- 1The Key Laboratory of Animal Disease and Human Health of Sichuan Province, College of Veterinary Medicine, Sichuan Agricultural University, Chengdu, Sichuan, China
- 2Sichuan Wolong National Natural Reserve Administration, Wenchuan, Sichuan, China
Corynebacterium pseudotuberculosis is a zoonotic pathogen that causes lymphadenitis in humans, livestock, and wildlife. In this study, C. pseudotuberculosis biovar equi strains were isolated from three alpacas. Antibiotic susceptibility tests and pathogenicity tests were also conducted. Moreover, one strain was sequenced using DNBSEQ and Oxford Nanopore technology. The three strains exhibited resistance to aztreonam, fosfomycin, and nitrofurantoin. The median lethal doses (LD50) of strains G1, S2 and BA3 in experimentally infected mice was 1.66 × 105 CFU, 3.78 × 105 CFU and 3.78 × 105 CFU, respectively. The sequencing of strain G1 resulted in the assembly of a chromosomal scaffold comprising 2,379,166 bp with a G + C content of 52.06%. Genome analysis of strain G1 revealed the presence of 48 virulence genes and 5 antibiotic resistance genes (ARGs). Comparative genomic analysis demonstrates a high degree of genetic similarity among C. pseudotuberculosis strains, in contrast to other Corynebacterium species, with a clear delineation between strains belonging to the two biovars (ovis and equi). The data of the present study contribute to a better understanding of the properties of C. pseudotuberculosis biovar equi strains and the potential risk they pose to alpacas and other livestock, as well as the necessity of ongoing surveillance and monitoring of infectious diseases in animals.
1. Introduction
Corynebacterium pseudotuberculosis is a facultative intracellular pathogen, which belongs to the Corynebacterium, Mycobacterium, Nocardia, and Rhodococcus (CMNR) group of the Actinomycetota (Mills et al., 1997; Dorella et al., 2006). It is a non-motile, non-capsulated, and non-sporulating Gram-positive bacterium, with fimbriae (Hard, 1969). This zoonotic pathogen is the main cause of caseous lymphadenitis (CLA) and can infect humans (Peel et al., 1997), livestock (Oliveira et al., 2014), and wildlife (Oliveira et al., 2016) worldwide through respiratory, digestive and wound transmission. C. pseudotuberculosis causes (Kinne, 2016) serious economic losses, including reduced reproductive efficiency and losses in wool, meat, and milk production, as well as increased expenses for the drugs and labor required for treatment (de Sá Guimarães et al., 2011). It is classified by host preference and nitrate reduction into two biovars: ovis (serotype I, nitrate-negative) causes lymphadenitis in small ruminants, mainly sheep and goats; equi (serotype II, nitrate-positive) results in ulcerative lymphangitis in equines (Bernardes et al., 2021) and oedematous skin disease (OSD) in buffaloes (Barakat et al., 1984; Moussa et al., 2016). However, C. pseudotuberculosis exhibits phenotypic intra-species variation, with certain strains isolated from sheep exhibiting nitrate reduction capability, and some equine isolates displaying nitrate negativity (Costa et al., 1998; Connor et al., 2000; Retamal et al., 2011). Considering the potential discrepancies due to mutations, genotypic tests have proven to be a more reliable and sensitive diagnostic tool compared to phenotypic tests (Asfour and Darwish, 2014; Almeida et al., 2017) The narG (nitrate reductase gene) gene was also included in PCR assays along with the 16S rRNA, pld (Phospholipase D) and rpoB (RNA polymerase beta subunit region) genes to improve the accuracy of multiplex PCR at the biovar level (Almeida et al., 2017).
CLA is one of the most serious infectious diseases in alpacas and has been reported in Germany (Sting et al., 2022), USA (Anderson et al., 2004), and Peru (Braga et al., 2006). In China, case of C. pseudotuberculosis infection have been documented in sheep (Liu, 2021), goats (Wang R. et al., 2021), Bactrian camels (Liu et al., 1986; Guo et al., 1988), buffaloes (Zhang et al., 1989), chickens (He et al., 1991), deer (Zhu and Lei, 2001), and humans (Du, 1997). Among these, the strains isolated from goats were identified as biovar ovis through phenotypic and genotypic tests, while strains isolated from other hosts were characterized only phenotypically, without fully determining their biovars. Strains isolated from chickens and deer were nitrate-negative, whereas strains isolated from humans were nitrate-positive. Both nitrate-positive and nitrate-negative strains have been reported in Bactrian camels. However, to the best of our knowledge, there have been no documented cases of CLA in alpacas in China.
The pathogenic mechanism of C. pseudotuberculosis is closely associated with its virulence factors, including several virulence genes, such as pld, iron uptake and regulation (fagA, fagB, fagC, fagD), mycolic acid, σ factor, and oligopeptide permease (oppA, oppB, oppC, oppD, oppF) (Billington et al., 2002). PLD is a highly potent exotoxin secreted by C. pseudotuberculosis, known for its necrotic and hemolytic activities (Sutherland et al., 1989). It is widely regarded as the most critical virulence factor produced by the bacterium, and the pld gene has been found to be widely detected in all C. pseudotuberculosis strains isolated from mammals (Songer, 1997; Aquino De Sá et al., 2013). Selective extracytoplasmic function (ECF) sigma factor family proteins, such as sigE, mainly regulate cell surface stress and control the expression of virulence-associated genes in various pathogenic bacteria (Kazmierczak et al., 2005). The Opp transporters are not only essential for nutrient accumulation but also contribute to the regulation of diverse intercellular signaling processes. One notable aspect of their regulatory function is the control they exert over the expression of virulence genes in pathogenic bacteria (Samen et al., 2004).
With the continuous advancement of sequencing technology, the number of available whole genome sequences for C. pseudotuberculosis has been growing. At present, a total of 125 C. pseudotuberculosis strains from 19 countries and regions have been fully sequenced (as of April 1, 2023). This abundance of genomic data provides a valuable resource for conducting comprehensive comparative genomics studies. Of these, only one isolate was yielded from camel (biovar ovis) and one isolate from llama (biovar equi), while the majority of C. pseudotuberculosis isolates were from goats (28%), sheep (24%), and horses (22%). Notably, the whole genome sequencing of C. pseudotuberculosis strain isolated from alpacas has not yet been reported (Data source: https://www.ncbi.nlm.nih.gov/genome/. The whole genome sequencing statistics of C. pseudotuberculosis are presented in Supplementary Table S1 and Supplementary Figure S1). All strains that infect sheep and goats are biovar ovis (strain PA02 was mistakenly labeled as biovar equi and is actually lacking the narG gene, as shown in Supplementary Figure S2). In contrast, all strains that infect horses and buffalo are biovar equi, while strains that infect cattle include both biovars ovis and equi. Seven C. pseudotuberculosis biovar equi strains that were isolated from equine were subjected to pan-genomic analysis, which revealed one of the smallest core genomes ever recorded and considerable genetic variability. There were no discernible genetic distinctions among the strains responsible for the three distinct types of infection, including those causing external abscesses, infections with abscess formation in the internal organs, and ulcerative lymphangitis (Barauna et al., 2017). The clustered regularly interspaced short palindromic repeats (CRISPR)-Cas (CRISPR-associated proteins) cluster is only seen in the biovar equi strains, while the methylation type III cluster is only found in biovar ovis strains (Parise et al., 2018).
The main objective of this study was to identify the causative bacteria responsible for the disease in three alpacas and investigate their biological characteristics, with a focus on pathogenicity assessed through mouse infection tests. Furthermore, it entailed the whole genome sequencing of the C. pseudotuberculosis biovar equi strain isolated from an alpaca, and this genome was compared to other C. pseudotuberculosis strains of both biovars ovis and equi.
2. Materials and methods
2.1. Sample collection
In November 2021, two diseased alpacas (G1 and S2) and a dead alpaca (BA3) from a local alpaca farm were brought to the Animal Hospital of Sichuan Agricultural University (Chengdu, Sichuan, China) for consultation. Tests for Brucella antibodies were negative for alpacas G1 and S2. Despite receiving careful medical attention, alpaca G1’s condition deteriorated, and it died on the third day of treatment. Necropsies were performed on the dead alpacas, and extremity pus from one diseased alpaca (S2) and liver abscesses from two dead alpacas (G1 and BA3) were aseptically collected.
2.2. Bacteriological examination
Samples were subjected to routine bacteriological examination using Luria-Bertani (LB) broth (Hopebio, Qingdao, China), including cultivation under both aerobic and anaerobic conditions. Three mono-colony isolates from one sick alpaca and two dead alpacas were marked as S2, G1, and BA3, respectively. The hemolytic ability of the strains was determined by inoculating them onto blood agar plates (Solarbio, Beijing, China) containing 5% sheep blood. Morphological and biochemical properties of the bacteria were characterized through gram staining and standard biochemical tests, according to the manufacturer’s instructions (Hopebio). The resulting isolates were stored at −80°C in 25% glycerol for subsequent experiments.
2.3. Molecular identification
The genomic DNA from the isolates was extracted using the Bacterial DNA Kit (Tiangen, Beijing, China) and stored at −80°C. For bacterial species identification, the partial 16S rRNA gene was amplified using the primers 27-F (5’-GATGGTCATAGGGATGAAGAGCTT-3′) and 1492-R (5’-AGGGATGAAGAGCTTCGGCTCTG-3′) (Fredriksson et al., 2013). Additionally, the narG gene was amplified using the primers F (5′-ACCCGTACTTGCACTCTTTC-3′) and R (5′-AGTCAGTACTTCCGCAGGTC′) (Almeida et al., 2017). Each PCR reaction (25 μL) contained 12.5 μL 2 × Pro Taq Master Mix (Accurate Biology, Hunan, China), 8.5 μL RNase-free H2O (Sangon Biotech, Shanghai, China), 1 μL forward and reverse primers (Sangon Biotech), and 2 μL template DNA. In the PCR test for amplifying the partial 16S rRNA gene, RNase-free water was used instead of template DNA as a negative control. For other PCR tests, Staphylococcus aureus ATCC 25923 was used as a negative control strain. The reaction conditions included predenaturation at 94°C for 5 min, followed by 35 cycles of 94°C for 1 min, annealing temperature for 50 s, and 72°C for 1 min, with a final extension at 72°C for 10 min. The PCR products were validated by 1% agarose gel electrophoresis with TS-GelRed (Tsingke Biotechnology, Beijing, China) and sent to Tsingke Biotechnology Co., Ltd. for Sanger sequencing. A homology search was performed in the GenBank database using the BLAST tool,1 and Clustal X2.02 for sequence alignment.
2.4. Antibiotic susceptibility testing
The Kirby-Bauer disk diffusion method (Oxoid, Basingstoke, UK) was utilized to test the antimicrobial susceptibility of the isolates. Representative drugs from 12 major classes of antibiotics, including β-lactams, carbapenems, polyphosphates, glycopeptides, aminoglycosides, tetracyclines, macrolides, amide alcohols, quinolones, sulfonamides, nitrofurans, and rifampicin, were selected. The bacterial solution cultured to 0.5 McFarland turbidity was evenly spread on Muller-Hinton (MH) solid medium (Hopebio) using a spreader stick. Disks were then placed on the medium and the culture was incubated at 37°C for 24–48 h. S. aureus ATCC 25923 was employed as the quality control strain in our study. The zone diameters for each drug were interpreted based on the Clinical and Laboratory Standards Institute (CLSI, 2019) and recorded as sensitive (S), intermediate (I), or resistant (R).
