
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 15 August 2023
Sec. Microbiotechnology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1220208
Sucrose synthase (SuSy, EC 2.4.1.13) is a unique glycosyltransferase (GT) for developing cost-effective glycosylation processes. Up to now, some SuSys derived from plants and bacteria have been used to recycle uridine 5′-diphosphate glucose in the reactions catalyzed by Leloir GTs. In this study, after sequence mining and experimental verification, a SuSy from Micractinium conductrix (McSuSy), a single-cell green alga, was overexpressed in Escherichia coli, and its enzymatic properties were characterized. In the direction of sucrose cleavage, the specific activity of the recombinant McSuSy is 9.39 U/mg at 37°C and pH 7.0, and the optimum temperature and pH were 60°C and pH 7.0, respectively. Its nucleotide preference for uridine 5′-diphosphate (UDP) was similar to plant SuSys, and the enzyme activity remained relatively high when the DMSO concentration below 25%. The mutation of the predicted N-terminal phosphorylation site (S31D) significantly stimulated the activity of McSuSy. When the mutant S31D of McSuSy was applied by coupling the engineered Stevia glycosyltransferase UGT76G1 in a one-pot two-enzyme reaction at 10% DMSO, 50 g/L rebaudioside E was transformed into 51.06 g/L rebaudioside M in 57 h by means of batch feeding, with a yield of 76.48%. This work may reveal the lower eukaryotes as a promising resource for SuSys of industrial interest.
Glycosylation of secondary metabolites with diverse structures, such as flavonoids and terpenoids, has a profound impact on their solubility, stability, or bioactivity (Xiao et al., 2014; Schwab et al., 2015). The majority of glycosylation reactions in nature are catalyzed by glycosyltransferases (GTs), and the representative Leloir GTs with exceptional regio- and stereoselectivity, as well as a broader substrate diversity, are considered the most promising biocatalysts for industrial applications (De Bruyn et al., 2015). A crucial prerequisite is to solve the limited availability of nucleotide-activated sugar donors.
Sucrose synthase (SuSy, EC 2.4.1.13) belongs to the glycosyltransferase-4 subfamily (GT-4), which can catalyze the reversible reaction of sucrose synthesis and cleavage (Stein and Granot, 2019). A large number of studies have shown that the activity of SuSy depended on pH value (Bungaruang et al., 2013; Gutmann and Nidetzky, 2016). At pH 7.5–9.5, it displays optimal activity in the direction of sucrose synthesis, while acidic pH promotes the reverse reaction and decomposes sucrose at pH 5.5–7.5 to produce nucleoside diphosphate (NDP) glucose and fructose. Recently, using sucrose to recover the “donor” uridine 5′-diphosphate (UDP) glucose (UDP-Glc) by combining SuSy with Leloir GT (SuSy-GT) has aroused considerable interest in the development of biocatalytic glycosylation process, because the glycosylation of most known conjugates by Leloir GT requires the participation of UDP-Glc (Gutmann et al., 2017; Schmölzer et al., 2017; Nidetzky et al., 2018). In the SuSy-GT cascade reaction, a UDP cycle is created using sucrose and SuSy which makes UDP-Glc continuously regenerated as an expedient donor for glucoside production. More than that, the removal of UDP, a product inhibitor of Leloir GT, will enhance the glycosylation efficiency (Terasaka et al., 2012).
It is known that SuSy has a broad substrate spectrum for different NDP “acceptors” (Kulmer et al., 2017). In the past five decades, more attention has been focused on plant SuSys with UDP preference, which is conducive to the production of UDP-Glc (Schmölzer et al., 2016; Zhang et al., 2019). Prokaryotic SuSys are diversified in nucleotide substrate preference, such as some characterized SuSys from Thermosynechococcus elongatus (TeSuSy), Nitrosomonas Europaea (NeSuSy), Acidithiobacillus caldus (AcSuSy), and Denitrovibrio acetiphilus (DaSuSy), which are more inclined to use adenosine 5′-diphosphate (ADP) as nucleotide (Figueroa et al., 2013; Diricks et al., 2015). But bacterial SuSys showed better thermostability than plant SuSys, which could be more suitable for application in large-scale industrial production by increasing reaction temperature to avoid microbial contamination (Desmet et al., 2012; Diricks et al., 2016). To obtain a bacterial SuSy variant suitable for UDP-Glc regeneration during glycosylation reactions, the affinity of AcSuSy for UDP has been significantly improved by introducing plant residues at positions of a putative nucleotide binding motif (QN motif) (Diricks et al., 2016). Comparison was made between the L637M-T640V double mutant of AcSuSy and the SuSy from Glycine max (GmSuSy). As a result, fitness in terms of kinetics, expressed by the relatively low Km values for UDP and sucrose, superseded enhanced thermostability in bacterial SuSys as the selection criterion, which made plant SuSys the strongly preferred choice (Gutmann et al., 2017).
However, most of the receptors to be glycosylated have poor water solubility, so dimethyl sulfoxide (DMSO) is added as cosolvent to increase the solubility of the substrate (Pei et al., 2019; Chu et al., 2021; Tao et al., 2023). The low stability and intolerance to organic cosolvents of plant SuSys have limited the further application of such SuSy-GT cascades (Schmölzer et al., 2016). Therefore, mining or engineering of SuSys for high robustness has become one of the important tasks for biocatalytic glycosylation. Undoubtedly, with the rapid development of sequencing technology and data mining algorithms, more SuSys would be uncovered from the ever-increasing numbers of sequences with improved properties (Zhang et al., 2019; Zhao et al., 2023). Herein, by sequence mining, we focused on SuSys from lower eukaryotes like green algae, and their characteristics are still little known. A candidate SuSy-encoding sequence derived from Micractinium conductrix (McSuSy) was overexpressed in Escherichia coli BL21 (DE3), and the recombinant SuSy was characterized. The site-directed mutagenesis was conducted at the predicted N-terminal phosphorylation site (S31) of McSuSy. Furthermore, the S31D mutant with enhanced activity and the engineered glycosyltransferase UGT76G1 (UGT76G1_S195Q) from Stevia rebaudiana were co-expressed in E. coli to construct a SuSy-GT reaction. In the biotransformation of rebaudioside E (RebE, PubChem CID 72710721) into rebaudioside M (RebM, PubChem CID 92023628) (Yu et al., 2022), the issue of low solubility of the intermediate product rebaudioside D (RebD, PubChem CID 71773169) was alleviated by addition of DMSO, to obtain an enhanced product yield.
Two SuSy sequences from Anabaena sp. PCC 7119 (AnSuSy, CAA09297) and Melioribacter roseus (MrSuSy, AFN74551) were used as templates for BLAST search in NCBI.1 The resulting 20,000 sequences were downloaded for further analysis (in April 2020). Multiple sequence alignment was performed using MAFFT-7.037 (Katoh et al., 2002) or ClustalW2 with default parameters. After removing redundancy, three putative SuSys from algae, including Micractinium conductrix (McSuSy, PSC73946) and Chara braunii (CbSuSy1, GBG73881; CbSuSy2, GBG70160), were selected from the sequences with conserved residues G302, G303, H438, R580, L581, K585, Q648, N654, E675, and E683 (Zheng et al., 2011; Diricks et al., 2015; Wu et al., 2015), and used as candidates in the subsequent experiments. The residue number refers to the sites of SuSy from Arabidopsis thaliana (AtSuSy1, CAA50317) in the multiple sequence alignment.
Solubility predictions from sequences were performed using Protein-sol3 (Hebditch et al., 2017). The translated protein sequences of McSuSy, CbSuSy1, and CbSuSy2 were used to construct a phylogenetic tree using MEGA 7.0 (Kumar et al., 2016) with the known SuSys from G. max (GmSuSy, AAC39323), A. thaliana (AtSuSy1,CAA50317; AtSuSy3,CAB80721), S. tuberosum (StSuSy1, AAA33841), D. acetiphilus (DaSuSy, ADD69694), A. caldus (AcSuSy, AIA55343), N. europaea (NeSuSy, CAD85125), M. roseus, T. elongatus (TeSuSy, BAC08600), and Anabaena sp. PCC 7119 by using the neighbor-joining method (Saitou and Nei, 1987). Motifs were found by MEME (Bailey and Elkan, 1994) according to the result of sequence alignment and displayed by WebLogo4 (Crooks et al., 2004).
To predict the phosphorylation sites of SuSys, protein sequences of McSuSy, GmSuSy, and the SuSy from Zea mays (ZmSuSy) were submitted to NetPhos 3.1 Server5 (Blom et al., 1999).
