- 1Department of Medical Laboratory, The General Hospital of Western Theater Command, Chengdu, China
- 2Department of Orthopaedics, The General Hospital of Western Theater Command, Chengdu, China
- 3Department of Clinical Pharmacy, The General Hospital of Western Theater Command, Chengdu, China
- 4School of Nursing, Chengdu Medical College, Chengdu, China
Enterococcus faecalis is a well-established resident of the human gastrointestinal tract and is also a major cause of human infections. Unfortunately, therapeutic options for E. faecalis infections remain limited, particularly with the emergence of vancomycin-resistant strains in hospital settings. Consequently, there has been a renewed interest in phage therapy as an alternative to antibiotics. In this study, we have isolated a bacteriophage, vB_EfaS-SFQ1, from hospital sewage, which effectively infects E. faecalis strain EFS01. Phage SFQ1 is a siphovirus and exhibits a relatively broad host range. Furthermore, it has a short latent period of approximately 10 min and a large burst size of about 110 PFU/cell at a multiplicity of infection (MOI) of 0.01, and it could effectively disrupt the biofilms formed by E. faecalis. Thus, this study provides a detailed characterization of E. faecalis phage SFQ1, which has great potential for treating E. faecalis infections.
Introduction
Enterococci are a group of low-GC Gram-positive cocci that comprise up to 60 different species (Cattoir, 2022). Among these species, Enterococcus faecalis and Enterococcus faecium are the two main species that have become a leading cause of human infections (Jabbari Shiadeh et al., 2019; Bright et al., 2020; Kakoullis et al., 2021). In recent years, the rise of vancomycin-resistant Enterococcus (VRE) has become a growing threat in hospital settings, and E. faecalis is the second most commonly isolated VRE species (Arias et al., 2011; Hayakawa et al., 2013). Moreover, E. faecalis could form biofilms that exist in the medical equipment, which makes the antibiotics ineffective against the bacteria in the biofilms (Venkateswaran et al., 2022). And the formation of biofilms makes it more difficult to cure infections (Ali and Neelakantan, 2022). Currently, the development of new antibacterial drugs is slow (Theuretzbacher et al., 2020). Consequently, there is an urgent need for novel therapeutic agents to combat multidrug-resistant bacterial infections (Gorski et al., 2016).
Phages, which are viruses that predate bacteria, can infect antibiotic-resistant bacteria, making them an attractive option for treating infections caused by multidrug-resistant bacteria (Dion et al., 2020; Strathdee et al., 2023). Phage therapy has already been successfully applied in various countries worldwide, including Belgium, Georgia, China, Germany, and the USA (Jault et al., 2018; Bao et al., 2020; Gelman et al., 2021; Leitner et al., 2021; Uyttebroek et al., 2022). Several clinical trials have been conducted to evaluate the safety and efficacy of phage therapy for various bacterial infections, including Pseudomonas aeruginosa infections in cystic fibrosis patients and Staphylococcus aureus infections in diabetic foot ulcers (Pires et al., 2020; Egido et al., 2021; Uyttebroek et al., 2022). While these studies have shown promising results, larger trials are needed to establish the safety and efficacy of phage therapy in a wider range of patients. On the other hand, more basic research is also needed to support clinical transitional research.
One of the key challenges facing phage therapy is the isolation of suitable phages for therapeutic use (Luong et al., 2020). The failure to isolate appropriate phages is common and can hinder rapid progress in phage therapy (Pires et al., 2015; Jault et al., 2018). Thus, isolation and characterization of phages are needed to perform phage therapy. In this study, we isolated a new lytic phage named vB_EfaS-SFQ1 from hospital sewage. Phage SFQ1 is a siphovirus and exhibits a relatively broad host range against E. faecalis, although it could not infect E. faecium. We provided a detailed biological and genomic description of phage SFQ1 and demonstrated its potential as a candidate for phage-based therapy against E. faecalis infections in vitro. Our findings contribute to the ongoing efforts to develop effective and safe phage-based therapies to combat multidrug-resistant bacterial infections.
Materials and methods
Strains and cultural conditions
Clinical strains of E. faecalis were isolated from the Department of Clinical Laboratory Medicine and subsequently preserved in our laboratory. To culture E. faecalis, brain heart infusion (BHI) medium was utilized and incubated aerobically with shaking at 37°C.
Isolation of phage that infects Enterococcus faecalis
The bacteriophage was isolated from hospital sewage using the previously described method (Duerkop et al., 2016). Briefly, 2 mL of hospital sewage was centrifuged at 10,000 × g for 2 min, and the resulting supernatant was filtered through a 0.22 μm filter. The filtrate was mixed with log-phase bacteria EFS01 and cultured overnight with shaking at 37°C. The mixture was then centrifuged and filtered using a 0.22 μm aseptic filter. Next, 10 μL of the resulting supernatant was mixed with 100 μL of EFS01 host bacteria and added to 5 mL semi-solid BHI before being poured onto agar plates. The plates were then incubated overnight at 37°C until plaques formed. A single plaque was selected and purified by performing the plaque assay three times.
Host range of SFQ1
To determine the host range of SFQ1, the double-layer agar method was conducted on a panel of clinically isolated E. faecalis and E. faecium strains (Table 1). The strains were grown in BHI medium and their susceptibility to lysis by SFQ1 was determined by the formation of clear plaques. The appearance of clear single plaques indicated sensitivity to the phage and the ability of SFQ1 to lyse the strain.
Transmission electron microscopy
Phage SFQ1 morphology was characterized by transmission electron microscopy (TEM) following previously established protocols (Chatterjee et al., 2019). In brief, the filtered phage lysate was loaded onto a copper grid and incubated for 10 min. The grid was then negatively stained with 10 μL of 2% phosphotungstic acid for an additional 5 min. After air-drying, the samples were visualized using a transmission electron microscope operating at 80 kV.
The optimal MOI of phage
To determine the optimal ratio of bacteriophages to bacteria that could produce the highest yield of progeny, the multiplicity of infection (MOI) needs to be determined. In this study, we investigated the optimal MOI for phage SFQ1 by mixing the phage and host bacteria at MOIs of 0.001, 0.01, 0.1, 1 and 10. The mixture was incubated at 37°C with shaking at 220 rpm for 5 h. The lysate was then filtered using a 0.45 μm filter, and the phage titer in each experiment was determined using the double-layer agar method. The experiment was repeated three times to ensure accuracy.
