
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol., 27 June 2023
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1221478
 Marie Verhaegen1
Marie Verhaegen1 Thomas Bergot1
Thomas Bergot1 Ernesto Liebana2
Ernesto Liebana2 Giuseppe Stancanelli2
Giuseppe Stancanelli2 Franz Streissl2
Franz Streissl2 Marie-Paule Mingeot-Leclercq3
Marie-Paule Mingeot-Leclercq3 Jacques Mahillon1
Jacques Mahillon1 Claude Bragard4*
Claude Bragard4*Despite growing attention, antibiotics (such as streptomycin, oxytetracycline or kasugamycin) are still used worldwide for the control of major bacterial plant diseases. This raises concerns on their potential, yet unknown impact on antibiotic and multidrug resistances and the spread of their genetic determinants among bacterial pathogens. Antibiotic resistance genes (ARGs) have been identified in plant pathogenic bacteria (PPB), with streptomycin resistance genes being the most commonly reported. Therefore, the contribution of mobile genetic elements (MGEs) to their spread among PPB, as well as their ability to transfer to other bacteria, need to be further explored. The only well-documented example of ARGs vector in PPB, Tn5393 and its highly similar variants (carrying streptomycin resistance genes), is concerning because of its presence outside PPB, in Salmonella enterica and Klebsiella pneumoniae, two major human pathogens. Although its structure among PPB is still relatively simple, in human- and animal-associated bacteria, Tn5393 has evolved into complex associations with other MGEs and ARGs. This review sheds light on ARGs and MGEs associated with PPB, but also investigates the potential role of antibiotic use in resistance selection in plant-associated bacteria.
Plant pathogenic bacteria (PPB) cause devastating losses of crops worldwide, notably in vineyards, pear and apple orchards, estimated from one billion dollars every year (Mansfield et al., 2012; Kannan et al., 2015) up to five billion euros (Sánchez et al., 2019; Schneider et al., 2020). To cite a few, PPB such as Erwinia amylovora (affecting mainly pear orchards), Xanthomonas oryzae (damaging rice cultures), Pseudomonas syringae pv. actinidiae (harmful to kiwi plantations) or Ralstonia solanacearum (impacting tomato yields) represent major concerns. Emerging bacterial plant diseases such as Candidatus Liberibacter sp. (also known as Huanglongbing on citrus) or Xylella fastidiosa, which is the causal agent of Pierce’s disease of grapevine, citrus variegated chlorosis, olive quick decline syndrome and many other plant diseases, are growing threats. Therefore, on a worldwide scale, farmers often resort to antibiotics as a simple and effective tool for the control of bacterial diseases.
Although the amount of antibiotics used in plant protection is considered to be very low compared to human and veterinary medicine (McGhee and Sundin, 2011; McManus, 2014; FAO, OIE, and WHO 2018; Sundin and Wang, 2018), it has been suggested that their use may be more widespread than previously thought (McManus et al., 2002; McManus, 2014; O’Neill, 2015; Taylor and Reeder, 2020). The amounts effectively applied on crops are difficult to assess due to the lack of precise monitoring in different parts of the world. Currently, antibiotics authorized as plant protection products (PPPs) are commonly grouped with fungicides, e.g., in the Food and Agriculture Organization (FAO) statistics, because specific antibiotics can also be antifungal agents. Presently, different legislations, such as in Europe and West Africa, do not authorize antibiotics as PPPs, while their use is, to some extent, allowed in the American and Asian continents. Five antibiotics are most regularly reported to be used in plant agriculture: streptomycin (the most used antibiotic worldwide), oxytetracycline, kasugamycin, oxolinic acid (OA) and gentamicin (McManus, 2014; Sundin and Wang, 2018; Miller et al., 2022).
The application of antibiotics to plants exerts selective pressure on plant-associated bacteria, which can lead to the development of antibiotic resistance. Four main mechanisms result in bacterial antibiotic resistances: (i) inactivation of the antibiotic itself, (ii) reduction of its penetration (e.g., alteration of the cell membrane resulting in decreased permeability) or active elimination via efflux pumps, (iii) modification of the antibiotic targets, and (iv) use of alternative pathways [for reviews on antibiotic resistance mechanisms, refer to (Alekshun and Levy, 2007; Van Hoek et al., 2011; Blair et al., 2015; Munita and Arias, 2016)]. Bacteria have a high potential to acquire resistance to antibiotics, either through chromosomal mutations or via horizontal gene transfer (HGT) of antibiotic resistance genes (ARGs).
On the one hand, spontaneous gene mutations associated with the mechanism of action of the antibiotic can lead to cell survival. A resistant subpopulation can emerge and become prominent as the initial susceptible population will die (Munita and Arias, 2016). On the other hand, genetic material can be horizontally transferred through three main mechanisms (Dagan, 2011; Gyles and Boerlin, 2014): (i) transformation, which happens when a recipient cell is able to integrate free DNA found in the extracellular medium, (ii) transduction, where a bacteriophage (a bacterial virus) is involved in the gene transfer, and (iii) conjugation, that requires direct contact between two living cells for the exchange of genetic material. Mobile genetic elements (MGEs) can serve as vectors for the transfer of genetic material, either as intracellular, moving from one location to another within the same genome (e.g., insertion sequences or transposons), or intercellular elements, transferred from one cell to another (e.g., conjugative plasmids or integrative and conjugative elements, aka ICEs; Partridge et al., 2018).
The extent to which the use of antibiotics in plant agriculture could impact the global antibiotic resistance issue is unknown. The present review focuses on ARGs and MGEs associated with PPB. It highlights the potential correlation between the use of antibiotics against bacterial plant pathogens and the appearance of resistance. It also gathers critical information to be taken into account for risk assessments performed during the authorization process of an antibiotic for plant protection purposes.
There are five major antibiotics most commonly used throughout the world against PPB (streptomycin, oxytetracycline, kasugamycin, oxolinic acid and gentamicin), although at least 15 have been identified as being used against plant diseases (Table 1; McManus, 2014; Sundin and Wang, 2018; Taylor and Reeder, 2020; Miller et al., 2022). They belong to eight different classes of antibiotics but the most prevalent are the aminoglycosides and β-lactams (Table 1). Among the top five, only kasugamycin is not used in human nor in veterinary medicine (Kumar et al., 2005; Aarestrup et al., 2008; McGhee and Sundin, 2011; McManus, 2014; World Health Organization, 2019). This is also the case of other antibiotics, such as ningnanmycin, validamycin or zhongshengmycin, used in China. Potentially, more antibiotics could be used in plant protection, but their use is not well monitored and/or their efficacy has not been demonstrated. In addition, in some parts of the world, antibiotics are applied to plants even though they are not authorized, due to poor control of antibiotic sales and/or lack of knowledge from producers (Chanvatik et al., 2019). Besides, there is a constant search for new antibiotics to fight plant diseases; for instance, penicillin shows great promises to combat citrus greening (Supplementary Table S1; Shin et al., 2016).
Globally, the use of antibiotics is permitted in the American and Asian continents but currently not approved in Europe (even if some limited derogations were accepted in the past) and West Africa (Figure 1; Supplementary Table S2). However, it is rather difficult to accurately list countries authorizing their use as PPPs, not only because available data are limited (Table 1), but also because the official lists of authorized pesticides are often available in the country’s official language only. Moreover, many countries suffer from a lack of monitoring of the use of antibiotics on plants. In fact, to the best of our knowledge, only three countries release information on the amounts used on crops: the United States, New Zealand and India.
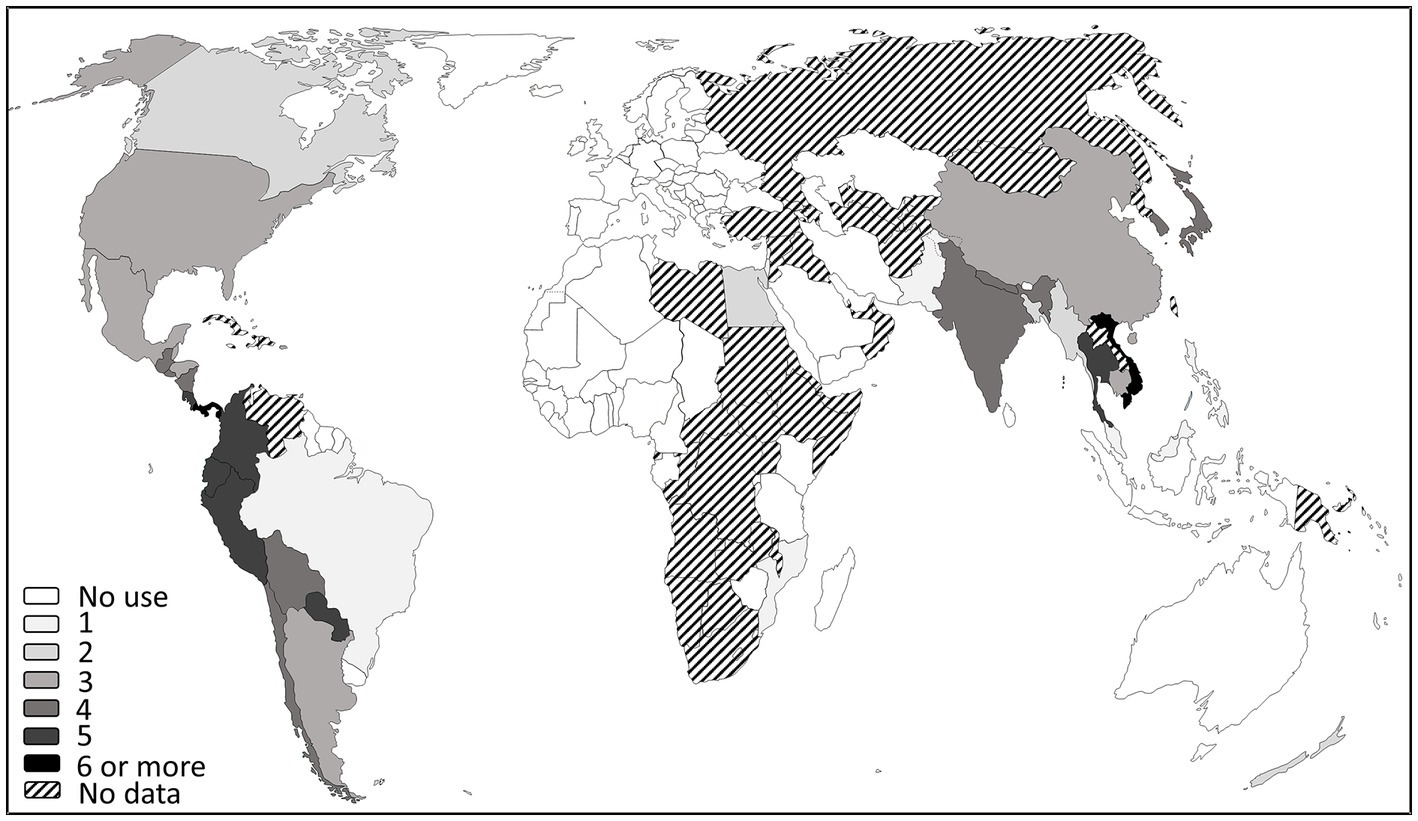
Figure 1. World map of countries where antibiotics are used as PPPs, compiled from scientific and grey literature searches screening the official list of authorized pesticides, when available, as of July 20, 2022 (list can be found in Supplementary Table S2). One hundred and ninety-five countries were considered, references could be found for more than 100 countries with around 70 with a clear indication that antibiotic use is not authorized in plants and around 30 countries with an indication of authorization. Numbers refer to the number of antibiotics used in each country. These data must be used with caution since legislations are subjected to rapid changes.
Several ARGs have been identified in PPB, mainly for the five main antibiotics used in plant protection. Also, the best documented ARGs are the streptomycin resistance genes, while the mechanisms involved in the other resistances are less known (Table 2; McManus, 2014; Sundin and Wang, 2018; Miller et al., 2022).