The homology model of McSuSy was constructed using the YASARA program (Krieger et al., 2002). The structures of UDP and sucrose were obtained from ZINC database.6 To construct the complex structure for evaluating the interaction between the protein and substrates, we tested molecular docking software such as LeDock and AutoDock Vina (Morris et al., 2009; Trott and Olson, 2010; Zhao and Caflisch, 2013) to dock the structure of AtSuSy1 (PDB ID: 3S27, chain A) with its substrates. The docking results obtained by LeDock have a relatively good reproducibility to the crystal structure of AtSuSy1, therefore, LeDock was further used to obtain the complex of McSuSy. UDP was first docked into the active site of McSuSy, resulting in the structure of McSuSy with UDP, which was then docked with sucrose. PyMOL (Version 2.4.1, Schrodinger LLC) was used to visualize and analyze the model structures generated, as well as to build illustrative figures.
After codon optimization for heterologous expression in E. coli, the coding region derived from the putative SuSy mentioned above (Supplementary Data) was synthesized and cloned into pRSFDuet-1 (Novagen) between the restriction endonuclease sites NcoI and EcoRI by GenScript (Nanjing, China). A 6-histidine tag was added at the C-terminus of SuSy. The generated plasmids were named as pRSF-McSuSy, pRSF-CbSuSy1, and pRSF-CbSuSy2, respectively.
The plasmid pRSF-McSuSy was used as the template for site-directed mutagenesis by a Mut Express® II Fast Mutagenesis Kit V2 (Vazyme Biotech Co., Ltd., Nanjing, China). The primers used in PCR to produce the plasmid mutants are listed in Supplementary Table 1.
The code-optimized genes coding for UGT76G1_S195Q and the S31D mutant of McSuSy were synthesized and cloned into the restriction endonuclease sites NdeI/XhoI and NcoI/EcoRI of the pRSFDuet-1, respectively (Yu et al., 2022). The obtained plasmid was named pRSF-S31D-S195Q. Then, the coding region of the McSuSy mutant was replaced by that of AtSuSy1 in pRSF-S31D-S195Q, giving another plasmid named pRSF-AtSuSy1-S195Q.
The aforementioned plasmids were, respectively transformed into E. coli BL21 (DE3) competent cells (TransGen Biotech, Beijing, China), resulting in the corresponding recombinant strain.
The recombined E. coli was first incubated in 5-mL Luria-Bertani medium containing 10 g/L tryptone, 10 g/L NaCl, 5 g/L yeast extract and 50 μg/mL kanamycin, and incubated overnight at 37°C with continuous shaking at 200 rpm. Then, 2% (v/v) of the overnight culture was incubated in shake flasks with 100-mL LB medium containing 50 μg/mL kanamycin to cultivate for about 2 h at 37°C. Isopropyl-β-D-thiogalactopyranoside in a final concentration of 0.1 mM was added when the culture turbidity (OD600) reached 0.5–0.6, and then the cultivation was continued at 16°C for another 24 h. The subsequent steps involving purification were performed at 4°C. Cells harvested by centrifugation at 5,289 g for 5 min, were resuspended in an appropriate lysis buffer [500 mM NaCl and 10% glycerine (v/v) in 20 mM sodium phosphate buffer, pH 8.0] and disrupted by sonication. After centrifuge twice at 6,665 g for 15 min, the 6 Histidine-tagged proteins in the supernatant (crude extract) were purified by immobilized metal-affinity chromatography (IMAC) using a Ni-charged resin (L00683, GenScript, Nanjing, China). The recombination proteins were eluted from the column by stepwise imidazole gradient. Fractions with SuSy activity were pooled and concentrated in an Amicon® Ultra-15 Centrifugal Filter Unit with an Ultracel-30 membrane (Merck Millipore Ltd., Ireland), and the buffer was exchanged to 50 mM HEPES (N-2-hydroxyethylpiperazine-N-ethane-sulphonicacid)-NaOH (pH 7.0). The protein expression and the purity of recombinant enzymes were analyzed using SDS-PAGE.
The activities of purified SuSys in the sucrose cleavage direction were measured with the standard reaction mixture containing 50 mM HEPES-NaOH (pH 7.0), 2 mM UDP, 200 mM sucrose, and appropriate amount of purified enzyme in a final volume of 50 μL. Reactions were carried out at 37°C for 5 min and stopped by heating at 95°C for 2 min, and control experiments were performed immediately to check the decomposition of sucrose by the heat treatment. The product fructose was determined by the reduction of NAD+ at 340 nm following the addition of a 150-μL solution that contained 50 mM HEPPS-NaOH (pH 7.0), 1 mM MgCl2, 1 mM NAD+, 1 mM ATP, 1 μg hexokinase, 1 μg P-glucose isomerase, and 1 μg glucose-6-P dehydrogenase (Figueroa et al., 2013). The SuSy activity of crude extract in the sucrose cleavage direction was measured by 3, 5-dinitrosalicylic acid (DNS) method (Miller, 1959). Appropriate amount of crude extract was added to 100 μL of assay solution containing 50 mM HEPES (pH 7.0), 200 mM sucrose and 5 mM UDP, and reactions were carried out at 37°C for 20 min and quenched by heating. Control experiments were performed to check decomposition of sucrose by the heat treatment. One unit of SuSy activity is defined as the amount of enzyme that releases 1 μmol of reducing sugars per minute under the specified conditions.
The pH optimum of SuSy activity in the cleavage direction was determined in the pH ranging from 5.0 to 8.5 at 0.5 pH unit intervals. Buffers used were 50 mM MES-HCl (pH 5.0–7.0) and HEPES-NaOH (pH 7.0–8.5). The initial concentrations of sucrose and UDP in the reaction mixture were 200 and 2 mM, respectively.
The temperature profiles were obtained by determining the SuSy activity in the direction of sucrose cleavage from 20 to 70°C. In the evaluation of thermal stability, the enzyme was pre-incubated in 50 mM HEPES buffer (pH 7.0) for 15 min from 30 to 60°C without any substrates, alternatively, with the addition of 200 mM sucrose. After the incubation, the residual activity in the sucrose cleavage direction was checked with the standard assay described above.
The influence of divalent metal ions on SuSy was investigated by measuring the activity in the presence of 2 mM of MgCl2, MnCl2, CaCl2, NiCl2, CuCl2, BaCl2, ZnCl2, or Ethylene diamine tetraacetic acid (EDTA). The influence of Dimethyl sulfoxide (DMSO) on SuSy was investigated by measuring the activity in the presence of different concentrations of DMSO.
The kinetic parameters for sucrose varying from 50 to 600 mM at a constant concentration of 2 mM UDP and for UDP varying from 0.05 to 5 mM at a constant concentration of 200 mM sucrose were measured at 37°C in 50 mM HEPES buffer (pH 7.0). Apparent Km and Vmax values were calculated by non-linear regression of the Michaelis–Menten equation using OriginPro Learning Edition.
All reactions were conducted in triplicate. The relative activity (%) was calculated in terms of that of the maximum activity (100%). Coupled enzymes used for SuSy activity assays were purchased from Shanghai yuanye Bio-Technology Co., Ltd., and all the other reagents were analytical grade and commercially available.
To explore the application potential of McSuSy, we established SuSy-GT reactions to convert RebE into RebM. The reaction mixtures (5 mL) contained 15, 20 or 30 g/L of RebE, 150, 200 or 300 g/L of sucrose, 0–30% (v/v) DMSO, potassium phosphate buffer (50 mM, pH 7.2), and appropriate amount of the crude extract prepared from E. coli BL21 (pRSF-S31D-S195Q), or E. coli BL21 (pRSF-AtSuSy1-S195Q), in which expression of two recombinant enzymes was under the same conditions as SuSys, except for the induction for 36 h. For the convenience of description, the reactions were named S31D-S195Q and AtSuSy1-S195Q, respectively. The reaction was incubated at 40°C and 200 rpm for 24 h, and the samples were taken and heated for 10 min at 95°C. In the fed-batch reactions, the powder of RebE and sucrose, which kept the mass ratio at 1:10, was added at the specified time. The enzyme activity of SuSy was measured by the DNS method as described above, but in potassium phosphate buffer (pH 7.2). The GT activity of UGT76G1_S195Q was measured as previously described using RebE and UDP-Glc as the substrates (Yu et al., 2022). After being properly diluted and filtered, the concentrations of RebD and RebM in the reaction mixtures were determined by HPLC as previously described (Yu et al., 2022).
