- 1Center for Marine Environmental Studies, Ehime University, Matsuyama, Japan
- 2Graduate School of Science and Engineering, Ehime University, Matsuyama, Japan
Marine bacteria are possible reservoirs of antibiotic-resistance genes (ARGs) originating not only from clinical and terrestrial hot spots but also from the marine environment. We report here for the first time a higher rate of the sulfonamide-resistance gene sul4 in marine bacterial isolates compared with other sul genes. Among four sulfonamide-resistance genes (sul1, sul2, sul3, and sul4), sul4 was most abundant (45%) in 74 sulfonamide-resistant marine isolates by PCR screening. The order of abundance was sul4 (33 isolates) >sul2 (6 isolates) >sul3 (5 isolates) >sul1 (1 isolate). Whole-genome sequencing of 23 isolates of sul4-expressing α- and γ-proteobacteria and bacilli revealed that sul4 was not accompanied by known mobile genetic elements. This suggests that sul4 in these marine isolates is clonally transferred and not horizontally transferable. Folate metabolism genes formed a cluster with sul4, suggesting that the cluster area plays a role in folate metabolism, at which sul4 functions as a dihydropteroate synthase. Thus, sul4 might be expressed in marine species and function in folate synthesis, but it is not a transferable ARG.
1. Introduction
Antibiotic-resistant bacteria (ARB) appeared almost immediately after the use of antibiotics began and now represents a major global health issue in human and animal chemotherapy. ARB develop via conservation of DNA mutations and the acquisition of antibiotic-resistance genes (ARGs) through horizontal gene transfer (HGT) (Levy and Marchall, 2004). HGT is thought to occur between bacteria via intra- and inter-species mechanisms. The One Health concept (G7 Presidency, 2015) involves a humans-animals-environment linkage in which HGT should be considered in relation to not only clinical but also environmental bacteria. ARGs could enter humans and animals from natural environmental bacteria.
Sulfonamides are synthetic antimicrobial agents and the first drugs used for chemotherapy of bacterial and protozoan infections in 1935 (Bickel, 1988). In developed countries, the use of sulfonamides in humans has been replaced by more-advanced antibiotics, but these drugs are still widely used in humans and animals in developing countries (Shimizu et al., 2013). During the long history of sulfonamides, many analogues and combination use with trimethoprim have been developed to overcome the development of resistant bacteria. Bacteria resistant to sulfonamides can be detected all over the world, and selective pressure (Na et al., 2014) and the stability of the compounds (Sukul and Spiteller, 2006; Zhang et al., 2013; Matsuura et al., 2021) have played a role in the development and dissemination of sulfonamide-resistant bacteria in the environment.
The main mechanism of sulfonamide resistance involves the acquisition of genes encoding enzymes analogous to dihydropteroate synthase (DHPS) (Yun et al., 2013). To date, four sulfonamide-resistance genes exhibiting transferable characteristics have been reported: sul1, sul2, sul3, and sul4 (Swedberg and Sköld, 1983; Rådström and Swedberg, 1988; Sundström et al., 1988; Perreten and Boerlin, 2003; Razavi et al., 2017). Among the four sul genes, sul1 and sul2 often form a gene cluster with integron integrase IntI1 on various plasmids, such as IncQ and pBP1 (Bean et al., 2009; Dungan and Bjorneberg, 2020). The gene cluster is transferred and spread among human and animal pathogens worldwide (Kerrn et al., 2002; Perreten and Boerlin, 2003; Antunes et al., 2005). Among clinically relevant bacteria, sul3 is of minor importance, but marine unculturable bacteria commonly harbor this gene (Suzuki et al., 2013). It was reported that sul3 is associated with non-classic class 1 integrons (Vinué et al., 2010). The sul4 gene was newly discovered in Chloroflexus isolated from river sediment (Razavi et al., 2017). Although the host range of sul4 remains unknown, gene sequences have been deposited in metagenome databases from aquatic environments of Asian and European countries, but no sequences from clinical specimens have been deposited (Razavi et al., 2017). The sul4 is linked to the transposase ISCR20, which may facilitate widespread distribution of the gene through aquatic environments (Razavi et al., 2017). These data indicate that sul genes are transferred among clinical and environmental bacteria through various mobile genetic elements (MGEs). This property could lead to the wider spread of sul genes.
Horizontal transfer of ARGs in the environment has been reported (Blahna et al., 2006; Aminov, 2011). A variety of MGEs, such as transposons, plasmids, and integrative and conjugative elements, play a role in HGT. Integrons coexisting with MGEs can uptake functional modules from other species and genera (Stokes et al., 2006; Stalder et al., 2012). The IntI1 gene is an essential part of class 1 integrons and has been reported as a candidate factor for monitoring anthropogenic pollution and ARGs in the environment (Gillings et al., 2015).
Several reports have described the presence of sul1, sul2, and sul3 in marine bacteria, predominantly culturable γ-proteobacteria (Wang et al., 2014; Jiang et al., 2019). In the present study, we focused on sulfonamide-resistant marine bacteria, including α- and γ-proteobacteria, which are major bacterial groups in the oceans (Sunagawa et al., 2015). As our results showed that sul4 is a common sulfonamide-resistance gene, we focused further studies on this gene. The transmissibility of sul4 and its formation of gene cluster with MGEs have not been reported to date. We conducted a whole-genome analysis using a nanopore long-read sequencing technique to identify the relative positioning of repetitive sequences of ARGs and MGEs in cultured sulfonamide-resistant marine bacteria. This is the first study to detect and determine the cluster structure of sul4 in marine bacteria.
2. Materials and methods
2.1. Bacterial strains
Sulfamethoxazole (SMX)-resistant bacteria used in this study were selected from isolates collected from surface seawater off the Takahama coast, Ehime Prefecture, Japan, in May and November, 2015. Ten liters of seawater was sampled each time, then 20 mL was employed for bacterial count experiment. Incubation was performed at 25°C. To select for SMX-resistant bacteria, low-nutrient seawater medium (peptone 0.01 g/L, yeast extract 0.005 g/L, agar 15 g/L in sterile seawater, pH 7.5) and nutrient seawater medium (peptone 5 g/L, yeast extract 1 g/L, agar 15 g/L in sterile seawater, pH 7.5) were used, with the addition of 64 μg/mL of SMX. A total of 74 SMX-resistant isolates (Supplementary Table S1) were obtained.
