
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 06 October 2023
Sec. Microbe and Virus Interactions with Plants
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1251698
This article is part of the Research TopicMicrobiome Associated With Plant Pathogens, Pathogenesis, and Their Applications in Developing Sustainable AgricultureView all 17 articles
Introduction: Tomato yellow leaf curl virus (TYLCV), which is a typical member of the genus Begomovirus, causes severe crop yield losses worldwide. RNA interference (RNAi) is an important antiviral defense mechanism in plants, but whether plant beneficial microbes used as biocontrol agents would modulate RNAi in defense against TYLCV remains unclear.
Methods: Here, we employed whole-transcriptome, bisulfite, and small RNA sequencing to decipher the possible role of Bacillus amyloliquefaciens Ba13 as a bacterial biocontrol agent against TYLCV in RNAi modulation.
Results: Potted tomato plants were exposed to whiteflies for natural viral infection 14 days after bacterial inoculation. Compared with non-inoculated controls, the abundance of TYLCV gene in the leaves of inoculated plants decreased by 70.1% at 28 days post-infection, which mirrored the pattern observed for plant disease index. The expression of the ARGONAUTE family genes (e.g., AGO3, AGO4, AGO5, and AGO7) involved in antiviral defense markedly increased by 2.44–6.73-fold following bacterial inoculation. The methylation level at CpG site 228 (in the open reading frame region of the RNA interference suppressing gene AV2) and site 461 (in the open reading frame regions of AV1 and AV2) was 183.1 and 63.0% higher in inoculated plants than in non-inoculated controls, respectively. The abundances of 10 small interfering RNAs matched to the TYLCV genome were all reduced in inoculated plants, accompanied by enhancement of photosystem and auxin response pathways.
Discussion: The results indicate that the application of Ba. amyloliquefaciens Ba13 enhances plant resistance to TYLCV through RNAi modulation by upregulating RNAi-related gene expression and enhancing viral genome methylation.
Tomato yellow leaf curl virus (TYLCV) is a single-stranded circular DNA virus of the genus Begomovirus (family Geminiviridae). Since it was first discovered in Israel, TYLCV has spread to many other places, including the Middle East, Mediterranean coast, Africa, and Asia (Lu et al., 2023). TYLCV is one of the most detrimental viruses to tomato cultivation and it causes severe crop yield losses in tropical and subtropical regions. Because TYLCV is transmitted through whiteflies (Bemisia tabaci) (Gafni, 2003), chemical pesticides are often used to control this virus disease (Palumbo et al., 2001). However, the long-term use of pesticides is harmful to the environment and easily induces resistance in B. tabaci (Cahill et al., 1996). Given its advantages such as low cost and environmental friendliness, the use of beneficial microbes as biocontrol strains has attracted increasing interest in the control of TYLCV disease.
Members of Bacillus (Kumar et al., 2016), Pseudomonas (Shen et al., 2014), and Trichoderma (Elsharkawy et al., 2013; Vitti et al., 2015) have been used to control plant diseases by improving plant systemic resistance. There are two main pathways of systemic resistance in plants (Wang et al., 2009). Systemic acquired resistance (SAR) is dependent on salicylic acid and the regulatory protein NPR1 (Mou et al., 2003), whereas induced systemic resistance (ISR) is modulated by hormones such as jasmonic acid and ethylene. It has been shown that plant beneficial microbes can activate disease resistance pathways by regulating the expression of resistance-related genes (Elsharkawy et al., 2012) and the biosynthesis of hormones (Niu et al., 2011), ultimately enhancing plant resistance to the virus.
In addition to systemic resistance, RNA interference (RNAi) is recognized as an important defense mechanism in plants against viral infection (Kanakala and Ghanim, 2016). RNAi can be divided into small interfering RNA (siRNA)-mediated post-transcriptional gene silencing (PTGS) and transcriptional gene silencing (TGS), depending on the site and mechanism of action (Arif et al., 2013). PTGS occurs in the cytoplasm to precisely target and degrade mRNA transcripts of specific genes, which has been widely demonstrated to play an essential role in plant resistance against viral infection (El-Sappah et al., 2021). This process is mainly mediated by 21–24 nt virus-derived siRNA cut by Dicer-like (DCL) nucleases (Zheng et al., 2019). RNA-directed DNA methylation (RdDM)-mediated TGS is another important mechanism of plant defense against geminiviral infection, and it inhibits virus replication by increasing the methylation level of cytosine on the viral genome (Raja et al., 2008). Among them, methylation of CpG sites is crucial for plant defense against virus invasion, and viruses even evade plant defense by reducing the frequency of CpG sites in the genome (de Pradena et al., 2020; Loew et al., 2020).
The occurrence of PTGS and TGS involves the ARGONAUTE (AGO) family proteins, which are effectors of RNAi in plants (Li Z. C. et al., 2019). A growing number of AGO proteins have been identified to participate in the antiviral defense processes in plants. In particular, AGO4 is a major component of TGS that has been widely studied. In the classical RNA polymerase IV-RdDM pathway, the siRNA binds to AGO4 to form an AGO4-siRNA complex. This complex then binds to polymerase IV to take part in methylation of virus genomic sequences that are homologous to the siRNA under the action of domains-rearranged methyltransferase 2 (Matzke et al., 2015). Additionally, AGO3 has been reported to play a vital role in abscisic acid (ABA)-mediated antiviral defense (Alazem et al., 2014). Evidence suggests that both AGO3 and AGO7 play antiviral roles during TCV infection (Zheng et al., 2019). However, it is still unclear whether inoculation with biocontrol strains could modulate AGOs to enhance plant antiviral defense.
A previous study has demonstrated the robust effects of a bacterial strain, Bacillus amyloliquefaciens Ba13, for plant growth promotion and TYLCV disease control in tomato (Guo et al., 2019). How the presence of Ba. amyloliquefaciens Ba13 modulates the RNAi-based antiviral system in the host plant remains unclear. Here we verified the biocontrol effects of Ba. amyloliquefaciens Ba13 against TYLCV and explored the underlying mechanisms from the RNAi perspective. We analyzed the changes in RNAi-related gene expression in TYLCV-infected tomato leaves, as well as the methylation rate of TYLCV genome and the abundance of antiviral siRNAs in plant cells, after bacterial inoculation. The aim of this study was to decipher the possible role of Ba. amyloliquefaciens Ba13 in the antiviral RNAi system against TYLCV. The results could provide new evidence for microbial control of viral plant diseases.
The biocontrol strain, Ba. amyloliquefaciens Ba13 (SUB3558699; GenBank: MG846076), was isolated from a stratified old manured loessial soil under a corn-wheat rotation system in Yangling, Shaanxi Province, China. The strain was preserved by the Resource and Environmental Biology Laboratory in the College of Resources & Environment, Northwest A&F University (Yangling, Shaanxi Province, China). Seeds of tomato (Solanum lycopersicum L.) cultivar “Jinpeng 1” were purchased from Xi’an Jinpeng Seedling Co., Ltd. (Xi’an, Shaanxi Province, China). To mimic natural viral infection, the TYLCV pathogen was sourced from viruliferous whiteflies (B. tabaci) collected in the field in Yangling.
The pot experiment was carried out in a greenhouse on campus of Northwest A&F University. Plump tomato seeds were selected and surface sterilized with 75% ethanol for 30 s, followed by at least five washes with sterile distilled water. The sterilized seeds were placed evenly onto a filter paper in sterile Petri dishes (∼50 seeds per dish). To each dish, 2 mL of sterile water was added and the seeds were incubated in the dark at 26°C until germination. The germinated seeds were evenly sown into potted trays (50 holes per tray) containing seedling substrate and cultivated for 21 days. When true leaves formed, tomato seedlings with uniform growth were selected and transplanted into 60 pots with a diameter of 18.3 cm and a height of 12 cm (one seedling per pot). Each pot contained fertile field soil collected from a wheat-maize rotation system in Yangling. The soil was classified as Eum-Orthic Anthrosols (Staff, 2010). The soil was passed through a 2-mm sieve before use. The experimental used a randomized complete block design with three blocks, and each block contained 20 pots. The pots in each block were randomly and equally divided into two groups. One group was inoculated with Ba. amyloliquefaciens Ba13 (treatment group), while the other group was not inoculated (control group).
Ba. amyloliquefaciens Ba13 was cultured in beef extract–peptone broth at 37°C for 48 h with a shaking frequency of 180 rpm. The bacterial culture was diluted to 1 × 108 colony-forming units (CFU) mL–1 with sterile phosphate-buffered saline. For the treatment group, 100 mL of diluted cell culture per pot was applied to the seedlings 7 days post-transplantation. The control group received 100 mL of sterile phosphate-buffered saline per pot. Natural viral infection was performed 14 days after bacterial inoculation. To do this, both non-inoculated controls and Ba. amyloliquefaciens Ba13-inoculated plants were placed in a greenhouse with whiteflies. Each block was covered with a 150-mesh high-quality nylon net of 1.2 m × 1.2 m × 1.5 m (length × width × height), and 1000 whiteflies were released under the net for infection over 4 weeks. The average temperature of the greenhouse was 28°C and the average light period per day was 12 h.
The infection rate and disease severity of tomato plants in each group were determined at 14 and 28 days post-viral infection (dpvi). The disease severity was scored as follows (Sun et al., 2016): 0 = healthy plant without with curling yellow leaves; 1 = mild dwarfing (∼4/5 height of healthy plant) with curling yellow leaves (<20% on the top); 2 = dwarfing (∼2/3 height of healthy plant) with leaf curling and yellowing (<40% on the top); 3 = obvious dwarfing (∼1/2 to 2/3 height of healthy plant) with typical curling yellow leaves (60%); and 4 = severe dwarfing (<1/2 height of healthy plant) with typical curling yellow leaves (60% to all), or early withering. The disease index and control rate were calculated as follows.
Plant height was measured 28 dpvi and leaf samples were taken for subsequent analyses. Three biological samples (repetitions) were collected from each group, and each biological repetition was a mixture of three leaf samples collected from the same part of three random plants in each block. The sample consisting of the first and second leaves from the apical growing point was used for virus quantification; the third leaf was used for whole-transcriptome RNA sequencing (RNA-seq), bisulfite sequencing, and small RNA-seq, as well as for quantitative validation of functional genes; and the fourth leaf was used for the analysis of plant hormone contents. The leaf samples destined for virus quantification and sequencing analyses were rinsed with distilled water and blotted dry, then immediately wrapped with tin foil and placed in liquid nitrogen. The samples for hormone analysis were weighed and then placed into liquid nitrogen. All samples were brought to the laboratory where they were kept at −80°C until used.
