
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 22 June 2023
Sec. Infectious Agents and Disease
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1204428
This article is part of the Research Topic Women in Infectious Agents and Disease: 2023 View all 10 articles
Staphylococcus aureus is one of the most common opportunistic human pathogens causing several infectious diseases. Ever since the emergence of the first methicillin-resistant Staphylococcus aureus (MRSA) strain decades back, the organism has been a major cause of hospital-acquired infections (HA-MRSA). The spread of this pathogen across the community led to the emergence of a more virulent subtype of the strain, i.e., Community acquired Methicillin resistant Staphylococcus aureus (CA-MRSA). Hence, WHO has declared Staphylococcus aureus as a high-priority pathogen. MRSA pathogenesis is remarkable because of the ability of this “superbug” to form robust biofilm both in vivo and in vitro by the formation of polysaccharide intercellular adhesin (PIA), extracellular DNA (eDNA), wall teichoic acids (WTAs), and capsule (CP), which are major components that impart stability to a biofilm. On the other hand, secretion of a diverse array of virulence factors such as hemolysins, leukotoxins, enterotoxins, and Protein A regulated by agr and sae two-component systems (TCS) aids in combating host immune response. The up- and downregulation of adhesion genes involved in biofilm formation and genes responsible for synthesizing virulence factors during different stages of infection act as a genetic regulatory see-saw in the pathogenesis of MRSA. This review provides insight into the evolution and pathogenesis of MRSA infections with a focus on genetic regulation of biofilm formation and virulence factors secretion.
Staphylococcus aureus is a normal microflora of the human nasal cavity and skin but may cause diseases such as skin and soft tissue infections, endocarditis, osteomyelitis, bacteremia, and lethal pneumonia on disruption of mucosal and cutaneous barriers due to surgical procedures, wounds, or chronic skin conditions. It is considered as one of the most common opportunistic human pathogens among immunocompromised patients, children, elderly, and patients with medical devices. Penicillin was extensively prescribed by doctors to cure S. aureus infections until Penicillin-resistant S. aureus was reported in the 1950's. To overcome this, in 1959 scientists developed a semisynthetic penicillin named methicillin which was resistant to penicillinase (Guo et al., 2020). Only after 2 years of the introduction of methicillin for clinical use, i.e., in 1961, a British scientist named Jevons isolated the first methicillin-resistant Staphylococcus aureus (MRSA) strain in Europe. MRSA possessed a gene mecA which was responsible for encoding the penicillin-binding protein 2a or 2′ (PBP2a or PBP2′). This gene was integrated into the chromosomal element (SCCmec) of methicillin-sensitive Staphylococcus aureus (MSSA). Since then, MRSA has been identified as one of the most notorious human pathogens across the world (Guo et al., 2020).
It is still debatable whether MRSA is more virulent than MSSA. Meta-analysis results of some of the epidemiological studies have indicated increased mortality and/or morbidity in the case of nosocomial MRSA infections such as those associated with surgery, pneumonia, and bloodstream, etc. as compared to MSSA infections (Cosgrove et al., 2003; Engemann et al., 2003; Gastmeier et al., 2005; Reed et al., 2005). Increased mortality by MRSA bacteremia as compared to MRSA pneumonia has been reported by Shurland et al. (2007). However, other studies reported no significant difference in mortality associated with nosocomial MRSA bacteremia (Cosgrove et al., 2005) or ventilator-associated pneumoniae (Zahar et al., 2005) as compared to those caused by MSSA. Another study comparing CA-MSSA and CA-MRSA skin infections also did not report any serious outcomes caused by the latter (Miller et al., 2007). To date, no study provides clear evidence that MRSA is more virulent than MSSA; however, treatment of invasive MRSA infections is challenging due to the lack of antibiotics and increased treatment costs (Gordon and Lowy, 2008).
Initially, MRSA was only associated with hospital outbreaks, more commonly termed as hospital-acquired methicillin-resistant Staphylococcus aureus (HA-MRSA). It was found to be responsible for 20–80% of secondary hospital infections (Krishnamurthy et al., 2014; Kemung et al., 2018). agr system, responsible for producing toxins for exhibiting virulence, is generally found to be either less active or mutated in HA-MRSA strains leading to high-level expression of adhesins, which aids in robust biofilm formation on implanted medical devices and are also more resistant to antibiotics (Painter et al., 2014; Suzuki et al., 2015).
Approximately 20 years after the first reported case of MRSA, community-acquired methicillin-resistant Staphylococcus aureus (CA-MRSA) was reported in Detroit, Michigan, USA in 1980 (Saravolatz et al., 1982). According to the Center for Disease Control and Prevention (CDC), CA-MRSA can be defined as an infection from an MRSA culture isolated within 48 h of admission of a patient in a hospital with no previous history of hospital admission or medical treatment with surgical procedures (Gorwitz et al., 2006). According to a CDC report of 2016, two out of 100 people are carriers of CA-MRSA, suggesting its ability to spread more easily than HA-MRSA. The presence of more mobile genetic elements viz., novel SCCmec elements, Panton-Valentine Leukocidin (PVL)-encoding genes, and ACME (arginine catabolite mobile element) in CA-MRSA is considered to be the plausible cause of more virulence observed in it as compared to HA-MRSA (Udo and Boswihi, 2017; Boswihi and Udo, 2018).
Recently, the evidence of the spread of MRSA from animals to humans led to the conclusion that animals are reservoirs as well as potent carriers of MRSA, which is a serious threat to human health (Voss et al., 2005; Loncaric et al., 2013; Van Boeckel et al., 2015). The plausible reason for the emergence of live-stock-associated methicillin-resistant Staphylococcus aureus (LA-MRSA) is the overuse of antibiotics for increasing the yield in live-stock production (Van Boeckel et al., 2015).
The entry of S. aureus into the endothelial tissues of the host begins when the bacterium, from an external source or from indigenous microflora, gains access to the bloodstream or underlying tissues leading to infections (Figure 1).
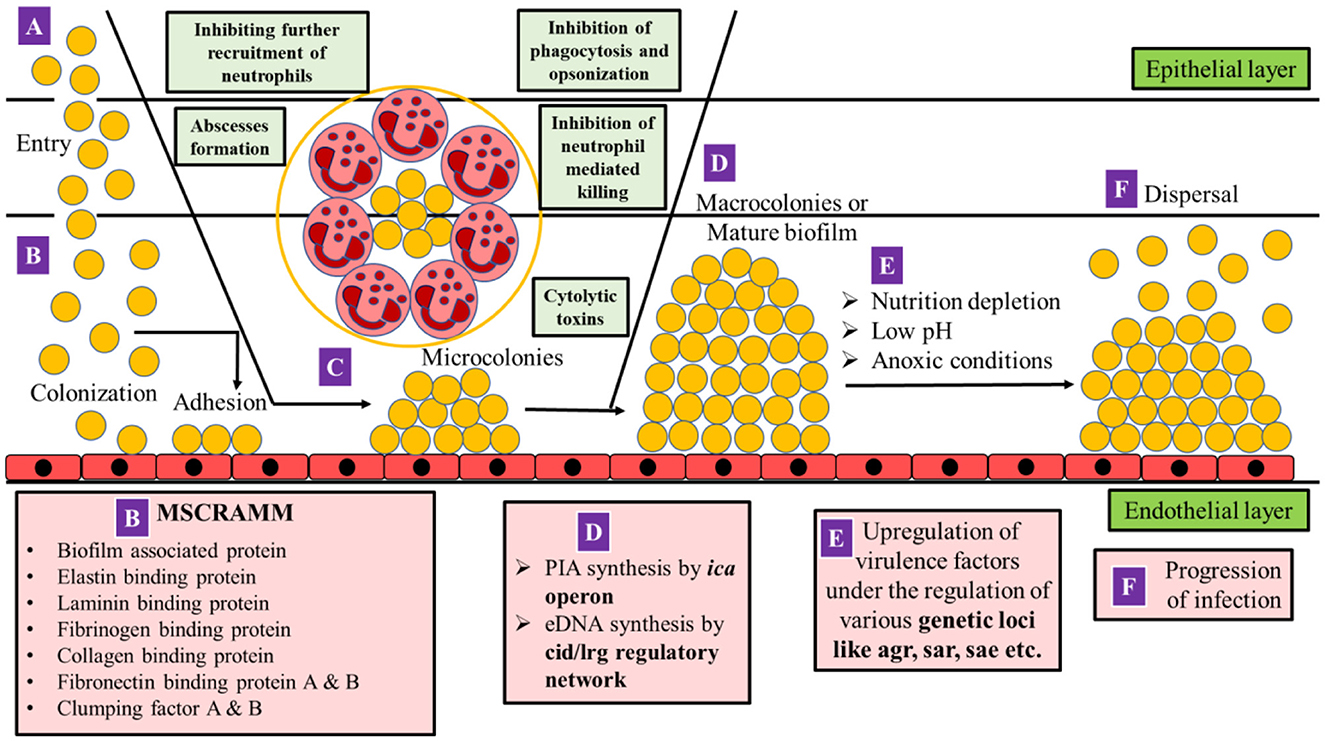
Figure 1. Mechanism of pathogenesis in MRSA. (A) S. aureus gains access to the bloodstream or underlying tissues due to a breach in cutaneous or mucosal barriers. (B) Colonization occurs on the endothelial layer by adhesion with the help of MSCRAMM. (C) As the S. aureus cells multiply, polymorphonuclear leukocytes (PMNs) are recruited, and a fibrin pseudo-capsule termed as abscess is formed with the help of coagulase proteins secreted by S. aureus which surrounds bacterial cells and recruited (PMNs). S. aureus can inhibit the further entry of PMNs at the site of infection, opsonization, phagocytosis, and neutrophil-mediated killing. The bacterium which is already phagocytosed by PMNs releases cytolytic toxins such as hemolysins, leukocidins such as Panton-Valentine Leukocidin (PVL) and PSM peptides to escape the host defense. The bacterial cells further multiply leading to the formation of microcolonies on the endothelial layer. (D) Synthesis of poly intracellular adhesin (PIA) and extracellular DNA (eDNA) is initiated by the ica operon and cid/lrg regulatory network, respectively, resulting in the formation of macrocolonies or a mature biofilm. (E) As biofilm matures, the core becomes anoxic, nutrient-deprived, and acidic in pH which triggers the upregulation of various virulence factors (agr, sar, and sae), which mediates (F) dispersal of biofilm as well as the progression of infection (adapted from Moormeier and Bayles, 2017; Sedarat and Taylor-Robinson, 2022).
The colonization of the mammalian cell's extracellular matrix proteins (fibrinogen, fibronectin, elastin, collagen, laminin, and vitronectin) occurs covalently or non-covalently, mediated by several molecules of S. aureus that are collectively termed as microbial surface components recognizing adhesive matrix molecules (MSCRAMMs; Table 1). Teichoic acids (TAs), which are common components of Gram-positive bacterial cell wall, also contribute significantly to the adherence to host cells (Qin et al., 2007). Approximately 15% of individuals (persistent carriers) have been reported to have continuous S. aureus colonization, whereas 70% of individuals have intermittent colonization (frequent S. aureus infection but immediate eradication), and the remaining 15% of individuals are non-carriers (Eriksen et al., 1995).
Apart from the host- and pathogen-associated factors, various other factors also play roles in S. aureus colonization. The nasal cavity is one of the most important sites of colonization for S. aureus as nose-picking can lead to the spread of bacteria to other body parts as well as to other hosts (Von Eiff et al., 2001; Wertheim et al., 2004, 2006). The nasal microbiota (Corynebacterium spp., Propionibacterium acnes, Staphylococcus epidermis, and Staphylococcus lugdunensis) and the invading pathogen S. aureus compete among themselves in various ways for colonization. Nutrition is one of the major limiting factors for colonization in the human nose and therefore S. aureus has been found to better adapt than coagulase-negative staphylococci due to the ability of the former to thrive in the low-nutrient environment (Lemon et al., 2010; Krismer et al., 2014; Liu et al., 2015; Lee et al., 2018).
The growth of S. aureus including MRSA has been found to be inhibited by the indigenous nasal bacterium Staphylococcus lugdunensis both in vitro and in vivo due to the production of an antimicrobial compound called lugdunin that can rapidly breakdown bacterial energy resources (Zipperer et al., 2016). The risk of colonization of S. aureus has been reported to be 6-fold lower in humans with Staphylococcus lugdunensis in their nasal cavities. However, nasal colonization with Staphylococcus lugdunensis has been reported in only 9–26% of the general population (Kaspar et al., 2016; Zipperer et al., 2016; Lee et al., 2018). S. aureus has been reported to compete with other commensal bacteria by the activation of host defense via upregulation of the production of antimicrobial proteins that are less harmful to it than to others (Krismer et al., 2017; Lee et al., 2018). Apart from the nasal cavity, other sites in the human body where S. aureus colonizes are armpits (8%), abdomen (15%), intestine (17–31%), perineum (22%), vagina (5%), and pharynx (4–64%; González-García et al., 2023). The virulence genes responsible for toxin production in S. aureus are downregulated during the colonization process.
The multiplication of bacteria after colonization on both biotic and abiotic surfaces leads to the formation of a three-dimensional complex community of bacteria within a layer of exopolysaccharide (EPS) termed as “biofilm” (Figure 1; Costerton et al., 1999; Sedarat and Taylor-Robinson, 2022). The accumulation of biofilm is facilitated by the formation of microcolonies due to the production of exopolysaccharide (a major component of biofilm), which comprises 97% water and 3% of structural and functional molecules (Guzmán-Soto et al., 2021). EPS of biofilm contains both positively and negatively charged groups as well as hydrophobic groups. The negatively charged groups of EPS include phosphates, sulfates, carboxyl groups, glutamic acid, and aspartic acid, whereas the positively charged groups include amino sugars such as polysaccharide intercellular adhesin (PIA). Despite positively charged PIA being a major component, the overall charge on the EPS surface is negative, serving as a better target site for positively charged drug candidates (Idrees et al., 2021).
The major EPS produced by S. aureus is polysaccharide intercellular adhesin (PIA), also known as poly-N-acetyl-glucosamine (PNAG; Mack et al., 1996). PIA/PNAG has a net positive charge and it promotes intercellular interactions by binding to the negatively charged surfaces of bacterial cells such as teichoic acids (O'Gara, 2007; Vergara-Irigaray et al., 2008). The multivalent electrostatic interaction of the cationic PIA polymer with the negatively charged wall teichoic acids on staphylococcal cells as revealed by single-cell force spectroscopy confirmed that the cationic nature of PIA is crucial for its attachment to the cell surface and intercellular adhesion (Formosa-Dague et al., 2016). PIA mutants exhibited a decreased ability to adhere to each other (Peng et al., 2022). There is no evidence for a covalent linkage of PIA to the cell surface (Cue et al., 2012). PIA is evident for biofilm formation under high-shear flow conditions such as those found inside catheters as compared to low-shear conditions such as those in subcutaneously implanted tissue, ocular infections, or platelet concentrate (Nguyen et al., 2020).
In addition to PIA/PNAG, biofilms contain bacterial proteins, eDNA, ions, and carbohydrates (Guzmán-Soto et al., 2021) as essential components with their ratios being variable. A number of staphylococcal strains exhibit PIA/PNAG-independent biofilm formation where the secreted proteins and extracellular DNA substitute for PIA/PNAG (Cue et al., 2012).
Extracellular DNA (eDNA) is another important structural component of biofilm matrix serving as a “glue” for the community due to its adhesive property. The destabilization of a matured biofilm by using DNAase I in Pseudomonas aerugionosa reported by Whitchurch et al. (2002) suggested the role of eDNA in biofilm formation in pathogenic bacteria. In a matured biofilm when there is a scarcity of nutrients, a subsequent population of damaged cells is eliminated in order to release nutrients for healthy cells. This self-destructive or suicidal act of cells is termed as autolysis or programmed cell death (PCD; Lewis, 2000). Along with the nutrients, the lysed cells also release genomic DNA in the form of eDNA. Rice et al. (2007) reported that only a small fraction of the bacterial population (< 1%) within the biofilm undergoes cell lysis to release a sufficient amount of eDNA required for biofilm stability (Bayles, 2007). eDNA can also confer antibiotic resistance mainly due to the horizontal gene transfer of eDNA to the healthy cells of biofilm (Molin and Tolker-Nielsen, 2003; Tetz et al., 2009).
Capsular polysaccharides (CPs) are long-chain polysaccharides attached covalently to the peptidoglycan layer of the cell wall. They aid in the colonization of the host as well as in biofilm formation, hence are indirectly involved in the progression of invasive diseases. However, the contrasting effect on the virulence of S. aureus is observed by the presence or absence of a capsule depending upon the type of infection (O'Riordan and Lee, 2004; Tuchscherr et al., 2010). CP enhances virulence in murine models of bacteremia (Thakker et al., 1998; Watts et al., 2005), septic arthritis (Nilsson et al., 1997), abscess formation (Portols et al., 2001), and surgical wound infection (McLoughlin et al., 2006). On the contrary, in mammary gland infections (Tuchscherr et al., 2005) and in catheter-induced endocarditis (Baddour et al., 1992; Nemeth and Lee, 1995), CP mutants are more virulent. This is likely because CP also inhibits the adherence of the underlying adhesins to their specific target molecule (Phlmann-Dietze et al., 2000; Risley et al., 2007). CP-negative S. aureus strains are frequently isolated from patients with osteomyelitis, mastitis, or cystic fibrosis, providing evidence that the loss of CP expression (due to mutations in any of the genes essential for CP biosynthesis or in the promoter region; Cocchiaro et al., 2006; Tuchscherr et al., 2010) may be advantageous for S. aureus during chronic infection (Herbert et al., 1997; Lattar et al., 2009; Tuchscherr et al., 2010). CPs also help bacteria in evading the phagocytic uptake by the host immune system and also protect the important bacterial cell wall constituents (Berni et al., 2020). Out of 13 serotypes of S. aureus (Berni et al., 2020), serotypes 1 and 2 (rarely reported among clinical isolates) produce mucoid colonies while the remaining serotypes produce non-mucoid colonies on a solid medium. Serotypes 5 and 8 (prevalent among clinical isolates as well as commensal sources) constitute ~25 and 50%, respectively, of the isolates recovered from humans from various geographic locations of the world (O'Riordan and Lee, 2004).
Teichoic acids are one of the major components of Gram-positive bacterial cell wall. These diverse anionic carbohydrate-based polymers are categorized into two classes: (1) lipoteichoic acids (LTAs), which are embedded in the lipid bilayer with the help of a diacylglycerol lipid anchor that can extend from the cell surface to the peptidoglycan layer, and (2) wall teichoic acids (WTAs), which are covalently attached to peptidoglycan matrix via a phosphodiester linkage to the C6 hydroxyl of the N-acetyl muramic acid sugars and can extend through and beyond the cell wall (Swoboda et al., 2010; Berni et al., 2020). WTAs comprise ~60% of the total cell mass of Gram-positive bacteria. S. aureus WTAs are essential for adhesion to host tissues as well as to artificial surfaces including glass and polystyrene (Gross et al., 2001). WTAs null mutants are defective in their ability to produce biofilm; however, the reduced production of PNAG (an inevitable component of biofilm) was not reported, suggesting an independent role of WTAs in biofilm formation (Vergara-Irigaray et al., 2008). WTA mutant S. aureus strains were also unable to adhere and colonize the nasal epithelial cells as well as endothelial tissues of the kidney and the spleen derived from cotton rats. D-alanylation, which takes place on LTA, was reported to be intact on these strains, implying that WTAs were independently involved in cell adhesion and eventually biofilm formation (Weidenmaier et al., 2005). The involvement of WTAs in host colonization, infection, and biofilm formation makes it an important virulence factor and the enzymes involved in its biosynthesis can be novel targets for the discovery of new antimicrobials (Weidenmaier and Peschel, 2008).
Signaling molecules such as cyclic AMP (cAMP) are not only linked with the regulation of carbon metabolism and stringent response but also with the expression of virulence genes and biofilm formation in Gram-positive bacteria (Schilcher and Horswill, 2020). S. aureus requires two molecules of ATP and the enzyme diadenylyl cyclase DacA to synthesize c-di-AMP. Phosphodiesterase GdpP is involved in the degradation of c-di-AMP (Corrigan et al., 2011). Screening for essential genes of S. aureus revealed that dacA disruption was not possible, indicating the importance of c-di-AMP formation for the viability of the bacterium (Chaudhuri et al., 2009). On the other hand, gdpP deletion in the S. aureus SEJ1 strain yielded a 3-fold increase in biofilm formation, highlighting the fact that high levels of c-di-AMP induced biofilm formation in this strain. However, similar experimental conditions did not replicate the results in S. aureus USA300 LAC or its corresponding gdpP mutant (Corrigan et al., 2011). Single nucleotide polymorphisms (SNPs) or deletions in gdpP in various homogenous oxacillin-resistant (HoR) S. aureus isolates exhibited a decrease in the expression of icaADBC and agr and an increase in the expression of penicillin-binding protein 2 (PBP2). This resulted in an absence of polysaccharide content in the biofilm and the formation of a more proteinaceous biofilm (Pozzi et al., 2012). Apart from this, gdpP mutants were also impaired in their ability to form eDNA suggesting that c-di-AMP is essential for eDNA release as well (DeFrancesco et al., 2017). The role of c-di-AMP in biofilm formation may be strain dependent as different studies yielded different biofilm phenotypes in gdpP mutants of S. aureus (Schilcher and Horswill, 2020).
S. aureus possesses small α-helical peptides called phenol soluble modulins (PSM) which act as surfactants and disrupt cell–cell interactions within the biofilm better than proteases, unlike other bacteria that commonly use nucleases and proteases for biofilm detachment (Otto, 2008). The detachment and dispersal of planktonic cells are mediated by changes conferred in pH, nutrition, waste accumulation, oxygen depletion, etc. (Otto, 2008).
Once colonization is established, the bacteria adhere and start multiplying on wounded tissues, resulting in the upregulation of virulence genes and the production of toxins that further aid in disease progression. The initially expressed adhesion genes are now downregulated (Novick, 2003; Foster et al., 2014).
During the onset of infection, 95% of iron is within host cells in the form of serum iron bound to host proteins. In such a situation of iron starvation, S. aureus secretes high-affinity iron-binding proteins such as staphyloferrin (Drechsel et al., 1993) and aureochelin (Courcol et al., 1997). S. aureus can also initiate transcription of Isd (iron surface determinants)-mediated heme-iron transport that facilitates the release of heme from hemoglobin, haptoglobin, and hemopexin. This free heme transports across the plasma membrane via the iron complex and free iron is released in the cytoplasm of bacterial cell as a result of oxidative degradation (Maresso and Schneewind, 2006). This helps the bacterium in survival along with evasion from host defense (Liu, 2009).
Gram-positive bacteria including S. aureus secrete a diverse array of chemo-attractants such as peptidoglycan (Dziarski and Gupta, 2005), N-terminal lipoylated structure of lipoproteins (Nguyen and Götz, 2016), formylated peptides (Krepel and Wang, 2019), and unmethylated CpG sequences in DNA (Hemmi et al., 2000). This gradient of chemo-attractants induces proinflammatory signaling and activation of local immune response by recruitment of neutrophils and macrophages to the site of infection via a process called chemotaxis (Kolaczkowska and Kubes, 2013). Moreover, PSMs secreted by S. aureus sheds lipoproteins that also act as neutrophil chemo-attractants (Hashimoto et al., 2006; Hanzelmann et al., 2016). These structures collectively termed as “pathogen-associated molecular patterns” (PAMPs) are specific to bacteria and hence are recognized by the host immune system which activates Toll-like receptors (TLRs; Kumar et al., 2011).
A fibrin pseudo-capsule termed as abscess is formed within 2–6 days of infection (Kobayashi et al., 2015) with the help of coagulase proteins secreted by S. aureus which surround bacterial cells and recruited polymorphonuclear leukocytes (PMNs). After recruitment, S. aureus further inhibits neutrophil extravasation, activation, and chemotaxis in various ways. The binding of the members of the staphylococcal superantigen-like protein (SSL) family viz. SSL2 and SSL4 to TLR2, SSL10 to CXCR4, and SSL5 to GPCRs, such as P-selectin glycoprotein ligand- 1 (PSGL-1; which normally binds to the P-selectin anchor on endothelial cells) on the leukocyte surface, subsequently blocking neutrophil adhesion and extravasation to the site of infection. SelX protein has also been reported to have a similar function as SSL5 (Cheung et al., 2021). CHIP (chemotaxis inhibitory protein) blocks neutrophil recognition of chemotactic factors while Eap (extracellular adhesion protein) prevents neutrophil binding to endothelial adhesion molecule (ICAM-1; Chavakis et al., 2002; de Haas et al., 2004). S. aureus protease staphopain degrades CXCR2 (which recognizes cytokines), leading to the inhibition of neutrophil migration toward cytokines (Laarman et al., 2012). Another Geh lipase has been reported that removes the pro-inflammatory lipoylated N- terminus of the bacterial lipoproteins, thereby disguising these PAMPs from neutrophils (Chen and Alonzo, 2019).
Phagocytosis is also inhibited by biofilm formation via PIA/PNAG production (refer Section 2.1.1), protective surface structures such as capsules (refer Section 2.1.1), and aggregation. The combined non-redundant activity of coagulase and von Willebrand factor-binding protein (vWbp) of S. aureus produces a protein called thrombi which binds to prothrombin (factor II of the coagulation process) and forms a complex called staphylothrombin (Friedrich et al., 2003; Cheng et al., 2010). In the absence of a vascular damage signal, staphylothrombin cleaves fibrinogen from the host cells and forms fibrin clots. Fibrinogen-binding proteins of S. aureus such as clumping factor A (ClfA) bind to these clots resulting in the formation of fibrin-containing bacterial aggregates (McAdow et al., 2011). FnBPA and FnBPB can also activate the aggregation of platelets (Fitzgerald et al., 2006). S. aureus cells that are already phagocytosed by PMNs fight for their survival by releasing cytolytic toxins which cause pore formation and eventually cell lysis (Figure 1; Peschel and Sahl, 2006; Chambers and DeLeo, 2009; Liu, 2009; Löffler et al., 2010; Spaan et al., 2013; Stapels et al., 2014).
The expression of the capsule, clumping factor A, Protein A, and various other complement inhibitors on the cell surface of the bacterium may help to overcome opsonophagocytosis, i.e., the marking of the bacterial pathogen by antibodies (immunoglobulins, Igs) or complement factors for efficient phagocytosis. The Igs bind to phagocytes by the Fc region and to the pathogen by the Fab region. The presence of Igs on the bacterial surface not only marks it for opsonization but also activates the classical pathway of complement fixation (Cheung et al., 2021). S. aureus produces three proteins to overcome opsonization by antibodies. (1) Surface protein A (SpA) non-specifically binds to the Fc region of IgG (Forsgren and Sjöquist, 1966) and Fab region of IgM, which acts as a B cell superantigen and causes B cell apoptosis. It also initiates the production of plasma B cells that specifically recognizes only protein A, thereby diverting the immune response away from other virulence factors (Goodyear and Silverman, 2003; Pauli et al., 2014). (2) Sbi (S. aureus binder of IgG) binds to the complement factor H and C3 apart from the Fc region of IgG (Zhang et al., 1998; Atkins et al., 2008). (3) SSL10 also binds to the Fc region of IgG and prevents receptor-mediated phagocytosis (Itoh et al., 2010).
