
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 04 September 2023
Sec. Microorganisms in Vertebrate Digestive Systems
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1237955
 Jian Wu1,2,3
Jian Wu1,2,3 Changxin Tian1,2,3,4*
Changxin Tian1,2,3,4* Jinzhen Jiao1,2,3
Jinzhen Jiao1,2,3 Qiongxian Yan1,2,3
Qiongxian Yan1,2,3 Chuanshe Zhou1,2,3
Chuanshe Zhou1,2,3 Zhiliang Tan1,2,3
Zhiliang Tan1,2,3Introduction: Feeding low protein (LP) diet to animals impose severe challenge to animals' immune homeostasis. However, limited knowledge about the underlying adaption mechanism of host and ruminal microbiota responding to LP diet were well understood. Herein, this study was performed to examine the changes in relative abundance of ruminal microbiota and host ruminal mucosal transcriptome profiles in response to a LP diet.
Methods: A total of twenty-four female Xiangdong balck goats with similar weight (20.64 ± 2.40 kg) and age (8 ± 0.3 months) were randomly assigned into two groups, LP (5.52% crude protein containing diet) and CON (10.77% crude protein containing diet) groups. Upon completion of the trial, all goats were slaughtered after a 16-hour fasting period in LiuYang city (N 28°15′, E 113°63′) in China. HE staining, free amino acids measurement, transcriptome analysis and microbiome analysis were applied to detect the morphology alterations, free amino acids profile alterations and the shift in host ruminal mucosal transcriptome and ruminal microbiota communities.
Results: Firstly, the results showed that feeding LP diet to goats decreased the rumen papilla width (P = 0.043), surface area (P = 0.013) and total ruminal free amino acids concentration (P = 0.016). Secondly, microbiome analysis indicated that 9 microbial genera, including Eubacterium and Prevotella, were enriched in LP group while 11 microbial genera, including Butyrivibrio and Ruminococcus, were enriched in CON group. Finally, in terms of immune-related genes, the expression levels of genes involved in tight junction categories (e.g., MYH11, PPP2R2C, and MYL9) and acquired immunity (e.g., PCP4 and CXCL13) were observed to be upregulated in the LP group when compared to the CON group.
Conclusion: Under the LP diet, the rumen exhibited increased relative abundance of pathogenic microbiota and VFA-degrading microbiota, leading to disruptions in immune homeostasis within the host's ruminal mucosa. These findings indicate that the ruminal microbiota interacts with host results in the disruption in animals' immune homeostasis under LP diet challenge.
Feeding low-protein (LP) diets to goats represent a common strategy for reducing the cost of breeding and nitrogen losses to the environment (Gebeyew et al., 2021). However, as the amino acids play a vital role in maintaining the procedure of animal growth and act as the basal component of the immunity system of the body, low-protein content in the feed imposes negative effects on ruminants' growth performance and homeostasis of the gastrointestinal tract (GIT) (Wang et al., 2021). Furthermore, inadequate protein supply for animals is closely linked to metabolic and immune disorders, such as inflammatory bowel diseases and cancer development (Lan et al., 2015; Soares et al., 2020).
Numerous microorganisms colonize the GIT of mammals and play vital roles in the procedures of extracting energy from diets, regulating the homeostasis of the immune system and producing hormones to regulate a series of body activities (Macpherson et al., 2017; Shanahan et al., 2017). Various factors, including diet composition, host genetics, and antibiotics, have been reported to impose significant effects on the diversity of microbiota in the GIT, and finally may impose severe challenges to host health (Gilbert et al., 2018). In humans and mice, a lower protein diet decreased the abundance of Bifidobacterium in gut digesta, which produced antimicrobial peptides to protect the gut from pathogenic bacteria infection and decreased the risk of gut inflammation (Liu et al., 2014; Feng et al., 2019). In addition to the microbiota residing in the digesta, the microbiota colonizing at the mucosal border is also vital to maintain host metabolic and immune homeostasis (Jiao et al., 2018). Despite the significance, its interactions with the host are still not well interpreted.
As the rumen represents the first site for interplay among ingested nutrients, host GIT, and the microbiota, maintaining metabolic, and immune homeostasis in the rumen is of great importance to ruminants. In lambs and beef steers, limited protein supply caused drastic alterations in bacteria diversity and fermentation parameters in the rumen digesta (Wang et al., 2017; Lv et al., 2020). Thus, we hypothesized that the ruminal mucosal microbiota of low-protein diets would be different from that of control diets, and such a variation could manipulate the molecular adaptation of the rumen. To validate this, we used a combination of 16S rRNA sequencing and mucosal transcriptome analysis to dissect bacterial diversity and the host responses in the rumen mucosa.
All procedures for animal experimentation were performed according to the protocol ISA-2019-0115 approved by the Animal Care Committee, Institute of Subtropical Agriculture, Chinese Academy of Sciences, Changsha, Hunan, China.
A total of 24 female Xiangdong black goats with similar weight (20.6 ± 2.4 kg) and age (8.0 ± 0.3 months) were used in this experiment (Gebeyew et al., 2021). These goats were randomly assigned into two groups, a control (CON) group and a low-protein (LP) group. The experimental diets were designed according to the feeding standards of goats in China (NY/T 816-2004) and met 1.3 times the maintenance requirement of metabolic energy (ME) on the basis of previous studies (Tang et al., 2019), and consisted of 70% of rice straw and 30% of concentrate. The detailed ingredients of experimental diets are shown in Supplementary Table 1. The low-protein diet was formulated to contain half of the CP content of the control diet, and this was achieved by replacing soybean meal with corn meal. The total feeding trial period was 70 days, including 25 days of adaptation and 45 days of experiments. These goats were maintained individually in metabolic cages with access to fresh water offered in equal portions at 07:00 and 17:00 h daily.
After the trial, all goats were slaughtered following a 16-h fasting period. Immediately after all the goats were killed by bleeding the jugular vein by a registered veterinarian in Liuyang city (N 28°15′, E 113°63′), China, ruminal tissues were collected from the ventral sac of the rumen and washed by cold PBS for three times. Afterward, the mucosa samples were separated from the underlying muscular layer, cut into 1 × 1 cm fragments, frozen in liquid nitrogen, and stored at −80°C for subsequent DNA extraction, transcriptome analysis, and free amino acid profile measurements. Moreover, two 2 × 2 cm mucosa samples from the ventral sac of the rumen were collected and fixated in formaldehyde for further HE staining analysis.
