
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 07 September 2023
Sec. Microbial Physiology and Metabolism
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1238482
 Yin Li
Yin Li Ming Ni*
Ming Ni*Klebsiella pneumoniae is an important Gram-negative opportunistic pathogen that is responsible for a variety of nosocomial and community-acquired infections. Klebsiella pneumoniae has become a major public health issue owing to the rapid global spread of extensively-drug resistant (XDR) and hypervirulent strains. Biofilm formation is an important virulence trait of K. pneumoniae. A biofilm is an aggregate of microorganisms attached to an inert or living surface by a self-produced exo-polymeric matrix that includes proteins, polysaccharides and extracellular DNA. Bacteria within the biofilm are shielded from antibiotics treatments and host immune responses, making it more difficult to eradicate K. pneumoniae-induced infection. However, the detailed mechanisms of biofilm formation in K. pneumoniae are still not clear. Here, we review the factors involved in the biofilm formation of K. pneumoniae, which might provide new clues to address this clinical challenge.
Klebsiella pneumoniae is an important pathogenic Gram-negative, nonmotile bacterium that is responsible for a variety of common infections including urinary tract infections (UTIs), pneumonia, bacteremia, purulent liver abscesses, and wound infection (Karyoute, 1989; Bouza et al., 2001; Luo et al., 2014; Mohamudha et al., 2016; Bart et al., 2021; D’Abbondanza and Shahrokhi, 2021; GBD 2019 Antimicrobial Resistance Collaborators, 2022). Given the rapid spread of extensively-drug resistant (XDR) (mainly carbapenem resistant K. pneumoniae [CRKP]) and hypervirulent strains worldwide, K. pneumoniae has become a major problem for public health. In 2020, the Antimicrobial Testing Leadership and Surveillance (ATLAS) program collected a total of 6,753 K. pneumoniae isolates from 57 countries across six regions worldwide. Of these isolates, 1,118 (16.6%) were CRKP strains (Lee et al., 2022). Because of the limitation of treatment options, CRKP is considered an urgent clinical threat. It was estimated that antibiotic resistant K. pneumoniae was responsible for more than 600,000 deaths globally in 2019 (Antimicrobial Resistance Collaborators, 2022). Hypervirulent K. pneumoniae (hvKP), a more virulent evolving variant of K. pneumoniae, is known to cause community-acquired, metastatic, and life-threatening infections such as pyogenic liver abscesses (PLA), central nervous system infection and endophthalmitis, which require rapid recognition and site-specific treatment (Russo and Marr, 2019). Moreover, via acquisition of carbapenem-resistant plasmids or hvKP-specific virulence determinants, XDR-hvKP strains are emerging (Han et al., 2022; Tian et al., 2022). The prevalence of the XDR-hvKP and its potential threat to human health are of concern.
Biofilm formation is an important virulence trait for K. pneumoniae (Khodadadian et al., 2018; Tang et al., 2020; Shadkam et al., 2021). A biofilm is a type of polymerization wherein microorganisms attach to inert or active surfaces through the extracellular polymeric substances (EPS) that they produce (Ashwath et al., 2022). The EPS is largely composed of polysaccharides, proteins, nucleic acids, lipids, and extracellular DNA (eDNA) (Bertoglio et al., 2018). The polysaccharides of K. pneumoniae biofilms are composed of mannose, glucose, their amines, and acetylated counterparts. The expression of the proteins in biofilms are different owing to the heterogeneity of the environment (Singh et al., 2019). Although the kinetics of biofilm formation vary among different strains, the process of biofilm formation is similar, including initial attachment, micro-colony formation, maturation, and dispersion (Bandeira et al., 2014; Koo et al., 2017; Al-Bayati and Samarasinghe, 2022). Each step requires many bacterial functions, such as exercise, adhesion, transportation, stress response, metabolic pathway activation, and extracellular matrix synthesis (Beloin et al., 2004; Domka et al., 2007).
The overall properties of biofilms are endowed by the biofilm matrix and can protect the resident cells against desiccation, chemical interference and other bacterial invasion. Moreover, biofilms help bacterial cells resist killing by the human phagocytic system, and they ensure that the biofilm community continues to adhere to the media surface (Yan and Bassler, 2019). It was estimated that the resistance of bacterial biofilm to antimicrobial agents might be 10–1,000-times higher than planktonic bacteria (de la Fuente-Núñez et al., 2016). Additionally, the bacteria in these fixed communities are often in close proximity to one another, which also increases the probability of chemical signal transduction and gene transfer between bacterial cells of the same or different species (Cook and Dunny, 2014; Koraimann and Wagner, 2014). This provides more convenient conditions for the spread of drug resistance genes. A previous report suggested that 65–80% of bacterial infections had some connection with biofilm formation (Al-Bayati and Samarasinghe, 2022). In patients with recurrent infections, it was observed that most serial isolates recovered from these patients were strong biofilm producers in vitro (Sanchez et al., 2013).
Nosocomial infections caused by K. pneumoniae are facilitated by the ability of the organism to form biofilms (Bandeira et al., 2014). It was reported that the majority of clinically isolated K. pneumoniae formed biofilms. Among the isolated K. pneumoniae strains, 54%, 29%, and 14% were strong, moderate and weak biofilm producers, respectively. The major sources of strong biofilm producers were isolated from urine, pus, and blood, whereas moderate biofilm producers were isolated from blood (Ashwath et al., 2022).
This review focuses on K. pneumoniae biofilms and summarizes various factors and genes affecting biofilm formation in K. pneumoniae.
The capsule affects different stages of biofilm formation of K. pneumoniae. It controls the initial adhesion through a series of behaviors, such as improving the regular initial spatial distribution and preventing bacterial interactions as much as possible (Balestrino et al., 2008). Davis et al. pointed out that the capsule was necessary for constructing an appropriate initial covering of mature biofilm structure (Davis and Brown, 2020). The expression of capsular polysaccharide also ensures that K. pneumoniae forms a typical three-dimensional mature biofilm structure (Balestrino et al., 2008).
The capsule is a toxic bacterial component. So far, researchers have identified 134 capsule synthesis loci (K loci) (Wyres et al., 2016). In a study using signature-tagged mutagenesis (STM) to screen a K. pneumoniae mutant library with unique characteristic markers to identify genes related to biofilm formation, the authors found that mutations in capsule gene cluster sites could lead to defects in biofilm formation (Boddicker et al., 2006). Mutants that insert transposons in the capsular wza and wzc loci show defective biofilm formation (Wu et al., 2011). Balestrino et al. (2008) discovered that the biofilm forming ability of K. pneumoniae ORF4 (wza homologous, transport of capsular polysaccharides) and ORF14 (glycosyl transferase, capsule biosynthesis) mutants on polyvinyl-chloride (PVC) was significantly reduced.
Further, treC and sugE have been shown to affect biofilm formation of K. pneumoniae by regulating the production of capsular polysaccharide (CPS). TreC encodes trehalose-6-phosphate hydrolase, and its deletion affects bacterial utilization of trehalose. TreC mutants show reduced mucus viscosity and produce less CPS, thereby reducing biofilm formation and preventing formation of advanced biofilm structures in K. pneumoniae. Adding glucose to the culture medium of K. pneumoniae treC mutant strains can restore CPS production and biofilm formation. However, a sugE (encoding an intima protein) mutant that increases biofilm formation in K. pneumoniae shows higher mucus viscosity and produces more CPS. It was suggested that the absence of sugE in K. pneumoniae would lead to changes in bacterial membrane structure and activate the downstream cascade, thus increasing CPS production during biofilm formation (Wu et al., 2011).
Biofilm formation in isolates containing magA (K1), rmpA+, rmpA2+, the virulence factors related to capsule production, is more obvious than in isolates with negative virulence factors. However, multivariate regression analysis showed that wcaG was the only independent risk factor for biofilm (Zheng et al., 2018). The wcaG positive genotype was involved in K1 and K54 capsular types, and was less associated with K16 and K58 capsular types (Turton et al., 2010). In addition, wcaG encodes the protein participating in the biosynthesis of fucose, and the deletion mutation of wcaG affects most capsule polysaccharide genes (Ho et al., 2011), thereby suggesting that wcaG may affect biofilm formation by changing the composition of capsule polysaccharides (Zheng et al., 2018).
One study also showed that the capsule could inhibit biofilm formation in K. pneumoniae. A recent study showed that strains lacking the wbaP gene, which was related to capsule production, formed stronger biofilms. While the strains with the super capsule containing wzc mutation could not form biofilms (Ernst et al., 2020). When carbohydrates were added to the medium, the biosynthesis of CPS increased, but the biofilm formation in K. pneumoniae decreased (Chen et al., 2020). A previous study showed that the expression of CPS in K. pneumoniae physically interrupted the function of type 1 fimbriae, hindered the biofilm formation mediated by fimbriae, and reduced the adhesion of bacteria to the surface (Schembri et al., 2005). In addition, CPS could inhibit bacterial surface interactions on non-biological substrates (dos Santos Goncalves et al., 2014). It was found that the capsule was costly in nutrient rich media, but it provided obvious adaptive advantages under conditions of malnutrition. Further, among strains forming more biofilms, the capsule often played a positive role in biofilm formation. The authors suggested that this was not because of the presence or absence of capsules, but was instead caused by the amount of capsule expressed by a given strain, which then affected biofilm formation. Moreover, the function of the capsule was not conservative in different isolates, but relied on other elements of the genome or serotype (Buffet et al., 1946).
The mechanism by which the capsule influences biofilm formation and the conditions under which it has positive or negative regulation on biofilm formation are still unclear. However, there seems to be a relationship with the O antigen, because the polysaccharide capsule is retained on the outer surface of the bacteria by interacting with the repeat sugar molecule of the lipopolysaccharide (LPS) molecule, namely “O antigen.” The waaL gene encodes a ligase involved in the connection of the LPS repeat O antigen to the LPS core, and inactivation of this gene is understood to lead to a significant reduction in capsule retention and an increase in biofilm formation (Singh et al., 2022).
The K. pneumoniae genome encodes for several types of fimbriae. Fimbriae are hair-like protein appendages extending from the cell surface (Wilksch et al., 2011). Fimbriae promote K. pneumoniae adhesion to non-biological surfaces, resulting in catheter related infections (Schroll et al., 2010). Type 1 and type 3 fimbriae, the most studied fimbriae, are encoded by fim and mrk gene clusters, respectively. In addition, ecp and kpa to kpg gene clusters found in recent years also encode fimbriae (Wu et al., 2010; Alcántar-Curiel et al., 2013).
