
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 11 July 2023
Sec. Microbe and Virus Interactions with Plants
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1227830
This article is part of the Research Topic Microbiomics in Food Security: Paradigm Shift in Omics View all 10 articles
 Priyanka Jha1*†
Priyanka Jha1*† Tamanna Kaur1†
Tamanna Kaur1† Ishita Chhabra2†
Ishita Chhabra2† Avirup Panja3
Avirup Panja3 Sushreeta Paul3
Sushreeta Paul3 Vijay Kumar1
Vijay Kumar1 Tabarak Malik4*
Tabarak Malik4*Endophytic fungi comprise host-associated fungal communities which thrive within the tissues of host plants and produce a diverse range of secondary metabolites with various bioactive attributes. The metabolites such as phenols, polyketides, saponins, alkaloids help to mitigate biotic and abiotic stresses, fight against pathogen attacks and enhance the plant immune system. We present an overview of the association of endophytic fungal communities with a plant host and discuss molecular mechanisms induced during their symbiotic interaction. The overview focuses on the secondary metabolites (especially those of terpenoid nature) secreted by endophytic fungi and their respective function. The recent advancement in multi-omics approaches paved the way for identification of these metabolites and their characterization via comparative analysis of extensive omics datasets. This study also elaborates on the role of diverse endophytic fungi associated with key agricultural crops and hence important for sustainability of agriculture.
Numerous microorganisms exist in mutualistic associations with plants, whereby their interaction results in a beneficial outcome for both entities. Microbes with such capabilities are ample in above-ground as well as in underground (edaphic) habitats. Epiphytes are microorganisms that are found on the external surface of plants, whereas endophytes are microorganisms that reside and establish themselves within plant tissues, specifically within the leaves or roots. Among these, the rhizosphere harbors a plethora of active microorganisms that enhance plant nutrition, growth, and development of plants. The rhizosphere can be defined as the soil zone in the proximity of plant roots that is affected by the presence of microorganisms and root exudates, including compounds secreted by the root system of the plants (Kanchan et al., 2020). The region is characterized by multifaceted plant-microbe interactions, encompassing mutualistic, symbiotic, or parasitic relationships. These interactions are capable of triggering the biosynthesis of secondary metabolites by the microbial cells, which can be advantageous for the plants by conferring tolerance to biotic and abiotic stresses contributing to the plant’s overall fitness (Koza et al., 2022). Endosymbiotic microorganisms, which are typically bacteria and/or fungi, establish mutually beneficial relationships with their plant host including improved host fitness. These organisms are considered to be non-pathogenic since they can complete a part of their life cycle or even the whole life cycle within their host without causing any symptoms of disease (Nair and Padmavathy, 2014). The endophytic community can be further divided into two subgroups which involve ‘obligate endophytes’ (endophytes that critically dependent on plant metabolism for their survival) and ‘facultative endophytes’ (endophytes that spend certain stages of their life cycle outside the host body and are mostly associated with plants present in their nearby soil environment) (Hardoim et al., 2008; Abreu-Tarazi et al., 2010).
Of all the microorganisms inhabiting the rhizosphere, endophytic fungi or fungal endophytes have garnered significant attention from researchers. This is owing to their ability to produce various bioactive molecules such as antibacterial compounds, and biostimulants, which can facilitate essential oil biosynthesis (Radic and Strukelj, 2012; Uzma et al., 2018; El Enshasy et al., 2019). Essential oils are the aromatic compounds that have high vapor pressure and low boiling point. Vanillin and 3-methoxy-4-hydroxytoluene was isolated from the roots of Zingiber officinale Rosc. infected with Streptomyces aureofaciens SMUAc130 (Taechowisan et al., 2005). The bacterial volatile compounds are significant in establishing interspecific and or intraspecific communication and role in defence mechanism against antagonistic microbes. Volatile organic compounds (VOCs) play extremely important role in antibiosis and signaling mechanism for microbes with symbiotic relation under competitive soil conditions (Polito et al., 2022). Endophytic fungi have been recognized for their multifaceted roles in plant-microbe interactions: nutrient solubilization in the plant rhizosphere, promoting plant growth, acting as biocontrol agents against pests and pathogens, inducing systemic resistance against biotic and abiotic stresses, and participating in the biosynthesis of secondary metabolites (Mehta et al., 2019; Poveda et al., 2020a; Cui et al., 2021; Poveda et al., 2021; Poveda and Baptista, 2021). Fungal secondary metabolites may act by causing alterations in the host plant’s morphology and physiology (Alam et al., 2021). Endophytic fungi establish symbiotic associations with host plants through a process that involves the enzymatic degradation of the host plant’s cell wall. To colonize the host plant tissue, endophytic fungi produce a range of cell wall-degrading enzymes including cellulase, laccase, pectinase, and xylanase. These enzymes facilitate the infiltration, colonization, and proliferation of endophytic fungi within the host plant tissue by altering the physical and structural properties of the host cell wall (Chow and Ting, 2019). Upon degradation of the host plant cell wall, host plant activates its defence mechanisms such as pattern-recognition receptors (PRRs) which provide innate immunity to the plant. In order to survive in plants, the endophytes require specific strategies to avoid being detected by the plant based immune system. One such strategies include, hiding of chitin by modifying it or oligomers derived from chitin employing chitin deacetylases (Cord-Landwehr et al., 2016). Apart from these, many fungal endophytes have been identified to secrete bioactive compounds known as secondary metabolites, which are helpful in coping with biotic and abiotic stresses. These secondary metabolites include mostly alkaloids, terpenoids, polyketides. Besides these major ones, certain secondary metabolites are also secreted such as flavonoids, saponins, phenols, and phenolic acids, aliphatic and chlorinated metabolites, peptides, and steroids. It was found that similar secondary metabolites are being produced by the fungal endophytes and even in larger concentrations as compared to the plants. Using gene clustering, transcription factors and horizontal gene transfer, we can study the biosynthesis pathways and genes encoding specific types of bioactive molecules.
Fungal endophytes are emerging source for terpenoids, an important class of secondary metabolites. Terpenoids are a diverse group of natural products, which consist of C5 isoprene units and are the largest class of natural products (Perveen, 2018). The structural diversity of terpenoids is substantial, with more than 80,000 terpenoids having been identified from both plant and microbial sources. The isoprene units required for terpenoid biosynthesis are typically synthesized through either the mevalonic acid pathway (MVA) or the methylerythritol phosphate pathway (MEP). Terpenoids exhibit a diverse range of pharmacological and nutritional activities, as well as utility in the food and cosmetic industries. Moreover, their therapeutic potential has been explored in the context of COVID-19 treatment due to their notable antiviral properties (Diniz et al., 2021). Due to these activities, there is increase in demand of terpenoids which imposes a strain on the terpenoid-producing plant species. A wide variety of terpenoids are produced by endophytic fungi but none of them is capable of producing terpenoids at commercial scale as their yield is low, especially after repeated subculturing.
Initially, with the advent of first generation sequencers alongwith amplicon based sequencing for variable regions of genomes was being employed for deciphering microbiome understanding. However, extensive studies on root morphology and root exudation in deciphering rhizobiomes can be done employing next generation sequencing (NGS). Metagenomics allows detailing of complete genomic information by assembling DNA sequences into genes. It also provides information about novel genes, bio based-products, biomolecules, interaction between microbial communities. Whereas, transcriptomics utilizes the NGS based information to identify and quantify the presence of the particular RNA molecule in biological samples. Metabolomics, on the other hand is a technique which is utilized to identify and analyze metabolite changes occurring due to overexpression or mutation in a desired gene (Chen et al., 2022). The combination of various omics-based approach can help in better understanding of plant-endophyte interaction mechanisms. This study focusses on the secondary metabolites (especially those of terpenoid nature) secreted by endophytic fungi. The study elaborates the role of endophytic fungi as potential source of terpenoid producer alongwith its host related molecular interactions. Concomitantly, a noteworthy discussion on recent multi-omics based approaches employed to have better understanding of host and endophytic fungi interaction for secondary metabolite production.
Endophytic fungi (EF), also known as fungal endophytes, are communities of fungi that form associations either inter- or intracellularly with host plant tissues while simultaneously providing benefits to the host and gaining benefits from it (Alam et al., 2021). These fungi establish a mutualistic or symbiotic relationship with the host plant, and are categorized into two groups based on this relationship. The first group is the Clavicipitaceous fungi or Balansiaceous group, which includes class I endophytes. This group comprises free-living symbiotic species that survive in cool season grasses (Poaceae), infect the ovules of host plants, and are transmitted vertically to progenies through host seeds. The living rhizomes and shoots of the host plant are colonized by these endophytes. The second group is the Non-Clavicipitaceous fungi or Non-Balansiaceous group, which includes class II, class III, and class IV endophytes. The Class I endophyte species are considered as obligate endophytes that offer protection to their host plant under conditions of drought stress, by secreting bioactive metabolites that exhibit defensive or supportive properties (Roberts and Lindow, 2014; Poveda et al., 2021). Non-Clavicipitaceous fungi or Non-Balansiaceous group (includes class II, class III and class IV endophytes) are non-grass host related groups having extensive biodiversity and distribution. These endophytes reside within plant tissues in a dormant or quiescent state without causing any apparent harm to the host plant, that is, they are not closely associated with the host plant. However, as soon as the chemical changes in the host plant occurs either through injuries or environmental stresses, these EFs then enter the host plant intracellularly through roots shoots or rhizomes (Carroll, 1988; Rodriguez et al., 2008; Mishra Y. et al., 2021). In Class II endophytes colonization of the host tissue occurs through roots, shoots and rhizomes whereas colonization occurs through shoots only and roots only in class III and class IV endophytes, respectively. The transmission occurs both vertically and horizontally in class II endophytes whereas only horizontally in class III and class IV endophytes (Rodriguez et al., 2009). Brief list of important crops and beneficial endophytic fungi with their respective functions have been provided in Table 1.
Endophytic fungi interact with the host plant via three types of interactions, that are, mutualistic, commensalistic, and pathogenic interactions (Figure 1). In mutualistic symbiosis, both host plants and EFs benefit from each other leading to evolutionary and ecological success. The EFs on colonizing the host plant tissue alters their metabolism improving plant’s tolerance to stresses such as heavy metal and drought, augmenting growth and development, nutrient acquisition. It can also protect the host plant from herbivore animals, pests, and pathogenic microorganisms whereas the host plant provides shelter and adequate amount of nutrients to the EFs for their proliferation and life cycle completion. In commensalistic or latent pathogenic relationship, EFs interact with the host plant and may or may not show any kind of beneficial effect on the plant physiology. According to various studies, EFs reside in the host plants as dormant or latent pathogen (Brown et al., 1998; Photita et al., 2004; Rodriguez and Redman, 2008; Gorzynska et al., 2018). In other words, during this stage the fungus is harmless and does not induce any symptom of the disease but as the environmental conditions become unfavorable, latent fungi becomes active and cause obvious pathogenic symptoms eventually leading to destruction of the host plant (Romero et al., 2001; Photita et al., 2004; Poveda et al., 2020b).
In the rhizosphere, plants engage in symbiotic interactions with a diverse range of microorganisms through pattern-recognition receptors (PRRs), which are cell surface proteins capable of detecting microbial- or pathogen-associated molecular patterns (MAMPs/PAMPs) produced by the interacting microbe. These PRRs are involved in triggering the first layer of plant innate immunity. Typically, during the formation of a mutualistic symbiotic relationship, the signaling pathways that hinder the expansion of endophytic proliferation, such as miRNA-mediated pathways involved in plant defence mechanisms, are suppressed (Plett and Martin, 2018). This extracellular recognition via MAMPs/PAMPs or damage-associated molecular patterns (DAMPs) has been identified to lead to first layer of innate immunity via triggered defences, and is coined as pattern-triggered immunity (PTI) (Tang et al., 2017; Saijo et al., 2018). Another PTI can be activated upon cellular disintegration through degradation of the plant cell wall compounds such as oligonucleotides, cellodextrins, and the compounds released under stress conditions (cutin monomers and small peptides), which in turn generates endogenous signals termed DAMPs that are also detected by PRRs. Upon infection of the host plant by a microorganism, the botrytis-induced kinase1 (BIK1) effector kinase of the pattern recognition receptors (PRRs) complex becomes activated. This activation leads to an increase in the cytosolic calcium (Ca2+) level mediated by cyclic nucleotide-gated channels (CNGCs). This elevation of Ca2+ is recognized as a crucial signal for triggering the activation of pathogen-associated molecular pattern (PAMP) signals involved in plant immunity during pathogen-associated molecular pattern-triggered immunity (PTI) (Tian et al., 2019). Establishment of successful infection by the pathogen by overcoming PTI via effector-triggered susceptibility (ETS) leads to activation of plants’ second immune system termed effector-triggered immunity (ETI) which is an amplified and robust defense system of the plant. Both Pathogen-Triggered Immunity (PTI) and Effector-Triggered Immunity (ETI) are plant defence mechanisms that respond to microbe invasion. These mechanisms alter the ionic balance, leading to increased cytosolic Ca2+ and apoplastic reactive oxygen species (ROS) levels. Additionally, they activate the mitogen-activated protein kinase (MAPK) pathway and cause the accumulation of nitric oxide (NO). This cascade results in the production and release of phytohormones such as ethylene (ET), jasmonic acid (JA), and salicylic acid (SA). Furthermore, it induces stomata closure and callose deposition, and initiates transcriptional and metabolic reprogramming to facilitate the plant’s defense response (Nguyen et al., 2021).