Streptomycin is used in plant protection against various PPB since the 1950s. The most common target of streptomycin is Erwinia amylovora, the causal agent of fire blight, which infects apple and pear trees, targeting leaves, flowers and shoots. Streptomycin is usually sprayed during bloom (McManus et al., 2002; Sundin and Wang, 2018).
Several mechanisms can be responsible for streptomycin resistance in PPB: (i) a point mutation of the rpsL gene, or (ii) the acquisition of strA-strB genes or aadA genes (Table 2). Other mechanisms have been suggested but they have not yet been confirmed and characterized. In E. amylovora, 95% of a total of 107 strains isolated in Mexico showed a mutation at codon 43 in rpsL, but the other 5% did not show rpsL mutation and did not carry strA-strB or aadA genes. The unknown resistance mechanism was not further investigated (de León Door et al., 2013). In Clavibacter michiganensis, another potential mechanism for streptomycin resistance was also suggested but needs further investigation (Lyu et al., 2019).
The rpsL gene encodes the ribosomal protein S12. A point mutation at codon 43 (mainly changing lysine (Lys) to arginine (Arg), rarely Lys to threonine or Lys to asparagine) prevents the binding of streptomycin to the ribosome. This mutation might be the most prevalent one since it retains a high environmental fitness even in the absence of streptomycin. Another less common mutation has been described at codon 88 (changing Lys to Arg). Both point mutations have been observed in various PPB (e.g., E. amylovora, C. michiganensis subsp. michiganensis, or in vitro in Xanthomonas oryzae pv. oryzicola or Erwinia carotovora, now named Pectobaterium carotovorum; Chiou and Jones, 1995b; Barnard et al., 2010; Zhang et al., 2011; Valenzuela et al., 2019; Escursell et al., 2021). In one streptomycin resistant strain of C. michiganensis, a point mutation at the 128th nucleotide of rpsL (Lys to Arg) was responsible for the resistance (Lyu et al., 2019). The rpsL chromosomal mutations might be the less worrying resistance mechanism because it is not prone to HGT.
Contrary to rpsL mutations, the acquisition of aadA or strA-strB genes requires more attention because of their presence on MGEs. Two alleles of aadA have been described, aadA1, the most prevalent allele, and aadA2. The aadA1 gene was found in isolates of X. oryzae pv. oryzae, whereas aadA2 conferred streptomycin resistance in a Pseudomonas strain (Schnabel and Jones, 1999; Xu et al., 2013). In X. oryzae pv. oryzae, three integrons carrying aadA1 have been described, which was the first report of resistance integrons in PPB (see sections 3.5 and 3.6; Xu et al., 2013).
The key actors in streptomycin resistance in PPB are the strA-strB genes, mainly found associated with Tn5393. This 6.7-kb transposon belongs to the Tn3 family. In its most simple forms, it encodes a putative transposase (TnpA) and a resolvase (TnpR), followed by a putative recombination site (res), an insertion sequence (IS) element (IS1133 in E. amylovora, IS6100 in Xanthomonas campestris), and by the strA-strB genes (Figure 2). These two genes are commonly encountered in human pathogens and are responsible for many infections associated with streptomycin resistant bacteria (Chiou and Jones, 1993; Sundin and Bender, 1995; Förster et al., 2015). They are widely disseminated among Gram-negative bacterial pathogens and commensals from humans, probably because of streptomycin use in clinical contexts. The strA-strB genes are usually found on small plasmids, such as RSF1010 in human pathogens, while they are most commonly encoded on large conjugative plasmids in plant pathogens (Förster et al., 2015).

Figure 2. Schematic representations of the different variants of Tn5393, based on GenBank accession numbers and references reported in Table 3. E. amylovora, P. syringae and X. campestris pv. vesicatoria are PPB, while Aeromonas sp., A. faecalis, S. enterica, Yersinia ruckeri and K. pneumoniae are human pathogenic and/or environmental bacteria. Tn5393 was first identified in E. amylovora and the successive variants discovered were then named alphabetically. It is however possible that Tn5393c, devoid of internal IS elements, compared to Tn5393, is the original version (Mindlin and Petrova, 2017). In this review, the original names of the different variants are used, but some publications have designated Tn5393c as Tn5393 and Tn5393a as an IS1133-bearing variant. Also note than Tn5393k was described twice but the two versions were not identical and are distinguished here as Tn5393k and Tn5393k’. Genes are not drawn to scale. Tc: truncated.
The strA-strB genes were identified in E. amylovora on plasmid pEA8.7, a plasmid very similar to the broad-host-range plasmid RSF1010, a 8.7-kb IncQ plasmid involved in streptomycin and sulfonamide resistances in bacterial human infections. It is non-conjugative but can be transferred horizontally through mobilization. Another RSF1010-like plasmid has also been observed in Erwinia herbicola in New Zealand, suggesting that IncQ plasmids of the RSF1010 family could be more widespread than previously thought (Palmer et al., 1997). It is hypothesized that the strA-strB sequence on RSF1010 originated from the insertion of Tn5393 into the ancestral plasmid from which RSF1010 evolved. Indeed, the right inverted repeat of Tn5393 (located downstream of strB) is conserved downstream of strB in RSF1010. This IR sequence would be the only vestige of Tn5393 on RSF1010. The strA-strB genes carried by RSF1010 diverged over time from the intact strA-strB genes encoded on Tn5393 from plant pathogens (Sundin, 2000).
Tn5393 is found on a variety of plasmids (Table 3). It was initially discovered on the large conjugative plasmid pEa34 (34 kb) in E. amylovora (Chiou, 1991; Chiou and Jones, 1993). The spread of streptomycin resistance genes among E. amylovora or other bacteria (pathogenic or not) could thus be facilitated by this conjugative plasmid via HGT. Tn5393 was also found on plasmid pEa29 from E. amylovora, where it can insert at several locations. This 29-kb plasmid is, unlike pEa34, unable to self-transfer to other strains, and is not related to pEa34. pEa29 is highly stable in E. amylovora and thus the integration of Tn5393 suggests the stable establishment of the resistance in E. amylovora populations.

Table 3. Organisms, main plasmid host and mobility, associated with the different variants of Tn5393 (Tn5393a to Tn5393l and Tn5393n).
Variants of Tn5393 have been described (Figure 2; Table 3). Tn5393a was identified in several strains of Pseudomonas syringae and Pseudomonas marginalis, and lacks the IS1133 of Tn5393 (Sundin and Bender, 1993, 1995; Han et al., 2004). It was also detected in isolates of E. amylovora, located on the conjugative plasmid pEU30 (30 kb; Foster et al., 2004; Förster et al., 2015). Interestingly, the conjugative machinery used by pEU30 most resembles the VirB system from pPSR1 of P. syringae (Foster et al., 2004). It could be hypothesized that the Tn5393a element detected in E. amylovora originated from P. syringae. Tn5393b was identified in X. campestris and contains IS6100 instead of IS1133. The presence of an insertion element increases the level of resistance in comparison to Tn5393a, which contains none (Sundin and Bender, 1995). IS6100 in Tn5393 from X. campestris is 100% identical to an element found in Mycobacterium fortuitum, Pseudomonas aeruginosa and Flavobacterium sp., suggesting its presence outside of PPB (Sundin and Bender, 1995). Also, the tnpR and res of Tn5393, Tn5393a and Tn5393b from E. amylovora, P. syringae and X. campestris, respectively, are identical (Sundin and Bender, 1995).
Other Tn5393 variants have been found in bacteria not pathogenic to plants (Figure 2; Table 3). Tn5393c, reported in the fish pathogen Aeromonas salmonicida (L’Abée-Lund and Sørum, 2000), does not carry any insertion sequence, like Tn5393a, and sequence comparison revealed that Tn5393c and Tn5393a are essentially the same. The presence of Tn5393c in A. salmonicida is surprising because streptomycin is not used to control A. salmonicida in Norway, where the transposon was first described in this bacterium. However, huge amounts of other antibiotics (including oxytetracycline and sulfonamides) have been used and could have selected for the transposon located on pRAS2 which also carries oxytetracycline and sulfonamide resistance determinants (L’Abée-Lund and Sørum, 2000).
Tn5393d was identified in a clinical strain of Alcaligenes faecalis, which can be responsible for human infections related to contamination of the medical equipment (Mantengoli and Rossolini, 2005). Tn5393d carries additional antibiotic resistance genes and is found on pFL424, a 44 -kb non-conjugative plasmid. This transposon seems to have originated from the consecutive insertion of two composite transposons containing aphA6b (conferring resistance to kanamycin, streptomycin and amikacin) and blaPER-1 (PER-1 extended spectrum beta-lactamase) genes (Mantengoli and Rossolini, 2005).
Tn5393e is a variant containing another transposon, Tn6023, and was found on the IncHI2 plasmid pSRC125. It was recovered from a multi-resistant Salmonella enterica serovar Typhimurium isolate of bovine origin. Tn6023 itself contains the aphA1b gene, conferring resistance to various aminoglycosides. Surprisingly, strains containing this transposon did not show resistance to streptomycin, even though the strA-strB genes present on the element did not carry any inactivating mutations, suggesting that these genes were not expressed (Cain and Hall, 2011). Tn5393f and Tn5393g were discovered in unidentified soil bacteria where manure had been applied. Both transposons were found on low GC conjugative plasmids carrying other ARGs such as sul2 or aac3, and Acinetobacter sp. was identified as the putative host (Heuer et al., 2009; Cain and Hall, 2011).
Tn5393h is derived from Tn5393a and includes Tn10, a tetracycline resistance transposon, inserted in the tnpA gene, which likely annihilates its mobility. Tn5393i contains IS1133, but this insertion element is not at the same location as in Tn5393a (namely in tnpR) (Cain and Hall, 2011). A variety of Tn5393 variants sequences are also available in GenBank (Cain and Hall, 2011). Tn5393j from Klebsiella pneumoniae carries another aminoglycoside resistance gene, aphA6, but its tnpR was not functional and its tnpA gene is missing (Espedido et al., 2013). Two variants were named Tn5393k but, in fact, they are not identical; for the sake of clarity, they are referred to as Tn5393k and Tn5393k’ (Figure 2; Table 3). Tn5393k is similar to Tn5393c and Tn5393f, and carries both IS26 and IS1. It is found in S. enterica sv. Typhimurium. The terminal inverted repeats (IRs) of Tn5393k exhibit 100% sequence identity with those in Tn5393, Tn5393c and Tn5393f (Yao et al., 2021). Tn5393k’ on the other hand is extremely similar to Tn5393d; the only difference is that it is lacking the ISPpu17 element (Adamczuk and Dziewit, 2017). Tn5393l carries the tetracycline resistance genes located in Tn10 and is also found in S. enterica sv. Typhimurium (Zhang et al., 2019). Finally, Tn5393n was found in Aeromonas caviae (Luo et al., 2022).
Tn5393 was also found in Corynebacterium striatum, Campylobacter jejuni, P. aeruginosa and Snodgrassella alvi (a honeybee gut symbiont; Sundin et al., 1995; Sundin, 2000; Ludvigsen et al., 2018). Similarly, Tn5393a was recently reported on a IncI1 type conjugative plasmid from S. enterica (Harmer, 2021) while Tn5393b was also found on the R64 plasmid of S. enterica sv. Typhimurium (Cain and Hall, 2011). Our recent BLAST run carried out on the Tn5393 sequence revealed its presence in many other bacteria, human pathogens or not. Among others, Tn5393 was found in Citrobacter koseri, Corynebacterium crudilactis, Enterobacter cancerogenus, Enterobacter cloacae, Enterobacter hormaechei, Escherichia coli, K. pneumoniae, Klebsiella quasipneumoniae, Klebsiella variicola and S. enterica, as well as in the plant root-associated bacterium Pseudomonas putida. However, this does not demonstrate that direct transfer of Tn5393 occurred between PPB and human pathogens but it certainly indicates that Tn5393 is much more widespread than previously thought. Similarly, the fact that this transposon was detected in diverse bacteria (Gram-negative as well as Gram-positive) in distinct geographical locations shows that it might be accessible to a large range of organisms.