In K. pneumoniae, biofilm formation is mainly mediated by type 3 fimbriae, and the Mrk protein is encoded by the operon containing the mrkABCDF gene (Allen et al., 1991). Type 3 fimbriae are mainly made up of the main fimbriae subunit, MrkA, which polymerizes to form a spiral fimbriae axis (Murphy and Clegg, 2012). In addition, ΔmrkA mutants are unable to attach to the abiotic surface to form biofilms (Di Martino et al., 2003; Jagnow and Clegg, 2003). Further, MrkA protein expression is significantly upregulated during biofilm thickening (Vuotto et al., 2017). MrkB and MrkC have sequence characteristics representing the periplasmic chaperone and usher translocatase, respectively. MrkD, present on the top of the fimbriae surface, also has adhesion characteristics of appendages, and determines specificity of fimbriae binding (Murphy and Clegg, 2012). The mrkA and mrkD genes play a key role in the biofilm formation of K. pneumoniae (Fang et al., 2021). The mrkA gene contributes to rapid biofilm formation while mrkD contributes to form dense K. pneumoniae biofilms (Ashwath et al., 2022). A gene cluster, mrkHIJ, adjacent to the type 3 fimbriae operon is related to the regulation of type 3 fimbriae expression. MrkH is a new transcription activator of the mrk gene cluster, which regulate mrkHI expression and contains the PilZ domain. MrkH binds to the region upstream of the mrkA promoter and activates the expression of the mrkABCDF operon. Therefore, mrkH is often referred to as a “biofilm switch” as it can initiate expression of genes involved in producing type 3 fimbriae (Wilksch et al., 2011; Tan et al., 2015). The biofilm formation capacity of K. pneumoniae carrying the mrkH box was clearly higher than strains without it (Fang et al., 2021). Wu et al. (2012) found that mrkHI transcription could be activated by MrkI. MrkI is a LuxR-like regulatory factor. The mrkI mutant reduces the mannose-resistant Klebsiella-like (MR/K) hemagglutinins (HA) activity and the number of type 3 fimbriae on the cell surface, leading to a significant reduction in biofilm formation, which can be rescued when providing wild type mrkI copies (Johnson et al., 2011; Wilksch et al., 2011). The expression of mrkHI is also actively regulated by Fur, which usually acts as a transcriptional activator to directly activate the transcription of mrkHI. The deletion of Fur reduces mrkH, mrkI, and mrkA transcription, thereby reducing type 3 fimbriae expression and biofilm formation (Wu et al., 2012). In addition, at least two components of pulmonary surfactant, phosphatidylcholine and cholesterol, promote the transcription of type 3 fimbriae genes and biofilm formation of K. pneumoniae (Willsey et al., 2018).
The type 3 fimbriae-dependent adhesion is probably the initial stage of K. pneumoniae colonization and biofilm formation on non-biological surfaces (Duguid, 1959). Type 3 fimbriae mediate the binding to the surface of damaged epithelium as they can bind to the extracellular matrix of urinary and respiratory tissues (Tamayo et al., 2007).
Type 3 fimbriae not only participate during the initial stages of K. pneumoniae biofilm formation, but also mediate the c-di-GMP dependent bacterial growth mode transformation from planktonic to biofilm. c-di-GMP is an important second messenger in bacteria (Tamayo et al., 2007). The activity of diguanylate cyclase (DGC) and phosphodiesterase (PDE) can regulate the intracellular concentration of c-di-GMP in bacteria (Simm et al., 2004; Hengge, 2009). The mrkHIJ gene cluster is associated with the regulation and sensing of c-di-GMP (Lin et al., 2016). When activated by c-di-GMP, MrkH recruits RNA polymerase to the mrkHI promoter to auto-activate mrkH expression. Increased MrkH production subsequently drives the expression of mrkABCDF, leading to type 3 fimbriae biosynthesis and biofilm formation (Tan et al., 2015). MrkJ encodes a hypothetical phosphodiesterase (PDE) which contains an EAL domain (the sequences encoding diguanylate cyclase and phosphodiesterase A share a lengthy consensus motif, comprising two adjacent domains termed GGDEF and EAL) mediating the hydrolysis of c-di-GMP (Tal et al., 1998; Johnson and Clegg, 2010). Because of intracellular accumulation of c-di-GMP, the absence of mrkJ leads to an increase in the production of type 3 fimbriae and biofilm formation (Johnson and Clegg, 2010; Wilksch et al., 2011). YjcC possesses PDE activity in the recombinant protein of its EAL domain. After receiving oxidative stress signal input, YjcC actively regulates oxidative stress responses by changing the level of c-di-GMP and has a negative impact on type 3 fimbriae expression and biofilm formation (Huang et al., 2013). YfiN harbors DGC domain plays a positive role in the expression of type 3 fimbriae (Wilksch et al., 2011). OmpR/EnvZ is a two-component system that senses osmotic signals and controls downstream gene expression in many species of Enterobacteriaceae. In response to osmotic stresses, the phosphorylated form of OmpR of K. pneumoniae regulates the expression of type 3 fimbriae to influence biofilm formation via modulating the level of intracellular c-di-GMP and MrkHIJ (Lin et al., 2018).
Type 1 fimbriae, also known as mannose sensitive fimbriae, can bind soluble mannose as a competitive inhibitor, as the name suggests. Type 1 fimbriae are encoded by the fim gene cluster, which is composed of eight genes (fimAICDFGHK) (Gomes et al., 2020). Regulation of fim gene expression is controlled by reversible DNA elements (fimS). Type 1 fimbriae are composed of a major fimbriae subunit, FimA, and a minor apical adhesion protein, FimH (Alcántar-Curiel et al., 2013). FimH is a component that promotes adhesion to the host surface and contains mannose (Sahly et al., 2008). The increased expression of the fimH gene plays an important role in the binding of bacteria to surfaces, leading to strong biofilm formation (Ashwath et al., 2022). The regulatory gene fimK constitutes the fim operon, and FimK has an EAL domain with PDE characteristics, which can regulate intracellular levels of c-di-GMP (Johnson and Clegg, 2010). Mutant strains that cannot produce FimK have a higher fimbriate shape than wild type K. pneumoniae and can be planted in the urinary tract of mice (Johnson and Clegg, 2010). FimK can reduce type 1 fimbriae, and inhibit biofilm formation and intracellular bacterial communities (Rosen et al., 2008). Type 1 fimbriae are key causes of UTIs (Struve et al., 2008) and they have high affinity of mannose residues on bladder cell surfaces (Rozen and Skaletsky, 2000), which can promote the adhesion and invasion of epithelial bladder cells, thus forming biofilm-like intracellular bacterial communities (Rosen et al., 2008). The regulation mechanisms of the biosynthesis of type 3 and type 1 fimbriae are shown in Figure 1.
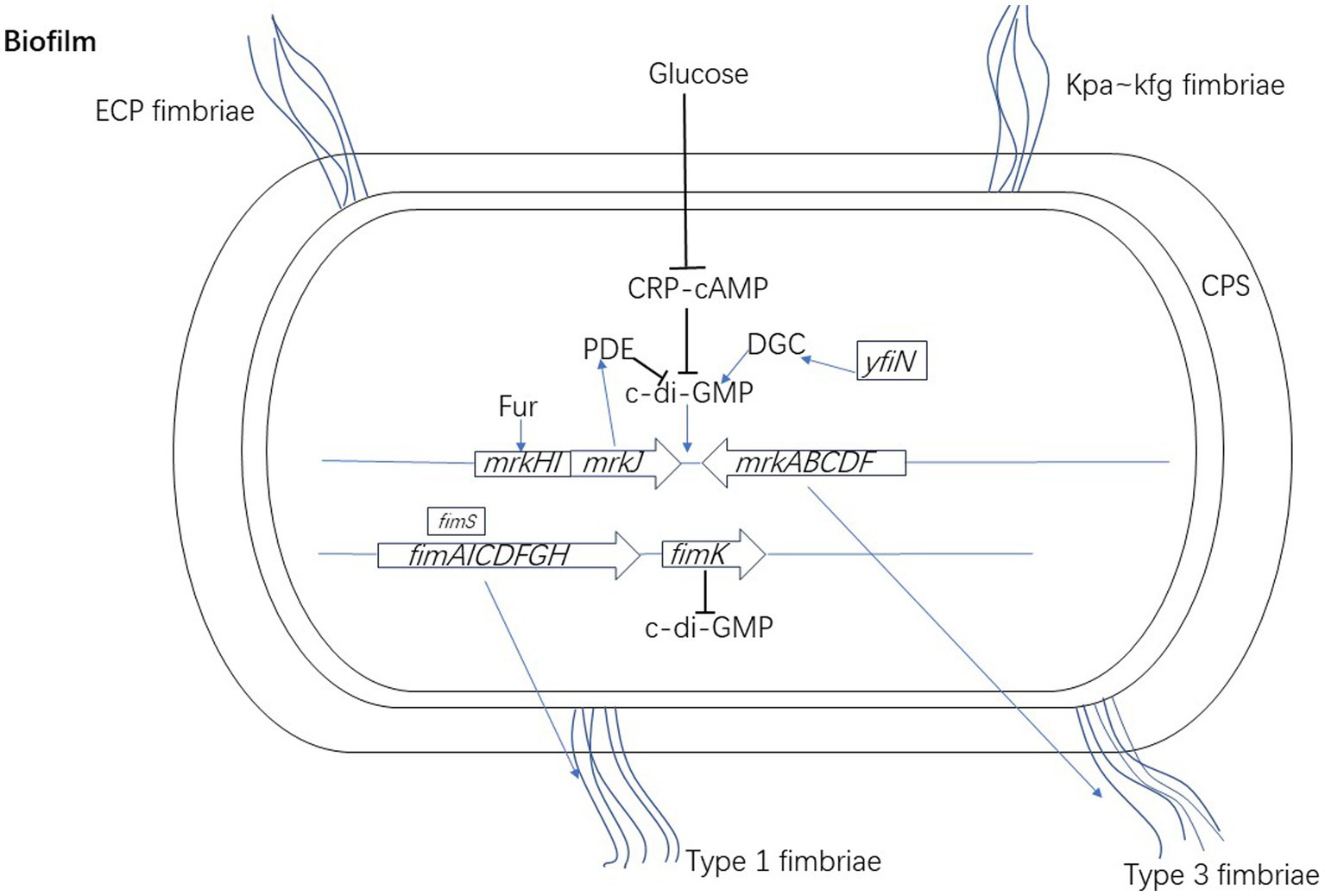
Figure 1. The regulation mechanisms of the biosynthesis of type 3 and type 1 fimbriae of K. pneumoniae. When activated by c-di-GMP, MrkH recruits RNA polymerase to the mrkHI promoter to auto-activate mrkH expression. Increased MrkH production subsequently drives the expression of mrkABCDF, leading to type 3 fimbriae biosynthesis and biofilm formation. The expression of mrkHI is also actively regulated by Fur, which usually acts as a transcriptional activator to directly activate the transcription of mrkHI. MrkJ encodes PDE which contains an EAL domain mediating the hydrolysis of c-di-GMP. YfiN harbors DGC domain plays a positive role in the expression of type 3 fimbriae by changing the level of c-di-GMP. CRP-cAMP indirectly regulates the expression of type 3 fimbriae through inhibiting the c-di-GMP signal pathway. Type 1 fimbriae are encoded by the fim gene cluster. The regulation of fim gene expression is controlled by fimS. The regulatory gene fimK constitutes the fim operon, and FimK can regulate intracellular levels of c-di-GMP.