In the context of fungal endophytes, many plants have chitin-specific receptors (PR-3) on their surface that recognize the chitin oligomers found on the fungal cell wall, resulting in the activation of the plant’s defence mechanism (Sanchez-Vallet et al., 2015). Besides these, plant pattern-triggered immune signaling, such as MIN7 and CAD1, are the important components in controlling the level and nurturing the endophytes present in the nearby soil environment so that host can survive easily in the microorganism rich environment (Chen et al., 2020). In addition to various pathways, plants utilize extracellular vesicles (EVs) to facilitate the transport of signaling lipids, proteins, RNA, and metabolites between cells. Research indicates that these EVs play a role in plant stress response by exhibiting antipathogenic activity. During stress, plant cells secrete EVs containing host-derived small interfering RNAs and microRNAs that are capable of silencing fungal genes and stress responses. This evidence suggests that plant EVs may mediate trans kingdom RNA interference (Rutter and Innes, 2018). Alternately, when fungal endophytes invades the host plant, they start producing chitin deacetylases, that leads to deacetylation of the chitosan oligomers, and these oligomers are not detected by the host plant chitin specific receptors (Cord-Landwehr et al., 2016). The formation of biofilms in endophytic bacteria aids in their ability to adhere to host plant tissues and facilitate communication among themselves, thereby enabling them to evade the host plant’s defence system. Endophytic microorganisms have evolved mechanisms to evade the oxidative stress generated by the plant host in response to pathogenic invasion. The plant’s defence system includes the production of reactive oxygen species (ROS) such as superoxide (O2-), hydrogen peroxide (H2O2), and hydroxyl radicals (OH), which are toxic to invading pathogens. To counteract this, endophytes have developed an array of antioxidative enzymes, such as superoxide dismutases (SODs), catalases (CatAs), peroxidases (PODs), alkyl hydroperoxide reductases (AhpCs), and glutathione-S-transferases (GSTs). These enzymes play a crucial role in the elimination of ROS and the protection of the endophyte from oxidative damage, allowing it to establish and maintain a symbiotic relationship with the host plant (Zeidler et al., 2004). Endophytic microbes possess genes that encode for molecules known as Microbe-Associated Molecular Patterns (MAMPs) inhibitors. MAMPs are recognized by plants and trigger their immune response (Figure 2).
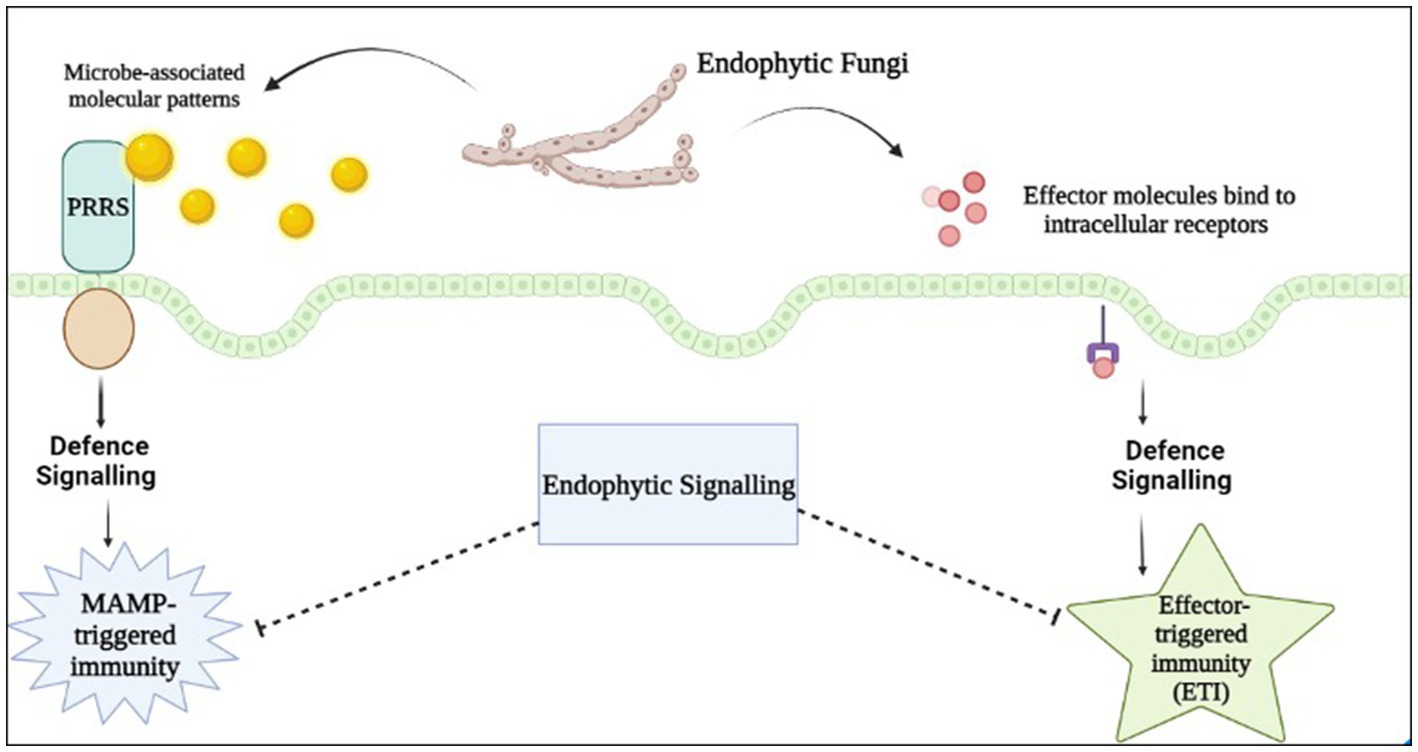
Figure 2. Schematic of plant immune system recognizes fungal signal molecules, resulting in two distinct defence mechanisms.
However, these inhibitors repress the MAMP-triggered immune response of the host plant. Endophytic communities can influence several physiological processes of the host plant, including the activation of silent gene clusters, leading to the synthesis of novel secondary metabolites. These secondary metabolites, which are also produced by the endophytes, play a crucial role in preventing the invasion of pathogenic microorganisms in the host plant, thereby preventing infections. The plant phenotype mostly depend upon the genetic arrangement within itself alongwith the biotic and abiotic stress and microbiome activity. The endophytic fungi stimulates the plant immune system through epigenetic events and hence, induce enhanced production of SMs leading to physiological and phenotypic change (Alam et al., 2021). Clustering of genes responsible for secondary metabolite production help in conserving the genetic arrangement. The encoding genes for these secondary metabolites are regulated by various factors such as horizontal gene transfer (HGT), transcription factors, the presence of effector molecules, and gene clustering.
Gene clustering can be defined as the phenomenon where the genes responsible for coding the biosynthesis of certain secondary metabolites tend to cluster together in close proximity to the telomeric or dynamic regions of the fungal chromosome. These clusters of genes can vary in two significant ways. Firstly, different clusters of genes can work together, resulting in the synthesis of highly complex secondary metabolites. Secondly, in some cases, different pathways for secondary metabolite production have gene clusters located adjacent to each other in the fungal genome (Chiang et al., 2016; Henke et al., 2016; Nützmann et al., 2018). Nevertheless, because they are involved in gene expression control, the mechanism of coding of gene clusters of secondary metabolites in unstable DNA regions remains unexplained (Osbourn, 2010a). The spatial arrangement of clustered genes is believed to be a fundamental necessity for the biosynthesis of bioactive products that are linked to a pathway. This is due to the fact that genes involved in a pathway are generally maintained in close proximity through genomic rearrangements (Deepika et al., 2016). The resulting genetic structure may allow favorable allele combinations to co-inherit at these multigene loci (Chu et al., 2011; Field et al., 2011). This may also regulate chromatin synchronization by rearranging the chromatins (Field and Osbourn, 2008; Osbourn and Field, 2009; Osbourn, 2010b).
Essentially correlated genes interspersed in higher plant genomes can form transcriptional clusters via the helix–loop–helix domain. In this, the formation of DNA loops results in the co-localization of cis-regulatory elements, leading to increased local concentrations of transcription factors at the corresponding gene’s transcription initiation sites, thereby initiating transcription. Any perturbations in the transcriptional regulation of clustered genes could result in the loss of their protein products and the accumulation of potentially harmful intermediates within the associated biochemical pathways. Many researchers showed that chromatin-level gene control is critical for the expression of secondary metabolic gene clusters (Osbourn, 2010a).
In the synthesis of secondary metabolites, clustered genes are regulated by two distinct groups of transcription factors, namely narrow domain transcription factors (NDTFs) and broad domain transcription factors (BDTFs). NDTFs exert their regulatory influence on the clustered genes directly, while BDTFs can act on the clustered genes at different gene locations than those of the NDTFs. This fact is exemplified by the renowned non-ribosomal peptide synthetase (NRPS) AflR, which is classified as a Zn2Cys6 transcription factor. AflR modulates the expression of the aflatoxin and sterigmatocystin gene clusters through its binding to the palindromic sequence 5′-TCG(N5)CGA-3′, an 11-bp motif that is present in the promoter regions of coding genes in selected Aspergillus species (Slot and Rokas, 2011). It also regulates three additional genes that are not associated with the aflatoxin metabolite gene cluster (Woloshuk et al., 1994; Yu et al., 1996; Cánovas et al., 2017). BDTFs, which are transcription factors that regulate the expression of multiple genes, function as high-level regulatory systems that respond to external stimuli that are not directly related to secondary biochemical gene clusters. Recent studies have revealed that ethylene-forming (EF) signals specifically disrupt transcription factors that are targeted by ethylene, indicating that these signals can influence gene expression in a variety of biological processes (Camehl et al., 2010). Thus secondary metabolites are synthesized by an organism through the interplay of developmental competence and environmental factors. These factors include, but are not limited to, nutrient supply, artificial light, pH, injury, infection, and developmental changes that occur during different stages of the host’s life cycle. It is widely accepted that a combination of these factors is essential for the efficient biosynthesis of secondary metabolites (Bayram and Braus, 2012; Xie et al., 2019). BDTFs are essential for transmitting environmental signals to the genome. They create and govern signaling transmission from environmental inputs to cellular responses during the creation of distinct SMs.
Several studies have investigated the molecular mechanisms underlying the relationship between Nucleosome-Dependent Transcription Factors (NDTFs) and Chromatin-Dependent Transcription Factors (BDTFs). These investigations aim to elucidate how BDTFs, or global transcription factors, perceive developmental or environmental cues, and subsequently transmit these signals to NDTFs through various biochemical pathways, such as chromatin and histone modifications, or through specific biochemical cascade reactions like methylation, phosphorylation, and acetylation. These responses are crucial in activating silent genes associated with specific secondary metabolites (SMs), which are required for specific cell metabolisms, growth stages, or environmental conditions. Furthermore, in addition to the plant’s genetic makeup, plant phenotypes are also influenced by the activity of the microbiome and environmental factors. This can lead to genetic changes in the appearance of endophytic fungi associated with the plant (Jia et al., 2016). It has been seen that the presence of EFs helps to improve host plant resistance to biotic and abiotic stresses (Rodriguez et al., 2008; Xie et al., 2019; Yan et al., 2019). While the precise processes are unknown, evidence shows that the presence of EFs alters plant genetic expression patterns (Mejia et al., 2014; Xie et al., 2019). Epigenetic interactions between the host and endophyte can modulate the host’s genomic expression. Endophyte-mediated alterations in DNA methylation and demethylation can enhance the host’s defence mechanisms in epigenetic processes (Geng et al., 2019). EFs strengthen plant immune systems and increase the quantity of SMs, which may cause physiological changes in infected host plants (Xie et al., 2019). Bailey et al. (2006) reported that exposure to EFs leads to changes in gene expression patterns in both the endophytic microorganisms and host organisms. These changes occur due to a complex system of genetic interactions between the EFs and hosts, resulting in altered genomic expressions during the interference process.
