- 1School of Biological Engineering, Henan University of Technology, Zhengzhou, Henan, China
- 2College of International Education, Henan University of Technology, Zhengzhou, Henan, China
Introduction: The biological contact oxidation reactor is an effective technology for the treatment of antibiotic wastewater, but there has been little research investigating its performance on the sulfamethazine wastewater treatment.
Methods: In this study, a novel two-stage biological contact oxidation reactor was used for the first time to explore the impact of sulfamethazine (SMZ) on the performance, microbial community, extracellular polymeric substances (EPS), and antibiotic-resistant genes (ARGs).
Results: The chemical oxygen demand (COD) and ammonia nitrogen (-N) removal efficiencies kept stable at 86.93% and 83.97% with 0.1–1 mg/L SMZ addition and were inhibited at 3 mg/L SMZ. The presence of SMZ could affect the production and chemical composition of EPS in the biofilm, especially for the pronounced increase in TB-PN yield in response against the threat of SMZ. Metagenomics sequencing demonstrated that SMZ could impact on the microbial community, a high abundance of Candidatus_Promineofilum, unclassified_c__Anaerolineae, and unclassified_c__Betaproteobacteria were positively correlated to SMZ, especially for Candidatus_Promineofilum.
Discussion: Candidatus_Promineofilum not only had the ability of EPS secretion, but also was significantly associated with the primary SMZ resistance genes of sul1 and sul2, which developed resistance against SMZ pressure through the mechanism of targeted gene changes, further provided a useful and easy-implement technology for sulfamethazine wastewater treatment.
1. Introduction
Antibiotics are widely used due to their good antibacterial or bactericidal effects (Harada, 2018), which are widespread in sewage treatment plants, sewers, landfill leachate, river runoff, and waste discharge from livestock farms (Zhang et al., 2022b). The widespread detection of antibiotic residues in various environments will accelerate the spread of antibiotic-resistant bacteria (ARB) and ARGs, further altering the microbial community and threatening ecological safety and human health (Bai et al., 2016; Ben et al., 2018). Among the various types of antibiotics, sulfonamides are detected with substantial frequency in the environment due to their broad antibacterial activity and high potency at a low cost (Acosta-Rangel et al., 2019). In particular, the typical short-acting sulfamethazine (SMZ) was considered the most emerging contaminant among other sulfur groups containing medicine, which was most frequently used in inhibiting the growth of gram-positive bacteria due to its two ionizable functional groups: the aniline amine and the amide moieties (Gao and Pedersen, 2005; Du et al., 2018). The levels of SMZ in the environment ranged from ng/L to μg/L (Li et al., 2019). Therefore, it is urgent to develop efficient approaches for removing SMZ from the environment (Fu et al., 2020).
In recent years, catalytic ozonation (Bai et al., 2016), Fenton (Zhuang and Wang, 2021), and photodegradation (Ouyang et al., 2020) have been used for SMZ wastewater treatment. However, it is limited due to large secondary pollution and economic costs. Our previous studies have confirmed that the biological contact oxidation reactor (BCOR) is a promising technology for the treatment of chloramphenicol wastewater due to its structural setup (Zhou et al., 2022), which is beneficial to the separation of microorganisms and diminishes interspecific competition, further removing the antibiotics step by step upon direct contact with wastewater. In this process, the microbial community in activated sludge played a crucial role in improving the performance of biotechnology in removing antibiotics from wastewater, although the antibiotics may cause stress to the microbes or even alter their community structure (Mulla et al., 2021). Furthermore, a shift in the associated microbial communities may disrupt the stability and performance of the reactor (Strous et al., 2006).
Previous studies have suggested that microbial resistance to SMZ may be related to the secretion of EPS (Zhang et al., 2019). EPS is not only an important reservoir for antibiotics but also contains active enzymes that can remove antibiotics, which was a response to the antibiotic stress to protect themselves and adhere to each other. Meanwhile, EPS also influenced the physicochemical and biological functions of activated sludge, especially for its main structures of tightly bound EPS (TB-EPS) and loosely bound EPS (LB-EPS) (Yu et al., 2023). As the key component of EPS (Wu et al., 2022), PN can transport small molecules and further facilitate the adsorption of antibiotics from wastewater through the amino and carboxyl groups (Song et al., 2023). These findings suggest that EPS might play an important role in the removal of SAs. However, less attention has been focused on investigating the effect of EPS on SMZ removal, especially the evolvement action roles of microorganisms and EPS against SMZ shocks during the SMZ wastewater treatment using BCOR, which is currently unclear.
