
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 19 June 2023
Sec. Microbe and Virus Interactions with Plants
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1200023
Our study aimed to identify potential biocontrol agents (BCAs) against major phytopathogens under in vitro conditions by screening the Freshwater Bioresources Culture Collection (FBCC), Korea. Of the identified 856 strains, only 65 exhibited antagonistic activity, among which only one representative isolation, Brevibacillus halotolerans B-4359 was selected based on its in vitro antagonistic activity and enzyme production. Cell-free culture filtrate (CF) and volatile organic compounds (VOCs) of B-4359 were shown to be effective against the mycelial growth of Colletotrichum acutatum. Notably, B-4359 was found to promote spore germination in C. acutatum instead of exhibiting a suppressive effect when the bacterial suspension was mixed with the spore suspension of C. acutatum. However, B-4359 showed an excellent biological control effect on the anthracnose of red pepper fruits. Compared to other treatments and untreated control, B-4359 played a more effective role in controlling anthracnose disease under field conditions. The strain was identified as B. halotolerans using BIOLOG and 16S rDNA sequencing analyses. The genetic mechanism underlying the biocontrol traits of B-4359 was characterized using the whole-genome sequence of B-4359, which was closely compared with related strains. The whole-genome sequence of B-4359 consisted of 5,761,776 bp with a GC content of 41.0%, including 5,118 coding sequences, 117 tRNA, and 36 rRNA genes. The genomic analysis identified 23 putative secondary metabolite biosynthetic gene clusters. Our results provide a deep understanding of B-4359 as an effective biocontrol agent against red pepper anthracnose for sustainable agriculture.
Plant pathogenic bacteria and fungi are responsible for major crop losses in terms of both the quality and quantity of agro-produce (Chakraborty and Newton, 2011). Approximately 10–15% of crop yield losses are caused by pathogenic infections globally (Peng et al., 2021). Phytopathogens produce toxins and enzymes that are harmful to host plants; even at low concentrations, these toxins are sufficient to destroy normal physiological functions in host plants (Thynne et al., 2017). Among bacterial phytopathogens, gram-negative bacteria in the genera, Xanthomonas, Pectobacterium, and Erwinia cause the most widespread and destructive losses in agricultural production (Mansfield et al., 2012).
Over the last two decades, the use of beneficial bacteria for the biological control of plant pathogens has become a major tool for protecting several crops (Samaras et al., 2021). Among several beneficial bacteria, species that can protect crop plants in several ways and promote plant growth either by direct or indirect mechanisms are commonly referred to as plant growth-promoting rhizobacteria (PGPR) (Gowtham et al., 2018). Plant diseases caused by fungi, bacteria, and viruses result in an estimated yield loss of 14% in diverse crops of agricultural importance, leading to an economic loss of USD 220 billion annually (Peng et al., 2021). Among these, plant pathogenic fungi are responsible for the majority of the devastating diseases, leading to famine and affecting food security worldwide (Fisher et al., 2012). Phytopathogenic fungi produce toxins that play a key role in the development of plant diseases, thereby adversely affecting host plants (Soyer et al., 2015). Many species of the genus Colletotrichum are associated with plant diseases commonly referred to as anthracnoses. Colletotrichum spp. can affect a wide range of hosts globally and cause crop loss in many staple foods (Prusky et al., 2000). Among the species of Colletotrichum, members of the Colletotrichum acutatum species complex have been well-documented causing diseases in a wide range of hosts (Talhinhas et al., 2005), including anthracnose in red pepper (Than et al., 2008).
Anthracnose in red peppers is associated with several Colletotrichum species, including C. acutatum (Harp et al., 2014), and may also infect other fruit and vegetable crops (Han et al., 2015). Red pepper anthracnose is typically characterized by dark brown to black, circular water-soaked spots with concentric rings of black acervuli developing beneath the skin of the fruit (Parisi et al., 2020); which often coalesce and cause softening and fruit rot (Montri et al., 2009). Anthracnose is typically controlled by the application of chemical fungicides under field conditions. The continuous use of chemical pesticides has increased public concern regarding the risks associated with hazardous residues in agricultural products and their adverse effects on ecosystem biodiversity (Meena et al., 2020). In addition, disease control using chemical pesticides is cost-effective for farmers in developing countries (Wesseling et al., 1997; Bordoh et al., 2020). Therefore, biological control using antagonistic microorganisms has emerged as an environment-friendly alternative for controlling plant diseases (Köhl et al., 2019; De la Lastra et al., 2021).
Brevibacillus spp. is one of the PGPR groups used as biofertilizers or biopesticides in different crops and against a variety of soil-borne and foliar pathogens (Nehra et al., 2016). Several studies have been conducted on antibiotic production by antagonistic microbes, such as Bacillus sp., Pseudomonas sp., and Streptomyces sp. (Beneduzi et al., 2012; Ngalimat et al., 2021). Bacillus spp. produce spores that can be used to develop an effective microbial biopesticide formulation in the form of a biocontrol agent (BCA) (Bonaterra et al., 2022). Several studies have demonstrated that secondary metabolites produced by antagonistic bacteria play key roles in controlling various phytopathogens (Han et al., 2015). In several cases, the control efficacy achieved by Bacillus strains has been associated with the production of antifungal and volatile organic compounds (VOCs) that induce plant defense reactions (Khalaf and Raizada, 2018). Whole-genome analysis of Bacillus species constitutes the basis for understanding the interactions between plants and other microorganisms (Magno-Perez-Bryan et al., 2015; Shaligram et al., 2016). Using genome mining, many antimicrobial compounds were discovered, such as brevicidine and antimicrobial cyclic lipopeptides, which are found in Brevibacillus laterosporus DSM 25 (Li Y. X. et al., 2018). Complete genome sequencing technology has good application prospects for the discovery of genome sequence information of unknown bacteria and the exploration of critical functional genes (Wang et al., 2020).
Considering these potential benefits, this study aimed to (i) screen in vitro antagonistic activity of various bacterial isolates obtained from the Freshwater Bioresources Culture Collection (FBCC), Korea, and identify effective antagonistic bacteria with inhibitory effects against major phytopathogens; (ii) evaluate the potential efficiency of the selected PGPR strain B-4359 as a BCA against red pepper anthracnose caused by C. acutatum under in vitro and field conditions and for plant growth promotion; and (iii) evaluate complete genome sequence of B-4359 and identify the divergent genomic characteristics among other Brevibacillus strains for comparative genome analysis.
All fungal pathogens tested in our study, including C. acutatum KACC 42403, Fusarium oxysporum f. sp. lycopersici KACC 40043, Colletotrichum coccodes KACC 48737, and Colletotrichum fructicola GYUN-11115 were procured from the Korean Agricultural Culture Collection (KACC), Korea. The antagonistic bacterium Brevibacillus halotolerans B-4359 was obtained from the FBCC, Korea. This bacterium was selected based on its antagonistic effects on various bacterial and fungal plant pathogens (provided in detail in the Supplementary material).
Brevibacillus halotolerans B-4359 was tested for in vitro antagonistic activity against five fungal pathogens, including C. acutatum KACC 42403, F. oxysporum f. sp. lycopersici KACC 40043, C. coccodes KACC 48737, and C. fructicola GYUN-11115, using a dual culture assay. Fungal pathogens were cultured onto potato dextrose agar (PDA) plates (90 mm diameter) at 25°C for 7 days. The bacteria B-4359 was cultured in tryptic soy agar (TSA) plates at 28°C for 2 d. A mycelial plug was removed from the fully grown PDA plate using a sterile cork borer (5 mm in diameter) and inoculated onto PDA medium supplemented with peptone (PDK) on one side 2 cm away from the edge, whereas the bacterium was streaked on the other side. Plates inoculated with fungal disks alone were used as controls. All the plates were incubated at 25°C for 10 d. The distance between B-4359 cells and the mycelium was measured as the inhibition of mycelial growth. The results are expressed as the percentage inhibition of the growth of fungal mycelia in the presence and absence of bacteria using the following equation (Montealegre et al., 2003).
The bacterial cells of B-4359 were cultured in tryptic soy broth (TSB) at 28°C under shaking conditions at 180 rpm for 24 h. The culture broth was centrifuged at 13,000 × g for 5 min at 4°C. The supernatant was collected and sterilized through a membrane filter (0.02 μm), which was used as a culture filtrate (CF). The mycelial plugs (5 mm diameter) of F. oxysporum f. sp. lycopersici KACC 40043, C. coccodes KACC 48737, C. fructicola GYUN-11115, and C. acutatum KACC 42403 were placed onto the center of PDK plates, and sterile paper discs (6 mm diameter) impregnated with 20 μL of the CF were placed onto the same plate at four different sides. Paper discs impregnated with TSB were used as untreated controls. Growth was measured 12 d after incubation at 25°C. Each treatment consisted of three replicates and the experiment was performed at least twice.
To determine the volatile organic compounds (VOCs) secreted by B. halotolerans B-4359 against the growth of fungal pathogens, exposure to B. halotolerans B-4359 VOCs was performed using the sandwich method described by Li N. et al. (2018). F. oxysporum f. sp. lycopersici KACC 40043, C. coccodes KACC 48737, C. fructicola GYUN-11115 and C. acutatum KACC 42403 were cultured on PDA plates at 25°C for 7 d. The antagonistic bacterium B-4359 was cultured on a TSA plate for 48 h. Mycelial plugs (5 mm diameter) of fungal pathogens were placed on the center of freshly prepared PDA plates, and bacteria were streaked onto TSA plates. Bacteria-inoculated plates were placed on top of PDA plates inoculated with mycelial plugs of the two fungal pathogens. Another set of fungal pathogen plates was sandwiched between TSA plates without streaked bacteria and used as a control group. All plates were sealed with a double layer of parafilm. Colony diameter was measured after incubating the C. acutatum KACC 42403 inoculated plates at 28°C for 12 d. The experiment was performed twice in triplicates.
For microscopic observation, a bacterial colony was picked with a sterile inoculation loop, placed in sterile distilled water (SDW) on a Formvar-coated copper grid, and stained with 2% uranyl acetate for negative staining. The preparation was examined using transmission electron microscopy (TEM; Hitachi, Japan). For bacterial endospore identification, the bacterium was streaked onto freshly prepared TSA plates and incubated at 28°C for 2 d. The bacterial colony was inoculated onto a glass slide and endospore formation was observed using a light microscope (Olympus BX43, Olympus, Tokyo, Japan).
To determine whether B-4359 is involved in inhibiting the conidia germination, the conidia from 7-d-old cultures at 25°C on PDA were harvested with SDW and adjusted to 105 spores/mL using a hemocytometer. The bacterial isolate B-4359 was cultured onto a TSA plate for 48 h at 28°C, and the cell suspensions were prepared in three different concentrations (106, 107, and 108 CFU/mL) in SDW. The conidial germination and appressorium formation of C. acutatum were tested on a covered glass surface treated with B-4359 bacterial suspensions using a previously described method (Kwak et al., 2012). Briefly, the conidial suspensions (10 μL) were mixed with bacterial suspensions (10 μL) in different concentrations and placed on a glass slide. Conidia suspensions without treating bacterial suspensions were considered as a control. The conidial germination and formation of appressorium and primary hyphae in the B-4359 treatment were assessed during incubation at 25°C and at different durations (0, 6, 18, 24, and 48 h) in Petri dishes containing moist paper. The glass slides were observed under a light microscope (Olympus BX43, Olympus, Tokyo, Japan). All the experiments were performed thrice in triplicate. The conidial germination inhibition rate (%) was calculated using the following formula:
To investigate the effect of B-4359 on anthracnose disease suppression in red peppers, fresh and healthy red pepper fruits of similar sizes were surface-sterilized in 1% NaOCl for 1 min and rinsed twice with SDW. The fungal pathogen C. acutatum GYUN-10586 causes anthracnose on red pepper and was isolated from the infected red pepper field, cultured onto PDA plates at 25°C, and used in our study for inoculation. Five wounds were made on one red pepper fruit using a sterile needle and B-4359 suspensions (10 μL) at 106 and 107 CFU/mL were dropped on each wound, allowed to dry, and the fruits were inoculated with 10 μL of fungal pathogen spore suspension (105 spore/mL) on each wound just by dropping. Pepper fruits treated with SDW served as a control. All fruits were placed on square plates (40 cm × 40 cm) containing moist paper. The disease severity (%) was recorded based on disease rating after incubating the plates at 28°C for 5 d. The disease severity (%) was calculated as follows: Based on the score, a disease index scale was established from 0 to 5 (0 = no symptoms, 1 = lesions with <10% disease incidence, 2 = symptoms with 11–20%, 3 = 21–40%, 4 = 41–70%, and 5 = symptoms with >71–100%. Disease severity (%) = [sum (class frequency × score of rating class)]/[(total number of fruits) × (maximal disease index)] × 100). The experiment was performed twice with five replicates.
