
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 24 May 2023
Sec. Microbe and Virus Interactions with Plants
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1201444
This article is part of the Research TopicBacterial Wilt: Pathogenic Mechanism, Disease Control, Bacteria-Plant and Bacteria-Environmental Microorganism InteractionsView all 6 articles
 Xue Ouyang1†
Xue Ouyang1† Jialan Chen1†
Jialan Chen1† Zhimao Sun2†
Zhimao Sun2† Rongbo Wang3†
Rongbo Wang3† Xuan Wu1
Xuan Wu1 Benjin Li3
Benjin Li3 Congfeng Song1*
Congfeng Song1* Peiqing Liu3*
Peiqing Liu3* Meixiang Zhang2*
Meixiang Zhang2*As one of the most destructive bacterial phytopathogens, Ralstonia solanacearum causes substantial annual yield losses of many important crops. Deciphering the functional mechanisms of type III effectors, the crucial factors mediating R. solanacearum-plant interactions, will provide a valuable basis for protecting crop plants from R. solanacearum. Recently, the NEL (novel E3 ligase) effector RipAW was found to induce cell death on Nicotiana benthamiana in a E3 ligase activity-dependent manner. Here, we further deciphered the role of the E3 ligase activity in RipAW-triggered plant immunity. We found that RipAWC177A, the E3 ligase mutant of RipAW, could not induce cell death but retained the ability of triggering plant immunity in N. benthamiana, indicating that the E3 ligase activity is not essential for RipAW-triggered immunity. By generating truncated mutants of RipAW, we further showed that the N-terminus, NEL domain and C-terminus are all required but not sufficient for RipAW-induced cell death. Furthermore, all truncated mutants of RipAW triggered ETI immune responses in N. benthamiana, confirming that the E3 ligase activity is not essential for RipAW-triggered plant immunity. Finally, we demonstrated that RipAW- and RipAWC177A-triggered immunity in N. benthamiana requires SGT1 (suppressor of G2 allele of skp1), but not EDS1 (enhanced disease susceptibility), NRG1 (N requirement gene 1), NRC (NLR required for cell death) proteins or SA (salicylic acid) pathway. Our findings provide a typical case in which the effector-induced cell death can be uncoupled with immune responses, shedding new light on effector-triggered plant immunity. Our data also provide clues for further in-depth study of mechanism underlying RipAW-induced plant immunity.
Plant diseases cause substantial yield losses of crops every year (Savary et al., 2019). Deciphering the molecular mechanisms underlying the plant-pathogen interactions will provide a valuable basis for protecting crops from these pathogens. During plant-pathogen interactions, pathogens usually secrete effectors as powerful weapons of invasion to facilitate their infection. However, after long-term confrontation with pathogens, plants have evolved effector-triggered immunity (ETI) accordingly to recognize pathogens, triggering hypersensitive response (HR) to restrict pathogen invasion (Jones and Dangl, 2006).
ETI is mediated by resistance protein (R protein) which is a kind of highly sensitive immune receptor (Jones and Dangl, 2006; Ngou et al., 2022a,b). Nucleotide-binding domain leucine-rich repeat receptors (NLRs), are the largest class of R proteins, and they are grouped into three categories according to their N-terminal signal transduction domains: Toll/interleukin-1 receptor/resistance protein (TIR) NLRs (TNLs), coiled-coil (CC) NLRs (CNLs) and RPW8-like CC domain (RPW8) NLRs (RNLs) (Jones et al., 2016). SGT1 (suppressor of G2 allele of skp1), an essential component of ETI, is required for plant immunity triggered by most NLRs by regulating their stability and activation (Azevedo et al., 2002; Muskett and Parker, 2003; Kadota et al., 2010). Except SGT1, EDS1 (enhanced disease susceptibility) also plays an important role in TNL-mediated immunity (Wiermer et al., 2005; Schultink et al., 2017). RNLs, such as NRG1 (N requirement gene 1) and NRC (NLR required for cell death), do not directly recognize pathogen effector proteins, but usually act downstream of TNLs and CNLs to activate immune signals and induce cell death, respectively (Peart et al., 2005; Wu et al., 2017), so they are called helper NLRs. The major defense hormone salicylic acid (SA) also plays an essential role in regulating plant immunity and programed cell death (Fu et al., 2012) and its signaling is well-integrated into NLR signaling pathways (Zhou and Zhang, 2020).
The bacterial pathogen Ralstonia solanacearum, one of the most destructive phytopathogens, can infect more than 200 plant species over a broad geographical range (Genin and Denny, 2012). The extensive genetic diversity of R. solanacearum strains causes serious bacterial wilt diseases, leading to great economic losses of many important crops worldwide every year (Qi et al., 2022). R. solanacearum secretes more than 70 type III effectors (T3Es) into plant cells as weapon to promote infection (Genin and Denny, 2012). Increasing studies showed that some R. solanacearum T3Es are endowed with enzymatic activities which function in its interaction with plants, such as RipTPS with trehalose-phosphate synthase activity (Poueymiro et al., 2014) and RipN with nudix hydrolase activity (Sun et al., 2019). Strikingly, 13 effectors in R. solanacearum have ubiquitin ligase structural domains and probably are endowed with ubiquitin ligase activity (Peeters et al., 2013). The presence of such a high abundance of ubiquitin ligases implies that ubiquitination may be an important mechanism by which R. solanacearum T3Es function to influence plant immunity.
Ubiquitination is a highly conserved eukaryote-specific post-translational modification of protein and plays an important role in regulating protein function (Rohde et al., 2007). The process of ubiquitin modification is mediated by a ubiquitin-activating enzyme (E1), a limited number of ubiquitin-conjugating enzymes (E2s) and lots of ubiquitin-ligating enzymes (E3s) (Rytkonen and Holden, 2007). E3 ubiquitin ligases are responsible for the specific recognition of target proteins and their subsequent ubiquitination, and therefore are considered as the most important component of ubiquitination (Buetow and Huang, 2016). According to the domains it contains and the way it binds to substrates, E3 ubiquitin ligases are classified into HECT (homologous to the E6-AP carboxyl terminus), U-box, RING (really interesting new gene), CRLs (cullin-RING ligases) and NEL (novel E3 ligase) (Vierstra, 2009). Among these, the NEL effector usually contains a typical LRR motif and cysteines that are essential for its ubiquitin ligase activity (Singer et al., 2013). Many effectors with NEL structural domains have been reported to affect plant immunity. For example, IpaH9.8 in Shigella dysenteriae degrades guanylate-binding protein (GBP) in host cells by ubiquitination, suppressing host immune response and hence promoting pathogen invasion (Li et al., 2017). RipV2 in R. solanacearum UW551 strain and RipAW in RS1000 strain depend on their NEL activities to inhibit plant PTI response and facilitate R. solanacearum infection in hosts (Cheng et al., 2021).
Recently, Niu et al. (2022) demonstrates that RipAW in R. solanacearum GMI1000 strain induces cell death in Nicotiana benthamiana and this process requires its E3 ligase activity. However, cell death is not always coupled with immune responses (Greenberg et al., 2000; Lapin et al., 2019; Yang et al., 2022). To further decipher the relationship of RipAW-triggered immunity with its NEL activity, we determined plant immunity triggered by RipAWC177A (the E3 ligase mutant) and RipAW’s truncated derivatives including RipAWN1-90, RipAWNEL, RipAWC322-448, RipAW ΔN and RipAW ΔC. We found that the E3 ligase activity was not essential for RipAW-triggered immunity. Furthermore, we explored the mechanism how RipAW triggered plant immunity by defining the roles of important immune regulators such as SGT1, EDS1, NRG1 and NRC in RipAW-triggered immunity. Our data provide basis for further in-depth study of the mechanisms behind RipAW-triggered plant immunity.
Wild-type N. benthamiana and eds1, nrg1 (Qi et al., 2018), nrc2/3/4 (Kourelis et al., 2022) mutants were grown in a walk-in chamber at 24°C under long-day conditions (16 h light/8 h dark). Escherichia coli strain DH5α and Agrobacterium tumefaciens strain GV3101 were used for cloning and gene expression, respectively. They were cultured on Luria-Bertani (LB) agar plates or in LB liquid medium with appropriate antibiotics at 37°C and 28°C, respectively. R. solanacearum strain CQPS-1 (Liu et al., 2017) was cultured at 28°C on Bacto-agar and glucose (BG) medium. Pseudomonas syringae strain DC3000 ΔhopQ1-1 (Wei et al., 2007) was cultured at 28°C on Luria-Marine (LM) agar medium plate.
The nucleic acid sequence and amino acid sequence of RipAW in R. solanacearum strain GMI1000 were obtained from NCBI1 and analyzed using SMART2. All the DNA fragments used to generate overexpression and silencing constructs were amplified by PCR from cDNA of N. benthamiana or genomic DNA of R. solanacearum. RipAW and its derivatives were cloned into pGWB505 or pCambia2300. The RipAWC177A mutant was generated by site-directed mutagenesis using corresponding primers. Fragments for virus-induced gene silencing (VIGS) were cloned into vector pTRV2. All the primers used for this experiment were listed in Supplementary Table S1.
