- 1Clinical Veterinary Laboratory, Institute of Animal Science and Technology, Ningxia University, Yinchuan, China
- 2Ningxia Xiaoming Agriculture and Animal Husbandry Co., Ltd., Yinchuan, China
- 3Ningxia Academy of Agriculture and Forestry Sciences, Yinchuan, China
Mycoplasma gallisepticum (MG) is one of the major causative agents of chronic respiratory diseases in poultry. The biofilms of MG are highly correlated to its chronic infection. However data on genes involved in biofilm formation ability are still scarse. MG strains with distinct biofilm intensity were screened by crystal violet staining morphotyped and characterized for the drug sensitivity. Two MG strains NX-01 and NX-02 showed contrasted ability to biofilm formation. The biofilm formation ability of NX-01 strain was significantly higher than that of NX-02 strain (p < 0.01). The drug sensitivity test showed that the stronger the ability of MG stain to form biofilms, the weaker its sensitivity to 17 antibiotic drugs. Moreover, putative key genes related to biofilm formation were screened by genome-wide analysis. A total of 13 genes and proteins related to biofilm formation, including ManB, oppA, oppD, PDH, eno, RelA, msbA, deoA, gapA, rpoS, Adhesin P1 precursor, S-adenosine methionine synthetase, and methionyl tRNA synthetase were identified. There were five major discrepancies between the two isolated MG strains and the five NCBI-published MG strains. These findings provide potential targets for inhibiting the formation of biofilm of MG, and lay a foundation for treating chronic infection.
1. Introduction
Mycoplasma gallisepticum (MG) is one of the important pathogens affecting poultry (Ghanem et al., 2017). It is widely distributed in all poultry countries worldwide, causing serious economic losses to the global poultry industry (Roussan et al., 2015; Feberwee et al., 2021; Marouf et al., 2022). MG mainly causes chronic respiratory diseases in chickens and other poultry and infectious sinusitis in turkeys (Sulyok et al., 2019). In addition, the infection of MG can cause low carcass grading and feed conversion efficiency of broiler chickens. With the increase of the weak brood rate, the egg production and egg hatching rate of laying hens tend to decrease (Beaudet et al., 2019).
Many therapeutic drugs have been developed for MG infection, such as macrolides, quinolones, tylomycin, and gentamicin (McAuliffe et al., 2006; Abd El-Hamid et al., 2019). However, the drug resistance of MG is becoming increasingly severe (Beylefeld et al., 2018), and biofilms may be one of the main causes. Bacterial biofilm plays an important role in the bacterial disease process, allowing bacteria to evade the host’s immune defenses, inducing drug resistance and increasing toxin accumulation (Sorci, 2013). The formation of biofilm poses significant clinical challenges and exacerbates the issues of drug resistance and bacterial virulence (Tassew et al., 2017; Feng et al., 2021). Studies have shown that the development of a biofilm by MG could contribute to persistent infections, rendering the eradication of MG challenging (Feng et al., 2020). The complete characterization of MG biofilm formation is key in assessing and combating resistance to infectious poultry diseases, particularly in commercial farming (Taylor-Robinson and Bébéar, 1997). Despite the wealth of information and corresponding treatment modalities available for biofilm formation in bacteria and other mycoplasma (Stewart, 2002; Sharma et al., 2019), there is a lack of research on MG biofilm.
Mycoplasma gallisepticum is a prokaryote intermediate between bacteria and viruses and lacks a cell wall (Ishfaq et al., 2020). A few key genes regulate the formation and attachment of bacterial biofilms to surfaces (Parker et al., 2009; Liu et al., 2022). For example, the formation of Streptococcus gordonii biofilm depends on arginine, which regulates the development of Streptococcus gordonii biofilm mainly by activating the regulatory pathway of the ArcR gene (Robinson et al., 2018). In addition, the ssrS gene of Haemophilus parasuis is associated with the antitoxin system, regulating biofilm formation and cell persistence (Jiang et al., 2021). Several genes that have a crucial role in the biofilm formation of various strains have been well-documented. However, there is a significant paucity of research on MG. So far, only a few articles have shown that ManB, VlhA, PDH, ABC transporter penetrase, ABC transporter ATP-binding protein, and other genes may be involved in forming MG biofilm (Chen et al., 2012; Wang et al., 2017). It is very important to study the regulatory genes of biofilm formation for controlling MG chronic infection. Genomic analyses can help elucidate genes functionalities in MG as recently documented (Semashko et al., 2022). Such approach could help to elucidate putative genes involved in biofilm formation ability and drug resistance.
The objective of this study was to screen MG strains with contrasted ability in terms of biofilm formation in Ningxia Hui Autonomous Region in China and study their drug sensitivity. It also aimed to gaining insights in the putative genes involved in biofilm formation ability using a genome-wide analysis on both strains having contrasted ability to biofilm formation.
2. Materials and methods
2.1. Main materials
The improved Frey culture medium was obtained from China Sea Biotechnology Co., LTD., Beijing, China. The bacterial genome DNA extraction kit was purchased from Tiangen Biochemical Technology Co., LTD., Beijing, China. Crystal violet was acquired from Chongming County Yuxi Reagent Factory. Ethanol (95%) and methanol were purchased from Guangnuo Chemical Technology Co., LTD., Shanghai, China. Glutaraldehyde fixative (2.5%, for electron microscopy) was from Proanti Biotechnology Development Co., LTD., Shaanxi, China. Ofloxacin, Ennofloxacin, Norfloxacin, Taymycin, Erythromycin, Talomycin, Temicoxacin, Spiramycin, Tetracycline Hydrochloride, Lincomycin, Gentamicin, Dakanamycin, Kanamycin, Streptomycin, Doxycycline, and Warnemylin were purchased from Golden Clonix Biotechnology Co., LTD., Beijing, China.
2.2. Bacterial strain isolation
Ethics approval and specific permission were not required for the study, as all samples were collected by the authors during necropsies with the consent of the owners. Air sacs of chickens suspected to be infected with M. gallisepticum were collected from poultry farms in Ningxia (Felice et al., 2020; Yadav et al., 2022). They were cut and placed in modified Frey’s liquid medium overnight at 4°C under sterile conditions. After filtration, they were inoculated into Frey liquid medium and incubated in a biochemical incubator at 37°C. The MG were purified by Kahya et al. (2010) and Sprygin et al. (2010), the harvested isolates were coated in the modified Frey’s solid medium and placed in a 5% CO2 incubator at 37°C for 3–7 days. Single colonies were selected and cultured in the modified Frey’s liquid medium, and the purified isolates were repeated three times to get the purified isolates. The colony morphology on the solid medium was observed with an inverted microscope.
Bacterial genomic DNA was extracted using the kit of Tiangen Biochemical Technology Company (Ahmad et al., 2020; Li L. et al., 2020). After the expanded culture, put 1 mL of fresh bacterial solution into a centrifuge tube and add 20 μL of protease K solution. Add 220 μL anhydrous ethanol and mix thoroughly. Then, the solution and precipitate are added to the adsorption Add 220 μL anhydrous ethanol and mix thoroughly. Then the solution and precipitate are added to the adsorption column, and 500 μL buffer GD is added to the column. Discard the waste liquid after centrifugation. Add 600 μL PW to the adsorption column, and the waste liquid was discarded after centrifugation. Add 50 μL eluent buffer TE in the middle of the adsorbed strain and let it stand at room temperature for 5 min. The solution was collected into the centrifuge tube to obtain the genome extract of MG strains. Meanwhile the isolated MG strain was amplified by PCR based on the OIE 16S rRNA and mgc2 primers to identify the MG strain in the bacterial solution (Table 1), nuclease-free water was used as a negative control in all PCR assays. Finally, the bacterial solution was mixed at a ratio of 1:1 to glycerol and stored at −80°C.
2.3. Determination of biofilm formation capacity
Modified tests using crystal violet staining were used to quantify biofilm formation (Hu et al., 2010). The suspension concentration of the prepared MG strains was adjusted to 1 × 107 CCU/mL, and 200 μL was inoculated on sterile 96-cell plates. Six replicates of the bacterial culture were prepared and subsequently incubated at 37°C for 12, 24, 36, and 48 h. Meanwhile, the crystal violet staining method was employed to measure the OD590 value of MG biofilm by enzyme marker, and the absorbance results were recorded. According to ODc value (ODc is equal to the average OD value of blank well), biofilm classification: OD ≤ ODc, no membrane capacity (−); ODc < OD ≤ 2ODc, weak biofilm forming ability (+); 2ODc < OD ≤ 4ODc, medium biofilm forming ability (++); and 4ODc < OD, strong biofilm forming ability (+++).
2.4. Determination of matrix growth curves of biofilm models in vitro
The growth curves of the biofilm matrix were measured by a semi-quantitative adhesion test (Liang et al., 2019). 200 μL of 1 × 106 CCU/mL bacterial suspension was inoculated on 96-cell plates with six replicates per strain, after which the plates were then incubated at 37°C for 0, 3, 6, 9, 12, 24, 36, 48, and 72 h. In the corresponding period time, crystal violet staining was used to detect the OD590 value of two MG biofilms. According to the absorbance results, the biofilm matrix growth curve was drawn.
2.5. Observation of biofilm morphology by scanning electron microscopy
Mycoplasma gallisepticum strains were inoculated into the liquid Frey medium for 12 h, and the suspension concentration was adjusted to 1 × 104 CCU/mL. The sterile six-well plate was used to hold the 14 mm round slip-plate, with 5 mL of bacterial suspension added to each well. The culture was incubated at 37°C for 24 h, and 1 mL 2.5% glutaraldehyde fixed solution (EM Grade) was added to fixed at 4°C overnight. The gradient elution was performed on each well was gradient eluted with ethanol of different concentration (30, 50, 70, 80, 90, and 95%). The obtained sample was sliced and photographed using an ISI-SX-40 scanning electron microscope (Shatila et al., 2021).
2.6. Drug sensitivity test
The drug sensitivity of the isolated stains were determined by Hannan’s microdilution method (Hannan, 2000). The suspension concentration of MG strains was adjusted to 1 × 104 CCU/mL. 40 μL 512 μg/mL of antibiotics were added to the first hole of the 96-well plate, and add 20 μL sterile PBS from well 2 to 12. Then 20 μL of liquid was discarded from well 1 to 2 and mixed, and the same operation was performed until well 12. Thus, the antibiotics were continuously diluted at multiple ratios. Finally, 180 μL diluted MG bacterial solution was added to well 1–12. Growth control wells (containing culture medium and bacterial solution) and negative control Wells (containing liquid culture medium) were also set up and each antibiotic were repeated three times, After being inoculated, the 96-well plates were transferred to a constant temperature incubator set at 37°C for a week. The color of the growth control well was yellow, and the negative control well was pink. The highest concentration of the drug with no color change in the test well was considered the lowest inhibitory concentration of the drug (Gautier-Bouchardon, 2018).
2.7. Genome-wide sequencing and quality control
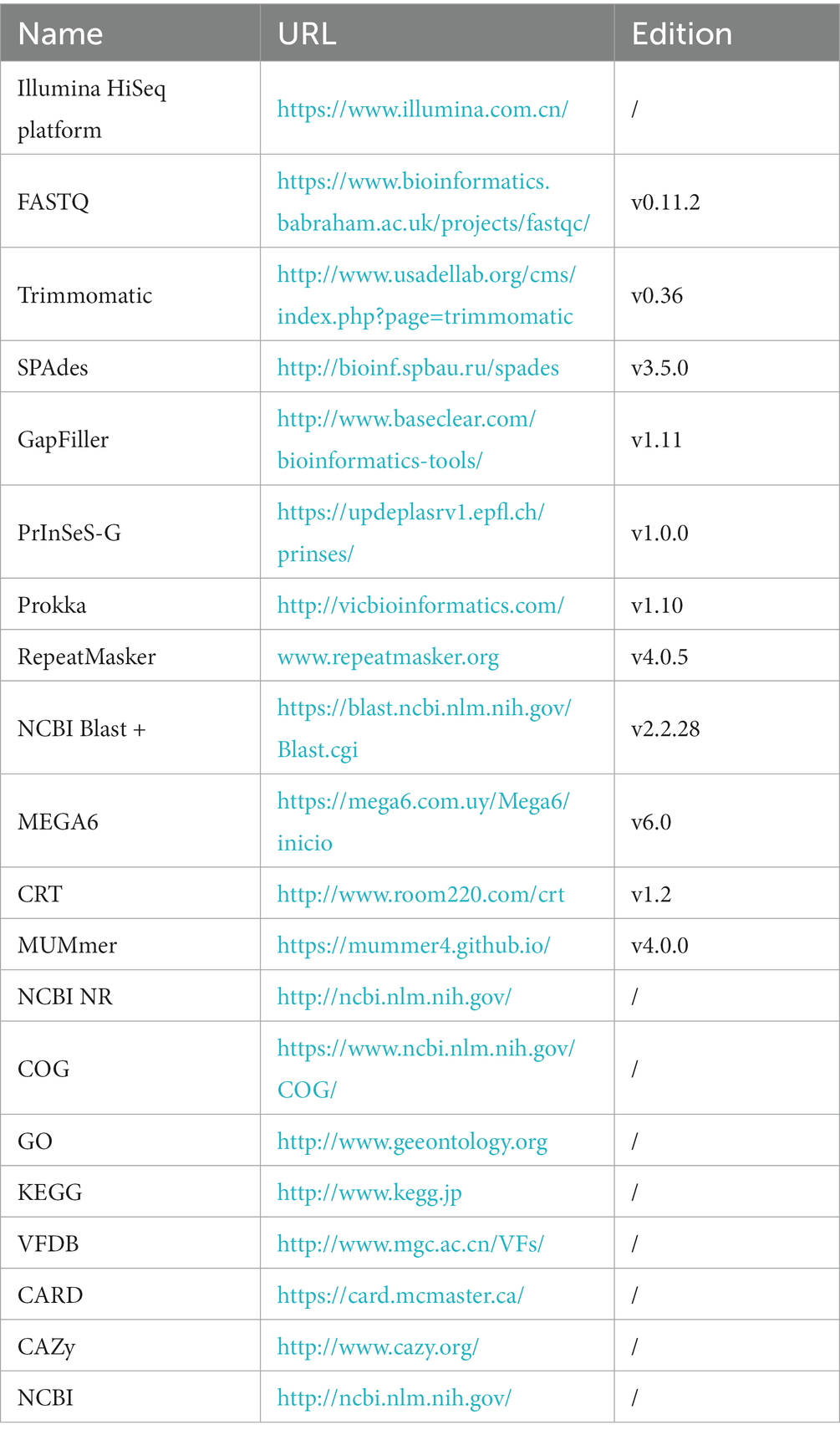
Sangon Bioengineering (Shanghai) Co., LTD. completed the genome-wide sequencing of the MG strains. MG complete genome sequences were obtained using Illumina HiSeq platform (Caporaso et al., 2012) for MG genome sequencing. The original image data file of the MG genome was processed by Base Calling and stored in FASTQ (Brown et al., 2017) file format. Next, the FastQC software was used to evaluate sequencing data quality. Using Trimmomatic (Bolger et al., 2014) was used to clear joint sequences and lower quality sequences to ensure that all the clean reads false discovery rate was <0.01.