In order to confirm the plant growth-promoting ability of B-4359, 3-week-old red-pepper seedlings were sown in plastic pots (10 cm diameter) containing horticultural soil (Baroker, Seoul Bio Co., Ltd., Eumseong, Korea). All the pots were treated with B-4359 bacterial suspensions at various concentrations (105, 106, and 107 CFU/mL) by soil drench (SD). Red pepper seedlings treated with SDW served as the control group. The plant growth (height) was measured after maintaining the plants at 25°C and 12 h/12 h (light/dark) for 20 d in a plant growth chamber.
To determine the growth curve of B-4359, a single bacterial colony of B-4359 cultured on TSA plates for 24 h was inoculated in TSB and incubated at 28°C for 72 h under shaking conditions (180 rpm). The optical density (OD) of culture broth was measured at 600 nm at 4 h intervals using a spectrophotometer, and 10 μL of the culture was inoculated onto TSA plates to check the number of colonies, from which CFU/mL value was calculated. The ability of B-4359 to produce IAA was also investigated. B-4359 was incubated at 180 rpm 28°C in TSB with tryptophan (500 μg/mL). The bacterial culture was centrifuged at 10,000 × g for 15 min, and 1 mL supernatant was transferred to a fresh tube to which 50 μL of 10 mM orthophosphoric acid and 2 mL of Salkowski reagent (1 mL of 0.5 M FeCl3 in 50 mL of 35% HClO4) were added. The mixture was incubated at room temperature (25°C) for 25 min and the absorbance of the pink color that developed was measured at 530 nm. The standard IAA solutions of 5, 10, 20, 50, and 100 μg/mL were prepared with culture medium, respectively. Absorbance was measured at 530 nm to obtain a standard curve.
To determine the effect of antagonistic B-4359 bacteria against red pepper anthracnose disease under field conditions, two-month-old healthy red pepper (cv. color king) seedlings were purchased from a local seedling market and transplanted into the field at a spacing of 40 × 100 cm. The experiment was conducted in a field at Andong National University, South Korea. The treatments were grouped as follows: control, chemical control, pyraclostrobin, pre-immersion with B-4359 (PI B-4359), foliar spray (FS) with B-4359 (FS B-4359), and foliar spray + soil drench with B-4359 (FS + SD B-4359). In the PI with B-4359 cell suspensions, the roots were pre-immersed in bacterial suspensions of B-4359 at 106 CFU/mL for 2 h before they were transplanted into the field. For the foliar spray treatment group, 250 mL/plant was used, whereas 500 mL/plant was used for the soil drench treatment group. After transplantation, all treatments were administered six times (10 d intervals) during the growth period of 60 d; the detailed treatment schedule is provided in Supplementary Table 1. Two months after transplantation, the first treatment was applied to red pepper plants. The disease rate (%) of anthracnose was calculated from disease-infected red pepper fruits after harvesting 2 weeks after the last treatment under field conditions. The disease rate (%) was calculated using the following formula: Disease rate (%) = Number of diseased fruits/total number of fruits × 100.
To investigate the effect of B-4359 on the expression of defense genes in red pepper plants, 2-month-old red pepper seedlings were treated with 50 mL of B-4359 cell suspensions (106 CFU/mL), 1 mM BTH (positive control) or SDW (negative control) using the foliar spray method. The leaves were sampled at 6 h and 24 h after treatment and stored at −80°C. Total RNA was isolated from red pepper leaf tissues using a RNeasy Plant Mini Kit (Qiagen, Hilden, Germany), following the manufacturer’s instructions. The cDNA synthesis was performed using iScript cDNA Synthesis Kit (Bio-Rad); real-time PCR was performed using SsoAdvanced Universal SYBR Green supermix (Bio-Rad) with housekeeping genes ACT F (5’-TGTTATGGTAGGGATGGGTC-3′), ACT R (5’-TTCTCTCTATTTGCCTTGGG-3′) (Wan et al., 2011), and pathogenesis-related genes CaPR1 F (5’-ACTTGCAATTATGATCC ACC-3′), CaPR1 R (5’-ACTCCAGTTACTGCACCATT-3′) (Yang et al., 2009). The thermocycler parameters were as follows: initial polymerase activation, 10 min at 95°C, and then 40 cycles of 30 s at 95°C, 60 s at 55°C and 30 s at 72°C. The expression level of the CaPR1 gene was compared by calculating relative transcript quantification using the ΔΔCT method.
Bacterial genomic DNA was extracted using a Solg Genomic DNA Prep Kit (Solgent, Daejeon, South Korea), following the manufacturer’s instructions. The concentration of the extracted DNA was determined using a Qubit 2.0 fluorometer (Invitrogen, Carlsbad, CA, United States). DNA contamination of the cultured strain was tested by sequencing the 16S rRNA gene using an ABI 3730 DNA sequencing machine (Applied Biosystems, Foster City, CA, USA). The integrity of the gDNA was verified using agarose gel electrophoresis and quantified using a Qubit 2.0 fluorometer (Invitrogen). Sequencing libraries were prepared according to the manufacturer’s instructions for 20 kb template preparation using the BluePippin Size-Selection System and PacBio DNA Template Prep Kit 1.0. Briefly, 10 μg of the gDNA was sheared to 20 kb using g-tubes (Covaris, Woburn, MA, United States); they were then purified, end-repaired, and the blunt-end SMARTbell adapters were ligated. The libraries were quantified using a Qubit 2.0 fluorometer (Invitrogen) and qualified using a DNA 12,000 chip (Agilent Technologies, Waldbronn, Germany). Subsequently, the libraries were sequenced using PacBio P6C4 chemistry in an 8-well SMART Cell v3 at PacBio RSII.
Five genes were selected based on the results of secondary metabolite analysis using antiSMASH. The PCR primers for the target genes from B-4359 and the housekeeping gene (16S rRNA) are shown in Table 1. Each primer name represents a gene corresponding to brevicidine for bre, macrobrevin for MAC, petrobactin for ptr6, basiliskamide for bask, bogorol A for bogA, and Brevibacillus 16S rRNA for br16. Total RNA from B-4359 cells was obtained from one-day-old cultures grown on TSA using the RNeasy Plus Mini kit (Qiagen, Valencia, CA, United States) according to the manufacturer’s instructions. The cDNA synthesis in 20 μL was performed using an iScript cDNA Synthesis Kit (Bio-Rad) following the manufacturer’s instructions. The SsoAdvanced Universal SYBR Green SuperMix (Bio-Rad) was used for real-time PCR analysis. Real-time PCR mixture in 10 μL of the total volume contained 5 μL of SYBR Green, 2 μL of each primer (5 pM), 2 μL of cDNA, and 1 μL of nuclease-free water. The thermal cycling program was set at 95°C for 3 min, 40 cycles at 95°C for 10 s, and 55°C for 30 s, followed by 55–95°C melting curve analysis.
Additional experiments, including a collection of fungal and bacterial pathogens, in vitro screening of antagonistic bacteria, in vitro enzyme activities by antagonistic bacteria, phosphate solubilization assays, siderophore production assays, and auxin and gibberellin detection assays, are described in detail in the Supplementary material.
Data were subjected to analysis of variance (ANOVA) using SAS JMP software (SAS Institute, Cary, NC, United States; SAS, 1995). Significant differences between treatment means were determined using the least significant difference (LSD) at p < 0.05. All the experiments were performed at least twice. For each experiment, data were analyzed separately. The results of a representative experiment are presented herein.
A total of 856 bacterial isolates were obtained from the FBCC, of which 57% were isolated from freshwater, 39% from the soil, and the remainder from sludge and plants. All the isolates were classified into 442 species and 182 genera (Table 2). Most of these genera come under the phylum Pseudomonadota, which accounts for the largest proportion of freshwater, soil, and other sources; while in the sludge, the phylum Bacillota was the most abundant (Supplementary Figure 1A). Among the strains isolated from freshwater, Flavobacterium sp. was the most common (9%), followed by Pseudomonas sp. and Rhodoferax sp., which accounted for 5% of the freshwater isolates. Of the strains isolated from the sludge, 50% were Bacillus spp., from which 20% were from the soil (Supplementary Figure 1B). Among all the isolates used for in vitro screening, only 65 strains exhibited antagonistic activity against at least one of the five pathogens used in our study, with a hit rate of 7.6%. Furthermore, the 65 strains were classified into 43 species and 25 genera. Of the isolates obtained from soil, 87% had no antagonistic ability, Bacillus sp., which accounted for 4%, and Pseudomonas sp., which accounted for 3%, showed the greatest antagonistic effect. In addition, 96% of strains from freshwater had no antagonistic ability and approximately 1% of Lactobacillus spp. showed the greatest antagonistic effect, and the other antagonists were less than 1% (data not shown).
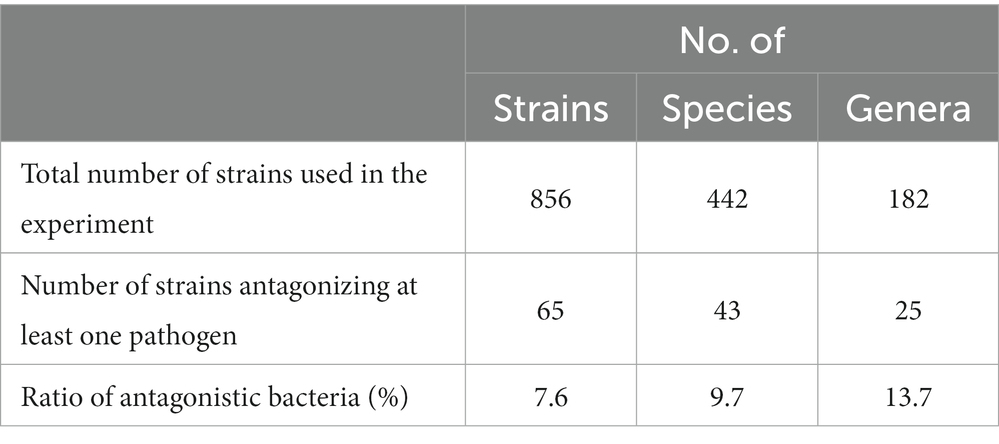
Table 2. Number of strains, species, and genera of the screened bacterial isolates used in this experiment.
As observed from the antifungal activity test, 44 strains inhibited the growth of the fungal pathogen C. acutatum with a hit rate of 5%; 15 strains inhibited the growth of the fungal pathogen F. oxysporum with a hit rate of 1.8% (Table 3). Antagonistic bacterial species, such as Streptomyces and Brevibacillus, were found to exhibit greater antifungal activity than other species in our study against C. acutatum and F. oxysporum (Supplementary Table 2). The strain Streptomyces sporoverrucosus B-1662 exhibited the greatest antagonistic activity against C. acutatum with an average inhibition percentage of 81, whereas the growth of the fungal pathogen F. oxysporum was strongly inhibited by the antagonistic strain S. virginiae B-4370, with an inhibition percentage of 71.1% (Supplementary Table 2). Three isolates, S. virginiae B-4371, Brevibacillus halotolerans B-4366, and S. sporoverrucosus B-1662, showed greater antagonistic activity against F. oxysporum with inhibition percentages of 68.3, 68.3, and 63.2, respectively (Supplementary Table 2).
