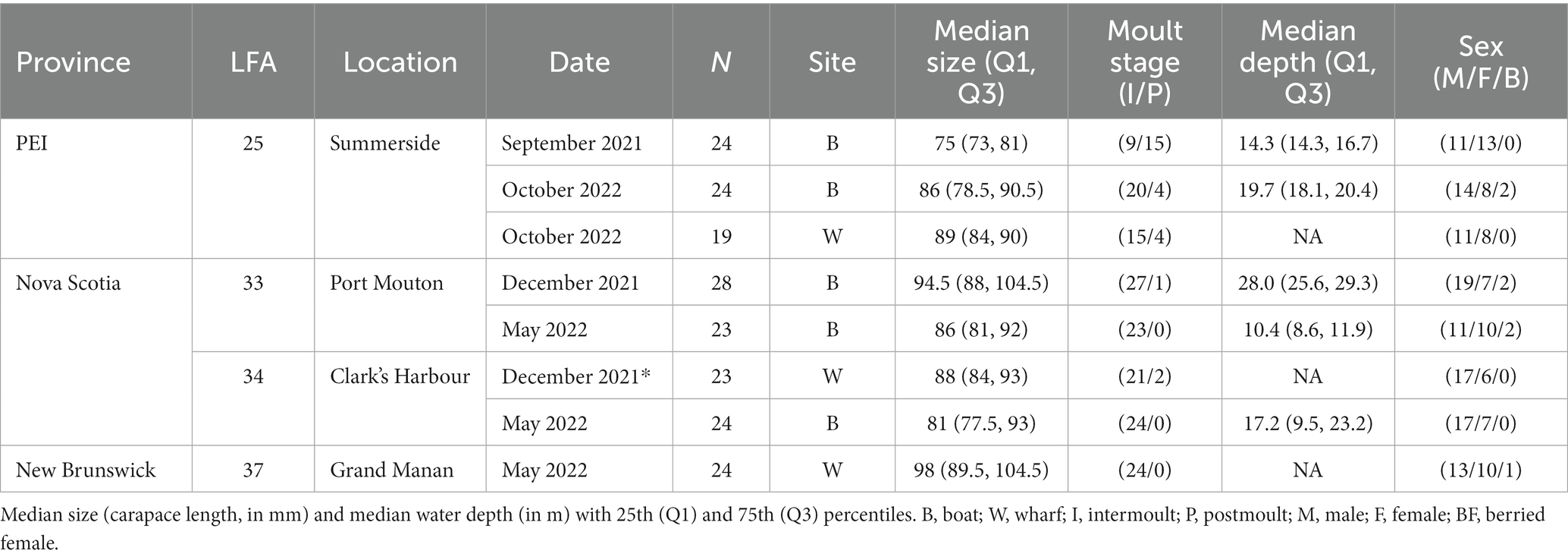
- 1Department of Health Management, Atlantic Veterinary College, University of Prince Edward Island, Charlottetown, PE, Canada
- 2Department of Animal Sciences and Aquaculture, Faculty of Agriculture, Dalhousie University, Bible Hill, NS, Canada
- 3Department of Computer and Information Sciences, University of Strathclyde, Glasgow, United Kingdom
The shell microbial community of lobsters—a key factor in the development of epizootic shell disease (ESD)—is still insufficiently researched in Atlantic Canada and many knowledge gaps remain. This study aimed to establish a baseline description and analysis of the shell microbiome of apparently healthy lobsters from four locations in the region. More than 180 lobster shell swab samples were collected from New Brunswick, Nova Scotia and Prince Edward Island (PEI). PacBio long-read 16S rDNA sequencing and bioinformatic analyses in QIIME2 identified the shell-associated bacteria. The shell microbiome of healthy lobsters consisted mainly of the bacterial classes Gammaproteobacteria, Saprospiria, Verrucomicrobiae, Alphaproteobacteria, Flavobacteriia, Acidimicrobiia and Planctomycetia. The microbial composition differed regionally and seasonally, with some classes showing decreased or increased relative abundances in the PEI samples as well as in the winter and spring samples in Nova Scotia. The core shell microbiome included potentially pathogenic as well as beneficial bacterial taxa, of which some were present only in certain regions. Bacterial taxa that have previously been associated with ESD were present on healthy lobsters in Atlantic Canada, but their frequency differed by location, sampling time, and moult stage. This study indicated that geographical and seasonal factors influenced the shell microbiome of apparently healthy lobsters more than host factors such as sex, size, and moult stage. Our results provide valuable reference microbial data from lobsters in a disease-free state.
1. Introduction
The American lobster (Homarus americanus) is an important species in the benthic ecosystem and supports Atlantic Canada’s most valuable fishery (DFO, 2021a). While lobster landings in Atlantic Canada are at an all-time high, lobster stocks in the southern range of the species have been threatened by ocean warming and disease outbreaks (Castro et al., 2012; Howell, 2012; DFO, 2021b).
Next-generation sequencing and faster computational methods have advanced microbial research in animals and humans over the past couple of decades. Multiple studies have highlighted the important influence of microbial communities on their host’s metabolism, growth, and immune functions (Berg et al., 2015; Bikel et al., 2015; Cornejo-Granados et al., 2017). Having a natural and healthy microbial community is thought to be beneficial for their hosts, as commensal bacteria can protect against invading pathogens, regulate the metabolism, synthesize vitamins, or help nutrient absorption (Ivanov et al., 2009; Venema, 2010; Bikel et al., 2015; Rajeev et al., 2021). The association between several chronic diseases and negative shifts in microbial communities—so-called dysbiosis—has changed the traditional view that a single pathogen is the cause of disease, towards a more holistic understanding where microbes and their host form one functional unit (Panda et al., 2014; Egan and Gardiner, 2016; Saraf et al., 2021).
Dysbiosis in the shell microbiome of American lobsters is hypothesized to be associated with the ecologically and economically important epizootic shell disease (ESD) (Meres et al., 2012). This syndrome is characterized by the formation of deep bacterial lesions on the carapace that spread rapidly over the whole lobster cuticle (Smolowitz et al., 2005; Cobb and Castro, 2006; Chistoserdov et al., 2012; Shields, 2013). Since its emergence in 1996, ESD has spread across most of the species’ range, most heavily affecting southern New England where lobster fisheries have decreased drastically since the emergence of ESD (Glenn and Pugh, 2006; Castro and Somers, 2012; Groner et al., 2018). While some bacterial species have been associated with ESD lesions (e.g., Aquimarina, Thalassobius) it is not clear whether these are causative agents or simply secondary invaders (Chistoserdov et al., 2012; Meres et al., 2012; Quinn et al., 2013, 2017). To determine ESD risk factors and why certain regions and lobster demographics are more susceptible to this disease, a general understanding of the lobster shell microbial community is necessary (Egan and Gardiner, 2016).
Previous research has identified the classes Acidimicrobiia, Alphaproteobacteria, Bacteroidota, Myxococcota (formerly: Deltaproteobacteria), Flavobacteriia, and Gammaproteobacteria as common members of the shell microbial community in lobsters unaffected by ESD. However, many former studies were restricted to small sample sizes often taken from lab-reared animals or did not use next-generation sequencing. Consequently, with no comprehensive studies of wild and healthy American lobsters, we lack comparable baseline data and are unable to determine which factors or microbial taxa impact the shell microbiome during disease onset and proliferation.
This study aimed to (1) describe the shell microbiome of apparently healthy lobsters sampled from four locations in Atlantic Canada, (2) determine which bacterial taxa make up the core shell microbiome and whether these differ by geographical, seasonal or host factors, and (3) ascertain whether ESD-associated taxa, in particular the genus Aquimarina, are present in the shell microbiome and, if so, whether there are risk factors for their frequency. As members of the Aquimarina genus have been linked to ESD in lobsters (Chistoserdov et al., 2012; Meres et al., 2012), it was also of interest how this genus was distributed in this study. Using third-generation long-read sequencing, we present a comprehensive description of the shell microbiome of lobsters from historically ESD-free regions. This dataset offers valuable baseline information on, what we define as, the “healthy” shell microbiome of lobsters in Atlantic Canada and the factors influencing it. It enhances our understanding of what roles certain microbes might play in the healthy lobster shell microbiome and how this may affect the development of ESD.
2. Materials and methods
2.1. Sample collection
Shell microbiome samples from freshly landed lobsters were collected from commercial fishing boats; once in New Brunswick [Grand Manan, Lobster Fishing Area (LFA) 37] and twice, respectively, in Nova Scotia (Clark’s Harbour, LFA 34; Port Mouton, LFA 33) and Prince Edward Island (Summerside, LFA 25) (Table 1 and Figure 1). The preferred way of sampling was to swab freshly trapped lobsters directly on fishing vessels. In cases where this was not possible, e.g., due to weather, samples were taken from freshly landed lobsters at the wharf. To determine any effect of these sampling methods on the shell microbiome, a direct comparison between boat and wharf sampling was implemented in Summerside (LFA 25) where during one sampling event (October 2022) lobsters were sampled on the boat directly after coming out of the water and again, at the wharf, after being in lobster crates for 6 h. Lobsters that showed signs of ESD were swabbed twice, once on shell disease lesions and once on an apparently healthy part of the carapace.
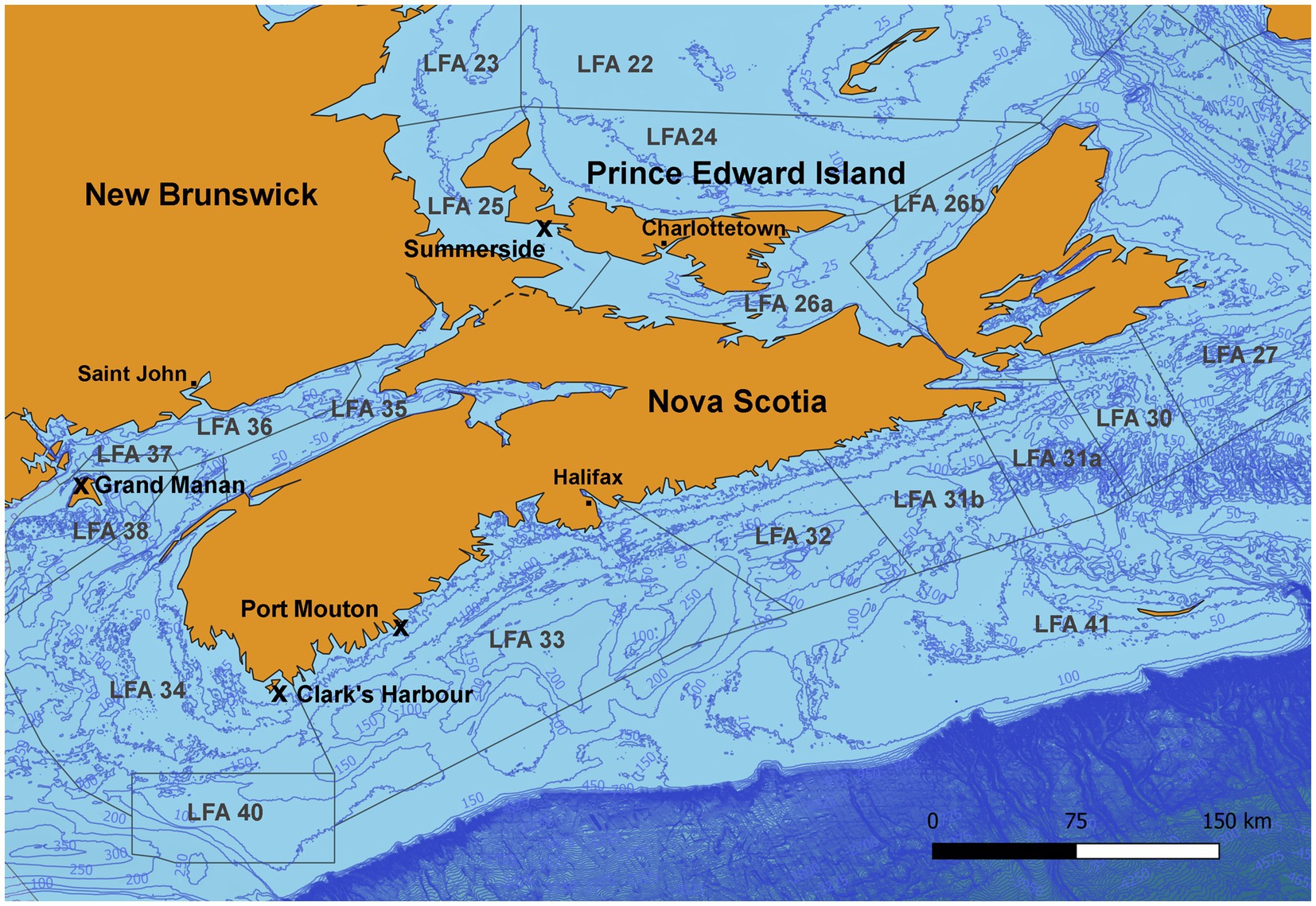
Figure 1. Map of the sampling areas in three Atlantic Canadian provinces and landing ports are indicated by an “x.” In Port Mouton (LFA33) and Clark’s Harbour (LFA 34), sampling occurred twice, once in December 2021 and again in May 2022. In Summerside (LFA 25), sampling occurred twice in September 2021 and October 2022).
