
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 19 September 2023
Sec. Microbe and Virus Interactions with Plants
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1226142
This article is part of the Research Topic Microbiome Associated With Plant Pathogens, Pathogenesis, and Their Applications in Developing Sustainable Agriculture View all 17 articles
Shihezi Reclamation Area is located at the southern edge of the Junggar Basin, with natural, soil, and climatic conditions unique to the production of peaches. In turn, peach orchards have accumulated rich microbial resources. As an important taxon of soil fungi, the diversity and community structure changes of yeast in the soil of peach orchards on spatial and temporal scales are still unknown. Here, we aimed to investigate the changes in yeast diversity and community structure in non-rhizosphere and rhizosphere soils of peach trees of different ages in the peach orchard and the factors affecting them, as well as the changes in the yeast co-occurrence network in the peach orchard at spatial and temporal scales. High-through put sequencing results showed that a total of 114 yeast genera were detected in all soil samples, belonging to Ascomycota (60 genera) and Basidiomycota (54 genera). The most dominant genus, Cryptococcus, was present in greater than 10% abundance in each sample. Overall, the differences in yeast diversity between non-rhizosphere and rhizosphere soil of peach trees at 3, 8 and 15 years were not significant. Principal coordinate analysis (PCoA) showed that differences in yeast community structure were more pronounced at the temporal scale compared to the spatial scale. The results of soil physical and chemical analysis showed that the 15-year-old peach rhizosphere soil had the lowest pH, while the OM, TN, and TP contents increased significantly. Redundancy analysis showed that soil pH and CO were key factors contributing to changes in soil yeast community structure in the peach orchard at both spatial and temporal scales. The results of co-occurrence network analysis showed that the peach orchard soil yeast network showed synergistic effects as a whole, and the degree of interactions and connection tightness of the 15-year-old peach orchard soil yeast network were significantly higher than the 3- and 8-year-old ones on the time scale. The results reveal the distribution pattern and mechanism of action of yeast communities in peach orchard soils, which can help to develop effective soil management strategies and improve the stability of soil microecology, thus promoting crop growth.
Yeast is known as the first “domesticated microorganism” in human history and is widely used in food fermentation, industry and agriculture, and pharmaceutical production because of its short growth cycle, high metabolic efficiency, and production of beneficial metabolites (Zhao et al., 2004). In recent years, yeasts have been shown to play a key role also in the biosorption of heavy metal ions in the environment, in building the balance of ecosystems, and in the prevention and control of polluted environments (He et al., 2022; Igwegbe et al., 2022). Yeast has an extremely wide distribution habitat, and since it prefers acidic and sugar-rich habitats, it is usually found in orchards, mainly from different organs of fruit trees such as leaves, flowers and fruits, as well as from orchard soil (Yang and Wang, 2009). Not only can novel yeast species be found in the orchard soil (Chen et al., 2010, 2012), but also many yeasts with excellent performance can be screened to add unique flavor to the fruit wine (Chen et al., 2018; Wang et al., 2022). There are also some orchard soil yeasts that can be used to control fruit tree diseases or to alleviate post-harvest diseases of fruits to extend the shelf life of fruit (Wang et al., 2016; Ferraz et al., 2019). In addition, some soil yeasts can indirectly promote plant root growth and development by enhancing colonization of arbuscular mycorrhizal (AM) fungal in host plants (Boby et al., 2008; Mirabal Alonso et al., 2008), and there are also yeasts that possess the ability to solubilize rock phosphate or produce plant growth regulators to improve plant growth directly (Nassar et al., 2005; El-Tarabily and Sivasithamparam, 2006; Kumla et al., 2020). Because of this, soil conditioners containing yeast have been developed to improve crop productivity (Ito and Ito, 2001). Research is also ongoing on the potential of soil yeast as a biofertilizer, which can also reduce the damage to the soil caused by conventional chemical fertilizers to some extent and rebuild the maintenance capacity of agroecosystems (Hernández-Fernández et al., 2021; Marques et al., 2021). Therefore, the study of yeast diversity in orchard soils is still of great importance and can provide rich strain resources for social production. In fact, in addition to exploring the function of soil yeasts in orchards, the search for relevant factors affecting the diversity and composition of yeast communities has also never stopped.
Soil yeast diversity and community structure are generally influenced by factors such as soil type, type of vegetation covered, climatic conditions, and geographic location (Vadkertiová et al., 2017). For instance, in vineyards, different grape types’ cultivated soils varied in their diversity of yeast composition and abundance of the same yeast species, and there was a significant relationship between some yeast species and particular grape varieties (Zhang et al., 2017). Moreover, distinct yeast strains obtained from various vineyard soils resulted in noticeably diverse fermentation flavors, providing directions for future screening of edible grape yeasts and genetic improvement of edible grapes (Wang et al., 2022). Yeast populations living in soil under various fruit tree species varied in species richness and evenness, with the highest species richness in soil next to apricot trees, according to a previous study of yeasts in 200 soil samples from five fruit tree species (apple, pear, plum, peach, and apricot) in two regions of Slovakia (Vadkertiová et al., 2019). Numerous earlier studies have demonstrated that the number of yeast cells tends to decrease as soil depth increases. This is because deeper soil contains less efficient nutrients and soil organic matter, which is unfavorable for the survival of yeast (Starmer and Lachance, 2011; Yurkov, 2018). As well, rhizosphere soil yeasts are more numerous than bulk soil due to their ease of uptake of simple organic carbon compounds secreted by plant roots and their ability to feed on spoilage fruits deposited in the soil (Cloete et al., 2009; Botha, 2011). Understanding the survival mechanism of yeast in orchard soil ecosystem is crucial for the growth and application of yeast resources.
Peach is rich in many essential substances for human body, including protein, crude fiber, various amino acids, carotenoids and minerals such as iron and phosphorus (Yin et al., 2017), and is known as the “first fruit of the world” (Zhou and Zhang, 2009). Shihezi, Xinjiang, is located in the middle of the northern foot of Tianshan Mountains and the southern edge of Junggar Basin (Fan et al., 2006). It has a dry climate, abundant light, heat and water sources, a wide day-night temperature range, long daylight hours, drought, and little rain during the fruiting season of fruit trees, all of which are very conducive to the accumulation of sugar and dry matter in fruits as well as the accumulation of rich yeast resources (Xu et al., 2015). Previous studies on microorganisms in peach orchards have focused either on the biological control of postharvest peach fruit diseases, on the structural analysis of peach leaf-attached yeast communities in peach orchards by a culture-independent method, or on the culturable yeast diversity of soil in peach orchards by a culture-dependent method (Yin et al., 2017; Liu et al., 2019; Wang et al., 2019). However, the yeast diversity and community composition in non-rhizosphere and rhizosphere soils of perennial peach trees containing both culturable and nonculturable yeasts, and whether they are influenced by annual (yearly) variation and soil factors, are not yet known, which will be a bottleneck for studying yeast adaptation mechanisms in peach orchards and their further development and utilization.
As a natural breeding ground for microorganisms, each gram of soil may contain millions of microbial species (Bunge et al., 2006), and most (>99%) are non-culturable microorganisms. Illumina MiSeq high-throughput sequencing technology allows for more comprehensive and accurate detection of species composition compared to traditional culture-dependent methods (Zebin et al., 2016). Here, we studied the diversity and composition of yeast communities associated with the non-rhizosphere and rhizosphere soil of peach trees of different ages based on Illumina MiSeq high-throughput sequencing technology in peach orchard of Shihezi, Xinjiang. The aim was to explore the differences in soil yeast diversity and community structure in the peach orchard at temporal and spatial scales, the correlation between soil factors and yeast communities, the variation of the soil yeast co-occurrence network in the peach orchard at spatial and temporal scales, and the mechanism of yeast action in the network. Our study provides supplemental information for a comprehensive understanding of peach orchard yeast resources, factors affecting soil yeast composition in perennial peach trees, and the mechanisms of action among yeasts in peach orchard soils, as well as some ideas for achieving sustainable agricultural development in the peach orchard.
Our soil samples were collected from a peach orchard in Shihezi 143rd Regiment (N44°28′, E85°82′, Altitude 450 m), Xinjiang. The soil type is gray desert soil with a medium loamy texture, cultivated at a depth of 80–100 cm with good permeability, and the peach trees are planted with a spacing of 5.0 m between plants and 6.0 m between rows, with a density of 330 plants per hectare (ha). The site is planted on 500 hectares and has year-round good irrigation conditions. All peach trees of the peach orchard are Amygdalus persica L. “Compressa.” The climate of the Shihezi region is typical of a temperate continental climate, with long and severe winters and short and hot summers (Han et al., 2008). The climate information of the sampling sites is shown in Table 1. The data of precipitation (PRECTP) and temperature (TEMP) from NOAA—Climate Prediction Center,1 land surface temperature (LST) and relative humidity (RH) from NASA GES DISC MERRA2—inst1_2d_asm_Nx,2 evaporation land (EVLAND) from NASA GES DISC MERRA2—tavg1_2d_lnd_Nx (see footnote 2).
Non-rhizosphere and rhizosphere soils were collected from peach trees of 3, 8 and 15 years of age, respectively. Non-rhizosphere soil samples, i.e., bulk soil samples (S3B, S8B, S15B) were collected at a distance of 1 m from the main trunk and at a depth of 30 cm, while rhizosphere soil samples (S3R, S8R, S15R) were collected at a distance of 30 cm from the main trunk and at a depth of 30 cm. Samples were collected using a five-point sampling method. Specifically, five peach trees each of three, eight, and fifteen years of age were randomly selected, and non-rhizosphere and rhizosphere soils were collected from each tree separately according to the previously described requirements, and plant residues and stones were removed with a shovel and sieve. Each of the five soil samples of non-rhizosphere and rhizosphere soils from the same tree age was then homogeneously mixed and divided into three equal parts, respectively. Both non-rhizosphere and rhizosphere soil samples have three replicates for each tree age, for a total of 18 samples. Samples were collected at the fruiting stage. Each sample was stored individually in sterile self-sealing bags and transported to the laboratory immediately, then filtered with a 2 mm sieve. They were divided into two parts: one part stored at room temperature for soil physicochemical analysis; the other part was stored in a −20°C refrigerator for subsequent DNA extraction.
PowerSoil® DNA Isolation Kit (MoBio Laboratories) was used to extract total DNA in triplicate from soil samples (0.25 g) by using manufacturer’s protocol. The DNA extractions from the same soil sample were combined, then quantified using a NanoDrop 2000 UV–Vis spectrophotometer (Thermo Scientific, United States). The integrity of the DNA was detected using 0.8% agarose gel electrophoresis. The purity and amount of DNA is shown in Supplementary Table S1. The region D1 of the LSU rRNA gene was amplified with a pair of specific primers with barcode NL1F (5′-GCATATCAATAAGCGGAGGAAAAG-3′) and NL2R (5′-CTTGTTCGCTATCGGTCTC-3′; Liu et al., 2018). The PCR reaction system were performed in 20 μL volume containing 5× FastPfu Buffer (4 μL), 5 μM forward primer and reverse primer (0.8 μL each), 2.5 mM dNTPs (2 μL), 0.4 μL FastPfu Polymerase, 0.2 μL BSA, and 10 ng of the template DNA. An equal amount of sterile water instead of template DNA was used as a negative control. Amplification was initiated with 5 min at 98°C, followed by 30 cycles of denaturation at 98°C for 30 s, primer annealing at 52°C for 30 s, extension at 72°C for 45 min, and final extension for 5 min at 72°C. Reactions, performed in triplicate, were combined. The PCR products were purified by using 2% agarose gel electrophoresis followed by the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, United States) and quantified using QuantiFluor™-ST (Promega, United States; Zhang et al., 2018). High-throughput sequencing with the Illumina MiSeq PE300 platform (Illumina, United States) was performed by using paired-end sequencing, which follows the instructions by Personal Biotechnology Co., Ltd. (Shanghai, China).
