- Pasteurien College, Suzhou Medical College of Soochow University, Suzhou, China
Staphylococcus aureus is an opportunistic foodborne pathogen occasionally isolated from diarrhea patients. In recent years, increasing studies have reported the detection of S. aureus in food poisoning incidents due to food contamination in the North and South of China. However, the epidemiology and genetic characteristics of S. aureus from food poisoning incidents in Eastern China remain unknown. The present study examined the genetic characteristics, antimicrobial resistance, and virulent factors of multidrug-resistant S. aureus isolated from 22 food poisoning incidents reported by the hospitals and health centers in Eastern China from 2011 to 2021. A total of 117 resistant and enterotoxigenic S. aureus isolates were collected and sequenced, among which 20 isolates were identified as methicillin resistant. Genetic analysis revealed 19 distinct CC/ST types, with CC6, CC22, CC59, CC88, and CC398 being the most frequent variants in methicillin-resistant S. aureus (MRSA). A considerable shift in CC types from CC1 to CC398 between 2011 and 2021 was observed in this study, indicating that CC398 may be the main epidemic strain circulating in the current food poisoning incidents. Additionally, genes for enterotoxins were detected in 55 isolates, with a prevalence of 27.8% (27/97) for methicillin-sensitive variants and 35.0% (7/20) for MRSA. The scn gene was detected in 59.0% of the isolates, demonstrating diverse contaminations of S. aureus among livestock-to-human transmission. Of the 117 isolates, only ten isolates displayed multi-drug resistance (MDR) to penicillin, tetracycline, and macrolides. None of the 117 foodborne S. aureus isolates tested positive for vanA in this study. Together, the present study provided phylogenetic characteristics of S. aureus from food poisoning incidents that emerged in Eastern China from 2011 to 2021. Our results suggested that these diarrhea episodes were hypotonic and merely transient low-MDR infections, however, further research for continued surveillance given the detection of virulence and antimicrobial resistance determinants is required to elucidate the genomic characteristics of pathogenic S. aureus in food poisoning incidents in the context of public health.
Introduction
Staphylococcus aureus is an opportunistic pathogen frequently detected in food as well as on the skin, nose and throat of humans (Argudín et al., 2010). It can cause acute food poisoning, which presents symptoms such as diarrhea, stomach pain, nausea, vomiting, and other related manifestations (Johler et al., 2015; Mahros et al., 2021). From 2005 to 2018, the European Centre for Disease Prevention and Control (ECDC) documented over 500,000 S. aureus bloodstream infections in European countries (Gagliotti et al., 2021). Similarly, over 241,000 illnesses of foodborne S. aureus infection were reported annually by the Centers for Disease Control and Prevention (CDC) in the United States (Kadariya et al., 2014). Previous research in China revealed frequent detection of S. aureus in animal meat, egg products, and dairy products, with a positive rate of 35.0% (Wu et al., 2018). In addition, a total of 1,150 S. aureus were isolated from 24 provinces in China, with 4.3% of retail foods being contaminated by S. aureus, and 7.9% of retail foods isolates being mecA positive. Moreover, 97.6% of S. aureus isolates were resistant to at least one antimicrobial compound, and 57.5% of these were multi drug resistant to penicillin (83.7%), linezolid (67.7%) and erythromycin (52.1%) (Wang et al., 2017).
Foodborne S. aureus is associated with various virulent factors, including genes responsible for producing enterotoxins, exfoliative toxins, toxic shock syndrome toxin (tsst-1), Panton-Valentine leucocidin, staphylococcal complement inhibitor, and hemolysins (Leung et al., 1993; Gouaux et al., 1997; Bhatia and Zahoor, 2007; Boyle-Vavra and Daum, 2007; Bukowski et al., 2010; Carrera et al., 2017; de Jong et al., 2018). Among these factors, enterotoxins are considered the primary culprits in causing staphylococcal food poisoning (SFP) (Bhatia and Zahoor, 2007). According to a recent study, Guo et al. reported an SFP outbreak caused by ST7 S. aureus strains in two kindergarten campuses in South China (Guo et al., 2023). Six antimicrobial resistance genes were detected including blaZ, ANT (4′)-Ib, tetK, lnuA, norA, and lmrS. Another study in South China analyzed the clonal complex (CC) of 62 distinct S. aureus strains and found that CC239 and CC3 were the dominant clones of food poisoning incidents (Xie et al., 2016). SCCmecIII-ST239 was the prevalent type, accounting for 43.4 to 79.5% of hospital and community-associated MRSA and harboring a series of virulence genes such as sea, seb, seh, eta, and pvl (Xie et al., 2016). Additionally, a total of 138 foodborne S. aureus were isolated from outbreaks in North China, with CC1, CC5, CC7, CC8, CC15, CC59, CC88, CC97, and CC398 as the predominant clones (Lv et al., 2021). However, there is a lack of research investigating the genetic characteristics of foodborne S. aureus in Eastern China during the past decade.
To identify S. aureus strains isolated from food poisoning incidents in Eastern China from 2011 to 2021, a combined bioinformatics approach was employed to study the genetic characteristics. In addition, the genotypic antimicrobial resistance profiles and virulent factor possession were examined for a comprehensive assessment of the potential risks associated with the presence of methicillin resistance in food poisoning incidents.
Materials and methods
Ethics approval
The experiment was strictly conducted according to the Guide for Care and Use from the Research Ethics Committee of Soochow University (20210220). All procedures involving human participants were performed to the ethical standards. Patients were given informed consent to participate in the study.
Description of food poisoning incidents
Information on the food poisoning incidents was gathered from the Center for Disease Control and Prevention of Suzhou, China. A total of 896 stool samples (n = 850) and vomit samples (n = 46) from diarrheal patients were collected from 22 hospitals and community health centers between 2011 and 2021 and forwarded for testing using the Chinese National Foodborne Disease Surveillance Manual (Li et al., 2018). These 22 facilities were dispersed among ten regions, between 30°93’N to 35°55’N and 120°21′E to 121°64′E in Eastern China. The collected samples were tested for pathogenic bacteria including Salmonella, Shigella, Staphylococcus aureus, Bacillus cereus, Vibrio parahaemolyticus, diarrhea-causing Escherichia coli, and Listeria monocytogenes. Food poisoning incidents with Staphylococcus aureus as the dominant pathogen were enrolled for further analysis. Detail descriptions of CC, ST types, isolation of year and geographic data of S. aureus were shown in Supplementary Table S1.
Bacterial isolation and identification
The present research was conducted during the period from 2011 to 2021. In the initial step, 10 g of stool samples and vomit samples were collected and transferred to the laboratory on ice. Then samples were homogenized in 0.1% peptone saline in a filter bag (Bkmam, Changde, China). After that, 100 ul were cultured onto Baird Parker agar (HopeBio 4,115, Beijing, China) and CHROMagar™ MRSA agar (Becton Dickinson, Franklin Lakes, NJ). The plates were incubated in the carbon dioxide incubator overnight at 37°C. Then, a loop full of bacterial culture from incubated tubes was streaked separately into the Baird Parker agar, and the plates were examined and studied carefully for the presence of characteristic colonies of S. aureus.
The S. aureus strains were identified by 16 s rRNA sequencing and MALDI-TOF MS (Bio-Merieux, Craponne, France). Generally, a 1.4-kb fragment of the 16S rRNA gene was amplified by PCR with universal primers (27F, 5’-AGAGTTTGATCMTGGCTCAG-3′, 1492R, 5’-TACGGYTACCTTGTTACGACTT-3′). The PCR product was purified and sequenced in both directions by use of conserved-region primers on the platform of Honsunbio company (Shanghai, China). Purified sequencing results were processed and edited by ABI 3100 (Applied Biosystems) and Sequencher (Gene Codes, Ann Arbor, Mich.), respectively. The altered sequences were identified by comparing them to GenBank via Blast. In the case of bacteria with a low identity, MALDI-TOF MS was enrolled for detection. Briefly, each sample was inoculated as triplicates on the target plate and covered by the freshly prepared matrix (Bruker Daltonik GmbH, Bremen, Germany). Mass spectra were compared with spectra obtained from the associated database (Hansen and Lee, 2017; Holzknecht et al., 2018).
Antimicrobial susceptibility testing
Susceptibility testing of confirmed S. aureus strains was performed according to the Clinical and Laboratory Standards Institute disk diffusion method (CLSI 2022). Staphylococcus aureus ATCC 25923 was used as a control strain. Disks from Oxoid were used. The following antimicrobial disks were tested: ampicillin (10 μg), penicillin (10 units), cefoxitin (30 μg), ceftazidime (30 μg), chloramphenicol (30 μg), clindamycin (2 μg), erythromycin (15 μg), gentamicin (10 μg), linezolid (30 μg), and tetracycline (30 μg). Cefoxitin was tested as a surrogate marker for the detection of methicillin resistance.
