
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 15 June 2023
Sec. Aquatic Microbiology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1203780
To understand the evolution of coscinodiscophycean diatoms, plastome sequences of six coscinodiscophycean diatom species were constructed and analyzed in this study, doubling the number of constructed plastome sequences in Coscinodiscophyceae (radial centrics). The platome sizes varied substantially in Coscinodiscophyceae, ranging from 119.1 kb of Actinocyclus subtilis to 135.8 kb of Stephanopyxis turris. Plastomes in Paraliales and Stephanopyxales tended to be larger than those in Rhizosoleniales and Coscinodiacales, which were due to the expansion of the inverted repeats (IRs) and to the marked increase of the large single copy (LSC). Phylogenomic analysis indicated that Paralia and Stephanopyxis clustered tightly to form the Paraliales-Stephanopyxales complex, which was sister to the Rhizosoleniales-Coscinodiscales complex. The divergence time between Paraliales and Stephanopyxales was estimated at 85 MYA in the middle Upper Cretaceous, indicating that Paraliales and Stephanopyxales appeared later than Coscinodiacales and Rhizosoleniales according to their phylogenetic relationships. Frequent losses of housekeeping protein-coding genes (PCGs) were observed in these coscinodiscophycean plastomes, indicating that diatom plastomes showed an ongoing reduction in gene content during evolution. Two acpP genes (acpP1 and acpP2) detected in diatom plastomes were found to be originated from an early gene duplication event occurred in the common progenitor after diatom emergence, rather than multiple independent gene duplications occurring in different lineages of diatoms. The IRs in Stephanopyxis turris and Rhizosolenia fallax-imbricata exhibited a similar trend of large expansion to the small single copy (SSC) and slightly small contraction from the LSC, which eventually led to the conspicuous increase in IR size. Gene order was highly conserved in Coscinodiacales, while multiple rearrangements were observed in Rhizosoleniales and between Paraliales and Stephanopyxales. Our results greatly expanded the phylogenetic breadth in Coscinodiscophyceae and gained novel insights into the evolution of plastomes in diatoms.
Diatoms are one of the most successful phytoplankton groups in contemporary oceans, accounting for roughly 20% of the primary productivity on Earth (Falkowski et al., 1998), as well as being the primary biological mediators of the silica cycle in oceans (Sumper and Brunner, 2008). Some diatom species are widely used as initial feeding in aquaculture, while others have demonstrated great potential to be important metabolites in the fields of biomedicine, bioenergy and biomaterials (e.g., Hemaiswarya et al., 2011; Kiatmetha et al., 2011; Nurachman et al., 2012). Some diatoms species proliferate rapidly and massively in regional waters, resulting in the development of harmful algal blooms (HABs) which have serious negative impact on economy and the ecological environment (Armbrust, 2009).
Diatoms are an extraordinarily diverse lineage in evolution and morphology, and it is believed that there are as many as 100,000 extant species around the world (Mann and Droop, 1996; Mann et al., 2021). Up to now, more than 18,200 species of diatoms have been effectively described in taxonomy around the world (Guiry and Guiry, 2023). There is no doubt that a large number of cryptic species have not yet been properly resolved due to their morphological similarities. Current classification system of diatoms based on molecular biological data, combined with morphological features, sexual reproduction, and fossil evidence (Medlin and Kaczmarska, 2004; Medlin and Desdevises, 2020), has revealed that the Bacillariophyta phylum could be divided into two subphyla, Bacillariophytina and Coscinodiscophytina. Bacillariophytina harbors two classes, Bacillariophyceae (pennates) and Mediophyceae (polar centrics), harboring 78.7 and 10.2% of diatom species, respectively. Coscinodiscophytina contains a single class Coscinodiscophyceae (radial centrics), which thus far has 1,548 species, occupying 8.5% (Yu et al., 2018; Guiry and Guiry, 2023).
Diatoms are unicellular photosynthetic heterokont algae that are globally distributed in marine and freshwater environments. Diatoms represent a lineage of photosynthetic heterokonts which acquired their chloroplasts from a red algal ancestor by secondary endosymbiosis that took place around one billion years ago (Bhattacharya et al., 2007), thus their chloroplasts are surrounded by four layers of membranes. Diatom nuclear genomes sequenced thus far harbored genes from a heterotrophic host cell, bacterial donors, and red algal endosymbionts, displaying mosaic nature of their genetic material (Armbrust et al., 2004; Bowler et al., 2008).
Nevertheless, in the latest 10 years, the number of diatom chloroplast genomes (plastid genomes, plastomes) increased rather rapidly driven by high throughput DNA sequencing technologies. As of 1st May 2023, more than 150 diatom plastomes from at least 130 species have been deposited in the GenBank database. The sequenced diatom plastomes display a canonical quadripartite organization with two inverted repeats (IRs) between a large single copy (LSC) region and a small single copy (SSC) region, and harbor a core set of 150 - 165 canonical genes (e.g., Kowallik et al., 1995; Oudot-Le Secq et al., 2007; Hamsher et al., 2019). Diatom plastomes show a reduced gene content in comparison with red algal plastomes (230–254), indicating that many chloroplast genes have been lost or transferred to the nuclear genome after secondary endosymbiosis (Lommer et al., 2010; Lang and Nedelcu, 2012). Meanwhile, some genes were acquired via lateral gene transfer from different sources (e.g., plasmids, bacteria), leading to the expansion of plastome sizes (Brembu et al., 2014; Ruck et al., 2014). Although they show a high degree of similarity in genome architecture and core gene set (Sabir et al., 2014; Ruck et al., 2017; Wang et al., 2022), diatom plastomes have undergone tremendous alterations in genome size, IR size and gene content, and gene order through evolutionary history of diatoms (Yu et al., 2018; Gastineau et al., 2021). Plastomes of some closely related intrageneric species or even intra-order species displayed highly conserved gene order and genome structure (Sabir et al., 2014; Hamsher et al., 2019; Xu et al., 2021; Zhang and Chen, 2022).
Thus far, only six plastomes from four genera in two orders (Rhizosoleniales and Coscinodiacales) in coscinodiscophycean diatoms have been sequenced (Table 1). Considering the rich morphological diversity of coscinodiscophycean diatom species, the important evolutionary status of this class which represents a basal lineage in the evolution of early diatoms, and their global distribution and important ecological significance, it is very necessary to fill this gap to understand their evolution trend and relationships. This study focused on the evolution of plastomes in coscinodiscophycean diatoms. Plastomes from six coscinodiscophycean diatom species including Guinardia delicatula, Guinardia striata, Actinocyclus sp., Coscinodiscus granii, Stephanopyxis turris, and Paralia sulcate, which were collected in the Jiaozhou Bay of China, were constructed in this study and compared with plastome sequences deposited in the GenBank database to ascertain their evolutionary dynamics.
Water samples were collected in Jiaozhou Bay (36°00′–36°12′N, 120°10′–120°24′E) of China from August 2020 to December 2021 onboard the R/V Chuangxin, which was operated by the Jiaozhou Bay National Marine Ecosystem Research Station. Single cells of each diatom species were isolated using single-cell capillary methods from water samples. Six unialgal diatom strains (CNS00558, CNS00513, CNS00114, CNS00554, CNS00378, and CNS00428) were successfully cultured in L1 medium with 1 ‰ volume fraction Na2SiO3. The culture was maintained at 18–20°C, 2,000–3,000 Lux in the photoperiod of 12 h light-12 h dark.
Total genomic DNA for each diatom strain was extracted using the DNAsecure Plant Kit (Tiangen Biotech, Beijing, China). After purification, genomic DNA samples were fragmented into a size of 350 bp using Covaris S220 ultrasonic crater (Covaris, USA) for library construction. The DNA libraries were sequenced using a NovaSeq 6000 platform (Illumina, San Diego, CA, USA), yielding about 5-Gb sequencing data of paired-end reads with 150 bp in length. Illumina sequencing raw data were trimmed using Trimmomatic v0.39 with the parameters: LEADING:3 TRAILING:3 SLIDING WINDOW:4:15 MINLEN:75 (Bolger et al., 2014).