The raw Illumina sequences were assigned to individual samples based on their unique barcodes. Raw sequence files were then demultiplexed, quality filtered and analyzed by merging Paired-end reads with FLASH and the Quantitative Insight Into Microbial Ecology (QIIME) software package, respectively (Caporaso et al., 2010; Magoč and Salzberg, 2011). Operational taxonomic unit (OTU) were clustered using UPARSE (version 7.1; http://drive5.com/uparse/) with a threshold of 97% pairwise identity (Edgar, 2013). UCHIME software was used to identify and remove the chimeric sequences. OTUs were classified taxonomically using the Ribosomal Database Project (RDP) classifier (version 2.2; http://sourceforge.net/pro-jects/rdp-classifier/; Wang et al., 2007) against the database of National Centre for Biotechnology Information (NCBI; National Centre for Biotechnology Information, https://www.ncbi.nlm.nih.gov/public/). Any OTUs representing non-yeast sequences were removed before down-stream analysis (Bokulich et al., 2013).
The soil water suspension was shaken for 30 min and then measured by a glass electrode meter for pH value. Conductivity (CO) was measured by electrode method after mixing a naturally dried soil sample with water at a ratio of 1:5 (M/V). Soil water content (SWC) was measured by the drying method, where a moist soil sample of known weight is dried in an oven and then weighed, and the moisture lost by heating represents the soil moisture in the moist sample. Organic matter (OM) was measured by titration with ferrous sulfate, using o-phenanthroline as the indicator. The total nitrogen (TN) was determined by the Kjeldahl method. Total phosphorus (TP) and total potassium (TK) were measured by acid solubilization (Bao, 2007).
The observed richness (Sobs), ACE index, Chao1 estimator, Shannon diversity index and Simpson diversity index of the samples were calculated using QIIME (Caporaso et al., 2010). And the rarefaction curve was plotted based on the diversity index using the “vegan” and “ggplot2” packages in R (v4.3.0). Alpha diversity and soil physical and chemical properties were compared between samples by SPSS Statistics v25.0 software (IBM, United States) based on Kruskal-Wallis test. All values are presented as mean ± standard error (mean ± SE). Differences were taken statistically significant at p < 0.05. The Venn diagram was drawn using the “VennDiagram” package and the community bar graph was plotted using “ggplot2” and “ggalluvial” packages in R (v4.2.1). Heatmap were created based on the “vegan” and “pheatmap” packages in R (v4.3.0) to analyze the differences in dominant genera across samples. Principal co-ordinate analysis (PCoA) was done based on Bray-Curtis at OTU level to analyze similarities or differences in the community composition of samples using “vegan” and “ape” packages in R (v4.3.0). Tests for differences between groups in PCoA were analyzed using ANOSIM (analysis of similarities) by vegan package in R. Redundancy analysis (RDA) was used to evaluate the relationships between soil factors and yeast communities, and the plot was drawn by the “vegan” and “ggplot2” packages in R (v4.3.0). Construct a co-occurrence network for each sample group based on the absolute abundance of OTUs. Network topological properties were calculated using the “igraph” package in R (v4.3.0). To reduce network complexity and ensure network reliability, the co-occurrence networks at the genus level were constructed by retaining OTUs with relative abundance ≥ 0.01% and Spearman’s correlation coefficient |r| ≥ 0.6, with significance p < 0.01. The co-occurrence network was visualized using Gephi (v.0.10.0; Barberán et al., 2012; Meng et al., 2022). The node data and edge data files for each sample used to generate the co-occurrence network graphs are shown in Supplementary Tables S2–S13.
According to the age of the peach trees and the sampling locations, we divided the 18 soil samples into 6 groups, named S3B (3-year-old non-rhizosphere), S3R (3-year-old rhizosphere), S8B (8-year-old non-rhizosphere), S8R (8-year-old rhizosphere), S15B (15-year-old non-rhizosphere), and S15R (15-year-old rhizosphere). Based on high-throughput sequencing of the D1 domain of the large subunit (LSU) rRNA gene, we obtained a total of 968,960 sequences from 18 soil samples after removing chimeras and sequences with low-quality reads, and 48,114 yeast sequence reads after excluding non-yeast sequence reads. All yeast sequence reads were clustered into 3,103 operational taxonomic units (OTUs) based on 97% similarity. The yeast rarefaction curves of all samples tended to be flat, indicating that the sequencing depth of the samples was sufficient and the sampling was reasonable (Figure 1).
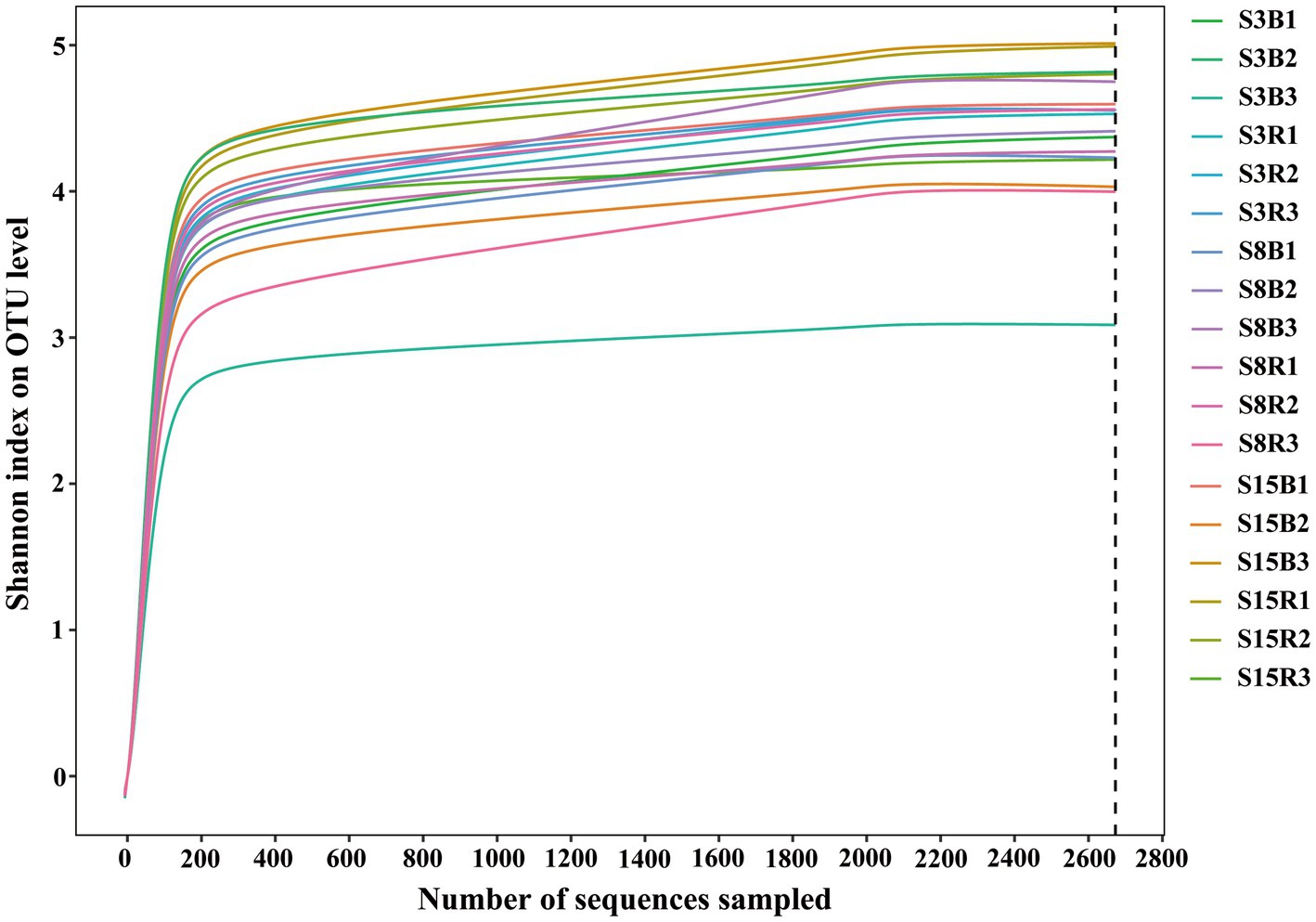
Figure 1. Rarefaction curves of all soil samples. Rarefaction curves of OTUs were clustered for a dissimilarity threshold of 3%. S3B, S3R, S8B, S8R, S15B, and S15R represent non-rhizosphere and rhizosphere soil samples from 3-year-old, 8-year-old, and 15-year-old peach trees, respectively. Each sample had three replicates.
The results of alpha diversity analysis showed that there were no significant differences in the observed species richness (Sobs), species richness (Chao1 and ACE indices) and species diversity (Shannon and Simpson indices; p > 0.05; Table 2). The OTUs of S3 (3-year rhizosphere and non-rhizosphere), S8 (8-year rhizosphere and non-rhizosphere) and S15 (15-year rhizosphere and non-rhizosphere) were 1,542, 1,568, and 1,581, respectively. We found that 454 OTUs shared between S3, S8 and S15; 700 OTUs are shared between S3 and S8, 755 OTUs between S3 and S15 and 587 OTUs between S8 and S15. The OTUs of non-rhizosphere (S3B, S8B and S15B) and rhizosphere (S3R, S8R, and S15R) were 2,095 and 2,115, respectively. Among them, the OTUs of S3B, S8B, and S15B were 1,037, 1,027, and 985, respectively; 261 OTUs shared between S3B, S8B, and S15B; 426 OTUs are shared between S3B and S8B, 442 OTUs between S3B and S15B and 347 OTUs between S8B and S15B. The OTUs of S3R, S8R, and S15R were 973, 970, and 1,037, respectively; 222 OTUs shared between S3R, S8R, and S15R; 359 OTUs are shared between S3R and S8R, 425 OTUs between S3R and S15R and 303 OTUs between S8R and S15R (Figure 2).
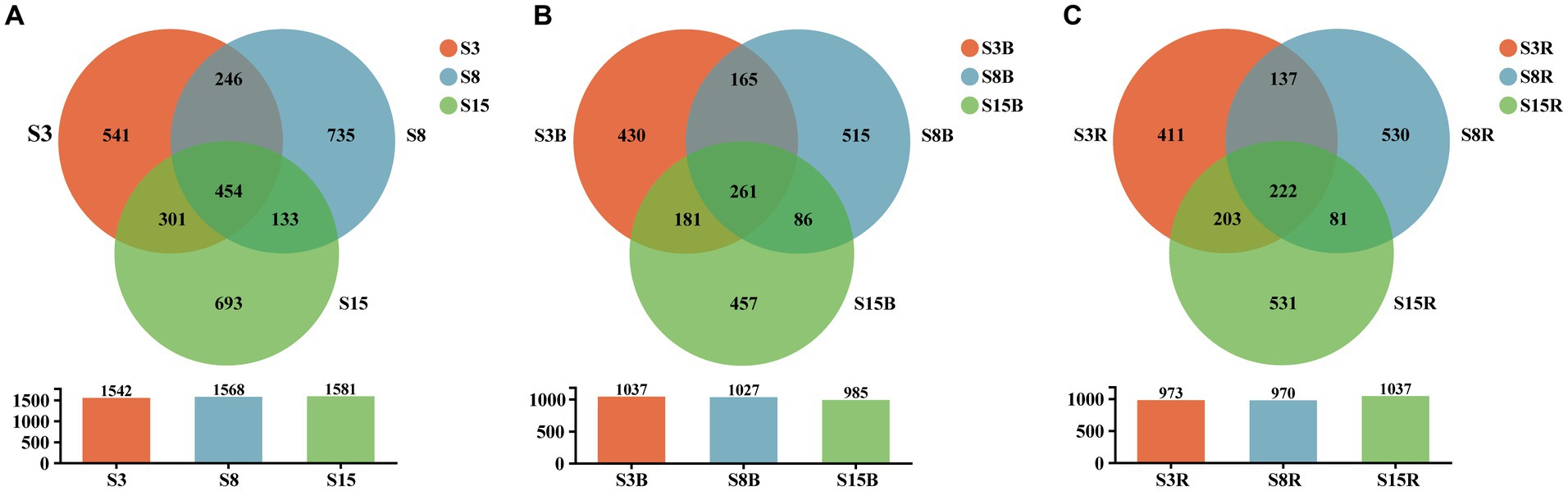
Figure 2. Venn diagram at the OTU level of soil samples from (A) non-rhizosphere and rhizosphere of 3-year-old (S3), 8-year-old (S8), and 15-year-old (S15) peach trees, (B) non-rhizosphere of 3-year-old (S3B), 8-year-old (S8B), and 15-year-old (S15B) peach trees and (C) rhizosphere of 3-year-old (S3R), 8-year-old (S8R), and 15-year-old (S15R) peach trees, respectively. Each circle with different colors in the diagram represents a group; middle core numbers represent the number of OTUs common to all groups. The shared and unique yeast OTUs were shown at a 0.03 dissimilarity distance after removing singletons.
