
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 31 August 2023
Sec. Terrestrial Microbiology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1208971
Soil bacteria are closely related to soil environmental factors, and their community structure is an important indicator of ecosystem health and sustainability. A large number of artificial grasslands have been established to control rocky desertification in the karst areas of southern China, but the influence of different use patterns on the soil bacterial community in artificial grasslands is not clear. In this study, three grassland use patterns [i.e., grazing (GG), mowing (MG), and enclosure (EG)] were used to investigate the effects of different use patterns on the soil bacterial community in artificial grassland by using 16S rDNA Illumina sequencing and 12 soil environmental indicators. It was found that, compared with EG, GG significantly changed soil pH, increased alkaline hydrolyzable nitrogen (AN) content (P < 0.05), and decreased soil total phosphorus (TP) content (P < 0.05). However, MG significantly decreased the contents of soil organic carbon (SOC), total phosphorus (TP), available nitrogen (AN), ammonium nitrogen (NH4+-N), β-1,4-glucosidase (BG), and N-acetyl-β-D-glucamosonidase (NAG) (P < 0.05). The relative abundance of chemoheterotrophy was significantly decreased by GG and MG (P < 0.05). GG significantly increased the relative abundance of Acidobacteria and Gemmatimonadota (P < 0.05) and significantly decreased the relative abundance of Proteobacteria (P < 0.05), but the richness index (Chao 1) and diversity index (Shannon) of the bacterial community in GG, MG, and EG were not significantly different (P > 0.05). The pH (R2 = 0.79, P = 0.029) was the main factor affecting the bacterial community structure. This finding can provide a scientific reference for ecological restoration and sustainable utilization of grasslands in the karst desertification areas.
Soil microorganisms, as the engine of the biogeochemical cycle, play a central role in regulating soil fertility, plant growth, and climate change (Nelson et al., 2016; He et al., 2022; Ma L. N. et al., 2022). They maintain the sustainable development of the soil ecosystem. Changes in the composition and abundance of soil microbial communities are important biological indicators for measuring soil fertility as they directly influence the composition and transformation of soil nutrients and indicate the level of ecosystem health (Baranova et al., 2019). Regulating the diversity and function of soil microbial can enhance terrestrial ecosystem services and promote the restoration of degraded ecosystems (Yu et al., 2022). Soil microbes play the role of “changer maker” in restoring degraded soil functions (Coban et al., 2022), so studying the links between microbial community functions and ecosystem changes has been a challenge for soil microbiology research (Fierer, 2017; Ma Q. X. et al., 2022). Because changes in microbial communities have an impact on community function and their sensitivity to adversity, it is unclear how microbial community structure responds to anthropogenic environmental perturbations, similar to different utilization patterns.
Soil bacteria are the most abundant and widely distributed among soil microorganisms, accounting for 70%−90% of all soil microorganisms, and their various metabolic activities are closely related to soil environmental factors and are extremely sensitive to environmental changes (Liu et al., 2015; Xue et al., 2017; Sui et al., 2019; Lu et al., 2022; Qiu et al., 2023). Soil bacterial communities are not only influenced by the climatic environment but also directly and indirectly affected by changes in vegetation (de Vries et al., 2017; Mod et al., 2021). There is evidence that bacterial communities respond directly or indirectly to anthropogenic changes in the soil environment (Hermans et al., 2020; Li et al., 2023). It was found that the composition of soil bacterial communities is closely related to the type of land use and that the type of land use can be correctly determined with an accuracy of up to 85% (Hermans et al., 2020). Some studies also found that heavily managed soils contain different bacterial communities compared to unmanaged soils (Drenovsky et al., 2010), and the type of land use correlates with changes in the composition of bacterial communities (Plassart et al., 2019). More specifically, management practices such as fertilization, establishment of artificial plants, or grazing have been shown to affect soil microbial communities (Fierer et al., 2012; Figuerola et al., 2015; Cassman et al., 2016). Overall, the composition of bacterial communities is strongly influenced by changes in the soil environment, many of which are a direct result of land-use change (Hermans et al., 2020). Therefore, by investigating the composition and variation of soil bacterial communities, we can better predict and control degraded ecosystem changes to realize the improvement and sustainable development of degraded ecosystems (Sun et al., 2022).
Karst landscapes account for ~15% of the world's total land area (Xiong et al., 2017; Garousi et al., 2021). Among them, South China Karst is one of the three major concentrated and continuous karst distribution areas in the world (Xiong et al., 2016; Zhang J. X. et al., 2021). Due to global climate change and unreasonable land use, the karst ecosystem in southern China has been severely degraded, and the problem of rocky desertification is prominent (Jiang et al., 2014), seriously threatening the ecological security and socioeconomic sustainable development of the region (Li S. L. et al., 2020). Desertification has long been a major economic, social, and environmental issue of concern to many countries and regions of the world (Oldroyd and Leyser, 2020). Karst desertification, which is similar to desertified landscapes, has attracted much attention from the international community (Xiong et al., 2002). It has been shown that the establishment of artificial grasslands is an important initiative to rapidly repair the damaged ecological environment of rocky desertification (Wang et al., 2019), which is of great significance to promote ecological reconstruction and economic development (Fang et al., 2018; Xin et al., 2020; Bai and Cotrufo, 2022). However, due to the different utilization of artificial grassland, desertification may change the microbial community structure formed by long-term evolution and then change the soil microbial–soil–plant nutrient relationship (Pei et al., 2021), ultimately having a profound effect on grassland ecosystems and their functions (Liu et al., 2022).
Since 1999, a large-scale special project of grain for green and vegetation restoration to control rocky desertification has been implemented in the karst areas of southern China. According to statistics, the area of ecological measures dominated by establishing artificial grassland has exceeded 10,000 km2 (Liu et al., 2021), and good social, economic, and ecological benefits have been achieved. Although many previous studies have been carried out on the response of grassland soil bacteria to land-use change in the karst desertification area, such as soil microbial communities (Li et al., 2018; Huang et al., 2020; Song et al., 2022; Barber et al., 2023), limitation of soil microbial resources (Chen et al., 2019; Sun et al., 2021; Qian et al., 2022; Wu et al., 2022; Soozandehfar et al., 2023), and community structures and functions (Wang D. et al., 2022). However, systematic studies on the response of soil bacterial communities in artificial grasslands to different use patterns are obviously lacking, especially for artificial grasslands in the karst desertification areas, which are at the stage of sporadic exploration. Therefore, there is an urgent need to clarify the response mechanisms of soil bacterial communities to different grassland utilization modes to help us comprehensively understand and predict the effects of external disturbances on grassland ecosystems and their future trends (Feng et al., 2022). In this study, it was hypothesized that different utilization patterns of artificial grassland in the karst desertification control area will change the structure, richness, and diversity of the soil bacterial community. To address the above hypotheses, this study analyzed the effects of different grassland use patterns on soil bacterial communities by using 16S rDNA Illumina sequencing and soil chemical properties analysis technology. The objectives were to (1) investigate changes in soil chemical index and soil enzyme activity under different grassland utilization patterns; (2) evaluate the influence of different use patterns on soil bacteria structure and diversity in artificial grassland; and (3) determine the relationship between soil bacterial community and environmental factors so as to provide theoretical support for ecological restoration and sustainable utilization of grassland in the karst desertification control areas in southern China.
The study area is located in Salaxi Town, Qixingguan District, Bijie City, Guizhou Province (105°02′01″-105°08′09″E, 27°11′36″-27°16′51″N), which is a typical karst plateau mountain area with light-to-moderate karst desertification. The karst desertification area is 55.931 km2, accounting for 64.93% of the total area of the demonstration area. The study area belongs to the subtropical monsoon humid climate, with an average altitude of 1,800 m, an annual average temperature of about 12°C, a frost-free period of 245 days, an average annual sunshine duration of 1,360 h, an annual average rainfall of 984.4 mm, and precipitation concentrated from June to September. The soil is zonal calcareous soil. The vegetation is dominated by Cyclobalanopsis glauca, Pyracantha fortuneana, Rhododendron simsii, Juglans regia, Rosa roxburghii, Artemisia lavandulaefolia, Chenopodium glaucum, Clinopodium chinense, Plantago asiatica, Stellaria media, Digitaria sanguinalis, and Polygonum hydropiper.
During the implementation of China's “13th Five-Year” National Key Research & Development Project, our research team established artificial grassland in the study area in mid-April 2012 to restore the damaged karst desertification ecosystem. The artificial grassland was sown with Lolium perenne+Trifolium repens+Dactylis glomerata, and it was planted according to the seed ratio of 2:2:1. After the establishment of artificial grassland, free grazing was the main practice, and the carrying capacity was sheep unit/600m2. To reveal the effect of artificial grassland on soil microbial bacterial communities under different utilization patterns, we set up grazing treatment (GG), mowing treatment (MG), and enclosure treatment (EG) for a comparative study in August 2019. The area of each treatment plot was ~3,000 m2, and three replicates were set. The average number of grazing animals on each plot was 5 (basically consistent with the local grazing situation). The grazing livestock were approximately 1-year-old Guizhou semi-fine wool sheep. Except for extreme weather, the grazing period was ~300 days per year. The stubble height in the mowing grassland was ~5 cm, and the mowing was carried out according to the normal forage phenology period or when the mowing height was reached. The enclosure grassland was not used in any way.
In mid-August 2021, three 10 m × 10 m sampling plots were set in each test site (nine sampling plots in total), and the distance between plots and their boundaries was >10 m. In each sampling plot, 15 sampling points (~3 cm away from the base of the plant) were evenly spaced using an “S”-shaped multi-point sampling method. After removing the litter layer from the surface, soil samples were collected from the surface layer (0–10 cm) using a soil auger. To reduce spatial heterogeneity, soil samples from 15 sampling points were mixed into 1 sample, and a total of 9 soil samples were obtained. The soil samples were removed from impurities and then divided into three parts. One part was placed in 15-ml sterilized centrifuge tubes and stored in liquid nitrogen for transport back to the laboratory, and these samples were stored in a refrigerator at −80°C for 16s rDNA analysis (Li H. N. et al., 2020; Sardar et al., 2021a,b). One part was placed in a sealed plastic bag and taken back to the laboratory for the determination of the soil enzyme activity. Another part of the soil samples was air-dried in the room and then passed through a 2-mm sieve to determine soil properties.