2.8. Genome splicing and component analysis and gene function annotation
To splice sequencing data, the SPAdes (Bankevich et al., 2012) was utilized (Table 2). In order to fill gaps in the resulting contigs, the GapFiller (Boetzer and Pirovano, 2012) was employed. The accuracy of the spliced sequences was improved using PrInSeS-G (Massouras et al., 2010) correction tool that can rectify splicing errors during editing and detect small insertion losses. The genetic elements were predicted using Prokka (Seemann, 2014), and repeated sequences in the genome were identified using RepeatMasker (Saha et al., 2008). The CRISPRs were predicted through CRT (Bland et al., 2007) analysis. To obtain functional annotation information, gene sequences were analyzed separately using NCBI NR (Yu and Zhang, 2013), COG (Tatusov et al., 2000), GO (The Gene Ontology, 2019), and KEGG (Kanehisa and Goto, 2000), VFDB (Chen et al., 2016), CARD (McArthur et al., 2013), and CAZy (Lombard et al., 2014), and compared using NCBI Blast + (Altschul et al., 1997).
2.9. Genome-wide comparative analysis
The annotation data of the genome-wide of the locally isolated MG strains were analyzed, including total gene length, GC content, coding gene, etc. Molecular evolutionary tree were constructed using N-J method and MEGA6 software (Tamura et al., 2013). The whole-genome sequences of MG strain were compared with those of the five MG strains available in the NCBI database. Subsequently, the linear analysis of genome-wide sequences of MG strains were conducted using the MUMmer (Delcher et al., 2003; Table 3). Finally, a plot was created to visualize the region of genetic differences.
3. Results
3.1. The bacterial strains showed contrasted ability in biofilm formation
According to strain isolation and identification, five MG strains NX-01, NX-02, NX-03, NX-04, and NX-05 were isolated during this study (Figures 1, 2). The capacity of biofilm formation showed that all 5 MG strains could form biofilms, with the following formation ability: NX-01 > NX-03 > NX-05 > NX-04 > NX-02 (Figure 3A), whereas the M. gallisepticum strains NX-01 and NX-03 showed strong biofilm forming ability (+++). Meanwhile, M. gallisepticum strain NX-05 showed medium biofilm-forming ability (++), and NX-04 and NX-02 were weak biofilm formers (+). The OD590 readings of the strong biofilm-forming strain NX-01 significantly differed from the medium and weak biofilm-forming strains at 12, 24, and 48 h of biofilm culture (p < 0.01). Additionally, at 36 h of biofilm culture, the OD590 readings of the M. gallisepticum strain NX-01 were significantly different from the other four strains (p < 0.01; Figure 3B). The results showed that NX-01 strain was the strongest strain and NX-02 strain was the weakest among the five MG strains. Therefore, these two strains were selected as the subsequent test bacteria.
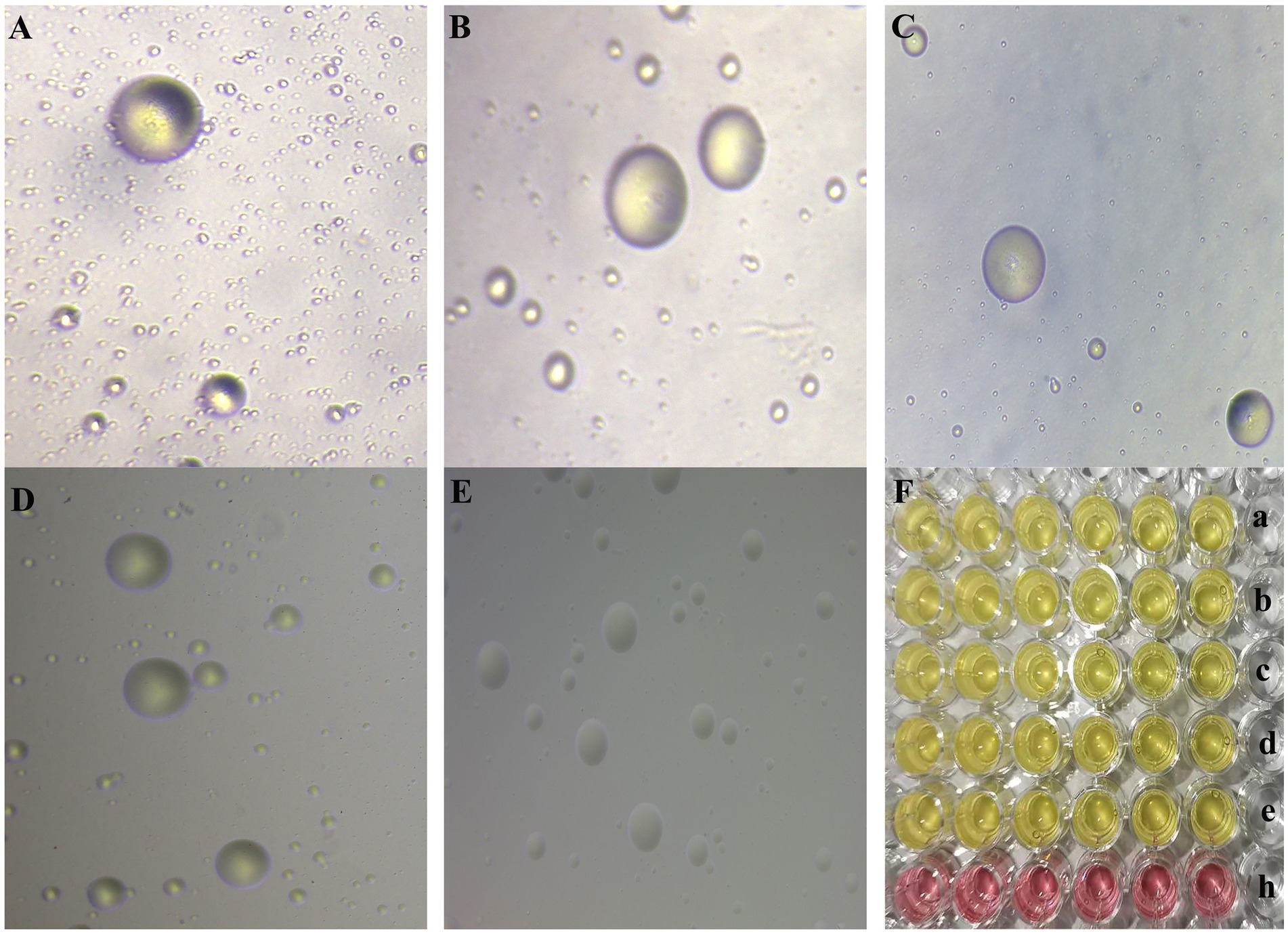
Figure 1. (A–E) The morphology of MG NX-01, NX-02, NX-03, NX-04, and NX-05 strains in modified solid medium under inverted microscope (10×), respectively. Consistent with the typical “fried egg” -like appearance of mycoplasma. (F) a–e was the color change of MG NX-01, NX-02, NX-03, NX-04, and NX-05 strains cultured in modified Frey liquid medium for 2–3 days, and h was the blank control group. Consistent with the characteristics of mycoplasma.
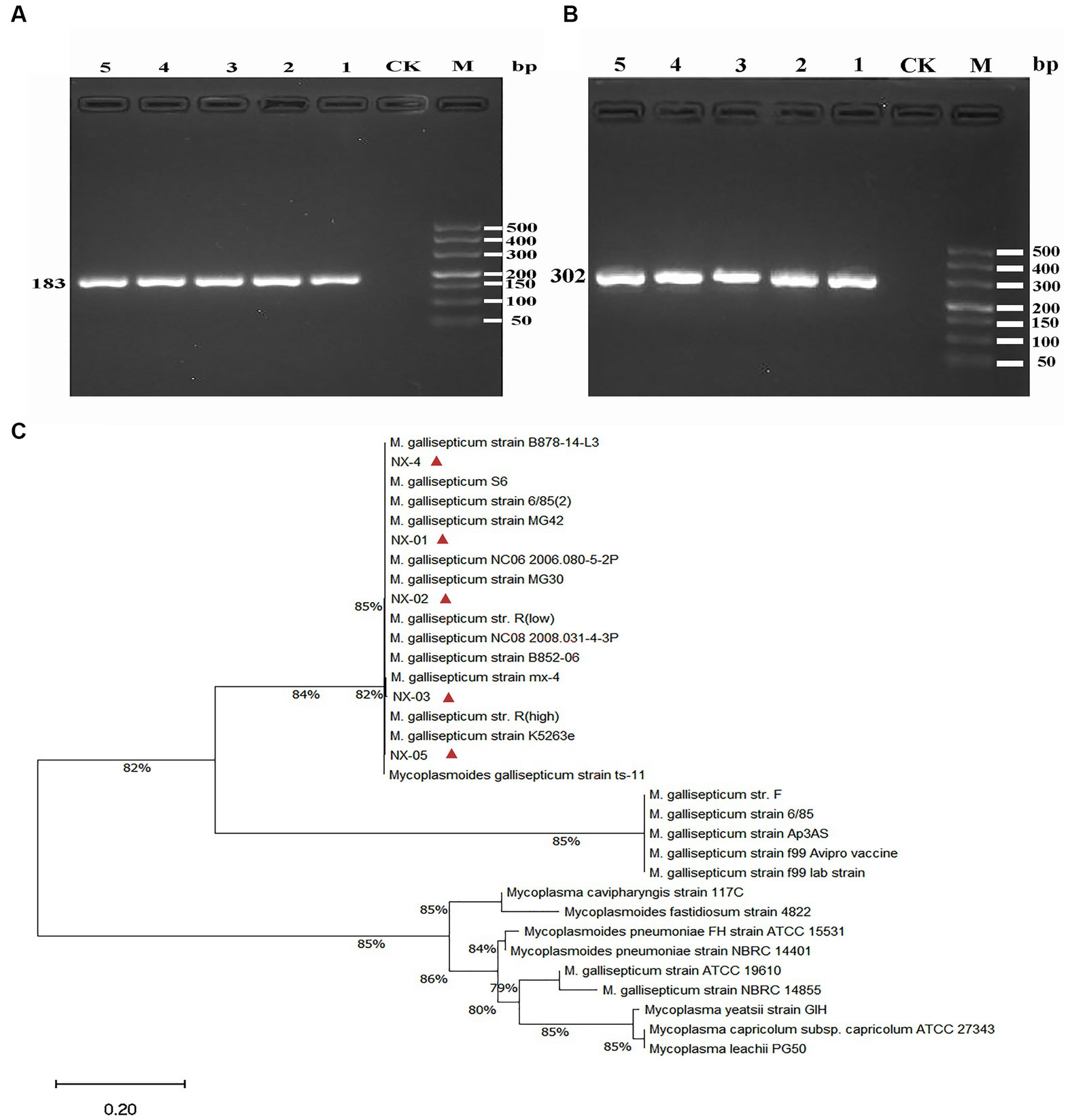
Figure 2. (A) The PCR results of MG 16S rRNA isolated from five strains. The fragment size was consistent with the expected size, indicating that the fragment was amplified correctly. (B) The PCR results of MG mgc2 isolated from five strains. The fragment size was consistent with the expected size, indicating that the fragment was amplified correctly. (C) The 16S rRNA phylogenetic tree of five MG isolates. The results showed that five MG strains were closely related to Mycoplasma gallisepticum uploaded by other NCBI. The above results indicated that the isolated strain was Mycoplasma gallisepticum.

Figure 3. (A) Biofilm formation in different strains of MG. The data are means from three independent experiments. The Mycoplasma gallisepticum strains NX-01 and NX-03 were determined to be strong biofilm producers. The M. gallisepticum strain NX-05 was determined to be a medium biofilm producer. The M. gallisepticum strains NX-02 and NX-04 were determined to be weak biofilm producers. (B) At 36 h of biofilm culture, the OD590 readings of the M. gallisepticum strain NX-01 were significantly different from the other four strains. (C) Biofilm growth curve of M. gallisepticum strain NX-01. (D) Biofilm growth curve of M. gallisepticum strain NX-02.
The matrix growth curve test of the biofilm model in vitro showed that the biofilm formation of the tested MG was divided into the initial adhesion stage, aggregation stage, and biofilm maturation stage. The two tested strains NX-01 and NX-02reached the initial adhesion stage after 12 h culture.
The growth rate of the NX-01 strain was the highest at 12–48 h, and the strain gathered to form microcolonies, corresponding to the aggregation stage of the biofilm. There was no significant change in the number of viable cells of stain at 48–72 h (p > 0.05), corresponding to the maturation stage of the biofilm, at which the microcolony continued to expand and formed a mature biofilm with a dense structure (Figure 3C).
The cells of the NX-02 strain adhered to each other and gathered to form microcolonies within 12–36 h, after which they rapidly multiplied and began to adhere to microplates. During the logarithmic phase of biofilm growth from 36 to 42 h, the formed strain microcolonies fused with each other and developed into biofilm with a mature three-dimensional structure. At 48–72 h, bacterial metabolic activity in the biofilm decreased (Figure 3D). The results showed that the membrane growth rate of strong membrane-forming MG strain NX-01 was faster than that of weak membrane-forming strain (p < 0.01), and the maintenance time of biofilm of NX-01 strain was longer than that of NX-02 strain (p < 0.05).
