
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 02 August 2023
Sec. Terrestrial Microbiology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1227297
Introduction: Conservation tillage is a widely used technique worldwide, but the effects of conservation tillage on bacterial community structure are poorly understood. We explored proportional alterations in the bacterial community under different tillage treatments.
Methodology: Hence, this study utilized high-throughput sequencing technique to investigate the structure and assembly processes of microbial communities in different tillage treatments.
Results and discussion: Tillage treatments included tillage no-straw retention (CntWt), no-tillage with straw retention (CntWntS), tillage with straw retention (CntWtS), no-tillage and no-straw retention (CntWnt). The influence of tillage practices on soil bacterial communities was investigated using Illumina MiSeq sequencing. Different tillage methods and straw retention systems significantly influenced soil parameters such as total potassium and pH were not affected by tillage practices, while straw retention significantly affected soil parameters including nitrogen content, available phosphorus and available potassium. Straw retention decreased bacterial diversity while increased bacterial richness. The effect of straw retention and tillage on bacterial communities was greater than with no tillage. Phylogenetic β-diversity analysis showed that deterministic homogeneous selection processes were dominated, while stochastic processes were more pronounced in tillage without straw retention. Ecological network analysis showed that microbial community correlation was increased in CntWntS and CntWnt. Straw retention treatment significantly increased the relative abundance of bacterial taxa Proteobacteria, Bacteroidetes, and OD1, while Nitrospirae, Actinobacteria, and Verrucomicrobia significantly decreased.
Conclusion: The conservation tillage practices significantly affect soil properties, bacterial composition, and assembly processes; however, further studies are required to investigate the impact of different crops, tillage practices and physiological characteristics on bacterial community structure and functions.
Agriculture is an important anthropogenic contributor to the variation of soil's physicochemical properties. These changes in soil properties proportionally affect the soil microbial community structure and composition (Kladivko, 2001). The relationship between soil and bacterial communities is an important sign of soil quality, sustainability of agriculture, and the ecosystem (Wang et al., 2016). Fertile soil in farmland is the prerequisite and foundation of sustainable agriculture development (Parikh and James, 2012). Tillage treatments affect soil biogeochemistry and help improve soil fertility (Frey et al., 1999; Gathala et al., 2011). No-tillage is an alternative to conventional tillage, and it usually increases the aggregation of soil organic matter (SOM) and soil biota (Six et al., 2000) and reduces soil erosion and production expenses by saving fuel, equipment, and labor (Uri, 1999; Huang et al., 2013). Soil microbial communities are more abundant in no-tillage soil than in conventional tillage (Govaerts et al., 2007; Helgason et al., 2009). Many Chinese farmers still use traditional tillage methods, especially in the Huang-Huai-Hai Plain in China, including deep plowing and rotational tillage (Chen et al., 2017). These practices are more costly, especially deep plowing (Wang et al., 2006). Nevertheless, tillage increases the bulk density of the soil up to a layer of 10–30 cm compared with no cultivation (Šíp et al., 2013). In addition, rotary tillage reduces the plow layer, enriches surface nutrients, and depletes deeper soil, causing soil infertility and preventing plant uptake of nutrients (Tian et al., 2012; LiangPeng et al., 2015).
Retention of crop straw stimulates microbial activity and biomass, reducing handling costs and time and preventing environmental pollution (Rodriguez et al., 2017). Studies have shown that availability of crop residues increased soil organic carbon (SOC), nitrogen (N) stocks (Brady and Weil, 1996; Dolan et al., 2006; Govaerts et al., 2006; Ke and Scheu, 2008), and microbial activity and decreased soil temperature (Sarrantonio and Gallandt, 2003; Wang et al., 2011). However, inappropriate straw incorporation methods could degrade soil structure and cause an imbalance in the nutrient supply (Kong, 2014; Chen et al., 2017). The fate of crop residues, which provide a diverse substrate, can significantly affect bacterial communities in the soil (Su et al., 2017). The impact of tillage on soil bacterial diversity and assemblages studied so far differs significantly between the different tillage methods (Souza et al., 2016; Dong et al., 2017).
With the recent advancement in deep sequence methods, several studies showed different findings about the impact of tillage and no-tillage practices on microbial community composition and structure. For example, several investigations have found consistent microbiome diversity in soil samples collected under different tillage practices (Hartman et al., 2018; Xia et al., 2019; Kraut-Cohen et al., 2020). Similarly, some studies indicated higher microbial alpha diversity in no-tillage soil than in tillage practices, and some also reported higher biomass richness in no-tillage samples (Feng et al., 2003; Sengupta and Dick, 2015; Wang et al., 2017; Schmidt et al., 2018). According to another research, appropriate post-harvest residue management practices can help restore soil nutrients by decomposing organic residues, thereby reducing the decline of microbial diversity in plantation soils (Chen and Xu, 2005; Xiong et al., 2021). To date, all research endeavors have investigated the impacts of diverse management practices on soil nutrients, long-term site sustainability, plant survival and growth, microbial composition, and several other pertinent factors. Nevertheless, prior research has not investigated the impact of distinct residue management and tillage practices on soil-related microbial community composition and assembly processes.
Numerous studies have shown that straw incorporation significantly affects nitrogen use and crop production (Duan et al., 2014). Both no-tillage and conventional tillage with residues applied had higher total nitrogen uptake and crop production than the no-residue methods applied in the lower middle Yangtze River Delta in China (Xu et al., 2010). In Central China, the yield was significantly higher under deep tillage with crop residues at a depth of 30 cm than without the incorporation of straw (Zhao et al., 2014; Chen et al., 2015). However, in northwest China, no significant difference in nitrogen uptake was found between treatments with and without straw return under no-tillage conditions (Zhang et al., 2009). The effects of tillage on crop production and nitrogen application differ depending on local climate, soil conditions, residue management, and crop rotation (Ponnamperuma, 1984). However, there is still a gap in knowledge about how straw retention affects soil microbial communities under both tillage and no-tillage. In contrast to microbial taxonomic composition, recent investigations have demonstrated that microorganisms' functional composition is more sensitive to environmental variables (Song et al., 2021). Therefore, it would be crucial to characterize the functional profiles of microbial communities to at least partially describe their metabolic properties (Gibbons et al., 2017). Xenobiotic substances are exotic substances that accumulate in the environment and endanger the biosphere. Xenobiotic substances cannot be biodegraded because they are extremely resistant. Although metagenomics technologies are widely used to reveal the activities of microbial communities, their widespread application is limited by their high cost and extremely difficult data processing (Louca et al., 2016). Tillage and residue management practices can impact the availability of nutrients such as carbon and nitrogen contents in soil (Kraut-Cohen et al., 2020). Therefore, in this study we performed PICRUSt, a system that predicts metagenomes using 16S data and a reference genome database using evolutionary modeling. Based on the relatively abundant bacterial sequences in all samples, most predicted metabolic pathways were classified into five functional groups: metabolism, environmental information processing, genetic information processing, cellular processes, and unclassified.
The tillage system produces soil disturbance at different levels, thus creating diverse soil ecosystems that affect bacterial community structure (Navarro-Noya et al., 2013; Sun et al., 2018). Soil crop cultivation conservation includes incorporating no-tillage, crop straw returning, and crop rotation, which is not well-defined and often interpreted diversely (Hobbs et al., 2008; Palm et al., 2014). In this study, we hypothesized that tillage decreases microbial diversity and richness, while straw retention increases microbial diversity and richness. Furthermore, microbial assembly processes were driven by tillage than straw retention in typical fluvo-aquatic soils. This study will help to interpret relationships between soil variables and changes in bacterial community structure and composition under different tillage practices.
We explained the experimental site and sampling strategy in our recently published study (Zhang et al., 2022). In brief, the long-term conservation field was commenced in 2006 and based on a completely randomized block design with three replications. Each treatment plot was 14 m × 6.5 m, and a maize–wheat rotation system was arranged randomly. Four treated soil samples are as follows: (1) tillage with no-straw retention (CntWt), (2) no-tillage with straw retention (CntWntS), (3) tillage with straw retention (CntWtS), and (4) no-tillage with no-straw retention (CntWnt) as control. Regarding tillage practice, soils were plowed to 20–22 cm depth with a moldboard plow. Regarding straw mulching, residues were crushed into 2–3 cm pieces for maize and 6–7 cm pieces for wheat and then were spread evenly on the soil surface as mulch. The amount of straw was the same in each plot. As for no-straw mulching treatments, all residues were removed from the plots. Each experimental plot was 14 m × 6.5 m in size. Three soil samples were randomly collected from each plot from the surface layer of soil (0–20 cm) using a sterile soil driller. The three samples from each plot were immediately mixed to form a composite soil sample. The composite samples were sieved through a 2-mm sieve to homogenize them and remove plant roots and stones before being transferred to the laboratory for storage at 4°C for measuring soil physicochemical properties and at −80°C for DNA extraction (Sparks et al., 1996).
According to the Fast-DNA SPIN kit protocol, the microbial genomic DNA was extracted from 0.5 g soil samples (MP Biomedical, Santa Ana, CA). DNA concentrations were quantified using a NanoDrop ND-2000 (NanoDrop2000, Thermo Scientific, and Wilmington, DE, USA) spectrophotometer according to the manufacturer's procedure.
The bacterial 16S rRNA gene was amplified with universal primers 515F (5′-GTAGTCTGTGCCAGCMGCCGCGG-3′) and 907R (5′-CCGTCAATTCMTTTRAGTTT-3′) that target the V4–V5 regions. Master Mix (20 μl) reaction mixture was prepared using a Taq enzyme (Takara, Japan). PCR amplification reaction conditions were as follows: initial denaturation at 95°C for 3 min, then 30 cycles of 95°C for the 30 s, 55°C for 30 s, 72°C for 1 min, with a final extension step of 72°C for 10 min. The amplified PCR product was verified by 1% agarose gel electrophoresis.
The high-throughput sequencing of 16S rRNA described by Caporaso et al. (2012) was used to find comprehensive coverage of bacterial community composition and diversity. The PCR products were sent to the Shanghai Personal Biotechnology Co., Ltd company. The amplified 16S rRNA gene was sequenced using the Illumina MiSeq platform. Complete data sets were deposited in the National Center for Biotechnology Information (NCBI) sequence read archive (SRA) with the accession number from SRR25082107 to SRR25082118.
We studied phylogenetic structure with null model-based analysis to test the assembly mechanisms of the bacterial populations (Kembel, 2010). In brief, phylogenetic turnover was calculated with 999 random sets, and the abundance-weighted mean nearest taxon distance (MNTD) was determined to analyze phylogenetic clustering in a single sample. Subsequently, to measure the standard effect size of the MNTD (ses.MNTD), the command “ses.mntd” in the “picante” package of R was applied (Webb et al., 2002; Stegen et al., 2012). Then, the β-nearest taxon index (βNTI), representing the difference between the βMNTD and the mean of the null distribution of βMNTD normalized by its standard deviation, was calculated to identify the community assembly processes further. The βMNTD and βNTI were calculated in R with the package MicEco using the command ses.comdist and ses.comdistnt, respectively (Zhang et al., 2019). Calculated βNTI values were used to examine phylogenetic turnover within a community to determine the role of deterministic and stochastic processes. If βNTI values are >2 or <-2, the deterministic processes shape bacterial community assembly processes across all treatments. Stochastic processes may play a significant role in community assembly processes when the values of βNTI are between −2 and 2. The βNTI > 2 showed significantly more phylogenetic turnover than predicted, often interpreted by chance as variable selections, while βNTI values <2 referred to less phylogenetic turnover than expected, i.e., homogeneous selection (Stegen et al., 2013). The Bray–Curtis-based Raup–Crick metric (RCbray) was further determined as described by Stegen et al. (2013) to quantify the role of specific stochastic processes. In brief, the obtained values of RCbray ranged between −1 and 1. We compared the βNTI values between 2 and−2 and RCbray values to further determine stochastic assembly processes. Thus, when βNTI values are between 2 and −2 and RCbray values > 0.95, the assembly process is dispersal limitation, when βNTI values are between 2 and −2 and RCbray values are between 0.95 and −0.95, the process is drift, and when βNTI values are between 2 and −2 and RCbray values <-0.95 is homogenizing dispersal (Liu et al., 2020).