Whole genome sequencing
After growing the isolates for 24 h at 37°C in Tryptone Soya Broth (AOBOX 02–049, Beijing, China), genomic DNA was extracted and purified using a HiPure Bacterial DNA Kit (D3146, Meiji Biotechnology Co., Ltd., Guangzhou, China). Library construction was performed with Vazyme TruePrep DNA Library Prep Kit TD501 (Vazyme, Nanjing, China). A Nanodrop ND-1000 spectrophotometer (Nanodrop Technologies, Wilmington, DE, United States) and a 1.0% (w/v) agarose gel were used to assess the quality of the extracted DNA. Purified DNA was whole-genome sequenced using Illumina HiSeq 4,000 platform (150 bp paired-end reads with ~200-fold average coverage) on the Honsunbio platform (Shanghai, China). The raw reads were trimmed, and genome was assembled by EToKi v1.0 (Zhou et al., 2020). The QUAST v2.3 was utilized to assess the quality of the genome assembly (Gurevich et al., 2013). The raw sequencing reads were uploaded to the China National GenBank with the accession number of CRA010922. The sequences can be accessed at https://bigd.big.ac.cn/gsa/browse/CRA010922.
Bioinformatics analysis
To classify STs into clonal complexes (CCs), the Illumina read files were subjected to multilocus sequence typing (MLST.20) and eBURST v3 analysis (Urwin and Maiden, 2003; Feil et al., 2004). A maximum likelihood core-genome phylogenetic tree was constructed using 117 strains based on 547 core genes (~96,496 SNPs). The S. aureus ATCC 25923 was employed as the reference genome for SNP analysis. The select minimum depth at SNP positions was set to 10x, while select minimum relative depth at SNP positions was set to 10%. The select minimum distance between SNPs (prune) was set to 10 bp and select minimum SNP quality was set to 30. The select minimum read mapping quality was set to 25 and the select minimum Z-score was set to 1.96 in CSI Phylogeny v1.4 (Kaas et al., 2014).
Antimicrobial-resistant genes and virulent factors were determined using the ResFinder, Mobile ElementFinder, and VFDB databases, with a minimum of 60% nucleotide identity retained in all algorithms (Liu et al., 2019; Bortolaia et al., 2020; Johansson et al., 2021). The detection of arg, icaA, icaB, icaC, icaD, icaR, luxS, ΦSa3 and ΦAVβ (SAAV_2008, SAAV_2009) genes was blast in MyDbFinder 2.0 with the select threshold of 98% and select minimum length of 60% on the platform of Centre for Genomic Epidemiology.1
Statistical analysis and visualization
The line chart and donut chart were made using GraphPad Prism 7, and statistical significance was assessed using One-way ANOVA with p < 0.05. The genotypic data were visualized in Grapetree and iTOL (Urwin and Maiden, 2003; Letunic and Bork, 2019).
Results and discussion
Description of the food poisoning incidents in eastern China from 2011 to 2021
In this investigation, the food poisoning incidents collected for this study were distributed between 30°93’N and 35°55’N and 120°21′E to 121°64′E (Figure 1). A total of 117 strains of S. aureus were isolated from 896 stool and vomit samples collected from diarrheal patients who registered to the 22 hospitals and community health centers between 2011 and 2021 in Eastern China. Among the 117 isolates, 20 isolates were identified as methicillin-resistant S. aureus (MRSA), with the majority of the strains collected from Region IV and Region II. Among them, six strains were isolated from vomit samples and the remaining 111 strains were detected from diarrheal stool. Further epidemiological analysis revealed that the food poisoning incidents in this study were divided sporadically into ten regions, with Suzhou (Region IV) and Kunshan (Region II) being the dominant regions of food poisoning incidents (Figure 1).
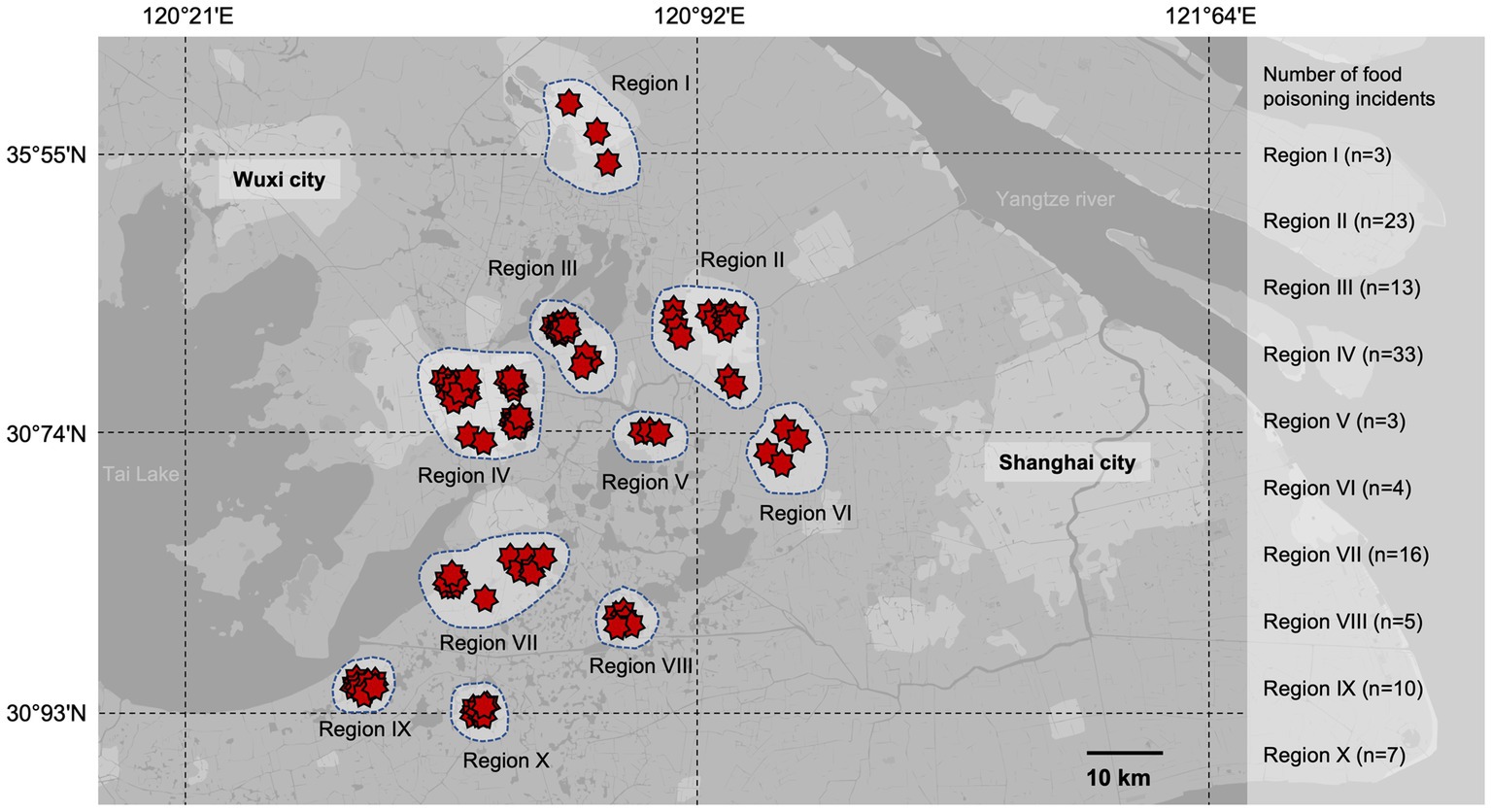
Figure 1. Geographic distribution of food poisoning incidents in Eastern China from 2011 to 2021. The food poisoning incidents collected for this study were distributed between 30°93’N and 35°55’N and 120°21′E to 121°64′E. A total of 117 strains of S. aureus were isolated and distributed to ten regions. The red stars represent the outbreaks of each food poisoning incident in the region.
Genetic diversity of Staphylococcus aureus isolates from the food poisoning incidents
The MLST typing of the 117 foodborne S. aureus genomes identified 19 unique CC/ST types, namely CC1, CC5, CC6, CC7, CC8, CC12, CC15, CC22, CC25, CC59, CC72, CC88, CC188, CC398, CC1281, ST672, ST944, ST1920, and ST2315 (Figure 2, Table 1). MRSA was detected in 20 isolates including CC6, CC22, CC59, CC88, and CC398, while MSSA isolates (n = 97) were widely distributed in the majority of the CC/ST types, indicating the diversity of S. aureus contamination in the current food poisoning incidents. Details of the housekeeping genes in MLST typing were described in Supplementary Table S2.
