
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 07 September 2023
Sec. Microbe and Virus Interactions with Plants
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1203796
This article is part of the Research Topic Beneficial Microbe-Plant Interactions Under Biotic/Abiotic Stress Conditions View all 23 articles
 Qian Wang1†
Qian Wang1† Juan Song1†
Juan Song1† Jinlian Zhang1
Jinlian Zhang1 Xiaojuan Qin1
Xiaojuan Qin1 Yihao Kang1
Yihao Kang1 Shilv Huang1
Shilv Huang1 Shengmao Zhou2*
Shengmao Zhou2* Tingsu Chen1*
Tingsu Chen1*Ginger is one of the important spice crops in the world. Due to the prevalence of ginger wilt disease and the lack of effective prevention and control methods, the planting area, total production and value have declined sharply, which have become a key factor restricting ginger industry development in China. Understanding the influence of microbial agents on the rhizosphere microbiota of ginger will facilitate developing novel technologies for the prevention and control of ginger wilt disease. In the new planting and continuous cropping ginger fields, using large-root ginger and microbial agents, two inoculation levels (inoculation and no inoculation) were designed, and high-throughput sequencing technology was used to study the bacterial community structure in the rhizosphere soil at mature stage of ginger. The results showed that newly planted ginger showed a significant yield advantage over continuous cropping ginger, with a yield increase of 39% to 56%, and the lowest ginger wilt disease index. The community structure at the phylum level of soil bacteria in each treatment was very similar to that in the control, but the abundance of some taxonomic units changed significantly. The four dominant phyla of bacteria in mature ginger rhizosphere soil were Proteobacteria, Actinobacteria, Chloroflexi, and Acidobacteria, accounting for 72.91% to 89.09% of the total. The microbial agent treatment of continuous cropping had beneficial microorganisms such as Acidobacteria and Gemmatimonadetes with abundances increased by 12.2% and 17.1%, respectively, compared to the control. The microbial inoculant treatment of newly planted ginger increased the abundance of Acidobacteria and Gemmatimonadetes by 34.4% and 10.7%, respectively, compared to the control. The composition of bacterial communities were affected by changes in soil properties. Redundancy analysis showed that the hydrolysable nitrogen, available phosphorus, available potassium, and organic matter were significantly related to the composition of soil bacterial communities. Therefore, the microbial agents can not only promote the proliferation of beneficial microorganisms in the continuous cropping soil but also further reshape the soil bacterial community structure by changing the soil physicochemical properties such as effective phosphorus. These results provided a reference for related research on the impact of ginger continuous cropping on soil environment and soil management improvement in ginger fields.
Ginger (Zingiber officinale Rosc.) is one of the most important spice crops in China, as well as in several other countries including India, Nepal, Indonesia, and Nigeria (Thekkan et al., 2020). Ginger is widely grown in China and has gradually developed into a large-scale and industrialized production, becoming an important source of income for farmers. However, ginger has a serious problem of continuous cropping obstacles, which leads to significant yield decrease and economic loss. The continuous cropping obstacle for ginger is mainly caused by ginger wilt disease caused by Ralstonia solanacarum, which does not occur when the soil and ginger seeds do not carry the pathogen. The incidence rate of ginger wilt disease is 60% when only the soil carries the pathogen, while it is 90% when both the soil and ginger seeds carry the pathogen (Wang et al., 2016). Currently, the methods for the prevention and control of ginger wilt disease mainly include strengthening field management, cultivating resistant varieties, using chemical agents, and applying biocontrol agents (Ohike et al., 2013; Thomas and Upreti, 2014). However, it has been shown that field management is difficult to effectively control the occurrence of ginger wilt disease, the breeding of resistant plants is time-consuming, and chemical control pollutes the environment, disrupts the ecological balance, and long-term use of chemical agents can lead to the pathogen developing resistance. Use of safe biological products is currently a trend (Guo et al., 2004). Therefore, use of beneficial microorganisms to replace harmful chemicals has aroused great interest (Kwon and Kim, 2014).
Microbial agents are microbial live preparations with a certain porous substance as a carrier, which can reproduce in soil or substrate and form an advantageous microbial community for plant growth (Hou et al., 2016). They can improve the soil microbial flora and metabolic activity, prevent and control soil-borne diseases, and thus have a significant promoting effect on plant growth. However, considering that R. solanacarum is a highly destructive soil-borne bacterium that can survive for a long time in soil, water, and plants, a single microbial agent has poor efficacy and is subject to problems such as poor persistence and strong environmental dependence, making it difficult to achieve ideal control effect (Huang et al., 2013). In contrast, use of composite microbial inoculant for biological control has become a new research hotspot, and the focus is the compound of beneficial bacteria. Reports on use of composite microbial agents to control R. solanacarum mainly focus on the combination of beneficial bacterial strains. For example, Wang et al. (2017) mixed two strains of Bacillus isolated from the healthy tomato rhizosphere, and the effect on controlling tomato bacterial wilt was significantly better than that of a single strain. Huang et al. (2015) mixed Bacillus amyloliquefaciens and Streptomyces sulfate to control tobacco bacterial wilt, achieving an efficacy of 65.85% and reducing the amount of chemical pesticides used by half. Therefore, it is important to improve the control effect of R. solanacarum on ginger by using composite microbial agents.
Arbuscular mycorrhiza (AM) fungi, Trichoderma spp., and plant growth-promoting rhizobacteria (PGPR), widely distributed, are important soil beneficial bacteria (Afaque et al., 2017; Che et al., 2019). AM fungi can form mutualistic associations with the roots of most terrestrial plants and promote the absorption and utilization of water and mineral elements, particularly nitrogen, phosphorus, and potassium, benefiting plant growth and development (Smith and Read, 2008; Sharifi, et al., 2007). PGPR includes a variety of soil microorganisms, such as nitrogen-fixing bacteria Azotobacter sp., and certain species of Pseudomonas and Bacillus sp. (Glick et al., 1998). Trichoderma spp. is a common type of fungus in soil. Studies have shown that Trichoderma has biocontrol effects, has the ability to dissolve insoluble inorganic phosphates and potassium in soil, and can promote the germination of various plant seeds and the growth of seedlings (Zhang et al., 2012). To date, many studies have reported the disease control effects of Trichoderma against tomato bacterial wilt and tobacco bacterial wilt (Zhu and Yao, 2014; Tan et al., 2011), but its effect on ginger disease control and the root soil microbial community has not received enough attention.
Rhizosphere microorganisms are important components of soil ecosystems (Yang et al., 2016), affecting energy flow and material conversion in soil environments and participating in many important biochemical reactions (Ling et al., 2014). Bacteria are one of the main components of microorganisms, and their stable community structure and functional diversity play an important role in maintaining soil microecological balance (Jacobsen and Hjelms, 2014). Therefore, studying soil microecosystem have important theoretical and practical significance in tackling challenges associated with continuous cropping (Xia et al., 2018). We speculate that, by applying microbial agents, the abundance, community composition, and functional diversity of rhizosphere microorganisms will be improved. Bacillus and Trichoderma as antagonists of plant pathogen are widely used to improve plant health and productivity, combining the use of biocontrol agents has potential synergistic effects (Huang et al., 2021). Our previous pot experiment results showed that the combination of AMF, Bacillus and Trichoderma had the best control effect than the single use, so the mixed bacteria were selected to carry out the study. In this study, a compound microbial agent with excellent promotion and disease prevention ability for ginger previously selected in a previous screening was applied under field conditions with both new planting and continuous cropping modes. The impact of the exogenous compound microbial agent on rhizosphere microorganisms was analyzed. High-throughput sequencing technology was used to explore the response of ginger rhizosphere bacterial communities to the compound microbial agent and the correlation between the changes in bacterial community structure and ginger disease prevention. This study provides a theoretical basis for exploring the field disease prevention mechanism of compound microbial agents on ginger.
The ginger variety used in the field experiment was Shandong Laiwu ginger, and the microbial agents applied were the experimental samples from Guangxi Academy of Agricultural Sciences, including a mix of arbuscular mycorrhizal (AM) agents in the ratio 1:1:1 (Glomus reticulatum LCGX-39, Glomus mosseae FSGX-1631, and Glomus versiforme LCGX-58), Bacillus velezensis KC-5, and Trichoderma viride BJM-11. There was no mutual inhibition among the strains.
The experiment was conducted at the Wu-Ming Li-Jian Research Station of Guangxi Academy of Agricultural Sciences (23°15′N, 108°02′E), and the tested soil was red soil. Before the experiment, the basic physical and chemical properties of the soil were as follows: available N 91.83 mg/kg, available P 81.86 mg/kg, available K 256.53 mg/kg, organic matter 23.67 g/kg, and pH 6.56.
The experimental area is in a subtropical monsoon climate zone with abundant light, heat, and water resources. The average annual temperature is 21.7°C, the average annual sunshine hours are 1,660, and the average annual rainfall is 1,300 mm.
The field for the continuous cropping experiment was used for planting ginger for two consecutive years, and the field for the new planting experiment was used for planting ginger for the first time.
There were four treatments in the experiment, including new planting plus microbial agent treatment (NT), new planting only control (NK), continuous cropping plus microbial agent treatment (CT), and continuous cropping only control (CK). Each treatment had three replicates, and a randomized complete block design was employed. The soil preparation was conducted on February 13, 2021, and the plastic film was removed on February 28. The ginger was planted on March 7. The AM agent was applied by spreading before planting ginger, with approximately 50 g of the agent per block (the spore count of the agent was approximately 85 spores per gram). The plants were irrigated with Bacillus velezensis (at a concentration of 108 CFU/mL) and Trichoderma viride (at a concentration of 106 CFU/mL) suspension since a month after planting for three times at April 7, May 7, and June 7, respectively, during the whole growth duration, with a rate of 200 kg/ha each time.
