- 1Department of Ecology and Environmental Science, Umeå University, Umeå, Sweden
- 2Department of Plant Physiology, Faculty of Agrobiology and Food Resources, Slovak University of Agriculture, Nitra, Slovakia
- 3Department of Marine Science, Bharathidasan University, Tiruchirappalli, Tamilnadu, India
- 4Department of Zoology, Goa University, Taleigão, Goa, India
- 5School of Biosciences and Veterinary Medicine, University of Camerino, Camerino, Italy
Climate change and the induced environmental disturbances is one of the major threats that have a strong impact on bacterial communities in the Antarctic environment. To cope with the persistent extreme environment and inhospitable conditions, psychrophilic bacteria are thriving and displaying striking adaptive characteristics towards severe external factors including freezing temperature, sea ice, high radiation and salinity which indicates their potential in regulating climate change’s environmental impacts. The review illustrates the different adaptation strategies of Antarctic microbes to changing climate factors at the structural, physiological and molecular level. Moreover, we discuss the recent developments in “omics” approaches to reveal polar “blackbox” of psychrophiles in order to gain a comprehensive picture of bacterial communities. The psychrophilic bacteria synthesize distinctive cold-adapted enzymes and molecules that have many more industrial applications than mesophilic ones in biotechnological industries. Hence, the review also emphasizes on the biotechnological potential of psychrophilic enzymes in different sectors and suggests the machine learning approach to study cold–adapted bacteria and engineering the industrially important enzymes for sustainable bioeconomy.
Introduction
Approximately 70% of the Earth’s surface is covered by ice which includes ice caps, ice sheets, glaciers, sea ice and high mountain ranges (Yusof et al., 2021). Antarctica, the least populated continent placed at the southernmost of the Earth, contains about 90% of the world’s ice and is continually blanketed in ice sheets. Around 15% part of the continent is covered by sea ice. However, the ongoing climate change and stratospheric ozone consistently impact the continent and the residing organisms by rapidly altering environmental factors including temperature, precipitation, UV radiation etc. (Yusof et al., 2021). Climate change is influenced by anthropogenic emissions of greenhouse gasses (GHGs), which result in an increase in global temperature of up to 1.5°C or even 2°C (IPCC, 2021). The increase in average temperature leads to the amplification of permafrost thawing, melting of the Antarctic ice sheet (Dietz and Koninx, 2022), and changes in ice mass (Stokes et al., 2022). An increase in Antarctic ice sheet loss enhances the penetration of light and energy levels into water that changes plankton productivity and composition, including microbes (Barnes et al., 2021). Hence, climate-induced disturbances are major threats to the Antarctic ecosystem, especially to microbial composition and functions (Gutt et al., 2021).
Microbes are important components that generate the base of polar food webs in both terrestrial and freshwater Antarctic ecosystems, hence affecting all trophic levels. They play a crucial role in the biogeochemical cycle (especially in carbon and nitrogen cycling) and functioning of an extreme Antarctic environment (Anesio et al., 2009). Microbes thriving in polar cryosphere, which comprises 14% of the Earth’s surface for more than 33 million kilometers (Malard and Pearce, 2018) are called psychrophilic sensu stricto and psychrotolerant organisms, based on the optimum growth temperature. The minimum, optimum and maximum growth temperatures for psychrophiles are <0, <15 and < 20°C, and for psychrotrophs >0, >20 and > 30°C, respectively (Moyer and Morita, 2007). To persist in extreme environment and inhospitable conditions, these microbes are thriving and displaying striking adaptive characteristics towards severe external factors including low humidity, precipitation, freezing temperature, sea ice, high radiation and salinity, nutrients limitation and strong winds (Feller and Gerday, 2003). This indicates the potential of polar microbes in regulating climate change’s environmental impacts and represents their distinctive adaptability of primitive life-forms. Therefore, it is crucial to find how Antarctic microorganisms adapt to different climate-induced environmental stresses.
Several previous studies reported that distinctive adaptive characteristics of psychrophiles are due to the specific protein adaptations and enzymes they secrete (Kohli et al., 2020). The different types of bonding (covalent and H-bond), amino acid composition, G + C content and folding pattern of proteins are responsible for the improved stability and adaptation of psychrophiles (Dutta and Chaudhuri, 2010). However, due to the difficulty in direct cultivation of psychrophiles from extreme Antarctic environment, the research on their adaptation is still on the elementary phase and most of the adaptation strategies and their underlying mechanisms (freezing tolerance/avoidance, regulation of protein synthesis and membrane fluidity) have not been fully understood.
In this review, we focus on the different adaptation strategies of Antarctic marine bacteria to environmental changes at morphological, physiological, and molecular level. Moreover, the review highlights the biotechnological applications of Antarctic bacteria and their enzymes and sourced proteins in different industries. To unlock “blackbox” of Antarctic bacteria and engineer cold adapted molecules including psychrozymes, in the last section of the review, we discussed the possible use of recent technologies such as omics and machine learning as an effective, fast and less labor- intensive approach. Additionally, we emphasize the existing research gaps and illustrate our view on future research directions on Antarctic microbes, an unexplored ecosystem.
Adaptation strategies of Antarctic bacteria
The adaptation of psychrophilic bacteria to the multitude of environmental stressors including low temperature is achieved via complex range of structural and physiological changes (Table 1) during long term evolution (Collins and Margesin, 2019). Several new emerging omics techniques including genomics, transcriptomics, metagenomics, metabolomics, etc. have increased our understanding of bacterial cold adaptation. These technologies demonstrated that psychrophilic bacteria employ various survival strategies to cope with the challenges confronted in their habitat. The following section includes the strategic tools adapted by bacteria at morphological, physiological and molecular level to enable life in the harsh Antarctic environment.
Structural adaptation
The cell envelope is a complex multi-layered (outer, peptidoglycan and inner membrane) structure of bacteria that provides protection from the unpredictable and hostile environment. Several studies reported the thickening of outer cell surfaces in particular, peptidoglycan layer (Figure 1) (gram positive) and lipopolysaccharide (gram negative bacteria) in cold adapted bacteria (Di Lorenzo et al., 2020). The thick cell surface strengthens the psychrophiles against cell disruption by ice formation, increased osmotic pressure and freezing/thawing at low/subzero temperature (Figure 1). Recent study on the structural elucidation of the highly heterogenous lipid A (LPS’ glycolipid moiety) in the three Antarctic bacteria strains Pseudoalteromonas tetraodonis SY174, Psychromonas arctica SY204b, and Psychrobacter cryohalolentis SY185 showed that structural alterations in the LPS’ glycolipid moiety increase the membrane flexibility and stability of these psychrophiles in Antarctica (Di Lorenzo et al., 2020). To tolerate the negative effect of temperature stress, bacteria alter the composition of cell membrane fatty acids (Hassan et al., 2020). Cold-adapted bacteria usually increases the polyunsaturated to saturated fatty acid ratio in membrane phospholipids to maintain optimal fluidity and membrane permeability (homeoviscous adaptation) (De Maayer et al., 2014). In this regard Králová (2017) studied the role of membrane fatty acids in adaptation of Flavobacterium sp. towards harsh Antarctic environment (Table 1). The study suggested that Antarctic Flavobacterium sp. mostly utilizes two mechanisms of homeoviscous adaptation in cold-adaptive response- unsaturation of fatty acids and synthesis of branched fatty acids (Králová, 2017). Similarly, Bajerski et al. (2017) investigated the biomembrane polar fatty acid adaptation of Chryseobacterium frigidisoli PB4T isolated from an Antarctic glacier in response to varying temperature (0°C to 20°C) and pH (5.5 to 8.5). Another strategy adopted by the Antarctic bacteria is the production of extracellular polymeric substances (EPSs) that protects the cell from sub-freezing temperature by forming a protective covering around the bacterial cell that act as a barrier to solutes diffusion and ice formation (Collins and Margesin, 2019) (Figure 1). In this context, Caruso et al. (2018) studied the EPS synthesis in four Antarctic sponge-associated bacteria (Shewanella sp. strain CAL606, Colwellia sp. strain GW185, and Winogradskyella sp. strains CAL384 and CAL396). EPS production by psychrophilic bacteria has an ecological role in cell adhesion to surfaces and cell protection due to their chemical composition (Lo Giudice et al., 2020). In addition, EPS also plays a vital role in biofilm formation that enhances access to nutrients and cell survivability. The adaptation of haloarchaea Halorubrum lacusprofundi in Deep Lake (Antarctica) is a good example of cold adaptation by biofilm (Liao et al., 2016). Moreover, EPS secreted from cold adapted microbes shows ice binding functions and ice recrystallization inhibitors (IRI) activity (Casillo et al., 2017). Additionally, cold adapted microbes produce a substantial amount of biosurfactants that play a potential role in their survival.
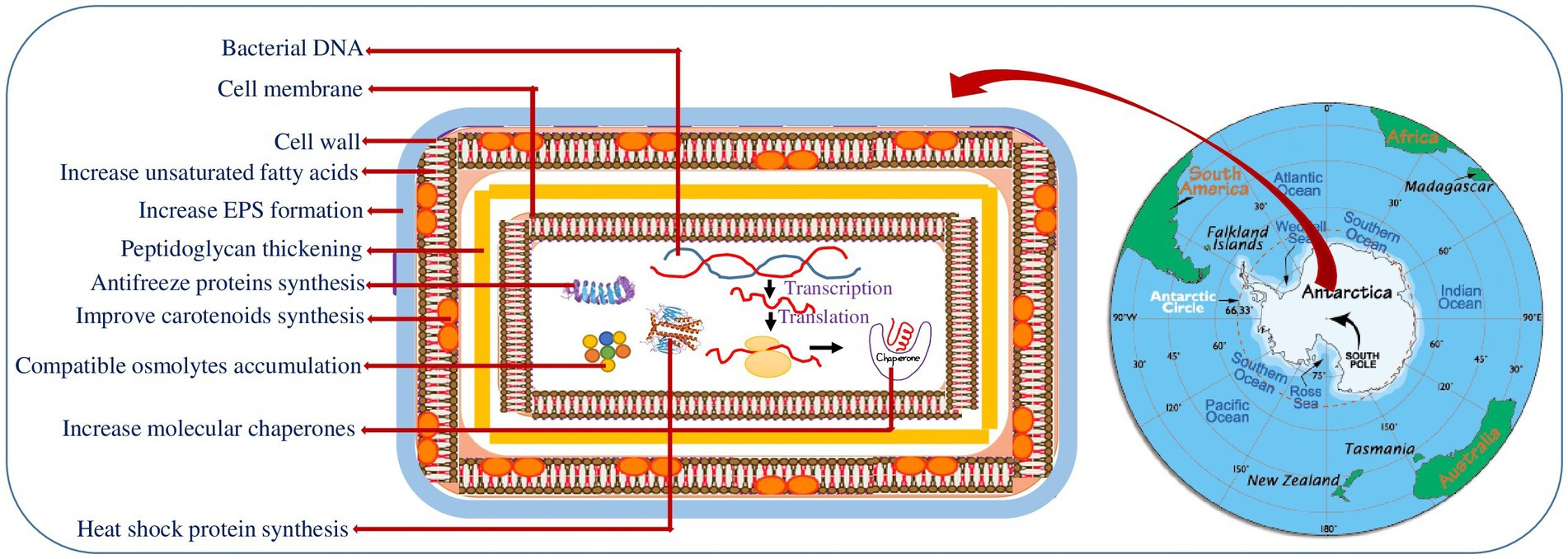
Figure 1. Schematic representation of the various cold adaptation strategies of Antarctic bacteria. Map of Antarctica retrieved from worldatlas.com.
Physiological adaptation
Psychrophilic bacteria developed several physiological adaptations such as metabolic alterations, synthesis of pigments, compatible osmolytes and ice binding proteins to optimize their metabolism in harsh Antarctic environment (Figure 1) (Collins and Margesin, 2019). Pigment formation is a common feature in cold-adapted microbes thriving in diverse habitat including glaciers, high altitude, marine water and ice cores (Pandey et al., 2018). Pigments, in particular polar carotenoids, have been suggested to serve in maintaining the membrane fluidity and rigidity. Moreover, these pigments act as a photoprotector, antioxidants, cryoprotectants, antimicrobials and light harvesters in psychrophilic microbes to counteract the low temperature and other environmental stresses (Pandey et al., 2018). The pigment production by cold adapted bacteria including Antarctic environment have been extensively reviewed by Sajjad et al. (2020). Accumulation of compatible osmolytes (glycine betaine, trehalose, glycerol, sucrose, mannitol etc.) is the other way of Antarctic bacteria to prevent cell shrinkage and water loss during sub-zero temperature (Goordial et al., 2016). Improved concentration of compatible osmolytes restore osmotic balance by reducing the freezing point of the cell cytoplasm (Fonseca et al., 2016). The osmolytes were also found to be involved in scavenging free radicals, counteracting protein aggregation, improving protein folding and stabilizing membranes and proteins at chilling temperatures (Collins and Margesin, 2019).
Additionally, psychrophiles synthesize several proteins namely ice binding, cold shock and heat shock proteins to sustain their physiological state in changing environment (Figure 1) (Kim et al., 2018). Ice binding proteins are antifreeze proteins (AFPs) that on binding to ice inhibits the growth of ice crystals and lower freezing temperature. The proteins were initially reported in Antarctic fish and in various cold organisms including bacteria. Bar Dolev et al. (2016) identified the multidomain ice adhesion AFP in the Antarctic bacterium Marinomonas primoryensis. AFPs causes thermal hysteresis (TH) in which the freezing temperature of water drops below the melting temperature. This ceases ice growth by creating a thermal hysteresis gap (Mangiagalli et al., 2017). Antifreeze proteins are also found to display ice recrystallisation inhibitors (IRI) activity. A study by Raymond et al. (2007) suggested the role of AFPs isolated from Antarctic Gram-negative bacteria Colwellia, strain SLW05 in ice recrystallization inhibition and ice-binding. The cold shock proteins are a family of widely distributed low molecular weight highly conserved proteins, synthesized under normal environment but strongly induced when exposed to cold conditions. The cold shock gene homologous to cspA of E. coli has been isolated in two isolates of Antarctic psychrotrophs- Gram-positive bacterium Arthrobacter protophormiae and Gram-negative Pseudomonas fluorescens which expressed constitutively at two different temperatures (4°C and 22°C) (Ray et al., 1994). Kim et al. (2007) characterized cold shock protein A of the Antarctic bacterium Streptomyces sp. AA8321 and depicted its role in inhibiting DNA replication during cold adaptation. Similarly, characterization and expression of three cold shock protein (CSP) genes under different stress conditions is studied in the Antarctic bacteria Psychrobacter sp. G (Song et al., 2012). A comparative proteomic study of cold-repressed proteins in the Pseudoalteromonas haloplanktis TAC125 located in Antarctic environment at 4°C and 18°C demonstrated that majority of these proteins expressed at 4°C were heat shock proteins associated to folding (Piette et al., 2011). Furthermore, the increase accretion of heat shock proteins has been observed in Antarctic psychrophiles during ocean warming and acidification. Recently, Yusof et al. (2021) characterized two Hsp70 genes in the Antarctic yeast, Glaciozyma antarctica PI12 that protect the functional activity of the yeast under different temperature stress (4°,15°, 25 ° and 37°C). The Antarctic ciliated protozoon E. focardii maintains a constitutive synthesis of some Hsp70 genes to preserve protein functions in the cold and induces high expression of other Hsp70 genes in response to the oxidative stress increased by the Antarctic ozone hole (Mozzicafreddo et al., 2021).
