- 1Department of Clinical Laboratory, The Affiliated Li Huili Hospital, Ningbo University, Ningbo, China
- 2Department of Clinical Laboratory, The First Affiliated Hospital of Wenzhou Medical University, Wenzhou, China
- 3Department of Clinical Laboratory, Ningbo No. 2 Hospital, Ningbo, China
Objective: The co-occurrence of colistin and tigecycline resistance genes in Klebsiella pneumoniae poses a serious public health problem. This study aimed to characterize a K. pneumoniae strain, K82, co-harboring a colistin resistance gene (CoRG) and tigecycline resistance gene (TRG), and, importantly, investigate the genetic characteristics of the plasmid with CoRG or TRG in GenBank.
Methods: K. pneumoniae strain K82 was subjected to antimicrobial susceptibility testing, conjugation assay, and whole-genome sequencing (WGS). In addition, comparative genomic analysis of CoRG or TRG-harboring plasmids from K82 and GenBank was conducted. K. pneumoniae strain K82 was resistant to all the tested antimicrobials including colistin and tigecycline, except for carbapenems.
Results: WGS and bioinformatic analysis showed that K82 belonged to the ST656 sequence type and carried multiple drug resistance genes, including mcr-1 and tmexCD1-toprJ1, which located on IncFIA/IncHI2/IncHI2A/IncN/IncR-type plasmid pK82-mcr-1 and IncFIB/IncFII-type plasmid pK82-tmexCD-toprJ, respectively. The pK82-mcr-1 plasmid was capable of conjugation. Analysis of the CoRG/TRG-harboring plasmid showed that mcr-8 and tmexCD1-toprJ1 were the most common CoRG and TRG of Klebsiella spp., respectively. These TRG/CoRG-harboring plasmids could be divided into two categories based on mash distance. Moreover, we found an IncFIB/IncHI1B-type plasmid, pSYCC1_tmex_287k, co-harboring mcr-1 and tmexCD1-toprJ1. To the best of our knowledge, this is the first report on the co-occurrence of mcr-1 and tmexCD1-toprJ1 on a single plasmid.
Conclusion: Our research expands the known diversity of CoRG and TRG-harboring plasmids in K. pneumoniae. Effective surveillance should be implemented to assess the prevalence of co-harboring CoRG and TRG in a single K. pneumoniae isolate or even a single plasmid.
1 Introduction
Klebsiella pneumoniae is one of the most important pathogens that can cause invasive hospital- and community-acquired infections, such as bacteremia, respiratory tract infections, and liver abscesses (Lee et al., 2017; David et al., 2019). With the extensive use of antibiotics, the rapid emergence of carbapenem-resistant hypervirulent K. pneumoniae (CR-hvKP) presents a severe challenge for clinical treatment (Shankar et al., 2018). In most cases, CR-hvKP is associated with an outbreak of infection in hospitals and high mortality rates (Yao et al., 2015). Therapeutic options for these species are mainly reliant on colistin and tigecycline, which are classified as critically important antimicrobials (He et al., 2019). Regrettably, the increasing use of colistin and tigecycline has inevitably resulted in the emergence of colistin and tigecycline-resistance isolates (Soliman et al., 2021).
Colistin resistance in K. pneumonia is commonly attributed to chromosomal mutations, including mgrB, phoP/phoQ, pmrA/pmrB, and crrA/crrB (Poirel et al., 2017). However, since the first discovery of mcr-1 in Escherichia coli in 2015 (Liu et al., 2016), nine variants of this gene (mcr-2 to mcr-10) have also been identified in E. coli and other Enterobacteriaceae strains (Gogry et al., 2021). In the persistent dissemination of colistin resistance, horizontal transfer of plasmid-borne mcr genes plays a significant role, which further worsens the severe situation (Wang et al., 2018). Until now, three plasmid-borne mcr genes, including mcr-1, mcr-7, and mcr-8, have been found in K. pneumonia (Xiaomin et al., 2020; Phetburom et al., 2021). In general, they led to the emergence of extensively drug-resistant K. pneumoniae, including colistin resistance along with other plasmid-mediated resistance genes, such as extended-spectrum beta-lactamase (ESBL) and carbapenem genes (Singh et al., 2021; Muraya et al., 2022).
Tigecycline resistance is a growing concern in gram-negative bacteria due to the emergence of plasmids containing mobile tigecycline-resistance genes, such as tet(X), tet(A), tet(K), and tet(M) variants, exacerbating transferable resistance between bacterial species (Linkevicius et al., 2016; Sun et al., 2019). New tet(X) variants have been identified from a variety of different bacterial species (Dong et al., 2022a; Lu et al., 2022, 2023). Among these tet(X) variants, the tet(X4) gene has been identified in a few studies in the Klebsiella pneumoniae strain, which poses a great threat to the clinical use of tigecycline (Li et al., 2022; Zhai et al., 2022). Recently, a novel resistance-nodulation-division (RND) efflux pump gene cluster, tmexCD1-toprJ1, encoded by plasmid was identified in K. pneumoniae isolates (Lv et al., 2020). Subsequently, its variants tmexCD2-toprJ2, tmexCD3-toprJ3, and tmexCD4-toprJ4, encoding tigecycline resistance, were revealed (Wang Y. et al., 2021; Gao et al., 2022). These highly transmissible resistance determinants are presenting a severe challenge for clinical management and treatment.
Worryingly, recent studies have found that tmexCD1-toprJ1 can be co-transferred with other mobile resistance genes, such as mcr-8.2, blaNDM − 1, and blakpc-2 in K. pneumoniae (Liu et al., 2022, 2023). These plasmid-mediated resistance determinants are highly transmissible, presenting a severe challenge for clinical management. Furthermore, the emergence of colistin and tigecycline resistance determinants in the endemic K. pneumoniae clone constitutes a true public threat. In this study, we characterize an ST656 multidrug-resistant K. pneumonia isolate, harboring colistin resistance gene (CoRG) mcr-1.1 and tigecycline resistance gene (TRG) tmexCD1-toprJ1, from the urine specimen of a bladder cancer patient. Moreover, we performed in silico typing and comparative analysis of CoRG or TRG-positive plasmids using the plasmids of Klebsiella species available in the NCBI RefSeq database. This study expands the diversity of known CoRG or TRG-carrying plasmids in K. pneumoniae strains and provides a basis for further prevention and control of the dissemination of such strains.
2 Materials and methods
2.1 Bacterial strains
The K. pneumoniae strain K82 was isolated from a urine sample from an 82-year-old man with a history of postoperative bladder cancer at Ningbo Medical Center, Li Huili Hospital in Zhejiang, China, in October 2016. The patient had a hospital-acquired urinary tract infection and was not treated with colistin or tigecycline. E. coli C600 (highly resistant rifampicin) were used as hosts for conjugal transfers. Strain K82 was initially identified by Vitek 2 Compact. Later, it was confirmed using matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS; bioMérieux, France). It was stored at −80°C in Luria-Bertani (LB) broth medium (Oxoid, UK) with 30% glycerol for further use.
2.2 Antimicrobial susceptibility testing
In vitro susceptibility tests of ceftazidime, cefepime, aztreonam, imipenem, meropenem, piperacillin/tazobactam, cefoperazone/sulbactam, amikacin, tobramycin, trimethoprim-sulfamethoxazole, levofloxacin, and minocycline were performed using the Vitek 2 Compact in N335 susceptibility cards (bioMérieux, France). The minimum inhibitory concentrations (MICs) of colistin and tigecycline were determined using the microdilution broth method, and the results were determined in accordance with the 2023 Clinical and Laboratory Standards Association (CLSI) guidelines (Clinical and Laboratory Standards Institute [CLSI], 2023). E. coli ATCC 25922 was used as the quality control strain.