Quantitative insights into the microbial ecology (QIIME; http://qiime.org/) (Caporaso et al., 2012) workflow was used to obtain quality-filtered sequences and high-quality reads. The reads were compared with the reference database (http://drive5.com/uchime/uchime_download.html), using the UCHIME algorithm to find chimeric sequences and remove them to obtain clean and effective reads. Alpha diversity was calculated using MOTHUR software (http://www.mothur.org). For alpha diversity, Chao1 and ACE indices were used to estimate richness, while we used Shannon and Simpson indices to estimate diversity. For β-diversity, principal coordinate analysis (PCoA) scaling was used for the clustering of different samples (Li et al., 2014). Functional gene predictions were performed using the PICRUSt (Langille et al., 2013). Heatmap was generated using ggplot2 in RStudio software. All statistical analyses were completed using SPSS package release 22.0 (SPSS Inc., Chicago, USA). Significance was accepted at a p-value of <0.05 unless otherwise noted. The microbial community network was built by calculating correlations using co-occurrence network (CoNet) inference. Using an ensemble method, the three metrics—Pearson's and Spearman's correlations and Bray–Curtis dissimilarities between paired OTUs—were merged. Spearman's correlation between microbial taxa was considered statistically robust when Spearman's correlation coefficient (r) is >0.6 and the p-value is <0.01. We calculated the relative abundance of OTUs in all samples and then identified the OTUs that were dominant with a relative abundance >0.5% in all samples. These dominant OTUs with relative abundance >0.5% were selected for network construction. We selected the most abundant bacterial taxa belonging to these OTUs at the phyla level in all samples for comparative analysis. The p-values for the four measures were integrated using the Brownian methodology and then corrected using the Benjamini–Hochberg method to lessen the probability of false-positive results. The network was seen using Cytoscape software. Different microbial taxa were represented by the nodes in the microbiome network (OTUs). The network edges showed physiologically or chemically relevant relationships and represented pairwise correlations between nodes. Groups of closely related nodes comprise modules (i.e., groups of coexisting or co-evolving microbes) (Shannon et al., 2003; Banerjee et al., 2016). Mantel tests with Pearson's correlation coefficient and 999 permutations were performed to analyze the correlation between βNTI values and Euclidean distances in environmental parameters.
The Illumina MiSeq sequencing produced 622,518 high-quality reads from four treatments. A total of 5,199 unique OTUs were identified from all the samples at 97% sequence similarity. Alpha diversity results of four treatments are shown in Table 1. The bacterial richness indices Chao1 and ACE showed higher richness in the CntWtS treatment, while lower bacterial richness was found in CntWnt. Shannon and Simpson diversity indices showed higher diversity in CntWt but lower diversity in the CntWntS treatment. The tillage with straw retention treatments increased soil bacterial richness, while no-tillage with straw retention decreased bacterial diversity. Therefore, no-tillage and straw retention practices significantly changed bacterial diversity.
The PCoA based on the OTUs level is shown in Figure 1. PCoA extracts coordinate axes that reflect the difference between samples. Samples containing similar community composition cluster together at the PCoA plot. The CntWntS and CntWtS had less diverse communities than CntWnt and CntWt, indicating that straw retention has a stronger influence on the bacterial population than tillage practices.
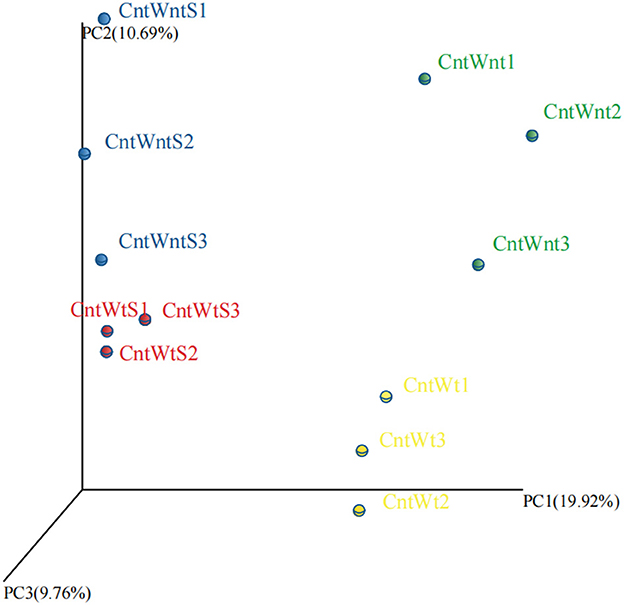
Figure 1. Principal Coordinates Analysis (PCoA) of unweighted UniFrac on 16S rRNA gene sequences from four treated soil samples with three replicates.
A total of 49 phyla were obtained from all samples using the MOTHUR program. The predominant phyla (≥1%) included Proteobacteria, Acidobacteria, Planctomycetes, Firmicutes, Actinobacteria, Bacteroidetes, Chloroflexi, Gemmatimonadetes, Crenarchaeota, WS3, Nitrospirae, Verrucomicrobia, and OD1 in all treatments (Supplementary Figure S1). The relative abundance of Proteobacteria and Bacteroidetes was higher in CntWtS compared with CntWnt treatment. Similarly, the relative abundance of Acidobacteria, Chloroflexi, Actinobacteria, and Verrucomicrobia was much higher in CntWt and Crenarchaeota, and OD1 and WS3 were higher in CntWntS.
The abundance of dominant (≥1%) classified and unclassified genera is shown in Supplementaary Table S1. In the CntWtS treatment, the relative abundance of unclassified genera Cytophagaceae, Sinobacteraceae, MND, envOPS12, Betaproteobacteria, Myxococcales, RB40, Xanthomonadaceae, and Piscirickettsiaceae was significantly higher than in other treatments. In the CntWntS treatment, the relative abundance of the classified genus Candidatus Nitrososphaera and the unclassified genera Sediment-1, Acidobacteria, CL500-15, agg27, and ABY1 was significantly higher than in other treatments. The relative abundance of unclassified genera within the Pirellulaceae, Rhodospirillaceae, Sva0725, Chitinophagaceae, and WD2101 was also higher in the CntWt treatment compared with treatments. Classified bacterial genera Lactococcus, Solibacillus, and Bacillus and unclassified genera iii1-15, RB41, Gemm-1 0319-6A21, mb2424, Haliangiaceae, and Syntrophobacteraceae had significantly higher abundance in the CntWnt treatment (Figure 2).
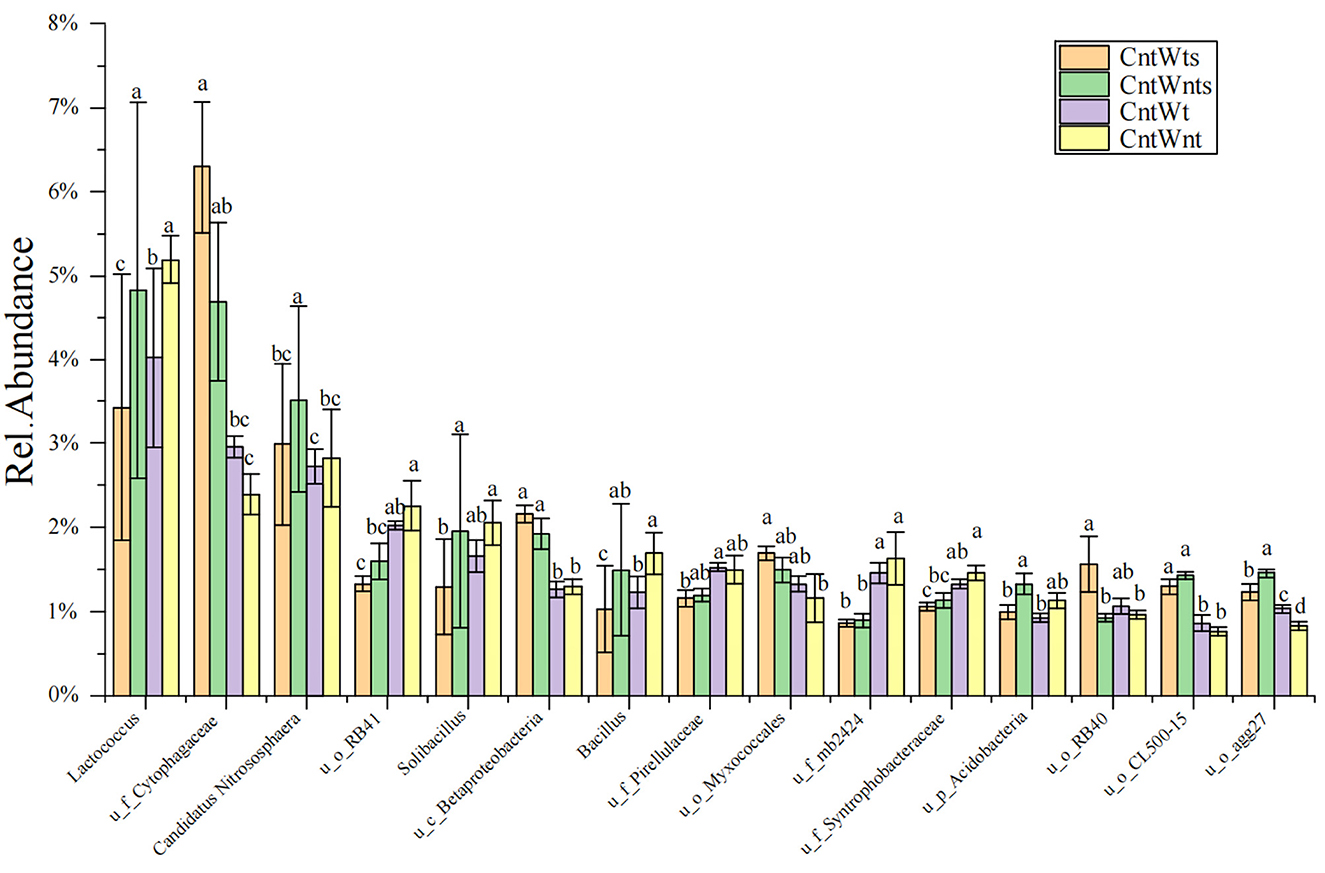
Figure 2. Relative abundances (≥ 1%) of dominant genera between four different treatments. “u” represents unclassified members at the genus level. Different lowercase letters show significant differences (p < 0.05).
