
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 14 September 2023
Sec. Microbial Physiology and Metabolism
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1235283
 Meichen Zhu1,2†
Meichen Zhu1,2† Yankun Liu1,2†
Yankun Liu1,2† Xuewei Yang1,2
Xuewei Yang1,2 Lirong Zhu1,2
Lirong Zhu1,2 Yanmei Shen1,2
Yanmei Shen1,2 Shipeng Duan1,2
Shipeng Duan1,2 Jinkui Yang1,2*
Jinkui Yang1,2*The p21-GTPase-activated protein kinases (PAKs) participate in signal transduction downstream of Rho GTPases, which are regulated by Rho GTPase-activating proteins (Rho-GAP). Herein, we characterized two orthologous Rho-GAPs (AoRga1 and AoRga2) and two PAKs (AoPak1 and AoPak2) through bioinformatics analysis and reverse genetics in Arthrobotrys oligospora, a typical nematode-trapping (NT) fungus. The transcription analyses performed at different development stages suggested that Aopaks and Aorga1 play a crucial role during sporulation and trap formation, respectively. In addition, we successfully deleted Aopak1 and Aorga1 via the homologous recombination method. The disruption of Aopak1 and Aorga1 caused a remarkable reduction in spore yield and the number of nuclei per cell, but did not affect mycelial growth. In ∆Aopak1 mutants, the trap number was decreased at 48 h after the introduction of nematodes, but nematode predatory efficiency was not affected because the extracellular proteolytic activity was increased. On the contrary, the number of traps in ∆Aorga1 mutants was significantly increased at 36 h and 48 h. In addition, Aopak1 and Aorga1 had different effects on the sensitivity to cell-wall-disturbing reagent and oxidant. A yeast two-hybrid assay revealed that AoPak1 and AoRga1 both interacted with AoRac, and AoPak1 also interacted with AoCdc42. Furthermore, the Aopaks were up-regulated in ∆Aorga1 mutants, and Aorga1 was down-regulated in ∆Aopak1 mutants. These results reveal that AoRga1 indirectly regulated AoPAKs by regulating small GTPases.
The p21-GTPase-activated protein kinase (PAK) family is present in all eukaryotes (Keniry and Sprague, 2003). The PAK proteins have a conserved N-terminal domain (Cdc42/Rac interactive binding, CRIB) required for binding to the Rho GTPases Cdc42 and Rac, and are regulated by an autoinhibitory mechanism involving the kinase and CRIB domains (Boyce and Andrianopoulos, 2011). This family is identified by sequence similarity with the kinase and CRIB domains, and divided into two groups (Cotteret et al., 2003; Keniry and Sprague, 2003; Molli et al., 2009). Saccharomyces cerevisiae contains three members of the PAK family, Ste20, Cla4, and Skm1 (Martin et al., 1997), each of which share some conserved functions and also perform distinct roles. The PAK family regulates cell and actin polarization throughout the cell cycle of S. cerevisiae (Holly and Blumer, 1999). Furthermore, the PAKs are required for the proper establishment of cell polarity in Schizosaccharomyces pombe (Qyang et al., 2002) and Cryptococcus neoformans (Nichols et al., 2004). In Talaromyces marneffei, PakA (Homologous protein of AoPak2) contributes to polarity establishment during conidial germination and polarization growth (Boyce and Andrianopoulos, 2007). In C. neoformans, Pak1 is involved in mating and virulence (Nichols et al., 2004). In addition, Ste20 protein kinases can trigger hyphal formation in the pathogenic fungus Candida albicans (Leberer et al., 1996). In Ustilago maydis, the deletion of cla4 causes defects in pathogenicity and the filamentous growth of the mating reaction (Leveleki et al., 2004). In Magnaporthe grisea, chm1 (Cla4 homolog in M. grisea) and mst20 (M. grisea Ste20 homolog) encode two PAK proteins with distinct functions, mst20 is dispensable for plant infection, and chm1 plays a critical role in appressorium formation and penetration in M. grisea (Li et al., 2004). Therefore, the PAK family play important roles in fungal conidial germination, pathogenicity, and spore production, especially in the establishment of polarity, and their specific functions vary by species.
The PAK family act as downstream effectors of the Rho GTPases in a variety of morphogenic processes. Rho GTPase family proteins include Rho1, Rho2, Rho3, Rho4, Rho5, Cdc42, and Rac (Mosaddeghzadeh and Ahmadian, 2021). Rho GTPases act as a molecular switch, toggling between an active GTP-bound form and an inactive GDP-bound form, and are involved in diverse cellular functions via the negative regulation of Rho GTPase activating proteins (Rho-GAPs; Bassilana et al., 2005; Ye et al., 2014). Three Rho-GAPs have been functionally studied in S. cerevisiae (Roumanie et al., 2001). Rho-GAPs are required for a variety of processes related to signal transduction in cell development. In M. oryzae, Morga2 to Morga7 (coding Rho-GAPs) are dispensable for conidiation, vegetative growth, appressorial formation and pathogenicity, but Molrg1 and Morga1 (homologous genes of rho-GAPs) are crucial for pathogenicity (Ye et al., 2014). In C. albicans, the deletion of the Cargd1 increases filamentous growth, and cells lacking Cargd1 present longer germ tubes, whereas the overexpression of rgd1 restricts hyphae growth (Ness et al., 2010). In addition, Cdc42 GAP (Cdc42 GTPase-activating protein, also known as p50RhoGAP or ARHGAP1) plays an important role in regulating mammalian cell genomic stability (Wang et al., 2007), and p200RhoGAP, a member of the Rho-GAP family, can mediate cross-talks between Ras- and Rho-regulated signaling pathways in cell growth regulation (Shang et al., 2007). Therefore, Rho-GAPs are required for mycelial development in various fungi, but the function and the mechanism underlying Rho-GAPs in Rho-mediated signaling pathways are still unknown in nematode-trapping (NT) fungi.
Plant-parasitic nematodes are a source of serious potential damage in agriculture and horticulture (Phani et al., 2021; Zhu et al., 2022a). Biological control stands out among the many nematode control methods, including chemical control and traditional practices of control (solarization, planting trap crops before sowing, in rotation, or after cultivation), with advantages of high efficiency, low toxicity and eco-friendliness (Philbrick et al., 2020; Sushil et al., 2022; Tapia-Vazquez et al., 2022). NT fungi are a promising source of biocontrol that can be employed to develop specialized structures called “traps” to capture, kill, and consume nematodes (Ji et al., 2020). Arthrobotrys oligospora, a representative NT fungus, has been sequenced for its whole genome and proteome (Yang et al., 2011). Recent studies have shown that signal transduction pathways play a vital role in hyphal growth and trap formation in A. oligospora. Rho-GAPs, small GTPases, and PAKs are involved in the transmission of signals between extracellular and intracellular regions, which is involved in the regulation of the cAMP-PKA pathway (Swaminathan et al., 2014; Yang et al., 2022) and mitogen-activated protein kinase (MAPK) cascades (Martin et al., 1997; Cansado et al., 2010). Recently, we have demonstrated that AoRac and AoCdc42 play a crucial role in hypha growth, lipid accumulation, DNA damage, sporulation, trap formation, pathogenicity, and stress response (Yang et al., 2022). Subsequently, the cAMP-PKA signaling pathway has been proven to be involved in hyphal growth, sporulation, trap morphogenesis, stress tolerance, and autophagy in A. oligospora (Chen et al., 2022; Zhu et al., 2022b). In addition, several genes are related to MAPK cascades, such as mkk1 (coding a MAP kinase kinase) and bck1 (coding a MAP kinase kinase kinase; Xie et al., 2021), ste7 (coding a MAP kinase kinase) and fus3 (coding a MAP kinase; Chen et al., 2021; Xie et al., 2023), hog1 (coding a MAP kinase; Kuo et al., 2020), and ime2 (coding a MAP kinase; Xie et al., 2020), which all participate in the development and pathogenicity of A. oligospora.
In this study, we identified two Rho-GAPs and two PAKs in A. oligospora using orthologous proteins in S. cerevisiae as reference sequences, and we have elucidated function and possible mechanisms of Aorga1 and Aopak1 in A. oligospora via phenotypic analysis, interaction protein verification, and expression pattern analysis. Our results indicate that PAKs are involved in the sporulation, pathogenicity, and stress resistance of A. oligospora under the indirect regulation of Rho-GAPs.
The wild-type (WT) strain A. oligospora (ATCC 24927) was maintained on potato dextrose agar (PDA) medium at 28°C, and the derived mutants were incubated on a PDA medium supplement with 200 μg/mL hygromycin. TGA (1% tryptone, 1% glucose, 2% agar) and TYGA (TGA with 0.5% yeast extracts and 1% molasses) media were used for analyzing the phenotypic traits. Sporulation and trap induction were performed on corn meal yeast extract (CMY) and water agar (WA) medium, respectively. The nematode Caenorhabditis elegans (strain N2) was maintained on oatmeal medium at room temperature for the bioassay. The mutant strains used in this study are listed in Supplementary Table S1.
The protein sequences of AoPak1 (AOL_s00004g340), AoPak2 (AOL_s00079g352), AoRga1 (AOL_s00110g81), and AoRga2 (AOL s00076g167) were identified in A. oligospora using the orthologs from the model fungus S. cerevisiae. The candidate sequences were chosen based on an E-value of ≤1e−10. Similarly, we retrieved homologous proteins from Magnaporthe oryzae, Neurospora crassa, Aspergillus nidulans, Beauveria bassiana and five NT fungi, including Dactylella cylindrospora, Drechslerella stenobrocha, Drechslerella brochopaga, Arthrobotrys flagrans, and Arthrobotrys entomopaga. The sequence similarities between orthologous PAKs and Rho-GAPs from different fungi were aligned with Geneious 4.8.5 software. The phylogenetic trees were constructed using MEGA 7.0 with the neighbor-joining method, the JTT + I + G substitution model with 1,000 bootstrap replicates. The phylogenetic trees were visualized using FigTree v1.4.2. The conserved domains were predicted using Pfam 35.0 (Zhu et al., 2022b).1
The targeted genes were disrupted using the homologous recombination method as described previously (Colot et al., 2006). Upstream and downstream 2000 bp fragments of the target genes were amplified from A. oligospora DNA. The selection marker gene hph was amplified from the pCSN44 plasmid. Three fragments were inserted into the pRS426 plasmid to form a fusion vector. The recombinant fragments were amplified by PCR and transformed into A. oligospora protoplasts in a PEG-mediated manner, as described previously (Tunlid et al., 1999; Zhu et al., 2023). Transformants were cultured on PDAS medium with 200 μg/mL hygromycin B and verified by PCR, and positive transformants were further confirmed by Southern blot analysis according to the instructions of the North2South Chemiluminescent Hybridization Detection Kit (Pierce, Rockford, United States). The primers are listed in Supplementary Table S4.
