- 1Jiangsu Co-Innovation Center for Prevention and Control of Important Animal Infectious Diseases and Zoonoses, College of Veterinary Medicine, Yangzhou University, Yangzhou, Jiangsu, China
- 2Institute of Comparative Medicine, Yangzhou University, Yangzhou, Jiangsu, China
- 3Zigong Center for Disease Control and Prevention, Zigong, Sichuan, China
- 4Center for Disease Control and Prevention of Sichuan Province, Chengdu, Sichuan, China
The fourth mobile sulfonamide resistance gene sul4 has been discovered in many metagenomic datasets. However, there is no reports of it in cultured bacteria. In this study, a sul4 positive clinical Salmonella enterica SC2020597 was obtained by conventional Salmonella isolation methods and characterized by species identification and antimicrobial susceptibility testing. Meanwhile, the genomic DNA was sequenced using both long-read and short-read methods. Following that, the complete genome was analyzed by bioinformatic methods. The sul4 gene in S. enterica SC2020597 differed from the sul4 identified in metagenomic data by one amino acid and could confer full resistance to sulfamethoxazole. Genetic location analysis showed that the sul4 in SC2020597 was carried by a complex chromosomally integrated hybrid plasmid. ISCR20-like was strongly associated with the mobilization of sul4 by core genetic context analysis. To the best of our knowledge, this is the first report of the emergence of sul4 in clinically cultured S. enterica. More important, the sul4 has the potential to spread to other bacteria with the help of mobile elements.
Introduction
Sulfonamides are bacteriostatic antimicrobials that inhibit bacterial cellular activity without directly killing the bacteria. They work by interfering with the synthesis of folic acid in bacteria, which is required for the formation of nucleic acids (Skold, 2000; Ovung and Bhattacharyya, 2021). In 1930’s, sulfonamides were firstly introduced for the treatment of human bacterial infections (Skold, 2000; Fernandez-Villa et al., 2019; Nunes et al., 2020). In the years that followed, over 150 sulfonamides and its derivatives were applied in human and veterinary medicine as antibacterial drugs (Baran et al., 2011). Given their extensive utilization, sulfonamide-subsisting bacteria were identified in 2008 for the first time (Dantas et al., 2008), and then many species of bacteria showing sulfonamides resistant were discovered (Deng et al., 2018; Ma et al., 2022). Currently, the most common mechanism of sulfonamide resistance in the majority of bacteria was plasmid-borne, highly mobilized sul1, sul2, and sul3, which encode dihydropteroate synthase (Wang et al., 2014; Nunes et al., 2020; Tang et al., 2022a; Venkatesan et al., 2023). In 2017, the fourth mobile sulfonamide resistance gene sul4 was identified in river sediment with amplicon metagenomic sequencing for the first time (Razavi et al., 2017). To date, many metagenomic analysis have revealed that sul4 is already present in a wide range of environmental samples from around the world (Razavi et al., 2017; Marathe et al., 2019; Hutinel et al., 2022). However, it has not yet been identified in cultured bacteria.
Salmonellosis caused by Salmonella enterica is a common foodborne diseases frequently occurred around the world (Newell et al., 2010; Jajere, 2019; Tang et al., 2022b). It is estimated that salmonellosis could cause 93.8 million foodborne illnesses and 155 thousand people deaths per year (Majowicz et al., 2010; Eng et al., 2015). The World Health Organization (WHO) has listed it as one of the global health concerns. Furthermore, an antibiotic resistance surveillance of foodborne pathogenic bacteria revealed that the prevalence of antibiotic resistance genes (ARGs) in Salmonella was serious, second only to Escherichia coli (Jajere, 2019). Many clinically critical ARGs, such as mcr-1 (Li et al., 2022), blaNDM-5 (Wang et al., 2020) and tet(X4) (Wang et al., 2021), have been identified in S. enterica in recently years. Therefore, the public health risk posed by antibiotics resistant S. enterica has increasingly arisen. In this study, we identified a fourth mobile sulfonamide resistance gene sul4 in clinical S. enterica for the first time, implying that Salmonella could be an important carrier of emerging ARGs.
Materials and methods
Bacterial isolate and antimicrobial susceptibility testing
According to previous method (Xia et al., 2009), the S. enterica SC2020597 was isolated from a hospital in Guangyuan, Sichuan province, China in 2020. Then, pure cultured SC2020597 was identified as S. enterica using matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF-MS) (Bruker, Bremen, Germany). Meanwhile, the species of isolated Salmonella was further confirmed by an online rMLST analysis (Jolley et al., 2018). Subsequently, the minimum inhibitory concentrations (MICs) of SC2020597 was tested by broth microdilution according to Clinical and Laboratory Standards Institute (CLSI) guidelines (https://clsi.org/). E. coli ATCC25922 was used for the quality control.
