
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 14 June 2023
Sec. Microbial Physiology and Metabolism
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1195137
This article is part of the Research TopicFunctional Insights into the Probiotic Mechanisms of Surface Protein ActionView all 6 articles
 Yujie Sha1,2*
Yujie Sha1,2* Qingyun Yan3
Qingyun Yan3 Jian Liu1,2*
Jian Liu1,2* Jiafeng Yu1,2
Jiafeng Yu1,2 Shicai Xu1,2
Shicai Xu1,2 Zhili He3
Zhili He3 Jing Ren1,2
Jing Ren1,2 Jie Qu1,2
Jie Qu1,2 Shiying Zheng1,2
Shiying Zheng1,2 Guomin Wang1
Guomin Wang1 Weiying Dong1
Weiying Dong1Clarifying mechanisms underlying the selective adhesion of probiotics and competitive exclusion of pathogens in the intestine is a central theme for shrimp health. Under experimental manipulation of probiotic strain (i.e., Lactiplantibacillus plantarum HC-2) adhesion to the shrimp mucus, this study tested the core hypothesis that homologous genes shared between probiotic and pathogen would affect the adhesion of probiotics and exclusion of pathogens by regulating the membrane proteins of probiotics. Results indicated that the reduction of FtsH protease activity, which significantly correlated with the increase of membrane proteins, could increase the adhesion ability of L. plantarum HC-2 to the mucus. These membrane proteins mainly involved in transport (glycine betaine/carnitine/choline ABC transporter choS, ABC transporter, ATP synthase subunit a atpB, amino acid permease) and regulation of cellular processes (histidine kinase). The genes encoding the membrane proteins were significantly (p < 0.05) up-regulated except those encoding ABC transporters and histidine kinases in L. plantarum HC-2 when co-cultured with Vibrio parahaemolyticus E1, indicating that these genes could help L. plantarum HC-2 to competitively exclude pathogens. Moreover, an arsenal of genes predicted to be involved in carbohydrate metabolism and bacteria-host interactions were identified in L. plantarum HC-2, indicating a clear strain adaption to host’s gastrointestinal tract. This study advances our mechanistic understanding of the selective adhesion of probiotics and competitive exclusion of pathogens in the intestine, and has important implications for screening and applying new probiotics for maintaining gut stability and host health.
Probiotics are defined as active microorganisms that are beneficial to the host, they can confer several benefits such as improving growth, digestion, and immunity (Parker, 1974; Zhang et al., 2018). Probiotics are widely used as alternatives to antibiotics. For example, Lactobacillus (e.g., L. casei, L. gasseri, L. helveticus, L. lactis, L. rhamnosus, L. reuteri, and L. acidophilus) and Lactiplantibacillus (L. plantarum and L. pentosus) have health-promoting effects (Kankainen et al., 2009; Zhang et al., 2018; Partrick et al., 2021; Wang et al., 2021), and are commonly accepted probiotics strains in the market.
Adhesion to the gut is important to the colonization of probiotics, which is conducive to the proliferation of probiotics but harmful to the pathogens in the intestine (Reid and Burton, 2002; Chen et al., 2007; Yadav et al., 2017). The molecules such as surface proteins, cell wall polysaccharides, peptidoglycans, bacteriocins, secreted proteins, and organic acids derived from probiotics Lactobacilli can mediate bacterial adhesion (Teame et al., 2020). The surface protein is one of the major adhesion molecules involved in probiotic mechanisms among different taxonomic groups of probiotics (Sanders et al., 2018). For example, L. casei surface layer protein could reduce the adhesion of pathogens to collagen and HT-29 cells, and thus decrease the pathogens-induced apoptosis of HT-29 cells (Meng et al., 2021). L. acidophilus surface layer protein also presented antibacterial effects against Staphylococcus aureus in combination with nisin (Wang et al., 2020).
Results from our previous studies suggested that the molecules located on the surface of L. plantarum HC-2 might participate in the competitive exclusion of Vibrio parahaemolyticus (Sha et al., 2016a), and the surface layer proteins of L. plantarum HC-2 were subsequently found to play an important role in its adhesion and colonization in the intestine of shrimp to competitively exclude the pathogens and promote beneficial bacteria growth (Du et al., 2019). However, whether other genes or proteins of L. plantarum HC-2 are involved in the adhesion and exclusion of pathogens remains unclear. Probiotics and pathogens compete for common cell receptors for binding to the intestine mucosa (Van den Abbeele et al., 2016; Singh et al., 2018). Thus, understanding the surface proteins of probiotics involved in host-probiotic interaction would provide new strategies for disrupting host-pathogen interaction.
Moreover, L. plantarum HC-2 could reduce the negative effects of aflatoxin by positively affecting the growth and resistance of P. vannamei (Fang et al., 2020). Thus, some other modes of action may exist for L. plantarum HC-2 to find a niche in the shrimp intestine. Different bacterial species may have unique biological activities, which may vary from one strain to another strain. So, the unique property is fundamental to understand and apply a specific probiotic strain (Minj et al., 2021). The characteristics of the genome and proteome could reflect the adaptability and function of the bacteria to specific niches, so that the probiotic properties of L. plantarum HC-2 can be predicted by analyzing its whole genome and proteome (O'sullivan et al., 2009; De Angelis et al., 2016).
This study mainly aimed to investigate which and how the genes and proteins of L. plantarum HC-2 affect the adhesion and exclusion of pathogens. We hypothesized that (i) the homologous genes shared between L. plantarum HC-2 and V. parahaemolyticus would affect the adhesion of L. plantarum HC-2 and competitive exclusion against pathogens in the mucus of shrimp by regulating the quantity of membrane proteins; (ii) the genome of L. plantarum HC-2 harbored key genes contributing to its adaptability to shrimp intestine niche. So, we first tested whether the shared genes between L. plantarum HC-2 and V. parahaemolyticus participated in adhesion of L. plantarum HC-2 to mucus, and examined the proteins involved in the adhesion of L. plantarum HC-2 to mucus and the response to V. parahaemolyticus exposure. Secondly, we analyzed the other probiotic characteristics of L. plantarum HC-2 based on its genome. We found that ftsH gene could affect the adhesion of L. plantarum HC-2 by regulating several membrane proteins, some of which play important roles in the competitive exclusion of pathogens. Arsenal genes in L. plantarum HC-2 genome are predicted to be involved in carbohydrate metabolism and bacteria-host interactions. These findings advance our mechanistic understanding of the selective adhesion of probiotics and competitive exclusion of pathogens in the intestine of shrimp, and have an important practical significance for the screening of probiotics, the optimization of feed additive formulations and the defense against diseases.
Lactiplantibacillus plantarum HC-2 was isolated from the hindgut of fish (Acanthogobius hasta) and stored at −80°C in Man, Rogosa, and Sharpe (MRS) broth (Qingdao Hope Biol-Technology Co., Ltd., Qingdao, China) containing 20% (v/v) glycerol. V. parahaemolyticus E1 was stored at −80°C in 2216E broth (Qingdao Hope Biol-Technology Co., Ltd., Qingdao, China) containing 20% (v/v) glycerol. L. plantarum HC-2 was cultured at 37°C under anaerobic conditions in MRS broth for 18 h, and V. parahaemolyticus E1 was cultured aerobically in 2216E broth at 28°C for 18 h as seed culture.
Lactiplantibacillus plantarum HC-2 was collected by centrifugation (12,000 g) at 4°C for 10 min. The DNA was extracted with a bacterial genomic DNA extraction kit (Tiangen, China) according to the manufacturer’s instructions. The quality of DNA was verified by 1% (w/v) agarose gel electrophoresis and quantified by Qubit® 2.0 Fluorometer (Thermo Scientific). The whole genome was sequenced by PacBio Sequel platform and Illumina NovaSeq PE150 at the Beijing Novogene Bioinformatics Technology Co., Ltd. Genomes were assembled using SMRT Link (v5.0.1). The low-quality reads (< 500 bp) were filtered, and the long reads (> 6,000 bp) were chosen as seed sequences. The sequences ranged from 500 to 6,000 bp were aligned to the seed sequence by Blasr to improve the accuracy. The obtained sequences were then corrected by the arrow algorithm in the variant Caller module of the SMRT Link software, which was then used as reference sequences to blast with Illumina data by bwa (filtered with base minimum mass value of 20, read depth of 4–1,000). The cyclization of chromosomal sequence depending on the overlap between the head and the tail, and the initial site was then corrected by blast with the DNA database. Finally, the plasmid database was used to blast the chromosome and plasmid sequences. The coding genes, transfer RNA genes, and ribosome RNA genes were predicted by GeneMarkSprogram, tRNAscan-SE, and rRNAmmer, respectively. Gene ontology (GO), Cluster of Orthologous Groups (COG) and Kyoto Encyclopedia of Genes and Genomes (KEGG) were used to predict the function of genes by blasting the whole genome (E-value of 1 × 10–5, minimal alignment length percentage of 40%).
A total of 29 genomes of V. parahaemolyticus were found at the 100% assembly level as of December 2018 from the National Center for Biotechnology Information (NCBI) genome database (the identifier for the genome assembly can be found in Supplementary Table S1). The multiple alignments of genomes between L. plantarum HC-2 and V. parahaemolyticus strains was performed using progressive Mauve option in software Mauve (Aaron et al., 2004). The consensus sequences existed in L. plantarum HC-2 and all V. parahaemolyticus strains were used as conserved genomic sequences for subsequent analysis.
A double-crossover mutagenesis was performed to construct the in-frame deletion of ftsH. In brief, two fragments (about 1 Kb) flanking ftsH were amplified independently with primers 464up-F-XhoI and 464up-R, or 464do-F and 464do-R-EcoRI. The obtained fragments were joined together by overlap PCR using primers 464do-F and 464up-R. The resulting fusion fragment was introduced into the temperature sensitive plasmid pG+host9 (Okano et al., 2009) through XhoI and EcoRI. The constructed plasmid was amplified in E. coli DH5α and transformed into L. plantarum HC-2 by electroporation. The transformants were selected by erythromycin (300 μg/mL) at 30°C.
The transformants were then separated as single clones at 37°C overnight. The integration of the plasmid to the genome was verified by primers Test464-F2 and Test464-R2 product length 1 Kb or 3 Kb. The integrated clones were inoculated in medium without erythromycin for 7–10 generations and spread as single clones on non-selective plates. The erythromycin resistance of these clones was confirmed through inoculation on plate with or without erythromycin (300 μg/mL). Clones loosing erythromycin resistance were checked by primers Test464-F2 and Test464-R2 and the resulting length 1 Kb indicated ΔftsH. The specific primers used in present study were listed in Supplementary Table S2.