The WT and mutant strains were activated on PDA medium for 5 days; then, colonies with a diameter of 9 mm were, respectively, inoculated on TG, PDA, and TYGA media for 7 days at 28°C. The colony diameter was recorded every 24 h, and photographs were taken on the seventh day to record morphology. For sporulation analysis, the activated WT and mutant strains were inoculated on CMY medium for 15 days at 28°C, sterile water (20 mL) was added to the cultures before they were shaken with glass beads to separate spores from mycelia, and then the cultures were filtered to obtain a conidial suspension. Then, the numbers of conidia per microliter of suspension were measured using a counter. The number of spores per cm2 medium can be used as an estimate of the conidial yield (Zhu et al., 2022b).
The suspensions containing 20,000 conidia of WT and mutants were inoculated separately on WA medium and incubated at 28°C for 3–4 days until the mycelium covered the entire plate. In total, 200\u00B0C. elegans were added to each plate to induce trap formation. The same number of nematodes as in the experimental group was added to the WA plate that was not inoculated with the strain to indicate the number of nematodes that died naturally, which was indicated by “uninfected.” We observed the trap morphology, and quantified the numbers of traps and nematode mortality at 12 h intervals.
The 9 mm colony discs of WT and mutant strains were added to PD broth medium with skimmed milk (8%) and incubated at 180 rpm and 28°C for 5 days. The fermentation solution was filtered under sterile conditions to obtain the liquid supernatant. The qualitative assessment of protease activity was carried out by adding fermentation broth to the casein skimmed milk plates, and the quantitative analysis of protease activity was carried out as previously described (Yang et al., 2021; Li et al., 2023). All experiments were performed with at least three repetitions.
For the staining of nuclei, 10 μg/mL DAPI (Sigma-Aldrich) was added to the 5-day-old mycelia of WT and mutants for 10 min, followed by the addition of 20 μL calcofluor white (CFW,10 μg/mL, Sigma-Aldrich). These were then photographed with a fluorescent microscope after 10 min of staining, followed by counting the number of nuclei in each cell using ImageJ.
The 9 mm fungal discs of activated WT and mutant strains were inoculated on TG plates supplemented with different concentrations of stress reagents, and the colony diameters were measured after 7 days of incubation at 28°C. The diameter of the colony cultured on TG medium without stress reagents was used as a control to calculate the relative growth inhibition (RGI; Zhen et al., 2018).
The mycelial samples used for the transcription analyses of genes related to proteases, sporulation, cell wall synthesis, and oxidative stress response were collected from PDA combined with nematodes, CMY, TG with Congo red (0.06 mg/mL) or menadione (0.07 mM), respectively. Total RNA was isolated using an RNA extraction kit (Axygen, Jiangsu, Suzhou) and reverse-transcribed cDNA using a PrimeScriptRT reagent kit (TaKaRa, Japan) according to the manufacturer’s instructions. The transcriptional levels of genes were detected using the LightCycler 480 SYBR green I master mix (Roche, Basel, Switzerland), and β-tubulin (AOL_s00076g640) was used as the internal control. The relative transcript level of each gene was calculated using the 2−∆∆CT method (Livak and Schmittgen, 2001). All primers used for RT-PCR are listed in Supplementary Table S4.
The coding sequences of AoRga1 and AoPak1 were cloned into the pGBKT7 vector as bait, respectively. The encoding sequences of small GTPases AoCdc42, AoRas, and AoRac were inserted separately into the pGADT7 vector as prey. The cDNA is used as the template for PCR amplification and the primers are listed in Supplementary Table S4. The pGBKT7 and pGADT7 fusion plasmids were co-transformed into the Y2H Gold (Weidi, Shanghai, China) competent cells and inoculated on synthetic dropout medium (SD/−Trp/−Leu and SD/−Trp/−Leu/–His/−Ade). The substrate X-α-gal was added to the SD/−Ade/–His/−Leu/−Trp medium to detect the α-galactosidase activity.
All experimental data have been presented as the mean ± standard deviation (SD) of three biological repetitions. The multiple T-test was performed using Prism 8.0 (GraphPad, San Diego, CA, United States) to statistically evaluate the differences between treatments. p < 0.05 was considered significant.
Based on the orthologous proteins in S. cerevisiae, we classified PAKs into Pak1 and Pak2, and Rho-GAP into Rga1, Rga2 and Rga3 (Figure 1). And two PAKs (AoPak1, AOL_s00004g340; AoPak2, AOL_s00079g352) and two Rho-GAPs (AoRga1, AOL_s00110g81; AoRga2, AOL s00076g167) were identified in A. oligospora. Bioinformatic analyses showed that AoPAKs were conserved between S. cerevisiae and filamentous fungi, and especially in five NT fungi, including A. flagrans, D. cylindrospora, A. entomopaga, D. brochopaga, and A. oligospora, wherein AoPak1 and AoPak2 showed high similarity (71.5–96.0% and 70.0–94.2%) with the orthologs from the other four NT fungi, respectively (Supplementary Table S2). Phylogenetic and domain analyses showed that A. flagrans is the closest evolutionary relative to A. oligospora, and the PAKs contain an STKc_PAK and a PBD domain in five NT fungi. In addition, AoPak2 also contains a PH_Cla4_Ste20 domain, and AoPak2 has a higher homology with Cla4 of S. cerevisiae, while AoPak1 is more similar to Ste20 (Figure 1A). Rho-GAPs contain a conserved Rho-GAP domain, and AoRga1 is conserved only in NT fungi with a high homology of 71.9–92.5% (Figure 1B; Supplementary Table S3).
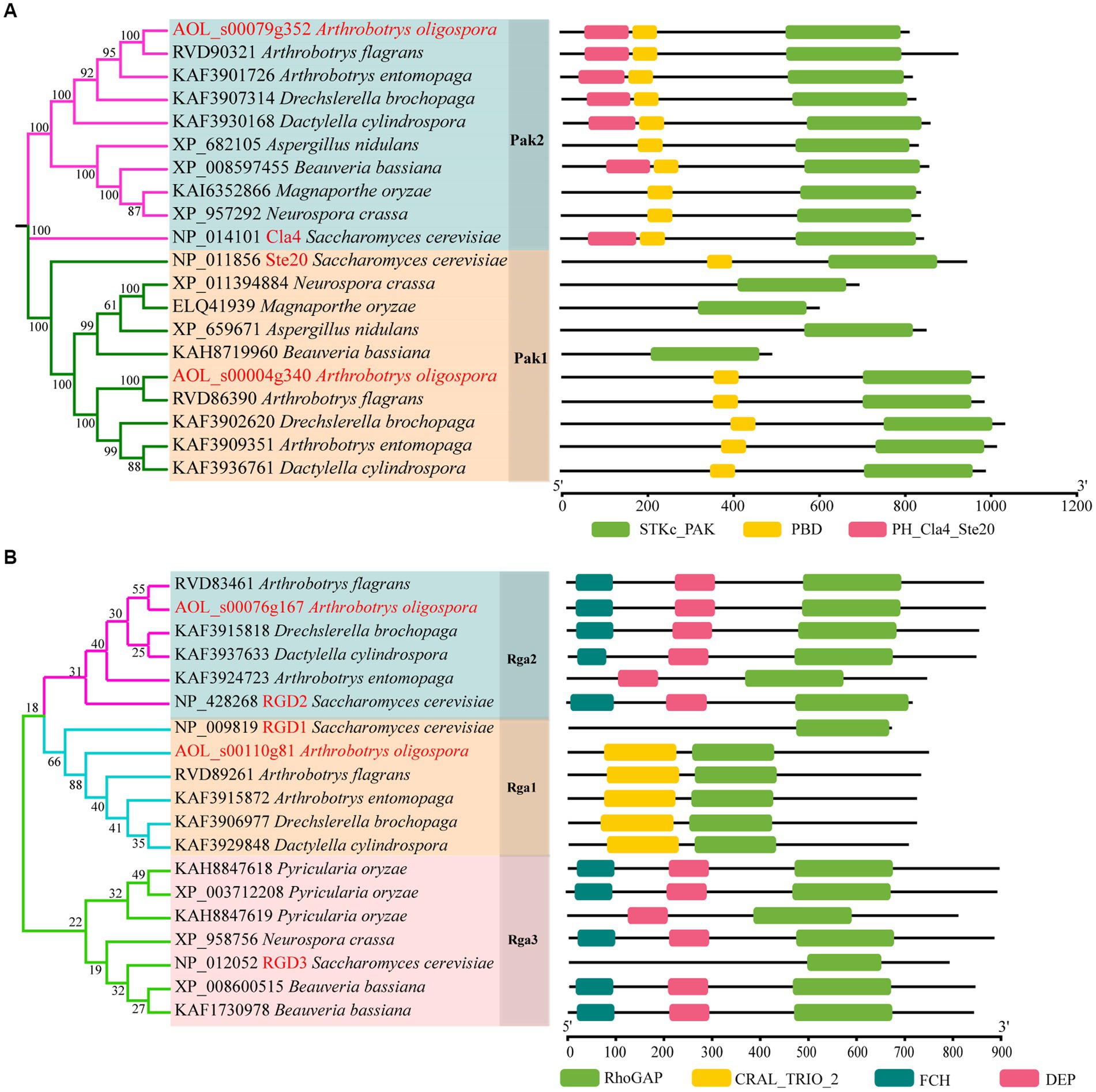
Figure 1. Phylogenetic analysis of Rho-GAPs and PAKs. (A, B) Phylogenetic and structure domain analyses of Rho-GAP and PAK orthologs from different fungi.
To determine the functions of Aopaks and Aorga1 in A. oligospora, we tried to delete the genes of Aopak1, Aopak2 and Aorga1 via homologous recombination. Finally, we successfully obtained ∆Aopak1 and ∆Aorga1 mutants with more than three positive transformants for each gene. The transformants were identified by genomic PCR amplification and Southern blotting analysis (Figures 2A,B). Because the individual transformants of each gene showed similar phenotypic traits, a single transformant for each gene was randomly selected for subsequent analysis.