2.3 Conjugation experiments
Conjugation experiments were performed with rifampicin-resistant E. coli C600 and azide-resistant E. coli J53 as the recipient, and the transferability of mcr-1 and tmexCD1-toprJ1 genes was investigated using strain K82 as a donor. The donor and recipient strains were grown in 3 mL LB broth overnight at 37°C. Subsequently, 50 μL of donor strain culture was mixed with 500 μL of recipient strain culture (v:v = 1:10) and 4.5 mL of fresh LB broth (Luo et al., 2022). In addition, 100 μL of the mixture was applied onto a cellulose filter membrane (pore size, 0.22 μm) already placed on an LB agar plate. After incubation at 37°C for 16 h to 18 h, the filter membrane was taken out and vortexed in 1 mL of LB broth. The vortex mixtures were plated on LB agar plates containing 2000 mg/L rifampicin or 200 mg/L sodium azide, together with 1 mg/L colistin or 2 mg/L tigecycline for the selection of the transconjugants, respectively. The conjugation frequency was calculated as the ratio of transconjugants over recipient cells.
2.4 Whole-genome sequencing assembly and annotation
The genomic DNA of K. pneumoniae K82 was extracted using a Qiagen Minikit (Qiagen, Hilden, Germany) based on the manufacturer’s recommendations. Whole-genome sequencing was performed using both the Illumina NovaSeq platform (Illumina, San Diego, CA, United States) and the long-read PacBio RS II platform (Pacific Biosciences, Menlo Park, CA, United States). De novo hybrid assembly of the Illumina and PacBio reads was performed using Unicycler v0.4.8 (Wick et al., 2017). The complete genome was annotated using prokka (Thirugnanasambandam et al., 2017). Antimicrobial resistance genes (ARGs) and virulence factors were identified using AMRFinderPlus and VFanalyzer, respectively (Liu et al., 2019; Feldgarden et al., 2021). Multilocus sequence typing (MLST) and capsular typing were performed using mlst and Kaptive, respectively. Plasmid replicons were analyzed with PlasmidFinder v2.1 (Carattoli et al., 2014). Insertion sequence (IS) elements were investigated through ISFinder (Couchoud et al., 2020).
2.5 Analysis of plasmids with colistin or tigecycline resistance determinants of genera Klebsiella
In total, 7,179 complete plasmid sequences of Klebsiella spp. were downloaded from the National Center for Biotechnology Information (NCBI) RefSeq GenBank. The ARG profiles of these plasmids were analyzed using AMRFinderPlus, and 119 plasmids carrying complete colistin or tigecycline resistance determinants were selected for further study (Supplementary Table S1). The ability of the mobilization and conjugation of plasmid was predicted using MOB-suite (Robertson and Nash, 2018). The neighbor-joining tree was constructed on the basis of the pairwise mash distances of the plasmids using Mashtree (Robertson and Nash, 2018). The tree was midpoint rooted and visualized using Interactive Tree of Life (iTOL).
2.6 Data availability
The complete genome sequences of K. pneumoniae strain K82 were deposited in GenBank with accession numbers CP124873-CP124878.
3 Results
3.1 General characteristics of Klebsiella pneumoniae K82
Klebsiella pneumoniae K82 was isolated from the urine of a patient after bladder cancer surgery. The strain was resistant to nearly all the tested antimicrobials (Table 1), including aminoglycoside, fluoroquinolone, colistin, tetracyclines, sulfonamide, and most β-lactams (aztreonam, ceftazidime, cefepime, piperacillin/tazobactam, cefoperazone/sulbactam), but remained susceptible to carbapenems (meropenem, imipenem).
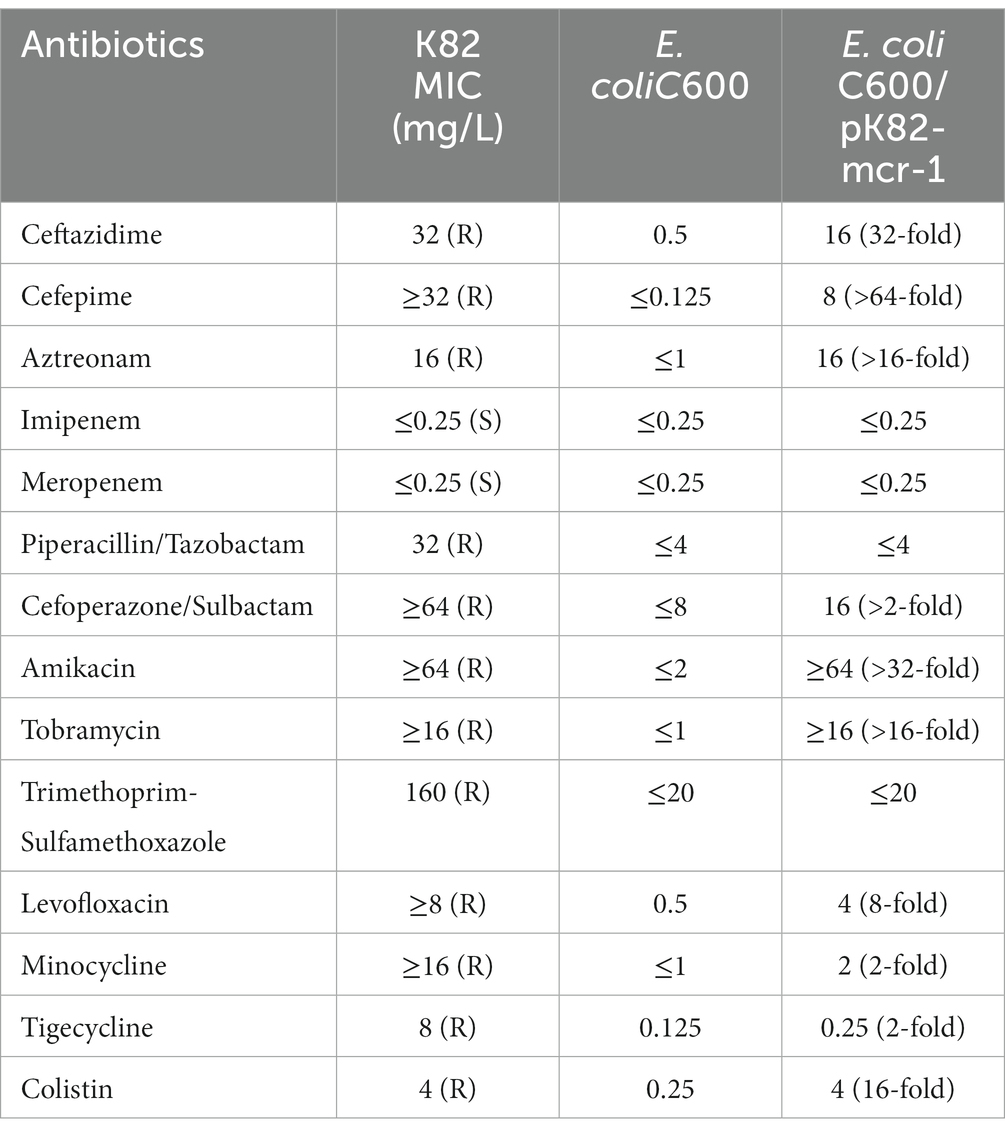
Table 1. Antimicrobial susceptibility of K. pneumoniae isolate K82, its transconjugants, and E. coli C600.
The results of WGS showed that the complete genome of K. pneumoniae K82 consisted of a 5.22-Mb chromosome and five plasmids. The genomic features are summarized in Table 2. In silico MLST and capsular typing showed that K. pneumoniae K82 belonged to ST656-KL23. A total of 20 ARGs and five metal resistance operons were identified in K. pneumoniae K82, and most of them were located on plasmids. The colistin-resistant and tigecycline-resistant determinants were carried by pK82-mcr-1 and pK82-tmexCD-toprJ, respectively.
