
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 08 September 2023
Sec. Food Microbiology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1238689
This article is part of the Research Topic Impact and Control of Food- and Waterborne Protozoan Parasites View all 5 articles
 Andrea Mancusi1
Andrea Mancusi1 Yolande T. R. Proroga1
Yolande T. R. Proroga1 Angela Giordano1
Angela Giordano1 Santa Girardi1
Santa Girardi1 Francescantonio D’Orilia2
Francescantonio D’Orilia2 Renato Pinto3
Renato Pinto3 Paolo Sarnelli3
Paolo Sarnelli3 Laura Rinaldi2,4
Laura Rinaldi2,4 Federico Capuano1
Federico Capuano1 Maria Paola Maurelli4*
Maria Paola Maurelli4*Toxoplasmosis, caused by the protozoan Toxoplasma gondii, is one of the main food-, water- and soil-borne zoonotic disease worldwide. Over the past 20 years many papers were published on the transmission of T. gondii by marine animals, including mollusks, which can concentrate the oocysts and release them. Sporulated oocysts may remain viable and infective for 18 months in seawater. Therefore, raw or undercooked bivalve mollusks pose a risk to humans. This study aimed to apply and validate for the first time a very sensitive digital droplet polymerase chain reaction (ddPCR) protocol to detect and quantify T. gondii DNA in mussels. Four concentration levels: 8000 genomic copies (gc)/μL, 800 gc/μL, 80 gc/μL, 8 gc/μL of a T. gondii reference DNA were tested. DNA was extracted from 80 pools of mussels (Mytilus galloprovincialis). Forty pools were contaminated with T. gondii reference DNA and used as positive controls, while 40 pools were used as negative controls. DdPCR reaction was prepared using a protocol, previously developed by the authors, for detection of T. gondii in meat. Amplification was obtained up 8 gc/μL. All infected replicates resulted positive, as well as no droplets were detected in negative controls. The droplets produced in the reaction ranged from 8,828 to 14,075 (average 12,627 droplets). The sensitivity and specificity of ddPCR were 100% (95%CI = 94.3–99.9). In addition, 100 pools of mussels collected in the Gulf of Naples were used to validate the protocol. Of these 16% were positive (95% CI = 9.7–25.0) for T. gondii. Samples were also tested by real-time PCR and no positive samples were found. Data obtained from ddPCR showed good identification of negative and positive samples with higher specificity and efficiency than real-time PCR. This tool could be very useful for a rapid sensitive detection of low DNA concentrations of T. gondii in mussels, reducing the risk of toxoplasmosis in humans.
Toxoplasmosis, caused by the protozoan Toxoplasma gondii, is one of the main food-, water- and soil-borne zoonotic disease in the world (FAO/WHO, 2014; EFSA Panel on Biological Hazards, 2018; Almeria and Dubey, 2021). The lifecycle of this parasite involves felids (definitive hosts) and all warm-blooded animals and humans (intermediate hosts) (Dubey, 2022; Marín-García et al., 2022). Oocysts, spread into the environment by definitive hosts, become infective after sporulation, representing a risk for intermediate hosts in which sporozoites contained in sporulated oocysts may develop in tachyzoites and tissue cysts (Hatam-Nahavandi et al., 2021). It is estimated that up to 30% of the human world’s population is affected by T. gondii (Nayeri et al., 2021). Human transmission can occur congenitally, with the passage of tachyzoites from mother to the fetus (Almeria and Dubey, 2021). Instead, the acquired toxoplasmosis in humans is mainly related to ingestion of raw/undercooked meat of infected animals containing tissue cysts or of other raw foods at risk (e.g., vegetable, fruit, mollusks) or water contaminated by sporulated oocysts (Caradonna et al., 2017; Ghozzi et al., 2017; EFSA European Centre for Disease Prevention and Control, 2021; Dámek et al., 2023). However, few food and waterborne outbreaks have been described worldwide, especially in Europe, as also reported in the last EFSA report (2021) and by Almeria and Dubey (2021). This may be due to several causes, mainly: (i) difficulty connecting infection with food consumption in immunocompetent individuals, where toxoplasmosis appears asymptomatic or mild symptomatic; and (ii) scarce and non-standardized procedures to detect T. gondii in food (Almeria and Dubey, 2021; EFSA European Centre for Disease Prevention and Control, 2021; Marín-García et al., 2022).