Immunoblot analysis indicated the accumulation of DnaK protein (homolog of eukaryotic Hsp 70 that plays vital role in several abiotic stresses, including thermal stress) in the Antarctic psychrotroph Shewanella sp. Ac10 at 24°C (Yoshimune et al., 2005). The study also suggested that recombinant SheDnaK gene facilitates the growth of mutant E. coli at 15°C. Similarly, García-Descalzo et al. (2014) studied the thermal adaptation in Antarctic bacteria Shewanella frigidimarina towards different temperature ranges, from 0°C to 30°C. The significant accumulation in heat shock and other stress proteins is observed in bacterial cells cultured at 28°C (García-Descalzo et al., 2014).
Moreover, studies have indicated that psychrophiles at chilling temperature alter their metabolic pathways to conserve energy and thrive in cold environment. For long term survival, psychrophiles either down regulate their primary metabolic pathways (glycolysis, tricarboxylic acid (TCA) cycle, electron transport chain, pentose phosphate pathway etc.) or substitute them with abridged alternative pathways (glyoxylate, methylglyoxal, acetate metabolism, ethanol oxidation pathway etc.) (Collins and Margesin, 2019).
Molecular adaptation
Molecular chaperones (RNA/DNA/protein) play an important role in stabilization of RNA/DNA and protein molecules at freezing temperature and in reducing protein aggregation and misfolding. These molecules are constitutively produced and expressed in psychrophiles as cold -adapted proteins. Several molecular chaperone proteins are identified from the Antarctic green alga Chlamydomonas sp. ICE-L shows high similarity with the genes of the psychrophilic bacterium Psychroflexus torquis and appears to be involved in tolerance to freezing, changing light and saline conditions in bacteria (Liu et al., 2016). Recently, Ijaq et al. (2022) investigated the functional role of hypothetical proteins from Pseudomonas sp. Lz4W, a Gram-negative psychrophilic bacterium adapted to survive in Antarctica. The study categorized two hypothetical proteins, HP AUB76544.1 and HP AUB76897, as chaperones that are important for correct folding and insertion of outer membrane proteins (HP AUB76544.1) as well as in sustaining the structural integrity and protein functions (HP AUB76897) (Ijaq et al., 2022). Furthermore, Solar Venero et al. (2022) studied the role of small non-coding RNAs (sRNAs) as genetic regulators in the adaptation of the Antarctic bacterium Pseudomonas extremaustralis towards changing climatic conditions and stress environment. The study showed the expression of novel sRNA, sRNA40 (identified by RNA-seq experiments) in response to different oxygen availability and oxidative stress and demonstrated that sRNA40 gene expression (associated to upregulation of selected secretory proteins) is triggered under aerobiosis/microaerobiosis conditions (Solar Venero et al., 2022).
Cold adapted enzymes are more flexible as compared to mesophilic enzymes due to the presence of relative low arginine content. Recently, one peroxidase named DyP (dye-decolorizing peroxidase) from an Antarctic bacteria Pseudomonas sp. AU10 was found to contain low arginine content (Cagide et al., 2023). Another study by Tang et al. (2019) characterized a cold-active alkaline pectate lyase from Antarctic bacterium Massilia eurypsychrophila with a low arginine content, suggesting that this is a distinctive property of cold adaptation. Petratos et al. (2020) studied the molecular dynamics of alcohol dehydrogenase (MoADH) from the cold-adapted bacterium Moraxella sp. TAE123 compared with the Escherichia coli (EcADH), Geobacillus stearothermophilus (GsADH), Thermus sp. ATN1 (ThADH). The cold adapted GH42 has also been identified in some microbes such as Halobacterium lacusprofundi, Arthrobacter sp. and Marinomonas sp. BSi20414 (Sheridan and Brenchley, 2000; Karan et al., 2013; Ding et al., 2017). Experimental study by Mangiagalli et al. (2021) on the cold adaptation of β-galactosidase from the psychrophilic Marinomonas ef1 shows the cold activity of this enzyme at 5°C and maintains stability up to 50°C, which suggests the origin of GH42 from the different evolutionary pathway. The biochemical and molecular features of GH42 and its biotechnological applications have been described in the recent review by Mangiagalli and Lotti (2021). Another study by Fan et al. (2016) reveals the structure of cold-active β-galactosidase from psychrotrophic Rahnella sp. R3. However, the glycoside hydrolase family (GH42) of other psychrophilic microbes is still poorly known. Thus, very little knowledge is available on the role of these enzymes in psychrophilic bacteria and adaptation to climate change in the Antarctic environment.
Another adaptation strategy by psychrophile bacteria is through horizontal gene transfer (HGT), which is considered as one of the important forces that regulates bacterial evolution (Martínez-Rosales et al., 2012). The occurrence of HGT was found in various Antarctic bacteria such as Marinomonas sp. ef1, Pseudomonas spp., Collimonas sp. (Martínez-Rosales et al., 2012; John et al., 2020; Hwang et al., 2021). Recently, Abe et al. (2020) used neural network called Batch Learning Self-Organizing Maps BLSOM method to detect HGTs in the genomes of two Antarctic bacteria (Sphingomonas sp. HMP6 and HMP9) compared with other continents.
Though, the various adaptation strategies have been studied in Antarctic psychrophiles in response to changing climate as evident from the previous studies stated above. However, the research is still on the elementary phase and there are many unexplored areas including the underlying mechanism of adaptation, molecular pathways and key components involved which will give better understanding of Antarctic psychrophiles adaptation towards ongoing changing environment. Upcoming research on cold -adapted bacteria needs to be emphasized on these aspects by using novel techniques such as metagenomics and machine learning. These methods could serve as a dynamic tool to decipher adaptation mechanisms in the Antarctic psychrophiles, as they are difficult to culture in laboratory conditions.
Industrial applications of Antarctic bacteria
Cold active enzymes
The last few decades witnessed extensive research on microbial diversity inhabiting harsh extreme environmental conditions. Bacteria inhabiting extreme environments, like in Antarctica regions, are undergoing several stress factors and to withstand such conditions they have developed numerous physiological and molecular strategies (Hamid et al., 2022). These psychrophilic bacteria can survive and grow at a temperature range from −2 to 20°C, and express cold-adapted enzymes with distinctive properties that allow higher catalytic efficiency, improved flexibility, and lower thermal stability (Bruno et al., 2019). The advantages of cold-adapted enzymes over mesophilic enzymes are illustrated in various research articles (Kuddus, 2015; Javed and Qazi, 2016). Cold-active enzymes isolated from microorganisms are classified into three groups.
Group 1: Heat-sensitive with other enzymatic characteristics similar to mesophilic enzymes.
Group 2: Heat-sensitive and relatively more active than mesophilic enzymes at a low temperature.
Group 3: Same thermostability as mesophilic enzymes but more active than mesophilic enzymes at a low temperature.
Since low temperatures are required for some industrial processes, there is a huge demand for cold-adapted enzymes in different biotechnological and industrial area, such as bioremediation, detergents, food and beverage processing, molecular biology and in textile industries (Figure 2) (Kuddus, 2018). Cold-active enzymes such as amylases, cellulases, lipases, pectinases, proteases, etc. from Antarctic bacteria establish an evident resource for several biotechnological applications. Many of the above enzymes are enormously used at commercial level, with special reference to hydrolases sourced from psychrophilic microorganisms (Hamid et al., 2022).
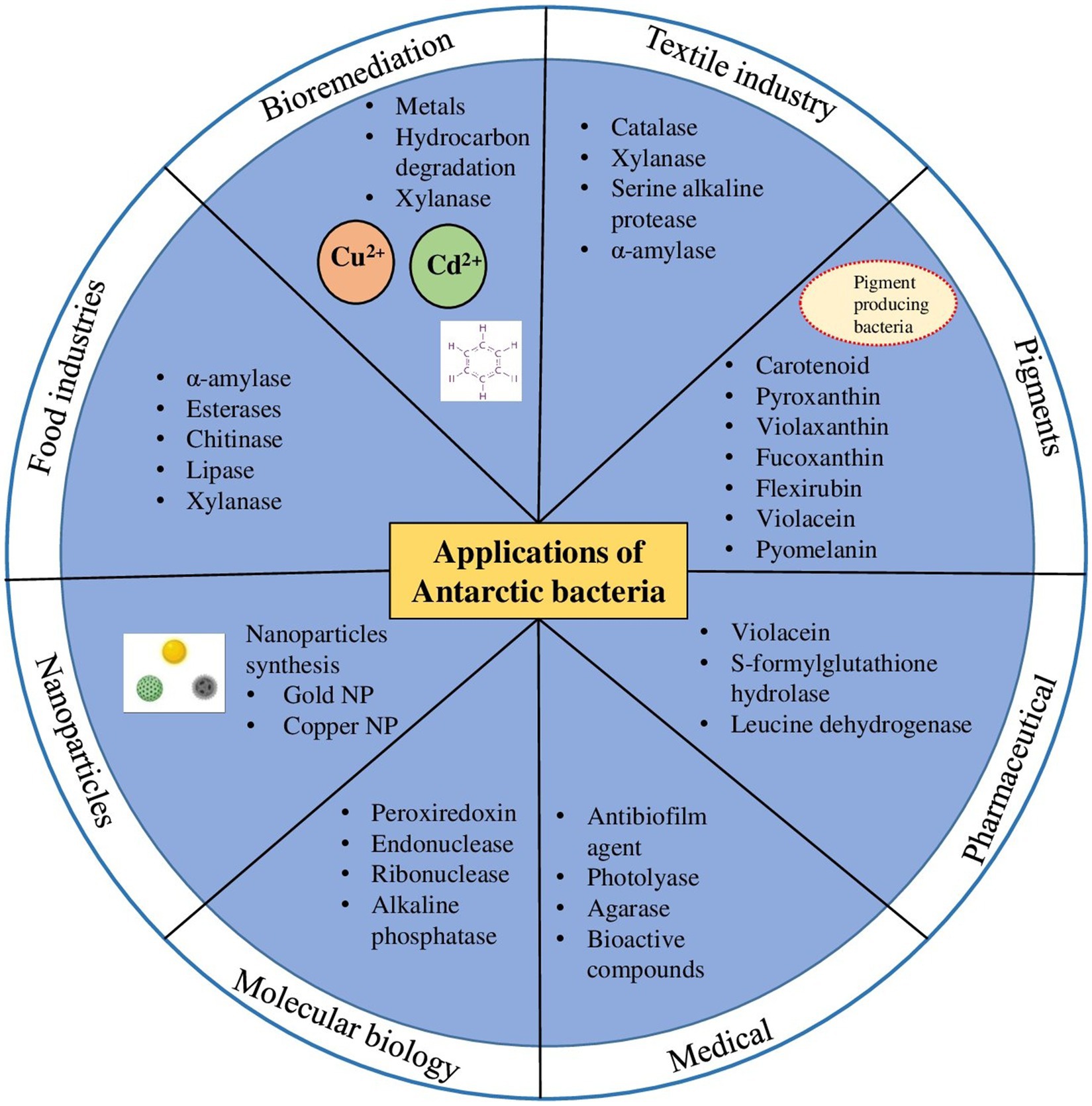
Figure 2. Biotechnological applications of cold-adapted molecules from Antarctic bacteria in different industrial sectors.
Food industry
Most of the Antarctic microbial enzymes have high catalytic competence at low and moderate temperatures when compared to mesophilic enzymes. This makes the cold-active enzymes unique in the food industry for their less biochemical requirements, reduction in process times, save energy costs and easy inactivation by gentle heat (Kuddus, 2018).
In dairy industries, the use of cold-active β-galactosidases is effectively operational in milk at lower temperatures that lead to lactose breakdown. Antarctic marine bacteria especially Pseudoalteromonas haloplanktis, has been proven to produce cold-active β-galactosidase with high efficacy in the hydrolysis of lactose under cold conditions (Cieśliński et al., 2005). Similarly, cold-active polygalacturonase isolated from P. haloplanktis can be used in juice manufacturing industries for degradation of pectin. Many other cold-active enzymes such as α-amylase, esterases, chitinases, and lipase have important applications in the food (Figure 2) and brewing industries, for cheese ripening, cheese flavoring, meat tenderizing, production of fatty acids and interesterification of fats, wheat bread making, improvement in food texture, etc. The gene encoding the cold active enzymes can be cloned into a suitable host to produce recombinant enzymes which are already proved to show high catalytic activity and stability at low temperatures (Table 2 and references therein). In case of food and brewage industries, low-temperature processing is highly favorable since it provides many advantages, for instances, prevention of bacterial contamination and occurrence of unwanted chemical responses, higher food quality, and persistence of flavor (Al-Maqtari et al., 2019).
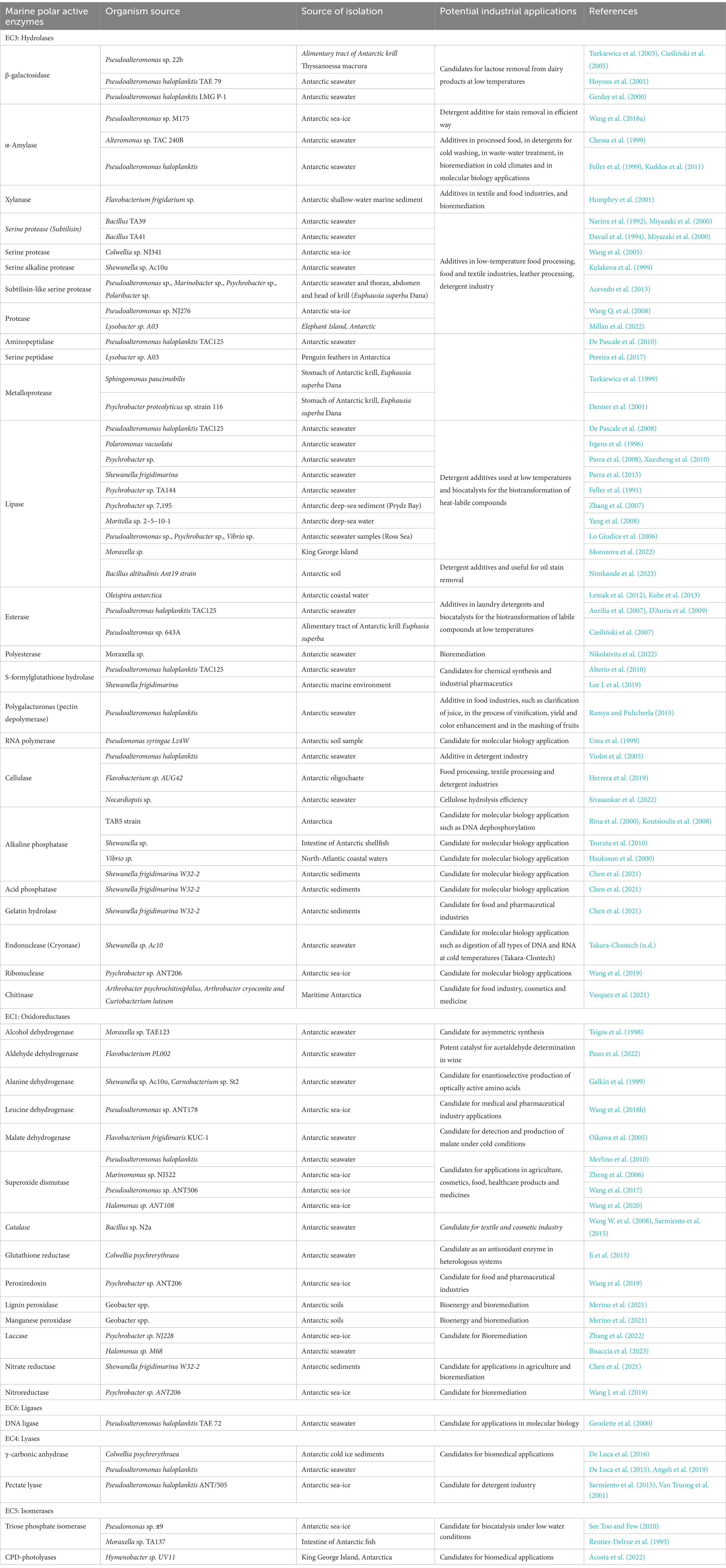
Table 2. Polar-active enzymes isolated from Antarctic bacteria and their potential industrial applications (Reproduced from Bruno et al. (2019) and updated until recent literature).
