
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 31 July 2023
Sec. Microorganisms in Vertebrate Digestive Systems
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1231837
Introduction: Gut microbes like segmented filamentous bacteria (SFB) play a key role in gut maturation during early life, as demonstrated in humans and mice. Our previous study demonstrated oral inoculation of ileum-spores containing SFB to chickens after hatch increases early SFB gut colonization, which increases immune maturation and resistance to bacteria, like Salmonella, as tested in vitro; however, more studies are needed for treatment optimization and in vivo testing. The objectives of this study were to (1) test a treatment that includes both spores and filamentous SFB, (2) validate antimicrobial ability of the treatment in layer hens in vivo, and (3) elucidate its molecular mechanism.
Methods: One-day-old specific pathogen-free layers (n = 12 per group) were orally treated with either PBS (CON) or SFB-based treatment (SFB). At 4 days post-inoculation (DPI), both CON and SFB groups were orally challenged with Salmonella Typhimurium. Total Enterobacteriaceae and Salmonella were examined by plating and enumeration in feces at 7,10 and 14 dpi; and in the ileum, cecum, and spleen at 16 dpi in euthanized birds. The presence and levels of SFB were determined from ilea scrapings via microscopy and qPCR, respectively. Relative gene expression of host-derived antimicrobial peptides and cytokines in the distal ileum was determined by RT-qPCR.
Results: At 10 and 14 dpi, a significant decrease in total Enterobacteriaceae was observed in the feces of the SFB group. At necropsy, the level of SFB was significantly higher in the SFB group than in the CON group, while a significant decrease in total Enterobacteriaceae and Salmonella was observed in the ceca of the SFB group. RT-qPCR revealed increased expression of β-defensin 14, and cytokines IL-10 and IFNγ.
Discussion: The introduction of SFB at hatch as a prophylactic treatment may benefit commercial partners as well as consumers by reducing the incidence of Enterobacteriaceae in food animals. Reduction of these bacteria in animals would, in turn, increase animal health, productivity, and safety for consumers. Studies to optimize the treatment for poultry industry applications are ongoing in our lab.
The gastrointestinal tract of animals serves as a reservoir for many bacterial species classified as gut symbionts that provide benefits for the host organism (Chow et al., 2010; Pudlo et al., 2015; Shang et al., 2018). These microorganisms often provide benefits to the host through alterations of the gut pH, production of secondary metabolites, and competitively excluding potentially pathogenic organisms (Redweik et al., 2020a). However, some symbionts have been shown to mechanistically impact the maturation of the gut immune system (Wickramasuriya et al., 2022). Of these microbes, segmented filamentous bacteria (SFB), or specifically Candidatus Arthromitus, are a gut-associated symbiont regularly found in numerous vertebrate species, including humans, pigs, mice, rats, turkey, and chickens (Klaasen et al., 1992; Redweik et al., 2020b). SFB are gram-positive, spore forming bacteria, closely related to Clostridia species that intimately bind to the host epithelium in the ileum of the gastrointestinal tract, as demonstrated in mice (Umesaki et al., 1995). SFB have drawn the attention of researchers owing to their unique morphology, lifecycle, and intimate relation with the host (Hedblom et al., 2018).
In contrast to enteric pathogens, which cause pathology by triggering an inflammatory response following binding to the gut epithelium, SFB drive the maturation of the gut immune system by influencing the production of antimicrobial peptides (AMPs) and the differentiation of naïve CD4+ T cells to form CD4+ TH17 cells (Ivanov and Littman, 2010). Induction of host immune maturation impacts the overall ability of both the innate and adaptive immune systems (Klaasen et al., 1992; Wickramasuriya et al., 2022). Triggering of adaptive immune maturation can provide extraintestinal protection and impact the immune system systemically (McAleer et al., 2016).
A significant amount of research has been accomplished utilizing defined mouse models and mice mono-associated with SFB (Klaasen et al., 1991; Umesaki et al., 1995; Atarashi et al., 2015; Schnupf et al., 2015, 2017; Shi et al., 2019). However, direct studies with SFB are hampered greatly due to the inconsistent ability to culture SFB readily in vitro in pure culture (Schnupf et al., 2015). In addition, SFB are host specific, and SFB isolated from mono-associated mice will not colonize any other hosts (Chen et al., 2017). This fact has greatly hindered translational studies to understand how SFB impact the immune systems of different host animals. Specifically, research on the relationship between SFB and the immune system of the chicken gut is severely lacking.
Recently, our lab has demonstrated the ability to prepare a chicken SFB-enriched inoculum that enables the SFB colonization of the ileum of layer chickens that would normally not receive SFB from their environment, due to their hatching in a clean environment and not in contact with hens (Redweik et al., 2020b). As tested in chicken ileum explants, this colonization resulted in measurable immunometabolic changes in the innate (Toll-like receptor, JAK–STAT), adaptive (T/B cell receptor, TH17 differentiation), and pathways like the mammalian target of rapamycin (mTOR) pathway in the gut immune system and increased ability to kill Salmonella in vitro (Redweik et al., 2020b). Therefore, in vivo testing is warranted, and the objectives of this study are to (1) prepare an optimized form of the SFB inoculum, (2) test the ability to increase gut immune maturation, and (3) antimicrobial activity against resident Enterobacteriaceae and Salmonella challenge in chickens in vivo.
Animal experiments were approved by Iowa State University Institutional Animal Care and Use Committee, Log #s 18–386 and 19–072. One-day-old male and female specific pathogen-free (SPF) White Leghorns (VALO; Adel, IA) were used in this study. Animal enrichments were added to open floor pens to minimize stress during experimental procedures. Animals were fed Purina Organic Starter Grower Feed with no antibiotic or probiotic supplementation. Euthanasia techniques (CO2 asphyxiation) follow the American Veterinary Medical Association Guidelines (2013) (AVMA guidelines for the euthanasia of animals, 2020).
Methods were described previously for the enrichment of ileal spores with minor modifications (Umesaki et al., 1995; Redweik et al., 2020b). Briefly, scrapings from the distal ileum of two-week-old commercial pullets (n = 10) were pooled in Phosphate-Buffered Saline (PBS) (3 mM EDTA). Pooled scrapings (n = 10) were then treated with chloroform (3% of total solution V/V). The inoculum was allowed to settle at room temperature (RT) for 10 min before the aqueous layer was transferred to a fresh tube. Residual chloroform was evaporated under CO2 (Wang et al., 2022). The entire solution was then pelleted at 4500 x g for 15 min. Solutions were not filtered to preserve SFB filaments in the preparations (Schnupf et al., 2015). The pellet was resuspended in a peptone-glycerol (1% peptone W/V, 15% glycerol V/V) solution and stored at −80°C until use in animal experiments. The suspensions were screened on blood agar to ensure the absence of culturable microbes.
Total DNA was extracted from intestinal scrapings utilizing a phenol-chloroform DNA extraction protocol (Gomes-Neto et al., 2017). SFB were enumerated by qPCR quantification of the 16S rRNA gene sequence using the primer pairs: SFB-specific F: 5’-AGGAGGAGTCTGCGGCACATTAGC-3′; and the universal R: 5’-TCCCCACTGCTGCCTCCCGTAG-3′ (Schnupf et al., 2015). The Maxima SYBR/Green master mix (Thermo Fischer, Waltham Massachusetts, United States, Catalog # K0223) was used with reaction conditions consisting of 0.1 mM primers, and 10 ng of template DNA. Samples for qPCR were run in duplicate utilizing a QuantStudio 3.0 thermocycler (Thermo Fischer, Waltham Massachusetts, United States). An SFB standard curve was created by ligating the SFB specific PCR amplicon into the pCR2.1 vector following manufacture instructions (Thermo Fischer, Waltham Massachusetts, United States, Catalog # K202040). The ligated pCR2.1:16SSFB vector was expressed in TOP10 E. coli, and plasmid DNA was extracted using the high-speed plasmid mini kit (IBI Scientific, Dubuque, Iowa, United States). The concentration of extracted plasmid DNA was determined using a Qubit 3.0 fluorometer (Thermo Fischer, Waltham Massachusetts, United States). The SFB standard curve was prepared by serially diluting the quantified plasmid stock in tenfold dilutions, and linear regression was performed using GraphPad Prism Software (Version 9.5.1; GraphPad, San Diego, CA). Calculations to convert the cycle threshold to colony forming unit per gram (CFU/g) were performed as described (Gomes-Neto et al., 2017).
