
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 24 July 2023
Sec. Virology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1202342
This article is part of the Research Topic Pathogen-Host Interaction in the Development of Viral Hepatitis View all 9 articles
Hepatitis B virus (HBV) antiviral Resistance-Associated Mutations (RAMs) in human immunodeficiency virus (HIV) coinfected patients undergoing highly active antiretroviral therapy (HAART) are complex and incompletely understood. We aimed to determine the prevalence of HBV coinfection, HBV genotypes, and RAMs in a cohort of people living with HIV (PLWH) in the northeastern region of Colombia. This cross-sectional study was carried out between February 2013 and February 2014. Virological, immunological and HAART data were collected from clinical records. In-house nested PCR and Sanger sequencing of the HBV pol gene were used to identify coinfections, genotypes, RAMs and HBV s antigen (HBsAg) escape mutants. Among 275 PLWH, HBV coinfection was confirmed in 32 patients (11.6%), of whom nine (28.2%) were HBsAg positive (active hepatitis B), and 23 (71.8%) were occult hepatitis B infections (OBI). All HBV sequences (n = 23) belonged to the genotype F3. Among HIV/HBV coinfections, 71.9% had CD4+ T cell counts above 200 cells/mm3 and 37.5% had undetectable HIV viral loads. The RAMs rtL80I, rtL180M, and rtM204V, which confer resistance to Lamivudine/Telbivudine and partially resistant to Entecavir, were found in all HBV isolates. An unknown rt236Y mutation to Tenofovir was also identified. Most patients under HAART received first-generation HBV antiviral therapy with a low genetic barrier to resistance. Antiviral Drug-associated Potential Vaccine-escape Mutations (ADAPVEMs) in the S gene were observed in all isolates ranging from 1–20 amino acid substitutions. However, no vaccine escape mutants were detected. In Conclusion, these findings highlight the importance of HBV molecular screening, antiviral resistance monitoring and new guidelines for PLWH to overcome RAMs and prevent HBV-related liver disease.
Human immunodeficiency virus (HIV) infection remains a major global public health problem. According to UNAIDS there were 38.4 million (33.9 million–43.8 million) people living with HIV (PLWH) in 2021 and 40.1 million (33.6 million–48.6 million) have died from acquired immunodeficiency syndrome (AIDS) related illnesses since the beginning of the epidemic (UNAIDS, 2023). Despite global efforts to prevent transmission, there is still no effective curative intervention or an effective vaccine to control the spread of the virus. The therapeutic approach against HIV focuses on inhibiting viral replication in CD4 + T cells through the use of highly active antiretroviral therapy (HAART) to allow partial recovery of the immune system and strengthen its ability to fight off opportunistic infections (Vittinghoff et al., 1999). HAART is a combination of three or more antiretroviral drugs and includes more than 30 molecules divided into six groups, according to their mechanism of action on HIV replication (Saag et al., 2018).
Widespread use of HAART in many countries has led to substantial reduction in AIDS-related deaths and increased life expectancy (Mocroft et al., 2003; Tanser et al., 2013). However, chronic hepatotropic virus infection is a major cause of morbidity and mortality in PLWH (Palella et al., 1998; Vallet-Pichard and Pol, 2004). Hepatitis B virus (HBV) is generally transmitted by percutaneous or mucosal exposure to infected blood and other body fluids. The mechanisms of transmission are the same as those for HIV, but HBV is 50–100 times more infectious (Thio et al., 2002). The World Health Organization (WHO) estimates the occurrence of 1.5 million new cases of HBV per year. In 2019, 296 million people were chronically infected and there were more than 820,000 HBV-related deaths worldwide (Platt et al., 2020; WHO, 2021). In addition, HIV/HBV co-infection is common in endemic areas with approximately 5–15% of PLWH chronically infected with hepatitis B (CHB) (Martín-Carbonero et al., 2011). CHB adversely affects the natural history of HIV and accelerates the progression to end-stage liver disease, including hepatic decompensation, cirrhosis, and hepatocellular carcinoma (HCC) (Martín-Carbonero et al., 2011).
Dual antiviral therapy is recommended for the treatment of HIV/HBV co-infection. HAART can therefore include two nucleoside/nucleotide reverse transcriptase inhibitors (NRTI) with anti-HBV activity, such as Tenofovir Disoproxil Fumarate (TDF), Tenofovir Alafenamide (TAF), Lamivudine (LAM), or Emtricitabine (FTC) (Shire and Sherman, 2005; Lampertico et al., 2017). However, inadequate suppression of viral replication in patients undergoing pharmacological treatment leads to in the development of NRTI-resistant mutations within the RT domain of the HBV pol gene (Archampong et al., 2017). Thus, antiviral therapy should include a correct diagnosis of HBV infection and antiviral susceptibility profile to prevent the emergence of drug-resistant mutations and resumption of active viral replication (He et al., 2015).
In Colombia, the number of PLWH has increased dramatically from 35,000 in 2012 to 134,636 in 2021, with a national prevalence of 0.3%. Despite public health policies and active screening programs in at-risk populations, approximately 30% of cases are underdiagnosed or unreported. In addition, the prevalence of HIV/HBV coinfection in the country was 4.77% of which 1.89% were CHB cases (CAC, 2021). However, coinfection increased threefold when the nucleic acid amplification testing was used, regardless of the HBV serologic profile, identifying 8.7% of cases as occult hepatitis B virus infection (OBI) (Bautista-Amorocho et al., 2014). OBI refers to the presence of HBV DNA in liver tissue or blood in the absence of detectable hepatitis B surface antigen (HBsAg) (Raimondo et al., 2019). Given the low sensitivity of HBsAg screening among PLWH, it is likely that a large proportion of HBV infections in this group remain undetected. In addition, the first-line HAART regimens often includes LAM in HIV mono-infected individuals, who are at increased risk for HBV NRTI-resistant mutations. In Colombia and Latin America, there is no evidence on the prevalence and genetic profile of HBV RAMs in co-infected PLWH. Therefore, we aimed to determine the prevalence of coinfection and the antiviral resistance patterns in the RT region of the HBV pol gene in a cohort of PLWH on HAART in the city of Bucaramanga, province of Santander.
We conducted a descriptive cross-sectional study between February 2013 and February 2014 in the city of Bucaramanga, province of Santander, located in the northeastern region of Colombia. Samples were collected using convenience sampling and included PLWH diagnosed at the IPS Medical Centre in the city. Blood (5 mL) was collected from each participant, along with demographic, clinical and laboratory parameters, including HBV serologic markers determined by enzyme-linked immunosorbent assay, HIV viral load measured with the Real Time HIV-1 Amplification Reagent Kit (Abbott, IL, USA) with a detection range of 50 to 10,000,000 copies/mL, CD4+ T-cell counts, and HAARTcombinations.