Type 1 and type 3 fimbriae can contribute to biofilm formation and compensate for each other (Stahlhut et al., 2012; Murphy et al., 2013; Ashwath et al., 2022). One study using the catheter bladder model, found that type 1 and type 3 fimbriae enhance biofilm formation on catheters (Stahlhut et al., 2012). Moreover, it was reported that gene clusters of type 3 and type 1 fimbriae have a cross regulatory effect, and the up-regulation of type 1 fimbriae can make up for the loss of type 3 fimbriae expression (Schroll et al., 2010). Type 3 fimbriae may have a more significant effect on biofilm formation than type 1 fimbriae (Fang et al., 2021). Bacterial strains that cannot produce type 1 fimbriae are as proficient as bacterial strains that can produce such fimbriae in biofilm formation (Clegg and Murphy, 2016).
Alcantar-Curiel et al. found that except for type 3 and type 1 fimbriae, the Escherichia coli common pilus (ECP) fimbriae gene cluster in the K. pneumoniae genome has an operon that is homologous to the E. coli ECP fimbriae. The ECP fimbriae gene cluster contains the ecpRABCDE gene and importantly, 90% of K. pneumoniae strains can produce ECP fimbriae. Ultrastructural and immunoassay analysis of K. pneumoniae showed that ECP can bind bacteria to each other, thus forming specific micro-colonies on cultured epithelial cells and stable biofilms on inert surfaces. ECP likely also plays an important role in cell adhesion, biofilm formation and several niche colonization, especially for isolates lacking MrkD adhesin or the entire type 3 fimbriae (Alcántar-Curiel et al., 2013).
Wu et al. (2010) found seven new fimbriae gene clusters in K. pneumoniae, namely kpa, kpb, kpc, kpd, kpe, kpf, and kpg. The loss of kpgC resulted in an obvious decrease in biofilm formation, adhesion to animal cells, and intestinal colonization in mice. Further, ΔkpaC and ΔkpeC mutants were also found to weaken biofilm formation and adhesion to Arabidopsis cells, respectively. The deletion of the kpjC usher-coding gene was shown to significantly reduce biofilm formation, while the loss of the kpaC usher gene was shown to only affect early and late stages of biofilm formation (Khater et al., 2015). The kpf gene cluster encodes type 1-like fimbriae, while the kpfR gene encoding the transcription inhibitor of the kpf gene cluster negatively regulates the expression of fimbriae. K. pneumoniae lacking the kpfR gene showed a hyperfimbriated phenotype and enhanced adhesion to epithelial host cells and biofilm formation (Gomes et al., 2020).
During biofilm formation, QS mediates inter-specific or intra-specific interactions through which bacterial cells communicate with each other (Bassler et al., 1993; Miller and Bassler, 2001). The QS system regulates the synthesis of fimbriae, exopolysaccharides, adhesins, and other substances through signaling molecules, thus affecting biofilm formation in bacteria (Yang et al., 2013; Gu et al., 2021). Depending on bacterial cell density, bacteria will produce and detect specific signaling molecules called auto inducers (AIs) to coordinate their gene expression (Bassler et al., 1993; Miller and Bassler, 2001). There are two main types of intercellular QS regulatory systems, namely type I and type II.
Type I QS is mainly used for intraspecific communication, which is usually related to the LuxI/LuxR system. LuxI synthetase produces N-acyl homoserine lactone (AHL) as an AI, and the LuxR transcription factor is their homologous receptor. However, K. pneumoniae does not produce AHL (Balestrino et al., 2005), rather encodes SdiA, which is an orphan LuxR receptor that reacts with exogenous AHL molecules produced by other bacteria (Pacheco et al., 2021). SdiA plays a repressive role in the expression of type 1 fimbriae in K. pneumoniae. Cells lacking SdiA regulator presents a hyperfimbriated phenotype that render the ΔsdiA mutant strain with a greater ability to form biofilm and agglutinate yeast cells (Pacheco et al., 2021).
Type II QS has an interspecific communication function, enabling bacteria to react not only to AI-2 produced by other species, but also to their own AI-2 (Chen et al., 2002; Zhang et al., 2020). De Araujo et al. observed that K. pneumoniae lacking AI-2 output (ΔtqsA) or input (ΔlsrCD) systems showed an increased surface coverage after growth in dynamic micro-fermentation but decreased biofilm thickness. In addition, production of AI-2 relies on the presence of luxS but the biofilm structure of ΔluxS mutants is different. In these mutants the surface coverage rate is lower, and fewer large colonies are formed. Mutations related to luxS and AI-2 transport systems both induce increased expression of wbbM and wzm in connection with LPS synthesis, which indicates that QS affects biofilm formation through LPS in K. pneumoniae (De Araujo et al., 2010). The regulation mechanisms of the QS in biofilm formation of K. pneumoniae is shown in Figure 2.
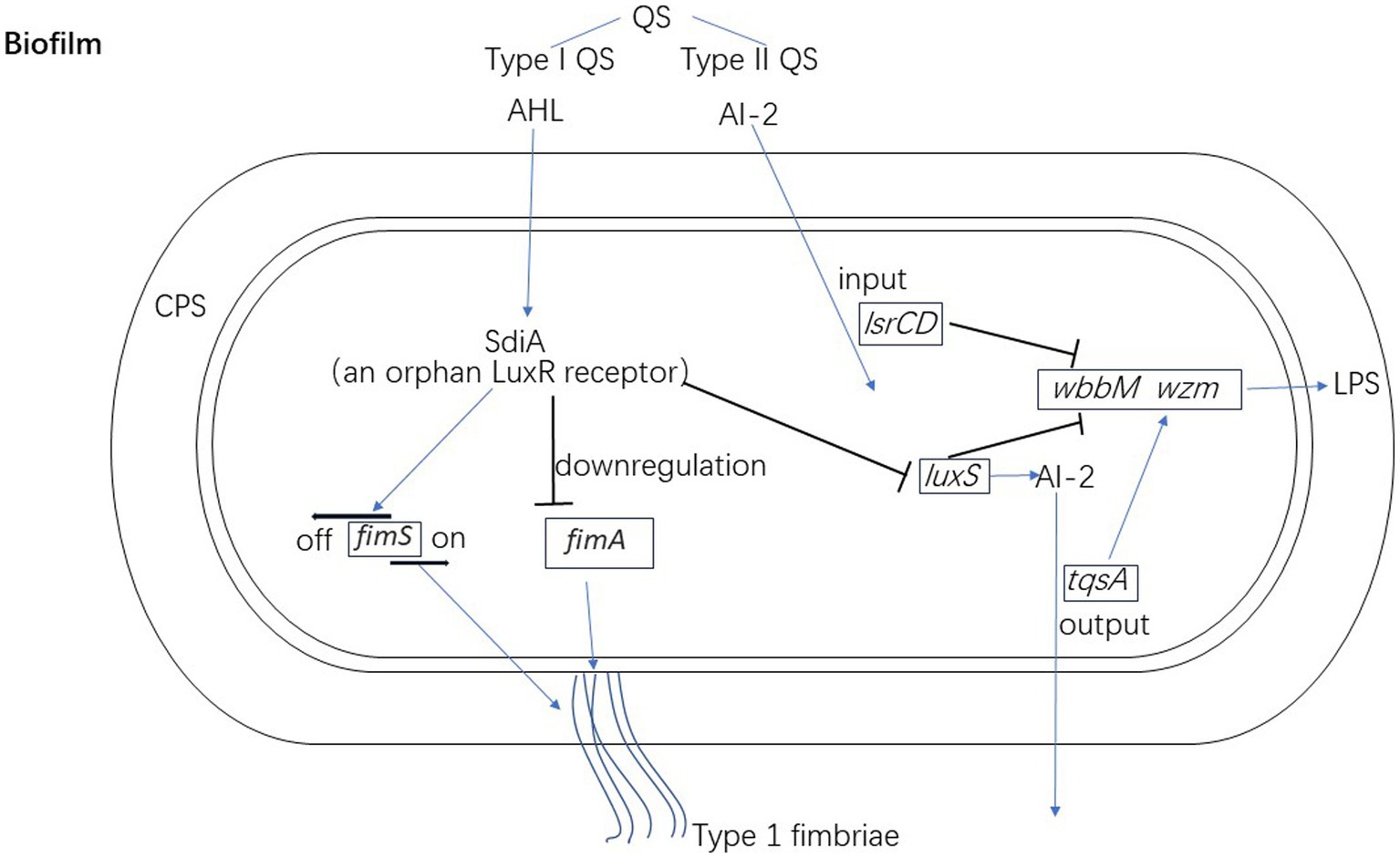
Figure 2. The regulation mechanisms of quorum qensing in biofilm formation of K. pneumoniae. Klebsiella pneumoniae encodes SdiA as an orphan LuxR receptor to down-regulate the expression of fimA (type 1 fimbriae) and luxS. SdiA also regulates the promoter region of the fimS at OFF orientation. AI-2 transport systems (tqsA and lsrCD) and luxS of K. pneumoniae both regulate the expression of wbbM and wzm in connection with LPS synthesis.
