- Department of Urology, Cancer Hospital of China Medical University, Liaoning Cancer Hospital and Institute, Shenyang, Liaoning, China
Aim: Recent observational and small-sample case-control studies have shown a relationship between gut microbiota composition and prostatic cancer (PCa). Nevertheless, the causal association between gut microbiota and PCa is still unclear. Herein, we used the Mendelian randomization (MR) method to explore the potential causal relationship between gut microbiota and PCa.
Methods: In this two-sample MR study, data were extracted from the summary statistics of gut microbiota from the largest available genome-wide association study meta-analysis conducted by the MiBioGen consortium (n = 14,306) and the Dutch Microbiome Project (n = 8,208). Summary statistics for PCa were obtained from the FinnGen consortium release data (n = 95,213). Inverse variance weighted (IVW), MR-Egger, strength test (F), and MR-PRESSO were used to examine the potential causal association between gut microbiota and PCa. Cochran's Q statistics were used to quantify the heterogeneity of instrumental variables.
Results: IVW estimates suggested that the relative abundance of Akkermansia muciniphila (odds ratio [OR] = 0.7926, 95% confidence interval [CI]: 0.6655–0.9440) and Bacteroides salyersiae (OR = 0.9023, 95% CI: 0.8262–0.9853) were negatively associated with the odds of PCa, while that of Eubacterium biforme (OR = 1.1629, 95% CI: 1.0110–1.3376) was positively associated with the odds of PCa. In addition, we explored these relationships among patients without other cancers and similarly found that the relative abundance of Akkermansia muciniphila, Bacteroides salyersiae, and Eubacterium biforme were linked to PCa (all P < 0.05).
Conclusion: Gut microbiota potentially influenced the occurrence of PCa. Our findings may provide some new ideas for researching the methods of PCa prevention. In addition, further studies are needed to explore the causal association and specific underlying mechanisms between gut microbiota and PCa.
Introduction
The role of gut microbiota in prostate cancer (PCa) has received a lot of attention in recent years (Rizzo et al., 2022), and the concept of “gut-prostate axis” has been proposed (Fujita et al., 2023; Matsushita et al., 2023). Previous basic studies suggested that the underlying mechanisms of gut microbiota's effects on the incidence and progression of PCa may involve modulating inflammation, oxidative stress, and immune function and interfering with lipid metabolism (Porter et al., 2018; Wheeler and Liss, 2019). Nevertheless, evidence from population-based studies on the relationship between gut microbiota and PCa remains limited, and most of the existing studies are small-sample case-control studies.
Studies have suggested that patients with PCa exhibit an increased relative abundance of the bacterial genera Veillonella, Bacteroides (Alanee et al., 2019), Streptococcus (Liss et al., 2018), Rikenellaceae, Alistipes, and Lachnospira (Matsushita et al., 2021). While in control groups, the relative abundance of Faecalibacterium prausnitzii and Eubacterium rectalie was higher than that in cases (Golombos et al., 2018). However, the firm conclusions for the causal relationship and its direction between gut microbiota and PCa are not yet enough to draw according to the existing observational studies' evidence. Inherent defects limited traditional observational studies, and thus the causal role of gut microbiota in the risk of PCa remained to be clarified (Tong et al., 2020). In spite of the fact that randomized controlled trial is recognized as a gold standard for determining causality, its application in clinical settings is impractical due to the long incubation period from certain microbiota exposure to oncogenesis (Spieth et al., 2016). Under the circumstances, a novel method for investigating the causal association between gut microbiota and PCa is warranted.
Mendelian randomization (MR) analysis has become a widely used approach that exploits single nucleotide polymorphisms (SNPs) as unconfounded instrumental variants (IVs) to explore the potential causal relationships between environmental exposures and diseases (Davey Smith and Hemani, 2014; Sekula et al., 2016). MR can avoid reverse causality inferences and reflect the long-term effects of exposures on outcomes. A recent two-sample MR analysis assessed the causal effect of gut microbiota on cancer risk and showed that the relative abundance of Alphaproteobacteria, Rhodospirillales, Adlercreutzia, and Coprobacter was associated with PCa (Wei et al., 2023). However, the findings of Wei's study were not in accordance with those of other epidemiological studies, and they also did not explore the causal associations between gut microbiota and cancer risk at the species level.