The land with a deep, loose, and fertile loamy and sandy soil layer and good drainage was selected, and the soil was well prepared. After application of base fertilizer, the land was ridged to a height of 60 cm and a width of 4.5 m. A drainage ditch with a width of 60 cm was built between each ridge. Before being planted, the pieces of ginger were first washed with tap water, then soaked in multi-bactericide (diluted at a ratio of 1:1,000) for 30 min before being dried at room temperature for 1–2 days. The disinfected ginger pieces were buried in sterile sand in a greenhouse for sprouting at 15°C–25°C and soil humidity 70%–80%. When the sprouts reached a length of 1 cm, they were transplanted to the field.
There were four ridges in total. Each ridge was 4.5 m wide and 70 m long divided into four small areas. The ginger is planted in two small beds with a 50 cm aisle in the middle. Each bed is 1.5 m wide, and two rows of ginger had a space of 25 cm × 45 cm between plants. The continuous cropping and new planting experiments were set with two treatments: inoculation and control (Figure 1).
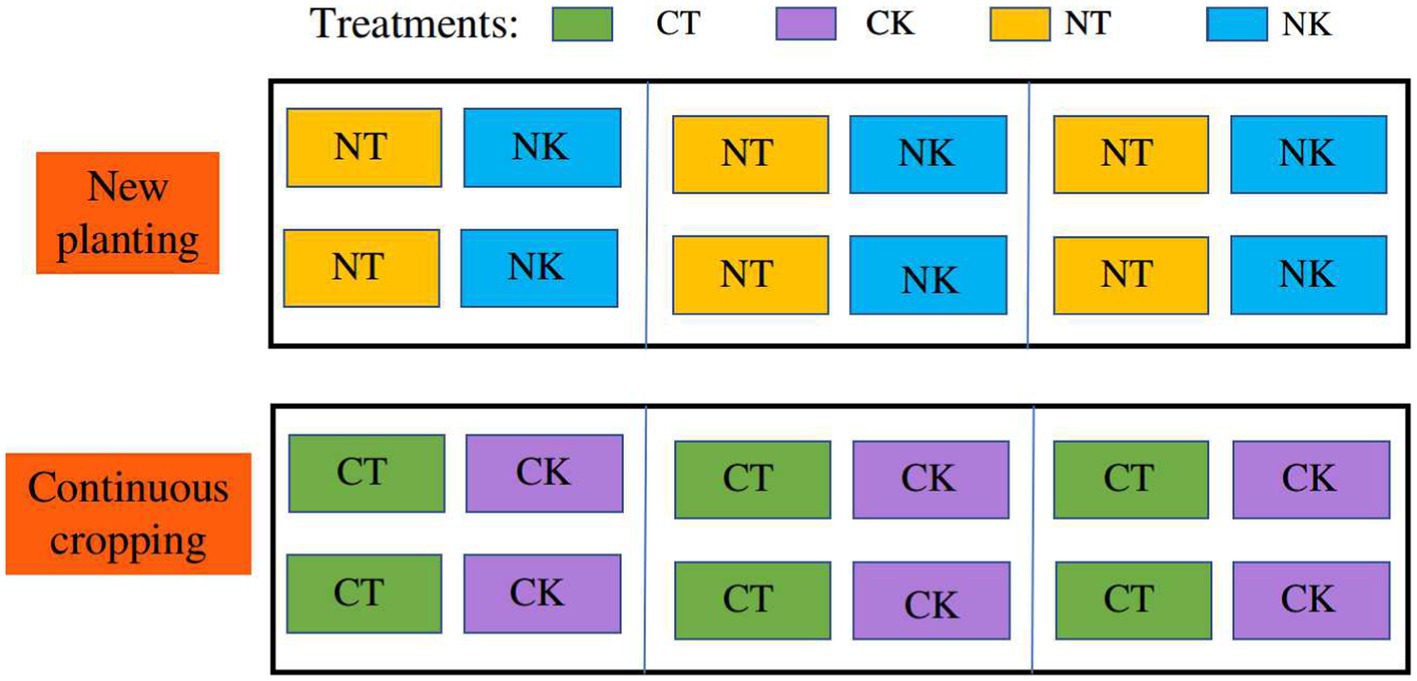
Figure 1. Field experiment design for microbial agent treatments in newly planted and continuous cropping ginger. NT, newly-planted microbial treatment; NK, newly-planted control; CT, continuous cropping microbial treatment; CK, continuous cropping control.
When gingers were harvested on December 10, 2021, the severity index and control efficacy of ginger wilt disease were scored according to NY/T1464.31 “Guidelines for Field Efficacy Trials of Pesticides Part 31: Fungicides for the Control of Ginger Wilt Disease.” The disease severity grading criteria are as follows: grade 0, asymptomatic; grade 1, slight yellowing of 10% of leaves, no obvious symptoms in fleshy stems; grade 3, yellowing of 11% to 30% of leaves, slight curling of leaf edges, water-stained spots appear on fleshy stems; grade 5, yellowing of 31% to 50% of leaves, curling of leaf edges, stunted plants, partial rotting of fleshy stems; grade 7, more than 51% of leaves are withered, plants wilt, and most of the fleshy stems are rot; grade 9, plants die, fleshy stems are rot or only fibrous vascular bundles are left. The disease severity index was calculated as follows: Disease severity index = Σ (Number of plants in each disease grade × Corresponding disease grade)/(Maximum disease grade × Total number of plants) × 100. The control efficacy (%) was calculated as (Disease severity index of control group − Disease severity index of treatment group)/Disease severity index of control group × 100.
The rhizosphere soil of ginger was collected by five-point sampling method at the harvest time in December 2021. The roots with soil were collected and put into a plastic bag, and kept on ice for transportation. In laboratory, the ginger roots were shaken to drop the loose surface soil that is the rhizosphere soil, which was then collected using a sterile brush (Feng et al., 2017). The soil was air-dried and sieved through a 20-mesh sieve, and 300 g was stored for determining the physical and chemical properties of the soil. The dried soil samples were used to measure soil pH using the water extraction method, soil organic matter content using the potassium dichromate heating method, available nitrogen using the alkaline hydrolysis diffusion method, available phosphorus using the molybdenum-antimony colorimetric method, and available potassium using the flame photometry method (Liang et al., 2009).
Genomic DNA was extracted from soil using the Fast DNA TM SPIN Kit (MP Biomedicals, Solon, OH, United States). After extraction, the DNA was checked for quality using 0.8% agarose gel electrophoresis and a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States).
The Ralstonia solanacearum in ginger rhizosphere soil of different treatments was determined by real-time fluorescence quantitative PCR. The flic gene of R. solanacearum was amplified using a primer pair flicF/flicR (5′-GAA CGC CAA CGG TGC GAA CT-3′/5′-GGC GGC CTT CAG GGA GGT C-3′) (Zhao et al., 2019). The plasmid containing the flic gene of R. solanacearum was diluted 10 times to prepare a standard curve of 103–109 copies/μL.
The SYBR® Premix Ex Taq™ real-time fluorescent PCR kit from TaKaRa company was used to run on the ABI 7500™ real-time fluorescent quantitative PCR detector. The amplification reaction system was 20 μL, SYBR® Premix Ex Taq™ (2 ×) 10 μL, ROX reference dye (50×) 0.4 μL, upstream and downstream primers (10 mmol L−1) 0.8 μL, DNA template 2 μL, double distilled water 6.0 μL. Three sample replicates were set in each group, and three replicates were measured. The copy number of flic gene in 1 g dry soil sample was calculated according to the measured Ct value, and the results were presented in logarithmic form l g (copies/g, dry soil).
The V3–V4 region of the bacterial 16S rRNA gene was amplified with PCR using 338F (5′-ACT CCT ACG GGA GGC AGC A-3′) and 806R (5′-GGA CTA CHV GGG TWT CTA AT-3′) primers (Yang et al., 2019). The PCR reaction mixture (25 μL) contained 0.25 μL of DNA polymerase, 5 μL of reaction buffer, 5 μL of high GC buffer, 2 μL of dNTP (10 mM), 2 μL of template DNA, 1 μL of each primer (10 μM), and 8.75 μL of ddH2O. The PCR conditions were as follows: 98°C for 30 s; 98°C for 15 s, 50°C for 30 s, 72°C for 30 s, 27 cycles; 72°C for 5 min. The PCR products were then used to prepare sequencing libraries using the TruSeq Nano DNA LT Library Prep Kit and sequenced on an Illumina MiSeq platform. Raw reads were deposited in the NCBI Sequence Read Archive (SRA) database (Accession Number: PRJNA977085).
The Illumina MiSeq raw paired-end sequencing data were in FASTQ format. The quality control was performed by removing sequences with a quality score less than Q20. The quality-filtered sequences were then assembled using FLASH with a minimum overlap of 10 bp and no mismatches. The sequences containing ambiguous bases or chimeras were removed using QIIME2 (Hugo et al., 2020). The filtered sequences were aligned to the 16S rRNA gene sequences in the GreenGenes database using the UCLUST algorithm implemented in QIIME2 with a 97% similarity threshold for operational taxonomic units (OTU) clustering.
