
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 29 June 2023
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1217270
This article is part of the Research Topic The Role of Metabolomics in Antimicrobial Drug Discovery and Development View all 6 articles
 Shixing Zhu1†
Shixing Zhu1† Jiali Yue1†
Jiali Yue1† Xintong Wang1†
Xintong Wang1† Jiayuan Zhang1
Jiayuan Zhang1 Mingming Yu1,2
Mingming Yu1,2 Yuanchao Zhan1*
Yuanchao Zhan1* Yuanqi Zhu3
Yuanqi Zhu3 Sherwin K. B. Sy4
Sherwin K. B. Sy4 Zhihua Lv1,2*
Zhihua Lv1,2*Introduction: The emergence of multidrug-resistant (MDR) Acinetobacter baumannii prompts clinicians to consider treating these infections with polymyxin combination.
Methods: Metabolomic analysis was applied to investigate the synergistic effects of polymyxin-B, amikacin and sulbactam combination therapy against MDR A. baumannii harboring OXA-23 and other drug resistant genes. The drug concentrations tested were based on their clinical breakpoints: polymyxin-B (2 mg/L), amikacin (16 mg/L), polymyxin-B/amikacin (2/16 mg/L), and polymyxin-B/amikacin/sulbactam (2/16/4 mg/L).
Results: The triple antibiotic combination significantly disrupted levels of metabolites involved in cell outer membrane structure including fatty acids, glycerophospholipids, nucleotides, amino acids and peptides as early as 15 min after administration. Amikacin and polymyxin-B alone perturbed a large number of metabolites at 15 min and 1 h, respectively, but the changes in metabolites were short-lived lasting for less than 4 h. In contrast, the combination treatment disrupted a large amount of metabolites beyond 4 h. Compared to the double-combination, the addition of sulbactam to polymyxin-B/amikacin combination produce a greater disorder in A. baumannii metabolome that further confer susceptibility of bacteria to the antibiotics.
Conclusion: The metabolomic analysis identified mechanisms responsible for the synergistic activities of polymyxin-B/amikacin/sulbactam against MDR A. baumannii.
Acinetobacter baumannii is a tenacious gram-negative nosocomial pathogen that can survive a wide range of temperatures, pH, and humidity (Gootz and Marra, 2008) and often causes hospital-acquired and ventilator-associated pneumonia (HAP/VAP), urinary tract infection, blood flow infection, meningitis, skin and soft tissue infections in critically ill patients (Clark et al., 2016; Freire et al., 2016). Over the past few decades, the number of community-acquired infections caused by A. baumannii has been steadily increasing, resulting in high morbidity and mortality (Dijkshoorn et al., 2007; Sengstock et al., 2010). Multidrug-resistant (MDR) strains have spread from areas with high antibiotic resistance to areas with low rates of antibiotic resistance (Perez et al., 2007).
Many broad-spectrum antibiotics are ineffective against MDR A. baumannii (Avedissian et al., 2019). Since early 2000s, polymyxins have been used to treat many infections caused by MDR A. baumannii (Li et al., 2006; Poirel et al., 2017). A. baumannii confers resistance to polymyxin-B through modification of lipid A or single nucleotide mutations leading to a complete loss of lipopolysaccharide (LPS) (Girardello et al., 2017). Resistance to colistin alone has been reported (Zhao et al., 2022). Monotherapy can no longer meet the clinical needs, and the combination of polymyxin-B and other antibiotics is needed to improve the survival rate of patients infected with MDR A. baumannii (Oo and Sy, 2018, 2020; Wang et al., 2023). There have been reports of improved results of polymyxin combination therapy in the clinic (Teo et al., 2015; Sharma et al., 2017; Hussein et al., 2019a). Even though sulbactam is primarily used as a β-lactamase inhibitor, it has inherent antibacterial activity against A. baumannii and is one of the main drugs to treat Acinetobacter infections, often in combination with other antibiotics (Penwell et al., 2015; Deng et al., 2022). In the absence of a partnering β-lactam, sulbactam can potentially enhance the activity of polymyxin-B by making the cell wall leaky.
We have previously shown that even though polymyxin-B/amikacin/sulbactam combination can exert synergistic activities against A. baumannii, patients can benefit from localized administration in addition to intravenous route to ensure sufficient drug concentrations at the site of infection such as the lung (Zhu et al., 2022a,b; Zhang et al., 2023). Due to the poor penetration of both amikacin and polymyxin-B into the lung, inhaled administration of these two drugs can improve probability of treatment success in HAP/VAP. As an extension of our previous work, we performed metabolomic analysis to understand mechanisms responsible for the synergistic activities of polymyxin-B in combination with amikacin and sulbactam against A. baumannii.
Polymyxin-B sulfate, amikacin, and sulbactam (Shanghai Macklin Biochemical Co., Ltd., Shanghai, China) solutions were prepared according to the Clinical and Laboratory Standards Institute (CLSI) guidelines (CLSI, 2020). The stock solutions of polymyxin-B sulfate, amikacin, and sulbactam were dissolved in pure water at a concentration of 5210 μg/mL for all three antibiotics and stored at −80°C. Working solutions were diluted in Millipore water and filtered before use. Clinical A. baumannii isolate 12 was obtained from the affiliated hospital of Qingdao University and grown in cation-adjusted Mueller-Hinton broth (CAMHB; LAND BRIDGE, Beijing, China). The quality control strain for antimicrobial susceptibility test was E. coli ATCC 25922 and A. baumannii ATCC 19606. The resistance genes carried by the clinical isolate 12 are shown in Table 1.
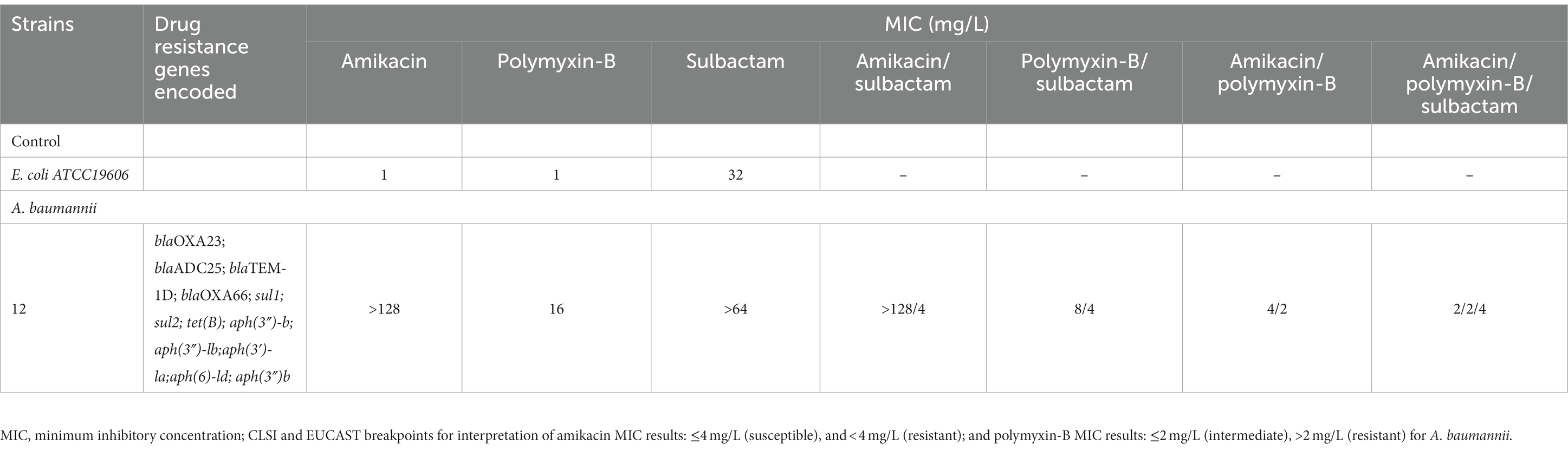
Table 1. Minimum inhibitory concentrations (MIC) of amikacin, polymyxin-B and sulbactam alone or in combination with or without sulbactam (4 mg/L) agains A. baumannii isolate 12, as well as drug resistance genes encoded.
The broth microdilution method was used to determine the MIC of various antibiotics against A. baumannii isolate 12, based on the CLSI guidelines (CLSI, 2020). Antibiotics were added in two-fold increment in each well of the sterile 96-well plate. The A. baumannii isolate 12 was cultured to a density of 0.5 McFarland and then diluted into each well of the 96-well plate to a final concentration of approximately 5 × 105 CFU/mL; the plate was then placed in a constant temperature incubator at 35 ± 2°C for 20 h. All MIC determination were performed in triplicate.
A single colony of A. baumannii isolate 12 was inoculated into 15 mL fresh MHB and incubated at 37°C overnight and then diluted 100-folds into 200 mL fresh MHB and shaking at 180 rpm (37°C). After the bacteria were cultured to logarithmic growth stage with an optical density (OD600) of approximately 0.5, antibiotics were added (Maifiah et al., 2017). The culture was divided into five treatment groups: (1) control without any antibiotics (AB-control); (2) polymyxin-B alone (2 μg/mL); (3) amikacin alone (16 μg/mL); (4) the combination of polymyxin-B and amikacin (2/16 μg/mL); and (5) the combination of polymyxin-B, amikacin, and sulbactam (2/16/4 μg/mL). Our previous study had shown that the triple combination could reduce MIC to or below the clinical breakpoints (Zhu et al., 2022a). The antibiotic concentrations were chosen based on CLSI and EUCAST breakpoints, which are associated with realistic pharmacokinetic concentrations in the clinic. In 2023, the CLSI lowered the Enterobacterales susceptibility breakpoint for amikacin from ≤16 mg/L to ≤4 mg/L (Sader et al., 2023). There is no susceptible breakpoint for polymyxin-B and the intermediate breakpoint was used (Satlin et al., 2020). Five replicates were prepared for each treatment. Samples were taken at three timepoints of 15 min, 1 h and 4 h and normalized to an OD600 of 0.5 before extraction (Zhu et al., 2022c).