Herein, in this two-sample MR study, we aimed to investigate the potential causal relationship between gut microbiota at different levels, especially at the species level, and PCa in order to provide some new ideas for exploring the methods for PCa prevention and treatment.
Methods
Data sources
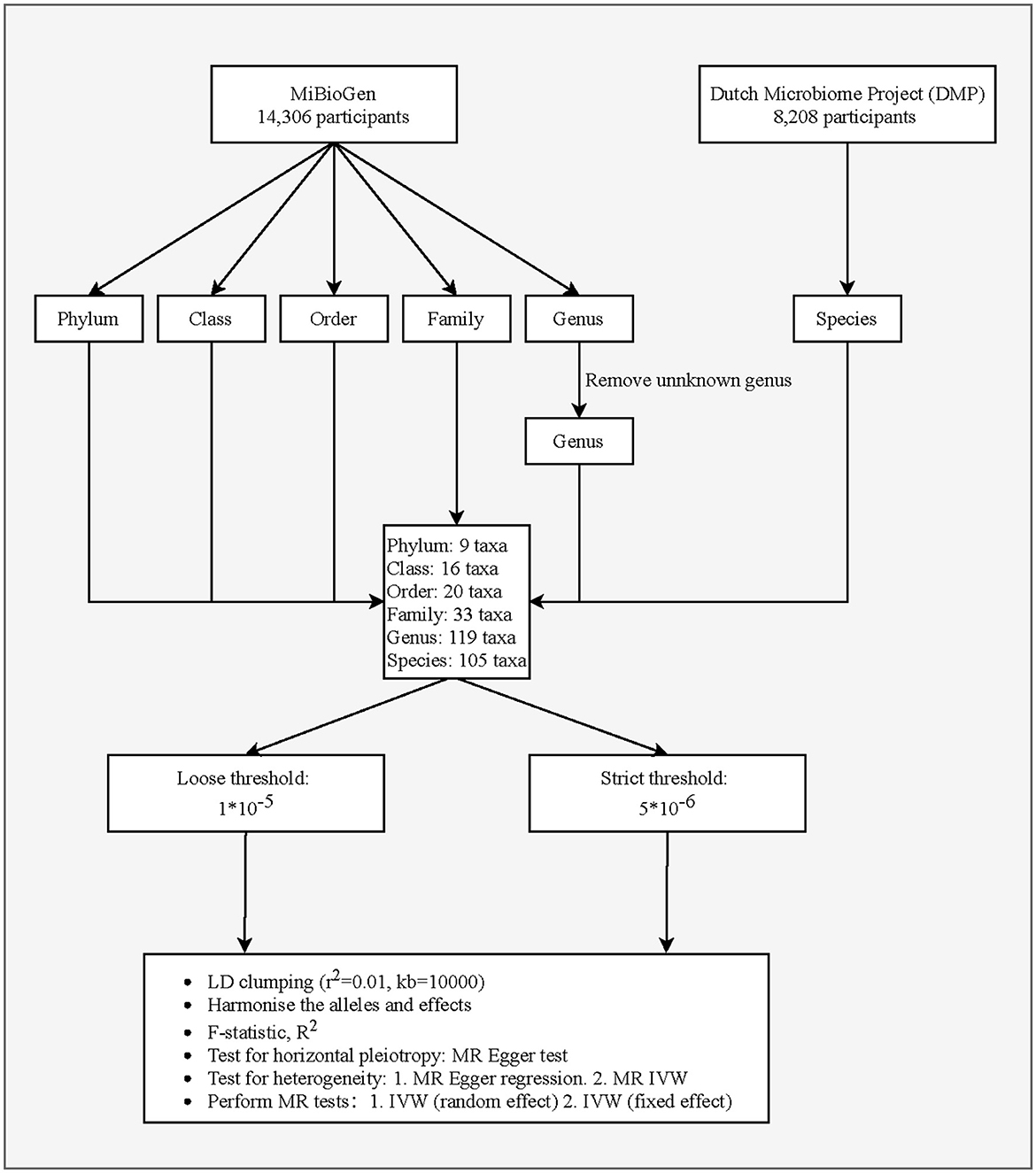
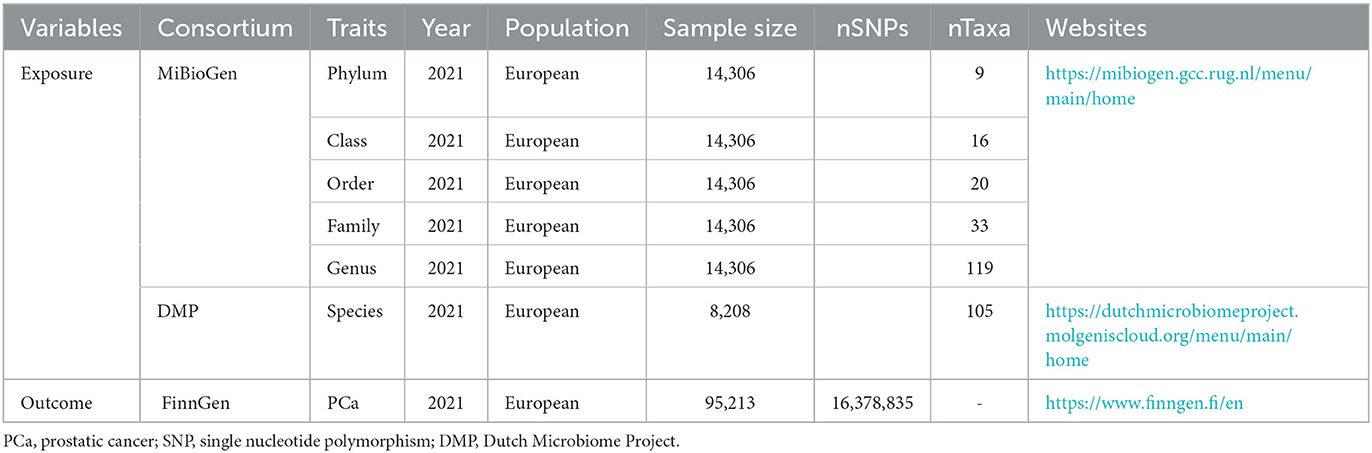
This study is a two-sample MR analysis. Data from the genome-wide association studies (GWASs) were extracted for gut microbiota and PCa. Figure 1 is the flowchart of this research procedure. Genetic variants of the gut microbiota were obtained from MiBioGen (Kurilshikov et al., 2021) and the Dutch Microbiome Project (DMP) (Lopera-Maya et al., 2022), and the sample size was 14,306 and 8,208, respectively. PCa cases (n = 6,311) and controls (n = 88,902) were obtained from the FinnGen consortium initially (Kurki et al., 2023). The detailed descriptions of exposure and outcome, including the data source, microbial taxa, race of population, sample size, the total number of SNPs, and website information, are presented in Table 1.
The study data from the databases are de-identified and publicly available. The informed consent of all participants (legal guardians for participants under 18 years old) has been obtained in each GWAS involved in this study and was ethically approved by the respective institutions. Therefore, no ethical approval of our agency's institutional review board was required.
Single nucleotide polymorphism selection
We extracted six levels of gut microbiota taxa, including phylum, class, order, family, genus, and species. SNPs significantly associated with gut microbiota were selected as potential instrument variables (IVs). We used two threshold standards to select the IVs, including a loose threshold (P < 1.0 × 10−5) and a strict threshold (P < 5.0 × 10−6). SNPs with a minor allele frequency of ≤ 0.01 were removed. The linkage disequilibrium threshold was set to be r2 = 0.01, clumping distance = 10,000 kb (for loose threshold), and r2 = 0.001 clumping distance = 10,000 kb (for strict threshold), respectively. We applied the MR-Egger regression test to monitor the potential horizontal pleiotropic effect (Burgess and Thompson, 2017), namely the confounding effect resulting from other diseases, which may violate the second assumption in MR analysis (only affect the outcome via the exposure). The intercept item of MR-Egger that was significant represents the existence of pleiotropy. In addition, palindromic SNPs were deleted due to the principle of MR to ensure that the same allele corresponds to the effects between SNPs and exposure and on the outcome.
The assumptions of MR analysis
For the purpose of minimizing the impact of bias on the results, MR must conform to three important assumptions. First, IVs must be independent of confounders related to exposure and outcome. Second, the IVs should be significantly linked to the exposure. The association strength between gut microbiota and IVs was estimated using the formula: F = β2/SE2 (Xie et al., 2023), where β was the regression coefficient for gut microbiota and IVs and SE was the standard error. A weak association between IVs and exposure is recognized when F < 10. Third, IVs influence outcomes through exposure only; that is, there is no horizontal pleiotropic effect of IVs on outcomes.
Statistical analysis
The statistical analyses were performed by R version 4.2.0 (Institute for Statistics and Mathematics, Vienna, Austria). The R package “TwoSampleMR” was used for MR analysis of the causal association between gut microbiota and PCs. P < 0.05 indicates the statistical significance of evidence for a potential causal relationship. The Wald ratio method was utilized to assess the role of individual IVs in the causal estimates. Calculation for the causal effect values was done using the inverse variance weighted (IVW) test, which is the primary method to obtain unbiased estimates when horizontal pleiotropy is absent. We used both fixed and random effects models for the IVW test. The effect size was expressed by odds ratios (ORs) with 95% confidence intervals (CIs). In addition, Bonferroni thresholds were also utilized to adjust for the P-value to control for multiple tests.
The test for heterogeneity was Cochrane's Q test, and IVs P < 0.05 were recognized as heterogeneous. The intercept of MR-Egger regression examined the potential pleiotropy in IVs, and P > 0.05 was deemed to be no horizontal pleiotropy. The examination of possible outliers was done using the MR-Pleiotropy RESidual Sum and Outlier (MR-PRESSO) test (R package “MR-PRESSO”) (Verbanck et al., 2018). Moreover, we performed the reverse causality analysis between gut microbiota and PCa.
