- 1College of Veterinary Medicine, Guangdong Laboratory for Lingnan Modern Agriculture, South China Agricultural University, Guangzhou, China
- 2Guangdong Provincial Key Laboratory of Zoonosis Prevention and Control, Guangzhou, China
- 3Guangdong Provincial Institution of Public Health, Guangdong Provincial Center for Disease Control and Prevention, Guangzhou, China
Waterfowl, such as ducks, are natural hosts for avian influenza viruses (AIVs) and act as a bridge for transmitting the virus to humans or susceptible chickens. Since 2013, chickens and ducks have been threatened by waterfowl-origin H5N6 subtype AIVs in China. Therefore, it is necessary to investigate the genetic evolution, transmission, and pathogenicity of these viruses. In this study, we determined the genetic characteristics, transmission, and pathogenicity of waterfowl-origin H5N6 viruses in southern China. The hemagglutinin (HA) genes of H5N6 viruses were classified into the MIX-like branch of clade 2.3.4.4h. The neuraminidase (NA) genes belonged to the Eurasian lineage. The PB1 genes were classified into MIX-like and VN 2014-like branches. The remaining five genes were clustered into the MIX-like branch. Therefore, these viruses belonged to different genotypes. The cleavage site of the HA proteins of these viruses was RERRRKR/G, a molecular characteristic of the H5 highly pathogenic AIV. The NA stalk of all H5N6 viruses contained 11 amino acid deletions at residues 58–68. All viruses contained 627E and 701D in the PB2 proteins, which were molecular characteristics of typical bird AIVs. Furthermore, this study showed that Q135 and S23 viruses could replicate systematically in chickens and ducks. They did not cause death in ducks but induced mild clinical signs in them. All the infected chickens showed severe clinical signs and died. These viruses were shed from the digestive and respiratory tracts and transmitted horizontally in chickens and ducks. Our results provide valuable information for preventing H5N6 avian influenza outbreaks.
1. Introduction
Avian influenza viruses (AIVs) can infect different types of animals, such as ducks, geese, chickens, quails, parrots, condors, pigs, horses, cats, dogs, tigers, seals, and humans (Buranathai et al., 2007; Jiao et al., 2012a,b,c; Long et al., 2019; Zhu et al., 2019; Zhuang et al., 2019; Puryear et al., 2023). As natural hosts for AIVs, ducks usually do not show clinical signs of AIV infection. Most AIV subtypes can be isolated from ducks, which facilitates the genetic recombination of the virus. Ducks often come into contact with terrestrial birds and even humans, which increases the chances of cross-host transmission of the AIVs (Cui et al., 2020).
In 1996, the first H5N1 subtype of highly pathogenic avian influenza virus (HPAIV) was discovered in domestic geese in Guangdong, south China (Xu et al., 1999). H5 HPAIVs have continued to circulate in birds in over 70 countries. The WHO/OIE/FAO classified the H5 AIV’s HA genes as Clades 0–9 (WHO, 2012). In 2006, clade 2 was subdivided into 2.1–2.5 (Neumann et al., 2010). In 2008, HA genes of clade 2 H5 virus were further divided into subclades 2.1.1–2.3.4. Since 2010, H5N1 HPAIV from clade 2.3.4 has recombined with many NA subtypes of viruses from waterfowl (including subtypes N2, N3, N5, N6, and N8) to produce several new viral subtypes. These viruses have been divided into four subclades, 2.3.4.1–2.3.4.4 (Bi et al., 2016; Du et al., 2017; Cui et al., 2020). The clade 2.3.4.4 was further separated into eight subclades, namely clades 2.3.4.4a–2.3.4.4h, owing to the evolution and spread of the virus worldwide (Gu et al., 2022). Clade 2.3.4.4a mainly refers to the H5N6 viruses that appeared in China in 2014. Clades 2.3.4.4b and 2.3.4.4c comprise H5N8 viruses that broke out in Eurasia and Africa between 2014 and 2018. Clade 2.3.4.4c includes the H5N8 AIVs that caused an avian flu outbreak in South Korea in 2014 and subsequently appeared in America and Europe and the H5N2 viruses that caused poultry outbreaks in the United States between 2014 and 2015. Clade 2.3.4.4e mainly refers to viruses that appeared in South Korea and Japan between 2016 and 2017. The viruses in Clade 2.3.4.4f and Clade 2.3.4.4 g were mainly H5N1 and H5N6 viruses found in China and the neighboring southwestern countries. H5N6 viruses of clades 2.3.4.4d and 2.3.4.4h were also isolated from samples in China (Cui et al., 2020; Li et al., 2020).
In April 2014, the first outbreak of H5N6 avian influenza in domestic poultry was recorded in Sichuan, China (Qu et al., 2019). Furthermore, H5N6 HPAIV has been widely concerned after a fatal human infection caused by a novel H5N6 virus in Sichuan, China, in 2014 (Pan et al., 2016). From 2014 to 2022, H5N6 AIVs caused 72 outbreaks in China, resulting in the death (or culling) of approximately 230,000 birds (Song et al., 2019; Siyu et al., 2023). The birds affected by the above outbreaks included chickens, ducks, geese, quails and wild birds such as wild geese, swans, mallards, northern shovelers, ferruginous pochards, great crested grebes, gadwalls and Eurasian wigeon (Cui et al., 2020). To prevent outbreaks of avian influenza outbreaks, we must continuously monitor the evolution of the H5N6 viruses and evaluate their pathogenicity and transmission.
Here, we isolated eight H5N6 AIVs from waterfowl during a routine surveillance of live poultry markets in southern China between 2017 and 2018. We analyzed the genetic characteristics of these viruses and selected the Q135 and S23 viruses to assess the pathogenicity and transmission of H5N6 AIVs in ducks and chickens.
2. Materials and methods
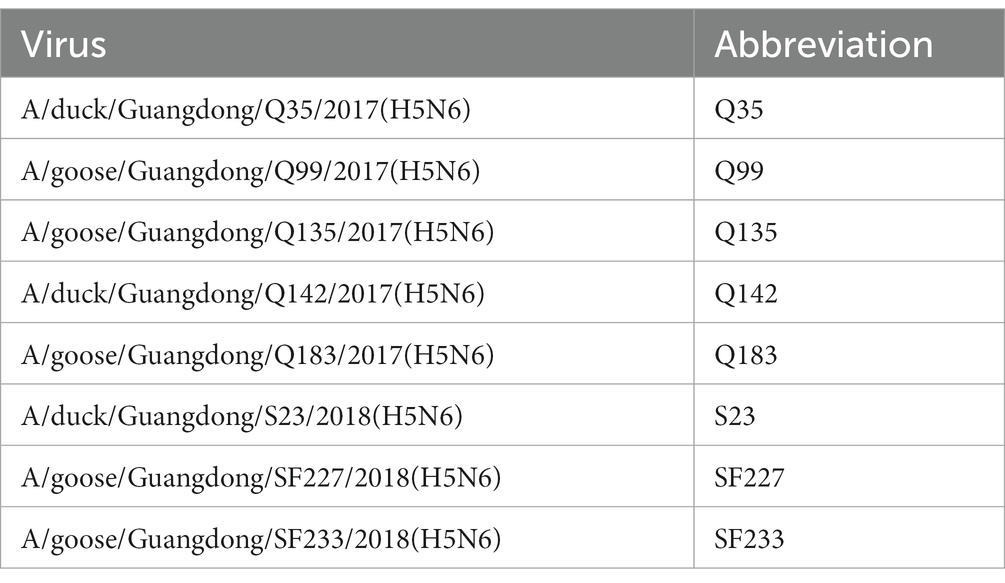
2.1. Viruses
As shown in Table 1, eight H5N6 HPAIVs (Q35, Q99, Q135, Q142, Q183, S23, SF227, and SF233) were isolated from waterfowl swabs from different live poultry markets of Guangdong Province, South China between 2017 and 2018. All swabs were collected in 1 mL of PBS, supplemented with penicillin and streptomycin. Subsequently, swab samples were shaken well and centrifuged at 10,000 g for 5 min. Then the supernatant was inoculated into 9-day-old SPF chicken embryo eggs that were cultured at 37°C for approximately 30 h to obtain viral allantoic fluid. The harvested allantoic fluid was centrifuged, dispensed into tubes, and stored at −80°C. Virus purification and 50% egg infective dose (EID50) determination were performed according to previous study (Thakur and Fezio, 1981; Jiao et al., 2018).
2.2. Phylogenetic analysis
Viral RNA was extracted from the allantoic fluid using the RNA extraction kit (Vazyme, China) and reverse transcribed using reverse transcriptase (M-MLV; Vazyme). Eight AIV genes were amplified using universal primers (Wu et al., 2020). Purification of polymerase chain reaction products was accomplished using the DNA purification kit (ThermoFisher Scientific, United States). The polymerase chain reaction products were sequenced by Tsingke Biotechnology Co., Ltd. Sequencing data were compiled using the SeqMan program from Lasergene 7 (DNASTAR, Inc.). Phylogenetic trees of all viral genes were constructed using MEGA10.2 software (Sinauer Associates, Inc.), using the neighbor-joining method and the maximum complex likelihood model. The trees were assessed for reliability using 1,000 bootstrap replicates (Qu et al., 2019).
The accession numbers for the sequences of our H5N6 viruses are available from NCBI GenBank (OQ829384–OQ829391, OQ829392–OQ829399, OQ829408–OQ829415, OQ829425–OQ829432, OQ829632–OQ829639, OQ830450–OQ830457, OQ830486–OQ830493, and OQ830564–OQ830571).
2.3. Duck experiments
Seven-day-old, healthy, nonimmune Muscovy ducks were purchased from a farm in Guangdong, China, and raised in isolators. The ducks were serologically negative for avian influenza virus, monitored using the hemagglutination-inhibition (HI) test once a week for 3 weeks. Three weeks later, 18 ducks were divided equally into two groups and labeled individually, and each inoculation group (six ducks) was inoculated intranasally with 108 EID50/0.1 mL of Q135 or S23 virus allantoic fluid. In addition, three ducks were inoculated with 0.1 mL of PBS as contact animals and were maintained with the inoculated ducks for 24 h after virus inoculation. On 3 DPI (day-post-inoculation), three ducks from the inoculated group were sacrificed, and the lungs, trachea, liver, spleen, kidneys, and brain were obtained. Each tissue sample (1 g) was cut and homogenized in 1 mL PBS containing antibiotics. The homogenate was centrifuged at 10,000 g for 5 min. After centrifugation, the virus titers of the supernatants were measured to check for viruses in the organs after inoculation into chicken embryos. The remaining ducks were observed for clinical signs up to 14 DPI. Additionally, oropharyngeal, and cloacal swabs were collected from all ducks at 3, 5, 7, 9, 11, and 13 DPI and suspended in 1 mL of PBS. The surviving ducks were euthanized after serum collection at 14 DPI. Seroconversion was confirmed using HI testing. Determination of HI antibodies in serum was performed as previously described (Jiao et al., 2018).
The supernatants of the tissue homogenates or swabs were 10-fold serially diluted, and 0.3 mL of each dilution was inoculated into three 9-day-old SPF chicken embryos (a rate of 0.1 mL/embryo). The embryos were cultured at incubator for 2 days. Positive allantoic fluid was determined using a hemagglutination test. Viral titers in the tissues and swabs were calculated according to a previous study(Thakur and Fezio, 1981; Jiao et al., 2018).
2.4. Chicken experiments
Four-week-old SPF chickens were purchased from Dahuanong Biotechnology Co., Ltd. in Guangdong, China, and raised in isolators. A total of 26 chickens were divided equally into two groups and labeled individually, and each inoculated group (eight chickens) was intranasally inoculated with 104 EID50/0.1 mL of Q135 or S23 virus allantoic fluid. In addition, five chickens were inoculated with 0.1 mL as contact animals and were maintained with the inoculated chickens for 24 h after virus inoculation. At 2 DPI, three chickens of each inoculated group were euthanized, and the lungs, trachea, liver, spleen, kidneys, and brain were collected from three chickens of each inoculated group. Each tissue sample (1 g) was cut and homogenized in 1 mL of PBS containing antibiotics. The homogenate was centrifuged at 10,000 g for 5 min. After centrifugation, the virus titers of the supernatants were measured to check for viruses in the organs after inoculation into chicken embryos. The remaining chickens were observed for clinical signs up to 14 DPI. Alternatively, oropharyngeal, and cloacal swabs were collected from all chickens at 3, 5, 7, 9, 11, and 13 DPI and suspended in 1 mL of PBS. The methods of inoculating the supernatants of tissue homogenates or swabs and virus titer determination were based on an infection study in ducks.