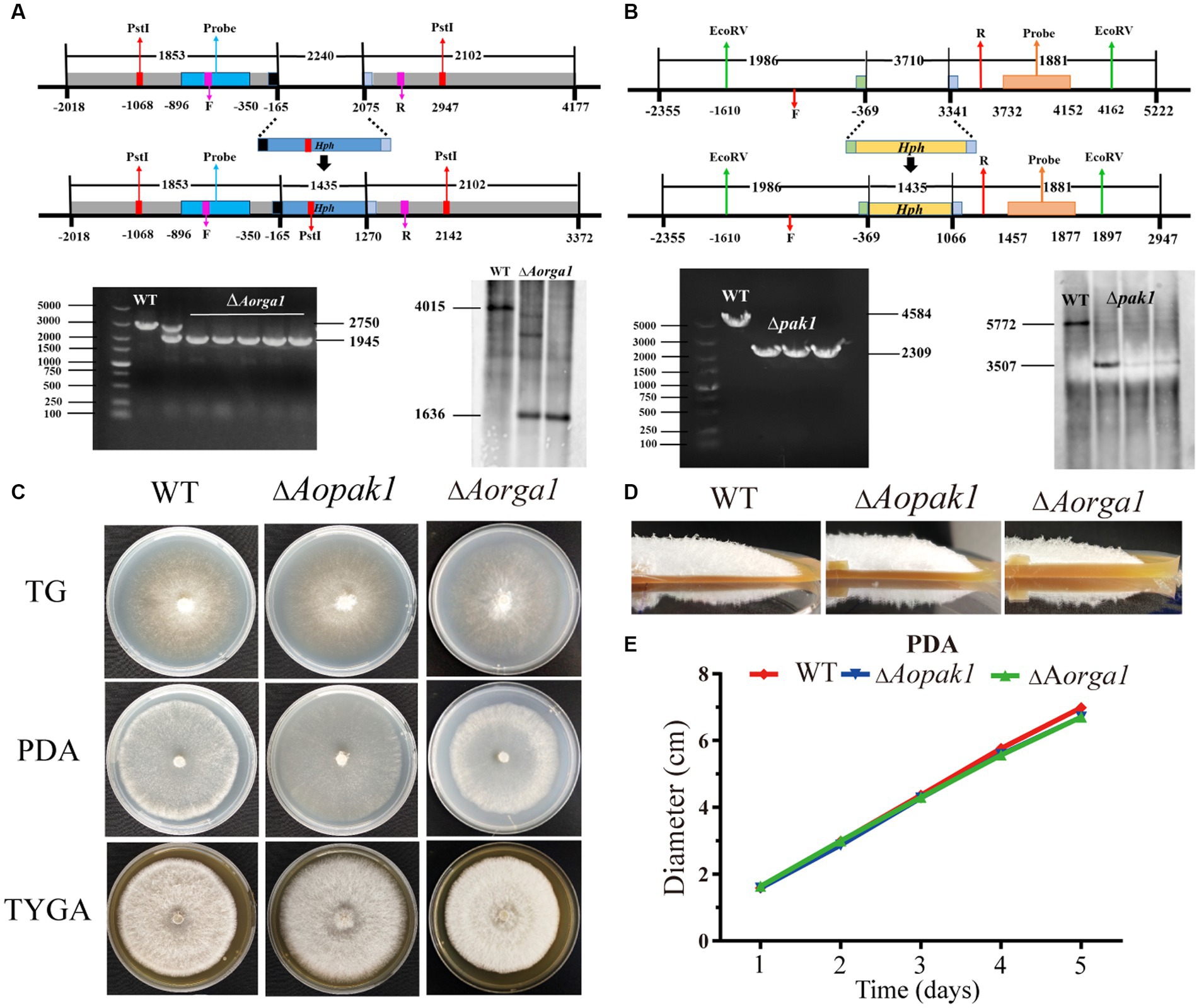
Figure 2. Comparison of the mycelial growth between WT and mutants (∆Aopak1 and ∆Aorga1). (A, B) Validation of Aorga1 and Aopak1 knockout strains using PCR and Southern blot. F, 5′ primer for validation of the transformants by PCR. R, 3′ primer for validation of the transformants by PCR. (C) Colony morphology of fungal strains cultured on TG, PDA and TYGA media for 5 days at 28°C. (D) The morphology of aerial hyphae incubated on TGYA medium for 5 days. (E) Comparison of mycelial growth on PDA medium.
WT and mutant strains were cultured on TG, PDA, and TYGA media for 5 days, and the colony morphology and mycelial growth between the WT and two mutant strains showed no obvious differences (Figure 2; Supplementary Figure S1). The mean colony diameters of the WT, ∆Aopak1 and ∆Aorga1 mutant strains cultured on TG media for 5 days were 6.65, 7.02, and 6.93 cm, respectively; these measurements were 6.98, 6.70, and 6.70 cm on PDA and 6.77, 7.05, and 6.75 cm on TYGA.
We added 200 nematodes to each plate of WT, ∆Aopak1, and ∆Aorga1 mutant strains to induce trap formation. The WT and mutants produced traps at 12 h post-induction (hpi), but there were differences in the ability of trap formation (Figure 3A). Compared with WT, the trap numbers of ∆Aorga1 mutants were significantly increased at 36 and 48 hpi, whereas those of ∆Aopak1 were decreased, most notably at 48 hpi (Figure 3B). However, the nematode predatory efficiency for ΔAopak1 and ∆Aorga1 was not significantly different from that of the WT at different time points (Figure 3C).
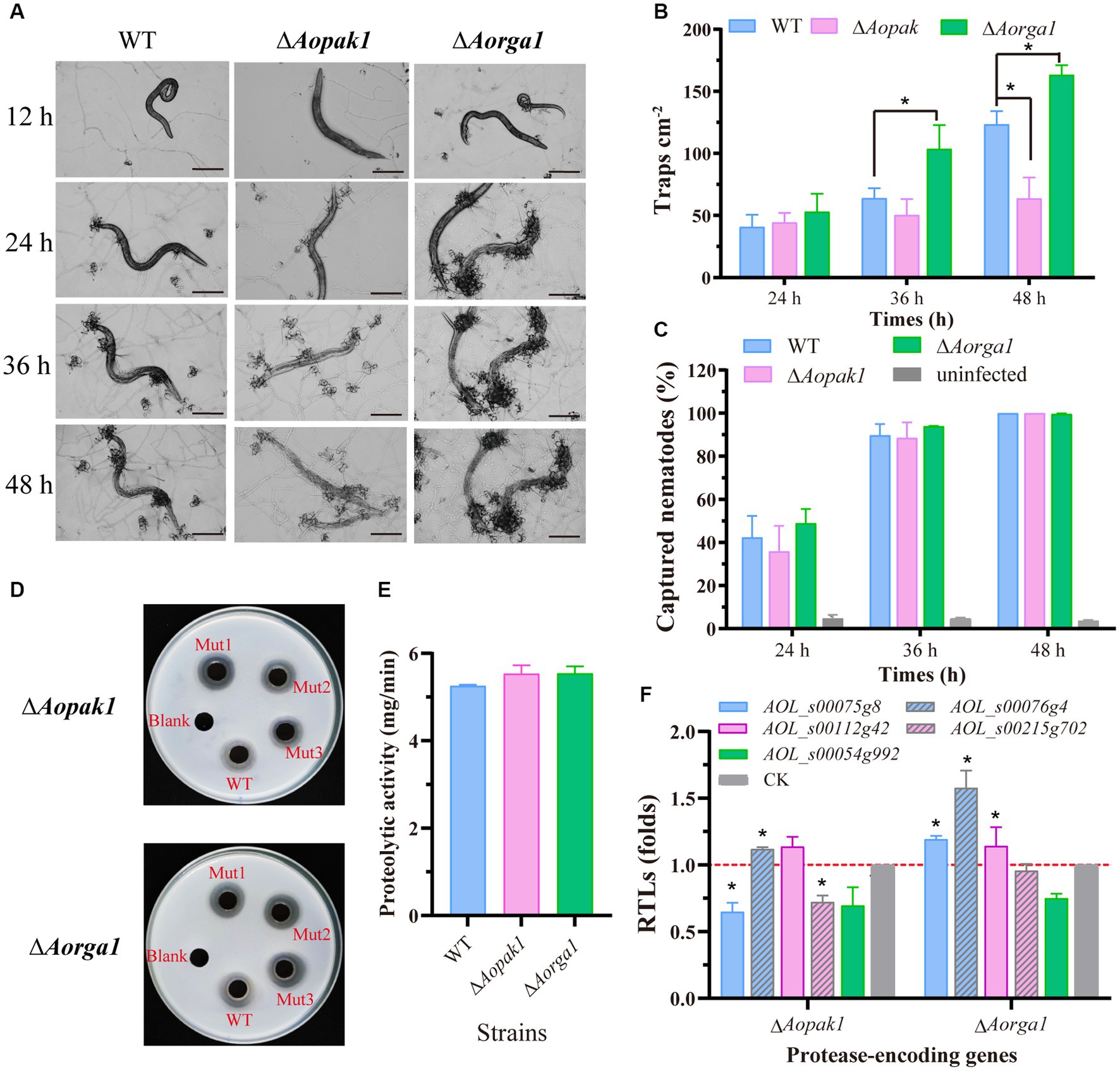
Figure 3. Comparison of trap formation and extracellular proteolytic activity in the WT and mutant strains. (A) The morphology of captured nematodes and traps at 12, 24, 36, and 48 h post-induction (hpi). Bar = 100 μm. (B) Number of traps at three different time points (24, 36, and 48 hpi). *, p < 0.05. (C) Comparison of captured nematodes in WT and mutants (∆Aopak1 and ∆Aorga1). (D, E) Qualitative and quantitative determination of extracellular proteolytic activity. (F) Relative transcript levels (RTLs) of protease-related genes between the WT strain and mutants (ΔAopak1 and ΔAorga1) cultured on PDA medium for 5 days, and then inducted by C. elegans for 24 h. * p < 0.05.
