
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
CORRECTION article
Front. Microbiol. , 08 August 2023
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1221779
This article is a correction to:
Pathogen genomics and phage-based solutions for accurately identifying and controlling Salmonella pathogens
 Angela V. Lopez-Garcia1
Angela V. Lopez-Garcia1 Manal AbuOun1
Manal AbuOun1 Javier Nunez-Garcia1
Javier Nunez-Garcia1 Janet Y. Nale2
Janet Y. Nale2 Edouard E. Gaylov3
Edouard E. Gaylov3 Preeda Phothaworn4
Preeda Phothaworn4 Chutikarn Sukjoi5
Chutikarn Sukjoi5 Parameth Thiennimitr5
Parameth Thiennimitr5 Danish J. Malik6
Danish J. Malik6 Sunee Korbsrisate4
Sunee Korbsrisate4 Martha R. J. Clokie3
Martha R. J. Clokie3 Muna F. Anjum1*
Muna F. Anjum1*A corrigendum on
Pathogen genomics and phage-based solutions for accurately identifying and controlling Salmonella pathogens
by Lopez-Garcia, A. V., AbuOun, M., Nunez-Garcia, J., Nale, J. Y., Gaylov, E. E., Phothaworn, P., Sukjoi, C., Thiennimitr, P., Malik, D. J., Korbsrisate, S., Clokie, M. R. J., and Anjum, M. F. (2023). Front. Microbiol. 14:1166615. doi: 10.3389/fmicb.2023.1166615
In the published article, there was an error in the Figure 3 and its legend as published. Figure 3B was removed from the manuscript; however, the associated legend text still needs to be removed and the key was not completely included in the original picture. The corrected Figure 3 and its caption appear below.
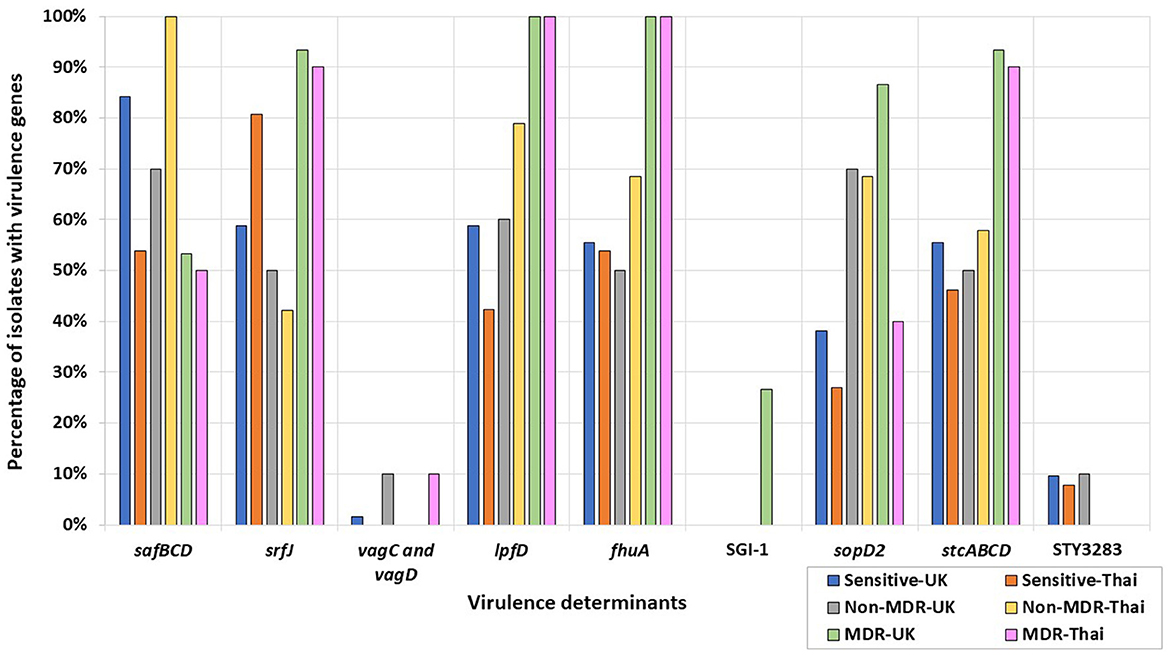
Figure 3. Percentages of UK and Thai MDR, non-MDR, and sensitive isolates harboring virulence determinants. MDR isolates from the UK (green), non-MDR isolates from the UK (grey), sensitive isolates from the UK (blue), MDR isolates from Thailand (pink), non-MDR isolates from Thailand (yellow), and sensitive isolates from Thailand (orange) have been included.
In the published article, there was an error in Table 1 as published. strA, strB and tetA(B) genes were misspelt as straA, straB and tet-AB. The corrected Table 1 and its caption appear below.