Total DNA was extracted from tomato plants using a modified hexadecyltrimethylammonium bromide (CTAB) method. PCR amplification was performed as described in the Supplementary Note 1, with previously reported primers for the full-length sequence of the complete TYLCV genome (Supplementary Table 1). The amplified products were sequenced to obtain the complete genome sequence. Then we determined the virus type by comparing the obtained sequence with available virus genotypes in the NCBI database. Gene sequence similarity analysis was performed using the MegAlign program in DNAstar (DNASTAR, Inc., Madison, WI, USA). The MEGA-X software (Kumar et al., 2018) and MUSCLE algorithm were used to align the nucleic acid sequences of multiple sequences, while the neighbor joining algorithm was applied to build phylogenetic trees for taxonomic analysis of the virus. There were 23 different TYLCV genotypes in the evolutionary tree (including the virus genotypes identified in this study). Their accession numbers in the NCBI database, origins, and the similarities to the genotype of the virus used in this study are listed in Supplementary Table 2. The search of virus open reading frame (ORF) was performed using the NCBI Open Reading Frame Finder.1
Quantification of TYLCV in tomato leaves was performed using the method of Sade et al. (2014). Total DNA was extracted from leaf samples using the M5 Plant Genomic DNA Kit (Mei5 Biotech Co., Ltd., Beijing, China). The concentration and purity of extracted DNA were measured using spectrophotometry with an ultra microplate (Epoch, BioTek, Winooski, VT, USA). The DNA integrity was detected using agarose gel electrophoresis. The DNA samples were subjected to real-time fluorescent quantitative PCR on an iQ5 PCR system (Bio-Rad, Hercules, CA, USA) as described in the Supplementary Note 1. The PCR amplification was performed using TYLCV-V1 as the primer (Supplementary Table 1) and the EF1 gene as the internal reference. There were three technical replicates for each sample. Data collection and analysis were performed using the Bio-Rad iQ5 software (v2.1.97). The relative expression level of the viral gene was calculated using the 2–ΔΔCT method (Schmittgen and Livak, 2008).
Frozen leaf samples were sent to BGI Biotech Co., Ltd. (Shenzhen, China) for transcriptome sequencing using the BGISEQ-500 platform (BGI Biotech). Total RNA was isolated from the leaf samples using Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Then mRNA libraries were constructed. Whole-transcriptome RNA-seq data processing was performed as described in Supplementary Note 2. The detection of significantly differentially expressed genes (DEGs) was performed following the method described by Wang et al. (2010). Genes with a | fold-change| > 2 and p-value ≤ 0.001 were defined as DEGs.
The DEGs were subjected to Gene Ontology (GO) and pathway enrichment analysis using the PANTHER classification system2 (Mi et al., 2005) and the Kyoto Encyclopedia of Genes and Genomes (KEGG),3 respectively (Kanehisa et al., 2008). The basic local alignment search tool (BLASTn) was adopted to identify homologous genes in the tomato genome.4 The raw sequences were deposited at the Sequence Read Archive in the National Center for Biotechnology Information (NCBI)5 under BioProject accession number: PRJNA553064.
The validation of functional genes was performed by reverse transcription quantitative real-time PCR (RT-qPCR) using six genes involved in gene silencing, plant growth, and disease resistance. The PCR protocols are provided in Supplementary Note 3. There were three technical replicates for each sample, and the primer sequences used are provided in Supplementary Table 1. The relative expression levels of the selected genes and small RNAs were calculated as described for virus quantification.
The content of ABA in tomato leaves was measured using a previously reported method (Pan et al., 2010) as described in Supplementary Data (Note S4). Briefly, the ABA was extracted from leaf samples with 80% methanol/water (v/v) followed by a mixture of 5% acetic acid/ethyl acetate and water (1:1, v/v). The levels of ABA in the extracts were measured using gas chromatography coupled to triple quadrupole mass spectrometry (Agilent, Santa Clara, CA, USA).
Gene-specific DNA methylation was assessed by a next generation sequencing-based BSP, according to previously published method (Gao et al., 2014). In brief, BSP primers were designed using the online MethPrimer software and listed in Supplementary Table 1. A total of 1 μg of genomic DNA was converted using the ZYMO EZ DNA Methylation-Gold Kit (Zymo Research, Irvine, CA, USA) and one twentieth of the elution products were used as templates for PCR amplification with 35 cycles using KAPA 2G Robust HotStart PCR Kit (Kapa Biosystems, Wilmington, MA, USA). For each sample, BSP products of multiple genes were pooled equally, 5′-phosphorylated, 3′-dA-tailed and ligated to barcoded adapter using T4 DNA ligase (NEB). Barcoded libraries from all samples were sequenced on Illumina platform. For the bisulfite sequencing reads of each sample, firstly, adapters and low-quality reads were removed using software Trimmomatic-0.36. After removing the adapter sequences and filtering out the low-quality reads, the clean sequencing reads were directly aligned to the target sequences using software Bsmap (v2.73) with the default parameters which combines genome hashing and bitwise masking to achieve fast and accurate bisulfite mapping. Methylation level of C base in TYLCV genome was calculated as follows.
Small RNA-seq was used to detected the changes in the types and abundances of siRNA in tomato leaves, which have lengths mainly concentrated in the range of 19–24 nt (Seo et al., 2013). For the sample preparation and library construction, small RNA-seq was performed using the BGISEQ-500 platform (BGI Biotech). Sample selection and total RNA extraction followed the same methods that were used for the whole-transcriptome RNA-seq. Small RNA libraries were then constructed and small RNA-seq data were processed as described in Supplementary Note 5. The siRNAs with a p-value ≤ 0.05 were considered significantly differentially expressed. The raw sequences were deposited at the Sequence Read Archive in the NCBI (see text footnote 5) under BioProject accession number: PRJNA553309.
Given that siRNAs can bind to viral mRNA for transcriptional and post-transcriptional inhibition of viral replication, we aligned all the differentially expressed (p < 0.05) siRNAs against the genome of the TYLCV-SJ which identified in this study. The alignment of sequences was performed using the SOAP2.22 short-read alignment software package (Li et al., 2009), with no mismatches allowed. For siRNAs that completely matched to the viral genome sequence, their name, sequence, matched site, abundance in the two treatment groups (Ba. amyloliquefaciens Ba13 and control), and corresponding viral genotype were recorded.
Data were statistically analyzed using DPS 4.0.6 The comparison of group means was performed using the Student’s t-test, except that the disease index values obtained at two time points were compared using the least-significant difference (LSD) test. A p-value of less than 0.05 was considered significant. The relative expression levels (fold-changes) of genes and small RNAs were plotted using Excel 2010 (Microsoft Corp., Redmond, WA, USA), and the relative quantities (chromatographic peaks) of plant hormones were drawn using Origin 2022b (OriginLab Corp., Northampton, MA, USA). The enrichment of DEGs in the biotic stress pathway was analyzed using Mapman (Thimm et al., 2004).
Based on PCR amplification and sequencing analysis, the genotype of the virus present in the tomato leaves was identified to be TYLCV virion DNA TYLCV-SJ (GenBank accession number: MN910280.1). The PCR results (Supplementary Figure 1) showed that the total length of the viral genome was 2781 nt, which encoded eleven ORFs (>150 bp). The starting and ending sites and coding proteins of corresponding genes and their functions are listed in Table 1. According to the phylogenetic tree (Supplementary Figure 2) and the comparison of virus genotypes, TYLCV-SJ (MN910280.1) had the closest genetic relationship with TYLCV-JSYC (MF590740) and TYLCV-FJFZ (KX885028). The sequence of TYLCV-SJ was 98.1% similar to that of the Israeli virus genotype TYLCV-IS (X15656), and therefore was deemed to be a relative of the Israeli virus in China.
In the pot experiment, the tomato plants presented typical symptoms of TYLCV disease such as leaf curling and yellowing after viral infection. Compared with the control group, the plant height of the group treated with Ba. amyloliquefaciens Ba13 increased by 15.4% (p < 0.05; Figure 1A), while the relative abundance of the TYLCV gene in tomato leaves decreased by 70.1% at 28 dpvi (p < 0.01; Figure 1B). Additionally, the disease index of the treatment group was significantly lower than that of the control group at 28 dpvi (p < 0.05). The corresponding control rates in the treatment group were 38.7 and 32.1% at 14 and 28 dpvi, respectively (Table 2).
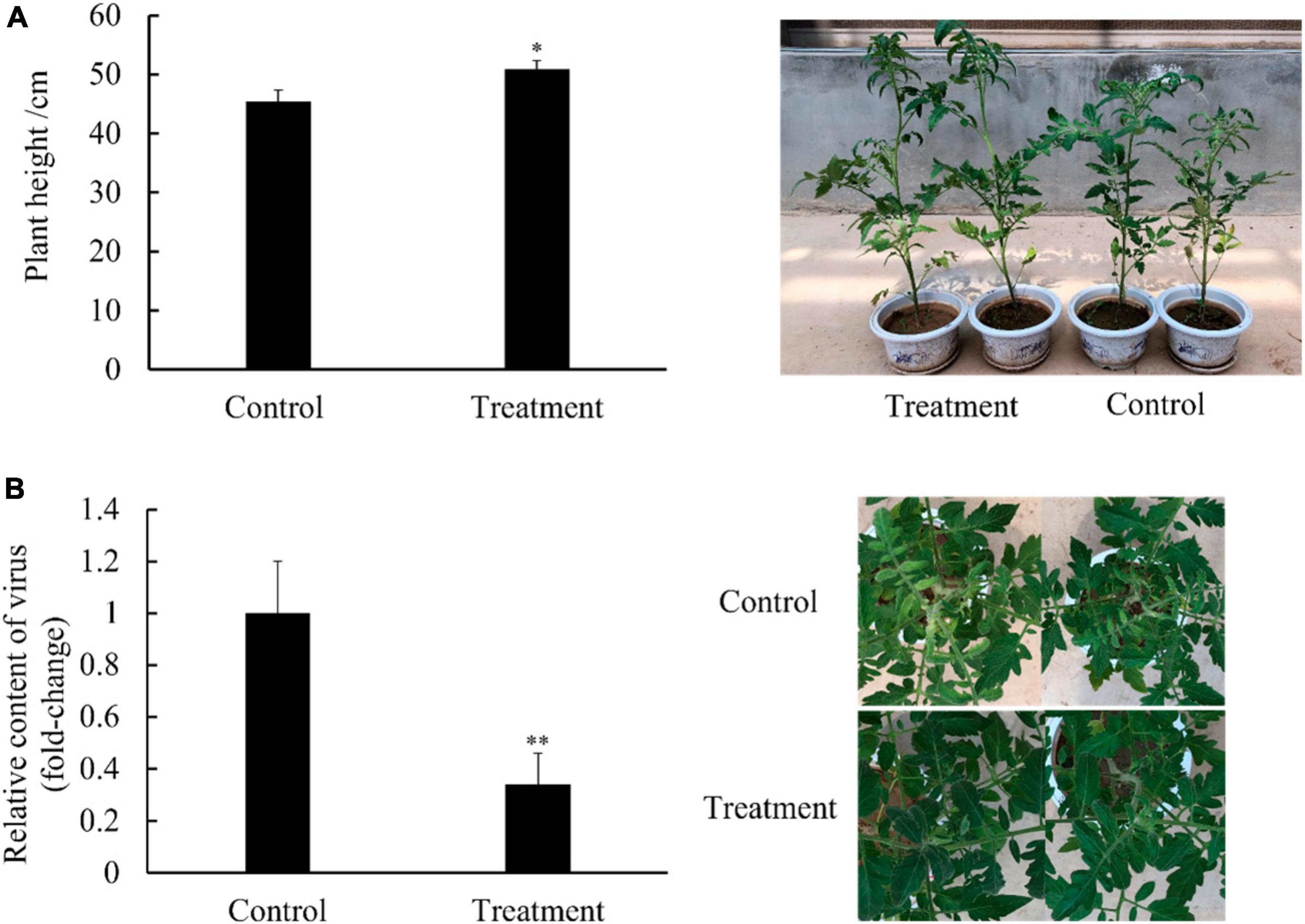
Figure 1. Effects of Bacillus amyloliquefaciens Ba13 in the promotion of tomato plant growth [(A); n = 30, error bars = standard deviation] and defense against tomato yellow leaf curl virus disease [(B); n = 3, error bars = standard deviation] at 28 days post-viral infection. **p < 0.01; *p < 0.05.