All three pathways (lectin, classical, and alternative) of the complement system possess the C3 convertase which cleaves C3 into C3a and C3b. The deposition of C3b on the bacteria marks it for opsonization. Other complement factors such as C5a (formed via the interaction of C3-C3b) act as a chemoattractant for the recruitment of more immune cells to the site of infection (Rooijakkers et al., 2005). Staphylococcal complement inhibitor (SCIN) inhibits C3 convertase, thus reducing the C3b deposition and C5a chemoattractant formation leading to the blockage of all three pathways of the complement system. Extracellular fibrinogen binding protein (Efb) of S. aureus binds to the C3 component via the C-terminus and fibrinogen via its N-terminus, covering the bacteria in a fibrinogen layer that inhibits the activation of the complement system as it fails to sense the surface-bound C3b (Ko et al., 2013). A homologous protein of Efb i.e., extracellular complement binding protein (Ecb), although lacking the fibrinogen binding activity, can inhibit the C3 convertase of the alternative pathway and all C5 convertases (Jongerius et al., 2010). Some S. aureus proteins such as collagen adhesion protein (Cna) inhibit the classical pathway (Kang et al., 2013), SdrE protein inhibits the alternative pathway (Sharp et al., 2012; Zhang et al., 2017) and the extracellular adherence protein (Eap) inhibits both lectin and classical pathways (Woehl et al., 2014). Finally, the SSL7 protein binds to the C5 component of the complement system as well as to the Fc region of the IgA antibody, inhibiting its recognition (Langley et al., 2005).
Proteolytic activity of various S. aureus proteins such as staphylococcal serine protease (V8 protease; SspA), cysteine protease (SspB), metalloprotease (aureolysin; Aur), and staphopain (Scp) have also been reported to inhibit opsonization (Dubin, 2002). Though the primary function of these proteases is nutrient acquisition, they may also destroy various immune defense proteins such as aureolysin cleaves C3 (Laarman et al., 2011) and V8 protease cleaves Igs (Rousseaux et al., 1983).
Despite the several mechanisms to evade phagocytosis, neutrophils can still manage to engulf S. aureus cells. Primary granules of neutrophil synthesize the enzyme Myeloperoxidase (MPO) which produces reactive oxygen species (ROS) and antimicrobial peptides (AMPs) such as defensins. Secondary granules of neutrophil secrete antimicrobial proteins such as lysozyme. S. aureus has evolved with various mechanisms to overcome both ROS and AMPs (Cheung et al., 2021). The orange pigment that gives S. aureus (aureus stands for golden) its name, Staphyloxanthin has been reported to scavenge the free radicals originating from ROS activity (Pelz et al., 2005; Clauditz et al., 2006). S. aureus synthesizes the enzyme superoxide dismutase which converts superoxide to less toxic H2O2 (Mandell, 1975; Clements et al., 1999). Furthermore, the catalase KatA and alkyl hydroperoxide reductase C AhpC detoxify the H2O2 by converting it into oxygen and water (Mandell, 1975; Cosgrove et al., 2007). MPO is directly inhibited by a staphylococcal peroxidase inhibitor (SPIN; De Jong et al., 2017).
Defensins and other AMPs are usually positively charged and hence are attracted to the negatively charged cell membrane and exhibit their bactericidal activity by forming pores in the bacterial membrane (Joo and Otto, 2015). The dlt operon of S. aureus esterifies hydroxyl groups in teichoic acids with alanyl residues which imparts an extra positive charge per alanine into the bacterial cell membrane, increasing the net charge and thereby inhibiting the binding of AMPs (Peschel et al., 1999). Moreover, a membrane-bound enzyme MrpF (multiple peptide resistance factor) adds Lys-PG on the outer layer of the cell membrane which also decreases the affinity of AMP binding (Peschel et al., 2001; Ernst et al., 2015). Finally, S. aureus secretes an enzyme named OatA, which acetylates the muramic acid residues of peptidoglycan reducing the efficacy of the neutrophil secreted antibacterial protein lysozyme which is otherwise very effective against other Gram-positive bacteria (Bera et al., 2005).
Neutrophil extracellular traps (NETs) are considered to be an integral component of the human immune system as they play a major role in host defenses during bacterial infections. NETs consist of activated neutrophils and DNA backbone along with proteins of various biological functions. NETs can ensnare but not kill S. aureus. The trapped bacteria from NETs can be released by degradation of the DNA backbone via S. aureus DNase leading to the persistence of the chronic infection. Eap, a protein secreted by S. aureus, can bind and aggregate linearized DNA, hindering the formation of NETs. Apart from this, the pathogen overcomes NET-mediated killing by expressing the surface protein FnBPB, which can neutralize the bactericidal activity of histones (Speziale and Pietrocola, 2021).
When encountered by neutrophils, S. aureus biofilms release the leukocidins such as LukAB and PVL (Panton-Valentine Leukocidin), which not only promotes S. aureus survival during phagocytosis but also induces the formation of NETs. Neutrophils and NETs can penetrate S. aureus biofilm but are unable to disrupt it (Malachowa et al., 2013). NETs can accumulate in organs and can cause tissue damage, as the infection progresses (Saffarzadeh et al., 2012; Weber, 2015). Therefore, the induction in the formation of NETs rather than blocking its antibacterial activity benefits the bacteria by favoring the colonization of deeper tissues, and thus providing better access to metabolic resources. This ensures the safer and optimal survival of bacteria in the host (Speziale and Pietrocola, 2021).
The abscess initially formed and matured within 6–14 days of infection accompanied by fibroblastic proliferation and tissue repair at the abscess margin and the formation of a fibrous capsule at the periphery (Kobayashi et al., 2015). The disruption of abscesses leads to the spread of S. aureus beneath the skin surface as well as in the bloodstream causing bacteremia. The bacterium can now adhere to endothelial surfaces and platelets causing endocarditis, metastatic abscesses, and bacterial uptake into endothelial cells where antibiotics and host defense molecules struggle to reach (Chavakis et al., 2005; Weidenmaier et al., 2005; Löffler et al., 2014). If endovascular spread cannot be controlled then systemic blood coagulation, massive production of microorganism-associated molecular pattern molecules (MAMP) and superantigen toxin-mediated cytokine storms can result in systemic inflammation, sepsis, multiple organ failure, and eventually death (Thomer et al., 2016; Lee et al., 2018).
S. aureus secretes a diverse array of virulence factors such as MSCRAMMs, hemolysins, leukotoxins, Protein A, exfoliative toxins, staphylococcal enterotoxins (SEs), and toxic-shock syndrome toxin-1 (TSST-1; refer Sections 3.1–3.6). Genes encoding these virulence factors (except MSCRAMMs) are located on the accessory genome, which comprises of mobile genetic elements (MGEs) such as plasmids, insertion sequences, pathogenicity islands, transposons, and prophages. These MGEs not only encode the genes for virulence factors production but also contain antibiotic-resistance determinants. Plasmids and transposons contain antibiotic-resistance genes while prophages and pathogenicity islands consist of toxins and other virulence determinants (Malachowa and DeLeo, 2010; Lindsay, 2019). The large family of staphylococcal pathogenicity islands (SaPIs) is widely known for enterotoxins and TSST. Toxins such as staphylococcal superantigen-like genes (SSLs), lipoprotein-like toxins (LPLs), α toxin, PSM peptides, leukocidin LukDE, and some enterotoxins are encoded on genomic islands and vary in their expression among different S. aureus isolates (Langley et al., 2010; Malachowa and DeLeo, 2010; Nguyen et al., 2015). Some other S. aureus toxins such as PVL, exfoliative toxins A and B, staphylokinase, immune evasion proteins such as CHIPS and SCIN, and several other enterotoxins are encoded on prophages (Cheung et al., 2021). β toxin encoding gene hlb has been reported to be non-functional in many S. aureus strains by the insertion of phage-encoding genes for CHIPS, SCIN, and staphylokinase (Carroll et al., 1993). This process is termed as “negative conversion” and it can be repaired by phage excision, which is important for infectious colonization (Katayama et al., 2013). Genomic islands such as vSAα, vSAβ, and vSAγ have also been reported to encode a diverse array of virulence factors. These MGEs have lost their ability to be transferred by non-MGE-specific mode of transfer and hence are now very stable and in fact are the characteristic of the entire species (Kläui et al., 2019). The genes encoding MSCRAMMs are located on the core genome, which is evident as they also exhibit general functions in the commensal lifestyle of S. aureus (Cheung et al., 2021).
S. aureus MSCRAMMs can bind to various proteins present in the extracellular matrix (ECM) of the host. Fibronectin-binding proteins, FnbPA and FnbPB, are responsible for bacterial attachment to fibronectin in vitro as well as binding to foreign bodies and plasma clots (Nizet and Bradley, 2011). Fibrinogen binding proteins or clumping factors, i.e., ClfA and ClfB (Ní Eidhin et al., 1998; O'Connell et al., 1998), are responsible for firm attachment of S. aureus to vascular thrombi in the situation of flow stress within the bloodstream (Fowler et al., 2000). Endocarditis studies in rats have reported reduced virulence in ClfA mutant S. aureus strains (Moreillon et al., 1995). Collagen-binding protein Cna is responsible for adherence to collagenous tissues such as cartilages (Patti et al., 1992). A Cna null mutant strain of S. aureus showed attenuated virulence in a murine septic arthritis model (Patti et al., 1994). Other important MSCRAMMs have been discussed in Table 1.
Hemolysins are toxins that lyse red blood cells as well as immune cells by binding to their specific receptors. Major classes of hemolysins include α, β, and γ hemolysins which are under the regulation of agr and sae locus. δ hemolysin (under the regulation of agr locus), also classified as Phenol Soluble Modulins (PSM), does not require a receptor to exhibit its hemolytic activity (Kong et al., 2016). α hemolysin/alpha toxin, encoded by the hla gene, is one of the major toxins under agr regulation. The alpha toxin, a 319 amino acid long pore-forming toxin, is shaped like a beta-barrel that can bind to disintegrin and metalloprotease 10 (ADAM10) receptor present on the cell membrane of the human host cell. α toxin can lyse red blood cells and leukocytes but is unable to neutralize neutrophils (Valeva et al., 1997). In animal models, hla mutant strains have been reported to cause less disease severity than the wild-type strains, suggesting the importance of hla toxin in staphylococcal infections (Wilke and Wardenburg, 2010; Berube and Bubeck Wardenburg, 2013). β hemolysin is a non-pore-forming toxin that can hydrolyze sphingomyelin, lyse erythrocytes (at low temperature), and monocytes but not lymphocytes and granulocytes (Walev et al., 1996). Its mode of action is still unclear, but it has been hypothesized that as the toxin acts as a sphingomyelinase, it may destabilize the bilipid layer of the plasma membrane of host cells, leading to an alteration in plasma membrane fluidity (Vandenesch et al., 2012). γ hemolysin is a bi-component toxin consisting of two polypeptide chains namely S (slow, HlgA, or HlgC) and F (fast, HlgB). The F component binds to the phosphatidylcholine of host cells, whereas the S component causes cell lysis by binding to the cell membrane of host cells (Meyer et al., 2009). Vandenesch et al. (2012) have reported the lysis of rabbit RBC as well as leukocytes such as macrophages, neutrophils, monocytes, and granulocytes by γ hemolysin. δ hemolysin/phenol soluble modulins (PSMs) are the only peptide toxins of S. aureus whose expression is under the direct control of AgrA and independent of RNAIII (Queck et al., 2008). δ toxin is small, amphipathic, and possesses a high affinity to lipids (Vandenesch et al., 2012). These multifunctional peptides produced by many S. aureus strains are hemolytic to erythrocytes, various organelles, bacterial protoplasts, and spheroplasts (Verdon et al., 2009). PSMα is a strong pro-inflammatory toxin that can lyse neutrophils post-phagocytosis and also contributes majorly to biofilm formation (Otto, 2014).
Leukotoxins such as LukDE, LukGH (LukAB), and Panton-Valentine Leukocidin (PVL) are pore-forming toxins under the regulation of sae and agr locus (Queck et al., 2008; Cheung et al., 2011; Alonzo and Torres, 2014). Alonzo et al. (2013) have reported lysis of phagocytic cells such as macrophages, dendritic cells, and lymphocytes by binding of LukDE toxin on the CCR5 chemokine receptor present in these immune cells. LukGH (LukAB), similar to PSMα peptide, binds to CD11b receptor in humans and causes lysis of immune cells after phagocytosis (DuMont et al., 2013). Panton-Valentine Leukocidin (PVL), commonly found in community-acquired methicillin-resistant Staphylococcus aureus (CA-MRSA), is a two-component toxin (LukS and LukF proteins that binds to TLR2 and TLR4, respectively, in animal models, and C5aR and C5L2 in humans) responsible for causing pore formation in the leukocyte cell membrane, which eventually results into cell lysis and tissue necrosis followed by skin and soft tissue infections (SSTIs; Rasigade et al., 2010). PVL has been reported to be 100-fold more potent than other leukotoxins (Kong et al., 2016).
Protein A (spa) is one of the major proteases among the agr downregulated surface proteins (Cheung et al., 2011). It can activate the TNFα receptor (TNFR1) in the lung parenchyma, resulting in a pro-inflammatory response (Gómez et al., 2004). Once released from the S. aureus envelope, Protein A can combat the host's humoral immune response by blocking Fc receptor-mediated opsonophagocytosis and may trigger apoptosis (refer Section 2.1.2; Peterson et al., 1977; Goodyear et al., 2006; Pauli et al., 2014; Le and Otto, 2015).
ETs are serine proteases (under the regulation of agr locus) that cleaves the protein desmoglein 1, which leads to the disruption of desmosomal cell linkages causing the detachment of skin epidermis, which results in a surge of infection (Eyre and Stanley, 1987; Hanakawa et al., 2002). ETs are also superantigens but less potent compared to other superantigens such as TSST-1 (Monday et al., 1999). ETs are responsible for a disease named staphylococcal scaled skin syndrome (SSSS), which majorly infects neonates and infants; however, adults who are immunocompromised or have renal impairment are also prone to it. The major symptoms include blistering of the skin, loss of superficial skin layers, dehydration, and other secondary infections (Bukowski et al., 2010).
Staphylococcal enterotoxins (SEs), secreted by entero-toxigenic S. aureus strains, are one of the most common causes of foodborne diseases as these toxins are heat stable. Based on antigenic heterogeneity, there are more than 24 different SEs identified that are under the regulation of agr and sae locus (Grispoldi et al., 2021). SEA to SEE, SEG to SEI, and SER to SET have reported emetic activity, while staphylococcal-like (SEl) proteins (SElL and SElQ) are not emetic in animal models, whereas SElJ, SElK, SElM to SElP, SElU, SElU2, and SElV are yet to be tested (Argudín et al., 2010). Ses induce cytokine release by T-cell activation and proliferation causing cell death via apoptosis and toxic shock syndrome (Balaban and Rasooly, 2000; Lin et al., 2010). SEF has been renamed as toxic shock syndrome toxin (TSST-1), widely known as a superantigen that can bind to HLA class II molecule on antigen-presenting cells and on T cell receptors leading to massive T-cell activation, proliferation, and release of cytokines (referred to as “cytokine storm”) viz., IL-8, MIP-3, IL-2, and TNFα inducing apoptosis and cell death due to lethal toxic shock. In lethal cases, TSST can lead to severe shock, organ dysfunction, and eventually death (Fraser and Proft, 2008; Otto, 2014; Stach et al., 2014). An epidemic was reported in the US from 2004 to 2014, which was found to be common among women using high-absorbency tampons in menstruation (mTSS). However, the incidence rate of mTSS was significantly reduced after the change in manufacturing and awareness regarding the use of tampons (Sharma et al., 2018). Currently, around 50% of TSS infections (such as skin and soft tissue infections) are due to non-menstrual toxic shock syndrome (nmTSS; Sharma et al., 2019).
The pathogenesis of S. aureus is largely dependent on the formation of biofilm and the production of virulence factors. The genetic regulatory network of biofilm and virulence in S. aureus is quite complex including a cross-regulation between different components viz. MSCRAMMs, ica locus, cid/lrg network, tar genes, cap operon, codY, SarA, and agr and sae TCS (Figure 2). During the onset of infection, the genetic systems responsible for initial colonization (viz. MSCRAMMs genes) and biofilm formation (viz., ica locus, cid/lrg network, and tar genes) are upregulated facilitating adherence to host tissue. As the infection progresses, there is a scarcity of nutrition and oxygen in the matured biofilm (due to an increase in bacterial cell density) that relieves the repression mediated by CodY (a global transcriptional regulator in S. aureus which represses the virulence genes with respect to nutrient availability and metabolism). Consequently, there is an upregulation of genetic systems responsible for the production of toxins (agr and sae TCS) that aid in acquiring nutrition, evading immune cells, and spread of infection. This suggests that apart from regulatory proteins, environmental factors such as nutrition, oxygen, pH, and reactive oxygen species (ROS) can also play a significant role. In this instance, the adhesion factors expressed initially are downregulated leading to the dispersal of biofilm. The dispersed planktonic cells can now adhere and colonize to other sites and spread the disease. The up- and downregulation of adhesion genes involved in biofilm formation and genes responsible for synthesizing virulence factors during different stages of infection act as a genetic regulatory see-saw in the pathogenesis of MRSA (Figure 3).
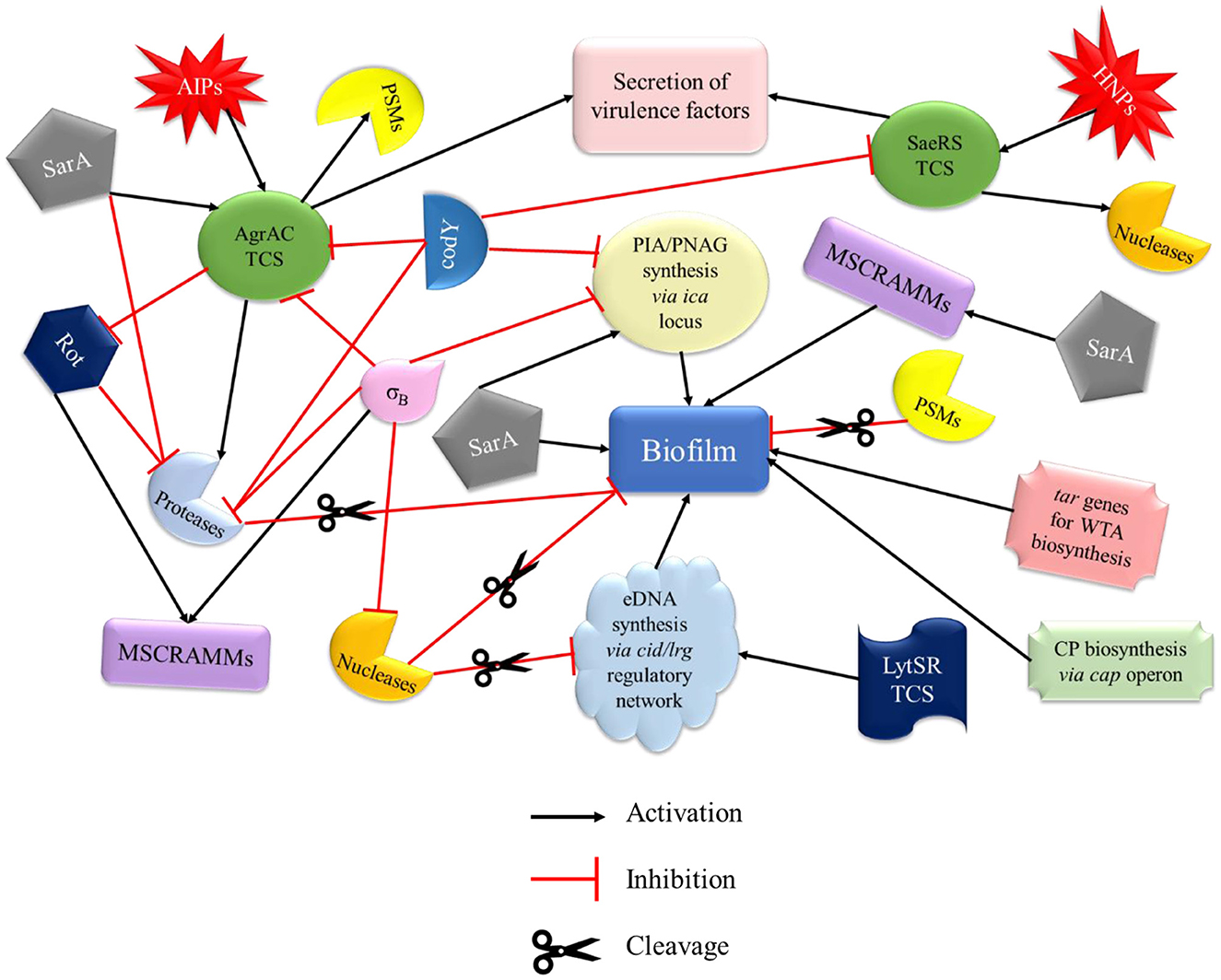
Figure 2. Regulatory network of biofilm and virulence in MRSA. The regulation of biofilm formation and virulence factors of MRSA involves a cross-talk of various systems that drive its pathogenesis. MSCRAMMs, PIA synthesis, CP, WTA biosynthesis genes, and SarA promote biofilm formation. PSMs and proteases (activated by AgrAC TCS) are directly involved in the disruption of biofilm, whereas nucleases (activated by SaeRS TCS and involved in the cleavage of eDNA), σB, and codY (by inhibiting PIA synthesis) play an indirect role. Agr and Sae TCSs, which are mainly involved in the secretion of virulence factors of MRSA, are primarily activated by AIP and HNP, respectively. AgrAC TCS can also be activated by SarA and repressed by σB and codY. codY is also involved in the downregulation of SaeRS TCS. MSCRAMMs, microbial surface components recognizing adhesive matrix molecules; PIA, polysaccharide intercellular adhesin; CP, capsule; WTA, wall teichoic acid; PSMs, phenol soluble modulins; AIP, autoinducing peptides; HNP, human neutrophil peptides; Rot, repressor of toxins.
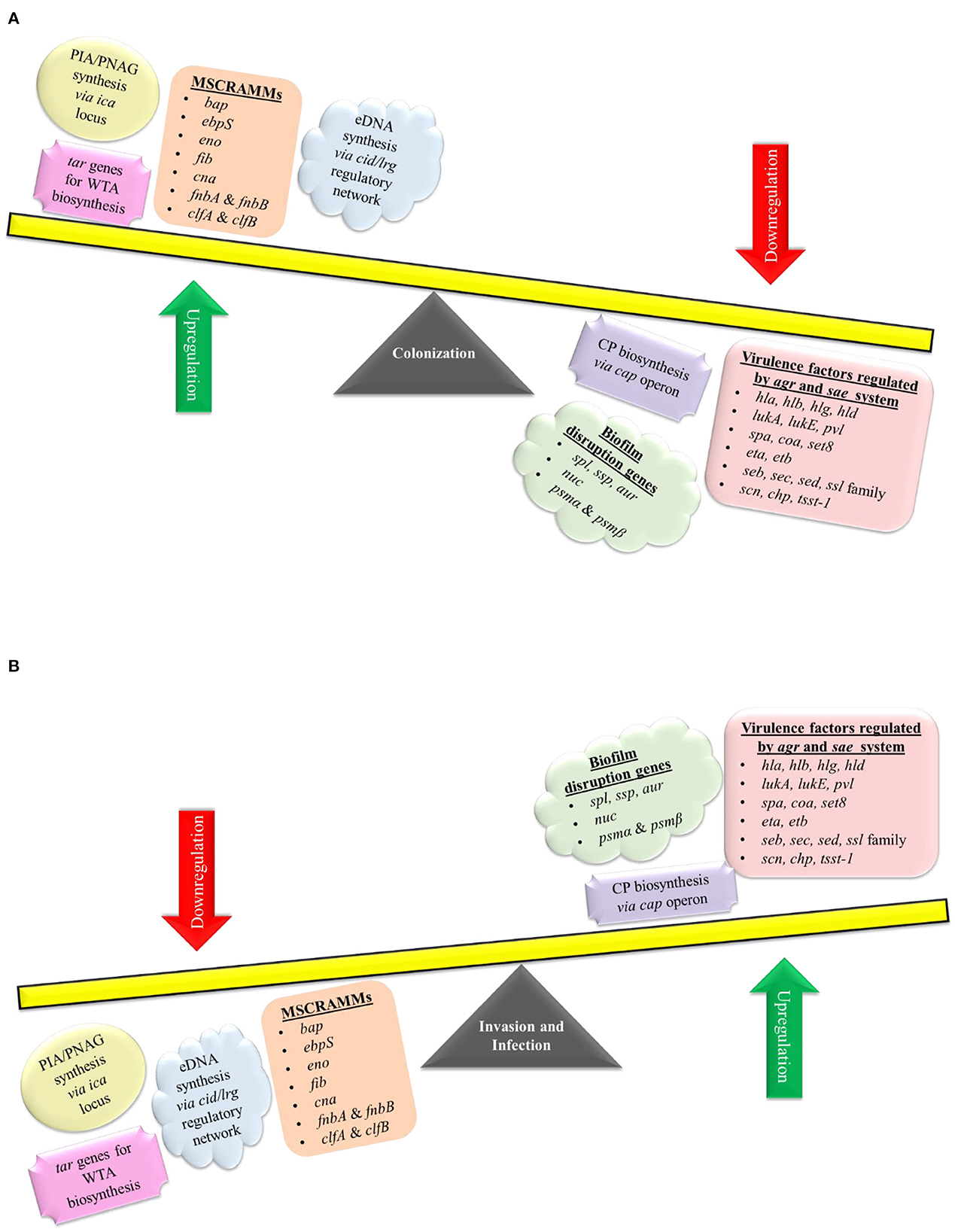
Figure 3. Genetic regulation of pathogenesis. S. aureus pathogenesis acts like a see-saw mechanism that involves an up- and downregulation of biofilm and virulence genes during different stages of infection. (A) For colonization, S. aureus upregulates ica locus, cid/lrg network, tar genes, and MSCRAMMs genes (bap, ebpS, eno, fib, cna, fnbA, fnbB, clfA, and clfB), which aids in adherence to host tissues as well as biofilm formation. Genes involved in biofilm disruption and virulence factor production are downregulated during colonization. As the infection progresses there is a scarcity of nutrition and oxygen in the matured biofilm, which leads to an (B) upregulation of biofilm disruption genes (spl, ssp, aur, nuc, psmα, and psmβ) and genetic systems such as cap operon, agr & sae TCS (which regulates genes such as hla, hlb, hlg, hld, lukA, lukB, pvl, spa, coa, set8, eta, etb, seb, sec, sed, ssl family genes, scn, chp, and tsst-1) that aid in acquiring nutrition, evading immune cells and spread of infection. Genes involved in colonization and biofilm formation are downregulated during the invasion and infection process.
The structural proteins, namely IcaA, IcaD, IcaB, and IcaC encoded by the ica operon synthesize PIA/PNAG. IcaA and IcaD are transmembrane proteins that simultaneously work as N-acetylglucosaminyltransferase-converting NAG monomers to PNAG oligomers of < 20 residues in length. The membrane-bound IcaC protein transports the PNAG oligomers across the cell membrane. Bacterial cell surface-associated IcaB protein deacetylates the PIA/PNAG oligomers that impart a positive charge to them and thus facilitates binding with the negatively charged bacterial cell surface (Cue et al., 2012).
The expression of the icaADBC operon is regulated by various direct and indirect factors. SarA and σB are global regulatory proteins of the operon, whereas some local proteins such as IcaR and TcaR regulate comparatively fewer genes. IcaR directly regulates the icaADBC operon, whereas proteins such as Rbf and Spx have an indirect effect (Cue et al., 2012; Figure 4).