Horizontal gene transfer (HGT) is the process by which genetic elements are transferred between isolated lineages without sexual reproduction. This unique evolutionary mechanism is considered a novel adaptive trait of effector proteins, enabling them to invade, degrade, and manipulate host organisms (Soanes and Richards, 2014). HGT can facilitate the acquisition of a new and complete metabolic pathway by transferring the metabolic cluster of an organism to another (Slot and Rokas, 2011). The mechanism of HGT remains to be debated as to how the expression is initiated in the recipient organism. Slot and Rokas (2011) reported the successful functional transfer of a gene cluster responsible for the biosynthesis of sterigmatocystin, a toxic secondary metabolite, from Aspergillus species to Podospora anserina. The presence of certain metabolites, such as djalonensone in various Alternaria fungi, and aureonitol in Chaetomium sp. and extracts of Helichrysum aureonitens, which originated from horizontal gene transfer or genetic recombination during the coevolution of hosts and endophytes, provide evidence for the development of genetic regulatory mechanisms of secondary metabolite biosynthesis (Aly et al., 2010; Kozyrovska, 2013).
Through the application of the aforementioned techniques are applied by human beings for secondary metabolite production, fungal endophytes are capable of synthesizing a diverse range of secondary metabolites, including alkaloids, terpenoids, polyketides, phenylpropanoids, lignins, flavonoids, saponins, phenols and phenolic acids, aliphatic and chlorinated metabolites, peptides, and steroids. These secondary metabolites play a critical role in enabling the host plant to combat a range of stress conditions, such as drought, salinity, nutrient deficiency, metal toxicity, and biotic stress caused by pathogenic microorganisms present in the plant’s immediate environment. The plant’s capacity to endure stressors protects it from harm and prevents destruction. Elicitors or elicitor factors can even induce the synthesis of secondary metabolites with potential applications in various industries, including but not limited to pharmaceuticals, agriculture, and cosmetics. These secondary metabolites may have commercially valuable properties such as antibiotic, anti-carcinogenic, cytotoxic, insecticidal, and allelopathic activities. Some specific examples of important secondary metabolites synthesized by elicitor factors are outlined in the table below (Table 2).
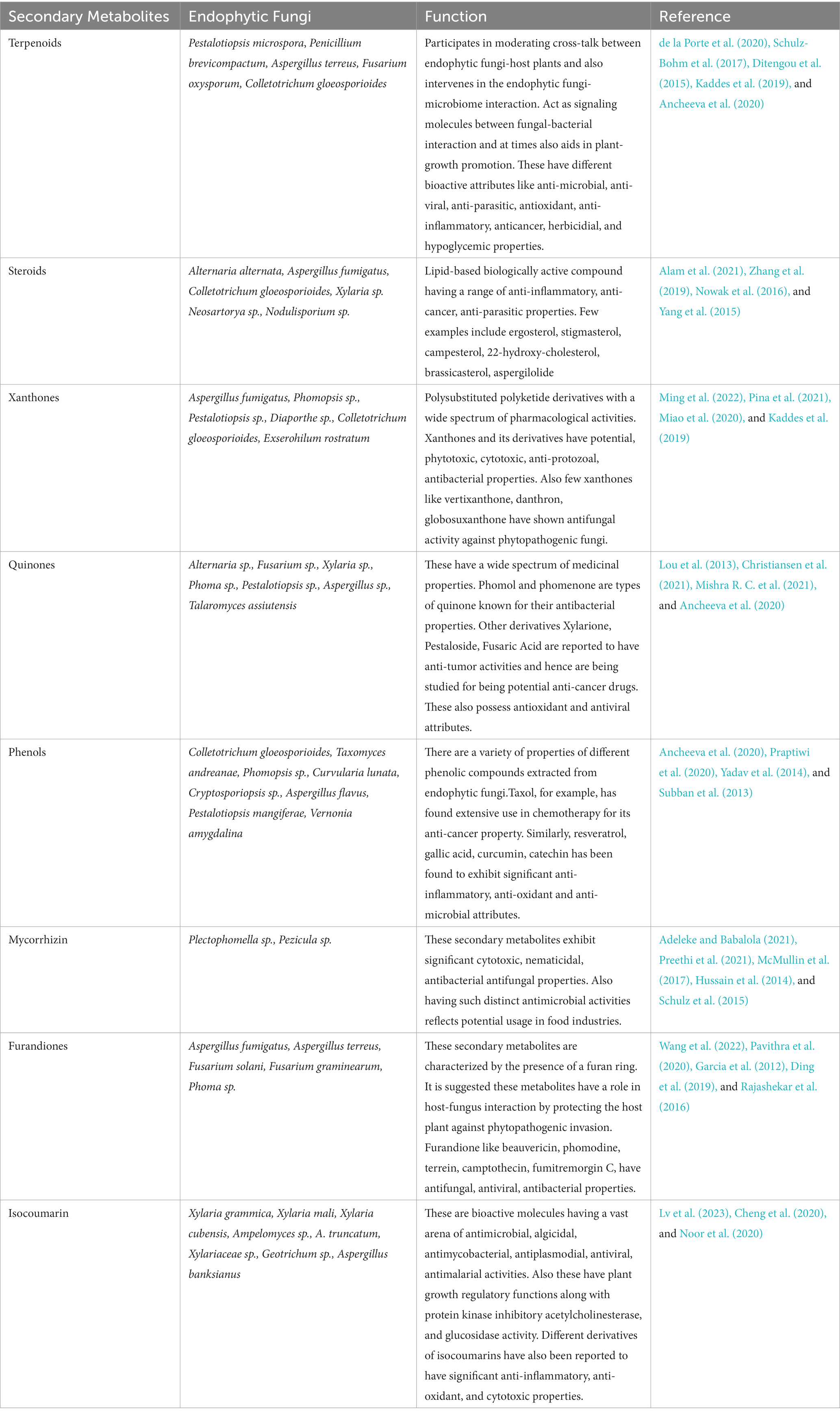
Table 2. Secondary metabolites produced by endophytic fungus with their respective structure and functions.
Endophytic fungi are capable of producing a diverse range of bioactive compounds that exhibit a wide array of biological activities including, but not limited to, insecticidal, antioxidant, antifungal, antiviral, antibacterial, and cytotoxic properties. These compounds belong to various chemical classes such as terpenoids, phenols, alkaloids, polyketides, quinones, steroids, enzymes, and peptides. The secretion of these bioactive compounds by endophytic fungi is known to host plant defence response, enabling it to better cope with both biotic and abiotic stressors. Methods to enhance the potential of endophytic fungi for producing these secondary metabolites include the activation of silent biosynthetic pathways, epigenetic modifications, and other techniques. Among these secondary metabolites, terpenoids have emerged as a particularly important class of molecules that have significant applications in human health and agriculture. Thus, endophytic fungi represent a promising novel source for the bioproduction of these valuable terpenoid compounds. The isoprene units are typically obtained through two pathways, the mevalonic acid pathway (MVA) and the methylerythritol phosphate pathway (MEP). Based on the number of carbon atoms in their skeletal structure [(C5)n], terpenoids can be classified into various subgroups, including hemi terpenoids (C5), monoterpenoids (C10), sesquiterpenoids (C15), diterpenoids (C20), sesterterpenoids (C25), triterpenoids (C30), and tetraterpenoids (C40) (Gershenzon and Dudareva, 2007).
The building blocks of terpenoids, which are a diverse group of natural products, are isopentenyl pyrophosphate and dimethylallyl pyrophosphate. These are synthesized through the mevalonate (MVA) and the methylerythritol phosphate (MEP) pathways, and are interconvertible by the action of isopentenyl pyrophosphate isomerase. Subsequently, prenyltransferases convert these shorter chain precursors to longer chain terpenoid skeletons, such as geranyl diphosphate, farnesyl diphosphate and geranylgeranyl diphosphate, which serve as the C10, C15, and C20 backbones of monoterpenoids, sesquiterpenoids and triterpenoids, diterpenoids and tetraterpenoids, respectively. Further structural diversification of terpenoids can be achieved by the introduction of functional groups, such as glycosyl, hydroxyl, ketone, carbonyl, and aldehyde, or by rearrangement of the carbon skeleton. These modifications can lead to the expression of various bioactivities associated with terpenoids (Sun et al., 2019). Numerous plant species are recognized for their capacity to biosynthesize terpenoids, including citral, menthol, camphor, salvinorin A (in the case of the plant Salvia divinorum), ginkgolide, and bilobalide (in the case of the plant Ginkgo biloba), and cannabinoids (in the case of the cannabis plant). Terpenoids are also commonly synthesized by tea, thyme, Spanish sage, and citrus fruits (such as lemon, orange, and mandarin). These terpenoids exhibit diverse functions in the food and cosmetic industries, as well as various pharmaceutical and nutritional applications. Furthermore, terpenoids have demonstrated good antiviral activity, and have therefore been investigated for their potential clinical use in the treatment of COVID-19. Due to these activities, there is an increase in demand of terpenoids which possess stress on the terpenoids producing plant species. In order to fulfil the increasing demand of terpenoids, other sources which are environmentally friendly and commercially fulfilling need to be discovered. The endophytes have the potential of synthesizing these terpenoids which have gained the attention for using the endophytes for alternate source of terpenoid-producing strains or terpenoid synthetic genes (Chen et al., 2021).
Endophytic fungi and its host plant undergo various complex interactions which enhance the plants growth and nutrition, also helps the plant in combating various biotic and abiotic stress conditions. Endophytes are known to exhibit a high degree of variability that is dependent on several factors such as the genotype of the host plant, growth stage, physiological state, tissue type, soil environment, and various agricultural practices (Gupta et al., 2020). These microorganisms have been shown to not only produce terpenoids themselves, but also stimulate their production in host plants through mechanisms such as horizontal gene transfer, the heterologous expression of terpenoid biosynthetic genes sourced from endophytes, biotransformation of terpenoids, and various signaling pathways that include elicitor recognition, signal transduction, integration with transcription factors, and gene activation (Zhai et al., 2017). Numerous scientific methodologies have been created to advance terpenoid biosynthesis research. These include metabolic engineering and synthetic biology, system biology approaches, modern metagenomic sequencing methods, and the de novo assembly of microbial genomes from metagenome data. These techniques enable the expression of the terpenoid biosynthetic pathway in industrial microbes, increase the potential gene pool involved in terpenoid biosynthesis from various environments, and offer effective strategies for discovering novel microbes and genes related to terpenoid biosynthesis in the endosphere and rhizosphere microbiomes, regardless of their culturability (Carrión et al., 2019; Belcher et al., 2020). Huperzine A (HupA) is a naturally occurring sesquiterpene alkaloid that is synthesized by members of the Huperziaceae family, which includes species such as Huperzia serrata. HupA has been found to possess potent anti-acetylcholinesterase activity, making it an effective treatment option for Alzheimer’s disease (AD). Furthermore, HupA’s ability to effectively inhibit acetylcholinesterase has been associated with minimal side effects, highlighting its potential as a safe and effective treatment option for AD (Zhao et al., 2013). The enzymes lysine decarboxylase and copper amine oxidase catalyze the first two steps of HupA biosynthetic pathway in which L-lysine is converted to 5-aminopentanal which is the precursor of HupA. Researchers successfully isolated two strains of endophytic fungi, EFs Shiraia sp. Slf14 (Zhu et al., 2010) and Cladosporium cladosporioides LF70 (Zhang et al., 2019), from the leaves of Huperzia serrata. These strains were found to produce Huperzine A (HupA) and were able to yield 142.6 μg/g and 39.61 μg/g dry mycelium, respectively. On sequencing the whole genome of Shiraia sp. Slf14, HupA biosynthetic gene cluster was identified, which is then expressed into Escherichia coli, which showed that genetically modified E. coli strain was able to convert cadaverine to 5-aminopentanal (Yang et al., 2016). Afterwards, some other strains of EFs were isolated, such as Colletotrichum gloeosporioides ES026, which were then sequenced and expressed for its yield of 5-aminopentanal which is a precursor of HupA.
