- Department of Laboratory Medicine, the Affiliated Hospital of Southwest Medical University, Luzhou, China
Colistin is highly promising against multidrug-resistant and extensively drug-resistant bacteria clinically. Bacteria are resistant to colistin mainly through mcr and chromosome-mediated lipopolysaccharide (LPS) synthesis-related locus variation. However, the current understanding cannot fully explain the resistance mechanism in mcr-negative colistin-resistant strains. Significantly, the contribution of efflux pumps to colistin resistance remains to be clarified. This review aims to discuss the contribution of efflux pumps and their related transcriptional regulators to colistin resistance in various bacteria and the reversal effect of efflux pump inhibitors on colistin resistance. Previous studies suggested a complex regulatory relationship between the efflux pumps and their transcriptional regulators and LPS synthesis, transport, and modification. Carbonyl cyanide 3-chlorophenylhydrazone (CCCP), 1-(1-naphthylmethyl)-piperazine (NMP), and Phe-Arg-β-naphthylamide (PAβN) all achieved the reversal of colistin resistance, highlighting the role of efflux pumps in colistin resistance and their potential for adjuvant development. The contribution of the efflux pumps to colistin resistance might also be related to specific genetic backgrounds. They can participate in colistin tolerance and heterogeneous resistance to affect the treatment efficacy of colistin. These findings help understand the development of resistance in mcr-negative colistin-resistant strains.
Introduction
The emergence and widespread occurrence of carbapenem-resistant gram-negative bacteria (CRGB: Enterobacteriaceae, Pseudomonas spp., Acinetobacter spp., etc.) poses a severe public health risk, with some metallo-β-lactamases-producing individuals also revealing inherent resistance to ceftazidime/avibactam. These CRGB acquire multiple antimicrobial resistance determinants under the action of mobile elements, promoting their resistance to tetracycline, quinolone, aminoglycosides, and other antibiotics. Additionally, energy-dependent efflux pumps, the classical pathway of bacterial resistance, play an essential role in multidrug resistance in bacteria. The efflux pumps can be divided into resistance-nodulation-cell division (RND), major facilitator superfamily (MFS), multidrug and toxic compound extrusion (MATE), small multidrug resistance (SMR), and ATP-binding cassette (ABC) super-families based on the differences in efflux transport proteins (Li and Nikaido, 2009). These transporters have a wide range of efflux substrates covering antibiotics for common clinical anti-infective treatments (β-lactams, tetracyclines, chloramphenicol, fluoroquinolones, aminoglycosides, sulfonamides, etc.), besides being involved in the elimination of bacterial intracellular metabolites. In the clinical infection setting, these efflux pumps also eliminate a wide range of chemical disinfectants favoring the long-term colonization of an abiotic surface by pathogens. These efflux pumps also respond quickly to enhance their viability when exposed to harsh environments (Fernández and Hancock, 2012). In particular, efflux pump transcription regulators are essential in regulating oxidative stress, physiological metabolism, and fitness (Holden and Webber, 2020). These results suggest that the efflux pumps are also an efficient means for bacteria to resist external environmental stress. Understanding their operation and development can help understand the evolutionary process of bacteria.
As carbapenems failed to treat CRGB, colistin (polymyxin B and E) was reintroduced to treat some refractory gram-negative infections. Colistin, as a cationic polypeptide compound, mainly acts on the outer membrane of bacteria carrying a negative charge. Colistin can destroy the stability of the outer membrane by displacing Ca2+ and Mg2+ ions, thus playing a bactericidal role (Falagas and Kasiakou, 2005). However, the lipid modification mediated by mutation and the inactivation of the two-component regulatory systems (TCS: PmrAB and PhoPQ) and mgrB are common causes of reduced susceptibility to colistin in bacteria. Plasmid-mediated horizontal transfer of mcr and its variants in bacteria promotes the rapid emergence of colistin resistance. In contrast, the role of efflux pumps in colistin resistance has received less attention. Previous studies have shown that efflux pump inhibitors (EPIs) CCCP and NMPs reverse colistin resistance, suggesting the role of efflux pumps in colistin resistance. Despite colistin’s much larger size than other common antibiotics and chemicals, the efflux of colistin compounds may be used as efflux substrates have been observed in many bacteria (Warner and Levy, 2010; Sundaramoorthy et al., 2019a). LPS modification and overexpression of efflux pumps and their regulators are also thought to be important components of the colistin resistance mechanisms in mcr-negative K. pneumoniae (Naha et al., 2022). In January 2023, we conducted a search on PubMed and Web of Science databases using keywords such as “colistin,” “efflux pump,” and “cationic antimicrobial peptide.” We screened over 100 publications summarized in this review, aiming to explore the role of the efflux pump in colistin resistance and provide a basis for elucidating the resistance mechanisms of mcr-negative colistin-resistant strains. The flow chart depicting the search and selection process is provided in Supplementary Figure S1.
General and novel colistin resistance mechanisms
The absence of the outer membrane limits the use of colistin against gram-positive bacteria. The colistin resistance mechanisms in gram-negative bacteria have been summarized in many previous reviews (Jeannot et al., 2017; Gogry et al., 2021). In general, the colistin resistance mechanism mainly includes two steps. First, point mutations in the TCS PmrAB, PhoPQ, CrrAB, and other lipid A modification coding genes located in chromosome loci produce more positively charged phosphoethanolamine to be added to the outer membrane lipids, resulting in weakened binding to colistin. Mutational inactivation and truncation of the insertion sequence of the PhoQ kinase inhibitor mgrB may also be effectively involved in this process. Second, phosphoethanolamine transferase encoded by mcr can also mediate the modification of the outer membrane (lipid A). More than 100 variants of mcr have been identified since it was first reported in 2015 (Liu et al., 2016).1 The mcr variants have completed horizontal transfer among different species with the help of mobile elements, posing a severe threat to public health. Compensatory mutations can effectively alleviate the fitness costs of mcr and promote the continued existence of plasmids carrying mcr (Yang et al., 2020). A recent study on A. hydrophila suggests that MlaA, the outer membrane lipoprotein-encoding gene, may be associated with high levels of colistin resistance (Liu et al., 2021). A recent study found that the RpoE stress system mediated colistin resistance in E. coli without disturbing the lipid A profile (Zeng et al., 2023). The discovery of colistin-degrading proteases also reveals the diversity of colistin resistance mechanisms (Lee et al., 2022).
Impacts of EPIs on colistin resistance
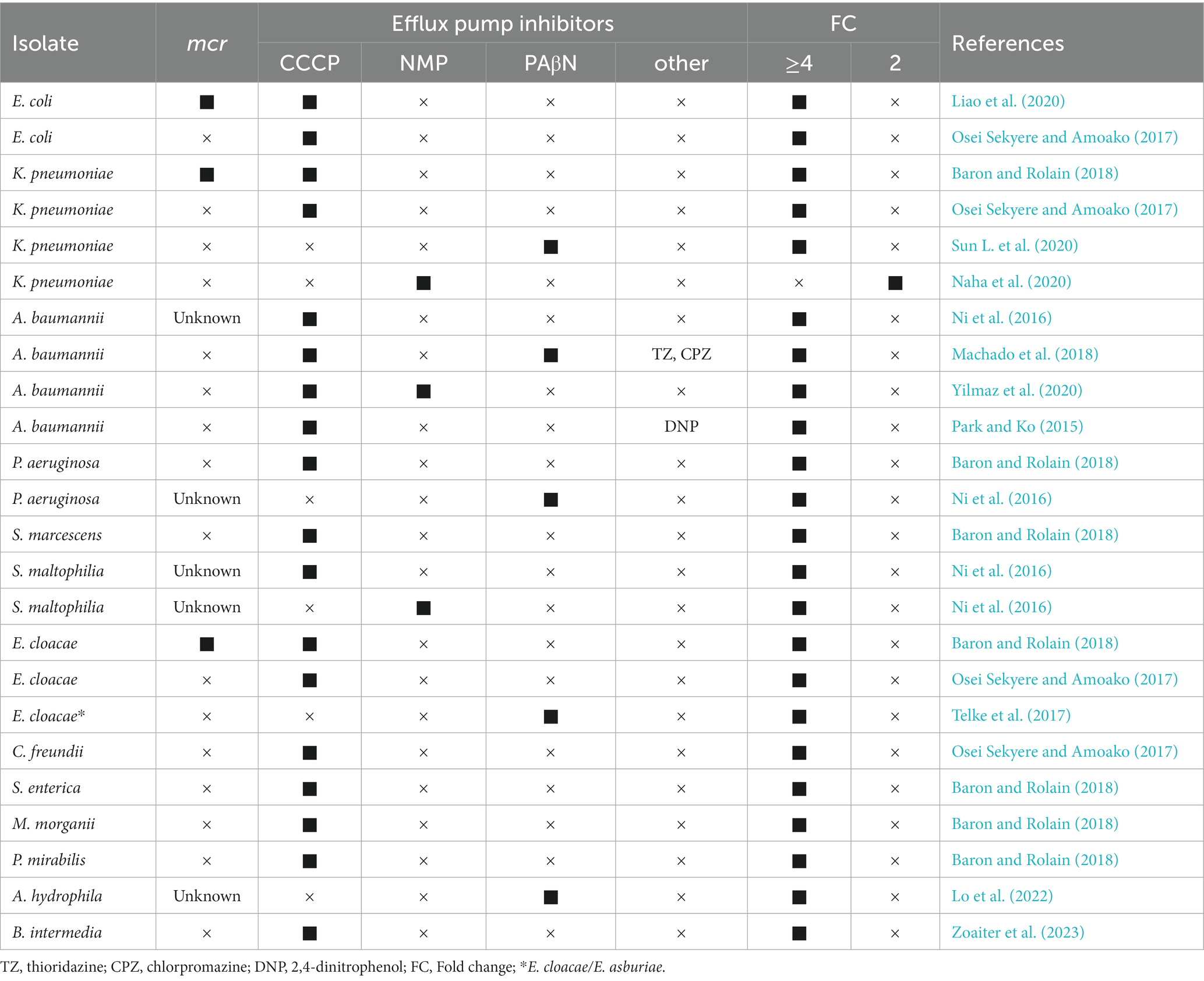
Although colistin is one of the few treatments available for multi-drug resistant pathogens, it does not seem effective in reducing patient mortality (Kelesidis and Falagas, 2015; Lee et al., 2020). Therefore, effective alternative treatment measures need to be developed. Current reviews of colistin resistance mechanisms have described less about the contribution of efflux pumps. Ni et al. (2016) successfully restored colistin susceptibility in colistin-resistant A. baumannii, P. aeruginosa, K. pneumoniae, and S. maltophilia using the efflux pump inhibitor CCCP (Table 1). Subsequent studies demonstrated that CCCP reversed colistin resistance in gram-negative bacteria that produced or did not produce mcr (Osei Sekyere and Amoako, 2017; Baron and Rolain, 2018). In contrast, other common EPIs (reserpine, verapamil, PAβN, and NMP) have more pronounced effects on the MICs of non-colistin-resistant strains (Table 1) (Ni et al., 2016; Osei Sekyere and Amoako, 2017). DNP, another proton-carrier inhibitor similar to CCCP, has apparent effects on colistin-resistant A. baumannii (Park and Ko, 2015). These results suggested that the differences in EPIs might be the main reason influencing the restoration of colistin susceptibility. The possible explanation is that CCCP-mediated electrochemical gradient depolarization can restore the negative charge of the outer membrane and lead to increased susceptibility to bacteria (Ni et al., 2016). Similarly, the MarR inhibitor salicylic acid can affect the negative cell surface charge of colistin-resistant E. coli and restore colistin susceptibility, but this effect is not apparent in colistin-sensitive E. coli (Sundaramoorthy et al., 2019b). Meanwhile, CCCP can reduce the metabolic activity of A. baumannii and increase its susceptibility to colistin (Park and Ko, 2015). A study in P. aeruginosa also showed that CCCP affected energy metabolism and thus decreased the colistin tolerance of P. aeruginosa biofilms (Pamp et al., 2008). In contrast, a previous study showed that CCCP contributed to the increase in polymyxin B resistance in wild-type and phop mutant in survival assays, which was mainly explained by the reduction of the proton gradient in the bacterial inner membrane (Alteri et al., 2011). Overall, more evidence is needed to confirm its impact on the cellular microenvironment and the potential involvement of other mechanisms.