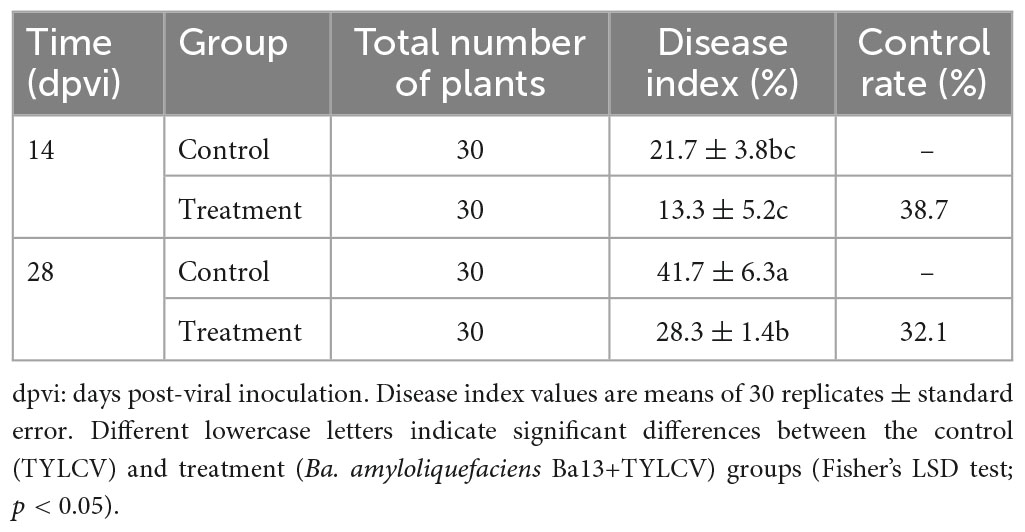
Table 2. The effect of Bacillus amyloliquefaciens Ba13 against tomato yellow leaf curl virus (TYLCV) disease.
Six leaf samples (three from each group) were analyzed using whole-transcriptome RNA-seq, which yielded 10.92 Gb of raw reads per sample. A total of 24,466 genes were detected, among which 1,897 genes (7.8%) were identified as DEGs with a | fold-change| > 2 and p < 0.001. There were 1352 DEGs upregulated and 545 DEGs downregulated in the group treated with Ba. amyloliquefaciens Ba13 relative to the control group. GO analysis showed that the DEGs were enriched in 2484 GO terms, 433 of which showed significant differences in expression. The 100 most significantly enriched GO terms, and the number of DEGs enriched in these terms, are provided in Supplementary Table 3. KEGG analysis revealed that a total of 126 pathways were enriched in the treatment group relative to the control group. Significant differences were found in 12 pathways including photosynthesis-antenna proteins, plant hormone signal transduction, and phenylpropanoid biosynthesis (p < 0.05; Figure 2).
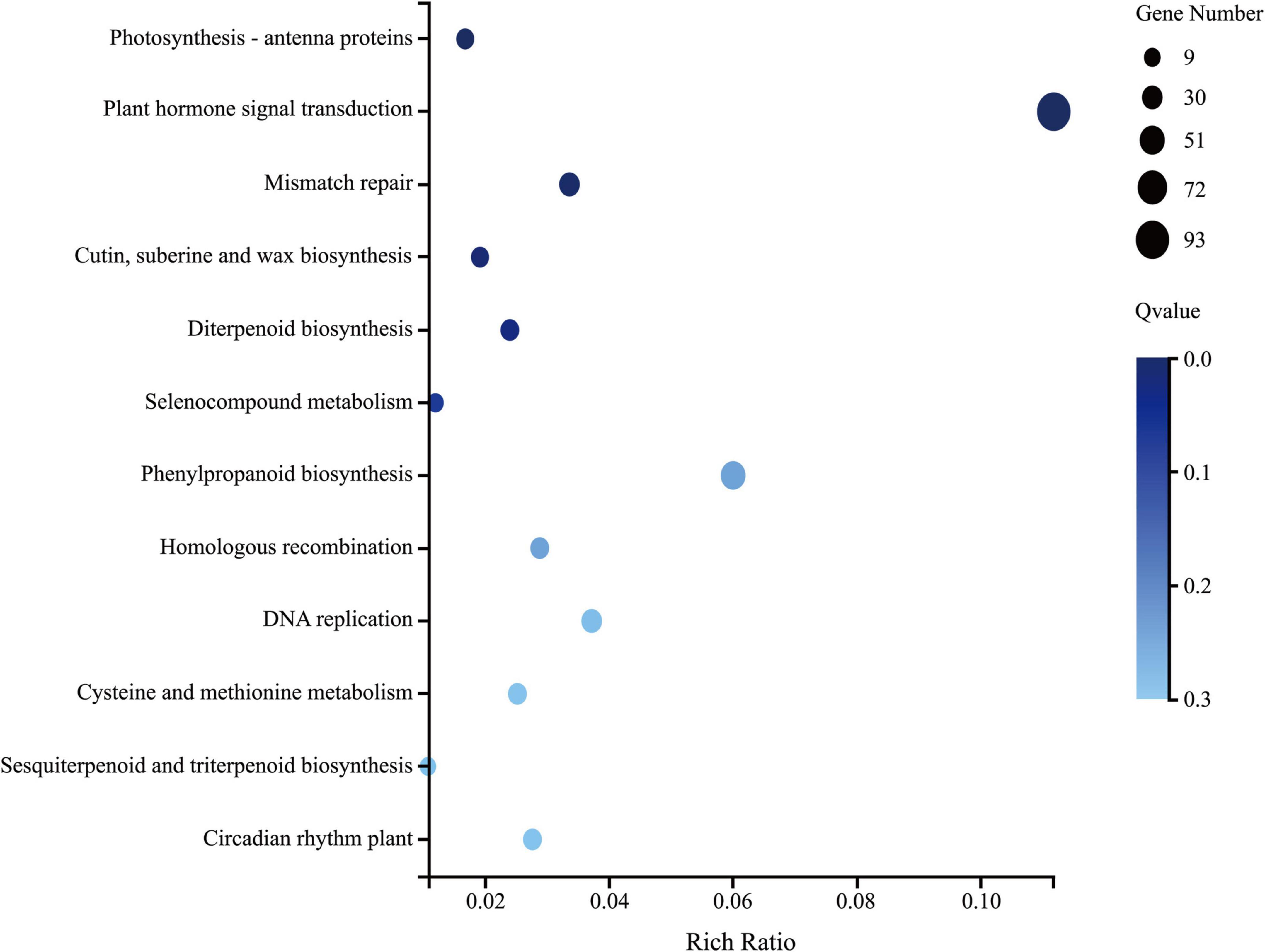
Figure 2. The 12 significantly enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways in tomato plants after treatment with Bacillus amyloliquefaciens Ba13.
We specifically analyzed gene silencing-related genes after treatment with Ba. amyloliquefaciens Ba13. GO analysis showed that a total of eight DEGs including AGO3, AGO4, AGO5, and AGO7 were enriched in the term gene silencing (GO: 0016458; Table 3). Multiple genes of the AGO family were upregulated in the RNAi system after treatment, with the fold-changes ranging from 2.03 to 6.73. The most pronounced upregulation was found in AGO4.
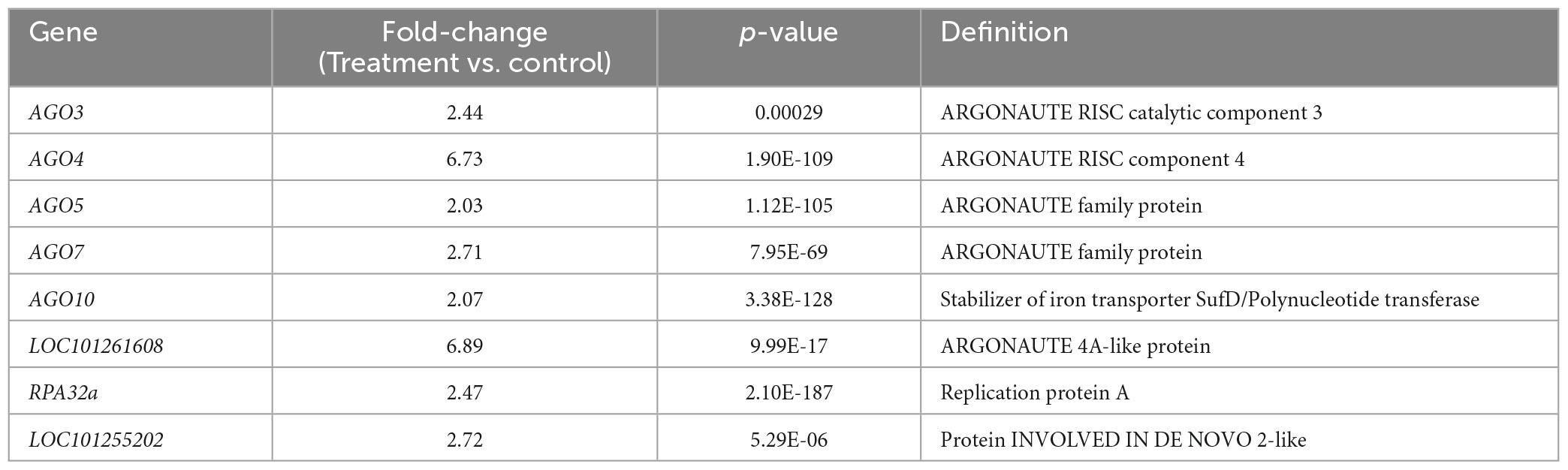
Table 3. Gene Ontology (GO) analysis of differentially expressed genes involved in gene silencing in tomato plants after treatment with Bacillus amyloliquefaciens Ba13.