The meat from different domestic and wild hosts has been investigated as source of T. gondii in humans, while environmental contamination is likely understudied and underestimated (Gabriël et al., 2022). In particular, a systematic and metanalysis review (López Ureña et al., 2022) recently published has highlighted that only in the last 20 years have been published papers on the transmission of T. gondii by fresh products and marine animals, including mollusks. This is due to increased knowledge also of other food- and water-borne zoonotic protozoa, e.g., Cryptosporidium spp., Giardia duodenalis, and Cyclospora cayetanensis (López Ureña et al., 2022).
Sporulated oocysts may remain viable and infective for 18 months in seawater (Lindsay and Dubey, 2009; Dubey, 2022). Studies on occurrence of T. gondii in the aquatic ecosystem started when lethal cases were reported due to this parasite in sea otters in California (Miller et al., 2004). The oocysts eliminated by felids are carried by freshwaters to coastal waters, where they can infect the marine animals (Fayer, 2004). In later years, T. gondii was found also in pinnipeds, mustelids and cetaceans, as well as in edible fishs and mollusks (Putignani et al., 2011; Zhang et al., 2014; Marino et al., 2019; Santoro et al., 2020; Nayeri et al., 2021; Dubey, 2022). Bivalve mollusks such as clams, cockles, mussels and oysters filter large volumes of water and can concentrate chemical and biological contaminants, including T. gondii oocysts. Moreover, mollusks can also release the parasite with feces after several days of ingestion from the water (Géba et al., 2021; Dubey, 2022). Therefore, raw or undercooked bivalve mollusks pose a risk to humans (Rosário et al., 2021).
Recently, we developed and validated a new droplet digital polymerase chain reaction (ddPCR) very sensitive (95.7%) and specific (100%) to detect T. gondii DNA in meat samples. This tool was compared with a real-time PCR protocol, which is usually the most used technique to detect T. gondii in food and environmental samples. The ddPCR showed a higher number of positive samples (7.6% of samples analyzed were positive by ddPCR vs 1.2% by qPCR) (Mancusi et al., 2022).
Therefore, in this study we applied and validated for the first time a ddPCR protocol on mussels, to obtain a more sensitive diagnostic tool to detect and quantify T. gondii DNA also in these marine animals.
To optimize ddPCR detection of T. gondii, a specific reference strain from the American Type Culture Collection (ATCC) was used. The T. gondii ATCC 50174D contained 2×105 genomic copies (gc)/μL in solution and was diluted with DNAse/RNAse free water to obtain four concentrations: 8000 gc/μL, 800 gc/μL, 80 gc/μL, and 8 gc/μL, to evaluate the amplification limit.
Internal tissue from 80 negative for T. gondii pools of marine mussels (Mytilus galloprovincialis) were homogenized by a stomacher. Each pool consisted of 10 mussels. The DNA was extracted from 25 mg of homogenized tissue for each pool, using a QIAamp DNA Mini kit (Qiagen, Hilden, Germany), according to the manufacturers’ instructions. Negativity was estimated using the real-time PCR protocol provided by the National Reference Centre for toxoplasmosis (Palermo, Italy). To evaluate the abovementioned concentrations, 40 negative DNA samples were contaminated with DNA extracted from the T. gondii ATCC, to obtain 10 replicates for each concentration. To the 40 samples used as negative controls only sterile water was added.
The ddPCR reaction was performed in a QX200 system (Bio-Rad, Hercules, CA, United States), using the protocol (i.e., mix preparation, primers and probe, Table 1) described by Mancusi et al. (2022) to amplify the 529 bp repeat element. Droplets were generated using a DG8 cartridge (Bio-Rad, Hercules, CA, United States), adding a volume of 70 μL of droplet generation oil for each well. Subsequently, the PCR amplification was carried out on a CFX96 instrument (Bio-Rad, Hercules, CA, USA), transferring 40 μL of droplet-partitioned samples to each well and following these thermal conditions: 96°C for 10 min followed by 45 cycles at 98°C for 30 s, 58.5°C for 1 min and a final stage at 98°C for 10 min. After thermocycling, the plate was read in the QX200 Droplet Reader and the QuantaSoft software was used to quantify the DNA target, expressed as the number of genomic copies/1 μL of reaction.

Table 1. Primers and probe to amplify the 529 bp repeat element by ddPCR (Mancusi et al., 2022).