Scanning electron microscopy (SEM) was used to visually detect the morphology of the biofilm adhesion stage of NX-01 and NX-02 strains. Bacterial aggregates of NX-01 strain was detected across the entire visual field at varying magnifications (Figure 4A). They formed the mature biofilm, which is closely adhered together after being wrapped by the extracellular polysaccharide and other substances secreted by pathogenic bacteria. The three-dimensional mushroom-like structure can even be observed at 50,000× magnification (Figure 4E). The distribution of the NX-02 strain was relatively dispersed in the adhesion stage (Figure 4B), and its cell gathered in some areas forming tiny colonies and organized mature biofilms. The results showed that both NX-01 and NX-02 strains could form biofilms. The biofilm formed by NX-01 strain was multilayer and denser than that of NX-02 strain, which was consistent with the results of the biofilm formation ability test (Figures 4C,D).
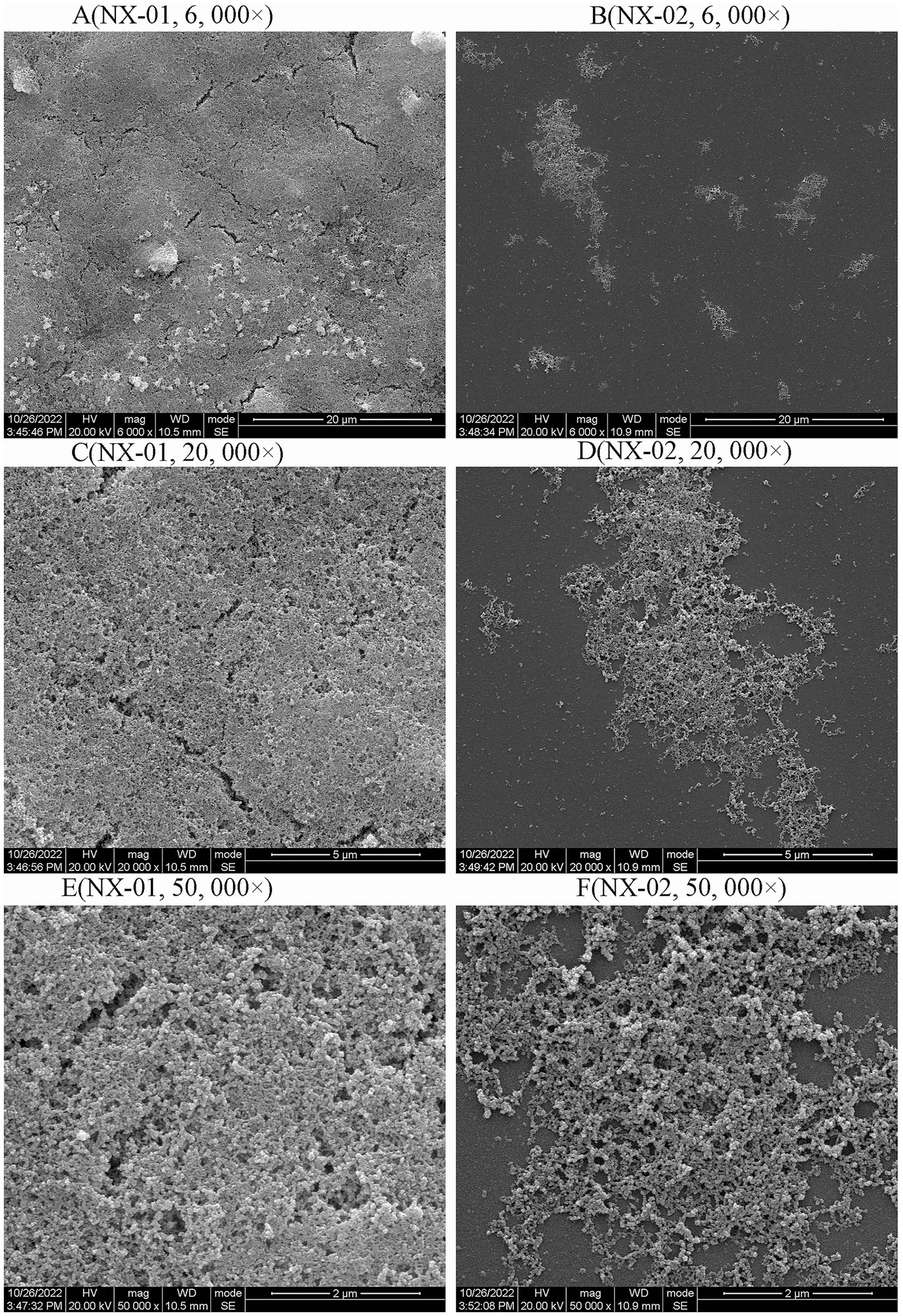
Figure 4. Scanning electron microscopy (SEM) analysis of the biofilm 0f NX-01 and NX-02 at gathering period. (A) MG NX-01(6,000×), (B) NX-02 (6,000×), (C) MG NX-01 (20,000×), (D) NX-02 (20,000×), (E) MG NX-01 (50,000×), and (F) NX-02 (50,000×).
3.2. Different drug sensitivity between NX-01 and NX-02
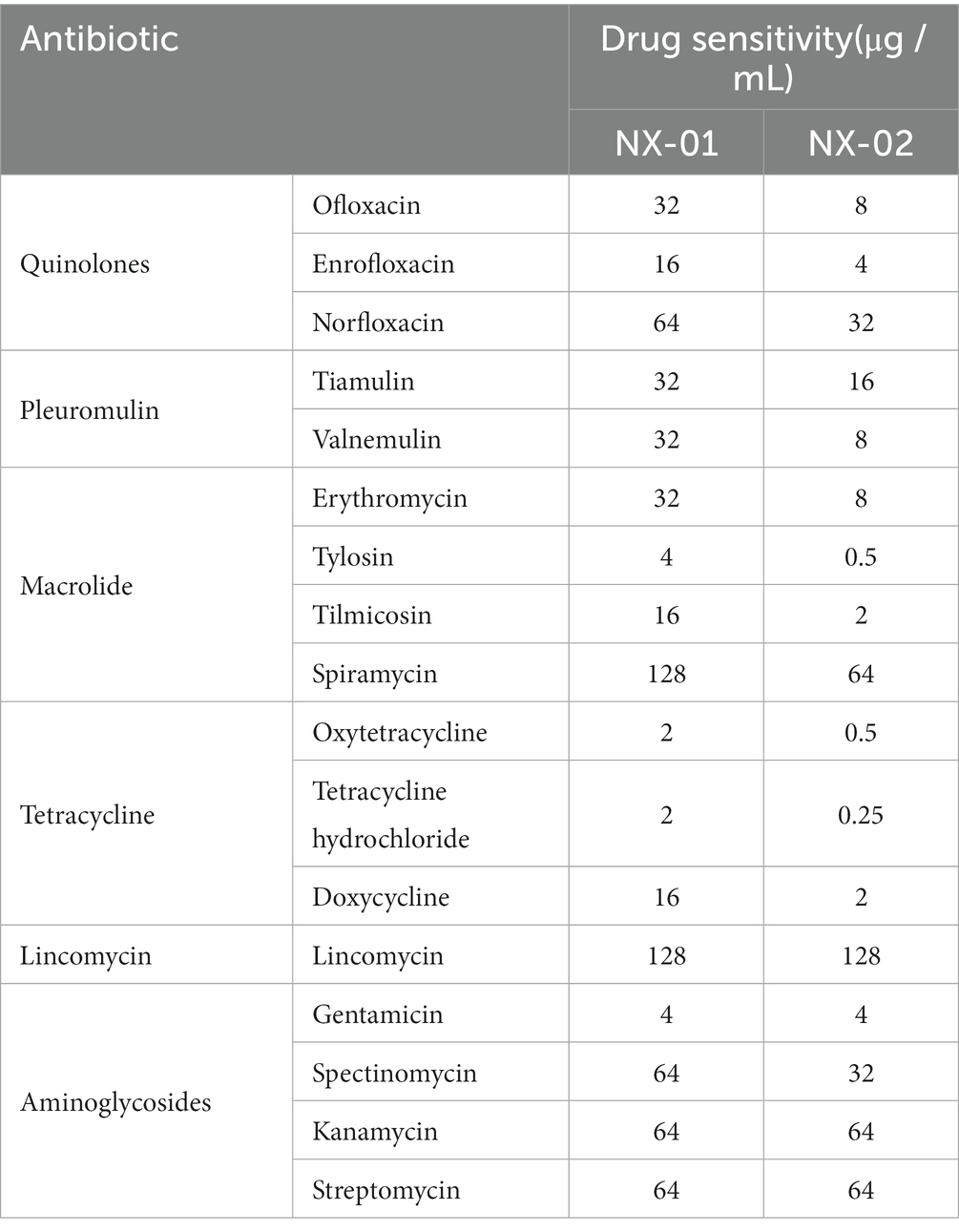
The sensitivity results of two MG stains isolated from Ningxia to 17 kinds of antibiotics showed that the order of sensitivity of NX-01 and NX-02 to six antibiotics was similar, which from strong to weak is tetracyclines, quinolones, macrolides, pleulopleuloides, aminoglycosides, and lincomycin (Table 4). The minimum inhibitory concentration of the NX-01 strain was 2 μg/mL, and that of the NX-02 strain was 0.25 μg/mL. The NX-02 strain was more sensitive to antibiotics than the NX-01 strain.
3.3. NX-01 and NX-02 harbors MG genomic charcteristics
DNA of NX-01 and NX-2 stains were, respectively, extracted and tested the integrity. All DNA sample for sequencing were not degrade in 0.7% nucleic acid gel electrophoresis.
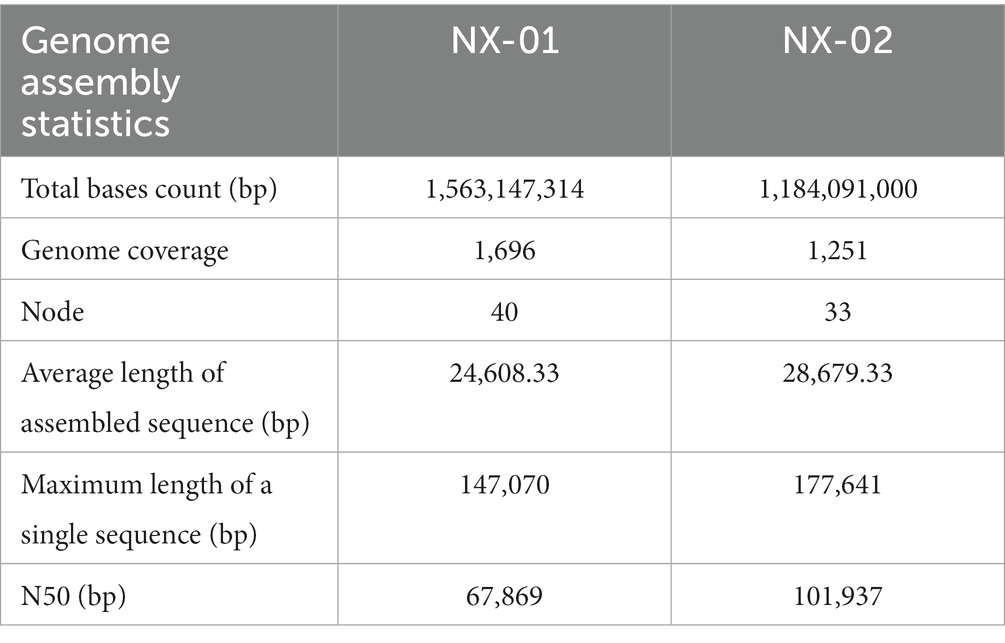
The genome-wide of two MG strains was sequenced using the Illumina HiSeq platform, and the total bases count of NX-01 and NX-02 strains were 1,563,147,314 and 1,184,091,000 bp, and the genome coverage was above 1,000× (Table 5). Compare and evaluate the assembly results after the initial assembly using software SPAdes, NX-01, and NX-02 strains were assembled to obtain 40 and 33 nodes, respectively. The average length of assembled sequences of them is 24608.33 and 28679.33 bp. The maximum length of a single sequences of MG strains NX-01 and NX-02 strains is 147,070 and 17,76,641 bp, respectively. The N50 lengths of them are 67,869 and 101,937 bp. NX-01 and NX-02 strains has been deposited in NCBI (Bio project PRJNA972534 and PRJNA972540).
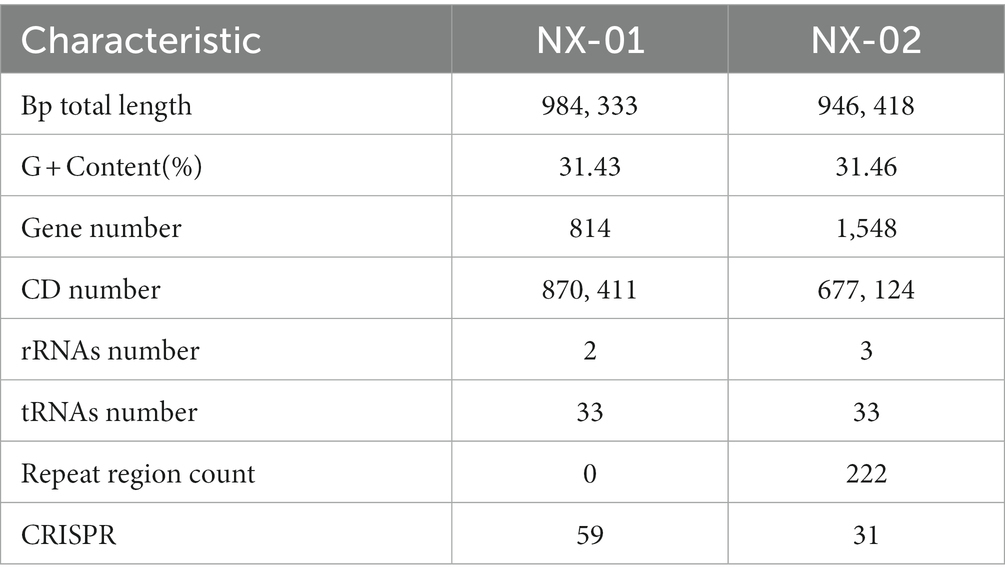
The genome-wide of the two MG strains showed that the genome size of NX-01 and NX-02 strains were 0.98 and 0.94 Mb, respectively (Table 6), and the GC content of their genome were 31.43 and 31.46%. The predicted CDS numbers in the genomes of NX-01 and NX-02 strains were 814 and 1,548 respectively, and predicted rRNAs were 2 and 3, respectively, and both had the tRNAs of 3.