2.2. Bacterial DNA extraction and classification
Total DNA was purified from cultured isolates using a QuickGene DNA Tissue S kit (Kurabo, Osaka). The 16S rRNA gene sequence (V3 region, 566 bp) was used for bacterial taxonomic identification. PCR mixtures contained 0.025 U of Ex Taq (Takara, Japan), 0.1 μM each of forward and reverse primers, 0.2 mM dNTP mixture (Takara, Japan), 1× Ex Taq buffer, and template DNA (approx. 10 ng). Primers and amplification condition are listed in Supplementary Table S2. PCR products were sequenced on a Genetic Analyzer 3130 (Thermo Fisher Scientific K.K., Japan) with BigDye Terminator and subsequently analyzed using the BLAST program. A sequence identity of ≥97% relative to database species was recognized as indicating the closest species. A phylogenetic tree of the isolates was constructed by Molecular Evolutionary Genetics Analysis 11 (MEGA11, https://www.megasoftware.net/) using the V3 region of the 16S rRNA gene sequence according to the maximum-likelihood algorithm and Jukes–Cantor model. The phylogenetic tree was visualized using version 5 of Interactive Tree Of Life (iTOL, https://itol.embl.de/) (Letunic and Bork, 2019).
2.3. Detection of ARGs and IntI1
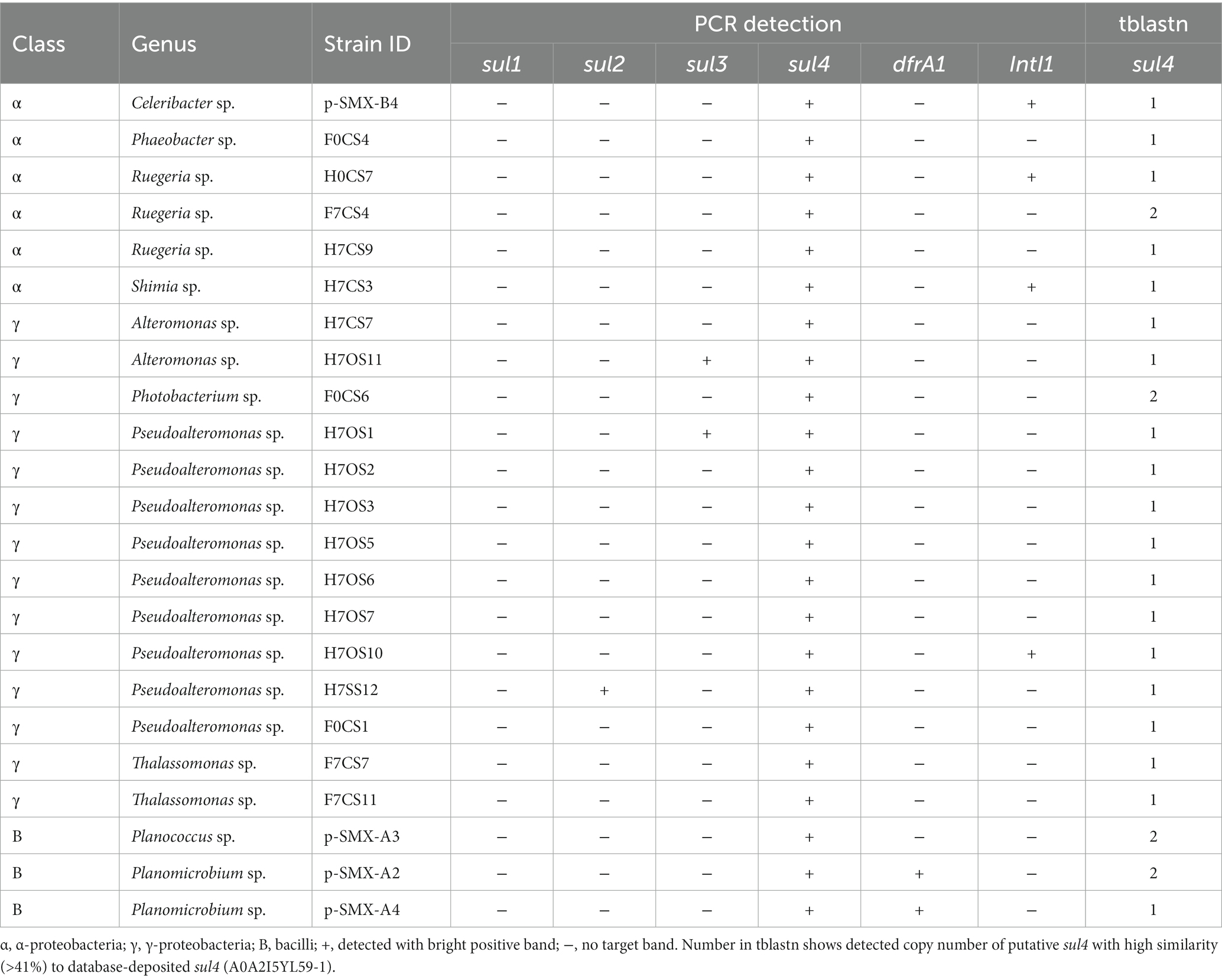
The sul1, sul2, sul3, sul4, dfrA1 (trimethoprim resistance), and IntI1 (class 1 integron integrase) among the 74 SMX-resistant isolates were detected using PCR. The dfrA1 gene plays a role in sulfonamide resistance in a different step of folate synthesis than sul (Huovinen et al., 1995). The IntI1 gene is frequently involved in the transfer of sul1 and sul2 (Koczura et al., 2016). The primers and cycle conditions are shown in Supplementary Table S2. PCR was carried out in a mixture of 0.025 U of Ex Taq (Takara, Japan), 0.5 μM each forward and reverse primer, 0.2 mM dNTP mixture (Takara, Japan), 1× Ex Taq buffer, and template DNA (50–100 ng). The amplification products were examined by electrophoresis on a 1.5% agarose gel with GelRed (Nacalai Tesque, Japan) staining. PCR product was confirmed based on band length and sequence.