Moreover, the intra-laboratory repeatability was assessed to verify the robustness of the ddPCR method, calculating the coefficient of variation (CV%) between the assays performed by two operators.
One hundred pool samples of marine mussels were collected from seven sites in the Gulf of Naples (Figure 1) and used for validation of the optimized ddPCR. The DNA extraction and ddPCR were performed according to the protocols above mentioned.
These 100 DNA samples were also analyzed by qPCR (Pepe et al., 2021), always amplifying the 529 bp repeat element, to compare the results obtained by ddPCR. Briefly, a final qPCR reaction volume of 20 μL was prepared containing: 1X PCR Master Mix (Bio-Rad, Hercules, CA, United States), 500 nM of each primer (AF1 and AF2 reported in Table 1), 250 nM of TaqMan probe described in Table 1 (Eurofins Genomics, Ebersberg, Germany) and 2 μL of template DNA. Moreover, Internal Amplification controls (IACs) (Applied Biosystems, Waltham, MA, United States), were used to verify the amplification reaction. The amplification conditions were as follows: 50°C for 2 min, 95°C for 10 min, followed by 40 cycles: 95°C for 15 s and 60°C for 1 min. A standard reference curve was prepared, using the four concentrations of T. gondii DNA (prepared as mentioned before) in triplicate. The quantified DNA target is expressed as the number of genomic copies/1 μL of reaction. Specificity of primers was evaluated in silico using the NCBI nucleotide BLAST tool and in lab using known positive samples for other two protozoa which can be found in mollusks, Giardia duodenalis and Cryptosporidium parvum.
The Standards for Reporting of Diagnostic Accuracy Studies (STARD) checklist (https://www.equator-network.org/reporting-guidelines/stard/) was used to improve our information on performances of techniques (Cohen et al., 2016).
Sensitivity, specificity, negative and positive predictive values (NPV and PPV) were calculated for ddPCR. The agreement between qPCR and ddPCR was calculated using Cohen’s κ statistic (Thrusfield, 2007).
The κ measure was interpreted as follows: 0, no agreement; 0.01–0.20, poor agreement; 0.21–0.40, fair agreement; 0.41–0.60, moderate agreement; 0.61–0.80, substantial agreement; and 0.81–1.0, nearly perfect agreement (Thrusfield, 2007).
The intra-assay coefficient of variation (CV) for each of the four dilution levels and overall were calculated using the following formula: [(CV% = standard deviation [SD]/mean value for each concentration level) × 100].
The 95% confidence intervals (95% CIs) were calculated using the free online software “Sample Size Calculator” (Creative Research Systems, CA, United States).
The ddPCR allowed DNA amplification up to 8 gc/μL (Figure 2). Samples which showed ≥ two droplets were considered positive. All infected replicates resulted positive, as well as no droplets were detected in negative controls. The droplets produced in the reaction ranged from 8,828 to 14,075 (average 12,627 droplets). The data obtained by ddPCR showed a good separation between negative and positive droplets with few interface droplets, highlighting the high specificity and efficiency of this technique. In fact, the sensitivity and specificity of ddPCR were 100% (95%CI = 94.3–99.9). An overall CV% = 14.8 was calculated for all the ddPCR positive replicates for all the concentration levels. The CV% for each concentration level were: 8000 gc/μL CV% = 7.8, 800 gc/μL CV% = 14.6, 80 gc/μL CV% = 16.6, 8 gc/μL CV% = 20.0.