2.5. Pathogenicity test
The animal study was reviewed and approved by the Sichuan Agricultural University Animal Ethical and Welfare Committee. Female Specific Pathogen Free (SPF) Kunming mice were obtained from Chengdu Dossy Experimental Animals CO., LTD. (Chengdu, China). The mice were housed in clean cages at 23 ± 2°C and a relative humidity of 50 ± 10%. The mice were 6 weeks old at the start of the study.
Preliminary experiments were conducted to determine the infection range of three C. pseudotuberculosis strains (6 × 104 to 107 CFU/mL) (CFU: Colony Forming Unit). Healthy mice (n = 78) were randomly divided into 13 groups of 6 mice each. Groups 1–4, 5–8, and 9–12 were intraperitoneally injected with 0.2 mL of G1, S2, and BA3 bacterial solutions (with normal saline and diluted to 6 × 104 to 107 CFU/mL), respectively. Group 13 served as the control group and received 0.2 mL of saline intraperitoneally.
Clinical signs and mortality were recorded every 2 h within 12 h post-injection and every day for 7 consecutive days. The median lethal dose (LD50) was calculated. Histopathological analysis was performed on the initial three mice that died following infection with strains G1, S2, and BA3, respectively. The liver, heart, spleen, lung, kidney, and intestines of dead mice were fixed in 10% formaldehyde for histopathological examination. On day 14, the organs of the remaining mice were collected and weighed to compare changes in organ index after infection with different concentrations of bacterial solutions.
2.6. Whole genome sequencing
2.6.1. Genome sequencing and assembly
The genome of a C. pseudotuberculosis strain was sequenced using a combination of the DNBSEQ platform (BGI, Shenzhen, China) and the Oxford Nanopore technology (ONT, Oxford, UK) at the Beijing Genomics Institute. The program Canu was utilized for self-correction. Draft genomic unitigs were assembled using Canu, a high-quality assembler that employs a corrected circular consensus sequence subread set. To improve the accuracy of the genome sequences, GATK3 was used to make single-base corrections.
2.6.2. Gene prediction and annotation
The functional annotation of the C. pseudotuberculosis strain genome for open reading frame (ORF) prediction was performed using glimmer34 with Hidden Markov models. tRNA, rRNA and sRNA recognition made use of tRNAscan-SE,5 RNAmmer, and the Rfam database. The tandem repeat annotation was obtained using the Tandem Repeat Finder,6 and the minisatellite DNA and microsatellite DNA were selected based on the number and length of repeat units. The Genomic Island Suite of Tools (GIST) was used for genomic island analysis7 with IslandPath DIOMB, SIGI-HMM, IslandPicker method. Prophage regions were predicted using the PHAge Search Tool (PHAST) web server8 and CRISPR-Cas identification using the CRISPRCasFinder.9
The best hit was extracted using the BLAST alignment tool for function annotation. Virulence factors and antibiotic resistance genes (ARGs) were identified based on the core datasets in the VFDB (Virulence Factors of Pathogenic Bacteria) and ARDB (Antibiotic Resistance Genes Database) databases. Additionally, general function annotation was performed using three databases: COG (Clusters of Orthologous Groups), GO (Gene Ontology), and KEGG (Kyoto Encyclopedia of Genes and Genomes).
2.6.3. Comparative genomics
(i) Structural Variation (Synteny). The genome sequence of the C. pseudotuberculosis strain was compared with that of the reference bacteria. (ii) FastANI (version 1.32) software was used to conduct the Average Nucleotide Identity (ANI) analysis. (iii) CD-HIT clustering analysis was conducted on the protein gene sets of the C. pseudotuberculosis strain investigated in this study and other Corynebacterium strains to obtain the core/specific/dispensable gene set. (iv) Gene families were constructed by pairwise aligning the protein sequences of multiple target genomes using the BLAST, eliminating redundancy with Solar (BGI), and clustering the alignment results using the Hcluster_sg software. (v) The phylogenetic tree was constructed by the TreeBeST10 using the method of neighbor-joining (NJ).
2.7. Statistical analysis
The in vitro experiment was performed in triplicate and the data were analyzed using SPSS software (IBM, Armonk, NY, USA). Statistical significance was determined by one-way ANOVA, and differences were considered significant at p < 0.05 and extremely significant at p < 0.01. The LD50 values were calculated by Probit analysis (Xiong et al., 2013). The difference in LD50 among the three C. pseudotuberculosis strains were examined by the Kruskal-Wallis test. All graphical presentations were generated by Adobe Illustrator 2020 (Adobe Inc., San Jose, CA, USA) and GraphPad Prism 9.0 (GraphPad Software, San Jose, CA, USA).
3. Results
3.1. Postmortem examination
All three alpacas exhibited purulent infections in the skin and subcutaneous tissue, with some abscesses that developed in “beaded” pattern. White, yellowish pus or caseous pus masses were observed after incision of the pustules. Additionally, skin ulceration and necrosis were observed (Supplementary Figure S3). Upon necropsy of dead alpaca (G1, BA3), multiple abscesses were found in subcutaneous tissue, anterior shoulder lymph nodes, and inguinal lymph nodes. Alpaca G1 was found to have abscesses in the liver and mesenteric lymph nodes, as well as hemorrhagic spots in the lung (Supplementary Figure S4).
3.2. Bacteriological examination
Dry, friable, and opaque porcelain white colonies were grown on LB solid medium under both anaerobic and aerobic conditions (Figure 1A). A narrow and transparent hemolytic ring formed on the blood plate, which is β-hemolysis (Figure 1B). Microscopic examination of Gram-stained samples showed a large number of blue-purple globular or rod-shaped Gram-positive bacteria that were arranged singly and sporadically (Figure 1C).
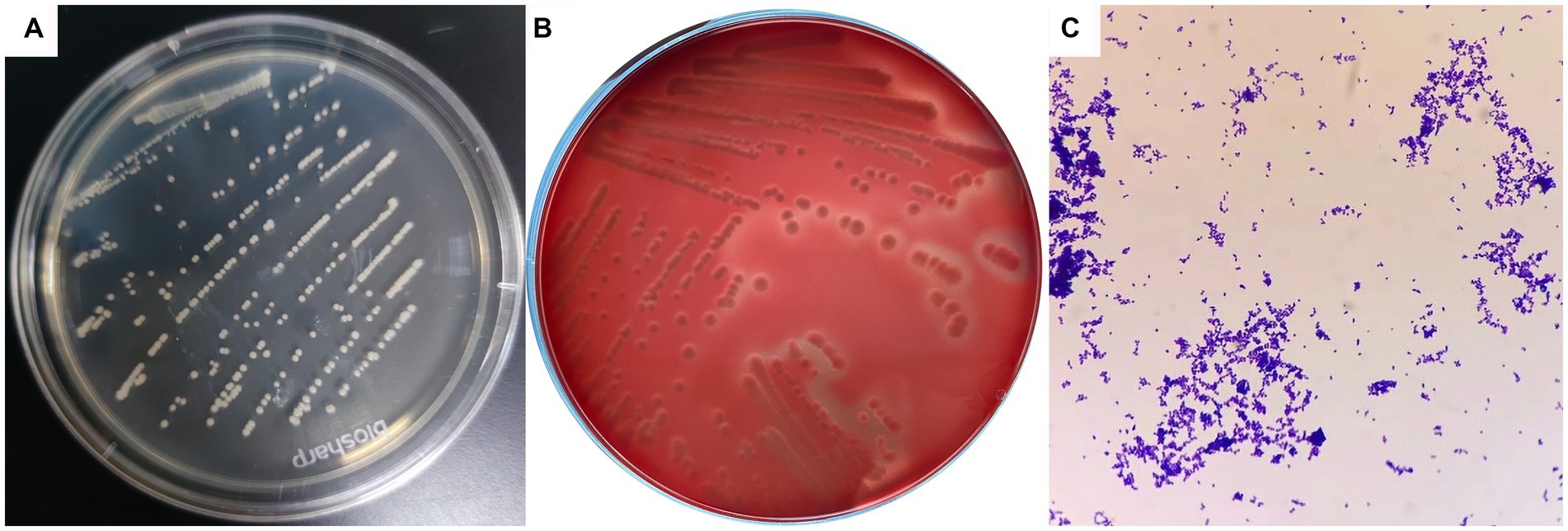
Figure 1. Colony morphology and bacterial morphology observation of C. pseudotuberculosis. (A) Colony morphology (LB solid medium). (B) Colony morphology (5% sheep blood). (C) Bacterial morphology (1000 ×).
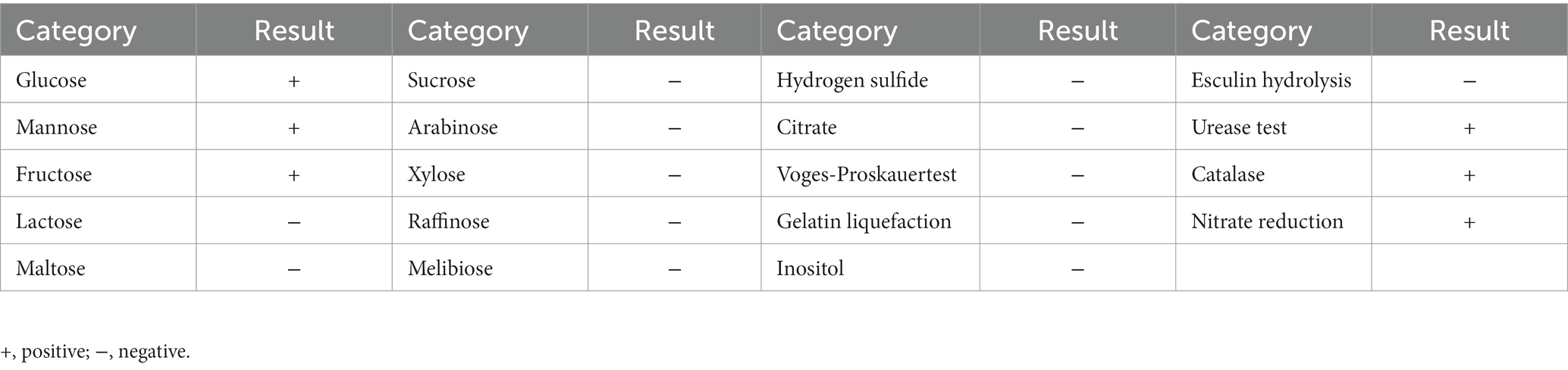
Biochemical identification was consistent for the three strains (Table 1), and was in accordance with the biochemical properties of C. pseudotuberculosis previously summarized (Dorella et al., 2006). These three strains were further identified as C. pseudotuberculosis biovar equi by nitrate reduction experiments (+).
3.3. Molecular identification
Identification analysis of the partial 16S rRNA gene confirmed that all three isolates were C. pseudotuberculosis (accession numbers OQ980206, OP903368 and OP903369). These sequences exhibited 100% similarity to the sequences MT649220 (dromedary, equi, India) and LC549531 (goat, ovis, Malaysia), suggesting a high degree of similarity between the three strains.