Inclusion criteria were as follows: HIV antibody positive confirmed by ELISA and Western Blot, duration of HIV infection over 6 months, and access to patient medical records. Exclusion criteria were as follows: pregnant and breastfeeding women, mental illness and patients under 18 years of age.
Active HBV infection was defined as positive of HBsAg serologic marker by enzyme immunoassay or rapid diagnostic tests in PLWH with or without viral DNA amplification by nested PCR. OBI was confirmed when HBV DNA was amplified by nested PCR in the absence of HBsAg and independent of other HBV serologic markers.
Total DNA was purified from 1 mL of serum using a QIAamp Ultra Sense Virus Kit (QIAgen, MD, USA) according to manufacturer’s instructions. Nucleic acid purity and concentration were determined by absorbance measurements 260/280 nm using a NanoDrop 2000C (Thermo Fisher, CA, USA). DNA aliquots were stored at −80°C until PCR processing.
Detection of HBV co-infection in HIV serum samples was carried out using an in-house nested PCR to amplify a 1,027 bp fragment of the pol gene containing the entire reverse transcriptase (RT) region using primers as described in Table 1. The first round of amplification was performed in a total volume of 25 μL, containing 0.2 μM of each primer, 11.5 μL of sample DNA, and 12.5 μL of enzyme OneTaq Hot Start 2X MM Polymerase (New England Biolabs, MA, USA) according to the manufacturer’s instructions. Cycling conditions were 94°C for 30 s, 35 cycles of 94°C for 15 s, 60°C for 15 s, and 68°C for 2 min, ending with 68°C for 10 min. Amplification was carried out using the ProFlex PCR System (Applied Biosystems, Thermo Fisher, CA, USA). The second round of PCR was performed under the same conditions, except that 1 μL of the first-round product was added to the reaction tubes. PCR products were separated by agarose gel electrophoresis and detected using SYBR Safe DNA Gel Stain and a 1 kb GeneRuler DNA Ladder (Thermo Scientific, CA, USA). Positive samples were confirmed by a second assay using the same conditions as above. Fragments were purified using the QIAquick PCR Purification Kit (QIAgen, MD, USA) and sequenced bidirectionally using the inner primers described in Table 1.
PCR fragments corresponding to the RT domain of the pol gene were sequenced using the Sanger platform 3730XL (Applied Biosystems, Thermo Fisher, CA, USA). Raw HBV DNA sequences from co-infected PLWH and two HBsAg (+) blood donors without prior NRTI treatment were automatically assembled using SeqMan Ultra (DNA Lasergene v17.1). The trimmed sequences were aligned to the consensus derived from representative HBV reference sequences from NCBI using Clustal W software. Local HBV isolates were deposited in the NCBI GenBank under the accession numbers: OQ262971-OQ262995.
HBV genotypes and subgenotypes were determined by phylogenetic inference using Molecular Evolutionary Genetics Analysis (MEGA, V.11) (Kumar et al., 2018). HBV isolates and sequences of reference genotypes and subgenotypes retrieved from GenBank were used for the analysis. The selection of the evolutionary model was based on the lower Bayesian Information Criterion (BIC) score, and the phylogenetic tree was generated by the Maximum Likelihood (ML) method, using the Subtree Pruning Regrafting (SPR) heuristic algorithm. The confidence level of the inferred tree was determined by bootstrapping with 1,000 replicates. The resulting tree was exported in Nowick’s format and edited using FigTree (v. 1.4.4).
HBV resistance and compensatory mutations to NRTIs were identified using the Geno2pheno [HBV] 2.0, drug resistance tool developed by the Max Planck Institute.1 HBV sequences from each co-infected individual were included in data collection. Mutations were assessed at 11 different positions (rtL80, rtI169, rtV173, rtL180, rtA181, rtS184, rtA194, rtS202, rtM204, rtN236, and rtM250), starting at codon 344 within the open reading frame (ORF) of the pol gene. Amino acid substitution profiles associated with INTR resistance were described according to the Clinical Practice Guidelines for the management of HBV infection (Lampertico et al., 2017). The ORF of the HBV envelope (S) gene was also analyzed to identify escape mutations in the “a determinant” within the Major Hydrophilic Region (MHR) and ADAPVEMs. Similarly, the deduced amino acid sequences for the P and S genes were aligned with the consensus sequences obtained from several annotations of the F genotype to exclude genotype-related polymorphisms and variations.
Demographic, clinical, and RAMs data were analyzed for all study participants. Continuous variables were presented as range, median and interquartile range for CD4+ cell counts and HIV viral loads, while categorical variables were presented as percentages. Comparisons between mono-infected and co-infected groups for categorical and continuous variables were assessed using Chi-square and Mann–Whitney tests, respectively. Statistical significance was set at value of p ≤ 0.05. Analyses were performed in Python using the pandas and SciPy Stats libraries.
The study was approved by the Ethics Committee of the Universidad de Santander (approval 015-2012), and written informed consent was obtained from all the participants before sampling and data collection. All procedures performed during the study were in accordance with the Declaration of Helsinki and the Colombian regulations on ethics in clinical research (Resolution 8430 1993).
A total of 275 PLWH were enrolled in the study, with a median age of 38 years (30–44) and 65.1% male. All HIV transmissions were sexually acquired. The majority of the participants were heterosexual (66.2%), followed by homosexual (15.6%) and bisexual (5.8%). HIV infection was classified according to the Centers for Disease Control and Prevention, with 32.4% of patients in C category (AIDS state), 34.5% in category B (symptomatic), and 30.2% in category A (asymptomatic). CD4+ T-cell counts showed that 24.7% of patients had less than 200 cells/mm3, 46.6% had between 200–500 cells/mm3, and 28.7% had more than 500 cells/mm3. Plasma HIV viral load was classified as undetectable (<50 copies/mL) in 43.2% of patients, low (51–10,000 copies/mL) in 34.2% and high (>100,000 copies/mL) in 7.3%. HBV co-infection was confirmed in 32 HIV samples (prevalence 11.6%), of which nine (28.2%) were HBsAg positive (active hepatitis B) and 23 (71.8%) were OBI. Among HIV/HBV co-infected patients, HBV serologic markers showed that 28.1% were HBsAg positive and 40.6% were total anti core antibodies (anti-HBc) positive. Surprisingly, 31.3% had antibodies against HBV S antigen (anti-HBs) and 9.4% were hepatitis B vaccinated. No significant differences were observed between HIV/HBV co-infected and HBV mono-infected patients, except for the positivity of the serological markers anti-HBc and HBsAg (p ≤ 0.05) (Table 2).
