) and extracellular polysaccharide synthesis (psl). ΔpelAΔpslA, which does not synthesize the extracellular polysaccharides Pel and Psl, showed reduced MV production in biofilms but not in planktonic conditions, suggesting that enhanced vesiculation is closely related to the synthesis of biofilm matrices in P. aeruginosa. Additionally, we found that blebbing occurred during bacterial attachment. Our findings indicate that biofilm-related factors are closely involved in enhanced MV formation in biofilms and that surface sensing facilitates vesiculation. Furthermore, this work expands the understanding of the infection strategy in P. aeruginosa biofilms.
Bacteria possess sophisticated tools to deliver their own substances to other cells. One of the mechanisms is the release of membrane vesicles (MVs), which range in size from 20 to 400 nm and are composed of bacterial outer surfaces such as membrane proteins, phospholipids, and lipopolysaccharides (LPS). MVs facilitate the transfer of nucleic acids and toxic components to other bacteria and host cells, and also play roles in the alteration of biofilm structure, resistance to phages, and release of unnecessary compounds (Tashiro et al., 2012; Toyofuku et al., 2015).
In Gram-negative bacteria, MVs caused by blebbing from the outer membrane (OM) are called outer membrane vesicles (OMVs), and several mechanisms of OMV formation have been identified in different species and culture conditions. Weakened OM-peptidoglycan cross-linking (Suzuki et al., 1978; Deatherage et al., 2009; Schwechheimer et al., 2015), accumulation of misfolded protein in the cell envelope (McBroom and Kuehn, 2007; Tashiro et al., 2009), and accumulation or modification of LPS and phospholipids in the outer leaflet of the OM (Elhenawy et al., 2016; Roier et al., 2016) cause blebbing of the OM. The Pseudomonas quinolone signal (PQS; 2-heptyl-3-hydroxy-4-quinolone), which is a quorum-sensing molecule produced by Pseudomonas aeruginosa (Pesci et al., 1999), is also known to stimulate OMV production in not only P. aeruginosa but also other bacteria (Mashburn and Whiteley, 2005; Tashiro et al., 2010a) by insertion into the outer leaflet and OM curvature (Schertzer and Whiteley, 2012). Explosive cell lysis (ECL) is another pathway of MV formation that is distinct from blebbing (Turnbull et al., 2016). ECL is triggered by endolysin, which degrades peptidoglycan, causing cell death and the release of large amounts of MVs.
The relationship between MVs and biofilms has been the focus of research for decades, as MVs are a component of biofilms formed under natural and experimental conditions (Beveridge et al., 1997; Schooling and Beveridge, 2006; Grande et al., 2015; Wang et al., 2015; Baeza and Mercade, 2020). However, research on MV biogenesis has been mainly conducted in a planktonic state. According to a report by the National Institutes of Health (NIH), among all microbial and chronic infections, 65 and 80%, respectively, are associated with biofilm formation (Jamal et al., 2018). Pathogenic bacteria cause infections by forming chronic biofilms that persist in mucus-rich environments (Costerton et al., 1999). Biofilms form in the lungs, cornea, and skin of individual animals, contributing to the initiation and spread of infection, as well as in the soil, hospital surfaces, and many other locations in the environment (Hall-Stoodley et al., 2004). Therefore, a comprehensive understanding of the function and formation mechanisms of MVs in biofilms is important for both clinical and environmental conditions.
The biofilm matrix, which helps protect biofilm-embedded cells from external environmental stress and threats (Hobley et al., 2015), is mainly composed of exopolysaccharides, extracellular proteins, eDNA, and MVs. Several studies have suggested that MVs promote biofilm formation in Helicobacter pylori (Yonezawa et al., 2009), Pseudomonas putida (Baumgarten et al., 2012), Vibrio cholerae (Altindis et al., 2014), and P. aeruginosa (Murphy et al., 2014), and also function to facilitate dispersion in Xylella fastidiosa (Ionescu et al., 2014) and P. aeruginosa (Esoda and Kuehn, 2019; Cooke et al., 2020). The MVs derived from planktonic and biofilm states are quite different in physical characteristics and proteomics in several bacteria, including P. aeruginosa (Schooling and Beveridge, 2006; Schooling et al., 2009; Toyofuku et al., 2012; Park et al., 2014b,2015; Couto et al., 2015), H. pylori (Grande et al., 2015), Lactobacillus reuteri (Grande et al., 2017), and Bordetella pertussis (Carriquiriborde et al., 2021). Based on these studies, the biogenesis and function of MVs in the biofilm state are considered to be different from those in the planktonic state.
Pseudomonas aeruginosa is a prominent opportunistic human pathogen that establishes chronic biofilm-based infections, such as cystic fibrosis, and is used as a model organism for biofilm formation. MV production by several bacteria, including P. aeruginosa, is increased in biofilms (Schooling and Beveridge, 2006; Grande et al., 2017), but the mechanisms have not been fully elucidated. P. aeruginosa induces host immune responses by releasing large amounts of MVs that retain antigens in the human body (Bauman and Kuehn, 2009; Bomberger et al., 2009; Ellis et al., 2010). Therefore, understanding the MV production mechanism under biofilm conditions leads to control of biofilm formation and bacterial infections.
Although biofilm growth mode is predominant in natural and disease environments, most studies on the function and mechanism of MV formation have been conducted using planktonic cultures. In this study, we aimed to unravel the molecular mechanism behind enhanced MV formation under biofilm conditions in P. aeruginosa. Liquid static biofilm was used for the biofilm model because MV formation was significantly promoted compared to planktonic conditions and is a suitable model for studying the relationship between attachment and MV formation. Transposon mutagenesis screens were performed to identify genes that affect enhanced MV formation in P. aeruginosa PAO1 biofilm. Many mutants lacking biofilm-related genes were identified as low-vesiculating mutants; some mutants (e.g., those related to flagellar motility and Psl synthesis) showed low-biofilm-forming abilities, whereas others showed low vesiculation even though biofilm formation was not repressed. We also showed that the initial attachment triggers vesiculation. These results provide novel insights into how bacteria produce MVs in clinically relevant biofilms.
The bacterial strains, plasmids, and primers used in this study are listed in Table 1 and Supplementary Table 1. P. aeruginosa and Escherichia coli were routinely grown at 37°C aerobically in Miller’s lysogeny broth (LB) (1% w/v tryptone, 0.5% w/v yeast extract, and 1% w/v NaCl). P. aeruginosa PAO1 and its mutants were inoculated to adjust an optical density at 600 nm of 0.02, and grown at 37°C for 12 h in either a shaking condition (planktonic condition: 100 ml LB medium in each flask at 200 rpm) or a static biofilm condition (10 ml LB medium in each petri dish without shaking). When required, the following antibiotics were used: 20 μg/ml tetracycline or 10 μg/ml gentamicin for E. coli, and 60 μg/ml tetracycline or 100 μg/ml gentamicin for P. aeruginosa. 2,6-Diaminopimelic acid (DAPA) was added to the medium at a concentration of 300 μM for the growth of E. coli β2163.
Vesicle extraction from P. aeruginosa culture was performed by ultracentrifugation of the filtered supernatant, as previously described (Takaki et al., 2020). P. aeruginosa was grown under shaking and static conditions overnight. In static biofilm conditions, the mucoid and viscous films attached to the bottom of petri dish were scratched using a scraper. The collected cultures were treated by sonication (38 kHz self-oscillation for 5 min) and vortex to isolate vesicles from the bacterial cell surfaces. After the removal of bacterial cells using centrifugation (6,000 × g for 15 min at 4°C), the supernatant was sequentially filtered through 0.45 and 0.20 μm pore-size membranes (CELLULOSE ACETATE, ADVANTEC) and ultracentrifuged at 150,000 × g for 2 h at 4°C using a Himac CP80WX and a P45AT angle rotor (Eppendorf Himac Technologies). The supernatant was removed, and the pellets were resuspended in 50 mM HEPES (pH 6.8) and 0.85% NaCl (HEPES-NaCl buffer).
Purification of extracted vesicles was conducted by density gradient ultracentrifugation using different layered concentrations of iodixanol, as previously described (Takaki et al., 2020). Briefly, the extracted vesicles were adjusted to 1 ml of 45% (wt/vol) iodixanol (OptiPrep; Axis-Shield Diagnostics Ltd.) in HEPES-NaCl, transferred to the bottom of ultracentrifuge tubes, and layered with iodixanol-HEPES-NaCl (2 ml of 40, 35, 30, and 25% and 1 ml of 20%). The samples were ultracentrifuged at 100,000 × g for 16 h at 4°C using a swinging bucket rotor (P40ST; Eppendorf Himac Technologies). The MV fraction was further ultracentrifuged at 200,000 × g for 1 h at 4°C using a P50A3 rotor, and the pellet was resuspended in HEPES-NaCl to remove excess iodixanol.
Quantification of vesicles using FM4-64 was followed by a previous report (Aktar et al., 2021). Briefly, to extract vesicles, the cell-free supernatant was ultracentrifuged at 200,000 × g for 1 h at 4°C using a P50A3 rotor. The extracted vesicles were incubated with 10 μg/ml of FM4-64 (Biotium, Inc.) for 30 min on a 96-well black plate at 37°C in the dark, and the fluorescence intensities were measured using a microplate reader (SpectraMax i3, Molecular Devices) at 506/750 nm (excitation/emission wavelength). Linoleic acid was used as a standard, and the value was presented as the phospholipid amount in vesicles normalized to the cell density.
Phase contrast, fluorescence microscopic observations were conducted using an Olympus IX73 microscope (Olympus), and images were captured with a scientific complementary metal oxide semiconductor (sCMOS) camera (Andor). The excitation light was emitted using a mercury lamp (U-HGLGPS, Olympus). The cell membrane images were recorded using the GWA filter set [Ex/Em (nm), 530–550/575–625], and the polysaccharide Psl images were recorded using the BNA filter set [Ex/Em (nm), 470–495/510–550]. The phase contrast and fluorescence channels were adjusted using the microscope imaging software CellSens Dimension (Olympus).
For phase contrast and fluorescence observation of bacterial cells attached to the glass, 18 × 18 mm coverslips were placed in a 6-well plate with 3 ml of the P. aeruginosa culture and OD600 adjusted to 0.1. Only half of the coverslips were then immersed. The cells were statically grown at 37°C for 2.5 h. Unattached cells were removed from coverslips after incubation by rinse with 1 ml of 1 × PBS. Psl was detected by labeling with 2 μg of HHA-FITC lectin (Hippeastrum Hybrid, EY Laboratories, Inc.) for 20 min at 25°C in the dark. Bacterial cells on coverslips were then stained with 10 μg/ml FM4-64 (SynaptoRed C2, Biotium, Inc.) for 10 min at 37°C in the dark. To observe free-living cells, planktonic bacterial cells in a 6-well plate were collected and centrifuged (6,000 × g for 2 min at room temperature), then cells were stained with FM4-64 as described.