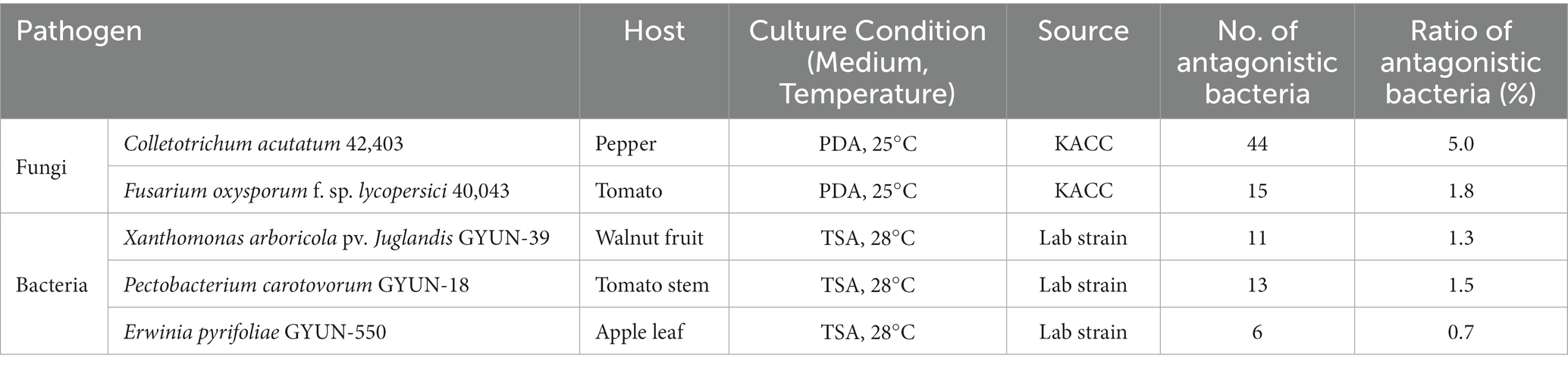
Table 3. List of the five phytopathogens tested in this study and their culture conditions, and a summary of the antagonistic activity results of all 856 isolates against pathogens.
In the case of the antibacterial activity test, 11, 13, and 6 strains showed antagonistic activity against the bacterial pathogen, Xanthomonas arboricola pv. juglandis GYUN-39; Pectobacterium carotovorum GYUN-18; and Erwinia pyrifoliae GYUN-550. The hit rate was determined to be 1.3, 1.5, and 0.7% for X. arboricola, P. carotovorum, and E. pyrifoliae, respectively (Table 3). Some species such as Paenibacillus terrae and Bacillus mycoides, exhibit antagonistic activity against X. arboricola. The three isolates of P. terrae showed different antagonistic activities against the three pathogenic bacteria. Among them, the isolate B-248 exhibited an antagonistic effect on C. acutatum, F. oxysporum, E. pyrifoliae, and X. arboricola but no activity against P. carotovorum (Supplementary Table 3). Two antagonistic strains, such as Lactobacillus plantarum subsp. argentoratensis and B. halotolerans exhibited greater antagonistic activity against P. carotovorum than the other strains. These three isolates were obtained from freshwater. In contrast, the other two strains, B-367 and 368, which were also isolated from the same area, showed no antagonistic effect against any of the five pathogens (data not shown). Similarly, antagonistic strains such as B. halotolerans and Chromobacterium alkanivorans exhibited the highest antagonistic activity against the bacterial pathogen E. pyrifoliae (Supplementary Table 3). From in vitro screening, nine strains exhibiting substantial antagonistic activity (in mm diameter) against phytopathogens were selected for further studies. The strain B-4359 was selected for further studies based on its antagonistic activity against several phytopathogens and various enzyme activities. The strain B-4359 was evaluated for the control of red pepper anthracnose because of the increased incidence of the disease in Korea.
Nine effective strains were evaluated for starch hydrolase, cellulase, protease, and chitinase activities, and each strain displayed different enzyme activities in vitro (Supplementary Figure 2). Strain B-4359 produced protease at a greater level and a lower level of chitinase, but two other enzymes, starch hydrolase, and cellulase, were not produced by the bacteria. The inhibition zones around the inoculation sites on the solid media in the Petri dishes were determined by the degradation of various substrates, such as soluble starch, CMS, proteins, and chitin, by amylase, cellulase, protease, and chitinase, respectively. In contrast, bacterium B-4359 is involved in the production of either siderophores or phosphate solubility (Supplementary Figure 2). This indicates that the mode of action of the bacterium might involve other mechanisms. Indole and gibberellin content were analyzed from our bacteria isolate B-4359, but it could not produce indole; while gibberellin was produced at 855.6 μg/mL (Supplementary Figure 3). This indicates that the strain may promote plant growth.
Morphological characteristics of the B. halotolerans isolate B-4359 obtained from FBCC were smooth, glossy, light brown, and flat colonies when cultured on TSA medium after 48 h incubation at 28°C (Figure 1A). When observed using a TEM, the bacterial cells were found to be rod-shaped, measuring approximately 0.84–1.49 μm × 2.08–4.10 μm (width × length; Figure 1B), motile possessing peritrichous flagella (Figure 1C). The bacterial cells were found to be gram-positive using Gram staining, and endospores were observed when the bacteria were cultured for 7 d in TSA (Figure 1D). Most of these characteristics are similar to those of Brevibacillus halotolerans sp. nov., as described by Song et al. (2017).
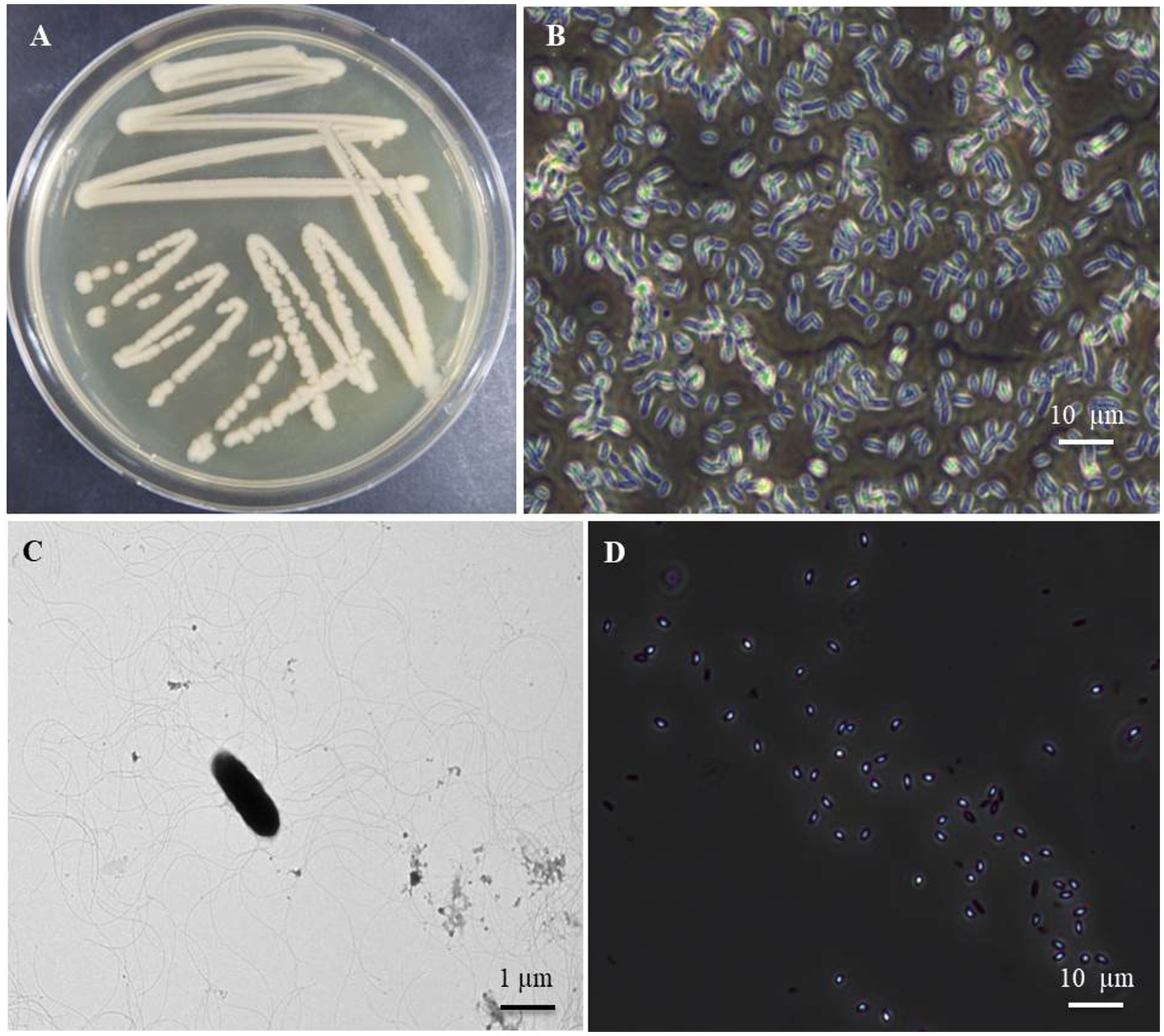
Figure 1. Cultural and morphological characteristics of the bacterial isolate B. halotolerans B-4359 using transmission electron microscopy (TEM) and light microscope. (A) Bacteria cultured at 28°C for 48 h showed smooth glossy and rod-shaped colonies, (B) Bacterial cells appeared rod-shaped under light microscopy with 400X magnification, and (C) rod-shaped bacteria appears with peritrichous flagella through TEM (Bar = 1 μm). Bar = 10 μm. (D) Endospores were observed in the 7-day-old cultured broth (Bar = 10 μm).
Bacterium B-4357 was tested for in vitro antagonistic activity against four fungal pathogens, including C. acutatum KACC 42403, F. oxysporum KACC 40043, C. coccodes KACC 48737, and C. fructicola GYUN-11115, which cause anthracnose, Fusarium wilt, leaf anthracnose, and apple bitter rot, respectively. Mycelial growth of all pathogenic fungi was inhibited by the B-4359 suspensions to a greater extent than that of the untreated control (Figure 2A). Among the four fungal pathogens, mycelial growth of C. coccodes KACC 48737 was greatly inhibited by B-4359 13 d after incubation at 25°C, when compared with the growth inhibition of other three pathogens. B-4359 showed inhibition percentages of 24.3, 27.2, 35.7, and 27.7 for C. acutatum KACC 42403, F. oxysporum KACC 40043, C. coccodes KACC 48737, and C. fructicola GYUN-11115, respectively.