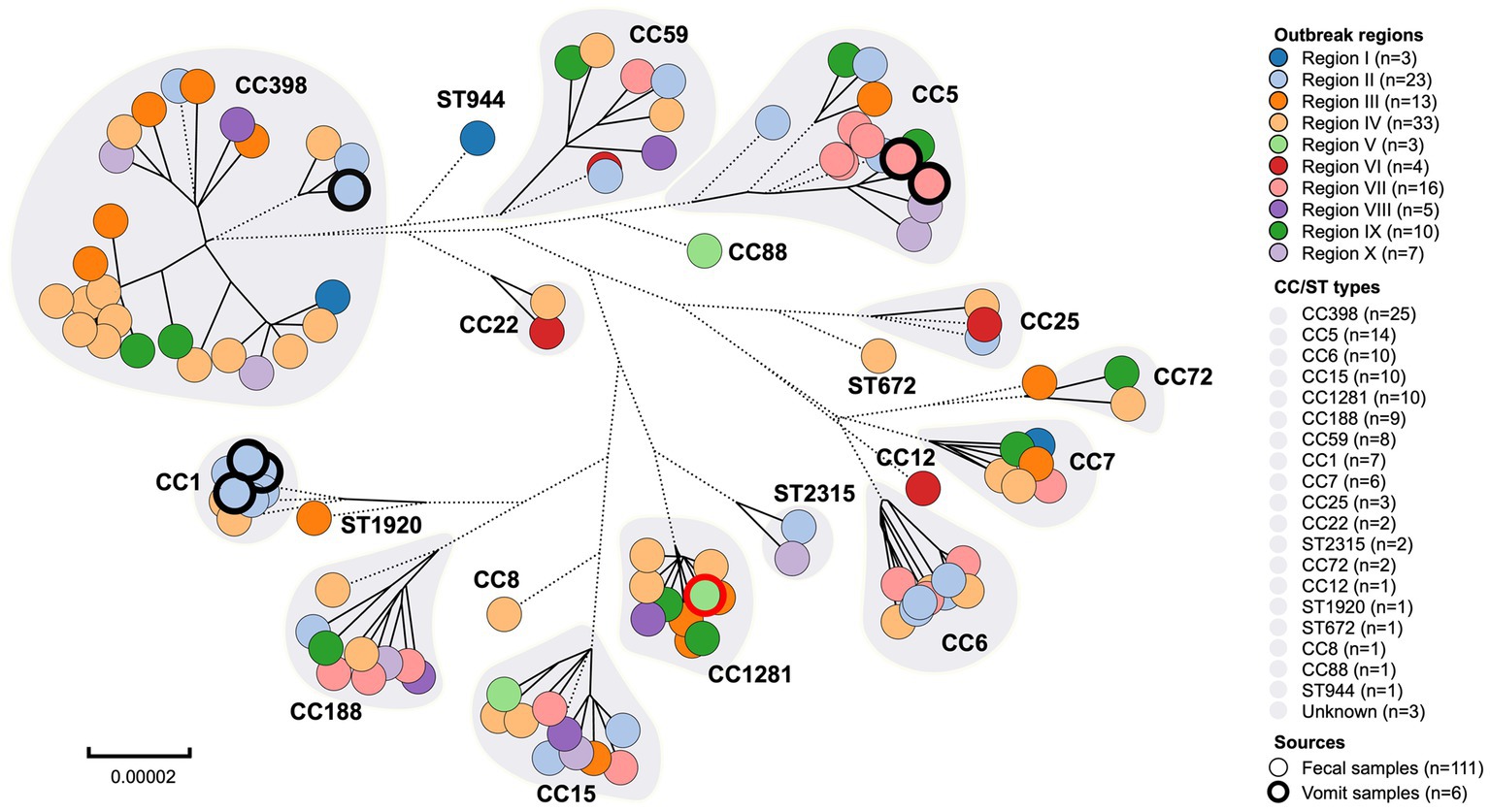
Figure 2. Clonal complexes (CC) and sequence types (ST) of S. aureus from food poisoning incidents in Eastern China. The dotted branches represented shorter distances longer than 0.00003, while the solid lines represented the genetic distance between strains.
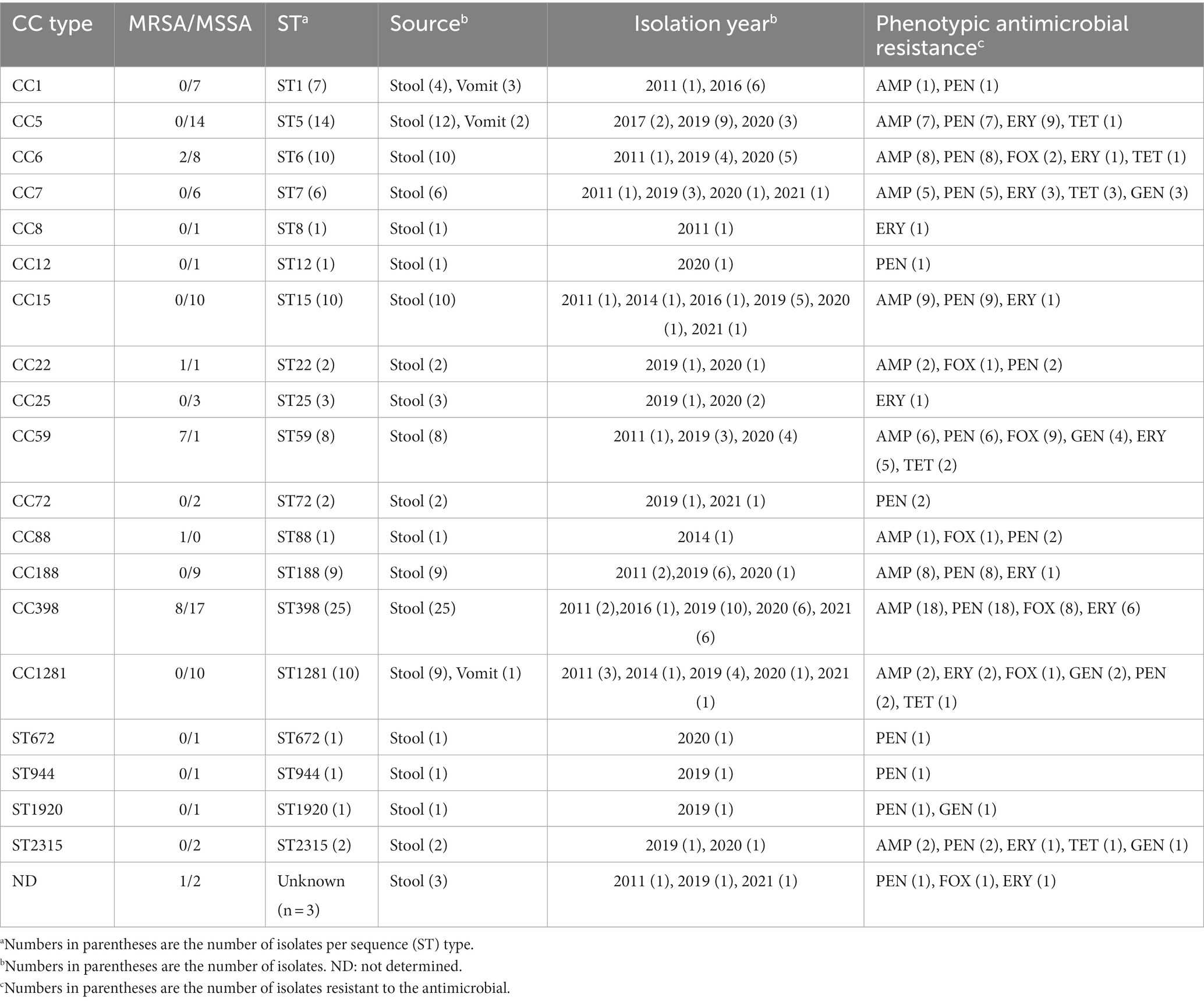
Table 1. CC, MRSA/MSSA, ST types and phenotypic antimicrobial resistance of foodborne S. aureus isolates from this study.
An intriguing finding from this study was the considerable shift in CC types from CC1 to CC398 between 2011 and 2021 (Figure 3B). Before 2018, food poisoning incidents were primarily linked to community-associated CC1. However, the subsequent five years witnessed an increase in the CC398 strains associated with livestock settings, indicating the intricate nature of foodborne S. aureus outbreaks.