Samples were taken as described in Koepper et al. (2023). Briefly, sterile cotton-tipped applicators were used to collect the lobster shell microbiome. The left side of the carapace was washed with 5 mL of sterile water (DNase/RNase/protein-free) filtered through a 0.22 μm filter mounted on a 60 mL syringe. Then the cotton swab was dipped in aliquoted Buffer A (20 mM Tris-HCl, 2 mM sodium EDTA, 1.2% Triton X-100, pH 8.0) (Birer et al., 2017) and used to swab the washed carapace region. To collect seawater controls, sterile cotton swabs were submerged in seawater at the respective sampling locations. Swabs were stored in 100% ethanol at room temperature until further processing.
2.2. Bacterial 16S DNA extraction and sequencing
Bacterial DNA was extracted using the Qiagen Blood and Tissue kit with an additional prior lysis step: swabs were placed in new 1.5 mL microcentrifuge tubes and incubated at 56°C until all ethanol evaporated. Then 300 μL of lysozyme reaction mix (1:10 lysozyme (11 mg/mL): lysozyme buffer (1 M NaCl, 0.5 M EDTA pH 8.0, 1 M Tris-HCL pH 8.0, sterile water) were added to each sample followed by a 30 min incubation at 37°C. The next steps followed the kit manufacturer’s instructions (starting at step 1a). Several extraction controls of molecular biology grade water were added right before sequencing. The quantity and quality of extracted DNA of the samples were checked by Nanodrop (Thermo Scientific, NanoDrop Spectrophotometer ND-1000). Extracted DNA was shipped to the Integrated Microbiome Resource Lab at Dalhousie University, Halifax, Nova Scotia for full-length 16S rRNA gene PacBio sequencing, where library preparation and subsequent sequencing took place according to their in-house protocols (Comeau et al., 2017a,b). In summary, the whole 16S rDNA region was amplified once prior to sequencing (40 cycles) with the full-length fusion primers 27F [AGRGTTYGATYMTGGCTCAG, (Paliy et al., 2009)], 1492R [RGYTACCTTGTTACGACTT, (Lane, 1991)] specific to the PacBio Sequel 2 system and PCR products were validated using a Coastal Genomics Analytical Gel. The PCR reactions were then cleaned and normalized using Charm Biotech Just-a-Plate 96-well Normalization Kit. Samples were pooled on plates (separate wells) and fluorometrically quantified to make one library. Lastly, single-molecule-real-time (SMRT) sequencing was performed by the PacBio Sequel 2 platform on 8 M chips with 240–320 Gb output (Comeau et al., 2017a,b).
2.3. Bioinformatic analysis
All bioinformatic steps were performed using the open-source bioinformatics platform QIIME2 via the Galaxy interface run through Docker on Windows (Bolyen et al., 2019; q2galaxy, 2023). The sequencing produced demultiplexed FASTQ files, which were processed in QIIME2 according to the PacBio CCS Amplicon standard operating procedure (Version 1) (Comeau et al., 2017b) after inspecting the read quality of the raw sequence. Briefly, forward and reverse primer sequences were trimmed with “cutadapt” (forward: 1,200, reverse: 18,000) and FASTQ read files were imported into QIIME2 via a manifest file. The DADA2 algorithm was used to denoise the reads and create amplicon sequence variants (ASVs) (Callahan et al., 2016). ASVs that appeared in only a single sample were removed. Taxonomy was assigned with a Bayesian-trained classifier (Greengenes 13_8 99%, McDonald et al., 2011) and ASVs of mitochondrial and eukaryotic origin were removed from the dataset. Although the seawater and extraction control samples were sequenced along with the swab samples, they failed the PacBio sequencing due to low DNA content and were subsequently removed during quality filtering.
2.4. Statistical analyses
To explore the microbial composition on several taxonomic levels, ASV frequencies were plotted on the phylum level (heat map), relative abundances on class level (bar graph) and the core microbiome and the frequencies of ESD-associated taxa were analyzed on the lowest possible taxonomic level. The relative abundances were calculated by merging ASVs by the respective taxonomic level and then dividing the abundance (number of reads) of one taxon by the total abundance of all taxa in a sample. The relative abundances per sample were subsequently plotted as a bar graph for the nine most abundant bacterial classes. The ASV phylum counts were Hellinger transformed by square rooting the relative abundances to plot the right skewed ASV frequencies in a heatmap. The web-based visualization and analysis tool Clustergrammer was used to create heatmaps (Fernandez et al., 2017).
The core microbiome was determined with the QIIME2 plugin “core-features” for each comparison group separately, e.g., for samples from male, female, and berried lobsters. Core taxa were defined as being present in at least 80% of the samples. The Venn diagrams presenting the amount of shared and unique core microbiome ASVs among sampling groups were created using the interactive web tool Venny (Oliveros, 2007).
To determine whether the frequencies of ESD-associated taxa were associated with any geographical, seasonal or host factors (sex, size, moult stage), initial descriptive statistics (occurrence, prevalence, mean and total number of reads, minimum and maximum counts of detected bacterial taxa) were summarized for the each of the previously identified ESD-associated taxa, according to literature [Aquimarina, Flavobacterium, Kiloniella, Polaribacter, Pseudoalteromonas, Sulfitobacter, Thalassobius and Vibrio; (Chistoserdov et al., 2012; Meres et al., 2012; Feinmann et al., 2017; Ishaq et al., 2023)]. The frequencies of these taxa were investigated, to assess their association with geographical, seasonal or host factors by building a regression model for count data. In initially fitted Poisson models, the variances of the bacterial frequencies were larger than the mean, suggesting overdispersion. Accordingly, the negative binomial model type with the best fit for the data was the NB1 negative binomial which assumes a constant mean. Subsequently, negative binomial regression (NB1) models were built for each of the taxa with the respective taxon frequency as the outcome and using LFA, sampling month, lobster sex, size and moult stage as explanatory variables. The models did not include an offset since there was no population at risk present. This analysis was conducted in Stata (Release 17, StatCorp, College Station, TX, United States, 2021). Forward model building was used, where variables that were associated with the outcome (bacterial taxon count) with a p-value <0.05 were kept in the multivariable final model. A p-value of 0.05 was considered significant for fixed effects and interactions. To gain a better understanding of the Aquimarina strains present on healthy lobster shells, respective ASVs of this genus were also assigned by the blast algorithm (Altschul et al., 1990). The above-described analysis of ESD-associated taxa was also performed on the rarefied dataset (normalized to 400 reads per sample, n = 185 samples). This was done as a sensitivity analysis to evaluate the use of rarefaction for these types of microbiome analysis.
3. Results
3.1. Descriptive statistics
Overall, the shell microbiome of 189 lobsters (19–28 lobsters per sampling event) was analyzed, of which 113 were male, 69 were female and 7 were egg-bearing females. Only one ESD-diseased lobster was sampled (LFA 33 in December), while all others were apparently healthy and showed no signs of shell disease. Lobster sizes ranged from 55 mm CL to 129 mm CL (median: 87 mm, Q1: 81 mm, Q3: 96 mm). Sampled male lobsters were slightly larger than female lobsters with a median of 88 mm CL compared to 87 mm CL. Berried female lobsters were the largest with 92 mm CL (median). 86% of lobsters (163 animals) were in the intermoult stage, whereas 14% (26 animals) were in postmoult stage. For samples collected on commercial fishing vessels, the fishing depth ranged from 7.5 m to 36.6 m (median: 15.2 m, Q1: 13.7, Q3: 22.0 m). For samples taken from the wharf, no water depth data was available. The summary statistics for each sampling event are shown in Table 1.
During denoising and quality filtering, an average of 9% of the reads were removed and the remaining forward and reverse reads were successfully merged into full denoised sequences. Of the total merged sequence variants, 21% were chimeric and consequently removed by the DADA2 algorithm. A summary of the number and percentage of reads passing the quality filtering steps is given in Supplementary Table S1. After filtering, the PacBio sequencing generated an average of 7,000 reads per sample, of which 5,173 ASVs (amplicon sequence variants) were classified. Overall, 326 taxonomic ranks were assigned to the ASVs, and the percentages assigned to each taxonomic level are shown in Figure 2. The full ASV table is included in the Supplementary Table S2.
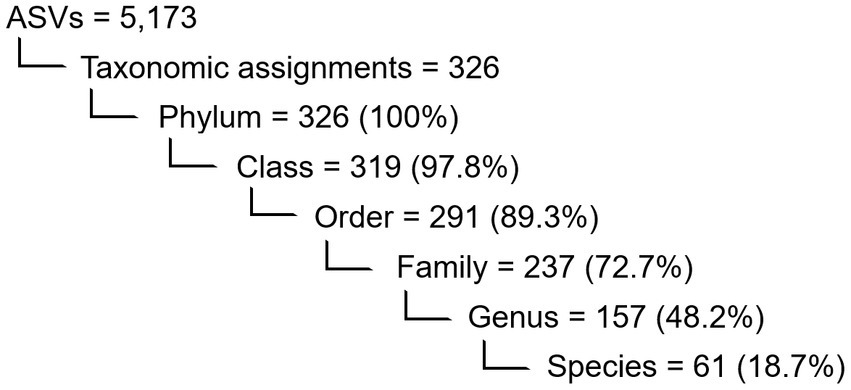
Figure 2. Numerical overview of the detected features in the shell microbiome of apparently healthy lobsters (N = 189) and the ASVs within which they were binned. Also indicated are the number and percentage (in brackets) of taxa assigned to each taxonomical level before rarefaction.
3.2. Shell microbial community
All ASVs were at least assigned to the phylum level; the most abundant bacterial phyla were Pseudomonadata (formerly: Proteobacteria, 46.5%), Bacteroidota (formerly: Bacteroidetes, 23.7%), Verrucomicrobia (12.4%) and Actinomycetota (formerly: Actinobacteria, 6.9%). The heatmap in Figure 3 also shows that the shell microbiome of lobsters from LFA 25 (PEI) was richer in Verrucomicrobia but lower in Actinomycetota and Planctomycetota (formerly: Planctomycetes). Of the less common phyla, Bacillota (formerly: Firmicutes), Mycoplasmatota (formerly: Tenericutes) and Fusobacteriota (formerly: Fusobacteria) were almost only present in LFA 34 in December (Supplementary Figures S1, S2 show heatmaps of bacterial phyla by sex and moult stage respectively).
