
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Microbiol. , 21 August 2023
Sec. Virology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1251705
This article is part of the Research Topic Respiratory RNA Viruses: Molecular Mechanisms of Viral Replication and Pathogenicity View all 18 articles
The main protease (Mpro) plays a crucial role in coronavirus, as it cleaves viral polyproteins and host cellular proteins to ensure successful replication. In this review, we discuss the preference in the recognition sequence of Mpro based on sequence-based studies and structural information and highlight the recent advances in computational and experimental approaches that have aided in discovering novel Mpro substrates. In addition, we provide an overview of the current understanding of Mpro host substrates and their implications for viral replication and pathogenesis. As Mpro has emerged as a promising target for the development of antiviral drugs, further insight into its substrate specificity may contribute to the design of specific inhibitors.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of the coronavirus disease 2019 (COVID-19) pandemic, is a positive-sense single-stranded RNA virus that utilizes its two cysteine proteases, nsp3/papain-like protease (PLpro), and nsp5/3-chymotrypsin-like protease (3CLpro), to cleave its polyproteins into functional viral proteins required for virus replication (Koudelka et al., 2021; Sabbah et al., 2021). Nsp3 cleaves three distinct sites of nsp1–nsp4, while nsp5 cleaves 11 distinct sites of nsp5–nsp16; thereby nsp5 is also referred to as the main protease (Mpro). Mpro is a conserved protease in the family Coronaviridae (Ullrich and Nitsche, 2020; Xiong et al., 2021). The mature Mpro is a dimeric cysteine protease and its catalytic dyad is formed by His41 and Cys145 (Ullrich and Nitsche, 2020; Hu et al., 2022). Besides viral polyproteins, viral proteases likewise cleave host proteins to hinder host immune responses and promote viral replication (Pablos et al., 2021). In this review, we first address the substrate specificity and further analyze the implication of Mpro cleavage on host substrates in various biological processes.
The substrate specificity of SARS-CoV Mpro has been previously investigated. The recombinant protein substrates with saturation mutagenesis at each of the P5 to P3’ positions were used to profile the sequence preference of Mpro substrates (Chuck et al., 2010). In addition, the 11 autoproteolytic cleavage site sequences in SARS-CoV-2 pp1ab and host substrates were applied to analyze the sequence logo of the cleavage site. Thus far, the consensus sequence motif of Mpro substrates is recognized as (L/F/M)-Q↓(S/A/G/N), where ↓ is the cleavage site. In brief, this motif is composed of a conserved P1 residue Gln flanked by a hydrophobic (Leu, Phe, or Val) at P2 and a small aliphatic amino acid (Ser, Asn, Gly, or Ala) at P1’ positions (Miczi et al., 2020; Koudelka et al., 2021; Moustaqil et al., 2021; Pablos et al., 2021; Zhang et al., 2021). The P1, P2, and P1’ residues are important to determine substrate specificity, whereas the less conserved P3, P4, and P3’ residues increase the recognition and binding stability of the substrates (Hu et al., 2022). P3 and P3’ positions prefer positively charged residues to negatively charged ones (Chuck et al., 2010). Although Mpro primarily prefers Gln, it has also been found to recognize non-canonical Met or His at the P1 residue (Koudelka et al., 2021; Pablos et al., 2021). The identification of new substrate sequences can aid in the design of specific inhibitors that can target Mpro activity with higher affinity and selectivity.
Computational and experimental methods are widely used for substrate identification. For computational methods, NetCorona 1.0, a publicly available web server originally designed to predict putative SARS-CoV Mpro cleavage sites, has been commonly used for identifying SARS-CoV-2 Mpro substrates (with a suggested threshold of 0.5), since the sequence of SARS-CoV-2 Mpro shares 96% identity with that of SARS-CoV Mpro (Miczi et al., 2020; Zhang et al., 2021; Scott et al., 2022). Another approach is to search for short stretches of homologous human-pathogen protein sequences (SSHHPS) using BLAST analysis, which is based on the principle that the cleavage site sequences found in the viral genome are identical to the cleavage sites on host cell substrates (Miczi et al., 2020). As to experimental methods, a commonly used screening procedure is the liquid chromatography–mass spectrometry (LC–MS)-based terminal amine isotopic labeling of substrates (TAILS) that not only identifies substrates but also their corresponding cleavage sites (Koudelka et al., 2021; Meyer et al., 2021; Pablos et al., 2021). Besides, Moustaqil et al. (2021) screened 71 human innate immune pathway proteins (HIIPs) using the cell-free Leishmania tarentolae protein expression system, which allows the direct visualization in SDS-PAGE of the target protein fused to GFP.
Table 1 lists the host proteins that have been identified as potential substrates for SARS-CoV-2 Mpro through computational or experimental methods, and further supported by the detection of cleaved products. Among the identified substrates, five proteins have available structure data in Protein Data Bank (PDB), while the rest were predicted by AlphaFold (Table 1). Through analysis of the structure information, we observed that the cleavage sites are commonly located in loops or loops connected to α-helixes or β-sheets (Figure 1), suggesting that most of the target sequences are accessible to Mpro. This implies that in addition to the prediction of cleavage sequences, structural analysis is also important for evaluation of the accessibility of putative cleavage sites (Miczi et al., 2020; Moustaqil et al., 2021).