Species identification of six coscinodiscophycean diatoms was performed based on comparative analysis of their full-length 18S rDNA sequences as well as their morphological characteristics (Figure 1). Full-length 18S rDNA sequences of these diatom strains were assembled using Illumina sequencing results using the GetOrganelle v1.7.4.1 (Jin et al., 2020) and SPAdes v3.14.0 (Bankevich et al., 2012). The 18S rDNA sequences of coscinodiscophycean diatoms downloaded from the GenBank database were used as reference sequences (Supplementary Table 1). Multiple sequence alignments of 18S rDNA sequences were conducted by using ClustalX v1.83 with the default settings (Thompson et al., 1997). Evolutionary relationships were evaluated based on the analysis of the similarity of the 18S rDNA sequences using MEGA v7.0 (Kumar et al., 2016). 18S rDNA sequences of strains CNS00558, CNS00513, CNS00554, CNS00378, and CNS00428 displayed very high similarities (>99.6%) with the reference sequences deposited in the GenBank database, respectively (Supplementary Table 2). According to molecular data and their morphological characteristics, these five strains were identified as Guinardia delicatula, Guinardia striata, Coscinodiscus granii, Stephanopyxis turris, and Paralia sulcate, respectively. Strain CNS00114 shared the highest similarity with Actinocyclus sp. (X85401), reaching 98.86% (Supplementary Table 2), suggesting that this strain represents an undescribed species in Actinocyclus.

Figure 1. Micrographs of six diatom species in class Coscinodiscophyceae. (A) Guinardia delicatula. (B) Guinardia striata. (C) Actinocyclus sp. (D) Coscinodiscus granii. (E) Stephanopyxis turris. (F) Paralia sulcate. Bar = 20 μm.
Clean reads were used to assemble complete plastome sequences using GetOrganelle v1.7.4.1 (Jin et al., 2020). To verify the completeness of these plastomes, GetOrganelle generally exported consistent assembly results with the same raw reads using different parameters, when a complete plastome was obtained (Jin et al., 2020). Whether an assembled plastome was circular was further confirmed by aligning short reads against the assembled plastome sequence using the MEM algorithm of BWA v0.7.17 (Li and Durbin, 2009) and SAMtools v1.10 (Li et al., 2009). Circular plastome was supported by perfect alignments of short reads along the entire length of assembled plastome sequence. The alignment was visualized using IGV v2.7.2 (Thorvaldsdottir et al., 2013). Annotation was conducted using MFannot1 and NCBI’s ORF Finder,2 which was further improved using NCBI’s Sequin v15.10. For best accuracy of comparative analysis, we had checked and re-annotated coscinodiscophycean diatom plastomes deposited in the GenBank database.
Due to the absence of four protein-coding genes (PCGs) (bas1, ycf88, ycf89, and ycf90) in Triparma laevis (Bolidophyceae, Ochrophyta) plastome when compared with coscinodiscophycean plastomes, a total of 116 PCGs from 12 coscinodiscophycean plastomes as well as Triparma laevis plastome (Tajima et al., 2016) were extracted and concatenated for phylogenomic analysis. The amino acid (AA) sequences of each PCG were individually aligned and checked using MAFFT v7.471 (Katoh and Standley, 2013), and ambiguously aligned regions were trimmed by trimAl v1.4 (Capella-Gutierrez et al., 2009). These AA datasets were concatenated using PhyloSuite v1.2.2 (Zhang et al., 2020). The best-fit model was tested and identified using ModelFinder (Kalyaanamoorthy et al., 2017). Phylogenomic tree was constructed by IQ-TREE v1.6.12 (Trifinopoulos et al., 2016) using default parameters with 5,000 ultrafast bootstrap analysis (Minh et al., 2013). T. laevis was used as the outgroup. The AA sequences of AcpP (AcpP1 and AcpP2) were aligned by ClustalX v1.83 with default settings (Thompson et al., 1997). The phylogenetic relationships were inferred with the Maximum Likelihood (ML) method based on the JTT matrix-based model (Jones et al., 1992) using MEGA v7.0 (Kumar et al., 2016). There were 113 positions in the final dataset of AcpP. Synteny analysis of plastomes was performed using the progressiveMauve software of the package Mauve v2.3.1 (Darling et al., 2010). Comparative illustration of coscinodiscophycean plastomes was conducted using circos v0.69 (Krzywinski et al., 2009).
Phylogenetic relationship and molecular dating were analyzed by calculating the codon evolution rate of nucleotide (nt) sequences of 109 PCGs which were shared by plastomes of selected diatom species and Ectocarpus siliculosus (Phaeophyceae, Ochrophyta). E. siliculosus was used as outgroup with its known fossil time. The nt sequences were aligned using MAFFT v7.471 and concatenated using PhyloSuite v1.2.2 (Zhang et al., 2020). The phylogenetic tree was constructed using IQ-TREE v1.6.12 (Trifinopoulos et al., 2016), and molecular dating was conducted using the PAML package v4.8a (Yang, 2007). Estimation of substitution rate was performed using baseml, and estimation of divergence times with the approximate likelihood method was carried out using mcmctree. The phylogenetic tree was displayed in Figtree v1.4.3 and visualized with 95% highest posterior density interval (HPD) for each node. Three calibrations of internal nodes, including E. siliculosus at 176–202 MYA (Matari and Blair, 2014), R. setigera at 90–93 MYA (Sinninghe-Damsté et al., 2004), and Thalassiosirales at 40–50 MYA (Sims et al., 2006), were conducted in divergence time estimation.
Complete plastome sequences of six coscinodiscophycean diatom species which exhibited rich diversity in cell morphology and cell size (Figure 1) were successfully assembled. These plastomes ranged in size from 120.5 kb in A. curvatulus to 135.8 kb in S. turris, and their G + C content was from 30.37% in S. turris to 32.39% in G. delicatula (Table 1), which were in the range of reported diatom plastomes (Ruck et al., 2017; Liu et al., 2021a,b; He et al., 2022; Wang et al., 2022). Similar to the reported diatom plastomes (e.g., Kowallik et al., 1995; Oudot-Le Secq et al., 2007; Tanaka et al., 2011; Hamsher et al., 2019), these newly assembled plastomes were mapped as canonical circular quadripartite structures with two IRs separating LSC and SSC. The conserved psbD-psbC overlapping region was detected in these coscinodiscophycean plastomes, but also in plastomes of Bacillariophyta and Ochrophyta (Kowallik et al., 1995; Liu et al., 2017). However, comparative analysis reveals that the psbD-psbC overlapping regions in the Stephanopyxis and Paralia plastomes reduced from the original 53 to 17 bp (GTGGAAACGCCCTTTAA), due to a single base mutation (T→C) which should have occurred in the common progenitor of Stephanopyxis and Paralia, and psbC started with GTG instead of ATG.
Combined the other six plastomes of coscinodiscophycean diatom species deposited in the GenBank database, we found that these 12 plastomes represented four orders in the class Coscinodiscophyceae (Table 1), which enabled us to obtain valuable evolutionary clues. The 135.8-kb Stephanopyxis plastome was the largest one found in Coscinodiscophyceae thus far, followed by the 132.2-kb Paralia plastome, and then the plastomes in Rhizosoleniales and Coscinodiacales. There is no significant difference in plastome sizes between Rhizosoleniales and Coscinodiacales (Table 1). Plastomes in Paraliales and Stephanopyxales tended to be larger than those in Rhizosoleniales and Coscinodiacales (Ruck et al., 2014; Sabir et al., 2014; Yu et al., 2018), which was caused not only by the expansion of the IR but also by the marked increase of the LSC. The size of LSC in Paraliales and Stephanopyxales is at least 8.0 kb larger than that in Rhizosoleniales and Coscinodiacales (Table 1). Numerous expanded intergenic regions were caused by accepting foreign DNA fragments in the Stephanopyxis and Paralia plastomes, which made them less compact when compared with plastomes in Rhizosoleniales and Coscinodiacales. However, it is difficult to trace the exact origin of some foreign fragments detected in diatom plastomes thus far, although a small fraction has been identified to be from diatom plasmids (Brembu et al., 2014; Ruck et al., 2014).
Phylogenomic analysis based on AA sequences of 116 PCGs shared by the 12 coscinodiscophycean plastomes as well as the T. laevis plastome (Tajima et al., 2016) as the outgroup revealed that the sampled coscinodiscophycean diatom species were grouped into four clades representing four orders (Figure 2). Guinardia and Rhizosolenia were recovered as the monophyletic clade Rhizosoleniales. In Rhizosoleniales, R. setigera was sister to Guinardia plus the left Rhizosolenia with high bootstrap support (100%), which shows that R. setigera may represent a hitherto undescribed genus (pseudo-Rhizosolenia) independent of Guinardia and Rhizosolenia. The controversial results that intraspecific genetic distance in G. striata (OM827251 and MG755796) exceeds interspecific genetic distance between G. striata and G. delicatula indicated that the G. striata strain (OM827251) we identified is not the same species as that (MG755796). More work needs to be carried out to reveal whether there are cryptic species in Guinardia. Actinocyclus and Coscinodiscus clustered together to form the clade Coscinodiacales which was sister to Rhizosoleniales. Paralia and Stephanopyxis, which represented two distinct orders, clustered tightly to form the Paraliales-Stephanopyxales complex. The Paraliales-Stephanopyxales complex was sister to Rhizosoleniales-Coscinodiscales complex with 100% bootstrap support (Figure 2).