After fixation in formaldehyde for 24 h, ruminal mucosa was embedded in paraffin wax (Sangon, Shanghai, China) and then successively sliced into 10 pieces of 4–5 μm thick sections followed by mounting onto poly-L-lysine-coated glass slides (Hailun, Changsha, China). The slides were stained with hematoxylin for 5 min followed by immersing the slides into hydrochloric acid and ammonia water for 10 s separately. After washing with running water for 1 h, all the slides were stained with eosin for 5 min. All the images were acquired under a 10× objective lens and a 10× ocular lens by a fluorescence microscope (Olympus, Tokyo, Japan) equipped with DP2-BSW software.
The free amino acid profiles of ruminal mucosa were determined using the procedures described in previous studies (Wu et al., 2020). In brief, 0.4 g of ruminal mucosa was weighed and ground into powder with liquid nitrogen. The free amino acids were extracted from the mucosa by adding 2 ml of 8% sulfosalicylic acid to the mucosa powder. After quiescence at 4°C for 12 h, the samples were centrifuged at 8,000 × g at 4°C for 10 min, and the supernatant fluid was collected and filtered through a 0.22-μm membrane. The samples were analyzed by an automatic amino acid analyzer (L-8900; Hitachi Global Inc., Hitachi, Japan).
Total RNA was extracted from ruminal mucosa following the instructions of the TaKaRa MiniBEST Universal RNA Extraction Kit (TaKaRa, Dalian, China; code no. 9767). The extracted RNA was reverse-transcribed to build the cDNA library using an mRNA-Seq Sample Preparation Kit (Illumina, San Diego, USA). After verifying and quantifying the cDNA, the libraries were sequenced on the Illumina HiSeq 4000 platform.
The procedures for filtering raw sequencing data were detailed in previous studies (Jiao et al., 2019), and the cleaned data were stored in FASTA format. HISAT2 (v2.04) was applied to map the clean reads to the reference genome (Kim et al., 2015). The clean reads were aligned to the reference coding gene set by Bowtie (Langmead and Salzberg, 2012), and the expression level was measured by RSEM (Li and Dewey, 2011). The analysis of differentially expressed genes (DEGs) was carried out using DESeq2, using thresholds with false discovery rate (FDR) <0.05 and an absolute value of fold change >1.2.
The total DNA of the ruminal mucosa was extracted using the bead-beating method, as detailed in our previous study (Jiao et al., 2015). The quantity of total DNA was measured by NanoDrop ND1000 (NanoDrop Technologies, Inc., Wilmington, DE, USA), and the quality of total DNA was observed by gel electrophoresis. Four samples (1/CON, 3/LP) exhibited significant degradation, failing to meet the standards for library sequencing. After amplifying the V3-V4 region of 16S rRNA (341F, ACTCCTACGGGAGGCAGCAG and 806R, GGACTACHVGGGTWTCTAAT) and running an agarose gel, the bands were purified with a QIAquick Gel Extraction Kit (Qiagen, Hilden, Germany), which was prior to sequencing by the Illumina MiSeq PE250 platform.
The QIIME (Quantitative Insights into Microbial Ecology) pipeline was utilized to perform quality control on the raw data, with a default quality threshold of 20 (Zhang et al., 2019). The quality control process involved a sliding window approach to identify low-quality reads alongside a length screening where reads longer than 200 bases were truncated to 200 bases. Afterward, the short reads were assembled into tags using FLASH, and tags were clustered into amplicon sequence variants (ASVs) of 99% similarity using the unoise3 command implemented in USEARCH (Edgar, 2010). The representative sequences were kept with a total sequencing count >3, and the ASVs were retained that were present in at least two sample replicates to mitigate incidental factors. Taxonomic assignments were performed against the RDP database with a 0.80 confidence threshold (Wang Q. et al., 2007). Alpha and beta diversities were analyzed using the QIIME pipeline. The Bray–Curtis similarity index was used to calculate the distance matrix for principal coordinate analysis (PCoA). The significance of grouping in the PCoA plots was tested by analysis of dissimilarity (ADONIS) with 999 permutations. To identify the differential microbial communities in the ruminal mucosa between these two groups, a linear discriminate analysis (LDA) effect size (LEfSe) method (Segata et al., 2011) was applied with an LDA threshold value of 2.0 using the R software (version 4.0.5).
Data were carried out with a one-way analysis of variance (ANOVA) using the R software (version 4.0.5). Student's t-test was applied to determine the effects of a low-protein diet on the ruminal mucosal morphology and free amino acid profiles, with a P-value of < 0.05 considered significant.
As shown in Table 1, feeding LP diet to goats significantly decreased ruminal papilla width (P = 0.043) and surface area (P = 0.013), while the length of ruminal papilla showed no significant difference (P = 0.268) between these two groups.
Feeding LP diet to goats significantly decreased the total amino acid concentration (P = 0.016). The concentration of glutamic acid (P = 0.011) and aspartic acid (P = 0.037) was significantly decreased in the ruminal mucosa under the LP diet. Similarly, feeding LP diet to goats significantly decreased the concentration of isoleucine (P = 0.033) and valine (P = 0.040). The concentration of leucine tended to decrease in the ruminal mucosa under the LP diet (P = 0.061) (Table 2).