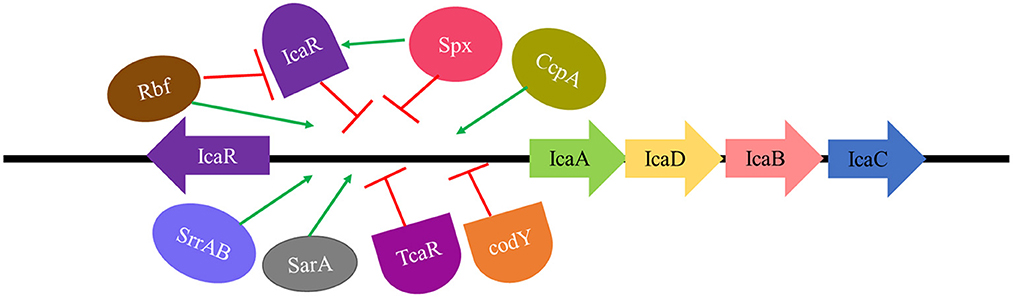
Figure 4. Regulation of ica locus. SarA, SrrAB TCS, and CcpA are direct activators, whereas Rbf exhibits an indirect effect in ica locus activation by repressing IcaR (repressor of ica locus). IcaR, TcaR, and codY are direct inhibitors whereas Spx exerts an indirect effect on ica locus repression by activating IcaR (repressor of ica locus).
(i) IcaR
IcaR repressor gene (which encodes the IcaR protein) is transcribed in a divergent manner from the other four genes (Conlon et al., 2002). The binding of IcaR to a DNA region immediately 5′ to the icaA gene and deletion of the short nucleotide sequence of 5 bp (TATTT) within the icaA-icaR intergenic region could significantly increase the expression of the icaADBC. icaR deletion can cause a 100-fold increase in ica locus expression and a 10-fold increase in PNAG/PIA production (Jefferson et al., 2004).
(ii) Sar A
SarA is a global regulatory protein as it affects the expression of many pathogenic genes in S. aureus, hence making it a major virulence factor. SarA protein can function as both activator and repressor of transcription (Bayer et al., 1996; Beenken et al., 2003; Weidenmaier et al., 2005; Oscarsson et al., 2006). SarA can directly bind to the ica promoter, enhancing PIA/PNAG production and, subsequently, biofilm formation (Valle et al., 2003; Jefferson et al., 2004; Tormo et al., 2005). The most important role that SarA plays in promoting biofilm formation is the repression of extracellular proteases and nucleases (Schilcher and Horswill, 2020). An elevated expression of proteases was reported in sarA mutants (Mrak et al., 2012; Zielinska et al., 2012), which also exhibited a decreased affinity for fibronectin binding (Blevins et al., 2002) and an inability to form static or flow cell biofilm (Beenken et al., 2003). Inhibition of all three classes of proteinases i.e., serine, cysteine, and metalloprotease (Tsang et al., 2008), or concurrent mutation of four extracellular proteases i.e., Aur, ScpA, SspA, and SspB can restore biofilm formation in sarA mutants (Loughran et al., 2014).
(iii) TcaR
TcaR is a member of the MarR family of transcription factors of S. aureus. Jefferson et al. (2004) demonstrated, via northern analysis, a 5-fold increase in transcription of the ica locus by the inactivation of the tcaR gene. The bacterial adherence and PIA/PNAG production were found to increase up to 500-fold in the icaR tcaR double mutant, indicating that tcaR is a negative regulator of the ica locus.
(iv) σB
σB is an alternative sigma factor found in S. aureus and other Gram-positive bacteria which is activated by signal transduction in response to environmental stress such as high temperature, extreme pH, high osmolarity, and use of antibiotics (Donegan and Cheung, 2009). Rachid et al. (2000) reported the role of σB in biofilm formation under high salinity and osmotic stress in an S. aureus mucosal isolate. σB has been reported to have a role in the expression of ica operon. Valle et al. (2003) reported that sarA- σBdouble mutants showed a decrease in icaADBC expression, but, on the other hand, also showed a significant increase in PIA/PNAG production and biofilm formation as compared to single sarA mutants. These results suggested the role of σB in the upregulation of a factor directly involved in the turnover of PIA/PNAG. An enhancement in PIA-dependent biofilm formation has been reported in σB mutants due to an increased accumulation of IcaC protein (Valle et al., 2019).
The available literature on σB-mediated regulation of PIA-dependent biofilm formation is contradictory. Cerca et al. (2008) studied icaADBC and icaR expressions in σB mutant SA113 and Newman strains of S. aureus. They concluded that σB was a positive regulator of both icaADBC and icaR transcripts. IcaR has been reported to be a weak repressor of icaADBC in S. aureus strains. Rachid et al. (2000) reported a decrease in the transcription of the ica locus in the sigB mutant of the MSSA MA12 strain. A biofilm-negative phenotype of the sigB mutant SH1000 and USA300 strain LAC (CA-MRSA isolate) was found to have no significant changes in the ica- dependent PIA formation but showed an enhanced Agr and protease activity (Lauderdale et al., 2009). However, both these strains were capable of forming PIA-independent biofilms (Boles and Horswill, 2008; Pozzi et al., 2012; Atwood et al., 2015). These contradictory results may be due to variations in some strain-specific properties, and the use of different media as well as biofilm models used in these studies (Schilcher and Horswill, 2020).
(v) Rbf
Rbf (regulator of biofilm), a member of the AraC/XylS family, is a transcriptional regulatory protein that plays a critical role in biofilm formation in S. aureus (Lim et al., 2004). Rbf positively regulates icaADBC transcription by inhibiting icaR expression (Gallegos et al., 1997). Cue et al. (2009) analyzed whether the activation of icaADBC mediated by Rbf is through direct binding to ica promoter or by repression of icaR. The results indicated that Rbf was unable to directly bind to the icaADBC promoter, suggesting that the expression of icaADBC by Rbf is indirect via inhibition of the expression of icaR.
(vi) Spx
Spx protein is a global transcriptional regulator that can act both as an activator as well as a repressor. It blocks transcription by directly binding to the α subunit of RNA polymerase, thereby inhibiting its interactions with target genes (Nakano et al., 2010). Spx mutants were found to exhibit increased transcription of icaADBC along with a decrease in icaR transcription. Thus, it can be concluded that Spx downregulates icaADBC expression by upregulating icaR expression. However, the exact mechanism of increased expression of icaR by Spx is still unclear (Pamp et al., 2006).
(vii) SrrAB
SrrAB is a two-component system (TCS) that prevents the phagocytosis of S. aureus by neutrophils in anaerobic conditions which is often found in the core region of a mature biofilm. Ulrich et al. (2007) demonstrated that phosphorylated SrrA protein can bind to 100 bp upstream of the promoter of ica operon resulting in an increase in its expression. Under anoxic conditions, the SrrAB mutant showed downregulation of icaA transcription and PIA/PNAG production. However, the SrrAB mutation did not exhibit any change in icaR expression, suggesting the direct activation of the icaADBC by SrrAB.
(viii) CcpA and CodY
The expression of icaADBC in S. aureus is also affected by changes in the levels of the TCA cycle intermediates with respect to the metabolic state of the cell (Vuong et al., 2005). The genes encoding for enzymes involved in the TCA cycle are usually repressed by CcpA in the presence of high glucose concentration among the majority of Gram-positive bacteria including S. aureus. High-intracellular levels of glucose-6 phosphate and fructose 1,6 bisphosphate regulate the activity of CcpA, which has been reported to be an activator of icaADBC. The synthesis of branched-chain amino acids is repressed due to the downregulation of the TCA cycle in high-glucose conditions. CodY transcriptional regulatory protein, which is responsive to branched-chain amino acids and a repressor of ica operon, is also downregulated, resulting in further activation of icaADBC and enhanced biofilm formation. When the biofilm matures and nutritional scarcity arises then CcpA activity is repressed, TCA cycle and branched chain amino acid synthesis genes are upregulated, resulting in the activation of codY and inhibition of icaADBC and, subsequently, biofilm formation. Thus, CodY and CcpA are both regulators of icaADBC expression in S. aureus (Seidl et al., 2008; Fujita, 2009). Mlynek et al. (2020) suggested that PIA synthesis in cells with low codY activity majorly contributes to biofilm formation. CodY regulates PIA-dependent biofilm formation and codY mutant strains exhibit different biofilm phenotypes. For instance, a transposon insertion in the codY gene of the clinical isolate S30 revealed reduced biofilm formation and PIA production (Tu Quoc et al., 2007). On the contrary, a codY allelic replacement mutation in two S. aureus clinical isolates SA564 and UAMS-1, reported an elevated biofilm formation, probably due to higher transcription of icaA and increased PIA synthesis during in vitro growth (Majerczyk et al., 2008). However, CodY-mediated regulation of the icaADBC locus is independent of IcaR (Majerczyk et al., 2008), SarA, and RNAIII (Majerczyk et al., 2010).
cidABC and lrgAB operons of S. aureus encodes bacteriophage such as holins and antiholins, respectively (Brunskill and Bayles, 1996b). These operons have also been reported to have a crucial role in staphylococcal murein hydrolase activity (Groicher et al., 2000; Rice et al., 2003). In bacteriophages, holins are responsible for the transport of murein hydrolases across the cytoplasmic membrane for cell lysis and antiholins have an inhibitory activity on the holins (Young, 1992). Similarly, in S. aureus the cell lysis and eDNA release are controlled by the antagonistic activity of cid and lrg gene products. Mutations in either cid or lrg operons lead to variation in biofilm formation which suggests that fine-tuning between survival and death is essential for a robust biofilm formation (Rice et al., 2007; Mann et al., 2009).
The cell death and lysis of S. aureus controlled by the cid/lrg system are under the regulation of another TCS, i.e., LytSR and transcriptional regulator cidR (Figure 5).
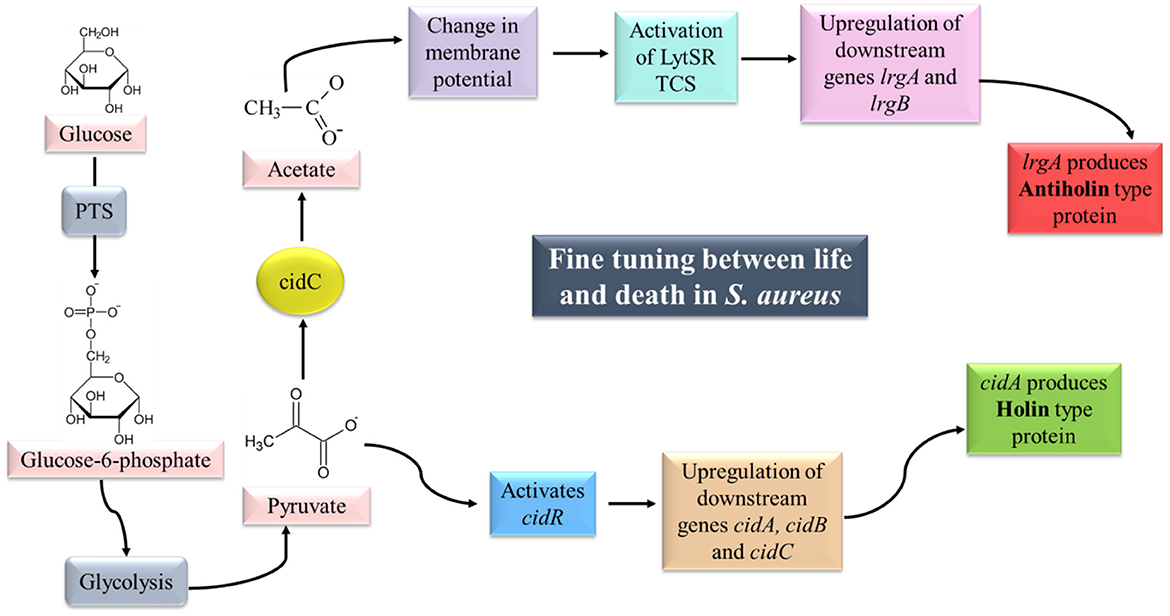
Figure 5. cid/lrg system and its regulation. LytSR TCS is activated by sensing a change in membrane potential (due to the conversion of pyruvate to acetate by CidC). Phosphorylated LytR results in upregulation of the lrgAB operon which is responsible for the production of Antiholin-like protein. Pyruvate on the other hand can activate cidR which can upregulate the cidABC operon, responsible for the production of Holin-like protein. Synthesis of Holin and Antiholin-like proteins maintain a fine tuning between life and death in S. aureus (adapted from Bayles, 2007; Sadykov and Bayles, 2012).
LytSR is a two-component regulatory system composed of a sensory histidine kinase protein (LytS) and a response regulator (LytR), which has been reported to control the expression of lrgAB operon (Brunskill and Bayles, 1996a; Sharma-Kuinkel et al., 2009). LytS senses the change in membrane potential of the cell which may be caused due to accumulation of acetate in the media under conditions of excessive glucose and oxygen (overflow metabolism; Rice et al., 2005; Patton et al., 2006). Activated LytS in turn phosphorylates the response regulator LytR, which can induce lrgAB expression. Mutation in lytSR genes has been reported to cause increased levels of autolysis due to increased murein hydrolase activity (Brunskill and Bayles, 1996a). Hence, LytSR-mediated control over the lrgAB operon is essential to regulate the autolysis phenomenon in biofilm.
cidR is a LysR type of transcriptional regulator (LTTR) responsible for DNA binding. Yang et al. (2005) reported two overlapping transcripts (cidABC and cidBC) of cid operon as revealed by northern blot analysis. The transcription of cidBC is induced by σB, while the transcription of cidABC is dependent on CidR and CcpA (Sadykov et al., 2019). The presence of excess glucose and/or acetic acid induces the transcription of cidABC. CidC-encoded pyruvate oxidase converts the end product of glycolysis, i.e., pyruvate into acetate (Patton et al., 2005). Pyruvate is also believed to serve as a co-inducer molecule for the activation of cidR (Schell, 1993).
In planktonic cells of S. aureus, the expression of cidABC is induced under excess glucose leading to a high rate of glycolysis which in turn inhibits aerobic respiration and diverts the carbon flow through fermentation pathways, known as the Crabtree effect. With enhanced murein hydrolase activity due to the activation of cidABC, holin-like proteins are produced that aid in the death and lysis of weakened cells. On the other hand, fermentative metabolism and CidC protein lead to the conversion of pyruvate into organic acids such as acetic acid. Acidification leads to disruption in the membrane potential of the cell which is sensed by LytSR TCS that further controls the expression of lrgAB. lrgAB transcription leads to the production of antiholin-like protein, thereby maintaining a fine balance between survival and death of S. aureus (Sadykov and Bayles, 2012).
The decrease in the expression of lrgA via induction of sigB by hyperglycemia-related factors such as advanced glycation end products (AGEs) secreted during diabetic foot infection (DFI) in S. aureus increases the release of eDNA, thereby enhancing biofilm formation (Xie et al., 2020). σB has also been reported to repress the expression of the secreted thermonuclease Nuc. Cell-free supernatants of a USA300 sigB mutant were found to inhibit the biofilm formation of different S. aureus strains. Subsequent fractionation and mass spectroscopy analysis revealed that Nuc was an active component in the supernatant which is responsible for the cleavage of eDNA, subsequently inhibiting biofilm formation. Kiedrowski et al. (2011) reported enhancement in biofilm formation in several S. aureus strains, including the USA300 lineage upon deletion of the nuc gene. The biofilm-negative phenotype of the sigB mutation has been observed to be partially repaired in a nuc-sigB double mutant.
CP5 and CP8 capsules (prevalent among S. aureus isolates) consist of repeating units of N-acetyl mannosaminuronic acid, N-acetyl L-fucosamine, and N-acetyl D-fucosamine. S. aureus have different serotypes due to the difference in glycosidic linkages between the sugars and the sites of O-acetylation of the mannosaminuronic acid residues of the capsule (O'Riordan and Lee, 2004; Kuipers et al., 2016). The pathway for capsule (CP) biosynthesis occurs in the cytoplasm via three distinct reactions, in which the universal cell envelope substrate UDP-D-N-acetylglucosamine (UDP-D-GlcNAc) is converted into the three different nucleotide-coupled sugars: UDP-N-acetyl-D-fucosamine (UDP-D-FucNAc), UDP-N-acetyl-L-fucosamine (UDP-L-FucNAc), and UDP-N-acetyl-D-mannosaminuronic acid (UDP-D-ManNAcA; Figure 6; Rausch et al., 2019).

Figure 6. Cascade of capsule biosynthesis. The pathway for capsule (CP) biosynthesis occurs in the cytoplasm via three distinct reactions in which the universal cell envelope substrate UDP-D-N-acetylglucosamine (UDP-D-GlcNAc) is converted into the three different nucleotide-coupled sugars: (1) UDP-N-acetyl- D-fucosamine (UDP-D-FucNAc) by the enzymes CapD and CapN, (2) UDP-N-acetyl-L-fucosamine (UDP-L-FucNAc) by the enzymes CapE, CapF, and CapG, and (3) UDP-N-acetyl-D-mannosaminuronic acid (UDP-D-ManNAcA) by the enzymes CapP and CapO. The enzymes CapK and CapJ translocate the modified precursor to the outer surface of the cell membrane where polymerization occurs (modified from Rausch et al., 2019).
The first reaction is catalyzed by the enzymes CapD and CapN that converts UDP-D-GlcNAc into the first soluble precursor UDP-D-FucNAc. The phosphosugar moiety of UDP-D-FucNAc is transferred to the membrane-anchored lipid carrier undecaprenyl-phosphate (C55P) by CapM, yielding lipid Icap (Li W. et al., 2014). The second reaction is catalyzed by the enzymes CapE, CapF, and CapG which convert UDP-D-GlcNAc into a second soluble precursor UDP-L-FucNAc. The transferase CapL further attaches L-FucNAc to lipid Icap, resulting in the formation of second CP lipid intermediate, lipid IIcap (Kneidinger et al., 2003). The third reaction is catalyzed by the epimerase CapP and the dehydrogenase CapO, which converts UDP-D-GlcNAc into a third soluble precursor UDP-D-ManNAcA (Kiser et al., 1999; Portols et al., 2001). The transmembrane protein CapI transfers the UDP-D-ManNAcA moiety to lipid IIcap, generating the final capsule precursor lipid IIIcap (Rausch et al., 2019). The modification of C55P coupled trisaccharide is carried out by acetyltransferase CapH, which catalyzes the O- acetylation of L-FucNAc residues at the C3 position in CP5 strains (Bhasin et al., 1998). The putative flippase CapK and the polymerase CapJ translocate the modified precursor to the outer surface of the cell membrane where polymerization occurs (Sau et al., 1997; O'Riordan and Lee, 2004). The attachment of CP precursors to the MurNAc (N-acetylmuramic acid) moiety of peptidoglycan occurs via an unknown mechanism that possibly involves a member of the LCP (LytR-CpsA-Psr) family of proteins (Kawai et al., 2011; Chan et al., 2014). This process has been assumed to release the lipid carrier C55P, which enters a new synthesis cycle (Rausch et al., 2019).
cap5 and cap8 are allelic gene loci that constitute a 17.5 kb region of the chromosomal DNA. Both loci consist of 16 linked genes [cap5A (cap8A) to cap5P (cap8P)], which are transcribed in one direction and are involved in biosynthesis, acetylation, transport, and regulation of capsule biosynthesis. In total, 12 out of these 16 genes are nearly identical in both loci (Sau et al., 1997; Rausch et al., 2019). The genes distinguishing CP5 and CP8 strains exhibit very little homology and are located in the central region of the loci (cap5H, cap5I, cap5J, and cap5K and similarly for cap8; O'Riordan and Lee, 2004). Wann et al. (1999) integrated cap5HIJK genes into the CP8 strain via homologous recombination resulting in a reciprocal loss of cap8HIJK. The recombinant strain started producing CP5, indicating that indeed cap5HIJK genes were responsible for the CP5 serotype.
cap8 and cap5 gene expression are both positively regulated by agr locus as well as sarA. Single mutants of agr and sarA as well as agr-sarA double mutant studies have reported agr locus to be a major regulator of cap8 gene expression. sarA gene was also found to be responsible for the activation of cap8 gene expression at the transcriptional level but its effect was minor as compared to agr (Luong et al., 2002). Positive regulation of cap5 gene expression by agr locus both in vitro and in vivo was reported in the rabbit endocarditis model. Similar to cap8 gene regulation, sarA was also found to exert a lesser positive impact on cap5 gene expression (van Wamel et al., 2002). Another global regulator mgr belonging to the MarR family of transcriptional regulators has been reported to upregulate CP8 biosynthesis and nuclease expression but downregulated the production of alpha toxin, protease, Protein A, and coagulase (Luong et al., 2003). The global repressor codY has also been reported to repress the cap operon under high-nutrient conditions, i.e., early and exponential growth phase (Pohl et al., 2009; Majerczyk et al., 2010). Phosphorylated SaeR (SaeRS TCS) binds to the promoter of the cap operon and represses both SigB- and SigA-dependent promoter activities (Keinhoerster et al., 2019). Environmental factors also play a major role in capsule expression both in vitro and in vivo. High-salt conditions, iron limitation, and growth on solid medium enhance the CP production, whereas high glucose, low oxygen, high CO2, alkaline conditions, and yeast extract repress the CP production. CP was reported to rarely express during ex vivo analysis of bacteria recovered from cystic fibrosis patients and only a few isolates were CP positive. This might be due to the high CO2 concentration in the lungs. Similarly, in cases of nasal colonization also, only a part of the S. aureus population was reported to be CP-positive (Keinhoerster et al., 2019).
WTA biosynthesis was first characterized in Bacillus subtilis 168, which makes poly(glycerol) phosphate, and hence the genes involved in its synthesis were known as tag genes (for teichoic acid glycerol). Bacillus subtilis W23 and S. aureus make poly(ribitol) phosphate and hence the genes involved in its synthesis were known as tar genes (for teichoic acid ribitol; Ward, 1981; Figure 7).
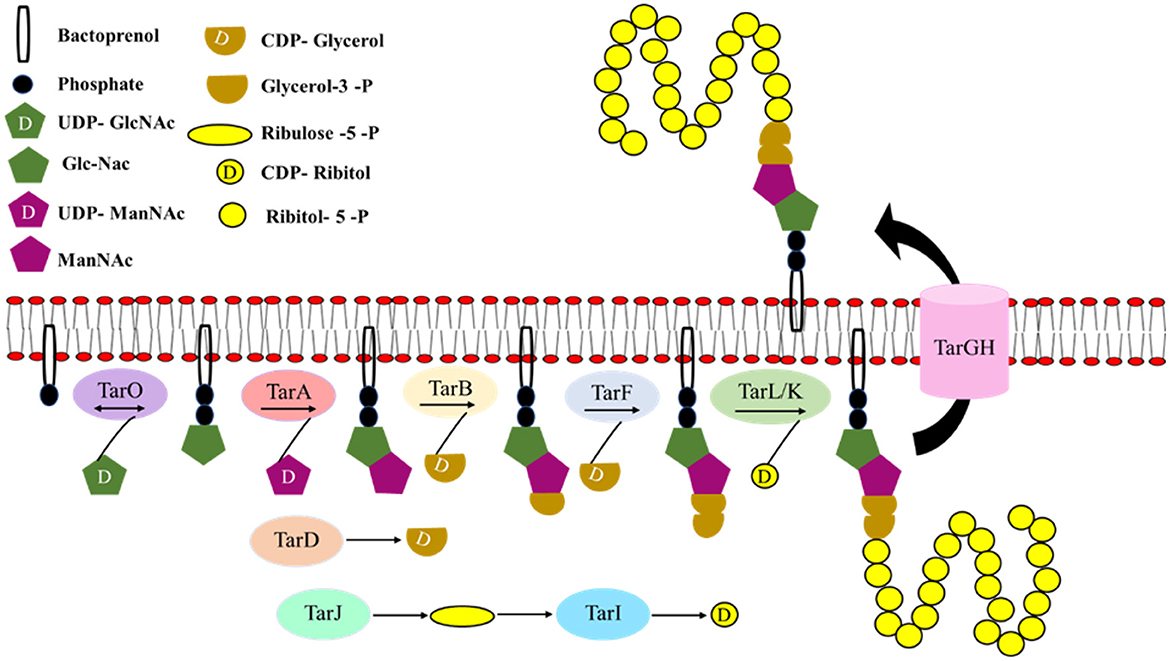
Figure 7. Cascade of wall teichoic acid biosynthesis. The first three steps of the WTA (wall teichoic acid) biosynthetic pathway are catalyzed by the enzymes TagO (TarO), TagA (TarA), and TagB (TarB), respectively leading to the formation of the linkage unit which comprises of a single phosphoglycerol unit from CDP-glycerol to the C4 hydroxyl of ManNAc-β1,4-GlcNAc disaccharide. TarF adds one additional glycerol phosphate unit to the linkage unit. The assembly of the poly(ribitol) phosphate main chain is carried out by one of the two bifunctional poly(ribitol) phosphate primases/polymerases known as TarK/TarL. Once the polymerization is completed in the cytoplasm, glycosylation occurs and the polymer is flipped out by an ABC-dependent transporter complex TarGH followed by which ligation to the cell wall occurs (modified from Swoboda et al., 2010).
The first three steps of the biosynthetic pathway are catalyzed by the enzymes TagO (TarO), TagA (TarA), and TagB (TarB), respectively. In the cytoplasm, N-acetylglucosamine phosphate (GlcNAc-1 phosphate) is transferred to an undecaprenyl phosphate (bactoprenyl phosphate) by a reversible phosphosugar transferase enzyme, TagO (TarO; Soldo et al., 2002; Price and Tsvetanova, 2007). Furthermore, TagA (TarA), an N-acetylmannosaminyl transferase, catalyzes the transfer of ManNAc from UDP-ManNAc to the C4 hydroxyl of GlcNAc residue to form ManNAc-β1,4-GlcNAc disaccharide (Ginsberg et al., 2006; Zhang et al., 2006). Finally, TagB (TarB), a glycerophosphotransferase, catalyzes the transfer of a single phosphoglycerol unit from CDP- glycerol (synthesized by TagD or TarD) to the C4 hydroxyl of ManNAc to complete the synthesis of linkage unit (Ginsberg et al., 2006; Bhavsar et al., 2007). The WTA linkage unit is highly conserved in all Gram-positive bacterial strains characterized so far. After these first three initial steps, the WTA pathways diverge (Brown et al., 2013).
In S. aureus, TarF (homolog of TagF that acts as a polymerase), which acts as a primase, adds one additional glycerol phosphate unit to the linkage unit (Swoboda et al., 2010). The assembly of the poly(ribitol) phosphate main chain is carried out by one of the two bifunctional poly(ribitol) phosphate primases/polymerases known as TarK/TarL. S. aureus TarL is a bifunctional enzyme having both primase and polymerase activities (Meredith et al., 2008). A cytidylyl transferase TarI and an alcohol dehydrogenase TarJ together synthesize the CDP-ribitol substrate that is utilized by TarL. TarL attaches more than 40 ribitol phosphates to complete the polymer synthesis (Brown et al., 2013). tarK gene is highly homologous to the tarL gene, suggesting that it might have the same enzymatic function. The reason for the presence of two homologous sets of tarIJL genes (tarI'J'K genes) with the same function (Qian et al., 2006) in S. aureus is still not clear; however, Meredith et al. (2008) have reported that tarK can nullify the loss of tarL and have an enzymatic function similar to tarL the gene product. However, strains that produced only TarL or TarK produced two electrophoretically distinct poly (ribitol) phosphate WTAs, known as L-WTA or K-WTA, respectively. K-WTA was found to be significantly shorter than L-WTA based on PAGE analysis. tarK gene expression is negatively regulated by agr quorum sensing system. As the expression of tarK can shorten K-WTA length by 50%, it is presumed that WTA chain length is dynamically regulated by S. aureus between pro-adhesion and low adhesion state to promote adhesion and dispersal of biofilm during different stages of infection. Once the polymerization is completed in the cytoplasm, glycosylation occurs and the polymer is flipped out by an ABC-dependent transporter complex TarGH followed by which ligation to the cell wall occurs (Swoboda et al., 2010).