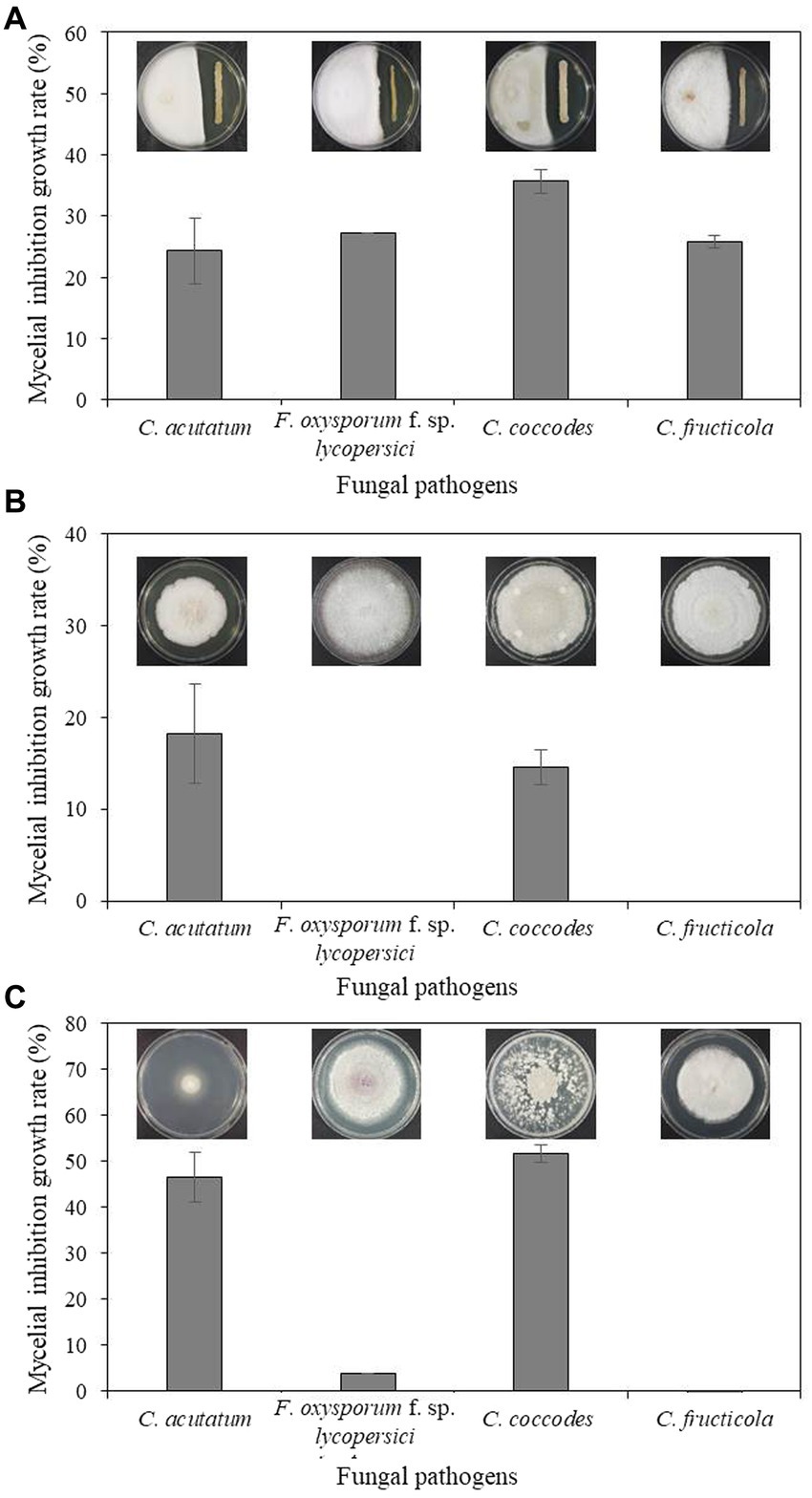
Figure 2. (A) In vitro antagonistic effect of B. halotolerans B-4359 against fungal plant pathogens. An in vitro antagonistic activity of B. halotolerans B-4359 against fungal pathogens (C. acutatum, F. oxysporum f. sp. lycopersici, C. coccodes, and C. fructicola) was tested using a confrontation plate assay by inoculating the mycelial plug at 2.0 cm away from one edge of the PDK plates and streaked with bacterial cultures at another edge on the same plate. The incubation days were 13, 10, 10, and 7 days for C. acutatum, F. oxysporum, f. sp. lycopersici, C. coccodes, and C. fructicola, respectively. (B) Inhibitory effect of culture filtrate (CF) of antagonistic bacterium B. halotolerans B-4359 on the fungal growth of C. acutatum. Paper discs impregnated with TSB were used as a non-treated control. The diameter of mycelial growth of fungal pathogens on PDK plates was recorded 12 d after incubation at 25°C. (C) The inhibitory effect of volatile organic compounds (VOCs) produced by B. halotolerans B-4359 against the growth of fungal pathogens was tested using a sandwich method. The experiment was performed twice with triplicates.
The cell-free CF of strain B-4359 was tested for the growth of C. acutatum. The colony diameter of the B-4359-CF-treated fungi was reduced relative to that of the untreated control (Figure 2B). The resulting inhibition rates of mycelial growth were 18.21 and 14.63% for C. acutatum KACC 42403 and C. coccodes KACC 48737, respectively, in the B-4359-CF treatment. This indicated that the presence of secondary metabolites in our antagonistic bacteria played a role in the growth inhibition of fungal pathogens. Furthermore, we tested whether VOCs produced by the antagonistic bacterium B. halotolerans B-4359 inhibited the growth of C. acutatum KACC 42403 in vitro (Figure 2C). The pathogen C. acutatum was inhibited by the VOCs produced by B. halotolerans B-4359 to a greater level using the sandwich method. The mycelial growth inhibition rates of C. acutatum KACC 42403 and C. coccodes KACC 48737 were 46.3 and 51.7%, respectively, by treatment with the presence of VOCs released from B. halotolerans isolate B-4359. This resulted in strain B-4359 having the ability to produce VOCs against the growth of C. acutaum. The mycelial growth of F. oxysporum KACC 40043 and C. fructicola GYUN-11115 was not inhibited by the VOCs produced by the B-4359 strain.
When the conidial spores of C. acutatum were treated with B-4359 bacterial suspensions in various concentrations under in vitro conditions, spore germination occurred faster than in the non-treated control (Figure 3A). No marked difference was observed in appressorium formation between the B-4359 treatment and control groups. B-4359-treated conidia at 107 CFU/mL resting on the hard glass surface started to germinate after the 6th hour of incubation, whereas water-treated conidia did not germinate. Similarly, the spore germination rate (%) had not exceeded 30% after the 18th h of incubation, whereas B-4359-treated conidia showed a germination rate of more than 70% at all concentrations (Figure 3B). These results suggest that B-4359 cell suspensions do not play a role in suppressing the growth of conidial spores of the fungal pathogen C. acutatum in vitro.
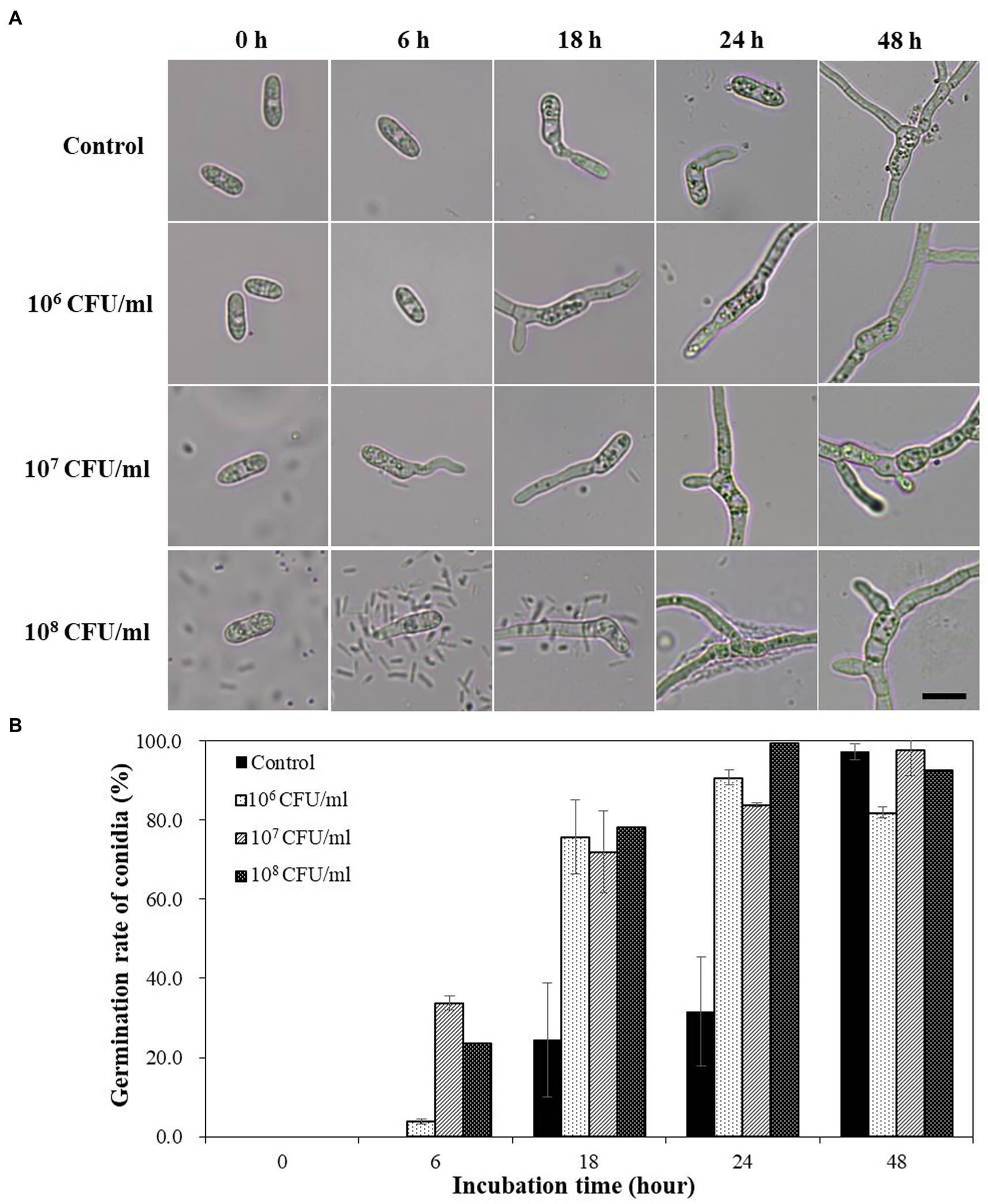
Figure 3. Effect of B-4359 cell suspensions on conidial germination rate and microscopic observation. (A) Microscopic observations of the fungal spore germination of C. acutatum GYUN-10586 after B. halotolerans B-4359 treatment during the incubation period from 0 to 48 h. (Bar = 10 μm). (B) The percentage of conidial germination rate by treatment with B-4359 cell suspensions was compared to the non-treated control. The experiment was repeated three times with three replicates per treatment producing similar results. The representative image is presented here.
B-4359 was tested for its preventive effect against the anthracnose pathogen C. acutatum GYUN-10586, a laboratory-owned strain isolated from our red pepper field at Andong National University. Disease severity (%) was reduced to a greater level at both concentrations of B-4359 suspensions in comparison with a non-treated control after 5 days of incubation (Figure 4). Disease severity was 37 and 58% in B-4359-treated plants at 106 and 107 CFU/mL, respectively, whereas it was 94% in non-treated control.
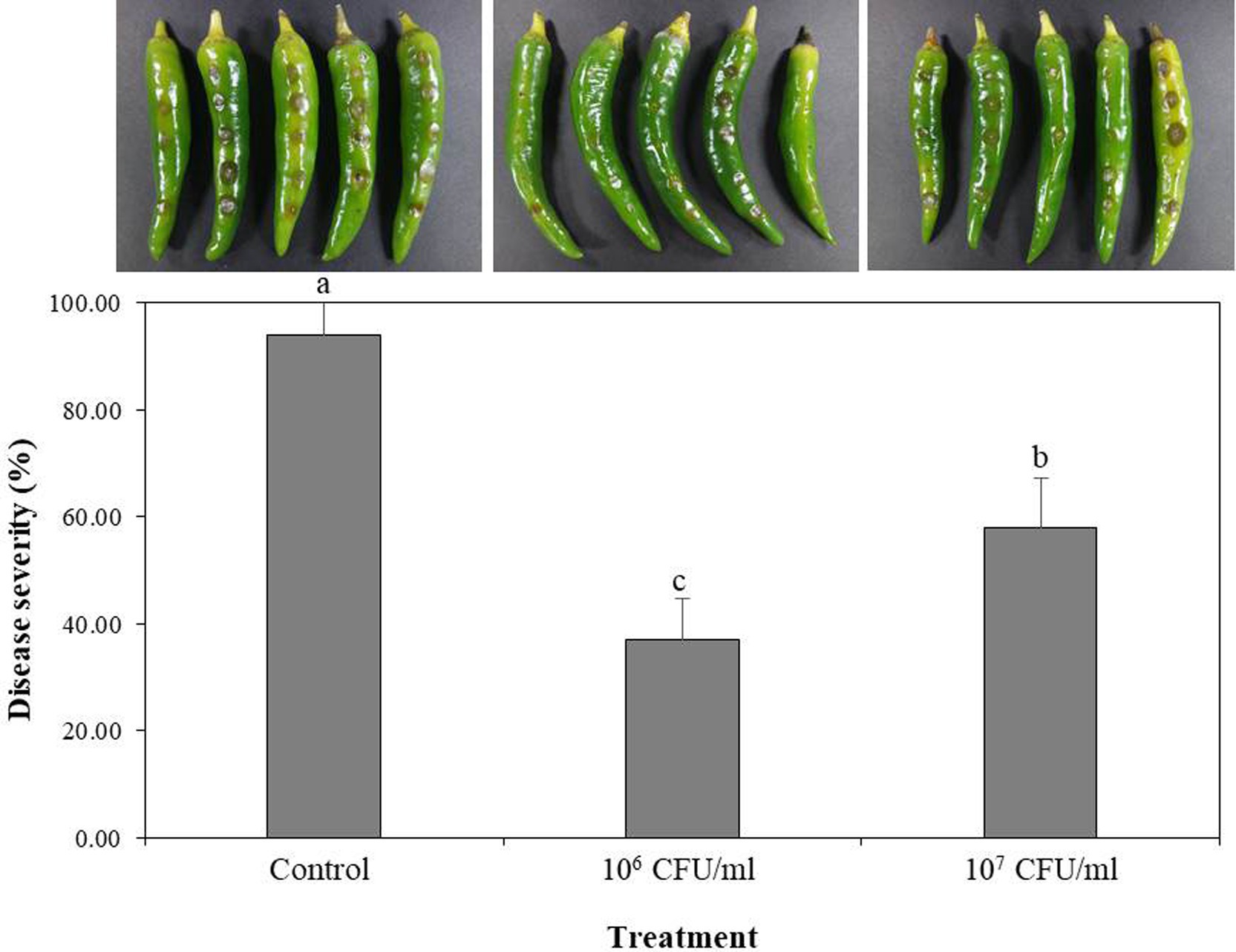
Figure 4. Effect of B. halotolerans B-4359 cell suspensions on suppression of disease severity (%) of anthracnose caused by C. acutatum GYUN-10586 on red-pepper fruits. Red-pepper fruits treated with SDW served as a control. The experiment was performed two times with five replicates. The results were compared with a non-treated control 3 days after incubation at 25°C. The experiment was performed two times with three replicates. Bars with the same letters do not differ from each other according to the least significant difference (LSD) (p < 0.05).