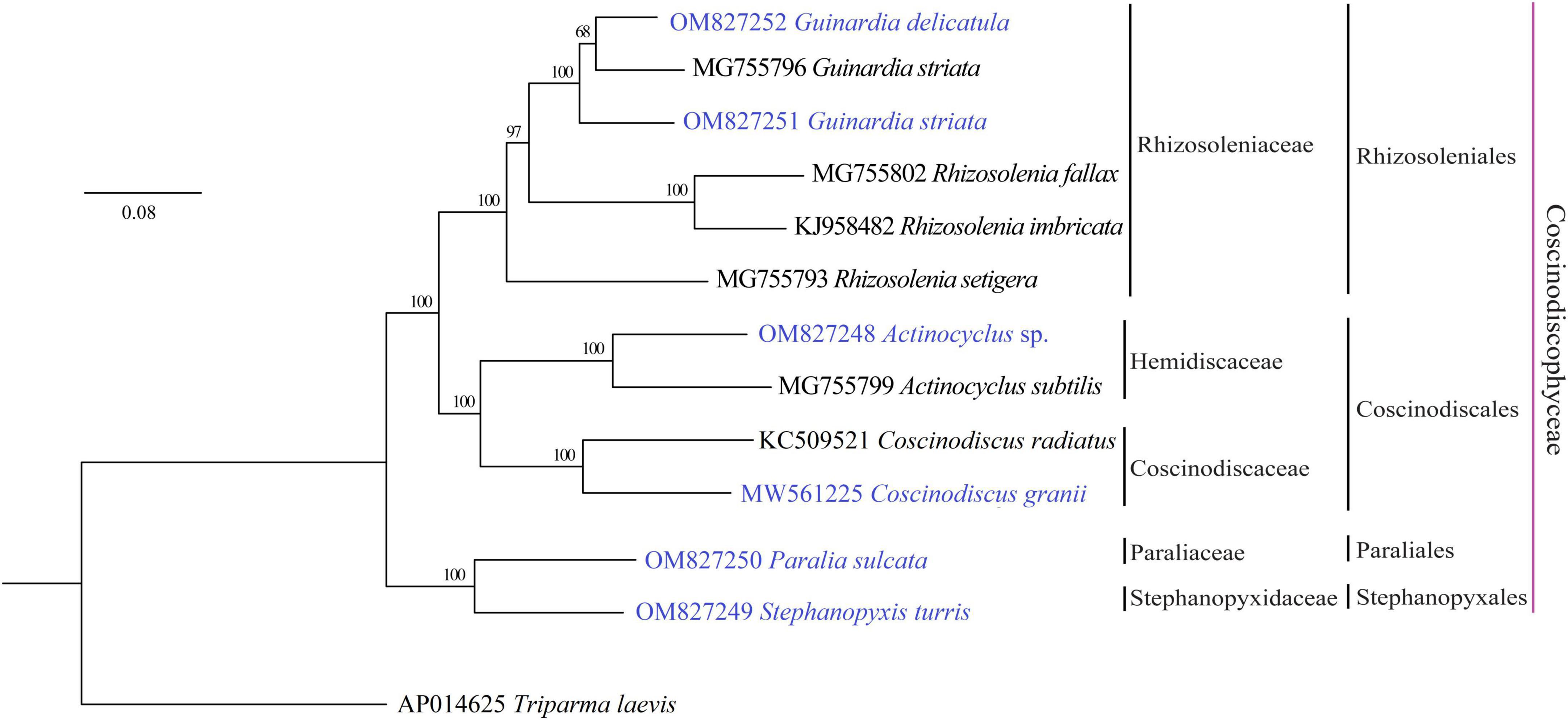
Figure 2. Maximum Likelihood (ML) phylogenomic tree of coscinodiscophycean diatom species based on concatenated amino acid sequences of 116 PCGs shared by plastomes of these diatoms as well as Triparma laevis as outgroup. Numbers at the branches represented bootstrap values. Branch lengths were proportional to the amount of sequence change, which were indicated by the scale bar below the trees.
A molecular dating tree was constructed to evaluate the divergence time in diatom lineages based on nt sequences of 109 PCGs shared by the plastomes of selected diatoms and E. siliculosus. Our results showed that ancient diatoms (Bacillariophyta) were estimated to appear at about 186 MYA, which was within the range of previous reports (Sorhannus, 2007; Medlin, 2015), while the Coscinodiscophyceae was separated from the Bacillariophyceae-Mediophyceae complex at about 142 MYA in the early Lower Cretaceous (Figure 3). These coscinodiscophycean diatom species were well recovered as a monophyletic clade (Figure 3). In Coscinodiscophyceae, the Paraliales-Stephanopyxales complex split at 124 MYA, followed by Coscinodiacales and Rhizosoleniales that split 114 MYA (Figure 3). The divergence time between Paraliales and Stephanopyxales was estimated at 85 MYA in the middle Upper Cretaceous, which was consistent with the time reported previously (Sorhannus, 2007). These results emphasize that Paraliales and Stephanopyxales appeared later than Coscinodiacales and Rhizosoleniales. In Coscinodiacales, Actinocyclus and Coscinodiscus split at about 101 MYA. In Rhizosoleniales, R. setigera emerged first at about 92 MYA and then the split between Guinardia and other Rhizosolenia occurred at 84 MYA.
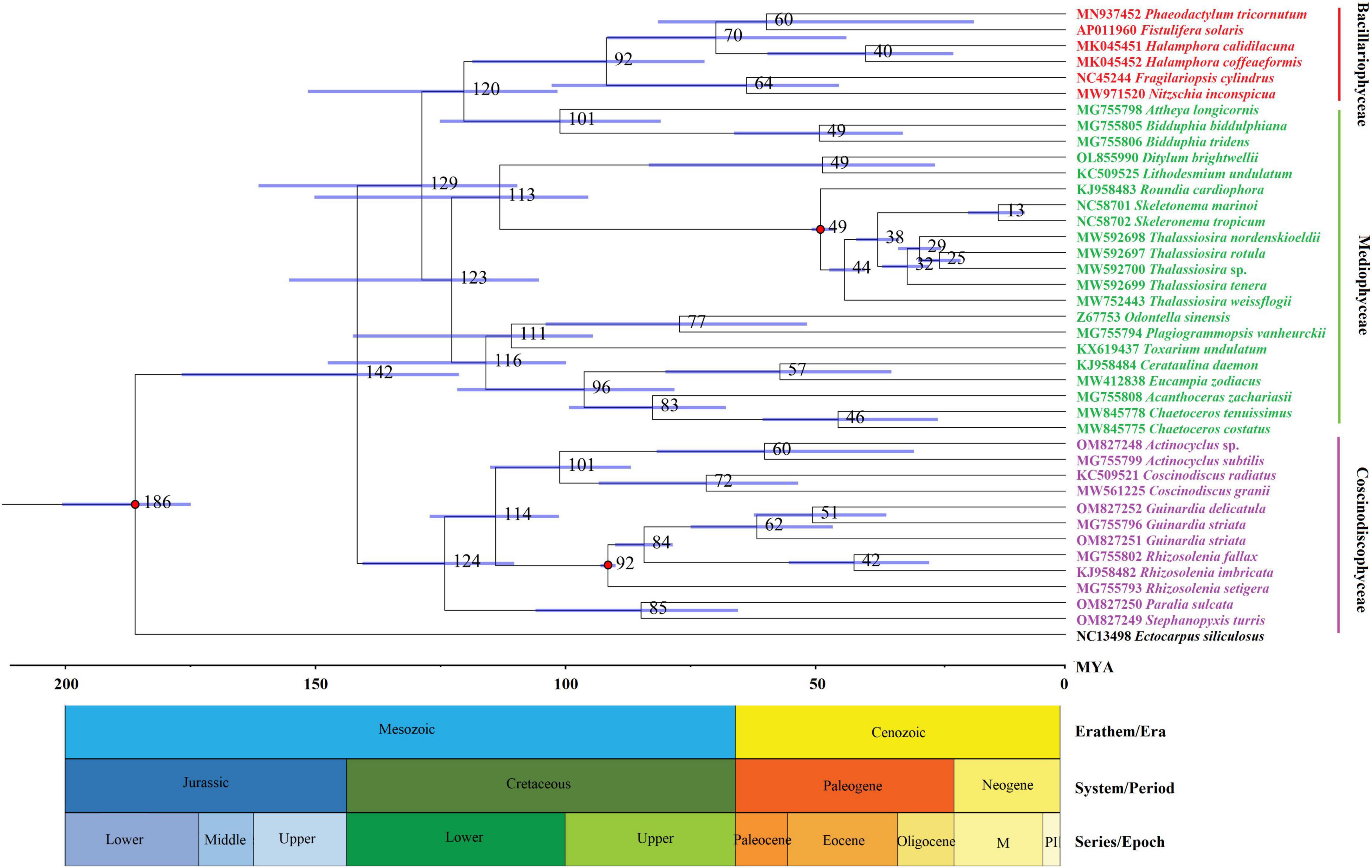
Figure 3. Time-calibrated phylogenetic analysis based on nucleotide sequences of 109 PCGs shared by the plastomes of selected diatom species as well as Ectocarpus siliculosus as outgroup. The fossil calibration taxa were indicated by red dots at corresponding nodes. Horizontal bars in blue color represented the 95% highest posterior density (HPD) values of the estimated divergence time.