For scanning electron microscope (SEM) observation, vesicles or bacterial cells were prepared on attached glasses. For vesicles and planktonic bacterial cells, samples were dropped onto glasses coated with 0.5% poly-L-lysine hydrobromide (Sigma-Aldrich). In the case of biofilms, two small coverslips were prepared: one was vertically stood and fixed with tape, and the other was laid at the bottom in the static culture. Fixation, dehydration through ascending acetone series, critical point drying, and osmium coating were conducted using previously reported methods (Tashiro et al., 2008). The samples were observed using SEM Regulus 8220 (Hitachi High-Tech).
Transmission electron microscope (TEM) observations of negatively stained vesicles and quick-freeze deep-etched (QFDE) bacterial cells were performed as previously described (Takaki et al., 2020) using a JEM-1010 (JEOL) equipped with a FastScan-F214 (T) CCD camera (TVIPS) or JEM-2100F at 80 kV with a CANTEGA G2 CCD camera (Olympus), or CANTEGA G2 (Olympus), respectively. To prepare samples for negative staining, Cu400 mesh grids (JEOL) were pretreated with 0.01% α-poly-L-lysine, vesicles were placed onto the grid, and samples were stained with 2% (NH4)6Mo7O24. For QFDE sample preparation, bacterial cells grown under shaking or static conditions were centrifuged; after washing, they were mixed with a rabbit lung slab and mica flakes. The mixture was placed on a paper disk attached to an aluminum disc. The samples were quickly frozen in liquid helium using a CryoPress (Valiant Instruments) and maintained at −180°C using a JFDV freeze-etching device (JEOL). The samples were freeze-fractured with a knife, freeze-dried, and coated with platinum and carbon. After floating in full-strength hydrofluoric acid and rinsing in water, the replica was cleaned with a commercial bleach containing sodium hypochlorite. The replica specimens were rinsed and placed on the grids.
The size of vesicles was measured using ZetaView PMX 120 (Particle Metrix). Polystyrene nanospheres (100 nm) were used to assess the measurement bias of size and concentration. Vesicles were diluted to allow for 50–200 particles in 1 frame.
Membrane vesicles were purified using the density gradient centrifugation method described above, except that a protein inhibitor was added to all solvents at appropriate concentrations in each step. Bacterial OMPs were isolated from lysed total cell proteins by Sarkosyl separation, as described previously (Tashiro et al., 2008). SDS-PAGE and peptide mass fingerprinting (PMF) were performed as reported previously (Takaki et al., 2020).
Transposon insertions in the PAO1 chromosome were generated as previously reported (Jacobs et al., 2003), with some modifications. Transposon insertions in the PAO1 chromosome were generated by mating P. aeruginosa PAO1 with β2163/pIT2 (ISlacZ/hah insertions).
Mating was performed for 4 h at 37°C on LB agar containing DAPA. Mutagenized cells were selected on LB agar containing 60 μg/ml tetracycline, 20 μg/ml chloramphenicol, and 40 μg/ml 5-bromo-4-chloro-3-indolyl-β-D-galactoside (X-gal; Wako) and used for screening.
The screening of strains with altered vesicle production was performed through two steps. In the first screening, P. aeruginosa wild-type strain and transposon mutants were inoculated in 200 μl of LB culture in 96-well plates and grown statically at 37°C for 16 h. To obtain colonies, cell cultures were plated on square LB agar plates containing 60 μg/ml tetracycline using a 96-pin microplate replicator and incubated overnight at 37°C. The colonies were temporarily stored at 4°C. After measuring OD600, cultures were transferred to 96-well PCR plates (FastGene Ultra Easy Cut; NIPPON Genetics) and centrifuged at 2,576 × g for 30 min at 4°C to remove cells. The supernatant (50 μl) was mixed with 50 μl of 10 μg/ml FM4-64 in fresh 96-well plates and incubated at 37°C for 30 min in the dark. Fluorescence was measured at 506/750 nm (excitation/emission) using a microplate reader. Fresh LB was used as the standard. Vesicle production was expressed as the amount of phospholipid normalized to the cell density. In the second screening, transposon mutants that showed alterations in MV production were used, and vesicle production was measured in triplicate as in the first screening.
Transposon insertion sites were determined according to a previous study (Jacobs et al., 2003) with some modifications. Two-stage PCR and sequencing were performed. For PCR experiments, KOD One PCR Master Mix (TOYOBO) and primer pairs (lacZ-211, CEKG2A, CEKG2B, CEKG2C for the first round, and lacZ-148 and CEKG4 for the second round) were used to amplify the DNA fragments. In the first round of PCR, the colony suspension was used as a template. In the second round, the PCR products from the first round were used as templates. The PCR products from the second round were cleaned up by using the FastGene Gel/PCR Extraction Kit (NIPPON Genetics Co., Ltd.) and used as sequencing templates with the primers shown in Supplementary Table 1. Sequencing analysis was performed by Eurofins.
A DNA fragment containing the open reading frame of pslA was amplified by PCR using primers pslA-F-Hind3 and pslA-R-Xba1. A broad-host-range vector pBBR1MCS-5 was digested with HinIII and XbaI, and the pslA DNA fragment was inserted into pBBR1MCS-5 by Gibson Assembly (New England Biolabs), yielding plasmid pPslA. Plasmids were transformed into PAO1 and ΔpslA.
To assess cell attachment, 96-well microtiter plates were inoculated with 200 μl of culture in LB at an OD600 of 0.02 and statically incubated at 37°C for 6 h. Biomass was quantified by crystal violet (CV) staining according as reported previously (O’Toole and Kolter, 1998). After measuring the OD600 using a microplate reader (SpectraMax i3, Molecular Devices), the supernatant was removed, and the wells were washed twice with 250 μl of distilled water. The plates were allowed to dry and 225 μl of 0.1% CV was added to each well. The plates were then incubated at room temperature. Following staining, the wells were washed thrice with 300 μl of distilled water to remove excess CV. After drying, 250 μl of a 4:1 ethanol:acetone solution was added to each well to solubilize the CV, and the absorbance was measured at 570 nm. The absorbance per OD600 was defined as the biofilm formation per cell.
PAO1 wild-type and transposon mutants were inoculated to 200 μl of LB at an initial OD600 of 0.02 in 96-well microtiter plates and grown at 37°C at 200 rpm. Growth curves were measured every hour, and their specific growth rates μ [h–1] were calculated as follows:
Xi and X0 are cell densities measured at 600 nm at each time. ti and t0 indicate the respective time [h].
For all experiments, results were calculated from more than three biological replicates and the average ± standard deviation (SD) is shown. Statistical analyses were conducted using Student’s t-tests or one-way ANOVA for two groups or three groups, respectively.
It is commonly recognized that MV formation increases under biofilm conditions in many bacteria. In this study, we used static culture, in which PAO1 was grown in LB liquid medium in a petri dish as a biofilm model, in contrast to a flask shaking culture as planktonic condition. After incubation overnight, the static culture medium was mucoid, and a viscous film was attached to the bottom. Whole cells and viscous films were scratched from the petri dish, and MVs contained in the supernatant were isolated by ultracentrifugation after cell removal. The ultracentrifuge pellet from the static culture was greater than that from the shaking culture (Figure 1A). Quantification of vesicles with lipid probe FM4-64 revealed that vesicle production was significantly enhanced in the static culture compared to that in the shaking culture (Figure 1B). These results indicate that vesiculation is enhanced in the static culture.
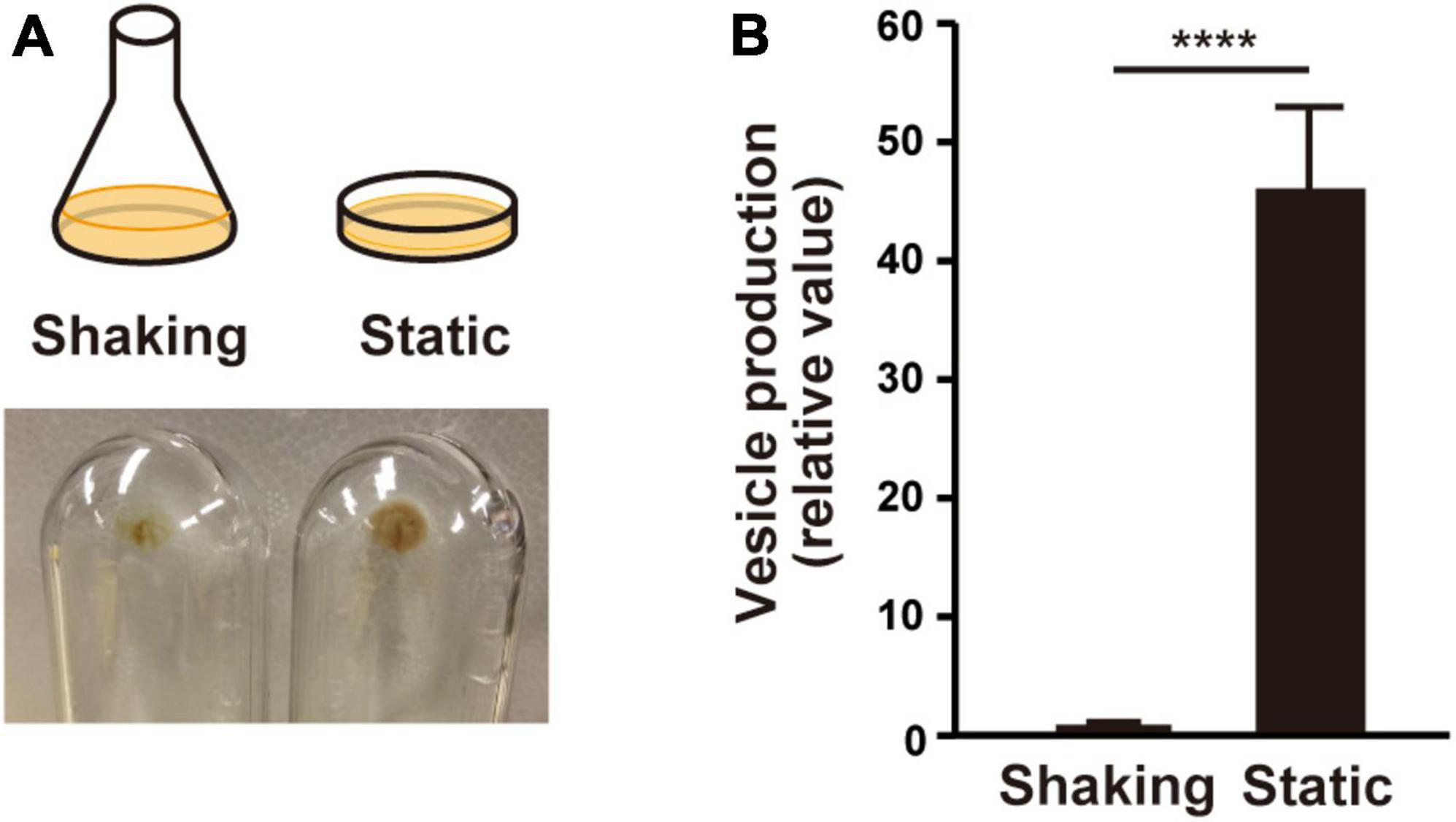
Figure 1. Vesicle formation is increased in liquid static culture in P. aeruginosa PAO1. (A) Graphical representation of culture conditions (upper) and visualization of ultracentrifuged pellets from the supernatant of the cultures from shaking (planktonic) and liquid static (biofilm) conditions after 12 h (lower). (B) Vesicle formation of PAO1. The amount of vesicles extracted from the supernatants was normalized to the cell density, and each value shown is relative to that of the shaking condition. The data are presented as the mean ± SD from three replicates. ****P < 0.0001 (t-tests).