In addition to the silencing-related genes, a number of the identified DEGs were found to be involved in disease resistance pathways. ABF, a key gene involved in ABA biosynthesis, was upregulated 2.40-fold. Regarding salicylic acid biosynthesis, NPR1, an important defense signal regulatory gene downstream of salicylic acid was upregulated 4.58-fold. For mitogen-activated protein kinase (MAPK), eight DEGs related to serine/threonine-protein kinase OXI1 (OX1), mitogen-activated protein kinase kinase kinase (MAPKKK) ANP1, and mitogen-activated protein kinase kinase 4/5 (MKK4/5) were all upregulated between 2.06- and 4.72-fold.
Some DEGs were also found to be involved in growth regulation pathways. Regarding photosynthesis-antenna proteins, 14 DEGs involved in the biosynthesis of four light-harvesting complex II (LHCII) proteins including Lhca4 and Lhcb1 were upregulated between 2.03- and 3.07-fold (Supplementary Figure 3). For plant hormone signal transduction, there were 16 DEGs related to early auxin-responsive proteins including auxin-responsive protein/indoleacetic acid (AUX/IAA), small auxin-up RNA (SAUR), and Gretchen Hagen 3 (GH3). Of these 16 DEGs, 15 were upregulated between 2.01- and 236.14-fold. Additionally, three DEGs related to the auxin response factor (ARF) were upregulated, with a maximum fold-change of 3.81. Moreover, the physiological functions of brassinosteroids are similar to those of auxin; in relation to brassinosteroid biosynthesis, 12 DEGs participating in the biosynthesis of the proteins brassinosteroid insensitive 1 (BRI1), brassinosteroid resistant 1/2 (BZR1/2), and BRI1 kinase inhibitor 1 (BKI1), were upregulated. Supplementary Table 4 summarizes the details (name, involved pathway, and expression change) of the above-mentioned genes.
The DEGs identified by whole-transcriptome RNA-seq were introduced into the corresponding biotic stress pathway in Mapman. Among the 1897 DEGs, 549 were enriched in this pathway. Following treatment with Ba. amyloliquefaciens Ba13, 97 of the 119 genes related to pathogenesis-related protein, auxin, and cell wall, were upregulated. Additionally, 67 of the 94 genes related to signaling and MAPK were upregulated (Figure 3). These results were consistent with the KEGG enrichment analysis of disease resistance-related genes.
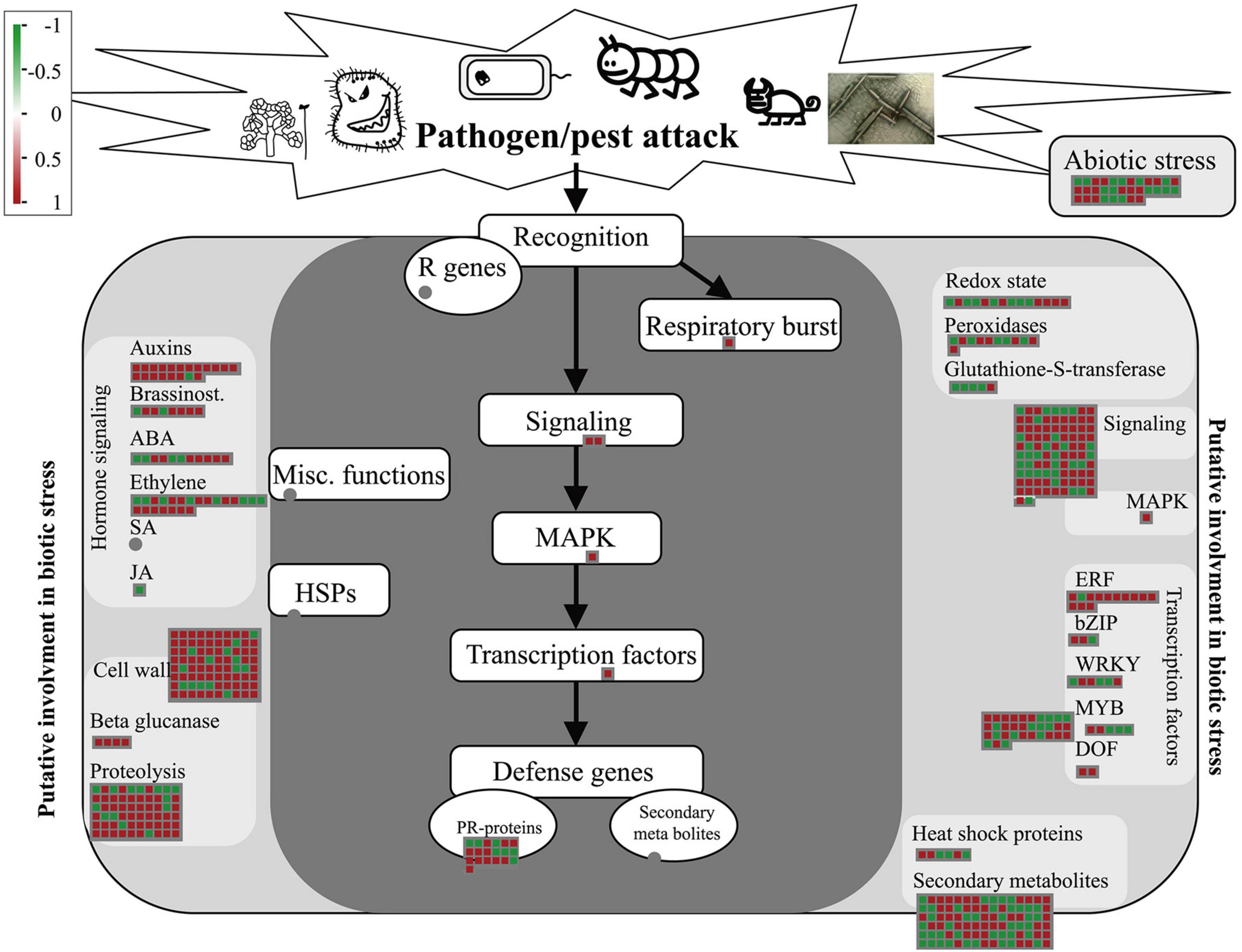
Figure 3. Gene expression changes of the biotic stress pathway in tomato plants after treatment with Bacillus amyloliquefaciens Ba13. Mapman was used to analyze the distribution of differentially expressed genes in the biotic stress pathway in the treatment group compared with the control group. Red squares indicate upregulated genes and green squares indicate downregulated genes.
The whole-transcriptome RNA-seq data were validated using RT-qPCR with six functional genes that were upregulated in the treatment group, with fold-changes of between 1.21 and 6.73. The PCR analysis revealed that all six genes increased their expression levels (between 1.61- and 4.34-fold) in the treatment group compared with the control group (Figure 4). The PCR results were generally consistent with the RNA-seq data. For example, according to the whole-transcriptome RNA-seq data, expression levels of AGO4 (an important component of TGS), FLS2 (a plant receptor kinase gene), and NPR1 (a regulatory gene in the resistance system) increased 6. 73-, 2. 13-, and 4.58-fold, respectively; their expression changes were 4. 34-, 1. 77-, and 4.55-fold based on the RT-qPCR analysis. These results demonstrated the reliability of the whole-transcriptome RNA-seq data.
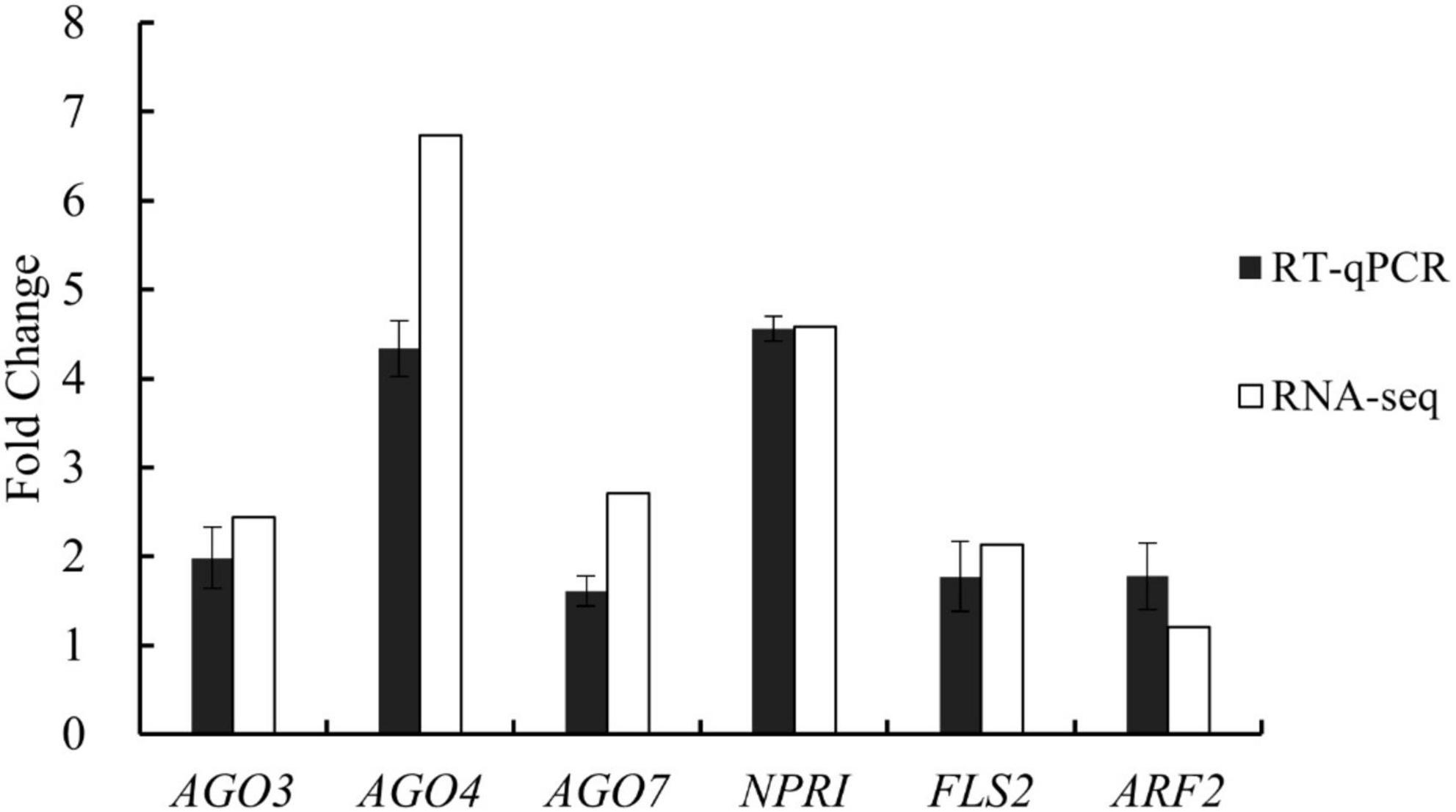
Figure 4. Expression changes of six genes involved in gene silencing, growth regulation, and disease resistance in tomato plants after treatment with Bacillus amyloliquefaciens Ba13 for validation of whole-transcriptome RNA sequencing (RNA-seq) data by reverse transcription quantitative real-time PCR (RT-qPCR).