Highly pathogenic CA-MRSA strains express the increased level of WTAs by upregulating the TarH ATPase subunit of the TarGH ABC transporter. This “WTA-high phenotype” is expressed in highly virulent strains in which the agr quorum sensing system is upregulated (van Dalen et al., 2020). Wanner et al. (2017) have reported increased skin abscess formation, T-cell proliferation, and IFN-γ production in mouse skin models infected with WTA-high phenotype strains. When agr is active, L-WTA is expressed resulting in long chains that may aid in colonization and infection (Meredith et al., 2008). Cross-regulation of WTA and CP biosynthesis is another important factor as both processes compete for C55P lipid carriers. CP production is thus very tightly regulated and only expressed in post exponential phase that too in a fraction of the population in order to ensure sufficient C55P lipid carrier for WTA production which is an inevitable component of the bacterial cell wall (Keinhoerster et al., 2019).
The agr locus (first reported by Peng et al., 1988) is 3.5 kb in size and consists of two divergent promoters P2 and P3 that generates the transcripts of RNAII and RNAIII, respectively. RNAII locus comprises of four genes namely, agrB, agrD, agrC, and agrA. agrD transcript encodes a 7–9 amino acid long autoinducing peptide (AIP), which also plays a role in extracellular quorum sensing signal in S. aureus (Ji et al., 1995). AgrB is a multifunctional endopeptidase that is responsible for maturation (thiolactone modification and C terminal cleavage) and the export of AIP across the cell membrane. AgrC and AgrA constitute the two-component signal transduction system, of which AgrC is a membrane-bound histidine kinase, which is auto-phosphorylated upon the binding of AIP. It then trans-phosphorylates the response regulator AgrA. Activated AgrA can bind to the P2 and P3 promoters of agr locus and can regulate the expression of downstream genes (Novick et al., 1995; Queck et al., 2008; Le and Otto, 2015; Figure 8).
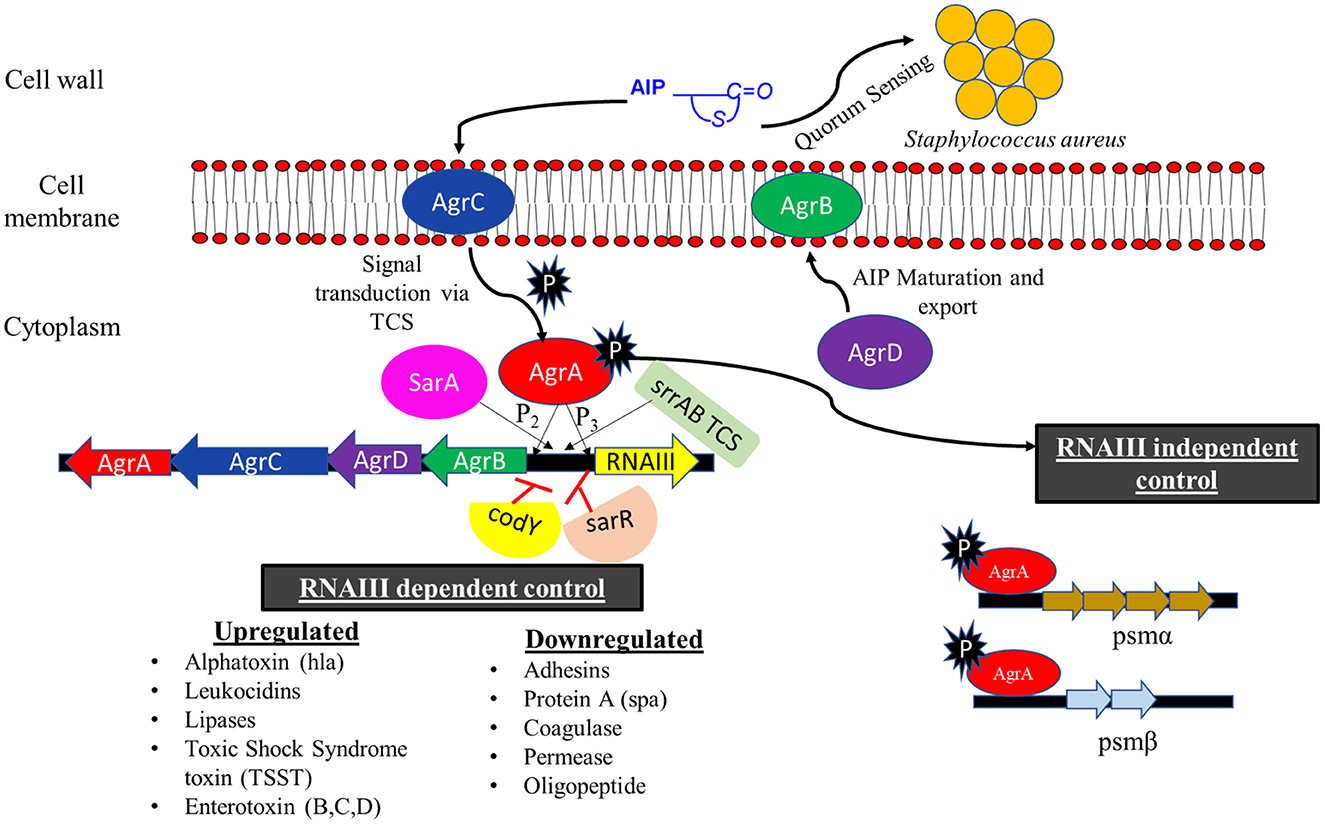
Figure 8. Regulation of agr (accessory gene regulator) two-component system. Autoinducing peptide (AIP) synthesized by AgrD is transported across the cell membrane by transmembrane protein AgrB. AIP acts as a signal for quorum sensing in S. aureus and can also activate the AgrAC TCS. The activated AgrA can bind to the P2 and P3 promoter of agr locus and initiate the transcription of agrACDB genes and RNAIII transcript, respectively. RNAIII transcript can mediate the expression of various virulence and biofilm disruption genes. The activated AgrA initiates the transcription of psmα and psmβ in RNAIII independent manner. SarA and SrrAB positively regulate the agr locus whereas codY and sarR exhibit a negative effect (modified from Le and Otto, 2015; Tan et al., 2018).
The majority of the virulence factors that are under the control of agr system are regulated by RNAIII. It is a messenger RNA that contains the hld gene for delta toxin or delta hemolysin (Janzon et al., 1989). It also activates the transcription of the hla gene for an alpha toxin or alpha hemolysin. RNAIII controls the expression of surface proteins, such as Protein A, coagulase, and fibrinogen binding protein, and repressor of toxin (Rot) protein by antisense base pairing with 5′ untranslated regions (5′ UTR) and forming RNA duplexes (Boisset et al., 2007). Rot protein binds to the promoter of many exoproteins (α, β, and γ hemolysin), enterotoxins (Toxic shock syndrome toxin), and leukocidins, inhibiting their transcription. The inhibition of Rot and surface proteins by RNA III leads to an upregulation of virulence factors along with the dispersal of biofilm. AgrA can also upregulate the transcription of phenol-soluble modulins psmα and psmβ operons in an RNAIII-independent manner (Queck et al., 2008; Le and Otto, 2015).
agr system is a global regulatory system of S. aureus for the upregulation of virulence factors, which aids S. aureus to cause several types of infections (Li S. et al., 2014; Tuchscherr and Löffler, 2016) and downregulation of surface proteins for disruption of biofilm. During the initial course of bacterial infection when the cell density is low, agr system is downregulated, resulting in an increased production of adhesins and surface proteins. Once colonization is established and nutrients become limited, the upregulation of degradative exoenzymes and toxins mediated by agr occurs, which helps not only to acquire nutrition for the cell but also to evade the host immune system (Fowler et al., 2004; Cheung et al., 2014). Yarwood and Schlievert (2003) have reported thicker and smoother biofilms on medical devices formed by agr mutant strains, leading to chronic and persisting infections in the host due to their inability to disseminate from the patient's body (Shopsin et al., 2010). The ability of Staphylococcus aureus to form biofilm in chronic relapsing infections associated with medical devices such as urinary catheters, intravenous catheters, and orthopedic prosthetics (Singh and Ray, 2014) shields the bacteria from the host immune system as well as antibiotics (Waters et al., 2016).
S. aureus strains can be classified into four agr groups (agrI, agrII, agrIII, and agrIV) based on the agrD gene (which encodes the synthesis of AIPs) and agrC gene (which encodes the receptor of agr TCS). The four AIP molecules are similar enough to bind with the AgrC receptor of the different groups but cannot activate the AgrA protein unless it is activated by the AIP of a similar group (Jabbari et al., 2012). Ikonomidis et al. (2009) and Khoramrooz et al. (2016) have reported that strains of agr group II and agr group III are more potent biofilm producers. Nichol et al. (2011) have also established a relationship between antibiotic resistance and agr groups in S. aureus. agr group I is widely associated with CA-MRSA genotypes, whereas agr group II is associated with HA-MRSA genotypes in humans.
(i) SarA family
It is encoded by the sar gene present in the sar locus (an important global virulence regulon that plays a major role in growth, biofilm formation, and production of toxins in S. aureus). It binds to P2–P3 intragenic region and activates agr transcription while another protein Sar R (which also binds to P2-P3 intragenic region) represses the transcription of agr (Reyes et al., 2011). sar locus possesses ~50,000 copies of the sarA gene per cell and it can regulate the expression of nearly 120 genes (encoding virulence, cell wall associated, and extracellular proteins) of S. aureus (Cheung et al., 2008; Fujimoto et al., 2009). It consists of three transcripts i.e., sarA, sarB, and sarC under the control of P1, P2, and P3 promoters, respectively (Cheung and Manna, 2005). SarA is a DNA binding protein that can bind to the P2 promoter of agr locus and upregulate the transcription of RNA III. Hence, the sar locus can directly activate the agr-mediated quorum sensing in S. aureus via the P2 promoter of agr locus (Rechtin et al., 1999; Sterba et al., 2003; Roberts et al., 2006). α toxin, phenol-soluble modulins, and Panton Valentine Leukocidin (PVL), which are responsible for causing critical infection in CA-MRSA, are regulated by SarA (Bronner et al., 2000; Dumitrescu et al., 2011; Zielinska et al., 2011). Production of extracellular proteases such as serine protease, cysteine protease, metalloprotease, and staphopain are also negatively regulated by SarA (Karlsson and Arvidson, 2002). It can bind to various promoters viz., sarA, mecA, and sarR under the same conditions and at the same time point as revealed by DNA affinity capture assay (DACA; Kim et al., 2022). Sar locus has been reported to play a major role in regulating antibiotic resistance mechanisms. Proteomic analysis revealed that mecA expression was reduced in sarA deficient mutant, the exact reason for which is still unknown. However, this might be a reason for antibiotic susceptibility in sarA deficient mutant strains as both genes are involved in biofilm formation and PBP2a expression (Kim et al., 2022). Also, the inactivation of sarA has led to a reduction in ciprofloxacin and vancomycin resistance (Lamichhane-Khadka et al., 2009). Sequence analysis of clinical strains of various lineage revealed that the sarA gene was highly conserved unlike the other global virulence regulator AgrA which had various mutations (Kim et al., 2022). SarA regulates the post-transcriptional expression of spa and collagen adhesion genes during the exponential growth phase by binding to the target mRNA. It changes the turnover as well as accounts for the stability of their transcripts (Roberts et al., 2006; Morrison et al., 2012). Unlike agr locus, whose transcription is initiated majorly in the exponential phase, SarA protein levels are constant throughout the growth phases of the bacterium (Cheung and Manna, 2005; Arya and Princy, 2013).
S. aureus has various SarA paralogs i.e., SarR, SarS, SarT, Rot, SarU, SarV, SarX, MgrA, and MarR, which are either inhibitor or stimulator of each other, and indirectly contributes to virulence, biofilm production, autolysis, antibiotic resistance, and metabolic processes (Trotonda et al., 2008; Ballal and Manna, 2009). Northern blot and transcriptional fusion analysis have confirmed that SarV, a 116-residue long polypeptide, which is a homolog of SarA and regulator of cell lysis, is regulated by sarA as well as mgrA. Various virulence and autolysis genes have been reported to be under the regulation of sarV (Cheung et al., 2008; Trotonda et al., 2009). SarR, a 13.6 kDa dimeric protein, is a repressor of SarA, which regulates agr expression directly by binding to the intragenic region of the P2–P3 promoters, where SarA also binds but with less affinity than SarR. SarR binds to sarA or the promoters of target genes, repressing the expression of sarA, agr, hla, hlb, and spa during the post-exponential growth phase (Cheung and Zhang, 2001; Manna and Cheung, 2001; Oscarsson et al., 2005; Arya and Princy, 2013).
SarS, a 250-residue-long polypeptide with 64% homology with SarA, is a repressor of hla transcription and activator of protein A (spa; Manna and Ray, 2007). Rot, a 166 amino acid long residue, is an additional regulatory protein that modulates the expression of sarS, a transcriptional regulator of virulence genes. Rot protein has been reported to negatively regulate the production of various toxins such as lipases, serine proteases, α toxin, β toxin, cysteine proteases, and several other proteases, suggesting that rot acts downstream of the agr locus and indirectly upregulates cell wall synthesis. Activation of agr results in the production of RNA III which inhibits the production of rot (Said-Salim et al., 2003; Schmidt et al., 2003). Treatment with protease inhibitors such as cysteine protease inhibitor E-64 or staphostatin SspC, a specific inhibitor of staphopain B, is necessary to restore biofilm formation in a rot mutant (Mootz et al., 2015). SarA and Rot both repress protease production and are thus important regulators of biofilm formation.
SarT, another 118-residue-long homologous protein of SarA that downregulates hla and RNA III expression, is negatively regulated by sarA (Manna and Cheung, 2003). SarU, a 247- residue-long protein with a molecular mass of 2.92 kDa, is adjoining to sarT but is transcribed in the opposite direction. Mutational analysis revealed an elevated expression of sarU in sarT mutants indicating that sarU is negatively regulated by sarT. sarU mutants exhibited a lower RNAII and RNAIII expression as compared to the parental strain as proved by transcriptional and northern blot analysis. This observation suggests that sarU indirectly activates agr locus by upregulating RNAIII expression and altering the expression of agr-mediated virulence genes (Liu et al., 2006; Arya and Princy, 2013). Finally, the global regulator MgrA (member of the SarA protein family) acts as a repressor of eight cell wall-anchored proteins. mgrA mutants exhibited an increased biofilm formation with a loss of bacterial clumping (Schilcher and Horswill, 2020).
(ii) SrrAB
SrrAB is a two-component system where SrrA is a 28 kDa, 241 amino acid long response regulator, and SrrB is a 66 kDa, 583 amino acid long histidine kinase. Low oxygen levels and redox environmental conditions such as pH serve as a signal for SrrAB TCS. Upon receiving the signal, the membrane-bound SrrB auto-phosphorylates at a conserved histidine residue. This phosphate group is then transferred to the aspartate residue of cytoplasmic SrrA. SrrA has been reported to bind to both P2 and P3 promoters of agr system, positively affecting its activity (Pragman et al., 2004; Tan et al., 2018).
(iii) CodY
The nutritional status of the cell greatly impacts biofilm formation and virulence factors production, which is regulated via CodY (global transcriptional regulator) in S. aureus (Schilcher and Horswill, 2020). CodY has been reported to strongly repress agr locus (Majerczyk et al., 2008) not by binding to the P2 and P3 promoter regions but rather with a region within the agrC gene (Majerczyk et al., 2010). However, this had no effect on agrA expression. On the contrary, Roux et al. (2014) reported in vitro binding of CodY to the P2 and P3 promoter of agr locus but with low affinity. Recently, CodY exhibited repression of rsaD, a small regulatory RNA (sRNA) responsible for causing cell death regulation during weak acid stress has been reported, which eventually causes eDNA release and biofilm formation (Augagneur et al., 2020).
The sae (staphylococcal accessory element) system was first identified by Giraudo et al. (1994) while studying a Tn551 insertional mutant in which the production of exoproteins such as nuclease, coagulase, α hemolysin, and β hemolysin was found to be altered. The sae operon consists of four genes, i.e., saeP, saeQ, saeR, and saeS, which are under the regulation of P1 and P3 promoters. The sae locus primarily comprises of a two-component system in which SaeR is the response regulator and SaeS is the histidine kinase (Giraudo et al., 1999). This system is activated by external stimuli such as H2O2 and α defensins and repressed under low pH and high NaCl concentrations, both resulting in membrane perturbation (Geiger et al., 2012; Haag and Bagnoli, 2015). The signal is sensed by SaeP which interacts with the extracellular linker peptide of SaeS leading to its autophosphorylation at His131 which then transphosphorylates SaeR at Asp51. SaeR can now bind to SBS (SaeR binding sequence), leading to the upregulation of saeR-mediated virulence genes (Figure 9).
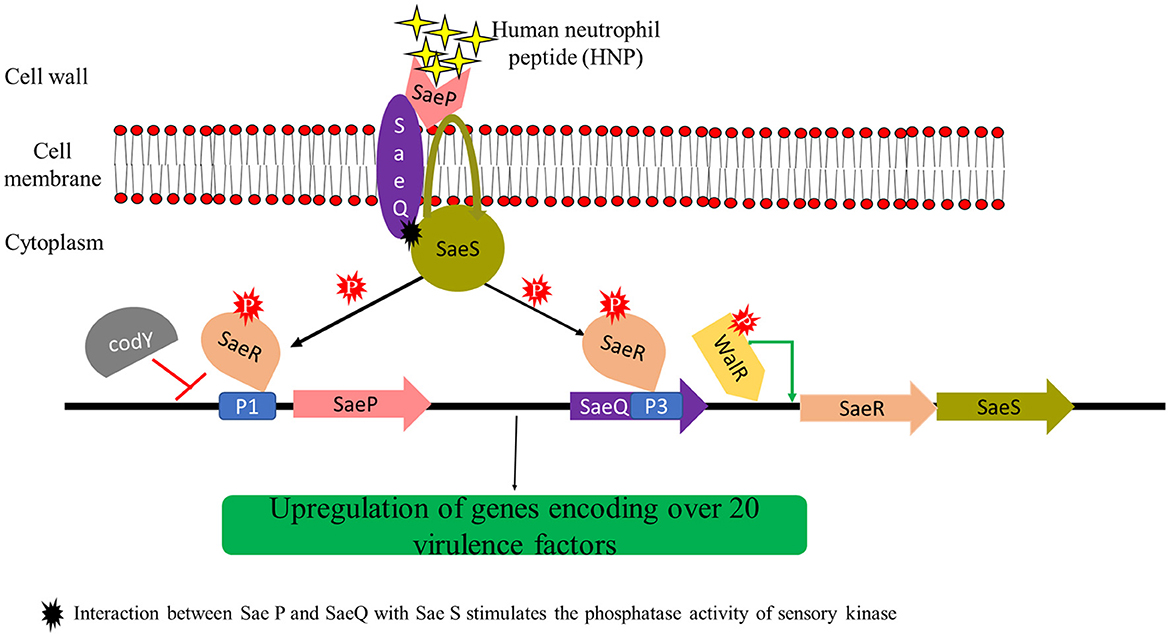
Figure 9. Regulation of sae (Staphylococcal accessory element) two-component system. Inducing molecules such as Human Neutrophil peptide 1 (HNP-1) are sensed by the extracellular membrane protein SaeP which interacts with the extracellular linker peptide of the cytoplasmic domain SaeS, activating the SaeRS TCS. Activated SaeR can induce the transcription of saePQRS (and genes encoding over 20 virulence factors) via P1 and P3 promoters. SaeP and SaeQ can form a protein complex with SaeS and induces the sensory kinase phosphatase activity resulting into the dephosphorylation of activated SaeR leading to the downregulation of virulence genes which are regulated by saeR. All these interactions are based on predictions (modified from Liu et al., 2016).
Very little was known about the role of auxiliary proteins SaeP and SaeQ in Sae TCS activity and disease progression. Collins et al. (2020) generated mutants of saeP, saeQ, and saeP-saeQ double mutant of the USA300 strain of S. aureus to analyze their function. The survival of S. aureus USA300ΔsaeP increased compared to the wild type USA300 strain post phagocytosis by neutrophils probably due to increased expression of bicomponent leukocidins by the former. The USA300ΔsaeQ also yielded a similar phenotype but the neutrophil interaction results were comparatively less significant. However, the saeP-saeQ double mutant strain (USA300ΔsaePsaeQ) exhibited a drastic increase in neutrophil survival and virulence during murine bacteremia as compared to the USA300ΔsaeP single mutant strain. These observations suggested the role of SaeP in the survival of bacterium after phagocytosis by neutrophils and the combined effect of SaeP and SaeQ in in vivo pathogenesis.
Flack et al. (2014) have reported that SaeS more specifically detects α-defensin 1/Human neutrophil peptide 1 (HNP-1) and human polymorphonuclear leucocytes (PMNs). SaeR and SaeS are transcribed via the P3 promoter located within the saeQ coding sequence. The P1 promoter is located upstream of the saeP gene and can transcribe all four genes of the locus. The P1 promoter has been reported to exhibit 2–30 times higher activity compared to the P3 promoter. Jeong et al. (2011) have reported that only basal level expression of saeRS genes from the P3 promoter is sufficient for activation of sae target genes and exoprotein production. P1 promoter has two saeR binding sites and thus it can be auto-induced by saeR transcripts produced by basal level expression of saeRS TCS via P3 promoter (Figure 7). The target genes of the sae system have been bifurcated into two classes: Class I target genes require high levels of phosphorylated SaeR for its activation, viz., fnbA, coa, P1 promoter, sae, and eap, while Class II target genes require basal/lower levels of phosphorylated SaeR for its activation, viz., hla and hlb genes (Mainiero et al., 2010; Haag and Bagnoli, 2015). SaeP and SaeQ have been reported to form a protein complex with SaeS and induce the sensory kinase phosphatase activity resulting in dephosphorylation of activated SaeR which downregulates the virulence genes regulated by saeR (Jeong et al., 2012; Haag and Bagnoli, 2015).
Depending on growth conditions, sae system can both positively (upregulation of biofilm forming genes, i.e., hla, hlb, coa, emp, eap, fnBPA, and fnBPB) and negatively (upregulation of biofilm dispersal factors such as nucleases and proteases) affect biofilm formation (Caiazza and O'Toole, 2003; Johnson et al., 2008; O'Neill et al., 2008; Huseby et al., 2010; McCourt et al., 2014; Zapotoczna et al., 2015; Liu et al., 2016). Moormeier et al. (2014) have reported that nuc transcription is positively regulated by the SaeRS system, as both sae and nuc mutant were unable to cause the dispersal of biofilms, which likely explains the degradation of eDNA during the dispersal stage of biofilm formation. Moreover, SaeS has been reported to exhibit polymorphism across S. aureus strains as a point mutation can lead to hyperactivation of SaeRS TCS leading to the inability of such strains to form robust biofilms (Mainiero et al., 2010; Cue et al., 2015). saeRS locus also acts synergistically with sarA to inhibit the production of extracellular proteases, resulting in an improved ability for biofilm formation (Arya and Princy, 2013).
SaeRS system positively regulates NETs (Berends et al., 2010; McDonald et al., 2012), SCIN, CHIPS (Rooijakkers et al., 2006), leukocidins (Münzenmayer et al., 2016), α hemolysin (Hla; Nygaard et al., 2013), pro-inflammatory cytokines, proteases, and toxins production (Watkins et al., 2011; Zurek et al., 2014; Cho et al., 2015). However, S. epidermis, which also possesses a SaeRS two-component system and is closely related to S. aureus, does not possess these virulence genes (Handke et al., 2008; Ravcheev et al., 2011). This indicates that S. aureus has acquired these virulence genes during evolution, which are being maintained by it under the control of the SaeRS two-component system (Liu et al., 2016).
Transcription of sae operon has been reported to be modulated by other global and local regulatory systems, which are mentioned as follows:
(i) agr system
The majority of toxins and exoproteins under the control of agr and sae operons are similar; however, there are several reports which suggest that agr and sae are independent of each other (Liu et al., 2016). sae operon does not possess AgrA or RNAIII binding sites, indicating that activation of agr operon should not have any significant effect on sae operon. Some of the virulence genes under the regulation of agr and sae are transcribed in opposite manner. coa and fnbA genes are repressed, whereas the cap gene is activated by agr operon. On the other hand, coa and fnbA genes are activated and the cap gene is repressed by the sae operon (Dassy et al., 1993; Wolz et al., 1996; Saravia-Otten et al., 1997; Luong et al., 2002; Steinhuber et al., 2003). Toxic shock syndrome toxin (TSST) is positively regulated by both agr and sae operon. However, agr mutants did not show impairment of TSST toxin production as observed in sae mutants (Baroja et al., 2016). Despite the positive regulation of hla expression via RNAIII activation by agr locus in vitro, sae operon has been reported to be inevitable for in vivo toxin production (Novick et al., 1993; Goerke et al., 2001).
(ii) σB
In the case of sae operon, σB has been reported to cause downregulation of saePQRS and sae target genes (hla, nuc, splABCDEF, and hlgABC; Mitchell et al., 2013) mostly via regulatory proteins or small non-coding RNAs which may be positively regulated by σB (Bischoff et al., 2004). No evidence of cross-talk between sae locus and σB has been reported, suggesting the fact that both are independent regulatory systems (Goerke et al., 2005; Liu et al., 2016).
(iii) CodY
Branched-chain amino acids (BCAA) viz., isoleucine, leucine, valine, and GTP are the key metabolites that activate CodY as a DNA-binding protein and bind to the sequence motif (AATTTTCWGAAAATT) of chromosomal DNA. CodY and SaeR can both bind to the sae P1 promoter in an opposite manner where one is the repressor and the other is the activator, respectively. CodY and SaeR have been reported to compete for binding to the saeP regulatory region. CodY is a stronger repressor of sae P1 promoter as compared to Rot. When nutrients are abundant, CodY inhibits the transcription of saeRS operon and sae dependent virulence genes but with the depletion of nutrients, the affinity of CodY to sae P1 promoter is lost resulting in an upregulation of sae operon. The subsequent increase in the production of cytotoxic factors helps S. aureus to combat the host immune system by destroying neutrophils. Pendleton et al. (2022) have reported that the cell membranes of codY mutant strains of S. aureus have a higher percentage of branched-chain fatty acids (BCFAs) as compared to the cell membranes of wild-type strains. This observation suggests the possibility of post-transcriptional regulation of the Sae system by the global repressor codY. Disruption of the lpdA gene which encodes dihydrolipoyl dehydrogenase (an important enzyme in BCFA synthesis) results in a reduction of SaeS kinase activity as well as the response regulator SaeR-P. This ultimately leads to a reduction in exotoxin secretion and attenuation of virulence. Thus, CodY acts as a nutritional checkpoint protein which ensures that the saePQRS operon is activated only when cytolytic and pore-forming toxins are to be secreted to acquire nutrition from the host and functions as a switch between commensal and invasive lifestyles of S. aureus (Mlynek et al., 2018).
(iv) WalRK
WalRK is one of the major two-component system of S. aureus which plays a role in cell wall metabolism and cell viability (Dubrac and Msadek, 2004; Dubrac et al., 2007). When the response regulator, WalR is produced in large amounts in its active form then the upregulation of sae operon has been observed. However, this upregulation was terminated when SaeRS two component system was deleted (Delauné et al., 2012). These results indicated that WalRK positively affects SaeRS TCS; however, the exact mechanism by which it occurs is still unknown (Liu et al., 2016).
(v) Fur regulon
Fur regulon of S. aureus is responsible for iron uptake, biofilm formation, and anti-oxidative stress response (Hantke, 1981; Litwin and Calderwood, 1993; Horsburgh et al., 2001; Johnson et al., 2005; Richardson et al., 2006; Lee and Helmann, 2007). saeRS transcription from both P1 and P3 promoters is downregulated in fur mutant as well as when iron from an exogenous source is supplied in media (fur is downregulated as iron is freely available), suggesting that Fur may be a positive regulator of saeRS (Johnson et al., 2011). However, the direct effect of Fur regulon on saeRS transcription is still unclear because sae operon does not possess Fur binding sites (Cho et al., 2015; Liu et al., 2016).