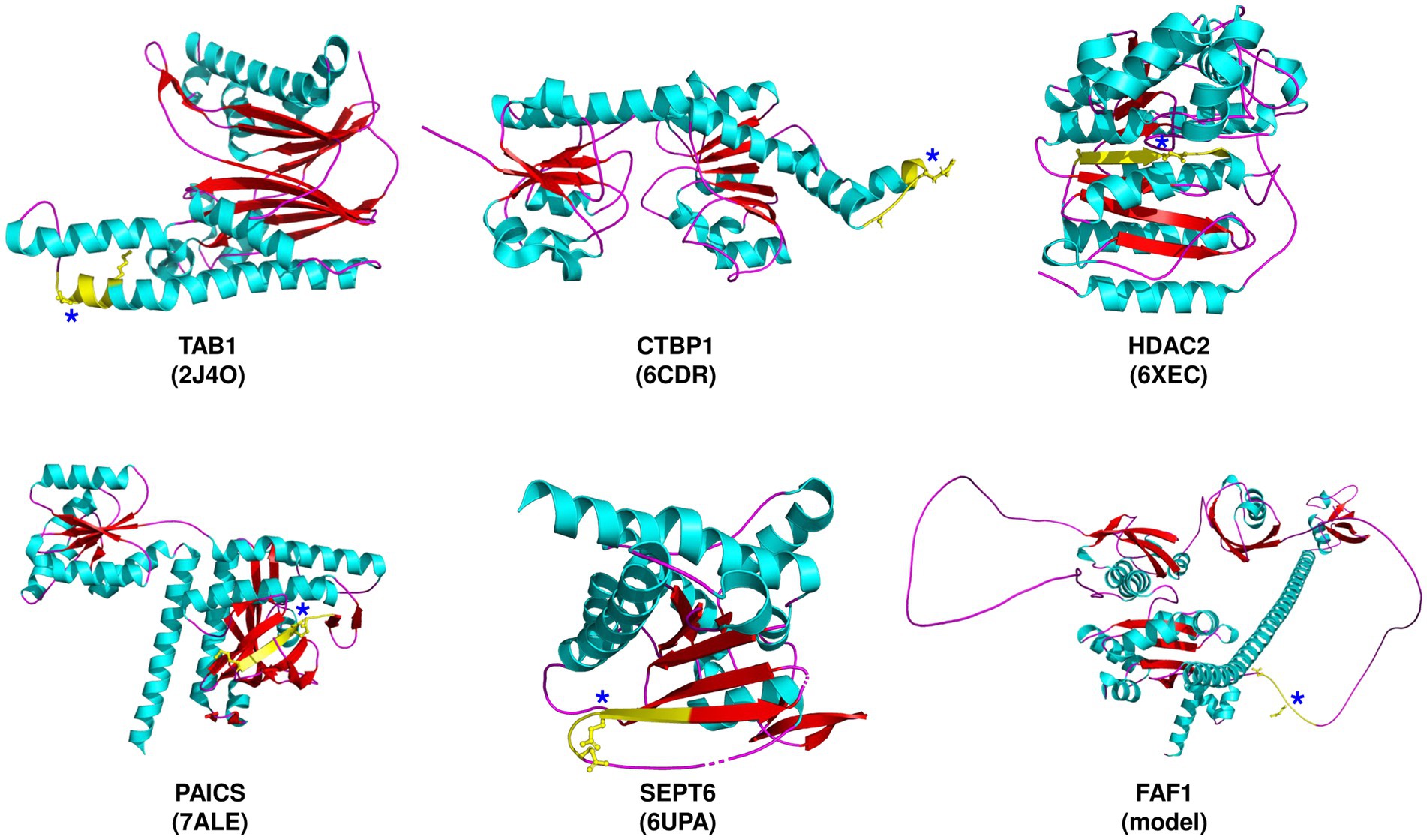
Figure 1. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Mpro cleavage sites in selected target proteins. The proteins are depicted along with the corresponding PDB ID, except for FAF1, which is predicted by AlphaFold. The predicted cleavage sequences (yellow) are shown, with P1 and P5 residues, and an asterisk denoting the P1-Gln residue.
Research on exploring the functional consequences of Mpro cleavage on host proteins is still ongoing. It is important to note that host proteins serve multiple functions, and their dysfunction may have implications for more than one biological process. The implications of Mpro cleavage, according to published information or the known biological function of the substrates, are discussed below.
The innate immune system releases inflammatory cytokines and chemokines as an immediate defense against invading pathogens. However, viruses can manipulate the innate immune response to evade the host’s antiviral defenses (Diamond and Kanneganti, 2022). Mpro was discovered to cleave interleukin-1 receptor-associated kinase 1 (IRAK1), a kinase involved in the regulation of the innate immune response (Miczi et al., 2020). Several viruses such as porcine epidemic diarrhea virus and borna disease virus 1 target IRAK1 to block IRAK1/TRAF6/NF-κB signaling pathway activation, consequently reducing the expression of the IFN-III subtypes, IFN-λ1, and -λ3 (Zhang et al., 2019; Zheng et al., 2022). Notably, inhibition of IRAK1 using pacritinib had effectively attenuated the pro-inflammatory cytokine release triggered by the GU-rich ssRNA sequence derived from the SARS-CoV-2 spike protein (Campbell et al., 2023). Similarly, the SARS-CoV-2 Mpro cleavage of the TAK1 binding protein (TAB1) results in decreased TAB1 protein levels in virus-infected cells and is proposed to inhibit cytokine production by disrupting the interaction between TAB1 and the transforming growth factor-β-activated kinase 1 (TAK1), which is necessary for constitutive activation of NF-κB (Jackson-Bernitsas et al., 2007; Moustaqil et al., 2021; Pablos et al., 2021). mRNA-decapping enzyme 1A (DCP1A), one of the interferon-stimulated genes (ISGs), was recently identified as an Mpro substrate (Song et al., 2023). Cleavage of DCP1A by porcine deltacoronavirus Mpro has been demonstrated to decrease antiviral activity (Zhu et al., 2020). It is conceivable that SARS-CoV-2 Mpro cleaves IRAK1, TAB1, and DCP1A to disturb the production of pro-inflammatory cytokines and attenuate the immune defense (Miczi et al., 2020).
