- 1School of Life Sciences, Guizhou Normal University, Guiyang, Guizhou, China
- 2Key Laboratory for Information System of Mountainous Areas and Protection of Ecological Environment of Guizhou Province, Guizhou Normal University, Guiyang, Guizhou, China
The microorganisms associated with a plant influence its growth and fitness. These microorganisms accumulate on the aerial and root surfaces of plants, as well as within the plants, as endophytes, although how the interaction between microorganisms protects the plant from pathogens is still little understood. In the current study, the impact of assembled the bacterial communities against the pathogenic fungus to promote Artemisia annua L. growths was investigated. We established a model of bacterium–fungus–plant system. Eight bacterial strains and a fungal pathogen Globisporangium ultimum (Glo) were isolated from wild A. annua roots and leaves, respectively. We assembled the six-bacteria community (C6: Rhizobium pusense, Paracoccus sp., Flavobacterium sp., Brevundimonas sp., Stenotrophomonas sp., and Bacillus sp.) with inhibition, and eight-bacteria community (C8) composing of C6 plus another two bacteria (Brevibacillus nitrificans and Cupriavidus sp.) without inhibition against Glo in individually dual culture assays. Inoculation of seedlings with C8 significantly reduced impact of Glo. The growth and disease suppression of A. annua seedlings inoculated with C8 + Glo were significantly better than those of seedlings inoculated with only Glo. C8 had more inhibitory effects on Glo, and also enhanced the contents of four metabolites in seedling roots compared to Glo treatment only. Additionally, the inhibitory effects of root extracts from A. annua seedlings showed that Glo was most sensitive, the degree of eight bacteria sensitivity were various with different concentrations. Our findings suggested that the non-inhibitory bacteria played a vital role in the bacterial community composition and that some bacterial taxa were associated with disease suppression. The construction of a defined assembled bacterial community could be used as a biological fungicide, promoting biological disease control of plants.
Introduction
The microorganisms living in close association with plants influence plant growth and fitness (Castrillo et al., 2017; Durán et al., 2018; Herrera et al., 2018; Finkel et al., 2020). In response to the plant and environmentally derived signals, these microorganisms accumulate on or within the plants (Fitzpatrick et al., 2018; Finkel et al., 2019; Thiergart et al., 2020). Antagonistic interactions between beneficial and pathogenic microorganisms are known to play a critical role in shaping the composition of the plant microbiota and in protecting plants from pathogens (Müller et al., 2016; Durán et al., 2018; Chen et al., 2020; Finkel et al., 2020). However, how the mechanisms involved are not fully understood.
A controllable microorganism-plant system can be established on aseptically grown plants which are then inoculated with a defined microbial community (McNulty et al., 2011; Bodenhausen et al., 2014); some research on such model systems has been reported (Berendsen et al., 2012; Mueller and Sachs, 2015; Wei et al., 2015). For example, leaves of the model plant Arabidopsis thaliana were inoculated with a controllable and defined bacterial community to characterize how some plant genes shaped the microbiota of the phyllosphere (Finkel et al., 2020). It has been reported that such simplified microbial communities colonizing the plants can protect the plant against invading pathogens. For example, a simplified and controllable synthetic community, made up of eight species of bacteria, could control maize seedling blight caused by Fusarium verticillioides by inhibiting fungal colonization and growth (Niu et al., 2017). Some biocontrol bacteria can protect plants from fungal disease: fluorescent pseudomonads can promote plant growth and produce antifungal chemicals to inhibit pathogenic fungi, whereas the rhizobacterium Variovorax boronicumulans produces the auxin plant growth regulator indole acetic acid to promote plant growth (Dieter and Geneviève, 2005). However, it is unclear whether different invading pathogenic fungi are controlled by different microbiota of plants.
Beneficial bacterial communities associated with plants can control invading pathogen fungi either directly or indirectly (Berendsen et al., 2012; Bai et al., 2015; Lebeis et al., 2015; Wagner et al., 2016; Thiergart et al., 2019; Shalev et al., 2022). Reports about how altering the composition of the bacterial community can serve as a barrier to protect the host plant from invading pathogen fungi, in which the invading pathogen fungi may not necessarily be killed, have been presented (Xue et al., 2015; Vale et al., 2016; Ventorino et al., 2016). For example, colonization by a simplified four-species bacterial community, composed of three high-abundance and one low-abundance bacterial species, not only protected Astragalus mongholicus from root rot disease, but also achieved plant-induced systemic resistance (Li et al., 2021). However, for most plants, our understanding of the mechanisms through which the bacterial microbiota protect host plants from pathogen fungi is limited because of the high complexity of bacteria microbiota–pathogen–plant systems, which include the interactions between the bacterial community and the pathogen, between the pathogen and the plant, and between bacterial community members and the plant.
Artemisia annua L., named Qinghao in Chinese, is an annual herbaceous member of the Asteraceae family and is also one of the most important traditional Chinese herbs (Committee of Chinese Pharmacopoeia, 2020), being the main source material of anti-malarial medicines based on artemisinin. A. annua is widely distributed all over the world and mainly grown in the southwest of China. In addition, A. annua plants can adapt to various conditions (Huang et al., 2010; Liu et al., 2013). A. annua is a well-known medicinal herb that has low toxicity and a very effective anti-malarial activity due to the presence of the sesquiterpene lactone artemisinin, the first-line anti-malarial drug recommended by the World Health Organization. There are many medicinal activities of A. annua (e.g., anticancer) associated with plant metabolites (including phenolics) (Ferreira et al., 2010; Huang et al., 2010; Lévesque and Peter, 2012). Recent studies of A. annua mainly focus on the artemisinin biosynthesis mechanism, with the goal of increasing the yield of artemisinin. A linkage has been reported between rhizosphere microbes and medicinal metabolites in A. annua (Luo et al., 2019a). Bacterial and fungal communities were markedly influenced by total organic carbon, available phosphorus, total nitrogen, available potassium, and water-soluble nitrogen (Luo et al., 2019b; Shi et al., 2022). Few studies of microbial influence on A. annua growth (e.g., phenotype, growth rate, and physiology) have been reported. Plant growth-promoting bacteria, (e.g., Sphingobium and Sphingomonas), and the fungal amplicon sequencing variants (ASVs) (e.g., of saprotrophs) were significantly enriched in the A. annua rhizosphere (Shi et al., 2022). Furthermore, A. annua could assemble the specific root-associated microbial communities with an increased abundance of plant growth-promoting microorganisms and build inter-kingdom co-occurrence networks in the rhizosphere (Shi et al., 2022). It has been reported that endophytic bacteria (43 strains), fungi (12 strains), and actinomycetes (43 strains) were isolated from A. annua roots (Tian et al., 2008). However, how the interaction between microbiota in the few studies on microbial influence on A. annua growth (e.g., phenotype, physiology, and secondary metabolism) have been reported. How the natural A. annua microbial assemblage can inhibit invading pathogens and promote the healthy growth of A. annua plants is still poorly understood.
In this study, we examined the inhibitory effect of a defined assembled bacterial community on A. annua against the fungal pathogen Globisporangium ultimum (OR416213), the cause of A. annua root rot. We studied the assembly of the assembled bacterial microbiota community of eight bacterial species, which was isolated from A. annua roots, and the response of colonized and non-colonized A. annua seedlings to G. ultimum infection was investigated. The questions which need to be resolved are: (1) Does the assembled bacterial microbiota have a direct inhibitory effect on the pathogen G. ultimum? (2) How does the bacterial microbiota affect G. ultimum in the disease-suppressive plant–bacterium interaction? (3) How does the A. annua root extract affect the growth of the fungal and bacterial isolates?
To answer these questions, we first isolated a major plant pathogen, G. ultimum from A. annua, and a number of bacterial strains associated with A. annua, and we analyzed the effects of the bacterial isolates against the plant pathogen based on screening and dual culture assays. Afterward, we constructed a six-species assembled bacterial microbiota (C6) in which each strain had an inhibitory effect on the pathogen G. ultimum. In addition, we also constructed another, eight-species assembled bacterial microbiota (C8) consisting of the six species described in C6 above plus two bacterial species having no inhibitory effects on the fungal pathogen, G. ultimum. We then compared the responses of A. annua plants to G. ultimum following inoculation with C8, C6 or no bacteria in vivo in a greenhouse pot experiment. Ultimately, the C8 assembled bacterial community was selected to examine the plant–bacteria–fungus interaction in promoting healthy growth of A. annua in the presence of G. ultimum.
This study provided evidence that colonization by the assembled bacterial microbiota can inhibit infection by the pathogen G. ultimum, to promote healthy growth of A. annua; more importantly, it provided an insight into the potential use of assembled bacterial microbiota as biological fungicide, promoting biological disease control of plants.
Materials and methods
Collection of A. annua plants
Healthy A. annua plants were collected from the wild (26°35′31.0″N, 106°43′09.6″E) by uprooting them randomly on 10 September 2021 in Guiyang, Guizhou Province, Southwest China. The samples were placed separately into sterile plastic bags, and transported to the laboratory at Guizhou Normal University, where they were used for isolation of pathogenic fungi, associated bacteria, and for plantlet regeneration.
Purification and infection of pathogenic fungi
Pathogenic fungi were isolated from mature leaves of A. annua plants according to a slight modification of the method of Melo et al. (2014). The leaf samples were rinsed in running water for 30 min to remove soil particles, etc., then sterilized by immersion in 75% (v/v) alcohol for 30 s, followed by transfer to 0.1% (w/v) HgCl2 for 6 min under shake conditions to ensure sufficient contact with the sterilant, then rinsed five times in sterile distilled water for 10 s each time. After the water on the leaf surface was dried using sterile filter paper, the sterilized leaves were cut into 1.5–2.0 cm pieces, and placed on potato dextrose agar (PDA), three leaf pieces per Petri dish. The PDA dishes were incubated at 28°C for 7 days in darkness. When the hyphal tips of the fungus protruded from the inner leaf segments on PDA, they were isolated and purified, then transferred to slants and stored at 4°C for use in subsequent experiments.
To screen for the most effective virulent fungus, infection experiments were performed by adding 1 ml fungal spore suspension liquid (106 CFU/ml) of each of the fungal isolates obtained from the wild A. annua plants into sterile soil containing sterile A. annua plantlets; after culture for 14 days, the pathogenicity rate of each fungus and the corresponding death rate of plantlets were recorded. The most consistently virulent strain was selected for use as the experimental pathogenic fungus.
