- 1Guizhou Institute of Technology, Guiyang, China
- 2Key Laboratory of Microbial Resources Collection and Preservation, Ministry of Agriculture and Rural Affairs, Beijing, China
Ethanol tolerance is crucial for the oenological yeasts. Rosa roxburghii Tratt, a Rosaceae plant native to China, is rich in nutritional and medicinal ingredients. In this study, ethanol-tolerant non-Saccharomyces yeasts were screened, and their oenological properties were further evaluated. Three ethanol-tolerant yeast strains (designated as C6, F112, and F15), which could tolerate 12% (v/v) ethanol treatment, were isolated from R. roxburghii, and identified as Candida tropicalis, Pichia guilliermondii, and Wickerhamomyces anomalus, respectively. The winemaking condition tolerances of these ethanol-tolerant yeast strains were similar to those of Saccharomyces cerevisiae X16. However, their growth, sugar metabolic performance and sulphureted hydrogen activities, were different. The β-glucosidase production ability of strain W. anomalus F15 was lower than that of S. cerevisiae X16, and strains of C. tropicalis C6 and P. guilliermondii F112 were similar to S. cerevisiae X16. Electronic sensory properties of the R. roxburghii wines fermented using ethanol-tolerant yeasts together with S. cerevisiae showed no significant differences. However, the mixed inoculation of the ethanol-tolerant yeast strains with S. cerevisiae could regulate the volatile aroma characteristics of the fermented R. roxburghii wine, enriching and enhancing the aroma flavor. Therefore, the selected ethanol-tolerant yeasts have the potential for application in the production of unique R. roxburghii wine.
Introduction
The flavor characteristics and quality of fruit wine are determined by various factors, including the type of fruit, the brewing process, and the metabolic activity of the selected yeast (Wei et al., 2019). Yeast can be classified into two categories based on their fermentation characteristics and physiological properties: Saccharomyces cerevisiae and non-Saccharomyces yeasts (Jolly et al., 2014). S. cerevisiae is preferred for its high fermentation activity and strong ethanol tolerance, making it a popular choice for fruit wine production, and it is readily available for purchase by producers (Parapouli et al., 2020). However, the commercial varieties of wine yeast are limited, leading to high similarity in the flavor characteristics of fermented fruit wines and a lack of complexity in taste and flavor. As a result, product homogenization is common, which does not meet the diverse needs of consumers for product diversity. Non-Saccharomyces yeast refers to a diverse group of yeast species that also play a crucial role in winemaking. This group includes Hanseniaspora uvarum (Pietrafesa et al., 2020), Wickerhamomyces anomalus (Padilla et al., 2018), Candida tropicalis (Egue et al., 2018), etc.
Research has shown that non-Saccharomyces yeast can metabolize a greater variety of compounds during fruit wine fermentation, resulting in more complex and aromatic wine characteristics that enhance the overall flavor quality (Morata et al., 2019). However, non-Saccharomyces yeast is typically more sensitive to ethanol, which accumulates during the fermentation process and can inhibit its growth and induce cell death, ultimately reducing fermentation efficiency (Contreras et al., 2014). Therefore, the screening of non-Saccharomyces yeast strains with higher ethanol tolerance is of great practical significance for the production of distinctive fruit wines.
Rosa roxburghii Tratt, a perennial plant belonging to the Rosaceae family and the Rosa genus, is widely distributed in southwestern China, such as Guizhou, Sichuan, and Yunnan (Liu X. et al., 2020). The fruit of R. roxburghii is rich in nutrients, such as vitamin C, polysaccharides, and carotenoids (Liu et al., 2021a). Moreover, it contains abundant bioactive substances, such as flavonoids, superoxide dismutase (SOD), and organic acids, which give it good medicinal value (Wang et al., 2021). However, due to its high content of phenolic and acidic compounds, the fresh fruit tastes sour and astringent in taste, making it unsuitable for consumption. Therefore, fermenting the fruit into R. roxburghii fruit wine is more appropriate (Liu et al., 2021b). Currently, the yeast strains used in R. roxburghii fruit wine production mostly come from the active dry yeast used in grape wine production rather than from the indigenous yeast strains of R. roxburghii. This leads to poor adaptability of the strains and serious homogenization of the resulting R. roxburghii wine. Therefore, screening and isolating excellent indigenous yeast strains of R. roxburghii with perfect brewing characteristics, especially non-Saccharomyces yeasts, will promote the healthy development of R. roxburghii fruit wine.
In our preliminary research, we used high-throughput sequencing technology to identify the diversity and population changes of non-Saccharomyces yeasts during the spontaneous fermentation process of R. roxburghii fruit (Liu X. Z. et al., 2020). Additionally, we isolated 80 cultivable non-Saccharomyces yeasts from the spontaneous fermentation broth of R. roxburghii fruit using culture-dependent approach (Liu X. Z. et al., 2020). In this study, ethanol-tolerant strains were screened from our previously isolated culturable non-Saccharomyces yeasts, and then species of these ethanol-tolerant yeasts were identified based on morphology and molecular approaches. In addition, we also analyzed brewing characteristics of these ethanol-tolerant yeasts. Moreover, aroma and quality characteristics of R. roxburghii fruit wines were further evaluated by co-inoculation of these non-Saccharomyces yeasts together with S. cerevisiae as fermentation starter. The results obtained from the present study were helpful to explore potential high-quality brewing strains for the production of characteristic R. roxburghii fruit wine.
Materials and methods
Yeast strains
The reference strain used in this study was the commercial S. cerevisiae X16 obtained from Laffort Company (France). A total of 80 strains of non-Saccharomyces yeasts, isolated from spontaneous fermentation of R. roxburghii were screened for ethanol-tolerant strains. All yeasts cells were cultured on yeast extract peptone dextrose (YEPD) solid medium (1% yeast extract, 2% peptone, 2% glucose, and 2% agar) containing 100 mg/L of Chloramphenicol at 28°C for 72 h and then stored at 4°C for later use.
Screening and of identification of ethanol-tolerant non-Saccharomyces yeast strains
Ethanol-tolerant strains were screened by culturing them in YEPD broth (1% yeast extract, 2% peptone, and 2% glucose) containing 12% (v/v) ethanol with the initial concentration of 108 cfu/ml, and the yeast cells were cultured at 28°C with shaking at 180 rpm for 36 h. The optical density (OD) values were measured at a wavelength of 600 nm using a spectrophotometer (Hitachi, Tokyo, Japan).
Yeast strains were identified using both morphological and molecular methods. Firstly, cells were scraped onto Wallerstein Laboratory nutrient agar and cultured for 72 h. The characteristics of the colony and cellular morphology were examined and photographed with a microscope (Olympus, Tokyo, Japan). Next, genomic DNA was extracted from three ethanol-tolerant strains (C6, F112, and F15) using a DNA extraction kit (B518257; Sangon Biotech, China) following the manufacturer’s instructions. The D1/D2 domain within the 26S rDNA was amplified using the polymerase chain reaction (PCR). The yeast species were then determined by comparing the 26S rDNA D1/D2 domain sequences in the GenBank database.
Growth curve detection and sugar metabolism analysis of ethanol-tolerant non-Saccharomyces yeast strains
The C6, F112, and F15 strains were inoculated into YEPD broth at a concentration of 108 cfu/ml and cultured under agitation at 180 rpm and 28°C for 48 h. The OD of the cultures was measured at the wavelength of 600 nm every 4 h, and a growth curve was plotted based on the time and OD600 nm values.
Strains C6, F112, and F15 were inoculated with a concentration of 108 cfu/ml into a 0.6% yeast powder solution containing 2% final concentration of glucose, sucrose, maltose, lactose, and galactose, respectively. The yeast powder solution was placed in test tubes containing Durham tubes and incubated at 28°C for 48 h. The formation of gas bubbles in the Durham tubes was observed. A positive reaction was recorded as “+” if bubbles formed. Otherwise, a negative reaction was recorded as “−.”
Analysis of winemaking tolerances of ethanol-tolerant non-Saccharomyces yeast strains
Strains of C6, F112, and F15 were inoculated in YEPD broth at a concentration of 108 cfu/ml with (1) glucose concentrations of 100, 150, 200, 250, or 300 g/L; (2) citric acid mass fractions of 1, 1.5, 2, 2.5, or 3% (w%); and (3) a sulfur dioxide contents of 50, 100, 150, 200, or 300 mg/L. All groups were cultured at 28°C and 180 rpm for 36 h with three replicates, and then OD600 nm values were measured.
Production capacity of hydrogen sulfide and β-glucosidase activity in ethanol-tolerant non-Saccharomyces yeast strains
The hydrogen sulfide (H2S) production activities of C6, F112, and F15 were investigated using BiGGY agar by comparing the depth of colony color (Caridi et al., 2022).
The ability of the strains to produce β-glucosidase was analyzed using the p-nitrophenyl-β-D-glucopyranoside (p-NPG) method. Strains C6, F112, and F15 were inoculated into YEPD medium and shaken at 180 rpm at 28°C for 72 h. The supernatant was obtained after centrifugation at 3,000 g for 10 min and used for the determination of enzyme activity. Enzyme activity units (U) were defined as the amount of enzyme required to produce 1 μmol of p-nitrophenol (p-NP) by hydrolyzing 1 μmol of p-NPG under conditions of pH 5.0 and 50°C for 1 min.