2.5. Statistical analysis
Statistically differences in viral titers were determined using the Student’s t-test using the GraphPad 9.0. An analysis was considered statistically significant at the value of p < 0.05.
2.6. Ethics statement
All animal experiments were approved via the Laboratory Animal Management and Ethics Committee of South China Agricultural University and were conducted in the ABSL-3 facility (SCAUABSL2019-004).
3. Results
3.1. Phylogenic study of H5N6 AIVs
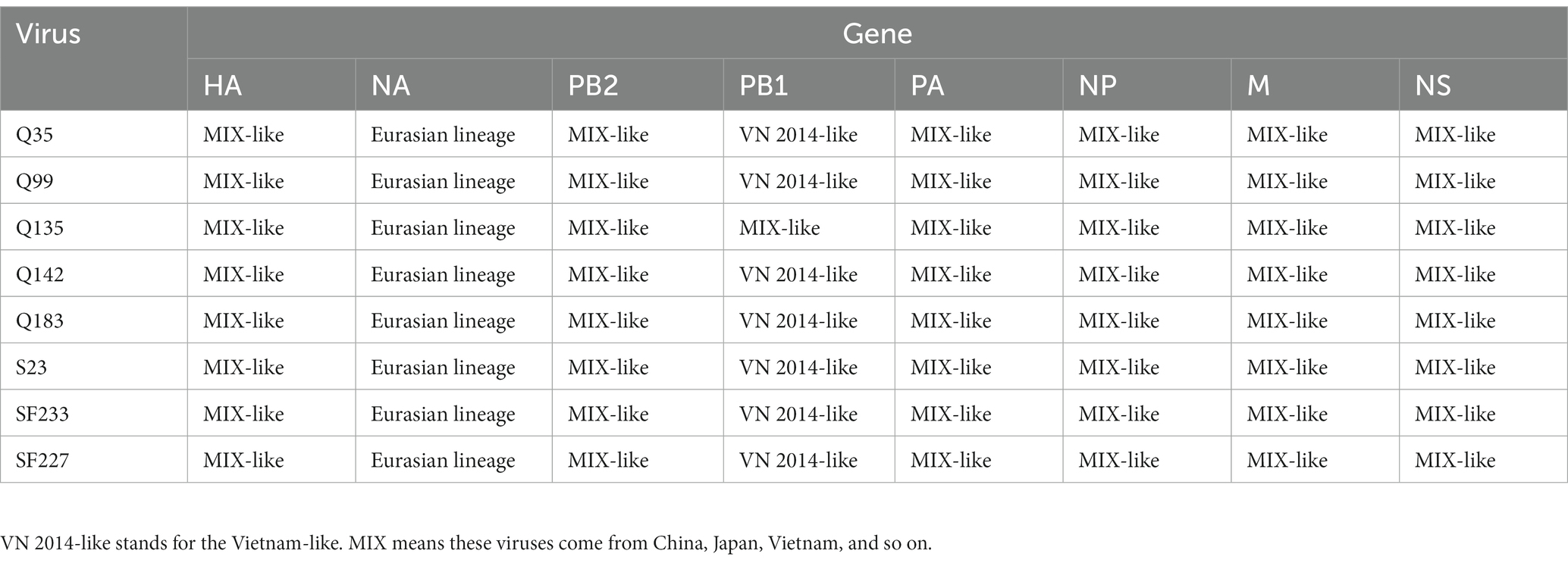
To resolve the genetic evolution of the eight H5N6 AIVs, viral genes were sequenced. We compared their sequences with those of representative viruses obtained from GenBank and performed a phylogenetic analysis. In this study, the HA genes of H5N6 AIVs were classified into MIX-like, Vietnam 2004-like (VN 2004-like), Vietnam 2014-like (VN 2014-like), Indonesia-like (IDN-like), Guiyang-like (GY-like), and Qinghai-like (QH-like). The HA genes of the H5N6 AIVs belonged to the MIX-like of Clade 2.3.4.4h (Figure 1). NA genes were divided into two branches: the Eurasian and North American lineages; the NA genes of our H5N6 AIVs were located in the Eurasian lineage (Figure 2). The PB2 genes belonged to the MIX-like branch (Figure 3). The PB1 genes were classified as MIX-like and VN 2014-like branches (Figure 4). The PA, NP, M, and NS genes were clustered into the MIX-like branch (Figures 5–8). In summary, the eight genes of the isolated AIVs belonged to different genotypes, indicating that their genetic evolution was diverse (Table 2).
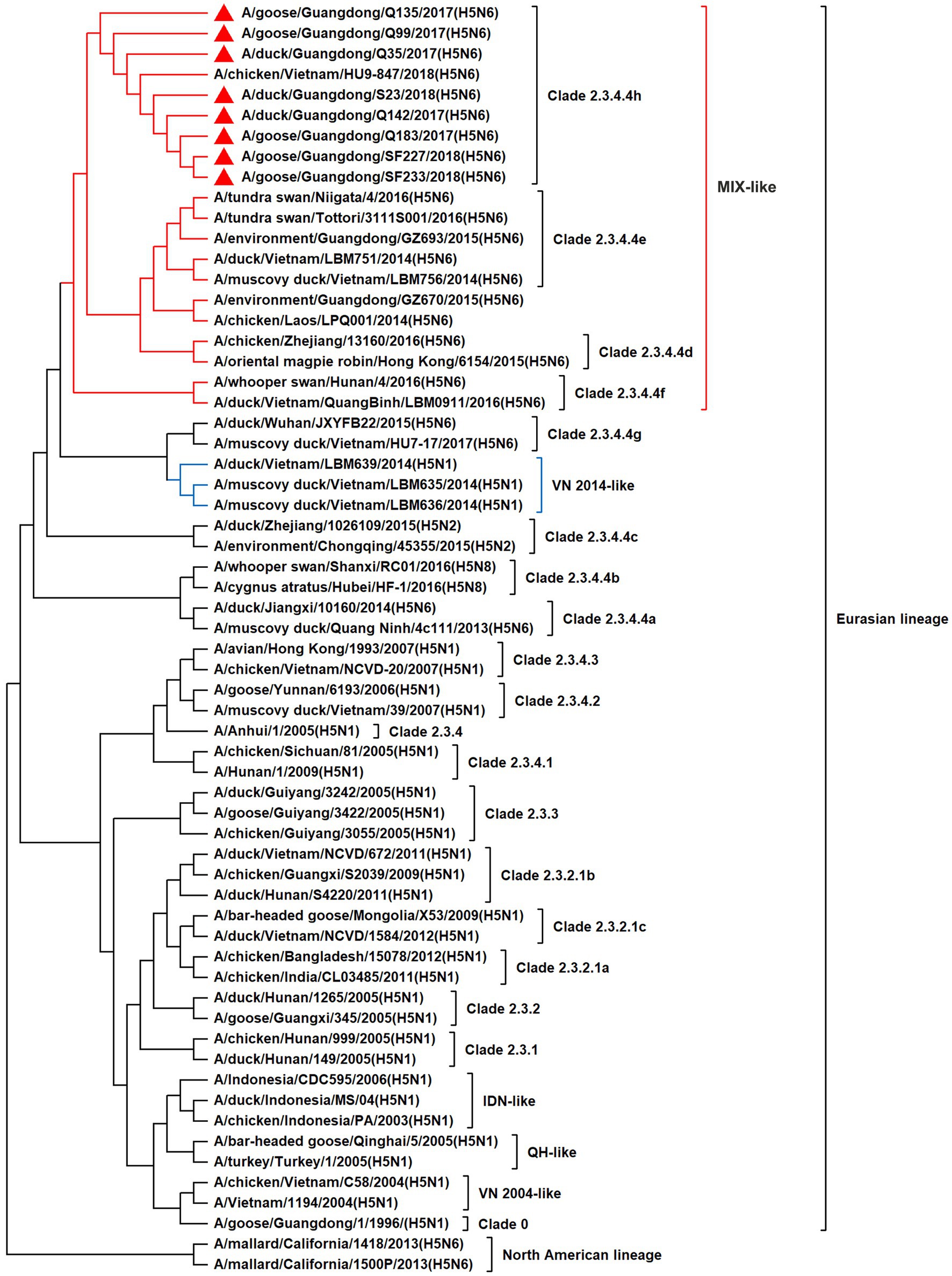
Figure 1. Phylogenetic relationships among H5 subtype AIVs based on their HA gene sequences (nucleotides 1–1,704). Our eight viruses have a triangle in front of the strain name. Qinghai, Guiyang, Vietnam, and Indonesia are abbreviated as QH, GY, VN, and IDN, respectively; MIX means these viruses come from China, Japan, Vietnam, and so on.
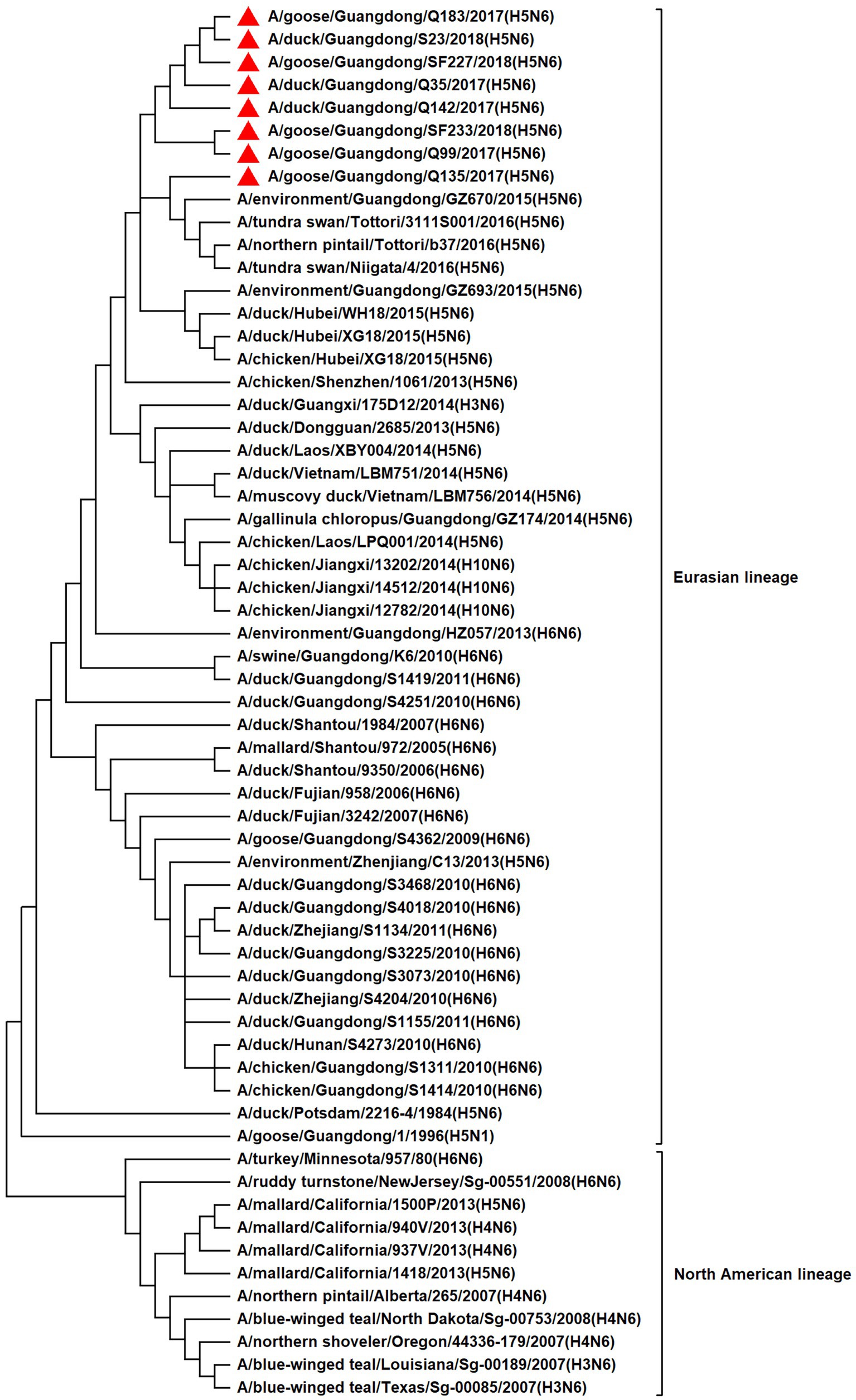
Figure 2. Phylogenetic relationships among N6 subtype AIVs based on their NA gene sequences (nucleotides 1–1,380). Our eight viruses have a triangle in front of the strain name.