Efflux pumps belonging to RND, MATE, SMR, and MFS families can use proton motive force to mediate efflux to various antibiotics (Lomovskaya and Watkins, 2001). Notably, CCCP reduces the activity of these multidrug-resistant efflux pumps by disrupting the proton motive force through interference with the transmembrane potential. A recent study found that CCCP, but not PAβN, reversed colistin resistance in a colistin-resistant K. pneumoniae strain that was mcr-negative and had no mutations in mgrB, phoPQ, pmrABCDK and reported efflux pump-related genes (ramAR, acrAB, kpnEF/GH, soxS, etc.) (Pu et al., 2023). It remains unclear whether other proton motive force-dependent and energy-driven efflux pumps contribute to colistin resistance (Figure 1). Additionally, the crrB mutant K. pneumoniae had a 4-fold decreased for colistin MIC in the presence of PAβN, suggesting a potential role of the efflux pump in colistin resistance (Sun L. et al., 2020). In the presence of NMP, the colistin MICs in colistin-resistant with mcr-negative A. baumannii/K have been observed. Pneumoniae lacking lipid A-related modification gene variation decreased significantly (Naha et al., 2020; Yilmaz et al., 2020). Li et al. also found that the colistin MICs in ST11-blaKPC-2 resistant lineage significantly reduced in the presence of NMP (Yang et al., 2021). Although these results suggest a potential pathway for the involvement of efflux pumps in colistin resistance, more substantial evidence is still needed. Considering the differences among species, we then summarized in detail the roles of the efflux pumps and their related genes in colistin resistance in different species.
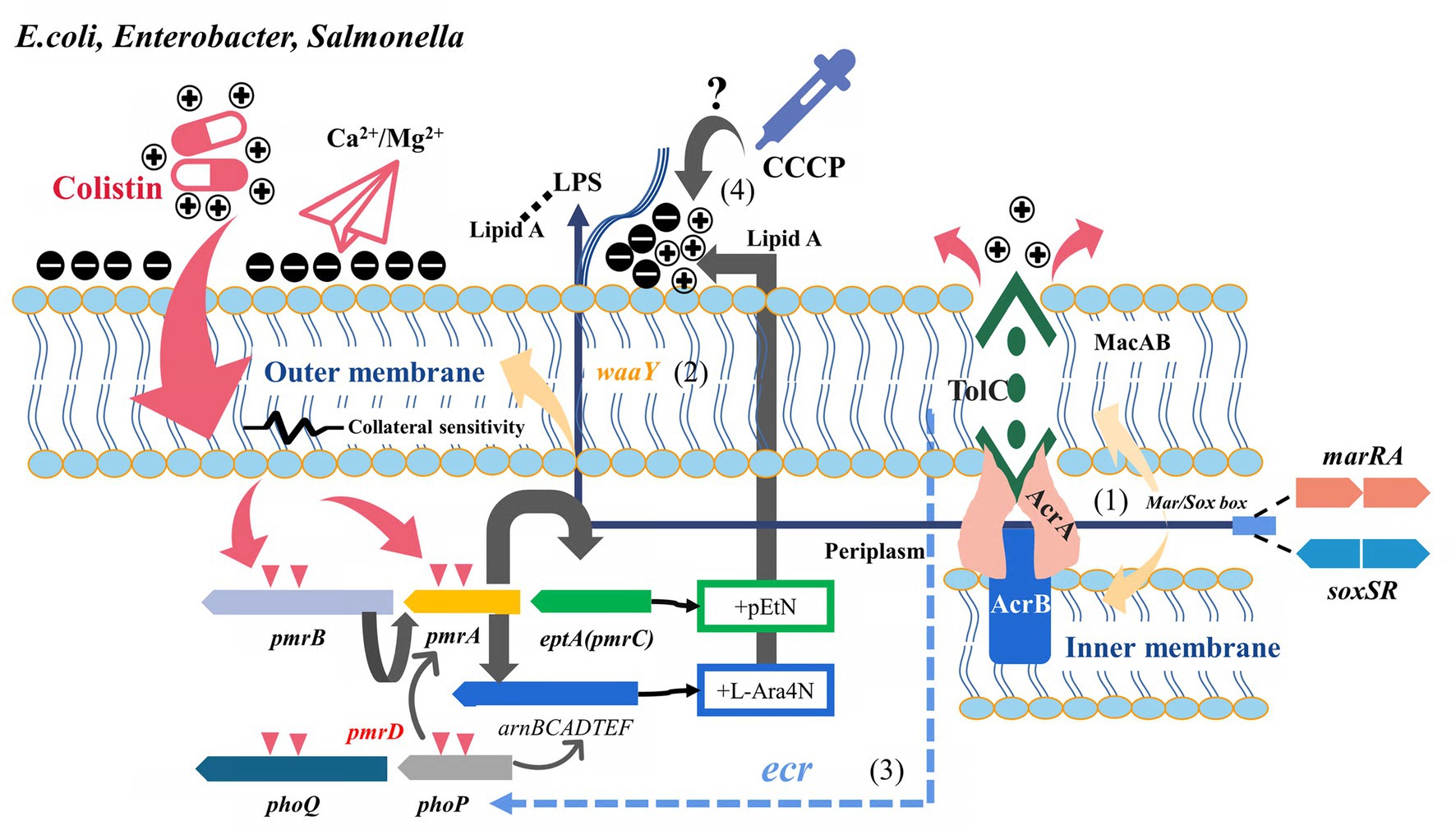
Figure 1. Potential relationship between multidrug resistance efflux pumps and their transcriptional regulatory factors and colistin resistance (E.coli, Enterobacter, and Salmonella). Two-component regulatory systems (PmrAB, PhoPQ, etc.) and related pathway variations are the classical pathways of mcr-negative colistin-resistant isolates. (1) Derepressed marA/soxS activated AcrAB-TolC or other RND efflux pumps involved in antibiotics efflux. (2) marA overexpression conferred collateral sensitivity to cationic antimicrobial peptides by up-regulating waaY. (3) Role of tolC in regulating PhoPQ via ecr in Enterobacter spp. remains to be confirmed. (4) Whether the effect of CCCP on the bacterial microenvironment mediates the change in colistin susceptibility is unclear.
Enterobacteriaceae
Escherichia coli, Enterobacter, and Salmonella
Colistin-resistant Enterobacteriaceae are widely identified in clinical infection settings and animal husbandry, and the status of antimicrobial resistance is a concern (Shen et al., 2019). As a transcription regulator commonly found in E. coli, K. pneumoniae, and other members of Enterobacteriaceae, marR played an essential role in regulating efflux pumps. Furthermore, marR mediated the inhibition of marA, while salicylic acid, antibiotic pressure, and endogenous amino acid replacement could release the disinhibition and activate marA expression under physiological conditions, thus upregulating the MDR efflux pump AcrAB-TolC (Grkovic et al., 2002). Activated marA could also upregulate the expression of gene encoding lipopolysaccharide core heptose (II) kinase, known as waay. This upregulation increases the negative charge of the bacterial outer membrane, subsequently enhancing its ability to bind cationic antimicrobial peptides (collateral sensitivity) (Lázár et al., 2018). Sundaramoorthy et al. (2019b) found that the negative charge of the outer membrane of colistin-resistant E. coli increased in the presence of the marR inhibitor salicylic acid, thereby restoring susceptibility to colistin. Another study found that the decreased polymyxin B susceptibility due to the marAB upregulation and marR mutant was observed, mainly attributed to the action of AcrAB and other TolC-dependent efflux pumps (Warner and Levy, 2010). These findings demonstrated the different pathways through which marRA is involved in colistin resistance. Another soxRS system, common in E. coli and Salmonella, plays an essential role in coping with superoxide, nitric oxide, and antibiotic stress. Increased soxS expression induced multidrug-resistant efflux pump AcrAB expression, decreased membrane permeability, and promoted the formation of multidrug-resistant phenotypes (Koutsolioutsou et al., 2005). The same binding sites exist in the promoters regulating Sox, Rob, and Mar systems (mar/sox box), resulting in a high degree of overlap between the genes (micF, acrAB, ompF, fumC, etc.) of soxRS and mar regulons (Figure 1) (Martin et al., 1999). Both soxRS and waaY could be activated by superoxide induction, but waaY transcription was SoxRS dependent (Figure 1) (Lee et al., 2009). Although the absence or overexpression of soxS in E. coli harboring the soxR mutation did not significantly affect CAMP susceptibility, the overexpression of SoxS in the tolC mutant contributed to increased susceptibility to polymyxin B (Warner and Levy, 2010). A recent study showed that not only the collateral susceptibility of antimicrobial resistance development to cationic polypeptides was associated with the changes in the regulation of LPS-related genes, but also a variety of physiological metabolism-related genes such as iron ion-binding proteins and transmembrane transport proteins played an important role (Grézal et al., 2023). This also reflected the pleiotropy of these global regulators involved in regulating multiple links of physiological metabolism. The disruption of tolC directly affected outer membrane integrity, and the intracellular accumulation of toxic substances activated marA, soxS, and rob to upregulate the efflux pumps (Rosner and Martin, 2009; Zgurskaya et al., 2011). Additionally, Rob might directly or indirectly increase marRAB expression by upregulating micF in response to colistin pressure, although the rob deficiency had little effect on CAMP susceptibility (Oh et al., 2000; Warner and Levy, 2010).
Heteroresistance may be an intermediate stage in the transition of susceptible bacteria to antimicrobial resistance, which is an essential reason for the failure of clinical anti-infective therapy (Falagas et al., 2008). Telke et al. found that soxRS-induced AcrAB-TolC efflux pump mediated heterogeneous resistance to colistin and could restore colistin susceptibility through PAβN in Enterobacter spp. (Telke et al., 2017). Unlike CCCP and DNP, PAβN could act directly as an inhibitor of AcrAB and AcrEF efflux pumps. Therefore, these results also suggested that AcrAB efflux pump was involved in colistin resistance (Misra et al., 2015). Colistin could mediate bactericidal effects by producing hydroxyl radicals or causing oxidative stress reactions as a class of bactericidal antibiotics (Bialvaei and Samadi Kafil, 2015; Yu et al., 2019). However, this bactericidal effect did not appear to influence the soxRS transcriptional levels in heterogeneous colistin-resistant Enterobacter isolates (Telke et al., 2017). The necessity of colistin heteroresistance by tolC (but not acrB) was further confirmed in E. cloacae, and tolC could activate the PhoQ-PhoP system by affecting the ecr (Figure 1) (Huang et al., 2019). The deletion of acrB is not sufficient to completely reverse colistin heterologous resistance in E. cloacae (Huang et al., 2019). Previous findings suggested efflux pumps are the primary mechanism for generating stable heterogeneous subpopulations (Manjunath et al., 2021). The fitness costs of these resistance mechanisms remain to be evaluated. Besides the co-regulation of tolC with Mar, Rob, and Sox, the expression of tolC, which carried multiple promoters, was also affected by EvgAS and PhoPQ, indicating that AcrAB-TolC had the potential to cope with different environments and responded quickly (Zhang et al., 2008; Pérez et al., 2012).
Transcriptome and proteomics analyses revealed that the expression of efflux pump AcrAB in E. coli and K. pneumoniae significantly increased after exposure to polymyxin B under experimental conditions (Ramos et al., 2016; Liu et al., 2020). The overexpression of cpxR in the absence of acrB affected the expression of TCS genes (phoP, phoQ, pmrB, pmrC, pmrH, and pmrD) and restored colistin susceptibility in Salmonella (Zhai et al., 2018). The variation in the acrB 620 site in Salmonella contributed to the restoration of colistin susceptibility, which could be explained by the defective macromolecular transport due to the altered conformation of the switch loop in this site (Cha et al., 2014; Kapach et al., 2020). Either tolC deletion alone or dual deletion of cpxR and tolC increased the susceptibility of S. enterica to colistin to varying degrees. The inactivation of tolC impaired the function of the entire RND efflux family and other tolC-dependent efflux pumps, not only the AcrAB pump (Zhang M. K. et al., 2021). These results highlighted the different contributions of acrB, soxRS, and marRA to colistin resistance in different backgrounds, the role of tolC mutations in regulating colistin resistance, and the complex regulatory network among the efflux pump, two-component system, and LPS modification.