Detergent industry
The removal of stains (lipids, polysaccharides, and proteins) by manual heating and beating of the clothes reduces the life of the fabrics and also decolorization (Kumar et al., 2021). On the other hand, harmful chemicals are most widely used in the chemical industries for removal of dirt which ultimately contaminates the environment. To overcome these issues, cold-active enzymes are the best alternative to the chemicals and moreover they increase the life of the fabrics, as manual heating is no longer required. The use of cold active enzymes such as lipases, amylases and proteases active at alkaline pH, with thermostability has solved the problems arising in the detergent industries. Switching mesophilic to cold-adaptive enzymes in the cleaning process ensures lowers wash temperatures and greater energy protection. A 10°C reduction in wash temperature created a 30% reduction in the consumption of electricity (Nielsen, 2005).
Proteases are found to be the most widely used enzymes in detergents and most of the cold-active proteases have shown remarkable stability and activity in a wide-ranging alkaline condition (Hamid et al., 2022). Cold-active proteases from Bacillus TA41, Colwellia sp. NJ341, Pseudoalteromonas sp. NJ276, P. haloplanktis etc. can be used as detergent additives for cold washing (Table 2 and references therein). The detergent manufacturers are seeking novel cold-active enzymes that may improve efficiency of detergents and retain the quality of fabrics (Al-Ghanayem and Joseph, 2020). Hence more studies on this aspect may lead to the discovery of many novel cold-active enzymes.
Molecular biology
Alkaline phosphatases which catalyze the hydrolysis of phosphate monoesters have a significant role in molecular cloning for dephosphorylation of DNA at the 5′ end to avoid its re-circularization. Based on this importance, New England Biolabs developed a recombinant alkaline phosphatase isolated from the Antarctic bacterial strain TAB5.1 In addition, cold active alkaline phosphatase from an Antarctic Vibrio sp., was reported to have a higher turnover number (kcat) and higher apparent Michaelis–Menten factor (Km) as compared with enzyme from E. coli (Hauksson et al., 2000). Similarly, to cold adapted alkaline phosphatases, uracil DNA N-glycosylases have been recently commercialized as a molecular biological tool by various companies (New England Biolabs Inc., Takara-Clontech, Affymetrix, Inc.) (Awazu et al., 2011; Muller-Greven et al., 2013).
DNA ligases are enzymes involved in DNA replication, DNA recombination and DNA repair. These enzymes are commonly used in molecular biology to catalyze the formation of a phosphodiester bond between adjacent 5′-phosphoryl and 3′-hydroxyl groups in double stranded DNA (Bruno et al., 2019). The well-established psychrophile P. haloplanktis TAE72, was also reported to produce DNA ligase and exhibit activity at temperatures as low as 4°C (Georlette et al., 2000).
Bioremediation
Bioremediation by mesophilic and thermophilic enzymes is ineffective in cold environmental conditions hence cold active enzymes are valuable tools for removal or biodegradation of pollutants. The use of cold-active enzymes could be more feasible and result oriented than the use of whole bacterial cells, since the whole cells requires multiple parameters of optimal growth (Kumar and Bharadvaja, 2019). Many cold adapted microorganisms, such as Pseudomonas sp., Rhodococcus sp., Oleispira antarctica and Sphingomonas spp. are proficient in degradation of petroleum hydrocarbons (Aislabie et al., 2006; Miri et al., 2019). Many cold active enzymes such as lipases, proteases, xylanase (Figure 2) etc. are regularly explored for various applications in bioremediation (Miri et al., 2019).
Bacterial pigments
Pigments of natural origin play an important role in the physiology and molecular processes of microorganisms because they act as a strategy of adaptation to various extreme environments, have a protective function against solar radiation, and are also involved in functional processes like photosynthesis (Sutthiwong et al., 2014). Bacteria also produce a wide range of pigments such as carotenoids, melanin, violacein, prodigiosin, pyocyanin, actinorhodin, and zeaxanthin (Venil et al., 2013). Antarctica environment is also well known for richness of bacterial species producing various pigments such as carotenoids, flexirubin, violaceins, tetrapyrroles, quinones, biochromes, etc. (Figure 2).
Antarctic bacteria are able to produce not only different kinds of carotenoids but other pigments as well. This is supposedly due to the harsh conditions to which they are exposed. Therefore, these microorganisms may be regarded as promising potential targets for further research on the growing market of biotechnological pigments for industrial applications, not only focusing on long established compounds, but also on unconventional pigments. Arthrobacter, Citricoccus, and Microbacterium from the phylum Actinobacteria, Chryseobacterium and Flavobacterium from the phylum Bacteroidetes, and Janthinobacterium, Pseudomonas, Lysobacter, and Serratia from the phylum Proteobacteria are among the main pigment-producing bacteria reported in Antarctic environments (Silva et al., 2021 and references therein).
The carotenoid content, pyroxanthin, violaxanthin, fucoxanthin, and nostoxanthine 3-sulfate from the Antarctic bacterium Pedobacter showed strong antioxidant capacity and protects the bacterium against oxidative damage caused by high levels of UV-B radiation (Correa-Llanten et al., 2012).
Microbial pigments are of great interest in cosmetic, dairy, food, pharmaceutical and textile industries (Figure 2), mainly due to its new chemical structures and most importantly they are natural. A recent report from the global food colorants market showed that natural products represent one third of the total colorants approximately, and three fourths of these natural colorants are used in food in beverages.
Other applications
Antarctic bacteria are a source of a wide range of applications in the medical sector due to their capacity of producing numerous compounds/metabolites (presence of diverse metabolic pathways resulting from evolutionary adaptation to subzero and nutrient deficient conditions) (Figure 2). Recently, a new anti-biofilm agent called “CATASAN” has been found in Antarctic bacteria Psychrobacter sp. TAE2020 which can be used against the human pathogen Staphylococcus epidermidis (D’Angelo et al., 2022). Similarly, another Antarctic bacteria Pseudomonas sp. TAE6080 is capable of inhibiting biofilm formation by the opportunistic pathogen Staphylococcus epidermidis (Riccardi et al., 2022). Several studies have shown that Antarctic microbes produce bioactive compounds to treat various diseases (Murray et al., 2021; Xiao et al., 2023). Quinones from orange-yellow pigmented Sphingomonas aerolata (Busse et al., 2003), isolated from the ice of Taylor dome and hydrocarbon-contaminated soils around Scott Base is used to treat Alzheimer’s, Huntington’s, Parkinson’s, and cardiovascular diseases (Nair and Abraham, 2020). Previous analysis showcased that the Antarctic pigments also have various biological activities such as antioxidant, antibacterial, antimalarial, antifungal, anticancer and many others (Silva et al., 2021). Experimental studies by Maeda et al. (2009) on fucoxanthin (characteristic carotenoid of brown algae) have revealed various applications of the compound in producing anti lymphangiogenic, antitumoral, neuroprotective, antidiabetic, anti-obesity, and anti-inflammatory effects. In addition, fucoxanthin prevents carcinogenesis and depressive behavior, such as the attenuation of bleomycin-induced lung fibrosis and ulcerative colitis (Wang Y. et al., 2019). Another pigment violaxanthin has proved to have antiproliferative and anti-inflammatory effects (Pasquet et al., 2011; Soontornchaiboon et al., 2012). Decaprenoxanthin from Arthrobacter psychrochitiniphilus strain 366 isolated from a biofilm formed on the surface of defrost water in Whalers Bay, Deception Island, Silva et al. (2019) has strong antioxidant properties. Apart from pigments, a variety of antimicrobial lipid-based substances have been isolated from Antarctic microorganisms with potential to be used in treatments of bacterial infections. Rhamnolipids, a special class of bacterial lipids purified form Antarctic marine sediment bacteria, Pseudomonas sp. BTN1 displayed antibacterial activity against Burkholderia cenocepacia (isolated from cystic fibrosis patient) and Staphylococcus aureus (Tedesco et al., 2016). Aminolipids, another category of microbial lipids purified from shallow-sea-sediment bacterium Aequorivita sp. is effective against methycilin-resistant Staphylococcus aureus (Chianese et al., 2018).
Besides bioactive compounds, Antarctic bacteria synthesizes metal nanoparticles (Figure 2) through biomineralization process. The biosynthesis of metal nanoparticles using Antarctic bacteria is a cost-effective, environmentally friendly process, without using toxic chemicals in the synthesis and purification steps. In recent years, synthesis of metal nanoparticles using cold adapted bacterial strains have gained attention due to the high stability (even at psychrophilic conditions) and diverse biomedical applications (Das et al., 2020; John et al., 2021, 2022). Das et al. (2020) biosynthesize gold nanoparticle (GNP) at different temperatures (4°, 10°, 25°, 30° and 37° C) using psychrotolerant Antarctic bacteria Bacillus sp. GL1.3. The synthesized gold nanoparticles exhibit antibacterial activity against sulfate-reducing bacteria (Desulfovibrio sp.) (Das et al., 2020). A similar study by Javani et al. (2015) identified four psychrophilic Antarctic bacteria namely Aeromonas salmonicida, Pseudomonas veronii, Psychrobacter sp. and Yersinia kristensenii that extracellularly biosynthesize nanosilver at 4°C and 30°C. The study demonstrated that the most active and stable nanoparticles with highest antibacterial activity were those prepared at 4°C. These nanoparticles possess high stability even after 10 months of incubation under light (Javani et al., 2015). An efficient novel approach is used by Plaza et al. (2016) to synthesize quantum dots (CdS and CdTe quantum dots) at room temperature by using heavy metal (cadmium and tellurite) resistant Antarctic bacteria, Pseudomonas, Psychrobacter and Shewanella. Recently, John et al. (2022) synthesizes silver nanoparticles (AgNPs) using three bacterial strains Rhodococcus, Brevundimonas and Bacillus isolated from an Antarctic consortium. Biosynthetic AgNPs show promising effects against common nosocomial pathogens and can be replaced with conventional antibiotics (John et al., 2022). Thus, despite being underexplored, Antarctic bacteria constitute a promising platform for biosynthesis of nanomaterials.
Advanced strategies to study Antarctic bacterial adaptation
Omics
To gain a comprehensive picture of bacterial communities, several “omics” approaches should be applied to reveal polar “blackbox” microbes (Figure 3). The development of genomic technologies in recent years has gained knowledge on microbial communities and their adaptation in the Antarctic ecosystem. The most common technique to reveal the taxonomical composition of cold adapted bacteria is 16S rRNA gene sequencing (Bowman and Ducklow, 2015), but the functional role of many other genes remains unknown. However, new technologies are being developed in high -throughput sequencing, which provides high quality data with short or long read sequences. To date, several genomes from psychrophilic bacteria and archaea have been sequenced (Pucciarelli et al., 2015; Ramasamy et al., 2019; John et al., 2020; Wang et al., 2021; Lee et al., 2022; Riccardi et al., 2022; Otur et al., 2023). The advantage of whole genome sequencing of Antarctic bacteria is to analyze and characterize the genes in the entire genome, especially genes coding industrially relevant enzymes using DNA sequencing methods and bioinformatics tools (assemble and analyze the structure and functions of specific gene). However, the whole genome is limited to cultivable bacteria which can grow as pure cultures in the laboratory conditions (Figure 3). Due to lack of cultivation methods in laboratory conditions, the majority of bacteria on our planet are uncultured and hence unidentified (Hug et al., 2016).
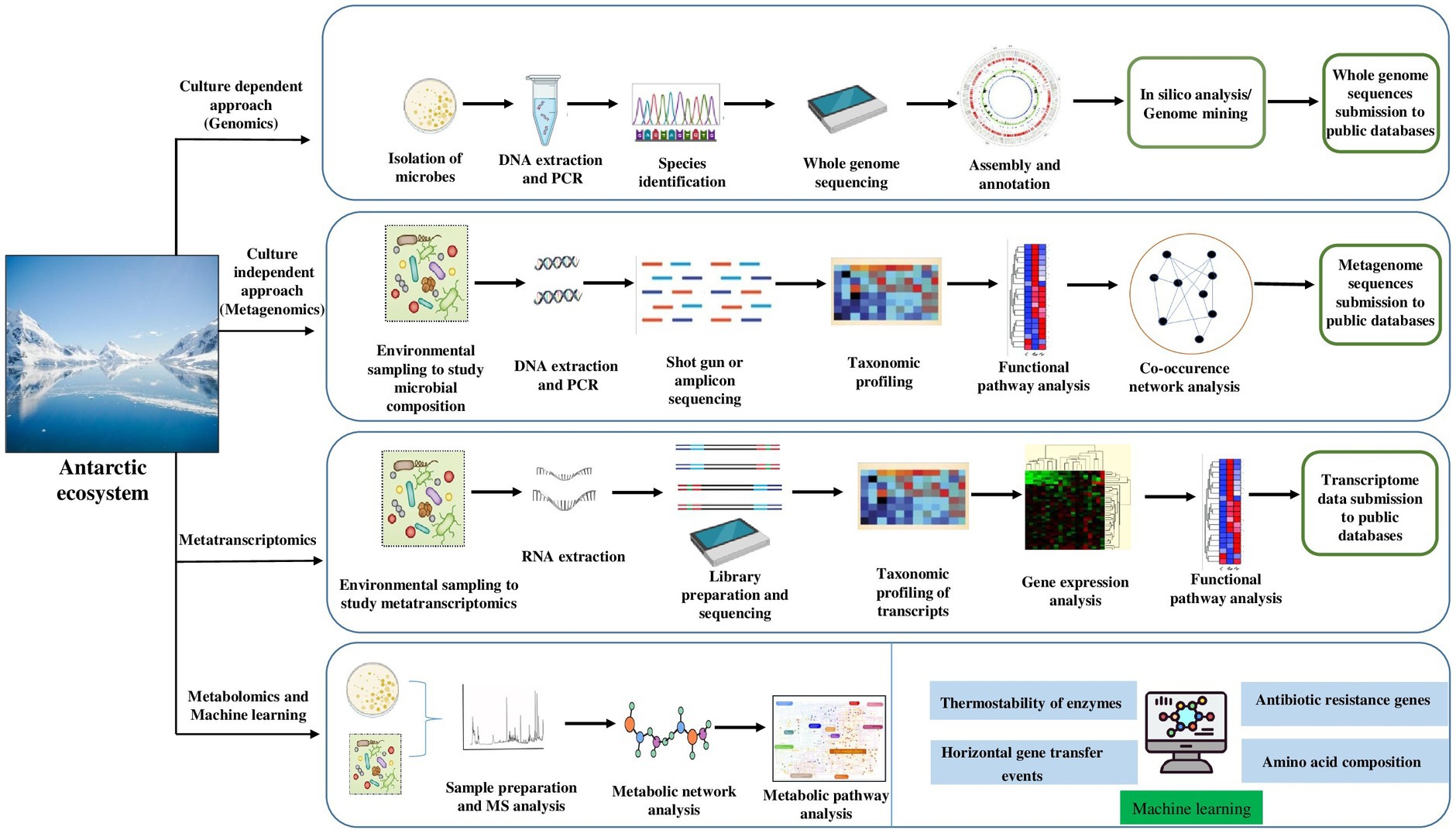
Figure 3. Advanced technologies – omics tools and machine learning approach to study the adaptation mechanism and potential of cold-adaptive molecules of Antarctic bacteria.