To test the effects of treatments on crop yield, soil physical and chemical properties, the least significant difference test was performed at 5% level using SPSS Statistics for Windows (SPSS 19.0) software package. The OTU table with 97% similarity was selected, and the bacterial community composition analysis and Venn analysis were performed using the R language (v.3.3.1) tool, and statistics and plotting were performed. Multivariate statistical methods such as principal coordinates analysis (PCoA) were used for analysis of differences in bacterial communities. The Metastats and LEfSe software was used to screen biomarker features of each group (Segata et al., 2011). To study the relationship between the soil physicochemical indices and samples and microbial communities, redundancy analysis (RDA) was used to correlate the biochemical indices as environmental factors with the sample communities.
In this study, soil properties, including alkaline nitrogen, available phosphorus, available potassium, organic matter, and pH in the ginger rhizosphere soils with four treatments were measured and the results were shown in Table 1. It can be seen from Table 1 that regardless of the microbial treatment (CT) or control (CK), continuous cropping of ginger significantly reduced the main physicochemical indicators of soil in the newly-planted microbial treatment (NT) compared to the control (NK) (p < 0.05). This indicates that continuous ginger cultivation led to more consumption of soil nutrients. There were also significant differences in soil physicochemical properties between the microbial treatment and control. The effective P, available K, and organic matter in NT were higher than those in NK, especially the organic matter, which was 13% higher in NT than in NK (p < 0.05). The alkaline nitrogen in CT was higher than that in CK, and the other indicators showed no significant difference. There was no significant difference in soil pH between the newly-planted and continuous cropping treatments.
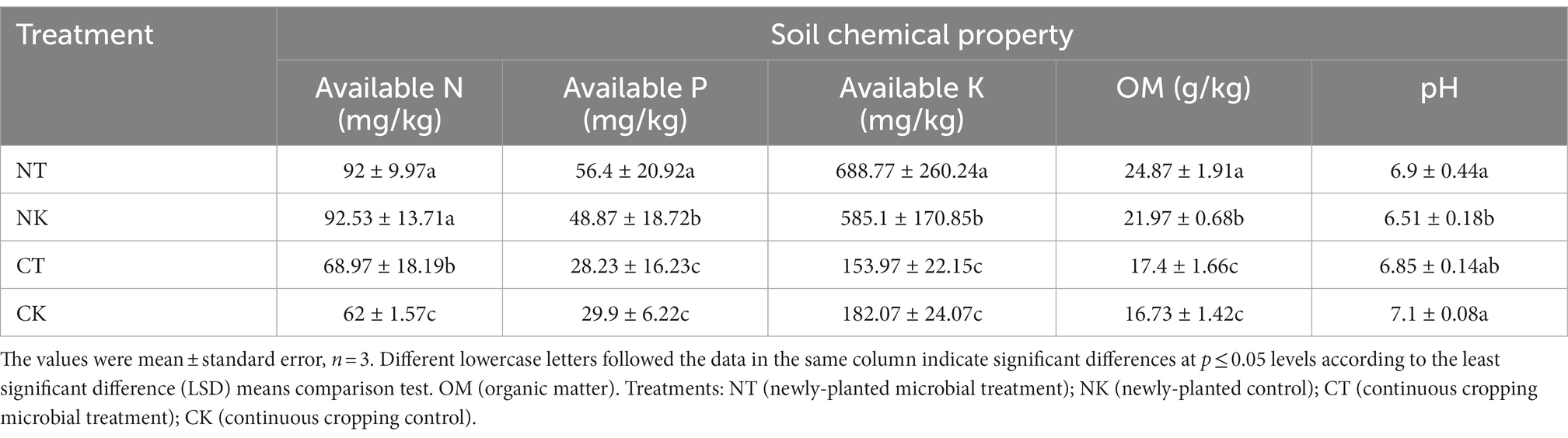
Table 1. Effects of different planting systems on the physicochemical and biological characteristics of the ginger rhizosphere soil.
As indicated in Table 2, the three treatments including NT, NK, and CT significantly reduced the disease index of ginger wilt (p < 0.05) compared to CK. Among them, NT had the lowest disease index (5.2) and the highest correctional efficiency (85.87%). NK had a disease index of 8.4 and a correctional efficiency of 77.17%, followed by CT with a disease index of 15.6 and a correctional efficiency of 57.61%. The flic gene abundance of R. solanacearum in the rhizosphere soil of NT treatment was 2.51 × 106 copies/g, dry soil, and the flic gene abundance of R. solanacearum in the rhizosphere soil of NK treatment was 10.62 × 106 copies/g, dry soil, it is 4.23 times that of NT. The flic gene abundance of R. solanacearum in rhizosphere soil of CT treatment was 15.23 × 106 copies/g, dry soil, and the flic gene abundance of R. solanacearum in rhizosphere soil of CK treatment was 38.47 × 106 copies/g, dry soil, it is 2.53 times that of CT. The abundance of flic gene in the rhizosphere of NT and CT was significantly lower than that of NK and CK (p < 0.05), respectively. Under the same microbial application conditions, the yield in NT was significantly increased by 39% compared to that in CT, and the yield in NK was significantly increased by 56% compared to that in CK.
The V3–V4 region of bacterial 16S rDNA gene was amplified, and the structure and composition of bacterial populations were analyzed by high-throughput sequencing. A total of 1,269,686 sequences with an average length of 416 bp were obtained from 12 soil samples using Illumina MiSeq. After quality control and chimera filtering, 634,843 high-quality sequences were obtained. Finally, 601,196 effective tags were detected in all the samples, accounting for 94.7% of the total quantified sequences. The quality reads of soil samples ranged from 53,462 to 69,760 in the 12 replicates (Supplementary Table S1). These sequences were clustered into operational taxonomic units (OTUs) at a 3% difference level and annotated with species names, resulting in a total of 23,872 OTUs. In the sparse curve analysis, all the curves tended to saturate (Supplementary Figure S1), and effective coverage of almost all the bacterial diversity was found at 97% sequence similarity by rank-abundance curve analysis (Supplementary Figure S2).
At the similarity level of 97%, OTUs were obtained for each treatment, and a Venn diagram was used to visualize the number of the shared and unique OTUs in different treatments. As shown in Figure 2, there were 2,059 shared OTUs among the four treatments, and the numbers of OTUs unique to NT, NK, CT, and CK were 319, 218, 273, and 248, respectively, indicating significant changes in the bacterial colony diversity in different treatments (Figure 2).
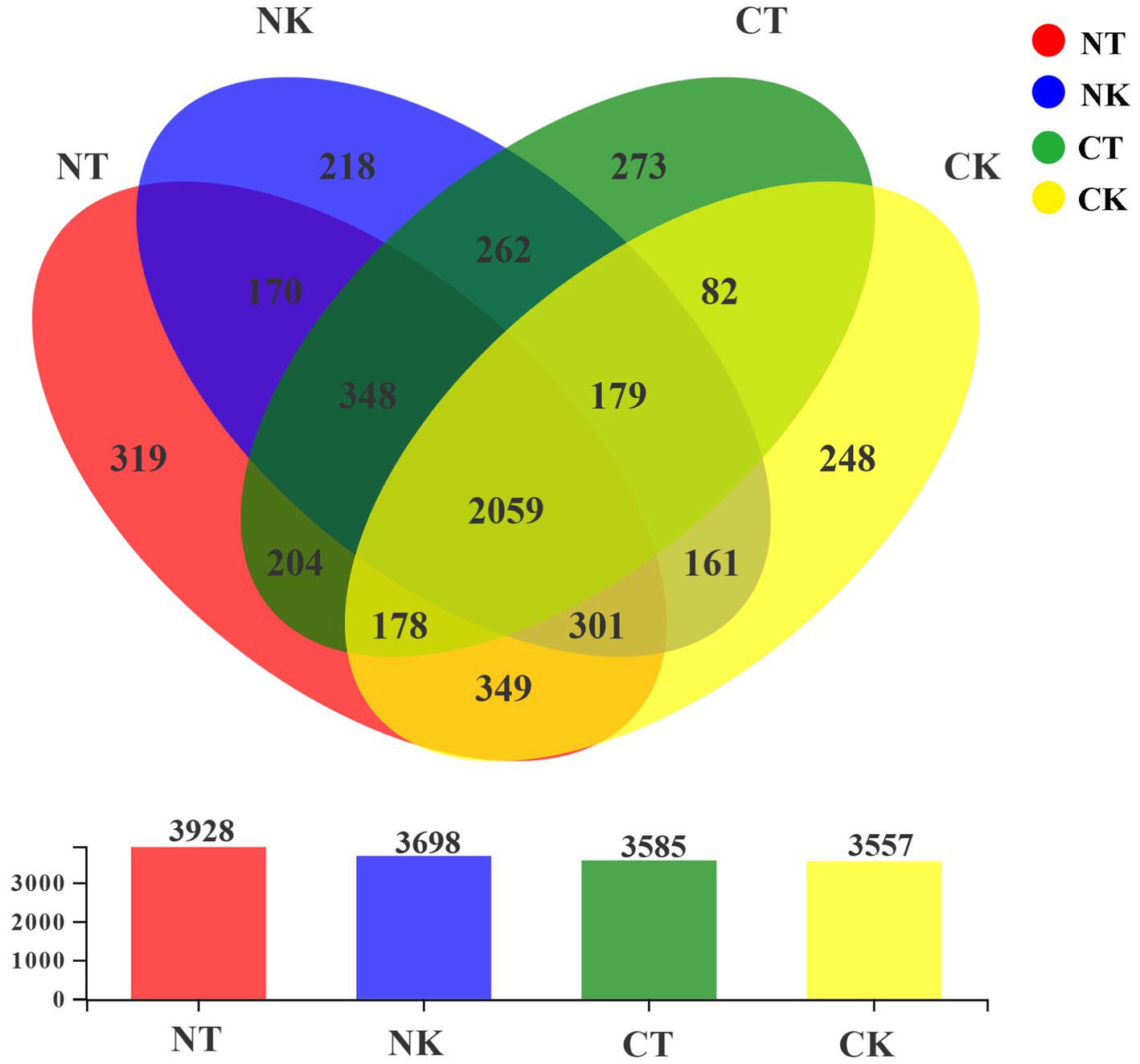
Figure 2. Venn diagram of different treatments (97% sequence similarity). NT, newly-planted microbial treatment; NK, newly-planted control; CT, continuous cropping microbial treatment; CK, continuous cropping control.