The mean ABA content of tomato leaves in the treatment group was 693.3 ng/g at 28 dpvi, which was 15% higher compared with that of the control group (p < 0.05). The retention time and content of ABA after Ba. amyloliquefaciens Ba13 treatment are shown in Figure 5.
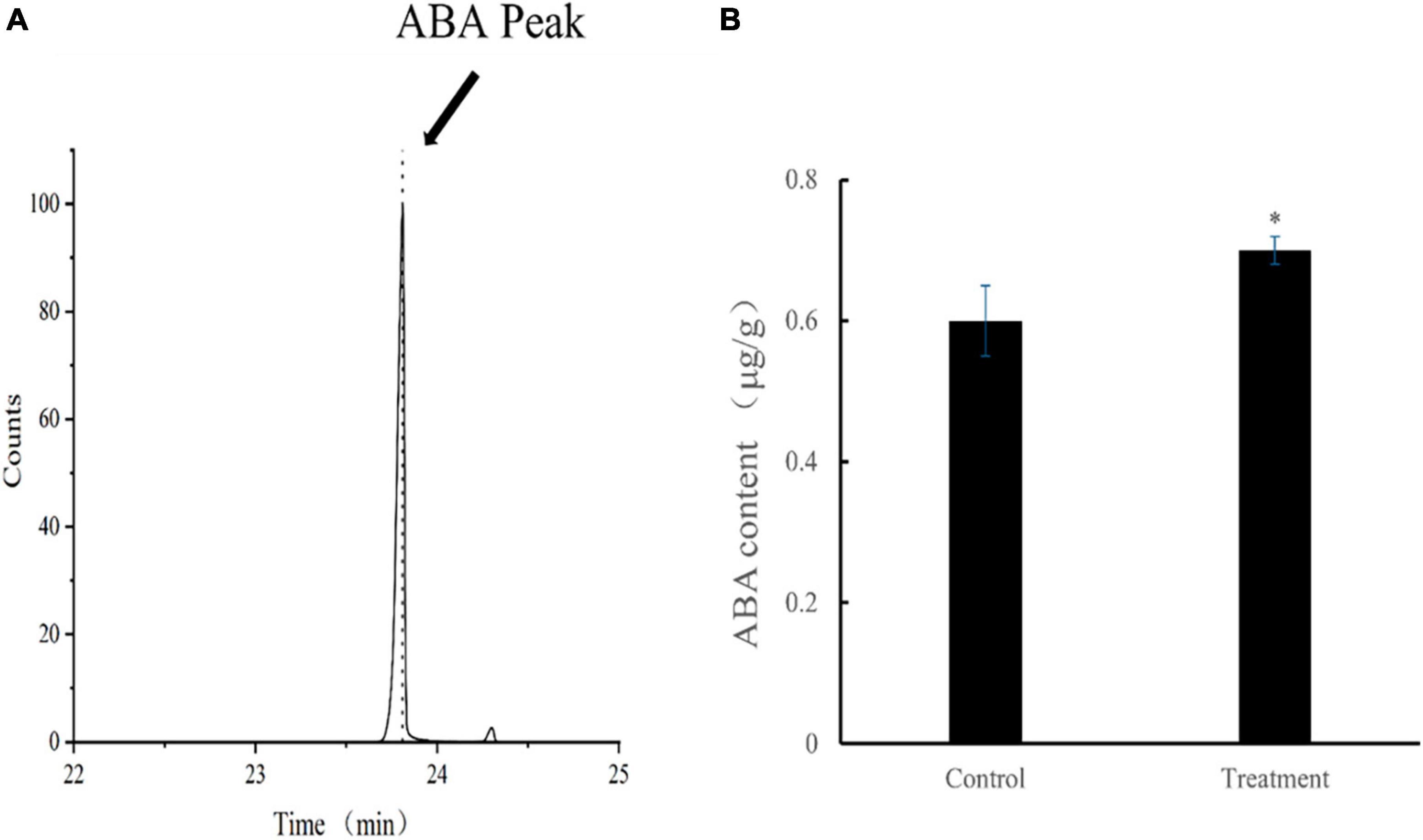
Figure 5. The content of abscisic acid (ABA) in leaves of tomato plants. (A) ABA standard retention time (23.787 min). (B) Changes in ABA content after Bacillus amyloliquefaciens Ba13 treatment compared with the control group. n = 3, error bar = standard deviation of ABA content in tomato leaves at 28 days post-viral infection. *p < 0.05.
Bisulfite sequencing detected methylation on 69 of 81 CpG sites in the TYLCV-SJ genome. The methylation level of sites 54, 228, 461, and 1244 ranged from 13.3 to 38.4%, which was significantly higher than that of other CpG sites (0.3%–1.8%; Figure 6). Among them, site 228 was located in the ORF region of AV1 and 461 sites were located in the ORF region of AV1 and AV2. After Ba. amyloliquefaciens Ba13 inoculation, the methylation level of sites 228 and 461 was 183.1% and 63.0% higher than that of the control group, respectively (p < 0.01). However, the methylation level of sites 54 and 1244 did not significantly differ between the two groups.
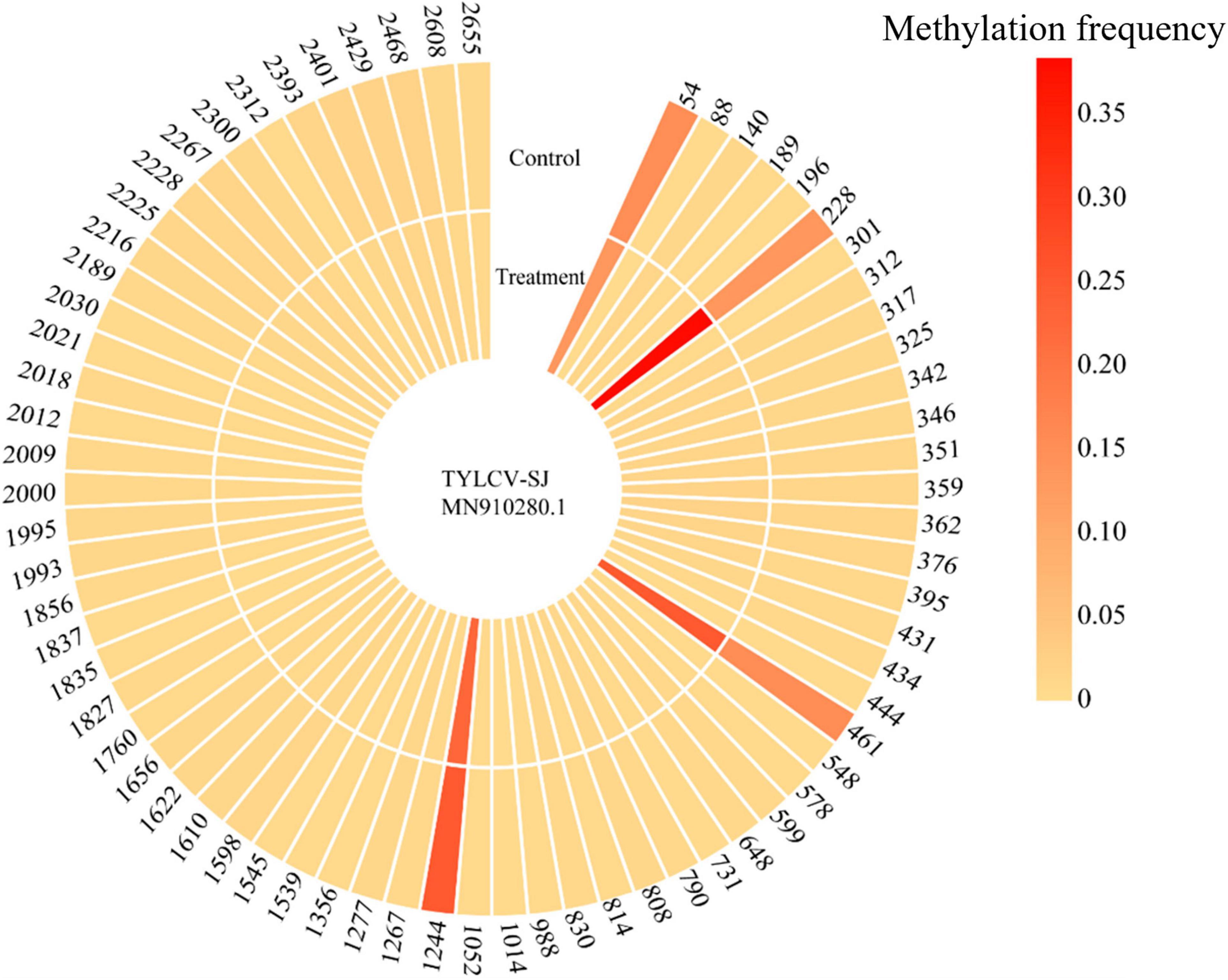
Figure 6. Changes in CpG methylation frequency of TYLCV genome after Bacillus amyloliquefaciens Ba13 treatment.