Results
Instrumental variables selection
After quality control, we identified 2,616 (P < 1.0 × 10−5) and 1,371 (P < 5.0 × 10−6) SNPs as IVs for 302 bacterial taxa, which comprised 9 phyla, 16 classes, 20 orders, 33 families, 119 genera, and 105 species. Then, we evaluated the horizontal pleiotropic effect at each taxa level. For both PCa and the five levels of gut microbiota, none of the IVs were outliers through the MR-PRESSO test. IVs in this analysis had no horizontal pleiotropy after removing pleiotropic SNPs identified using the MR-PRESSO outlier test and MR-Egger regression (both MR-PRESSO global test P > 0.05 and MR-Egger regression P > 0.05).
Two-sample MR analysis
Figure 2 shows the relationship between 53 bacterial taxa and PCa. Whether the IVW estimates used a loose threshold (P < 1.0 × 10−5) or a strict threshold (P < 5.0 × 10−6), the relative abundance of seven bacterial taxa was all significantly associated with the odds of PCa. To be specific, the increased relative abundance of Melainabacteria (at class level), Gastranaerophilales (at order level), and Prevotellaceae (at family level) was negatively associated with the odds of PCa, while that of Acidaminococcaceae (at family level), Ruminococcus torques group, Lachnospiraceae UCG-008 (at genus level), and Eubacterium biforme (at species level) had positive relationships.
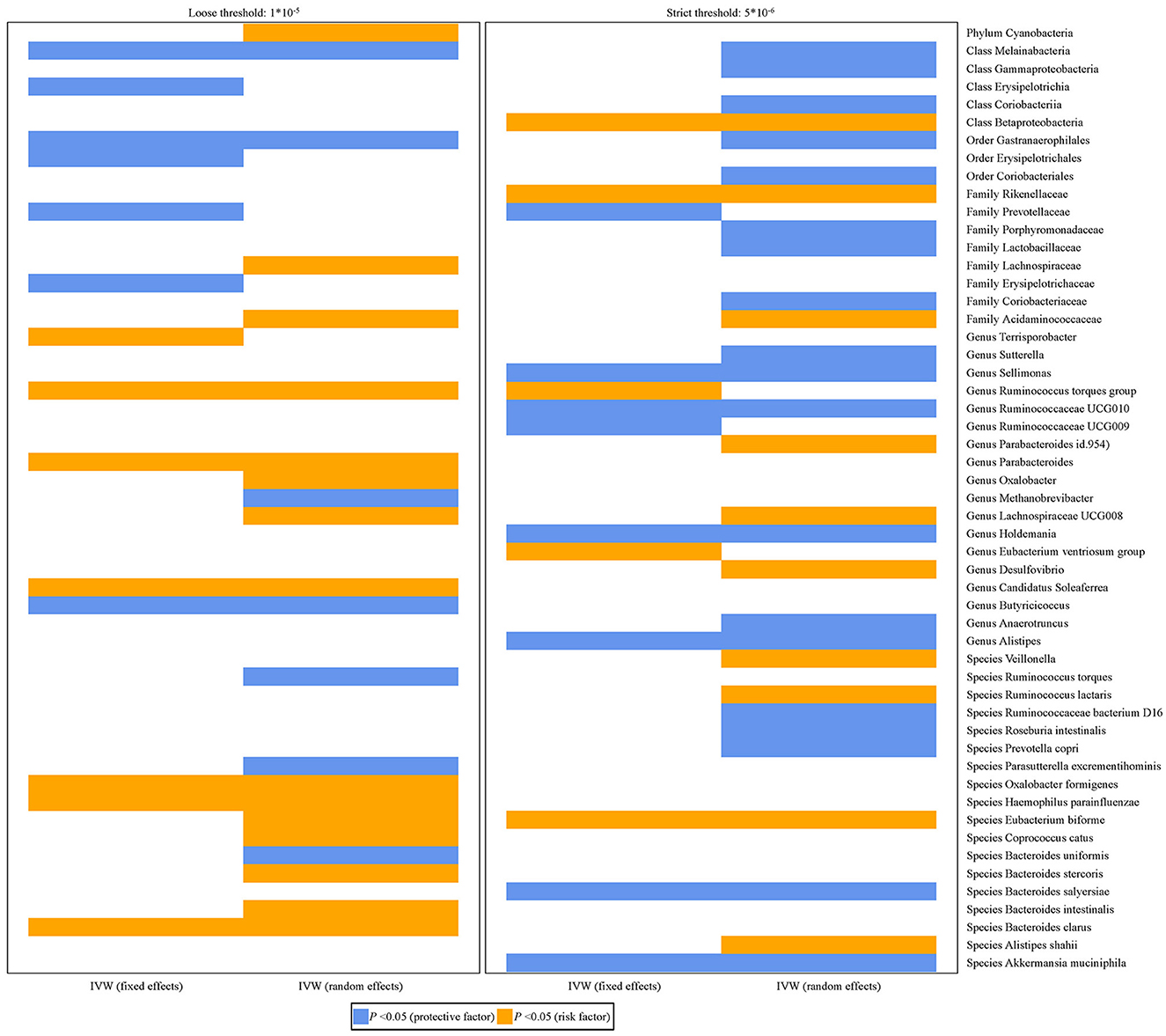
Figure 2. Histograms for the potential causal association between gut microbiota and PCa. The blue color represents OR < 1 while orange color represents OR > 1.