Conjugation assay and electroporation experiment
In order to investigate the transfer ability of sul4, both conjugation assays and electroporation experiments were performed. For the conjugation assay, we used SC2020597 as donor strains and E. coli C600 as recipients. The donor and recipient strains were cultured into the logarithmic growth phase with an OD600 value of 0.4 in LB broth, then mixed at a ratio of 1:1 and cultured overnight on LB agar plates. The transconjugants were screened on LB agar plates containing rifampin (300 mg/L) and trimethoprim/sulfamethoxazole (4/76 mg/L). For the electroporation experiment, the genomic DNA of SC2020597 was used as donor DNA, and electrocompetent cells of S. enterica ATCC13076 were used as recipients. Electroporation conditions were 200 Ω, 1.8 kV and 25 uF. The transconjugants were screened on LB agar plates containing trimethoprim/sulfamethoxazole (4/76 mg/L). Then, all transconjugants were confirmed by PCR methods targeted at sul4 and 16S rDNA genes.
Genomic DNA extraction, sequencing, and cyclic plasmid detection
The genomic DNA of SC2020597 was extracted using FastPure Bacteria DNA Isolation Mini Kit (Vazyme™, China) following the protocol descripting in the manufacturer. The purity and quality of extracted genomic DNA were evaluated using NanoDrop (Thermo ScientificTM) and using a dsDNA High Sensitivity (HS) Assay kit on the Qubit 4 Fluorometer, respectively. Then, 200 μg genomic DNA was sent to GENEWIZ (Suzhou China) to subject short-read sequencing with PE150 strategy on Illumina Hiseq 2,500 platform. Meanwhile, long-read genomic sequencing of SC2020597 was conducted at Oxford Nanopore Technologies MinION platform in our laboratory. Briefly, the long-read sequencing library was prepared using the SQK-RBK109 1D Rapid Barcoding genomic DNA kit according to the user handbook. Then, the prepared library was sequenced with R9.4 flow cells on MinION and the sequencing process was managed with MinKNOW.
The cyclic plasmid form of the chromosomally integrated plasmid was detected using the inverse PCR method with primers cir_F: TTCAGACGGACTGGACATCG and cir_R: GCGGAATTGTTCAGGGGGTA. The genomic DNA of SC2020597 was used as template DNA. The master mix for application in long fragments was used for PCR amplification reaction system.
Data analysis
The short-read raw reads were filtered to remove low-quality base and adapters using fastp with default parameters (Chen et al., 2018). Then, the complete genome of SC2020597 was generated with a hybrid assembly strategy combining of clean short-read data and long-read data using Unicycler (Wick et al., 2017). Functional annotation of the complete genome was performed by a web-based RAST annotation engine (Aziz et al., 2008; Overbeek et al., 2014; Brettin et al., 2015). Antibiotic resistance genes (ARGs), plasmid replicon genes and insertion sequences (ISs) were identified using abricate tool (https://github.com/tseemann/abricate) based on AMRFinderPlus (Feldgarden et al., 2019), PlasmidFinder (Carattoli et al., 2014) and ISFinder (Siguier et al., 2006) databases, respectively. Multilocus sequence typing (MLST) of the complete bacterial genome was performed using mlst tool (https://github.com/tseemann/mlst). Using Salmonella genomes as input data, the web-based application SISTR was used to identify the Salmonella serovar (Yoshida et al., 2016). Plasmid comparisons and genetic context comparisons visualization were performed with BRIG (Alikhan et al., 2011) and Easyfig (Sullivan et al., 2011) tools.
Functional confirmation of sul4
To confirm the resistance function of the mutated sul4, TA-cloning was performed using a 5 min TA/Blunt-Zero Cloning Kit developed by Vazyme (Vazyme, China). Briefly, the sul4 gene and its predicted promoter were amplified by PCR using primers sul4_F: TGCCTGCAGGTCGACTCTAGAACCCAAAAGTCTGTAGCCCAAA, sul4_R: ACGGCCAGTGAATTGAGCTCTGGTCTAGTICAAAATCGATCATGT, and then cloned into pUC19 vector. Meanwhile, in order to verify the effect of the base mutation on the function of sul4, an unmutated sul4 recombinant expression plasmid was constructed. Subsequently, the recombinant plasmids were introduced chemically into E. coli DH5α. At last, we tested the resistance phenotype of the transconjugants using broth microdilution.
Data availability
The genome sequences of SC2020597 were deposited into the National Center for Biotechnology information (NCBI) under BioProject PRJNA946266.