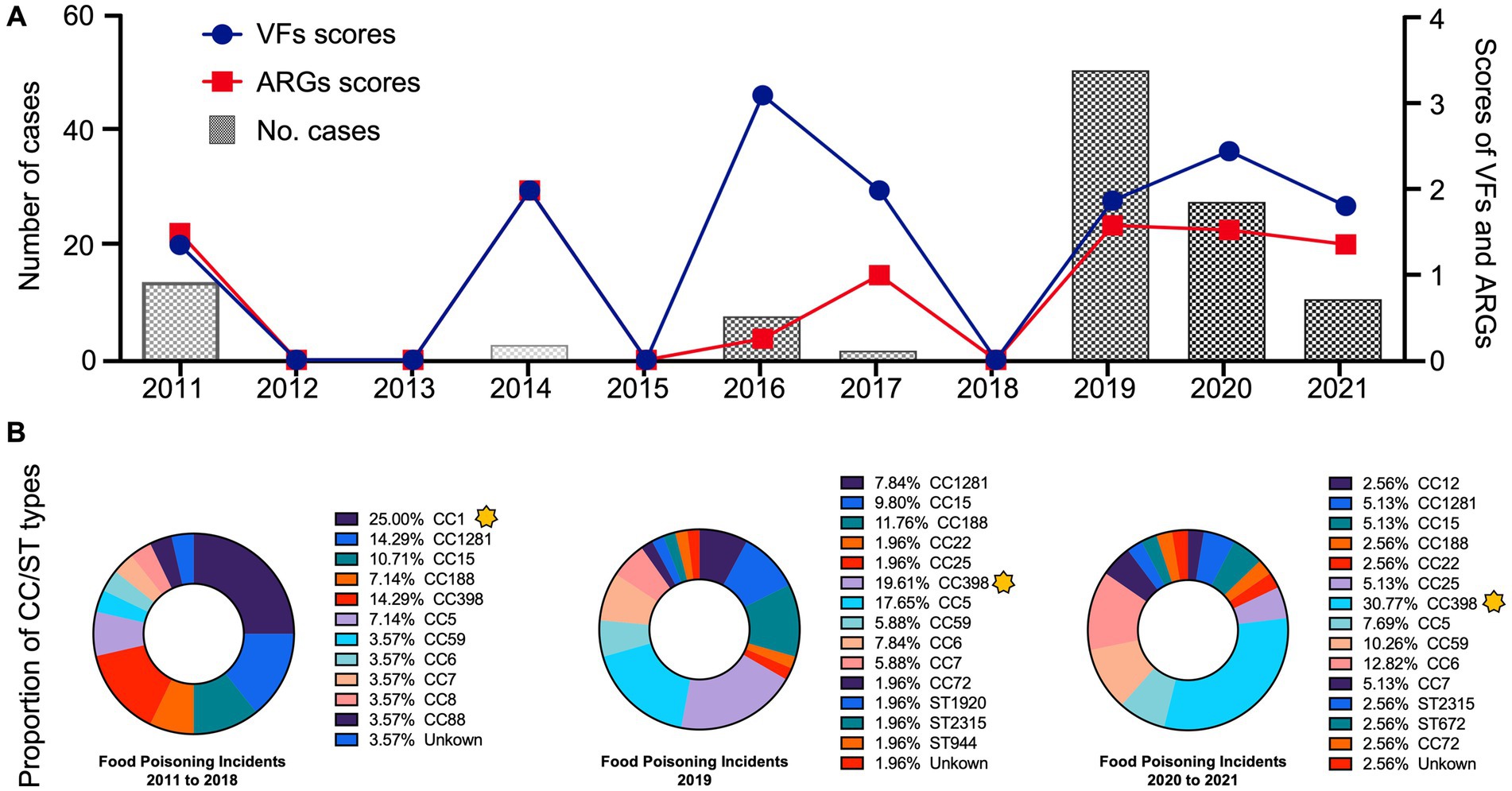
Figure 3. (A) The line chart represented the total number of ARGs and VFs from 117 S. aureus strains between 2011 to 2021. (B) The donut chart represented the proportion of CC and ST types of S. aureus strains in the current food poisoning incidents. The yellow stars marked the dominant types, which showed a considerable shift in CC types from CC1 to CC398 between 2011 and 2021.
Region IV and Region II were identified as the most prevalent regions among the current food poisoning incidents, with CC398 (n = 25), CC5 (n = 14), CC6 (n = 10), and CC15 (n = 10) being the dominant types (Figure 2). Historically, S. aureus CC8, CC15, and CC45 had been the predominant strains causing foodborne diarrhea in European countries (Baumgartner et al., 2014). Analysis of 1850 food products from China revealed that S. aureus CC1 (10.7%) was the most common type, followed by CC7 (10.6%) and CC5 (4.8%), suggesting a different major variation compared to the food poisoning incidents we found in this study (Wu et al., 2018). This disparity could potentially be attributed to differences in Chinese and Western food cultures, as well as variations in cooking practices.
CC398 was identified as the most predominant clonal complex in this study (Figure 2, Table 1). Previous retrospective analysis revealed that human and animal infections with S. aureus CC398 occurred in nations throughout West Europe and Eastern Asia, with the initial cases harboring the strain being identified in the Netherlands (Stegger et al., 2010; Laumay et al., 2021). Since then, the annual growth in the detection rate of CC398 had been increasing, reaching 20.0% in 2006 in Europe (Laumay et al., 2021). In China, the positive rate for CC398 strains in livestock-associated food products varied between 4.6 to 33.3% depending on the region (Wu et al., 2018; Li W. et al., 2021). Similarly, CC398 was detected in meat products with a prevalence of 11 to 31%, indicating that CC398 had entered the food chain and performed as the primary epidemic strain circulating over the world (Verkade and Kluytmans, 2014).
S. aureus CC5 was a common community-associated MRSA lineage circulating in poultry (Aires-de-Sousa, 2017). Studies identified that the majority of S. aureus isolated from poultry belonged to the avian-associated spectrum of CC5, which emerged from a human-to-poultry host jump and was characterized by numerous signatures of adaptation to the avian host including carriage of the ΦAVβ prophage genes (Matuszewska et al., 2020). In the present study, a local blast was performed to detect the ΦAVβ genes (SAAV_2008, SAAV_2009) in S. aureus strains (Lowder et al., 2009; Tang et al., 2017). All CC5 isolates carried ΦAVβ genes, which was consistent with the identification of these isolates in poultry products from current food poisoning incidents (Figure 2, Table 1).
According to previous studies, CC6 was identified as a prevalent pathogen causing food poisoning in adults. Previous study indicated that a total of 868 S. aureus isolates were collected from meat products in China, of which 47 strains were CC6, accounting for about 5.4% (Wu et al., 2018). There was evidence that S. aureus CC6 variations were isolated from diarrhea, and virulence varied between enterotoxin-encoding mobile genetic elements in Eastern Asia (Suzuki et al., 2014). In this study, CC6 isolates carried antimicrobial resistance genes such as aadD1, blaZ, mecA, ermB, and tetM, indicating a multidrug-resistant (MDR) spectrum in China (Figure 4, Table 1). As a result, it is imperative to develop tools for accurate screening for MDR S. aureus in foods.