The A. tumefaciens cells harboring the corresponding plasmids were suspended in an infiltration buffer containing 150 mM acetosyringone, 10 mM MgCl2 and 10 mM MES (pH 5.6), and adjusted to an OD600 of 0.5. The inoculum preparations were incubated at 28°C for 2 h. The inoculum preparations were then infiltrated into leaves of five-week-old N. benthamiana using a needless syringe. The infiltrated leaves were photographed with a digital camera at 48 or 72 h post infiltration to visualize cell death triggered by effectors.
Ralstonia solanacearum strain CQPS-1 and P. syringae strain DC3000 ΔhopQ1-1 were grown in BG and LM liquid medium, respectively, at 28°C overnight. Bacterial cells were collected by centrifugation, washed with sterile water and adjusted to a final OD600 of 0.001 for CQPS-1 and 0.00002 for DC3000 ΔhopQ1-1. RipAW or its derivatives including RipAWC177A, RipAWN1-90, RipAWNEL, RipAWC322-448, RipAW ΔN and RipAW ΔC was transiently expressed in N. benthamiana leaves. LTI6b, a small intrinsic protein not related to plant immunity (Martinière et al., 2011), was commonly used as the control in plant immunity study (Lee et al., 2020). Therefore, LTI6b was transiently expressed as a control. The leaves were inoculated with CQPS-1 or DC3000 ΔhopQ1-1 48 h post-infiltration, and the bacterial titers were measured at 2–3 days post-inoculation (dpi).
Total RNA was extracted using the RNAsimple Total RNA Extraction Kit (TIANGEN), and quantitative expression of the defense-related genes NbHIN1 and NbPR1a was determined by qRT-PCR with SYBR Green PCR Master Mix as previously described (An et al., 2022). NbEF1α was selected as an internal control. The primers used for amplification were listed in Supplementary Table S1.
Total soluble proteins were extracted from N. benthamiana leaves expressing RipAW, RipE1, INF1, NahG or RipAW’s derivatives. Proteins were separated by SDS-PAGE and transferred onto PVDF membranes (Thermo Fisher) which were then blocked using 5% defatted milk in TBST buffer (1× TBS containing 0.05% Tween 20) for 60 min at room temperature. The membranes were incubated with anti-FLAG (1:8000, Sigma), anti-HA (1:10000, Abmart) or anti-GFP (1:8000, ABclonal) at room temperature for 1 h and then with a horseradish peroxidase-conjugated secondary antibody (1:10000, Sigma) for another 45 min. Signals were detected and photographed using the SuperSignal West Pico Chemiluminescent substrate (Thermo Scientific) and the Bio-Rad ChemiDoc Touch Imaging System.
The extent of plant cell death can be quantified by electrolyte leakage according to Sang et al. (2020). After 36 h of transient expression of effectors in N. benthamiana leaves, leaf discs were collected with a hole punch (1.2 cm in diameter) and immersed in 5 mL of sterile water. After 1 h of gentle shaking at room temperature, the ion conductivity of the water measured using a conductivity meter Waterproof Pocket Tester (Oakton). Three replicates were set up in parallel for each group of experiments.
VIGS assay using TRV (tobacco rattle virus) vectors in N. benthamiana was carried out according to Senthil-Kumar and Mysore (2014). For TRV-mediated gene silencing, A. tumefaciens cultures expressing TRV2::NbSGT1 and those expressing TRV1 were mixed at 1:1 ratio to a final OD600 of 1.0, and infiltrated into the first two true leaves of two-week-old N. benthamiana. After being covered with plastic film to keep high humidity for 2 days, plants were cultured under normal conditions. TRV2::GFP was generated as control plants. Three-week-old plants after VIGS were used for corresponding assays.
All experiments were repeated for at least three times with similar results. Statistical analysis was performed using Student’s t-test along with analysis of variance to discriminate statistical differences between treatments.
RipAW is a new member of the NEL family (Nakano et al., 2017), and it has been reported that its E3 ligase activity is required for RipAW-induced cell death in N. benthamiana (Niu et al., 2022). Here, RipAW in R. solanacearum strain GMI1000 induced cell death in N. benthamiana, but its E3 ligase mutant RipAWC177A could not (Figure 1A). Western blot analysis using the anti-FLAG antibody confirmed the expression of RipAW and RipAWC177A proteins (Figure 1B). These results confirmed the essential role of its E3 ligase activity in RipAW-induced cell death. To further define the role of E3 ligase activity in RipAW-triggered immunity, we detected the expression of two famous defense-related genes NbHIN1 and NbPR1a (Delaney et al., 1994; Gopalan et al., 1996) in N. benthamiana leaves expressing RipAW or RipAWC177A. Compared to the control (LTI6b), RipAW expression extremely increased the expression level of NbHIN1 and NbPR1a (Figure 1C). Unexpectedly, expressing RipAWC177A also significantly promoted expression of NbHIN1 and NbPR1a, although not as high as RipAW did. In addition, we evaluated whether the overexpression of RipAWC177A affected plant resistance to pathogens. After expression of RipAWC177A in N. benthamiana, tobacco leaves were inoculated with R. solanacearum strain CQPS-1 or P. syringae strain DC3000 ΔhopQ1-1. The result showed that RipAWC177A expression significantly restricted the growth of CQPS-1 (Figure 1D) and DC3000 ΔhopQ1-1 (Figure 1E). These results indicate that the E3 ligase activity of RipAW is not essential for induction of plant immunity.
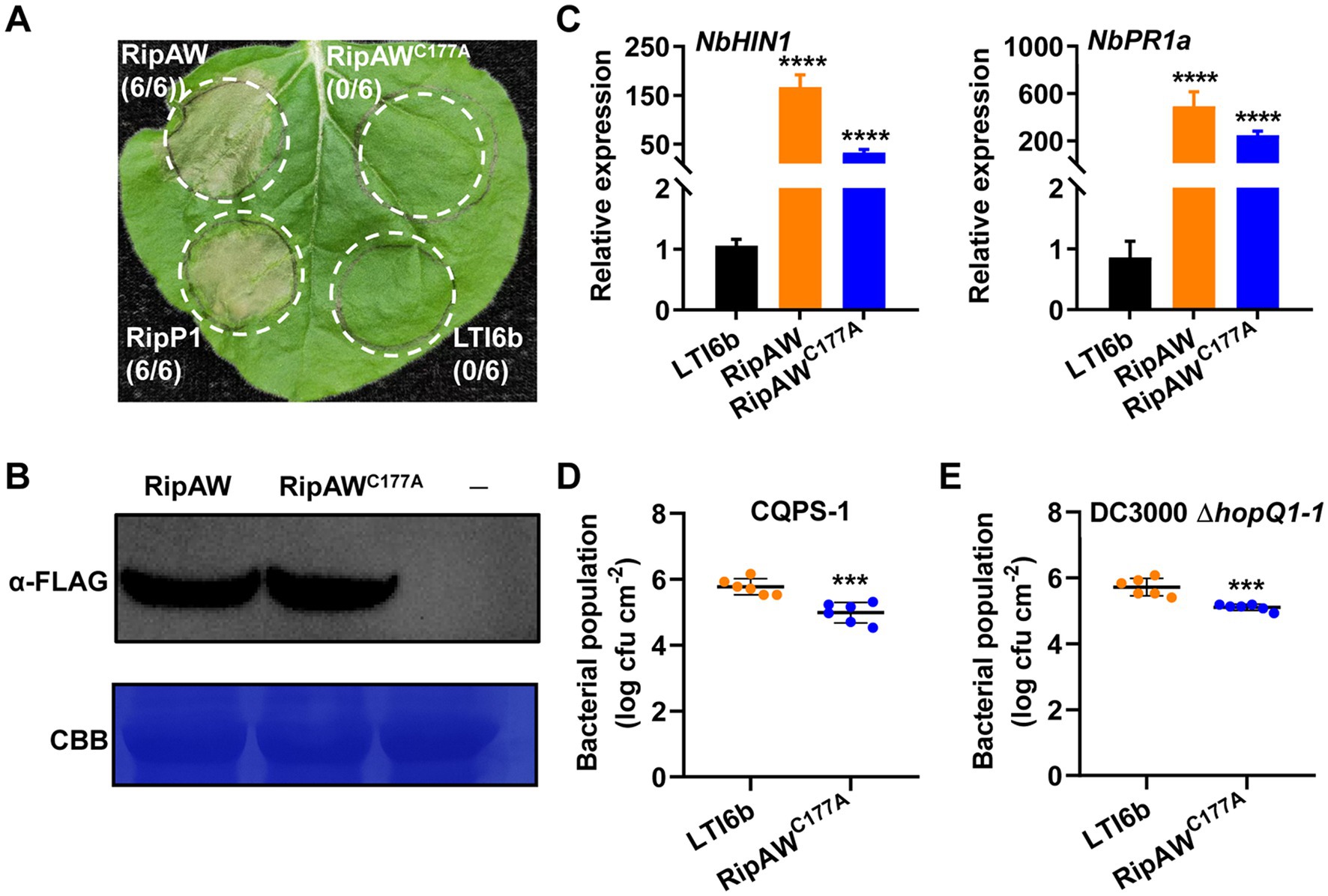
Figure 1. Plant immunity triggered by RipAWC177A in Nicotiana benthamiana. (A) The RipAWC177A mutant failed to induce cell death in N. benthamiana. RipAW and RipAWC177A were expressed in the same leaf of N. benthamiana. RipP1 and LTI6b were the positive control and negative control, respectively. Photographs were taken 2 days post inoculation (dpi). Circles indicate the infiltrated areas. The fractions in brackets represent the number of leaves displaying HR over the total number of infiltrated leaves. (B) Detection of RipAW and RipAWC177A protein expression by western blot using anti-FLAG antibody. Total protein was extracted from the leaves 2 days after agroinfiltration and detected by western blot. The blot was stained with Coomassie brilliant blue (CBB) to confirm equal loading. (C) Expression of defense-related genes NbHIN1 and NbPR1a in N. benthamiana leaves expressing LTI6b (control), RipAW, or RipAWC177A by qRT–PCR. (D,E) Growth of Ralstonia solanacearum CQPS-1 (D) and Pseudomonas syringae DC3000 ΔhopQ1-1 (E) in N. benthamiana leaves expressing RipAWC177A and LTI6b. RipAWC177A and LTI6b were agroinfiltrated into N. benthamiana leaves. 48 h later, CQPS-1 or DC3000 ΔhopQ1-1 was infiltrated into the same areas. The bacterial population was determined at 2 dpi for CQPS-1 and 3 dpi for DC3000 ΔhopQ1-1. Values are means ± standard errors (SEs) from six biological replicates. *** and **** indicate significant differences at p ≤ 0.001 and p ≤ 0.0001, respectively.