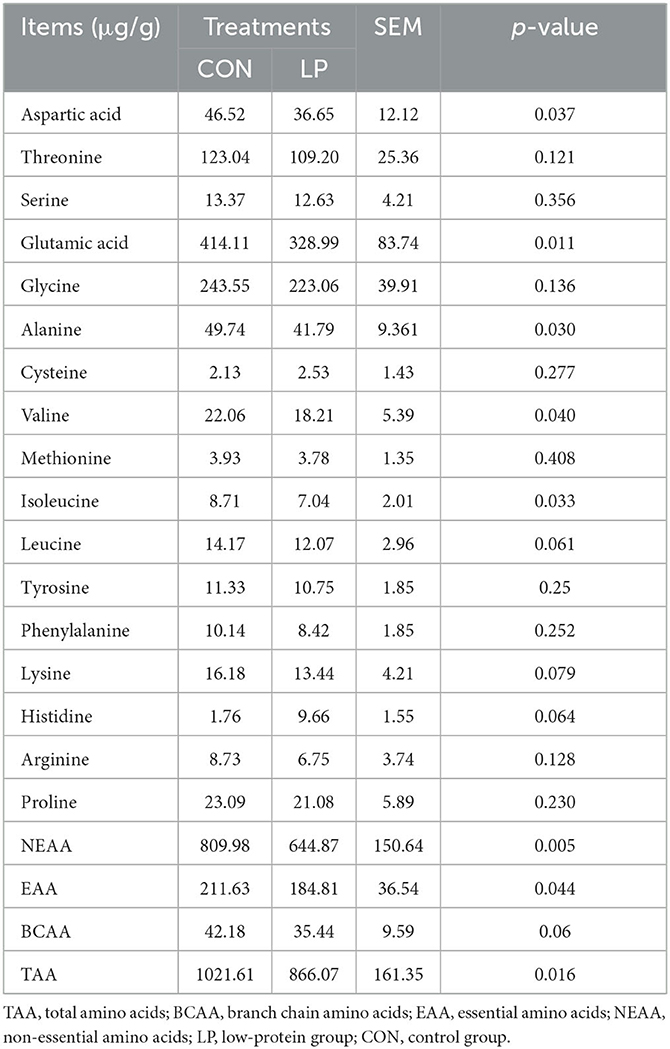
Table 2. Effects of different dietary protein levels on rumen mucosa and free amino acid profiles in goats.
The transcriptome results showed that 41 differentially expressed (DE) genes were observed between the LP and CON groups, of which the expression of 28 genes was upregulated, while 13 genes were downregulated in the LP group in comparison with the CON group. The majority of DEGs were related to immune and muscle categories. For immune-related genes, the expression of genes involved in the categories of tight junction such as MYH11, PPP2R2C, and MYL9, cancer development such as TAGLN and CALD1, and acquired immunity such as PCP4 and CXCL13 was upregulated in the LP group when compared with that in the CON group (Table 3). For muscle-related genes, the expression of genes involved in the categories of muscle contraction such as LMOD1, SYNM, CNN1, TPM2, MYH11, MYL9, MYLK, and TPM1, muscle cell proliferation such as PDLIM3 and RBPMS2, and muscle development such as LDB3 and DMPK was upregulated in the LP group compared with that in the CON group (Table 4). The expression of gene encoding desmin, which acts as a biomarker of muscle, was downregulated in the LP group when compared with that in the CON group.
The alpha diversity analysis in ruminal mucosa showed lower ACE (P = 0.013), Chao1 (P = 0.015), and Shannon (P = 0.031) diversity indexes in the LP group than in the CON group, while the Simpson index showed no significant differences (P = 0.098, Table 5). Beta diversity analysis showed a clear distance separation between the LP and CON groups, and the major variance observed between these two groups was 14.8% (Figure 1). To explore these findings, we performed a LEfSe analysis to identify the differential microbial taxa between the LP and CON groups. The results showed that 9 microbial genera, namely, Intestinimonas, Atopobium, Sphingomonas, Fretibacterium, Eubacterium, Treponema, Prevotella, Desulfobulbus, and Campylobacter were enriched in the LP group while 11 microbial genera, namely, Desulfovibrio, Fibrobacter, Succiniclastium, Saccharofermentans, Butyrivibrio, Ruminococcus, Ruminobacter, Succinimonas, Clostridium-XIVa, Pseudobutyrivibrio, and Pyramidobacter were enriched in the CON group (Figure 2, LDA score >2).
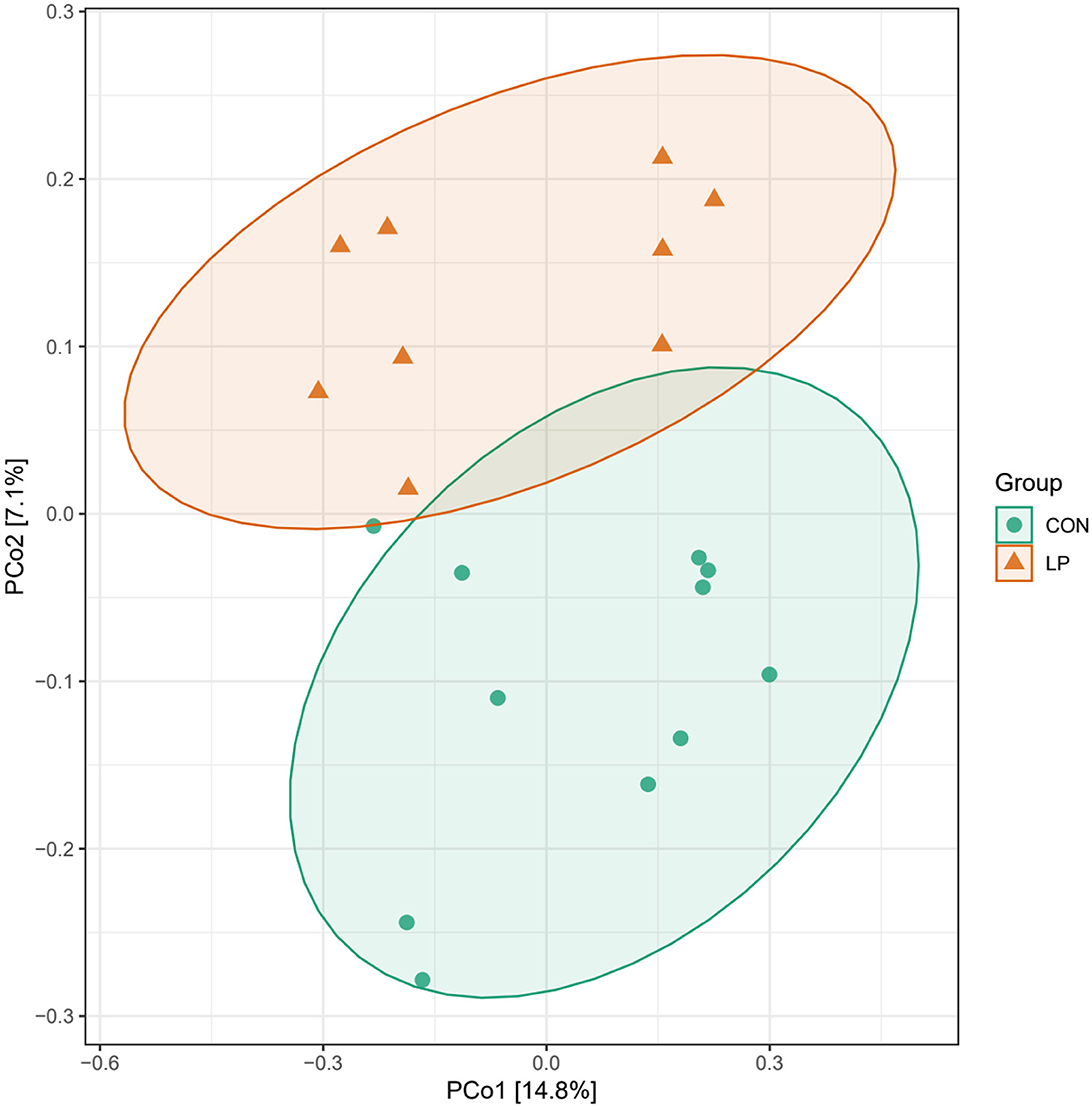
Figure 1. Principal coordinate analysis of ruminal mucosal microbiota of goats fed with LP vs. CON diets. LP, low-protein group; CON, control group.