Next, 3,103 OTUs were identified as two phyla (Ascomycota and Basidiomycota) and 114 genera. Ascomycota contained 60 genera accounting for 44.78% of all yeast sequences, and Basidiomycota had 54 genera accounting for 55.22% (Tables 3, 4). These include 19 dominant genera that accounted for greater than 1% were Cryptococcus (21.40%), Pichia (9.615%), Clavispora (9.249%), Tausonia (5.491%), Zygosaccharomyces (4.849%), Solicoccozyma (4.527%), Udeniomyces (3.912%), Candida (3.294%), Filobasidium (3.284%), Trigonopsis (3.124%), Aureobasidium (3.024%), Papiliotrema (2.822%), Saturnispora (2.752%), Rhodotorula (2.060%), Saitozyma (2.058%), Cyniclomyces (1.567%), Goffeauzyma (1.814%), Naganishia (1.594%), and Cyberlindnera (1.122%). The 19 dominant genera accounted for 87.558% of all yeast sequences. In addition, a total of 15 rare yeast genera (genus with less than 10% frequency of occurrence) were included in all soil samples, accounting for approximately 13.16% of all yeast genera. 33 yeast genera were shared by 18 soil samples, and 24 yeast genera were significantly different among S3B, S3R, S8B, S8R, S15B, and S15R groups (p < 0.05; Tables 3, 4).
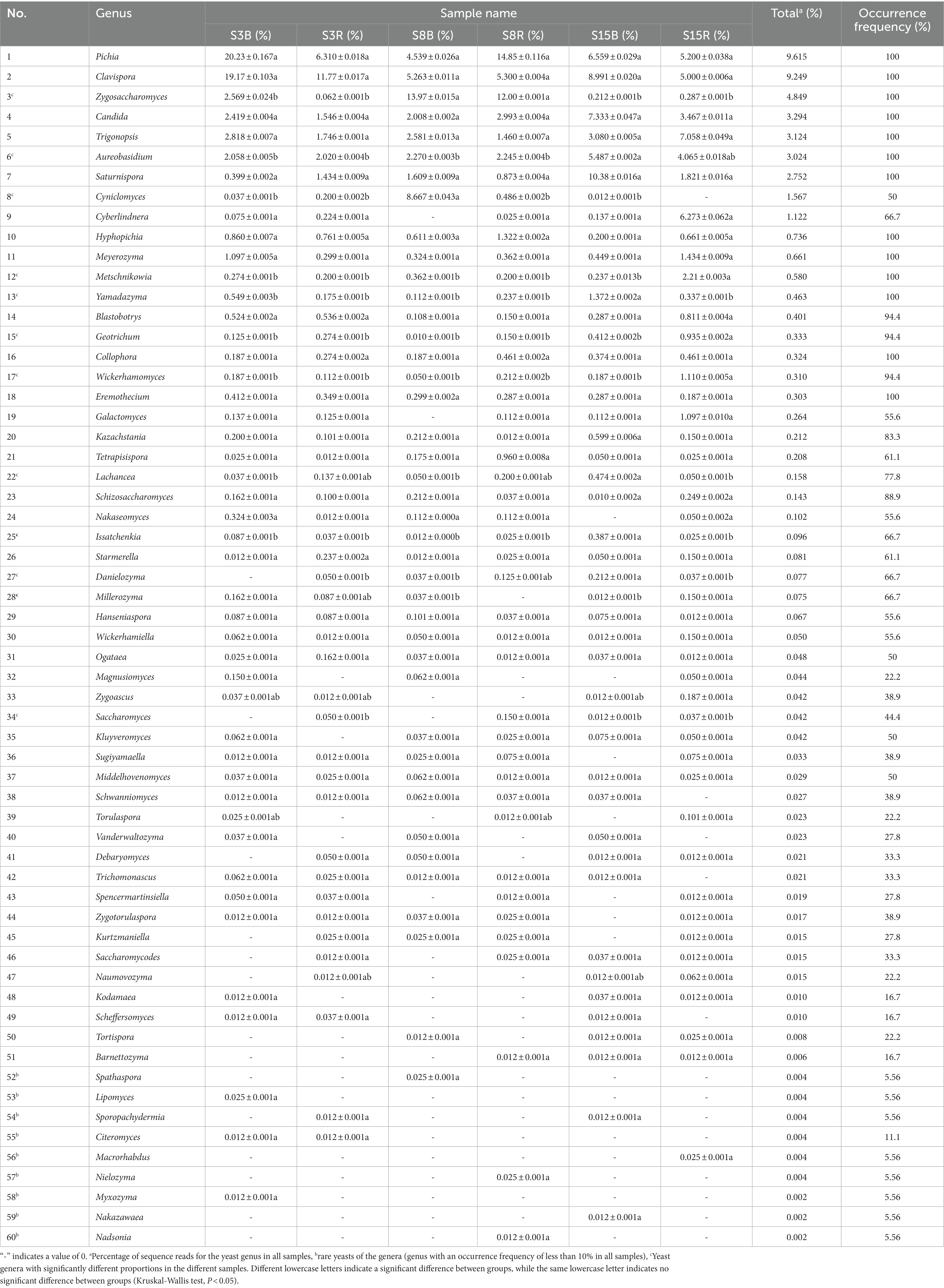
Table 3. The percentage and frequency of occurrence of Ascomycetous yeasts (accounted for 44.78%) in all samples.
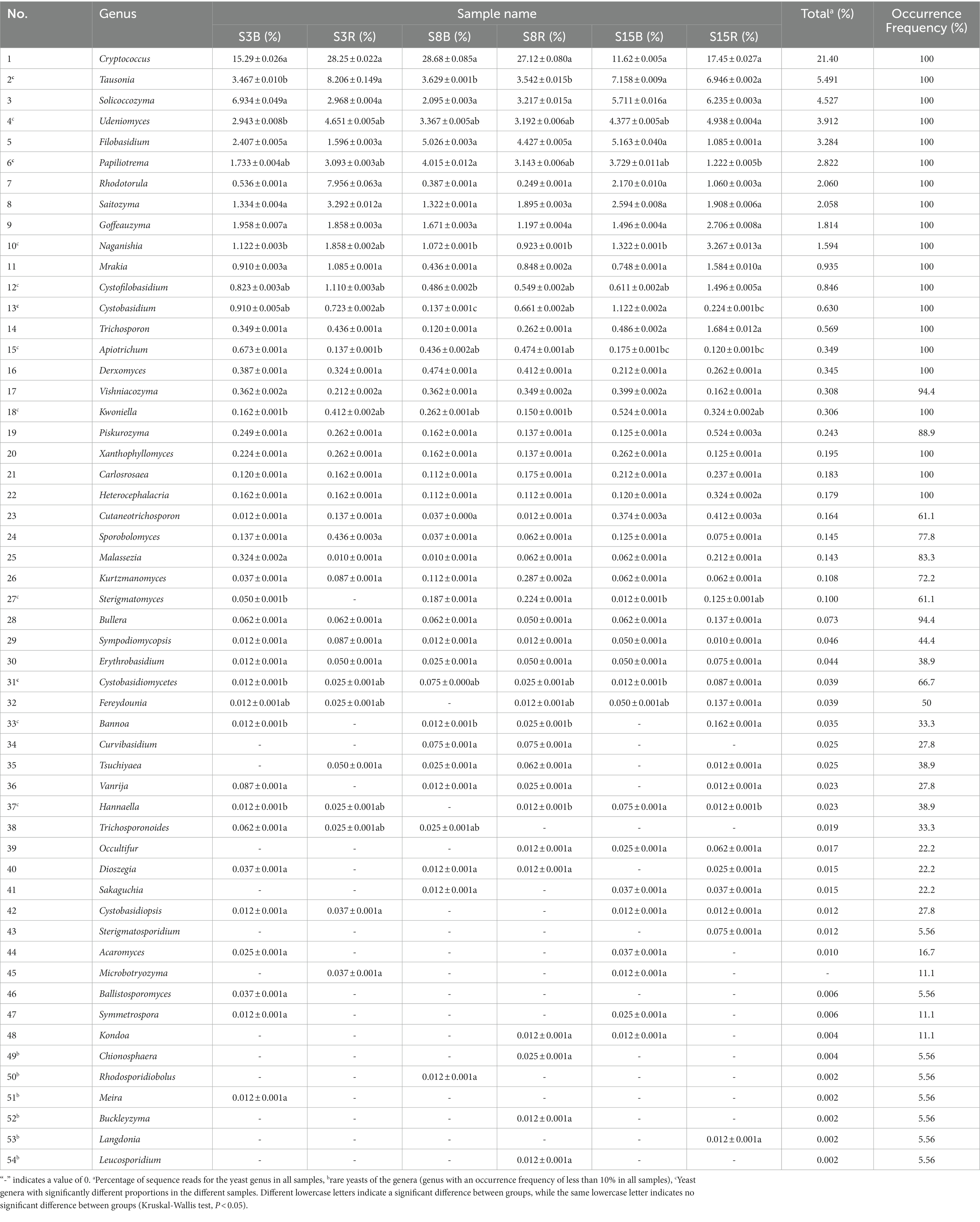
Table 4. The percentage and frequency of occurrence of Basidiomycetes yeasts (accounted for 55.22%) in all samples.
After that, we show the proportions of 19 dominant genera in each group of soil samples to reveal the differences in yeast community composition between non-rhizosphere and rhizosphere soils of peach trees at different ages (Figure 3). The results showed that the top three dominant genera in S3 were Cryptococcus (21.77%), Clavispora (15.47%) and Pichia (13.27%) in that order; Cryptococcus (27.90%), Zygosaccharomyces (12.98%) and Pichia (9.696%) in S8; and Cryptococcus (14.53%), Tausonia (7.052%), and Clavispora (6.996%) in S15 (Figure 3A). The top three dominant genera in non-rhizosphere soil samples were Cryptococcus (18.53%), Clavispora (11.14%), and Pichia (10.44%) in that order; and Cryptococcus (24.27%), Pichia (8.787%), and Clavispora (7.358%) were found in the rhizosphere soil samples (Figure 3B). Further analysis showed that the top three dominant genera in the soil samples of S3B group were Pichia (20.23%), Clavispora (19.17%), and Cryptococcus (15.29%), in S3R group were Cryptococcus (28.25%), Clavispora (11.77%), and Tausonia (8.206%), in S8B group were Cryptococcus (28.68%), Zygosaccharomyces (13.97%), and Cyniclomyces (8.667%), in S8R group were Cryptococcus (27.12%), Pichia (14.85%), and Zygosaccharomyces (12.00%), S15B soils were Cryptococcus (11.62%), Saturnispora (10.38%), and Clavispora (8.991%), and the top three dominant genera in S15R soils were Cryptococcus (17.45%), Trigonopsis (7.058%), and Tausonia (6.946%), respectively (Figure 3C). And the relative abundances of 7 of the 19 dominant genera differed significantly among the S3B, S3R, S8B, S8R, S15B, and S15R groups, namely Tausonia, Zygosaccharomyces, Udeniomyces, Aureobasidium, Papiliotrema, Cyniclomyces, and Naganishia (p < 0.05). Tausonia and Udeniomyces were mainly present in S3R, S15B and S15R, Zygosaccharomyces was mainly present in S8B and S8R, Aureobasidium was mainly found in S15B and S15R, Papiliotrema and Cyniclomyces were mainly detected in S8B and Cyniclomyces could not be detected in S15R, and Naganishia was mainly found in S3R and S15R (Figure 4; Tables 3, 4). In addition, the heat map results largely show that soil samples from the same tree age are clustered together (Figure 4).