Except the NEL domain, Niu et al. (2022) reported that the N terminus of RipAW is also required for induction of cell death in N. benthamiana. To further determine the functional domains in RipAW, we constructed a series of truncated mutants of RipAW. The NEL domain of RipAW is located at amino acids 97–306 (Nakano et al., 2017). We generated mutants including RipAWN1-90, RipAWNEL, RipAWC322-448, N-terminal and C-terminal deletion mutants of RipAW, namely RipAW ΔN and RipAW ΔC, which deleted the N terminus and C terminus of RipAW, respectively (Figure 2A). The above derivatives, as well as wild-type RipAW and RipAWC177A, were expressed in the leaves of N. benthamiana. We found that, except for the full-length RipAW which induced strong cell death, none of its derivatives could induce cell death in N. benthamiana at 3 dpi (Figure 2B). Electrolyte leakage measurement showed that only full-length RipAW induced higher ion leakage, but its derivatives did not (Figure 2C), confirming the cell death phenotypes. Western blot analysis confirmed the expression of RipAW and its derivatives in N. benthamiana, except RipAWN1-90 which was too small to be detected (Figure 1D). These results indicate that not only the N terminus, the NEL domain but also the C terminus is required for RipAW-induced cell death.
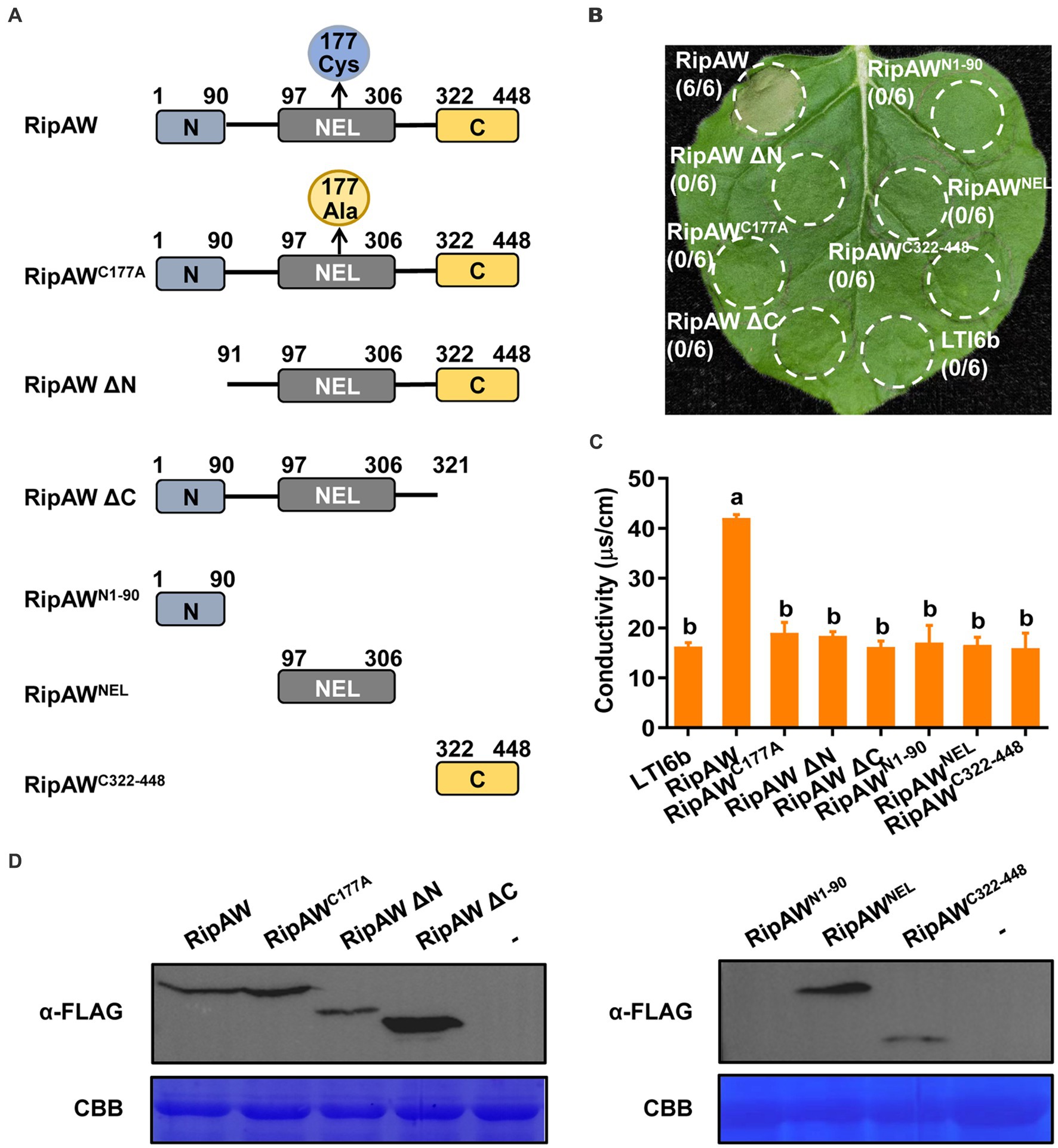
Figure 2. Induction of cell death by RipAW and its mutants. (A) Schematic illustration of RipAW and its derivatives RipAWC177A (cysteine at position 177 of RipAW is substituted by alanine), RipAW ΔN, RipAW ΔC, RipAWN1-90, RipAWNEL and RipAWC322-448. The N-terminus, NEL domain and C-terminus of RipAW are indicated as blue, gray and yellow boxes, respectively. The positions of amino acid residues are indicated by numbers. (B) RipAW induces cell death in N. benthamiana, but all its derivatives do not. RipAW and its derivatives were expressed in the same leaf of N. benthamiana. LTI6b was the negative control. Photographs were taken at 3 dpi. Circles indicate the infiltrated areas. The fractions in brackets represent the number of leaves displaying HR over the total number of infiltrated leaves. (C) Cell death in (B) was evaluated by the degree of ion leakage. The degree of ion leakage from the leaf discs was measured 44 h after agroinfiltration using a conductivity meter. Values are means ± SEs from three biological replicates. Different letters indicate significant differences at p ≤ 0.01. (D) Detection of RipAW and its derivatives with western blot using anti-FLAG antibody. Total protein was extracted from the leaves 2 days after agroinfiltration. The blot was stained with Coomassie brilliant blue (CBB) to confirm equal loading.
The E3 ligase mutant RipAWC177A still triggered plant immunity (Figure 1), indicating that some other regions also contribute to RipAW-triggered immunity. We expressed RipAW derivatives individually in N. benthamiana, and investigated their effects on plant resistance to CQPS-1 and DC3000 ΔhopQ1-1. Results showed that, compared with the control, expression of RipAWN1-90, RipAWNEL, RipAWC322-448, RipAW ΔN and RipAW ΔC all significantly suppressed the colonization of both CQPS-1 (Figure 3A) and DC3000 ΔhopQ1-1 (Figure 3B) in N. benthamiana. These results indicate that the N- and C-terminal regions of RipAW can also trigger plant immunity. Meanwhile, we examined the expression of the plant immune-related gene NbHIN1 and found that it was up-regulated significantly by these RipAW derivatives (Figure 3C). This result further confirmed that RipAW-induced plant immunity is not completely dependent on the E3 ubiquitin ligase activity of RipAW.