Nutritional conditions are also an important factor for biofilm formation. Excess nutrition may promote the planktonic growth model, while malnutrition environments are more favorable for the biofilm growth model (Stanley and Lazazzera, 2004).
Previous studies have found that high concentration of sugars (such as glucose) prohibit biofilm formation in K. pneumoniae and E. coli (Jackson et al., 2002; Sutrina et al., 2015). Glucose-rich medium inhibits the production of cyclic AMP (cAMP), a well-known second messenger that has important effects on gene regulation (Rickenberg, 1974). Furthermore, cAMP forms a homodimer (CRP-cAMP) with its signal transduction target, cAMP receptor protein (CRP), and then combines with the CRP binding site in the DNA promoter region to regulate mRNA transcription. External glucose inhibits the function of CRP-cAMP in K. pneumoniae. CRP indirectly regulates the expression of type 3 fimbriae through the c-di-GMP signal pathway (Lin et al., 2016). In addition, CRP mediates catabolite repression. The absence of CRP increases the concentration of c-di-GMP and reduces the activity of PDE in cells. The expression of mrkHI depends on c-di-GMP which in turn increases the expression of MrkH and MrkI, leading to the high expression of type 3 fimbriae. It was reported that inserting an open-reading frame containing CRP-activation domain into K. pneumoniae resulted in biofilm deficiency (Boddicker et al., 2006). However, other studies found that crp mutant K. pneumoniae strains could not express MrkA, the major subunit of the fimbrial shaft, which indicated that CRP was required for fimbriae production and biofilm formation (Ou et al., 2017; Panjaitan et al., 2019). These studies indicate the important regulating role of CRP in biofilm formation of K. pneumoniae.
Cellobiose also affects biofilm formation in K. pneumoniae. It has previously been shown that the celB deletion mutation, which leads to cellobiose deficiency, clearly decreased biofilm formation in K. pneumoniae. Moreover, celB encodes the cellobiose-specific subunit IIC of enzyme II (EIIC) of a carbohydrate phosphotransferase system (PTS, a sugar transport system in bacteria) (Wu et al., 2012). Horng et al. (2018) showed that a non-characteristic enzyme II complex homolog of PTS in K. pneumoniae actively regulated biofilm formation by enhancing eDNA and capsular polysaccharide production.
Different carbon sources can also affect the biofilm formation of K. pneumoniae. The isolates formed more robust biofilms when grown with fucose as the sole carbon source than with glucose or glycerol. It was related to the positive modulates of fucose to hypermucoviscosity of K. pneumoniae (Hudson et al., 2022).
The presence of bile salts can stimulate biofilm formation in K. pneumoniae, which is related to the production of poly-β-1,6-N-acetyl-d-glucosamine (PNAG) (Chen et al., 2014). PNAG is a common bacterial surface polysaccharide and a significant component of the biofilm EPS (Chen et al., 2020). PAGA, which is encoded by pgaABCD (Cywes-Bentley et al., 2013), mediates the intercellular binding of bacterial species and surface adhesion. The biofilm formation in pgaA mutants was shown to be significantly decreased (Wu et al., 2011). The loss of pgaC in K. pneumoniae reduces PNGA production, and significantly affects the enhancement of 1% bile salt mixture on K. pneumoniae biofilm (Chen et al., 2014).
Iron is indispensable in K. pneumoniae growth and virulence factor expression (Chhibber et al., 2013; Chen et al., 2020). A study showed that a certain concentration of iron (0.16 mM FeCl2) could promote biofilm formation in K. pneumoniae by inhibiting succinic acid. This may be due to a reduction in protein and polysaccharide expression in the biofilm EPS since succinic acid participates in pyruvate metabolism and amino acid synthesis (Liu et al., 2022). Chen et al. observed that biofilm formation was strongest when K. pneumoniae was cultured in LB broth supplemented with 50 μM iron. When the strain was cultured with an iron chelator, biofilm formation decreased (Chen et al., 2020). Chhibber et al. studied the biofilm formation of K. pneumoniae in the presence of Co [II] (iron antagonist ions) and depolymerase producing phage (degrading extracellular polysaccharides on biofilm structure). A significant reduction was observed in the growth of younger biofilms (1–3 days old) when 500 μM CoSO4 and 10 μM FeCl3 supplemented medium was used. Moreover, a complete eradication of the younger biofilms was observed when both elements were present (Chhibber et al., 2013).
The use of some drugs will instead promote biofilm formation. In the presence of sub-MICs of cefotaxime, the biomass increased and was positively related to the antibiotic concentration (Hennequin et al., 2012). When CRKP strain was under antibiotic pressure, the expression of the Psp and Pho family genes [PspB-PspC complex is a pressure receptor that plays a role in molecular switch during the process of biofilm pressure response (Flores-Kim and Darwin, 2016)] was induced, thus further mediating the downstream stress responses, and compensating for the adsorption, colonization, and biofilm formation (Bowen et al., 2021). Cadavid et al. (2018) found that in K. pneumoniae, hydrochlorothiazide and acetaminophen could promote biofilm formation.
Antibiotic-resistant genes in special plasmids can regulate the biofilm formation of K. pneumoniae (Maeyama et al., 2004). Multidrug-resistant (MDR) K. pneumoniae often forms stronger biofilms than non-MDR strains (Shadkam et al., 2021). The plasmid encoding cephalosporin enzyme was shown to obtain a transcription factor, namely AmpR, which was involved in upregulating capsule synthesis and antiserum killing, regulating the expression of type 3 fimbriae and biofilm formation (Hennequin et al., 2012). In addition, compared to the control strains, strong biofilm formation was found in NDM-1 producing K. pneumoniae. Moreover, the resistance genes blaNDM-1 of K. pneumoniae were observed to be maximally up-regulated in 24 h-biofilms (Al-Bayati and Samarasinghe, 2022).
Bacterial efflux pumps are made up of transmembrane proteins, which export a variety of harmful substances including different types of antibiotics from the intracellular environment to the external environment. This process is one of the causes of MDR (Du et al., 2018). The role of efflux pumps in biofilm formation is still controversial. Tang et al. found that the efflux pump inhibitor, CCCP, had a dose-dependent effect on biofilm formation (Knight et al., 2018). In another study, researchers found that the up-regulation of the AcrAB multidrug efflux system was observed only in XDR strains with biofilm growth that could be considered an essential factor in the biofilm-forming ability in K. pneumoniae (Vuotto et al., 2017). In turn, biofilms have been shown to up-regulate K. pneumoniae efflux pump genes acrA, emrB, oqxA, and qacEΔ1 (Tang et al., 2020). However, some studies have suggested that there is no correlation between the expression of efflux pump genes (acrA, kexD, kdeA, kpnEF and ketM) and biofilm formation (Türkel et al., 2018).
The physical environment of bacteria will affect biofilm formation. Some physical and chemical properties of the surface on which bacteria grow may interfere with biofilm formation by damaging the initial bacterial attachment to the surface (Bos et al., 1999; Li and Logan, 2004). Biofilm production of K. pneumoniae decreased at 37°C compared to at 30°C, but the difference was not significant (Hostacká et al., 2010). In another experiment with 17 CRKP isolates, biofilm formation was greater at 37° C than at 25° C (Gual-de-Torrella et al., 2022). An increase in the pH of culture medium led to an increase in biofilm formation, and in K. pneumoniae this increased by 151–319% at pH 8.5 and by 113–177% at pH 7.5, respectively compared to at pH 5.5 (Hostacká et al., 2010).
Klebsiella pneumoniae growing under simulated microgravity (SMG) conditions formed a thicker biofilm than those growing under normal gravity conditions. Moreover, under SMG conditions, the cellulose production and expression of type 3 fimbriae of K. pneumoniae were enhanced. Therefore, K. pneumoniae isolated from orbital spacecrafts poses a potential threat to the health of astronauts (Wang et al., 2016).
Recently, the CRISPR-Cas9 technique has been implemented to eliminate certain bacteria by use of bacteriophages or bacterial conjugation. This technique allows targeted editing of genomes by inducing double-stranded-DNA breaks (DSBs). However, a novel type of biofilm (“R-biofilm”) was found in clinical isolates of K. pneumoniae after DSBs. R-biofilms are mainly made up of extracellular proteins and/or DNA, which may be released by dead bacteria. In addition to bacterial SOS reaction (severe DNA damage in cells will result in SOS reaction), new signaling pathways also participate in the formation of R-biofilms. Furthermore, R-biofilms form a fixed ring or disk shape with better ductility, which can protect living bacterial cells in the body from harmful conditions such as exposure to ethanol, hydrogen peroxide, and ultraviolet radiation. The discovery of R-biofilms indicated the limited effect of the current popular Cas9-mediated sterilization tools, because the resulting DSBs may facilitate the formation of these new protective biofilms (Liu et al., 2020).
In the past decades we have gained considerable knowledge about the molecular mechanisms involved in the biofilm formation of K. pneumoniae. Similar to other bacteria, biofilm formation of K. pneumoniae is an adaptive response to various stressors such as nutritional deficiency, physical environment change, and drugs (especially antibiotics). Biofilm formation is not a precisely conserved process, the pattern of biofilm formation of K. pneumoniae is similar to other Gram-negative bacteria (Ruhal and Kataria, 2021). For example, O antigen of LPS is related to the production of capsule polysaccharide of K. pneumoniae and influences biofilm formation, which is common in Gram-negative bacteria (Fedtke et al., 2007; Lee et al., 2016). In general, all flagellated bacteria approach by motility and condition the surface by the secretion of polysaccharides to help cells adhere. As mentioned previously, K. pneumoniae use type 1 and type 3 fimbriae to adhere to surfaces (Schroll et al., 2010). Pseudomonas aeruginosa also use flagellar motility to reach surfaces and subsequently use type IV pili motility to crawl on surface (Zhao et al., 2013). Regulation by the two-component system via c-di-GMP are involved in the biofilm formation of most Gram-negative bacteria, including K. pneumoniae (Ruhal and Kataria, 2021). QS plays significant roles in biofilm formation and dispersal (Solano et al., 2014). The QS molecules are various in different bacteria. K. pneumoniae encodes SdiA as an AI to inhibit biofilm formation (Pacheco et al., 2021). However, P. aeruginosa produce AHL as an AI to influence biofilm formation (Pesci et al., 1997). Owing to the rapid spread of CRKP and hvKP, the most interesting aspect of K. pneumoniae biofilm formation is the impact of carbapenem-resistant plasmids or hvKP-specific virulence plasmids on biofilm formation.