One-step growth curve
The one-step phage growth process was conducted following the previously described method with some modifications. Initially, 5 mL of E. faecalis EFS01 in the exponential growth phase was mixed with phage SFQ1 at an MOI of 0.01. After 5 min of incubation, the mixtures were diluted by 200 times. The culture was further incubated at 37°C, and samples were collected at 10 min intervals. The phage titers in each sample were determined using the double-layer agar method.
Stability of phage SFQ1
The stability of phage SFQ1 was evaluated using various assays. Firstly, pH sensitivity was tested by mixing 10 μL of concentrated phage stock solution (2 × 1010 PFU/mL) with 990 μL of BHI medium, adjusted with HCl or NaOH to pH values ranging from 2.0 to 13.0. The mixture was then incubated at 37°C for 60 min, and the phage titer was determined using the double agar layer method. The thermal stability of phage SFQ1 was tested by incubating the phage at temperatures ranging from 4°C to 70°C for 1 h, followed by determining the survival phage titer using the double-layer agar method. Additionally, the chloroform sensitivity assay was conducted by incubating phage SFQ1 (108 PFU/mL) with varying ratios of chloroform (10%, 30%, 50%, 70%, 90%) at 37°C with shaking for 10 min. The mixture was then centrifuged, and the phage titer in the upper layer was calculated using the double-layer agar method.
Phage genome extraction
Extraction of the phage SFQ1 genome was conducted following a previously described method (Han et al., 2022). Initially, DNase I and RNase A were added to the phage stocks to attain a final concentration of 1 μg/mL, which facilitated the removal of contaminated DNA and RNA. The mixture was then incubated at 37°C for 60 min. Subsequently, the phage genomic DNA was extracted using the phenol and chloroform method (Duerkop et al., 2016). The DNA was finally dissolved in sterile water and preserved at a temperature of −20°C until genome sequencing.
Phage genome sequencing
The genome of phage SFQ1 was sequenced using the Illumina Hiseq 2500 platform. The reads were processed using Fastp to remove any errors and trimmed for quality control. Then, CLC software from QIAGEN (Germany) was applied to assemble the phage genome and annotated with RAST (Overbeek et al., 2014), an online tool that predicted the open reading frames (ORFs). Each ORF’s DNA and protein sequences were manually searched for homologs by BlastN and BlastP. Finally, the phage genome was visualized using the GraphPad Prism 8.0.2, and the sequence was deposited in NCBI under accession number OQ831052. The antibiotic resistance genes were predicted by the online prediction platform ResFinder1 and the virulence factors were predicted using the VirulenceFinder,2 respectively.
Phylogenetic and comparative analysis of phage genome
To investigate the genetic relationships between phage SFQ1 and other E. faecalis phages, a phylogenetic tree was constructed using complete genome sequences of 21 phages, including SFQ1. The analysis was carried out with MEGA 7 software, which allowed for the assessment of similarities and differences in their genetic traits. Additionally, intergenomic similarities between SFQ1 and other E. faecalis phages were calculated using VIRIDIC (Moraru et al., 2020) to gain a deeper understanding of their relationships.
Biofilm assays
The methodology for the biofilm assay was adapted from a previous study (Goodarzi et al., 2022). To establish a biofilm, fill each well of a 96-well plate with 100 μL of BHI medium containing an overnight culture of E. faecalis. The plates were incubated at 37°C for 24 h, 48 h, or 72 h to allow the biofilm to form. After removing the planktonic cells, 100 μL of phage with a titer of 108 PFU/mL was added to each well, while control wells were added with 100 μL of SM buffer. Incubation was continued at 37°C for 6 h. Then, plates were washed with PBS and the biofilm biomass was stained with crystal violet for 15 min. To quantify the results, crystal violet was solubilized in 0.2 mL of 95% ethanol, and the OD600 was determined using a SpectraMax M3 multimode microplate reader. Each sample was examined in triplicate, and BHI medium alone served as the negative control.
Statistical analysis
Each experiment was repeated three times to ensure the reliability of the results. The statistical analysis was carried out using GraphPad Prism 8.0.2, and student’s t-test was used where applicable. A p-value less than 0.05 was considered statistically significant.
Results
Isolation of Enterococcus faecalis bacteriophage SFQ1
Bacteriophage that specifically targets E. faecalis strain EFS01 was obtained from the sewage of the hospital. Phage plaques were first observed on a top agar lawn containing the clinical strain EFS01. The plaque was purified three times, resulting in the isolation of phage SFQ1. This phage formed clear plaques that were approximately 3 mm in diameter on a double-layered agar plate, as shown in Figure 1A. Electron microscopy revealed that phage SFQ1 had an icosahedral head with a diameter of around 50 nm and a long tail (Figure 1B). Thus, phage SFQ1 is a siphovirus and was designated vB_EFaS_SFQ1.