In the sequence mining, the prokaryote-derived SuSy templates AnSuSy and MrSuSy were used for sequence collection and the sequences without conservative residues G302, G303, H438, R580, L581, K585, and E675 (Zheng et al., 2011), and residues Q648, N654, and E683 that contribute to UDP-Glc binding were removed (the residue number refers to AtSuSy1 in the multiple sequence alignment) (Diricks et al., 2015; Wu et al., 2015). As a result, only 14 sequences from lower eukaryote sources like green algae remained together with a large number of the putative plant SuSys. These 14 sequences fall into Helicosporidium sp. (KDD76488), M. conductrix (PSC73946), and C. braunii (GBG70160, GBG73779, GBG73781, GBG73784, GBG73881, GBG76990, GBG86653, GBG89666, GBG89682, GBG92531, GBG92534, and GBG92536). The sequence from Helicosporidium sp. was ruled out for heterologous expression due to the low predicted protein solubility (0.389). Two sequences (GBG73881, GBG70160) with 69.48% identity (called CbSuSy1 and CbSuSy2, respectively) from those putative SuSys from C. braunii, along with the one from M. conductrix (McSuSy) was selected and synthesized for expression in E. coli. Enzyme activities of the crude extracts containing McSuSy and CbSuSy2 were around 15.5 and 5.5 mU/mg total protein, respectively. However, it was difficult to detect the SuSy activity of CbSuSy1. Therefore, McSuSy was chosen for further study of enzymatic properties.
McSuSy fused with a C-terminal histidine-tag that was overexpressed in E. coli BL21 (pRSF-McSuSy), was purified by Ni-NTA affinity chromatography (Supplementary Table 2). In SDS-PAGE, it corresponded to a band close to 97.2 kDa (Supplementary Figure 1). The specific activity of recombinant McSuSy is 9.39 U/mg at 37°C and pH 7.0. The kinetic constants for the cleavage reaction were determined: Km (UDP) 0.14 mM; Km (sucrose) 90 mM (Supplementary Figure 2).
According to the pH profile (Figure 1A), McSuSy reached its maximum activity at pH 7.0 and showed high enzymatic activity (>70% of maximum value) between pH 7 and pH 7.5. From pH 6.0–7.5, its activity was still higher than 40% of the maximum value, while the activity was undetectable at pH 8.5. The optimal temperature of McSuSy was 60°C ranging from 20 to 70°C (Figure 1B). After incubating the enzymes for 15 min between 30 and 60°C with or without sucrose, the thermostability of McSuSy was determined by measuring the residual activity. The enzyme remained stable up to 42°C after 15 min of incubation without substrates, but its activity sharply decayed beyond 50°C (Figure 1C). It is worth mentioning that sucrose is known to act as a stabilizing agent, and the result found that sucrose plays a positive role in maintaining enzyme activity. Addition of 200 mM sucrose enhanced enzyme activity by about 2 U/mg at the lower incubation temperature (30, 37°C) compared to the case without sucrose.
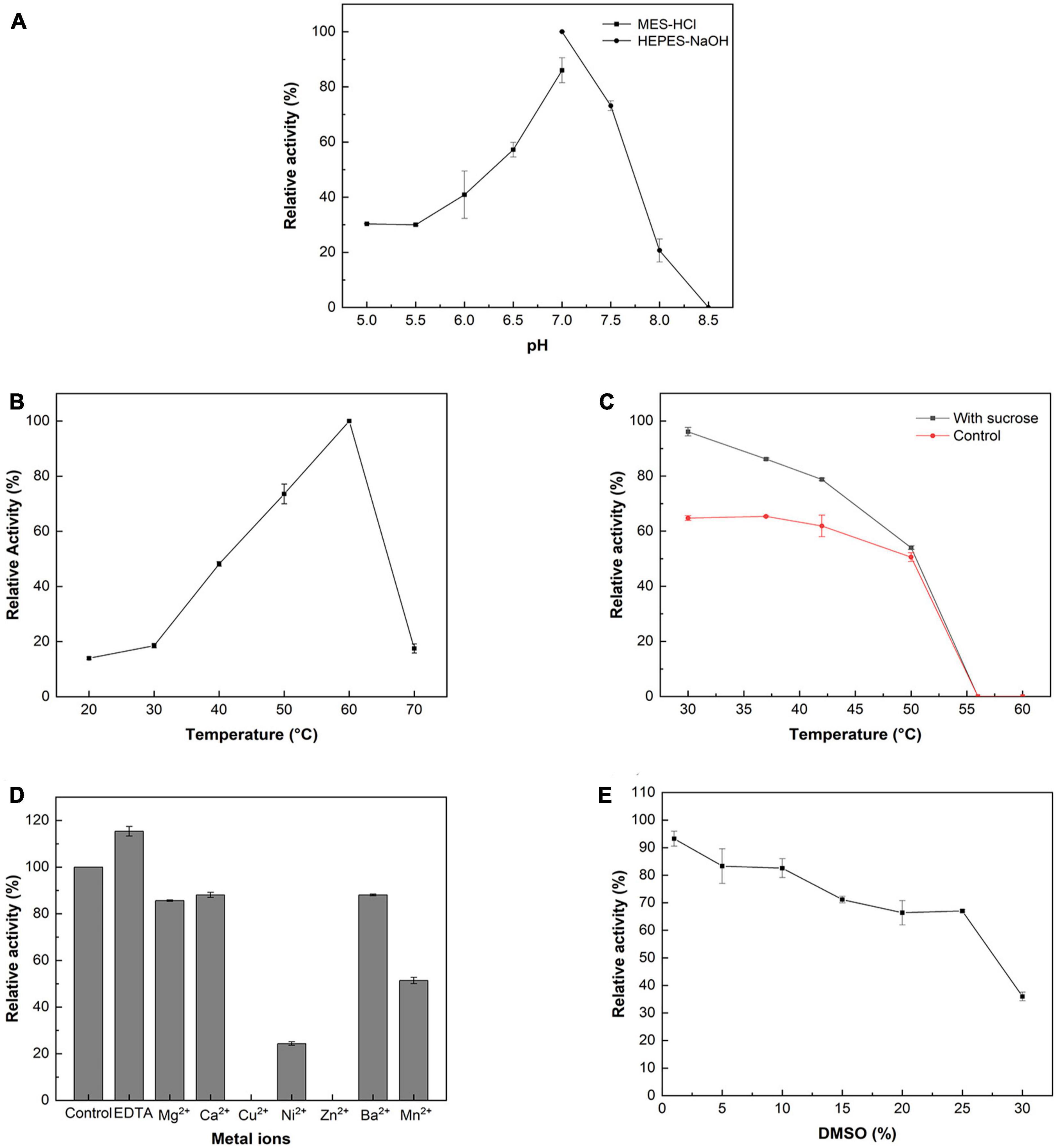
Figure 1. Enzymatic properties of the recombinant McSuSy. (A) pH profile; (B) Temperature profile; (C) Thermal stability; (D) Effect of metal ions on the activity of McSuSy; (E) Effect of DMSO on the activity of McSuSy. The data are presented as the means ± standard deviation of triplicates. The relative activity (%) was calculated in terms of that of the maximum activity (100%).
In terms of the effect of divalent metal ions, adding 2 mM of Mg2+, Ca2+, Cu2+, Ni2+, Zn2+, Ba2+, and Mn2+, the activities of McSuSy were observed to decrease (Figure 1D). It was especially strongly inhibited by Cu2+ and Zn2+, resulting in undetectable activity, since these ions may influence the interaction with clusters of histidine on the protein surface (Elling, 1995). In the presence of ethylene diamine tetraacetic acid (EDTA), a chelating agent, the activity of McSuSy was promoted by removing trace metal ions.
The effect of DMSO on McSuSy activity was investigated by adding different concentrations of DMSO (v/v) in the assay mixtures (Figure 1E). When the DMSO concentration increased from 1 to 25%, the enzyme activity decreased gradually. At 10% DMSO, McSuSy maintained a high activity (>80% of maximum value), and while adding 30% DMSO reduced enzyme activity to 36%.
SuSys, like StSuSy (S. tuberosum L) and SuSyNe (N. europaea) show high flexibility for nucleoside diphosphates in the cleavage reaction (Römer et al., 2004; Diricks et al., 2015). Plant SuSys preferentially utilize UDP as an acceptor nucleotide, while bacterial SuSys prefer ADP (Table 1). The Km value of McSuSy for UDP is 0.14 mM, indicating that McSuSy has a higher affinity for UDP. And it was difficult to determine the enzyme activity under the same conditions when ADP was the glycosyl receptor. Homology modeling was carried out using the crystal structure of AtSuSy1 (PDB ID: 3S27) as a template, which has 55.17% sequence identity with McSuSy, and the complex was obtained by substrate docking using LeDock. The observed secondary structure of McSuSy is very similar to that of the AtSuSy1 monomer (Figure 2A). Two sequence fragments, residues 333 to 345 and residues 683 to 718, were found in the active site of McSuSy, which are highly conserved in plant SuSys (Figure 2B). To be specific, the residues 333 to 345 of McSuSy (light blue), corresponding to the residues 300 to 312 of AtSuSy1, participate in the binding of fructose and G336 (G303 in AtSuSy1) also interacts with β-phosphate of UDP by forming hydrogen bonds (Figures 2C, D; Zheng et al., 2011; Wu et al., 2015). The residues 683 to 718 of McSuSy (pink) corresponding to the residues 648 to 683 of AtSuSy1 belong to the nucleotide-binding domain, which contains the “QN” motif, playing a significant role in the nucleotide preference of SuSy (Wu et al., 2015; Diricks et al., 2016). In particular, the two amino acids Q683 and N689 (Q648 and N654 in AtSuSy1) are highly conserved in plant SuSys, while in bacteria the residues are highly variable (Diricks et al., 2016). For example, R636 and A642 in the N. europaea create a more spacious binding site for the preference toward the bulkier ADP substrate (Wu et al., 2015). As shown in Figures 2C, D, the UDP moieties bind of McSuSy are the same way as AtSuSy1, especially in the indicated “QN” motif, which also implies a similar preference for nucleotide bases (Zheng et al., 2011).