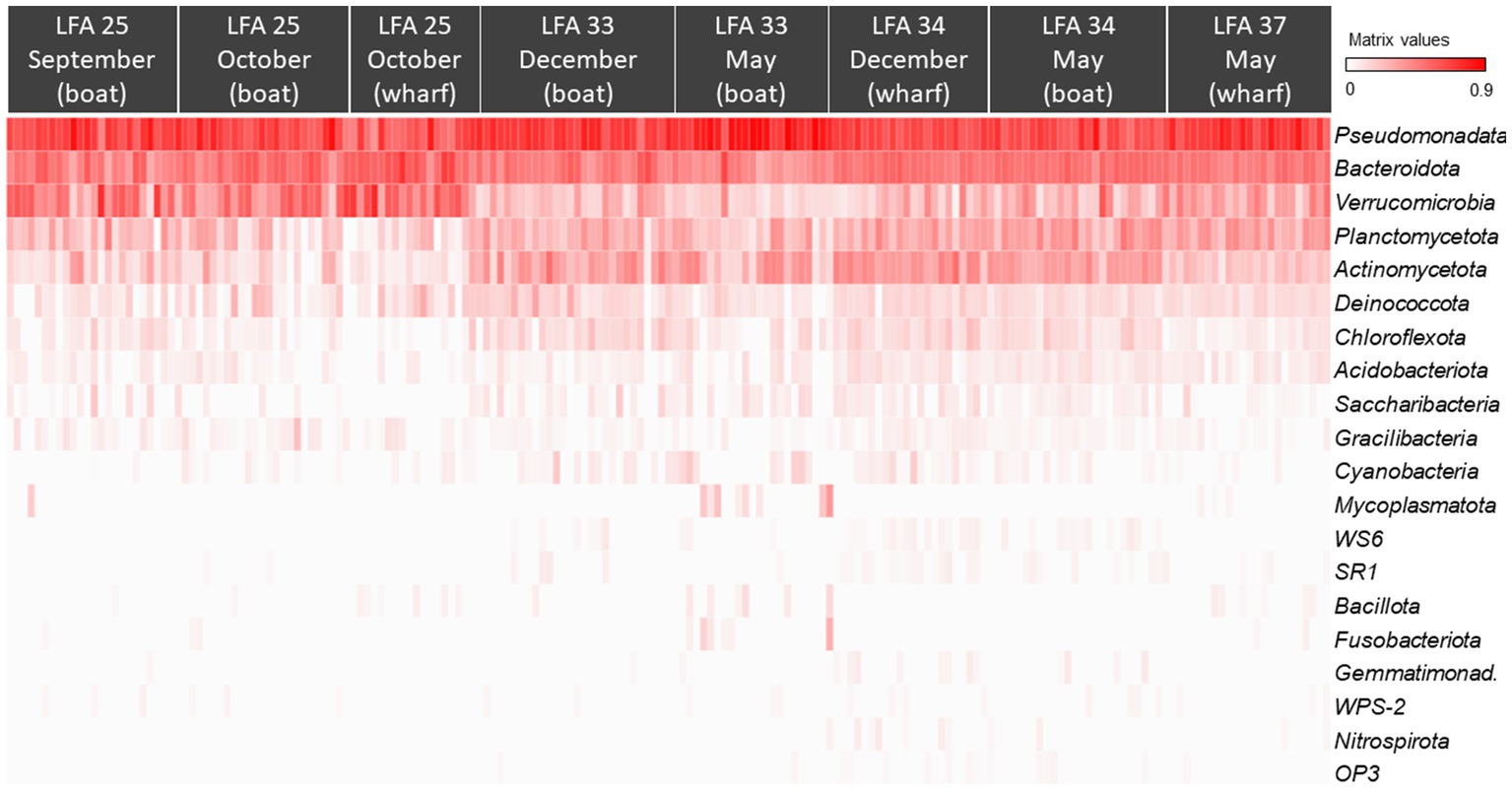
Figure 3. The Hellinger root transformed ASV frequencies per phyla presented in a heat map grouped by sampling location (LFA) and time (month) (N = 189). Red colour indicates the highest frequencies of the respective phylum and white the lowest.
The most abundant classes in the lobster shell microbiome were Gammaproteobacteria, Saprospiria, Verrucomicrobiae, Alphaproteobacteria, Flavobacteriia, Acidimicrobiia, Planctomycetia, Deltaproteobacteria and Deinococci (Table 2). Their relative abundances are shown in the bar graph in Figure 4 and together they made up 96.7% of the total bacterial community. The relative abundance of some bacterial classes differed between samples from different regions (LFAs) and months. The proportions of Deinococci, Deltaproteobacteria and Flavobacteriia were similar across all sampling regions and times. Acidimicrobiia had the lowest relative abundances in LFA 25 sampled in 2022 (1.1%) whereas Alphaproteobacteria (16.8%, only in September 2021) and Verrucomicrobiae (22.8% in 2021, 28.1% in 2022) had by far the highest proportions there. Gammaproteobacteria were highest in LFA 34 in December (51.3%) but lowest in LFA 25 in October 2022 (22.9%). Planctomycetia were most abundant in samples from LFA 33, 34, and 37 in May (8.1%–10.7%) and least abundant in LFA 25 (2.0%–4.0%). Saprospiria occurred with the highest frequencies in LFA 25 in 2022 (18.8%). Male and female lobsters had similar proportions of most bacterial classes, except Deinococci and Saprospiria which were more abundant in females and Acidimicobiia which was more abundant in males. Berried females (N = 7) had less Gammaproteobacteria (27.4%) than males (34.1%) and unberried females (33.1%) but more Planctomycetia. Female lobsters had a lower frequency of Acidimicrobiia (6.3%) than males (8.6%) and berried females (9.0%). Between the moult stages, Alphaproteobacteria and Verrucomicrobiae were more abundant in postmoult lobsters, and Acidimicrobiia, Gammaproteobacteria and Planctomycetia in intermoult lobsters (Supplementary Tables S3, S4).
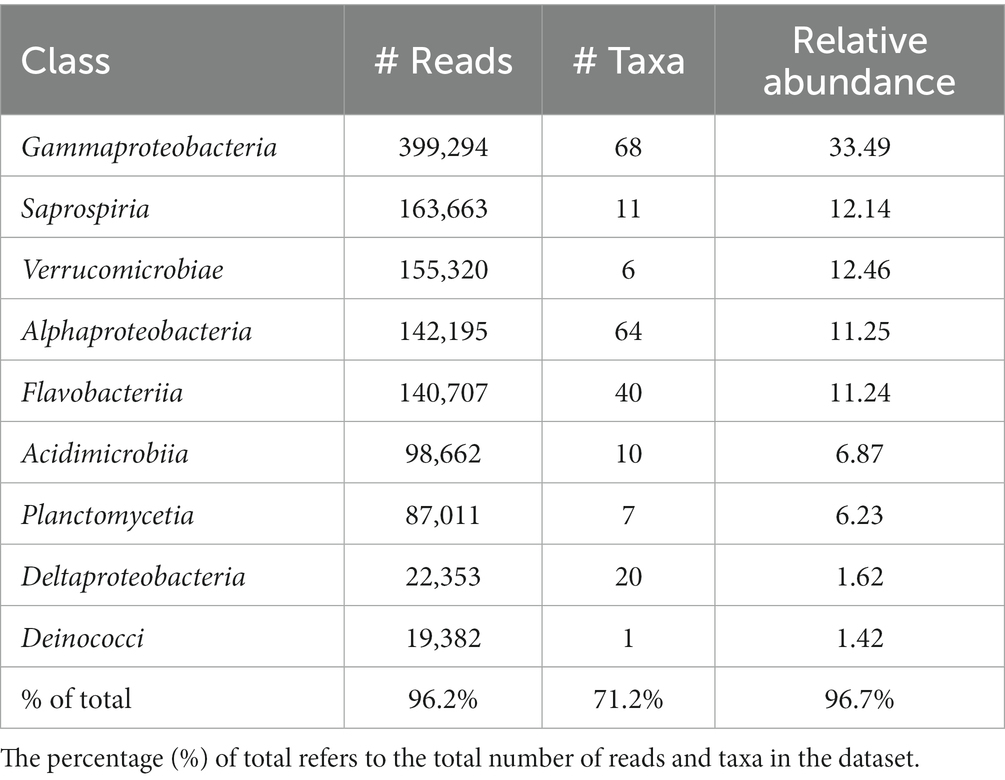
Table 2. Number of total reads, number of taxa and the relative abundance of the nine most abundant bacterial classes in the shell microbiome of apparently healthy lobsters (N = 189).
Figure 4. Bar plot showing the relative abundance of the nine most common bacterial classes in the shell microbiome of American lobster (on average these account for just over 96% of the total abundance) N = 189.
The most abundant families were Saprospiraceae (11.9%), Verrucomicrobiaceae (12.4%) Flavobacteriaceae (10.0%), and Thiotrichaceae (8.7%). Together they made up 42.9% of the shell microbiome on family level. The Vibrionaceae family was only represented by six ASVs with a relative abundance of 0.004%. The most abundant genera >1% were Rubritalea (12.0%, one ASV), Cocleimonas (7.9%, one ASV), Candidatus Endobugula (2.2%, one ASV), Loktanella (1.6%), Octadecabacter (1.8%) and Maribacter (1.2%). The genus Aquimarina was represented with a relative abundance of 0.4%.
3.3. Core microbiome
Venn diagrams show the number and proportion of shared and unique ASVs between sampling groups (Figure 5). Overall, the core microbiome of lobsters was comprised of a total of 24 bacterial taxa and Table 3 summarizes which bacterial taxa differed between sampling groups.
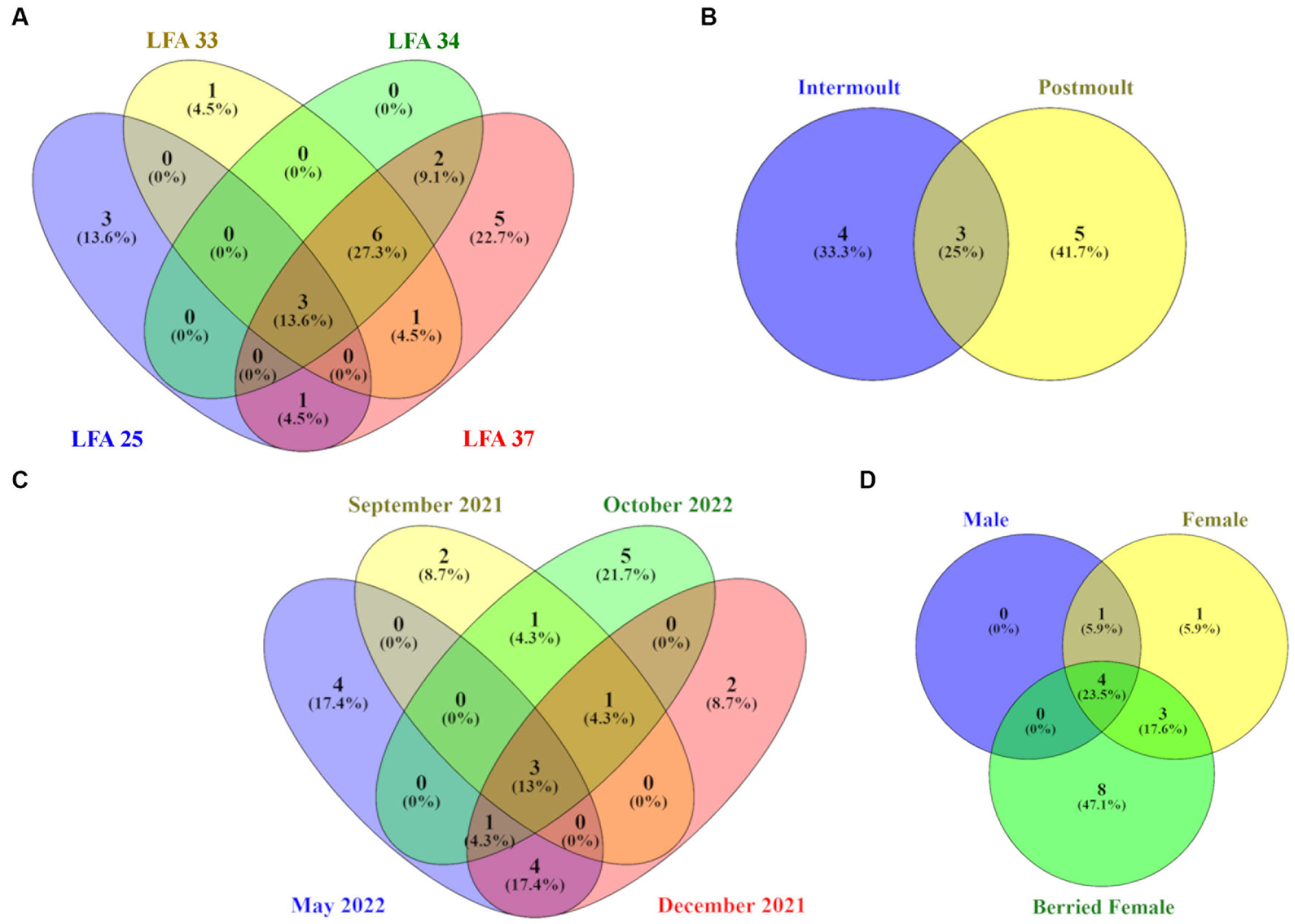
Figure 5. Venn diagrams showing the number and percentage of unique and shared ASVs on the lobster shell microbiome by (A) LFA, (B) sampling month, (C) moult stage and (D) sex. Core ASVs were calculated for each category per sampling group respectively, so percentages in the circles refer to the total ASVs identified for each of the four sampling groups.