2.4. Genome analysis
Long-read MinION (Oxford Nanopore Technologies, United Kingdom) was used to search for the sul4 among the genomes of 33 isolates that were sul4-positive in PCR screening and to determine the gene structure of the surrounding regions. Total DNA was extracted using a Wizard Genomic DNA Purification kit (Promega, United States) according to the protocol for Gram-negative bacteria. DNA quality was checked using e-spect (Malcom, Japan), and DNA was quantified using a QuantiFluor® dsDNA system (Promega). We prepared the DNA libraries according to the Oxford Nanopore Technologies’ protocol for environmental bacteria. A NEBNext Ultra II End Repair/dA-Tailing Module (New England Biolabs, United States) was used to generate 5′ phosphorylated and 3′ dA-tailed (end-repaired) DNA samples, which were subsequently purified using AMPure XP beads (Beckman Coulter, United Kingdom) and eluted using nuclease-free water. Native Barcoding Expansions (EXP-NBD104 or EXP-NBD114) (Oxford Nanopore Technologies, United Kingdom) were ligated to the end-repaired DNA using Blunt/TA Ligase Master Mix (New England Biolabs). After purification using AMPure XP beads and elution with nuclease-free water, individual barcoded DNA samples were pooled and ligated to sequencing adaptor molecules in Adapter Mix II using NEBNext Quick T4 DNA Ligase (New England Biolabs). The ligation products were washed twice using long fragmentation buffer and eluted with elution buffer. The multiplexed barcoded samples were prepared as sequencing libraries and loaded into a FLO-MIN106D flow cell R9.4.1. Sequencing was carried out on a MinION Mk1B with MinKNOW software (21.10.4) for 24 h using standard settings. Base-calling of FAST5 raw sequencing data was performed using the high-accuracy (Qscore >9) option with Nanopore Guppy software v.6.0.7 (ONT, United Kingdom). A higher Qscore may result in reduced detection sensitivity, but it also reduces false positives. Adapter sequences were demultiplexed using Porechop v.0.2.4,1 and read quality was checked using Nanoplot with default settings. A summary of the runs is shown in Supplementary Table S3. The reads for each isolate were assembled using Unicycler v.0.4.9 (Wick et al., 2017) with default settings, and the resulting contigs were visualized using Bandage v.0.8.1 (Wick et al., 2017). The gene sul4 was determined based on amino acid identity relative to deposited sequences in the Comprehensive Antibiotic Resistance Database (CARD) (Jia et al., 2017, https://card.mcmaster.ca/). We searched for high amino acid sequence similarity (E-value <1.0, identity >41%, coverage >80%) relative to database-deposited sul4 gene products (ARO:3004361). MGEs were identified using the reference center for bacterial insertion sequences ISfinder database (ISfinder: https://www-is.biotoul.fr/). All sul genes and MGEs were confirmed using tblastn 2.12.0. The threshold values in previous reports were >40 coverage by nucleotide identity (Dai et al., 2022), whereas ours is higher due to longer sequence was targeted. Thus our identification should be reasonable in long-read method.
3. Results and discussion
3.1. Profile of sul genes harbored by SMX-resistant isolates
Occurrence rate of SMX-resistant bacteria on nutrient seawater medium was 27.0% (May) and 8.6% (November), whereas those on low nutrient seawater medium was 39.1% (May) and 97.4% (November). Since marine bacteria generally grow well on low nutrient medium (Carlucci et al., 1986), high rate in low nutrient medium should be reflected by the property.
A phylogenetic tree of SMX-resistant isolates based on the 16S rRNA gene and detection of the sul1, sul2, sul3, sul4, dfrA1, and IntI1 genes by PCR are shown in Figure 1. A total of 41 sul-positive isolates (55%) were identified among the 74 SMX-resistant isolates. The sul-negative isolates should have other mechanisms, such as efflux pumps (Su et al., 2015). Of 37 α-proteobacteria isolates, 16 were sul4-positive, 1 was sul1-positive, and 1 was sul3-sul4-double positive. dfrA1 was detected in 10 of the 74 SMX-resistant isolates. Three isolates showed the combination sul4 and dfrA1. Of 34 γ-proteobacteria, 14 isolates were sul4-positive, 6 were sul2-positive, and 4 were sul3-positive. Two isolates had dfrA1 but not sul genes. All 3 of the bacilli were sul4-positive, and 2 were also dfrA1-positive. The frequency of sul genes among all 74 SMX-resistant isolates was ranked sul4 > sul2 > sul3 > sul1, showing that sul4 is most common. Most past studies of aquatic bacteria have reported that sul1 or sul2 is dominant in cultured bacteria (Rosser and Young, 1999; Stoll et al., 2012; Wang et al., 2014). PCR-based analyses using aquatic environmental DNA also showed that sul1 or sul2 is dominant (Muziasari et al., 2014; Suzuki et al., 2019, 2021), which suggests that sul1 and sul2 are distributed worldwide among a variety of bacterial communities. On the other hand, Suzuki et al. (2013) detected sul3 at a higher abundance than sul1 and sul2 from a non-culturable community in Manila Bay, Philippines. Taken together with previous reports regarding sul3, the high detection rates in the present study suggest that sul3 and sul4 might be more common in the marine environment than other environments.
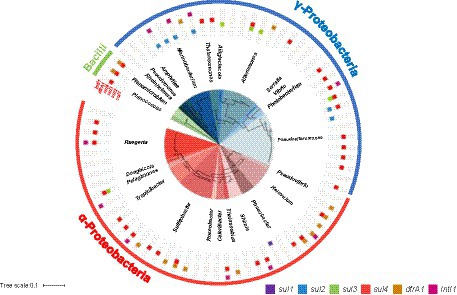
Figure 1. Phylogenetic tree of 74 SMX-resistant isolates based on the 16S rRNA gene (center circle), and detection of target genes by PCR.
The gene sul4 was first discovered in river sediments of India in 2017, and it has since been found in environmental metagenomes from several countries (Razavi et al., 2017). However, the host range and distribution are unknown. In the present study, we detected sul4 by PCR, and the results indicated that sul4 is dominant among culturable SMX-resistant bacteria from seawater, with an occupation of 45% among sul-positive isolates. All sul4-positive α-proteobacteria isolates were classified in the family Rhodobacteraceae (Figure 1). Bacteria belonging to the Rhodobacteraceae are abundant key members of coastal seawater communities that form biofilms (Elifantz et al., 2013). In aquatic environments, biofilms are considered hotspots for horizontal transfer of ARGs (Abe et al., 2020), suggesting there is a high potential for HGT among sul4-positive Rhodobacteraceae in ocean waters. A ballast water study reported that major ARGs, including sul1 and sul2, were detected in Rhodobacteraceae (Lv et al., 2020). In contrast to sul4-positive α-proteobacteria, sul4-positive γ-proteobacteria were classified into four families (Alteromonadaceae, Vibrionaceae, Pseudoalteromonadaceae, Colwelliaceae), suggesting sul4 has a wide host range in this phylum.