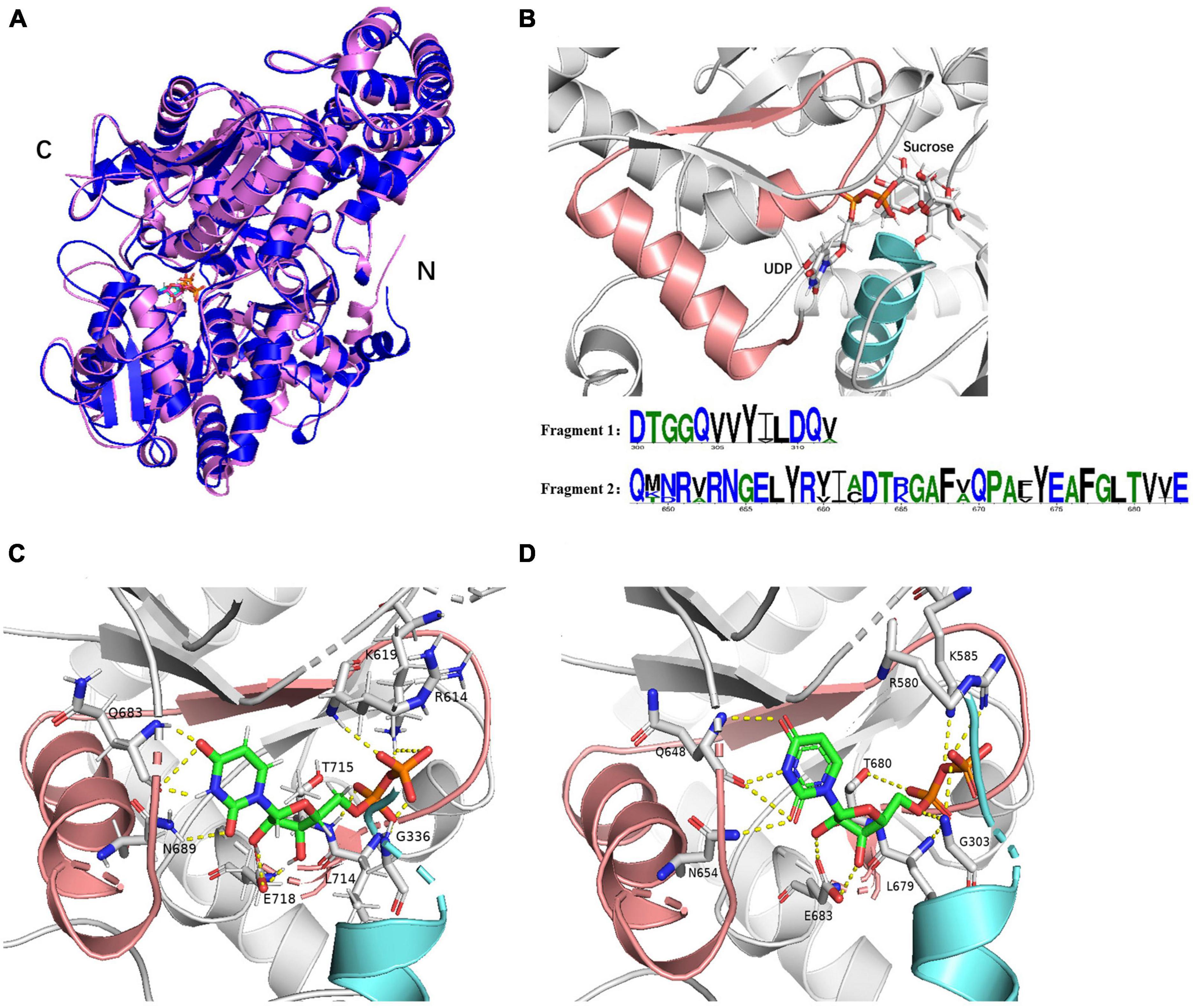
Figure 2. The structure models of McSuSy and AtSuSy1. (A) Ribbon drawing of the structure of McSuSy (blue) aligned with that of AtSuSy1 (purple). (B) The McSuSy complex with two conserved sequence fragments: residues 300–312 (light blue) and residues 648–683 (pink). The residue number refers to the sites of AtSuSy1 in the multiple sequence alignment. (C,D) The UDP binding sites of McSuSy and AtSuSy1, respectively.
Studies have demonstrated that phosphorylation affected the catalytic activities of SuSys in sucrose cleavage, which may adjust the apparent affinity of the enzyme for sucrose and UDP to activate the formation of UDP-Glc and fructose from sucrose plus UDP (Huber et al., 1996; Takeda et al., 2017). Three residues including S7, T22, and S31 at the N-terminus of McSuSy (Supplementary Table 3), which were predicted reliably as phosphorylation sites by NetPhos 3.1 Server, were mutated into two different acidic amino acid residues Asp (D) or Glu (E), respectively. As shown in Figure 3, the enzyme activity of crude extracts from the S7D, S7E, and T22D mutants declined slightly, and those from both S31D and S31E mutants increased significantly (more than 40% compared with wild-type McSuSy). After purification, the kinetic parameter of the S31D mutant (McSuSy_S31D) was determined (Supplementary Figure 3). The Km values of McSuSy_S31D were 0.092 mM and 88 mM for UDP and sucrose, respectively. A Km drop of more than 34.3% indicated an increased affinity for UDP compared with the wild type of McSuSy (Table 1).
The engineered glucosyltransferase UGT76G1_S195Q derived from S. rebaudiana, was recently reported that can efficiently convert RebE into RebM via a two-step continuous glycosylation (Figure 4; Yu et al., 2022). In the present study, McSuSy_S31D and UGT76G1_S195Q prepared from E. coli BL21 (pRSF-S31D-S195Q) were coupled to form a SuSy-GT system (S31D-S195Q). A control experiment (AtSuSy1-S195Q) was performed under the same conditions using UGT76G1_S195Q and AtSuSy1, which was widely applied in various glycosylation reactions (Chen T. Y. et al., 2021; Chu et al., 2021; Ping et al., 2022). The cascade reactions were carried out at pH 7.2 and 40°C, and the mass ratio of RebE to sucrose was set to 1:10 (Yu et al., 2022). The GT activity in AtSuSy1-S195Q was apparently higher than that in S31D-S195Q (Supplementary Table 5), suggesting the expression difference of UGT76G1_S195Q existed when SuSys were different, although the genes encoding GT and SuSy were cloned into the same sites of plasmid. The SuSy activity in AtSuSy1-S195Q was 1.85 folds of that in S31D-S195Q (Supplementary Table 5). The reaction mixture changed from clear to milky white after reaction of 3 h, and the intermediate product RebD started to accumulate as visible white precipitate. After 24 h of reaction (Figure 5A), the products RebD and RebM synthesized by S31D-S195Q were 8.43 and 9.48 g/L, respectively, where the RebM yield was 35.5%. The concentration of RebM in AtSuSy1-S195Q was 7.75 g/L, which was 18.2% lower than that in S31D-S195Q. Considering that UDP-Glc and UDP from the cell lysates are in limited amounts and prone to degradation for a long time of reaction (Baroja-Fernández et al., 2012; Gutmann and Nidetzky, 2016), an extra 1 mM UDP was added to the initial solutions, which promoted RebM production in both reactions. The concentrations of RebM in S31D-S195Q and AtSuSy1-S195Q were 10.52 and 11.87 g/L, respectively. The RebD concentrations in both reactions kept almost the same (around 7 g/L).