Cellular metabolites of A. baumannii were extracted by previously reported methods (Hussein et al., 2019b; Zhao et al., 2021). Firstly, samples were centrifuged at 3,220 × g at 4°C for 10 min, then the bacterial pellets were washed with cold saline twice. 500 μL cold chloroform-methanol–water (CMW) (1:3:1 by volume) solution containing 1 μM of each of the internal standards (CHAPS, CAPS, PIPES, and Tris) was added. The samples were flash frozen in liquid nitrogen and thawed on ice, and then fully vortexed. The samples were then centrifuged for 10 min at 3,220 × g at 4°C to remove cell debris, and then 300 μL of the supernatants were added to 1.5 mL Eppendorf tubes for centrifugation. After centrifugation at 14,000 × g at 4°C for 10 min, 200 μL of supernatant was transferred into injection vials for analysis by liquid chromatography-high resolution mass spectrometry (LC–MS). Quality control (QC) samples were obtained by pooling the samples, and their cellular metabolites were extracted as described above.
The LC–MS method was established based on a previously reported method with modifications (Zhu et al., 2022c). Samples were analyzed by LC–MS equipped with an Ultimate 3,000 ultra-high-performance liquid chromatography (UHPLC) system (Thermo Scientific, CA, United States) and Q-Exactive Orbitrap mass spectrometer (Thermo Scientific, CA, United States) operated in both positive and negative electrospray ionization (ESI) modes (polarity switching) at 35,000 resolution with a detection range of m/z 50 to 1,275 Da. The separation was performed on a HILIC column (2.1 × 100 mm, 1.7 μm; ACE 1.7 μm, HILIC-A, United Kingdom) coupled with guard column (ACE UHPLC pre-column filter) operated at 40°C. The mobile phase which consisted of 10 mM ammonium acetate in water (solvent A), and acetonitrile (solvent B) was delivered at a flow rate of 0.3 mL/min. The gradient elution mode started from 80 to 20% over 15 min, then to 5% of solvent B at 18 min, followed by a wash with 5% B for 3 min, and re-equilibration for 8 min with 80% B. The injection volume was 10 μL.
The raw data file obtained by LC–MS analysis was analyzed by Progenesis QI program (Waters, Mass, United States). The workflow was established for importing data, automatic alignment, peak selection based on the minimum intensity of 50,000, and subsequent identification of metabolites. Metabolites were identified by LC retention time and accurate m/z value. The maximum retention time shift for peak alignment was 0.2 min, and the mass tolerance was 5 ppm. Metabolites were normalized by the mean, log10-transformed and auto scaling. Statistical analysis was performed using MetaboAnalyst 5.0 web portal.1 Principal component analysis (PCA) was performed for all groups at each time point. Student’s t-test was used to determine significantly altered metabolites (p < 0.05). False discovery rate (FDR) <0.05 and fold change (FC) ≥2 (log2FC ≥ 1 or ≤ −1) were also evaluated by the algorithm. Metabolites and pathway analysis were searched using KEGG (Kyoto Encyclopedia of Genes and Genomes) and HMDB (The Human Metabolome Database) databases.
Table 2 lists the in vitro susceptibility results of A. baumannii isolate 12 to commonly used antibiotics. This isolate is resistant to common antibiotics used in the clinic, including amikacin and polymyxin-B (Table 1). The addition of 4 μg/mL sulbactam did not reduce the MIC of amikacin monotherapy, whereas the polymyxin-B MIC was reduced from 16 to 8 μg/mL in the presence of 4 μg/mL sulbactam. The MIC values of each drug in amikacin/polymyxin-B and amikacin/polymyxin-B/sulbactam combination had significant reduction from their monotherapy MICs.
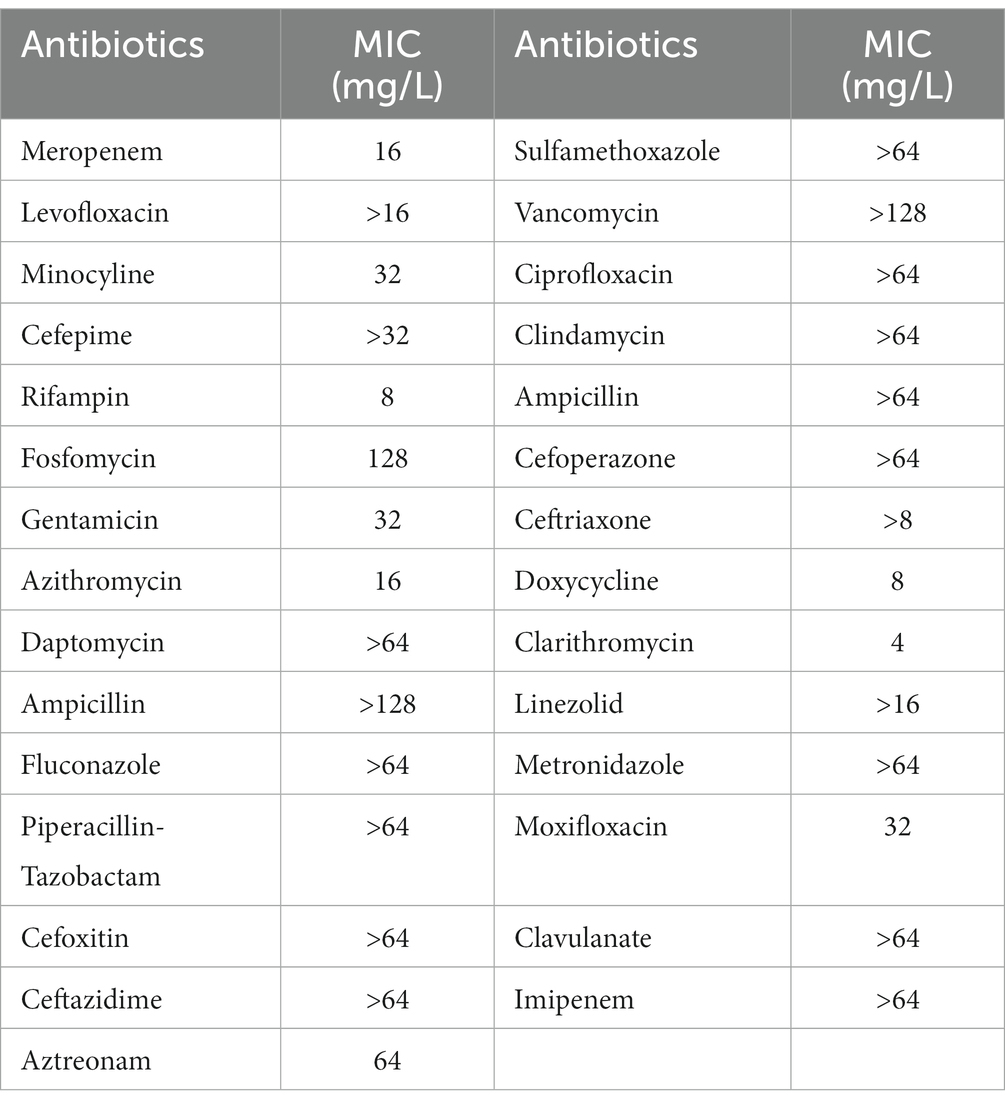
Table 2. Minimum inhibitory concentrations (MIC) of commonly used clinical antibiotics against Acinetobacter baumannii isolate 12.
In the present study, a total of 555 metabolites in bacteria extract were identified by KEGG and HMDB databases. Among them, 73 were associated with known metabolic pathways. PCA was performed to delineate putative metabolites that were affected by differential effects of drugs on the A. baumannii isolate (Figure 1). After 15 min of treatment, polymyxin-B, amikacin, and the combination groups were clustered and were significantly distanced from the control group. However, there was an overlap between polymyxin-B/amikacin and polymyxin-B/amikacin/sulbactam combination groups. Even though there were differences characterized by the distance between the monotherapies and control group after 1 h and 4 h treatment, their distances started to shrink, whereas the clusters representing the two combination therapies were quite distinct from the control group. At 4 h treatment, there was a clear separation between polymyxin-B/amikacin and polymyxin-B/amikacin/sulbactam treatment groups.
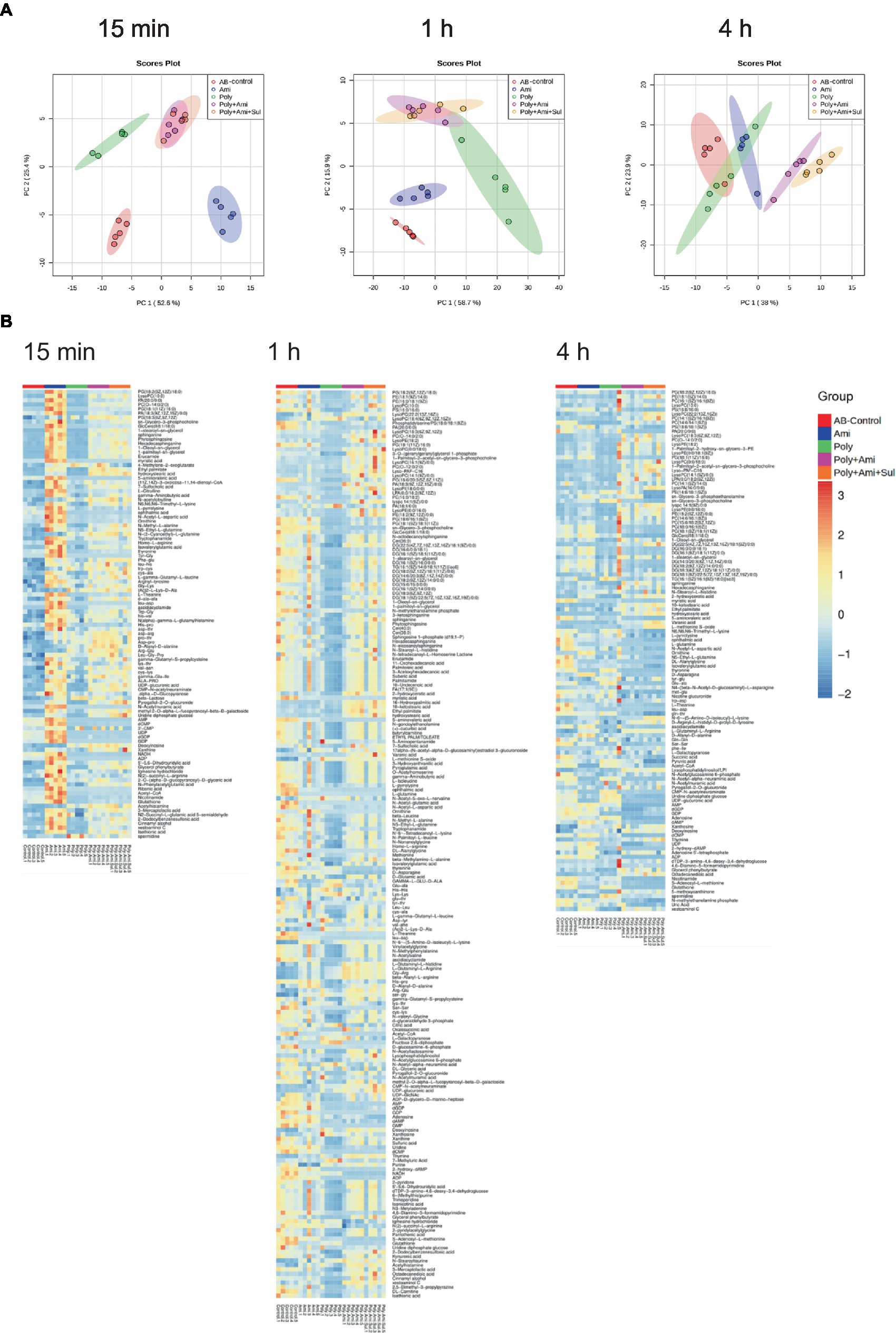
Figure 1. Principal component analysis (A) and heatmap (B) profiles of MDR A. baumannii treated with amikacin and polymyxin-B as monotherapy and in combination with and without sulbactam at 15 min, 1 h and 4 h.