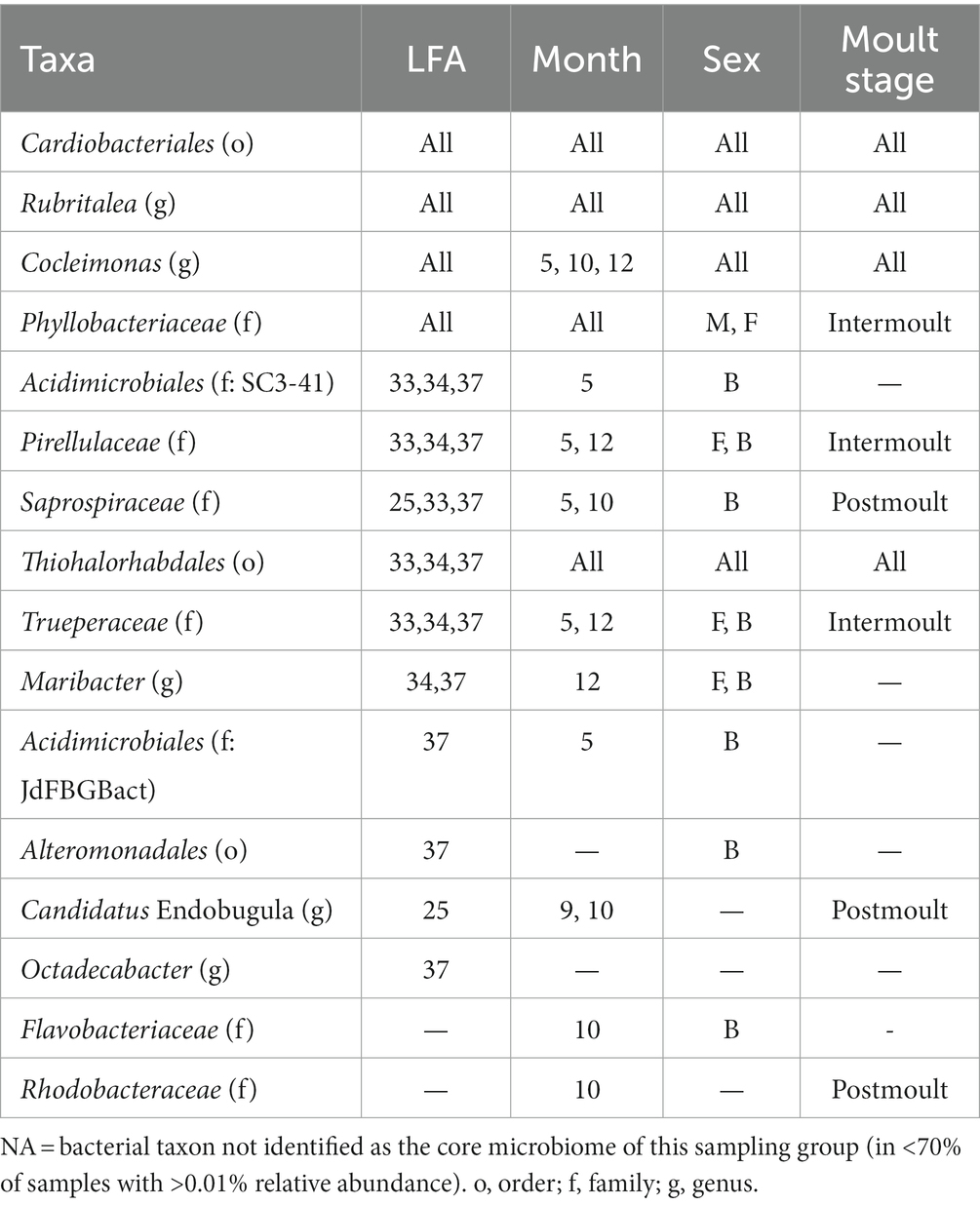
Table 3. Table of bacterial taxa (several ASVs may have been assigned to one taxonomic group) comprising the core microbiome and their occurrence by lobster fishing area (LFA), sampling month (5 = May, 9 = September, 10 = October, 12 = December), Sex (B, berried female; F, female; M, male) and moult stage.
On a geographical scale, the LFAs 33, 34 and 37 shared the highest proportion of ASVs (27.3%), while LFA 25 (PEI) and LFA 37 (New Brunswick) samples had the highest proportions of unique ASVs (13.6%, 22.7%). Only 13.6% of ASVs were shared among all LFAs. Seasonally, samples taken in October had the highest number of unique ASVs (21.7%) but the shell microbial community was most similar in May and December with 17.4% of shared features, such as members of the Pirellulaceae and the Trueperaceae family. Samples from September 2021 and October 2022 were both collected in LFA 25, but only 4.3% of ASVs were shared between the sampling events compared to ASVs that were unique to either sampling event. The shell microbiome of intermoult and postmoult lobsters was not very similar and almost 75% of ASVs were unique to either moult stage. Regarding lobster sex, the highest proportion, with 47.1% of ASVs, was unique to berried females alone, while 23.5% of core ASVs were shared between all three sex groups. Males and females shared only 5.9% of core microbial ASVs.
The core-shell microbiome consisted, for the most part, of the phyla Bacteroidota, Planctomycetota and Pseudomonadata. Of the 16 core taxa identified, only Cardiobacteriales and Rubritalea were present in all LFAs and sampling months and moult stages. Cocleimonas was detected as a core bacterium in all months, sexes, moult stages and months except September. Thiohalorhabdales was detected as a core bacterium in all months, sexes, moult stages and LFAs except LFA 25. Maribacter and Trueperaceae occurred in both berried, and unberried females as a core bacterium.
3.4. ESD-associated taxa
Both two most prominent ESD-associated taxa Aquimarina (class: Flavobacteriia) and Thalassobius (class: Alphaproteobacteria) were detected in this study. One species of the genera Aquimarina, Polaribacter, Pseudoalteromonas and Thalassobius, two species of the genus Flavobacterium and three species of the genera Sulfitobacter and Vibrio were identified, respectively, in the shell microbiome of Canadian lobsters. Aquimarina had the highest mean frequency (8.50, max = 202) and occurred with a prevalence of 47%, whereas Thalassocius had the lowest mean frequency (0.05, max = 86) and was found in only 9% of the samples. Of the ESD-associated taxa which have been reported by Chistoserdov et al. (2005, 2012), Bell et al. (2012), Meres et al. (2012) and Ishaq et al. (2023), only Aquimarina, Polaribacter and Sulfitobacter were detected in sufficient numbers to model explanatory factors for their frequency (Table 4).
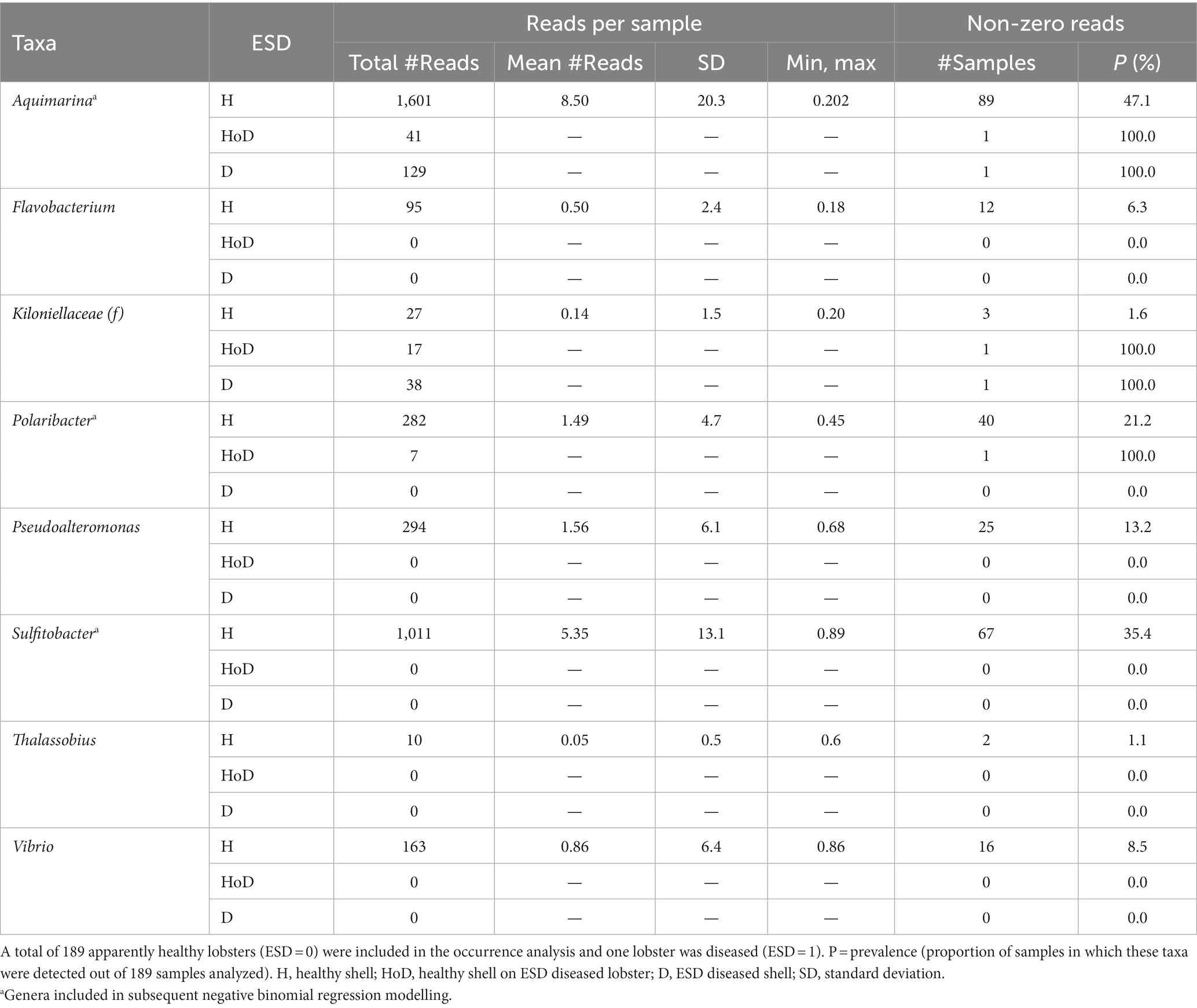
Table 4. Distribution of taxa that have been previously associated with epizootic shell disease grouped by disease status.
Negative binomial regression models were fitted for all ESD-associated taxa, but only for Aquimarina, Polaribacter and Sulfitobacter the number of reads differed significantly by the explanatory variables. For Aquimarina, LFA and sampling month (e.g., sampling events) as well as the moult stage were significant factors during the forward model building process and were subsequently included in the final NB1 model (sampling event: p = 0.000; moult stage: p = 0.029, Wald-test). This indicated that the frequency of Aquimarina depended on the sampling area and sampling season and whether lobsters were in inter- or postmoult. The predicted counts for Aquimarina were highest in LFA 37 (New Brunswick) with an estimated 26.4 reads per sample, and more than double in postmoult lobsters (16.3 estimated reads per sample) compared to intermoult lobsters (7.7 reads per sample). For Sulfitobacter, LFA and sampling month (p = 0.000, Wald test)—combined as sampling event—and moult stage (p = 0.005, Wald test) were significant predictors in the final NB1 model. The predicted Sulfitobacter counts were highest in LFA 25 with an estimated 10.7 reads per sample (September 2021) and in postmoult lobsters with 9.0 reads as compared to intermoult lobsters (4.0). For Polaribacter, lobster sex was the only significant factor in the final NB1 model, and berried females had the highest estimated reads per sample with 3.37. The negative binomial regression model parameters for all analyzed taxa are shown in Table 5 and the mean Sulfitobacter and Aquimarina counts per sampling events and moult stage are shown in Figures 6, 7. The results for the rarefied dataset within the framework of the sensitivity analysis are presented in the Supplementary Tables S5, S6 and Supplementary Figure S3. Briefly, the overall patterns of the estimates from the negative binomial regression models did not differ much between the rarefied and non-rarefied dataset. Aquimarina counts were highest in LFA 37 and Sulfitobacter counts were highest in LFA 25 in post moult lobsters in both approaches. However, when using the rarefied dataset, Polaribacter occurrence did not differ significantly by lobster sex, and Aquimarina occurrence did not differ significantly by moult stage. This information loss could be due to the implemented rarefaction step, which led us to present the raw data as our main results.