In this study, 12 indices of soil samples were determined, namely pH, soil organic carbon (SOC), total nitrogen (TN), total phosphorus (TP), available nitrogen (AN), available phosphorus (AP), nitrate nitrogen (NO3−-N), ammonium nitrogen (NH4+-N), β-1,4-glucosidase (BG), N-acetyl-β -D-glucosaminidase (NAG), acid phosphatase (LCP), and leucine aminopeptidase (LAP). Soil pH was determined in suspension with a water-to-soil ratio of 2.5:1 using a pH meter (PHC-3C, Leici, Shanghai, China). SOC, TN, TP, AN, and AP were determined using the method described by Bao (2000). SOC and TN were determined using an automatic elemental analyzer (FlashSmart, Thermo Fisher, USA). TP, AN, and AP were determined on a continuous flow analyzer (Flowsys, Systea, Italy). Nitrogen and ammonium nitrogen in the soil were extracted with potassium chloride solution according to ISO standards and determined with a continuous flow analyzer (Flowsys, Systea, Italy). Soil enzyme activity was analyzed for BG, NAG, LCP, and LAP according to the method of Jiao et al. (2022) and determined with UV-visible spectrophotometer (Specord 200 Plus, Analytik, Germany), with active units expressed as IUg−1 units.
The DNA was extracted with the TGuide S96 Magnetic Soil DNA Kit (Tiangen Biotech Beijing Co., Ltd.) according to manufacturer instructions. The DNA concentration of the samples was measured with the Qubit dsDNA HS Assay Kit and Qubit 4.0 Fluorometer (Invitrogen, Thermo Fisher Scientific, Oregon, USA). The 338F: 5′- ACTCCTACGGGAGGCAGCA-3′ and 806R: 5′- GGACTACHVGGGTWTCTAAT-3′ universal primer set was used to amplify the V3-V4 region of 16S rRNA gene from the genomic DNA extracted from each sample. Both the forward and reverse 16S primers were tailed with sample-specific Illumina index sequences to allow for deep sequencing. The PCR was performed in a total reaction volume of 10 μl: DNA template 5-50 ng, *Vn F (10μM) 0.3 μl, *Vn R (10μM) 0.3 μl, KOD FX Neo Buffer 5 μl, dNTP (2 mM each) 2 μl, KOD FX Neo 0.2 μl, and ddH2O up to 10 μl. Vn F and Vn R are selected according to the amplification area: initial denaturation at 95°C for 5 min, followed by 25 cycles of denaturation at 95°C for 30 s, annealing at 50°C for 30 s, and extension at 72°C for 40 s, and a final step at 72°C for 7 min. A total of PCR amplicons were purified with Agencourt AMPure XP beads (Beckman Coulter, Indianapolis, IN) and quantified using the Qubit dsDNA HS Assay Kit and Qubit 4.0 Fluorometer (Invitrogen, Thermo Fisher Scientific, Oregon, USA). After the individual quantification step, amplicons were pooled in equal amounts. For the constructed library, use Illumina NovaSeq 6000 (Illumina, Santiago CA, USA) for sequencing.
According to the quality of single nucleotide, raw data were primarily filtered by Trimmomatic (Version 0.33) (Edgar, 2013). The identification and removal of primer sequences were processed by Cutadapt (Version 1.9.1) (Callahan et al., 2016). PE reads obtained from previous steps were assembled by USEARCH (Version 10) (Segata et al., 2011) and followed by chimera removal using UCHIME (Version 8.1) (Quast et al., 2013). The high-quality reads generated from the above steps were used in the following analysis. Sequences with similarity ≥ 97% were clustered into the same operational taxonomic unit (OTU) by USEARCH (Version 10.0) (Edgar, 2013), and the OTUs with a relative abundance of < 0.005% were filtered. Taxonomy annotation of the OTUs was performed based on the Naive Bayes classifier in QIIME2 (Bolyen et al., 2019) using the SILVA database (release 132) (Quast et al., 2013) with a confidence threshold of 70%. The alpha diversity was calculated and displayed by the QIIME2 and R software, respectively. Beta diversity was determined to evaluate the degree of similarity of microbial communities from different samples using QIIME.
The one-way analysis of variance (ANOVA) and multiple comparisons (Tukey's pairwise comparisons) were used to analyze the effects of different use patterns on soil properties, relative abundance of bacterial phylum, and bacterial community richness and diversity indices (SPSS 19.0, Chicago, IL, USA). The data were log-transformed to fit the normal distribution of the data. The Chao 1 and Shannon indices of alpha diversity were calculated using the Mothur software based on OTU information, which was used to reflect the richness and diversity of soil microbial communities. The common and unique OTUs and soil bacterial structure at the phylum level among grassland samples under different utilization patterns were presented using the Vegan software package in R (Chen and Boutros, 2011). PCoA analysis and similarity test (ANOSIM) based on Bray–Curtis distance measure were used to compare the beta diversity of soil bacteria in grassland under different use patterns.
The variance inflation factor (VIF) was used to filter the multicollinearity of environmental factors when VIF >10. Redundancy analysis (RDA) was used to analyze the correlation between the selected environmental factors and soil microbial communities. To test the correlation of environmental factors with bacterial community composition, we performed a Monte Carlo permutation test of soils using the R “ecodist” package, and the Bray–Curtis index served as the dissimilarity metric. The relationships between environmental factors and soil bacterial structure at the phylum level were analyzed using the Spearman correlation heatmap of the Vegan software package in R. The Functional Annotation of Prokaryotic Taxa database 12 (FAPROTAX, v1.2.3) was used to predict the ecological function of soil bacteria (Sansupa et al., 2021).
Different use patterns have different effects on soil chemical properties and soil enzyme activity in grassland (Figure 1). Compared to EG, GG significantly changed soil pH, increased the soil AN content (P < 0.05), and decreased the soil TP content and NAG content (P < 0.05). MG significantly decreased the contents of SOC, TP, AN, NH4+-N, BG, and NAG (P < 0.05). However, GG and MG had no significant effect on pH, SOC, AP, NO3−-N, ACP, and LAP (P > 0.05).
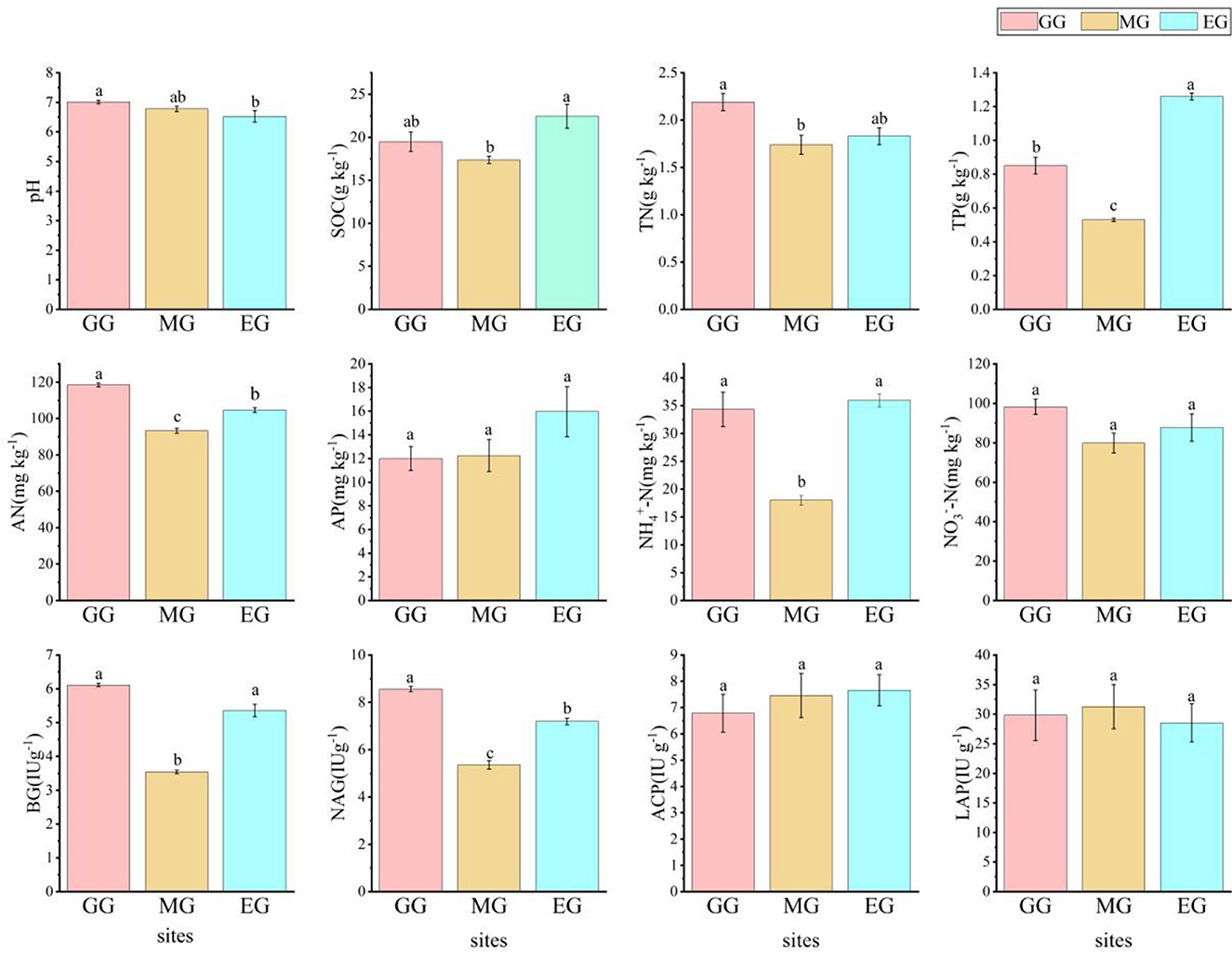
Figure 1. Soil chemical properties and soil enzyme activity under different grassland use patterns [different lowercase letters indicate the significant differences between different treatments (P < 0.05, n = 3)]. SOC, soil organic carbon; TN, total nitrogen; TP, total phosphorus; AN, available nitrogen; AP, available phosphorus; NO3−-N, nitrate nitrogen; NH4+-N, ammonium nitrogen; BG, β-1,4-glucosidase; NAG, N-acetyl-β -D-glucosaminidase; LCP, acid phosphatase; LAP, leucine aminopeptidase; GG, grazing grassland; MG, mowing grassland; EG, enclosure grassland.