Klebsiella pneumoniae
Multicenter studies from China have shown that the insertion inactivation of mgrB is the primary mechanism of colistin resistance in K. pneumoniae (Li et al., 2023). However, acrR insertion by IS26 is widespread in ST11-blaKPC-2-producing PR-CRKP (Polymyxin-resistant carbapenem-resistant K. pneumonia), which may be related to the low level of colistin resistance in these high-risk clones (Yang et al., 2021; Li et al., 2023). ramRA is a widely described global regulator in K. pneumoniae compared with E. coli and Salmonella, which mediates resistance to antibiotics such as tigecycline, nitrofurantoin, and beta-lactams by regulating AcrAB and OqxAB (Hao et al., 2022). The expression of soxS, ramA, and acrAB-tolC significantly upregulated in mcr-negative colistin-resistant K. pneumoniae lacking specific TCS-related gene variants, suggesting the role of soxS, ramA, and efflux pumps in colistin resistance (Naha et al., 2020). Similar to marA and soxS, ramA can also affect LPS modification. ramA directly binds to and activates the genes involved in lipid A biosynthesis: lpxC, lpxL-2, and lpxO, thereby modifying lipid A and resulting in decreased colistin susceptibility and increased anti-serum phagocytosis under the condition of increased ramA expression (Figure 2) (De Majumdar et al., 2015). Li et al. demonstrated in vitro that ramR variants can mediate polymyxin resistance, possibly related to derepressed ramA-mediated LPS alterations and multidrug resistance efflux pump overexpression (AcrAB) (Li et al., 2023). Another survey in China showed that the transcription level of ramA in colistin-resistant K. pneumoniae was not statistically significantly different from that in non-colistin-resistant strains, but the study lacked the main antimicrobial resistant and hypervirulent lineages ST11 and ST23 prevalent in China (Wang et al., 2017). However, ramA overexpression and variation were common in some colistin-resistant K. pneumoniae ST11 and ST147 isolates (Naha et al., 2020; Lv et al., 2021; Bolourchi et al., 2021a,b). Cationic antimicrobial peptides are also common in the human environment. Thus, whether these high-risk clones contribute to the co-evolution of hypervirulence and antimicrobial resistance by activating the efflux pump is not clear. A previous study showed that ΔacrB K. pneumoniae mutant had increased susceptibility to colistin and that LPS and CPS production were not significantly affected, revealing a role for AcrAB in the antimicrobial peptides resistance (Padilla et al., 2010). A less-described class of multidrug efflux RND transporter, KexD, is also present in K. pneumoniae and can be expressed constitutively with E. coli TolC or K. pneumoniae KocC (Ogawa et al., 2012). KexD has been proven to contribute to colistin resistance, and both of them are more conducive to high-level colistin resistance in K. pneumoniae under the induction of its neighbor gene crrB (Figure 2) (Cheng et al., 2018; Pantel et al., 2023). The specific diversity of the crrBAC-kexD cluster in the K. pneumoniae ST11 group suggests that ST11, a closely related lineage with carbapenem resistance and hypervirulence, is at risk of further developing colistin resistance (Kim et al., 2022). Overall, the colistin resistance mechanisms in mcr-negative colistin-resistant pathogens may be accumulated, highlighting the synergistic involvement of multiple mechanisms. Notably, the homolog of mexCD-oprJ efflux, tmexCD-toprJ, can be plasmid-mediated to acquire resistance to tigecycline in Enterobacteriaceae rapidly (Lv et al., 2020). In recent years, it has been identified that tmexCD-toprJ and mcr coexist in the same host of mobile elements, declaring the failure of last-line antibiotics (tigecycline and colistin) treatment and suggesting the threat of the rapid emergence of superbugs (Sun S. et al., 2020; Dong et al., 2022).
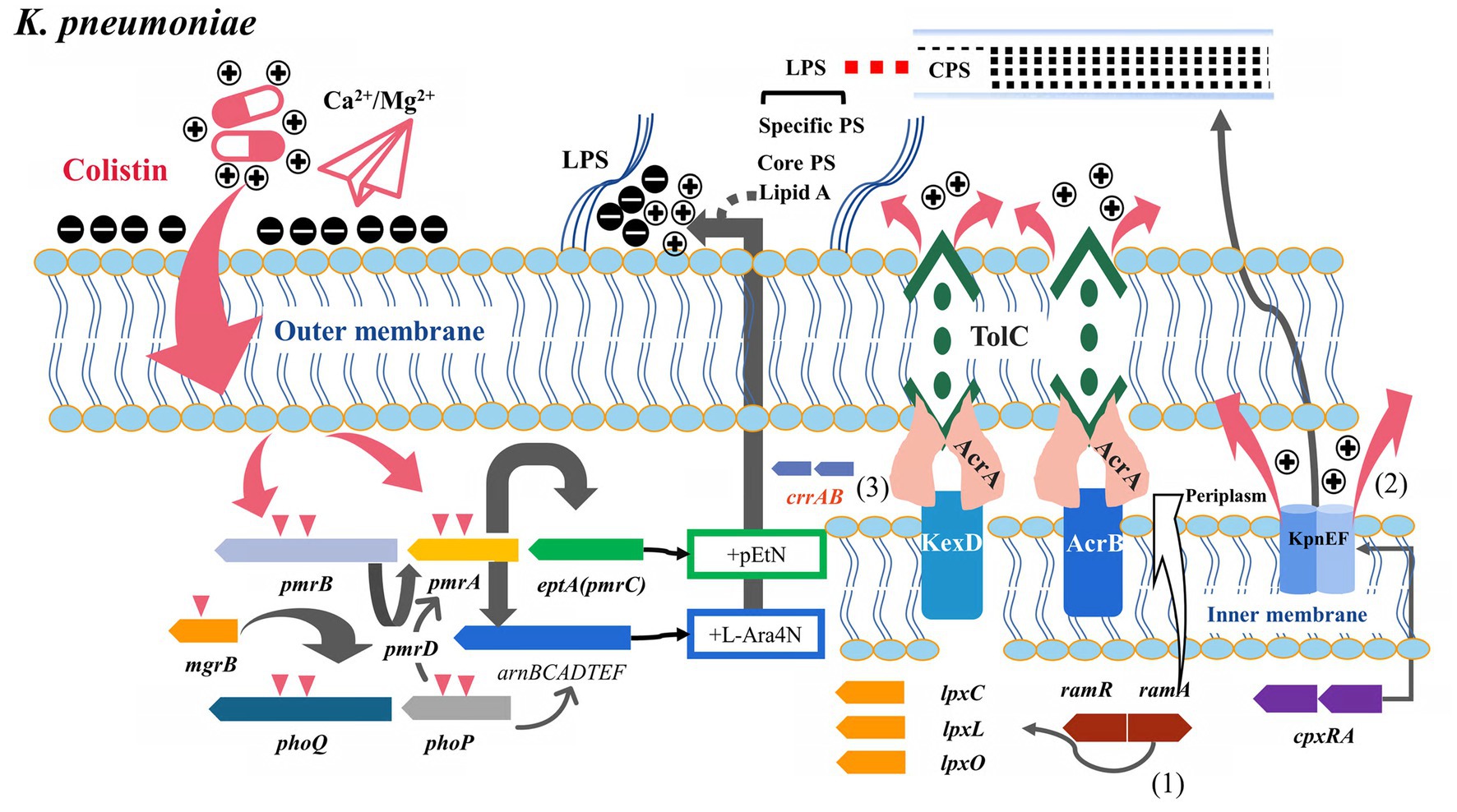
Figure 2. Potential relationship between multidrug resistance efflux pumps and their transcriptional regulatory factors and colistin resistance (K. pneumoniae). (1) ramAR was the dominant efflux pump transcriptional regulator in K. pneumoniae, binding to lpxC/X/O and participating in lipopolysaccharide synthesis. (2) KpnEF efflux pump was regulated by cpxRA and involved in colistin efflux and capsule synthesis. (3) Co-expression of CrrAB with the adjacent KexD efflux pump promoted the development of high levels of colistin resistance.
A study showed that several mcr-negative colistin-resistant K. pneumoniae strains isolated from CC15 and CC101 had different degrees of overexpression of acrAB, ramA, kpnEF (SMR pump), and kpnGH (MFS pump) (Naha et al., 2022). KpnEF efflux pumps belong to the small MDR family. The ΔKpnEF mutants showed increased susceptibility to various cationic antimicrobial peptides such as colistin, whereas KpnEF expression differs in the cpxAR mutant background (Figure 2) (Srinivasan and Rajamohan, 2013). The kpnGH is homologous to emrAB and belongs to the MFS efflux pump. In vitro, ΔkpnGH mutant susceptibility to cephalosporins, imipenem, polymyxin B, chlorhexidine, and other antimicrobial agents increased (Srinivasan et al., 2014).
Acinetobacter baumannii
The antimicrobial resistance surveillance from China showed that the colistin resistance rate of A. baumannii was still relatively low, but more than half of the isolates showed carbapenem-resistant A. baumannii (CRAB) (Gao et al., 2017; Zhang et al., 2020). A. baumannii had intrinsic resistance to multiple antibiotics. Hence, colistin has become one of the few options for treating CRAB. In the clinical application of colistin in anti-infection therapy, it is easy to induce A. baumannii to lead to colistin resistance by modifying phosphoethanolamine mediated by the two-component regulatory system PmrAB (Qureshi et al., 2015). In A. baumannii, three significant efflux pumps, AdeAB, AdeIJK, and AdeFGH, are involved in expelling antibiotics such as tetracycline, beta-lactam, and quinolones (Ayoub Moubareck and Hammoudi Halat, 2020). After A. baumannii was exposed to colistin, the upregulation of Ade cluster encoding genes was common; emrB and macAB overexpression were also observed (Cheah et al., 2016; Hua et al., 2017; Boinett et al., 2019). In E. coli, MacA demonstrated high affinity and specificity for the core LPS, suggesting that MacAB-TolC could be involved in LPS transport (Lu and Zgurskaya, 2013). phoP inhibited macAB transcription, and macAB deletion attenuated Salmonella’s virulence more than the tolC mutant (Nishino et al., 2006). Despite the upregulation of MDR efflux pumps, the laboratory-induced colistin-resistant isolates showed restoration of susceptibility to cefepime, azithromycin, and teicoplanin compared with the parental isolates (Moffatt et al., 2010; Li et al., 2015). Hua et al. also found that colistin induced partial restoration of antibiotic susceptibility in resistant strains but they mainly focused on β-lactams (Hua et al., 2017). Increased susceptibility to bacitracin, vancomycin, and beta-lactams was also observed in A. baumannii with high permeability of the outer membrane (Leus et al., 2018). These phenomena suggested that efflux pumps might be more involved in the transport of toxic compounds rather than in the efflux of the dominant antibiotics in colistin-resistant strains with increased outer membrane permeability due to LPS loss (Henry et al., 2012; Henry et al., 2015). Such collateral sensitivity could be masked by multiple β-lactamases and other plasmid-mediated resistance determinants in some clinical isolates of colistin-resistant A. baumannii.
Machado et al. (2018) identified several heterogeneous colistin-resistant strains that lacked TCS gene variants (lpxACD and pmrCAB). The EPIs CCCP, NMP, and PAβB were found to reverse polymyxin resistance to varying degrees in these strains. Furthermore, these strains’ efflux pump genes adeB, adeJ, adeG, craA, amvA, abeS, and abeM were overexpressed after colistin exposure (Machado et al., 2018). Colistin resistance mediated by the mutations in the pmr operon is generally expensive for A. baumannii (Geisinger and Isberg, 2017). Colistin heteroresistant A. baumannii isolates with only lpxACD mutations and overexpression of adeAB, adeG, and adeIJK were also observed in other studies, highlighting the role of efflux pumps in colistin heteroresistance (Chen et al., 2020). Besides participating in antibiotics efflux, the overexpression of these efflux pumps may be associated with fitness advantages mediated by these clinical isolates at specific sites of infection (Yoon et al., 2016). Another study also demonstrated that the resistance of A. baumannii to colistin could be reversed by NMP. AdeRS mutations mediating AdeAB or other RND-type efflux system overexpression were suggested to be the possible cause of colistin resistance in these isolates (Yilmaz et al., 2020). Additionally, increased expression of efflux transporter proteins (AdeABC and HlyD family) after colistin exposure was also observed in another study (Cheah et al., 2016). However, neither AdeRS nor AdeAB was shown to affect the colistin MIC in vitro (Richmond et al., 2016). These findings also suggested that efflux pump overexpression played a role in the excretion of toxic compounds and maintaining outer membrane integrity besides antibiotic efflux. The role of efflux pumps in antimicrobial resistance is also influenced by specific physiological settings (Leus et al., 2018). Additionally, different biological effects were found in AdeB deletion strains with different genetic backgrounds, suggesting that the function of the AdeAB efflux pump might be heterogeneous in other individuals (Richmond et al., 2016). These results indicated that the AdeAB pump could target different substrates under different living environments and selective pressures.