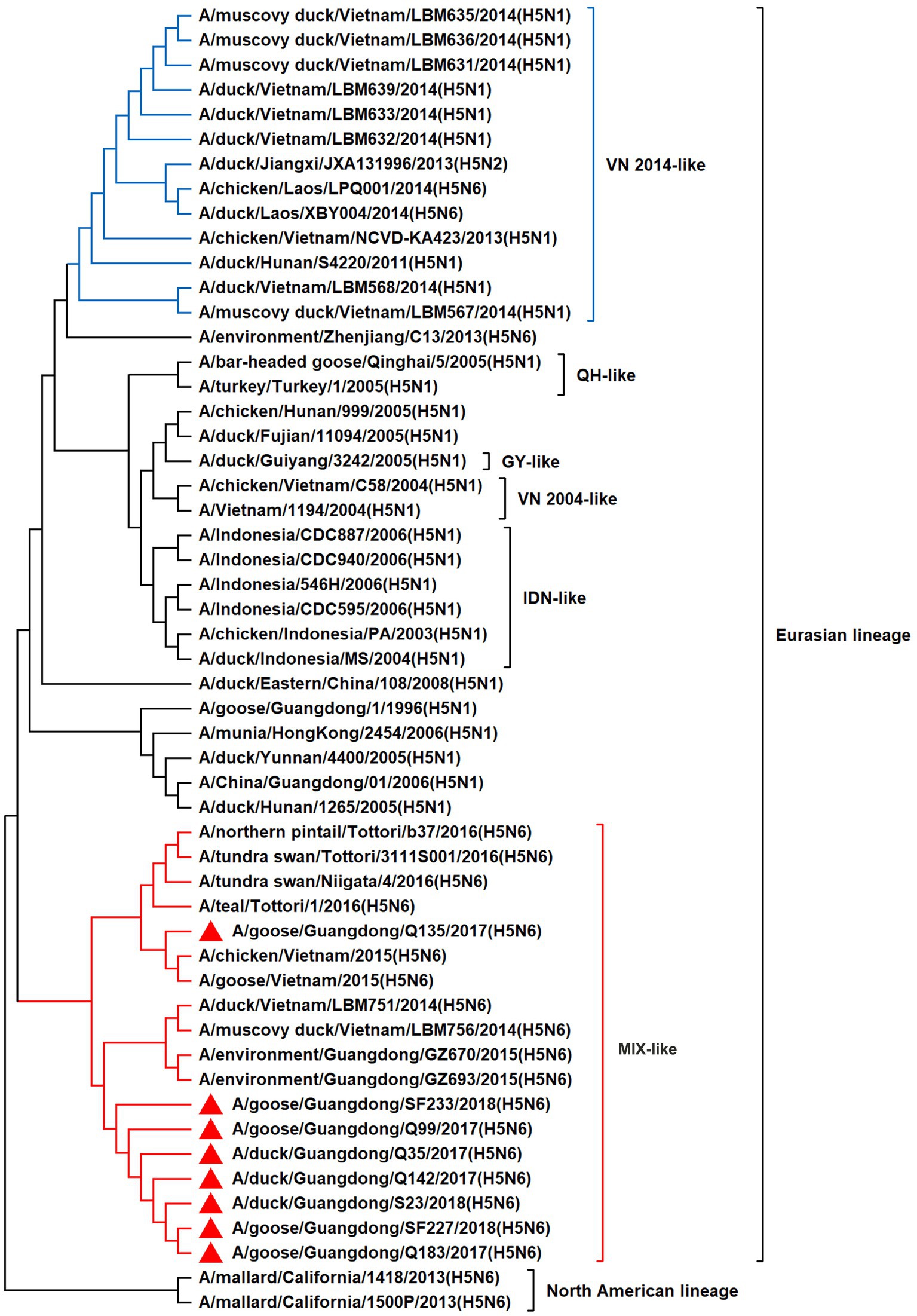
Figure 3. Phylogenetic relationships among H5 subtype AIVs based on their PB2 gene sequences (nucleotides 1–2,280). Our eight viruses have a triangle in front of the strain name. Qinghai, Guiyang, Vietnam, and Indonesia are abbreviated as QH, GY, VN, and IDN, respectively.
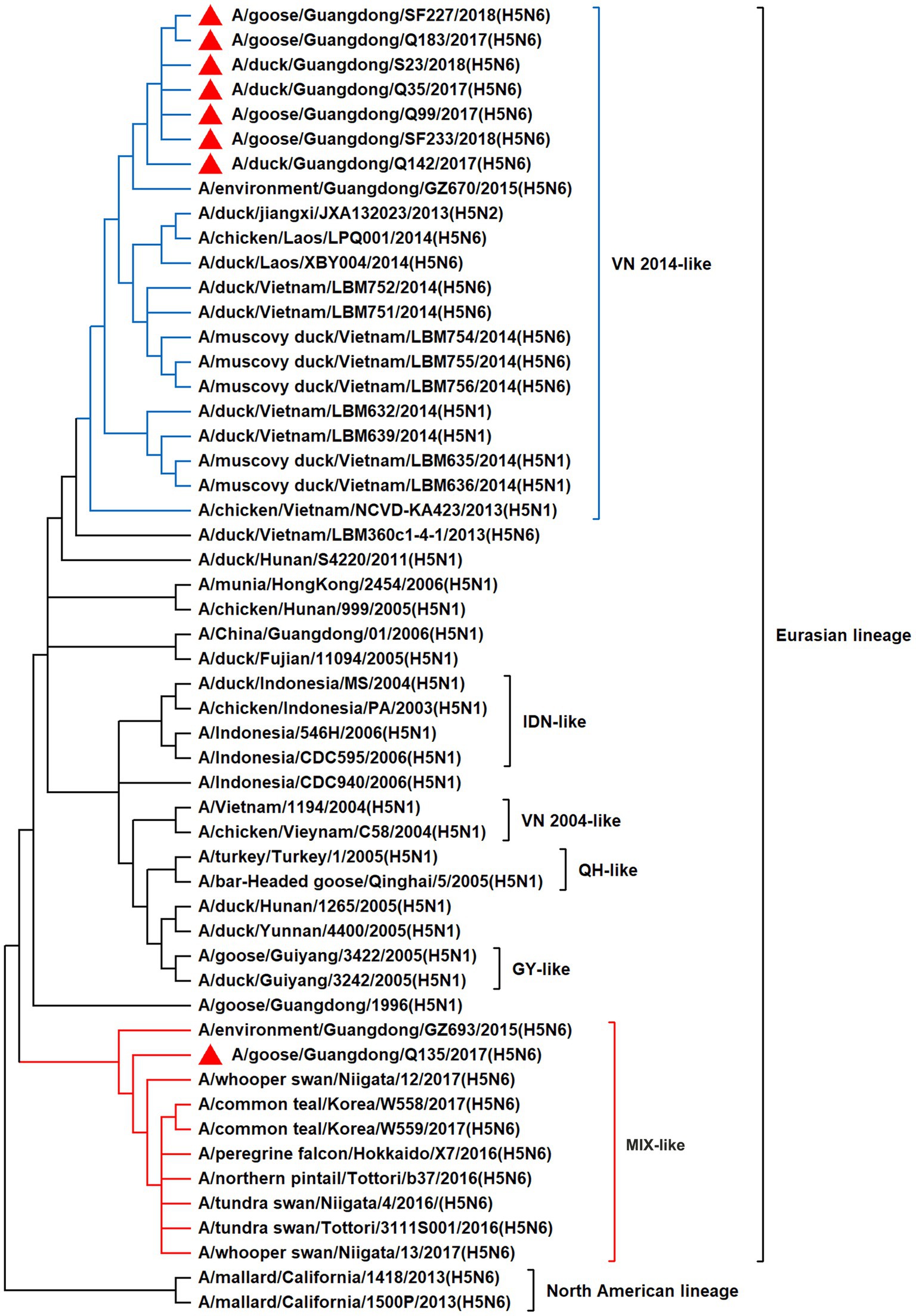
Figure 4. Phylogenetic relationships among H5 subtype AIVs based on their PB1 gene sequences (nucleotides 1–2,274). Our eight viruses have a triangle in front of the strain name. Qinghai, Guiyang, Vietnam, and Indonesia are abbreviated as QH, GY, VN, and IDN, respectively; MIX means these viruses come from China, Japan, Vietnam, and so on.
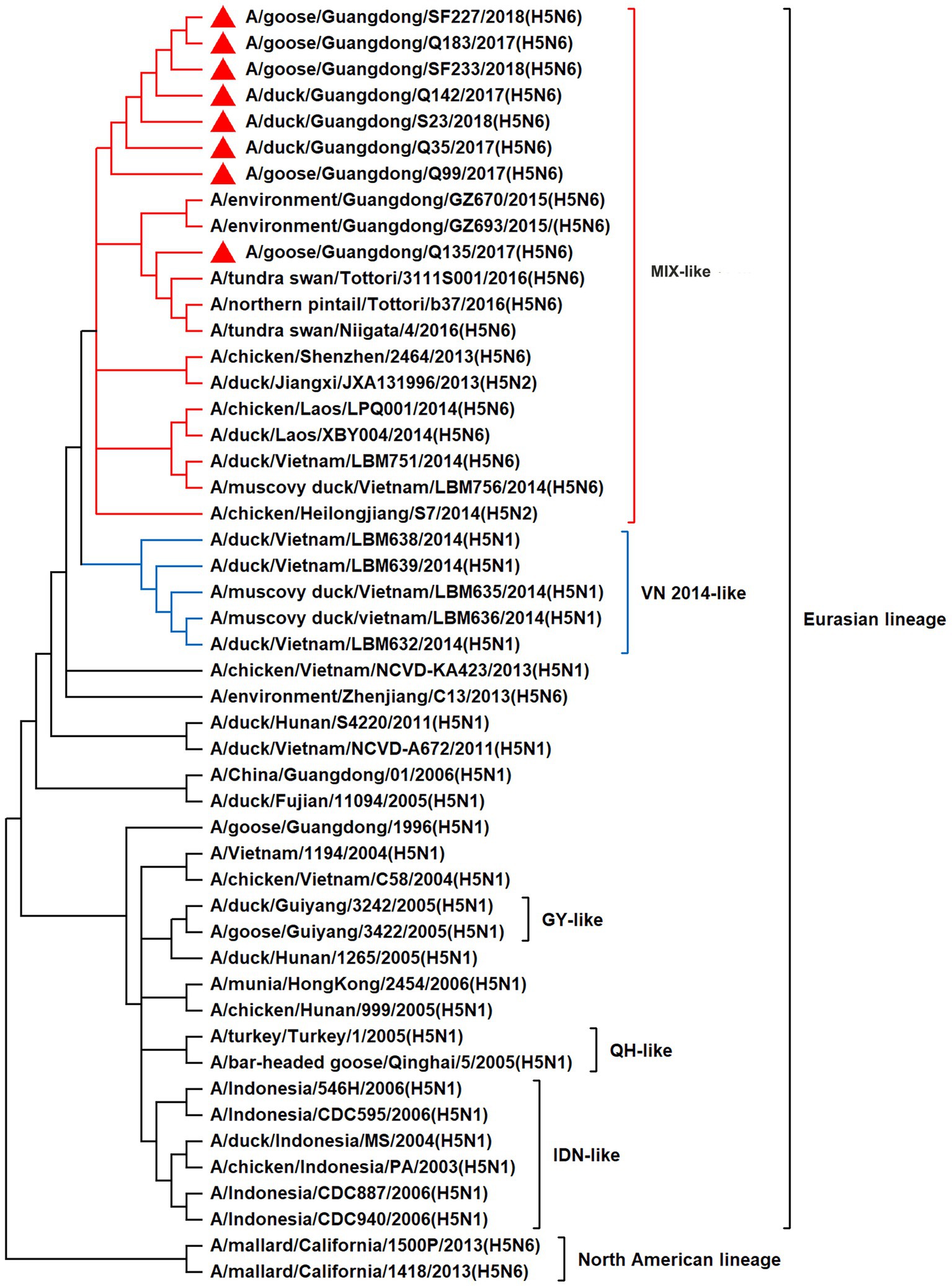
Figure 5. Phylogenetic relationships among H5 subtype AIVs based on their PA gene sequences (nucleotides 1–2,151). Our eight viruses have a triangle in front of the strain name. Qinghai, Guiyang, Vietnam, and Indonesia are abbreviated as QH, GY, VN, and IDN, respectively; MIX means these viruses come from China, Japan, Vietnam, and so on.