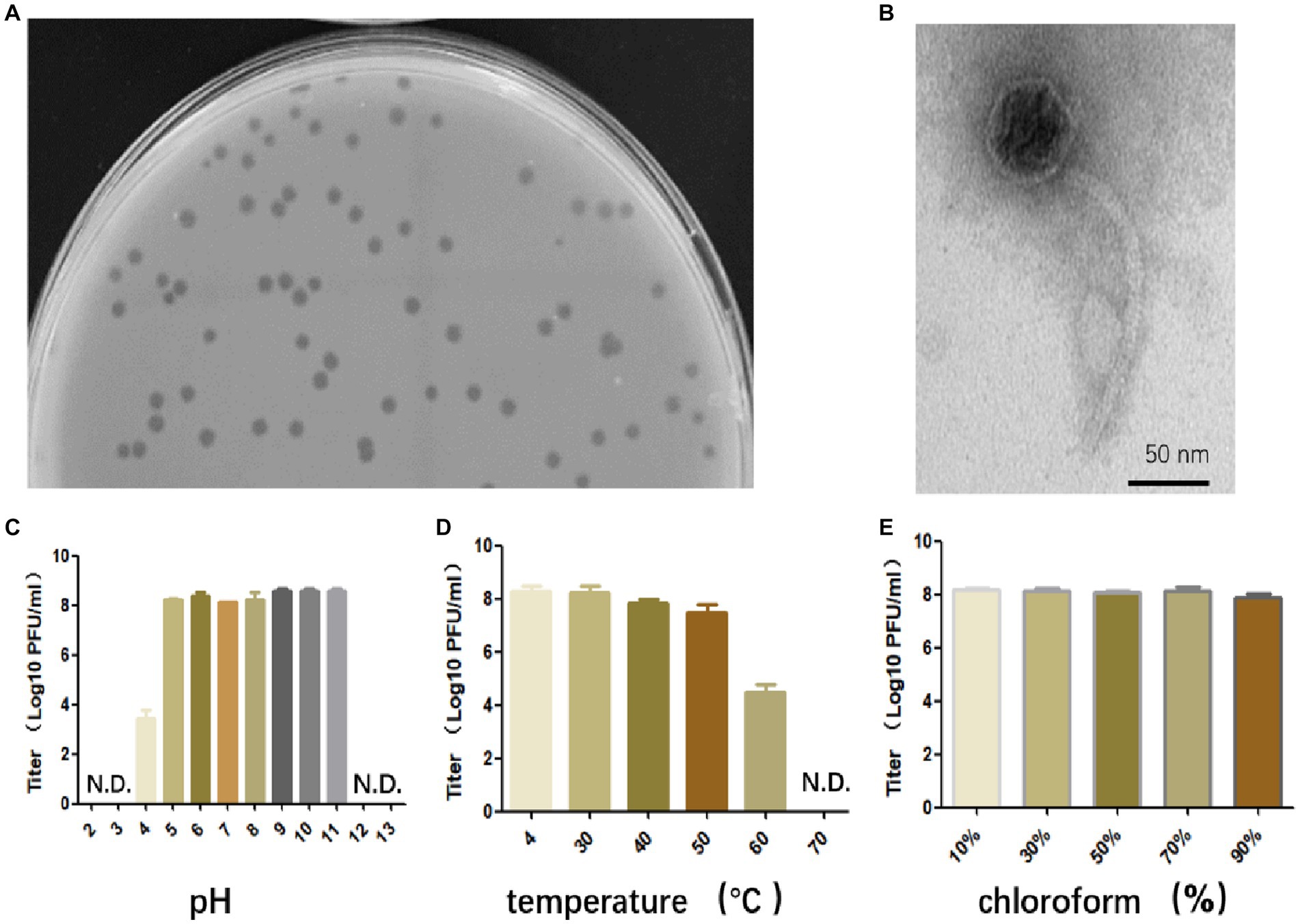
Figure 1. The biological characteristics of phage SFQ1. (A) The plaques of phage SFQ1 formed on a double-layered agar plate. (B) The transmission electron microscopy image displays the morphology of the phage SFQ1, with a scale bar of 50 nm. (C) Phage SFQ1 is stable over a broad range of pH values from 5 to 11. (D) SFQ1 could tolerate 50°C treatment but was completely inactivated at 70°C. (E) SFQ1 is completely resistant to chloroform treatment, and its titer is not changed for various concentrations of chloroform treatment.
Stability of bacteriophage
Phage SFQ1’s stability was evaluated under different pH, temperature, and chloroform treatments. The results revealed that SFQ1 remained viable within a pH range of 5 to 11 (Figure 1C). Additionally, SFQ1 was able to survive at 50°C but rapidly became inactivated at 60°C (Figure 1D). However, after chloroform treatment, the titer of SFQ1 is not changed using various concentrations of chloroform, indicating that it is completely resistant to chloroform (Figure 1E). These findings suggest that SFQ1 can withstand moderate acid and alkali conditions, high temperatures, and chloroform treatment.
Biological characterization of bacteriophage
The optimal MOI was determined by mixing phage with bacteria at different ratios, and when the MOI was 0.01, it produced the maximum titer of particles (1.44 × 1010 PFU/mL), indicating that 0.01 is the optimal MOI of SFQ1 (Figure 2A).
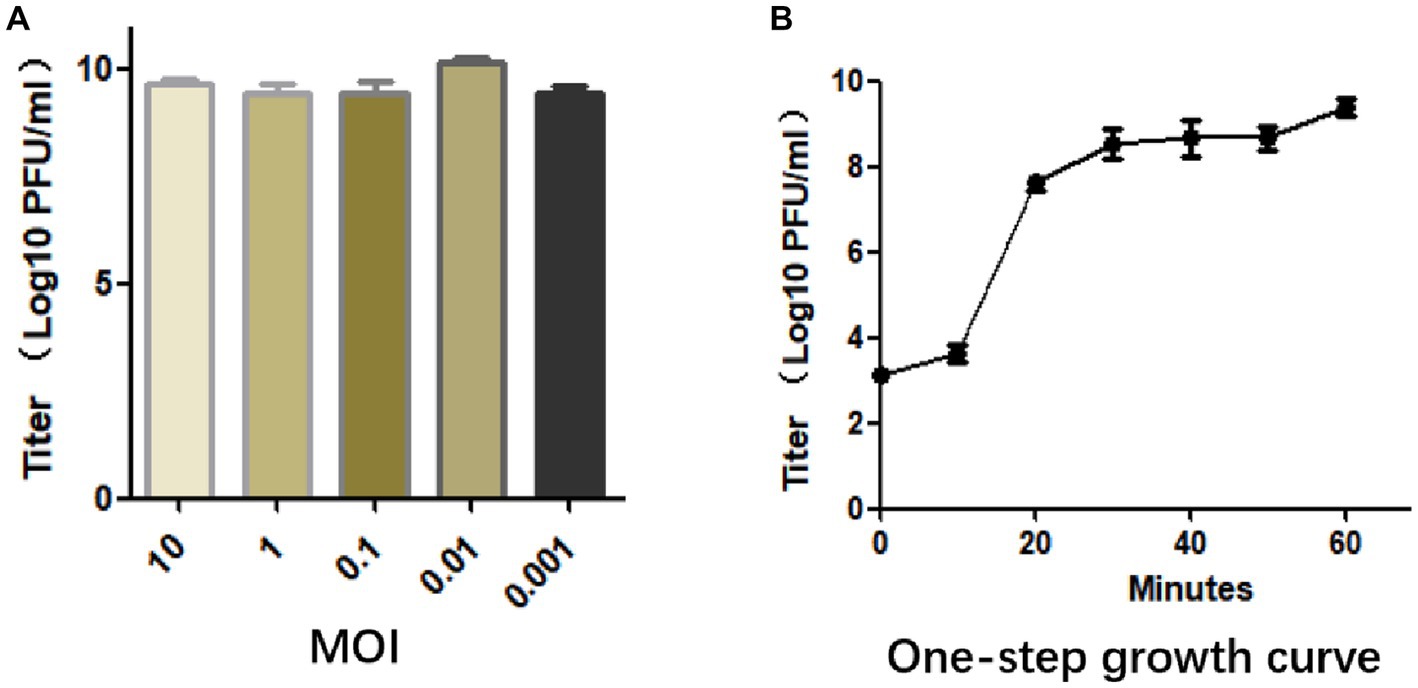
Figure 2. (A) The multiplicity of infection assay indicates that SFQ1 produced the most progenies when the phage and host bacteria were mixed at MOIs of 0.01. (B) The one-step growth curve for bacteriophage SFQ1 indicates that the phage titer increased after 10 min of infection and plateaued at 30 min, with a lysis time of approximately 30 min.