Bacterial growth curve analysis of B-4359 in liquid culture revealed that the growth has increased rapidly from 12 h onwards. This was determined by measuring the absorbance of the culture broth at an optical density (OD) of 600 nm using a spectrophotometer (Figure 5A). The growth of bacteria remained in the stationary phase from 24 to 72 h. Bacterial colonies were found to be the highest after 24 h of incubation on TSA medium at 108 CFU/mL, and the number of bacterial colonies decreased until 56 h of incubation and thereafter started to increase again. Isolate, B-4359 was further assayed for IAA production in association with plant growth-promoting activity. Strain B-4359 was more effective in IAA detection than the untreated control (Figure 5B). The IAA production detected on the sixth day by B-4359 cells was about 3-fold increased at 27.3 μg/mL, whereas on the third day, IAA was detected at 9.4 μg/mL. This indicated that B-4359 synthesis high amounts of IAA, depending on exogenous tryptophan. IAA detection in B-4359 effectively promoted plant growth.
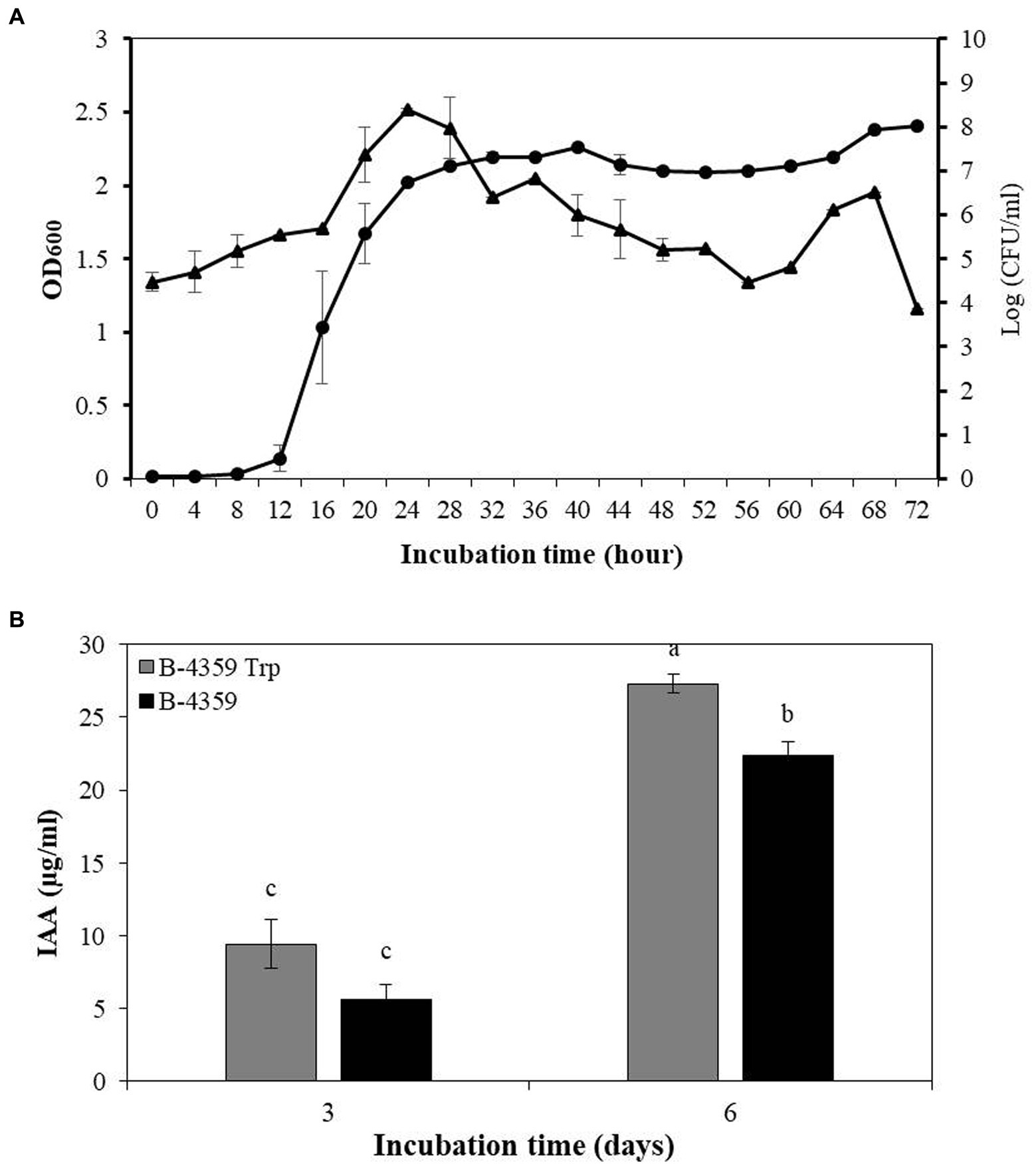
Figure 5. Bacterial growth curve and IAA production of B. halotolerans B-4359. (A) Bacterial growth curve of B. halotolerans B-4359 incubated in TSB at 28°C and 180 rpm. The optical density (OD) was measured at 600 nm in every 4 h interval. The plot represents as OD600 (●) and Log (CFU/mL) (▲). (B) IAA production by B. halotolerans B-4359. The absorbance of pink color development at 530 nm was measured. B-4359 was incubated in TSB with tryptophan (500 μg/mL) at 28°C and 180 rpm. An IAA standard curve was used to quantify the absorbance of the pink produced at 530 nm.
Bacterium B-4359 was tested for its ability to suppress red pepper anthracnose caused by C. acutatum under field conditions. The B-4359 treatment showed a significant (p < 0.05) effect on the reduction of anthracnose, with a disease rate of 0.4% by pre-immersion, compared with the foliar spray or foliar spray + soil drench, with 41.1 and 27.8%, respectively, 2 weeks after the last treatment. When red pepper seedlings were treated with B-4359 by pre-immersion for the first time, followed by chemical treatments exhibited a greater reduction of anthracnose disease than when plants were treated with various chemical fungicides at all six times without any pre-immersion. Overall, the treatment with B-4359 by foliar spray or foliar spray + soil drench showed better performance in controlling disease incidence (%) than the chemical treatment (pyraclostrobin) and a non-treated control (Figure 6). For all treatments, the disease rate (%) in the first experiment was lower than that in the second experiment. This may be due to the development of resistance at an earlier stage of bacterial treatment.
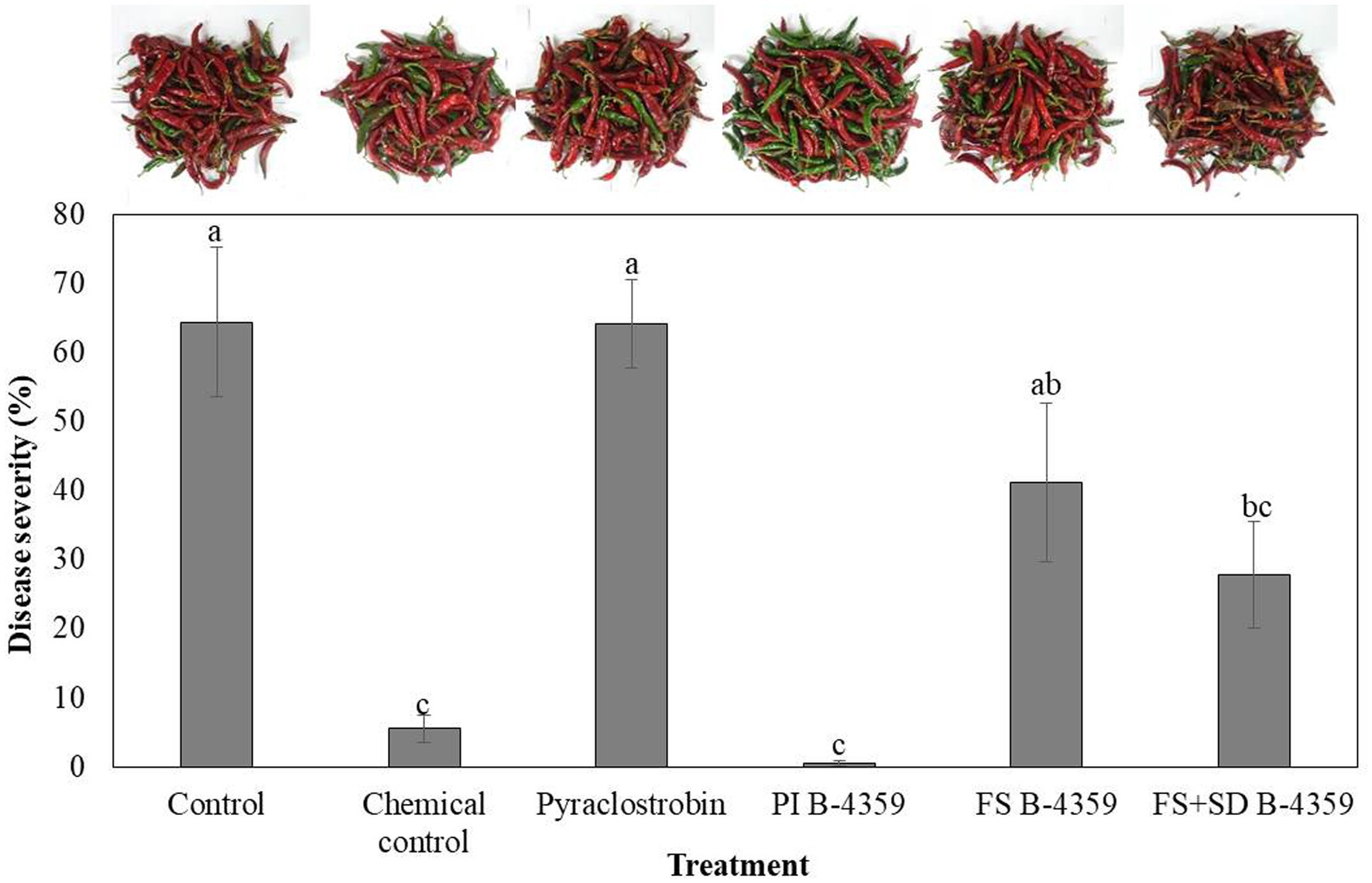
Figure 6. Suppression of anthracnose by treatment with B-4359 under field conditions. After transplanting two-month-old red-pepper seedlings in the field, the plants were treated with B-4359 by soil drench or foliar spray or mixing of both, control (water), chemical control, pyraclostrobin (negative control) six times in 60 d. Disease rate (%) was recorded from disease-infected red-pepper fruits after 10 days of the last treatment. The data for B-4359 were compared with the control. For all the treatments, three different plots with 20 replicates (plants) were used. Differences in letters on bars indicate statistically significant between the treated and the control according to LSD (p < 0.05). PI: pre-immersion, FS: foliar spray, SD: soil drench.