3.2 The characteristics of plasmid pK82-mcr-1
The plasmid pK82-mcr-1 was 315,303-bp in length with an average G + C content of 47.57% (Figure 1A). A total of five replication proteins were identified on the plasmid, which belonged to the IncFIA(HI1), IncHI2, IncHI2A, IncN, and IncR family incompatibility groups. The result of the BLASTn search against the NCBI database showed that pK82-mcr-1 shared high similarity (>70% coverage) with four mcr-1-carrying plasmids, the highest of which was pSLK172-1 (accession number CP017632) from the E. coli strain. The four plasmids belonged to the IncHI2/IncHI2A family and the backbone structure of the IncHI2/IncHI2A type plasmid, including replication, conjugative transfer system, maintenance, and stability functional regions, were conserved in these plasmids as well as pK82-mcr-1. The mcr-1 gene of pK82-mcr-1 was located between the ISApl1 and pap2. The structure (ISApl1-mcr-1-pap2) resulted from Tn6330 losing one copy of ISApl1, which was common in other mcr-1-carrying plasmids (Snesrud et al., 2018). A total of 12 ARGs, including mcr-1, three copies of mercury resistance gene clusters, and a tellurium resistance gene cluster, were identified in the MDR region of pK82-mcr-1. Moreover, we observed that the complete backbone sequence (from vagC gene to umuD gene) of the IncR-type plasmid was presented in the variable region of pK82-mcr-1. The results of the conjugation experiment showed that pK82-mcr-1 could be successfully transferred to the recipient E. coli C600 at a frequency of 1.18 × 10−4 cells per recipient cell. The transconjugant acquired most of the antimicrobial resistance of the donor strain K82, except for piperacillin/tazobactam, cefoperazone/sulbactam, trimethoprim-sulfamethoxazole, and tetracyclines (Table 1).
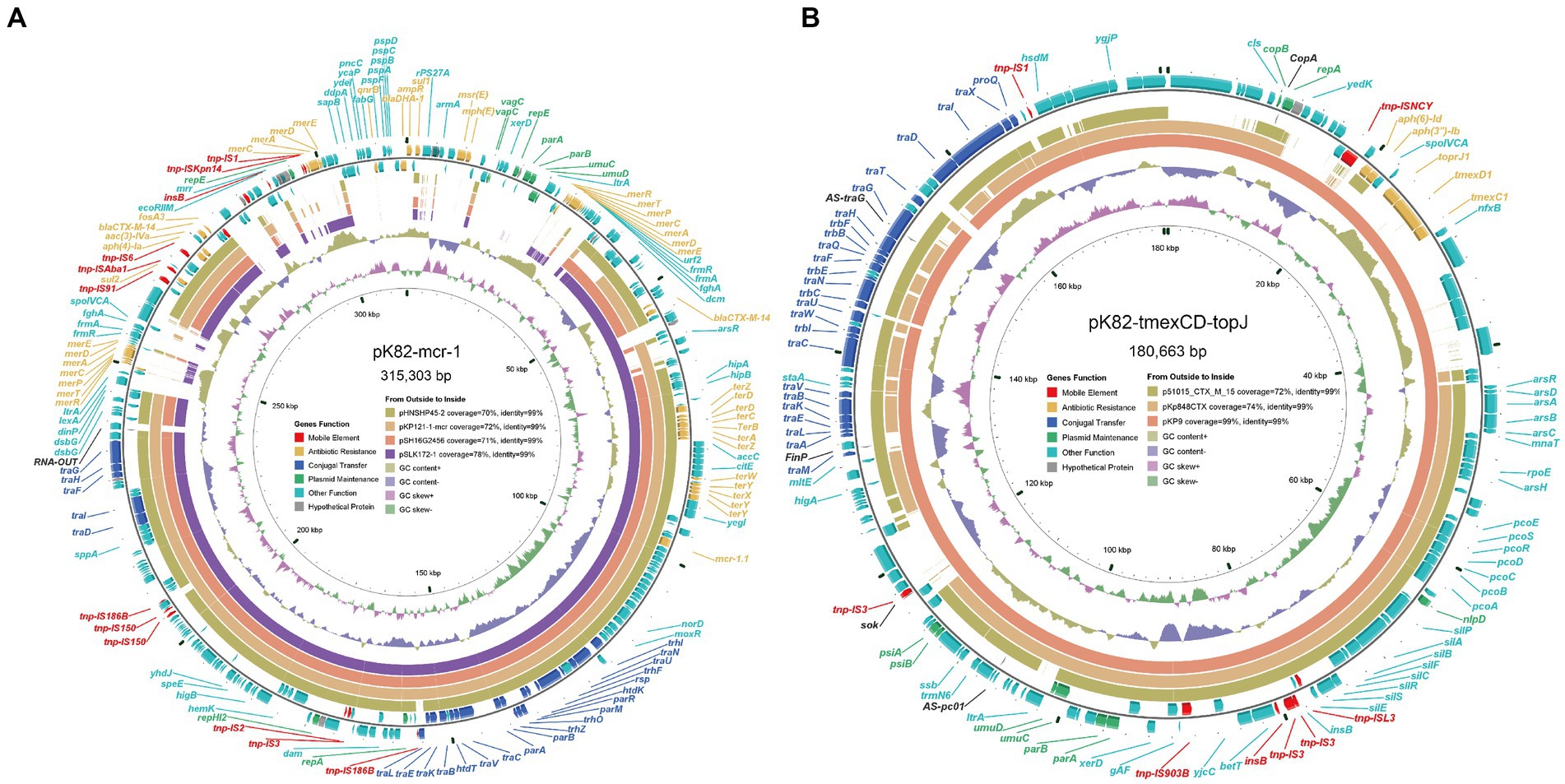
Figure 1. Diagram of plasmids pK82-mcr-1 and pK82-tmexCD-toprJ. (A) The plasmid map of pK82-mcr-1; (B) The plasmid map of pK82-tmexCD-toprJ.
3.3 Comparative analysis of plasmid pK82-tmexCD-toprJ and tmexCD-toprJ-harboring plasmids
pK82-tmexCD-toprJ was a 180,663-bp IncFIB(K)/IncFII(K) type plasmid with an average G + C content of 51.84% (Figure 1B). pK82-tmexCD-toprJ exhibits the highest similarity (99% coverage and 99% identity) with the tmexCD-toprJ-harboring plasmid pKP9 (accession number MZ690484), followed by the tmexCD-toprJ-negative plasmid pKp845CTX (accession number NC_024992). In addition to tmexCD1-toprJ1, we also found two ARGs and three metal resistance gene clusters conferring resistance to arsenic, copper, and silver (Figure 1B). Although conjugation transfer-related genes were identified on the pK82-tmexCD-toprJ, we could not transfer it into E. coli C600 or E. coli J53 by conjugation, which was consistent with the previous reports (Dong et al., 2022b).
To investigate the evolution of pK82-tmexCD-toprJ, we constructed a mash distance-based phylogenetic tree of plasmids carrying tmexCD-toprJ or its variants and aforementioned tmexCD-toprJ-negative plasmid pKp845CTX (Supplementary Figure S1). The results showed that pK82-tmexCD-toprJ was in the same branch, with six tmexCD1-toprJ1-harboring plasmids and pKp845CTX. All these plasmids were from K. pneumoniae (Supplementary Figure S1). Comparative plasmid analysis of pK82-tmexCD-toprJ with these seven plasmids showed that the plasmid backbone region, including maintaining stability (parB, umuCD), the conjugative transfer region (tra), and the metal resistance region (sil, pco, ars genes) were conserved among these plasmids (Supplementary Figure S2). The results indicate that these plasmids might have evolved from the same ancestor. In addition, we observed that plasmids MZ690482 and MZ690487 acquired more resistance genes, including tetA and blaTEM-1, which suggests that this type of plasmid has higher plasticity and aggravates the possibility of further spread.
3.4 Characteristics of plasmids with colistin or tigecycline resistance determinants of Klebsiella spp.
Totals of 119 plasmids of Klebsiella spp. carrying colistin or tigecycline resistance genes were obtained from GenBank for subsequent analysis together with pK82-mcr-1 and pK82-tmexCD-toprJ. Among these plasmids, 67 carried CoRG, 55 carried TRG, and one plasmid, pSYCC1_tmex_287k (CP113179), carried the two resistance determinants simultaneously (Figure 2A). Most of these plasmids were predicted to be capable of conjugative transfer. The types of CoRG and TRG carried by these plasmids were five and three, respectively. The most common CoRG was mcr-8, while the most common TRG was tmexCD1-toprJ1 (Figure 2B). A total of 16 and 15 Inc. groups were identified on plasmids carrying CoRG and TRG, respectively. The most common Inc. group of the former was IncFII, and the most common Inc. group of the latter was IncHI1B (Figure 2C). Notably, more than half of CoRG-harboring (52.2%, 35/67) and TRG-harboring (63.6%, 35/55) plasmids were hybrid plasmid types, in which IncFIA/IncFII and IncFIB/IncHI1B were the most common hybrid groups for CoRG-harboring and TRG-harboring plasmids, respectively (Figure 2D).