On the other hand, hyperinflammation, characterized by cytokine storm, is a significant contributor to severe cases of COVID-19 (Diamond and Kanneganti, 2022). SARS-CoV-2 Mpro specifically cleaved Nod-like receptor protein 12 (NLRP12), as evidenced by significant reductions of NLRP12 protein levels in SARS-CoV-2 infected cells (Moustaqil et al., 2021). Its cleavage is proposed to enhance pro-inflammatory cytokine and chemokine production via NF-κB signaling, and perturb the NLRP3 inflammasome assembly to trigger the cleavage of pro-caspase-1, thereby enhancing the release of IL-1β, all associated with the hyperinflammation observed in severe COVID-19. Another ISG cleaved by Mpro is the solute carrier family 25 member 22 (SLC25A22; Zhang et al., 2021). Knockout of SLC25A22, a mitochondrial glutamate carrier, has been associated with decreased immunosuppressive function in colorectal cancer (Yoo et al., 2020; Zhou et al., 2021), implying its involvement in immune response activation.
Fas-associated factor 1 (FAF1) is a positive regulator of type I interferon (IFN) signaling and is involved in the activation of the Fas-mediated pathway of apoptosis. However, there are contrasting results on the role of FAF1 in regulating the antiviral immune response. FAF1 is suggested to reduce virus-induced type I IFN activation by inhibiting nuclear translocation of the transcription factor IRF3 (Song et al., 2016). In contrast, FAF1 is hypothesized to bind competitively to NLRX1 to free the mitochondrial antiviral signaling protein (MAVS) upon RNA virus infection, which subsequently interacts with the retinoic acid-inducible gene (RIG)-I to initiate type I IFN signaling (Kim et al., 2017). Furthermore, virus infection is postulated to prevent aggregation of FAF1, which inhibits FAF1-dependent suppression of MAVS and then activates antiviral immunity (Dai et al., 2018). More studies are needed to confirm the role of FAF1 cleavage in virus infection.
Viruses can affect host gene expression at the transcriptional level. In addition, since viruses lack functional ribosomes, they attempt to usurp the host’s translational apparatus by competing with cellular mRNA to achieve successful replication. For instance, RNA polymerase II-associated protein 1 (RPAP1), which is crucial to bridging RNA polymerase II with gene-enhancer elements to increase transcription, and the polypyrimidine tract-binding protein (PTBP1), essential for pre-mRNA splicing and mRNA export, are both cleaved by Mpro. Proteolysis of PTBP1 after SARS-CoV-2 infection leads to the redistribution of PTBP1 from the nucleus to the cytoplasm (Pablos et al., 2021). In polioviruses, proteolysis of PTBP1 is speculated to switch viral translation to replication (Back et al., 2002). Thus, Mpro might target RPAP1 and PTBP1 to divert transcription and translation machineries from host to virus.
Pinin (PNN), a multifunctional nuclear phosphoprotein involved in the regulation of transcription and alternative RNA splicing, has also been identified as a substrate of Mpro (Meyer et al., 2021). Depletion of PNN has been demonstrated to result in apoptosis in vitro and early lethality in vivo (Leu et al., 2012). Furthermore, PNN binds to the transcriptional co-repressor C-terminal binding protein 1 (CTBP1). The interaction of PNN and CTBP1 alters CTBP1 silencing function (Alpatov et al., 2004). The overlapping pathways enriched in PNN-KD and CTBP1-KD cells include the TNFα-induced canonical NFκB signaling pathway and the IFN response pathway (Zhang et al., 2016). CTBP1-mutated neuronal cells were more susceptible to West Nile virus than control cells, consistent with the lower expression of IFN-response genes in CTBP1-mutated cells (Vijayalingam et al., 2020). Cleavage of PNN and CTBP1 by Mpro is suggested to alter the transcription of host antiviral response genes and induce apoptosis (Miczi et al., 2020; Meyer et al., 2021). Furthermore, Mpro cleaves Histone deacetylase 2 (HDAC2), which primarily regulates gene transcription by modifying histones and is also required for ISG transcriptional elongation (Chang et al., 2004). In consequence, the cleavage of HDAC2 by Mpro results in the impairment of ISG expression (Song et al., 2023).
Yes-associated protein 1 (YAP1), a transcriptional co-activator, participates in Hippo pathway. Since YAP negatively regulated an antiviral immune response via inhibiting the translocation of IRF3 to the nucleus, cleavage of YAP1 is presumed to enhance innate immunity (Wang et al., 2017). The kinase activity of mitogen-activated kinase-kinase-kinase-kinase 5 (MAP4K5), another Hippo pathway regulator, can be inactivated by Mpro cleavage. cAMP response element binding protein 1 (CREB1) is a transcription factor that dimerizes with ATF1 to regulate the transcription of anti-apoptotic and cell proliferation genes. Besides, CREB1 binds YAP1 and forms a positive feedback loop with each other (Chen et al., 2018). Mpro cleavages of YAP1, MAP4K5, and CREB1 indicate that SARS-CoV-2 can hijack the Hippo-YAP signaling pathway (Pablos et al., 2021) for mediating a variety of cellular processes, including cell proliferation, differentiation, apoptosis, and immune response.