A total of 720,323 raw reads were detected in all samples, generating an average of 716,180 clean reads and 695,633 effective reads, and the average sequence length was 420.44 bp (Supplementary Table S1). After species annotation, the average sequence obtained from all samples was 56,723, ranging from 49,679 to 60,248 (Supplementary Table S2). A total of 2,427 OTUs were obtained, which were divided into 26 phyla, 54 classes, 153 orders, 288 families, 505 genera, and 575 species (Supplementary Table S3). We illustrated the similarities and differences between OTUs from different treatments using the Venn diagram (Figure 2). The numbers of unique OTUs in the GG, MG, and EG were 47, 61, and 51, respectively, and the number of shared OTUs was 2,042. The Chao 1 and Shannon indices represent the richness and diversity of bacterial communities, which are used to study the alpha diversity of soil microbial bacteria in grasslands under different utilization patterns (Table 1). There was no significant difference in bacterial community richness (Chao 1) and diversity (Shannon) among treatments. This indicated that grazing and mowing did not significantly change the richness and diversity of the bacterial communities compared to EG.
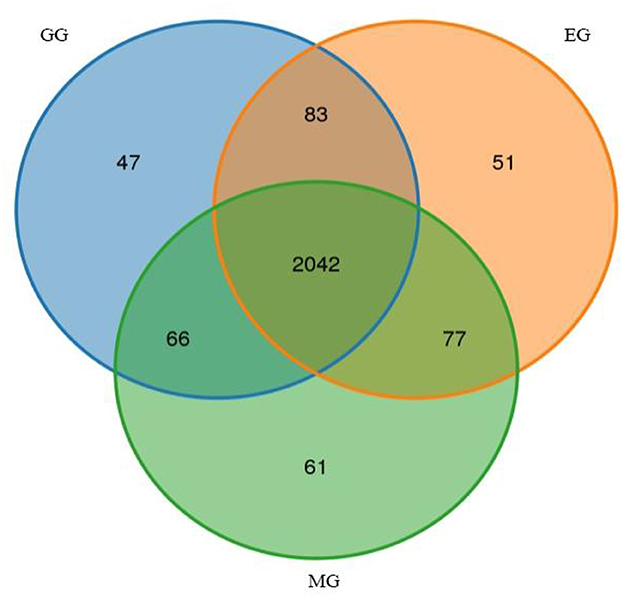
Figure 2. Venn diagram of unique and shared bacterial OTUs under different grassland use patterns (at the 3% evolutionary distance). GG, grazing grassland; MG, mowing grassland; EG, enclosure grassland.
The PCoA of soil bacterial communities under different use patterns was analyzed using the Bray–Curtis distance metric. The PCoA results showed that the bacterial communities in different treatments were largely separated (Figure 3), and the ANOSIM test results based on the Bray–Curtis distance metric also confirmed this finding (R2 = 0.513, P = 0.001) (Figure 4), which indicated that different utilization patterns significantly changed the soil bacterial community structure in grassland.
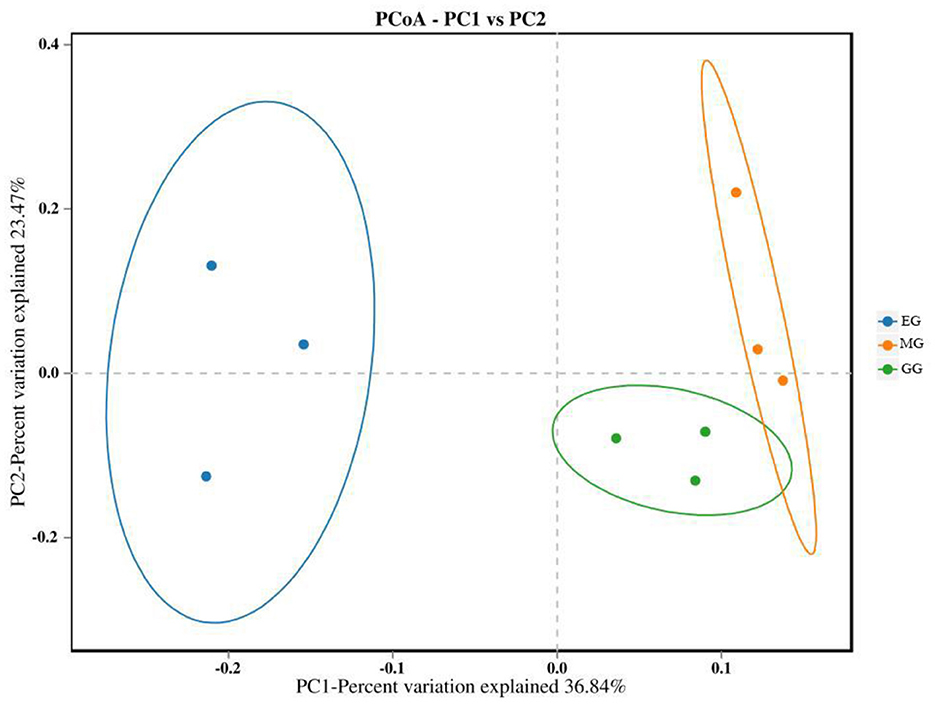
Figure 3. PCoA of bacterial community composition under different grassland use patterns. GG, grazing grassland; MG, mowing grassland; EG, enclosure grassland.
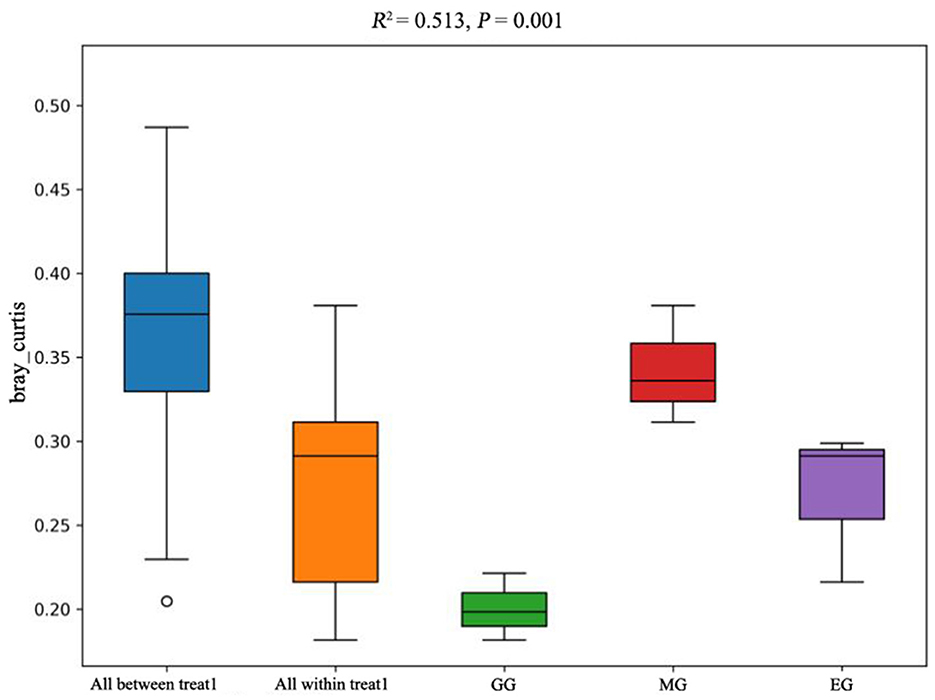
Figure 4. ANOSIM of soil bacterial community composition under different grassland use patterns based on the Bray–Curtis distance metric. GG, grazing grassland; MG, mowing grassland; EG, enclosure grassland.
At the phylum level, a total of 10 soil bacterial phyla with a relative abundance of >1% were obtained (Figure 5). The order was as follows: Proteobacteria (29.88%), Acidobacteriota (29.46%), Actinobacteriota (7.61%), Bacteroidota (6.13%), Chloroflexi (4.97%), Gemmatimonadota (3.83%), unclassified_Bacteria (3.71%), Verrucomicrobiota (3.25%), Myxococcota (3.25%), and Nitrospirota, (2.33%). Among them, Proteobacteria and Acidobacteriota were the dominant bacteria. The results of ANOVA showed that different use patterns had a significant effect on the relative abundance of Proteobacteria, Acidobacteriota, Gemmatimonadota, and Nitrospirota (P < 0.05) but no significant effect on other phyla (P > 0.05). Compared with EG, the GG significantly increased the relative abundance of Acidobacteriota and Gemmatimonadota (P < 0.05) but significantly decreased the relative abundance of Proteobacteria (P < 0.05) and had no significant difference with MG (P > 0.05). Compared to GG, MG significantly increased the relative abundance of Nitrospirota (P < 0.05) but had no significant difference with EG (P > 0.05). However, there were no significant differences between the three treatments at the family and genus levels (Supplementary Table S4).