Furthermore, the disruption of the Aopak1 and Aorga1 genes resulted in a slight increase in proteolytic activity compared to WT strains (Figures 3D,E), which is consistent with the nematode-digesting capacity of the strains. To further explore the regulation of Aopak1 and Aorga1 in terms of serine proteases, the relative transcript levels (RTLs) of five protease-related genes were determined by RT-PCR in WT, ∆Aopak1, and ∆Aorga1 strains. The RTL of 76 g4 was significantly up-regulated (p < 0.05) in both ∆Aopak1 and ∆Aorga1 mutants, and the RTL of 54 g992 was significantly down-regulated (p < 0.05) in the Aopak1 and Aorga1 disruption strains. The opposite expression trend of 75 g8 was observed in the ∆Aopak1 and ∆Aorga1 mutants (Figure 3F).
The deletion of both Aopak1 and Aorga1 resulted in a remarkable reduction in conidiophores. Particularly in the ΔAopak1 mutants, not only did the conidiophores become more sparse, but the number of spores on each conidiophore was also remarkably decreased (Figure 4A). On the contrary, the conidia attached on the conidiophores of the ΔAorga1 mutant strains showed multiple whorls (Figure 4B). As a result, the spore yields of the mutants were significantly reduced compared to the WT, and the conidia yields of WT, ΔAopak1, and ΔAorga1 strains were 2.23, 0.40, and 1.59 × 105 conidia/cm2, respectively (Figure 4C). Following this, the transcriptional levels of most sporulation-related genes were down-regulated in the ΔAopak1 mutant, especially AovosA, AovelB, AoveA, AomedA, Aohyp1, and AobrlA. However, these genes were up-regulated in the ΔAorga1 mutant, except for AomedA and Aohyp1 (Figure 4D).
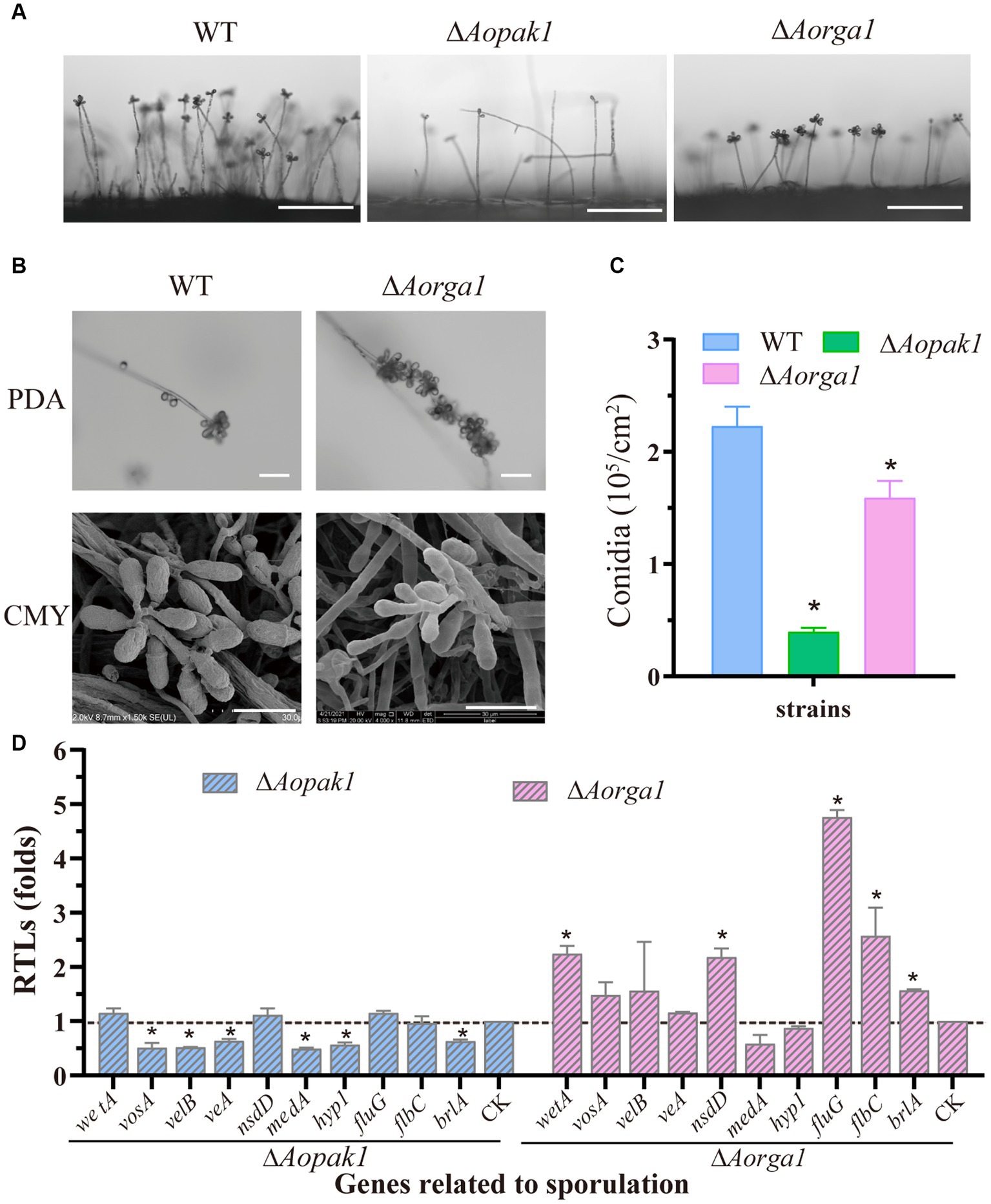
Figure 4. Comparison of conidiation and the transcript level analysis of sporulation-related genes between the WT and mutant strains. (A) Conidiophore observation in WT and mutants (ΔAopak1 and ΔAorga1) cultured on PDA for 3 days. Bar = 100 μm. (B) Comparison of conidiophore morphology between the WT and ΔAorga1 mutant strains cultured on CMY for 7 days. Bar = 20 μm. (C) Spore yield statistics for WT and mutants cultured on CMY for 15 days. * p < 0.05. (D) Relative transcription levels (RTLs) of sporulation-related genes between the WT and mutant strains. The expression of the corresponding genes in WT was normalized and used as CK. * p < 0.05.
To probe the roles of Aopak1 and Aorga1 in stress response, we treated strains with a cell-wall-disturbing reagent (Congo red) and oxidant (menadione). The results show that the ΔAopak1 mutant was more sensitive to the cell wall and oxidative stress reagents, as the RGI value of the ΔAopak1 mutant was significantly higher than that of the WT strain. However, the sensitivity of the ΔAorga1 mutant to the cell wall and oxidative stress reagents was decreased, and the RGI values here were significantly lower than those of WT (Figures 5A,B). Aohex (coding hexokinase), a gene related to cell wall biosynthesis, was significantly down-regulated in the ΔAopak1 mutant, whereas the RTLs of three cell-wall-synthesis-related genes (Aochs-1, coding chitin synthases; Aoglu, coding β-glucosidase; and Aogls, coding 1,3-β-glucan synthase) were remarkably up-regulated in the ΔAorga1 mutant (Figure 5C). Similarly, five oxidative-stress-response-related genes, including Aothi (coding thioredoxin), Aoper (coding peroxidase), Aoglr (glutathione reductase), Aogld (coding glutathione dehydrogenase), and Aocat-1 (coding catalase), were significantly up-regulated in the ΔAorga1 mutant (Figure 5D).
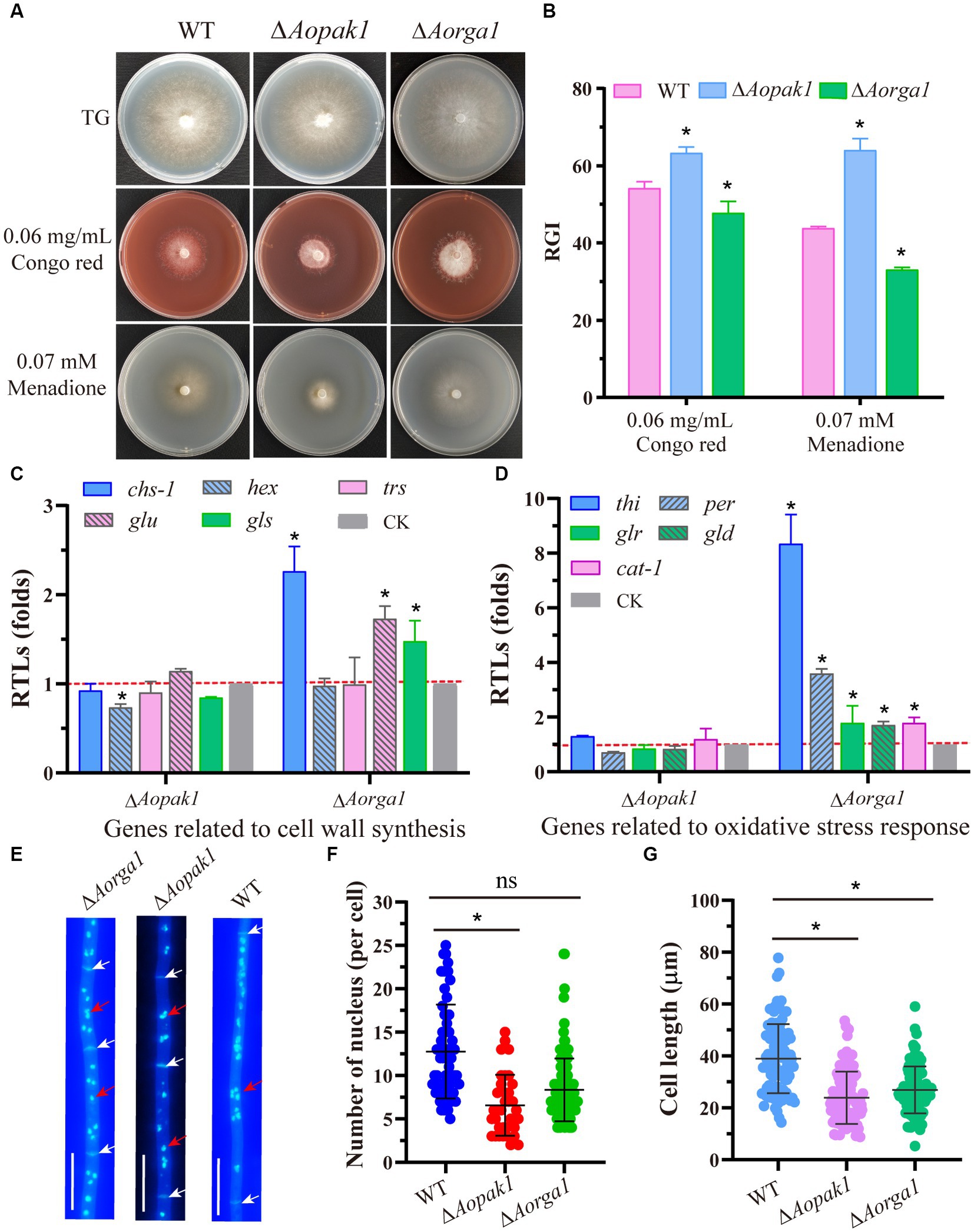
Figure 5. Comparison of stress responses and the numbers of nuclei. (A) Colony morphology of WT and mutants cultured on TG plates supplemented with 0.06 mg/mL Congo red and 0.07 mM menadione, respectively. (B) Relative growth inhibition (RGI) rate of WT and mutants cultured on (A). * p < 0.05. (C, D) Relative transcription levels (RTLs) of genes associated with cell wall synthesis and oxidative stress response in the mutants compared with the WT strain. * p < 0.05. (E) Representative images of nuclei stained with DAPI and visualized using a fluorescent microscope. White arrow, septum. Red arrow, nuclei. Bar = 10 μm. (F) Statistical analysis of the number of nuclei. ns, not statistically significant, * p < 0.05. (G) Statistical analysis of cell length. ns, not statistically significant, * p < 0.05.