To maintain homeostasis, cells undergo two types of programmed cell death (PCD)-apoptosis and autophagy (Kennedy, 2015). Inhibition of these PCDs by SARS-CoV-2 aids the virus to avoid elimination in the cells and ensure viable cells for viral replication, while induction may benefit the virus by the regulation of immune response and virus release (Li et al., 2020, 2021, 2022). Moreover, SARS-CoV-2 exploits autophagy to prevent virus degradation (Chen et al., 2020). Several proteins involved in apoptosis and autophagy have been identified to be targeted by Mpro.
Baculoviral IAP repeat-containing protein 6 (BIRC6) functions as an inhibitor of apoptosis and autophagy by ubiquitinating pro-apoptotic factors and LC3B, leading to their proteasomal degradation (Ehrmann et al., 2022). Mpro cleavage of BIRC6 may promote apoptosis and autophagy, in line with the induction of apoptosis and autophagy upon SARS-CoV-2 infection (Li et al., 2020, 2021). Transactive response DNA binding protein 43 kDa (TDP-43) is critical in RNA regulation, including the expression of viral RNA (reviewed in Rahic et al., 2023). Cleavage of TDP-43 by Mpro induced cytotoxicity in neurons, which could contribute to the pathogenicity of SARS-CoV-2 in the nervous system (Yang et al., 2023).
Galectin-8 (LGALS8) is involved in the regulation of immune responses and directly binds to Spike S1 glycans and the autophagy adaptor NDP52 (Pablos et al., 2021). LGALS8 is proposed to sense the glycosylated Spike S1 protein and activate xenophagy, a type of selective autophagy targeting invading pathogens to lysosomes, to reduce SARS-CoV-2 infection (Pablos et al., 2021). Furthermore, the autophagy adaptor protein FYVE and the coiled coil domain containing 1 (FYCO1) has been identified as a candidate COVID-19 susceptibility and severity gene and is believed to be the key mediator that connects double-membrane vesicles (the main site of coronavirus replication) from the endoplasmic reticulum to the microtubule network in host cells (Reggiori et al., 2011; Parkinson et al., 2020; Lee et al., 2021; Jahanafrooz et al., 2022). The elimination of FYCO1 resulted in the accumulation of early autophagosomes (Pankiv et al., 2010). Mpro cleavage of LGALS8 and FYCO1 possibly enables SARS-CoV-2 to escape antiviral xenophagy (Pablos et al., 2021) and induce incomplete autophagy.
SARS-CoV-2 infection alters host cell metabolism (Andrade Silva et al., 2021; Mullen et al., 2021). In fact, proteins that play roles in cell metabolism were found to be substrates of Mpro. Cleavage of Ring finger protein 20 (RNF20) destabilizes the RNF20/RNF40 complex, which is essential for their ubiquitin E3 ligase activity. As a result, this blocks the degradation of the sterol regulatory element binding protein 1 (SREBP1), and subsequently increasing the lipid metabolism for promoting SARS-CoV-2 replication (Zhang et al., 2021).
Phosphoribosylaminoimidazole succinocarboxamide synthetase (PAICS), a de novo purine biosynthetic enzyme was previously identified to be crucial in influenza virus replication (Karlas et al., 2010; Generous et al., 2014). PAICS is proposed to be a candidate for a noncanonical route for SARS-CoV-2 infection in human placentas (Constantino et al., 2021). SARS-CoV-2 infection has been reported to promote de novo purine synthesis through nsp9 (Qin et al., 2022). Silencing of PAICS reduced virus titers (~10-fold), suggesting that cleavage of PAICS by Mpro results in altered function of PAICS (Meyer et al., 2021), which may influence the de novo purine synthesis.
Insulin receptor substrate 2 (IRS2) regulates insulin signaling and the control of glucose homeostasis. Hepatitis C virus infection downregulates IRS2 expression by upregulating the suppressor of cytokine signaling (SOCS) and by activating the mTOR/S6K1 signaling pathway, resulting in insulin resistance (Kawaguchi et al., 2004; Pazienza et al., 2007; Bose et al., 2012). Notably, new-onset hyperglycemia has been associated with SARS-CoV-2 because non-diabetic COVID-19 patients were found to have increased risk of insulin resistance (Chen et al., 2021; Wihandani et al., 2023), which may be associated with Mpro cleavage of IRS2 (Pablos et al., 2021).
The intracellular transport system and cytoskeletons are essential for viral infections, particularly for transporting viral components to specific subcellular compartment sites of translation, replication, and secretion. The Golgi apparatus is an integral component of the viral life cycle. SARS-CoV-2 remodels the Golgi structure for viral release, hence, Mpro cleavage of Golgin subfamily A member 3 (GOLGA3), which is involved in the organization of the Golgi apparatus and its associated vesicles (Meyer et al., 2021; Pablos et al., 2021), may also be linked to this modulation (Zhang et al., 2022). Moreover, GOLGA3 has been associated with COVID-19 and has been identified to interact with nsp13 (Gordon et al., 2020; Deng et al., 2021). Mpro cleavage of GOLGA3 may play a role in reconfiguring the endoplasmic reticulum to facilitate Golgi trafficking during virus assembly.
Although RNA viruses replicate in the cytoplasm, they also exploit the nucleocytoplasmic trafficking system to inhibit the host immune response (Sajidah et al., 2021), which may explain why SARS-CoV-2 Mpro cleaves the nuclear pore complex 107 kDa subunit (NUP107) and Importin subunit alpha-4 (IMA4), which are both important members of nuclear pore transport (Pablos et al., 2021). IMA4, also known as karyopherin subunit alpha-3 (KPNA3), has been shown to be targeted by the Japanese encephalitis virus NS5 protein to hinder the nuclear import of its cargo molecules IFN regulatory factor 3 and NF-κB, thereby subsequently inhibiting type 1 IFN production (Ye et al., 2017).