In this study, a novel two-stage biological contact oxidation reactor was constructed to (1) investigate the long-term impacts of increasing SMZ stress on the performance; (2) explore the production and chemical composition of EPS produced by the activated sludge against SMZ shocks; and (3) reveal the shift of the microbial community and the abundance of ARGs with the gradient concentrations of SMZ by the metagenomics sequencing technology. The results highlight the crucial roles of microbial EPS in responding to different concentrations of SMZ shocks during biological wastewater treatment.
2. Materials and methods
2.1. Experimental setup
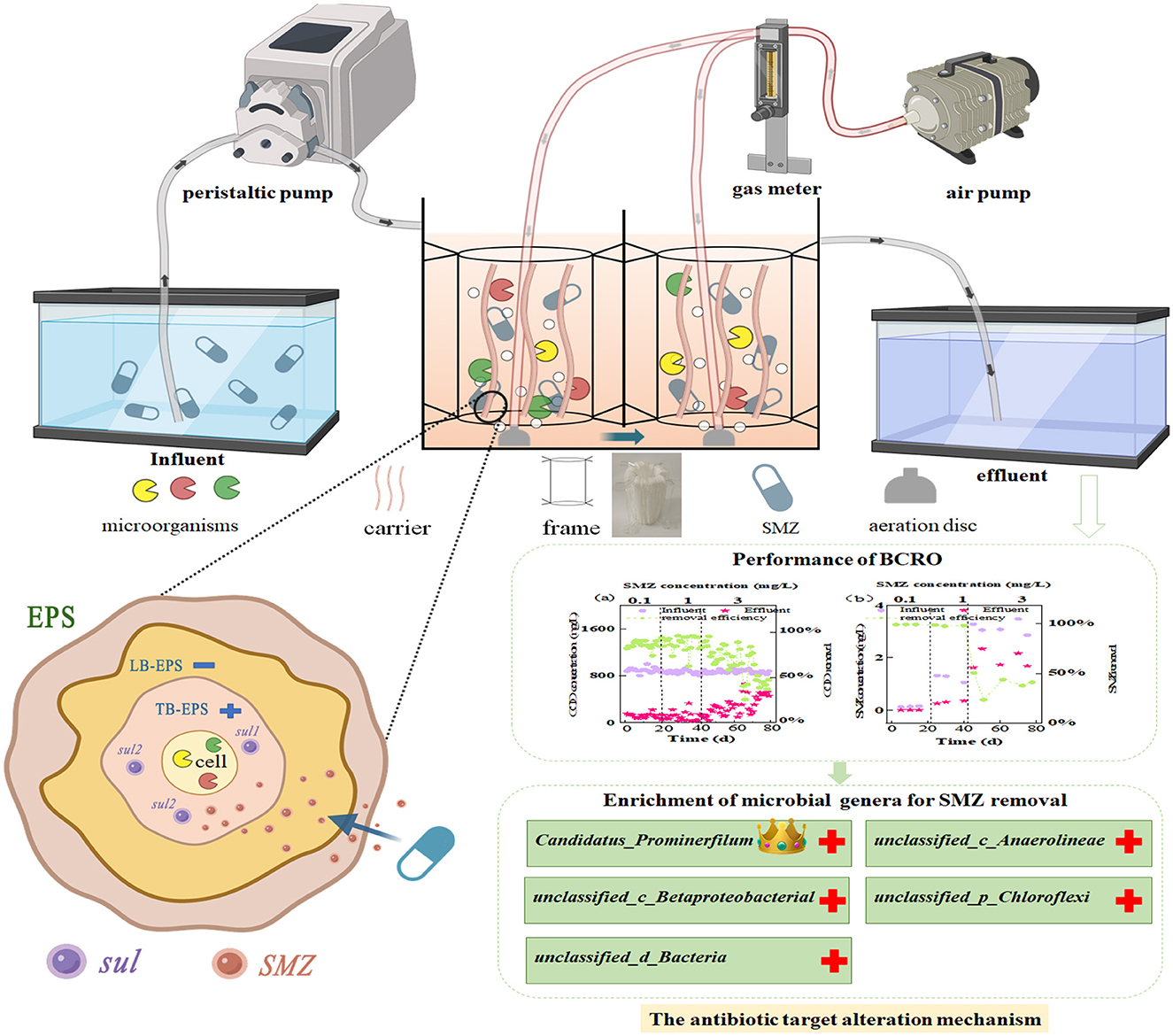
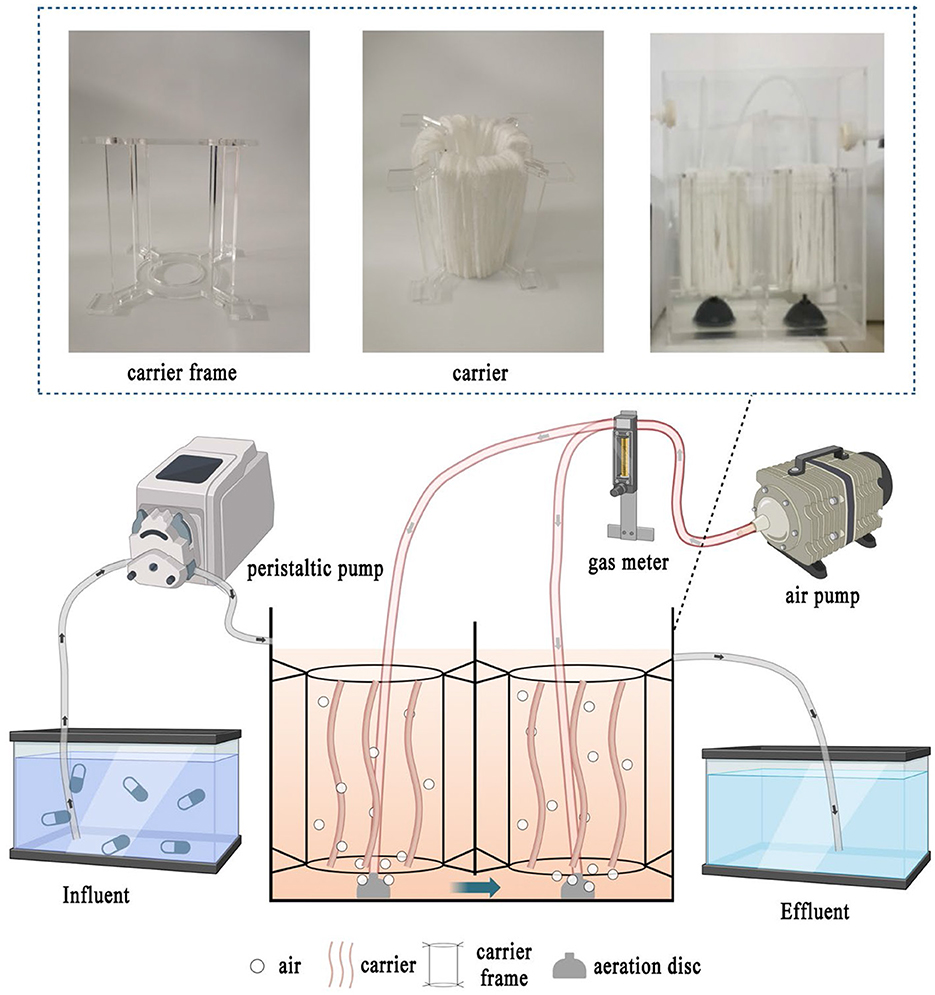
A two-stage biological contact oxidation reactor with an effective volume of ~6 L was set up in Figure 1, which is composed of two identical rectangular chambers (125 × 125 × 200 mm). The influent was pumped (BT101s, Lead Fluid, China) into the inlet and flowed to the outlet downward and upwardly through the overflow ports on both sides of the bottom in the middle baffle of the two chambers. The microporous aeration disc (LZB-3WB, Xiangjin, China) was fixed at the bottom of each chamber with dissolved oxygen (DO) of 3–4 mg/L. The acrylic zigzag circle frame with top and bottom inner diameters of 50 and 60 mm was suspended and fixed at 50–100 mm above the aeration disc in each chamber through its four serrations. Furthermore, the mixed polyethylene carrier (Teijin, Japan), which has a compact and porous fiber structure with a filling capacity yield of 3.5 g/L carriers, was wrapped around the zigzag circle frame to serve as a filter that absorbed the activated sludge and formed an efficient biofilm.
2.2. Biofilm culturing
Seed sludge was gathered from the secondary sedimentation tank of the Wulongkou Municipal Wastewater Treatment Plant (MWTP). The original mixed liquor-suspended solids (MLSS) of the two chambers were maintained at ~4.0 g/L. After aerating for 3 days, the sludge was almost adsorbed onto the carrier surface, and the liquid was clarified. Afterward, the microorganisms were inhabited at a high density in the activated sludge, which is beneficial to forming a biofilm and realizing the selective distribution and natural differentiation of microorganisms in space and function through synergy and antagonism.
2.3. Wastewater components
Initially, 0.05 g of standard SMZ was dissolved in a small amount of 50 mL of acetonitrile (HPLC grade) and then stored in amber-colored vials at −20°C. After the completion of biofilm incubation, the synthetic SMZ wastewater was prepared based on the previous study (Shi et al., 2018; Wan et al., 2018), with the main composition of CH3COONa 1.15 g/L, NH4Cl 0.34 g/L, K2HPO4 0.10 g/L, and trace elements of 1 mL/L (1.50 g/L FeCl3·6H2O, 0.15 g/L H3BO3, 0.03 g/L CuSO4·5H2O, 0.12 g/L MnCl4·H2O, 0.06 g/L NaMoO4·2H2O, 0.12 g/L ZnSO4·7H2O, 0.15 g/L CoCl2·6H2O). It was pumped into the reactor continuously at room temperature (25–30°C) for 80 days. The flow rate of the peristaltic pump was 4.25 mL/min to maintain the hydraulic retention time (HRT) of 24 h. The operation process could be generally divided into three phases based on SMZ shocks in the influent, which gradually increased from 0.10 to 1 mg/L and 3 mg/L, according to previous studies (Yang et al., 2015; Li et al., 2019).
2.4. Analysis of a wastewater sample
The influent and effluent samples were collected daily and centrifuged at 10,000 rpm for 10 min, and the supernatant was taken to determine COD, -N, and levels as previously described (Zhou et al., 2022). The DO and pH values were measured using a DO meter (YSI 550A, USA) and a pH meter (HQ30d, HACH, America). The supernatant samples were filtrated using a 0.22-μm filter membrane (PTFE) for SMZ quantification using an analytical liquid chromatograph (LC-2030C, Shimadzu, Japan), equipped with the SB-C18 chromatographic column (Agilent ZORBAX, 150 × 4.6 mm, 5 μm) at a detection wavelength of 270 nm, an injection volume of 20 μL, and a flow rate of 1 mL/min. A combination of two mobile stages was programmed with pure water (A) and acetonitrile (B), and the mobile stage elution gradient was 0–2 min 60% A, 2–4 min 70% A, and 4–6 min 75% A.
2.5. EPS extraction and component analysis
The seed sludge (CK) biofilm samples obtained from the carrier in the reactor at the end of stages 2 (Z1) and 3 (Z3) were used to extract the EPS using thermal extraction. First, 25 mL of the sludge mixture was centrifuged at 4,000 r/min for 5 min at 4°C to remove the supernatant. Then, the sludge was replenished to its original volume with PBS buffer (NaCl 8.5%, Na2HPO4 2.2%, NaH2PO4 0.4%, distilled water 100 mL, pH 7), heated to 70°C, mixed well, and centrifuged at 4,000 r/min for 10 min at 4°C to obtain the supernatant (LB-EPS). The supernatant was then supplemented with PBS buffer to its original volume and heated in a water bath at 70°C for 30 min. Then, it was centrifuged at 4,000 r/min for 15 min at 4°C to obtain the supernatant (TB-EPS). Afterward, solutions of LB-EPS and TB-EPS were filtered through a 0.45-μm membrane.