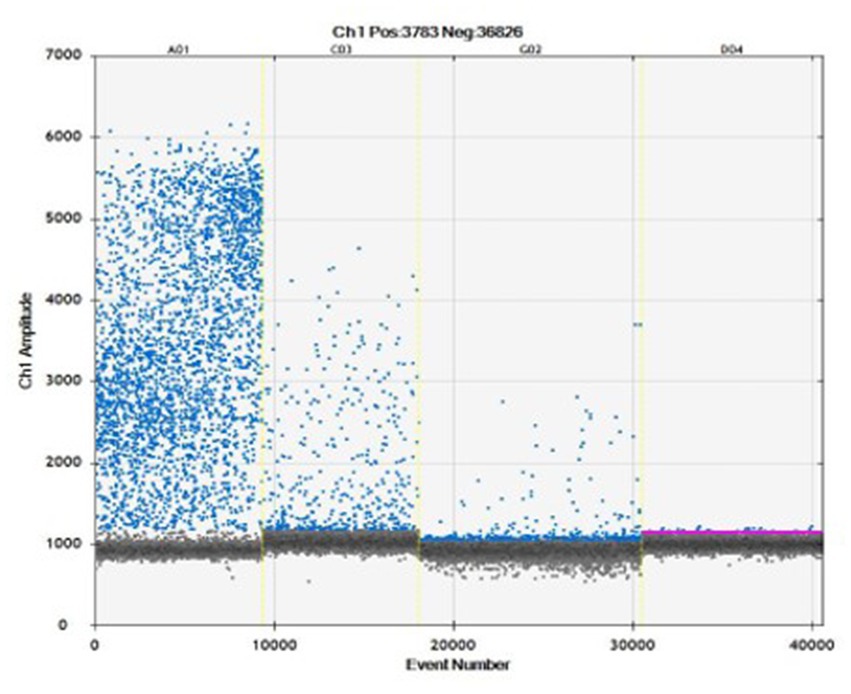
Figure 2. Amplification plot obtained to evaluate four concentrations: 8000 gc/μL (A01), 800 gc/μL (C03), 80 gc/μL (G02), 8 gc/μL (D04) in ddPCR.
No significant intra-laboratory variation in results was reported (CV% < 0.1).
Of the 100 mussel pools (composed by 10 individual samples) examined, the ddPCR detected 16 positive samples (16.0%; 95%CI = 9.7–25.0), with concentrations ranging from 0.1 to 1.9 gc/μL. No positive samples at T. gondii were detected by the qPCR reference method. Therefore, no agreement was found between ddPCR and qPCR (κ = 0). Samples examined showed no inhibitory effects on qPCR, as demonstrated by the results obtained using the IACs. No cross-reactions with G. duodenalis or C. parvum amplification were found in silico or in lab, showing a specificity of 100% by ddPCR.
All performance of the ddPCR has been summarized in Table 2.
Although, T. gondii is one of the most widespread zoonotic foodborne protozoa, no systematic food controls are carried out for this parasite, caused mainly by the lack of specific regulations and standardized methods to detect the parasite in food matrices (Pinto-Ferreira et al., 2019; Gabriël et al., 2022).
Recent outbreaks due to contamination with T. gondii oocysts in the environment have highlighted significant risks to public health. Indeed, several studies showed that oocysts are very resistant for months and years in the environment, including the marine environment (López Ureña et al., 2022). Oocysts were found in wild and commercial bivalve mollusks (clams, oysters, and mussels) with detection rates varied between 2.1% (Fung et al., 2021) and 6.6% (Ghozzi et al., 2017) in clams from Canada and Tunisia, respectively; from 1.3% (Silva et al., 2020) to 31.0% (Marquis et al., 2019) in oysters from Brazil and United States, respectively; from 1.4% (Shapiro et al., 2015) to 46.3% (Staggs et al., 2015) in different species of mussels from United States (López Ureña et al., 2022). T gondii DNA has been found also in different fish species ranging from 2.9 to 100% (depending on fish species; Marino et al., 2019), in sea otters (54.8%) and cetaceans (30.9%) (Ahmadpour et al., 2022).
Unfortunately, it is not easy to compare different studies for the detection of T. gondii in marine animals including mollusks, because several molecular methods and protocols are used (i.e., singleplex PCR, (semi-)nested, PCR–restriction fragment length polymorphism, qPCR), characterized by different performance (sensitivity, specificity, accuracy, precision, repeatability, detection limit) (Nayeri et al., 2021; Marín-García et al., 2022). The most common targets employed are the B1 gene or the 529 bp DNA repeat element which are multi-copy loci that increase the sensitivity of T. gondii detection (López Ureña et al., 2022). In particular, the 529 bp repeat element resulted more sensitive and accurate of B1 also at low concentrations of T. gondii DNA (Edvisson et al., 2006; Sağlam et al., 2021). However, more performant tools are in continuous development. In this study better results were obtained by ddPCR than qPCR (16.0% of positive samples for T. gondii obtained by ddPCR vs 0% by qPCR), confirming that this technique is very useful to increase the sensitivity, accuracy and precision for detection and quantification of small amounts of DNA of the pathogens to be recognized, preserving all the advantages of the qPCR (Kao et al., 2021). This ddPCR protocol was previously successfully used for the detection of T. gondii in meat, showing 7.6% of positive sample vs 1.2% by qPCR (Mancusi et al., 2022).