The OTUs were classified into 32 bacterial phyla and 94 classes. At the phylum level (Figure 3A), the top 13 phyla were Proteobacteria (24.16% to 29.71%), Actinobacteria (25.05% to 28%), Chloroflexi (13.73% to 16.32%), Acidobacteria (9.97% to 15.06%), Firmicutes (3.14% to 5.92%), Gemmatimonadetes (2.47% to 4.07%), Bacteroidetes (1.42% to 3.82%), Cyanobacteria (0.51% to 2.89%), Nitrospirae (0.93% to 1.79%), Rokubacteria (0.74% to 2.34%), Planctomycetes (0.88% to 1.03%), Patescibacteria (0.49% to 1.15%), and Verrucomicrobia (0.56% to 1.04%). Proteobacteria and Actinobacteria were the dominant phyla, accounting for 49.21% to 57.71% of the total bacteria, indicating their high abundance. The phylum-level abundance of Proteobacteria, Actinobacteria, Firmicutes, and Bacteroidetes showed significant changes between the new planting and continuous cropping treatments. No matter new planting or continuous cropping, the application of microbial agents increased the abundance of Chloroflexi, Acidobacteria, Gemmatimonadetes, Cyanobacteria, and Nitrospirae. Therefore, the microbial agents significantly altered the relative abundance of dominant bacterial taxa, affecting the composition and structure of the bacterial community.
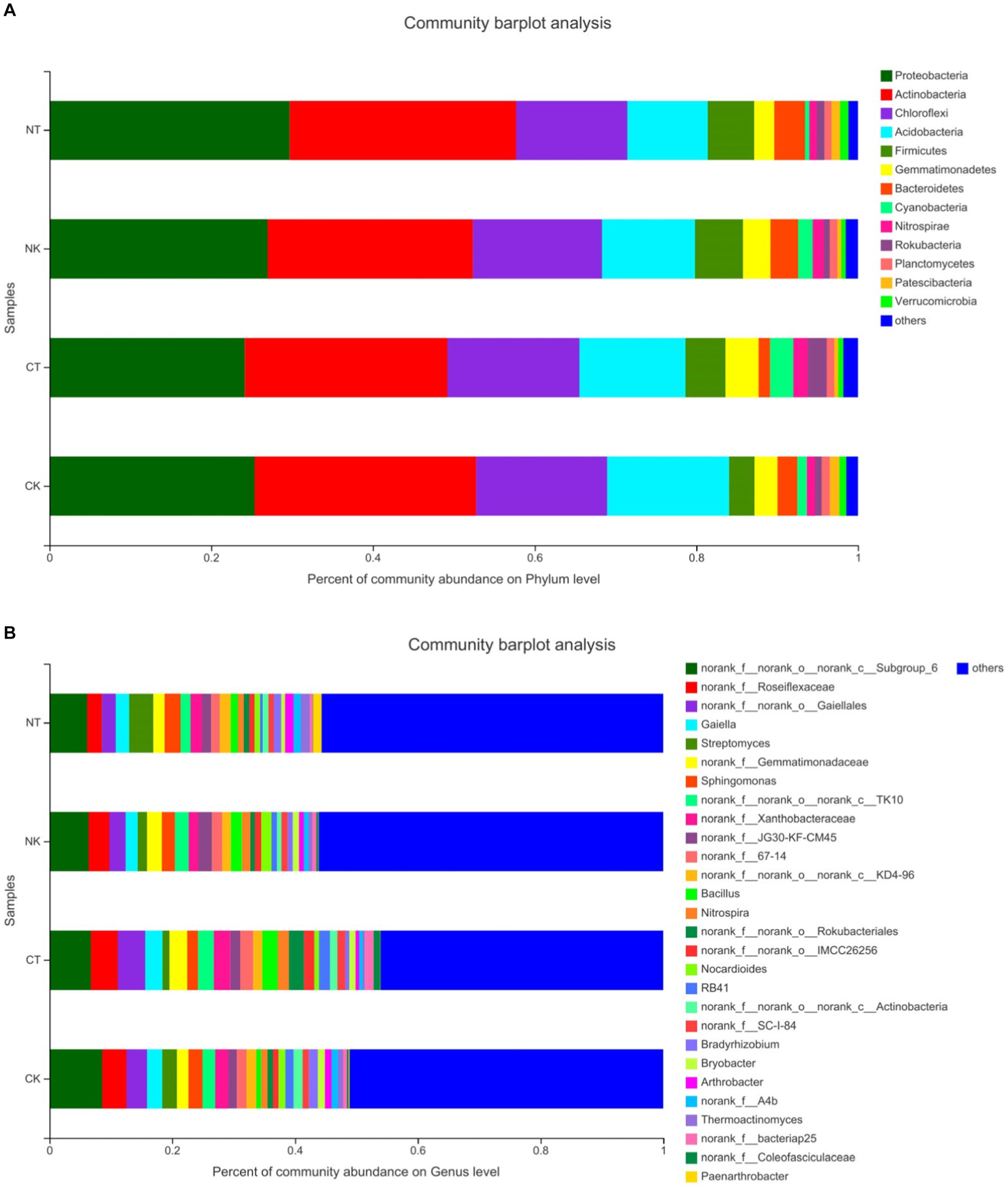
Figure 3. Main community composition of bacteria at phylum level (A) and genus level (B) under different treatments. NT, newly-planted microbial treatment; NK, newly-planted control; CT, continuous cropping microbial treatment; CK, continuous cropping control.
From Figure 3B, it can be seen that the top 10 genera in terms of abundance were Roseiflexaceae, Gaiellales, Gaiella, Streptomyces, Gemmatimonadaceae, Sphingomonas, Xanthobacteraceae, Bacillus, Nitrospira, and Rokubacteriales, followed by Nocardioides, Bradyrhizobium, Bryobacter, and Arthrobacter. The abundance of Roseiflexaceae, Gaiellales, and Gaiella in the continuous cropping group (CK and CT) was higher than that in the new planting group (NK and NT), while the abundance of Streptomyces in the new planting group (NK and NT) was higher than that in the continuous cropping group (CK and CT). There was little difference in the abundance of bacterial agents and control treatments in the continuous cropping group, but in the new planting group, the abundance NT was significantly higher at 3.97% compared to NK at 1.52%. Bacillus is a potential beneficial bacterium that can increase plant disease resistance. In the continuous cropping group, the abundance of Bacillus in CT was 2.5%, which is significantly lower in CK at 0.8%. However, in the new planting group, there was little difference in the abundance of Bacillus between NT and NK. These data suggest that the microbial agents can significantly change the relative abundance of dominant bacterial genera in the community.
Principal co-ordinates analysis (PCoA) of sequencing showed significantly separate clustering of the soil microbiota community between each pair of control and microbial agent treated groups (p = 0.001), with the main principal component (PC) scores: PC1 = 27.5%, PC2 = 14.22%, demonstrating the effects of the microbial agent treatments on the indigenous bacterial community in the rhizosphere soil of field ginger (Figure 4).