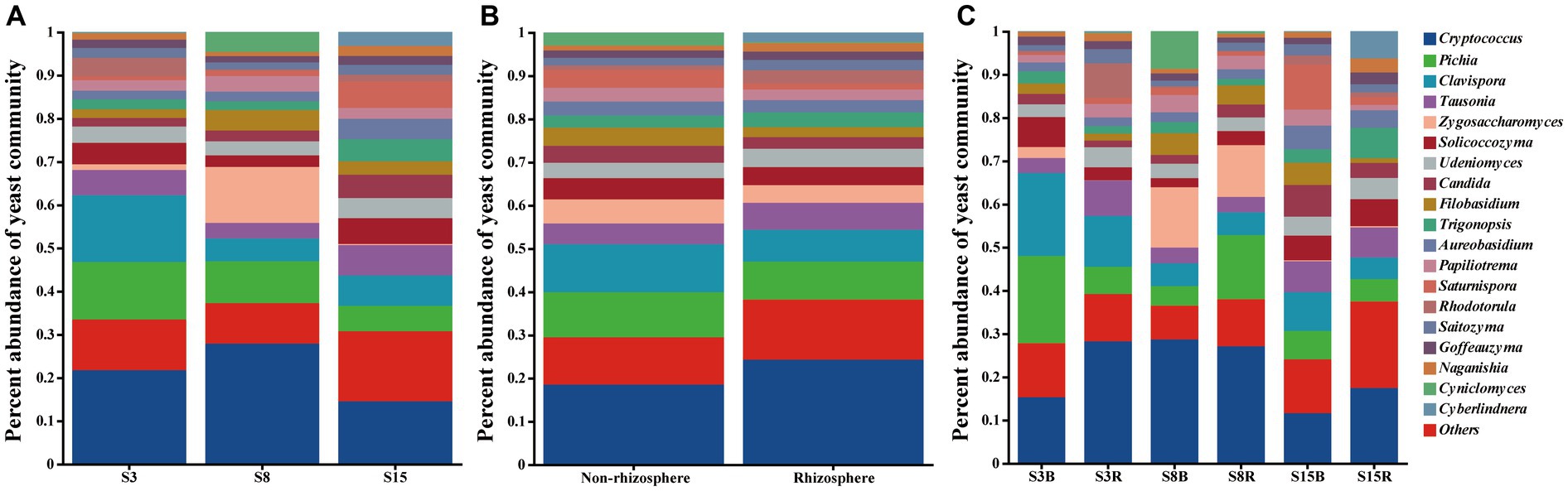
Figure 3. Proportion of dominant yeast genera in (A) peach trees of different ages soil samples, (B) non-rhizosphere and rhizosphere soil samples, and (C) non-rhizosphere and rhizosphere soil samples from peach trees of different ages. Others indicated that yeast genera accounted for less than 1%. Each sample had three replicates (Replicates are not specifically shown in the legend, but have been involved in the analysis). Sample abbreviations are same as presented in Figures 1, 2.
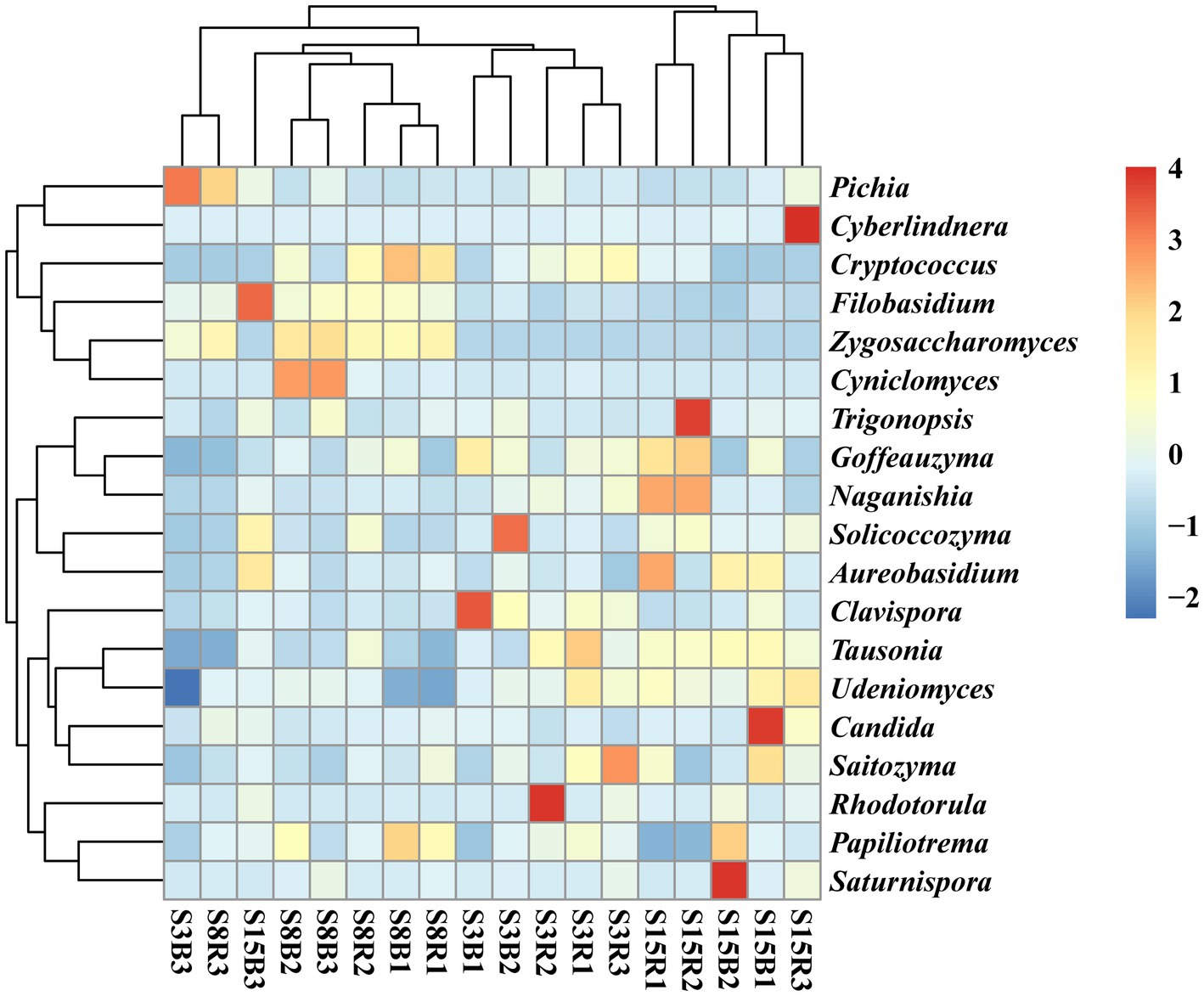
Figure 4. Heatmap of the distribution of the top 19 dominant yeast genera among the different soil samples. The normalized relative abundance of each genus is indicated by a gradient of color from blue (low abundance) to red (high abundance). Sample abbreviations are same as presented in Figure 1.
We further evaluated the similarity of the yeast community composition of 18 soil samples based on PCoA (Figure 5). The results showed that the variance explained by the first principal axis (PCoA1) alone was 43.92%, and the variance explained by the second principal axis (PCoA2) alone was 14.16%. In general, the 18 samples were first clustered together according to tree age, followed by clustering according to rhizosphere or non-rhizosphere criteria, only with an overlapping between the samples from the S3 and S15 groups on the score plots, indicating significant differences in community composition between groups, and this result can also be proved by both R-value (0.4315) and p-value (p = 0.001). S3 and S15 were more similar in community composition for intergroups, and there were significant differences in community composition between rhizosphere or non-rhizosphere samples at the same tree age.
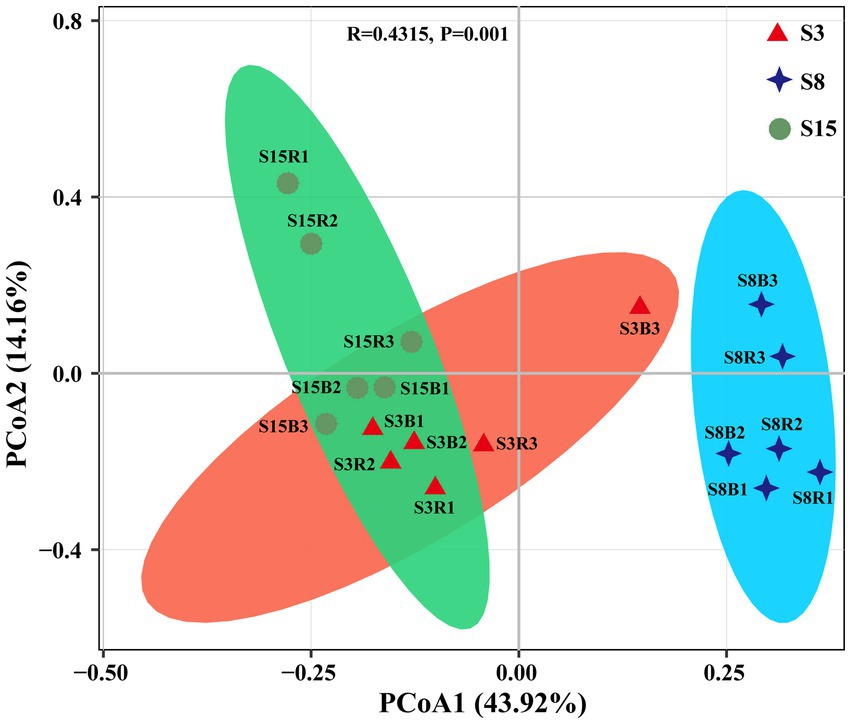
Figure 5. Principal Coordinates analysis (PCoA) based on Bray-Curtis distance method at the OTU level. Red triangles, blue diamonds and green circles represent samples from non-rhizosphere and rhizosphere of 3-year-old (S3), 8-year-old (S8), and 15-year-old (S15) peach trees, respectively. Sample abbreviations are same as presented in Figures 1, 2.
In order to clarify the relationship between soil sample similarity, yeast community structure and soil factors, we first examined the soil physical and chemical properties that including soil pH, conductivity (CO), soil water content (SWC), organic matter (OM), total nitrogen (TN), total phosphorus (TP) and total potassium (TK). The analysis of soil physical and chemical properties showed that the pH values were significantly higher in S3B and S8R and significantly lower in S15R than in the rest of the samples; SWC values were significantly higher in S3B and significantly lower in S8R and S15B than in other samples; while the OM, TN and TK contents were significantly higher in S15R than in other samples (p < 0.05); the CO and TK levels were not significantly different in all samples (p > 0.05; Table 5).
Moving on, redundancy analysis (RDA) based on yeast genera and soil physical and chemical properties in non-rhizosphere soil samples showed that the first and second RDA components explained 36.1% and 20.3% of the variation, respectively, for a total of 56.4% of the total variation (Figure 6A). The degree of influence of soil factors on yeast communities in non-rhizosphere soils was in the following order: CO (F-ratio: 2.06, p values: 0.04) > TK (F-ratio: 1.90, p-values: 0.086) > SWC (F-ratio: 1.18, p-values: 0.348) > pH (F-ratio: 1.08, p-values: 0.40) > TP (F-ratio: 1.08, p-values: 0.42) > TN (F-ratio: 0.33, p-values: 0.826) > OM (F-ratio: 1.29, p-values: 0.454). Among them, CO was significantly associated with the yeast communities in non-rhizosphere soils (p < 0.05). CO was positively correlated with Pichia but negatively correlated with Cyniclomyces, Cryptococcus, Filobasidium, and Papiliotrema.