The combination of sul4 and IntI1 was found in α-proteobacteria (Sulfitobacter sp., Ruegeria sp., and Thalassobius aestuarii) and γ-proteobacteria (Photobacterium), although not at a high frequency (Figure 1). IntI1 is associated with mobility of ARGs and has been linked to genes conferring resistance to antibiotics and anthropogenic pollutants (Gillings et al., 2015). Some ARGs detected in human and animal pathogens were initially detected in aquatic bacteria (Cabello et al., 2013), suggesting the possibility of transfer of marine-originating ARGs to humans via integrons with IntI1. We further examined the primary structure of the peripheral region of sul4 in the genome to determine whether sul4 could form a transfer unit.
3.2. Genomic analysis of sul4-positive isolates
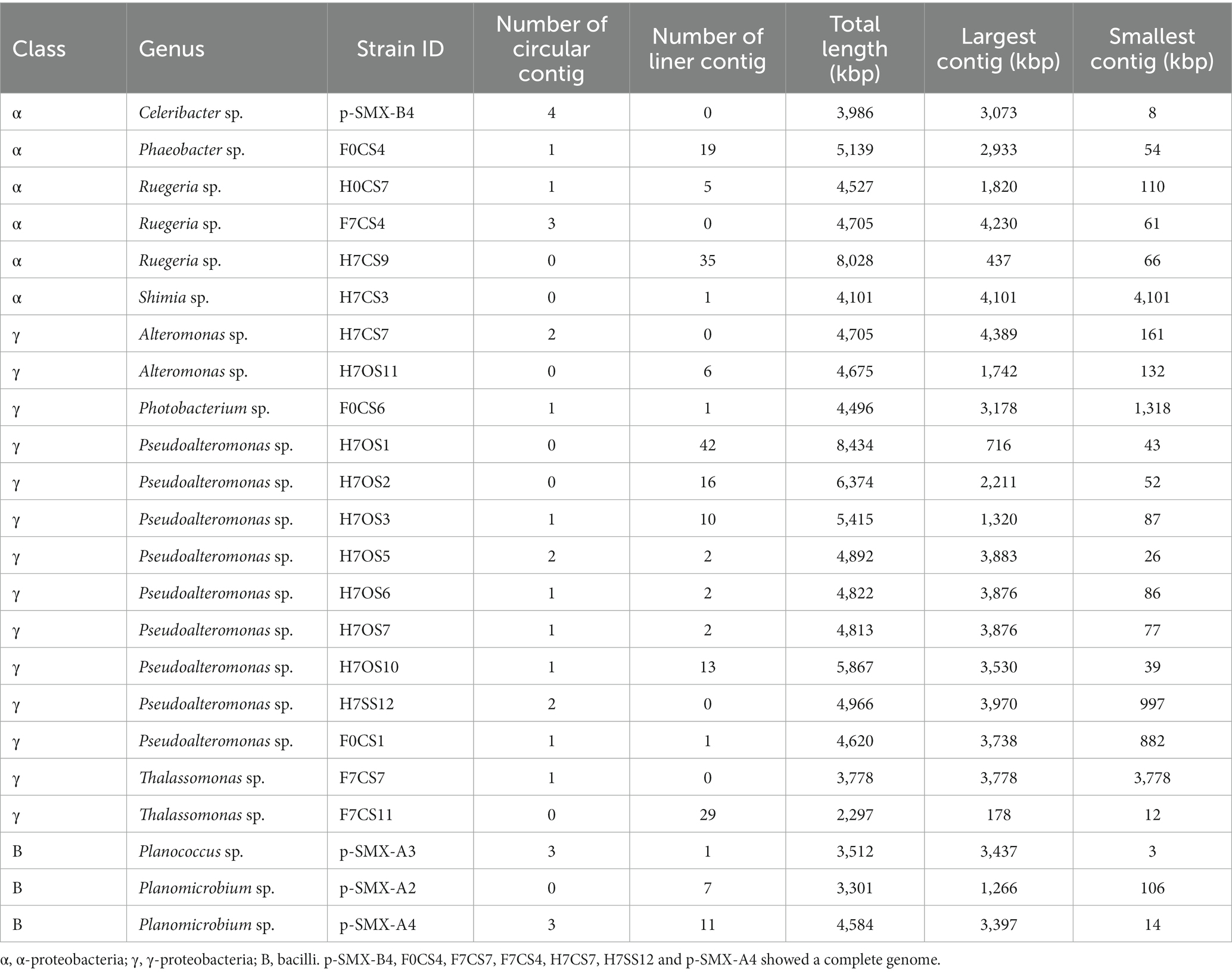
MinION sequencing data for each isolate gave 2,519–45,720 reads, with an average read length of 5,022–24,011 bp and an average read quality of 11–13 (Supplementary Table S3). The genome assembly is summarized in Table 1. After trimming of barcodes and adapters, closed circular contigs were obtained for five isolates via de novo assembly, which indicated the genome size was similar to the standard deposited in NCBI. These five isolates were Celeribacter sp. (pSMX.B4) and Ruegeria sp. (F7CS4) from the α-proteobacteria; and Alteromonas sp. (H7CS7), Pseudoalteromonas sp. (H7SS12), and Thalassomonas sp. (F7CS7) from the γ-proteobacteria (Table 1). Although other isolates gave linear contigs, the total lengths were close to the genome size in the database (Table 1), suggesting that the sequence data covered almost the whole genome. Only Thalassomonas sp. F7CS11 showed a small genome size (2,297 kbp). The genomes of related species are reportedly larger than 7 Mbp (Olonade et al., 2015), suggesting that the whole genome sequence was not obtained for F7CS11. However, as we did identify the sul4-encoding region in this isolate, it was included for further analysis.