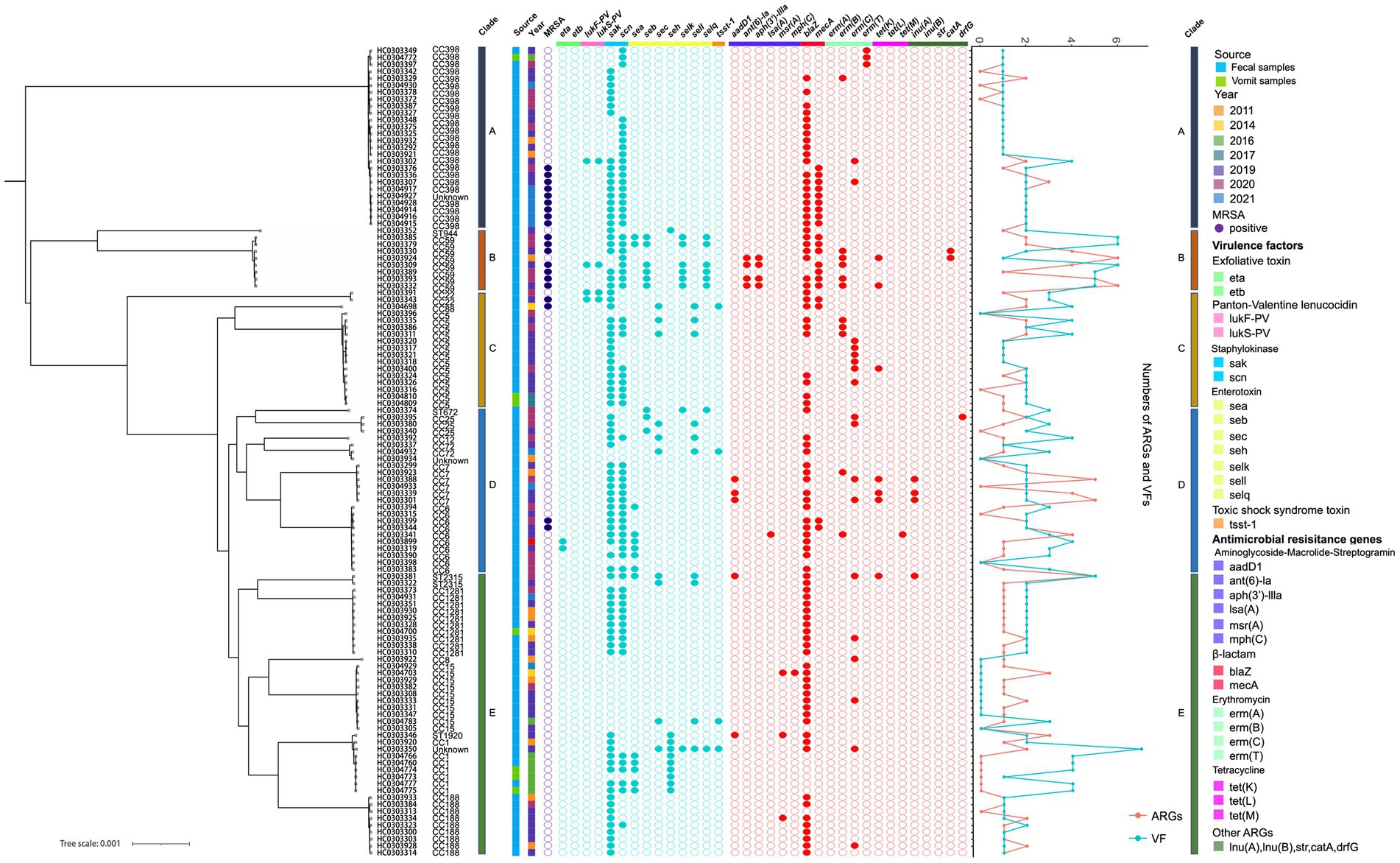
Figure 4. Genetic characteristics of 117 foodborne S. aureus strains. The clade, source, year, antimicrobial and virulence genes are depicted by colored squares. Filled or empty circles indicate the presence (filled) or absence (empty) of MRSA (violet), antimicrobial (red) and virulence (turquoise) genes in the 117 foodborne S. aureus strains. The line graph represents the total number of ARGs and VFs in the 117 strains.
The final predominant strain that caused diarrhea in food poisoning incidents was S. aureus CC15. Previous studies reported that a total of 8 CC types and 12 ST types of S. aureus were isolated from food poisoning incidents, with CC15 accounting for 5% of the total and carrying numerous enterotoxin genes such as sec, sed, and see (Lv et al., 2021). However, CC15 isolated in this study was not found to carry any enterotoxin genes, nor as lukF-PV and lukS-PV, suggesting a low pathogenicity but a high prevalence of hypotonic S. aureus contamination in food poisoning incidents in Eastern China (Figure 4, Table 1).
Virulence factors of Staphylococcus aureus isolates from the food poisoning incidents
One of the significant causes of diarrhea and other gastrointestinal problems was S. aureus enterotoxin genes (SEs). Genes for enterotoxins were found in 55 isolates in this investigation, with a prevalence of 27.8% (27/97) for MSSA and 3.5% (7/20) for MRSA (Table 2). CC1, CC5, CC25, and CC59 commonly had part of the sea, seb, sec, seh, selk, sell, and selq genes, however, neither clade A nor clade D contained any enterotoxin genes, except for five sea genes present in CC6 (Figure 4, Table 2).
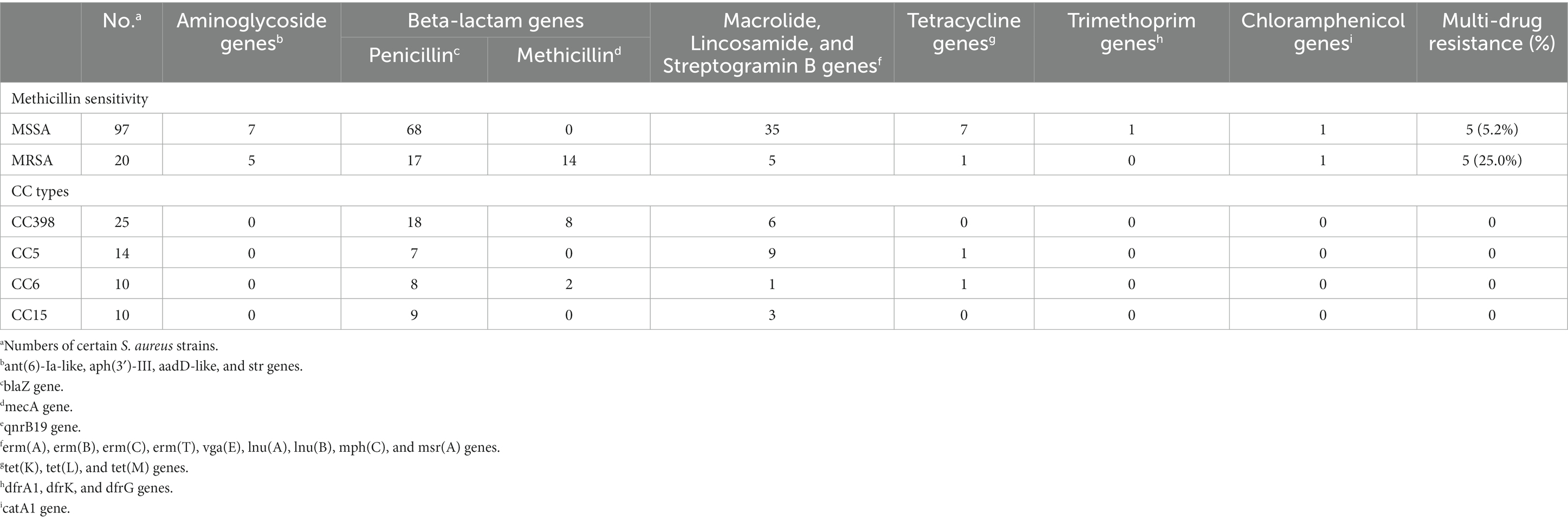
Table 2. Distribution of antimicrobial resistance genes according to sensitivity toward methicillin (MSSA vs. MRSA) and CC types.
The carrier rate of foodborne S. aureus enterotoxins varied greatly. A study examining the rate of classical staphylococcus enterotoxins revealed that 39.3% of the foodborne isolates were enterotoxin-positive, with sec and sea as the dominant genes (Zhang et al., 2022). Moreover, a correlation was observed between the prevalence of specific enterotoxin genes, e.g., sea and seh, with the severity of the associated illness (Argudín et al., 2010). This finding underscores the importance of monitoring the prevalence of enterotoxins in foodborne S. aureus and its relevance to outbreak tracing.
The pathogenicity of these S. aureus isolates differed from that of other virulent factors. A total of 14 virulent factors, including the functional components Panton-Valentine leucocidin, toxic shock syndrome toxin, staphylokinase, exfoliative toxin, and enterotoxin, were examined among 117 foodborne S. aureus strains. Of the 117 isolates, only two exfoliative toxin-positive genes (eta) and four tsst-1-positive genes were detected, showing that S. aureus isolates from the foodborne epidemic was hypotonic and enterotoxins were the predominant pathogenicity in the context of food safety (Figure 4, Table 2).
The pvl gene was detected only in four isolates (4/117, 3.4%) in this study, which was surpassed by prior research that found Panton-Valentine leucocidin to be detectable in MRSA at a rate of 24.1% (Wu et al., 2019). Such a high PVL carriage rate among MRSA isolates was observed for food in China, suggesting considerable diversity in the frequency of PVL across foodborne S. aureus strains.
Furthermore, a total of 69 isolates were confirmed to contain scn genes, including 18 isolates from the MRSA group (Figure 4, Table 2). As a recognized marker of the immune evasion cluster (IEC), the scn gene was found in high frequency in human hosts, indicating that the gene may be utilized to differentiate strains that are transferred to humans from environments and animals (de Jong et al., 2018). The scn gene appeared to be more prevalent in the MSSA group. An earlier study conducted in China found an 81.3% (52/64) prevalence of scn in MSSA isolates from the intestinal tracts of adult patients (Li et al., 2022). In our study, the scn gene was detected in more than 59.0% of the isolates, demonstrating that the causes of infections in food poisoning incidents were widespread, and included both human-to-human transmission and environmental and livestock-associated contaminations.