The narG gene was successfully amplified and identified in all three isolates as C. pseudotuberculosis biovar equi (accession numbers OQ817706, OQ817707 and OQ817708). These sequences displayed 100% identity to both the sequences CP003652 (strain Cp162, equi, camel) and CP017384 (strain I37, equi, cattle).
3.4. Antibiotic susceptibility testing
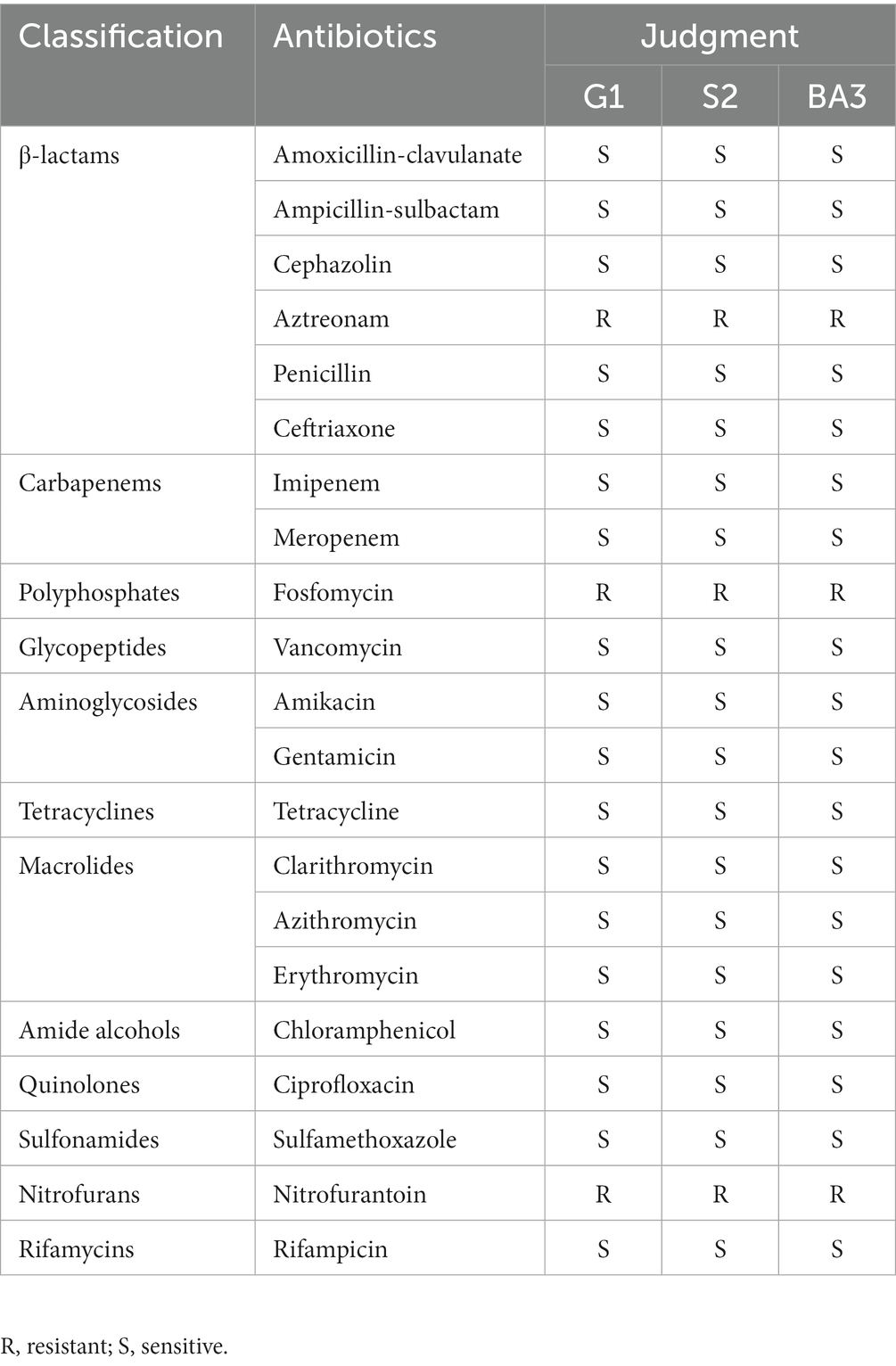
The antibiotic susceptibility test showed that the three C. pseudotuberculosis strains were sensitive to all antibiotics tested except aztreonam, fosfomycin and nitrofurantoin. Specific results are shown in Table 2.
3.5. Pathogenicity test
After artificial infection with C. pseudotuberculosis for 5–6 h, the mice began to develop abnormal symptoms, including disheveled coat, depression, accelerated respiration, slow reaction, slow movement, and inappetence. Survival curves were plotted according to the time of death in each group (Figure 2). The LD50 values for mice infected with strains G1, S2 and BA3 were 1.66 × 105 CFU, 3.78 × 105 CFU and 3.78 × 105 CFU, respectively (p > 0.05). The LD50 of strain G1 was less than that of the strains S2 and BA3, but the difference is not significant (p > 0.05).
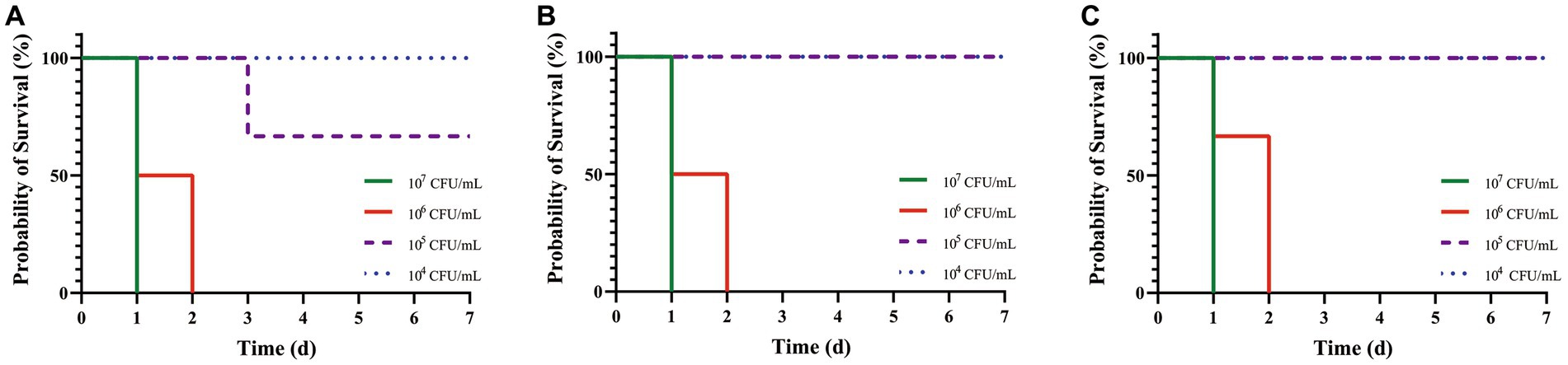
Figure 2. Survival curves of mice challenged with C. pseudotuberculosis strains at different concentrations. (A) G1 strain. (B) S2 strain. (C) BA3 strain.
Necropsy of dead mice (107 CFU/mL group) showed dilatation of the gastrointestinal tract with significant fluid accumulation, hemorrhagic spots in the liver, and a congested and enlarged spleen. The dead mice did not show typical CLA or suppuration symptoms, which may be explained by acute death of mice post-injection and the fact that the infected organs lacked the time to develop the organ lesions. Sterile collection was performed on the liver, spleen, and kidneys of the dead mice, and after streaking and incubation, C. pseudotuberculosis was isolated from all organs. The pathological sections revealed that there was pronounced central vein congestion (red arrow) and diffuse hepatic cell lipid degeneration (black arrow) in the liver tissue (Figure 3A). Heart tissue (Figure 3B) showed no apparent abnormalities. Spleen tissue (Figure 3C) showed irregularities in the white pulp, along with numerous small vacuoles (black arrow) and a large area of extramedullary hematopoietic foci in the red pulp (red arrow), with a significant amount of neutrophil infiltration (blue arrow). Lung tissue (Figure 3D) exhibited extensive alveolar interstitial congestion and expansion (red arrow), with only a few lymphocyte infiltrates observed around blood vessels (blue arrow). Kidney tissue (Figure 3E) displayed a small number of epithelial cells that underwent cellular hydropic degeneration and cytoplasmic relaxation (black arrow), and a large portion of renal tubular interstitial congestion and expansion (red arrow). Intestine tissue (Figure 3F) showed a considerable amount of villous epithelial cells that experienced necrosis and shedding (yellow arrow), and there was also necrosis or atrophy of the basal intestinal glands in the lamina propria, resulting in a reduced structure (orange arrow), together with a small amount of lymphocyte infiltration (blue arrow).
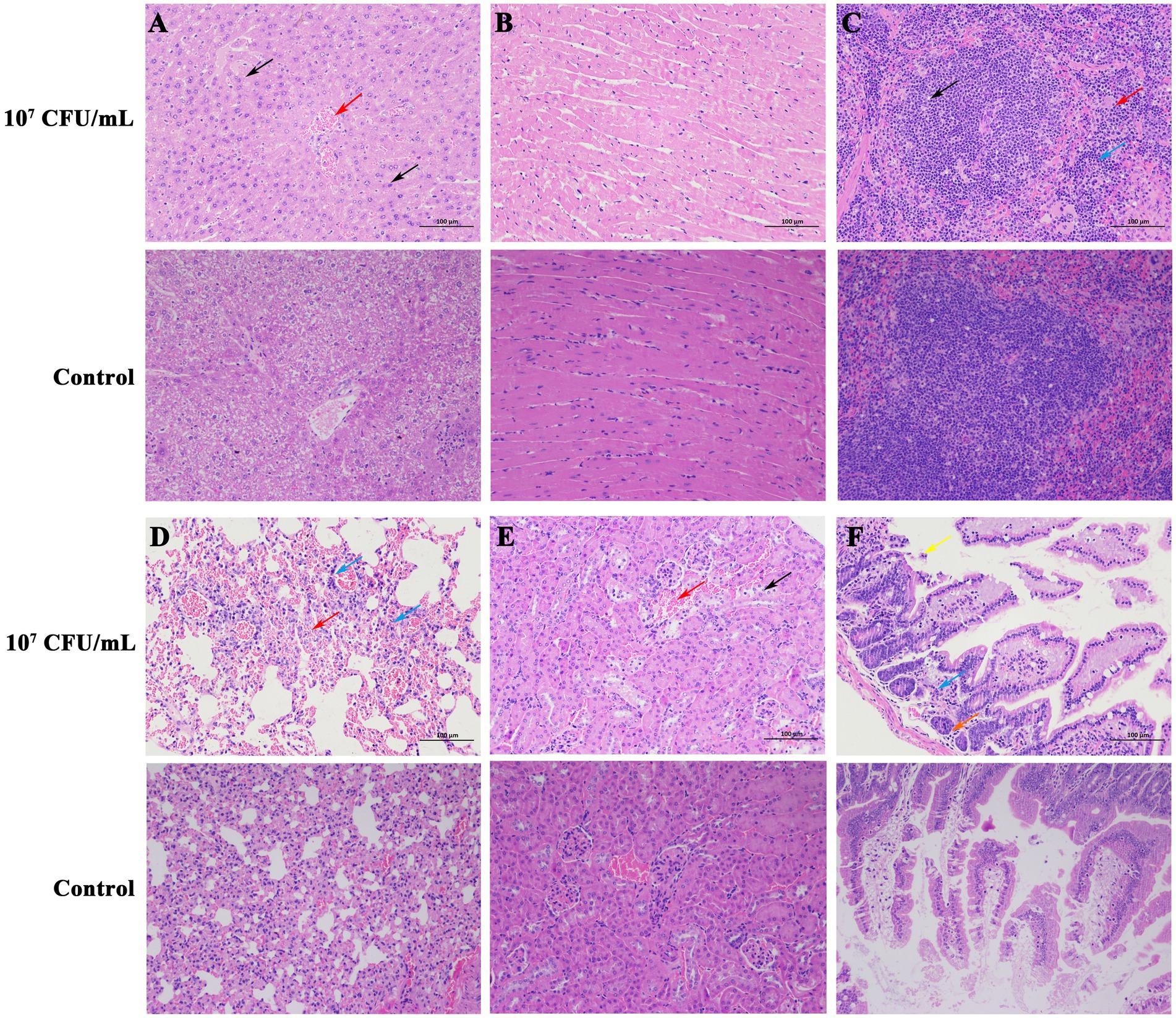
Figure 3. Histopathologic changes in mice killed by C. pseudotuberculosis (200×). (A) Liver. (B) Heart. (C) Spleen. (D) Lung. (E) Kidney. (F) Intestine.