PacBio RSII NGS equipment was used at a sequencing depth of 179.344× to obtain complete sequence data for B-4359. The generated raw data were assembled using the HGAP2 protocol to obtain a FASTA file consisting of a single contig. The genome of B-4359 strain consisted of two circular chromosomes of 5,761,776 bp (contig 1, 414,359 bp and contig 2, 5,347,417 bp), with 5,118 predicted protein-coding sequences (CDSs), 36 rRNA genes, 117 tRNA genes, and an average G + C content of 41% based on NCBI Prokaryotic Genomes Automatic Annotation Pipeline (PGAAP) analysis. The first outer circle in the gray region of the genome map represents one contig, second circle represents the forward strand, third circle represents the CDSs on the reverse strand, and fourth circle indicates the tRNA and rRNA positions. The fifth circle represents the GC skew used as a reference point. Values higher than this are indicated in green, whereas lower values are indicated in red. The sixth circle is the GC ratio metric, where values greater than the average GC ratio are expressed in blue and lower values are represented in yellow; the GC skew and GC ratio are expressed at 10 kb intervals (Figure 7A).
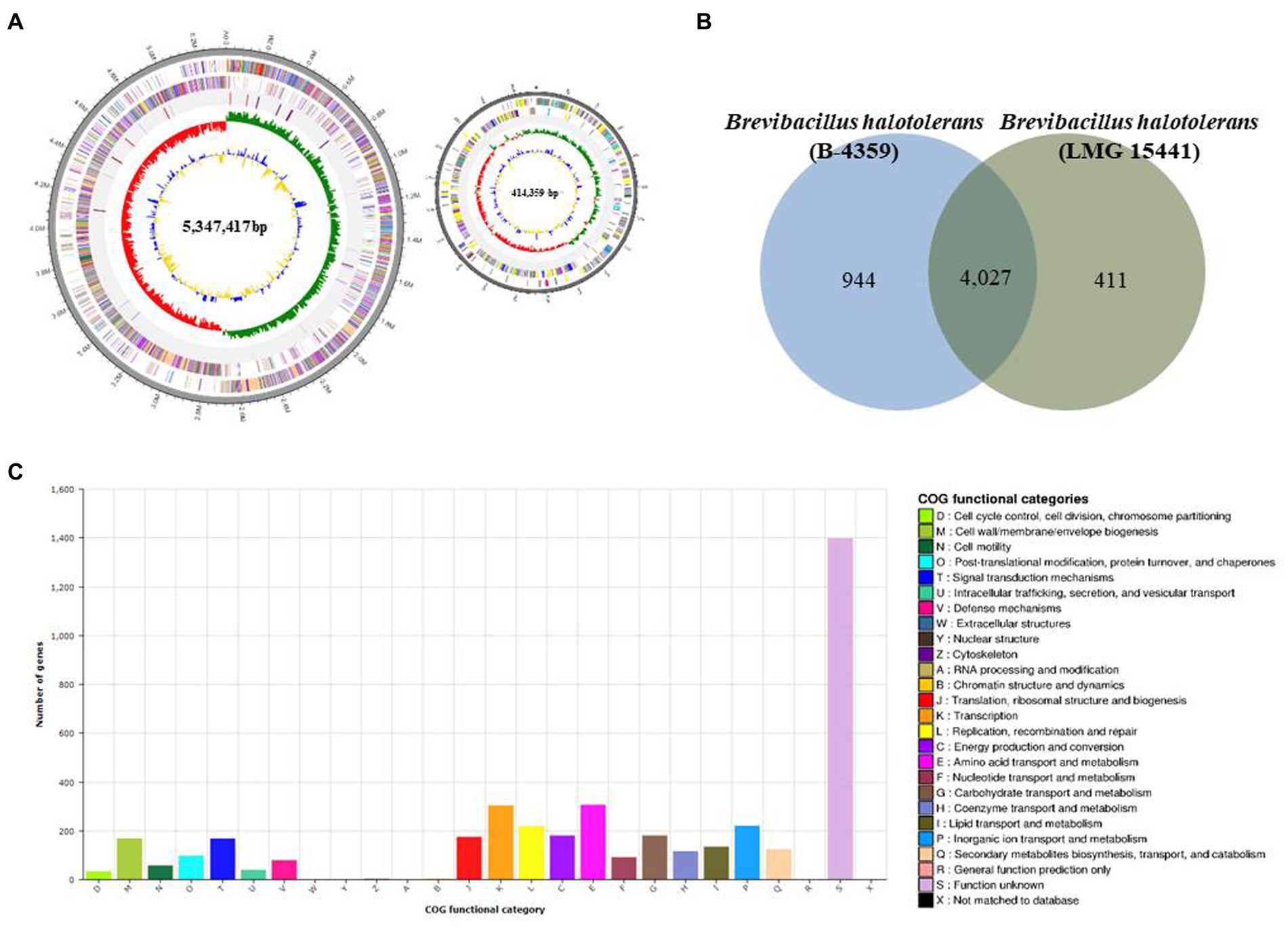
Figure 7. Whole-genome map and COG annotation of Brevibacillus halotolerans B-4359. (A) Marked characteristics are shown from the outside to the center; coding sequence (CDS) on the forward strand, CDS on the reverse strand, tRNA, rRNA, guanine-cytosine (GC)-content, and GC skew. CDS genome consists of a single circular chromosome that is 5.7 mb in size, and represents one contig from the outer part of the circle; the second circle represents forward, the third represents reverse strain, and the fourth circle represents tRNA and rRNA positions. (B) Distribution of orthologous genes in the B-4359 and B. halotolerans LMG 15441. The Venn diagram shows the summary of unique SNPs from the total genes of the B-4359. This analysis exploits all CDS of the genomes and is not restricted to the core genome. (C) The clusters of orthologous genes (COG) function annotation of B-4359, and distribution of genes in different COG function categories.
The Venn diagram in Figure 7B compares the common CDSs between B. halotolerans B-4359 and B. halotolerans LMG 15441. This section summarizes the unique protein-encoding genes among all B-4359 genes in total. In total, 4,027 common high-expression gene families were shared between B-4359 and LMG 15441 (Figure 7B), whereas 944 and 411 genes were unique to B-4359 and LMG 15441, respectively. All predicted CDSs of B-4359 were compared with the clusters of orthologous genes (COG) databases to identify homologous amino acid sequences. Each functionally annotated protein was assigned a COG number, representing a class of proteins; then, the proteins were subjected to functional clustering analysis according to the COG function. Totally, 123 and 81 genes were associated with secondary metabolites biosynthesis, transport, catabolism, and defense mechanisms, respectively (Figure 7C). Furthermore, secondary metabolite biosynthesis genes of B-4359 were detected using antiSMASH. A total of 23 biosynthetic gene clusters were identified, of which 8 secondary metabolite gene clusters encoding NRP biosynthesis were identified (Supplementary Table 4). A genome search revealed that strain B-4359 produces several antimicrobial compounds that suppress the growth of fungal pathogens. Five secondary metabolites with >95% similarity were identified using antiSMASH. In gene expression for five secondary metabolite gene clusters using real-time PCR, basiliskamide, and macrobrevin were expressed at a higher level than the other gene clusters (Supplementary Figure 4).
From the in vitro screening performed in our study, nine antagonistic bacterial strains were selected from 856 isolates obtained from FBCC, Korea. From these nine isolates, based on in vitro antagonistic activity against three fungal pathogens (C. acutatum, F. oxysporum, and R. solani) and three bacterial pathogens (X. arboricola, P. carotovorum, and E. pyrifoliae), and in vitro enzyme activity, only one representative isolate B. halotolerans B-4359 was selected for further studies. Brevibacillus can be found in a variety of environments including soil, seawater, and animal intestines, and it produces various metabolites with antifungal activity that can control plant diseases as BCAs (Yang and Yousef, 2018). Many biological and chemical agents have been used to control bacterial and fungal pathogens in the field (Lee et al., 2017). In recent years, the demand for safe and eco-friendly agricultural products has been increasing (Huh and Kim, 2010). Biological control practices are in high demand as alternatives to synthetic pesticides. Biological control practices are particularly important in organic crop production (Cook, 1993; Yang et al., 2002; Kim and Yun, 2011). Although Brevibacillus spp. possessed fewer antagonistic effects on plant pathogens than Bacillus spp. and Pseudomonas spp., they perform biological control functions against fungal plant pathogens and pests (Joo et al., 2005; Prasanna et al., 2013; Le Han et al., 2022). B. halotolerans has recently been reported as a new species with larvicidal activity (Katak et al., 2021). In addition, the abundance of bacteria in the genera Bacillus, Microbacterium, and Pseudomonas have been reported to be effective for biological control, with a positive correlation with Brevibacillus sp. (Li et al., 2021). This study describes the biological control effects and characterization of B. halotolerans B-4359.
Secondary metabolites, including volatile and non-volatile compounds produced by different bacterial species, play different roles in plants (Rao et al., 2016; Elnahal et al., 2022). The VOCs derived from bacterial sources contribute to disease suppression (Schulz-Bohm et al., 2017). Several Bacillus species produce VOCs that exhibit antifungal activities against various phytopathogenic fungi (Zhao et al., 2019; Calvo et al., 2020). Similarly, many other soil bacteria have been displayed to produce VOCs with antifungal effects that contribute to the inhibition of conidia (Garbeva et al., 2011). VOCs produced by Streptomyces spp. exhibit antifungal properties against Rhizoctonia solani, resulting in plant disease suppression (Cordovez et al., 2015); while VOCs from the tomato rhizosphere bacterium, Pseudomonas donghuensis P482, exhibit strong antifungal activity (Ossowicki et al., 2017). These reports are in agreement with our result, where the VOCs derived from strain B-4359 were effective in inhibiting the growth of fungal mycelia of C. acutatum using the sandwich plate method. However, the cell-free CF of B-4359 inhibited pathogenic mycelial growth to some extent compared to a non-treated control, but not to a significant level. In support of this, Wang et al. (2009) previously reported that the CFs from Bacillus spp. exhibited remarkable antifungal activity against several fungal phytopathogens, whereas some other studies reported that the CFs of B. amyloliquefaciens AK-0 inhibited the growth and spore germination of Cylindrocarpon destructans (Kim et al., 2016).
Our strain B-4359 did not exhibit an inhibitory effect on conidial germination when mixed with conidial suspensions of the fungal pathogen; however, treatment with B-4359 cell suspensions promoted spore germination in comparison with that of non-treated control. But the general phenomenon is that when new biocontrol products are to be developed, strains are mostly selected based on their ability to inhibit our target pathogens under in vitro conditions via antagonistic effects or in vitro suppression of spore germination. Strains with no in vitro effect are often eliminated without further testing in planta. Our results are supported by a previous report (Besset-Manzoni et al., 2019), where the BCAs have not exhibited antagonistic effects in vitro but had good efficacy in planta. In contrast, a previous study by Han et al. (2015) stated that the conidial germination of C. gloeosporioides was inhibited by four B. atrophaeus strains, of which the B. atrophaeus strain HM03 suppressed the conidial germination of C. gloeosporioides and C. acutatum. This antagonistic effect may be owing to the secretion of certain antifungal metabolites by Bacillus spp. (Fan et al., 2017). Of note, B-4359 displayed a disease-control effect against anthracnose in red pepper fruits. Brevibacillus is known to have a germination inhibitory effect on spores of other fungal phytopathogens, such as Alternaria sp., Fusarium sp., and Colletotrichum sp. (Saber et al., 2003; Chandel et al., 2010; Ahmed, 2017), but such an inhibitory effect was not observed with B-4359 treatment.