At baseline, 89.4% (n = 246) of PLWH were receiving HAART and most combinations (99.6%) included an NRTI. In 156/246 (56.7%) regimens contained NRTI+NNRTI (non-nucleos(t)ides reverse transcriptase inhibitors), of which 62.2% received LAM, Zidovudine (ZDV), and Efavirenz (EFV). The NRTI + PI (protease inhibitors) family of drugs were administered in 56/246 individuals (22.7%). Of these, LAM, ZDV, Lopinavir (LPV), and Ritonavir (RTV) were the most commonly used in 42.9% of patients. The results also highlighted 23 patients receiving more than two molecules from the same family of NRTIs and ten subjects treated with drugs from three different HAART regimens, such as NRTI+NNRTI+PI. Only one participant benefited from the integrase inhibitor Raltegravir (RAL) with the protease inhibitor LPV. In summary, the data show that 38 different HAART regimens are being used in PLWH from Colombia (Table 3).
A total of 24 HIV samples tested positive for HBV using nested PCR, with only one sample showing positive results for HBsAg (patient ID 0119-077). High quality DNA was successfully extracted from 23 samples, allowing the collection of DNA data based on electropherogram profiles for subsequent phylogenetic analyses. Based on evolutionary clustering, all patients were infected with the HBV genotype F3, with clade support after 1,000 bootstrap replicates of 98 and 75%, respectively (Figure 1).
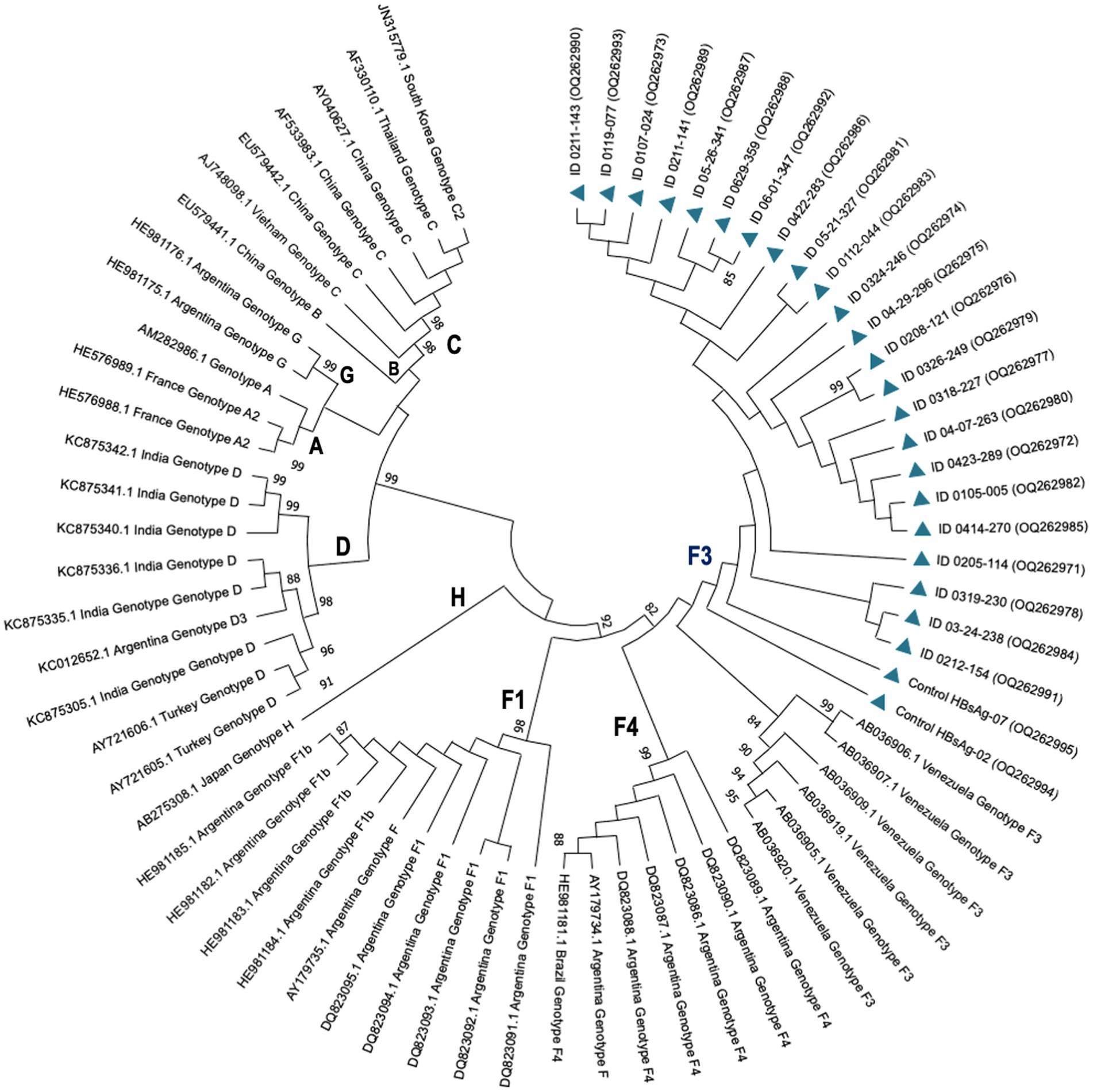
Figure 1. Phylogenetic tree showing HBV genotypes and subgenotypes by maximum likelihood inference. HBV isolates from PLWH in Colombia are represented by codes OQ262971-OQ262995 (blue triangles). Reference strains are represented by their accession numbers as they appear in GenBank followed by the country of isolation and their corresponding genotypes. Probability values ≥0.80 are indicated at the nodes of the tree.
Inferred amino acid sequences at position 27–344 of the HBV RT domain of the pol gene and 21–136 of the envelope (S) gene (partially overlapping pol gene within the MHR) were used for NRTI and HBsAg mutant scape analyses. All HBV sequences (n = 23) showed resistance and/or compensatory mutations that were typically associated with prolonged NRTI treatment. The most common amino acid substitution profiles are shown in Table 4 and are as follows: rt80I, rt180M, rt204V in 91.3% and rt180M, rt204V in 4.3%, both profiles associated with cross-resistance to LAM and Telbivudine (LdT) and partial resistance to Entecavir (ETV) (Figure 2). In addition, rt236Y was detected as an unknown mutation in TDF. ADAPVEMs in the envelope (S) gene were observed in all isolates ranging from 1–20 amino acid substitutions (Figure 3). However, no vaccine escape mutants were detected.