Figure 2. Differential microbe of ruminal mucosa in goats fed with CON vs. LP at the genus level. LP, low-protein group; CON, control group.
We investigated the correlations between differentially expressed genes (DEGs) and the prominent microbes identified by the LEfSe analysis, in order to gain insight into the interaction between the host and microbiota in the rumen (Figure 3). Using Spearman correlations, we identified a strong negative association between the majority of immune and muscle-related DEGs and the prominent microbes, such as Prevotella and Eubacterium.
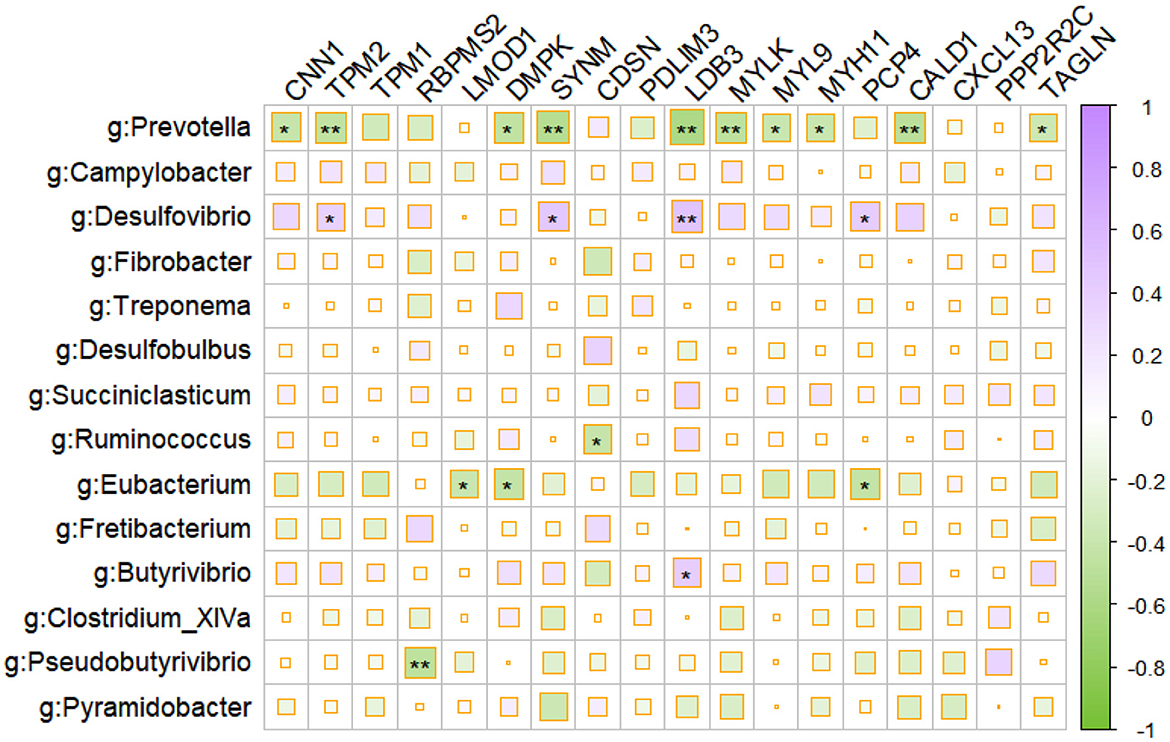
Figure 3. Correlation plot depicting gene–microbe correlations. Color and size of the squares indicate the magnitude of the correlation, asterisks indicate the significance of the correlation (**indicates a p-value of < 0.05, and *indicates a p-value of < 0.1).
It is usually considered that the protein ingested from the diet is converted into NH3-N in the rumen and then utilized by the ruminal microbiota to synthesize microbial protein (MCP). In addition to MCP, peptides and free amino acids digested from the dietary protein and synthesized by the ruminal microbiota (Russell et al., 2009) could be absorbed in the rumen mucosa (Poole et al., 2003). Not surprisingly, low-protein content in the diet induced a decline in the ruminal mucosal free amino acid concentrations, especially branched-chain amino acids (BCAAs, such as valine and isoleucine) and non-essential amino acids (NEAAs, such as glutamic acid and aspartic acid). It has been reported that the scanty of glutamate and BCAAs could induce the suppression of cell proliferation and protein synthesis (Sancak et al., 2010; Li et al., 2016). Hence, it is reasonable to observe a decline in the morphology of the rumen papilla.