The preparation of shrimp mucus and adherence experiments were performed as previously study (Sha et al., 2016b). In briefly, the experimental shrimp were obtained after being fed for 4 h from Ruizi Seafood Development Co.Ltd. (Qingdao, China). Crude mucus was obtained from the intestines of shrimp by gentle scraping and was suspended in cold HEPES-Hanks’s Buffer (H-H Buffer). Then, the solution was centrifuged to remove non-soluble material and was diluted with H-H Buffer to a final concentration of 1 mg/mL. 100 μL prepared shrimp mucus was added into the wells of a 96-well micro-titer plate and incubated overnight at 4°C. Then, the wells were washed three times with sterile PBS to remove the non-immobilized mucus. Subsequently, the ΔftsH mutant, ΔftsH-C and wild type bacteria were added to the immobilized mucus and incubated at 37°C for 1 h, respectively. Wells were washed five times with sterile PBS and treated with 0.05% trypsin (100 μL) for 10 min at room temperature to liberate the bacteria. The liberated bacteria were then serially diluted (103 ~ 105) and plated on MRS plates, which incubated at 37°C under anaerobic conditions for 24 h. The number of colony-forming units (CFUs) was counted, and the adhesion ability of each type strain was calculated by normalizing the liberated bacteria CFUs to total CFUs before adhesion.
Cells from L. plantarum HC-2 wild type and mutant were collected at the stationary phase for differential proteome analysis. The bacterial pellets were collected by centrifugation (12,000 g) at 4°C for 10 min, and then transferred to 1.5 mL centrifuge tube with SDT lysis buffer (4% SDS, 10 mM DL-DTT, 100 mM TEAB) before ultrasonication for 5 min on ice. The lysate was centrifuged at 12,000 g for 15 min at 4°C for collecting the supernatant, which was then reduced with 10 mM DTT for 1 h at 56°C. Sufficient iodoacetamide was added to alkylate for 1 h at room temperature in the dark. Subsequently, four volumes pre-cold acetone were added and mixed completely, and the mixture were incubated at −20°C overnight. The protein precipitation was washed with 1 mL pre-cold acetone and then dissolved in dissolution buffer (8 M Urea, 100 mM TEAB, pH = 8.5). Protein concentration of the lysate was determined by Bradford method (Bradford, 1976). For each sample, 20 μg proteins were loaded on the 12% SDS-PAGE. After a concentration with 80 V for 20 min, the protein was separated by 120 V for 90 min, and then visualized using coomassie brilliant blue R-250. The trypsin treatment and desalted were performed as previous study (Zhang et al., 2016).
Peptides were analyzed using an EASY-nLC™ 1,200 nano UHPLC system connected to a Q Exactive™ HF-X mass spectrometer (Thermo Fisher) with ion source of Nanospray Flex™ (ESI). In data dependent mode, the mass spectrometer automatically switched between MS1 and MS2 spectra. MS1 spectra had a full scan mass-to-charge (m/z) range from 350 to 1,500 using a maximum injection time of 20 ms and an automatic gain control (AGC) target value of 3 × 106. Top 40 abundant precursors were isolated, fragmented by higher energy collisional dissociation (HCD) using 27% normalized collision energy (NCE) and analyzed at a resolution of 15,000 with a scan range from 200 to 2,000 m/z with an AGC target value of 1 × 105. The raw data of MS detection was acquired by set the intensity threshold of 2.2 × 104 and the dynamic exclusion parameter of 20 s.
Peptide/protein identification and quantification were performed with Proteome Discover 2.2 (PD2.2, Thermo) by searching the raw data against database consisting of 22,110 sequences obtained from the zip file named Match_result-X101SC21061420_Z01_J001_B1_43 in ProteomeXchange Consortium with the dataset identifier PXD036035. The search parameters were as follows: mass tolerance for precursor ion and product ion was 10 ppm and 0.02 Da, respectively; fixed modification with carbamidomethyl; dynamic modification with methionine oxidation; N-Terminal modification with acetylation; allowance of 2 missed cleavage; identified peptides spectrum matches with a credibility of more than 99% and at least one unique peptide; false discovery rate set as 0.01. The mutant strains with fold changes ≥1.5 or ≤ 0.67, and p value ≤0.05 (t-test) were considered as significantly differentially expressed proteins.
GO and InterPro (IPR) functional analysis were performed by the interproscan software against the non-redundant database (including Pfam, PRINTS, ProDom, SMART, ProSite, PANTHER). The protein family and pathway were analyzed by COG and KEGG. Subcellular localization for each protein was predicted according to GO annotation by UniProt.1 The protein–protein interactions were predicted using the STRING-DB software.2
Establishment of the mono-and co-culture system for L. plantarum HC-2 and V. parahaemolyticus E1 were performed as described previously (Sha et al., 2016a). In briefly, equal volumes of MRS and 2216E broth were mixed, and 150 mL of mixed medium was inoculated with 150 μL of the L. plantarum HC-2 and V. parahaemolyticus E1 seed cultures and incubated at 28°C for 36 h. Monocultures of both strains were prepared as a control. Samples were collected to monitor the adhesion-related gene expression at t = 3, 6, 9, 12, 24, and 36 h, respectively. After the samples were withdrawn, an equal volume of the mixed medium was added to ensure that the volume of the cultures remained at 150 mL.
The methods of RNA isolation, reverse transcription, and quantitative real-time PCR (qPCR) were performed according to previous study (Sha et al., 2016b). In briefly, RNA isolation was performed from bacteria using an RNA fast extraction kit according to the manufacturer’s protocol (Fastagen, China). 2 μg of RNA was used for reverse transcription using a TransScript® One-Step gDNA Removal and cDNA Synthesis Kit according to the manufacturer’s protocol (TransGen Biotech Co., Ltd., China). qPCR was performed using TransStar Top Green qPCR Supermix according to the manufacturer’s protocol (TransGen Biotech Co., Ltd., China) with three replicates of each sample under the following two steps: denaturation at 94°C for 30 s and then 40 cycles of 94°C for 5 s and 60°C for30 s. The specificity of PCR products were confirmed by the dissociation curve analysis at the end of qPCR. Microsoft Excel was used to analyze the data and the expression changes were determined with the 2−ΔΔCt method. MA1 and pvuA were selected as the reference genes for L. plantarum HC-2 and V. parahaemolyticus E1, respectively. Specific primers used in present study were listed in Supplementary Table S2.
All statistical analyses were performed with SPSS (version 20), and student’s t-test was used to analyze differences between two compared groups with a significant level at p ≤ 0.05. PCA analysis was performed with the R package vegan. The graphic works were performed with origin 8.0 or R software.
The raw sequencing data and assembly sequence of L. plantarum HC-2 genome can be found at the National Center for Biotechnology Information (NCBI) (BioProject: PRJNA868485; Sequence Read Archive (SRA): SRR21010423). The mass spectrometry proteomics data of L. plantarum HC-2 have been deposited to the ProteomeXchange Consortium via the PRIDE (PRoteomics IDEntification Database) partner repository with the dataset identifier PXD036035.
The complete genome of L. pentousus HC-2 consists of 3,364,894 nucleotides (one chromosome and two plasmids) with an average GC content of 45.41%. In silico analyses revealed the presence of 3,259 open reading frames (ORFs), resulting in a coding percentage of 84.12%. One or more protein families (PFAM) are attributed to 66.68% of these ORFs; 72.72% of them have similarities (> 97%) to at least one COG; 1.17% of the ORFs remain unknown and 79.78% of them show similarities (> 97%) to defined genes.
All of these circles are fairly symmetrical in the genome by observing the predicted origin of replication through GC-skew analysis and the ORF orientation shift (Supplementary Figure S1, circles 2 to 5 and 8). The five rRNA loci are located in the GC-content spikes (average GC content of 52.66%) (Supplementary Figure S1, circle 6) and all rRNA loci are in the same direction as the DNA replication (Supplementary Figure S1, circle 7). Sixty-four tRNAs are identified in the genome, representing all 20 amino acids, with redundant tRNAs for all amino acids except tryptophan and cysteine.
Nearly 20,000 sequences were obtained which were homologous between L. plantarum HC-2 and at least one of V. parahaemolyticus strains. In order to ensure that the sequences were conserved in both L. plantarum HC-2 and V. parahaemolyticus strains, the presence of homologous sequences in all studies strains was selected as the filter. Seventeen sequences were filtered out which were homologous between L. plantarum HC-2 and all V. parahaemolyticus strains, and then were assigned to 11 genes based on their location in L. plantarum HC-2 genome (Table 1). These genes mainly attribute to the essential cellular metabolic processes such as transcription, translation, glycolysis, and protein stability (Table 1).