For these reasons, the ddPCR could be very useful for a rapid sensitive detection of low DNA concentrations of T. gondii, in order to perform a standardized food inspection on several matrices, reducing the public health risk of this parasite.
However, our preliminary results need to be confirmed by further studies on field samples.
Finally, this study confirms the presence of T. gondii in mussels in Campania region, as previously showed by Santoro et al., 2020, but the prevalence of T. gondii found in our study in mussels collected in Gulf of Naples is higher (16% vs 10.2% in Santoro et al., 2020). Moreover, this occurrence is higher also than other previous studies on clams, mussels and oyster collected in other two Italian regions: Apulia and Sardinia (Putignani et al., 2011; Tedde et al., 2019), highlighting the need of ministerial regulations to prevent toxoplasmosis infection.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The manuscript presents research on animals that do not require ethical approval for their study.
AM, YP, AG, and SG performed sampling and laboratory analyses. AM, LR, FC, and MM conceived the study. FD’O, RP, PS, LR, FC, and MM supervised the study. MM and AM wrote the manuscript. All authors contributed to manuscript revision, read and approved the submitted version.
This study was supported by the Regional Project “Control and reduction of toxoplasmosis in animals and humans – ToxoCamp,” Campania Region, Italy.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Ahmadpour, E., Rahimi, M. T., Ghojoghi, A., Rezaei, F., Hatam-Nahavandi, K., Oliveira, S. M. R., et al. (2022). Toxoplasma gondii infection in marine animal species, as a potential source of food contamination: a systematic review and Meta-analysis. Acta Parasitol. 67, 592–605. doi: 10.1007/s11686-021-00507-z
Almeria, S., and Dubey, J. P. (2021). Foodborne transmission of toxoplasma gondii infection in the last decade. An overview. Res. Vet. Sci. 135, 371–385. doi: 10.1016/j.rvsc.2020.10.019
Caradonna, T., Marangi, M., Del Chierico, F., Ferrari, N., Reddel, S., Bracaglia, G., et al. (2017). Detection and prevalence of protozoan parasites in ready-to-eat packaged salads on sale in Italy. Food Microbiol. 67, 67–75. doi: 10.1016/j.fm.2017.06.006
Cohen, J. F., Korevaar, D. A., Altman, D. G., Bruns, D. E., Gatsonis, C. A., Hooft, L., et al. (2016). STARD 2015 guidelines for reporting diagnostic accuracy studies: explanation and elaboration. BMJ Open 6:e012799. doi: 10.1136/bmjopen-2016-012799
Dámek, F., Swart, A., Waap, H., Jokelainen, P., le Roux, D., Deksne, G., et al. (2023). Systematic review and modelling of age-dependent prevalence of toxoplasma gondii in livestock, wildlife and felids in Europe. Pathogens 12:97. doi: 10.3390/pathogens12010097
Dubey, J. P. (2022). Toxoplasmosis of animals and humans. 3rd, (Boca Raton, Florida, USA: CRC press Taylor & Francis Group).
Edvisson, B., Lappalainen, M., and Evengard, B., ESCMID Study Group for Toxoplasmosis (2006). Real-time PCR targeting a 529-bp repeat element for diagnosis of toxoplasmosis. Clin. Microbiol. Infect. 12, 131–136. doi: 10.1111/j.1469-0691.2005.01332.x
EFSA European Centre for Disease Prevention and Control (2021). The European Union one health 2020 Zoonoses report. EFSA J. Sci. Opin. 19:6971. doi: 10.2903/j.efsa.2021.6971
EFSA Panel on Biological Hazards (2018). Public health risks associated with food-borne parasites. EFSA J. Sci. Opin. 16:5495. doi: 10.2903/j.efsa.2018.5495
FAO/WHO (2014). Multicriteria-based ranking for risk management of food-borne parasites. Report of a joint FAO/WHO expert meeting, 3–7 September 2012, FAO Headquarters, Rome, Italy.