The various factors and genes affecting biofilm formation in K. pneumoniae are shown in Figure 3 and Table 1. Some controversy remains regarding certain factors in different studies. Moreover, the mechanisms by which the above factors affect the biofilm formation of K. pneumoniae requires further study. For instance, the role and the molecular mechanisms of the capsule of K. pneumoniae in biofilm formation is still unclarified. In summary, realizing the commonality and specifics of biofilm formation between K. pneumoniae and other bacteria will lead to a deep understanding of bacterial interactions within natural or host infection environment. Furthermore, it would be helpful to develop new therapeutic strategies for K. pneumoniae biofilm.
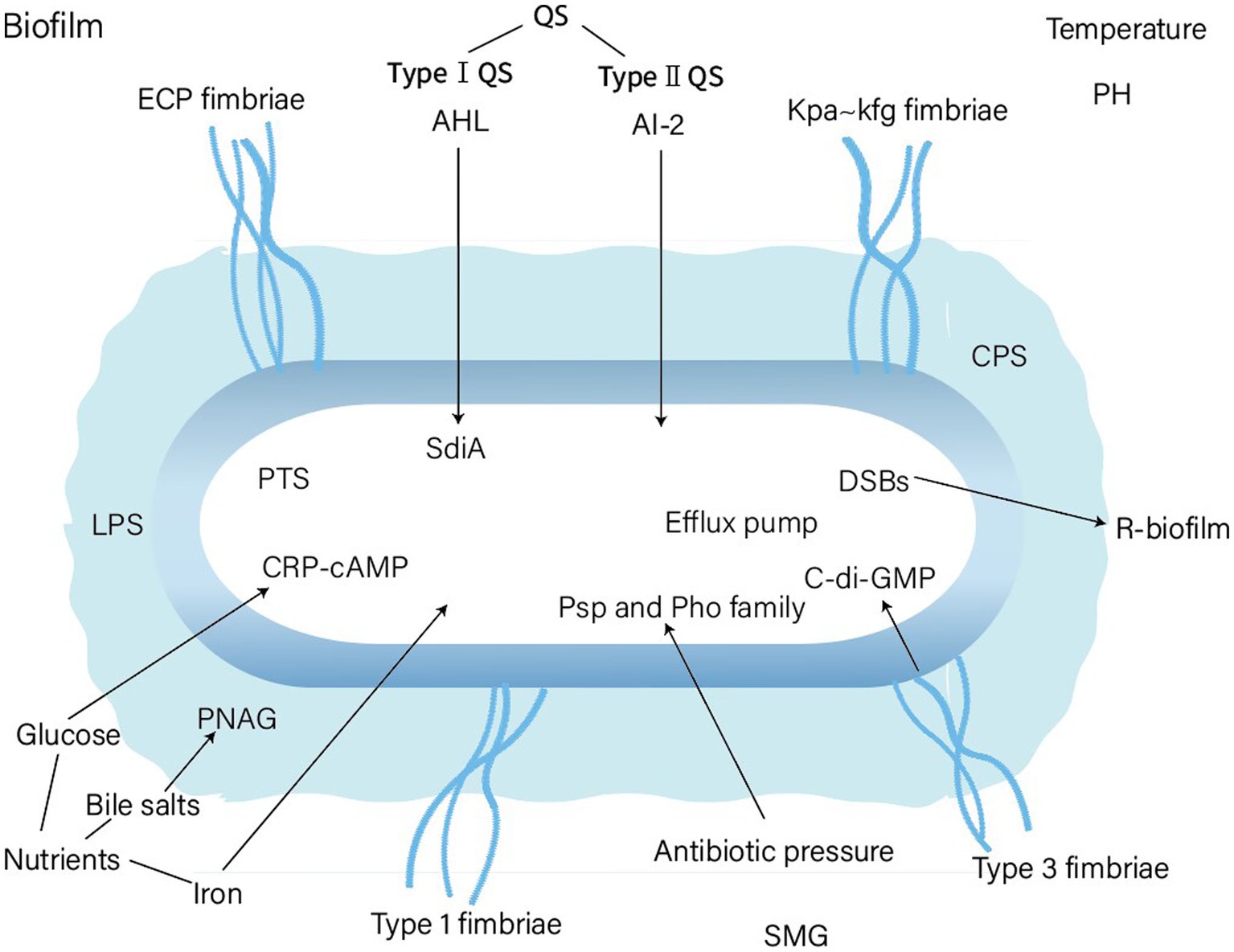
Figure 3. Factors affecting biofim formation of Klebsiella pneumoniae. PTS, phosphotransferase system; PNAG, Poly-β-1,6-N-acetyl-d-glucosamine; DSBs, double strand DNA breaks; SMG, simulated microgravity.
YL wrote the manuscript and searched for references. MN developed the concept and added valuable insights into the manuscript. All authors have read and agreed to the published version of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Al-Bayati, M., and Samarasinghe, S. (2022). Biofilm and gene expression characteristics of the Carbapenem-resistant Enterobacterales, Escherichia coli IMP, and Klebsiella pneumoniae NDM-1 associated with common bacterial infections. Int. J. Environ. Res. Public Health 19:4788. doi: 10.3390/ijerph19084788
Alcántar-Curiel, M. D., Blackburn, D., Saldaña, Z., Gayosso-Vázquez, C., Iovine, N., de la Cruz, M. A., et al. (2013). Multi-functional analysis of Klebsiella pneumoniae fimbrial types in adherence and biofilm formation. Virulence 4, 129–138. doi: 10.4161/viru.22974
Allen, B. L., Gerlach, G. F., and Clegg, S. (1991). Nucleotide sequence and functions of mrk determinants necessary for expression of type 3 fimbriae in Klebsiella pneumoniae. J. Bacteriol. 173, 916–920. doi: 10.1128/jb.173.2.916-920.1991
Antimicrobial Resistance Collaborators (2022). Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399, 629–655. doi: 10.1016/S0140-6736(21)02724-0
Ashwath, P., Deekshit, V. K., Rohit, A., Dhinakaran, I., Karunasagar, I., Karunasagar, I., et al. (2022). Biofilm formation and associated gene expression in multidrug-resistant Klebsiella pneumoniae isolated from clinical specimens. Curr. Microbiol. 79:73. doi: 10.1007/s00284-022-02766-z
Balestrino, D., Ghigo, J. M., Charbonnel, N., Haagensen, J. A. J., and Forestier, C. (2008). The characterization of functions involved in the establishment and maturation of Klebsiella pneumoniae in vitro biofilm reveals dual roles for surface exopolysaccharides. Environ. Microbiol. 10, 685–701. doi: 10.1111/j.1462-2920.2007.01491.x
Balestrino, D., Haagensen, J. A. J., Rich, C., and Forestier, C. (2005). Characterization of type 2 quorum sensing in Klebsiella pneumoniae and relationship with biofilm formation. J. Bacteriol. 187, 2870–2880. doi: 10.1128/JB.187.8.2870-2880.2005
Bandeira, M., Carvalho, P. A., Duarte, A., and Jordao, L. (2014). Exploring dangerous connections between Klebsiella pneumoniae biofilms and healthcare-associated infections. Pathogens 3, 720–731. doi: 10.3390/pathogens3030720
Bart, S. M., Rubin, D., Kim, P., Farley, J. J., and Nambiar, S. (2021). Trends in hospital-acquired and ventilator-associated bacterial pneumonia trials. Clin. Infect. Dis. 73, e602–e608. doi: 10.1093/cid/ciaa1712
Bassler, B. L., Wright, M., Showalter, R. E., and Silverman, M. R. (1993). Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol. Microbiol. 9, 773–786. doi: 10.1111/j.1365-2958.1993.tb01737.x
Beloin, C., Valle, J., Latour-Lambert, P., Faure, P., Kzreminski, M., Balestrino, D., et al. (2004). Global impact of mature biofilm lifestyle on Escherichia coli K-12 gene expression. Mol. Microbiol. 51, 659–674. doi: 10.1046/j.1365-2958.2003.03865.x
Bertoglio, F., Bloise, N., Oriano, M., Petrini, P., Sprio, S., Imbriani, M., et al. (2018). Treatment of biofilm communities: an update on new tools from the Nanosized world. Appl. Sci. 8:845. doi: 10.3390/app8060845
Boddicker, J. D., Anderson, R. A., Jagnow, J., and Clegg, S. (2006). Signature-tagged mutagenesis of Klebsiella pneumoniae to identify genes that influence biofilm formation on extracellular matrix material. Infect. Immun. 74, 4590–4597. doi: 10.1128/IAI.00129-06
Bos, R., van der Mei, H. C., and Busscher, H. J. (1999). Physico-chemistry of initial microbial adhesive interactions – its mechanisms and methods for study. FEMS Microbiol. Rev. 23, 179–229. doi: 10.1016/S0168-6445(99)00004-2
Bouza, E., Juan, R. S., Muñoz, P., Voss, A., and Kluytmans, J. (2001). A European perspective on nosocomial urinary tract infections II. Report on incidence, clinical characteristics and outcome (ESGNI-004 study). European study group on nosocomial infection. Clin. Microbiol. Infect. 7, 532–542. doi: 10.1046/j.1198-743x.2001.00324.x
Bowen, T., Yingang, X., Junhong, L., Hongbin, T., and Fengming, W. (2021). Pressure response of carbapenems Klebsiella pneumoniae under antibiotic stress. Infect. Genet. Evol. 92:104915. doi: 10.1016/j.meegid.2021.104915
Buffet, A., Rocha, E. P. C., and Rendueles, O. (1946). Nutrient conditions are primary drivers of bacterial capsule maintenance in Klebsiella. Proc. Biol. Sci. 2021:20202876. doi: 10.1098/rspb.2020.2876
Cadavid, E., Robledo, S. M., Quiñones, W., and Quiñones, W. (2018). Induction of biofilm formation in Klebsiella Pneumoniae ATCC 13884 by several drugs: the possible role of quorum sensing modulation. Antibiotics 7:103. doi: 10.3390/antibiotics7040103
Chen, K. M., Chiang, M. K., Wang, M., Ho, H. C., Lu, M. C., and Lai, Y. C. (2014). The role of pgaC in Klebsiella pneumoniae virulence and biofilm formation. Microb. Pathog. 77, 89–99. doi: 10.1016/j.micpath.2014.11.005
Chen, T., Dong, G., Zhang, S., Zhang, X., Zhao, Y., Cao, J., et al. (2020). Effects of iron on the growth, biofilm formation and virulence of Klebsiella pneumoniae causing liver abscess. BMC Microbiol. 20:36. doi: 10.1186/s12866-020-01727-5
Chen, X., Schauder, S., Potier, N., van Dorsselaer, A., Pelczer, I., Bassler, B. L., et al. (2002). Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415, 545–549. doi: 10.1038/415545a
Chen, L., Wilksch, J. J., Liu, H., Zhang, X., Torres, V. V. L., Bi, W., et al. (2020). Investigation of LuxS-mediated quorum sensing in Klebsiella pneumoniae. J. Med. Microbiol. 69, 402–413. doi: 10.1099/jmm.0.001148
Chhibber, S., Nag, D., and Bansal, S. (2013). Inhibiting biofilm formation by Klebsiella pneumoniae B5055 using an iron antagonizing molecule and a bacteriophage. BMC Microbiol. 13:174. doi: 10.1186/1471-2180-13-174
Clegg, S., and Murphy, C. N. (2016). Epidemiology and virulence of Klebsiella pneumoniae. Microbiol. Spectr. 4:10.1128. doi: 10.1128/microbiolspec.UTI-0005-2012
Cook, L. C. C., and Dunny, G. M. (2014). The influence of biofilms in the biology of plasmids. Microbiol. Spectr. 2:0012. doi: 10.1128/microbiolspec.PLAS-0012-2013
Cywes-Bentley, C., Skurnik, D., Zaidi, T., Roux, D., Deoliveira, R. B., Garrett, W. S., et al. (2013). Antibody to a conserved antigenic target is protective against diverse prokaryotic and eukaryotic pathogens. Proc. Natl. Acad. Sci. U. S. A. 110, E2209–E2218. doi: 10.1073/pnas.1303573110
D'Abbondanza, J. A., and Shahrokhi, S. (2021). Burn infection and burn Sepsis. Surg. Infect. 22, 58–64. doi: 10.1089/sur.2020.102
Davis, R. T., and Brown, P. D. (2020). spoT-mediated stringent response influences environmental and nutritional stress tolerance, biofilm formation and antimicrobial resistance in Klebsiella pneumoniae. APMIS 128, 48–60. doi: 10.1111/apm.13006