Fluorescent in situ hybridization (FISH) was performed based on methods described previously (Haroon et al., 2013; Batani et al., 2019) with minor modifications. Briefly, stored ilea scrapings were pelleted at 13,000 x g for 5 min and washed 2 times with PBS. After the second wash, pellets were resuspended in 50 mL transformation buffer (100 mM CaCl2, 30 mM MnCl2, 20 mM MgCl2, 100 mM potassium acetate, and 10% glycerol V/V). Ilea scrapings were then supplemented with 2,000 ng/mL of the SFB-specific probe (GGG TAC TTA TTG CGT TTG CGA CGG CAC) corresponding to position 801–827 in the universal 16S rRNA gene sequence of SFB (Accession number: X87244) (Liao et al., 2012). The SFB-specific probe was 6-Carboxyfluorescein (6-FAM) labeled (Thermo Fischer, Waltham Massachusetts, United States). After hybridization and washing, FISH-tagged SFB were resuspended in PBS and stored at 4°C for microscopy (Batani et al., 2019). Fluorescence microscopy was performed utilizing a Leica DM4 B fluorescent microscope with L5 filter cube (excitation 480/40 nm) for the detection of the SFB probe (Leica, Wetzlar, Germany).
Day-old birds were distributed into 2 pens (n = 18 birds/pen) in the same room. Immediately after placement, birds were orally inoculated with sodium bicarbonate and either 50 μL chloroform-treated scrapings (SFB) or 50 μL PBS (CON). The SFB inoculum contained 104 SFB as determined by qPCR. Food and water were provided 30 min after treatment. Feces from three birds randomly in each pen were collected at days 11 and 14 post SFB treatment to monitor levels of total Enterobacteriaceae. Birds (n = 6) were euthanized on 2-, 5-, and 16-days post inoculation via CO2 asphyxiation. To assess levels of SFB, scrapings of the distal ileum were performed, as before, without chloroform treatment and stored in peptone-glycerol at −80°C for future experiments.
Results from the pilot study showed SFB cross-contamination of the CON group through the environment. To address this issue, in this second experiment, birds were distributed into 2 pens (n = 12 birds/pen) in separate rooms. Immediately after placement, birds were orally inoculated with sodium bicarbonate and either 50 μL of chloroform-treated scrapings (104 CFU SFB) or 50 μL PBS (CON). At day 4, the birds were orally challenged with 200 μL of nalidixic acid resistant χ3761 (107 CFUs) in PBS. Feces were collected aseptically on 7-, 10-, and 14-days post inoculation (dpi), resuspended in PBS, and plated on MacConkey agar with and without nalidixic acid to track χ3761 and total Enterobacteriaceae, respectively. At 16 dpi all birds were euthanized. Intestinal tissues, including contents (ileum and cecum), and extraintestinal tissue (spleen) were aseptically collected, homogenized in PBS, and plated on MacConkey agar with and without nalidixic acid for bacterial enumeration. The limit of detection (LOD) was calculated for each sample as described (Ott et al., 2020). Samples that did not show growth were enriched overnight in 500 μL Luria Bertani (LB) broth with 0.1% glucose at 37°C and re-plated. If no growth was detected after enrichment, these values are displayed as 0 CFU/g tissue. Levels of SFB in the distal ileum were assessed via qPCR as in the pilot study.
Total RNA was extracted from flash-frozen ilea tissues using RNAzol® RT according to manufacture instructions (Molecular Research Center, Cincinnati, OH, United States, Catalog # RN190) and RNA concentration and quality were assessed via a Nanodrop 2000 (Thermo Fischer, Waltham, Massachusetts, USA). Reverse transcription assays were performed via the High-Capacity cDNA Reverse Transcription Kit according to manufactures instructions (Thermo Fischer, Waltham Massachusetts, United States, Catalog # 4374966). The RT-qPCR assays were performed using a QuantStudio 3.0 thermocycler (Thermo Fischer, Waltham Massachusetts, USA) and cycling conditions as described (Redweik et al., 2021). Genes encoding host-derived AMPs (b-defensins 12 and 14, cathelicidin b1 and Fowlicidins 1–3) and cytokines (IL-2, 6, 10, 17, and IFNg) were assessed using primers described in Table 1. Differences in gene expression were determined via 2-▵▵method using the gene encoding glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as a stably expressed housekeeping control gene.
Statistical analyses of the data were performed using the GraphPad Prism software (Version 9.5.1; GraphPad, San Diego, CA). Differences in total Enterobacteriaceae in feces or SFB quantification in ilea scrapings were measured at each timepoint using Student’s t-test. Analyses of differences between either Salmonella or total Enterobacteriaceae were determined in each organ tested using Student’s t-test with Holm-Šidák’s correction. Mann–Whitney t-tests were utilized to analyze all RT-qPCR data with Holm-Šidák’s correction.
Through gram-staining and FISH microscopy, we detected SFB filaments in only SFB birds at 5 days post inoculation (dpi). Small holdfast-like morphologies were detected in both SFB and CON groups 2 dpi. Large filaments were found in all birds at 16 dpi (Figure 1). qPCR investigation of the levels of ilea-associated SFB, demonstrated low levels of SFB in all birds at 2 dpi. At 5 dpi, 50% (3/6) of SFB birds demonstrated a large expansion of SFB. At 16 dpi, all birds demonstrated colonization with SFB with no differences between CON and SFB birds (Figure 2A). In the Salmonella challenge study, housing CON and SFB birds in different rooms resulted in the colonization of SFB in treated birds only. Gram-strain and FISH microscopy of scrapings from the distal ileum in SFB birds at 16 dpi demonstrated an abundance of filamentous SFB (Figure 3). CON birds showed a complete lack of SFB. Furthermore, SFB-specific qPCR demonstrated a significantly higher (p < 0.0001) amount of SFB in SFB birds compared to CON (Figure 2B). Specifically, all CON birds demonstrated a lack of SFB amplification in qPCR assays.