Furthermore, disrupting Aopak1 and Aorga1 resulted in a decrease in the nuclei number of per cell and cell length. Statistical analysis showed that each cell has an average of 13, 7, and 8 nuclei in the WT, ΔAopak1, and ΔAorga1 mutants, respectively (Figures 5E,F). The mean cell length of ΔAopak1 and ΔAorga1 mutants was significantly shorter than that of WT (Figure 5G).
To probe the regulation mechanism of Aopak and Aorga1 in A. oligospora, we determined the transcription patterns of three genes at different developmental stages. The transcription level of Aopak2 in the sporulation stage was significantly higher than that in the vegetative growth stage, and the transcriptional level of Aorga1 was remarkably down-regulated at 24 h after the induction of C. elegans (Figure 6A). Previous studies showed that small GTPases can bind to the CRIB domain of PAKs and are regulated by Rho-GAPs (Boyce and Andrianopoulos, 2011; Ye et al., 2014). Therefore, we detected the transcription of three genes encoding the Rho GTPases in ΔAorga1 mutants, and we found that Aorac was significantly down-regulated (Figure 6B). Moreover, the Aorga1 was significantly up-regulated in ΔAocdc42, ΔAorho2, and ΔAorac mutants, whereas Aopak1 was down-regulated in three mutants (Figure 6C). Interestingly, Aorga1 was down-regulated in ΔAopak1 mutants, while Aopak1 and Aopak2 were both up-regulated in ΔAorga1 mutants (Figures 6D,E). In addition, Y2H analysis showed that AoPak1 cannot interact with AoRga1, but they can both interact with AoRac, and AoPak1 can also interact with AoCdc42 (Figure 6F; Supplementary Figure S2). These results demonstrate that AoRga1 indirectly regulates AoPAK by regulating smallGTPases.
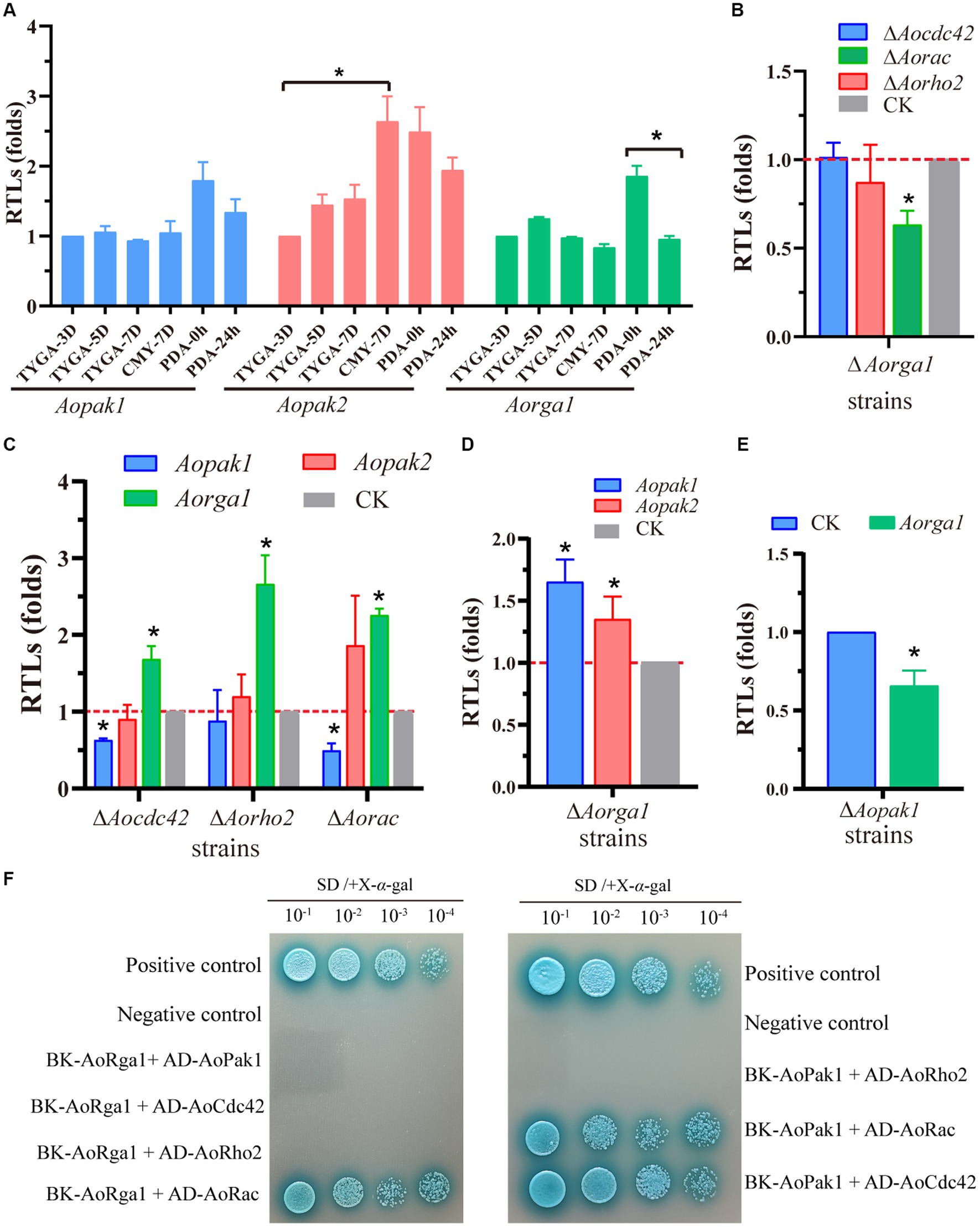
Figure 6. AoRho-GAP indirectly regulates AoPAKs by regulating Rho GTPases. (A) Transcription patterns of Aopak1, Aopak2, and Aorga1 in different developmental stages of WT. * p < 0.05. (B–E) Relative transcription levels (RTLs) of Aopak1, Aopak2, Aorga1, Aocdc42, Aorho2, and Aorac in different mutants. The expression of the corresponding genes in WT was normalized and used as CK. * p < 0.05. (F) Verification of the interaction relationship between AoPak1, AoRga1, and Rho GTPases (AoCdc42, AoRho2, and AoRac) by Y2H assay. The interaction of pGBKT7-53 with pGADT7-T was used as a positive control, and the interaction of pGBKT7-lam with ADT7-T was used as a negative control.
Signaling pathways play a crucial role in the vegetative growth and development in fungi, as they can sense alterations in various physical and chemical stimuli in the environment and translate them into intracellular signals (Yang et al., 2011; Zhu et al., 2022a). PAKs function upstream of the MAPK cascades and are regulated by small GTPases, which are negatively regulated by Rho-GAPs (Dan et al., 2001; Boyce and Andrianopoulos, 2011; Rawat and Chernoff, 2015). PAKs participate in multiple phenotypes in fungi, including polarized morphogenesis (Leveleki et al., 2004; Nichols et al., 2004), cell morphogenesis (Boyce et al., 2009), conidial germination (Boyce and Andrianopoulos, 2007), virulence and hyphal formation (Leberer et al., 1997), and pathogenicity (Li et al., 2004; Rolke and Tudzynski, 2008). In addition, Rho-GAPs play a vital role in cell proliferation (Shang et al., 2007), pathogenicity (Ye et al., 2014), and growth (Yang et al., 2003). Here, we found that AoPak1 and AoRga1 were also involved in multiple biological processes in the NT fungus A. oligospora, such as sporulation, trap formation, and stress response.
The disruption of Aopak1 or Aorga1 had no effect on hyphal growth (Figure 2). Similarly, the deletion of skm1 (coding a PAK protein) manifested no detectable phenotype under laboratory conditions, but the overexpression of skm1, ste20 or cla4 lacking an N-terminus led to growth arrest in S. cerevisiae (Martin et al., 1997). However, the deletion of Molrg1 resulted in a dramatic decrease in the growth rate of aerial hyphae in M. oryzae, while the six other Rho-GAP-domain containing genes did not impact vegetative growth due to functional redundancy (Ye et al., 2014). In M. oryzae, ∆mst20 mutants were reduced during aerial hyphae growth (Li et al., 2004). The presence of ∆cla4 mutants completely removed the ability to form filaments (Szabo, 2001). Based on these results we speculate functional redundancy between Aopak1 and Aopak2, meaning the deletion of Aopak1 does not affect mycelial growth.
