
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 28 July 2023
Sec. Microorganisms in Vertebrate Digestive Systems
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1203678
This article is part of the Research TopicExploring the Oral-Gut Microbiome Interactions: Pathways to Therapeutic Strategies and Implications for Systemic HealthView all 9 articles
 Lingming Hu1†
Lingming Hu1† Zhaojun Ni1†
Zhaojun Ni1† Kangqing Zhao1
Kangqing Zhao1 Xiangxue Li1
Xiangxue Li1 Xuejiao Gao1
Xuejiao Gao1 Yulin Kang2
Yulin Kang2 Zhoulong Yu1
Zhoulong Yu1 Ying Qin3
Ying Qin3 Jingwen Zhao3
Jingwen Zhao3 Wenjuan Peng3
Wenjuan Peng3 Lin Lu1,4,5
Lin Lu1,4,5 Hongqiang Sun1*
Hongqiang Sun1*Introduction: The relationship between oral and gut microbiota in alcohol dependence (AD) is not well understood, particularly the effects of oral microbiota on the intestinal microbiota. The current study aimed to explore the association between oral and gut microbiota in AD to clarify whether oral microbiota could ectopically colonize into the gut.
Methods: 16S rRNA sequence libraries were used to compare oral and gut microbial profiles in persons with AD and healthy controls (HC). Source Tracker and NetShift were used to identify bacteria responsible for ectopic colonization and indicate the driver function of ectopic colonization bacteria.
Results: The α-diversity of oral microbiota and intestinal microbiota was significantly decreased in persons with AD (all p < 0.05). Principal coordinate analysis indicated greater similarity between oral and gut microbiota in persons with AD than that in HC, and oral-gut overlaps in microbiota were found for 9 genera in persons with AD relative to only 3 genera in HC. The contribution ratio of oral microbiota to intestinal microbiota composition in AD is 5.26% based on Source Tracker,and the AD with ectopic colonization showed the daily maximum standard drinks, red blood cell counts, hemoglobin content, and PACS scores decreasing (all p < 0.05).
Discussion: Results highlight the connection between oral-gut microbiota in AD and suggest novel potential mechanistic possibilities.
Alcohol dependence (AD) causes physical and psychological damage to patients and contributed to more than 3 million deaths in 2016, accounting for 5.3 percent of total global mortality (World Health Organization, 2018). Alcohol dependence was the most prevalent substance use disorder in China with a lifetime prevalence of 1.3% and a 12-month prevalence of 0.7% as of 2015 (Huang et al., 2019). The pathogenesis of AD remains unclear, although recent studies have implicated dysfunction of the oral or gut microbiota correlated with alcohol dependence (Leclercq et al., 2014; Dubinkina et al., 2017). Previous study suggested that oral and gut microbiota as well as their products may have an impact on nutritional metabolism, immune protection, and neuro-regulation in individuals from infancy to senescence (Hehemann et al., 2010; Adak and Khan, 2019). Meanwhile, gut microbiota has a profound influence on the progression of AD and animal studies have shown that fecal microbiota transplantation from AD into germ-free mice interferes with recipient cognitive function, induces anxiety/depression behavior decreases expression of brain-derived neurotrophic factor (BDNF), and the function of mGluR1/PKC ε pathway in the brain (Zhao et al., 2020). In addition, oral bacteria, such as Neisseria, are known to participate in the metabolism of nitrate and acetaldehyde, perhaps contributing to AD-related cognitive impairment (Fan et al., 2018; Rosier et al., 2020). And increased levels of P. gingivalis, resulting from a disturbed balance of the oral microbiome, may impair the function of the blood–brain barrier (BBB) and elicit neuroinflammation (Panza et al., 2019).
Both oral and gut microflora may thus contribute to the pathogenesis of AD, but little is known about the association between the two bacterial populations. Healthy people generate and swallow about 1.5 L of saliva per day containing an enormous number of oral microbiota and metabolites (Humphrey and Williamson, 2001; Atarashi et al., 2017). Normal intestinal barrier function would prevent cross-colonization of oral and gut microbiota in healthy individuals, but the loss of intestinal integrity has been shown to cause invasion of oral bacteria into the intestine, like gastrointestinal diseases or in germ-free animal models (Humphrey and Williamson, 2001; Bull-Otterson et al., 2013; Atarashi et al., 2017; Park et al., 2021). The phenomenon of oral bacteria found in the gut is one type of ectopic colonization in bacteria that has been observed in many disease states already (Strandwitz, 2018; Rashidi et al., 2021). More oral microbiota has been found colonized in the gut of irritable bowel syndrome (IBS), inflammatory bowel disease (IBD), and colorectal cancer (CRC), compared with healthy individuals (Gevers et al., 2014; Iwauchi et al., 2019; Pittayanon et al., 2019; Thomas et al., 2019). Such ectopic colonization might aggravate the disturbed gut microbiota and immunoreactivity. Effects of oral microbiota on gut microbiota have been found to contribute to microglia-mediated neuroinflammation in major depression disorder (MDD) (Scassellati et al., 2021), and heavy alcohol consumption is known to impair gut barrier function and cause dysfunction of oral and gut microbiota (Forsyth et al., 2009; Leclercq et al., 2017). Therefore, the association between oral and gut microbiota seems to affect the progression of AD, but this association is still unclear.
The current study investigated whether oral microbiota could be ectopically colonized into the gut and whether those ectopic colonization bacteria are related to clinical features in AD. Findings from this study may contribute to a better understanding of oral-gut microbiota in AD, expose potential mechanisms and inform therapeutic strategies of AD in clinical practice.
Thirty-three male patients diagnosed with alcohol dependence were recruited from the Second People’s Hospital of Guizhou Province during July and September 2019. alcohol dependence was diagnosed using the Diagnostic and Statistical Manual of Mental Disorders, 4th edition (DSM-IV). Other than nicotine dependence, as determined by the Mini International Neuropsychiatric Interview (M.I.N.I), patients did not present with other substance use disorders or other DSM-VI mental health or Axis I disorders. Twenty-one male healthy controls (HC) without somatic and psychiatric illnesses were also recruited. HC did not have a current or lifetime history of alcohol use or other substance use disorders. The inclusion and exclusion criteria are shown below.
The inclusion and exclusion criteria are shown below. The inclusion criteria for the alcohol dependence (AD) group were as follows: (1) diagnosis of AD according to the Diagnostic and Statistical Manual of Mental Disorders, 4th edition (DSM-IV); (2) Han Chinese males aged 18–60 years. The exclusion criteria for the AD group were as follows: (1) had or having the infectious disease; (2) had or having heart, brain, liver, kidney, oral and other serious diseases; (3) had or having metabolic diseases that can lead to the abnormal immune system; (4) had or having neurodegenerative diseases; (5) used antibiotics, steroids or other microbiota-modulating medications within 2 months before enrollment; (6) previous or current DSM-IV diagnosis of schizophrenia, depression, anxiety disorder, bipolar disorder, mental retardation, dementia (excluding mild cognitive function), and substance dependence other than alcohol and nicotine; (7) had irregular eating habits that affected oral flora (except alcohol) in recent 2 months; (8) alcohol abstinence longer than 1 week. The inclusion criteria for the HC group were as follows: Han Chinese males aged 18–60 years. The exclusion criteria for the HC group were as follows: (1)–(8) exclusion criteria for patients with alcohol dependence but (6) patients with a previous or current DSM-IV diagnosis of schizophrenia, depression, anxiety disorder, bipolar disorder, mental retardation, dementia (excluding MCI), and substance dependence, including alcohol (excluding nicotine); (9) drinking alcohol during sample collection.
G*Power3.1.9.7 software was used to calculate the sample size, and it was assumed that the test standard α = 0.05 (bilateral), the effect value was 0.5, the beta value was 0.20, and the degree of certainty (1-β) was 0.80. It was calculated that the minimum sample size required was 18.
All subjects gave written informed consent for the collection of saliva and feces and participation in the study.
All experiments were conducted by the Helsinki Declaration of 1975 and the ethical standards of the home institution Committee on Human Experimentation. Ethical approval was granted by the ethics committees of Peking University Sixth Hospital (2019, ethical review No. 6) and the Second People’s Hospital of Guizhou Province.
Clinical features of AD participants were indexed, including withdrawal days, dependence years, first drinking age (FD age), frequency of drinking weekly (FREW), frequency of drinking daily (FRED), average standard drinks per day (ASD), maximum standard drinks per day (MSD), smoking status, body mass index (BMI), aspartate aminotransferase (AST), alanine transaminase (ALT), gamma-glutamyl transpeptidase (GGT), total bilirubine (TBIL), red blood cell (RBC), hemoglobin (HB), mean corpuscular volume (MCV), triiodothyronine (TT3), tetraiodothyronine (TT4), thyroid stimulating hormone (TSH); alcohol withdrawal syndrome (evaluated by Clinical Institute Withdrawal Assessment for Alcohol-Revised, CIWA-Ar), alcohol craving (evaluated by Pennsylvania Alcohol Craving Scale, PACS), cognitive function (evaluated by Montreal Cognitive Assessment, MoCA), nicotine consumption (evaluated by Fagerstrom test for nicotine dependence, FTND), sleeping patterns (evaluated by Pittsburg Sleep Quality Index, PSQI) and symptoms of anxiety /depression (evaluated by Hamilton Anxiety Scale and Hamilton Depression scale, HAMA and HAMD) were recorded. Control subjects were assessed for cognitive function, depression and anxiety, and sleeping patterns.