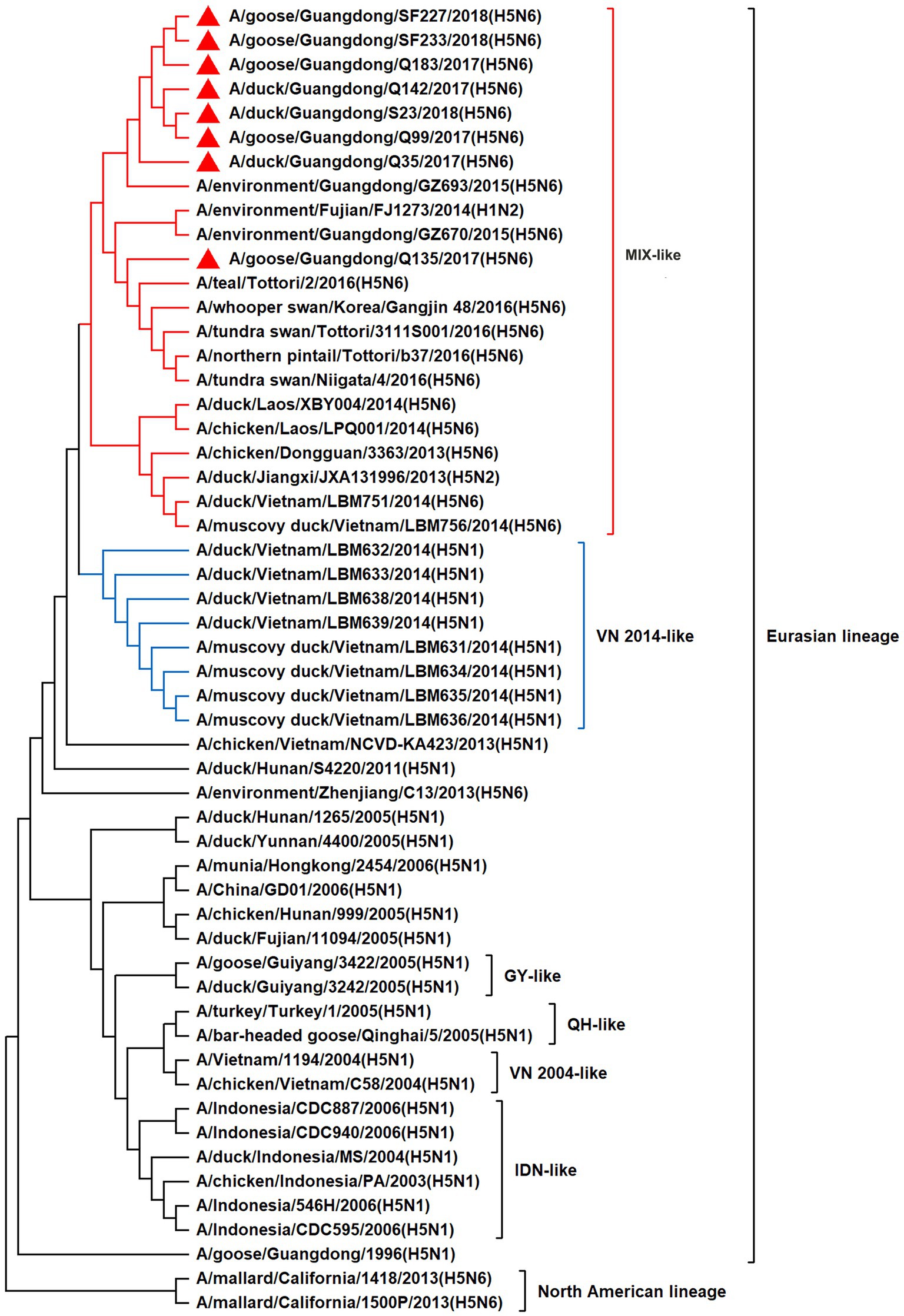
Figure 6. Phylogenetic relationships among H5 subtype AIVs based on their NP gene sequences (nucleotides 1–1,497). Our eight viruses have a triangle in front of the strain name. Qinghai, Guiyang, Vietnam, and Indonesia are abbreviated as QH, GY, VN, and IDN, respectively; MIX means these viruses come from China, Japan, Vietnam, and so on.
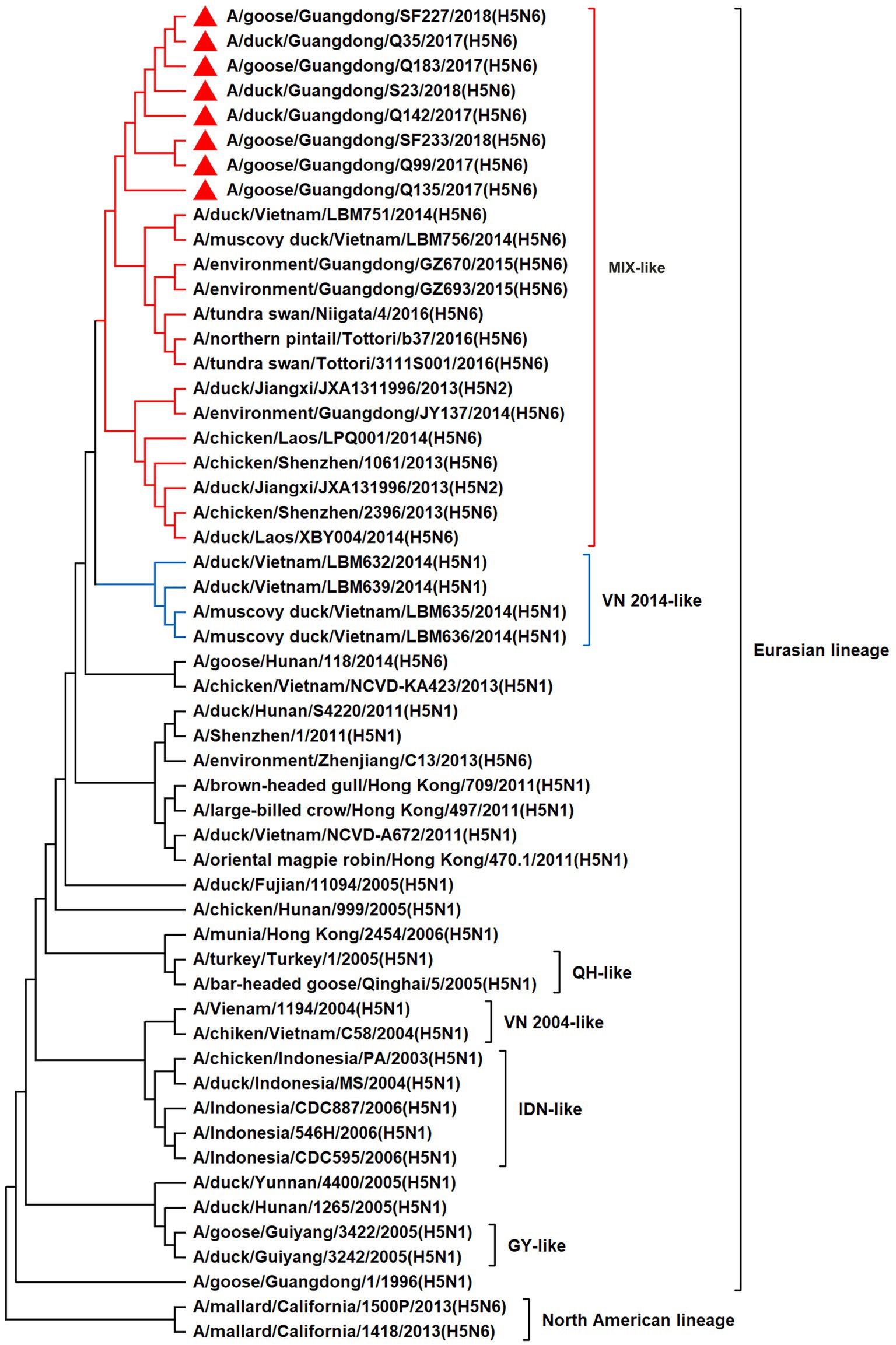
Figure 7. Phylogenetic relationships among H5 subtype AIVs based on their M gene sequences (nucleotides 1–982). Our eight viruses have a triangle in front of the strain name. Qinghai, Guiyang, Vietnam, and Indonesia are abbreviated as QH, GY, VN, and IDN, respectively; MIX means these viruses come from China, Japan, Vietnam and so on.
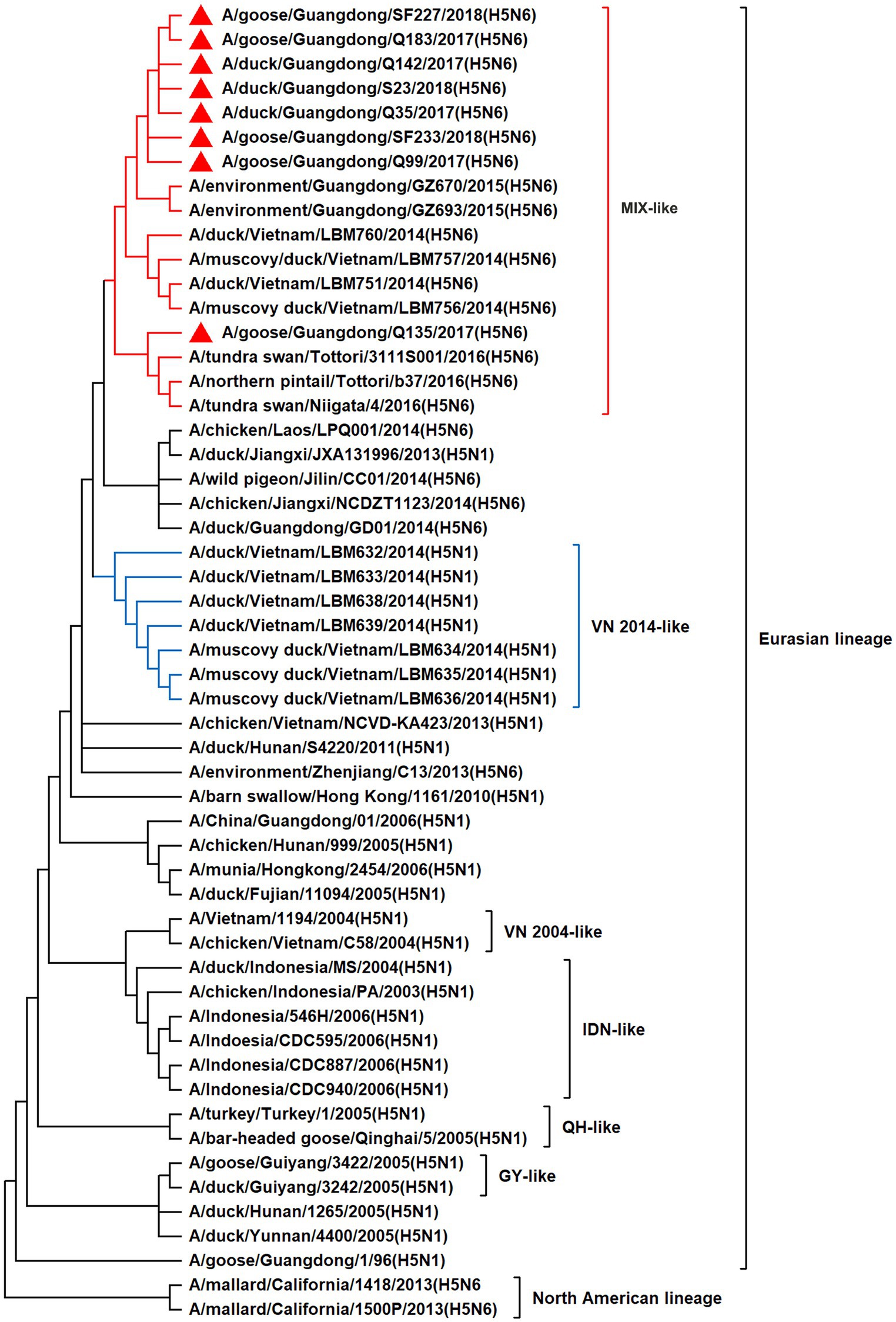
Figure 8. Phylogenetic relationships among H5 subtype AIVs based on their NS gene sequences (nucleotides 1–823). Our eight viruses have a triangle in front of the strain name. Qinghai, Guiyang, Vietnam, and Indonesia are abbreviated as QH, GY, VN, and IDN, respectively; MIX means these viruses come from China, Japan and Vietnam, and so on.