The traps are important infectious structures of NT fungi, and mature trap formation is essential to their pathogenicity (Zhu et al., 2022a). The disruption of Aopak1 resulted in a reduction in trap number at 48 hpi, while the numbers of traps were increased in ∆Aorga1 mutants at 36 and 48 hpi (Figure 3B). However, the nematode predation abilities of ΔAopak1 and ΔAorga1 mutants were similar to that of WT (Figure 3C). NT fungi can secrete cuticle-degrading serine proteases, which act as key mediators of virulence against nematodes, and many related gens have been cloned (Yang et al., 2005, 2017; Liang et al., 2011; Tzean et al., 2016). For example, serine protease PII was first identified in A. oligospora; it can immobilize the free-living nematode Panagrellus redivivus, and degrade the nematode cuticle (Tunlid et al., 1994). Herein, we analyzed the expressions of serine protease-related genes and found that most genes were differently expressed in ΔAopak1 and ΔAorga1 mutants, and 76 g4 (encoding cuticle-degrading protease PII) was remarkably up-regulated in ΔAopak1 and ΔAorga1 mutants, suggesting that Aopak1 and Aorga1 play crucial roles in the regulation of extracellular proteolytic activity. In addition, previous studies have showed that cla4 is required for pathogenesis in C. albicans (Leberer et al., 1997), B. maydis (Kitade et al., 2019), U. maydis (Leveleki et al., 2004), C. purpurea (Rolke and Tudzynski, 2008), and V. dahliae (Tian et al., 2015). Meanwhile, mst20 plays a key role in the pathogenicity of M. grisea (Li et al., 2004), U. maydis (Smith et al., 2004), and C. neoformans (Nichols et al., 2004), but has a negligible effect on pathogenicity in B. maydis (Kitade et al., 2019). Moreover, the deletion of Molrg1 resulted in a complete loss of pathogenicity in M. oryzae, but the appressorial formation and pathogenicity of six genes’ (coding Rho-GAP proteins) mutants were similar to those of WT (Ye et al., 2014). These results indicate that the roles of PAKs and Rho-GAPs in pathogenicity vary among fungal species, and there is functional redundancy among homologous genes. Similarly, Aopak and Aorga1 have been shown to be involved in trap formation in A. oligospora, and Aopak2 may complement the functional defects caused by the knockout of Aopak1. In contrast to studies in other species, the knockout of Aorga1 here resulted in an increase in the number of traps and an accelerated rate of nematode digestion. The specific mechanisms involved here need to be further explored.
The main component of biocontrol agents is conidia, and the ability to produce spores is one of the key factors at play in the fecundity and fitness of biocontrol fungi (Zhang et al., 2019). Studies have shown that the deletion of Morga1 results in a high percentage of larger or gherkin-shaped conidia and decreases in conidiation (Ye et al., 2014). In M. grisea, conidiation was reduced in both ∆chm1 and ∆mst20 mutants, most notably in the former, which showed more conidia with abnormal morphologies (Li et al., 2004). The conidiation of ∆ste20 mutants was similar to that of WT, while ∆cla4 strains showed more severe defects than the WT strain in Bipolaris maydis (Kitade et al., 2019). Consistently with the results for M. grisea, the deletion of Aopak1 and Aorga1 resulted in a significantly decrease in spore yield (Figure 4C), and the conidiophore morphology of ΔAorga1 mutants was abnormal (Figure 4B). The transcriptions of several sporulation-related genes (Bai et al., 2023b), including AovosA, AovelB, AoveA, AomedA, Aohyp1, and AobrlA, were significantly down-regulated in the ΔAopak1 mutant, while AowetA, AonsdD, AofluG, AoflbC, and AobrlA were up-regulated in the ΔAorga1 mutant (Figure 4D). Except for brlA, the differentially expressed genes regulated by Aopak1 and Aorga1 were different, and these genes were regulated in opposite patterns in the ΔAopak1 and ΔAorga1 mutants, suggesting that AoPak1 and AoRga1 play different roles in the regulation of conidia formation.
Stress response is essential to fungi’s sense of their environment, and enables them to make timely adjustments to adapt to changes. In this study, we found that the deletion of Aopak1 led to increased sensitivity to oxidant and cell-wall-disturbing reagents, whereas the disruption of Aorga1 resulted in an increased resistance to these stressors (Figure 5B). The alterations in the transcriptional levels of related genes were consistent with the corresponding phenotypes, especially in the ΔAorga1 mutant, where most of the related genes associated with cell wall synthesis and oxidative stress response were significantly up-regulated (Figures 4C,D). These results suggest that Aopak1 and Aorga1 play critical and diverse roles in stress response. Previous studies have shown that PAKs, upstream of MAPK cascades, are indirectly regulated by Rho-GAPs (Bassilana et al., 2005; Boyce and Andrianopoulos, 2011; Ye et al., 2014). In S. cerevisiae, activated Ste20 phosphorylates the MAPK kinase. The kinase Ste11 can activate the MAPK pathways regulating cell wall integrity and filamentous growth (Wu et al., 1995; van Drogen et al., 2000). In A. oligospora, Bck1, Mkk1, and Slt2 signaling cascade has been proved to be involved in multi-stress tolerance (Zhen et al., 2018; Xie et al., 2021). In addition, some transcription factors downstream of MAPK cascades have been confirmed to be involved in stress resistance, such as Ste12 (Bai et al., 2023a), RlmA (Yang et al., 2023), and Swi6 (Xie et al., 2022). Therefore, AoPaks and AoRho-GAPs may influence the stress response by regulating the downstream effectors, including the MAPK cascades and related transcription factors.
The transcription pattern analysis and Y2H results show that Rho GTPases link AoPAK and AoRho-GAP, enabling regulation between them, and there is negative feedback between Aopak and Aorga1 (Figure 6). Studies have confirmed that small GTPases are negatively regulated by Rho-GAP (Etienne-Manneville and Hall, 2002; Burridge and Wennerberg, 2004). In this study, Aopak1 was shown to be positively regulated by the phenotype of A. oligospora, similarly to the effect seen in other species, such as C. albicans (Leberer et al., 1997), B. maydis (Kitade et al., 2019), U. maydis (Leveleki et al., 2004), C. purpurea (Rolke and Tudzynski, 2008), V. dahliae (Tian et al., 2015), M. grisea (Li et al., 2004), and C. neoformans (Nichols et al., 2004). On the other hand, the deletion of Aorga1 resulted in alterations in the number of traps, nematode digestion efficiency, and stress response, which is a novel finding compared to other species. This may be related to the homology of the protein sequence in filamentous fungi. These results can be partly explained by alterations in the transcription pattern of the related genes in each mutant strain, and deserve more detailed study. Combined with the results of this study, we see that AoPAKs regulate the sporulation, trap formation, stress resistance, and number of nuclei via the indirect regulation of AoRho-GAP in A. oligospora (Figure 7). Our study elucidates the mechanisms involved in signal transduction pathways regulating conidia and trap formation, and it highlights the roles played by AoPAKs and AoRho-GAPs in improving the qualities of biocontrol fungi.
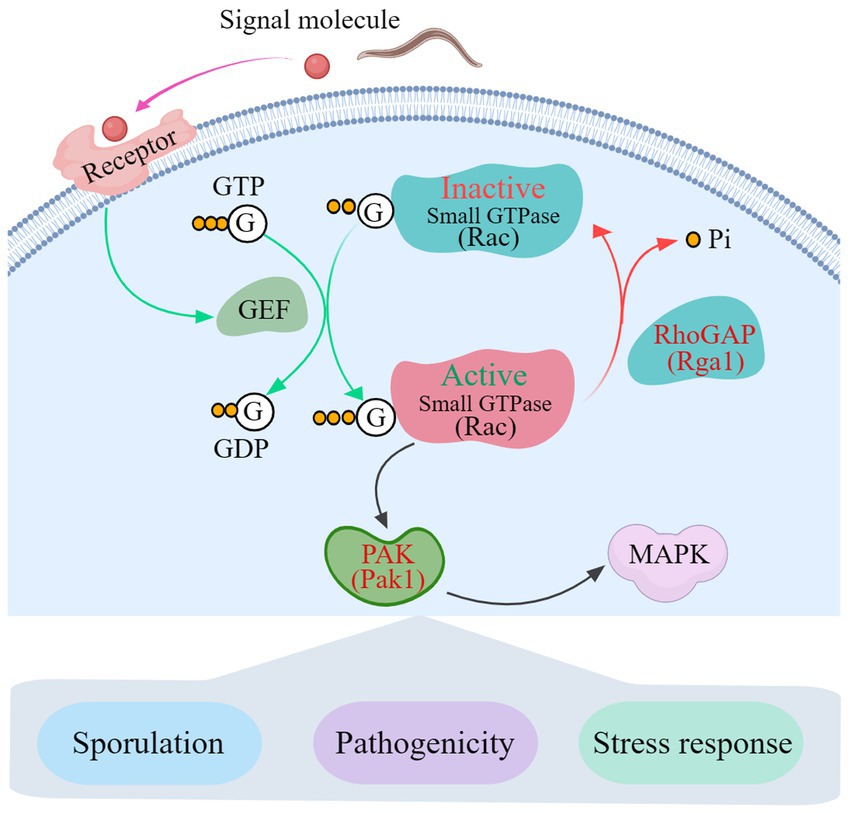
Figure 7. A proposed interaction model between Rho-GAP and PAKs in A. oligospora. GTP, Guanosine triphosphate; GDP, Guanosine-5′-diphosphate; GEF, guanine-nucleotide exchange factor; RhoGAP, Rho GTPase-activating protein; PAK, p21-GTPase-activated protein kinase; MAPK, mitogen-activated protein kinase.
We identified and characterized a Rho GAP and two PAK-coding genes, Aorga1, Aopak1 and Aopak2, from the NT fungus A. oligospora. Our results show that Aopak1 and Aorga1 play crucial roles in conidiation, trap formation, and response to oxidant and cell-wall-disturbing reagents. In particular, Aorga1 negatively regulates trap formation and nematode digestion, which is a novel finding in the context of other fungi. Our findings provide new insights into the Rho-GAP- and PAKs-mediated signaling pathways that regulate trap formation, conidiation, and stress resistance in NT fungi.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
JY conceived and designed the study, and revised the manuscript. MZ wrote the manuscript. MZ and YL conducted the experiments. XY, LZ, YS, and SD analyzed the data. All authors contributed to the article and approved the submitted version.
This study was provided by the Scientific Research Fund Project of Yunnan Provincial Department of Education (2022Y019).