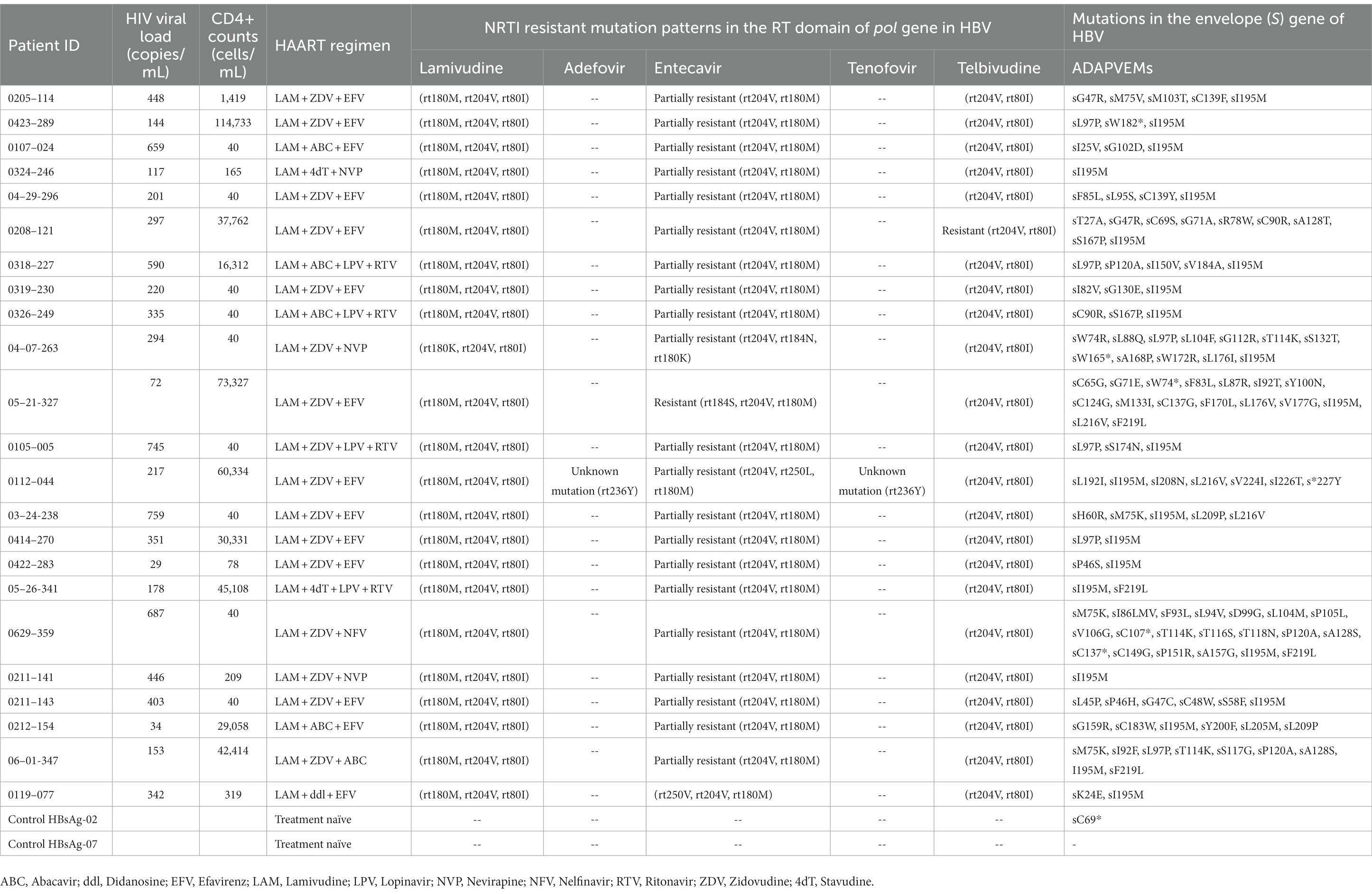
Table 4. NRTI resistance and/or compensatory mutations in the RT domain and ADAPVEMs in HBV isolates of PLWH in the Colombian cohort.
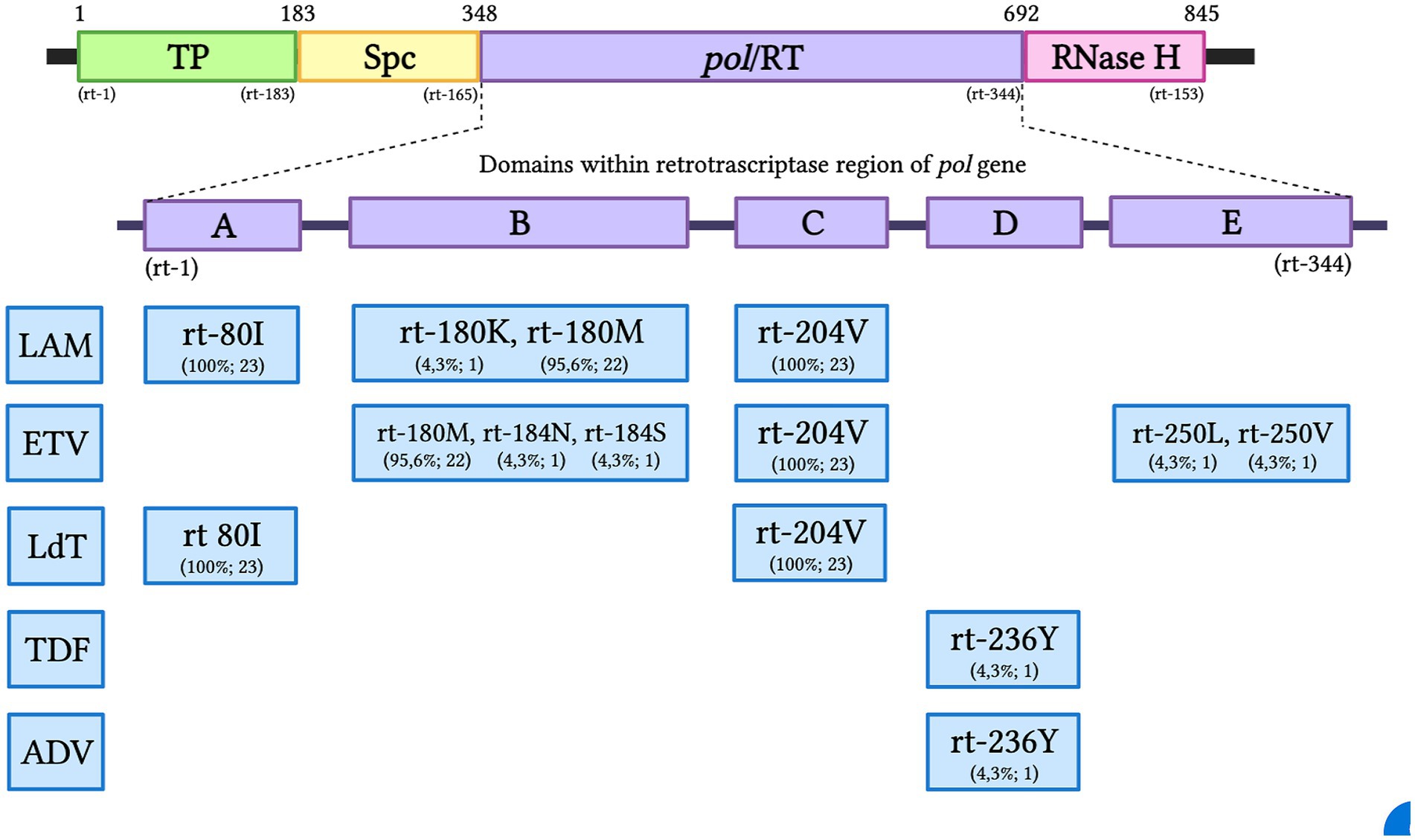
Figure 2. The HBV pol gene with the RT region and the A-E domains of Antiviral Resistance-Associated Mutations (RAMs). The resistance pattern found for each NRTI is presented as a percentage and the total number of subjects in the Colombian HIV/HBV cohort: TP, Terminal Profile; SPC, Space region; Pol/RT, Polymerase/Reverse Transcriptase; LAM, Lamivudine; ETV, Entecavir; LdT, Telbivudine; TDF, Tenofovir; ADV, Adefovir.