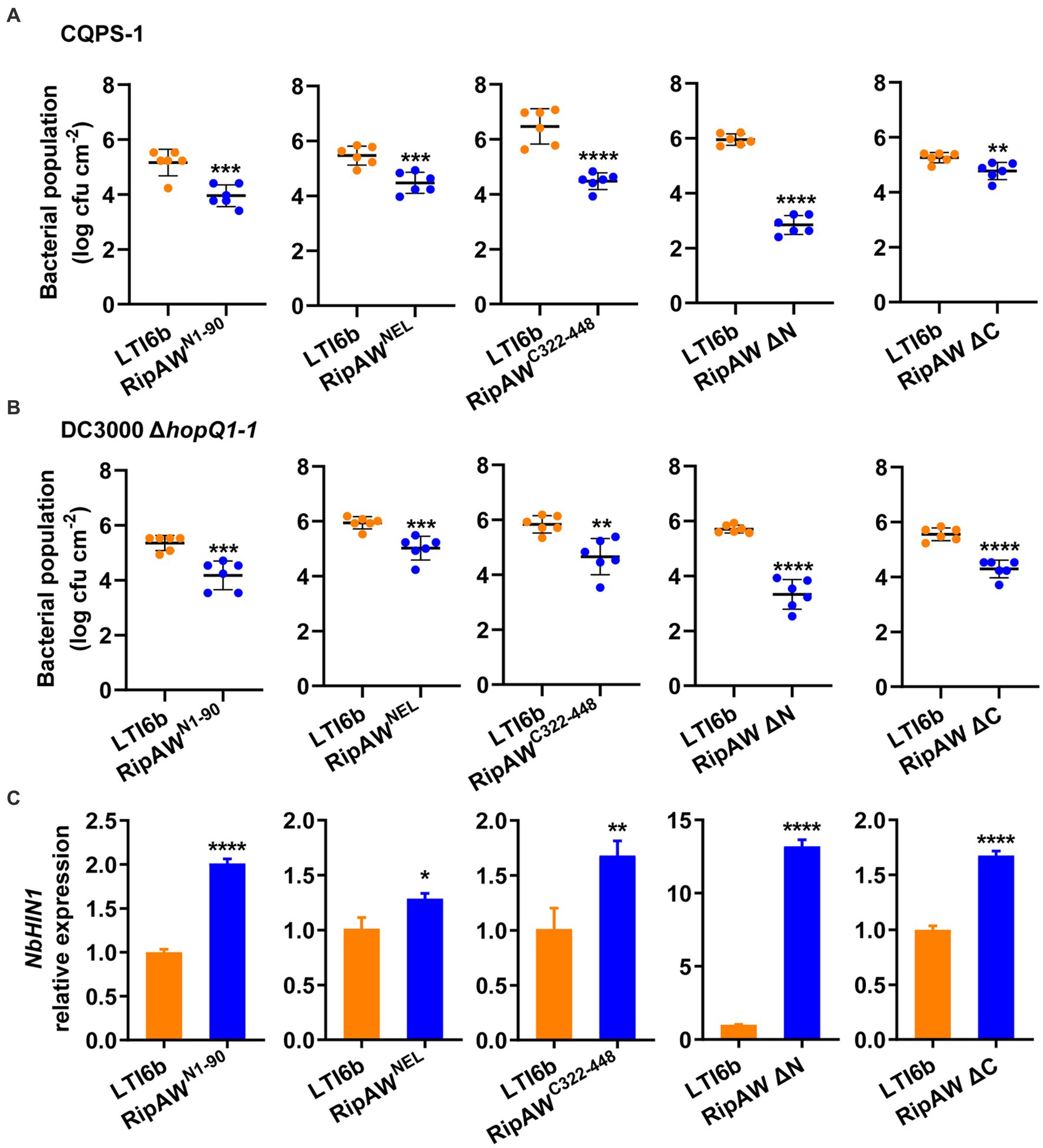
Figure 3. Plant immunity triggered by RipAW mutants. (A,B) Bacterial population of R. solanacearum CQPS-1 (A) and P. syringae DC3000 ΔhopQ1-1 (B) in N. benthamiana leaves expressing corresponding RipAW derivatives. Agrobacterial cells harboring RipAW mutants or LTI6b were infiltrated into N. benthamiana leaves. 48 h later, CQPS-1 or DC3000 ΔhopQ1-1 was infiltrated into the same areas. The bacterial population was determined at 2 dpi for CQPS-1 and 3 dpi for DC3000 ΔhopQ1-1. (C) NbHIN1 expression in N. benthamiana leaves expressing LTI6b (control) or RipAW’s derivatives by qRT–PCR. Values are means ± SEs from three biological replicates. *, **, ***, and **** indicate significant differences at p ≤ 0.05, p ≤ 0.01, p ≤ 0.001 and p ≤ 0.0001, respectively.
SGT1 is a major ETI regulator (Azevedo et al., 2002; Kadota et al., 2010), and RipAW-induced cell death has been reported to be SGT1-dependent (Niu et al., 2022). We wonder whether RipAW-triggered plant immunity is also SGT1-dependent. We silenced SGT1 in N. benthamiana by VIGS and tested RipAW- and RipAWC177A-induced immune responses in the NbSGT1-silenced plants. Results showed that, similar to INF1 whose cell death induction is dependent on SGT1 (Bos et al., 2006), RipAW could not induce cell death in the NbSGT1-silenced N. benthamiana (Figure 4A). The ubiquitin E3 ligase mutant RipAWC177A could not induce cell death in both control and the NbSGT1-silenced plants. The ion leakage results confirmed the results of cell death (Figure 1B). qRT-PCR analysis showed that NbSGT1 expression in TRV2::NbSGT1 N. benthamiana was only 18.81% of that in TRV2::GFP plants (Figure 4C), indicating that NbSGT1 was successfully silenced. Expression of RipAWC177A significantly decreased CQPS-1’s population in TRV2::GFP plants, but this effect was abolished in NbSGT1-silenced plants (Figure 4D). Expression of RipAW also failed to promote plant resistance to CQPS-1 in NbSGT1-silenced plants (Figure 4D). These results indicate that not only RipAW-induced cell death but also RipAW-triggered immunity is dependent on NbSGT1. We further analyzed the expression of defense-related gene NbHIN1 and found that both RipAW- and RipAWC177A-enhanced NbHIN1 expression was significantly impaired in the NbSGT1-silenced plants compared with that in control (TRV2::GFP) plants (Figure 4E), confirming the important role of NbSGT1 in RipAW and RipAWC177A-triggered plant immunity. Western blot analysis showed the successful expression of RipAW, RipAWC177A and INF1 proteins (Figure 4F).
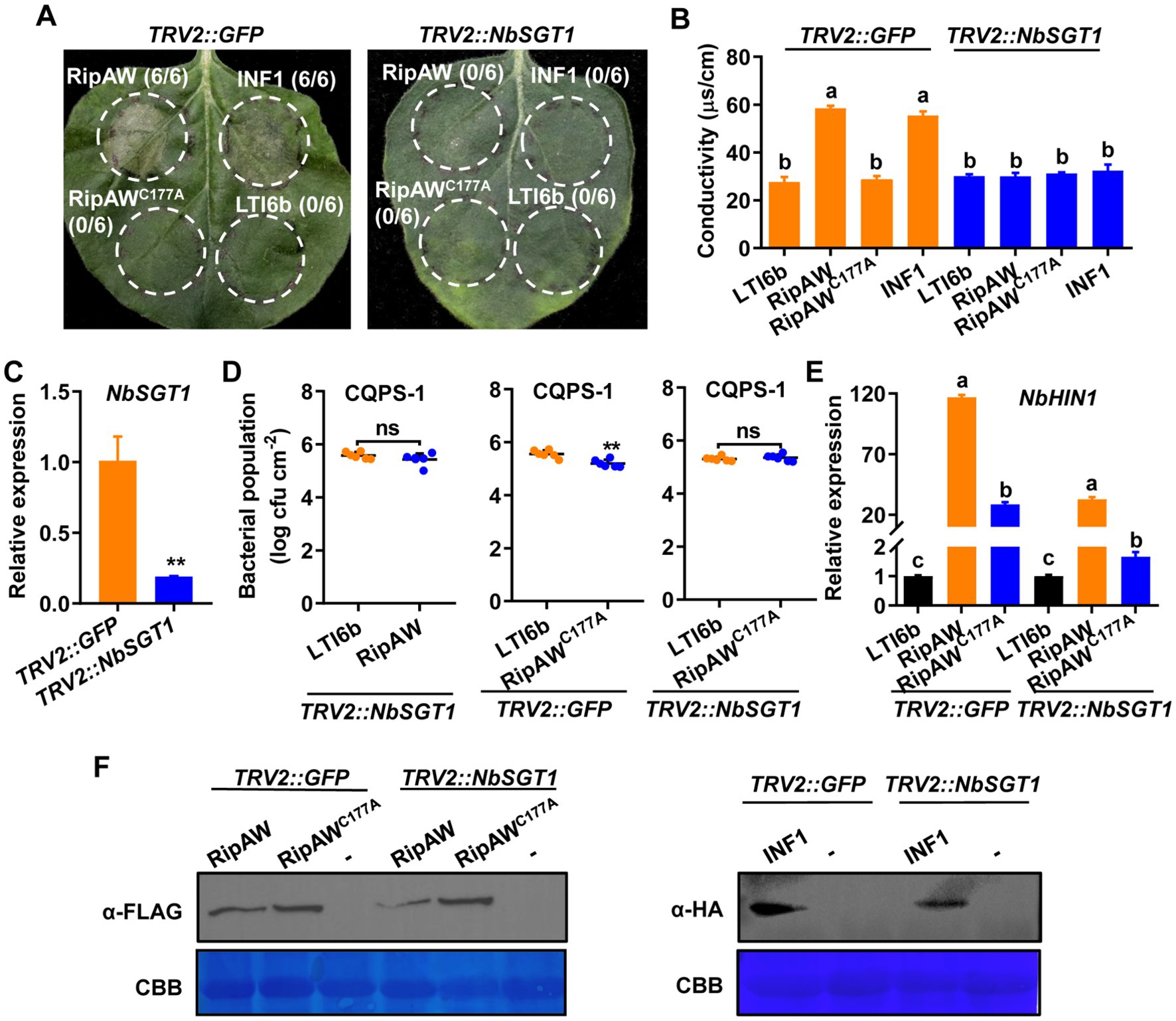
Figure 4. Silencing NbSGT1 impairs defense triggered by RipAW and RipAWC177A. (A) Both RipAW and RipAWC177A cannot induce cell death in NbSGT1-silenced N. benthamiana leaves. Photographs were taken at 3 days after infiltration. Circles indicate the infiltrated areas. The fractions in brackets represent the number of leaves displaying HR over the total number of infiltrated leaves. (B) Cell death in (A) was evaluated by the degree of ion leakage. The degree of ion leakage from the leaf discs was measured 2 days after agroinfiltration using a conductivity meter. (C) NbSGT1 expression in leaves of TRV2::NbSGT1 and TRV2::GFP N. benthamiana. (D) Growth of R. solanacearum CQPS-1 in TRV2::NbSGT1 and TRV2::GFP N. benthamiana leaves expressing RipAW and RipAWC177A. LTI6b, RipAW and RipAWC177A were agroinfiltrated into TRV2::NbSGT1 and TRV2::GFP N. benthamiana leaves, respectively. 48 h later, CQPS-1 was infiltrated into the same areas. The bacterial population was determined at 2 dpi. (E) NbHIN1 expression in TRV2::NbSGT1 and TRV2::GFP N. benthamiana leaves expressing LTI6b (control), RipAW or RipAWC177A by qRT–PCR. Values in (B–E) are means ± SEs from at least three biological replicates. Different small letters in (B,E) and ** in panels (C,D) indicate significant differences at p ≤ 0.001. ns indicates no significant difference. (F) Detection of RipAW, RipAWC177A and INF1 in TRV2::NbSGT1 and TRV2::GFP N. benthamiana by western blot using anti-FLAG or anti-HA antibody. Total protein was extracted from the leaves 2 days after agroinfiltration. The blot was stained with Coomassie brilliant blue (CBB) to confirm equal loading.