(vi) Rot
Rot, a member of the SarA protein family, is known to repress the expression of toxins (such as hla) in S. aureus (McNamara et al., 2000; Li and Cheung, 2008). Li and Cheung (2008) have reported that Rot can bind to the P1 promoter of the sae locus and repress it suggesting that Rot represses hla transcription via the P1 promoter of the sae locus. However, as the P1 promoter is not involved in the transcription of sae target genes (Jeong et al., 2011), the observations of Li and Cheung (2008) seem to be unlikely (Liu et al., 2016).
(vii) Fak system
Fatty acid kinase (fakA and fakB1/fakB2) mutants of S. aureus have been reported to exhibit a decrease in α hemolysin production as well as other sae target genes, indicating positive regulation of SaeRS TCS by the Fak system. It is believed that the acyl-PO4 group of FakB may donate the phosphoryl group to SaeR leading to its activation (Parsons et al., 2014).
(viii) ScrA
A novel protein ScrA acts via the SaeRS TCS to regulate virulence gene expression in S. aureus. ScrA protein acts as an intermediate between ArlRS and SaeRS systems (Wittekind et al., 2022). ArlRS TCS regulates genes involved in adhesion (ebh and sdrD), virulence (nuc, lukA, and esxA), and transcriptional factors (sarV and mgrA; Crosby et al., 2020). scrA expression is increased directly via the ArlR response regulator or indirectly via other regulators such as mgrA. ScrA has been hypothesized to stimulate SaeS kinase activity. This activates the SaeRS TCS which upregulates the production of adhesins and hemolysins and downregulates proteases which in turn increases cellular aggregation, biofilm formation, and hemolysis. The activated SaeRS TCS acts as a feedback inhibitor and directly or indirectly represses scrA (Wittekind et al., 2022).
S. aureus being a normal microflora of humans will always coexist with mankind. The extensive use of antibiotics across the world has led to the emergence of more resistant strains such as MRSA requiring novel antibiotics and treatment strategies. The ease of infection, high mortality rates, lack of suitable animal models, and increase in antibiotic resistance in MRSA have proved to be major hurdles to advances in clinical research. S. aureus pathogenesis is more complex as it is not dependent on a single major virulence factor that leads to disease progression. Secretion of a diverse array of virulence factors during its course of infection poses a major challenge in both drug and vaccine development. Though humans have made outstanding achievements in understanding the pathogenesis of MRSA, there are still gaps in knowledge and some important challenges to overcome. First, the adaptive immune evasion mechanisms post S. aureus infections remain unknown. Second, a vaccine targeting multiple factors against S. aureus is yet not successfully developed. Apart from these, a better understanding of bacterial pathogenesis, prevention of transmission and infection, and advancement in diagnosis requires focused concentration by researchers, policymakers, funding agencies, and well-coordinated multidisciplinary approaches that may help control the transmission of this highly successful pathogen.
HP has written the article. SR has designed the concept and edited the article. All authors contributed to the article and approved the submitted version.
The authors would like to thank School of Life Sciences, Central University of Gujarat for providing the facilities for this study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Alonzo III, F., Kozhaya, L., Rawlings, S. A., Reyes-Robles, T., DuMont, A. L., Myszka, D. G., et al. (2013). CCR5 is a receptor for Staphylococcus aureus leukotoxin ED. Nature 493, 51–55. doi: 10.1038/nature11724
Alonzo III, F., and Torres, V. J. (2014). The bicomponent pore-forming leucocidins of Staphylococcus aureus. Microbiol. Mol. Biol. Rev. 78, 199–230. doi: 10.1128/MMBR.00055-13
Argudín, M. Á., Mendoza, M. C., and Rodicio, M. R. (2010). Food poisoning and Staphylococcus aureus enterotoxins. Toxins 2, 1751–1773. doi: 10.3390/toxins2071751
Arya, R., and Princy, S. A. (2013). An insight into pleiotropic regulators Agr and Sar: Molecular probes paving the new way for antivirulent therapy. Fut. Microbiol. 8, 1339–1353. doi: 10.2217/fmb.13.92
Atkins, K. L., Burman, J. D., Chamberlain, E. S., Cooper, J. E., Poutrel, B., Bagby, S., et al. (2008). S. aureus IgG-binding proteins SpA and Sbi: Host specificity and mechanisms of immune complex formation. Mol. Immunol. 45, 1600–1611. doi: 10.1016/j.molimm.2007.10.021
Atwood, D. N., Loughran, A. J., Courtney, A. P., Anthony, A. C., Meeker, D. G., Spencer, H. J., et al. (2015). Comparative impact of diverse regulatory loci on Staphylococcus aureus biofilm formation. Microbiologyopen 4, 436–451. doi: 10.1002/mbo3.250
Augagneur, Y., King, A. N., Germain-Amiot, N., Sassi, M., Fitzgerald, J. W., Sahukhal, G. S., et al. (2020). Analysis of the CodY RNome reveals RsaD as a stress-responsive riboregulator of overflow metabolism in Staphylococcus aureus. Mol. Microbiol. 113, 309–325. doi: 10.1111/mmi.14418
Baddour, L., Lowrance, C., Albus, A., Lowrance, J., Anderson, S., and Lee, J. (1992). Staphylococcus aureus microcapsule expression attenuates bacterial virulence in a rat model of experimental endocarditis. J. Infect. Dis. 165, 749–753. doi: 10.1093/infdis/165.4.749
Balaban, N., and Rasooly, A. (2000). Staphylococcal enterotoxins. Int. J. Food Microbiol. 61, 1–10. doi: 10.1016/S0168-1605(00)00377-9
Ballal, A., and Manna, A. C. (2009). Expression of the sarA family of genes in different strains of Staphylococcus aureus. Microbiology 155, 2342. doi: 10.1099/mic.0.027417-0
Baroja, M. L., Herfst, C. A., Kasper, K. J., Xu, S. X., Gillett, D. A., Li, J., et al. (2016). The SaeRS two-component system is a direct and dominant transcriptional activator of toxic shock syndrome toxin 1 in Staphylococcus aureus. J. Bacteriol. 198, 2732–2742. doi: 10.1128/JB.00425-16
Bayer, M. G., Heinrichs, J. H., and Cheung, A. L. (1996). The molecular architecture of the sar locus in Staphylococcus aureus. J. Bacteriol. 178, 4563–4570. doi: 10.1128/jb.178.15.4563-4570.1996
Bayles, K. W. (2007). The biological role of death and lysis in biofilm development. Nat. Rev. Microbiol. 5, 721–726. doi: 10.1038/nrmicro1743
Beenken, K. E., Blevins, J. S., and Smeltzer, M. S. (2003). Mutation of sarA in Staphylococcus aureus limits biofilm formation. Infect. Immunity 71, 4206–4211. doi: 10.1128/IAI.71.7.4206-4211.2003
Bera, A., Herbert, S., Jakob, A., Vollmer, W., and Götz, F. (2005). Why are pathogenic staphylococci so lysozyme resistant? The peptidoglycan O-acetyltransferase OatA is the major determinant for lysozyme resistance of Staphylococcus aureus. Mol. Microbiol. 55, 778–787. doi: 10.1111/j.1365-2958.2004.04446.x
Berends, E. T., Horswill, A. R., Haste, N. M., Monestier, M., Nizet, V., and von Köckritz-Blickwede, M. (2010). Nuclease expression by Staphylococcus aureus facilitates escape from neutrophil extracellular traps. J. Innate Immunity 2, 576–586. doi: 10.1159/000319909
Berni, F., Enotarpi, J., Voskuilen, T., Li, S., van der Marel, G. A., and Codée, J. D. (2020). Synthetic carbohydrate-based cell wall components from Staphylococcus aureus. Drug Discov. Today 38, 35–43. doi: 10.1016/j.ddtec.2021.01.003
Berube, B. J., and Bubeck Wardenburg, J. (2013). Staphylococcus aureus α-toxin: Nearly a century of intrigue. Toxins 5, 1140–1166. doi: 10.3390/toxins5061140
Bhasin, N., Albus, A., Michon, F., Livolsi, P. J., Park, J. S., and Lee, J. C. (1998). Identification of a gene essential for O-acetylation of the Staphylococcus aureus type 5 capsular polysaccharide. Mol. Microbiol. 27, 9–21. doi: 10.1046/j.1365-2958.1998.00646.x
Bhavsar, A. P., D'Elia, M. A., Sahakian, T. D., and Brown, E. D. (2007). The amino terminus of Bacillus subtilis TagB possesses separable localization and functional properties. J. Bacteriol. 189, 6816–6823. doi: 10.1128/JB.00910-07
Bischoff, M., Dunman, P., Kormanec, J., Macapagal, D., Murphy, E., Mounts, W., et al. (2004). Microarray-based analysis of the Staphylococcus aureus σB regulon. J. Bacteriol. 186, 4085–4099. doi: 10.1128/JB.186.13.4085-4099.2004
Blevins, J. S., Beenken, K. E., Elasri, M. O., Hurlburt, B. K., and Smeltzer, M. S. (2002). Strain-dependent differences in the regulatory roles of sarA and agr in Staphylococcus aureus. Infect. Immunity 70, 470–480. doi: 10.1128/IAI.70.2.470-480.2002
Boisset, S., Geissmann, T., Huntzinger, E., Fechter, P., Bendridi, N., Possedko, M., et al. (2007). Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev. 21, 1353–1366. doi: 10.1101/gad.423507
Boles, B. R., and Horswill, A. R. (2008). Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 4, e1000052. doi: 10.1371/journal.ppat.1000052
Boswihi, S. S., and Udo, E. E. (2018). Methicillin-resistant Staphylococcus aureus: An update on the epidemiology, treatment options and infection control. Curr. Med. Res. Practice 8, 18–24. doi: 10.1016/j.cmrp.2018.01.001
Bronner, S., Stoessel, P., Gravet, A., Monteil, H., and Prévost, G. (2000). Variable expressions of Staphylococcus aureus bicomponent leucotoxins semiquantified by competitive reverse transcription-PCR. Appl. Environ. Microbiol. 66, 3931–3938. doi: 10.1128/AEM.66.9.3931-3938.2000
Brown, S., Santa Maria Jr, J. P., and Walker, S. (2013). Wall teichoic acids of gram-positive bacteria. Ann. Rev. Microbiol. 67, 313–336. doi: 10.1146/annurev-micro-092412-155620
Brunskill, E. W., and Bayles, K. W. (1996a). Identification and molecular characterization of a putative regulatory locus that affects autolysis in Staphylococcus aureus. J. Bacteriol. 178, 611–618. doi: 10.1128/jb.178.3.611-618.1996
Brunskill, E. W., and Bayles, K. W. (1996b). Identification of LytSR-regulated genes from Staphylococcus aureus. J. Bacteriol. 178, 5810–5812. doi: 10.1128/jb.178.19.5810-5812.1996
Bukowski, M., Wladyka, B., and Dubin, G. (2010). Exfoliative toxins of Staphylococcus aureus. Toxins 2, 1148–1165. doi: 10.3390/toxins2051148
Burke, F. M., Di Poto, A., Speziale, P., and Foster, T. J. (2011). The A domain of fibronectin-binding protein B of Staphylococcus aureus contains a novel fibronectin binding site. FEBS J. 278, 2359–2371. doi: 10.1111/j.1742-4658.2011.08159.x
Caiazza, N. C., and O'Toole, G. A. (2003). Alpha-toxin is required for biofilm formation by Staphylococcus aureus. J. Bacteriol. 185, 3214–3217. doi: 10.1128/JB.185.10.3214-3217.2003
Carneiro, C. R., Postol, E., Nomizo, R., Reis, L. F., and Brentani, R. R. (2004). Identification of enolase as a laminin-binding protein on the surface of Staphylococcus aureus. Microbes Infect. 6, 604–608. doi: 10.1016/j.micinf.2004.02.003
Carroll, J., Cafferkey, M., and Coleman, D. (1993). Serotype F double-and triple-converting phage insertionally inactivate the Staphylococcus aureus β-toxin determinant by a common molecular mechanism. FEMS Microbiol. Lett. 106, 147–155. doi: 10.1016/0378-1097(93)90073-B
Cerca, N., Brooks, J. L., and Jefferson, K. K. (2008). Regulation of the intercellular adhesin locus regulator (icaR) by SarA, σB, and IcaR in Staphylococcus aureus. J. Bacteriol. 190, 6530–6533. doi: 10.1128/JB.00482-08
Chambers, H. F., and DeLeo, F. R. (2009). Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 7, 629–641. doi: 10.1038/nrmicro2200
Chan, Y. G.-Y., Kim, H. K., Schneewind, O., and Missiakas, D. (2014). The capsular polysaccharide of Staphylococcus aureus is attached to peptidoglycan by the LytR-CpsA-Psr (LCP) family of enzymes. J. Biol. Chem. 289, 15680–15690. doi: 10.1074/jbc.M114.567669
Chaudhuri, R. R., Allen, A. G., Owen, P. J., Shalom, G., Stone, K., Harrison, M., et al. (2009). Comprehensive identification of essential Staphylococcus aureus genes using Transposon-Mediated Differential Hybridisation (TMDH). BMC Genom. 10, 1–18. doi: 10.1186/1471-2164-10-291
Chavakis, T., Hussain, M., Kanse, S. M., Peters, G., Bretzel, R. G., Flock, J.-I., et al. (2002). Staphylococcus aureus extracellular adherence protein serves as anti-inflammatory factor by inhibiting the recruitment of host leukocytes. Nat. Med. 8, 687–693. doi: 10.1038/nm728
Chavakis, T., Wiechmann, K., Preissner, K. T., and Herrmann, M. (2005). Staphylococcus aureus interactions with the endothelium. Thromb. Haemostasis 94, 278–285. doi: 10.1160/TH05-05-0306
Chen, X., and Alonzo III, F. (2019). Bacterial lipolysis of immune-activating ligands promotes evasion of innate defenses. Proc. Natl. Acad. Sci. U. S. A. 116, 3764–3773. doi: 10.1073/pnas.1817248116
Cheng, A. G., McAdow, M., Kim, H. K., Bae, T., Missiakas, D. M., and Schneewind, O. (2010). Contribution of coagulases towards Staphylococcus aureus disease and protective immunity. PLoS Pathog. 6, e1001036. doi: 10.1371/journal.ppat.1001036
Cheung, A. L., and Manna, A. C. (2005). Role of the distal sarA promoters in SarA expression in Staphylococcus aureus. Infect. Immunity 73, 4391–4394. doi: 10.1128/IAI.73.7.4391-4394.2005
Cheung, A. L., Nishina, K., and Manna, A. C. (2008). SarA of Staphylococcus aureus binds to the sarA promoter to regulate gene expression. J. Bacteriol. 190, 2239–2243. doi: 10.1128/JB.01826-07
Cheung, A. L., and Zhang, G. (2001). Are the structures of SarA and SarR similar? Trends Microbiol. 9, 570–573. doi: 10.1016/S0966-842X(01)02253-3
Cheung, G. Y., Bae, J. S., and Otto, M. (2021). Pathogenicity and virulence of Staphylococcus aureus. Virulence 12, 547–569. doi: 10.1080/21505594.2021.1878688
Cheung, G. Y., Kretschmer, D., Duong, A. C., Yeh, A. J., Ho, T. V., Chen, Y., et al. (2014). Production of an attenuated phenol-soluble modulin variant unique to the MRSA clonal complex 30 increases severity of bloodstream infection. PLoS Pathog. 10, e1004298. doi: 10.1371/journal.ppat.1004298
Cheung, G. Y., Wang, R., Khan, B. A., Sturdevant, D. E., and Otto, M. (2011). Role of the accessory gene regulator agr in community-associated methicillin-resistant Staphylococcus aureus pathogenesis. Infect. Immunity 79, 1927–1935. doi: 10.1128/IAI.00046-11
Cho, H., Jeong, D.-W., Liu, Q., Yeo, W.-S., Vogl, T., Skaar, E. P., et al. (2015). Calprotectin increases the activity of the SaeRS two component system and murine mortality during Staphylococcus aureus infections. PLoS Pathog. 11, e1005026. doi: 10.1371/journal.ppat.1005026
Clauditz, A., Resch, A., Wieland, K.-P., Peschel, A., and Gtz, F. (2006). Staphyloxanthin plays a role in the fitness of Staphylococcus aureus and its ability to cope with oxidative stress. Infect. Immunity 74, 4950–4953. doi: 10.1128/IAI.00204-06
Clements, M. O., Watson, S. P., and Foster, S. J. (1999). Characterization of the major superoxide dismutase of Staphylococcus aureus and its role in starvation survival, stress resistance, and pathogenicity. J. Bacteriol. 181, 3898–3903. doi: 10.1128/JB.181.13.3898-3903.1999
Cocchiaro, J. L., Gomez, M. I., Risley, A., Solinga, R., Sordelli, D. O., and Lee, J. C. (2006). Molecular characterization of the capsule locus from non-typeable Staphylococcus aureus. Mol. Microbiol. 59, 948–960. doi: 10.1111/j.1365-2958.2005.04978.x
Collins, M. M., Behera, R. K., Pallister, K. B., Evans, T. J., Burroughs, O., Flack, C., et al. (2020). The accessory gene saeP of the SaeR/S two-component gene regulatory system impacts Staphylococcus aureus virulence during neutrophil interaction. Front. Microbiol. 11, 561. doi: 10.3389/fmicb.2020.00561
Conlon, K. M., Humphreys, H., and O'Gara, J. P. (2002). icaR encodes a transcriptional repressor involved in environmental regulation of ica operon expression and biofilm formation in Staphylococcus epidermidis. J. Bacteriol. 184, 4400–4408. doi: 10.1128/JB.184.16.4400-4408.2002
Corrigan, R. M., Abbott, J. C., Burhenne, H., Kaever, V., and Gründling, A. (2011). c-di-AMP is a new second messenger in Staphylococcus aureus with a role in controlling cell size and envelope stress. PLoS Pathog. 7, e1002217. doi: 10.1371/journal.ppat.1002217
Cosgrove, K., Coutts, G., Jonsson, I.-M., Tarkowski, A., Kokai-Kun, J. F., Mond, J. J., et al. (2007). Catalase (KatA) and alkyl hydroperoxide reductase (AhpC) have compensatory roles in peroxide stress resistance and are required for survival, persistence, and nasal colonization in Staphylococcus aureus. J. Bacteriol. 189, 1025–1035. doi: 10.1128/JB.01524-06
Cosgrove, S. E., Qi, Y., Kaye, K. S., Harbarth, S., Karchmer, A. W., and Carmeli, Y. (2005). The impact of methicillin resistance in Staphylococcus aureus bacteremia on patient outcomes: Mortality, length of stay, and hospital charges. Infect. Control Hospit. Epidemiol. 26, 166–174. doi: 10.1086/502522
Cosgrove, S. E., Sakoulas, G., Perencevich, E. N., Schwaber, M. J., Karchmer, A. W., and Carmeli, Y. (2003). Comparison of mortality associated with methicillin-resistant and methicillin-susceptible Staphylococcus aureus bacteremia: A meta-analysis. Clin. Infect. Dis. 36, 53–59. doi: 10.1086/345476
Costerton, J. W., Stewart, P. S., and Greenberg, E. P. (1999). Bacterial biofilms: A common cause of persistent infections. Science 284, 1318–1322. doi: 10.1126/science.284.5418.1318
Courcol, R. J., Trivier, D., Bissinger, M.-C., Martin, G. R., and Brown, M. (1997). Siderophore production by Staphylococcus aureus and identification of iron-regulated proteins. Infect. Immunity 65, 1944–1948. doi: 10.1128/iai.65.5.1944-1948.1997
Crosby, H. A., Tiwari, N., Kwiecinski, J. M., Xu, Z., Dykstra, A., Jenul, C., et al. (2020). The Staphylococcus aureus ArlRS two-component system regulates virulence factor expression through MgrA. Mol. Microbiol. 113, 103–122. doi: 10.1111/mmi.14404
Cucarella, C., Solano, C., Valle, J., Amorena, B., Lasa, I., and Penads, J. R. (2001). Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J. Bacteriol. 183, 2888–2896. doi: 10.1128/JB.183.9.2888-2896.2001
Cue, D., Junecko, J. M., Lei, M. G., Blevins, J. S., Smeltzer, M. S., and Lee, C. Y. (2015). SaeRS-dependent inhibition of biofilm formation in Staphylococcus aureus Newman. PLoS ONE 10, e0123027. doi: 10.1371/journal.pone.0123027
Cue, D., Lei, M. G., and Lee, C. Y. (2012). Genetic regulation of the intercellular adhesion locus in staphylococci. Front. Cell. Infect. Microbiol. 2, 38. doi: 10.3389/fcimb.2012.00038
Cue, D., Lei, M. G., Luong, T. T., Kuechenmeister, L., Dunman, P. M., O'Donnell, S., et al. (2009). Rbf promotes biofilm formation by Staphylococcus aureus via repression of icaR, a negative regulator of icaADBC. J. Bacteriol. 191, 6363–6373. doi: 10.1128/JB.00913-09
Dassy, B., Hogan, T., Foster, T. J., and Fournier, J.-M. (1993). Involvement of the accessory gene regulator (agr) in expression of type 5 capsular polysaccharide by Staphylococcus aureus. Microbiology 139, 1301–1306. doi: 10.1099/00221287-139-6-1301
de Haas, C. J., Veldkamp, K. E., Peschel, A., Weerkamp, F., Van Wamel, W. J., Heezius, E. C., et al. (2004). Chemotaxis inhibitory protein of Staphylococcus aureus, a bacterial antiinflammatory agent. J. Exp. Med. 199, 687–695. doi: 10.1084/jem.20031636
De Jong, N. W., Ramyar, K. X., Guerra, F. E., Nijland, R., Fevre, C., Voyich, J. M., et al. (2017). Immune evasion by a staphylococcal inhibitor of myeloperoxidase. Proc. Natl. Acad. Sci. U. S. A. 114, 9439–9444. doi: 10.1073/pnas.1707032114
DeFrancesco, A. S., Masloboeva, N., Syed, A. K., DeLoughery, A., Bradshaw, N., Li, G.-W., et al. (2017). Genome-wide screen for genes involved in eDNA release during biofilm formation by Staphylococcus aureus. Proc. Natl. Acad. Sci. U. S. A. 114, E5969–E5978. doi: 10.1073/pnas.1704544114
Delauné, A., Dubrac, S., Blanchet, C., Poupel, O., Mäder, U., Hiron, A., et al. (2012). The WalKR system controls major staphylococcal virulence genes and is involved in triggering the host inflammatory response. Infect. Immunity 80, 3438–3453. doi: 10.1128/IAI.00195-12
Donegan, N. P., and Cheung, A. L. (2009). Regulation of the mazEF toxin-antitoxin module in Staphylococcus aureus and its impact on sigB expression. J. Bacteriol. 191, 2795–2805. doi: 10.1128/JB.01713-08
Drechsel, H., Freund, S., Nicholson, G., Haag, H., Jung, O., Zähner, H., et al. (1993). Purification and chemical characterization of staphyloferrin B, a hydrophilic siderophore from staphylococci. Biometals 6, 185–192. doi: 10.1007/BF00205858
Dubin, G. (2002). Extracellular Proteases of Staphylococcus spp. 383, 1075–1086. doi: 10.1515/BC.2002.116
Dubrac, S., Boneca, I. G., Poupel, O., and Msadek, T. (2007). New insights into the WalK/WalR (YycG/YycF) essential signal transduction pathway reveal a major role in controlling cell wall metabolism and biofilm formation in Staphylococcus aureus. J. Bacteriol. 189, 8257–8269. doi: 10.1128/JB.00645-07
Dubrac, S., and Msadek, T. (2004). Identification of genes controlled by the essential YycG/YycF two-component system of Staphylococcus aureus. Am. Soc. Microbiol. 186, 1175–1181. doi: 10.1128/JB.186.4.1175-1181.2004
Dumitrescu, O., Choudhury, P., Boisset, S., Badiou, C., Bes, M., Benito, Y., et al. (2011). β-Lactams interfering with PBP1 induce panton-valentine leukocidin expression by triggering sarA and rot global regulators of Staphylococcus aureus. Antimicrob. Agents Chemother. 55, 3261–3271. doi: 10.1128/AAC.01401-10
DuMont, A. L., Yoong, P., Surewaard, B. G., Benson, M. A., Nijland, R., van Strijp, J. A., et al. (2013). Staphylococcus aureus elaborates leukocidin AB to mediate escape from within human neutrophils. Infect. Immunity 81, 1830–1841. doi: 10.1128/IAI.00095-13
Dziarski, R., and Gupta, D. (2005). Peptidoglycan recognition in innate immunity. J. Endotoxin Res. 11, 304–310. doi: 10.1177/09680519050110050801
Engemann, J. J., Carmeli, Y., Cosgrove, S. E., Fowler, V. G., Bronstein, M. Z., Trivette, S. L., et al. (2003). Adverse clinical and economic outcomes attributable to methicillin resistance among patients with Staphylococcus aureus surgical site infection. Clin. Infect. Dis. 36, 592–598. doi: 10.1086/367653
Eriksen, N. R., Espersen, F., Rosdahl, V. T., and Jensen, K. (1995). Carriage of Staphylococcus aureus among 104 healthy persons during a 19-month period. Epidemiol. Infect. 115, 51–60. doi: 10.1017/S0950268800058118
Ernst, C. M., Kuhn, S., Slavetinsky, C. J., Krismer, B., Heilbronner, S., Gekeler, C., et al. (2015). The lipid-modifying multiple peptide resistance factor is an oligomer consisting of distinct interacting synthase and flippase subunits. MBio 6, e02340–e02314. doi: 10.1128/mBio.02340-14