We are grateful to Microbial Library of the Germplasm Bank of Wild Species from Southwest China for preserving and providing experimental strains.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1235283/full#supplementary-material
Bai, N., Xie, M., Liu, Q., Wang, W., Liu, Y., and Yang, J. (2023a). AoSte12 is required for mycelial development, conidiation, trap morphogenesis, and secondary metabolism by regulating hyphal fusion in nematode-trapping fungus Arthrobotrys oligospora. Microbiol. Spectr. 11:e0395722. doi: 10.1128/spectrum.03957-22
Bai, N., Xie, M., Liu, Q., Zhu, Y., Yang, X., Zhang, K. Q., et al. (2023b). AoMedA has complex regulatory relationship with AoBrlA, AoAbaA, and AoWetA in conidiation, trap formation, and secondary metabolism in the nematode-trapping fungus Arthrobotrys oligospora. Appl. Environ. Microbiol. e0098323. doi: 10.1128/aem.00983-23 [Online ahead of print].
Bassilana, M., Hopkins, J., and Arkowitz, R. A. (2005). Regulation of the Cdc42/Cdc24 GTPase module during Candida albicans hyphal growth. Eukaryot. Cell 4, 588–603. doi: 10.1128/EC.4.3.588-603.2005
Boyce, K. J., and Andrianopoulos, A. (2007). A p21-activated kinase is required for conidial germination in penicillium marneffei. PLoS Pathog. 3, e162–e1569. doi: 10.1371/journal.ppat.0030162
Boyce, K. J., and Andrianopoulos, A. (2011). Ste20-related kinases: effectors of signaling and morphogenesis in fungi. Trends Microbiol. 19, 400–410. doi: 10.1016/j.tim.2011.04.006
Boyce, K. J., Schreider, L., and Andrianopoulos, A. (2009). In vivo yeast cell morphogenesis is regulated by a p21-activated kinase in the human pathogen penicillium marneffei. PLoS Pathog. 5:e1000678. doi: 10.1371/journal.ppat.1000678
Burridge, K., and Wennerberg, K. (2004). Rho and Rac take center stage. Cells 116, 167–179. doi: 10.1016/s0092-8674(04)00003-0
Cansado, J., Soto, T., Gacto, M., and Perez, P. (2010). Rga4, a rho-GAP from fission yeast: finding specificity within promiscuity. Commun. Integr. Biol. 3, 436–439. doi: 10.4161/cib.3.5.12284
Chen, S.-A., Lin, H.-C., and Hsueh, Y.-P. (2022). The cAMP-PKA pathway regulates prey sensing and trap morphogenesis in the nematode-trapping fungus Arthrobotrys oligospora. G3. 12:jkac217. doi: 10.1093/g3journal/jkac217
Chen, S.-A., Lin, H.-C., Schroeder, F. C., and Hsueh, Y.-P. (2021). Prey sensing and response in a nematode-trapping fungus is governed by the MAPK pheromone response pathway. Genetics 217:iyaa008. doi: 10.1093/genetics/iyaa008
Colot, H. V., Park, G., Turner, G. E., Ringelberg, C., Crew, C. M., Litvinkova, L., et al. (2006). A high-throughput gene knockout procedure for neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. U. S. A. 103, 10352–10357. doi: 10.1073/pnas.0601456103
Cotteret, S., Jaffer, Z. M., Beeser, A., and Chernoff, J. (2003). p21-activated kinase 5 (Pak5) localizes to mitochondria and inhibits apoptosis by phosphorylating BAD. Mol. Cell. Biol. 23, 5526–5539. doi: 10.1128/MCB.23.16.5526-5539.2003
Dan, I., Watanabe, N. M., and Kusumi, A. (2001). The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell Biol. 11, 220–230. doi: 10.1016/s0962-8924(01)01980-8
Etienne-Manneville, S., and Hall, A. (2002). Rho GTPases in cell biology. Nature 420, 629–635. doi: 10.1038/nature01148
Holly, S. P., and Blumer, K. J. (1999). PAK-family kinases regulate cell and actin polarization throughout the cell cycle of Saccharomyces cerevisiae. J. Cell Biol. 147, 845–856. doi: 10.1083/jcb.147.4.845
Ji, X., Yu, Z., Yang, J., Xu, J., Zhang, Y., Liu, S., et al. (2020). Expansion of adhesion genes drives pathogenic adaptation of nematode-trapping fungi. iScience 23:101057. doi: 10.1016/j.isci.2020.101057
Keniry, M. E., and Sprague, G. F. (2003). Identification of p21-activated kinase specificity determinants in budding yeast: a single amino acid substitution imparts Ste20 specificity to Cla4. Mol. Cell. Biol. 23, 1569–1580. doi: 10.1128/MCB.23.5.1569-1580.2003
Kitade, Y., Sumita, T., Izumitsu, K., and Tanaka, C. (2019). Cla4 PAK-like kinase is required for pathogenesis, asexual/sexual development and polarized growth in Bipolaris maydis. Curr. Genet. 65, 1229–1242. doi: 10.1007/s00294-019-00977-9
Kuo, C.-Y., Chen, S.-A., and Hsueh, Y.-P. (2020). The high osmolarity glycerol (HOG) pathway functions in osmosensing, trap morphogenesis and conidiation of the nematode-trapping fungus Arthrobotrys oligospora. J. Fungi 6:191. doi: 10.3390/jof6040191
Leberer, E., Harcus, D., Broadbent, I. D., Clark, K. L., Dignard, D., Ziegelbauer, K., et al. (1996). Signal transduction through homologs of the Ste20p and Ste7p protein kinases can trigger hyphal formation in the pathogenic fungus Candida albicans. Proc. Natl. Acad. Sci. U. S. A. 93, 13217–13222. doi: 10.1073/pnas.93.23.13217
Leberer, E., Ziegelbauer, K., Schmidt, A., Harcus, D., Dignard, D., Ash, J., et al. (1997). Virulence and hyphal formation of Candida albicans require the Ste20p-like protein kinase CaCla4p. Curr. Biol. 7, 539–546. doi: 10.1016/s0960-9822(06)00252-1
Leveleki, L., Mahlert, M., Sandrock, B., and Bolker, M. (2004). The PAK family kinase Cla4 is required for budding and morphogenesis in Ustilago maydis. Mol. Microbiol. 54, 396–406. doi: 10.1111/j.1365-2958.2004.04296.x
Li, L., Xue, C. Y., Bruno, K., Nishimura, M., and Xu, J. R. (2004). Two PAK kinase genes, CHM1 and MST20, have distinct functions in Magnaporthe grisea. Mol. Plant-Microbe Interact. 17, 547–556. doi: 10.1094/MPMI.2004.17.5.547
Li, X., Zhu, M., Liu, Y., Yang, L., and Yang, J. (2023). Aoatg11 and Aoatg33 are indispensable for mitophagy, and contribute to conidiation, the stress response, and pathogenicity in the nematode-trapping fungus Arthrobotrys oligospora. Microbiol. Res. 266:127252. doi: 10.1016/j.micres.2022.127252
Liang, L., Yang, J., Li, J., Mo, Y., Li, L., Zhao, X., et al. (2011). Cloning and homology modeling of a serine protease gene (PrC) from the nematophagous fungus Clonostachys rosea. Ann. Microbiol. 61, 511–516. doi: 10.1007/s13213-010-0166-5
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Martin, H., Mendoza, A., RodriguezPachon, J. M., Molina, M., and Nombela, C. (1997). Characterization of SKM1, a Saccharomyces cerevisiae gene encoding a novel Ste20/PAK-like protein kinase. Mol. Microbiol. 23, 431–444. doi: 10.1046/j.1365-2958.1997.d01-1870.x
Molli, P., Li, D., Murray, B., Kayala, S., and Kumar, R. (2009). PAK signaling in oncogenesis. Oncogene 28, 2545–2555. doi: 10.1038/onc.2009.119
Mosaddeghzadeh, N., and Ahmadian, M. (2021). The RHO family GTPases: mechanisms of regulation and signaling. Cells 10:1831. doi: 10.3390/cells10071831
Ness, F., Prouzet-Mauleon, V., Vieillemard, A., Lefebvre, F., Noel, T., Crouzet, M., et al. (2010). The Candida albicans Rgd1 is a RhoGAP protein involved in the control of filamentous growth. Fungal Genet. Biol. 47, 1001–1011. doi: 10.1016/j.fgb.2010.07.007
Nichols, C. B., Fraser, J. A., and Heitman, J. (2004). PAK kinases Ste20 and Pak1 govern cell polarity at different stages of mating in Cryptococcus neoformans. Mol. Biol. Cell 15, 4476–4489. doi: 10.1091/mbc.e04-05-0370
Phani, V., Khan, M. R., and Dutta, T. K. (2021). Plant-parasitic nematodes as a potential threat to protected agriculture: current status and management options. Crop Prot. 144:105573. doi: 10.1016/j.cropro.2021.105573
Philbrick, A. N., Adhikari, T. B., Louws, F. J., and Gorny, A. M. (2020). Meloidogyne enterolobii, a major threat to tomato production: current status and future prospects for its management. Front. Plant Sci. 11:606395. doi: 10.3389/fpls.2020.606395
Qyang, Y. B., Yang, P. R., Du, H. Y., Lai, H., Kim, H. W., and Marcus, S. (2002). The p21-activated kinase, Shk1, is required for proper regulation of microtubule dynamics in the fission yeast, Schizosaccharomyces pombe. Mol. Microbiol. 44, 325–334. doi: 10.1046/j.1365-2958.2002.02882.x
Rawat, S. J., and Chernoff, J. (2015). Regulation of mammalian Ste20 (Mst) kinases. Trends Biochem. Sci. 40, 149–156. doi: 10.1016/j.tibs.2015.01.001
Rolke, Y., and Tudzynski, P. (2008). The small GTPase Rac and the p21-activated kinase Cla4 in Claviceps purpurea: interaction and impact on polarity, development and pathogenicity. Mol. Microbiol. 68, 405–423. doi: 10.1111/j.1365-2958.2008.06159.x
Roumanie, O., Weinachter, C., Larrieu, I., Crouzet, M., and Doignon, F. (2001). Functional characterization of the Bag7, Lrg1 and Rgd2 RhoGAP proteins from Saccharomyces cerevisiae. FEBS Lett. 506, 149–156. doi: 10.1016/s0014-5793(01)02906-4