Traditionally utilized medicinal plants Nothapodytes nimmoniana and Campotheca acuminata contain campothecin which belongs to the class of monoterpene indole alkaloids and functions as potent topoisomerase inhibitor in DNA replication. Reports from various studies suggest that Entrophospora infrequens and Neurospora crassa isolated from N. nommoniana produce campothecin (Puri et al., 2005; Rehman et al., 2008). Kusari et al. (2009) suggested that Fusarium solani isolated from the barks of C. acuminata are potential source of campothecin production. The endophytic strain of F. solani utilizes enzymes encoded by its gene in presence of strictosidine synthase, one of the majorly important plant enzyme for the campothecin biosynthesis. Another report by Yan et al. (2019) suggested that ginsenosides Rh2 and Rg3 are produced by strain PDA-2 which is closely related to Agrobacterium rhizogenes which was isolated from Panax ginseng. The ginsenosides Rh2 and Rg3 have major roles in inhibiting tumor cell proliferation and induce apoptosis. In a study by Palem et al. (2015), authors have suggested biosynthesis of vincristine and vinblastine by endophytic fungus Talaromyces radicus isolated from various tissues of Catharanthus roseus. The study screened 22 endophytic fungi for presence of gene encoding tryptophan decarboxylase which could only be amplified in T. radicus. The gene tryptophan decarboxylase is one of the most important enzyme terpenoid indole alkaloid biosynthetic pathway. Maximum yield of 670 μg/L and 70 μg/L of vincristine and vinblastine from modified M2 medium and PDB medium was obtained, respectively. Parthasarathy et al. (2020) recorded the vinblastine production Curvularia verruculosa isolated from leaves of C. roseus. Maximum yield of 182 μg/L of vinblastine was obtained from endophytic fungi C. verruculosa.
Stress can be defined as any substance, microorganism or unfavorable condition which directly or indirectly affects the plant’s growth and development, metabolism or nutrition. Stress on vegetation depends on factors such as, temperature, mineral deficiency, long rainy periods, desiccation problems, insects and pathogens, herbicides and pesticides, pollutants, climatic conditions like drought or floods, increased UV radiations and so on. Based on the LICHTENTHALER model of stress in plants (Lichtenthaler, 1998), stress can be divided into several distinct phases. The first phase is the response phase, which is characterized by an alarm reaction and marks the beginning of stress. This is followed by the restitution phase, where the plant enters a stage of resistance and continues to experience stress. The third phase is the terminal phase, where the plant enters a stage of exhaustion after long-term stress. Finally, the regeneration phase allows the plant to recover and rejuvenate after experiencing stress. These phases of stress describe in detail the changes that occur in the plant during the unfavorable or stress conditions. Initially, there is decline in physiological functions which leads to decrease in metabolic activities and decline in growth, development and vitality of the plant. In response, the plant’s defence system gets activated leading to hardening of the plant by increasing the plant’s resistance against stressors by reaching plant resistance maximum. If the stressors are removed before senescence the plant survives via regeneration but if the stressors are not being removed then the plant leads to cell death which ultimately causes plant death (Lichtenthaler, 1998). Plant stressors can be divided into three types, that is, natural or abiotic stressors such as high light, heat, drought, mineral deficiency, low temperature, chilling, wounding, UV-A and UV-B, biotic stressors such as insects, pathogens, elicitors, bacteria, fungi and virus, anthropogenic stressors such as, herbicides, air pollutants, peroxyacyl nitrates, free radicals, acid rain, acid fog and heavy metal load. Plants sense these stressors and act accordingly via activation of signal transduction pathways after signal perception further leading to gene expression or metabolic responses in the plant. Under stress conditions, plants synthesize bioactive molecules such as phytohormones and secondary metabolites which not only have the tendency to overcome the stress conditions but also have antifungal, anticarcinogenic and various other medicinal properties which can be utilized to treat various diseases. EFs have the capability of producing the secondary metabolites (terpenoids, alkaloids, phenols, quinones) that would help in fighting the biotic and abiotic stress in plants as well as can be utilized in pharmaceutical industry due to its medicinal properties (Gershenzon and Dudareva, 2007; Christiansen et al., 2021).
When endophytic fungi interact with their host plant, the plant’s defence system is activated to counteract the presence of the endophytic fungi which are perceived as potential pathogens. However, endophytic fungi are capable of evading these defence mechanisms and colonizing the host plant. Subsequently, endophytic fungi produce bioactive compounds, such as terpenoids, phenols, alkaloids, steroids, quinones, poly-ketones, and peptides, which exhibit inhibitory effects on the growth of pathogens and herbivores invading the host plant in the presence of endophytic fungi (Lu et al., 2021). This phenomenon provides a protective mechanism to the host plant from pathogenic microorganisms. Various terpenoids are synthesized by endophytes which will help host plants to combat biotic and abiotic stresses. Examples of these terpenoids include indole diterpenoids, synthesized in endophytic infected grasses, are neurotoxins, responsible for intoxication, helpful in protecting the grass from cattle grazing (Hardoim et al., 2015) multi-cyclic indolosesquiterpene synthesized by Streptomyces sp. HKI0595 in Kandelia candel has antibacterial activity (Maier et al., 2011), cadinane sesquiterpene derivatives synthesized by Phomopsis cassia in Cassia spectabilis has antifungal activity against phytopatogenic fungi Cladosporium cladosporioids and C. sphaerospermum. Persicaria minor also known as kesum is herbaceous plant widely found in south-east Asia and is mainly important for flavonoid and terpenoid synthesis. Study by Samad et al. (2019) reported that six miRNAs of Persicaria minor post-transcriptionally regulated the terpenoid biosynthesis induced by Fusarium oxysporum.
A large diversity of microorganisms is present in the soil that interacts with the plants growing in that soil. These interactions are known as plant-microbe interaction. These interactions play a very important role in making a sustainable balanced ecosystem. Plants synthesize various organic and inorganic nutrients which makes the soil nutrient enriched that is very beneficial for the microbial consortia growing in that soil. Plants house endophytes which are one of the examples of plant-microbe interaction. Such interactions between plants and various microbes such as bacteria and fungi are beneficial and attract the interest of various researchers due to their potential for combatting biotic and abiotic stresses along with its application in agriculture. Throughout their life cycle, plants engage in the synthesis of two distinct categories of metabolites: primary and secondary metabolites. Primary metabolites, which are essential for the plant’s growth, development, and nourishment, remain constant across all plant species. In contrast, secondary metabolites function as signaling molecules that play a crucial role in signal transduction. These metabolites are typically secreted by plants in response to herbivore, pathogen, or environmental stress but in low quantities. However, research suggests that endophytes, particularly EFs, have the capability to produce these secondary metabolites in large quantities when compared to plants. There are several researchers working for the discovery of these secondary metabolites as these are potential source for depleting the use of chemical pesticides, insecticides or herbicides and these discoveries are categorized as: (a) metabolites that are unknown, (b) metabolites with known function but unknown structure, (c) metabolites with unknown function but known structures (Luo et al., 2022).
Researchers face several difficulties in the discovery of these secondary metabolites which include (1) metabolites are secreted by the plants in very low concentrations, (2) the metabolites that are being secreted and synthesized are being actively metabolized as well, (3) they have diverse physical and chemical properties which require specific assays to determine their functions and structures, (4) these metabolites are secreted at specific stages of the plant’s life which makes it difficult for scientists to discover them (Luo et al., 2022). To surmount the hurdles in the study of plant secondary metabolites, it is imperative to adopt interdisciplinary methods. A multi-omics approach, for instance, has been instrumental in the identification of these metabolites via the methodical comparative analysis of extensive datasets by researchers. In a broader aspect, “omics” are the scientific fields that are related to measuring biological molecules in high throughput methods. This includes various biological fields such as genomics, proteomics, metagenomics, metabolomics, phenomics, epigenomics, transcriptomics and so on. When all these fields are studied in an integrated manner for discovery of some biological molecule, then it is called a multi-omics approach. This review utilizes a multi-omics approach to investigate a diverse range of secondary metabolites synthesized by endophytes. By employing one or multiple omics approaches, a scientific experiment can be designed to align with the function and structure of the metabolite. Each omics approach has its unique characteristics and can complement the limitations of the other omics approach, thereby providing a comprehensive understanding of the endophytic secondary metabolite’s synthesis and its potential applications.
Genomics is the field of biology that deals with the study of the whole genome (including structure, function, evolution, mapping and editing of genome) of the organism. Genome is the whole sequence of the DNA set of the organism. The fundamental genetic material, DNA, contains essential information about the regulation of gene expression, including promoter regions, untranslated regulatory regions, and splicing sites, as well as the protein-coding sequence that determines the function of a given gene within an organism (Chu et al., 2020). The field of genomics is driven by advanced technologies such as high-throughput DNA sequencing techniques, such as Illumina HiSeq, PacBio, and Nanopore sequencing, as well as single nucleotide polymorphism (SNP) chips that enable the identification and analysis of genetic variations among individuals. The whole genome sequencing analysis would help us understand that which genes are responsible for growth and development, defence system, nutrient acquisition, production of secondary metabolites, and various other processes of the organism which we are studying. This approach of whole genome sequencing would help in determining the genes that are directly or indirectly influencing the various bioactivities or metabolic activities of the endophyte or plant which is synthesizing the secondary metabolites.
In the context of the interactions between endophytes and plants, various biotic and abiotic stresses may occur during their interactions leading to the production of secondary metabolites as a defence mechanism. Genomics approaches utilizing high-throughput sequencing techniques offer a powerful tool for identifying the specific genes responsible for the activation of signaling pathways leading to the production of secondary metabolites (Alam et al., 2021). Additionally, such approaches can be used to investigate plant growth promotion, endophytic secretory systems, surface attachment and insertion elements, transport systems, and other related metabolic mechanisms. This detailed knowledge about the genes and its functions would provide us with better understanding about the ecology and evolution of the endophyte and we could accordingly extract the gene responsible for secondary metabolite production and amplify it using PCR for its further utilization. Compounds like alkaloids, flavonoids, minerals, polyphenols and vitamins are mostly synthesized by endophytes have positive influence on adjusting in adverse conditions and hence, promoting plant health (Balestrini et al., 2021). These molecules/compounds can be detected and analyzed employing techniques such as gas chromatography–mass spectroscopy (GC–MS), Fourier transform infrared (FT-IR), nuclear magnetic resonance (NMR) spectroscopy, metabolite fingerprinting, time-of-flight mass spectrometry (TOF-MS), Orbitrap Mass Spectrometer (Orbitrap-MS), flash chromatography. The signaling pathway initiation and colonization factors due to host cell and endophytic interactions can be evaluated on the basis of proteomics studies. However, there are new omics tools available for deciphering the fungal genome and the metabolites produced by them through epigenomics, ionomics, fluxomics, lipidomics, nutrigenomics, toxicogenomics. By integrating these various techniques, the secondary metabolite production can be verified and analyzed at every level (Table 3; Shankar and Sharma, 2022). The host endophyte interaction from recognition phase to stress resistance development can be thoroughly studied via transcriptomic analysis.
During the investigation of the metabolic output of the colocalization phenomenon of prenyltransferase (Polyprenyl synt (PT)) and terpene synthase (Terpene synth C (TPS)) domains in plant genomes, the biosynthesis of sesterterpenes, a rare type of terpenoid, was discovered to occur in both plants and fungi (Huang et al., 2017, 2018). P. indica, has the potential of acting as plant probiotic agent, that has been revealed during its genome sequence analysis (Qiang et al., 2012). EFs of order sebacinales helps the host plant in enhancing its growth, development and stress tolerance potential, is revealed by this approach (Weiss et al., 2011). Several bioinformatic tools, such as plantiSMASH (Kautsar et al., 2017), phyto cluster (Töpfer et al., 2017), and clusterfinder (Schläpfer et al., 2017; Chavali and Rhee, 2018), have been developed to predict plant biosynthetic gene clusters (BGCs) using plant genomic sequences, protein annotations, and gene expression profiles. This facilitates the identification of plant secondary metabolites, which can have important implications in fields such as drug discovery and agriculture.