Genes associated with the biofilm formation were predicted in the S. aureus strains. Interestingly, the icaC, icaD, icaR, and capsule genes (capA to capP) were found in majority of the 117 isolates, whereas icaB was found in 107 strains (Supplementary Table S4, Supplementary Figure S1). Furthermore, arg and luxS genes were found in all isolates, which were linked to pathogenicity, such as Quorum Sensing, demonstrating the potential ability to form biofilm in connection to pathogenicity in S. aureus (Reading and Sperandio, 2006) (see Table 3).
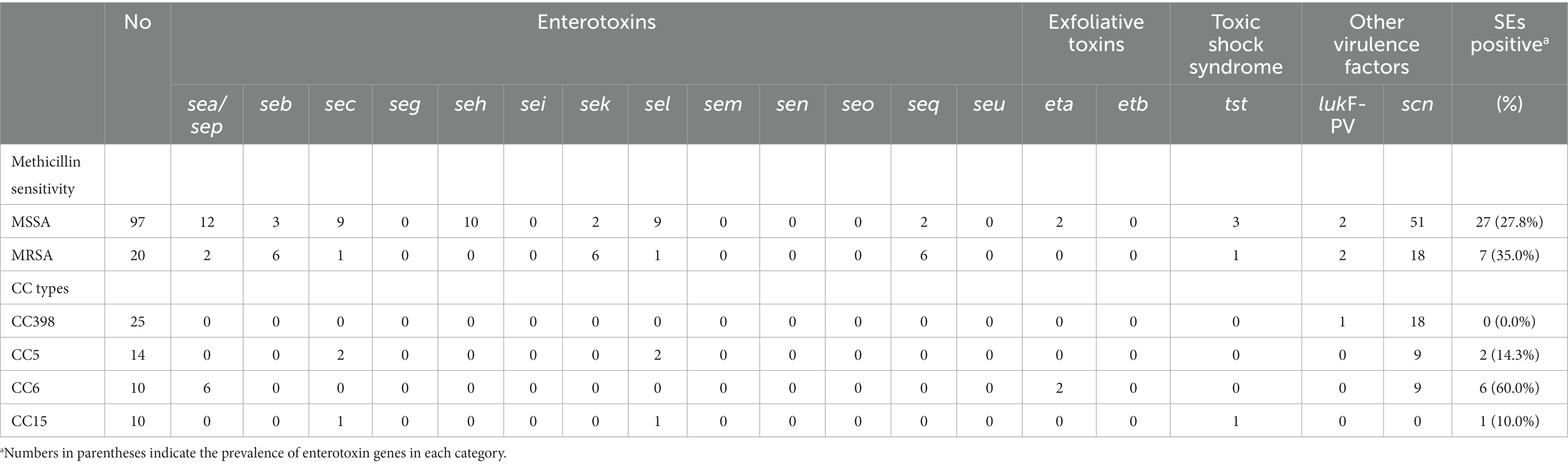
Table 3. Distribution of virulence factors according to sensitivity toward methicillin (MSSA vs. MRSA) and CC types.
Finally, prophage ΦSa3 and ΦAVβ were identified in this study (Supplementary Table S4, Supplementary Figure S1). The majority of human-associated S. aureus harbored β-hemolysin negative-converting bacteriophages, which are classified as ΦSa3 (Goerke et al., 2009). In this study, prophage ΦSa3 was found in 69 isolates, which was consistent with the findings of the scn genes and indicated that S. aureus has jumped its host from animals to humans. Additionally, ΦAVβ prophage genes were the indicators of the human-to-poultry host jump (Lowder et al., 2009). A total of 14 isolates were found to harbor the ΦAVβ genes (SAAV_2008, SAAV_2009) in CC5 strains, which showed that chicken items may have been a source of contamination for the current food poisoning incidents. However, further research is required to elucidate the bacteriophages of pathogenic S. aureus in food poisoning incidents.
Antimicrobial resistance of Staphylococcus aureus isolates from food poisoning incidents
The antimicrobial susceptibility pattern observed in S. aureus isolates from various outbreaks revealed that only ten isolates displayed resistance to at least one antimicrobial, including penicillin, tetracycline, and macrolides (Figure 4, Table 2, Supplementary Table S5). Antimicrobial resistance indicators were substantially more prevalent in CC398 isolates than in other CC types. Macrolide, Lincosamide, and Streptogramin B genes were found in 36.8% of MSSA isolates and 25.0% of MRSA isolates, respectively. Similar to previous research, we found that 93.0% of the S. aureus isolates in our investigation were penicillin-resistant (Table 2) (Li et al., 2022).
The frequency of antimicrobial resistance genes from S. aureus in diarrhea patients had been extensively investigated. For instance, a total of 187 S. aureus clinical isolates were collected in South China from 2010 to 2016 (Liang et al., 2019). Among them, 103 isolates were identified as MRSA with resistance to erythromycin (64.1%), clindamycin (48.5%), gentamicin (36.9%) and ciprofloxacin (34.0%). These findings demonstrated that the therapeutic management of hypervirulent MRSA infection may be complicated by MDR isolates colonizing livestock.
Intriguingly, tetracycline resistance is a hallmark of the CC398 clade, and tetracycline accumulation in CC398 has been reported to be associated with rapid radiation from humans to livestock (Price et al., 2012). However, all the CC398 strains were tetracycline susceptible in this study. This may be explained by the fact that the current strains were all isolated from human infection instead of livestock origination. In a prior study, we conducted a sampling of pork in Beijing and found that the livestock-associated CC398 harbored a high number of tetracycline resistance genes including tetK, and tetM (Li H. et al., 2021). Altogether, our findings support the differential carriage of tetracycline resistance in S. aureus between humans and livestock reported by Price et al.
Furthermore, CC59 was found to harbor a bunch of antimicrobial resistance and virulence genes including eta, etb, scn, sea,seb,selk, selq, ant(6)-la, blaZ, erm(B), tet(K) and others in this study. CC59 was a predominant clonal lineage of community-acquired S. aureus circulating in Asia (Pang et al., 2020). Previous research described two distinct clones of the ST59 sequence type, PVL-negative/SAK-positive and PVL-positive/SAK-negative (Hung et al., 2016). In this study, a similar mode of distinct clones was observed that seven CC59 strains were PVL-negative/SAK-positive and the rest one was PVL-positive/SAK-negative. Moreover, CC59 isolated from food chain were reported to be resistant to ampicillin, penicillin, erythromycin, tetracycline and others (Pang et al., 2020), which was consistent with the results in this study.
In addition, vancomycin-resistant S. aureus strains were reportedly isolated from food products in Egypt, with a detection rate of 64.7% for vanA and 29.4% for vanB (Saber et al., 2022). However, none of the 117 foodborne S. aureus isolates in this study tested positive for vanA (Supplementary Table S3). The S. aureus virulence factors linked to staphylococcal food poisoning are of importance in terms of food safety. In this study, the enterotoxin A, B, and C genes (sea/seb/s) were shown to be the most frequent enterotoxin genes in the current foodborne S. aureus isolates. These enterotoxins have been linked to staphylococcal food poisoning as well as toxic shock syndrome in humans (Benkerroum, 2018). Although our results suggested that these diarrhea episodes were hypotonic and merely transient low-MDR infections, however, further research for continued surveillance given the detection of virulence and antimicrobial resistance determinants is required to elucidate the genomic characteristics of pathogenic S. aureus in food poisoning incidents in the context of public health.
This study has some limitations. This investigation primarily focused on the events produced by S. aureus due to the range of pathogenic bacteria leading to food poisoning incidents. Nevertheless, diarrhea caused by Salmonella and Shigella was more frequent in most cases (Kotloff, 2022). Additionally, the sample size and strain counts were limited and only covered the period from 2011 to 2021 in terms of food poisoning incidents in Eastern China. To enhance the utilization of genetics in food safety research and manufacturing, it is crucial to gather additional data on foodborne S. aureus in future studies.