Annotation results of NR database showed that more than 90% of the sequenced genes of the two isolates strains were derived from genome of reported MG, which proved that both of them were MG.
3.4. Functional genes differ between NX-01 and NX-02 genomes
The functional annotation of COG genes showed (Figures 5A,B) that the number of functional genomes of NX-01 and NX-02 strains were 487 and 648, respectively. The predicted types of functional genes of these two MG strains were identical, whereas the number of certain functional genes differed significantly. The number of the two functional genes, including ribosome structure and biogenesis, and DNA replication and repair, was 40 less in the NX-01 strain than in the NX-02 strain. The number of cell wall or membrane or envelope-related genes of NX-01 strain was higher than that of NX-02 strain. The number of other functional genes of NX-01 was less or equal to that of NX-02. S-adenosylmethionine synthetase and methionyl tRNA synthetase were annotated in the genome-wide of NX-01 and NX-02 strains, both of which are biofilm-related enzymes. A biofilm-related protein, adhesin P1 precursor, was only noted in the genome-wide of the NX-01 strain.
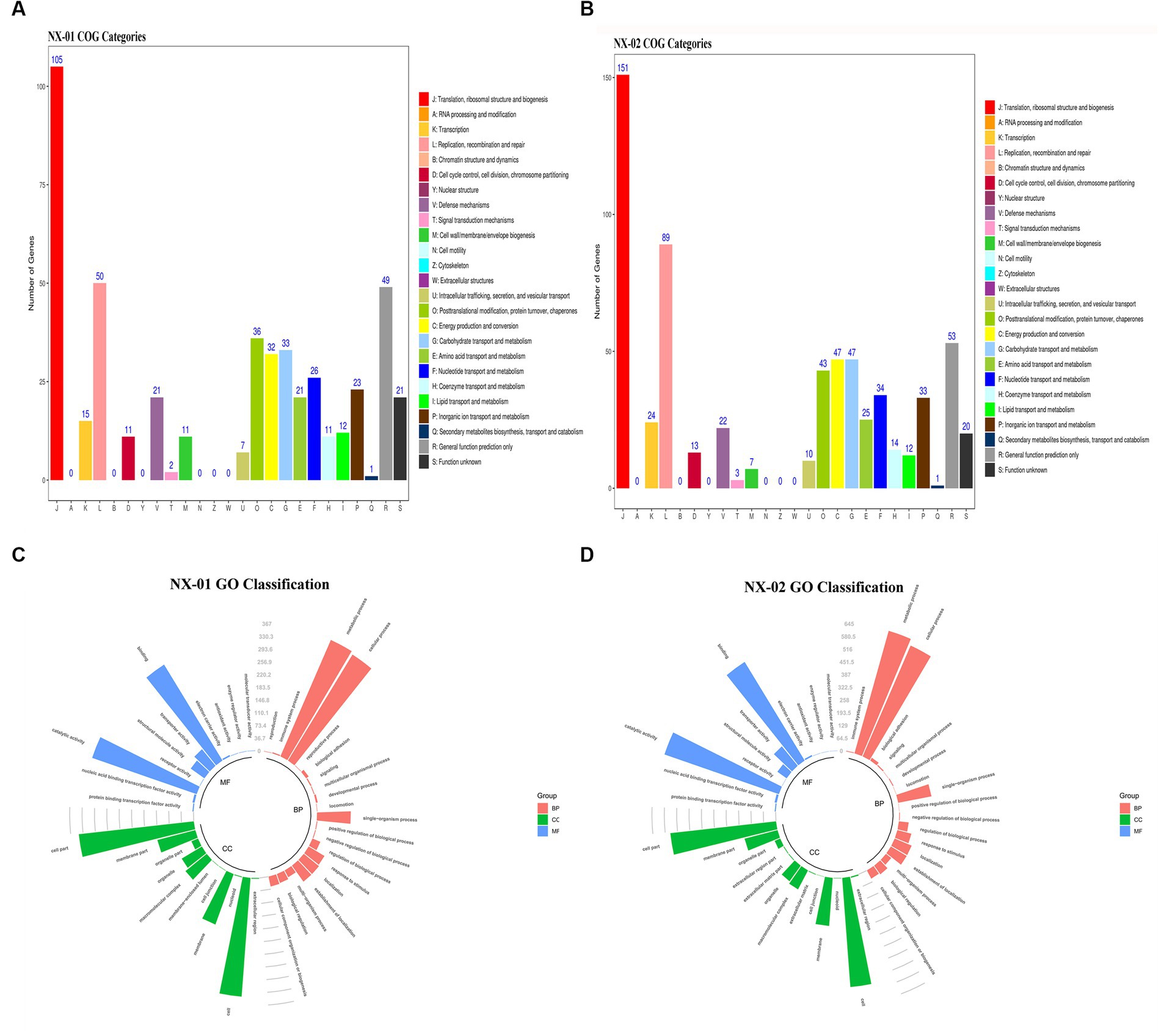
Figure 5. Annotation statistics of the genomes of Mycoplasma gallisepticum strains NX-01 and NX-02 in databases. (A) Functional annotation results of COG database gene of M. gallisepticum strains NX-01. (B) Functional annotation results of COG database gene of M. gallisepticum strain NX-02. (C) GO database gene classification annotation results of M. gallisepticum strain NX-01. (D) GO database gene classification annotation results of M. gallisepticum strain NX-02.
GO database annotation showed (Figures 5C,D) that the functional genes of the two MG strains could be divided into biological process, molecular function, and cell component. NX-01 and NX-02 strains annotated 537 and 947 functional genes, respectively. The same gene may have multiple functions, the total number of genes of NX-01 and NX-02 strains involved in biological processes were 781 and 1,920, involved in molecular function were 1,127 and 1,431, involved in cell components were 1,133 and 1,785, respectively. The most annotated genes of two MG strains,in biological processes category belong to metabolic processes, including 367 genes in NX-01strain and 645 genes in NX-02 strain. The most annotated genes of two MG strains in the category of molecular functions belong to catalytic activity, including 327 genes in NX-01strain and 629 genes in NX-02 strain. The most annotated genes of two MG strains in the category of cell components belong to cells and cell parts, including 343 genes in NX-01 strain and 554 in NX-02 strain. These results indicated that the number of functional genes of NX-01 was significantly different than that of NX-02.
The results of KEGG database annotation results showed that the genes of two MG strains could be divided into five branches, including cell process, environmental information processing, genetic information processing, metabolism, and organism system (Figures 6A,B). The gene number of the NX-01 strain was 302, and that of the NX-02 strain was 502. Metabolism accounted for the highest proportion of genes in the two strains. Metabolism-related genes of NX-01 strain account for 62.3% of the total genes, including energy metabolism, cofactor, and vitamin metabolism, while which of NX-02 strain lack glycan biosynthesis and metabolism genes. The number of metabolism-related genes in NX-02 strain accounted for 55.9% of the total genes. The genes related to genetic information processing accounted for 26.1% in NX-01 strain compared 27.5% in NX-02 strain. Cell process genes account for the least proportion of total genes, with only 0.3% of genes in NX-01 and 0.1% in NX-02. Five genes, oppA, oppD, pdhA, eno, and msbA were annotated in the environmental information processing pathway of two MG strains. Seven genes, manB, pdhB, pdhC, pdhD, RelA, deoA, and gapA, were annotated in the metabolic pathway. One genes, ropS, was annotated in the basic human pathway. All of these genes are biofilm-related gene, and among them, PDH, eno, and gapA participate in multiple pathways at the same time.
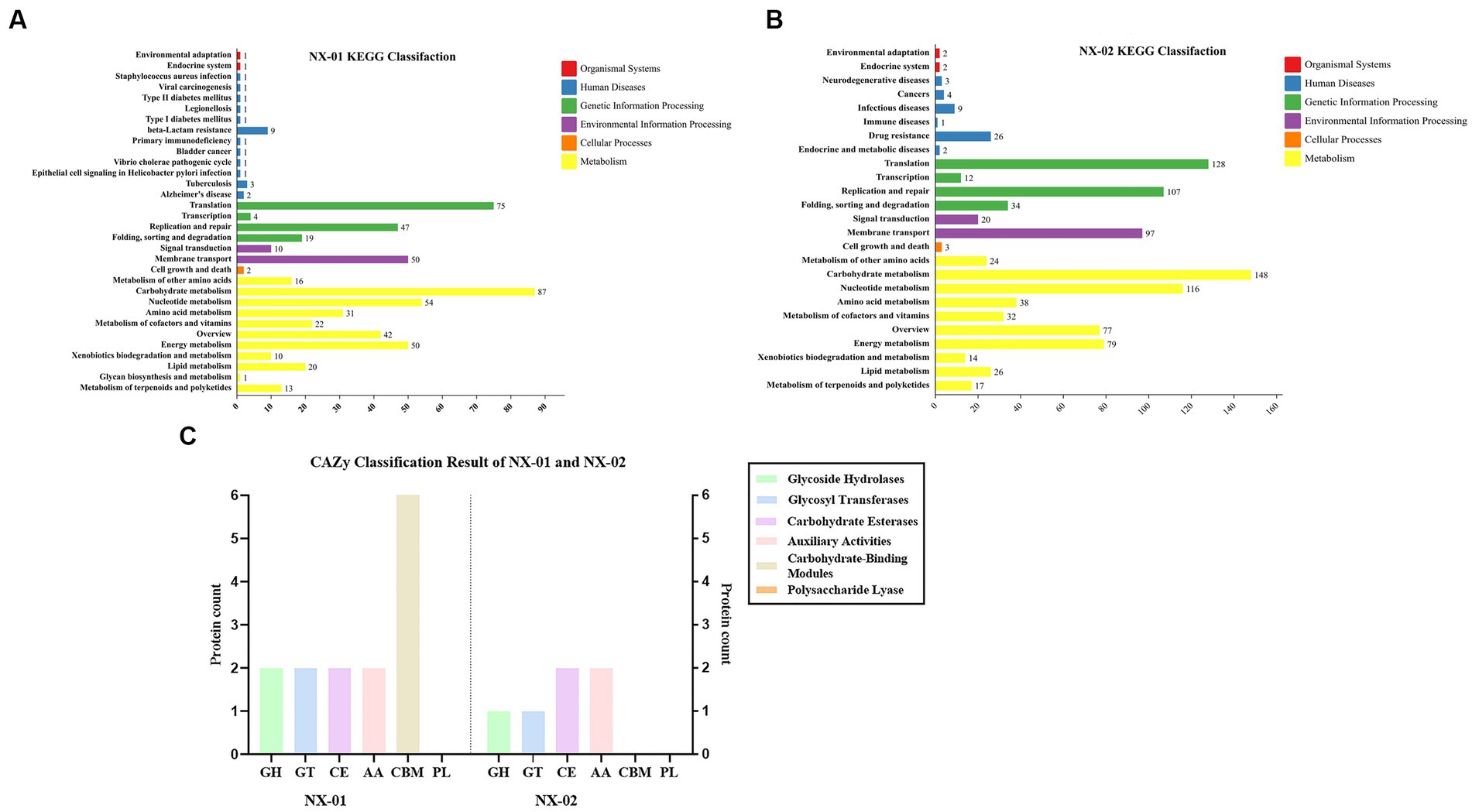
Figure 6. (A) Annotation results of KEGG database gene function classification of Mycoplasma gallisepticum strain NX-01. (B) Annotation results of KEGG database gene function classification of M. gallisepticum strain NX-02. (C) The CAZy database gene annotation results of M. gallisepticum strains NX-01 and NX-02.
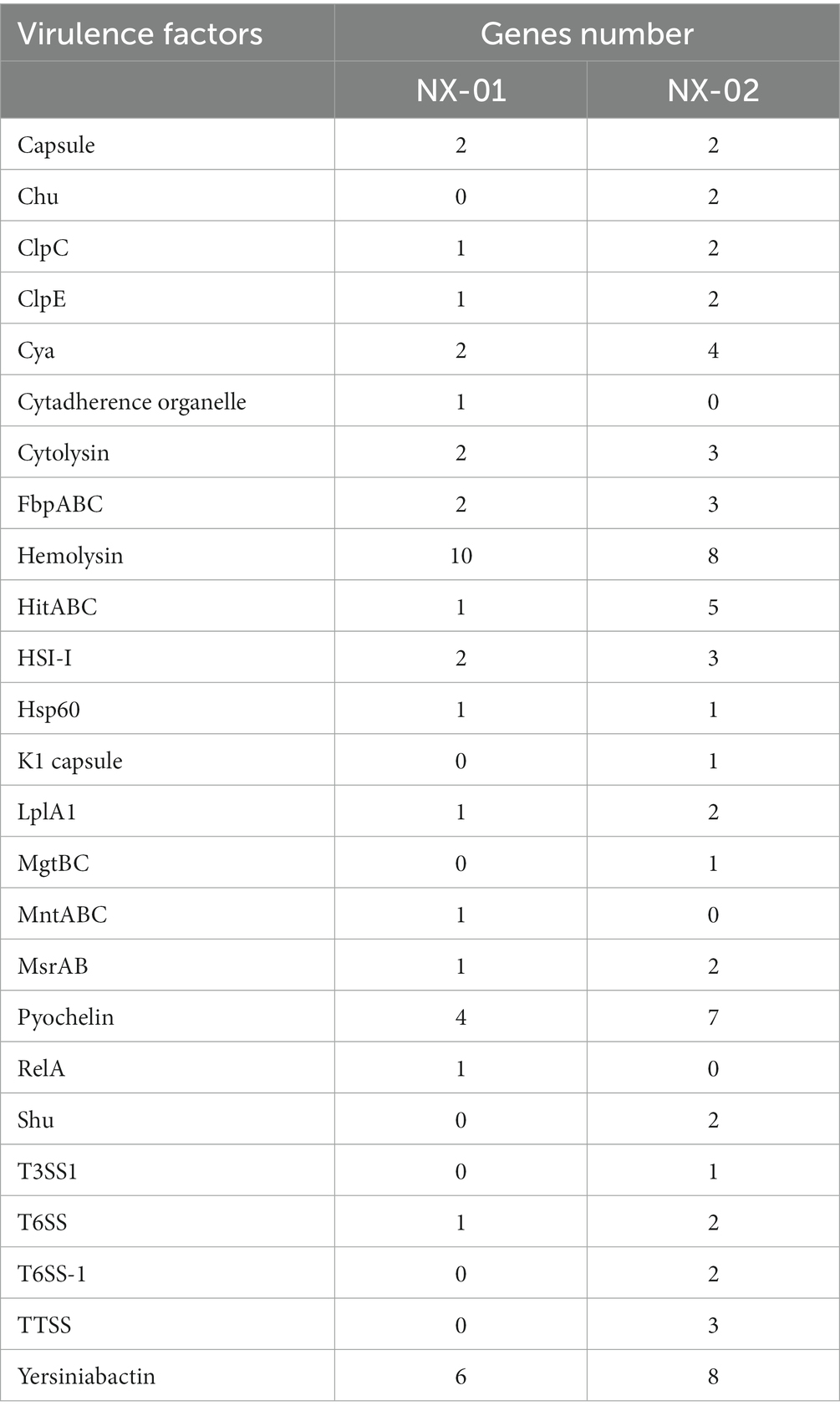
The annotation of the VFDB database showed that NX-01 strain has 18 species virulence factors while NX-02 has 22 species (Table 7). NX-01 strain is characterized by Cytadherence organelle, MntA transporter, and RelA virulence factor. Seven toxic factors, including Clp-type ATPase chaperone protein, type III secretion system ATPase VscN, ATPase EscN, and K1 capsule, were unique to the NX-02 strain. Hemolysin factor was the most annotated MG factor in these two strains, which was cytotoxic to various cell types. From the number of noted virulence factors, it was found that the MG strain NX-02 had more virulence factors than the NX-01 strain, which may lead to stronger virulence of the MG strain NX-02 than NX-01.