All altered metabolites were grouped by metabolic pathways, which were assigned to lipids, amino sugars, nucleotides, amino acids and central carbon metabolism. The affected metabolic pathways were primarily involved in the metabolisms of lipids, nucleotides and amino acids.
A number of lipid molecules were affected by the combination therapy, as illustrated in Figures 2A,B. Among them, sn-glycero-3-phosphocholine, sn-glycero-3-phosphoethanolamine and N-methylethanolamine phosphate were three important intermediates for outer membrane structure. The level of sn-glycero-3-phosphocholine (log2FC = −3.5 to1.1) was remarkably affected by polymyxin-B/amikacin/sulbactam at the three time points. The level of N-methylethanolamine phosphate was significantly depleted at 1 h and 4 h (log2FC = −4.2 to −3.2). However, the level of sn-glycero-3-phosphoethanolamine (log2FC = −1.1) was reduced only at 4 h.
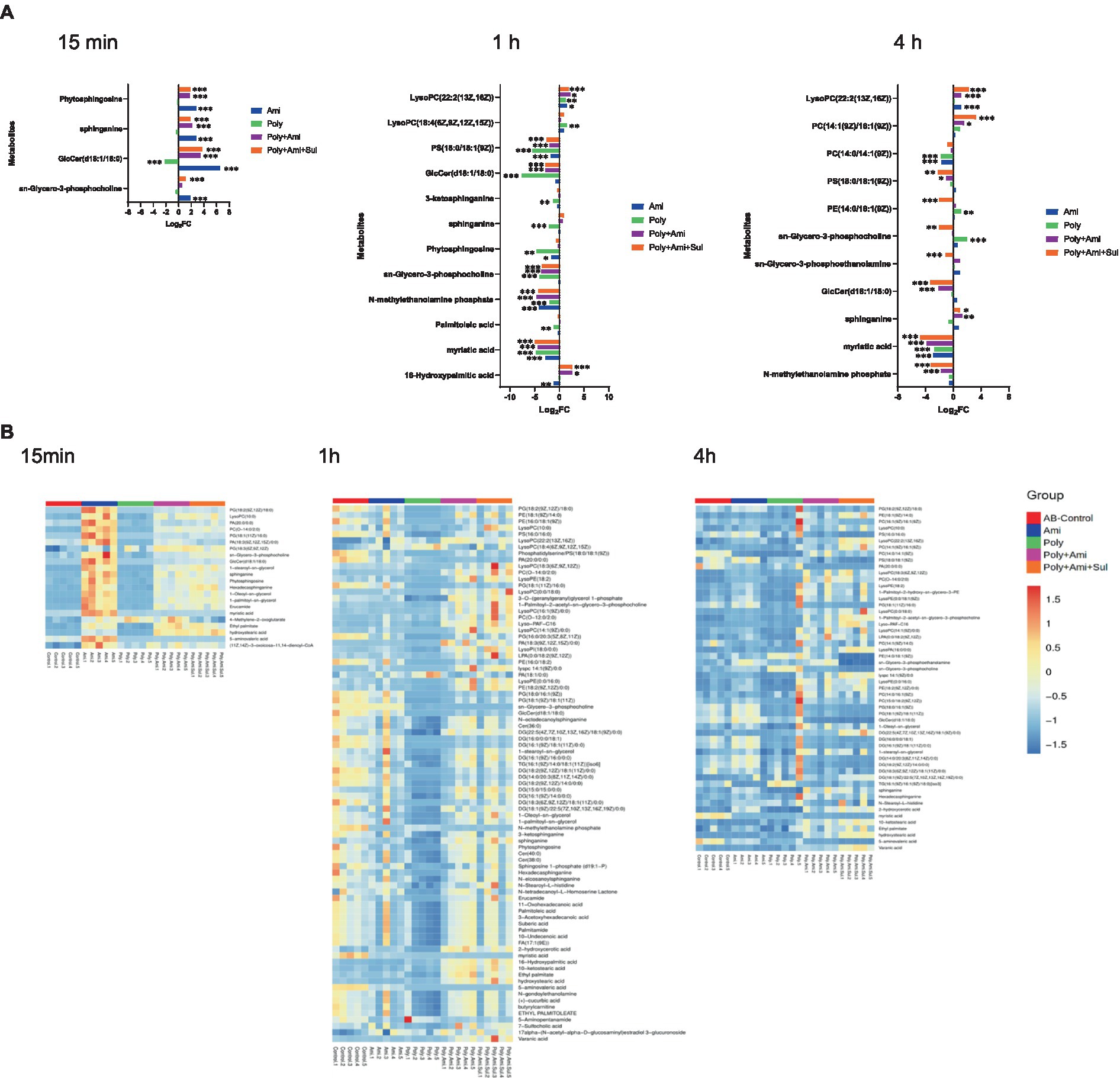
Figure 2. Perturbations of lipid metabolites in A. baumannii treated with antibiotics alone and in combination at 15 min, 1 h and 4 h. (A) Bar charts show the log2FC values of metabolites associated with lipid pathway. (B) Heatmap shows the normalized relative intensity of the significantly disrupted metabolites involved in lipid metabolism (p value < 0.05, log2FC ≤ −1 or log2FC ≥ 1, FDR < 0.05). *p < 0.05; **p < 0.01; ***p < 0.001.
In addition, the polymyxin-B/amikacin/sulbactam combination perturbed the levels of sphinganine and glucosylceramide (d18:1/18:0; log2FC = −3.3 to 3.8), which were two important intermediates associated with the sphingolipid pathway, at 15 min and 4 h, respectively. Perturbation of metabolites associated with the fatty acid pathway occurred after 1 h in all four treatment conditions. Myristic acid and 16-hydroxypalmitic acid were two intermediates in the fatty acid pathway that were affected. The level of myristic acid (log2FC = −4.9 to −4.8) was reduced by polymyxin-B/amikacin/sulbactam combination at 1 h and 4 h. In addition, the level of 16-hydroxypalmitic acid (log2FC = 2.6) was also increased at 1 h. In contrast, the level of palmitoleic acid (log2FC = −1.2) was only depleted by polymyxin-B monotherapy at 1 h.
The results showed that amikacin and polymyxin-B alone caused lipid metabolism disorder at an early stage, but the effect was weakened after 4 h. While the disturbance induced by polymyxin-B/amikacin treatment was similar to that of polymyxin-B/amikacin/sulbactam treatment at 1 h, the effect was weaker in the polymyxin-B/amikacin group than that in the polymyxin-B/amikacin/sulbactam combination at 4 h (Figures 2A,B).
The polymyxin-B/amikacin/sulbactam combination significantly perturbed the levels of UDP-glucuronic acid (UDP-GlcA), CMP-N-acetylneuraminate (CMP-Neu5Ac), and uridine diphosphate glucose (UDP-Glc) (log2FC = −4.0 to 3.9), which are the three essential intermediates involved in amino sugars, lipopolysaccharides and peptidoglycans pathways (Figures 3, 4). The levels of D-glucosamine-6-phosphate (GlcN-6P, log2FC = −2.9) were reduced at 1 h.