Gene repertoires in coscinodiscophycean plastomes showed substantial variations, ranging from 154 genes in R. fallax to 163 genes in two Coscinodiscus plastomes (Table 1 and Supplementary Table 3). Comparative analysis revealed that 120 PCGs, three ribosomal RNA genes (rRNAs), 27 transfer RNA genes (tRNAs) and two regulatory small RNA genes (sRNAs) were shared by these coscinodiscophycean plastomes, with 13 PCGs lost to various degrees in different lineages of Coscinodiscophyceae (Figure 4 and Table 2), which is most likely due to their horizontal transfer to corresponding nuclear genomes (Ruck et al., 2014; Liu et al., 2021a,b). Twenty-seven tRNAs were sufficient for messenger RNA translation in diatom plastomes which contained one more tRNA (trnR3) than the plastomes of brown algae (Liu et al., 2017). In addition to two copies of trnN(guu) located at IRa and IRb, respectively, another perfect copy of trnN(guu) which shared the same sequence as the formers was situated in SSC of the Stephanopyxis plastome, but was absent in other coscinodiscophycean plastomes, indicating that the duplication of trnN(guu) occurred only in the Stephanopyxis plastome. Two sRNAs, ffs and ssrA, were conservatively located in the adjacent upstream (5′) region of psbX and adjacent downstream (3′) region of trnR3(ccg), respectively. ffs is a signal-recognition particle RNA that participates in the transmembrane transport of nuclear-encoded genes with plastid localization, and ssrA is a small regulatory RNA that interacts with stalled ribosomes to resume translation (Galachyants et al., 2012).
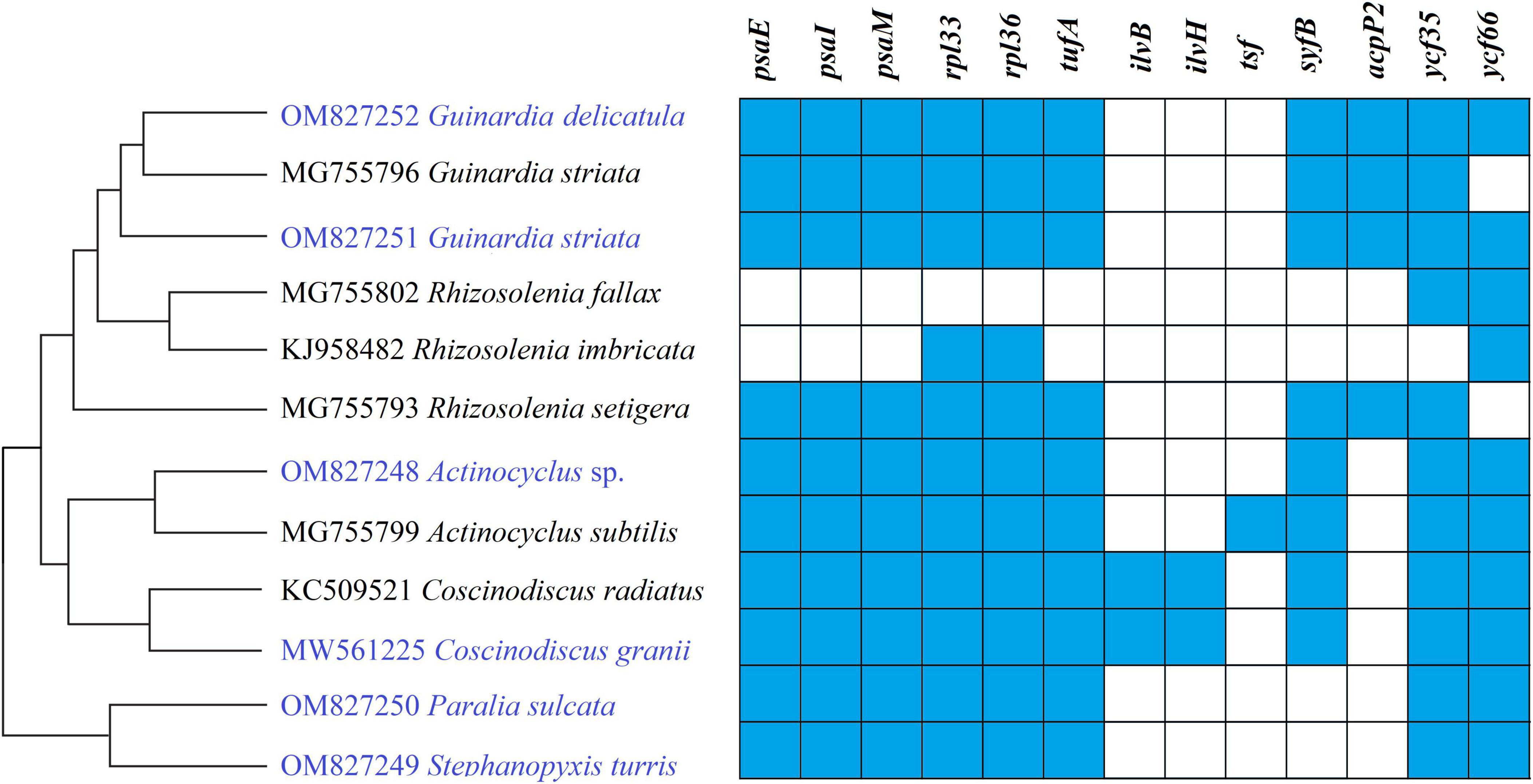
Figure 4. Changes in gene content of PCGs in the 12 coscinodiscophycean plastomes. The Maximum Likelihood (ML) tree on the left showed their phylogenetic relationships, and the matrix on the right showed the presence (blue) and absence (white) of 16 PCGs.
All these coscinodiscophycean cpDNAs belong to intron-less plastomes. The vast majority of sequenced diatom plastomes contain no intron, and only several diatom plastomes harbor a small number of introns. So far, plastid introns were detected in six housekeeping genes including atpB, groEL, petB, petD, psaA, and rnl among sequenced diatom plastomes (Brembu et al., 2014; Ruck et al., 2017; Yu et al., 2018; Hamsher et al., 2019; Gastineau et al., 2021; Górecka et al., 2021; Liu et al., 2021a). Overall, diatom plastomes exhibit an essential characteristic of being uncontaminated by intron. This characteristic is relatively similar in chloroplasts originating from secondary endosymbiosis (e.g., Phaeophyceae) (Liu et al., 2017), but significantly different from chloroplasts originating from primary endosymbiosis (e.g., Ulvophyceae and land plants), which typically contain more group I or/and group II introns in housekeeping genes (Liu et al., 2023).
Frequent loss of housekeeping PCGs were observed in these coscinodiscophycean plastomes, suggesting that diatom plastomes showed an ongoing reduction in gene content during evolution. Five PCGs including ilvB, ilvH, tsf, syfB, and acpP2 were absent from plastomes of both Paralia and Stephanopyxis. Considering their phylogenetic relationship in Coscinodiscophyceae (Figure 4), it is most likely that the loss of these five PCGs happened after the split of the Paraliales-Stephanopyxales complex from the Rhizosoleniales-Coscinodiscales complex. Three PCGs including ilvB, ilvH, and tsf were missing in all of six Rhizosoleniales plastomes, suggesting that the loss of these three genes might occur earlier than the loss of other 10 PCGs in Rhizosoleniales. The plastomes of Rhizosolenia fallax-imbricata lost more PCGs compared with the other lineages (Yu et al., 2018). Especially, some housekeeping PCGs associated with photosystem I (psaE, psaI, and psaM) and ribosome (rpl33 and rpl36) were lost. Combined with phylogenetic analysis, it can be seen that the loss of these genes in the R. fallax-imbricata plastomes should be recent events. Considering the important function of these genes, they are most likely transferred into the nuclear genome through endosymbiotic gene transfer. Similar events have been identified in other diatoms. For example, endosymbiotic gene transfers of petF into nuclear genomes were identified to occur in the plastomes of Thalassiosira and Skeletonema species (Liu et al., 2021a,b). The transfer of petF from plastome to nuclear genome is linked to ecological success of Thalassiosira oceanica (Lommer et al., 2010). In Coscinodiscophyceae, two PCGs, ilvB and ilvH, were only present in Coscinodiscus plastomes, while tsf were found only in A. subtilis plastome. These three PCGs have been lost multiple times in the evolution of diatoms, and nowadays are rarely scattered in plastomes of specific species in different lineages of diatoms (Yu et al., 2018). It was found that tsf was present in nuclear genomes of some diatoms (e.g., Phaeodactylum and Thalassiosira species) with tsf-lacking plastomes (Ruck et al., 2014).