De Araujo, C., Balestrino, D., Roth, L., Charbonnel, N., and Forestier, C. (2010). Quorum sensing affects biofilm formation through lipopolysaccharide synthesis in Klebsiella pneumoniae. Res. Microbiol. 161, 595–603. doi: 10.1016/j.resmic.2010.05.014
de la Fuente-Núñez, C., Cardoso, M. H., de Souza Cândido, E., Franco, O. L., and Hancock, R. E. W. (2016). Synthetic antibiofilm peptides. Biochim. Biophys. Acta 1858, 1061–1069. doi: 10.1016/j.bbamem.2015.12.015
Di Martino, P., Cafferini, N., Joly, B., and Darfeuille-Michaud, A. (2003). Klebsiella pneumoniae type 3 pili facilitate adherence and biofilm formation on abiotic surfaces. Res. Microbiol. 154, 9–16. doi: 10.1016/S0923-2508(02)00004-9
Domka, J., Lee, J., Bansal, T., and Wood, T. K. (2007). Temporal gene-expression in Escherichia coli K-12 biofilms. Environ. Microbiol. 9, 332–346. doi: 10.1111/j.1462-2920.2006.01143.x
dos Santos Goncalves, M., Delattre, C., Balestrino, D., Charbonnel, N., Elboutachfaiti, R., Wadouachi, A., et al. (2014). Anti-biofilm activity: a function of Klebsiella pneumoniae capsular polysaccharide. PLoS One 9:e99995. doi: 10.1371/journal.pone.0099995
Du, D., Wang-Kan, X., Neuberger, A., van Veen, H. W., Pos, K. M., Piddock, L. J. V., et al. (2018). Multidrug efflux pumps: structure, function and regulation. Nat. Rev. Microbiol. 16, 523–539. doi: 10.1038/s41579-018-0048-6
Duguid, J. P. (1959). Fimbriae and adhesive properties in Klebsiella strains. J. Gen. Microbiol. 21, 271–286. doi: 10.1099/00221287-21-1-271
Ernst, C. M., Braxton, J. R., Rodriguez-Osorio, C. A., Zagieboylo, A. P., Li, L., Pironti, A., et al. (2020). Adaptive evolution of virulence and persistence in carbapenem-resistant Klebsiella pneumoniae. Nat. Med. 26, 705–711. doi: 10.1038/s41591-020-0825-4
Fang, R., Liu, H., Zhang, X., Dong, G., Li, J., Tian, X., et al. (2021). Difference in biofilm formation between carbapenem-resistant and carbapenem-sensitive Klebsiella pneumoniae based on analysis of mrkH distribution. Microb. Pathog. 152:104743. doi: 10.1016/j.micpath.2021.104743
Fedtke, I., Mader, D., Kohler, T., Moll, H., Nicholson, G., Biswas, R., et al. (2007). A Staphylococcus aureus ypfP mutant with strongly reduced lipoteichoic acid (LTA) content: LTA governs bacterial surface properties and autolysin activity. Mol. Microbiol. 65, 1078–1091. doi: 10.1111/j.1365-2958.2007.05854.x
Flores-Kim, J., and Darwin, A. J. (2016). Interactions between the cytoplasmic domains of PspB and PspC silence the Yersinia enterocolitica phage shock protein response. J. Bacteriol. 198, 3367–3378. doi: 10.1128/JB.00655-16
GBD 2019 Antimicrobial Resistance Collaborators (2022). Global mortality associated with 33 bacterial pathogens in 2019: a systematic analysis for the global burden of disease study 2019. Lancet 400, 2221–2248. doi: 10.1016/S0140-6736(22)02185-7
Gomes, A. E. I., Pacheco, T., Dos Santos, C. D. S., Pereira, J. A., Ribeiro, M. L., Darrieux, M., et al. (2020). Functional insights from KpfR, a new transcriptional regulator of Fimbrial expression that is crucial for Klebsiella pneumoniae pathogenicity. Front. Microbiol. 11:601921. doi: 10.3389/fmicb.2020.601921
Gu, Y., Tian, J., Zhang, Y., Wu, R., Li, L., Zhang, B., et al. (2021). Dissecting signal molecule AI-2 mediated biofilm formation and environmental tolerance in Lactobacillus plantarum. J. Biosci. Bioeng. 131, 153–160. doi: 10.1016/j.jbiosc.2020.09.015
Gual-de-Torrella, A., Delgado-Valverde, M., Pérez-Palacios, P., Oteo-Iglesias, J., Rojo-Molinero, E., Macià, M. D., et al. (2022). Prevalence of the fimbrial operon mrkABCD, mrkA expression, biofilm formation and effect of biocides on biofilm formation in carbapenemase-producing Klebsiella pneumoniae isolates belonging or not belonging to high-risk clones. Int. J. Antimicrob. Agents 60:106663. doi: 10.1016/j.ijantimicag.2022.106663
Han, Y. L., Wen, X. H., Zhao, W., Cao, X. S., Wen, J. X., Wang, J. R., et al. (2022). Epidemiological characteristics and molecular evolution mechanisms of carbapenem-resistant hypervirulent Klebsiella pneumoniae. Front. Microbiol. 13:1003783. doi: 10.3389/fmicb.2022.1003783
Hengge, R. (2009). Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 7, 263–273. doi: 10.1038/nrmicro2109
Hennequin, C., Aumeran, C., Robin, F., Traore, O., and Forestier, C. (2012). Antibiotic resistance and plasmid transfer capacity in biofilm formed with a CTX-M-15-producing Klebsiella pneumoniae isolate. J. Antimicrob. Chemother. 67, 2123–2130. doi: 10.1093/jac/dks169
Hennequin, C., and Forestier, C. (2009). oxyR, a LysR-type regulator involved in Klebsiella pneumoniae mucosal and abiotic colonization. Infect. Immun. 77, 5449–5457. doi: 10.1128/IAI.00837-09
Hennequin, C., Robin, F., Cabrolier, N., Bonnet, R., and Forestier, C. (2012). Characterization of a DHA-1-producing Klebsiella pneumoniae strain involved in an outbreak and role of the AmpR regulator in virulence. Antimicrob. Agents Chemother. 56, 288–294. doi: 10.1128/AAC.00164-11
Ho, J.-Y., Lin, T. L., Li, C. Y., Lee, A., Cheng, A. N., Chen, M. C., et al. (2011). Functions of some capsular polysaccharide biosynthetic genes in Klebsiella pneumoniae NTUH K-2044. PLoS One 6:e21664. doi: 10.1371/journal.pone.0021664
Horng, Y.-T., Dewi Panjaitan, N. S., Chang, H. J., Wei, Y. H., Chien, C. C., Yang, H. C., et al. (2022). A protein containing the DUF1471 domain regulates biofilm formation and capsule production in Klebsiella pneumoniae. J. Microbiol. Immunol. Infect. 55, 1246–1254. doi: 10.1016/j.jmii.2021.11.005
Horng, Y. T., Wang, C. J., Chung, W. T., Chao, H. J., Chen, Y. Y., and Soo, P. C. (2018). Phosphoenolpyruvate phosphotransferase system components positively regulate Klebsiella biofilm formation. J. Microbiol. Immunol. Infect. 51, 174–183. doi: 10.1016/j.jmii.2017.01.007
Hostacká, A., Ciznár, I., and Stefkovicová, M. (2010). Temperature and pH affect the production of bacterial biofilm. Folia Microbiol. 55, 75–78. doi: 10.1007/s12223-010-0012-y
Huang, C. J., Wang, Z. C., Huang, H. Y., Huang, H. D., and Peng, H. L. (2013). YjcC, a c-di-GMP phosphodiesterase protein, regulates the oxidative stress response and virulence of Klebsiella pneumoniae CG43. PLoS One 8:e66740. doi: 10.1371/journal.pone.0066740
Hudson, A. W., Barnes, A. J., Bray, A. S., Ornelles, D. A., and Zafar, M. A. (2022). Klebsiella pneumoniae l-Fucose metabolism promotes gastrointestinal colonization and modulates its virulence determinants. Infect. Immun. 90:e0020622. doi: 10.1128/iai.00206-22
Huertas, M. G., Zárate, L., Acosta, I. C., Posada, L., Cruz, D. P., Lozano, M., et al. (2014). Klebsiella pneumoniae yfiRNB operon affects biofilm formation, polysaccharide production and drug susceptibility. Microbiology 160, 2595–2606. doi: 10.1099/mic.0.081992-0
Jackson, D. W., Simecka, J. W., and Romeo, T. (2002). Catabolite repression of Escherichia coli biofilm formation. J. Bacteriol. 184, 3406–3410. doi: 10.1128/JB.184.12.3406-3410.2002
Jagnow, J., and Clegg, S. (2003). Klebsiella pneumoniae MrkD-mediated biofilm formation on extracellular matrix-and collagen-coated surfaces. Microbiology 149, 2397–2405. doi: 10.1099/mic.0.26434-0
Johnson, J. G., and Clegg, S. (2010). Role of MrkJ, a phosphodiesterase, in type 3 fimbrial expression and biofilm formation in Klebsiella pneumoniae. J. Bacteriol. 192, 3944–3950. doi: 10.1128/JB.00304-10
Johnson, J. G., Murphy, C. N., Sippy, J., Johnson, T. J., and Clegg, S. (2011). Type 3 fimbriae and biofilm formation are regulated by the transcriptional regulators MrkHI in Klebsiella pneumoniae. J. Bacteriol. 193, 3453–3460. doi: 10.1128/JB.00286-11
Karyoute, S. M. (1989). Burn wound infection in 100 patients treated in the burn unit at Jordan University hospital. Burns 15, 117–119. doi: 10.1016/0305-4179(89)90142-3.