Up till now the whole genome sequence of Antarctic bacteria is available for a few taxa. The advancement in sequencing methods and bioinformatic approach could provide the understanding of their physiological and metabolic roles. The portable sequencers of Oxford Nanopore Technologies MinION could be used as an in situ sequencing tool for community composition and functional profiling of microbes thrive in Antarctic environment. Although, the high error rate of Nanopore sequencing compared to the amplicon sequencing technology by the Illumina platform is gradually decreasing, the hybrid assembly strategy (i.e., Illumina short reads assembled together with Nanopore long reads) is considered the best to cover the novel taxa and their functions in various Antarctic microbial communities.
The use of long-read metagenomic sequencing by Waschulin et al. (2022) revealed the biosynthetic potential of uncultured bacterial phyla such as Acidobacteriota, Verrucomicrobiota and Gemmatimonadota. Additionally, the uncultivable bacteria and their genetic functions can be explored through the metagenomic approach (genetic material of mixed community directly recovered from natural environment without obtaining the pure culture) which can either be sequence based, including high-throughput sequencing and bioinformatic analysis (high quality metagenome-assembled genomes, obtained through combination of binning approaches), or function based by involving functional expression of metagenomic libraries to identify target genes/gene clusters (Figure 3) (Alneberg et al., 2018).
Uncultivated microbial clades (candidate phyla) belonging to Genome Taxonomy Database (GTDB) in the Antarctic Ace Lake may play an important role in nutrient cycling (Williams et al., 2022). Similar studies have been reported by Williams et al. (2021) on microbial “dark matter” i.e. Candidatus bacterial phyla of metagenome-assembled genomes (MAGs) obtained from an Antarctic Lake. Recently, Fonseca et al. (2022) identified the bacterial family Woeseiaceae for the first time in Antarctic sediments, but the cellular and molecular adaptation of this family to the cold environment is unknown. Although the metagenomic approach reveals the taxonomic composition, to deeply understand the expression of genes in the microbes to environmental changes, the metatranscriptome provides the information about microbial functions associated with the environment (Sutherland et al., 2022). The functional diversity of the microbial communities has been recently investigated under the Antarctic ice shelf using multiomics approach such as metagenomics, metatranscriptomics, single-cell genomics by Martínez-Pérez et al. (2022).
A study by Médigue et al. (2005) sequenced the genome of the Antarctic bacterium Pseudoalteromonas haloplanktis TAC125 and using in silico analysis revealed the composition of the proteome for cold adaptation. Similarly, Fondi et al. (2015) investigated several metabolic features of Pseudoalteromonas haloplanktis TAC125 and variations in cellular metabolic fluxes through in silico modeling.
Recent developments in the omics era will open a remarkable milestone in structural and functional metagenomics (Prayogo et al., 2020). In fact, metagenome mining is applied to bacterial communities to screen novel classes of cold adapted enzymes for biotechnological applications (Kumar et al., 2021). A recent study by Blázquez-Sánchez et al. (2022) found Antarctic bacteria Moraxella sp. strain TA144 (Mors1) and Oleispira antarctica RB-8 (OaCut) hydrolyze aliphatic and aromatic polyesters at moderate temperatures. In addition, metagenome analysis revealed the members of the Moraxellaceae family harbors candidate genes for polyethylene terephthalate (PET) hydrolases (Blázquez-Sánchez et al., 2022). The majority of bacteria living in the cold expresses certain genes as adaptation strategies. Dall Agnol et al. (2014), using transcriptomes (Figure 3) and proteomes, unveiled global gene expression in response to thermal adaptation of E. antarcticum B7 at different temperatures (0°C and 37°C). In a recent study by Ijaq et al. (2022), Pseudomonas sp. Lz4W genome analysis revealed 743 CDS annotated as hypothetical proteins, including 61 hypothetical proteins at translational level. However, with the current global climate crisis, there is the urgent need to understand all the adaptation mechanisms to the changing environmental conditions, particularly in the polar region. Recent developments in proteomics along with gene expression profiling have been coupled for the discovery of various biomolecules in psychrophilic bacteria. Garcia-Lopez et al. (2021) presented a review of the current knowledge on psychrophiles for their biomolecules and metabolic pathways. Understanding the adaptation strategies of Antarctic bacteria using the omics approach will help to reveal their metabolic changes (Figure 3) to future climate change scenario.
Machine learning approach
Machine learning (ML) is a branch of artificial intelligence (AI) that designs mathematical models to execute certain tasks from assembled information in less time and cost. Generally, two ML models – supervised (also known as predictive) and unsupervised (descriptive)- have been extensively used in most research areas in the field of microbiology (Goodswen et al., 2021; Greener et al., 2022). The supervised model requires to be trained using the training data set which includes text, images, and alphanumeric data. The most used supervised model algorithms are classification and regression. While the unsupervised algorithm uses unlabeled data which involves clustering and association rule mining (Goodswen et al., 2021). The ML approach has evolved rapidly in recent years to understand microbial and molecular processes using high-throughput data. Considering the various applications of this approach in biology, very scarce studies are available for psychrophilic microbes, especially bacteria. Previously, Lee C. et al. (2019) used ML method (classification and regression tree machine learning algorithms called CART) to study the bacterial communities and geochemical variables. The study provides a clue to unravel the bacterial communities link to changing environmental conditions using ML approach in other habitats of the Antarctic environment. Later, the mobilized colistin resistance (mcr) gene, that is a type of antibiotic resistance gene (ARG), has been identified using machine learning tools (Figure 3) in polar Psychrobacter (Cuadrat et al., 2020). Similarly, Arango-Argoty et al. (2018) developed a tool called DeepARG using a deep learning approach for the prediction of antibiotic resistance genes from metagenomic data. Recent study by Marcoleta et al. (2022) employ DeepARG tool to detect ARG from North Antarctic soils microbial communities (Pseudomonas, Streptomyces, Gemmatimonas, Paenibacillus, and Polaromonas). Yet, sparse information is available for antibiotic resistance genes (ARG) of resistome profile of Antarctic bacteria. Moreover, genomic, and metabolic pathways of novel taxa of bacteria are poorly known. The ML is a promising approach and can be widely used in omics data to explore the presence of ARGs in Antarctic habitats.
Furthermore, studies based on machine learning have been reported on thermophilic and mesophilic bacterial proteins, however little is known about the application of ML approach in psychrophilic enzymes. Recent study on psychrophilic amino acid composition (AAC) using machine learning (ML) algorithm showed that psychrophiles proteins contains high frequency of Ala, Gly, Ser, and Thr, compared to Glu, Lys, Arg, Ile, Val, and Leu amino acids (Huang et al., 2023a). The support vector machine learning model (SVM) in combination with molecular dynamics (MD) is employed to study the thermostability of psychrophilic alpha-amylase (Figure 3) (exhibited high activity at low temperature) isolated from Pseudoalteromonas haloplanktis (Li et al., 2020). The study revealed the presence of two single point mutations (S255K and S340P) and one double mutation (S255K/S340P) at non-conserved residues that enhanced thermostability of enzyme without altering its catalytic activity (Li et al., 2020). Similarly, to understand the antifreeze peptides and proteins interactions to ice crystals, several computational predictors have been used (Jiang et al., 2022). Recently, ML-guided robotic strain isolation platform for the isolation of diverse microbes from human feces have been used by Huang et al. (2023b). These ML approaches can be applicable to bridge the knowledge gap on genes functions in bacterial genome in response to climate change adaptation (Figure 3). Hence, we think this algorithm can be extendable to the Antarctic ecosystem in the upcoming studies to develop the understanding on bacterial dark matter and their adaptation.
ML has the potential to accelerate HIV drug discovery by narrowing down the number of antiviral compounds selected for in vitro and in vivo testing.
Conclusion and future perspectives
The cold-adapted Antarctic psychrophilic bacteria represent excellent model organisms to study climate change induced stress adaptation. These bacteria are thriving in harsh and inhospitable Antarctic environment and displaying immense potential of regulating climate change factors. Therefore, the comprehensive review on Antarctic bacteria highlighted the adaptation strategies of psychrophiles at various levels (structural, physiological and molecular) in response to the changing environment. Many of these adaptation tools including biosurfactants, EPS, PUFA, membrane pigments, molecular chaperones and the underlying mechanisms are superficially studied due to the difficulty in some bacterial cultivation. The aid of ML and omics approach, particularly metagenomics, could provide insight into how psychrophilic bacteria adapt to cope with cellular and molecular mechanisms as survival strategies. Extensive research using these approaches needs to be done to better understand the bacterial adaptation in Antarctic environment and implementing this knowledge for improving the tolerance ability of other bacteria, for an environmentally sustainable future. The multi-omics tools coupled with ML algorithms might explore the industrial potential of biomolecules from Antarctic bacteria effectively in less time and labor. This is another aspect of upcoming research that needs to be emphasized. In recent years, the exploitation of psychrophilic enzymes (amylase, lipase, protease, hydrolase, pectinase, cellulase etc.) has been increased in different biotechnological industries due to their improved catalytic efficiency, flexibility, and low thermal stability. The cold-active enzymes are the best eco-friendly alternative to synthetic chemicals and moreover it increases the shelf life of the products (fabrics, foods etc.). Therefore, the manufacturers are seeking novel cold-active enzymes that further improve the efficiency and quality of products (particularly in the detergents industry). Hence, this is an additional aspect of research that needs to be explored. Overall, the review discusses the most recent studies on Antarctic bacterial adaptation as future climate model and suggested novel approaches for upcoming research in this direction.
Author contributions
KPR and LM conceptualized the idea and constructed figures. RR and KPR constructed tables. KPR, LM, RR, and KR wrote the original draft. CM and SP revised the manuscript. KPR wrote the final draft with all other authors. All authors contributed to the article and approved the submitted version.
Funding
The authors are thankful to Umeå University for the funding provided for Open access fee.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Abe, T., Akazawa, Y., Toyoda, A., Niki, H., and Baba, T. (2020). Batch-learning self-organizing map identifies horizontal gene transfer candidates and their origins in entire genomes. Front. Microbiol. 11:1486. doi: 10.3389/fmicb.2020.01486
Acevedo, J. P., Rodriguez, V., Saavedra, M., Munoz, M., Salazar, O., Asenjo, J. A., et al. (2013). Cloning, expression and decoding of the cold adaptation of a new widely represented thermolabile subtilisin-like protease. J. Appl. Microbiol. 114, 352–363. doi: 10.1111/jam.12033
Acosta, S., Canclini, L., Marizcurrena, J. J., Castro-Sowinski, S., and Hernández, P. (2022). Photo-repair effect of a bacterial Antarctic CPD-photolyase on UVC-induced DNA lesions in human keratinocytes. Environ. Toxicol. Pharmacol. 96:104001. doi: 10.1016/j.etap.2022.104001
Aislabie, J., Saul, D. J., and Foght, J. M. (2006). Bioremediation of hydrocarbon-contaminated polar soils. Extremophiles 10, 171–179. doi: 10.1007/s00792-005-0498-4
Al-Ghanayem, A. A., and Joseph, B. (2020). Current prospective in using cold-active enzymes as eco-friendly detergent additive. Appl. Microbiol. Biotechnol. 104, 2871–2882. doi: 10.1007/s00253-020-10429-x
Al-Maqtari, Q. A., Waleed, A. A., and Mahdi, A. A. (2019). Cold-active enzymes and their applications in industrial fields-a review. Int. J. Res. Agric. Sci. 6, 2348–3997.
Alneberg, J., Karlsson, C. M., Divne, A. M., Bergin, C., Homa, F., Lindh, M. V., et al. (2018). Genomes from uncultivated prokaryotes: a comparison of metagenome-assembled and single-amplified genomes. Microbiome 6, 1–14. doi: 10.1186/s40168-018-0550-0
Alterio, V., Aurilia, V., Romanelli, A., Parracino, A., Saviano, M., D’Auria, S., et al. (2010). Crystal structure of an S-formylglutathione hydrolase from Pseudoalteromonas haloplanktis TAC125. Biopolymers 93, 669–677. doi: 10.1002/bip.21420
Anesio, A. M., Hodson, A. J., Fritz, A., Psenner, R., and Sattler, B. (2009). High microbial activity on glaciers: importance to the global carbon cycle. Glob. Chang. Biol. 15, 955–960. doi: 10.1111/j.1365-2486.2008.01758.x
Angeli, A., Del Prete, S., Osman, S. M., AlOthman, Z., Donald, W. A., Capasso, C., et al. (2019). Activation studies of the γ-carbonic anhydrases from the Antarctic marine bacteria Pseudoalteromonas haloplanktis and Colwellia psychrerythraea with amino acids and amines. Mar. Drugs 17:238. doi: 10.3390/md17040238
Arango-Argoty, G., Garner, E., Pruden, A., Heath, L. S., Vikesland, P., and Zhang, L. (2018). Deep ARG: a deep learning approach for predicting antibiotic resistance genes from metagenomic data. Microbiome 6, 1–15. doi: 10.1186/s40168-018-0401-z
Aurilia, V., Parracino, A., Saviano, M., and D’Auria, S. (2007). The psychrophilic bacterium Pseudoalteromonas halosplanktis TAC125 possesses a gene coding for a cold-adapted feruloyl esterase activity that shares homology with esterase enzymes from γ-proteobacteria and yeast. Gene 397, 51–57. doi: 10.1016/j.gene.2007.04.004
Ausuri, J., Dell’Anno, F., Vitale, G. A., Palma Esposito, F., Funari, V., Franci, G., et al. (2022). Bioremediation of Multiple Heavy Metals Mediated by Antarctic Marine Isolated Dietzia psychralcaliphila JI1D. JMSE 10:1669. doi: 10.3390/jmse10111669
Awazu, N., Shodai, T., Takakura, H., Kitagawa, M., Mukai, H., and Kato, I. (2011). U.S. patent no. 8, 034, 597. Washington, DC: U.S. patent and trademark office.