To further characterized phage SFQ1, a one-step growth curve experiment was conducted, which showed that phage SFQ1 had a 10 min latent period and plateaued 30 min after phage infection (Figure 2B). Thus, the phage lysis time is about 30 min, and the burst size is about 110 PFU/cell based on the curve.
The host range of SFQ1 was estimated by plaque assays. Ten clinically isolated E. faecalis strains and five E. faecium strains were tested for their sensitivity against SFQ1. Six E. faecalis strains could be lysed by SFQ1, indicating a modest host range, while none of the E. faecium strains could be infected by SFQ1 (Table 1).
Genomic characterization of phage SFQ1
The genome structure of SFQ1, a newly isolated E. faecalis phage, was investigated in this study. The phage genome was shown to be linear double-stranded DNA with a length of 40,787 base pairs and a G + C content of 35%. The genomic similarity of SFQ1 to E. faecalis Phage (NC_042126.1) was found to be 94.2%, based on sequence alignment. The SFQ1 genome encodes 63 predicted open reading frames (ORFs), with 17 of them considered functional genes and the remaining 46 annotated as hypothetical proteins (Figure 3). The complete genome sequence of SFQ1 is available at GenBank (accession no. OQ831052).
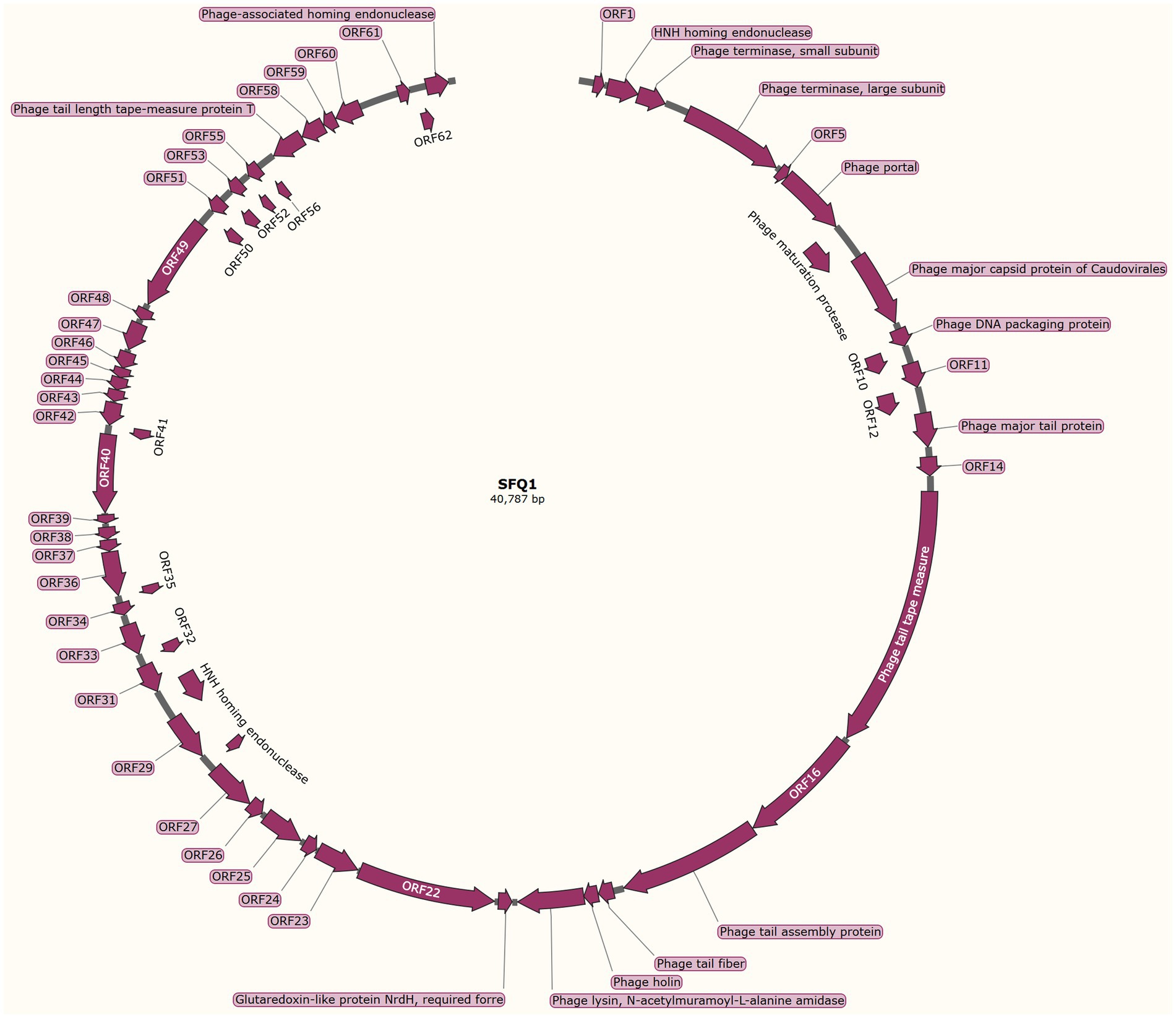
Figure 3. The genome characterization of phage SFQ1. The circular genome map of SFQ1 was generated using GraphPad Prism 8.0.2, and the annotation of ORFs was performed using RAST and the BLASTP database. The function of each predicted ORFs of phage SFQ1 are shown, and 17 out of the 63 predicted ORFs are functionally annotated.