Shang, X., Moon, S. Y., and Zheng, Y. (2007). p200 RhoGAP promotes cell proliferation by mediating cross-talk between Ras and rho signaling pathways. J. Biol. Chem. 282, 8801–8811. doi: 10.1074/jbc.M609375200
Smith, D. G., Garcia-Pedrajas, M. D., Hong, W., Yu, Z. Y., Gold, S. E., and Perlin, M. H. (2004). An ste20 homologue in Ustilago maydis plays a role in mating and pathogenicity. Eukaryot. Cell 3, 180–189. doi: 10.1128/EC.3.1.180-189.2004
Sushil, S. N., Joshi, D., Roy, S., Rao, G. P., and Pathak, A. D. (2022). Plant quarantine regulations with reference to sugarcane in India: strengths and challenges. Sugar Tech 24, 1319–1329. doi: 10.1007/s12355-022-01125-3
Swaminathan, K., Muller-Taubenberger, A., Faix, J., Rivero, F., and Noegel, A. A. (2014). A Cdc42-and Rac-interactive binding (CRIB) domain mediates functions of coronin. Proc. Natl. Acad. Sci. U. S. A. 111, E25–E33. doi: 10.1073/pnas.1315368111
Szabo, R. (2001). Cla4 protein kinase is essential for filament formation and invasive growth of Yarrowia lipolytica. Mol. Gen. Genomics. 265, 172–179. doi: 10.1007/s004380000405
Tapia-Vazquez, I., Montoya-Martinez, A. C., De los Santos-Villalobos, S., Ek-Ramos, M. J., Montesinos-Matias, R., and Martinez-Anaya, C. (2022). Root-knot nematodes (Meloidogyne spp.) a threat to agriculture in Mexico: biology, current control strategies, and perspectives. World J. Microbiol. Biotechnol. 38:26. doi: 10.1007/s11274-021-03211-2
Tian, H., Zhou, L., Guo, W. Z., and Wang, X. Y. (2015). Small GTPase Rac1 and its interaction partner Cla4 regulate polarized growth and pathogenicity in verticillium dahliae. Fungal Genet. Biol. 74, 21–31. doi: 10.1016/j.fgb.2014.11.003
Tunlid, A., Ahman, J., and Oliver, R. P. (1999). Transformation of the nematode-trapping fungus Arthrobotrys oligospora. FEMS Microbiol. Lett. 173, 111–116. doi: 10.1111/j.1574-6968.1999.tb13491.x
Tunlid, A., Rosen, S., Ek, B., and Rask, L. (1994). Purification and characterization of an extracellular serine-protease from the nematode-trapping fungus Arthrobotrys oligospora. Microbiology (Reading) 140, 1687–1695. doi: 10.1099/13500872-140-7-1687
Tzean, Y., Chou, T.-H., Hsiao, C.-C., Shu, P.-Y., Walton, J. D., and Tzean, S.-S. (2016). Cloning and characterization of cuticle-degrading serine protease from nematode-trapping fungus Arthrobotrys musiformis. Mycoscience 57, 136–143. doi: 10.1016/j.myc.2015.12.003
van Drogen, F., O’Rourke, S. M., Stucke, V. M., Jaquenoud, M., Neiman, A. M., and Peter, M. (2000). Phosphorylation of the MEKK Ste11p by the PAK-like kinase Ste20p is required for MAP kinase signaling in vivo. Curr. Biol. 10, 630–639. doi: 10.1016/s0960-9822(00)00511-x
Wang, L., Yang, L., Debidda, M., Witte, D., and Zheng, Y. (2007). Cdc42 GTPase-activating protein deficiency promotes genomic instability and premature aging-like phenotypes. Proc. Natl. Acad. Sci. U. S. A. 104, 1248–1253. doi: 10.1073/pnas.0609149104
Wu, C. L., Whiteway, M., Thomas, D. Y., and Leberer, E. (1995). Molecular characterization of Ste20p, a potential mitogen-activated protein or extracellular signal-regulated kinase kinase (MEK) kinase kinase from Saccharomyces cerevisiae. J. Biol. Chem. 270, 15984–15992. doi: 10.1074/jbc.270.27.15984
Xie, M., Bai, N., Yang, J., Jiang, K., Zhou, D., Zhao, Y., et al. (2020). Protein kinase Ime2 is required for mycelial growth, conidiation, osmoregulation, and pathogenicity in nematode-trapping fungus Arthrobotrys oligospora. Front. Microbiol. 10:3065. doi: 10.3389/fmicb.2019.03065
Xie, M., Bai, N., Yang, X., Liu, Y., Zhang, K. Q., and Yang, J. (2023). Fus3 regulates asexual development and trap morphogenesis in the nematode-trapping fungus Arthrobotrys oligospora. iScience 26:107404. doi: 10.1016/j.isci.2023.107404
Xie, M., Ma, N., Bai, N., Yang, L., Yang, X., Zhang, K.-Q., et al. (2022). PKC-SWI6 signaling regulates asexual development, cell wall integrity, stress response, and lifestyle transition in the nematode-trapping fungus Arthrobotrys oligospora. Sci. China Life Sci. 65, 2455–2471. doi: 10.1007/s11427-022-2118-0
Xie, M., Yang, J., Jiang, K., Bai, N., Zhu, M., Zhu, Y., et al. (2021). AoBck1 and AoMkk1 are necessary to maintain cell wall integrity, vegetative growth, conidiation, stress resistance, and pathogenicity in the nematode-trapping fungus Arthrobotrys oligospora. Front. Microbiol. 12:649582. doi: 10.3389/fmicb.2021.649582
Yang, J. K., Huang, X. W., Tian, B. Y., Sun, H., Duan, J. X., Wu, W. P., et al. (2005). Characterization of an extracellular serine protease gene from the nematophagous fungus Lecanicillium psalliotae. Biotechnol. Lett. 27, 1329–1334. doi: 10.1007/s10529-005-0482-1
Yang, L., Li, X., Bai, N., Yang, X., Zhang, K.-Q., and Yang, J. (2022). Transcriptomic analysis reveals that rho GTPases regulate trap development and lifestyle transition of the nematode-trapping fungus Arthrobotrys oligospora. Microbiol Spectr 10:e0175921. doi: 10.1128/spectrum.01759-21
Yang, L., Li, X., Xie, M., Bai, N., Yang, J., Jiang, K., et al. (2021). Pleiotropic roles of Ras GTPases in the nematode-trapping fungus Arthrobotrys oligospora identified through multi-omics analyses. iScience 24:102820. doi: 10.1016/j.isci.2021.102820
Yang, P. R., Qyang, Y., Bartholomeusz, G., Zhou, X., and Marcus, S. (2003). The novel rho GTPase-activating protein family protein, Rga8, provides a potential link between Cdc42/p21-activated kinase and rho signaling pathways in the fission yeast, Schizosaccharomyces pombe. J. Biol. Chem. 278, 48821–48830. doi: 10.1074/jbc.M306819200
Yang, L.-Q., Sang, P., Zhang, R.-P., and Liu, S.-Q. (2017). Substrate-induced changes in dynamics and molecular motions of cuticle-degrading serine protease PL646: a molecular dynamics study. RSC Adv. 7, 42094–42104. doi: 10.1039/c7ra07797a
Yang, J., Wang, L., Ji, X., Feng, Y., Li, X., Zou, C., et al. (2011). Genomic and proteomic analyses of the fungus Arthrobotrys oligospora provide insights into nematode-trap formation. PLoS Pathog. 7:e1002179. doi: 10.1371/journal.ppat.1002179
Yang, J., Wang, W., Liu, Y., Xie, M., and Yang, J. (2023). The MADS-box transcription factor AoRlmA is involved in the regulation of mycelium development, conidiation, cell-wall integrity, stress response, and trap formation of Arthrobotrys oligospora. Microbiol. Res. 268:127299. doi: 10.1016/j.micres.2022.127299
Ye, W., Chen, X., Zhong, Z., Chen, M., Shi, L., Zheng, H., et al. (2014). Putative RhoGAP proteins orchestrate vegetative growth, conidiogenesis and pathogenicity of the rice blast fungus Magnaporthe oryzae. Fungal Genet. Biol. 67, 37–50. doi: 10.1016/j.fgb.2014.03.008
Zhang, A.-X., Mouhoumed, A.-Z., Tong, S.-M., Ying, S.-H., and Feng, M.-G. (2019). BrlA and AbaA govern virulence-required dimorphic switch, conidiation, and pathogenicity in a fungal insect pathogen. msystems 4, e00140–e00119. doi: 10.1128/mSystems.00140-19
Zhen, Z., Xing, X., Xie, M., Yang, L., Yang, X., Zheng, Y., et al. (2018). MAP kinase Slt2 orthologs play similar roles in conidiation, trap formation, and pathogenicity in two nematode-trapping fungi. Fungal Genet. Biol. 116, 42–50. doi: 10.1016/j.fgb.2018.04.011
Zhu, M.-C., Li, X.-M., Zhao, N., Yang, L., Zhang, K.-Q., and Yang, J.-K. (2022a). Regulatory mechanism of trap formation in the nematode-trapping fungi. J. Fungi 8:406. doi: 10.3390/jof8040406
Zhu, M.-C., Zhao, N., Liu, Y.-K., Li, X.-M., Zhen, Z.-Y., Zheng, Y.-Q., et al. (2022b). The cAMP-PKA signalling pathway regulates hyphal growth, conidiation, trap morphogenesis, stress tolerance, and autophagy in Arthrobotrys oligospora. Environ. Microbiol. 24, 6524–6538. doi: 10.1111/1462-2920.16253
Keywords: p21-activated kinase, Rho GTPase-activating protein, conidiation, trap formation, Arthrobotrys oligospora
Citation: Zhu M, Liu Y, Yang X, Zhu L, Shen Y, Duan S and Yang J (2023) p21-activated kinase is involved in the sporulation, pathogenicity, and stress response of Arthrobotrys oligospora under the indirect regulation of Rho GTPase-activating protein. Front. Microbiol. 14:1235283. doi: 10.3389/fmicb.2023.1235283
Received: 06 June 2023; Accepted: 04 September 2023;
Published: 14 September 2023.
Edited by:
Hector Riveros-Rosas, National Autonomous University of Mexico, MexicoReviewed by:
Parul Singh, National Heart, Lung, and Blood Institute (NIH), United StatesCopyright © 2023 Zhu, Liu, Yang, Zhu, Shen, Duan and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinkui Yang, amlua3VpOTYwQHludS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.