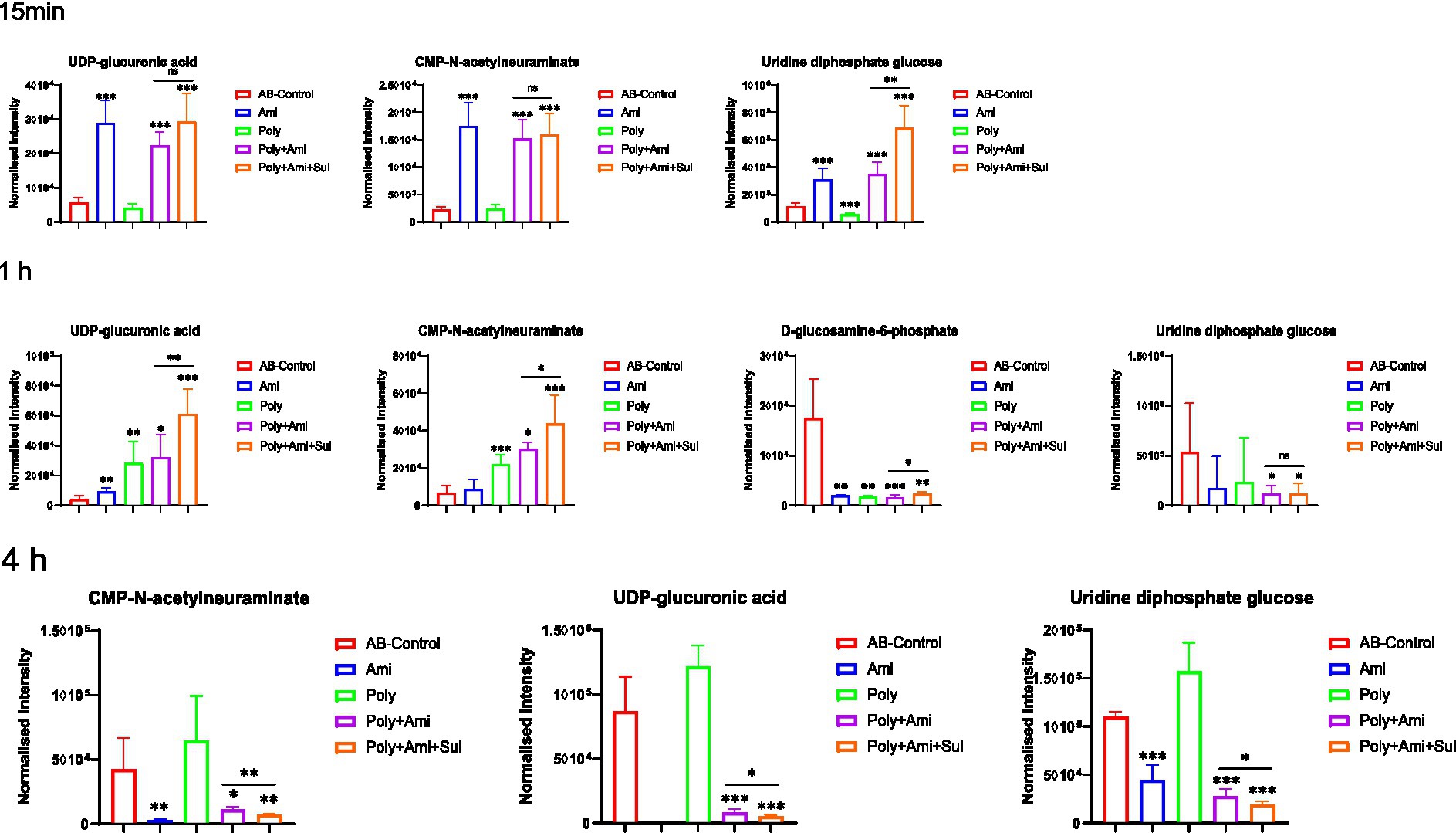
Figure 3. Perturbations of metabolites associated with LPS, peptidoglycan and amino sugar pathway in A. baumannii treated with antibiotics alone and the combination at 15 min, 1 h and 4 h. Box plots show the differences of normalized relative intensity of the metabolites involved in pathways (p value < 0.05, log2FC ≤ −1 or log2FC ≥ 1, FDR < 0.05). *p < 0.05; **p < 0.01; ***p < 0.001.
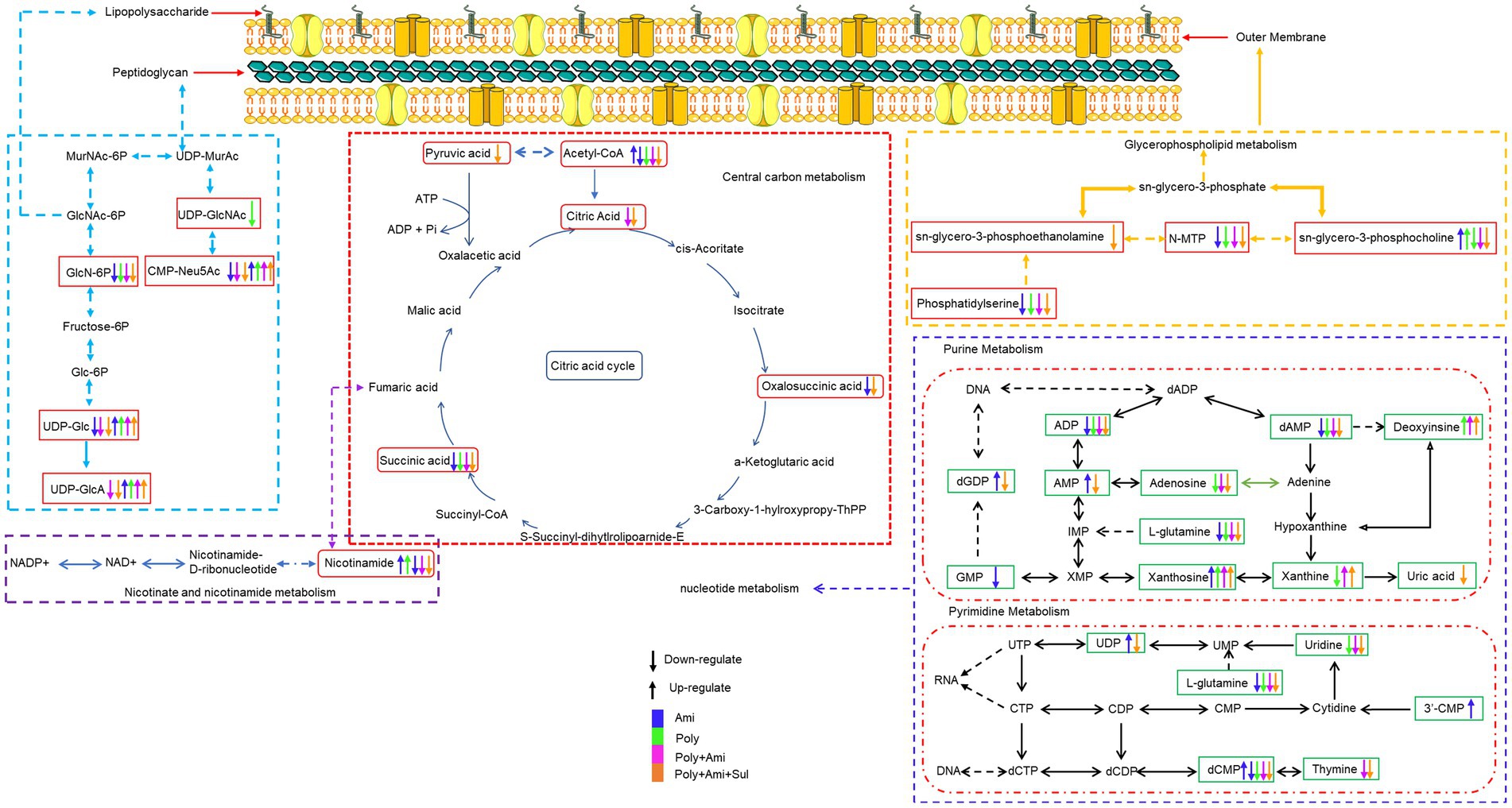
Figure 4. Illustration of affected metabolomic pathways in A. baumannii treated with antibiotics as monotherapy and combination therapy.
The disturbances induced by polymyxin-B/amikacin combination were similar but weakened compared to the polymyxin-B/amikacin/sulbactam combination (Figure 3). For monotherapy, amikacin-induced metabolite disorder was generally weaker than that in the combination treatments at 1 h after administration. Polymyxin-B alone caused metabolic changes primarily at 1 h (Figure 3).
The central carbon metabolism (CCM) pathway is slightly affected at the three timepoints but combination therapies showed greater differences compared with the monotherapies (Figure 5). The triple-combination treatment significantly reduced the level of acetyl-CoA (log2FC = −6.8 to −2.1), which is an essential intermediate for CCM, fatty acid and glutathione pathways. In addition, polymyxin-B/amikacin/sulbactam combination therapy disturbed the levels of citric acid/oxalosuccinic acid (log2FC = 1.2/1.6) and succinic acid/pyruvic acid (log2FC = −2.5/ −2.1) at 1 h and 4 h, respectively. The changes induced by the polymyxin-B/amikacin combination and monotherapy treatments were weaker than that of polymyxin-B/amikacin/sulbactam combination in the CCM pathway (Figure 5).
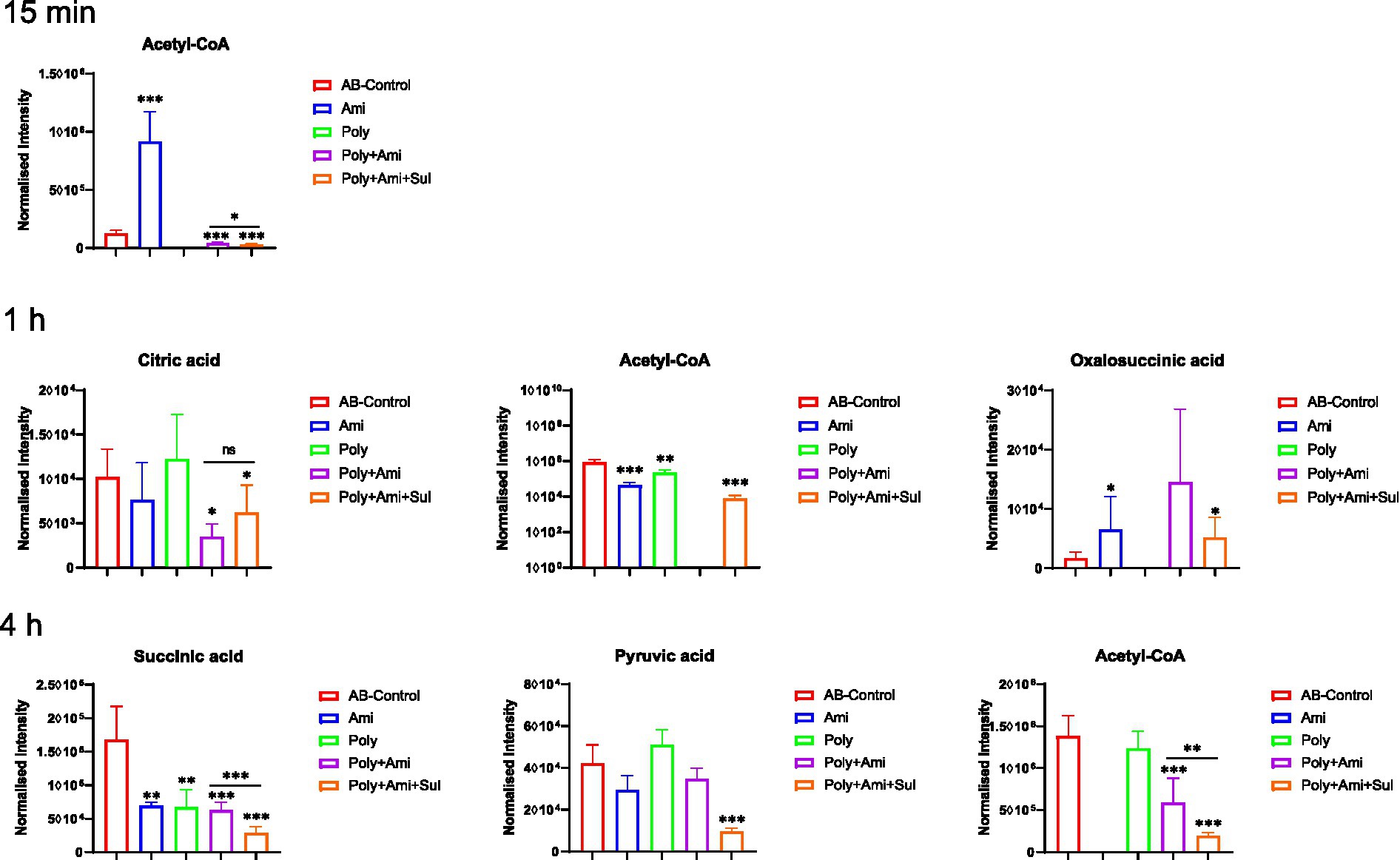
Figure 5. Perturbations of central carbon metabolism in A. baumannii treated with antibiotics alone and the combination at 15 min, 1 h and 4 h. Box plots show the differences of normalized relative intensity of the metabolites involved in CCM pathways (p-value < 0.05, log2FC ≤ −1 or log2FC ≥ 1, FDR < 0.05). *p < 0.05; **p < 0.01; ***p < 0.001.