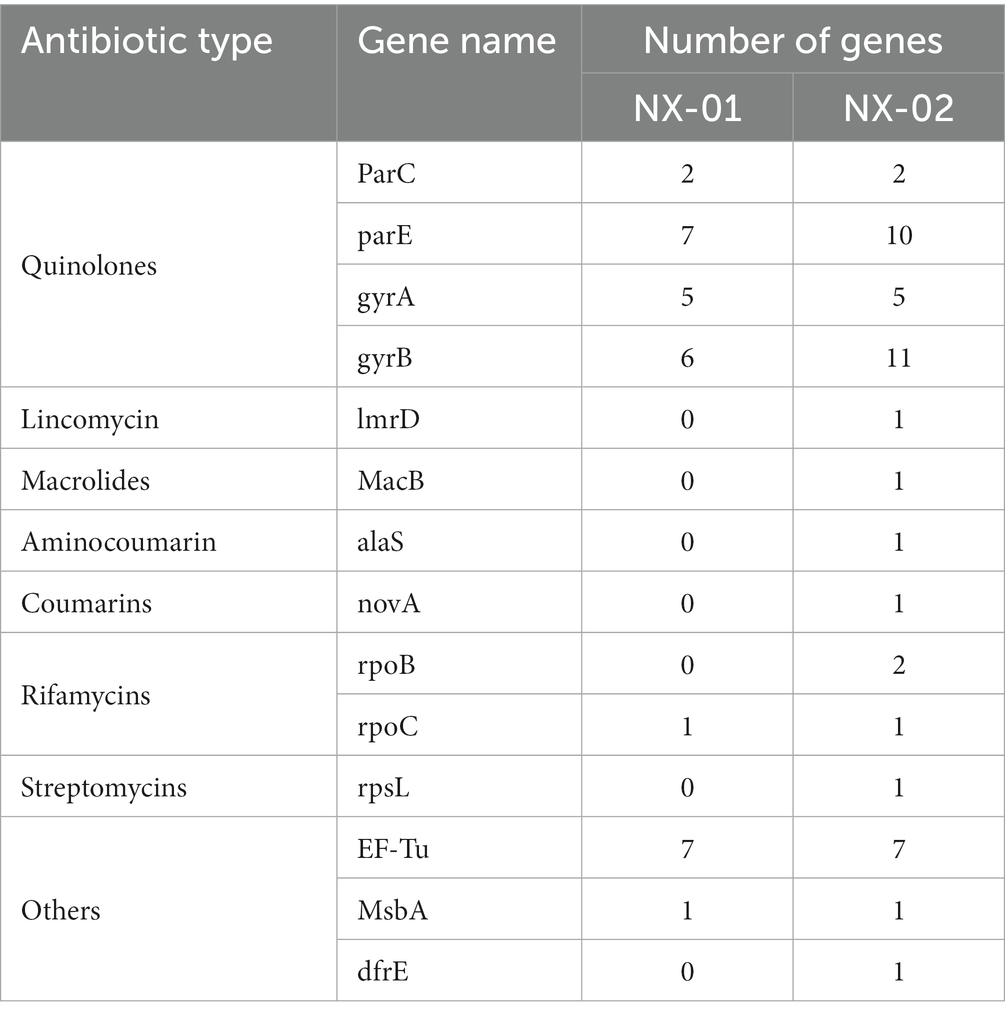
The annotation of the CARD database showed seven kinds of drug-resistant genes in the M. gallisepticum strain NX-01 and 14 kinds in the MG strain NX-02 (Table 8). Quinolone resistance genes is the most frequent occurring drug resistance gene in two MG strains, which NX-01 strain had 20 quinolone resistance genes, and NX-02 strain had 28. Additional resistance genes were also detected, among which the immunogenic protein EF-Tu accounted for the largest proportion which range from 77.7% to 87.5%. Others genes including specific lincomycin resistance factor ImrD, macrolide resistance factor MacB, aminocoumarin resistance factor alaS, coumarin resistance factor novA, rifamycin resistance factor rpoB, and streptomycin resistance factor rpsL, occurs in the NX-02 strain. The results showed that NX-02 strain annotated more drug resistance factors than NX-01 strain, and they may have different degrees of resistance to antibiotics.
The annotation of the CAZy database showed (Figure 6C) that M. gallisepticum strain NX-01 was annotated to five carbohydrate enzymes (gene number 14), including glycoside hydrolase (GH), glycosyltransferase (GT), carbohydrate esterase (CE), helper activity enzyme (AA), and carbohydrate-binding module (CBM). NX-02 strain had one less carbohydrate-binding module (CBM) and six genes. The number of related enzymes annotated by NX-01 strain was higher than that of the NX-02 strain, especially the carbohydrate-binding module, which may indicate different metabolic capacities.
3.5. NX-01 and NX-02 belongs to a divergent clade
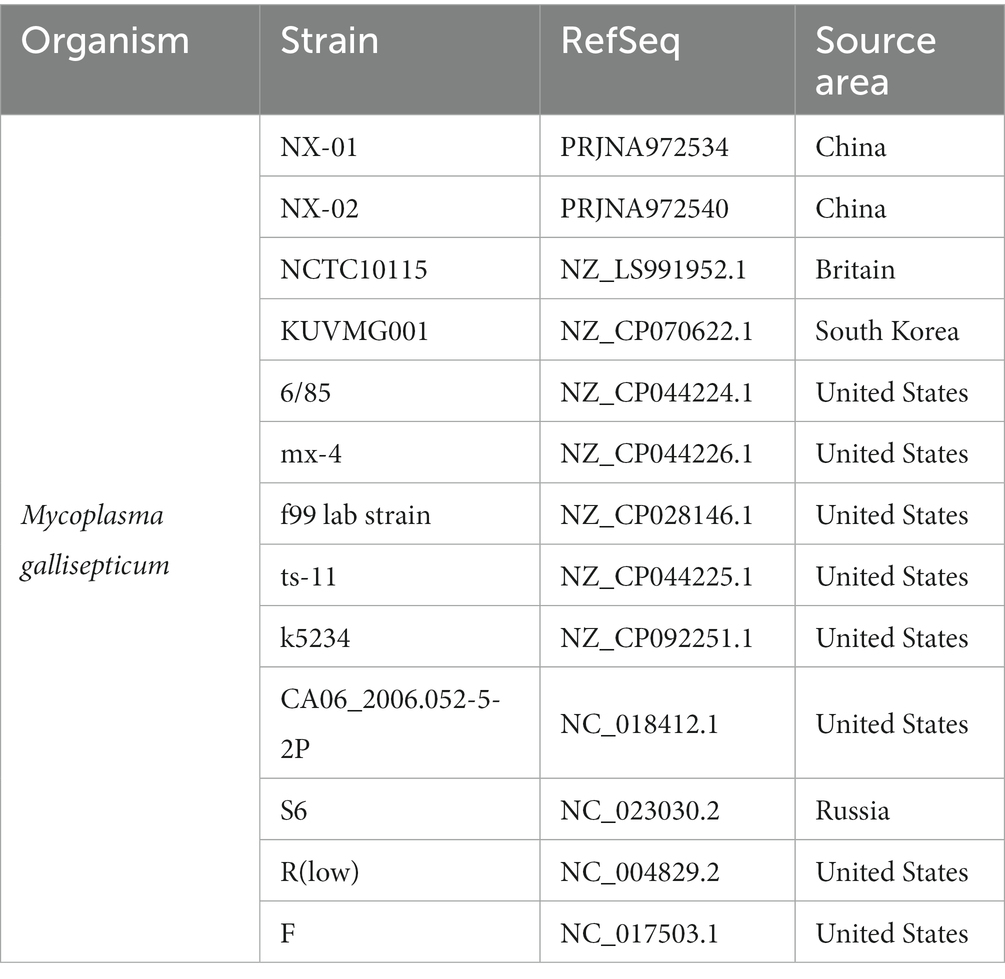
A phylogenetic tree based on genome-wide construction is shown in Figure 7A. NX-01 and NX-02 strains had the closest homology and were in the same group. They were slightly distant from other MG strains, such as S6, ts-11, 6/85, R(low), NCTC10115, and so on, and were on different evolutionary branches. They were related to M. gallisepticum strains mx-4 and CA06_2006-052-5-2p, and furthest related to M. gallisepticum strains KUVMG01 and faa lab. The results showed that the 12 MG strains formed three large branches without clear distinctions based on geographic location. The two Ningxia MG isolates were closely related to M. gallisepticum Russian strain S6, suggesting that the two Ningxia MG isolates and MG strain S6 may derive from a common ancestor.
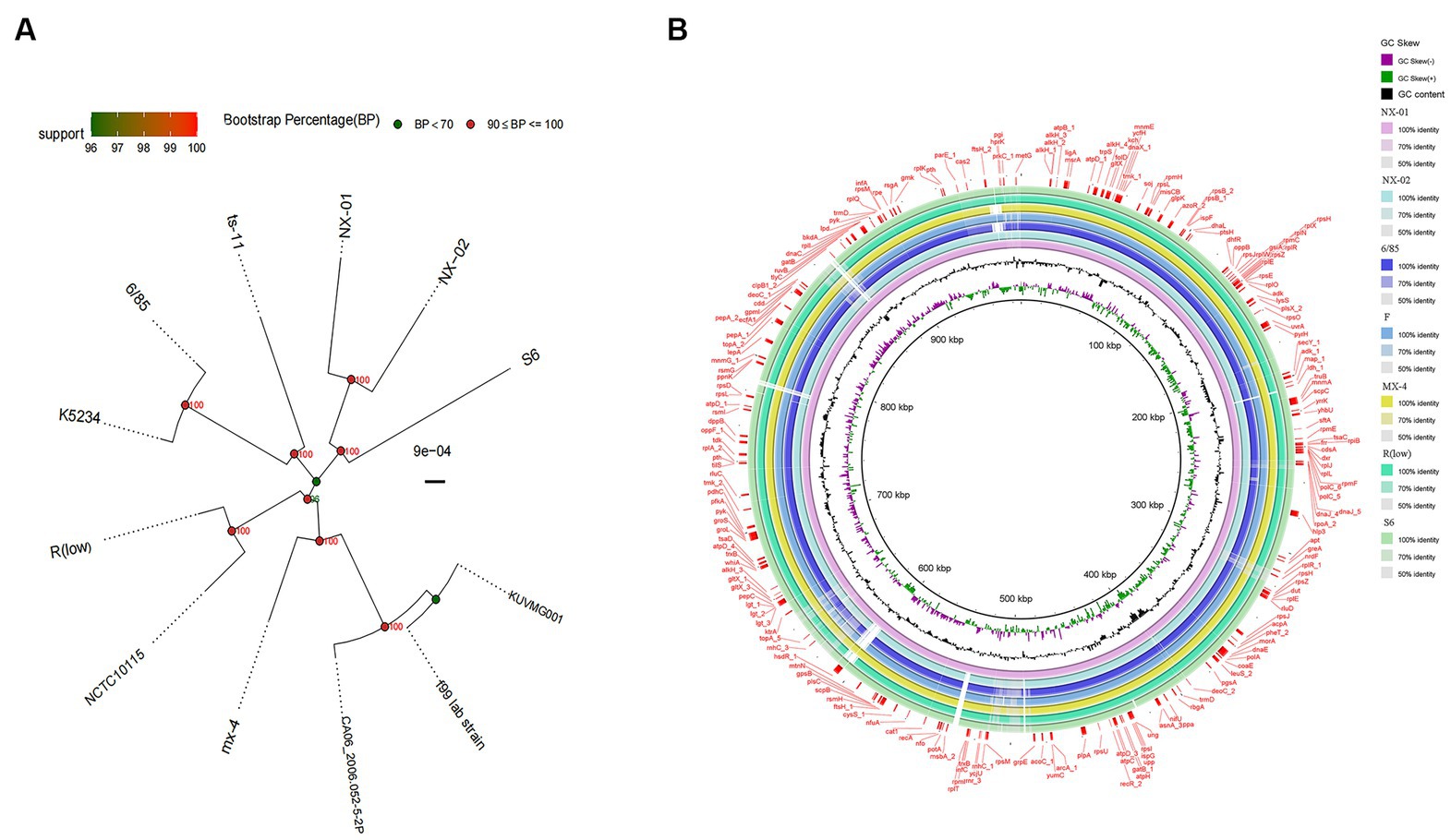
Figure 7. (A) Phylogenetic tree based on different MG genome-wide sequences. NX-01 and NX-02 strains had the closest homology and were in the same group. It was furthest related to Mycoplasma gallisepticum strains KUVMG01 and faa lab. (B) Genome circle of seven MG strains. On laps 1, 2, and 3 (from inside out), the GC skew is black. Green is skew+ and purple is skew−. In circles 4–10 are the full genomes of seven MG strains, namely M. gallisepticum strains NX-01, NX-02, 6/85, F, MX-4, R(LOW), and S6.