The protein (PN) and polysaccharide (PS) concentrations were determined using the Bradford method and the phenol-sulfuric acid method (Peng et al., 2022). The organic compositions and fluorescence characteristics in EPS were determined using three-dimensional excitation and emission matrix fluorescence (3D-EEM). The excitation wavelength (Ex) was scanned from 200 to 400 nm, and the emission wavelength (Em) was scanned from 300 to 500 nm, both in 5 nm increments (Zhang et al., 2022a).
2.6. Metagenomics sequencing
The seed sludge (CK) and the biofilm sludge samples collected at the end of stages 2 (Z1) and 3 (Z3) were used for metagenomics analysis on an Illumina HiSeq 4000 platform (Majorbio, China). The raw reads were used to generate clean reads using fastp (Chen et al., 2018) on the Majorbio Cloud Platform (cloud.majorbio.com). A non-redundant gene catalog was constructed using CD-HIT (Fu et al., 2012). The species abundance was calculated from the sum of gene abundances corresponding to the species. Representative sequences from the non-redundant gene catalog were annotated by Blastp based on the NCBI NR database, using DIAMOND for taxonomic annotation with an e-value cutoff of 1e−5. Antibiotic resistance gene annotation was conducted using DIAMOND (Buchfink et al., 2015) against the CARD database with an e-value cutoff of 1e−5. Network analysis was used to explore the potential correlations between bacteria and ARGs based on Spearman's correlation coefficient > 0.6 (P < 0.05). The data were deposited into the NCBI Sequence Read Archive (SRA) database under accession number PRJNA838398.
3. Results
3.1. Reactor performance
The performance of the two-stage biological contact oxidation reactor for the treatment of SMZ wastewater is shown in Figure 2. The SMZ shocks in stages 1–2 barely influenced the performance of BCOR in COD removal (Figure 2A), and the average COD removal efficiency was 86.93 and 88.04%, respectively. However, the obvious fluctuation of COD removal efficiency appeared in stage 3, and it reduced to 66.39%, mainly attributed to the increasing SMZ stress. In addition, NH4+-N removal efficiency kept relatively stable at 83.87–90.03% when the SMZ concentration increased from 0.10 to 1 mg/L (Figure 2B), indicating that the nitrifying bacteria still had strong activity even under the increasing SMZ pressure. However, the NH4+-N removal efficiency illustrated a decreasing tendency when the SMZ concentration in the influent increased to 3 mg/L. The high SMZ concentration (3 mg/L) in the present research could impact the removal of organic matter and the process of ammonia oxidization.
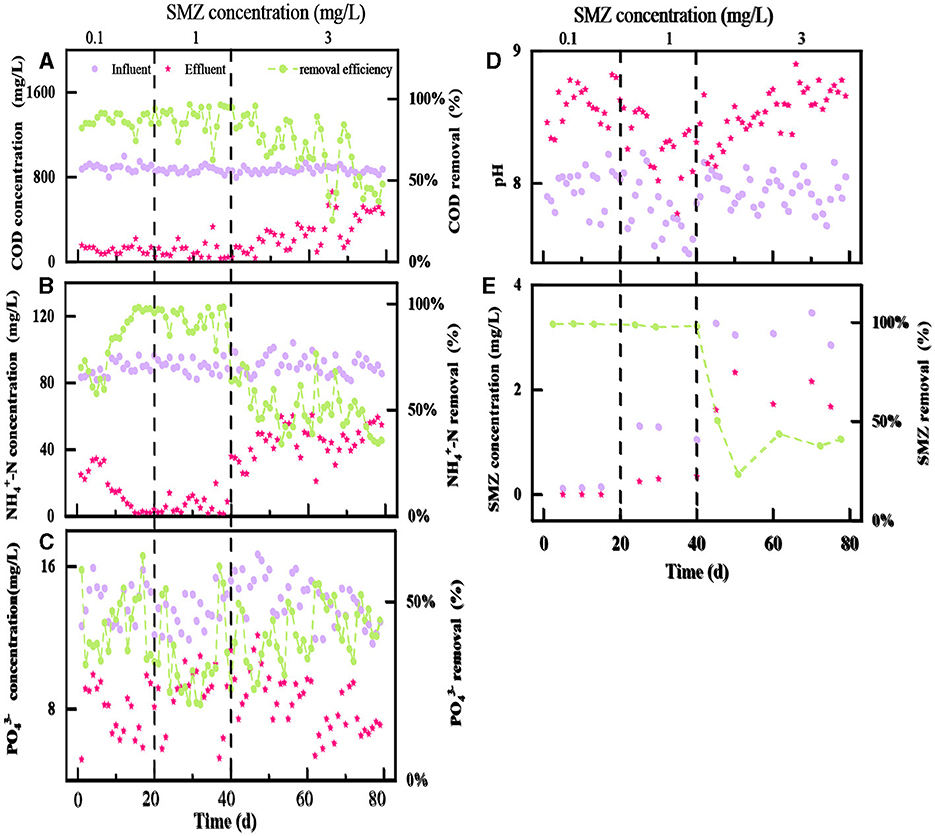
Figure 2. The variations of COD (A), -N (B), (C), pH (D), and SMZ (E) concentrations in influent and effluent at different stages.
Moreover, there was no significant difference in the effect of different SMZ concentrations on removal efficiency (Figure 2C), which always fluctuated around 42.12%. The pH of the effluent is higher than that of the influent (Figure 2D). It has been reported that pH affects phosphorus removal by the microbial community. With effluent pH exceeding 8, phosphorus precipitation tends to form in the system (Wang et al., 2022), affecting phosphorus utilization by microorganisms and reducing phosphorus removal efficiency. Overall, the two-stage biological contact oxidation reactor had a limited effect on the removal of during the SMZ wastewater treatment.
Afterward, SMZ was undetectable in the effluents during stage 1 (Figure 2E), indicating that activated sludge completely adsorbed or degraded the dosed SMZ. Afterward, the SMZ removal efficiency was above 90% in stage 2. In stage 3, the average removal efficiency of SMZ decreased to 39.36% with 3 mg/L SMZ added to the influent. The gradient concentrations of SMZ resulted in a significant reduction in the performance of the two-stage biological contact oxidation reactor during SMZ wastewater treatment.
3.2. Response of EPS production to SMZ shocks
Biofilms are complex structures composed of microorganisms, and EPS, as the main component of biofilms, is a key factor affecting the performance of reactors (Xiong et al., 2020). EPS content is strongly influenced by SMZ in Figure 3. The addition of SMZ to the influent inhibited the secretion of LB-EPS (Figure 3A), which was reduced from 73.53 mg/(g VSS) in CK to 42.04 mg/(g VSS) in Z3. On the contrary, the production of TB-EPS substantially increased after each SMZ shock to 44.33 mg/(g VSS) in Z1 and 53.79 mg/(g VSS) in Z3, respectively (Figure 3B), which was significantly higher than the 10.34 mg/(g VSS) in CK. It has been reported that EPS can resist antibiotic stress by the adsorption of antibiotics through the hydrophobic region and functional groups of PN (Fan et al., 2021). The pronounced increase in PN yield mainly contributed to the elevated TB-EPS production, reaching 41.97 mg/(g VSS) and 47.79 mg/(g VSS) in Z1 and Z3, respectively. Its production was promoted to protect microbial cells against SMZ shocks. In particular, SMZ is positively correlated with TB-EPS and TB-PN content and negatively correlated with LB-EPS content, while TB-EPS is also positively correlated with PN content (Figure 3).