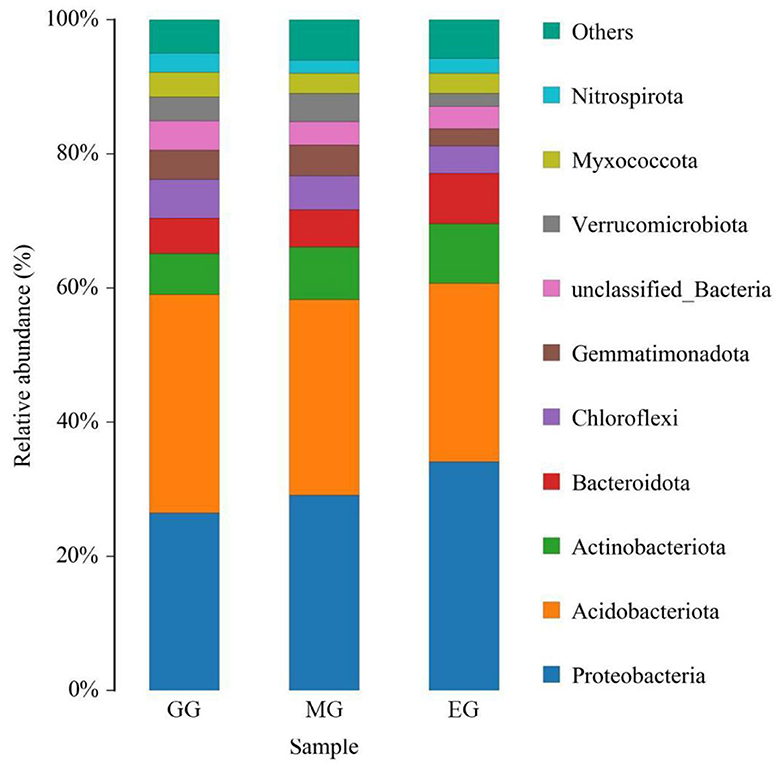
Figure 5. Relative abundance of soil bacterial phylum under different grassland use patterns (relative abundance>1%). GG, grazing grassland; MG, mowing grassland; EG, enclosure grassland.
The variance inflation factor (VIF) was used to filter the multicollinearity of environmental factors (Supplementary Table S5). Redundancy analysis of soil microbial structure at the phylum level and soil properties indicated that the first and second axes explain 38.93% and 24.94% of the variation in the bacterial community, respectively (Figure 6). Further analysis by Monte Carlo test showed that pH (R2 = 0.79, p = 0.029) had a higher correlation with the bacterial community composition than other environmental factors, which was the main factor affecting the bacterial community structure (Table 2). The related heatmap was used to reveal the relationship between environmental factors and soil microbial structure at the phylum level (relative abundance> 1%) (Figures 7A, B). The results showed that there was a significantly positive correlation between pH and Acidobacteriota (P < 0.05) but a significantly negative correlation with Actinobacteria (P < 0.05). SOC and TP have a significantly negative correlation with Gemmatimonadota (P < 0.05). AP and TP have a significantly negative correlation with Verrucomicrobiota (P < 0.05). TN, AN, and BG have a significantly positive correlation with Nitrospirota (P < 0.05). TN has a significantly positive correlation with Myxococcota (P < 0.05). ACP has a significantly positive correlation with Bacteroidota (P < 0.05) but a significantly negative correlation with Acidobacteriota (P < 0.05).
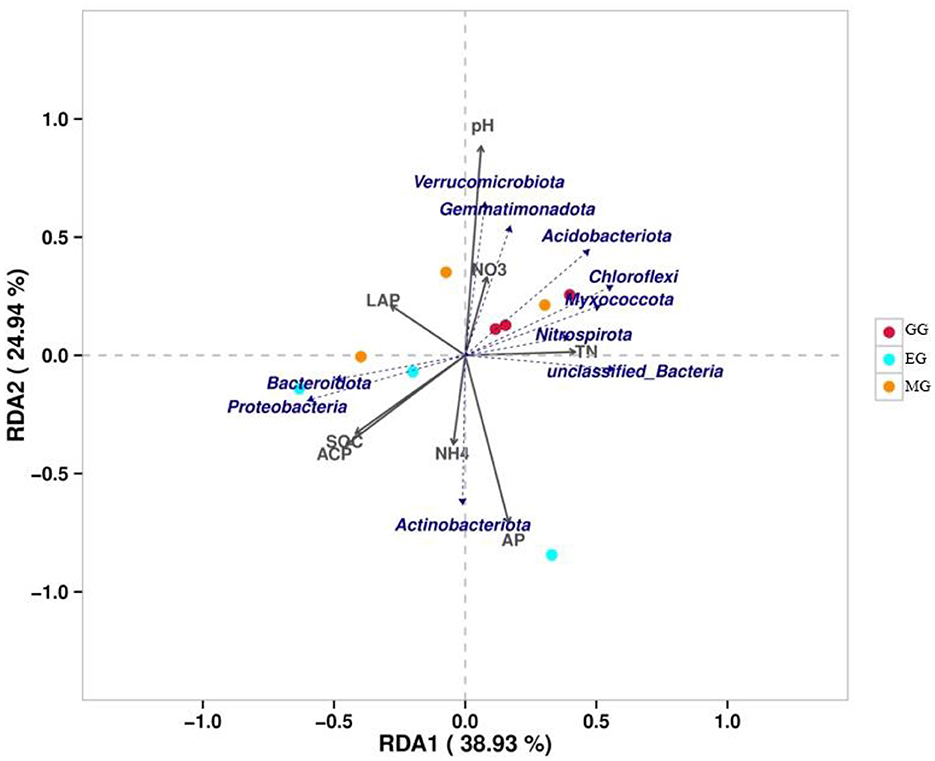
Figure 6. Redundancy analysis of soil properties (gray arrows) and soil microbial structure (blue arrows) at the phylum level. SOC, soil organic carbon; TN, total nitrogen; AP, available phosphorus; NO3, nitrate nitrogen; NH4, ammonium nitrogen; LCP, acid phosphatase; LAP, leucine aminopeptidase; GG, grazing grassland; MG, mowing grassland; EG, enclosure grassland.
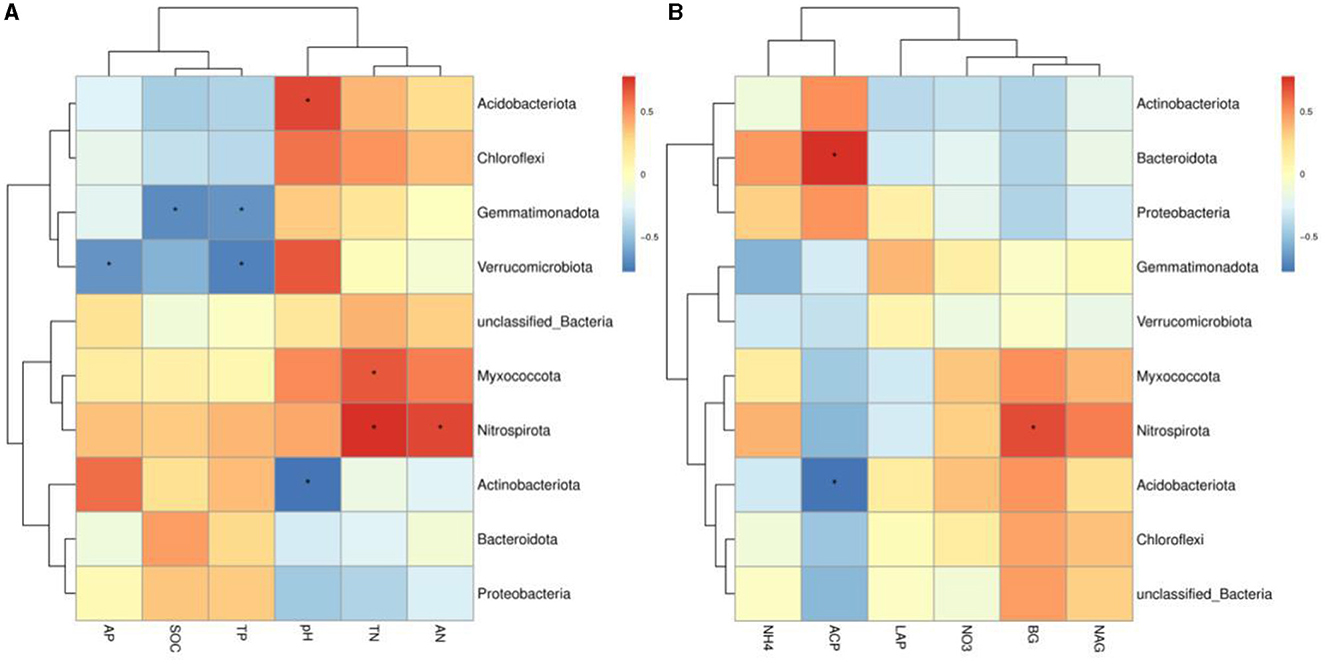
Figure 7. (A, B) Heatmap of the correlation between environmental factors and soil bacterial community at the phylum level (relative abundance >1%). Spearman's correlation coefficients are shown in different colors, and the legend on the right shows the color range of the different correlation coefficients (P-values: * < 0.05). SOC, soil organic carbon; TN, total nitrogen; TP, total phosphorus; AN, available nitrogen; AP, available phosphorus; NO3−-N, nitrate nitrogen; NH4+-N, ammonium nitrogen; BG, β-1,4-glucosidase; NAG, N-acetyl-β -D-glucosaminidase; LCP, acid phosphatase; LAP, leucine aminopeptidase; GG, grazing grassland; MG, mowing grassland; EG, enclosure grassland.
FAPROTAX software was used to predict the ecological function of soil bacteria in different utilization patterns (Figure 8), and a total of 46 functional groups were obtained. In all the ecological function groups, the relative abundance of chemoheterotrophy, aerobic chemoheterotrophy, ureolysis decomposition, and fermentation bacteria was relatively high, with an average abundance of 34.14%, 28.80%, 4.07%, and 3.34%, respectively. The results of ANOVA showed that GG and MG significantly decreased the relative abundance of chemoheterotrophy compared to EG (P < 0.05), but there was no significant difference between GG and MG (P > 0.05). There was no significant difference in the relative abundance of aerobic chemoheterotrophy, ureolysis, and fermentation bacteria between GG, MG, and EG (P > 0.05).