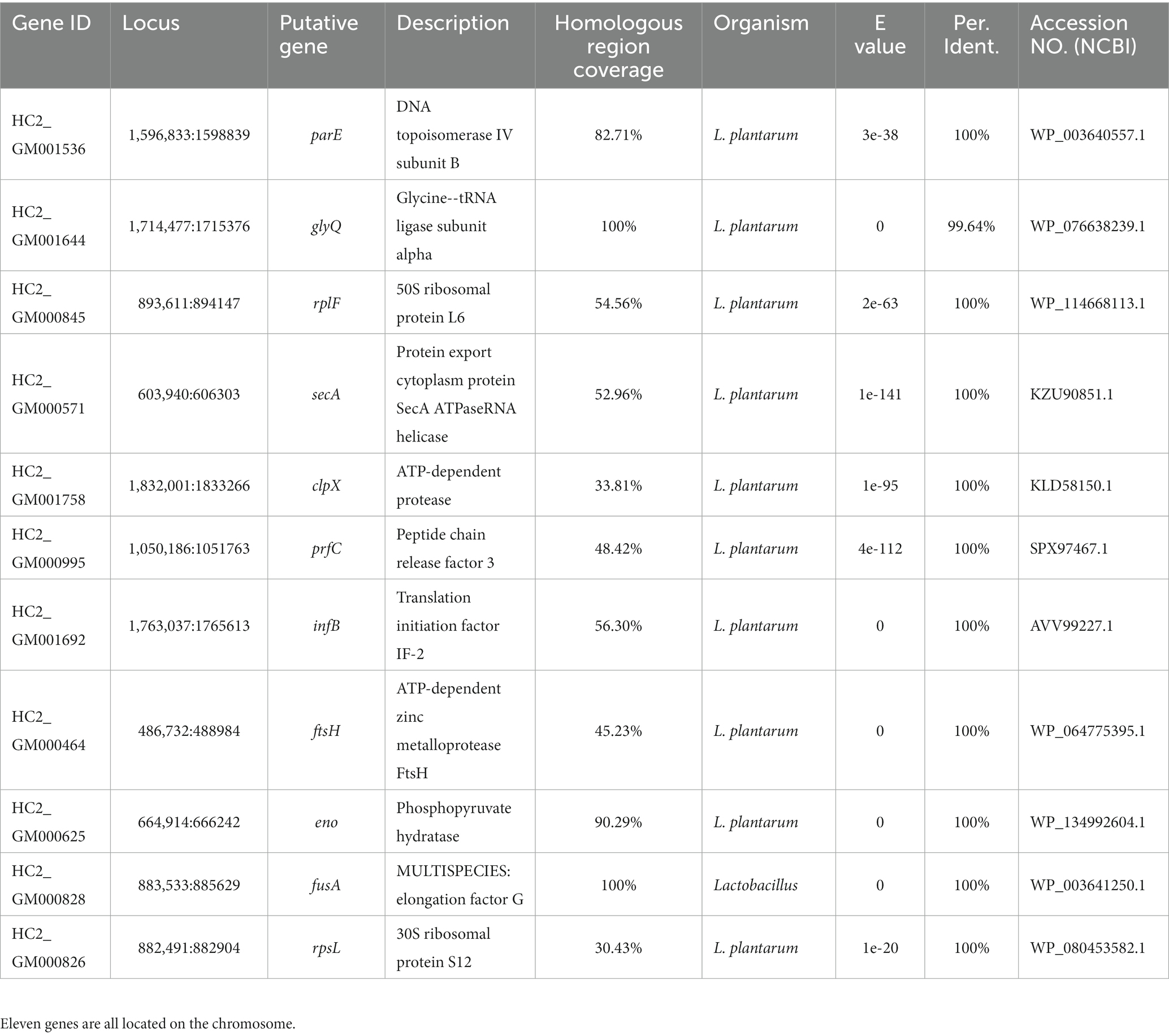
Table 1. Eleven genes of Lactiplantibacillus plantarum HC-2 with homology to all investigated Vibrio parahaemolyticus genomes.
In order to elucidate the role of ftsH in the adhesion of L. plantarum HC-2 to shrimp mucus, a mutant strain was constructed by double-crossover mutagenesis. PCR results confirmed the absence of ftsH in L. plantarum HC-2 genome (data not shown). One mutant was screened out from nearly 5,000 mutants. The ΔftsH mutant exhibited a significantly (p < 0.05) higher adhesion rate than the wild type (more than 2 times) (Figure 1A). When complemented with the ftsH, the adhesion rate nearly restored the level of the wild type (Figure 1A). The results provide evidence that a strong connection between the adhesion ability and FtsH role does exist in L. plantarum.
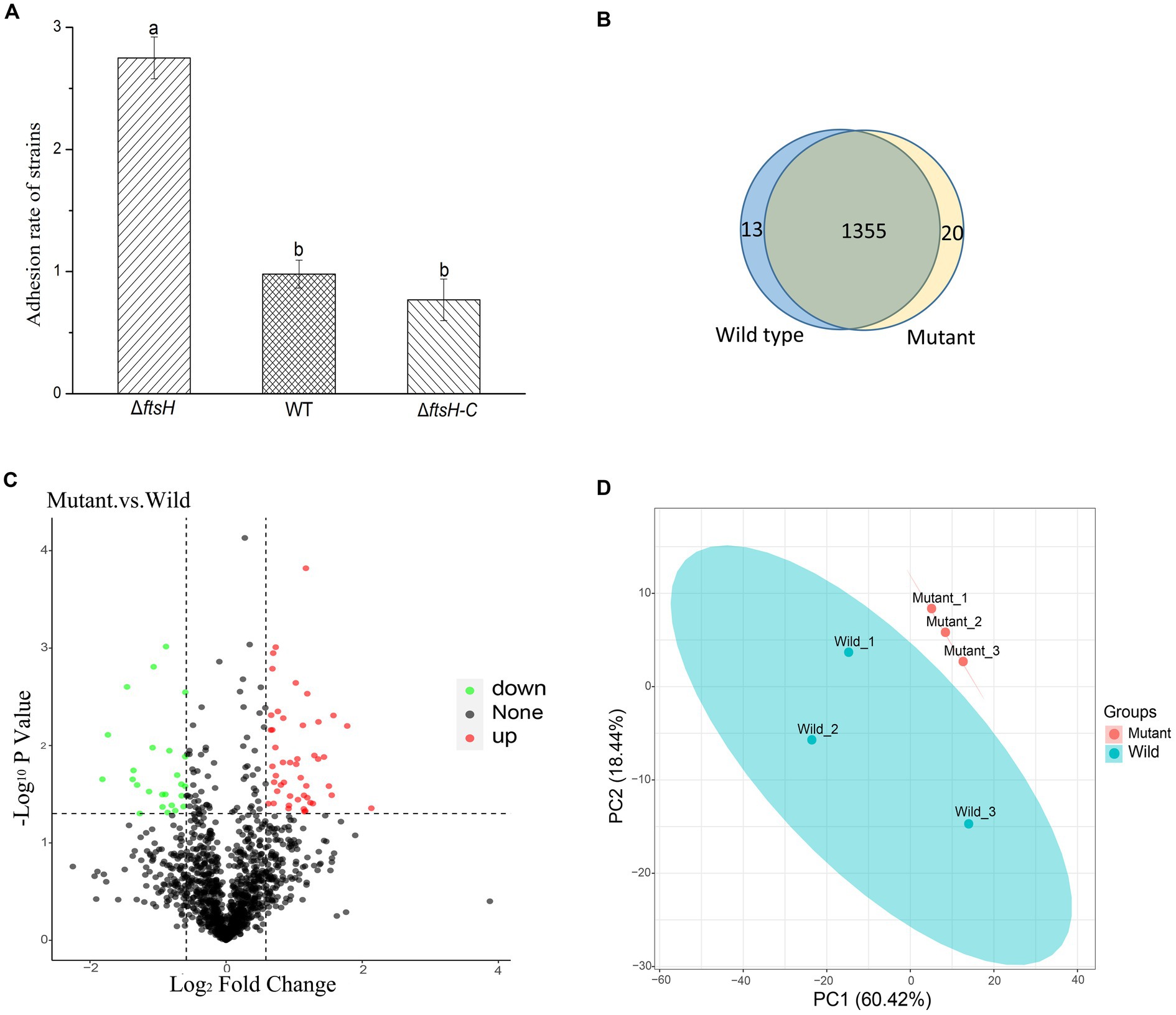
Figure 1. The adhesion of Lactiplantibacillus plantarum HC-2 is affected by FtsH expression. (A) The adhesion rate of wild type (WT), ftsH mutant (ΔftsH), and ftsH mutant complemented with the ftsH (ΔftsH-C). (B) The Veen diagram shows the unique and shared proteins between wild type and mutant groups. (C) Volcano plot of changes at the protein level of L. plantarum HC-2 after FtsH expression decreased. (D) Principal coordinates analysis based on the Unifrac distance.
Because of the regulatory protease activity of FtsH (Bove et al., 2012), we sought to ascertain whether inactivation of ftsH could alter the surface proteins of L. plantarum. A total of 1,369 and 1,376 proteins were detected in the wild type and mutant, respectively (Figure 1B). In addition, we identified that 26 proteins were significantly down-expressed and 54 proteins were significantly up-expressed (Figure 1C). Although the deletion of ftsH inhibited the expression of several genes, the expression of more proteins was up-regulated. The up-regulated proteins mainly belonged to the following categories: (i) translation, ribosomal structure and biogenesis, (ii) general function, (iii) amino acid transport and metabolism, (iv) carbohydrate transport and metabolism, and (v) replication, recombination and repair (Figure 2). The down-regulated proteins were also involved in mostly of the functions indicated above, but the numbers were less than the up-regulated proteins. Global proteomic profiles of the wild type and mutant strains were compared revealing a distinct separation between these two types of strains, suggesting that ftsH deletion induced the expression changes of proteins which may contribute to the increased adhesion ability of L. plantarum HC-2 (Figure 1D). Among the differentially expressed proteins, six proteins were considered to be related to the adhesion of L. plantarum HC-2 due to their higher expression in the membrane of mutant strains (Table 2) and were selected as adhesion-related biomakers.