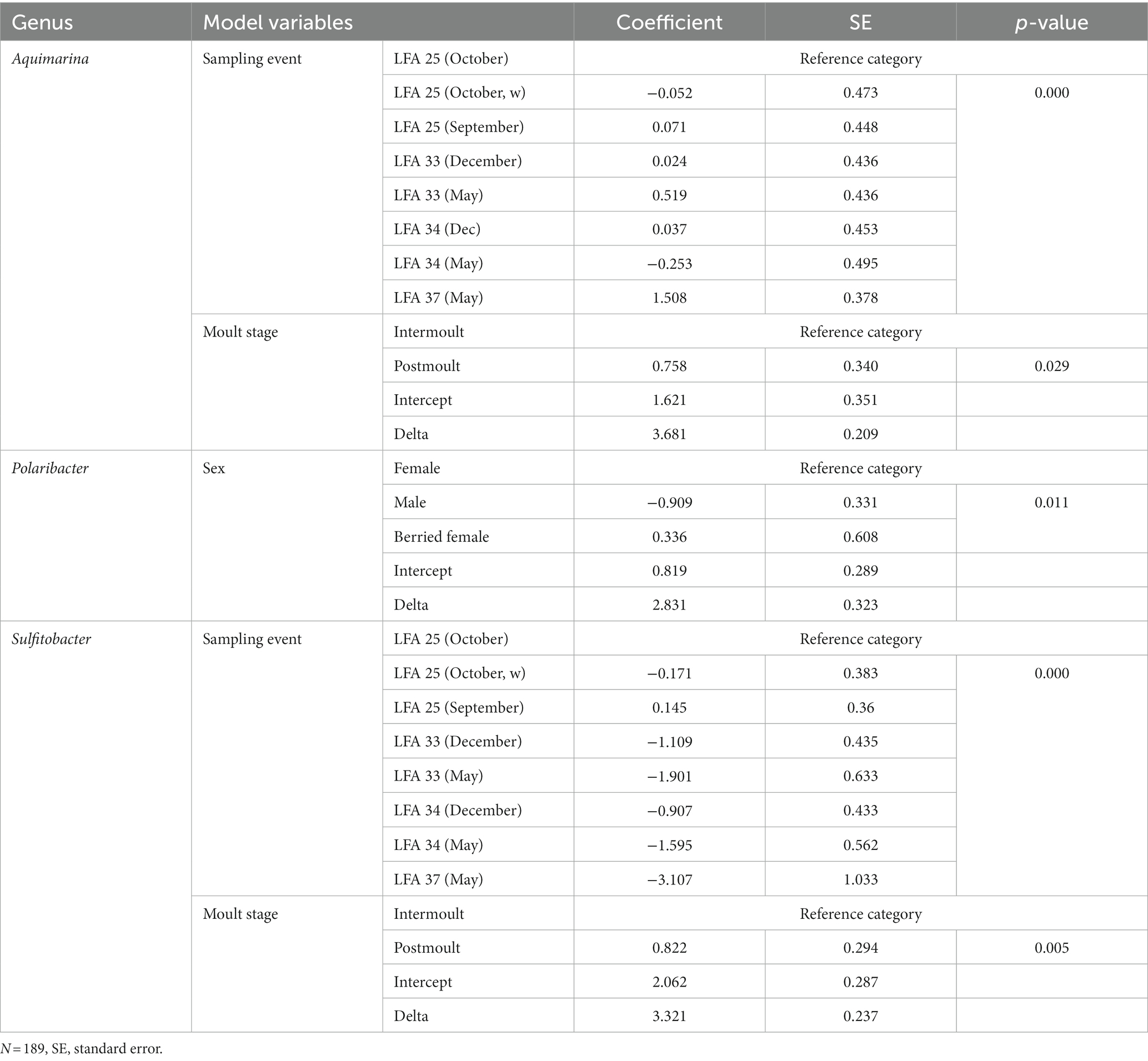
Table 5. The factor variables and respective outputs of the negative binomial regression models fitted to the frequency of each bacterial genus.
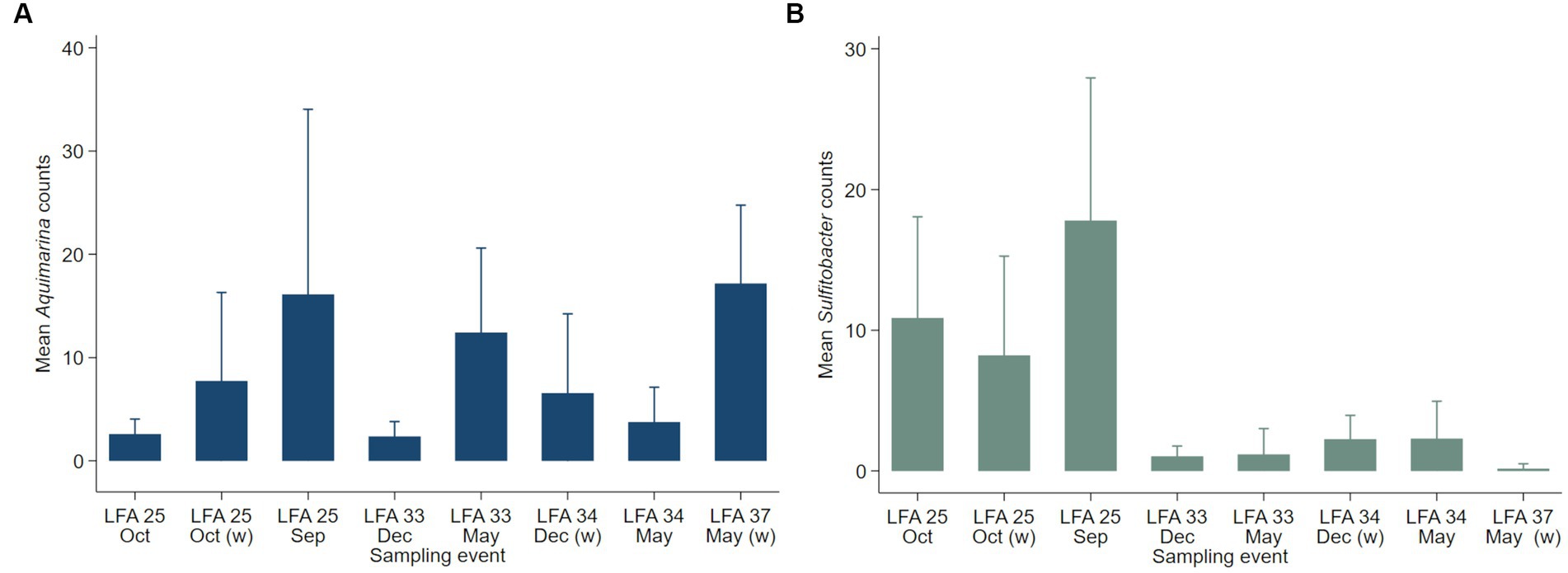
Figure 6. Mean counts (with 95% confidence intervals) of genera (A) Aquimarina and (B) Sulfitobacter per LFA and sampling month (N = 189). Only the shell microbial data from healthy lobsters were considered for negative binomial analysis.
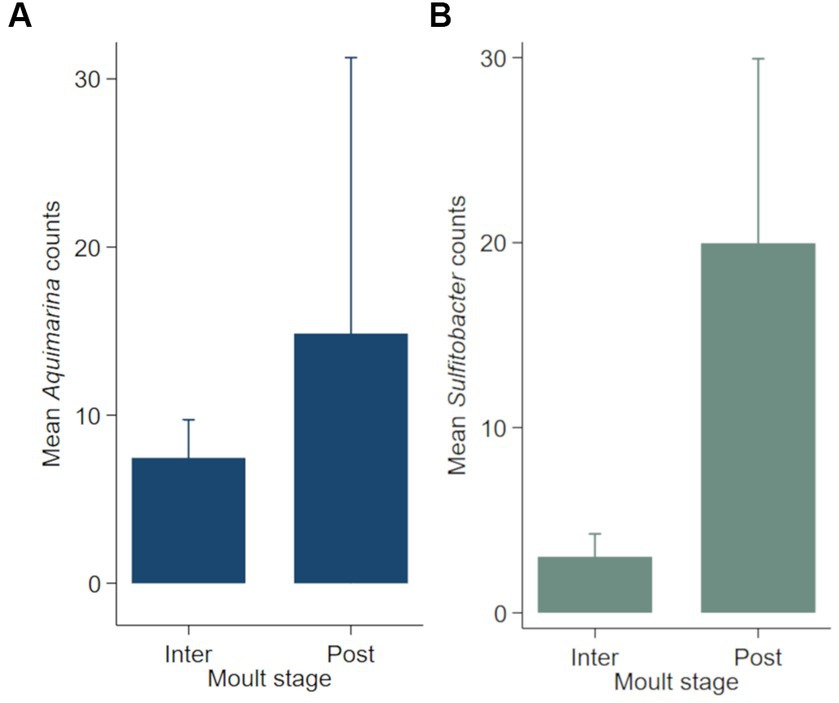
Figure 7. Mean counts (with 95% confidence intervals) of genera (A) Aquimarina and (B) Sulfitobacter per moult stage (N = 189). Only the shell microbial data from healthy lobsters were considered for negative binomial analysis.
The Blast assignment identified 12 different strains of Aquimarina (Table 6) and A. algiphila A. amphilecti, A. macrocephali, A. muelleri and A. pacifica were identified to species level. Aquimarina strain AQ103 and Aquimarina algiphila had the highest prevalences with 18.0% and 15.9%, respectively.
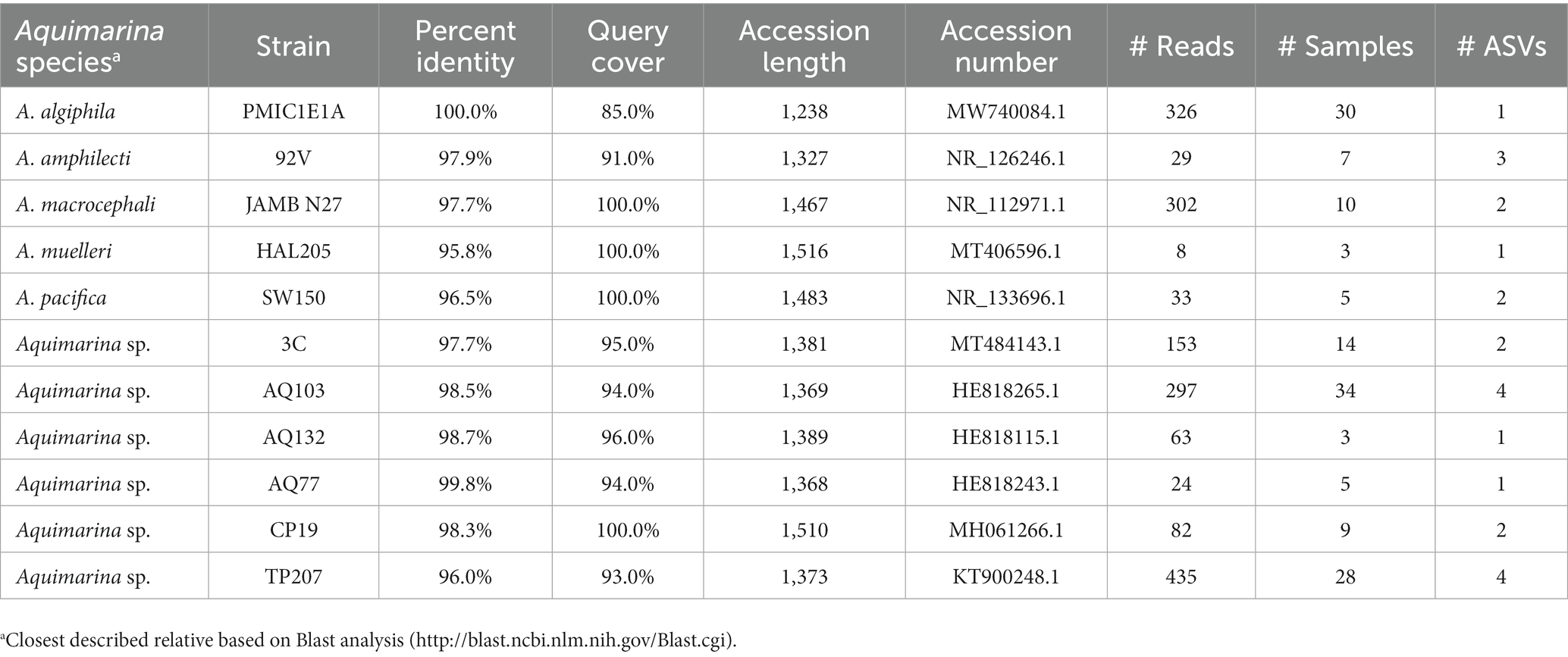
Table 6. Aquimarina species and strains identified by a Blast analysis, including the accession numbers of the closest match in the database as well as the number of reads and ASV the respective taxa had and in how many samples they were detected.
4. Discussion
This study collected and analyzed shell microbial samples from over 180 lobsters from Nova Scotia, New Brunswick and Prince Edward Island; within the most important lobster fishing areas in Canada (LFA 33 and 34). As almost all animals were free of shell disease and sampled from areas with reportedly low to no ESD prevalence, these results provide new insights into the shell microbial community of apparently healthy lobsters. We detected that sampling time and sampling region, as well as to some extent moult stage, rather than lobster sex or size, influence the shell microbiome; that potential pathogens, as well as potential beneficial bacterial taxa, are present in the core microbiome; and that the frequency of taxa previously associated with ESD depends on geographic and seasonal factors. Due to the large sample size as well as the standardized sampling and processing protocols, the presented microbiome data can also serve as a relevant baseline reference for future studies.