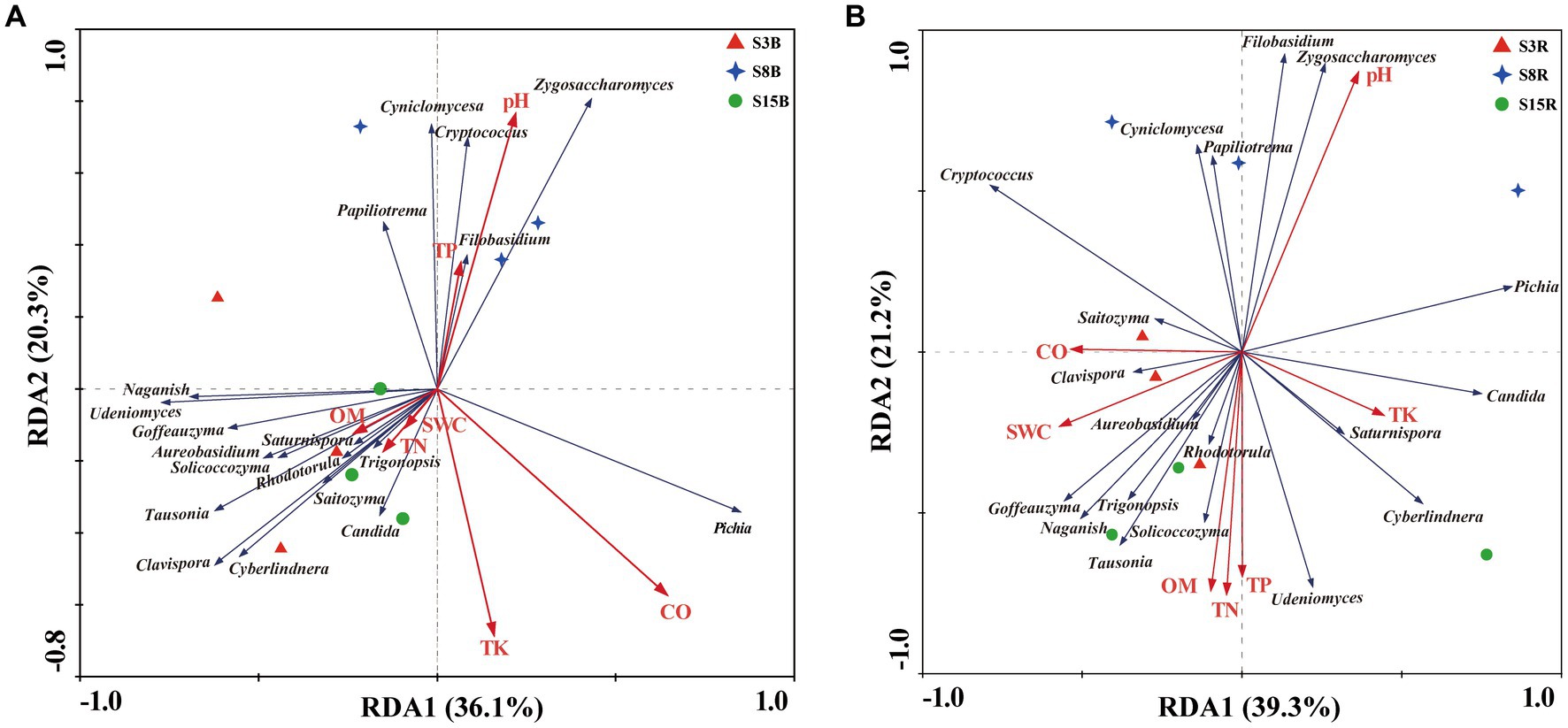
Figure 6. Redundancy analysis (RDA) of the correlation between the dominant yeast genera and soil physicochemical properties in (A) non-rhizosphere and (B) rhizosphere soil samples from peach trees of different ages. Red, blue, and green symbols in (A) and (B) represent non-rhizosphere and rhizosphere soil samples from 3-year-old (S3), 8-year-old (S8), and 15-year-old (S15) peach trees, respectively. Red and blue arrows represent the soil physical and chemical properties and genera, respectively. Soil physicochemical properties: pH, Conductivity (CO), Soil water content (SWC), Organic matter (OM), Total nitrogen (TN), Total phosphorus (TP), Total potassium (TK). Sample abbreviations are same as presented in Figure 1.
Immediately after, we performed a RDA of the correlation between the yeast genera and soil physical and chemical properties in rhizosphere soil samples. The results show that the first and second RDA components explained 39.3% and 21.2% of the variation, respectively, for a total of 60.5% of the total variation (Figure 6B). The degree of influence of soil factors on yeast communities in rhizosphere soils was in the following order: pH (F-ratio: 1.97, p-values: 0.036) > TN (F-ratio: 1.54, p-values: 0.224) > CO (F-ratio: 1.27, p-values: 0.260) > TK (F-ratio: 1.70, p-values: 0.178) > SWC (F-ratio: 1.20, p-values: 0.344) > TP (F-ratio: 0.80, p-values: 0.548) > OM (F-ratio: 0.56, p-values: 0.650). pH was the soil factors that has significant effects on the distribution of yeast communities in rhizosphere soils (p < 0.05). pH was positively correlated with Zygosaccharomyces, Filobasidium, Cyniclomyces, and Papiliotrema but negatively correlated with Clavispora, Trigonopsis, Tausonia, Solicoccozyma, Udeniomyces, Goffeauzyma, and Naganishia.
The above results indicate there is a correlation between the yeast communities and soil physical and chemical properties, particularly CO and pH levels in the soil, and that soil chemical properties are important factors influencing the appearance of differences in yeast community structure in non-rhizosphere and rhizosphere soil samples.
To investigate the potential interactions of yeast communities and changes in co-occurrence networks at temporal and spatial scales, we constructed yeast co-occurrence networks based on random matrix theory in all samples, in samples of each tree age, and in non-rhizosphere and rhizosphere samples, respectively (Figure 7). The topological properties of the network indicate that the ALL co-occurrence network consists of 126 nodes and 153 edges with an average degree of 2.429, implying that each node is directly connected to approximately two other nodes. The average degree reveals the degree of connectivity of the components in the yeast network, and the higher the average degree, the higher the degree of network interactions. Secondly, the network average clustering coefficient (ACC), average path length (APL), and density of the ALL co-occurrence network are 0.644, 3.769, and 0.019, respectively. These three together reveal the tightness of each component of the network, where the smaller the APL is, the higher the network tightness. The modularity of the network was 0.809 (Values > 0.4 indicate that the network have modular structures), indicating a high degree of modularity and representing a high degree of classification of the community structure and function of yeast. Overall, complex relationships existed in the soil yeast community of the peach orchard (Table 6).
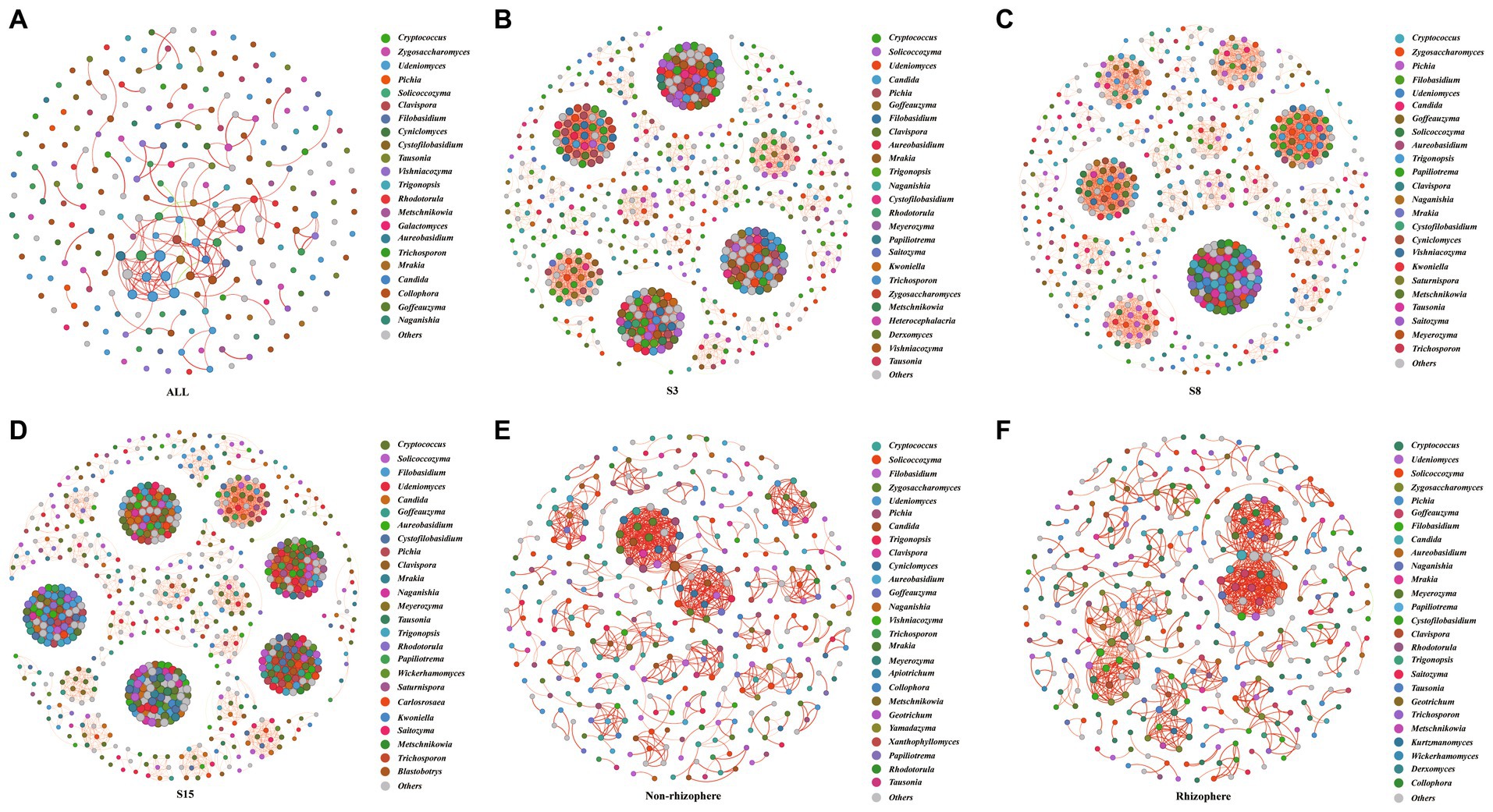
Figure 7. Network of co-occurring 90% cutoff OTUs based on correlation analysis. A connection stands for a strong (Spearman’s |r| > 0.6) and significant (p-value <0.01) correlation. Nodes in the network represent different genera (OTUs belonging to the same genera are grouped into the same color); the size of each node is proportional to the number of connections (that is, degree). A red edge represents a positive interaction, and a green edge represents a negative interaction. The thickness of the line is proportional to the correlation coefficient between OTUs. The greater the number of lines indicates the more closely related that OTU is to the others. Others indicated that yeast genera accounted for less than 1%. (A) ALL: All soil samples in this study. The remaining sample abbreviations (B) S3, (C) S8 (D) S15 (E) Non-rhizosphere (F) Rhizosphere are same as presented in Figures 1, 2.
On the time scale, the degree of interactions and tightness of connections among components in the S15 yeast network were significantly higher than those in S3 and S8. In the case that the network average clustering coefficients (ACC) and the average path lengths (APL) of the three network graphs were consistent, the S15 network had the highest number of nodes (731) and edges (11679) and the highest average degree (31.95), density (0.44), and modularity (0.845). The next highest degree and tightness of network interactions was S3, and S8 was the lowest. At the spatial scale, the average degree (4.944), APL (1.506), and density (0.014) of the yeast co-occurrence network of rhizosphere soils were slightly higher, and the ACC (0.886) and modularity (0.842) were slightly lower than those of the non-rhizosphere, indicating a slightly higher degree of interactions and a slightly lower degree of tightness and modularity in the rhizosphere soil network compared to the non-rhizosphere soil. The number of positively correlated edges in the network was greater than the number of negatively correlated edges, both at the overall level and at the temporal and spatial scales, indicating greater synergy and less antagonism among yeast communities. The largest synergistic effect of yeast network was found in the S15 on the time scale, with more than 99.78% of the positive correlation edges, and in the rhizosphere network on the spatial scale, with 100% of the positive correlation edges.