3.2. Molecular characteristics of H5N6 AIVs
To determine the molecular characteristics of the eight H5N6 viruses, their amino acid sequences were analyzed with reference to the genomic sequence of H5N1 AIV (A/goose/Guangdong/1/1996, GGD1-96). Herein, the cleavage sites in the HA protein of the eight H5N6 AIVs were RERRRKR/G, consistent with the characteristic of HPAIV. Compared with the reference strain GGD1-96, these viruses had amino acid deletions at positions 142 and 345. The amino acids related to the HA protein receptor-binding specificity were 190E, 225G, and 226-228QRG/QHG (the amino acid positions listed refer to H3 numbering). The HA protein of the GGD1-96 virus has seven potential glycosylation sites (26, 27, 39, 181, 302, 500, and 559). All seven potential glycosylation sites in our viruses were conserved; however, they added one potential glycosylation site at position 140 (NHT). NA proteins had five potential glycosylation sites (70, 86, 146, 201, and 402), but Q135 virus has an additional potential glycosylation site at 49 (NMS). The NA proteins had a deletion of 11 amino acids residues 58–68 in the stalk region of all isolated viruses. In the viral NA proteins, the amino acids were 119E, 152R, 275H, 293R, and 295 N, which correlated with the affinity of the active site and the neuraminidase inhibitor. The PB2 proteins in our viruses were the 627E and 701D. The PB1 proteins in our viruses were the 13P. The PA protein of the S23 virus was 383E, whereas that of the other seven viruses were 383D. A 479F amino acid residue of the NP proteins was observed in our viruses. The M1 proteins were found to have amino acid residues of 30D and 215A in all eight viruses. The amino acids at the amantadine/rimantadine resistance-associated sites of the M2 proteins were 26 L, 27 V, 30A, 31S, and 34G. Compared with the GGD1-96 NS1 proteins, our isolated viruses had these mutations (42S, 92E, 103F, 106 M, and 149A) and a five-amino-acid deletion at positions 83–87. In addition, their binding motif of the PDZ domain of the NS1 proteins was ESEV.
3.3. Pathogenicity and replication of the H5N6 HPAIVs isolated from waterfowl in ducks
To investigate the pathogenicity of H5N6 HPAIVs in ducks, we inoculated two groups of ducks with 108 EID50/0.1 mL of Q135 and S23 viruses, respectively. At 3 DPI, three ducks from each inoculated group were euthanized to test whether the virus could cause a systemic infection. During the 14 DPI, all ducks were observed for clinical signs.
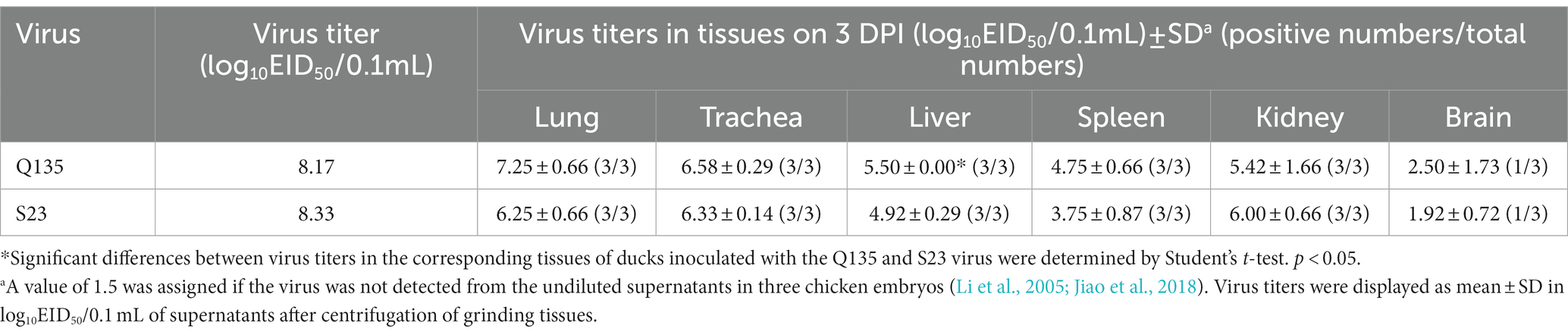
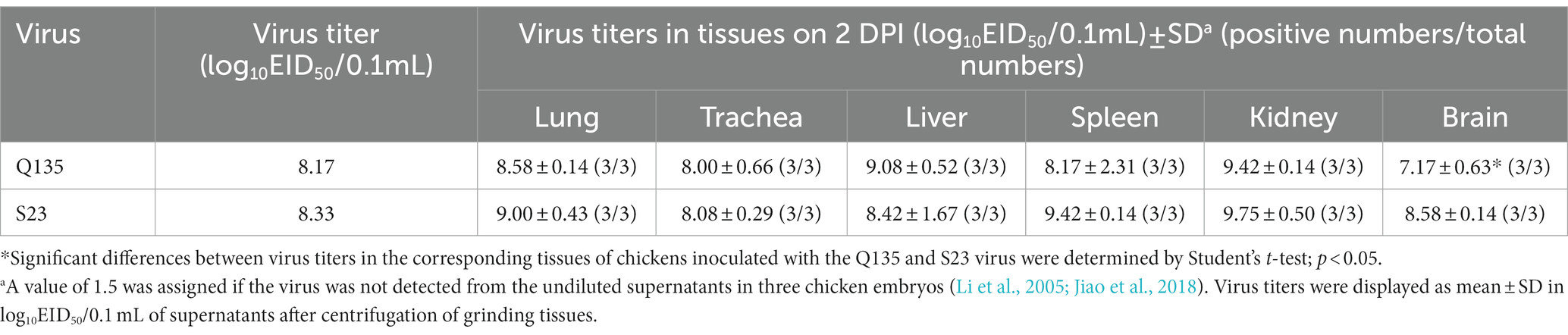
No ducks died after inoculation with the Q135 or S23 virus, but they presented mild clinical symptoms, such as low activity and loss of appetite, from 3 to 6 DPI. All human-infected ducks seroconverted at the 14 DPI, and HI antibody titers of the inoculated groups were 6–8 log2, which proved that all the inoculated ducks were infected with viruses. As shown in Table 3, we inoculated each grinding supernatant of the organs into chicken embryos and tested the viral replication titers of the Q135 in the lungs, trachea, liver, spleen, kidneys, and brain, which were 7.25, 6.58, 5.50, 4.75, 5.42, and 2.50 log10EID50/0.1 mL, respectively. The virus titers of S23 in the corresponding organs were determined using the same method and were 6.25, 6.33, 4.92, 3.75, 6.00, and 1.92 log10EID50/0.1 mL, respectively. Overall, the two viruses replicated in the respiratory, digestive, urinary, immune and nervous systems of the inoculated ducks. The virus titers of the Q135 virus were higher than those of S23 in the organs of the ducks, except in the kidneys.
These results indicated that Q135 and S23 viruses caused systemic infection and induced mild clinical signs, but do not cause death in inoculated and naturally infected ducks.
3.4. Transmission of the H5N6 HPAIVs isolated from waterfowl in ducks
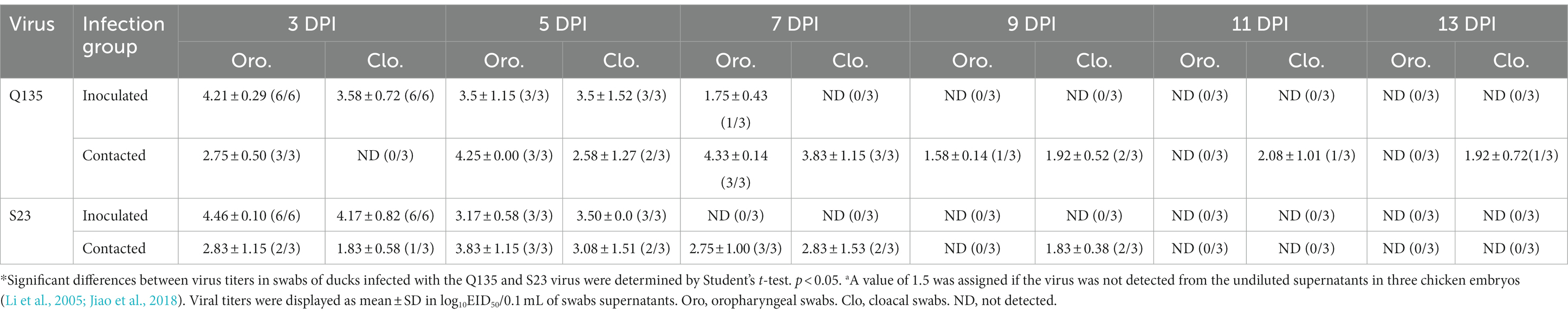
To explore the virus shedding in infected ducks and the horizontal transmission of the H5N6 HPAIVs in the ducks, three ducks were intranasally inoculated with 0.1 mL of PBS, raised alongside the inoculated groups at 1 DPI, and designated as the contacted group. We obtained oropharyngeal and cloacal swabs from all ducks at 3, 5, 7, 9, 11, and 13 DPI. Then, we inoculated chicken embryos with the supernatants of the swabs from the inoculated and contacted ducks to determine their virus titers.