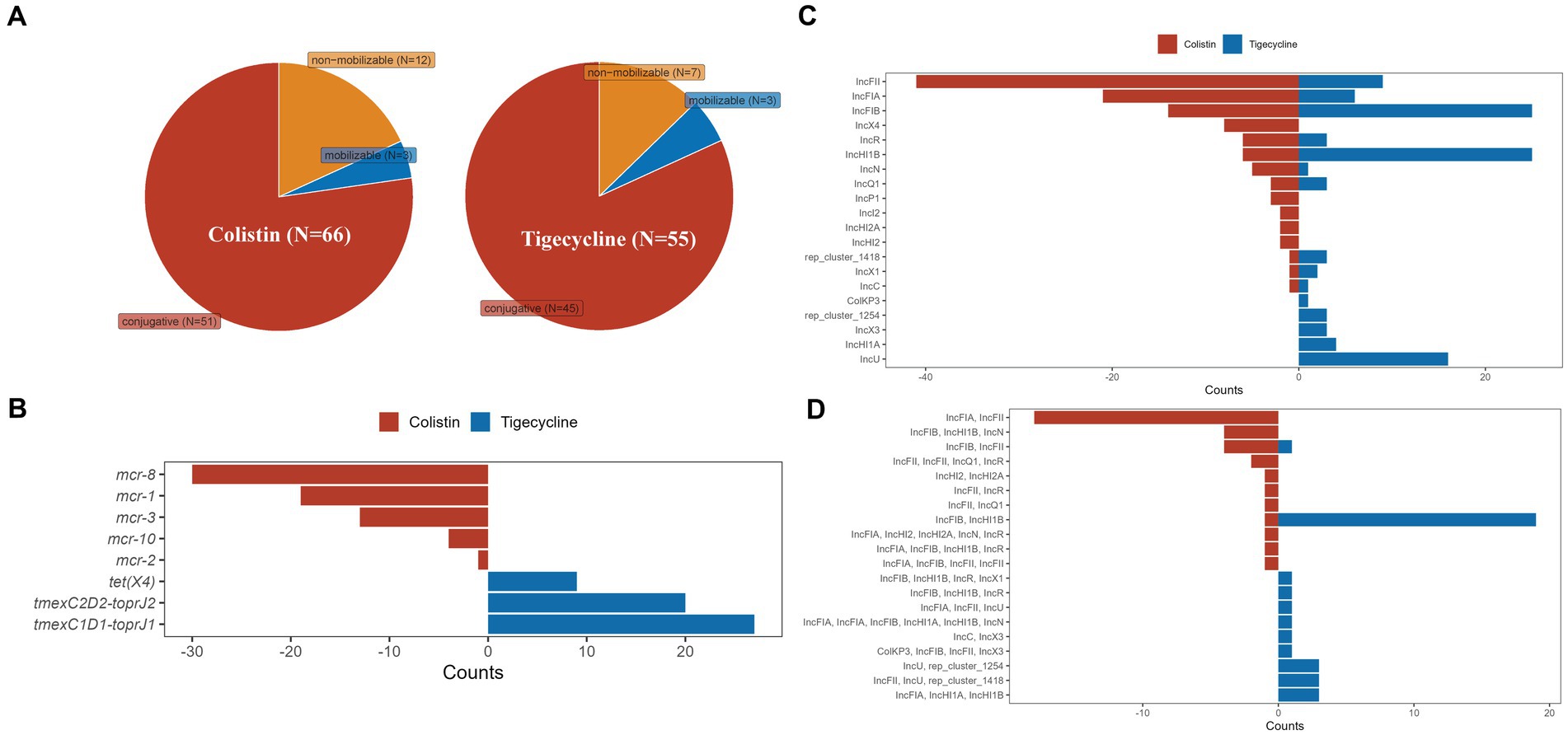
Figure 2. Characteristics of plasmids with colistin or tigecycline resistance determinants of Klebsiella spp. (A) The predicted transferability of the plasmids carrying CoRG or TRG; (B) The types of CoRG and TRG carried by the plasmids; (C) The Inc. group types of the plasmids carrying CoRG or TRG; (D) The Inc. group types of the hybrid plasmids carrying CoRG or TRG.
We further analyzed the antimicrobial resistance genotypes of these plasmids (Figure 3). ARGs carried by these plasmids could confer resistance to 14 classes of antimicrobials, and up to 11 classes of antimicrobial resistance could be provided by a single plasmid. Resistance to β-lactam was the most common phenotype (61.16%, 74/121) of these TRG/CoRG-harboring plasmids, some of which (16.53%, 20/121) even exhibit resistance to carbapenems. The counts of the resistance class of TRG-harboring plasmids (median = 8) were higher than that of CoRG-harboring plasmids (median = 2). The results of the mash clustering tree showed that these TRG/CoRG-harboring plasmids could be divided into two main categories (Cluster-TRG and Cluster-CoRG), which were highly in consistent with their phenotypes (Figure 3). However, we noticed that some CoRG-harboring plasmids were closely related to TRG-harboring plasmids, suggesting the possibility that a single plasmid exhibited both tigecycline and colistin resistance.
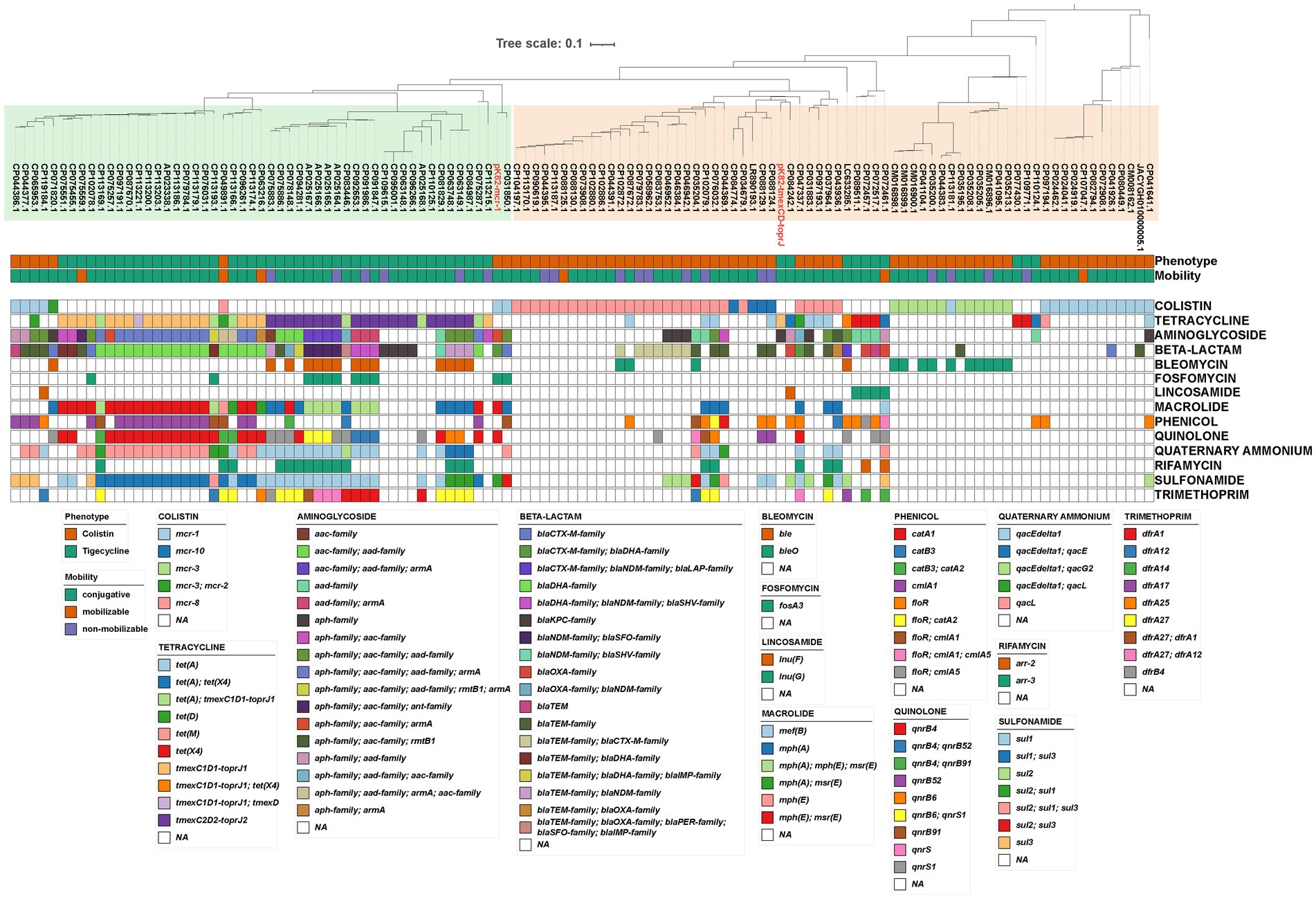
Figure 3. Phylogenetic analysis and antimicrobial resistance gene of plasmids carrying CoRG or TRG. Mash-based evolutionary relationships, plasmid characteristics, and antimicrobial resistance genes are shown from top to bottom, respectively. The meaning of the colored squares of each row is shown in the legends on the outside, and white means not present.