Results and discussion
Characteristic of the Salmonella enterica isolate SC2020597
The isolate SC2020597 was recovered from a clinical patient. We identified it as S. enterica using MALDI-TOF-MS and confirmed by Ribosomal Multilocus Sequence Typing (rMLST) analysis. MLST analysis showed that isolate SC2020597 belonged to ST26 S. enterica. Serovar analysis classified the isolate as S. enterica subsp. enterica serovar Thompson, which is one of the most frequent Salmonella serovars involved in human infection (Eun et al., 2019). According to previous investigations, the prevalence of S. Thompson in clinical patients and food in China was 3.9 and 5.4%, respectively (Wang et al., 2017; Fan et al., 2020). The serovar has the potential to cause outbreaks of Salmonella infection. Antimicrobial susceptibility testing showed that the isolate was resistant to multiple antibiotics including kanamycin, ampicillin, tetracycline, sulfamethoxazole and trimethoprim/sulfomethoxazole, but sensitive to aztreonam, meropenem, ciprofloxacin, colistin and enrofloxacin (Supplementary Table S1).
Functional analysis of sul4
The draft genome of SC2020597 was obtained by short-read genome assembly. Many ARGs, plasmid replicon genes and ISs were identified in the draft genome. Of note, we found a sul4 gene in isolate SC2020597, which had previously only been found in metagenomes (Razavi et al., 2017; Marathe et al., 2019; Hutinel et al., 2022). The sul4 gene in SC2020597 showed 100% coverage and 99.88% nucleic acid identity to sul4 (NG_056174). The one nucleic acid substitution of sul4 causes one amino acid change (W120R). To verify the function of the novel sul4, the intact sul4 gene and its promoter were cloned into pUC19 vector and introduced into E. coli DH5α. The sul4 positive transconjugants were resistant to trimethoprim/sulfomethoxazole and had a MIC for trimethoprim/sulfomethoxazole that was more than 16-fold higher than E. coli DH5 with an empty vector (≤ 1/19 mg/L to >32/608 mg/L). Meanwhile, unmutated sul4-bearing E. coli DH5 also showed full resistance to trimethoprim/sulfomethoxazole (>32/608 mg/L). This demonstrated that W120R substitution had no effect to the function of sul4.
Genomic feature of SC2020597
To decipher the genomic structure feature of SC2020597, long-read sequencing was performed. Then, the complete genome of SC2020597 was generated by a hybrid assembly strategy using short-read and long-read data. The isolate harbored one chromosome with a length of 5, 035, 375 bp and two plasmids, pSC2020597_48k with a length of 48, 530 bp, and pSC2020597_5k with a length of 5, 754 bp. Plasmid replicon analysis showed that pSC2020597_48k was an IncFII type plasmid and pSC2020597_5k was a Col type small plasmid. Of note, no ARG was found in the two plasmids. Online blastn analysis showed that many plasmids from Salmonella were similar to pSC2020597_48k, indicating that such plasmids were common in Salmonella. Plasmid pSC2020597_5k belonged to a group of small plasmids with a broad host range that were widely distributed in Enterobacteriaceae.
A total of 22 ARGs, including sul4, were detected in SC2020597, and all of them were located on chromosome (Supplementary Table S2). In addition, three plasmid replicon genes were discovered on the chromosome of SC2020597. Further analysis found that a multi-drugs resistant plasmid was integrated into chromosome of SC2020597. The chromosomally integrated plasmid was 330, 232 bp in length and was designated as PSC2020597-sul4-330 k-c (Figure 1A). The phenomenon that plasmid was integrated into chromosome have been frequently reported (Muthuirulandi Sethuvel et al., 2021; Shigemura et al., 2021; Chang et al., 2022), especially in Salmonella, and is commonly mediated by homologous recombination of ISs. We found that PSC2020597-sul4-330 k-c was flanked by IS1F on chromosome of SC2020597. Meanwhile, a 7 bp direct repeat sequence was found around PSC2020597-sul4-330 k-c (Figure 1B). These findings provided strong evidences that PSC2020597-sul4-330 k-c was integrated into chromosome via the homologous recombination of IS1R. The precursor of PSC2020597-sul4-330 k-c was a hybrid plasmid with complex structure. Half of PSC2020597-sul4-330 k-c was composed by a typical IncHI2/HI2A plasmid. Another half was made up of flexible genetic arrays. Blastn analysis with NCBI nr database showed that PSC2020597-sul4-330 k-c was most closely related to pSIn_quan12 (GenBank: ON960352.1), which had 66% coverage and 99.98% identity to PSC2020597-sul4-330 k-c (Figure 1A). Meanwhile, many other plasmids similar to PSC2020597-sul4-330 k-c were also found in nr database, and the complete structure of PSC2020597-sul4-330 k-c could almost be covered by the genetic array of these plasmids (Figure 1A). Hence, PSC2020597-sul4-330 k-c was most likely formed through the recombination of diversity of plasmids and then integrated into chromosome. The discovery of sul4 in a complex chromosomally integrated plasmid implied that sul4 gene had already appeared and spread in plasmids. We should take notice to monitor the spread of sul4 by plasmids or other mobile genetic elements.