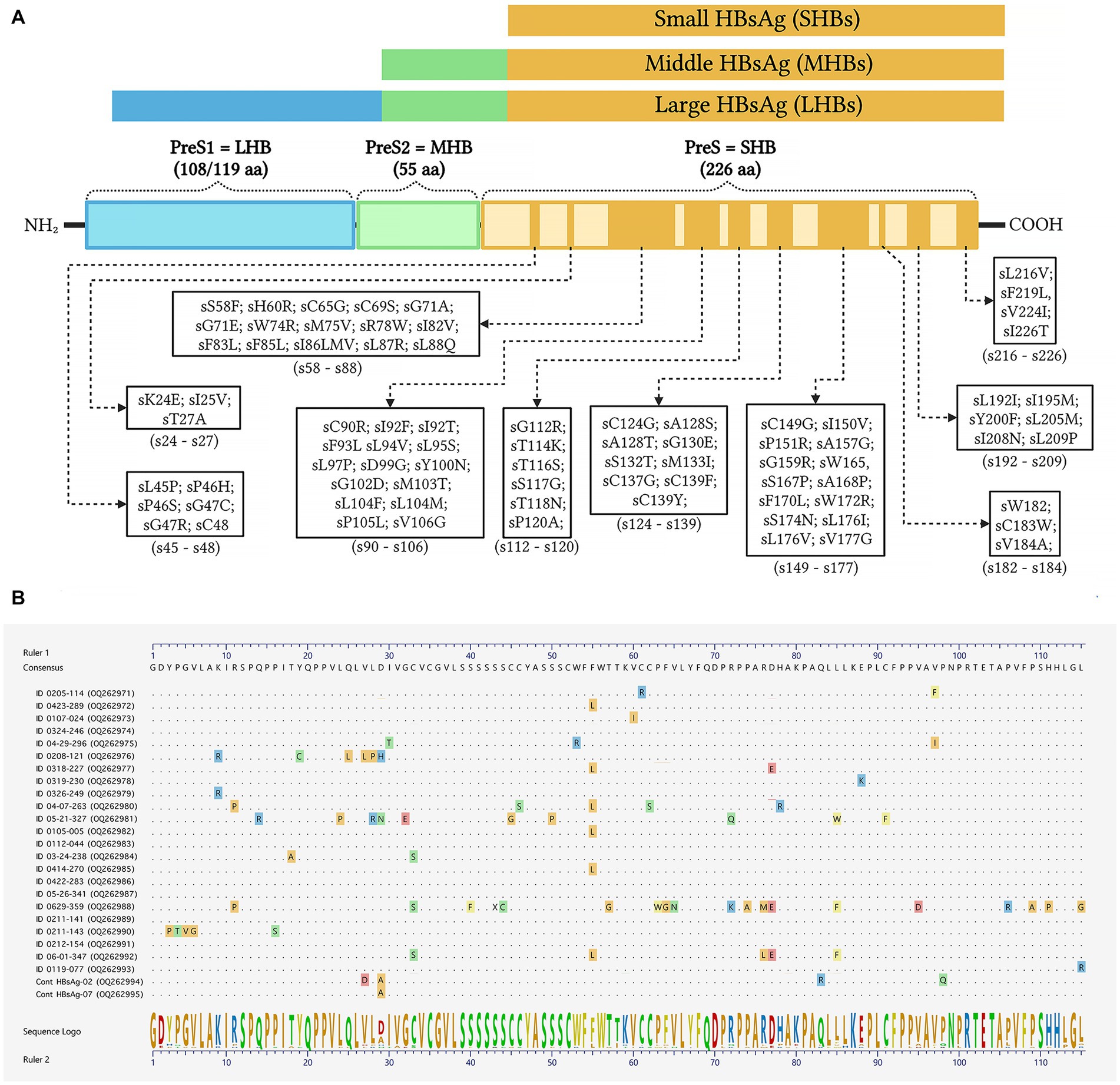
Figure 3. Schematic representation of amino acid substitutions in the HBV envelope/surface proteins. (A) Color bars show the large HBV surface antigen (LHBs), middle surface antigen (MHBs), and small surface antigen (SHBs) with antiviral drug-associated potential vaccine-escape mutations (ADAPVEMs). Each ADAPVEM found in the present study is plotted along the SHBs according to the output of the geno2pheno system outputs. (B) Clustal W alignments of the SHBs at amino acid positions 21–136. The sequences confirmed all amino acid substitutions found in PLWH co-infected with HBV (GenBank accession numbers OQ262971-OQ262995). The consensus amino acid sequence was derived from the same F genotype strains used to construct the phylogenetic tree in Figure 1.
In this cross-sectional study, we found that 11.6% of PLWH in the city of Bucaramanga (Colombia) were co-infected with HBV when the nucleic acid testing was performed on all specimens, regardless of HBV serologic profile. The local prevalence in the northeastern region of Colombia was higher than previously reported in the country (between 2.9 and 3.3%), probably because only the HBsAg test was included in the serological screening (Tengan et al., 2017). Nevertheless, the high frequency of HIV/HBV co-infection reported in the present study was similar to that reported in Latin American countries, such as Brazil and Cuba with10.3% (Corredor et al., 2005; Soares Sampaio et al., 2009) or in the Caribbean with 13.55% in Haiti (Schweitzer et al., 2015). The regional prevalence of HBV in the Americas may have increased in the last five years, as the Pan American Health Organization estimates that the annual infections with the HIV virus rose by 21% from 2010 through 2021 (Pan American Health Organization, 2021).