The annotated genes in SFQ1’s genome were genes involved in DNA replication and modification, phage genome packaging, structural proteins, and host lysis proteins modules. ORF2 was predicted to encode the HNH homing endonuclease, which is involved in phage DNA packing by cooperation with terminase, which is encoded by ORF3 and ORF4. While the phage structural proteins, such as tail fiber and capsid, are encoded in a specific region. ORF13 encodes the phage major tail protein, ORF15 encodes the phage tail tape measure protein, ORF17 encodes the phage tail assembly protein. The phage lysin, which includes a N-acetylmuramoyl-L-alanine amidase, is also predicted close to the tail fiber gene. But, most of the genes are unknown proteins, such as the genes that are used to control host metabolism are not annotated. And none of the ORFs encoded virulence factors or antibiotic resistance genes, and the genome of SFQ1 did not encode lysogenic modules, such as integrase or repressor proteins. These data indicate that SFQ1 is a strictly lytic phage and can be safely used to treat E. faecalis infections.
Phylogenetic analysis
To better understand the evolution and relationship of phage SFQ1 with other E. faecalis phages, the genome of SFQ1 was compared with that of 20 E. faecalis phages, which were downloaded from NCBI that have homology with SFQ1.
Based on the whole genome sequences, the phylogenetic tree showed that IME278 was grouped into a clade with siphovirus, which is consistent with the results of the whole genome similarity analysis (Figure 4). Phage vB_EfaS_AL3 was highly similar to phage SFQ1. Then, we used VIRIDIC to calculate the intergenomic similarities, showing the similarity between phages SFQ1 and NC_042126.1 was 84.2%, while that with NC_042023.1 was 80.1% (Figure 5).
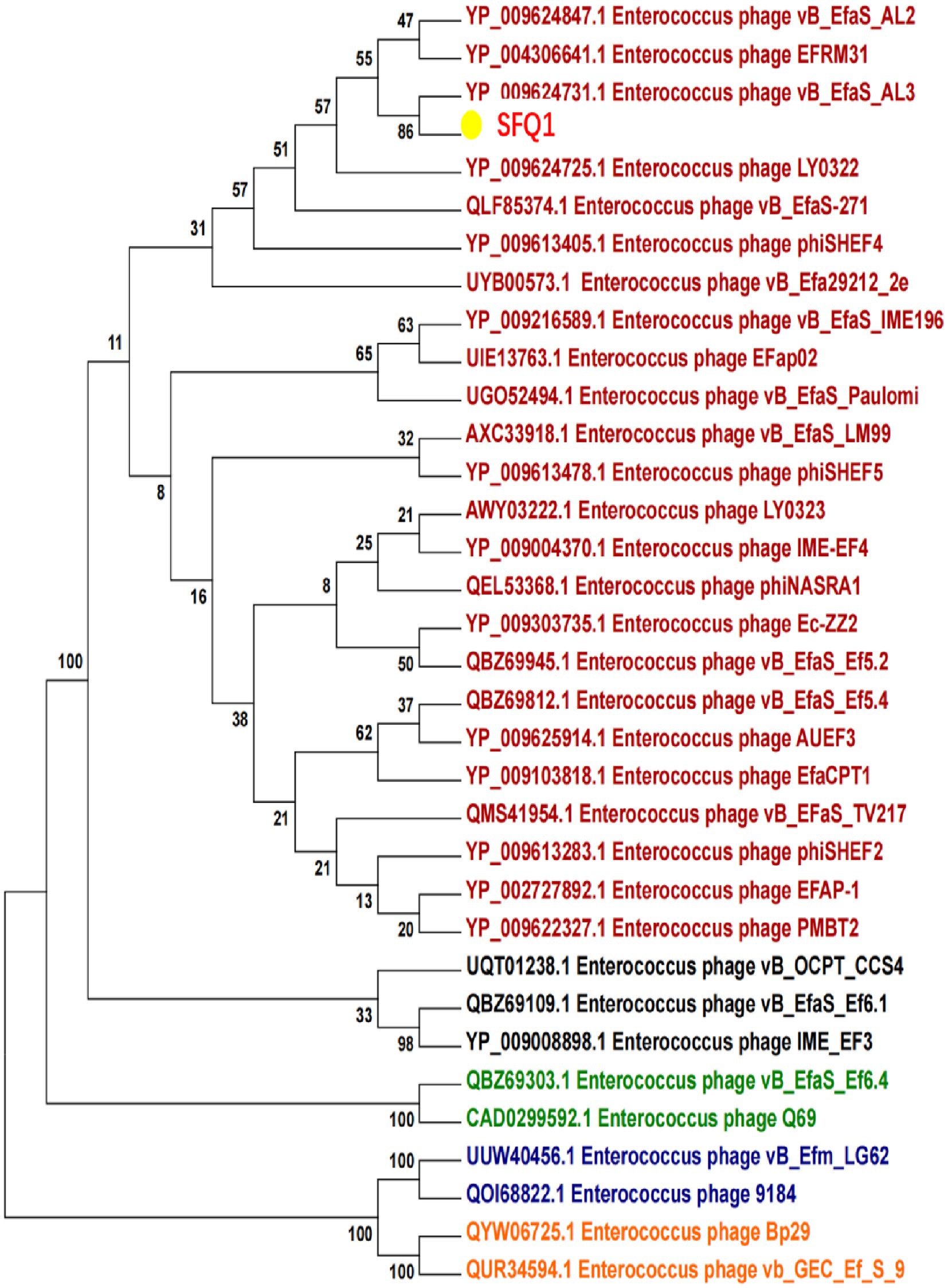
Figure 4. Phylogenetic analysis of SFQ1 and various E. faecalis bacteriophages based on the similarity of the whole genome sequences. SFQ1 was labeled in red in a clade with phage vB_EfaS_AL3.