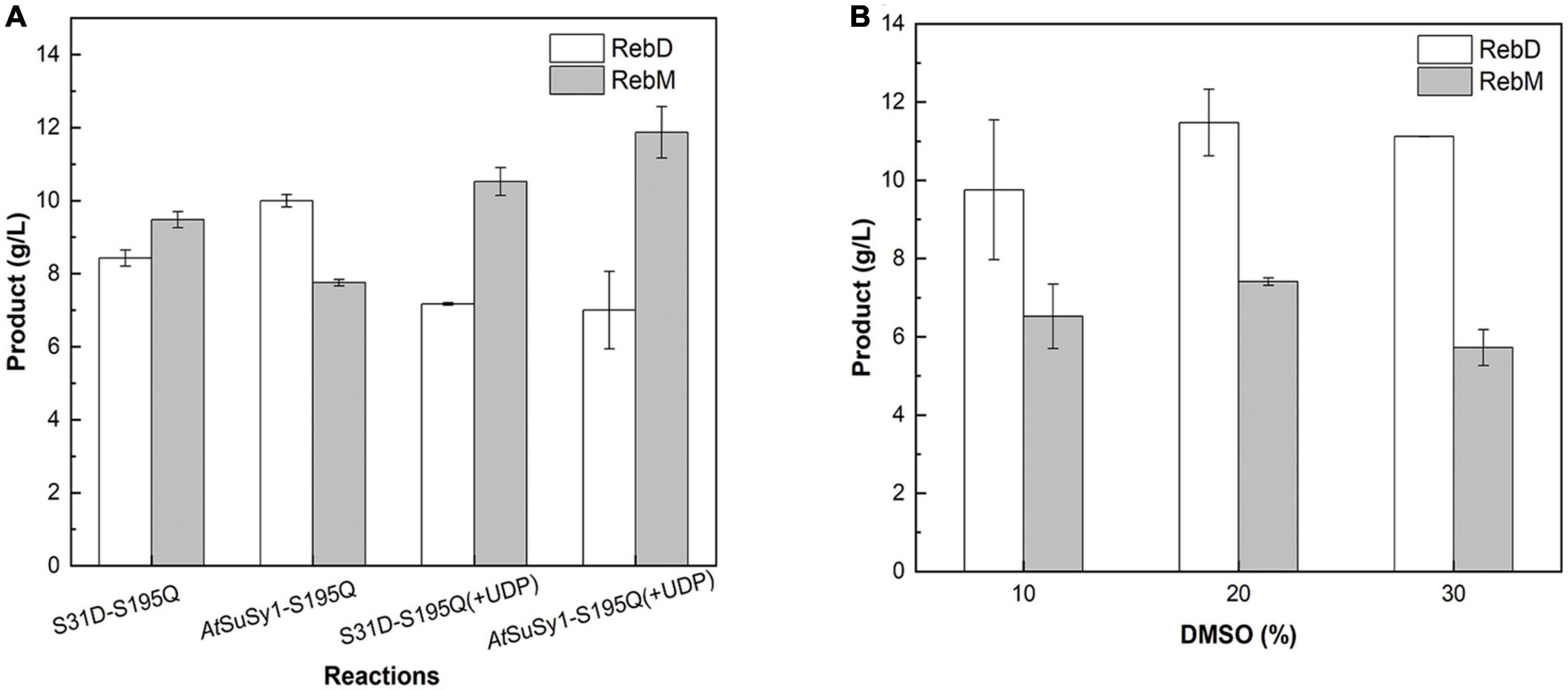
Figure 5. Production of RebM from RebE in the SuSy-UGT reactions. (A) The reactions catalyzed by AtSuSy1-S195Q and S31D-S195Q at 40°C for 24 h, with the crude extract (1.21 U/mL UGT in ∼6 mg/mL of total protein), 20 g/L RebE, and 200 g/L sucrose, with or without 1 mM UDP. (B) The reactions catalyzed by S31D-S195Q at 40°C for 8 h, with the crude extract (6 mg/mL of total protein), 20 g/L RebE, 200 g/L sucrose, 1 mM UDP, and different concentrations of DMSO. Data are plotted as means ± standard deviation of duplicates.
The poor water solubility of RebD (Zhang et al., 2021) may lead to low RebD concentration in the aqueous solutions, which would limit the rate of the second glycosylation reaction that RebD was converted into RebM by UGT76G1_S195Q. Since addition of DMSO would improve the RebD solubility, the influence of DMSO concentrations on the activities of McSuSy_S31D and AtSuSy1 of the crude extracts were investigated (Supplementary Figure 4). In the range from 1 to 30% DMSO, McSuSy_S31D remained higher residual activity than that of AtSuSy1. Therefore, 10, 20, and 30% (v/v) DMSO was added to the S31D-S195Q reactions (Figure 5B). After reaction of 8 h, the total yield of RebD and RebM was the highest in the reaction mixtures containing 20% DMSO, and the RebD concentration kept more than 10 g/L in all the systems. In view that McSuSy maintained more that 80% of maximum activity at 10% DMSO (Figure 1E and Supplementary Figure 4), the subsequent RebM synthesis from 30 g/L RebE was performed at 10% DMSO (Figure 6). Compared with RebM (22.90 g/L) in the 24-h batch production (D), the fed-batch reactions (A, B, and C) obtained more RebM (28.17, 28.95, and 29.62 g/L, respectively), reaching the yields of above 70%. Subsequently, 20 g/L RebE and 200 g/L sucrose were used as the initial substrates, and equivalent to 10 g/L RebE and 10-fold sucrose were fed at 10, 24, and 37 h. Incubation at 40°C continued for another 20 h. The resultant product of interest in the 57-h reaction mixture was 51.06 g/L, with a RebM yield of 76.48%.
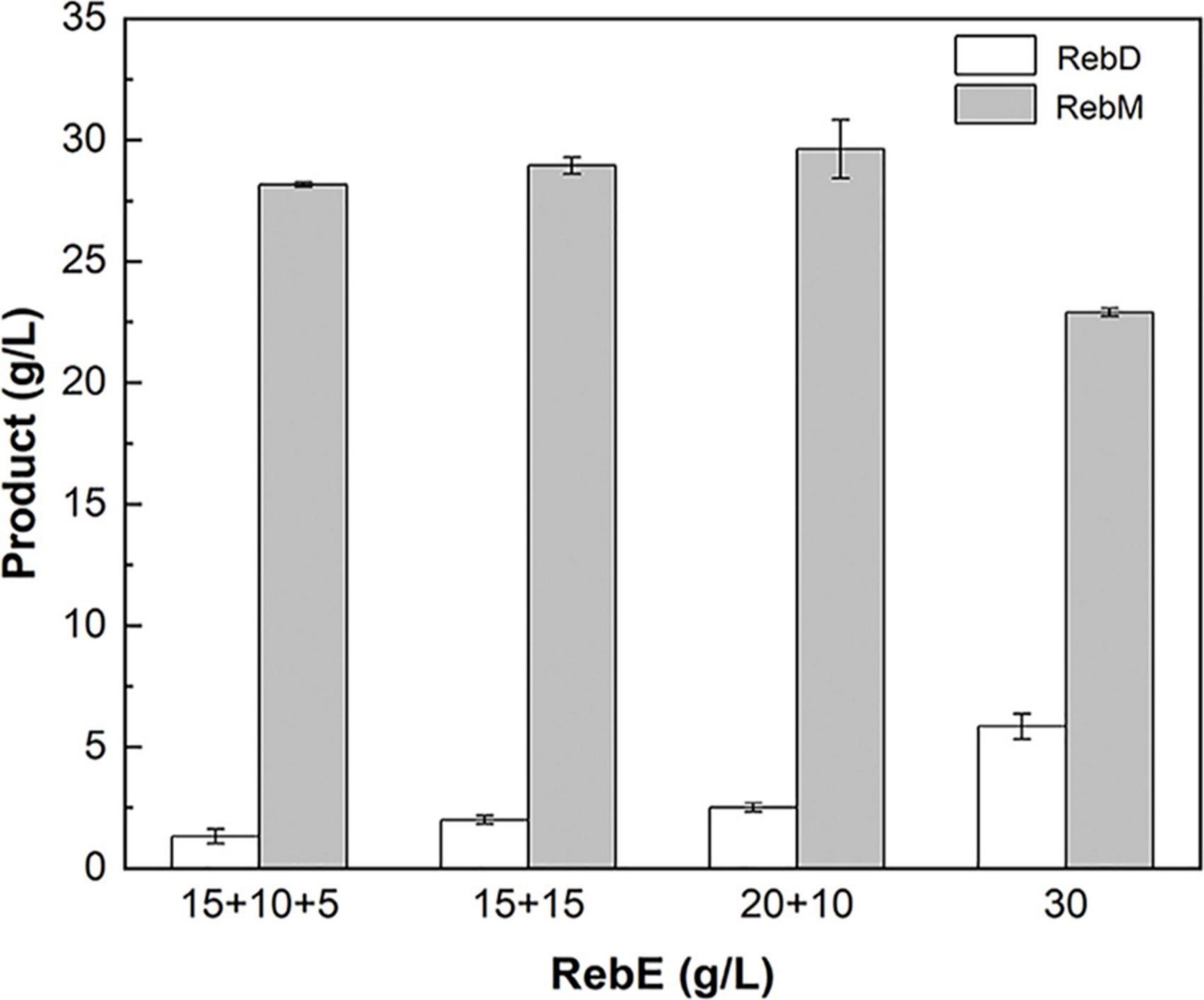
Figure 6. Fed-batch synthesis of RebM catalyzed by S31D-S195Q. The reactions were carried out at 40°C for 24 h, with the crude extract (2.14 U/mL UGT and 3.93 U/mL SuSy in 10 mg/mL of total protein), RebE, sucrose (10 times of RebE), 10% DMSO, and 1 mM UDP. A: the initial concentration of RebE was 15 g/L, and the RebE powder equivalent to 10 and 5 g/L, was added at 6 and 10 h, respectively; B:the initial concentration of RebE was 15 g/L, and the RebE powder equivalent to 15 g/L was added at 10 h; C: the initial concentration of RebE was 20 g/L, and the RebE powder equivalent to 10 g/L was added at 10 h; D: the initial concentration of RebE was 30 g/L. When feeding, sucrose was added along with RebE to keep the constant radio of 10:1. Data are plotted as means ± standard deviation of duplicates.