The genome-wide circle map of the seven strains of MG (Figure 7B) showed that the genome-wide of NX-01 strain was significantly different from that of the other six strains. Mycoplasma gallisepticum strain NX-01 had fewer gene deletions and had the least difference from M. gallisepticum Russian strain S6. The MG gene differed less between NX-02 and the other five strains were less than NX-01. Compared with NX-01 strain, NX-02 strain had little genetic difference in MG with five strains from other regions. In addition, NX-02 strain has less genome-wide differences from F and R (low) strains in the United States. Five major differences were found in the comparative genome of seven MG strains, which were 490–500, 525–540, 600–615, 782–792, and 882–892 kb fragment regions. The results indicated that the genetic information of MG isolates from Ningxia had rich regional specificity.
3.6. Evidence of differences of NX-01 and NX-02 with other MG epidemic strains
The two MG were from Ningxia were different from the five endemic M. gallisepticum strains [6/85, F, MX-4, R(low), and S6] in five regions of the world, and the two MG isolates were different from M. gallisepticum strain F, but little from M. gallisepticum strains R(low) and S6.
In the fragment region of 490–500 kb (Figure 8A), the NX-01 and NX-02 strains showed more long fragment deletion than the other five strains. In addition, NX-01 and NX-02 strains showed translocation of MG recombinase Polymerase recA, rpmA, and nfo compared with other five MG strains. The direction of NX-01 and NX-02 strains were consistent with S6, R(low), and 6/85 gene fragments. Compared with MX-4 and F strains, NX-01 and NX-02 strains had similar inversions of larger gene segments.
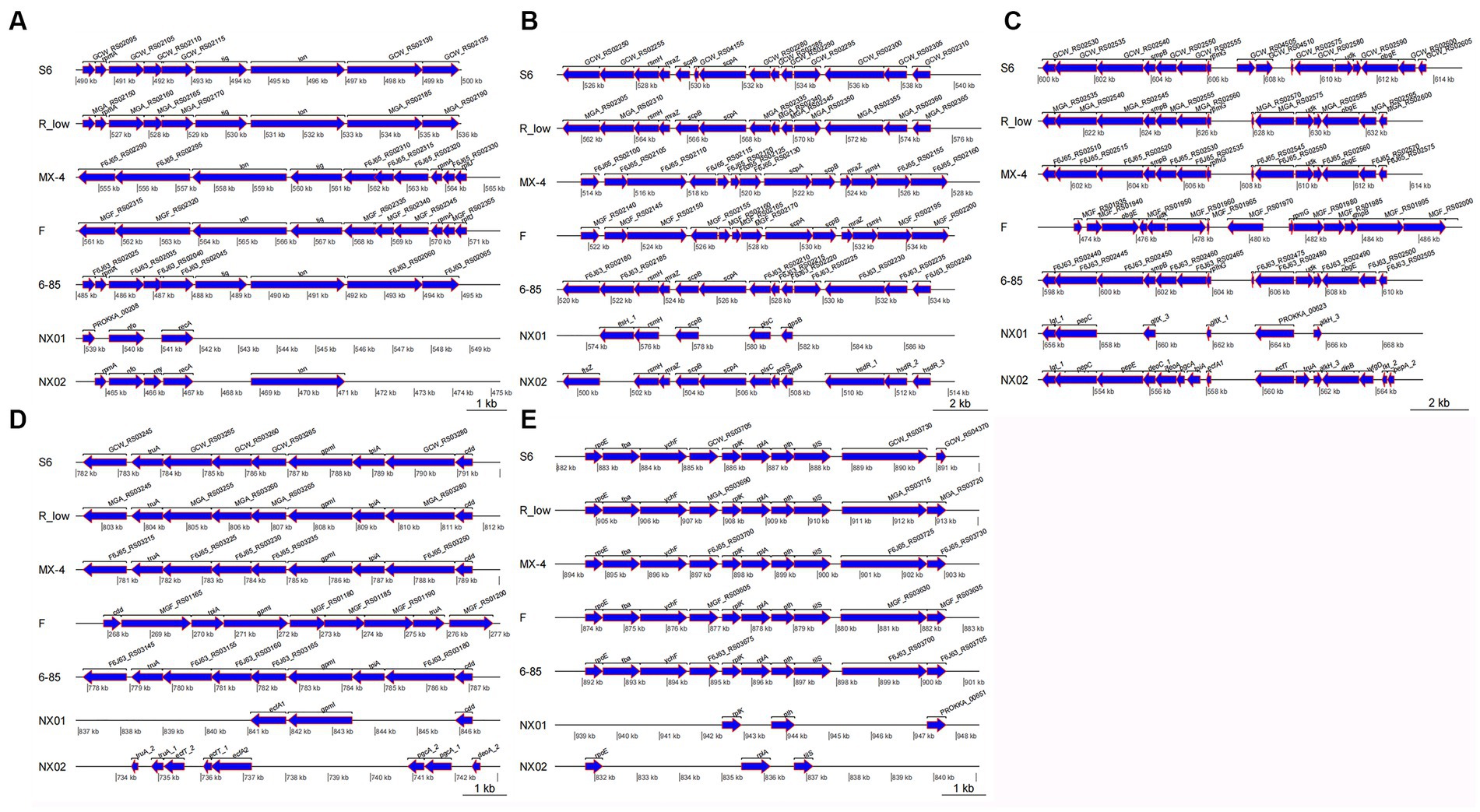
Figure 8. (A) Differences of 490–500 kb in seven MG genomes. (B) Differences of 520–540 kb in seven MG genomes. (C) Differences of 600–615 kb in seven MG genomes. (D) Differences of 782–792 kb in seven MG genomes. (E) Differences of 882–892 kb in seven MG genomes.
In the 520–540 kb fragment (Figure 8B), the two MG strains still had longer fragment deletion than the other five MG strains, and the same genes, such as rsmH, scpB, and plsC, were displaced. Mycoplasma gallisepticum NX-02 strain-specific fssZ can cause host immunogenicity. Two MG strains were in the same direction as M. gallisepticum strains S6, R(low), and 6/85, and the whole segment was inverted with M. gallisepticum strain MX-4 and F.
The 600–615 kb sequences were compared (Figure 8C). Besides removing longer fragments in two of the MG strains compared to the other five, the NX-01 strain is distinguished by the absence of pepE, geoA, and pgcA genes as opposed to the M. gallisepticum strain NX-02. Notably, the pepE was associated with the adhesion process of mycoplasma. Mycoplasma gallisepticum strain NX-02 had more deoA, pgcA, and tpiA genes than the other six strains. The two MG and M. gallisepticum strain F showed an inversion of the entire segment, and the direction of the gene segments of the other four MG strains was the same.
The 782–792 kb sequences were compared (Figure 8D), which revealed found that NX-01 strain had more deletion of MG fragments than the other six strains. NX-01 strain showed more ecfA1 gene than the other five MG isolates from different regions, and the similar genes cdd and gpml were displaced. The ecfT, pgcA, and deoA genes were unique to the NX-02 strain, and the similar gene truA was translocated. The direction of the segments of two MG strains was consistent with that of 6/85, R(low), and S6 strains, and the whole segment was inverted with F strain.
In the 882–892 kb difference region (Figure 8E), the direction of MG gene fragments in two strains was consistent with that in the other five strains. However, fragment deletion was the largest among the five different regions. The same genes rplK, pth, and rpoE of the two MG strains and the other five MG strains were shifted to different degrees. The length of the tilS in M. gallisepticum strain NX-02 was shorter and more translocated than in the other five MG strains.
4. Discussion
Due to the absence of cell walls and defense mechanisms, mycoplasma typically has a limited lifespan in the external environment (Justice-Allen et al., 2010). The formation of biofilms helps mycoplasma survive in the environment outside of its host (Beko et al., 2022). Thus far, only a limited number of studies have demonstrated the ability of certain Mycoplasma gallisepticum strains to develop biofilms, and subsequent investigations in this area are needed (Perez et al., 2020). The present study measured the biofilm-forming ability of five strains of MG isolated from chicken farms in Ningxia, China. Our findings revealed that the MG strains could create biofilms. Yet, proficiency in doing so exhibited notable variations. NX-01 strain was found to have the strongest biofilm formation capacity, while NX-02 strain had the weakest biofilm formation ability.
At the same time, scanning electron microscopy also confirmed that M. gallisepticum strain NX-01 could form a more mature and dense multilayer biofilm. On the other hand, the MG strain NX-02 could only form flaky biofilm clusters in some areas.
Biofilms provide a “safe haven” for persistent bacteria to escape antibiotics, dramatically increasing antibiotic resistance (Cepas et al., 2018; Yan and Bassler, 2019). In the drug sensitivity test of two MG strains, the MG strain NX-01, which exhibited robust biofilm formation, displayed greater antibiotic resistance than the NX-02 strain, thus demonstrating weaker biofilm formation. In general, both strains exhibited poor drug sensitivity. Currently, there is no globally accepted criterion for antimicrobial resistance in MG. Nonetheless, our results showed that the two Ningxia MG isolates had a certain degree of insensitivity to antibiotics commonly used to treat mycoplasma, which was especially evident for the M. gallisepticum strain NX-01, probably due to its strong biofilm formation capacity.
Two MG strains were predicted to contain multiple quinolone-resistant genes, such as Parc, ParE, gyrA, and gyrB. The drug sensitivity test also verified that the two MG strains were not highly sensitive to norfloxacin, which may be related to the frequent use of quinolone antibiotics in treating MG infection. Two MG-specific RecA genes were found in the comparative genome, and they mainly mediated SOS response. The SOS response induces resistance to antibiotic agents by repairing DNA damage caused by antibiotic agents. Quinolones are effective inducers of SOS expression(Pribis et al., 2019). The drug sensitivity test also verified the insensitivity of the two MG strains to quinolones. The NX-02 strain exhibited more specific resistance genes, such as lmrD, MacB, and alaS, among others, than the NX-01 strain. Despite this, the NX-02 strain demonstrated greater overall sensitivity in the drug sensitivity test than the NX-01 strain. It was observed that both biofilm and drug-resistance genes impacted the drug resistance of the bacteria. However, the mechanism underlying this phenomenon should be further explored.
By genome-wide analysis, 10 genes related to biofilm formation were predicted in both MG strains: ManB, oppA, oppD, PDH, eno, RelA, msbA, deoA, gapA, and rpoS. Phosphomanose mutase (ManB) is major in regulating extracellular polysaccharide synthesis of Pseudomonas aeruginosa biofilms, designed to maintain biofilm structure and antibiotic resistance (Ghafoor et al., 2011; Rachmawati et al., 2022). oppA and oppD belong to the ATP-dependent transporter family. They exert different roles in bacterial transport and have been shown to influence biofilm formation (Lee et al., 2004; Li H. et al., 2020). PdhA, pdhB, pdhC, and pdhD were all predicted as genes encoding pyruvate dehydrogenase complex (PDH). PDH can convert pyruvate into acetyl coenzyme A, providing energy for bacterial growth and metabolism (Matic et al., 2003). Monica also found that PDH operons affect the formation of Streptococcus mutans biofilms (Busuioc et al., 2010). Vania reported that pdhA regulates the formation of Staphylococcus epidermidis biofilms (Gaio et al., 2021). Enolase encoded by eno is a key enzyme in the glycolysis pathway and a key gene in forming Staphylococcus aureus biofilm (Didiasova et al., 2019; Pant et al., 2022). The ribosome-related enzyme RelA has been shown to induce metabolic resistance in persistent biofilm cells, used to maintain bacterial biofilms and resist antibiotics (Hall et al., 2020). MsbA is a multidrug-resistant protein gene that can affect bacterial adhesion and biofilm formation (Brown et al., 2014; Padayatti et al., 2019). DeoA is one of the key enzymes used by deoxyribonucleoside. Previous studies have shown that the microcolony of Mycoplasma enters a state of energy starvation, where deoxyribonucleoside is preferred. It helps them survive under pressure (Fisunov et al., 2022). GapA is an essential protein for MG host cell adhesion and virulence and may be involved in biofilm formation (Goh et al., 1998; Ruger et al., 2022). Quorum-sensing phenomenon (QS) is interdependent with biofilm formation, and rpoS is considered the major positive modulator of QS response. Previous studies have shown that rpoS is closely related to the formation of multiple bacterial biofilms (Mika and Hengge, 2014; Hall et al., 2018; Zhang et al., 2021). However, the mechanism of MG biofilm reaction with QS remains unclear.
In addition, three proteins or enzymes associated with biofilm formation were predicted: Adhesin P1 precursor, S-adenosine methionine synthetase, and methionyl tRNA synthetase. In particular, the Adhesin P1 precursor was a differential protein only predicted in the genome-wide NX-01 strain. Adhesin P1 precursor is related to adhesion protein P1, the most important membrane surface adhesion protein of Mycoplasma pneumoniae. It was found that adhesion protein P1 was involved in the biofilm formation of Mycoplasma pneumoniae and Streptococcus mutans (Kornspan et al., 2011; Tang et al., 2016). Therefore, the Adhesin P1 precursor might be significantly related to the distinct biofilm formation capacity of the two MG strains. However, as the genome-wide study was insufficient, further verification is necessary. In both strains of MG, the genome was analyzed for the presence of S-adenosine methionine synthetase and methionyl tRNA synthetase, where the former has a crucial role in protein methylation and various metabolic pathways in bacteria, making it a significant metabolite. It is also a precursor of QS, producing signal molecules related to inducing biofilm formation(Kleiner et al., 2019; Renard et al., 2021). Methionyl tRNA synthetase is necessary for bacteria to make proteins. Ulrike’s research has revealed that this synthetase also has a crucial role in the modulation of virulence and biofilm formation in Acinetobacter baumannii (Blaschke et al., 2021).