The sequence of Tn5393a also retrieved numerous matches in the BLAST analysis, mostly in antibiotic resistant human pathogens, and in two PPB, P. syringae pv. actinidiae (plasmid pMG2_SR198) and Agrobacterium tumefaciens (plasmids pTi and pAt). Finally, Tn5393d displays strong similarities with plasmid p17-84_OXA of Acinetobacter baumannii, a critical multi-resistant pathogen in human health for which new antibiotics are needed according to the WHO (World Health Organization, 2017), and with pOXA58_100004 from Acinetobacter pitii. Tn5393e was found in other human bacteria, among which some are pathogenic.
In relation with human health implications, several important findings are worth mentioning. First, a tigecycline resistance mechanism called “resistance-nodulation-division” (RND) family efflux pump (tmexCD1-toprJ1 pump) was identified in K. pneumoniae isolates from humans and chickens. The corresponding gene tmexCD1-toprJ1 appeared to be found in Tn5393 and the authors suggest that it originated from the chromosome of Aeromonas spp. through Tn5393-mediated translocation (Sun et al., 2020). Second, a tigecycline resistance gene, tet(Y), was identified on a plasmid of a multi-resistant clinical isolate of A. baumannii. This plasmid carried numerous ARGs, including strA, strB, aac(6′)-Ib3, msr(E), mph(E), floR, ARR-3, sul1, dfrA19, tet(39) and tet(Y). Interestingly, the tet(Y) gene was located inside the sequence of Tn5393 and the authors also concluded that Tn5393 played a role in its transmission from Aeromonas spp. (Wang Z. et al., 2021). Lastly, in Australia, a multi-resistant A. baumannii strain that contains a fragment of Tn5393, was isolated, showing again its potential of ARGs spreading (Post and Hall, 2009). It is interesting to note that some species of Aeromonas or Acinetobacter can also be found in the environment and genetic transfers in these genera should be carefully considered.
Recently, the genomic evolution of Tn5393 and kin was examined and different steps were proposed on how Tn5393 could have acquired various MGEs and became more complex.1 It is also suggested that other variants of Tn5393 could exist (Lima-Mendez et al., 2020; Ross et al., 2021). Additionally, when comparing the different TnpA sequences from Tn5393 and its variants, it is rather complicated to establish their exact filiation and to determine how exchanges between bacterial species have shaped their structures. Still, the complexity of Tn5393 is apparently greater among human or animal pathogens than in PPB (Figure 2; Table 3).
Oxytetracycline belongs to the tetracycline class of antibiotics and is the second most used antibiotic in plant agriculture, mainly against fire blight (E. amylovora) in the case of streptomycin resistance reports. However, it is only used as a second line of defense because it is considered as less effective than streptomycin (McManus et al., 2002; Sundin and Wang, 2018). Various genes can be involved in tetracycline resistance: (i) expression of efflux pumps, (ii) enzymatic inactivation of the antibiotic or (iii) ribosomal protein protecting from the action of tetracyclines (Chopra and Roberts, 2001).
Concerning PPB, cases of oxytetracycline resistance are rarely reported. To the best of our knowledge, oxytetracycline resistance has not been observed yet in field isolates of E. amylovora, although, resistant strains can be obtained in vitro. In fact, the RP1 plasmid carrying an oxytetracycline resistance gene can be transferred from both E. herbicola and P. syringae pv. syringae to E. amylovora. Nevertheless, after pathogenesis, the plasmid phenotype could only be recovered in 0.03% of the isolates, suggesting that it may not be stable in that species (Lacy et al., 1984).
Oxytetracycline resistance cases have been reported in P. syringae (da Silva and Lopes, 1995; Spotts and Cervantes, 1995; Hwang et al., 2005), Xanthomonas arboricola pv. pruni (Herbert et al., 2022) and A. tumefaciens (Luo and Farrand, 1999). The resistance mechanisms were not described, except in X. arboricola pv. pruni, where it was shown that the tetracycline resistance gene tetC was carried by a plasmid, coding for an efflux pump (Herbert et al., 2022). Given the plethora of known tetracycline resistance genes (36 efflux pump genes, 13 ribosomal protection protein genes, 13 enzyme genes, 1 other gene conferring an unknown mechanism of resistance and 11 mosaic genes reported so far; Chopra and Roberts, 2001; Van Hoek et al., 2011; Roberts, 2021), others could be present in PPB but they have not been described yet. When searching for identity while running BLAST on PPB genomes with the sequence of tetC mentioned above, several strains present sequences with high identity percentage, such as X. arboricola pv. pruni, Burkholderia cepacia, Agrobacterium fabrum and several Streptomyces, which could be an indication that tetracycline resistance genes are present in other PPB.
Kasugamycin is an aminoglycoside antibiotic used only in plants, not in human nor veterinary medicine. In E. coli, point mutations in various genes (ksgA, ksgB, ksgC and ksgD) were shown to confer resistance (Sparling, 1970; Sparling et al., 1973; Yoshikawa et al., 1975; Fouts and Barbour, 1981; Yoshii et al., 2012). In PPB, different mechanisms have been identified (Table 2). The presence of aac(2′)-IIa gene in Burkholderia glumae and Acidovorax avenae ssp. avenae was shown to cause the resistance to kasugamycin (Yoshii et al., 2012). The Aac(2′)-IIa acyltransferase inactivates kasugamycin via acetylation of the 2′-amino residue of the antibiotic. In the chromosome of B. glumae, this acetyltransferase is encoded on the IncP genomic island, suggesting that this kasugamycin resistance gene might have been acquired by HGT and that the gene could hypothetically spread among PPB (Yoshii et al., 2012). The aac(2′)-IIa gene was later found on a conjugative IncP-1β plasmid, pAAA83, in an A. avenae ssp. avenae strain, raising even more concern about the potential transmission of this resistance gene (Yoshii et al., 2015). In E. amylovora, resistance to kasugamycin and blasticidin S is induced by the in vitro deletion of both permease genes opp and dpp, suggesting that Opp and Dpp act synergistically to transport these antibiotics. This deletion has not been observed in field isolates, so the relevance of such mechanism in the environment remains to be assessed (Ge et al., 2018). Similarly, in vitro mutation of the ksgA gene in E. amylovora confers kasugamycin resistance but it requires a two-step mutational process and these mutants displayed a considerably reduced fitness (McGhee and Sundin, 2011; Ge et al., 2018).
In PPB, the exact mechanisms involved in OA resistance have not been described in great details (Table 2). In vitro mutants of B. glumae resistant to the OA quinolone were obtained from strains isolated in rice fields in Japan, where the antibiotic is used as PPP. GyrA83 mutation (substitution of serine by arginine or isoleucine at position 83) seemed to be involved in the resistance mechanism (Maeda et al., 2007a,b). Others have also demonstrated that OA resistant B. glumae could be obtained in vitro, however such mutants were unable to survive in paddy fields (Hikichi et al., 1998, 2001).
In Israel, OA was introduced in 1997 when the use of streptomycin was abandoned. About 2 years later, OA resistant E. amylovora and B. glumae were reported. Yet, the exact mechanisms leading to OA resistance in these strains remained unknown, although it was suggested that it might consist into chromosomal mutations rather than gene acquisition through HGT (Manulis et al., 2003; Kleitman et al., 2005).
Resistance to the gentamicin aminoglycoside was only reported in X. oryzae pv. oryzae, where the acquisition of the integron-borne aacA3 gene was incriminated. As indicated in Table 2, aacA3 encodes an aminoglycoside-3’-N-acetyltransferase enzyme that deactivates gentamicin (Xu et al., 2013).
The aadA1 gene was found in X. oryzae pv. oryzae on three integrons and associated with other antibiotic resistance gene cassettes such as aacA3 (conferring resistance to tobramycin, kanamycin, gentamicin and netilmicin) or arr3 (conferring resistance to rifampicin; Xu et al., 2013). Both aadA1 and aadA2 also confer spectinomycin resistance. The co-occurrence of streptomycin and gentamicin resistance is therefore possible and it is then reasonable to speculate that these gene cassettes could be co-transferred under the selective pressure of streptomycin or gentamicin.
In a recent study conducted in South Carolina (United States), strains of X. arboricola pv. pruni were isolated and characterized for their resistance to both streptomycin and oxytetracycline. They carried a plasmid encoding tetC, tetR and strA-strB, with a region similar to Tn5393 (Herbert et al., 2022). It was previously described that, when isolating epiphytic bacteria (not PPB) from Michigan apple orchards, almost all tetracycline resistant strains were also streptomycin resistant and that tetracycline resistance genes were found on plasmids that also carried Tn5393 (Schnabel and Jones, 1999). Similarly, several plasmid-borne tetracycline resistance elements have been found in various epiphytic bacteria and usually the tetracycline resistance was associated with transposons, mainly Tn5393 (carried by an uncharacterized plasmid; Schnabel and Jones, 1999). These data indicate that PPB could become resistant to different antibiotics through the acquisition of resistance genes from other plant-associated bacteria if the genes are present on MGEs.
In 2005, several P. syringae were isolated and assessed for their antibiotic resistance to six antibiotics. One strain of P. syringae pv. syringae was resistant to kanamycin and tetracycline. Eight isolates were resistant to streptomycin, 16 isolates were resistant to rifampicin and 36 to chloramphenicol. Fifty-five strains in total were resistant to ampicillin. Some of these strains were resistant to several antibiotics, and although the resistance mechanisms were not characterized, this study further highlights the possibility of ARGs co-occurrence (Hwang et al., 2005).
Along the same lines, an OA resistant B. glumae was reported to display cross-resistance with other quinolones such as ciprofloxacin (Hikichi et al., 2001), while another isolated B. glumae was resistant to polymyxin B (Paz-Carrasco et al., 2018).
Finally, it is worth mentioning three studies that have addressed resistance to antibiotics mainly used in China. First, it was possible to obtain X. oryzae pv. oryzae strains resistant to phenazine-1-carboxylic acid (PCA), also called shenqinmycin in China, after in vitro exposition to increasing concentrations of the antibiotic (Pan et al., 2018). Second, zhongshengmycin resistant strains of X. oryzae were obtained in vitro and the resistance was related to increasing fatty acid biosynthesis. The exact underlying mechanisms are not known yet (Wang Q. et al., 2021). Third, bismerthiazol resistant mutants of X. oryzae pv. oryzae could also be induced, both in vivo and in vitro (Zhu et al., 2013). However, these mutants were all obtained in vitro or induced, so the natural occurrence of such resistance remains to be demonstrated. Nevertheless, this indicates that X. oryzae pv. oryzae is highly adaptable and may easily develop resistances.
This section mostly focuses on streptomycin, which has been used the longest in plant agriculture, compared to other antibiotics, and remains the most used antibiotic in this context today. Scientific evidence that can directly link the use of streptomycin and the abundance of streptomycin resistance genes or streptomycin resistant strains are relatively scarce. In fact, although several studies suggest that the application of streptomycin in fields does not influence the abundance of resistance genes in PPB and other surrounding bacteria, opposite conclusions have been reached by other studies.
Several studies reported that the application of streptomycin on orchards did not, or not adversely, affect the bacterial population of the soil (Walsh et al., 2013; Shade et al., 2013a) or described minimal alterations with a slight decrease in phylogenetic diversity of bacterial communities of apple tree flowers (Shade et al., 2013b), on the short-term. Another study indicated a decrease of diversity in streptomycin-treated soil (Tolba et al., 2002). When looking at the bacterial communities of apple leaves, a higher frequency of streptomycin resistant bacteria was observed in samples that were not treated with streptomycin (Yashiro and McManus, 2012). However, they only focused on apple leaves and not on the other bacterial communities (soil or roots) that could be affected differently. This work also dealt with the overall bacterial community, and not with specific species that could become resistant without significantly increasing the overall abundance of resistance. Moreover, as indicated before, streptomycin application could select for other ARGs.