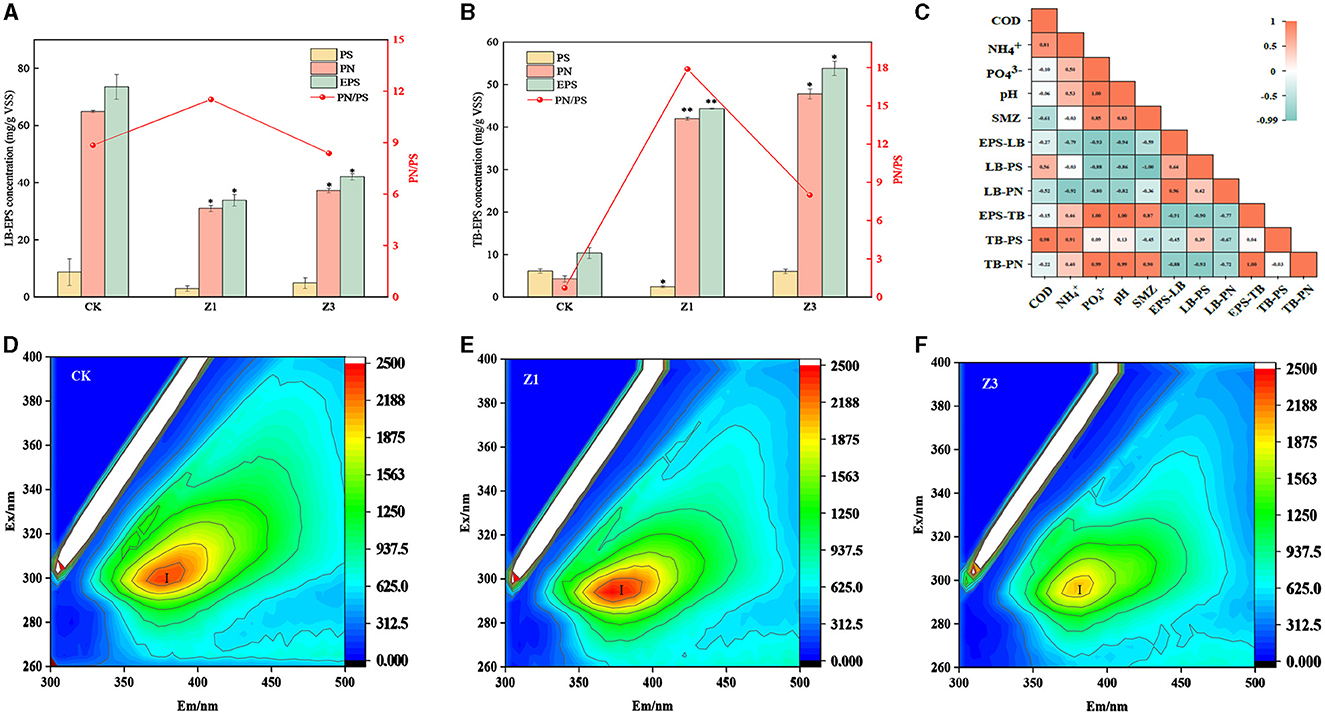
Figure 3. Variations of LB-EPS (A) and TB-EPS (B) concentration at different stages (CK: seeded sludge; Z1 and Z3, indicated the sludge collected at the end of stage 2, 3); correlation analysis of extracellular polymers with environmental factors (C); excitation-emission matrix fluorescence spectra for EPS in CK (D), Z1(E), and Z3 (F).
The chemical composition of EPS was further investigated at different SMZ concentrations by the 3D-EEM fluorescence spectra in Figures 3D–F. The I-peak (250 nm < Ex < 400 nm, 200 nm < Em < 380 nm) was associated with aromatic protein-like substances, according to Zhu et al. (2022). The I-peak appeared at all SMZ concentrations, while its intensity significantly decreased with increasing SMZ concentrations. Compared to 0 mg/L SMZ, the location of the I-peak in TB-EPS was shifted by 20/20 nm and 25/25 nm along the excitation/emission scale at 1 and 3 mg/L SMZ, respectively. Additionally, the fluorescence shift further illustrated that SMZ could affect the chemical composition of EPS.
3.3. Effect of SMZ on the bacterial community
The impact of SMZ pressure on the microbial composition and distribution was investigated by the metagenomics sequencing technology in Figure 4. The bacterial community composition and abundance differed significantly with an increase in SMZ concentration, and the flora with poor environmental adaptability were gradually eliminated. At the phylum level, the abundance of Chloroflexi increased from 19.25% (CK) to 20.12% (Z1) and 41.75% (Z3) with an increase in SMZ concentration (Figure 4A). Moreover, the abundance of Proteobacteria reached 39.54% in Z1, while it decreased to 28.62% in Z3. In contrast, the abundance of Actinobacteria had an obvious decrease from 44.22% (CK) to 8% (Z3) under the selection pressure of SMZ in the environment.
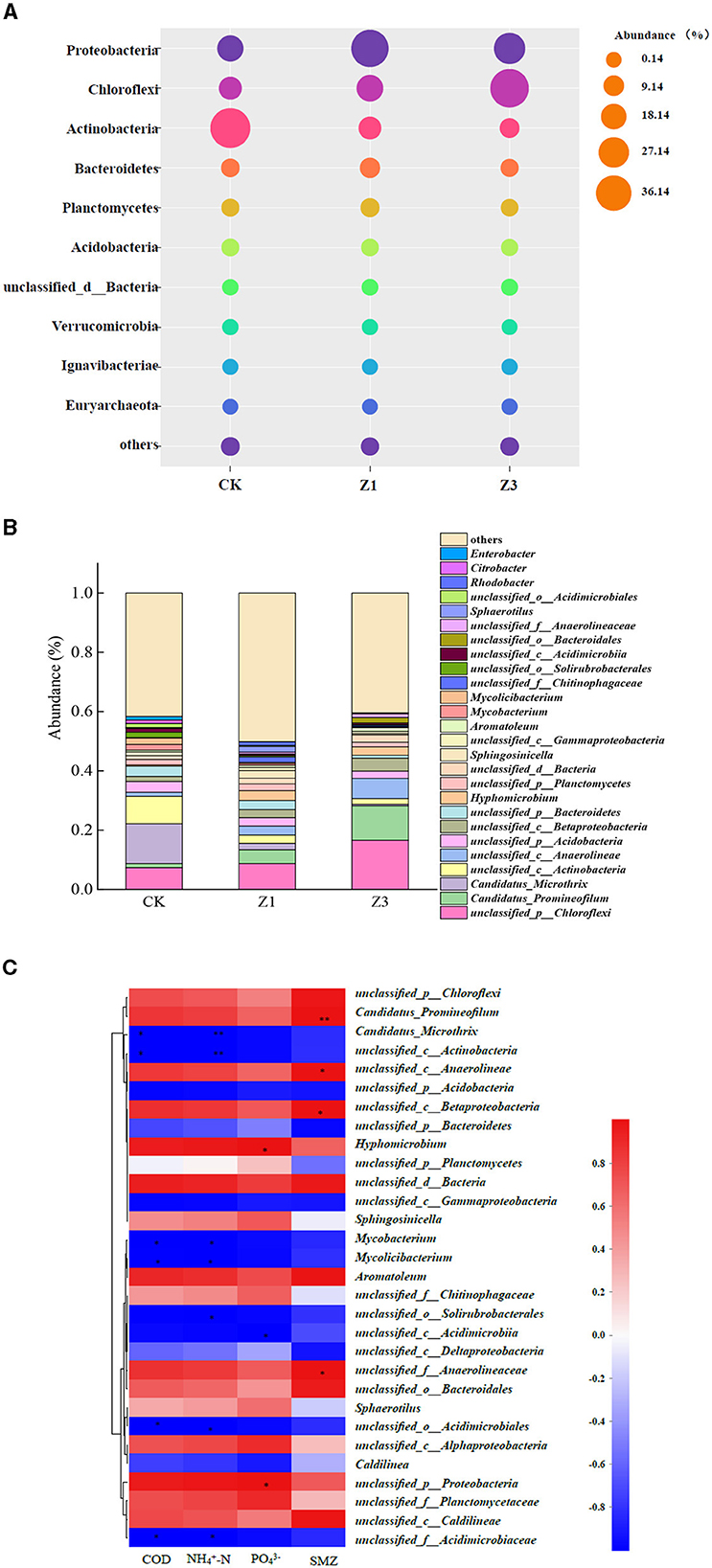
Figure 4. Phylum-level classification of bacterial communities (the relative abundance of <1% was defined as “others”) (A); percentage of bacterial community abundance on genus level (the relative abundance of <1% was defined as “others”) (B); a correlation heatmap of the top 30 genera related to environmental factors (C). *Represents P less than 0.05, **represents P less than 0.01.