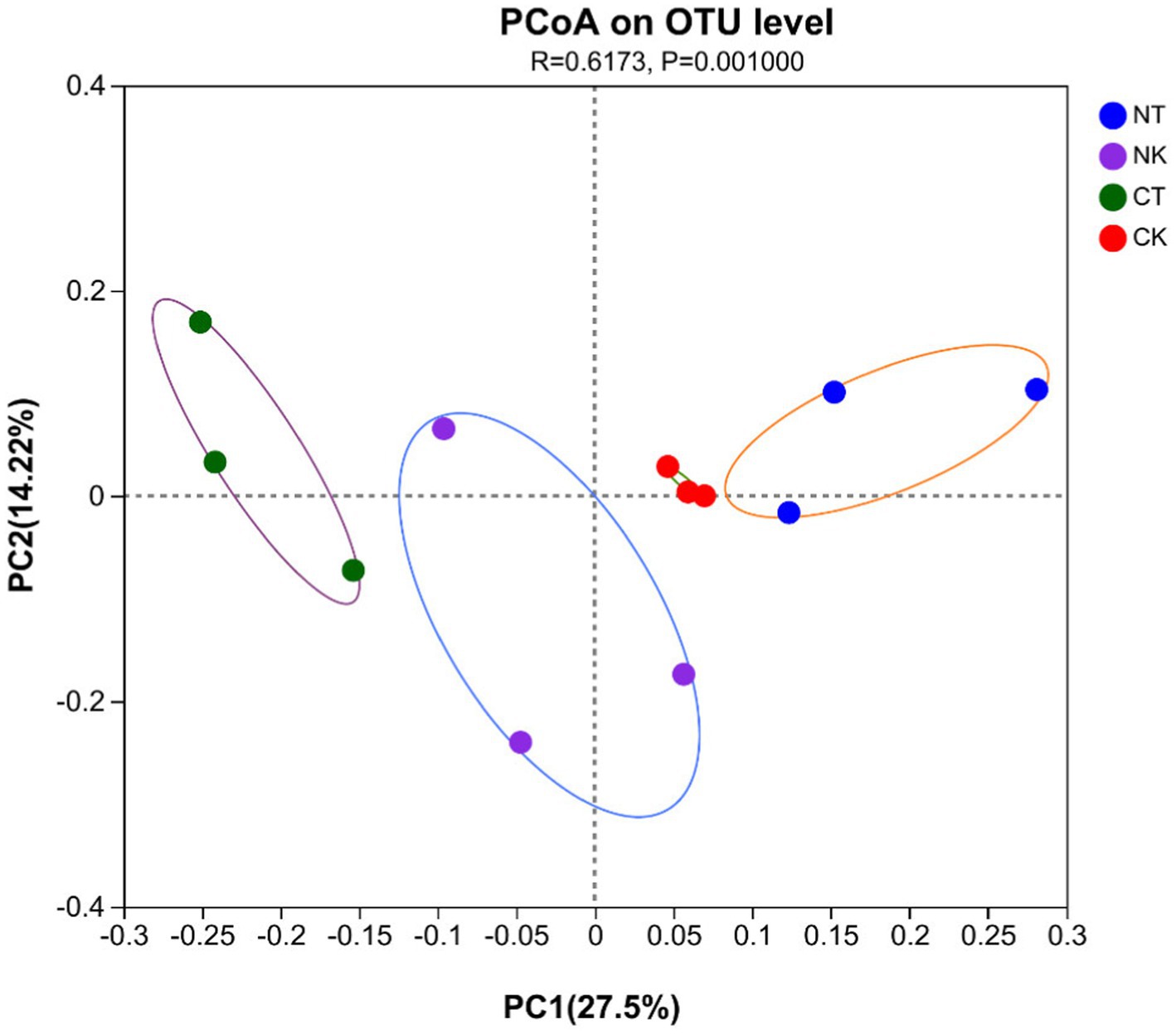
Figure 4. Principal coordinate analysis (PCoA) of bacterial community structure based on the Bray–Curtis distance metric in all soil samples. NT, newly-planted microbial treatment; NK, newly-planted control; CT, continuous cropping microbial treatment; CK, continuous cropping control.
LEfSe analysis was conducted with the LDA threshold of 3.5 to identify the microbial biomarkers that differed significantly between the microbial agents. The results showed that Streptomyces and Streptomycetales were significantly enriched in NT compared to NK, while Oxyphotobacteria and Chloroflexia were significantly enriched in NK (Figure 5C). In addition, Rokubacteria and Bacillaceae were enriched in CT, while Streptomycetaceae and Pseudonocardiales were enriched in CK (Figure 5D). When comparing NT and CT, Alphaproteobacteria and Streptomycetales were significantly enriched in NT, while Gaiellales and Blastocatellia were significantly enriched in CT (Figure 5B). Finally, when comparing NK and CK, Thermomicrobiales and Burkholderiaceae were significantly enriched in NK, while Streptomycetaceae and Streptomycetales were significantly enriched in CK (Figure 5A). The results suggest that different cropping systems have a significant impact on the composition of microbial communities in the ginger rhizosphere soil.
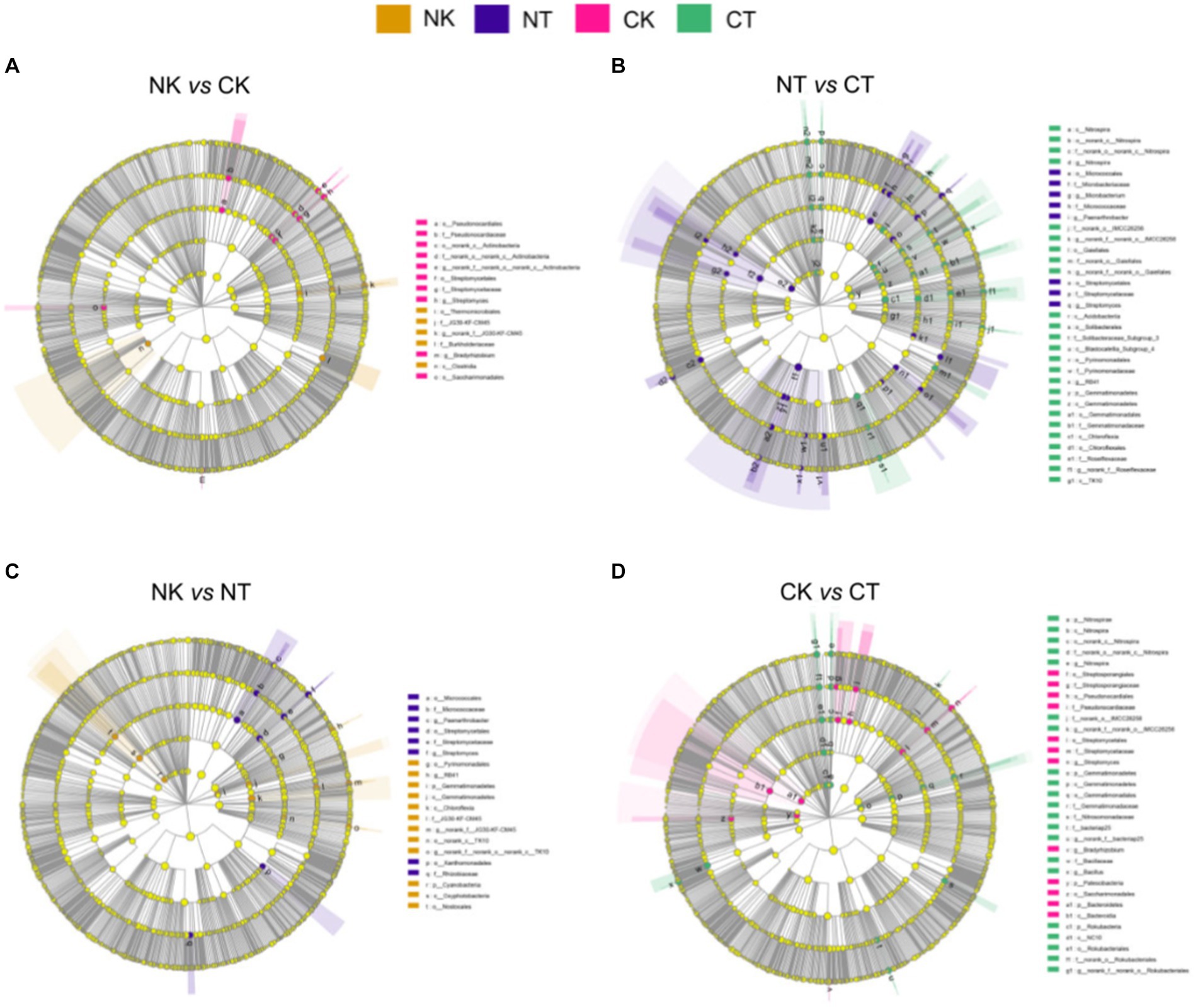
Figure 5. Cladograms plotted from LEfSe comparison analysis indicating the taxonomic representation of statistically and biologically consistent differences of identified biomarkers among different cropping systems. NT, newly-planted microbial treatment; NK, newly-planted control; CT, continuous cropping microbial treatment; CK, continuous cropping control.
The results of RDA indicated that soil properties, including hydrolyzable nitrogen (R2 = 0.6913**), available phosphorus (R2 = 0.4981**), available potassium (R2 = 0.5821**), and organic matter (R2 = 0.5715**), were the significant predictors of bacterial community composition at the genus level in the ginger rhizosphere soil (Supplementary Table S2). The magnitude of the effects of soil physicochemical properties on the bacterial community composition followed the order of hydrolyzable nitrogen, available potassium, organic matter, and available phosphorus. Available phosphorus was positively correlated with hydrolyzable nitrogen, available potassium, and organic matter, but negatively correlated with pH (Figure 6).