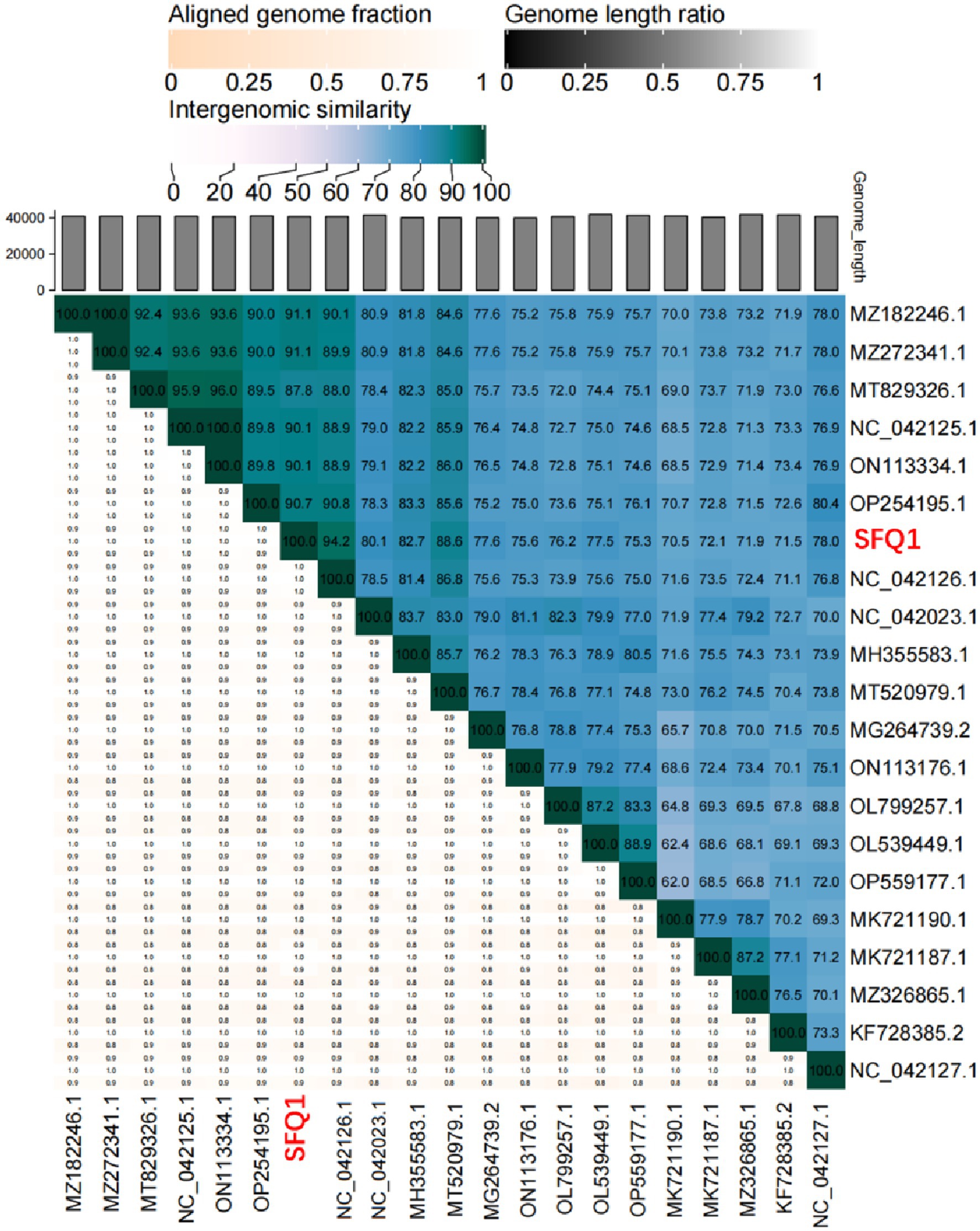
Figure 5. Heatmap of the average nucleotide identity values between SFQ1 and 20 most similar bacteriophages. According to a matrix of Hadamard values of pairwise alignment coverage and the percentage identity, the ANI values were generated and SFQ1 was marked in red.
Biofilm disruption
The efficiency of phage SFQ1 in destroying existing biofilms of E. faecalis EFS01 was assessed under various biofilm-forming conditions (Figure 6). Biofilm experiments were conducted thrice with duplicate readings, and the biofilm was grown to three-time points of 24 h, 48 h, and 72 h. At each stage, phage SFQ1 significantly reduced the E. faecalis EFS01 biofilm within 6 h, and the reduction was statistically significant (p < 0.05) (Figure 6), indicating that SFQ1 can disrupt the biofilm efficiently.
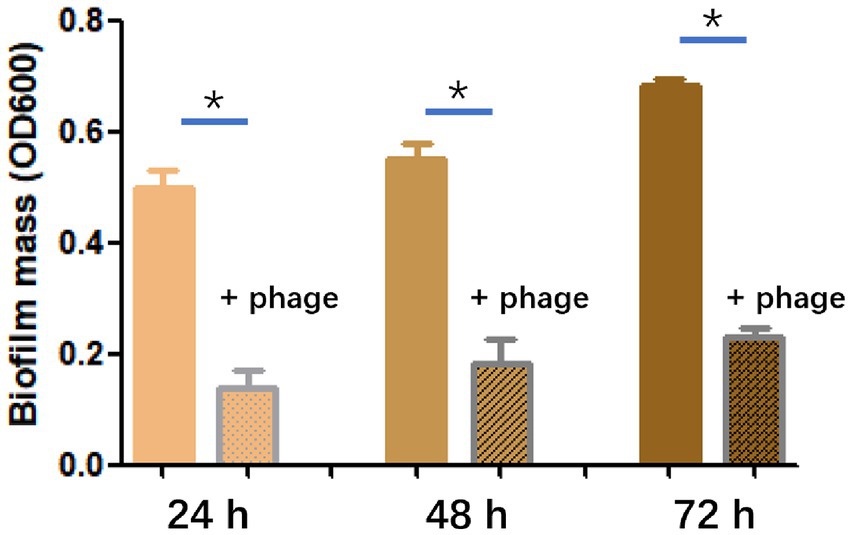
Figure 6. The effect of phage SFQ1 on biofilms formed by E. faecalis EFS01 was evaluated by measuring biofilm using crystal violet assay before and after phage treatment. Biofilms were formed for 24 h, 48 h, or 72 h, and were treated with phage SFQ1 for 6 h. The results showed that phage SFQ1 had a significant effect on biofilms formed for all three-time points. The asterisks indicate statistical significance (p < 0.05).
Discussion
Phage therapy has been widely recognized as a promising strategy for treating infectious diseases caused by various bacteria, including E. faecalis (El Haddad et al., 2019). Thus, more and more E. faecalis phages were identified that targets different parts of the E. faecalis cell wall, and the cocktail with different phages could reduce the likelihood of phage resistance development (Banla et al., 2018; Al-Zubidi et al., 2019; Chatterjee et al., 2019). One study identified a cocktail of phages that were effective in killing antibiotic-resistant E. faecalis strains in vitro but not effective in the gut of mice (Buttimer et al., 2022). In addition to the well-known phage therapy, phages also play a vital role in treating many other diseases. For instance, (Duan et al., 2019) discovered phages that specifically target cytolysin-positive E. faecalis strains, which can cause hepatocyte lysis under the alcohol condition and lead to alcoholic hepatitis. The use of these phages has shown promising results in treating alcoholic hepatitis caused by E. faecalis. The development of phages with specificity against different E. faecalis strains has the potential to revolutionize the treatment of infectious diseases caused by this bacterium. Moreover, the characterization of E. faecalis phages can also shed light on their potential role in the gut microbiome and provide new therapeutic opportunities for gut microbiome-associated diseases. Therefore, further research and characterization of E. faecalis phages are essential for developing effective therapies against E. faecalis infections and other microbiome-associated diseases.