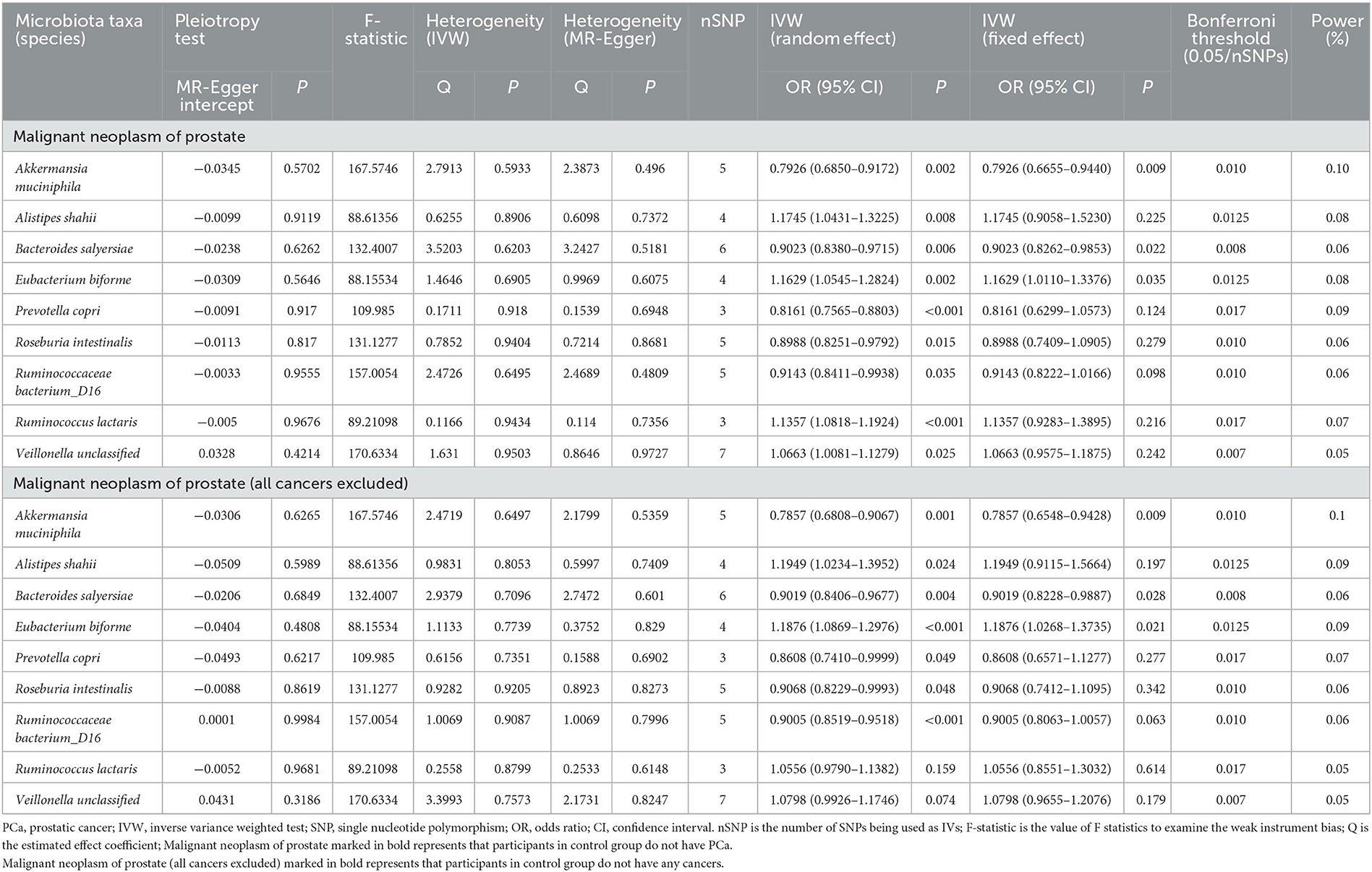
When focused on the role of bacterial taxa at the species level in PCa (fixed effect), the relative abundance of Akkermansia muciniphila (OR = 0.7926, 95% CI: 0.6655–0.9440) and Bacteroides salyersiae (OR = 0.9023, 95% CI: 0.8262–0.9853) was both negatively associated with the odds of PCa, while that of Eubacterium biforme was positively associated with the odds of PCa (OR = 1.1629, 95% CI: 1.0110–1.3376) (Table 2).
We further explored these associations among patients without any other cancers and similarly found the negative relationship between the relative abundance of Akkermansia muciniphila (OR = 0.7857, 95% CI: 0.6548–0.9428) and Bacteroides salyersiae (OR = 0.9019, 95% CI: 0.8228–0.9887) and PCa, and the positive relationship between the relative abundance of Eubacterium biforme (OR = 1.1876, 95% CI: 1.0268–1.3735) and PCa.
Table 2 also shows the pleiotropy and heterogeneity test results. We indicated the impact of comparatively accurate MR results in three species of gut microbiota on PCa by sensitivity analysis. No horizontal pleiotropy was observed in Akkermansia muciniphila (P = 0.5702), Bacteroides salyersiae (P = 0.6262), and Eubacterium biforme (P = 0.5646) for PCa. Furthermore, there was no heterogeneity in Akkermansia muciniphila (IVW: P = 0.5933; MR-Egger: P = 0.4960), Bacteroides salyersiae (IVW: P = 0.6203; MR-Egger: P = 0.5181), and Eubacterium biforme (IVW: P = 0.6905; MR-Egger: P = 0.6075) for PCa. Results of IVW were comparatively reliable when heterogeneity and pleiotropy were absent, indicating the potential causal relationships between these three species of gut microbiota and PCa were comparatively steady.
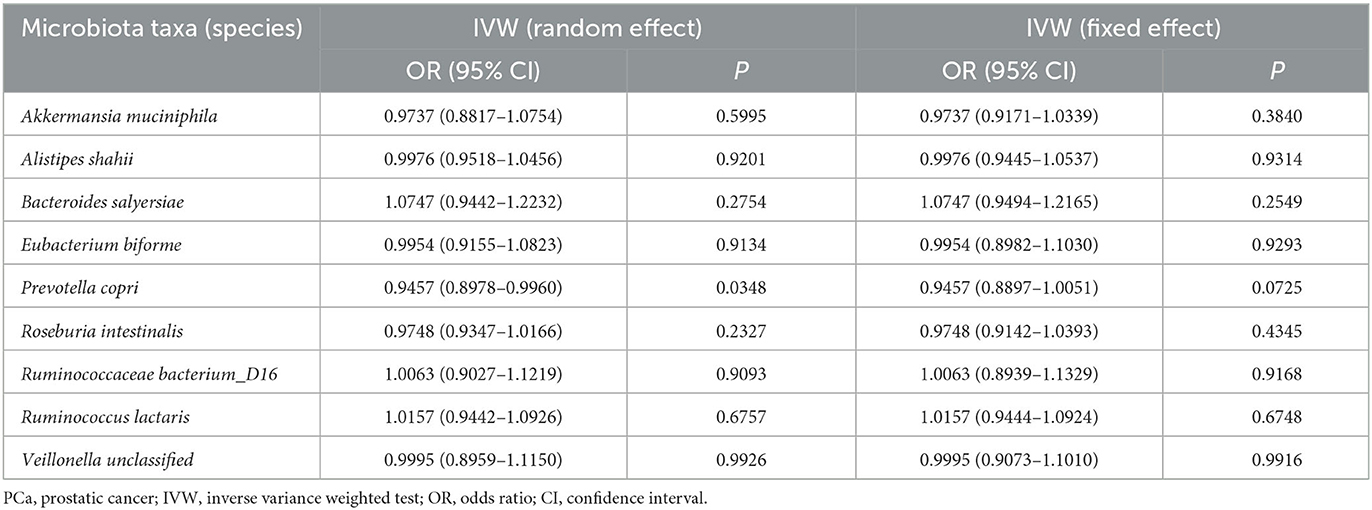
In addition, we used the Bonferroni threshold to assess these relationships and found that only high a relative abundance of Akkermansia muciniphila (P = 0.010) was associated with low odds of PCa among all participants or those without any other cancers. The results of reverse causality analysis also showed that there was no reverse causality relationship between gut microbiota and PCa (fixed effect) (Table 3).