Furthermore, BCAs substituted for chemicals in modern agriculture require the ability to stimulate plant growth and ameliorate diseases (Kamil et al., 2018). The plant growth-promoting ability of B-4359 was tested, and the B-4359 suspension treatment promoted plant growth. Plant growth-promoting bacteria with biocontrol abilities can produce secondary metabolites such as antibiotics and phytohormones (Backer et al., 2018). Our results showed that B-4359 produced IAA in addition to effectively inhibiting pathogenic fungi, resulting in promoting plant growth. The ability of bacteria to produce IAA is an important factor that influences plant growth (Chandra et al., 2018). These compounds stimulate root growth, resulting in a larger root surface area that enables plants to access more nutrients from the soil, thereby promoting plant growth (Souza et al., 2013). The application of B-4359 in the field showed an excellent biological control effect in the second investigation. Disease incidence under field conditions is affected by a variety of parameters, such as the nature of pathogenicity, environmental factors, and soil microbes (Wei et al., 2015). Strain B-4359 did not directly antagonize the pathogen in vitro or inhibit spore germination; it did induce the expression of the plant systemic resistance marker gene CaPR1 and the housekeeping gene actin (ACT).
Our study further reports the whole-genome sequence of strain B-4359, which consists of chromosome 5,761,776 bp. The B-4359 genome was compared with those of similar species of the same genus using whole-genome analysis. The non-ribosomal peptide (NRP) gene clusters of Brevibacillus spp. encode antimicrobial cyclic peptides such as bacillibactin, brevicidine, and brevicidine B (Li Y. X. et al., 2018; Zhao and Kuipers, 2021). In particular, biocontrol-related genes and gene clusters involved in antibiotic resistance could result in differences in biocontrol targets and efficacy between B-4359 and other Brevibacillus strains. These results suggest that all other strains could prevent disease through the genes involved in the synthesis of secondary metabolites. The genome of strain B-4359 was investigated for the presence of secondary metabolites using antiSMASH and eight secondary metabolite gene clusters encoding NRP biosynthesis were identified. Among them, siderophore (petrobacterin) and three antibacterial peptides (bogorol A, macrobrevin, brevicidine) were identified as 100% in contig 2; they play a role in the suppression of pathogens by inducing systemic resistance (Schneider et al., 2007). Similarly, in a recent report by Le Han et al. (2022), genomic analysis of the strain B. halotolerans 7WMA2 isolated from marine sediments revealed 23 putative biosynthetic secondary metabolite gene clusters responsible for NRP, polyketides, and siderophores.
In conclusion, strain B-4359 obtained from FBCC exhibited remarkable antagonistic activity against several fungal pathogens, including C. acutatum, which causes anthracnose in red peppers. B-4359 cell suspensions suppressed red pepper anthracnose in vitro despite promoting conidial germination. This strain produced IAA and showed growth-promoting effects on red pepper seedlings. The B-4359 suspension treatment has been demonstrated to control red pepper anthracnose under field conditions in Andong, Gyeongbuk Province, South Korea. Whole-genome sequencing revealed that the core genome of B-4359 was 98.6% similar to that of B. halotolerans LMG 15441. The strain was identified as B. halotolerans B-4359 based on the whole-genome sequence analysis. The whole-genome sequencing of strain B-4359 revealed antimicrobial secondary metabolites such as bogorol A, basiliskamide A/B, macrobrevin, brevicidine, and tauramamide. Petrobactin siderophore-coding regions have also been identified. Our results indicate that B-4359 may be a promising BCA for controlling phytopathogens in an eco-friendly manner. Future studies will apply proteomic and transcriptomic methods to investigate the signaling pathways involved in the antagonistic effects of secondary metabolites.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
HK and YL contributed to design and perform the experiments. Y-JH and M-HL were involved in the field experiments. YL performed the RNA sequencing experiment and supervised the project. KB and YJ analyzed the data and wrote the manuscript. All authors contributed to the manuscript and approved the submitted version.
This work was supported by the Korea Environment Industry & Technology Institute (KEITI) through a project to make multi-ministerial national biological research resources a more advanced program funded by the Korea Ministry of Environment (MOE) (2021003420002).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1200023/full#supplementary-material
Ahmed, A. I. (2017). Biological control of potato brown leaf spot disease caused by Alternaria alternata using Brevibacillus formosus strain DSM 9885 and Brevibacillus brevis strain NBRC 15304. J. Plant Pathol. Microbiol. 08, 1–8. doi: 10.4172/2157-7471.1000413
Backer, R., Rokem, J. S., Ilangumaran, G., Lamont, J., Praslickova, D., Ricci, E., et al. (2018). Plant growth-promoting Rhizobacteria: context, mechanisms of action, and roadmap to commercialization of biostimulants for sustainable agriculture. Front. Plant Sci. 9:1473. doi: 10.3389/fpls.2018.01473
Beneduzi, A., Ambrosini, A., and Passaglia, L. M. (2012). Plant growth-promoting rhizobacteria (PGPR): their potential as antagonists and biocontrol agents. Gen. Mol. Biol. 35, 1044–1051. doi: 10.1590/S1415-47572012000600020
Besset-Manzoni, Y., Joly, P., Brutel, A., Gerin, F., Soudière, O., Langin, T., et al. (2019). Does in vitro selection of biocontrol agents guarantee success in planta? A study case of wheat protection against fusarium seedling blight by soil bacteria. PLoS One 14:e0225655. doi: 10.1371/journal.pone.0225655
Bonaterra, A., Badosa, E., Daranas, N., Francés, J., Roselló, G., and Montesinos, E. (2022). Bacteria as biological control agents of plant diseases. Microorganisms 10:1759. doi: 10.3390/microorganisms10091759
Bordoh, P. K., Ali, A., Dickinson, M., Siddiqui, Y., and Romanazzi, G. (2020). A review on the management of postharvest anthracnose in dragon fruits caused by Colletotrichum spp. Crop Prot. 130:105067. doi: 10.1016/j.cropro.2019.105067
Calvo, H., Mendiara, I., Arias, E., Gracia, A. P., Blanco, D., and Venturini, M. E. (2020). Antifungal activity of the volatile organic compounds produced by Bacillus velezensis strains against postharvest fungal pathogens. Postharvest Biol. Technol. 166:111208. doi: 10.1016/j.postharvbio.2020.111208
Chakraborty, S., and Newton, A. C. (2011). Climate change, plant diseases and food security: an overview. Plant Pathol. 60, 2–14. doi: 10.1111/j.1365-3059.2010.02411.x
Chandel, S., Allan, E. J., and Woodward, S. (2010). Biological control of fusarium oxysporum f. sp. lycopersici on tomato by Brevibacillus brevis. J. Phytopathol. 158, 470–478. doi: 10.1111/j.1439-0434.2009.01635.x
Chandra, S., Askari, K., and Kumari, M. (2018). Optimization of indole acetic acid production by isolated bacteria from Stevia rebaudiana rhizosphere and its effects on plant growth. J. Gen. Eng. Biotech. 16, 581–586. doi: 10.1016/j.jgeb.2018.09.001
Cook, R. J. (1993). Making greater use of introduced microorganisms for biological control of plant pathogens. Annu. Rev. Phytopathol. 31, 53–80. doi: 10.1146/annurev.py.31.090193.000413
Cordovez, V., Carrion, V. J., Etalo, D. W., Mumm, R., Zhu, H., van Wezel, G. P., et al. (2015). Diversity and functions of volatile organic compounds produced by Streptomyces from a disease-suppressive soil. Front. Microbiol. 6:1081. doi: 10.3389/fmicb.2015.01081
De la Lastra, E., Camacho, M., and Capote, N. (2021). Soil bacteria as potential biological control agents of fusarium species associated with asparagus decline syndrome. Appl. Sci. 11:8356. doi: 10.3390/app11188356
Elnahal, A. S., El-Saadony, M. T., Saad, A. M., Desoky, E. S. M., El-Tahan, A. M., Rady, M. M., et al. (2022). The use of microbial inoculants for biological control, plant growth promotion, and sustainable agriculture: a review. Euro. J. Plant Pathol. 1:34.
Fan, H., Ru, J., Zhang, Y., Wang, Q., and Li, Y. (2017). Fengycin produced by Bacillus subtilis 9407 plays a major role in the biocontrol of apple ring rot disease. Microbiol. Res. 199, 89–97. doi: 10.1016/j.micres.2017.03.004
Fisher, M. C., Henk, D., Briggs, C. J., Brownstein, J. S., Madoff, L. C., McCraw, S. L., et al. (2012). Emerging fungal threats to animal, plant and ecosystem health. Nature 484, 186–194. doi: 10.1038/nature10947
Garbeva, P., Hol, W. H. G., Termorshuizen, A. J., Kowalchuk, G. A., and de Boer, W. (2011). Fungistasis and general soil biostasis - a new synthesis. Soil Biol. Biochem. 43, 469–477. doi: 10.1016/j.soilbio.2010.11.020
Gowtham, H. G., Murali, M., Singh, S. B., Lakshmeesha, T. R., Murthy, K. N., Amruthesh, K. N., et al. (2018). Plant growth promoting rhizobacteria Bacillus amyloliquefaciens improves plant growth and induces resistance in chilli against anthracnose disease. Biol. Control 126, 209–217. doi: 10.1016/j.biocontrol.2018.05.022
Han, J. H., Shim, H., Shin, J. H., and Kim, K. S. (2015). Antagonistic activities of Bacillus spp. strains isolated from tidal flat sediment towards anthracnose pathogens Colletotrichum acutatum and C. gloeosporioides in South Korea. Plant Pathol. J. 31, 165–175. doi: 10.5423/PPJ.OA.03.2015.0036
Harp, T., Kuhn, P., Roberts, P. D., and Pernezny, K. L. (2014). Management and cross-infectivity potential of Colletotrichum acutatum causing anthracnose on bell pepper in Florida. Phytoparasitica 42, 31–39. doi: 10.1007/s12600-013-0334-9
Huh, E. J., and Kim, J. W. (2010). Consumer knowledge and attitude to spending on environment-friendly agricultural products. Kor. J. Human Ecol. 19, 883–896. doi: 10.5934/KJHE.2010.19.5.883
Joo, G. J., Kim, Y. M., Kim, J. T., Rhee, I. K., Kim, J. H., and Lee, I. J. (2005). Gibberellins-producing rhizobacteria increase endogenous gibberellins content and promote growth of red peppers. J. Microbiol. 43, 510–515.
Kamil, F. H., Saeed, E. E., El-Tarabily, K. A., and Abu Qamar, S. F. (2018). Biological control of mango dieback disease caused by Lasiodiplodia theobromae using streptomycete and non-streptomycete actinobacteria in the United Arab Emirates. Front. Microbiol. 9:829. doi: 10.3389/fmicb.2018.00829
Katak, R. M., Rocha, E. M., Oliveira, J. C., Muniz, V. A., Oliveira, M. R., Ferreira, F. A. S., et al. (2021). Larvicidal activities against Aedes aegypti of supernatant and pellet fractions from cultured Bacillus spp. isolated from Amazonian microenvironments. Trop. Med. Infect. Dis. 6:104. doi: 10.3390/tropicalmed6020104
Khalaf, E. M., and Raizada, M. N. (2018). Bacterial seed endophytes of domesticated cucurbits antagonize fungal and oomycete pathogens including powdery mildew. Front. Microbiol. 9:42. doi: 10.3389/fmicb.2018.00042
Kim, Y. S., Balaraju, K., and Jeon, Y. H. (2016). Biological characteristics of Bacillus amyloliquefaciens AK-0 and suppression of ginseng root rot caused by Cylindrocarpon destructans. J. Appl. Microbiol. 122, 166–179. doi: 10.1111/jam.13325
Kim, S. T., and Yun, S. C. (2011). Biocontrol activity of Myxococcus sp. KYC 1126 against Phytophthora blight on hot pepper. Res. Plant Dis. 17, 121–128. doi: 10.5423/RPD.2011.17.2.121
Köhl, J., Kolnaar, R., and Ravensberg, W. J. (2019). Mode of action of microbial biological control agents against plant diseases: relevance beyond efficacy. Front. Plant Sci. 10:845. doi: 10.3389/fpls.2019.00845
Kwak, Y. K., Kim, I. S., Cho, M. C., Lee, S. C., and Kim, S. (2012). Growth inhibition effect of environment-friendly farm materials in Colletotrichum acutatum in vitro. J. Bio Environ. Contr. 21, 127–133.