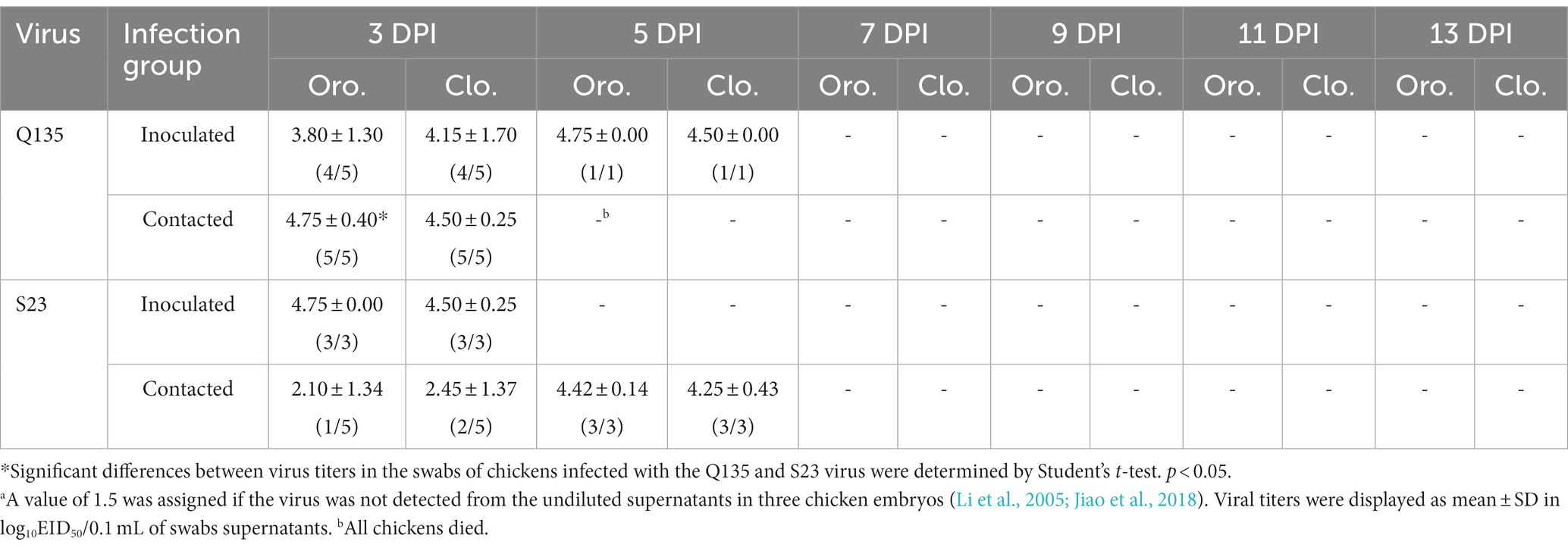
For the inoculated ducks, as shown in Table 4, Q135 virus titers ranged from 1.75–4.21 log10EID50/0.1 mL in the oropharyngeal swabs at the 3, 5, and 7 DPI, and 3.50–3.58 log10EID50/0.1 mL in the cloacal swab at the 3 and 5 DPI. S23 virus titers ranged from 3.17–4.46 log10EID50/0.1 mL in the oropharyngeal swabs at the 3 and 5 DPI, and 3.50–4.17 log10EID50/0.1 mL in the cloacal swabs at the 3 and 5 DPI. Q135-and S23-infected ducks shed the virus by 7 and 5 DPI, respectively. Despite the poor sustained shedding time of S23, monitoring data at 3 DPI showed that oropharyngeal and cloacal swabs had higher viral titers than those of Q135, suggesting that S23 was rapidly cleared from the ducks after infection. For the contacted ducks, two viruses were shed through the oropharynx until 9 DPI or 7 DPI (Q135 virus infection group was 1.58–4.33 log10EID50/0.1 mL and S23 virus infection group was 2.75–3.83 log10EID50/0.1 mL). The Q135 virus was tested using cloacal swabs at 3, 6, 9, 11, and 13 DPI (1.92–3.83 log10EID50/0.1 mL). Additionally, the S23 virus was tested using cloacal swabs at 3, 6, and 9 DPI (1.83–3.08 log10EID50/0.1 mL). Q135-infected ducks shed the virus by 13 DPI, with an infection peak infection at 7 DPI. By contrast, S23-infected ducks shed the undetectable virus on 11 DPI, with the peak of infection 2 days earlier than Q135. All contacted ducks survived and were seroconverted at the 14 DPI, and HI antibody titers of the contacted groups were 5–7 log2.
Therefore, both the Q135 and S23 viruses can be transmitted among ducks by direct contact. In particularly, the Q135 virus had a longer shedding time in contacted ducks than the S23 virus did.
3.5. Pathogenicity and replication of the H5N6 HPAIVs isolated from waterfowl in chickens
To understand the pathogenicity of the viruses in chickens, we inoculated two groups of chickens with 104 EID50/0.1 mL of Q135 and S23 viruses. At 2 DPI, three chickens from each inoculated group were euthanized to test whether the virus could cause a systemic infection.
All chickens in the two groups displayed typical clinical symptoms before death, including decreased water and food intake, chicken claw edema, bleeding foot scales, oropharyngeal bleeding, greenish diarrhea, and even nervous signs. For Q135 virus, 3, 3, 1, and 1 inoculated chickens died at 2, 3, 4, and 5 DPI, respectively; 2 and 3 contacted chickens died at 3 and 4 DPI, respectively. For S23 virus, 5 and 3 inoculated chickens died at 2 and 3 DPI, respectively; 2, 2, and 1 contacted chickens died at 4, 5, and 6 DPI, respectively. As shown in Table 5, we inoculated each grinding supernatant of the organs into chicken embryos and tested the virus replication titers of Q135 in the lungs, trachea, liver, spleen, kidneys, and brain, and were 8.58, 8.00, 9.08, 8.17, 9.42, and 7.17 log10EID50/0.1 mL, respectively. Furthermore, the virus titers of S23 in the corresponding organs were determined using the same method, which were 9.00, 8.08, 8.42, 9.42, 9.75, and 8.58 log10EID50/0.1 mL, respectively. Overall, Q135 and S23 viruses replicated in the respiratory, digestive, urinary, immune, and nervous systems of the inoculated chickens. The viral titers of S23 were higher than those of Q135 in most chicken organs of chickens, except in the liver.
Hence, Q135 and S23 viruses caused severe systemic infections and 100% mortality in artificially inoculated and naturally infected chickens.
3.6. Transmission of the H5N6 HPAIVs isolated from waterfowl in chickens
The methods of assessing the viral shedding in infected chickens and the horizontal transmission of the H5N6 HPAIVs in the chickens were based on an infection study in ducks. As shown in Table 6, we found that virus titers were 3.80 log10EID50/0.1 mL and 4.15 log10EID50/0.1 mL in the oropharyngeal and cloacal swabs, respectively, from Q135-infected chickens at 3 DPI. Virus titers were 4.75 log10EID50/0.1 mL and 4.50 log10EID50/0.1 mL in the oropharyngeal and cloacal swabs, respectively, from Q135-infected chickens at 5 DPI. Virus titers were 4.75 log10EID50/0.1 mL and 4.50 log10EID50/0.1 mL in the oropharyngeal and cloacal swabs, respectively, from S23-infected chickens at 3 DPI. Additionally, the virus titers of oropharyngeal and cloacal swabs from contacted chicken were determined using the same method and were 4.75 log10EID50/0.1 mL and 4.50 log10EID50/0.1 mL, respectively, from Q135-infected chickens at 3 DPI. Virus titers were 2.10 log10EID50/0.1 mL, 2.45 log10EID50/0.1 mL in the oropharyngeal and cloacal swabs, respectively, from S23-infected chickens at 3 DPI. Virus titers were 4.42 log10EID50/0.1 mL and 4.25 log10EID50/0.1 mL in the oropharyngeal and cloacal swabs, respectively, from S23-infected chickens at 5 DPI.
Thus, we demonstrated that Q135 and S23 were shed via the respiratory and digestive tracts and directly transmitted among chickens.
4. Discussion
H5 subtype viruses have accumulated over 27 genotypes through mutations in HA genes and have acquired NA genes from H3N2, H6N2, H4N6, H6N6, and H3N8 viruses and internal genes from H9N2, H7N9, and other unknown viruses (Zhuang et al., 2019; Cui et al., 2020). Since 2013, emerging H5N6 HPAIVs have spread from Asia to other countries via western and eastern routes, causing diseases in birds and humans (Bi et al., 2016). From 2014 to 2022, Clade 2.3.4.4a–h H5N6 HPAIVs caused the death (or culling) of approximately 230,000 birds in China (Song et al., 2019; Cui et al., 2020; Siyu et al., 2023). In addition, there were 65 cases of human infection with the H5N6 virus in China from 2014 to 2021, 64 of which occurred in southern China. These viruses belonged to clade 2.3.4.4a, 2.3.4.4b, 2.3.4.4d, 2.3.4.4g, and 2.3.4.4h (Zhu et al., 2022). In this study, eight H5N6 HPAIVs were isolated from domestic waterfowl from 2017 to 2018, in southern China. The HA genes of these viruses belonged to the MIX-like branch of clade 2.3.4.4h, whereas the NA genes belonged to the Eurasian lineage. The PB2 genes were also categorized into the MIX-like branch, and the PB1 genes were classified as MIX-like and VN 2014-like branches. The PA, NP, M, and NS genes of the H5N6 viruses were clustered into a MIX-like branch (Figures 1–8). This suggests that Clade 2.3.4.4h H5N6 HPAIV in south China underwent genetic recombination.
Since their initial emergence, H5N6 HPAIVs have undergone further genetic evolution owing to the accumulation of mutations. These mutations could lead to changes in the receptor-binding properties, virulence, and antigenicity of the virus, thereby affecting its ability to infect and spread between hosts. In the present study, the isolated strains had RERRRKR/G sequences at the HA cleavage sites and they were 100% lethal to chickens, which indicated they were HPAIVs (Huang et al., 2021). In our strains, the amino acid residues related to HA protein receptor-binding site were 190E, 225G, and 226–228QRG/QHG, indicating that the viruses tended to bind to the α-2-3-galactoside sialic acid receptor. All our viruses were mutated to a potential glycosylation site at position 140 (NHT) of HA proteins and the Q135 virus added one potential glycosylation site at position 49 (NMS) of the NA protein, which may lead to immune escape by interfering with antibody recognition(Wagner et al., 2000; Zhao et al., 2017). The deletion of 11 amino acids from residues 58–68 in the stalk region of NA proteins has been associated with increased viral virulence in mammals (Matsuoka et al., 2009; Fan et al., 2014). Fortunately, their NA and M2 proteins did not occur resistant mutation of neuraminidase inhibitors or Amantadine (Fan et al., 2014; Moorthy et al., 2014). The 627 K and 701 N mutations in PB2 enhance viral replication and pathogenicity in mammals(Hatta et al., 2001; Li et al., 2005). However, all our viruses were E at the 627 site and D at the 701 site of the PB2 proteins, which are molecular characteristics of typical AIVs from birds. The L13P of the PB1 proteins, D383K of PA, L479F substitution in the NP, and 30D and 215A of M1 were present in all our viruses, and these mutations helped to enhance their viral pathogenicity in mammals (Gabriel et al., 2008; Long et al., 2008; Fan et al., 2009; Song et al., 2011, 2015; Yang et al., 2021). The NS1 proteins have these mutations P42S, D92E, L103F, I106M, and V149A and a five-amino-acid deletion at positions 83–87 compared with GGD1-96 (Long et al., 2008), which are the molecular characterization that enhance viral virulence (Donelan et al., 2003; Seo et al., 2004; Banet-Noach et al., 2007; Jiao et al., 2008). In addition, their binding motif of the PDZ domain of the NS1 proteins is ESEV, indicating that they were avian-origin AIVs (Obenauer et al., 2006; Jackson et al., 2008). Thus, the H5N6 viruses in this study have some molecular characteristics contributing to enhanced pathogenicity in avians or mammals.
Ducks are natural hosts of AIVs, and almost all AIV subtypes can be isolated from ducks (Chen et al., 2006; Deng et al., 2013). However, no AIVs, including the highly pathogenic H5 strain, were known to cause disease symptoms or death in ducks before 2002 (Webster et al., 1992; Chen et al., 2004). In the late 20th century, researchers discovered that AIVs do not cause disease in ducks but mainly infect monolayer columnar epithelial cells in their colon, using virus titers as high as 107–8 EID50/mL (Webster et al., 1978; Kida et al., 1980). Additionally, early H5N1 AIVs could not replicate systemically or induce strong immune responses in ducks (Burggraaf et al., 2014; Kuchipudi et al., 2014). However, the H5N1 virus, isolated from wild waterfowl in 2002, caused severe neurological symptoms in ducks, ultimately killing them (Sturm-Ramirez et al., 2004). From April to June 2005, there was a large outbreak of H5N1 infection among migratory waterfowl in Qinghai Lake, western China (Chen et al., 2005; Liu et al., 2005). The H5N1 viruses isolated in 2004, 2008, and 2012, and the H5N6 virus isolated between 2014 and 2016 could replicate in multiple organs of ducks, such as lungs, trachea, liver, spleen, kidneys, pancreas, intestines, and brain and even cause death (Wei et al., 2013; Yuan et al., 2014; Uchida et al., 2019;Wu et al., 2019; Huang et al., 2021). Moreover, Wei found that H5N1 highly pathogenic viruses induce an excessive immune response resulting in immune damage, multiple system organ failure, and disease exacerbation in ducks (Wei et al., 2013). In our study, we selected Q135 and S23 viruses for the pathogenicity study based on the sequencing results of eight viral genes, differences in time and host of the virus isolated. The two H5N6 HPAIVs (Q135 and S23) replicated in the lungs, trachea, liver, pancreas, intestine, spleen, cloacal bursa, trachea, kidneys, and brain, but were not lethal to ducks; it exhibited mild clinical signs for 3–6 DPI. Overall, the two viruses replicated in the respiratory, digestive, urinary, immune, and nervous systems of the inoculated ducks. Additionally, there were differences in the horizontal transmission patterns of viruses isolated at different stages in the duck populations. In the 1980s, ducks infected with AIV shed the virus only through the cloaca. The virus did not spread widely among the duck population (Webster et al., 1978; Kida et al., 1980). In 2002, H5N1 viruses were isolated from the drinking water of diseased and contacted ducks, demonstrating the effective transmission of the virus (Sturm-Ramirez et al., 2004). Recent studies have shown that infected ducks shed H5N6 AIVs through their digestive and respiratory tracts (Wu et al., 2019; Huang et al., 2021). In this study, inoculated and contacted ducks shed the H5N6 virus through the oropharynx and cloaca. These two viruses are also horizontally transmitted in ducks through contact. Ducks have a long shedding time when infected with H5N6 HPAIVs; this is concerning, since they could be a potentially effective source of avian influenza transmission.