Eyre, R. W., and Stanley, J. R. (1987). Human autoantibodies against a desmosomal protein complex with a calcium-sensitive epitope are characteristic of pemphigus foliaceus patients. J. Exp. Med. 165, 1719–1724. doi: 10.1084/jem.165.6.1719
Fitzgerald, J. R., Loughman, A., Keane, F., Brennan, M., Knobel, M., Higgins, J., et al. (2006). Fibronectin-binding proteins of Staphylococcus aureus mediate activation of human platelets via fibrinogen and fibronectin bridges to integrin GPIIb/IIIa and IgG binding to the FcγRIIa receptor. Mol. Microbiol. 59, 212–230. doi: 10.1111/j.1365-2958.2005.04922.x
Flack, C. E., Zurek, O. W., Meishery, D. D., Pallister, K. B., Malone, C. L., Horswill, A. R., et al. (2014). Differential regulation of staphylococcal virulence by the sensor kinase SaeS in response to neutrophil-derived stimuli. Proc. Natl. Acad. Sci. U. S. A. 111, E2037–E2045. doi: 10.1073/pnas.1322125111
Formosa-Dague, C., Feuillie, C., Beaussart, A., Derclaye, S., Kucharikova, S., Lasa, I., et al. (2016). Sticky matrix: adhesion mechanism of the staphylococcal polysaccharide intercellular adhesin. ACS Nano 10, 3443–3452. doi: 10.1021/acsnano.5b07515
Forsgren, A., and Sjöquist, J. (1966). “Protein a” from S. aureus: I. Pseudo-immune reaction with human γ-globulin. J. Immunol. 97, 822–827. doi: 10.4049/jimmunol.97.6.822
Foster, T. J., Geoghegan, J. A., Ganesh, V. K., and Höök, M. (2014). Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat. Rev. Microbiol. 12, 49–62. doi: 10.1038/nrmicro3161
Fowler Jr, V. G., Sakoulas, G., McIntyre, L. M., Meka, V. G., Arbeit, R. D., Cabell, C. H., et al. (2004). Persistent bacteremia due to methicillin-resistant Staphylococcus aureus infection is associated with agr dysfunction and low-level in vitro resistance to thrombin-induced platelet microbicidal protein. J. Infect. Dis. 190, 1140–1149. doi: 10.1086/423145
Fowler, T., Wann, E. R., Joh, D., Johansson, S., Foster, T. J., and Höök, M. (2000). Cellular invasion by Staphylococcus aureus involves a fibronectin bridge between the bacterial fibronectin-binding MSCRAMMs and host cell β1 integrins. Eur. J. Cell Biol. 79, 672–679. doi: 10.1078/0171-9335-00104
Fraser, J. D., and Proft, T. (2008). The bacterial superantigen and superantigen-like proteins. Immunol. Rev. 225, 226–243. doi: 10.1111/j.1600-065X.2008.00681.x
Friedrich, R., Panizzi, P., Fuentes-Prior, P., Richter, K., Verhamme, I., Anderson, P. J., et al. (2003). Staphylocoagulase is a prototype for the mechanism of cofactor-induced zymogen activation. Nature 425, 535–539. doi: 10.1038/nature01962
Fujimoto, D. F., Higginbotham, R. H., Sterba, K. M., Maleki, S. J., Segall, A. M., Smeltzer, M. S., et al. (2009). Staphylococcus aureus SarA is a regulatory protein responsive to redox and pH that can support bacteriophage lambda integrase-mediated excision/recombination. Mol. Microbiol. 74, 1445–1458. doi: 10.1111/j.1365-2958.2009.06942.x
Fujita, Y. (2009). Carbon catabolite control of the metabolic network in Bacillus subtilis. Biosci. Biotechnol. Biochem. 73, 245–259. doi: 10.1271/bbb.80479
Gallegos, M.-T., Schleif, R., Bairoch, A., Hofmann, K., and Ramos, J. L. (1997). Arac/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61, 393–410. doi: 10.1128/.61.4.393-410.1997
Gastmeier, P., Sohr, D., Geffers, C., Behnke, M., Daschner, F., and Rüden, H. (2005). Mortality risk factors with nosocomial Staphylococcus aureus infections in intensive care units: results from the German Nosocomial Infection Surveillance System (KISS). Infection 33, 50–55. doi: 10.1007/s15010-005-3186-5
Geiger, T., Francois, P., Liebeke, M., Fraunholz, M., Goerke, C., Krismer, B., et al. (2012). The stringent response of Staphylococcus aureus and its impact on survival after phagocytosis through the induction of intracellular PSMs expression. PLoS Pathog. 8, e1003016. doi: 10.1371/journal.ppat.1003016
Ginsberg, C., Zhang, Y.-H., Yuan, Y., and Walker, S. (2006). In vitro reconstitution of two essential steps in wall teichoic acid biosynthesis. ACS Chem. Biol. 1, 25–28. doi: 10.1021/cb0500041
Giraudo, A. T., Calzolari, A., Cataldi, A. A., Bogni, C., and Nagel, R. (1999). The sae locus of Staphylococcus aureus encodes a two-component regulatory system. FEMS Microbiol. Lett. 177, 15–22. doi: 10.1111/j.1574-6968.1999.tb13707.x
Giraudo, A. T., Raspanti, C. G., Calzolari, A., and Nagel, R. (1994). Characterization of a Tn 551-mutant of Staphylococcus aureus defective in the production of several exoproteins. Can. J. Microbiol. 40, 677–681. doi: 10.1139/m94-107
Goerke, C., Fluckiger, U., Steinhuber, A., Bisanzio, V., Ulrich, M., Bischoff, M., et al. (2005). Role of Staphylococcus aureus global regulators sae and σB in virulence gene expression during device-related infection. Infect. Immunity 73, 3415–3421. doi: 10.1128/IAI.73.6.3415-3421.2005
Goerke, C., Fluckiger, U., Steinhuber, A., Zimmerli, W., and Wolz, C. (2001). Impact of the regulatory loci agr, sarA and sae of Staphylococcus aureus on the induction of α-toxin during device-related infection resolved by direct quantitative transcript analysis. Mol. Microbiol. 40, 1439–1447. doi: 10.1046/j.1365-2958.2001.02494.x
Gómez, M. I., Lee, A., Reddy, B., Muir, A., Soong, G., Pitt, A., et al. (2004). Staphylococcus aureus protein A induces airway epithelial inflammatory responses by activating TNFR1. Nat. Med. 10, 842–848. doi: 10.1038/nm1079
González-García, S., Hamdan-Partida, A., Valdez-Alarcón, J. J., Bustos-Hamdan, A., and Bustos-Martínez, J. (2023). Main Factors of Staphylococcus aureus Associated With the Interaction to the Cells for Their Colonization and Persistence. IntechOpen. doi: 10.5772/intechopen.107974
Goodyear, C. S., and Silverman, G. J. (2003). Death by a B cell superantigen: in vivo VH-targeted apoptotic supraclonal B cell deletion by a staphylococcal toxin. J. Exp. Med. 197, 1125–1139. doi: 10.1084/jem.20020552
Goodyear, C. S., Sugiyama, F., and Silverman, G. J. (2006). Temporal and dose-dependent relationships between in vivo B cell receptor-targeted proliferation and deletion-induced by a microbial B cell toxin. J. Immunol. 176, 2262–2271. doi: 10.4049/jimmunol.176.4.2262
Gordon, R. J., and Lowy, F. D. (2008). Pathogenesis of methicillin-resistant Staphylococcus aureus infection. Clin. Infect. Dis. 46(Suppl.5), S350–S359. doi: 10.1086/533591
Gorwitz, R. J., Jernigan, D. B., and Jernigan, J. A. (2006). Strategies for Clinical Management of MRSA in the Community; Summary of an Experts' Meeting Convened by the Centers for Disease Control and Prevention. Available online at: https://stacks.cdc.gov/view/cdc/12423
Grispoldi, L., Karama, M., Armani, A., Hadjicharalambous, C., and Cenci-Goga, B. T. (2021). Staphylococcus aureus enterotoxin in food of animal origin and staphylococcal food poisoning risk assessment from farm to table. Ital. J. Anim. Sci. 20, 677–690. doi: 10.1080/1828051X.2020.1871428
Groicher, K. H., Firek, B. A., Fujimoto, D. F., and Bayles, K. W. (2000). The Staphylococcus aureus lrgAB operon modulates murein hydrolase activity and penicillin tolerance. J. Bacteriol. 182, 1794–1801. doi: 10.1128/JB.182.7.1794-1801.2000
Gross, M., Cramton, S. E., Gtz, F., and Peschel, A. (2001). Key role of teichoic acid net charge in Staphylococcus aureus colonization of artificial surfaces. Infect. Immunity 69, 3423–3426. doi: 10.1128/IAI.69.5.3423-3426.2001
Guo, Y., Song, G., Sun, M., Wang, J., and Wang, Y. (2020). Prevalence and therapies of antibiotic-resistance in Staphylococcus aureus. Front. Cell. Infect. Microbiol. 10, 107. doi: 10.3389/fcimb.2020.00107
Guzmán-Soto, I., McTiernan, C., Gonzalez-Gomez, M., Ross, A., Gupta, K., Suuronen, E. J., et al. (2021). Mimicking biofilm formation and development: Recent progress in in vitro and in vivo biofilm models. Iscience 24, 102443. doi: 10.1016/j.isci.2021.102443
Haag, A. F., and Bagnoli, F. (2015). The role of two-component signal transduction systems in Staphylococcus aureus virulence regulation. Staphylococcus aureus 2015, 145–198. doi: 10.1007/82_2015_5019
Hanakawa, Y., Schechter, N. M., Lin, C., Garza, L., Li, H., Yamaguchi, T., et al. (2002). Molecular mechanisms of blister formation in bullous impetigo and staphylococcal scalded skin syndrome. J. Clin. Investig. 110, 53–60. doi: 10.1172/JCI0215766
Handke, L., Rogers, K., Olson, M., Somerville, G. A., Jerrells, T., Rupp, M. E., et al. (2008). Staphylococcus epidermidis saeR is an effector of anaerobic growth and a mediator of acute inflammation. Infect. Immunity 76, 141–152. doi: 10.1128/IAI.00556-07
Hantke, K. (1981). Regulation of ferric iron transport in Escherichia coli K12: isolation of a constitutive mutant. Mol. General Genet. 182, 288–292. doi: 10.1007/BF00269672
Hanzelmann, D., Joo, H.-S., Franz-Wachtel, M., Hertlein, T., Stevanovic, S., Macek, B., et al. (2016). Toll-like receptor 2 activation depends on lipopeptide shedding by bacterial surfactants. Nat. Commun. 7, 12304. doi: 10.1038/ncomms12304
Hashimoto, M., Tawaratsumida, K., Kariya, H., Kiyohara, A., Suda, Y., Krikae, F., et al. (2006). Not lipoteichoic acid but lipoproteins appear to be the dominant immunobiologically active compounds in Staphylococcus aureus. J. Immunol. 177, 3162–3169. doi: 10.4049/jimmunol.177.5.3162
Hemmi, H., Takeuchi, O., Kawai, T., Kaisho, T., Sato, S., Sanjo, H., et al. (2000). A Toll-like receptor recognizes bacterial DNA. Nature 408, 740–745. doi: 10.1038/35047123
Herbert, S., Worlitzsch, D., Dassy, B., Boutonnier, A., Fournier, J.-M., Bellon, G., et al. (1997). Regulation of Staphylococcus aureus capsular polysaccharide type 5: CO2 inhibition in vitro and in vivo. J. Infect. Dis. 176, 431–438. doi: 10.1086/514061
Horsburgh, M. J., Ingham, E., and Foster, S. J. (2001). in Staphylococcus aureus, Fur is an interactive regulator with PerR, contributes to virulence, and is necessary for oxidative stress resistance through positive regulation of catalase and iron homeostasis. J. Bacteriol. 183, 468–475. doi: 10.1128/JB.183.2.468-475.2001
Huseby, M. J., Kruse, A. C., Digre, J., Kohler, P. L., Vocke, J. A., Mann, E. E., et al. (2010). Beta toxin catalyzes formation of nucleoprotein matrix in staphylococcal biofilms. Proc. Natl. Acad. Sci. U. S. A. 107, 14407–14412. doi: 10.1073/pnas.0911032107
Idrees, M., Sawant, S., Karodia, N., and Rahman, A. (2021). Staphylococcus aureus biofilm: Morphology, genetics, pathogenesis and treatment strategies. Int. J. Environ. Res. Public Health 18, 7602. doi: 10.3390/ijerph18147602
Ikonomidis, A., Vasdeki, A., Kristo, I., Maniatis, A. N., Tsakris, A., Malizos, K. N., et al. (2009). Association of biofilm formation and methicillin-resistance with accessory gene regulator (agr) loci in Greek Staphylococcus aureus clones. Microbial Pathogen. 47, 341–344. doi: 10.1016/j.micpath.2009.09.011
Itoh, S., Hamada, E., Kamoshida, G., Yokoyama, R., Takii, T., Onozaki, K., et al. (2010). Staphylococcal superantigen-like protein 10 (SSL10) binds to human immunoglobulin G (IgG) and inhibits complement activation via the classical pathway. Mol. Immunol. 47, 932–938. doi: 10.1016/j.molimm.2009.09.027
Jabbari, S., King, J. R., and Williams, P. (2012). Cross-strain quorum sensing inhibition by Staphylococcus aureus. Part 2: A spatially inhomogeneous model. Bullet. Math. Biol. 74, 1326–1353. doi: 10.1007/s11538-011-9702-0
Janzon, L., Löfdahl, S., and Arvidson, S. (1989). Identification and nucleotide sequence of the delta-lysin gene, hld, adjacent to the accessory gene regulator (agr) of Staphylococcus aureus. Mol. General Genet. 219, 480–485. doi: 10.1007/BF00259623
Jefferson, K. K., Pier, D. B., Goldmann, D. A., and Pier, G. B. (2004). The teicoplanin-associated locus regulator (TcaR) and the intercellular adhesin locus regulator (IcaR) are transcriptional inhibitors of the ica locus in Staphylococcus aureus. J. Bacteriol. 186, 2449–2456. doi: 10.1128/JB.186.8.2449-2456.2004
Jeong, D.-W., Cho, H., Lee, H., Li, C., Garza, J., Fried, M., et al. (2011). Identification of the P3 promoter and distinct roles of the two promoters of the SaeRS two-component system in Staphylococcus aureus. J. Bacteriol. 193, 4672–4684. doi: 10.1128/JB.00353-11
Jeong, D. W., Cho, H., Jones, M. B., Shatzkes, K., Sun, F., Ji, Q., et al. (2012). The auxiliary protein complex SaePQ activates the phosphatase activity of sensor kinase SaeS in the SaeRS two-component system of Staphylococcus aureus. Mol. Microbiol. 86, 331–348. doi: 10.1111/j.1365-2958.2012.08198.x
Ji, G., Beavis, R. C., and Novick, R. P. (1995). Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc. Natl. Acad. Sci. U. S. A. 92, 12055–12059. doi: 10.1073/pnas.92.26.12055
Johnson, M., Cockayne, A., and Morrissey, J. A. (2008). Iron-regulated biofilm formation in Staphylococcus aureus Newman requires ica and the secreted protein Emp. Infect. Immunity 76, 1756–1765. doi: 10.1128/IAI.01635-07
Johnson, M., Cockayne, A., Williams, P. H., and Morrissey, J. A. (2005). Iron-responsive regulation of biofilm formation in Staphylococcus aureus involves fur-dependent and fur-independent mechanisms. J. Bacteriol. 187, 8211–8215. doi: 10.1128/JB.187.23.8211-8215.2005
Johnson, M., Sengupta, M., Purves, J., Tarrant, E., Williams, P. H., Cockayne, A., et al. (2011). Fur is required for the activation of virulence gene expression through the induction of the sae regulatory system in Staphylococcus aureus. Int. J. Medical Microbiol. 301, 44–52. doi: 10.1016/j.ijmm.2010.05.003
Jongerius, I., Garcia, B. L., Geisbrecht, B. V., van Strijp, J. A., and Rooijakkers, S. H. (2010). Convertase inhibitory properties of Staphylococcal extracellular complement-binding protein. J. Biol. Chem. 285, 14973–14979. doi: 10.1074/jbc.M109.091975
Joo, H.-S., and Otto, M. (2015). Mechanisms of resistance to antimicrobial peptides in staphylococci. Biochimica et Biophysica Acta 1848, 3055–3061. doi: 10.1016/j.bbamem.2015.02.009
Kang, M., Ko, Y.-P., Liang, X., Ross, C. L., Liu, Q., Murray, B. E., et al. (2013). Collagen-binding microbial surface components recognizing adhesive matrix molecule (MSCRAMM) of Gram-positive bacteria inhibit complement activation via the classical pathway. J. Biol. Chem. 288, 20520–20531. doi: 10.1074/jbc.M113.454462
Karlsson, A., and Arvidson, S. (2002). Variation in extracellular protease production among clinical isolates of Staphylococcus aureus due to different levels of expression of the protease repressor sarA. Infect. Immunity 70, 4239–4246. doi: 10.1128/IAI.70.8.4239-4246.2002
Kaspar, U., Kriegeskorte, A., Schubert, T., Peters, G., Rudack, C., Pieper, D. H., et al. (2016). The culturome of the human nose habitats reveals individual bacterial fingerprint patterns. Environ. Microbiol. 18, 2130–2142. doi: 10.1111/1462-2920.12891
Katayama, Y., Baba, T., Sekine, M., Fukuda, M., and Hiramatsu, K. (2013). Beta-hemolysin promotes skin colonization by Staphylococcus aureus. J. Bacteriol. 195, 1194–1203. doi: 10.1128/JB.01786-12
Kawai, Y., Marles-Wright, J., Cleverley, R. M., Emmins, R., Ishikawa, S., Kuwano, M., et al. (2011). A widespread family of bacterial cell wall assembly proteins. EMBO J. 30, 4931–4941. doi: 10.1038/emboj.2011.358
Keinhoerster, D., George, S. E., Weidenmaier, C., and Wolz, C. (2019). Function and regulation of Staphylococcus aureus wall teichoic acids and capsular polysaccharides. Int. J. Medical Microbiol. 309, 151333. doi: 10.1016/j.ijmm.2019.151333
Kemung, H. M., Tan, L. T.-H., Khan, T. M., Chan, K.-G., Pusparajah, P., Goh, B.-H., et al. (2018). Streptomyces as a prominent resource of future anti-MRSA drugs. Front. Microbiol. 9, 2221. doi: 10.3389/fmicb.2018.02221
Khoramrooz, S. S., Mansouri, F., Marashifard, M., Hosseini, S. A. A. M., Chenarestane-Olia, F. A., Ganavehei, B., et al. (2016). Detection of biofilm related genes, classical enterotoxin genes and agr typing among Staphylococcus aureus isolated from bovine with subclinical mastitis in southwest of Iran. Microbial Pathog. 97, 45–51. doi: 10.1016/j.micpath.2016.05.022
Kiedrowski, M. R., Kavanaugh, J. S., Malone, C. L., Mootz, J. M., Voyich, J. M., Smeltzer, M. S., et al. (2011). Nuclease modulates biofilm formation in community-associated methicillin-resistant Staphylococcus aureus. PLoS ONE 6, e26714. doi: 10.1371/journal.pone.0026714
Kim, B., Lee, H.-J., Jo, S.-H., Kim, M.-G., Lee, Y., Lee, W., et al. (2022). Study of SarA by DNA affinity capture assay (DACA) employing three promoters of key virulence and resistance genes in methicillin-resistant Staphylococcus aureus. Antibiotics 11, 1714. doi: 10.3390/antibiotics11121714
Kiser, K. B., Bhasin, N., Deng, L., and Lee, J. C. (1999). Staphylococcus aureus cap5P encodes a UDP-N-acetylglucosamine 2-epimerase with functional redundancy. J. Bacteriol. 181, 4818–4824. doi: 10.1128/JB.181.16.4818-4824.1999
Kläui, A. J., Boss, R., and Graber, H. U. (2019). Characterization and comparative analysis of the Staphylococcus aureus genomic island v Saβ: An in silico approach. J. Bacteriol. 201, e00777–e00718. doi: 10.1128/JB.00777-18
Kneidinger, B., O'Riordan, K., Li, J., Brisson, J.-R., Lee, J. C., and Lam, J. S. (2003). Three highly conserved proteins catalyze the conversion of UDP-N-acetyl-d-glucosamine to precursors for the biosynthesis of O antigen in Pseudomonas aeruginosa O11 and capsule in Staphylococcus aureus type 5: Implications for the UDP-N-acetyl-l-fucosamine biosynthetic pathway. J. Biol. Chem. 278, 3615–3627. doi: 10.1074/jbc.M203867200
Ko, Y.-P., Kuipers, A., Freitag, C. M., Jongerius, I., Medina, E., van Rooijen, W. J., et al. (2013). Phagocytosis escape by a Staphylococcus aureus protein that connects complement and coagulation proteins at the bacterial surface. PLoS Pathog. 9, e1003816. doi: 10.1371/journal.ppat.1003816
Kobayashi, S. D., Malachowa, N., and DeLeo, F. R. (2015). Pathogenesis of Staphylococcus aureus abscesses. Am. J. Pathol. 185, 1518–1527. doi: 10.1016/j.ajpath.2014.11.030
Kolaczkowska, E., and Kubes, P. (2013). Neutrophil recruitment and function in health and inflammation. Nat. Rev. Immunol. 13, 159–175. doi: 10.1038/nri3399
Kong, C., Neoh, H. M., and Nathan, S. (2016). Targeting Staphylococcus aureus toxins: A potential form of anti-virulence therapy. Toxins 8, 72. doi: 10.3390/toxins8030072
Krepel, S. A., and Wang, J. M. (2019). Chemotactic ligands that activate G-protein-coupled formylpeptide receptors. Int. J. Mol. Sci. 20, 3426. doi: 10.3390/ijms20143426
Krishnamurthy, A., Nagel, S., and Orlov, D. (2014). Sizing up repo. J. Fin. 69, 2381–2417. doi: 10.1111/jofi.12168
Krismer, B., Liebeke, M., Janek, D., Nega, M., Rautenberg, M., Hornig, G., et al. (2014). Nutrient limitation governs Staphylococcus aureus metabolism and niche adaptation in the human nose. PLoS Pathog. 10, e1003862. doi: 10.1371/journal.ppat.1003862
Krismer, B., Weidenmaier, C., Zipperer, A., and Peschel, A. (2017). The commensal lifestyle of Staphylococcus aureus and its interactions with the nasal microbiota. Nat. Rev. Microbiol. 15, 675–687. doi: 10.1038/nrmicro.2017.104
Kuipers, A., Stapels, D. A., Weerwind, L. T., Ko, Y.-P., Ruyken, M., Lee, J. C., et al. (2016). The Staphylococcus aureus polysaccharide capsule and Efb-dependent fibrinogen shield act in concert to protect against phagocytosis. Microbiology 162, 1185. doi: 10.1099/mic.0.000293
Kumar, H., Kawai, T., and Akira, S. (2011). Pathogen recognition by the innate immune system. Int. Rev. Immunol. 30, 16–34. doi: 10.3109/08830185.2010.529976
Laarman, A. J., Mijnheer, G., Mootz, J. M., Van Rooijen, W. J., Ruyken, M., Malone, C. L., et al. (2012). Staphylococcus aureus Staphopain A inhibits CXCR2-dependent neutrophil activation and chemotaxis. EMBO J. 31, 3607–3619. doi: 10.1038/emboj.2012.212
Laarman, A. J., Ruyken, M., Malone, C. L., van Strijp, J. A., Horswill, A. R., and Rooijakkers, S. H. (2011). Staphylococcus aureus metalloprotease aureolysin cleaves complement C3 to mediate immune evasion. J. Immunol. 186, 6445–6453. doi: 10.4049/jimmunol.1002948
Lamichhane-Khadka, R., Cantore, S. A., Riordan, J. T., Delgado, A., Norman, A. E., Duenas, S., et al. (2009). sarA inactivation reduces vancomycin-intermediate and ciprofloxacin resistance expression by Staphylococcus aureus. Int. J. Antimicrobial Agents 34, 136–141. doi: 10.1016/j.ijantimicag.2009.01.018
Langley, R., Patel, D., Jackson, N., Clow, F., and Fraser, J. D. (2010). Staphylococcal superantigen super-domains in immune evasion. Crit. Rev. Immunol. 30, 40. doi: 10.1615/CritRevImmunol.v30.i2.40
Langley, R., Wines, B., Willoughby, N., Basu, I., Proft, T., and Fraser, J. D. (2005). The staphylococcal superantigen-like protein 7 binds IgA and complement C5 and inhibits IgA-FcαRI binding and serum killing of bacteria. J. Immunol. 174, 2926–2933. doi: 10.4049/jimmunol.174.5.2926
Lattar, S. M., Tuchscherr, L. P., Caccuri, R. L., Centrón, D., Becker, K., Alonso, C. A., et al. (2009). Capsule expression and genotypic differences among Staphylococcus aureus isolates from patients with chronic or acute osteomyelitis. Infect. Immunity 77, 1968–1975. doi: 10.1128/IAI.01214-08
Lauderdale, K. J., Boles, B. R., Cheung, A. L., and Horswill, A. R. (2009). Interconnections between Sigma B, agr, and proteolytic activity in Staphylococcus aureus biofilm maturation. Infect. Immunity 77, 1623–1635. doi: 10.1128/IAI.01036-08
Le, K. Y., and Otto, M. (2015). Quorum-sensing regulation in staphylococci—An overview. Front. Microbiol. 6, 1174. doi: 10.3389/fmicb.2015.01174
Lee, A. S., de Lencastre, H., Garau, J., Kluytmans, J., Malhotra-Kumar, S., Peschel, A., et al. (2018). Methicillin-resistant Staphylococcus aureus. Nat. Rev. Dis. Prim. 4, 18033. doi: 10.1038/nrdp.2018.33
Lee, J.-W., and Helmann, J. D. (2007). Functional specialization within the Fur family of metalloregulators. Biometals 20, 485–499. doi: 10.1007/s10534-006-9070-7
Lemon, K. P., Klepac-Ceraj, V., Schiffer, H. K., Brodie, E. L., Lynch, S. V., and Kolter, R. (2010). Comparative analyses of the bacterial microbiota of the human nostril and oropharynx. MBio 1, e00129–e00110. doi: 10.1128/mBio.00129-10
Lewis, K. (2000). Programmed death in bacteria. Microbiol. Mol. Biol. Rev. 64, 503–514. doi: 10.1128/MMBR.64.3.503-514.2000
Li, D., and Cheung, A. (2008). Repression of hla by rot is dependent on sae in Staphylococcus aureus. Infect. Immunity 76, 1068–1075. doi: 10.1128/IAI.01069-07