The two-component system and efflux pumps can be activated in bacteria by pH, nutrients, redox state, osmotic pressure, quorum signaling, and antibiotics (Worthington et al., 2013; Bazyleu and Kumar, 2014). One reported that the expression of AdeRS, AdeABC, and AdeFGH promoters did not show significant differences under sub-MIC colistin concentration and different growth conditions (Gil et al., 2021). However, the activation of the efflux pumps adeB, adeG, adeJ, adeH, and autoinducer synthase (abaI) by subinhibitory colistin has been observed in some clinical isolates (Sato et al., 2018; Shenkutie et al., 2022). Multiple biological effects on bacteria are observed at sublethal antibiotic concentrations, one of which can directly engage or interfere with quorum sensing systems (QS) (Figure 3) (Andersson and Hughes, 2014). As a signal molecule in the QS system, abaI is essential in regulating biofilms and can be used as a substrate for the AdeFGH efflux pump (He et al., 2015). A. baumannii isolates in a biofilm state has higher minimal biofilm inhibition concentrations (MBICs), which helps reduce the therapeutic effect of colistin (Kim et al., 2015). However, AdeRS did not always exist in A. baumannii isolates (Montaña et al., 2015). The restoration of colistin susceptibility by EPIs was observed in some colistin-resistant isolates without adeRS, suggesting the existence of other regulatory pathways and efflux pumps involved in colistin resistance (Yilmaz et al., 2020). Lin et al. observed that the transcription of emrB and several emrB- like genes were upregulated in colistin resistance-induced A. baumannii. Furthermore, ΔemrB mutants had increased susceptibility to colistin, demonstrating the contribution of the EmrAB pump to colistin resistance in A. baumannii (Lin et al., 2017). EmrAB was previously described mainly in E. coli and could mediate increased resistance to nalidixic acid, thiolactamycin, nitroquinoline, and hydrophobic proton uncouplers, with relatively little information available in A. baumannii (Yousefian et al., 2021). In another study, significant transcriptional changes were observed in the MATE (ydhE), MFS (mdfA), and SMR (ynfA and sugE) efflux pumps of colistin-resistant A. baumannii upon exposure to subinhibitory colistin concentrations (Paul et al., 2020). The overexpression of the MATE family efflux pump and mdfA contributes to the efflux of cationic compounds, but their contribution to colistin resistance remains confirmed (He et al., 2011; Li et al., 2015). A putative solvent/toluene-tolerant efflux ABC transporter protein, Ttg2C, may be essential in high-level colistin resistance in A. baumannii (Thi Khanh Nhu et al., 2016).
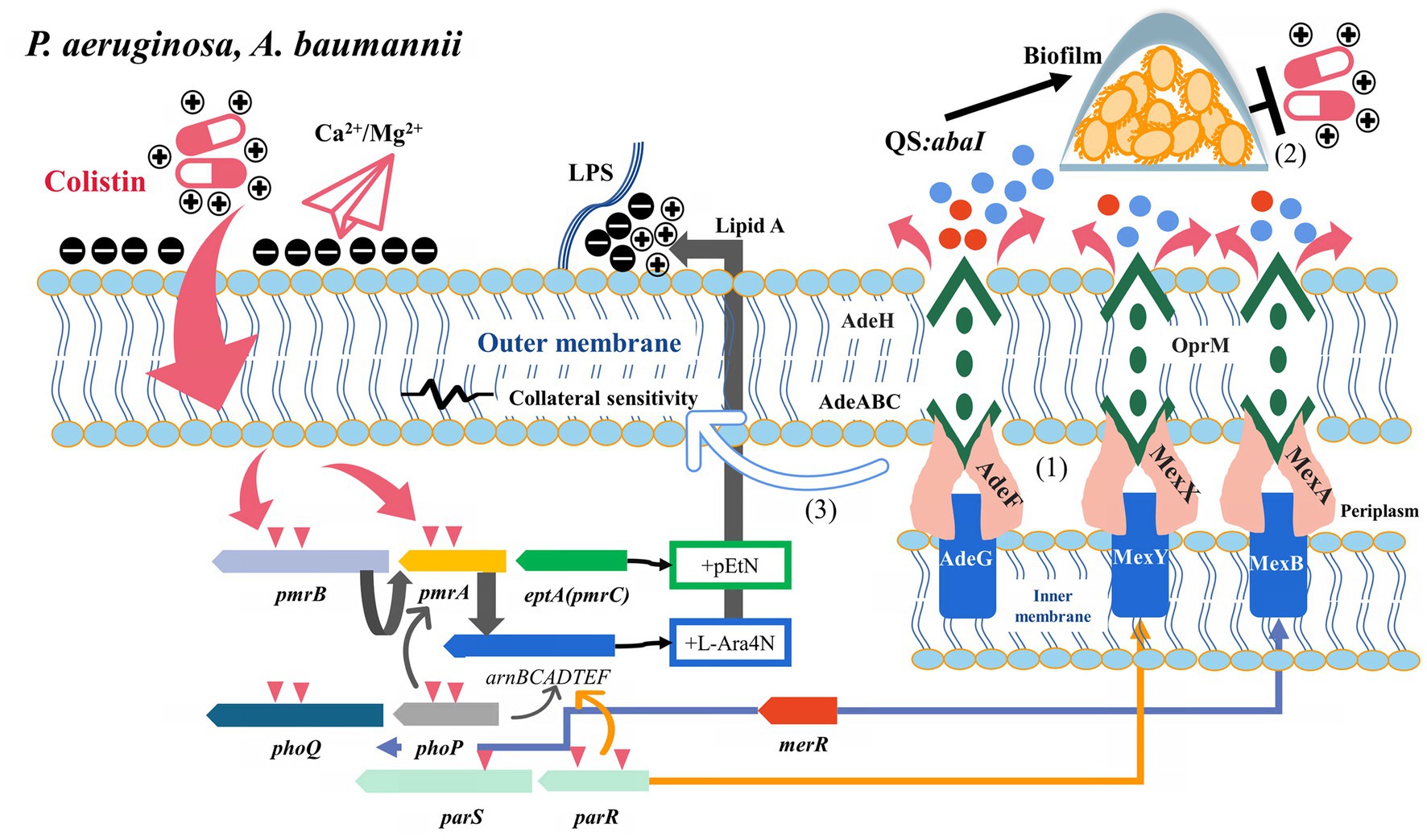
Figure 3. Potential relationship between multidrug resistance efflux pumps and their transcriptional regulatory factors and colistin resistance (P. aeruginosa and A. baumannii). (1) AdeRS, merR, and parRS activated AdeAB, MexXY-OprM, and MexAB-OprM to efflux antibiotics, respectively. (2) Efflux pumps were involved in colistin tolerance by transporting signaling molecules to activate the quorum sensing system. (3) Some colistin-resistant A. baumannii strains with efflux pump overexpression exhibited increased susceptibility to multiple antibiotics, which is associated with the disruption of LPS.
Pseudomonas aeruginosa
Pseudomonas aeruginosa is a common pathogen causing burn infection and cystic fibrosis, exhibiting intrinsic resistance to different antibiotics. MexAB-OprM, MexXY-OprM, and MexCD-OprJ are widely described RND family efflux pumps in P. aeruginosa with various efflux substrates: β-lactams, aminoglycosides, quinolones, tetracyclines, tigecycline, macrolides, amphenicols, novobiocin, sulfonamides, and trimethoprim (Li et al., 2015). MexXY is an induced efflux system, often characterized by concentration-dependent induction by ribosomal inhibitors (such as chloramphenicol, tetracyclines, macrolides, and aminoglycosides) (Jeannot et al., 2005). However, the overexpression of MexXY under exposure to ribosome-targeting antimicrobial agents was inversely correlated with colistin susceptibility and accompanied by the downregulation of the arn operon (Poole et al., 2015). Meanwhile, the heterogeneity of MexXY expression was observed in clinically isolated colistin-resistant P. aeruginosa isolates with different resistance levels, suggesting the possibility of intervention by other mechanisms (Goli et al., 2016). In an experiment on chlorhexidine (cationic polypeptide compounds) induced resistance, several mutants with decreased colistin susceptibility were observed, and these mutants exhibited MexY overexpression. While adding chlorpromazine significantly reduced the chlorhexidine MICs in the resistant mutants (Tag ElDein et al., 2021). Under colistin selective pressure, the two-component regulator ParR-ParS activated the arnBCADTEF operon, promoted mexY overexpression and inhibited oprD to reduce susceptibility to colistin (Fernández et al., 2010; Muller et al., 2011). mexXY deletion in ParRS-dependent pathways increased colistin tolerance by upregulating arnA and pmrA expression. The simultaneous overexpression of the arn opern and mexXY induced by the dual activation of PmrAB and ParRS contributed to the high-level resistance of the pmrB mutants to colistin, suggesting that the synergistic effect of efflux pumps and LPS modification promoted the development of colistin resistance (Puja et al., 2020) (Figure 3). In another study on the development of cross-resistance to colistin by exposing P. aeruginosa to chlorhexidine, proteomics revealed that the upregulation of MexA expression might be related to colistin resistance (Hashemi et al., 2019). Zhang W. et al. (2021) also observed significant upregulation of mexAB-oprM in laboratory-induced colistin-resistant P. aeruginosa. These results were similar to the previous findings of Pamp et al., who found that MexAB was involved in colistin tolerance, especially in adapting different subpopulations of bacteria to colistin in P. aeruginosa biofilms (Pamp et al., 2008). The efflux pump activator MerR induced mexAB-oprM and mexEF-oprN to participate in biofilm tolerance and acted as a repressor of phoPQ to participate in colistin resistance (Chambers and Sauer, 2013). In P. aeruginosa, the efflux pump is involved in the transport of signal molecules, and its expression can also be affected by the QS system. The genes related to QS were upregulated when P. aeruginosa was exposed to subinhibitory colistin concentration (Figure 3) (Cummins et al., 2009). However, whether this pathway can promote colistin resistance by activating efflux pumps or biofilms remains to be confirmed.
rsmA is a post-transcriptional regulatory protein involved in regulating various virulence-related genes, and its deletion causes overexpression of the MexEF-OprN pump and downregulation of type III secretion (Burrowes et al., 2006). However, the disruption of type III via secretion rsmA is associated with the overexpression of MexCD-OprJ or MexEF-OprN (Linares et al., 2005; Mlynarcik and Kolar, 2019). On exposure to different membrane-targeted drugs, these strains can mobilize various genetic determinants, such as pmr operons and efflux pumps, in response to environmental stresses (Chiang et al., 2012). The MexAB-oprM, MexCD-oprJ, and MuxABC-opmB efflux pumps all contribute to colistin-tolerant subpopulations (Chiang et al., 2012).
Stenotrophomonas maltophilia
Stenotrophomonas maltophilia is widespread in the natural environment and exhibits natural resistance to numerous antibiotics, significantly limiting clinical use options. Despite colistin’s in vitro antimicrobial activity, the assessment of colistin susceptibility in S. maltophilia is influenced by various factors in vitro. (Martínez-Servat et al., 2018). Lin et al. found that MacABCsm had a broader substrate spectrum on macrolides, aminoglycosides, and polymyxin than its counterpart in E. coli, and the deletion of MacAB resulted in a significant decrease in the colistin MIC of S. maltophilia isolates (Lin et al., 2014). Meanwhile, MacABCsm was stably expressed in S. maltophilia, associated with intrinsic resistance (Lin et al., 2014). Although multiple genes encoding efflux pumps have been identified in colistin-resistant S. maltophilia, their specific contribution to colistin resistance remains elucidated (Li et al., 2019).
Aeromonas hydrophila
Aeromonas hydrophila is a common group of opportunistic pathogens in the Aeromonas genus associated with aquatic environments. The AheABC efflux pump regulates the efflux of cefoperazone, cefuroxime, erythromycin, pristinamycin, and trityltin in A. hydrophila and participates in its MDR phenotype (Hernould et al., 2008). A recent study showed that the expression levels of three putative RND efflux pump genes, AHA0021, AHA1320, and AheB, significantly increased in MDR A. hydrophila. Also, PAβN significantly reduced the MIC of piperacillin/tazobactam, imipenem, erythromycin, and polymyxin B, suggesting the contribution of the RND efflux pump to colistin resistance (Lo et al., 2022).
Efflux pumps are associated with intrinsic colistin resistance in other gram-negative bacteria
Serratia marcescens and Proteus mirabilis
Proteus and S. marcescens are also common but easily overlooked opportunistic pathogens in clinical infections, often showing inherent resistance to colistin. LPS modification mediated by L-Ara4N in lipid A, Kdo residues, and arnBCADTEF operon contributes to the intrinsic resistance of P. mirabilis and S. marcescens to colistin (Olaitan et al., 2014). A recent study showed that the ABC transporter MacAB contributed to the intrinsic colistin resistance of S. marcescens, which was previously thought to be associated with the efflux of macrolide antibiotics and could be constitutively expressed with tolC (Shirshikova et al., 2021). In Salmonella, phoP inhibited macAB transcription, and macAB deletion attenuated the virulence of Salmonella more than the tolC mutant (Nishino et al., 2006). LPS played a role in colistin resistance and was a critical virulence factor in pathogenesis. However, whether macAB in S. marcescens is associated with PhoPQ or other TCSs remains to be investigated. The MFS efflux pump family SmvA was overexpressed in K. pneumoniae with increased resistance to multiple cationic biocides (chlorhexidine and octenidine) (Wand et al., 2019). Another study on P. mirabilis demonstrated that SmvA expression was insufficient to explain the differences in polymyxin B MIC of these intrinsically resistant isolates (Pelling et al., 2019).