The nodes in the yeast co-occurrence network were divided by genus level, and to explore the variation of core species in the peach orchard soil yeast co-occurrence network, we counted the degree of all genera in each network (Supplementary Tables S14–S19) and enumerated the top five ranked hub genera (Table 7). The analysis showed that nodes in the ALL, S3, S8, S15, non-rhizosphere, and rhizosphere networks belonged to 40, 74, 74, 82, 62, and 68 genera, respectively. Hub genera in the ALL yeast network were, in order, Zygosaccharomyces (39.87%), Cryptococcus (9.80%), Pichia (9.15%), Udeniomyces (6.54%), and Clavispora (5.88%). The composition and proportion of hub genera in each network changed with spatial and temporal changes. For example, Pichia accounted for 11.55% and 15.03% in the S3 and S8 networks, respectively, and only 6.92% in S15. The unique hub genera in the S3 and S15 networks were Clavispora and Filobasidium, respectively, and the unique hub genera in S8 were Zygosaccharomyces and Trigonopsis. The non-rhizosphere compared with the rhizosphere network each had two unique hub genera, Pichia and Cyniclomyces for the non-rhizosphere and Goffeauzyma and Filobasidium for the rhizosphere.
Based on high-throughput sequencing technology we obtained a total of 3,103 yeast OTUs from peach orchard soil, identified as 114 genera belonging to Ascomycota and Basidiomycota (Tables 3, 4). The species richness and species diversity of the soil samples in this study (Table 2) were at a high level compared to other soil types (Yurkov et al., 2016; Vadkertiová et al., 2019). This suggests that peach orchard soils are rich in yeast resources, which is consistent with previous studies obtained using culturable methods (Wang et al., 2019). But compared to traditional culture methods, high-throughput sequencing technology is significantly more advantageous and can provide more comprehensive detection of species composition in habitats (Zebin et al., 2016). Also, the results of the Alpha diversity index among groups showed no significant differences in yeast community diversity between non-rhizosphere and rhizosphere soils at ages 3, 8, and 15 years (Table 2), which indicated that the overall yeast distribution in the peach orchard soil was relatively stable. It validates the conclusions obtained by previous studies that the composition of the soil microbial community under fruit trees is generally more stable than that of annual crops, as it is less likely to be disturbed by management practices (Vadkertiová et al., 2017; Mercado-Blanco et al., 2018).
In our study, Ascomycetous yeast genera were more numerous than Basidiomycetes yeast genera, which further validates the idea that Ascomycetous yeasts are usually more frequent and abundant in agricultural soils, orchards, and grasslands (Sláviková and Vadkertiová, 2003; Yurkov et al., 2012). In addition, we found that 33 yeast genera were detected in both non-rhizosphere and rhizosphere soils of 3-, 8-, and 15-year-old peach trees in the peach orchard, suggesting that these genera may be resident yeasts in peach orchard soils (Tables 3, 4). Among them, Cryptococcus, Pichia, Candia, Rhodotorula, and Hanseniaspora can be found in most soil types, but their diversity and abundance of these species vary from one habitat to another (Poliakova et al., 2001; Wang, 2007; Xu, 2009), this is supported by our findings. Apart from that, Saitozyma, Solicoccozyma, and Goffeauzyma are dominant yeasts in our study and are reported to be equally dominant in other soil types (França et al., 2016; Groenewald et al., 2018; Yurkov, 2018). In fact, not every yeast isolated from soil is a native soil dweller but may come from sources other than soil (Phaff et al., 1978; Phaff and Starmer, 1987). For example, some species of the Ascomycetous genera Aureobasidium, Hanseniaspora, Metschnikowia, Saccharomyces, and Pichia, as well as the Basidiomycetes genera Rhodotorula, Cystobasidium, Vishniacozyma, and Sporobolomyces that were detected in this study, are usually dominant species isolated from the above-ground vegetative organs (leaves, flowers, and fruits) of the plant (SlÁviková et al., 2009; Sipiczki, 2016; Yurkov, 2018). This may be due to the fact that our sampling time was at the peak of the peach tree’s fruiting season, and there would be epiphytic yeast entering the soil with the fallen peaches or leaves. Furthermore, the rare yeast genera discovered in this study accounted for approximately 13.16% of the yeast genera in all soil samples (Tables 3, 4), which is significantly lower than the proportion of rare yeasts found in other orchards (Vadkertiová et al., 2019), forest (França et al., 2016), grassland, and shrub soils (Yurkov et al., 2016). Previous studies have shown that reduced precipitation leads to increased populations of rare species in soil habitats (Yurkov et al., 2016). In contrast, the field management pattern of the peach orchard in this study provided sufficient water, so this may be more favorable for yeast survival.
Although there were no significant differences in yeast diversity among the groups, their community structure showed significant variation at temporal and spatial scales, particularly at the temporal scale (Figure 5). The yeast community was most evenly distributed at 15 years compared to the soil yeast community composition at 3 and 8 years (Figure 3; Tables 3, 4). This indicates that the soil yeast community was already more stable at 15 years. Previous studies have shown that an increase in shared species diversity can improve the stability of microbial communities (Wang et al., 2013). The variation in the number of shared yeast OTUs among the three ages in this study also proved this (Figure 2A). And, the stability of the yeast community also contributes to the resistance of peach trees. In addition, the abundance of the shared dominant genera Zygosaccharomyces and Aureobasidium increased significantly in 8- and 15-year-old peach soils, respectively. Zygosaccharomyces has been reported to be involved in the solubilization of soil insoluble phosphate, which may be related to the high phosphorus demand of 8-year-old peach trees (Gizaw et al., 2017; Petkova et al., 2022). Moreover, 8-year-old peach trees may accumulate pathogenic fungi with increasing age, and Zygosaccharomyces also has the ability to produce siderophore compounds (iron (III) ion compounds) that inhibit the growth of fungal phytopathogens (Hider and Kong, 2010; Petkova et al., 2022). Aureobasidium has been shown to be effective against postharvest fruit pathogens (Di Francesco et al., 2020; Podgórska-Kryszczuk, 2023).
In terms of spatial scale, we found that the total number of yeast OTUs in peach rhizosphere soil was higher than that in non-rhizosphere soil. In agreement with a previous study, rhizosphere microorganisms have better abundance and diversity than non-rhizosphere microorganisms (Yue et al., 2018). In addition, the number of shared OTUs in non-rhizosphere soil samples among different tree ages was higher than the number of shared OTUs in their rhizosphere samples (Figure 2). This indicates that the yeast community is more specific in the rhizosphere soil of different tree ages, possibly influenced by factors such as the rhizosphere secretion of peach trees. It has been reported that root secretions have a selective role in shaping the rhizosphere microbial community structure, which is unique of different plants (Paterson et al., 2007). For example, Candida, Geotrichum, Rhodotorula, and Meyerozyma were all detected in all rhizosphere samples in this study, and their representative species are thought to be associated with nitrification in the soil, where nitrite and phosphate are solubilized in vitro to nitrate (Chen et al., 2012; Nakayan et al., 2013). It is worth noting that the abundance of the yeast genera Metschnikowia, Wickerhamomyces, Geotrichum, and Torulaspora was significantly increased in the 15-year peach rhizosphere soil samples compared to the 3- and 8-year samples. The results of the present study suggest that the increase in abundance of the first three may be caused by the accumulation of a large number of pathogens due to the increase in the number of years of colonization of peach trees. Some representative species of Metschnikowia (Sipiczki, 2020; Wang et al., 2021), Wickerhamomyces (Lanhuang et al., 2022), and Geotrichum (Kawtharani et al., 2022) have been shown to be antagonistic to various pathogens and can be widely used as biocontrol agents in organic agriculture. T. delbrueckii in the genus Torulaspora has been reported to be able to produce phytase under certain conditions, increasing the nutritional content of peach and improving the absorption of trace elements in peach by humans (Kaur et al., 2007). All these results indicate that the rhizosphere microbial community and plant growth and development are mutually influential. In summary, the changes in yeast community structure in the soil of the peach orchard in this study were consistent with previous findings that soil microbial diversity has certain spatial and temporal characteristics (Kowalchuk et al., 2002; Yue et al., 2018).
The drivers of yeast community assembly in soils are more complex and mainly include the effects of environmental conditions and vegetation (Mašínová et al., 2017), among which soil physicochemical properties are the key factors. Because the soil physical and chemical properties reflect both the growth state of plants and the survival conditions of yeast in the soil. In this study, there were significant differences in soil physicochemical properties among the samples, especially in pH, organic matter (OM), total nitrogen (TN), and total phosphorus (TP; Table 5). The results of the RDA analysis clearly revealed that conductivity (CO) and pH were the main factors influencing the structure of the yeast community (Figure 6). The same conclusion was reached in a previous study of the diversity of soil yeasts isolated from South Victoria Land, Antarctica (Connell et al., 2008). The soil conductivity CO reflects the amount of salt in the soil water solution. Generally speaking, the higher the CO value of the soil within a suitable range, the more fast-acting nutrients are available to the plant (Zhang et al., 2009). We found a significant positive correlation between pichia and CO, suggesting that pichia may be beneficial to plant growth. In addition, soil pH is also one of the main factors influencing the composition of the soil yeast community. It has been described as the “master soil variable” that influences a myriad of soil biological, chemical, and physical properties and processes and affects plant growth and biomass production (Minasny et al., 2016). The pH of the soil samples collected for this study ranged from 7.76 to 8.41, with an overall weak alkalinity. We found a negative correlation with pH for most of the yeast genera, indicating that overall the yeast community still prefers an acidic environment, which is a common characteristic of yeasts (Chen, 2012). In contrast, the genera Zygosaccharomyces, Filobasidium, Cyniclomyces, and Papiliotrema in this study showed a positive correlation with pH, indicating that these three yeast genera prefer alkaline environments for survival.
In this study, we found that 15-year-old peach inter-root soils had the lowest pH but significantly higher levels of OM, TN, and TP than the other samples (p < 0.05; Table 5). This indicates that the 15-year rhizosphere soil fertility was higher. Because pH can affect soil function and plant nutrition effectiveness by influencing the chemical solubility and availability of essential plant nutrients, pesticide performance, and organic matter decomposition (McCauley et al., 2009). Furthermore, Tausonia, Solicoccozyma, Trigonopsis, and Goffeauzyma in this study showed positive correlations with OM, TN, and TP, indicating that they grow in abundance in nutrient-rich environments and can be used to indicate soil fertility. In summary, soil CO and pH play an important role in coordinating crop growth and soil yeast community structure. Tracing the factors that contribute to differences in the structure of soil yeast communities helps us better provide solutions to improve soil ecology and thus contribute to the sustainable development of fruit trees.
To further understand the survival mechanisms of yeast communities in peach orchard soils, we conducted a co-occurrence network analysis of yeast communities in soils in multiple dimensions: overall, temporal, and spatial (Figure 7). Co-occurrence network analysis has now been widely used in the field of microbiology. It measures the interactions between different microbial taxa by correlating the abundance of microbial taxa across multiple soil samples and extracting simple patterns from complex interactions to identify cooperative or competitive relationships between species and further infer community assembly and evolutionary mechanisms (Goberna and Verdú, 2022; Guseva et al., 2022). By calculating the degree of the network nodes, the central node microorganisms of the network can be screened for microorganisms with potential ecological functions (Shetty et al., 2017). The results of this study show that there is significant spatial and temporal specificity in soil yeast community interactions in the peach orchard, with particularly pronounced variation on the temporal scale. This is consistent with the changes in yeast community structure. The degree of interactions and connection tightness of the 15-year-old yeast community were higher than those of the 3-year-old and 8-year-old ones, while there were obvious yeast network modularity, core yeast genera, and network connection nodes, indicating the reliability of the interactions among the 15-year-old soil yeasts. This indicates that the yeast community has acquired certain structural and functional stability in its long-term evolution with increasing age. In addition, there were more positive connections than negative connections in the peach orchard soil yeast network, indicating that yeasts in peach orchard soil prefer to coexist in a synergistic mutualistic manner, and the strongest synergistic effect among the three tree-aged soil yeast networks was found among 15-year-old soil yeasts, which further supports the strong stability of their network structure and function.