3.5 Co-occurrence of mcr-1 and tmexCD-toprJ in one plasmid
Surprisingly, we found an mcr-1 with a frameshift mutation encoding a truncated MCR-1 protein on the tmexCD-toprJ-harboring plasmid pSYCC1_tmex_287k. pSYCC1_tmex_287k was 287,882-bp in length, and its replicon belonged to the IncFIB/IncHI1B Inc. group, which was the most common Inc. group of hybrid TRG-harboring plasmid. pSYCC1_tmex_287k shared high similarity (coverage >95% and identity >99%) with seven tmexCD-toprJ-carrying plasmids, while intact or truncated mcr-1 was not found on them (Figure 4A). Comparative genomic analysis revealed that pSYCC1_tmex_287k had an insertion of an mcr-1-harboring fragment between the dcm gene and ecoRIIR gene compared to these similar sequences (Figure 4B). The genetic context of mcr-1 in pSYCC1_tmex_287k was almost identical to that of Tn6330, while ISApl1 downstream of pap2 was disrupted by ISEc33 insertion. The results indicated that the plasmid pSYCC1_tmex_287k might be generated by the insertion of mcr-harboring Tn6330 into a tmexCD-toprJ-carrying plasmid of the IncFIB/IncHI1B Inc. group.
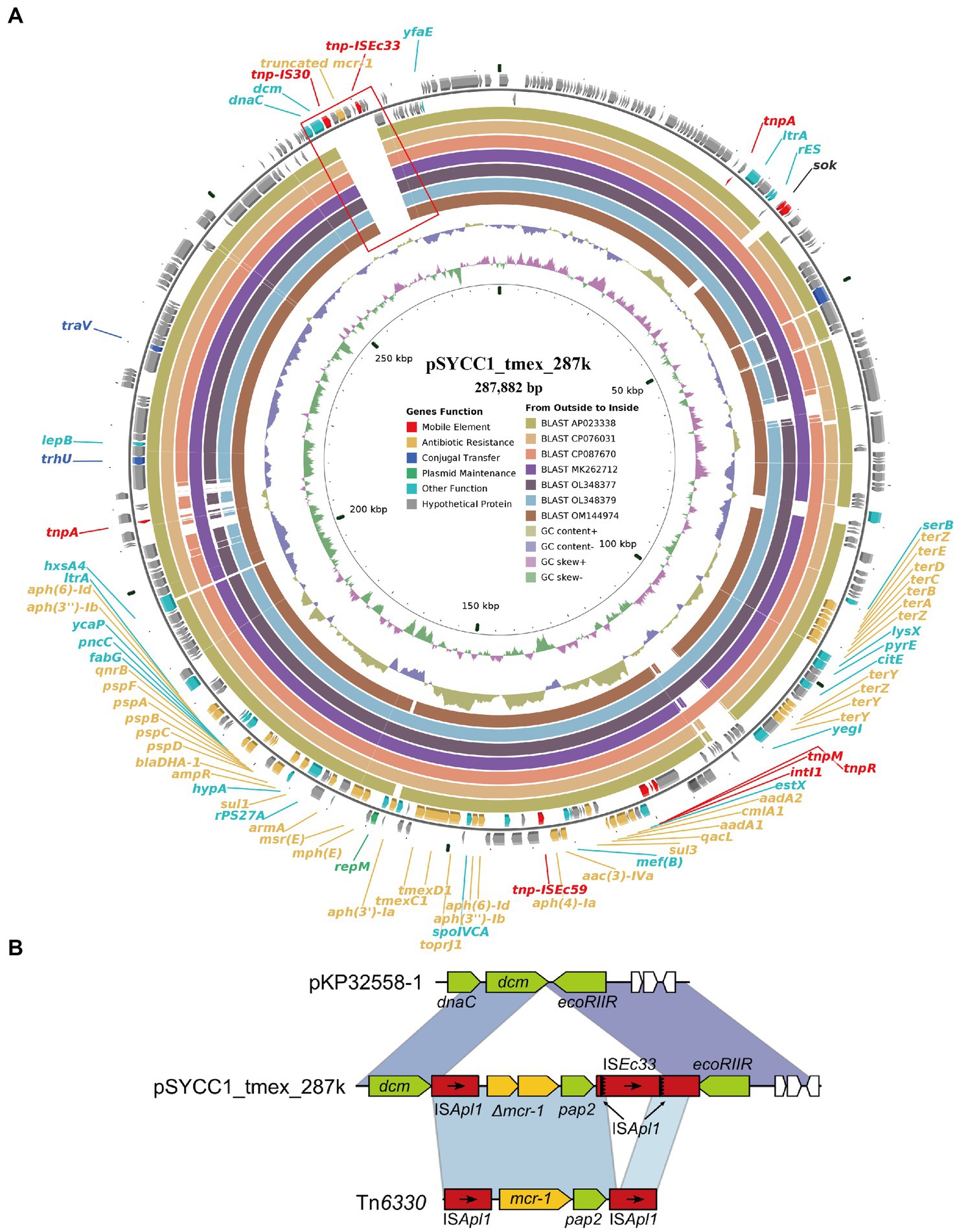
Figure 4. Diagram of plasmids pSYCC1_tmex_287k. (A) The plasmid map of pSYCC1_tmex_287k; (B) The genetic context of mutated mcr-1 in pSYCC1_tmex_287k.
4 Discussion
Colistin and tigecycline are important treatment options for multidrug-resistant and pan-drug-resistant Gram-negative bacteria, especially carbapenemase-producing K. pneumoniae in patients with severe infection (Karakonstantis et al., 2020). The emergence of colistin and tigecycline-resistant strains presents a difficult problem for clinical treatment. In this study, we identified a multidrug-resistant K. pneumoniae isolate carrying the plasmid-encoded CoRG mcr-1.1 and TRG tmexCD1-toprJ1 simultaneously. Moreover, we characterized all plasmids with CoRG or TRG in GenBank. The findings of this study will provide a new perspective on multiple resistance mechanisms to the last-resort antimicrobials. Of note, this represents the rare report of co-occurrence of mcr-1 and tmexCD1-toprJ1 on a single plasmid.