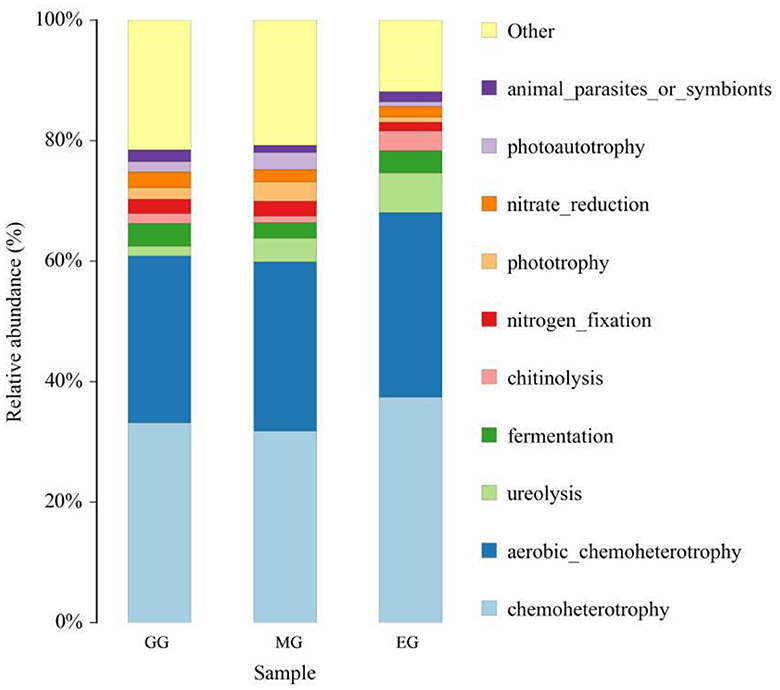
Figure 8. Relative abundance of soil bacterial community for ecological function under different grassland use patterns. GG, grazing grassland; MG, mowing grassland; EG, enclosure grassland.
Artificial grassland is an ecosystem under human disturbance, and its quality and function were good or bad, which often cannot be evaluated from macroscopic perspectives such as vegetation cover (Wang et al., 2020), while soil chemical properties and soil microorganisms can better evaluate the quality of grassland under different use patterns (Baptistella et al., 2020; Song et al., 2022). In this study, there were significant differences in the soil chemistry of artificial grassland under different use patterns after 2 years. It has been shown that grazing can increase the soil pH of artificial grassland (Wei et al., 2022), which is consistent with the results of this study. This may be related to the excreta of grazing livestock. As the excreta mature, the soil animal community increases and the cation cycle in the grassland ecosystem accelerates, thus offsetting soil acidification and resulting in a trend toward weak alkalinity of the soil. As MG and EG were not disturbed by livestock, the difference in soil pH was not significant. In this study, compared with EG, GG significantly increased soil TN and AN content (P < 0.05) but significantly decreased TP content (P < 0.05), which is consistent with the results of Zheng et al. (2022) and Guan et al. (2022). This may be due to nitrogen enrichment in the grassland ecosystem caused by grazing and increased soil phosphorus supply capacity through activation of mineral-bound phosphorus, but long-term grazing leads to depletion of the soil phosphorus pool (Wang R. Z. et al., 2022). Compared with EG, MG significantly decreased the content of soil SOC, TP, AN, NH4+-N, NO3−-N, BG, and NAG (P < 0.05), indicating that MG was not conducive to the accumulation of TN in soil, and at the same time, the available carbon and nitrogen sources by microorganisms were reduced, which decreased the activity of microorganisms and subsequently decreased the enzyme activity, which is consistent with the research results of Wang et al. (2023). GG and MG had no significant effect on soil AP, ACP, and LAP (P > 0.05). This indicates that the available phosphorus did not obviously respond to GG and MG, but the AP content in EG was relatively higher, which may be related to its higher SOC and more complete decomposition.
As an important component of the soil system, the structure and diversity of soil microorganisms play an important role in the stability of the grassland ecosystem (Sennett et al., 2023). The dominant taxa in soil bacteria have important functions in grassland ecosystems (Ou et al., 2023). In this study, the results of PCoA showed that there were significant differences among the groups tested by ANOSIM (R2 = 0.513, P = 0.001), and the soil bacterial communities of grasslands under different utilization patterns were basically separated, indicating that the different use patterns significantly changed the bacterial community structure. The results of ANOVA showed that the relative abundance of Proteobacteria, Acidobacteriota, Gemmatimonadota, and Nitrospirota was significantly affected by the different use patterns (P < 0.05). Proteobacteria, Acidobacteriota, Gemmatimonadota, and Nitrospirota are considered potential decomposer nutrients in the soil (Berlemont and Martiny, 2013). Proteobacteria can make full use of soil carbon and grow rapidly under the condition of sufficient soil carbon (Zhang Y. T. et al., 2020). They can also use nutrients produced by the decomposition of organic matter for growth and metabolism (Huang et al., 2022). Gemmatimonadota are believed to promote the rapid decomposition of organic matter such as litter, animal, and plant remains in the soil by decomposing organic matter (Lozupone and Knight, 2008). Proteobacteria and Gemmatimonadota can promote microbial phosphate mineralization (Tan et al., 2013). In this study, MG and GG decreased SOC content and changed the developmental environment of Proteobacteria compared with EG. Most Acidobacteriota are oligotrophic and can maintain basal activity and participate in geochemical cycles by using fewer carbon and nitrogen sources in low-nutrient stress environments (Rime et al., 2015; Sun et al., 2021; Venkatachalam et al., 2021). Compared with EG, the relative abundance of Acidobacteriota in GG and MG increased, indicating that they can adapt to the disturbance of grazing and mowing so that the relative abundance of Acidobacteriota that efficiently utilized nutrient resources increased, and this result is consistent with Li et al. (2023). In this study, Proteobacteria and Acidobacteriota are the dominant phylum, which is consistent with the results of previous studies (Gao et al., 2017; Zhang C. et al., 2021; Song et al., 2022; Zhu et al., 2022).
The alpha diversity of soil microbes is used to describe the microbial community of an individual or a group habitat (Lozupone and Knight, 2008). The Chao1 index and Shannon index can be used to evaluate the richness and diversity of bacterial communities, and the higher the index value, the higher the richness and diversity of bacterial communities (Xue et al., 2023). In this study, the richness index (Chao1) and diversity index (Shannon) of the soil bacterial community had no significant difference among the different utilization patterns, indicating that GG and MG did not significantly change the richness and diversity of the bacterial communities compared with EG. The Venn diagram showed that the unique OTUS in MG was the largest, indicating that the unique microbial community was the most in EG, which is consistent with the study results of Zheng et al. (2022). However, compared with EG and GG, MG had no significant effect on the diversity of the soil bacterial community, indicating that external disturbance had some but limited effect on the diversity of the soil bacterial community.
Changes in land use will affect the soil structure and nutrient status, and the structure of soil bacterial communities will also change, while soil environmental factors are important influences on the structure of soil bacterial communities (Saleem, 2015; Melo et al., 2020). The RDA showed that soil properties explained 63.87% of the variation in the bacterial communities. This indicated that the variation in microbial community composition was highly correlated with soil properties. Previous studies have shown that soil pH is one of the key factors affecting the structure of soil microbial communities (Fierer and Jackson, 2006; Griffiths et al., 2011; Ling et al., 2016; Yun et al., 2016). In our study, the Monte Carlo test showed that pH was one of the key factors affecting the soil microbial community under different use patterns, which is consistent with the results of previous studies (Fan et al., 2019; Wang et al., 2021; Song et al., 2022). This may be due to the fact that most of the bacterial groups in this study showed a relatively narrow growth tolerance and poor growth environment in the karst area (Xue et al., 2017). In this study, pH was positively associated with Acidobacteriota, which is consistent with many studies (Dai et al., 2017; Zhang J. Y. et al., 2020; Song et al., 2022). This suggests that Acidobacteriota is closely related to pH under different grassland use patterns. Verrucomicrobiota has a sparsely linked non-ribosomal peptide synthetase system, and its gene cluster has the potential to synthesize antibiotics (Bergmann et al., 2011), which indicated that external disturbance may lead to a reduction in antibiotic synthesis in the soil, which is not conducive to plant disease resistance (Fu et al., 2022). This result confirms the finding of this study, namely that the relative abundance of Verrucomicrobiota in GG and MG was decreased compared to EG. Soil AP and TP were closely related to Verrucomicrobiota, indicating that soil phosphorus played an important role in the establishment of Verrucomicrobiota. Gemmatimonadota can convert various sugar molecules into vitamins (Xu et al., 2020), and the relative abundance of Gemmatimonadota in EG was decreased in this study.
The functional diversity of microorganisms is fundamental to ensuring that all basic soil functions perform as expected (Eduardo et al., 2015; Ma et al., 2021). The results of this study showed that the dominant bacterial group in the soil ecological function group was mainly chemoheterotrophy and aerobic chemoheterotrophy, which is consistent with Zhang et al. (2018) and Wu et al. (2021). The results of ecological function prediction in soil bacteria showed that the relative abundance of aerobic chemoheterotrophic was the highest in EG and the lowest in MG. This indicated that a large number of litter were not easily used by plants in EG, and a large number of chemoheterotrophic bacteria were produced, but the litter could decompose in time in MG, which decreased the relative abundance of aerobic chemoheterotrophic.