Based on the distribution pattern of two acpP genes (acpP1 and acpP2) in diatom plastomes, acpP was previously considered to have experienced multiple independent duplications in the acpP1/2-containning plastomes, meanwhile it was believed to have undergone multiple losses in the acpP-lacking plastomes during the evolution of diatoms (Yu et al., 2018). Our phylogenetic analysis based on the AA sequences of AcpP1 and AcpP2 in the plastomes of diatoms and T. laevis revealed that the acpP genes were clearly grouped into two clades representing acpP1 and acpP2 lineages, respectively (Figure 5). Each clade harbored related members from Bacillariophyceae, Mediophyceae and Coscinodiscophyceae. These results indicated that acpP1 and acpP2 in diatom plastomes were not derived from multiple gene duplications occurring in different lineages of diatoms, instead they originated from an early gene duplication event occurred in the common progenitor after diatom emergence, followed by frequent loss of one or both genes (acpP1 or/and acpP2) in different diatom lineages (Ruck et al., 2014).
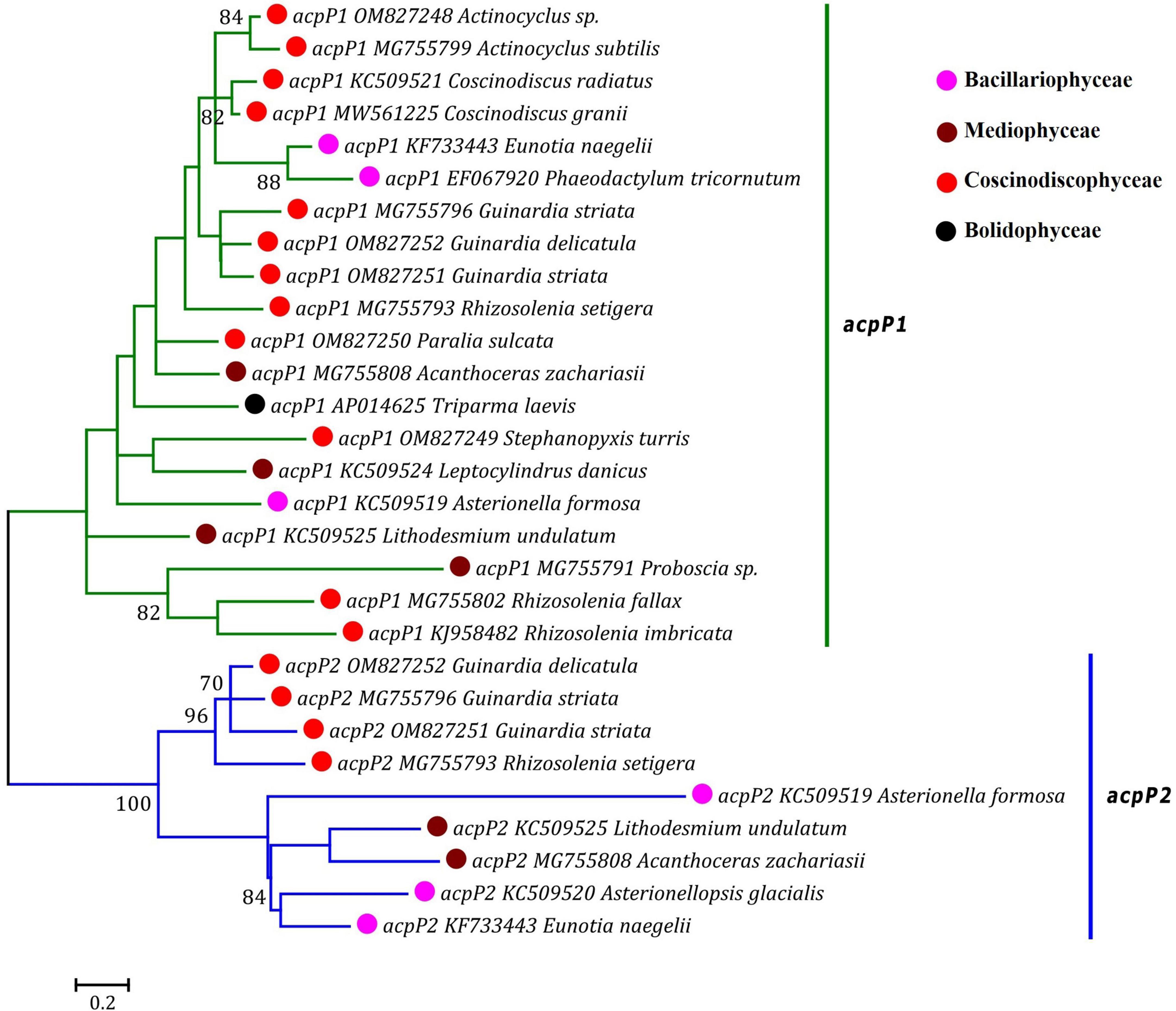
Figure 5. Unrooted Maximum Likelihood (ML) phylogenetic tree based on the amino acid sequences of AcpP1 and AcpP2 found in plastomes of diatoms and Triparma laevis. Numbers at the branches represented bootstrap values. The bootstrap support values greater than 70% were displayed at branches. Branch lengths were proportional to the amount of sequence change, which were indicated by the scale bar below the trees.
Size changes in these coscinodiscophycean plastomes were mainly caused by variations of IRs, which ranged from 11.3 kb in A. curvatulus to 20.1 kb in S. turris. Similar to most of the IR-containing plastomes, diatom IRs were composed of the conserved rns-trnI-trnA-rnl-rrn5 gene block and additional genes that flanked the ribosomal gene operon. However, IRs of these coscinodiscophycean plastomes shared a much larger conserved gene block containing nine genes, i.e., acpP-trnP-ycf89- rns-trnI-trnA-rnl-rrn5-psbA. At the intrageneric level of Guinardia, Actinocyclus and Coscinodiscus, the structure and size of IRs show little fluctuations (Figure 6), suggesting that IRs were generally conserved among closely related species. However, IRs in S. turris and R. fallax-imbricata exhibited a similar trend of large expansion to SSCs and slightly small contraction from LSCs, which eventually led to the conspicuous increase in IR sizes. The most obvious expansion of IRs was observed in Stephanopyxis plastome with the 20.1-kb IR which harbored 23 genes and was much longer than those in other coscinodiscophycean lineages (Figure 6).