In this study, we characterized an effective E. faecalis phage SFQ1, it is a dsDNA phage with a large burst size, and lyse the host effectively. Moreover, its genome was sequenced and did not encode any lysogenic gene, antibiotic resistance gene, or virulent gene. Thus, SFQ1 is an effective and safe candidate for phage therapy.
Biofilms are communities of bacteria that can be highly resistant to antibiotics and can contribute to persistent infections (Khalifa et al., 2015; Sanz et al., 2017). Thus, researchers have investigated the potential of using phages to treat biofilm-related E. faecalis infections. A study found that a phage cocktail was effective in reducing E. faecalis biofilm formation in vitro, suggesting its potential use as a therapeutic agent (Khalifa et al., 2015). In this study, we also test the effect of SFQ1 on disrupting the existing biofilm, and we found that all three stage biofilms could be significantly disrupted by phage SFQ1, thus in clinical therapy, SFQ1 could be effective when combined with the use of antibiotics.
In conclusion, our findings highlight the potential of SFQ1 as a promising candidate for phage therapy against E. faecalis infections. Future studies should focus on identifying its specific receptor, which is critical for developing effective phage cocktails and minimizing the risk of phage resistance. Overall, this study suggested the importance of exploring the potential of phages as alternative therapies against antibiotic-resistant bacteria.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author contributions
WG and XW conceived and designed the experiments. FS, JS, JT, and HX performed the experiments. FS and JS analyzed the data. WG and XW wrote the paper. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by Natural Science Foundation of Sichuan Province of China (2023NSFSC1902).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Ali, I. A. A., and Neelakantan, P. (2022). Antibiofilm activity of phytochemicals against Enterococcus faecalis: a literature review. Phytother. Res. 36, 2824–2838. doi: 10.1002/ptr.7488
Al-Zubidi, M., Widziolek, M., Court, E. K., Gains, A. F., Smith, R. E., Ansbro, K., et al. (2019). Identification of novel bacteriophages with therapeutic potential that target Enterococcus faecalis. Infect. Immun. 87:e00512. doi: 10.1128/IAI.00512-19
Arias, C. A., Panesso, D., McGrath, D. M., Qin, X., Mojica, M. F., Miller, C., et al. (2011). Genetic basis for in vivo daptomycin resistance in enterococci. N. Engl. J. Med. 365, 892–900. doi: 10.1056/NEJMoa1011138
Banla, I. L., Kommineni, S., Hayward, M., Rodrigues, M., Palmer, K. L., Salzman, N. H., et al. (2018). Modulators of Enterococcus faecalis cell envelope integrity and antimicrobial resistance influence stable colonization of the mammalian gastrointestinal tract. Infect. Immun. 86:e00381. doi: 10.1128/IAI.00381-17
Bao, J., Wu, N., Zeng, Y., Chen, L., Li, L., Yang, L., et al. (2020). Non-active antibiotic and bacteriophage synergism to successfully treat recurrent urinary tract infection caused by extensively drug-resistant Klebsiella pneumoniae. Emerg. Microbes. Infect. 9, 771–774. doi: 10.1080/22221751.2020.1747950
Bright, M., Parfitt, E. C., Pasquill, K., Steele, L., and Laupland, K. B. (2020). Occurrence and determinants of enterococcal bloodstream infections: a population-based study. Infect. Dis. 52, 638–643. doi: 10.1080/23744235.2020.1774074
Buttimer, C., Sutton, T., Colom, J., Murray, E., Bettio, P. H., Smith, L., et al. (2022). Impact of a phage cocktail targeting Escherichia coli and Enterococcus faecalis as members of a gut bacterial consortium in vitro and in vivo. Front. Microbiol. 13:936083. doi: 10.3389/fmicb.2022.936083
Cattoir, V. (2022). The multifaceted lifestyle of enterococci: genetic diversity, ecology and risks for public health. Curr. Opin. Microbiol. 65, 73–80. doi: 10.1016/j.mib.2021.10.013
Chatterjee, A., Johnson, C. N., Luong, P., Hullahalli, K., McBride, S. W., Schubert, A. M., et al. (2019). Bacteriophage resistance alters antibiotic-mediated intestinal expansion of enterococci. Infect. Immun. 87:e00085. doi: 10.1128/IAI.00085-19
Dion, M. B., Oechslin, F., and Moineau, S. (2020). Phage diversity, genomics and phylogeny. Nat. Rev. Microbiol. 18, 125–138. doi: 10.1038/s41579-019-0311-5
Duan, Y., Llorente, C., Lang, S., Brandl, K., Chu, H., Jiang, L., et al. (2019). Bacteriophage targeting of gut bacterium attenuates alcoholic liver disease. Nature 575, 505–511. doi: 10.1038/s41586-019-1742-x
Duerkop, B. A., Huo, W., Bhardwaj, P., Palmer, K. L., and Hooper, L. V. (2016). Molecular basis for lytic bacteriophage resistance in enterococci. mBio 7:e01304. doi: 10.1128/mBio.01304-16
Egido, J. E., Costa, A. R., Aparicio-Maldonado, C., Haas, P. J., and Brouns, S. J. J. (2021). Mechanisms and clinical importance of bacteriophage resistance. FEMS Microbiol. Rev. 46:fuab048. doi: 10.1093/femsre/fuab048
El Haddad, L., Harb, C. P., Gebara, M. A., Stibich, M. A., and Chemaly, R. F. (2019). A systematic and critical review of bacteriophage therapy against multidrug-resistant ESKAPE organisms in humans. Clin. Infect. Dis. 69, 167–178. doi: 10.1093/cid/ciy947
Gelman, D., Yerushalmy, O., Alkalay-Oren, S., Rakov, C., Ben-Porat, S., Khalifa, L., et al. (2021). Clinical phage microbiology: a suggested framework and recommendations for the in-vitro matching steps of phage therapy. Lancet Microbe. 2, e555–e563. doi: 10.1016/S2666-5247(21)00127-0