All subjects fasted overnight and did not brush their teeth. Fresh saliva and feces were collected at 7:00 to 8:00 am into sterile EP tubes. Saliva was collected after patients had rinsed the mouth with water 5 min beforehand and 1–2 ml of naturally secreted saliva was removed by Pap dropper into an EP tube. The procedure was repeated once. Feces were collected after patients had urinated to avoid contamination and defecated into a clean bedpan, from which 1–2 g stool, which had not been exposed to air, was removed with a sampling spoon. The procedure was repeated once. All samples were frozen in liquid nitrogen for 1 min and stored at −80°C. All samples were collected by procedures specified in the Human Microbiome Project (HMP) version 12 standard process (Integrative HMP Research Network Consortium, 2019).
DNA was extracted from the oral and gut microbial community using NEBnext microbiome DNA enrichment kit (New England Biolabs, Ipswich, MA, US). The detailed procedure following the manufacturer’s instructions, including 600 μl Buffer with magnetic beads +20 μl Proteinase K + 5 μl RNase A was added to 96-well deep plates before washing three times and adding 100 μl Elution Buffer. 100–200 mg sample was transferred to the centrifuge tube with grinding beads and 1 ml Buffer (ATL/PVP-10) added and the sample was ground and incubated at 65°C for 20 min before centrifugation at 14000 × g for 5 min. 0.6 ml Buffer PCI was added with mixing by vortex for 15 s and centrifugation at 12000 × g for 10 min. The supernatant was transferred to a deep well plate with magnetic beads binding solution and DNA was transferred to a 1.5 ml centrifuge tube. DNA concentration and purity were assessed by NanoDrop 2000 micro-ultraviolet spectrophotometer and quantified with a Qubit Fluorometer using Qubit® dsDNA BR Assay kit (Invitrogen, USA). DNA quality was checked by running an aliquot on 1% agarose gel.
Variable regions, V3 ~ V4, of the bacterial 16S rRNA gene, were amplified with degenerate PCR primers, 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5′-GGAC TACHVGGGTWT-CTAAT-3′) and products purified by Agencourt AMPure XP beads and eluted with elution buffer. Agilent Technologies 2,100 bioanalyzer qualified the Libraries and sequenced using an Illumina HiSeq 2,500 platform (BGI, Shenzhen, China), following the manufacturer’s instructions. 2 × 250 bp paired-end reads were generated. Raw reads were filtered and paired-end reads were tagged for clustering into operational taxonomic units (OTUs). OTU representative sequences were taxonomically classified using Ribosomal Database Project (RDP) Classifier v.2.2 with a minimum confidence threshold of 0.6 and trained on the Green-genes database v201305 by QIIME 2 (Bolyen et al., 2019). Alpha and beta diversity were estimated by MOTHUR (v1.31.2) and QIIME 2, respectively (Schloss et al., 2009; Wade and Prosdocimi, 2020).
Different classification levels were plotted with R package v3.4.1 and R package “gg plots” (Shamsheer et al., 2022). Principal coordinate analysis (PCoA) of OTUs was conducted by R package “ade4” (Jombart, 2008). Wilcoxon or Kruskal–Wallis’ testing was used to ascertain significant species or functions by R (v3.4.1) software.
DNA extraction, 16S rRNA sequencing, and analyses were performed by BGI Co., LTD. Shenzhen, China.
Source Tracker is a Bayesian approach to allow the estimation of the proportion of contaminants in a given community that come from possible source environments (Knights et al., 2011). Oral microbiota was considered source environments and gut microbiota sink environments for calculation of the contribution rate of oral microbiota to gut microbiota. Source Tracker R script was downloaded from https://github.com/danknights/sourcetracker. NetShift was used to quantify oral and gut community changes between the AD and HC to identify the microbial taxa which serve as “driver-bacteria,” promoting the change from the healthy to the diseased state. The correlation network was established based on the Pearson coefficients between bacteria communities in AD and HC. Each genus was regarded as a point in the network and the genus with a significant correlation relationship was assigned an edge (Kuntal et al., 2019). The Netshift application was used to elucidate the drivers of HC to the AD network. NetShift algorithm could be downloaded from NetShift was downloaded from https://web.niapps.net/netshift.
Statistical analyses were performed with IBM SPSS 23.0, R-4.2.0, and Graphpad prism 8.0.2. Differences in numerical variables between AD and HC, continuous variables consistent with normal distribution and homogeneity of variance were assessed by a two-tailed Student’s t-test and results expressed as the mean ± standard deviation, other continuous variables assessed by nonparametric test and results expressed as the median (Q25, Q75). Discontinuous variables were analyzed by the chi-square test. The α-diversity index and β-diversity index were calculated and compared between HC and AD microbiota. Principal coordinate analysis (PCoA) was used to evaluate the species differences in the microbiota between different sample groups. Based on the Wilcoxon test, calculated the characteristics of composition in oral/gut microbiota between AD and HC from phylum to genus. Oral-gut overlap microbiota based on common OTUs number by VeenDiagram package in R. Comparison of clinical features between ectopic colonization (EC) patients and no ectopic colonization (NE) patients by Mann–Whitney U test. Correlational analyses were assessed by the Spearman test. The value of p < 0.05 was considered statistically significant.
No significant differences were present in age, occupation, educational status, marital status, annual income, BMI, or smoking status. No significant difference in FTND was found, but HAMD, HAMA, and PSQI scores were higher for AD than for HC (p < 0.001), and MoCA scores were lower for AD than for HC (p < 0.05). Demographic and clinical characteristics of the study population are summarized in Table 1.
The α-diversity of oral microbiota (Shannon index) and intestinal microbiota (Sob index, Chao1 index, ACE index, and Coverage index) was significantly decreased in AD, compared with HC in Figures 1A,B (all p < 0.05). Based on weighted Unifrac, the β-diversity of oral microbiota between AD and HC exhibited statistical difference (p < 0.01); and based on weighted Unifrac, the β-diversity of gut microbiota between AD and HC exhibited statistical difference (p < 0.001) in Figures 1C,D. Compared with HC, the characteristics of composition in the oral and gut microbiota of AD from phylum to genus were summarized based on the Wilcoxon test (all p < 0.05), in Supplementary Figures S1, S2.
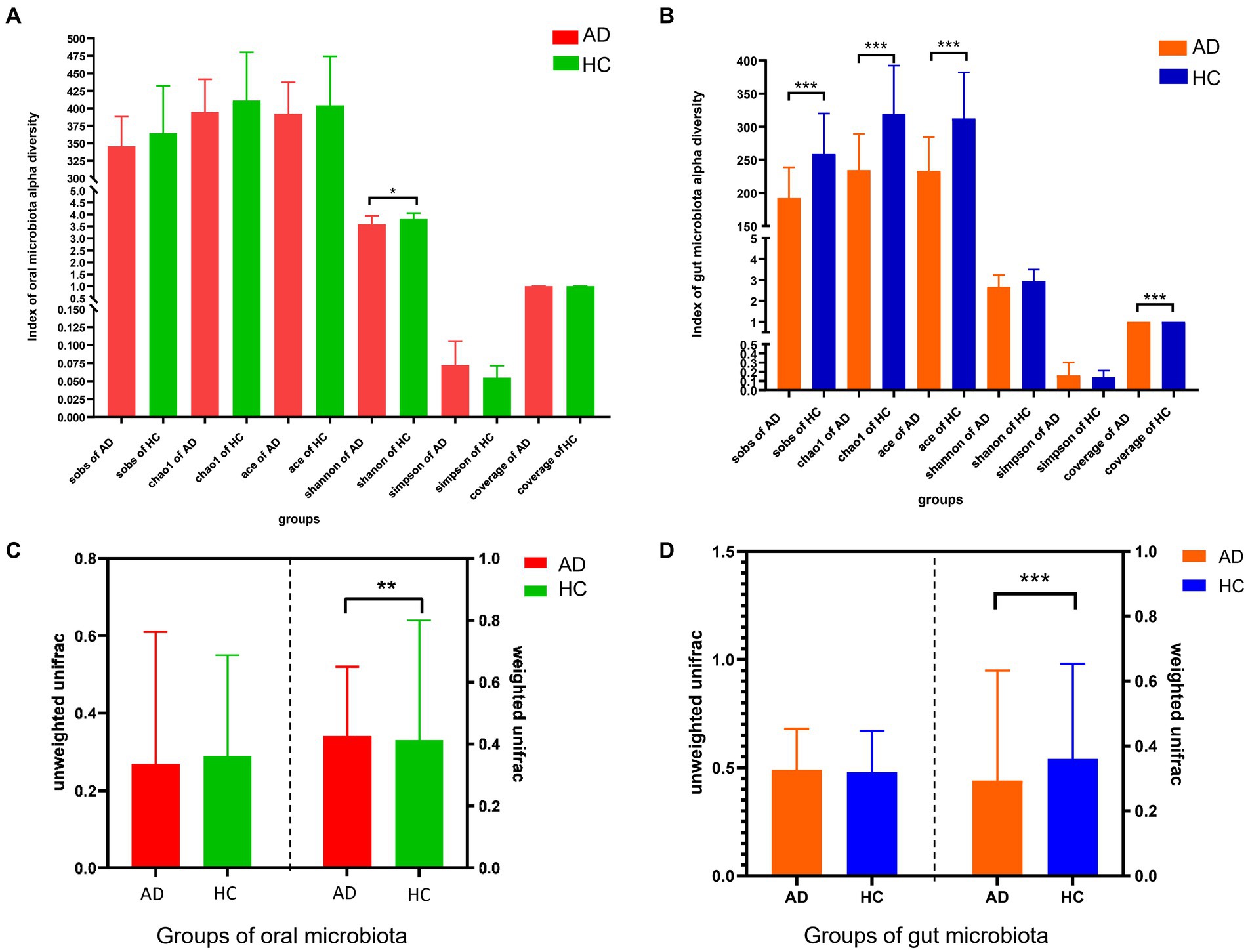
Figure 1. Differences in alpha and beta diversity between alcohol dependence and healthy controls. (A) α-diversity analysis showed that compared with the HC, the Shannon index of oral flora in the AD was lower, p < 0.05; no statistical difference was seen between AD and HC in the Sobs, Chao 1, ACE, Simpson, and Good coverage indices. (B) α-diversity analysis showed that compared with the HC, the intestinal flora observed species, Chao1, ACE and Good Coverage indices of the AD were lower, all p < 0.001; no statistical difference between AD and HC in the Shannon, Simpson indices. (C) Unweighted similarity analysis of β-diversity analysis showed that there was no statistical difference in β-diversity between the AD and HC, when sample sequence abundance was not considered; the results of weighted similarity analysis showed that there was a statistical difference in β-diversity between AD and HC, p < 0.01, when sample sequence abundance was considered. (D) Unweighted similarity analysis showed no statistical difference in β-diversity between AD and HC, when the sample sequence abundance was not considered; the results of the weighted similarity analysis showed a statistical difference in β-diversity between AD and HC, p < 0.001, when the sample sequence abundance was considered. All p-values are Bonferroni adjusted. * Indicates p < 0.05, ** indicates p < 0.01, *** indicates p < 0.001. AD indicates alcohol dependence, HC indicates healthy controls.