We named the MVs formed under shaking and static culture conditions planktonic MVs (p-MVs) and biofilm MVs (b-MVs), respectively. p-MVs and b-MVs were purified using gradient ultracentrifugation from crude MVs and visualized by TEM and SEM. The results showed no significant changes in their appearance (Figures 2A, B). Nanoparticle tracking analysis (NTA) also showed that the average diameters and distributions between p-MVs and b-MVs were not significantly different (Figure 2C). These results indicate no significant differences in size and shape between p-MVs and b-MVs.
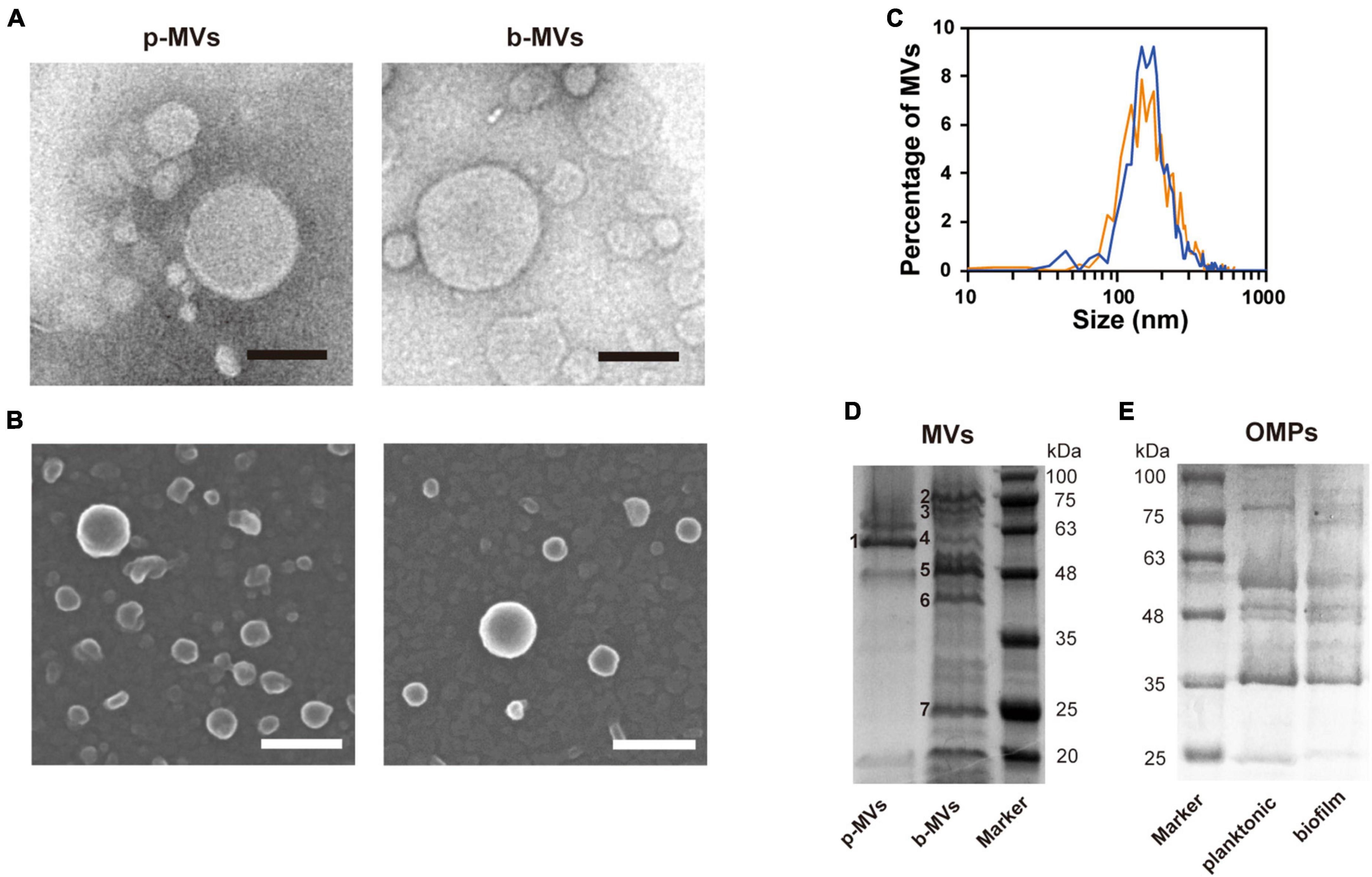
Figure 2. Characteristics and protein patterns of p-MVs and b-MVs. (A) TEM observation of negatively stained purified MVs. Bar, 100 nm. (B) SEM observation of purified MVs. Bar, 100 nm. (C) Nano-tracking analysis of p-MVs (orange) and b-MVs (blue). Protein patterns of MVs (D) and OMPs (E) separated by SDS-PAGE with Coomassie brilliant blue. Inset numbers indicate the identified bands shown in Table 2.
To further investigate whether different growth conditions alter the components of MVs, purified p-MVs and b-MVs with protease inhibitors were separated by SDS-PAGE, and the protein profiles were compared. Some major bands were excised and identified using PMF. Although a major component of p-MVs was leucine aminopeptidase PaAP, consistent with previous reports (Bauman and Kuehn, 2006; Tashiro et al., 2010b), the protein pattern of b-MVs was significantly different (Figure 2D). FptA, PA3923, Esp, OprD, OpdQ, and PA4824 were the major protein components of b-MVs (Table 2). However, no significant differences in the OMPs between planktonic and biofilm cells were observed (Figure 2E), suggesting that the different distribution of proteins in b-MVs is not due to altered OMP profiles. These results indicate that a specific mechanism may induce MV production in the static biofilm condition.
It was of interest to investigate what is the state of the bacteria and how b-MVs interact with bacterial cells in the static culture. We prepared two glasses in the liquid static culture; one was placed vertically, and the other was placed horizontally at the bottom of the culture, and after incubation overnight were visualized using SEM (Figure 3A). On the glass placed vertically, thick biofilms were observed at the air-liquid interface (Figure 3B and Supplementary Figures 1A–D) and the bottom edge of the glass (Figure 3C and Supplementary Figures 1E–G). Fibrous structures formed connections between cells, and b-MVs were observed on both the glass and fibrous structures (Figures 3B, C). These fibrous structures are considered as Psl exopolysaccharides and flagella in PAO1 biofilms according to a previous report (Byrd et al., 2010). A thick biofilm was also observed on the edge of the glass placed horizontally (Figure 3D and Supplementary Figures 1H, I), and b-MVs were intertwined with fibrous structures (Figure 3D). On the other hand, that fibrous structures were not observed on the cell surfaces grown in shaking culture (Supplementary Figure 1J). These results indicate that high MV production is related to biofilm formation in static culture and that MVs function as biofilm components.
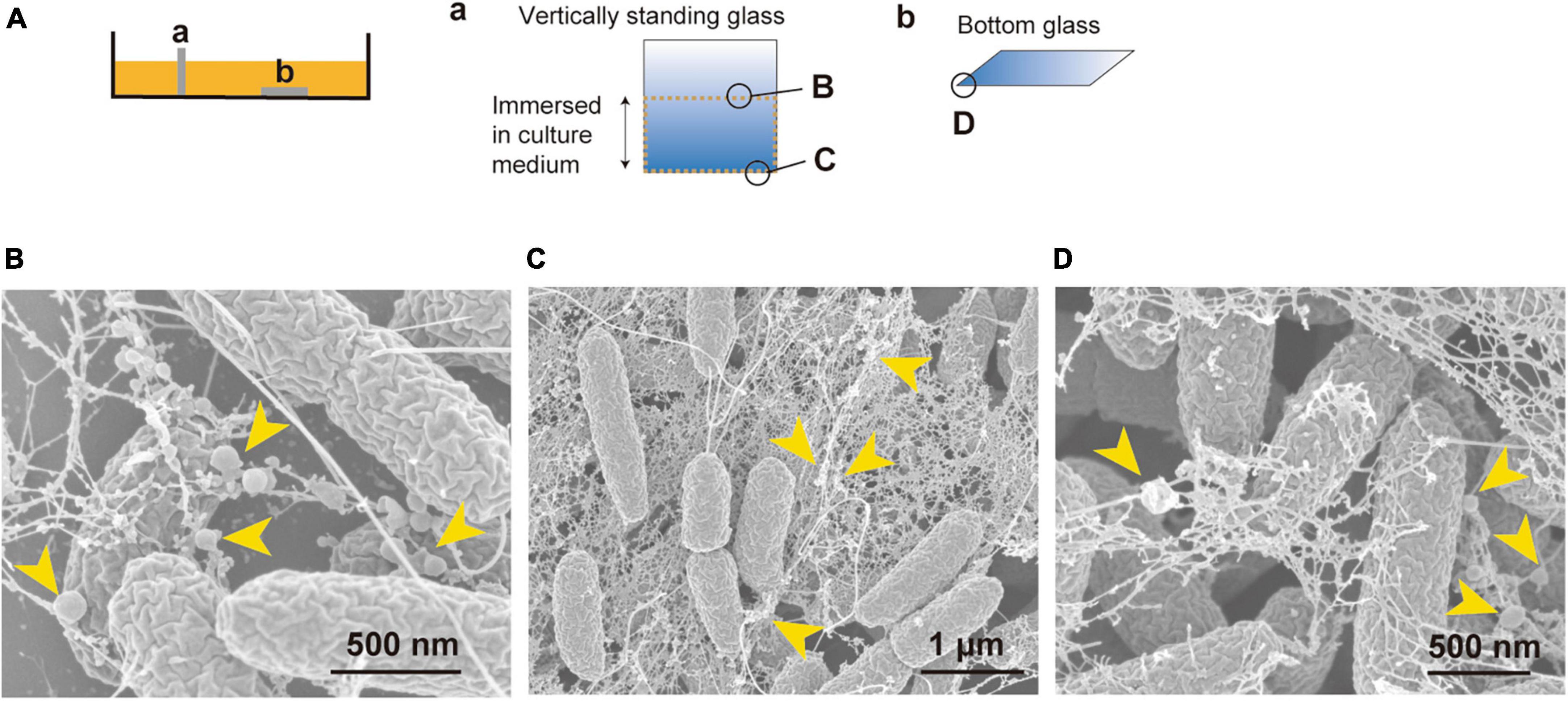
Figure 3. Vesicles are a biofilm component and are associated with filamentous structures in PAO1 liquid static culture. (A) Cover glasses were either vertically standing (a) or placed at the bottom of the static culture (b). Observation areas of B, C, and D are shown in circles. (B–D) Bacterial cells inside biofilm visualized by SEM. The air-liquid interface (B) and the bottom (C) of the vertically standing glass, and the edge of the glass placed at the bottom of the static culture (D) are shown. Vesicles are attached to filamentous structures and are present as a component of the biofilm (yellow arrows). Additional images are shown in Supplementary Figure 1.