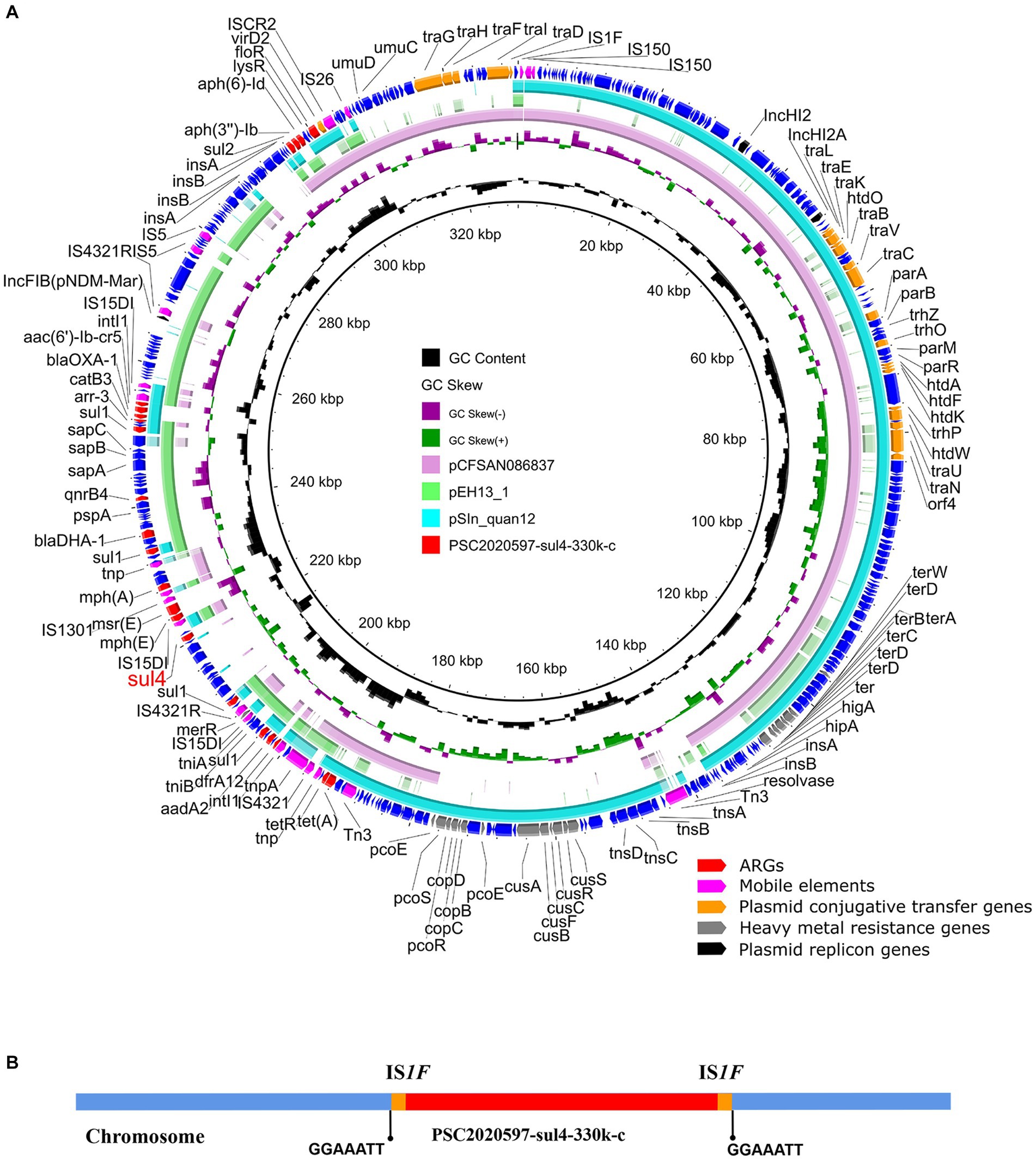
Figure 1. Structure analysis of sul4-bearing chromosomally integrated plasmid in Salmonella enterica. (A) Comparative analysis of chromosomally integrated plasmid PSC2020597-sul4-330 k-c in this study with other similar plasmids including pSIn_quan12 (ON960352.1), pEH13_1 (CP089098.1), and pCFSAN086837 (CP039438.1). (B) The integration sites of chromosomal plasmids PSC2020597-sul4-330 k-c.