Neisseria spp.
Among Neisseria spp., N. meningitidis and N. gonorrhoeae are closely associated with clinical infections and causative agents of bacterial meningitis and gonorrhea, respectively. The MtrCDE efflux system in Neisseria is thought to be the leading cause of the low-level intrinsic resistance to colistin (Moffatt et al., 2019). MtrCDE is Neisseria’s most widely explored RND pump, contributing to resistance to β-lactams, macrolides, rifampicin, detergents, bile salts, and cationic polypeptides. The contribution of MtrCDE to colistin resistance has been demonstrated in vitro (Tzeng et al., 2005). The epistatic effects between mtrD and mtr promoter region promote the formation of the multidrug resistance phenotype (Wadsworth et al., 2018).
Yersinia spp.
Yersinia comprises dozens of species, and only Y. pestis, Y. pseudotuberculosis, and Y. enterocolitica are closely related to clinical infection. Significantly different from other bacteria, the resistance of Yersinia to cationic peptides (including colistin) is affected by temperature as well as individual differences, which may be related to the successful adaptation of these pathogens at different sites (Bengoechea et al., 1996, 1998). Bengoechea et al. found that the rosAB locus encoded a temperature-regulated efflux pump and could participate in colistin efflux in response to antibiotic pressure (Bengoechea and Skurnik, 2000).
Burkholderia
Like S. maltophilia, Burkholderia is widespread in the environment and exhibits inherent resistance to various antibiotics, including colistin. The primary mechanism of intrinsic resistance of Burkholderia to colistin is still Ara4N synthesis and Ara4N transfer to lipid A (Loutet and Valvano, 2011). A previously suggested norM belonging to MATE transporters contributes to colistin resistance, but it is mainly associated with the presence of tetracycline (Fehlner-Gardiner and Valvano, 2002). Another class of yej operons (yejA1, yejA2, yejB, yejE, and yejF) belonging to ABC transporters was also found to be directly activated by colistin and conferred colistin resistance. To further clarify the contribution of the efflux pump to colistin resistance, Zoaiter et al. (2023) found that CCCP restored susceptibility in Burkholderia isolates, while VRP, PAβN, and RSP did not. The genomic analysis showed that the efflux pump genes YejABEF, LolCDE, and NorM were widely present in these isolates (Zoaiter et al., 2023). This result suggested that multiple efflux pumps might be involved in the intrinsic colistin resistance of Burkholderia. Another study explored the interaction of the Amrab-OprA, BpeEF-OprC, and BpeAB-OprB efflux pumps with the outer membrane permeability and showed that only the simultaneous presence of Amrab-OprA inactivation and hyperporination contributed to the increased susceptibility to colistin (Krishnamoorthy et al., 2019).
Future perspectives
Compared with the two-component regulatory system PmrAB/PhoPQ and mcr-mediated colistin resistance, relatively little information is available on the role played by efflux pumps in this regard. Although these findings suggest the possibility of EPIs reversing polymyxin resistance, EPI application to reverse colistin resistance still needs to be confirmed. CCCP has advantages over other EPIs in reversing colistin resistance, and its cytotoxicity limits its clinical application. Therefore, identifying these atypical resistant strains and developing suitable EPIs are crucial. Whether EPIs can reverse polymyxin resistance mediated by polymyxin-degrading protease and RpoE stress response pathways not involving structural changes in lipid A is uncertain. On the contrary, various reports showed that the bactericidal effect of CCCP on colistin was different. Therefore, the impact of CCCP on the bacterial intracellular microenvironment remains to be clarified. Other efflux pumps that rely on proton power to mediate colistin resistance remain to be identified. Since efflux pumps and associated transcription factors play a role in response to environmental stress, their activation can provide an adaptive advantage for these antibiotics-resistant strains in specific environments (Holden and Webber, 2020). However, the fitness effects of efflux pumps and related transcription factors mediating colistin resistance are still less explored compared with those of colistin resistance caused by LPS-related locus variants. The role of efflux pumps and their regulators in LPS synthesis, transport, and outer membrane integrity in colistin-resistant strains with different genetic and living backgrounds remains to be determined.
Conclusion
The efflux pumps’ structure, function, and regulation have been previously summarized in detail in many comprehensive reviews (Piddock, 2006; Puzari and Chetia, 2017; Nishino et al., 2021). We specifically examined the effects of efflux pumps, their regulators, and EPIs on colistin susceptibility. Efflux pumps play a role in the classical pathway of antibiotic resistance and participate in the efflux of various metabolites and signaling molecules. The correlation between efflux pumps and their transcriptional regulators and LPS modification/transport indicates colistin resistance mechanisms’ complexity. The observation that efflux pumps and their regulator (ramR, KpnEF, KexD, etc.) independently mediate colistin susceptibility in some isolates indicates colistin resistance mechanisms’ diversity. The effect of efflux pumps on colistin susceptibility is also mediated through multiple pathways, such as heteroresistance and tolerance. These findings indicate that the multidrug resistance efflux pumps may participate in different stages of occurrence and development of colistin resistance. Overall, the contribution of efflux pumps and their regulators to colistin resistance is multi-pathway, including outer membrane permeability, LPS modification, and environmental adaptation, besides the direct involvement in efflux. For developing adjuvants acting as EPIs, it is critical to identify colistin-resistant strains that are mcr-negative and lack specific mutations related to LPS modification.
Author contributions
YD, JH, WX, and ZZ contributed to the conception and design of the study. WX, XX, and CJ collected and explored literature. JH, MT, and GL performed the diagrams visualization. YD, ZZ, CY, JH, and JL prepared and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Sichuan Science and Technology Program (2020YFQ0045 and 2021YFS0329) and School-level scientific research project of Southwest Medical University (2019ZQN017).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1207441/full#supplementary-material
Footnotes
References
Alteri, C. J., Lindner, J. R., Reiss, D. J., Smith, S. N., and Mobley, H. L. (2011). The broadly conserved regulator PhoP links pathogen virulence and membrane potential in Escherichia coli. Mol. Microbiol. 82, 145–163. doi: 10.1111/j.1365-2958.2011.07804.x
Andersson, D. I., and Hughes, D. (2014). Microbiological effects of sublethal levels of antibiotics. Nat. Rev. Microbiol. 12, 465–478. doi: 10.1038/nrmicro3270
Ayoub Moubareck, C., and Hammoudi Halat, D. (2020). Insights into Acinetobacter baumannii: a review of microbiological, virulence, and resistance traits in a threatening nosocomial pathogen. Antibiotics (Basel) 9:119. doi: 10.3390/antibiotics9030119
Baron, S. A., and Rolain, J. M. (2018). Efflux pump inhibitor CCCP to rescue colistin susceptibility in mcr-1 plasmid-mediated colistin-resistant strains and gram-negative bacteria. J. Antimicrob. Chemother. 73, 1862–1871. doi: 10.1093/jac/dky134
Bazyleu, A., and Kumar, A. (2014). Incubation temperature, osmolarity, and salicylate affect the expression of resistance-nodulation-division efflux pumps and outer membrane porins in Acinetobacter baumannii ATCC19606T. FEMS Microbiol. Lett. 357, 136–143. doi: 10.1111/1574-6968.12530
Bengoechea, J. A., Díaz, R., and Moriyón, I. (1996). Outer membrane differences between pathogenic and environmental Yersinia enterocolitica biogroups probed with hydrophobic permeants and polycationic peptides. Infect. Immun. 64, 4891–4899. doi: 10.1128/iai.64.12.4891-4899.1996
Bengoechea, J. A., Lindner, B., Seydel, U., Ramón, D., and Ignacio, M. (1998). Yersinia pseudotuberculosis and Yersinia pestis are more resistant to bactericidal cationic peptides than Yersinia enterocolitica. Microbiology (Read.) 144, 1509–1515. doi: 10.1099/00221287-144-6-1509
Bengoechea, J. A., and Skurnik, M. (2000). Temperature-regulated efflux pump/potassium antiporter system mediates resistance to cationic antimicrobial peptides in yersinia. Mol. Microbiol. 37, 67–80. doi: 10.1046/j.1365-2958.2000.01956.x
Bialvaei, A. Z., and Samadi Kafil, H. (2015). Colistin, mechanisms and prevalence of resistance. Curr. Med. Res. Opin. 31, 707–721. doi: 10.1185/03007995.2015.1018989
Boinett, C. J., Cain, A. K., Hawkey, J., Do Hoang, N. T., Khanh, N. N. T., Thanh, D. P., et al. (2019). Clinical and laboratory-induced colistin-resistance mechanisms in Acinetobacter baumannii. Microb. Genom. 5:e000246. doi: 10.1099/mgen.0.000246
Bolourchi, N., Shahcheraghi, F., Giske, C. G., Nematzadeh, S., Noori Goodarzi, N., Solgi, H., et al. (2021a). Comparative genome analysis of colistin-resistant OXA-48-producing Klebsiella pneumoniae clinical strains isolated from two Iranian hospitals. Ann. Clin. Microbiol. Antimicrob. 20:74. doi: 10.1186/s12941-021-00479-y
Bolourchi, N., Shahcheraghi, F., Giske, C. G., Nematzadeh, S., Solgi, H., and Badmasti, F. (2021b). Genome analysis of an OXA-48-producing carbapenem- and colistin-resistant Klebsiella pneumoniae sequence type 11 clone isolated from an inpatient. Gene Rep. 25:101394. doi: 10.1016/j.genrep.2021.101394
Burrowes, E., Baysse, C., Adams, C., and O'Gara, F. (2006). Influence of the regulatory protein RsmA on cellular functions in Pseudomonas aeruginosa PAO1, as revealed by transcriptome analysis. Microbiology (Read.) 152, 405–418. doi: 10.1099/mic.0.28324-0
Cha, H. J., Müller, R. T., and Pos, K. M. (2014). Switch-loop flexibility affects transport of large drugs by the promiscuous AcrB multidrug efflux transporter. Antimicrob. Agents Chemother. 58, 4767–4772. doi: 10.1128/aac.02733-13
Chambers, J. R., and Sauer, K. (2013). The MerR-like regulator BrlR impairs Pseudomonas aeruginosa biofilm tolerance to colistin by repressing PhoPQ. J. Bacteriol. 195, 4678–4688. doi: 10.1128/jb.00834-13
Cheah, S. E., Johnson, M. D., Zhu, Y., Tsuji, B. T., Forrest, A., Bulitta, J. B., et al. (2016). Polymyxin resistance in Acinetobacter baumannii: genetic mutations and transcriptomic changes in response to clinically relevant dosage regimens. Sci. Rep. 6:26233. doi: 10.1038/srep26233
Chen, L., Lin, J., Lu, H., Zhang, X., Wang, C., Liu, H., et al. (2020). Deciphering colistin heteroresistance in Acinetobacter baumannii clinical isolates from Wenzhou, China. J. Antibiot. (Tokyo) 73, 463–470. doi: 10.1038/s41429-020-0289-2