The affected nucleotide metabolic pathway mainly included pyrimidine and purine. For the pyrimidine metabolism, three key affected metabolites by polymyxin-B/amikacin/sulbactam (log2FC = −4.3/−2.1/1.2 at 1 h and log2FC = −2.9/−1.2/2.3 at 4 h, respectively) and polymyxin-B/amikacin (log2FC = −5.6/−2.6/1.3 at 1 h) combinations were dCMP, uridine and thymine. The magnitude of metabolite changes due to polymyxin-B/amikacin was smaller compared to that of polymyxin-B/amikacin/sulbactam, especially at 15 min and 4 h.
For monotherapy, only amikacin perturbed the levels of dCMP, 3’-CMP and UDP (log2FC = 1.4 to 1.9) at 15 min. Polymyxin-B did not induce obvious disturbance of metabolites related to the pyrimidine metabolic pathway.
Both monotherapies and combination therapies affected the purine metabolic pathway, which primarily occurred after 1 h of treatment. The affected metabolites due to combination therapies were adenosine, AMP, GDP, dGDP, dAMP, ADP, deoxyinosine, uric acid, xanthosine; these changes were of varying degrees depending on time and the type of combination treatment.
For monotherapy, amikacin decreased the levels of NADH, GDP and GMP (log2FC = −1.3 to −3.1), whereas polymyxin-B significantly reduced the levels of 8 metabolites, including NADH, adenosine, dAMP, xanthine, ADP, sulfuric acid, deoxyinosine and xanthosine (log2FC = −5.7 to 2.2) at 1 h. At 4 h, the effect of monotherapy is significantly reduced (Figure 6).
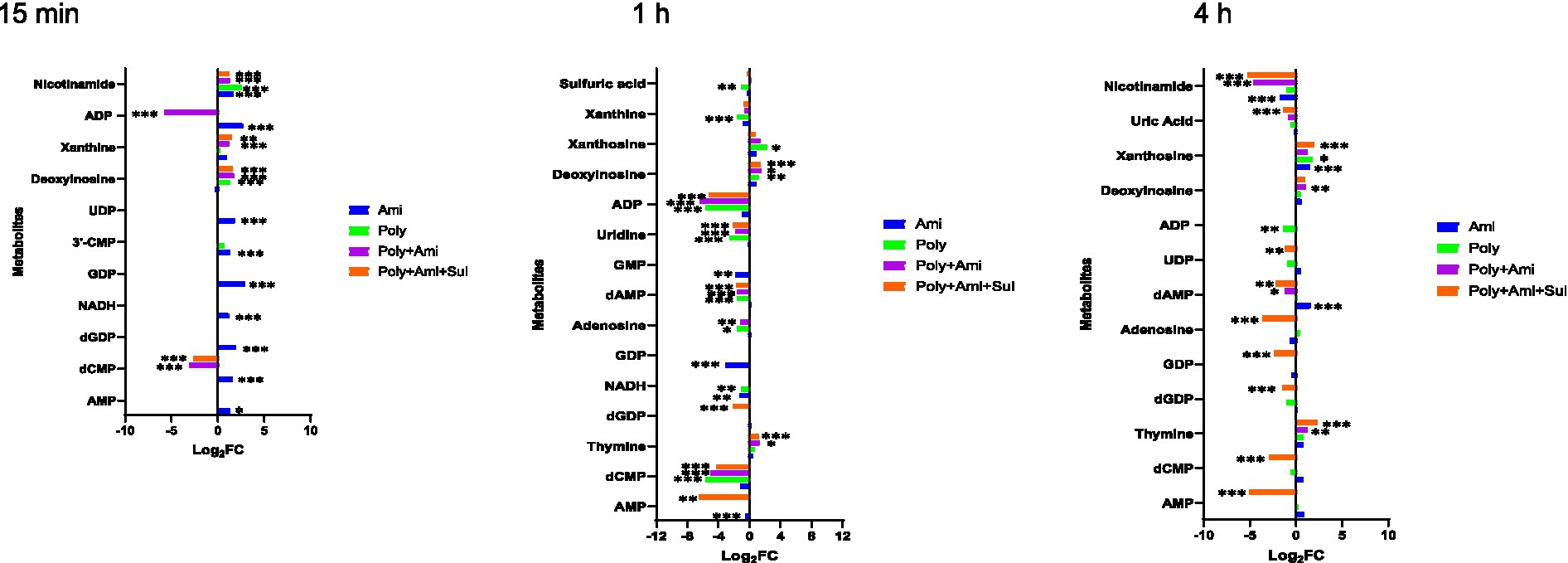
Figure 6. Perturbations of metabolites associated with nucleotide and derivative metabolism in A. baumannii treated with antibiotics alone and the combination at 15 min, 1 h and 4 h. Bar charts show the log2FC values of metabolites associated with pathway (p value < 0.05, log2FC ≤ −1 or log2FC ≥ 1, FDR < 0.05). *p < 0.05; **p < 0.01; ***p < 0.001.
The combination treatments also remarkably reduced the level of nicotinamide (log2FC = −5.2/−4.6), which is a key metabolite involved in nicotinate and nicotinamide metabolic pathway (Figures 4, 5).
A large number of amino acids and peptides were affected by both monotherapies and combination therapies (Figure 7A). The metabolic pathways of the affected metabolites include arginine biosynthesis, glutathione, alanine, aspartate, and glutamate metabolism (Figure 7B).
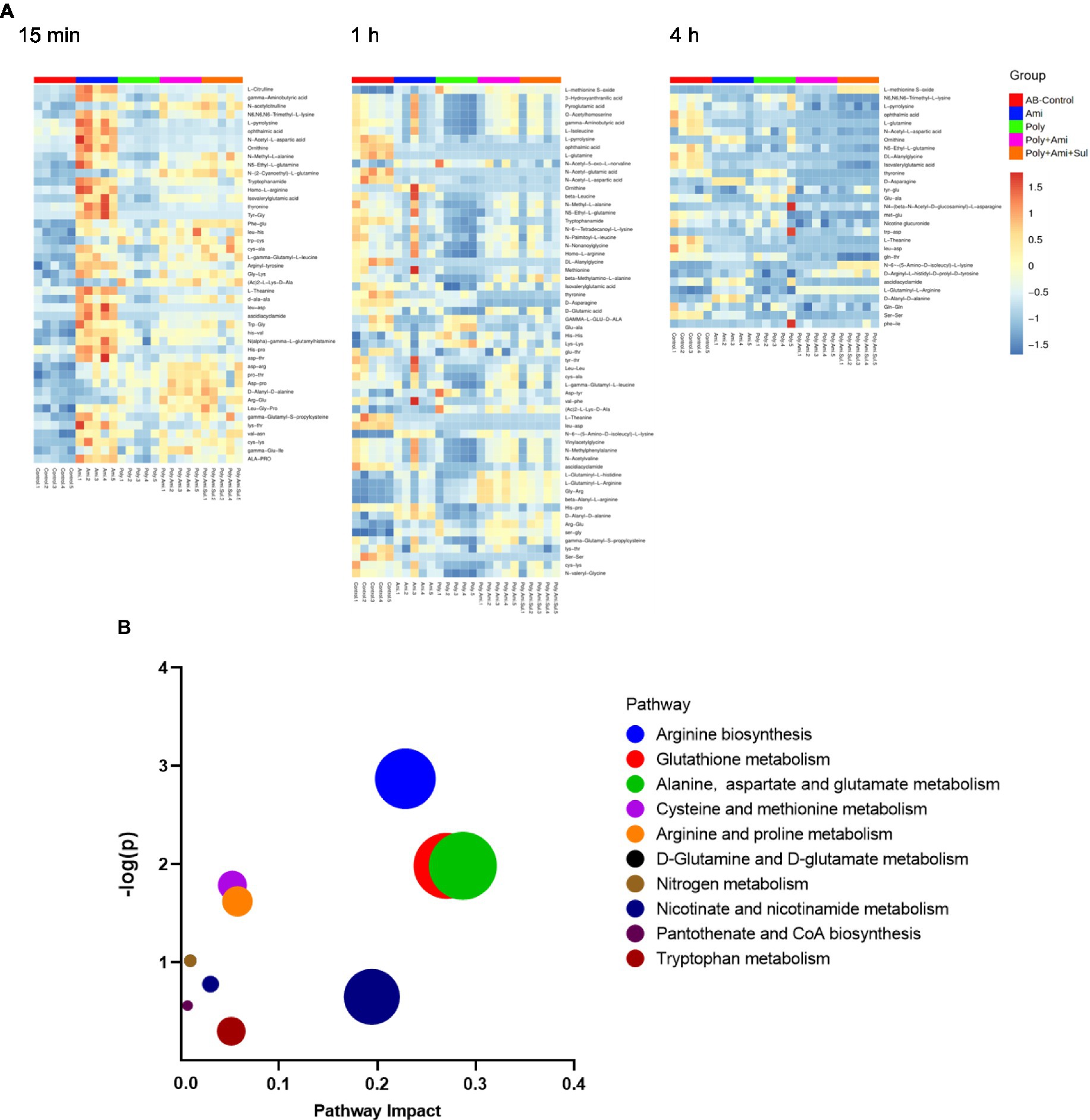
Figure 7. Heat map (A) profiles of normalized relative intensity significantly perturbed metabolites in amino acid and peptide in A. baumannii treated with antibiotics alone and the combination at 15 min, 1 h and 4 h (p value < 0.05, log2FC ≤ −1 or log2FC ≥ 1 and FDR < 0.05). Enrichment analysis (B) of amino acid metabolism pathway.