4.1. Boat vs. wharf sampling
Although the sampling protocol for this study was standardized as much as possible, differences in sample collection—either from the boat or from the wharf—persisted. Hence, a direct comparison of the effect of these methods on the resulting shell microbiome was implemented in LFA 25. There were differences in the relative abundance of some bacterial taxa, for example, the class Gammaproteobacteria had lower relative abundance in samples taken from the wharf. Still, these observations were in line with the overall trend of shell microbial samples from LFA 25 having a lower representation of this class than other regions. Furthermore, the heatmaps and relative abundance bar plots do not show obvious differences in boat and wharf samples from the same sampling event. Therefore, we assume that both sampling methods yield similar outputs and that results are comparable across this study.
4.2. The overall shell microbial community
With around 5,000 ASVs detected, this study shows a similar microbial richness as others that have analyzed shell microbiota in American lobster (Zha et al., 2019; Schaubeck et al., 2023) and other crustaceans (Zha et al., 2019; Kraemer et al., 2020) in the wild. In some studies of lab-reared lobsters, up to 18,000 taxonomic units (similar to ASVs) were detected (Feinmann et al., 2017). The higher richness in shell samples from tank-based compared to wild animals is potentially linked to higher bacterial load in tank environments. While the opposite was observed in wild and captive eagle rays (Clavere-Graciette et al., 2022) and tank-acclimated horseshoe crabs (Friel et al., 2020), there is limited data on the microbial difference between wild and tank-based crustaceans.
The most common phyla detected on shells of apparently healthy lobsters are comparable to what has been observed on other crustaceans. Actinomycetota, Bacteroidota, Planctomycetota, and Pseudomonadota were previously found on healthy American lobsters (Chistoserdov et al., 2012; Feinmann et al., 2017). In addition to the aforementioned phyla, Feinmann et al. (2017) also detected Deferribacteraceae in apparently healthy lab-reared American lobsters, which were not present in our dataset. Bacillota, Fusobacteriota and Mycoplasmatota were less abundant on lobsters in this study, but they are common marine taxa and have been previously recorded on shells of lobsters and brown crabs (Payne et al., 2008; Bell et al., 2012; Meres et al., 2012; Quinn et al., 2013; Bergen et al., 2022). Bell et al. (2012) showed that, among others, Bacillota, Fusobacteriota and Mycoplasmatota were over-represented in healthy compared to shell-diseased American lobsters. Here, we saw a higher frequency of these three taxa in LFA 34 in December compared to the same region in spring or to LFA 33 in December. In addition to disease state (not analyzed in this study), it appears that geographical and seasonal factors play a role in the distribution of these phyla.
In this study, we confirmed previous observations that the most common bacterial classes on lobster carapaces are Acidimicrobiia, Alphaproteobacteria, Flavobacteriia, Gammaproteobacteria, Saprospiria and Verrucomicrobiae (Chistoserdov et al., 2012; Meres et al., 2012; Quinn et al., 2013; Ishaq et al., 2023). Some of these classes showed differential distributions between sampling sites and times. While there are studies on how the bacterial community differs by shell disease state on lobsters, significantly less is known about the effect of spatiotemporal and host factors on bacterial community. For example, Verrucomicrobiae had higher relative abundance in healthy compared to shell-diseased spiny lobsters in New Zealand (Zha et al., 2019) and Alphaproteobacteria levels were higher in American lobsters without ESD (Meres et al., 2012). We observed that Verrucomicrobiae and Alphaproteobacteria abundances were highest in LFA 25 in September and in postmoult lobsters. At the same time, LFA 25 had the highest proportion of postmoult lobsters, so it is difficult to discern whether a higher abundance of these taxa can be attributed to location or to the moult stage. A recent study found that on rock crabs (Cancer irrotatus) carapaces, Verrucomicrobiae also had the highest abundance in the Northumberland Straight (close proximity to LFA 25) compared to other regions in Atlantic Canada, which also points towards geographical effects on the lobster shell microbial community (Koepper et al., 2023). Saprospiria has only been isolated from lobster shells once (Andrews, 2018, unpublished thesis), but colonized the gut and hemolymph of spiny lobsters (Ooi et al., 2017, 2019). In the present study, Saprospiria was the least common in LFA 34 in December but had almost 10% higher abundance during May of the following year. Members of the Saprospiria class are sensitive to environmental factors such as temperature and CO2 in benthic communities (Raulf et al., 2015; Hicks et al., 2018). If conditions are less favourable for this class during winter, this could explain the seasonal difference in their relative abundance in the lobster shell microbiome.
Planctomycetia, which were the least abundant in LFA 25 (both years), are a dominant bacterial class in coastal sediments (Feinmann et al., 2017; Hicks et al., 2018). They have also been found to be dominant in healthy shrimp and absent in shell-diseased brown crabs (Chen et al., 2017; Bergen et al., 2022). Lastly, members of the Acidimicrobiia were previously detected in healthy (not shell-diseased) lobsters (Chistoserdov et al., 2012; Quinn et al., 2013; Feinmann et al., 2017; Zha et al., 2019) and brown crabs (Bergen et al., 2022).
4.3. The core microbial composition
Of the 16 bacterial taxa identified as core microbes on lobster shells, all have previously been detected on crustacean carapaces except Acidimicrobiales (family: SC3-41, JdFBGBact), Alteromonadales and Trueperaceae. The families SC3-41 and JdFBGBact of the Acidimicrobiales and Pirellulaceae have been detected before in water samples from salmon farms and on the seaweed Sargassum muticum as an epibiont (Rud et al., 2017; Serebryakova et al., 2018). While members of the Trueperaceae have not yet been found on lobster carapaces, Ooi et al. (2019) detected Trueperaceae in the hemolymph of lab-reared spiny lobsters (Panulirus ornatus) where the taxa occurred at higher frequencies in control compared to thermally stressed hosts.
Most other core bacterial taxa found in our study have been previously associated with shell disease in American lobsters and other crustaceans. Two of the four taxa that were positively linked to shell disease by previous research and present here as core bacteria were shared between most sampling groups: Cardiobacteriales and Thiohalorhabdales. Members of the order Cardiobacteriales have only been found in shell disease lesions and never on healthy carapaces of American lobsters albeit with overall low frequency (Quinn et al., 2013). Here, this taxon was observed in the core microbiome in all sampling groups (LFAs, months, sexes and moult stages). Similarly, the order Thiohalorhabdales has been associated with tail fan necrosis lesions in spiny lobsters previously (Zha et al., 2019). While the genus Maribacter occurs both on lobster shells free of ESD and on ESD lesions, Chistoserdov et al. (2012) and Schaubeck et al. (2023) reported a higher frequency of this genus in ESD lesions (Quinn et al., 2012, 2013). Here, Maribacter is a core bacterium on lobster shells sampled from LFA 33 and 37 and in berried and unberried females. Flavobacteriaceae are a ubiquitous family in crustacean shell microbiomes, but they contain genera that have previously been associated with ESD and other shell diseases (Aquimarina) and, as such, Flavobacteriaceae abundance increases in shell disease lesions (Chistoserdov et al., 2005; Zha et al., 2019). Here, members of Flavobacteriaceae were core bacteria only in October and in berried females.
Three taxa previously associated with non-shell diseased crustaceans, and present here as core bacteria, were shared between all sampling groups: Cocleimonas, Saprospiraceae and Rubritalea. Cocleimonas previously occurred in both shell-diseased and healthy brown crabs (Bergen et al., 2022), but only colonized non-ESD American lobster (Quinn et al., 2013; Feinmann et al., 2017). Saprospiraceae were most commonly identified from non-ESD lobsters and from cuticles unaffected by shell disease (Feinmann et al., 2017; Andrews, 2018; Zha et al., 2019). The genus Rubritalea is a common epibiont on marine animals and has been associated with healthy (non-ESD) lobster shells in previous research (Feinmann et al., 2017; Ishaq et al., 2023). This genus produces carotenoids, which may have antimicrobial functions and interestingly, ESD-diseased lobsters have fewer carotenoids on their shells (Prince and Bayer, 2005). Therefore, Rubritalea could be a beneficial shell colonizer for their hosts. Furthermore, two known marine symbionts were part of the core microbiome only in one LFA respectively: Candidatus Endobugula in LFA 25 and Octadecabacter in LFA 37. Mathew and Lopanik (2014) detected the symbiotic relationship between members of the genus Candidatus Endobugula and its bryozoan host, but in lobsters, this microbe was majorly isolated from shells that were transitioning into shell disease (Feinmann et al., 2017). Octadecabacter is a dominant subcuticular symbiont in brittle stars (Morrow et al., 2018), but in American lobsters, this genus has been detected with an increased presence in shell-diseased lobsters and in ESD lesions (Chistoserdov et al., 2012; Quinn et al., 2012; Andrews, 2018; Zha et al., 2019).
Another core microbial taxon of the family Phyllobacteriaceae was shared by all sampling regions and times in this study. Phyllobacteriaceae were both isolated from healthy cuticles and ESD lesions in American lobsters, and an ubiquitous distribution of this family is coherent (Quinn et al., 2012; Feinmann et al., 2017). The Rhodobacteraceae family is ubiquitous on lobster carapaces and has been detected in healthy lobsters as well as in shell disease lesions (Chistoserdov et al., 2012; Quinn et al., 2013; Feinmann et al., 2017). In shell diseased brown crabs, Bergen et al. (2022) detected a shift toward this bacterial family. Here, Rhodobacteraceae were part of the core microbiome only in October (LFA 25) in postmoult lobsters. This could indicate their role as early colonizers of the new shell, as they commonly dominate marine biofilms (Elifantz et al., 2013).
4.4. Differential frequency of ESD-associated taxa
The data of this study provided us with a unique opportunity to demonstrate that the distribution of shell microbial taxa previously associated with ESD is influenced by location and sampling time as well as by host factors.
The genera considered to be associated with ESD Aquimarina, Polaribacter, Pseudoalteromonas, Sulfitobacter, Thalassobius and Vibrio have been found in the shell microbiome of American lobsters from Atlantic Canada before, but not the genera Flavobacterium and Kiloniella (Quinn et al., 2013). In our samples of apparently healthy American lobsters, all the above-mentioned genera were present. However, some occurred with a frequency too low to evaluate factors associated with their distribution. With a high number of sampled lobsters and several sampling events, both taxa most commonly associated with ESD were detected, although with low relative frequencies (Aquimarina: 0.1%, Thalassobius: 0.001%). In a previous analysis of 75 lobsters (ESD diseased and healthy) sampled from the northern and southern Gulf of Maine (US), Thalassobius did not occur in the shell microbiome (Ishaq et al., 2023). Quinn et al. (2013) have reported that the distribution of ESD-associated taxa in Atlantic Canada differs from Southern New England populations. This may be because these disease-associated taxa would be less common in ESD-free lobster populations or in regions of the Gulf of Maine and Gulf of St. Lawrence compared to Southern New England.
Although only one diseased lobster was collected in this study, ESD disease state affected the frequency of the two ESD-associated taxa when this lobster was included in the analyses. If not, the time and location of the sampling event, as well as lobster moult stage significantly affected the distribution of Aquimarina and Sulfitobacter in our models. The occurrence of Aquimarina was highest in LFA 37, a fishing area close in proximity to LFA 35 in the Bay of Fundy where ESD prevalence in Atlantic Canada is anecdotally the highest (Petritchenko, personal communication, 2023). A seasonal difference in Aquimarina counts was seen in LFA 33, where they were higher in May than December. ESD has been associated with higher temperatures and the ESD peak in the Gulf of Maine is observed during May and June (Reardon et al., 2018). Aquimarina counts were also significantly higher in postmoult lobsters compared to intermoult lobsters. Postmoult lobsters have younger shells, however, ESD has been associated with individuals with older shells (Groner et al., 2018). Lower Aquimarina counts in older shells, which seem to be a risk factor for ESD, was an unexpected result and may emphasize the opportunistic nature of this genus on healthy lobsters. For Sulfitobacter our model indicated that both the LFA and the sampling month, as well as moult stages were significantly associated with its frequency in the samples. Predicted Sulfitobacter counts were highest in LFA 25 and in postmoult lobsters. As most of the postmoult lobsters were sampled in LFA 25, it is likely that this taxon is more common in this fishing region, where more postmoult animals happened to be sampled. Large, female lobsters are reportedly more susceptible to ESD as they usually moult only every 2 years (Glenn and Pugh, 2006; Castro and Somers, 2012; Hoenig et al., 2017). Lobsters with a longer intermoult phase have a longer exposure to changes in their shell microbiome and potentially to pathogens. The negative binomial models developed here indicate that lobster sex and size do not predict the frequency of ESD-associated taxa Aquimarina and Sulfitobacter. While it has been shown that males, females and berried females react differently to environmental stressors and prefer different habitats according to their size (Koepper et al., 2021), this may not have a large influence on their shell microbial community in healthy individuals. The genus Polaribacter was significantly more abundant on shells of berried female lobsters, although only a small number (N = 7) of this population group was sampled. While berried females are reportedly more susceptible to ESD (Groner et al., 2018), our results should be interpreted with caution as our study only considered healthy lobsters.