Isolation of endophytic bacteria
Endophytic bacteria were isolated from the roots of A. annua seedlings, which were regenerated from the wild-harvested A. annua plants collected in Guiyang and grown in the humus soil collected from Changbai Mountains, Jilin Province, China (41°35′N, 127°40′E). After separating from the seedling aerial parts, the roots were shaken vigorously to remove loose soil, then rinsed with sterile water until the adhering soil was completely removed, and sterilized as described earlier for leaves. The sterilized roots were cut into pieces, and then extracted in 0.5 mmol/L phosphate-buffered saline (PBS) by ultrasonic vibration for 10 min. The root extract in PBS was diluted into 10–5, 10–6, and 10–7 concentrations. A 0.1 ml aliquot of each concentration was added and spread onto nutrient agar (NA, containing 5.0 g peptone, 18 g agar, 5.0 g NaCl, 1,000 ml distilled water, pH 7.0–7.2), with five replicate plates for each concentration. All NA plates were incubated at 37°C for 3 days; endogenous bacteria were purified and stored at −18°C prior to the assembled community construction stage.
Dual culture assays and assembly of bacterial communities
Following the method of Gkarmiri et al. (2015), with minor modifications, dual culture assays of the bacterium–fungus interactions were established in 9-cm diameter Petri dishes containing PDA medium. A 7-mm diameter plug cut from the edge of an actively growing colony of each test fungus was inoculated in the middle of each Petri dish. Fresh cells of endophytic bacteria were streaked in 3-cm long parallel lines on either side of the fungal plug; the concentration of each bacterial suspension was 106 CFU/ml. Control plates with the fungus only were also conducted. After all treatments were incubated at 28°C in darkness for 7 days, fungal hyphae challenged with each endophytic bacterial isolate and from control plates were photographed, and the sizes of the inhibition zones were measured.
Based on the fungal inhibitory effects, a six-species assembled bacterial community (C6) was constructed, to consider the synergistic effect among the bacteria. Two bacterial strains without significant inhibitory effect against the pathogenic fungus was added into C6 to construct an eight-species assembled bacterial community (C8), which was composed of the following eight bacteria strains isolated from A. annua roots: Brevundimonas sp. Yang 2023, G16 Flavobacterium sp., 0410ARD7G4 Paracoccus sp., CA34 Rhizobium pusense, YFCC6454 Stenotrophomonas sp., DE024 Bacillus sp., S186B Brevibacillus nitrificans, and NBRC102508 Cupriavidus sp. The same volume and concentration (106 CFU/ml) of each isolate was mixed together (“C6” and “C8” treatments), and “Glo” represented the pathogenic fungus; other treatments included “C8 + Glo” and “C6 + Glo.” We compared the inhibitory effects of the two bacterial communities (C6 and C8) against the pathogenic fungus by soil pot experiments, and selected the bacterial assembled community with the greater antifungal inhibitory effect for the subsequent disease-control pot experiments.
DNA extraction, amplification, and sequencing
After the fungal and bacterial isolates were purified and screened for pathogenicity and antifungal effects, respectively, we submitted each to Sangon Biotechnology Co., Ltd. (Shanghai, China) for sequencing. Isolates were transported in an expanded polystyrene foam box containing dry ice to the laboratory of Sangon Biotechnology. The DNA was extracted from bacteria using Ezup Column Bacteria Genomic DNA Purification Kit (SK8255, Sangon Biotechnology, Shanghai, China), and the DNA was extracted from fungus with Ezup Column Fungi Genomic DNA Purification Kit (SK8259, Sangon Biotechnology, Shanghai, China), according to the manufacturer’s protocol. The barcoded primers 7F (5′-CAGAGTTTGATCCTGGCT-3′) and 1540R (5′-AGGAGGTGATCCAGCCGCA-3′) were used to amplify the V3–V4 region 16S rDNA of bacteria; the length of the PCR product was 1,500 bp; the barcoded primers ITS1 (5′-TCCGTAGTGACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) were used to amplify the internal transcribed spacer (ITS) V1–V2 region of fungal rDNA; the length of the PCR product was 600 bp. The PCR amplification procedure was as follows: pre-denaturation at 95°C for 4 min, followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 57°C for 30 s, extension at 72°C for 90 s, repair and extension at 72°C for 10 min, and termination at 4°C. The purified PCR products were sequenced on the Illumina MiSeq platform. The sequence alignment of bacterial 16S rDNA was conducted in the GenBank database,1 and the sequence alignment of fungal ITS was conducted by BLAST of the NCBI website (see text footnote 1), and submit the sequence data to the Genbank of NCBI,2 at last the accession numbers were got. Phylogenetic trees were constructed by the adjacency method using MEGA11 software, and the bootstrap replication number was set to 1,000.
Soil preparation and physicochemical properties
Control of the pathogenic fungus G. ultimum by the bacterial assembled community was tested in vivo using humus soil, which was collected from Changbai Mountains. The organic matter, available nitrogen, available phosphorus, available potassium concentrations, and pH of the soil were 81.75 g/kg, 115.47 mg/kg, 69.92 mg/kg, 86.51 mg/kg, and 6.83, respectively. The soil sampling field had no history of A. annua growing there before the soil was collected.
A tissue-culture glass container (height 120 mm and diameter 80 mm) was filled with 100 g soil, and sterilized in an autoclave at 105°C for 1 h. After autoclaving, the soil medium was allowed to cool, a 0.1 g sub-sample of the soil was diluted with sterilized deionized water into different concentration (10–2, 10–3, 10–4, 10–5, 10–6, and 10–7), and then 10–5, 10–6, and 10–7 soil solutions were spread on NA (for culturing bacteria) and PDA (for fungi) plates, and incubated at 28°C for 7 days; if no microbial growth occurred, it meant that the soil was sterilized completely and could be used for pot experiment of disease control. The soil that was not sterilized (natural soil) was used for the disease-control treatment. Before subsequent experiments, all soil medium was supplemented with the same volume of sterile deionized water.
Preparation of fungal and bacterial suspension liquid
After a fungal colony had grown robustly on a PDA plate, the fungal hyphae was transferred into a 150-ml flask containing 50 ml sterile deionized water, and then incubated at 28°C with constant agitation at 200 rpm for 6 h, a method which was conducive to the spores falling from the mycelium and being suspended in the water, forming the fungal spore suspension, which was diluted with sterile deionized water to achieve a concentration of 106 CFU/ml; this was stored at 4°C for use in the disease-control experiment.
After activation was performed at 25°C in darkness, bacterial colonies on NA plates were each transferred individually with an inoculation loop into a 150-ml flask with 50 ml NA broth, and then incubated at 37°C with constant agitation at 200 rpm for 6 h; eventually, the bacterial NA broth was diluted using sterile deionized water to achieve the concentration of 106 CFU/ml and stored at 4°C for use in the disease-control experiment.
Soil pot experiment of the effect of a bacterial assembled community against fungal disease
Regenerated tissue of A. annua, which was cultured according to the method of Wang et al. (2022), was transplanted into the sterilized soil in containers to get many sterile seedlings, then, the sterile seedlings were transplanted to containers with the sterile humus soil, one seedling per container, and the plants were allowed to grow for 3 days. Subsequently, each container with one sterile seedling was inoculated with 10 ml Glo, C8, or Glo + C8 solutions (106 CFU/ml), with control seedlings being treated with 10 ml of sterile deionized water; every treatment was replicated 15 times, with all the plants being grown at 24°C (12-h light/12-h darkness photoperiod). Fungal disease incidence and plant mortality were recorded every 7 days. The antagonistic capacity of C8 toward the pathogenic fungus was assessed in terms of plant growth, concentrations of chlorophyll a and b, malondialdehyde (MDA), and proline, and activities of the antioxidant enzymes superoxide dismutase (SOD) and peroxidase (POD). Moreover, the concentrations of metabolites (artemisinin, scopoletin, chrysosplenol-D, and chrysosplenetin) of A. annua roots were determined after the seedlings were cultured for 7 days after inoculation with Glo.
Inhibitory effect of A. annua extract on microbial isolates
To examine whether A. annua affects the growth of the fungal or bacterial isolates used in this study, the inhibitory experiment was conducted. A sample (400 g fresh weight) of A. annua roots was cut into pieces which were put into a 1,000-ml flask, to which were added 400 ml methanol; the flask was then sealed and placed on a shaker at 25°C at 200 rpm for 12 h. After ultrasonic extraction for 30 min, the extract was filtered into a round-bottomed flask, and evaporated using a rotary vacuum evaporator and stored in a brown bottle at 4°C.
Artemisia annua extract, which was adjusted to a density of 1 mg/ml, was sterile filtered by passage through a bacterial filter membrane into PDA at 50∼60°C into plates and into NA broth into rigid test-tubes (diameter 20 mm, height 200 mm), respectively, to final concentrations of A. annua extract in the medium of 0 (control, CK), 3, 6, 12, and 24 mg/ml. After activation was performed, a 7-mm diameter plug cut from the edge of an actively growing colony of the pathogenic fungus G. ultimum was inoculated onto a PDA plate containing A. annua extract, incubated at 28°C, and the diameter of the fungus colony was recorded after culture for 24 and 48 h. To determine the impact on bacterial growth, 1 ml bacterial suspension (106 CFU/ml) was inoculated into NA broth containing A. annua extract, incubated at 37°C with constant agitation at 200 rpm for 24 h, and then the optical density (OD) at 600 nm was determined using spectrophotometry, repeated three times for each concentration.
Statistical analysis
Excel 2016 was used to calculate the mean values of each treatment in dual culture assays, soil pot incubation experiments and inhibitory experiments of the A. annua extract. SPSS 23.0 software (SPSS Inc., Chicago, IL, USA) was used to carry out statistical analysis on experimental data, by two-way ANOVA, with Duncan’s multiple range test being used to determine significant differences among treatments at P ≤ 0.5.
Results
Isolation and pathogenicity of fungi isolated from A. annua
Five different endogenous fungal strains were isolated from A. annua leaves (Figure 1A). Through sequence alignment of fungal ITS sequences, they were clustered into three different genera and the accession numbers inquired in Genbank at NCBI website: Th1 Cerrena unicolor (OR416211) (M1), QC11 Fusarium tricinctum (OR416212) (M2), and CBS398.51 G. ultimum (OR416213) (M3) (Figure 1B). According to the inoculation experiment results, G. ultimum caused the highest disease incidence (100%) and plant death rate (100%) (Figure 1C). After plants inoculated with G. ultimum were cultured for 14 days, A. annua plants displayed typical symptoms: all seedings became chlorotic and died; in addition, the surfaces of the soil in the glass container ware covered with flocculent white hyphae (Figure 1D). Therefore, G. ultimum was used as the pathogen in the following experiments.