The effects of streptomycin and kasugamycin application on bacteria in the apple phyllosphere have been investigated. While the use of streptomycin did not result in any increase of streptomycin resistant E. amylovora, it was correlated with an increase in resistance in other epiphytic bacteria, such as Pantoea agglomerans or Pseudomonas spp. No kasugamycin resistant bacteria could be isolated, however the antibiotic application changed the microbial spectrum in the orchard (Tancos and Cox, 2017). Along the same lines, there was no influence on the abundance of culturable oxytetracycline and gentamicin resistant bacteria when these antibiotics where applied, as compared to untreated soils (Rodríguez-Sánchez et al., 2008). On the opposite, kasugamycin resistant Gram-negative bacteria (mainly residents of the plant phyllosphere or colonists of apple, such as P. agglomerans, Pseudomonas graminis, P. syringae and Stenotrophomonas spp.) could be isolated in another study from orchard soil, apple flowers and leaves treated with kasugamycin (McGhee and Sundin, 2011).
Sundin et al. (1995) observed the distribution of streptomycin resistance transposon Tn5393 in the apple phylloplane and soil of ornamental pear (that had received previous applications of streptomycin) and tomato (no prior exposure to streptomycin; Sundin et al., 1995). The recovery of streptomycin resistant bacteria was generally higher in soil samples, but the highest occurrence was from phylloplane samples from an orchard where streptomycin had been applied. They showed that the use of streptomycin for plant protection can also select for streptomycin resistance in non-target commensal bacteria that inhabit either plant surfaces or the surrounding soil (Chiou and Jones, 1993; Sundin et al., 1995). They investigated the risk of co-selection of other antibiotic resistance genes (tetracycline), but found no evidence that the use of streptomycin increased tetracycline resistance (Sundin et al., 1995). One should however note that streptomycin and tetracycline resistance genes were recently found on the same plasmid (Herbert et al., 2022).
In a bioinformatic analysis, where a total of 127 genomes of E. amylovora from different geographic regions were examined, the greatest number of streptomycin resistant isolates was observed in Western North America (in particular in British Columbia, Canada), where streptomycin was used, at least during the survey (1993–1998; Sholberg et al., 2001; Parcey et al., 2020). Several additional studies showed that the orchards where streptomycin use is the greatest were usually those where the highest numbers of streptomycin resistant bacteria were detected (Chiou, 1991; Norelli et al., 1991; Burr et al., 1993).
Regarding oxytetracycline resistance, the finding of a tetracycline resistant isolate of A. tumefaciens could not be linked to oxytetracycline application (Luo and Farrand, 1999). On the opposite, antibiotic sprays of streptomycin and oxytetracycline were positively correlated with resistance in P. syringae pv. syringae (Spotts and Cervantes, 1995). In epiphytic bacteria, the use of oxytetracycline generally resulted in less abundant bacterial populations than when streptomycin was used, but streptomycin resistant isolates were more common than the tetracycline resistant ones. Moreover, tetracycline resistant strains could be observed in orchards where tetracycline had not been applied, although higher numbers of tetracycline resistant bacteria could be detected in another orchard where oxytetracycline was applied (Schnabel and Jones, 1999).
In Israel, the introduction of OA to replace streptomycin led to the isolation of OA resistant E. amylovora and B. glumae. However, the incidence of OA resistance in E. amylovora was sporadic and irrespective of the number of sprays applied and the severity of the disease (Kleitman et al., 2005). In another study also conducted in Israel after introduction of OA, several OA resistant strains were isolated from different orchards but none was resistant to both streptomycin and OA (Manulis et al., 2003). Concerning gentamicin resistance, the only report available in X. oryzae pv. oryzae in China has not been linked to the use of gentamicin (Xu et al., 2013).
The abundance of streptomycin and tetracycline resistance genes in flowers, leaves and soil samples from orchards treated with streptomycin in Switzerland was analyzed using multiplex qPCR (Duffy et al., 2014). Three orchards that had not been treated with streptomycin prior to the assays were used, and samples were collected over a three-year period. By using qPCR to assess the abundance of ARGs, this study overcame the intrinsic problem linked to culture-dependent assays where unculturable bacteria are overlooked. This method allows for a qualitative assessment of ARGs in the complete apple tree ecosystem but does not identify the species that contain the resistance genes. The results indicated no consistent increase in streptomycin resistance genes in streptomycin-treated samples. Tetracycline resistance genes were also quantified in this study, and no increase was reported with streptomycin use. It is however important to note that due to the nature of the technique used, streptomycin resistance genes, notably strA-strB and aadA, had to be selected for the screening. Other potential resistance mechanisms might have been ignored.
Similarly, the abundance of various tetracycline resistance genetic determinants and several gentamicin resistance genes was investigated under field conditions with five applications of oxytetracycline and gentamicin during 16 months. This study indicated that the occurrence of these genes was not related to the application of antibiotics (Rodríguez-Sánchez et al., 2008).
Little is known about the resilience of an ecosystem to antibiotic resistances, years after the discontinuation of the antibiotic use.
In Israel, streptomycin was removed from the list of approved antibiotics for plant agriculture in 1997 and has not been used since. A decline in the incidence of streptomycin resistance in E. amylovora was observed in the subsequent years (from 57% in 1998 to 15% in 2001). Surprisingly, within 4 years, the resistance had almost disappeared (Manulis et al., 2003).
Other studies have been conducted in a few American states. In California, streptomycin resistance among bacteria from orchards declined from 1973 to 1977. However, streptomycin resistant E. amylovora strains could still be isolated, even though streptomycin had not been applied in these orchards since 1971 (Schroth et al., 1979). In Washington state, streptomycin resistant E. amylovora strains could still be isolated 5 years after termination of streptomycin use (Loper et al., 1991). Schnabel and Jones (1999) showed that streptomycin resistance did not diminish at their study site over the course of 2 years in the absence of the selection pressure (Schnabel and Jones, 1999). In Michigan, a decline in the incidence of streptomycin resistance was observed from 1991 to 1992 when oxytetracycline was used rather than streptomycin. However, streptomycin resistance in E. amylovora quickly reoccurred in 1993 when the use of streptomycin was resumed (McManus and Jones, 1994). This could suggest that streptomycin resistant strains have lower fitness than streptomycin sensitive strains, although Loper et al. (1991) found streptomycin resistant strains where this antibiotic had never been applied, suggesting similar fitness capabilities under no selective pressure (Loper et al., 1991).
It was shown that there is a high presence of streptomycin resistance in agricultural sites without streptomycin treatment history, and that HGT of strA and strB occurred regardless of soil treatment with antibiotics (Tolba et al., 2002). It has also been shown that strA gene is more prevalent in soils from compost, forest or agriculture samples than from vegetable gardens, apple orchards or mixed fruit orchards (Popowska et al., 2012). However, in the same study, strB was less prevalent in compost, forest or agricultural soils than in apple orchards or mixed fruit orchards. The aadA gene followed the same pattern as strB but was only detected in agricultural soil at very low rates, while absent in compost or forest soils. It was rather prevalent in vegetable gardens, apple orchards or mixed fruit orchards.
It is also worth mentioning that the streptomycin resistant transposon Tn5393 was detected in bacteria with no prior exposure to streptomycin. It seems that this transposable element is indigenous to both phylloplane and soil microbial communities (Sundin et al., 1995). In fact, strA-strB and Tn5393 are both present in non-target bacteria (Chiou and Jones, 1993; Sundin et al., 1995). Gentamicin resistance genes are also commonly found in environments associated with sewage effluent or farm animals (Heuer et al., 2002).
There are many studies reporting the presence of antibiotic resistant bacteria and ARGs in orchards. This does not establish, per se, a link with the use of antibiotics. Research on the relationship between antibiotic use and antibiotic resistance in PPB is largely incomplete. Many other factors could drive the emergence of resistant strains, such as cross-resistance to other PPPs, e.g., fungicides or copper, the use of manure, irrigation water, sewage sludge or antibiotic resistance from animal or human uses. As of now, there are not enough data to support either view with confidence. More large-scale and long-term research in fields with non-treated controls are needed to be able to have statistically relevant data on the issue.
What is known for sure is that streptomycin resistance is mainly mediated by strA-strB found on the Tn5393 transposon, which seems to be rather widespread. The strA-strB genes found in PPB are broadly disseminated in human pathogens, although the alleles of the genes are different, suggesting diverse routes of acquisition (Sundin and Bender, 1996; Sundin, 2000, 2002). Detection of similar strA-strB genes in PPB and in human pathogens does not automatically mean that transfer occurred directly between these organisms. In the clinical context, the main problem with Tn5393 is its ability to translocate and mobilize other ARGs rather than streptomycin resistance per se. Strains of two major human pathogens (S. enterica and K. pneumoniae) have been found to carry Tn5393 variants, as well as several species of Aeromonas, which could indicate that the transposon is also circulating in this bacterial genus. The selection of Tn5393 in plant-associated bacteria driven by extensive streptomycin use in agriculture could create a reservoir that may negatively influence the antibiotic resistance crisis. The fact that the same Tn5393 can be found in PPB and in S. enterica is a worrying fact.
By using antibiotics on plants, not only are the associated resistance genes selected, but also the MGEs that carry these ARGs, which is often underestimated. Tn5393 is the only well-documented example, so far, of MGE associated with antibiotic resistance in PPB, but it cannot be excluded that other “mobile” resistance genes are yet to be discovered or to emerge. Besides, it is known that the use of antibiotics selects not only for the resistance genes, but also contributes to the evolution of complex vectors (MGEs) encoding several ARGs (O’Brien, 2002), which is a plausible phenomenon also in PPB.
The translocation of ARGs among plasmids thanks to transposons also potentially accelerates the spread of ARGs (Yao et al., 2022). The fact that Tn5393 can translocate to other plasmids also suggests the risk of insertion into other conjugative plasmids, which could lead to further spread of the resistance genes (Falkenstein et al., 1989; McManus et al., 2002; Llop et al., 2006). This suggests Tn5393 is evolving and that IS or other transposons have inserted into its structure resulting into more complex, better-fit elements, potentially carrying other ARGs. Tn5393 structures are still relatively simple in PPB (Figure 2; Table 3), but in human or animal pathogens, variants of the transposon show the potential complexity that could be attained in PPB within a few years, with the formation of complex transposons simultaneously carrying several ARGs.
Regarding the other antibiotics used in plant protection, less information is yet available regarding the resistance risk that they entail. Since the beginning of its use in plant protection, there have been very few reports of tetracycline resistance in PPB. It could therefore be assumed that oxytetracycline resistance is not an issue on the short-term. However, risks related to the transfer of tetracycline resistance genes from epiphytic bacteria to human pathogens or PPB cannot be excluded. Besides, the uncertainty linked to the lack of studies cannot be underestimated. When tetracycline resistance is present, it is typically encoded on plasmids associated with Tn5393, which indicates a potential link between streptomycin and tetracycline resistance and the potential dissemination of both resistances at the same time (Schnabel and Jones, 1999; Herbert et al., 2022).
A better understanding of the extent to which epiphytic bacteria can serve as reservoirs of ARGs for PPB or human pathogens is crucial. In fact, it is very likely that strA-strB and Tn5393 were first transferred from non-target bacteria to plant pathogens, resulting in streptomycin resistant PPB (Chiou and Jones, 1993; Sundin et al., 1995). Regarding the case of oxytetracycline resistance potential acquisition, the availability of tetracycline resistance genes in the bacterial populations exposed to tetracycline and their ability to transfer to E. amylovora are clear factors contributing to the risk of tetracycline resistance selection in this pathogen. Even though tetracycline resistance in E. amylovora by chromosomal mutation does not easily occur (Lacy et al., 1984), the use of tetracycline could potentially lead to the selection of resistant strains of E. amylovora or other PPB through the acquisition of tetracycline resistance genes from other plant (or soil) bacteria if the genes are present on MGEs (Chiou and Jones, 1993; Sundin et al., 1995).