Table 2 shows isolates with PCR and tblastn search results for the whole-genome sequence data. Among 33 sul4-positives with PCR (Figure 1), 23 isolates were finally confirmed as harboring sul4 via tblastn search: 6 α-proteobacteria, 14 γ-proteobacteria, and 3 bacilli (Table 2). Sequences of the 23 sul4-harboring isolates had hits for sulfonamide-resistance genes in the CARD, which contained efflux pumps, sul1, sul2, sul3, sul4, and dfrA1. Those genes annotated to sul4 exhibited amino acid identity of 41%–51% relative to reported sul4 gene products (ARO:3004361). These data confirmed that the isolates examined in the present study harbor putative sul4. In contrast to the above 23 isolates, sul4 was not detected in the whole genome sequence of the remaining 10 isolates. This false-negative detection may have been due to a high sequencing error rate associated with the nanopore long-read sequencing method. These errors could not be sufficiently corrected, despite our setting of a high Qscore of >9.
Primary structures within the 5-kbp area around sul4 are shown in Figure 2. Four types of gene arrangement were observed. In type 1, sul4 was followed by the phosphoglucosamine mutase gene, and this arrangement was observed only in the γ-proteobacteria. In type 2, sul4 was located immediately after the dihydroneopterin aldolase gene (folB), followed by the phosphoglucosamine mutase and permease of the drug/metabolite transporter (DMT) superfamily genes. This type was found in 5 of the α-proteobacteria isolates. Type 3 was similar to type 2; however, the permease DMT superfamily gene was not present. This type was found in α-proteobacteria, Celeribacter sp. (pSMX.B4). In type 4, found in bacilli, sul4 was followed by folB and 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase (folK). In this manner, the four types of gene arrangement differed among the bacterial classes. Interestingly, none of the types were accompanied by MGEs such as transposons or insertion sequences within the 5-kbp region, suggesting sul4 is not located on an MGE. IntI1 was detected in four isolates (Table 2) at a location distant from sul4; the distance from sul4 in p.SMX.B4 and H7CS3 was 1.2 Mbp, and that in H7OS10 was 870 kbp. H0CS7 harbored IntI1 and sul4 on different contigs. This distant positioning suggests that IntI1 is not connected to sul4. No other integrase or transposon sequences were found in the sul4 area (Figure 2), suggesting sul4 in these marine bacteria might not be a mobilizable gene.
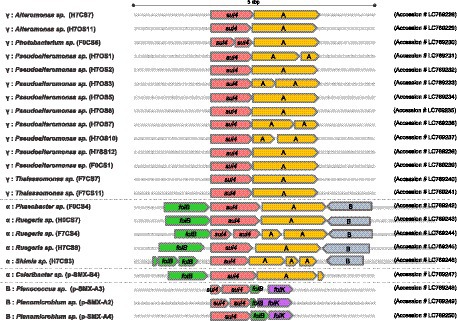
Figure 2. sul4 and the peripheral region within a 5 kbp area. The gene cluster around sul4 was classified into 4 types. A: Phosphoglucosamine mutase; B: Permease of the drug/metabolite transporter (DMT) superfamily. Accession number of each cluster is indicated at the right.
The first report of sul4 (Razavi et al., 2017) indicated the gene was accompanied by an attI site, followed by the complete qacE gene, or with a transposase ISCR20 belonging to the insertion sequence common region (ISCR) family. These data suggest sul4 has been widely distributed via transferable MGEs (Razavi et al., 2017). In contrast, sul4 was not accompanied by an MGE in the present study, suggesting that marine bacterial sul4 acquired independently of Chloroflexus sul4, which was accompanied by an ISCR.
The gene sul4 in our study showed another interesting characteristic. The peripheral area of sul4 encodes multiple folate metabolism (fol) genes. The folB gene was found in types 2, 3, and 4, and folK was found in type 4 (Figure 2). The genes encoding the folate synthesis machinery are reportedly clustered on the genome (de Crécy-Lagard et al., 2007), suggesting that fol genes around putative sul4 function in the folate synthesis system. The gene sul4 possibly encodes a native DHPS in the marine bacteria in our study. The sul1 and sul2 genes showed lower Km values for p-aminobenzoate and higher affinity for sulfonamides than native DHPS (Sköld, 2000). The sul4 gene product might function as an essential DHPS in the host.
In Pseudoalteromonas sp. (H7SS12), sul2 was found in an area with transposase (Supplementary Figure S1). The sul2 was suggested to be capable of horizontal transfer between human and animal Escherichia coli (Trobos et al., 2009). The gene sul2 of the isolate H7SS12 is encoded with the aminoglycoside-resistance genes strB and strA and florfenicol efflux gene floR on a transposon area (Supplementary Figure S1), suggesting the presence of a mobilizable ARG-unit with multidrug resistance properties. The resistance to sulfonamide, aminoglycoside, and florfenicol has also been reported in Vibrio cholerae (ICDC-VC4210, CDC-VC0143) (Wang et al., 2016) and V. cholerae O139 (MO10) (Waldor et al., 1996). The identities of the transposon areas of Pseudoalteromonas sp. (H7SS12) relative to these Vibrio species were 45% (ICDC-VC4210), 77% (CDC-VC0143), and 68% (MO10) (Supplementary Figure S1) when compared with the complete transposase set. These data suggest that marine bacteria have acquired multidrug resistance via recombination. Furthermore, the concentration site of strB, strA, and sul2 was observed on the plasmid pKP27-MCR1 (CP041641.1) of Klebsiella pneumoniae (PIMB15ND2KP27) (Mbelle et al., 2019) and on the mobile plasmid of Salmonella typhimurium TST207 (Tamamura et al., 2013). This cluster was also found in an Antarctic ice core (1,200 to 2,800 years before present) (Okubo et al., 2019). The multidrug resistance cluster of sulfonamides and aminoglycosides might have been formed via transfer between pathogenic and marine bacteria thousands of years ago.
Some metagenomic studies have reported detection of ARGs in ocean waters (Port et al., 2014; Hatosy and Martiny, 2015; Cuadrat et al., 2020), indicating that efflux channels and tetracycline-resistance genes are abundant (Cuadrat et al., 2020). The results of the present study suggest that metabolic systems of marine bacteria might be capable of playing a role in resistance to antimicrobials, which could then evolute to ARGs that are disseminated to pathogenic bacteria. Marine bacteria could thus be considered an ancestral origin of ARGs.