Cheng, Y. H., Lin, T. L., Lin, Y. T., and Wang, J. T. (2018). A putative RND-type efflux pump, H239_3064, contributes to colistin resistance through CrrB in Klebsiella pneumoniae. J. Antimicrob. Chemother. 73, 1509–1516. doi: 10.1093/jac/dky054
Chiang, W. C., Pamp, S. J., Nilsson, M., Givskov, M., and Tolker-Nielsen, T. (2012). The metabolically active subpopulation in Pseudomonas aeruginosa biofilms survives exposure to membrane-targeting antimicrobials via distinct molecular mechanisms. FEMS Immunol. Med. Microbiol. 65, 245–256. doi: 10.1111/j.1574-695X.2012.00929.x
Cummins, J., Reen, F. J., Baysse, C., Mooij, M. J., and O'Gara, F. (2009). Subinhibitory concentrations of the cationic antimicrobial peptide colistin induce the pseudomonas quinolone signal in Pseudomonas aeruginosa. Microbiology (Read.) 155, 2826–2837. doi: 10.1099/mic.0.025643-0
De Majumdar, S., Yu, J., Fookes, M., McAteer, S. P., Llobet, E., Finn, S., et al. (2015). Elucidation of the RamA regulon in Klebsiella pneumoniae reveals a role in LPS regulation. PLoS Pathog. 11:e1004627. doi: 10.1371/journal.ppat.1004627
Dong, N., Zeng, Y., Wang, Y., Liu, C., Lu, J., Cai, C., et al. (2022). Distribution and spread of the mobilised RND efflux pump gene cluster tmexCD-toprJ in clinical gram-negative bacteria: a molecular epidemiological study. Lancet Microbe 3, e846–e856. doi: 10.1016/s2666-5247(22)00221-x
Falagas, M. E., and Kasiakou, S. K. (2005). Colistin: the revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clin. Infect. Dis. 40, 1333–1341. doi: 10.1086/429323
Falagas, M. E., Makris, G. C., Dimopoulos, G., and Matthaiou, D. K. (2008). Heteroresistance: a concern of increasing clinical significance? Clin. Microbiol. Infect. 14, 101–104. doi: 10.1111/j.1469-0691.2007.01912.x
Fehlner-Gardiner, C. C., and Valvano, M. A. (2002). Cloning and characterization of the Burkholderia vietnamiensis norM gene encoding a multi-drug efflux protein. FEMS Microbiol. Lett. 215, 279–283. doi: 10.1111/j.1574-6968.2002.tb11403.x
Fernández, L., Gooderham, W. J., Bains, M., McPhee, J. B., Wiegand, I., and Hancock, R. E. (2010). Adaptive resistance to the "last hope" antibiotics polymyxin B and colistin in Pseudomonas aeruginosa is mediated by the novel two-component regulatory system ParR-ParS. Antimicrob. Agents Chemother. 54, 3372–3382. doi: 10.1128/aac.00242-10
Fernández, L., and Hancock, R. E. (2012). Adaptive and mutational resistance: role of porins and efflux pumps in drug resistance. Clin. Microbiol. Rev. 25, 661–681. doi: 10.1128/cmr.00043-12
Gao, L., Lyu, Y., and Li, Y. (2017). Trends in drug resistance of Acinetobacter baumannii over a 10-year period: Nationwide data from the China surveillance of antimicrobial resistance program. Chin. Med. J. 130, 659–664. doi: 10.4103/0366-6999.201601
Geisinger, E., and Isberg, R. R. (2017). Interplay between antibiotic resistance and virulence during disease promoted by multidrug-resistant bacteria. J. Infect. Dis. 215, S9–s17. doi: 10.1093/infdis/jiw402
Gil, S. P. M., Tajuelo, A., López-Siles, M., and McConnell, M. J. (2021). Subinhibitory concentrations of clinically-relevant antimicrobials affect resistance-nodulation-division family promoter activity in Acinetobacter baumannii. Front. Microbiol. 12:780201. doi: 10.3389/fmicb.2021.780201
Gogry, F. A., Siddiqui, M. T., Sultan, I., and Haq, Q. M. R. (2021). Current update on intrinsic and acquired colistin resistance mechanisms in bacteria. Front. Med. (Lausanne) 8:677720. doi: 10.3389/fmed.2021.677720
Goli, H. R., Nahaei, M. R., Ahangarzadeh Rezaee, M., Hasani, A., Samadi Kafil, H., and Aghazadeh, M. (2016). Emergence of colistin resistant Pseudomonas aeruginosa at Tabriz hospitals, Iran. IJM 8, 62–69. Available at: https://pubmed.ncbi.nlm.nih.gov/27092226/
Grézal, G., Spohn, R., Méhi, O., Dunai, A., Lázár, V., Bálint, B., et al. (2023). Plasticity and stereotypic rewiring of the transcriptome upon bacterial evolution of antibiotic resistance. Mol. Biol. Evol. 40:msad020. doi: 10.1093/molbev/msad020
Grkovic, S., Brown, M. H., and Skurray, R. A. (2002). Regulation of bacterial drug export systems. Microbiol. Mol. Biol. Rev. 66, 671–701. doi: 10.1128/mmbr.66.4.671-701.2002
Hao, M., Ye, F., Jovanovic, M., Kotta-Loizou, I., Xu, Q., Qin, X., et al. (2022). Structures of class I and class II transcription complexes reveal the molecular basis of RamA-dependent transcription activation. Adv. Sci. (Weinh) 9:e2103669. doi: 10.1002/advs.202103669
Hashemi, M. M., Holden, B. S., Coburn, J., Taylor, M. F., Weber, S., Hilton, B., et al. (2019). Proteomic analysis of resistance of gram-negative bacteria to chlorhexidine and impacts on susceptibility to colistin, antimicrobial peptides, and Ceragenins. Front. Microbiol. 10:210. doi: 10.3389/fmicb.2019.00210
He, X., Lu, F., Yuan, F., Jiang, D., Zhao, P., Zhu, J., et al. (2015). Biofilm formation caused by clinical Acinetobacter baumannii isolates is associated with overexpression of the AdeFGH efflux pump. Antimicrob. Agents Chemother. 59, 4817–4825. doi: 10.1128/aac.00877-15
He, G. X., Zhang, C., Crow, R. R., Thorpe, C., Chen, H., Kumar, S., et al. (2011). SugE, a new member of the SMR family of transporters, contributes to antimicrobial resistance in Enterobacter cloacae. Antimicrob. Agents Chemother. 55, 3954–3957. doi: 10.1128/aac.00094-11
Henry, R., Crane, B., Powell, D., Deveson Lucas, D., Li, Z., Aranda, J., et al. (2015). The transcriptomic response of Acinetobacter baumannii to colistin and doripenem alone and in combination in an in vitro pharmacokinetics/pharmacodynamics model. J. Antimicrob. Chemother. 70, 1303–1313. doi: 10.1093/jac/dku536
Henry, R., Vithanage, N., Harrison, P., Seemann, T., Coutts, S., Moffatt, J. H., et al. (2012). Colistin-resistant, lipopolysaccharide-deficient Acinetobacter baumannii responds to lipopolysaccharide loss through increased expression of genes involved in the synthesis and transport of lipoproteins, phospholipids, and poly-β-1,6-N-acetylglucosamine. Antimicrob. Agents Chemother. 56, 59–69. doi: 10.1128/aac.05191-11
Hernould, M., Gagné, S., Fournier, M., Quentin, C., and Arpin, C. (2008). Role of the AheABC efflux pump in Aeromonas hydrophila intrinsic multidrug resistance. Antimicrob. Agents Chemother. 52, 1559–1563. doi: 10.1128/aac.01052-07
Holden, E. R., and Webber, M. A. (2020). MarA, RamA, and SoxS as mediators of the stress response: survival at a cost. Front. Microbiol. 11:828. doi: 10.3389/fmicb.2020.00828
Hua, X., Liu, L., Fang, Y., Shi, Q., Li, X., Chen, Q., et al. (2017). Colistin resistance in Acinetobacter baumannii MDR-ZJ06 revealed by a multiomics approach. Front. Cell. Infect. Microbiol. 7:45. doi: 10.3389/fcimb.2017.00045
Huang, L., Feng, Y., and Zong, Z. (2019). Heterogeneous resistance to colistin in Enterobacter cloacae complex due to a new small transmembrane protein. J. Antimicrob. Chemother. 74, 2551–2558. doi: 10.1093/jac/dkz236
Jeannot, K., Bolard, A., and Plésiat, P. (2017). Resistance to polymyxins in gram-negative organisms. Int. J. Antimicrob. Agents 49, 526–535. doi: 10.1016/j.ijantimicag.2016.11.029
Jeannot, K., Sobel, M. L., El Garch, F., Poole, K., and Plésiat, P. (2005). Induction of the MexXY efflux pump in Pseudomonas aeruginosa is dependent on drug-ribosome interaction. J. Bacteriol. 187, 5341–5346. doi: 10.1128/jb.187.15.5341-5346.2005
Kapach, G., Nuri, R., Schmidt, C., Danin, A., Ferrera, S., Savidor, A., et al. (2020). Loss of the periplasmic chaperone Skp and mutations in the efflux pump AcrAB-TolC play a role in acquired resistance to antimicrobial peptides in Salmonella typhimurium. Front. Microbiol. 11:189. doi: 10.3389/fmicb.2020.00189
Kelesidis, T., and Falagas, M. E. (2015). The safety of polymyxin antibiotics. Expert Opin. Drug Saf. 14, 1687–1701. doi: 10.1517/14740338.2015.1088520
Kim, S. J., Cho, H., and Ko, K. S. (2022). Comparative analysis of the colistin resistance-regulating gene cluster in klebsiella species. J. Microbiol. 60, 461–468. doi: 10.1007/s12275-022-1640-z
Kim, H. A., Ryu, S. Y., Seo, I., Suh, S. I., Suh, M. H., and Baek, W. K. (2015). Biofilm formation and colistin susceptibility of Acinetobacter baumannii isolated from Korean nosocomial samples. Microb. Drug Resist. 21, 452–457. doi: 10.1089/mdr.2014.0236
Koutsolioutsou, A., Peña-Llopis, S., and Demple, B. (2005). Constitutive soxR mutations contribute to multiple-antibiotic resistance in clinical Escherichia coli isolates. Antimicrob. Agents Chemother. 49, 2746–2752. doi: 10.1128/aac.49.7.2746-2752.2005
Krishnamoorthy, G., Weeks, J. W., Zhang, Z., Chandler, C. E., Xue, H., Schweizer, H. P., et al. (2019). Efflux pumps of Burkholderia thailandensis control the permeability barrier of the outer membrane. Antimicrob. Agents Chemother. 63:e00956-19. doi: 10.1128/aac.00956-19
Lázár, V., Martins, A., Spohn, R., Daruka, L., Grézal, G., Fekete, G., et al. (2018). Antibiotic-resistant bacteria show widespread collateral sensitivity to antimicrobial peptides. Nat. Microbiol. 3, 718–731. doi: 10.1038/s41564-018-0164-0
Lee, D. H., Cha, J. H., Kim, D. W., Lee, K., Kim, Y. S., Oh, H. Y., et al. (2022). Colistin-degrading proteases confer collective resistance to microbial communities during polymicrobial infections. Microbiome 10:129. doi: 10.1186/s40168-022-01315-x
Lee, J. H., Lee, K. L., Yeo, W. S., Park, S. J., and Roe, J. H. (2009). SoxRS-mediated lipopolysaccharide modification enhances resistance against multiple drugs in Escherichia coli. J. Bacteriol. 191, 4441–4450. doi: 10.1128/jb.01474-08
Lee, Y. T., Sun, J. R., Wang, Y. C., Chiu, C. H., Kuo, S. C., Chen, T. L., et al. (2020). Multicentre study of risk factors for mortality in patients with Acinetobacter bacteraemia receiving colistin treatment. Int. J. Antimicrob. Agents 55:105956. doi: 10.1016/j.ijantimicag.2020.105956
Leus, I. V., Weeks, J. W., Bonifay, V., Smith, L., Richardson, S., and Zgurskaya, H. I. (2018). Substrate specificities and efflux efficiencies of RND efflux pumps of Acinetobacter baumannii. J. Bacteriol. 200:e00049-18. doi: 10.1128/jb.00049-18
Li, J., Liu, S., Fu, J., Yin, J., Zhao, J., Zhong, C., et al. (2019). Co-occurrence of colistin and meropenem resistance determinants in a Stenotrophomonas strain isolated from sewage water. Microb. Drug Resist. 25, 317–325. doi: 10.1089/mdr.2018.0418
Li, Z., Liu, X., Lei, Z., Li, C., Zhang, F., Wu, Y., et al. (2023). Genetic diversity of polymyxin-resistance mechanisms in clinical isolates of carbapenem-resistant Klebsiella pneumoniae: a multicenter study in China. Microbiol. Spectr. 11:e0523122. doi: 10.1128/spectrum.05231-22
Li, X. Z., and Nikaido, H. (2009). Efflux-mediated drug resistance in bacteria: an update. Drugs 69, 1555–1623. doi: 10.2165/11317030-000000000-00000