We hypothesized that the surface structures in static culture are different from those in the shaking condition, and that these alterations induce b-MV formation. To investigate the spatial structure of the bacterial envelope, we applied quick-freeze deep-etch and replica electron microscopy (QFDE-EM) method to visualize the cellular surface with high spatial and sub-millisecond time resolutions. When cells were grown under shaking conditions, the intracellular compartments, including the presumptive OM, peptidoglycan (PG) and inner membrane (IM) were clearly visualized in the freeze-fractured sections (Figure 4A and Supplementary Figures 2B, C). Cell sections from the static condition showed different surface characteristics; some curvatures of the OM were observed at the surface (Figure 4B and Supplementary Figures 2E, F), suggesting that there are vesicles released from the protrusion of the OM in static biofilm conditions. Furthermore, the OM and IM were separated, and large periplasmic spaces were observed in some cells (Figure 4C and Supplementary Figure 2F); these vesicles are therefore considered to be composed of the OM as a result of blebbing. b-MVs observed under this condition seemed to be released due to alteration of cell envelope structure.
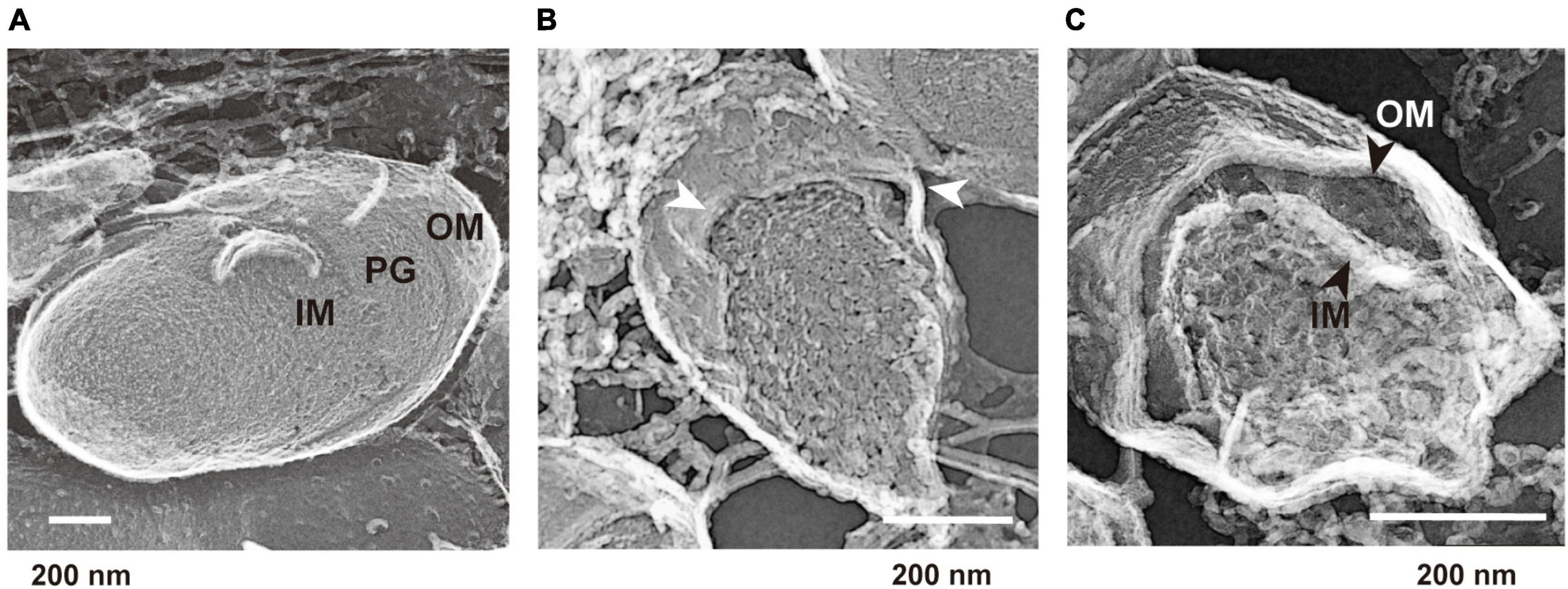
Figure 4. Quick-freeze deep-etch and replica electron micrographs of P. aeruginosa PAO1 cells. (A) An image of cells grown under the shaking condition for 12 h. Outer membrane (OM), inner membrane (IM), and peptidoglycan (PG) are shown at the cross sections. (B,C) Images of cells grown under the static condition for 12 h. The protrusion of the OM is shown by white arrows (B). A broad space between the OM and IM (black arrows) was observed in some cells (C). Additional images are shown in Supplementary Figure 2.
To identify the factors involved in increased MV formation in biofilms, we constructed a P. aeruginosa mutant library using transposon T8 derived from Tn5. The 8,023 transformants obtained were statically grown in LB medium with microtiter plates, and MV production was analyzed with the lipid probe FM4-64. After the first and second screenings, 66 mutants were identified as low-vesiculation strains (2/3 decrease relative to wild type), whereas no mutant was obtained that produced more MVs (twofold increase). The transposon insertion sites of low MV-producing mutants were determined by sequencing and are shown in Table 3.
Many of the genes identified in this screen were involved in the biofilm lifecycle: flagella synthesis and motility (flgG, H, I, J, K, L, fliD, I, K, R, A, motC, D, Y, A, and fleN, R, Q), exopolysaccharide synthesis (pslE, I, K, L, and algC), cyclic diguanylate (c-di-GMP) synthesis (siaC and D), quorum sensing (lasR), and cis-2-decenoic acid synthesis (dspI). In addition, multiple transposon insertions were identified in genes related to metabolic pathways, including the leucine/isovalerate catabolic pathway (liuB and C) and purine metabolism pathway (purC, D, E, L, and M).
To elucidate the interplay between biofilm and MV biogenesis, biofilm formation of the low MV-producing mutants on microtiter plates was quantified using a CV assay. This assay allowed us to identify the factors associated with the initial attachment in biofilm formation. Mutants related to flagella motility (flg genes, fli genes, fle genes, and mot genes) and polysaccharide synthesis (psl genes) showed significantly decreased biofilm formation, as well as MV production (Table 3), suggesting that those biofilm-related factors are also involved in increased MV formation. However, algC::T8, which are involved in exopolysaccharide synthesis, showed high biofilm-forming ability for unknown reasons. Mutants of genes related to purine metabolism (purM, C, L, D, and E) commonly showed decreased MV formation, but no common feature in growth or biofilm formation was observed (Table 3), suggesting that decreased MV formation in static culture is not due to differences in biofilm formation capacity in purine metabolism-related mutants. There were also several strains that seemed to impair growth and lack the energy to produce MVs.
To gain further insight into the effects of biofilm-related factors on MV production, we extracted MVs and quantified MV production per bacterial cell in WT and several mutants under shaking (planktonic) and static (biofilm) conditions. The flagellar synthesis-defective mutant (flgK::T8) and the motility-defective mutant (motA::T8) showed similar MV formation compared to that in the WT under planktonic conditions (Figure 5A), but were significantly repressed under biofilm conditions (Figure 5B), indicating that the induction of MV formation by flagellar motility is a biofilm-specific process.
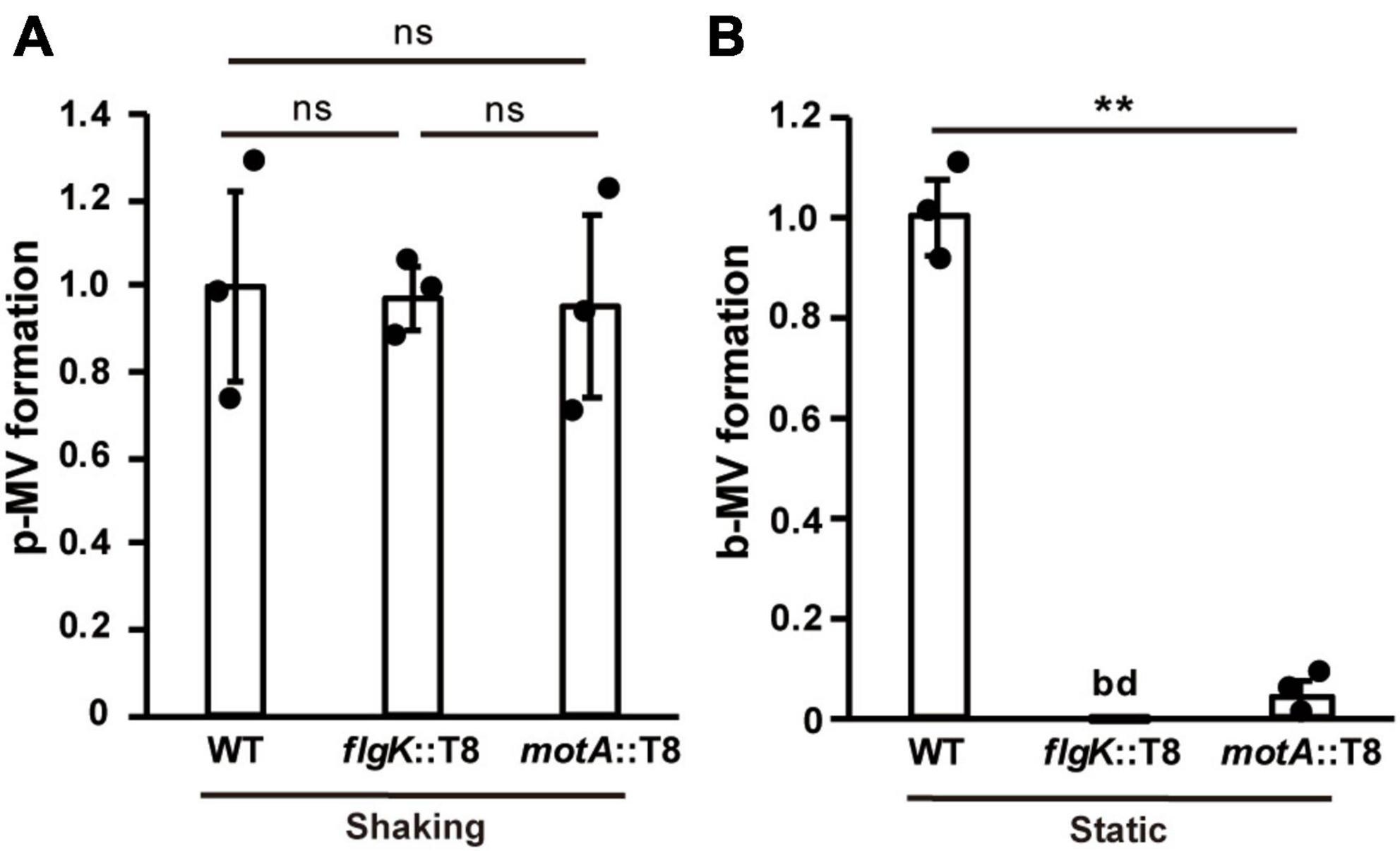
Figure 5. Flagella motility enhances MV formation in biofilms. Planktonic MV (p-MV) and biofilm MV (b-MV) formations of PAO1 wild type (WT), flgK::T8, and motA::T8 were examined under shaking (A) and static biofilm (B) conditions, respectively. The amount of vesicles extracted from the supernatants was normalized to the cell density, and each value shown is relative to that of WT. The data are presented as the mean ± SD from three replicates. **P < 0.01; ns, not significant; bd, below the limit of detection. One-way ANOVA followed by Turkey’s multiple comparison tests was used.