At the genus level (Figure 4B), Candidatus_Microthrix and unclassified_c__Actinobacteria were the most prevalent genera in CK, accounting for 22.6% of all observed sequences. However, their abundances showed a downward trend and reduced to 0.5 and 1.9% in Z3, respectively, with the increased SMZ concentration, probably because the SMZ concentrations exceeded these bacteria' tolerance levels. In contrast, the abundances of unclassified_d__Bacteria, unclassified_c__Betaproteobacteria, unclassified_c__Anaerolineae, Candidatus_Promineofilum, and unclassified_p__Chloroflexi increased with the increased SMZ concentration, especially for Candidatus_Promineofilum, whose abundance increased from 1.4% in CK to 11.6% in Z3, indicating higher levels of adaptation for SMZ. Furthermore, the SMZ was significantly correlated with Candidatus_Promineofilum (P < 0.05), followed by unclassified_c__Anaerolineae, unclassified_c__Betaproteobacteria, and unclassified_f__Anaerolineaceae (P < 0.05) (Figure 4C). Meanwhile, the relative abundances of genera Candidatus_Promineofilum, unclassified_c__Betaproteobacteria, and unclassified_f__Anaerolineaceae exhibited an increasing trend in the SMZ wastewater treatment system (Figure 4B), further demonstrating their powerful tolerance to SMZ.
3.4. Co-occurrence associations between the microbial community and ARGs
The presence of SMZ not only affects the microbial community but also leads to the development, migration, and transformation of diverse ARGs. Therefore, the correlation between the abundance of ARGs and the microbial community has been further explored in Figure 5. During the SMZ wastewater treatment system, the types of antibiotics with higher abundance in the sludge samples from CK, Z1, and Z3 were bacteriocins and sulfonamides. The abundance of sulfonamide antibiotics increased substantially from 12% (CK) to 31% (Z1) and 56% (Z3), respectively (Figure 5A). This phenomenon indicated that the SMZ concentration in the influent serves as a crucial factor for enriching its corresponding sulfonamide antibiotics. Among these ARGs (Figure 5B), bacA, sul1, and sul2 were the top three dominant genes across the three samples. sul1 and sul2 are the sulfonamide resistance genes, and their abundances increased with the increasing SMZ concentration, which enhanced the propagation and transformation of ARGs among bacteria. Afterward, AAN60217, ABG36700, and ACJ39691 associated with sul1, and YP_001969930 associated with sul2 also exhibited an increasing trend during the SMZ wastewater treatment (Figure 5C), further revealing that the selection pressure imposed by increased SMZ promoted the occurrence of these associated bacteria. The co-occurrence pattern of sul1, sul2, and microbial communities revealed that the 19 bacterial genera were speculated as possible hosts (Figure 5D). The microorganisms positively associated with sul1 and sul2 were mainly attributed to Chloroflexi and Proteobacteria, especially Candidatus_Promineofilum. It was not only strongly correlated with sul1 and sul2, but it was also the most enriched genus associated with the removal of SMZ in this system.
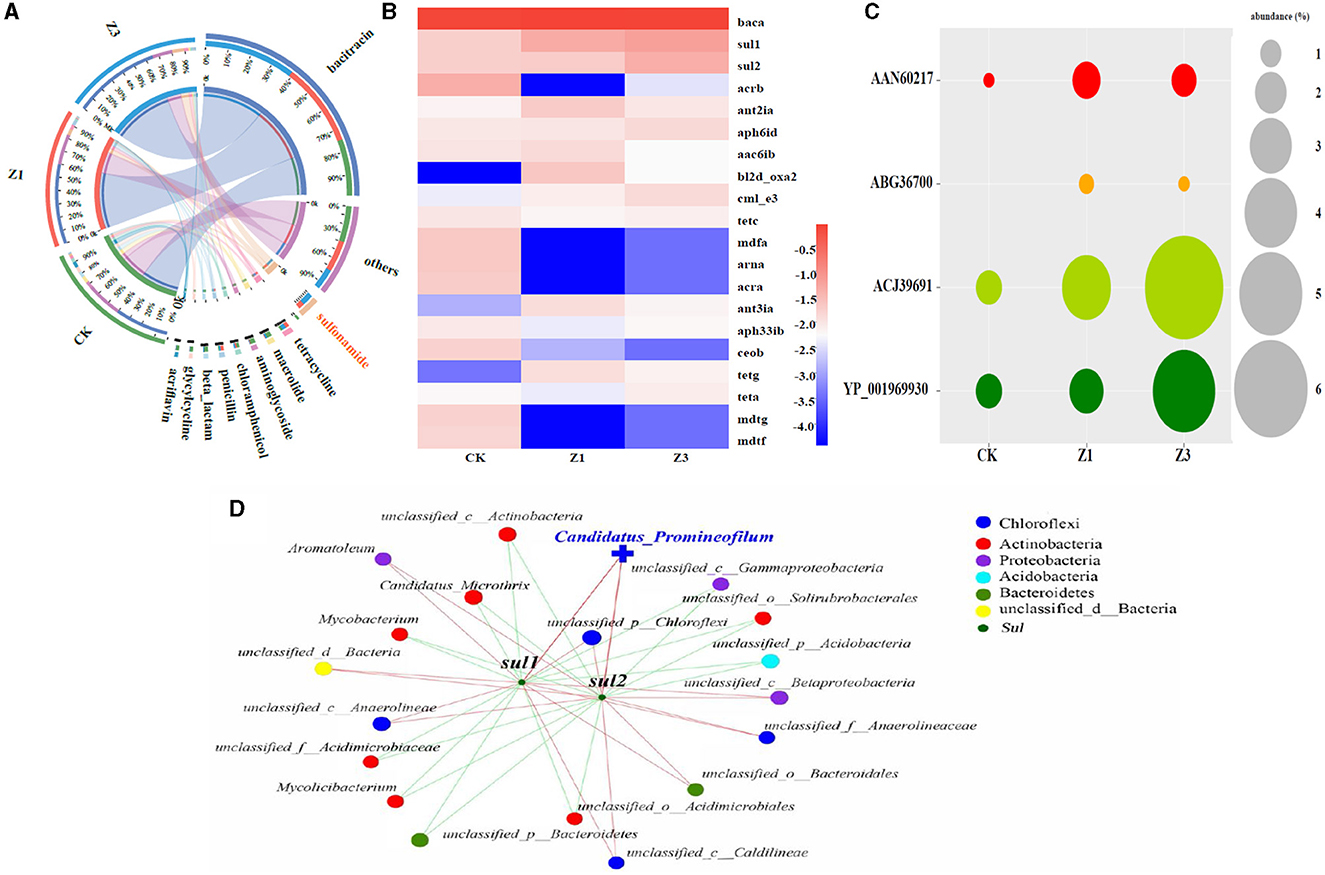
Figure 5. Circos analysis between the biofilm samples and antibiotic type (others: abundance < 3%) (A); heatmap analysis of the top 20 ARGs types in each sample (B); the abundance of ARGs associated with SMZ (C); network analysis revealing the co-occurrence patterns among the ARGs types associated with SMZ and the top 20 genera (D).
3.5. Antibiotic resistance mechanism
Antibiotics exert selective pressure on microbial communities and promote the emergence of ARGs. Sulfonamide antibiotics were dissipated and driven by microbes mainly through the initial ipso-hydroxylation (Chen et al., 2022). To analyze the factors that contribute to the mechanism of resistance of biota to SMZ, it is crucial to understand the mechanism of action of antibiotics. During the SMZ wastewater treatment system, the antibiotic target alteration was persistently prevalent in different SMZ concentrations (Figure 6A), which was considered a primary resistance mechanism for bacterial resistance to SMZ, among which the relative contribution of Candidatus_Promineofilum, unclassified_f__Anaerolineaceae, Hyphomicrobium, unclassified_c__Anaerolineae, and unclassified_p__Chloroflexi to this resistance mechanism gradually increased with the increased SMZ concentration (Figure 6B), indicating that the members of these genera could resist the adverse environment of SMZ pressure through antibiotic target alteration mechanisms (Figure 6C).