Based on unweighted Unifrac and weighted Unifrac to explore the relationship between oral and gut microbiota found that β-diversity between oral and gut microbiota in AD (p < 0.001, p < 0.001) and HC (p < 0.05, p < 0.001) exhibited significant difference in Figures 2A,B. Additionally, the β-diversity heatmap and principal coordinate analysis (PCoA) both indicate that the difference between oral and gut microbiota in AD was decreased compared to HC in Supplementary Figures S3A–C and Figures 2C,D. At the phylum level, significant differences between oral and gut microbiota were found in the top 10 species of HC (all p < 0.05), but four species had no significant differences between oral and gut microbiota in the top 10 species of AD in Figures 2E,F.
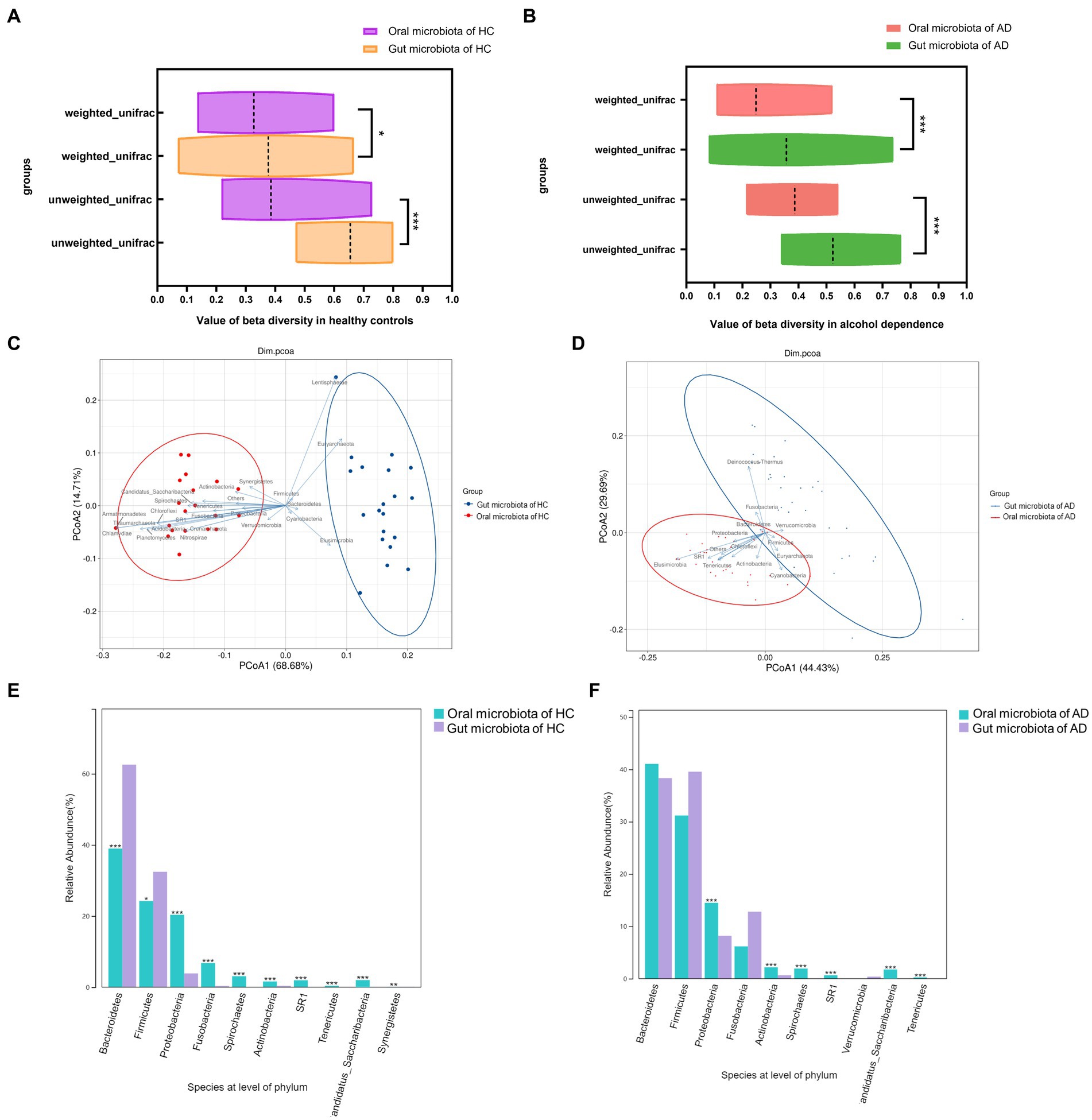
Figure 2. Relationships between oral and gut microbiota in alcohol dependence and healthy controls. (A) Difference in β-diversity between oral and gut microbiota in HC. Weight unifrac, p < 0.05; unweight unifrac, p < 0.001. (B) Difference in β-diversity between oral and gut microbiota in AD. Weight unifrac, p < 0.001; unweight unifrac, p < 0.001. (C) Distribution of oral and gut microbiota in HC analyzed by Principal Co-ordinates Analysis (PCoA). (D) Distribution of oral and gut microbiota in AD analyzed by Principal Co-ordinates Analysis (PCoA). (E) Significant differences between oral and gut in the top 10 species at the level of phylum in HC (all p < 0.05). (F) Bacteroidetes, Firmicutes, Fusobacteria, and Verrucomicrobia have no differences between oral and gut top 10 species at the level of phylum in AD. *Wilcoxon test indicated significant differences between different groups. All p-values are Bonferroni adjusted. * Indicates p < 0.05, ** indicates p < 0.01, *** indicates p < 0.001. AD indicates alcohol dependence, HC indicates healthy controls.
Analysis of species composition indicated that overlaps between oral and gut microbiota were found for 9 genera in AD, including the genus of Prevotella, Streptococcus, Veillonella, Neisseria, Fusobacterium, Haemophilus, Alloprevotella, Megasphaera, and Bacteroides. And the genus of Fusobacterium, Alloprevotella, and Veillonella also are the oral/gut microbiota overlap for HC. Therefore, there were 6 AD oral/gut overlap genera, Prevotella, Streptococcus, Neisseria, Haemophilus, Megasphaera, and Bacteroides, that were not found in the HC oral/gut overlap microbiota (Figures 3A,B).