Septin (SEPT) is recognized as a component of the cytoskeleton (Mostowy and Cossart, 2012). Septin polymerizes into filaments at the cell cortex or in association with other cytoskeletal proteins, such as actin or microtubules. Mpro cleaves several septin proteins, including SEPT2, SEPT6, and SEPT9, to affect the septin complex, causing an unstable filament structure and inducing cilia dysfunction (Lee et al., 2023).
With the help of computational and experimental methods, scientists have gained valuable insights into the substrates of Mpro. NetCorona analysis is widely used for substrate prediction. Intriguingly, some of the identified substrates have low NetCorona scores (Table 1), implying other issues should be considered. Further information, like binding affinity, may improve the original algorithm. The steric effects on substrate specificity also play an important role for the assessment. Notably, the cleavage sites of HDAC2 and PAICS are buried in the structure, warranting further study regarding the mechanism of Mpro cleavage of these two proteins. Deep learning of sequenced-based prediction and structural analysis can likewise improve the accuracy of prediction.
Identification of viral host substrates helps determine specific virus-host interactions, including the cellular pathways involved, and the mechanisms of viral replication and pathogenesis. Consequently, researchers can gain valuable insights into how viruses cause diseases and develop strategies to control or treat viral infections. After COVID-19 infection, certain individuals developed post-acute sequelae of SARS-CoV-2 infection (PASC), known as long COVID. The persistence of viral RNA or proteins for weeks in these patients implies the presence of an impaired immune response. Exploring the potential role of Mpro in this aspect would be valuable. Besides, identifying the specific sequences of host substrates targeted by Mpro can have significant implications in developing peptidomimetic protease inhibitors. Discovering new substrate sequences can enhance the design of effective antiviral strategies. Continued research is essential to improve our understanding of Mpro function and develop potent antiviral therapies against coronaviruses.
W-CS conceived and supervised the review topic. IM, Y-CL, and W-CS participated in the writing and preparation of the manuscript. All authors contributed to the article and approved the submitted version.
This work is supported by the grant (MOST 111-2320-B-039-060) from the National Science and Technology Council, Taiwan, and the grant (CMU111-MF-29) from China Medical University.
Y-CL was employed by the company Sinphar Pharmaceutical Co, Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Alpatov, R., Munguba, G. C., Caton, P., Joo, J. H., Shi, Y., Shi, Y., et al. (2004). Nuclear speckle-associated protein Pnn/Drs binds to the transcriptional corepressor Ctbp and relieves Ctbp-mediated repression of the E-cadherin gene. Mol. Cell. Biol. 24, 10223–10235. doi: 10.1128/MCB.24.23.10223-10235.2004
Andrade Silva, M., Da Silva, A., Do Amaral, M. A., Fragas, M. G., and Câmara, N. O. S. (2021). Metabolic alterations in Sars-Cov-2 infection and its implication in kidney dysfunction. Front. Physiol. 12:624698. doi: 10.3389/fphys.2021.624698
Back, S. H., Kim, Y. K., Kim, W. J., Cho, S., Oh, H. R., Kim, J.-E., et al. (2002). Translation of polioviral mRNA is inhibited by cleavage of polypyrimidine tract-binding proteins executed by polioviral 3cPro. J. Virol. 76, 2529–2542. doi: 10.1128/jvi.76.5.2529-2542.2002
Bose, S. K., Shrivastava, S., Meyer, K., Ray, R. B., and Ray, R. (2012). Hepatitis C virus activates the Mtor/S6k1 signaling pathway in inhibiting Irs-1 function for insulin resistance. J. Virol. 86, 6315–6322. doi: 10.1128/JVI.00050-12
Campbell, G. R., Rawat, P., and Spector, S. A. (2023). Pacritinib inhibition of Irak1 blocks aberrant Tlr8 signalling by Sars-Cov-2 and Hiv-1-derived RNA. J. Innate Immun. 15, 96–106. doi: 10.1159/000525292
Chang, H. M., Paulson, M., Holko, M., Rice, C. M., Williams, B. R., Marie, I., et al. (2004). Induction of interferon-stimulated gene expression and antiviral responses require protein deacetylase activity. Proc. Natl. Acad. Sci. U. S. A. 101, 9578–9583. doi: 10.1073/pnas.0400567101
Chen, L., Feng, P., Peng, A., Qiu, X., Zhu, X., He, S., et al. (2018). Camp response element-binding protein and yes-associated protein form a feedback loop that promotes neurite outgrowth. J. Cell. Mol. Med. 22, 374–381. doi: 10.1111/jcmm.13324