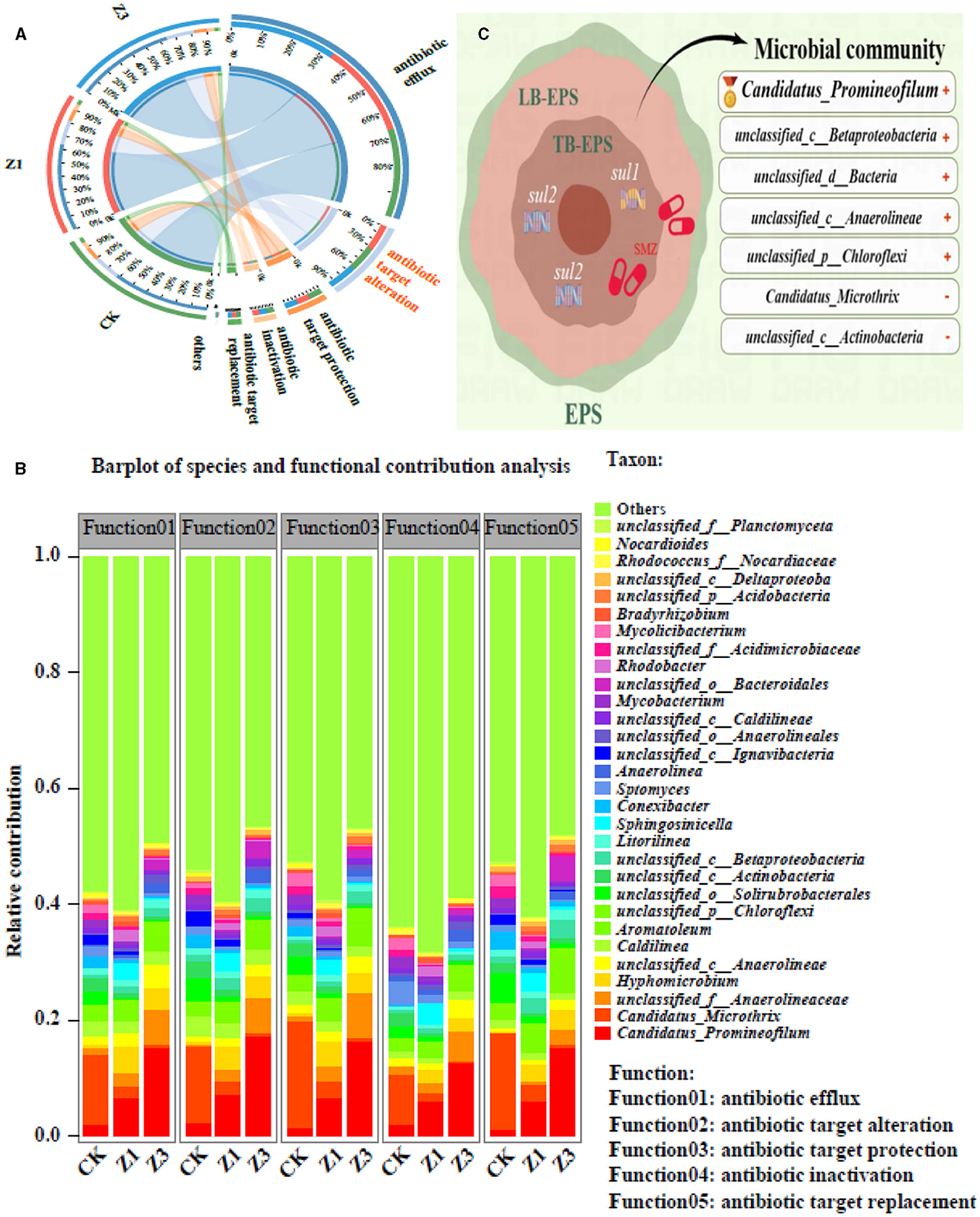
Figure 6. Circos analysis between the biofilm samples and resistance mechanism (others: abundance < 1%) (A); relative contributions for bacterial genera (top 30 genera) to resistance mechanisms (B); the mechanisms of tolerance and removal of SMZ (C).
4. Discussion
The two-stage biological contact oxidation reactor exhibited excellent performance in removing low concentrations (0.1–1 mg/L) of SMZ with a removal rate higher than 90% (Figure 2E), which was higher than the 56.0% removal rate reported by Yang et al. (2015) for the treatment of 1 mg/L SMZ using the lab-scale sequencing batch reactor. However, the SMZ, COD, and -N removal efficiencies all had a decreasing trend when the SMZ concentration in the influent increased to 3 mg/L. This phenomenon was similar to Yang et al. (2015) in that SMZ was not completely removed by the acclimatized activated sludge. However, it was lower than the 95.4% removal under 0.1–5 mg/L SMZ during the MBR system (Shi et al., 2018). There are huge differences in the biodegradation of SMZ, which might be due to the differences in study conditions (including activated sludge) and also from place to place. The addition of the antibiotic SMZ affects the structure of a bacterial community in this system. The biofilm is subjected to greater environmental stress, and the species abundance is affected by the increase in time and SMZ concentration (Ciofu et al., 2022).
The EPS is essential for providing stability and adhesion of biofilms to the carrier in the reactor (Das, 2021). The TB-EPS contents kept an increasing tendency with an increase in SMZ concentration (Figure 3B), indicating that SMZ stimulated the microbial secretion of TB-EPS in the biofilm, especially given the pronounced increase in PN yield. PN plays a key role in the response to changes in environmental conditions. However, the variation trend in TB-EPS was opposite to that in LB-EPS, which may be because SMZ could migrate into the activated sludge by dynamically complexing with EPS. TB-EPS plays a central role in the protection of microbial cells, which can bind with antibiotics and secrete active enzymes for the removal of antibiotics, which then can alleviate cellular osmotic pressure imbalance (Sun et al., 2022). Furthermore, as the first barrier of microbial cells, EPS also contacts directly with SMZ in aqueous environments (Guo et al., 2020) and plays a key role in the biosorption of SMZ (Wang et al., 2018), further reducing it to undetectable levels in the effluent (Xu and Sheng, 2020). Consequently, microorganisms in the biofilm produced the EPS to form a network structure outside the cells, owing to the self-protection strategy against the toxicity of SMZ (Zhang et al., 2016).
The performance of BCOR in the treatment of SMZ wastewater is closely related to the key bacteria associated with SMZ biodegradation in activated sludge. Chloroflexi and Proteobacteria were enriched with an increase in SMZ concentration and became the dominant phylum (Figure 4A). Furthermore, Chloroflexi is involved in pathways for the complete hydrolytic or oxidative degradation of various recalcitrant organic matter, including aromatic compounds, polyaromatic hydrocarbons, polychlorobiphenyls, and organochlorine compounds (Liu et al., 2022). Proteobacteria play an important role in the degradation or transformation of sulfonamide compounds (Reis et al., 2018). Meanwhile, the abundance of unclassified_d__Bacteria, unclassified_c__Betaproteobacteria, unclassified_c__Anaerolineae, Candidatus_Promineofilum, and unclassified_p__Chloroflexi exhibited an increasing trend with the gradient concentrations of SMZ, especially for Candidatus_Promineofilum belonging to Chloroflexi. It was not only highly correlated with SMZ (Figure 4C) but also had the strongest correlation with the primary resistance genes to sulfonamides: sul1 and sul2 (Figure 5B), which may be their potential hosts. Previous studies have reported that Candidatus_Promineofilum is one of the most abundant populations in activated sludge (McIlroy et al., 2016), which contributed predominantly to aromatic degradation, quinoline degradation, and denitrification (Tian et al., 2023). Moreover, Candidatus_Promineofilum plays essential roles in EPS secretion, containing the exopolysaccharide biosynthesis genes (glmS and wbpA) and amino acid biosynthesis genes (ltaE and metH) (Dong et al., 2023). This provides an opportunity to protect and regulate itself by producing the EPS under the SMZ stress, and the increased levels of EPS further facilitate the SMZ adsorption.
Bacteria will adopt appropriate strategies to break the inhibitory effect of antibiotics according to the antibacterial mechanism of antibiotics, including mutation of the drug target (Garcia-Bustos et al., 2022), secretion of hydrolase (Pulingam et al., 2022), and excretion of antibiotics from cells through an efflux pump (Zhou et al., 2023). Previous studies have reported that bacterial resistance to sulfonamides is mainly associated with mutations in the gene encoding dihydrofolate synthetase (DHPs) in the chromosome, which reduces the affinity between DHPs and sulfonamides. Sul, as an alternative gene to DHPs (Aslam et al., 2012), has a much lower affinity for sulfanilamide and is the most common way to generate a sulfonamide resistance mechanism (Figure 6B). Therefore, in the microbial community, unclassified_d__Bacteria, unclassified_c__Betaproteobacteria, unclassified_c__Anaerolineae, Candidatus_Promineofilum, and unclassified_p__Chloroflexi in the SMZ-treated wastewater system mainly resist SMZ stress through targeted gene changes (Figure 6A).