Li, S., Huang, H., Rao, X., Chen, W., Wang, Z., and Hu, X. (2014). Phenol-soluble modulins: novel virulence-associated peptides of staphylococci. Fut. Microbiol. 9, 203–216. doi: 10.2217/fmb.13.153
Li, W., Ulm, H., Rausch, M., Li, X., O'Riordan, K., Lee, J. C., et al. (2014). Analysis of the Staphylococcus aureus capsule biosynthesis pathway in vitro: characterization of the UDP-GlcNAc C6 dehydratases CapD and CapE and identification of enzyme inhibitors. Int. J. Medical Microbiol. 304, 958–969. doi: 10.1016/j.ijmm.2014.06.002
Lim, Y., Jana, M., Luong, T. T., and Lee, C. Y. (2004). Control of glucose-and NaCl-induced biofilm formation by rbf in Staphylococcus aureus. J. Bacteriol. 186, 722–729. doi: 10.1128/JB.186.3.722-729.2004
Lin, C.-F., Chen, C.-L., Huang, W.-C., Cheng, Y.-L., Hsieh, C.-Y., Wang, C.-Y., et al. (2010). Different types of cell death induced by enterotoxins. Toxins 2, 2158–2176. doi: 10.3390/toxins2082158
Lindsay, J. (2019). Staphylococci: Evolving genomes. Microbiol. Spectr. 7, GPP3-0071-2019. doi: 10.1128/microbiolspec.GPP3-0071-2019
Litwin, C. M., and Calderwood, S. B. (1993). Cloning and genetic analysis of the Vibrio vulnificus fur gene and construction of a fur mutant by in vivo marker exchange. J. Bacteriol. 175, 706–715. doi: 10.1128/jb.175.3.706-715.1993
Liu, C. M., Price, L. B., Hungate, B. A., Abraham, A. G., Larsen, L. A., Christensen, K., et al. (2015). Staphylococcus aureus and the ecology of the nasal microbiome. Sci. Adv. 1, e1400216. doi: 10.1126/sciadv.1400216
Liu, G. Y. (2009). Molecular pathogenesis of Staphylococcus aureus infection. Pediatr. Res. 65, 71–77. doi: 10.1203/PDR.0b013e31819dc44d
Liu, Q., Yeo, W.-S., and Bae, T. (2016). The SaeRS two-component system of Staphylococcus aureus. Genes 7, 81. doi: 10.3390/genes7100081
Liu, Y., Manna, A. C., Pan, C.-H., Kriksunov, I. A., Thiel, D. J., Cheung, A. L., et al. (2006). Structural and function analyses of the global regulatory protein SarA from Staphylococcus aureus. Proc. Natl. Acad. Sci. U. S. A. 103, 2392–2397. doi: 10.1073/pnas.0510439103
Löffler, B., Hussain, M., Grundmeier, M., Brück, M., Holzinger, D., Varga, G., et al. (2010). Staphylococcus aureus panton-valentine leukocidin is a very potent cytotoxic factor for human neutrophils. PLoS Pathog. 6, e1000715. doi: 10.1371/journal.ppat.1000715
Löffler, B., Tuchscherr, L., Niemann, S., and Peters, G. (2014). Staphylococcus aureus persistence in non-professional phagocytes. Int. J. Medical Microbiol. 304, 170–176. doi: 10.1016/j.ijmm.2013.11.011
Loncaric, I., Brunthaler, R., and Spergser, J. (2013). Suspected goat-to-human transmission of methicillin-resistant Staphylococcus aureus sequence type 398. J. Clin. Microbiol. 51, 1625–1626. doi: 10.1128/JCM.03052-12
Loughran, A. J., Atwood, D. N., Anthony, A. C., Harik, N. S., Spencer, H. J., Beenken, K. E., et al. (2014). Impact of individual extracellular proteases on Staphylococcus aureus biofilm formation in diverse clinical isolates and their isogenic sarA mutants. Microbiologyopen 3, 897–909. doi: 10.1002/mbo3.214
Luong, T., Sau, S., Gomez, M., Lee, J. C., and Lee, C. Y. (2002). Regulation of Staphylococcus aureus capsular polysaccharide expression by agr and sarA. Infect. Immunity 70, 444–450. doi: 10.1128/IAI.70.2.444-450.2002
Luong, T. T., Newell, S. W., and Lee, C. Y. (2003). Mgr, a novel global regulator in Staphylococcus aureus. J. Bacteriol. 185, 3703–3710. doi: 10.1128/JB.185.13.3703-3710.2003
Mack, D., Fischer, W., Krokotsch, A., Leopold, K., Hartmann, R., Egge, H., et al. (1996). The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear beta-1, 6-linked glucosaminoglycan: Purification and structural analysis. J. Bacteriol. 178, 175–183. doi: 10.1128/jb.178.1.175-183.1996
Mainiero, M., Goerke, C., Geiger, T., Gonser, C., Herbert, S., and Wolz, C. (2010). Differential target gene activation by the Staphylococcus aureus two-component system saeRS. J. Bacteriol. 192, 613–623. doi: 10.1128/JB.01242-09
Majerczyk, C. D., Dunman, P. M., Luong, T. T., Lee, C. Y., Sadykov, M. R., Somerville, G. A., et al. (2010). Direct targets of CodY in Staphylococcus aureus. J. Bacteriol. 192, 2861–2877. doi: 10.1128/JB.00220-10
Majerczyk, C. D., Sadykov, M. R., Luong, T. T., Lee, C., Somerville, G. A., and Sonenshein, A. L. (2008). Staphylococcus aureus CodY negatively regulates virulence gene expression. J. Bacteriol. 190, 2257–2265. doi: 10.1128/JB.01545-07
Malachowa, N., and DeLeo, F. R. (2010). Mobile genetic elements of Staphylococcus aureus. Cell. Mol. Life Sci. 67, 3057–3071. doi: 10.1007/s00018-010-0389-4
Malachowa, N., Kobayashi, S. D., Freedman, B., Dorward, D. W., and DeLeo, F. R. (2013). Staphylococcus aureus leukotoxin GH promotes formation of neutrophil extracellular traps. J. Immunol. 191, 6022–6029. doi: 10.4049/jimmunol.1301821
Mandell, G. (1975). Catalase, superoxide dismutase, and virulence of Staphylococcus aureus. In vitro and in vivo studies with emphasis on staphylococcal–leukocyte interaction. J. Clin. Investig. 55, 561–566. doi: 10.1172/JCI107963
Mann, E. E., Rice, K. C., Boles, B. R., Endres, J. L., Ranjit, D., Chandramohan, L., et al. (2009). Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLoS ONE 4, e5822. doi: 10.1371/journal.pone.0005822
Manna, A., and Cheung, A. L. (2001). Characterization of sarR, a modulator of sar expression in Staphylococcus aureus. Infect. Immunity 69, 885–896. doi: 10.1128/IAI.69.2.885-896.2001
Manna, A. C., and Cheung, A. L. (2003). sarU, a sarA homolog, is repressed by SarT and regulates virulence genes in Staphylococcus aureus. Infect. Immunity 71, 343–353. doi: 10.1128/IAI.71.1.343-353.2003
Manna, A. C., and Ray, B. (2007). Regulation and characterization of rot transcription in Staphylococcus aureus. Microbiology 153, 1538–1545. doi: 10.1099/mic.0.2006/004309-0
Maresso, A., and Schneewind, O. (2006). Iron acquisition and transport in Staphylococcus aureus. Biometals 19, 193–203. doi: 10.1007/s10534-005-4863-7
McAdow, M., Kim, H. K., DeDent, A. C., Hendrickx, A. P., Schneewind, O., and Missiakas, D. M. (2011). Preventing Staphylococcus aureus sepsis through the inhibition of its agglutination in blood. PLoS Pathog. 7, e1002307. doi: 10.1371/journal.ppat.1002307
McCourt, J., O'Halloran, D. P., McCarthy, H., O'Gara, J. P., and Geoghegan, J. A. (2014). Fibronectin-binding proteins are required for biofilm formation by community-associated methicillin-resistant Staphylococcus aureus strain LAC. FEMS Microbiol. Lett. 353, 157–164. doi: 10.1111/1574-6968.12424
McDonald, B., Urrutia, R., Yipp, B. G., Jenne, C. N., and Kubes, P. (2012). Intravascular neutrophil extracellular traps capture bacteria from the bloodstream during sepsis. Cell Host Microbe 12, 324–333. doi: 10.1016/j.chom.2012.06.011
McLoughlin, R. M., Solinga, R. M., Rich, J., Zaleski, K. J., Cocchiaro, J. L., Risley, A., et al. (2006). CD4+ T cells and CXC chemokines modulate the pathogenesis of Staphylococcus aureus wound infections. Proc. Natl. Acad. Sci. U. S. A. 103, 10408–10413. doi: 10.1073/pnas.0508961103
McNamara, P. J., Milligan-Monroe, K. C., Khalili, S., and Proctor, R. A. (2000). Identification, cloning, and initial characterization of rot, a locus encoding a regulator of virulence factor expression in Staphylococcus aureus. J. Bacteriol. 182, 3197–3203. doi: 10.1128/JB.182.11.3197-3203.2000
Meredith, T. C., Swoboda, J. G., and Walker, S. (2008). Late-stage polyribitol phosphate wall teichoic acid biosynthesis in Staphylococcus aureus. J. Bacteriol. 190, 3046–3056. doi: 10.1128/JB.01880-07
Meyer, F., Girardot, R., Piémont, Y., Prévost, G., and Colin, D. A. (2009). Analysis of the specificity of Panton-Valentine leucocidin and gamma-hemolysin F component binding. Infect. Immunity 77, 266–273. doi: 10.1128/IAI.00402-08
Miller, L. G., Quan, C., Shay, A., Mostafaie, K., Bharadwa, K., Tan, N., et al. (2007). A prospective investigation of outcomes after hospital discharge for endemic, community-acquired methicillin-resistant and-susceptible Staphylococcus aureus skin infection. Clin. Infect. Dis. 44, 483–492. doi: 10.1086/511041
Mitchell, G., Fugère, A., Pépin Gaudreau, K., Brouillette, E., Frost, E. H., Cantin, A. M., et al. (2013). SigB is a dominant regulator of virulence in Staphylococcus aureus small-colony variants. PLoS ONE 8, e65018. doi: 10.1371/journal.pone.0065018
Mlynek, K. D., Bulock, L. L., Stone, C. J., Curran, L. J., Sadykov, M. R., Bayles, K. W., et al. (2020). Genetic and biochemical analysis of CodY-mediated cell aggregation in Staphylococcus aureus reveals an interaction between extracellular DNA and polysaccharide in the extracellular matrix. J. Bacteriol. 202, e00593–e00519. doi: 10.1128/JB.00593-19
Mlynek, K. D., Sause, W. E., Moormeier, D. E., Sadykov, M. R., Hill, K. R., Torres, V. J., et al. (2018). Nutritional regulation of the Sae two-component system by CodY in Staphylococcus aureus. J. Bacteriol. 200, e00012–00018. doi: 10.1128/JB.00012-18
Molin, S., and Tolker-Nielsen, T. (2003). Gene transfer occurs with enhanced efficiency in biofilms and induces enhanced stabilisation of the biofilm structure. Curr. Opin. Biotechnol. 14, 255–261. doi: 10.1016/S0958-1669(03)00036-3
Monday, S. R., Vath, G. M., Ferens, W. A., Deobald, C., Rago, J. V., Gahr, P. J., et al. (1999). Unique superantigen activity of staphylococcal exfoliative toxins. J. Immunol. 162, 4550–4559. doi: 10.4049/jimmunol.162.8.4550
Moormeier, D., Bose, J., Horswill, A., and Bayles, K. (2014). Temporal and stochastic control of Staphylococcus aureus biofilm development. MBio 5, e01341–01314. doi: 10.1128/mBio.01341-14
Moormeier, D. E., and Bayles, K. W. (2017). Staphylococcus aureus biofilm: A complex developmental organism. Mol. Microbiol. 104, 365–376. doi: 10.1111/mmi.13634
Mootz, J. M., Benson, M. A., Heim, C. E., Crosby, H. A., Kavanaugh, J. S., Dunman, P. M., et al. (2015). R ot is a key regulator of Staphylococcus aureus biofilm formation. Mol. Microbiol. 96, 388–404. doi: 10.1111/mmi.12943
Moreillon, P., Entenza, J. M., Francioli, P., McDevitt, D., Foster, T. J., Francois, P., et al. (1995). Role of Staphylococcus aureus coagulase and clumping factor in pathogenesis of experimental endocarditis. Infect. Immunity 63, 4738–4743. doi: 10.1128/iai.63.12.4738-4743.1995
Morrison, J. M., Anderson, K. L., Beenken, K. E., Smeltzer, M. S., and Dunman, P. M. (2012). The staphylococcal accessory regulator, SarA, is an RNA-binding protein that modulates the mRNA turnover properties of late-exponential and stationary phase Staphylococcus aureus cells. Front. Cell. Infect. Microbiol. 2012, 26. doi: 10.3389/fcimb.2012.00026
Mrak, L. N., Zielinska, A. K., Beenken, K. E., Mrak, I. N., Atwood, D. N., Griffin, L. M., et al. (2012). saeRS and sarA act synergistically to repress protease production and promote biofilm formation in Staphylococcus aureus. PLoS ONE 7, e38453. doi: 10.1371/journal.pone.0038453
Münzenmayer, L., Geiger, T., Daiber, E., Schulte, B., Autenrieth, S. E., Fraunholz, M., et al. (2016). Influence of Sae-regulated and Agr-regulated factors on the escape of Staphylococcus aureus from human macrophages. Cell. Microbiol. 18, 1172–1183. doi: 10.1111/cmi.12577
Nakano, M. M., Lin, A., Zuber, C. S., Newberry, K. J., Brennan, R. G., and Zuber, P. (2010). Promoter recognition by a complex of Spx and the C-terminal domain of the RNA polymerase α subunit. PLoS ONE 5, e8664. doi: 10.1371/journal.pone.0008664
Ní Eidhin, D., Perkins, S., Francois, P., Vaudaux, P., Höök, M., and Foster, T.J. (1998). Clumping factor B (ClfB), a new surface-located fibrinogen-binding adhesin of Staphylococcus aureus. Mol. Microbiol. 30, 245–257. doi: 10.1046/j.1365-2958.1998.01050.x
Nemeth, J., and Lee, J. C. (1995). Antibodies to capsular polysaccharides are not protective against experimental Staphylococcus aureus endocarditis. Infect. Immunity 63, 375–380. doi: 10.1128/iai.63.2.375-380.1995
Nguyen, H. T., Nguyen, T. H., and Otto, M. (2020). The staphylococcal exopolysaccharide PIA–Biosynthesis and role in biofilm formation, colonization, and infection. Comput. Struct. Biotechnol. J. 18, 3324–3334. doi: 10.1016/j.csbj.2020.10.027
Nguyen, M. T., and Götz, F. (2016). Lipoproteins of Gram-positive bacteria: Key players in the immune response and virulence. Microbiol. Mol. Biol. Rev. 80, 891–903. doi: 10.1128/MMBR.00028-16
Nguyen, M. T., Kraft, B., Yu, W., Demicrioglu, D. D., Hertlein, T., Burian, M., et al. (2015). The νSaα specific lipoprotein like cluster (lpl) of S. aureus USA300 contributes to immune stimulation and invasion in human cells. PLoS Pathog. 11, e1004984. doi: 10.1371/journal.ppat.1004984
Nichol, K. A., Adam, H. J., Hussain, Z., Mulvey, M. R., McCracken, M., Mataseje, L. F., et al. (2011). Comparison of community-associated and health care-associated methicillin-resistant Staphylococcus aureus in Canada: Results of the CANWARD 2007–2009 study. Diagnost. Microbiol. Infect. Dis. 69, 320–325. doi: 10.1016/j.diagmicrobio.2010.10.028
Nilsson, I.-M., Lee, J. C., Bremell, T., Ryden, C., and Tarkowski, A. (1997). The role of staphylococcal polysaccharide microcapsule expression in septicemia and septic arthritis. Infect. Immunity 65, 4216–4221. doi: 10.1128/iai.65.10.4216-4221.1997
Nizet, V., and Bradley, J. S. (2011). “Staphylococcal infections,” in Infectious Diseases of the Fetus and Newborn (WB Saunders), 489–515.
Novick, R., Projan, S., Kornblum, J., Ross, H., Ji, G., Kreiswirth, B., et al. (1995). Theagr P2 operon: An autocatalytic sensory transduction system in Staphylococcus aureus. Mol. General Genet. 248, 446–458. doi: 10.1007/BF02191645
Novick, R. P. (2003). Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48, 1429–1449. doi: 10.1046/j.1365-2958.2003.03526.x
Novick, R. P., Ross, H., Projan, S., Kornblum, J., Kreiswirth, B., and Moghazeh, S. (1993). Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 12, 3967–3975. doi: 10.1002/j.1460-2075.1993.tb06074.x
Nygaard, T. K., Pallister, K. B., Zurek, O. W., and Voyich, J. M. (2013). The impact of α-toxin on host cell plasma membrane permeability and cytokine expression during human blood infection by CA-MRSA USA300. J. Leukocyte Biol. 94, 971–979. doi: 10.1189/jlb.0213080
O'Brien, L. M., Walsh, E. J., Massey, R. C., Peacock, S. J., and Foster, T. J. (2002). Staphylococcus aureus clumping factor B (ClfB) promotes adherence to human type I cytokeratin 10: Implications for nasal colonization. Cell. Microbiol. 4, 759–770. doi: 10.1046/j.1462-5822.2002.00231.x
O'Connell, D. P., Nanavaty, T., McDevitt, D., Gurusiddappa, S., Hk, M., and Foster, T. J. (1998). The fibrinogen-binding MSCRAMM (clumping factor) of Staphylococcus aureus has a Ca2+-dependent inhibitory site. J. Biol. Chem. 273, 6821–6829. doi: 10.1074/jbc.273.12.6821
O'Gara, J. P. (2007). ica and beyond: biofilm mechanisms and regulation in Staphylococcus epidermidis and Staphylococcus aureus. FEMS Microbiol. Lett. 270, 179–188. doi: 10.1111/j.1574-6968.2007.00688.x
O'Neill, E., Pozzi, C., Houston, P., Humphreys, H., Robinson, D. A., Loughman, A., et al. (2008). A novel Staphylococcus aureus biofilm phenotype mediated by the fibronectin-binding proteins, FnBPA and FnBPB. J. Bacteriol. 190, 3835–3850. doi: 10.1128/JB.00167-08
O'Riordan, K., and Lee, J. C. (2004). Staphylococcus aureus capsular polysaccharides. Clin. Microbiol. Rev. 17, 218–234. doi: 10.1128/CMR.17.1.218-234.2004
Oscarsson, J., Harlos, C., and Arvidson, S. (2005). Regulatory role of proteins binding to the spa (protein A) and sarS (staphylococcal accessory regulator) promoter regions in Staphylococcus aureus NTCC 8325-4. Int. J. Medical Microbiol. 295, 253–266. doi: 10.1016/j.ijmm.2005.05.003
Oscarsson, J., Kanth, A., Tegmark-Wisell, K., and Arvidson, S. (2006). SarA is a repressor of hla (α-hemolysin) transcription in Staphylococcus aureus: Its apparent role as an activator of hla in the prototype strain NCTC 8325 depends on reduced expression of sarS. J. Bacteriol. 188, 8526–8533. doi: 10.1128/JB.00866-06
Otto, M. (2008). Staphylococcal biofilms. Bacterial Biofilms 207–228. doi: 10.1007/978-3-540-75418-3_10
Otto, M. (2014). Staphylococcus aureus toxins. Curr. Opin. Microbiol. 17, 32–37. doi: 10.1016/j.mib.2013.11.004
Painter, K. L., Krishna, A., Wigneshweraraj, S., and Edwards, A. M. (2014). What role does the quorum-sensing accessory gene regulator system play during Staphylococcus aureus bacteremia? Trends Microbiol. 22, 676–685. doi: 10.1016/j.tim.2014.09.002
Palma, M., Shannon, O., Quezada, H. C., Berg, A., and Flock, J.-I. (2001). Extracellular fibrinogen-binding protein, Efb, from Staphylococcus aureus blocks platelet aggregation due to its binding to the α-chain. J. Biol. Chem. 276, 31691–31697. doi: 10.1074/jbc.M104554200
Pamp, S. J., Frees, D., Engelmann, S., Hecker, M., and Ingmer, H. (2006). Spx is a global effector impacting stress tolerance and biofilm formation in Staphylococcus aureus. J. Bacteriol. 188, 4861–4870. doi: 10.1128/JB.00194-06
Park, P. W., Roberts, D., Grosso, L., Parks, W., Rosenbloom, J., Abrams, W., et al. (1991). Binding of elastin to Staphylococcus aureus. J. Biol. Chem. 266, 23399–23406. doi: 10.1016/S0021-9258(18)54510-5
Parsons, J. B., Broussard, T. C., Bose, J. L., Rosch, J. W., Jackson, P., Subramanian, C., et al. (2014). Identification of a two-component fatty acid kinase responsible for host fatty acid incorporation by Staphylococcus aureus. Proc. Natl. Acad. Sci. U. S. A. 111, 10532–10537. doi: 10.1073/pnas.1408797111
Patti, J. M., Boles, J. O., and Hook, M. (1993). Identification and biochemical characterization of the ligand binding domain of the collagen adhesin from Staphylococcus aureus. Biochemistry 32, 11428–11435. doi: 10.1021/bi00093a021
Patti, J. M., Bremell, T., Krajewska-Pietrasik, D., Abdelnour, A., Tarkowski, A., Rydén, C., et al. (1994). The Staphylococcus aureus collagen adhesin is a virulence determinant in experimental septic arthritis. Infect. Immunity 62, 152–161. doi: 10.1128/iai.62.1.152-161.1994
Patti, J. M., Jonsson, H., Guss, B., Switalski, L. M., Wiberg, K., Lindberg, M., et al. (1992). Molecular characterization and expression of a gene encoding a Staphylococcus aureus collagen adhesin. J. Biol. Chem. 267, 4766–4772. doi: 10.1016/S0021-9258(18)42898-0
Patton, T. G., Rice, K. C., Foster, M. K., and Bayles, K. W. (2005). The Staphylococcus aureus cidC gene encodes a pyruvate oxidase that affects acetate metabolism and cell death in stationary phase. Mol. Microbiol. 56, 1664–1674. doi: 10.1111/j.1365-2958.2005.04653.x
Patton, T. G., Yang, S. J., and Bayles, K. W. (2006). The role of proton motive force in expression of the Staphylococcus aureus cid and lrg operons. Mol. Microbiol. 59, 1395–1404. doi: 10.1111/j.1365-2958.2006.05034.x
Pauli, N. T., Kim, H. K., Falugi, F., Huang, M., Dulac, J., Henry Dunand, C., et al. (2014). Staphylococcus aureus infection induces protein A–mediated immune evasion in humans. J. Exp. Med. 211, 2331–2339. doi: 10.1084/jem.20141404
Pelz, A., Wieland, K.-P., Putzbach, K., Hentschel, P., Albert, K., and Gotz, F. (2005). Structure and biosynthesis of staphyloxanthin from Staphylococcus aureus. J. Biol. Chem. 280, 32493–32498. doi: 10.1074/jbc.M505070200
Pendleton, A., Yeo, W.-S., Alqahtani, S., DiMaggio Jr, D. A., Stone, C. J., Li, Z., et al. (2022). Regulation of the sae two-component system by branched-chain fatty acids in Staphylococcus aureus. Mbio 13, e01472–e01422. doi: 10.1128/mbio.01472-22
Peng, H.-L., Novick, R., Kreiswirth, B., Kornblum, J., and Schlievert, P. (1988). Cloning, characterization, and sequencing of an accessory gene regulator (agr) in Staphylococcus aureus. J. Bacteriol. 170, 4365–4372. doi: 10.1128/jb.170.9.4365-4372.1988
Peng, Q., Tang, X., Dong, W., Sun, N., and Yuan, W. (2022). A review of biofilm formation of Staphylococcus aureus and its regulation mechanism. Antibiotics 12, 12. doi: 10.3390/antibiotics12010012
Peschel, A., Jack, R. W., Otto, M., Collins, L. V., Staubitz, P., Nicholson, G., et al. (2001). Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MprF is based on modification of membrane lipids with l-lysine. J. Exp. Med. 193, 1067–1076. doi: 10.1084/jem.193.9.1067
Peschel, A., Otto, M., Jack, R. W., Kalbacher, H., Jung, G., and Gtz, F. (1999). Inactivation of the dlt Operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J. Biol. Chem. 274, 8405–8410. doi: 10.1074/jbc.274.13.8405
Peschel, A., and Sahl, H.-G. (2006). The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat. Rev. Microbiol. 4, 529–536. doi: 10.1038/nrmicro1441
Peterson, P. K., Verhoef, J., Sabath, L. D., and Quie, P. G. (1977). Effect of protein A on staphylococcal opsonization. Infect. Immunity 15, 760–764. doi: 10.1128/iai.15.3.760-764.1977
Phlmann-Dietze, P., Ulrich, M., Kiser, K. B., Dring, G., Lee, J. C., Fournier, J. M., et al. (2000). Adherence of Staphylococcus aureus to endothelial cells: influence of capsular polysaccharide, global regulator agr, and bacterial growth phase. Infect. Immunity 68, 4865–4871. doi: 10.1128/IAI.68.9.4865-4871.2000
Pietrocola, G., Nobile, G., Gianotti, V., Zapotoczna, M., Foster, T. J., Geoghegan, J. A., et al. (2016). Molecular interactions of human plasminogen with fibronectin-binding protein B (FnBPB), a fibrinogen/fibronectin-binding protein from Staphylococcus aureus. J. Biol. Chem. 291, 18148–18162. doi: 10.1074/jbc.M116.731125
Pohl, K., Francois, P., Stenz, L., Schlink, F., Geiger, T., Herbert, S., et al. (2009). CodY in Staphylococcus aureus: A regulatory link between metabolism and virulence gene expression. J. Bacteriol. 191, 2953–2963. doi: 10.1128/JB.01492-08
Portols, M., Kiser, K. B., Bhasin, N., Chan, K. H., and Lee, J. C. (2001). Staphylococcus aureus Cap5O has UDP-ManNAc dehydrogenase activity and is essential for capsule expression. Infect. Immunity 69, 917–923. doi: 10.1128/IAI.69.2.917-923.2001
Pozzi, C., Waters, E. M., Rudkin, J. K., Schaeffer, C. R., Lohan, A. J., Tong, P., et al. (2012). Methicillin resistance alters the biofilm phenotype and attenuates virulence in Staphylococcus aureus device-associated infections. PLoS Pathog. 8, e1002626. doi: 10.1371/journal.ppat.1002626
Pragman, A. A., Yarwood, J. M., Tripp, T. J., and Schlievert, P. M. (2004). Characterization of virulence factor regulation by SrrAB, a two-component system in Staphylococcus aureus. J. Bacteriol. 186, 2430–2438. doi: 10.1128/JB.186.8.2430-2438.2004
Price, N. P. J., and Tsvetanova, B. (2007). Biosynthesis of the tunicamycins: a review. J. Antibiot. 60, 485–491.