Discussion
We conducted an MR analysis to explore the potential causal relationship between gut microbiota and PCa. The results showed that Melainabacteria (class level), Gastranaerophilales (order level), Prevotellaceae (family level), Acidaminococcaceae (family level), Ruminococcus torques group (genus level), Lachnospiraceae UCG-008 (genus level), Akkermansia muciniphila (species level), Bacteroides salyersiae (species level), and Eubacterium biforme (species level) were associated with PCa. To be specific, the relative abundance of Akkermansia muciniphila and Bacteroides salyersiae was both negatively associated with the odds of PCa, while that of Eubacterium biforme was positively associated with the odds of PCa. Besides, no reverse causality has been found between them.
The firm conclusions for the causal relationship between gut microbiota and PCa are not yet enough to draw according to the evidence from existing observational studies. In recent years, only a few studies have explored the association between gut microbiota and the risk of cancer using MR analyses, which is a widely used approach to explore the potential causal relationships between environmental exposures and diseases. For example, a univariable and multivariable MR study by Wei et al. (2023) assessed the causal effect of gut microbiota on five common cancers, including breast, endometrial, lung, ovarian, and PCa. Their results showed that a higher abundance of class Alphaproteobacteria was associated with a lower risk of PCa. Differently, in our study, we focused on the relationship between gut microbiota and PCa only and observed gut microbiota at phylum, class, order, family, genus, and species levels. We found that the relative abundance of Akkermansia muciniphila, Bacteroides salyersiae, and Eubacterium biforme was all associated with PCa. Our study detailed the group of gut microbiotas and found three specific species, which may provide some references for further basic and prospective research on the causal relationship between gut microbiota and PCa. In Wei's study, the associations between SNPs and PCa were obtained from the GWAS study from the Prostate Cancer Association Group to Investigate Cancer-Associated Alterations in the Genome (PRACTICAL) Consortium, which consists of cases diagnosed with PCa and controls of European descent. Another two-sample MR study by Long et al. (2023), also based on the population from the PRACTICAL Consortium, examined the causal relationship between gut microbiota and cancer. Although we used the GWAS study from the Finngen Consortium, the race of the study population was similar. Long's results showed that the genus Ruminococcustorquesgroup, class Verrucomicrobiae, family Verrucomicrobiaceae, order Verrucomicrobiales, genus Terrisporobacter, genus Roseburia, and class Alphaproteobacteria were causally associated with PCa. The different gut microbiota we explored may complement previous studies.
In fact, the underlying mechanisms of these gut microbiota and PCa are complex and unclear. The class Melainabacteria has been identified as an accurate biomarker of zinc (Zn) status in the human body (Chen et al., 2021). As Zn plays a growth-modulatory role in PCa, Melainabacteria may influence the occurrence and development of PCa by modulating Zn levels (To et al., 2020; Zhang et al., 2022). Melainabacteria exist in groundwater, wastewater treatment plants, and herbivorous mammal and human guts and have the function of synthesizing vitamins B and K, suggesting they are beneficial bacteria to their hosts (Di Rienzi et al., 2013). Similarly to the previous studies, our findings showed that an increased abundance of Melainabacteria was associated with low odds of PCa. Gastranaerophilales is one of the probiotics with impaired abundance in colitis (Wang et al., 2021; Wu et al., 2021). Inflammation is a risk factor for prostate carcinogenesis, with diet, chemical injury, and an altered microbiome being causally implicated (de Bono et al., 2020). However, no study has been conducted to describe the potential mechanisms by which Gastranaerophilales play a protective role in prostate carcinogenesis, which needs further basic research for clarification. Prevotellaceae is also a probiotic that plays an important role in colitis and some cancers (Zhang L. et al., 2019; Qu et al., 2021). Li et al. (2022) performed an analysis on alterations of gut microbiota diversity, composition, and metabonomics in benign prostatic hyperplasia rats and showed that there was a strong correlation between Prevotellaceae and differential metabolites. However, the specific mechanism of the potential protective role of Prevotellaceae in PCa has not been clear. According to previous studies, we speculated that Prevotellaceae may moderate inflammation (Qu et al., 2021), oxidative stress (Cui et al., 2018), and metabolic disorders (Li et al., 2022) and further influence PCa development.
The high relative abundance of the family Acidaminococcaceae, genus Ruminococcus torques group, and genus Lachnospiraceae UCG-008 was found to be associated with a high risk of PCa in this study. Acidaminococcaceae clustered into one group with the Veillonellaceae, which has been reported as a marker of dysbacteriosis and plays a possible role in carcinogenesis (Yan et al., 2015; Kasai et al., 2016). A bidirectional, two-sample MR study on the association between Graves' disease and the gut microbiome showed that the Ruminococcus torques group was identified as a risk factor (Cao et al., 2023). Ruminococcus torques group is a genera derived from the genus Mediterraneibacter from the family Lachnospiraceae and is identified as a butyrate-producing bacterium (Salyers et al., 1977). Butyrate-producing bacteria were found to promote fat deposition because they are able to convert dietary fiber to butyrate by fermentation (Yang et al., 2010; Blaut, 2015). The abundance of Ruminococcus torques group is thus supposed to be positively related to fat accumulation. Excess fat can accelerate the growth of prostate tumors by inducing inflammation (Hayashi et al., 2018). In addition, altered lipid metabolism, especially the excessive accumulation of cholesterol and fatty acids, promotes the malignant transformation of PCa via the formation of cholesteryl esters (Wang et al., 2022). Lachnospiraceae UCG-008 is considered a potential harmful genus in human colonic microbiota (Huang et al., 2019). Some medicines improve gut microbial dysbiosis by increasing beneficial bacteria and decreasing harmful bacteria, including Lachnospiraceae UCG-008 in order to relieve diseases (Huang et al., 2019; Yin et al., 2021). In conclusion, how gut microbiota influence the development of PCa by disturbing the metabolism is still unclear; further studies are needed to clarify the specific biological mechanisms.
We additionally explored the relationships between gut microbiota and the risk of PCa at the species level. Our findings indicated that the high relative abundance of Akkermansia muciniphila and Bacteroides salyersiae was both associated with a low risk of PCa, while that of Eubacterium biforme was positively associated with the risk of PCa. Akkermansia muciniphila is a Gram-negative anaerobic bacterium that contributes to homeostasis maintenance and barrier integrity in the gastrointestinal tract (Zhai et al., 2019; Zhang T. et al., 2019). Recently, it has been reported that intravenous injection of Akkermansia muciniphila-derived extracellular vesicles in immune-competent mice reduced the tumor burden of PCa without inducing obvious toxicity in normal tissues, indicating a potential association between the abundance of Akkermansia muciniphila and PCa (Luo et al., 2021). Similarly, our MR study supplemented and suggested a potential causal relationship between the high relative abundance of Akkermansia muciniphila and the low risk of PCa. Wang et al. (2020) found the membrane protein from Akkermansia muciniphila, namely Amuc_1100, was associated with tumourigenesis of colorectal cancer through modulation of CD8 T cells in mice. Gu et al. (2021) also suggested that Akkermansia muciniphila and its outer protein Amuc_1100 regulated the tryptophan metabolism in colitis. Another animal study also showed the effect of improved Amuc_1100 from Akkermansia muciniphila on metabolism in obese and diabetic mice (Plovier et al., 2017). As the mechanism of PCa development mentioned previously, Akkermansia muciniphila and Amuc_1100 may also influence PCa by improving inflammation and regulating glucose metabolism and lipid metabolism, but further mechanistic exploration is needed. Bacteroides salyersiae is a beneficial commensal with Akkermansia muciniphila and plays a similar role in the host (Derosa et al., 2020). Our study considered that the application of probiotics in the prevention and treatment of PCa was very promising. Gut microflora evolves with a complex polysaccharide-rich diet and dietary fermentation, resulting in the production of short-chain fatty acids such as butyrate, which represent a primary energy source for colonic epithelial cells and preserve them from inflammation (Ahmad et al., 2000; Atarashi et al., 2013). Eubacterium biforme is one of them. Aside from this, a lack of fiber in diets is associated with a low concentration of short-chain fatty acids (Pituch-Zdanowska et al., 2015). Daily attention to dietary fiber consumption may be the key to preventing inflammatory diseases by regulating intestinal flora. Daily consumption of bifidobacteria-rich foods such as yogurt may be a viable way to regulate probiotics; however, whether a high relative abundance of Eubacterium biforme is associated with a high risk of PCa is not determinate.