The effects of the four treatments varied with time. Amikacin was driving the most changes in metabolites at 15 min. The levels of 10 key metabolites including L-citrulline, 5-aminovaleric acid, gamma-aminobutyric acid, N6,N6,N6-trimethyl-L-lysine, L-pyrrolysine, glutathione, ophthalmic acid, N-acetyl-L-aspartic acid, 3-mercaptolactic acid, and N2-succinyl-L-glutamic acid 5-semialdehyde (log2FC = 1.2 to 2.8) were significantly disturbed. At this time point, polymyxin-B/amikacin/sulbactam and polymyxin-B/amikacin treatments induced perturbations of 6 and 5 metabolites (log2FC = −2.3 to 2.5/ log2FC = −2.4 to 2.2), respectively.
At 1 h, polymyxin-B caused changes in a total of 18 metabolites including 5-aminovaleric acid, L-methionine S-oxide, 3-hydroxyanthranilic acid, pyroglutamic acid, O-acetylhomoserine, gamma-aminobutyric acid, L-isoleucine, L-pyrrolysine, ophthalmic acid, L-glutamine, N-acetyl-5-oxo-L-norvaline, N-acetyl-glutamic acid, N-acetyl-L-aspartic acid, pantothenic acid, S-adenosyl-L-methionine, glutathione, kynurenic acid and 3-mercaptolactic acid (log2FC = −6.8 to 1.2). For combination therapies, 10 and 11 key metabolites (log2FC = −5.8 to 1.1/log2FC = −6.3 to 1.0) were significantly affected by polymyxin-B/amikacin and polymyxin-B/amikacin/sulbactam, respectively (Table 3).
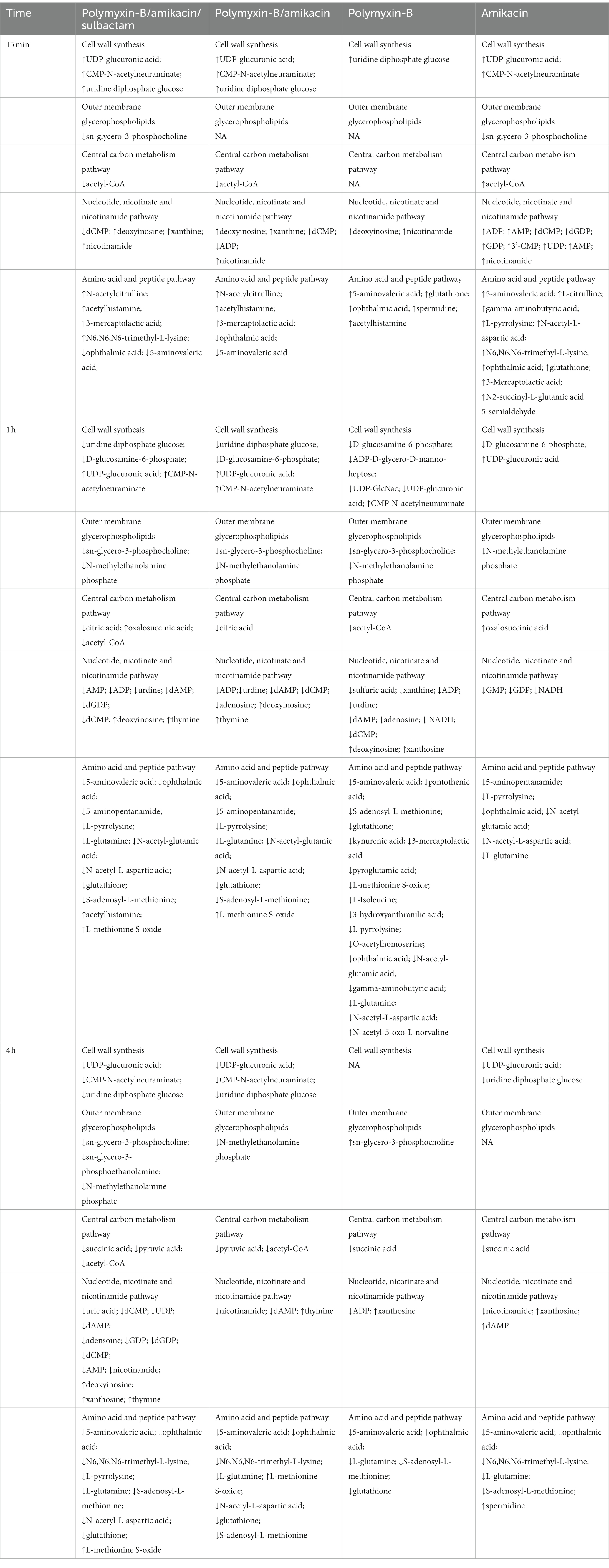
Table 3. Sequence of metabolomic changes in A. baumannii after polymyxin-B and amikacin treatments as monotherapy and in combination with and without sulbactam.
At 4 h, the combination of polymyxin-B/amikacin/sulbactam induced the most disturbance for a total of 9 metabolites including 5-aminovaleric acid, L-methionine-S-oxide, N6,N6,N6-trimethyl-L-lysine, L-pyrrolysine, ophthalmic acid, L-glutamine, N-acetyl-L-aspartic acid, S-adenosyl-L-methionine and glutathione (log2FC = −8.5 to 2.7). The treatment of polymyxin-B/amikacin perturbed 8 metabolites (Table 3). At this time point, the effects of amikacin and polymyxin-B alone, which induced disturbances in the levels of 6 and 5 key metabolites (log2FC = −5.2 to −1.2/log2FC = −2.5 to 1.4), respectively (Table 3) were weaker than that during 15 min and 1 h post-treatment.
This clinical isolate carried OXA-23, aph(3′)-la and other resistance genes, which made it resistant to both sulbactam and amikacin (Umezawa, 1979; Yang et al., 2019). The monotherapy treatment against this MDR isolate would have required a large plasma amikacin exposure. However, the risk of human toxicity increases when the steady-state trough amikacin concentration is greater than 10 mg/L (Dijkstra et al., 2014; Xu et al., 2022). In this isolate, the combination of polymyxin-B/amikacin reduced amikacin MIC value from >128 μg/mL in monotherapy to 2–4 μg/mL with and without sulbactam, respectively (Table 1). Combination therapy confers susceptibility in this MDR isolate to each of the antimicrobial agents in the combination.
In a previous study, we evaluated the time course of bacterial dynamics of several A. baumannii isolates in response to amikacin, polymyxin-B alone and in combination with sulbactam (4 mg/L fixed) using time-kill assays (Zhu et al., 2022a). The results showed that amikacin/polymyxin-B combined with or without sulbactam could inhibit bacterial growth; the bactericidal effect was enhanced after adding sulbactam (4 mg/L). Bacterial growth were inhibited by 4 h in the antibiotic combinations (Zhu et al., 2022a).
Metabolomics was used to elucidate the pathways that resulted in synergistic effects of polymyxin-B and amikacin with or without sulbactam. Disequilibrium of key metabolic pathways was induced by polymyxin-B/amikacin/sulbactam; altered metabolites occurred with lipid, nucleotide, and amino acid metabolisms, as well as CCM and cell wall synthesis. The sequence of events is summarized in Table 3.
Lipopolysaccharide and peptidoglycan are the main components of cell wall. The resistance mechanism of A. baumannii to colistin is mainly due to the modification of lipoprotein A or the loss of LPS; polymyxin-B exerts its effect through electrostatic interaction with LPS. To counteract the effect of polymyxins, A. baumannii can increase its own cell survival by upregulating arginine biosynthesis to weaken the electrostatic interaction between colistin-LPS. Metabolomics and transcriptomics results showed that polymyxin-B monotherapy can reduce the synthesis rate of peptidoglycan and lipoprotein A (Han et al., 2019; Zhu et al., 2019; Zhao et al., 2021).
The effect of combination therapy on lipopolysaccharide and peptidoglycan metabolic pathways occurred earlier than that in monotherapy (Table 3). The combination of polymyxin-B/amikacin/sulbactam reduced the level of glucosamine-6-phosphate (GlcN6P), UDP-Glc and UDP-GlcA, which are important metabolites and precursors of glycosylated protein and lipid biosynthesis and related to cell wall maintenance (Zhao et al., 2014; Warth et al., 2015; Kuang et al., 2016). This may be due to the effect of sulbactam, since sulbactam can disintegrate the cell wall of A. baumannii, allowing companion antibiotics to enter the bacterial cell (Horii et al., 2002; Penwell et al., 2015). This shows that sulbactam in the combination therapy has the advantage of destabilizing cell wall synthesis.
The integrity and stability of the cell outer membrane play an important role in the normal growth of bacteria (Zhang and Rock, 2008). Polymyxin-B can disrupt the outer membrane of gram-negative bacteria, as well as phospholipid exchange, resulting in an increased cell permeability (Velkov et al., 2013; Zhao et al., 2021). However, due to the drug resistance in this isolate, polymyxin-B alone did not have much effect on the metabolites related to the outer membrane pathway. Interestingly, the combination of polymyxin-B/amikacin/sulbactam significantly affected the metabolisms of sn-glycero-3-phosphocholine, sn-glycero-3-phosphoethanolamine and N-methylethanolamine phosphate, which are three essential intermediates involved in glycerophospholipid pathway (Zhang and Rock, 2008; McHenry et al., 2012). This result demonstrated that polymyxin-B/amikacin/sulbactam combination can destabilize bacterial outer membrane to overcome the drug resistance caused by cell membrane modification (Loutet and Valvano, 2011).