Although presently kasugamycin resistance does not appear as a major problem in plant agriculture, the recent emergence of a transmissible kasugamycin resistance gene constitutes a considerable threat for the effective control of the diseases involved, especially because the resistance gene might spread to other bacteria through HGT via MGEs. Dissemination of kasugamycin resistance among PPB is therefore a possibility. Kasugamycin is often considered as an interesting alternative to the other antibiotics used in plant protection because of its non-use in human or veterinary medicine. In fact, it was shown to be efficient for the management of fire blight when streptomycin resistant strains were present (McGhee and Sundin, 2011). Conversely, in this same study, Enterobacteriaceae and Pseudomonas spp. resistant to both kasugamycin and streptomycin were isolated, which is concerning because of the potential resistance transfer to E. amylovora and the potential link between these two ARGs. The authors dismissed the potential cross-resistance among singly resistant spontaneous mutants for either kasugamycin or streptomycin, because kasugamycin resistant E. amylovora strains were sensitive to streptomycin and vice versa. However, this does not mean that the double resistance could not arise in the future. An increasing number of antibiotics restricted to plant protection are proposed to control bacterial diseases, but cross-resistances and selection of antibiotic resistance vectors should be investigated.
Nowadays, very little information is known about gentamicin resistance in PPB. However, the only gentamicin resistance report was due to a gene found on an integron, which may then be able to spread through HGT. Rare cases of resistance to OA have been observed in Israel in isolates of E. amylovora and B. glumae, where the antibiotic is used as PPP, but the exact resistance mechanisms were not characterized, though it seems that they only involved chromosomal mutations so far (Manulis et al., 2003; Kleitman et al., 2005), such as the GyrA83 mutation observed in vitro (Maeda et al., 2007b).
Because of the main way of application of antibiotics on plants (spraying), it was suggested that it might result in a limited selection for resistance, due to photodegradation, soil adsorption or deactivation and the substantial dilution (McManus, 2014). However, contrarily to animal and human medicine, where the use of antibiotics is rather controlled, when used in plant health, antibiotics are sprayed on a large scale and with relatively high doses, probably notably because of the factors cited above, which is also a source of great concerns due to the large environmental exposure. Besides, it is well-known that antibiotic concentrations already well below the minimal inhibitory concentrations (MICs) select for antibiotic resistant bacteria (Bengtsson-Palme and Larsson, 2016), both pathogenic and commensal, which can then become vectors of ARGs. It is also worth noting that, in this review, the purity of the antibiotic, the potential role of the excipients (Killiny et al., 2020), the way of use (spraying or injection) as well as the type of crops were not taken into consideration. Yet, they all play a role in the amount of antibiotic that is effectively applied and the potential subsequent selection pressure on bacteria.
Efficient and safe alternative control measures are urgently needed to manage bacterial diseases in plant health, to avoid resorting to antibiotics in the first place. Reducing the need to use antibiotics would be the safest way to avoid the selection and emergence of antibiotic resistance. Many innovative control measures are being explored as new potential alternatives to antibiotics, notably antagonistic bacteria or competitive fungi (Poveda et al., 2021; Poveda and Baptista, 2021), bacteriophages (Buttimer et al., 2017; Grace et al., 2021) or for the control of animal vectors of PPB (Di Serio et al., 2019; Vicente-Díez et al., 2021). In the European Union, despite the absence of authorization of antibiotic use, fire blight is not so much of a problem because other strategies are in place to control this bacterial disease. In other locations of the world, using antibiotics is often the choice of convenience, but this strategy also hampers the development and application of alternative methods.
Even though the use of antibiotics in plant protection is considered relatively low in comparison to the use for human and veterinary medicine, the impact it could potentially have on the phytobiome cannot be overlooked, with potential unintended side effects such as the development of antibiotic resistance. It must also be stressed that PPB are not the only bacteria and microorganisms associated with plants (e.g., fungi, that can also be controlled through antibiotics with antifungal properties, as well as soil bacteria, might also develop resistance). Another aspect not developed in this paper is the use of biocontrol agents to control PPB or fungi, that might themselves carry ARGs. They are also sometimes spread heavily on plants, and are currently approved in Europe and other countries, such as Streptomyces lydicus WYEC 108 [US Environmental Protection Agency Office of Pesticide Programs, 2005; European Food Safety Authority (EFSA), 2013; European Food Safety Authority (EFSA) et al., 2020]. They could also represent a source of ARGs and MGEs, potentially interacting with the microorganisms associated with plants.
The contribution of MGEs to the spread of antibiotic resistance in PPB, as well as their ability to transfer to other bacteria, need to be further investigated carefully. In fact, the only well-studied example of ARGs vector in PPB, Tn5393, is concerning because of its occurrence outside of PPB and its structure evolving into complex associations of MGEs and ARGs. The effect of antibiotic use on non-pathogenic plant-associated bacteria is not well studied and largely unknown, still they represent another piece of the puzzle that could allow MGEs to travel from crops to the environment and to the human consumers. Whether the use of antibiotics in plant health has an impact on the global problematic of antibiotic resistance remains unanswered.
The extent of the risk of resistance that goes along with antibiotic use in plant protection cannot be excluded given the current lack of data. To reduce the resistance risk, the adoption of strong antimicrobial stewardship practices (Miller et al., 2022) is essential. The development of surveillance programs practical and achievable also by low- and middle-income countries that are harmonized and collect quantitative data on the use and sales of antibiotics, as well as the crops and area of their application is necessary to better understand the situation and assess the risks of antibiotic resistance selection and spread. Accurate data on the amounts of antibiotic used in different crops are crucial to better identify and quantify the related risk of development of antibiotic resistance.
Unravelling the risks potentially associated with the use of antibiotics and potential gene transfers among PPB species via MGEs is urgent, which is why surveillance in plants and soils needs to be improved. In order to assess the potential spread of these ARGs, it is also of uttermost importance to develop and use new detection methods (modern tools to carry out genetic analyses). Whole genome sequencing (WGS) of resistant field strains and metagenomics of field samples could clarify where the genes are located and how they can be transferred (presence on MGEs), as well as contribute greatly to resistance prediction (Arango-Argoty et al., 2018). In addition, rapid and inexpensive tests and/or tools are needed to facilitate the identification of PPB and to characterize their resistomes.
MV, TB, and CB carried out the original literature screening under the supervision of CB, JM, and M-PM-L. MV drafted the manuscript with inputs from JM and CB. MV, EL, GS, FS, M-PM-L, JM, and CB edited, revised and approved the final version of the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the European Food Safety Authority (EFSA, grant to MV and TB) under Agreement Number GP/EFSA/ALPHA/2020/02.
The authors are grateful to the team of the Laboratory of Food and Environmental Microbiology for their critical inputs throughout the writing of this article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors EL, GS, and FS are employed by the European Food Safety Authority (EFSA). However, the present article is published under the sole responsibility of the authors and may not be considered as an EFSA scientific output. The positions and opinions presented in this article are those of the authors alone and do not necessarily represent the views/any official position or scientific works of EFSA. To know about the views or scientific outputs of EFSA, please consult its website “www.efsa.europa.eu”.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1221478/full#supplementary-material
1. ^https://tncentral.ncc.unesp.br/TnPedia/index.php/Transposons_families/Tn3_family#The_Tn163_Clade
Aarestrup, F. M., Wegener, H. C., and Collignon, P. (2008). Resistance in bacteria of the food chain: epidemiology and control strategies. Expert Rev. Anti-Infect. Ther. 6, 733–750. doi: 10.1586/14737140.6.5.733
Adamczuk, M., and Dziewit, L. (2017). Genome-based insights into the resistome and mobilome of multidrug-resistant Aeromonas sp. ARM81 isolated from wastewater. Arch. Microbiol. 199, 177–183. doi: 10.1007/s00203-016-1285-6
Alekshun, M. N., and Levy, S. B. (2007). Molecular mechanisms of antibacterial multidrug resistance. Cells 128, 1037–1050. doi: 10.1016/j.cell.2007.03.004
Arango-Argoty, G., Garner, E., Pruden, A., Heath, L. S., Vikesland, P., and Zhang, L. (2018). DeepARG: a deep learning approach for predicting antibiotic resistance genes from metagenomic data. Microbiome 6, 1–15. doi: 10.1186/s40168-018-0401-z
Barbosa, R. A., Santini, P. T., and Guilherme, L. R. G. (2018). Kasugamycin influence on bacterial blight of coffee and on green coffee beans physicochemical quality. Coffee Sci. 13, 98–103. doi: 10.25186/cs.v13i1.1384
Barnard, A. M. L., Simpson, N. J. L., Lilley, K. S., and Salmond, G. P. C. (2010). Mutations in rpsL that confer streptomycin resistance show pleiotropic effects on virulence and the production of a carbapenem antibiotic in Erwinia carotovora. Microbiology 156, 1030–1039. doi: 10.1099/mic.0.034595-0
Bengtsson-Palme, J., and Larsson, D. G. J. (2016). Concentrations of antibiotics predicted to select for resistant bacteria: proposed limits for environmental regulation. Environ. Int. 86, 140–149. doi: 10.1016/j.envint.2015.10.015
Bian, C., Duan, Y., Wang, J., Xiu, Q., Wang, J., Hou, Y., et al. (2020). Validamycin A induces broad-spectrum resistance involving salicylic acid and jasmonic acid/ethylene signaling pathways. Mol. Plant-Microbe Interact. 33, 1424–1437. doi: 10.1094/MPMI-08-20-0211-R
Blair, J. M. A., Webber, M. A., Baylay, A. J., Ogbolu, D. O., and Piddock, L. J. V. (2015). Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 13, 42–51. doi: 10.1038/nrmicro3380
Burr, T. J., Norelli, J. L., Reid, C. L., Capron, L. K., Nelson, L. S., Aldwinckle, H. S., et al. (1993). Streptomycin-resistant bacteria associated with fire blight infections. Plant Dis. 77, 63–66. doi: 10.1094/PD-77-0063
Buttimer, C., McAuliffe, O., Ross, R. P., Hill, C., O’Mahony, J., and Coffey, A. (2017). Bacteriophages and bacterial plant diseases. Front. Microbiol. 8, 1–15. doi: 10.3389/fmicb.2017.00034
Cain, A. K., and Hall, R. M. (2011). Transposon Tn5393e carrying the aphA1-containing transposon Tn6023 upstream of strAB does not confer resistance to streptomycin. Microb. Drug Resist. 17, 389–394. doi: 10.1089/mdr.2011.0037
Cameron, A., and Sarojini, V. (2014). Pseudomonas syringae pv. actinidiae: chemical control, resistance mechanisms and possible alternatives. Pseudomonas Syringae pv. actinidiae, 63, 1–11. doi: 10.1111/ppa.12066
Chanvatik, S., Donnua, S., Lekagul, A., Kaewkhankhaeng, W., Vongmongkol, V., Athipunyakom, P., et al. (2019). Antibiotic use in mandarin production (Citrus reticulata Blanco) in major mandarin-producing areas in Thailand: a survey assessment. PLoS One 14, 1–14. doi: 10.1371/journal.pone.0225172
Chiou, C.-S. (1991). The analysis of plasmid-mediated streptomycin resistance in Erwinia amylovora. Phytopathology 81:710. doi: 10.1094/phyto-81-710
Chiou, C.-S., and Jones, A. L. (1993). Nucleotide sequence analysis of a transposon (Tn5393) carrying streptomycin resistance genes in Erwinia amylovora and other Gram-negative bacteria. J. Bacteriol. 175, 732–740. doi: 10.1128/jb.175.3.732-740.1993
Chiou, C.-S., and Jones, A. L. (1995a). Expression and identification of the strA-strB gene pair from streptomycin-resistant Erwinia amylovora. Gene 152, 47–51. doi: 10.1016/0378-1119(94)00721-4
Chiou, C.-S., and Jones, A. L. (1995b). Molecular analysis of high-level streptomycin resistance in Erwinia amylovora. Phytopathology 85, 324–328. doi: 10.1094/Phyto-85-324
Chopra, I., and Roberts, M. (2001). Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65, 232–260. doi: 10.1128/MMBR.65.2.232
da Silva, V. L., and Lopes, C. A. (1995). Pseudomonas Syringae pv. tomato resistant to streptomycin and oxytetracycline in tomato plants treated or not with agricultural antibiotics. Fitopatol. Bras. 20, 80–84.