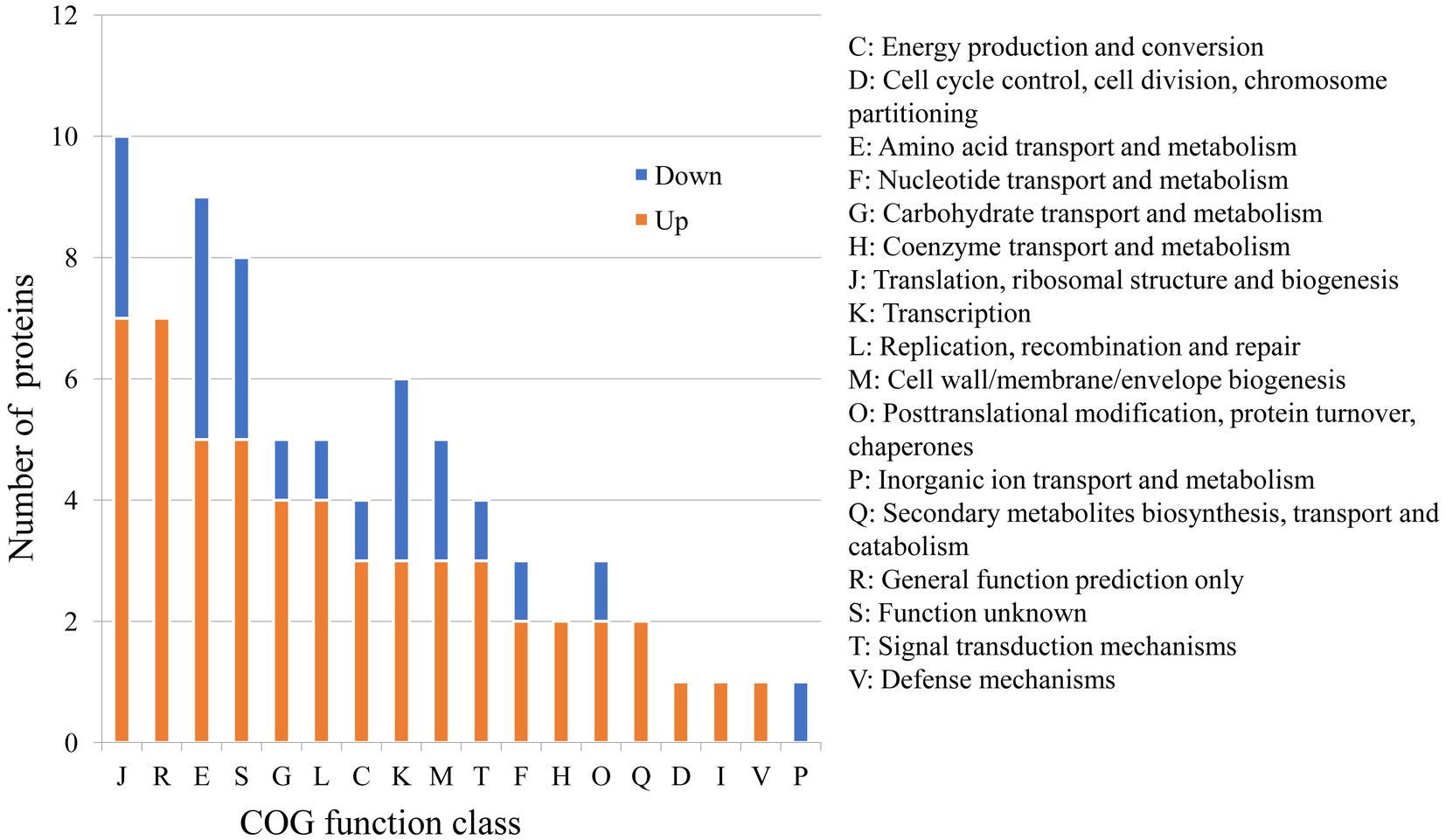
Figure 2. Predicted functions of differentially expressed proteins between wild type and mutant strains based on the Cluster of Orthologous Groups (COG) analysis.
As shown in Figure 3, the expression of genes coding A0A241RSP9 was significantly up-regulated at the presence of V. parahaemolyticus E1 for 3 h. The expression of genes coding G0M1M7, I9L3P8, A0A2S9W1Y1, and A0A241RSP9 were significantly up-regulated when co-cultured with V. parahaemolyticus E1 for 6 h. However, the expression of gene coding I9L3P8 was 8-fold higher than that of the control group. At 12 h, three membrane protein-coding genes (I9L3P8, G0M1M7, A0A2S9W1Y1) were up-regulated, and the expression of I9L3P8 and A0A2S9W1Y1 were 4-fold higher than that of the control group. Thus, the presence of pathogenic bacteria can induce the expression of L. plantarum HC-2 surface-associated proteins.
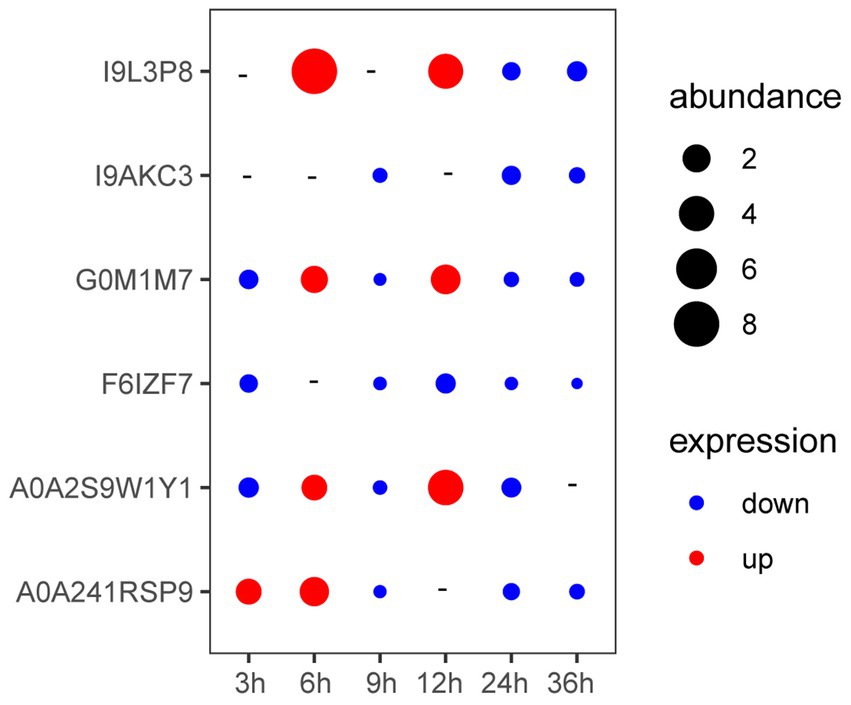
Figure 3. The expression of genes encoding membrane proteins in Lactiplantibacillus plantarum HC-2 when co-cultured with Vibrio parahaemolyticus E1 at different time points. “-” means no change. “abundance” means the expression fold-changes of the target gene relative to the reference gene.
Lactiplantibacillus plantarum HC-2 has been shown to be able to ferment in vitro a variety of carbohydrates (Sha, 2016). Among them, Methyl-α-D-Mannopyranoside, N-acetylglucosamine, amygdalin, Arbutin, Esculin ferric citrate, salicin, D-lactose, D-melibiose and D-melezitose could be utilized by L. plantarum HC-2 as the only carbon source, but the related genes were not found in the genome of L. plantarum HC-2, which need to be further investigated. In addition, in silico analysis of the annotated genome sequence of L. plantarum HC-2 also predicted its capacity to ferment mannitol, inositol and glycogen.
Seven percent of the identified genes in the L. plantarum HC-2 genome were involved in carbohydrate metabolism. The analysis of BlastKOALA predicted that L. plantarum HC-2 possessed three complete carbohydrate metabolism pathways involving EMP, pentose phosphate pathway and galactose metabolism (Figure 4), these pathways formed the central core of carbohydrate metabolism of L. plantarum HC-2. In addition, 15 carbohydrate utilization pathways were predicted in the genome of L. plantarum HC-2. As such, the wide repertoire of enzymes involved in carbohydrate metabolism are found in its genome, which are also corroborated by the abundant number of genes for the phosphoenolpyruvate- (PEP) dependent sugar phosphotransferase system (PTS) (67 genes) and the presence of specific genes or gene clusters involved in carbohydrate utilization by L. plantarum HC-2 (Figure 5).
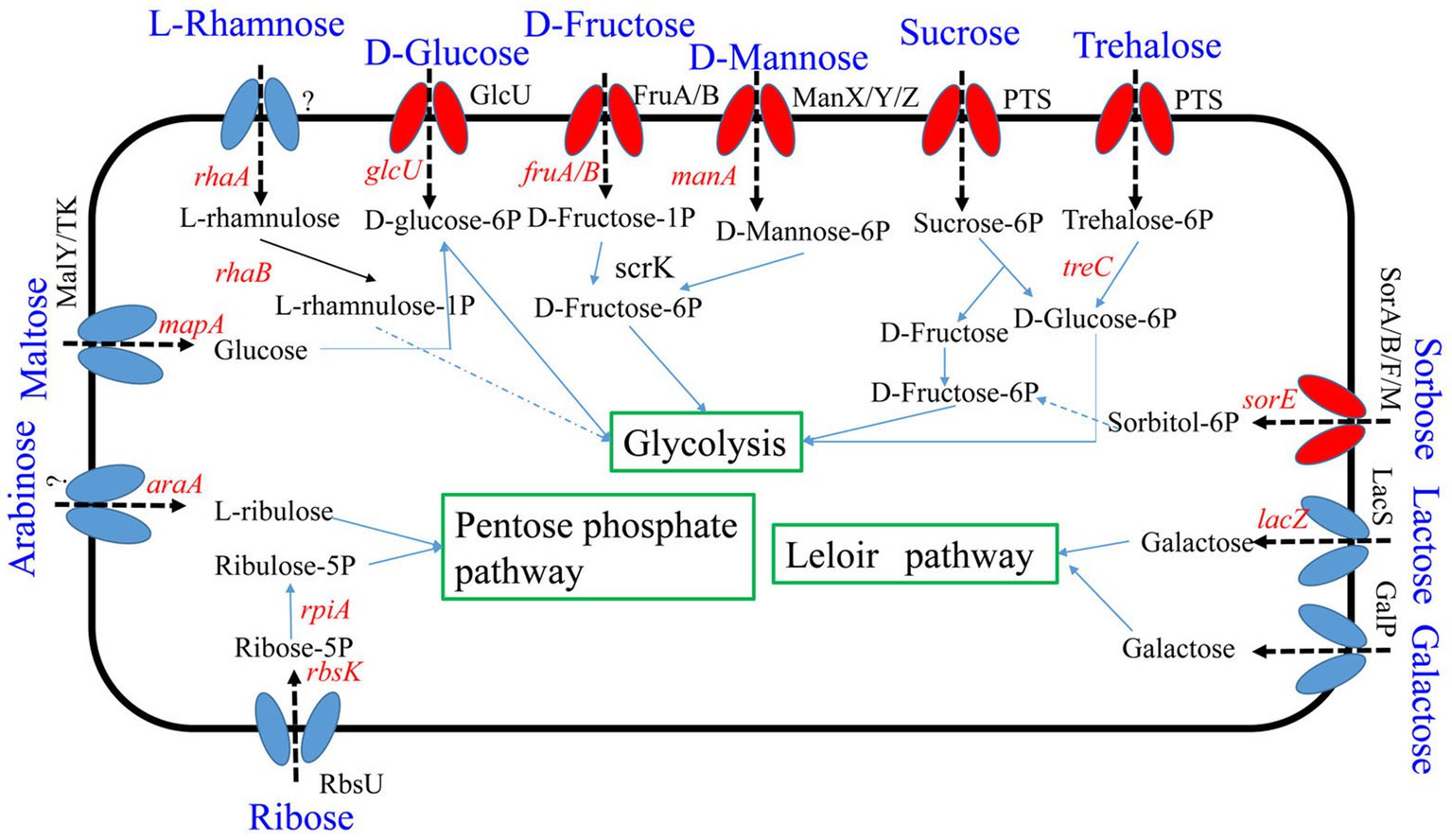
Figure 4. Predicted carbohydrate metabolism pathways of Lactiplantibacillus plantarum HC-2. Red: phosphotransferase system; blue: permease protein.