Li, X. Z., Plésiat, P., and Nikaido, H. (2015). The challenge of efflux-mediated antibiotic resistance in gram-negative bacteria. Clin. Microbiol. Rev. 28, 337–418. doi: 10.1128/cmr.00117-14
Liao, W., Lin, J., Jia, H., Zhou, C., Zhang, Y., Lin, Y., et al. (2020). Resistance and Heteroresistance to colistin in Escherichia coli isolates from Wenzhou, China. Infect. Drug Resist. 13, 3551–3561. doi: 10.2147/idr.S273784
Lin, Y. T., Huang, Y. W., Liou, R. S., Chang, Y. C., and Yang, T. C. (2014). MacABCsm, an ABC-type tripartite efflux pump of Stenotrophomonas maltophilia involved in drug resistance, oxidative and envelope stress tolerances and biofilm formation. J. Antimicrob. Chemother. 69, 3221–3226. doi: 10.1093/jac/dku317
Lin, M. F., Lin, Y. Y., and Lan, C. Y. (2017). Contribution of EmrAB efflux pumps to colistin resistance in Acinetobacter baumannii. J. Microbiol. 55, 130–136. doi: 10.1007/s12275-017-6408-5
Linares, J. F., López, J. A., Camafeita, E., Albar, J. P., Rojo, F., and Martínez, J. L. (2005). Overexpression of the multidrug efflux pumps MexCD-OprJ and MexEF-OprN is associated with a reduction of type III secretion in Pseudomonas aeruginosa. J. Bacteriol. 187, 1384–1391. doi: 10.1128/jb.187.4.1384-1391.2005
Liu, J., Huang, Z., Ruan, B., Wang, H., Chen, M., Rehman, S., et al. (2020). Quantitative proteomic analysis reveals the mechanisms of polymyxin B toxicity to Escherichia coli. Chemosphere 259:127449. doi: 10.1016/j.chemosphere.2020.127449
Liu, J., Xiao, G., Zhou, W., Yang, J., Wang, Y., Wu, Y., et al. (2021). Various novel colistin resistance mechanisms interact to facilitate adaptation of Aeromonas hydrophila to complex colistin environments. Antimicrob. Agents Chemother. 65:e0007121. doi: 10.1128/aac.00071-21
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/s1473-3099(15)00424-7
Lo, C. C., Liao, W. Y., Chou, M. C., Wu, Y. Y., Yeh, T. H., and Lo, H. R. (2022). Overexpression of resistance-nodulation-division efflux pump genes contributes to multidrug resistance in Aeromonas hydrophila clinical isolates. Microb. Drug Resist. 28, 153–160. doi: 10.1089/mdr.2021.0084
Lomovskaya, O., and Watkins, W. J. (2001). Efflux pumps: their role in antibacterial drug discovery. Curr. Med. Chem. 8, 1699–1711. doi: 10.2174/0929867013371743
Loutet, S. A., and Valvano, M. A. (2011). Extreme antimicrobial peptide and polymyxin B resistance in the genus burkholderia. Front. Microbiol. 2:159. doi: 10.3389/fmicb.2011.00159
Lu, S., and Zgurskaya, H. I. (2013). MacA, a periplasmic membrane fusion protein of the macrolide transporter MacAB-TolC, binds lipopolysaccharide core specifically and with high affinity. J. Bacteriol. 195, 4865–4872. doi: 10.1128/jb.00756-13
Lv, L., Wan, M., Wang, C., Gao, X., Yang, Q., Partridge, S. R., et al. (2020). Emergence of a plasmid-encoded resistance-nodulation-division efflux pump conferring resistance to multiple drugs, including tigecycline, in klebsiella pneumoniae. mBio. 11. doi: 10.1128/mBio.02930-19
Lv, F., Cai, J., He, Q., Wang, W., Luo, Y., Wang, X., et al. (2021). Overexpression of efflux pumps mediate Pan resistance of Klebsiella pneumoniae sequence type 11. Microb. Drug Resist. 27, 1405–1411. doi: 10.1089/mdr.2020.0395
Machado, D., Antunes, J., Simões, A., Perdigão, J., Couto, I., McCusker, M., et al. (2018). Contribution of efflux to colistin heteroresistance in a multidrug resistant Acinetobacter baumannii clinical isolate. J. Med. Microbiol. 67, 740–749. doi: 10.1099/jmm.0.000741
Manjunath, A., Thumu, S. C. R., Kumar, S., and Halami, P. M. (2021). Bacterial heteroresistance: an evolving novel way to combat antibiotics. Biologia (Bratisl.) 76, 3029–3041. doi: 10.1007/s11756-021-00820-y
Martin, R. G., Gillette, W. K., Rhee, S., and Rosner, J. L. (1999). Structural requirements for marbox function in transcriptional activation of mar/sox/rob regulon promoters in Escherichia coli: sequence, orientation and spatial relationship to the core promoter. Mol. Microbiol. 34, 431–441. doi: 10.1046/j.1365-2958.1999.01599.x
Martínez-Servat, S., Yero, D., Huedo, P., Marquez, R., Molina, G., Daura, X., et al. (2018). Heterogeneous colistin-resistance phenotypes coexisting in Stenotrophomonas maltophilia isolates influence colistin susceptibility testing. Front. Microbiol. 9:2871. doi: 10.3389/fmicb.2018.02871
Misra, R., Morrison, K. D., Cho, H. J., and Khuu, T. (2015). Importance of real-time assays to distinguish multidrug efflux pump-inhibiting and outer membrane-destabilizing activities in Escherichia coli. J. Bacteriol. 197, 2479–2488. doi: 10.1128/jb.02456-14
Mlynarcik, P., and Kolar, M. (2019). Molecular mechanisms of polymyxin resistance and detection of mcr genes. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc Czech Repub. 163, 28–38. doi: 10.5507/bp.2018.070
Moffatt, J. H., Harper, M., and Boyce, J. D. (2019). Mechanisms of polymyxin resistance. Adv. Exp. Med. Biol. 1145, 55–71. doi: 10.1007/978-3-030-16373-0_5
Moffatt, J. H., Harper, M., Harrison, P., Hale, J. D., Vinogradov, E., Seemann, T., et al. (2010). Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob. Agents Chemother. 54, 4971–4977. doi: 10.1128/aac.00834-10
Montaña, S., Vilacoba, E., Traglia, G. M., Almuzara, M., Pennini, M., Fernández, A., et al. (2015). Genetic variability of AdeRS two-component system associated with tigecycline resistance in XDR-Acinetobacter baumannii isolates. Curr. Microbiol. 71, 76–82. doi: 10.1007/s00284-015-0829-3
Muller, C., Plésiat, P., and Jeannot, K. (2011). A two-component regulatory system interconnects resistance to polymyxins, aminoglycosides, fluoroquinolones, and β-lactams in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 55, 1211–1221. doi: 10.1128/aac.01252-10
Naha, S., Sands, K., Mukherjee, S., Dutta, S., and Basu, S. (2022). A 12 year experience of colistin resistance in Klebsiella pneumoniae causing neonatal sepsis: two-component systems, efflux pumps, lipopolysaccharide modification and comparative phylogenomics. J. Antimicrob. Chemother. 77, 1586–1591. doi: 10.1093/jac/dkac083
Naha, S., Sands, K., Mukherjee, S., Roy, C., Rameez, M. J., Saha, B., et al. (2020). KPC-2-producing Klebsiella pneumoniae ST147 in a neonatal unit: clonal isolates with differences in colistin susceptibility attributed to AcrAB-TolC pump. Int. J. Antimicrob. Agents 55:105903. doi: 10.1016/j.ijantimicag.2020.105903
Ni, W., Li, Y., Guan, J., Zhao, J., Cui, J., Wang, R., et al. (2016). Effects of efflux pump inhibitors on colistin resistance in multidrug-resistant gram-negative bacteria. Antimicrob. Agents Chemother. 60, 3215–3218. doi: 10.1128/aac.00248-16
Nishino, K., Latifi, T., and Groisman, E. A. (2006). Virulence and drug resistance roles of multidrug efflux systems of Salmonella enterica serovar typhimurium. Mol. Microbiol. 59, 126–141. doi: 10.1111/j.1365-2958.2005.04940.x
Nishino, K., Yamasaki, S., Nakashima, R., Zwama, M., and Hayashi-Nishino, M. (2021). Function and inhibitory mechanisms of multidrug efflux pumps. Front. Microbiol. 12:737288. doi: 10.3389/fmicb.2021.737288
Ogawa, W., Onishi, M., Ni, R., Tsuchiya, T., and Kuroda, T. (2012). Functional study of the novel multidrug efflux pump KexD from Klebsiella pneumoniae. Gene 498, 177–182. doi: 10.1016/j.gene.2012.02.008
Oh, J. T., Cajal, Y., Skowronska, E. M., Belkin, S., Chen, J., Van Dyk, T. K., et al. (2000). Cationic peptide antimicrobials induce selective transcription of micF and osmY in Escherichia coli. Biochim. Biophys. Acta 1463, 43–54. doi: 10.1016/s0005-2736(99)00177-7
Olaitan, A. O., Morand, S., and Rolain, J. M. (2014). Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front. Microbiol. 5:643. doi: 10.3389/fmicb.2014.00643
Osei Sekyere, J., and Amoako, D. G. (2017). Carbonyl cyanide m-Chlorophenylhydrazine (CCCP) reverses resistance to colistin, but not to carbapenems and tigecycline in multidrug-resistant Enterobacteriaceae. Front. Microbiol. 8:228. doi: 10.3389/fmicb.2017.00228
Padilla, E., Llobet, E., Doménech-Sánchez, A., Martínez-Martínez, L., Bengoechea, J. A., and Albertí, S. (2010). Klebsiella pneumoniae AcrAB efflux pump contributes to antimicrobial resistance and virulence. Antimicrob. Agents Chemother. 54, 177–183. doi: 10.1128/aac.00715-09
Pamp, S. J., Gjermansen, M., Johansen, H. K., and Tolker-Nielsen, T. (2008). Tolerance to the antimicrobial peptide colistin in Pseudomonas aeruginosa biofilms is linked to metabolically active cells, and depends on the pmr and mexAB-oprM genes. Mol. Microbiol. 68, 223–240. doi: 10.1111/j.1365-2958.2008.06152.x
Pantel, L., Juarez, P., Serri, M., Boucinha, L., Lessoud, E., Lanois, A., et al. (2023). Missense mutations in the CrrB protein mediate Odilorhabdin derivative resistance in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 65:e00139-21. doi: 10.1128/aac.00139-21
Park, Y. K., and Ko, K. S. (2015). Effect of carbonyl cyanide 3-chlorophenylhydrazone (CCCP) on killing Acinetobacter baumannii by colistin. J. Microbiol. 53, 53–59. doi: 10.1007/s12275-015-4498-5
Paul, D., Mallick, S., Das, S., Saha, S., Ghosh, A. K., and Mandal, S. M. (2020). Colistin induced assortment of antimicrobial resistance in a clinical isolate of Acinetobacter baumannii SD01. Infect. Disord. Drug Targets 20, 501–505. doi: 10.2174/1871526519666190426153258
Pelling, H., Bock, L. J., Nzakizwanayo, J., Wand, M. E., Denham, E. L., MacFarlane, W. M., et al. (2019). De-repression of the smvA efflux system arises in clinical isolates of Proteus mirabilis and reduces susceptibility to chlorhexidine and other biocides. Antimicrob. Agents Chemother. 63:e01535-19. doi: 10.1128/aac.01535-19
Pérez, A., Poza, M., Fernández, A., Fernández Mdel, C., Mallo, S., Merino, M., et al. (2012). Involvement of the AcrAB-TolC efflux pump in the resistance, fitness, and virulence of Enterobacter cloacae. Antimicrob. Agents Chemother. 56, 2084–2090. doi: 10.1128/aac.05509-11
Piddock, L. J. (2006). Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin. Microbiol. Rev. 19, 382–402. doi: 10.1128/cmr.19.2.382-402.2006
Poole, K., Lau, C. H., Gilmour, C., Hao, Y., and Lam, J. S. (2015). Polymyxin susceptibility in Pseudomonas aeruginosa linked to the MexXY-OprM multidrug efflux system. Antimicrob. Agents Chemother. 59, 7276–7289. doi: 10.1128/aac.01785-15
Pu, D., Zhao, J., Lu, B., Zhang, Y., Wu, Y., Li, Z., et al. (2023). Within-host resistance evolution of a fatal ST11 hypervirulent carbapenem-resistant Klebsiella pneumoniae. Int. J. Antimicrob. Agents 61:106747. doi: 10.1016/j.ijantimicag.2023.106747
Puja, H., Bolard, A., Noguès, A., Plésiat, P., and Jeannot, K. (2020). The efflux pump MexXY/OprM contributes to the tolerance and acquired resistance of Pseudomonas aeruginosa to colistin. Antimicrob. Agents Chemother. 64:e02033-19. doi: 10.1128/aac.02033-19