14 days after intraperitoneal injection of C. pseudotuberculosis, enlarged spleens of the mice were observed at necropsy. Changes in organ indices were consistent with necropsy findings. After intraperitoneal injection of strain G1, the spleen organ index of mice in 105 CFU/mL group was significantly higher than in the control group and 104 CFU/mL group (p < 0.01), and 104 CFU/mL group was significantly higher than the control group (p < 0.05). After intraperitoneal injection of S2 or BA3 strains, the spleen organ index of mice in 105 CFU/mL group was significantly higher than the control group (p < 0.01) and 104 CFU/mL group (p < 0.05), while there was no significant difference between 104 CFU/mL group and the control group (p > 0.05). There was no significant difference in the organ index of other organs (liver, heart, lung and kidney) (p > 0.05) (Figure 4). The spleen is an important immune organ in the host, and when exposed to external infections, immune cell infiltration, hypersplenism and portal hypertension can cause splenomegaly (Wang X. et al., 2021).
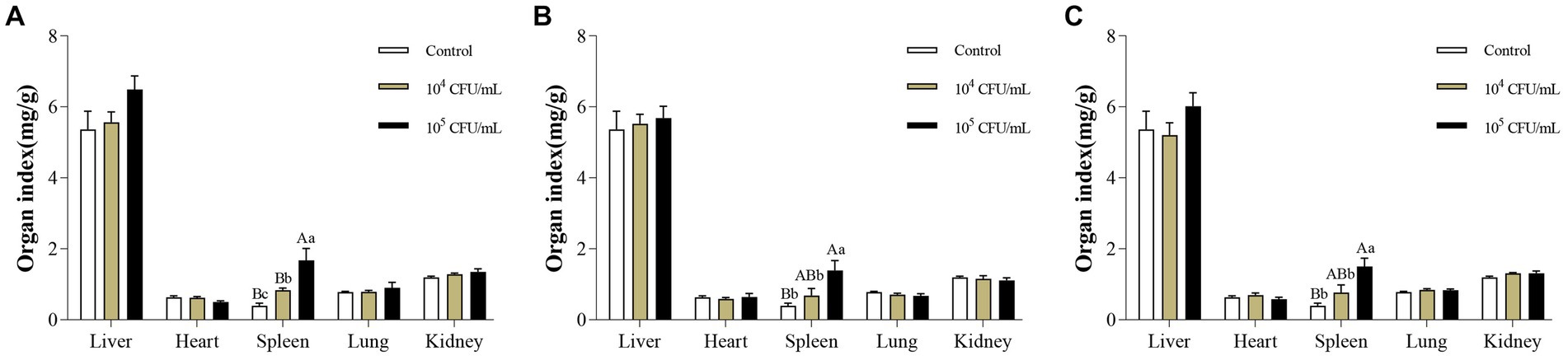
Figure 4. Changes of organ index in mice infected with different concentrations of C. pseudotuberculosis. (A) G1 strain. (B) S2 strain. (C) BA3 strain. Significant differences were observed among the different letter marks. Uppercase letters indicate p < 0.01, while lowercase letters indicate p < 0.05. The absence of any letter mark, or the presence of the same letter mark, indicates no significant difference (p > 0.05).
3.6. Whole genome sequencing
Based on the consistent biochemical properties, biovars identification, and antibiotic sensitivity test results of these three C. pseudotuberculosis strains, and no significant difference in LD50 among them, only one strain was specifically chosen for whole genome sequencing and comparative genomics analysis. The strain G1 was chosen due to its lower LD50 compared to the other two strains.
3.6.1. Genome sequencing and assembly of Corynebacterium pseudotuberculosis strain G1
Initially, a total of 1,314 Mb of raw data were produced from the DNBSEQ sequencing platform, from 8,764,210 reads (Supplementary Table S2). After single-molecule nanopore DNA sequencing and data clean-up, 2,331,063,777 bp of sequence data were obtained, with reads mean length of 6,908 bp (Supplementary Table S3). The final assembly was composed of a chromosomal scaffold of 2,379,166 bp (52.06% G + C) (Supplementary Table S4; Supplementary Figure S5) without any plasmid. The genome has been uploaded to the NCBI (National Center for Biotechnology Information) database with accession number CP121342.
3.6.2. Gene prediction and annotation of Corynebacterium pseudotuberculosis strain G1
A total of 49 tRNA-encoding genes and 4 rRNA operons (four 5S rRNAs, four 16S rRNAs and four 23S rRNAs) were identified in the chromosome (Supplementary Table S5). The results of tandem repeat prediction are presented in Supplementary Table S6. Phages were not predicted in this study. Three different CRISPR clusters were detected in the chromosome of C. pseudotuberculosis strain G1, and details are shown in Supplementary Table S7.
After comparing the ORFs to the ARDB database, we annotated that the strain G1 contains five ARGs, including rpoB, rbpA, EF-Tu mutant, thyA, and murA (identity >60%), and is predicted to be resistant to rifamycin, elfamycin, salicylic acid, and fosfomycin. In addition, we compared the strain G1 with the VFDB database and annotated 43 genes related to virulence factors, including adherence (such as spaC and spaI), exotoxin (pld), immune modulation (rfbB and ndk), nutritional/metabolic factor (such as fagA-D and ciuA-E), and stress survival (such as sigE, sigH, and sigA/rpoV). Specific virulence genes are shown in Supplementary Table S8 (identity >60%). Furthermore, we aligned the strain G1 with the I37 strain (accession number: CP017384) to annotate the virulence factor-related genes oppA, oppB, oppC, oppD, and oppF (identity >98%).
After comparing the predicted gene set with the COG database, the homologous gene annotation was completed, and the COG functional clustering was obtained. The genome of strain G1 has been annotated with 1538 genes that have known functions and 55 genes that have unknown functions. The highest number of genes is related to metabolism (n = 896), followed by cellular and information. There are 98 genes predicted for general function prediction only (Supplementary Figure S6A). Through the annotation in the GO database, we can determine the potential function of a gene based on its annotation in various categories. Supplementary Figure S6B displays the statistical results of the annotation findings for the strain G1 in the three categories (cellular component, molecular function, and biological process) of the GO database. The KEGG database divides biological pathways into eight categories, with each category further subdivided and labeled with the relevant genes. This allows for easy identification of all annotated genes associated with a specific function. Supplementary Figure S6C shows the histogram resulting from the KEGG secondary classification statistics of the strain G1.
3.6.3. Comparative genomics
As shown in Figure 5, strain G1 clusters with the other 12 strains of C. pseudotuberculosis, with intraspecies ANI values greater than 98.6. The highest ANI values (> 99.94) were observed between strain G1 and strain Cp162 (equi, camel), followed by >99.93 with strain I37 (equi, cattle). ANI values were greater than 98.9 between strains of the same biovars of C. pseudotuberculosis, and ranged from 98.6 to 98.9 between strains of different biovars of C. pseudotuberculosis. The interspecies ANI values between C. pseudotuberculosis and C. ulcerans ranged from 84.8 to 85.0. Interspecies ANI values were less than 80 for strains of C. pseudotuberculosis versus C. diphtheriae,C. glutamicum and C. pseudopelargi.
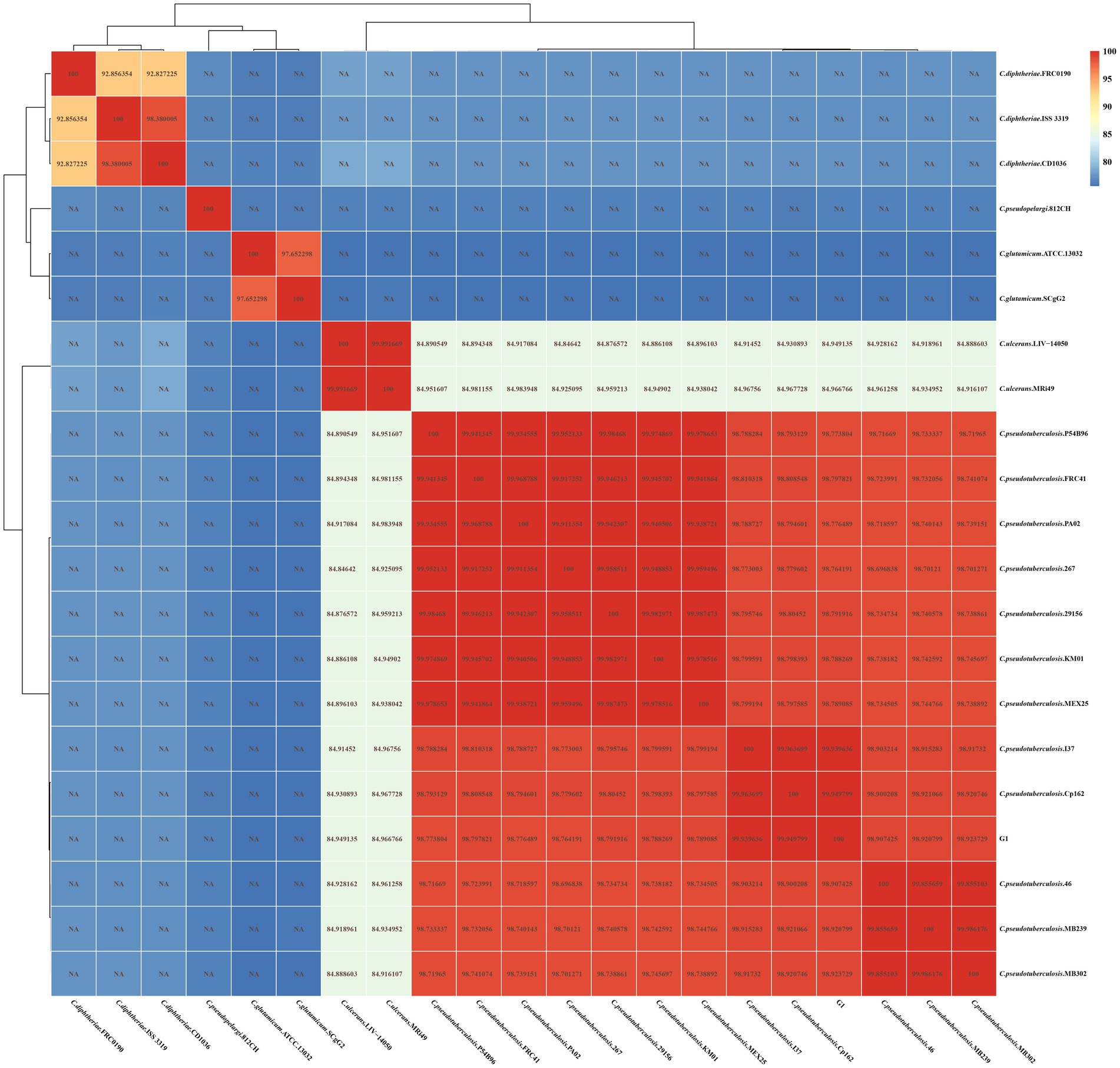
Figure 5. ANI heat map of 21 Corynebacterium strains. The horizontal and vertical axes represent the strain names. Each value in the heat map represents the ANI value.