Goodarzi, F., Hallajzadeh, M., Sholeh, M., Talebi, M., Pirhajati Mahabadi, V., and Amirmozafari, N. (2022). Anti-biofilm activity of a lytic phage against vancomycin-resistant Enterococcus faecalis. Iran. J. Pathol. 17, 285–294. doi: 10.30699/ijp.2022.541855.2760
Gorski, A., Międzybrodzki, R., Weber-Dąbrowska, B., Fortuna, W., Letkiewicz, S., Rogóż, P., et al. (2016). Phage therapy: combating infections with potential for evolving from merely a treatment for complications to targeting diseases. Front. Microbiol. 7:9. doi: 10.3389/fmicb.2016.01515
Han, K., Dong, Y., An, X., Song, L., Li, M., Fan, H., et al. (2022). Potential application of a newly isolated phage BUCT609 infecting Stenotrophomonas maltophilia. Front. Microbiol. 13:1001237. doi: 10.3389/fmicb.2022.1001237
Hayakawa, K., Marchaim, D., Palla, M., Gudur, U. M., Pulluru, H., Bathina, P., et al. (2013). Epidemiology of vancomycin-resistant Enterococcus faecalis: a case-case-control study. Antimicrob. Agents Chemother. 57, 49–55. doi: 10.1128/AAC.01271-12
Jabbari Shiadeh, S. M., Pormohammad, A., Hashemi, A., and Lak, P. (2019). Global prevalence of antibiotic resistance in blood-isolated Enterococcus faecalis and Enterococcus faecium: a systematic review and meta-analysis. Infect. Drug Resist. 12, 2713–2725. doi: 10.2147/IDR.S206084
Jault, P., Leclerc, T., Jennes, S., Pirnay, J. P., Que, Y. A., Resch, G., et al. (2018). Efficacy and tolerability of a cocktail of bacteriophages to treat burn wounds infected by Pseudomonas aeruginosa (PhagoBurn): a randomised, controlled, double-blind phase 1/2 trial. Lancet Infect. Dis. 19, 35–45. doi: 10.1016/S1473-3099(18)30482-1
Kakoullis, L., Papachristodoulou, E., Chra, P., and Panos, G. (2021). Mechanisms of antibiotic resistance in important gram-positive and gram-negative pathogens and novel antibiotic solutions. Antibiotics 10:415. doi: 10.3390/antibiotics10040415
Khalifa, L., Brosh, Y., Gelman, D., Coppenhagen-Glazer, S., Beyth, S., Poradosu-Cohen, R., et al. (2015). Targeting Enterococcus faecalis biofilms with phage therapy. Appl. Environ. Microbiol. 81, 2696–2705. doi: 10.1128/AEM.00096-15
Leitner, L., Ujmajuridze, A., Chanishvili, N., Goderdzishvili, M., Chkonia, I., Rigvava, S., et al. (2021). Intravesical bacteriophages for treating urinary tract infections in patients undergoing transurethral resection of the prostate: a randomised, placebo-controlled, double-blind clinical trial. Lancet Infect. Dis. 21, 427–436. doi: 10.1016/S1473-3099(20)30330-3
Luong, T., Salabarria, A. C., Edwards, R. A., and Roach, D. R. (2020). Standardized bacteriophage purification for personalized phage therapy. Nat. Protoc. 15, 2867–2890. doi: 10.1038/s41596-020-0346-0
Moraru, C., Varsani, A., and Kropinski, A. M. (2020). VIRIDIC-A novel tool to calculate the Intergenomic similarities of prokaryote-infecting viruses. Viruses 12:1268. doi: 10.3390/v12111268
Overbeek, R., Olson, R., Pusch, G. D., Olsen, G. J., Davis, J. J., Disz, T., et al. (2014). The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res. 42, D206–D214. doi: 10.1093/nar/gkt1226
Pires, D. P., Costa, A. R., Pinto, G., Meneses, L., and Azeredo, J. (2020). Current challenges and future opportunities of phage therapy. FEMS Microbiol. Rev. 44, 684–700. doi: 10.1093/femsre/fuaa017
Pires, D. P., Vilas Boas, D., Sillankorva, S., and Azeredo, J. (2015). Phage therapy: a step forward in the treatment of Pseudomonas aeruginosa infections. J. Virol. 89, 7449–7456. doi: 10.1128/JVI.00385-15
Sanz, M., Beighton, D., Curtis, M. A., Cury, J. A., Dige, I., Dommisch, H., et al. (2017). Role of microbial biofilms in the maintenance of oral health and in the development of dental caries and periodontal diseases. Consensus report of group 1 of the joint EFP/ORCA workshop on the boundaries between caries and periodontal disease. J. Clin. Periodontol. 44, S5–S11. doi: 10.1111/jcpe.12682
Strathdee, S. A., Hatfull, G. F., Mutalik, V. K., and Schooley, R. T. (2023). Phage therapy: from biological mechanisms to future directions. Cells 186, 17–31. doi: 10.1016/j.cell.2022.11.017
Theuretzbacher, U., Outterson, K., Engel, A., and Karlén, A. (2020). The global preclinical antibacterial pipeline. Nat. Rev. Microbiol. 18, 275–285. doi: 10.1038/s41579-019-0288-0
Uyttebroek, S., Chen, B., Onsea, J., Ruythooren, F., Debaveye, Y., Devolder, D., et al. (2022). Safety and efficacy of phage therapy in difficult-to-treat infections: a systematic review. Lancet Infect. Dis. 22, e208–e220. doi: 10.1016/S1473-3099(21)00612-5
Keywords: bacteriophage, Enterococcus faecalis , antibiotic resistance, phage therapy, vancomycin-resistant
Citation: Song F, Sheng J, Tan J, Xie H, Wang X and Guo W (2023) Characterization of an Enterococcus faecalis bacteriophage SFQ1 as a potential therapeutic agent. Front. Microbiol. 14:1210319. doi: 10.3389/fmicb.2023.1210319
Edited by:
Bing Liu, Xi’an Jiaotong University, ChinaReviewed by:
Mengzhe Li, Beijing University of Chemical Technology, ChinaMengjun Cheng, Fudan University, China
Copyright © 2023 Song, Sheng, Tan, Xie, Wang and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoyu Wang, c2NuY3d4eUAxNjMuY29t; Wenqiong Guo, Mjk5ODA4NzJAcXEuY29t
 Fuqiang Song1
Fuqiang Song1 Wenqiong Guo
Wenqiong Guo