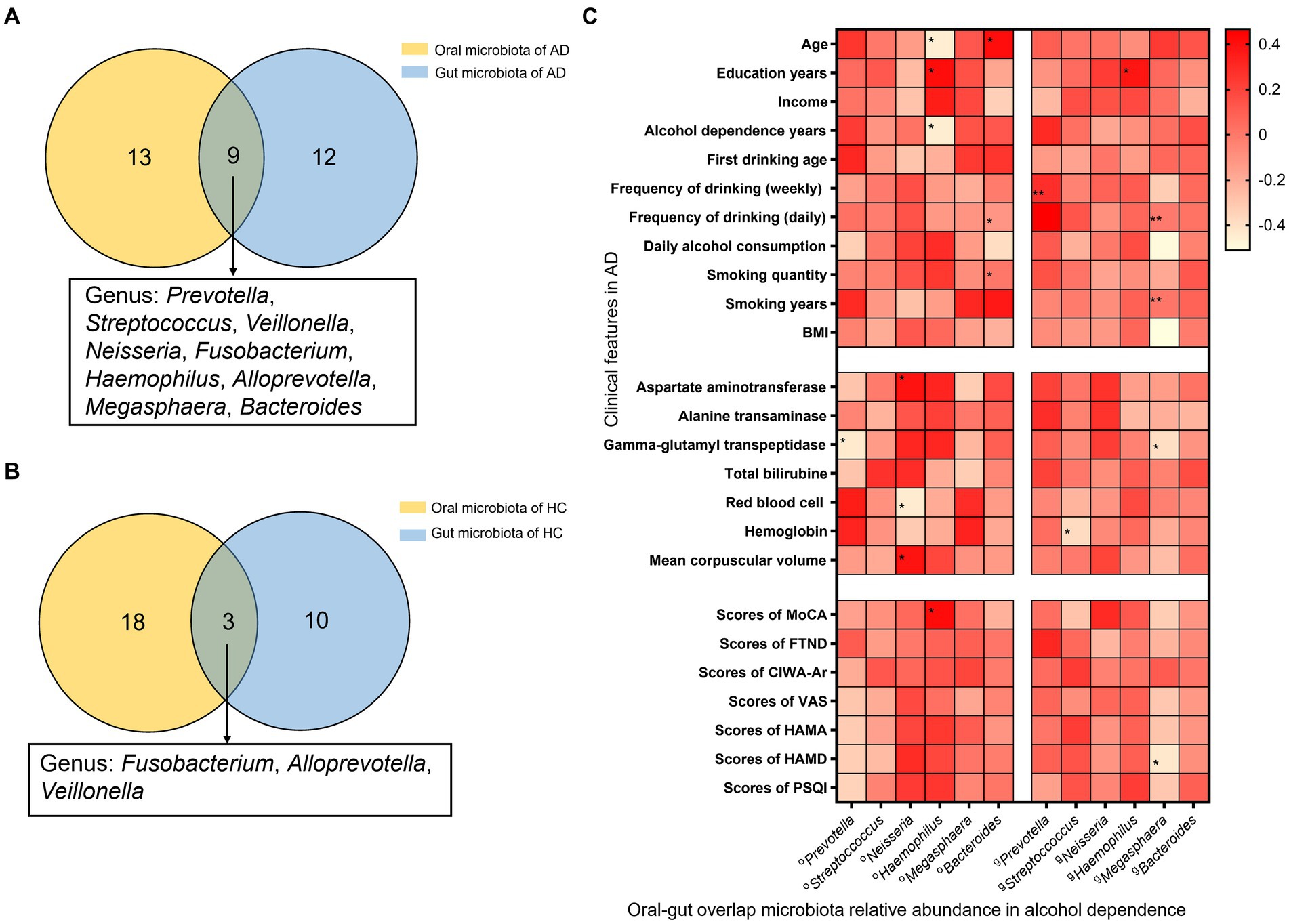
Figure 3. Oral and gut overlap microbiota and correlation with clinical features in AD. (A) There are 9 genera were oral-gut overlap bacteria, Prevotella, Streptococcus, Veillonella, Neisseria, Fusobacterium, Haemophilus, Alloprevotella, Megasphaera, and Bacteroides, in AD. (B) There are 3 genera were oral-gut overlap bacteria, Fusobacterium, Alloprevotella, and Veillonella, in HC. (C) Heat map correlation between oral/gut overlap bacteria and clinical features in AD. *Spearman test indicated the association between different groups. * Indicates p < 0.05, ** indicates p < 0.01. AD indicates alcohol dependence, HC indicates healthy controls.
The correlation analysis between relative abundance of oral-gut microbiota and clinical features found that, in the relative abundance of oral microbiota, Prevotella is negatively correlated with GGT and scores of PACS (r = −0.43, p = 0.02; r = −0.39, p = 0.03); Neisseria is positively correlated with AST, MCV, and TSH (r = 0.39, p = 0.03; r = 0.39, p = 0.03; r = 0.38, p = 0.06), and negatively correlated with RBC (r = −0.44, p = 0.01); Haemophilus is positively correlated with education years, scores of MoCA (r = 0.42, p = 0.02; r = 0.42, p = 0.02), and negatively correlated with age, alcohol dependence years (r = −0.45, p = 0.01; r = −0.45, p = 0.01); Bacteroides is positively correlated with age, smoking years (r = 0.41, p = 0.03; r = 0.37, p = 0.04), and negatively correlated with daily alcohol consumption (r = −0.38, p = 0.04). And in the relative abundance of gut microbiota, found that Prevotella is positively correlated with frequency of drinking (daily) (r = 0.49, p = 0.01); Streptococcus is negatively correlated with HB (r = −0.38, p = 0.04) and positively correlated with TT3 (r = 0.39, p = 0.04); Haemophilus is positively correlated with education years (r = 0.38, p = 0.04); Megasphaera is negatively correlated with daily alcohol consumption, BMI, GGT, scores of PACS, scores of HAMD (r = −0.50, p = 0.01; r = −0.51, p = 0.00; r = −0.39, p = 0.03; r = −0.43, p = 0.02) (Figure 3C).
Source Tracker analysis explored the ectopic colonization of oral microbiota in the gut, showing five AD, No. 3, 7, 9, 16, and 21, had a fraction of their gut microbiota which had originated from the oral cavity, but no positive results in HC. The specific oral bacteria involved in ectopic colonization, Abiotrophia, Actinomyces, Alloprevotella, Bacteroidales, Blautia, and Campylobacter…, are shown in Table 2 and Supplementary Figure S4. The contribution ratio of oral microbiota to intestinal microbiota composition in AD is 5.26%, and in HC is 0.00% based on Source Tracker. Analyze of microbiota composition between ectopic colonization and non-ectopic colonization patients, the ratio of Firmicutes to Bacteroidetes (F\B) is 1.23 for ectopic colonization and 1.06 for non-ectopic colonization compared with 0.61 for HC at the level of phylum in Supplementary Figures S5A–C. According to the Mann–Whitney U test, in contrast to AD patients without ectopic colonization, the patients with ectopic colonization showed the daily maximum standard drinks, red blood cell counts, hemoglobin content, and PACS scores decreasing (all p < 0.05) in Figures 4A–D. Three predominant driver bacteria, Prevotella, Neisseria, and Saccharibacteria in the level of the genus were found by Netshift analysis, and they were also belonging to ectopic colonization bacteria in Figure 4E and Table 2.
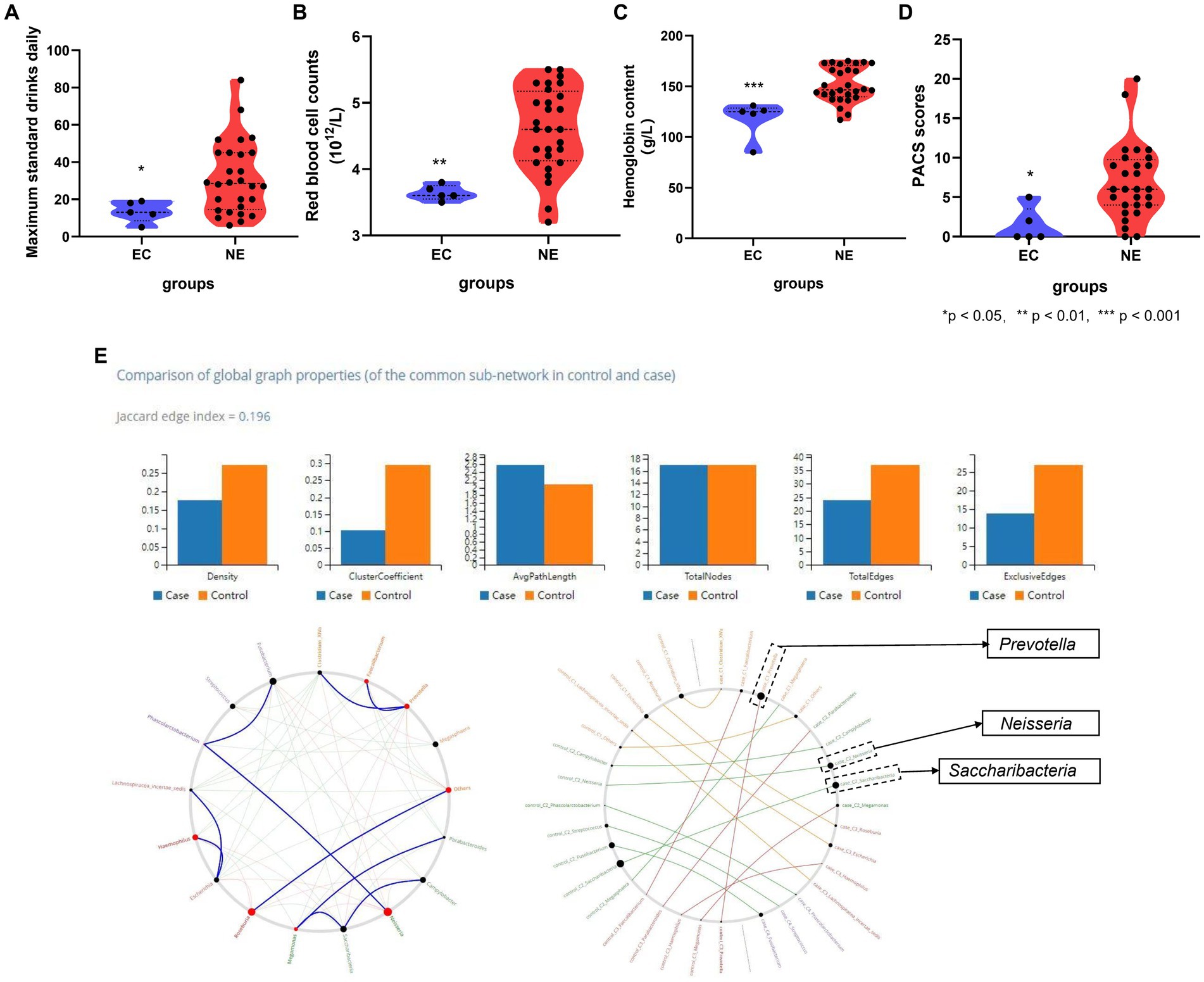
Figure 4. The association between clinical features and ectopic colonization bacteria in AD. (A) Ectopic colonization patients had lower maximum standard drinking daily (p < 0.05); (B) ectopic colonization patients had lower red blood cell counts (p < 0.01); (C) ectopic colonization patients had lower hemoglobin content (p < 0.001); (D) ectopic colonization patients had lower PACS scores (p < 0.05). (E) Genera that drive healthy states to alcohol dependence, Prevotella, Neisseria, and Saccharibacteria, are indicated by bigger black circles from NetShift analysis. *Mann–Whitney test indicated significant differences between different groups. *Indicates p < 0.05, ** indicates p < 0.01, *** indicates p < 0.001. Data are expressed as mean ± SEM (n = 21–33). EC indicates ectopic colonization patients, NE indicates no ectopic colonization patients, PACS indicates Pennsylvania alcohol craving scale, “Case” indicates a relative abundance of gut genera in alcohol dependence, “Control” indicates a relative abundance of gut genera in healthy controls.