Although most NLRs require SGT1 to function, EDS1 and helper NLR NRG1 also play critical roles in some TIR-NLRs (Wiermer et al., 2005; Bos et al., 2006; Schultink et al., 2017; Ngou et al., 2022b) and a specific subset of CC-NLRs requires another class of helper NLRs termed NRC proteins (Wu et al., 2017). To determine whether RipAW-triggered plant immunity depends on these important ETI components, we expressed RipAW/RipAWC177A in eds1, nrg1 and nrc2/3/4 mutants, respectively. We found that RipAW-induced cell death was not affected in eds1, nrg1 and nrc2/3/4 mutants (Figure 5A). Bacterial population assay further showed that RipAWC177A maintained its function to restrict CQPS-1 growth in eds1, nrg1 and nrc2/3/4 mutants (Figure 5B). RipAW and RipAWC177A proteins were successfully expressed in both WT and tested mutants (Figure 5C). These results indicate that RipAW- and RipAWC177A-triggered plant immunity do not depend on EDS1 and helper NLRs NRG1 and NRCs.
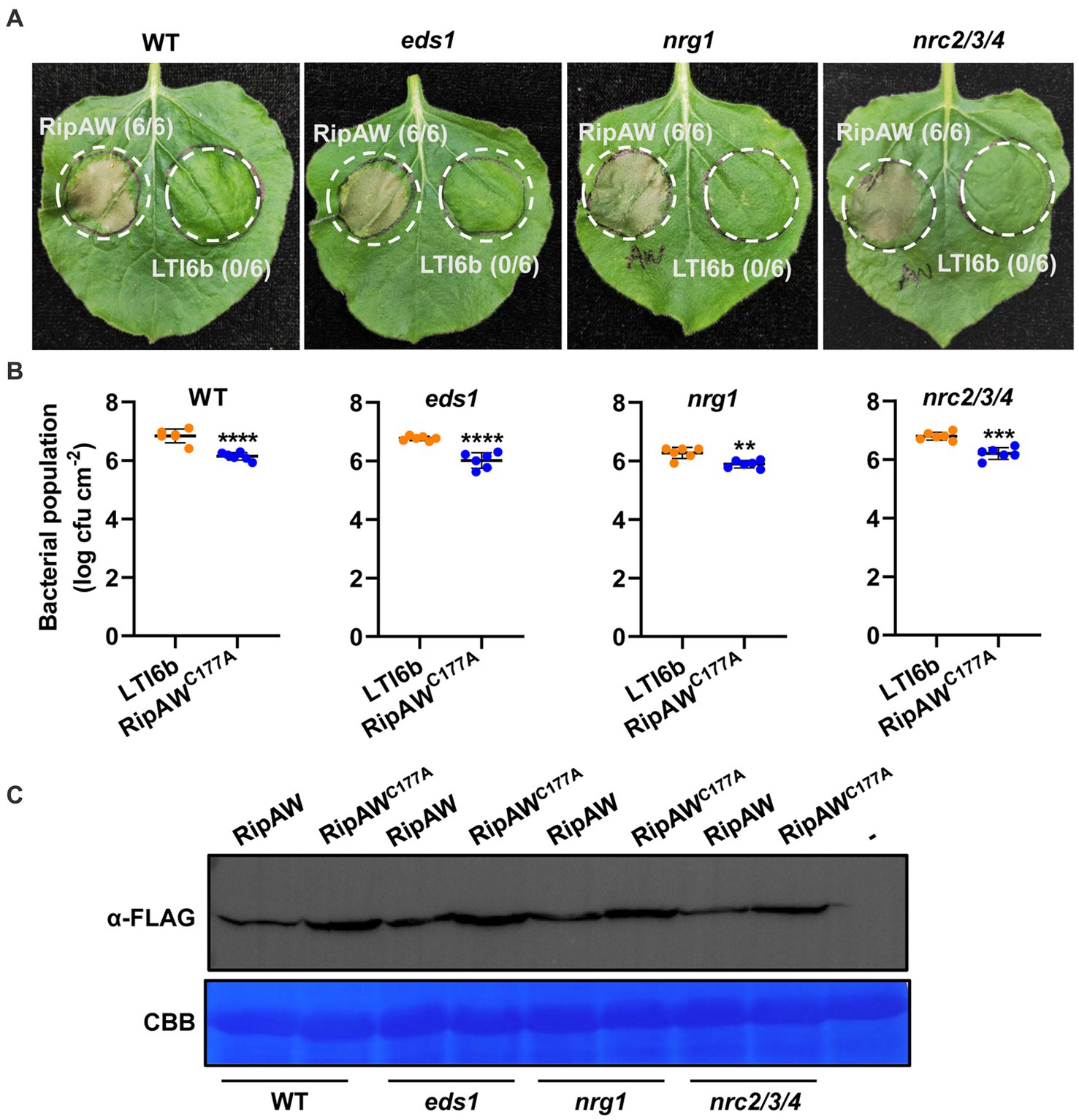
Figure 5. RipAWC177A-triggered immunity in eds1 and mutants of Helper NLRs. (A) RipAW-induced cell death in wild-type (WT) N. benthamiana and eds1, nrg1, nrc2/3/4 mutants. RipAW and LTI6b (control) were expressed in the same leaf of wild-type N. benthamiana, eds1, nrg1 and nrc2/3/4 mutants, respectively. Photographs were taken at 3 dpi. Circles indicate the infiltrated areas. The fractions in brackets represent the number of leaves displaying HR over the total number of infiltrated leaves. (B) Growth of R. solanacearum CQPS-1 in WT and mutant leaves expressing LTI6b or RipAWC177A. LTI6b and RipAWC177A were agroinfiltrated into WT and mutant leaves. 48 h later, CQPS-1 was infiltrated into the same areas. The bacterial population was determined at 2 dpi. Values are means ± SEs from six biological replicates. ** and *** indicate significant differences at p ≤ 0.01 and p ≤ 0.001, respectively. (C) Detection of RipAW and RipAWC177A in WT and mutants by western blot using anti-FLAG antibody. Total protein was extracted from the leaves 2 days after agroinfiltration and subjected to western blot analysis. The blot was stained with Coomassie brilliant blue (CBB) to confirm equal loading.