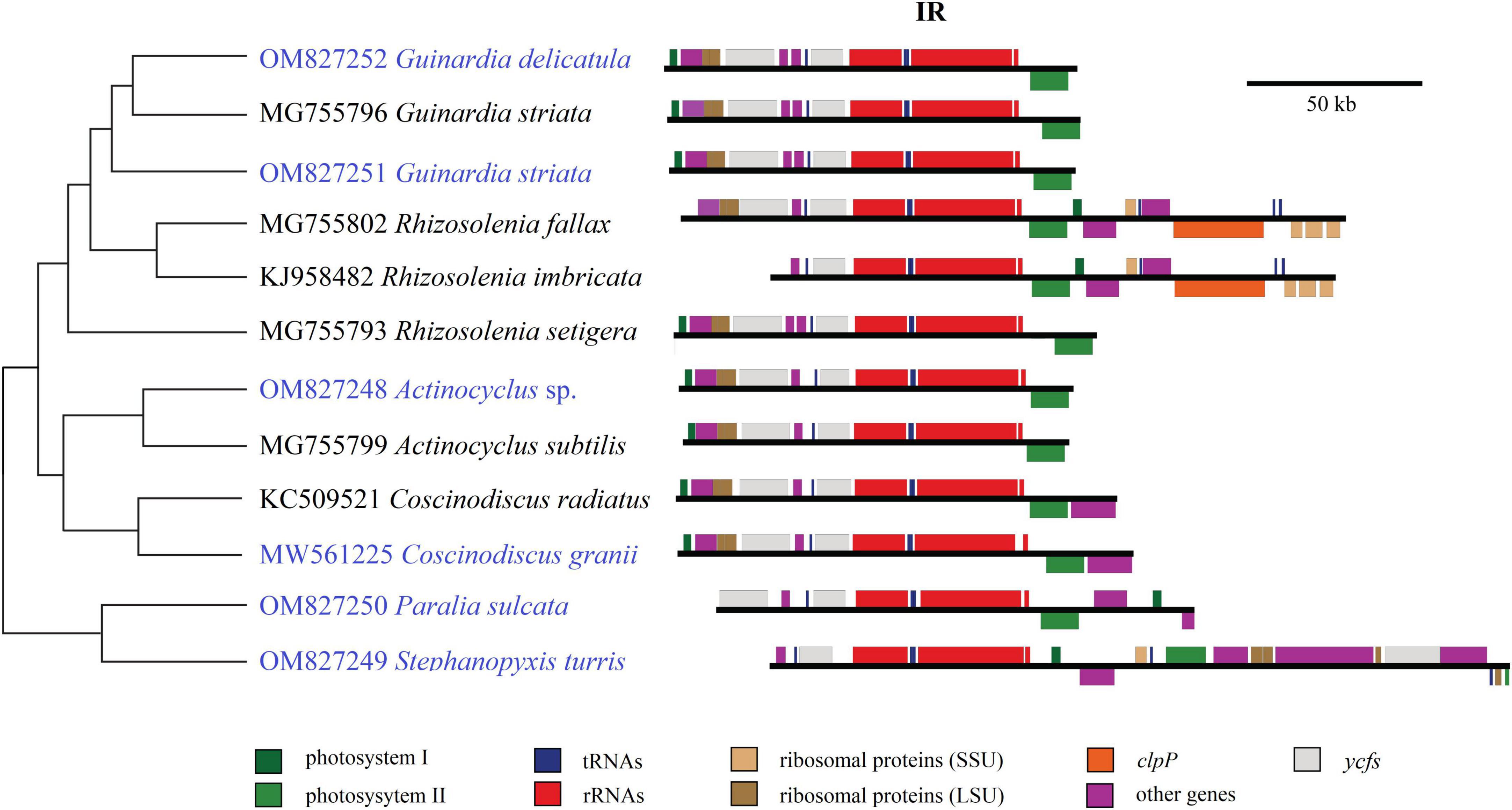
Figure 6. Comparison of inverted repeats (IRs) in the 12 coscinodiscophycean plastomes. The Maximum Likelihood (ML) tree on the left showed their phylogenetic relationships, and the photo on the right showed the structure and size of IRs. Annotated genes were colored according to their functional categories.
In the evolution of diatoms, the contraction and expansion of IRs occurred frequently in many lineages, which resulted in a huge variations of IR sizes, ranging from 6.8 kb in Synedra acus to 79.0 kb in Climaconeis cf. scalaris, an eleven-fold difference (Galachyants et al., 2012; Yu et al., 2018; Gastineau et al., 2021). Interestingly, it was found that the circular Pseudo-nitzschia multiseries plastome (KR709240) could lack IR (Cao et al., 2016; He et al., 2022), which may represent a new trend in the evolution of diatom plastomes if the assembly was correct. As found in the green algae (e.g., Ulva), IR is not an essential feature of plastomes (Liu and Melton, 2021; Liu et al., 2023), and it would change considerably in different eukaryotic photosynthetic lineages.
Mauve alignments showed that Actinocyclus and Coscinodiscus plastomes shared the same gene order and showed high collinearity at the intra-order level of Coscinodiacales (Supplementary Figure 1). At the intrageneric level of Rhizosolenia, multiple inversion and translocation events were identified (Supplementary Figure 1). Within the genus Guinardia, we observed that one of IR gene clusters composing of 14 genes was inverted in the G. striata plastome (MG755796) (Yu et al., 2018), which resulted in two IR gene clusters being arranged in a forward direction instead of a reverse direction (Figure 7 and Supplementary Figure 2). If this was not due to assembly issues, the emergence of this unusual IR rearrangement represents new evolutionary features that are significantly different from our newly sequenced Guinardia plastomes (OM827251 and OM827252) which maintain a usual arrangement of IRs.
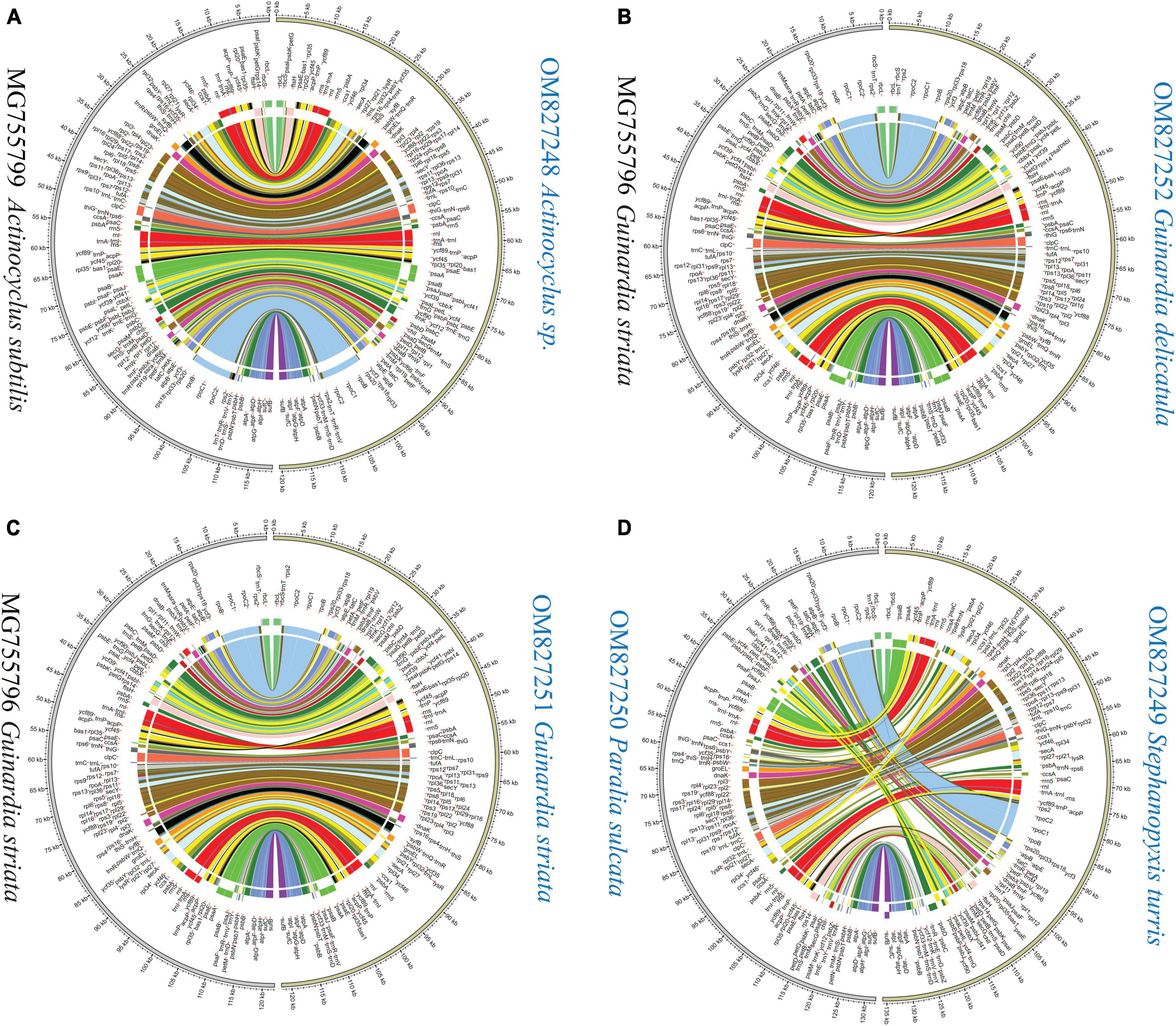
Figure 7. Pair-wise comparison of gene order and genome rearrangement between coscinodiscophycean plastomes. (A) Comparison of Actinocyclus sp. (OM827248) and Actinocyclus subtilis (MG755799). (B) Comparison of Guinardia delicatula (OM827252) and Guinardia striata (MG755796). (C) Comparison of Guinardia striata (OM827251) and Guinardia striata (MG755796). (D) Comparison of Stephanopyxis turris (OM827249) and Paralia sulcate (OM827250).