Dafny-Yelin, M., Moy, J. C., Mairesse, O., Silberstein, M., Sapir, G., and Michaeli, D. (2021). Efficacy of fire blight management in pome fruit in northern Israel: copper agents and their effect on yield parameters. J. Plant Pathol. 103, 151–161. doi: 10.1007/s42161-020-00665-5
Dagan, T. (2011). Phylogenomic networks. Trends Microbiol. 19, 483–491. doi: 10.1016/j.tim.2011.07.001
de León Door, A. P., Romo Chacón, A., and Acosta Muñiz, C. (2013). Detection of streptomycin resistance in Erwinia amylovora strains isolated from apple orchards in Chihuahua, Mexico. Eur. J. Plant Pathol. 137, 223–229. doi: 10.1007/s10658-013-0241-4
Di Serio, F., Bodino, N., Cavalieri, V., Demichelis, S., Di Carolo, M., Dongiovanni, C., et al. (2019). Collection of data and information on biology and control of vectors of Xylella fastidiosa. EFSA supporting publication, 1628E, 102. doi: 10.2903/sp.efsa.2019.EN-1628
Duffy, B., Holliger, E., and Walsh, F. (2014). Streptomycin use in apple orchards did not increase abundance of mobile resistance genes. FEMS Microbiol. Lett. 350, 180–189. doi: 10.1111/1574-6968.12313
Escursell, M. M., Roschi, A., Smits, T. H. M., and Rezzonico, F. (2021). Characterization and direct molecular discrimination of rpsL mutations leading to high streptomycin resistance in Erwinia amylovora. J. Plant Pathol. 103, 99–108. doi: 10.1007/s42161-020-00600-8
Espedido, B. A., Steen, J. A., Ziochos, H., Grimmond, S. M., Cooper, M. A., Gosbell, I. B., et al. (2013). Whole genome sequence analysis of the first Australian OXA-48-producing outbreak-associated Klebsiella pneumoniae isolates: the resistome and in vivo evolution. PLoS One 8, 1–6. doi: 10.1371/journal.pone.0059920
European Food Safety Authority (EFSA) (2013). Conclusion on the peer review of the pesticide risk assessment of the active substance Streptomyces lydicus WYEC 108. EFSA J. 11, 1–30. doi: 10.2903/j.efsa.2013.3425
European Food Safety Authority (EFSA)Anastassiadou, M., Bernasconi, G., Brancato, A., Carrasco Cabrera, L., Ferreira, L., et al. (2020). Review of the existing maximum residue levels for Streptomyces lydicus strain WYEC 108 according to article 12 of regulation (EC) no 396/2005. EFSA J. 18, 1–11. doi: 10.2903/j.efsa.2020.6241
Falkenstein, H., Zeller, W., and Geider, K. (1989). The 29 kb plasmid, common in strains of Erwinia amylovora, modulates development of fireblight symptoms. Microbiology 135, 2643–2650. doi: 10.1099/00221287-135-10-2643
FAO, OIE, and WHO (2018). Monitoring Global Progress on Addressing Antimicrobial Resistance. Available at: https://www.who.int/publications/i/item/monitoring-global-progress-on-addressing-antimicrobial-resistance. (Accessed 28 October 2022).
Förster, H., McGhee, G. C., Sundin, G. W., and Adaskaveg, J. E. (2015). Characterization of streptomycin resistance in isolates of Erwinia amylovora in California. Phytopathology 105, 1302–1310. doi: 10.1094/PHYTO-03-15-0078-R
Foster, G. C., McGhee, G. C., Jones, A. L., and Sundin, G. W. (2004). Nucleotide sequences, genetic organization, and distribution of pEU30 and pEL60 from Erwinia amylovora. Appl. Environ. Microbiol. 70, 7539–7544. doi: 10.1128/AEM.70.12.7539-7544.2004
Fouts, K. E., and Barbour, S. D. (1981). Transductional mapping of ksgB and a new Tn5-induced kasugamycin resistance gene, ksgD, in Escherichia coli K-12. J. Bacteriol. 145, 914–919. doi: 10.1128/jb.145.2.914-919.1981
Fricke, W. F., McDermott, P. F., Mammel, M. K., Zhao, S., Johnson, T. J., Rasko, D. A., et al. (2009). Antimicrobial resistance-conferring plasmids with similarity to virulence plasmids from avian pathogenic Escherichia coli strains in Salmonella enterica serovar Kentucky isolates from poultry. Appl. Environ. Microbiol. 75, 5963–5971. doi: 10.1128/AEM.00786-09
Ge, Y., Lee, J. H., Hu, B., and Zhao, Y. (2018). Loss-of-function mutations in the Dpp and Opp permeases render Erwinia amylovora resistant to kasugamycin and blasticidin S. Mol. Plant-Microbe Interact. 31, 823–832. doi: 10.1094/MPMI-01-18-0007-R
Grace, E. R., Rabiey, M., Friman, V. P., and Jackson, R. W. (2021). Seeing the forest for the trees: use of phages to treat bacterial tree diseases. Plant Pathol. 70, 1987–2004. doi: 10.1111/ppa.13465
Gyles, C., and Boerlin, P. (2014). Horizontally transferred genetic elements and their role in pathogenesis of bacterial disease. Vet. Pathol. 51, 328–340. doi: 10.1177/0300985813511131
Han, H. S., Koh, Y. J., Hur, J.-S., and Jung, J. S. (2004). Occurrence of the strA-strB streptomycin resistance genes in Pseudomonas species isolated from kiwifruit plants. J. Microbiol. 42, 365–368.
Harmer, C. J. (2021). HI1 and I1 resistance plasmids from Salmonella enterica serovar Typhimurium strain SRC27 are epidemic. Microb. Drug Resist. 27, 1495–1504. doi: 10.1089/mdr.2020.0579
Herbert, A., Hancock, C. N., Cox, B., Schnabel, G., Moreno, D., Carvalho, R., et al. (2022). Oxytetracycline and streptomycin resistance genes in Xanthomonas arboricola pv. pruni, the causal agent of bacterial spot in peach. Front. Microbiol. 13, 1–13. doi: 10.3389/fmicb.2022.821808
Heuer, H., Kopmann, C., Binh, C. T. T., Top, E. M., and Smalla, K. (2009). Spreading antibiotic resistance through spread manure: characteristics of a novel plasmid type with low %G+C content. Environ. Microbiol. 11, 937–949. doi: 10.1111/j.1462-2920.2008.01819.x
Heuer, H., Krögerrecklenfort, E., Wellington, E. M. H., Egan, S., Van Elsas, J. D., Van Overbeek, L., et al. (2002). Gentamicin resistance genes in environmental bacteria: prevalence and transfer. FEMS Microbiol. Ecol. 42, 289–302. doi: 10.1016/S0168-6496(02)00342-2
Hijaz, F., Nehela, Y., Al-Rimawi, F., Vincent, C. I., and Killiny, N. (2020). The role of the xylem in oxytetracycline translocation within citrus trees. Antibiotics 9, 1–10. doi: 10.3390/antibiotics9100691
Hijaz, F., Nehela, Y., Batuman, O., and Killiny, N. (2021). Detection of oxytetracycline in citrus phloem and xylem saps using europium-based method. Antibiotics 10:1036. doi: 10.3390/antibiotics10091036
Hikichi, Y., Egami, H., Oguri, Y., and Okuno, T. (1998). Fitness for survival of Burkholderia glumae resistant to oxolinic acid in rice plants. Japanese J. Phytopathol. 64, 147–152. doi: 10.3186/jjphytopath.64.147
Hikichi, Y., Tsujiguchi, K., Maeda, Y., and Okuno, T. (2001). Development of increased oxolinic acid-resistance in Burkholderia glumae. J. Gen. Plant Pathol. 67, 58–62. doi: 10.1007/pl00012988
Hwang, M. S. H., Morgan, R. L., Sarkar, S. F., Wang, P. W., and Guttman, D. S. (2005). Phylogenetic characterization of virulence and resistance phenotypes of Pseudomonas syringae. Appl. Environ. Microbiol. 71, 5182–5191. doi: 10.1128/AEM.71.9.5182-5191.2005
Kannan, V. R., Bastas, K. K., and Sangeethadevi, R. (2015). “Scientific and economic impact of plant pathogenic bacteria” in Sustainable approaches to controlling plant pathogenic bacteria, eds. V. R. Kannan and K. K. Bastas (CRC Press (Taylor & Francis Group, USA)), 369–392. doi: 10.1201/b18892-16
Killiny, N., Hijaz, F., Gonzalez-blanco, P., Jones, S. E., Pierre, M. O., and Vincent, C. I. (2020). Effect of adjuvants on oxytetracycline uptake upon foliar application in citrus. Antibiotics 9, 1–16. doi: 10.3390/antibiotics9100677
Kleitman, F., Shtienberg, D., Blachinsky, D., Oppenheim, D., Zilberstaine, M., Dror, O., et al. (2005). Erwinia amylovora populations resistant to oxolinic acid in Israel: prevalence, persistence and fitness. Plant Pathol. 54, 108–115. doi: 10.1111/j.1365-3059.2005.01146.x
Kumar, K., Gupta, S. C., Chander, Y., and Singh, A. K. (2005). Antibiotic use in agriculture and its impact on the terrestrial environment. Adv. Agron. 87, 1–54. doi: 10.1016/S0065-2113(05)87001-4
L’Abée-Lund, T. M., and Sørum, H. (2000). Functional Tn5393-like transposon in the R plasmid pRAS2 from the fish pathogen Aeromonas salmonicida subspecies salmonicida isolated in Norway. Appl. Environ. Microbiol. 66, 5533–5535. doi: 10.1128/AEM.66.12.5533-5535.2000
Lacy, G. H., Stromberg, V. K., and Cannon, N. P. (1984). Erwinia amylovora mutants and in planta-derived transconjugants resistant to oxytetracycline. Can. J. Plant Pathol. 6, 33–39. doi: 10.1080/07060668409501588
Lee, Y. S., Kim, G. H., Song, Y. R., Oh, C. S., Koh, Y. J., and Jung, J. S. (2020). Streptomycin resistant isolates of Pseudomonas syringae pv. actinidiae in Korea. Res. Plant Dis. 26, 44–47. doi: 10.5423/RPD.2020.26.1.44
Liang, X., Yu, X., Pan, X., Wu, J., Duan, Y., Wang, J., et al. (2018). A thiadiazole reduces the virulence of xanthomonas oryzae pv. oryzae by inhibiting the histidine utilization pathway and quorum sensing. Mol. Plant Pathol. 19, 116–128. doi: 10.1111/mpp.12503
Lima-Mendez, G., Alvarenga, D. O., Ross, K., Hallet, B., Van Melderen, L., Varani, A. M., et al. (2020). Toxin-antitoxin gene pairs found in Tn3 family transposons appear to be an integral part of the transposition module. MBio 11, 1–19. doi: 10.1128/mBio.00452-20
Llop, P., Donat, V., Rodríguez, M., Cabrefiga, J., Ruz, L., Palomo, J. L., et al. (2006). An indigenous virulent strain of Erwinia amylovora lacking the ubiquitous plasmid pEA29. Phytopathology 96, 900–907. doi: 10.1094/PHYTO-96-0900