We found no statistically significant differences in HIV viral load and CD4+ cell counts between HIV/HBV co-infected and mono-infected patients. This observation has been replicated in previous studies (Wondimeneh et al., 2013; Sarkar et al., 2016) but other authors have demonstrated lower baseline CD4+ cell counts, increased HIV viral load, and AIDS-related comorbidities in HIV-HBV co-infected patients [Idoko et al., 2009; Thio et al., 2013; The Canadian Observational Cohort (CANOC) Collaboration et al., 2019]. Lower CD4 counts have been associated with higher levels of HBV viral replication (HBV DNA > 200,000 IU/mL) and may increase the risk of developing HCC (Clifford et al., 2008). In Colombia, hepatopathies are a major cause of morbidity and mortality among PLWH. Future longitudinal studies in a large cohort will allow investigation of the etiology and progression of chronic liver disease.
Here, we reported genotype F3 in all co-infected patients. These results are limited to Colombia, where our group and others have described that genotype F3 was predominant (>75%) in different populations such as blood donors, PLWH, patients with cirrhosis and HCC, followed in a lesser proportion by genotypes F1b, A, and G (Alvarado Mora et al., 2011; Bautista-Amorocho et al., 2014; Duque-Jaramillo et al., 2017). The results also showed that most PLWH were receiving HAART (89.3%), and of these, 56.7% were receiving a combination of NRTI + NNRTI, followed by NRTI + PI (22.8%). In both combinations, 96.7% had LAM as an NRTI drug and none had TDF.
To the best of our knowledge, this is the first study in Colombia to investigate antiviral resistance mutation profiles of the HBV pol gene associated with NRTI treatment. Of the 12 known codons that can confer drug resistance along the conserved RT domain of HBV, seven were identified in the present study. All HBV sequences (n = 23) from PLWH were on HAART and showed mutations conferring clinical resistance to different NRTIs. The most common were the triple amino acid substitutions rt80I, rt180M, and rt204V, which are associated with LAM and LdT resistance and were found in 91.3% of the cases. Interestingly, we also observed the triple combination of antiviral resistance rt80I, rt180M, and rt204A, described as compensatory or secondary mutations in patients undergoing ETV therapy. This pattern of amino acid substitutions can restore RT activity, as a consequence of defects caused by primary resistance (Delaney et al., 2003; Deng and Tang, 2011; Lee et al., 2012). Thus, the replication potential of HBV could persist due to compensatory mutation. A high prevalence of LAM resistance mutations has also been reported in previous studies in South Africa in treatment-naïve and treatment-experienced individuals with CHB and OBI mostly infected with genotype A1 (Selabe et al., 2007). In France, 90% of rt180M and rt204V LAM resistance patterns were estimated in PLWH after 48 months of treatment (Benhamou et al., 1999). Similarly, in an international multi-cohort study, three different resistance mutation patterns (rt204V/I, rt180M plus rt204V/I, and rt173L plus rt180M plus rt204V/I) were observed in 50 and 94% of patients after 2 years and 4 years of LAM-based HAART, respectively (Matthews et al., 2006). Finally, in India, double (rt180M, rt204V) and triple (rt173L, rt180M, rt204V) mutations associated with genotype D were found in HIV/HBV coinfected patients after 24–48 months of antiviral therapy (Pal et al., 2015). In Latin America, there are no previous reports of RAMs to LAM or other NRTIs in HIV/HBV co-infected patients on HAART.
Due to the organization of the HBV genome, the S gene which encodes the envelope proteins (small, medium and large), is completely overlapped by the pol gene. Therefore, mutations in the pol ORF associated with antiviral RAMs, may result in changes in the HBsAg that interfere with the vaccine-derived protection (Torresi, 2002). In patients where LAM has been widely used continuously for several years, scape mutants and/or ADAPVEMs with nucleotide variation within the S gene are likely to occur. The current results show ADAPVEMs in all HIV/HBV co-infections associated with LAM and LdT treatment. However, no vaccine escape mutants were observed. In the present study HBV vaccine coverage was relatively low (17.1%); therefore, NRTI treatment as part of the HIV HAART, has the potential to cause the emergence of ADAPTVEMs and probably spread escape mutants in HBV-infected individuals.
Our study had several limitations. First, HBV viral load and alanine aminotransferase (ALT) were not measured due to insufficient sample volumes. Second, adherence to HAART or duration of treatment could not be confirmed in mono-and co-infected PLWH. Third, a cohort of HIV/HBV antiretroviral treatment-naïve and CHB mono-infected individuals should be included in future studies to characterize natural drug resistance mutations associated with genotype F circulation in Colombia.
The prevalence of HIV/HBV coinfection in Colombian patients was high, mainly associated with OBI cases. Therefore, nucleic acid amplification testing is recommended for HBV screening regardless of HBsAg, anti-HBs and anti-HBc serologic status. Most NRTI-resistant mutant strains correspond to low genetic barrier antivirals such as Lamivudine an Telbivudine. The American Association for the Study of Liver Diseases (AASLD) discourages the use of LAM as the sole antiviral drug to treat HBV infection within the HIV HAART regimen, due to its low potency and the high risk of mutations that confer cross-resistance to other anti-HBV drugs such as Emtricitabine, Telbivudine, and Entecavir (Terrault et al., 2016). Therefore, a high genetic barrier HBV antiviral such as Tenofovir Disoproxil Fumarate or Tenofovir Alafenamide plus Lamivudine is recommended as first-line of HAART in HIV co-infection (Thio et al., 2015; Wandera et al., 2022).
The datasets presented in this study were uploaded in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/genbank/, OQ262971-OQ262995.
The studies involving human participants were reviewed and approved by Ethics Committee of the Universidad de Santander. The patients/participants provided their written informed consent to participate in this study.
HB-A designed the study, recruited the patients, performed the sequencing, bioinformatics analysis, and drafted the manuscript. HB-A, JP-V, and JAS-S performed the experiments and analyzed the data. JAS-S prepared the tables, performed the statistics, and revised the manuscript. JP-V designed the figures. All authors have read and approved the final version of the submitted manuscript.
The authors would like to thank to the staff involved in collecting the blood samples and the volunteer patients enrolled in this study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
This study was funded by Ministerio de Ciencia y Tecnología (MinCiencias) of Colombia, Grant 758-2018 code number 129977757637 and Vicerrectoría de Investigaciones de la Universidad de Santander (UDES).