As mentioned before, MR is a relatively good study design to clarify the causal effect of potential risk factors on diseases of interest. By exploring the gut microbiota that modulate PCa risk, MR studies facilitate the recommendation of public health policies and clinical interventions that effectively reduce the incidence and social burden of PCa. With the rapid development of omics technologies in recent years, including genomics, proteomics, transcriptomics, metabolomics, and epigenomics, researchers can use a large number of novel exposures/intermediate phenotypes generated in observational studies to assess associations with clinical endpoints. The MR approaches summarize the results from published GWAS studies facilitately and do not need a separate study to carry out MR analyses (Sekula et al., 2016). Compared with observational studies, which commonly suffer from biased results due to confounding, MR can deal with any confounding by design as long as a valid genetic instrumental variable is available. However, two major limitations also influence the judgment of causation through MR approaches. First, no valid instrument for every research question is available because of a lack of knowledge about MR, and the publicly available sources of data to provide information on the associations of interest are not always available. Second, MR studies have potential limitations in the statistical power of the study design, which depends on several aspects, such as the proportion of variance in the exposure explained by the genetic instrumental variable and the magnitude of the causal association between exposure and outcome (Brion et al., 2013; Burgessm, 2014). Besides, in recent years, artificial intelligence has been explored to improve the diagnosis of PCa, including radiological and histological diagnoses (Ström et al., 2020; Mata et al., 2021). Ramírez-Mena et al. (2023) combined gene expression and AI for the detection and screening of PCa and found it can decrease the misclassification rates of anatomopathological analysis, thus reducing the need for repeated biopsies. Their algorithm may be applied to urine or blood samples rather than traditional living tissue, which could be a part of the liquid biopsy strategy in PCa in the future. To the best of our knowledge, AI has not been applied in the calculation of causal association between the gut microbiota and the risk of PCa, and we believe the potential causal relationship indicated by MR approaches can provide some references for further studies that focus on the underlying mechanisms.
There are some strengths and limitations in this study. The current study refined the taxonomic units of gut microbiota to the species level and explored their relationship with the risk of PCa, which may be more comprehensive compared to the previous studies. Nevertheless, there were two major limitations in this study. Our study data were extracted from the MiBioGen consortium, DMP, and FinnGen consortium, which may cause the selection bias. Furthermore, these databases only contain the European population, so the potential causal relationship between gut microbiota and the risk of PCa is limited, and whether the results are generalizable to other populations requires further studies to clarify. Another limitation was that the locations of the SNPs we used (whether they are present in the enhancer/promoter/coding region of the genome) are not available, which limited the explanation for the mechanisms of the potential causal association between gut microbiota and PCa.
Conclusion
Our study suggested a potential causal relationship between gut microbiota and the risk of PCa. Further studies are warranted to elucidate the causal association and specific underlying mechanisms of gut microbiota and the development of PCa.
Data availability statement
Publicly available datasets were analyzed in this study. This data can be found here: the GWAS Catalog, https://www.ebi.ac.uk/gwas/.
Author contributions
QX and BH designed the study, collected, analyzed, and interpreted the data. QX wrote the manuscript. BH critically reviewed, edited, and approved the manuscript. Both authors read and approved the final manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ahmad, M. S., Krishnan, S., Ramakrishna, B. S., Mathan, M., Pulimood, A. B., and Murthy, S. N. (2000). Butyrate and glucose metabolism by colonocytes in experimental colitis in mice. Gut 46, 493–499. doi: 10.1136/gut.46.4.493
Alanee, S., El-Zawahry, A., Dynda, D., Dabaja, A., McVary, K., Karr, M., et al. (2019). A prospective study to examine the association of the urinary and fecal microbiota with prostate cancer diagnosis after transrectal biopsy of the prostate using 16sRNA gene analysis. Prostate 79, 81–87. doi: 10.1002/pros.23713
Atarashi, K., Tanoue, T., Oshima, K., Suda, W., Nagano, Y., Nishikawa, H., et al. (2013). Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 500, 232–236. doi: 10.1038/nature12331
Blaut, M. (2015). Gut microbiota and energy balance: role in obesity. Proc. Nutr. Soc. 74, 227–234. doi: 10.1017/S0029665114001700
Brion, M. J., Shakhbazov, K., and Visscher, P. M. (2013). Calculating statistical power in Mendelian randomization studies. Int. J. Epidemiol. 42, 1497–1501. doi: 10.1093/ije/dyt179
Burgess, S., and Thompson, S. G. (2017). Interpreting findings from Mendelian randomization using the MR-Egger method. Eur. J. Epidemiol. 32, 377–389. doi: 10.1007/s10654-017-0255-x
Burgessm, S. (2014). Sample size and power calculations in Mendelian randomization with a single instrumental variable and a binary outcome. Int. J. Epidemiol. 43, 922–929. doi: 10.1093/ije/dyu005
Cao, J., Wang, N., Luo, Y., Ma, C., Chen, Z., Chenzhao, C., et al. (2023). A cause-effect relationship between Graves' disease and the gut microbiome contributes to the thyroid-gut axis: A bidirectional two-sample Mendelian randomization study. Front. Immunol. 14, 977587. doi: 10.3389/fimmu.2023.977587