Bajerski, F., Wagner, D., and Mangelsdorf, K. (2017). Cell membrane fatty acid composition of Chryseobacterium frigidisoli PB4T, isolated from Antarctic glacier forefield soils, in response to changing temperature and pH conditions. Front. Microbiol. 8:677. doi: 10.3389/fmicb.2017.00677
Bar Dolev, M., Bernheim, R., Guo, S., Davies, P. L., and Braslavsky, I. (2016). Putting life on ice: bacteria that bind to frozen water. J. R. Soc. Interface 13:20160210. doi: 10.1098/rsif.2016.0210
Barnes, D. K., Sands, C. J., Paulsen, M. L., Moreno, B., Moreau, C., Held, C., et al. (2021). Societal importance of Antarctic negative feedbacks on climate change: blue carbon gains from sea ice, ice shelf and glacier losses. Sci. Nat. 108, 1–14. doi: 10.1007/s00114-021-01748-8
Bisaccia, M., Binda, E., Rosini, E., Caruso, G., Dell’Acqua, O., Azzaro, M., et al. (2023). A novel promising laccase from the psychrotolerant and halotolerant Antarctic marine Halomonas sp. M68 strain. Front. Microbiol. 14:1078382. doi: 10.3389/fmicb.2023.1078382
Blázquez-Sánchez, P., Engelberger, F., Cifuentes-Anticevic, J., Sonnendecker, C., Griñén, A., Reyes, J., et al. (2022). Antarctic polyester hydrolases degrade aliphatic and aromatic polyesters at moderate temperatures. Appl. Environ. Microbiol. 88, e01842–e01821. doi: 10.1128/aem.01842-21
Bowman, J. S., and Ducklow, H. W. (2015). Microbial communities can be described by metabolic structure: a general framework and application to a seasonally variable, depth-stratified microbial community from the coastal West Antarctic peninsula. PLoS One 10:e0135868. doi: 10.1371/journal.pone.0135868
Bruno, S., Coppola, D., di Prisco, G., Giordano, D., and Verde, C. (2019). Enzymes from marine polar regions and their biotechnological applications. Mar. Drugs 17:544. doi: 10.3390/md17100544
Busse, H. J., Denner, E. B., Buczolits, S., Salkinoja-Salonen, M., Bennasar, A., and Kampfer, P. (2003). Sphingomonas aurantiaca sp. nov., Sphingomonas aerolata sp. nov. and Sphingomonas faeni sp. nov., air-and dustborne and Antarctic, orange-pigmented, psychrotolerant bacteria, and emended description of the genus Sphingomonas. Int. J. Syst. Evol. Microbiol. 53, 1253–1260. doi: 10.1099/ijs.0.02461-0
Cabrera, M. Á., Márquez, S. L., and Pérez-Donoso, J. M. (2022). Comparative genomic analysis of Antarctic Pseudomonas isolates with 2, 4, 6-trinitrotoluene transformation capabilities reveals their unique features for xenobiotics degradation. Genes 13:1354. doi: 10.3390/genes13081354
Cabrera, M. Á., Márquez, S. L., Quezada, C. P., Osorio, M. I., Castro-Nallar, E., González-Nilo, F. D., et al. (2020). Biotransformation of 2, 4, 6-trinitrotoluene by Pseudomonas sp. TNT3 isolated from deception island, Antarctica. Environ. Pollut. 262:113922. doi: 10.1016/j.envpol.2020.113922
Cagide, C., Marizcurrena, J. J., Vallés, D., Alvarez, B., and Castro-Sowinski, S. (2023). A bacterial cold-active dye-decolorizing peroxidase from an Antarctic Pseudomonas strain. Appl. Microbiol. Biotechnol. 107, 1707–1724. doi: 10.1007/s00253-023-12405-7
Caruso, C., Rizzo, C., Mangano, S., Poli, A., Di Donato, P., Finore, I., et al. (2018). Production and biotechnological potential of extracellular polymeric substances from sponge-associated Antarctic bacteria. Appl. Environ. Microbiol. 84, e01624–e01617. doi: 10.1128/aem.01624-17
Casillo, A., Parrilli, E., Sannino, F., Mitchell, D. E., Gibson, M. I., Marino, G., et al. (2017). Structure-activity relationship of the exopolysaccharide from a psychrophilic bacterium: a strategy for cryoprotection. Carbohydr. Polym. 156, 364–371. doi: 10.1016/j.carbpol.2016.09.037
Cheng, Z., Shi, C., Gao, X., Wang, X., and Kan, G. (2022). Biochemical and Metabolomic responses of Antarctic bacterium Planococcus sp. O5 induced by copper ion. Toxics 10:302. doi: 10.3390/toxics10060302
Chen, Y., Gao, P., Tang, X., and Xu, C. (2021). Characterisation and bioactivities of an exopolysaccharide from an Antarctic bacterium Shewanella frigidimarina W32–2. Aquaculture 530:735760. doi: 10.1016/j.aquaculture.2020.735760
Chessa, J. P., Feller, G., and Gerday, C. (1999). Purification and characterization of the heat-labile α-amylase secreted by the psychrophilic bacterium TAC 240B. Can. J. Microbiol. 45, 452–457. doi: 10.1139/w99-021
Chianese, G., Esposito, F. P., Parrot, D., Ingham, C., De Pascale, D., and Tasdemir, D. (2018). Linear aminolipids with moderate antimicrobial activity from the Antarctic gram-negative bacterium Aequorivita sp. Mar. Drugs 16:187. doi: 10.3390/md16060187
Cieśliński, H., Białkowska, A. M., Długołęcka, A., Daroch, M., Tkaczuk, K. L., Kalinowska, H., et al. (2007). A cold-adapted esterase from psychrotrophic Pseudoalteromas sp. strain 643A. Arch. Microbiol. 188, 27–36. doi: 10.1007/s00203-007-0220-2
Cieśliński, H., Kur, J., Białkowska, A., Baran, I., Makowski, K., and Turkiewicz, M. (2005). Cloning, expression, and purification of a recombinant cold-adapted β-galactosidase from Antarctic bacterium Pseudoalteromonas sp. 22b. Protein Expr. Purif. 39, 27–34. doi: 10.1016/j.pep.2004.09.002
Collins, T., and Margesin, R. (2019). Psychrophilic lifestyles: mechanisms of adaptation and biotechnological tools. Appl. Microbiol. Biotechnol. 103, 2857–2871. doi: 10.1007/s00253-019-09659-5
Correa-Llanten, D. N., Amenabar, M. J., and Blamey, J. M. (2012). Resistance to hypoosmotic shock of liposomes containing novel pigments from an Antarctic bacterium. Microbiol. Biotechnol. Lett. 40, 215–219. doi: 10.4014/kjmb.1205.05018
Cuadrat, R. R., Sorokina, M., Andrade, B. G., Goris, T., and Davila, A. M. (2020). Global Ocean resistome revealed: exploring antibiotic resistance gene abundance and distribution in TARA oceans samples. Gigascience 9:giaa046. doi: 10.1093/gigascience/giaa046
D’Angelo, C., Casillo, A., Melchiorre, C., Lauro, C., Corsaro, M. M., Carpentieri, A., et al. (2022). CATASAN is a new anti-biofilm agent produced by the marine Antarctic bacterium Psychrobacter sp. TAE2020. Mar. Drugs 20:747. doi: 10.3390/md20120747
D’Auria, S., Aurilia, V., Marabotti, A., Gonnelli, M., and Strambini, G. (2009). Structure and dynamics of cold-adapted enzymes as investigated by phosphorescence spectroscopy and molecular dynamics studies. 2. The case of an esterase from Pseudoalteromonas haloplanktis. J. Phys. Chem. B 113, 13171–13178. doi: 10.1021/jp9043286
Dall Agnol, H. P., Baraúna, R. A., de Sá, P. H., Ramos, R. T., Nóbrega, F., Nunes, C. I., et al. (2014). Omics profiles used to evaluate the gene expression of Exiguobacterium antarcticum B7 during cold adaptation. BMC Genomics 15, 1–12. doi: 10.1186/1471-2164-15-986
Darham, S., Syed-Muhaimin, S. N., Subramaniam, K., Zulkharnain, A., Shaharuddin, N. A., Khalil, K. A., et al. (2021). Optimisation of Various Physicochemical Variables Affecting Molybdenum Bioremediation Using Antarctic Bacterium, Arthrobacter sp. Strain AQ5-05. Water 13:2367. doi: 10.3390/w13172367
Das, K. R., Tiwari, A. K., and Kerkar, S. (2020). Psychrotolerant Antarctic bacteria biosynthesize gold nanoparticles active against sulphate reducing bacteria. Prep. Biochem. Biotechnol. 50, 438–444. doi: 10.1080/10826068.2019.1706559
Davail, S., Feller, G., Narinx, E., and Gerday, C. (1994). Cold adaptation of proteins. Purification, characterization, and sequence of the heat-labile subtilisin from the Antarctic psychrophile Bacillus TA41. J. Biol. Chem. 269, 17448–17453.
De Lemos, E. A., Procópio, L., Da Mota, F. F., Jurelevicius, D., Rosado, A. S., and Seldin, L. (2023). Molecular characterization of Paenibacillus antarcticus IPAC21, a bioemulsifier producer isolated from Antarctic soil. Front. Microbiol. 14:1142582. doi: 10.3389/fmicb.2023.1142582
De Luca, V., Vullo, D., Del Prete, S., Carginale, V., Osman, S. M., AlOthman, Z., et al. (2016). Cloning, characterization and anion inhibition studies of a γ-carbonic anhydrase from the Antarctic bacterium Colwellia psychrerythraea. Bioorg. Med. Chem. 24, 835–840. doi: 10.1016/j.bmc.2016.01.005
De Luca, V., Vullo, D., Del Prete, S., Carginale, V., Scozzafava, A., Osman, S. M., et al. (2015). Cloning, characterization and anion inhibition studies of a new γ-carbonic anhydrase from the Antarctic bacterium Pseudoalteromonas haloplanktis. Bioorg. Med. Chem. 23, 4405–4409. doi: 10.1016/j.bmc.2015.06.021
De Maayer, P., Anderson, D., Cary, C., and Cowan, D. A. (2014). Some like it cold: understanding the survival strategies of psychrophiles. EMBO Rep. 15, 508–517. doi: 10.1002/embr.201338170
Denner, E. B., Mark, B., Busse, H. J., Turkiewicz, M., and Lubitz, W. (2001). Psychrobacter proteolyticus sp. nov., a psychrotrophic, halotolerant bacterium isolated from the Antarctic krill Euphausia superba Dana, excreting a cold-adapted metalloprotease. Syst. Appl. Microbiol. 24, 44–53. doi: 10.1078/0723-2020-00006
De Pascale, D., Cusano, A. M., Autore, F., Parrilli, E., Di Prisco, G., Marino, G., et al. (2008). The cold-active Lip1 lipase from the Antarctic bacterium Pseudoalteromonas haloplanktis TAC125 is a member of a new bacterial lipolytic enzyme family. Extremophiles 12, 311–323. doi: 10.1007/s00792-008-0163-9
De Pascale, D., Giuliani, M., De Santi, C., Bergamasco, N., Amoresano, A., Carpentieri, A., et al. (2010). PhAP protease from Pseudoalteromonas haloplanktis TAC125: gene cloning, recombinant production in E. coli and enzyme characterization. Pol. Sci. 4, 285–294. doi: 10.1016/j.polar.2010.03.009
Dietz, S., and Koninx, F. (2022). Economic impacts of melting of the Antarctic ice sheet. Nat. Commun. 13:5819. doi: 10.1038/s41467-022-33406-6
Di Lorenzo, F., Crisafi, F., La Cono, V., Yakimov, M. M., Molinaro, A., and Silipo, A. (2020). The structure of the lipid a of gram-negative cold-adapted Bacteria isolated from Antarctic environments. Mar. Drugs 18:592. doi: 10.3390/md18120592
Ding, H., Zeng, Q., Zhou, L., Yu, Y., and Chen, B. (2017). Biochemical and structural insights into a novel thermostable β-1, 3-galactosidase from Marinomonas sp. BSi20414. Mar. Drugs 15:13. doi: 10.3390/md15010013
Dutta, A., and Chaudhuri, K. (2010). Analysis of tRNA composition and folding in psychrophilic, mesophilic and thermophilic genomes: indications for thermal adaptation. FEMS Microbiol. Lett. 305, 100–108. doi: 10.1111/j.1574-6968.2010.01922.x
Effendi, D. B., Sakamoto, T., Ohtani, S., Awai, K., and Kanesaki, Y. (2022). Possible involvement of extracellular polymeric substrates of Antarctic cyanobacterium Nostoc sp. strain SO-36 in adaptation to harsh environments. J. Plant Res. 135, 771–784. doi: 10.1007/s10265-022-01411-x
Fan, Y., Yi, J., Hua, X., Feng, Y., Yang, R., and Zhang, Y. (2016). Structure analysis of a glycosides hydrolase family 42 cold-adapted β-galactosidase from Rahnella sp R3. RSC Adv. 6, 37362–37369. doi: 10.1039/c6ra04529d
Feller, G., d’Amic, D., and Gerday, C. (1999). Thermodynamic stability of a cold-active α-amylase from the Antarctic bacterium Alteromonas haloplanctis. Biochemistry 38, 4613–4619. doi: 10.1021/bi982650+
Feller, G., and Gerday, C. (2003). Psychrophilic enzymes: hot topics in cold adaptation. Nat. Rev. Microbiol. 1, 200–208. doi: 10.1038/nrmicro773
Feller, G., Thiry, M., Arpigny, J. L., and Gerday, C. (1991). Cloning and expression in Escherichia coli of three lipase-encoding genes from the psychrotrophic Antarctic strain Moraxella TA144. Gene 102, 111–115. doi: 10.1016/0378-1119(91)90548-p
Fondi, M., Maida, I., Perrin, E., Mellera, A., Mocali, S., Parrilli, E., et al. (2015). Genome-scale metabolic reconstruction and constraint-based modelling of the Antarctic bacterium Pseudoalteromonas haloplanktis TAC 125. Environ. Microbiol. 17, 751–766. doi: 10.1111/1462-2920.12513
Fonseca, F., Meneghel, J., Cenard, S., Passot, S., and Morris, G. J. (2016). Determination of intracellular vitrification temperatures for unicellular microorganisms under conditions relevant for cryopreservation. PLoS One 11:e0152939. doi: 10.1371/journal.pone.0152939
Fonseca, V. G., Kirse, A., Giebner, H., Vause, B. J., Drago, T., Power, D. M., et al. (2022). Metabarcoding the Antarctic peninsula biodiversity using a multi-gene approach. ISME Commun. 2:37. doi: 10.1038/s43705-022-00118-3
Galkin, A., Kulakova, L., Ashida, H., Sawa, Y., and Esaki, N. (1999). Cold-adapted alanine dehydrogenases from two Antarctic bacterial strains: gene cloning, protein characterization, and comparison with mesophilic and thermophilic counterparts. Appl. Environ. Microbiol. 65, 4014–4020. doi: 10.1128/aem.65.9.4014-4020.1999
García-Descalzo, L., García-López, E., Alcázar, A., Baquero, F., and Cid, C. (2014). Proteomic analysis of the adaptation to warming in the Antarctic bacteria Shewanella frigidimarina. Biochim Biophys Acta 1844, 2229–2240. doi: 10.1016/j.bbapap.2014.08.006
García-Descalzo, L., Lezcano, M. Á., Carrizo, D., and Fairén, A. G. (2023). Changes in membrane fatty acids of a halo-psychrophile exposed to magnesium perchlorate and low temperatures: implications for Mars habitability. Front. Astron. Space Sci. 10:65. doi: 10.3389/fspas.2023.1034651
Garcia-Lopez, E., Alcazar, P., and Cid, C. (2021). Identification of biomolecules involved in the adaptation to the environment of cold-loving microorganisms and metabolic pathways for their production. Biomol. Ther. 11:1155. doi: 10.3390/biom11081155
Georlette, D., Jonsson, Z. O., Van Petegem, F., Chessa, J. P., Van Beeumen, J., Hübscher, U., et al. (2000). A DNA ligase from the psychrophile Pseudoalteromonas haloplanktis gives insights into the adaptation of proteins to low temperatures. Eur. J. Biochem. 267, 3502–3512. doi: 10.1046/j.1432-1327.2000.01377.x
Gerday, C., Aittaleb, M., Bentahir, M., Chessa, J. P., Claverie, P., Collins, T., et al. (2000). Cold-adapted enzymes: from fundamentals to biotechnology. Trends Biotechnol. 18, 103–107. doi: 10.1016/s0167-7799(99)01413-4
Goodswen, S. J., Barratt, J. L., Kennedy, P. J., Kaufer, A., Calarco, L., and Ellis, J. T. (2021). Machine learning and applications in microbiology. FEMS Microbiol. Rev. 45:fuab015. doi: 10.1093/femsre/fuab015
Goordial, J., Raymond-Bouchard, I., Zolotarov, Y., de Bethencourt, L., Ronholm, J., Shapiro, N., et al. (2016). Cold adaptive traits revealed by comparative genomic analysis of the eurypsychrophile Rhodococcus sp. JG3 isolated from high elevation McMurdo Dry Valley permafrost, Antarctica. FEMS Microbiol. Ecol. 92:fiv154. doi: 10.1093/femsec/fiv154
Greener, J. G., Kandathil, S. M., Moffat, L., and Jones, D. T. (2022). A guide to machine learning for biologists. Nat. Rev. Mol. Cell Biol. 23, 40–55. doi: 10.1038/s41580-021-00407-0
Gushgari-Doyle, S., Lui, L. M., Nielsen, T. N., Wu, X., Malana, R. G., Hendrickson, A. J., et al. (2022). Genotype to ecotype in niche environments: adaptation of Arthrobacter to carbon availability and environmental conditions. ISME Commun. 2:32. doi: 10.1038/s43705-022-00113-8
Gutt, J., Isla, E., Xavier, J. C., Adams, B. J., Ahn, I. Y., Cheng, C. H. C., et al. (2021). Antarctic ecosystems in transition–life between stresses and opportunities. Biol. Rev. 96, 798–821. doi: 10.1111/brv.12679
Hamid, B., Bashir, Z., Yatoo, A. M., Mohiddin, F., Majeed, N., Bansal, M., et al. (2022). Cold-active enzymes and their potential industrial applications—a review. Molecules 27:5885. doi: 10.3390/molecules27185885
Hassan, N., Anesio, A. M., Rafiq, M., Holtvoeth, J., Bull, I., Haleem, A., et al. (2020). Temperature driven membrane lipid adaptation in glacial psychrophilic bacteria. Front. Microbiol. 11:824. doi: 10.3389/fmicb.2020.00824
Hauksson, J. B., Andrésson, Ó. S., and Ásgeirsson, B. (2000). Heat-labile bacterial alkaline phosphatase from a marine Vibrio sp. Enzym. Microb. Technol. 27, 66–73. doi: 10.1016/s0141-0229(00)00152-6
Herrera, L. M., Braña, V., Fraguas, L. F., and Castro-Sowinski, S. (2019). Characterization of the cellulase-secretome produced by the Antarctic bacterium Flavobacterium sp. AUG42. Microbiol. Res. 223, 13–21. doi: 10.1016/j.micres.2019.03.009
He, Y., Qu, C., Zhang, L., and Miao, J. (2021). DNA photolyase from Antarctic marine bacterium Rhodococcus sp. NJ-530 can repair DNA damage caused by ultraviolet. 3. Biotech 11, 1–9. doi: 10.1007/s13205-021-02660-8
Hoyoux, A., Jennes, I., Dubois, P., Genicot, S., Dubail, F., François, J. M., et al. (2001). Cold-adapted β-galactosidase from the Antarctic psychrophile Pseudoalteromonas haloplanktis. Appl. Environ. Microbiol. 67, 1529–1535. doi: 10.1128/aem.67.4.1529-1535.2001
Huang, A., Lu, F., and Liu, F. (2023a). Discrimination of psychrophilic enzymes using machine learning algorithms with amino acid composition descriptor. Front. Microbiol. 14:1130594. doi: 10.3389/fmicb.2023.1130594
Huang, Y., Sheth, R. U., Zhao, S., Cohen, L. A., Dabaghi, K., Moody, T., et al. (2023b). High-throughput microbial culturomics using automation and machine learning. Nat. Biotechnol., 1–10. doi: 10.1038/s41587-023-01674-2
Hug, L. A., Baker, B. J., Anantharaman, K., Brown, C. T., Probst, A. J., Castelle, C. J., et al. (2016). A new view of the tree of life. Nat. Microbiol. 1, 1–6. doi: 10.1038/nmicrobiol.2016.48
Humphry, D. R., George, A., Black, G. W., and Cummings, S. P. (2001). Flavobacterium frigidarium sp. nov., an aerobic, psychrophilic, xylanolytic and laminarinolytic bacterium from Antarctica. Int. J. Syst. Evol. Microbiol. 51, 1235–1243. doi: 10.1099/00207713-51-4-1235
Hussain, A., and Ray, M. K. (2022). DEAD box RNA helicases protect Antarctic Pseudomonas syringae Lz4W against oxidative stress. Infect. Genet. Evol. 106:105382. doi: 10.1016/j.meegid.2022.105382
Hwang, K., Choe, H., Nasir, A., and Kim, K. M. (2021). Complete genome of Polaromonas vacuolata KCTC 22033T isolated from beneath Antarctic Sea ice. Mar. Genomics 55:100790. doi: 10.1016/j.margen.2020.100790
Ijaq, J., Chandra, D., Ray, M. K., and Jagannadham, M. V. (2022). Investigating the functional role of hypothetical proteins from an Antarctic bacterium Pseudomonas sp. Lz4W: emphasis on identifying proteins involved in cold adaptation. Front. Genet. 13:825269. doi: 10.3389/fgene.2022.825269
IPCC (2021). Climate Change 2021: The Physical Science Basis. Contribution of Working Group I to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change (Cambridge: Cambridge University Press).