In total, 15,448 siRNA sequences were obtained by small RNA-seq, 8480 of which had significant differences in their expression (p < 0.05). There were 4578 siRNA upregulated and 3902 siRNA downregulated in the treatment group compared with the control group. Ten siRNAs completely matched to the genome of the TYLCV genotype used in this study (MN910280.1), and their virus binding sites and sequences are provided in Table 4. All the 10 siRNAs were downregulated in the Ba. amyloliquefaciens Ba13 treatment group compared with the control group, with 21–22 nt possibly participating in RNAi. For example, siRNA5429 was matched to the ORF region of AV1, and siRNA5327, siRNA12690, siRNA16430, and siRNA15617 were matched to the ORF region of C1. Their abundances in the treatment group were reduced by 47.9, 39.7, 60.3, 99.7, and 62.7%, respectively.
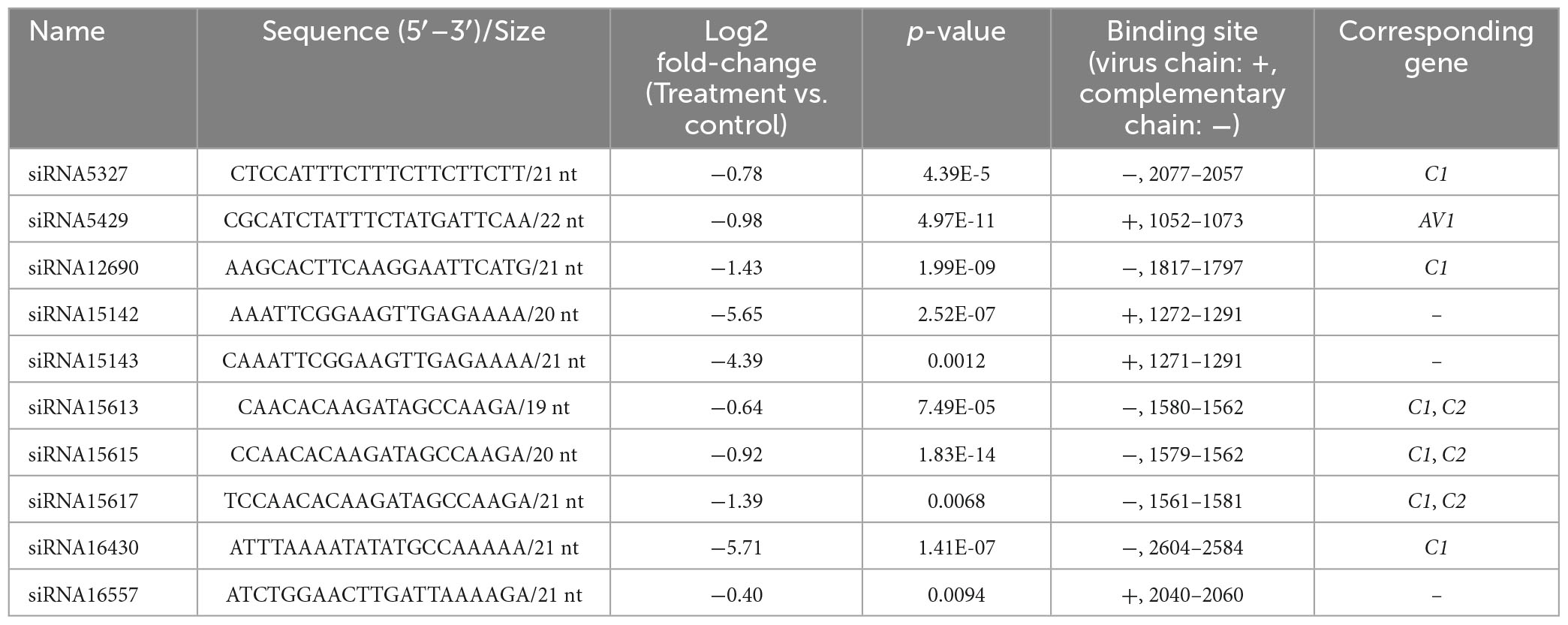
Table 4. Changes in the abundance of 10 virus-derived small interfering RNAs (siRNAs) in the leaves of tomato plants after treatment with Bacillus amyloliquefaciens Ba13.
RNAi is an antiviral system which allow plants to directly act on viruses. The application of Ba. amyloliquefaciens Ba13 as a biocontrol strain against TYLCV necessitates a holistic understanding of how this strain modulates the RNAi-based antiviral system in its host plants. In the present study, we found that inoculation with Ba. amyloliquefaciens Ba13 increased the expression of RNAi-related genes (e.g., AGO3, AGO4, AGO5, AGO7) in tomato leaves, with improved levels of viral genome methylation and ABA content. These results indicate that the tomato plants might have initiated RNAi-related pathways after TYLCV invasion, whereas the application of Ba. amyloliquefaciens Ba13 could improve plant resistance by enhancing RNAi.
As an integral part of RNAi, DNA methylation-mediated TGS is considered to be an important pathway for plant defense against infection by geminiviruses (Mei et al., 2020). For DNA viruses, AGO4 is the main effector protein in the antiviral silencing machinery. Based on the whole-transcriptome RNA-seq data, we observed upregulation of AGO4 expression in tomato leaves upon Ba. amyloliquefaciens Ba13 inoculation. Taking into account the role of AGO4–siRNA complex in viral genome methylation, we expected that Ba. amyloliquefaciens Ba13 mediated increased methylation level of the TYLCV genome. This hypothesis was supported by the higher methylation levels at CpG sites in AV1 and AV2 following Ba. amyloliquefaciens Ba13 inoculation. Previous study has found that after infection with the tomato leaf curl New Delhi virus, the methylation level of the viral intergenic spacer in a resistant cultivar was substantially higher than in a sensitive cultivar after infection. Additionally, Brough et al. (1992) introduced the cytosine methylated DNA of tomato golden mosaic virus into tobacco protoplasts, leading to a reduction in viral replication to 1/20–1/5 of the original level. These observations indicate that Ba. amyloliquefaciens Ba13 inoculation increased the methylation level of viral DNA through upregulation of AGO4 expression, which in turn inhibited AV1 and AV2 expression and enhanced plant resistance to TYLCV. It has been shown that the methylation frequency of CpG sites in the plant genome markedly increases during virus invasion. CpG methylation frequency is a key factor in inhibiting geminivirus replication and reducing plant symptoms (Rodriguez-Negrete et al., 2013). Further study is needed to ascertain whether inoculation with Ba. amyloliquefaciens Ba13 can affect the frequency of plant genome methylation, thereby improving tomato resistance to TYLCV.
In addition to AGO4, other AGO family members (e.g., AGO3, AGO5, AGO7) play non-negligible roles in plant antiviral defense. AGO3 has been reported to take part in ABA-mediated antiviral defense processes (Alazem et al., 2014). Promoters of AGO genes contain several ABA responsive elements, and their expression levels are affected by exogenous ABA (Alazem et al., 2017). Therefore, ABA can enhance the expression of AGO3—which is necessary for plants to resist Bamboo mosaic virus (Alazem et al., 2017). In the case of TYLCV infection, Ba. amyloliquefaciens Ba13 inoculation increased AGO3 expression in tomato plants. To clarify whether the upregulation of AGO3 expression is related to ABA, we looked at ABA biosynthesis related genes and ABA content in tomato leaves. We observed a remarkable upregulation in the expression of ABF upon Ba. amyloliquefaciens Ba13 inoculation, along with an increase in the ABA content. These results provide strong evidence that the application of Ba. amyloliquefaciens Ba13 improved ABA biosynthesis and enhanced the expression of AGO3, thereby facilitating plant defense against TYLCV.
With regard to AGO5 and AGO7 expression, we observed a distinct upregulation in tomato leaves upon Ba. amyloliquefaciens Ba13 inoculation. AGO5 can bind to small RNAs derived from viruses or viroids. Potato virus X (PVX) infection induces AGO5 expression, which is essential for Arabidopsis to limit PVX infection (Brosseau and Moffett, 2015). AGO7 is also involved in PTGS and thus plays a role in plant defense against viruses (Garcia-Ruiz et al., 2015). For example, AGO7 has been shown to collaborate with AGO1 in clearing viral RNA with different levels of secondary structure (Qu et al., 2008). AGO7 additionally participates in plant defense against Turnip crinkle virus, and together with AGO3, it becomes the major contributor to virus clearance in the leaves (Zheng et al., 2019). The collective results allow us to conclude that the application of Ba. amyloliquefaciens Ba13 upregulated the expression of AGO5 and AGO7, which synergistically acted to improve the efficiency of clearing viral RNAs in tomato plants.
While the expression levels of multiple AGO genes were upregulated in tomato plants inoculated with Ba. amyloliquefaciens Ba13, plant resistance to TYLCV was enhanced. It has been reported that an increase in AGO gene expression enhances plant resistance to different viruses such as Bamboo mosaic virus, Potato virus X, Turnip crinkle virus (Qu et al., 2008; Brosseau and Moffett, 2015; Alazem et al., 2017). This leads us to posit that Ba. amyloliquefaciens Ba13 is likely to regulate plant defense against other viruses by upregulating AGO expression. We additionally found that Streptomyces pactum Act12 can increase tomato resistance to TYLCV by upregulating AGO gene expression (Li Y. L. et al., 2019). This suggests that other biocontrol bacteria may perform similar functions as observed in Ba. amyloliquefaciens Ba13. More experiments are required to verify whether the effects and mechanisms of Ba. amyloliquefaciens Ba13 are universal for other viruses and biocontrol strains.
RNAi plays a role in disease control via siRNAs, which target viral mRNAs with corresponding sequences at specific sites for degradation. We compared the obtained siRNA sequences against the TYLCV genotype (MN910280.1) and found 10 siRNAs completely matched to our obtained viral sequence. The siRNA matcher site in the ORF were corresponding to AV1, C1, and C2. AV1 and C1 are, respectively, involved in coding virus coat protein and replication-related proteins, which help TYLCV growth and spread. C2 can revert TGS and decrease DNA methylation in plants by interfering with the methyl cycle (Buchmann et al., 2009). The complete matching of these siRNAs to the functional gene regions of TYLCV indicates that the siRNAs may interfere with viral protein synthesis by complementary binding and degrading mRNA of gene transcription. This result illustrates that RNAi-related pathways were initiated in tomato plants in response to TYLCV infection.