SA plays a vital role in the activation of immune responses (Zhou and Zhang, 2020). RipE1, another T3E in R. solanacearum, has been reported to trigger plant immunity by regulating SA-dependent responses (Sang et al., 2020). Here, we co-expressed RipAW or RipE1 (as a positive control) with NahG which blocks SA accumulation in plant (Delaney et al., 1994). Results showed that co-expression of NahG obviously impaired RipE1-induced cell death but did not affect RipAW-induced cell death (Figure 6A) which was further confirmed by the ion leakage result (Figure 6B). NahG, RipAW and RipE1 were all expressed successfully in our experiment (Figure 6C). These results indicate that, different with RipE1, RipAW-triggered plant immunity is probably independent of SA signaling pathway. To confirm this, we investigated the effect of NahG co-expression on RipAW and RipAWC177A-induced NbHIN1 gene expression. We found that RipAW and RipAWC177A-induced NbHIN1 gene expression was not influenced by NahG (Figures 6D,E). In addition, NahG co-expression also did not affect RipAWC177A-restricted CQPS-1 growth in N. benthamiana (Figure 6F). These results confirm that RipAW-triggered cell death and immunity is independent on SA pathway.
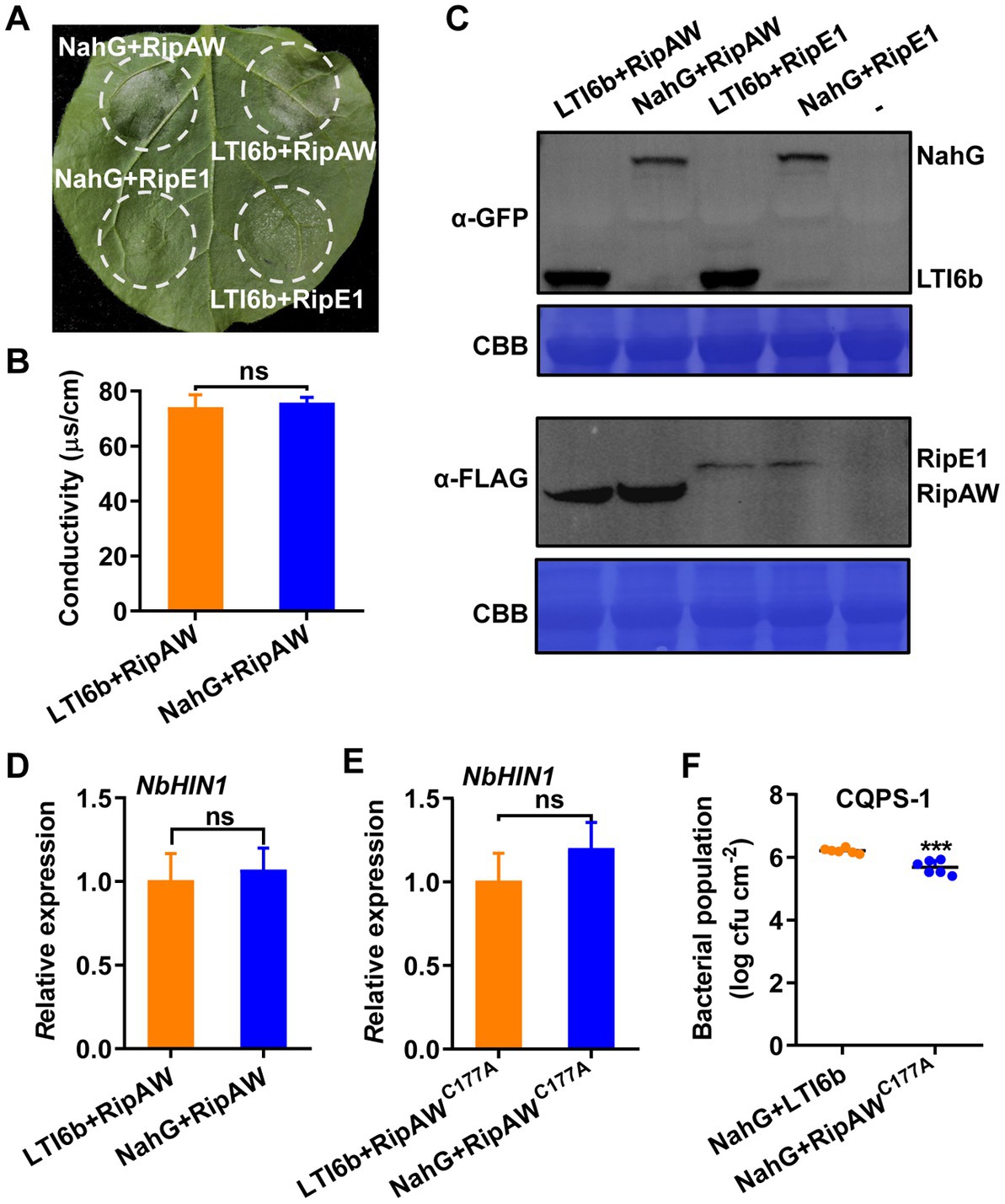
Figure 6. RipAWC177A-triggered immunity is independent of SA pathway. (A) RipAW induces cell death when co-expressed with NahG. RipAW and RipE1 were co-expressed with LTI6b (control) or NahG in the same leaf. Photographs were taken at 3 dpi. Circles indicate the infiltrated areas. The fractions in brackets represent the number of leaves displaying HR over the total number of infiltrated leaves. (B) Cell death in (A) was quantified by the degree of ion leakage. The degree of ion leakage from the leaf discs was measured 2 days after agroinfiltration using a conductivity meter. (C) Detection of proteins in N. benthamiana by western blot using anti-FLAG (for RipAW and RipE1) or anti-GFP (for LTI6 and NahG) antibody. Total protein was extracted from the leaves 2 days after agroinfiltration. The blot was stained with Coomassie brilliant blue (CBB) to confirm equal loading. (D) NbHIN1 expression in N. benthamiana leaves co-expressing LTI6b (control) or NahG with RipAW by qRT–PCR. (E) NbHIN1 expression in N. benthamiana leaves co-expressing LTI6b (control) or NahG with RipAWC177A by qRT–PCR. (F) Growth of R. solanacearum CQPS-1 in N. benthamiana leaves co-expressing NahG with LTI6b (control) or RipAWC177A. LTI6b and RipAWC177A were agroinfiltrated into NahG-expressed N. benthamiana leaves, respectively. 24 h later, CQPS-1 was infiltrated into the same leaf areas. The bacterial population was determined at 2 dpi. Values are means ± SEs from six biological replicates. *** indicates significant differences at p ≤ 0.001 and ns indicates no significant difference.
As one of the most destructive bacterial phytopathogens, R. solanacearum is endowed with an exceptionally large repertoire of effectors that functions in its interaction with plants (Mansfield et al., 2012; Coll and Valls, 2013). Since ubiquitination is important in the regulation of many cellular processes of plants (Rytkonen and Holden, 2007), it is not surprising that some phytopathogens secrete effectors to interfere extensively with ubiquitination pathways. For example, P. syringae type III effector AvrPtoB depends on its E3 ligase activity to degrade NPR1 (Non-expresser of PR genes 1) and hence represses SA-dependent immune responses (Chen et al., 2017). NEL is a novel class of E3 ligase which is distinct from typical RING- and HECT-types (Ashida and Sasakawa, 2016). Type III effector RipAW in R. solanacearum strain RS1000 was found to have a NEL domain and suppress plant PTI responses in a E3 ligase activity-dependent manner (Nakano et al., 2017). Niu et al. (2022) further demonstrated that RipAW in R. solanacearum strain GMI1000 induced cell death in N. benthamiana which requires its E3 ligase activity. These previous studies demonstrate the importance of the E3 ligase activity for the function of RipAW in the plant-R. solanacearum interactions. Here, we further defined the role of E3 ligase activity in RipAW-triggered plant immunity, providing new insights into the functional mechanism of RipAW in R. solanacearum.
RipAWC177A, the E3 ligase mutant of RipAW, lost the ability to induce cell death but retained that to trigger plant immunity in N. benthamiana (Figure 1), indicating that the E3 ligase is not essential for RipAW-triggered immunity. All truncated mutants of RipAW, including RipAWN1-90, RipAWNEL, RipAWC322-448, RipAW ΔN and RipAW ΔC, showed the ability to trigger plant immunity without inducing cell death (Figures 2, 3), confirming that the NEL contributes to but is not essential for RipAW-triggered immunity. Niu et al. (2022) demonstrated that both the N-terminal region and NEL domain are required for RipAW-induced cell death. Here, we confirmed Niu et al. (2022)‘s finding and further provided evidence that the C-terminal region is also required for RipAW-induced cell death in N. benthamiana. Although being required, none of them alone is sufficient to trigger cell death in N. benthamiana.
Early in 2000, researchers found that cell death associated with HR to CaMV was uncoupled genetically from plant resistance (Greenberg et al., 2000). Several following studies also showed that ETI-induced cell death was sometimes segregated from plant immunity (Glocova et al., 2005; Menna et al., 2015; Pitsili et al., 2020; Yang et al., 2022). Recently, the Xanthomonas effector XopQ-triggered immune responses were found to be separated from ETI-induced cell death in N. benthamiana (Prautsch et al., 2023). In the present study, RipAWC177A and RipAW’s derivatives triggered plant immunity to restrict bacterial infection without inducing ETI-triggered cell death, which is consistent with these previous studies. These results indicate that different ETI outputs are not always coupled together and plant ETI responses may be triggered without inducing cell death. Therefore, except cell death, it would be better to simultaneously consider some other immune responses as outputs when screening for potential effectors in pathogens that are recognized by plants.