Irgens, R. L., Gosink, J. J., and Staley, J. T. (1996). Polaromonas vacuolata gen. Nov., sp. nov., a psychrophilic, marine, gas vacuolate bacterium from Antarctica. Int. J. Syst. Bacteriol. 46, 822–826. doi: 10.1099/00207713-46-3-822
Javani, S., Marín, I., Amils, R., and Abad, J. P. (2015). Four psychrophilic bacteria from Antarctica extracellularly biosynthesize at low temperature highly stable silver nanoparticles with outstanding antimicrobial activity. Colloids Surf. A Physicochem. Eng. Asp. 483, 60–69. doi: 10.1016/j.colsurfa.2015.07.028
Javed, A., and Qazi, J. I. (2016). Psychrophilic microbial enzymes implications in coming biotechnological processes. Am. Sci. Res. J. Eng. Technol. Sci. 23, 103–120.
Jiang, W., Yang, F., Chen, X., Cai, X., Wu, J., Du, M., et al. (2022). Molecular simulation-based research on antifreeze peptides: advances and perspectives. J. Future Food 2, 203–212. doi: 10.1016/j.jfutfo.2022.06.002
Ji, M., Barnwell, C. V., and Grunden, A. M. (2015). Characterization of recombinant glutathione reductase from the psychrophilic Antarctic bacterium Colwellia psychrerythraea. Extremophiles 19, 863–874. doi: 10.1007/s00792-015-0762-1
John, M. S., Nagoth, J. A., Ramasamy, K. P., Ballarini, P., Mozzicafreddo, M., Mancini, A., et al. (2020). Horizontal gene transfer and silver nanoparticles production in a new Marinomonas strain isolated from the Antarctic psychrophilic ciliate Euplotes focardii. Sci. Rep. 10, 1–14. doi: 10.1038/s41598-020-66878-x
John, M. S., Nagoth, J. A., Ramasamy, K. P., Mancini, A., Giuli, G., Miceli, C., et al. (2022). Synthesis of bioactive silver nanoparticles using new bacterial strains from an Antarctic consortium. Mar. Drugs 20:558. doi: 10.3390/md20090558
John, M. S., Nagoth, J. A., Zannotti, M., Giovannetti, R., Mancini, A., Ramasamy, K. P., et al. (2021). Biogenic synthesis of copper nanoparticles using bacterial strains isolated from an Antarctic consortium associated to a psychrophilic marine ciliate: characterization and potential application as antimicrobial agents. Mar. Drugs 19:263. doi: 10.3390/md19050263
Karan, R., Capes, M. D., DasSarma, P., and DasSarma, S. (2013). Cloning, overexpression, purification, and characterization of a polyextremophilic β-galactosidase from the Antarctic haloarchaeon Halorubrum lacusprofundi. BMC Biotechnol. 13, 1–11. doi: 10.1186/1472-6750-13-3
Kim, E. J., Kim, J. E., Hwang, J. S., Kim, I. C., Lee, S. G., Kim, S., et al. (2019). Increased productivity and antifreeze activity of ice-binding protein from Flavobacterium frigoris PS1 produced using Escherichia coli as bioreactor. Appl. Biochem. Microbiol. 55, 489–494. doi: 10.1134/S0003683819050077
Kim, J., Ha, S., and Park, W. (2018). Expression and deletion analyses of cspE encoding cold-shock protein E in Acinetobacter oleivorans DR1. Res. Microbiol. 169, 244–253. doi: 10.1016/j.resmic.2018.04.011
Kim, M. J., Lee, Y. K., Lee, H. K., and Im, H. (2007). Characterization of cold-shock protein a of Antarctic Streptomyces sp. AA8321. Protein J. 26, 51–59. doi: 10.1007/s10930-006-9044-1
Kloska, A., Cech, G. M., Sadowska, M., Krause, K., Szalewska-Pałasz, A., and Olszewski, P. (2020). Adaptation of the marine bacterium Shewanella baltica to low temperature stress. Int. J. Mol. Sci. 21:4338. doi: 10.3390/ijms21124338
Kohli, I., Joshi, N. C., Mohapatra, S., and Varma, A. (2020). Extremophile–an adaptive strategy for extreme conditions and applications. Curr. Genomics 21, 96–110. doi: 10.2174/1389202921666200401105908
Koutsioulis, D., Wang, E., Tzanodaskalaki, M., Nikiforaki, D., Deli, A., Feller, G., et al. (2008). Directed evolution on the cold adapted properties of TAB5 alkaline phosphatase. Protein Eng. Des. Sel. 21, 319–327. doi: 10.1093/protein/gzn009
Králová, S. (2017). Role of fatty acids in cold adaptation of Antarctic psychrophilic Flavobacterium spp. Syst. Appl. Microbiol. 40, 329–333. doi: 10.1016/j.syapm.2017.06.001
Krucoń, T., Ruszkowska, Z., Pilecka, W., Szych, A., and Drewniak, Ł. (2023). Bioprospecting of the Antarctic Bacillus subtilis strain for potential application in leaching hydrocarbons and trace elements from contaminated environments based on functional and genomic analysis. Environ. Res. 227:115785. doi: 10.1016/j.envres.2023.115785
Kube, M., Chernikova, T. N., Al-Ramahi, Y., Beloqui, A., Lopez-Cortez, N., Guazzaroni, M. E., et al. (2013). Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica. Nat. Commun. 4:2156. doi: 10.1038/ncomms3156
Kuddus, M. (2015). Cold-active microbial enzymes. Biochem. Physiol. 4:e132. doi: 10.4172/2168-9652.1000e132
Kuddus, M. (2018). Cold-active enzymes in food biotechnology: an updated mini review. J. Appl. Biol. 6, 58–63. doi: 10.7324/jabb.2018.60310
Kuddus, M., Arif, J. M., and Ramteke, P. W. (2011). An overview of cold-active microbial a-amylase: adaptation strategies and biotechnological potentials. Biotechnology 10, 246–258. doi: 10.3923/biotech.2011.246.258
Kulakova, L., Galkin, A., Kurihara, T., Yoshimura, T., and Esaki, N. (1999). Cold-active serine alkaline protease from the psychrotrophic bacterium Shewanella strain Ac10: gene cloning and enzyme purification and characterization. Appl. Environ. Microbiol. 65, 611–617. doi: 10.1128/aem.65.2.611-617.1999.5
Kumar, A., Mukhia, S., and Kumar, R. (2021). Industrial applications of cold-adapted enzymes: challenges, innovations and future perspective. Biotech 11, 1–18. doi: 10.1007/s13205-021-02929-y
Kumar, L., and Bharadvaja, N. (2019). “Enzymatic bioremediation: a smart tool to fight environmental pollutants” in Smart Bioremediation Technologies (Cambridge: Academic Press), 99–118.