Laboratory-scale fermentation of R. roxburghii fruit wine
Fresh, mature, and non-rotten R. roxburghii (Supplementary Figure 1) was juiced with juice extractor (Midea, WJE2802D, China) and then treated with 100 mg/L of potassium metabisulfite and 20 mg/L of pectinase at room temperature for 12 h. The juice was then adjusted to 24°Brix with crystalline sucrose and divided into four groups, with each group replicated in triplicate in 2 L sterile triangular flasks. For the C6 + S. cerevisiae X16 group, F112 + S. cerevisiae X16 group, and F15 + S. cerevisiae X16 group, each group was inoculated with 108 cfu/ml of the C6, F112, or F15 strain and 107 cfu/ml of S. cerevisiae X16, with a control group that was only inoculated with 107 cfu/ml of S. cerevisiae X16. The flasks were left to ferment statically at 26°C until fermentation was completed. After fermentation, the R. roxburghii fruit wine from each group was centrifuged at 4,000 rpm for 10 min, and the supernatant was used for the determination of the quality indicators of R. roxburghii fruit wine.
Analysis of flavor and quality characteristics of R. roxburghii fruit wine
The alcohol content, total sugar, total acidity, and volatile acid content of the R. roxburghii wine were determined following the methods described by Liu et al. (2021b). The sensory characteristics of the R. roxburghii fruit wine were analyzed using an electronic tongue system. For this, 80 ml of each group of R. roxburghii fruit wine was taken and added to a dedicated beaker for the electronic tongue system. The electronic tongue system was used according to the instructions in the user manual to test each group of R. roxburghii fruit wine. The sampling time was 120 s, the sampling speed was 1/s, each sample was measured in triplicate, and each replicate was collected four times.
The headspace solid-phase microextraction-gas chromatography-mass spectrometry system (TQ8040, Agilent, USA) was used to analyze the aroma characteristics of the R. roxburghii fruit wine. The aroma components of R. roxburghii fruit wine were extracted at 40°C for 30 min, with cyclohexanone used as the internal standard for determining the aroma components of R. roxburghii fruit wine. The odor activity value (OAV) of each aroma component was calculated by referring to the threshold values of each volatile aroma component.
Statistical analysis
Data results were presented as mean ± SD. Principal component analysis (PCA) and one-way ANOVA were performed using SPSS 21.0 to test for significant differences among the groups. A p-value of less than 0.05 was considered statistically significant. Each experiment was repeated in triplicate.
Results
Screening of ethanol-tolerant non-Saccharomyces yeast strains
When the native non-Saccharomyces yeasts isolated from R. roxburghii were treated with 12% (v/v) ethanol, most of them died. However, three yeast strains (designated as C6, F112, and F15) exhibited satisfactory growth with OD600nm values of 0.56 ± 0.02, 0.47 ± 0.01, and 0.48 ± 0.01, respectively. Therefore, C6, F112, and F15 were selected as ethanol-tolerant strains for further analysis.
Identification of ethanol-tolerant non-Saccharomyces yeast strains
The identification of the three ethanol-tolerant yeast strains was initially based on morphological characteristics on WL agar. As shown in Figure 1, the colony color of C6, F112, and F15 was white, and their colony topography was convex and opaque.
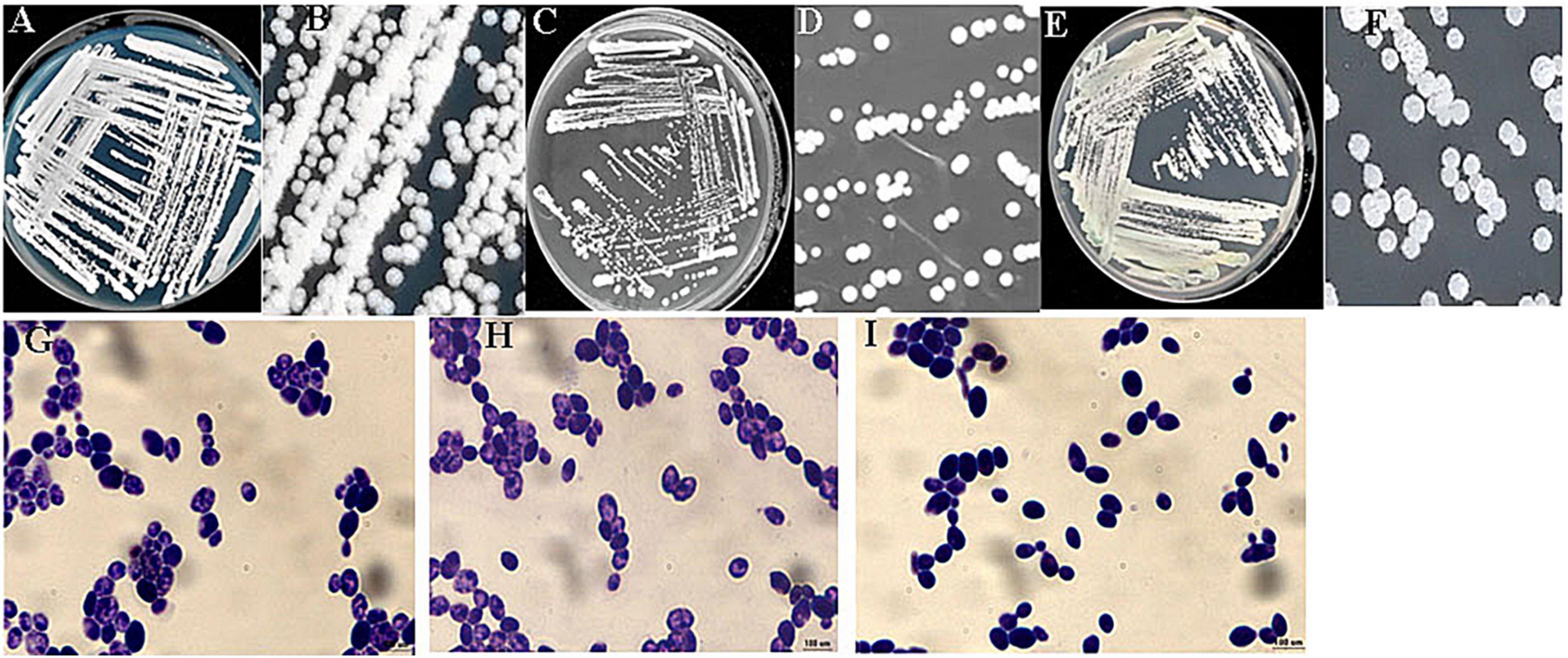
Figure 1. Colony and cell morphologies of ethanol-tolerant non-Saccharomyces yeasts isolated from R. Roxburghii. (A,B) Colony morphology of C6 on WL medium; (C,D) colony morphology of F112 on WL medium; (E,F) colony morphology of F15 on WL medium; (G) cell morphology of C6 following crystal violet staining (100×); (H) cell morphology of F112 following crystal violet staining (100×); and (I) cell morphology of F15 following crystal violet staining (100×).
To confirm the identity of the ethanol-tolerant yeast strains, their 26S rDNA D1/D2 domain sequences were compared. The analysis revealed that the 26S rDNA sequences of C6, F112, and F15 had the highest similarity to C. tropicalis, Pichia guilliermondii, and W. anomalus, respectively. Therefore, these three strains of ethanol-tolerant yeasts (C6, F112, and F15) were identified and named C. tropicalis C6, P. guilliermondii F112, and W. anomalus F15 based on the results of morphological characteristics and sequence alignment.