Next, we examined whether the effect of exopolysaccharides on MV formation is altered by culture conditions. P. aeruginosa produces Pel and Psl exopolysaccharides (Ryder et al., 2007). Pel is a glucose-rich, cellulose-like polymer associated with pellicle formation at air-liquid interfaces. Psl is rich in mannose and galactose and is involved in initial attachment (Ghafoor et al., 2011). Biofilm formation in the PAO1 strain is mainly regulated by Psl polysaccharide (Ghafoor et al., 2011; Colvin et al., 2012), but Pel polysaccharide has some influence in the absence of Psl (Tashiro et al., 2014). Therefore, we compared MV formation using a mutant strain ΔpelAΔpslA, in which both genes were deleted in a frame. Biofilm formation by this mutant was significantly repressed (Figure 6A). While MV formation in ΔpelAΔpslA was slightly increased compared to that in WT under shaking conditions (Figure 6B), it was significantly decreased under static conditions (Figure 6C), indicating that exopolysaccharides play a critical role in biofilm-specific MV formation. When we compared the effect of Pel and Psl using the single mutants, both biofilm formation and b-MV formation were decreased in ΔpslA to the same level as the double mutant ΔpelAΔpslA, but not in ΔpelA (Supplementary Figures 3A, B), suggesting that increased MV formation in PAO1 biofilm is dependent on Psl. Complementation analysis showed that MV formation of ΔpslA expressing plasmid-based pslA was at the same level as the control (Supplementary Figure 3C).
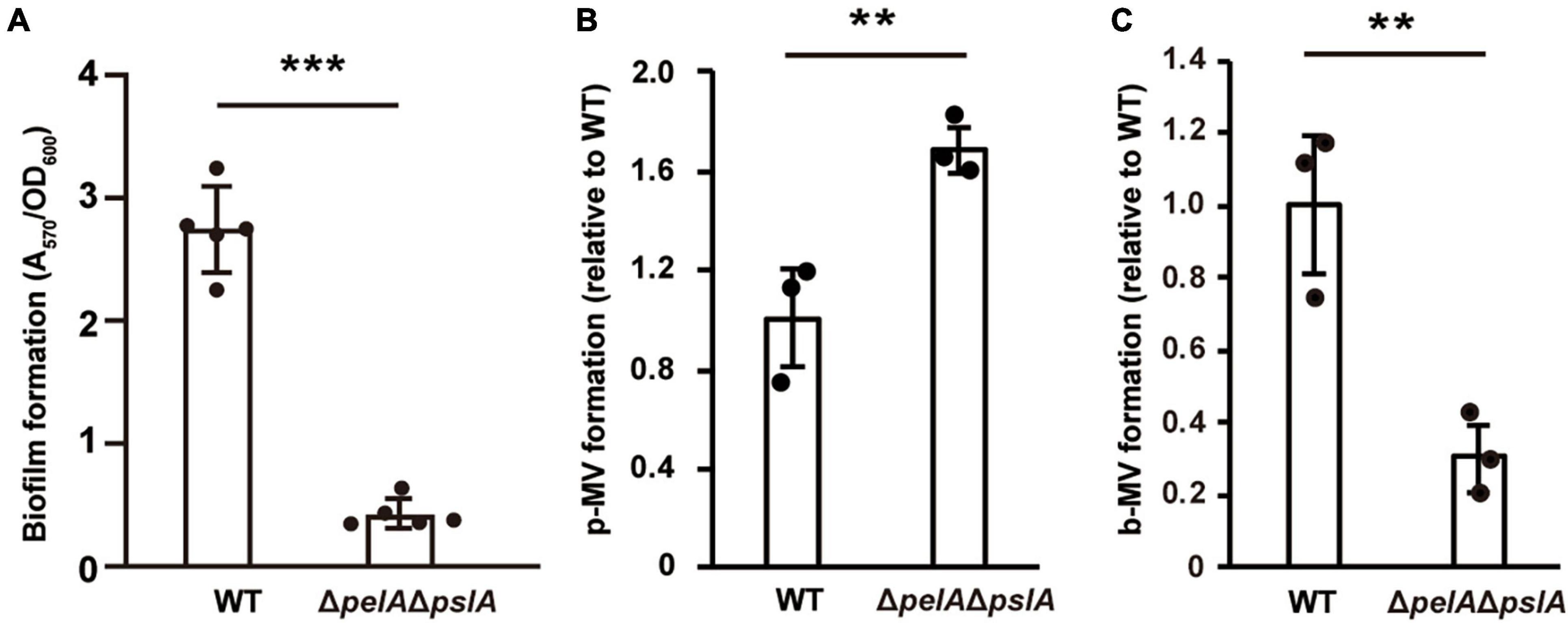
Figure 6. Exopolysaccharides enhance MV formation in static culture. (A) Biofilm formation of WT and ΔpelAΔpslA on microtiter plates. The amount of biofilm was normalized to the cell density. The data are presented as the mean ± SD from five replicates. ***P < 0.001. Planktonic MV (p-MV) and biofilm MV (b-MV) formations of WT and ΔpelAΔpslA were examined under shaking (B) and biofilm (C) conditions, respectively. The amount of vesicles extracted from the supernatants was normalized to cell densities, and each value shown is relative to that of WT. The data are presented as the mean ± SD from three replicates. **P < 0.01 (t-tests).
To further investigate the effect of flagella motility and exopolysaccharides on MV production, we focused on the early stage of biofilm formation. WT and each mutant were grown for 2.5 h under static conditions, in which coverslips were vertically stood, and glass-attached cells were observed by fluorescence microscopy. Bacterial attachments were repressed in low-biofilm forming mutants (flgK::T8, motA::T8, and ΔpelAΔpslA), and staining Psl-specific HHA-lectin revealed that Psl was actively formed in WT, but not synthesized in flgK::T8, as well as ΔpelAΔpslA (Supplementary Figure 4). The Psl was detected in the glass-attached cells of motA::T8, but the signal level was significantly lower than that of WT. When bacterial cells attached to glass were stained with FM4-64, active MV production was observed on the coverslips in both WT and mutants (Figure 7). MVs on the cellular surface were rarely observed in free-living cells, not attached to the glass, in any of the samples (Figure 7). These results suggest that flagellar motility and Psl indirectly enhance MV formation as a result of increasing contact between bacterial cells and surfaces, and surface attachment activates MV blebbing.
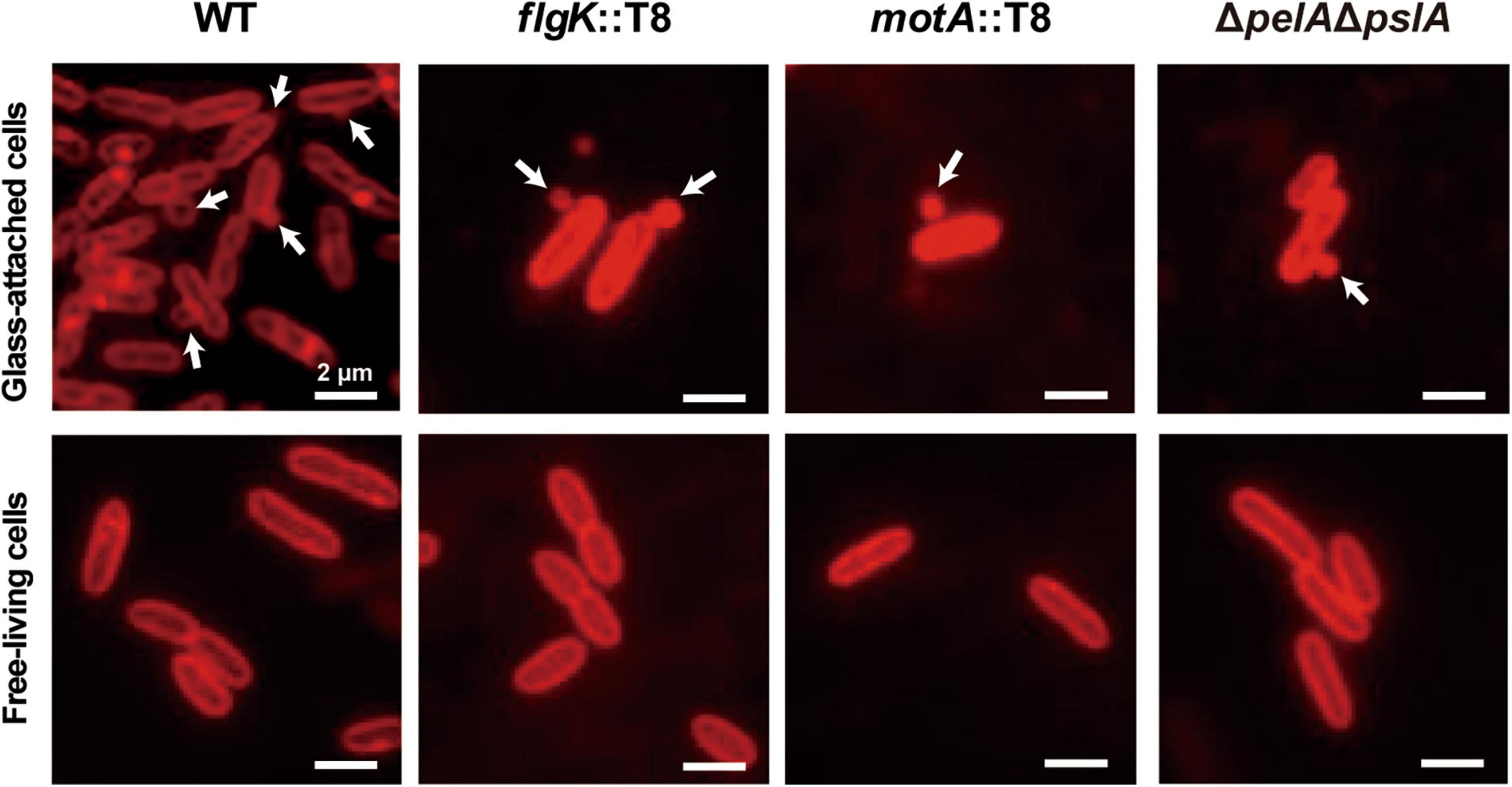
Figure 7. Surface sensing facilitates MV formation. Bacterial cells (PAO1 WT, flgK::T8, motA::T8, and ΔpelAΔpslA) were grown in a static biofilm condition for 2.5 h, and half of coverslips were immersed in each culture during incubation. Glass-attached cells on coverslips (upper row) and free-living cells in cultures (lower row) were stained with FM4-64. Released vesicles are shown by white arrows. Bar, 2 μm.