Based on the Spearman correlation test, to explore the association between ectopic colonization bacteria in the relative abundance of gut microbiota and clinical features in AD. Alloprevotella is positively correlated with first drinking age and ALT (r = 0.41, p = 0.03; r = 0.36, p = 0.04), Escherichia is negatively correlated with AST (r = −0.38, p = 0.04), Fusobacterium is positively correlated with MCV (r = −0.39, p = 0.03). Lachnospiracea_incertae_sedis is negatively correlated with frequency of drinking (weekly), AST, ALT, GGT, and TSH (r = −0.53, p = 0.00; r = −0.54, p = 0.00; r = −0.55, p = 0.00; r = −0.44, p = 0.01; r = −0.45, p = 0.0). Megasphaera is negatively correlated with average standard drinks per day, scores of PACS, scores of HAMD, and BMI (r = −0.46, p = 0.01; r = −0.42, p = 0.02; r = −0.42, p = 0.02; r = −0.52, p = 0.00). Prevotella is positively correlated with frequency of drinking (daily) (r = 0.47, p = 0.00). Saccharibacteria is negatively correlated with TBiL (r = −0.39, p = 0.01). Streptococcus is negatively correlated with HB (r = −0.37, p = 0.04) in Table 3.
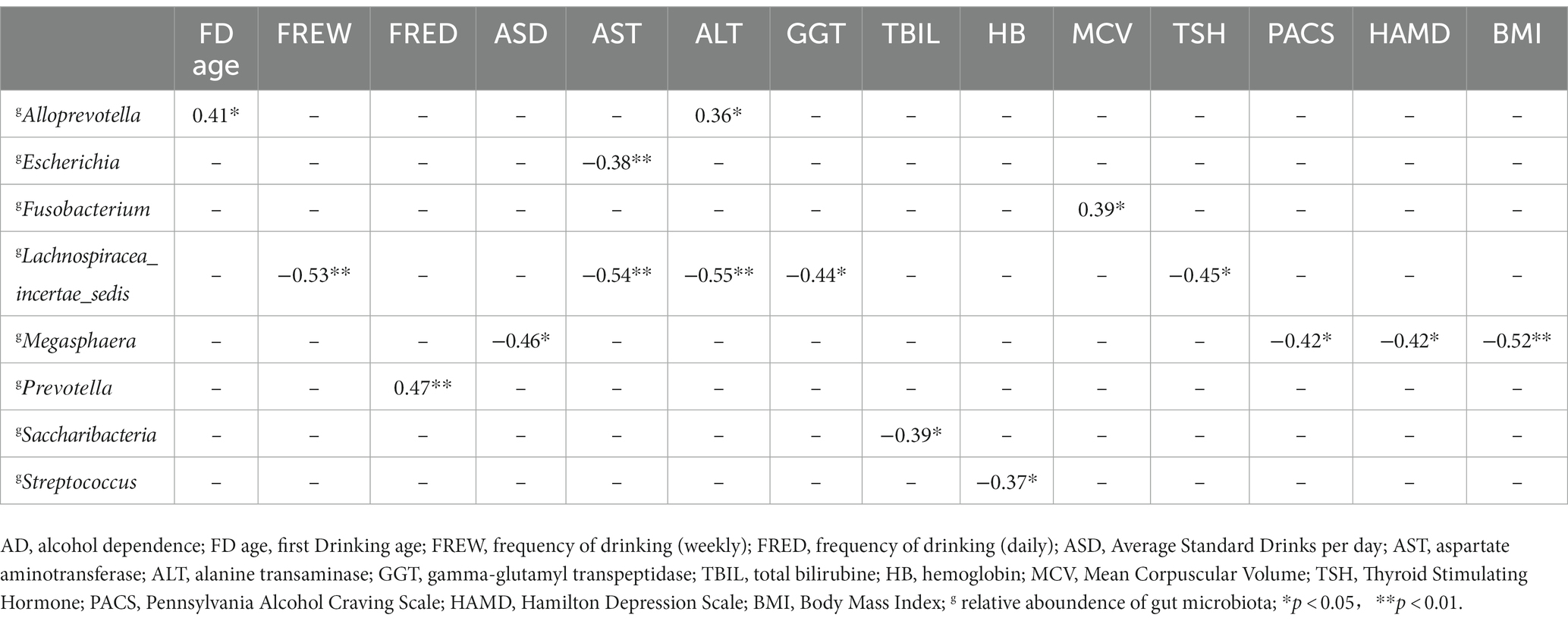
Table 3. The correlation analysis between ectopic colonization bacteria and clinical features in alcohol dependence.
The exploration of the relationship between oral and gut microbiota revealed that differences between oral and gut microbiota decreased and the oral/gut microbiota overlap increased in AD, compared with HC. Genera involved in ectopic colonization of oral microbiota to gut were shown connected with AD symptom severity based on Source Tracker and NetShift analysis. Compared with previous studies which only focused on single niche, the current study emphasized the oral-gut microbiota axis and the connection between oral and gut microbiota in AD (Liao et al., 2022; Zhao et al., 2023). To the best of our knowledge, this is the first study to explore the ectopic colonization in AD combined with methods of Source Tracker and NetShift, along with other methods, this will help to enrich the etiological hypothesis of the oral-gut microbiota axis in AD.
Heavy alcohol consumption is likely to cause disturbances of oral-gut microbiota. In current study, the α-diversity of oral and intestinal microbiota decreased in AD, this change may predispose patients to inflammatory responses and metabolic disorders (Karkman et al., 2017). At level the of phylum, SR1 decreased and Firmicutes and Proteobacteria increased in the oral microbiota of AD, and at the level of genus, Prevotella decreased, and Bacteroides and Fusobacterium increased in the gut microbiota of AD; such compositional change may be pro-inflammatory (Brandsma et al., 2019; Ballini et al., 2020). In summary, alcohol affects the ecological niches of both oral and gut, these could provide prerequisites for ectopic colonization of oral bacteria to the gut (Li et al., 2019).
In this study, the distances between oral and gut microbiota in AD are shortened based on PCoA, which indicated that oral and gut microbiota share a greater similarity in AD. This phenomenon also has been found in elderly people, which is believed an enhanced tendency for oral bacteria to invade the gut (Iwauchi et al., 2019). Then, the current study also found 6 oral/gut overlap bacteria, Prevotella, Streptococcus, Neisseria, Haemophilus, Megasphaera, and Bacteroides in AD. A previous study found 7 oral/gut overlap bacteria, Actinomyces, Bifidobacterium, Dialister, Granulicatella, Lactobacillus, Megasphaera, and Veillonella in alcohol use disorder (AUD) (Kobyliak et al., 2018; Ames et al., 2020; Maitre et al., 2020). And the difference in specific bacteria might be related to alcohol consumption, race, and dietary habits.
However, the question of what causes changes in the association between oral and gut microbiota and whether oral microbiota can ectopically colonize the gut in AD needs to be answered by the approaches of Source Tracker and NetShift. The contribution ratio of oral microbiota to the composition of intestinal microbiota in AD is 5.26%, and no ectopic colonization was found in HC. Among the current study, those showing ectopic colonization often suffered lower red blood cell counts and hemoglobin content, meanwhile, their daily maximum standard drinks and craving for alcohol were also lower than non-ectopic colonization. This reduction in craving seems to confirm the prevailing hypothesis of the work that ectopic bacteria from the oral cavity to the gut contribute to the pathological state, because low red blood cell counts and hemoglobin content represent the poor state of nutrition in AD, and impaired nutritional status often with lower levels of ghrelin and alcohol craving, and consumed high quantity of ultra-processed food had higher obsessive thoughts about alcohol (Addolorato et al., 2006; Amadieu et al., 2021). Thus, ectopic colonization in AD might be regarded as a sign of deterioration in diseases (Ley et al., 2008; Wade, 2013). Previous studies have indicated that oral microbiota is a reservoir of pathogenic bacteria (Lim et al., 2017). Colonization of the gut from oral bacteria may precipitate intestinal microbiota disturbance and barrier damage, resulting in low levels of systemic inflammation and distal spread (Bull-Otterson et al., 2013; Imai et al., 2021). In addition, HC had a proportion of the intestinal microbiota which derived from the oral microbiota of less than 1% according to the Chinese study, which similar to the results of current study (Guo et al., 2022).