Growth characteristics of ethanol-tolerant non-Saccharomyces yeast strains
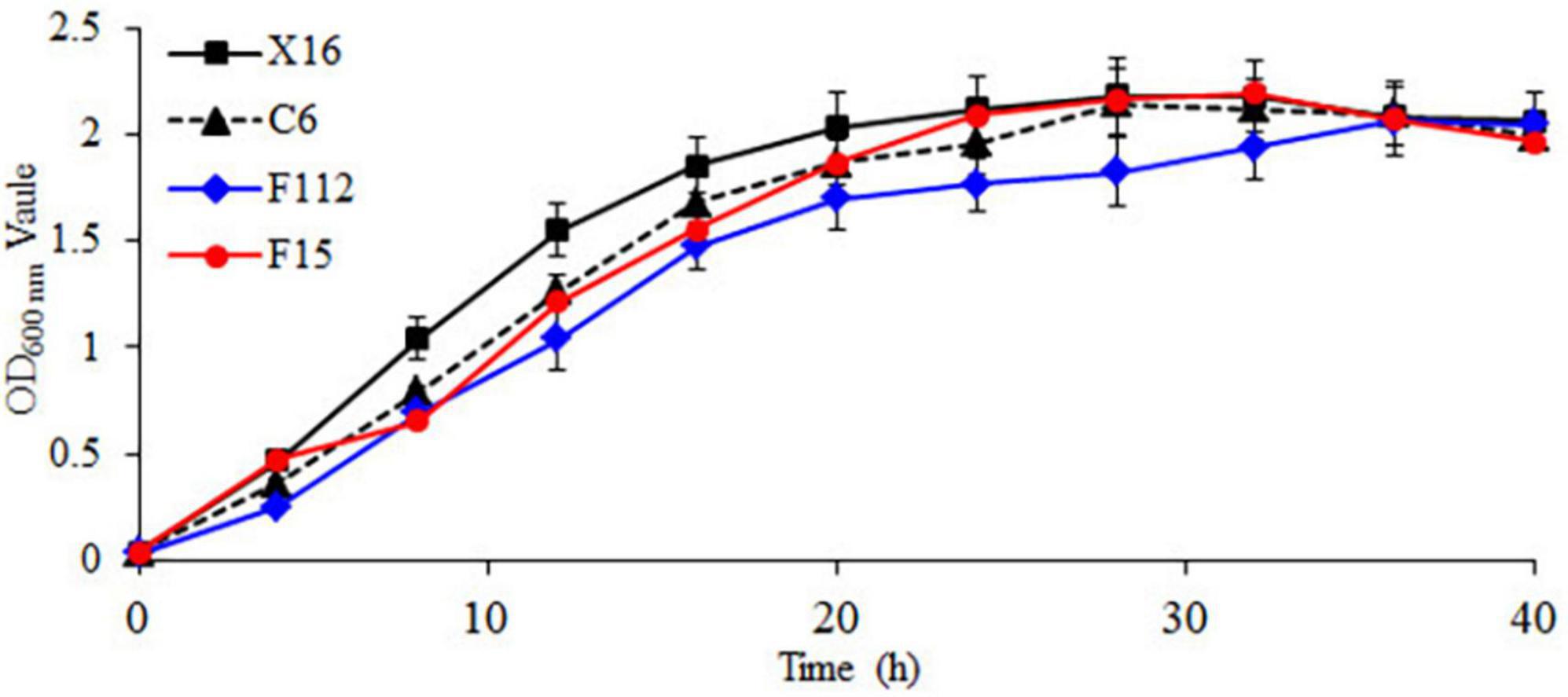
The growth curves of the strains are shown in Figure 2, with a lag phase from 0 to 4 h, a logarithmic growth phase from 4 to 20 h, and a stationary phase after 20 h. During the logarithmic growth phase, the OD600 nm values of C. tropicalis C6, P. guilliermondii F112, and W. anomalus F15 were all lower than those of the commercial S. cerevisiae X16. During the stationary phase, the OD600 nm of P. guilliermondii F112 was lower than that of S. cerevisiae X16. Throughout the entire growth period, the growth of P. guilliermondii F112 was lower than that of S. cerevisiae X16, while the growth of C. tropicalis C6 and W. anomalus F15 was basically consistent with that of S. cerevisiae X16 in the later stages of the stationary phase.
Winemaking condition tolerances of ethanol-tolerant non-Saccharomyces yeast strains
To assess the tolerance of the selected yeasts to winemaking conditions, their OD600 nm values were measured after exposure to different concentrations of glucose, SO2, and citric acid. Results demonstrated that all three ethanol-tolerant yeast strains exhibited excellent sugar tolerance, as they were able to grow in all glucose concentrations tested (100–300 mg/L) (Figure 3A). Furthermore, C. tropicalis C6, P. guilliermondii F112, and W. anomalus F15 displayed similar sulfur dioxide and acid tolerance to S. cerevisiae X16 within the tested ranges of sulfur dioxide (50–300 mg/L) and citric acid (2.5–4.0%) concentrations, respectively (Figures 3B, C). Therefore, C. tropicalis C6, P. guilliermondii F112, and W. anomalus F15 showed perfect tolerance to the winemaking environment.
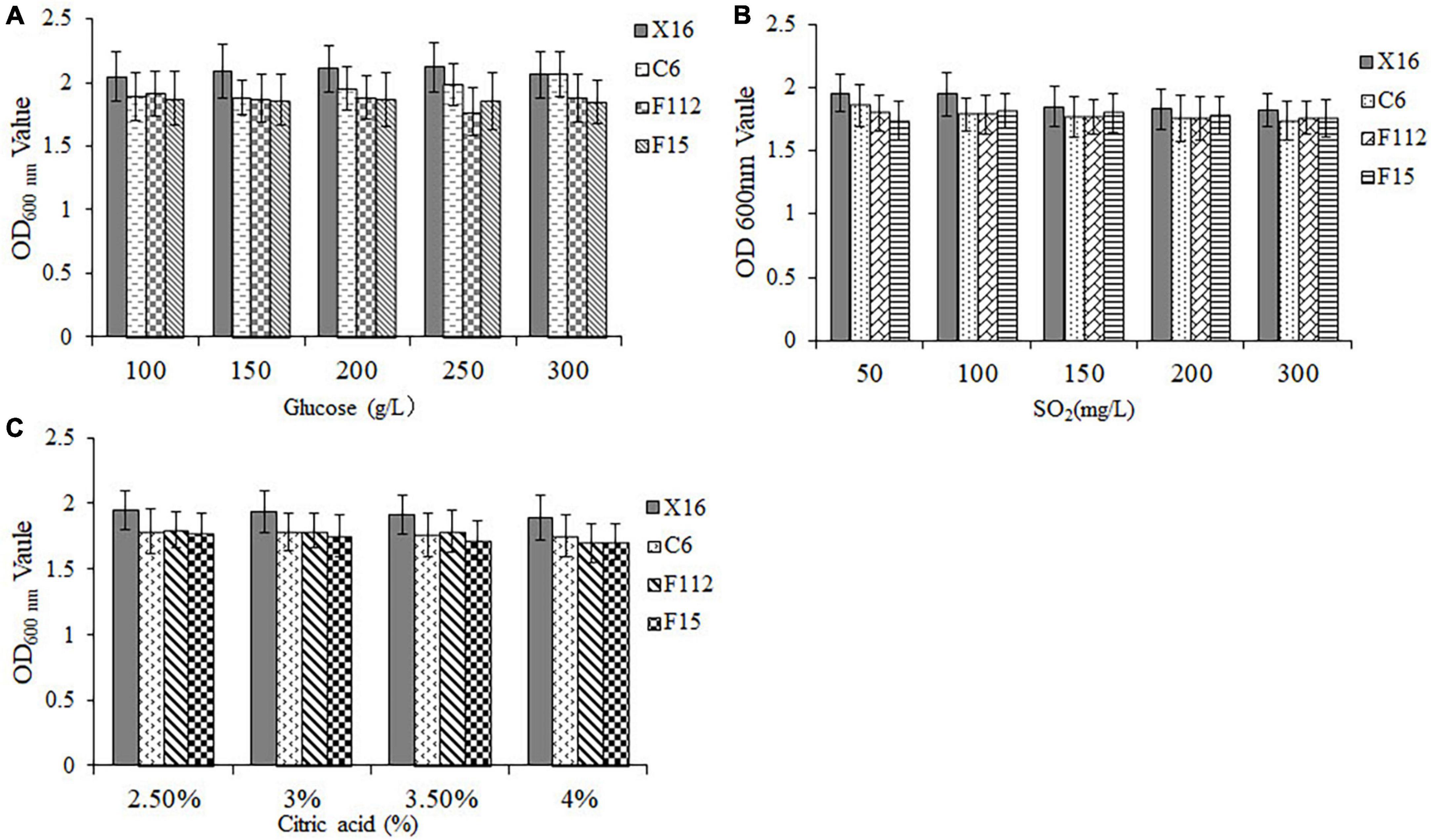
Figure 3. Winemaking condition tolerances of ethanol-tolerant non-Saccharomyces yeasts isolated from R. roxburghii. (A) Glucose tolerance; (B) SO2 tolerance; and (C) citric acid tolerance.
Sugar metabolic performance of ethanol-tolerant non-Saccharomyces yeast strains
As shown in Table 1, different strains have different utilization characteristics for different sugars. W. anomalus F15 can only metabolize glucose with the least number of sugars that it can utilize. On the other hand, C. tropicalis C6 can metabolize all types of sugars except for galactose, with the broadest range of sugar utilization. P. guilliermondii F112 can utilize three types of sugars (glucose, sucrose, and maltose). Therefore, C. tropicalis C6 has the widest range of sugar metabolism, and its sugar utilization characteristics are similar to those of S. cerevisiae X16, except for galactose.