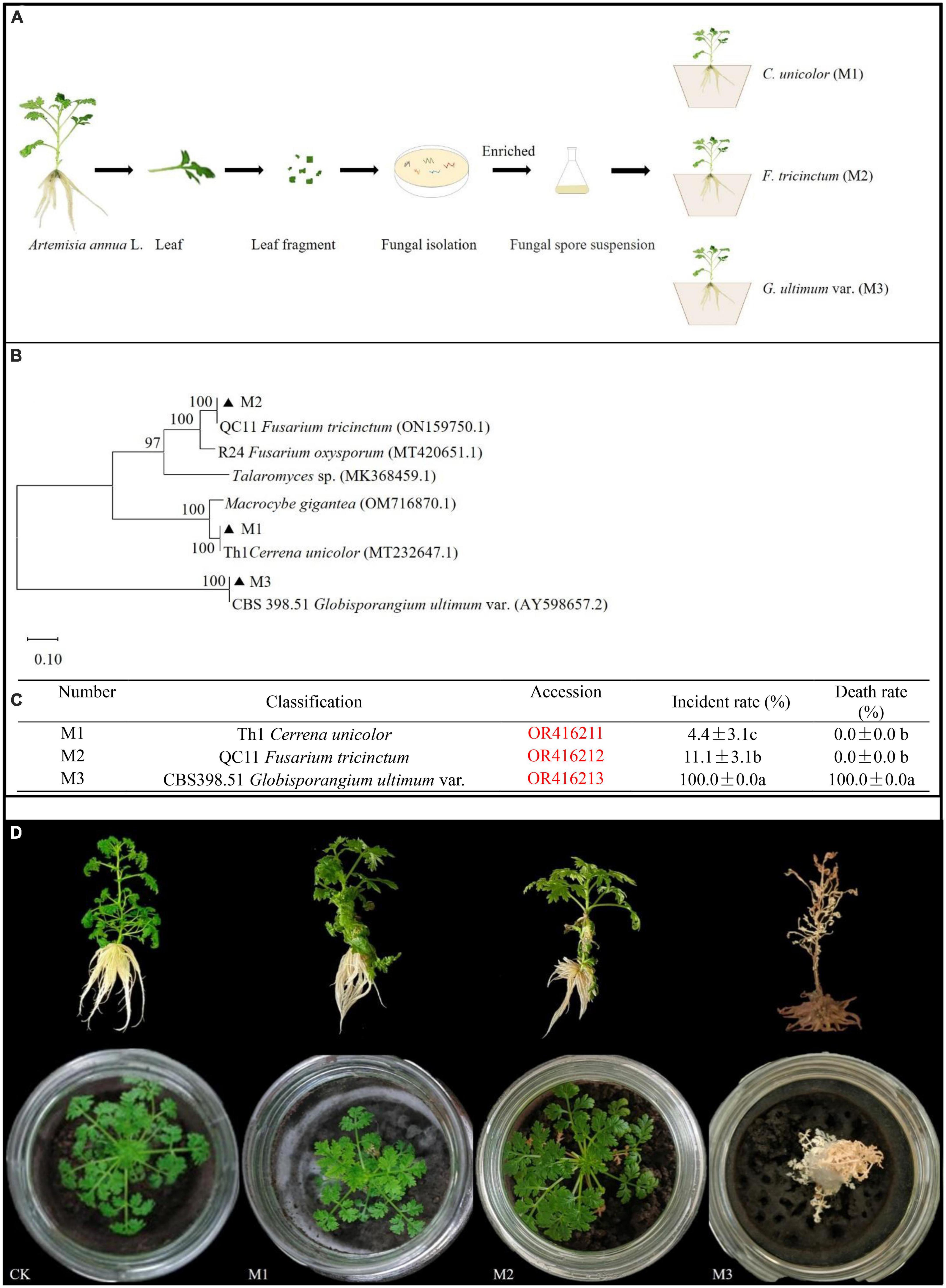
Figure 1. Phylum of fungal isolates from A. annua leaves and pathogenic capacity of endogenously fungi pathogens. (A) The separation process of pathogenic fungi. (B) Phylum of endogenous fungi isolates. (C) Plant disease incident rate and death rate in A. annua with fungi pathogenic infection, different lower-case letters indicate significant differences among fungi pathogen (p < 0.05), same lower-case letters mean no differences. (D) The symptoms of plants.
Screening of antifungal bacteria
To screen as many bacterial isolates as possible, more sterile roots were cut into pieces in PBS buffer solution, and spread on NA plates (Figure 2A), and at last 60 endogenous bacterial strains (Supplementary Table 1) were isolated from A. annua roots, named N1–N60, After the DNA of each strain was sequenced and the 16S rDNA sequences were aligned, the bacterial strains were clustered into eighteen different genera (Supplementary Figure 1) and the accession numbers inquired in Genbank at NCBI website. Because the similarity of N16 and CHNTR43 Brevundimonas sp. is 96.28%, N16 is named as Brevundimonas sp. Yang 2023 (OR415840). Dual culture assay of every bacterium–fungus interaction was conducted on PDA plate. Six bacterial taxa, which could each inhibit the growth of the fungus G. ultimum (Figure 2B), were selected to be candidates for inclusion in construction of the community of antifungal bacteria (“assembled bacterial community”). The order of inhibitory effect toward the fungus was as follows: CA34 R. pusense (OR415843) > 0410ARD7G 4 Paracoccus sp. (OR415841) > G16 Flavobacterium sp. (OR415847) > Brevundimonas sp. Yang 2023 (OR415840) > YFCC6454 Stenotrophomonas sp. (OR415842) > DE024 Bacillus sp. (OR415844) (Table 1). Although there was no obvious inhibitory zone around S186B B. nitrificans (OR415845) and NBRC102508 Cupriavidus sp. (OR415846), the white hyphae of the fungus reduced obviously (Figure 2B), and it was believed to have an inhibitory effect on the fungus.
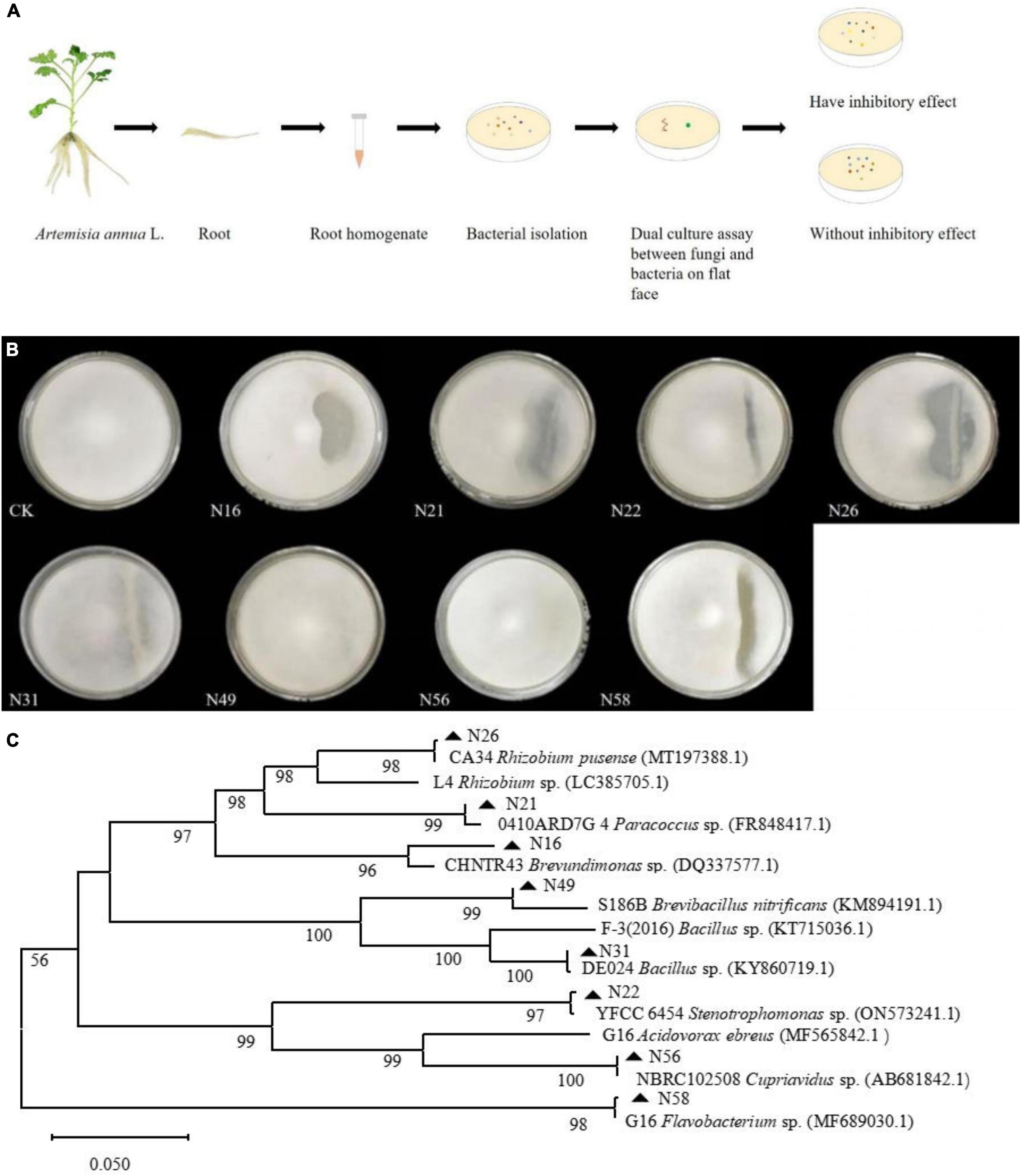
Figure 2. Dual culture assay of every bacterial-fungal interaction and phylum of endogenous bacteria isolates. (A) Isolation of endogenous bacteria and dual culture assay. (B) Inhibitory effects of eight candidates composing bacteria against on G. ultimum var. (C) Phylum of endogenous bacteria isolates. CK, Brevundimonas sp. Yang 2023, G16 Flavobacterium sp., 0410ARD7G4 Paracoccus sp., CA34 Rhizobium pusense, YFCC6454 Stenotrophomonas sp., DE024 Bacillus sp., S186B Brevibacillus nitrificans, and NBRC102508 Cupriavidus sp., respectively.
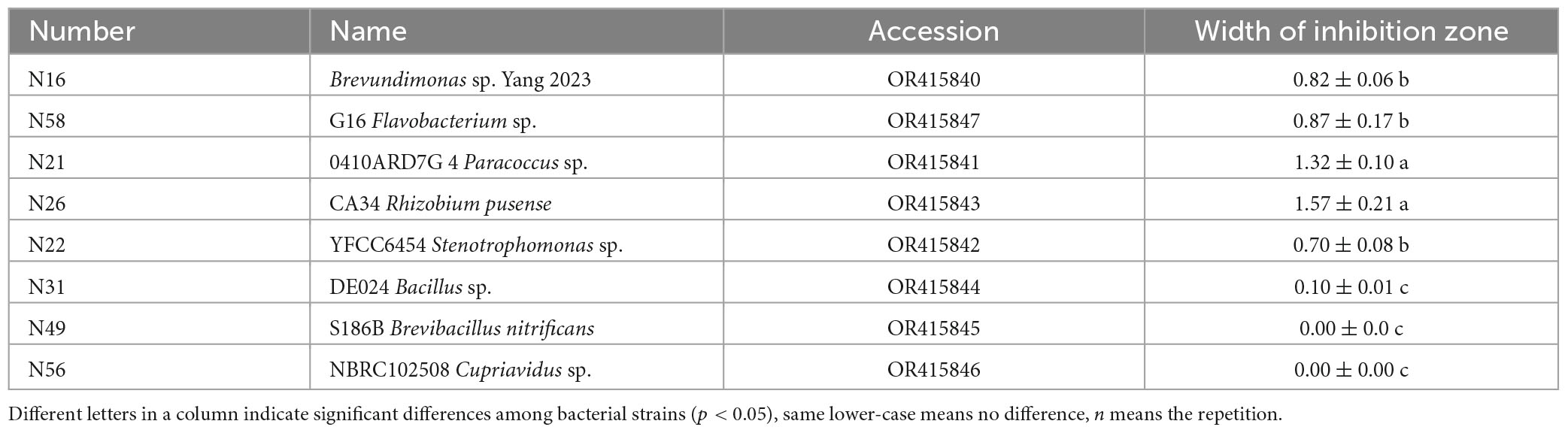
Table 1. Inhibition of differently disease-resistant bacteria on Globisporangium ultimum var. (mean ± standard deviation, n = 3).