Chen, S.-W., Himeno, M., Koui, Y., Sugiyama, M., Nishitsuji, H., Mizokami, M., et al. (2020). Modulation of hepatitis B virus infection by epidermal growth factor secreted from liver sinusoidal endothelial cells. Sci. Rep. 10:14349. doi: 10.1038/s41598-020-71453-5
Chen, M., Zhu, B., Chen, D., Hu, X., Xu, X., Shen, W. J., et al. (2021). Covid-19 may increase the risk of insulin resistance in adult patients without diabetes: a 6-month prospective study. Endocr. Pract. 27, 834–841. doi: 10.1016/j.eprac.2021.04.004
Chuck, C. P., Chong, L. T., Chen, C., Chow, H. F., Wan, D. C., and Wong, K. B. (2010). Profiling of substrate specificity of Sars-Cov 3cl. PLoS One 5:E13197. doi: 10.1371/journal.pone.0013197
Constantino, F. B., Cury, S. S., Nogueira, C. R., Carvalho, R. F., and Justulin, L. A. (2021). Prediction of non-canonical routes for Sars-Cov-2 infection in human placenta cells. Front. Mol. Biosci. 8:614728. doi: 10.3389/fmolb.2021.614728
Dai, T., Wu, L., Wang, S., Wang, J., Xie, F., Zhang, Z., et al. (2018). Faf1 regulates antiviral immunity by inhibiting mavs but is antagonized by phosphorylation upon viral infection. Cell Host Microbe 24, 776–790.E5. doi: 10.1016/j.chom.2018.10.006
Deng, H., Yan, X., and Yuan, L. (2021). Human genetic basis of coronavirus disease 2019. Signal Transduct. Target. Ther. 6:344. doi: 10.1038/s41392-021-00736-8
Diamond, M. S., and Kanneganti, T.-D. (2022). Innate immunity: the first line of defense against Sars-Cov-2. Nat. Immunol. 23, 165–176. doi: 10.1038/s41590-021-01091-0
Ehrmann, J. F., Grabarczyk, D. B., Heinke, M., Deszcz, L., Kurzbauer, R., Hudecz, O., et al. (2022). Structural basis of how the Birc6/SMAC complex regulates apoptosis and autophagy. Biorxiv [Preprint]. doi: 10.1126/science.ade8873
Generous, A., Thorson, M., Barcus, J., Jacher, J., Busch, M., and Sleister, H. (2014). Identification of putative interactions between swine and human influenza a virus nucleoprotein and human host proteins. Virol. J. 11:228. doi: 10.1186/s12985-014-0228-6
Gordon, D. E., Jang, G. M., Bouhaddou, M., Xu, J., Obernier, K., White, K. M., et al. (2020). A Sars-Cov-2 protein interaction map reveals targets for drug repurposing. Nature 583, 459–468. doi: 10.1038/s41586-020-2286-9
Hu, Q., Xiong, Y., Zhu, G. H., Zhang, Y. N., Zhang, Y. W., Huang, P., et al. (2022). The Sars-Cov-2 main protease (M(pro)): structure, function, and emerging therapies for Covid-19. Medcomm 3:E151. doi: 10.1002/mco2.151
Jackson-Bernitsas, D. G., Ichikawa, H., Takada, Y., Myers, J. N., Lin, X. L., Darnay, B. G., et al. (2007). Evidence that Tnf-Tnfr1-Tradd-Traf2-rip-Tak1-Ikk pathway mediates constitutive Nf-Κb activation and proliferation in human head and neck squamous cell carcinoma. Oncogene 26, 1385–1397. doi: 10.1038/sj.onc.1209945
Jahanafrooz, Z., Chen, Z., Bao, J., Li, H., Lipworth, L., and Guo, X. (2022). An overview of human proteins and genes involved in Sars-Cov-2 infection. Gene 808:145963. doi: 10.1016/j.gene.2021.145963
Karlas, A., Machuy, N., Shin, Y., Pleissner, K.-P., Artarini, A., Heuer, D., et al. (2010). Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature 463, 818–822. doi: 10.1038/nature08760
Kawaguchi, T., Yoshida, T., Harada, M., Hisamoto, T., Nagao, Y., Ide, T., et al. (2004). Hepatitis C virus down-regulates insulin receptor substrates 1 and 2 through up-regulation of suppressor of cytokine signaling 3. Am. J. Pathol. 165, 1499–1508. doi: 10.1016/S0002-9440(10)63408-6
Kennedy, P. G. E. (2015). Viruses, apoptosis, and neuroinflammation—a double-edged sword. J. Neuro-Oncol. 21, 1–7. doi: 10.1007/s13365-014-0306-y
Kim, J.-H., Park, M.-E., Nikapitiya, C., Kim, T.-H., Uddin, M. B., Lee, H.-C., et al. (2017). Fas-associated factor-1 positively regulates type i interferon response to RNA virus infection by targeting Nlrx1. PLoS Pathog. 13:E1006398. doi: 10.1371/journal.ppat.1006398
Koudelka, T., Boger, J., Henkel, A., Schonherr, R., Krantz, S., Fuchs, S., et al. (2021). N-terminomics for the identification of in vitro substrates and cleavage site specificity of the Sars-Cov-2 main protease. Proteomics 21:2000246. doi: 10.1002/pmic.202000246
Lee, A. R., Kweon, Y. C., Lee, S. M., and Park, C. Y. (2023). Human coronavirus 3cl proteases cleave septins and disrupt hedgehog signaling. J. Med. Virol. 95:E28618. doi: 10.1002/jmv.28618
Lee, J.-W., Lee, I.-H., Sato, T., Kong, S. W., and Iimura, T. (2021). Genetic variation analyses indicate conserved Sars-Cov-2–host interaction and varied genetic adaptation in immune response factors in modern human evolution. Develop. Growth Differ. 63, 219–227. doi: 10.1111/dgd.12717