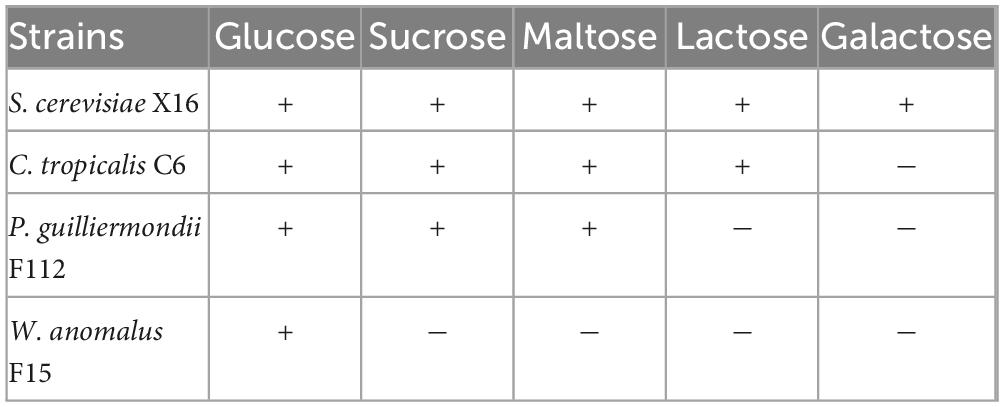
Table 1. Sugar utilization characteristics of ethanol-tolerant non-Saccharomyces yeasts isolated from R. Roxburghii.
Sulphureted hydrogen and β-glucosidase production abilities of ethanol-tolerant non-Saccharomyces yeasts strains
The ability of ethanol-tolerant yeasts to produce H2S production ability was evaluated by observing the color depth on BiGGY agar (Caridi et al., 2022). As shown in Figure 4, C. tropicalis C6 had a similar H2S production ability to the control (S. cerevisiae X16), while P. guilliermondii F112 and W. anomalus F15 exhibited stronger H2S production abilities than the control, as evidenced by the colony depth color on the filter paper.
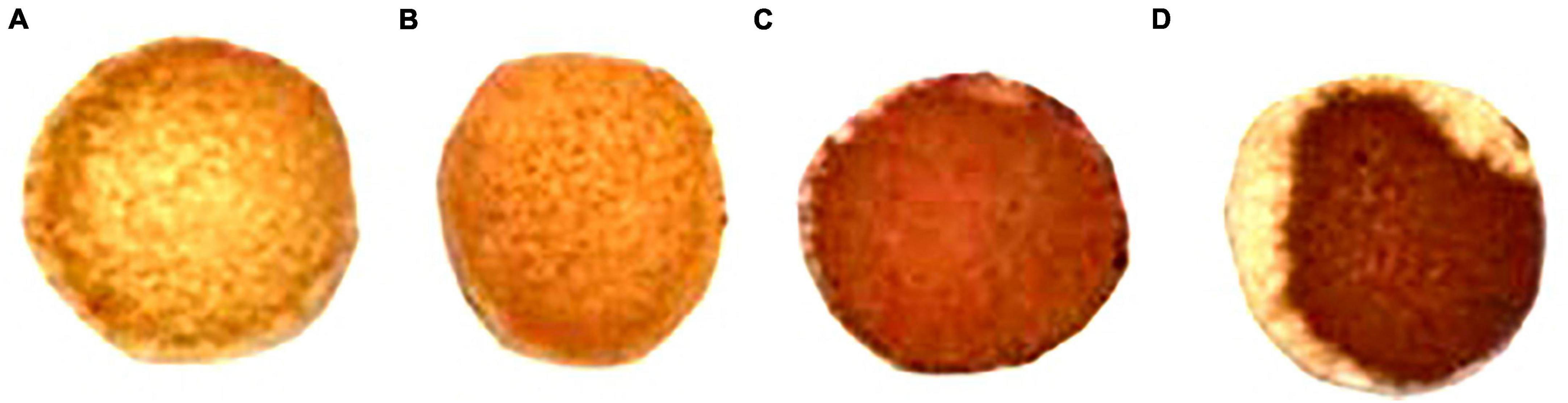
Figure 4. Hydrogen sulfide production ability of C6, F112, and F15 strains on BiGGY agar. (A) S. cerevisiae X16; (B) strain of C. tropicalis C6; (C) strain of P. guilliermondii F112; and (D) strain of W. anomalus F15.
Flavor compounds in fruit are often present in the form of glycoconjugate, making them flavorless (Gueguen et al., 1996). β-Glucosidase are enzymes that could hydrolyze these glycosyl bonds, thereby releasing the flavor compounds to wines (Haslbeck et al., 2017). To investigate the β-glucosidases of the selected strains, namely, C. tropicalis C6, P. guilliermondii F112, and W. anomalus F15, p-NPG colorimetry was used. The result showed that the β-glucosidase production abilities of C. tropicalis C6 and P. guilliermondii F112 were similar to those of S. cerevisiae X16. However, the strain of W. anomalus F15 exhibited significantly lower β-glucosidase production ability than S. cerevisiae X16 (Table 2).
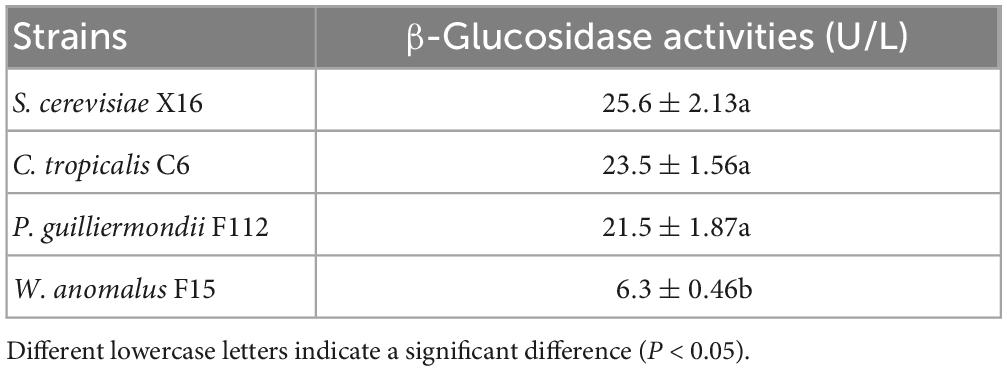
Table 2. β-Glucosidase production capacity of ethanol-tolerant non-Saccharomyces yeasts isolated from R. Roxburghii.
Winemaking properties of the ethanol-tolerant non-Saccharomyces yeasts in laboratory-scale
The combination of non-saccharomyces yeasts with S. cerevisiae as fermentation starters has been widely studied and accepted in wine production (Comitini et al., 2011). To further analyze the fermentative properties of the ethanol-tolerant yeast strains, R. roxburghii wine was fermented by co-inoculating the C. tropicalis C6, P. guilliermondii F112, or W. anomalus F15 together with S. cerevisiae X16. Dynamic changes of the non-Saccharomyces yeasts population during R. roxburghii wine fermentation were monitored by colony counting method, and the results showed that the proportion of C. tropicalis C6, P. guilliermondii F112, and W. anomalus F15 gradually decreased, in contrast, the proportion of S. cerevisiae X16 gradually increased and dominate at the middle and later periods of fermentation (Supplementary Figure 2).
The physicochemical parameters of the fermented R. roxburghii wines are listed in Table 3. Ethanol degrees of wines fermented by P. guilliermondii F112 or W. anomalus F15 were lower than the wine produced by S. cerevisiae X16 alone, while the C. tropicalis C6 fermented wine was similar to that produced by S. cerevisiae X16. The pH and volatile acidity parameters of the four groups of R. roxburghii wines were similar, with no differences found among them. The total acidity were lower in the C. tropicalis C6 and P. guilliermondii F112 groups compared to the S. cerevisiae X16 group.
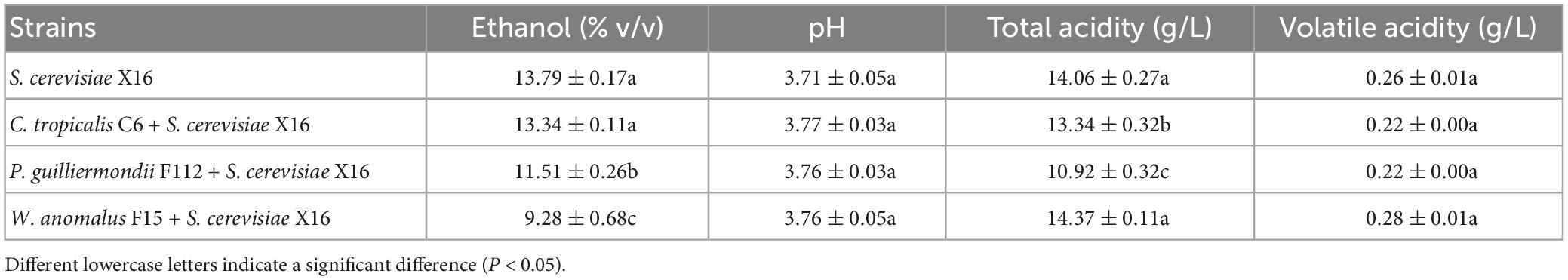
Table 3. Oenological parameters of R. roxburghii wine fermented with ethanol-tolerant non-Saccharomyces yeasts in combination with S. cerevisiae.
In addition, an electronic tongue system was used to perform sensory analysis and differentiate the sensory characteristics of the R. roxburghii wines fermented with different yeast strains. However, no significant differences in the sensory characteristics including in sourness, bitterness, astringency, aftertaste-A, aftertaste-B, umami, richness, and saltiness were found among the three types of R. roxburghii wines (Figure 5).