5. Conclusions
The two-stage biological contact oxidation reactor displayed stable and efficient removal of SMZ (90%) at 0.1–1 mg/L SMZ, which was strongly correlated with the composition of EPS, especially for TB-PN played an important role in responding to the threat of SMZ. Moreover, SMZ has an obvious effect on the bacterial community. Candidatus_Promineofilum, with the ability to secrete EPS, was enriched mostly with the gradient SMZ concentrations, which accounted for 11.6% of the overall microbial community and became the predominant SMZ-degrading bacteria genus in BCOR biofilm. Furthermore, Candidatus_Promineofilum was not only highly correlated with SMZ but also had the strongest correlation with the primary sulfonamide resistance genes of sul1 and sul2. It could fight against SMZ stress through the mechanism of targeted gene changes.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author contributions
TC: writing—original draft. JC: methodology and investigation. YC: methodology, resources, and supervision. SZ: project administration. J-HQ: visualization and writing—review and editing. ZW: conceptualization. JP: formal analysis. LF: supervision. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (42107139), the Doctoral Scientific Research Startup Foundation from Henan University of Technology (2019BS046), Key Scientific Research Projects in Henan Higher Education Institutions (22A180014), the Innovative Funds Plan of Henan University of Technology (2021ZKCJ15), and the Cultivation Programme for Young Backbone Teachers in Henan University of Technology (21421206).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Acosta-Rangel, A., Sánchez-Polo, M., Rozalen, M., Rivera-Utrilla, J., Polo, A., and Mota, A. (2019). Comparative study of the oxidative degradation of different 4-aminobenzene sulfonamides in aqueous solution by sulfite activation in the presence of Fe (0), Fe (II), Fe (III), Or Fe (VI). Water 11, 2332. doi: 10.3390/w11112332
Aslam, M., Checkley, S., Avery, B., Chalmers, G., Bohaychuk, V., Gensler, G., et al. (2012). Phenotypic and genetic characterization of antimicrobial resistance in Salmonella serovars isolated from retail meats in Alberta, Canada. Food Microbiol. 32, 110–117. doi: 10.1016/j.fm.2012.04.017
Bai, Z., Yang, Q., and Wang, J. (2016). Catalytic ozonation of sulfamethazine using Ce0.1Fe0.9OOH as catalyst: Mineralization and catalytic mechanisms. Chem. Eng. J. 300, 169–176. doi: 10.1016/j.cej.2016.04.129
Ben, W., Zhu, B., Yuan, X., Zhang, Y., Yang, M., and Qiang, Z. (2018). Occurrence, removal and risk of organic micropollutants in wastewater treatment plants across China: comparison of wastewater treatment processes. Water Res. 130, 38–46. doi: 10.1016/j.watres.2017.11.057
Buchfink, B., Xie, C., and Huson, D. H. (2015). Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60. doi: 10.1038/nmeth.3176
Chen, J., Yang, Y., Ke, Y., Chen, X., Jiang, X., Chen, C., et al. (2022). Sulfonamide-metabolizing microorganisms and mechanisms in antibiotic-contaminated wetland sediments revealed by stable isotope probing and metagenomics. Environ. Int. 165, 107332. doi: 10.1016/j.envint.2022.107332
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, 884–890. doi: 10.1093/bioinformatics/bty560
Ciofu, O., Moser, C., Jensen, P. Ø., and Høiby, N. (2022). Tolerance and resistance of microbial biofilms. Nat. Rev. Microbiol. 20, 621–635. doi: 10.1038/s41579-022-00682-4
Das, S. (2021). Structural and mechanical characterization of biofilm-associated bacterial polymer in the emulsification of petroleum hydrocarbon. 3 Biotech 11, 239. doi: 10.1007/s13205-021-02795-8
Dong, K., Qiu, Y., Wang, X., Yu, D., Yu, Z., Feng, J., et al. (2023). Towards low carbon demand and highly efficient nutrient removal: establishing denitrifying phosphorus removal in a biofilm-based system. Bioresour. Technol. 372, 128658. doi: 10.1016/j.biortech.2023.128658
Du, L., Cheng, S., Hou, Y., Sun, X., Zhou, D., and Liu, B. (2018). Influence of sulfadimethoxine (SDM) and sulfamethazine (SM) on anammox bioreactors: performance evaluation and bacterial community characterization. Bioresour. Technol. 267, 84–92. doi: 10.1016/j.biortech.2018.05.067
Fan, N. S., Fu, J. J., Huang, D. Q., Ma, Y. L., Lu, Z. Y., Jin, R. C., et al. (2021). Resistance genes and extracellular proteins relieve antibiotic stress on the anammox process. Water Res. 202, 117453. doi: 10.1016/j.watres.2021.117453
Fu, C., Yi, X., Liu, Y., and Zhou, H. (2020). Cu2+ activated persulfate for sulfamethazine degradation. Chemosphere 257, 127294. doi: 10.1016/j.chemosphere.2020.127294
Fu, L., Niu, B., Zhu, Z., Wu, S., and Li, W. (2012). CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152. doi: 10.1093/bioinformatics/bts565
Gao, J., and Pedersen, J. A. (2005). Adsorption of sulfonamide antimicrobial agents to clay minerals. Environ. Sci. Technol. 39, 9509–9516. doi: 10.1021/es050644c
Garcia-Bustos, V., Cabañero-Navalón, M. D., and Lletí, M. S. (2022). Resistance to beta-lactams in Gram-negative bacilli: relevance and potential therapeutic alternatives. Rev. Española Quimioterapia 35, 1. doi: 10.37201/req/s02.01.2022
Guo, H., Chen, Z., Lu, C., Guo, J., Li, H., Song, Y., et al. (2020). Effect and ameliorative mechanisms of polyoxometalates on the denitrification under sulfonamide antibiotics stress. Bioresour. Technol. 305, 123073. doi: 10.1016/j.biortech.2020.123073
Harada, K. (2018). Antibiotic residue in environmental water in Vietnam. Yakugaku Zasshi 138, 271–275. doi: 10.1248/yakushi.17-00177-1
Li, N., Liu, Q., Zhou, G. Q., Dai, M. X., and Kong, Q. (2019). Contaminant removal and microorganism response of activated sludge in sulfamethazine wastewater treatment. Int. Biodeterior. Biodegradation. 143, 104705. doi: 10.1016/j.ibiod.2019.05.022
Liu, R., Wei, X., Song, W., Wang, L., Cao, J., Wu, J., et al. (2022). Novel Chloroflexi genomes from the deepest ocean reveal metabolic strategies for the adaptation to deep-sea habitats. Microbiome 10, 1–17. doi: 10.1186/s40168-022-01263-6
McIlroy, S. J., Karst, S. M., Nierychlo, M., Dueholm, M. S., Albertsen, M., Kirkegaard, R. H., et al. (2016). Genomic and in situ investigations of the novel uncultured Chloroflexi associated with 0092 morphotype filamentous bulking in activated sludge. ISME J. 10, 2223–2234. doi: 10.1038/ismej.2016.14
Mulla, S. I., Bagewadi, Z. K., Faniband, B., Bilal, M., Chae, J. C., Bankole, P. O., et al. (2021). Various strategies applied for the removal of emerging micropollutant sulfamethazine: a systematic review. Environ. Sci. Pollut. Res. 30, 1–15. doi: 10.1007/s11356-021-14259-w
Ouyang, Z., Yang, C., He, J., Yao, Q., Zhang, B., Wang, H., et al. (2020). Homogeneous photocatalytic degradation of sulfamethazine induced by Fe (III)-carboxylate complexes: kinetics, mechanism and products. Chem. Eng. J. 402, 126122. doi: 10.1016/j.cej.2020.126122
Peng, T., Wang, Y., Wang, J., Fang, F., Yan, P., and Liu, Z. (2022). Effect of different forms and components of EPS on sludge aggregation during granulation process of aerobic granular sludge. Chemosphere 303, 135116. doi: 10.1016/j.chemosphere.2022.135116