The bacteria that drive state from health to AD, including Prevotella, Neisseria, and Saccharibacteria, were also belonging to ectopic colonization of oral bacteria based on NetShift analysis. Previous studies found that, Prevotella is an opportunistic pathogen that may increase branched-chain amino acid (BCAA) synthesis in the gut and lead to insulin resistance (Wu et al., 2011; Wang et al., 2021). Prevotella and Neisseria were also enriched in the gut of Crohn’s disease (CD) (Zhang et al., 2020). Bacteroides and Prevotella have both been shown to contribute to metabolic endotoxemia in a high-fat fed mouse model (Ding et al., 2020). Saccharibacteria may improve renal function through a symbiotic relationship with other host bacteria, which could decrease the concentration of serum creatinine (−0.011 [95% CI –0.019, −0.003], p = 0.007) and increase the estimated glomerular filtration rate (eGFR) (0.012 [95% CI 0.004, 0.020], p = 0.003) (Xu et al., 2020), and was found to reduce inflammation in a mouse model of periodontitis (Chipashvili et al., 2021).
Ectopic colonization bacteria correlated with the clinical feature in AD, such as first drinking age, frequency of drinking (weekly and daily), average standard drinks per day, BMI, AST, ALT, GGT, TBIL, HB, MCV, TSH, PACS scores and HAMD scores. Therefore, the presence of the ectopic genera may be an objective indicator of disease severity. Meanwhile, the current study also compared the species’ relative abundance of 6 oral/gut overlap bacteria with clinical characteristics, further combining the results in Figure 3C and Table 3 revealing that the ectopic colonization of the oral flora to the intestine could be one of the reasons for the increased number of oral-gut overlap bacteria in alcohol dependent, and oral ectopic colonized bacteria might play a role in the intestine and correlate with the severity of clinical symptoms. In terms of mechanism about ectopic colonization, previous research found that ectopic colonization bacteria from the oral cavity, such as Fusobacterium nucleatum, Porphyromonas gingivalis, and Klebsiella spp., aggravate gut dysbiosis and favor pro-inflammatory bacteria (Arimatsu et al., 2014), and activation of the NF-ĸB-PPAR-I FABP/Angptl4 pathway decreases lipid accumulation and induces intestinal apoptosis (Nakajima et al., 2015; Olsen and Yamazaki, 2019). Furthermore, P. gingivalis activates the MAL-PI3K pathway, and Klebsiella spp. resulted in potent Th1 cell differentiation in the gut (Maekawa et al., 2014; Atarashi et al., 2017; Scassellati et al., 2021). Meanwhiles, amounts of bacteria and their products invade the blood, activating B cells and inflammatory response, increasing fibrinogen production, and vascular endothelial injury in the liver (Wiest et al., 2014; Sansores-España et al., 2021). Thrombotic occlusion of capillaries and excessive interleukin-1β (IL-1β) and interleukin-6 (IL-6) secretion may follow (Leclercq et al., 2017; Imai et al., 2021). Low-grade inflammation in peripheral tissues and vagus nerve fibers (IL-1β) in the gut-brain axis may weaken the blood–brain barrier (BBB) (Maekawa et al., 2014; Acharya et al., 2017; Leclercq et al., 2017; Sansores-España et al., 2021). Inflammatory responses in the CNS may cause activated microglia to produce tumor necrosis factor (TNF), IL-1β, and IL-6 and neurotrophic factor (BDNF) from astroglia. Inflammation damages the limbic system and dorsal lateral prefrontal cortex (DLPFC) of the brain, exacerbating the cognitive impairment and affective disturbance of AD (Lee et al., 2005; Liu et al., 2016; Pascual et al., 2021) (Supplementary Figure S6). We could deduce that ectopic colonization of oral microbiota could be involved in intestinal and brain dysfunction in AD through the microbiota-gut-brain axis from the network above (Figure 5).
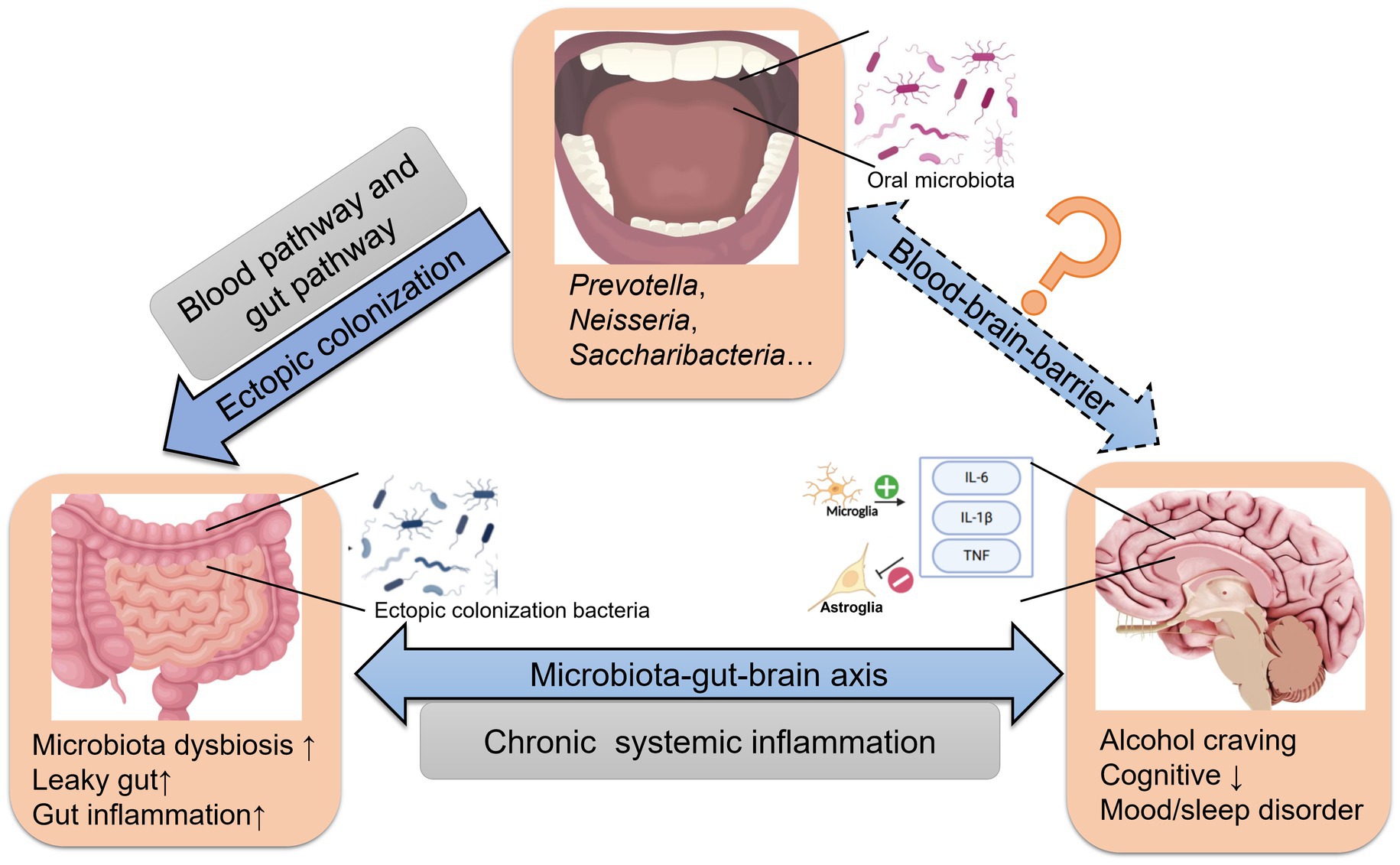
Figure 5. The effect of ectopically colonized bacteria on the microbiota-gut-brain axis in AD. Oral microbiota arrive at the gut via the blood and/or gut pathway and colonize the distal intestine. Then aggravate gastrointestinal microbiota dysbiosis, barrier injury and inflammation, ultimately affecting alcohol craving in AD.
We acknowledge some limitations to the current study. Ectopic colonization in AD was investigated using data analysis of Source Tracker, thus it is still not enough to conclude that there is ectopic colonization in the gut by specific oral bacteria, and experimental verification is necessary. This study only included male subjects, lack of representation of females as one of the limitations of the study.
The decreased divergence between oral and gut microbiota and increased oral-gut overlap bacteria were found in AD. Ectopic colonization bacteria were only found in AD based on Source Tracker. Connections between oral-gut microbiota and mechanisms of AD progression are highlighted and could be regard as the potential biomarker for oral-gut microbiota mechanisms in AD development.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA895096; https://www.ncbi.nlm.nih.gov/, PRINA905862.
The studies involving human participants were reviewed and approved by the Helsinki Declaration of 1975 and the ethical standards of the home institution Committee on Human Experimentation. Ethical approval was granted by the ethics committees of Peking University Sixth Hospital (2019, ethical review No. 6) and the Second People’s Hospital of Guizhou Province. The patients/participants provided their written informed consent to participate in this study.