It is estimated that more than one-third of the world's soils are in degraded conditions (Wall and Six, 2015). This study was conducted on the effects of different grassland utilization patterns on soil bacterial communities in a special land-degraded landscape (karst desertification) area in the subtropical monsoon zone of China. We investigated the effects of grazing, mowing, and enclosure on the soil properties and the structure and diversity of soil bacterial communities in the artificial grassland. We drew the following conclusions: (1) Compared with EG, GG significantly changed soil pH, increased alkaline hydrolyzable nitrogen content, and decreased soil total phosphorus content. However, MG significantly decreased the contents of soil organic carbon, total phosphorus, available nitrogen, ammonium nitrogen, β-1,4-glucosidase, and N-acetyl-β-D-glucamosonidase (P < 0.05). (2) Different grassland use patterns significantly changed the structure of the soil bacterial community but did not significantly change the richness and diversity of the soil bacterial community. (3) Proteobacteria (29.88%) and Acidobacteriota (29.46%) were the dominant phyla. EG significantly decreased the relative abundance of Acidobacteriota and Gemmatimonadota and significantly increased the relative abundance of Proteobacteria and chemoheterotrophy. (4) pH was the main factor affecting the bacterial community structure, which is consistent with the results of the study conducted by Bahram et al. (2018) on a global scale. In this study, different grassland utilization patterns significantly altered the structure of soil bacterial communities but did not significantly alter the richness and diversity of soil bacterial communities, which may be related to the short observation period. Future studies should continue long-term observations to identify suitable grassland use patterns in the karst desertification control areas and to provide theoretical support for the ecological restoration of the karst desertification environment and the sustainable development of grassland ecosystems.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA957490.
YC, KX, and SS performed the sampling, contributed to the experimental design, analyzed the data, and wrote the manuscript. KX provided the study site and funded the study. All authors contributed to the article and approved the submitted version.
This research was funded by the Key Science and Technology Program of Guizhou Provence: Poverty Alleviation Model and Technology demonstration for Ecoindustries Derivated from the Karst Desertification Control (No. 5411 2017 QianKehe Pingtai Rencai), the National Technical Innovation Center for Karst Desertification Control and Green Development (Qiankehe Zhongyindi [2023]05), Natural Science Research Project of Education Department of Guizhou Province [Qianjiaohe KY Zi (2022) 157], and Academic New Seedling Fund Project of Guizhou Normal University (Qianshi Xinmiao B15).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1208971/full#supplementary-material
Bahram, M., Hildebrand, F., Forslund, S. K., Anderson, J. L., Soudzilovskaia, N. A., Bodegom, P. M., et al. (2018). Structure and function of the global topsoil microbiome. Nature 560, 233–237. doi: 10.1038/s41586-018-0386-6
Bai, Y. F., and Cotrufo, M. F. (2022). Grassland soil carbon sequestration: current understanding, challenges, and solutions. Science 377, 603–608. doi: 10.1126/science.abo23
Baptistella, J. L. C., Andrade, d. e., Favarin, S. A. L., and Mazzafera, J. L. P. (2020). Urochloa in tropical agroecosystems. Front. Sustain. Food Syst. 4, 119. doi: 10.3389/fsufs.2020.00119
Baranova, A., Oldeland, J., Wang, S. L., and Schickhoff, U. (2019). Grazing impact on forage quality and macronutrient content of rangelands in Qilian Mountains, NW China. J. Mount. Sci. 16, 43–53. doi: 10.1007/s11629-018-5131-y
Barber, N. A., Klimek, D. M., Bell, J. K., and Swingley, W. D. (2023). Restoration age and reintroduced bison may shape soil bacterial communities in restored tallgrass prairies. FEMS. Microbiol. Ecol. 99, fiad007. doi: 10.1093./femsec/fiad007
Bergmann, G. T., Bates, S. T., Eilers, K. G., Lauber, C. L., Caporaso, J. G., Walters, W. A., et al. (2011). The under-recognized dominance of Verrucomicrobia in soil bacterial communities. Soil Biol. Biochem. 43, 1450–1465. doi: 10.1016/j.soilbio.03012
Berlemont, R., and Martiny, A. C. (2013). Phylogenetic distribution of potential cellulases in bacteria. Appl. Environ. Microbiol. 79, 1545–1554. doi: 10.1128/AEM.03305-12
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A.W., Johnson, A.J.A., and Holmes, S.P. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Cassman, N. A., Leite, M. F., Pan, Y., Hollander, d. e., van Veen, M., Kuramae, J. A. E. E., et al. (2016). Plant and soil fungal but not soil bacterial communities are linked in long-term fertilized grassland. Sci. Rep. 6, 23680. doi: 10.1038/srep23680
Chen, H., Li, D. J., Mao, Q., Xiao, K., and Wang, K. (2019). Resource limitation of soil microbes in karst ecosystems. Sci. Total Environ. 650, 241–248. doi: 10.1016/j.scitotenv.09036
Chen, H. B., and Boutros, P. C. (2011). Venn Diagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics. 12, 35. doi: 10.1186/1471-2105-12-35
Coban, O., Deyn, G. B. D., and Ploeg, M. V. D. M. (2022). Soil microbiota as game-changers in restoration of degraded lands. Science. 375, abe0725. doi: 10.1126./science.abe0725
Dai, Y. T., Yan, Z. J., Xie, J. H., Wu, H. X., Xu, L. B., Hou, X. Y., et al. (2017). Soil Bacteria diversity in rhizosphere under two types of vegetation restoration based on high throughput sequencing. Acta Pedologica Sin. 54, 735–748. doi: 10.11766/trxb201603150062
de Vries, F. T., Griffiths, R. I., Bailey, M., Craig, H., Girlanda, M., Gweon, H. S., et al. (2017). Soil bacterial networks are less stable under drought than fungal networks. Nat. Commun. 9, 3033. doi: 10.1038/s41467-018-05516-7
Drenovsky, R. E., Steenwerth, K. L., Jackson, L. E., and Scow, K. M. (2010). Land use and climatic factors structure regional patterns in soil microbial communities. Glob. Ecol. Biogeogr. J. 19, 27–39. doi: 10.1111/j.1466-8238.2009.00486.x
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. methods 10, 996. doi: 10.1038/NMETH.2604
Eduardo, P., Marta, G., and Miguel, V. (2015). Phylogenetic structure of soil bacterial communities predicts ecosystem functioning. FEMS microbiol. ecol. 91, fiv031. doi: 10.1093./femsec/fiv031
Fan, Z. Z., Lu, S. Y., Liu, S., Guo, H., Wang, T., Zhou, J. X., et al. (2019). Changes in plant rhizosphere microbial communities under different vegetation restoration patterns in karst and non-karst ecosystems. Sci. Rep. 9, 8761. doi: 10.1038/s41598-019-44985-8
Fang, J. Y., Jing, H. C., Zhang, W. H., Gao, S. Q., Duan, Z. Y., Wang, H. S., et al. (2018). The concept of “Grass-based Livestock Husbandry” and its practice in Hulun Buir, Inner Mongolia. Chin. Sci. Bull. 63, 1619–1631. doi: 10.1360/N972018-00042
Feng, Y., Zhang, J., Berdugo, M., Guirado, E., Guerra, C. A., Egidi, E., et al. (2022). Temperature thresholds drive the global distribution of soil fungal decomposers. Global Change Biol. 28, 2779–2789. doi: 10.1111/gcb.16096
Fierer, N. (2017). Embracing the unknown: disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 15, 579–590. doi: 10.1038/nrmicro.2017.87
Fierer, N., and Jackson, R. B. (2006). The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. U.S.A. 103, 626–631. doi: 10.1073/pnas.0507535103
Fierer, N., Lauber, C. L., Ramirez, K. S., Zaneveld, J., Bradford, M. A., Knight, R., et al. (2012). Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients. ISME J. 6, 1007–1017. doi: 10.1038/ismej.2011.159
Figuerola, E. L. M., Guerrero, L. D., Türkowsky, D., Wall, L. G., and Erijman, L. (2015). Crop monoculture rather than agriculture reduces the spatial turnover of soil bacterial communities at a regional scale. Environ. Microbiol. 17, 678–88. doi: 10.1111/1462-2920.12497
Fu, B. Y., Zhang, J. H., Yang, M. X., Xu, H. S., Peng, Z. P., Xue, C., et al. (2022). Effects of nitrogen topdressing timing on microbial community structure in strong gluten wheat rhizosphere soil. J. Agr. Univ. Hebei. 45, 1–8. doi: 10.13320/j.cnki.jauh.2022.0036
Gao, X. F., Han, G. D., and Zhang, G. G. (2017). Soil microbial community structure and composition of Stipa Breviflora on the desert steppe. Acta Ecol. Sin. 37, 5129–5136. doi: 10.5846/stxb201605100902
Garousi, F., Shan, Z. J., Ni, K., Yang, H., Shan, J., Cao, J. H., et al. (2021). Decreased inorganic N supply capacity and turnover in calcareous soil under degraded rubber plantation in the tropical karst region. Geoderma. 381, 114754. doi: 10.1016/j.geoderma.2020.114754
Griffiths, R. I., Thomson, B. C., James, P., Bell, T., Bailey, M., and Whiteley, A. S. (2011). The bacterial biogeography of British soils. Environ. Microbiol. 13, 1642–1654. doi: 10.1111/j.1462-2920.2011.02480.x
Guan, W. T., Zheng, Z. R., Diao, Z. Y., Ma, P., Jin, S. L., Wang, X., et al. (2022). Stoichiometric characteristics and their storage of soil carbon, nitrogen and phosphorus in the temperate meadow steppe under different disturbances. Acta Agrestia Sin. 30, 2959–2966. doi: 10.11733/j.issn.1007-0435.11012
He, M., Fang, K., Chen, L. Y., Feng, X. H., Qin, S. Q., Kou, D., et al. (2022). Depth-dependent drivers of soil microbial necromass carbon across Tibetan alpine grasslands. Global Change Biol. 8, 936–949. doi: 10.1111/gcb.15969
Hermans, S. M., Buckley, H. L., Case, B. S., Curran-Cournane, F., Taylor, M., Lear, G., et al. (2020). Using soil bacterial communities to predict physico-chemical variables and soil quality. Microbiome. 8, 79. doi: 10.1186/s40168-020-00858-1
Huang, L. B., Bai, J. H., Wang, J. J., Zhang, G. L., Wang, W., Wang, X., et al. (2022). Different stochastic processes regulate bacterial and fungal community assembly in estuarine wetland soils. Soil Biol. Biochem. 167, 108586. doi: 10.1016/j.soilbio.2022.108586
Huang, Y. F., Shu, Y. G., Xiao, S. Y., and Chen, M. J. (2020). Quantification of soil nutrient levels and enzyme activities in different grassland cate-gories in karst mountains. Acta Prataculturae Sin. 29, 93–104. doi: 10.11686/cyxb2019519
Jiang, Z. C., Lian, Y. Q., and Qin, X. Q. (2014). Rocky desertification in Southwest China: impacts, causes, and restoration. Earth Sci. Rev. 132, 1–12. doi: 10.1016/j.earscirev.01005
Jiao, P. Y., Guo, W., Chen, Z. L., Liu, X., Hu, Y. L., Wang, Y. Z., et al. (2022). Soil enzyme stoichiometric characteristics of pinus massoniana plantations at different stand ages in mid-subtropical areas. Environ. Sci. 43, 1059–1068. doi: 10.13227/j.hjkx.202107043
Li, B. Y., Liu, G. H., Gu, Z., Liu, X. D., and Shu, L. F. (2023). Effects of fire disturbance on soil bacterial community in Pinus tabulaeformis forest. Chin. J. Ecol. 3, 1–13.