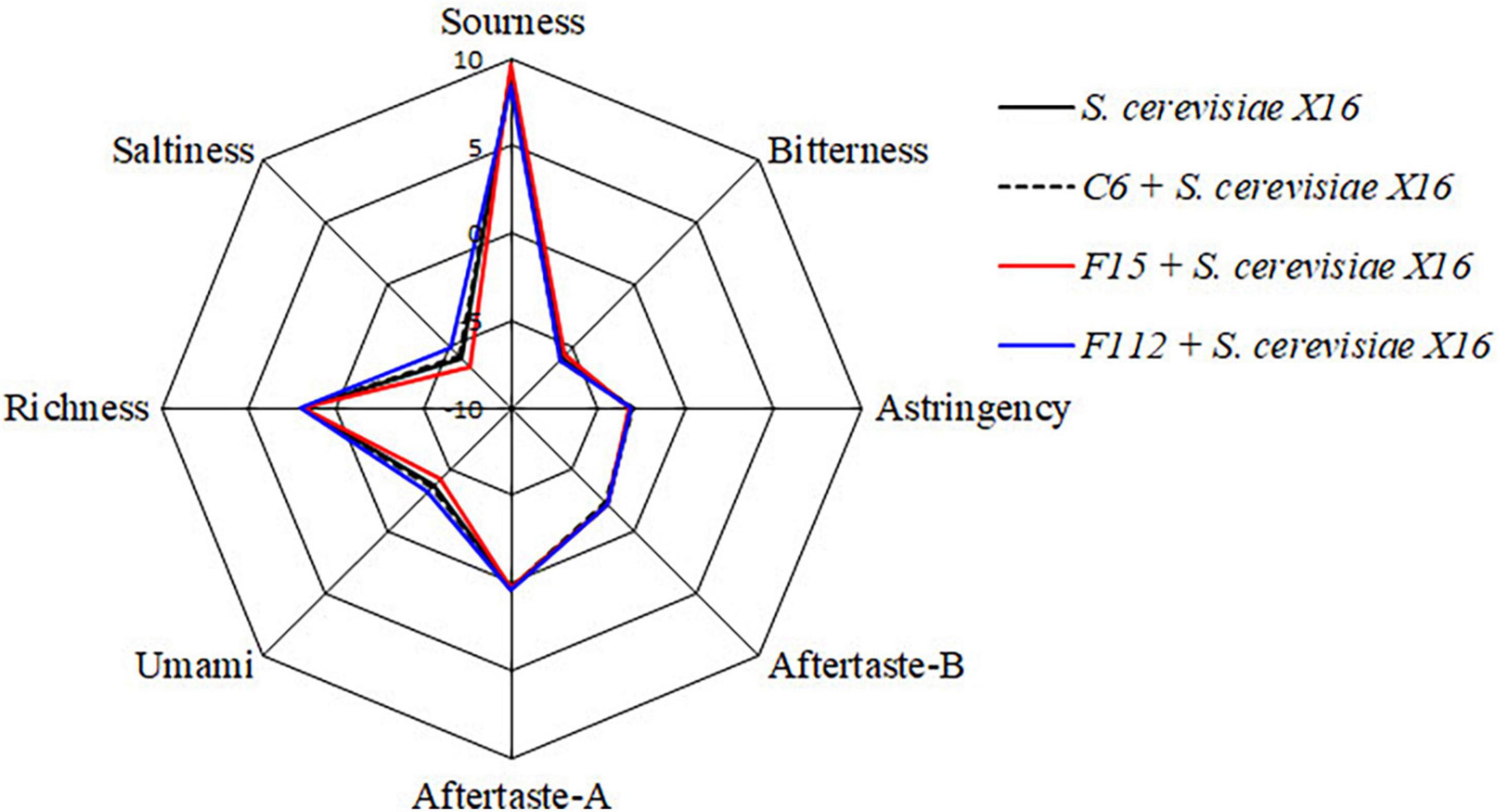
Figure 5. Radar chart of taste attribute of R. Roxburghii wine fermented with ethanol-tolerant non-Saccharomyces yeasts in combination with S. cerevisiae.
The volatile aroma profiles of the R. roxburghii wines fermented with the selected yeast strains were further examined by using GC-MS analysis. A total of 66 volatile compounds, including 32 volatile esters, 10 volatile alcohols, 6 volatile acids, 3 volatile aldoketones, and 15 other volatile chemicals, were identified in the four groups of fermented R. roxburghii wines (Table 4). The R. roxburghii wines co-fermented with the three ethanol-tolerant yeast strains contained 50, 46, and 55 volatile compounds, respectively, whereas only 44 volatile compounds were detected in the R. roxburghii wine inoculated with S. cerevisiae X16 alone. Additionally, 22 chemicals, including 10 esters, 1 alcohol, 1 acid, and 10 other compounds, were specifically detected in the R. roxburghii wines inoculated with the ethanol-tolerant yeasts. On the other hand, octyl acetate, decanoic acid, 1-nonanal, and 2,4-di-tert-butylphenol were specifically discovered in the S. cerevisiae X16 group. Overall, the co-inoculation of these ethanol-tolerant yeast strains isolated from R. roxburghii along with S. cerevisiae increased the types of volatile compounds in the wine (Supplementary Figure 3).
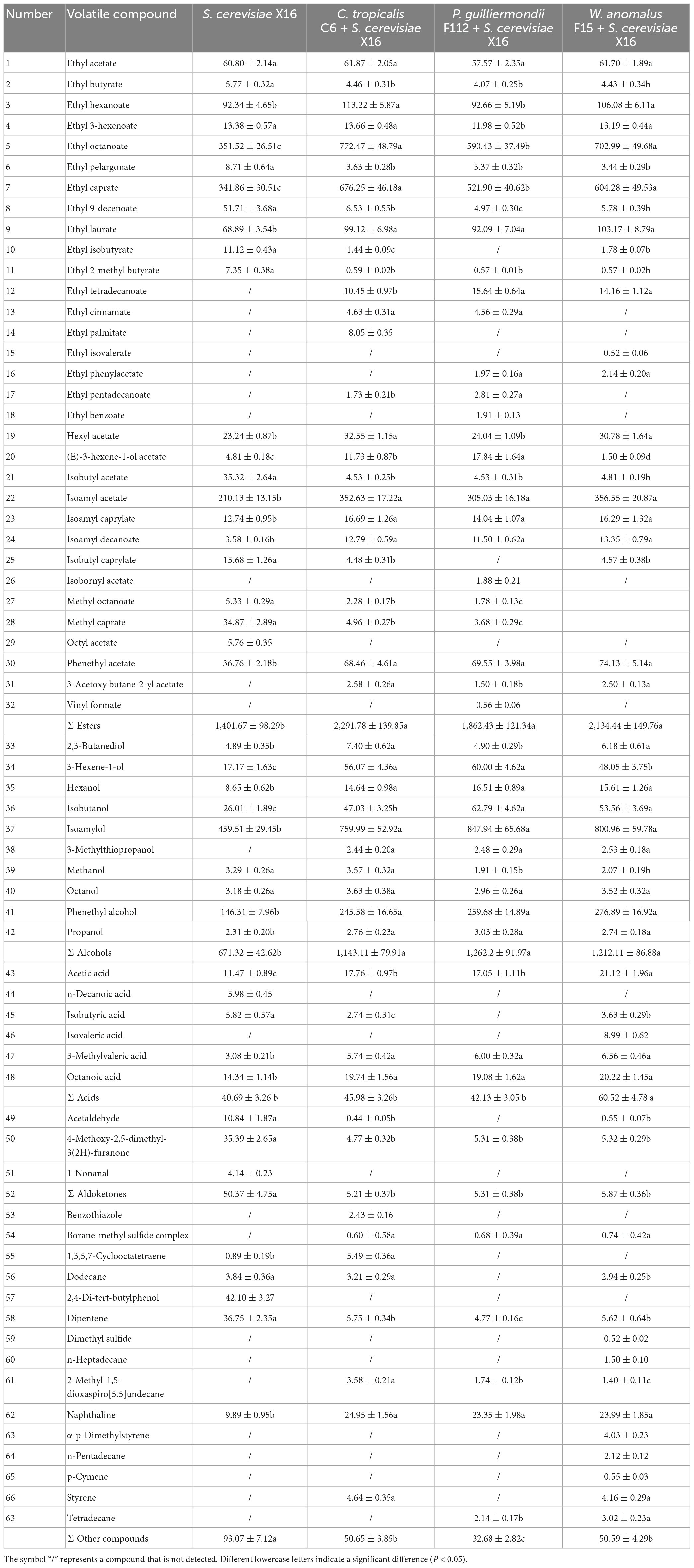
Table 4. Volatile compounds (mg/L) in R. roxburghii wines fermented with ethanol-tolerant non-Saccharomyces yeasts combined with S. cerevisiae.