Another notable change in response to the LP diet was genes involved in the muscle, which was likely the result of cells from the muscle layer that remained adhered to the dissected mucosal samples. Previous studies indicated that feeding a low-protein diet to mammals induces a decline in muscle layer thickness and muscle destruction in the GIT of animals (Peng et al., 2017). Although TMP1, TMP2, DMPK, PDLIM3, RBPMS2, and LDB3, which were reported to engage in muscle repairment (Jauvin et al., 2017), muscle cell proliferation (Notarnicola et al., 2012; Yin et al., 2020), and muscle regeneration (Knight et al., 2003), were upregulated in LP diet, the upregulation of MYLK, MYH11, and MYL9 may induce muscle relaxation and GIT movement retention (Iwasaki et al., 2001) and finally impose negative effects on nutrient digestion and immune homeostasis (Kashyap et al., 2013). Muscle repairment and muscle regeneration are closely related processes. Muscle repairment refers to the physiological mechanisms involved in restoring damaged muscle tissue, while muscle regeneration refers to the process by which new muscle cells are formed to replace damaged or lost muscle tissue (Notarnicola et al., 2012; Yin et al., 2020). Movement retention is linked to both muscle repairment and muscle regeneration. When muscles are properly repaired and regenerated, movement retention is improved, allowing individuals to maintain their ability to perform various physical activities (Iwasaki et al., 2001). Additionally, the decline in muscle layer thickness and muscle destruction in the rumen may account for the downregulation of DES, which was considered the major component of muscle fibers in the LP group (Cizkova et al., 2009). The LMOD1, which promoted muscle relaxation (Nanda et al., 2017), was also upregulated in the ruminal mucosa and involved in the procedures of rumen contraction shunt.
In addition to the function of muscle contraction, the MYH11 also plays vital roles in a series of biological processes, such as intracellular signaling transduction and cell adhesion (Derycke et al., 2011; Bowers et al., 2012). It has been reported that the promotion of MYH11 expression was related to cell mobility and would cause the disruption of organ integrity and barrier function (Poninska et al., 2022). As anticipated, we observed a decline in the width and surface area of the ruminal papilla. Synchronously, LP inclusion resulted in the upregulation of PCP4, which played a vital role in the procedure of pathological microbiota infection resistance (Ge, 2003; Sui et al., 2021). It is not surprising to find that CALD1 and TAGLN, which promoted inflammation, were upregulated in the LP group, implying an elevated inflammation risk in the GIT (Shen et al., 2010; Liu et al., 2021). Notably, the procedures of antigen presentation to immune cells were upregulated via the upregulation of CXCL13 (Ohmatsu et al., 2007; Wang X. B. et al., 2007), to enhance the immune system and finally alleviate the negative effect of limited protein supply for goats.
Feeding LP diet to goats decreased the ruminal microbiota diversity, which was in line with previous reports in beef steers (Wang et al., 2017). It is noteworthy that several pathogenic bacteria genera propagated in ruminal mucosa during LP inclusion, namely, Fretibacterium, Treponema, Intestinimonas, Sphingomonas, and Campylobacter (Brooks et al., 2017; Gubert et al., 2020; Fan and Pedersen, 2021). These pathogens possessed the capacity to produce toxic substances such as endotoxin in the GIT (Zhao et al., 2013; Fan and Pedersen, 2021), thereby aggravating the above-mentioned disruption of immune homeostasis in the ruminal mucosa. In addition, the higher proportion of starch as a primary carbohydrate source in the LP group may provide a more suitable growth and metabolic environment for Fretibacterium and Campylobacter. This could potentially lead to a relatively higher abundance of Fretibacterium in the LP group.
Additionally, the results of our analysis revealed a significant negative association between DEGs related to immune response and muscle function and the prominent microbes, such as Prevotella and Eubacterium, in the rumen, suggesting that these microbial taxa may have a suppressive effect on the immune system and muscle development of the host. Meanwhile, the surge of H2S-producing Desulfobulbus in the mucosal microbiota during LP intervention (Cabrera et al., 2006) might also contribute to the disruption of immune stability in the ruminal mucosa. Intriguingly, LP inclusion declined the abundances of generally accepted carbohydrate-degrading bacteria in the ruminal mucosa, namely, fiber-degrading Fibrobacter and Ruminococcus (Yeoman et al., 2021), acetate producers Desulfovibrio and Saccharofermentans (Rettenmaier et al., 2021), succinate producers Succinimonas and Pyramidobacter (Gilbert et al., 2018), propionate producers Succiniclastium, and butyrate producers Butyrivibrio and Pseudobutyrivibrio (Pidcock et al., 2021). These highlighted the significance of ruminal microbiota in short-chain fatty acid (SCFA) production for energy supply and indicated a decline of carbohydrate fermentation in ruminal mucosa during LP intervention. Since the SCFAs not only act as the energy source for ruminants but also act as vital signaling molecules in maintaining immune homeostasis (Yao et al., 2022), the decline in the abundance of VFA-producing microbiota in the LP diet may also contribute to the host mucosal immune disruption.
Feeding LP diets to goats significantly decreased NEAA and BCAA concentration in the rumen mucosa, restrained the ruminal papilla growth, altered the gene expression related to muscle contraction, and depressed the gene expression related to immune homeostasis. Moreover, the low-protein diet also induced a decline in the relative abundance of carbohydrate-degrading microbiota while the promotion induced the abundance of pathogenic microbiota in the ruminal mucosa. Taken together, feeding LP diets to goats imposed severe challenges to goats as a result of the combination of pathogenic microbiota enrichment and the disruption of ruminal immune homeostasis.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://ngdc.cncb.ac.cn/gsa—CRA011528 and CRA011517.
The animal studies were approved by the Animal Care Committee, Institute of Subtropical Agriculture, Chinese Academy of Sciences. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
JW and ZT designed the research. CT and JJ performed the research and analyzed the samples. CZ and CT contributed intellectually to the analysis and interpretation of the data. CT, JW, and ZT wrote the manuscript. All authors read and approved the final version of the manuscript.