Loper, J. E., Henkels, M. D., Roberts, R. G., Grove, G. G., Willett, M. J., and Smith, T. J. (1991). Evaluation of streptomycin, oxytetracycline, and copper resistance of Erwinia amylovora isolated from pear orchards in Washington state. Plant Dis. 75, 287–290. doi: 10.1094/PD-75-0287
Ludvigsen, J., Amdam, G. V., Rudi, K., and L’Abée-Lund, T. M. (2018). Detection and characterization of streptomycin resistance (strA-strB) in a honeybee gut symbiont (Snodgrassella alvi) and the associated risk of antibiotic resistance transfer. Microb. Ecol. 76, 588–591. doi: 10.1007/s00248-018-1171-7
Luo, Z. Q., and Farrand, S. K. (1999). Cloning and characterization of a tetracycline resistance determinant present in Agrobacterium tumefaciens C58. J. Bacteriol. 181, 618–626. doi: 10.1128/jb.181.2.618-626.1999
Luo, X., Mu, K., Zhao, Y., Zhang, J., Qu, Y., Hu, D., et al. (2022). Emergence of blaNDM–1-carrying Aeromonas caviae K433 isolated from patient with community-acquired pneumonia. Front. Microbiol. 13, 1–9. doi: 10.3389/fmicb.2022.825389
Lyu, Q., Bai, K., Kan, Y., Jiang, N., Thapa, S. P., Coaker, G., et al. (2019). Variation in streptomycin resistance mechanisms in Clavibacter michiganensis. Phytopathology 109, 1849–1858. doi: 10.1094/PHYTO-05-19-0152-R
Maeda, Y., Horita, M., Shinohara, H., Kiba, A., Ohnishi, K., Tsushima, S., et al. (2007a). Analysis of sources of oxolinic acid-resistant field strains of Burkholderia glumae based on rep-PCR analysis and nucleotide sequences of gyrB and rpoD. J. Gen. Plant Pathol. 73, 46–52. doi: 10.1007/s10327-006-0313-9
Maeda, Y., Kiba, A., Ohnishi, K., and Hikichi, Y. (2007b). Amino acid substitutions in GyrA of Burkholderia glumae are implicated in not only oxolinic acid resistance but also fitness on rice plants. Appl. Environ. Microbiol. 73, 1114–1119. doi: 10.1128/AEM.02400-06
Mansfield, J., Genin, S., Magori, S., Citovsky, V., Sriariyanum, M., Ronald, P., et al. (2012). Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 13, 614–629. doi: 10.1111/j.1364-3703.2012.00804.x
Mantengoli, E., and Rossolini, G. M. (2005). Tn5393d, a complex Tn5393 derivative carrying the PER-1 extended-spectrum β-lactamase gene and other resistance determinants. Antimicrob. Agents Chemother. 49, 3289–3296. doi: 10.1128/AAC.49.8.3289-3296.2005
Manulis, S., Kleitman, F., Shtienberg, D., Shwartz, H., Oppenheim, D., Zilberstaine, M., et al. (2003). Changes in the sensitivity of Erwinia amylovora populations to streptomycin and oxolinic acid in Israel. Plant Dis. 87, 650–654. doi: 10.1094/PDIS.2003.87.6.650
McGhee, G. C., and Sundin, G. W. (2011). Evaluation of kasugamycin for fire blight management, effect on nontarget bacteria, and assessment of kasugamycin resistance potential in Erwinia amylovora. Phytopathology 101, 192–204. doi: 10.1094/PHYTO-04-10-0128
McManus, P. S. (2014). Does a drop in the bucket make a splash? Assessing the impact of antibiotic use on plants. Curr. Opin. Microbiol. 19, 76–82. doi: 10.1016/j.mib.2014.05.013
McManus, P. S., and Jones, A. (1994). Epidemiology and genetic analysis of streptomycin-resistant Erwinia amylovora from Michigan and evaluation of oxytetracycline for control. Phytopathology 84, 627–633. doi: 10.1094/Phyto-84-627
McManus, P. S., Stockwell, V. O., Sundin, G. W., and Jones, A. L. (2002). Antibiotic use in plant agriculture. Annu. Rev. Phytopathol. 40, 443–465. doi: 10.1146/annurev.phyto.40.120301.093927
McVay, J., Sun, X., Jones, D., Urbina, H., Aldeek, F., Cook, J. M., et al. (2019). Limited persistence of residues and metabolites in fruit and juice following penicillin trunk infusion in citrus affected by Huanglongbing. Crop Prot. 125:104753. doi: 10.1016/j.cropro.2019.03.001
Miller, S. A., Ferreira, J. P., and Lejeune, J. T. (2022). Antimicrobial use and resistance in plant agriculture: a one health perspective. Agric. 12, 1–27. doi: 10.3390/agriculture12020289
Mindlin, S. Z., and Petrova, M. A. (2017). On the origin and distribution of antibiotic resistance: permafrost bacteria studies. Mol. Genet. Microbiol. Virol. 32, 169–179. doi: 10.3103/S0891416817040048
Munita, J. M., and Arias, C. A. (2016). Mechanisms of antibiotic resistance. Microbiol. Spectr. 6:34. doi: 10.3389/fmicb.2015.00034
Naue, C. R., Barbosa, M. A. G., Batista, D. D. C., De Souza, E. B., and Mariano, R. D. L. R. (2014). Effet of treatment of “red globe” vine cuttings on the control of bacterial canker caused by Xanthomonas campestris pv. viticola. Rev. Bras. Frutic. 36, 853–861. doi: 10.1590/0100-2945-435/13
Németh, J. (2004). Practice of applying streptomycin to control fireblight in Hungary. EPPO Bull. 34, 381–382. doi: 10.1111/j.1365-2338.2004.00764.x
Norelli, J. L., Burr, T. J., Lo Cicero, A. M., Gilbert, M. T., and Katz, B. H. (1991). Homologous streptomycin resistance gene present among diverse gram-negative bacteria in New York state apple orchards. Appl. Environ. Microbiol. 57, 486–491. doi: 10.1128/aem.57.2.486-491.1991
O’Brien, T. F. (2002). Emergence, spread, and environmental effect of antimicrobial resistance: how use of an antimicrobial anywhere can increase resistance to any antimicrobial anywhere else. Clin. Infect. Dis. 34, S78–S84. doi: 10.1086/340244
O’Neill, J. (2015). Antimicrobials in Agriculture and the Environment: Reducing Unnecessary Use and Waste - the Review on Antimicrobial Resistance. Available at: https://amr-review.org/sites/default/files/Antimicrobials%20in%20agriculture%20and%20the%20environment%20-%20Reducing%20unnecessary%20use%20and%20waste.pdf (Accessed 13 February 2023)..
Palmer, E. L., Teviotdale, B. L., and Jones, A. L. (1997). A relative of the broad-host-range plasmid RSF1010 detected in Erwinia amylovora. Appl. Environ. Microbiol. 63, 4604–4607. doi: 10.1128/aem.63.11.4604-4607.1997
Pan, X., Xu, S., Wu, J., Luo, J., Duan, Y., Wang, J., et al. (2018). Screening and characterization of Xanthomonas oryzae pv. oryzae strains with resistance to pheazine-1-carboxylic acid. Pestic. Biochem. Physiol. 145, 8–14. doi: 10.1016/j.pestbp.2017.12.003
Parcey, M., Gayder, S., Morley-Senkler, V., Bakkeren, G., Úrbez-Torres, J. R., Ali, S., et al. (2020). Comparative genomic analysis of Erwinia amylovora reveals novel insights in phylogenetic arrangement, plasmid diversity, and streptomycin resistance. Genomics 112, 3762–3772. doi: 10.1016/j.ygeno.2020.04.001
Partridge, S. R., Kwong, S. M., Firth, N., and Jensen, S. O. (2018). Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 31, e00088–17. doi: 10.1128/CMR.00088-17
Paz-Carrasco, L. C., Intriago-Mendoza, L. D., Basso, M. F., and Celi-Herán, R. E. (2018). Prevalence of Burkholderia glumae in rice crops in Ecuador. J. Agric. Univ. Puerto Rico 102, 65–78. doi: 10.46429/jaupr.v102i1-2.17532
Popowska, M., Rzeczycka, M., Miernik, A., Krawczyk-Balska, A., Walsh, F., and Duffy, B. (2012). Influence of soil use on prevalence of tetracycline, streptomycin, and erythromycin resistance and associated resistance genes. Antimicrob. Agents Chemother. 56, 1434–1443. doi: 10.1128/AAC.05766-11
Post, V., and Hall, R. M. (2009). AbaR5, a large multiple-antibiotic resistance region found in Acinetobacter baumannii. Antimicrob. Agents Chemother. 53, 2667–2671. doi: 10.1128/AAC.01407-08
Poveda, J., and Baptista, P. (2021). Filamentous fungi as biocontrol agents in olive (Olea europaea L.) diseases: mycorrhizal and endophytic fungi. Crop Prot. 146:105672. doi: 10.1016/j.cropro.2021.105672
Poveda, J., Roeschlin, R. A., Marano, M. R., and Favaro, M. A. (2021). Microorganisms as biocontrol agents against bacterial citrus diseases. Biol. Control 158:104602. doi: 10.1016/j.biocontrol.2021.104602
Rao, G. P. (2021). Our understanding about phytoplasma research scenario in India. Springer India 74, 371–401. doi: 10.1007/s42360-020-00303-1
Roberts, M. (2021). Mechanism of resistance for characterized tet and otr genes. Available at: https://faculty.washington.edu/marilynr/tetweb1.pdf (Accessed January 12, 2023).
Rodríguez, C., Lang, L., Wang, A., Altendorf, K., García, F., and Lipski, A. (2006). Lettuce for human consumption collected in Costa Rica contains complex communities of culturable oxytetracycline- and gentamicin-resistant bacteria. Appl. Environ. Microbiol. 72, 5870–5876. doi: 10.1128/AEM.00963-06
Rodríguez-Sánchez, C., Altendorf, K., Smalla, K., and Lipski, A. (2008). Spraying of oxytetracycline and gentamicin onto field-grown coriander did not affect the abundance of resistant bacteria, resistance genes, and broad host range plasmids detected in tropical soil bacteria. Biol. Fertil. Soils 44, 589–596. doi: 10.1007/s00374-007-0242-6
Ross, K., Varani, A. M., Snesrud, E., Huang, H., Alvarenga, D. O., Zhang, J., et al. (2021). TnCentral: a prokaryotic transposable element database and web portal for transposon analysis. MBio 12, 1–15. doi: 10.1128/mBio.02060-21
Sánchez, B., Mosbach-Schulz, O., Rodríguez Cerezo, E., Barreiro Hurle, J., Soto Embodas, I., Baker, R., et al. (2019). Estimating the Economic, Social and Environmental Impacts of EU Priority Pests: A Joint EFSA and JRC Project with a Focus on Xylella fastidiosa. In 2nd European Conference on Xylella fastidiosa 29th–30th Oct 2019, 1–17. Available at: https://www.efsa.europa.eu/sites/default/files/event/191029-xylella/S6.P4 SANCHEZ MOSBACH.pdf (Accessed 18 January 2023).