As the cell wall and membrane stability of bacteria are destroyed, the permeability and integrity of the outer membrane are no longer stable, allowing for more drugs to enter the bacteria. Amikacin antibacterial mechanism is via the inhibition of protein synthesis that leads to cell death (Taber et al., 1987; Kotra et al., 2000). This mechanism of action is consistent with our result that a large number of amino acids and peptides were affected. Glutathione was significantly decreased by polymyxin-B/amikacin/sulbactam. It is an important marker of oxidative stress response. Previous studies have shown that polymyxins may induce oxidative stress through the formation of hydroxyl radicals and reactive oxygen species, which target DNA, RNA and lipids (Cabiscol et al., 2000; Sampson et al., 2012). The decrease in glutathione level is also an early activation signal of apoptosis; the subsequent generation of oxygen free radicals promotes cell death (Armstrong et al., 2002). Acetyl-CoA, an important intermediate in the synthesis of amino acids and fatty acids (Gest, 1987; Murima et al., 2014; Wolfe, 2015; Hussein et al., 2019b), was significantly disrupted by the triple-antibiotic combination. In addition, the triple-antibiotic combination perturbed some metabolites, which are all critical intermediates in the CCM pathway including tricarboxylic acid (TCA) cycle, pentose phosphate pathway and gluconeogenesis/glycolysis pathway, such as citric acid, oxalosuccinic acid, succinic acid and pyruvic acid (Table 3 and Figure 5).
Nucleotide metabolism pathway is essential for energy, lipid and protein biosynthesis (Moffatt and Ashihara, 2002; Armenta-Medina et al., 2014; Zhao et al., 2021). The triple-antibiotic combination disturbs the stability of pyrimidine and purine metabolism over a longer period of time. This will have a greater impact on the stability and normal growth of bacteria. In addition, nicotinamide as an ATP reserve pool (Hussein et al., 2019a), was also significantly affected by the combination of polymyxin-B/amikacin/sulbactam, suggesting that the energy balance in the bacteria was disoriented. The action of combination therapy was more sustained than that of single-agent administration. The addition of sulbactam provided stronger disruption than that of polymyxin-B/amikacin combination.
Our previous study demonstrated the synergistic effect of another triple-combination therapy consisting of polymyxin-B/meropenm/sulbactam (Menegucci et al., 2019; Fedrigo et al., 2021; Zhang et al., 2022; Zhu et al., 2022b) and demonstrated disruption of the metabolomic profile of a clinical A. baumannii isolate (Zhu et al., 2022c). The metabolites involved in the cell wall synthesis, outer membrane integrity and central carbon metabolism were primarily affected by polymyxin-B/meropenem or polymyxin-B/meropenem/sulbactam combinations. The addition of sulbactam to the double combination induced metabolic disruption at an earlier time. In contrast, the present study replaced meropenem with amikacin; this triple-antibiotic combination with amikacin also disrupted metabolisms of the nucleotides, amino acids and peptides. There is a stark difference in treatment effects for the two triple-antibiotic combinations that may explain why the combination with amikacin resulted in a better bactericidal effect and inhibition of mutant selection for their clinical regimens based on model-simulated parameters (Zhang et al., 2022; Zhu et al., 2022a).
In conclusion, the present study elucidated the effects of combination therapy on a clinical A. baumannii isolate. The metabolic pathway analysis showed that the effect of the combination is both enhanced and sustained. At the early stage, polymyxin-B disturbs the stability of cell membrane, making it easier for amikacin and sulbactam to enter the bacteria. Sulbactam further disrupts cell wall synthesis allowing amikacin to penetrate the cell and alter protein synthesis.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Armenta-Medina, D., Segovia, L., and Perez-Rueda, E. (2014). Comparative genomics of nucleotide metabolism: a tour to the past of the three cellular domains of life. BMC Genomics 15:800. doi: 10.1186/1471-2164-15-800
Armstrong, J. S., Steinauer, K. K., Hornung, B., Irish, J. M., Lecane, P., Birrell, G. W., et al. (2002). Role of glutathione depletion and reactive oxygen species generation in apoptotic signaling in a human B lymphoma cell line. Cell Death Differ. 9, 252–263. doi: 10.1038/sj.cdd.4400959
Avedissian, S. N., Liu, J., Rhodes, N. J., Lee, A., Pais, G. M., Hauser, A. R., et al. (2019). A review of the clinical pharmacokinetics of polymyxin B. Antibiotics 8:31. doi: 10.3390/antibiotics8010031
Cabiscol, E., Tamarit, J., and Ros, J. (2000). Oxidative stress in bacteria and protein damage by reactive oxygen species. Int. Microbiol. 3, 3–8.
Clark, N. M., Zhanel, G. G., and Lynch, J. P. 3rd (2016). Emergence of antimicrobial resistance among Acinetobacter species: a global threat. Curr. Opin. Crit. Care 22, 491–499. doi: 10.1097/MCC.0000000000000337
CLSI (2020). Performance standards for antimicrobial susceptibility testing-thirty edition: M100. Wayne, PA: Clinical and Laboratory Standards Institute.
Deng, Y., Chen, L., Yue, M., Huang, X., Yang, Y., and Yu, H. (2022). Sulbactam combined with tigecycline improves outcomes in patients with severe multidrug-resistant Acinetobacter baumannii pneumonia. BMC Infect. Dis. 22:795. doi: 10.1186/s12879-022-07778-5
Dijkshoorn, L., Nemec, A., and Seifert, H. (2007). An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 5, 939–951. doi: 10.1038/nrmicro1789
Dijkstra, J. A., Sturkenboom, M. G. G., Van Hateren, K., Koster, R. A., Greijdanus, B., and Alffenaar, J. W. C. (2014). Quantification of amikacin and kanamycin in serum using a simple and validated LC-MS/MS method. Bioanalysis 6, 2125–2133. doi: 10.4155/bio.14.191
Fedrigo, N. H., Shinohara, D. R., Mazucheli, J., Nishiyama, S. A. B., Carrara-Marroni, F. E., Martins, F. S., et al. (2021). Pharmacodynamic evaluation of suppression of in vitro resistance in Acinetobacter baumannii strains using polymyxin B-based combination therapy. Sci. Rep. 11:11339. doi: 10.1038/s41598-021-90709-2
Freire, M. P., De Oliveira Garcia, D., Garcia, C. P., Campagnari Bueno, M. F., Camargo, C. H., Kono Magri, A. S. G., et al. (2016). Bloodstream infection caused by extensively drug-resistant Acinetobacter baumannii in cancer patients: high mortality associated with delayed treatment rather than with the degree of neutropenia. Clin. Microbiol. Infect. 22, 352–358. doi: 10.1016/j.cmi.2015.12.010
Gest, H. (1987). Evolutionary roots of the citric acid cycle in prokaryotes. Biochem. Soc. Symp. 54, 3–16.
Girardello, R., Visconde, M., Cayo, R., Figueiredo, R. C., Mori, M. A., Lincopan, N., et al. (2017). Diversity of polymyxin resistance mechanisms among Acinetobacter baumannii clinical isolates. Diagn. Microbiol. Infect. Dis. 87, 37–44. doi: 10.1016/j.diagmicrobio.2016.10.011
Gootz, T. D., and Marra, A. (2008). Acinetobacter baumannii: an emerging multidrug-resistant threat. Expert Rev. Anti-Infect. Ther. 6, 309–325. doi: 10.1586/14787210.6.3.309
Han, M. L., Zhu, Y., Creek, D. J., Lin, Y. W., Gutu, A. D., Hertzog, P., et al. (2019). Comparative metabolomics and transcriptomics reveal multiple pathways associated with polymyxin killing in Pseudomonas aeruginosa. mSystems 4:e00149-18. doi: 10.1128/mSystems.00149-18
Horii, T., Mase, K., Suzuki, Y., Kimura, T., Ohta, M., Maekawa, M., et al. (2002). Antibacterial activities of beta-lactamase inhibitors associated with morphological changes of cell wall in Helicobacter pylori. Helicobacter 7, 39–45. doi: 10.1046/j.1523-5378.2002.00054.x
Hussein, M., Han, M. L., Zhu, Y., Zhou, Q., Lin, Y. W., Hancock, R. E. W., et al. (2019a). Metabolomics study of the synergistic killing of polymyxin B in combination with amikacin against polymyxin-susceptible and -resistant Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 64:e01587-19. doi: 10.1128/AAC.01587-19
Hussein, M., Han, M. L., Zhu, Y., Zhou, Q., and Velkov, T. (2019b). Metabolomics study of the synergistic killing of polymyxin B in combination with amikacin against polymyxin-susceptible and-resistant Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 64:e01587-19. doi: 10.1128/AAC.01587-19
Kotra, L. P., Haddad, J., and Mobashery, S. (2000). Aminoglycosides: perspectives on mechanisms of action and resistance and strategies to counter resistance. Antimicrob. Agents Chemother. 44, 3249–3256. doi: 10.1128/AAC.44.12.3249-3256.2000
Kuang, B., Zhao, X., Zhou, C., Zeng, W., Ren, J., Ebert, B., et al. (2016). Role of UDP-glucuronic acid decarboxylase in xylan biosynthesis in arabidopsis. Mol. Plant 9, 1119–1131. doi: 10.1016/j.molp.2016.04.013
Li, J., Nation, R. L., Turnidge, J. D., Milne, R. W., Coulthard, K., Rayner, C. R., et al. (2006). Colistin: the re-emerging antibiotic for multidrug-resistant gram-negative bacterial infections. Lancet Infect. Dis. 6, 589–601. doi: 10.1016/s1473-3099(06)70580-1
Loutet, S. A., and Valvano, M. A. (2011). Extreme antimicrobial peptide and polymyxin B resistance in the genus burkholderia. Front. Microbiol. 2:159. doi: 10.3389/fmicb.2011.00159
Maifiah, M. H., Creek, D. J., Nation, R. L., Forrest, A., Tsuji, B. T., Velkov, T., et al. (2017). Untargeted metabolomics analysis reveals key pathways responsible for the synergistic killing of colistin and doripenem combination against Acinetobacter baumannii. Sci. Rep. 7:45527. doi: 10.1038/srep45527