Lee, C. W., Yoo, W., Park, S. H., Le, L. T. H. L., Jeong, C. S., Ryu, B. H., et al. (2019). Structural and functional characterization of a novel cold-active S-formylglutathione hydrolase (SfSFGH) homolog from Shewanella frigidimarina, a psychrophilic bacterium. Microb. Cell Factories 18, 1–13. doi: 10.1186/s12934-019-1190-1
Lee, G. L. Y., Zakaria, N. N., Futamata, H., Suzuki, K., Zulkharnain, A., Shaharuddin, N. A., et al. (2022). Metabolic pathway of phenol degradation of a cold-adapted Antarctic Bacteria, Arthrobacter sp. Catalysts 12:1422. doi: 10.3390/catal12111422
Lee, J., Cho, J., Cho, Y. J., Cho, A., Woo, J., Lee, J., et al. (2019). The latitudinal gradient in rock-inhabiting bacterial community compositions in Victoria land, Antarctica. Sci. Total Environ. 657, 731–738. doi: 10.1016/j.scitotenv.2018.12.073
Lemak, S., Tchigvintsev, A., Petit, P., Flick, R., Singer, A. U., Brown, G., et al. (2012). Structure and activity of the cold-active and anion-activated carboxyl esterase OLEI01171 from the oil-degrading marine bacterium Oleispira antarctica. Biochem. J. 445, 193–203. doi: 10.1042/bj20112113
Liao, Y., Williams, T. J., Ye, J., Charlesworth, J., Burns, B. P., Poljak, A., et al. (2016). Morphological and proteomic analysis of biofilms from the Antarctic archaeon, Halorubrum lacusprofundi. Sci. Rep. 6, 1–17. doi: 10.1038/srep37454
Li, J., Xiao, X., Zhou, M., and Zhang, Y. (2023). Strategy for the adaptation to stressful conditions of the novel isolated conditional Piezophilic strain Halomonas titanicae ANRCS81. Appl. Environ. Microbiol. 89, e01304–e01322. doi: 10.1128/aem.01304-22
Li, Q., Yan, Y., Liu, X., Zhang, Z., Tian, J., and Wu, N. (2020). Enhancing thermostability of a psychrophilic alpha-amylase by the structural energy optimization in the trajectories of molecular dynamics simulations. Int. J. Biol. Macromol. 142, 624–633. doi: 10.1016/j.ijbiomac.2019.10.004
Liu, C., Wang, X., Wang, X., and Sun, C. (2016). Acclimation of Antarctic Chlamydomonas to the sea-ice environment: a transcriptomic analysis. Extremophiles 20, 437–450. doi: 10.1007/s00792-016-0834-x
Lo Giudice, A., Michaud, L., De Pascale, D., De Domenico, M., Di Prisco, G., Fani, R., et al. (2006). Lipolytic activity of Antarctic cold-adapted marine bacteria (Terra Nova Bay, Ross Sea). J. Appl. Microbiol. 101, 1039–1048. doi: 10.1111/j.1365-2672.2006.03006.x
Lo Giudice, A., Poli, A., Finore, I., and Rizzo, C. (2020). Peculiarities of extracellular polymeric substances produced by Antarctic bacteria and their possible applications. Appl. Microbiol. Biotechnol. 104, 2923–2934. doi: 10.1007/s00253-020-10448-8
Maeda, H., Hosokawa, M., Sashima, T., Murakami-Funayama, K., and Miyashita, K. (2009). Anti-obesity and anti-diabetic effects of fucoxanthin on diet-induced obesity conditions in a murine model. Mol. Med. Rep. 2, 897–902. doi: 10.3892/mmr_00000189
Malard, L. A., and Pearce, D. A. (2018). Microbial diversity and biogeography in Arctic soils. Environ. Microbiol. Rep. 10, 611–625. doi: 10.1111/1758-2229.12680
Mangiagalli, M., Bar-Dolev, M., Tedesco, P., Natalello, A., Kaleda, A., Brocca, S., et al. (2017). Cryo-protective effect of an ice-binding protein derived from Antarctic bacteria. FEBS J. 284, 163–177. doi: 10.1111/febs.13965
Mangiagalli, M., Lapi, M., Maione, S., Orlando, M., Brocca, S., Pesce, A., et al. (2021). The co-existence of cold activity and thermal stability in an Antarctic GH42 β-galactosidase relies on its hexameric quaternary arrangement. FEBS J. 288, 546–565. doi: 10.1111/febs.15354
Mangiagalli, M., and Lotti, M. (2021). Cold-active β-galactosidases: insight into cold adaptation mechanisms and biotechnological exploitation. Mar. Drugs 19:43. doi: 10.3390/md19010043
Marcoleta, A. E., Arros, P., Varas, M. A., Costa, J., Rojas-Salgado, J., Berríos-Pastén, C., et al. (2022). The highly diverse Antarctic peninsula soil microbiota as a source of novel resistance genes. Sci. Total Environ. 810:152003. doi: 10.1016/j.scitotenv.2021.152003
Marizcurrena, J. J., Herrera, L. M., Costábile, A., Morales, D., Villadóniga, C., Eizmendi, A., et al. (2019). Validating biochemical features at the genome level in the Antarctic bacterium Hymenobacter sp. strain UV11. FEMS Microbiology Letters 366. doi: 10.1093/femsle/fnz177
Martínez-Pérez, C., Greening, C., Bay, S. K., Lappan, R. J., Zhao, Z., De Corte, D., et al. (2022). Phylogenetically and functionally diverse microorganisms reside under the Ross ice shelf. Nat. Commun. 13:117. doi: 10.1038/s41467-021-27769-5
Martínez-Rosales, C., Fullana, N., Musto, H., and Castro-Sowinski, S. (2012). Antarctic DNA moving forward: genomic plasticity and biotechnological potential. FEMS Microbiol. Lett. 331, 1–9. doi: 10.1111/j.1574-6968.2012.02531.x
Médigue, C., Krin, E., Pascal, G., Barbe, V., Bernsel, A., Bertin, P. N., et al. (2005). Coping with cold: the genome of the versatile marine Antarctica bacterium Pseudoalteromonas haloplanktis TAC125. Genome Res. 15, 1325–1335. doi: 10.1101/gr.4126905
Merino, C., Kuzyakov, Y., Godoy, K., Jofré, I., Nájera, F., and Matus, F. (2021). Iron-reducing bacteria decompose lignin by electron transfer from soil organic matter. Sci. Total Environ. 761:143194. doi: 10.1016/j.scitotenv.2020.143194
Merlino, A., Krauss, I. R., Castellano, I., De Vendittis, E., Rossi, B., Conte, M., et al. (2010). Structure and flexibility in cold-adapted iron superoxide dismutases: the case of the enzyme isolated from Pseudoalteromonas haloplanktis. J. Struct. Biol. 172, 343–352. doi: 10.1016/j.jsb.2010.08.008
Millan, G. C. L., Veras, F. F., Stincone, P., Pailliè-Jiménez, M. E., and Brandelli, A. (2022). Biological activities of whey protein hydrolysate produced by protease from the Antarctic bacterium Lysobacter sp. A03. Biocatalysis and agricultural. Biotechnology 43:102415. doi: 10.1016/j.bcab.2022.102415
Miri, S., Naghdi, M., Rouissi, T., Kaur Brar, S., and Martel, R. (2019). Recent biotechnological advances in petroleum hydrocarbons degradation under cold climate conditions: a review. Crit. Rev. Environ. Sci. Technol. 49, 553–586. doi: 10.1080/10643389.2018.1552070
Miyazaki, K., Wintrode, P. L., Grayling, R. A., Rubingh, D. N., and Arnold, F. H. (2000). Directed evolution study of temperature adaptation in a psychrophilic enzyme. J. Mol. Biol. 297, 1015–1026. doi: 10.1006/jmbi.2000.3612
Morozova, O. V., Andreeva, I. S., Zhirakovskiy, V. Y., Pechurkina, N. I., Puchkova, L. I., Saranina, I. V., et al. (2022). Antibiotic resistance and cold-adaptive enzymes of Antarctic culturable bacteria from King George Island. Pol. Sci. 31:100756. doi: 10.1016/j.polar.2021.100756
Moyer, C. L., and Morita, R. Y. (2007). Psychrophiles and psychrotrophs. Encyclopedia of Life Science, 1. John Wiley & Sons, Ltd. Hoboken, NJ
Mozzicafreddo, M., Pucciarelli, S., Swart, E. C., Piersanti, A., Emmerich, C., Migliorelli, G., et al. (2021). The macronuclear genome of the Antarctic psychrophilic marine ciliate Euplotes focardii reveals new insights on molecular cold adaptation. Sci. Rep. 11:18782. doi: 10.1038/s41598-021-98168-5
Muller-Greven, J. C., Post, M. A., and Kubu, C. J. (2013). U.S. patent no. 8,486,665. Washington, DC: U.S. patent and trademark office
Murray, A. E., Lo, C. C., Daligault, H. E., Avalon, N. E., Read, R. W., Davenport, K. W., et al. (2021). Discovery of an Antarctic ascidian-associated uncultivated Verrucomicrobia with antimelanoma palmerolide biosynthetic potential. Msphere 6, e00759–e00721. doi: 10.1128/mSphere.00759-21
Nair, S., and Abraham, J. (2020). “Natural products from actinobacteria for drug discovery” in Advances in Pharmaceutical Biotechnology: Recent Progress and Future Applications. eds. J. K. Patra, A. C. Shukla, and G. Das (Singapore: Springer), 333–363.
Narinx, E., Davail, S., Feller, G., and Gerday, C. (1992). Nucleotide and derived amino acid sequence of the subtilisin from the Antarctic psychrotroph Bacillus TA39. Biochim. Biophys. Acta 1131, 111–113. doi: 10.1016/0167-4781(92)90108-c
Nikolaivits, E., Taxeidis, G., Gkountela, C., Vouyiouka, S., Maslak, V., Nikodinovic-Runic, J., et al. (2022). A polyesterase from the Antarctic bacterium Moraxella sp. degrades highly crystalline synthetic polymers. J. Hazard. Mater. 434:128900. doi: 10.1016/j.jhazmat.2022.128900
Nimkande, V. D., Sivanesan, S., and Bafana, A. (2023). Screening, identification, and characterization of lipase-producing halotolerant Bacillus altitudinis Ant19 from Antarctic soil. Arch. Microbiol. 205:113. doi: 10.1007/s00203-023-03453-8
Oikawa, T., Yamamoto, N., Shimoke, K., Uesato, S., Ikeuchi, T., and Fujioka, T. (2005). Purification, characterization, and overexpression of psychrophilic and thermolabile malate dehydrogenase of a novel Antarctic psychrotolerant, Flavobacterium frigidimaris KUC-1. Biosci. Biotechnol. Biochem. 69, 2146–2154. doi: 10.1271/bbb.69.2146
Orellana-Saez, M., Pacheco, N., Costa, J. I., Mendez, K. N., Miossec, M. J., Meneses, C., et al. (2019). In-Depth Genomic and Phenotypic Characterization of the Antarctic Psychrotolerant Strain Pseudomonas sp. MPC6 Reveals Unique Metabolic Features, Plasticity, and Biotechnological Potential. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.01154
Orlando, M., Pucciarelli, S., and Lotti, M. (2020). Endolysins from Antarctic Pseudomonas display lysozyme activity at low temperature. Mar. Drugs 18:579. doi: 10.3390/md18110579
Otur, Ç., Okay, S., and Kurt-Kızıldoğan, A. (2023). Whole genome analysis of Flavobacterium aziz-sancarii sp. nov., isolated from Ardley Island (Antarctica), revealed a rich resistome and bioremediation potential. Chemosphere 313:137511. doi: 10.1016/j.chemosphere.2022.137511
Pandey, N., Jain, R., Pandey, A., and Tamta, S. (2018). Optimisation and characterisation of the orange pigment produced by a cold adapted strain of Penicillium sp. (GBPI_P155) isolated from mountain ecosystem. Mycology 9, 81–92. doi: 10.1080/21501203.2017.1423127
Parra, L. P., Espina, G., Devia, J., Salazar, O., Andrews, B., and Asenjo, J. A. (2015). Identification of lipase encoding genes from Antarctic seawater bacteria using degenerate primers: expression of a cold-active lipase with high specific activity. Enzym. Microb. Technol. 68, 56–61. doi: 10.1016/j.enzmictec.2014.10.004
Parra, L. P., Reyes, F., Acevedo, J. P., Salazar, O., Andrews, B. A., and Asenjo, J. A. (2008). Cloning and fusion expression of a cold-active lipase from marine Antarctic origin. Enzym. Microb. Technol. 42, 371–377. doi: 10.1016/j.enzmictec.2007.11.003
Pasquet, V., Morisset, P., Ihammouine, S., Chepied, A., Aumailley, L., Berard, J. B., et al. (2011). Antiproliferative activity of violaxanthin isolated from bioguided fractionation of Dunaliella tertiolecta extracts. Mar. Drugs 9, 819–831. doi: 10.3390/md9050819
Paun, V. I., Banciu, R. M., Lavin, P., Vasilescu, A., Fanjul-Bolado, P., and Purcarea, C. (2022). Antarctic aldehyde dehydrogenase from Flavobacterium PL002 as a potent catalyst for acetaldehyde determination in wine. Sci. Rep. 12:17301. doi: 10.1038/s41598-022-22289-8
Pereira, J. Q., Ambrosini, A., Passaglia, L. M. P., and Brandelli, A. (2017). A new cold-adapted serine peptidase from Antarctic Lysobacter sp. A03: insights about enzyme activity at low temperatures. Int. J. Biol. Macromol. 103, 854–862. doi: 10.1016/j.ijbiomac.2017.05.142
Petratos, K., Gessmann, R., Daskalakis, V., Papadovasilaki, M., Papanikolau, Y., Tsigos, I., et al. (2020). Structure and dynamics of a thermostable alcohol dehydrogenase from the Antarctic Psychrophile Moraxella sp. TAE123. ACS. Omega 5, 14523–14534. doi: 10.1021/acsomega.0c01210
Piette, F., D’Amico, S., Mazzucchelli, G., Danchin, A., Leprince, P., and Feller, G. (2011). Life in the cold: a proteomic study of cold-repressed proteins in the Antarctic bacterium Pseudoalteromonas haloplanktis TAC125. Appl. Environ. Microbiol. 77, 3881–3883. doi: 10.1128/aem.02757-10
Plaza, D. O., Gallardo, C., Straub, Y. D., Bravo, D., and Pérez-Donoso, J. M. (2016). Biological synthesis of fluorescent nanoparticles by cadmium and tellurite resistant Antarctic bacteria: exploring novel natural nanofactories. Microb. Cell Factories 15, 1–11. doi: 10.1186/s12934-016-0477-8
Prayogo, F. A., Budiharjo, A., Kusumaningrum, H. P., Wijanarka, W., Suprihadi, A., and Nurhayati, N. (2020). Metagenomic applications in exploration and development of novel enzymes from nature: a review. J. Genet. Eng. Biotechnol. 18, 1–10. doi: 10.1186/s43141-020-00043-9
Pucciarelli, S., Devaraj, R. R., Mancini, A., Ballarini, P., Castelli, M., Schrallhammer, M., et al. (2015). Microbial consortium associated with the Antarctic marine ciliate Euplotes focardii: an investigation from genomic sequences. Microb. Ecol. 70, 484–497. doi: 10.1007/s00248-015-0568-9
Ramasamy, K. P., Telatin, A., Mozzicafreddo, M., Miceli, C., and Pucciarelli, S. (2019). Draft genome sequence of a new Pseudomonas sp. strain, ef1, associated with the psychrophilic Antarctic ciliate Euplotes focardii. Microbiol. Resour. Announc. 8, e00867–e00819. doi: 10.1128/mra.00867-19
Ramya, L. N., and Pulicherla, K. K. (2015). Molecular insights into cold active polygalacturonase enzyme for its potential application in food processing. J. Food Sci. Technol. 52, 5484–5496. doi: 10.1007/s13197-014-1654-6
Ray, M. K., Sitaramamma, T., Ghandhi, S., and Shivaji, S. (1994). Occurrence and expression of cspA, a cold shock gene, in Antarctic psychrotropic bacteria. FEMS Microbiol. Lett. 116, 55–60. doi: 10.1111/j.1574-6968.1994.tb06675.x
Raymond, J. A., Fritsen, C., and Shen, K. (2007). An ice-binding protein from an Antarctic Sea ice bacterium. FEMS Microbiol. Ecol. 61, 214–221. doi: 10.1111/j.1574-6941.2007.00345.x
Rentier-Delrue, F., Mande, S. C., Moyens, S., Terpstra, P., Mainfroid, V., Goraj, K., et al. (1993). Cloning and overexpression of the triosephosphate isomerase genes from psychrophilic and thermophilic bacteria: structural comparison of the predicted protein sequences. J. Mol. Biol. 229, 85–93. doi: 10.1006/jmbi.1993.1010
Riccardi, C., Calvanese, M., Ghini, V., Alonso-Vásquez, T., Perrin, E., Turano, P., et al. (2023). Metabolic robustness to growth temperature of a cold-adapted marine bacterium. Msystems 8, e01124–e01122. doi: 10.1128/msystems.01124-22
Riccardi, C., D’Angelo, C., Calvanese, M., Ricciardelli, A., Tutino, M. L., Parrilli, E., et al. (2022). Genome analysis of a new biosurfactants source: the Antarctic bacterium Psychrobacter sp. TAE2020. Mar. Genomics 61:100922. doi: 10.1016/j.margen.2021.100922
Ricciardelli, A., Casillo, A., Vergara, A., Balasco, N., Corsaro, M. M., Tutino, M. L., et al. (2019). Environmental conditions shape the biofilm of the Antarctic bacterium Pseudoalteromonas haloplanktis TAC125. Microbiol. Res. 218, 66–75. doi: 10.1016/j.micres.2018.09.010