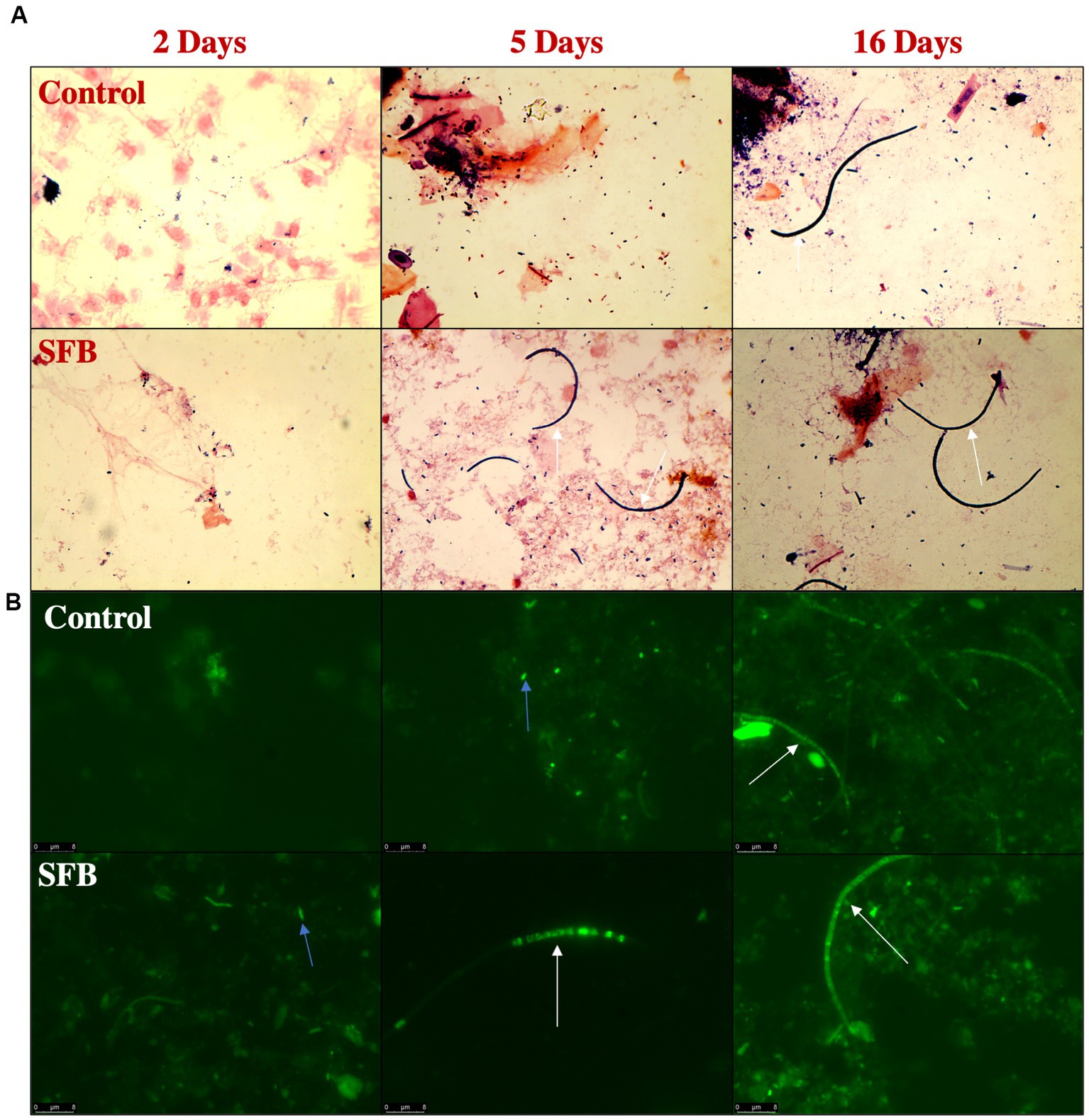
Figure 1. Detection of Segmented Filamentous Bacteria (SFB) via Gram-Staining and FISH Microscopy in the Pilot Inoculation Experiment. Representative images of ilea scrapings analyzed by gram-stain (A) and FISH microscopy (B) demonstrate the inability to detect SFB filaments in Control birds prior to 16-days post inoculation (dpi) and presence of filaments in SFB treated birds after 5 dpi. Arrows indicate intracellular offspring (blue) or filamentous SFB (white).
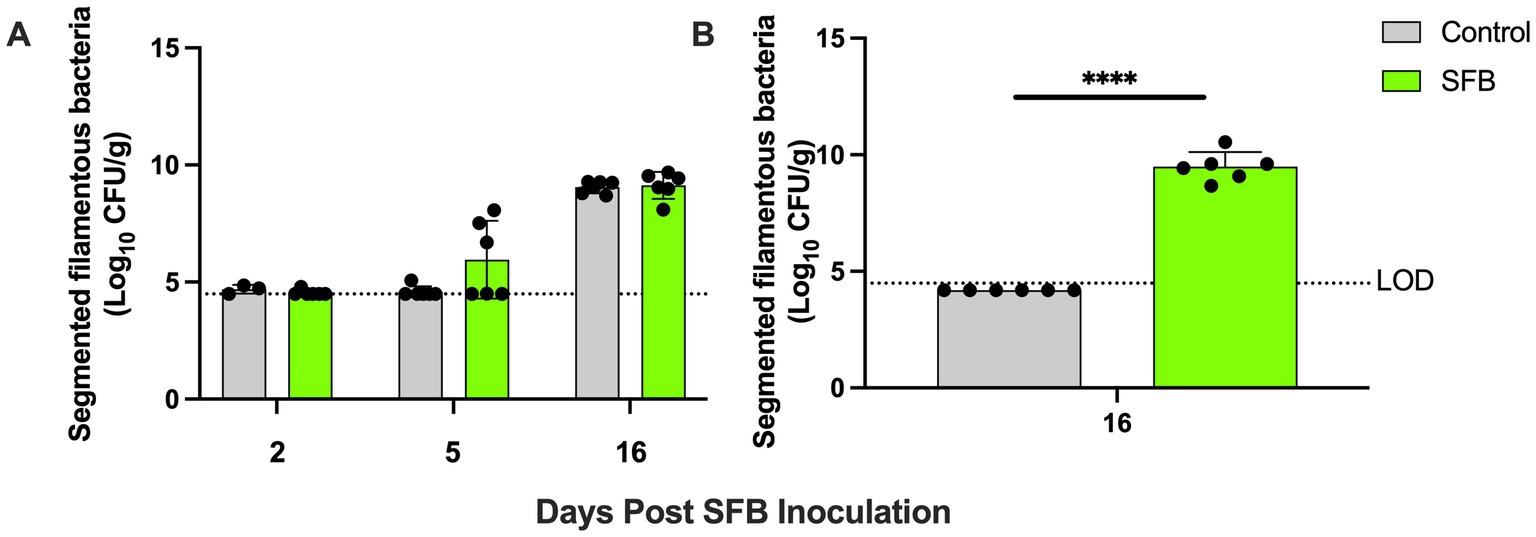
Figure 2. Ilea-associated Segmented Filamentous Bacteria (SFB) Quantification. Log10 CFU/g of SFB detected in ilea scrapings calculated by qPCR in (A) the pilot inoculation experiment at respective 2-, 5-, and 16- days post inoculation (dpi) and (B) Salmonella challenge study at final necropsy (16 dpi). ****, p < 0.0001. Dots indicate individual birds. The horizontal dashed line represents the limit of detection (LOD). Samples that did not demonstrate amplification in qPCR assays were set to half of the LOD.