Khater, F., Balestrino, D., Charbonnel, N., Dufayard, J. F., Brisse, S., and Forestier, C. (2015). In silico analysis of usher encoding genes in Klebsiella pneumoniae and characterization of their role in adhesion and colonization. PLoS One 10:e0116215. doi: 10.1371/journal.pone.0116215
Khodadadian, R., Rahdar, H. A., Javadi, A., Safari, M., and Khorshidi, A. (2018). Detection of VIM-1 and IMP-1 genes in Klebsiella pneumoniae and relationship with biofilm formation. Microb. Pathog. 115, 25–30. doi: 10.1016/j.micpath.2017.12.036
Knight, D. B., Rudin, S. D., Bonomo, R. A., and Rather, P. N. (2018). Defining the role of efflux pumps in resistance to antimicrobial therapy, surface motility, and biofilm formation. Front. Microbiol. 9:1902. doi: 10.3389/fmicb.2018.01902
Koo, H., Allan, R. N., Howlin, R. P., Stoodley, P., and Hall-Stoodley, L. (2017). Targeting microbial biofilms: current and prospective therapeutic strategies. Nat. Rev. Microbiol. 15, 740–755. doi: 10.1038/nrmicro.2017.99
Koraimann, G., and Wagner, M. A. (2014). Social behavior and decision making in bacterial conjugation. Front. Cell. Infect. Microbiol. 4:54. doi: 10.3389/fcimb.2014.00054
Lee, Y. L., Ko, W. C., and Hsueh, P. R. (2022). Geographic patterns of global isolates of carbapenem-resistant Klebsiella pneumoniae and the activity of ceftazidime/avibactam, meropenem/vaborbactam, and comparators against these isolates: results from the antimicrobial testing leadership and surveillance (ATLAS) program, 2020. Int. J. Antimicrob. Agents 60:106679. doi: 10.1016/j.ijantimicag.2022.106679
Lee, K. J., Lee, M. A., Hwang, W., Park, H., and Lee, K. H. (2016). Deacylated lipopolysaccharides inhibit biofilm formation by gram-negative bacteria. Biofouling 32, 711–723. doi: 10.1080/08927014.2016.1193595
Li, B., and Logan, B. E. (2004). Bacterial adhesion to glass and metal-oxide surfaces. Colloids Surf. B Biointerfaces 36, 81–90. doi: 10.1016/j.colsurfb.2004.05.006
Lin, T.-H., Chen, Y., Kuo, J. T., Lai, Y. C., Wu, C. C., Huang, C. F., et al. (2018). Phosphorylated OmpR is required for type 3 fimbriae expression in Klebsiella pneumoniae under hypertonic conditions. Front. Microbiol. 9:2405. doi: 10.3389/fmicb.2018.02405
Lin, D., Fan, J. M., Wang, J., Liu, L., Xu, L., Li, F., et al. (2018). The fructose-specific phosphotransferase system of Klebsiella pneumoniae is regulated by global regulator CRP and linked to virulence and growth. Infect. Immun. 86:e00340-18. doi: 10.1128/IAI.00340-18
Lin, C. T., Lin, T. H., Wu, C. C., Wan, L., Huang, C. F., and Peng, H. L. (2016). CRP-cyclic AMP regulates the expression of type 3 fimbriae via cyclic di-GMP in Klebsiella pneumoniae. PLoS One 11:e0162884. doi: 10.1371/journal.pone.0162884
Liu, Y., Pan, C., Ye, L., Si, Y., Bi, C., Hua, X., et al. (2020). Nonclassical biofilms induced by DNA breaks in Klebsiella pneumoniae. mSphere 5:e00336-20. doi: 10.1128/mSphere.00336-20
Liu, K., Tan, S., Ye, W., Hou, L., and Fang, B. (2022). Low-concentration iron promotes Klebsiella pneumoniae biofilm formation by suppressing succinic acid. BMC Microbiol. 22:95. doi: 10.1186/s12866-022-02518-w
Luo, Y., Wang, Y., Ye, L., and Yang, J. (2014). Molecular epidemiology and virulence factors of pyogenic liver abscess causing Klebsiella pneumoniae in China. Clin. Microbiol. Infect. 20, O818–O824. doi: 10.1111/1469-0691.12664
Maeyama, R., Mizunoe, Y., Anderson, J. M., Tanaka, M., and Matsuda, T. (2004). Confocal imaging of biofilm formation process using fluoroprobed Escherichia coli and fluoro-stained exopolysaccharide. J. Biomed. Mater. Res. A 70, 274–282. doi: 10.1002/jbm.a.30077
Miller, M. B., and Bassler, B. L. (2001). Quorum sensing in bacteria. Annu. Rev. Microbiol. 55, 165–199. doi: 10.1146/annurev.micro.55.1.165
Mohamudha, P. R., Belgode, N. H., Laura, M., and Michael, R. M. (2016). Carbapenem resistance mechanisms among blood isolates of Klebsiella pneumoniae and Escherichia coli. Afr. J. Microbiol. Res. 10, 45–53. doi: 10.5897/AJMR2015.7802
Murphy, C. N., and Clegg, S. (2012). Klebsiella pneumoniae and type 3 fimbriae: nosocomial infection, regulation and biofilm formation. Future Microbiol. 7, 991–1002. doi: 10.2217/fmb.12.74
Murphy, C. N., Mortensen, M. S., Krogfelt, K. A., and Clegg, S. (2013). Role of Klebsiella pneumoniae type 1 and type 3 fimbriae in colonizing silicone tubes implanted into the bladders of mice as a model of catheter-associated urinary tract infections. Infect. Immun. 81, 3009–3017. doi: 10.1128/IAI.00348-13
Ou, Q., Fan, J., Duan, D., Xu, L., Wang, J., Zhou, D., et al. (2017). Involvement of cAMP receptor protein in biofilm formation, fimbria production, capsular polysaccharide biosynthesis and lethality in mouse of Klebsiella pneumoniae serotype K1 causing pyogenic liver abscess. J. Med. Microbiol. 66, 1–7. doi: 10.1099/jmm.0.000391
Pacheco, T., Gomes, A. É. I., Siqueira, N. M. G., Assoni, L., Darrieux, M., Venter, H., et al. (2021). SdiA, a quorum-sensing regulator, suppresses fimbriae expression, biofilm formation, and quorum-sensing signaling molecules production in Klebsiella pneumoniae. Front. Microbiol. 12:597735. doi: 10.3389/fmicb.2021.597735
Pal, S., Verma, J., Mallick, S., Rastogi, S. K., Kumar, A., and Ghosh, A. S. (2019). Absence of the glycosyltransferase WcaJ in Klebsiella pneumoniae ATCC13883 affects biofilm formation, increases polymyxin resistance and reduces murine macrophage activation. Microbiology 165, 891–904. doi: 10.1099/mic.0.000827
Panjaitan, N. S. D., Horng, Y. T., Cheng, S. W., Chung, W. T., and Soo, P. C. (2019). EtcABC, a putative EII complex, regulates type 3 fimbriae via CRP-cAMP signaling in Klebsiella pneumoniae. Front. Microbiol. 10:1558. doi: 10.3389/fmicb.2019.01558
Pesci, E. C., Pearson, J. P., Seed, P. C., and Iglewski, B. H. (1997). Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 179, 3127–3132. doi: 10.1128/jb.179.10.3127-3132.1997
Rickenberg, H. V. (1974). Cyclic AMP in prokaryotes. Annu. Rev. Microbiol. 28, 353–369. doi: 10.1146/annurev.mi.28.100174.002033
Rosen, D. A., Pinkner, J. S., Jones, J. M., Walker, J. N., Clegg, S., and Hultgren, S. J. (2008). Utilization of an intracellular bacterial community pathway in Klebsiella pneumoniae urinary tract infection and the effects of FimK on type 1 pilus expression. Infect. Immun. 76, 3337–3345. doi: 10.1128/IAI.00090-08
Rozen, S., and Skaletsky, H. (2000). Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132, 365–386. doi: 10.1385/1-59259-192-2:365
Ruhal, R., and Kataria, R. (2021). Biofilm patterns in gram-positive and gram-negative bacteria. Microbiol. Res. 251:126829. doi: 10.1016/j.micres.2021.126829
Russo, T. A., and Marr, C. M. (2019). Hypervirulent Klebsiella pneumoniae. Clin. Microbiol. Rev. 32:3. doi: 10.1128/CMR.00001-19
Sahly, H., Navon-Venezia, S., Roesler, L., Hay, A., Carmeli, Y., Podschun, R., et al. (2008). Extended-spectrum beta-lactamase production is associated with an increase in cell invasion and expression of fimbrial adhesins in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 52, 3029–3034. doi: 10.1128/AAC.00010-08
Sanchez, C. J., Mende, K., Beckius, M. L., Akers, K. S., Romano, D. R., Wenke, J. C., et al. (2013). Biofilm formation by clinical isolates and the implications in chronic infections. BMC Infect. Dis. 13:47. doi: 10.1186/1471-2334-13-47
Schembri, M. A., Blom, J., Krogfelt, K. A., and Klemm, P. (2005). Capsule and fimbria interaction in Klebsiella pneumoniae. Infect. Immun. 73, 4626–4633. doi: 10.1128/IAI.73.8.4626-4633.2005
Schroll, C., Barken, K. B., Krogfelt, K. A., and Struve, C. (2010). Role of type 1 and type 3 fimbriae in Klebsiella pneumoniae biofilm formation. BMC Microbiol. 10:179. doi: 10.1186/1471-2180-10-179