We determined the β NTI for paired samples to assess whether neutral or niche-based mechanisms better explain the assembly of the bacterial community in different soil with different tillage treatments (Figure 3A). Our results indicated that deterministic processes assembled 87.88% of the bacterial community, and βNTI scores below −2 showed homogeneous selection as the dominant mechanism shaping community assemblage. In comparison, 12.12% community exhibited stochastic assembly processes and the βNTI values were below 2 and above −2. Quantitative assembly mechanism analysis further revealed that in CntWt treatments, homogeneous selection dominated assembly processes followed by homogeneous dispersal (Figure 3B). Interestingly, our study showed community assembly dynamic was not driven by variable selection in all treatments. The Mantel test was performed for βMNTD and βNTI values and measured environmental variables for further validations. Here, we applied both matrices and the results indicated that measured environmental variables and βMNTD and βNTI values were non-significantly correlated with community assembly processes (Supplementary Figure S3).
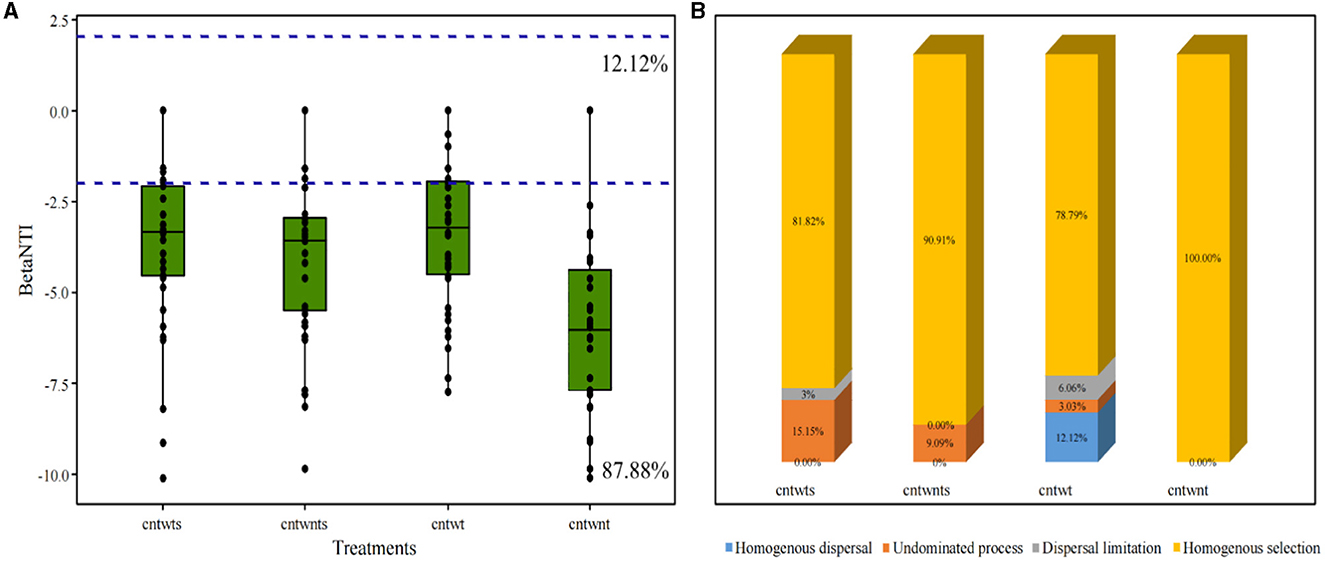
Figure 3. Boxplot of βNTI values grouped by treatment of the soil. (A) Horizontal blue dashed lines indicate lower and upper betaNTI significant thresholds (−2 and +2) respectively. (B) Percentage of bacterial community assembly governing processes.
The relationship between the dominant genera and environmental factors analyzed by the redundancy analysis (RDA) method (Figure 4) shows that bacterial community structure is affected by various environmental factors under conservation tillage treatments. The phylum Proteobacteria positively correlated with TN and VP and negatively correlated with TP. Bacteroidetes were positively correlated with AN, VP, and VK and negatively correlated with TP. Actinobacteria negatively correlated with AN and VK. The phyla WS3 positively correlated with TN and VK. Nitrospirae negatively correlated with OC and AN. OD1 was positively correlated with OC, TN, AN, and VK (Supplementary Table S4). Unclassified, such as Sinobacteraceae, Betaproteobacteria, Myxococcales, Acidobacteria, Cytophagaceae, and Sediment-1, positively correlated with total nitrogen. The available nitrogen concentration positively correlated with the abundance of Betaproteobacteria, Cytophagaceae, and Sediment-1 and negatively correlated with the abundance of unclassified genera RB41, mb2424, and 031F9-6A21. Unclassified Acidobacteria positively correlated with organic carbon. Classified genus Lactococcus and unclassified Sinobacteraceae, MND1, Myxococcales, and Cytophagaceae were positively associated with available phosphorus, while other Betaproteobacteria were negatively associated with total phosphorus. Soil-available potassium positively correlated with unclassified Betaproteobacteria, Myxococcales, mb2424, Acidobacteria, Cytophagaceae, and Sediment-1 and was not correlated with total potassium. Soil pH was negatively correlated with classified Lactococcus and unclassified Xanthomonadaceae (Supplementary Table S3).
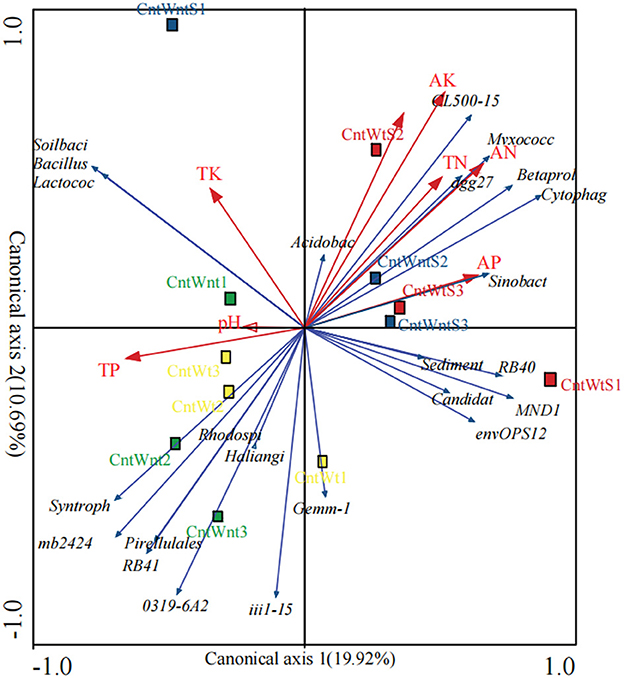
Figure 4. Redundancy analysis of bacterial communities in different residue and tillage treatment soils as affected by environmental properties (b). AP, Available Phosphorus; TP, Total Phosphorus; TK, Total Potassium; SOC, Soil Organic Carbon; AK, Available Potassium; AN, Available Nitrogen, TN: Total Nitrogen. The percentage indicates the proportion of variation elucidated by the first and second canonical axes.
We constructed microbial co-occurrence networks based on the correlation analysis of taxonomic profiles in different tillage practices to further understand the impact of different tillage practices on microbial composition (Figure 5). Overall, 49, 50, 46, and 50 nodes were inter-linked 173 (57.80% positive and 42.2% negative), 185 (83.78% positive and 16.21% negative), 120 (95% positive and 5% negative), and 194 (70.10% positive and 29.90% negative) in CntWts, CntWntS, CntWt, and CntWnt, respectively (Table 2). The significant and robust associations between taxa (average degree) were higher in CntWnt than CntWntS, CntWtS, and CntWt (7.18, 7.14, 6.93, and 4.36, respectively). However, in CntWt and CntWntS, the positive connections were higher at 95 and 83.78%, respectively, while lower in CntWts at 75.80%, showing that the connections between OTUs were disturbed during different tillage practices. Interacting microbial networks clustered into modules can be examined to find significant modular relationships. Our results indicate that bacterial networks can all be broken down into smaller interconnected modules, with individual nodes exhibiting different roles in the microbial network. The bacterial taxa in the network consisted mainly of p__Proteobacteria, p__Planctomycetes, p__Acidobacteria, p__Bacteroidetes, p__Firmicutes, and p__Gemmatimonadetes. In the CntWtS network, Sinobacteraceae;g__(p__Proteobacteria) was detected as the most critical taxa (strongest interaction). In CntWntS network, iii1-15;g__(p__Acidobacteria) and MND1;g__ (p__Proteobacteria) were similarly designated as key taxa. The relatively few and unimportant key taxa in CntWt are Cytophagaceae;g__(p__Bacteroidetes). The key taxa in CntWnt are iii1-15;g__(p__Acidobacteria) and Cytophagaceae;g__(p__Bacteroidetes).
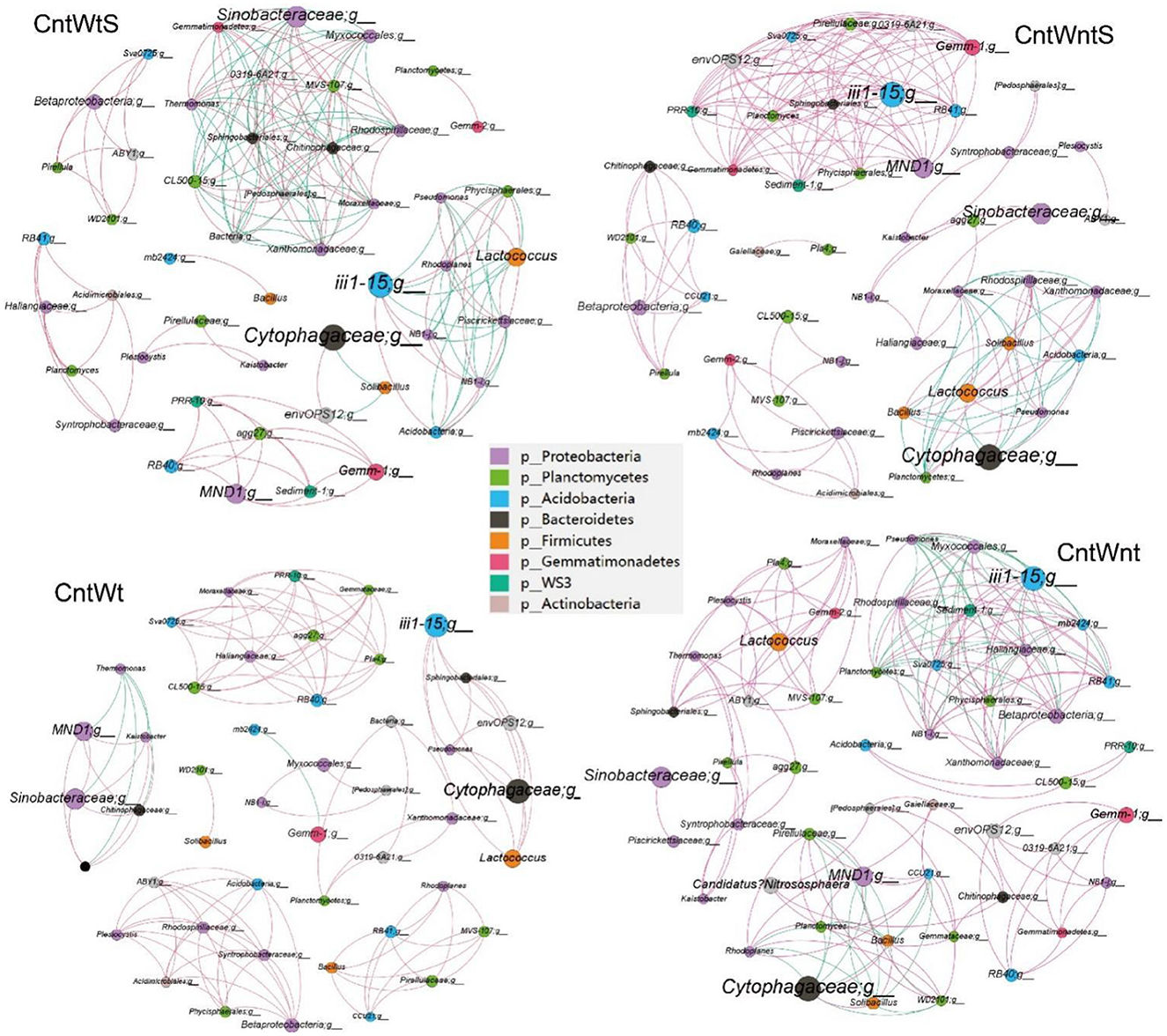
Figure 5. The bacterial co-occurrence networks under the CntWtS and CntWntS, CntWt and CntWnt treatments based on correlation analysis in the field experiment. A connection stands for a strong (Spearman's R > 0.9) and significant (P value < 0.01) correlation for different treatments. Each node name represents a taxon at the bacterial genus level, with unidentified genus names ending in; g_The size of each node is proportional to the number of links (degree), The networks in the field experiment are colored by the bacterial phyla. Each bacterial network consists of different closely related bacterial modules to identify keystone taxa (module hubs). The size of each node is proportional to the number of relative abundances. The red edges indicate positive interactions between two bacterial nodes, while green edges indicate negative interactions.