K. pneumoniae K82 belongs to ST656, a rare clone endemic in China (Wang et al., 2012). Recently, carbapenem-resistant ST656 strains have been reported worldwide (Roberts et al., 2022). In addition to mcr-1 and tmexCD1-toprJ1, K82 also carried multiple ARGs, including ESBL, as well as various metal resistance operons conferring resistance to multiple drug classes. Most of these ARGs located on IncFIA/IncHI2/IncHI2A/IncN/IncR-type plasmid pK82-mcr-1 and IncFIB/IncFII-type plasmid pK82-tmexCD-toprJ were found to be associated with mobile genetic elements (MGEs), which play important roles in the resistance gene transfer (He et al., 2022). The genetic context of mcr-1 was similar to Tn6330, which, apart from ISApl1 downstream of pap2, was disrupted by ISEc33 insertion. This insertion event may be of great importance in the transferability and evolution of resistance genes (Wang et al., 2017). It has been reported that tmexCD1-toprJ1 has disseminated among K. pneumoniae strains from different sources, such as poultry, food markets, and patients, and is located on plasmids or chromosomes (Sun et al., 2023). Its orthologous variants, tmexCD2-toprJ2, tmexCD3-toprJ3, and tmexCD4-toprJ4, were reported in various species of Enterobacteriaceae, while the plasmid types were different from pK82-tmexCD-toprJ. The tmexCD2-toprJ2 cluster was mainly located on IncHI1B plasmids, while the tmexCD3-toprJ3 gene cluster was located on SXT/R391 ICE (Wang Q. et al., 2021; Wang Y. et al., 2021). The TmexCD4-toprJ4 cluster was identified on untypeable plasmids, which was closely related (92 to 99% amino acid identity) to tmexCD1-toprJ1, tmexCD2-toprJ2, and tmexCD3-toprJ3 (Gao et al., 2022). Interestingly, recently, an ST656 K. pneumoniae strain carrying the mcr-1 and tmexCD1-toprJ1 genes was also reported, whereas the drug resistance genes were located on the IncX1 and IncR/IncN plasmids, respectively (Wang et al., 2023). These results indicate that ST656 K. pneumoniae is a potential high-risk clone that acquires resistance genes through various MGEs to produce resistance to almost all available antimicrobials. It is urgently vital to enhance the global surveillance.
In silico typing based on plasmid replicons of Klebsiella spp. showed that CoRG and TRG were diverse among plasmids, yet multiple genes were widely distributed across the plasmids, which is consistent with the previous reports (Algarni et al., 2022). The types of CoRG carried by plasmid replicons successively were mcr-8, mcr-1, mcr-3, mcr-10, and mcr-2, while the types of TRG were tmexCD1-toprJ1, tmexCD2-toprJ2, and tet(X4). As the above result shows, mcr-8 and tmexCD1-toprJ1 are the most common CoRG and TRG, respectively. The role of plasmid harboring mcr-8 in colistin-resistant K. pneumoniae should be highlighted, as it indicates that the genetic context of mcr-8 is heterogeneous and diverse (Wu et al., 2020). The crucial Inc. group was IncF, which is present in 56% of all multi-replicons, along with IncH, IncR, and IncU replicons (Douarre et al., 2020). The IncF-type plasmids are widely distributed in clinically relevant Enterobacteriaceae isolates (Johnson and Nolan, 2009), which is consistent with our report. These disparate plasmids contribute to the bacteria of antibiotic resistance gene dissemination among bacterial pathogens (Von Wintersdorff et al., 2016). Furthermore, there were too many ARGs carried by these plasmids, which is similar to pK82-mcr-1. Some horizontally transmitted accessory genes located on transposons and plasmids can be acquired by these ARGs carried by different plasmids. Worse still, the ARGs shuttle between resistant and sensitive strains, which increases resistance in different classes of antimicrobials (Acman et al., 2022). Furthermore, it poses a significant threat to global public health, especially when the genes are carried by a single plasmid among Enterobacteriaceae (Liu Z. et al., 2021). In our report, we noticed that some CoRG-harboring plasmids were closely related to TRG-harboring plasmids, suggesting the possibility that a single plasmid exhibits both tigecycline and colistin resistance, which presents a great threat to public health. Therefore, our study indicated that CoRG or TRG-carrying plasmids in K. pneumoniae strains are full of diversity, which is worth exploring more broadly.
Conjugative plasmids have facilitated the spread of antimicrobial resistance genes among clinically important pathogens (Matamoros et al., 2017). Recently, the rapid emergence of plasmid-mediated resistance genes mcr-8 and tmexCD1-toprJ1 confers transferable resistance to both tigecycline and colistin, which has attracted intense attention (Sun et al., 2020). Interestingly, in our study, we found an mcr-1 with a frameshift mutation on the tmexCD-toprJ-harboring plasmid pSYCC1_tmex_287k (CP113179), which indicated the co-occurrence of mcr-1 and tmexCD1-toprJ1 on a single plasmid of the IncFIB/IncHI1B Inc. group. The coexistence of mcr-1 and other TRGs in the same strain makes clinical treatment more challenging, and this coexistence phenomenon has been described many times (Liu Y. et al., 2021; Lu et al., 2022). However, a report showed that mcr-1 coexisted with other TRGs on an IncHI2-type single plasmid in E. coli (Xu et al., 2021), which increases the challenges of controlling antibiotic resistance. It was reported that mcr-1 and tet(X4)-coharboring plasmids could evolve into a plasmid with lower fitness costs, which would accelerate the transmission of mcr-1 and TRGs globally (Lu et al., 2021). Our research extends the known diversity and mechanistic insights of CoRG and TRG-harboring plasmids in K. pneumonia, which might benefit the development of new antibacterial agents.
5 Conclusion
The present study documents a clinically isolated ST656 K. pneumoniae isolate co-harboring plasmid-encoded resistance gene mcr-1 and tmexCD1-toprJ1 from the urine specimen of a bladder cancer patient. These resistance determinants are located on distinct plasmids. In addition, the analysis of plasmids with colistin or tigecycline resistance determinants of genera Klebsiella reveals that the tmexCD-toprJ-harboring plasmid pSYCC1_tmex_287k contains a frameshift mutation mcr-1, which represents the first report of the coexistence of mcr-1 and tmexCD-toprJ in one plasmid in China. This will inevitably accelerate the horizontal transmission of K. pneumonia resistance to colistin and tigecycline among Enterobacteriaceae species. Measures must be implemented to strengthen reasonable monitoring to avoid the spread of colistin- and tigecycline-resistant strains in China.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Ethics statement
The studies involving humans were approved by Medical Ethics Committee of Ningbo Medical Center Li Huili Hospital. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author contributions
QM contributed to the conception and design of the study. YZ, CQ, JY, QL, RZ, LQ, and QM participated and acquired the data. YZ conducted the experiments and drafted the manuscript. CQ analyzed and interpreted the data. All authors contributed to the article and approved the submitted version.
Funding
This study was funded by the Zhejiang Province Medical and Health Technology Project (No. 2023KY245) and Ningbo Medical Science and Technology Plan Project (No. 2022Y03).
Acknowledgments
We would like to give special thanks to all colleagues in the Laboratory of Clinical Microbiology for their assistance.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1221428/full#supplementary-material
References
Acman, M., Wang, R., Van Dorp, L., Shaw, L. P., Wang, Q., Luhmann, N., et al. (2022). Role of mobile genetic elements in the global dissemination of the carbapenem resistance gene Bla(NDM). Nat. Commun. 13:1131. doi: 10.1038/s41467-022-28819-2
Algarni, S., Han, J., Gudeta, D. D., Khajanchi, B. K., Ricke, S. C., Kwon, Y. M., et al. (2022). In silico analyses of diversity and dissemination of antimicrobial resistance genes and mobile genetics elements, for plasmids of enteric pathogens. Front. Microbiol. 13:1095128. doi: 10.3389/fmicb.2022.1095128
Carattoli, A., Zankari, E., García-Fernández, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/aac.02412-14
Clinical and Laboratory Standards Institute [CLSI] (2023). “Performance standards for antimicrobial susceptibility testing” in CLSI Supplement M100. 33rd ed (Wayne, PA: CLSI).