Chen, L., Wang, Z., Wang, P., Yu, X., Ding, H., Wang, Z., et al. (2021). Effect of long-term and short-term imbalanced Zn manipulation on gut microbiota and screening for microbial markers sensitive to zinc status. Microbiol. Spectr. 9, e0048321. doi: 10.1128/Spectrum.00483-21
Cui, H. X., Hu, Y. N., Li, J. W., and Yuan, K. (2018). Hypoglycemic mechanism of the berberine organic acid salt under the synergistic effect of intestinal flora and oxidative stress. Oxid. Med. Cell Longev. 2018, 8930374. doi: 10.1155/2018/8930374
Davey Smith, G., and Hemani, G. (2014). Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum. Mol. Genet. 23, R89–98. doi: 10.1093/hmg/ddu328
de Bono, J. S., Guo, C., Gurel, B., De Marzo, A. M., Sfanos, K. S., Mani, R. S., et al. (2020). Prostate carcinogenesis: inflammatory storms. Nat. Rev. Cancer 20, 455–469. doi: 10.1038/s41568-020-0267-9
Derosa, L., Routy, B., Fidelle, M., Iebba, V., Alla, L., Pasolli, E., et al. (2020). Gut bacteria composition drives primary resistance to cancer immunotherapy in renal cell carcinoma patients. Eur. Urol. 78, 195–206. doi: 10.1016/j.eururo.2020.04.044
Di Rienzi, S. C., Sharon, I., Wrighton, K. C., Koren, O., Hug, L. A., Thomas, B. C., et al. (2013). The human gut and groundwater harbor non-photosynthetic bacteria belonging to a new candidate phylum sibling to Cyanobacteria. Elife 2, e01102. doi: 10.7554/eLife.01102.024
Fujita, K., Matsushita, M., De Velasco, M. A., Hatano, K., Minami, T., Nonomura, N., et al. (2023). The gut-prostate axis: a new perspective of prostate cancer biology through the gut microbiome. Cancers (Basel) 15, 1375. doi: 10.3390/cancers15051375
Golombos, D. M., Ayangbesan, A., O'Malley, P., Lewicki, P., Barlow, L., Barbieri, C. E., et al. (2018). The role of gut microbiome in the pathogenesis of prostate cancer: a prospective, pilot study. Urology 111, 122–128. doi: 10.1016/j.urology.2017.08.039
Gu, Z., Pei, W., Shen, Y., Wang, L., Zhu, J., Zhang, Y., et al. (2021). Akkermansia muciniphila and its outer protein Amuc_1100 regulates tryptophan metabolism in colitis. Food Funct. 12, 10184–10195. doi: 10.1039/D1FO02172A
Hayashi, T., Fujita, K., Nojima, S., Hayashi, Y., Nakano, K., Ishizuya, Y., et al. (2018). High-fat diet-induced inflammation accelerates prostate cancer growth via IL6 signaling. Clin. Cancer Res. 24, 4309–4318. doi: 10.1158/1078-0432.CCR-18-0106
Huang, J., Wang, Q., Xu, Q., Zhang, Y., Lin, B., Guan, X., et al. (2019). In vitro fermentation of O-acetyl-arabinoxylan from bamboo shavings by human colonic microbiota. Int. J. Biol. Macromol. 125, 27–34. doi: 10.1016/j.ijbiomac.2018.12.024
Kasai, C., Sugimoto, K., Moritani, I., Tanaka, J., Oya, Y., Inoue, H., et al. (2016). Comparison of human gut microbiota in control subjects and patients with colorectal carcinoma in adenoma: Terminal restriction fragment length polymorphism and next-generation sequencing analyses. Oncol. Rep. 35, 325–333. doi: 10.3892/or.2015.4398
Kurilshikov, A., Medina-Gomez, C., Bacigalupe, R., Radjabzadeh, D., Wang, J., Demirkan, A., et al. (2021). Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat. Genet. 53, 156–165. doi: 10.1038/s41588-020-00763-1
Kurki, M. I., Karjalainen, J., Palta, P., Sipila, T. P., Kristiansson, K., Donner, K. M., et al. (2023). FinnGen provides genetic insights from a well-phenotyped isolated population. Nature 613, 508–518. doi: 10.1038/s41586-022-05473-8
Li, L. Y., Han, J., Wu, L., Fang, C., Li, W. G., Gu, J. M., et al. (2022). Alterations of gut microbiota diversity, composition and metabonomics in testosterone-induced benign prostatic hyperplasia rats. Mil. Med. Res. 9, 12. doi: 10.1186/s40779-022-00373-4
Liss, M. A., White, J. R., Goros, M., Gelfond, J., Leach, R., Johnson-Pais, T., et al. (2018). Metabolic biosynthesis pathways identified from fecal microbiome associated with prostate cancer. Eur. Urol. 74, 575–582. doi: 10.1016/j.eururo.2018.06.033
Long, Y., Tang, L., Zhou, Y., Zhao, S., and Zhu, H. (2023). Causal relationship between gut microbiota and cancers: a two-sample Mendelian randomisation study. BMC Med. 21, 66. doi: 10.1186/s12916-023-02761-6
Lopera-Maya, E. A., Kurilshikov, A., van der Graaf, A., Hu, S., Andreu-Sanchez, S., Chen, L., et al. (2022). Effect of host genetics on the gut microbiome in 7,738 participants of the dutch microbiome project. Nat. Genet. 54, 143–151. doi: 10.1038/s41588-021-00992-y
Luo, Z. W., Xia, K., Liu, Y. W., Liu, J. H., Rao, S. S., Hu, X. K., et al. (2021). Extracellular vesicles from akkermansia muciniphila elicit antitumor immunity against prostate cancer via modulation of CD8(+) T Cells and macrophages. Int. J. Nanomed. 16, 2949–2963. doi: 10.2147/IJN.S304515
Mata, L. A., Retamero, J. A., Gupta, R. T., García, Figueras,. R., and Luna, A. (2021). Artificial intelligence-assisted prostate cancer diagnosis: radiologic-pathologic correlation. Radiographics 41, 1676–1697. doi: 10.1148/rg.2021210020
Matsushita, M., Fujita, K., Hatano, K., De Velasco, M. A., Tsujimura, A., Uemura, H., et al. (2023). Emerging relationship between the gut microbiome and prostate cancer. World J. Mens Health. 41, 759–768. doi: 10.5534/wjmh.220202
Matsushita, M., Fujita, K., Motooka, D., Hatano, K., Fukae, S., Kawamura, N., et al. (2021). The gut microbiota associated with high-Gleason prostate cancer. Cancer Sci. 112, 3125–3135. doi: 10.1111/cas.14998
Pituch-Zdanowska, A., Banaszkiewicz, A., and Albrecht, P. (2015). The role of dietary fibre in inflammatory bowel disease. Prz. Gastroenterol. 10, 135–141. doi: 10.5114/pg.2015.52753
Plovier, H., Everard, A., Druart, C., Depommier, C., Van Hul, M., Geurts, L., et al. (2017). A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat. Med. 23, 107–113. doi: 10.1038/nm.4236
Porter, C. M., Shrestha, E., Peiffer, L. B., and Sfanos, K. S. (2018). The microbiome in prostate inflammation and prostate cancer. Prostate Cancer Prostatic Dis. 21, 345–354. doi: 10.1038/s41391-018-0041-1