Fayer, R. (2004). Cryptosporidium: a water-borne zoonotic parasite. Vet. Parasitol. 126, 37–56. doi: 10.1016/j.vetpar.2004.09.004
Fung, R., Manore, A. J. W., Harper, S. L., Sargeant, J. M., Shirley, J., Caughey, A., et al. (2021). Clams and potential foodborne toxoplasma gondii in Nunavut Canada. Zoonoses Public Health 68, 277–283. doi: 10.1111/zph.12822
Gabriël, S., Dorny, P., Saelens, G., and Dermauw, V. (2022). Foodborne parasites and their complex life cycles challenging food safety in different food chains. Foods 12:142. doi: 10.3390/foods12010142
Géba, E., Rousseau, A., Le Guernic, A., Escotte-Binet, S., Favennec, L., La Carbona, S., et al. (2021). Survival and infectivity of toxoplasma gondii and Cryptosporidium parvum oocysts bioaccumulated by Dreissena polymorpha. J. Appl. Microbiol. 130, 504–515. doi: 10.1111/jam.14802
Ghozzi, K., Marangi, M., Papini, R., Lahmar, I., Challouf, R., Houas, N., et al. (2017). First report of Tunisian coastal water contamination by protozoan parasites using mollusk bivalves as biological indicators. Mar. Pollut. Bull. 117, 197–202. doi: 10.1016/j.marpolbul.2017.01.057
Hatam-Nahavandi, K., Calero-Bernal, R., Rahimi, M. T., Pagheh, A. S., Zarean, M., Dezhkam, A., et al. (2021). Toxoplasma gondii infection in domestic and wild felids as public health concerns: a systematic review and meta-analysis. Sci. Rep. 4;11(1):9509 11:9509. doi: 10.1038/s41598-021-89031-8
Kao, Y. F., Peake, B., Madden, R., Cowan, S. R., Scimeca, R. C., Thomas, J. E., et al. (2021). A probe-based droplet digital polymerase chain reaction assay for early detection of feline acute cytauxzoonosis. Vet. Parasitol. 292:109413. doi: 10.1016/j.vetpar.2021.109413
Lindsay, D. S., and Dubey, J. P. (2009). Long-term survival of toxoplasma gondii sporulated oocysts in seawater. J. Parasitol. 95, 1019–1020. doi: 10.1645/GE-1919.1
López Ureña, N. M., Chaudhry, U., Calero Bernal, R., Cano Alsua, S., Messina, D., Evangelista, F., et al. (2022). Contamination of soil, water, fresh produce, and bivalve mollusks with toxoplasma gondii oocysts: A systematic review. Microorganisms 10:517. doi: 10.3390/microorganisms10030517
Mancusi, A., Giordano, A., Bosco, A., Girardi, S., Proroga, Y. T. R., Morena, L., et al. (2022). Development of a droplet digital polymerase chain reaction tool for the detection of Toxoplasma gondii in meat samples. Parasitol. Res. 121, 1467–1473. doi: 10.1007/s00436-022-07477-9
Marín-García, P.-J., Planas, N., and Llobat, L. (2022). Toxoplasma gondii in foods: prevalence, control, and safety. Foods 11:2542. doi: 10.3390/foods11162542
Marino, A. M. F., Giunta, R. P., Salvaggio, A., Castello, A., Alfonzetti, T., Barbagallo, A., et al. (2019). Toxoplasma gondii in edible fishes captured in the Mediterranean basin. Zoonoses Public Health 66, 826–834. doi: 10.1111/zph.12630
Marquis, N. D., Bishop, T. J., Record, N. R., Countway, P. D., and Fernández, R. (2019). Molecular Epizootiology of toxoplasma gondii and Cryptosporidium parvum in the eastern oyster (Crassostrea virginica) from Maine (USA). Pathogens 8:125. doi: 10.3390/pathogens8030125
Miller, M. A., Grigg, M. E., Kreuder, C., James, E. R., Melli, A. C., Crosbie, P. R., et al. (2004). An unusual genotype of Toxoplasma gondii is common in California sea otters (Enhydra lutris nereis) and is a cause of mortality. Int. J. Parasitol. 34, 275–284. doi: 10.1016/j.ijpara.2003.12.008
Nayeri, T., Sarvi, S., and Daryani, A. (2021). Toxoplasma gondii in mollusks and cold-blooded animals: a systematic review. Parasitology 148, 895–903. doi: 10.1017/S0031182021000433