Gene family statistics for the 21 strains of Corynebacterium are presented in Supplementary Table S9 and Supplementary Figure S8. The analysis revealed a total of 1,080 homologous gene families, with 762 Single Copy Orthologs identified among all 20 strains of Corynebacterium. Notably, the strain G1 showed 8 unique paralogs and 2 unique families.
The protein coding DNA sequences (CDSs) of strain G1, 12 strains of C. pseudotuberculosis and 8 strains of other Corynebacterium were clustered, and the number of pan genes was 6224, the number of core genes was 931, and the number of dispensable genes was 3277 (Figure 6A). Analyses of the core gene set and pan gene set of different numbers of strain combinations, resulted in the core gene and pan gene dilution curves (Figure 6B). According to the distribution of dispensable genes in different samples, a heat map was drawn to show the clustering among samples (Figure 6C). Different species of Corynebacterium were clustered separately, but different biovars of C. pseudotuberculosis were not clustered.
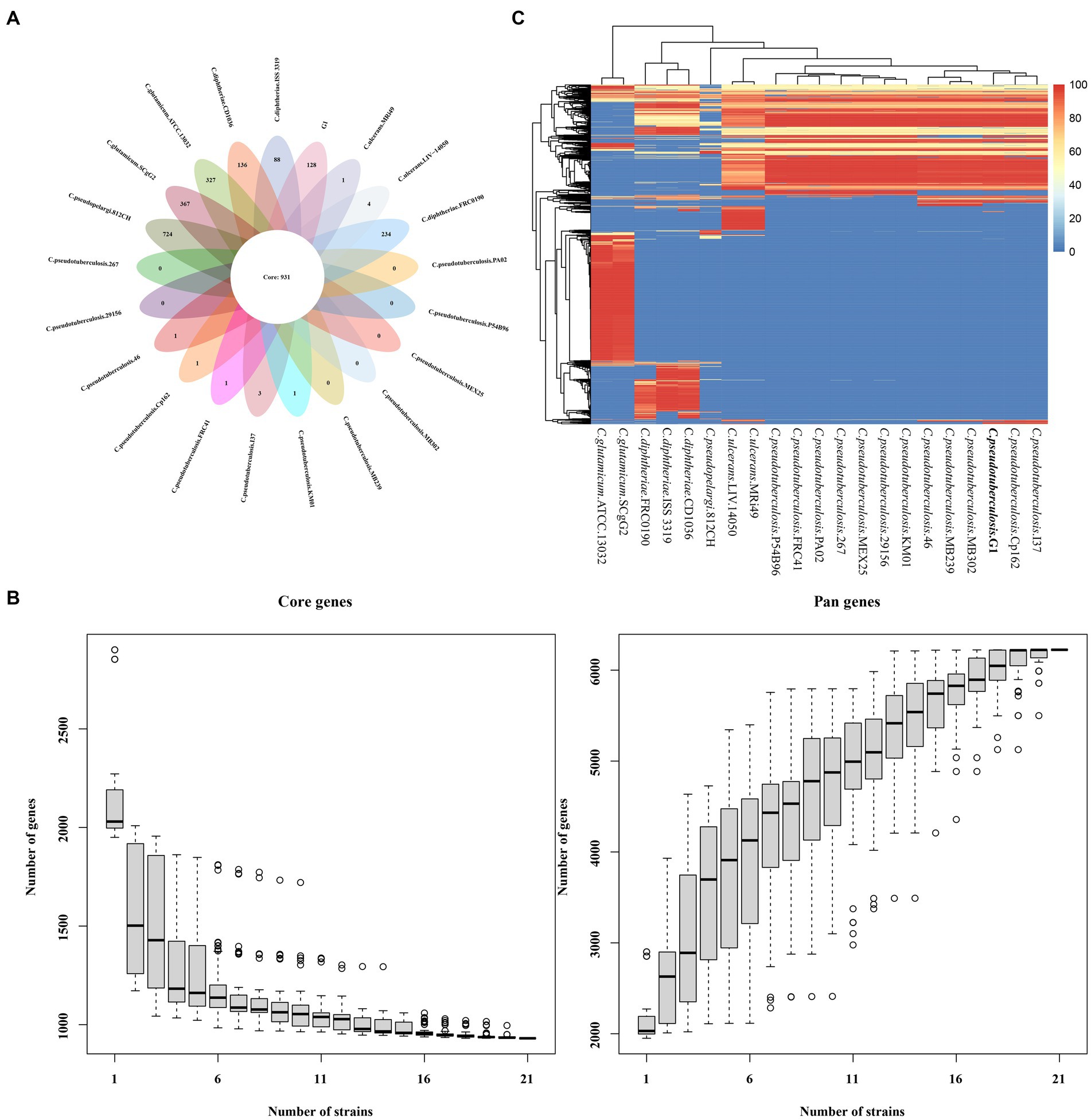
Figure 6. The core and pan-genome of 21 Corynebacterium strains. (A) Venn plots showing the number of core genes and strain-specific genes in 21 the Corynebacterium strains. (B) Dilution curve of the pan-genome and core genome of the 21 Corynebacterium strains. (C) Heat map displaying the dispensable genes.
Mapped sequence library genes of the core genes, dispensable genes, and specific genes to the corresponding functions of the COG database are shown in Supplementary Figure S7. Of the 128 specific genes in C. pseudotuberculosis strain G1, only one gene (Cell wall/membrane/envelope biogenesis) matched the COG database. In the core and dispensable gene sequence libraries of the 21 Corynebacterium strains, 850 and 1934 genes, respectively, matched the COG database, with the majority of core genes (91.30%, n = 931) and dispensable genes (59.02%, n = 3277) having COG classification information. Translation, ribosomal structure and biogenesis genes had the largest proportion among the core genes (16.00%), while inorganic ion transport and metabolism genes had the highest proportion among the dispensable genes (7.93%).
To infer the phylogenetic relationships of strain G1 and 20 strains of Corynebacterium, we constructed phylogenetic trees based on gene family and core-pan genome (Figure 7).
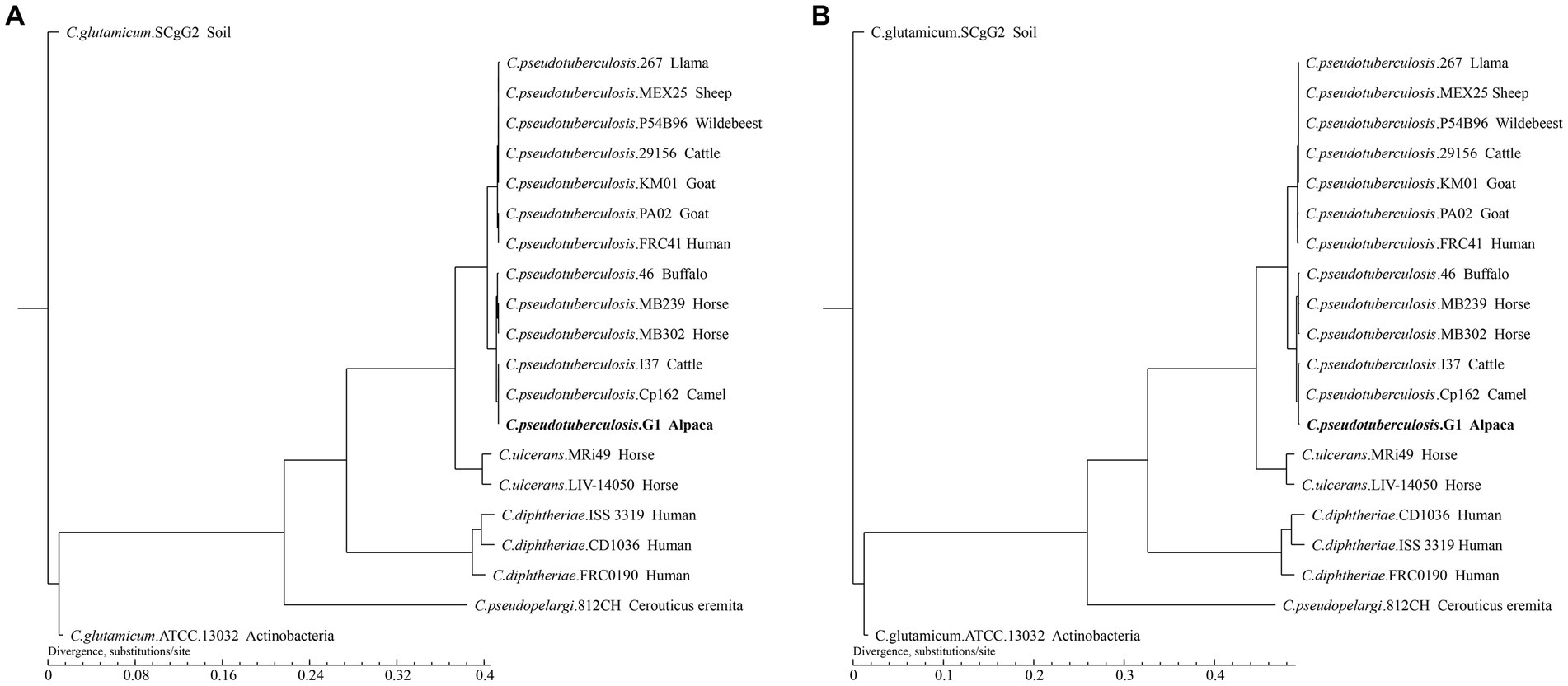
Figure 7. Phylogenetic tree of the 21 Corynebacterium strains. The tree was constructed using TreeBeST and PHYML based on (A) Core-pan gene. (B) Gene family. The numbers on the branches represent percent bootstrapping values from 1000 replicates. Each sequence is identified by its species, strain name, and host. Sequences obtained in this study are shown in bold.
The phylogenetic tree shows that different species of Corynebacterium clustered separately, while 13 C. pseudotuberculosis strains were divided into two biovars with clear demarcation, and strain G1 with C. pseudotuberculosis biovar equi clustered in the same clade. On the phylogenetic tree, C. pseudotuberculosis strain G1 was the closest relative to Cp162 and I37, and this result was consistent with the ANI analysis. The phylogenetic trees constructed based on the results of the two analyses showed the same branching structure, indicating good conservation of core-pan genes and gene families among these Corynebacterium strains.