The transfer ability of sul4
By the genetic analysis, we found that sul4 was located on a chromosomally integrated plasmid. However, a previous study demonstrated that chromosomally integrated plasmids could also be transferred to other recipients by conjugation assay (Chang et al., 2022). Here, we verified that there is a circle form of chromosomally integrated plasmid PSC2020597-sul4-330 k-c in some clones of SC2020597 using the inverse PCR method and confirmed by Sanger sequencing (Supplementary Figure S1). This phenomenon implied that the chromosomally integrated plasmid PSC2020597-sul4-330 k-c had potential to horizontal transfer by conjugation. Subsequently, we used E. coli C600 as recipients to test the transfer of sul4 by conjugation assay. The sul4 gene could not be transferred into E. coli C600 after multiple tries. Then, we attempted to verify the transfer ability of sul4 using electroporation experiments with S. enterica ATCC13076 as recipients. The sul4-positive S. enterica ATCC13076 was successfully screened and showed resistance to trimethoprim/sulfomethoxazole.
The core genetic structure of sul4
Mobile elements played a key role in the dissemination of ARGs (Partridge et al., 2018). Previous research discovered an ISCR family transposase ISCR20-like (GneBank: MG649402.1) downstream of sul4 that could be mobilized along with their adjacent genes via rolling-circle transposition without the assistance of any other transposase protein (Razavi et al., 2017). Therefore, sul4-ISCR20-like was considered to be mobilizable as an entire integrin (Razavi et al., 2017). Subsequently, we investigated the core genetic structure of sul4 in PSC2020597-sul4-330 k-c. We also found an ISCR20-like in the downstream of sul4, which was consistent with previous structure of sul4 in metagenome (GneBank: MG649402.1) (Figure 2). In addition, other sul4-bearing genetic contexts associated with ISCR20-like or truncated ISCR20-like were also found in other bacterial chromosome or plasmid in NCBI nr database (Figure 2). These findings demonstrated that the dissemination of sul4 was probably driven by ISCR20-like. Although the transfer of sul4-ISCR20-like has not been verified by experiment, other similar transfer events have been confirmed, such as tet(X4)-ISCR2 (He et al., 2019). Hence, the mobilization of sul4-ISCR20-like was likely to happen and promoted the dissemination of sul4. Of note, sul4 was found to be integrated into a class 1 integron in the plasmid of Aeromonas sp. FDAARGOS 1402 (Figure 2). It means that novel mobile genetic structure harboring sul4 is emerging, which may accelerate the propagation of sul4. Therefore, we should take more attention to the surveillance of sul4 in order to slow its spread.
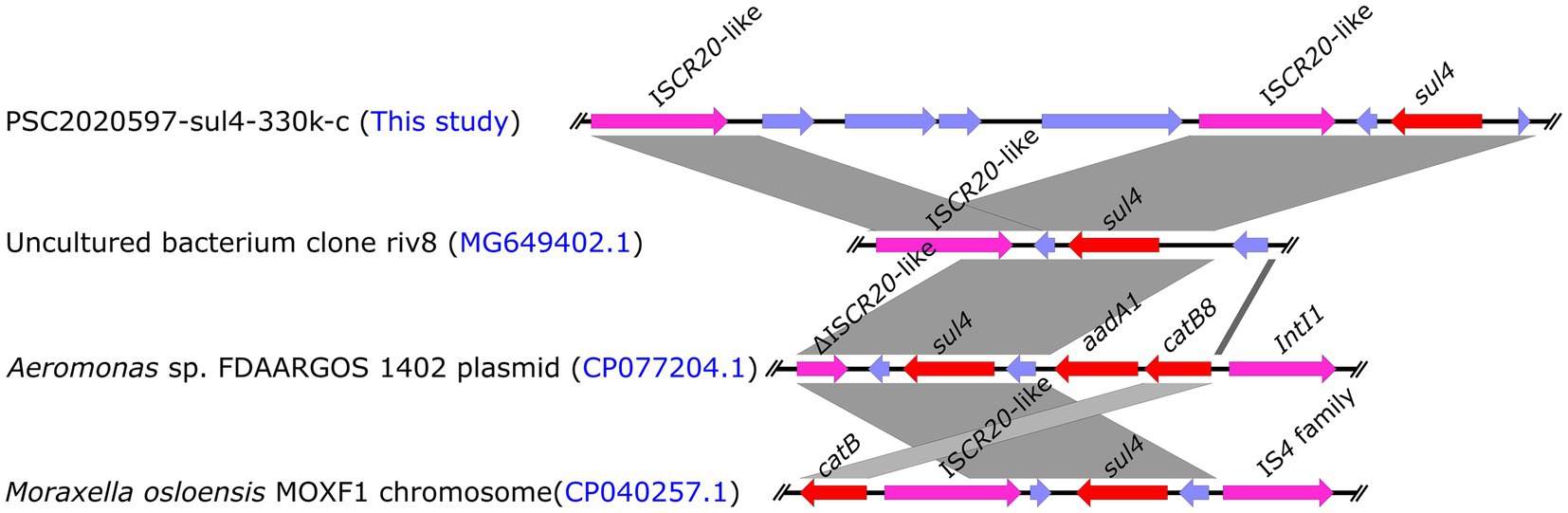
Figure 2. Linear comparison of the core genetic structures of sul4 gene. Comparative analysis of the genetic context of sul4 in PSC2020597-sul4-330 k-c with the genetic contexts of sul4 in uncultured riv8 (MG649402.1), isolates FDAARGOS 1402 (CP077204.1), and MOXF1 (CP040257.1).