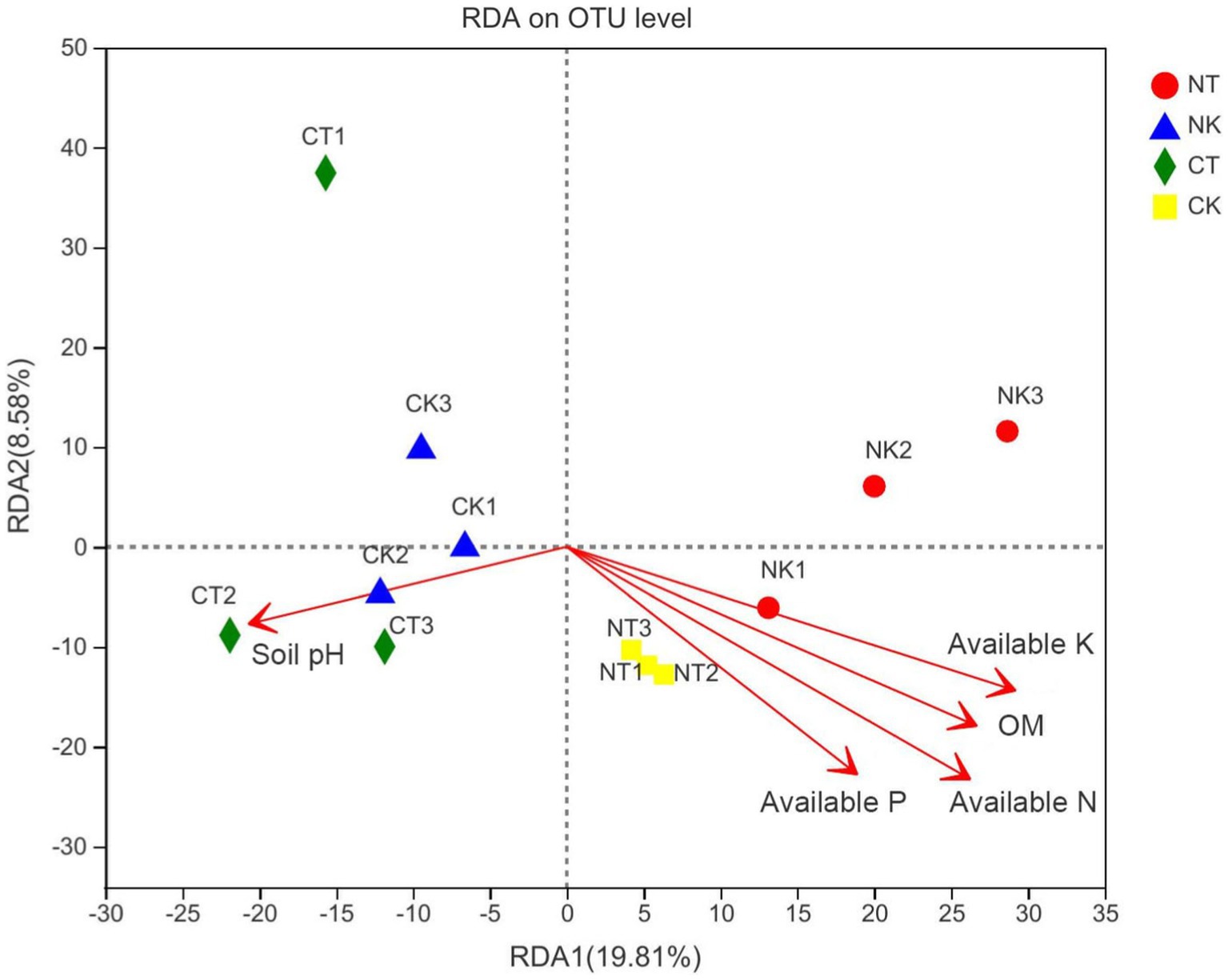
Figure 6. Redundancy analysis (RDA) of soil bacterial community structure associated with soil properties. NT, newly-planted microbial treatment; NK, newly-planted control; CT, continuous cropping microbial treatment; CK, continuous cropping control.
By calculating the Spearman correlation between the environmental factors and bacterial genera, it was found that the abundance of 12 genera was significantly and positively correlated with the environmental factors, while the abundance of 5 genera was significantly and negatively correlated with the soil physicochemical factors. Specifically, norank_f_JG30-KF-CM45, Marmoricia, RB41, and norank_f_norank_o_11–24 were significantly and negatively correlated with pH; Bradyrhizobium, Mycobacterium, norank_f_JG30-KF-AS9, norank_f_norank_o_S085, and Kribbella were significantly and positively correlated with available phosphorus, and norank_f_JG30-KF-CM45 was significantly and negatively correlated with available phosphorus; Bradyrhizobium, Mycobacterium, norank_f_norank_o_Saccharimonadales, norank_f_norank_o_S085, and Streptomyces were significantly and positively correlated with hydrolysable nitrogen, while norank_f_norank_o_norank_c_TK10 was significantly and negatively correlated with hydrolysable nitrogen; Bradyrhizobium, Mycobacterium, norank_f_norank_o_Saccharimonadales, norank_f_norank_o_S085, Kribbella, Streptomyces, Dongia, and Acidbacter were significantly and positively correlated with available potassium, while norank_f_JG30-KF-CM45, Marmoricia, norank_f_bacteriap25, and MND1 were significantly and negatively correlated with available potassium; Bradyrhizobium, Mycobacterium, norank_f_norank_o_Saccharimonadales, norank_f_norank_o_S085, and Streptomyces were significantly and positively correlated with organic matter, while norank_f_norank_o_norank_c_TK10 and norank_f_JG30-KF-CM45 were significantly and negatively correlated with organic matter (Figure 7).

Figure 7. Heatmap correlation between dominant bacteria genara and soil physico-chemical properties.
The results of soil physicochemical properties showed that regardless of the cultivation method or inoculant treatment, the changes in soil pH were not significant, and the impact on the rhizosphere microorganisms was also weak. This is likely due to the short-term effects of continuous cropping, the buffering effect of the soil on pH, and the complexity of the field environment. It has been reported that different crop cultivation methods result in different nutrient utilization and transformation efficiencies in the soil, which in turn lead to different fertility properties (Xun et al., 2015; Liu et al., 2019). The results of this study showed that after 1 year of continuous cropping, the content of available nitrogen, available phosphorus, available potassium, and organic matter in the rhizosphere soil of ginger significantly decreased compared to the newly planted ginger. Therefore, continuous cropping may lead to nutrient loss in soil.
Based on the view of sustainability of agricultural production, one of the most effective measures is to use one or several beneficial bacteria as microbial fertilizers for crop cultivation. These microbial fertilizers not only interact with the crops, but also directly affect the soil physicochemical properties such as soil nutrients and pH, which in turn have feedback effects on the microorganisms and crops (Liu, et al., 2017). In this study, after inoculation of microbial agents in newly planted ginger, the contents of available phosphorus, available potassium, and organic matter in the soil were significantly increased. This is similar to the conclusion of Zhang et al. (2019) that in the first year of planting Panax notoginseng (Burk.) Chen, the soil nitrogen content remained basically unchanged, while the phosphorus content increased significantly, and the potassium content decreased significantly. In the present study, however, the microbial agent inoculation treatment in ginger under continuous cropping condition did not cause significant changes in soil physicochemical properties, which may be related to the factors such as soil type, crop species, application cycle, and complexity of the field environment.
Soil microbial community structure and composition are closely related to the soil environment, and planting patterns can cause changes in the soil physical and chemical properties, thus changing the microbial community structure. Proteobacteria, Acidobacteria, Bacteriodetes and Actinobacteria are the dominant groups in the soil bacteria in farmland. Microbial agents change the relative abundance of Proteobacteria, Bacteriodetes, and Acidobacteria to some extent (Li et al., 2014). In this study, Proteobacteria were the most abundant group in the soil for all the four treatments, with a relative abundance of 24.16% to 29.71%, which is consistent with the results of previous studies (Shen et al., 2015; Zhu et al., 2017). The abundance of bacterial phyla in the soil differed between the continuous cropping and the new planting treatments. Among them, the changes in the relative abundance of Proteobacteria, Actinobacteria, Firmicutes and Bacteriodetes were the most obvious, and their abundance in the continuous cropping treatment was significantly lower than that in the new planting treatment. Therefore, different cropping patterns significantly affect the relative abundance of dominant bacterial groups and the composition of bacterial community structure in soil. Based on the results of this study, continuous ginger cropping will reduce the diversity of the rhizosphere soil bacterial community, degrade the structure of the bacterial community, cause an imbalance in the rhizosphere microecology, and thus might trigger the continuous cropping diseases.
The abundance of R. solanacearum is an important factor leading to the incidence of ginger (Prameela and Suseela, 2020). Inoculation of microbial agents can significantly reduce the abundance of R. solanacearum in the rhizosphere soil of ginger (p < 0.05), inhibit the growth of R. solanacearum to a certain extent, delay the development of ginger bacterial wilt and reduce its disease index. Inoculation of microbial agents resulted in a decrease in the number of R. solanacearum in the soil. It is speculated that there is a complex interaction between microbial agents, R. solanacearum and soil bacterial communities. Microbial agents can inhibit the growth of R. solanacearum by regulating soil bacterial communities.
Microbial agents (bio-fertilizers, organic fertilizers) can improve soil microbial community structure and control soil-borne diseases by increasing the abundance of beneficial bacteria in the soil (Zhao et al., 2011). Our research found that regardless of whether it was newly planted or continuously cropped, the bacterial inoculant treatment (NT and CT) could increase the abundance of Chloroflexi, Acidobacteria, Gemmatimonadetes, Cyanobacteria and Nitrospirae. In particular, Acidobacteria and Gemmatimonadetes, which have strong stress resistance and are beneficial to plants, were found to be significantly increased by 12.2% and 17.1%, respectively, in CT compared to CK in the continuously cropped group. In the newly planted group, the treatment NT increased the abundance of Acidobacteria and Gemmatimonadetes by 34.4% and 10.7%, respectively, compared to NK. Acidobacteria and Gemmatimonadetes can improve soil environment, promote ginger growth, and produce antibacterial substances to inhibit the pathogenic bacteria that settle in plant roots, thereby reducing the occurrence of soil-borne diseases (Liu et al., 2021). Bacillus is a group of potential beneficial bacteria that can increase plant disease resistance. This study found that the abundance of Bacillus sp. in the microbial agent treatment was significantly higher than that in CK in the continuous cropping group. This may be due to the good colonization of Bacillus velezensis KC-5 in the soil. B. velezensis KC-5 was isolated and screened from the rhizosphere soil of ginger. After applying B. velezensis KC-5 to the rhizosphere soil of ginger, the specific root exudates of ginger would promote the growth and reproduction of the bacteria. However, there was little difference in the abundance of Bacillus between NT and NK in the new planting. This may be due to the newly planted experimental area shows a milder occurrence of bacteria wilt, the new treatment with B. velezensis KC-5 shows colonization in the new area (NT). As for the new control (NK), even though there is no colonization of KC-5, the rhizosphere soil may contain indigenous B. velezensis, resulting in no significant difference. However, in the continuous cropping experimental area, the occurrence of bacteria wilt is severe. After applying the microbial agent, the continuous cropping treatment (CT) shows colonization of B. velezensis KC-5. On the other hand, the continuous cropping control (CK) not only lacks colonization of KC-5 but also has a reduced abundance of beneficial bacteria in the rhizosphere due to the severe occurrence of bacteria wilt, resulting in significant differences when compared with the continuous cropping control (CK). Nitrospira are the ammonia-oxidizing bacteria that contribute to soil nitrification (Saikia et al., 2018). Their abundance being significantly increased in the microbial agent treatment may have promoted the plant growth. Our research confirmed that the bio-bacterial agents can improve the soil bacterial community structure. Therefore, applying beneficial bacteria to regulate the soil microbial community composition can help prevent ginger wilt disease.