Synteny analysis was conducted between strain G1 and other C. pseudotuberculosis strains. The analysis revealed that strain G1 has a chromosomal inversion compared with strain Cp162 (equi, camel), a chromosomal translocation with four biovar equi (I37, MB302, MB239, and 46) and three biovar ovis (267, FRC41, and PA02) strains, and four chromosomal translocations with two biovar ovis (29,156 and MEX25) strains (Figure 8). No host or biovar-specific patterns were observed.
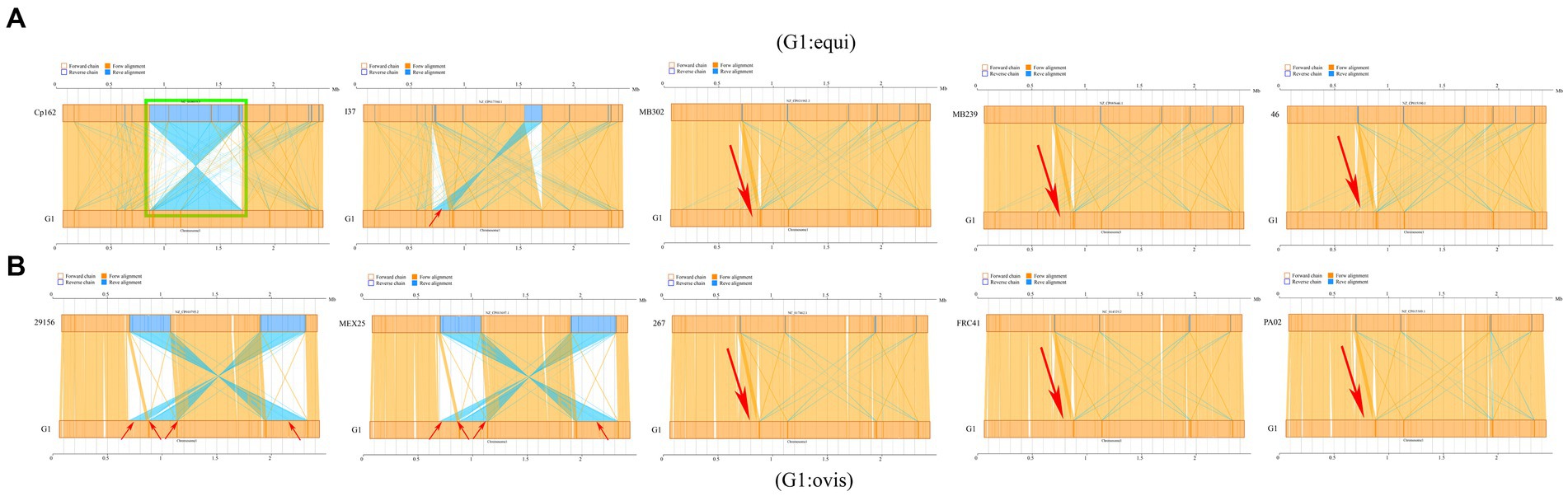
Figure 8. Synteny analysis. (A) Comparison of strain G1 with C. pseudotuberculosis biovar equi strains (Cp162 - camel, I37 - cattle, MB302 - horse, MB239 - horse, 46 - buffalo). (B) Comparison of strain G1 with C. pseudotuberculosis biovar ovis strains (29156 - cattle, MEX25 - sheep, 267 - llama, FRC41 - human, PA02 - goat). Green box indicates chromosome inversion, while red arrow represents chromosome translocation.
4. Discussion
CLA, caused by C. pseudotuberculosis, is one of the most important bacterial infectious diseases in camelids. This disease is characterized by abscesses of one or more superficial lymph nodes, and it can also infect internal organs such as the liver, lungs, and mammary glands, causing great losses to the breeding industry and attracting the attention of many scholars (Kinne, 2016). CLA has been reported in Old World Camels (Bactrian camel and dromedary camel) from several countries, such as China (Mullali et al., 2017), Russia, India (Purohit et al., 1985), Iran (Esterabadi et al., 1975), Saudi Arabia (Radwan et al., 1989), United Arab Emirates (Afzal et al., 1996), Oman, Jordan (Hawari, 2008), Kazakhstan (Samartsev, 1950), Egypt (Nashed and Mahmoud, 1987), Ethiopia (Domenech, 1980), Kenya (Wernery, 2012), Spain (Tejedor-Junco et al., 2008), as well as in Australia (Wernery et al., 2014). CLA has also been reported in New World Camels (alpaca and llama) from the USA (Lopes et al., 2012), Peru (Braga et al., 2006), Sweden, Germany (Sting et al., 2022) and Italy. Specifically, there have been reports of a llama in USA (Lopes et al., 2012) and 130 alpacas in Germany (Sting et al., 2022) being infected with biovar ovis strains. CLA by C. pseudotuberculosis biovar equi has not previously been reported in alpacas. Previous reports have solely documented the infection of goats with C. pseudotuberculosis biovar ovis strains in Sichuan Province, China (Zhang et al., 2015). While the alpacas in this study were imported from Chile and the Netherlands, it is worth noting that in Chile, cases of C. pseudotuberculosis biovar equi strains infection in horses have been reported (Cavalcante et al., 2015). Therefore, the pathogenic bacteria isolated in this study may have originated from the country from where the alpacas were introduced. Horses can only be infected by C. pseudotuberculosis biovar equi strains, while cattle and camelids can be infected by both biovars. It is crucial to be vigilant, as the outbreak of this pathogen in Chinese farms would pose a significant threat to the cattle, horse, and camelid breeding industries, resulting in economic losses such as death of farmed animals and reduced production. Therefore, effective prevention and control measures must be taken to prevent the spread and infection of this pathogen.
The genome of strain G1 is approximately 2.379 Mb, which is smaller than the genomes of biovar equi strains isolated from buffaloes (strains 31–36, 38, 39, and 48) and camel (strain 267). The GC content is 52.06%, which is lower than that of previously sequenced whole genomes of C. pseudotuberculosis strains (Supplementary Table S1). Three CRISPR clusters were detected on the chromosome of the strain G1, which were only detected in biovar equi strains (Parise et al., 2018). The gene set was subjected to COG functional annotation, and the categorical statistics were similar to those of six biovar equi strains (MEX1, MEX9, MEX25, MEX29, MEX30, and MEX31) isolated from equines in Mexico, except for strain MEX30 which harbors a gene related to chromatin structure and dynamics gene (Parise et al., 2018).
The widespread prevalence and significant economic impact of this disease have prompted investigations into the molecular mechanisms of virulence in this pathogen (Dorella et al., 2009). In sheep and goats, the virulence factor-related genes pld and fagA, B were detected in 100% of isolates, and fagC, D genes were detected in more than 95% (Aquino De Sá et al., 2013). Notably, all fagD-negative isolates were obtained from superficial abscesses, suggesting variations in the virulence potential of the clinical isolates (Aquino De Sá et al., 2013). A mutation in fagB (C) can reduce the pathogenicity of C. pseudotuberculosis in goat infections (Billington et al., 2002). To study the role of Opp in C. pseudotuberculosis, an OppD-deficient strain was created, and it was observed that the mutant strain had impaired growth when exposed to the toxic glutathione peptide, similar to the wild-type strain. The ΔoppD strain also showed a reduced ability to adhere to and infect macrophages compared to the wild-type strain, but both strains had similar potential to colonize spleens and cause injury and death to infected mice (Moraes et al., 2014).The above ten virulence genes (pld, fagA-D, oppA-F) were detected in the strain LY20 isolated from a goat in China (Wang R. et al., 2021). Whole genome sequencing of C. pseudotuberculosis strain I37 isolated from cattle (accession number CP017384, Israel) also revealed the presence of these ten virulence genes. At present, there are few reports related to C. pseudotuberculosis from alpaca, and the results of whole genome sequencing of C. pseudotuberculosis isolated from llama showed the presence of the pld and fagA-D (Lopes et al., 2012). In this study, we annotated 48 genes related to virulence factors in five categories (adherence, exotoxin, immune modulation, nutritional/metabolic factor, and stress survival). Among them, pld is the only exotoxin-related virulence gene. Other genes include σ factors (sigE, sigH, sigA/rpoV), iron uptake and regulation-related genes (fagA-D, ciuA-E), oligopeptide permease (oppA-F), minor pilin proteins (spaC, spaI), and superoxide dismutase (sodA).
The success of antimicrobial treatment to CLA is poor (Kinne, 2016). Performing resistance testing of isolates can provide guidance for the use of antibiotics. At present, there have been limited studies on antibiotic susceptibility of C. pseudotuberculosis in alpaca, with most research focusing on horse (Foley et al., 2004; Rhodes et al., 2015), sheep (Gallardo et al., 2019), and goat (Connor et al., 2000; Abebe and Sisay, 2015) isolates. The minimum inhibitory concentration (MIC) of the antimicrobial agent showed no significant differences between the isolates from horses and cows compared to those from sheep and goats, except in the case of amikacin (Costa et al., 1998). The MIC of equine C. pseudotuberculosis isolates demonstrated that several commonly used antimicrobials are effective against C. pseudotuberculosis in vitro. Abscess location was not associated with different MIC patterns in cultured isolates (Rhodes et al., 2015). It has been determined that 59 C. pseudotuberculosis isolates from sheep and goats were primarily susceptible to norfloxacin (77.97%), doxycycline HCl (72.88%), and kanamycin (72.88%), while mainly resistant to ampicillin (28.81%), clindamycin (25.42%), and doxycycline HCl (22.04%) (Abebe and Sisay, 2015). Strain XH02 (Boer goat, China) exhibited high sensitivity to 13 antibacterial agents, including chloromycetin, tetracycline, norfloxacin, minocycline, cefoxitin, clarithromycin, roxithromycin, and ceftriaxone, but completely resistant to nitrofurantoin and furazolidone (Zhou et al., 2016). However, the susceptibility of five alpaca isolates to gentamicin, sulfonamide, oxacillin, neomycin, and ceftiofur varied (Anderson et al., 2004). In this study, 21 antibiotics in 12 categories commonly used in clinical practice were selected for antibiotic susceptibility testing. The three strains were sensitive to penicillin, cefotaxime, meropenem, erythromycin and other antibiotics, resistant to aztreonam, fosfomycin, and nitrofurantoin. Combined with the antibiotic sensitivity results and clinical manifestations of the sick alpaca (S2), meropenem with high sensitivity was selected for treatment, and the sick alpaca symptoms improved, and gradually recovered.