SGT1 acts as a molecular chaperone for NLR proteins and has been proved as a key signaling component in ETI triggered by several effectors, such as Phytophthora infestans RXLR effector AVR3a (Bos et al., 2006), R. solanacearum effector RipB (Nakano and Mukaihara, 2019) and RipE1 (Jeon et al., 2020). RipAW-induced cell death also requires SGT1 (Niu et al., 2022). Here, using NbSGT1-silenced plants, we further showed that RipAW- and RipAWC177A-triggered plant immunity which is likely uncoupled from cell death is also dependent on SGT1 (Figure 4). Except SGT1, EDS1 and some helper NLRs such as NRG1 and NRCs are also well-known downstream regulators in plant ETI (Peart et al., 2005; Wiermer et al., 2005; Schultink et al., 2017; Wu et al., 2017). However, in this study, both RipAW-induced cell death and RipAWC177A-induced resistance to bacterial growth retained in eds1, nrg1 and nrc2/3/4 mutants (Figure 5), suggesting that RipAW-triggered plant immunity is independent of EDS1, NRG1 and NRC2/3/4. NahG blocks SA accumulation in plants (Delaney et al., 1994). RipE1 was found to trigger cell death partially through SA signaling pathway by co-expressing with NahG (Sang et al., 2020). However, NahG did not affect RipAW-induced cell death and RipAWC177A-induced resistance, implying that SA signaling pathway is not essential for RipAW and RipAWC177A-triggered plant immunity.
In summary, our data demonstrates that the E3 ligase activity is not essential for plant immunity triggered by the NEL effector RipAW in R. solanacearum. The N-terminus, C-terminus and NEL domain all contribute to RipAW-triggered immunity in N. benthamiana, and they are jointly required for RipAW-induced cell death. RipAW- and RipAWC177A-triggered immunity in N. benthamiana requires SGT1, but not EDS1, NRG1, NRC proteins or SA pathway. Our findings shed new light on effector-triggered immunity and provide clues for further in-depth study of mechanism underlying RipAW-induced plant immunity.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
MZ, PL, and CS conceived and designed the experiments. XO, JC, ZS, and RW performed the experiments. XO, JC, XW, BL, and MZ analyzed the data. PL, CS, and MZ drafted and modified the manuscript. All authors read and approved the manuscript.
This research was supported by the National Natural Science Foundation of China (32072399 and 32272641), the Fundamental Research Funds for the Central Universities (GK202201017), and the Program of Fujian Key Laboratory for Monitoring and Integrated Management of Crop Pests (MIMCP-202203).
We thank Wei Ding from Southwest University for providing the CQPS-1 strain.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1201444/full#supplementary-material
An, Y. Y., Chen, J. L., Xu, Z. Y., Ouyang, X., Cao, P., Wang, R. B., et al. (2022). Three amino acid residues are required for the recognition of Ralstonia solanacearum RipTPS in Nicotiana tabacum. Front. Plant Sci. 13:1040826. doi: 10.3389/fpls.2022.1040826
Ashida, H., and Sasakawa, C. (2016). Shigella IpaH family effectors as a versatile model for studying pathogenic bacteria. Front. Cell. Infect. Microbiol. 5:100. doi: 10.3389/fcimb.2015.00100
Azevedo, C., Sadanandom, A., Kitagawa, K., Freialdenhoven, A., Shirasu, K., and Schulze-Lefert, P. (2002). The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science 295, 2073–2076. doi: 10.1126/science.1067554
Bos, J. I. B., Kanneganti, T. D., Young, C., Cakir, C., Huitema, E., Win, J., et al. (2006). The C-terminal half of Phytophthora infestans RXLR effector AVR3a is sufficient to trigger R3a-mediated hypersensitivity and suppress INF1-induced cell death in Nicotiana benthamiana. Plant J. 48, 165–176. doi: 10.1111/j.1365-313X.2006.02866.x
Buetow, L., and Huang, D. T. (2016). Structural insights into the catalysis and regulation of E3 ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 17, 626–642. doi: 10.1038/nrm.2016.91
Chen, H., Chen, J., Li, M., Chang, M., Xu, K. M., Shang, Z. H., et al. (2017). A bacterial type III effector targets the master regulator of salicylic acid signaling, NPR1, to subvert plant immunity. Cell Host Microbe 22, 777–788.e7. doi: 10.1016/j.chom.2017.10.019
Cheng, D., Zhou, D., Wang, Y. D., Wang, B. S., He, Q., Song, B. T., et al. (2021). Ralstonia solanacearum type III effector RipV2 encoding a novel E3 ubiquitin ligase (NEL) is required for full virulence by suppressing plant PAMP-triggered immunity. Biochem. Biophys. Res. Commun. 550, 120–126. doi: 10.1016/j.bbrc.2021.02.082
Coll, N. S., and Valls, M. (2013). Current knowledge on the Ralstonia solanacearum type III secretion system. Microb. Biotechnol. 6, 614–620. doi: 10.1111/1751-7915.12056
Delaney, T. P., Uknes, S., Vernooij, B., Friedrich, L., Weymann, K., Negrotto, D., et al. (1994). A central role of salicylic acid in plant disease resistance. Science 266, 1247–1250. doi: 10.1126/science.266.5188.1247
Fu, Z. Q., Yan, S. P., Saleh, A., Wang, W., Ruble, J., Oka, N., et al. (2012). NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 486, 228–232. doi: 10.1038/nature11162
Genin, S., and Denny, T. P. (2012). Pathogenomics of the Ralstonia solanacearum species complex. Annu. Rev. Phytopathol. 50, 67–89. doi: 10.1146/annurev-phyto-081211-173000
Glocova, I., Thor, K., Roth, B., Babbick, M., Pfitzner, A. J. P., and Pfitzner, U. M. (2005). Salicylic acid (SA)-dependent gene activation can be uncoupled from cell death-mediated gene activation: the SA-inducible NIMIN-1 and NIMIN-2 promoters, unlike the PR-1a promotor, do not respond to cell death signals in tobacco. Mol. Plant Pathol. 6, 299–314. doi: 10.1111/J.1364-3703.2005.00288.X
Gopalan, S., Wei, W., and He, S. Y. (1996). Hrp gene-dependent induction of hin1: a plant gene activated rapidly by both harpins and the avrPto gene-mediated signal. Plant J. 10, 591–600. doi: 10.1046/j.1365-313X.1996.10040591.x
Greenberg, J. T., Silverman, F. P., and Liang, H. (2000). Uncoupling salicylic acid-dependent cell death and defense-related responses from disease resistance in the Arabidopsis mutant acd5. Genetics 156, 341–350. doi: 10.1093/genetics/156.1.341
Jeon, H., Kim, W., Kim, B., Lee, S., Jayaraman, J., Jung, G., et al. (2020). Ralstonia solanacearum type III effectors with predicted nuclear localization signal localize to various cell compartments and modulate immune responses in Nicotiana spp. Plant Pathol. J. 36, 43–53. doi: 10.5423/Ppj.Oa.08.2019.0227
Jones, J. D. G., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1146/annurev-phyto-081211-173000
Jones, J. D. G., Vance, R. E., and Dangl, J. L. (2016). Intracellular innate immune surveillance devices in plants and animals. Science 354:aaf6395. doi: 10.1126/science.aaf6395
Kadota, Y., Shirasu, K., and Guerois, R. (2010). NLR sensors meet at the SGT1-HSP90 crossroad. Trends Biochem. Sci. 35, 199–207. doi: 10.1016/j.tibs.2009.12.005
Kourelis, J., Contreras, M. P., Harant, A., Pai, H., Lüdke, D., Adachi, H., et al. (2022). The helper NLR immune protein NRC3 mediates the hypersensitive cell death caused by the cell-surface receptor Cf-4. PLoS Genet. 18:e1010414. doi: 10.1371/journal.pgen.1010414
Lapin, D., Kovacova, V., Sun, X. H., Dongus, J. A., Bhandari, D., von Born, P., et al. (2019). A coevolved EDS1-SAG101-NRG1 module mediates cell death signaling by TIR-domain immune receptors. Plant Cell 31, 2430–2455. doi: 10.1105/tpc.19.00118
Lee, D., Lal, N. K., Lin, Z. D., Ma, S., Liu, J., Castro, B., et al. (2020). Regulation of reactive oxygen species during plant immunity through phosphorylation and ubiquitination of RBOHD. Nat. Commun. 11:1838. doi: 10.1038/s41467-020-15601-5