Qu, Y., Li, X., Xu, F., Zhao, S., Wu, X., Wang, Y., et al. (2021). Kaempferol alleviates murine experimental colitis by restoring gut microbiota and inhibiting the LPS-TLR4-NF-kappaB Axis. Front. Immunol. 12, 679897. doi: 10.3389/fimmu.2021.679897
Ramírez-Mena, A., Andrés-León, E., Alvarez-Cubero, M. J., Anguita-Ruiz, A., Martinez-Gonzalez, L. J., and Alcala-Fdez, J. (2023). Explainable artificial intelligence to predict and identify prostate cancer tissue by gene expression. Comput. Methods Programs Biomed. 240, 107719. doi: 10.1016/j.cmpb.2023.107719
Rizzo, A., Santoni, M., Mollica, V., Fiorentino, M., Brandi, G., and Massari, F. (2022). Microbiota and prostate cancer. Semin. Cancer Biol. 86, 1058–1065. doi: 10.1016/j.semcancer.2021.09.007
Salyers, A. A., West, S. E., Vercellotti, J. R., and Wilkins, T. D. (1977). Fermentation of mucins and plant polysaccharides by anaerobic bacteria from the human colon. Appl. Environ. Microbiol. 34, 529–533. doi: 10.1128/aem.34.5.529-533.1977
Sekula, P., Del Greco, M. F., Pattaro, C., and Kottgen, A. (2016). Mendelian randomization as an approach to assess causality using observational data. J. Am. Soc. Nephrol. 27, 3253–3265. doi: 10.1681/ASN.2016010098
Spieth, P. M., Kubasch, A. S., Penzlin, A. I., Illigens, B. M., Barlinn, K., and Siepmann, T. (2016). Randomized controlled trials - a matter of design. Neuropsychiatr. Dis. Treat 12, 1341–1349. doi: 10.2147/NDT.S101938
Ström, P., Kartasalo, K., Olsson, H., Solorzano, L., Delahunt, B., Berney, D. M., et al. (2020). Artificial intelligence for diagnosis and grading of prostate cancer in biopsies: a population-based, diagnostic study. Lancet Oncol. 21, 222–232. doi: 10.1016/S1470-2045(19)30738-7
To, P. K., Do, M. H., Cho, J. H., and Jung, C. (2020). Growth modulatory role of zinc in prostate cancer and application to cancer therapeutics. Int. J. Mol. Sci. 21, 2991. doi: 10.3390/ijms21082991
Tong, J., Zhang, X., Fan, Y., Chen, L., Ma, X., Yu, H., et al. (2020). Changes of intestinal microbiota in ovarian cancer patients treated with surgery and chemotherapy. Cancer Manag. Res. 12, 8125–8135. doi: 10.2147/CMAR.S265205
Verbanck, M., Chen, C. Y., Neale, B., and Do, R. (2018). Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 50, 693–698. doi: 10.1038/s41588-018-0099-7
Wang, L., Tang, L., Feng, Y., Zhao, S., Han, M., Zhang, C., et al. (2020). A purified membrane protein from Akkermansia muciniphila or the pasteurised bacterium blunts colitis associated tumourigenesis by modulation of CD8(+) T cells in mice. Gut 69, 1988–1997. doi: 10.1136/gutjnl-2019-320105
Wang, X., Cheng, K., Liu, Z., Sun, Y., Zhou, L., Xu, M., et al. (2021). Bioactive constituents of Mosla chinensis-cv. Jiangxiangru ameliorate inflammation through MAPK signaling pathways and modify intestinal microbiota in DSS-induced colitis mice. Phytomedicine 93, 153804. doi: 10.1016/j.phymed.2021.153804
Wang, X., Sun, B., Wei, L., Jian, X., Shan, K., He, Q., et al. (2022). Cholesterol and saturated fatty acids synergistically promote the malignant progression of prostate cancer. Neoplasia 24, 86–97. doi: 10.1016/j.neo.2021.11.004
Wei, Z., Yang, B., Tang, T., Xiao, Z., Ye, F., Li, X., et al. (2023). Gut microbiota and risk of five common cancers: A univariable and multivariable Mendelian randomization study. Cancer Med. 12, 10393–10405. doi: 10.1002/cam4.5772
Wheeler, K. M., and Liss, M. A. (2019). The microbiome and prostate cancer risk. Curr. Urol. Rep. 20, 66. doi: 10.1007/s11934-019-0922-4
Wu, F., Lei, H., Chen, G., Chen, C., Song, Y., Cao, Z., et al. (2021). In vitro and in vivo studies reveal that hesperetin-7-o-glucoside, a naturally occurring monoglucoside, exhibits strong anti-inflammatory capacity. J. Agric. Food Chem. 69, 12753–12762. doi: 10.1021/acs.jafc.1c05793
Xie, J., Huang, H., Liu, Z., Li, Y., Yu, C., Xu, L., et al. (2023). The associations between modifiable risk factors and nonalcoholic fatty liver disease: A comprehensive Mendelian randomization study. Hepatology 77, 949–964. doi: 10.1002/hep.32728
Yan, X., Yang, M., Liu, J., Gao, R., Hu, J., Li, J., et al. (2015). Discovery and validation of potential bacterial biomarkers for lung cancer. Am. J. Cancer Res. 5, 3111–3122.
Yang, X., Zhang, B., Guo, Y., Jiao, P., and Long, F. (2010). Effects of dietary lipids and Clostridium butyricum on fat deposition and meat quality of broiler chickens. Poult. Sci. 89, 254–260. doi: 10.3382/ps.2009-00234
Yin, R., Liu, S., Jiang, X., Zhang, X., Wei, F., and Hu, J. (2021). The qingchangligan formula alleviates acute liver failure by regulating galactose metabolism and gut microbiota. Front. Cell Infect. Microbiol. 11, 771483. doi: 10.3389/fcimb.2021.771483
Zhai, Q., Feng, S., Arjan, N., and Chen, W. (2019). A next generation probiotic, Akkermansia muciniphila. Crit. Rev. Food Sci. Nutr. 59, 3227–3236. doi: 10.1080/10408398.2018.1517725
Zhang, L., Liu, Y., Zheng, H. J., and Zhang, C. P. (2019). The oral microbiota may have influence on oral cancer. Front. Cell Infect. Microbiol. 9, 476. doi: 10.3389/fcimb.2019.00476
Zhang, T., Li, Q., Cheng, L., Buch, H., and Zhang, F. (2019). Akkermansia muciniphila is a promising probiotic. Microb. Biotechnol. 12, 1109–1125. doi: 10.1111/1751-7915.13410
Keywords: gut microbiota, prostatic cancer, Mendelian randomization study, endometriosis, causal inference
Citation: Xie Q and Hu B (2023) Effects of gut microbiota on prostatic cancer: a two-sample Mendelian randomization study. Front. Microbiol. 14:1250369. doi: 10.3389/fmicb.2023.1250369
Received: 30 June 2023; Accepted: 26 September 2023;
Published: 06 November 2023.
Edited by:
George Grant, University of Aberdeen, United KingdomReviewed by:
Mudasir Rashid, Howard University Hospital, United StatesIgor Łoniewski, Pomeranian Medical University, Poland
Copyright © 2023 Xie and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bin Hu, aHViaW51cm9sb2d5JiN4MDAwNDA7b3V0bG9vay5jb20=
 Qingpeng Xie
Qingpeng Xie Bin Hu
Bin Hu