Almost all waterfowl-origin AIVs are weakly pathogenic to ducks. However, these waterfowl-origin viruses are highly pathogenic when transmitted from waterfowl to chickens. In chickens, these viruses cause severe inflammation and tissue damage to most organs, including the respiratory and digestive systems. The clinical signs of infected chickens include respiratory distress, lethargy, reduced egg production, and sudden death. The severity of avian influenza in chickens depends on various factors, including age, breed, immune status, and the virus strain (Webster et al., 1992; Yoon et al., 2014). Most strains of the H5N6 virus are highly pathogenic, causing a mortality rate of up to 100% in infected flocks. Since 1996, there has been a trend toward increased pathogenicity of H5 HPAIV in chickens. A low dose (103 EID50/0.1 mL) of the H5N1 virus GGD1-96 strain did not cause death in inoculated chickens; however, the same dose of H5N1 HPAIV isolated in 2012 caused illness and death in inoculated chickens (Yuan et al., 2014). The H5 subtype HPAIV induces the overexpression of cytokines such as IFN-I, IL-6, OAS, IL-8, and Mx1 in infected chickens, causing immune damage and an excessive inflammatory response, which accelerates the death of infected chickens (Wei et al., 2013; Song et al., 2019; Chen et al., 2020). In this study, two H5N6 HPAIVs (Q135 and S23) replicated in all tested tissues of chickens. Overall, both viruses replicated in the respiratory, digestive, urinary, immune, and nervous systems of the inoculated chickens. Infected chickens exhibited typical avian influenza symptoms from 2 DPI, and all inoculated and contacted chickens died within 3–5 DPI. In addition to strain differences, the horizontal transmission of H5 subtype viruses among contacted chickens is related to the dose of the infected virus in the inoculated chickens. When chickens were inoculated with H5N1 HPAIV at 103 EID50/0.1 mL, they showed mild clinical signs and undetectable viral shedding. When chickens were inoculated with H5N1 or H5N6 HPAIV at 106 EID50/0.1 mL, those in the direct contact group began shedding the virus at 2 DPI and eventually died (Yuan et al., 2014; Jiao et al., 2016). However, some H5N6 AIVs do not kill chickens but spread silently through flocks (Mei et al., 2019; Uchida et al., 2019). The H5N6 HPAIVs that have become prevalent in recent years are mainly transmitted in bird populations through direct contact with droppings or through the oral and nasal secretions of infected birds (Qu et al., 2019; Chen et al., 2020). The H5N6 virus can also be transmitted via air and aerosols in crowded flocks, such as in crowded live-bird markets (Zhou et al., 2016). In this study, inoculated and contacted chickens shed viruses via the oropharynx and cloaca, confirming that both H5N6 HPAIVs were transmitted horizontally in chickens.
In summary, waterfowl-origin clade 2.3.4.4h H5N6 HPAIVs displayed genetic diversity in their phylogenetic analysis. Our H5N6 viruses replicated systematically, causing severe and mild clinical symptoms in chickens and ducks, respectively. These viruses are shed from the infected poultry and transmitted horizontally in chickens and ducks. These findings provide a basis for preventing H5N6 avian influenza outbreaks.
Data availability statement
The data presented in the study are deposited in the NCBI GenBank repository, accession numbers OQ829384–OQ829391, OQ829392–OQ829399, OQ829408–OQ829415, OQ829425–OQ829432, OQ829632–OQ829639, OQ830450–OQ830457, OQ830486–OQ830493 and OQ830564–OQ830571.
Ethics statement
The animal study was reviewed and approved by the Laboratory Animal Management and Ethics Committee of South China Agricultural University.
Author contributions
ZH, XW, and PJ designed and performed the main experiments. YL, SF, XH, LZ, JZ, YD, and WL performed the experiments. ZH, PJ, and RY drafted the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the National Key Research and Development Program of China (2021YFD1800202), the National Natural Science Foundation of China (32072844), and the Laboratory of Lingnan Modern Agriculture Project (NT2021007).
Acknowledgments
The authors would like to thank the Key Laboratory of Animal Vaccine Development, Ministry of Agriculture and Rural Affairs.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Banet-Noach, C., Panshin, A., Golender, N., Simanov, L., Rozenblut, E., Pokamunski, S., et al. (2007). Genetic analysis of nonstructural genes (Ns1 and Ns2) of H9n2 and H5n1 viruses recently isolated in Israel. Virus Genes 34, 157–168. doi: 10.1007/s11262-006-0057-9
Bi, Y., Chen, Q., Wang, Q., Chen, J., Jin, T., Wong, G., et al. (2016). Genesis, evolution and prevalence of H5n6 avian influenza viruses in China. Cell Host Microbe 20, 810–821. doi: 10.1016/j.chom.2016.10.022
Buranathai, C., Amonsin, A., Chaisigh, A., Theamboonlers, A., Pariyothorn, N., and Poovorawan, Y. (2007). Surveillance activities and molecular analysis of H5n1 highly pathogenic avian influenza viruses from Thailand, 2004-2005. Avian Dis. 51, 194–200. doi: 10.1637/7594-040306R.1
Burggraaf, S., Karpala, A. J., Bingham, J., Lowther, S., Selleck, P., Kimpton, W., et al. (2014). H5n1 infection causes rapid mortality and high cytokine levels in chickens compared to ducks. Virus Res. 185, 23–31. doi: 10.1016/j.virusres.2014.03.012
Chen, H., Deng, G., Li, Z., Tian, G., Li, Y., Jiao, P., et al. (2004). The evolution of H5n1 influenza viruses in ducks in southern China. Proc. Natl. Acad. Sci. U. S. A. 101, 10452–10457. doi: 10.1073/pnas.0403212101
Chen, H., Li, Y., Li, Z., Shi, J., Shinya, K., Deng, G., et al. (2006). Properties and dissemination of H5n1 viruses isolated during an influenza outbreak in migratory waterfowl in Western China. J. Virol. 80, 5976–5983. doi: 10.1128/JVI.00110-06
Chen, H., Smith, G. J., Zhang, S. Y., Qin, K., Wang, J., Li, K. S., et al. (2005). Avian flu: H5n1 virus outbreak in migratory waterfowl. Nature 436, 191–192. doi: 10.1038/nature03974
Chen, Z., Wang, Z., Zhao, X., Guan, Y., Xue, Q., Li, J., et al. (2020). Pathogenicity of different H5n6 highly pathogenic avian influenza virus strains and host immune responses in chickens. Vet. Microbiol. 246:108745. doi: 10.1016/j.vetmic.2020.108745
Cui, Y., Li, Y., Li, M., Zhao, L., Wang, D., Tian, J., et al. (2020). Evolution and extensive Reassortment of H5 influenza viruses isolated from wild birds in China over the past decade. Emerg. Microbes Infect. 9, 1793–1803. doi: 10.1080/22221751.2020.1797542
Deng, G., Tan, D., Shi, J., Cui, P., Jiang, Y., Liu, L., et al. (2013). Complex Reassortment of multiple subtypes of avian influenza viruses in domestic ducks at the Dongting Lake region of China. J. Virol. 87, 9452–9462. doi: 10.1128/JVI.00776-13
Donelan, N. R., Basler, C. F., and Garcia-Sastre, A. (2003). A recombinant influenza a virus expressing an Rna-binding-defective Ns1 protein induces high levels of Beta interferon and is attenuated in mice. J. Virol. 77, 13257–13266. doi: 10.1128/JVI.77.24.13257-13266.2003
Du, Y., Chen, M., Yang, J., Jia, Y., Han, S., Holmes, E. C., et al. (2017). Molecular evolution and emergence of H5n6 avian influenza virus in Central China. J. Virol. 91:e00143-17. doi: 10.1128/JVI.00143-17
Fan, Z., Ci, Y., Ma, Y., Liu, L., Wang, D., Ma, J., et al. (2014). Phylogenetic analysis of a novel H6n6 avian influenza virus isolated from a green peafowl in China and its pathogenic potential in mice. Infect. Genet. Evol. 28, 107–112. doi: 10.1016/j.meegid.2014.09.008
Fan, S., Deng, G., Song, J., Tian, G., Suo, Y., Jiang, Y., et al. (2009). Two amino acid residues in the matrix protein M1 contribute to the virulence difference of H5n1 avian influenza viruses in mice. Virology 384, 28–32. doi: 10.1016/j.virol.2008.11.044
Gabriel, G., Herwig, A., and Klenk, H. D. (2008). Interaction of polymerase subunit Pb2 and Np with Importin Alpha1 is a determinant of host range of influenza a virus. PLoS Pathog. 4:E11. doi: 10.1371/journal.ppat.0040011
Gu, W., Shi, J., Cui, P., Yan, C., Zhang, Y., Wang, C., et al. (2022). Novel H5n6 Reassortants bearing the clade 2.3.4.4b ha gene of H5n8 virus have been detected in poultry and caused multiple human infections in China. Emerg. Microbes Infect. 11, 1174–1185. doi: 10.1080/22221751.2022.2063076
Hatta, M., Gao, P., Halfmann, P., and Kawaoka, Y. (2001). Molecular basis for high virulence of Hong Kong H5n1 influenza a viruses. Science 293, 1840–1842. doi: 10.1126/science.1062882
Huang, J., Wu, S., Wu, W., Liang, Y., Zhuang, H., Ye, Z., et al. (2021). The biological characteristics of novel H5n6 highly pathogenic avian influenza virus and its pathogenesis in ducks. Front. Microbiol. 12:628545. doi: 10.3389/fmicb.2021.628545
Jackson, D., Hossain, M. J., Hickman, D., Perez, D. R., and Lamb, R. A. (2008). A new influenza virus virulence determinant: the Ns1 protein four C-terminal residues modulate pathogenicity. Proc. Natl. Acad. Sci. U. S. A. 105, 4381–4386. doi: 10.1073/pnas.0800482105
Jiao, P., Song, Y., Huang, J., Xiang, C., Cui, J., Wu, S., et al. (2018). H7n9 avian influenza virus is efficiently transmissible and induces an antibody response in chickens. Front. Immunol. 9:789. doi: 10.3389/fimmu.2018.00789
Jiao, P., Song, H., Liu, X., Song, Y., Cui, J., Wu, S., et al. (2016). Pathogenicity, transmission and antigenic variation of H5n1 highly pathogenic avian influenza viruses. Front. Microbiol. 7:635. doi: 10.3389/fmicb.2016.00635
Jiao, P., Song, Y., Yuan, R., Wei, L., Cao, L., Luo, K., et al. (2012a). Complete genomic sequence of an H5n1 influenza virus from a parrot in southern China. J. Virol. 86, 8894–8895. doi: 10.1128/JVI.01243-12
Jiao, P., Tian, G., Li, Y., Deng, G., Jiang, Y., Liu, C., et al. (2008). A single-amino-acid substitution in the Ns1 protein changes the pathogenicity of H5n1 avian influenza viruses in mice. J. Virol. 82, 1146–1154. doi: 10.1128/JVI.01698-07
Jiao, P., Wei, L., Yuan, R., Gong, L., Cao, L., Song, Y., et al. (2012b). Complete genome sequence of an H5n2 avian influenza virus isolated from a parrot in southern China. J. Virol. 86, 8890–8891. doi: 10.1128/JVI.01189-12
Jiao, P., Yuan, R., Song, Y., Wei, L., Ren, T., Liao, M., et al. (2012c). Full genome sequence of a recombinant H5n1 influenza virus from a condor in southern China. J. Virol. 86, 7722–7723. doi: 10.1128/JVI.01043-12
Kida, H., Yanagawa, R., and Matsuoka, Y. (1980). Duck influenza lacking evidence of disease signs and immune response. Infect. Immun. 30, 547–553. doi: 10.1128/iai.30.2.547-553.1980
Kuchipudi, S. V., Tellabati, M., Sebastian, S., Londt, B. Z., Jansen, C., Vervelde, L., et al. (2014). Highly pathogenic avian influenza virus infection in chickens but not ducks is associated with elevated host immune and pro-inflammatory responses. Vet. Res. 45:118. doi: 10.1186/s13567-014-0118-3
Li, Z., Chen, H., Jiao, P., Deng, G., Tian, G., Li, Y., et al. (2005). Molecular basis of replication of duck H5n1 influenza viruses in a mammalian mouse model. J. Virol. 79, 12058–12064. doi: 10.1128/JVI.79.18.12058-12064.2005
Li, Y., Li, M., Li, Y., Tian, J., Bai, X., Yang, C., et al. (2020). Outbreaks of highly pathogenic avian influenza (H5n6) virus subclade 2.3.4.4h in swans, Xinjiang, Western China, 2020. Emerg. Infect. Dis. 26, 2956–2960. doi: 10.3201/eid2612.201201
Liu, J., Xiao, H., Lei, F., Zhu, Q., Qin, K., Zhang, X. W., et al. (2005). Highly pathogenic H5n1 influenza virus infection in migratory birds. Science 309:1206. doi: 10.1126/science.1115273
Long, J. S., Mistry, B., Haslam, S. M., and Barclay, W. S. (2019). Host and viral determinants of influenza a virus species specificity. Nat. Rev. Microbiol. 17, 67–81. doi: 10.1038/s41579-018-0115-z
Long, J. X., Peng, D. X., Liu, Y. L., Wu, Y. T., and Liu, X. F. (2008). Virulence of H5n1 avian influenza virus enhanced by a 15-nucleotide deletion in the viral nonstructural gene. Virus Genes 36, 471–478. doi: 10.1007/s11262-007-0187-8
Matsuoka, Y., Swayne, D. E., Thomas, C., Rameix-Welti, M. A., Naffakh, N., Warnes, C., et al. (2009). Neuraminidase stalk length and additional glycosylation of the Hemagglutinin influence the virulence of influenza H5n1 viruses for mice. J. Virol. 83, 4704–4708. doi: 10.1128/JVI.01987-08
Mei, K., Guo, Y., Zhu, X., Qu, N., Huang, J., Chen, Z., et al. (2019). Different pathogenicity and transmissibility of goose-origin H5n6 avian influenza viruses in chickens. Viruses 11:612. doi: 10.3390/v11070612
Moorthy, N. S., Poongavanam, V., and Pratheepa, V. (2014). Viral M2 Ion Channel protein: a promising target for anti-influenza drug discovery. Mini Rev. Med. Chem. 14, 819–830.