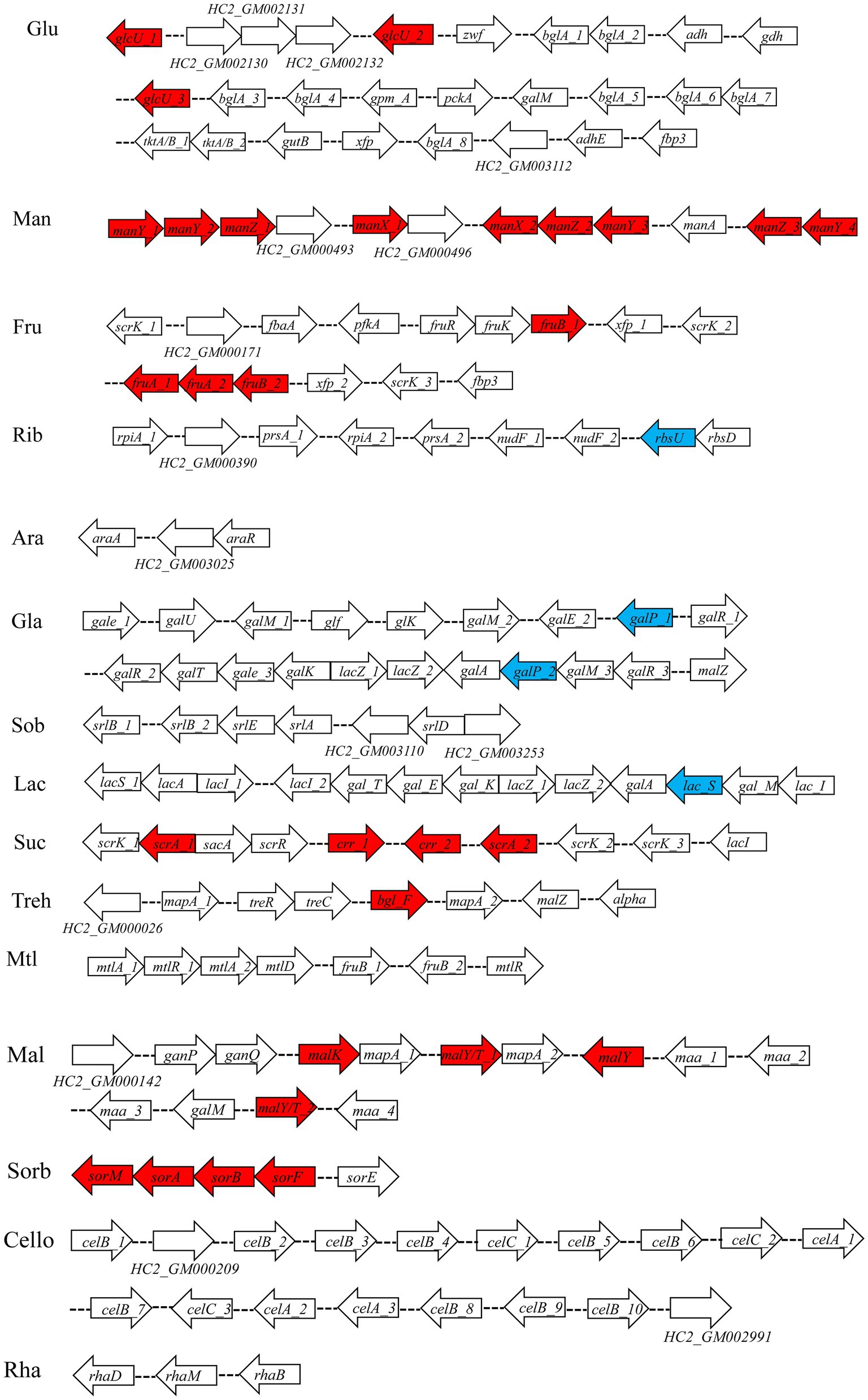
Figure 5. Predicted gene clusters involved in major carbohydrate metabolism. Red: phosphotransferase system; blue: permease protein. Glu, glucose; Man, mannose; Fru, fructose; Rib, ribose; Ara, arabinose; Gla, galactose; Sob, sorbitol; Lac, lactose; Suc, sucrose; Treh, treholose; Mtl, mannitol; Mal, maltose; Sorb, Sorbose; Cello, cellobiose; Rha, rhamnose.
The adhesion genes of probiotic L. plantarum HC-2 were also analyzed, combined with annotation and published studies (Van Tassell and Miller, 2011; Abriouel et al., 2017). A total of 34 adhesion genes were predicted in the genome of L. plantarum HC-2 (Table 3).
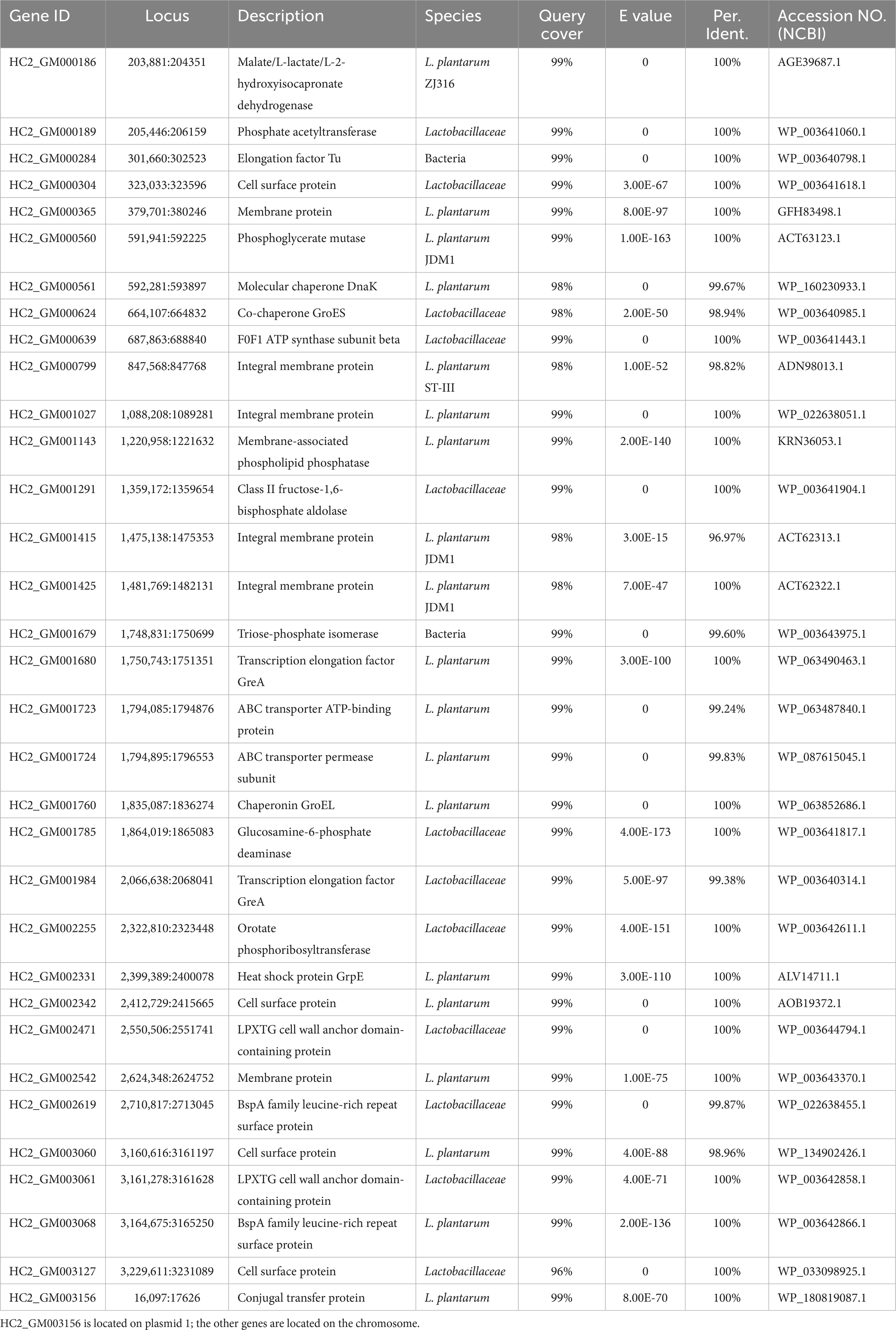
Table 3. Predicted genes encoding proteins with roles in adhesion or interaction with the host in the genome of Lactiplantibacillus plantarum HC-2.
Probiotics are widely used as a kind of alternatives to antibiotics (Parker, 1974; Zhang et al., 2018). Adhesion of probiotics to the gut mucus is the first step to exert different effects against the activity of pathogens (e.g., competition, exclusion, displacement, and repression) (Reid and Burton, 2002; Chen et al., 2007; Yadav et al., 2017). This study clarified the genes and proteins of L. plantarum HC-2 involved in adhesion to shrimp intestine and competitive exclusion against pathogenic V. parahaemolyticus. The results generally supported our core hypothesis that the shared ftsH gene could influence the adhesion of L. plantarum HC-2 to the shrimp mucus by regulating membrane proteins. Also, the genome of L. plantarum HC-2 harbors many genes relating to carbohydrate utilization and probiotic-host interactions.