Schnabel, E. L., and Jones, A. L. (1999). Distribution of tetracycline resistance genes and transposons among phylloplane bacteria in Michigan apple orchards. Appl. Environ. Microbiol. 65, 4898–4907. doi: 10.1128/aem.65.11.4898-4907.1999
Schneider, K., van der Werf, W., Cendoya, M., Mourits, M., Navas-Cortés, J. A., Vicent, A., et al. (2020). Impact of Xylella fastidiosa subspecies pauca in European olives. Proc. Natl. Acad. Sci. U. S. A. 117, 9250–9259. doi: 10.1073/pnas.1912206117
Schroth, M., Thomson, S. V., and Moller, W. J. (1979). Streptomycin resistance in Erwinia amylovora. Phytopathology 69, 565–568. doi: 10.1094/Phyto-69-565
Shade, A., Klimowicz, A. K., Spear, R. N., Linske, M., Donato, J. J., Hogan, C. S., et al. (2013a). Streptomycin application has no detectable effect on bacterial community structure in apple orchard soil. Appl. Environ. Microbiol. 79, 6617–6625. doi: 10.1128/AEM.02017-13
Shade, A., McManus, P. S., and Handelsman, J. (2013b). Unexpected diversity during community succession in the apple flower microbiome. mBio 4, e00602–12. doi: 10.1128/mBio.00602-12
Shin, K., Ascunce, M. S., Narouei-Khandan, H. A., Sun, X., Jones, D., Kolawole, O. O., et al. (2016). Effects and side effects of penicillin injection in huanglongbing affected grapefruit trees. Crop Prot. 90, 106–116. doi: 10.1016/j.cropro.2016.08.025
Sholberg, P. L., Bedford, K. E., Haag, P., and Randall, P. (2001). Survey of Erwinia amylovora isolates from British Columbia for resistance to bactericides and virulence on apple. Can. J. Plant Pathol. 23, 60–67. doi: 10.1080/07060660109506910
Sparling, P. F. (1970). Kasugamycin resistance: 30S ribosomal mutation with an unusual location on the Escherichia coli chromosome. Science 167, 56–58. doi: 10.1126/science.167.3914.56
Sparling, P. F., Ikeya, Y., and Elliot, D. (1973). Two genetic loci for resistance to kasugamycin in Escherichia coli. J. Bacteriol. 113, 704–710. doi: 10.1128/jb.113.2.704-710.1973
Spotts, R. A., and Cervantes, L. A. (1995). Copper, oxytetracycline, and streptomycin resistance of Pseudomonas syringae pv. syringae strains from pear orchards in Oregon and Washington. Plant Dis. 79, 1132–1135. doi: 10.1094/PD-79-1132
Stockwell, V. O., and Duffy, B. (2012). Use of antibiotics in plant agriculture. Rev. sci. tech. Off. int. Epiz. 31, 199–210. doi: 10.20506/rst.31.1.2104
Sun, S., Gao, H., Liu, Y., Jin, L., Wang, R., Wang, X., et al. (2020). Co-existence of a novel plasmid-mediated efflux pump with colistin resistance gene mcr in one plasmid confers transferable multidrug resistance in Klebsiella pneumoniae. Emerg. Microbes Infect. 9, 1102–1113. doi: 10.1080/22221751.2020.1768805
Sundin, G. W. (2000). Examination of base pair variants of the strA-strB streptomycin resistance genes from bacterial pathogens of humans, animals and plants. J. Antimicrob. Chemother. 46, 848–849. doi: 10.1093/jac/46.5.848
Sundin, G. W. (2002). Distinct recent lineages of the strA-strB streptomycin-resistance genes in clinical and environmental bacteria. Curr. Microbiol. 45, 63–69. doi: 10.1007/s00284-001-0100-y
Sundin, G. W., and Bender, C. L. (1993). Ecological and genetic analysis of copper and streptomycin resistance in Pseudomonas syringae pv. syringae. Appl. Environ. Microbiol. 59, 1018–1024. doi: 10.1128/aem.59.4.1018-1024.1993
Sundin, G. W., and Bender, C. L. (1995). Expression of the strA-strB streptomycin resistance genes in Pseudomonas syringae and Xanthomonas campestris and characterization of IS6100 in X. campestris. Appl. Environ. Microbiol. 61, 2891–2897. doi: 10.1128/aem.61.8.2891-2897.1995
Sundin, G. W., and Bender, C. L. (1996). Dissemination of the strA-strB streptomycin-resistance genes among commensal and pathogenic bacteria from humans, animals and plants. Mol. Ecol. 5, 133–143. doi: 10.1111/j.1365-294X.1996.tb00299.x
Sundin, G. W., Mayfield, C. T., Zhao, Y., Gunasekera, T. S., Foster, G. L., and Ullrich, M. S. (2004). Complete nucleotide sequence and analysis of pPSR1 (72,601 bp), a pPT23A-family plasmid from Pseudomonas syringae pv. syringae A2. Mol. Genet. 270, 462–476. doi: 10.1007/s00438-003-0945-9
Sundin, G. W., Monks, D. E., and Bender, C. L. (1995). Distribution of the streptomycin resistance transposon Tn5393 among phylloplane and soil bacteria from managed agricultural habitats. Can. J. Microbiol. 41, 792–799. doi: 10.1139/m95-109
Sundin, G. W., and Wang, N. (2018). Antibiotic resistance in plant-pathogenic bacteria. Annu. Rev. Phytopathol. 56, 161–180. doi: 10.1136/pgmj.48.558.216
Tancos, K. A., and Cox, K. D. (2017). Effects of consecutive streptomycin and kasugamycin applications on epiphytic bacteria in the apple phyllosphere. Plant Dis. 101, 158–164. doi: 10.1094/PDIS-06-16-0794-RE
Taylor, P., and Reeder, R. (2020). Antibiotic use on crops in low and middle-income countries based on recommendations made by agricultural advisors. CABI Agriculture and Bioscience 1, 1–14. doi: 10.1186/s43170-020-00001-y
Tolba, S., Egan, S., Kallifidas, D., and Wellington, E. M. H. (2002). Distribution of streptomycin resistance and biosynthesis genes in streptomycetes recovered from different soil sites. FEMS Microbiol. Ecol. 42, 269–276. doi: 10.1016/S0168-6496(02)00379-3
US Environmental Protection Agency Office of Pesticide Programs (2005). Streptomyces lydicus WYEC 108, USA. Available at: http://www.epa.gov/opp00001/methods/atmpmethods/QC-23-01.pdf. (Accessed 28 February 2023).
Valenzuela, M., Méndez, V., Montenegro, I., Besoain, X., and Seeger, M. (2019). Streptomycin resistance in Clavibacter michiganensis subsp. michiganensis strains from Chile is related to an rpsL gene mutation. Plant Pathol. 68, 426–433. doi: 10.1111/ppa.12971
Van Hoek, A. H. A. M., Mevius, D., Guerra, B., Mullany, P., Roberts, A. P., and Aarts, H. J. M. (2011). Acquired antibiotic resistance genes: an overview. Front. Microbiol. 2, 1–27. doi: 10.3389/fmicb.2011.00203
Vicente-Díez, I., Blanco-Pérez, R., González-Trujillo, M. D. M., Pou, A., and Campos-Herrera, R. (2021). Insecticidal effect of entomopathogenic nematodes and the cell-free supernatant from their symbiotic bacteria against Philaenus spumarius (Hemiptera: Aphrophoridae) nymphs. Insects 12, 1–12. doi: 10.3390/insects12050448
Vidaver, A. K. (2002). Uses of antimicrobials in plant agriculture. Clin. Infect. Dis. 34, S107–S110. doi: 10.1086/340247
Vincent, C., Pierre, M., Li, J., and Wang, N. (2019). Implications of heat treatment and systemic delivery of foliar-applied oxytetracycline on citrus physiological management and therapy delivery. Front. Plant Sci. 10, 1–7. doi: 10.3389/fpls.2019.00041
Walsh, F., Pelludat, C., Duffy, B., Smith, D. P., Owens, S. M., Frey, J. E., et al. (2014). Impact of streptomycin applications on antibiotic resistance in apple orchards. Agrar. Schweiz 5, 300–305.
Walsh, F., Smith, D. P., Owens, S. M., Duffy, B., and Frey, J. E. (2013). Restricted streptomycin use in apple orchards did not adversely alter the soil bacteria communities. Front. Microbiol. 4, 1–8. doi: 10.3389/fmicb.2013.00383
Wang, Z., Li, H., Zhang, J., Wang, X., Zhang, Y., and Wang, H. (2021). Identification of a novel plasmid-mediated tigecycline resistance-related gene, tet(Y), in Acinetobacter baumannii. J. Antimicrob. Chemother. 77, 58–68. doi: 10.1093/jac/dkab375
Wang, Q., Lin, M., Shen, P., and Guan, Y. (2021). Elevation of fatty acid biosynthesis metabolism contributes to zhongshengmycin resistance in Xanthomonas oryzae. Antibiotics 10, 1–13. doi: 10.3390/antibiotics10101166
Welch, T. J., Fricke, W. F., McDermott, P. F., White, D. G., Rosso, M. L., Rasko, D. A., et al. (2007). Multiple antimicrobial resistance in plague: an emerging public health risk. PLoS One 2:e309. doi: 10.1371/journal.pone.0000309
World Health Organization (2017). WHO Publishes List of Bacteria for Which New Antibiotics are Urgently Needed. Available at: https://www.who.int/news-room/detail/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (Accessed January 19, 2023).
World Health Organization (2019). Critically Important Antimicrobials for Human Medicine, 6th Revision. Geneva: World Health Organization.
Xu, Y., Luo, Q., and Zhou, M. (2013). Identification and characterization of integron-mediated antibiotic resistance in the phytopathogen Xanthomonas oryzae pv. oryzae. PLoS One 8:8. doi: 10.1371/journal.pone.0055962
Yang, T., Zhang, T., Zhou, X., Wang, P., Gan, J., Song, B., et al. (2021). Dysregulation of ClpP by small-molecule activators used against Xanthomonas oryzae pv. oryzae infections. J. Agric. Food Chem. 69, 7545–7553. doi: 10.1021/acs.jafc.1c01470
Yao, L., Ding, Y., Ding, M., Yan, X., Zhang, F., Zhang, Z., et al. (2021). Characterization of a novel class 1 integron InSW39 and a novel transposon Tn5393k identified in an imipenem-nonsusceptible Salmonella Typhimurium strain in Sichuan, China. Diagn. Microbiol. Infect. Dis. 99:115263. doi: 10.1016/j.diagmicrobio.2020.115263
Yao, Y., Maddamsetti, R., Weiss, A., Ha, Y., Wang, T., Wang, S., et al. (2022). Intra- and interpopulation transposition of mobile genetic elements driven by antibiotic selection. Nat. Ecol. Evol. 6, 555–564. doi: 10.1038/s41559-022-01705-2
Yashiro, E., and McManus, P. S. (2012). Effect of streptomycin treatment on bacterial community structure in the apple phyllosphere. PLoS One 7:e37131. doi: 10.1371/journal.pone.0037131
Yoshii, A., Moriyama, H., and Fukuhara, T. (2012). The novel kasugamycin 2’-N-acetyltransferase gene aac(2′)-IIa, carried by the IncP Island, confers kasugamycin resistance to rice-pathogenic bacteria. Appl. Environ. Microbiol. 78, 5555–5564. doi: 10.1128/AEM.01155-12
Yoshii, A., Omatsu, T., Katayama, Y., Koyama, S., Mizutani, T., Moriyama, H., et al. (2015). Two types of genetic carrier, the IncP genomic island and the novel IncP-1β plasmid, for the aac(2′)-IIa gene that confers kasugamycin resistance in Acidovorax avenae ssp. avenae. Mol. Plant Pathol. 16, 288–300. doi: 10.1111/mpp.12182
Yoshikawa, M., Okuyama, A., and Tanaka, N. (1975). A third kasugamycin resistance locus, ksgC, affecting ribosomal protein S2 in Escherichia coli K-12. J. Bacteriol. 122, 796–797. doi: 10.1128/jb.122.2.796-797.1975
Zhang, Y., Chen, Y., Zhu, X., Xu, Y., Hou, Y., Gao, T., et al. (2011). A molecular mechanism of resistance to streptomycin in Xanthomonas oryzae pv. oryzicola. Phytoparasitica 39, 393–401. doi: 10.1007/s12600-011-0172-6
Zhang, D., Zhao, Y., Feng, J., Hu, L., Jiang, X., Zhan, Z., et al. (2019). Replicon-based typing of IncI-complex plasmids, and comparative genomics analysis of IncIγ/K1 plasmids. Front. Microbiol. 10, 1–8. doi: 10.3389/fmicb.2019.00048
Keywords: antibiotic resistance, Erwinia amylovora , horizontal gene transfer, one health, plant pathogenic bacteria, strA-strB , streptomycin, Tn5393
Citation: Verhaegen M, Bergot T, Liebana E, Stancanelli G, Streissl F, Mingeot-Leclercq M-P, Mahillon J and Bragard C (2023) On the use of antibiotics to control plant pathogenic bacteria: a genetic and genomic perspective. Front. Microbiol. 14:1221478. doi: 10.3389/fmicb.2023.1221478
Received: 12 May 2023; Accepted: 08 June 2023;
Published: 27 June 2023.
Edited by:
Elisabeth Grohmann, Berlin Technical University of Applied Sciences, GermanyReviewed by:
Alexander N. Ignatov, Peoples’ Friendship University of Russia, RussiaCopyright © 2023 Verhaegen, Bergot, Liebana, Stancanelli, Streissl, Mingeot-Leclercq, Mahillon and Bragard. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Claude Bragard, Y2xhdWRlLmJyYWdhcmRAdWNsb3V2YWluLmJl
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.