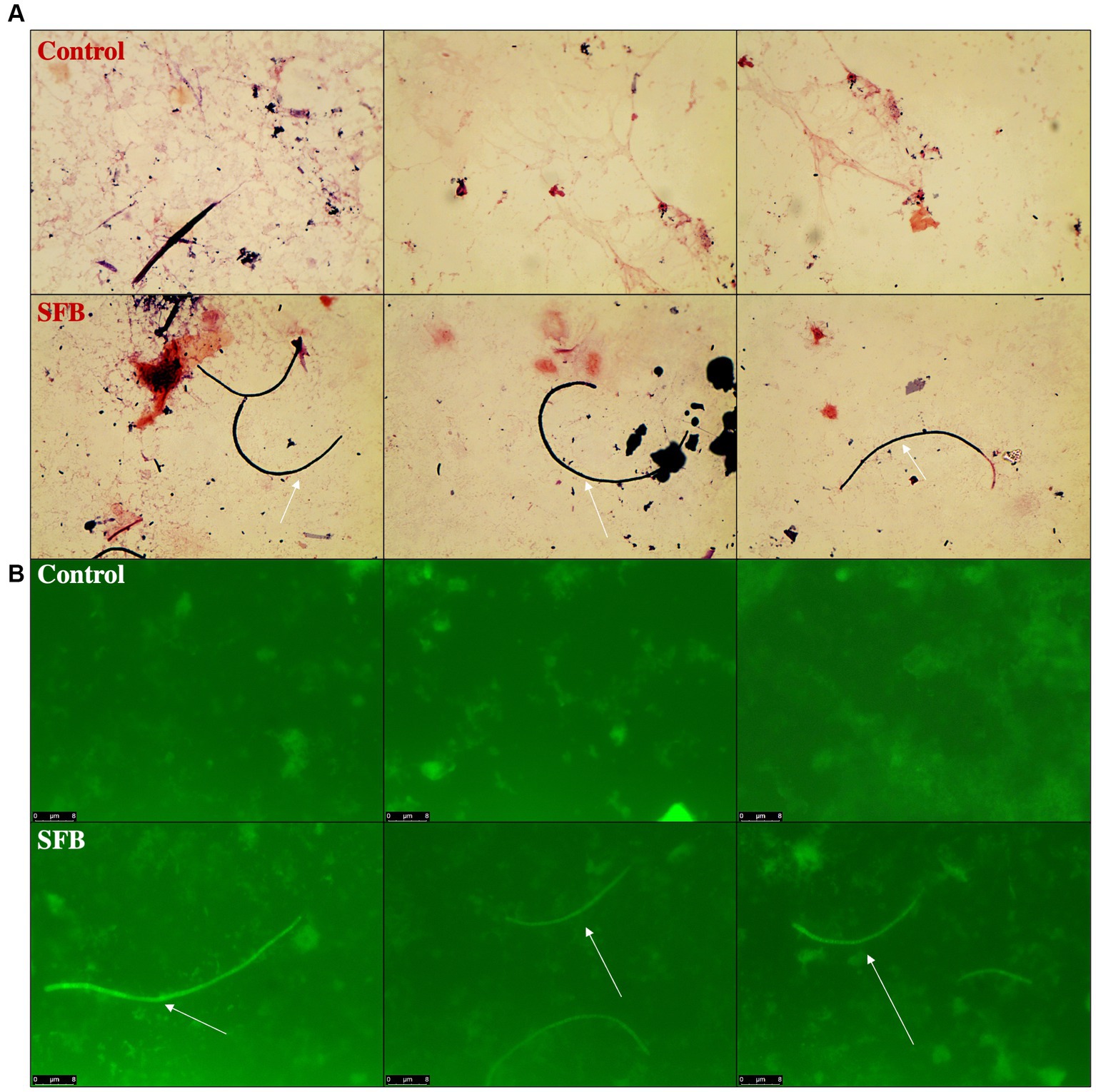
Figure 3. Detection of Segmented Filamentous Bacteria (SFB) via Gram-Staining and FISH Microscopy in Salmonella Challenge Study. Representative images of ilea scrapings analyzed by gram-stain (A) and FISH microscopy (B) demonstrate the inability to detect SFB in Control birds and presence of SFB in SFB treated birds at final necropsy (16-days post inoculation). White arrows indicate filamentous SFB.
In the pilot study, at days 11 and 14 post-inoculation, SFB birds demonstrated significantly lower levels of Enterobacteriaceae in feces compared to CON birds tested (p < 0.01; Figure 4A). This was further confirmed in the Salmonella challenge, where the introduction of SFB significantly reduced the amount of Enterobacteriaceae detected in the feces (p < 0.05) at 10 and 14 dpi (Figure 4B). At the final necropsy (16 dpi) Salmonella χ3761 was detected in all organs tested (Figure 5). The CFU/g tissue of Salmonella χ3761 was significantly reduced (p < 0.001) in the ceca of SFB birds compared to CON (Figure 5A). Furthermore, the CFU/g tissue of total Enterobacteriaceae were reduced in the ceca (p < 0.01) of SFB birds compared to CON (Figure 6A).
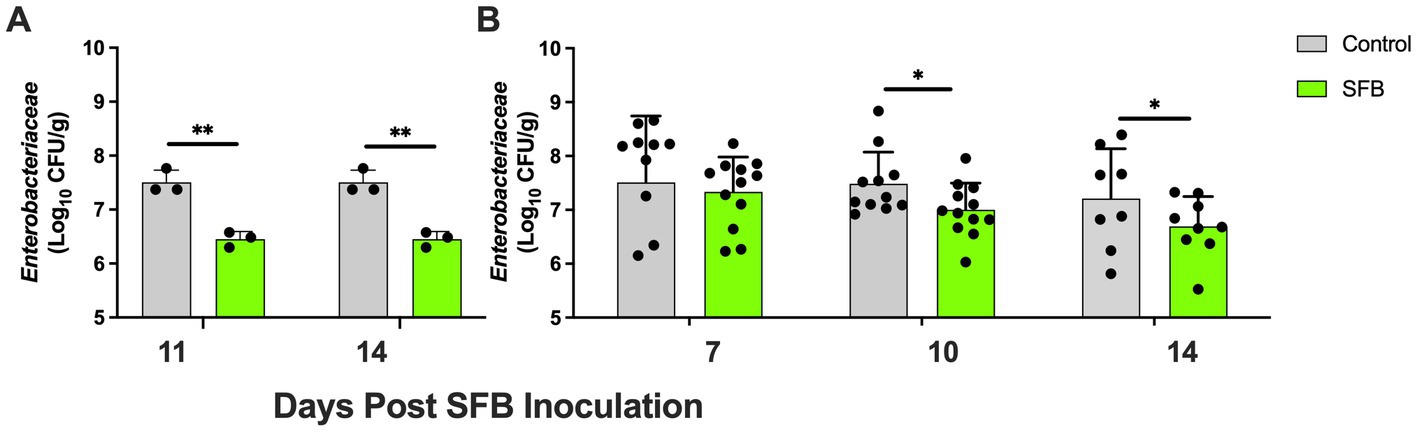
Figure 4. Fecal Tracking of Total Enterobacteriaceae. Total Enterobacteriaceae in feces in the pilot inoculation experiment at 11- and 14-days post inoculation (dpi) (A) and in the Salmonella challenge at 7-, 10-, and 14-dpi (B). *, p < 0.05; **, p < 0.01. Each dot represents a separate bird tested.
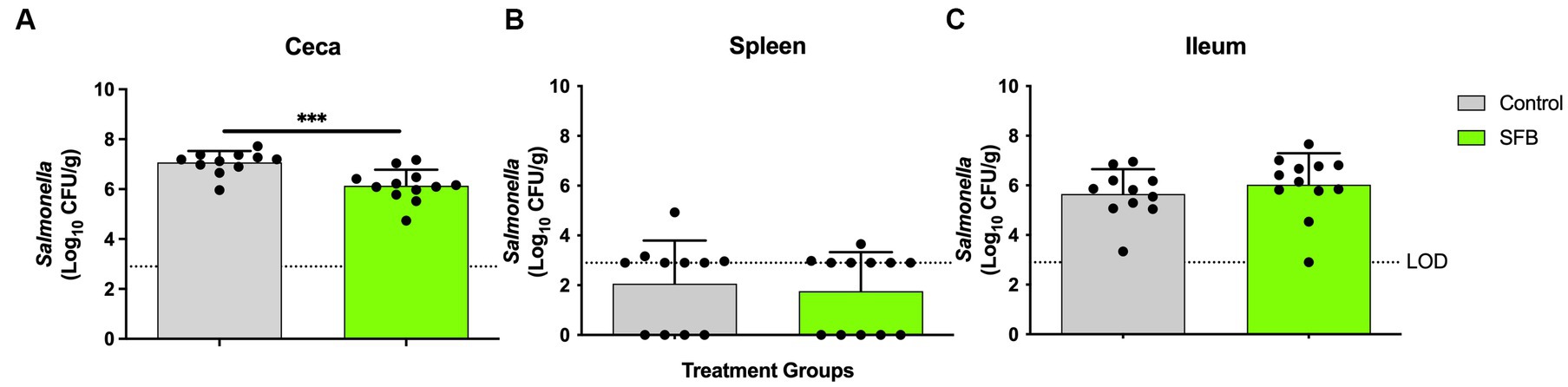
Figure 5. Salmonella Enumeration at Final Necropsy. Total Salmonella enumerated in the (A) ceca (B) spleen and (C) ileum at final necropsy, 16-days post inoculation. ***, p < 0.001. Dots indicate individual birds. The horizontal dashed line represents the limit of detection (LOD). Samples that remained negative after enrichment were assigned a 0 value.