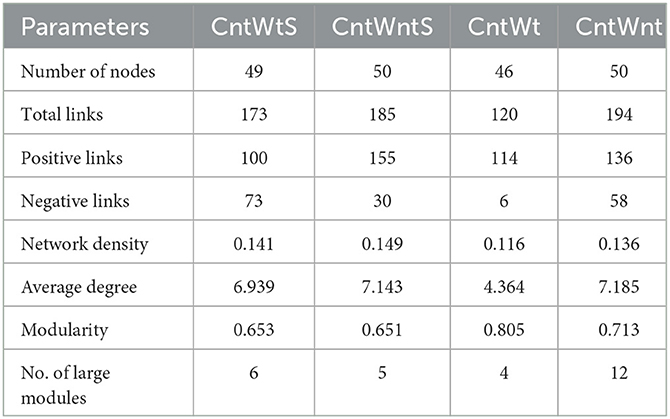
Table 2. Topological properties of a correlation network diagram of soil bacterial communities at the genus level in soils.
The functional prediction was carried out using the KEGG database. It was classified into seven major categories and 41 sub-categories (Figure 6A). Functional prediction indicated that metabolisms of xenobiotic biodegradation, nucleotide, lipids, energy, amino acids, and carbohydrate were significantly higher in tillage without straw retention treatment. At the same time, they were significantly lower in no-tillage and straw retention treatments. These findings suggest that tillage treatment significantly increases metabolic activities. Tillage with straw retention treatments showed significantly increased metabolism compared to no-tillage and straw retention treatment (Figure 6B). Our findings suggest that among all treatments, no-tillage and straw retention significantly decreased overall metabolic activity potential compared to control and other treatments.
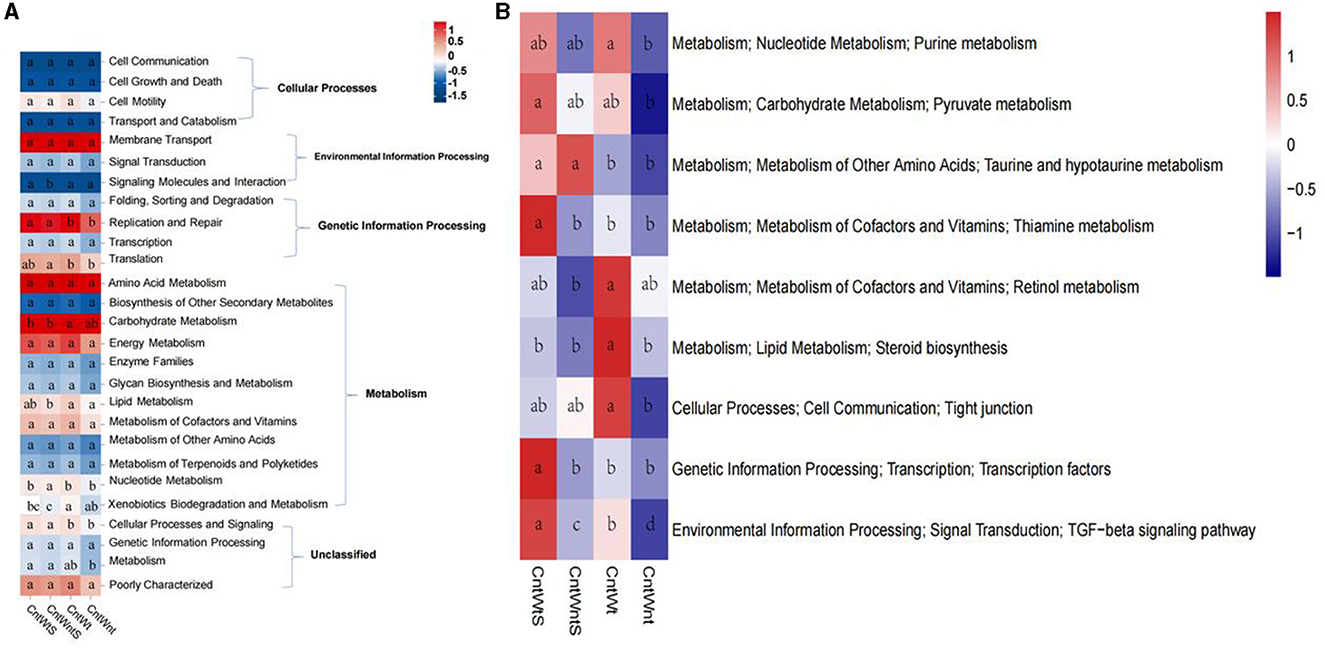
Figure 6. Predicted function of bacteria in four treatments. Values of each functional gene (row) was z-zero transformed. (A) KEGG level I and II. (B) Heatmap shows the third level of KEGG. The significant tests of gene distribution between groups was completed using ANOVA p < 0.05.
In our experiment, bacterial richness and diversity were lower in no-tillage treatments with or without crop straw retention. The no-tillage practices benefit the formation of soil micro-aggregates, which have different O2 concentrations inside and outside of the aggregates (Zhang X. et al., 2018). Some studies reported decreased bacterial diversity following organic material alteration, which helps the enrichment of copiotrophic taxa in soil (Fernandez, 2015; Lian et al., 2016). Our results indicate that tillage with straw retention treatment (CntWtS) increased bacterial richness and decreased bacterial diversity (Table 1), which is consistent with the previous study (Zhang Y. et al., 2018). While some studies reported decreased bacterial diversity following organic material amendment, ascribed to the incorporated material favoring the copiotrophic type microbial taxa (Ceja-Navarro et al., 2010; Fernandez, 2015). Soil disturbance caused by tillage and crop residues has been demonstrated as an important component affecting the microbial community (Jansa et al., 2002; Six et al., 2006; Cookson et al., 2008). A recent study showed that the soil aggregates and tillage practices affected soil bacterial communities compared with no-residue treatment (Zhang X. et al., 2018). The connection between tillage and soil bacterial communities may be complex, as tillage affects the soil's physiochemical properties and has a mutually dependent relationship with soil bacterial communities (Trivedi et al., 2017). Another study has shown that leaving crop residue on the soil surface increases soil OC, and crop residues with more cellulose or lignin and less N will decompose more slowly (Benbi and Khosa, 2014; Panettieri et al., 2015). The deposition of N significantly affected the relative abundances of bacterial phyla but reduced the bacterial diversity and changed the bacterial composition at low-level taxonomic levels (Frey et al., 2014; Freedman and Zak, 2015). According to Jiao et al. (2004), tillage practices affect the supply of soil nutrients, which include soil moisture content, OM decomposition, and porosity of the soil.
Several studies on microbial community ecology focus on the relative contribution of deterministic and stochastic processes (Stegen et al., 2013; Zhang et al., 2016; Zhou and Ning, 2017). In this study, the dominance of homogeneous selection in community assembly processes was confirmed by quantifying various ecological mechanisms in community assembly (Figure 3). Bacterial community assembly processes showed different trends, such as higher stochastic processes (15.15% undominated and 3% dispersal limitation) associated with tillage practices with no-straw retention treatments. On the other hand, no-tillage with straw retention was more associated with deterministic homogeneous selection. Finally, no-tillage with no-straw retention showed 100% homogeneous selection. To better understand microbial community assembly processes, we have established a conceptual model (Figure 7). This shows that ecological processes can occur in the following forms: (i) under weak selection (CntWt), microbial community assembly processes show the contribution of stochastic processes; (ii) under medium selection (CntWts), environmental selection leads to deterministic processes dominating the microbial communities; and (iii) under strong selection (CntWts), microbial diversity is reduced, which ultimately induces the deterministic processes. Our findings are contrary to previous studies indicating that tillage practices impact bacterial assembly processes (Wang et al., 2020; Li et al., 2021), while in line with Zhuxiu et al. (2021), who found that both stochastic and deterministic processes play a significant role in bacterial community assembly processes under conservation tillage practices.
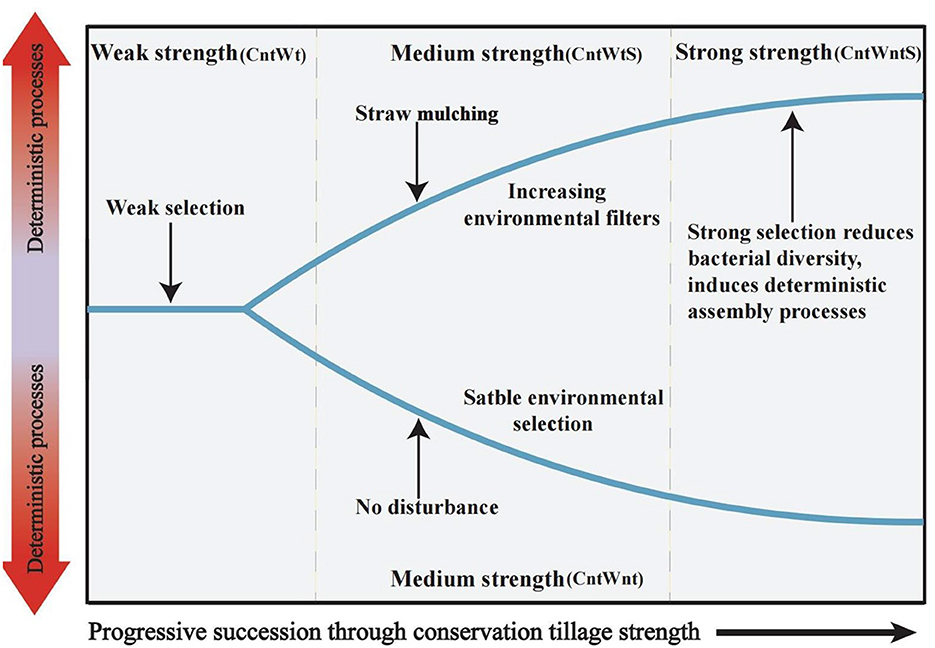
Figure 7. Conceptual model showing how ecological selection determines microbial community assembly processes by different tillage treatments. Weak strength in samples with tillage and no straw retention (CntWt); Medium strength in samples of no-tillage with no straw retention (CntWnt) and tillage with straw retention (CntWtS); Strong strength in samples with no-tillage with straw retention (CntWntS).