Due to the rarity of Aquimarina, this genus is considered to belong to the microbial rare biosphere. In this study, Aquimarina was less rare (relative abundance = 0.4%) than in a recent survey where the relative abundance was mostly below 0.1% over several marine environments (Silva et al., 2022). In contrast, on healthy lobster shells sampled from Long Island Sound, a region with historically high rates of ESD, the relative abundance of Aquimarina was around 4% (Schaubeck et al., 2023). The blast analysis showed that several Aquimarina species and strains are present on healthy American lobster shells. A. muelleri and A. macrocephali have previously been detected on ESD lesions in Atlantic Canada and on black spot lesions of brown crabs, although A. macrocephali was also isolated from healthy crabs in the same study (Quinn et al., 2013; Bergen et al., 2022). Although not yet isolated from ESD lesions, one of the most prevalent Aquimarina species detected here, A. algiphila (prevalence = 15.9%) can degrade chitin (Nedashkovskaya et al., 2018) and is closely related to A. pacifica. The strain that has most commonly been associated with ESD lesions, A. “homaria” strain I32.4 was not found in this study (Chistoserdov et al., 2012; Ranson et al., 2018). Accordingly, this strain seems to be rare in healthy lobsters in Atlantic Canada. However, other strains may also have the capability to act opportunistically when encountering a suitable host, and successively could be involved in ESD progression. So far, no work has been carried out on the distribution of these taxa in healthy lobsters in Atlantic Canada and no consistent ESD monitoring is currently being implemented in the region. This makes it difficult to determine whether differences in the frequency of these taxa stem from geographic variation alone or if a higher frequency could be linked to lobsters being more likely to develop ESD.
5. Limitations
As sampling for this study took place on commercial lobster fishing vessels, sampling times were restricted to the respective fishing seasons. This is the reason not all regions could be sampled at the same time, potentially introducing seasonal differences in the microbial community that are difficult to account for. For example, the shell microbial community in LFA 25 in September showed differences in microbial composition when compared to samples taken in other regions during May. However, it was not possible to sample LFA 25 in May for better spatial comparison as the fishing season there only lasts from mid-August until mid-October. While we analyzed all sampling events by LFA and months separately to account for this limitation, future studies could profit from a more balanced sampling design. Retrieving samples from commercial lobster boats also led to targeting more intermoult lobsters compared to freshly moulted (postmoult) animals, because fishing usually takes place well after the moulting season. Only in LFA 25, where samples were taken not long after the lobster moult, more postmoult lobsters were represented in the samples. If postmoult lobsters have a distinct shell microbial community, when compared to intermoult lobsters, this could have caused bias, as it is difficult to determine whether the microbial differences stem from a geographic influence or because more postmoult lobsters were present in the samples. Additionally, not many berried female lobsters were sampled, potentially because they are usually not targeted by the fishery. Having roughly equal numbers of males, females and berried females would enable a better microbial comparison to be made between those groups, given that berried females have also been shown to be more susceptible to shell disease. Furthermore, no seawater controls or blanks could be sequenced as these samples contained too little DNA for PacBio sequencing. This inhibited the control for any potential contaminating ASVs in the shell samples. Nevertheless, the consistency in sample collection, processing and analysis makes this a valuable dataset for a first description of healthy lobster microbiomes and for crustacean microbiome studies in the future.
6. Conclusion
This comprehensive analysis of the shell microbial community of American lobster highlights differences in relative abundances of the most common bacterial phyla and classes. These differences are more pronounced when comparing samples from different LFAs and at different times of the year, than by the host factors (such as lobster sex, size or moult condition). The marine epibiotic community is highly variable and depends on environmental factors such as temperature and salinity. Epibionts can also provide the host with important metabolic products (Wahl et al., 2012). LFA 25 stands out with the highest abundance of Verrucomicrobiae and the lowest abundance of Acidimicrobiia and Planctomycetia, and while Planctomycetia has been associated with healthy crustaceans, it is unknown in what way this may affect the local lobster populations, if at all. Of 16 core microbial taxa, only two occurred in all LFAs and sampling months, and four occurred in both males and (berried) females and in intermoult and postmoult lobsters. This indicates a certain degree of variation in the distribution of the core microbiome and could mean that bacterial taxa play a distinct role in the microbial community depending on where the host is located as well as the time of year. Some potential pathogens (e.g., Cardiobacteriales and Thiohalorhabdales) and beneficial bacteria (e.g., Rubritalea) were shared among regions and sampling times. However, a potential symbiont Candidatus Endobugula was found only in the core microbiome in shell samples from LFA 25, whereas Flavobacteriaceae, which are enriched on shell-diseased lobsters, were only identified as core epibionts in samples from October (LFA 25) and berried females. Furthermore, it was shown that the occurrence of ESD-associated taxa, Aquimarina in particular, depends on the geographic area, sampling time and the lobster moult stage. The unique composition of the core microbiome, together with distinct distributions of the ESD-associated taxa, highlights the differences in the microbial community on healthy lobster shells from Atlantic Canada. It has yet to be determined whether this is concomitant with lobsters being more predisposed to shell disease in areas where ESD-associated taxa frequency is highest or where the core microbiome features certain potential pathogens. In this study, we provided a detailed overview of the Aquimarina species and strains detected in the dataset, which will be a useful tool for ESD research in Canadian water and beyond. For future studies, we encourage a holistic approach in shell disease monitoring, combined with microbiome assessment of healthy and diseased lobsters, to elucidate the risk factors for this important disease.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1026623/.
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
SK, KFC, KKT, and CWR designed the research. SK collected the data and wrote the manuscript. SK and HS analyzed the data. SK, KFC, JTM, CWR, HS, and KKT revised and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This project received funding via the OFI Phase-2 One Ocean Health Grant (Ocean Frontiers Institute, Dalhousie University, Nova Scotia, Canada) through the Canada First Research Excellence Fund. Additional funding was received from the Atlantic Veterinary College Research Fund (University of Prince Edward Island, Prince Edward Island, Canada).
Acknowledgments
The Prince Edward Island Fishermen’s Association (PEIFA) and the Fishermen and Scientist Research Association (FSRS) in Nova Scotia assisted in organizing and conducting sample collection and the authors are grateful for the fishers who accommodated our sample collection on their vessels. The authors would also like to thank members of the Clark Lab at the Dalhousie University’s Faculty of Agriculture for their help in processing the samples as well as the Integrated Microbiome Resource Lab (IMR) for their support in sequencing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1245818/full#supplementary-material
References
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Andrews, J. (2018). Environmental effects on the microbial community of Homarus americanus afflicted with the epizootic shell disease. Available at: http://jbox.gmu.edu/handle/1920/11647
Bell, S. L., Allam, B., McElroy, A., Dove, A., and Taylor, G. T. (2012). Investigation of epizootic shell disease in American lobsters (Homarus americanus) from Long Island sound:1. Characterization of associated microbial communities. J. Shellfish Res. 31, 473–484. doi: 10.2983/035.031.0207
Berg, G., Rybakova, D., Grube, M., and Köberl, M. (2015). The plant microbiome explored: implications for experimental botany. J. Exp. Bot. 67, 995–1002. doi: 10.1093/jxb/erv466
Bergen, N., Krämer, P., Romberg, J., Wichels, A., Gerlach, G., and Brinkhoff, T. (2022). Shell disease syndrome is associated with reduced and shifted epibacterial diversity on the carapace of the crustacean Cancer pagurus. Microbiol. Spectr. 10:e0341922. doi: 10.1128/spectrum.03419-22
Bikel, S., Valdez-Lara, A., Cornejo-Granados, F., Rico, K., Canizales-Quinteros, S., Soberón, X., et al. (2015). Combining metagenomics, metatranscriptomics and viromics to explore novel microbial interactions: towards a systems-level understanding of human microbiome. Comput. Struct. Biotechnol. J. 13, 390–401. doi: 10.1016/j.csbj.2015.06.001
Birer, C., Tysklind, N., Zinger, L., and Duplais, C. (2017). Comparative analysis of DNA extraction methods to study the body surface microbiota of insects: a case study with ant cuticular bacteria. Mol. Ecol. Resour. 17, e34–e45. doi: 10.1111/1755-0998.12688
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Callahan, B. J., Mcmurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Castro, K., Cobb, J., Gomez-Chiarri, M., and Tlusty, M. (2012). Epizootic shell disease in American lobsters Homarus americanus in southern New England: past, present and future. Dis. Aquat. Org. 100, 149–158. doi: 10.3354/dao02507
Castro, K. M., and Somers, B. A. (2012). Observations of epizootic shell disease in American lobsters, Homarus americanus, in southern New England. J. Shellfish Res. 31, 423–430. doi: 10.2983/035.031.0202
Chen, W., Ng, T., Wu, J., Chen, J., and Wang, H.-C. (2017). Microbiome dynamics in a shrimp grow-out pond with possible outbreak of acute hepatopancreatic necrosis disease. Sci. Rep. 7:12. doi: 10.1038/s41598-017-09923-6
Chistoserdov, A., Quinn, R., Gubbala, S., and Smolowitz, R. (2012). Bacterial communities associated with lesions of shell disease in the American lobster, Homarus americanus. J. Shellfish Res. 31, 449–462. doi: 10.2983/035.031.0205
Chistoserdov, A., Smolowitz, R., Mirasol, F., and Hsu, A. (2005). Culture-dependent characterization of the microbial community associated with epizootic shell disease lesions in American lobster, Homarus americanus. J. Shellfish Res. 24, 741–747. doi: 10.2983/0730-8000(2005)24[741:CCOTMC]2.0.CO;2
Clavere-Graciette, A. G., McWhirt, M. E., Hoopes, L. A., Bassos-Hull, K., Wilkinson, K. A., Stewart, F. J., et al. (2022). Microbiome differences between wild and aquarium whitespotted eagle rays (Aetobatus narinari). Anim Microbiome 4, 34–15. doi: 10.1186/S42523-022-00187-8
Cobb, J. S., and Castro, K. M. (2006). Shell disease in lobsters: a synthesis. Rhode Island: University of Rhode Island.
Comeau, A., Douglas, G., and Langille, M. (2017a). Integrated microbiome research (IMR): protocols. Available at: https://imr.bio/protocols.html (Accessed January 10, 2023)
Comeau, A. M., Douglas, G. M., and Langille, M. G. I. (2017b). Microbiome helper: a custom and streamlined workflow for microbiome research. mSystems :2. doi: 10.1128/msystems.00127-16
Cornejo-Granados, F., Lopez-Zavala, A. A., Gallardo-Becerra, L., Mendoza-Vargas, A., Sánchez, F., Vichido, R., et al. (2017). Microbiome of Pacific Whiteleg shrimp reveals differential bacterial community composition between wild, aquacultured and AHPND/EMS outbreak conditions. Sci. Rep. 7:11783. doi: 10.1038/s41598-017-11805-w, 7
DFO (2021a). Canada’s Fisheries Fast Facts 2021. Available at: https://waves-vagues.dfo-mpo.gc.ca/library-bibliotheque/41039634.pdf?
DFO (2021b). Stock status update for American lobster (Homarus americanus) in lobster fishing area 34 for 2020.