Probiotic bacteria adhering to the gut may compete the colonized niches with pathogens, and thus prevent pathogen infestation (Servin, 2004; Fukuda, 2017). The major driving forces for enhancing bacterial adhesion are protein–protein and protein-carbohydrate interactions. The bacteria mainly share molecular mechanisms of initial adhesion to the host, and the presence of common adhesion proteins on the surface of bacteria could affect the adhesion-related molecules (Westerlund and Korhonen, 1993; Pizarro-Cerdá and Cossart, 2006; Kline et al., 2009; Iniguez-Palomares et al., 2011; Singh et al., 2018). L. plantarum HC-2 and V. parahaemolyticus can be adhered to the intestine of Litopenaeus vannamei (Sha et al., 2016a), thus the adhesion-related proteins encoded by homologous genes of these strains may compete for adhesion niches in the intestine as they may play a common functional role (Edgar and Batzoglou, 2006). In this study, homologous sequences obtained among L. plantarum HC-2 and all V. parahaemolyticus genomes were finally assigned to 11 genes which can attribute to the essential cellular metabolic processes. Several of them show multiple functions in different subcellular localization, like proteins related to transcription and translation which not only attribute to the essential cellular metabolic processes in cells but also act as adhesins when emerged on the cell surface of bacteria, such as genes numbered HC2_GM000845 (rplF), HC2_GM000995 (prfC), and HC2_GM000828 (fusA). Moreover, the gene numbered HC2_GM000464 was annotated as ftsH and was found that there was only one copy in this genome. The ftsH gene was firstly found in Escherichia coli and then proved in almost all bacteria to perform cellular roles for FtsH (Schumann, 1999). FtsH is a ubiquitous membrane-integrated protein, an ATP-and Zn2+-dependent metalloprotease, which belongs to ATPase associated with different cellular activities (AAA) protein family and in responsible for regulating the protein stability of specific and critical regulators to control cellular processes. Given the critical role of FtsH in cellular processes, the ftsH gene was selected as the target gene for mining adhesion-related biomarkers among 11 genes. In this study, the double-crossover mutagenesis was performed and obtained nearly 5,000 mutants one ftsH mutant. The number of mutants showed desired genotype only 0.02% due to the fact that the second homologous crossover is random. Thus, it is difficult to obtain the desired genotype at the absence of selection pressure, and a suitable method should be developed to get target mutagenesis of L. plantarum.
FtsH protease is very important for controlling the membrane protein, heat shock response, lipopolysaccharide biosynthesis, viral infection, and mediating colicin activity in E. coli (Schumann, 1999), and it could also regulate the cytoplasmic substrates in Caulobacter crescentus and Bacillus subtilis (Langklotz et al., 2012). However, the FtsH is dispensable for normal growth of bacteria except that survival upon stress conditions like heat shock or osmotic stress (Beier et al., 1997; Deuerling et al., 1997; Fischer et al., 2002; Lithgow et al., 2004; Fiocco et al., 2009; Langklotz et al., 2012). In L. plantarum, the FtsH mutant reduced the capacity to form biofilms and changed the physico-chemical surface properties, which possibly linked to the alterations of surface charge and/or architecture (Bove et al., 2012). Given that in present study, the bacteria were washed three times with phosphate buffer solution before being used to test their adhesion to mucus, and there was basically no difference in charge on the surface of cells. However, we found that the FtsH mutant increased the adhesion ability to the mucus (Garrett et al., 2008). This is consistent with previous studies, which identified that the down-expression of FtsH could promote biofilm formation in L. plantarum and E. coli (Langklotz et al., 2012; Gu et al., 2021). Therefore, the absence of ftsH in L. plantarum HC-2 might mainly alter the membrane architecture depending on the regulatory protease activity of FtsH (Garrett et al., 2008; Langklotz et al., 2012; Gu et al., 2021).
The obtained mutant strains increased the adhesion ability to mucus of shrimp, and comparative proteomic analysis was further performed to create a molecular map of the adhesion regulation processes. 1355 proteins were present in both groups, while 13 and 20 proteins were uniquely identified in wild type and mutant strains, respectively (Figure 1B). There was basically no change in the number of total proteins expressed, but the number of uniquely expressed proteins exceeded 1% in each group. A total of 80 differential expressed proteins were found, including 54 up-regulated proteins. Many increased functional activities in the mutant strains reveal that the improvement of L. plantarum HC-2 adhesion requires the mobilization of its more basic biological and cellular processes. In addition, the proteomic results showed that the expression of FtsH was still existed in the mutant strains with 10.56% down-regulation (data not shown) and showed no significant difference from the wild type strain, suggesting that the mutants were partially repaired. This speculation could be supported by the elevated abundance of proteins for reducing damage such as proteins involved in enzyme metabolism and DNA repair pathways (Figure 2). L. acidophilus also responded to oxidative stress in the similarly way (Calderini et al., 2017). However, further studies are needed to clarify how the missing ftsH gene is repaired. A 10.56% decrease in FtsH expression already altered the adhesion ability of this group, so the results in this study can be used for subsequent analysis to test our hypotheses.
Surface proteins play an important role in the adhesion of bacteria to mucus in two ways. The first one is that surface proteins are bound to the membrane by hydrophobic transmembrane domain(s) and lipoprotein, and the second is that surface proteins are attached to the cell surface by noncovalent or recognizing some cell-wall-binding domains (e.g., LPXTG, CWBD1, and GW proteins) (Deepika and Charalampopoulos, 2010). We found that the differentially expressed proteins were predicted to be located in the cytoplasm and membrane rather than the cell wall, suggesting that FtsH mainly affects the adhesion of L. plantarum by regulating the expression of the membrane proteins.
Cell division inhibitor MinD (protein ID: G0M1M7) participates in the division of cell, and the overexpression of MinD could result in filamentation of cell by enhancing MinC-mediated inhibition of cell division (Nguyen et al., 2013). However, the minC gene was identified as antimicrobial activities other than being involved in cell division in L. acidophilus (Nguyen, 2012; Nguyen et al., 2013). Thus, the members of cell division inhibitors may play different roles in different strains. In this study, the increased MinD in membrane may result in filamentation of cell which may participate in the adhesion as an anchoring site for other adhesion molecules. The protein I9L3P8 encoded by choS was reported to have a compatible solutes glycine betaine/carnitine/choline transport system (Rossi et al., 2016). Moreover, choline was found to be a direct precursor of phosphatidylcholine, which was a major lipid membrane component in the Rhizobiaceae family (Dupont et al., 2004). The choS was also involved in tolerance to stress from the external environment (Rossi et al., 2016), so the higher expression of gene encoding for I9L3P8 may be a response to protect itself against pathogens and enhance adhesion to the host. The A0A2S9W1Y1 is annotated as ATP synthase subunit a, which is a membrane-bound stator subunit of the ATP synthase and is essential for proton translocation (Ishmukhametov et al., 2017). These three proteins were significantly up-regulated in L. plantarum HC-2 when it cocultured with V. parahaemolyticus E1 for 6 h and 12 h while down-regulated at other time points. Biological membranes are a mix of membrane proteins and their lipid partners. One potential consequence of membrane crowding is that this conformational change could cause a change in free energy that is associated with the area available to the rest of the proteins, resulting in depletion forces (Phillips et al., 2009). These changes play a critical role in sensory and signaling pathways. Therefore, during this experiment, it was observed that the expression of genes can at times be up or down. The similar gene-expression patterns were also found in Pseudomonas.syringae pv. actinidiae during the first five days after infection of kiwifruit (McAtee et al., 2018), in which the expression of several genes from the plasmid-borne putative aromatic pathway showed a fluctuating pattern over the infection time course. In addition, compared with V. parahaemolyticus E1 after incubation alone, aphA in V. parahaemolyticus E1 was also observed to be up-regulated at 2 h and 6 h but down-regulated at 4 h and 8 h when V. parahaemolyticus E1 was cocultured with the same number of its own cells at 28°C (Sha et al., 2016a). The probiotics and pathogens share similar nutritional substrates, especially the amino acids. Thus, the gene encoding amino acid permeases (A0A241RSP9) was induced at earlier time. As above mentioned, four proteins (G0M1M7, I9L3P8, A0A2S9W1Y1, A0A241RSP9) may be involved in the competitive exclusion against pathogens in the mucus of shrimp by regulating their quantity of proteins on the surface of L. plantarum HC-2.
The application of strains is benefit from their adaption to the host intestinal environment and interaction with the host. As a kind of facultatively heterofermentative LAB, L. plantarum HC-2 genome possesses genes for both the phosphoketolase and Embden-Meyerhof pathways (EMP). Thus, it mainly ferments carbohydrate via EMP by using glucose. However, in the absence of six-carbon sugars, it can also ferment five-carbon carbohydrates such as arabinose or ribose via the phosphoketolase pathway (Bustos et al., 2005; Abriouel et al., 2017). As animals have limited evolved abilities to digest complex polysaccharides (Cantarel et al., 2012), the enzymes produced by intestinal microorganisms are needed to metabolize carbohydrate (e.g., xylan, β-glucan, cellulose, and chitosan). The genes encoding carbohydrate-modifying enzymes could be used to predict the possible adaption and enrichment of bacteria in the intestine (Abriouel et al., 2017). In present study, many enzymes involved in degrading of xylan and β-glucan are found in L. plantarum HC-2 genome. As the accessory enzymes for the degradation of xylan, acetyl xylan esterase (four genes, Table 3), glycerol-3-phosphate cytidylyltransferase, eukaryotic-like serine/threonine-protein kinase, xylan endo-1,3-beta-xylosidase activity and xylanase were predicted in L. plantarum HC-2 genome sequence, which can remove the terminal acetate residues from xylan polymers and act in the xylan catabolic pathway (Kim et al., 2020). Furthermore, 12 genes coding for enzymes involved in the degradation of beta-glucan were also predicted in L. plantarum HC-2 (Supplementary Table S3). Xylan occurs in nearly all plants and some marine algae, and could act as immune modulators to enhance animal immunity (Yang et al., 2019). However, as a kind of soluble non-starch polysaccharides (SNPs), the presence of xylan in diet could also interfere with feed utilization and negatively affect animal performance (Sinha et al., 2011; Yao et al., 2019). Beta-glucan often used as a feed additive in aquaculture, which can enhance fish and shrimp innate immune responses (Chang et al., 2003; Yamamoto et al., 2018), but it is also a kind of SNPs. Therefore, the presence of these enzymes is predicted to contribute to the degradation of xylan and beta-glucan in the intestine. Interestingly, the L. plantarum HC-2 genome has 22 genes responsible for the cellulose synthesis, including cellulose synthase (Supplementary Table S3). The cellulose synthase (17 genes) could accumulate cellulose on the cell wall surface as an extracellular matrix for adhesion of cell and formation of biofilm for protecting the bacteria (Abriouel et al., 2017). Over 7% (229 of 3,259 genes) of the identified genes in L. plantarum HC-2 are involved in carbohydrate metabolism, which is similar to the genome of L. pentosus MP-10 (Abriouel et al., 2017). The abundance of carbohydrate metabolism genes is important for L. plantarum HC-2 to adapt the intestinal environment, which may enhance its possibilities of survival and persistence and competitiveness to pathogens within the intestine of host.