Comparatively, although the rhizosphere network was more interactions than the non-rhizosphere, the degree of connectivity tightness and modularity were lower than the non-rhizosphere. It indicates that the interactions between rhizosphere soil yeasts are more random and do not have stability and reliability. And the fact that yeast synergistic effect is lower in the rhizosphere network than in the non-rhizosphere network also illustrates the same issue. Unlike the previous conclusions obtained that a more stable microbial network exists in rhizosphere soils compared to non-rhizosphere soils of wheat (Fan et al., 2018). This may be due to the fact that yeast is more sensitive to environmental changes in soil only as a taxon of fungi, while yeast in inter-rhizosphere soil is more susceptible to the influence of plant roots compared to non-rhizosphere yeast, in addition to the influence of the environment (Paterson et al., 2007; Fan et al., 2018).
At the same time, our observation of each network graph revealed that OTU nodes in low-abundance yeast genera are also likely to have a high degree (Figure 7), which reaffirms the important role of low-abundance microbial genera in maintaining the stability and function of microbial communities (Guo et al., 2022). Furthermore, we found that each network differed in the composition of the hub yeast genera, which may also be a response of the peach orchard soil yeast community to temporal and spatial changes. Hubs in the network are usually defined as keystone species because if these taxa are removed, the network may also split; thus, they play a crucial role in the network structure and can be identified as targets for microbial regulation to improve crop productivity (Olesen et al., 2007). In this study, we can take the hub yeast genera in the network as a reference and then use its related soil factor as a condition for the improvement of peach orchard soil to improve the quality of soil microbiology and finally achieve the purpose of maintaining the health of peach trees and promoting the quality growth of peach trees.
In this study, we found for the first time the living strategies of soil yeasts at the spatial and temporal scales of perennial peach trees. Unlike soil yeast diversity in the peach orchard, yeast community structure varies significantly on spatial and temporal scales. Soil factors such as CO and pH were the main factors influencing the differences in yeast community structure. This study reveals the changes in the diversity and community structure of non-rhizosphere and rhizosphere soil yeast at different ages in the peach orchard and the factors affecting them, as well as the spatio-temporal response of the soil yeast network in peach orchards, providing new insights into the role of soil yeast resources in achieving sustainable agricultural development in peach orchards and its spatio-temporal adaptation mechanisms.
The data of high throughput sequencing in this project has been deposited in the Sequence Read Archive (SRA) of the National Center for Biotechnology Information (NCBI) under the accession number PRJNA992790 (https://www.ncbi.nlm.nih.gov/sra/?term=PRJNA992790).
Ethical approval is not applicable in the case of the study. However, the collection of the soil samples from the peach orchard was verbally permitted by the farm owner.
SSZ designed and performed the experiments, analyzed the data, and drafted the manuscript. YLC helped design experiments, analyzed the data, and drafted the manuscript. YL performed sample collection, DNA extraction, PCR amplification and analyzed part of the data. JX performed sample collection and soil chemical property analysis. YHL performed sample collection, DNA extraction, and PCR amplification. YFS designed and performed the experiments and analyzed data. All authors contributed to the article and approved the submitted version.
The work was funded by the National Natural Science Foundation of China (project number: 31860003).
The authors would like to thank all the reviewers who participated in the review.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1226142/full#supplementary-material
OTU, Operational taxonomic units; CO, Conductivity; SWC, Soil water content; OM, Organic matter; TN, Total nitrogen; TP, Total phosphorus; TK, Total potassium; PRECTP, Average annual precipitation; TEMP, Average annual temperature; LST, Average annual land surface temperature; RH, Average annual relative humidity; EVLAND, Average annual evaporation land; PCoA, Principal coordinates analysis; RDA, Redundancy analysis.
Barberán, A., Scott, T., Bates-Casamayor, K., and Fierer, N. (2012). Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 6, 343–351. doi: 10.1038/ismej.2011.119
Boby, V., Balakrishna, A., and Bagyaraj, D. (2008). Interaction between Glomus mosseae and soil yeasts on growth and nutrition of cowpea. Microbiol. Res. 163, 693–700. doi: 10.1016/j.micres.2006.10.004
Bokulich, N. A., Subramanian, S., Faith, J. J., Gevers, D., Gordon, J. I., Knight, R., et al. (2013). Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 10, 57–59. doi: 10.1038/nmeth.2276
Botha, A. (2011). The importance and ecology of yeasts in soil. Soil Biol. Biochem. 43, 1–8. doi: 10.1016/j.soilbio.2010.10.001
Bunge, J., Epstein, S. S., and Peterson, D. G. (2006). Comment on "computational improvements reveal great bacterial diversity and high metal toxicity in soil". Science 313:918. doi: 10.1126/science.1126593
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Chen, R. (2012) Study on soil microbial diversity and new species of yeasts in apple orchards. Ph.D. thesis. Shandong Agricultural University.
Chen, R., Jiang, Y.-M., Wei, S.-C., and Wang, Q.-M. (2012). Kwoniella shandongensis sp. nov., a basidiomycetous yeast isolated from soil and bark from an apple orchard. Int. J. Syst. Evol. Microbiol. 62, 2774–2777. doi: 10.1099/ijs.0.039172-0
Chen, R., Wei, S.-C., Jiang, Y.-M., Wang, Q.-M., and Bai, F.-Y. (2010). Kazachstania taianensis sp. nov., a novel ascomycetous yeast species from orchard soil. Int. J. Syst. Evol. Microbiol. 60, 1473–1476. doi: 10.1099/ijs.0.016501-0
Chen, Z. J., Yang, X. C., Zhao, J., Hu, J. X., and Yuan, W. Y. (2018). Breeding of Saccharomyces cerevisiae with high ethanol tolerance and its application in kiwi wine. Sci. Technol. Food Ind. 39, 141–145,151. doi: 10.13386/j.issn1002-0306.2018.02.027
Cloete, K. J., Valentine, A. J., Stander, M. A., Blomerus, L. M., and Botha, A. (2009). Evidence of symbiosis between the soil yeast Cryptococcus laurentii and a sclerophyllous medicinal shrub, Agathosma betulina (berg.) Pillans. Microb. Ecol. 57, 624–632. doi: 10.1007/s00248-008-9457-9
Connell, L., Redman, R., Craig, S., Scorzetti, G., Iszard, M., and Rodriguez, R. (2008). Diversity of soil yeasts isolated from South Victoria land, Antarctica. Microb. Ecol. 56, 448–459. doi: 10.1007/s00248-008-9363-1
Di Francesco, A., Di Foggia, M., and Baraldi, E. (2020). Aureobasidium pullulans volatile organic compounds as alternative postharvest method to control brown rot of stone fruits. Food Microbiol. 87:103395. doi: 10.1016/j.fm.2019.103395
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
El-Tarabily, K. A., and Sivasithamparam, K. (2006). Potential of yeasts as biocontrol agents of soil-borne fungal plant pathogens and as plant growth promoters. Mycoscience 47, 25–35. doi: 10.1007/s10267-005-0268-2
Fan, L. H., Cui, Y. J., He, Q., and Pan, X. L. (2006). Analysis on the climate change characteristics in the Shihezi region, Xinjiang in recent 40 years. Arid Zone Res. 23, 334–338. doi: 10.13866/j.azr.2006.02.024
Fan, K., Gilbert, P., Jack, A., and Chu, H. (2018). Wheat rhizosphere harbors a less complex and more stable microbial co-occurrence pattern than bulk soil. Soil Biol. Biochem. 125, 251–260. doi: 10.1016/j.soilbio.2018.07.022
Ferraz, P., Cássio, F., and Lucas, C. (2019). Potential of yeasts as biocontrol agents of the phytopathogen causing cacao witches’ broom disease: is microbial warfare a solution? Front. Microbiol. 10:1766. doi: 10.3389/fmicb.2019.01766
França, L., Sannino, C., Turchetti, B., Buzzini, P., and Margesin, R. (2016). Seasonal and altitudinal changes of culturable bacterial and yeast diversity in alpine forest soils. Extremophiles 20, 855–873. doi: 10.1007/s00792-016-0874-2
Gizaw, B., Tsegay, Z., Tefera, G., and Aynalem, E. (2017). Phosphate solubilizing yeast isolated and characterized from teff rhizosphere soil collected from gojam; Ethiopia. J. Bacteriol. Mycol. 5, 218–223. doi: 10.15406/jbmoa.2017.05.00120
Goberna, M., and Verdú, M. (2022). Cautionary notes on the use of co-occurrence networks in soil ecology. Soil Biol. Biochem. 166:108534. doi: 10.1016/j.soilbio.2021.108534
Groenewald, M., Lombard, L., de Vries, M., Lopez, A. G., Smith, M., and Crous, P. W. (2018). Diversity of yeast species from Dutch garden soil and the description of six novel Ascomycetes. FEMS Yeast Res. 18:7. doi: 10.1093/femsyr/foy076
Guo, B., Zhang, L., Sun, H., Gao, M., Yu, N., Zhang, Q., et al. (2022). Microbial co-occurrence network topological properties link with reactor parameters and reveal importance of low-abundance genera. NPJ Biofilms Microbiomes 8:3. doi: 10.1038/s41522-021-00263-y
Guseva, K., Darcy, S., Simon, E., Alteio, L. V., Montesinos-Navarro, A., and Kaiser, C. (2022). From diversity to complexity: microbial networks in soils. Soil Biol. Biochem. 169:108604. doi: 10.1016/j.soilbio.2022.108604
Han, C. G., Ding, J. L., Pu, Y. J., Liu, H. T., and Wei, J. B. (2008). The change characteristics of the temperatures and precipitation in Shihezi, Xinjiang in the recent 41 years. J Arid Land Resourc Environ 22, 50–54.
He, M., Xu, Y., Qiao, Y., Zhang, Z., Liang, J., Peng, Y., et al. (2022). A novel yeast strain Geotrichum sp. CS-67 capable of accumulating heavy metal ions. Ecotoxicol. Environ. Saf. 236:113497. doi: 10.1016/j.ecoenv.2022.113497
Hernández-Fernández, M., Cordero-Bueso, G., Ruiz-Muñoz, M., and Cantoral, J. M. (2021). Culturable yeasts as biofertilizers and biopesticides for a sustainable agriculture: a comprehensive review. Plan. Theory 10:822. doi: 10.3390/plants10050822
Hider, R. C., and Kong, X. (2010). Chemistry and biology of siderophores. Nat. Prod. Rep. 27, 637–657. doi: 10.1039/b906679a
Igwegbe, C. A., Obiora-Okafo, I. A., Iwuozor, K. O., Ghosh, S., Kurniawan, S. B., Rangabhashiyam, S., et al. (2022). Treatment technologies for bakers’ yeast production wastewater. Environ. Sci. Pollut. Res. Int. 29, 11004–11026. doi: 10.1007/s11356-021-17992-4
Kaur, P., Kunze, G., and Satyanarayana, T. (2007). Yeast phytases: present scenario and future perspectives. Crit. Rev. Biotechnol. 27, 93–109. doi: 10.1080/07388550701334519
Kawtharani, H., Beaufort, S., Anson, P., Taillandier, P., Mathieu, F., and Snini, S. P. (2022). Impact of the inoculation method of Geotrichum candidum, used as biocontrol agent, on T-2 toxin produced by fusarium sporotrichioides and F. langsethiae during the malting process. Toxins 14:239. doi: 10.3390/toxins14040239
Kowalchuk, G. A., Buma, D. S., De Boer, W., Klinkhamer, P. G., and Van Veen, J. A. (2002). Effects of above-ground plant species composition and diversity on the diversity of soil-borne microorganisms. Antonie Van Leeuwenhoek 81, 509–520. doi: 10.1023/a:1020565523615
Kumla, J., Nundaeng, S., Suwannarach, N., and Lumyong, S. (2020). Evaluation of multifarious plant growth promoting trials of yeast isolated from the soil of Assam tea (Camellia sinensis var. assamica) plantations in northern Thailand. Microorganisms 8:1168. doi: 10.3390/microorganisms8081168
Lanhuang, B., Yang, Q., Godana, E. A., and Zhang, H. (2022). Efficacy of the yeast wickerhamomyces anomalus in biocontrol of gray mold decay of tomatoes and study of the mechanisms involved. Foods 11:720. doi: 10.3390/foods11050720
Liu, K. P., Wei, Y. M., Xiong, J., Sun, Y. F., and Lei, Y. H. (2018). Analysis of diversity of endobiotic yeasts isolated from Locusta migratoria manilensis and Ischiodon scutellaris Fabricius. Guangdong Agric Sci 9, 90–95. doi: 10.16768/j.issn.1004-874X.2018.09.015
Liu, H., Zhang, Y., Wang, C., Sun, Y. F., and Lei, Y. H. (2019). Community structure analysis of yeast attached to flat peach leaves in Xinjiang. Ecol Sci 38, 29–34. doi: 10.14108/j.cnki.1008-8873.2019.04.005
Magoč, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Marques, A. R., Resende, A. A., Gomes, F. C., Santos, A. R. O., Rosa, C. A., Duarte, A. A., et al. (2021). Plant growth–promoting traits of yeasts isolated from the tank bromeliad Vriesea minarum LB Smith and the effectiveness of Carlosrosaea vrieseae for promoting bromeliad growth. Braz. J. Microbiol. 52, 1417–1429. doi: 10.1007/s42770-021-00496-1
Mašínová, T., Bahnmann, B. D., Větrovský, T., Tomšovský, M., Merunková, K., and Baldrian, P. (2017). Drivers of yeast community composition in the litter and soil of a temperate forest. FEMS Microbiol. Ecol. 93:223. doi: 10.1093/femsec/fiw223
McCauley, A., Jones, C., and Jacobsen, J. (2009). Soil pH and organic matter. Nutr Manage Module 8, 1–12.