Alvarado Mora, M. V., Romano, C. M., Gomes-Gouvêa, M. S., Gutierrez, M. F., Botelho, L., Carrilho, F. J., et al. (2011). Molecular characterization of the Hepatitis B virus genotypes in Colombia: a Bayesian inference on the genotype F. Infect. Genet. Evol. 11, 103–108. doi: 10.1016/j.meegid.2010.10.003
Archampong, T. N., Boyce, C. L., Lartey, M., Sagoe, K. W., Obo-Akwa, A., Kenu, E., et al. (2017). HBV Genotypes and drug resistance mutations in antiretroviral treatment-naive and treatment-experienced HBV–HIV-coinfected patients. Antivir. Ther. 22, 13–20. doi: 10.3851/IMP3055
Bautista-Amorocho, H., Castellanos-Domínguez, Y. Z., Rodríguez-Villamizar, L. A., Velandia-Cruz, S. A., Becerra-Peña, J. A., and Farfán-García, A. E. (2014). Epidemiology, risk factors and genotypes of HBV in HIV-infected patients in the Northeast Region of Colombia: high prevalence of occult Hepatitis B and F3 Subgenotype Dominance. PLoS One 9:e114272. doi: 10.1371/journal.pone.0114272
Benhamou, Y., Bochet, M., Thibault, V., di Martino, V., Caumes, E., Bricaire, F., et al. (1999). Long-term incidence of hepatitis B virus resistance to lamivudine in human immunodeficiency virus-infected patients. Hepatology 30, 1302–1306. doi: 10.1002/hep.510300525
Clifford, G. M., Rickenbach, M., Polesel, J., Dal Maso, L., Steffen, I., Ledergerber, B., et al. (2008). Influence of HIV-related immunodeficiency on the risk of hepatocellular carcinoma. AIDS 22, 2135–2141. doi: 10.1097/QAD.0b013e32831103ad
Corredor, B., Marité, L. L., de Los, A. R., Moreno, A. G., Frómeta, S. S., MCM, V., et al. (2005). Detection of hepatitis B and hepatitis C markers in HIV positive patients, 2000-2004. Rev. Cubana Med. Trop. 57, 212–213.
CAC. (2021). Fondo Colombiano de Enfermedades de Alto Costo. In: Situación Del VIH y Sida En Colombia 2021. Available at: https://cuentadealtocosto.org/site/publicaciones/situacion-del-vih-y-sida-en-colombia-2021/ (Accessed June 25, 2023).
Delaney, W. E., Yang, H., Westland, C. E., Das, K., Arnold, E., Gibbs, C. S., et al. (2003). The Hepatitis B virus polymerase Mutation rt V173L is selected during lamivudine therapy and enhances viral replication in vitro. J. Virol. 77, 11833–11841. doi: 10.1128/JVI.77.21.11833-11841.2003
Deng, L., and Tang, H. (2011). Hepatitis B virus drug resistance to current nucleos(t)ide analogs: mechanisms and mutation sites. Hepatol. Res. 41, 1017–1024. doi: 10.1111/j.1872-034X.2011.00873.x
Duque-Jaramillo, A., Rendón, J., Cortés-Mancera, F., Correa, G., Restrepo, J. C., and Navas, M. C. (2017). Infección oculta por el virus de la hepatitis B en pacientes sometidos a trasplante hepático. Rev. Colomb. Gastroenterol. 31:347. doi: 10.22516/25007440.109
He, X., Wang, F., Huang, B., Chen, P., and Zhong, L. (2015). Detection and analysis of resistance mutations of hepatitis B virus. Int. J. Clin. Exp. Med. 8, 9630–9639.
Idoko, J., Meloni, S., Muazu, M., Nimzing, L., Badung, B., Hawkins, C., et al. (2009). Impact of hepatitis b virus infection on human immunodeficiency virus response to antiretroviral therapy in Nigeria. Clin. Infect. Dis. 49, 1268–1273. doi: 10.1086/605675
Lee, Y. S., Chung, Y. H., Kim, J. A., Jin, Y. J., Park, W. H., Kim, S. E., et al. (2012). rt L180M mutation of hepatitis B virus is closely associated with frequent virological resistance to adefovir dipivoxil therapy. J. Gastroenterol. Hepatol. 27, 300–305. doi: 10.1111/j.1440-1746.2011.06853.x
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lampertico, P., Agarwal, K., Berg, T., Buti, M., Janssen, H. L. A., Papatheodoridis, G., et al. (2017). EASL 2017 clinical practice guidelines on the management of hepatitis B virus infection. J. Hepatol. 67, 370–398. doi: 10.1016/j.jhep.2017.03.021
Martín-Carbonero, L., Teixeira, T., Poveda, E., Plaza, Z., Vispo, E., González-Lahoz, J., et al. (2011). Clinical and virological outcomes in HIV-infected patients with chronic hepatitis B on long-term nucleos(t) ide analogues. AIDS 25, 73–79. doi: 10.1097/QAD.0b013e328340fde2
Matthews, G. V., Bartholomeusz, A., Locarnini, S., Ayres, A., Sasaduesz, J., Seaberg, E., et al. (2006). Characteristics of drug resistant HBV in an international collaborative study of HIV-HBV-infected individuals on extended lamivudine therapy. AIDS 20, 863–870. doi: 10.1097/01.aids.0000218550.85081.59
Mocroft, A., Ledergerber, B., Katlama, C., Kirk, O., Reiss, P., d'Arminio Monforte, A., et al. (2003). Decline in the AIDS and death rates in the EuroSIDA study: an observational study. Lancet 362, 22–29. doi: 10.1016/S0140-6736(03)13802-0
Pal, A., Sarkar, N., Saha, D., Guha, S. K., Saha, B., Chakrabarti, S., et al. (2015). High incidence of lamivudine-resistance-associated vaccine-escape HBV mutants among HIV-coinfected patients on prolonged antiretroviral therapy. Antivir. Ther. 20, 545–554. doi: 10.3851/IMP2942
Palella, F. J., Delaney, K. M., Moorman, A. C., Loveless, M. O., Fuhrer, J., Satten, G. A., et al. (1998). Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. N. Engl. J. Med. 338, 853–860. doi: 10.1056/NEJM199803263381301
Pan American Health Organization. (2021). HIV epidemic and response in Latin America and the Caribbean. Tech Reports. Available at: https://www.paho.org/en/documents/hiv-epidemic-and-response-latin-america-and-caribbean-october-2022 (Accessed June 25, 2023).