Pulingam, T., Parumasivam, T., Gazzali, A. M., Sulaiman, A. M., Chee, J. Y., Lakshmanan, M., et al. (2022). Antimicrobial resistance: prevalence, economic burden, mechanisms of resistance and strategies to overcome. Eur. J. Pharm. Sci. 170, 106103. doi: 10.1016/j.ejps.2021.106103
Reis, P. J., Homem, V., Alves, A., Vilar, V. J., Manaia, C. M., and Nunes, O. C. (2018). Insights on sulfamethoxazole bio-transformation by environmental Proteobacteria isolates. J. Hazard. Mater. 358, 310–318. doi: 10.1016/j.jhazmat.2018.07.012
Shi, B. J., Wang, Y., Geng, Y. K., Liu, R. D., Pan, X. R., Li, W. W., et al. (2018). Application of membrane bioreactor for sulfamethazine-contained wastewater treatment. Chemosphere 193, 840–846. doi: 10.1016/j.chemosphere.2017.11.051
Song, T., Zhang, X., Li, J., Xie, W., Dong, W., and Wang, H. (2023). Sulfamethoxazole impact on pollutant removal and microbial community of aerobic granular sludge with filamentous bacteria. Bioresour. Technol. 379, 128823. doi: 10.1016/j.biortech.2023.128823
Strous, M., Pelletier, E., Mangenot, S., Rattei, T., Lehner, A., Taylor, M. W., et al. (2006). Deciphering the evolution and metabolism of an anammox bacterium from a community genome. Nature 440, 790–794. doi: 10.1038/nature04647
Sun, Z., Li, Y., Li, M., Wang, N., Liu, J., Guo, H., et al. (2022). Steel pickling rinse wastewater treatment by two-stage MABR system: reactor performance, extracellular polymeric substances (EPS) and microbial community. Chemosphere 299, 134402. doi: 10.1016/j.chemosphere.2022.134402
Tian, H., Li, Y., Chen, H., Zhang, J., Hui, M., Xu, X., et al. (2023). Aerobic biodegradation of quinoline under denitrifying conditions in membrane-aerated biofilm reactor. Environ. Pollut. 326, 121507. doi: 10.1016/j.envpol.2023.121507
Wan, X. P., Gao, M. M., Ye, M. S., Wang, Y. K., Xu, H., Wang, M. Y., et al. (2018). Formation, characteristics and microbial community of aerobic granular sludge in the presence of sulfadiazine at environmentally relevant concentrations. Bioresour. Technol. 250, 486–494. doi: 10.1016/j.biortech.2017.11.071
Wang, L., Li, Y., Wang, L., Zhang, H., Zhu, M., Zhang, P., et al. (2018). Extracellular polymeric substances affect the responses of multi-species biofilms in the presence of sulfamethizole. Environ. Pollut. 235, 283–292. doi: 10.1016/j.envpol.2017.12.060
Wang, Y., Kuntke, P., Saakes, M., Weijden, R. D., Buisman, C. J. N., and Lei, Y. (2022). Electrochemically mediated precipitation of phosphate minerals for phosphorus removal and recovery: progress and perspective. Water Res. 209, 117891. doi: 10.1016/j.watres.2021.117891
Wu, X., Zhang, L., Lv, Z., Xin, F., Dong, W., Liu, G., et al. (2022). N-acyl-homoserine lactones in extracellular polymeric substances from sludge for enhanced chloramphenicol-degrading anode biofilm formation in microbial fuel cells. Environ. Res. 207, 112649. doi: 10.1016/j.envres.2021.112649
Xiong, F., Zhao, X., Wen, D., and Li, Q. (2020). Effects of N-acyl-homoserine lactones-based quorum sensing on biofilm formation, sludge characteristics, and bacterial community during the startup of bioaugmented reactors. Sci. Total Environ. 735, 139449. doi: 10.1016/j.scitotenv.2020.139449
Xu, J., and Sheng, G. P. (2020). Microbial extracellular polymeric substances (EPS) acted as a potential reservoir in responding to high concentrations of sulfonamides shocks during biological wastewater treatment. Bioresour. Technol. 313, 123654. doi: 10.1016/j.biortech.2020.123654
Yang, N., Wan, J. F., Zhao, S. J., and Wang, Y. (2015). Removal of concentrated sulfamethazine by acclimatized aerobic sludge and possible metabolic products. PeerJ 3, e1359. doi: 10.7717/peerj.1359
Yu, J., Xiao, K., Xu, H., Li, Y., Xue, Q., Xue, W., et al. (2023). Spectroscopic fingerprints profiling the polysaccharide/protein/humic architecture of stratified extracellular polymeric substances (EPS) in activated sludge. Water Res. 235, 119866. doi: 10.1016/j.watres.2023.119866
Zhang, Q. Q., Bai, Y. H., Wu, J., Zhu, W. Q., Tian, G. M., Zheng, P., et al. (2019). Microbial community evolution and fate of antibiotic resistance genes in anammox process under oxytetracycline and sulfamethoxazole stresses. Bioresour. Technol. 293, 122096. doi: 10.1016/j.biortech.2019.122096
Zhang, W., Zhou, X., Cao, X., and Li, S. (2022a). Accelerating anammox nitrogen removal in low intensity ultrasound-assisted ASBBR: performance optimization, EPS characterization and microbial community analysis. Sci. Total Environ. 817, 152989. doi: 10.1016/j.scitotenv.2022.152989
Zhang, Y., Cheng, D., Xie, J., Zhang, Y., Wan, Y., Zhang, Y., and Shi, X. (2022b). Impacts of farmland application of antibiotic-contaminated manures on the occurrence of antibiotic residues and antibiotic resistance genes in soil: a meta-analysis study. Chemosphere 300, 134529. doi: 10.1016/j.chemosphere.2022.134529
Zhang, Y., Geng, J., Ma, H., Ren, H., Xu, K., and Ding, L. (2016). Characterization of microbial community and antibiotic resistance genes in activated sludge under tetracycline and sulfamethoxazole selection pressure. Sci. Total Environ. 571, 479–486. doi: 10.1016/j.scitotenv.2016.07.014
Zhou, J., Chen, Y., Li, W. X., Qu, J. H., Chen, T., Wang, Y. P., et al. (2023). Deciphering the microbial community tolerance mechanism and alteration of antibiotic resistance genes during chloramphenicol wastewater treatment. Int. Biodeterior. Biodegrad. 178, 105546. doi: 10.1016/j.ibiod.2022.105546
Zhou, J., Chen, Y., Qu, J. H., Wang, Y. K., Mai, W. N., Wan, D. J., et al. (2022). Responses of microbial community and antibiotic resistance genes to co-existence of chloramphenicol and salinity. Appl. Microbiol. Biotechnol. 106, 7683–7697. doi: 10.1007/s00253-022-12188-3
Zhu, J., You, H., Ng, H. Y., Li, Z., Xie, B., Chen, H., et al. (2022). Impacts of bio-carriers on the characteristics of cake layer and membrane fouling in a novel hybrid membrane bioreactor for treating mariculture wastewater. Chemosphere. 300, 134593. doi: 10.1016/j.chemosphere.2022.134593
Keywords: sulfadimethoxine, biological contact oxidation technology, microbial community, extracellular polymeric substances, antibiotic-resistant genes
Citation: Zhou J, Chen T, Cui J, Chen Y, Zhao S, Qu J-H, Wang Z, Pan J and Fan L (2023) Responses of the microbial community and the production of extracellular polymeric substances to sulfamethazine shocks in a novel two-stage biological contact oxidation system. Front. Microbiol. 14:1240435. doi: 10.3389/fmicb.2023.1240435
Received: 15 June 2023; Accepted: 01 August 2023;
Published: 29 August 2023.
Edited by:
Alejandro Rodriguez-Sanchez, University of Granada, SpainReviewed by:
Aqiang Ding, Chongqing University, ChinaShiyang Zhang, Wuhan University of Technology, China
Copyright © 2023 Zhou, Chen, Cui, Chen, Zhao, Qu, Wang, Pan and Fan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian-Hang Qu, amhxdUBoYXV0LmVkdS5jbg==
 Jia Zhou1
Jia Zhou1 Jian-Hang Qu
Jian-Hang Qu Zitong Wang
Zitong Wang