This work was supported by grants from Strategic Priority Research Program of the Chinese Academy of Sciences (No. XDA26040304) and the National Natural Science Foundation of China (No. 31730092).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1237955/full#supplementary-material
AAs, amino acids; BCAAs, branch-chain amino acids; CALD1, Caldesmon; CXCL13, C-X-C motif chemokine 13; CNN1, calponin-1; DEG, differential expression genes; FDMPK, Myotonin-protein kinase isoform X1; DSG1, Desmoglein-1-alpha; EAA, essential amino acids; GIT, gastrointestinal tract; LDB3, LIM domain-binding protein 3 isoform X1; MCP, Microbial protein; MYH11, Myosin-11 isoform X2; MYL9, Myosin regulatory light polypeptide 9 isoform X1; MYLK, Myosin light chain kinase, SM, smooth muscle; MYH11, Myosin-11; MYL9, Myosin regulatory light polypeptide 9; PCP4, Purkinje cell protein; PDLIM3, PDZ and LIM domain protein 3 isoform X1; PPP1R1A, Protein phosphatase 1 regulatory subunit 1A; PPP1R12B, Protein phosphatase 1 regulatory subunit 12B; PPP2R2C, serine/threonine-protein phosphatase 2A 55 kDa regulatory subunit B gamma isoform; RBPMS2, RNA-binding protein with multiple splicing 2; SYNM, synemin isoform X1; TAGLN, Transgelin; TPM1, Tropomyosin alpha-1 chain; TAA, total amino acids; VFA, volatile fatty acids.
Bowers, R. R., Manevich, Y., Townsend, D. M., and Tew, K. D. (2012). Sulfiredoxin redox-sensitive interaction with S100A4 and non-muscle myosin IIA regulates cancer cell motility. Biochemistry 51, 7740–7754. doi: 10.1021/bi301006w
Brooks, P. T., Brakel, K. A., Bell, J. A., Bejcek, C. E., Gilpin, T., Brudvig, J. M., et al. (2017). Transplanted human fecal microbiota enhanced Guillain Barre syndrome autoantibody responses after Campylobacter jejuni infection in C57BL/6 mice. Microbiome 5, 1–22. doi: 10.1186/s40168-017-0284-4
Cabrera, G., Perez, R., Gomez, J. M., Abalos, A., and Cantero, D. (2006). Toxic effects of dissolved heavy metals on Desulfovibrio vulgaris and Desulfovibrio sp strains. J. Hazar. Mater. 135, 40–46. doi: 10.1016/j.jhazmat.2005.11.058
Cizkova, D., Soukup, T., and Mokry, J. (2009). Expression of nestin, desmin and vimentin in intact and regenerating muscle spindles of rat hind limb skeletal muscles. Histochem. Cell Biol. 131, 197–206. doi: 10.1007/s00418-008-0523-7
Derycke, L., Stove, C., Vercoutter-Edouart, A. S., De Wever, O., Dolle, L., Colpaert, N., et al. (2011). The role of non-muscle myosin IIA in aggregation and invasion of human MCF-7 breast cancer cells. Int. J. Dev. Biol. 55, 835–840. doi: 10.1387/ijdb.113336ld
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Fan, Y., and Pedersen, O. (2021). Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol. 19, 55–71. doi: 10.1038/s41579-020-0433-9
Feng, Y. Q., Duan, Y. F., Xu, Z. J., Lyu, N., Liu, F., Liang, S. H., et al. (2019). An examination of data from the American Gut Project reveals that the dominance of the genus Bifidobacterium is associated with the diversity and robustness of the gut microbiota. Microbiol. Open 8, 939. doi: 10.1002/mbo3.939
Ge, L. (2003). Identification of the delta-6 desaturase of human sebaceous glands: expression and enzyme activity. J. Invest. Dermatol. 121, 434–434. doi: 10.1046/j.1523-1747.2003.12123.x
Gebeyew, K., Chen, W. X., Yan, Q. X., He, Z. X., and Tan, Z. L. (2021). Growth of pancreas and intestinal enzyme activities in growing goats: influence of a low-protein diet. Agric. Basel 11, 1155. doi: 10.3390/agriculture11111155
Gilbert, J. A., Blaser, M. J., Caporaso, J. G., Jansson, J. K., Lynch, S. V., and Knight, R. (2018). Current understanding of the human microbiome. Nat. Med. 24, 392–400. doi: 10.1038/nm.4517
Gubert, C., Kong, G., Renoir, T., and Hannan, A. J. (2020). Exercise, diet and stress as modulators of gut microbiota: Implications for neurodegenerative diseases. Neurobiol. Dis. 134, 104621. doi: 10.1016/j.nbd.2019.104621
Iwasaki, T., Murata-Hori, M., Ishitobi, S., and Hosoya, H. (2001). Diphosphorylated MRLC is required for organization of stress fibers in interphase cells and the contractile ring in dividing cells. Cell Struct. Funct. 26, 677–683. doi: 10.1247/csf.26.677
Jauvin, D., Chretien, J., Pandey, S. K., Martineau, L., Revillod, L., Bassez, G., et al. (2017). Targeting DMPK with antisense oligonucleotide improves muscle strength in myotonic dystrophy type 1 mice. Molec. Ther. Nucl. Acids 7, 465–474. doi: 10.1016/j.omtn.2017.05.007
Jiao, J., Wu, J., Wang, M., Zhou, C., Zhong, R., and Tan, Z. (2018). Rhubarb supplementation promotes intestinal mucosal innate immune homeostasis through modulating intestinal epithelial microbiota in goat kids. J. Agric. Food Chem. 66, 1047–1057. doi: 10.1021/acs.jafc.7b05297
Jiao, J., Zhang, X., Wang, M., Zhou, C., Yan, Q., and Tan, Z. (2019). Linkages between epithelial microbiota and host transcriptome in the ileum during high-grain challenges: implications for gut homeostasis in goats. J. Agric. Food Chem. 67, 551–561. doi: 10.1021/acs.jafc.8b05591