The 16S rRNA sequencing indicated that at the phylum level, bacterial community composition was very similar in all treatments, dominated by Proteobacteria, Acidobacteria, Planctomycetes, Firmicutes, Bacteroidetes, Chloroflexi, Gemmatimonadetes, Actinobacteria, Nitrospirae, and Crenarchaeota (Supplementary Figure S1). This result supports several other studies (Navarro-Noya et al., 2013; Wang et al., 2016). The relative abundance of Proteobacteria and Bacteroidetes significantly increased in tillage with straw retention treatments, which are responsible for labile carbon sources and increased in crop residue retention treatment (Fierer et al., 2007; Navarro-Noya et al., 2013). Many studies revealed that an application of crop residue retention supported Proteobacteria, particularly for bacteria at order levels of this phylum, such as Myxococcales, Pseudomonadales, and Sphingomonadales (Wallenstein et al., 2007; de Gannes et al., 2013; De la Cruz-Barrón et al., 2017; Su et al., 2017). The relative abundance of Betaproteobacteria and Myxococcales was significantly higher in crop straw retention treatments (Figure 2), Myxococcales are well-known to be able to decompose macromolecules, and some members can decompose cellulose (Bernard et al., 2007; Ceja-Navarro et al., 2010; Negassa et al., 2015). A previous study showed that Betaproteobacteria growth was positively linked with the soil's quality and amount of nutrients (Lin et al., 2010). Tillage alters the surrounding environment for Pseudomonas, Rudaea, Bacillus, and other bacteria and then changes the decay process of residue (Xia et al., 2019). The relative abundance of the family Cytophagaceae was significantly higher in straw retention treatments but was not significantly influenced by either depth or tillage treatments (McBride et al., 2014; Cania et al., 2019). Members of Cytophagaceae are aerobic and cellulolytic, which help to regulate the carbon cycle (Monserrate et al., 2001).
Applying crop residues into the soil leads to a shift in the relative abundances of various phyla, favoring the prevalence of syntrophic microorganisms such as Actinobacteria and Firmicutes (Ramirez-Villanueva et al., 2015). Another study stated that tillage with crop rotation increased the relative abundance of Acidobacteria than in a no-tillage (Yin et al., 2010). However, in this study, the relative abundance of phyla Acidobacteria and Firmicutes decreased with the application of straw retention in tillage and no-tillage treatment (Supplementary Figure S1). The reduction in acidobacteria might be due to the high level of available carbon sources as it is reported that the relative abundance of acidobacteria was higher in the low level of carbon (Fierer et al., 2007), while in this study, total organic carbon was relatively higher with straw retention application (Zhang et al., 2022). The relative abundance of the genus Lactococcus, Bacillus, and Solibacillus (Firmicutes) and unclassified _o_iii1-15 (Actinobacteria) decreased in tillage with straw retention treatment (Figure 2). Our findings are contrary to previous studies reported that the relative abundances of the plowed and no-tilled communities underwent significant changes during the growth stage, albeit with distinct trends. Actinobacteria are typically characterized as bacteria that thrive in nutrient-rich environments. These contrary findings might be due to experimental design, sampling size (Xue et al., 2021), soil types (Bickel and Or, 2020), and availability of nutrients (Bastida et al., 2021), which are predominant factors affecting microbial distribution. For example, a previous study showed that Actinobacteria appeared in oligotrophic conditions with low microbial activity (Jenkins et al., 2010; Che et al., 2016). Previous research also showed that Lactococcus represented 2.72% of the soil bacterial community and that the relative abundance of Bacillus, Lactococcus, Streptococcus, and Enterococcus in farmland, grassland, and woodland is significantly higher than in cropland (Cheng et al., 2018; Liang et al., 2019). This study also showed that the relative abundance of members of the phylum Crenarchaeota and class Thermoplasmata was higher in the 10–30 cm layer under conventional tillage with straw retention conditions, while tillage without straw retention increased the abundance of phylum Crenarchaeota's genus Candidatus Nitrososphaera (Barber et al., 1996). The significant correlation of phylum Planctomycetes with biomass and various pools of soil was noted, which is accountable for the anaerobic oxidation of ammonium (Buckley et al., 2006; Kuenen, 2008; Humbert et al., 2010).
However, the PCoA based on the unweighted UniFrac distance matrixes showed that straw retention and tillage system significantly affected the bacterial communities more than the control. These findings are in agreement with previous studies that reported that tillage practices and straw retention impact soil microbial community composition (Wang et al., 2017; Lu et al., 2019; Fernandez-Gnecco et al., 2022). The RDA showed that changes related to the different treatment and environmental factors were generated in bacterial communities (Negassa et al., 2015; de Graaff et al., 2019). To some extent, the bacterial community structure was affected by various environmental factors. TP and AK have a more significant impact on the bacterial community in treatments such as straw retention and tillage treatment (Figure 4).
In tillage with straw retention treatment, part of the metabolism (i.e., nucleotide, carbohydrate, and amino acid metabolism) significantly increased. Many studies have revealed that carbohydrate metabolism is an enzymatic process that changes external substrates into metabolic precursors, such as hydrolyzed polymers into monomers and d-fructose-6-phosphate and acetyl-CoA and pyruvate (Bräsen et al., 2014). Enzymes such as alpha-aminoadipic semialdehyde synthase and S-adenosylmethionine synthetase are involved in amino acid degradation (Guo et al., 2015; Gao et al., 2016). Yang et al. (2014) found that anaerobic microbes are involved in carbohydrate decay and energy conversion. In the amino acid metabolism which is a central process adopted by soil microbial communities in the presence of constant moisture stress is higher than normal environmental conditions (Fierer et al., 2012; Monard et al., 2012). The co-occurrence network further exhibited that total links are lower in no-tillage with straw retention and higher in tillage treatment, indicating that straw retention has a different effect on bacterial relationship. Tillage with no-straw retention impacts bacteria and demonstrates that no-tillage can make the network of soil biota more compact (Chaffron et al., 2010). No-tillage and straw retention can significantly promote the positive connection of dominant bacterial networks. The more compact network topology may be caused by several factors, which may be related to the no-tillage reduction in human microbial disturbances, and the return of straw to the bacteria to bring enough nitrogen and carbon sources, as well as appropriate habitat and breeding environment (Zaneveld et al., 2010). Another study reported that most studies consider the soil biota to be a black box, and little is known about the entire soil network (Schloss et al., 2009). In general, crop type (Fyles et al., 1988; Kaiser and Heinemeyer, 1993; Grayston et al., 1998), crop rotation (McGill et al., 1986; Dick, 1992), soil type (Girvan et al., 2003), sampling season (Spedding et al., 2004; Zhang et al., 2012), and agriculture management drive bacterial distributions in soils. In our study, straw retention treatments decreased soil bacterial diversity, and no-tillage treatments reduced the number of OTUs and bacterial diversity compared to tillage treatments, which differ from previous work in several significant ways. The contrary findings might be due to crop rotation because the wheat and maize straw return mode significantly changed bacterial diversity in soil and showed a downward trend (Zhang et al., 2023). Another reason might be an increase in risk disease pathogens, which may relate to a change in soil electrical conductivity and impact on soil microbial diversity, as reported in a previous study that maize straw reduces fungal diversity and fungal pathogens were abundant in maize straw-treated soil (Su et al., 2020).
We observed clear variations in soil physicochemical properties and microbial community structure by different tillage treatments. Different tillage practices significantly influenced soil properties except for pH and total potassium, while straw retention treatments significantly alter soil properties. Bacterial communities responded differently to different tillage treatments such as the relative abundance of the phyla Proteobacteria, Bacteroidetes, WS3, OD1, Acidobacteria, Actinobacteria, Nitrospirae, Verrucomicrobia, Cytophagaceae, and Betaproteobacteria was significantly affected by the addition of straw retention in tillage and no-tillage treatments. On the other hand, tillage practices with or without straw retention also changed some bacterial relative abundance, such as Planctomycetes, Firmicutes, Gemmatimonadetes, Crenarchaeota, Chloroflexi, and Lactococcus. Tillage without straw retention significantly affected soil bacterial diversity as compared to other treatments. Based on bacterial network analysis, no-tillage treatment can increase the co-occurrence between dominant bacterial genera. The relationship between bacterial genera with straw retention treatment was found to have different effects on tillage and no-tillage treatments. The function prediction analysis indicated that tillage treatment significantly increased the potential of metabolic activities. We can conclude that tillage practices and straw retention treatments are essential in soil microbial community structure and microbial metabolic activities. Current results revealed that we can alter microbial community structure in agricultural soil to improve soil fertility by managing tillage practices. We conducted this study in a controlled environment, and further research is needed on agricultural soils and open fields to determine the effect of long-term conservation tillage practices on soil bacterial community structure, functions, and their dynamics. Further experiments must consider testing different crop rotation and their impact on soil microbial community composition at a larger scale.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
HC and SH: conceptualization, supervision, and project administration. MuzK: methodology, formal analysis, resources, and writing—original draft preparation. HL: software. HC, SH, and MuzK: validation. AZ: investigation. SH: data curation. HL and MudK: writing—review and editing. HC: visualization and funding acquisition. All authors have read and agreed to the published version of the manuscript.
This study was financially supported by the National Natural Science Foundation of China (42077026 and 4207070252) and the Jiangsu Postdoctoral Excellent Program (2022ZB155).
We thank Dr. Jie Hu for her valuable advice on data analysis.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1227297/full#supplementary-material
Banerjee, S., Kirkby, C. A., Schmutter, D., Bissett, A., Kirkegaard, J. A., Richardson, A. E., et al. (2016). Network analysis reveals functional redundancy and keystone taxa amongst bacterial and fungal communities during organic matter decomposition in an arable soil. Soil Biol. Biochem. 97, 188–198. doi: 10.1016/j.soilbio.2016.03.017
Barber, R., Orellana, M., Navarro, F., Diaz, O., and Soruco, M. (1996). Effects of conservation and conventional tillage systems after land clearing on soil properties and crop yield in Santa Cruz, Bolivia. Soil Tillage Res. 38, 133–152. doi: 10.1016/0167-1987(96)01012-4
Bastida, F., Eldridge, D. J., García, C., Kenny Png, G., Bardgett, R. D., Delgado-Baquerizo, M., et al. (2021). Soil microbial diversity–biomass relationships are driven by soil carbon content across global biomes. ISME J. 15, 2081–2091. doi: 10.1038/s41396-021-00906-0
Benbi, D., and Khosa, M. (2014). Effects of temperature, moisture, and chemical composition of organic substrates on C mineralization in soils. Commun. Soil Sci. Plant Anal. 45, 2734–2753. doi: 10.1080/00103624.2014.950423
Bernard, L., Mougel, C., Maron, P. A., Nowak, V., Lévêque, J., Henault, C., et al. (2007). Dynamics and identification of soil microbial populations actively assimilating carbon from 13C-labelled wheat residue as estimated by DNA-and RNA-SIP techniques. Environ. Microbiol. 9, 752–764. doi: 10.1111/j.1462-2920.2006.01197.x
Bickel, S., and Or, D. (2020). Soil bacterial diversity mediated by microscale aqueous-phase processes across biomes. Nat. Commun. 11, 116. doi: 10.1038/s41467-019-13966-w
Brady, N. C., and Weil, R. R. (1996). The Nature and Properties of Soils. Hoboken, NJ: Prentice-Hall Inc.