Li, D. J., Liu, J., Chen, H., Zheng, L., and Wang, K. L. (2018). Soil microbial community responses to forage grass cultivation in degraded karst soils, Southwest China. Land Degrad. Dev. 29, 4262–4270. doi: 10.1002/ldr.3188
Li, H. N., Zhang, Z. G., Duan, J. T., Li, N., Li, B. X., Song, T. T., et al. (2020). Electrochemical disinfection of secondary effluent from a wastewater treatment plant: removal efficiency of ARGs and variation of antibiotic resistance in surviving bacteria. Chem. Eng. J. 392, 123674. doi: 10.1016/j.cej.2019.123674
Li, S. L., Xu, S., Wang, T. J., Yue, F. J., Peng, T., Zhong, J., et al. (2020). Effects of agricultural activities coupled with karst structures on riverine biogeochemical cycles and environmental quality in the karst region. Agr. Ecosyst. Environ. 303, 107120. doi: 10.1016/j.agee.2020.107120
Ling, N., Zhu, C., Xue, C., Chen, H., Duan, Y. H., Peng, C., et al. (2016). Insight into how organic amendments can shape the soil microbiome in long-term field experiments as revealed by network analysis. Soil Biol. Biochem. 99, 137–149. doi: 10.1016/j.soilbio.05.005
Liu, M., Bai, X. Y., Tan, Q., Luo, G. J., Zhao, C. W., Wu, L. H., et al. (2022). Monitoring impacts of ecological engineering on ecosystem services with geospatial techniques in karst areas of SW China. Geocarto Int. 7, 5091–5115. doi: 10.1080/10106049.2021.1903580
Liu, W. X., Liu, L. L., Yang, X., Deng, M. F., Wang, Z., Wang, P. D., et al. (2021). Long-term nitrogen input alters plant and soil bacterial, but not fungal beta diversity in a semiarid grassland. Global Change Biol. 37, 3939–3950. doi: 10.1111/gcb.15681
Liu, X., Wang, S. J., Liu, X. M., Huang, T. Z., and Li, Y. (2015). Compositional characteristics and variations of soil microbial community in karst area of Puding County, Guizhou Province, China. Earth Envion. 43, 490–497. doi: 10.14050/j.cnki.1672-9250.05002
Lozupone, C. A., and Knight, R. (2008). Species divergence and the measurement of microbial diversity. FEMS Microbiol. Rev. 32, 557–578. doi: 10.1111/j.1574-6976.2008.00111.x
Lu, Z. X., Wang, P., Ou, H. B., Wei, S. X., Wu, L. C., Jiang, Y. Liu, Z. M., et al. (2022). Effects of different vegetation restoration on soil nutrients, enzyme activities, and microbial communities in degraded karst landscapes in southwest China. Forest Ecol. Manag. 508, 120002. doi: 10.1016/j.foreco.2021.120002
Ma, L. N., Zhang, C. X., Xu, X. F., Wang, C. W., Liu, G. F., Liang, C. Z., et al. (2022). Different facets of bacterial and fungal communities drive soil multifunctionality in grasslands spanning a 3,500 km transect. Funct. Ecol. 36, 3120–3133. doi: 10.1111/1365-2435.14220
Ma, Q. X., Xu, M., Liu, M. J., Cao, X. C., Hill, P. W., Chadwick, D. R., et al. (2022). Organic and inorganic sulfur and nitrogen uptake by co-existing grassland plant species competing with soil microorganisms. Soil Biol. Biochem. 168, 108627. doi: 10.1016/j.soilbio.2022.108627
Ma, X., Luo, Z. Z., Zhang, Y. Q., Liu, J. H., Niu, Y. N., Cai, L. Q., et al. (2021). Distribution characteristics and ecological function predictions of soil bacterial communities in rainfed alfalfa fields on the Loess Plateau. Acta Prataculturae Sin. 30, 54–67. doi: 10.11686/cyxb2020381
Melo, V. F., Barros, L. S., Silva, M. C. S., Veloso, T. G. R., Senwo, Z. N., Matos, K. S., et al. (2020). Soil bacterial diversities and response to deforestation, land use and burning in North Amazon, Brazil. Appl. Soil Ecol. 158, 103775. doi: 10.1016/j.apsoil.2020.103775
Mod, H. K., Buri, A., Yashiro, E. Guex, N., Malard, L., Pinto-Figueroa, E., Pagni, M., et al. (2021). Predicting spatial patterns of soil bacteria under current and future environmental conditions. ISME J. 15, 2547–2560. doi: 10.1038/s41396-021-00947-5
Nelson, M. B., Martiny, A. C., and Martiny, J. B. H. (2016). Global biogeography of microbial nitrogen-cycling traits in soil. P. Natl. Acad. Sci. USA. 113, 8033–8040. doi: 10.1073/pnas.1601070113
Oldroyd, G. E. D., and Leyser, O. (2020). A plant's diet, surviving in a variable nutrient environment. Science 368, eaba0196. doi: 10.1126./science.aba0196
Ou, J., Hao, J., Wang, B., Ren, X. Y., Mi, J., Dong, K. H., et al. (2023). Effects of mowing intensity on soil bacterial community structure in salinized grassland of Northern Shanxi Province. J. Shanxi Univ. 3, 1–12. doi: 10.13451./j.sxu.ns.2023011
Pei, G. T., Sun, J. F., He, T. X., and Hu, B. Q. (2021). Effects of long-term human disturbances on soil microbial diversity and community structure in a karst grassland ecosystem of northwestern Guangxi, China. Chin. J. Plant Ecol. 45, 74–84. doi: 10.17521/cjpe.2020.0316
Plassart, P., Prévost-Bouré, N. C., Uroz, S., Dequiedt, S., Stone, D., Creamer, R., et al. (2019). Soil parameters, land use, and geographical distance drive soil bacterial communities along a European transect. Sci. Rep. 9, 605. doi: 10.1038/s41598-018-36867-2
Qian, Q., Zhang, R. Y., Liu, K. P., Liang, Y. M., Zhang, W., Pan, F. J., et al. (2022). Soil phosphorus characteristics of different land use types and ecological restoration modes in karst region. Chin. J. Ecol. 4, 2128–2136. doi: 10.13292/j.1000-4890.202208.003
Qiu, J., Zhang, J. F., Zhang, X., Wei, Y. P., Chen, A., et al. (2023). Effects of graphene on soil enzyme activities and microbial diversity in rhizosphere of Cunninghamia lanceolata Seedlings. Acta Agri. Univ. Jiangxiensis. 4, 1–10.