We observed Ba. amyloliquefaciens Ba13-mediated downregulation of all 10 siRNAs matched to the TYLCV genotype in tomato leaves, consistent with the pattern observed in a previous study. Rodriguez-Negrete et al. (2009) investigated the recovery of plants (with less susceptible symptoms) after infection with pepper golden mosaic virus. They found a lower total abundance of virus-derived siRNAs but a higher DNA methylation level of the viral genome in the recovered tissue than in the diseased tissue of pepper plants. Additionally, we found that the disease symptoms of Ba. amyloliquefaciens Ba13-inoculated tomato plants were alleviated compared with those of non-inoculated controls. The decrease in siRNA abundances could be related to the decrease in virus quantity in tomato plants treated with Ba. amyloliquefaciens Ba13. Ba. amyloliquefaciens Ba13 inoculation enhanced plant defense against TYLCV, allowing faster recovery from viral infection.
Our results indicated that the expression levels of genes related to SA, ABA, and RNAi pathways were upregulated in tomato leaves after inoculation with Ba. amyloliquefaciens Ba13. In fact, the defense processes of plants against viruses involve the participation and collaboration of multiple mechanisms, such as RNAi, dominant resistance, and SAR (Nicaise, 2014). Thus, inoculation with Ba. amyloliquefaciens Ba13 is likely to induce multifaceted plant response through various defense systems. Although both SAR and RNAi play a role in plant defense against viruses, RNAi has higher specificity for viruses compared with the broad-spectrum resistance mechanism of SAR and can target degradation of viruses. While this study has focused on RNAi modulation by Ba. amyloliquefaciens Ba13, the relationship between RNAi and other defense mechanisms in inoculated plants should be clarified by further studies.
Furthermore, we found that the application of Ba. amyloliquefaciens Ba13 strongly affected the expression of genes related to photosystem and auxin response pathways. A total of 12 DEGs related to the biosynthesis of LHCII chlorophyll a/b binding protein 1 (LHCb1), LHCb3, and LHCb7 in the LHCII pathway (the most abundant light harvester in plants) were upregulated in tomato leaves upon Ba. amyloliquefaciens Ba13 inoculation. Additionally, 13 of the 14 DEGs related to auxin/indoleacetic acid and small auxin-up RNA, as well as two DEGs related to GH3, were upregulated in tomato leaves of inoculated plants. Montasser et al. (2012) found that the content of photosynthesis related chemical components (e.g., chlorophyll a, b) decreased in tomato leaves infected with TYLCV compared with normal leaves. Wu et al. (2012) showed that auxin/indoleacetic acid proteins were significantly expressed in tomato plants during root and stem development. Small auxin-up RNA plays a major role in modulating cell elongation and positively regulates cell expansion by modulating the transport of auxin (Chae et al., 2012). Furthermore, GH-like genes mediate the interaction between auxin and salicylic acid signaling pathways. Overexpression of GH3 can enhance the systemic disease resistance in plants (Zhang et al., 2007). Taken together, these results indicate that the presence of Ba. amyloliquefaciens Ba13 could attenuate the negative effects of viral infection on leaf photosynthetic efficiency, promote plant growth, and as such, enhance plant defense against TYLCV.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, MN910280.1, https://www.ncbi.nlm.nih.gov/, PRJNA553064, https://www.ncbi.nlm.nih.gov/, PRJNA553309.
HL conceived the study and proposed the experimental approaches. QG, YS, and CJ conducted the experiments. QG, YS, ZK, ZL, and YZL contributed to data processing and analysis. QG, YS, and YLL wrote the manuscript. All authors read, commented on, and approved the final version of the manuscript.
This research was financially supported by the Qinchuangyuan Scientific and Technological Innovation Funds (No. 2021ZDZXNY-0005), the Science and Technology Major Project of Ordos (2022EEDSKJZDZX019), the Science and Technology Planning Project of Inner Mongolia Autonomous Region (2022YFHH0114), the Natural Science Foundation of Shaanxi Province (No. 2021JQ-151), and the Youth Project of National Natural Science Foundation of China (31600407).
We thank Chaofeng Lin (Transcend Envirotech Consulting Co., Ltd.) for improving the English and Lu Wang (Plant Science Facility of the Institute of Botany, Chinese Academy of Sciences) for technical assistance with GC-MS analysis.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1251698/full#supplementary-material
Alazem, M., He, M., Moffett, P., and Lin, N. (2017). Abscisic acid induces resistance against bamboo mosaic virus through Argonaute 2 and 3. Plant Physiol. 174, 339–355. doi: 10.1104/pp.16.00015
Alazem, M., Lin, K., and Lin, N. (2014). The abscisic acid pathway has multifaceted effects on the accumulation of Bamboo mosaic virus. Mol. Plant Microbe. Interact. 27, 177–189. doi: 10.1094/MPMI-08-13-0216-R
Arif, M. A., Frank, W., and Khraiwesh, B. (2013). Role of RNA interference (RNAi) in the Moss Physcomitrella patens. Int. J. Mol. Sci. 14, 1516–1540. doi: 10.3390/ijms14011516
Brosseau, C., and Moffett, P. (2015). Functional and genetic analysis identify a role for Arabidopsis ARGONAUTE5 in antiviral RNA silencing. Plant Cell 27, 1742–1754. doi: 10.1105/tpc.15.00264
Brough, C. L., Gardiner, W. E., Inamdar, N. M., Zhang, X. Y., Ehrlich, M., and Bisaro, D. M. (1992). DNA methylation inhibits propagation of tomato golden mosaic virus DNA in transfected protoplasts. Plant Mol. Biol. 18, 703–712. doi: 10.1007/BF00020012
Buchmann, R. C., Asad, S., Wolf, J. N., Mohannath, G., and Bisaro, D. M. (2009). Geminivirus AL2 and L2 proteins suppress transcriptional gene silencing and cause genome-wide reductions in cytosine methylation. J. Virol. 83, 5005–5013. doi: 10.1128/JVI.01771-08
Cahill, M., Jarvis, W., Gorman, K., and Denholm, I. (1996). Resolution of baseline responses and documentation of resistance to buprofezin in Bemisia tabaci (Homoptera: Aleyrodidae). Bull. Entomol. Res. 86, 117–122. doi: 10.1017/s0007485300052342
Chae, K., Isaacs, C. G., Reeves, P. H., Maloney, G. S., Muday, G. K., Nagpal, P., et al. (2012). Arabidopsis small auxin up RNA63 promotes hypocotyl and stamen filament elongation. Plant J. 71, 684–697. doi: 10.1111/j.1365-313X.2012.05024.x
de Pradena, A., Jimenez, A., San Leon, D., Simmonds, P., Garcia, J., and Valli, A. (2020). Plant virus genome is shaped by specific dinucleotide restrictions that influence viral infection. Mbio 11, e2818–e2819. doi: 10.1128/mBio.02818-19
El-Sappah, A. H., Yan, K., Huang, Q. L., Islam, M. M., Li, Q. Z., Wang, Y., et al. (2021). Comprehensive mechanism of gene silencing and its role in plant growth and development. Front. Plant. Sci. 12:705249. doi: 10.3389/fpls.2021.705249
Elsharkawy, M. M., Shimizu, M., Takahashi, H., and Hyakumachi, M. (2012). Induction of systemic resistance against cucumber mosaic virus by Penicillium simplicissimum GP17-2 in Arabidopsis and tobacco. Plant Pathol. 61, 964–976. doi: 10.1111/j.1365-3059.2011.02573.x
Elsharkawy, M. M., Shimizu, M., Takahashi, H., Ozaki, K., and Hyakumachi, M. (2013). Induction of systemic resistance against cucumber mosaic virus in Arabidopsis thaliana by trichoderma asperellum SKT-1. Plant Pathol. J. 29, 193–200. doi: 10.5423/ppj.si.07.2012.0117
Gafni, Y. (2003). Tomato yellow leaf curl virus, the intracellular dynamics of a plant DNA virus. Mol. Plant Pathol. 4, 9–15. doi: 10.1046/j.1364-3703.2003.00147.x
Gao, F., Zhang, J. Y., Jiang, P. P., Gong, D. S., Wang, J. W., Xia, Y. D., et al. (2014). Marked methylation changes in intestinal genes during the perinatal period of preterm neonates. BMC Genomics 15:716. doi: 10.1186/1471-2164-15-716
Garcia-Ruiz, H., Carbonell, A., Hoyer, J. S., Fahlgren, N., Gilbert, K. B., Takeda, A., et al. (2015). Roles and programming of Arabidopsis ARGONAUTE proteins during Turnip Mosaic Virus infection. Plos Pathog. 11:e1004755. doi: 10.1371/journal.ppat.1004755
Gong, P., Tan, H., Zhao, S. W., Li, H., Liu, H., Ma, Y., et al. (2021). Geminiviruses encode additional small proteins with specific subcellular localizations and virulence function. Nat. Commun. 12:4278. doi: 10.1038/s41467-021-24617-4
Guo, Q., Li, Y. L., Lou, Y., Shi, M. D., Jiang, Y., Zhou, J., et al. (2019). Bacillus amyloliquefaciens Ba13 induces plant systemic resistance and improves rhizosphere microecology against tomato yellow leaf curl virus disease. Appl. Soil Ecol. 137, 154–166. doi: 10.1016/j.apsoil.2019.01.015
Kanakala, S., and Ghanim, M. (2016). RNA interference in insect vectors for plant viruses. Viruses Basel 8:329. doi: 10.3390/v8120329
Kanehisa, M., Araki, M., Goto, S., Hattori, M., Hirakawa, M., Itoh, M., et al. (2008). KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36, 480–484. doi: 10.1093/nar/gkm882
Kumar, S., Chauhan, P. S., Agrawal, L., Raj, R., Srivastava, A., Gupta, S., et al. (2016). Paenibacillus lentimorbus inoculation enhances tobacco growth and extenuates the virulence of cucumber mosaic virus. PLoS ONE 11:e0149980. doi: 10.1371/journal.pone.0149980
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Li, R. Q., Yu, C., Li, Y. R., Lam, T. W., Yiu, S. M., Kristiansen, K., et al. (2009). SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics 25, 1966–1967. doi: 10.1093/bioinformatics/btp336
Li, Y. L., Guo, Q., Li, Y. Z., Sun, Y. F., Xue, Q. H., and Lai, H. X. (2019). Streptomyces pactum Act12 controls tomato yellow leaf curl virus disease and alters rhizosphere microbial communities. Biol. Fertil. Soils 55, 149–169. doi: 10.1007/s00374-019-01339-w
Li, Z. C., Li, W. Q., Guo, M. X., Liu, S. M., Liu, L., Yu, Y., et al. (2019). Origin, evolution and diversification of plant ARGONAUTE proteins. Plant J. 109, 1086–1097. doi: 10.1111/tpj.15615
Loew, L., Goonawardane, N., Ratcliff, J., Nguyen, D., and Simmonds, P. (2020). Use of a small DNA virus model to investigate mechanisms of CpG dinucleotide-induced attenuation of virus replication. J. Gen. Virol. 101, 1202–1218. doi: 10.1099/jgv.0.001477
Lu, D. Y. H., Liao, J. Y., Fajar, A., Chen, J. B., Wei, Y., Zhang, Z. H., et al. (2023). Co-infection of TYLCV and ToCV increases cathepsin B and promotes ToCV transmission by Bemisia tabaci MED. Front. Microbiol. 14:1107038. doi: 10.3389/fmicb.2023.1107038
Matzke, M. A., Kanno, T., and Matzke, A. J. M. (2015). RNA-directed DNA methylation: The evolution of a complex epigenetic pathway in flowering plants. In: Merchant SS ed. Annu. Rev. Plant Biol. 66, 243–267. doi: 10.1146/annurev-arplant-043014-114633
Mei, Y., Wang, Y., Li, F., and Zhou, X. (2020). The C4 protein encoded by tomato leaf curl Yunnan virus reverses transcriptional gene silencing by interacting with NbDRM2 and impairing its DNA-binding ability. PLoS Pathol. 16:1008829. doi: 10.1371/journal.ppat.1008829
Mi, H. Y., Lazareva-Ulitsky, B., Loo, R., Kejariwal, A., Vandergriff, J., Rabkin, S., et al. (2005). The panther database of protein families, subfamilies, functions and pathways. Nucleic Acids Res. 33, D284–D288. doi: 10.1093/nar/gki078
Montasser, M. S., Al-Own, F. D., Haneif, A. M., and Afzal, M. (2012). Effect of Tomato yellow leaf curl bigeminivirus (TYLCV) infection on tomato cell ultrastructure and physiology. Can. J. Plant Pathol. 34, 114–125. doi: 10.1080/07060661.2012.661767
Mou, Z., Fan, W. H., and Dong, X. N. (2003). Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113, 935–944. doi: 10.1016/s0092-8674(03)00429-x
Nicaise, V. (2014). Crop immunity against viruses: Outcomes and future challenges. Front. Plant Sci. 21:660. doi: 10.3389/fpls.2014.00660
Niu, D. D., Liu, H. X., Jiang, C. H., Wang, Y. P., Wang, Q. Y., Jin, H. L., et al. (2011). The plant growth-promoting rhizobacterium Bacillus cereus AR156 induces systemic resistance in Arabidopsis thaliana by simultaneously activating salicylate- and jasmonate/ethylene-dependent signaling pathways. Mol. Plant Microbe Interact. 24, 533–542. doi: 10.1094/MPMI-09-10-0213
Padmanabhan, C., Zheng, Y., Shamimuzzaman, M., Fei, Z., and Ling, K. S. (2017). Tomato yellow leaf curl virus C4 protein is a determinant of disease phenotype in tomato. Phytopathology 107, 91–91.