The development of a sustainable agriculture is of utter importance. Microorganisms like the endophytic fungi represent an intriguing area of study in the field of plant-microbe interactions in this context. The unique ability of these fungi to inhabit and colonize host plant tissues and thereby stimulate the production of wide range of secondary metabolites with diverse biological activities, including antifungal, antibacterial, anticancer, and immunomodulatory properties are of extreme importance. These attributes enhance the plants disease resistance ability, growth and nutrient uptake abilities. Most of these secondary metabolites produced by these fungi are valuable sources of antimicrobial, antitumor, and antiviral agents, among other bioactive compounds, making them a vital component in the development of new drugs. Multi-omics technologies can further help in deciphering the physiological development of endophytes in host plant. Concomitantly, generation of more high-throughput data will provide updated information on the core areas of study, thus discovering unknown genes, metabolites, and microbial species for better harnessing of beneficial aspects from endophytes. Thus, in conclusion it can be well understood that the overall, investigation of endophytic fungi and their metabolic products has gained significant attention in recent years, and this approach holds great promise for sustainable agriculture. With the increasing demand for eco-friendly and non-toxic agricultural practices, endophytic fungi present an attractive alternative to chemical pesticides and fertilizers. Further studies on these fungi will not only provide a better understanding of their interactions with host plants but also contribute to the development of sustainable agricultural practices and the discovery of novel bioactive compounds for various applications.
PJ and TM: conceptualization, editing, and supervision. TK and IC: literature review and drafting of original manuscript. AP, SP, and VK: writing and editing. All the authors have read and approved the final version of the manuscript.
IC was employed by Metropolis Healthcare Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abreu-Tarazi, M. F., Navarrete, A. A., Andreote, F. D., Almeida, C. V., Tsai, S. M., and Almeida, M. (2010). Endophytic bacteria in long-term in vitro cultivated “axenic” pineapple microplants revealed by PCR–DGGE. World J. Microb. Biot. 26, 555–560. doi: 10.1007/s11274-009-0191-3
Adeleke, B. S., and Babalola, O. O. (2021). Pharmacological potential of fungal endophytes associated with medicinal plants: a review. J. Fungi. 7:147. doi: 10.3390/jof7020147
Ahmad, I., del Mar Jiménez-Gasco, M., Luthe, D. S., Shakeel, S. N., and Barbercheck, M. E. (2020). Endophytic Metarhizium robertsii promotes maize growth, suppresses insect growth, and alters plant defense gene expression. Biol. Control 144:104167. doi: 10.1016/j.biocontrol.2019.104167
Alam, B., Liˇ, J., Gě, Q., Khan, M. A., Gōng, J., Mehmood, S., et al. (2021). Endophytic fungi: from symbiosis to secondary metabolite communications or vice versa? Front. Plant Sci. 12:791033. doi: 10.3389/fpls.2021.791033
Aly, A. H., Debbab, A., Kjer, J., and Proksch, P. (2010). Fungal endophytes from higher plants: a prolific source of phytochemicals and other bioactive natural products. Fungal Divers. 41, 1–16. doi: 10.1007/s13225-010-0034-4
Ancheeva, E., Daletos, G., and Proksch, P. (2020). Bioactive secondary metabolites from endophytic fungi. Curr. Med. Chem. 27, 1836–1854. doi: 10.2174/0929867326666190916144709
Avinash, K. S., Ashwini, H. S., Babu, H. N., and Krishnamurthy, Y. L. (2015). Antimicrobial potential of crude extract of Curvularia lunata, an endophytic fungi isolated from Cymbopogon caesius. J. Mycol. 2015:185821. doi: 10.1155/2015/185821
Bagy, H. M. K., Hassan, E. A., Nafady, N. A., and Dawood, M. F. (2019). Efficacy of arbuscular mycorrhizal fungi and endophytic strain Epicoccum nigrum ASU11 as biocontrol agents against blackleg disease of potato caused by bacterial strain Pectobacterium carotovora subsp. atrosepticum PHY7. Biol. Control 134, 103–113. doi: 10.1016/j.biocontrol.2019.03.005
Bailey, B. A., Bae, H., Strem, M. D., Roberts, D. P., Thomas, S. E., Crozier, J., et al. (2006). Fungal and plant gene expression during the colonization of cacao seedlings by endophytic isolates of four Trichoderma species. Planta 224, 1449–1464. doi: 10.1007/s00425-006-0314-0
Bajaj, R., Huang, Y., Gebrechristos, S., Mikolajczyk, B., Brown, H., Prasad, R., et al. (2018). Transcriptional responses of soybean roots to colonization with the root endophytic fungus Piriformospora indica reveals altered phenylpropanoid and secondary metabolism. Sci. Rep. 8:10227. doi: 10.1038/s41598-018-26809-3
Balestrini, R., Brunetti, C., Cammareri, M., Caretto, S., Cavallaro, V., Cominelli, E., et al. (2021). Strategies to modulate specialized metabolism in mediterranean crops: from molecular aspects to field. Int. J. Mol. Sci. 22:2887. doi: 10.3390/ijms22062887
Bayram, Ö., and Braus, G. H. (2012). Coordination of secondary metabolism and development in fungi: the velvet family of regulatory proteins. FEMS Microbiol. Rev. 36, 1–24. doi: 10.1111/j.1574-6976.2011.00285.x
Belcher, M. S., Mahinthakumar, J., and Keasling, J. D. (2020). New frontiers: harnessing pivotal advances in microbial engineering for the biosynthesis of plant-derived terpenoids. Curr. Opin. Biotechnol. 65, 88–93. doi: 10.1016/j.copbio.2020.02.001
Bilal, S., Shahzad, R., Imran, M., Jan, R., Kim, K. M., and Lee, I. J. (2020). Synergistic association of endophytic fungi enhances Glycine max L. resilience to combined abiotic stresses: heavy metals, high temperature and drought stress. Ind. Crop. Prod. 143:111931. doi: 10.1016/j.indcrop.2019.111931
Brown, K. B., Hyde, K. D., and Guets, D. I. (1998). Preliminary studies on endophytic fungal communities of Musa acuminata species complex in Hong Kong and Australia. Fungal Divers. 1, 27–51.
Camehl, I., Sherameti, I., Venus, Y., Bethke, G., Varma, A., Lee, J., et al. (2010). Ethylene signalling and ethylene-targeted transcription factors are required to balance beneficial and nonbeneficial traits in the symbiosis between the endophytic fungus Piriformospora indica and Arabidopsis thaliana. New Phytol. 185, 1062–1073. doi: 10.1111/j.1469-8137.2009.03149.x
Cánovas, D., Studt, L., Marcos, A. T., and Strauss, J. (2017). High-throughput format for the phenotyping of fungi on solid substrates. Sci. Rep. 7:4289. doi: 10.1038/s41598-017-03598-9
Carrión, V. J., Perez-Jaramillo, J., Cordovez, V., Tracanna, V., De Hollander, M., Ruiz-Buck, D., et al. (2019). Pathogen-induced activation of disease-suppressive functions in the endophytic root microbiome. Science 366, 606–612. doi: 10.1126/science.aaw9285
Carroll, G. (1988). Fungal endophytes in stems and leaves: from latent pathogen to mutualistic symbiont. Ecology 69, 2–9. doi: 10.2307/1943154
Chavali, A. K., and Rhee, S. Y. (2018). Bioinformatics tools for the identification of gene clusters that biosynthesize specialized metabolites. Brief. Bioinform. 19, 1022–1034. doi: 10.1093/bib/bbx020
Chen, Y., Hu, B., Xing, J., and Li, C. (2021). Endophytes: the novel sources for plant terpenoid biosynthesis. Appl. Microbiol. Biotechnol. 105, 4501–4513. doi: 10.1007/s00253-021-11350-7
Chen, T., Nomura, K., Wang, X., Sohrabi, R., Xu, J., Yao, L., et al. (2020). A plant genetic network for preventing dysbiosis in the phyllosphere. Nature 580, 653–657. doi: 10.1038/s41586-020-2185-0
Chen, X., Sun, M., Chong, S., Si, J., and Wu, L. (2022). Transcriptomic and metabolomic approaches deepen our knowledge of plant–endophyte interactions. Front. Plant Sci. 12:700200. doi: 10.3389/fpls.2021.700200
Cheng, M. J., Wu, M. D., Aung, T., Chang, C. T., Hsieh, S. Y., and Chen, J. J. (2020). A new isocoumarin from the fungus Xylaria Mali. Chem. Nat. Compd. 56, 221–223. doi: 10.1007/s10600-020-02992-6
Cheng, X. F., Xie, M. M., Li, Y., Liu, B. Y., Liu, C. Y., Wu, Q. S., et al. (2022). Effects of field inoculation with arbuscular mycorrhizal fungi and endophytic fungi on fruit quality and soil properties of Newhall navel orange. Appl. Soil Ecol. 170:104308. doi: 10.1016/j.apsoil.2021.104308
Chiang, Y. M., Ahuja, M., Oakley, C. E., Entwistle, R., Asokan, A., Zutz, C., et al. (2016). Development of genetic dereplication strains in aspergillus nidulans results in the discovery of aspercryptin. Angew. Chem. 55, 1662–1665. doi: 10.1002/anie.201507097
Chow, Y. Y., and Ting, A. S. Y. (2019). Influence of fungal infection on plant tissues: FTIR detects compositional changes to plant cell walls. Fungal Ecol. 37, 38–47. doi: 10.1016/j.funeco.2018.10.004
Christiansen, J. V., Isbrandt, T., Petersen, C., Sondergaard, T. E., Nielsen, M. R., Pedersen, T. B., et al. (2021). Fungal quinones: diversity, producers, and applications of quinones from aspergillus, Penicillium, Talaromyces, fusarium, and Arthrinium. Appl. Microbiol. Biotechnol. 105, 8157–8193. doi: 10.1007/s00253-021-11597-0
Chu, L., Huang, J., Muhammad, M., Deng, Z., and Gao, J. (2020). Genome mining as a biotechnological tool for the discovery of novel marine natural products. Crit. Rev. Biotechnol. 40, 571–589. doi: 10.1080/07388551.2020.1751056
Chu, H. Y., Wegel, E., and Osbourn, A. (2011). From hormones to secondary metabolism: the emergence of metabolic gene clusters in plants. Plant J. 66, 66–79. doi: 10.1111/j.1365-313X.2011.04503.x
Colla, G., Rouphael, Y., Di Mattia, E., El-Nakhel, C., and Cardarelli, M. (2015). Co-inoculation of Glomus intraradices and Trichoderma atroviride acts as a biostimulant to promote growth, yield and nutrient uptake of vegetable crops. J. Sci. Food Agric. 95, 1706–1715. doi: 10.1002/jsfa.6875
Contreras-Cornejo, H. A., Macías-Rodríguez, L., Del-Val, E., and Larsen, J. (2018). The root endophytic fungus Trichoderma atroviride induces foliar herbivory resistance in maize plants. Appl. Soil Ecol. 124, 45–53. doi: 10.1016/j.apsoil.2017.10.004
Cord-Landwehr, S., Melcher, R. L. J., Kolkenbrock, S., and Moerschbacher, B. M. (2016). A chitin deacetylase from the endophytic fungus Pestalotiopsis sp. efficiently inactivates the elicitor activity of chitin oligomers in rice cells. Sci. Rep. 6:38018. doi: 10.1038/srep38018
Cui, R., Lu, X., Chen, X., Malik, W. A., Wang, D., Wang, J., et al. (2021). A novel raffinose biological pathway is observed by symbionts of cotton Verticillium dahliae to improve salt tolerance genetically on cotton. J. Agron. Crop Sci. 207, 956–969. doi: 10.1111/jac.12556
de la Porte, A., Schmidt, R., Yergeau, É., and Constant, P. (2020). A gaseous milieu: extending the boundaries of the rhizosphere. Trends Microbiol. 28, 536–542. doi: 10.1016/j.tim.2020.02.016
Deepika, V. B., Murali, T. S., and Satyamoorthy, K. (2016). Modulation of genetic clusters for synthesis of bioactive molecules in fungal endophytes: a review. Microbiol. Res. 182, 125–140. doi: 10.1016/j.micres.2015.10.009
Deja-Sikora, E., Kowalczyk, A., Trejgell, A., Szmidt-Jaworska, A., Baum, C., Mercy, L., et al. (2020). Arbuscular mycorrhiza changes the impact of potato virus Y on growth and stress tolerance of Solanum tuberosum L. in vitro. Front. Microbiol. 10:2971. doi: 10.3389/fmicb.2019.02971
Ding, L., Xu, P., Li, T., Liao, X., He, S., and Xu, S. (2019). Asperfurandiones a and B, two antifungal furandione analogs from a marine-derived fungus aspergillus versicolor. Nat. Prod. Res. 33, 3404–3408. doi: 10.1080/14786419.2018.1480622
Diniz, L. R. L., Perez-Castillo, Y., Elshabrawy, H. A., Filho, C. D. S. M. B., and de Sousa, D. P. (2021). Bioactive terpenes and their derivatives as potential SARS-CoV-2 proteases inhibitors from molecular modeling studies. Biomol. Ther. 11:74. doi: 10.3390/biom11010074
Ditengou, F. A., Müller, A., Rosenkranz, M., Felten, J., Lasok, H., Van Doorn, M. M., et al. (2015). Volatile signalling by sesquiterpenes from ectomycorrhizal fungi reprogrammes root architecture. Nat. Commun. 6:6279. doi: 10.1038/ncomms7279
El Enshasy, H. A., Hanapi, S. Z., Malek, R. A., Abdelgalil, S. A., and Leng, O. M. (2019) in Endophytic fungi: The desired biostimulants for essential oil production in advances in endophytic fungal research: Present status and future challenges. ed. B. P. Singh (Cham: Springer International Publishing), 211–232.