Meng, L., Xu, C., Wu, F., and Huhe, S. (2022). Microbial co-occurrence networks driven by low-abundance microbial taxa during composting dominate lignocellulose degradation. Sci. Total Environ. 845:157197. doi: 10.1016/j.scitotenv.2022.157197
Mercado-Blanco, J., Abrantes, I., Barra Caracciolo, A., Bevivino, A., Ciancio, A., Grenni, P., et al. (2018). Belowground microbiota and the health of tree crops. Front. Microbiol. 9:1006. doi: 10.3389/fmicb.2018.01006
Minasny, B., Hong, S. Y., Hartemink, A. E., Kim, Y. H., and Kang, S. S. (2016). Soil pH increase under paddy in South Korea between 2000 and 2012. Agr Ecosyst Environ 221, 205–213. doi: 10.1016/j.agee.2016.01.042
Mirabal Alonso, L., Kleiner, D., and Ortega, E. (2008). Spores of the mycorrhizal fungus Glomus mosseae host yeasts that solubilize phosphate and accumulate polyphosphates. Mycorrhiza 18, 197–204. doi: 10.1007/s00572-008-0172-7
Nakayan, P., Hameed, A., Singh, S., Young, L.-S., Hung, M.-H., and Young, C.-C. (2013). Phosphate-solubilizing soil yeast Meyerozyma guilliermondii CC1 improves maize (Zea mays L.) productivity and minimizes requisite chemical fertilization. Plant and Soil 373, 301–315. doi: 10.1007/s11104-013-1792-z
Nassar, A. H., El-Tarabily, K. A., and Sivasithamparam, K. (2005). Promotion of plant growth by an auxin-producing isolate of the yeast Williopsis saturnus endophytic in maize (Zea mays L.) roots. Biol. Fertil. Soils 42, 97–108. doi: 10.1007/s00374-005-0008-y
Olesen, J. M., Bascompte, J., Dupont, Y. L., and Jordano, P. (2007). The modularity of pollination networks. Proc. Natl. Acad. Sci. U. S. A. 104, 19891–19896. doi: 10.1073/pnas.0706375104
Paterson, E., Gebbing, T., Abel, C., Sim, A., and Telfer, G. (2007). Rhizodeposition shapes rhizosphere microbial community structure in organic soil. New Phytol. 173, 600–610. doi: 10.1111/j.1469-8137.2006.01931.x
Petkova, M., Petrova, S., Spasova-Apostolova, V., and Naydenov, M. (2022). Tobacco plant growth-promoting and antifungal activities of three endophytic yeast strains. Plan. Theory 11:751. doi: 10.3390/plants11060751
Phaff, H. J., Miller, M. W., and Mrak, E. M. (1978). The life of yeasts, vol. 59. Cambridge, Mass: Harvard University Press, 553.
Phaff, H. J., and Starmer, W. (1987). Yeasts associated with plants, insects and soil. London: Academic Press, 123–179.
Podgórska-Kryszczuk, I. (2023). Biological control of aspergillus flavus by the yeast Aureobasidium pullulans in vitro and on tomato fruit. Plan. Theory 12:236. doi: 10.3390/plants12020236
Poliakova, A., Chernov, I., and Panikov, N. S. (2001). Yeast biodiversity in hydromorphic soils with reference to grass-Sphagnum swamp in Western Siberia and the hammocky tundra region (Barrow, Alaska). Mikrobiologiia 70, 714–720.
Shetty, S. A., Hugenholtz, F., Lahti, L., Smidt, H., and de Vos, W. M. (2017). Intestinal microbiome landscaping: insight in community assemblage and implications for microbial modulation strategies. FEMS Microbiol. Rev. 41, 182–199. doi: 10.1093/femsre/fuw045
Sipiczki, M. (2016). Overwintering of vineyard yeasts: survival of interacting yeast communities in grapes mummified on vines. Front. Microbiol. 7:212. doi: 10.3389/fmicb.2016.00212
Sipiczki, M. (2020). Metschnikowia pulcherrima and related pulcherrimin-producing yeasts: fuzzy species boundaries and complex antimicrobial antagonism. Microorganisms 8:1029. doi: 10.3390/microorganisms8071029
Sláviková, E., and Vadkertiová, R. (2003). The diversity of yeasts in the agricultural soil. J. Basic Microbiol. 43, 430–436. doi: 10.1002/jobm.200310277
SlÁviková, E., Vadkertiová, R., and Vránová, D. (2009). Yeasts colonizing the leaves of fruit trees. Ann. Microbiol. 59, 419–424. doi: 10.1007/BF03175125
Vadkertiová, R., Dudášová, H., and Balaščáková, M. (2017). “Yeasts in agricultural and managed soils,” in Yeasts in Natural Ecosystems: Diversity, eds. P. Buzzini, M. A. Lachance, and A. M. Yurkov (Switzerland: Springer) 117–144.
Vadkertiová, R., Dudášová, H., Stratilová, E., and Balaščáková, M. (2019). Diversity of yeasts in the soil adjacent to fruit trees of the Rosaceae family. Yeast 36, 617–631. doi: 10.1002/yea.3430
Wang, Q. G. (2007). Studies on species diversity and application of wild yeast in Xinjiang. M.D. thesis. Shangdong Institute of Light Industry, Jinan, China.
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Wang, R., Jing, M., Zhao, H., Xu, X., and Shao, Y. (2016). Isolation and screening of antagonism yeast against postharvest mango anthracnose. Food Res Dev 37, 180–183. doi: 10.3969/j.issn.1005-6521.2016.07.044
Wang, X., Wang, Z., and Feng, T. (2022). Screening of yeast in various vineyard soil and study on its flavor compounds from brewing grape wine. Molecules 27:512. doi: 10.3390/molecules27020512
Wang, Q. Q., Zhang, Y., Lei, Y. H., Feng, L., Xiong, J., and Sun, Y. F. (2019). Diversity of culturable yeast and screening of functional yeast in soils of flat peach trees of different ages in Shihezi. Biotic Resourc 9, 44–52. doi: 10.14188/j.ajsh.2019.01.007
Wang, S., Zhang, H., Ruan, C., Yi, L., Deng, L., and Zeng, K. (2021). Metschnikowia citriensis FL01 antagonize Geotrichum citri-aurantii in citrus fruit through key action of iron depletion. Int. J. Food Microbiol. 357:109384. doi: 10.1016/j.ijfoodmicro.2021
Wang, H.-D., Zhang, L.-L., and Zhu, Z.-H. (2013). Effects of clipping and fertilizing on the relationships between species diversity and ecosystem functioning and mechanisms of community stability in alpine meadow. Chin J Plant Ecol 37, 279–295. doi: 10.3724/SP.J.1258.2013.00028
Xu, M. X. (2009). Species diversity of wild yeast collected in the east part of Qinhai Province of China. M.D. thesis. Shangdong Institute of Light Industry, Jinan, China.
Xu, Y. N., Li, Q., Liu, Q. P., Xiao, J., Zheng, X. J., and Shi, X. W. (2015). Research Progress of non-saccharomyces yeasts in Xinjian. Food Ind. 36, 266–270. doi: 10.13414/j.cnki.zwpp.2019.01.012
Yang, Q., and Wang, Z. (2009). Distribution and function of yeast in eco-environments. Environ. Sci. Technol. 32, 86–91. doi: 10.3969/j.issn.1003-6504.2009.04.021
Yin, X. H., Wang, Q. G., Zhang, C., and Shi, J. Y. (2017). Isolation and identification of antagonistic bacteria against peach brown rot and its antibacterial activity. Sci. Technol. Food Ind. 38, 128–132. doi: 10.13386/j.issn1002-0306.2017.09.016
Yue, C., Guanghui, L., and Yan, L. (2018). Soil microbial functional diversity of rhizosphere and non-rhizosphere of three dominant herbaceous plants in the Dushanzi District. Acta Ecol. Sin. 38, 3110–3117. doi: 10.5846/stxb201703070382
Yurkov, A. (2018). Yeasts of the soil—obscure but precious. Yeast 35, 369–378. doi: 10.1002/yea.3310
Yurkov, A., Kemler, M., and Begerow, D. (2012). Assessment of yeast diversity in soils under different management regimes. Fungal Ecol. 5, 24–35. doi: 10.1016/j.funeco.2011.07.004
Yurkov, A. M., Röhl, O., Pontes, A., Carvalho, C., Maldonado, C., and Sampaio, J. P. (2016). Local climatic conditions constrain soil yeast diversity patterns in Mediterranean forests, woodlands and scrub biome. FEMS Yeast Res. 16:fov103. doi: 10.1093/femsyr/fov103
Zebin, C., Tiyuan, X., Dingkang, W., Guoxing, W., Jiani, L., Zhang, Y., et al. (2016). Diversity analysis of endophytic Fungi in Mentha using Illumina MiSeq high-throughput sequencing technology. Medicinal Plant 7. 412:127042. doi: 10.1016/j.surfcoat.2021.127042
Zhang, J. E., Gao, A. X., Xu, H. Q., and Luo, M. Z. (2009). Effects of maize/peanut in tercropping on rhizosphere soil microbes and nutrient contents. Chin. J. Appl. Ecol. 20, 1597–1602. doi: 10.13287/j.1001-9332.2009.0253
Zhang, H., Wang, Y., Chen, S., Zhao, Z., Feng, J., Zhang, Z., et al. (2018). Water bacterial and fungal community compositions associated with urban lakes, Xi’an, China. Int. J. Environ. Res. Public Health 15:469. doi: 10.3390/ijerph15030469
Zhang, J. J., Yang, X., Guo, C., Li, K., Zhang, W. Y., and Liu, C. H. (2017). The composition and differences of yeasts from the soils cultivating different grape varieties. Liquor Making 44, 76–81. doi: 10.3969/j.issn.1002-8110.2017.01.018
Zhao, L. F., Xu, Y. J., and Zhou, H. J. (2004). Study progress on comprehensive utilization of yeast. Food Res Dev 25, 29–32. doi: 10.3969/j.issn.1005-6521.2004.04.009
Keywords: yeast community, spatio-temporal scales, high-throughput sequencing, co-occurrence network, orchard soil
Citation: Zhu S, Cai Y, Li Y, Xiong J, Lei Y and Sun Y (2023) Effects of temporal and spatial scales on soil yeast communities in the peach orchard. Front. Microbiol. 14:1226142. doi: 10.3389/fmicb.2023.1226142
Received: 20 May 2023; Accepted: 31 August 2023;
Published: 19 September 2023.
Edited by:
Dilfuza Egamberdieva, Leibniz Center for Agricultural Landscape Research (ZALF), GermanyReviewed by:
Mahendra Vikram Singh Rajawat, Prabhat Fertilizers and Chemical Works, IndiaCopyright © 2023 Zhu, Cai, Li, Xiong, Lei and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: YongHui Lei, NDk3OTc2QHFxLmNvbQ==; YanFei Sun, ODE3MTEzMDhAcXEuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.