Ginger’s susceptibility to bacteria wilt is influenced by various factors. Wang et al. (2017) demonstrated that temperature and pH are crucial soil factors affecting the pathogenicity of R. solanacarum. Other studies (Mahmood and Bashir, 2011) have suggested that severe occurrences of bacteria wilt may be primarily attributed to soil compaction, imbalanced pH levels, and deteriorated physicochemical properties. In this study, the rhizosphere soil of the newly planted experimental area (with less bacteria wilt) showed significantly higher levels of available nitrogen (AN), available phosphorus (AP), and available potassium (AK) compared to the continuous cropping soil (experiencing severe bacteria wilt). This indicates that increasing the content of AN, AP, and AK in the soil plays a vital role in reducing the incidence of bacteria wilt. Many studies have shown the importance of soil physicochemical properties on soil microbial community, and the increase in soil organic matter and available nitrogen content can promote the formation of bacterial diversity (Liu et al., 2014; Li et al., 2018). With the accumulation of soil nutrients, the relative abundance of Actinobacteria increases (Zeng et al., 2017). Through correlation analysis, this study found that the abundance of Bradyrhizobium and Mycobacterium was significantly correlated with the hydrolysable nitrogen, available phosphorus, available potassium, and organic matter in the soil; Streptomyces was significantly correlated with the available potassium and organic matter content. More experimental results demonstrated that cropping patterns, soil physicochemical properties, and soil microbial community structure are correlated each other (Ofek-Lalzar et al., 2014; Pang et al., 2017). The above-mentioned research results indicate that different planting patterns and soil chemical properties have inconsistent effects on various soil bacterial communities. As this research focused on analyzing the composition of the bacteria in the rhizosphere soil of mature ginger, further research is needed to investigate the impact of bacterial agents and newly planted ginger on the bacterial community structure at other stages of ginger growth.
Interestingly, high-throughput sequencing results showed that the application of microbial agents did not affect the abundance of Ralstonia, so in addition to directly inhibiting pathogens, microbial agents may also reduce the incidence of bacterial wilt through some indirect ways (Tan et al., 2016). Therefore, the specific ways of microbial agents to inhibit pathogens need to be explored in the future.
High-throughput sequencing results showed that application of microbial inoculant altered the bacterial community structure in ginger rhyzosphere soil. The dominant phyla in the four treatments were Proteobacteria, Actinobacteria, Chloroflexi and Acidobacteria, which accounted for 72.91% to 89.09%. The abundance of bacteria at phyla level of Proteobacteria, Actinobacteria, Firmicutes, and Bacteroidetes in the new planting treatment was significantly higher than that in the continuous cropping treatment. The microbial agent treatment improved the abundance of the potential probiotics such as Acidobacteria and Gemmatimonadetes in the soil, which helps optimize the soil microecological balance. The difference in bacterial community structure between the continuous cropping and new planting conditions was greater than that between the microbial and non-microbial inoculant treatments. Correlation analysis showed that the cropping pattern and microbial agent treatments caused significant changes in the physical and chemical properties of the soil, and hydrolyzable N, available P, available K, and organic matter were the significant factors affecting the composition of genus-level bacteria communities in ginger rhizosphere soil. In the future, transcriptome and metabolome analyses associated with specific microbial taxa are required to further explore the potential mechanism, safety, and application strategy of the microbial agents.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
QW: validation and visualization. QW and JS: investigation and manuscript writing and editing. JS, YK, and JZ: data curation. XQ and SH: project administration. TC and SZ: funding acquisition. All authors contributed to the article and approved the submitted version.
This work was supported in part by Guangxi Key Research and Development Project (AB21238002), Guangxi Nature Science Fund (2019GXNSFDA245013), and Guangxi Academy of Agricultural Sciences Project (2023YM96 and 2022JM59).
We thank Shangtao Jiang of Nanjing Agricultural University for providing guidance on the data analyses. We would like to thank Yangrui Li of Guangxi Academy of Agricultural Sciences, Nanning, Guangxi for helping to improve the manuscript, and appreciate the help of Haiying Liang of Department of Genetic and Biochemistry, Clemson University in editing the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1203796/full#supplementary-material
Afaque, M., Zahoor, A., John, S. A., Shukla, P. K., and Ramteke, P. W. (2017). Suppression of bacterial wilt of tomato (Lycopersicon esculentum mill.) by plant growth promoting rhizobacteria (article). Vegetos 30, 429–434. doi: 10.5958/2229-4473.2017.00114.8
Che, Y. M., Zhao, F. G., Chen, T. J., Zhu, Y., Zhang, W., and Liu, X. (2019). Study on growth effects of co-inoculation with different AM fungi, Trichoderma harzianum sp. and growth-promoting rhizobacteria. J. Qingdao. Agric. Univ. 36, 95–102.
Feng, S., Liu, X. L., Wu, X. L., Jiang, S. J., Zhou, Z. F., Zhang, W., et al. (2017). Effects of water conditions on microbial community in maize rhizosphere. Crops 2017, 127–134. doi: 10.16035/j.issn.1001-7283.2017.01.023
Glick, B. R., Penrose, D. M., and Li, J. (1998). A model for the lowering of plant ethylene concentrations by plant growth promoting bacteria. J. Theor. Biol. 190, 63–68. doi: 10.1006/jtbi.1997.0532
Guo, J. H., Qi, H. Y., Guo, Y. H., Ge, H. L., Gong, L. Y., Zhang, L. X., et al. (2004). Biocontrol of tomato wilt by plant growth-promoting rhizobacteria. Biol. Control 29, 66–72. doi: 10.1016/S1049-9644(03)00124-5
Hou, L. M., Meng, R. Q., Nie, L. C., and Qi, Y. B. (2016). Effects of different microbial agents on substrate enzyme activities and tomato yield and quality. Chin. J. Appl. Ecol. 27, 2520–2526. doi: 10.13287/j.1001-9332.201608.015
Huang, M. Y., Gu, W. J., Zhang, F. B., Lu, Y. S., Xie, K. Z., and Jiang, R. P. (2013). Stability test of ferment filtrate by Arthrobacter YB6 strain and its control effect on Ralstonia solanacearum. Guangdong Agric. Sci. 18, 75–78. doi: 10.16768/j.issn.1004-874x.2013.18.036
Huang, X. Q., Liu, Y., Zhang, L., Zhou, X. Q., Wu, W. X., et al. (2015). Screening Synergistic Bactericide Combined with Bacillus against Tobacco Bacterial Wilt[J]. Agrochemicals, 54:848–851. doi: 10.16820/j.cnki.1006-0413.2015.11.021
Huang, Z. H., Liu, B. W., Yin, Y., Liang, F., Xie, D. S., Han, T. T., et al. (2021). Impact of biocontrol microbes on soil microbial diversity in ginger (Zingiber officinale Roscoe). Pest Manag. Sci. 77, 5537–5546. doi: 10.1002/ps.6595
Hugo, M., Ayako, M., Takayuki, N., Chiho, M., and Toshinori, O. (2020). Amplicon sequence variant-based oral microbiome analysis using QIIME2. J. Osaka Dent. Univ. 54, 273–281. doi: 10.18905/JODU.54.2_273
Jacobsen, C. S., and Hjelms, M. H. (2014). Agricultural soils, pesticides and microbial diversity. Curr. Opin. Biotechnol. 27, 15–20. doi: 10.1016/j.copbio.2013.09.003
Kwon, J. W., and Kim, S. D. (2014). Characterization of an antibiotic produced by Bacillus subtilis JW-1 that suppresses Ralstonia solanacearum. J. Microbiol. Biotechnol. 24, 13–18. doi: 10.4014/jmb.1308.08060
Liang, C. C., Li, T., Xiao, Y. P., Liu, M. J., Zhang, H. B., and Zhao, Z. W. (2009). Effects of inoculation with arbuscular mycorrhizal fungi on maize grown in multi-metal contaminated soils. Int. J. Phytoremediation 11, 692–703. doi: 10.1080/15226510902787310
Li, J., Wen, Y., Li, X., Li, Y. T., Yang, X. D., Lin, Z., et al. (2018). Soil labile organic carbon fractions and soil organic carbon stocks as affected by long-term organic and mineral fertilization regimes in the North China Plain. Soil Tillage Res. 175, 281–290. doi: 10.1016/j.still.2017.08.008
Ling, N., Deng, K. Y., Song, Y., Wu, Y. C., Zhao, J., Raza, W., et al. (2014). Variation of rhizosphere bacterial community in watermelon continuous mono-cropping soil by long-term application of a novel bioorganic fertilizer. Microbiol. Res. 169, 570–578. doi: 10.1016/j.micres.2013.10.004
Liu, D. H., Qiao, Y., Li, S. L., Zhang, Z., Li, F., Xu, W., et al. (2021). Effects of different planting patterns on soil nutrients and microbial community of Foshou yam. Hubei Agric. Sci. 60, 79–84. doi: 10.14088/j.cnki.issn0439-8114.2021.23.017
Liu, J. J., Sui, Y. Y., Yu, Z. H., Shi, Y., Chu, H. Y., Jin, J., et al. (2014). High throughput sequencing analysis of biogeographical distribution of bacterial communities in the black soils of Northeast China. Soil Biol. Biochem. 70, 113–122. doi: 10.1016/j.soilbio.2013.12.014
Liu, J. J., Yu, Z. H., Yao, Q., Hu, X. J., Zhang, W., Mi, G., et al. (2017). Distinct soil bacterial communities in response to the cropping system in a Mollisol of Northeast China. Appl. Soil Ecol. 119, 407–416. doi: 10.1016/j.apsoil.2017.07.013
Liu, T. Y., Huang, Y. L., Wang, X. R., Lin, J. Q., et al. (2006). Studies on techniques for soil improvement in Sanming tobacco planting area[J]. Chinese Tobacco Science, 27: 10–15. doi: 10.13496/j.issn.1007-5119.2006.03.003
Liu, Z., Liu, J. J., Xu, Y., Zhang, W., Mi, G., Yao, Q., et al. (2019). Effects of continuous cropping years of soybean on the bacterial community structure in black soil. Acta Ecol. Sin. 39, 4337–4346. doi: 10.5846/stxb201801270212
Li, X. G., Ding, C. F., Zhang, T. L., and Wang, X. X. (2014). Fungal pathogen accumulation at the expense of plant-beneficial fungi as a consequence of consecutive peanut monoculturing. Soil Biol. Biochem. 72, 11–18. doi: 10.1016/j.soilbio.2014.01.019
Mahmood, R., and Bashir, U. (2011). Relationship between soil physicochemical characteristics and soil-borne diseases[J]. Mycopath, 9: 87–93.