Probiotic lactobacilli can express adhesion-related proteins to mimic the colonization process of pathogens, resulting in several probiotic effects. The identification and characterization of these proteins, often strain-specific, involved in interaction with the host are necessary to evaluate a priori the probiotic potential of Lactobacillus sp. Candidates (Abriouel et al., 2017). In this study, the possible interaction of L. plantarum HC-2 with the host was bioinformatically predicted from the genome sequence. Several adhesion-related proteins were predicted, such as cell surface proteins, LPXTG cell wall anchor domain-containing proteins and elongation factors. However, the role of these proteins in L. plantarum HC-2 has not yet been determined, requiring further mutation or proteomic studies for this purpose.
In summary, this study identified and clarified the underlying mechanisms of genes and proteins of L. plantarum HC-2 involved in adhesion to intestine and competitive exclusion against pathogenic V. parahaemolyticus. The findings advance our mechanistic understanding of the selective adhesion of probiotics and competitive exclusion of pathogens. Also, the repertoire of genes identified in the L. plantarum HC-2 genome have an important implications for its applications in other animals as a potential probiotic.
The strain HC-2 was previously identified as Lactobacillus pentosus (now NCBI changed the name of Lactobacillus pentosus to Lactiplantibacillus pentosus) based on the analysis of partial 16S rRNA sequence which was more similar to L. pentosus JCM 1558, and therefore named as L. pentosus HC-2. However, now the complete genome of strain HC-2 was obtained, and the phylogenetic tree was constructed based on the genomes of strain HC-2, 5 strains from L. plantarum and 5 strains from L. pentosus (Supplementary Figure S2). The result showed that HC-2 clustered well with L. plantarum strains, so the strain HC-2 was renamed as L. plantarum HC-2 from now on. This result was also confirmed by calculating ANI (Average Nucleotide Identity)3 of strain HC-2 with L. plantarum or L. pentosus and estimating DNA–DNA relatedness using the Genome-to-Genome Distance Calculator (GGDC) 3.0 provided by the DSMZ4 (Supplementary Table S4).
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, SRR21010423. http://www.proteomexchange.org/, PXD036035. https://www.ncbi.nlm.nih.gov/, PRJNA868485.
The animal study was reviewed and approved by Science and Technology Ethics Committee of Dezhou University (Dezhou University, Dezhou, China).
YS: conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, resources, supervision, visualization, writing—original draft, and writing—review and editing. QY: formal analysis, writing—original draft, and visualization. JL: methodology and funding acquisition. JY: conceptualization and writing—review and editing. SX: writing—review and editing. ZH: writing—review and editing. JR: conceptualization and writing—review and editing. JQ: conceptualization and writing—review and editing. SZ: writing—review and editing. GW: data curation and methodology. WD: data curation and methodology. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (31802309), the Natural Science Foundation of Shandong Province (CN) (ZR2017BC027), Research Foundation for Advanced Talents of Dezhou University (2019xgrc35).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1195137/full#supplementary-material
Aaron, C. E., Darling, B. M., Blattner, F. R., and Perna, N. T. (2004). Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403. doi: 10.1101/
Abriouel, H., Perez Montoro, B., Casimiro-Soriguer, C. S., Perez Pulido, A. J., Knapp, C. W., Caballero Gomez, N., et al. (2017). Insight into potential probiotic markers predicted in Lactobacillus pentosus MP-10 genome sequence. Front. Microbiol. 8:891. doi: 10.3389/fmicb.2017.00891
Beier, D., Spohn, G., Rappuoli, R., and Scarlato, V. (1997). Identification and characterization of an operon of Helicobacter pylori that is involved in motility and stress adaptation. J. Bacteriol. 179, 4676–4683. doi: 10.1128/jb.179.15.4676-4683.1997
Bove, P., Capozzi, V., Garofalo, C., Rieu, A., Spano, G., and Fiocco, D. (2012). Inactivation of the ftsH gene of Lactobacillus plantarum wcfs1: effects on growth, stress tolerance, cell surface properties and biofilm formation. Microbiol. Res. 167, 187–193. doi: 10.1016/j.micres.2011.07.001
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Bustos, G., Moldes, A. B., Cruz, J. M., and Domínguez, J. M. (2005). Influence of the metabolism pathway on lactic acid production from hemicellulosic trimming vine shoots hydrolyzates using Lactobacillus pentosus. Biotechnol. Prog. 21, 793–798. doi: 10.1021/bp049603v
Calderini, E., Celebioglu, H. U., Villarroel, J., Jacobsen, S., Svensson, B., and Pessione, E. (2017). Comparative proteomics of oxidative stress response of Lactobacillus acidophilus NCFM reveals effects on DNA repair and cysteine de novo synthesis. Proteomics 17:178. doi: 10.1002/pmic.201600178
Cantarel, B. L., Lombard, V., and Henrissat, B. (2012). Complex carbohydrate utilization by the healthy human microbiome. PLoS One 7:e28742. doi: 10.1371/journal.pone.0028742
Chang, C.-F., Su, M.-S., Chen, H.-Y., and Liao, I.-C. (2003). Dietary β-1, 3-glucan effectively improves immunity and survival of Penaeus monodon challenged with white spot syndrome virus. Fish Shellfish Immunol. 15, 297–310. doi: 10.1016/S1050-4648(02)00167-5
Chen, X., Xu, J., Shuai, J., Chen, J., Zhang, Z., and Fang, W. (2007). The s-layer proteins of Lactobacillus crispatus strain zj001 is responsible for competitive exclusion against Escherichia coli o157: H7 and Salmonella typhimurium. Int. J. Food Microbiol. 115, 307–312. doi: 10.1016/j.ijfoodmicro.2006.11.007
De Angelis, M., Calasso, M., Cavallo, N., Di Cagno, R., and Gobbetti, M. (2016). Functional proteomics within the genus Lactobacillus. Proteomics 16, 946–962. doi: 10.1002/pmic.201500117
Deepika, G., and Charalampopoulos, D. (2010). Surface and adhesion properties of lactobacilli. Adv. Appl. Microbiol. 70, 127–152. doi: 10.1016/s0065-2164(10)70004-6
Deuerling, E., Mogk, A., Richter, C., Purucker, M., and Schumann, W. (1997). The ftsH gene of Bacillus subtilis is involved in major cellular processes such as sporulation, stress adaptation and secretion. Mol. Microbiol. 23, 921–933. doi: 10.1046/j.1365-2958.1997.2721636.x
Du, Y., Zhou, S., Liu, M., Wang, B., Jiang, K., Fang, H., et al. (2019). Understanding the roles of surface proteins in regulation of Lactobacillus pentosus HC-2 to immune response and bacterial diversity in midgut of Litopenaeus vannamei. Fish Shellfish Immunol. 86, 1194–1206. doi: 10.1016/j.fsi.2018.12.073
Dupont, L., Garcia, I., Poggi, M. C., Alloing, G., Mandon, K., and Le Rudulier, D. (2004). The Sinorhizobium meliloti abc transporter cho is highly specific for choline and expressed in bacteroids from Medicago sativa nodules. J. Bacteriol. 186, 5988–5996. doi: 10.1128/JB.186.18.5988-5996.2004
Edgar, R. C., and Batzoglou, S. (2006). Multiple sequence alignment. Curr. Opin. Struct. Biol. 16, 368–373. doi: 10.1016/j.sbi.2006.04.004
Fang, H., Wang, B., Jiang, K., Liu, M., and Wang, L. (2020). Effects of Lactobacillus pentosus HC-2 on the growth performance, intestinal morphology, immune-related genes and intestinal microbiota of Penaeus vannamei affected by aflatoxin B1. Aquaculture 525:735289. doi: 10.1016/j.aquaculture.2020.735289
Fiocco, D., Collins, M., Muscariello, L., Hols, P., Kleerebezem, M., Msadek, T., et al. (2009). The Lactobacillus plantarum ftsH gene is a novel member of the CtsR stress response regulon. J. Bacteriol. 191, 1688–1694. doi: 10.1128/JB.01551-08
Fischer, B., Rummel, G., Aldridge, P., and Jenal, U. (2002). The FtsH protease is involved in development, stress response and heat shock control in Caulobacter crescentus. Mol. Microbiol. 44, 461–478. doi: 10.1046/j.1365-2958.2002.02887.x
Fukuda, K. (2017). Is it feasible to control pathogen infection by competitive binding of probiotics to the host? Virulence 8, 1502–1505. doi: 10.1080/21505594.2017.1382798
Garrett, T. R., Bhakoo, M., and Zhang, Z. (2008). Bacterial adhesion and biofilms on surfaces. Prog. Nat. Sci. 18, 1049–1056. doi: 10.1016/j.pnsc.2008.04.001
Gu, Y., Tian, J., Zhang, Y., Wu, R., Li, L., Zhang, B., et al. (2021). Dissecting signal molecule AI-2 mediated biofilm formation and environmental tolerance in Lactobacillus plantarum. J. Biosci. Bioeng. 131, 153–160. doi: 10.1016/j.jbiosc.2020.09.015
Iniguez-Palomares, C., Jimenez-Flores, R., Vazquez-Moreno, L., Ramos-Clamont-Montfort, G., and Acedo-Felix, E. (2011). Protein-carbohydrate interactions between Lactobacillus salivarius and pig mucins. J. Anim. Sci. 89, 3125–3131. doi: 10.2527/jas.2010-2996
Ishmukhametov, R. R., DeLeon-Rangel, J., Zhu, S., and Vik, B. S. (2017). Analysis of an N-terminal deletion in subunit a of the Escherichia coli ATP synthase. J. Bioenerg. Biomembr. 49, 171–181. doi: 10.1007/s10863-017-9694-z