Platt, L., French, C. E., McGowan, C. R., et al. (2020). Prevalence and burden of HBV co-infection among people living with HIV: A global systematic review and meta-analysis. J. Viral Hepat. 27, 294–315. doi: 10.1111/jvh.13217
Raimondo, G., Locarnini, S., Pollicino, T., Levrero, M., Zoulim, F., and Lok, A. S. (2019). Taormina workshop on occult HBV infection faculty members update of the statements on biology and clinical impact of occult hepatitis B virus infection. J. Hepatol. 71, 397–408. doi: 10.1016/j.jhep.2019.03.034
Saag, M. S., Benson, C. A., Gandhi, R. T., Hoy, J. F., Landovitz, R. J., Mugavero, M. J., et al. (2018). Antiretroviral drugs for treatment and prevention of HIV infection in adults: 2018 recommendations of the international antiviral society-USA panel. JAMA 320, 379–396. doi: 10.1001/jama.2018.8431
Sarkar, J., Saha, D., Bandyopadhyay, B., Saha, B., Kedia, D., Mazumder, D. N. G., et al. (2016). Baseline characteristics of HIV & amp; hepatitis B virus (HIV/HBV) co-infected patients from Kolkata, India. Indian J. Med. Res. 143:636. doi: 10.4103/0971-5916.187113
Schweitzer, A., Horn, J., Mikolajczyk, R. T., Krause, G., and Ott, J. J. (2015). Estimations of worldwide prevalence of chronic hepatitis B virus infection: a systematic review of data published between 1965 and 2013. Lancet 386, 1546–1555. doi: 10.1016/S0140-6736(15)61412-X
Selabe, S. G., Lukhwareni, A., Song, E., Leeuw, Y. G. M., Burnett, R. J., and Mphahlele, M. J. (2007). Mutations associated with lamivudine-resistance in therapy-naïve hepatitis B virus (HBV) infected patients with and without HIV co-infection: Implications for antiretroviral therapy in HBV and HIV co-infected South African patients. J. Med. Virol. 79, 1650–1654. doi: 10.1002/jmv.20974
Shire, N. J., and Sherman, K. E. (2005). Management of HBV/HIV-Coinfected Patients. Semin. Liver Dis. 25, 48–57. doi: 10.1055/s-2005-915646
Soares Sampaio, A., Arraes de Alencar, L. C., Moura, P. M. M. F. D., Barros Correia, J. D., Barros Barreto, S. D., and Castelo, A. (2009). Prevalencia de la co-infección con hepatitis B y C en pacientes HIV positivos y factores de riesgo asociados. Actual. SIDA 12–17.
Tanser, F., Baernighausen, T., Graspa, E., Zaidi, J., and Newell, M.-L. (2013). High coverage of ART associated with decline in risk of HIV acquisition in rural Kwa Zulu-Natal, South Africa. Science (80-) 339, 966–972. doi: 10.1126/science.1228160
Tengan, F. M., Abdala, E., Nascimento, M., Bernardo, W. M., and Barone, A. A. (2017). Prevalence of hepatitis B in people living with HIV/AIDS in Latin America and the Caribbean: a systematic review and meta-analysis. BMC Infect. Dis. 17:587. doi: 10.1186/s12879-017-2695-z
Terrault, N. A., Bzowej, N. H., Chang, K. M., Hwang, J. P., Jonas, M. M., and Murad, M. H. (2016). AASLD guidelines for treatment of chronic hepatitis B. Hepatology 63, 261–283. doi: 10.1002/hep.28156
The Canadian Observational Cohort (CANOC) CollaborationRana, U., Driedger, M., Sereda, P., Pan, S., Ding, E., et al. (2019). Characteristics and outcomes of antiretroviral-treated HIV-HBV co-infected patients in Canada. BMC Infect. Dis. 19:982. doi: 10.1186/s12879-019-4617-8
Thio, C. L., Seaberg, E. C., Skolasky, R. Jr., Phair, J., Visscher, B., Muñoz, A., et al. (2002). HIV-1, hepatitis B virus, and risk of liver-related mortality in the Multicenter Cohort Study (MACS). Lancet 360, 1921–1926. doi: 10.1016/S0140-6736(02)11913-1
Thio, C. L., Smeaton, L., Hollabaugh, K., Saulynas, M., Hwang, H., Saravanan, S., et al. (2015). Comparison of HBV-active HAART regimens in an HIV-HBV multinational cohort: outcomes through 144 weeks. AIDS 29, 1173–1182. doi: 10.1097/QAD.0000000000000686
Thio, C. L., Smeaton, L., Saulynas, M., Hwang, H., Saravan, S., Kulkarni, S., et al. (2013). Characterization of HIV–HBV coinfection in a multinational HIV-infected cohort. AIDS 27, 191–201. doi: 10.1097/QAD.0b013e32835a9984
Torresi, J. (2002). The virological and clinical significance of mutations in the overlapping envelope and polymerase genes of hepatitis B virus. J. Clin. Virol. 25, 97–106. doi: 10.1016/S1386-6532(02)00049-5
UNAIDS. (2023) Global HIV & AIDS statistics — Fact sheet. 1–5. Available at: https://www.unaids.org/en/resources/fact-sheet
Vallet-Pichard, A., and Pol, S. (2004). Hepatitis viruses and human immunodeficiency virus co-infection: pathogenesis and treatment. J. Hepatol. 41, 156–166. doi: 10.1016/j.jhep.2004.05.001
Vittinghoff, E., Scheer, S., O’Malley, P., Colfax, G., Holmberg, S. D., and Buchbinder, S. P. (1999). Combination antiretroviral therapy and recent declines in AIDS incidence and mortality. J. Infect. Dis. 179, 717–720. doi: 10.1086/314623
Wandera, B. O., Onyango, D. M., and Musyoki, S. K. (2022). Hepatitis B virus genetic multiplicity and the associated HBV lamivudine resistance mutations in HBV/HIV co-infection in Western Kenya: a review article. Infect. Genet. Evol. 98:105197. doi: 10.1016/j.meegid.2021.105197
WHO (2021). Global progress report on HIV, viral hepatitis and sexually transmitted infections. Available at: https://www.who.int/publications/i/item/9789240027077 (Accessed December 4, 2021)
Keywords: HIV, HBV, coinfection, NRTI, antiviral, resistance, mutation
Citation: Bautista-Amorocho H, Silva-Sayago JA and Picón-Villamizar J (2023) High frequency of Lamivudine and Telbivudine resistance mutations in hepatitis B virus isolates from human immunodeficiency virus co-infected patients on highly active antiretroviral therapy in Bucaramanga, Colombia. Front. Microbiol. 14:1202342. doi: 10.3389/fmicb.2023.1202342
Received: 08 April 2023; Accepted: 07 June 2023;
Published: 24 July 2023.
Edited by:
Min Yao, Nantong University, ChinaReviewed by:
Lorenzo Piermatteo, University of Rome Tor Vergata, ItalyCopyright © 2023 Bautista-Amorocho, Silva-Sayago and Picón-Villamizar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Henry Bautista-Amorocho, aGVucnliYXUzM0BnbWFpbC5jb20=
†ORCID: Henry Bautista-Amorocho, http://orcid.org/0000-0001-7746-6376
Jorge Alexander Silva-Sayago, http://orcid.org/0000-0001-8147-6374
Jirehl Picón-Villamizar, https://orcid.org/0009-0000-3292-0264
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.