In the present study, the prokaryotic AnSuSy and MrSuSy were used as templates for sequence collection, and the homology, as well as the active site of the reported SuSys, were also considered in the sequence screening process. Particularly, the sequences that have the conserved residues contributing to UDP-Glc binding, corresponding to Q648, N654, and E683 in AtSuSy1 remained (Diricks et al., 2015; Wu et al., 2015). The phylogenetic tree shows the classification and evolutionary relationship of three selected sequences (McSuSy, CbSuSy1, and CbSuSy2) from the algae and several other characterized SuSys from plants and bacteria (Supplementary Figure 5). They are close to those from plants, falling in the Eukaryotic group, and share the common conserved active site residues in retaining GT-B glycosyltransferases (Supplementary Table 4), which was known from the multiple sequence alignment. What we focused on was McSuSy, which was heterologously expressed in E. coli with higher activity than CbSuSy1 and CbSuSy2. Lower pH values are known to promote the cleavage reaction of SuSys, yielding NDP-glucose and fructose, and with the increasing of the pH, NDP-glucose synthesis is disfavored (Bungaruang et al., 2013; Gutmann and Nidetzky, 2016; Schmölzer et al., 2016). While the McSuSy displayed the highest activity at pH 7.0 in sucrose degradation (Figure 1A), which is different from other sources of SuSys preferring to hydrolyze sucrose at acidic pH. And it is suitable to apply in SuSy-GT cascade reactions where Leloir GT having the optimal neutral or slightly basic pH.
In addition, the plant SuSy is a known phosphoserine-containing enzyme (Komina et al., 2002). One distinctive characteristic feature of SuSys is that phosphorylation of the N-terminus at the major phosphorylation site in plants contributes to the fine-tuning of enzyme activity and may be responsible for changes in membrane binding (Komina et al., 2002; Hardin et al., 2004). In contrast, interestingly, the N-terminal sequence alignment of prokaryotic SuSys shows that a highly conserved motif was found in cyanobacteria SuSys as a putative phosphoacceptor, but for non-cyanobacteria SuSys, there is no definite motif to distinguish (Schmölzer et al., 2016). Previous studies have shown that phosphorylation or introducing the negative charge at the N-terminal phosphorylation site of plant SuSys, such as at S15 of ZmSuSy and S11 of GmSuSy and StSuSy1, has affected their catalytic activities in sucrose cleavage (Komina et al., 2002; Hardin et al., 2004; Sauerzapfe et al., 2008). In the N-terminal sequence alignment of four SuSys involving McSuSy, GmSuSy, StSuSy1, and ZmSuSy, the reported phosphorylation site is conserved (Figure 7) in McSuSy (S31), which is identical to the predicted results obtained from NetPhos 3.1 Server (Supplementary Table 3). S31D mutation of McSuSy showed a nearly 1.2-fold increase in the enzyme activity, which suggest that introduction of the negative charge at S31, like phosphorylation, may affect the N-terminal conformation and the interactions between adjacent region, thus stimulating the catalytic activity of McSuSy (Hardin et al., 2004; Zheng et al., 2011). Low Km values for UDP are beneficial for in vitro recycling of UDP-Glc in SuSy-GT coupled systems due to favored sucrose cleavage, and the product can be synthesized with endogenesis UDP. The Km of McSuSy for UDP (0.14 mM) is comparable to that of most plant SuSys (Table 1). Its S31D mutant reached an even lower Km at 0.092 mM, the lowest in those of SuSys expressed in E. coli, as listed in Table 1. But both Kcat values for UDP were much the same (17.3 s–1 vs. 19 s–1). Although the catalytic efficiency (Kcat/Km) of the S31D mutant was increased by over 67% compared with the wild type, it was still inferior to that of the two Arabidopsis-derived SuSys (Table 1). The affinity for sucrose, indicated by the Km values (∼90 mM), was much worse than that of plants, which implies the demand for high-concentration sucrose in the reactions and that McSuSy would not be inhibited by high concentrations of sucrose as well. However, the catalytic efficiency of McSuSy_S31D was higher than all of the reported prokaryotic SuSys (Table 1).
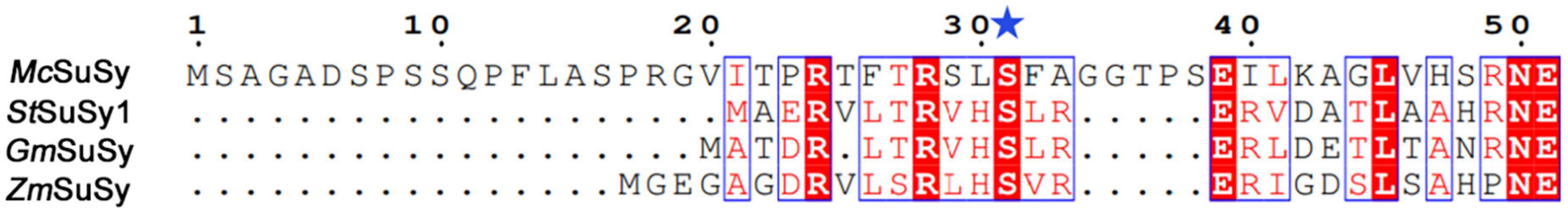
Figure 7. Sequence alignment of the selected SuSys at N-terminus. The site corresponding to S31 in McSuSy, S11 in StSuSy1 and GmSuSy, and S15 in ZmSuSy is marked with a blue star.
RebM, originally identified from S. rebaudiana, is a next-generation of non-calorie sweetener in the approved list of food additivities (Sun et al., 2021). With the growing demands for sustainability, people have aroused great interest in obtaining safe and “natural” products through microbial production. The SuSy-GT cascade reactions have previously been used in enzymatic glycosylation of steviol glycosides to produce RebA, RebE, and RebD (Wang et al., 2016; Chen et al., 2018, 2020; Chen L. L. et al., 2021), and now to produce RebM (Guo et al., 2022; Yu et al., 2022). The strategies of co-immobilizing or fusing the glycosyltransferases have also been applied (Wang et al., 2021, 2023). Guo et al. (2022) used a variant UGT76G1-T284S/M88L/L200A coupled with sucrose synthase AtSuSy1 in a cascade reaction that produced 23.37 g/L RebM from RebD with a yield of 90.5%. In this study, RebM was synthesis from RebE via a two-step continuous glycosylation as previously reported (Yu et al., 2022). With the aid of SuSy, the sugar donor UDPG for GT can be regenerated from sucrose and the trace UDP (usually 1–2 mM) (Mao et al., 2006) in the crude extract that was prepared from the recombinant cells. In case that the UDP/UDP-Glc in the cell lysate was used for UDP-Glc regeneration, RebM yield in S31D-S195Q was higher than that in AtSuSy1-S195Q, suggesting the better UDP affinity of McSuSy_S31D than that of AtSuSy1 (Table 1). When additional 1 mM UDP was supplemented, AtSuSy1 with higher activity (Supplementary Table 5) showed its advantages; the RebM concentration in S31D-S195Q was only increased by 1 g/L, while that in AtSuSy1-S195Q was increased by 4.1 g/L, exceeding the production of RebM by S31D-S195Q. However, the bottleneck limiting the product yield in such reactions was not the enzyme activities but the poor solubility of RebD. As the intermediate product, it formed quickly at high RebE concentrations, and mainly exist as the insoluble precipitate when the reaction speed of further conversion to RebM was limited by the low RebD concentration in the solution. At 10% DMSO, the production of RebM was enhanced (Figure 6), probably due to the improved solubility of RebD. Based on the addition of 10% DMSO, we kept the initial RebE concentration (20 g/L) as used in Figure 6 and increased the number of feeding (three times) as well as the reaction time (total 57 h). Finally, 51.06 g/L RebM was generated from 50 g/L RebE. The results also indicated that both enzymes have good stability under the above conditions. As far as we know, this is the highest concentration of RebM obtained by biotransformation ever reported. The fed-batch synthesis of RebM can be further optimized to achieve a higher yield. RebE, instead of RebA, used as raw material for RebM production, may have an impact on Stevia breeding and cultivation.