Farhat, H., Urooj, F., Irfan, M., Sohail, N., Majeed, S., Ullah, S., et al. (2023). Biological control potential of endophytic fungi with amelioration of systemic resistance in sunflower and GC–MS metabolic profiling of Talaromyces assiutensis. Curr. Microbiol. 80:61. doi: 10.1007/s00284-022-03161-4
Field, B., Fiston-Lavier, A. S., Kemen, A., Geisler, K., Quesneville, H., and Osbourn, A. E. (2011). Formation of plant metabolic gene clusters within dynamic chromosomal regions. Proc. Natl. Acad. Sci. U. S. A. 108, 16116–16121. doi: 10.1073/pnas.1109273108
Field, B., and Osbourn, A. E. (2008). Metabolic diversification–independent assembly of operon-like gene clusters in different plants. Science 320, 543–547. doi: 10.1126/science.1154990
Garcia, A., Rhoden, S. A., Bernardi-Wenzel, J., Orlandelli, R. C., Azevedo, J. L., and Pamphile, J. A. (2012). Antimicrobial activity of crude extracts of endophytic fungi isolated from medicinal plant Sapindus saponaria L. J. Appl. Pharm. Sci. 2, 035–040. doi: 10.7324/JAPS.2012.21007
Geng, S., Kong, X., Song, G., Jia, M., Guan, J., Wang, F., et al. (2019). DNA methylation dynamics during the interaction of wheat progenitor Aegilops tauschii with the obligate biotrophic fungus Blumeria graminis f. sp. tritici. New Phytol. 221, 1023–1035. doi: 10.1111/nph.15432
Gershenzon, J., and Dudareva, N. (2007). The function of terpene natural products in the natural world. Nat. Chem. Biol. 3, 408–414. doi: 10.1038/nchembio.2007.5
Gorzynska, K., Slachetka, M., Ryszka, P., Turnau, K., Plachno, B. J., and Lembicz, M. (2018). Incidence, identification, and mycoparasitic activity of Clonostachys epichloë, a hyperparasite of the fungal endophyte Epichloëtyphina. Plant Dis. 102, 1973–1980. doi: 10.1094/PDIS-02-18-0320-RE
Gupta, S., Chaturvedi, P., Kulkarni, M. G., and Van Staden, J. (2020). A critical review on exploiting the pharmaceutical potential of plant endophytic fungi. Biotechnol. Adv. 39:107462. doi: 10.1016/j.biotechadv.2019.107462
Hamayun, M., Hussain, A., Khan, S. A., Kim, H. Y., Khan, A. L., Waqas, M., et al. (2017). Gibberellins producing endophytic fungus Porostereum spadiceum AGH786 rescues growth of salt affected soybean. Front. Microbiol. 8:686. doi: 10.3389/fmicb.2017.00686
Hardoim, P. R., Van Overbeek, L. S., Berg, G., Pirttilä, A. M., Compant, S., Campisano, A., et al. (2015). The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 79, 293–320. doi: 10.1128/MMBR.00050-14
Hardoim, P., Van-Overbeek, L., and Van-Elsas, J. (2008). Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol. 16, 463–471. doi: 10.1016/j.tim.2008.07.008
Henke, M. T., Soukup, A. A., Goering, A. W., McClure, R. A., Thomson, R. J., and Keller, N. P. (2016). New aspercryptins, lipopeptide natural products, revealed by HDAC inhibition in aspergillus nidulans. ACS Chem. Biol. 11, 2117–2123. doi: 10.1021/acschembio.6b00398
Huang, A. C., Hong, Y. J., Bond, A. D., Tantillo, D. J., and Osbourn, A. (2018). Diverged plant terpene synthases reroute the carbocation cyclization path towards the formation of unprecedented 6/11/5 and 6/6/7/5 sesterterpene scaffolds. Angew. Chem. Int. Ed. 57, 1291–1295. doi: 10.1002/anie.201711444
Huang, A. C., Kautsar, S. A., Hong, Y. J., Medema, M. H., Bond, A. D., Tantillo, D. J., et al. (2017). Unearthing a sesterterpene biosynthetic repertoire in the Brassicaceae through genome mining reveals convergent evolution. Proc. Natl. Acad. Sci. U. S. A. 114, E6005–E6014. doi: 10.1073/pnas.1705567114
Hussain, H., Kliche-Spory, C., Al-Harrasi, A., Al-Rawahi, A., Abbas, G., Green, I. R., et al. (2014). Antimicrobial constituents from three endophytic fungi. Asian Pac J Trop Med 7, S224–S227. doi: 10.1016/S1995-7645(14)60236-4
Ismail, A. H., Mehmood, A., Qadir, M., Husna, A. I., Hamayun, M., and Khan, N. (2020). Thermal stress alleviating potential of endophytic fungus Rhizopus oryzae inoculated to sunflower (Helianthus annuus L.) and soybean (Glycine max L.). Pak. J. Bot. 52, 1857–1865. doi: 10.30848/PJB2020-5(10)
Jia, M., Chen, L., Xin, H. L., Zheng, C. J., Rahman, K., Han, T., et al. (2016). A friendly relationship between endophytic fungi and medicinal plants: a systematic review. Front. Microbiol. 7:906. doi: 10.3389/fmicb.2016.00906
Kaddes, A., Fauconnier, M. L., Sassi, K., Nasraoui, B., and Jijakli, M. H. (2019). Endophytic fungal volatile compounds as solution for sustainable agriculture. Molecules 24:1065. doi: 10.3390/molecules24061065
Kanchan, V., Kumar, N., Shandilya, C., Mohapatra, S., Bhayana, S., and Varma, A. (2020). Revisiting plant-microbe interactions and microbial consortia application for enhancing sustainable agriculture: a review. Front. Microbiol. 11:560406. doi: 10.3389/fmicb.2020.560406
Kautsar, S. A., Suarez Duran, H. G., Blin, K., Osbourn, A., and Medema, M. H. (2017). plantiSMASH: automated identification, annotation and expression analysis of plant biosynthetic gene clusters. Nucleic Acids Res. 45, W55–W63. doi: 10.1093/nar/gkx305
Khan, A. L., Hamayun, M., Khan, S. A., Kang, S.-M., Shinwari, Z.-K., and Kamran, M. (2012). Pure culture of Metarhizium anisopliae LHL07 reprograms soybean to higher growth and mitigates salt stress. World J. Microbiol. Biotechnol. 28, 1483–1494. doi: 10.1007/s11274-011-0950-9
Kisaakye, J., Fourie, H., Coyne, D., Cortada, L., Khamis, F. M., Subramanian, S., et al. (2023). Endophytic fungi improve management of the burrowing nematode in banana (Musa spp.) through enhanced expression of defence-related genes. Nematology 25, 427–442. doi: 10.1163/15685411-bja10229
Koza, N. A., Adedayo, A. A., Babalola, O. O., and Kappo, A. P. (2022). Microorganisms in plant growth and development: roles in abiotic stress tolerance and secondary metabolites secretion. Microorganisms 10:1528. doi: 10.3390/microorganisms10081528
Kozyrovska, N. O. (2013). Crosstalk between endophytes and a plant host within information-processing networks. Biopolym. Cell. 29, 234–243. doi: 10.7124/bc.00081D
Kusari, S., Zühlke, S., and Spiteller, M. (2009). An endophytic fungus from Camptotheca acuminata that produces camptothecin and analogues. J. Nat. Prod. 72, 2–7. doi: 10.1021/np800455b
Li, D., Bodjrenou, D. M., Zhang, S., Wang, B., Pan, H., Yeh, K. W., et al. (2021). The endophytic fungus Piriformospora indica reprograms banana to cold resistance. Int. J. Mol. Sci. 22:4973. doi: 10.3390/ijms22094973
Li, R., Duan, W., Ran, Z., Chen, X., Yu, H., Fang, L., et al. (2023). Diversity and correlation analysis of endophytes and metabolites of Panax quinquefolius L. in various tissues. BMC Plant Biol. 23:275. doi: 10.1186/s12870-023-04282-z
Lichtenthaler, H. K. (1998). The stress concept in plants: an introduction. Ann. N. Y. Acad. Sci. 851, 187–198. doi: 10.1111/j.1749-6632.1998.tb08993.x
Liu, X., Du, Y., Na, X., Wang, M., Qu, Y., Ge, L., et al. (2023). Integrative transcriptome and metabolome revealed the molecular mechanism of Bacillus megaterium BT22-mediated growth promotion in Arabidopsis thaliana. J. Plant Physiol. 285:153995. doi: 10.1016/j.jplph.2023.153995
Lopez, D. C., and Sword, G. A. (2015). The endophytic fungal entomopathogens Beauveria bassiana and Purpureocillium lilacinum enhance the growth of cultivated cotton (Gossypium hirsutum) and negatively affect survival of the cotton bollworm (Helicoverpa zea). Biol. Control 89, 53–60. doi: 10.1016/j.biocontrol.2015.03.010
Lou, J., Fu, L., Peng, Y., and Zhou, L. (2013). Metabolites from Alternaria fungi and their bioactivities. Molecules 18, 5891–5935. doi: 10.3390/molecules18055891
Lu, C., Liu, H., Jiang, D., Wang, L., Jiang, Y., Tang, S., et al. (2019). Paecilomyces variotii extracts (ZNC) enhance plant immunity and promote plant growth. Plant Soil 441, 383–397. doi: 10.1007/s11104-019-04130-w
Lu, H., Wei, T., Lou, H., Shu, X., and Chen, Q. (2021). A critical review on communication mechanism within plant-endophytic fungi interactions to cope with biotic and abiotic stresses. J. Fungi. 7:719. doi: 10.3390/jof7090719
Luo, F., Yu, Z., Zhou, Q., and Huang, A. (2022). Multi-omics-based discovery of plant signaling molecules. Meta 12:76. doi: 10.3390/metabo12010076
Lv, L. X., Mo, T. X., Liang, M., Huang, L. L., Li, B. C., Qin, X. Y., et al. (2023). Secondary metabolites from the endophytic fungus Xylaria grammica and their anti-inflammatory activities. Chem. Nat. Compd. 59, 154–156. doi: 10.1007/s10600-023-03940-w
Maier, A., Fiebig, H. H., Lin, W. H., Peschel, G., and Hertweck, C. (2011). Kandenols A-E, eudesmenes from an endophytic Streptomyces sp. of the mangrove tree Kandelia candel. J. Nat. Prod. 75, 2223–2227. doi: 10.1021/np300387n
Malinich, E. A., Wang, K., Mukherjee, P. K., Kolomiets, M., and Kenerley, C. M. (2019). Differential expression analysis of Trichoderma virens RNA reveals a dynamic transcriptome during colonization of Zea mays roots. BMC Genomics 20:289. doi: 10.1186/s12864-019-5651-z
McMullin, D. R., Green, B. D., Prince, N. C., Tanney, J. B., and Miller, J. D. (2017). Natural products of Picea endophytes from the Acadian forest. J. Nat. Prod. 80, 1475–1483. doi: 10.1021/acs.jnatprod.6b01157
Mehmood, A., Hussain, A., Irshad, M., Hamayun, M., Iqbal, A., and Khan, N. (2019). In vitro production of IAA by endophytic fungus aspergillus awamori and its growth promoting activities in Zea mays. Symbiosis 77, 225–235. doi: 10.1007/s13199-018-0583-y
Mehta, P., Sharma, R., Putatunda, C., and Walia, A. (2019). “Endophytic fungi: role in phosphate solubilization” in Advances in endophytic fungal research: Present status and future challenges. ed. B. P. Singh (Cham: Springer International Publishing), 183–209.