Membrane vesicles are a component of the biofilm matrix, and their close relationship has long been the focus of attention (Schooling and Beveridge, 2006; Wang et al., 2015). Previous reports showed that the production of MVs is significantly increased under biofilm conditions in several bacteria (Schooling and Beveridge, 2006; Yonezawa et al., 2009; Grande et al., 2017), and MVs regulate biofilm morphology (Esoda and Kuehn, 2019; Cooke et al., 2020); however, the mechanisms of increased MVs in biofilms are not fully understood. In this study, we comprehensively identified the factors involved in enhanced MV production in static biofilms in LB medium by screening a transposon mutant library from the P. aeruginosa PAO1 strain. We identified 55 genes required for enhanced MV formation in static culture where biofilm is formed.
The liquid static culture used in this study showed a significant enhancement in MV formation compared to the shaking culture conditions (Figure 1). Furthermore, this static condition was suitable for screening altered MV-forming mutants and the relationship between initial attachment and MV formation, in contrast to the colony-biofilm. Observation of the static culture showed thick biofilm formation with MVs at the pellicle and edge of the cover glass (Figure 3). Based on a previous report, it is estimated that the fibrous substance entangling cells and MVs is Psl (Byrd et al., 2010), the main component of the biofilm matrix in P. aeruginosa. Psl appears to be used by cells to aggregate and adhere to surfaces.
Notably, the observation of the bacterial envelope in the biofilm state by QFDE-EM revealed several curvatures of OM and the separation of OM and IM (Figures 4B, C and Supplementary Figures 2E, F). Such bacterial plasmolysis, which causes a large gap in the periplasmic space, has been observed in a hypervesiculating E. coli mutant (ΔmlaEΔnlpI) (Ojima et al., 2021) and under hypervesiculating conditions (presence of 1% glycine) (Aktar et al., 2021), suggesting that plasmolysis is one of the key factors for increased MV formation in biofilms. Furthermore, the MV protein composition is significantly different between p-MVs and b-MVs (Figure 2D and Table 2), and biofilm-specific regulation drives enhanced MV formation.
Using a transposon mutant library of PAO1, we isolated 66 low-MV producing mutants in P. aeruginosa biofilms, but no high-MV producing mutant strain was obtained, suggesting that the static biofilm is high-vesiculating condition in P. aeruginosa (Table 3). This result is different from a previous study that screened altered MV-forming E. coli mutants; only a few low-vesiculation mutants were obtained, although there were many hypervesiculating mutants which lack components of the OM, peptidoglycan synthesis, or the σE envelope stress pathways (McBroom et al., 2006). In the present study, we found that mutations in flagellar motility, exopolysaccharide synthesis, metabolic pathways, two-component systems, and signal synthesis reduced MV production in static biofilms. Examining the effect of these genes constructing mutations with non-polar effect will yield further insight regarding the contribution of specific factors to MV formation under biofilm conditions.
As both flagellar motility and exopolysaccharide (Psl) synthesis are involved in the initial attachment of biofilm formation (Byrd et al., 2009; Leid et al., 2009), we hypothesized that surface attachment triggers MV formation under biofilm conditions. To test this hypothesis, fluorescence microscopy observations of the early stage of WT and each mutant biofilm cells with membrane staining was conducted. It revealed that cells attached to coverslips produced MVs more actively than free-living cells in liquid static culture (Figure 7). Flagella motility and exopolysaccharide synthesis-defective mutants, which showed low MV production in static culture, also produced MVs on coverslips, suggesting that surface sensing is closely related to MV production. This observation is consistent with a previous result by Byrd et al. (2010); protrusions approximately 100 nm in diameter were observed on the membrane of adherent P. aeruginosa PAO1, but a similar structure was not found in its Δpsl mutant. During initial attachment, bacterial cells sense surface attachment through various pathways, including pili, signal receptions, two-component regulatory systems, and envelope stress on the cell wall (O’Toole and Wong, 2016; Chang, 2017). Although the details of the mechanism are unclear, it appears that these factors are intimately involved in the induction of OM curvature and blebbing.
We also found that several mutants related to biofilm formation and maintenance showed low vesiculation in static culture. Two mutants with disruptions in siaC and siaD, which are genes important for c-di-GMP synthesis (Chen et al., 2020), were identified with low vesiculation and low biofilm formation. c-di-GMP is an intracellular signaling molecule and a key signal for P. aeruginosa to switch from planktonic to attachment states during the early stage of biofilm formation, and extracellular polysaccharide synthesis is enhanced with increased c-di-GMP levels (Romling et al., 2013). While the direct relationship has not been fully investigated, the intracellular level of c-di-GMP may be associated with MV production in biofilms.
In contrast, mutations in lasR and dspI resulted in low MV formation, although the ability to form biofilms were not significantly different from WT. Las is one of the quorum sensing systems using N-(3-oxo-dodecanoyl) L-homoserine lactone (3-oxo-C12-HSL), and the LasR and 3-oxo-C12-HSL complex activates transcription related to virulence factors (Parsek and Greenberg, 2000). DspI is a putative enoyl-coenzyme A (CoA) hydratase/isomerase that is involved in the synthesis of cis-2-decenoic acid (CDA), which regulates biofilm dispersion and the transcription of many genes related to motility and virulence (Amari et al., 2013; Rahmani-Badi et al., 2015). Since these mutants showed low production of MVs without suppressing biofilm formation, it is possible that the genes are involved in the induction of MV formation in biofilms and should be analyzed in the future.
There were six low-vesiculation mutants of purine nucleotide metabolism and their growth was impaired. The de novo purine is essential for both DNA and RNA production, with purines playing an important role in energy generation (ATP and GTP) and signal molecule synthesis (c-di-GMP, cAMP, and (p)ppGpp) (Jewett et al., 2009). Energy storage and signaling or global changes in metabolism are likely to be involved in MV production in biofilms.
Furthermore, three low-vesiculation mutants of the leucine/isovalerate utilization (Liu) pathway had no effect on growth. Acetyl-CoA, the end product of the Liu pathway, is used for the tricarboxylic acid cycle and glyoxylate cycle (Hoschle et al., 2005; Diaz-Perez et al., 2018). Leucine is one of the biofilm-promoting amino acids in P. aeruginosa (Bernier et al., 2011), but the effect of these genes on biofilm formation was not significant in this study. These factors are also potential candidates for direct involvement in MV formation in biofilms.
Biofilm formation is the optimal growth mode for P. aeruginosa when chronically infecting host epithelial cells, and enhanced MV formation in this state is a sophisticated infection strategy to efficiently deliver virulence factors to the host. Indeed, FptA, EprS, OprD, and OprQ were abundant in b-MVs, while PaAP was a major component of p-MVs. OprD, which is an important porin for the entry of carbapenem and imipenem antibiotics into cells, is abundant in PAO1 biofilm cells and clinical P. aeruginosa isolates (Park et al., 2014a), and is also rich in MVs derived from colony biofilms (Toyofuku et al., 2012; Couto et al., 2015; Park et al., 2015). OprD functions as a receptor for laminin, a major component of the lung extracellular matrix, and in P. aeruginosa it strongly adheres to basal lamina (Paulsson et al., 2019), suggesting that MVs contribute to stabilizing biofilm formation in lung cells during infection. EprS is an autotransporter (AT) protein with serine protease activity that is secreted by the type V secretion system (T5SS) and induces inflammatory responses (Kida et al., 2013). Thus, b-MVs possess a high abundance of proteins that contribute to cell adhesion and inflammation, respectively, and their pathogenicity is crucial to chronic infection.
Although the underlying mechanisms of enhanced MV production in bacterial biofilms are not fully understood, this study revealed significant findings using P. aeruginosa static biofilms. Plasmolysis and OM curvature occur in biofilm cells, and it seems that blebbing is one of the key factors to release MVs actively from OM in static biofilms. In addition, we identified factors involved in enhanced MV production in biofilms. Flagella motility and Psl synthesis indirectly control MV formation in biofilms, and the attachment during the early stage of biofilm formation may be the switch for MV blebbing. Future work will seek to elucidate the mechanisms and factors that directly influence MV production in biofilms.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
MK and YT contributed to the conception and prepared the manuscript. MK, TS, SU, MS, KK, and YT designed the experiments. MK, TS, and SU performed the analyses of vesicle. MK, SU, MT, KH, YOT, MM, and RN contributed the microscopic observations. TS, KH, and MM contributed the protein analyses. MK performed the construction and analyses of transposon mutants. MS, KK, HF, and YT supervised the work. All authors reviewed the manuscript and approved the submitted version.
This work was supported by JST PRESTO (grant number JPMJPR19H8) to YT, JSPS KAKENHI (grant numbers JP19H02920, JP19KK0394, and JP22K19915) to YT, and JST CREST (grant number JPMJCR19S5) to MM. MK was supported by JST, the establishment of university fellowships toward the creation of science technology innovation (grant number JPMJFS2119).