4. Conclusion
Our results showed that sul4 is present in marine α- and γ-proteobacteria and bacilli without taxonomic bias in the harboring bacteria. This is the first study to detect sul4 in defined marine bacteria. In addition, sul4 was not located on an MGEs, suggesting it is not mobile. We found that sul4 was flanked by genes related to folate synthesis, suggesting that sul4 codes for a housekeeping DHPS in the host bacteria and is clonally inherited within species. The sul2 in the sul4-harboring isolate was clustered with a transposon, suggesting the presence of a unit for transfer to other bacteria.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
SaS contributed to conception and design of the study and funding. SuS and AK performed molecular experiments and analysis of data. RK isolated bacteria. KW contributed data curation and manuscript review. SuS and SaS wrote the first draft of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by KAKENHI 16H01782 and 20H00633, JSPS.
Acknowledgments
The authors thank Joeselle Serrana and Ken Aquino Bongulto for their consulting MinION operation.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1230548/full#supplementary-material
Footnotes
References
Abe, K., Nomura, N., and Suzuki, S. (2020). Biofilms: hot spots of horizontal gene transfer (HGT) in aquatic environments, with a focus on a new HGT mechanism. FEMS Microbiol. Ecol. 96:fiaa031. doi: 10.1093/femsec/fiaa031
Aminov, R. I. (2011). Horizontal gene exchange in environmental microbiota. Front. Microbiol. 2:158. doi: 10.3389/fmicb.2011.00158
Antunes, P., Machado, J., Sousa, J. C., and Peixe, L. (2005). Dissemination of sulfonamide resistance genes (sul1, sul2, and sul3) in Portuguese Salmonella enterica strains and relation with integrons. Antimicrob. Agents Chemother. 49, 836–839. doi: 10.1128/AAC.49.2.836-839.2005
Bean, D. C., Livermore, D. M., and Hall, L. M. C. (2009). Plasmids imparting sulfonamide resistance in Escherichia coli: implications for persistence. Antimicrob. Agents Chemother. 53, 1088–1093. doi: 10.1128/AAC.00800-08
Bickel, M. H. (1988). The development of sulfonamides (1932–1938) as a focal point in the history of chemotherapy. Gesnerus 45, 67–86. doi: 10.1163/22977953-04501006
Blahna, M. T., Zalewski, C. A., Reuer, J., Kahlmeter, G., Foxman, B., and Marrs, C. F. (2006). The role of horizontal gene transfer in the spread of trimethoprim-sulfamethoxazole resistance among uropathogenic Escherichia coli in Europe and Canada. J. Antimicrob. Chemother. 57, 666–672. doi: 10.1093/jac/dkl020
Cabello, F. C., Godfrey, H. P., Tomova, A., Ivanova, L., Dölz, H., Millanao, A., et al. (2013). Antimicrobial use in aquaculture re-examined: its relevance to antimicrobial resistance and to animal and human health. Environ. Microbiol. 15, 1917–1942. doi: 10.1111/1462-2920.12134
Carlucci, A. F., Shmp, S. L., and Craven, D. B. (1986). Growth characteristics of low-nutrient bacteria from the north-east and central Pacific Ocean. FEMS Microbiol. Lett. 38, 1–10. doi: 10.1111/j.1574-6968.1986.tb01932.x
Cuadrat, R. R. C., Sorokina, M., Andrade, B. G., Goris, T., and Avila, A. M. R. D. (2020). Global ocean resistome revealed: exploring antibiotic resistance gene abundance and distribution in TARA oceans samples. Gigascience 9:giaa046. doi: 10.1093/gigascience/giaa046
Dai, D., Brown, C., Bürgmann, H., Larsson, D. G. J., Nambi, I., Zhang, T., et al. (2022). Long-read metagenomic sequencing reveals shifts in associations of antibiotic resistance genes with mobile genetic elements. Microbiome 10:20. doi: 10.1186/s40168-021-01216-5
de Crécy-Lagard, V., El Yacoubi, B., de la Garza, R., Noiriel, A., and Hanson, A. (2007). Comparative genomics of bacterial and plant folate synthesis and salvage: predictions and validations. BMC Genomics 8:245. doi: 10.1186/1471-2164-8-245
Dungan, R. S., and Bjorneberg, D. L. (2020). Antibiotic resistance genes, class 1 integrons, and IncP-1/IncQ-1 plasmids in irrigation return flows. Environ. Pollut. 257:113568. doi: 10.1016/j.envpol.2019.113568
Elifantz, H., Horn, G., Ayon, M., Cohen, Y., and Minz, D. (2013). Rhodobacteraceae are the key members of the microbial community of the initial biofilm formed in Eastern Mediterranean coastal seawater. FEMS Microbiol. Ecol. 85, 348–357. doi: 10.1111/1574-6941.12122
G7 Presidency, (2015). Final report by the Federal Government on the G7 presidency 2015. Available at: https://www.mofa.go.jp/files/000084020.pdf
Gillings, M. R., Gaze, W. H., Pruden, A., Smalla, K., Tiedje, J. M., and Zhu, Y. G. (2015). Using the class 1 integron-integrase gene as a proxy for anthropogenic pollution. ISME J. 9, 1269–1279. doi: 10.1038/ismej.2014.226
Hatosy, S. M., and Martiny, A. C. (2015). The ocean as a global reservoir of antibiotic resistance genes. Appl. Environ. Microbiol. 81, 7593–7599. doi: 10.1128/AEM.00736-15
Heuer, H., and Smalla, K. (2007). Manure and sulfadiazine synergistically increased bacterial antibiotic resistance in soil over at least two months. Environ. Microbiol. 9, 657–666. doi: 10.1111/j.1462-2920.2006.01185.x
Huovinen, P., Sundström, L., Swedberg, G., and Sköld, O. (1995). Trimethoprim and sulfonamide resistance. Antimicrob. Agents Chemother. 39, 279–289. doi: 10.1128/aac.39.2.279
Jia, B., Raphenya, A. R., Alcock, B., Waglechner, N., Guo, P., Tsang, K. K., et al. (2017). CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 45, D566–D573. doi: 10.1093/nar/gkw1004
Jiang, H., Cheng, H., Liang, Y., Yu, S., Yu, T., Fang, J., et al. (2019). Diverse mobile genetic elements and conjugal transferability of sulfonamide resistance genes (sul1, sul2, and sul3) in Escherichia coli isolates from Penaeus vannamei and pork from large markets in Zhejiang, China. Front. Microbiol. 10:1787. doi: 10.3389/fmicb.2019.01787