When checking the volatile compound contents of R. roxburghii wines with the different ethanol-tolerant yeast strains, variations in the levels of volatile esters, alcohols, acids, aldoketones, and other chemicals were observed (Table 4). Co-inoculation with the three ethanol-tolerant yeasts resulted in an increase in volatile esters and alcohols, as well as a decrease in volatile aldoketones and other compounds (Table 4). In addition, R. roxburghii wines co-fermented with W. anomalus F15 and S. cerevisiae X16 exhibited higher levels of volatile acids compared to those produced with S. cerevisiae X16 alone.
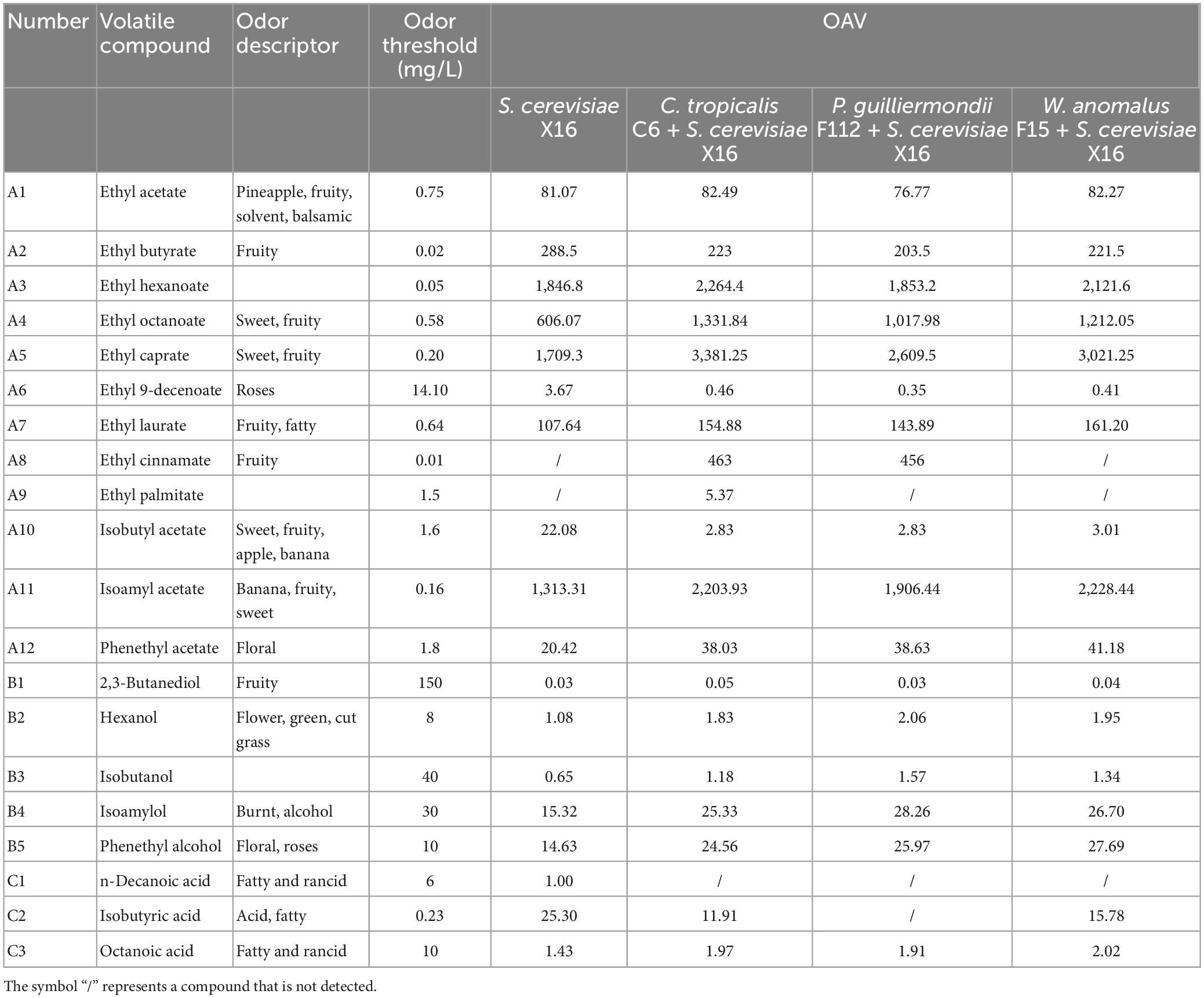
The OAV was used to further evaluate the contribution of the main aromatic compounds to the aromatic characteristics of R. roxburghii wine. Compounds with OAV greater than 1 were considered to have a significant impact on the aroma, while those with OAV less than 1 were considered less important. Table 5 shows the calculated OAVs of twenty aromatic compounds in R. roxburghii wine. Thirteen compounds had OAVs greater than 1, while only 1 had an OAV less than 1 across all 4 groups of R. roxburghii wine. Specifically, ethyl 9-decenoate had an OAV greater than 1 only in the S. cerevisiae X16 group, whereas isobutanol had the only OAV less than 1 in the same group. The OAVs of ethyl caprate, isoamyl acetate, ethyl hexanoate, and ethyl octanoate were high in all four groups, suggesting that these compounds strongly contribute to the aroma of R. roxburghii wine.
Principal component analysis was used to further assess the impact of the main aromatic compounds on the characteristics of R. roxburghii wine. As depicted in Figure 6, the three principal components, PC1, PC2, and PC3, accounted for 62.84, 25.02, and 12.14% of the total variance, respectively, explaining 100.00% of the total variance. Most of the compounds were clustered in the positive axis of PC1 and PC2, and significant differences in distribution were observed among the four groups of fermented R. roxburghii wine. Ethyl palmitate might be closely related to the mixed fermentation of C. tropicalis C6 and S. cerevisiae X16, while W. anomalus F15 and S. cerevisiae X16 produced wines were characterized by compounds located in the positive PC1 and PC2, such as fruity and rosy chemicals (e.g., isobutyl acetate and ethyl 9-decenoate), which may contribute to the aroma of S. cerevisiae X16 fermented wine. However, it was challenging to identify the main volatile characteristics of P. guilliermondii F112 and S. cerevisiae X16 fermented R. roxburghii wines. Moreover, the mixed fermented wine using W. anomalus F15 and S. cerevisiae X16 was closely clustered to many esters, alcohols and acids, such as ethyl laurate, phenethyl alcohol, and octanoic acid, which may endow the wine more complex aroma characteristics.
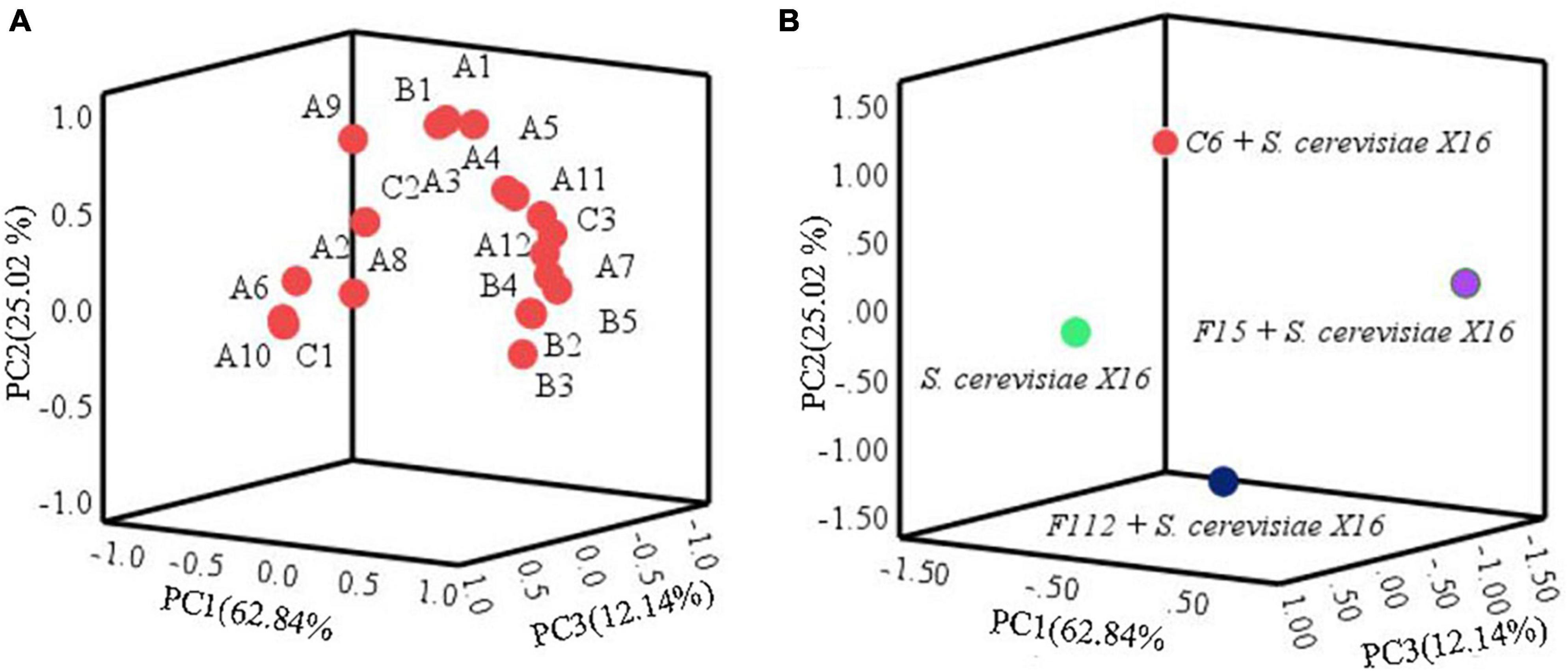
Figure 6. Principal component analysis of R. roxburghii wine fermentation with ethanol-tolerant non-Saccharomyces yeasts in combination with S. cerevisiae. (A) Principal component load plot of volatile aroma compounds; and (B) principal component score of volatile aroma compounds. A1, ethyl acetate; A2, ethyl butyrate; A3, ethyl hexanoate; A4, ethyl octanoate; A5, ethyl caprate; A6, ethyl 9-decenoate; A7, ethyl laurate; A8, ethyl cinnamate; A9, ethyl palmitate; A10, isobutyl acetate; A11, isoamyl acetate; A12, phenethyl acetate; B1, 2,3-butanediol; B2, hexanol; B3, isobutanol; B4, isoamylol; B5, phenethyl alcohol; C1, n-decanoic acid; C2, isobutyric acid; C3, octanoic acid.