Conclusion
In conclusion, a combined bioinformatics approach was employed to study the genetic characteristics of S. aureus strains isolated from food poisoning incidents in Eastern China from 2011 to 2021. A number of 19 unique CC/ST types were identified among the foodborne S. aureus genomes, with CC398, CC5, CC6, and CC15 being the dominant types, respectively. Genes for enterotoxins were found in 55 isolates, while the rest virulence factors were merely detected, showing that S. aureus isolates from the foodborne epidemic were hypotonic and enterotoxins were the predominant pathogenicity. Antimicrobial resistance indicators were substantially more prevalent in CC398 isolates, however, only ten isolates displayed multi-drug resistance in the present study, suggesting that these diarrhea episodes may not pose a major clinical risk for treatment-resistance infections among the food poisoning incidents that occurred in Eastern China between 2011 to 2021.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: CRA010922 (https://ngdc.cncb.ac.cn/gsa/browse/CRA010922).
Ethics statement
The studies involving humans were approved by Research Ethics Committee of Soochow University. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
SY, YZ, DF, QJ, and TL performed the analysis. HL and ZZ wrote the main manuscript text and GJ prepared the figures. All authors contributed to the article and approved the submitted version.
Funding
The project was supported by the National Natural Science Foundation of China (No. 32170003 to ZZ, No. 82202465 to HL). This work was supported by agricultural innovation grants from Suzhou Science and Technology Project (N316460121 to HL) and college students’ innovation and entrepreneurship training program of Soochow University (202310285177Y to SY).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1225472/full#supplementary-material
Footnotes
References
Aires-de-Sousa, M. (2017). Methicillin-resistant Staphylococcus aureus among animals: current overview. Clin. Microbiol. Infect. 23, 373–380. doi: 10.1016/j.cmi.2016.11.002
Argudín, M. Á., Mendoza, M. C., and Rodicio, M. R. (2010). Food poisoning and Staphylococcus aureus enterotoxins. Toxins (Basel). 2, 1751–1773. doi: 10.3390/toxins2071751
Baumgartner, A., Niederhauser, I., and Johler, S. (2014). Virulence and resistance gene profiles of Staphylococcus aureus strains isolated from ready-to-eat foods. J. Food Prot. 77, 1232–1236. doi: 10.4315/0362-028X.JFP-14-027
Benkerroum, N. (2018). Staphylococcal enterotoxins and enterotoxin-like toxins with special reference to dairy products: an overview. Crit. Rev. Food Sci. Nutr. 58, 1943–1970. doi: 10.1080/10408398.2017.1289149
Bhatia, A., and Zahoor, S. (2007). Staphylococcus aureus enterotoxins: a review. J. Clin. Diagn. Res. 3, 188–197.
Bortolaia, V., Kaas, R. S., Ruppe, E., Roberts, M. C., Schwarz, S., Cattoir, V., et al. (2020). ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 75, 3491–3500. doi: 10.1093/jac/dkaa345
Boyle-Vavra, S., and Daum, R. S. (2007). Community-acquired methicillin-resistant Staphylococcus aureus: the role of Panton-valentine leukocidin. Lab. Investig. 87, 3–9. doi: 10.1038/labinvest.3700501
Bukowski, M., Wladyka, B., and Dubin, G. (2010). Exfoliative toxins of Staphylococcus aureus. Toxins (Basel). 2, 1148–1165. doi: 10.3390/toxins2051148
Carrera, M., Böhme, K., Gallardo, J. M., Barros-Velázquez, J., Cañas, B., and Calo-Mata, P. (2017). Characterization of foodborne strains of Staphylococcus aureus by shotgun proteomics: functional networks, virulence factors and species-specific peptide biomarkers. Front. Microbiol. 8:2458. Published 2017 Dec 11. doi: 10.3389/fmicb.2017.02458
de Jong, N. W. M., Vrieling, M., Garcia, B. L., Koop, G., Brettmann, M., Aerts, P. C., et al. (2018). Identification of a staphylococcal complement inhibitor with broad host specificity in equid Staphylococcus aureus strains. J. Biol. Chem. 293, 4468–4477. doi: 10.1074/jbc.RA117.000599
Feil, E. J., Li, B. C., Aanensen, D. M., Hanage, W. P., and Spratt, B. G. (2004). eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186, 1518–1530. doi: 10.1128/JB.186.5.1518-1530.2004
Gagliotti, C., Högberg, L. D., Billström, H., Eckmanns, T., Giske, C. G., Heuer, O. E., et al. (2021). Staphylococcus aureus bloodstream infections: diverging trends of meticillin-resistant and meticillin-susceptible isolates, EU/EEA, 2005 to 2018. Euro Surveill. 26:2002094. doi: 10.2807/1560-7917.ES.2021.26.46.2002094
Goerke, C., Pantucek, R., Holtfreter, S., Schulte, B., Zink, M., Grumann, D., et al. (2009). Diversity of prophages in dominant Staphylococcus aureus clonal lineages. J. Bacteriol. 191, 3462–3468. doi: 10.1128/JB.01804-08
Gouaux, E., Hobaugh, M., and Song, L. (1997). Alpha-Hemolysin, gamma-hemolysin, and leukocidin from Staphylococcus aureus: distant in sequence but similar in structure. Protein Sci. 6, 2631–2635. doi: 10.1002/pro.5560061216
Guo, Y., Yu, X., Wang, J., Hua, D., You, Y., Wu, Q., et al. (2023). A food poisoning caused by ST7 staphylococcal aureus harboring sea gene in Hainan province, China. Front. Microbiol. 14:1110720. Published 2023 Mar 16. doi: 10.3389/fmicb.2023.1110720
Gurevich, A., Saveliev, V., Vyahhi, N., and Tesler, G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075. doi: 10.1093/bioinformatics/btt086
Hansen, R. L., and Lee, Y. J. (2017). Overlapping MALDI-mass spectrometry imaging for in-parallel MS and MS/MS data acquisition without sacrificing spatial resolution. J. Am. Soc. Mass Spectrom. 28, 1910–1918. doi: 10.1007/s13361-017-1699-7
Holzknecht, B. J., Dargis, R., Pedersen, M., Pinholt, M., and Christensen, J. J., Danish Enterococcal Study Group (2018). Typing of vancomycin-resistant enterococci with MALDI-TOF mass spectrometry in a nosocomial outbreak setting. Clin. Microbiol. Infect. 24, 1104.e1–1104.e4. doi: 10.1016/j.cmi.2018.03.020
Hung, W. C., Wan, T. W., Kuo, Y. C., Yamamoto, T., Tsai, J. C., Lin, Y. T., et al. (2016). Molecular evolutionary pathways toward two successful community-associated but multidrug-resistant st59 methicillin-resistant staphylococcus aureus lineages in Taiwan: dynamic modes of mobile genetic element salvages. PLoS One 11:e0162526. doi: 10.1371/journal.pone.0162526
Johansson, M. H. K., Bortolaia, V., Tansirichaiya, S., Aarestrup, F. M., Roberts, A. P., and Petersen, T. N. (2021). Detection of mobile genetic elements associated with antibiotic resistance in Salmonella enterica using a newly developed web tool: MobileElementFinder. J. Antimicrob. Chemother. 76, 101–109. doi: 10.1093/jac/dkaa390
Johler, S., Giannini, P., Jermini, M., Hummerjohann, J., Baumgartner, A., and Stephan, R. (2015). Further evidence for staphylococcal food poisoning outbreaks caused by egc-encoded enterotoxins. Toxins (Basel). 7, 997–1004. Published 2015 Mar 20. doi: 10.3390/toxins7030997
Kaas, R. S., Leekitcharoenphon, P., Aarestrup, F. M., and Lund, O. (2014). Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS One 9:e104984. Published 2014 Aug 11. doi: 10.1371/journal.pone.0104984
Kadariya, J., Smith, T. C., and Thapaliya, D. (2014). Staphylococcus aureus and staphylococcal food-borne disease: an ongoing challenge in public health. Biomed. Res. Int. 2014:827965. doi: 10.1155/2014/827965
Kotloff, K. L. (2022). Bacterial diarrhoea. Curr. Opin. Pediatr. 34, 147–155. doi: 10.1097/MOP.0000000000001107
Laumay, F., Benchetrit, H., Corvaglia, A. R., van der Mee-Marquet, N., and François, P. (2021). The Staphylococcus aureus CC398 lineage: an evolution driven by the acquisition of prophages and other mobile genetic elements. Genes (Basel) 12:1752. Published 2021 Oct 30. doi: 10.3390/genes12111752