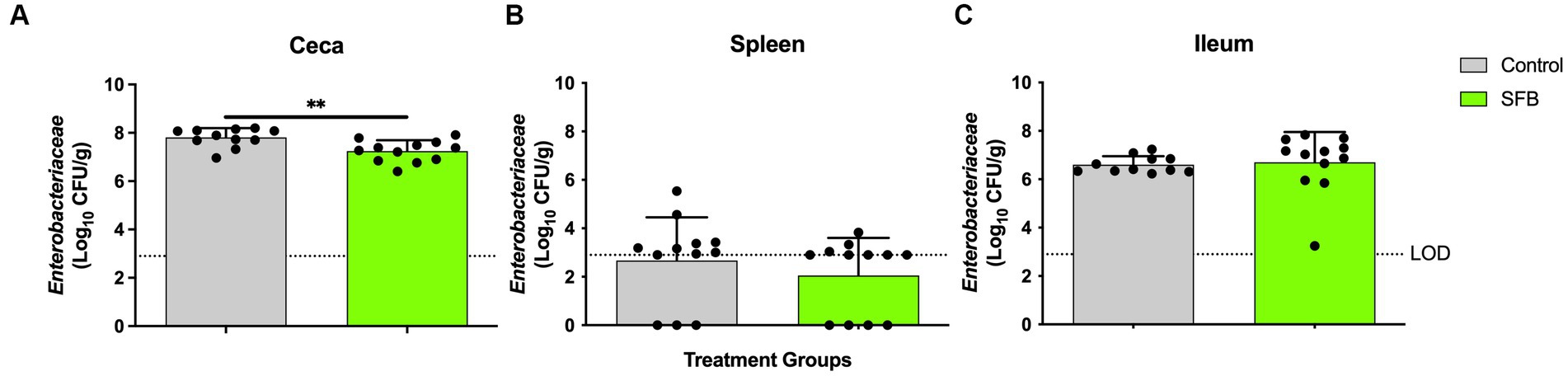
Figure 6. Total Enterobacteriaceae Enumeration at Final Necropsy. Total Enterobacteriaceae enumerated in the ceca (A), spleen (B), and ileum (C) at 16-days post inoculation (dpi). **, p < 0.01. Dots indicate individual birds. The horizontal dashed line represents the limit of detection (LOD). Samples that remained negative after enrichment were assigned a 0 value.
To determine if treatment with SFB preparations results in transcriptional changes in genes of key host-derived AMPs and cytokines, the Log2 fold changes in gene expression were measured via RT-qPCR. Increased gene expression of the AMP β-defensin 14 (p < 0.01; Figure 7A), regulatory cytokine IL-10 (p < 0.0001; Figure 7B), and cytokine IFNγ (p < 0.05; Figure 7B) was observed in the SFB group compared to CON (Figure 7A). There were no significant differences detected in any other AMP or cytokine tested (Figures 7A,B).
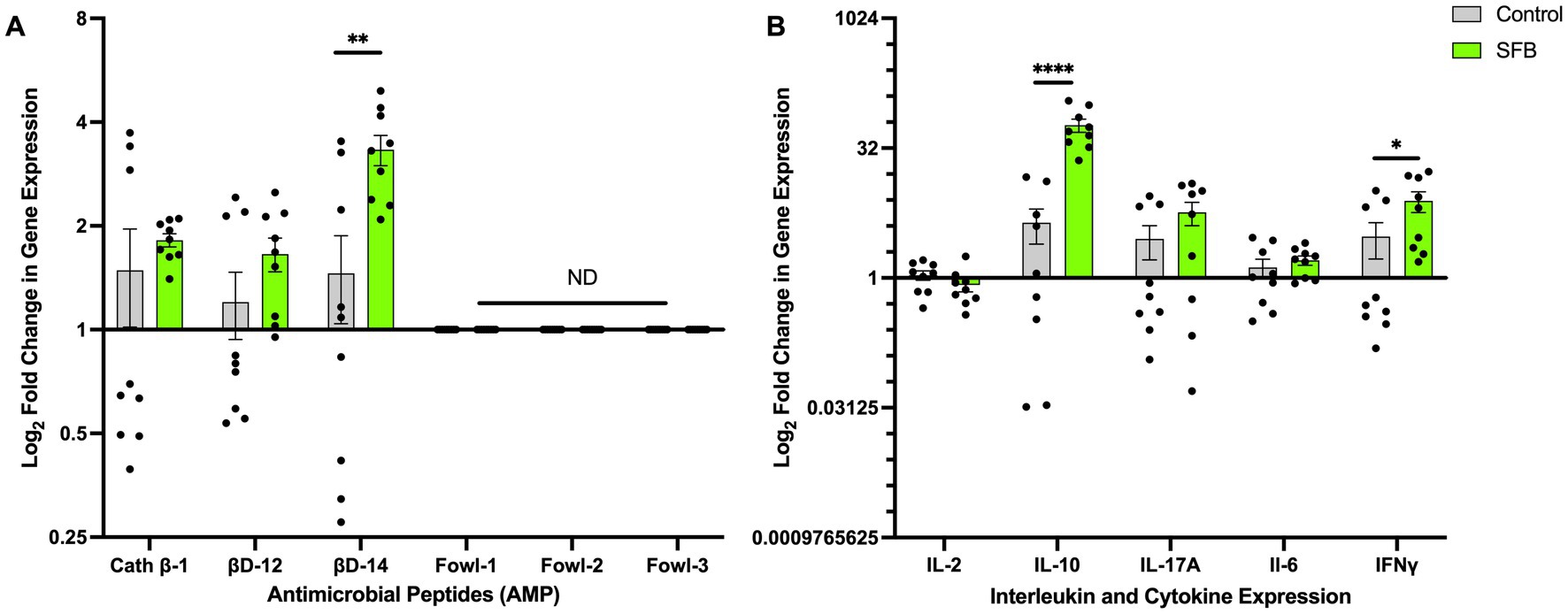
Figure 7. Changes in Antimicrobial Peptide and Interleukin Gene Expression. Log2 fold change in gene expression of select antimicrobial peptides (AMPs) (A) and interleukins (B). *, p < 0.05; **, p < 0. 01; ****, p < 0.0001. ND denotes no amplification detected.
Although the lifecycle of SFB and its interaction with the host have been studied in mice, limited studies have been reported on the role of these bacteria in agriculturally relevant animals, such as chickens. A substantial amount of research in mice has demonstrated the key role that SFB play in the maturation of the mouse gut immune system in both the innate and adaptive immune responses (Schnupf et al., 2013; Atarashi et al., 2015; Farkas et al., 2015). Because of the host-specificity of SFB (Chen et al., 2017), our lab has previously studied the interaction of chicken SFB with their host and demonstrated the ability of our chloroform-treated inoculum to promote colonization of the distal ileum with SFB (Redweik et al., 2020b). Identification of the specific role that SFB play in the limitation of pathogen colonization of the gut of chickens will facilitate possible probiotic treatment options in the place of antibiotic intervention.