Horizontal gene transfer (HGT) of drug-resistance genes leads to global infections of multi-drug resistant (MDR) microorganisms in hospitals and communities, posing a serious potential hazard to public health safety (Warnes et al., 2012). PCR amplification of the ARGs in C. pseudotuberculosis strains obtained from goats and sheep from Egypt indicated the presence of the β-lactams resistance gene (bla) in 40% of the isolates, and the aminoglycoside resistance gene (aadA2) in 42% of the isolates (Tawab et al., 2019). Upon analyzing the complete genome sequence of the C. pseudotuberculosis strain Cp267 isolated from llama (accession number CP003407, USA), we discovered that it carries 12 ARGs, mainly expressed as MRD protein (marR, norM, and emrB), lincomycin resistance protein (lmrB), bleomycin resistance protein and glycopeptide antibiotic resistance protein, and no β-lactam resistance gene was found (Lopes et al., 2012). In this study, the complete genome sequence of the C. pseudotuberculosis strain G1 isolated from alpaca was annotated with five ARGs, including rifamycin resistance genes rpoB and rbpA, elfamycin antibiotic gene EF-Tu mutant, fosfomycin antibiotic gene murA, and pyrazine antibiotic gene pncA. Our findings indicate that these ARGs exhibited a low correlation with the resistance phenotype, suggesting that detection of ARGs cannot replace routine antibiotic susceptibility testing. The expression of ARGs is influenced by various factors, including gene expression level, drug resistance mechanism, and bacterial adaptability to the environment. Therefore, the presence of a certain ARG does not necessarily imply that the bacterium will exhibit resistance to the corresponding antibiotic. Lastly, the difference in the carriage of resistance genes between the strains analyzed in this study and previous studies may be attributed to the variation in antibiotic usage in different regions to combat bacterial infectious diseases.
Several different serological tests (haemagglutination, haemagglutination inhibition, and ELISAs) have been tried for the diagnosis of CLA but, with the exception of ELISA, are rarely used on camelids (Kinne, 2016). An in-house ELISA was developed based on commercially available ELITEST, using protein A substituted anti-goat/sheep conjugate, is currently routinely used to detect CLA antibodies in dromedaries with good results (Berlin et al., 2015). Serologic testing was performed on 232 alpacas using a commercially available ELISA based on PLD as antigen and an in-lab ELISA based on whole cell antigens (WCA), and showed a substantial degree of agreement of 89.5% for both tests (Sting et al., 2022). Further comparative studies showed that the immunoblot had a sensitivity superior to both ELISAs (Sting et al., 2022). In conclusion, serological testing using ELISA combined with validation by immunoblot should be considered as critical methods to control CLA in alpaca herds.
5. Conclusion
To our knowledge, this is the first report of infection with C. pseudotuberculosi from alpaca in China, and the first whole genome sequencing of C. pseudotuberculosis biovar equi strain in alpaca. Comparative genomics analysis based on ANI value, gene families, core-pan genome, and synteny analysis demonstrated a high degree of genetic similarity among C. pseudotuberculosis strains, in contrast to other Corynebacterium species, with a clear delineation between strains belonging to the two biovars (ovis and equi). This study provides fundamental information for further exploration of the pathogenic mechanisms of C. pseudotuberculosis in alpaca and practical guidance for the prevention, diagnosis, and treatment of CLA in this species.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Ethics statement
The animal study was reviewed and approved by the Sichuan Agricultural University Animal Ethical and Welfare Committee. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
WM: methodology, data analysis and visualization, and writing – original draft. SC: clinical case handling. LH: writing – original draft. JY: data analysis and visualization. WZ: investigation. ZhZ and ZiZ: resources. HL and HF: writing – review and editing. TH: project administration. GP: conceptualization and supervision. All authors have read and approved the manuscript.
Funding
This research was supported by Sichuan Wolong National Nature Reserve Administration (510000-02-064387), and the State Forestry and Grassland Administration (2022-2222219002).
Acknowledgments
The authors thank Zhiyou Dong and Qianlan Li for their invaluable assistance in the feeding and management of experimental animals. Additionally, we extend our heartfelt appreciation to Cao Deng for his expert guidance in the partial data analysis of the whole genome.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1206187/full#supplementary-material
Footnotes
1. ^http://www.ncbi.nlm.nih.gov/BLAST
3. ^https://www.broadinstitute.org/gatk/
4. ^http://www.cbcb.umd.edu/software/glimmer/
6. ^http://tandem.bu.edu/trf/trf.html
7. ^http://www5.esu.edu/cpsc/bioinfo/software/GIST/
8. ^http://phast.wishartlab.com/
9. ^https://crisprcas.i2bc.paris-saclay.fr//CrisprCasFinder/Index
10. ^http://treesoft.svn.sourceforge.net/viewvc/treesoft/trunk/treebest/
References
Abebe, D., and Sisay, T. T. (2015). Determination of Corynebacterium pseudotuberculosis prevalence and antimicrobial susceptibility pattern of isolates from lymph nodes of sheep and goats at an organic export abattoir, Modjo. Ethiopia. Lett Appl. Microbiol. 61, 469–476. doi: 10.1111/lam.12482
Afzal, M., Sakir, M., and Hussain, M. M. (1996). Corynebacterium pseudotuberculosis infection and lymphadenitis (taloa or mala) in the camel. Trop. Anim. Hlth. Prod. 28, 158–162. doi: 10.1007/BF02299568
Almeida, S., Dorneles, E. M. S., Diniz, C., Abreu, V., Sousa, C., Alves, J., et al. (2017). Quadruplex PCR assay for identification of Corynebacterium pseudotuberculosis differentiating biovar Ovis and Equi. BMC Vet. Res. 13:290. doi: 10.1186/s12917-017-1210-5
Almeida, S., Sousa, C., Abreu, V., Diniz, C., Dorneles, E. M. S., Lage, A. P., et al. (2017). Exploration of nitrate Reductase metabolic pathway in Corynebacterium pseudotuberculosis. Int. J. Genomics 2017, 1–12. doi: 10.1155/2017/9481756
Anderson, D. E., Rings, D. M., and Kowalski, J. (2004). Infection with Corynebacterium pseudotuberculosis in five alpacas. J. Am. Vet. Med. Assoc. 225, 1743–1747. doi: 10.2460/javma.2004.225.1743
Aquino De Sá, M. D. C., Gouveia, G. V., Krewer, C. D. C., Veschi, J. L. A., de Mattos-Guaraldi, A. L., and Da Costa, M. M. (2013). Distribution of PLD and FagA, B, C and D genes in Corynebacterium pseudotuberculosis isolates from sheep and goats with caseus lymphadenitis. Genet. Mol. Biol. 36, 265–268. doi: 10.1590/S1415-47572013005000013
Asfour, H. A. E., and Darwish, S. F. (2014). Evaluation of phenotypic methods versus molecular methods for differentiation of coagulase positive staphylococci causing bovine mastitis with a special reference to atypical Staphylococcus aureus. Int. J. Curr. Microbiol. App. Sci. 3, 543–558.
Barakat, A. A., Selim, S. A., Atef, A., Saber, M. S., Nafie, E. K., and El-Edeeby, A. A. (1984). Two serotypes of Corynebacterium pseudotuberculosis isolated from different animal species. Rev. Sci. Tech. 3, 151–163. doi: 10.20506/rst.3.1.147
Barauna, R. A., Ramos, R. T., Veras, A. A., Pinheiro, K. C., Benevides, L. J., Viana, M. V., et al. (2017). Assessing the genotypic differences between strains of Corynebacterium pseudotuberculosis biovar equi through comparative genomics. PLoS One 12:e170676. doi: 10.1371/journal.pone.0170676
Berlin, M., Joseph, M., Jose, S., Raghavan, R., and Wernery, U. (2015). Production of a caseous lymphadenitis vaccine for dromedaries. J. Camel. Pract. Res. 22:163. doi: 10.5958/2277-8934.2015.00026.0
Bernardes, J. S., Eberle, R. J., Vieira, F. R. J., and Coronado, M. A. (2021). A comparative pan-genomic analysis of 53 C. pseudotuberculosis strains based on functional domains. J. Biomol. Struct. Dyn. 39, 6974–6986. doi: 10.1080/07391102.2020.1805017
Billington, S. J., Esmay, P. A., Songer, J. G., and Jost, B. H. (2002). Identification and role in virulence of putative iron acquisition genes from Corynebacterium pseudotuberculosis. FEMS Microbiol. Lett. 208, 41–45. doi: 10.1111/j.1574-6968.2002.tb11058.x
Braga, W. U., Chavera, A., and Gonzalez, A. (2006). Corynebacterium pseudotuberculosis infection in highland alpacas (Lama pacos) in Peru. Vet. Rec. 159, 23–24. doi: 10.1136/vr.159.1.23
Cavalcante, A. L. Q., Dias, L. M., Alves, J. T. C., Veras, A. A. O., Guimarães, L. C., Rocha, F. S., et al. (2015). Complete genome sequence of Corynebacterium pseudotuberculosis strain E19, isolated from a Horse in Chile. Genome Announc. 3, e1315–e1385. doi: 10.1128/genomeA.01385-15
CLSI (2019). Performance Standards for Antimicrobial Susceptibility Testing. 29th ed. CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute.
Connor, K. M., Quirie, M. M., Baird, G., and Donachie, W. (2000). Characterization of United Kingdom isolates of Corynebacterium pseudotuberculosis using pulsed-field gel electrophoresis. J. Clin. Microbiol. 38, 2633–2637. doi: 10.1128/JCM.38.7.2633-2637.2000
Costa, L. R. R., Spier, S. J., and Hirsh, D. C. (1998). Comparative molecular characterization of Corynebacterium pseudotuberculosis of different origin. Vet. Microbiol. 62, 135–143. doi: 10.1016/S0378-1135(98)00202-8
de Sá Guimarães, A., Carmo, F. B. D., Pauletti, R. B., Seyffert, N., Ribeiro, D., Lage, A. P., et al. (2011). Caseous lymphadenitis: epidemiology, diagnosis, and control. IIOAB J. 2, 33–43.
Domenech, J. (1980). Etube bactériologique de Corynebacterium pseudotuberculosis et de Corynebacterium pyogenes isolés chez le deomadaire en Ethiopie. Revue d'elevage et de medecine veterinaire des pays tropicaux. 33, 123–126. doi: 10.19182/remvt.8220
Dorella, F. A., Pacheco, L. G. C., Oliveira, S. C., Miyoshi, A., and Azevedo, V. (2006). Corynebacterium pseudotuberculosis: microbiology, biochemical properties, pathogenesis and molecular studies of virulence. Vet. Res. 37, 201–218. doi: 10.1051/vetres:2005056
Dorella, F. A., Pacheco, L. G., Seyffert, N., Portela, R. W., Meyer, R., Miyoshi, A., et al. (2009). Antigens of Corynebacterium pseudotuberculosis and prospects for vaccine development. Expert Rev. Vaccines 8, 205–213. doi: 10.1586/14760584.8.2.205
Du, Y. (1997). A case of thyroid abscess induced by Corynebacterium pseudotuberculosis. Labor Med. 12:123.
Esterabadi, A. H., Entessar, F., Hedayati, H., Narimani, A. A., and Sadri, M. (1975). Isolation of Corynebacterium pseudotuberculosis from camel in Iran. Arch Razi Inst. 27, 61–66. doi: 10.22092/ARI.1975.108778
Foley, J. E., Spier, S. J., Mihalyi, J., Drazenovich, N., and Leutenegger, C. M. (2004). Molecular epidemiologic features of Corynebacterium pseudotuberculosis isolated from horses. Am. J. Vet. Res. 65, 1734–1737. doi: 10.2460/ajvr.2004.65.1734
Fredriksson, N. J., Hermansson, M., and Britt-Marie, W. (2013). The choice of PCR primers has great impact on assessments of bacterial community diversity and dynamics in a wastewater treatment plant. PLoS One 8:e76431. doi: 10.1371/journal.pone.0076431
Gallardo, A. A., Toledo, R. A., González Pasayo, R. A., Azevedo, V., Robles, C., Paolicchi, F. A., et al. (2019). Corynebacterium pseudotuberculosis biovar ovis: evaluación de la sensibilidad antibiótica in vitro. Rev. Argent. Microbiol. 51, 334–338. doi: 10.1016/j.ram.2018.12.001
Guo, Z., Sun, J., Cheng, J., Zheng, L., and Bao, Z. (1988). Diagnosis of Corynebacterium pseudotuberculosis in camels. Vet. Orien., 101–103.