Egan, S., and Gardiner, M. (2016). Microbial dysbiosis: rethinking disease in marine ecosystems. Front. Microbiol. 7:8. doi: 10.3389/fmicb.2016.00991
Elifantz, H., Horn, G., Ayon, M., Cohen, Y., and Minz, D. (2013). Rhodobacteraceae are the key members of the microbial community of the initial biofilm formed in eastern Mediterranean coastal seawater. FEMS Microbiol. Ecol. 85, 348–357. doi: 10.1111/1574-6941.12122
Feinmann, S., Martínez, A., Bowen, J., and Tlusty, M. (2017). Fine-scale transition to lower bacterial diversity and altered community composition precedes shell disease in laboratory-reared juvenile American lobster. Dis. Aquat. Org. 124, 41–54. doi: 10.3354/dao03111
Fernandez, N. F., Gundersen, G. W., Rahman, A., Grimes, M. L., Rikova, K., Hornbeck, P., et al. (2017). Clustergrammer, a web-based heatmap visualization and analysis tool for high-dimensional biological data. Sci. Data 4, 1–12. doi: 10.1038/sdata.2017.151
Friel, A. D., Neiswenter, S. A., Seymour, C. O., Bali, L. R., McNamara, G., Leija, F., et al. (2020). Microbiome shifts associated with the introduction of wild Atlantic horseshoe crabs (Limulus polyphemus) into a touch-tank exhibit. Front. Microbiol. 11, 1–16. doi: 10.3389/FMICB.2020.01398
Glenn, R. P., and Pugh, T. L. (2006). Epizootic shell disease in American lobster (Homarus americanus) in Massachusetts coastal waters: interactions of temperature, maturity, and intermolt duration. J. Crustac. Biol. 26, 639–645. doi: 10.1651/S-2754.1
Groner, M. L., Shields, J. D., Landers, D. F., Swenarton, J., and Hoenig, J. M. (2018). Rising temperatures, molting phenology, and epizootic shell disease in the American lobster. Am. Nat. 192, 163–177. doi: 10.5061/dryad.n102b1s
Hicks, N., Liu, X., Gregory, R., Kenny, J., Lucaci, A., Lenzi, L., et al. (2018). Temperature driven changes in benthic bacterial diversity influences biogeochemical cycling in coastal sediments. Front. Microbiol. 9:1730. doi: 10.3389/fmicb.2018.01730
Hoenig, J. M., Groner, M. L., Smith, M. W., Vogelbein, W. K., Taylor, D. M., Landers, D. F., et al. (2017). Impact of disease on the survival of three commercially fished species. Ecol. Appl. 27, 2116–2127. doi: 10.1002/eap.1595
Howell, P. (2012). The status of the southern New England lobster stock. J. Shellfish Res. 31, 573–579. doi: 10.2983/035.031.0217
Ishaq, S. L., Turner, S. M., Lee, G., Scarlett Tudor, M., Macrae, J. D., Hamlin, H., et al. (2023). Water temperature and disease alters bacterial diversity and cultivability from American lobster (Homarus americanus) shells. IScience 26:106606. doi: 10.1016/j.isci.2023.106606
Ivanov, I. I., Atarashi, K., Manel, N., Brodie, E. L., Shima, T., Karaoz, U., et al. (2009). Induction of intestinal Th17 cells by segmented filamentous bacteria. Cells 139, 485–498. doi: 10.1016/J.CELL.2009.09.033
Koepper, S., Kelley, S., Thakur, K. K., and Clark, K. F. (2023). Interspecies and spatial differences in the shell microbiome of Atlantic rock crab Cancer irrotatus and European green crab Carcinus maenas from Atlantic Canada. Front. Mar. Sci. 10:1152544. doi: 10.3389/fmars.2023.1152544
Koepper, S., Revie, C. W., Stryhn, H., Clark, K. F., Scott-Tibbetts, S., and Thakur, K. K. (2021). Spatial and temporal patterns in the sex ratio of American lobsters (Homarus americanus) in southwestern Nova Scotia, Canada. Sci. Rep. 11, 24100–24111. doi: 10.1038/s41598-021-03233-8
Kraemer, P., Gerlach, G., and Brinkhoff, T. (2020). Diversity and flexibility of the bacterial communities on Cancer pagurus at different temperatures. J. Sea Res. 156:101827. doi: 10.1016/j.seares.2019.101827
Lane, D. (1991). “16S/23S rRNA sequencing” in Nucleic acid techniques in bacterial systematics (New York, NY: Wiley)
Mathew, M., and Lopanik, N. B. (2014). Host differentially expressed genes during association with its defensive endosymbiont. Biol. Bull. 226, 152–163. doi: 10.1086/BBLv226n2p152
McDonald, D., Price, M. N., Goodrich, J., Nawrocki, E. P., Desantis, T. Z., Probst, A., et al. (2011). An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6, 610–618. doi: 10.1038/ismej.2011.139
Meres, N., Ajuzie, C., Sikaroodi, M., Vemulapalli, M., Shields, J. D., and Gillevet, P. (2012). Dysbiosis in epizootic shell disease of the American lobster (Homarus americanus). J. Shellfish Res. 31, 463–472. doi: 10.2983/035.031.0206
Morrow, K. M., Tedford, A. R., Pankey, M. S., and Lesser, M. P. (2018). A member of the Roseobacter clade, Octadecabacter sp., is the dominant symbiont in the brittle star Amphipholis squamata. FEMS Microbiol. Ecol. 94:30. doi: 10.1093/femsec/fiy030
Nedashkovskaya, O., Kim, S.-G., Stenkova, A. M., and Kukhlevskiy, A. (2018). Aquimarina algiphila sp. nov., a chitin degrading bacterium isolated from the red alga Tichocarpus crinitus. Int. J. Syst. Evol. Microbiol. 68, 892–898. doi: 10.1099/ijsem.0.002606
Oliveros, J. (2007). Venny. an interactive tool for comparing lists with Venn’s diagrams. Available at: https://bioinfogp.cnb.csic.es/tools/venny/index.html.
Ooi, M. C., Goulden, E. F., Smith, G. G., and Bridle, A. R. (2019). Haemolymph microbiome of the cultured spiny lobster Panulirus ornatus at different temperatures. Sci. Rep. 9, 1677–1613. doi: 10.1038/s41598-019-39149-7
Ooi, M. C., Goulden, E. F., Smith, G. G., Nowak, B. F., and Bridle, A. R. (2017). Developmental and gut-related changes to microbiomes of the cultured juvenile spiny lobster Panulirus ornatus. FEMS Microbiol. Ecol. 93:159. doi: 10.1093/femsec/fix159
Paliy, O., Kenche, H., Abernathy, F., and Michail, S. (2009). High-throughput quantitative analysis of the human intestinal microbiota with a phylogenetic microarray. Appl. Environ. Microbiol. 75, 3572–3579. doi: 10.1128/AEM.02764-08
Panda, S., Guarner, F., and Manichanh, C. (2014). Structure and functions of the gut microbiome. Endocr. Metab. Immune Disord. Drug Targets 14, 290–299. doi: 10.2174/1871530314666140714120744
Payne, M. S., Høj, L., Wietz, M., Hall, M. R., Sly, L., and Bourne, D. G. (2008). Microbial diversity of mid-stage Palinurid phyllosoma from great barrier reef waters. J. Appl. Microbiol. 105, 340–350. doi: 10.1111/J.1365-2672.2008.03749.X
Prince, D., and Bayer, R. (2005). “Are all lobsters created equal? Understanding the role of host susceptibility in the development of shell disease in Homarus americanus” in Lobster shell disease workshop (Boston: New England Aquarium), 58–67.
q2galaxy . q2galaxy: An interface for generating Galaxy tool descriptions automatically from QIIME2 actions (2023). Available at: https://github.com/qiime2/q2galaxy.
Quinn, R., Cawthorn, R., Summerfield, R., Smolowitz, R., and Chistoserdov, A. (2013). Bacterial communities associated with lesions of two forms of shell disease in the American lobster (Homarus americanus, Milne Edwards) from Atlantic Canada. Can. J. Microbiol. 59, 380–390. doi: 10.1139/cjm-2012-0679
Quinn, R. A., Hazra, S., Smolowitz, R., and Chistoserdov, A. Y. (2017). Real-time PCR assay for Aquimarina macrocephali subsp. homaria and its distribution in shell disease lesions of Homarus americanus, Milne-Edwards, 1837, and environmental samples. J. Microbiol. Methods 139, 61–67. doi: 10.1016/J.MIMET.2017.04.001
Quinn, R., Metzler, A., Tlusty, M., Smolowitz, R., Leberg, P., and Chistoserdov, A. (2012). Lesion bacterial communities in American lobsters with diet-induced shell disease. Dis. Aquat. Org. 98, 221–233. doi: 10.3354/dao02446
Rajeev, R., Adithya, K. K., Kiran, G. S., and Selvin, J. (2021). Healthy microbiome: a key to successful and sustainable shrimp aquaculture. Rev. Aquac. 13, 238–258. doi: 10.1111/RAQ.12471
Ranson, H. J., LaPorte, J., Spinard, E., Chistoserdov, A. Y., Gomez-Chiarri, M., Nelson, D. R., et al. (2018). Draft genome sequence of the putative marine pathogen Aquimarina sp. strain I32.4. Genome Announc. 6, 7–8. doi: 10.1128/genomeA.00313-18
Raulf, F. F., Fabricius, K., Uthicke, S., de Beer, D., Abed, R. M. M., and Ramette, A. (2015). Changes in microbial communities in coastal sediments along natural CO2 gradients at a volcanic vent in Papua New Guinea. Environ. Microbiol. 17, 3678–3691. doi: 10.1111/1462-2920.12729
Reardon, K. M., Wilson, C. J., Gillevet, P. M., Sikaroodi, M., and Shields, J. D. (2018). Increasing prevalence of epizootic shell disease in American lobster from the nearshore Gulf of Maine. Bull. Mar. Sci. 94, 903–921. doi: 10.5343/bms.2017.1144
Rud, I., Kolarevic, J., Holan, A. B., Berget, I., Calabrese, S., and Terjesen, B. F. (2017). Deep-sequencing of the bacterial microbiota in commercial-scale recirculating and semi-closed aquaculture systems for Atlantic salmon post-smolt production. Aquac. Eng. 78, 50–62. doi: 10.1016/j.aquaeng.2016.10.003
Saraf, V. S., Sheikh, S. A., Ahmad, A., Gillevet, P. M., Bokhari, H., and Javed, S. (2021). Vaginal microbiome: normalcy vs dysbiosis. Arch. Microbiol. 203, 3793–3802. doi: 10.1007/S00203-021-02414-3
Schaubeck, A., Cao, D., and Cavaleri, V. (2023). Carapace microbiota in American lobsters (Homarus americanus) associated with epizootic shell disease and the green gland. Front. Mar. Sci. 14:1093312. doi: 10.3389/fmicb.2023.1093312
Serebryakova, A., Aires, T., Viard, F., Serrão, E. A., and Engelen, A. H. (2018). Summer shifts of bacterial communities associated with the invasive brown seaweed Sargassum muticum are location and tissue dependent. PLoS One 13, e0206734–e0206718. doi: 10.1371/journal.pone.0206734
Shields, J. D. (2013). Complex etiologies of emerging diseases in lobsters (Homarus americanus) from Long Island Sound 1. Can. J. Fish. Aquat. Sci. 70, 1576–1587. doi: 10.1139/cjfas-2013-0050
Silva, S. G., Paula, P., da Silva, J. P., Mil-Homens, D., Teixeira, M. C., Fialho, A. M., et al. (2022). Insights into the antimicrobial activities and metabolomes of Aquimarina (Flavobacteriaceae, Bacteroidetes) species from the rare marine biosphere. Mar. Drugs 20:423. doi: 10.3390/MD20070423/S1
Smolowitz, R., Chistoserdov, A., and Hsu, A. (2005). Epizootic shell disease in the American lobster, Homarus americanus. State of Lobster Science: Lobster Shell Disease Workshop (Boston, MA), 4–11.
Venema, K. (2010). Role of gut microbiota in the control of energy and carbohydrate metabolism. Curr. Opin. Clin. Nutr. Metab. Care 13, 432–438. doi: 10.1097/MCO.0b013e32833a8b60
Wahl, M., Goecke, F., Labes, A., Dobretsov, S., and Weinberger, F. (2012). The second skin: ecological role of epibiotic biofilms on marine organisms. Front. Microbiol. 3:292. doi: 10.3389/fmicb.2012.00292
Keywords: microbial ecology, core microbiome, epizootic shell disease, 16S rRNA gene, PacBio, Aquimarina
Citation: Koepper S, Clark KF, McClure JT, Revie CW, Stryhn H and Thakur KK (2023) Long-read sequencing reveals the shell microbiome of apparently healthy American lobsters Homarus americanus from Atlantic Canada. Front. Microbiol. 14:1245818. doi: 10.3389/fmicb.2023.1245818
Edited by:
Svetlana Yurgel, Agricultural Research Service (USDA), United StatesReviewed by:
Tina Keller-Costa, University of Lisbon, PortugalAndrew Winters, Wayne State University, United States
Copyright © 2023 Koepper, Clark, McClure, Revie, Stryhn and Thakur. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Svenja Koepper, c2tvcHBlckB1cGVpLmNh
 Svenja Koepper
Svenja Koepper K. Fraser Clark
K. Fraser Clark J. Trenton McClure
J. Trenton McClure Crawford W. Revie
Crawford W. Revie Henrik Stryhn1
Henrik Stryhn1 Krishna K. Thakur
Krishna K. Thakur