It is suggested that the lifecycle of SFB is propagated through the adhesion of spores or holdfast cells to the host epithelium in the ileum (Chase and Erlandsen, 1976; Pamp et al., 2012). The transmission of SFB in mammalian hosts is done, in part, thanks to vertical transmission from the mother. However, in the chicken production environment, the eggs are separated from the layer hens and disinfected, thus negating the opportunity for this vertical transmission (Smith, 2019). Our inoculation strategy demonstrates the ability to introduce chicks at day-of-hatch with SFB that propagate to full length filaments in the distal ileum. Furthermore, our data show that birds that do not receive SFB directly via inoculation can receive them through shed SFB that are spread throughout the environment. However, in the pilot study, filamentous SFB were only detected in SFB-treated birds at 5 dpi. Birds in the Control group that did not initially receive inoculation of SFB likely began to acquire SFB through environmental spreading at this time point. Based on the lifecycle of SFB, the filamentous form at 5 dpi suggests that the SFB had bound to the host epithelium and progressed through their lifecycle (Pamp et al., 2012). Early SFB colonization is important for the maturation of the gut immune system as although both groups were eventually colonized with SFB in the pilot study, the reduction of Enterobacteriaceae was only detected in the SFB-treated birds. When birds were segregated into separate rooms, birds that did not receive SFB on day of hatch remained SFB-negative throughout the entire experiment. These results demonstrate the importance of oral treatment of birds with SFB.
The presence of SFB causes shifts in the gut microbiota in humans, including a reduction of some Enterobacteriaceae (Chen et al., 2018). In addition, SFB enhance the production of AMPs from intestinal epithelial cells in vitro and in SFB-inoculated germ-free mice (Schnupf et al., 2015). AMPs prevent bacterial pathogens from inducing attachment derived inflammation and lesions (Gubatan et al., 2021; Nazeer et al., 2021). Production of AMPs also prevents SFB from overgrowing and causing pathology in the host (Flannigan and Denning, 2018). Our RT-qPCR data demonstrate that the colonization of SFB in the distal ileum of layer hens triggers the production of AMPs in the ileum. Specifically, we establish that colonization of SFB triggered a log2 fold increase in gene expression of β-defensin 14. Recent in vitro testing on chicken-derived cathelicidins show significant bactericidal effects against various Enterobacteriaceae (Veldhuizen et al., 2013). Avian β-defensins like β-defensin 12 and 14 possess broad-spectrum antimicrobial, LPS neutralizing, and chemotactic properties (Shao et al., 2016). We observed an upregulation of AMP expression in ilea tissue, which may partly elucidate the mechanism by which the reduction of Enterobacteriaceae was observed in SFB treated birds.
The up-regulation of IL-10 in the distal ileum of SFB colonized birds suggests that SFB are promoting an anti-inflammatory environment in the gut while synergistically eliminating Salmonella. Normally, the expression of IL-10 is linked to continued systemic infection in chickens (Haghighi et al., 2008; Shanmugasundaram et al., 2015; Lalsiamthara and Lee, 2017). However, we did not detect any differences in Salmonella (Figures 5B,C) or Enterobacteriaceae (Figures 6B,C) load in the ileum and the spleen of CON or SFB birds. In addition, our recent study in chickens, demonstrated that SFB-associated inflammation was non-pathological (Redweik et al., 2020a,b), similar to what has been previously shown in mice (Davis and Savage, 1974; Flannigan and Denning, 2018; Shi et al., 2019). Although we did not observe a significant increase in the expression of IL-17A in SFB-treated birds, only one time point was used for gene expression evaluation. The colonization of SFB has been demonstrated to be directly linked to the differentiation of naïve CD4+ T cells to activated TH17 cells in germ-free mice (Gaboriau-Routhiau et al., 2009; Lécuyer et al., 2014; Flannigan et al., 2017; Schnupf et al., 2017; Flannigan and Denning, 2018; Shi et al., 2019). TH17 cells play a dual role in regulating host inflammation while providing antimicrobial activity against pathogens in the intestine (Blaschitz and Raffatellu, 2010; Crhanova et al., 2011; Huber et al., 2012). Future experiments will aim to track the production of these cytokines throughout the colonization of SFB.
Further, RT-qPCR revealed an increased gene expression of the immunostimulatory cytokine IFNγ in the distal ileum of SFB-treated birds. IFNγ is a cytokine produced by T lymphocytes and natural killer cells throughout the chicken immune system (St Paul et al., 2012; Bagheri et al., 2022). Increased expression of IFNγ in the intestine of Salmonella enterica infected chickens has been correlated to a robust T-cell response that results in the rapid clearance of Salmonella (Penha Filho et al., 2012; Onuigbo et al., 2018; Ibe et al., 2019). This increase of IFNγ caused by the colonization of SFB is correlated to a significant reduction in Salmonella in the ceca. We also demonstrated the reduction of total Enterobacteriaceae in the ceca and shed in the feces, which indicates a broad protection against multiple bacteria. As intestinal Salmonella was only measured at one timepoint (16 dpi), a continuation of the study may exhibit decreased shedding of Salmonella due to this relation with IFNγ expression.
This study demonstrates the ability of an SFB-enriched inoculum to colonize the distal ileum and induce modulations in the innate and adaptive immune responses. Furthermore, we identify the impact the colonization of SFB plays on Enterobacteriaceae in the gastrointestinal tract of chickens. Finally, the colonization of SFB indicated protective capabilities against continued Salmonella Typhimurium infection in the ceca of layers. By enhancing the ability of layers to clear Salmonella infection, SFB may serve as a beneficial organism for egg producers. Current efforts are underway to obtain a pure culture of SFB in vitro to optimize dosage in vivo. Other studies are underway to assess the protective abilities of SFB against other foodborne pathogens common in the chicken gastrointestinal tract and the overall impact on the complex gut microbiota.
The original contributions presented in the study are included in the article, further inquiries can be directed to the corresponding author.
The animal study was reviewed and approved by Iowa State University Institutional Animal Care and Use Committee.
MM conceived and designed the experiments and contributed reagents, materials, and analysis tools. JM-J, LO, and MM performed the experiments and revised the manuscript. JM-J and MM analyzed the data and wrote the manuscript. All authors contributed to the article and approved the submitted version.
This research was supported by the United States Department of Agriculture (USDA) National Institute of Food and Agriculture (NIFA) project #026895–0000, USDA- Hatch projects (IOW05700-NC1202 and IOW04202), and Kent corp. to MM. The funders had no role in study design, data collection, and interpretation, or the decision to submit the work for publication. Mention of commercial products is for the sole purpose of providing specific information, not a recommendation or endorsement by the funders.
The authors thank Graham Redweik for the generation of the pCR2.1:16SSFB vector expressing Top10 E. coli.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Achanta, M., Sunkara, L. T., Dai, G., Bommineni, Y. R., Jiang, W., and Zhang, G. (2012). Tissue expression and developmental regulation of chicken cathelicidin antimicrobial peptides. J. Anim.l Sci. Biotechnol. 3:15. doi: 10.1186/2049-1891-3-15
Atarashi, K., Tanoue, T., Ando, M., Kamada, N., Nagano, Y., Narushima, S., et al. (2015). Th17 cell induction by adhesion of microbes to intestinal epithelial cells. Cells 163, 367–380. doi: 10.1016/j.cell.2015.08.058
AVMA guidelines for the euthanasia of animals . American Veterinary Medical Association. (2020) Available at: https://www.avma.org/resources-tools/avma-policies/avma-guidelines-euthanasia-animals (Accessed April 11, 2023).