Letunic, I., and Bork, P. (2019). Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259. doi: 10.1093/nar/gkz239
Leung, D. Y., Meissner, H. C., Fulton, D. R., Murray, D. L., Kotzin, B. L., and Schlievert, P. M. (1993). Toxic shock syndrome toxin-secreting Staphylococcus aureus in Kawasaki syndrome. Lancet 342, 1385–1388. doi: 10.1016/0140-6736(93)92752-F
Li, W, Wu, S, Fu, P, Liu, J, Han, H, Bai, L, et al. (2018). National molecular tracing network for foodborne disease surveillance in China. Food Control. 88, 28–32. doi: 10.1016/j.foodcont.2017.12.032
Li, W., Cui, Q., Bai, L., Fu, P., Han, H., Liu, J., et al. (2021). Application of whole-genome sequencing in the national molecular tracing network for foodborne disease surveillance in China. Foodborne Pathog. Dis. 18, 538–546. doi: 10.1089/fpd.2020.2908
Li, Y., Tang, Y., Jiang, Z., Wang, Z., Li, Q., and Jiao, X. (2022). Molecular characterization of methicillin-sensitive Staphylococcus aureus from the intestinal tracts of adult patients in China. Pathogens 11:978. Published 2022 Aug 26. doi: 10.3390/pathogens11090978
Li, H., Tang, T., Stegger, M., Dalsgaard, A., Liu, T., and Leisner, J. J. (2021). Characterization of antimicrobial-resistant Staphylococcus aureus from retail foods in Beijing. China. Food Microbiol. 93:103603. doi: 10.1016/j.fm.2020.103603
Liang, Y., Tu, C., Tan, C., el-Sayed Ahmed, M. A. E. G., Dai, M., Xia, Y., et al. (2019). Antimicrobial resistance, virulence genes profiling and molecular relatedness of methicillin-resistant Staphylococcus aureus strains isolated from hospitalized patients in Guangdong Province, China. Infect Drug Resist. 12, 447–459. Published 2019 Feb 25. doi: 10.2147/IDR.S192611
Liu, B., Zheng, D., Jin, Q., Chen, L., and Yang, J. (2019). VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 47, D687–D692. doi: 10.1093/nar/gky1080
Lowder, B. V., Guinane, C. M., Zakour, N. L. B., Weinert, L. A., Conway-Morris, A., Cartwright, R. A., et al. (2009). Recent human-to-poultry host jump, adaptation, and pandemic spread of Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A. 106, 19545–19550. doi: 10.1073/pnas.0909285106
Lv, G., Jiang, R., Zhang, H., Wang, L., Li, L., Gao, W., et al. (2021). Molecular characteristics of Staphylococcus aureus from food samples and food poisoning outbreaks in Shijiazhuang, China. Front. Microbiol. 12:652276. Published 2021 Jun 22. doi: 10.3389/fmicb.2021.652276
Mahros, M. A., Abd-Elghany, S. M., and Sallam, K. I. (2021). Multidrug-, methicillin-, and vancomycin-resistant Staphylococcus aureus isolated from ready-to-eat meat sandwiches: an ongoing food and public health concern. Int. J. Food Microbiol. 346:109165. doi: 10.1016/j.ijfoodmicro.2021.109165
Matuszewska, M., Murray, G. G. R., Harrison, E. M., Holmes, M. A., and Weinert, L. A. (2020). The evolutionary genomics of host specificity in Staphylococcus aureus. Trends Microbiol. 28, 465–477. doi: 10.1016/j.tim.2019.12.007
Pang, R., Wu, S., Zhang, F., Huang, J., Wu, H., Zhang, J., et al. (2020). The genomic context for the evolution and transmission of community-associated staphylococcus aureus st59 through the food chain. Front. Microbiol. 11:422. doi: 10.3389/fmicb.2020.00422
Price, L. B., Stegger, M., Hasman, H., Aziz, M., Larsen, J., Andersen, P. S., et al. (2012). Staphylococcus aureus CC398: host adaptation and emergence of methicillin resistance in livestock. MBio 3, e00305–e00311. doi: 10.1128/mBio.00305-11
Reading, N. C., and Sperandio, V. (2006). Quorum sensing: the many languages of bacteria. FEMS Microbiol. Lett. 254, 1–11. doi: 10.1111/j.1574-6968.2005.00001.x
Saber, T., Samir, M., el-Mekkawy, R. M., Ariny, E., el-Sayed, S. R., Enan, G., et al. (2022). Methicillin- and vancomycin-resistant Staphylococcus aureus from humans and ready-to-eat meat: characterization of antimicrobial resistance and biofilm formation ability. Front. Microbiol. 12:735494. Published 2022 Feb 8. doi: 10.3389/fmicb.2021.735494
Stegger, M., Lindsay, J. A., Sørum, M., Gould, K. A., and Skov, R. (2010). Genetic diversity in CC398 methicillin-resistant Staphylococcus aureus isolates of different geographical origin. Clin. Microbiol. Infect. 16, 1017–1019. doi: 10.1111/j.1469-0691.2009.03003.x
Suzuki, Y., Omoe, K., Hu, D. L., Sato'o, Y., Ono, H. K., Monma, C., et al. (2014). Molecular epidemiological characterization Ofstaphylococcus aureusisolates originating from food poisoning outbreaks that occurred in Tokyo, Japan. Jpn. J. Microbiol. 58, 570–580. doi: 10.1111/1348-0421.12188
Tang, Y., Larsen, J., Kjeldgaard, J., Andersen, P. S., Skov, R., and Ingmer, H. (2017). Methicillin-resistant and -susceptible Staphylococcus aureus from retail meat in Denmark. Int. J. Food Microbiol. 249, 72–76. doi: 10.1016/j.ijfoodmicro.2017.03.001
Urwin, R., and Maiden, M. C. (2003). Multi-locus sequence typing: a tool for global epidemiology. Trends Microbiol. 11, 479–487. doi: 10.1016/j.tim.2003.08.006
Verkade, E., and Kluytmans, J. (2014). Livestock-associated Staphylococcus aureus CC398: animal reservoirs and human infections. Infect. Genet. Evol. 21, 523–530. doi: 10.1016/j.meegid.2013.02.013
Wang, W., Baloch, Z., Jiang, T., Zhang, C., Peng, Z., Li, F., et al. (2017). Enterotoxigenicity and antimicrobial resistance of Staphylococcus aureus isolated from retail food in China. Front. Microbiol. 8:2256. doi: 10.3389/fmicb.2017.02256
Wu, S., Huang, J., Wu, Q., Zhang, J., Zhang, F., Yang, X., et al. (2018). Staphylococcus aureus isolated from retail meat and meat products in China: incidence, antibiotic resistance and genetic diversity. Front. Microbiol. 9:2767. Published 2018 Nov 15. doi: 10.3389/fmicb.2018.02767
Wu, S., Huang, J., Zhang, F., Wu, Q., Zhang, J., Pang, R., et al. (2019). Prevalence and characterization of food-related methicillin-resistant Staphylococcus aureus (MRSA) in China. Front. Microbiol. 10:304. Published 2019 Feb 20. doi: 10.3389/fmicb.2019.00304
Xie, X., Bao, Y., Ouyang, N., Dai, X., Pan, K., Chen, B., et al. (2016). Molecular epidemiology and characteristic of virulence gene of community-acquired and hospital-acquired methicillin-resistant Staphylococcus aureus isolates in sun Yat-sen memorial hospital, Guangzhou, southern China. BMC Infect. Dis. 16:339. Published 2016 Jul 22. doi: 10.1186/s12879-016-1684-y
Zhang, J., Wang, J., Jin, J., Li, X., Zhang, H., Shi, X., et al. (2022). Prevalence, antibiotic resistance, and enterotoxin genes of Staphylococcus aureus isolated from milk and dairy products worldwide: a systematic review and meta-analysis. Food Res. Int. 162:111969. doi: 10.1016/j.foodres.2022.111969
Keywords: Staphylococcus aureus , food poison, diarrhea, antimicrobial resistance, virulence
Citation: Yu S, Zhou Y, Feng D, Jiang Q, Li T, Jiang G, Zhou Z and Li H (2023) Whole genome sequence-based characterization of virulence and antimicrobial resistance gene profiles of Staphylococcus aureus isolated from food poisoning incidents in eastern China. Front. Microbiol. 14:1225472. doi: 10.3389/fmicb.2023.1225472
Edited by:
Bo Yang, Jiangnan University, ChinaReviewed by:
Valentine Usongo, Health Canada, CanadaLili Zhang, Northeast Agricultural University, China
Changyong Cheng, Zhejiang A & F University, China
Copyright © 2023 Yu, Zhou, Feng, Jiang, Li, Jiang, Zhou and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Heng Li, aGxpQHN1ZGEuZWR1LmNu
†These authors have contributed equally to this work
‡Present Address: Heng Li, Soochow University, Suzhou, Jiangsu, China
 Shuyang Yu
Shuyang Yu Yuxuan Zhou†
Yuxuan Zhou† Zhemin Zhou
Zhemin Zhou Heng Li
Heng Li