Assembly of the community of antifungal bacteria
Because the hyphae of the fungus G. ultimum grew very quickly, they reached the 3-cm long parallel lines of antifungal bacteria after only 24 h culture, at which point, some of the bacterial colonies did not grow out of the medium surface, which would affect the measurement of the inhibition zone; after 48 h culture, the hyphae of the fungus G. ultimum covered the plate, so that the inhibition zones caused by antifungal bacteria were obvious, and could be measured. The six bacterial isolates with inhibitory effects were mixed in equal volumes of bacterial suspensions (106 CFU/ml) to generate an assembled bacterial community (C6). To investigate whether two bacteria without antifungal activity could contribute to preventing fungal pathogens from infecting A. annua plants, we also selected two bacterial isolates, which did not cause visible inhibitory zones in the dual assay, and six bacterial isolates with antifungal activity to construct another assembled bacterial community (C8) (Figure 2B and Supplementary Table 3). We then mixed all the eight species in equal volumes of bacterial suspensions (106 CFU/ml) and constructed assembled bacterial community (C8). A typical phylum picture of the eight bacterial isolates is displayed in Figure 2C.
Screening of assembled bacterial communities against a pathogenic fungus
After A. annua plants were treated with G. ultimum and either or none of the two assembled communities (C6, C8, or CK) in an in vivo pot experiment for 14 days, those plants inoculated with C6 or neither community (CK) all died, although the six bacterial species in C6 each had an inhibitory effect on the pathogenic fungus in vitro. In contrast, the plants inoculated with the eight bacterial species of C8 grew vigorously in the presence of G. ultimum. So, the C8 community was screened for the subsequent pot experiment (Supplementary Figure 2).
Effects of the assembled bacterial community against fungal disease
After sterile seedlings were inoculated with G. ultimum for 7 or 14 days, the disease incidence rate were 100 and 100% (Figures 3A, C), and the average death rates were 24.5 and 100%, respectively (Figures 3B, D). In addition, after culture for 7 days, the soil surfaces were covered with a large amount of white hypha, with some hyphae climbing up the aerial parts of the seedlings (Supplementary Figure 2), until the seedling was all entwined by the hyphae and could not stand; at this point, the leaves of the seedling dehydrated and became yellow, and seedling growth was obviously inhibited. After culture for 14 days, the roots rotted and the aerial parts became completely dry (Figure 3E). However, after sterile seedlings were inoculated with the assembled community C8 and G. ultimum simultaneously for 7 and 14 days, the average disease incidence rates decreased by 91 and 73.3%, respectively (Figures 3A, C), and the average death rates declined by 24.5 and 89.9%, respectively (Figures 3B, D). At the same time, the plants inoculated with the eight-species assembled bacterial community (C8) grew similarly to the control (CK) seedlings (Figure 3E).
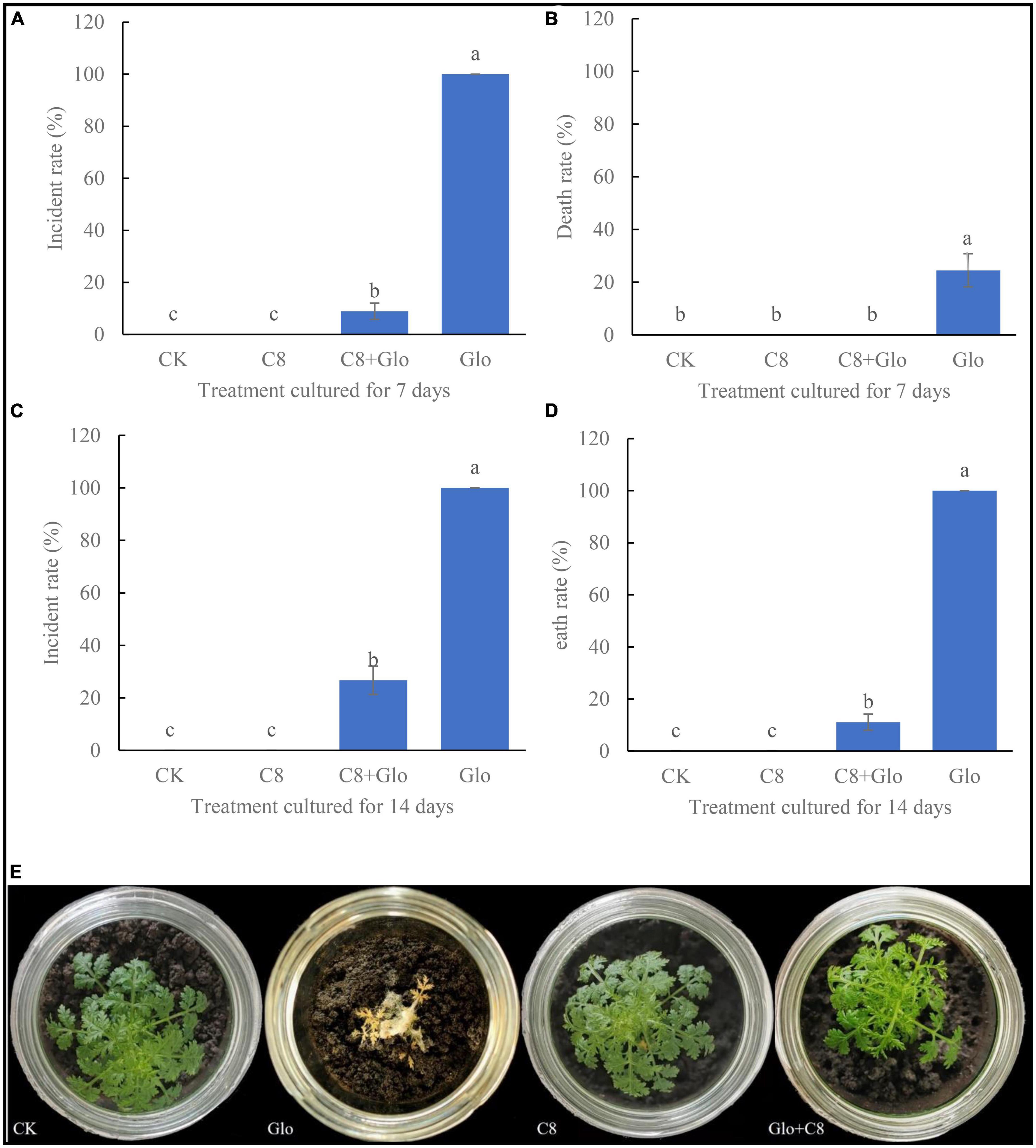
Figure 3. (A) Incidental rate of seedling treated with sterile water CK, G. ultimum var. Glo, assembled community C8 and C8 + Glo after cultured for 7 days. (B) Death rate cultured for 7 days. (C) Incidental rate cultured for 14 days. (D) Death rate cultured for 14 days. Different letters in a column indicate significant differences among bacterial strains (p < 0.05), same lower-case means no difference. (E) The pictures of seedling cultured for 14 days in the treatments.
The growth of seedlings colonized by an assembled bacterial community in response to fungal infection
After A. annua plants were inoculated with G. ultimum for 14 days, the mean dry weight, fresh weight, plant height, and root length significantly decreased (relative to CK) by 77.8, 65.4, 60.6, and 28.8%, respectively (p < 0.05) (Figures 4A-C, E). However, when the plants were inoculated with C8 + Glo, the average number of the plant roots increased significantly (relative to CK), whereas the other indexes of plant growth were not significantly different from those in the CK treatment. When the plants were inoculated with C8 only, they grew better than CK seedlings, although the only significant difference was with respect to the mean number of roots (Figure 4D). These results showed that the assembled bacterial community C8 could also improve the growth of non-diseased A. annua plants.
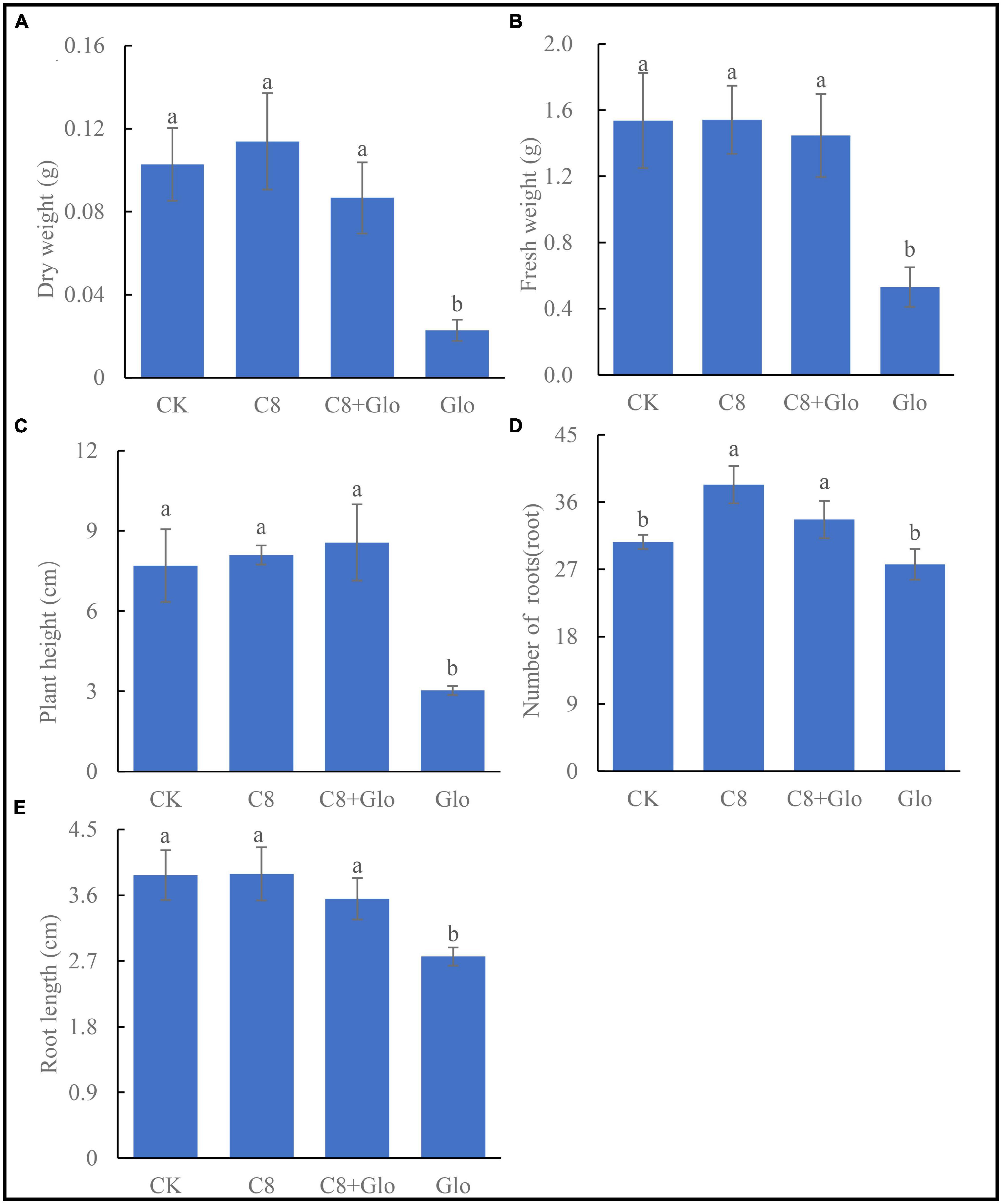
Figure 4. The seedling growth of bacteria assembled community against fungal disease. (A) Dry height, (B) fresh weight, (C) plant height, (D) number of roots, and (E) root length of A. annua plants cultured for 14 days. Different letters in a bar diagram indicate significant differences among strains (p < 0.05), same lower-case means no difference.