Kerrn, M. B., Klemmensen, T., Frimodt-Møller, N., and Espersen, F. (2002). Susceptibility of Danish Escherichia coli strains isolated from urinary tract infections and bacteraemia, and distribution of sul genes conferring sulphonamide resistance. J. Antimicrob. Chemother. 50, 513–516. doi: 10.1093/jac/dkf164
Koczura, R., Mokracka, J., Taraszewska, A., and Łopacinska, N. (2016). Abundance of class 1 integron-integrase and sulfonamide resistance genes in river water and sediment is affected by anthropogenic pressure and environmental factors. Microb. Ecol. 72, 909–916. doi: 10.1007/s00248-016-0843-4
Lane, D. J., Pace, B., Olsen, G. J., Stahl, D. A., Sogin, M. L., and Pace, N. R. (1985). Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc. Natl. Acad. Sci. U. S. A. 82, 6955–6959. doi: 10.1073/pnas.82.20.6955
Letunic, I., and Bork, P. (2019). Interactive Tree of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259. doi: 10.1093/nar/gkz239
Levy, S. B., and Marchall, B. (2004). Antibacterial resistance worldwide: causes, challenges and responses. Nat. Med. 10, S122–S129. doi: 10.1038/nm1145
Lv, B., Cui, Y., Tian, W., Wei, H., Chen, Q., Liu, B., et al. (2020). Vessel transport of antibiotic resistance genes across oceans and its implication for ballast water management. Chemosphere 253:126697. doi: 10.1016/j.chemosphere.2020.126697
Matsuura, R., Kanehara, R., Kadoya, A., and Suzuki, S. (2021). Adsorption of sulfonamides to marine diatoms and arthropods. Environ. Toxicol. Pharmacol. 82:103557. doi: 10.1016/j.etap.2020.103557
Mbelle, N. M., Feldman, C., Sekyere, J. O., Maningi, N. E., Modipane, L., and Essack, S. Y. (2019). The resistome, mobilome, virulome and phylogenomics of multidrug-resistant Escherichia coli clinical isolates from Pretoria, South Africa. Sci. Rep. 9:16457. doi: 10.1038/s41598-019-52859-2
Muyzer, G., de Waal, E. C., and Uitterlinden, A. G. (1993). Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59, 695–700. doi: 10.1128/aem.59.3.695-700.1993
Muziasari, W. I., Managaki, S., Pärnänen, K., Karkman, A., Lyra, C., Tamminen, M., et al. (2014). Sulphonamide and trimethoprim resistance genes persist in sediments at Baltic Sea aquaculture farms but are not detected in the surrounding environment. PLoS One 9:e92702. doi: 10.1371/journal.pone.0092702
Na, G., Zhang, W., Zhou, S., Gao, H., Lu, Z., Wu, X. L., et al. (2014). Sulfonamide antibiotics in the northern Yellow Sea are related to resistant bacteria: implications for antibiotic resistance genes. Mar. Pollut. Bull. 84, 70–75. doi: 10.1016/j.marpolbul.2014.05.039
Okubo, T., Ae, R., Noda, J., Iizuka, Y., Usui, M., and Tamura, Y. (2019). Detection of the sul2-str A-str B gene cluster in an ice core from Dome Fuji Station, East Antarctica. J. Glob. Antimicrob. Resist. 17, 72–78. doi: 10.1016/j.jgar.2018.11.005
Olonade, I., Joaquim, L. J., and Trindade, M. (2015). Draft genome sequences of marine isolates of Thalassomonas viridans and Thalassomonas actiarum. Genome Announc. 3, e00297–e00215. doi: 10.1128/genomeA.00297-15
Perreten, V., and Boerlin, P. (2003). A new sulfonamide resistance gene (sul3) in Escherichia coli is widespread in the pig population of Switzerland. Antimicrob. Agents Chemother. 47, 1169–1172. doi: 10.1128/aac.47.3.1169-1172.2003
Port, J. A., Cullen, A. C., Wallace, J. C., Smith, M. N., and Faustman, E. M. (2014). Metagenomic frameworks for monitoring antibiotic resistance in aquatic environments. Environ. Health Perspect. 122, 222–228. doi: 10.1289/ehp.1307009
Rådström, P., and Swedberg, G. (1988). RSF1010 and conjugative plasmid contain sul1, one of two known genes for plasmid-borne sulfonamide resistance dihydropteroate syntetase. Antimicrob. Agents Chemother. 32, 1684–1692. doi: 10.1128/AAC.32.11.1684
Razavi, M., Marathe, N. P., Gillings, M. R., Flach, C. F., Kristiansson, E., and Larsson, D. G. J. (2017). Discovery of the fourth mobile sulfonamide resistance gene. Microbiome 5:160. doi: 10.1186/s40168-017-0379-y
Rosser, S., and Young, H. K. (1999). Identification and characterization of class 1 integrons in bacteria from an aquatic environment. J. Antimicrob. Chemother. 44, 11–18. doi: 10.1093/jac/44.1.11
Shah, S. Q. A., Cabello, F. C., L’Abée-Lund, T. M., Tomova, A., Godfrey, H. P., Buschmann, A. H., et al. (2014). Antimicrobial resistance and antimicrobial resistance genes in marine bacteria from salmon aquaculture and non-aquaculture sites. Environ. Microbiol. 16, 1310–1320. doi: 10.1111/1462-2920.12421
Shimizu, A., Takada, H., and Koike, T. (2013). Ubiquitous occurrence of sulfonamides in tropical Asian waters. Sci. Total Environ. 452-453, 108–115. doi: 10.1016/j.scitotenv.2013.02.027
Sköld, O. (2000). Sulfonamide resistance: mechanisms and trends. Drug Resist. Updates 3, 155–160. doi: 10.1054/drup.2000.0146
Stalder, T., Barraud, O., Casellas, M., Dagot, C., and Ploy, M. C. (2012). Integron involvement in environmental spread of antibiotic resistance. Front. Microbiol. 3:119. doi: 10.3389/fmicb.2012.00119
Stokes, H. W., Nesbo, C. L., Holley, M., Bahl, M. I., Gillings, M. R., and Boucher, Y. (2006). Class 1 integrons potentially predating the association with tn402-like transposition genes are present in a sediment microbial community. J. Bacteriol. 188, 5722–5730. doi: 10.1128/JB.01950-05
Stoll, C., Sidhu, J. P. S., Tiehm, A., and Toze, S. (2012). Prevalence of clinically relevant antibiotic resistance genes in surface water samples collected from Germany and Australia. Environ. Sci. Technol. 46, 9716–9726. doi: 10.1021/es302020s
Su, C.-C., Bolla, J. R., Kumar, N., Radhakrishnan, A., Long, F., Delmar, J. A., et al. (2015). Structure and function of Neisseria gonorrhoeae MtrF illuminates a class of antimetabolite efflux pumps. Cell Rep. 11, 61–70. doi: 10.1016/j.celrep.2015.03.003
Sukul, P., and Spiteller, M. (2006). Sulfonamides in the environment as veterinary drugs. Rev. Environ. Contam. Toxicol. 187, 67–101.