Mchenry, A. J., Sciacca, M. F. M., Brender, J. R., and Ramamoorthy, A. (2012). Does cholesterol suppress the antimicrobial peptide induced disruption of lipid raft containing membranes? BBA-Biomembranes 1818, 3019–3024. doi: 10.1016/j.bbamem.2012.07.021
Menegucci, T. C., Fedrigo, N. H., Lodi, F. G., Albiero, J., Nishiyama, S. A. B., Mazucheli, J., et al. (2019). Pharmacodynamic effects of sulbactam/meropenem/polymyxin-B combination against extremely drug resistant Acinetobacter baumannii using checkerboard information. Microb. Drug Resist. 25, 1266–1274. doi: 10.1089/mdr.2018.0283
Moffatt, B. A., and Ashihara, H. (2002). Purine and pyrimidine nucleotide synthesis and metabolism. Arabidopsis Book 1:e0018. doi: 10.1199/tab.0018
Murima, P., Mckinney, J. D., and Pethe, K. (2014). Targeting bacterial central metabolism for drug development. Chem. Biol. 21, 1423–1432. doi: 10.1016/j.chembiol.2014.08.020
Oo, C., and Sy, S. K. B. (2018). Fixed-dose combinations: a potential means to boost drug development for selected drugs. Drug Discov. Today 23, 457–459. doi: 10.1016/j.drudis.2017.11.001
Oo, C., and Sy, S. K. B. (2020). Learning and augmenting natural processes: potential means of combating antimicrobial resistance from a drug R&D perspective. Drug Discov. Today 25, 1–3. doi: 10.1016/j.drudis.2019.07.012
Penwell, W. F., Shapiro, A. B., Giacobbe, R. A., Gu, R. F., Gao, N., Thresher, J., et al. (2015). Molecular mechanisms of sulbactam antibacterial activity and resistance determinants in Acinetobacter baumannii. Antimicrob. Agents Chemother. 59, 1680–1689. doi: 10.1128/AAC.04808-14
Perez, F., Hujer, A. M., Hujer, K. M., Decker, B. K., Rather, P. N., and Bonomo, R. A. (2007). Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 51, 3471–3484. doi: 10.1128/AAC.01464-06
Poirel, L., Jayol, A., and Nordmann, P. (2017). Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin. Microbiol. Rev. 30, 557–596. doi: 10.1128/CMR.00064-16
Sader, H. S., Mendes, R. E., Kimbrough, J. H., Kantro, V., and Castanheira, M. (2023). Impact of the recent clinical and laboratory standards institute breakpoint changes on the antimicrobial Spectrum of aminoglycosides and the activity of Plazomicin against multidrug-resistant and carbapenem-resistant enterobacterales from United States medical centers. Open Forum Infect. Dis. 10:ofad058. doi: 10.1093/ofid/ofad058
Sampson, T. R., Liu, X., Schroeder, M. R., Kraft, C. S., Burd, E. M., and Weiss, D. S. (2012). Rapid killing of Acinetobacter baumannii by polymyxins is mediated by a hydroxyl radical death pathway. Antimicrob. Agents Chemother. 56, 5642–5649. doi: 10.1128/AAC.00756-12
Satlin, M. J., Lewis, J. S., Weinstein, M. P., Patel, J., Humphries, R. M., Kahlmeter, G., et al. (2020). Clinical and laboratory standards institute and European committee on antimicrobial susceptibility testing position statements on polymyxin B and colistin clinical breakpoints. Clin. Infect. Dis. 71, e523–e529. doi: 10.1093/cid/ciaa121
Sengstock, D. M., Thyagarajan, R., Apalara, J., Mira, A., Chopra, T., and Kaye, K. S. (2010). Multidrug-resistant Acinetobacter baumannii: an emerging pathogen among older adults in community hospitals and nursing homes. Clin. Infect. Dis. 50, 1611–1616. doi: 10.1086/652759
Sharma, R., Patel, S., Abboud, C., Diep, J., Ly, N. S., Pogue, J. M., et al. (2017). Polymyxin B in combination with meropenem against carbapenemase-producing Klebsiella pneumoniae: pharmacodynamics and morphological changes. Int. J. Antimicrob. Agents 49, 224–232. doi: 10.1016/j.ijantimicag.2016.10.025
Taber, H. W., Mueller, J. P., Miller, P. F., and Arrow, A. S. (1987). Bacterial uptake of aminoglycoside antibiotics. Microbiol. Rev. 51, 439–457. doi: 10.1128/mr.51.4.439-457.1987
Teo, J., Lim, T. P., Hsu, L. Y., Tan, T. Y., Sasikala, S., Hon, P. Y., et al. (2015). Extensively drug-resistant Acinetobacter baumannii in a Thai hospital: a molecular epidemiologic analysis and identification of bactericidal polymyxin B-based combinations. Antimicrob. Resist. Infect. Control 4:2. doi: 10.1186/s13756-015-0043-x
Umezawa, H. (1979). Studies on aminoglycoside antibiotics: enzymic mechanism of resistance and genetics. Jpn. J. Antibiot. 32, S1–S14.
Velkov, T., Roberts, K. D., Nation, R. L., Thompson, P. E., and Li, J. (2013). Pharmacology of polymyxins: new insights into an 'old' class of antibiotics. Future Microbiol. 8, 711–724. doi: 10.2217/fmb.13.39
Wang, J., Stegger, M., Moodley, A., and Yang, M. (2023). Drug combination of ciprofloxacin and polymyxin B for the treatment of multidrug-resistant Acinetobacter baumannii infections: a drug pair limiting the development of resistance. Pharmaceutics 15:720. doi: 10.3390/pharmaceutics15030720
Warth, B., Siegwart, G., Lemmens, M., Krska, R., Adam, G., and Schuhmacher, R. (2015). Hydrophilic interaction liquid chromatography coupled with tandem mass spectrometry for the quantification of uridine diphosphate-glucose, uridine diphosphate-glucuronic acid, deoxynivalenol and its glucoside: in-house validation and application to wheat. J. Chromatogr. A 1423, 183–189. doi: 10.1016/j.chroma.2015.10.070
Wolfe, A. J. (2015). Glycolysis for microbiome generation. Microbiol. Spectr. 3:14. doi: 10.1128/microbiolspec.MBP-0014-2014
Xu, L., Cheng, X., Zhu, G., Hu, J., Li, Q., and Fan, G. (2022). Therapeutic drug monitoring of amikacin: quantification in plasma by liquid chromatography-tandem mass spectrometry and work experience of clinical pharmacists. Eur. J. Hosp. Pharm. 29, e77–e82. doi: 10.1136/ejhpharm-2021-003049
Yang, Y. X., Xu, Q. Y., Li, T. T., Fu, Y., Shi, Y., Lan, P., et al. (2019). OXA-23 is a prevalent mechanism contributing to sulbactam resistance in diverse Acinetobacter baumannii clinical strains. Antimicrob. Agents Chemother. 63:e01676-18. doi: 10.1128/AAC.01676-18
Zhang, J., Diao, S., Liu, Y., Wang, H., Liu, Y., Zhu, S., et al. (2022). The combination effect of meropenem/sulbactam/polymyxin-B on the pharmacodynamic parameters for mutant selection windows against carbapenem-resistant Acinetobacter baumannii. Front. Microbiol. 13:1024702. doi: 10.3389/fmicb.2022.1024702
Zhang, Y. M., and Rock, C. O. (2008). Membrane lipid homeostasis in bacteria. Nat. Rev. Microbiol. 6, 222–233. doi: 10.1038/nrmicro1839
Zhang, J., Song, C., Wu, M., Yue, J., Zhu, S., Zhu, P., et al. (2023). Physiologically-based pharmacokinetic modeling to inform dosing regimens and routes of administration of rifampicin and colistin combination against Acinetobacter baumannii. Eur. J. Pharm. Sci. 185:106443. doi: 10.1016/j.ejps.2023.106443
Zhao, Y., Chen, H., Du, F., Yasmeen, A., Dong, J., Cui, X., et al. (2014). Signal amplification of glucosamine-6-phosphate based on ribozyme glmS. Biosens. Bioelectron. 62, 337–342. doi: 10.1016/j.bios.2014.06.067
Zhao, J., Han, M. L., Zhu, Y., Lin, Y. W., Wang, Y. W., Lu, J., et al. (2021). Comparative metabolomics reveals key pathways associated with the synergistic activity of polymyxin B and rifampicin combination against multidrug-resistant Acinetobacter baumannii. Biochem. Pharmacol. 184:114400. doi: 10.1016/j.bcp.2020.114400
Zhao, J., Zhu, Y., Lin, Y. W., Yu, H., Wickremasinghe, H., Han, J., et al. (2022). Polymyxin dose tunes the evolutionary dynamics of resistance in multidrug-resistant Acinetobacter baumannii. Clin. Microbiol. Infect. 28:1026.e1. doi: 10.1016/j.cmi.2022.02.043
Zhu, S., Song, C., Zhang, J., Diao, S., Heinrichs, T. M., Martins, F. S., et al. (2022a). Effects of amikacin, polymyxin-B, and sulbactam combination on the pharmacodynamic indices of mutant selection against multi-drug resistant Acinetobacter baumannii. Front. Microbiol. 13:1013939. doi: 10.3389/fmicb.2022.1013939
Zhu, S., Zhang, J., Lv, Z., Zhu, P., Oo, C., Yu, M., et al. (2022b). Prediction of tissue exposures of meropenem, colistin, and sulbactam in pediatrics using physiologically based pharmacokinetic modeling. Clin. Pharmacokinet. 61, 1427–1441. doi: 10.1007/s40262-022-01161-y
Zhu, S., Zhang, J., Song, C., Liu, Y., Oo, C., Heinrichs, M. T., et al. (2022c). Metabolomic profiling of polymyxin-B in combination with meropenem and sulbactam against multi-drug resistant Acinetobacter baumannii. Front. Microbiol. 13:1013934. doi: 10.3389/fmicb.2022.1013934
Zhu, Y., Zhao, J., Maifiah, M. H. M., Velkov, T., Schreiber, F., and Li, J. (2019). Metabolic responses to Polymyxin treatment in Acinetobacter baumannii ATCC 19606: integrating transcriptomics and metabolomics with genome-scale metabolic modeling. mSystems 4:e00157-18. doi: 10.1128/mSystems.00157-18
Keywords: Acinetobacter baumannii , metabolomics, polymyxin-B, amikacin, sulbactam significance
Citation: Zhu S, Yue J, Wang X, Zhang J, Yu M, Zhan Y, Zhu Y, Sy SKB and Lv Z (2023) Metabolomics revealed mechanism for the synergistic effect of sulbactam, polymyxin-B and amikacin combination against Acinetobacter baumannii. Front. Microbiol. 14:1217270. doi: 10.3389/fmicb.2023.1217270
Received: 16 May 2023; Accepted: 13 June 2023;
Published: 29 June 2023.
Edited by:
Je Chul Lee, Kyungpook National University, Republic of KoreaReviewed by:
Chulhee Choi, Chungnam National University, Republic of KoreaCopyright © 2023 Zhu, Yue, Wang, Zhang, Yu, Zhan, Zhu, Sy and Lv. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuanchao Zhan, emhhbnl1YW5jaGFvQG91Yy5lZHUuY24=; Zhihua Lv, bHZ6aGlodWFAb3VjLmVkdS5jbg==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.