Different diatom lineages showed significantly different evolution rates in gene order. Coscinodiacales and Rhizosoleniales appeared earlier than Paraliales and Stephanopyxales in the evolution of diatoms (Figure 3). Gene order in Coscinodiacales were highly conserved at the intra-order level, while multiple rearrangements were observed in Rhizosoleniales and between Paraliales and Stephanopyxales (Figure 7 and Supplementary Figure 1). Considering all instances of IR variations, it seems that variations of gene orders matched well with variations of IRs in these diatom plastomes, consistent with previous understanding that. IR regions might play an important role in stabilizing the architecture of plastomes (Turmel and Lemieux, 2018). It is reasonable to assume that accelerating IR changes involves frequent genome recombination and rearrangement (Figure 7 and Supplementary Figure 1), when considering substantial variation of the additional genes that flanked the ribosomal gene operon (rns-trnI-trnA-rnl-rrn5). However, despite frequent plastome rearrangements in these diatoms, the affiliation of gene clusters has not changed, suggesting that rearrangements were strictly restricted within LSC and SSC. This is different from what was observed in the IR-losing plastomes in green algae where gene clusters have undergone a larger range of frequent changes (Liu and Melton, 2021; Liu et al., 2023).
Construction of six plastomes of diatom species in the class Coscinodiscophyceae in our work substantially boosted the number of assembled plastomes in this class, which in turn helped to reveal new evolutionary trends and details of diatom plastomes. The coscinodiscophycean plastomes in different lineages exhibited different evolutionary trends. The plastomes in Coscinodiacales were highly conserved in genome size, gene content, IR structure and gene order at the intra-order level, but plastomes in Rhizosoleniales and Paraliales-Stephanopyxales complex displayed multiple changes in the above aspects, even at the intrageneric level (e.g., Rhizosolenia). Comparative analysis revealed that 120 protein-coding genes (PCGs), three ribosomal RNA genes (rRNAs), 27 transfer RNA genes (tRNAs) and two regulatory small RNA genes (sRNAs) were shared by these coscinodiscophycean plastomes, but the other 13 PCGs were lost to varying degrees in different coscinodiscophycean lineages. The lost housekeeping PCGs could have been transferred into the nuclear genome via endosymbiotic gene transfer. Frequent loss of housekeeping PCGs were observed in these coscinodiscophycean plastomes, suggesting that diatom plastomes showed an ongoing reduction in gene content during evolution. The IRs in S. turris and R. fallax-imbricata showed a trend of expansion to the SSC and contraction from the LSC, which eventually led to the conspicuous increase in IR size. Different lineages of diatoms show significantly different evolution rates in gene order. As more genomic data accumulate in future, we will be able to depict more comprehensive evolutionary pathways and reveal evolution mechanism of species diversity in diatoms.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in this article/Supplementary material.
NC designed the study. YW, HH, and FL performed the experiments. FL and YW performed the analysis. FL wrote the manuscript with contributions from coauthors. All authors have read and approved the final version of the manuscript.
This work was financially supported by the National Natural Science Foundation of China (Nos. 42176162 and 42276133), the Strategic Priority Research Program of Chinese Academy of Sciences (Nos. XDB42000000, XDA23050302, and XDA23050403), the Science and Technology Basic Resources Investigation Program of China (No. 2018FY100200), the Chinese Academy of Sciences Pioneer Hundred Talents Program (to NC), and the Taishan Scholar Project Special Fund (to NC).
We are grateful to colleagues from the Jiaozhou Bay National Marine Ecosystem Research Station for their help in field sampling, and all staffs of marine ecological environment genomics research group in Institute of Oceanology, Chinese Academy of Sciences. Statistical analyses were supported by Oceanographic Data Center, Institute of Oceanology, Chinese Academy of Sciences (IOCAS).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1203780/full#supplementary-material
Armbrust, E. V. (2009). The life of diatoms in the world’s oceans. Nature 459, 185–192. doi: 10.1038/nature08057
Armbrust, E. V., Berges, J. A., Bowler, C., Green, B. R., Martinez, D., Putnam, N. H., et al. (2004). The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science 306, 79–86.
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bhattacharya, D., Archibald, J. M., Weber, A. P., and Reyes-Prieto, A. (2007). How do endosymbionts become organelles? Understanding early events in plastid evolution. Bioessays 29, 1239–1246. doi: 10.1002/bies.20671
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bowler, C., Allen, A. E., Badger, J. H., Grimwood, J., Jabbari, K., Kuo, A., et al. (2008). The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456, 239–244.
Brembu, T., Winge, P., Tooming-Klunderud, A., Nederbragt, A. J., Jakobsen, K. S., and Bones, A. M. (2014). The chloroplast genome of the diatom Seminavis robusta: new features introduced through multiple mechanisms of horizontal gene transfer. Mar. Genomics 16, 17–27. doi: 10.1016/j.margen.2013.12.002
Cao, M., Yuan, X.-L., and Bi, G. (2016). Complete sequence and analysis of plastid genomes of pseudo-nitzschia multiseries (bacillariophyta). Mitochondrial DNA Part A 27, 2897–2898. doi: 10.3109/19401736.2015.1060428
Capella-Gutierrez, S., Silla-Martinez, J. M., and Gabaldon, T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Darling, A. E., Mau, B., and Perna, N. T. (2010). ProgressiveMauve: multiple genome alignment with gene gain. Loss and rearrangement. PLoS One 5, e11147. doi: 10.1371/journal.pone.0011147
Falkowski, P. G., Barber, R. T., and Smetacek, V. (1998). Biogeochemical controls and feedbacks on ocean primary production. Science 281, 200–206. doi: 10.1126/science.281.5374.200
Galachyants, Y. P., Morozov, A. A., Mardanov, A. V., Beletsky, A. V., Ravin, N. V., Petrova, D. P., et al. (2012). Complete Chloroplast genome sequence of freshwater Araphid pennate diatom alga Synedra acus from lake baikal. International Journal of Biology. 4, 27–35. doi: 10.5539/ijb.v4n1p27
Gastineau, R., Davidovich, N., Davidovich, O., Lemieux, C., Turmel, M., Wróbel, R., et al. (2021). Extreme enlargement of the inverted repeat region in the plastid genomes of diatoms from the genus climaconeis. Int. J. Mol. Sci. 22, 7155. doi: 10.3390/ijms22137155
Górecka, E., Gastineau, R., Davidovich, N., Davidovich, O., Ashworth, M., Sabir, J., et al. (2021). Mitochondrial and plastid genomes of the Monoraphid diatom Schizostauron trachyderma. Int. J. Mol. Sci. 22, 11139. doi: 10.3390/ijms222011139
Guiry, M. D., and Guiry, G. M. (2023). AlgaeBase. World-wide electronic publication. Galway: National University of Ireland.
Hamsher, S. E., Keepers, K. G., Pogoda, C. S., Stepanek, J. G., Kane, N. C., and Kociolek, J. P. (2019). Extensive chloroplast genome rearrangement amongst three closely related Halamphora spp. (Bacillariophyceae), and evidence for rapid evolution as compared to land plants. PLoS One 14:e0217824. doi: 10.1371/journal.pone.0217824
He, Z., Chen, Y., Wang, Y., Liu, K., Xu, Q., Li, Y., et al. (2022). Comparative analysis of pseudo-nitzschia chloroplast genomes revealed extensive inverted region variation and pseudo-nitzschia speciation. Front. Mar. Sci. 9:784579. doi: 10.3389/fmars.2022.784579
Hemaiswarya, S., Raja, R., Kumar, R. R., Ganesan, V., and Anbazhagan, C. (2011). Microalgae: a sustainable feed source for aquaculture. World J. Microbiol. Biotechnol. 27, 1737–1746.
Jin, J.-J., Yu, W.-B., Yang, J.-B., Song, Y., dePamphilis, C. W., Yi, T.-S., et al. (2020). GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21, 241. doi: 10.1186/s13059-020-02154-5
Jones, D. T., Taylor, W. R., and Thornton, J. M. (1992). The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8, 275–282.
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A., and Jermiin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kiatmetha, P., Siangdang, W., Bunnag, B., Senapin, S., and Withyachumnarnkul, B. (2011). Enhancement of survival and metamorphosis rates of Penaeus monodon larvae by feeding with the diatom Thalassiosira weissflogii. Aquac. Int. 19, 599–609.
Kowallik, K. V., Stoebe, B., Schaffran, I., Kroth-Pancic, P., and Freier, U. (1995). The chloroplast genome of a chlorophyll a+c-containing alga, Odontella sinensis. Plant Mol. Biol. Rep. 13, 336–342. doi: 10.1016/0014-5793(96)00658-8
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lang, B. F., and Nedelcu, A. M. (2012). “Plastid genomes of algae,” in Advances in photosynthesis and respiration including bioenergy and related processes: genomics of chloroplasts and mitochondria, eds R. Bock and V. Knoop (Dordrecht: Springer), 59–87. doi: 10.1007/978-94-007-2920-9_3
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Liu, F., and Melton, J. T. (2021). Chloroplast genomes of the green-tide forming alga Ulva compressa: comparative chloroplast genomics in the genus Ulva (Ulvophyceae, Chlorophyta). Front. Mar. Sci. 8, 668542. doi: 10.3389/fmars.2021.668542
Liu, F., Chen, N., Wang, H., Li, J., Wang, J., and Qu, F. (2023). Novel insights into chloroplast genome evolution in the green macroalgal genus Ulva (Ulvophyceae, Chlorophyta). Front. Plant Sci. 14:1126175.