Bräsen, C., Esser, D., Rauch, B., and Siebers, B. (2014). Carbohydrate metabolism in Archaea: current insights into unusual enzymes and pathways and their regulation. Microbiol. Mol. Biol. Rev. 78, 89–175. doi: 10.1128/MMBR.00041-13
Buckley, D. H., Huangyutitham, V., Nelson, T. A., Rumberger, A., and Thies, J. E. (2006). Diversity of planctomycetes in soil in relation to soil history and environmental heterogeneity. Appl. Environ. Microbiol. 72, 4522–4531. doi: 10.1128/AEM.00149-06
Cania, B., Vestergaard, G., Krauss, M., Fliessbach, A., Schloter, M., Schulz, S., et al. (2019). A long-term field experiment demonstrates the influence of tillage on the bacterial potential to produce soil structure-stabilizing agents such as exopolysaccharides and lipopolysaccharides. Environ. Microbiomes 14, 1–14. doi: 10.1186/s40793-019-0341-7
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Huntley, J., Fierer, N., et al. (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624. doi: 10.1038/ismej.2012.8
Ceja-Navarro, J. A., Rivera, F. N., Patiño-Zúñiga, L., Govaerts, B., Marsch, R., Vila-Sanjurjo, A., et al. (2010). Molecular characterization of soil bacterial communities in contrasting zero tillage systems. Plant Soil 329, 127–137. doi: 10.1007/s11104-009-0140-9
Chaffron, S., Rehrauer, H., Pernthaler, J., and von Mering, C. (2010). A global network of coexisting microbes from environmental and whole-genome sequence data. Genome Res. 20, 947–959. doi: 10.1101/gr.104521.109
Che, R., Wang, W., Zhang, J., Nguyen, T. T. N., Tao, J., Wang, F., et al. (2016). Assessing soil microbial respiration capacity using rDNA-or rRNA-based indices: a review. J. Soils Sediments 16, 2698–2708. doi: 10.1007/s11368-016-1563-6
Chen, C. R., and Xu, Z. H. (2005). Soil carbon and nitrogen pools and microbial properties in a 6-year-old slash pine plantation of subtropical Australia: impacts of harvest residue management. For. Ecol. Manage. 206, 237–247. doi: 10.1016/j.foreco.2004.11.005
Chen, J., Tang, Y., Yin, Y., Pang, D., Cui, Z., Zheng, M., et al. (2015). Effects of straw returning plus nitrogen fertilizer on nitrogen utilization and grain yield in winter wheat. Acta Agron. Sin. 41, 160–167. doi: 10.3724/SP.J.1006.2015.00160
Chen, J., Zheng, M.-j., Pang, D.-w., Yin, Y.-p., Han, M.-m., Li, Y.-x., et al. (2017). Straw return and appropriate tillage method improve grain yield and nitrogen efficiency of winter wheat. J. Integr. Agric. 16, 1708–1719. doi: 10.1016/S2095-3119(16)61589-7
Cheng, Z., Chen, Y., and Zhang, F. (2018). Effect of reclamation of abandoned salinized farmland on soil bacterial communities in arid northwest China. Sci. Total Environ. 630, 799–808. doi: 10.1016/j.scitotenv.2018.02.259
Cookson, W. R., Murphy, D. V., and Roper, M. M. (2008). Characterizing the relationships between soil organic matter components and microbial function and composition along a tillage disturbance gradient. Soil Biol. Biochem. 40, 763–777. doi: 10.1016/j.soilbio.2007.10.011
de Gannes, V., Eudoxie, G. W., and Hickey, J. (2013). Prokaryotic successions and diversity in composts as revealed by 454-pyrosequencing. Bioresour. Technol. 133, 573–580. doi: 10.1016/j.biortech.2013.01.138
de Graaff, M. A., Hornslein, N., Throop, H., Kardol, P., and van Diepen, L. T. (2019). Effects of Agricultural Intensification on Soil Biodiversity and Implications for Ecosystem Functioning: A Meta-analysis Advances in Agronomy. Cambridge, MA: Academic Press Inc. doi: 10.1016/bs.agron.2019.01.001
De la Cruz-Barrón, M., Cruz-Mendoza, A., Navarro–Noya, Y. E., Ruiz-Valdiviezo, V. M., Ortíz-Gutiérrez, D., Ramírez-Villanueva, D. A., et al. (2017). The bacterial community structure and dynamics of carbon and nitrogen when maize (Zea mays L.) and its neutral detergent fibre were added to soil from Zimbabwe with contrasting management practices. Microb. Ecol. 73, 135–152. doi: 10.1007/s00248-016-0807-8
Dick, R. P. (1992). A review: long-term effects of agricultural systems on soil biochemical and microbial parameters. Agric. Ecosyst. Environ. 40, 25–36. doi: 10.1016/0167-8809(92)90081-L
Dolan, M., Clapp, C., Allmaras, R., Baker, J., and Molina, J. (2006). Soil organic carbon and nitrogen in a Minnesota soil as related to tillage, residue and nitrogen management. Soil Tillage Res. 89, 221–231. doi: 10.1016/j.still.2005.07.015
Dong, W., Liu, E., Yan, C., Tian, J., Zhang, H., Zhang, Y., et al. (2017). Impact of no tillage vs. conventional tillage on the soil bacterial community structure in a winter wheat cropping succession in northern China. Eur. J. Soil Biol. 80, 35–42. doi: 10.1016/j.ejsobi.2017.03.001
Duan, Y., Xu, M., Gao, S., Yang, X., Huang, S., Liu, H., et al. (2014). Nitrogen use efficiency in a wheat–corn cropping system from 15 years of manure and fertilizer applications. Field Crops Res. 157, 47–56. doi: 10.1016/j.fcr.2013.12.012
Feng, Y., Motta, A. C., Reeves, D. W., Burmester, C. H., van Santen, E., Osborne, J. A., et al. (2003). Soil microbial communities under conventional-till and no-till continuous cotton systems. Soil Biol. Biochem. 35, 1693–1703. doi: 10.1016/j.soilbio.2003.08.016
Fernandez, A. (2015). Effects of Cover Crop and Fertilizer Incorporation on the Structure and Function of Microbial Communities in soils Under Long-Term Organic Management. University of Minnesota Digital Conservancy. Available online at: https://hdl.handle.net/11299/181726
Fernandez-Gnecco, G., Covacevich, F., Consolo, V. F., Behr, J. H., Sommermann, L., Moradtalab, N., et al. (2022). Effect of long-term agricultural management on the soil microbiota influenced by the time of soil sampling. Front. Soil Sci. 2, 837508. doi: 10.3389/fsoil.2022.837508
Fierer, N., Bradford, M. A., and Jackson, R. B. (2007). Toward an ecological classification of soil bacteria. Ecology 88, 1354–1364. doi: 10.1890/05-1839
Fierer, N., Leff, J. W., Adams, B. J., Nielsen, U. N., Bates, S. T., Lauber, C. L., et al. (2012). Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc. Nat. Acad. Sci. 109, 21390–21395. doi: 10.1073/pnas.1215210110
Freedman, Z. B., and Zak, D. R. (2015). Atmospheric N deposition alters connectance, but not functional potential among saprotrophic bacterial communities. Mol. Ecol. 24, 3170–3180. doi: 10.1111/mec.13224
Frey, S., Elliott, E., and Paustian, K. (1999). Bacterial and fungal abundance and biomass in conventional and no-tillage agroecosystems along two climatic gradients. Soil Biol. Biochem. 31, 573–585. doi: 10.1016/S0038-0717(98)00161-8
Frey, S. D., Ollinger, S., Nadelhoffer, K., Bowden, R., Brzostek, E., Burton, A., et al. (2014). Chronic nitrogen additions suppress decomposition and sequester soil carbon in temperate forests. Biogeochemistry 121, 305–316. doi: 10.1007/s10533-014-0004-0
Fyles, I., Juma, N., and Robertson, J. (1988). Dynamics of microbial biomass and faunal populations in long-term plots on a Gray Luvisol. Can. J. Soil Sci. 68, 91–100. doi: 10.4141/cjss88-008
Gao, J., Liu, G., Li, H., Xu, L., Du, L., Yang, B., et al. (2016). Predictive functional profiling using marker gene sequences and community diversity analyses of microbes in full-scale anaerobic sludge digesters. Bioprocess Biosyst. Eng. 39, 1115–1127. doi: 10.1007/s00449-016-1588-7
Gathala, M. K., Ladha, J., Saharawat, Y. S., Kumar, V., Kumar, V., Sharma, P. K., et al. (2011). Effect of tillage and crop establishment methods on physical properties of a medium-textured soil under a seven-year rice– wheat rotation. Soil Sci. Soc. Am. J. 75, 1851–1862. doi: 10.2136/sssaj2010.0362
Gibbons, S. M., Lekberg, Y., Mummey, D. L., Sangwan, N., Ramsey, P. W., Gilbert, J. A., et al. (2017). Invasive plants rapidly reshape soil properties in a grassland ecosystem. mSystems 2, e00178–00116. doi: 10.1128/mSystems.00178-16
Girvan, M. S., Bullimore, J., Pretty, J. N., Osborn, A. M., and Ball, A. S. (2003). Soil type is the primary determinant of the composition of the total and active bacterial communities in arable soils. Appl. Environ. Microbiol. 69, 1800–1809. doi: 10.1128/AEM.69.3.1800-1809.2003
Govaerts, B., Mezzalama, M., Sayre, K. D., Crossa, J., Nicol, J. M., Deckers, J., et al. (2006). Long-term consequences of tillage, residue management, and crop rotation on maize/wheat root rot and nematode populations in subtropical highlands. Appl. Soil Ecol. 32, 305–315. doi: 10.1016/j.apsoil.2005.07.010