Mejia, L. C., Herre, E. A., Sparks, J. P., Winter, K., Garcia, M. N., and Van Bael, S. A. (2014). Pervasive effects of a dominant foliar endophytic fungus on host genetic and phenotypic expression in a tropical tree. Front. Microbiol. 5:479. doi: 10.3389/fmicb.2014.00479
Miao, C., Wang, J., Huang, R., Liu, S., Zheng, K., Liu, C., et al. (2020). Antifungal xanthones produced by the endophytic fungus Paraconionthyrium sp. YM 311593. Folia Microbiol. 65, 567–572. doi: 10.1007/s12223-019-00762-8
Ming, Q., Li, Y., Jiang, X., Huang, X., He, Y., Qin, L., et al. (2022). Xanthones and benzophenones isolated from the endophytic fungus Penicillium sp. ct-28 of Corydlis tomentella and their cytotoxic activity. Fitoterapia 157:105127. doi: 10.1016/j.fitote.2022.105127
Mishra, R. C., Kalra, R., Dilawari, R., Deshmukh, S. K., Barrow, C. J., and Goel, M. (2021). Characterization of an endophytic strain Talaromyces assiutensis, CPEF04 with evaluation of production medium for extracellular red pigments having antimicrobial and anticancer properties. Front. Microbiol. 12:665702. doi: 10.3389/fmicb.2021.665702
Mishra, Y., Sharma, L., Dhiman, M., and Sharma, M. M. (2021). “Endophytic fungal diversity of selected medicinal plants and their bio-potential applications” in Fungi bio-prospects in sustainable agriculture, environment and Nano-Technology, chap. 10. eds. V. K. Sharma, M. P. Shah, S. Parmar, and A. Kumar (Cambridge, MA: Academic Press), 227–283.
Nair, D. N., and Padmavathy, S. (2014). Impact of endophytic microorganisms on plants, environment and humans. Sci. World J. 2014:250693. doi: 10.1155/2014/250693
Nguyen, Q. M., Iswanto, A., Son, G. H., and Kim, S. H. (2021). Recent advances in effector-triggered immunity in plants: new pieces in the puzzle create a different paradigm Int. J. Mol. Sci. 22:4709. doi: 10.3390/ijms22094709
Nicoletti, R. (2019). Endophytic fungi of citrus plants. Agriculture 9:247. doi: 10.3390/agriculture9120247
Noor, A. O., Almasri, D. M., Bagalagel, A. A., Abdallah, H. M., Mohamed, S. G. A., Mohamed, G. A., et al. (2020). Naturally occurring isocoumarins derivatives from endophytic fungi: sources, isolation, structural characterization, biosynthesis, and biological activities. Molecules 25:395. doi: 10.3390/molecules25020395
Nowak, R., Drozd, M., Mendyk, E., Lemieszek, M., Krakowiak, O., Kisiel, W., et al. (2016). A new method for the isolation of ergosterol and peroxyergosterol as active compounds of Hygrophoropsis aurantiaca and in vitro antiproliferative activity of isolated ergosterol peroxide. Molecules 21:946. doi: 10.3390/molecules21070946
Nützmann, H. W., Scazzocchio, C., and Osbourn, A. (2018). Metabolic gene clusters in eukaryotes. Annu. Rev. Genet. 52, 159–183. doi: 10.1146/annurev-genet-120417-031237
Osbourn, A. (2010a). Gene clusters for secondary metabolic pathways: an emerging theme in plant biology. Plant Physiol. 154, 531–535. doi: 10.1104/pp.110.161315
Osbourn, A. (2010b). Secondary metabolic gene clusters: evolutionary toolkits for chemical innovation. Trends Genet. 26, 449–457. doi: 10.1016/j.tig.2010.07.001
Osbourn, A. E., and Field, B. (2009). Operons. Cell. Mol. Life Sci. 66, 3755–3775. doi: 10.1007/s00018-009-0114-3
Palem, P. P. C., Kuriakose, G. C., and Jayabaskaran, C. (2015). An endophytic fungus, Talaromyces radicus, isolated from Catharanthus roseus, produces vincristine and vinblastine, which induce apoptotic cell death. PLoS One 10:e0144476. doi: 10.1371/journal.pone.0144476
Pappas, M. L., Liapoura, M., Papantoniou, D., Avramidou, M., Kavroulakis, N., Weinhold, A., et al. (2018). The beneficial endophytic fungus fusarium solani strain K alters tomato responses against spider mites to the benefit of the plant. Front. Plant Sci. 9:1603. doi: 10.3389/fpls.2018.01603
Parthasarathy, R., Shanmuganathan, R., and Pugazhendhi, A. (2020). Vinblastine production by the endophytic fungus Curvularia verruculosa from the leaves of Catharanthus roseus and its in vitro cytotoxicity against HeLa cell line. Anal. Biochem. 593:113530. doi: 10.1016/j.ab.2019.113530
Pavithra, G., Bindal, S., Rana, M., and Srivastava, S. (2020). Role of endophytic microbes against plant pathogens: a review. Asian J. Plant Sci. 19, 54–62. doi: 10.3923/ajps.2020.54.62
Perveen, S. (2018). Introductory chapter: terpenes and Terpenoids. IntechOpen. doi: 10.5772/intechopen.79683
Photita, W., Lumyong, S., Lumyong, P., McKenzie, E. H. C., and Hyde, K. D. (2004). Are some fungi isolated as endophytes of Musa acuminata latent pathogens? Fungal Divers. 16, 131–140.
Pina, J. R. S., Silva-Silva, J. V., Carvalho, J. M., Bitencourt, H. R., Watanabe, L. A., Fernandes, J. M. P., et al. (2021). Antiprotozoal and antibacterial activity of ravenelin, a xanthone isolated from the endophytic fungus Exserohilum rostratum. Molecules 26:3339. doi: 10.3390/molecules26113339
Plett, J. M., and Martin, F. M. (2018). Know your enemy, embrace your friend: using omics to understand how plants respond differently to pathogenic and mutualistic microorganisms. Plant J. 93, 729–746. doi: 10.1111/tpj.13802
Polito, G., Semenzato, G., Del Duca, S., Castronovo, L. M., Vassallo, A., Chioccioli, S., et al. (2022). Endophytic bacteria and essential oil from Origanum vulgare ssp. vulgare share some VOCs with an antibacterial activity. Microorganisms. 10:1424. doi: 10.3390/microorganisms10071424
Potshangbam, M., Devi, S. I., Sahoo, D., and Strobel, G. A. (2017). Functional characterization of endophytic fungal community associated with Oryza sativa L. and Zea mays L. Front. Microbiol. 8:325. doi: 10.3389/fmicb.2017.00325
Poveda, J., Abril-Urias, P., and Escobar, C. (2020a). Biological control of plant-parasitic nematodes by filamentous fungi inducers of resistance: Trichoderma, mycorrhizal and endophytic fungi. Front. Microbiol. 11:992. doi: 10.3389/fmicb.2020.00992
Poveda, J., and Baptista, P. (2021). Filamentous fungi as biocontrol agents in olive (Olea europaea L.) diseases: mycorrhizal and endophytic fungi. Crop Prot. 146:105672. doi: 10.1016/j.cropro.2021.105672
Poveda, J., Eugui, D., Abril-Urías, P., and Velasco, P. (2021). Endophytic fungi as direct plant growth promoters for sustainable agricultural production. Symbiosis 85, 1–19. doi: 10.1007/s13199-021-00789-x
Poveda, J., Zabalgogeazcoa, I., Soengas, P., Rodríguez, V. M., Cartea, M. E., Abilleira, R., et al. (2020b). Brassica oleracea var. acephala (kale) improvement by biological activity of root endophytic fungi. Sci. Rep. 10:20224. doi: 10.1038/s41598-020-77215-7
Praptiwi, P., Fathoni, A., and Ilyas, M. (2020). Diversity of endophytic fungi from Vernonia amygdalina, their phenolic and flavonoid contents and bioactivities. Biodiversitas J. Biol. Divers. 21:202. doi: 10.13057/biodiv/d210202
Preethi, K., Manon Mani, V., and Lavanya, N. (2021). “Endophytic fungi: a potential source of bioactive compounds for commercial and therapeutic applications”, in Endophytes, eds R. H. Patil, V. L. Maheshwari (Singapore: Springer). doi: 10.1007/978-981-15-9371-0_12
Puri, S. C., Verma, V., Amna, T., Qazi, G. N., and Spiteller, M. (2005). An endophytic fungus from Nothapodytes foetida that produces camptothecin. J. Nat. Prod. 68, 1717–1719. doi: 10.1021/np0502802
Qiang, X., Ding, J., Lin, W., Li, Q., Xu, C., Zheng, Q., et al. (2019). Alleviation of the detrimental effect of water deficit on wheat (Triticum aestivum L.) growth by an indole acetic acid-producing endophytic fungus. Plant Soil 439, 373–391. doi: 10.1007/s11104-019-04028-7
Qiang, X., Weiss, M., Kogel, K. H., and Schafer, S. (2012). Piriformospora indica- a mutualistic basidiomycete with an exceptionally large plant host range. Mol. Plant Pathol. 13, 508–518. doi: 10.1111/j.1364-3703.2011.00764.x
Radic, N., and Strukelj, B. (2012). Endophytic fungi: the treasure chest of antibacterial substances. Phytomed 19, 1270–1284. doi: 10.1016/j.phymed.2012.09.007
Rajashekar, Y., Tonsing, N., Shantibala, T., and Manjunath, J. R. (2016). 2, 3-Dimethylmaleic anhydride (3, 4-Dimethyl-2, 5-furandione): a plant derived insecticidal molecule from Colocasia esculenta var. esculenta (L.) Schott. Sci. Rep. 6, 1–7. doi: 10.1038/srep20546
Rehman, S., Shawl, A. S., Kour, A., Andrabi, R., Sudan, P., Sultan, P., et al. (2008). An endophytic Neurospora sp. from Nothapodytes foetida producing camptothecin. Appl. Biochem. Microbiol. 44, 203–209. doi: 10.1134/S0003683808020130
Renuka, S., and Ramanujam, B. (2016). Fungal endophytes from maize (Zea mays L.): isolation, identification and screening against maize stem borer, Chilo partellus (Swinhoe). J Pure Appl. Microbiol. 10, 523–529.
Roberts, E., and Lindow, S. (2014). Loline alkaloid production by fungal endophytes of fescue species select for particular epiphytic bacterial microflora. ISME J. 8, 359–368. doi: 10.1038/ismej.2013.170
Rodriguez, R. J., Henson, J., Van Volkenburgh, E., Hoy, M., Wright, L., Beckwith, F., et al. (2008). Stress tolerance in plants via habitat-adapted symbiosis. ISME J. 2, 404–416. doi: 10.1038/ismej.2007.106
Rodriguez, R., and Redman, R. (2008). More than 400 million years of evolution and some plants still can’t make it on their own: plant stress tolerance via fungal symbiosis. J. Exp. Bot. 59, 1109–1114. doi: 10.1093/jxb/erm342
Rodriguez, R. J., White, J. F. Jr., Arnold, A. E., and Redman, R. S. (2009). Fungal endophytes: diversity and functional roles. New Phytol. 182, 314–330. doi: 10.1111/j.1469-8137.2009.02773.x
Romero, A., Carrion, G., and Rico-Gray, V. (2001). Fungal latent pathogens and endophytes from leaves of Parthenium hysterophorus (Asteraceae). Fungal Divers. 7, 81–87.