Neumann, G., Chen, H., Gao, G. F., Shu, Y., and Kawaoka, Y. (2010). H5n1 influenza viruses: outbreaks and biological properties. Cell Res. 20, 51–61. doi: 10.1038/cr.2009.124
Obenauer, J. C., Denson, J., Mehta, P. K., Su, X., Mukatira, S., Finkelstein, D. B., et al. (2006). Large-scale sequence analysis of avian influenza isolates. Science 311, 1576–1580. doi: 10.1126/science.1121586
Pan, M., Gao, R., Lv, Q., Huang, S., Zhou, Z., Yang, L., et al. (2016). Human infection with a novel, highly pathogenic avian influenza a (H5n6) virus: Virological and clinical findings. J. Infect. 72, 52–59. doi: 10.1016/j.jinf.2015.06.009
Puryear, W., Sawatzki, K., Hill, N., Foss, A., Stone, J. J., Doughty, L., et al. (2023). Highly pathogenic avian influenza a(H5n1) virus outbreak in New England seals, United States. Emerg. Infect. Dis. 29, 786–791. doi: 10.3201/eid2904.221538
Qu, N., Zhao, B., Chen, Z., He, Z., Li, W., Liu, Z., et al. (2019). Genetic characteristics, pathogenicity and transmission of H5n6 highly pathogenic avian influenza viruses in southern China. Transbound. Emerg. Dis. 66, 2411–2425. doi: 10.1111/tbed.13299
Seo, S. H., Hoffmann, E., and Webster, R. G. (2004). The Ns1 gene of H5n1 influenza viruses circumvents the host anti-viral cytokine responses. Virus Res. 103, 107–113. doi: 10.1016/j.virusres.2004.02.022
Siyu, F., Yating, S., Xiaowu, H., Junsheng, Z., Sumei, S., Luxiang, Z., et al. (2023). H5 subtype highly pathogenic avian influenza epidemic characteristics in China, 2020-2022. Chin J Anim Infect Dis, 1–7. doi: 10.19958/j.cnki.cn31-2031/s.20230103.003
Song, J., Feng, H., Xu, J., Zhao, D., Shi, J., Li, Y., et al. (2011). The pa protein directly contributes to the virulence of H5n1 avian influenza viruses in domestic ducks. J. Virol. 85, 2180–2188. doi: 10.1128/JVI.01975-10
Song, Y., Li, W., Wu, W., Liu, Z., He, Z., Chen, Z., et al. (2019). Phylogeny, pathogenicity, transmission, and host immune responses of four H5n6 avian influenza viruses in chickens and mice. Viruses 11:1048. doi: 10.3390/v11111048
Song, J., Xu, J., Shi, J., Li, Y., and Chen, H. (2015). Synergistic effect of S224p and N383d substitutions in the pa of H5n1 avian influenza virus contributes to mammalian adaptation. Sci. Rep. 5:10510. doi: 10.1038/srep10510
Sturm-Ramirez, K. M., Ellis, T., Bousfield, B., Bissett, L., Dyrting, K., Rehg, J. E., et al. (2004). Reemerging H5n1 influenza viruses in Hong Kong in 2002 are highly pathogenic to ducks. J. Virol. 78, 4892–4901. doi: 10.1128/JVI.78.9.4892-4901.2004
Thakur, A. K., and Fezio, W. L. (1981). A computer program for estimating Ld50 and its confidence limits using modified Behrens-reed-Muench Cumulant method. Drug Chem. Toxicol. 4, 297–305. doi: 10.3109/01480548109018136
Uchida, Y., Mine, J., Takemae, N., Tanikawa, T., Tsunekuni, R., and Saito, T. (2019). Comparative pathogenicity of H5n6 subtype highly pathogenic avian influenza viruses in chicken, Pekin duck and Muscovy duck. Transbound. Emerg. Dis. 66, 1227–1251. doi: 10.1111/tbed.13141
Wagner, R., Wolff, T., Herwig, A., Pleschka, S., and Klenk, H. D. (2000). Interdependence of Hemagglutinin glycosylation and neuraminidase as regulators of influenza virus growth: a study by reverse genetics. J. Virol. 74, 6316–6323. doi: 10.1128/JVI.74.14.6316-6323.2000
Webster, R. G., Bean, W. J., Gorman, O. T., Chambers, T. M., and Kawaoka, Y. (1992). Evolution and ecology of influenza a viruses. Microbiol. Rev. 56, 152–179. doi: 10.1128/mr.56.1.152-179.1992
Webster, R. G., Yakhno, M., Hinshaw, V. S., Bean, W. J., and Murti, K. G. (1978). Intestinal influenza: replication and characterization of influenza viruses in ducks. Virology 84, 268–278. doi: 10.1016/0042-6822(78)90247-7
Wei, L., Jiao, P., Song, Y., Cao, L., Yuan, R., Gong, L., et al. (2013). Host immune responses of ducks infected with H5n1 highly pathogenic avian influenza viruses of different Pathogenicities. Vet. Microbiol. 166, 386–393. doi: 10.1016/j.vetmic.2013.06.019
WHO (2012). Continued evolution of highly pathogenic avian influenza a (H5n1): updated nomenclature. Influenza Other Respi. Viruses 6, 1–5. doi: 10.1111/j.1750-2659.2011.00298.x
Wu, S., Huang, J., Huang, Q., Zhang, J., Liu, J., Xue, Q., et al. (2020). Host innate immune response of geese infected with clade 2.3.4.4 H5n6 highly pathogenic avian influenza viruses. Microorganisms 8:224. doi: 10.3390/microorganisms8020224
Wu, S., Zhang, J., Huang, J., Li, W., Liu, Z., He, Z., et al. (2019). Immune-related gene expression in ducks infected with waterfowl-origin H5n6 highly pathogenic avian influenza viruses. Front. Microbiol. 10:1782. doi: 10.3389/fmicb.2019.01782
Xu, X., Subbarao Cox, N. J., and Guo, Y. (1999). Genetic characterization of the pathogenic influenza a/goose/Guangdong/1/96 (H5n1) virus: similarity of its Hemagglutinin gene to those of H5n1 viruses from the 1997 outbreaks in Hong Kong. Virology 261, 15–19. doi: 10.1006/viro.1999.9820
Yang, F., Xiao, Y., Liu, F., Yao, H., Wu, N., and Wu, H. (2021). Molecular characterization and antigenic analysis of Reassortant H9n2 subtype avian influenza viruses in eastern China in 2016. Virus Res. 306:198577. doi: 10.1016/j.virusres.2021.198577
Yoon, S. W., Webby, R. J., and Webster, R. G. (2014). Evolution and ecology of influenza a viruses. Curr. Top. Microbiol. Immunol. 385, 359–375. doi: 10.1007/82_2014_396
Yuan, R., Cui, J., Zhang, S., Cao, L., Liu, X., Kang, Y., et al. (2014). Pathogenicity and transmission of H5n1 avian influenza viruses in different birds. Vet. Microbiol. 168, 50–59. doi: 10.1016/j.vetmic.2013.10.013
Zhao, D., Liang, L., Wang, S., Nakao, T., Li, Y., Liu, L., et al. (2017). Glycosylation of the Hemagglutinin protein of H5n1 influenza virus increases its virulence in mice by exacerbating the host immune response. J. Virol. 91:e02215-16. doi: 10.1128/JVI.02215-16
Zhou, J., Wu, J., Zeng, X., Huang, G., Zou, L., Song, Y., et al. (2016). Isolation of H5n6, H7n9 and H9n2 avian influenza a viruses from air sampled at live poultry markets in China, 2014 and 2015. Euro Surveill. 21:21. doi: 10.2807/1560-7917.ES.2016.21.35.30331
Zhu, H., Damdinjav, B., Gonzalez, G., Patrono, L. V., Ramirez-Mendoza, H., Amat, J. A. R., et al. (2019). Absence of adaptive evolution is the Main barrier against influenza emergence in horses in Asia despite frequent virus interspecies transmission from wild birds. PLoS Pathog. 15:E1007531. doi: 10.1371/journal.ppat.1007531
Zhu, W., Li, X., Dong, J., Bo, H., Liu, J., Yang, J., et al. (2022). Epidemiologic, clinical, and genetic characteristics of human infections with influenza a(H5n6) viruses, China. Emerg. Infect. Dis. 28, 1332–1344. doi: 10.3201/eid2807.212482
Keywords: H5N6 avian influenza virus, genetic evolution, pathogenicity, transmission, duck, chicken
Citation: He Z, Wang X, Lin Y, Feng S, Huang X, Zhao L, Zhang J, Ding Y, Li W, Yuan R and Jiao P (2023) Genetic characteristics of waterfowl-origin H5N6 highly pathogenic avian influenza viruses and their pathogenesis in ducks and chickens. Front. Microbiol. 14:1211355. doi: 10.3389/fmicb.2023.1211355
Edited by:
Zhixun Xie, Guangxi Veterinary Research Institute, ChinaCopyright © 2023 He, Wang, Lin, Feng, Huang, Zhao, Zhang, Ding, Li, Yuan and Jiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Runyu Yuan, Y2VjaWxpYV95cnlAaG90bWFpbC5jb20=; Peirong Jiao, cHJqaWFvQHNjYXUuZWR1LmNu
 Zhuoliang He
Zhuoliang He Xia Wang1
Xia Wang1 Junsheng Zhang
Junsheng Zhang Weiqiang Li
Weiqiang Li Runyu Yuan
Runyu Yuan Peirong Jiao
Peirong Jiao