Palumbo, J. C., Horowitz, A. R., and Prabhaker, N. (2001). Insecticidal control and resistance management for Bemisia tabaci. Crop Prot. 20, 739–765. doi: 10.1016/s0261-2194(01)00117-x
Pan, X. Q., Welti, R., and Wang, X. M. (2010). Quantitative analysis of major plant hormones in crude plant extracts by high-performance liquid chromatography-mass spectrometry. Nat. Protoc. 5, 986–992. doi: 10.1038/nprot.2010.37
Qu, F., Ye, X. H., and Morris, T. J. (2008). Arabidopsis DRB4, AGO1, AGO7, and RDR6 participate in a DCL4-initiated antiviral RNA silencing pathway negatively regulated by DCL1. Proc. Natl. Acad. Sci. U.S.A. 105, 14732–14737. doi: 10.1073/pnas.0805760105
Raja, P., Sanville, B. C., Buchmann, R. C., and Bisaro, D. M. (2008). Viral genome methylation as an epigenetic defense against geminiviruses. J. Virol 82, 8997–9007. doi: 10.1128/JVI.00719-08
Ramesh, S. V., Mishra, A. K., and Praveen, S. (2007). Hairpin RNA-Mediated strategies for silencing of Tomato leaf curl virus AC1 and AC4 genes for effective resistance in plants. Oligonucleotides 17, 251–257. doi: 10.1089/oli.2006.0063
Rodriguez-Negrete, E., Carrillo-Tripp, J., and Rivera-Bustamante, R. F. (2009). RNA silencing against geminivirus: Complementary action of posttranscriptional gene silencing and transcriptional gene silencing in host recovery. J. Virol. 83, 1332–1340. doi: 10.1128/jvi.01474-08
Rodriguez-Negrete, E., Lozano-Duran, R., Piedra-Aguilera, A., Cruzado, L., Bejarano, E. R., and Castillo, A. G. (2013). Geminivirus Rep protein interferes with the plant DNA methylation machinery and suppresses transcriptional gene silencing. New Phytol. 199, 464–475. doi: 10.1111/nph.12286
Sade, D., Sade, N., Shriki, O., Lerner, S., Gebremedhin, A., Karavani, A., et al. (2014). Water balance, hormone homeostasis, and sugar signaling are all involved in tomato resistance to tomato yellow leaf curl virus. Plant Physiol. 165, 1684–1697. doi: 10.1104/pp.114.243402
Schmittgen, T. D., and Livak, K. J. (2008). Analyzing real-time PCR data by the comparative C-T method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Seo, J. K., Wu, J. G., Lii, Y. F., Li, Y., and Jin, H. L. (2013). Contribution of small RNA pathway components in plant immunity. Mol. Plant Microbe Interact. 26, 617–625. doi: 10.1094/mpmi-10-12-0255-ia
Shen, L. L., Wang, F. L., Yang, J. G., Qian, Y. M., Dong, X. W., and Zhan, H. X. (2014). Control of tobacco mosaic virus by Pseudomonas fluorescens CZ powder in greenhouses and the field. Crop Prot. 56, 87–90. doi: 10.1016/j.cropro.2013.11.020
Staff, S. S. (2010). Keys to Soil Taxonomy, 11th Edn. Washington, DC: USDA-Natural Resources Conservation Service.
Sun, S., Kang, X. P., Xing, X. J., Chen, Z. F., Zheng, S. W., and Xing, G. M. (2014). Transient expression of siRNA targeted against the TYLCV AV1, AC1 and AC3 genes for high resistance in tomato. Sci. Hortic. 179, 321–327. doi: 10.1016/j.scienta.2014.10.002
Sun, W. J., Lv, W. J., Li, L. N., Yin, G., Hang, X. F., Xue, Y. F., et al. (2016). Eugenol confers resistance to tomato yellow leaf curl virus (TYLCV) by regulating the expression of SlPer1 in tomato plants. Nat. Biotechnol. 33, 345–354. doi: 10.1016/j.nbt.2016.01.001
Thimm, O., Blasing, O., Gibon, Y., Nagel, A., Meyer, S., Kruger, P., et al. (2004). MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37, 914–939. doi: 10.1111/j.1365-313X.2004.02016.x
van Wezel, R., Dong, X. L., Liu, H. T., Tien, P., Stanley, J., and Hong, Y. G. (2002). Mutation of three cysteine residues in Tomato yellow leaf curl virus-China C2 protein causes dysfunction in pathogenesis and posttranscriptional gene-silencing suppression. Mol. Plant Microbe Interact. 15, 203–208. doi: 10.1094/MPMI.2002.15.3.203
Vitti, A., La Monaca, E., Sofo, A., Scopa, A., Cuypers, A., and Nuzzaci, M. (2015). Beneficial effects of Trichoderma harzianum T-22 in tomato seedlings infected by cucumber mosaic virus (CMV). Biocontrol 60, 135–147. doi: 10.1007/s10526-014-9626-3
Wang, L. K., Feng, Z. X., Wang, X., Wang, X. W., and Zhang, X. G. (2010). DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26, 136–138. doi: 10.1093/bioinformatics/btp612
Wang, S. A., Wu, H. J., Qiao, J. Q., Ma, L. L., Liu, J., Xia, Y. F., et al. (2009). Molecular mechanism of plant growth promotion and induced systemic resistance to tobacco mosaic virus by Bacillus spp. J. Microbiol. Biotechnol. 19, 1250–1258. doi: 10.4014/jmb.0901.008
Wu, J., Peng, Z., Liu, S. Y., He, Y. J., Cheng, L., Kong, F. L., et al. (2012). Genome-wide analysis of Aux/IAA gene family in Solanaceae species using tomato as a model. Mol. Genet. Genomics 287, 295–311. doi: 10.1007/s00438-012-0675-y
Wu, J., Zhang, J., and Zhou, X. (2010). AV2 protein encoded by tomato yellow leaf curl China virus is an RNA silencing suppressor. Phytopathology 100, S139–S139.
Yaakov, N., Levy, Y., Belausov, E., Gaba, V., Lapidot, M., and Gafni, Y. (2011). Effect of a single amino acid substitution in the NLS domain of tomato yellow leaf curl virus-Israel (TYLCV-IL) capsid protein (CP) on its activity and on the virus life cycle. Virus Res. 158, 8–11. doi: 10.1016/j.virusres.2011.02.016
Zhang, Z. Q., Li, Q., Li, Z. M., Staswick, P. E., Wang, M. Y., Zhu, Y., et al. (2007). Dual regulation role of GH3.5 in salicylic acid and auxin signaling during Arabidopsis-Pseudomonas syringae interaction. Plant Physiol. 145, 450–464. doi: 10.1104/pp.107.106021
Zhao, S., Gong, P., Ren, Y., Liu, H., Li, H., Li, F., et al. (2022). The novel C5 protein from tomato yellow leaf curl virus is a virulence factor and suppressor of gene silencing. Stress Biol. 2:19. doi: 10.1007/s44154-022-00044-3
Keywords: Bacillus amyloliquefaciens, RNA interference, ARGONAUTE family, plant viral disease, antiviral defense
Citation: Guo Q, Sun Y, Ji C, Kong Z, Liu Z, Li Y, Li Y and Lai H (2023) Plant resistance to tomato yellow leaf curl virus is enhanced by Bacillus amyloliquefaciens Ba13 through modulation of RNA interference. Front. Microbiol. 14:1251698. doi: 10.3389/fmicb.2023.1251698
Received: 02 July 2023; Accepted: 18 September 2023;
Published: 06 October 2023.
Edited by:
Jian-Wei Guo, Kunming University, ChinaReviewed by:
Gaurav Raikhy, Louisiana State University Health Shreveport, United StatesCopyright © 2023 Guo, Sun, Ji, Kong, Liu, Li, Li and Lai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hangxian Lai, bGFpaGFuZ3hpYW5AMTYzLmNvbQ==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.