Rutter, B. D., and Innes, R. W. (2018). Extracellular vesicles as key mediators of plant–microbe interactions. Curr. Opin. Plant Biol. 44, 16–22. doi: 10.1016/j.pbi.2018.01.008
Sadeghi, F., Samsampour, D., Seyahooei, M. A., Bagheri, A., and Soltani, J. (2019). Diversity and spatiotemporal distribution of fungal endophytes associated with Citrus reticulata cv. Siyahoo. Curr. Microbiol. 76, 279–289. doi: 10.1007/s00284-019-01632-9
Saijo, Y., Loo, E. P., and Yasuda, S. (2018). Pattern recognition receptors and signaling in plant–microbe interactions. Plant J. 93, 592–613. doi: 10.1111/tpj.13808
Samad, A. F. A., Rahnamaie-Tajadod, R., Sajad, M., Jani, J., Murad, A. M. A., Noor, N. M., et al. (2019). Regulation of terpenoid biosynthesis by miRNA in Persicaria minor induced by fusarium oxysporum. BMC Genomics 20:586. doi: 10.1186/s12864-019-5954-0
Sanchez-Vallet, A., Mesters, J. R., and Thomma, B. P. (2015). The battle for chitin recognition in plant-microbe interactions. FEMS Microbiol. Rev. 39, 171–183. doi: 10.1093/femsre/fuu003
Saxena, J., Chandra, S., and Nain, L. (2013). Synergistic effect of phosphate solubilizing rhizobacteria and arbuscular mycorrhiza on growth and yield of wheat plants. J. Soil Sci. Plant Nutr. 13, 511–525. doi: 10.4067/S0718-95162013005000040
Schläpfer, P., Zhang, P., Wang, C., Kim, T., Banf, M., Chae, L., et al. (2017). Genome-wide prediction of metabolic enzymes, pathways, and gene clusters in plants. Plant Physiol. 173, 2041–2059. doi: 10.1104/pp.16.01942
Schulz, B., Haas, S., Junker, C., Andrée, N., and Schobert, M. (2015). Fungal endophytes are involved in multiple balanced antagonisms. Curr. Sci. 109, 39–45.
Schulz-Bohm, K., Geisen, S., Wubs, E. R., Song, C., de Boer, W., and Garbeva, P. (2017). The prey’s scent–volatile organic compound mediated interactions between soil bacteria and their protist predators. ISME J. 11, 817–820. doi: 10.1038/ismej.2016.144
Shahabivand, S., Parvaneh, A., and Aliloo, A. A. (2017). Root endophytic fungus Piriformospora indica affected growth, cadmium partitioning and chlorophyll fluorescence of sunflower under cadmium toxicity. Ecotoxicol. Environ. Saf. 145, 496–502. doi: 10.1016/j.ecoenv.2017.07.064
Shankar, A., and Sharma, K. K. (2022). Fungal secondary metabolites in food and pharmaceuticals in the era of multi-omics. Appl. Microbiol. Biotechnol. 106, 3465–3488. doi: 10.1007/s00253-022-11945-8
Sinno, M., Ranesi, M., Gioia, L., d’Errico, G., and Woo, S. L. (2020). Endophytic fungi of tomato and their potential applications for crop improvement. Agriculture 10:587. doi: 10.3390/agriculture10120587
Slot, J. C., and Rokas, A. (2011). Horizontal transfer of a large and highly toxic secondary metabolic gene cluster between fungi. Curr. Biol. 21, 134–139. doi: 10.1016/j.cub.2010.12.020
Soanes, D., and Richards, T. A. (2014). Horizontal gene transfer in eukaryotic plant pathogens. Annu. Rev. Phytopathol. 52, 583–614. doi: 10.1146/annurev-phyto-102313-050127
Subban, K., Subramani, R., and Johnpaul, M. (2013). A novel antibacterial and antifungal phenolic compound from the endophytic fungus Pestalotiopsis mangiferae. Nat. Prod. Res. 27, 1445–1449. doi: 10.1080/14786419.2012.722091
Sun, W., Qin, L., Xue, H., Yu, Y., Ma, Y., Wang, Y., et al. (2019). Novel trends for producing plant triterpenoids in yeast. Crit. Rev. Biotechnol. 39, 618–632. doi: 10.1080/07388551.2019.1608503
Taechowisan, T., Lu, C., Shen, Y., and Lumyong, S. (2005). Secondary metabolites from endophytic Streptomyces aureofaciens CMUAc130 and their antifungal activity. Microbiology 151, 1691–1695. doi: 10.1099/mic.0.27758-0
Tang, M. J., Lu, F., Yang, Y., Sun, K., Zhu, Q., Xu, F. J., et al. (2022). Benefits of endophytic fungus Phomopsis liquidambaris inoculation for improving mineral nutrition, quality, and yield of rice grains under low nitrogen and phosphorus condition. J. Plant Growth Regul. 41, 2499–2513. doi: 10.1007/s00344-021-10462-8
Tang, D., Wang, G., and Zhou, J. M. (2017). Receptor kinases in plant-pathogen interactions: more than pattern recognition. Plant Cell 29, 618–637. doi: 10.1105/tpc.16.00891
Tian, W., Hou, C., Ren, Z., Wang, C., Zhao, F., Dahlbeck, D., et al. (2019). A calmodulin-gated calcium channel links pathogen patterns to plant immunity. Nature 572, 131–135. doi: 10.1038/s41586-019-1413-y
Töpfer, N., Fuchs, L. M., and Aharoni, A. (2017). The PhytoClust tool for metabolic gene clusters discovery in plant genomes. Nucleic Acids Res. 45, 7049–7063. doi: 10.1093/nar/gkx404
Uzma, F., Mohan, C. D., Hashem, A., Konappa, N. M., Rangappa, S., Kamath, P. V., et al. (2018). Endophytic fungi-alternative sources of cytotoxic compounds: a review. Front. Pharmacol. 9:309. doi: 10.3389/fphar.2018.00309
Wang, Z., Wang, L., Pan, Y., Zheng, X., Liang, X., Sheng, L., et al. (2022). Research advances on endophytic fungi and their bioactive metabolites. Bioprocess Biosyst. Eng. 46, 165–170. doi: 10.1007/s00449-022-02840-7
Waqas, M., Khan, A. L., Hamayun, M., Shahzad, R., Kang, S. M., Kim, J. G., et al. (2015). Endophytic fungi promote plant growth and mitigate the adverse effects of stem rot: an example of Penicillium citrinum and aspergillus terreus. J. Plant Interact. 10, 280–287. doi: 10.1080/17429145.2015.1079743
Weiss, M., Sýkorová, Z., Garnica, S., Riess, K., Martos, F., Krause, C., et al. (2011). Sebacinales everywhere: previously overlooked ubiquitous fungal endophytes. PLoS One 6:e16793. doi: 10.1371/journal.pone.0016793
Woloshuk, C. P., Foutz, K. R., Brewer, J. F., Bhatnagar, D., Cleveland, T. E., and Payne, G. A. (1994). Molecular characterization of aflR, a regulatory locus for aflatoxin biosynthesis. Appl. Environ. Microbiol. 60, 2408–2414. doi: 10.1128/aem.60.7.2408-2414.1994
Xie, W., Hao, Z., Yu, M., Wu, Z., Zhao, A., Li, J., et al. (2019). Improved phosphorus nutrition by arbuscular mycorrhizal symbiosis as a key factor facilitating glycyrrhizin and liquiritin accumulation in Glycyrrhiza uralensis. Plant Soil 439, 243–257. doi: 10.1007/s11104-018-3861-9
Yadav, M., Yadav, A., and Yadav, J. P. (2014). In vitro antioxidant activity and total phenolic content of endophytic fungi isolated from Eugenia jambolana lam. Asian Pac J Trop Med 7, S256–S261. doi: 10.1016/S1995-7645(14)60242-X
Yan, H., Jin, H., Fu, Y., Yin, Z., and Yin, C. (2019). Production of rare ginsenosides Rg3 and Rh2 by endophytic bacteria from Panax ginseng. J. Agric. Food Chem. 67, 8493–8499. doi: 10.1021/acs.jafc.9b03159
Yang, H., Peng, S., Zhang, Z., Yan, R., Wang, Y., Zhan, J., et al. (2016). Molecular cloning, expression, and functional analysis of the copper amine oxidase gene in the endophytic fungus Shiraia sp. Slf14 from Huperzia serrata. Protein Expr. Purif. 128, 8–13. doi: 10.1016/j.pep.2016.07.013
Yang, H., Tong, J., Lee, C. W., Ha, S., Eom, S. H., and Im, Y. J. (2015). Structural mechanism of ergosterol regulation by fungal sterol transcription factor Upc2. Nat. Commun. 6:6129. doi: 10.1038/ncomms7129
Yu, J. H., Butchko, R. A., Fernandes, M., Keller, N. P., Leonard, T. J., and Adams, T. H. (1996). Conservation of structure and function of the aflatoxin regulatory gene aflR from aspergillus nidulans and A. flavus. Curr. Genet. 29, 549–555. doi: 10.1007/BF02426959
Yuan, Y., Feng, H., Wang, L., Li, Z., Shi, Y., Zhao, L., et al. (2017). Potential of endophytic fungi isolated from cotton roots for biological control against verticillium wilt disease. PLoS One 12:e0170557. doi: 10.1371/journal.pone.0170557
Zakaria, L., and Aziz, W. N. W. (2018). Molecular identification of endophytic fungi from banana leaves (Musa spp.). Trop. Life Sci. Res. 29:201. doi: 10.21315/tlsr2018.29.2.14
Zeidler, D., Zahringer, U., Gerber, I., Dubery, I., Hartung, T., Bors, W., et al. (2004). Innate immunity in Arabidopsis thaliana: lipopolysaccharides activate nitric oxide synthase (NOS) and induce defense genes. Proc. Natl. Acad. Sci. U. S. A. 101, 15811–15816. doi: 10.1073/pnas.0404536101
Zhai, X., Jia, M., Chen, L., Zheng, C. J., Rahman, K., Han, T., et al. (2017). The regulatory mechanism of fungal elicitor-induced secondary metabolite biosynthesis in medical plants. Crit. Rev. Microbiol. 43, 238–261. doi: 10.1080/1040841X.2016.1201041
Zhang, Y., Li, Y., Chen, X., Meng, Z., and Guo, S. (2020). Combined metabolome and transcriptome analyses reveal the effects of mycorrhizal fungus Ceratobasidium sp. AR2 on the flavonoid accumulation in Anoectochilus roxburghii during different growth stages. Int. J. Mol. Sci. 21:564. doi: 10.3390/ijms21020564
Zhang, P. L., Wang, G., Xu, F. Q., Liu, J. S., Wang, J. T., Zhang, R., et al. (2019). Aspergilolide, a steroid lactone produced by an endophytic fungus aspergillus sp. MBL1612 isolated from Paeonia ostii. Nat. Prod. Res. 33, 2133–2138. doi: 10.1080/14786419.2018.1488706
Zhao, X., Wang, Z., Shu, S., Wang, W., Xu, H., Ahn, Y., et al. (2013). Ethanol and methanol can improve huperzine a production from endophytic Colletotrichum gloeosporioides ES026. PLoS One 8:e61777. doi: 10.1371/journal.pone.0061777
Zhu, D., Wang, J., Zeng, Q., Zhang, Z., and Yan, R. (2010). A novel endophytic huperzine A–producing fungus, Shiraia sp. Slf14, isolated from Huperzia serrata. J. Appl. Microbiol. 109:1469–1478. doi: 10.1111/j.1365-2672.2010.04777.x
Keywords: secondary metabolites, terpenoids, symbiosis, alkaloids, endophytic fungi
Citation: Jha P, Kaur T, Chhabra I, Panja A, Paul S, Kumar V and Malik T (2023) Endophytic fungi: hidden treasure chest of antimicrobial metabolites interrelationship of endophytes and metabolites. Front. Microbiol. 14:1227830. doi: 10.3389/fmicb.2023.1227830
Received: 23 May 2023; Accepted: 26 June 2023;
Published: 11 July 2023.
Edited by:
Anukool Vaishnav, Agroscope, SwitzerlandReviewed by:
Bishun Deo Prasad, Dr. Rajendra Prasad Central Agricultural University, IndiaCopyright © 2023 Jha, Kaur, Chhabra, Panja, Paul, Kumar and Malik. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tabarak Malik, dGFiYXJhay5tYWxpa0BqdS5lZHUuZXQ=; Priyanka Jha, cHJpeWFua2EuYnQuamhhQGdtYWlsLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.