LH, ZN, LL, and HS were responsible for the study concept and design. KZ contributed to the acquisition of human data. LH, YK, and ZY performed the gut and oral microbiota analysis. YK and XG assisted with data analysis and interpretation of findings. XL provided critical revision of the manuscript for important intellectual content. All authors critically reviewed the content and approved the final version for publication.
Our study got support from the National Natural Science Foundation of China (nos. 81971235, 81771429, and 81571297) and the National Clinical Research Center for Mental Disorders (Peking University Sixth Hospital) NCRC2020M09.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1203678/full#supplementary-material
AD, alcohol dependence; BMI, body mass index; CIWA-Ar, Revised Clinic Institute Alcohol Withdrawal Syndrome Assessment Scale; DSM-IV, 4th edition of the Diagnostic and Statistical Manual of Mental Disorders; F/B, The Ratio of Firmicutes to Bacteroidetes; FTND, Fagerstrom Test of Nicotine Dependence; HAMA, Hamilton Anxiety Scale; HAMD, Hamilton Depression scale; M.I.N.I, Mini-International Neuropsychiatric Interview; MoCA, Montreal cognitive assessment; OTUs, operational taxonomic units; PACS, Pennsylvania Alcohol Craving Scale; PCoA, Principal Co-ordinates Analysis; PSQI, Pittsburg Sleep Quality Index.
Acharya, C., Sahingur, S. E., and Bajaj, J. S. (2017). Microbiota, cirrhosis, and the emerging oral-gut-liver axis. JCI Insight 2:e94416. doi: 10.1172/jci.insight.94416
Adak, A., and Khan, M. R. (2019). An insight into gut microbiota and its functionalities. Cell. Mol. Life Sci. 76, 473–493. doi: 10.1007/s00018-018-2943-4
Addolorato, G., Capristo, E., Leggio, L., Ferrulli, A., Abenavoli, L., Malandrino, N., et al. (2006). Relationship between ghrelin levels, alcohol craving, and nutritional status in current alcoholic patients. Alcohol. Clin. Exp. Res. 30, 1933–1937. doi: 10.1111/j.1530-0277.2006.00238.x
Amadieu, C., Leclercq, S., Coste, V., Thijssen, V., Neyrinck, A. M., Bindels, L. B., et al. (2021). Dietary fiber deficiency as a component of malnutrition associated with psychological alterations in alcohol use disorder. Clin Nutr. 40, 2673–2682. doi: 10.1016/j.clnu.2021.03.029
Ames, N. J., Barb, J. J., Schuebel, K., Mudra, S., Meeks, B. K., Tuason, R. T. S., et al. (2020). Longitudinal gut microbiome changes in alcohol use disorder are influenced by abstinence and drinking quantity. Gut Microbes 11, 1608–1631. doi: 10.1080/19490976.2020.1758010
Arimatsu, K., Yamada, H., Miyazawa, H., Minagawa, T., Nakajima, M., Ryder, M. I., et al. (2014). Oral pathobiont induces systemic inflammation and metabolic changes associated with alteration of gut microbiota. Sci. Rep. 4:4828. doi: 10.1038/srep04828
Atarashi, K., Suda, W., Luo, C., Kawaguchi, T., Motoo, I., Narushima, S., et al. (2017). Ectopic colonization of oral bacteria in the intestine drives T(H)1 cell induction and inflammation. Science (New York, N.Y.) 358, 359–365. doi: 10.1126/science.aan4526
Ballini, A., Dipalma, G., Isacco, C. G., Boccellino, M., Di Domenico, M., Santacroce, L., et al. (2020). Oral microbiota and immune system crosstalk: a translational research. Biology 9:131. doi: 10.3390/biology9060131
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Brandsma, E., Kloosterhuis, N. J., Koster, M., Dekker, D. C., Gijbels, M. J. J., van der Velden, S., et al. (2019). A proinflammatory gut microbiota increases systemic inflammation and accelerates atherosclerosis. Circ. Res. 124, 94–100. doi: 10.1161/CIRCRESAHA.118.313234
Bull-Otterson, L., Feng, W., Kirpich, I., Wang, Y., Qin, X., Liu, Y., et al. (2013). Metagenomic analyses of alcohol induced pathogenic alterations in the intestinal microbiome and the effect of Lactobacillus rhamnosus GG treatment. PLoS One 8:e53028. doi: 10.1371/journal.pone.0053028
Chipashvili, O., Utter, D. R., Bedree, J. K., Ma, Y., Schulte, F., Mascarin, G., et al. (2021). Episymbiotic Saccharibacteria suppresses gingival inflammation and bone loss in mice through host bacterial modulation. Cell Host Microbe 29, 1649–1662.e7. doi: 10.1016/j.chom.2021.09.009
Ding, N., Zhang, X., Zhang, X. D., Jing, J., Liu, S. S., Mu, Y. P., et al. (2020). Impairment of spermatogenesis and sperm motility by the high-fat diet-induced dysbiosis of gut microbes. Gut 69, 1608–1619. doi: 10.1136/gutjnl-2019-319127
Dubinkina, V. B., Tyakht, A. V., Odintsova, V. Y., Yarygin, K. S., Kovarsky, B. A., Pavlenko, A. V., et al. (2017). Links of gut microbiota composition with alcohol dependence syndrome and alcoholic liver disease. Microbiome 5:141. doi: 10.1186/s40168-017-0359-2
Fan, X., Peters, B. A., Jacobs, E. J., Gapstur, S. M., Purdue, M. P., Freedman, N. D., et al. (2018). Drinking alcohol is associated with variation in the human oral microbiome in a large study of American adults. Microbiome 6:59. doi: 10.1186/s40168-018-0448-x
Forsyth, C. B., Farhadi, A., Jakate, S. M., Tang, Y., Shaikh, M., and Keshavarzian, A. (2009). Lactobacillus GG treatment ameliorates alcohol-induced intestinal oxidative stress, gut leakiness, and liver injury in a rat model of alcoholic steatohepatitis. Alcohol (Fayetteville, NY) 43, 163–172. doi: 10.1016/j.alcohol.2008.12.009
Gevers, D., Kugathasan, S., Denson, L. A., Vázquez-Baeza, Y., Van Treuren, W., Ren, B., et al. (2014). The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe 15, 382–392. doi: 10.1016/j.chom.2014.02.005
Guo, X. J., Jiang, T., Ma, X. X., Hu, X. J., Huang, J. B., Cui, L. T., et al. (2022). Relationships between diurnal changes of tongue coating microbiota and intestinal microbiota. Front. Cell. Infect. Microbiol. 12:813790. doi: 10.3389/fcimb.2022.813790
Hehemann, J. H., Correc, G., Barbeyron, T., Helbert, W., Czjzek, M., and Michel, G. (2010). Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature 464, 908–912. doi: 10.1038/nature08937
Huang, Y., Wang, Y., Wang, H., Liu, Z., Yu, X., Yan, J., et al. (2019). Prevalence of mental disorders in China: a cross-sectional epidemiological study. Lancet Psychiatry 6, 211–224. doi: 10.1016/S2215-0366(18)30511-X
Humphrey, S. P., and Williamson, R. T. (2001). A review of saliva: normal composition, flow, and function. J. Prosthet. Dent. 85, 162–169. doi: 10.1067/mpr.2001.113778
Imai, J., Kitamoto, S., and Kamada, N. (2021). The pathogenic oral-gut-liver axis: new understandings and clinical implications. Expert. Rev. Clin. Immunol. 17, 727–736. doi: 10.1080/1744666X.2021.1935877
Integrative HMP Research Network Consortium (2019). Integrative human microbiome project. Nature 569, 641–648. doi: 10.1038/s41586-019-1238-8
Iwauchi, M., Horigome, A., Ishikawa, K., Mikuni, A., Nakano, M., Xiao, J. Z., et al. (2019). Relationship between oral and gut microbiota in elderly people. Immun. Inflam. Disease 7, 229–236. doi: 10.1002/iid3.266
Jombart, T. (2008). Adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics (Oxford, England) 24, 1403–1405. doi: 10.1093/bioinformatics/btn129
Karkman, A., Lehtimäki, J., and Ruokolainen, L. (2017). The ecology of human microbiota: dynamics and diversity in health and disease. Ann. N. Y. Acad. Sci. 1399, 78–92. doi: 10.1111/nyas.13326
Knights, D., Kuczynski, J., Charlson, E. S., Zaneveld, J., Mozer, M. C., Collman, R. G., et al. (2011). Bayesian community-wide culture-independent microbial source tracking. Nat. Methods 8, 761–763. doi: 10.1038/nmeth.1650
Kobyliak, N., Abenavoli, L., Mykhalchyshyn, G., Kononenko, L., Boccuto, L., Kyriienko, D., et al. (2018). A multi-strain probiotic reduces the fatty liver index, cytokines and aminotransferase levels in NAFLD patients: evidence from a randomized clinical trial. J. Gastrointestinal Liver Diseases 27, 41–49. doi: 10.15403/jgld.2014.1121.271.kby
Kuntal, B. K., Chandrakar, P., Sadhu, S., and Mande, S. S. (2019). 'NetShift': a methodology for understanding 'driver microbes' from healthy and disease microbiome datasets. ISME J. 13, 442–454. doi: 10.1038/s41396-018-0291-x