We would like to thank Takako Otsuka, Tomomi Sugaya, and Satomi Aiba (Shizuoka University) for their technical support. We also thank Hirotaka Kobayashi (National Institute of Infectious Diseases) for help with SEM analysis, Junko Shiomi (Osaka Metropolitan University) for QFDE-EM analysis, and Aya Takamori (Osaka Metropolitan University) for PMF analysis. Professor Colin Manoil (University of Washington) provided the plasmid pIT2.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1252155/full#supplementary-material
Aktar, S., Okamoto, Y., Ueno, S., Tahara, Y. O., Imaizumi, M., Shintani, M., et al. (2021). Incorporation of plasmid DNA into bacterial membrane vesicles by peptidoglycan defects in Escherichia coli. Front. Microbiol. 12:747606. doi: 10.3389/fmicb.2021.747606
Altindis, E., Fu, Y., and Mekalanos, J. J. (2014). Proteomic analysis of Vibrio cholerae outer membrane vesicles. Proc. Natl. Acad. Sci. U. S. A. 111, E1548–E1556. doi: 10.1073/pnas.1403683111
Amari, D. T., Marques, C. N. H., and Davies, D. G. (2013). The putative Enoyl-Coenzyme A hydratase DspI is required for production of the Pseudomonas aeruginosa biofilm dispersion autoinducer cis-2-decenoic acid. J. Bacteriol. 195, 4600–4610. doi: 10.1128/JB.00707-13
Baeza, N., and Mercade, E. (2020). Relationship between membrane vesicles, extracellular ATP and biofilm formation in antarctic Gram-negative bacteria. Microbial Ecol. 81, 645–656. doi: 10.1007/s00248-020-01614-6
Bauman, S. J., and Kuehn, M. J. (2006). Purification of outer membrane vesicles from Pseudomonas aeruginosa and their activation of an IL-8 response. Microbes Infect. 8, 2400–2408. doi: 10.1016/j.micinf.2006.05.001
Bauman, S. J., and Kuehn, M. J. (2009). Pseudomonas aeruginosa vesicles associate with and are internalized by human lung epithelial cells. BMC Microbiol. 9:26. doi: 10.1186/1471-2180-9-26
Baumgarten, T., Sperling, S., Seifert, J., Von Bergen, M., Steiniger, F., Wick, L. Y., et al. (2012). Membrane vesicle formation as a multiple-stress response mechanism enhances Pseudomonas putida DOT-T1E cell surface hydrophobicity and biofilm formation. Appl. Environ. Microbiol. 78, 6217–6224. doi: 10.1128/AEM.01525-12
Bernier, S. P., Ha, D.-G., Khan, W., Merritt, J. H., and O’toole, G. A. (2011). Modulation of Pseudomonas aeruginosa surface-associated group behaviors by individual amino acids through c-di-GMP signaling. Res. Microbiol. 162, 680–688. doi: 10.1016/j.resmic.2011.04.014
Beveridge, T. J., Makin, S. A., Kadurugamuwa, J. L., and Li, Z. (1997). Interactions between biofilms and the environment. FEMS Microbiol. Rev. 20, 291–303. doi: 10.1111/j.1574-6976.1997.tb00315.x
Bomberger, J. M., Maceachran, D. P., Coutermarsh, B. A., Ye, S., O’toole, G. A., and Stanton, B. A. (2009). Long-distance delivery of bacterial virulence factors by Pseudomonas aeruginosa outer membrane vesicles. PLoS Pathog. 5:e1000382. doi: 10.1371/journal.ppat.1000382
Byrd, M. S., Pang, B., Mishra, M., Swords, W. E., and Wozniak, D. J. (2010). The Pseudomonas aeruginosa exopolysaccharide Psl facilitates surface adherence and NF-kappaB activation in A549 cells. mBio 1:e00140-10. doi: 10.1128/mBio.00140-10
Byrd, M. S., Sadovskaya, I., Vinogradov, E., Lu, H., Sprinkle, A. B., Richardson, S. H., et al. (2009). Genetic and biochemical analyses of the Pseudomonas aeruginosa Psl exopolysaccharide reveal overlapping roles for polysaccharide synthesis enzymes in Psl and LPS production. Mol. Microbiol. 73, 622–638. doi: 10.1111/j.1365-2958.2009.06795.x
Carriquiriborde, F., Martin Aispuro, P., Ambrosis, N., Zurita, E., Bottero, D., Gaillard, M. E., et al. (2021). Pertussis vaccine candidate based on outer membrane vesicles derived from biofilm culture. Front. Immunol. 12:730434. doi: 10.3389/fimmu.2021.730434
Chang, C. Y. (2017). Surface sensing for biofilm formation in Pseudomonas aeruginosa. Front. Microbiol. 8:2671. doi: 10.3389/fmicb.2017.02671
Chen, G., Gan, J., Yang, C., Zuo, Y., Peng, J., Li, M., et al. (2020). The SiaA/B/C/D signaling network regulates biofilm formation in Pseudomonas aeruginosa. EMBO J. 39:e103412. doi: 10.15252/embj.2019103412
Colvin, K. M., Irie, Y., Tart, C. S., Urbano, R., Whitney, J. C., Ryder, C., et al. (2012). The Pel and Psl polysaccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix. Environ. Microbiol. 14, 1913–1928. doi: 10.1111/j.1462-2920.2011.02657.x
Cooke, A. C., Florez, C., Dunshee, E. B., Lieber, A. D., Terry, M. L., Light, C. J., et al. (2020). Pseudomonas quinolone signal-induced outer membrane vesicles enhance biofilm dispersion in Pseudomonas aeruginosa. mSphere 5:e01109-20. doi: 10.1128/mSphere.01109-20
Costerton, J., Stewart, P., and Greenberg, E. (1999). Bacterial biofilms: A common cause of persistent infections. Science 284, 1318–1322. doi: 10.1126/science.284.5418.1318
Couto, N., Schooling, S. R., Dutcher, J. R., and Barber, J. (2015). Proteome profiles of outer membrane vesicles and extracellular matrix of Pseudomonas aeruginosa biofilms. J. Proteome Res. 14, 4207–4222. doi: 10.1021/acs.jproteome.5b00312
Deatherage, B. L., Lara, J. C., Bergsbaken, T., Barrett, S. L. R., Lara, S., and Cookson, B. T. (2009). Biogenesis of bacterial membrane vesicles. Mol. Microbiol. 72, 1395–1407. doi: 10.1111/j.1365-2958.2009.06731.x
Demarre, G. L., GueìRout, A.-M., Matsumoto-Mashimo, C., Rowe-Magnus, D. A., MarlieÌRe, P., and Mazel, D. (2005). A new family of mobilizable suicide plasmids based on broad host range R388 plasmid (IncW) and RP4 plasmid (IncPα) conjugative machineries and their cognate Escherichia coli host strains. Res. Microbiol. 156, 245–255. doi: 10.1016/j.resmic.2004.09.007
Diaz-Perez, A. L., Nunez, C., Meza Carmen, V., and Campos-Garcia, J. (2018). The expression of the genes involved in leucine catabolism of Pseudomonas aeruginosa is controlled by the transcriptional regulator LiuR and by the CbrAB/Crc system. Res. Microbiol. 169, 324–334. doi: 10.1016/j.resmic.2018.05.004
Elhenawy, W., Bording-Jorgensen, M., Valguarnera, E., Haurat, M. F., Wine, E., and Feldman, M. F. (2016). LPS remodeling triggers formation of outer membrane vesicles in Salmonella. mBio 7:e00940-16. doi: 10.1128/mBio.00940-16
Ellis, T. N., Leiman, S. A., and Kuehn, M. J. (2010). Naturally produced outer Membrane vesicles from Pseudomonas aeruginosa elicit a potent innate immune response via combined sensing of both lipopolysaccharide and protein components. Infect. Immun. 78, 3822–3831. doi: 10.1128/IAI.00433-10
Esoda, C. N., and Kuehn, M. J. (2019). Pseudomonas aeruginosa leucine aminopeptidase influences early biofilm composition and structure via vesicle-associated antibiofilm activity. mBio 10:e02548-19. doi: 10.1128/mBio.02548-19
Ghafoor, A., Hay, I. D., and Rehm, B. H. A. (2011). Role of exopolysaccharides in Pseudomonas aeruginosa biofilm formation and architecture. Appl. Environ. Microbiol. 77, 5238–5246. doi: 10.1128/AEM.00637-11
Grande, R., Celia, C., Mincione, G., Stringaro, A., Di Marzio, L., Colone, M., et al. (2017). Detection and physicochemical characterization of membrane vesicles (MVs) of Lactobacillus reuteri DSM 17938. Front. Microbiol. 8:1040. doi: 10.3389/fmicb.2017.01040
Grande, R., Di Marcantonio, M. C., Robuffo, I., Pompilio, A., Celia, C., Di Marzio, L., et al. (2015). Helicobacter pylori ATCC 43629/NCTC 11639 outer membrane vesicles (OMVs) from biofilm and planktonic phase associated with extracellular DNA (eDNA). Front. Microbiol. 6:1369. doi: 10.3389/fmicb.2015.01369
Hall-Stoodley, L., Costerton, J., and Stoodley, P. (2004). Bacterial biofilm: From the nature environment to infectious diseases. Nat. Rev. Microbiol. 2, 95–108. doi: 10.1038/nrmicro821
Hobley, L., Harkins, C., Macphee, C. E., and Stanley-Wall, N. R. (2015). Giving structure to the biofilm matrix: An overview of individual strategies and emerging common themes. FEMS Microbiol. Rev. 39, 649–669. doi: 10.1093/femsre/fuv015
Holloway, B. W., Krishnapillai, V., and Morgan, A. F. (1979). Chromosomal genetics of Pseudomonas. Microbiol. Rev. 43, 73–102. doi: 10.1128/mr.43.1.73-102.1979
Hoschle, B., Gnau, V., and Jendrossek, D. (2005). Methylcrotonyl-CoA and geranyl-CoA carboxylases are involved in leucine/isovalerate utilization (Liu) and acyclic terpene utilization (Atu), and are encoded by liuB/liuD and atuC/atuF, in Pseudomonas aeruginosa. Microbiology 151, 3649–3656. doi: 10.1099/mic.0.28260-0
Ionescu, M., Zaini, P. A., Baccari, C., Tran, S., Da Silva, A. M., and Lindow, S. E. (2014). Xylella fastidiosa outer membrane vesicles modulate plant colonization by blocking attachment to surfaces. Proc. Natl. Acad. Sci. U. S. A. 111, E3910–E3918. doi: 10.1073/pnas.1414944111
Jacobs, M. A., Alwood, A., Thaipisuttikul, I., Spencer, D., Haugen, E., Ernst, S., et al. (2003). Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 100, 14339–14344. doi: 10.1073/pnas.2036282100
Jamal, M., Ahmad, W., Andleeb, S., Jalil, F., Imran, M., Nawaz, M. A., et al. (2018). Bacterial biofilm and associated infections. J. Chin. Med. Assoc. 81, 7–11. doi: 10.1016/j.jcma.2017.07.012
Jewett, M. C., Miller, M. L., Chen, Y., and Swartz, J. R. (2009). Continued protein synthesis at low [ATP] and [GTP] enables cell adaptation during energy limitation. J. Bacteriol. 191, 1083–1091. doi: 10.1128/JB.00852-08