Kankainen, M., Paulin, L., Tynkkynen, S., von Ossowski, I., Reunanen, J., Partanen, P., et al. (2009). Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human-mucus binding protein. P. Natl. A. Sci. 106, 17193–17198. doi: 10.1073/pnas.0908876106
Kim, M.-J., Jang, M.-U., Nam, G.-H., Shin, H., Song, J.-R., and Kim, T.-J. (2020). Functional expression and characterization of acetyl xylan esterases ce family 7 from Lactobacillus antri and Bacillus halodurans. J. Microbiol. Biotechnol. 30, 155–162. doi: 10.4014/jmb.2001.01004
Kline, K. A., Fälker, S., Dahlberg, S., Normark, S., and Henriques-Normark, B. (2009). Bacterial adhesins in host-microbe interactions. Cell Host Microbe 5, 580–592. doi: 10.1016/j.chom.2009.05.011
Langklotz, S., Baumann, U., and Narberhaus, F. (2012). Structure and function of the bacterial AAA protease FtsH. Biochim. Biophys. Acta 1823, 40–48. doi: 10.1016/j.bbamcr.2011.08.015
Lithgow, J. K., Ingham, E., and Foster, S. J. (2004). Role of the hprT–ftsH locus in Staphylococcus aureus. Microbiology 150, 373–381. doi: 10.1099/mic.0.26674-0
McAtee, P., Brian, L., Curran, B., Linden, O., Nieuwenhuizen, J., and Chen, X. (2018). Re-programming of Pseudomonas syringae pv. Actinidiae gene expression during early stages of infection of kiwifruit. BMC Genomics 19:822. doi: 10.1186/s12864-018-5197-5
Meng, J., Wang, Y.-Y., and Hao, Y.-P. (2021). Protective function of surface layer protein from Lactobacillus casei fb05 against intestinal pathogens in vitro. Biochem. Biophys. Res. Commun. 546, 15–20. doi: 10.1016/j.bbrc.2021.01.101
Minj, J., Chandra, P., Paul, C., and Sharma, R. K. (2021). Bio-functional properties of probiotic Lactobacillus: current applications and research perspectives. Crit. Rev. Food Sci. Nutr. 61, 2207–2224. doi: 10.1080/10408398.2020.1774496
Nguyen, T. (2012). Cell division inhibitor minC from Lactobacillus acidophilus VTCC-B-871 and detection of antimicrobial activities in functional study. Afr. J. Pharm. Pharmaco. 6, 3293–3298. doi: 10.5897/ajpp12.1476
Nguyen, T. H., Doan, V. T., Ha, L. D., and Nguyen, H. N. (2013). Molecular cloning, expression of mind gene from Lactobacillus acidophilus VTCC-B-871 and analyses to identify Lactobacillus rhamnosus pn04 from Vietnam hottuynia cordata thunb. Indian J. Microbiol. 53, 385–390. doi: 10.1007/s12088-013-0384-1
Okano, K., Zhang, Q., Shinkawa, S., Yoshida, S., Tanaka, T., Fukuda, H., et al. (2009). Efficient production of optically pure D-lactic acid from raw corn starch by using a genetically modified L-lactate dehydrogenase gene-deficient and α-amylase-secreting Lactobacillus plantarum strain. Appl. Environ. Microbiol. 75, 462–467. doi: 10.1128/AEM.01514-08
O'sullivan, O., O'Callaghan, J., Sangrador-Vegas, A., McAuliffe, O., Slattery, L., Kaleta, P., et al. (2009). Comparative genomics of lactic acid bacteria reveals a niche-specific gene set. BMC Microbiol. 9, 1–9. doi: 10.1186/1471-2180-9-50
Partrick, K. A., Rosenhauer, A. M., Auger, J., Arnold, A. R., Ronczkowski, N. M., Jackson, L. M., et al. (2021). Ingestion of probiotic (Lactobacillus helveticus and Bifidobacterium longum) alters intestinal microbial structure and behavioral expression following social defeat stress. Sci. Rep. 11, 1–12. doi: 10.1038/s41598-021-83284-z
Phillips, R., Ursell, T., Wiggins, P., and Sens, P. (2009). Emerging roles for lipids in shaping membrane-protein function. Nature 459, 379–385. doi: 10.1038/nature08147
Pizarro-Cerdá, J., and Cossart, P. (2006). Bacterial adhesion and entry into host cells. Cells 124, 715–727. doi: 10.1016/j.cell.2006.02.012
Reid, G., and Burton, J. (2002). Use of Lactobacillus to prevent infection by pathogenic bacteria. Microb. Infect. 4, 319–324. doi: 10.1016/S1286-4579(02)01544-7
Rossi, F., Zotta, T., Iacumin, L., and Reale, A. (2016). Theoretical insight into the heat shock response (HSR) regulation in Lactobacillus casei and L. rhamnosus. J. Theor. Biol. 402, 21–37. doi: 10.1016/j.jtbi.2016.04.029
Sanders, M. E., Benson, A., Lebeer, S., Merenstein, D. J., and Klaenhammer, T. R. (2018). Shared mechanisms among probiotic taxa: implications for general probiotic claims. Curr. Opin. Biotechnol. 49, 207–216. doi: 10.1016/j.copbio.2017.09.007
Schumann, W. (1999). FtsH – a single-chain charonin? FEMS Microbiol. Rev. 23, 1–11. doi: 10.1016/S0168-6445(98)00024-2
Servin, A. L. (2004). Antagonistic activities of lactobacilli and Bifidobacteria against microbial pathogens. FEMS Microbiol. Rev. 28, 405–440. doi: 10.1016/j.femsre.2004.01.003
Sha, Y. (2016) Studies on probiotic mechannism of lactic acid bacteria in Litopenaeus vannamei. [dissertation/doctoral thesis]. Institute of Oceanology, Chinese Acadamy of sciences.
Sha, Y., Wang, B., Liu, M., Jiang, K., and Wang, L. (2016a). Interaction between Lactobacillus pentosus HC-2 and Vibrio parahaemolyticus E1 in Litopenaeus vannamei in vivo and in vitro. Aquaculture 465, 117–123. doi: 10.1016/j.aquaculture.2016.09.007
Sha, Y., Wang, L., Liu, M., Jiang, K., Xin, F., and Wang, B. (2016b). Effects of lactic acid bacteria and the corresponding supernatant on the survival, growth performance, immune response and disease resistance of Litopenaeus vannamei. Aquaculture 452, 28–36. doi: 10.1016/j.aquaculture.2015.10.014
Singh, K. S., Kumar, S., Mohanty, A. K., Grover, S., and Kaushik, J. K. (2018). Mechanistic insights into the host-microbe interaction and pathogen exclusion mediated by the mucus-binding protein of Lactobacillus plantarum. Sci. Rep. 8:14198. doi: 10.1038/s41598-018-32417-y
Sinha, A. K., Kumar, V., Makkar, H. P., De Boeck, G., and Becker, K. (2011). Non-starch polysaccharides and their role in fish nutrition–a review. Food Chem. 127, 1409–1426. doi: 10.1016/j.foodchem.2011.02.042
Teame, T., Wang, A., Xie, M., Zhang, Z., Yang, Y., Ding, Q., et al. (2020). Paraprobiotics and postbiotics of probiotic lactobacilli, their positive effects on the host and action mechanisms: a review. Front. Nutr. 7:570344. doi: 10.3389/fnut.2020.570344
Van den Abbeele, P., Marzorati, M., Derde, M., De Weirdt, R., Joan, V., Possemiers, S., et al. (2016). Arabinoxylans, inulin and Lactobacillus reuteri 1063 repress the adherent-invasive Escherichia coli from mucus in a mucosa-comprising gut model. NPJ Biofilms Microbi. 2:16016. doi: 10.1038/npjbiofilms.2016.16
Van Tassell, M. L., and Miller, M. J. (2011). Lactobacillus adhesion to mucus. Nutrients 3, 613–636. doi: 10.3390/nu3050613
Wang, H., Niu, Y., Pan, J., Li, Q., and Lu, R. (2020). Antibacterial effects of Lactobacillus acidophilus surface-layer protein in combination with nisin against Staphylococcus aureus. LWT 124:109208. doi: 10.1016/j.lwt.2020.109208
Wang, J., Zhang, J., Liu, W., Zhang, H., and Sun, Z. (2021). Metagenomic and metatranscriptomic profiling of Lactobacillus casei Zhang in the human gut. NPJ Biofilms Microbi. 7:55. doi: 10.1038/s41522-021-00227-2
Westerlund, B., and Korhonen, T. (1993). Bacterial proteins binding to the mammalian extracellular matrix. Mol. Microbiol. 9, 687–694. doi: 10.1111/j.1365-2958.1993.tb01729.x
Yadav, A. K., Tyagi, A., Kumar, A., Panwar, S., Grover, S., Saklani, A. C., et al. (2017). Adhesion of lactobacilli and their anti-infectivity potential. Crit. Rev. Food Sci. Nutr. 57, 2042–2056. doi: 10.1080/10408398.2014.918533
Yamamoto, F. Y., Yin, F., Rossi, W. Jr., Hume, M., and Gatlin, D. M. III (2018). β-1, 3 glucan derived from Euglena gracilis and Algamune™ enhances innate immune responses of red drum (Sciaenops ocellatus L.). Fish Shellfish Immunol. 77, 273–279. doi: 10.1016/j.fsi.2018.04.003
Yang, P., Hu, H., Li, Y., Ai, Q., Zhang, W., Zhang, Y., et al. (2019). Effect of dietary xylan on immune response, tight junction protein expression and bacterial community in the intestine of juvenile turbot (Scophthalmus maximus L.). Aquaculture 512:734361. doi: 10.1016/j.aquaculture.2019.734361
Yao, W., Li, X., Chowdhury, M. K., Wang, J., and Leng, X. (2019). Dietary protease, carbohydrase and micro-encapsulated organic acid salts individually or in combination improved growth, feed utilization and intestinal histology of pacific white shrimp. Aquaculture 503, 88–95. doi: 10.1016/j.aquaculture.2018.12.064
Zhang, H., Liu, T., Zhang, Z., Payne, S. H., Zhang, B., McDermott, J. E., et al. (2016). Integrated proteogenomic characterization of human high-grade serous ovarian cancer. Cells 166, 755–765. doi: 10.1016/j.cell.2016.05.069
Keywords: Lactiplantibacillus plantarum , probiotics, mucus, adhesion, competitive exclusion
Citation: Sha Y, Yan Q, Liu J, Yu J, Xu S, He Z, Ren J, Qu J, Zheng S, Wang G and Dong W (2023) Homologous genes shared between probiotics and pathogens affect the adhesion of probiotics and exclusion of pathogens in the gut mucus of shrimp. Front. Microbiol. 14:1195137. doi: 10.3389/fmicb.2023.1195137
Received: 28 March 2023; Accepted: 29 May 2023;
Published: 14 June 2023.
Edited by:
Hector Riveros-Rosas, National Autonomous University of Mexico, MexicoReviewed by:
Xiangli Tian, Ocean University of China, ChinaCopyright © 2023 Sha, Yan, Liu, Yu, Xu, He, Ren, Qu, Zheng, Wang and Dong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yujie Sha, c3lqbHc4OUAxNjMuY29t; Jian Liu, bGl1amlhbkBkenUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.