Le Han, H. L., Jiang, L., Tran, T. N. T., Muhammad, N., Kim, S. G., Pham, V. P. T., et al. (2022). Whole-genome analysis and secondary metabolites production of a new strain Brevibacillus halotolerans 7WMA2: a potential biocontrol agent against fungal pathogens. Chemosphere 307:136004. doi: 10.1016/j.chemosphere.2022.136004
Lee, G., Lee, S. H., Kim, K. M., and Ryu, C. M. (2017). Colonizing yeast Pseudozyma churashimaensis elicits systemic defense of pepper against bacterial and viral pathogens. Sci. Rep. 7:39432. doi: 10.1038/srep39432
Li, N., Alfiky, A., Wang, W., Islam, M., Nourollahi, K., Liu, X., et al. (2018). Volatile compound-mediated recognition and inhibition between Trichoderma biocontrol agents and fusarium oxysporum. Front. Microbiol. 9:2614. doi: 10.3389/fmicb.2018.02614
Li, C., Shi, W., Wu, D., Tian, R., Wang, B., Lin, R., et al. (2021). Biocontrol of potato common scab by Brevibacillus laterosporus BL12 is related to the reduction of pathogen and changes in soil bacterial community. Biol. Control 153:104496. doi: 10.1016/j.biocontrol.2020.104496
Li, Y. X., Zhong, Z., Zhang, W. P., and Qian, P. Y. (2018). Discovery of cationic nonribosomal peptides as gram-negative antibiotics through global genome mining. Nat. Commun. 9, 1–9. doi: 10.1038/s41467-018-05781-6
Magno-Perez-Bryan, M. C., Martinez-Garcia, P. M., Hierrezuelo, J., Rodriguez-Palenzuela, P., Arrebola, E., Ramos, C., et al. (2015). Comparative genomics within the Bacillus genus reveal the singularities of two robust Bacillus amyloliquefaciens biocontrol strains. Mol. Plant Microbe Inter. 28, 1102–1116. doi: 10.1094/MPMI-02-15-0023-R
Mansfield, J., Genin, S., Magori, S., Citovsky, V., Sriariyanum, M., Ronald, P., et al. (2012). Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 13, 614–629. doi: 10.1111/j.1364-3703.2012.00804.x
Meena, R. S., Kumar, S., Datta, R., Lal, R., Vijayakumar, V., Brtnicky, M., et al. (2020). Impact of agrochemicals on soil microbiota and management: a review. Land 9:34. doi: 10.3390/land9020034
Montealegre, J. R., Reyes, R., Pérez, L. M., Herrera, R., Silva, P., and Besoain, X. (2003). Selection of bioantagonistic bacteria to be used in biological control of Rhizoctonia solani in tomato. Electron. J. Biotechnol. 6, 115–127. doi: 10.2225/vol6-issue2-fulltext-8
Montri, P., Taylor, P. W. J., and Mongkolporn, O. (2009). Pathotypes of Colletotrichum capsici, the causal agent of chili anthracnose, in Thailand. Plant Dis. 93, 17–20. doi: 10.1094/PDIS-93-1-0017
Nehra, V., Saharan, B. S., and Choudhary, M. (2016). Evaluation of Brevibacillus brevis as a potential plant growth promoting rhizobacteria for cotton (Gossypium hirsutum) crop. Springerplus 5, 1–10. doi: 10.1186/s40064-016-2584-8
Ngalimat, M. S., Mohd Hata, E., Zulperi, D., Ismail, S. I., Ismail, M. R., Mohd Zainudin, N. A. I., et al. (2021). Plant growth-promoting bacteria as an emerging tool to manage bacterial rice pathogens. Microorganisms 9:682. doi: 10.3390/microorganisms9040682
Ossowicki, A., Jafra, S., and Garbeva, P. (2017). The antimicrobial volatile power of the rhizospheric isolate Pseudomonas donghuensis P482. PLoS One 12:e0174362. doi: 10.1371/journal.pone.0174362
Parisi, M., Alioto, D., and Tripodi, P. (2020). Overview of biotic stresses in pepper (Capsicum spp.): sources of genetic resistance, molecular breeding and genomics. Inter. J. Mol. Sci. 21:2587. doi: 10.3390/ijms21072587
Peng, Y., Li, S. J., Yan, J., Tang, Y., Cheng, J. P., Gao, A. J., et al. (2021). Research progress on phytopathogenic fungi and their role as biocontrol agents. Front. Microbiol. 12:670135. doi: 10.3389/fmicb.2021.670135
Prasanna, L., Eijsink, V. G., Meadow, R., and Gåseidnes, S. (2013). A novel strain of Brevibacillus laterosporus produces chitinases that contribute to its biocontrol potential. Appl. Microbiol. Biotechnol. 97, 1601–1611. doi: 10.1007/s00253-012-4019-y
Prusky, D., Freeman, S., and Dickman, M. B. (2000). Colletotrichum: Host specificity, pathology, and host-pathogen interaction (BOOK). St. Paul APS Press.
Rao, A. P., Agbo, B. E., Ikpoh, I. S., Udoekong, N. S., and Etuk, H. A. (2016). Biological control mechanisms against plant-based pathogens. J. Biopest. Environ. 2, 1–11.
Saber, M. M., Abdou, Y. A., El-Gantiry, S. M., and Ahmed, S. S. (2003). Biocontrol of anthracnose disease of soybean caused by Colletotrichum dematium. Egypt. J. Phytopathol. 31, 17–29.
Samaras, A., Nikolaidis, M., Antequera-Gómez, M. L., Cámara-Almirón, J., Romero, D., Moschakis, T., et al. (2021). Whole genome sequencing and root colonization studies reveal novel insights in the biocontrol potential and growth promotion by Bacillus subtilis MBI 600 on cucumber. Front. Microbiol. 11:600393. doi: 10.3389/fmicb.2020.600393
SAS Institute. (1995). JMP statistics and graphics guide, version 3. SAS Institute, Cary,North:Carolina United States of America. 65–95.
Schneider, K., Chen, X. H., Vater, J., Franke, P., Nicholson, G., Borris, R., et al. (2007). Macrolactin is the polyketide biosynthesis product of the pks2 cluster of Bacillus amyloliquefaciens FZB42. J. Nat. Prod. 70, 1417–1423. doi: 10.1021/np070070k
Schulz-Bohm, K., Martín-Sánchez, L., and Garbeva, P. (2017). Microbial volatiles: small molecules with an important role in intra and inter-kingdom interactions. Front. Microbiol. 8:2484. doi: 10.3389/fmicb.2017.02484
Shaligram, S., Kumbhare, S. V., Dhotre, D. P., Muddeshwar, M. G., Kapley, A., Joseph, N., et al. (2016). Genomic and functional features of the biosurfactant producing Bacillus sp. AM13. Funct. Integr. Genom. 16, 557–566. doi: 10.1007/s10142-016-0506-z
Song, J., Wang, Y., Song, Y., Zhao, B., Wang, H., Zhou, S., et al. (2017). Brevibacillus halotolerans sp. nov., isolated from saline soil of a paddy field. Inter. J. Syst. Evol. Microbiol. 67, 772–777. doi: 10.1099/ijsem.0.001579
Souza, R., Beneduzi, A., Ambrosini, A., Costa, P. B., Meyer, J., Vargas, L. K., et al. (2013). The effect of plant growth-promoting rhizobacteria on the growth of rice (Oryza sativa L.) cropped in southern Brazilian fields. Plant Soil 366, 585–603. doi: 10.1007/s11104-012-1430-1
Soyer, J. L., Hamiot, A., Ollivier, B., Balesdent, M. H., Rouxel, T., and Fudal, I. (2015). The APSES transcription factor LmStuA is required for sporulation, pathogenic development and effector gene expression in Leptosphaeria maculans. Mol. Plant Pathol. 16, 1000–1005. doi: 10.1111/mpp.12249
Talhinhas, P., Sreenivasaprasad, S., Neves-Martins, J., and Oliveira, H. (2005). Molecular and phenotypic analyses reveal association of diverse Colletotrichum acutatum groups and a low level of C. gloeosporioides with olive anthracnose. Appl. Environ. Microbiol. 71, 2987–2998. doi: 10.1128/AEM.71.6.2987-2998.2005
Than, P. P., Prihastuti, H., Phoulivong, S., Taylor, P. W., and Hyde, K. D. (2008). Chilli anthracnose disease caused by Colletotrichum species. J Zhejiang Univ Sci B 9, 764–778. doi: 10.1631/jzus.B0860007
Thynne, E., Saur, I. M., Simbaqueba, J., Ogilvie, H. A., Gonzalez-Cendales, Y., Mead, O., et al. (2017). Fungal phytopathogens encode functional homologues of plant rapid alkalinization factor (RALF) peptides. Mol. Plant Pathol. 18, 811–824. doi: 10.1111/mpp.12444
Wan, H., Yuan, W., Ruan, M., Ye, Q., Wang, R., Li, Z., et al. (2011). Identification of reference genes for reverse transcription quantitative real-time PCR normalization in pepper (Capsicum annuum L.). Biochem. Biophys. Res. Commu. 416, 24–30. doi: 10.1016/j.bbrc.2011.10.105
Wang, H., Wen, K., Zhao, X., Wang, X., Li, A., and Hong, H. (2009). The inhibitory activity of endophytic Bacillus sp. strain CHM1 against plant pathogenic fungi and its plant growth-promoting effect. Crop Prot. 28, 634–639. doi: 10.1016/j.cropro.2009.03.017
Wang, K., Zhao, Y., Wang, X., Qu, C., and Miao, J. (2020). Complete genome sequence of Bacillus sp. N1-1, a κ-selenocarrageenan degrading bacterium isolated from the cold seep in the South China Sea. Mar. Genomics 54:100771. doi: 10.1016/j.margen.2020.100771
Wei, Z., Yang, T., Friman, V. P., Xu, Y., Shen, Q., and Jousset, A. (2015). Trophic network architecture of root-associated bacterial communities determines pathogen invasion and plant health. Nat. Commun. 6:8413. doi: 10.1038/ncomms9413
Wesseling, C., McConnell, R., Partanen, T., and Hogstedt, C. (1997). Agricultural pesticide use in developing countries: health effects and research needs. Inter. J. Health Serv. 27, 273–308. doi: 10.2190/E259-N3AH-TA1Y-H591
Yang, H. S., Sohn, H. B., and Chung, Y. R. (2002). Biological control of pythium damping-off of cucumber by Bacillus stearothermophilus YC4194. In Proceedings of the Korean Society of Plant Pathology Conference The Korean Society of Plant Pathology.
Yang, X., and Yousef, A. E. (2018). Antimicrobial peptides produced by Brevibacillus spp.: structure, classification and bioactivity: a mini review. World J. Microbiol. Biotechnol. 34, 1–10. doi: 10.1007/s11274-018-2437-4
Yang, J. W., Yu, S. H., and Ryu, C. M. (2009). Priming of defense-related genes confers root-colonizing bacilli-elicited induced systemic resistance in pepper. Plant Pathol. J. 25, 389–399. doi: 10.5423/PPJ.2009.25.4.389
Zhao, X., and Kuipers, O. P. (2021). BrevicidineB, a new member of the brevicidine family, displays an extended target specificity. Front. Microbiol. 12:693117. doi: 10.3389/fmicb.2021.693117
Keywords: phytopathogen, Brevibacillus halotolerans B-4359, Colletotrichum acutatum , red pepper anthracnose, sustainable agriculture, whole-genome sequencing, biocontrol agent
Citation: Kim H, Lee Y, Hwang Y-J, Lee M-H, Balaraju K and Jeon Y (2023) Identification and characterization of Brevibacillus halotolerans B-4359: a potential antagonistic bacterium against red pepper anthracnose in Korea. Front. Microbiol. 14:1200023. doi: 10.3389/fmicb.2023.1200023
Received: 04 April 2023; Accepted: 31 May 2023;
Published: 19 June 2023.
Edited by:
Motaher Hossain, Bangabandhu Sheikh Mujibur Rahman Agricultural University, BangladeshReviewed by:
Farjana Sultana, International University of Business Agriculture and Technology, BangladeshCopyright © 2023 Kim, Lee, Hwang, Lee, Balaraju and Jeon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongho Jeon, eW9uZ2JhY0BhbmRvbmcuYWMua3I=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.