Discussion
In order to evaluate yeast strains for winemaking, ethanol tolerance is an essential property (Novo et al., 2014). Researchers have made numerous efforts to isolate ethanol-tolerant yeasts from various sources, including fruits and fermentation conditions, for industrial purposes (Osho, 2005; Tikka et al., 2013). In a recent study, ethanol-tolerant yeast flora was isolated, identified, and screened from the Indian cashew apple, and seven strains of ethanol-tolerant yeasts were identified as Candida spp. (Desai et al., 2012). Besides, mutational breeding techniques were also applied to produce ethanol-tolerant yeasts. For example, the Pichia terricola H5 strain, which initially displayed 8% ethanol tolerance, was subjected to ultraviolet irradiation and diethyl sulfate mutagenesis to increase its ethanol tolerance (Gao et al., 2022). As a result, two mutant strains (UV5 and UV8) that demonstrated high tolerance to ethanol were obtained, and modified aroma profiles were evident in the fermentation samples exposed to these strains. In the present study, ethanol-tolerant non-Saccharomyces yeasts were screened for the first time from R. roxburghii. Three strains of yeasts displaying high ethanol tolerance were successfully obtained and subsequently identified as C. tropicalis, P. guilliermondii, and W. anomalus (Figure 1).
While S. cerevisiae is the most commonly isolated ethanol-tolerant yeast and widely used for wine making (Alexandre et al., 2004), non-Saccharomyces yeasts have been considered sensitive to ethanol and are typically dominant in the early stage of spontaneous wine fermentation (Liu et al., 2021c). However, some non-Saccharomyces species were proved to be tolerant to ethanol. For example, three strains of Candida spp. yeast isolated from Indian cashew apple were able to tolerate up to 10% (v/v) ethanol (Desai et al., 2012). Besides, a strain of Candida stellata was found to produce ethanol levels up to 13.48% (v/v) during the fermentation of Macabeo grape must, indicating a high tolerance to ethanol (Clemente-Jimenez et al., 2004). In this study, we obtained a strain of C. tropicalis that exhibited robust growth in YEPD broth containing 12% (v/v) of ethanol (Figure 1). These results indicated that Candida species may be a better source for screening ethanol-tolerant yeast.
Among non-Saccharomyces yeasts, W. anomalus has gained increasing attention in recent years due to its unique physiological characteristics and metabolic features (Liu et al., 2021d). These yeast species have been reported to tolerate various extreme environmental conditions such as high/low pH, high osmotic pressure, and anaerobic conditions (Schneider et al., 2012). In our previous study, a fruity aroma-producing strain of W. anomalus C11 was isolated from R. roxburghii, which was capable of withstanding 9% (v/v) ethanol treatment (Liu et al., 2021b). In the present study, we isolated another strain of W. anomalus F15 from R. roxburghii, which displayed a higher ethanol tolerance of up to 12% (v/v) than W. anomalus C11. Moreover, W. anomalus F15 was also found to be tolerant to glucose, sulfur dioxide, and citric acid, suggesting that this strain of W. anomalus may have a better potential for application in winemaking (Figure 3).
Aroma characteristic is an important parameter in assessing wine quality (Styger et al., 2011). Combining non-Saccharomyces starters with S. cerevisiae during winemaking to enhance the richness and complexity of wine has been widely accepted by researchers and wine producers (Contreras et al., 2015). In this study, R. roxburghii wines were fermented by co-inoculating ethanol-tolerant non-Saccharomyces yeasts with S. cerevisiae. The results showed that the levels of volatile esters and alcohol compounds significantly increased in the three mixed-fermentation wines compared to those fermented with S. cerevisiae alone. On the other hand, the levels of aldoketones and other compounds significantly decreased in mixed-fermentation wines (Table 4 and Supplementary Figure 2). Therefore, co-inoculating ethanol-tolerant non-Saccharomyces yeasts with S. cerevisiae can regulate the aromatic characteristics of R. roxburghii wine, contributing to the enrichment of different types of R. roxburghii wine.
Ester compounds are a type of metabolite generated during alcohol metabolism (Sumby et al., 2021). Most ester compounds exhibit floral and fruity aromatic characteristics and are important contributors to the aroma profiles of various fermented wines (Rojas et al., 2001). Our study found that using native non-Saccharomyces yeasts of R. roxburghii in combination with commercial S. cerevisiae can increase the diversity and concentration of volatile esters compounds (Table 4 and Supplementary Figure 2). For example, the levels of ethyl octanoate, ethyl caprate, and ethyl laurate in non-Saccharomyces yeasts-fermented wines were higher than those in R. roxburghii wine fermented with S. cerevisiae alone (Table 4). Additionally, seven types of ethyl eater chemicals, including ethyl tetradecanoate, ethyl cinnamate, and ethyl palmitate, were specifically detected in the three types of R. roxburghii wines produced with the native non-Saccharomyces yeasts of R. roxburghii (Table 4).
Conclusion
This study represents the first report on the screening and oenological property analysis of ethanol-tolerant non-Saccharomyces yeasts isolated from R. roxburghii. We obtained three strains of ethanol-tolerant yeasts designated as C6, F112, and F15, which were identified as C. tropicalis, P. guilliermondii, and W. anomalus, respectively, after treating them with 12% (v/v) of ethanol. These strains showed similar winemaking condition tolerances to S. cerevisiae X16, but their growth, sugar metabolic performance, and activities of sulphureted hydrogen production were different. The β-glucosidase production ability of strain W. anomalus F15 was lower than that of S. cerevisiae X16, and strains of C. tropicalis C6 and P. guilliermondii F112 were similar to S. cerevisiae X16. Mixed inoculation of these ethanol-tolerant yeast strains with S. cerevisiae regulated the volatile aroma characteristics of the fermented R. roxburghii wine, enriching and enhancing its aroma flavor. Therefore, the selected ethanol-tolerant yeasts exhibit potential applications in the production of unique R. roxburghii wine.
Data availability statement
The original contributions presented in this study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
YL and XL wrote the original draft manuscript. PD, XT, WZ, and MK conducted the experiments. XL and MH conceived and designed the experiments. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the Guizhou Provincial Science and Technology Foundation [ZK (2023)137], Key Laboratory of Microbial Resources Collection and Preservation, Ministry of Agriculture and Rural Affairs (KLMRCP2021-02), National Natural Science Foundation of China (32260641), and Guizhou Fruit Wine Brewing Engineering Research Center [Qianjiaoji (2022)050].