Conclusion
To our best knowledge, this is the first report of the emergence of sul4 in clinically cultured S. enterica. The genetic location analysis showed that sul4 had already emerged in common and continually evolving hybrid plasmids of Enterobacteriaceae. Meanwhile, we found that ISCR20-like played an important role in the spread of sul4. These findings demonstrated that sul4 has the potential to be prevalent in various bacteria, and further reduce the effect of sulfonamide.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA946266.
Author contributions
KP, JD, and NZ: conception and design. KP, JD, NZ, and XS: methodology. WH: collection and assembly of data. KP and WH: data analysis and interpretation. KP: writing—original draft. RL and XY: writing—reviewing and editing. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Sichuan Science and Technology Program (2022ZDZX0017), Postgraduate Research & Practice Innovation Program of Jiangsu Province (SJCX21_1631) and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1242369/full#supplementary-material
References
Alikhan, N. F., Petty, N. K., Ben Zakour, N. L., and Beatson, S. A. (2011). BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402
Aziz, R. K., Bartels, D., Best, A. A., Dejongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genom. 9:75. doi: 10.1186/1471-2164-9-75
Baran, W., Adamek, E., Ziemianska, J., and Sobczak, A. (2011). Effects of the presence of sulfonamides in the environment and their influence on human health. J. Hazard. Mater. 196, 1–15. doi: 10.1016/j.jhazmat.2011.08.082
Brettin, T., Davis, J. J., Disz, T., Edwards, R. A., Gerdes, S., Olsen, G. J., et al. (2015). RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 5:8365. doi: 10.1038/srep08365
Carattoli, A., Zankari, E., Garcia-Fernandez, A., Larsen, M. V., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Chang, M. X., Zhang, J., Zhang, J. F., Ding, X. M., Lu, Y., Zhang, J., et al. (2022). Formation, transmission, and dynamic evolution of a multidrug-resistant chromosomally integrated plasmid in Salmonella Spp. Front. Microbiol. 13:846954. doi: 10.3389/fmicb.2022.846954
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Dantas, G., Sommer, M. O., Oluwasegun, R. D., and Church, G. M. (2008). Bacteria subsisting on antibiotics. Science 320, 100–103. doi: 10.1126/science.1155157
Deng, Y., Li, B., and Zhang, T. (2018). Bacteria that make a meal of sulfonamide antibiotics: blind spots and emerging opportunities. Environ. Sci. Technol. 52, 3854–3868. doi: 10.1021/acs.est.7b06026
Eng, S. K., Pusparajah, P., Ab Mutalib, N. S., Ser, H. L., Chan, K. G., and Lee, L. H. (2015). Salmonella: A review on pathogenesis, epidemiology and antibiotic resistance. Front. Life Sci. 8, 284–293. doi: 10.1080/21553769.2015.1051243
Eun, Y., Jeong, H., Kim, S., Park, W., Ahn, B., Kim, D., et al. (2019). A large outbreak of Salmonella enterica serovar Thompson infections associated with chocolate cake in Busan Korea. Epidemiol. Health 41:e2019002. doi: 10.4178/epih.e2019002
Fan, J., Zhang, L., He, J., Zhao, M., Loh, B., Leptihn, S., et al. (2020). Plasmid dynamics of mcr-1-positive Salmonella spp. in a general Hospital in China. Front. Microbiol. 11:604710. doi: 10.3389/fmicb.2020.604710
Feldgarden, M., Brover, V., Haft, D. H., Prasad, A. B., Slotta, D. J., Tolstoy, I., et al. (2019). Validating the AMRFinder tool and resistance gene database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrob. Agents Chemother. 63:e00483-19. doi: 10.1128/AAC.00483-19
Fernandez-Villa, D., Aguilar, M. R., and Rojo, L. (2019). Folic acid antagonists: antimicrobial and Immunomodulating mechanisms and applications. Int. J. Mol. Sci. 20:4996. doi: 10.3390/ijms20204996