Leclercq, S., De Saeger, C., Delzenne, N., de Timary, P., and Stärkel, P. (2014). Role of inflammatory pathways, blood mononuclear cells, and gut-derived bacterial products in alcohol dependence. Biol. Psychiatry 76, 725–733. doi: 10.1016/j.biopsych.2014.02.003
Leclercq, S., de Timary, P., Delzenne, N. M., and Stärkel, P. (2017). The link between inflammation, bugs, the intestine and the brain in alcohol dependence. Transl. Psychiatry 7:e1048. doi: 10.1038/tp.2017.15
Lee, J., Fukumoto, H., Orne, J., Klucken, J., Raju, S., Vanderburg, C. R., et al. (2005). Decreased levels of BDNF protein in Alzheimer temporal cortex are independent of BDNF polymorphisms. Exp. Neurol. 194, 91–96. doi: 10.1016/j.expneurol.2005.01.026
Ley, R. E., Hamady, M., Lozupone, C., Turnbaugh, P. J., Ramey, R. R., Bircher, J. S., et al. (2008). Evolution of mammals and their gut microbes. Science (New York, N.Y.) 320, 1647–1651. doi: 10.1126/science.1155725
Li, B., Ge, Y., Cheng, L., Zeng, B., Yu, J., Peng, X., et al. (2019). Oral bacteria colonize and compete with gut microbiota in gnotobiotic mice. Int. J. Oral Sci. 11:10. doi: 10.1038/s41368-018-0043-9
Liao, Y., Tong, X. T., Jia, Y. J., Liu, Q. Y., Wu, Y. X., Xue, W. Q., et al. (2022). The effects of alcohol drinking on oral microbiota in the Chinese population. Int. J. Environ. Res. Public Health 19:5729. doi: 10.3390/ijerph19095729
Lim, Y., Totsika, M., Morrison, M., and Punyadeera, C. (2017). Oral microbiome: a new biomarker reservoir for oral and oropharyngeal cancers. Theranostics 7, 4313–4321. doi: 10.7150/thno.21804
Liu, P., Liu, Y., Ye, W., Ma, J., and Gao, D. (2016). Flower-like N-doped MoS2 for photocatalytic degradation of RhB by visible light irradiation. Nanotechnology 27:225403. doi: 10.1088/0957-4484/27/22/225403
Maekawa, T., Krauss, J. L., Abe, T., Jotwani, R., Triantafilou, M., Triantafilou, K., et al. (2014). Porphyromonas gingivalis manipulates complement and TLR signaling to uncouple bacterial clearance from inflammation and promote dysbiosis. Cell Host Microbe 15, 768–778. doi: 10.1016/j.chom.2014.05.012
Maitre, Y., Micheneau, P., Delpierre, A., Mahalli, R., Guerin, M., Amador, G., et al. (2020). Did the brain and oral microbiota talk to each other? Review of the literature. J. Clin. Med. 9:3876. doi: 10.3390/jcm9123876
Nakajima, M., Arimatsu, K., Kato, T., Matsuda, Y., Minagawa, T., Takahashi, N., et al. (2015). Oral Administration of P. gingivalis induces dysbiosis of gut microbiota and impaired barrier function leading to dissemination of Enterobacteria to the liver. PLoS One 10:e0134234. doi: 10.1371/journal.pone.0134234
Olsen, I., and Yamazaki, K. (2019). Can oral bacteria affect the microbiome of the gut? J. Oral Microbiol. 11:1586422. doi: 10.1080/20002297.2019.1586422
Panza, F., Lozupone, M., Solfrizzi, V., Watling, M., and Imbimbo, B. P. (2019). Time to test antibacterial therapy in Alzheimer's disease. Brain 142, 2905–2929. doi: 10.1093/brain/awz244
Park, S. Y., Hwang, B. O., Lim, M., Ok, S. H., Lee, S. K., Chun, K. S., et al. (2021). Oral-gut microbiome Axis in gastrointestinal disease and Cancer. Cancers 13:2124. doi: 10.3390/cancers13092124
Pascual, M., Calvo-Rodriguez, M., Núñez, L., Villalobos, C., Ureña, J., and Guerri, C. (2021). Toll-like receptors in neuroinflammation, neurodegeneration, and alcohol-induced brain damage. IUBMB Life 73, 900–915. doi: 10.1002/iub.2510
Pittayanon, R., Lau, J. T., Yuan, Y., Leontiadis, G. I., Tse, F., Surette, M., et al. (2019). Gut microbiota in patients with irritable bowel syndrome-a systematic review. Gastroenterology 157, 97–108. doi: 10.1053/j.gastro.2019.03.049
Rashidi, A., Ebadi, M., Weisdorf, D. J., Costalonga, M., and Staley, C. (2021). No evidence for colonization of oral bacteria in the distal gut in healthy adults. Proc. Natl. Acad. Sci. U. S. A. 118:e2114152118. doi: 10.1073/pnas.2114152118
Rosier, B. T., Buetas, E., Moya-Gonzalvez, E. M., Artacho, A., and Mira, A. (2020). Nitrate as a potential prebiotic for the oral microbiome. Sci. Rep. 10:12895. doi: 10.1038/s41598-020-69931-x
Sansores-España, L. D., Melgar-Rodríguez, S., Olivares-Sagredo, K., Cafferata, E. A., Martínez-Aguilar, V. M., Vernal, R., et al. (2021). Oral-gut-brain axis in experimental models of periodontitis: associating gut dysbiosis with neurodegenerative diseases. Front. Aging 2:781582. doi: 10.3389/fragi.2021.781582
Scassellati, C., Marizzoni, M., Cattane, N., Lopizzo, N., Mombelli, E., Riva, M. A., et al. (2021). The complex molecular picture of gut and Oral microbiota-brain-depression system: what we know and what we need to know. Front. Psych. 12:722335. doi: 10.3389/fpsyt.2021.722335
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing Mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Shamsheer, B., Riaz, N., Yousaf, Z., Hyder, S., Aftab, A., Iqbal, R., et al. (2022). Genetic diversity analysis for wild and cultivated accessions of Cymbopogon citratus (D.C.) Stapf using phytochemical and molecular markers. PeerJ 10:e13505. doi: 10.7717/peerj.13505
Strandwitz, P. (2018). Neurotransmitter modulation by the gut microbiota. Brain Res. 1693, 128–133. doi: 10.1016/j.brainres.2018.03.015
Thomas, A. M., Manghi, P., Asnicar, F., Pasolli, E., Armanini, F., Zolfo, M., et al. (2019). Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat. Med. 25, 667–678. doi: 10.1038/s41591-019-0405-7
Wade, W. G. (2013). The oral microbiome in health and disease. Pharmacol. Res. 69, 137–143. doi: 10.1016/j.phrs.2012.11.006
Wade, W. G., and Prosdocimi, E. M. (2020). Profiling of oral bacterial communities. J. Dent. Res. 99, 621–629. doi: 10.1177/0022034520914594
Wang, D. D., Nguyen, L. H., Li, Y., Yan, Y., Ma, W., Rinott, E., et al. (2021). The gut microbiome modulates the protective association between a Mediterranean diet and cardiometabolic disease risk. Nat. Med. 27, 333–343. doi: 10.1038/s41591-020-01223-3
Wiest, R., Lawson, M., and Geuking, M. (2014). Pathological bacterial translocation in liver cirrhosis. J. Hepatol. 60, 197–209. doi: 10.1016/j.jhep.2013.07.044
World Health Organization. Global status report on alcohol and health (2018) [EB/OL]. Available at: https://www.who.int/substanceabuse/publications/globalalcoholreport/en/
Wu, G. D., Chen, J., Hoffmann, C., Bittinger, K., Chen, Y. Y., Keilbaugh, S. A., et al. (2011). Linking long-term dietary patterns with gut microbial enterotypes. Science (New York, N.Y.) 334, 105–108. doi: 10.1126/science.1208344
Xu, F., Fu, Y., Sun, T. Y., Jiang, Z., Miao, Z., Shuai, M., et al. (2020). The interplay between host genetics and the gut microbiome reveals common and distinct microbiome features for complex human diseases. Microbiome 8:145. doi: 10.1186/s40168-020-00923-9
Zhang, T., Kayani, M. U. R., Hong, L., Zhang, C., Zhong, J., Wang, Z., et al. (2020). Dynamics of the salivary microbiome during different phases of Crohn's disease. Front. Cell. Infect. Microbiol. 10:544704. doi: 10.3389/fcimb.2020.544704
Zhao, W., Hu, Y., Li, C., Li, N., Zhu, S., Tan, X., et al. (2020). Transplantation of fecal microbiota from patients with alcoholism induces anxiety/depression behaviors and decreases brain mGluR1/PKC ε levels in mouse. BioFactors (Oxford, England) 46, 38–54. doi: 10.1002/biof.1567
Keywords: alcohol dependence, oral microbiota, gut microbiota, ectopic colonization, Source Tracker
Citation: Hu L, Ni Z, Zhao K, Li X, Gao X, Kang Y, Yu Z, Qin Y, Zhao J, Peng W, Lu L and Sun H (2023) The association between oral and gut microbiota in male patients with alcohol dependence. Front. Microbiol. 14:1203678. doi: 10.3389/fmicb.2023.1203678
Received: 12 April 2023; Accepted: 17 July 2023;
Published: 28 July 2023.
Edited by:
Elena Ilina, Federal Research and Clinical Center of Physical-Chemical Medicine, RussiaReviewed by:
Naile Dame-Teixeira, University of Brasília, BrazilCopyright © 2023 Hu, Ni, Zhao, Li, Gao, Kang, Yu, Qin, Zhao, Peng, Lu and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongqiang Sun, c3VuaHFAYmptdS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.