The primary invasion of host cells by mycoplasma is adhesion, which also seems necessary for forming mycoplasma biofilms (Sachse et al., 1996). Genome-wide analysis also annotated two MG adhesion-related genes. For example, heat shock proteins Hsp60, Clpc, and ClpE are important virulence factors mediating cell apoptosis, adhesion, invasion, virulence, and reproduction (Garduño et al., 1998). NX-01 strain is characterized by Cytadherence organelle, which is related to Mycoplasma’s cellular adherence and invasiveness (Romero-Arroyo et al., 1999). Among the specific virulence genes of the NX-02, TTSS, T3SS, T6SS system, and K1 Capsule had a key role in host bacterial infection (Francis et al., 2001; Schwarz et al., 2010). In addition, a specific non-catalytic carbohydrate-binding module (CBM) of the NX-01 strain was discovered from the CAZy database. CBMs attach to proteins on the surface of bacterial cells and act as carbohydrate-specific adhesins (Higgins et al., 2011). The regulatory mechanisms of MG biofilms and adhesins have not yet been fully explored; however, both were found to be closely associated with severe chronic mycoplasma infection.
The comparative genome showed that NX-01 and NX-02 strains were closely related to the Russian strain S6, which is far away from MG endemic strains in other areas. The results indicated that the two MG Ningxia isolates had regional specificity. The genome-wide of the NX-01 and NX-02 strains differed from each other. It is worth noting that NX-01 and S6 strains slightly differed, just as NX-02 with F and R(low) strains. Previous research has demonstrated that S6 strain exhibits robust biofilm formation capacity, whereas F and R(low) strains exhibit relatively poor biofilm formers. In this study, the genomic association analysis of NX-01 and S6 showed that the genomes of NX-01 and S6 strains were similar in size, 984,333 and 985,433 bp, respectively, and the CG content were also similar, 31.4 and 31.5%, with both strains containing 33 tRNAs. The P1 homologous adhesion gene was discovered in S6 strain (GenBank: U44804.1). The P1 homologous adhesin gene was discovered in strain S6 (GenBank: U44804.1), and the P1 homologous adhesin gene sequence of S6 strain was compared with the P1 adhesin precursor of NX-01 strain at NCBI Blast. The results showed that the Query Cover was 85%, the E-value was 0, and the per identity was 98.28%. This indicates that the gene sequences of the two MG strains have high homology. Only the strong biofilm formation NX-01 strain was found to contain the P1 mucoadhesive precursor, and it has been shown that the P1 mucoadhesive in Mycoplasma pneumoniae is associated with the formation of its biofilm.
Whether these findings are related to the ability of MG biofilm formation needs to be further investigated. We found five big differences between the two MG strains and the five MG strains from other areas, which contained homologous genes and differential genes. Also, some homologous genes were inverted, which may affect the biological characteristics of MG.
5. Conclusion
Mycoplasma gallisepticum predicted key genes that have been shown to regulate biofilm formation in other strains, and these genes may also have a regulatory role in MG biofilm formation. MG biofilm formation capacity has a direct influence on antibiotic drugs. We speculated that Adhesin P1 precursor might significantly differ from MG biofilm formation ability; however, further in vivo verification, such as gene knockout or use of transposon-mutated mycoplasma clones, is needed. In summary, our results provide a theoretical basis for the molecular mechanism of MG biofilm formation and a potential target for inhibiting the formation of MG biofilm.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/bioproject/; Two genomic information involved in this paper are PRJNA972534 and PRJNA972540.
Author contributions
SH and FY designed the research and analyzed data. XM and LW cultured Mycoplasma gallisepticum biofilm. LG and YG performed genome annotation. XM performed writing—original draft preparation. SH and JL wrote, reviewed, and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work is supported by the Technology Innovation Team Construction Project of Ningxia Hui Autonomous Region (Grant No. 2022BSB03107), the School-Enterprise Joint Innovation Project of Yinchuan, Ningxia (Grant No. 2022XQ009), and the Demonstration Base Construction Project of Production-Education Integration Graduate Student Joint Training of Ningxia University.
Acknowledgments
The authors thank all the teachers and peers who helped with our experiment, and we also thank all the authors of this paper for their hard work.
Conflict of interest
LG was employed by Ningxia Xiaoming Agriculture and Animal Husbandry Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1196747/full#supplementary-material
References
Abd El-Hamid, M. I., Awad, N. F. S., Hashem, Y. M., Abdel-Rahman, M. A., Abdelaziz, A. M., Mohammed, I. A. A., et al. (2019). In vitro evaluation of various antimicrobials against field mycoplasma gallisepticum and mycoplasma synoviae isolates in Egypt. Poult. Sci. 98, 6281–6288. doi: 10.3382/ps/pez576
Ahmad, T., Ullah, S., Moosa, A., Liu, Y., Chengrong, N., and Shujian, H. (2020). First report of pre-harvest soft rot of peach fruit (Prunus persica) caused by Enterobacter mori in China. Plant Dis. doi: 10.1094/PDIS-06-20-1285-PDN
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Beaudet, J., Tulman, E. R., Pflaum, K., Canter, J. A., Silbart, L. K., and Geary, S. J. (2019). Immunologic pathways in protective versus maladaptive host responses to attenuated and pathogenic strains of Mycoplasma gallisepticum. Infect. Immun. 87:e00613. doi: 10.1128/IAI.00613-18
Beko, K., Nagy, E. Z., Grozner, D., Kreizinger, Z., and Gyuranecz, M. (2022). Biofilm formation and its impact on environmental survival and antibiotic resistance of Mycoplasma anserisalpingitidis strains. Acta Vet. Hung. doi: 10.1556/004.2022.00029
Beylefeld, A., Wambulawaye, P., Bwala, D. G., Gouws, J. J., Lukhele, O. M., Wandrag, D. B. R., et al. (2018). Evidence for multidrug resistance in nonpathogenic mycoplasma species isolated from south African poultry. Appl. Environ. Microbiol. 84:e01660-18. doi: 10.1128/AEM.01660-18
Bland, C., Ramsey, T. L., Sabree, F., Lowe, M., Brown, K., Kyrpides, N. C., et al. (2007). CRISPR recognition tool (CRT): a tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinformatics 8:209. doi: 10.1186/1471-2105-8-209
Blaschke, U., Skiebe, E., and Wilharm, G. (2021). Novel genes required for surface-associated motility in Acinetobacter baumannii. Curr. Microbiol. 78, 1509–1528. doi: 10.1007/s00284-021-02407-x
Boetzer, M., and Pirovano, W. (2012). Toward almost closed genomes with GapFiller. Genome Biol. 13:R56. doi: 10.1186/gb-2012-13-6-r56
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Brown, N. A., Dos Reis, T. F., Goinski, A. B., Savoldi, M., Menino, J., Almeida, M. T., et al. (2014). The aspergillus nidulans signalling mucin MsbA regulates starvation responses, adhesion and affects cellulase secretion in response to environmental cues. Mol. Microbiol. doi: 10.1111/mmi.12820
Brown, J., Pirrung, M., and McCue, L. A. (2017). FQC dashboard: integrates FastQC results into a web-based, interactive, and extensible FASTQ quality control tool. Bioinformatics 33, 3137–3139. doi: 10.1093/bioinformatics/btx373
Busuioc, M., Buttaro, B. A., and Piggot, P. J. (2010). The pdh operon is expressed in a subpopulation of stationary-phase bacteria and is important for survival of sugar-starved Streptococcus mutans. J. Bacteriol. 192, 4395–4402. doi: 10.1128/JB.00574-10
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Huntley, J., Fierer, N., et al. (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624. doi: 10.1038/ismej.2012.8
Cepas, V., Lopez, Y., Munoz, E., Rolo, D., Ardanuy, C., Marti, S., et al. (2018). Relationship between biofilm formation and antimicrobial resistance in gram-negative bacteria. Microb. Drug Resist. 25, 72–79. doi: 10.1089/mdr.2018.0027
Chen, H., Yu, S., Hu, M., Han, X., Chen, D., Qiu, X., et al. (2012). Identification of biofilm formation by Mycoplasma gallisepticum. Vet. Microbiol. 161, 96–103. doi: 10.1016/j.vetmic.2012.07.013
Chen, L., Zheng, D., Liu, B., Yang, J., and Jin, Q. (2016). VFDB 2016: hierarchical and refined dataset for big data analysis—10 years on. Nucleic Acids Res., 44, D694–D697. doi: 10.1093/nar/gkv1239
Delcher, A. L., Salzberg, S. L., and Phillippy, A. M. (2003). Using MUMmer to identify similar regions in large sequence sets. Curr. Protoc. Bioinformatics 10:Unit 10.3. doi: 10.1002/0471250953.bi1003s00
Didiasova, M., Schaefer, L., and Wygrecka, M. (2019). When place matters: shuttling of Enolase-1 across cellular compartments. Front. Cell Dev. Biol. 7:61. doi: 10.3389/fcell.2019.00061
Feberwee, A., de Wit, S., and Dijkman, R. (2021). Clinical expression, epidemiology, and monitoring of Mycoplasma gallisepticum and Mycoplasma synoviae: an update. Avian Pathol. 51, 2–18. doi: 10.1080/03079457.2021.1944605
Felice, V., Lupini, C., Mescolini, G., Silveira, F., Guerrini, A., Catelli, E., et al. (2020). Molecular detection and characterization of Mycoplasma gallisepticum and Mycoplasma synoviae strains in backyard poultry in Italy. Poult. Sci. 99, 719–724. doi: 10.1016/j.psj.2019.12.020
Feng, M., Burgess, A. C., Cuellar, R. R., Schwab, N. R., and Balish, M. F. (2021). Modelling persistent Mycoplasma pneumoniae biofilm infections in a submerged BEAS-2B bronchial epithelial tissue culture model. J. Med. Microbiol. 70. doi: 10.1099/jmm.0.001266
Feng, M., Schaff, A. C., and Balish, M. F. (2020). Mycoplasma pneumoniae biofilms grown in vitro: traits associated with persistence and cytotoxicity. Microbiology 166, 629–640. doi: 10.1099/mic.0.000928
Fisunov, G. Y., Pobeguts, O. V., Ladygina, V. G., Zubov, A. I., Galyamina, M. A., Kovalchuk, S. I., et al. (2022). Thymidine utilisation pathway is a novel phenotypic switch of Mycoplasma hominis. J. Med. Microbiol. 71:001468. doi: 10.1099/jmm.0.001468
Francis, M. S., Lloyd, S. A., and Wolf-Watz, H. (2001). The type III secretion chaperone LcrH co-operates with YopD to establish a negative, regulatory loop for control of Yop synthesis in Yersinia pseudotuberculosis. Mol. Microbiol. 42, 1075–1093. doi: 10.1046/j.1365-2958.2001.02702.x
Gaio, V., Lopes, N., Cerca, N., and Franca, A. (2021). codY and pdhA expression is induced in Staphylococcus epidermidis biofilm and planktonic populations with higher proportions of viable but non-culturable cells. Front. Cell. Infect. Microbiol. 11:771666. doi: 10.3389/fcimb.2021.771666
Garduño, R. A., Garduño, E., and Hoffman, P. S. (1998). Surface-associated hsp60 chaperonin of Legionella pneumophila mediates invasion in a HeLa cell model. Infect. Immun. 66, 4602–4610. doi: 10.1128/IAI.66.10.4602-4610.1998
Gautier-Bouchardon, A. V. (2018). Antimicrobial Resistance in Mycoplasma spp. Microbiol. Spectr. 6. doi: 10.1128/microbiolspec.ARBA-0030-2018
Ghafoor, A., Hay, I. D., and Rehm, B. H. (2011). Role of exopolysaccharides in Pseudomonas aeruginosa biofilm formation and architecture. Appl. Environ. Microbiol. 77, 5238–5246. doi: 10.1128/AEM.00637-11
Ghanem, M., Wang, L., Zhang, Y., Edwards, S., Lu, A., Ley, D., et al. (2017). Core genome multilocus sequence typing: a standardized approach for molecular typing of Mycoplasma gallisepticum. J. Clin. Microbiol. 56:e01145-17. doi: 10.1128/JCM.01145-17
Goh, M. S., Gorton, T. S., Forsyth, M. H., Troy, K. E., and Geary, S. J. (1998). Molecular and biochemical analysis of a 105 kDa Mycoplasma gallisepticum cytadhesin (GapA). Microbiology 144, 2971–2978.