In the published article, the reference for the virulence genes safBCD, srfJ, lpfD, and fhuA were present in both MDR S. Kentucky isolates, which were identified as clones of a ST198 S. Kentucky global lineage (Martínez and Baquero, 2002; Beceiro et al., 2013) was incorrectly written as (Martínez and Baquero, 2002; Beceiro et al., 2013). It should be deleted.
In the published article, there was an error to Materials and methods, Phylogenetic and SNP analysis. The reference name should not contain commas.
This sentence previously stated:
“Snippy version v4.6.0 (Seemann, 2015) was used to detect SNPs in the core genome of S. Kentucky isolates BL700 and BL800, and two S. Kentucky MDR ST198 isolates from earlier research [SAMN08784244 and SAMN08784253; (Hawkey et al., 2019)], aligning them against the reference 201,001,922 (CP028357).”
The corrected sentence appears below:
“Snippy version v4.6.0 (Seemann, 2015) was used to detect SNPs in the core genome of S. Kentucky isolates BL700 and BL800, and two S. Kentucky MDR ST198 isolates from earlier research [SAMN08784244 and SAMN08784253; (Hawkey et al., 2019)], aligning them against the reference 201001922 (CP028357).”
In the published article, there was an error in Results, Antimicrobial resistance characterization. The Thai S.1,4,12:i:- isolate does not present resistance to streptomycin.
This sentence previously stated:
“One Thai isolate, serotyped as S. 1,4,12:i:-, was resistant to seven antimicrobial classes (ampicillin, chloramphenicol, ciprofloxacin-nalidixic acid, gentamicin-streptomycin, sulfamethoxazole, tetracycline, and trimethoprim).”
The corrected sentence appears below:
“One Thai isolate, serotyped as S. 1,4,12:i:-, was resistant to seven antimicrobial classes (ampicillin, chloramphenicol, ciprofloxacin-nalidixic acid, gentamicin, sulfamethoxazole, tetracycline, and trimethoprim).”
In the published article, there was an error in Results, Characterization of MDR isolates with distinct virulence profiles, 3rd Paragraph. The tetA(B) gene was misspelt.
This sentence previously stated:
“From the resolved genome of MDR isolate BL708 S. 1,4,[5],12:i:-, we identified SGI-4 in the chromosome with genes showing resistance to copper, arsenic, mercury, and antimicrobials [blaTem−1b, sul2, tet(AB), strA, strB].”
The corrected sentence appears below:
“From the resolved genome of MDR isolate BL708 S. 1,4,[5],12:i:-, we identified SGI-4 in the chromosome with genes showing resistance to copper, arsenic, mercury, and antimicrobials [blaTEM−1B, sul2, tetA(B), strA, strB].”
In the published article, there was an error with the percentage of MDR isolates reported in the Author's Summary.
The sentence previously stated:
“The results indicated genes harboring resistance to antimicrobials differed between the countries, possibly due to differing farming practices; however, 17–18% of all isolates were multidrug resistant.”
The corrected sentence appears below:
“The results indicated genes harboring resistance to antimicrobials differed between the countries, possibly due to differing farming practices; however, 14%−15% of all isolates were multidrug resistant.”
In the published article, there was an error in Supplementary Table S5. The tet-AB gene was misspelt and should be replaced by tetA(B). The supplementary table with the error typo amended has been submitted.
The authors apologize for these errors and state that this does not change the scientific conclusions of the article in any way. The original article has been updated.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Hawkey, J., Le Hello, S., Doublet, B., Granier, S. A., Hendriksen, R. S., Fricke, W. F., et al. (2019). Global phylogenomics of multidrug-resistant Salmonella enterica serotype Kentucky ST198. Microb. Genom. 5. doi: 10.1099/mgen.0.000269
Keywords: antimicrobial resistance, Salmonella, virulence genes, genomics, bacteriophages, serovar
Citation: Lopez-Garcia AV, AbuOun M, Nunez-Garcia J, Nale JY, Gaylov EE, Phothaworn P, Sukjoi C, Thiennimitr P, Malik DJ, Korbsrisate S, Clokie MRJ and Anjum MF (2023) Corrigendum: Pathogen genomics and phage-based solutions for accurately identifying and controlling Salmonella pathogens. Front. Microbiol. 14:1221779. doi: 10.3389/fmicb.2023.1221779
Received: 12 May 2023; Accepted: 26 July 2023;
Published: 08 August 2023.
Approved by:
Frontiers Editorial Office, Frontiers Media SA, SwitzerlandCopyright © 2023 Lopez-Garcia, AbuOun, Nunez-Garcia, Nale, Gaylov, Phothaworn, Sukjoi, Thiennimitr, Malik, Korbsrisate, Clokie and Anjum. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Muna F. Anjum, TXVuYS5Bbmp1bUBhcGhhLmdvdi51aw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.