Leu, S., Lin, Y.-M., Wu, C.-H., and Ouyang, P. (2012). Loss of PNN expression results in mouse early embryonic lethality and cellular apoptosis through Srsf1-mediated alternative expression of Bcl-Xs and Icad. J. Cell Sci. 125, 3164–3172. doi: 10.1242/jcs.100859
Li, F., Li, J., Wang, P. H., Yang, N., Huang, J., Ou, J., et al. (2021). Sars-Cov-2 spike promotes inflammation and apoptosis through autophagy by Ros-suppressed Pi3k/Akt/Mtor signaling. Biochim. Biophys. Acta Mol. basis Dis. 1867:166260. doi: 10.1016/j.bbadis.2021.166260
Li, S., Zhang, Y., Guan, Z., Li, H., Ye, M., Chen, X., et al. (2020). Sars-Cov-2 triggers inflammatory responses and cell death through Caspase-8 activation. Signal Transduct. Target. Ther. 5:235. doi: 10.1038/s41392-020-00334-0
Li, X., Zhang, Z., Wang, Z., Gutiérrez-Castrellón, P., and Shi, H. (2022). Cell deaths: involvement in the pathogenesis and intervention therapy of Covid-19. Signal Transduct. Target. Ther. 7:186. doi: 10.1038/s41392-022-01043-6
Meyer, B., Chiaravalli, J., Gellenoncourt, S., Brownridge, P., Bryne, D. P., Daly, L. A., et al. (2021). Characterising proteolysis during Sars-Cov-2 infection identifies viral cleavage sites and cellular targets with therapeutic potential. Nat. Commun. 12:5553. doi: 10.1038/s41467-021-25796-w
Miczi, M., Golda, M., Kunkli, B., Nagy, T., Tozser, J., and Motyan, J. A. (2020). Identification of host cellular protein substrates of Sars-Cov-2 main protease. Int. J. Mol. Sci. 21:9523. doi: 10.3390/ijms21249523
Mostowy, S., and Cossart, P. (2012). Septins: the fourth component of the cytoskeleton. Nat. Rev. Mol. Cell Biol. 13, 183–194. doi: 10.1038/nrm3284
Moustaqil, M., Ollivier, E., Chiu, H. P., Van Tol, S., Rudolffi-Soto, P., Stevens, C., et al. (2021). Sars-Cov-2 proteases Plpro and 3clpro cleave Irf3 and critical modulators of inflammatory pathways (NLRP12 and TAB1): implications for disease presentation across species. Emerg. Microbes Infect. 10, 178–195. doi: 10.1080/22221751.2020.1870414
Mullen, P. J., Garcia, G., Purkayastha, A., Matulionis, N., Schmid, E. W., Momcilovic, M., et al. (2021). Sars-Cov-2 infection rewires host cell metabolism and is potentially susceptible to Mtorc1 inhibition. Nat. Commun. 12:1876. doi: 10.1038/s41467-021-22166-4
Pablos, I., Machado, Y., De Jesus, H. C. R., Mohamud, Y., Kappelhoff, R., Lindskog, C., et al. (2021). Mechanistic insights into Covid-19 by global analysis of the Sars-Cov-2 3cl(pro) substrate degradome. Cell Rep. 37:109892. doi: 10.1016/j.celrep.2021.109892
Pankiv, S., Alemu, E. A., Brech, A., Bruun, J. A., Lamark, T., Overvatn, A., et al. (2010). Fyco1 is a Rab7 effector that binds to Lc3 and Pi3p to mediate microtubule plus end-directed vesicle transport. J. Cell Biol. 188, 253–269. doi: 10.1083/jcb.200907015
Parkinson, N., Rodgers, N., Head Fourman, M., Wang, B., Zechner, M., Swets, M. C., et al. (2020). Dynamic data-driven meta-analysis for prioritisation of host genes implicated in COVID-19. Sci. Rep. 10:22303. doi: 10.1038/s41598-020-79033-3
Pazienza, V., Clément, S., Pugnale, P., Conzelman, S., Foti, M., Mangia, A., et al. (2007). The hepatitis C virus core protein of genotypes 3a and 1b downregulates insulin receptor substrate 1 through genotype-specific mechanisms. Hepatology 45, 1164–1171. doi: 10.1002/hep.21634
Qin, C., Rao, Y., Yuan, H., Wang, T. Y., Zhao, J., Espinosa, B., et al. (2022). Sars-Cov-2 couples evasion of inflammatory response to activated nucleotide synthesis. Proc. Natl. Acad. Sci. U. S. A. 119:E2122897119. doi: 10.1073/pnas.2122897119
Rahic, Z., Buratti, E., and Cappelli, S. (2023). Reviewing the potential links between viral infections and Tdp-43 proteinopathies. Int. J. Mol. Sci. 24:1581. doi: 10.3390/ijms24021581
Reggiori, F., De Haan, C. A. M., and Molinari, M. (2011). Unconventional use of Lc3 by coronaviruses through the alleged subversion of the erad tuning pathway. Viruses 3, 1610–1623. doi: 10.3390/v3091610
Sabbah, D. A., Hajjo, R., Bardaweel, S. K., and Zhong, H. A. (2021). An updated review on Sars-Cov-2 main proteinase (M(pro)): protein structure and small-molecule inhibitors. Curr. Top. Med. Chem. 21, 442–460. doi: 10.2174/1568026620666201207095117
Sajidah, E. S., Lim, K., and Wong, R. W. (2021). How Sars-Cov-2 and other viruses build an invasion route to hijack the host nucleocytoplasmic trafficking system. Cells 10:1424. doi: 10.3390/cells10061424
Scott, B. M., Lacasse, V., Blom, D. G., Tonner, P. D., and Blom, N. S. (2022). Predicted coronavirus Nsp5 protease cleavage sites in the human proteome. BMC Genom Data 23:25. doi: 10.1186/s12863-022-01044-y