Bagheri, S., Paudel, S., Wijewardana, V., Kangethe, R. T., Cattoli, G., Hess, M., et al. (2022). Production of interferon gamma and interleukin 17A in chicken T-cell subpopulations hallmarks the stimulation with live, irradiated and killed avian pathogenic Escherichia coli. Dev. Comp.Immunol. 133:104408. doi: 10.1016/j.dci.2022.104408
Batani, G., Bayer, K., Böge, J., Hentschel, U., and Thomas, T. (2019). Fluorescence in situ hybridization (FISH) and cell sorting of living bacteria. Sci. Rep. 9:18618. doi: 10.1038/s41598-019-55049-2
Blaschitz, C., and Raffatellu, M. (2010). Th17 cytokines and the gut mucosal barrier. J. Clin. Immunol. 30, 196–203. doi: 10.1007/s10875-010-9368-7
Chase, D. G., and Erlandsen, S. L. (1976). Evidence for a complex life cycle and endospore formation in the attached, filamentous, segmented bacterium from murine ileum. J. Bacteriol. 127, 572–583. doi: 10.1128/jb.127.1.572-583.1976
Chen, B., Chen, H., Shu, X., Yin, Y., Li, J., Qin, J., et al. (2018). Presence of segmented filamentous Bacteria in human children and its potential role in the modulation of human gut immunity. Front. Microbiol. 9:1403. doi: 10.3389/fmicb.2018.01403
Chen, H., Yin, Y., Wang, Y., Wang, X., and Xiang, C. (2017). Host specificity of Flagellins from segmented filamentous Bacteria affects their patterns of interaction with mouse Ileal mucosal proteins. Appl. Environ. Microbiol. 83, e01061–e01017. doi: 10.1128/AEM.01061-17
Chow, J., Lee, S. M., Shen, Y., Khosravi, A., and Mazmanian, S. K. (2010). Host-bacterial symbiosis in health and disease. Adv. Immunol. 107, 243–274. doi: 10.1016/B978-0-12-381300-8.00008-3
Crhanova, M., Hradecka, H., Faldynova, M., Matulova, M., Havlickova, H., Sisak, F., et al. (2011). Immune response of chicken gut to natural colonization by gut microflora and to Salmonella enterica Serovar Enteritidis infection. Infect. Immun. 79, 2755–2763. doi: 10.1128/IAI.01375-10
Davis, C. P., and Savage, D. C. (1974). Habitat, succession, attachment, and morphology of segmented, filamentous microbes indigenous to the murine gastrointestinal tract. Infect. Immun. 10, 948–956. doi: 10.1128/iai.10.4.948-956.1974
Farkas, A. M., Panea, C., Goto, Y., Nakato, G., Galan-Diez, M., Narushima, S., et al. (2015). Induction of Th17 cells by segmented filamentous bacteria in the murine intestine. J. Immunol. Methods 421, 104–111. doi: 10.1016/j.jim.2015.03.020
Flannigan, K. L., and Denning, T. L. (2018). Segmented filamentous bacteria-induced immune responses: a balancing act between host protection and autoimmunity. Immunology 154, 537–546. doi: 10.1111/imm.12950
Flannigan, K. L., Ngo, V. L., Geem, D., Harusato, A., Hirota, S. A., Parkos, C. A., et al. (2017). IL-17A-mediated neutrophil recruitment limits expansion of segmented filamentous bacteria. Mucosal Immunol. 10, 673–684. doi: 10.1038/mi.2016.80
Gaboriau-Routhiau, V., Rakotobe, S., Lécuyer, E., Mulder, I., Lan, A., Bridonneau, C., et al. (2009). The key role of segmented filamentous Bacteria in the coordinated maturation of gut helper T cell responses. Immunity 31, 677–689. doi: 10.1016/j.immuni.2009.08.020
Gomes-Neto, J. C., Mantz, S., Held, K., Sinha, R., Segura Munoz, R. R., Schmaltz, R., et al. (2017). A real-time PCR assay for accurate quantification of the individual members of the altered Schaedler Flora microbiota in gnotobiotic mice. J. Microbiol. Methods 135, 52–62. doi: 10.1016/j.mimet.2017.02.003
Gubatan, J., Holman, D. R., Puntasecca, C. J., Polevoi, D., Rubin, S. J., and Rogalla, S. (2021). Antimicrobial peptides and the gut microbiome in inflammatory bowel disease. World J. Gastroenterol. 27, 7402–7422. doi: 10.3748/wjg.v27.i43.7402
Haghighi, H. R., Abdul-Careem, M. F., Dara, R. A., Chambers, J. R., and Sharif, S. (2008). Cytokine gene expression in chicken cecal tonsils following treatment with probiotics and Salmonella infection. Vet. Microbiol. 126, 225–233. doi: 10.1016/j.vetmic.2007.06.026
Haroon, M. F., Skennerton, C. T., Steen, J. A., Lachner, N., Hugenholtz, P., and Tyson, G. W. (2013). “Chapter one - in-solution fluorescence in situ hybridization and fluorescence-activated cell sorting for single cell and population genome recovery” in Methods in enzymology microbial metagenomics, Metatranscriptomics, and Metaproteomics. ed. E. F. DeLong (Cambridge, MA, (USA): Academic Press), 3–19.
Hedblom, G. A., Reiland, H. A., Sylte, M. J., Johnson, T. J., and Baumler, D. J. (2018). Segmented filamentous Bacteria – metabolism meets immunity. Front. Microbiol. 9:1991. doi: 10.3389/fmicb.2018.01991
Hong, Y. H., Song, W., Lee, S. H., and Lillehoj, H. S. (2012). Differential gene expression profiles of β-defensins in the crop, intestine, and spleen using a necrotic enteritis model in 2 commercial broiler chicken lines. Poult. Sci. 91, 1081–1088. doi: 10.3382/ps.2011-01948
Huber, S., Gagliani, N., and Flavell, R. A. (2012). Life, death, and miracles: Th17 cells in the intestine. Eur. J. Immunol. 42, 2238–2245. doi: 10.1002/eji.201242619
Ibe, M. I., Odimegwu, D. C., and Onuigbo, E. B. (2019). Alginate-coated chitosan microparticles encapsulating an oral plasmid-cured live Salmonella enterica serovar Gallinarum vaccine cause a higher expression of interferon-gamma in chickens compared to the parenteral live vaccine. Avian Pathol. 48, 423–428. doi: 10.1080/03079457.2019.1616673
Ivanov, I. I., and Littman, D. R. (2010). Segmented filamentous bacteria take the stage. Mucosal Immunol. 3, 209–212. doi: 10.1038/mi.2010.3
Klaasen, H. L. B. M., Koopman, J. P., Poelma, F. G. J., and Beynen, A. C. (1992). Intestinal, segmented, filamentous bacteria. FEMS Microbiol. Rev. 88, 165–180. doi: 10.1111/j.1574-6968.1992.tb04986.x
Klaasen, H. L. B. M., Koopman, J. P., Van Den Brink, M. E., Van Wezel, H. P. N., and Beynen, A. C. (1991). Mono-association of mice with non-cultivable, intestinal, segmented, filamentous bacteria. Arch. Microbiol. 156:148. doi: 10.1007/bf00290989
Lalsiamthara, J., and Lee, J. H. (2017). Immunization with Salmonella Enteritidis secreting mucosal adjuvant labile toxin confers protection against wild type challenge via augmentation of CD3+CD4+ T-cell proliferation and enhancement of IFN-γ, IL-6 and IL-10 expressions in chicken. Vaccine 35, 767–773. doi: 10.1016/j.vaccine.2016.12.042
Lécuyer, E., Rakotobe, S., Lengliné-Garnier, H., Lebreton, C., Picard, M., Juste, C., et al. (2014). Segmented filamentous bacterium uses secondary and tertiary lymphoid tissues to induce gut IgA and specific T helper 17 cell responses. Immunity 40, 608–620. doi: 10.1016/j.immuni.2014.03.009