Physiological indexes of seedlings inoculated with an assembled bacterial community and G. ultimum
Compared with control (CK) uninoculated seedlings, the chlorophyll a (Chla) concentration of A. annua plants inoculated with C8 and C8 + Glo increased, whereas the concentration in seedlings inoculated with G. ultimum (Glo) decreased although there no significant difference between the CK and inoculation treatments, and the Chla concentrations of seedlings inoculated with C8 and C8 + Glo were significantly higher than those inoculated with only Glo (Figure 5A). The chlorophyll b (Chlb) concentration also varied with different treatments, but there were no significant differences among the CK, C8, and Glo treatments (Figure 5B). Surprisingly, the concentration of MDA, which reflects damage due to oxidative stress, was lower in Glo than in C8 + Glo (Figure 5C). However, compared with CK, the activities of the antioxidant enzymes SOD and especially POD treated with Glo increased significantly 160.04 U⋅g–1FW⋅h–1 and 13,330.30 U⋅g–1⋅min–1, respectively; the SOD activities treated with C8 and C8 + Glo decreased 49.51 and 201.51 U⋅g–1FW⋅h–1, respectively, whereas POD activities treated with C8 and C8 + Glo increased significantly 2,181.64 and 3,285.27 U⋅g–1⋅min–1 (Figures 5D, E). The proline concentrations varied similarly to POD activities, significantly increasing 81.02, 18.11, and 17.15 μg⋅g–1 FW (Figure 5F).
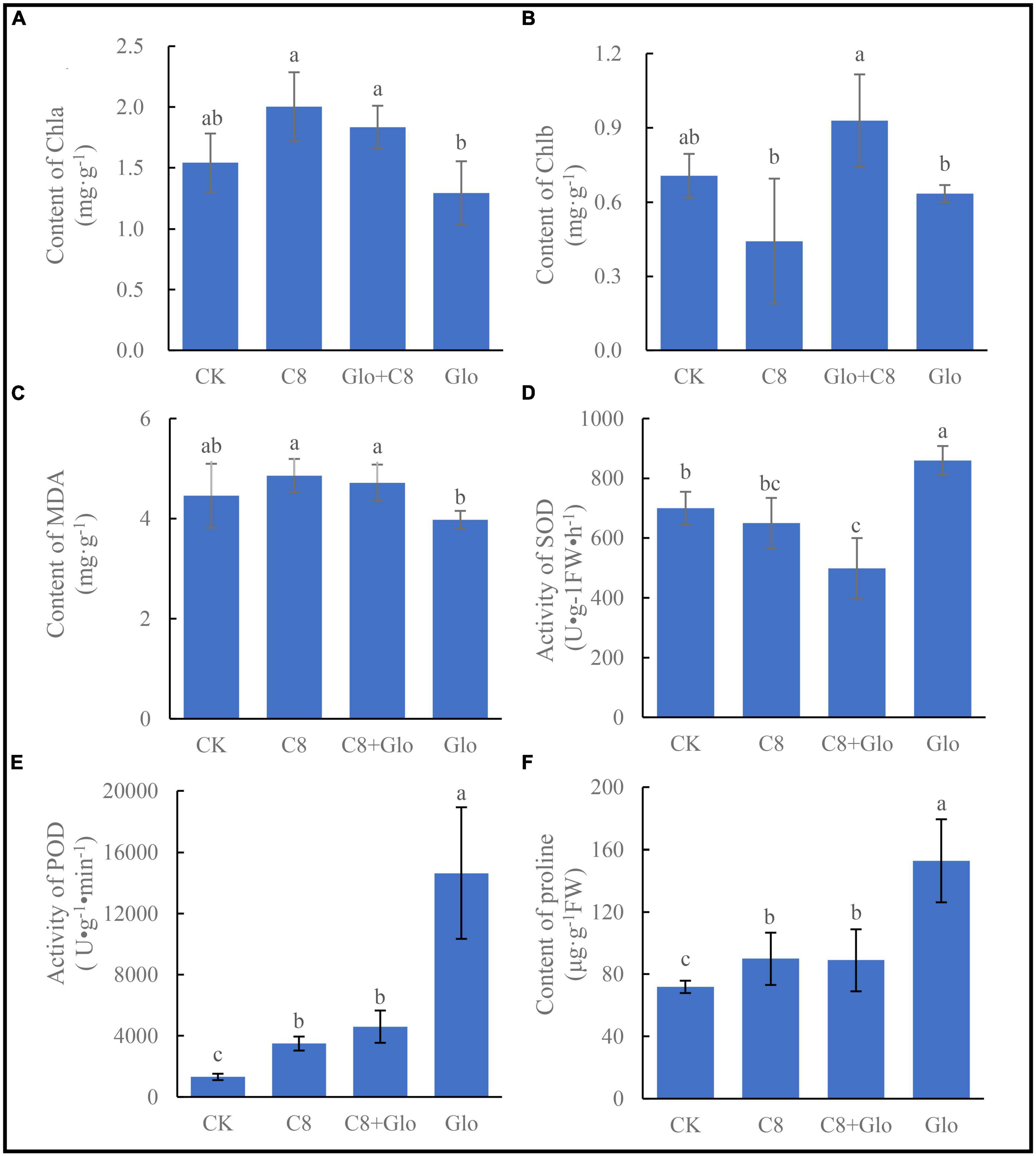
Figure 5. The seedling physiological indexes of bacteria assembled community against fungal disease. (A) The chlorophyll a (Chla) content, (B) the chlorophyll b (Chlb) content, (C) content of MDA, (D) activity of SOD, (E) activity of POD, and (F) the proline contents. Different letters in a bar diagram indicate significant differences among bacterial strains (p < 0.05), same lower-case means no difference.
Metabolite concentrations in seedlings inoculated with the assembled bacterial community and G. ultimum
A study was carried out on the concentrations of the main metabolites to determine whether colonization by the assembled bacterial community affected the main metabolites; the metabolite concentrations of A. annua roots were measured after the plants were cultured for 7 days. The results showed that the concentrations of the four tested metabolites of seedlings treated with C8, C8 + Glo, and Glo were significantly lower than those in CK seedlings, except for chrysosplenetin and artemisinin treated by C8 + Glo. Compared with Glo, the concentrations of the four metabolites in seedlings inoculated with C8 + Glo increased significantly, and the metabolite concentrations in seedlings inoculated with C8 increased except for artemisinin (Figures 6A-D).
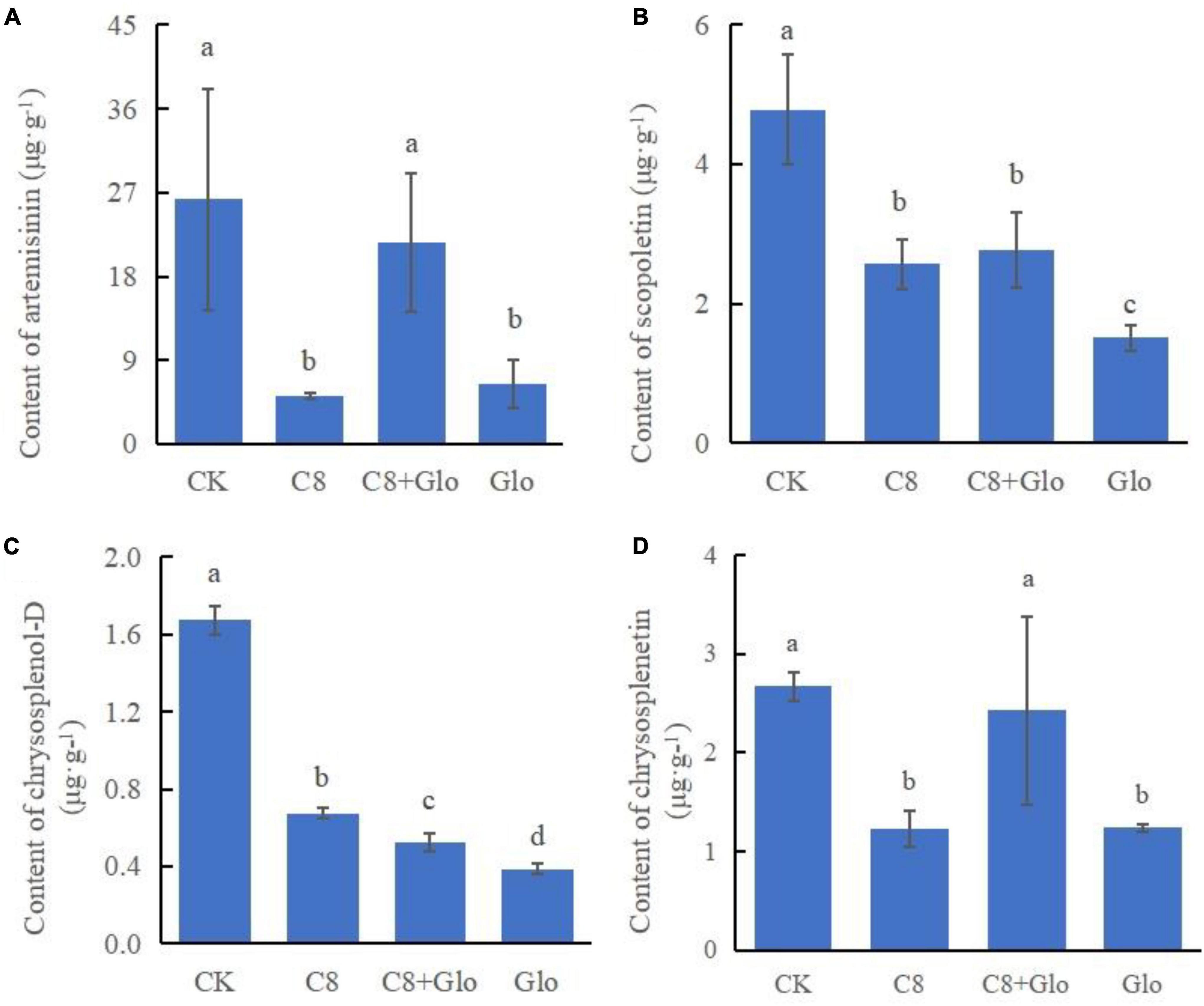
Figure 6. Metabolite contents of the seedling in bacteria assembled community against fungal disease. The contents of (A) artemisinin, (B) scopoletin, (C) chrysosplenol-D, and (D) chrysosplenetin of A. annua roots after the seedlings were cultured for 7 days. Different letters in a bar diagram indicate significant differences among strains (p < 0.05), same lower-case means no difference.