Liu, F., Jin, Z., Wang, Y., Bi, Y., and Melton, J. T. (2017). Plastid genome of Dictyopteris divaricata (Dictyotales, Phaeophyceae): understanding the evolution of plastid genomes in brown algae. Mar. Biotech. 19, 627–637. doi: 10.1007/s10126-017-9781-5
Liu, K., Chen, Y., Cui, Z., Liu, S., Xu, Q., and Chen, N. (2021a). Comparative analysis of chloroplast genomes of thalassiosira species. Front. Mar. Sci. 8:788307. doi: 10.3389/fmars.2021.788307
Liu, K., Xu, Q., Liu, K., Zhao, Y., and Chen, N. (2021b). Chloroplast genomes for five skeletonema species: comparative and phylogenetic analysis. Front. Plant Sci. 12:2962. doi: 10.3389/fpls.2021.774617
Lommer, M., Roy, A.-S., Schilhabel, M., Schreiber, S., Rosenstiel, P., and LaRoche, J. (2010). Recent transfer of an iron-regulated gene from the plastid to the nuclear genome in an oceanic diatom adapted to chronic iron limitation. BMC Genomics 11:718. doi: 10.1186/1471-2164-11-718
Mann, D. G., and Droop, S. J. M. (1996). Biodiversity, biogeography and conservation of diatoms. Hydrobiologia 336, 19–32.
Mann, D. G., Trobajo, R., Sato, S., Li, C., Witkowski, A., Rimet, F., et al. (2021). Ripe for reassessment: a synthesis of available molecular data for the speciose diatom family Bacillariaceae. Mol. Phylogenetics Evol. 158, 106985. doi: 10.1016/j.ympev.2020.106985
Matari, N. H., and Blair, J. E. (2014). A multilocus timescale for oomycete evolution estimated under three distinct molecular clock models. BMC Evol. Biol. 14:101. doi: 10.1186/1471-2148-14-101
Medlin, L. K. (2015). A timescale for diatom evolution based on four molecular markers: reassessment of ghost lineages and major steps defining diatom evolution. Vie Milieu-Life Environ. 65, 219–238.
Medlin, L. K., and Desdevises, Y. (2020). Phylogenetic reconstruction of diatoms using a seven-gene dataset, multiple outgroups, and morphological data for a total evidence approach. Phycologia 59, 422–436.
Medlin, L. K., and Kaczmarska, I. (2004). Evolution of the diatoms: V. Morphological and cytological support for the major clades and a taxonomic revision. Phycologia 43, 245–270.
Minh, B. Q., Nguyen, M. A. T., and von Haeseler, A. (2013). Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 30, 1188–1195.
Nurachman, Z., Anita, S., Anward, E. E., Novirani, G., Mangindaan, B., et al. (2012). Oil productivity of the tropical marine diatom Thalassiosira sp. Bioresour. Technol. 108, 240–244.
Oudot-Le Secq, M.-P., Grimwood, J., Shapiro, H., Armbrust, E. V., Bowler, C., and Green, B. R. (2007). Chloroplast genomes of the diatoms Phaeodactylum tricornutum and Thalassiosira pseudonana: comparison with other plastid genomes of the red lineage. Mol. Genet. Genomics. 277, 427–439. doi: 10.1007/s00438-006-0199-4
Ruck, E. C., Linard, S. R., Nakov, T., Theriot, E. C., and Alverson, A. J. (2017). Hoarding and horizontal transfer led to an expanded gene and intron repertoire in the plastid genome of the diatom, Toxarium undulatum (Bacillariophyta). Curr. Genet. 63, 499–507. doi: 10.1007/s00294-016-0652-9
Ruck, E. C., Nakov, T., Jansen, R. K., Theriot, E. C., and Alverson, A. J. (2014). Serial gene losses and foreign DNA underlie size and sequence variation in the plastid genomes of diatoms. Genome Biol. Evol. 6, 644–654. doi: 10.1093/gbe/evu039
Sabir, J. S. M., Yu, M., Ashworth, M. P., Baeshen, N. A., Baeshen, M. N., Bahieldin, A., et al. (2014). Conserved gene order and expanded inverted repeats characterize plastid genomes of Thalassiosirales. PLoS One 9:e107854. doi: 10.1371/journal.pone.0107854
Sims, P. A., Mann, D. G., and Medlin, L. K. (2006). Evolution of the diatoms: insights from fossil, biological and molecular data. Phycologia 45, 361–402.
Sinninghe-Damsté, J. S., Muyzer, G., Abbas, B., Rampen, S. W., Masse, G., Allard, W. G., et al. (2004). The rise of the rhizosolenid diatoms. Science 304, 584–587. doi: 10.1126/science.1096806
Sorhannus, U. (2007). A nuclear-encoded small-subunit ribosomal RNA timescale for diatom evolution. Marine Micropaleontol. 65, 1–12.
Sumper, M., and Brunner, E. (2008). Silica biomineralisation in diatoms: the model organism Thalassiosira pseudonana. ChemBioChem 9, 1187–1194. doi: 10.1002/cbic.200700764
Tajima, N., Saitoh, K., Sato, S., Maruyama, F., Ichinomiya, M., Yoshikawa, S., et al. (2016). Sequencing and analysis of the complete organellar genomes of parmales, a closely related group to Bacillariophyta (diatoms). Curr. Genet. 62, 887–896. doi: 10.1007/s00294-016-0598-y
Tanaka, T., Fukuda, Y., Yoshino, T., Maeda, Y., Muto, M., Matsumoto, M., et al. (2011). High-throughput pyrosequencing of the chloroplast genome of a highly neutral-lipid-producing marine pennate diatom, Fistulifera sp. strain JPCC DA0580. Photosynth. Res. 109, 223–229. doi: 10.1007/s11120-011-9622-8
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (1997). The ClustalX windows interface flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Thorvaldsdottir, H., Robinson, J. T., and Mesirov, J. P. (2013). Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14, 178–192. doi: 10.1093/bib/bbs017
Trifinopoulos, J., Lam-Tung, N., von Haeseler, A., and Minh, B. Q. (2016). W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44, W232–W235. doi: 10.1093/nar/gkw256
Turmel, M., and Lemieux, C. (2018). Evolution of the plastid genome in green algae. Adv. Bot. Res. 85, 157–193.
Wang, Y., Wang, J., Chen, Y., Liu, S., Zhao, Y., and Chen, N. (2022). Comparative analysis of Bacillariophyceae chloroplast genomes uncovers extensive genome rearrangements associated with speciation. Int. J. Environ. Res. Public Health 19, 10024. doi: 10.3390/ijerph191610024
Xu, Q., Cui, Z., and Chen, N. (2021). Comparative analysis of chloroplast genomes of seven chaetoceros species revealed variation hotspots and speciation time. Front. Microbiol. 12:742554. doi: 10.3389/fmicb.2021.742554
Yang, Z. (2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591.
Yu, M., Ashworth, M. P., Hajrah, N. H., Khiyami, M. A., Sabir, M. J., Alhebshi, A. M., et al. (2018). Evolution of the plastid genomes in diatoms. Adv. Bot. Res. 85, 129–155.
Zhang, D., Gao, F., Jakovlic, I., Zou, H., Zhang, J., Li, W. X., et al. (2020). PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 20, 348–355. doi: 10.1111/1755-0998.13096
Keywords: diatom, plastome, Coscinodiscophyceae, gene duplication, inverted repeats, phylogenomic analysis
Citation: Liu F, Wang Y, Huang H and Chen N (2023) Evolutionary dynamics of plastomes in coscinodiscophycean diatoms revealed by comparative genomics. Front. Microbiol. 14:1203780. doi: 10.3389/fmicb.2023.1203780
Received: 11 April 2023; Accepted: 01 June 2023;
Published: 15 June 2023.
Edited by:
Ondrej Prasil, Academy of Sciences of the Czech Republic (ASCR), CzechiaReviewed by:
Tore Brembu, Norwegian University of Science and Technology, NorwayCopyright © 2023 Liu, Wang, Huang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Feng Liu, bGl1ZmVuZ0BxZGlvLmFjLmNu; Nansheng Chen, Y2hlbm5zQHFkaW8uYWMuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.