Thus, McSuSy is the first characterized SuSy derived from eukaryotic algae, with UDP preference and DMSO tolerance and provides a desirable SuSy candidate to construct the cost-effective SuSy-GT cascade reactions for glycosylation of compounds with low solubility, especially some hydrophobic receptors.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in this article/Supplementary material.
YL and HJ conceived and designed the experiment. KC, LL, and RM performed all the research. YL, RM, and LL wrote the manuscript with input from all authors who reviewed the final manuscript. HP, JD, and YT extracted and analyzed the data. YL and LL revised the manuscript. All authors contributed to the article and approved the submitted version.
This work was financial supported by the National Key R&D Program of China (2021YFC2101500), NSFC (21878155) and the Jiangsu Synergetic Innovation Center for Advanced Bio-manufacture, and PAPD.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1220208/full#supplementary-material
Bailey, T. L., and Elkan, C. (1994). Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2, 28–36.
Baroja-Fernández, E., Muñoz, F. J., Li, J., Bahaji, A., Almagro, G., Montero, M., et al. (2012). sucrose synthase activity in the sus1/sus2/sus3/sus4 Arabidopsis mutant is sufficient to support normal cellulose and starch production. Proc. Natl. Acad. Sci. U.S.A. 109:321. doi: 10.1073/pnas.1117099109
Blom, N., Gammeltoft, S., and Brunak, S. (1999). Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 294, 1351–1362. doi: 10.1006/jmbi.1999.3310
Bungaruang, L., Gutmann, A., and Nidetzky, B. (2013). Leloir glycosyltransferases and natural product glycosylation: Biocatalytic synthesis of the C-glucoside nothofagin, a major antioxidant of Redbush Herbal Tea. Adv. Synth. Catal. 355, 2757–2763. doi: 10.1002/adsc.201300251
Chen, L. L., Cai, R. X., Weng, J. Y., Li, Y., Jia, H. H., Chen, K. Q., et al. (2020). Production of rebaudioside D from stevioside using a UGTSL2 Asn358Phe mutant in a multi-enzyme system. Microb. Biotechnol. 13, 974–983. doi: 10.1111/1751-7915.13539
Chen, L. L., Pan, H. Y., Cai, R. X., Li, Y., Jia, H. H., Chen, K. Q., et al. (2021). Bioconversion of stevioside to rebaudioside E using glycosyltransferase UGTSL2. Appl. Biochem. Biotechnol. 193, 637–649. doi: 10.1007/s12010-020-03439-y
Chen, L. L., Sun, P., Zhou, F., Li, Y., Chen, K. Q., Jia, H. H., et al. (2018). Synthesis of rebaudioside D, using glycosyltransferase UGTSL2 and in situ UDP-glucose regeneration. Food Chem. 259, 286–291. doi: 10.1016/j.foodchem.2018.03.126
Chen, T. Y., Chen, Z. Y., Wang, N., Chu, J. L., Fan, B., Cheng, C., et al. (2021). Highly regioselective and efficient biosynthesis of polydatin by an engineered UGTBL1 – AtSuSy cascade reaction. J. Agric. Food Chem. 69, 8695–8702. doi: 10.1021/acs.jafc.1c02518
Chu, J. L., Yue, J. H., Qin, S., Li, Y. Q., Wu, B., and He, B. F. (2021). Biocatalysis for rare ginsenoside Rh2 production in high level with co-immobilized UDP-glycosyltransferase Bs-Yjic mutant and sucrose synthase Atsusy. Catalysts 11:132. doi: 10.3390/catal11010132
Crooks, G. E., Hon, G., Chandonia, J. M., and Brenner, S. E. (2004). WebLogo: A sequence logo generator. Genome Res. 14, 1188–1190. doi: 10.1101/gr.849004
Curatti, L., Porchia, A. C., Herrera-Estrella, L., and Salerno, G. L. (2000). A prokaryotic sucrose synthase gene (susA) isolated from a filamentous nitrogen-fixing cyanobacterium encodes a protein similar to those of plants. Planta 211, 729–735. doi: 10.1007/s004250000343
De Bruyn, F., Maertens, J., Beauprez, J., Soetaert, W., and De Mey, M. (2015). Biotechnological advances in UDP-sugar based glycosylation of small molecules. Biotechnol. Adv. 33, 288–302. doi: 10.1016/j.biotechadv.2015.02.005
Desmet, T., Soetaert, W., Bojarová, P., Křen, V., Dijkhuizen, L., Eastwick-Field, V., et al. (2012). Enzymatic glycosylation of small molecules: Challenging substrates require tailored catalysts. Chem. Eur. J. 18, 10786–10801. doi: 10.1002/chem.201103069
Diricks, M., De Bruyn, F., Van Daele, P., Walmagh, M., and Desmet, T. (2015). Identification of sucrose synthase in nonphotosynthetic bacteria and characterization of the recombinant enzymes. Appl. Microbiol. Biotechnol. 99, 8465–8474. doi: 10.1007/s00253-015-6548-7
Diricks, M., Gutmann, A., Debacker, S., Dewitte, G., Nidetzky, B., and Desmet, T. (2016). Sequence determinants of nucleotide binding in sucrose synthase: Improving the affinity of a bacterial sucrose synthase for UDP by introducing plant residues. Protein Eng. Des. Sel. 30, 141–148. doi: 10.1093/protein/gzw048
Elling, L. (1995). Effect of metal ions on sucrose synthase from rice grains-a study on enzyme inhibition and enzyme topography. Glycobiology 5, 201–206. doi: 10.1093/glycob/5.2.201
Figueroa, C. M., Asencion Diez, M. D., Kuhn, M. L., McEwen, S., Salerno, G. L., Iglesias, A. A., et al. (2013). The unique nucleotide specificity of the sucrose synthase from Thermosynechococcus elongatus. FEBS Lett. 587, 165–169. doi: 10.1016/j.febslet.2012.11.011
Guo, B. D., Deng, Z. W., Meng, F., Wang, Q. F., Zhang, Y., Yuan, Z. B., et al. (2022). Enhancement of rebaudioside M production by structure-guided engineering of glycosyltransferase UGT76G1. J. Agric. Food Chem. 70, 5088–5094. doi: 10.1021/acs.jafc.2c01209
Gutmann, A., and Nidetzky, B. (2016). Unlocking the potential of leloir glycosyltransferases for applied biocatalysis: Efficient synthesis of uridine 5’-diphosphate-glucose by sucrose synthase. Adv. Synth. Catal. 358, 3600–3609. doi: 10.1002/adsc.201600754
Gutmann, A., Lepak, A., Diricks, M., Desmet, T., and Nidetzky, B. (2017). Glycosyltransferase cascades for natural product glycosylation: Use of plant instead of bacterial sucrose synthases improves the UDP-glucose recycling from sucrose and UDP. Biotechnol. J. 12:1600557. doi: 10.1002/biot.201600557
Hardin, S. C., Winter, H., and Huber, S. C. (2004). Phosphorylation of the amino terminus of maize sucrose synthase in relation to membrane association and enzyme activity. Plant Physiol. 134, 1427–1438. doi: 10.1104/pp.103.036780
Hebditch, M., Carballo-Amador, M. A., Charonis, S., Curtis, R., and Warwicker, J. (2017). Protein-sol: A web tool for predicting protein solubility from sequence. Bioinformatics 33, 3098–3100. doi: 10.1093/bioinformatics/btx345
Huber, S. C., Huber, J. L., Liao, P. C., Gage, D. A., McMichael, R. W., Chourey, P. S., et al. (1996). Phosphorylation of serine-15 of maize leaf sucrose synthase. Occurrence in vivo and possible regulatory significance. Plant Physiol. 112, 793–802. doi: 10.1104/pp.112.2.793
Katoh, K., Misawa, K., Kuma, K., and Miyata, T. (2002). MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066. doi: 10.1093/nar/gkf436
Komina, O., Zhou, Y., Sarath, G., and Chollet, R. (2002). In vivo and in vitro phosphorylation of membrane and soluble forms of soybean nodule sucrose synthase. Plant Physiol. 129, 1664–1673. doi: 10.1104/pp.002360
Krieger, E., Koraimann, G., and Vriend, G. (2002). Increasing the precision of comparative models with YASARA NOVA–a self-parameterizing force field. Proteins Struct. Funct. Genet. 47, 393–402. doi: 10.1002/prot.10104