Song, S., Lee, J. J., Kim, H. J., Lee, J. Y., Chang, J., and Lee, K. J. (2016). FAS-associated factor 1 negatively regulates the antiviral immune response by inhibiting translocation of interferon regulatory factor 3 to the nucleus. Mol. Cell. Biol. 36, 1136–1151. doi: 10.1128/MCB.00744-15
Song, L., Wang, D., Abbas, G., Li, M., Cui, M., Wang, J., et al. (2023). The main protease of Sars-Cov-2 cleaves histone deacetylases and Dcp1a, attenuating the immune defense of the interferon-stimulated genes. J. Biol. Chem. 299:102990. doi: 10.1016/j.jbc.2023.102990
Ullrich, S., and Nitsche, C. (2020). The Sars-Cov-2 main protease as drug target. Bioorg. Med. Chem. Lett. 30:127377. doi: 10.1016/j.bmcl.2020.127377
Vijayalingam, S., Ezekiel, U. R., Xu, F., Subramanian, T., Geerling, E., Hoelscher, B., et al. (2020). Human IPSC-derived neuronal cells from Ctbp1-mutated patients reveal altered expression of neurodevelopmental gene networks. Front. Neurosci. 14:562292. doi: 10.3389/fnins.2020.562292
Wang, S., Xie, F., Chu, F., Zhang, Z., Yang, B., Dai, T., et al. (2017). Yap antagonizes innate antiviral immunity and is targeted for lysosomal degradation through Ikkɛ-mediated phosphorylation. Nat. Immunol. 18, 733–743. doi: 10.1038/ni.3744
Wihandani, D. M., Purwanta, M. L. A., Mulyani, W. R. W., Putra, I., and Supadmanaba, I. G. P. (2023). New-onset diabetes in Covid-19: the molecular pathogenesis. Biomedicine 13, 3–12. doi: 10.37796/2211-8039.1389
Xiong, M., Su, H., Zhao, W., Xie, H., Shao, Q., and Xu, Y. (2021). What coronavirus 3c-like protease tells us: from structure, substrate selectivity, to inhibitor design. Med. Res. Rev. 41, 1965–1998. doi: 10.1002/med.21783
Yang, J., Li, Y., Wang, S., Li, H., Zhang, L., Zhang, H., et al. (2023). The Sars-Cov-2 main protease induces neurotoxic Tdp-43 cleavage and aggregates. Signal Transduct. Target. Ther. 8:109. doi: 10.1038/s41392-023-01386-8
Ye, J., Chen, Z., Li, Y., Zhao, Z., He, W., Zohaib, A., et al. (2017). Japanese encephalitis virus Ns5 inhibits type I interferon (IFN) production by blocking the nuclear translocation of IFN regulatory factor 3 and Nf-Κb. J. Virol. 91, E00039–E00017. doi: 10.1128/JVI.00039-17
Yoo, H. C., Yu, Y. C., Sung, Y., and Han, J. M. (2020). Glutamine reliance in cell metabolism. Exp. Mol. Med. 52, 1496–1516. doi: 10.1038/s12276-020-00504-8
Zhang, X., Guo, Y., Xu, X., Tang, T., Sun, L., Wang, H., et al. (2019). Mir-146a promotes Borna disease virus 1 replication through Irak1/Traf6/Nf-Κb signaling pathway. Virus Res. 271:197671. doi: 10.1016/j.virusres.2019.197671
Zhang, J., Kennedy, A., Xing, L., Bui, S., Reid, W., Joppich, J., et al. (2022). Sars-Cov-2 triggers golgi fragmentation via down-regulation of Grasp55 to facilitate viral trafficking. Biorxiv [Preprint]. doi: 10.1101/2022.03.04.483074
Zhang, Y., Kwok, J. S.-L., Choi, P.-W., Liu, M., Yang, J., Singh, M., et al. (2016). Pinin interacts with C-terminal binding proteins for RNA alternative splicing and epithelial cell identity of human ovarian cancer cells. Oncotarget 7, 11397–11411. doi: 10.18632/oncotarget.7242
Zhang, S., Wang, J., and Cheng, G. (2021). Protease cleavage of Rnf20 facilitates coronavirus replication via stabilization of Srebp1. Proc. Natl. Acad. Sci. U. S. A. 118:e2107108118. doi: 10.1073/pnas.2107108118
Zheng, H.-Q., Li, C., Zhu, X.-F., Wang, W.-X., Yin, B.-Y., Zhang, W.-J., et al. (2022). Mir-615 facilitates porcine epidemic diarrhea virus replication by targeting Irak1 to inhibit type iii interferon expression. Front. Microbiol. 13:1071394. doi: 10.3389/fmicb.2022.1071394
Zhou, Q., Peng, Y., Chen, L.-S., Chen, H., Kang, W., Nie, Y., et al. (2021). Iddf2021-Abs-0183 Slc25a22 drives immune suppression in kras-mutant colorectal cancer. Gut 70, A53–A55. doi: 10.1136/gutjnl-2021-IDDF.51
Keywords: SARS-CoV-2, main protease, substrate, virus-host interaction, virus pathogenesis, viral replication
Citation: Melano I, Lo Y-C and Su W-C (2023) Characterization of host substrates of SARS-CoV-2 main protease. Front. Microbiol. 14:1251705. doi: 10.3389/fmicb.2023.1251705
Received: 11 July 2023; Accepted: 31 July 2023;
Published: 21 August 2023.
Edited by:
Qiang Ding, Tsinghua University, ChinaReviewed by:
Sourish Ghosh, Indian Institute of Chemical Biology (CSIR), IndiaCopyright © 2023 Melano, Lo and Su. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wen-Chi Su, dDIzNTE0QG1haWwuY211aC5vcmcudHc=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.