Kida, Y., Taira, J., Yamamoto, T., Higashimoto, Y., and Kuwano, K. (2013). EprS, an autotransporter protein of Pseudomonas aeruginosa, possessing serine protease activity induces inflammatory responses through protease-activated receptors. Cell Microbiol. 15, 1168–1181. doi: 10.1111/cmi.12106
Kovach, M. E., Elzer, P. H., Steven Hill, D., Robertson, G. T., Farris, M. A., Roop, R. M., et al. (1995). Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166, 175–176. doi: 10.1016/0378-1119(95)00584-1
Leid, J. G., Kerr, M., Selgado, C., Johnson, C., Moreno, G., Smith, A., et al. (2009). Flagellum-mediated biofilm defense mechanisms of Pseudomonas aeruginosa against host-derived lactoferrin. Infect. Immun. 77, 4559–4566. doi: 10.1128/IAI.00075-09
Mashburn, L. M., and Whiteley, M. (2005). Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature 437, 422–425. doi: 10.1038/nature03925
McBroom, A., Johnson, A., Vemulapalli, S., and Kuehn, M. (2006). Outer membrane vesicle production by Escherichia coli is independent of membrane instability. J. Bacteriol. 188, 5385–5392. doi: 10.1128/JB.00498-06
McBroom, A. J., and Kuehn, M. J. (2007). Release of outer membrane vesicles by Gram-negative bacteria is a novel envelope stress response. Mol. Microbiol. 63, 545–558. doi: 10.1111/j.1365-2958.2006.05522.x
Murphy, K., Park, A. J., Hao, Y., Brewer, D., Lam, J. S., and Khursigara, C. M. (2014). Influence of O polysaccharides on biofilm development and outer membrane vesicle biogenesis in Pseudomonas aeruginosa PAO1. J. Bacteriol. 196, 1306–1317. doi: 10.1128/JB.01463-13
Ojima, Y., Sawabe, T., Nakagawa, M., Tahara, Y. O., Miyata, M., and Azuma, M. (2021). Aberrant membrane structures in hypervesiculating Escherichia coli strain ΔmlaE ΔnlpI visualized by electron microscopy. Front. Microbiol. 12:706525. doi: 10.3389/fmicb.2021.706525
O’Toole, G. A., and Kolter, R. (1998). Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30, 295–304. doi: 10.1046/j.1365-2958.1998.01062.x
O’Toole, G. A., and Wong, G. C. (2016). Sensational biofilms: Surface sensing in bacteria. Curr. Opin. Microbiol. 30, 139–146. doi: 10.1016/j.mib.2016.02.004
Park, A. J., Murphy, K., Krieger, J. R., Brewer, D., Taylor, P., Habash, M., et al. (2014a). A temporal examination of the planktonic and biofilm proteome of whole cell Pseudomonas aeruginosa PAO1 using quantitative mass spectrometry. Mol. Cell Proteomics 13, 1095–1105. doi: 10.1074/mcp.M113.033985
Park, A. J., Surette, M. D., and Khursigara, C. M. (2014b). Antimicrobial targets localize to the extracellular vesicle-associated proteome of Pseudomonas aeruginosa grown in a biofilm. Front. Microbiol. 5:464. doi: 10.3389/fmicb.2014.00464
Park, A. J., Murphy, K., Surette, M. D., Bandoro, C., Krieger, J. R., Taylor, P., et al. (2015). Tracking the dynamic relationship between cellular systems and extracellular subproteomes in Pseudomonas aeruginosa biofilms. J. Proteome Res. 14, 4524–4537. doi: 10.1021/acs.jproteome.5b00262
Parsek, M. R., and Greenberg, E. P. (2000). Acyl-homoserine lactone quorum sensing in Gram-negative bacteria: A signaling mechanism involved in associations with higher organisms. Proc. Natl. Acad. Sci. U. S. A. 97, 8789–8793. doi: 10.1073/pnas.97.16.8789
Paulsson, M., Su, Y. C., Ringwood, T., Udden, F., and Riesbeck, K. (2019). Pseudomonas aeruginosa uses multiple receptors for adherence to laminin during infection of the respiratory tract and skin wounds. Sci. Rep. 9:18168. doi: 10.1038/s41598-019-54622-z
Pesci, E., Milbank, J., Pearson, J., Mcknight, S., Kende, A., Greenberg, E., et al. (1999). Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 96, 11229–11234. doi: 10.1073/pnas.96.20.11229
Rahmani-Badi, A., Sepehr, S., Fallahi, H., and Heidari-Keshel, S. (2015). Dissection of the cis-2-decenoic acid signaling network in Pseudomonas aeruginosa using microarray technique. Front. Microbiol. 6:383. doi: 10.3389/fmicb.2015.00383
Roier, S., Zingl, F. G., Cakar, F., Durakovic, S., Kohl, P., Eichmann, T. O., et al. (2016). A novel mechanism for the biogenesis of outer membrane vesicles in Gram-negative bacteria. Nat. Commun. 7:10515. doi: 10.1038/ncomms10515
Romling, U., Galperin, M. Y., and Gomelsky, M. (2013). Cyclic di-GMP: The first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. 77, 1–52. doi: 10.1128/MMBR.00043-12
Ryder, C., Byrd, M., and Wozniak, D. (2007). Role of polysaccharides in Pseudomonas aeruginosa biofilm development. Curr. Opin. Microbiol. 10, 644–648. doi: 10.1016/j.mib.2007.09.010
Schertzer, J. W., and Whiteley, M. (2012). A bilayer-couple model of bacterial outer membrane vesicle biogenesis. mBio 3:e00297-11. doi: 10.1128/mBio.00297-11
Schooling, S., and Beveridge, T. (2006). Membrane vesicles: An overlooked component of the matrices of biofilms. J. Bacteriol. 188, 5945–5957. doi: 10.1128/JB.00257-06
Schooling, S., Hubley, A., and Beveridge, T. (2009). Interactions of DNA with biofilm-derived membrane vesicles. J. Bacteriol. 191, 4097–4102. doi: 10.1128/JB.00717-08
Schwechheimer, C., Rodriguez, D. L., and Kuehn, M. J. (2015). NlpI-mediated modulation of outer membrane vesicle production through peptidoglycan dynamics in Escherichia coli. MicrobiologyOpen 4, 375–389. doi: 10.1002/mbo3.244
Suzuki, H., Nishimura, Y., Yasuda, S., Nishimura, A., Yamada, M., and Hirota, Y. (1978). Murein-lipoprotein of Escherichia coli: A protein involved in the stabilization of bacterial cell envelope. Mol. Gen. Genet. 167, 1–9. doi: 10.1007/BF00270315
Takaki, K., Tahara, Y. O., Nakamichi, N., Hasegawa, Y., Shintani, M., Ohkuma, M., et al. (2020). Multilamellar and multivesicular outer membrane vesicles produced by a Buttiauxella agrestis tolB mutant. Appl. Environ. Microbiol. 86:e01131-20. doi: 10.1128/AEM.01131-20
Tashiro, Y., Ichikawa, S., Nakajima-Kambe, T., Uchiyama, H., and Nomura, N. (2010a). Pseudomonas quinolone signal affects membrane vesicle production in not only Gram-negative but also Gram-positive bacteria. Microbes Environ. 25, 120–125. doi: 10.1264/jsme2.ME09182
Tashiro, Y., Ichikawa, S., Shimizu, M., Toyofuku, M., Takaya, N., Nakajima-Kambe, T., et al. (2010b). Variation of physiochemical properties and cell association activity of membrane vesicles with growth phase in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 76, 3732–3739. doi: 10.1128/AEM.02794-09
Tashiro, Y., Inagaki, A., Ono, K., Inaba, T., Yawata, Y., Uchiyama, H., et al. (2014). Low concentrations of ethanol stimulate biofilm and pellicle formation in Pseudomonas aeruginosa. Biosci. Biotechnol. Biochem. 78, 178–181. doi: 10.1080/09168451.2014.877828
Tashiro, Y., Nomura, N., Nakao, R., Senpuku, H., Kariyama, R., Kumon, H., et al. (2008). Opr86 is essential for viability and is a potential candidate for a protective antigen against biofilm formation by Pseudomonas aeruginosa. J. Bacteriol. 190, 3969–3978. doi: 10.1128/JB.02004-07
Tashiro, Y., Sakai, R., Toyofuku, M., Sawada, I., Nakajima-Kambe, T., Uchiyama, H., et al. (2009). Outer membrane machinery and alginate synthesis regulators control membrane vesicle production in Pseudomonas aeruginosa. J. Bacteriol. 191, 7509–7519. doi: 10.1128/JB.00722-09
Tashiro, Y., Uchiyama, H., and Nomura, N. (2012). Multifunctional membrane vesicles in Pseudomonas aeruginosa. Environ. Microbiol. 14, 1349–1362. doi: 10.1111/j.1462-2920.2011.02632.x
Toyofuku, M., Roschitzki, B., Riedel, K., and Eberl, L. (2012). Identification of proteins associated with the Pseudomonas aeruginosa biofilm extracellular matrix. J. Proteome Res. 11, 4906–4915. doi: 10.1021/pr300395j
Toyofuku, M., Tashiro, Y., Hasegawa, Y., Kurosawa, M., and Nomura, N. (2015). Bacterial membrane vesicles, an overlooked environmental colloid: Biology, environmental perspectives and applications. Adv. Colloid Interface Sci. 226, 65–77. doi: 10.1016/j.cis.2015.08.013
Turnbull, L., Toyofuku, M., Hynen, A. L., Kurosawa, M., Pessi, G., Petty, N. K., et al. (2016). Explosive cell lysis as a mechanism for the biogenesis of bacterial membrane vesicles and biofilms. Nat. Commun. 7:11220. doi: 10.1038/ncomms11220
Wang, W., Chanda, W., and Zhong, M. (2015). The relationship between biofilm and outer membrane vesicles: A novel therapy overview. FEMS Microbiol. Lett. 362:fnv117. doi: 10.1093/femsle/fnv117
Keywords: membrane vesicles, biofilm, Pseudomonas aeruginosa, Psl, flagellar motility
Citation: Kanno M, Shiota T, Ueno S, Takahara M, Haneda K, Tahara YO, Shintani M, Nakao R, Miyata M, Kimbara K, Futamata H and Tashiro Y (2023) Identification of genes involved in enhanced membrane vesicle formation in Pseudomonas aeruginosa biofilms: surface sensing facilitates vesiculation. Front. Microbiol. 14:1252155. doi: 10.3389/fmicb.2023.1252155
Received: 03 July 2023; Accepted: 13 November 2023;
Published: 01 December 2023.
Edited by:
Dipshikha Chakravortty, Indian Institute of Science (IISc), IndiaReviewed by:
Paula Maria Tribelli, National Scientific and Technical Research Council (CONICET), ArgentinaCopyright © 2023 Kanno, Shiota, Ueno, Takahara, Haneda, Tahara, Shintani, Nakao, Miyata, Kimbara, Futamata and Tashiro. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yosuke Tashiro, dGFzaGlyby55b3N1a2VAc2hpenVva2EuYWMuanA=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.