Govaerts, B., Mezzalama, M., Unno, Y., Sayre, K. D., Luna-Guido, M., Vanherck, K., et al. (2007). Influence of tillage, residue management, and crop rotation on soil microbial biomass and catabolic diversity. Appl. Soil Ecol. 37, 18–30. doi: 10.1016/j.apsoil.2007.03.006
Grayston, S. J., Wang, S., Campbell, C. D., and Edwards, A. C. (1998). Selective influence of plant species on microbial diversity in the rhizosphere. Soil Biol. Biochem. 30, 369–378. doi: 10.1016/S0038-0717(97)00124-7
Guo, J., Peng, Y., Ni, B.-J., Han, X., Fan, L., Yuan, Z., et al. (2015). Dissecting microbial community structure and methane-producing pathways of a full-scale anaerobic reactor digesting activated sludge from wastewater treatment by metagenomic sequencing. Microb. Cell Fact. 14, 33. doi: 10.1186/s12934-015-0218-4
Hartman, K., van der Heijden, M. G. A., Wittwer, R. A., Banerjee, S., Walser, J.-C., Schlaeppi, K., et al. (2018). Correction to: Cropping practices manipulate abundance patterns of root and soil microbiome members paving the way to smart farming. Microbiome 6, 74. doi: 10.1186/s40168-018-0456-x
Helgason, B. L., Walley, F. L., and Germida, J. J. (2009). Fungal and bacterial abundance in long-term no-till and intensive-till soils of the Northern Great Plains. Soil Sci. Soc. Am. J. 73, 120–127. doi: 10.2136/sssaj2007.0392
Hobbs, P. R., Sayre, K., and Gupta, R. (2008). The role of conservation agriculture in sustainable agriculture. Philos. Trans. R. Soc. B Biol. Sci. 363, 543–555. doi: 10.1098/rstb.2007.2169
Huang, M., Jiang, L., Zou, Y., Xu, S., and Deng, G. (2013). Changes in soil microbial properties with no-tillage in Chinese cropping systems. Biol. Fertil. Soils 49, 373–377. doi: 10.1007/s00374-013-0778-6
Humbert, S., Tarnawski, S., Fromin, N., Mallet, M.-P., Aragno, M., Zopfi, J., et al. (2010). Molecular detection of anammox bacteria in terrestrial ecosystems: distribution and diversity. ISME J. 4, 450. doi: 10.1038/ismej.2009.125
Jansa, J., Mozafar, A., Anken, T., Ruh, R., Sanders, I., Frossard, E., et al. (2002). Diversity and structure of AMF communities as affected by tillage in a temperate soil. Mycorrhiza 12, 225–234. doi: 10.1007/s00572-002-0163-z
Jenkins, S. N., Rushton, S. P., Lanyon, C. V., Whiteley, A. S., Waite, I. S., Brookes, P. C., et al. (2010). Taxon-specific responses of soil bacteria to the addition of low level C inputs. Soil Biol. Biochem. 42, 1624–1631. doi: 10.1016/j.soilbio.2010.06.002
Jiao, Y., Hendershot, W. H., and Whalen, J. K. (2004). Agricultural practices influence dissolved nutrients leaching through intact soil cores. Soil Sci. Soc. Am. J. 68, 2058–2068. doi: 10.2136/sssaj2004.2058
Kaiser, E.-A., and Heinemeyer, O. (1993). Seasonal variations of soil microbial biomass carbon within the plough layer. Soil Biol. Biochem. 25, 1649–1655. doi: 10.1016/0038-0717(93)90166-9
Ke, X., and Scheu, S. (2008). Earthworms, Collembola and residue management change wheat (Triticum aestivum) and herbivore pest performance (Aphidina: Rhophalosiphum padi). Oecologia 157, 603–617. doi: 10.1007/s00442-008-1106-y
Kembel, S. W. (2010). Disentangling niche and neutral influences on community assembly: assessing the performance of community phylogenetic structure tests. Ecol. Lett. 12, 949–960. doi: 10.1111/j.1461-0248.2009.01354.x
Kladivko, E. J. (2001). Tillage systems and soil ecology. Soil Tillage Res. 61, 61–76. doi: 10.1016/S0167-1987(01)00179-9
Kong, L. (2014). Maize residues, soil quality, and wheat growth in China. A review. Agron. Sustain. Dev. 34, 405–416. doi: 10.1007/s13593-013-0182-5
Kraut-Cohen, J., Zolti, A., Shaltiel-Harpaz, L., Argaman, E., Rabinovich, R., Green, S. J., et al. (2020). Effects of tillage practices on soil microbiome and agricultural parameters. Sci. Total Environ. 705, 135791. doi: 10.1016/j.scitotenv.2019.135791
Kuenen, J. G. (2008). Anammox bacteria: from discovery to application. Nat. Rev. Microbiol. 6, 320. doi: 10.1038/nrmicro1857
Langille, M. G., Zaneveld, J., Caporaso, J. G., McDonald, D., Knights, D., Reyes, J. A., et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814. doi: 10.1038/nbt.2676
Li, X., Rui, J., Mao, Y., Yannarell, A., and Mackie, R. (2014). Dynamics of the bacterial community structure in the rhizosphere of a maize cultivar. Soil Biol. Biochem. 68, 392–401. doi: 10.1016/j.soilbio.2013.10.017
Li, Y., Li, T., Zhao, D., Wang, Z., and Liao, Y. (2021). Different tillage practices change assembly, composition, and co-occurrence patterns of wheat rhizosphere diazotrophs. Sci. Total Environ. 767, 144252. doi: 10.1016/j.scitotenv.2020.144252
Lian, T., Wang, G., Yu, Z., Li, Y., Liu, X., Jin, J., et al. (2016). Carbon input from 13 C-labelled soybean residues in particulate organic carbon fractions in a Mollisol. Biol. Fertil. Soils 52, 331–339. doi: 10.1007/s00374-015-1080-6
Liang, H., Wang, X., Yan, J., and Luo, L. (2019). Characterizing the intra-vineyard variation of soil bacterial and fungal communities. Front. Microbiol. 10, 1239. doi: 10.3389/fmicb.2019.01239
LiangPeng, N., LiWei, G., HaiYan, N., Jie, W., ZengJia, L., and TangYuan, N. (2015). Effects of rotational tillage on tilth soil structure and crop yield and quality in maize–wheat cropping system. Acta Agron. Sin. 41, 468–478. doi: 10.3724/SP.J.1006.2015.00468
Lin, Y.-T., Huang, Y.-J., Tang, S.-L., Whitman, W. B., Coleman, D. C., Chiu, C.-Y., et al. (2010). Bacterial community diversity in undisturbed perhumid montane forest soils in Taiwan. Microb. Ecol. 59, 369–378. doi: 10.1007/s00248-009-9574-0
Liu, J., Zhu, S., Liu, X., Yao, P., Ge, T., Zhang, X.-H., et al. (2020). Spatiotemporal dynamics of the archaeal community in coastal sediments: assembly process and co-occurrence relationship. ISME J. 14, 1463–1478. doi: 10.1038/s41396-020-0621-7
Louca, S., Parfrey, L. W., and Doebeli, M. (2016). Decoupling function and taxonomy in the global ocean microbiome. Science 353, 1272–1277. doi: 10.1126/science.aaf4507
Lu, J., Qiu, K., Li, W., Wu, Y., Ti, J., Chen, F., et al. (2019). Tillage systems influence the abundance and composition of autotrophic CO2-fixing bacteria in wheat soils in North China. Eur. J. Soil Biol. 93, 103086. doi: 10.1016/j.ejsobi.2019.103086
McBride, M. J., Liu, W., Lu, X., Zhu, Y., and Zhang, W. (2014). “The family cytophagaceae,” in The Prokaryotes: Other Major Lineages of Bacteria and The Archaea, eds E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin: Springer), 577–593. doi: 10.1007/978-3-642-38954-2_382
McGill, W., Cannon, K., Robertson, J., and Cook, F. (1986). Dynamics of soil microbial biomass and water-soluble organic C in Breton L after 50 years of cropping to two rotations. Can. J. Soil Sci. 66, 1–19. doi: 10.4141/cjss86-001
Monard, C., Mchergui, C., Nunan, N., Martin-Laurent, F., and Vieuble-Gonod, L. (2012). Impact of soil matric potential on the fine-scale spatial distribution and activity of specific microbial degrader communities. FEMS Microbiol. Ecol. 81, 673–683. doi: 10.1111/j.1574-6941.2012.01398.x
Monserrate, E., Leschine, S. B., and Canale-Parola, E. (2001). Clostridium hungatei sp. nov., a mesophilic, N2-fixing cellulolytic bacterium isolated from soil. Int. J. Syst. Evol. Microbiol. 51, 123–132. doi: 10.1099/00207713-51-1-123
Navarro-Noya, Y. E., Gómez-Acata, S., Montoya-Ciriaco, N., Rojas-Valdez, A., Suárez-Arriaga, M. C., Valenzuela-Encinas, C., et al. (2013). Relative impacts of tillage, residue management and crop-rotation on soil bacterial communities in a semi-arid agroecosystem. Soil Biol. Biochem. 65, 86–95. doi: 10.1016/j.soilbio.2013.05.009
Negassa, W. C., Guber, A. K., Kravchenko, A. N., Marsh, T. L., Hildebrandt, B., Rivers, M. L., et al. (2015). Properties of soil pore space regulate pathways of plant residue decomposition and community structure of associated bacteria. PLoS ONE 10, e0123999. doi: 10.1371/journal.pone.0123999
Palm, C., Blanco-Canqui, H., DeClerck, F., Gatere, L., and Grace, P. (2014). Conservation agriculture and ecosystem services: an overview. Agric. Ecosyst. Environ. 187, 87–105. doi: 10.1016/j.agee.2013.10.010
Panettieri, M., Berns, A. E., Knicker, H., Murillo, J., and Madejón, E. (2015). Evaluation of seasonal variability of soil biogeochemical properties in aggregate-size fractioned soil under different tillages. Soil Tillage Res. 151, 39–49. doi: 10.1016/j.still.2015.02.008
Parikh, S. J., and James, B. R. (2012). Soil: the foundation of agriculture. Nat. Educ. Knowl. 3, 2.
Ponnamperuma, F. (1984). Straw as a source of nutrients for wetland rice. Organic Matter Rice 117, 136.