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1202440/full#supplementary-material
References
Alexandre, H., Costello, P. J., Remize, F., Guzzo, J., and Guilloux-Benatier, M. (2004). Saccharomyces cerevisiae–Oenococcus oeni interactions in wine: Current knowledge and perspectives. Int. J. Food Microbiol. 93, 141–154. doi: 10.1016/j.ijfoodmicro.2003.10.013
Caridi, A., Sidari, R., Pulvirenti, A., Blaiotta, G., and Ritieni, A. (2022). Clonal selection of wine yeasts with differential adsorption activities towards phenolics and ochratoxin A. Food Biotechnol. 36, 22–37. doi: 10.1080/08905436.2021.2006064
Clemente-Jimenez, J. M., Mingorance-Cazorla, L., Martıìnez-Rodrıìguez, S., Francisco Heras-Vázquez, J. L., and Rodrıìguez-Vico, F. (2004). Molecular characterization and oenological properties of wine yeasts isolated during spontaneous fermentation of six varieties of grape must. Food Microbiol. 21, 149–155. doi: 10.1016/S0740-0020(03)00063-7
Comitini, F., Gobbi, M., Domizio, P., Romani, C., Lencioni, L., Mannazzu, I., et al. (2011). Selected non-Saccharomyces wine yeasts in controlled multistarter fermentations with Saccharomyces cerevisiae. Food Microbiol. 28, 873–882. doi: 10.1016/j.fm.2010.12.001
Contreras, A., Hidalgo, C., Henschke, P. A., Chambers, P. J., Curtin, C., and Varela, C. (2014). Evaluation of non-Saccharomyces yeasts for the reduction of alcohol content in wine. Appl. Environ. Microbiol. 80, 1670–1678. doi: 10.1128/AEM.03780-13
Contreras, A., Hidalgo, C., Schmidt, S., Henschke, P. A., Curtin, C., and Varela, C. (2015). The application of non-Saccharomyces yeast in fermentations with limited aeration as a strategy for the production of wine with reduced alcohol content. Int. J. Food Microbiol. 205, 7–15. doi: 10.1016/j.ijfoodmicro.2015.03.027
Desai, M. V., Dubey, K. V., Vakil, B. V., and Ranade, V. V. (2012). Isolation, identification and screening of the yeast flora from Indian cashew apple for sugar and ethanol tolerance. Int. J. Biotechnol. Wellness Ind. 1, 259–265.
Egue, L. A. N., Bouatenin, J. P. K. M., N’guessan, F. K., and Koussemon-Camara, M. (2018). Virulence factors and determination of antifungal susceptibilities of Candida species isolated from palm wine and sorghum beer. Microb. Pathog. 124, 5–10. doi: 10.1016/j.micpath.2018.08.007
Gao, J., He, X., Huang, W., You, Y., and Zhan, J. (2022). Enhancing ethanol tolerance via the mutational breeding of Pichia terricola H5 to improve the flavor profiles of wine. Fermentation 8:149. doi: 10.3390/fermentation8040149
Gueguen, Y., Chemardin, P., Janbon, G., Arnaud, A., and Galzy, P. (1996). A very efficient β-glucosidase catalyst for the hydrolysis of flavor precursors of wines and fruit juices. J. Agric. Food Chem. 44, 2336–2340. doi: 10.1021/jf950360j
Haslbeck, K., Jerebic, S., and Zarnkow, M. (2017). Characterization of the unfertilized and fertilized hop varieties progress and hallertauer tradition-Analysis of free and glycosidic-bound flavor compounds and β-glucosidase activity. Brew. Sci. 70, 148–158. doi: 10.23763/BRSC17-15HASLBECK
Jolly, N. P., Varela, C., and Pretorius, I. S. (2014). Not your ordinary yeast: Non-Saccharomyces yeasts in wine production uncovered. FEMS Yeast Res. 14, 215–237. doi: 10.1111/1567-1364.12111
Liu, X. Z., Li, Y. F., Yu, Z. H., Hardie, W. J., and Huang, M. Z. (2020). Biodiversity of non-Saccharomyces yeasts during natural fermentation of Rosa roxburghii. Acta Microbiol. Sin. 60, 1696–1708. doi: 10.13343/j.cnki.wsxb.20190523
Liu, X., Li, Y., Yu, Z., Liu, X., Hardie, W. J., and Huang, M. (2020). Screening and characterisation of β-glucosidase production strains from Rosa roxburghii Tratt. Int. J. Food Eng. 17, 1–9. doi: 10.1515/ijfe-2020-0152
Liu, X., Li, Y., Zhao, H., Yu, Z., and Huang, M. (2021a). Oenological property analysis of selected Hanseniaspora uvarum isolated from Rosa roxburghii Tratt. Int. J. Food Eng. 17, 445–454. doi: 10.1515/ijfe-2020-0331
Liu, X., Li, Y., Zhao, H., Yu, Z., Hardie, W. J., and Huang, M. (2021b). Identification and fermentative properties of an indigenous strain of Wickerhamomyces anomalus isolated from Rosa roxburghii Tratt. Brit. Food J. 123, 4069–4081. doi: 10.1108/BFJ-11-2020-0993
Liu, X., Li, Y., Zhang, Y., Zeng, S., and Huang, M. (2021c). Yeast diversity investigation of ‘Beihong’ (V. vinifera× V. amurensis) during spontaneous fermentation from Guiyang region, Guizhou, China. Food Sci Technol. Res. 27, 887–896. doi: 10.3136/fstr.27.887
Liu, X., Li, Y., Zhou, J., and Huang, M. (2021d). Effects of co-inoculation and sequential inoculation of Wickerhamomyces anomalus and Saccharomyces cerevisiae on the physicochemical properties and aromatic characteristics of longan (Dimocarpus longan Lour.) wine. Qual. Assur. Saf. Crop. 13, 56–66. doi: 10.15586/qas.v13i2.893
Morata, A., Escott, C., Bañuelos, M. A., Loira, I., Fresno, J. M., González, C., et al. (2019). Contribution of non-Saccharomyces yeasts to wine freshness. A review. Biomolecules 10:34. doi: 10.3390/biom10010034
Novo, M., Gonzalez, R., Bertran, E., Martínez, M., Yuste, M., and Morales, P. (2014). Improved fermentation kinetics by wine yeast strains evolved under ethanol stress. LWT-Food Sci. Technol. 58, 166–172. doi: 10.1016/j.lwt.2014.03.004
Osho, A. (2005). Ethanol and sugar tolerance of wine yeasts isolated from fermenting cashew apple juice. Afr. J. Biotechnol. 4, 660–662. doi: 10.5897/AJB2005.000-3119
Padilla, B., Gil, J. V., and Manzanares, P. (2018). Challenges of the non-conventional yeast Wickerhamomyces anomalus in winemaking. Fermentation 4:68. doi: 10.3390/fermentation4030068
Parapouli, M., Vasileiadis, A., Afendra, A. S., and Hatziloukas, E. (2020). Saccharomyces cerevisiae and its industrial applications. AIMS Microbiol. 6, 1–31. doi: 10.3934/microbiol.2020001
Pietrafesa, A., Capece, A., Pietrafesa, R., Bely, M., and Romano, P. (2020). Saccharomyces cerevisiae and Hanseniaspora uvarum mixed starter cultures: Influence of microbial/physical interactions on wine characteristics. Yeast 37, 609–621. doi: 10.1002/yea.3506
Rojas, V., Gil, J. V., Piñaga, F., and Manzanares, P. (2001). Studies on acetate ester production by non-Saccharomyces wine yeasts. Int. J. Food Microbiol. 70, 283–289. doi: 10.1016/S0168-1605(01)00552-9
Schneider, J., Rupp, O., Trost, E., Jaenicke, S., Passoth, V., Goesmann, A., et al. (2012). Genome sequence of Wickerhamomyces anomalus DSM 6766 reveals genetic basis of biotechnologically important antimicrobial activities. FEMS Yeast Res. 12, 382–386. doi: 10.1111/j.1567-1364.2012.00791.x
Styger, G., Prior, B., and Bauer, F. F. (2011). Wine flavor and aroma. J. Ind. Microbiolo. Biotechnol. 38, 1145–59. doi: 10.1007/s10295-011-1018-4
Sumby, K. M., Grbin, P. R., and Jiranek, V. (2021). Microbial modulation of aromatic esters in wine: Current knowledge and future prospects. Food Chem. 121, 1–16. doi: 10.1016/j.foodchem.2009.12.004
Tikka, C., Osuru, H. P., Atluri, N., Raghavulu, P. C. V., Yellapu, N. K., Mannur, I. S., et al. (2013). Isolation and characterization of ethanol tolerant yeast strains. Bioinformation 9, 421–425. doi: 10.6026/97320630009421
Wang, L., Lv, M., An, J., Fan, X., Dong, M., Zhang, S., et al. (2021). Botanical characteristics, phytochemistry and related biological activities of Rosa roxburghii Tratt fruit, and its potential use in functional foods: A review. Food Funct. 12, 1432–1451. doi: 10.1039/D0FO02603D
Keywords: ethanol tolerance, non-Saccharomyces yeast, Rosa roxburghii Tratt, fruit wine, volatile aroma
Citation: Li Y, Ding P, Tang X, Zhu W, Huang M, Kang M and Liu X (2023) Screening and oenological property analysis of ethanol-tolerant non-Saccharomyces yeasts isolated from Rosa roxburghii Tratt. Front. Microbiol. 14:1202440. doi: 10.3389/fmicb.2023.1202440
Received: 08 April 2023; Accepted: 17 May 2023;
Published: 01 June 2023.
Edited by:
Carlo Giuseppe Rizzello, Sapienza University of Rome, ItalyReviewed by:
Severino Zara, University of Sassari, ItalyGianluca Bleve, National Research Council (CNR), Italy
Copyright © 2023 Li, Ding, Tang, Zhu, Huang, Kang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mingzheng Huang, aHVhbmdtaW5nemhlbmdAZ2l0LmVkdS5jbg==; Xiaozhu Liu, bGl1eGlhb3podV84NDA5MTRAMTYzLmNvbQ==
 Yinfeng Li1
Yinfeng Li1 Xiaozhu Liu
Xiaozhu Liu