He, T., Wang, R., Liu, D., Walsh, T. R., Zhang, R., Lv, Y., et al. (2019). Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat. Microbiol. 4, 1450–1456. doi: 10.1038/s41564-019-0445-2
Hutinel, M., Larsson, D. G. J., and Flach, C. F. (2022). Antibiotic resistance genes of emerging concern in municipal and hospital wastewater from a major Swedish city. Sci. Total Environ. 812:151433. doi: 10.1016/j.scitotenv.2021.151433
Jajere, S. M. (2019). A review of Salmonella enterica with particular focus on the pathogenicity and virulence factors, host specificity and antimicrobial resistance including multidrug resistance. Vet World 12, 504–521. doi: 10.14202/vetworld.2019.504-521
Jolley, K. A., Bray, J. E., and Maiden, M. C. J. (2018). Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 3:124. doi: 10.12688/wellcomeopenres.14826.1
Li, R., Peng, K., Huang, W., Sun, X., Huang, Y., Lei, G., et al. (2022). The genomic epidemiology of mcr-positive Salmonella enterica in clinical patients from 2014 to 2017 in Sichuan, China and global epidemiological features. J. Infect. 85, 702–769. doi: 10.1016/j.jinf.2022.08.042
Ma, J., Zhou, W., Wu, J., Liu, X., Lin, J., Ji, X., et al. (2022). Large-scale studies on antimicrobial resistance and molecular characterization of Escherichia coli from food animals in developed areas of eastern China. Microbiol. Spectr. 10:e0201522. doi: 10.1128/spectrum.02015-22
Majowicz, S. E., Musto, J., Scallan, E., Angulo, F. J., Kirk, M., O'brien, S. J., et al. (2010). The global burden of Nontyphoidal Salmonella gastroenteritis. Clin. Infect. Dis. 50, 882–889. doi: 10.1086/650733
Marathe, N. P., Berglund, F., Razavi, M., Pal, C., Droge, J., Samant, S., et al. (2019). Sewage effluent from an Indian hospital harbors novel carbapenemases and integron-borne antibiotic resistance genes. Microbiome 7:97. doi: 10.1186/s40168-019-0710-x
Muthuirulandi Sethuvel, D. P., Anandan, S., Murugan, D., Asokan, K., Vasudevan, K., Jacob, J. J., et al. (2021). Hybrid genome assembly of Shigella sonnei reveals the novel finding of chromosomal integration of an IncFII plasmid carrying a mphA gene. Access Microbiol. 3:000189
Newell, D. G., Koopmans, M., Verhoef, L., Duizer, E., Aidara-Kane, A., Sprong, H., et al. (2010). Food-borne diseases - the challenges of 20 years ago still persist while new ones continue to emerge. Int. J. Food Microbiol. 139, S3–S15. doi: 10.1016/j.ijfoodmicro.2010.01.021
Nunes, O. C., Manaia, C. M., Kolvenbach, B. A., and Corvini, P. F. (2020). Living with sulfonamides: a diverse range of mechanisms observed in bacteria. Appl. Microbiol. Biotechnol. 104, 10389–10408. doi: 10.1007/s00253-020-10982-5
Overbeek, R., Olson, R., Pusch, G. D., Olsen, G. J., Davis, J. J., Disz, T., et al. (2014). The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res. 42, D206–D214. doi: 10.1093/nar/gkt1226
Ovung, A., and Bhattacharyya, J. (2021). Sulfonamide drugs: structure, antibacterial property, toxicity, and biophysical interactions. Biophys. Rev. 13, 259–272. doi: 10.1007/s12551-021-00795-9
Partridge, S. R., Kwong, S. M., Firth, N., and Jensen, S. O. (2018). Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 31:e00088-17. doi: 10.1128/CMR.00088-17
Razavi, M., Marathe, N. P., Gillings, M. R., Flach, C. F., Kristiansson, E., and Joakim Larsson, D. G. (2017). Discovery of the fourth mobile sulfonamide resistance gene. Microbiome 5:160. doi: 10.1186/s40168-017-0379-y
Shigemura, H., Maeda, T., Nakayama, S., Ohishi, A., Carle, Y., Ookuma, E., et al. (2021). Transmission of extended-spectrum cephalosporin-resistant Salmonella harboring a blaCMY-2-carrying IncA/C2 plasmid chromosomally integrated by ISEcp1 or IS26 in layer breeding chains in Japan. J. Vet. Med. Sci. 83, 1345–1355. doi: 10.1292/jvms.21-0085