Ofek-Lalzar, M., Sela, N., Goldman-Voronov, M., Stefan, J. G., Yitzhak, H., and Dror, M. (2014). Niche and host-associated functional signatures of the root surface microbiome. Nat. Commun. 5, 1–9. doi: 10.1038/ncomms5950
Ohike, T., Makuni, K., Okanami, M., and Ano, T. (2013). Screening of endophytic bacteria against fungal plant pathogens. J. Environ. Sci. 25, S122–S126. doi: 10.1016/S1001-0742(14)60640-9
Pang, G., Cai, F., Li, R. X., Zhao, Z., Li, R., Gu, X. L., et al. (2017). Trichoderma-enriched organic fertilizer can mitigate microbiome degeneration of monocropped soil to maintain better plant growth. Plant Soil 416, 181–192. doi: 10.1007/s11104-017-3178-0
Prameela, T. P., and Suseela, B. R. (2020). Bacterial wilt of ginger (Zingiber officinale Rosc.) incited by Ralstonia pseudosolanacearum-a review based on pathogen diversity, diagnostics and management. J. Plant Pathol. 102, 709–719. doi: 10.1007/s42161-020-00487-5
Saikia, J., Sarma, R. K., Dhandia, R., Yadav, A., Bharali, R., Gupta, V. K., et al. (2018). Alleviation of drought stress in pulse crops with ACC deaminase producing rhizobacteria isolated from acidic soil of Northeast India. Sci. Rep. 8:3560. doi: 10.1038/s41598-018-21921-w
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12:R60. doi: 10.1186/gb-2011-12-6-r60
Sharifi, M., Ghorbanli, M., and Ebrahimzadeh, H. (2007). Improved growth of salinity-stressed soybean after inoculation with salt pre-treated mycorrhizal fungi. J. Hytopathol. 164, 1144–1151. doi: 10.1016/j.jplph.2006.06.016
Shen, B. J., Zhu, Z. K., Yuan, H. C., Ge, T. D., Wang, J. R., Chen, M. L., et al. (2015). Effects of different plantation type on the abundance and diversity of soil microbes in subtropical red soils. Environ. Sci. 36, 3839–3844. doi: 10.13227/j.hjkx.2015.10.037
Tan, H., Zhou, S., Deng, Z., He, M., and Cao, L. X. (2011). Ribosomal-sequence-directed selection for endophytic streptomycete strains antagonistic to Ralstonia solanacearum to control tomato bacterial wilt. Biol. Control 59, 245–254. doi: 10.1016/j.biocontrol.2011.07.018
Tan, S., Gu, Y., Yang, C., Dong, Y., Mei, X., Shen, Q., et al. (2016). Bacillus amyloliquefaciens T-5 may prevent Ralstonia solanacearum infection through competitive exclusion. Biol. Fertil. Soils 52, 341–351. doi: 10.1007/s00374-015-1079-z
Thekkan, P. P., and Rajamma, S. B. (2020). Bacterial wilt of ginger (Zingiber officinale Rosc.) incited by Ralstonia pseudosolanacearum-A review based on pathogen diversity, diagnostics and management[J]. Journal of Plant Pathology, 102: 709–719. doi: 10.1007/s42161-020-00487-5
Thomas, P., and Upreti, R. (2014). Testing of bacterial endophytes from non-host sources as potential antagonistic agents against tomato wilt pathogen Ralstonia solanacearum. Adv. Microbiol. 4, 656–666. doi: 10.4236/aim.2014.410071
Wang, L. L., Zhou, X. D., Li, G. A., Yao, H. Y., and Wang, F. (2017). Screening and identifying of solanacearum and the control antagonistic bacteria against Ralstonia effects on tomato bacterial wilt in the field. Plant Prot. 43, 182–185.
Wang, Q., Zhang, J. L., Long, Y. Y., Li, D. P., Chen, T. S., and Che, J. L. (2016). Resources of arbuscular mycorrhizal fungi in ginger fields of Liujiang, Guangxi. Southwest China J. Agric. Sci. 29, 115–119. doi: 10.16213/j.cnki.scjas.2016.01.023
Xia, T. Y., Chen, Z. B., Su, Y., Liu, J. N., Yu, L., Wang, D. K., et al. (2018). Microbial diversity of tobacco rhizosphere soil in different growth stages of marigold-tobacco intercropping system. Southwest China J. Agric. Sci. 31, 680–686. doi: 10.16213/j.cnki.scjas.2018.4.007
Xun, W. B., Huang, T., Zhao, J., Ran, W., Wang, B. R., Shen, Q. R., et al. (2015). Environmental conditions rather than microbial inoculum composition determine the bacterial composition, microbial biomass and enzymatic activity of reconstructed soil microbial communities. Soil Biol. Biochem. 90, 10–18. doi: 10.1016/j.soilbio.2015.07.018
Yang, C., Liu, N., and Zhang, Y. (2019). Soil aggregates regulate the impact of soil bacterial and fungal communities on soil respiration. Geoderma 337, 444–452. doi: 10.1016/j.geoderma.2018.10.002
Yang, R. P., Mo, Y. L., Liu, C. M., Wang, Y. Q., Ma, J. X., Zhang, Y., et al. (2016). The effects of cattle manure and garlic rotation on soil under continuous cropping of watermelon (Citrullus lanatus L.). PLoS One 11, 1–15. doi: 10.1371/journal.pone.0156515
Zeng, Q. C., An, S. S., and Liu, Y. (2017). Soil bacterial community response to vegetation succession after fencing in the grassland of China. Sci. Total Environ. 609, 2–10. doi: 10.1016/j.scitotenv.2017.07.102
Zhang, M. M., Deng, C. S., Ma, J. F., Zhang, Y. R., Geng, B. L., and Shun, P. (2012). Screening and identification of multi-functional Trichoderma spp. J. Agro-Environ. Sci. 31, 1571–1575.
Zhang, Y., Zheng, Y., Xia, P., Xun, L. L., and Liang, Z. S. (2019). Impact of continuous Panax notoginseng plantation on soil microbial and biochemical properties. Sci. Rep. 9:13205. doi: 10.1038/s41598-019-49625-9
Zhao, M. L., Yuan, J., Shen, Z. Z., Dong, M., Liu, H. J., Wen, T., et al. (2019). Predominance of soil vs root effect in rhizosphere microbiota reassembly. FEMS Microbiol. Ecol. 95:fiz139. doi: 10.1093/femsec/fiz139
Zhao, Q., Dong, C., Yang, X., Mei, X. L., Ran, W., Shen, Q. R., et al. (2011). Biocontrol of fusarium wilt disease for Cucumis melo melon using bio-organic fertilizer. Appl. Soil Ecol. 47, 67–75. doi: 10.1016/j.apsoil.2010.09.010
Zhu, H. H., and Yao, Q. (2004). Localized and systemic increase of phenols in tomato roots induced by Glomus versiforme inhibits Ralstonia solanacearum. J. Phytopathol. 152, 537–542. doi: 10.1111/j.1439-0434.2004.00892
Keywords: ginger, ginger wilt disease, microbial agents, rhizosphere, bacterial community
Citation: Wang Q, Song J, Zhang J, Qin X, Kang Y, Huang S, Zhou S and Chen T (2023) Effects of microbial agent application on the bacterial community in ginger rhizosphere soil under different planting years. Front. Microbiol. 14:1203796. doi: 10.3389/fmicb.2023.1203796
Received: 11 April 2023; Accepted: 21 August 2023;
Published: 07 September 2023.
Edited by:
Lin Chen, Chinese Academy of Forestry, ChinaReviewed by:
Caixia Dong, Nanjing Agricultural University, ChinaCopyright © 2023 Wang, Song, Zhang, Qin, Kang, Huang, Zhou and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shengmao Zhou, TWFvbWFvemhvdTcwQGd4YWFzLm5ldA==; Tingsu Chen, Q2hlbnRzMDAxQGd4YWFzLm5ldA==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.