Jiao, J. Z., Huang, J. Y., Zhou, C. S., and Tan, Z. L. (2015). Taxonomic identification of ruminal epithelial bacterial diversity during rumen development in goats. Appl. Environ. Microbiol. 81, 3502–3509. doi: 10.1128/AEM.00203-15
Kashyap, P. C., Marcobal, A., Ursell, L. K., Larauche, M., Duboc, H., Earle, K. A., et al. (2013). Complex interactions among diet, gastrointestinal transit, and gut microbiota in humanized mice. Gastroenterology 144, 967–977. doi: 10.1053/j.gastro.2013.01.047
Kim, D., Landmead, B., and Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–U121. doi: 10.1038/nmeth.3317
Knight, R. F., Bader, D. M., and Backstrom, J. R. (2003). Membrane topology of Bves/Pop1A, a cell adhesion molecule that displays dynamic changes in cellular distribution during development. J. Biol. Chem. 278, 32872–32879. doi: 10.1074/jbc.M301961200
Lan, A., Andriamihaja, M., Blouin, J. M., Liu, X., Descatoire, V., Desclee de Maredsous, C., et al. (2015). High-protein diet differently modifies intestinal goblet cell characteristics and mucosal cytokine expression in ileum and colon. J. Nutr. Biochem. 26, 91–98. doi: 10.1016/j.jnutbio.2014.09.007
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–U354. doi: 10.1038/nmeth.1923
Li, B., and Dewey, C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 12, 323. doi: 10.1186/1471-2105-12-323
Li, X. G., Sui, W. G., Gao, C. Q., Yan, H. C., Yin, Y. L., Li, H. C., et al. (2016). L-Glutamate deficiency can trigger proliferation inhibition via down regulation of the mTOR/S6K1 pathway in pig intestinal epithelial cells. J. Animal Sci. 94, 1541–1549. doi: 10.2527/jas.2015-9432
Liu, X. X., Blouin, J. M., Santacruz, A., Lan, A., Andriamihaja, M., Wilkanowicz, S., et al. (2014). High-protein diet modifies colonic microbiota and luminal environment but not colonocyte metabolism in the rat model: the increased luminal bulk connection. Am. J. Physiol. Gastrointest. Liver Physiol. 307, G459–G470. doi: 10.1152/ajpgi.00400.2013
Liu, Y. X., Xie, S. H., Zhu, K. Y., Guan, X. L., Guo, L., and Lu, R. Q. (2021). CALD1 is a prognostic biomarker and correlated with immune infiltrates in gastric cancers. Heliyon 7, e07257. doi: 10.1016/j.heliyon.2021.e07257
Lv, Cui, K., Qi, M. L., Wang, S. Q., Diao, Q. Y., and Zhang, N. F. (2020). Ruminal microbiota and fermentation in response to dietary protein and energy levels in weaned lambs. Animals 10, 109. doi: 10.3390/ani10010109
Macpherson, A. J., de Aguero, M. G., and Ganal-Vonarburg, S. C. (2017). How nutrition and the maternal microbiota shape the neonatal immune system. Nat. Rev. Immunol. 17, 508–517. doi: 10.1038/nri.2017.58
Nanda, V., Wang, T., Iyer, D., Pjanic, M., Liu, B. X., Nguyen, T., et al. (2017). Functional regulatory mechanism of smooth muscle cell-restricted LMOD1 coronary artery disease locus. Circulation 136, A21021–A21021. doi: 10.1161/circ.136.suppl_1.21021
Notarnicola, C., Rouleau, C., Le Guen, L., Virsolvy, A., Richard, S., Faure, S., et al. (2012). The RNA-Binding protein RBPMS2 regulates development of gastrointestinal smooth muscle. Gastroenterology 143, 687–U204. doi: 10.1053/j.gastro.2012.05.047
Ohmatsu, H., Sugaya, M., Kadono, T., and Tamaki, K. (2007). CXCL13 and CCL21 are expressed in ectopic lymphoid follicles in cutaneous Lymphoproliferative disorders. J. Investigat. Dermatol. 127, 2466–2468. doi: 10.1038/sj.jid.5700873
Peng, Y., Yu, K. F., Mu, C. L., Hang, S. Q., Che, L. Q., and Zhu, W. Y. (2017). Progressive response of large intestinal bacterial community and fermentation to the stepwise decrease of dietary crude protein level in growing pigs. Appl. Microbiol. Biotechnol. 101, 5415–5426. doi: 10.1007/s00253-017-8285-6
Pidcock, S. E., Skvortsov, T., Santos, F. G., Courtney, S. J., Sui-Ting, K., Creevey, C. J., et al. (2021). Phylogenetic systematics of Butyrivibrio and Pseudobutyrivibrio genomes illustrate vast taxonomic diversity, open genomes and an abundance of carbohydrate- active enzyme family isoforms. Microbial. Genom. 7, 638. doi: 10.1099/mgen.0.000638
Poninska, J. K., Bilinska, Z. T., Truszkowska, G., Michalak, E., Podgorska, A., Stepien-Wojno, M., et al. (2022). Good performance of the criteria of American College of Medical Genetics and Genomics/Association for Molecular Pathology in prediction of pathogenicity of genetic variants causing thoracic aortic aneurysms and dissections. J. Translat. Med 20, 42. doi: 10.1186/s12967-022-03251-8
Poole, C. A., Wong, E. A., McElroy, A. P., Veit, H. P., and Webb, K. E. (2003). Ontogenesis of peptide transport and morphological changes in the ovine gastrointestinal tract. Small Ruminant Res. 50, 163–176. doi: 10.1016/S0921-4488(03)00103-2
Rettenmaier, R., Thieme, N., Streubel, J., Di Bello, L., Kowollik, M. L., Huang, L. R., et al. (2021). Variimorphobacter saccharofermentans gen. nov., sp. nov., a new member of the family Lachnospiraceae, isolated from a maize- fed biogas fermenter. Int. J. System. Evolut. Microbiol. 71, 5044. doi: 10.1099/ijsem.0.005044
Russell, J. B., Muck, R. E., and Weimer, P. J. (2009). Quantitative analysis of cellulose degradation and growth of cellulolytic bacteria in the rumen. Fems Microbiol. Ecol. 67, 183–197. doi: 10.1111/j.1574-6941.2008.00633.x