Sunagawa, S., Coelho, L. P., Chaffron, S., Kultima, J. R., Labadie, K., Salazar, G., et al. (2015). Structure and function of the global ocean microbiome. Science 348:1261359. doi: 10.1126/science.1261359
Sundström, L., Rådström, P., Swedberg, G., and Sköld, O. (1988). Site-specific recombination promotes linkage between trimethoprim- and sulfonamide resistance genes. Sequence characterization of dhfrV and sulI and a recombination active locus of Tn21. Mol. Gen. Genet. 213, 191–201. doi: 10.1007/BF00339581
Suzuki, S., Nakanishi, S., Tamminen, M., Yokokawa, T., Sato-Takabe, Y., Ohta, K., et al. (2019). Occurrence of sul and tet(M) genes in bacterial community in Japanese marine aquaculture environment throughout the year: profile comparison with Taiwanese and Finnish aquaculture waters. Sci. Total Environ. 669, 649–656. doi: 10.1016/j.scitotenv.2019.03.111
Suzuki, S., Ogo, M., Miller, T. W., Shimizu, A., Takada, H., and Sringan, M. A. T. (2013). Who possesses drug resistance genes in the aquatic environment?: sulfamethoxazole (SMX) resistance genes among the bacterial community in water environment of Metro-Manila, Philippines. Front. Microbiol. 4:102. doi: 10.3389/fmicb.2013.00102
Suzuki, S., Ogo, M., Takada, H., Seki, K., Mizukawa, K., Kadoya, A., et al. (2021). Contamination of antibiotics and sul and tet(M) genes in veterinary wastewater, river, and coastal sea in Thailand. Sci. Total Environ. 791:148423. doi: 10.1016/j.scitotenv.2021.148423
Swedberg, G., and Sköld, O. (1983). Plasmid-borne sulfonamide resistance determinants studied by restriction enzyme analysis. J. Bacteriol. 153, 1228–1237. doi: 10.1128/jb.153.3.1228-1237.1983
Tamamura, Y., Tanaka, K., Akiba, M., Kanno, T., Hatama, S., Ishihara, R., et al. (2013). Complete nucleotide sequences of virulence-resistance plasmids carried by emerging multidrug-resistant Salmonella enterica serovar Typhimurium isolated from cattle in Hokkaido, Japan. PLos One 8:e77644. doi: 10.1371/journal.pone.0077644
Trobos, M., Christensen, H., Sunde, M., Nordentoft, S., Agersø, Y., Simonsen, G. S., et al. (2009). Characterization of sulphonamide-resistant Escherichia coli using comparison of sul2 gene sequences and multilocus sequence typing. Microbiology 155, 831–836. doi: 10.1099/mic.0.024190-0
Vinué, L., Sáenz, Y., Rojo-Bezares, B., Olarte, I., Undabeitia, E., Somalo, S., et al. (2010). Genetic environment of sul genes and characterisation of integrons in Escherichia coli isolates of blood origin in a Spanish hospital. Int. J. Antimicrob. Agents 35, 492–496. doi: 10.1016/j.ijantimicag.2010.01.012
Waldor, M. K., Tschape, H., and Mekalanos, J. J. (1996). A new type of conjugative transposon encodes resistance to sulfamethoxazole, trimethoprim, and streptomycin in Vibrio cholerae O139. J. Bacteriol. 178, 4157–4165. doi: 10.1128/jb.178.14.4157-4165.1996
Waldron, L. S., and Gillings, M. R. (2015). Screening foodstuffs for class 1 integrons and gene cassettes. J. Vis. Exp. 100:e52889. doi: 10.3791/52889
Wang, N., Yang, X., Jiao, S., Zhang, J., Ye, B., and Gao, S. (2014). Sulfonamide-resistant bacteria and their resistance genes in soils fertilized with manures from Jiangsu province, Southeastern China. PLoS One 9:e112626. doi: 10.1371/journal.pone.0112626
Wang, R., Yu, D., Yue, J., and Kan, B. (2016). Variations in SXT elements in epidemic Vibrio cholerae O1 El Tor strains in China. Sci. Rep. 6:22733. doi: 10.1038/srep22733
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595
Yun, M.-K., Wu, Y., Li, Z., Waddel, M. B., Ferreira, A. M., Lee, R. E., et al. (2013). Catalysis and sulfa drug resistance in dihydropteroate synthase. Science 335, 1110–1114. doi: 10.1126/science.1214641
Keywords: sul4 , sulfonamide resistance, dihydropteroate synthase, marine bacteria, α-proteobacteria
Citation: Shindoh S, Kadoya A, Kanechi R, Watanabe K and Suzuki S (2023) Marine bacteria harbor the sulfonamide resistance gene sul4 without mobile genetic elements. Front. Microbiol. 14:1230548. doi: 10.3389/fmicb.2023.1230548
Edited by:
Kristina Kadlec, Independent Researcher, Wunstorf, GermanyReviewed by:
Atte Johannes Von Wright, University of Eastern Finland, FinlandBijit Bhowmik, Croda Inc, United States
Copyright © 2023 Shindoh, Kadoya, Kanechi, Watanabe and Suzuki. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Satoru Suzuki, d3JpdGVyc29oQGdtYWlsLmNvbQ==
 Suzune Shindoh1
Suzune Shindoh1 Kozo Watanabe
Kozo Watanabe Satoru Suzuki
Satoru Suzuki