Shadkam, S., Goli, H. R., Mirzaei, B., Gholami, M., and Ahanjan, M. (2021). Correlation between antimicrobial resistance and biofilm formation capability among Klebsiella pneumoniae strains isolated from hospitalized patients in Iran. Ann. Clin. Microbiol. Antimicrob. 20:13. doi: 10.1186/s12941-021-00418-x
Simm, R., Morr, M., Kader, A., Nimtz, M., and Römling, U. (2004). GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol. Microbiol. 53, 1123–1134. doi: 10.1111/j.1365-2958.2004.04206.x
Singh, S., Wilksch, J. J., Dunstan, R. A., Mularski, A., Wang, N., Hocking, D., et al. (2022). LPS O antigen plays a key role in Klebsiella pneumoniae capsule retention. Microbiol. Spectr. 10:e0151721. doi: 10.1128/spectrum.01517-21
Singh, A. K., Yadav, S., Chauhan, B. S., Nandy, N., Singh, R., Neogi, K., et al. (2019). Classification of clinical isolates of Klebsiella pneumoniae based on their in vitro biofilm forming capabilities and elucidation of the biofilm matrix chemistry with special reference to the protein content. Front. Microbiol. 10:669. doi: 10.3389/fmicb.2019.00669
Solano, C., Echeverz, M., and Lasa, I. (2014). Biofilm dispersion and quorum sensing. Curr. Opin. Microbiol. 18, 96–104. doi: 10.1016/j.mib.2014.02.008
Stahlhut, S. G., Struve, C., Krogfelt, K. A., and Reisner, A. (2012). Biofilm formation of Klebsiella pneumoniae on urethral catheters requires either type 1 or type 3 fimbriae. FEMS Immunol. Med. Microbiol. 65, 350–359. doi: 10.1111/j.1574-695X.2012.00965.x
Stanley, N. R., and Lazazzera, B. A. (2004). Environmental signals and regulatory pathways that influence biofilm formation. Mol. Microbiol. 52, 917–924. doi: 10.1111/j.1365-2958.2004.04036.x
Struve, C., Bojer, M., and Krogfelt, K. A. (2008). Characterization of Klebsiella pneumoniae type 1 fimbriae by detection of phase variation during colonization and infection and impact on virulence. Infect. Immun. 76, 4055–4065. doi: 10.1128/IAI.00494-08
Sutrina, S. L., Daniel, K., Lewis, M., Charles, N. T., Anselm, C. K., Thomas, N., et al. (2015). Biofilm growth of Escherichia coli is subject to cAMP-dependent and cAMP-independent inhibition. J. Mol. Microbiol. Biotechnol. 25, 209–225. doi: 10.1159/000375498
Tal, R., Wong, H. C., Calhoon, R., Gelfand, D., Fear, A. L., Volman, G., et al. (1998). Three cdg operons control cellular turnover of cyclic di-GMP in Acetobacter xylinum: genetic organization and occurrence of conserved domains in isoenzymes. J. Bacteriol. 180, 4416–4425. doi: 10.1128/JB.180.17.4416-4425.1998
Tamayo, R., Pratt, J. T., and Camilli, A. (2007). Roles of cyclic diguanylate in the regulation of bacterial pathogenesis. Annu. Rev. Microbiol. 61, 131–148. doi: 10.1146/annurev.micro.61.080706.093426
Tan, J. W., Wilksch, J. J., Hocking, D. M., Wang, N., Srikhanta, Y. N., Tauschek, M., et al. (2015). Positive autoregulation of mrkHI by the cyclic di-GMP-dependent MrkH protein in the biofilm regulatory circuit of Klebsiella pneumoniae. J. Bacteriol. 197, 1659–1667. doi: 10.1128/JB.02615-14
Tang, M., Wei, X., Wan, X., Ding, Z., Ding, Y., and Liu, J. (2020). The role and relationship with efflux pump of biofilm formation in Klebsiella pneumoniae. Microb. Pathog. 147:104244. doi: 10.1016/j.micpath.2020.104244
Tian, D., Liu, X., Chen, W., Zhou, Y., Hu, D., Wang, W., et al. (2022). Prevalence of hypervirulent and carbapenem-resistant Klebsiella pneumoniae under divergent evolutionary patterns. Emerg. Microbes Infect. 11, 1936–1949. doi: 10.1080/22221751.2022.2103454
Türkel, İ., Yıldırım, T., Yazgan, B., Bilgin, M., and Başbulut, E. (2018). Relationship between antibiotic resistance, efflux pumps, and biofilm formation in extended-spectrum beta-lactamase producing Klebsiella pneumoniae. J. Chemother. 30, 354–363. doi: 10.1080/1120009X.2018.1521773
Turton, J. F., Perry, C., Elgohari, S., and Hampton, C. V. (2010). PCR characterization and typing of Klebsiella pneumoniae using capsular type-specific, variable number tandem repeat and virulence gene targets. J. Med. Microbiol. 59, 541–547. doi: 10.1099/jmm.0.015198-0
Vuotto, C., Longo, F., Pascolini, C., Donelli, G., Balice, M. P., Libori, M. F., et al. (2017). Biofilm formation and antibiotic resistance in Klebsiella pneumoniae urinary strains. J. Appl. Microbiol. 123, 1003–1018. doi: 10.1111/jam.13533
Wang, H., Yan, Y., Rong, D., Wang, J., Wang, H., Liu, Z., et al. (2016). Increased biofilm formation ability in Klebsiella pneumoniae after short-term exposure to a simulated microgravity environment. Microbiology 5, 793–801. doi: 10.1002/mbo3.370
Wilksch, J. J., Yang, J., Clements, A., Gabbe, J. L., Short, K. R., Cao, H., et al. (2011). MrkH, a novel c-di-GMP-dependent transcriptional activator, controls Klebsiella pneumoniae biofilm formation by regulating type 3 fimbriae expression. PLoS Pathog. 7:e1002204. doi: 10.1371/journal.ppat.1002204
Willsey, G. G., Ventrone, S., Schutz, K. C., Wallace, A. M., Ribis, J. W., Suratt, B. T., et al. (2018). Pulmonary surfactant promotes virulence gene expression and biofilm formation in Klebsiella pneumoniae. Infect. Immun. 86:e00135-18. doi: 10.1128/IAI.00135-18
Wu, M. C., Chen, Y. C., Lin, T. L., Hsieh, P. F., and Wang, J. T. (2012). Cellobiose-specific phosphotransferase system of Klebsiella pneumoniae and its importance in biofilm formation and virulence. Infect. Immun. 80, 2464–2472. doi: 10.1128/IAI.06247-11
Wu, C. C., Huang, Y. J., Fung, C. P., and Peng, H. L. (2010). Regulation of the Klebsiella pneumoniae Kpc fimbriae by the site-specific recombinase KpcI. Microbiology 156, 1983–1992. doi: 10.1099/mic.0.038158-0
Wu, C. C., Lin, C. T., Cheng, W. Y., Huang, C. J., Wang, Z. C., and Peng, H. L. (2012). Fur-dependent MrkHI regulation of type 3 fimbriae in Klebsiella pneumoniae CG43. Microbiology 158, 1045–1056. doi: 10.1099/mic.0.053801-0
Wu, M. C., Lin, T. L., Hsieh, P. F., Yang, H. C., and Wang, J. T. (2011). Isolation of genes involved in biofilm formation of a Klebsiella pneumoniae strain causing pyogenic liver abscess. PLoS One 6:e23500. doi: 10.1371/journal.pone.0023500
Wyres, K. L., Wick, R. R., Gorrie, C., Jenney, A., Follador, R., Thomson, N. R., et al. (2016). Identification of capsule synthesis loci from whole genome data. Microb. Genom. 2:e000102. doi: 10.1099/mgen.0.000102
Yan, J., and Bassler, B. L. (2019). Surviving as a community: antibiotic tolerance and persistence in bacterial biofilms. Cell Host Microbe 26, 15–21. doi: 10.1016/j.chom.2019.06.002
Yang, Y., Yao, F., Zhou, M., Zhu, J., Zhang, X., Bao, W., et al. (2013). F18ab Escherichia coli flagella expression is regulated by acyl-homoserine lactone and contributes to bacterial virulence. Vet. Microbiol. 165, 378–383. doi: 10.1016/j.vetmic.2013.04.020
Zhang, L., Li, S., Liu, X., Wang, Z., Jiang, M., Wang, R., et al. (2020). Sensing of autoinducer-2 by functionally distinct receptors in prokaryotes. Nat. Commun. 11:5371. doi: 10.1038/s41467-020-19243-5
Zhao, K., Tseng, B. S., Beckerman, B., Jin, F., Gibiansky, M. L., Harrison, J. J., et al. (2013). Psl trails guide exploration and microcolony formation in Pseudomonas aeruginosa biofilms. Nature 497, 388–391. doi: 10.1038/nature12155
Keywords: Klebsiella pneumoniae , biofilm, capsule, fimbriae, factors
Citation: Li Y and Ni M (2023) Regulation of biofilm formation in Klebsiella pneumoniae. Front. Microbiol. 14:1238482. doi: 10.3389/fmicb.2023.1238482
Received: 11 June 2023; Accepted: 23 August 2023;
Published: 07 September 2023.
Edited by:
Nityananda Chowdhury, Medical University of South Carolina, United StatesReviewed by:
Norma Velazquez-Guadarrama, Federico Gómez Children’s Hospital, MexicoCopyright © 2023 Li and Ni. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ming Ni, bmltaW5nQHRqaC50am11LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.