Rina, M., Pozidis, C., Mavromatis, K., Tzanodaskalaki, M., Kokkinidis, M., and Bouriotis, V. (2000). Alkaline phosphatase from the Antarctic strain TAB5: properties and psychrophilic adaptations. Eur. J. Biochem. 267, 1230–1238. doi: 10.1046/j.1432-1327.2000.01127.x
Sajjad, W., Din, G., Rafiq, M., Iqbal, A., Khan, S., Zada, S., et al. (2020). Pigment production by cold-adapted bacteria and fungi: colorful tale of cryosphere with wide range applications. Extremophiles 24, 447–473. doi: 10.1007/s00792-020-01180-2
Sarmiento, F., Peralta, R., and Blamey, J. M. (2015). Cold and hot extremozymes: industrial relevance and current trends. Front. Bioeng. Biotechnol. 3:148. doi: 10.3389/fbioe.2015.00148
See Too, W. C., and Few, L. L. (2010). Cloning of triose phosphate isomerase gene from an Antarctic psychrophilic Pseudomonas sp. by degenerate and splinkerette PCR. World J. Microbiol. Biotechnol. 26, 1251–1259. doi: 10.1007/s11274-009-0295-9
Sheridan, P. P., and Brenchley, J. E. (2000). Characterization of a salt-tolerant family 42 β-galactosidase from a psychrophilic Antarctic Planococcus isolate. Appl. Environ. Microbiol. 66, 2438–2444. doi: 10.1128/AEM.66.6.2438-2444.2000
Silva, T. R. E., Silva, L. C. F., de Queiroz, A. C., Alexandre Moreira, M. S., de Carvalho Fraga, C. A., de Menezes, G. C. A., et al. (2021). Pigments from Antarctic bacteria and their biotechnological applications. Crit. Rev. Biotechnol. 41, 809–826. doi: 10.1080/07388551.2021.1888068
Silva, T. R., Tavares, R. S., Canela-Garayoa, R., Eras, J., Rodrigues, M. V., Neri-Numa, I. A., et al. (2019). Chemical characterization and biotechnological applicability of pigments isolated from Antarctic bacteria. Mar. Biotechnol. 21, 416–429. doi: 10.1007/s10126-019-09892-z
Sivasankar, P., Poongodi, S., Sivakumar, K., Al-Qahtani, W. H., Arokiyaraj, S., and Jothiramalingam, R. (2022). Exogenous production of cold-active cellulase from polar Nocardiopsis sp. with increased cellulose hydrolysis efficiency. Arch. Microbiol. 204:218. doi: 10.1007/s00203-022-02830-z
Solar Venero, E. C., Matera, G., Vogel, J., López, N. I., and Tribelli, P. M. (2022). Small RNAs in the Antarctic bacterium Pseudomonas extremaustralis responsive to oxygen availability and oxidative stress. Environ. Microbiol. Rep. 14, 604–615. doi: 10.1111/1758-2229.13084
Song, W., Lin, X., and Huang, X. (2012). Characterization and expression analysis of three cold shock protein (CSP) genes under different stress conditions in the Antarctic bacterium Psychrobacter sp. G. Polar Biology 35, 1515–1524. doi: 10.1007/s00300-012-1191-6
Soontornchaiboon, W., Joo, S. S., and Kim, S. M. (2012). Anti-inflammatory effects of violaxanthin isolated from microalga Chlorella ellipsoidea in RAW 264.7 macrophages. Biol. Pharm. Bull. 35, 1137–1144. doi: 10.1248/bpb.b12-00187
Stokes, C. R., Abram, N. J., Bentley, M. J., Edwards, T. L., England, M. H., Foppert, A., et al. (2022). Response of the East Antarctic ice sheet to past and future climate change. Nature 608, 275–286. doi: 10.1038/s41586-022-04946-0
Styczynski, M., Rogowska, A., Gieczewska, K., Garstka, M., Szakiel, A., and Dziewit, L. (2020). Genome-based insights into the production of carotenoids by Antarctic bacteria, Planococcus sp. ANT_H30 and Rhodococcus sp. ANT_H53B. Molecules 25:4357. doi: 10.3390/molecules25194357
Styczynski, M., Rogowska, A., Nyabayo, C., Decewicz, P., Romaniuk, F., Pączkowski, C., et al. (2022). Heterologous production and characterization of a pyomelanin of Antarctic Pseudomonas sp. ANT_H4: a metabolite protecting against UV and free radicals, interacting with iron from minerals and exhibiting priming properties toward plant hairy roots. Microb. Cell Factories 21, 1–17. doi: 10.1186/s12934-022-01990-3
Sutherland, B. J., Finke, J. F., Saunders, R., Warne, S., Schulze, A. D., Strohm, J. H., et al. (2022). Metatranscriptomics reveals a shift in microbial community composition and function during summer months in a coastal marine environment. Environ. DNA. doi: 10.1002/edn3.353
Sutthiwong, N., Fouillaud, M., Valla, A., Caro, Y., and Dufossé, L. (2014). Bacteria belonging to the extremely versatile genus Arthrobacter as novel source of natural pigments with extended hue range. Food Res. Int. 65, 156–162. doi: 10.1016/j.foodres.2014.06.024
Takara-Clontech. Available at: https://www.takarabio.com/products/cloning/modifying-enzymes/nucleases/cryonasecold-active-nuclease
Tang, Y., Wu, P., Jiang, S., Selvaraj, J. N., Yang, S., and Zhang, G. (2019). A new cold-active and alkaline pectate lyase from Antarctic bacterium with high catalytic efficiency. Appl. Microbiol. Biotechnol. 103, 5231–5241. doi: 10.1007/s00253-019-09803-1
Tedesco, P., Maida, I., Palma Esposito, F., Tortorella, E., Subko, K., Ezeofor, C. C., et al. (2016). Antimicrobial activity of monoramnholipids produced by bacterial strains isolated from the Ross Sea (Antarctica). Mar. Drugs 14:83. doi: 10.3390/md14050083
Tengku-Mazuki, T. A., Subramaniam, K., Zakaria, N. N., Convey, P., Abdul Khalil, K., Lee, G. L. Y., et al. (2020). Optimization of phenol degradation by Antarctic bacterium Rhodococcus sp. Antarctic Science 32, 486–495. doi: 10.1017/s0954102020000358
Teoh, C. P., Lavin, P., Lee, D. J. H., González-Aravena, M., Najimudin, N., Lee, P. C., et al. (2021). Genomics and transcriptomics analyses provide insights into the cold adaptation strategies of an Antarctic bacterium, Cryobacterium sp. SO1. Polar. Biol. 44, 1305–1319. doi: 10.1007/s00300-021-02883-8
Terashima, M., Ohashi, K., Takasuka, T. E., Kojima, H., and Fukui, M. (2019). Antarctic heterotrophic bacterium Hymenobacter nivis P3T displays light-enhanced growth and expresses putative photoactive proteins. Environ. Microbiol. Rep. 11, 227–235. doi: 10.1111/1758-2229.12702
Tribelli, P. M., Pezzoni, M., Brito, M. G., Montesinos, N. V., Costa, C. S., and López, N. I. (2020). Response to lethal UVA radiation in the Antarctic bacterium Pseudomonas extremaustralis: polyhydroxybutyrate and cold adaptation as protective factors. Extremophiles 24, 265–275. doi: 10.1007/s00792-019-01152-1
Tsigos, I., Velonia, K., Smonou, I., and Bouriotis, V. (1998). Purification and characterization of an alcohol dehydrogenase from the Antarctic psychrophile Moraxella sp. TAE123. Eur. J. Biochem. 254, 356–362. doi: 10.1046/j.1432-1327.1998.2540356.x
Tsuruta, H., Mikami, B., Higashi, T., and Aizono, Y. (2010). Crystal structure of cold-active alkaline phosphatase from the psychrophile Shewanella sp. Biosci. Biotechnol. Biochem. 74, 69–74. doi: 10.1271/bbb.90563
Turkiewicz, M., Gromek, E., Kalinowska, H., and Zielińska, M. (1999). Biosynthesis and properties of an extracellular metalloprotease from the Antarctic marine bacterium Sphingomonas paucimobilis. J. Biotechnol. 70, 53–60. doi: 10.1016/s0168-1656(99)00057-7
Turkiewicz, M., Kur, J., Białkowska, A., Cieśliński, H., Kalinowska, H., and Bielecki, S. (2003). Antarctic marine bacterium Pseudoalteromonas sp. 22b as a source of cold-adapted β-galactosidase. Biomol. Eng. 20, 317–324. doi: 10.1016/s1389-0344(03)00039-x
Uma, S., Jadhav, R. S., Seshu Kumar, G., Shivaji, S., and Ray, M. K. (1999). A RNA polymerase with transcriptional activity at 0 °C from the Antarctic bacterium Pseudomonas syringae. FEBS Lett. 453, 313–317. doi: 10.1016/s0014-5793(99)00660-2
Van Truong, L., Tuyen, H., Helmke, E., Binh, L. T., and Schweder, T. (2001). Cloning of two pectate lyase genes from the marine Antarctic bacterium Pseudoalteromonas haloplanktis strain ANT/505 and characterization of the enzymes. Extremophiles 5, 35–44. doi: 10.1007/s007920000170
Vasquez, Y. M. S. C., Gomes, M. B., E Silva, T. R., Duarte, A. W. F., Rosa, L. H., and de Oliveira, V. M. (2021). Cold-adapted chitinases from Antarctic bacteria: taxonomic assessment and enzyme production optimization. Biocatal. Agric. Biotechnol. 34:102029. doi: 10.1016/j.bcab.2021.102029
Venil, C. K., Zakaria, Z. A., and Ahmad, W. A. (2013). Bacterial pigments and their applications. Process Biochem. 48, 1065–1079. doi: 10.1016/j.procbio.2013.06.006
Violot, S., Aghajari, N., Czjzek, M., Feller, G., Sonan, G. K., Gouet, P., et al. (2005). Structure of a full length psychrophilic cellulase from Pseudoalteromonas haloplanktis revealed by X-ray diffraction and small angle X-ray scattering. J. Mol. Biol. 348, 1211–1224. doi: 10.1016/j.jmb.2005.03.026
Wang, J., Ma, Y., Yang, J., Jin, L., Gao, Z., Xue, L., et al. (2019). Fucoxanthin inhibits tumour-related lymphangiogenesis and growth of breast cancer. J. Cell. Mol. Med. 23, 2219–2229. doi: 10.1111/jcmm.14151
Wang, Q. F., Hou, Y. H., Xu, Z., Miao, J. L., and Li, G. Y. (2008). Purification and properties of an extracellular cold-active protease from the psychrophilic bacterium Pseudoalteromonas sp. NJ276. Biochem. Eng. J. 38, 362–368. doi: 10.1016/j.bej.2007.07.025
Wang, Q. F., Miao, J. L., Hou, Y. H., Ding, Y., Wang, G. D., and Li, G. Y. (2005). Purification and characterization of an extracellular cold-active serine protease from the psychrophilic bacterium Colwellia sp. NJ341. Biotechnol. Lett. 27, 1195–1198. doi: 10.1007/s10529-005-0016-x
Wang, Q., Nie, P., Hou, Y., and Wang, Y. (2020). Purification, biochemical characterization and DNA protection against oxidative damage of a novel recombinant superoxide dismutase from psychrophilic bacterium Halomonas sp. ANT108. Protein Expr. Purif. 173:105661. doi: 10.1016/j.pep.2020.105661
Wang, W., Mu, H., Ren, X., Ouyang, Q., and Li, J. (2023). Genome-based classification of Pedobacter polysacchareus sp. nov., isolated from Antarctic soil producing exopolysaccharide. FEMS Microbiol. Lett. 370, fnad031. doi: 10.1093/femsle/fnad031
Wang, W., Sun, M., Liu, W., and Zhang, B. (2008). Purification and characterization of a psychrophilic catalase from Antarctic Bacillus. Can. J. Microbiol. 54, 823–828. doi: 10.1139/w08-066
Wang, X., He, Y., Deng, Y., Zuo, Z., Li, D., Chen, F., et al. (2022). A diguanylate cyclase regulates biofilm formation in Rhodococcus sp. NJ-530 from Antarctica. Biotech 12, 1–10. doi: 10.1007/s13205-021-03093-z
Wang, X., Kan, G., Ren, X., Yu, G., Shi, C., Xie, Q., et al. (2018a). Molecular cloning and characterization of a novel α-amylase from Antarctic Sea ice bacterium Pseudoalteromonas sp. M175 and its primary application in detergent. Biomed. Res. Int. 2018:3258383. doi: 10.1155/2018/3258383
Wang, Y., Han, H., Cui, B., Hou, Y., Wang, Y., and Wang, Q. (2017). A glutathione peroxidase from Antarctic psychrotrophic bacterium Pseudoalteromonas sp. ANT506: cloning and heterologous expression of the gene and characterization of recombinant enzyme. Bioengineered 8, 742–749. doi: 10.1080/21655979.2017.1373534
Wang, Y., Hou, Y., Nie, P., Wang, Y., Ren, X., Wei, Q., et al. (2019). A novel cold-adapted and salt-tolerant RNase R from Antarctic Sea-ice bacterium Psychrobacter sp. Ant206. Molecules 24:2229. doi: 10.3390/molecules24122229
Wang, Y., Hou, Y., Wang, Y., Zheng, L., Xu, X., Pan, K., et al. (2018b). A novel cold-adapted leucine dehydrogenase from Antarctic Sea-ice bacterium Pseudoalteromonas sp. ANT178. Mar. Drugs 16:359. doi: 10.3390/md16100359
Wang, Y., Ma, L., He, J., Liu, Z., Weng, S., Wang, L., et al. (2021). Whole genome sequencing and comparative genomic analyses of Planococcus alpniumensis MSAK28401T, a new species isolated from Antarctic krill. BMC Microbiol. 21, 1–15. doi: 10.1186/s12866-021-02347-3
Waschulin, V., Borsetto, C., James, R., Newsham, K. K., Donadio, S., Corre, C., et al. (2022). Biosynthetic potential of uncultured Antarctic soil bacteria revealed through long read metagenomic sequencing. ISME J. 16, 101–111. doi: 10.1038/s41396-021-01052-3
Williams, T. J., Allen, M. A., Ivanova, N., Huntemann, M., Haque, S., Hancock, A. M., et al. (2021). Genome analysis of a verrucomicrobial endosymbiont with a tiny genome discovered in an Antarctic Lake. Front. Microbiol. 12:674758. doi: 10.3389/fmicb.2021.674758
Williams, T. J., Allen, M. A., Panwar, P., and Cavicchioli, R. (2022). Into the darkness: the ecologies of novel ‘microbial dark matter’phyla in an Antarctic Lake. Environ. Microbiol. 24, 2576–2603. doi: 10.1111/1462-2920.16026
Xiao, Y., Yan, F., Cui, Y., Du, J., Hu, G., Zhai, W., et al. (2023). A symbiotic bacterium of Antarctic fish reveals environmental adaptability mechanisms and biosynthetic potential towards antibacterial and cytotoxic activities. Front. Microbiol. 13:1085063. doi: 10.3389/fmicb.2022.1085063
Xuezheng, L., Shuoshuo, C., Guoying, X., Shuai, W., Ning, D., and Jihong, S. (2010). Cloning and heterologous expression of two cold-active lipases from the Antarctic bacterium Psychrobacter sp. G. Polar Res. 29, 421–429. doi: 10.1111/j.1751-8369.2010.00189.x
Yang, X., Lin, X., Fan, T., Bian, J., and Huang, X. (2008). Cloning and expression of lipP, a gene encoding a cold-adapted lipase from Moritella sp. 2-5-10-1. Curr. Microbiol. 56, 194–198. doi: 10.1007/s00284-007-9051-2
Yoshimune, K., Galkin, A., Kulakova, L., Yoshimura, T., and Esaki, N. (2005). Cold-active DnaK of an Antarctic psychrotroph Shewanella sp. Ac10 supporting the growth of dnaK-null mutant of Escherichia coli at cold temperatures. Extremophiles 9, 145–150. doi: 10.1007/s00792-004-0429-9
Yusof, N. A., Hashim, N. H. F., and Bharudin, I. (2021). Cold adaptation strategies and the potential of psychrophilic enzymes from the Antarctic yeast, Glaciozyma antarctica PI12. J. Fungi 7:528. doi: 10.3390/jof7070528
Zhang, A., Hou, Y., Wang, Q., and Wang, Y. (2022). Characteristics and polyethylene biodegradation function of a novel cold-adapted bacterial laccase from Antarctic Sea ice psychrophile Psychrobacter sp. NJ228. J. Hazard. Mater. 439:129656. doi: 10.1016/j.jhazmat.2022.129656
Zhang, J., Lin, S., and Zeng, R. (2007). Cloning, expression, and characterization of a cold-adapted lipase gene from an Antarctic deep-sea psychrotrophic bacterium, Psychrobacter sp. 7195. J. Microbiol. Biotechnol. 17, 604–610.
Keywords: Antarctic bacteria, climate change, psychrophiles, omics, machine learning, biotechnological applications
Citation: Ramasamy KP, Mahawar L, Rajasabapathy R, Rajeshwari K, Miceli C and Pucciarelli S (2023) Comprehensive insights on environmental adaptation strategies in Antarctic bacteria and biotechnological applications of cold adapted molecules. Front. Microbiol. 14:1197797. doi: 10.3389/fmicb.2023.1197797
Edited by:
Prashant Kumar Singh, Mizoram University, IndiaReviewed by:
Alysson Wagner Fernandes Duarte, Federal University of Alagoas, BrazilGarvita Singh, University of Delhi, India
Copyright © 2023 Ramasamy, Mahawar, Rajasabapathy, Rajeshwari, Miceli and Pucciarelli. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kesava Priyan Ramasamy, a2VzYXZhLnJhbWFzYW15QHVtdS5zZQ==
 Kesava Priyan Ramasamy
Kesava Priyan Ramasamy Lovely Mahawar
Lovely Mahawar Raju Rajasabapathy
Raju Rajasabapathy Kottilil Rajeshwari4
Kottilil Rajeshwari4 Cristina Miceli
Cristina Miceli Sandra Pucciarelli
Sandra Pucciarelli