Hall, C. W., Hinz, A. J., Gagnon, L. B., Zhang, L., Nadeau, J. P., Copeland, S., et al. (2018). Pseudomonas aeruginosa biofilm antibiotic resistance gene ndvB expression requires the RpoS stationary-phase sigma factor. Appl. Environ. Microbiol. 84:e02762-17. doi: 10.1128/AEM.02762-17
Hall, D. C. Jr., Król, J. E., Cahill, J. P., Ji, H. F., and Ehrlich, G. D. (2020). The development of a pipeline for the identification and validation of small-molecule RelA inhibitors for use as anti-biofilm drugs. Microorganisms 8:1310. doi: 10.3390/microorganisms8091310
Hannan, P. C. (2000). Guidelines and recommendations for antimicrobial minimum inhibitory concentration (MIC) testing against veterinary mycoplasma species. International research programme on comparative Mycoplasmology. Vet. Res. 31, 373–395. doi: 10.1051/vetres:2000100
Higgins, M. A., Ficko-Blean, E., Meloncelli, P. J., Lowary, T. L., and Boraston, A. B. (2011). The overall architecture and receptor binding of pneumococcal carbohydrate-antigen-hydrolyzing enzymes. J. Mol. Biol. 411, 1017–1036. doi: 10.1016/j.jmb.2011.06.035
Hu, Q., Han, X., Zhou, X., Ding, S., Ding, C., and Yu, S. (2010). Characterization of biofilm formation by Riemerella anatipestifer. Vet. Microbiol. 144, 429–436. doi: 10.1016/j.vetmic.2010.02.023
Ishfaq, M., Zhang, W., Ali Shah, S. W., Wu, Z., Wang, J., Ding, L., et al. (2020). The effect of Mycoplasma gallisepticum infection on energy metabolism in chicken lungs: through oxidative stress and inflammation. Microb. Pathog. 138:103848. doi: 10.1016/j.micpath.2019.103848
Jiang, R., Xiang, M., Chen, W., Zhang, P., Wu, X., Zhu, G., et al. (2021). Biofilm characteristics and transcriptomic analysis of Haemophilus parasuis. Vet. Microbiol. 258:109073. doi: 10.1016/j.vetmic.2021.109073
Justice-Allen, A., Trujillo, J., Corbett, R., Harding, R., Goodell, G., and Wilson, D. (2010). Survival and replication of Mycoplasma species in recycled bedding sand and association with mastitis on dairy farms in Utah. J. Dairy Sci. 93, 192–202. doi: 10.3168/jds.2009-2474
Kahya, S., Temelli, S., Eyigor, A., and Carli, K. T. (2010). Real-time PCR culture and serology for the diagnosis of Mycoplasma gallisepticum in chicken breeder flocks. Vet. Microbiol. 144, 319–324. doi: 10.1016/j.vetmic.2010.01.012
Kanehisa, M., and Goto, S. (2000). KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. doi: 10.1093/nar/28.1.27
Kleiner, D., Shmulevich, F., Zarivach, R., Shahar, A., Sharon, M., Ben-Nissan, G., et al. (2019). The interdimeric interface controls function and stability of Ureaplasma urealiticum methionine S-adenosyltransferase. J. Mol. Biol. 431, 4796–4816. doi: 10.1016/j.jmb.2019.09.003
Kornspan, J. D., Tarshis, M., and Rottem, S. (2011). Adhesion and biofilm formation of Mycoplasma pneumoniae on an abiotic surface. Arch. Microbiol. 193, 833–836. doi: 10.1007/s00203-011-0749-y
Lee, E. M., Ahn, S. H., Park, J. H., Lee, J. H., Ahn, S. C., and Kong, I. S. (2004). Identification of oligopeptide permease (Opp) gene cluster in Vibrio fluvialis and characterization of biofilm production by oppA knockout mutation. FEMS Microbiol. Lett. 240, 21–30. doi: 10.1016/j.femsle.2004.09.007
Li, L., Li, H., Shi, Y., Chai, A. L., Xie, X., and Li, B. (2020). First report of bacterial leaf spot of cucurbita pepo caused by erwinia persicina in China. Plant Dis. doi: 10.1094/PDIS-06-20-1241-PDN
Li, H., Qiao, Y., Du, D., Wang, J., and Ma, X. (2020). Deletion of the oligopeptide transporter Lmo2193 decreases the virulence of Listeria monocytogenes. J. Vet. Sci. 21:e88. doi: 10.4142/jvs.2020.21.e88
Liang, Z., Qi, Y., Guo, S., Hao, K., Zhao, M., and Guo, N. (2019). Effect of AgWPA nanoparticles on the inhibition of Staphylococcus aureus growth in biofilms. Food Control 100, 240–246. doi: 10.1016/j.foodcont.2019.01.030
Liu, Z., Li, H., Li, L., Ma, Q., Fang, Z., Wang, H., et al. (2022). Gene-trait matching analysis reveals putative genes involved in Bifidobacterium spp. biofilm formation. Gene 826:146449. doi: 10.1016/j.gene.2022.146449
Lombard, V., Golaconda Ramulu, H., Drula, E., Coutinho, P. M., and Henrissat, B. (2014). The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, D490–D495. doi: 10.1093/nar/gkt1178
Marouf, S., Khalf, M. A., Alorabi, M., El-Shehawi, A. M., El-Tahan, A. M., El-Hack, M. E. A., et al. (2022). Mycoplasma gallisepticum: a devastating organism for the poultry industry in Egypt. Poult. Sci. 101:101658. doi: 10.1016/j.psj.2021.101658
Massouras, A., Hens, K., Gubelmann, C., Uplekar, S., Decouttere, F., Rougemont, J., et al. (2010). Primer-initiated sequence synthesis to detect and assemble structural variants. Nat. Methods 7, 485–486. doi: 10.1038/nmeth.f.308
Matic, J. N., Wilton, J. L., Towers, R. J., Scarman, A. L., Minion, F. C., Walker, M. J., et al. (2003). The pyruvate dehydrogenase complex of Mycoplasma hyopneumoniae contains a novel lipoyl domain arrangement. Gene 319, 99–106. doi: 10.1016/S0378-1119(03)00798-4
McArthur, A. G., Waglechner, N., Nizam, F., Yan, A., Azad, M. A., Baylay, A. J., et al. (2013). The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 57, 3348–3357. doi: 10.1128/AAC.00419-13
McAuliffe, L., Ellis, R. J., Miles, K., Ayling, R. D., and Nicholas, R. A. J. (2006). Biofilm formation by mycoplasma species and its role in environmental persistence and survival. Microbiology 152, 913–922. doi: 10.1099/mic.0.28604-0
Mika, F., and Hengge, R. (2014). Small RNAs in the control of RpoS, CsgD, and biofilm architecture of Escherichia coli. RNA Biol. 11, 494–507. doi: 10.4161/rna.28867
Padayatti, P. S., Lee, S. C., Stanfield, R. L., Wen, P. C., Tajkhorshid, E., Wilson, I. A., et al. (2019). Structural insights into the lipid a transport pathway in MsbA. Structure 27, 1114–1123. doi: 10.1016/j.str.2019.04.007
Pant, N., Miranda-Hernandez, S., Rush, C., Warner, J., and Eisen, D. P. (2022). Effect of savirin in the prevention of biofilm-related Staphylococcus aureus prosthetic joint infection. Front. Pharmacol. 13:989417. doi: 10.3389/fphar.2022.989417
Parker, D., Soong, G., Planet, P., Brower, J., Ratner, A. J., and Prince, A. (2009). The NanA neuraminidase of Streptococcus pneumoniae is involved in biofilm formation. Infect. Immun. 77, 3722–3730. doi: 10.1128/IAI.00228-09
Perez, K., Mullen, N., Canter, J. A., Ley, D. H., and May, M. (2020). Phenotypic diversity in an emerging mycoplasmal disease. Microb. Pathog. 138:103798. doi: 10.1016/j.micpath.2019.103798
Pribis, J. P., Garcia-Villada, L., Zhai, Y., Lewin-Epstein, O., Wang, A. Z., Liu, J., et al. (2019). Gamblers: an antibiotic-induced evolvable cell subpopulation differentiated by reactive-oxygen-induced general stress response. Mol. Cell 74:e7. doi: 10.1016/j.molcel.2019.02.037
Rachmawati, D., Fahmi, M. Z., Abdjan, M. I., Wasito, E. B., Siswanto, I., Mazlan, N., et al. (2022). In vitro assessment on designing novel antibiofilms of pseudomonas aeruginosa using a computational approach. Molecules 27:8935. doi: 10.3390/molecules27248935
Renard, A., Diene, S. M., Courtier-Martinez, L., Gaillard, J. B., Gbaguidi-Haore, H., Mereghetti, L., et al. (2021). 12/111phiA prophage domestication is associated with autoaggregation and increased ability to produce biofilm in Streptococcus agalactiae. Microorganisms 9:1112. doi: 10.3390/microorganisms9061112
Robinson, J. C., Rostami, N., Casement, J., Vollmer, W., Rickard, A. H., and Jakubovics, N. S. (2018). ArcR modulates biofilm formation in the dental plaque colonizer Streptococcus gordonii. Mol Oral Microbiol 33, 143–154. doi: 10.1111/omi.12207
Romero-Arroyo, C. E., Jordan, J., Peacock, S. J., Willby, M. J., Farmer, M. A., and Krause, D. C. (1999). Mycoplasma pneumoniae protein P30 is required for cytadherence and associated with proper cell development. J. Bacteriol. 181, 1079–1087. doi: 10.1128/JB.181.4.1079-1087.1999
Roussan, D. A., Khawaldeh, G., and Shaheen, I. A. (2015). A survey of Mycoplasma gallisepticum and Mycoplasma synovaie with avian influenza H9 subtype in meat-type chicken in Jordan between 2011-2015. Poult. Sci. 94, 1499–1503. doi: 10.3382/ps/pev119
Ruger, N., Szostak, M. P., and Rautenschlein, S. (2022). The expression of GapA and CrmA correlates with the Mycoplasma gallisepticum in vitro infection process in chicken TOCs. Vet. Res. 53:66. doi: 10.1186/s13567-022-01085-2
Sachse, K., Grajetzki, C., Rosengarten, R., Hänel, I., Heller, M., and Pfützner, H. (1996). Mechanisms and factors involved in Mycoplasma bovis adhesion to host cells. Zentralbl Bakteriol 284, 80–92. doi: 10.1016/S0934-8840(96)80157-5
Saha, S., Bridges, S., Magbanua, Z. V., and Peterson, D. G. (2008). Empirical comparison of ab initio repeat finding programs. Nucleic Acids Res. 36, 2284–2294. doi: 10.1093/nar/gkn064
Schwarz, S., West, T. E., Boyer, F., Chiang, W. C., Carl, M. A., Hood, R. D., et al. (2010). Burkholderia type VI secretion systems have distinct roles in eukaryotic and bacterial cell interactions. PLoS Pathog. 6:e1001068. doi: 10.1371/journal.ppat.1001068
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Semashko, T. A., Arzamasov, A. A., Evsyutina, D. V., Garanina, I. A., Matyushkina, D. S., Ladygina, V. G., et al. (2022). Role of DNA modifications in Mycoplasma gallisepticum. PLoS One 17:e0277819. doi: 10.1371/journal.pone.0277819
Sharma, D., Misba, L., and Khan, A. U. (2019). Antibiotics versus biofilm: an emerging battleground in microbial communities. Antimicrob. Resist. Infect. Control 8:76. doi: 10.1186/s13756-019-0533-3
Shatila, F., Yasa, I., and Yalcin, H. T. (2021). Biofilm formation by Salmonella enterica strains. Curr. Microbiol. 78, 1150–1158. doi: 10.1007/s00284-021-02373-4
Sorci, G. (2013). Immunity, resistance and tolerance in bird-parasite interactions. Parasite Immunol. 35, 350–361. doi: 10.1111/pim.12047
Sprygin, A. V., Andreychuk, D. B., Kolotilov, A. N., Volkov, M. S., Runina, I. A., Mudrak, N. S., et al. (2010). Development of a duplex real-time TaqMan PCR assay with an internal control for the detection of Mycoplasma gallisepticum and Mycoplasma synoviae in clinical samples from commercial and backyard poultry. Avian Pathol. 39, 99–109. doi: 10.1080/03079451003604621
Stewart, P. S. (2002). Mechanisms of antibiotic resistance in bacterial biofilms. Int. J. Med. Microbiol. 292, 107–113. doi: 10.1078/1438-4221-00196
Sulyok, K. M., Kreizinger, Z., Beko, K., Forro, B., Marton, S., Banyai, K., et al. (2019). Development of molecular methods for rapid differentiation of mycoplasma gallisepticum vaccine strains from field isolates. J. Clin. Microbiol. 57:e01084-18. doi: 10.1128/JCM.01084-18
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tang, W., Bhatt, A., Smith, A. N., Crowley, P. J., Brady, L. J., and Long, J. R. (2016). Specific binding of a naturally occurring amyloidogenic fragment of Streptococcus mutans adhesin P1 to intact P1 on the cell surface characterized by solid state NMR spectroscopy. J. Biomol. NMR 64, 153–164. doi: 10.1007/s10858-016-0017-1
Tassew, D. D., Mechesso, A. F., Park, N. H., Song, J. B., Shur, J. W., and Park, S. C. (2017). Biofilm formation and determination of minimum biofilm eradication concentration of antibiotics in Mycoplasma hyopneumoniae. J. Vet. Med. Sci. 79, 1716–1720. doi: 10.1292/jvms.17-0279
Tatusov, R. L., Galperin, M. Y., Natale, D. A., and Koonin, E. V. (2000). The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28, 33–36. doi: 10.1093/nar/28.1.33
Taylor-Robinson, D., and Bébéar, C. (1997). Antibiotic susceptibilities of mycoplasmas and treatment of mycoplasmal infections. J. Antimicrob. Chemother. 40, 622–630. doi: 10.1093/jac/40.5.622
The Gene Ontology (2019). The gene ontology resource: 20 years and still GOing strong. Nucleic Acids Res. 47, D330–D338. doi: 10.1093/nar/gky1055
Wang, Y., Yi, L., Zhang, F., Qiu, X., Tan, L., Yu, S., et al. (2017). Identification of genes involved in Mycoplasma gallisepticum biofilm formation using mini-Tn4001-SGM transposon mutagenesis. Vet. Microbiol. 198, 17–22. doi: 10.1016/j.vetmic.2016.11.021
Yadav, J. P., Singh, Y., Jindal, N., and Mahajan, N. K. (2022). Rapid and specific detection of Mycoplasma gallisepticum and Mycoplasma synoviae infection in poultry using single and duplex PCR assays. J. Microbiol. Methods 192:106365. doi: 10.1016/j.mimet.2021.106365
Yan, J., and Bassler, B. L. (2019). Surviving as a community: antibiotic tolerance and persistence in bacterial biofilms. Cell Host Microbe 26, 15–21. doi: 10.1016/j.chom.2019.06.002
Yu, K., and Zhang, T. (2013). Construction of customized sub-databases from NCBI-nr database for rapid annotation of huge metagenomic datasets using a combined BLAST and MEGAN approach. PLoS One 8:e59831. doi: 10.1371/journal.pone.0083943
Keywords: Mycoplasma gallisepticum, biofilm, formation, drug sensitivity, whole-genome analysis
Citation: Ma X, Wang L, Yang F, Li J, Guo L, Guo Y and He S (2023) Drug sensitivity and genome-wide analysis of two strains of Mycoplasma gallisepticum with different biofilm intensity. Front. Microbiol. 14:1196747. doi: 10.3389/fmicb.2023.1196747
Edited by:
Mohamed Hijri, Montreal University, CanadaReviewed by:
Mangesh Vasant Suryavanshi, Cleveland Clinic, United StatesBidyut Mohapatra, The University of the West Indies, Cave Hill, Barbados
Adolphe Zeze, Félix Houphouët-Boigny National Polytechnic Institute, Côte d'Ivoire
Copyright © 2023 Ma, Wang, Yang, Li, Guo, Guo and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shenghu He, aGVzaGVuZ2h1MzA4QDE2My5jb20=
 Xiaoyan Ma
Xiaoyan Ma Li Wang1
Li Wang1