Ramirez-Villanueva, D. A., Bello-López, J. M., Navarro-Noya, Y. E., Luna-Guido, M., Verhulst, N., Govaerts, B., et al. (2015). Bacterial community structure in maize residue amended soil with contrasting management practices. Appl. Soil Ecol. 90, 49–59. doi: 10.1016/j.apsoil.2015.01.010
Rodriguez, C., Alaswad, A., Benyounis, K. Y., and Olabi, A. G. (2017). Pretreatment techniques used in biogas production from grass. Renew. Sust. Energ. Rev. 68, 1193–1204. doi: 10.1016/j.rser.2016.02.022
Sarrantonio, M., and Gallandt, E. (2003). The role of cover crops in North American cropping systems. J. Crop Prod. 8, 53–74. doi: 10.1300/J144v08n01_04
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Schmidt, R., Gravuer, K., Bossange, A. V., Mitchell, J., and Scow, K. (2018). Long-term use of cover crops and no-till shift soil microbial community life strategies in agricultural soil. PLoS ONE 13, e0192953. doi: 10.1371/journal.pone.0192953
Sengupta, A., and Dick, W. A. (2015). Bacterial community diversity in soil under two tillage practices as determined by pyrosequencing. Microb. Ecol. 70, 853–859. doi: 10.1007/s00248-015-0609-4
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Šíp, V., Vavera, R., Chrpová, J., Kusá, H., and Ružek, P. (2013). Winter wheat yield and quality related to tillage practice, input level and environmental conditions. Soil Tillage Res. 132, 77–85. doi: 10.1016/j.still.2013.05.002
Six, J., Elliott, E., and Paustian, K. (2000). Soil macroaggregate turnover and microaggregate formation: a mechanism for C sequestration under no-tillage agriculture. Soil Biol. Biochem. 32, 2099–2103. doi: 10.1016/S0038-0717(00)00179-6
Six, J., Frey, S., Thiet, R., and Batten, K. (2006). Bacterial and fungal contributions to carbon sequestration in agroecosystems. Soil Sci. Soc. Am. J. 70, 555–569. doi: 10.2136/sssaj2004.0347
Song, X., Zeng, J., Zhou, Y., Chen, Q., Yang, H., Shou, L., et al. (2021). Partial function prediction of sulfate-reducing bacterial community from the rhizospheres of two typical coastal wetland plants in China. J. Oceanol. Limnol. 39, 185–197. doi: 10.1007/s00343-019-9177-7
Souza, R. C., Mendes, I. C., Reis-Junior, F. B., Carvalho, F. M., Nogueira, M. A., Vasconcelos, A. T. R., et al. (2016). Shifts in taxonomic and functional microbial diversity with agriculture: how fragile is the Brazilian Cerrado? BMC Microbiol. 16, 42. doi: 10.1186/s12866-016-0657-z
Sparks, D. L., Page, A. L., Helmke, P. A., Loeppert, R. H., Soltanpour, P. N., Tabatabai, M. A., et al. (1996). Methods of Soil Analysis. Part 3 - Chemical Methods. doi: 10.2136/sssabookser5.3
Spedding, T., Hamel, C., Mehuys, G., and Madramootoo, C. (2004). Soil microbial dynamics in maize-growing soil under different tillage and residue management systems. Soil Biol. Biochem. 36, 499–512. doi: 10.1016/j.soilbio.2003.10.026
Stegen, J. C., Lin, X., Konopka, A. E., and Fredrickson, J. K. (2012). Stochastic and deterministic assembly processes in subsurface microbial communities. ISME J. 6, 1653–1664. doi: 10.1038/ismej.2012.22
Stegen, J. C., Lin, X., Fredrickson, J. K., Chen, X., Kennedy, D. W., Murray, C. J., et al. (2013). Quantifying community assembly processes and identifying features that impose them. ISME J. 7, 2069–2079. doi: 10.1038/ismej.2013.93
Su, P., Lou, J., Brookes, P. C., Luo, Y., He, Y., Xu, J., et al. (2017). Taxon-specific responses of soil microbial communities to different soil priming effects induced by addition of plant residues and their biochars. J. Soils Sediments 17, 674–684. doi: 10.1007/s11368-015-1238-8
Su, Y., Yu, M., Xi, H., Lv, J., Ma, Z., Kou, C., et al. (2020). Soil microbial community shifts with long-term of different straw return in wheat-corn rotation system. Sci. Rep. 10, 6360. doi: 10.1038/s41598-020-63409-6
Sun, R., Li, W., Dong, W., Tian, Y., Hu, C., Liu, B., et al. (2018). Tillage changes vertical distribution of soil bacterial and fungal communities. Front. Microbiol. 9, 699. doi: 10.3389/fmicb.2018.00699
Tian, S., Ning, T., Zhao, H., Wang, B., Li, N., Han, H., et al. (2012). Response of CH4 and N2O emissions and wheat yields to tillage method changes in the North China Plain. PLoS ONE 7, e51206. doi: 10.1371/journal.pone.0051206
Trivedi, P., Delgado-Baquerizo, M., Jeffries, T. C., Trivedi, C., Anderson, I. C., Lai, K., et al. (2017). Soil aggregation and associated microbial communities modify the impact of agricultural management on carbon content. Environ. Microbiol. 19, 3070–3086. doi: 10.1111/1462-2920.13779
Uri, N. D. (1999). Factors affecting the use of conservation tillage in the United States. Water Air Soil Pollut. 116, 621–638. doi: 10.1023/A:1005168928627
Wallenstein, M. D., McMahon, S., and Schimel, J. (2007). Bacterial and fungal community structure in Arctic tundra tussock and shrub soils. FEMS Microbiol. Ecol. 59, 428–435. doi: 10.1111/j.1574-6941.2006.00260.x
Wang, X.-B., Cai, D.-X., Hoogmoed, W. B., Oenema, O., and Perdok, U. D. (2006). Potential effect of conservation tillage on sustainable land use: a review of global long-term studies. Pedosphere 16, 587–595. doi: 10.1016/S1002-0160(06)60092-1
Wang, Y., Li, C., Tu, C., Hoyt, G. D., DeForest, J. L., Hu, S., et al. (2017). Long-term no-tillage and organic input management enhanced the diversity and stability of soil microbial community. Sci. Total Environ. 609, 341–347. doi: 10.1016/j.scitotenv.2017.07.053
Wang, Y., Tu, C., Cheng, L., Li, C., Gentry, L. F., Hoyt, G. D., et al. (2011). Long-term impact of farming practices on soil organic carbon and nitrogen pools and microbial biomass and activity. Soil Tillage Res. 117, 8–16. doi: 10.1016/j.still.2011.08.002
Wang, Z., Li, Y., Li, T., Zhao, D., and Liao, Y. (2020). Tillage practices with different soil disturbance shape the rhizosphere bacterial community throughout crop growth. Soil Tillage Res. 197, 104501. doi: 10.1016/j.still.2019.104501
Wang, Z., Liu, L., Chen, Q., Wen, X., and Liao, Y. (2016). Conservation tillage increases soil bacterial diversity in the dryland of northern China. Agron. Sustain. Dev. 36, 1–9. doi: 10.1007/s13593-016-0366-x
Webb, C. O., Ackerly, D. D., McPeek, M. A., and Donoghue, M. J. (2002). Phylogenies and community ecology. Annu. Rev. Ecol. Syst. 33, 475–505. doi: 10.1146/annurev.ecolsys.33.010802.150448
Xia, X., Zhang, P., He, L., Gao, X., Li, W., Zhou, Y., et al. (2019). Effects of tillage managements and maize straw returning on soil microbiome using 16S rDNA sequencing. J. Integr. Plant Biol. 61, 765–777. doi: 10.1111/jipb.12802
Xiong, Q., Li, L., Luo, X., He, X., Zhang, L., Pan, K., et al. (2021). Driving forces for recovery of forest vegetation after harvesting a subalpine oak forest in eastern Tibetan Plateau. Environ. Sci. Pollut. Res. 28, 67748–67763. doi: 10.1007/s11356-021-15367-3
Xu, Y., Nie, L., Buresh, R. J., Huang, J., Cui, K., Xu, B., et al. (2010). Agronomic performance of late-season rice under different tillage, straw, and nitrogen management. Field Crops Res. 115, 79–84. doi: 10.1016/j.fcr.2009.10.005
Xue, R., Zhao, K., Yu, X., Stirling, E., Liu, S., Ye, S., et al. (2021). Deciphering sample size effect on microbial biogeographic patterns and community assembly processes at centimeter scale. Soil Biol. Biochem. 156, 108218. doi: 10.1016/j.soilbio.2021.108218
Yang, Y., Yu, K., Xia, Y., Lau, F. T., Tang, D. T., Fung, W. C., et al. (2014). Metagenomic analysis of sludge from full-scale anaerobic digesters operated in municipal wastewater treatment plants. Appl. Microbiol. Biotechnol. 98, 5709–5718. doi: 10.1007/s00253-014-5648-0
Yin, C., Jones, K. L., Peterson, D. E., Garrett, K. A., Hulbert, S. H., Paulitz, T. C., et al. (2010). Members of soil bacterial communities sensitive to tillage and crop rotation. Soil Biol. Biochem. 42, 2111–2118. doi: 10.1016/j.soilbio.2010.08.006
Zaneveld, J. R., Lozupone, C., Gordon, J. I., and Knight, R. (2010). Ribosomal RNA diversity predicts genome diversity in gut bacteria and their relatives. Nucleic Acids Res. 38, 3869–3879. doi: 10.1093/nar/gkq066
Zhang, B., He, H., Ding, X., Zhang, X., Zhang, X., Yang, X., et al. (2012). Soil microbial community dynamics over a maize (Zea mays L.) growing season under conventional-and no-tillage practices in a rainfed agroecosystem. Soil Tillage Res. 124, 153–160. doi: 10.1016/j.still.2012.05.011
Zhang, C., Liu, H., Liu, S., Hussain, S., Zhang, L., Yu, X., et al. (2022). Response of fungal sub-communities in a maize-wheat rotation field subjected to long-term conservation tillage management. Front. Microbiol. 13, 829152. doi: 10.3389/fmicb.2022.829152
Zhang, C.-J., Pan, J., Duan, C.-H., Wang, Y.-M., Liu, Y., Sun, J., et al. (2019). Prokaryotic diversity in mangrove sediments across southeastern china fundamentally differs from that in other biomes. mSystems 4, e00442–00419. doi: 10.1128/mSystems.00442-19
Zhang, S., Li, M., Cui, X., and Pan, Y. (2023). Effect of different straw retention techniques on soil microbial community structure in wheat–maize rotation system. Front. Microbiol. 13, 1069458. doi: 10.3389/fmicb.2022.1069458
Zhang, W., Li, Y., Wang, C., Wang, P., Hou, J., Yu, Z., et al. (2016). Modeling the biodegradation of bacterial community assembly linked antibiotics in river sediment using a deterministic–stochastic combined model. Environ. Sci. Technol. 50, 8788–8798. doi: 10.1021/acs.est.6b01573
Zhang, X., Xin, X., Zhu, A., Yang, W., Zhang, J., Ding, S., et al. (2018). Linking macroaggregation to soil microbial community and organic carbon accumulation under different tillage and residue managements. Soil Tillage Res. 178, 99–107. doi: 10.1016/j.still.2017.12.020
Zhang, Y., Zhang, M., Tang, L., Che, R., Chen, H., Blumfield, T., et al. (2018). Long-term harvest residue retention could decrease soil bacterial diversities probably due to favouring oligotrophic lineages. Microb. Ecol. 76, 771–781. doi: 10.1007/s00248-018-1162-8
Zhang, Y. X., Yang, J. L., Liu, W., Li, L., Gao, Y. J., Xiao, L. L., et al. (2009). Effects of nitrogen rate on nitrogen uptake and utilization of winter wheat and residual nitrate in soil under straw mulch. Agric. Res. Arid Areas 27, 189–193.
Zhao, Y., Guo, H., Xue, Z., Mu, X., and Li, C. (2014). Effects of tillage and straw returning on biomass and water use efficiency in a winter wheat and summer maize rotation system. Acta Agron. Sin. 40, 1797–1807. doi: 10.3724/SP.J.1006.2014.01797
Zhou, J., and Ning, D. (2017). Stochastic community assembly: does it matter in microbial ecology? Microbiol. Mol. Biol. Rev. 81, e00002–00017. doi: 10.1128/MMBR.00002-17
Keywords: conservation tillage, soil properties, soil bacterial community structure, Illumina sequencing, KEGG pathway, bacterial network analysis
Citation: Khan MH, Liu H, Zhu A, Khan MH, Hussain S and Cao H (2023) Conservation tillage practices affect soil microbial diversity and composition in experimental fields. Front. Microbiol. 14:1227297. doi: 10.3389/fmicb.2023.1227297
Received: 23 May 2023; Accepted: 06 July 2023;
Published: 02 August 2023.
Edited by:
Ruibo Sun, Anhui Agricultural University, ChinaReviewed by:
Talat Bilal Yasoob, Ghazi University, PakistanCopyright © 2023 Khan, Liu, Zhu, Khan, Hussain and Cao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hui Cao, aGNhb0BuamF1LmVkdS5jbg==; Sarfraz Hussain, MjAyMTA5MzZAaGh1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.