Couchoud, C., Bertrand, X., Valot, B., and Hocquet, D. (2020). Deciphering the role of insertion sequences in the evolution of bacterial epidemic pathogens with panISa software. Microb. Genom. 6:e000356. doi: 10.1099/mgen.0.000356
David, S., Reuter, S., Harris, S. R., Glasner, C., Feltwell, T., Argimon, S., et al. (2019). Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nat. Microbiol. 4, 1919–1929. doi: 10.1038/s41564-019-0492-8
Dong, N., Zeng, Y., Cai, C., Sun, C., Lu, J., Liu, C., et al. (2022a). Prevalence, transmission, and molecular epidemiology of tet(X)-positive bacteria among humans, animals, and environmental niches in China: An epidemiological, and genomic-based study. Sci. Total Environ. 818:151767. doi: 10.1016/j.scitotenv.2021.151767
Dong, N., Zeng, Y., Wang, Y., Liu, C., Lu, J., Cai, C., et al. (2022b). Distribution and spread of the mobilised RND efflux pump gene cluster tmexCD-toprJ in clinical gram-negative bacteria: a molecular epidemiological study. Lancet Microbe 3, e846–e856. doi: 10.1016/s2666-5247(22)00221-x
Douarre, P. E., Mallet, L., Radomski, N., Felten, A., and Mistou, M. Y. (2020). Analysis of COMPASS, a new comprehensive plasmid database revealed prevalence of multireplicon and extensive diversity of IncF plasmids. Front. Microbiol. 11:483. doi: 10.3389/fmicb.2020.00483
Feldgarden, M., Brover, V., Gonzalez-Escalona, N., Frye, J. G., Haendiges, J., Haft, D. H., et al. (2021). AMRFinderPlus and the reference gene catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci. Rep. 11:12728. doi: 10.1038/s41598-021-91456-0
Gao, X., Wang, C., Lv, L., He, X., Cai, Z., He, W., et al. (2022). Emergence of a novel plasmid-mediated Tigecycline resistance gene cluster, tmexCD4-toprJ4, in Klebsiella quasipneumoniae and Enterobacter roggenkampii. Microbiol. Spectr. 10:e0109422. doi: 10.1128/spectrum.01094-22
Gogry, F. A., Siddiqui, M. T., Sultan, I., and Haq, Q. M. R. (2021). Current update on intrinsic and acquired Colistin resistance mechanisms in Bacteria. Front. Med. 8:677720. doi: 10.3389/fmed.2021.677720
He, J., Du, X., Zeng, X., Moran, R. A., Van Schaik, W., Zou, Q., et al. (2022). Phenotypic and genotypic characterization of a Hypervirulent Carbapenem-resistant Klebsiella pneumoniae ST17-KL38 clinical isolate harboring the Carbapenemase IMP-4. Microbiol. Spectr. 10:e0213421. doi: 10.1128/spectrum.02134-21
He, T., Wang, R., Liu, D., Walsh, T. R., Zhang, R., Lv, Y., et al. (2019). Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat. Microbiol. 4, 1450–1456. doi: 10.1038/s41564-019-0445-2
Johnson, T. J., and Nolan, L. K. (2009). Pathogenomics of the virulence plasmids of Escherichia coli. Microbiol. Mol. Biol. Rev. 73, 750–774. doi: 10.1128/mmbr.00015-09
Karakonstantis, S., Kritsotakis, E. I., and Gikas, A. (2020). Treatment options for K. pneumoniae, P. Aeruginosa and A. baumannii co-resistant to carbapenems, aminoglycosides, polymyxins and tigecycline: an approach based on the mechanisms of resistance to carbapenems. Infection 48, 835–851. doi: 10.1007/s15010-020-01520-6
Lee, C. R., Lee, J. H., Park, K. S., Jeon, J. H., Kim, Y. B., Cha, C. J., et al. (2017). Antimicrobial resistance of Hypervirulent Klebsiella pneumoniae: epidemiology, Hypervirulence-associated determinants, and resistance mechanisms. Front. Cell. Infect. Microbiol. 7:483. doi: 10.3389/fcimb.2017.00483
Li, Y., Li, Y., Bu, K., Wang, M., Wang, Z., and Li, R. (2022). Antimicrobial resistance and genomic epidemiology of tet(X4)-bearing Bacteria of pork origin in Jiangsu, China. Genes (Basel) 14:36. doi: 10.3390/genes14010036
Linkevicius, M., Sandegren, L., and Andersson, D. I. (2016). Potential of tetracycline resistance proteins to evolve Tigecycline resistance. Antimicrob. Agents Chemother. 60, 789–796. doi: 10.1128/aac.02465-15
Liu, C., Du, P., Yang, P., Zheng, J., Yi, J., Lu, M., et al. (2023). Emergence and inter- and Intrahost evolution of Pandrug-resistant Klebsiella pneumoniae Coharboring tmexCD1-toprJ1, Bla(NDM-1), and Bla(KPC-2). Microbiol. Spectr. 11:e0278622. doi: 10.1128/spectrum.02786-22
Liu, Z., Hang, X., Xiao, X., Chu, W., Li, X., Liu, Y., et al. (2021). Co-occurrence of Bla (NDM-1) and mcr-9 in a conjugative IncHI2/HI2A plasmid from a bloodstream infection-causing Carbapenem-resistant Klebsiella pneumoniae. Front. Microbiol. 12:756201. doi: 10.3389/fmicb.2021.756201
Liu, Y., Lin, Y., Wang, Z., Hu, N., Liu, Q., Zhou, W., et al. (2021). Molecular mechanisms of Colistin resistance in Klebsiella pneumoniae in a tertiary care teaching hospital. Front. Cell. Infect. Microbiol. 11:673503. doi: 10.3389/fcimb.2021.673503
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/s1473-3099(15)00424-7
Liu, C., Wu, Y., Fang, Y., Sang, Z., Huang, L., Dong, N., et al. (2022). Emergence of an ST1326 (CG258) multi-drug resistant Klebsiella pneumoniae Co-harboring mcr-8.2, ESBL genes, and the resistance-nodulation-division efflux pump gene cluster tmexCD1-toprJ1 in China. Front. Microbiol. 13:800993. doi: 10.3389/fmicb.2022.800993
Liu, B., Zheng, D., Jin, Q., Chen, L., and Yang, J. (2019). VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 47, D687–d692. doi: 10.1093/nar/gky1080
Lu, X., Du, Y., Peng, K., Zhang, W., Li, J., Wang, Z., et al. (2022). Coexistence of tet(X4), mcr-1, and Bla(NDM-5) in ST6775 Escherichia coli isolates of animal origin in China. Microbiol. Spectr. 10:e0019622. doi: 10.1128/spectrum.00196-22
Lu, X., Xiao, X., Liu, Y., Li, R., and Wang, Z. (2021). Emerging opportunity and Destiny of mcr-1- and tet(X4)-Coharboring plasmids in Escherichia coli. Microbiol. Spectr. 9:e0152021. doi: 10.1128/Spectrum.01520-21
Lu, X., Zhang, L., Peng, K., Wang, Q., Liu, R., Wang, Z., et al. (2023). Characterisation of a novel Tigecycline resistance gene tet(X22) and its coexistence with Bla(NDM-1) in a Pseudomonas caeni isolate. Int. J. Antimicrob. Agents 62:106961. doi: 10.1016/j.ijantimicag.2023.106961
Luo, X., Yu, L., Feng, J., Zhang, J., Zheng, C., Hu, D., et al. (2022). Emergence of extensively drug-resistant ST170 Citrobacter portucalensis with plasmids pK218-KPC, pK218-NDM, and pK218-SHV from a tertiary hospital, China. Microbiol. Spectr. 10:e0251022. doi: 10.1128/spectrum.02510-22
Lv, L., Wan, M., Wang, C., Gao, X., Yang, Q., Partridge, S. R., et al. (2020). Emergence of a plasmid-encoded resistance-nodulation-division efflux pump conferring resistance to multiple drugs, including Tigecycline, in Klebsiella pneumoniae. MBio 11:e02930-19. doi: 10.1128/mBio.02930-19
Matamoros, S., Van Hattem, J. M., Arcilla, M. S., Willemse, N., Melles, D. C., Penders, J., et al. (2017). Global phylogenetic analysis of Escherichia coli and plasmids carrying the mcr-1 gene indicates bacterial diversity but plasmid restriction. Sci. Rep. 7:15364. doi: 10.1038/s41598-017-15539-7
Muraya, A., Kyany'a, C., Kiyaga, S., Smith, H. J., Kibet, C., Martin, M. J., et al. (2022). Antimicrobial resistance and virulence characteristics of Klebsiella pneumoniae isolates in Kenya by whole-genome sequencing. Pathogens 11:545. doi: 10.3390/pathogens11050545