Qian, Z., Yin, Y., Zhang, Y., Lu, L., Li, Y., and Jiang, Y. (2006). Genomic characterization of ribitol teichoic acid synthesis in Staphylococcus aureus: Genes, genomic organization and gene duplication. BMC Genom. 7, 1–12. doi: 10.1186/1471-2164-7-74
Qin, Z., Ou, Y., Yang, L., Zhu, Y., Tolker-Nielsen, T., Molin, S., et al. (2007). Role of autolysin-mediated DNA release in biofilm formation of Staphylococcus epidermidis. Microbiology 153, 2083–2092. doi: 10.1099/mic.0.2007/006031-0
Queck, S. Y., Jameson-Lee, M., Villaruz, A. E., Bach, T.-H. L., Khan, B. A., Sturdevant, D. E., et al. (2008). RNAIII-independent target gene control by the agr quorum-sensing system: Insight into the evolution of virulence regulation in Staphylococcus aureus. Mol. Cell 32, 150–158. doi: 10.1016/j.molcel.2008.08.005
Rachid, S., Ohlsen, K., Wallner, U., Hacker, J., Hecker, M., and Ziebuhr, W. (2000). Alternative transcription factor ςB is involved in regulation of biofilm expression in a Staphylococcus aureus mucosal isolate. J. Bacteriol. 182, 6824–6826. doi: 10.1128/JB.182.23.6824-6826.2000
Rasigade, J.-P., Laurent, F., Lina, G., Meugnier, H., Bes, M., Vandenesch, F., et al. (2010). Global distribution and evolution of Panton-Valentine leukocidin-positive methicillin-susceptible Staphylococcus aureus, 1981–2007. J. Infect. Dis. 201, 1589–1597. doi: 10.1086/652008
Rausch, M., Deisinger, J. P., Ulm, H., Müller, A., Li, W., Hardt, P., et al. (2019). Coordination of capsule assembly and cell wall biosynthesis in Staphylococcus aureus. Nat. Commun. 10, 1404. doi: 10.1038/s41467-019-09356-x
Ravcheev, D. A., Best, A. A., Tintle, N., DeJongh, M., Osterman, A. L., Novichkov, P. S., et al. (2011). Inference of the transcriptional regulatory network in Staphylococcus aureus by integration of experimental and genomics-based evidence. J. Bacteriol. 193, 3228–3240. doi: 10.1128/JB.00350-11
Rechtin, T. M., Gillaspy, A. F., Schumacher, M. A., Brennan, R. G., Smeltzer, M. S., and Hurlburt, B. K. (1999). Characterization of the SarA virulence gene regulator of Staphylococcus aureus. Mol. Microbiol. 33, 307–316. doi: 10.1046/j.1365-2958.1999.01474.x
Reed, S. D., Friedman, J. Y., Engemann, J. J., Griffiths, R. I., Anstrom, K. J., Kaye, K. S., et al. (2005). Costs and outcomes among hemodialysis-dependent patients with methicillin-resistant or methicillin-susceptible Staphylococcus aureus bacteremia. Infect. Control Hospit. Epidemiol. 26, 175–183. doi: 10.1086/502523
Reyes, D., Andrey, D. O., Monod, A., Kelley, W. L., Zhang, G., and Cheung, A. L. (2011). Coordinated regulation by AgrA, SarA, and SarR to control agr expression in Staphylococcus aureus. J. Bacteriol. 193, 6020–6031. doi: 10.1128/JB.05436-11
Rice, K. C., Firek, B. A., Nelson, J. B., Yang, S.-J., Patton, T. G., and Bayles, K. W. (2003). The Staphylococcus aureus cidAB operon: Evaluation of its role in regulation of murein hydrolase activity and penicillin tolerance. J. Bacteriol. 185, 2635–2643. doi: 10.1128/JB.185.8.2635-2643.2003
Rice, K. C., Mann, E. E., Endres, J. L., Weiss, E. C., Cassat, J. E., Smeltzer, M. S., et al. (2007). The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc. Natl. Acad. Sci. U. S. A. 104, 8113–8118. doi: 10.1073/pnas.0610226104
Rice, K. C., Nelson, J. B., Patton, T. G., Yang, S.-J., and Bayles, K. W. (2005). Acetic acid induces expression of the Staphylococcus aureus cidABC and lrgAB murein hydrolase regulator operons. J. Bacteriol. 187, 813–821. doi: 10.1128/JB.187.3.813-821.2005
Richardson, A. R., Dunman, P. M., and Fang, F. C. (2006). The nitrosative stress response of Staphylococcus aureus is required for resistance to innate immunity. Mol. Microbiol. 61, 927–939. doi: 10.1111/j.1365-2958.2006.05290.x
Risley, A. L., Loughman, A., Cywes-Bentley, C., Foster, T. J., and Lee, J. C. (2007). Capsular polysaccharide masks clumping factor A-mediated adherence of Staphylococcus aureus to fibrinogen and platelets. J. Infect. Dis. ases 196, 919–927. doi: 10.1086/520932
Roberts, C., Anderson, K. L., Murphy, E., Projan, S. J., Mounts, W., Hurlburt, B., et al. (2006). Characterizing the effect of the Staphylococcus aureus virulence factor regulator, SarA, on log-phase mRNA half-lives. J. Bacteriol. 188, 2593–2603. doi: 10.1128/JB.188.7.2593-2603.2006
Roche, F. M., Downer, R., Keane, F., Speziale, P., Park, P. W., and Foster, T. J. (2004). The N-terminal A domain of fibronectin-binding proteins A and B promotes adhesion of Staphylococcus aureus to elastin. J. Biol. Chem. 279, 38433–38440. doi: 10.1074/jbc.M402122200
Rooijakkers, S. H., Ruyken, M., Roos, A., Daha, M. R., Presanis, J. S., Sim, R. B., et al. (2005). Immune evasion by a staphylococcal complement inhibitor that acts on C3 convertases. Nat. Immunol. 6, 920–927. doi: 10.1038/ni1235
Rooijakkers, S. H., Ruyken, M., Van Roon, J., Van Kessel, K. P., Van Strijp, J. A., and Van Wamel, W. J. (2006). Early expression of SCIN and CHIPS drives instant immune evasion by Staphylococcus aureus. Cell. Microbiol. 8, 1282–1293. doi: 10.1111/j.1462-5822.2006.00709.x
Rousseaux, J., Rousseaux-Prévost, R., Bazin, H., and Biserte, G. (1983). Proteolysis of rat IgG subclasses by Staphylococcus aureus V8 proteinase. Biochimica et Biophysica Acta 748, 205–212. doi: 10.1016/0167-4838(83)90296-0
Roux, A., Todd, D. A., Velázquez, J. V., Cech, N. B., and Sonenshein, A. L. (2014). Cody-mediated regulation of the Staphylococcus aureus. Agr system integrates nutritional and population density signals. J. Bacteriol. 196, 1184–1196. doi: 10.1128/JB.00128-13
Sadykov, M. R., and Bayles, K. W. (2012). The control of death and lysis in staphylococcal biofilms: A coordination of physiological signals. Curr. Opin. Microbiol. 15, 211–215. doi: 10.1016/j.mib.2011.12.010
Sadykov, M. R., Windham, I. H., Widhelm, T. J., Yajjala, V. K., Watson, S. M., Endres, J. L., et al. (2019). CidR and CcpA synergistically regulate Staphylococcus aureus cidABC expression. J. Bacteriol. 201, e00371–e00319. doi: 10.1128/JB.00371-19
Saffarzadeh, M., Juenemann, C., Queisser, M. A., Lochnit, G., Barreto, G., Galuska, S. P., et al. (2012). Neutrophil extracellular traps directly induce epithelial and endothelial cell death: A predominant role of histones. PLoS ONE 7, e32366. doi: 10.1371/journal.pone.0032366
Said-Salim, B., Dunman, P., McAleese, F., Macapagal, D., Murphy, E., McNamara, P., et al. (2003). Global regulation of Staphylococcus aureus genes by Rot. J. Bacteriol. 185, 610–619. doi: 10.1128/JB.185.2.610-619.2003
Saravia-Otten, P., Müller, H., and Arvidson, S. (1997). Transcription of Staphylococcus aureus fibronectin binding protein genes is negatively regulated by agr and an agr-independent mechanism. J. Bacteriol. 179, 5259–5263. doi: 10.1128/jb,.179.17.5259-5263.1997
Saravolatz, L. D., Markowitz, N., Arking, L., Pohlod, D., and Fisher, E. (1982). Methicillin-resistant Staphylococcus aureus: Epidemiologic observations during a community-acquired outbreak. Ann. Internal Med. 96, 11–16. doi: 10.7326/0003-4819-96-1-11
Sau, S., Bhasin, N., Wann, E. R., Lee, J. C., Foster, T. J., and Lee, C. Y. (1997). The Staphylococcus aureus allelic genetic loci for serotype 5 and 8 capsule expression contain the type-specific genes flanked by common genes. Microbiology 143, 2395–2405. doi: 10.1099/00221287-143-7-2395
Schell, M. A. (1993). Molecular biology of the LysR family of transcriptional regulators. Ann. Rev. Microbiol. 47, 597–627. doi: 10.1146/annurev.mi.47.100193.003121
Schilcher, K., and Horswill, A. R. (2020). Staphylococcal biofilm development: Structure, regulation, and treatment strategies. Microbiol. Mol. Biol. Rev. 84, e00026–e00019. doi: 10.1128/MMBR.00026-19
Schmidt, K. A., Manna, A. C., and Cheung, A. L. (2003). SarT influences sarS expression in Staphylococcus aureus. Infect. Immunity 71, 5139–5148. doi: 10.1128/IAI.71.9.5139-5148.2003
Sedarat, Z., and Taylor-Robinson, A. W. (2022). Biofilm formation by pathogenic bacteria: Applying a Staphylococcus aureus model to appraise potential targets for therapeutic intervention. Pathogens 11, 388. doi: 10.3390/pathogens11040388
Seidl, K., Goerke, C., Wolz, C., Mack, D., Berger-Bchi, B., and Bischoff, M. (2008). Staphylococcus aureus CcpA affects biofilm formation. Infect. Immunity 76, 2044–2050. doi: 10.1128/IAI.00035-08
Sharma, H., Smith, D., Turner, C. E., Game, L., Pichon, B., Hope, R., et al. (2018). Clinical and molecular epidemiology of staphylococcal toxic shock syndrome in the United Kingdom. Emerg. Infect. Dis. 24, 258. doi: 10.3201/eid2402.170606
Sharma, H., Turner, C. E., Siggins, M. K., El-Bahrawy, M., Pichon, B., Kearns, A., et al. (2019). Toxic shock syndrome toxin 1 evaluation and antibiotic impact in a transgenic model of staphylococcal soft tissue infection. Msphere 4, e00665–e00619. doi: 10.1128/mSphere.00665-19
Sharma-Kuinkel, B. K., Mann, E. E., Ahn, J. S., Kuechenmeister, L. J., Dunman, P. M., and Bayles, K. W. (2009). The Staphylococcus aureus LytSR two-component regulatory system affects biofilm formation. J. Bacteriol. 191, 4767–4775. doi: 10.1128/JB.00348-09
Sharp, J. A., Echague, C. G., Hair, P. S., Ward, M. D., Nyalwidhe, J. O., Geoghegan, J. A., et al. (2012). Staphylococcus aureus surface protein SdrE binds complement regulator factor H as an immune evasion tactic. PLoS ONE 7, e38407. doi: 10.1371/journal.pone.0038407
Shopsin, B., Eaton, C., Wasserman, G. A., Mathema, B., Adhikari, R. P., Agolory, S., et al. (2010). Mutations in agr do not persist in natural populations of methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 202, 1593–1599. doi: 10.1086/656915
Shurland, S., Zhan, M., Bradham, D. D., and Roghmann, M.-C. (2007). Comparison of mortality risk associated with bacteremia due to methicillin-resistant and methicillin-susceptible Staphylococcus aureus. Infect. Control Hospit. Epidemiol. 28, 273–279. doi: 10.1086/512627
Siboo, I. R., Cheung, A. L., Bayer, A. S., and Sullam, P. M. (2001). Clumping factor A mediates binding of Staphylococcus aureus to human platelets. Infect. Immunity 69, 3120–3127. doi: 10.1128/IAI.69.5.3120-3127.2001
Singh, R., and Ray, P. (2014). Quorum sensing-mediated regulation of staphylococcal virulence and antibiotic resistance. Fut. Microbiol. 9, 669–681. doi: 10.2217/fmb.14.31
Soldo, B., Lazarevic, V., and Karamata, D. (2002). tagO is involved in the synthesis of all anionic cell-wall polymers in Bacillus subtilis 168. Microbiology 148, 2079–2087. doi: 10.1099/00221287-148-7-2079
Spaan, A. N., Surewaard, B. G., Nijland, R., and Van Strijp, J. A. (2013). Neutrophils versus Staphylococcus aureus: A biological tug of war. Ann. Rev. Microbiol. 67, 629–650. doi: 10.1146/annurev-micro-092412-155746
Speziale, P., and Pietrocola, G. (2021). Staphylococcus aureus induces neutrophil extracellular traps (NETs) and neutralizes their bactericidal potential. Comput. Struct. Biotechnol. J. 19, 3451–3457. doi: 10.1016/j.csbj.2021.06.012
Stach, C. S., Herrera, A., and Schlievert, P. M. (2014). Staphylococcal superantigens interact with multiple host receptors to cause serious diseases. Immunol. Res. 59, 177–181. doi: 10.1007/s12026-014-8539-7
Stapels, D. A., Ramyar, K. X., Bischoff, M., von Köckritz-Blickwede, M., Milder, F. J., Ruyken, M., et al. (2014). Staphylococcus aureus secretes a unique class of neutrophil serine protease inhibitors. Proc. Natl. Acad. Sci. U. S. A. 111, 13187–13192. doi: 10.1073/pnas.1407616111
Steinhuber, A., Goerke, C., Bayer, M. G., Dring, G., and Wolz, C. (2003). Molecular architecture of the regulatory locus sae of Staphylococcus aureus and its impact on expression of virulence factors. J. Bacteriol. 185, 6278–6286. doi: 10.1128/JB.185.21.6278-6286.2003
Sterba, K. M., Mackintosh, S. G., Blevins, J. S., Hurlburt, B. K., and Smeltzer, M. S. (2003). Characterization of Staphylococcus aureus SarA binding sites. J. Bacteriol. 185, 4410–4417. doi: 10.1128/JB.185.15.4410-4417.2003
Suzuki, N., Ohtaguro, N., Yoshida, Y., Hirai, M., Matsuo, H., Yamada, Y., et al. (2015). A compound inhibits biofilm formation of Staphylococcus aureus from Streptomyces. Biol. Pharmaceut. Bullet. 38, 889–892. doi: 10.1248/bpb.b15-00053
Swoboda, J. G., Campbell, J., Meredith, T. C., and Walker, S. (2010). Wall teichoic acid function, biosynthesis, and inhibition. Chembiochem 11, 35–45. doi: 10.1002/cbic.200900557
Tan, L., Li, S. R., Jiang, B., Hu, X. M., and Li, S. (2018). Therapeutic targeting of the Staphylococcus aureus accessory gene regulator (agr) system. Front. Microbiol. 9, 55. doi: 10.3389/fmicb.2018.00055
Tetz, G. V., Artemenko, N. K., and Tetz, V. V. (2009). Effect of DNase and antibiotics on biofilm characteristics. Antimicrob. Agents Chemother. 53, 1204–1209. doi: 10.1128/AAC.00471-08
Thakker, M., Park, J.-S., Carey, V., and Lee, J. C. (1998). Staphylococcus aureus serotype 5 capsular polysaccharide is antiphagocytic and enhances bacterial virulence in a murine bacteremia model. Infect. Immunity 66, 5183–5189. doi: 10.1128/IAI.66.11.5183-5189.1998
Thomer, L., Schneewind, O., and Missiakas, D. (2016). Pathogenesis of Staphylococcus aureus bloodstream infections. Ann. Rev. Pathol. 11, 343. doi: 10.1146/annurev-pathol-012615-044351
Tormo, M. A. A. N., Mart, M., Valle, J., Manna, A. C., Cheung, A. L., Lasa, I., et al. (2005). SarA is an essential positive regulator of Staphylococcus epidermidis biofilm development. J. Bacteriol. 187, 2348–2356. doi: 10.1128/JB.187.7.2348-2356.2005
Trotonda, M. P., Tamber, S., Memmi, G., and Cheung, A. L. (2008). MgrA represses biofilm formation in Staphylococcus aureus. Infect. Immunity 76, 5645–5654. doi: 10.1128/IAI.00735-08
Trotonda, M. P., Xiong, Y. Q., Memmi, G., Bayer, A. S., and Cheung, A. L. (2009). Role of mgrA and sarA in methicillin-resistant Staphylococcus aureus autolysis and resistance to cell wall-active antibiotics. J. Infect. Dis. 199, 209–218. doi: 10.1086/595740
Tsang, L. H., Cassat, J. E., Shaw, L. N., Beenken, K. E., and Smeltzer, M. S. (2008). Factors contributing to the biofilm-deficient phenotype of Staphylococcus aureus sarA mutants. PLoS ONE 3, e3361. doi: 10.1371/journal.pone.0003361
Tu Quoc, P. H., Genevaux, P., Pajunen, M., Savilahti, H., Georgopoulos, C., Schrenzel, J., et al. (2007). Isolation and characterization of biofilm formation-defective mutants of Staphylococcus aureus. Infect. Immunity 75, 1079–1088. doi: 10.1128/IAI.01143-06
Tuchscherr, L., and Löffler, B. (2016). Staphylococcus aureus dynamically adapts global regulators and virulence factor expression in the course from acute to chronic infection. Curr. Genet. 62, 15–17. doi: 10.1007/s00294-015-0503-0
Tuchscherr, L., Löffler, B., Buzzola, F. R., and Sordelli, D. O. (2010). Staphylococcus aureus adaptation to the host and persistence: role of loss of capsular polysaccharide expression. Fut. Microbiol. 5, 1823–1832. doi: 10.2217/fmb.10.147
Tuchscherr, L. P., Buzzola, F. R., Alvarez, L. P., Caccuri, R. L., Lee, J. C., and Sordelli, D. O. (2005). Capsule-negative Staphylococcus aureus induces chronic experimental mastitis in mice. Infect. Immunity 73, 7932–7937. doi: 10.1128/IAI.73.12.7932-7937.2005
Udo, E. E., and Boswihi, S. S. (2017). Antibiotic resistance trends in methicillin-resistant Staphylococcus aureus isolated in Kuwait hospitals: 2011-2015. Medical Principl. Practice 26, 485–490. doi: 10.1159/000481944
Ulrich, M., Bastian, M., Cramton, S. E., Ziegler, K., Pragman, A. A., Bragonzi, A., et al. (2007). The staphylococcal respiratory response regulator SrrAB induces ica gene transcription and polysaccharide intercellular adhesin expression, protecting Staphylococcus aureus from neutrophil killing under anaerobic growth conditions. Mol. Microbiol. 65, 1276–1287. doi: 10.1111/j.1365-2958.2007.05863.x
Valeva, A., Walev, I., Pinkernell, M., Walker, B., Bayley, H., Palmer, M., et al. (1997). Transmembrane β-barrel of staphylococcal α-toxin forms in sensitive but not in resistant cells. Proc. Natl. Acad. Sci. U. S. A. 94, 11607–11611. doi: 10.1073/pnas.94.21.11607
Valle, J., Echeverz, M., and Lasa, I. (2019). σB inhibits poly-N-acetylglucosamine exopolysaccharide synthesis and biofilm formation in Staphylococcus aureus. J. Bacteriol. 201, e00098–e00019. doi: 10.1128/JB.00098-19
Valle, J., Toledo-Arana, A., Berasain, C., Ghigo, J. M., Amorena, B., Penadés, J. R., et al. (2003). SarA and not σB is essential for biofilm development by Staphylococcus aureus. Mol. Microbiol. 48, 1075–1087. doi: 10.1046/j.1365-2958.2003.03493.x
Van Boeckel, T. P., Brower, C., Gilbert, M., Grenfell, B. T., Levin, S. A., Robinson, T. P., et al. (2015). Global trends in antimicrobial use in food animals. Proc. Natl. Acad. Sci. U. S. A. 112, 5649–5654. doi: 10.1073/pnas.1503141112
van Dalen, R., Peschel, A., and van Sorge, N. M. (2020). Wall teichoic acid in Staphylococcus aureus host interaction. Trends Microbiol. 28, 985–998. doi: 10.1016/j.tim.2020.05.017
van Wamel, W., Xiong, Y. Q., Bayer, A., Yeaman, M., Nast, C., and Cheung, A. (2002). Regulation of Staphylococcus aureus type 5 capsular polysaccharides by agr and sarA in vitro and in an experimental endocarditis model. Microbial Pathog. 33, 73–79. doi: 10.1006/mpat.2002.0513
Vandenesch, F., Lina, G., and Henry, T. (2012). Staphylococcus aureus hemolysins, bi-component leukocidins, and cytolytic peptides: A redundant arsenal of membrane-damaging virulence factors? Front. Cell. Infect. Microbiol. 2, 12. doi: 10.3389/fcimb.2012.00012
Verdon, J., Girardin, N., Lacombe, C., Berjeaud, J. M., and Héchard, Y. (2009). δ-hemolysin, an update on a membrane-interacting peptide. Peptides 30, 817–823. doi: 10.1016/j.peptides.2008.12.017
Vergara-Irigaray, M., Maira-Litrán, T., Merino, N., Pier, G. B., Penadés, J. R., and Lasa, I. (2008). Wall teichoic acids are dispensable for anchoring the PNAG exopolysaccharide to the Staphylococcus aureus cell surface. Microbiology 154, 865–877. doi: 10.1099/mic.0.2007/013292-0
Von Eiff, C., Becker, K., Machka, K., Stammer, H., and Peters, G. (2001). Nasal carriage as a source of Staphylococcus aureus bacteremia. N. Engl. J. Med. 344, 11–16. doi: 10.1056/NEJM200101043440102
Voss, A., Loeffen, F., Bakker, J., Klaassen, C., and Wulf, M. (2005). Methicillin-resistant Staphylococcus aureus in pig farming. Emerg. Infect. Dis. 11, 1965. doi: 10.3201/eid1112.050428
Vuong, C., Kidder, J. B., Jacobson, E. R., Otto, M., Proctor, R. A., and Somerville, G. A. (2005). Staphylococcus epidermidis polysaccharide intercellular adhesin production significantly increases during tricarboxylic acid cycle stress. J. Bacteriol. 187, 2967–2973. doi: 10.1128/JB.187.9.2967-2973.2005
Walev, I., Weller, U., Strauch, S., Foster, T., and Bhakdi, S. (1996). Selective killing of human monocytes and cytokine release provoked by sphingomyelinase (beta-toxin) of Staphylococcus aureus. Infect. Immunity 64, 2974–2979. doi: 10.1128/iai.64.8.2974-2979.1996
Wann, E. R., Dassy, B., Fournier, J.-M., and Foster, T. J. (1999). Genetic analysis of the cap5 locus of Staphylococcus aureus. FEMS Microbiol. Lett. 170, 97–103. doi: 10.1111/j.1574-6968.1999.tb13360.x
Wann, E. R., Gurusiddappa, S., and Hk, M. (2000). The fibronectin-binding MSCRAMM FnbpA of Staphylococcus aureus is a bifunctional protein that also binds to fibrinogen. J. Biol. Chem. 275, 13863–13871. doi: 10.1074/jbc.275.18.13863
Wanner, S., Schade, J., Keinhörster, D., Weller, N., George, S. E., Kull, L., et al. (2017). Wall teichoic acids mediate increased virulence in Staphylococcus aureus. Nat. Microbiol. 2, 1–12. doi: 10.1038/nmicrobiol.2016.257
Ward, J. (1981). Teichoic and teichuronic acids: Biosynthesis, assembly, and location. Microbiol. Rev. 45, 211–243. doi: 10.1128/mr.45.2.211-243.1981
Waters, E. M., Rowe, S. E., O'Gara, J. P., and Conlon, B. P. (2016). Convergence of Staphylococcus aureus persister and biofilm research: Can biofilms be defined as communities of adherent persister cells? PLoS Pathog. 12, e1006012. doi: 10.1371/journal.ppat.1006012
Watkins, R. L., Pallister, K. B., and Voyich, J. M. (2011). The SaeR/S gene regulatory system induces a pro-inflammatory cytokine response during Staphylococcus aureus infection. PLoS ONE 6, e19939. doi: 10.1371/journal.pone.0019939
Watts, A., Ke, D., Wang, Q., Pillay, A., Nicholson-Weller, A., and Lee, J. C. (2005). Staphylococcus aureus strains that express serotype 5 or serotype 8 capsular polysaccharides differ in virulence. Infect. Immunity 73, 3502–3511. doi: 10.1128/IAI.73.6.3502-3511.2005
Weber, C. (2015). Neutrophil extracellular traps mediate bacterial liver damage. Nat. Rev. Gastroenterol. Hepatol. 12, 251–251. doi: 10.1038/nrgastro.2015.60
Weidenmaier, C., and Peschel, A. (2008). Teichoic acids and related cell-wall glycopolymers in Gram-positive physiology and host interactions. Nat. Rev. Microbiol. 6, 276–287. doi: 10.1038/nrmicro1861
Weidenmaier, C., Peschel, A., Xiong, Y. Q., Kristian, S. A., Dietz, K., Yeaman, M. R., et al. (2005). Lack of wall teichoic acids in Staphylococcus aureus leads to reduced interactions with endothelial cells and to attenuated virulence in a rabbit model of endocarditis. J. Infect. Dis. 191, 1771–1777. doi: 10.1086/429692
Wertheim, H. F., Van Kleef, M., Vos, M. C., Ott, A., Verbrugh, H. A., and Fokkens, W. (2006). Nose picking and nasal carriage of Staphylococcus aureus. Infect. Control Hospit. Epidemiol. 27, 863–867. doi: 10.1086/506401
Wertheim, H. F., Vos, M. C., Ott, A., van Belkum, A., Voss, A., Kluytmans, J. A., et al. (2004). Risk and outcome of nosocomial Staphylococcus aureus bacteraemia in nasal carriers versus non-carriers. Lancet 364, 703–705. doi: 10.1016/S0140-6736(04)16897-9
Whitchurch, C. B., Tolker-Nielsen, T., Ragas, P. C., and Mattick, J. S. (2002). Extracellular DNA required for bacterial biofilm formation. Science 295, 1487–1487. doi: 10.1126/science.295.5559.1487
Wilke, G. A., and Wardenburg, J. B. (2010). Role of a disintegrin and metalloprotease 10 in Staphylococcus aureus α-hemolysin–mediated cellular injury. Proc. Natl. Acad. Sci. U. S. A. 107, 13473–13478. doi: 10.1073/pnas.1001815107
Wittekind, M. A., Frey, A., Bonsall, A. E., Briaud, P., Keogh, R. A., Wiemels, R. E., et al. (2022). The novel protein ScrA acts through the SaeRS two-component system to regulate virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 117, 1196–1212. doi: 10.1111/mmi.14901
Woehl, J. L., Stapels, D. A., Garcia, B. L., Ramyar, K. X., Keightley, A., Ruyken, M., et al. (2014). The extracellular adherence protein from Staphylococcus aureus inhibits the classical and lectin pathways of complement by blocking formation of the C3 proconvertase. J. Immunol. 193, 6161–6171. doi: 10.4049/jimmunol.1401600
Wolz, C., McDevitt, D., Foster, T. J., and Cheung, A. L. (1996). Influence of agr on fibrinogen binding in Staphylococcus aureus Newman. Infect. Immunity 64, 3142–3147. doi: 10.1128/iai.64.8.3142-3147.1996
Xie, X., Liu, X., Li, Y., Luo, L., Yuan, W., Chen, B., et al. (2020). Advanced glycation end products enhance biofilm formation by promoting extracellular DNA release through sigB upregulation in Staphylococcus aureus. Front. Microbiol. 11, 1479. doi: 10.3389/fmicb.2020.01479
Yang, S.-J., Rice, K. C., Brown, R. J., Patton, T. G., Liou, L. E., Park, Y. H., et al. (2005). A LysR-type regulator, CidR, is required for induction of the Staphylococcus aureus cidABC operon. J. Bacteriol. 187, 5893–5900. doi: 10.1128/JB.187.17.5893-5900.2005
Yarwood, J. M., and Schlievert, P. M. (2003). Quorum sensing in Staphylococcus infections. J. Clin. Investig. 112, 1620–1625. doi: 10.1172/JCI200320442
Young, R. (1992). Bacteriophage lysis: Mechanism and regulation. Microbiol. Rev. 56, 430–481. doi: 10.1128/mr.56.3.430-481.1992
Zahar, J.-R., Clec'h, C., Tafflet, M., Garrouste-Orgeas, M., Jamali, S., Mourvillier, B., et al. (2005). Is methicillin resistance associated with a worse prognosis in Staphylococcus aureus ventilator-associated pneumonia? Clin. Infect. Dis. 41, 1224–1231. doi: 10.1086/496923
Zapotoczna, M., McCarthy, H., Rudkin, J. K., O'Gara, J. P., and O'Neill, E. (2015). An essential role for coagulase in Staphylococcus aureus biofilm development reveals new therapeutic possibilities for device-related infections. J. Infect. Dis. 212, 1883–1893. doi: 10.1093/infdis/jiv319
Zhang, L., Jacobsson, K., Vasi, J., Lindberg, M., and Frykberg, L. (1998). A second IgG-binding protein in Staphylococcus aureus. Microbiology 144, 985–991. doi: 10.1099/00221287-144-4-985
Zhang, Y., Wu, M., Hang, T., Wang, C., Yang, Y., Pan, W., et al. (2017). Staphylococcus aureus SdrE captures complement factor H's C-terminus via a novel ‘close, dock, lock and latch'mechanism for complement evasion. Biochem. J. 474, 1619–1631. doi: 10.1042/BCJ20170085
Zhang, Y. H., Ginsberg, C., Yuan, Y., and Walker, S. (2006). Acceptor substrate selectivity and kinetic mechanism of Bacillus subtilis TagA. Biochemistry 45, 10895–10904. doi: 10.1021/bi060872z
Zielinska, A. K., Beenken, K. E., Joo, H. S., Mrak, L. N., Griffin, L. M., Luong, T. T., et al. (2011). Defining the strain-dependent impact of the Staphylococcal accessory regulator (sarA) on the alpha-toxin phenotype of Staphylococcus aureus. J. Bacteriol. 193, 2948–2958. doi: 10.1128/JB.01517-10
Zielinska, A. K., Beenken, K. E., Mrak, L. N., Spencer, H. J., Post, G. R., Skinner, R. A., et al. (2012). sarA-mediated repression of protease production plays a key role in the pathogenesis of Staphylococcus aureus USA 300 isolates. Mol. Microbiol. 86, 1183–1196. doi: 10.1111/mmi.12048
Zipperer, A., Konnerth, M. C., Laux, C., Berscheid, A., Janek, D., Weidenmaier, C., et al. (2016). Human commensals producing a novel antibiotic impair pathogen colonization. Nature 535, 511–516. doi: 10.1038/nature18634
Keywords: methicillin-resistant Staphylococcus aureus (MRSA), pathogenesis, biofilm, virulence, two-component system (TCS)
Citation: Patel H and Rawat S (2023) A genetic regulatory see-saw of biofilm and virulence in MRSA pathogenesis. Front. Microbiol. 14:1204428. doi: 10.3389/fmicb.2023.1204428
Received: 12 April 2023; Accepted: 30 May 2023;
Published: 22 June 2023.
Edited by:
Alina Maria Holban, University of Bucharest, RomaniaReviewed by:
Minh-Thu Nguyen, University Hospital Münster, GermanyCopyright © 2023 Patel and Rawat. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Seema Rawat, c2VlbWEucmF3YXRAY3VnLmFjLmlu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.