Liao, N., Yin, Y., Sun, G., Xiang, C., Liu, D., Yu, H. D., et al. (2012). Colonization and distribution of segmented filamentous bacteria (SFB) in chicken gastrointestinal tract and their relationship with host immunity. FEMS Microbiol. Ecol. 81, 395–406. doi: 10.1111/j.1574-6941.2012.01362.x
McAleer, J. P., Nguyen, N. L. H., Chen, K., Kumar, P., Ricks, D. M., Binnie, M., et al. (2016). Pulmonary Th17 anti-fungal immunity is regulated by the gut microbiome. J. Immunol. 197, 97–107. doi: 10.4049/jimmunol.1502566
Nazeer, N., Uribe-Diaz, S., Rodriguez-Lecompte, J. C., and Ahmed, M. (2021). Antimicrobial peptides as an alternative to relieve antimicrobial growth promoters in poultry. Br. Poult. Sci. 62, 672–685. doi: 10.1080/00071668.2021.1919993
Onuigbo, E., Iseghohimhen, J., Chah, K., Gyang, M., and Attama, A. (2018). Chitosan/alginate microparticles for the oral delivery of fowl typhoid vaccine: innate and acquired immunity. Vaccine 36, 4973–4978. doi: 10.1016/j.vaccine.2018.05.087
Ott, L. C., Stromberg, Z. R., Redweik, G. A. J., Wannemuehler, M. J., and Mellata, M. (2020). Mouse genetic background affects transfer of an antibiotic resistance plasmid in the gastrointestinal tract. mSphere 5:5. doi: 10.1128/mSphere.00847-19
Pamp, S. J., Harrington, E. D., Quake, S. R., Relman, D. A., and Blainey, P. C. (2012). Single-cell sequencing provides clues about the host interactions of segmented filamentous bacteria (SFB). Genome Res. 22, 1107–1119. doi: 10.1101/gr.131482.111
Penha Filho, R. A. C., Moura, B. S., de Almeida, A. M., Montassier, H. J., Barrow, P. A., and Berchieri Junior, A. (2012). Humoral and cellular immune response generated by different vaccine programs before and after Salmonella Enteritidis challenge in chickens. Vaccine 30, 7637–7643. doi: 10.1016/j.vaccine.2012.10.020
Pudlo, N. A., Urs, K., Kumar, S. S., German, J. B., Mills, D. A., and Martens, E. C. (2015). Symbiotic human gut Bacteria with variable metabolic priorities for host mucosal Glycans. MBio 6, e01282–e01215. doi: 10.1128/mBio.01282-15
Redweik, G. A. J., Jochum, J., and Mellata, M. (2020a). Live bacterial prophylactics in modern poultry. Front. Vet. Sci. 7:826. doi: 10.3389/fvets.2020.592312
Redweik, G. A. J., Kogut, M. H., Arsenault, R. J., Lyte, M., and Mellata, M. (2021). Reserpine improves Enterobacteriaceae resistance in chicken intestine via neuro-immunometabolic signaling and MEK1/2 activation. Commun. Biol. 4:1359. doi: 10.1038/s42003-021-02888-3
Redweik, G. A. J., Kogut, M. H., Arsenault, R. J., and Mellata, M. (2020b). Oral treatment with Ileal spores triggers Immunometabolic shifts in chicken gut. Front. Vet. Sci. 7:629. doi: 10.3389/fvets.2020.00629
Schnupf, P., Gaboriau-Routhiau, V., and Cerf-Bensussan, N. (2013). Host interactions with segmented filamentous Bacteria: an unusual trade-off that drives the post-natal maturation of the gut immune system. Semin. Immunol. 25, 342–351. doi: 10.1016/j.smim.2013.09.001
Schnupf, P., Gaboriau-Routhiau, V., Gros, M., Friedman, R., Moya-Nilges, M., Nigro, G., et al. (2015). Growth and host interaction of mouse segmented filamentous bacteria in vitro. Nature 520, 99–103. doi: 10.1038/nature14027
Schnupf, P., Gaboriau-Routhiau, V., Sansonetti, P. J., and Cerf-Bensussan, N. (2017). Segmented filamentous bacteria, Th17 inducers and helpers in a hostile world. Curr. Opin. Microbiol. 35, 100–109. doi: 10.1016/j.mib.2017.03.004
Shang, Y., Kumar, S., Oakley, B., and Kim, W. K. (2018). Chicken gut microbiota: importance and detection technology. Front. Vet. Sci. 5:254. doi: 10.3389/fvets.2018.00254
Shanmugasundaram, R., Kogut, M. H., Arsenault, R. J., Swaggerty, C. L., Cole, K., Reddish, J. M., et al. (2015). Effect of Salmonella infection on cecal tonsil regulatory T cell properties in chickens. Poult. Sci. 94, 1828–1835. doi: 10.3382/ps/pev161
Shao, Y., Wang, Z., Tian, X., Guo, Y., and Zhang, H. (2016). Yeast β-d-glucans induced antimicrobial peptide expressions against Salmonella infection in broiler chickens. Int. J. Biol. Macromol. 85, 573–584. doi: 10.1016/j.ijbiomac.2016.01.031
Shi, Z., Zou, J., Zhang, Z., Zhao, X., Noriega, J., Zhang, B., et al. (2019). Segmented filamentous Bacteria prevent and cure rotavirus infection. Cells 179, 644–658.e13. doi: 10.1016/j.cell.2019.09.028
Smith, J. A. (2019). Broiler production without antibiotics: United States field perspectives. Anim. Feed Sci. Technol. 250, 93–98. doi: 10.1016/j.anifeedsci.2018.04.027
St Paul, M., Barjesteh, N., Paolucci, S., Pei, Y., and Sharif, S. (2012). Toll-like receptor ligands induce the expression of interferon-gamma and interleukin-17 in chicken CD4+ T cells. BMC. Res. Notes 5:616. doi: 10.1186/1756-0500-5-616
Umesaki, Y., Okada, Y., Matsumoto, S., Imaoka, A., and Setoyama, H. (1995). Segmented filamentous bacteria are indigenous intestinal bacteria that activate intraepithelial lymphocytes and induce MHC class II molecules and fucosyl asialo GM1 glycolipids on the small intestinal epithelial cells in the ex-germ-free mouse. Microbiol. Immunol. 39, 555–562. doi: 10.1111/j.1348-0421.1995.tb02242.x
Veldhuizen, E. J. A., Brouwer, E. C., Schneider, V. A. F., and Fluit, A. C. (2013). Chicken Cathelicidins display antimicrobial activity against multiresistant Bacteria without inducing strong resistance. PLoS One 8:e61964. doi: 10.1371/journal.pone.0061964
Wang, H., Yu, R., Webb, J., Dollar, P., and Freedman, D. L. (2022). Anaerobic biodegradation of chloroform and dichloromethane with a Dehalobacter enrichment culture. Appl. Environ. Microbiol. 88, e01970–e01921. doi: 10.1128/aem.01970-21
Weerts, E. A. W. S., Matthijs, M. G. R., Bonhof, J., van Haarlem, D. A., Dwars, R. M., Gröne, A., et al. (2021). The contribution of the immune response to enhanced colibacillosis upon preceding viral respiratory infection in broiler chicken in a dual infection model. Vet. Immunol. Immunopathol. 238:110276. doi: 10.1016/j.vetimm.2021.110276
Keywords: segmented filamentous bacteria, Enterobacteriaceae , antimicrobial peptides, cytokines, layer chickens
Citation: Meinen-Jochum J, Ott LC and Mellata M (2023) Segmented filamentous bacteria-based treatment to elicit protection against Enterobacteriaceae in Layer chickens. Front. Microbiol. 14:1231837. doi: 10.3389/fmicb.2023.1231837
Received: 31 May 2023; Accepted: 12 July 2023;
Published: 31 July 2023.
Edited by:
Thi Thu Hao Van, RMIT University, AustraliaReviewed by:
Sunil Banskar, University of Arizona, United StatesCopyright © 2023 Meinen-Jochum, Ott and Mellata. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Melha Mellata, bW1lbGxhdGFAaWFzdGF0ZS5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.