Hard, G. C. (1969). Electron microscopic examination of Corynebacterium ovis. J. Bacteriol. 97, 1480–1485. doi: 10.1128/jb.97.3.1480-1485.1969
Hawari, A. D. (2008). Corynebacterium pseudotuberculosis infection (Caseous lymphadenitis) in camels (Camelus Dromedarius) in Jordan. Am. J. Anim. Vet. Sci. 3, 68–72. doi: 10.3844/ajavsp.2008.68.72
He, W., Liu, H., and Wei, D. (1991). Diagnosis report of Pseudotuberculosis in chicken. Chin. J. Vet. Med. 17:16.
Kazmierczak, M. J., Wiedmann, M., and Boor, K. J. (2005). Alternative sigma factors and their roles in bacterial virulence. Microbiol. Mol. Biol. Rev. 69, 527–543. doi: 10.1128/mmbr.69.4.527-543.2005
Kinne, U. W. A. J. (2016). Caseous lymphadenitis (Pseudotuberculosis) in Camelids: a review. Austin J. Vet. Sci. Anim. Husb. 3:1022.
Liu, M. (2021). Isolation of Corynebacterium pseudotuberculosis and its specific bacteriophage. Master Northwest A&F University, Xianyang
Liu, H., Zhang, Y., Huang, Z., Wang, Y., and Shen, M. (1986). Study on Corynebacterium pseudotuberculosis from camels. J. Ningxia Agric. For Sci. Technol. 46–48.
Lopes, T., Silva, A., Thiago, R., Carneiro, A., Dorella, F. A., Rocha, F. S., et al. (2012). Complete genome sequence of Corynebacterium pseudotuberculosis strain Cp267, isolated from a Llama. J. Bacteriol. 194, 3567–3568. doi: 10.1128/JB.00461-12
Mills, A. E., Mitchell, R. D., and Lim, E. K. (1997). Corynebacterium pseudotuberculosis is a cause of human necrotising granulomatous lymphadenitis. Pathology 29, 231–233. doi: 10.1080/00313029700169944
Moraes, P. M. R. O., Seyffert, N., Silva, W. M., Castro, T. L. P., Silva, R. F., Lima, D. D., et al. (2014). Characterization of the Opp peptide transporter of Corynebacterium pseudotuberculosis and its role in virulence and pathogenicity. Biomed. Res. Int. 2014, 1–7. doi: 10.1155/2014/489782
Moussa, I. M., Ali, M. S., Hessain, A. M., Kabli, S. A., Hemeg, H. A., and Selim, S. A. (2016). Vaccination against Corynebacterium pseudotuberculosis infections controlling caseous lymphadenitis (CLA) and oedematousskin disease. Saudi J. Biol. Sci. 23, 718–723. doi: 10.1016/j.sjbs.2016.06.005
Mullali,, Wulsbai, Y. W., Uang, B., and Hazibai, H. (2017). Diagnosis and treatment of camel impetigo (Corynebacterium pseudotuberculosis). Vet. Clin. Sci. 10:68. doi: 10.3969/j.issn.2096-3637.2017.10.060
Nashed, S. M., and Mahmoud, A. Z. (1987). Microbiological and histopathological studies for rare cases of Corynebacterium infection in camel. AVMJ 18, 82–85.
Oliveira, M., Barroco, C., Mottola, C., Santos, R., Lemsaddek, A., Tavares, L., et al. (2014). First report of Corynebacterium pseudotuberculosis from caseous lymphadenitis lesions in Black Alentejano pig (Sus scrofa domesticus). BMC Vet. Res. 10:218. doi: 10.1186/s12917-014-0218-3
Oliveira, A., Teixeira, P., Azevedo, M., Jamal, S. B., Tiwari, S., Almeida, S., et al. (2016). Corynebacterium pseudotuberculosis may be under anagenesis and biovar Equi forms biovar Ovis: a phylogenic inference from sequence and structural analysis. BMC Microbiol. 16:100. doi: 10.1186/s12866-016-0717-4
Parise, D., Parise, M. T. D., Viana, M. V. C., Muñoz-Bucio, A. V., Cortés-Pérez, Y. A., Arellano-Reynoso, B., et al. (2018). First genome sequencing and comparative analyses of Corynebacterium pseudotuberculosis strains from Mexico. Stand. Genomic Sci. 13:21. doi: 10.1186/s40793-018-0325-z
Peel, M. M., Palmer, G. G., Stacpoole, A. M., and Kerr, T. G. (1997). Human lymphadenitis due to Corynebacterium pseudotuberculosis: report of ten cases from Australia and review. Clin. Infect. Dis. 24, 185–191. doi: 10.1093/clinids/24.2.185
Purohit, N., Chouhan, D., and Choudhary, R. (1985). Lymphangitis in the camel. Agri. Pract. 6, 23–24.
Radwan, A. I., El-Magawry, S., Hawari, A., Al-Bekairi, S. I., and Rebleza, R. M. (1989). Corynebacterium pseudotuberculosis infection in camels (Camelus dromedarius) in Saudi Arabia. Trop. Anim. Health Prod. 21, 229–230.
Retamal, P., Ríos, M., Cheuquepán, F., Abalos, P., Pizarro-Lucero, J., Borie, C., et al. (2011). Host associated polymorphisms in the Corynebacterium pseudotuberculosis rpoB gene sequence. Vet. Microbiol. 151, 400–403. doi: 10.1016/j.vetmic.2011.03.012
Rhodes, D. M., Magdesian, K. G., Byrne, B. A., Kass, P. H., Edman, J., and Spier, S. J. (2015). Minimum inhibitory concentrations of equine Corynebacterium pseudotuberculosis isolates (1996-2012). J. Vet. Intern. Med. 29, 327–332. doi: 10.1111/jvim.12534
Samartsev, A. (1950). Infectious pustular dermatitis in camels. Proc. Kazakh Res. Vet. Inst. 5, 190–197.
Samen, U., Gottschalk, B., Eikmanns, B. J., and Reinscheid, D. J. (2004). Relevance of peptide uptake systems to the physiology and virulence of Streptococcus agalactiae. J. Bacteriol. 186, 1398–1408. doi: 10.1128/JB.186.5.1398-1408.2004
Songer, J. G. (1997). Bacterial phospholipases and their role in virulence. Trends Microbiol. 5, 156–161. doi: 10.1016/S0966-842X(97)01005-6
Sting, R., Geiger, C., Rietschel, W., Blazey, B., Schwabe, I., Rau, J., et al. (2022). Corynebacterium pseudotuberculosis infections in Alpacas (Vicugna pacos). Animals 12:1612. doi: 10.3390/ani12131612
Sutherland, S. S., Speijers, E. J., and Andres, B. (1989). Comparison of the exotoxins of four strains of Corynebacterium pseudotuberculosis. Res. Vet. Sci. 47, 190–194.
Tawab, A. A. A. E., Rizk, A. M., Afifi, S. E., and Mohamed, S. R. (2019). Corynebacterium pseudotuberculosis infection in small ruminant and molecular study of virulence and resistance genes in Beni-Suef governorate. Benha Vet. Med. J. 37, 122–127. doi: 10.21608/BVMJ.2019.18710.1125
Tejedor-Junco, M. T., Lupiola, P., Schulz, U., and Gutierrez, C. (2008). Isolation of nitrate-reductase positive Corynebacterium pseudotuberculosis from dromedary camels. Trop. Anim. Health Prod. 40, 165–167. doi: 10.1007/s11250-007-9077-2
Wang, X., Li, J., and Wang, L. (2021). Effect of Mycobacterium tuberculosis infection on the expression of IL-12 in mouse spleen and its correlation with body mass ratio and organ index of mice. Int. J. Clin. Exp. Med. 20, 1132–1135. doi: 10.3969/j.issn.1671-4695.2021.11.004
Wang, R., Lin, C., Chi, X. L., Chen, S., and Zhang, X. (2021). Isolation and identification of Corynebacterium pseudotuberculosis from goats and histopathological observation of infected mice. Chin. J. Vet. Sci. 41, 413–422. doi: 10.16303/j.cnki.1005-4545.2021.12.18
Warnes, S. L., Highmore, C. J., and Keevil, C. W. (2012). Horizontal transfer of antibiotic resistance genes on abiotic touch surfaces: implications for public health. MBio 3, e412–e489. doi: 10.1128/mBio.00489-12
Wernery, U. (2012). Caseous lymphadenitis (Pseudotuberculosis) in camelids. J. Camel Pract. Res. 19, 21–27.
Xiong, H., Wei, B., Wei, R., Yang, N., and Qi, M. (2013). Calculation of median lethal dose (LD50) for Yersinia pestis by SPSS package. Chin. J. Zoonoses. 29, 1127–1130. doi: 10.3969/cjz.j.issn.1002-2694.2013.11.020
Zhang, M., Li, H., and Zhou, Z. (2015). Diagnosis and drug sensitivity test of Corynebacterium pseudotuberculosis in Goats in Dazhou, Sichuan. Chin. J. Vet. Med. 51, 54–56. doi: 10.3969/j.issn.0529-6005.2015.12.019
Zhang, Z., Teng, Z., Ye, B., Yang, Z., and Zhu, C. (1989). Diagnosis of cutaneous Corynebacterium pseudotuberculosis infection in Buffalo. Chin. J. Pre. Vet. 1, 20–21.
Zhou, Z., Li, H., Zhang, M., Wang, Z., Zhou, R., Hu, S., et al. (2016). Genome sequence of Corynebacterium pseudotuberculosis strain XH02 isolated from a Boer Goat in Xuanhan, China. Genome Announc. 4, e1316–e1329. doi: 10.1128/genomeA.01329-16
Keywords: alpaca, Corynebacterium pseudotuberculosis, biovar equi, virulence, antibiotic resistance, pathogenicity, genomic sequencing
Citation: Meng W, Chen S, Huang L, Yang J, Zhang W, Zhong Z, Zhou Z, Liu H, Fu H, He T and Peng G (2023) Isolation, characterization, and pathogenicity assessment of Corynebacterium pseudotuberculosis biovar equi strains from alpacas (Vicugna pacos) in China. Front. Microbiol. 14:1206187. doi: 10.3389/fmicb.2023.1206187
Edited by:
Lei Deng, Harvard Medical School, United StatesReviewed by:
Michael John Calcutt, University of Missouri, United StatesYuwei Gao, Chinese Academy of Agricultural Sciences (CAAS), China
Hualei Wang, Jilin University, China
Copyright © 2023 Meng, Chen, Huang, Yang, Zhang, Zhong, Zhou, Liu, Fu, He and Peng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tingmei He, 248109505@qq.com; Guangneng Peng, pgn.sicau@163.com
†These authors have contributed equally to this work
 Wanyu Meng1†
Wanyu Meng1†