Pepe, P., Bosco, A., Capuano, F., Baldi, L., Giordano, A., Mancusi, A., et al. (2021). Towards an integrated approach for monitoring toxoplasmosis in southern Italy. Animals 11, 1949–1961. doi: 10.3390/ani11071949
Pinto-Ferreira, F., Caldart, E. T., Pasquali, A. K. S., Mitsuka-Breganó, R., Freire, R. L., and Navarro, I. T. (2019). Patterns of transmission and sources of infection in outbreaks of human toxoplasmosis. Emerg. Infect. Dis. 25, 2177–2182. doi: 10.3201/eid2512.181565
Putignani, L., Mancinelli, L., Del Chierico, F., Menichella, D., Adlerstein, D., Angelici, M. C., et al. (2011). Investigation of toxoplasma gondii presence in farmed shellfish by nested-PCR and real-time PCR fluorescent amplicon generation assay (FLAG). Exp. Parasitol. 127, 409–417. doi: 10.1016/j.exppara.2010.09.007
Rosário, M. K. S. D., Silva, J., Melo, H. F. R., Monteiro, T. R. M., Costa, D. R. D., Scofield, A., et al. (2021). Molecular investigation of toxoplasma gondii in oysters (Crassostrea spp.) sold on beaches in the state of Pará Brazil. Rev. Bras. Parasitol. Vet. 12:e023320. doi: 10.1590/S1984-29612021004
Sağlam, T., Düşen, S., Mete, E., and Karaman, U. (2021). Comparative evaluation of re 529-Bp sequence and B1 gene in the detection of toxoplasma gondii through PCR in water samples of Denizli Turkey. Acta Parasitol. 67, 555–559. doi: 10.1007/s11686-021-00494-1
Santoro, M., Viscardi, M., Boccia, F., Borriello, G., Lucibelli, M. G., Auriemma, C., et al. (2020). Parasite load and STRs genotyping of toxoplasma gondii isolates from Mediterranean mussels (Mytilus galloprovincialis) in southern Italy. Front. Microbiol. 11:355. doi: 10.3389/fmicb.2020.00355
Shapiro, K., VanWormer, E., Aguilar, B., and Conrad, P. A. (2015). Surveillance for Toxoplasma gondii in California mussels (Mytilus californianus) reveals transmission of atypical genotypes from land to sea. Environ. Microbiol. 17, 4177–4188. doi: 10.1111/1462-2920.12685
Silva, C. M., Silva, A. L. P., Watanabe, K. F. C., Chaves Bezerra, N. P., Bezerra, D. C., Gomes, H. M., et al. (2020). First report of detection of Toxoplasma gondii DNA in oysters (Crassostrea sp.) in the state of Maranhão. Rev. Bras. Parasitol. 29:e003720. doi: 10.1590/s1984-29612020050
Staggs, S. E., Keely, S. P., Ware, M. W., Schable, N., See, M. J., Gregorio, D., et al. (2015). The development and implementation of a method using blue mussels (Mytilus spp.) as biosentinels of Cryptosporidium spp. and Toxoplasma gondii contamination in marine aquatic environments. Parasitol. Res. 114, 4655–4667. doi: 10.1007/s00436-015-4711-9
Tedde, T., Marangi, M., Papini, R., Salza, S., Normanno, G., Virgilio, S., et al. (2019). Toxoplasma gondii and other zoonotic protozoans in Mediterranean mussel (Mytilus galloprovincialis) and blue mussel (Mytilus edulis): A food safety concern? J. Food Prot. 82, 535–542. doi: 10.4315/0362-028X.JFP-18-157
Keywords: droplet digital polymerase chain reaction, mollusks, mussels, Toxoplasma gondii , toxoplasmosis
Citation: Mancusi A, Proroga YTR, Giordano A, Girardi S, D’Orilia F, Pinto R, Sarnelli P, Rinaldi L, Capuano F and Maurelli MP (2023) First application of a droplet digital PCR to detect Toxoplasma gondii in mussels. Front. Microbiol. 14:1238689. doi: 10.3389/fmicb.2023.1238689
Received: 12 June 2023; Accepted: 29 August 2023;
Published: 08 September 2023.
Edited by:
Panagiotis Karanis, University of Nicosia, CyprusReviewed by:
Hamed Mirjalali, Shahid Beheshti University of Medical Sciences, IranCopyright © 2023 Mancusi, Proroga, Giordano, Girardi, D’Orilia, Pinto, Sarnelli, Rinaldi, Capuano and Maurelli. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maria Paola Maurelli, bWFyaWFwYW9sYS5tYXVyZWxsaUB1bmluYS5pdA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.