In vitro effects of A. annua root extract on the pathogenic fungus and beneficial bacteria
To examine whether the metabolites of A. annua affect the fungal and bacterial strains, five concentrations of the root extracts were added to the fungal and bacterial culture media, then inoculated with the fungus and the bacterial isolates, respectively, and cultured for 24 h. The results showed that the inhibitory effects of the root extract on G. ultimum increased with increasing extract concentrations (Figure 7A), and the diameters of the fungus were negatively correlated with the extract concentrations, and the regression equation was y = 7.07 − 0.24x, R2 = 0.97, p < 0.05. In contrast to G. ultimum, the bacterial isolates were differentially sensitive to the root extract concentration; isolates 0410ARD7G4 Paracoccus sp., DE024 Bacillus sp., S186B B. nitrificans, NBRC102508 Cupriavidus sp., Brevundimonas sp. Yang 2023, and CA34 R. pusense began to be significantly inhibited at 3.0, 6.0, 6.0, 6.0, 12.0, and 24.0 mg/ml extract concentrations, respectively (Figures 7D, G-I, B, E, respectively), although it is worth noting that YFCC6454 Stenotrophomonas sp. and G16 Flavobacterium sp. were not affected by all extract concentrations (Figures 7F, C). The order of the degree of sensitivity of the microbial strains to the extracts was G. ultimum ∼ DE024 Bacillus sp. > 0410ARD7G4 Paracoccus sp. ∼ S186B B. nitrificans ∼ NBRC102508 Cupriavidus sp. > Brevundimonas sp. Yang 2023 > CA34 R. pusense > G16 Flavobacterium sp. ∼ YFCC6454 Stenotrophomonas sp. (Figures 7A, G, D, H, I, B, E, C, F, respectively).
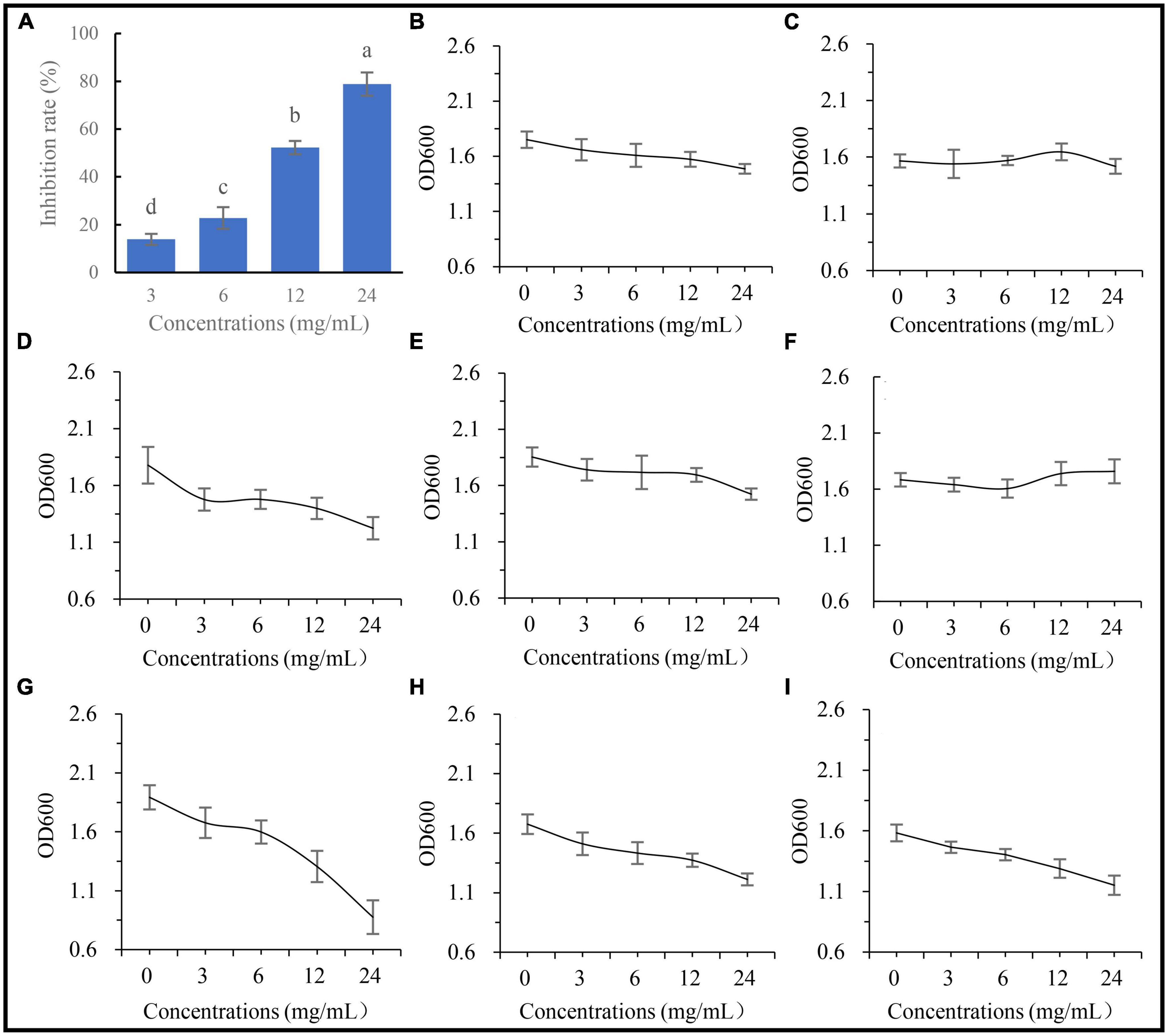
Figure 7. Affection of extracting solution from A. annua on pathogenic fungus and bacteria. (A) Inhibition rates of extracting solution on G. ultimum var. The OD600 of extracting solution against (B) Brevundimonas sp. Yang 2023, (C) G16 Flavobacterium sp., (D) 0410ARD7G4 Paracoccus sp., (E) CA34 Rhizobium pusense, (F) YFCC6454 Stenotrophomonas sp., (G) DE024 Bacillus sp., (H) S186B Brevibacillus nitrificans, and (I) NBRC102508 Cupriavidus sp.
Discussion
Existence and isolation of the A. annua pathogenic fungus
Under natural conditions, all plants suffer from abiotic and biotic stresses, such as drought, waterlogging, nutrient deficiency, nematodes, and pathogenic fungi (Farh et al., 2018; Yuan et al., 2018). A. annua is a pioneer plant, the only source of the antimalarial drug artemisinin (Committee of Chinese Pharmacopoeia, 2020), and is tolerant to soil nutrient deficiency (Luo et al., 2019b). The current study is the first report that A. annua plants are affected by pathogenic microorganisms (Figure 1). There are microorganisms pathogenic on A. annua, such as G. ultimum, but possibly natural associations between A. annua and bacterial communities achieve suppression of such pathogens.
Isolation and assembly complex of antifungal bacteria associated with the plants achieve adaptation to the environment
The composition of the microbial community in the rhizosphere of plants, such as maize, paddy rice, wheat, and so on, has been extensively studied (Edwards et al., 2015; Schlemper et al., 2018; Walters et al., 2018). Some members of the bacterial community which directly influenced A. mongholicus growth and improved the adaptation of the plants to the environment have been reported (Li et al., 2021). However, it is unclear how A. annua plants associate with protective microbes to improve their adaptation to different environments. Because we discovered that the sterile plantlets, which were regenerated from A. annua collected in Guiyang, could not grow directly in sterile humus soil from Changbai Mountains when inoculated with G. ultimum, it may be that geographic differences cause such results. When we transferred the sterile plantlets to glass containers with non-sterile humus soil inoculated with G. ultimum and cultured them for a period of time, the plants were alive and grew vigorously, indicating that the soil microbes were beneficial to the growth of A. annua plants; a previous study had reported that plant root systems have a strong selective effect on microorganisms in the surrounding soil (Dibbern et al., 2014), supporting our strategy of isolating bacteria from the roots of A. annua plants grown in natural humus soil from Changbai Mountains. After screening of bacterium–fungus interactions with the dual culture assay, bacterial isolates with antifungal activity were selected.
A previous study had reported that a simplified four-species bacterial community, which was composed of three high-abundance bacteria (Stenotrophomonas sp., Rhizobium sp., and Ochrobactrum sp.) and one low-abundance bacterium (Advenella sp.), had inhibitory effects on a root rot disease of A. mongholicus (Li et al., 2021). A disease-induced assembly of a three-species bacterial community (Microbacterium sp., Stenotrophomonas sp., and Xanthomonas sp.) interacted in biofilm formation to reduce disease incidence (Berendsen et al., 2018). Investigation of bacteria and fungi abundances by amplicon sequencing revealed that the abundance of Sphingomonas and Sphingobium (both plant growth-promoting bacteria) and saprotrophic fungi in the rhizosphere soil of A. annua plants increased dramatically (Shi et al., 2022), but specific isolates were not purified. In our study, 60 bacterial strains were obtained from A. annua roots belonging to 18 bacterial genera, from which eight bacterial genera were selected to make up an antifungal assembled bacterial community with activity against Glo. We compared the seedlings inoculated with C8 + Glo and C6 + Glo; the C6 community was composed of six antifungal bacterial isolates individually inhibiting the pathogenic fungus Glo, and the C8 community was composed of C6 plus another two bacterial isolates, S186B B. nitrificans and NBRC102508 Cupriavidus sp., which had no inhibitory effect on Glo. The results showed that the incidence of disease and the death rate of the seedlings inoculated with C8 + Glo after culture 14 days were significantly lower than in C6 + Glo (Supplementary Figure 2). The results explained that antifungal and non-antifungal bacteria in the assembled community synergistically enhanced plant disease-suppressive activity.