Siguier, P., Perochon, J., Lestrade, L., Mahillon, J., and Chandler, M. (2006). ISfinder: the reference Centre for bacterial insertion sequences. Nucleic Acids Res. 34, D32–D36. doi: 10.1093/nar/gkj014
Skold, O. (2000). Sulfonamide resistance: mechanisms and trends. Drug Resist. Updat. 3, 155–160. doi: 10.1054/drup.2000.0146
Sullivan, M. J., Petty, N. K., and Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Tang, B., Chang, J., Chen, Y., Lin, J., Xiao, X., Xia, X., et al. (2022a). Escherichia fergusonii, an underrated repository for antimicrobial resistance in food animals. Microbiol. Spectr. 10:e0161721. doi: 10.1128/spectrum.01617-21
Tang, B., Elbediwi, M., Nambiar, R. B., Yang, H., Lin, J., and Yue, M. (2022b). Genomic characterization of antimicrobial-resistant Salmonella enterica in duck, chicken, and pig farms and retail Markets in Eastern China. Microbiol Spectr 10:e0125722. doi: 10.1128/spectrum.01257-22
Venkatesan, M., Fruci, M., Verellen, L. A., Skarina, T., Mesa, N., Flick, R., et al. (2023). Molecular mechanism of plasmid-borne resistance to sulfonamide antibiotics. Nat. Commun. 14:4031. doi: 10.1038/s41467-023-39778-7
Wang, Y., Cao, C., Alali, W. Q., Cui, S., Li, F., Zhu, J., et al. (2017). Distribution and antimicrobial susceptibility of foodborne Salmonella Serovars in eight provinces in China from 2007 to 2012 (except 2009). Foodborne Pathog. Dis. 14, 393–399. doi: 10.1089/fpd.2016.2237
Wang, Z., He, J., Li, Q., Tang, Y., Wang, J., Pan, Z., et al. (2020). First detection of NDM-5-positive Salmonella enterica Serovar typhimurium isolated from retail pork in China. Microb. Drug Resist. 26, 434–437. doi: 10.1089/mdr.2019.0323
Wang, Y. N., Liu, F., Xu, X. B., Huang, H., Lyu, N., Ma, S. F., et al. (2021). Detection of plasmid-mediated Tigecycline resistance gene tet(X4) in a Salmonella enterica Serovar Llandoff isolate. Infect. Microb. Dis. 3, 198–204. doi: 10.1097/IM9.0000000000000077
Wang, N., Yang, X., Jiao, S., Zhang, J., Ye, B., and Gao, S. (2014). Sulfonamide-resistant bacteria and their resistance genes in soils fertilized with manures from Jiangsu Province Southeastern China. PLoS One 9:e112626. doi: 10.1371/journal.pone.0112626
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595
Xia, S., Hendriksen, R. S., Xie, Z., Huang, L., Zhang, J., Guo, W., et al. (2009). Molecular characterization and antimicrobial susceptibility of Salmonella isolates from infections in humans in Henan Province, China. J. Clin. Microbiol. 47, 401–409. doi: 10.1128/JCM.01099-08
Yoshida, C. E., Kruczkiewicz, P., Laing, C. R., Lingohr, E. J., Gannon, V. P., Nash, J. H., et al. (2016). The Salmonella in silico typing resource (SISTR): an open web-accessible tool for rapidly typing and subtyping draft Salmonella genome assemblies. PLoS One 11:e0147101. doi: 10.1371/journal.pone.0147101
Keywords: sulfonamide resistance, sul4, chromosomally integrated plasmid, Salmonella enterica, clinical
Citation: Peng K, Deng J, Zou N, Sun X, Huang W, Li R and Yang X (2023) Emergence of the fourth mobile sulfonamide resistance gene sul4 in clinical Salmonella enterica. Front. Microbiol. 14:1242369. doi: 10.3389/fmicb.2023.1242369
Edited by:
Biao Tang, Zhejiang Academy of Agricultural Sciences, ChinaReviewed by:
Chang-Wei Lei, Sichuan University, ChinaLili Zhang, Jiangsu Academy of Agricultural Sciences (JAAS), China
Bao-Tao Liu, Qingdao Agricultural University, China
Copyright © 2023 Peng, Deng, Zou, Sun, Huang, Li and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ruichao Li, cmNobDg4QHl6dS5lZHUuY24=; Xiaorong Yang, eWFuZ3lhbmd4ckAxNjMuY29t
†These authors have contributed equally to this work
 Kai Peng
Kai Peng Jianping Deng3†
Jianping Deng3† Ruichao Li
Ruichao Li Xiaorong Yang
Xiaorong Yang