Kulmer, S. T., Gutmann, A., Lemmerer, M., and Nidetzky, B. (2017). Biocatalytic cascade of polyphosphate kinase and sucrose synthase for synthesis of nucleotide-activated derivatives of glucose. Adv. Synth. Catal. 359, 292–301. doi: 10.1002/adsc.201601078
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Mao, Z. C., Shin, H. D., and Chen, R. R. Z. (2006). Engineering the E. coli UDP-glucose synthesis pathway for oligosaccharide synthesis. Biotechnol. Progr. 22, 369–374. doi: 10.1021/bp0503181
Miller, G. L. (1959). Use of dinitrosalicilic acid reagent for determination of reducing sugars. Anal. Chem. 31, 426–428. doi: 10.1021/ac60147a030
Morris, G. M., Huey, R., Lindstrom, W., Sanner, M. F., Belew, R. K., Goodsell, D. S., et al. (2009). AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 30, 2785–2791. doi: 10.1002/jcc.21256
Nidetzky, B., Gutmann, A., and Zhong, C. (2018). Leloir glycosyltransferases as biocatalysts for chemical production. ACS Catal. 8, 6283–6300. doi: 10.1021/acscatal.8b00710
Pei, J. J., Chen, A. N., Zhao, L. G., Cao, F. L., and Xiao, W. (2019). Synergistic catalysis of glycosyltransferase and sucrose synthase to produce isoquercitrin through glycosylation of quercetin. Chem. Nat. Compd. 55, 453–457. doi: 10.1007/s10600-019-02712-9
Ping, Q., Yang, L. F., Jiang, J. J., Yuan, J. C., Ai, S., Sun, S. Q., et al. (2022). Efficient synthesis of rebaudioside D2 through UGT94D1-catalyzed regio-selective glycosylation. Carbohydr. Res. 522:108687. doi: 10.1016/j.carres.2022.108687
Römer, U., Schrader, H., Günther, N., Nettelstroth, N., Frommer, W. B., and Elling, L. (2004). Expression, purification and characterization of recombinant sucrose synthase 1 from Solanum tuberosum L. for carbohydrate engineering. J. Biotechnol. 107, 135–149. doi: 10.1016/j.jbiotec.2003.10.017
Saitou, N., and Nei, M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Sauerzapfe, B., Engels, L., and Elling, L. (2008). Broadening the biocatalytic properties of recombinant sucrose synthase 1 from potato (Solanum tuberosum L.) by expression in Escherichia coli and Saccharomyces cerevisiae. Enzyme Microb. Technol. 43, 289–296. doi: 10.1016/j.enzmictec.2008.04.001
Schmölzer, K., Gutmann, A., Diricks, M., Desmet, T., and Nidetzky, B. (2016). Sucrose synthase: A unique glycosyltransferase for biocatalytic glycosylation process development. Biotechnol. Adv. 34, 88–111. doi: 10.1016/j.biotechadv.2015.11.003
Schmölzer, K., Lemmerer, M., Gutmann, A., and Nidetzky, B. (2017). Integrated process design for biocatalytic synthesis by a Leloir glycosyltransferase: UDP-glucose production with sucrose synthase. Biotechnol. Bioeng. 114, 924–928. doi: 10.1002/bit.26204
Schwab, W., Fischer, T., and Wüst, M. (2015). Terpene glucoside production: Improved biocatalytic processes using glycosyltransferases. Eng. Life Sci. 15, 376–386. doi: 10.1002/elsc.201400156
Stein, O., and Granot, D. (2019). An overview of sucrose synthases in plants. Front. Plant Sci. 10:95. doi: 10.3389/fpls.2019.00095
Sun, L. C., Xin, F. J., and Alper, H. S. (2021). Bio-synthesis of food additives and colorants-a growing trend in future food. Biotechnol. Adv. 47:107694. doi: 10.1016/j.biotechadv.2020.107694
Takeda, H., Niikura, M., Narumi, A., Aoki, H., Sasaki, T., and Shimada, H. (2017). Phosphorylation of rice sucrose synthase isoforms promotes the activity of sucrose degradation. Plant Biotechnol. 34, 107–113. doi: 10.5511/plantbiotechnology.17.0326a
Tao, Y. H., Xu, J. J., Shao, J. L., He, X. Y., Cai, R. X., Chen, K., et al. (2023). Three glycosyltransferase mutants in a one-pot multi-enzyme system with enhanced efficiency for biosynthesis of quercetin-3,4’-O-diglucoside. J. Agric. Food Chem. 71, 6662–6672. doi: 10.1021/acs.jafc.3c01043
Terasaka, K., Mizutani, Y., Nagatsu, A., and Mizukami, H. (2012). In situ UDP-glucose regeneration unravels diverse functions of plant secondary product glycosyltransferases. FEBS Lett. 586, 4344–4350. doi: 10.1016/j.febslet.2012.10.045
Trott, O., and Olson, A. J. (2010). AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461. doi: 10.1002/jcc.21334
Wang, X. Q., Li, J. Q., Li, L. N., Zhu, L. P., and Huang, F. (2023). Fusion glycosyltransferase design for enhanced conversion of rebaudioside A into rebaudioside M in cascade. Mol. Catal. 547:113317. doi: 10.1016/j.mcat.2023.113317
Wang, Y., Chen, L. L., Li, Y., Li, Y. Y., Yan, M., Chen, K. Q., et al. (2016). Efficient enzymatic production of rebaudioside A from stevioside. Biosci. Biotech. Bioch. 80, 67–73. doi: 10.1080/09168451.2015.1072457
Wang, Z. Y., Liu, W. B., Liu, W., Ma, Y. Y., Li, Y. T., Wang, B. Q., et al. (2021). Co-immobilized recombinant glycosyltransferases efficiently convert rebaudioside A to M in cascade. RSC Adv. 11, 15785–15794. doi: 10.1039/d0ra10574k
Wu, R., Asencion Diez, M. D., Figueroa, C. M., Machtey, M., Iglesias, A. A., Ballicora, M. A., et al. (2015). The crystal structure of Nitrosomonas europaea sucrose synthase reveals critical conformational changes and insights into the sucrose metabolism in prokaryotes. J. Bacteriol. 197, 2734–2746. doi: 10.1128/JB.00110-15
Xiao, J. B., Muzashvili, T. S., and Georgiev, M. I (2014). Advances in the biotechnological glycosylation of valuable flavonoids. Biotechnol. Adv. 32, 1145–1156. doi: 10.1016/j.biotechadv.2014.04.006
Yu, J., Tao, Y. H., Pan, H. Y., Lin, L., Sun, J. Y., Ma, R. Q., et al. (2022). Mutation of stevia glycosyltransferase UGT76G1 for efficient biotransformation of rebaudioside E into rebaudioside M. J. Funct. Foods 92:105033. doi: 10.1016/j.jff.2022
Zhang, L., Gao, Y. N., Liu, X. F., Guo, F., Ma, C. X., Liang, J. H., et al. (2019). Mining of sucrose synthases from Glycyrrhiza uralensis and their application in the construction of an efficient UDP-recycling system. J. Agric. Food Chem. 67, 11694–11702. doi: 10.1021/acs.jafc.9b05178
Zhang, T. T., Myint, K. Z., Xia, Y. M., and Wua, J. (2021). A comparative study on physicochemical and micellar solubilization performance between monoglucosyl rebaudioside A and rebaudioside A. J. Sci. Food Agric. 102, 2651–2659. doi: 10.1002/jfa.11604
Zhao, H., and Caflisch, A. (2013). Discovery of ZAP70 inhibitors by high-throughput docking into a conformation of its kinase domain generated by molecular dynamics. Bioorg. Med. Chem. Lett. 23, 5721–5726. doi: 10.1016/j.bmcl.2013.08.009
Zhao, L. T., Ma, Z. B., Wang, Q., Hu, M. F., Zhang, J. X., Chen, L., et al. (2023). Engineering the thermostability of sucrose synthase by reshaping the subunit interaction contributes to efficient UDP-glucose production. J. Agric. Food Chem. 71, 3832–3841. doi: 10.1021/acs.jafc.2c08642
Keywords: biotransformation, glycosylation, glycosyltransferase, Micractinium conductrix, rebaudioside M, sucrose synthase
Citation: Chen K, Lin L, Ma R, Ding J, Pan H, Tao Y, Li Y and Jia H (2023) Identification of sucrose synthase from Micractinium conductrix to favor biocatalytic glycosylation. Front. Microbiol. 14:1220208. doi: 10.3389/fmicb.2023.1220208
Received: 10 May 2023; Accepted: 11 July 2023;
Published: 15 August 2023.
Edited by:
Mingtao Huang, South China University of Technology, ChinaReviewed by:
Shrameeta Shinde, Miami University, United StatesCopyright © 2023 Chen, Lin, Ma, Ding, Pan, Tao, Li and Jia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan Li, bGl5YW5Abmp0ZWNoLmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.