Li, P., Jiang, W., Yu, Q., Liu, W., Zhou, P., Li, J., et al. (2017). Ubiquitination and degradation of GBPs by a Shigella effector to suppress host defence. Nature 551, 378–383. doi: 10.1038/nature24467
Liu, Y., Tang, Y. M., Qin, X. Y., Yang, L., Jiang, G. F., Li, S. L., et al. (2017). Genome sequencing of Ralstonia solanacearum CQPS-1, a phylotype I strain collected from a highland area with continuous cropping of tobacco. Front. Microbiol. 8:974. doi: 10.3389/fmicb.2017.00974
Mansfield, J., Genin, S., Magori, S., Citovsky, V., Sriariyanum, M., Ronald, P., et al. (2012). Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 13, 614–629. doi: 10.1111/j.1364-3703.2012.00804.x
Martinière, A., Shvedunova, M., Thomson, A. J., Evans, N. H., Penfield, S., Runions, J., et al. (2011). Homeostasis of plasma membrane viscosity in fluctuating temperatures. New Phytol. 192, 328–337. doi: 10.1111/j.1469-8137.2011.03821.x
Menna, A., Nguyen, D., Guttman, D. S., and Desveaux, D. (2015). Elevated temperature differentially influences effector-triggered immunity outputs in Arabidopsis. Front. Plant Sci. 6:995. doi: 10.3389/fpls.2015.00995
Muskett, P., and Parker, J. (2003). Role of SGT1 in the regulation of plant R gene signalling. Microbes Infect. 5, 969–976. doi: 10.1016/S1286-4579(03)00183-7
Nakano, M., and Mukaihara, T. (2019). The type III effector RipB from Ralstonia solanacearum RS1000 acts as a major avirulence factor in Nicotiana benthamiana and other Nicotiana species. Mol. Plant Pathol. 20, 1237–1251. doi: 10.1111/mpp.12824
Nakano, M., Oda, K., and Mukaihara, T. (2017). Ralstonia solanacearum novel E3 ubiquitin ligase (NEL) effectors RipAW and RipAR suppress pattern-triggered immunity in plants. Microbiology 163, 992–1002. doi: 10.1099/mic.0.000495
Ngou, B. P. M., Ding, P. T., and Jones, J. D. G. (2022a). Thirty years of resistance: Zig-zag through the plant immune system. Plant Cell 34, 1447–1478. doi: 10.1093/plcell/koac041
Ngou, B. P. M., Jones, J. D. G., and Ding, P. T. (2022b). Plant immune networks. Trends Plant Sci. 27, 255–273. doi: 10.1016/j.tplants.2021.08.012
Niu, Y., Fu, S. Y., Chen, G., Wang, H. J., Wang, Y. S., Hu, J. X., et al. (2022). Different epitopes of Ralstonia solanacearum effector RipAW are recognized by two Nicotiana species and trigger immune responses. Mol. Plant Pathol. 23, 188–203. doi: 10.1111/mpp.13153
Peart, J. R., Mestre, P., Lu, R., Malcuit, I., and Baulcombe, D. C. (2005). NRG1, a CC-NB-LRR protein, together with N, a TIR-NB-LRR protein, mediates resistance against tobacco mosaic virus. Curr. Biol. 15, 968–973. doi: 10.1016/j.cub.2005.04.053
Peeters, N., Guidot, A., Vailleau, F., and Valls, M. (2013). Ralstonia solanacearum, a widespread bacterial plant pathogen in the post-genomic era. Mol. Plant Pathol. 14, 651–662. doi: 10.1111/mpp.12038
Pitsili, E., Phukan, U. J., and Coll, N. S. (2020). Cell death in plant immunity. Cold Spring Harb. Perspect. Biol. 12:a036483. doi: 10.1101/cshperspect.a036483
Poueymiro, M., Cazale, A. C., Francois, J. M., Parrou, J. L., Peeters, N., and Genin, S. (2014). A Ralstonia solanacearum type III effector directs the production of the plant signal metabolite trehalose-6-phosphate. MBio 5, e02065–e02014. doi: 10.1128/mBio.02065-14
Prautsch, J., Erickson, J. L., Özyürek, S., Gormanns, R., Franke, L., Lu, Y., et al. (2023). Effector XopQ-induced stromule formation in Nicotiana benthamiana depends on ETI signaling components ADR1 and NRG1. Plant Physiol. 191, 161–176. doi: 10.1093/plphys/kiac481
Qi, P. P., Huang, M. L., Hu, X. H., Zhang, Y., Wang, Y., Li, P. Y., et al. (2022). Aralstonia solanacearum effector targets TGA transcription factors to subvert salicylic acid signaling. Plant Cell 34, 1666–1683. doi: 10.1093/plcell/koac015
Qi, T., Seong, K., Thomazella, D. P. T., Kim, J. R., Pham, J., Seo, E., et al. (2018). NRG1 functions downstream of EDS1 to regulate TIR-NLR-mediated plant immunity in Nicotiana benthamiana. Proc. Natl. Acad. Sci. U. S. A. 115, E10979–E10987. doi: 10.1073/pnas.1814856115
Rohde, J. R., Breitkreutz, A., Chenal, A., Sansonetti, P. J., and Parsot, C. (2007). Type III secretion effectors of the IpaH family are E3 ubiquitin ligases. Cell Host Microbe 1, 77–83. doi: 10.1016/j.chom.2007.02.002
Rytkonen, A., and Holden, D. W. (2007). Bacterial interference of ubiquitination and deubiquitination. Cell Host Microbe 1, 13–22. doi: 10.1016/j.chom.2007.02.003
Sang, Y. Y., Yu, W. J., Zhuang, H. Y., Wei, Y. L., Derevnina, L., Yu, G., et al. (2020). Intra-strain elicitation and suppression of plant immunity by Ralstonia solanacearum type-III effectors in Nicotiana benthamiana. Plant Commun. 1:100025. doi: 10.1016/j.xplc.2020.100025
Savary, S., Willocquet, L., Pethybridge, S. J., Esker, P., Mcroberts, N., and Nelson, A. (2019). The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 3, 430–439. doi: 10.1038/s41559-018-0793-y
Schultink, A., Qi, T. C., Lee, A., Steinbrenner, A. D., and Staskawicz, B. (2017). Roq1 mediates recognition of the Xanthomonas and Pseudomonas effector proteins XopQ and HopQ1. Plant J. 92, 787–795. doi: 10.1111/tpj.13715
Senthil-Kumar, M., and Mysore, K. S. (2014). Tobacco rattle virus-based virus-induced gene silencing in Nicotiana benthamiana. Nat. Protoc. 9, 1549–1562. doi: 10.1038/nprot.2014.092
Singer, A. U., Schulze, S., Skarina, T., Xu, X. H., Cui, H., Eschen-Lippold, L., et al. (2013). A pathogen type III effector with a novel E3 ubiquitin ligase architecture. PLoS Pathog. 9:e1003121. doi: 10.1371/journal.ppat.1003121
Sun, Y. H., Li, P., Shen, D., Wei, Q. L., He, J. G., and Lu, Y. J. (2019). The Ralstonia solanacearum effector RipN suppresses plant PAMP-triggered immunity, localizes to the endoplasmic reticulum and nucleus, and alters the NADH/NAD+ ratio in Arabidopsis. Mol. Plant Pathol. 20, 533–546. doi: 10.1111/mpp.12773
Vierstra, R. D. (2009). The ubiquitin-26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 10, 385–397. doi: 10.1038/nrm2688
Wei, C. F., Kvitko, B. H., Shimizu, R., Crabill, E., Alfano, J. R., Lin, N. C., et al. (2007). A Pseudomonas syringae pv. Tomato DC3000 mutant lacking the type III effector HopQ1-1 is able to cause disease in the model plant Nicotiana benthamiana. Plant J. 51, 32–46. doi: 10.1111/j.1365-313X.2007.03126.x
Wiermer, M., Feys, B. J., and Parker, J. E. (2005). Plant immunity: the EDS1 regulatory node. Curr. Opin. Plant Biol. 8, 383–389. doi: 10.1016/j.pbi.2005.05.010
Wu, C. H., Abd-el-Haliem, A., Bozkurt, T. O., Belhaj, K., Terauchi, R., Vossen, J. H., et al. (2017). NLR network mediates immunity to diverse plant pathogens. Proc. Natl. Acad. Sci. 114, 8113–8118. doi: 10.1073/pnas.1702041114
Yang, K., Chen, C., Wang, Y., Li, J. L., Dong, X. H., Cheng, Y., et al. (2022). Nep1-like proteins from the biocontrol agent Pythium oligandrum enhance plant disease resistance independent of cell death and reactive oxygen species. Front. Plant Sci. 13:830636. doi: 10.3389/fpls.2022.830636
Keywords: RipAW, Ralstonia solanacearum , cell death, plant immunity, SGT1
Citation: Ouyang X, Chen J, Sun Z, Wang R, Wu X, Li B, Song C, Liu P and Zhang M (2023) Ubiquitin E3 ligase activity of Ralstonia solanacearum effector RipAW is not essential for induction of plant defense in Nicotiana benthamiana. Front. Microbiol. 14:1201444. doi: 10.3389/fmicb.2023.1201444
Received: 06 April 2023; Accepted: 04 May 2023;
Published: 24 May 2023.
Edited by:
Yong Zhang, Southwest University, ChinaReviewed by:
Yunhao Sun, Zhongkai University of Agriculture and Engineering, ChinaCopyright © 2023 Ouyang, Chen, Sun, Wang, Wu, Li, Song, Liu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Congfeng Song, Y2Zzb25nQG5qYXUuZWR1LmNu;; Peiqing Liu, bGl1cGVpcWluZzExQDE2My5jb20=;; Meixiang Zhang, bWVpeGlhbmd6aGFuZ0Bzbm51LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.