Phetburom, N., Boueroy, P., Chopjitt, P., Hatrongjit, R., Akeda, Y., Hamada, S., et al. (2021). Klebsiella pneumoniae Complex harboring mcr-1, mcr-7, and mcr-8 isolates from slaughtered pigs in Thailand. Microorganisms 9:2436. doi: 10.3390/microorganisms9122436
Poirel, L., Jayol, A., and Nordmann, P. (2017). Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin. Microbiol. Rev. 30, 557–596. doi: 10.1128/cmr.00064-16
Roberts, L. W., Hoi, L. T., Khokhar, F. A., Hoa, N. T., Giang, T. V., Bui, C., et al. (2022). Genomic characterisation of multidrug-resistant Escherichia coli, Klebsiella pneumoniae, and Acinetobacter baumannii in two intensive care units in Hanoi, Viet Nam: a prospective observational cohort study. Lancet Microbe 3, e857–e866. doi: 10.1016/s2666-5247(22)00181-1
Robertson, J., and Nash, J. H. E. (2018). MOB-suite: software tools for clustering, reconstruction and typing of plasmids from draft assemblies. Microb. Genom. 4. doi: 10.1099/mgen.0.000206
Shankar, C., Veeraraghavan, B., Nabarro, L. E. B., Ravi, R., Ragupathi, N. K. D., and Rupali, P. (2018). Whole genome analysis of hypervirulent Klebsiella pneumoniae isolates from community and hospital acquired bloodstream infection. BMC Microbiol. 18:6. doi: 10.1186/s12866-017-1148-6
Singh, S., Pathak, A., Rahman, M., Singh, A., Nag, S., Sahu, C., et al. (2021). Genetic characterisation of Colistin resistant Klebsiella pneumoniae clinical isolates from North India. Front. Cell. Infect. Microbiol. 11:666030. doi: 10.3389/fcimb.2021.666030
Snesrud, E., Mcgann, P., and Chandler, M. (2018). The birth and demise of the ISApl1-mcr-1-ISApl1 composite transposon: the vehicle for transferable Colistin resistance. MBio 9:e02381-17. doi: 10.1128/mBio.02381-17
Soliman, A. M., Ramadan, H., Zarad, H., Sugawara, Y., Yu, L., Sugai, M., et al. (2021). Coproduction of Tet(X7) conferring high-level Tigecycline resistance, Fosfomycin FosA4, and Colistin Mcr-1.1 in Escherichia coli strains from chickens in Egypt. Antimicrob. Agents Chemother. 65:e02084-20. doi: 10.1128/AAC.02084-20
Sun, J., Chen, C., Cui, C. Y., Zhang, Y., Liu, X., Cui, Z. H., et al. (2019). Plasmid-encoded tet(X) genes that confer high-level tigecycline resistance in Escherichia coli. Nat. Microbiol. 4, 1457–1464. doi: 10.1038/s41564-019-0496-4
Sun, S., Gao, H., Liu, Y., Jin, L., Wang, R., Wang, X., et al. (2020). Co-existence of a novel plasmid-mediated efflux pump with colistin resistance gene mcr in one plasmid confers transferable multidrug resistance in Klebsiella pneumoniae. Emerg. Microbes Infect. 9, 1102–1113. doi: 10.1080/22221751.2020.1768805
Sun, L., Wang, H., Meng, N., Wang, Z., Li, G., Jiao, X., et al. (2023). Distribution and spread of the mobilized RND efflux pump gene cluster tmexCD-toprJ in Klebsiella pneumoniae from different sources. Microbiol. Spectr. 11:e0536422. doi: 10.1128/spectrum.05364-22
Thirugnanasambandam, R., Inbakandan, D., Abraham, L. S., Kumar, C., Sundaram, S. M., Subashni, B., et al. (2017). De novo assembly and annotation of the whole genomic analysis of Vibrio campbellii RT-1 strain, from infected shrimp: Litopenaeus vannamei. Microb. Pathog. 113, 372–377. doi: 10.1016/j.micpath.2017.11.016
Von Wintersdorff, C. J., Penders, J., Van Niekerk, J. M., Mills, N. D., Majumder, S., Van Alphen, L. B., et al. (2016). Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front. Microbiol. 7:173. doi: 10.3389/fmicb.2016.00173
Wang, X., Chen, J., Kang, Y., Jiang, N., An, S., and Gao, Z. J. C. M. J. (2012). Prevalence and characterization of plasmid-mediated blaESBL with their genetic environment in Escherichia coli and Klebsiella pneumoniae in patients with pneumonia. Chin Med J 125, 894–900. doi: 10.3760/cma.j.issn.0366-6999.2012.05.029
Wang, Q., Peng, K., Liu, Y., Xiao, X., Wang, Z., and Li, R. (2021). Characterization of TMexCD3-TOprJ3, an RND-type efflux system conferring resistance to Tigecycline in Proteus mirabilis, and its associated integrative conjugative element. Antimicrob. Agents Chemother. 65:e0271220. doi: 10.1128/aac.02712-20
Wang, Q., Sun, J., Ding, Y., Li, X. P., Liu, Y. H., and Feng, Y. (2017). Genomic insights into mcr-1-positive plasmids carried by Colistin-resistant Escherichia coli isolates from inpatients. Antimicrob. Agents Chemother. 61:e00361-17. doi: 10.1128/aac.00361-17
Wang, R., Van Dorp, L., Shaw, L. P., Bradley, P., Wang, Q., Wang, X., et al. (2018). The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat. Commun. 9:1179. doi: 10.1038/s41467-018-03205-z
Wang, Y., Zhou, J., Liu, H., Wang, Q., Zhang, P., Zhu, J., et al. (2023). Emergence of high-level colistin resistance mediated by multiple determinants, including mcr-1.1, mcr-8.2 and crrB mutations, combined with tigecycline resistance in an ST656 Klebsiella pneumoniae. Front. Cell. Infect. Microbiol. 13:1122532. doi: 10.3389/fcimb.2023.1122532
Wang, Y., Zhu, B., Liu, M., Dong, X., Ma, J., Li, X., et al. (2021). Characterization of IncHI1B plasmids encoding efflux pump TmexCD2-ToprJ2 in Carbapenem-resistant Klebsiella variicola, Klebsiella quasipneumoniae, and Klebsiella michiganensis strains. Front. Microbiol. 12:759208. doi: 10.3389/fmicb.2021.759208
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595
Wu, B., Wang, Y., Ling, Z., Yu, Z., Shen, Z., Zhang, S., et al. (2020). Heterogeneity and diversity of mcr-8 genetic context in chicken-associated Klebsiella pneumoniae. Antimicrob. Agents Chemother. 65:e01872-20. doi: 10.1128/aac.01872-20
Xiaomin, S., Yiming, L., Yuying, Y., Zhangqi, S., Yongning, W., and Shaolin, W. (2020). Global impact of mcr-1-positive Enterobacteriaceae bacteria on "one health". Crit. Rev. Microbiol. 46, 565–577. doi: 10.1080/1040841x.2020.1812510
Xu, Y., Liu, L., Zhang, H., and Feng, Y. (2021). Co-production of Tet(X) and MCR-1, two resistance enzymes by a single plasmid. Environ. Microbiol. 23, 7445–7464. doi: 10.1111/1462-2920.15425
Yao, B., Xiao, X., Wang, F., Zhou, L., Zhang, X., and Zhang, J. (2015). Clinical and molecular characteristics of multi-clone carbapenem-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in a tertiary hospital in Beijing, China. Int. J. Infect. Dis. 37, 107–112. doi: 10.1016/j.ijid.2015.06.023
Keywords: colistin, MCR-1, tigecycline, tmexCD1-toprJ1, plasmid
Citation: Zhao Y, Qian C, Ye J, Li Q, Zhao R, Qin L and Mao Q (2024) Convergence of plasmid-mediated Colistin and Tigecycline resistance in Klebsiella pneumoniae. Front. Microbiol. 14:1221428. doi: 10.3389/fmicb.2023.1221428
Edited by:
Xia Xiao, Yangzhou University, ChinaReviewed by:
Jiyun Li, Hunan Agricultural University, ChinaTao He, Jiangsu Academy of Agricultural Sciences (JAAS), China
Copyright © 2024 Zhao, Qian, Ye, Li, Zhao, Qin and Mao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qifeng Mao, bWFvcWlmZW5nMjE3QDE2My5jb20=
 Yujie Zhao
Yujie Zhao Changrui Qian
Changrui Qian Jianzhong Ye
Jianzhong Ye Qingcao Li1
Qingcao Li1 Qifeng Mao
Qifeng Mao