Effect on plants of assembled bacterial community against G. ultimum
An earlier study reported that an assembled bacterial community influenced the root growth of model plant Arabidopsis, with one single bacterial genus, Variovorax, of the assembled community manipulating plant hormone levels to achieve the positive effects of the assembled root community on root growth (Finkel et al., 2020). In the current study, the eight-species community had a robust effect on A. annua plants infected by the pathogen G. ultimum. Similarly, the concentrations of four metabolites of A. annua roots inoculated with C8 + Glo were significantly higher than in those inoculated with Glo, with the concentrations of artemisinin and scopoletin being significantly higher with C8 + Glo than with C8, suggesting that the eight-species assembled bacterial community enhanced the levels of A. annua metabolites under disease-suppressive condition. It has been reported that artemisinin has anti-fungal and anti-bacterial activities in addition to anti-malarial activity (Huang et al., 1993; Appalasamy et al., 2014), whereas scopoletin, chrysosplenol-D and chrysosplenol all had antioxidant activities to reduce the impact of oxidative stress (Lai et al., 2007; Brisibe et al., 2009; Yang et al., 2009; Ferreira et al., 2010), which could suggest that they would contribute to increased disease suppression in A. annua.
The increased disease resistance of C8-treated A. annua was also demonstrated by physiological indexes, with the concentrations of Chla and Chlb treated with C8 + Glo being markedly higher than when treated with Glo alone (Figures 5A, B), suggesting that the eight-species community had a robust beneficial effect on plant chlorophyll levels under disease attack. However, the POD and SOD activities were higher in Glo than in C8 + Glo (Figures 5D, E), possibly reflecting that the plants were responding to the increased oxidative stress associated with disease; unexpectedly, the concentration of the end-product of the stress, MDA, was lower in diseased plants.
Mechanism supposition of assembled bacteria community against fungi pathogen
After inoculating sterile seedlings with C8 + Glo in sterile soil and culturing for 7 days, the eight bacteria were traced by 16S RNA sequencing. For more sensitivity and specificity, ASVs were used to quantified the abundances of the endogenetic bacteria in roots (Callahan et al., 2016, 2019). ASV values ranged from 1.00 to 6,952.33 in C8 treatment, NBRC102508 Cupriavidus sp. was the highest, followed by CA34 R. pusense; and ASV varied from 1.67 to 7,454.33 in C8 + Glo treatment, but CA34 R. pusense was the highest, followed by NBRC102508 Cupriavidus sp., and G16 Flavobacterium sp. was the lowest in both treatments (Supplementary Table 2). It was discovered that eight bacteria could colonized in endosphere of plant roots, although the abundances of the endogenetic bacteria were different. In addition, the crude extract from C8 culture solution and dual cultural assay exhibited that C8 had inhibitory effect on Glo (Supplementary Figures 3, 4), which suggested that secretions from eight bacteria complex prevented pathogen Glo from growth. It could be seen that such functionally assembled community could be used as biological fungicides to help the growth of plants.
Different effects of A. annua root extract against fungi and bacteria
Other plant metabolites, such as coumarins and phenolic acids (Brown, 1992; Rice-Evans et al., 1996; Ferreira et al., 2010) have antioxidant activities, whereas some volatile components have anti-microbial activities (Juteau et al., 2002; Wu et al., 2007). To test whether such components affect the growth of fungal and bacterial isolates, five different concentrations of root extracts were tested for activity against bacterial and fungal growth in vitro. After culture for 24 h, the extract was significantly more inhibitory toward Glo than toward any one of the eight bacterial isolates (Figures 7B–I).
In all, the assembled bacterial community from roots of A. annua plants protected the plants against the Glo pathogen. In turn, the root extract of A. annua plants had inhibitory activity toward the pathogenic fungus, while the concentrations of some potentially anti-fungal and anti-stress metabolites were lower in C8 + Glo than in Glo. These findings suggested that A. annua plants were associated with a natural root bacterial community which suppressed the effects of the pathogen and this effect could be replicated by the assembled community of the A. annua root eight-bacteria. The specific mechanism of this bacteria–plant interactions deserve to be studied in more detail in the future.
Conclusion
The assemblage of six bacterial species with antifungal activity and two bacterial species without effectively protected the plants against Glo as an assembled community applied to sterile plants. These findings suggested that microbes with various ecological functions could be assembled into assembled communities for plant disease suppression. Such functional complex communities could be applied for use as biological fungicides, promoting biological disease control of plants.
Data availability statement
The original contributions presented in this study are included in this article/Supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
YW conducted the experiments, calculated the data, and prepared the figures with direction of Z-nY and S-qL. S-qL wrote and edited the manuscript. Z-nY reviewed and edited the final manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the Science and Technology Plan Project of Guizhou Province, China (Qiankehe Basics [2020]1Y179) and Project of Guizhou Provincial Characteristic Key Laboratory (QJHKY [2021]002). The funder played no role in the design of the work, analysis and interpretation of data or in writing the manuscript.
Acknowledgments
We thank Sangon Biotechnology Co., Ltd. (Sangon Biotech, Shanghai, China) for gene sequence, and thank the NCBI website for sequence alignment. Thanks for Shunxing Yang and Kaifeng Wang helping YW during the experiment period.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1218474/full#supplementary-material
Abbreviations
PDA, potato dextrose agar; NA, nutrient agar; OD, opacity degree; MDA, malondialdehyde; SOD, superoxide dismutase; POD, peroxidase; PBS, phosphate buffered saline; C6, six-bacteria assembled community; C8, eight-bacteria assembled community; Glo, Globisporangium ultimum var.; C8 + Glo, eight-bacteria synthetic community and G. ultimum var.
Footnotes
- ^ https://submit.ncbi.nlm.nih.gov/
- ^ https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome
References
Appalasamy, S., Lo, K. Y., Ch’ng, S. J., Nornadia, K., Othman, S. A., and Chan, L. (2014). Antimicrobial activity of artemisinin and precursor derived from in vitro plantlets of Artemisia annua L. Biomed. Res. Int. 2014:215872. doi: 10.1155/2014/215872
Bai, Y., Müller, D. B., Srinivas, G., Garrido-Oter, R., Potthoff, E., Rott, M., et al. (2015). Functional overlap of the Arabidopsis leaf and root microbiota. Nature 528, 364–363. doi: 10.1038/nature16192
Berendsen, R. L., Pieterse, C. M., and Bakker, P. A. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486. doi: 10.1016/j.tplants.2012.04.001
Berendsen, R. L., Vismans, G., Yu, K., Song, Y., de Jonge, R., Burgman, W. P., et al. (2018). Disease-induced assemblage of a plant-beneficial bacterial consortium. ISME J. 12, 1496–1507. doi: 10.1038/s41396-018-0093-1
Bodenhausen, N., Bortfeld-Miller, M., Ackermann, M., and Vorholt, J. A. (2014). A synthetic community approach reveals plant genotypes affecting the phyllosphere microbiota. PLoS Genet. 10:e1004283. doi: 10.1371/journal.pgen.1004283
Brisibe, E. A., Umoren, U. E., Brisibe, F., Magalhaees, P. M., Ferreira, J. F. S., Luthria, D., et al. (2009). Nutritional characterisation and antioxidant capacity of different tissues of Artemisia annua L. Food Chem. 115, 1240–1246. doi: 10.1016/j.foodchem.2009.01.033
Brown, G. D. (1992). Two new compounds from Artemisia annua. J. Nat. Prod. 55, 1756–1760. doi: 10.1021/np50090a006
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J., and Holmes, S. P. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Callahan, B. J., Wong, J., Heiner, C., Oh, S., Theriot, C. M., Gulati, A. S., et al. (2019). High-throughput amplicon sequencing of the full-length 16S rRNA gene with single-nucleotide resolution. Nucleic Acids Res. 47:e103. doi: 10.1093/nar/gkz569
Castrillo, G., Teixeira, P. J. P. L., Paredes, S. H., Law, T. F., de Lorenzo, L., Feltcher, M. E., et al. (2017). Root microbiota drive direct integration of phosphate stress and immunity. Nature 543, 513–518. doi: 10.1038/nature21417
Chen, T., Nomura, K., Wang, X., Sohrabi, R., Xu, J., Yao, L., et al. (2020). A plant genetic network for preventing dysbiosis in the phyllosphere. Nature 580, 653–657. doi: 10.1038/s41586-020-2185
Committee of Chinese Pharmacopoeia (2020). Chinese pharmacopoeia, 1st Edn. Beijing: China Medical Science and Technology Press.
Dibbern, D., Schmalwasser, A., Lueders, T., and Totsche, K. U. (2014). Selective transport of plant root-associated bacterial populations in agricultural soils upon snowmelt. Soil Biol. Biochem. 69, 187–196. doi: 10.1016/j.soilbio.2013.10.040
Dieter, H., and Geneviève, D. (2005). Biological control of soil-borne pathogens by Fluorescent pseudomonads. Nat. Rev. Microbiol. 3, 307–319. doi: 10.1038/nrmicro1129
Durán, P., Thiergart, T., Garrido-Oter, R., Agler, M., Kemen, E., Schulze-Lefert, P., et al. (2018). Microbial interkingdom interactions in roots promote Arabidopsis survival. Cell 175, 973–983.e14. doi: 10.1016/j.cell.2018.10.020
Edwards, J., Johnson, C., Santos-Medellin, C., Lurie, E., Podishetty, N. K., Bhatnagar, S., et al. (2015). Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. U.S.A. 112, E911–E920. doi: 10.1073/pnas.1414592112
Farh, M. E., Kim, Y., Kim, Y. J., and Yang, D. (2018). Cylindrocarpon destructans/Ilyonectria radicicola-species complex: Causative agent of ginseng root-rot disease and rusty symptoms. J. Ginseng Res. 42, 9–15.