Sancak, Y., Bar-Peled, L., Zoncu, R., Markhard, A. L., Nada, S., and Sabatini, D. M. (2010). Ragulator-rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141, 290–303. doi: 10.1016/j.cell.2010.02.024
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12, R60. doi: 10.1186/gb-2011-12-6-r60
Shanahan, F., van Sinderen, D., O'Toole, P. W., and Stanton, C. (2017). Feeding the microbiota: transducer of nutrient signals for the host. Gut 66, 1709–1717. doi: 10.1136/gutjnl-2017-313872
Shen, J., Yang, M., Ju, D., Jiang, H., Zheng, J. P., Xu, Z., et al. (2010). Disruption of SM22 promotes inflammation after artery injury via nuclear factor kappaB activation. Circ. Res. 106, 1351–1362. doi: 10.1161/CIRCRESAHA.109.213900
Soares, M. H., Rodrigues, G. A., Barbosa, L. M. R., Valente Junior, D. T., Santos, F. C., Rocha, G. C., et al. (2020). Effects of crude protein and lactose levels in diets on growth performance, intestinal morphology, and expression of genes related to intestinal integrity and immune system in weaned piglets. Animal Sci. J. 91, e13429. doi: 10.1111/asj.13429
Sui, B. K., Chen, D., Liu, W., Tian, B., Lv, L., Pei, J., et al. (2021). Comparison of lncRNA and mRNA expression in mouse brains infected by a wild-type and a lab-attenuated Rabies lyssavirus. J. General Virol.102, 1538. doi: 10.1099/jgv.0.001538
Tang, S. X., He, Y., Zhang, P. H., Jiao, J. Z., Han, X. F., Yan, Q. X., et al. (2019). Nutrient digestion, rumen fermentation and performance as ramie (Boehmeria nivea) is increased in the diets of goats. Animal Feed Sci. Technol. 247, 15–22. doi: 10.1016/j.anifeedsci.2018.10.013
Wang, C., Liu, Q., Guo, G., Huo, W. J., Liang, Y., Pei, C. X., et al. (2017). Effects of different dietary protein levels and rumen-protected folic acid on ruminal fermentation, degradability, bacterial populations and urinary excretion of purine derivatives in beef steers. J. Agric. Sci. 155, 1477–1486. doi: 10.1017/S0021859617000533
Wang, N., Wu, C. F., Qin, Y., Liu, S. M., and Zhang, R. (2021). Multi-Angle Investigation of the Fractal Characteristics of Nanoscale Pores in the Lower Cambrian Niutitang Shale and Their Implications for CH4 Adsorption. J. Nanosci. Nanotechnol. 21, 156–167. doi: 10.1166/jnn.2021.18463
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Wang, X. B., Yuling, H., Yanping, J., Xinti, T., Yaofang, Y., Feng, Y., et al. (2007). CCL19 and CXCL13 synergistically regulate interaction between B cell acute lymphocytic leukemia CD23(+) CD5(+) B cells and CD8(+) T cells. J. Immunol. 179, 2880–2888. doi: 10.4049/jimmunol.179.5.2880
Wu, J., Zhang, X. L., Wang, R., Wang, M., He, Z. X., Tan, Z. L., et al. (2020). Replacing corn grain with corn gluten feed: Effects on the rumen microbial protein synthesis, functional bacterial groups and epithelial amino acid chemosensing in growing goats. Animal Feed Sci. Technol. 270, 114684. doi: 10.1016/j.anifeedsci.2020.114684
Yao, Y., Cai, X. Y., Fei, W. D., Ye, Y. Q., Zhao, M. D., and Zheng, C. H. (2022). The role of short-chain fatty acids in immunity, inflammation and metabolism. Crit. Rev. Food Sci. Nutr. 62, 1–12. doi: 10.1080/10408398.2020.1854675
Yeoman, C. J., Fields, C. J., Lepercq, P., Ruiz, P., Forano, E., White, B. A., et al. (2021). In Vivo competitions between fibrobacter succinogenes, ruminococcus flavefaciens, and ruminoccus albus in a gnotobiotic sheep model revealed by multi-omic analyses. Mbio 12, e03533. doi: 10.1128/mBio.03533-20
Yin, H. D., Zhao, J., He, H. R., Chen, Y. Q., Wang, Y., Li, D. Y., et al. (2020). Gga-miR-3525 TargetsPDLIM3through the MAPK signaling pathway to regulate the proliferation and differentiation of skeletal muscle satellite cells. Int. J. Molec. Sci. 21, 5573. doi: 10.3390/ijms21155573
Zhang, X. L., Wu, J., Han, X. F., Tan, Z. L., and Jiao, J. Z. (2019). Effects of rumen-protected glucose on ileal microbiota and genes involved in ileal epithelial metabolism and immune homeostasis in transition dairy cows. Animal Feed Sci. Technol. 254, 114199. doi: 10.1016/j.anifeedsci.2019.06.003
Zhao, F. J., Zhang, X. H., Liu, S. Q., Zeng, T. B., Yu, J., Gu, W. M., et al. (2013). Assessment of the immune responses to Treponema pallidum Gpd DNA vaccine adjuvanted with IL-2 and chitosan nanoparticles before and after Treponema pallidum challenge in rabbits. Sci. China-Life Sci. 56, 174–180. doi: 10.1007/s11427-012-4434-4
Keywords: immune homeostasis, low protein diet, microbiome, transcriptome, goat
Citation: Wu J, Tian C, Jiao J, Yan Q, Zhou C and Tan Z (2023) The epithelial transcriptome and mucosal microbiota are altered for goats fed with a low-protein diet. Front. Microbiol. 14:1237955. doi: 10.3389/fmicb.2023.1237955
Received: 10 June 2023; Accepted: 14 August 2023;
Published: 04 September 2023.
Edited by:
Benoit St-Pierre, South Dakota State University, United StatesReviewed by:
Jun Zhang, Northwest A&F University, ChinaCopyright © 2023 Wu, Tian, Jiao, Yan, Zhou and Tan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Changxin Tian, NTA1NjMyNDMzQHFxLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.