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–6. doi: 10.1093/nar/gks1219
Rime, T., Hartmann, M., Brunner, I., Widmer, F., and Zeyer, J. Frey, B. (2015). Vertical distribution of the soil microbiota along a successional gradient in a glacier forefield. Mol. Ecol. 24, 1091–1108. doi: 10.1111/mec.13051
Sansupa, C., Wahdan, S. F. M., Hossen, S., Disayathanoowat, T., Wubet, T., Purahong, W., et al. (2021). Can we use functional annotation of prokaryotic taxa (faprotax) to assign the ecological functions of soil bacteria? Appl. Sci. 11, 688. doi: 10.3390/app11020688
Sardar, M. F., Zhu, C. X., Geng, B., Ahmad, H. R., Song, T. T., Li, H. N., et al. (2021b). The fate of antibiotic resistance genes in cow manure composting: shaped by temperature-controlled composting stages. Bioresource Technol. 320, 124403. doi: 10.1016/j.biortech.2020.124403
Sardar, M. F., Zhu, C. X., Geng, B., Huang, Y. L., Abbasi, B., Zhang, Z. G., et al. (2021a). Enhanced control of sulfonamide resistance genes and host bacteria during thermophilic aerobic composting of cow manure. Environ. Pollut. 275, 16587. doi: 10.1016/j.envpol.2021.116587
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Huttenhower, G. C., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12, r60. doi: 10.1186/gb-2011-12-6-r60
Sennett, L. B., Goyer, C., Burton, D. L., Zebarth, B. J., and Whitney, S. (2023). Chemical fumigation combined with soil amendments of contrasting carbon availability alters soil bacterial and fungal community diversity. Appl. Soil Ecol. 188, 104858. doi: 10.1016/j.apsoil.2023.104858
Song, S. Z., Xiong, K. N., Chi, Y. K., He, C., Fang, J. Z., He, S. Y., et al. (2022). Effect of cultivated pastures on soil bacterial communities in the karst rocky desertification area. Front. Microbiol. 13, 922989. doi: 10.3389/fmicb.2022.922989
Soozandehfar, H., MacColl, K., and Maherali, H. (2023). Grassland restoration increases mutualistic benefits that soil biota provide to plants. Int. J. Plant Sci. 4, 224. doi: 10.1086./724224
Sui, X., Zhang, R., Frey, B., Frey, B., Yang, L. B., Li, M. H., et al. (2019). Land use change effects on diversity of soil bacterial, acidobacterial and fungal communities in wetlands of the sanjiang plain, northeastern china. Sci. Rep. 9, 18535. doi: 10.1038/s41598-019-55063-4
Sun, C. L., Wang, Y. W., Wang, C. J., Li, Q. J., Wu, Z. H., Yuan, D. S., et al. (2021). Effects of land use conversion on soil extracellular enzyme activity and its stoichiometric characteristics in karst mountainous areas. Acta Ecol. Sin. 4110, 4140–4149. doi: 10.5846/stxb202007161864
Sun, H. F., Li, X. L., Jin, L. Q., Zhao, Y. R., Li, C. Y., Zhang, J., et al. (2022). Changes in soil bacterial community diversity in degraded patches of alpine meadow in the source area of the yellow river. Environ. Sci. 43, 4662–4673. doi: 10.13227/j.hjkx.202112106
Tan, H., Barret, M., Mooij, M. J., Rice, O., Morrissey, J., Dobson, A., et al. (2013). Long-term phosphorus fertilisation increased the diversity of the total bacterial community and the phod phosphorus mineraliser group in pasture soils. Biol. Fertil. Soils. 49, 661–672. doi: 10.1007/s00374-012-0755-5
Venkatachalam, S., Kannan, V. M., Saritha, V. N., Loganathachetti, D. S., Mohan, M., Krishnan, K. P., et al. (2021). Bacterial diversity and community structure along the glacier foreland of Midtre Lovénbreen, Svalbard, Arctic. Ecol. Indic. 126, 107704. doi: 10.1016/j.ecolind.2021.107704
Wall, D. H., and Six, J. (2015). Give soils their due. Science 347, 695. doi: 10.1126/science.aaa8493
Wang, D., Zhou, H., Zuo, J., Chen, P., She, Y., Yao, B., et al. (2022). Responses of soil microbial metabolic activity and community structure to different degraded and restored grassland gradients of the Tibetan Plateau. Front. Plant Sci. 13, 770315. doi: 10.3389/fpls.2022.770315
Wang, K. L., Yue, Y. M., Chen, H. S., Wu, X. B., Xiao, J., Qi, X. K., et al. (2019). The comprehensive treatment of karst rocky desertification and its regional restoration effects. Acta Ecol. Sin. 39, 7432–7440. doi: 10.5846/stxb201909051849
Wang, K. L., Yue, Y. M., Chen, H. S., and Zeng, F. P. (2020). Mechanisms and realization pathways for integration of scientifific poverty alleviation and ecosystem services enhancement. Bull. Chin. Acad. Sci. 35, 1264–1272. doi: 10.16418/j.issn.1000-3045.20200729002
Wang, R. Z., Yang, J. J., Liu, H. Y., Sardans, J., Zhang, Y. H., et al. (2022). Nitrogen enrichment buffers phosphorus limitation by mobilizing mineral-bound soil phosphorus in grasslands. Ecology. 103, e3616. doi: 10.1002/ecy.3616
Wang, Z. Q., Zhang, J. X., Yang, X. L., Huang, X., Chen, S. L., and Qiao, Y. (2021). Characteristics of soil microbial diversity in different patches of alpine meadow. Acta Agrestia Sin. 29,1916-1926. doi: 10.11733/j.issn.1007-0435.09007
Wang, Z. T., Liu, T. X., Tong, X., Duan, L. M., Li, D. F., Liu, X. Y., et al. (2023). Changes in vegetation characteristics and soil enzyme activities under different treatments in semi-arid meadow grassland. Acta Prataculturae Sinica 3, 1–15.
Wei, W., Zhou, J. J., Liu, Y. F., Baima, G. W., and Renzeng, W. D. (2022). Impact of different grazing utilization on vegetation and soil characteritics of Kobresia tibetica swamp alpine meadow. Grassland and Turf. 42, 55–61. doi: 10.13817/j.cnki.cyycp.04008
Wu, M. H., Xue, K., Wei, P. J., Jia, Y. L., Zhang, Y., Chen, S. Y., et al. (2022). Soil microbial distribution and assembly are related to vegetation biomass in the alpine permafrost regions of the Qinghai-Tibet Plateau. Sci. Total Environ.. 834, 155259. doi: 10.1016/j.scitotenv.2022.155259
Wu, X. H., Wang, R., Gao, C. Q., Gao, S., Du, L. L., Khan, A., et al. (2021). Variations of soil properties effect on microbial community structure and functional structure under land uses. Acta Ecol. Sin. 41, 7989–8002. doi: 10.5846/stxb202012133169
Xin, X. P., Jin, D. Y., Ge, Y., Wang, J. H., Chen, J. Q., Qi, J. G., et al. (2020). Climate change dominated long-term soil carbon losses of Inner Mongolian grasslands. Global Biogeochem. Cy. 34, e2020GB.006559. doi: 10.1029/2020GB006559
Xiong, K. N., Chi, Y. K., and Shen, X. Y. (2017). Research on photosynthetic leguminous forage in the karst rocky desertification regions of Southwestern China. Pol. J. Environ. Stud. 26, 2319–2329. doi: 10.15244/pjoes/69472
Xiong, K. N., Li, P., Zhou, Z. F., An, Y. L., Lv, T., Lan, A. J., et al. (2002). Remote sensing of Karst Rocky Desertification: A Typical GIS Study—Taking Guizhou Province as an Example. Beijing: Beijing Geological Pres.
Xiong, K. N., Zhu, D. Y., Peng, T., Yu, L. F., Xue, J. H., Li, P., et al. (2016). Study on ecological industry technology and demonstration for karst rocky desertification control of the karst plateau-gorge. Acta Ecol. Sin. 36, 7109–7113. doi: 10.5846/stxb201610172104
Xu, J. Y., Zhou, J. R., Wu, J., Wang, J., Cheng, S. Y., Zhao, G., et al. (2020). Medium-and long-term effects of the veterinary antibiotic sulfadiazine on soil microorganisms in a rice field. J. Agro-Environ. Sci. 39, 1757–1766. doi: 10.11654/jaes.2020-0123
Xue, G. P., Bai, H. M., Du, J. W., Zhu, C. X., Fu, C. Y., Jiang, W., et al. (2023). Effects of different treatments on structure and diversity of soil bacterial community in pepper continuous cropping soil. China Vegetables. 4, 1–7. doi: 10.19928./j.cnki.1000-6346.2023.2014
Xue, L., Ren, H., Li, S., Leng, X., and Yao, X. (2017). Soil bacterial community structure and co-occurrence pattern during vegetation restoration in karst rocky desertification area. Front. Microbiol. 8, 2377. doi: 10.3389/fmicb.2017.02377
Yu, J., Yin, Q., Niu, J. M., Yan, Z. J., Wang, H., Wang, Y. Q., et al. (2022). Consistent effects of vegetation patch type on soil microbial communities across three successional stages in a desert ecosystem. Land Degrad. Dev. 33, 1552–1563. doi: 10.1002/ldr.4194
Yun, Y., Wang, H., Man, B., Xiang, X., Zhou, J. P., Qiu, X., et al. (2016). The relationship between ph and bacterial communities in a single karst ecosystem and its implication for soil acidification. Front. Microbiol. 7, 1955. doi: 10.3389/fmicb.2016.01955
Zhang, C., Yan, R. R., Liang, Q. W., Na, R. S., Li, T., Yang, X. F., et al. (2021). Study on soil physical and chemical properties and carbon and nitrogen sequestration of grassland under different utilization modes. Acta Prataculturae Sin. 30, 90–98. doi: 10.11686/cyxb2020278
Zhang, J. X., Chen, H. S., Fu, Z. Y., and Wang, K. L. (2021). Effects of vegetation restoration on soil properties along an elevation gradient in the karst region of southwest China. Agr. Ecosyst. Environ. 320, 107572. doi: 10.1016/j.agee.2021.107572
Zhang, J. Y., Yang, X. M., Song, Y. H., Liu, H. F., Wang, G. L., Xue, S., et al. (2020). Revealing the nutrient limitation and cycling for microbes under forest management practices in the Loess Plateau - Ecological stoichiometry. Geoderma 361, 114108. doi: 10.1016/j.geoderma.2019.114108
Zhang, X. Y., Hu, B. X., Ren, H. J., and Zhang, J. (2018). Composition and functional diversity of microbial community across a mangrove-inhabited mudflat as revealed by 16S rDNA gene sequences. Sci. Total Environ. 633, 518–528. doi: 10.1016/j.scitotenv.03158
Zhang, Y. T., Gao, X. L., Hao, X. Y., Alexander, T. W., Shi, X. J., Jin, L., et al. (2020). Heavy grazing over 64 years reduced soil bacterial diversity in the foothills of the Rocky Mountains, Canada. Appl. Soil Ecol. 147, 103361. doi: 10.1016/j.apsoil.09011
Zheng, J. H., Zhao, M. L., Wang, Q., Zhang, F., Zhang, B., and Zhang, J. (2022). Effects of management regime on soil microbial community structure and diversity of Stipa grandis grassland. Acta Ecol. Sin. 42, 4998–5008. doi: 10.5846/stxb202103290818
Keywords: artificial grassland, soil microorganism, sustainable utilization, karst desertification, Illumina sequencing
Citation: Chi Y, Song S and Xiong K (2023) Effects of different grassland use patterns on soil bacterial communities in the karst desertification areas. Front. Microbiol. 14:1208971. doi: 10.3389/fmicb.2023.1208971
Received: 20 April 2023; Accepted: 07 August 2023;
Published: 31 August 2023.
Edited by:
Yongchun Li, Zhejiang Agriculture and Forestry University, ChinaReviewed by:
Xiangchao Cui, Xinyang Normal University, ChinaCopyright © 2023 Chi, Song and Xiong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kangning Xiong, eGlvbmdrbkBnem51LmVkdS5jbg==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.