Ferreira, J. F. S., Luthria, D. L., Sasaki, T., and Heyerick, A. (2010). Flavonoids from Artemisia annua L. as antioxidants and their potential synergism with artemisinin against malaria and cancer. Molecules 15, 3135–3170. doi: 10.3390/molecules15053135
Finkel, O. M., Salas-González, I., Castrillo, G., Conway, J. M., Law, T. F., Teixeira, P. J. P. L., et al. (2020). A single bacterial genus maintains root growth in a complex microbiome. Nature 587, 103–108. doi: 10.1038/s41586-020-2778-7
Finkel, O. M., Salas-González, I., Castrillo, G., Spaepen, S., Law, T. F., Teixeira, P. J. P. L., et al. (2019). The effects of soil phosphorus content on plant microbiota are driven by the plant phosphate starvation response. PLoS Biol. 17:e3000534. doi: 10.1371/journal.pbio.3000534
Fitzpatrick, C. R., Copeland, J., Wang, P. W., Guttman, D. S., Kotanen, P. M., and Johnson, M. T. J. (2018). Assembly and ecological function of the root microbiome across angiosperm plant species. Proc. Natl. Acad. Sci. U.S.A. 115, E1157–E1165. doi: 10.1073/pnas.1717617115
Gkarmiri, K., Finlay, R. D., Alström, S., Thomas, E., Cubeta, M. A., and Högberg, N. (2015). Transcriptomic changes in the plant pathogenic fungus Rhizoctonia solani AG-3 in response to the antagonistic bacteria Serratia proteamaculans and Serratia plymuthica. BMC Genomics 16:630. doi: 10.1186/s12864-015-1758-z
Herrera, P. S., Gao, T., Law, T. F., Finkel, O. M., Mucyn, T., Teixeira, P. J. P. L., et al. (2018). Design of synthetic bacterial communities for predictable plant phenotypes. PLoS Biol. 16:e2003962. doi: 10.1371/journal.pbio.2003962
Huang, L., Liu, J. F., Liu, L. X., Li, D. F., Zhang, Y., Nui, H. Z., et al. (1993). Study on the antipyretic and anti-inflammatory effects of Artemisia annua L. China. J. Chin. Mater. Med. 18, 44–48.
Huang, L., Xie, C., Duan, B., and Chen, S. (2010). Mapping the potential distribution of high artemisinin-yielding Artemisia annua L. (Qinghao) in China with a geographic information system. Chin. Med. 5:18. doi: 10.1186/1749-8546-5-18
Juteau, F., Masotti, V., Bessière, J. M., Dherbomez, M., and Viano, J. (2002). Antibacterial and antioxidant activities of Artemisia annua essential oil. Fitoterapia 73, 532–535. doi: 10.1016/s0367-326x(02)00175-2
Lai, J. P., Lim, Y. H., Su, J., Shen, H., and Ong, C. N. (2007). Identification and characterization of major flavonoids and caffeoylquinic acids in three Compositae plants by LC/DAD-APCI/MS. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 848, 215–225.
Lebeis, S. L., Paredes, S. H., Lundberg, D. S., Breakfield, N., Gehring, J., McDonald, M., et al. (2015). Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science 349, 860–864. doi: 10.1126/science.aaa8764
Lévesque, F., and Peter, H. S. (2012). Continuous-flflow synthesis of the antimalaria drug artemisinin. Angew. Chem. Int. Edit. 51, 1706–1709. doi: 10.1002/anie.201107446
Li, Z., Bai, X., Jiao, S., Li, Y., Li, P., Yang, Y., et al. (2021). A simplified synthetic community rescues Astragalus mongholicus from root rot disease by activating plant-induced systemic resistance. Microbiome 9:217. doi: 10.1186/s40168-021-01169-9
Liu, H., Tian, X., Zhang, Y., Wang, C., and Jiang, H. (2013). The discovery of Artemisia annua L. in the Shengjindian cemetery, Xinjiang, China and its implications for early uses of traditional Chinese herbal medicine qinghao. J. Ethnopharmacol. 146, 278–286. doi: 10.1016/j.jep.2012.12.044
Luo, S., Zhao, C., Yang, Z., Di, S., Zheng, Z., and Hu, J. (2019b). Correlation analysis of nutrients, enzymes, and microbial biomass in soils phenolics of Artemisia annua L. Pak. J. Agri. Sci. 56, 171–178. doi: 10.21162/PAKJAS/19.5605
Luo, S., Zhao, C., Yang, Z., Hu, J., and Di, S. (2019a). Soil microbes and medical metabolites of Artemisia annua L. along altitudinal gradient in Guizhou Karst terrains of China. J. Plant Interact. 14, 167–176. doi: 10.1080/17429145.2019.1602886
McNulty, N. P., Yatsunenko, T., Hsiao, A., Faith, J. J., Muegge, B. D., Goodman, A. L., et al. (2011). The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Sci. Transl. Med. 3:106ra106. doi: 10.1126/scitranslmed.3002701
Melo, I. S., Santos, S. N., Rosa, L. H., Parma, M. M., Silva, L. J., Queiroz, S. C. N., et al. (2014). Isolation and biological activities of an endophytic Mortierella alpina strain from the Antarctic moss Schistidium antarctici. Extremophiles 18, 15–23.
Mueller, U. G., and Sachs, J. L. (2015). Engineering microbiomes to improve plant and animal health. Trends Microbiol. 23, 606–617. doi: 10.1016/j.tim.2015.07.009
Müller, D. B., Vogel, C., Bai, Y., and Vorholt, J. A. (2016). The plant microbiota: Systems-level insights and perspectives. Annu. Rev. Genet. 50, 211–223. doi: 10.1146/annurev-genet-120215-034952
Niu, B., Paulson, J. N., Zheng, X., and Kolter, R. (2017). Simplified and representative bacterial community of maize roots. Proc. Natl. Acad. Sci. U.S.A. 114, E2450–E2459. doi: 10.1073/pnas.1616148114
Rice-Evans, C. A., Miller, N. J., and Paganga, G. (1996). Structure-antioxidant activity relationships of flavonoids and phenolic acids. Free Radic. Biol. Med. 20, 933–956. doi: 10.1016/0891-5849(95)02227-9
Schlemper, T. R., van Veen, J. A., and Kuramae, E. E. (2018). Co-variation of bacterial and fungal communities in different sorghum cultivars and growth stages is soil dependent. Microb. Ecol. 76, 205–214. doi: 10.1007/s00248-017-1108-6
Shalev, O., Karasov, T. L., Lundberg, D. S., Ashkenazy, H., Ayutthaya, P. P. N., and Weigel, D. (2022). Commensal Pseudomonas strains facilitate protective response against pathogens in the host plant. Nat. Ecol. Evol. 6, 383–396. doi: 10.1038/s41559-022-0167
Shi, Y., Pan, Y., Li, X., Zhu, Z., Fu, W., Hao, G., et al. (2022). Assembly of rhizosphere microbial communities in Artemisia annua: Recruitment of plant growth-promoting microorganisms and inter-kingdom interactions between bacteria and fungi. Plant Soil 470, 127–139. doi: 10.1007/s11104-021-04829-9
Thiergart, T., Durán, P., Ellis, T., Vannier, N., Garrido-Oter, R., Kemen, E., et al. (2020). Root microbiota assembly and adaptive differentiation among European Arabidopsis populations. Nat. Ecol. Evol. 4, 122–131. doi: 10.1038/s41559-019-1063-3
Thiergart, T., Zgadzaj, R., Bozsóki, Z., Garrido-Oter, R., Radutoiu, S., and Schulze-Lefert, P. (2019). Lotus japonicus symbiosis genes impact microbial interactions between symbionts and multiking domcommensal communities. mBio 10, e01833–19. doi: 10.1128/mBio.01833-19
Tian, X.-M., Wu, Y., and Yu, Z. (2008). Isolation and screening of endophytes from Artemisia annua and their antagonistic activity to plant pathogens. Acta Agric. Boreali-occidentalis Sin. 17, 186–190.
Vale, P. F., McNally, L., Doeschl-Wilson, A., King, K. C., Popat, R., Domingo-Sananes, M. R., et al. (2016). Beyond killing: Can we find new ways to manage infection? Evol. Med. Public. Health 2016, 148–157. doi: 10.1093/emph/eow012
Ventorino, V., Parillo, R., Testa, A., Viscardi, S., Espresso, F., and Pepe, O. (2016). Chestnut green waste composting for sustainable forest management: Microbiota dynamics and impact on plant disease control. J. Environ. Manag. 166, 168–177. doi: 10.1016/j.jenvman.2015.10.018
Wagner, M. R., Lundberg, D. S., del Rio, T. G., Tringe, S. G., Dangl, J. L., and Mitchell-Olds, T. (2016). Host genotypeand age shapethe leaf and root microbiomes of a wild perennial plant. Nat. Commun. 7:12151. doi: 10.1038/ncomms12151
Walters, W. A., Jin, Z., Youngblut, N., Wallace, J. G., Sutter, J., Zhang, W., et al. (2018). Largescale replicated field study of maize rhizosphere identifies heritable microbes. Proc. Natl. Acad. Sci. U.S.A. 115, 7368–7373. doi: 10.1073/pnas.1800918115
Wang, Y., Yang, Z., Luo, S., Ding, Q., Yang, S., and Wang, K. (2022). Establishment of tissue culture regeneration system of Artemisia Annua L. Mol. Plant Breed. 13:43.
Wei, Z., Yang, T., Friman, V., Xu, Y., Shen, Q., and Jousset, A. (2015). Trophic network architecture of root-associated bacterial communities determines pathogen invasion and plant health. Nat. Commun. 6:8413. doi: 10.1038/ncomms9413
Wu, J., Ding, W., Zhang, Y., and Guo, W. (2007). Antifungal activities of primary extracts of Artemisia annua L. against two fungi. Agrochemicals 46, 713–718. doi: 10.3969/j.issn.1006-0413.2007.10.024
Xue, C., Penton, C. R., Shen, Z., Zhang, R., Huang, Q., Li, R., et al. (2015). Manipulating the banana rhizosphere microbiome for biological control of Panama disease. Sci. Rep. 5:11124. doi: 10.1038/srep11124
Yang, G., Li, B., Zhang, X., Wang, Y., Li, Q., Zhang, W., et al. (2009). Studies on flavonoids and their antioxidant Activities of Artemisia annua. Chin. Herb. Med. 33, 1684–1688.
Keywords: Artemisia annua, assembled bacterial community, pathogenic fungus, plant–bacterium interaction, Globisporangium ultimum
Citation: Wang Y, Yang Z-n and Luo S-q (2023) An assembled bacterial community associated with Artemisia annua L. causes plant protection against a pathogenic fungus. Front. Microbiol. 14:1218474. doi: 10.3389/fmicb.2023.1218474
Received: 07 May 2023; Accepted: 12 September 2023;
Published: 09 October 2023.
Edited by:
Trevor Carlos Charles, University of Waterloo, CanadaReviewed by:
Walaa K. Mousa, Mansoura University, EgyptFangdong Zhan, Yunnan Agricultural University, China
Copyright © 2023 Wang, Yang and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shi-qiong Luo, c2hpcWlvbmdsdW9AMTYzLmNvbQ==; Zhan-nan Yang, eWFuZ3poYW5uYW5AMTYzLmNvbQ==
†These authors have contributed equally to this work
 Yu Wang
Yu Wang Zhan-nan Yang
Zhan-nan Yang Shi-qiong Luo
Shi-qiong Luo