Puzari, M., and Chetia, P. (2017). RND efflux pump mediated antibiotic resistance in gram-negative bacteria Escherichia coli and Pseudomonas aeruginosa: a major issue worldwide. World J. Microbiol. Biotechnol. 33:24. doi: 10.1007/s11274-016-2190-5
Qureshi, Z. A., Hittle, L. E., O'Hara, J. A., Rivera, J. I., Syed, A., Shields, R. K., et al. (2015). Colistin-resistant Acinetobacter baumannii: beyond carbapenem resistance. Clin. Infect. Dis. 60, 1295–1303. doi: 10.1093/cid/civ048
Ramos, P. I., Custódio, M. G., Quispe Saji, G. D., Cardoso, T., da Silva, G. L., Braun, G., et al. (2016). The polymyxin B-induced transcriptomic response of a clinical, multidrug-resistant Klebsiella pneumoniae involves multiple regulatory elements and intracellular targets. BMC Genomics 17:737. doi: 10.1186/s12864-016-3070-y
Richmond, G. E., Evans, L. P., Anderson, M. J., Wand, M. E., Bonney, L. C., Ivens, A., et al. (2016). The Acinetobacter baumannii two-component system AdeRS regulates genes required for multidrug efflux, biofilm formation, and virulence in a strain-specific manner. MBio 7, e00430–e00416. doi: 10.1128/mBio.00430-16
Rosner, J. L., and Martin, R. G. (2009). An excretory function for the Escherichia coli outer membrane pore TolC: upregulation of marA and soxS transcription and rob activity due to metabolites accumulated in tolC mutants. J. Bacteriol. 191, 5283–5292. doi: 10.1128/jb.00507-09
Sato, Y., Unno, Y., Ubagai, T., and Ono, Y. (2018). Sub-minimum inhibitory concentrations of colistin and polymyxin B promote Acinetobacter baumannii biofilm formation. PLoS One 13:e0194556. doi: 10.1371/journal.pone.0194556
Shen, Y., Lv, Z., Yang, L., Liu, D., Ou, Y., Xu, C., et al. (2019). Integrated aquaculture contributes to the transfer of mcr-1 between animals and humans via the aquaculture supply chain. Environ. Int. 130: 104708. doi: 10.1016/j.envint.2019.03.056
Shenkutie, A. M., Zhang, J., Yao, M., Asrat, D., Chow, F. W. N., and Leung, P. H. M. (2022). Effects of sub-minimum inhibitory concentrations of imipenem and colistin on expression of biofilm-specific antibiotic resistance and virulence genes in Acinetobacter baumannii sequence type 1894. Int. J. Mol. Sci. 23:12705. doi: 10.3390/ijms232012705
Shirshikova, T. V., Sierra-Bakhshi, C. G., Kamaletdinova, L. K., Matrosova, L. E., Khabipova, N. N., Evtugyn, V. G., et al. (2021). The ABC-type efflux pump MacAB is involved in protection of Serratia marcescens against aminoglycoside antibiotics, polymyxins, and oxidative stress. Clin. Vaccine Immunol. 6:e00033-21. doi: 10.1128/mSphere.00033-21
Srinivasan, V. B., and Rajamohan, G. (2013). KpnEF, a new member of the Klebsiella pneumoniae cell envelope stress response regulon, is an SMR-type efflux pump involved in broad-spectrum antimicrobial resistance. Antimicrob. Agents Chemother. 57, 4449–4462. doi: 10.1128/aac.02284-12
Srinivasan, V. B., Singh, B. B., Priyadarshi, N., Chauhan, N. K., and Rajamohan, G. (2014). Role of novel multidrug efflux pump involved in drug resistance in Klebsiella pneumoniae. PLoS One 9:e96288. doi: 10.1371/journal.pone.0096288
Sun, S., Gao, H., Liu, Y., Jin, L., Wang, R., Wang, X., et al. (2020). Co-existence of a novel plasmid-mediated efflux pump with colistin resistance gene mcr in one plasmid confers transferable multidrug resistance in Klebsiella pneumoniae. Emerg. Microbes Infect. 9, 1102–1113. doi: 10.1080/22221751.2020.1768805
Sun, L., Rasmussen, P. K., Bai, Y., Chen, X., Cai, T., Wang, J., et al. (2020). Proteomic changes of Klebsiella pneumoniae in response to colistin treatment and crrB mutation-mediated colistin resistance. Antimicrob. Agents Chemother. 64:e02200-19. doi: 10.1128/aac.02200-19
Sundaramoorthy, N. S., Mohan, H. M., Subramaniam, S., Raman, T., Selva Ganesan, S., Sivasubamanian, A., et al. (2019a). Ursolic acid inhibits colistin efflux and curtails colistin resistant Enterobacteriaceae. AMB Express 9:27. doi: 10.1186/s13568-019-0750-4
Sundaramoorthy, N. S., Suresh, P., Selva Ganesan, S., GaneshPrasad, A., and Nagarajan, S. (2019b). Restoring colistin sensitivity in colistin-resistant E. coli: combinatorial use of MarR inhibitor with efflux pump inhibitor. Sci. Rep. 9:19845. doi: 10.1038/s41598-019-56325-x
Tag ElDein, M. A., Yassin, A. S., El-Tayeb, O., and Kashef, M. T. (2021). Chlorhexidine leads to the evolution of antibiotic-resistant Pseudomonas aeruginosa. Eur. J. Clin. Microbiol. Infect. Dis. 40, 2349–2361. doi: 10.1007/s10096-021-04292-5
Telke, A. A., Olaitan, A. O., Morand, S., and Rolain, J. M. (2017). soxRS induces colistin hetero-resistance in Enterobacter asburiae and Enterobacter cloacae by regulating the acrAB-tolC efflux pump. J. Antimicrob. Chemother. 72, 2715–2721. doi: 10.1093/jac/dkx215
Thi Khanh Nhu, N., Riordan, D. W., do Hoang Nhu, T., Thanh, D. P., Thwaites, G., Huong Lan, N. P., et al. (2016). The induction and identification of novel colistin resistance mutations in Acinetobacter baumannii and their implications. Sci. Rep. 6:28291. doi: 10.1038/srep28291
Tzeng, Y. L., Ambrose, K. D., Zughaier, S., Zhou, X., Miller, Y. K., Shafer, W. M., et al. (2005). Cationic antimicrobial peptide resistance in Neisseria meningitidis. J. Bacteriol. 187, 5387–5396. doi: 10.1128/jb.187.15.5387-5396.2005
Wadsworth, C. B., Arnold, B. J., Sater, M. R. A., and Grad, Y. H. (2018). Azithromycin resistance through interspecific Acquisition of an Epistasis-Dependent Efflux Pump Component and Transcriptional Regulator in Neisseria gonorrhoeae. MBio 9. doi: 10.1128/mBio.01419-18
Wand, M. E., Jamshidi, S., Bock, L. J., Rahman, K. M., and Sutton, J. M. (2019). SmvA is an important efflux pump for cationic biocides in Klebsiella pneumoniae and other Enterobacteriaceae. Sci. Rep. 9:1344. doi: 10.1038/s41598-018-37730-0
Wang, X., Liu, Y., Qi, X., Wang, R., Jin, L., Zhao, M., et al. (2017). Molecular epidemiology of colistin-resistant Enterobacteriaceae in inpatient and avian isolates from China: high prevalence of mcr-negative Klebsiella pneumoniae. Int. J. Antimicrob. Agents 50, 536–541. doi: 10.1016/j.ijantimicag.2017.05.009
Warner, D. M., and Levy, S. B. (2010). Different effects of transcriptional regulators MarA, SoxS and rob on susceptibility of Escherichia coli to cationic antimicrobial peptides (CAMPs): rob-dependent CAMP induction of the marRAB operon. Microbiology (Read.) 156, 570–578. doi: 10.1099/mic.0.033415-0
Worthington, R. J., Blackledge, M. S., and Melander, C. (2013). Small-molecule inhibition of bacterial two-component systems to combat antibiotic resistance and virulence. Future Med. Chem. 5, 1265–1284. doi: 10.4155/fmc.13.58
Yang, Q. E., MacLean, C., Papkou, A., Pritchard, M., Powell, L., Thomas, D., et al. (2020). Compensatory mutations modulate the competitiveness an dynamics of plasmid-mediated colistin resistance in Escherichia coli clones. ISME J. 14, 861–865. doi: 10.1038/s41396-019-0578-6
Yang, Y., Yang, Y., Chen, G., Lin, M., Chen, Y., He, R., et al. (2021). Molecular characterization of carbapenem-resistant and virulent plasmids in Klebsiella pneumoniae from patients with bloodstream infections in China. Emerg. Microbes. Infect. 10, 700–709. doi: 10.1080/22221751.2021.1906163
Yilmaz, Ş., Hasdemir, U., Aksu, B., Altınkanat Gelmez, G., and Söyletir, G. (2020). Alterations in AdeS and AdeR regulatory proteins in 1-(1-naphthylmethyl)-piperazine responsive colistin resistance of Acinetobacter baumannii. J. Chemother. 32, 286–293. doi: 10.1080/1120009x.2020.1735118
Yoon, E. J., Balloy, V., Fiette, L., Chignard, M., Courvalin, P., and Grillot-Courvalin, C. (2016). Contribution of the Ade resistance-nodulation-cell division-type efflux pumps to fitness and pathogenesis of Acinetobacter baumannii. MBio 7:e00697-16. doi: 10.1128/mBio.00697-16
Yousefian, N., Ornik-Cha, A., Poussard, S., Decossas, M., Berbon, M., Daury, L., et al. (2021). Structural characterization of the EmrAB-TolC efflux complex from E. coli. Biochim. Biophys. Acta Biomembr. 1863:183488. doi: 10.1016/j.bbamem.2020.183488
Yu, Z., Zhu, Y., Fu, J., Qiu, J., and Yin, J. (2019). Enhanced NADH metabolism involves colistin-induced killing of Bacillus subtilis and Paenibacillus polymyxa. Molecules 24:387. doi: 10.3390/molecules24030387
Zeng, X., Hinenoya, A., Guan, Z., Xu, F., and Lin, J. (2023). Critical role of the RpoE stress response pathway in polymyxin resistance of Escherichia coli. J. Antimicrob. Chemother. 78, 732–746. doi: 10.1093/jac/dkad003
Zgurskaya, H. I., Krishnamoorthy, G., Ntreh, A., and Lu, S. (2011). Mechanism and function of the outer Membrane Channel TolC in multidrug resistance and physiology of enterobacteria. Front. Microbiol. 2:189. doi: 10.3389/fmicb.2011.00189
Zhai, Y. J., Huang, H., Liu, J., Sun, H. R., He, D., Pan, Y. S., et al. (2018). CpxR overexpression increases the susceptibility of acrB and cpxR double-deleted Salmonella enterica serovar typhimurium to colistin. J. Antimicrob. Chemother. 73, 3016–3024. doi: 10.1093/jac/dky320
Zhang, A., Rosner, J. L., and Martin, R. G. (2008). Transcriptional activation by MarA, SoxS and rob of two tolC promoters using one binding site: a complex promoter configuration for tolC in Escherichia coli. Mol. Microbiol. 69, 1450–1455. doi: 10.1111/j.1365-2958.2008.06371.x
Zhang, W., Yuan, Y., Li, S., Deng, B., Zhang, J., and Li, Z. (2021). Comparative transcription analysis of resistant mutants against four different antibiotics in Pseudomonas aeruginosa. Microb. Pathog. 160:105166. doi: 10.1016/j.micpath.2021.105166
Zhang, M. K., Zhang, M. Y., Liu, S. B., Yang, Y. Y., Zhai, Y. J., He, D. D., et al. (2021). Double deletion of cpxR and tolC significantly increases the susceptibility of Salmonella enterica serovar typhimurium to colistin. J. Antimicrob. Chemother. 76, 3168–3174. doi: 10.1093/jac/dkab332
Zhang, H., Zhang, G., Zhang, J., Duan, S., Kang, Y., Yang, Q., et al. (2020). Antimicrobial activity of colistin against contemporary (2015 - 2017) P. aeruginosa and A. baumannii isolates from a Chinese surveillance program. Front. Microbiol. 11:1966. doi: 10.3389/fmicb.2020.01966
Keywords: efflux pump, colistin resistance, lipopolysaccharide, efflux pump inhibitors, collateral susceptibility, heteroresistance
Citation: Ding Y, Hao J, Xiao W, Ye C, Xiao X, Jian C, Tang M, Li G, Liu J and Zeng Z (2023) Role of efflux pumps, their inhibitors, and regulators in colistin resistance. Front. Microbiol. 14:1207441. doi: 10.3389/fmicb.2023.1207441
Edited by:
Mona I. Shaaban, Mansoura Universiy, EgyptReviewed by:
Yuan Liu, Yangzhou University, ChinaCong Shen, Guangzhou University of Chinese Medicine, China
Copyright © 2023 Ding, Hao, Xiao, Ye, Xiao, Jian, Tang, Li, Liu and Zeng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinbo Liu, bGl1amI3MjAzQHN3bXUuZWR1LmNu; Zhangrui Zeng, emVuZ3poYW5ncnVpQHN3bXUuZWR1LmNu
†These authors have contributed equally to this work and share first authorship
 Yinhuan Ding†
Yinhuan Ding† Jingchen Hao
Jingchen Hao Weijia Xiao
Weijia Xiao Xue Xiao
Xue Xiao Jinbo Liu
Jinbo Liu